- 1Department of Bioenergy Science and Technology, Chonnam National University, Gwangju, South Korea
- 2Plant Systems Engineering Research Center, Korea Research Institute of Bioscience and Biotechnology, Deajeon, South Korea
- 3Department of Biosystems and Bioengineering, KRIBB School of Biotechnology, Korea University of Science and Technology, Daejeon, South Korea
The cuticle of land plants is the first physical barrier to protect their aerial parts from biotic and abiotic stresses. DEWAX, an AP2/ERF-type transcription factor, negatively regulates cuticular wax biosynthesis. In this study, we investigated the resistance to Botrytis cinerea in Arabidopsis thaliana and Camelina sativa overexpressing DEWAX and in Arabidopsis dewax mutant. Compared to wild type (WT) leaves, Arabidopsis DEWAX OX and dewax leaves were more and less permeable to toluidine blue dye, respectively. The ROS levels increased in DEWAX OX leaves, but decreased in dewax relative to WT leaves. Compared to WT, DEWAX OX was more resistant, while dewax was more sensitive to B. cinerea; however, defense responses to Pseudomonas syringae pv. tomato DC3000:GFP were inversely modulated. Microarray and RT-PCR analyses indicated that the expression of defense-related genes was upregulated in DEWAX OX, but downregulated in dewax relative to WT. Transactivation assay showed that DEWAX upregulated the expression of PDF1.2a, IGMT1, and PRX37. Chromatin immunoprecipitation assay revealed that DEWAX directly interacts with the GCC-box motifs of PDF1.2a promoter. In addition, ectopic expression of DEWAX increased the tolerance to B. cinerea in C. sativa. Taken together, we suggest that increased ROS accumulation and DEWAX-mediated upregulation of defense-related genes are closely associated with enhanced resistance to B. cinerea in Arabidopsis and C. sativa.
Introduction
The hydrophobic cuticle covering the aboveground parts of land plants forms a physical and chemical barrier to infection by bacterial and fungal pathogens, as well as drought stress (Nawrath, 2006; Pollard et al., 2008; Beisson et al., 2012; Li-Beisson et al., 2013; Yeats and Rose, 2013). The cuticle is composed of a cutin polyester layer, an intracuticular wax layer embedded in the cutin layer, and an outermost epicuticular wax layer (Bernard and Joubes, 2013; Yeats and Rose, 2013). The structural polymer cutin is cross-linked via ester bonds among glycerols and C16 and C18 polyhydroxy, epoxy, ω-hydroxy, and α,ω-dicarboxylic fatty acid components (Pollard et al., 2008; Beisson et al., 2012). Cuticular wax is a mixture of very-long-chain fatty acids and their derivatives, such as alkanes, aldehydes, ketones, primary and secondary alcohols, and wax esters (Kunst and Samuels, 2009; Li-Beisson et al., 2013; Lee and Suh, 2015). The cutin and wax components are synthesized from C16 and C18 fatty acid precursors produced in the plastids by the endoplasmic reticulum (ER)-localized enzymes and exported to the extracellular space through the cytoplasmic ATP-binding cassette transporters (Pighin et al., 2004; Bird et al., 2007; Panikashvili et al., 2007, 2011; Kunst and Samuels, 2009; McFarlane et al., 2010; Bessire et al., 2011; Bernard and Joubes, 2013; Li-Beisson et al., 2013; Lee and Suh, 2015).
When pathogenic fungi come in contact with a host plant, they first secrete cutinase or lipase to break down the cutin layer for penetration into the plant body (Kolattukudy, 1985; Kolattukudy et al., 1995). Degraded cutin is recognized by pathogen and promotes disease development, whereas as a counterprogram, it is recognized by the plant immune system and acts as a signal transmitter to induce an immune response (Woloshuk and Kolattukudy, 1986; Schweizer et al., 1996). Overexpression of a fungal cutinase in Arabidopsis plants enhanced solute permeability and led to increased resistance to the necrotrophic fungus Botrytis cinerea (Chassot et al., 2007). Several Arabidopsis mutants with increased cuticular permeability, including lacerate (lcr), bodyguard (bdg), long-chain acyl-CoA synthetase-2 (lacs-2) and -3, fiddlehead (fdh), permeable cuticle1 (fec1), and myeloblast transcription factor 96 (myb96), also display enhanced resistance to B. cinerea (Yephremov et al., 1999; Wellesen et al., 2001; Kurdyukov et al., 2006; Bessire et al., 2007, 2011; Chassot et al., 2007; Voisin et al., 2009; Seo et al., 2011). However, some Arabidopsis mutants with defective cuticle, such as glycerol-3-phosphate acyltransferase4 gpat8 (gpat4 gpat8), acyl-CoA binding protein1 (acbp1), and glycosylphosphatidyl-anchored lipid transfer protein1 (ltpg1), are more susceptible to fungal pathogens, including B. cinerea and Alternaria brassicicola (Li et al., 2007; Lee et al., 2009; Xue et al., 2014). Xia et al. (2009) showed that an intact cuticle layer is involved in the systemically acquired resistance (SAR) response, and cuticle-mediated SAR is independent of salicylic acid- or jasmonic acid-mediated defense signaling. Disruption of acbp3, 4, and 6 increased the cuticle permeability and susceptibility to bacterial and fungal pathogens due to defective SAR signal generation (Xia et al., 2012). Therefore, it is important to investigate the roles of the cuticle in plant immunity.
Cuticular wax biosynthesis is controlled by various environmental stress conditions, and several transcription factors involved in its regulation have been reported (Jenks et al., 2001; Cameron et al., 2006; Shepherd and Griffiths, 2006; Raffaele et al., 2007; Kosma et al., 2009; Seo et al., 2011; Lee and Suh, 2013, 2015; Go et al., 2014; Kim et al., 2017). Among them, MYB30 modulates hypersensitive cell death-related lipid signaling by the upregulation of very-long-chain fatty acid synthesis (Raffaele et al., 2007). MYB96 activation-tagging lines with increased total wax load showed resistance to the virulent pathogen, Pseudomonas syringae pv. tomato DC3000, by inducing salicylic acid biosynthesis (Seo and Park, 2010). The APETALA2/Ethylene-Responsive Factor (AP2/ERF)-type transcription factor DEWAX represses cuticular wax biosynthesis via downregulation of the cuticular wax biosynthetic genes CER1, LACS2, ACLA2, and ECR (Go et al., 2014). However, defense responses of DEWAX overexpression (OX) lines, which display a permeable cuticle, against pathogen attacks have not yet been reported.
Camelina sativa, which belongs to the Brassicaceae family, has recently gained increasing interest for its potential non-food usages, including in cosmetics, lubricants, and as biofuels (Moser, 2010; Waraich et al., 2013). C. sativa has a short life cycle (100–120 days), and can be grown on marginal agricultural lands owing to its characteristics, such as tolerance to cold stress, relatively good growth under nutrient-poor soil conditions, and high levels of polyunsaturated fatty acids in the seed oils (Putnam et al., 1993; Zubr, 1997; Enjalbert et al., 2013). To date, transgenic C. sativa plants with high seed-oil content or enhanced drought resistance have been developed via Agrobacterium-mediated transformation (Liu et al., 2012; Lee et al., 2014; An and Suh, 2015; Kim et al., 2016). However, only few reports on genetically improved C. sativa with enhanced tolerance to pathogens are available (Zakharchenko et al., 2013).
In this study, we observed that DEWAX OX plants were more resistant to the fungal pathogen, B. cinerea, but were more susceptible to the bacterial pathogen, Pto DC3000 compared with wild type (WT). To understand the molecular mechanisms underlying the defense responses of Arabidopsis and C. sativa plants overexpressing DEWAX to B. cinerea, further analyses were carried out. Finally, we revealed that DEWAX plays as a transcriptional activator that upregulates the expression of the defense-related genes such as PDF1.2a, IGMT1, and PRX37 via binding to their gene promoters. The results suggest that DEWAX-mediated upregulation of defense-related genes in addition to an increase in ROS levels is important in the enhanced tolerance to B. cinerea in Arabidopsis and C. sativa overexpressing DEWAX.
Materials and Methods
Plant Materials and Growth Conditions
DEWAX overexpression lines (OX1 and OX2) and dewax knockout mutant were described in Go et al. (2014). A. thaliana (ecotype Columbia-0) seeds were surface-sterilized with 75% (v/v) ethanol and 0.05% (v/v) Triton X-100, washed with 100% ethanol, dried on filter paper, and germinated on half MS (Murashuge and Skoog, 1962) medium containing 1% sucrose and 0.6% phytoagar (Duchefa), which was adjusted to pH 5.7 using KOH. Seven-day-old seedlings were transplanted to mixed soil (soil/vermiculite/perlite, 3:2:1, v/v/v) and grown in a growth room under long-day conditions (16-h-light and 8-h-dark cycle) at 24 ± 2°C. C. sativa ‘CAME’ was grown on mixed soil (soil/vermiculite/perlite, 3:2:1, v/v/v) at 25 ± 3°C under a 16-h-light/8-h-dark cycle (Lee et al., 2014).
RNA Isolation and Quantitative RT-PCR Analysis
Total RNA was isolated from 4-week-old Col-0, dewax, DEWAX lines (OX1 and OX2), and 6-week-old transgenic C. sativa lines (T1 generation) using the NucleoSpin RNA Plant Extraction Kit (Macherey-Nagel) according to the instructions of the manufacturer. The total RNA was reverse-transcribed with GoScriptTM Reverse Transcription System (Promega), and the cDNA was used in RT-PCR analysis. RT-PCR was performed using the Access QuickTM RT-PCR system (Promega, Madison, WI, United States) with gene-specific primers (Supplementary Table S1). Arabidopsis EIF4 (At3g13920) and C. sativa ACTIN11 genes (Hutcheon et al., 2010) were used in RT-PCR analysis as a control for cDNA quality and quantity. Quantitative RT-PCR (qRT-PCR) was executed using SYBR® FAST Universal 2x qPCR Master Mix (KAPA Biosystems, Wilmington, MA, United States) in a final volume of 20 μL. PCR was performed according to the manufacturer’s protocols using gene specific primers described in Supplementary Table S1. PP2A (At1g13320) and CsACT11 genes were used to determine the RNA quality and quantity.
Toluidine Blue Staining
Four-week-old Arabidopsis rosette leaves were stained at room temperature for 8 min without shaking using freshly prepared staining solution containing 0.05% toluidine blue and 0.01% Tween-20. After staining, leaves were rinsed five times with distilled water and photographed.
ROS Assay
For in situ detection of hydrogen peroxide, Arabidopsis leaves were immersed in 10 mM MES (pH 6.5) containing 0.1% 3,3′-diaminobenzidine tetrahydrochloride (DAB, Sigma-Aldrich), gently vacuum-infiltrated, and incubated for 30 min as described in Piisilä et al. (2015). After staining, the leaves were de-stained by boiling in ethanol-lactophenol (2:1) for 5 min, washed twice with 50% ethanol, rinsed once with distilled water, and then photographed. For fluorometric measurements of ROS levels, Arabidopsis leaves were incubated in 10% MS salt solution containing 20 μM 2′,7′-dichlorofluorescein diacetate (H2DCFDA, Sigma-Aldrich) and 0.1% Tween-20 in the dark for 30 min. DCF fluorescence in aliquots taken from the incubation solution was measured using a Synergy H1 Hybrid Multi-Mode Microplate Reader (BioTek) at 488 and 525 nm. Leaf fresh weights were used to normalize the values (Umbach et al., 2012). For measurement of hydrogen peroxide (H2O2) content, 5-week-old leaves (200 mg) were incubated in 1.5 mL of 5 mM morpholineethanesulfonic acid (MES)-KOH buffer (pH 6.7) for 7 h (Horemans et al., 2007). Peroxidase reaction was started by adding 3,5-dichloro-2-hydroxyvenxenesulfonic acid (DCHBS), 4-aminoantipyrine (4-AAP), and peroxidase based on the manufacturer’s instructions (Megazyme) and the reaction mixture was incubated for 1 h at 25°C. Absorbance for the quinoneimine dye was spectrophotometrically measured using a Synergy H1 Hybrid Multi-Mode Microplate Reader (BioTek) at 520 nm. H2O2 content was estimated using a standard curve prepared similarly with 0–3 μM H2O2.
Pathogen Inoculation
Botrytis cinerea strain B05.10 was kindly provided by Dr. Mengiste (Purdue University, United States) and was cultured on 2× V8 tomato agar (36% V8 juice, 0.2% CaCO3, and 2% Bacto-agar). Harvesting and inoculation of conidia were carried out according to Veronese et al. (2006). Detached Arabidopsis leaves were laid out on 3MM paper with water and inoculated with 5-μl droplets of conidial suspension (5 × 105 spores/mL), kept under a sealed transparent cover to maintain high humidity and transferred to a growth chamber (21 ± 2°C, 16 h light/8 h dark). C. sativa plants were sprayed with B. cinerea conidial suspension (2 × 105 spores/mL). After inoculation, the plants were transferred to growth chambers (21 ± 2°C, 16 h light/8 h dark) and covered with a sealed clear lid to maintain low light intensity and high humidity. Quantification of B. cinerea biomass in C. sativa was carried out according to Gachon and Saindrenan (2004).
A Pto DC3000 strain harboring pDSK-GFPuv (Pto DC3000:GFP, Wang et al., 2007) was used for dip inoculation (Katagiri et al., 2002). Briefly, 3-week-old Arabidopsis plants grown in meshed pots were dipped in 50 ml of bacterial suspensions (optical density at 600 nm of 0.1) containing 0.05% Silwet L-77 (Lehle Seeds, Round Rock, TX, United States) for few seconds. After inoculation, the plants were covered with a clear lid and transferred to a growth room (23 ± 2°C, 16 h light/8 h dark). Inoculated plants were photographed under ultraviolet light to determine PtoDC3000:GFP proliferation using GFP signals and bacterial populations were measured by serial dilution assays (Katagiri et al., 2002). Colony forming unit (CFU) was normalized as CFU/mg using total weights of the inoculated leaves.
Transactivation Assay in Yeast
For the transcactivation assay in yeast, DEWAX-F (603 bp for full-length), DEWAX-N (312 bp for N-terminal region), and DEWAX-C (291 bp for C-terminal region) DNA fragments were amplified via PCR using DEWAX cDNA and DEWAX-specific primers (Supplementary Table S1). SmaI and SacI-digested PCR fragments were cloned into the pGBKT7 vector (BD Biosciences Clontech). The recombinant vector (pAtDEWAX-F, pAtDEWAX-N, and pAtDEWAX-C) were transformed into yeast strain Y190 (MATa, HIS3, lacZ, trp1, leu2, cyhr2) according to the manufacturer’s instructions (BD Biosciences Clontech). Transformants were selected on selective medium (SD-Trp) supplemented with 25 mM 3-amino-1, 2, 4-aminotriazole (3-AT). Transformants were cultured on selective medium (SD-Trp-His) for 1 day at 30°C and filter-lift assays were performed for 16 h at 37°C, as described in Breeden and Nasmyth (1985).
Transcriptional Activation Assay
To generate the reporter constructs, DNA fragments corresponding to the PDF1.2a (∼1.3 kb), IGMT (∼1.5 kb), PRX37 (∼1.2 kb) promoter regions were amplified using gene-specific primers and cloned into the SalI and SpeI sites of the GAL4-LUC binary vector. The recombinant pPZP212 vector (Hajdukiewicz et al., 1994) harboring Arabidopsis DEWAX driven by the CaMV35S promoter, reported by Go et al. (2014), was used as an effector construct. Protoplast isolation was carried out according to Yoo et al. (2007).
The reporter and effector constructs were isolated using a Qiagen Plasmid Midi Kit and co-introduced into tobacco protoplasts (Miura et al., 2007). After incubation of the transfected tobacco protoplasts in the dark at 23°C for 16 h, the protoplasts were subjected to enzymatic assays to quantify luciferase and β-glucuronidase activities using a dual-luciferase assay system (Promega). Luciferase gene expression was normalized to that of the β-glucuronidase gene.
ChIP Assay
Chromatin immunoprecipitation (ChIP) assay was performed using 35S:MYC-DEWAX transgenic plants, which were previously reported (Go et al., 2014). Genomic DNA was isolated from 10-day-old 35S:MYC-DEWAX transgenic seedlings and immune-precipitated with an anti-myc antibody (Santa Cruz Biotechnology). Quantitative RT-PCR was performed as described previously (Gendrel et al., 2005). Arabidopsis actin7 (At5g09810) was used to control cDNA quantity and quality.
Construction of a Binary Vector and Transformation of C. sativa
To generate transgenic C. sativa plants overexpressing Arabidopsis DEWAX, the full-length DEWAX gene was amplified from cDNA converted from RNA isolated from 3-week-old leaves with DEWAX F1 and DEWAX R1 primers (Supplementary Table S1). The PCR product was digested with Sac1 and Sma1 and cloned into the binary vector pBA002. The binary vectors were transformed into Agrobacterium strain GV3101 by the freeze-thaw method (An, 1987).
Agrobacterium harboring the 35S:MYC-DEWAX construct (Go et al., 2014) was inoculated in YEP medium (containing 50 μg/mL rifampicin and 50 μg/mL spectinomycin) and cultured at 30°C overnight with shaking. The culture was centrifuged at 3,000 rpm for 20 min and the pellet was resuspended in transformation solution (5% sucrose and 0.05% Silwet L-77) at an OD600 of 0.8. Five- to six-week-old C. sativa plants were transformed with Agrobacterium by a modified floral dip method (Lu and Kang, 2008; Liu et al., 2012). T1 transgenic seeds were sowed on mixed soil and selected by spraying of 0.03% (v/v) BASTA herbicide.
Genomic DNA Isolation and PCR Analysis
Genomic DNA was isolated from leaves of 3-week old non-transgenic and transgenic C. sativa plants (T1 generation) using extraction buffer (200 mM Tris-HCl, pH 7.5, 250 mM NaCl, 25 mM EDTA, and 0.5% sodium dodecyl sulfate) and subjected to genomic DNA PCR analysis with CaMV35S promoter F and DEWAX CR1 primers (Supplementary Table S1). Thermal cycles included 32 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and elongation at 72°C for 60 s.
Results
Overexpression of DEWAX Increased Cuticle Permeability and ROS Accumulation in Arabidopsis
To investigate the cuticle permeability of WT, dewax, and two DEWAX OX lines, rosette leaves of 4-week-old plants were stained with 0.05% toluidine blue. As shown in Figure 1A, compared to WT, Arabidopsis DEWAX OX and dewax leaves were more and less permeable to toluidine blue dye, respectively. Based on the previous reports that Arabidopsis bdg and lacs2 mutants with a permeable cuticle showed increased ROS levels (L’Haridon et al., 2011), ROS assay was performed in the leaves of WT, dewax, and DEWAX OX lines. Both in situ assay of hydrogen peroxide and fluorometric measurements of ROS levels showed that the levels of ROS were lower in dewax leaves, but higher in DEWAX OX1 and OX2 leaves than in WT leaves (Figures 1B,C). When hydrogen peroxide content was spectrophotometrically measured after leaves of WT, dewax, and DEWAX OX lines were preadapted in incubation buffer, the levels of hydrogen peroxide were approximately twofold higher in DEWAX OX1 and OX2 leaves than in WT leaves, but no remarkable differences were detected between WT and dewax leaves (Figure 1D).
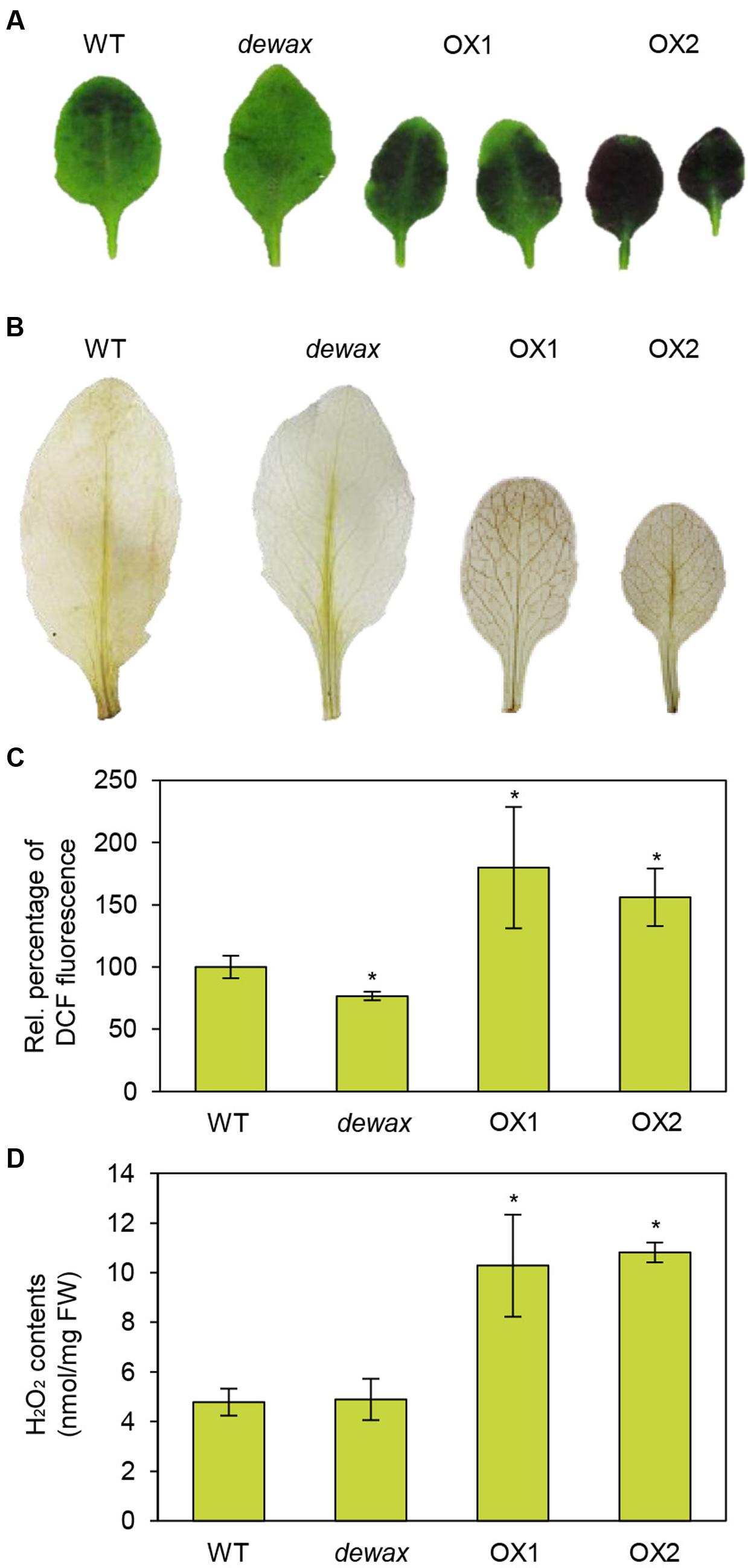
FIGURE 1. Overexpression of DEWAX increased cuticle permeability and ROS accumulation in Arabidopsis. (A) Cuticle permeability analysis. Leaves of WT, dewax, and DEWAX overexpression lines were stained with 0.05% toluidine blue for 8 min. (B) In situ detection of hydrogen peroxide. Leaves of WT, dewax, and DEWAX overexpression lines were incubated in 10 mM MES (pH 6.5) containing 0.1% DAB for 30 min, de-stained by boiling in ethanol-lactophenol (2:1) for 5 min, washed with 50% ethanol, rinsed with distilled water, and then photographed. (C) Measurements of ROS levels. Leaves of WT, dewax, and DEWAX overexpression lines were incubated in 10% MS salt solution containing 20 μM H2DCFDA and 0.1% Tween-20 for 30 min. DCF fluorescence in aliquots taken from the extracts was measured at 488 and 525 nm. Leaf fresh weights were used to normalize the values. Data were statistically analyzed using Student’s t-test (∗P < 0.05). Error bars indicate ± SD from triplicate experiments. (D) Measurements of hydrogen peroxide content. Five-week-old WT, dewax, OX1, and OX2 leaves (200 mg) were incubated in 1.5 mL of 5 mM MES buffer (pH 6.7) for 7 h. Peroxidase reaction was started by adding DCHBS, 4-AAP, and peroxidase (Megazyme) and the reaction mixture was incubated for 1 h at 25°C. Absorbance for the quinoneimine dye was spectrophotometrically measured at 520 nm. H2O2 content was estimated using a standard curve. Data were statistically analyzed using Student’s t-test (∗P < 0.01). Error bars indicate ± SD from triplicate experiments.
Overexpression of DEWAX Enhanced Resistance, While Disruption of DEWAX Increased Susceptibility to B. cinerea
To investigate the response of WT, dewax, and two DEWAX OX lines to the necrotrophic pathogen B. cinerea and the hemibiotrophic bacterial pathogen Pto DC3000, first, we examined visible disease symptoms 72 h after inoculation of B. cinerea on the adaxial surface of the leaves. In a spot inoculation assay, significantly larger lesions were observed on the leaves of dewax mutant, while smaller lesions were formed on DEWAX OX as compared with WT leaves (Figures 2A,B), indicating that DEWAX OX shows enhanced resistance, and dewax enhanced susceptibility, to B. cinerea. On the other hand, compared to WT, DEWAX OX was more susceptible to Pto DC3000:GFP, while dewax was more resistant to this pathogen (Figures 2C,D). These results indicate that DEWAX is involved in the disease resistance responses of Arabidopsis and plays a positive role in the defense to fungal pathogens, but a negative role in the defense to bacterial pathogens.
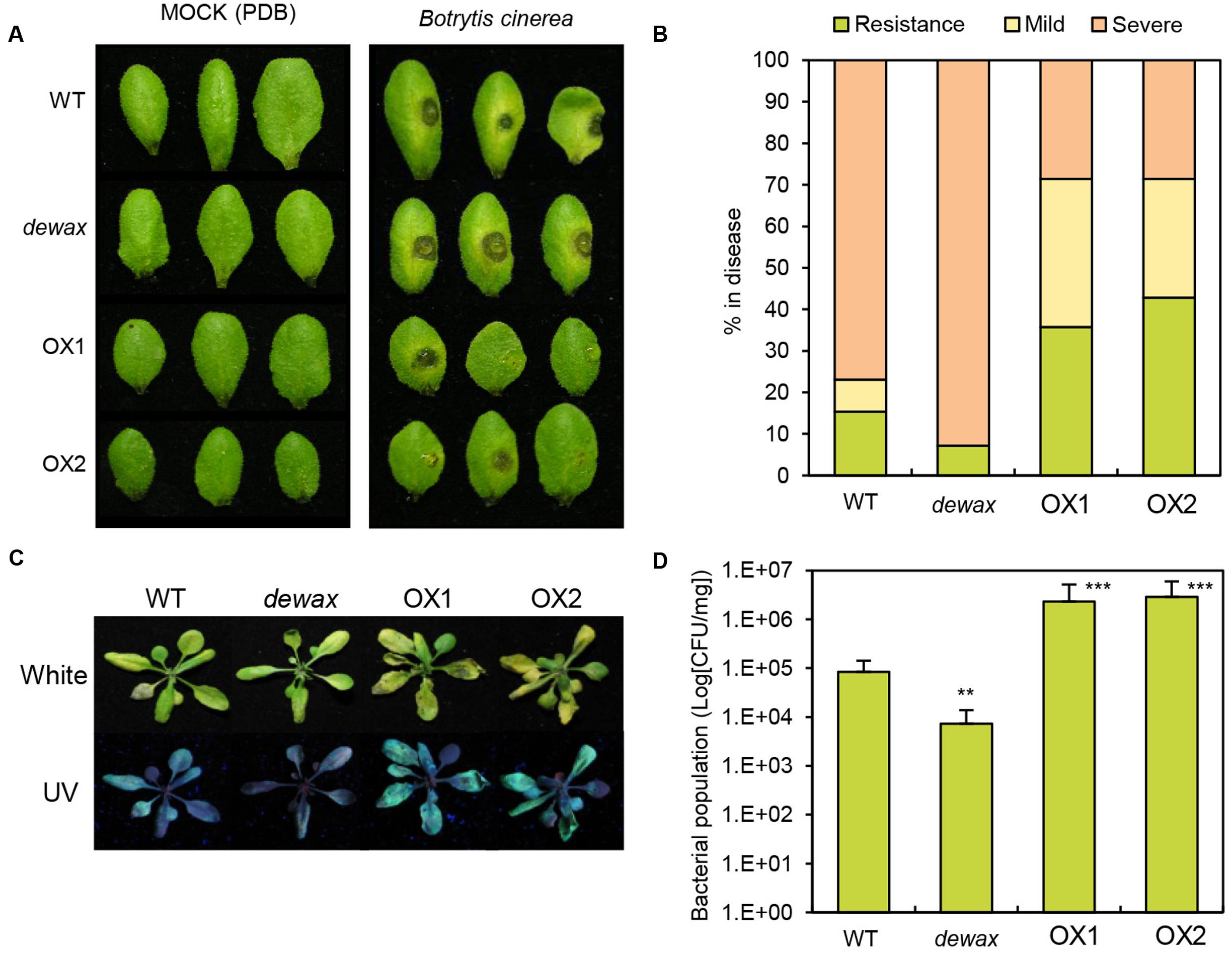
FIGURE 2. Overexpression of DEWAX confers tolerance to Botrytis cinerea and susceptibility to Pto DC3000:GFP in transgenic Arabidopsis. (A) Macroscopic symptoms on leaves of WT, dewax, and DEWAX overexpression lines at 6 days after inoculation with potato dextrose broth (Mock) or B. cinerea. (B) Disease symptom scores at 6 days after inoculation with B. cinerea (0 = non-infection, 1 = lesions at the point of inoculation, 2 = larger lesions surrounded by extensive chlorosis). (C) Disease phenotypes of WT, dewax and DEWAX overexpression lines after dip-inoculation with Pto DC3000:GFP bacteria. Photographs were taken 3 dpi under white and UV lights. (D) Bacterial populations of Pto DC3000:GFP in WT, dewax and DEWAX overexpression lines were measured at 3 dpi. Colony forming unit (CFU) was normalized as CFU/mg using total weights of the inoculated leaves. The results are representative of three independent experiments. Error bars indicate SE. Asterisks indicate a statistically significant difference compared with wild type (WT) using Student’s t-test (∗∗P < 0.01, ∗∗∗P < 0.001).
Expression of DEWAX-Regulated Genes in DEWAX OX Lines and the dewax Mutant
To obtain further insight in the DEWAX-mediated defense responses of Arabidopsis, comparative transcriptome analysis of WT and DEWAX OX was conducted (Go et al., 2014; E-MEXP-3781 at http://www.ebi.ac.uk/arrayexpress). Interestingly, the expression of genes involved in pathogen defense and ROS production was clearly upregulated in the stems of DEWAX OX as compared to those of WT (Penninckx et al., 2003; Almagro et al., 2009; Pedreira et al., 2011). Genes that were upregulated 10-fold and more in DEWAX OX2 than in WT are listed in Figure 3A. RT-PCR was carried out to confirm their transcript levels in 4-week-old WT, dewax, and DEWAX OX1 and OX2 leaves. The expression levels of PDF1.2a and PDF1.2b, involved in plant defense, were remarkably induced in DEWAX OX1 and OX2 leaves, but reduced in dewax as compared to WT leaves. The transcript levels of indole glucosinolate methyltransferase (IGMT1) and peroxidase C2 precursor (PRX37 and PRX38) were increased in DEWAX OX1 and OX2 leaves, while no significant downregulation was observed in dewax relative to WT. No noticeable differences in the expression of At1g67810 and At4g16260, encoding SULFUR (Stimulate CpNifS Cysteine Desulfurase Activity) and a β-1,3-glucanase precursor, respectively, were observed in dewax, and DEWAX OX1 and OX2 leaves (Figure 3B and Supplementary Figure S1).
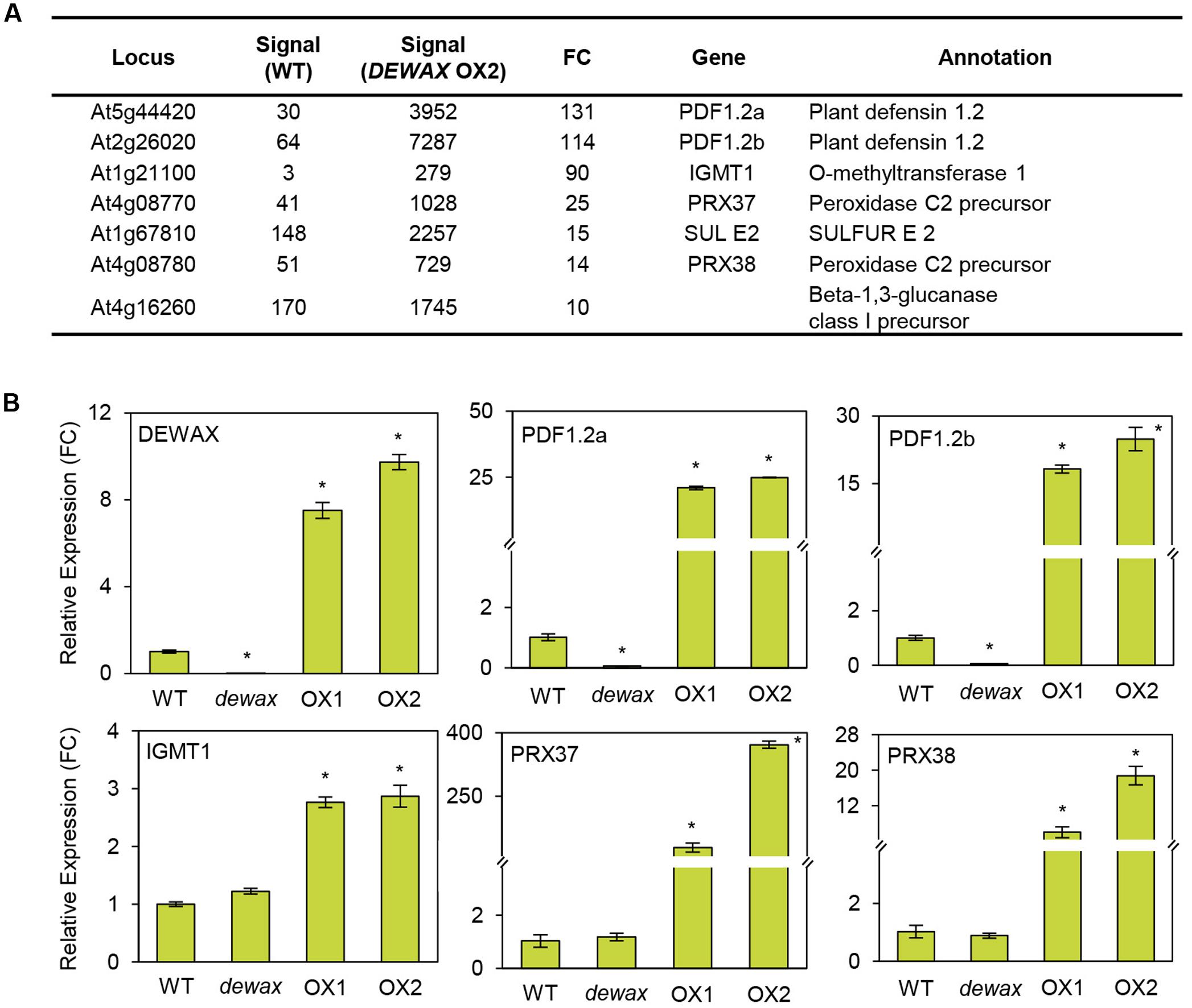
FIGURE 3. Microarray (A) and qRT-PCR (B) analyses of genes activated by DEWAX overexpression. (A) List of genes that were upregulated in stems of DEWAX OX2 relative to WT. Total RNA was isolated from stems of DEWAX OX2 and WT plants and subjected to microarray analysis using Arabidopsis ATH1 gene chips. FC, fold change. (B) Quantitative RT-PCR (qRT-PCR) analysis of genes upregulated in leaves of DEWAX OX2 as compared to WT. Total RNA was isolated from leaves of 4-week-old WT, dewax, and DEWAX overexpression lines (OX1 and OX2) and subjected to qRT-PCR analysis. The PP2A (At1g13320) gene was used as a reference to determine the RNA quality and quantity. Data were statistically analyzed using Student’s t-test (∗P < 0.01). Error bars indicate ± SD from triplicate experiments.
DEWAX Functions as a Transcriptional Activator
DEWAX has been reported to function as a transcriptional repressor in cuticular wax biosynthesis (Go et al., 2014). However, upregulation of defense-related genes in DEWAX OX and their downregulation in dewax prompted us to examine whether DEWAX acts as a transcriptional activator in tobacco protoplasts and yeast cells. A reporter construct, GAL4-LUC, harbors a luciferase gene driven by a promoter including a GAL4 motif. DEWAX was translationally ligated to the C-terminus of the GAL4 DNA-binding domain under the control of CaMV35S promoter in pBI221 vector (Lee et al., 2013) and used as an effector construct, GAL4DB:DEWAX. Tobacco protoplasts were transformed with a GAL4-LUC reporter construct only or co-transformed with a GAL4-LUC construct and an effector construct, GAL4DB or GAL4DB:DEWAX. Strong luciferase activities were observed in protoplasts transformed with GAL4-LUC reporter and a GAL4DB:DEWAX effector constructs as compared to those transformed with GAL4-LUC reporter and GAL4DB effector constructs (Figure 4A).
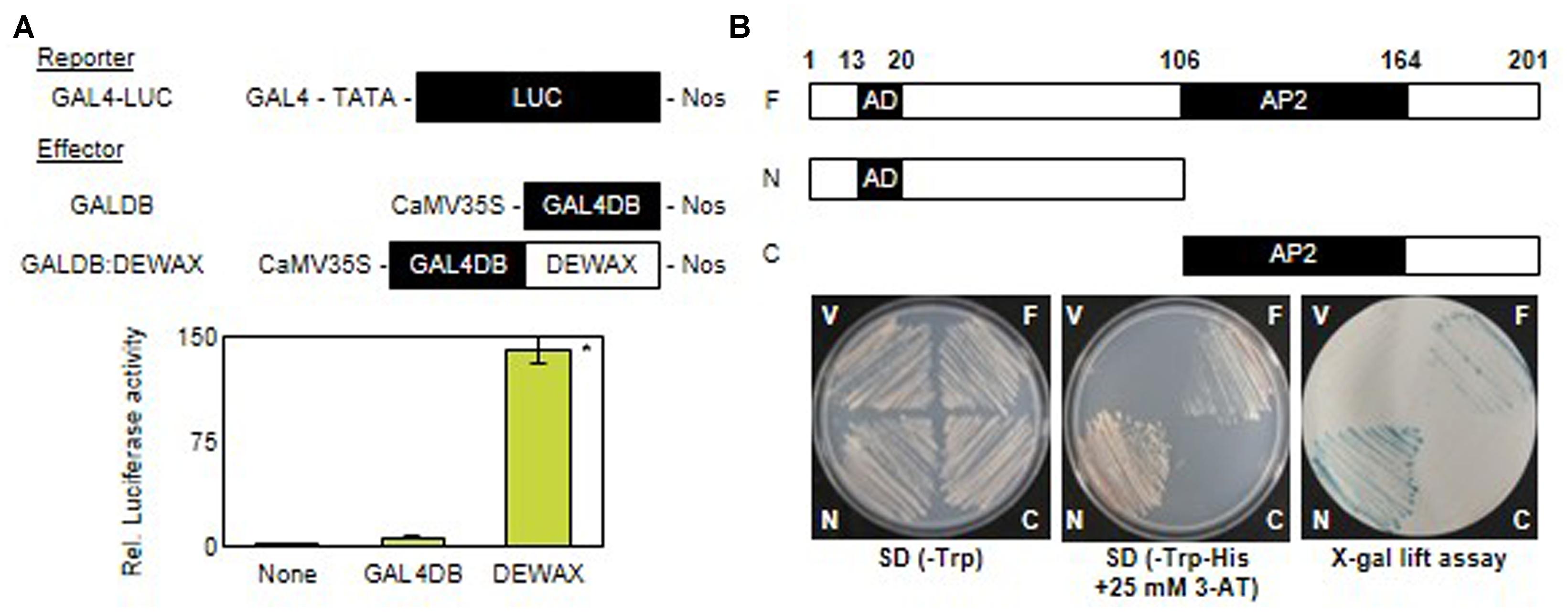
FIGURE 4. Transcriptional activation assay of DEWAX gene in tobacco protoplast and yeast. (A) Relative luciferase activities in tobacco protoplasts co-transfected with reporter and effector plasmids. The luciferase activities are expressed relative to values obtained with the reporter construct alone (none). GAL4, galactose-inducible gene promoter; LUC, luciferase; GAL4DB, GAL4 DNA-binding domain; Nos, nopaline synthase terminator; CaMV35, cauliflower mosaic virus 35S promoter. Biological triplicates were averaged and statistically analyzed using Student’s t-test (∗P < 0.01). Error bars indicate ± SD of the means. (B) Growth and β-galactosidase (X-Gal) assay of yeast transformed with F, N, or C construct on SD/Trp- and SD-His- plus 25 mM 3-AT media; F, full-length of the DEWAX present in pGBKT7; N, N-terminal domain of DEWAX present in pGBKT7; C, C-terminal domain of DEWAX present in pGBKT7; V, pGBKT7 plasmid without DEWAX as a control; AD, activation domain; AP, AP2 domain of the DEWAX required for DNA binding.
In addition, the full-length (F, amino acids 1 to 201), N-terminal (N, amino acids 1 to 106), and C-terminal (C, amino acids 107 to 201) regions of DEWAX were cloned downstream of the GAL4-binding domain in the pGBKT7 vector. Yeast Y190 cells, which possess the HIS3 and lacZ genes driven by the promoter including GAL4-responsive elements, were transformed with the constructed vectors. Yeast cells that were able to survived on tryptophan-deficient selective medium (SD/-Trp) were further selected on tryptophan- and histidine-deficient medium (SD/-Trp-His) supplemented with 10 mM 3-amino-1, 2, 4-aminotriazole (3-AT) and subsequently subjected to an X-gal filter-lifting assay. Yeast cells transformed with the F or N construct survived on SD/-Trp-His medium supplemented with 10 mM 3-AT, and generated blue colonies by the β-galactosidase activities in X-gal lift assay. However, yeast cells transformed with the pGBKT7 vector or the C construct did not survive on the selective medium, and showed no β-galactosidase activity. These results indicate that DEWAX is able to act as a transcriptional activator, and amino acid residues 1 to 106 of DEWAX are involved in transcriptional activation (Figure 4B).
DEWAX Promotes the Expression of PDF1.2 via Direct Binding to Conserved Sequence Motifs in its Promoter
To examine whether DEWAX is able to activate the expression of the putative target genes, PDF1.2a, IGMT1, and PRX37, their promoter regions were transcriptionally ligated to the luciferase gene (LUC) in the Gal4-LUC binary vector, and used as reporter constructs (Figure 5A). p35S and p35S-DEWAX as effector constructs were used in transactivation assay of tobacco protoplasts (Go et al., 2014). After tobacco protoplasts were co-transformed with each reporter construct containing LUC, the effector construct, and the internal control construct harboring the GUS gene, luciferase (LUC) and β-glucuronidase (GUS) activities were measured, and the LUC activity was normalized to the GUS activity. When DEWAX was expressed, LUC activity was upregulated in the protoplasts transformed with PDF1.2a, IGMT1, and PRX37 reporter constructs by approximately 2.3, 4.2, and 6.3-fold, respectively, but was downregulated in the protoplasts transformed with CER1 by approximately twofold relative to the control co-transformed with p35S (Figure 5B).
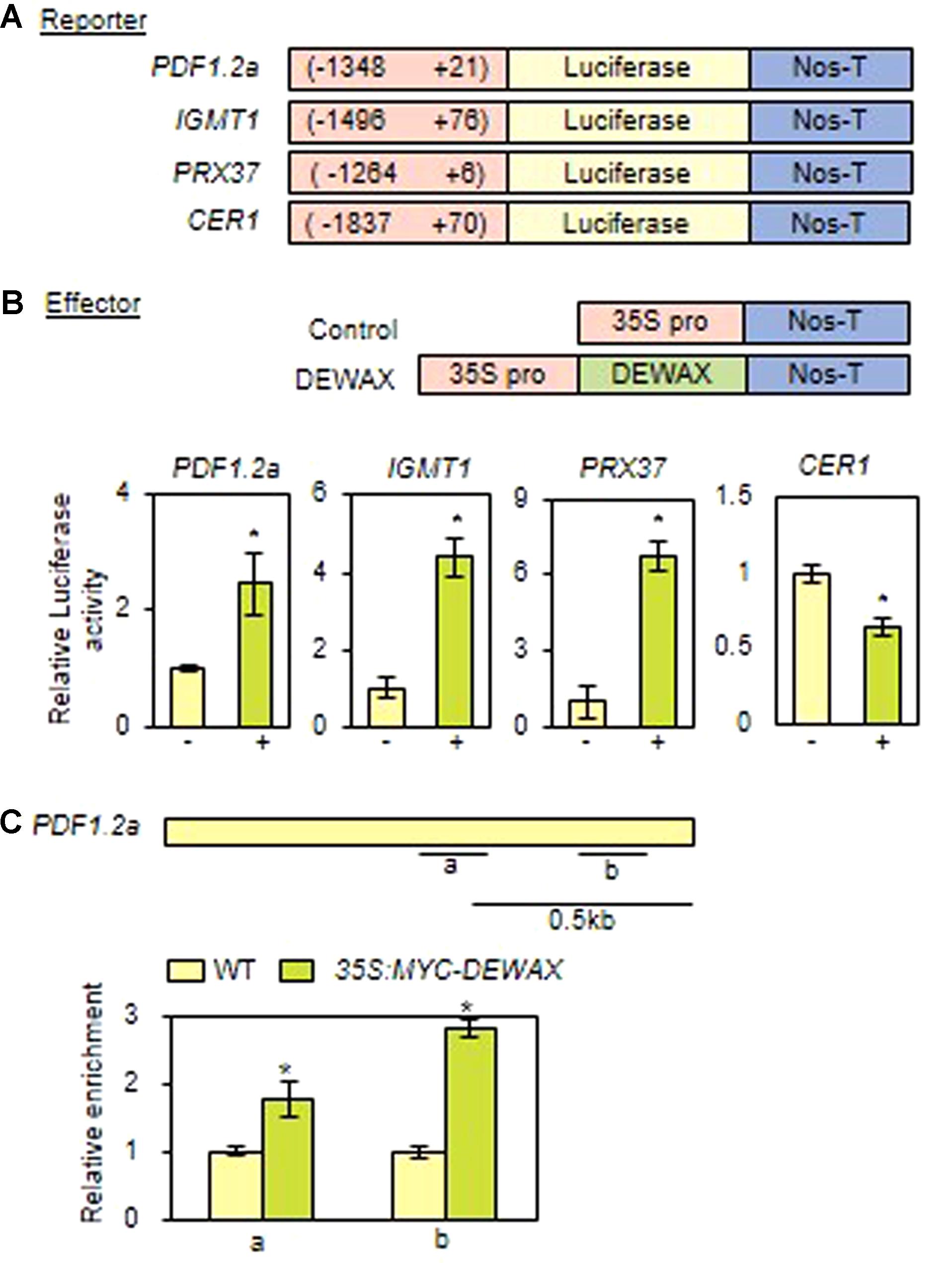
FIGURE 5. DEWAX upregulates the expression of the PDF1.2 gene by direct binding to conserved sequence motifs in its gene promoter. (A) Schematic diagrams of reporter and effector constructs used for the transcriptional activation assay. In the reporter constructs, the promoter regions of DEWAX-activated genes were fused to the luciferase gene. In the effector constructs, DEWAX was cloned between the CaMV35S promoter and the terminator of the nopaline synthase gene (Nos-T). (B) Transcriptional activation assay of DEWAX-activated genes in tobacco protoplasts. The LUC activity was normalized to the GUS activity, which was from the internal control construct harboring the GUS gene. Mean fold change in relative luciferase activity was calculated by dividing the normalized luciferase activity obtained from the protoplasts transformed with the control effector construct. Data were statistically analyzed using Student’s t-test (∗P < 0.01). Error bars indicate ± SD of the mean from triplicate experiments. –, P35S; +, P35S-DEWAX. (C) Description of the promoter region of the PDF1.2a gene and chromatin immunoprecipitation (ChIP) assay. a and b indicate regions including consensus GCC-box motifs that were used in ChIP assay. In each qRT-PCR measurement, the value for WT was set to 1 after normalization against actin7. Data were statistically analyzed using Student’s t-test (∗P < 0.01). Error bars indicate ± SD of the mean from triplicate samples.
We further investigated whether DEWAX directly interacts with the consensus GCC-box motifs of PDF1.2a promoter region using 35S:MYC-DEWAX transgenic plants (Go et al., 2014). Quantitative real-time ChIP-PCR assay showed that DEWAX binds directly to the promoters of the PDF1.2a gene in planta (Figure 5C). This observation indicates that DEWAX activates the expression of PDF1.2a gene by direct interaction with its promoter region.
Overexpression of DEWAX Enhanced the Resistance to B. cinerea in C. sativa
As DEWAX overexpression in Arabidopsis led to strong resistance to B. cinerea, we next evaluated how overexpression of DEWAX in a Brassica crop, C. sativa, affects disease resistance. The 35SP-DEWAX construct harboring DEWAX gene under the control of CaMV 35S promoter was used in the transformation of C. sativa (Figure 6A). Transgenic plants were screened through spraying of 0.03% BASTA herbicide (Figure 6B). Genomic DNA PCR analysis of non-transgenic (NT) and 16 transgenic plants (TO) using gene-specific primers (Supplementary Table S1) showed that Arabidopsis DEWAX was integrated in all transgenic plants tested (Supplementary Figure S2A). RT-PCR analysis revealed that the DEWAX gene was overexpressed in the leaves of all transgenic plants tested (Supplementary Figure S2B and Table S1).
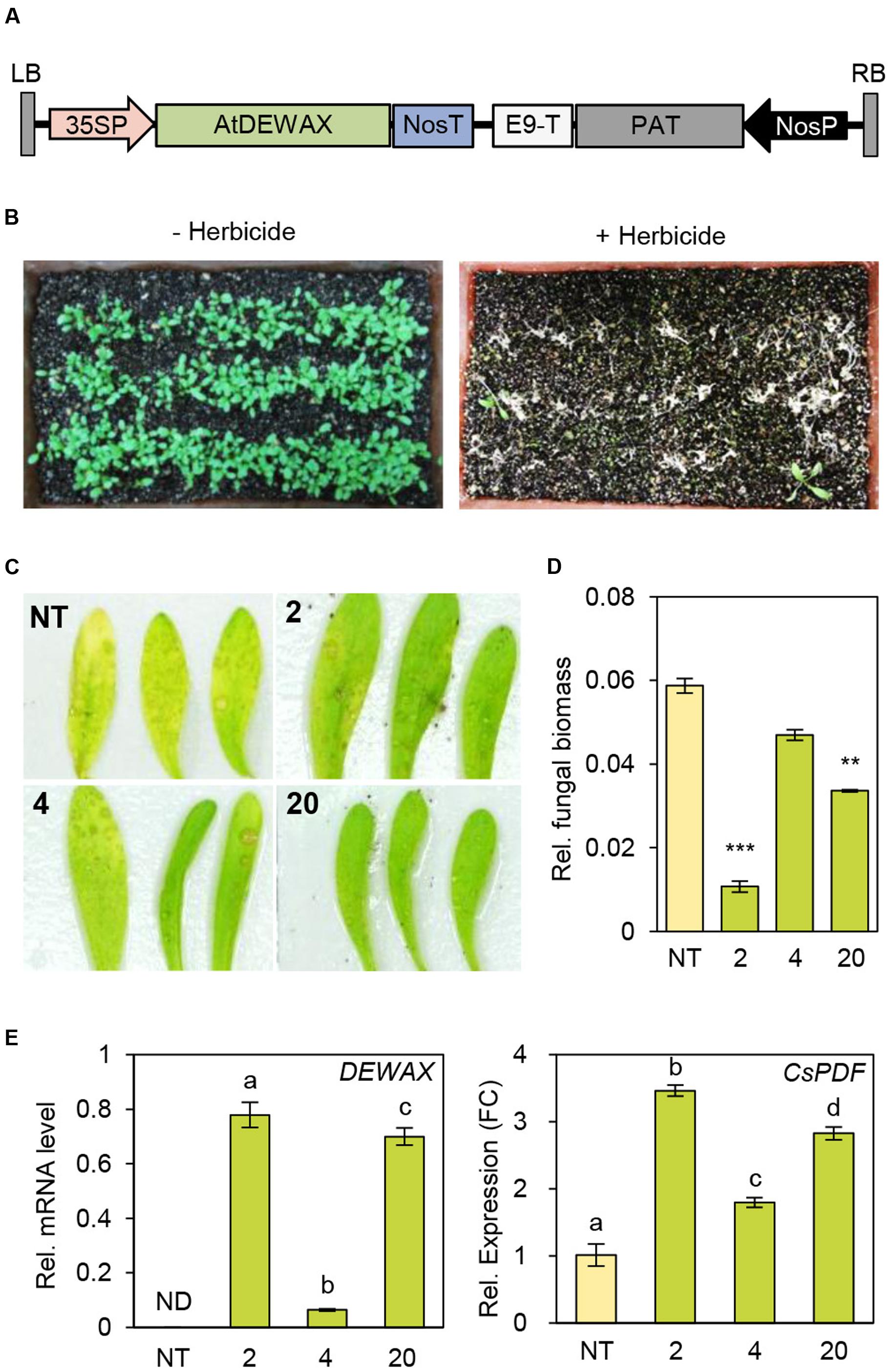
FIGURE 6. Generation of transgenic Camelina sativa plants overexpressing DEWAX. (A) Schematic diagram of the 35SP-DEWAX construct. LB, left border; 35SP, cauliflower mosaic virus 35S promoter; NosT, nopaline synthase terminator; E9-T, pea rbcs-E9 terminator; PAT, phosphinothricin acetyltransferase; NosP, nopaline synthase promoter; RB, right border. (B) Selection of transgenic C. sativa lines resistant to the herbicide BASTA. (C) Macroscopic symptoms on leaves of non-transgenic (NT) and DEWAX C. sativa OX plants (2, 4, and 20) at 4 days after inoculation with B. cinerea. (D) Quantification of B. cinerea biomass in NT and DEWAX C. sativa OX plants. The infected plants were collected at 4 days post inoculation, and genomic DNA was isolated. B. cinerea biomass was quantified by real-time quantitative PCR with B. cinerea cutinase A-specific primers and normalized to C. sativa iASK gene expression. All data represent the average of at least three plants. Error bars represent ± SD of the means. Data were statistically analyzed using Student’s t-test (∗∗P < 0.01, ∗∗∗P < 0.001). (E) qRT-PCR analysis of DEWAX and C. sativa PDF (CsPDF) genes. Total RNA was isolated from leaves of 5-week-old NT and DEWAX C. sativa OX plants and subjected to qRT-PCR analysis. The CsActin11 gene (Hutcheon et al., 2010) was used as a reference to determine the RNA quality and quantity. Data were statistically analyzed using Student’s t-test (∗∗P < 0.01). Error bars indicate ± SD from triplicate experiments.
Next, we examined the defense responses of NT and TO plants to B. cinerea. Visible disease symptoms, B. cinerea biomass, and the expression of Arabidopsis DEWAX were investigated in leaves of NT and TO-2, TO-4, TO-17, and TO-20 lines 4 days after inoculation with B. cinerea. Yellowing or chlorosis occurred more quickly in NT than in TO leaves (Figure 6C). To measure B. cinerea biomass, genomic DNA was isolated from the leaves 4 days after inoculation with B. cinerea and subjected to real-time quantitative PCR using B. cinerea-specific cutinase A-specific gene primers (Supplementary Table S1), while C. sativa GSK3/shaggy-like (iASK) gene was used for normalization. Compared to NT plants, all TO lines tested had lower B. cinerea biomass (Figure 6D). Subsequently quantitative RT-PCR analysis showed that the levels of C. sativa PDF increased by 2- to 3.5-fold in DEWAX OX lines compared with NT (Figure 6E). These results indicated that overexpression of Arabidopsis DEWAX confers tolerance to B. cinerea in transgenic C. sativa.
Discussion
Plant immune responses are essential for not only growth and development, but also productivity and yields of crops. In this study, we investigated a plant defense mechanism related to the cuticle, which is the first defensive barrier of the aerial parts of plants. We revealed that overexpression of DEWAX increased cuticle permeability and ROS accumulation in Arabidopsis. DEWAX is a transcriptional activator that upregulates the expression of the defense-related genes, PDF1.2a, IGMT1, and PRX37, by binding to their gene promoters. In particular, DEWAX directly interacts with the GCC motifs in the PDF1.2a promoter. Overexpression of DEWAX conferred enhanced tolerance to necrotrophic fungal pathogen, B. cinerea in Arabidopsis and C. sativa. Therefore, convergent line of evidence supports that increased ROS levels as well as upregulation of defense-related genes might be related to the resistance to B. cinerea in Arabidopsis and C. sativa overexpressing DEWAX.
Go et al. (2014) reported the AP/ERF transcription factor DEWAX as a transcriptional repressor involved in cuticular wax biosynthesis during daily light/dark cycles. However, we revealed that DEWAX is also able to act as a transcriptional activator that induces defense-related genes in this study. AP2/ERF transcription factors are known to function as both negative and positive regulators (Ohta et al., 2001; Aharoni et al., 2004; Yant et al., 2010). The N-terminus of DEWAX harbors the “EDLL” motif, which is an acidic-type transcriptional activation domain of AP2/ERF transcription factors (Tiwari et al., 2012), which is consistent with our yeast transactivation assay results. Direct binding of DEWAX as well as binding of OCTADECANOID-RESPONSIVE ARABIDOPSIS (ORA)59 and ERF96 with the “EDLL” motif to the two GCC motifs in the PDF1.2 promoter region, is essential for the expression of PDF1.2 (Solano et al., 1998; Zarei et al., 2011; Catinot et al., 2015). Although it remains unclear how exactly the bifunctional DEWAX regulates the expression of its target genes positively or negatively, possibly, DEWAX interacts with a co-activator, such as MED25 of the mediator complex, followed by the recruitment of transcriptional machinery including histone acetyltransferase, or with a co-repressor that interacts with histone deacetylase (Cevik et al., 2012; Huang and Geng, 2017).
The expression of the PDF1.2a, PDF1.2b, IGMT1, PRX37, and PRX38 genes was upregulated in Arabidopsis overexpressing DEWAX. The Arabidopsis mutant iop1 displaying induced expression of PDF1.2 shows enhanced resistance against necrotrophic fungal pathogens, including A. brassicicola, B. cinerea, and Plectosphaerella cucumerina (Penninckx et al., 2003). Overexpression of wasabi defensin, a small and cysteine-rich protein, in rice, potato, and orchid, enhances their resistance to B. cinerea and Magnaporthe grisea (Lay and Anderson, 2005). In addition, mechanical wounding rapidly activates the expression of apoplastic class III peroxidase genes PRX37 and PRX38 and the release of peroxidases to the apoplast, where they generate ROS and oxidize phenolic substrates, resulting in increased cross-linking of the cell wall (Almagro et al., 2009; Pedreira et al., 2011; Minibayeva et al., 2015), suggesting that upregulation of PRX37 and PRX38 by DEWAX may be involved in elevating ROS accumulation in DEWAX OX lines. Moreover, IGMT1 catalyzes the transmethylation of 1-hydroxy-indol-3-yl-methyl glucosinolate intermediates and 4-hydroxy-indol-3-yl-methyl to 1-methoxy-indol-3-yl-methyl and 4-methoxyindol-3-ylmethylglucosinolate, respectively, which are activated by β-thioglucoside glucohydrolase for antifungal defense (Bednarek et al., 2009; Clay et al., 2009). Therefore, the upregulation of PDF1.2, PRX37, PRX38, and IGMT1 by DEWAX might be important for plant innate immunity.
Some Arabidopsis mutants with increased cuticular permeability exhibit resistance to pathogen infection (Nawrath, 2006; Serrano et al., 2014). For example, lcr/lacerate/cyp86A8, bdg, lacs-2 and -3, fdh, fec1, and myb96 showed enhanced resistance to B. cinerea, similar to our observations in DEWAX OX lines (Yephremov et al., 1999; Wellesen et al., 2001; Bessire et al., 2007, 2011; Chassot et al., 2007; Voisin et al., 2009; Seo et al., 2011). We also observed increased hydrogen peroxide levels in DEWAX OX leaves (Figure 1B), which is consistent with the results in the Arabidopsis mutants, bdg and lacs2 (L’Haridon et al., 2011), although we could not exclude the possibility that the higher levels of hydrogen peroxide in DEWAX OX leaves may be attributed, to some extent, to smaller DEWAX OX leaves. Transgenic lines overexpressing oxalate decarboxylase and thus, having higher levels of oxalic acid, which inhibits ROS production, showed decreased innate immune response (Cessna et al., 2000; L’Haridon et al., 2011). Therefore, elevated ROS production in Arabidopsis overexpressing DEWAX might be involved in the resistance to B. cinerea, although the molecular mechanisms underlying immune responses of mutants with permeable cuticle to B. cinerea are still largely unknown.
The expression of PDF1.2 was significantly upregulated in Arabidopsis seedlings treated with jasmonic acid (JA) and ethephon, a form of liquid ethylene (Lorenzo et al., 2003; Pre et al., 2008). The JA- and ethylene-responsive expression of PDF1.2 depended on ERF1 and ORA59 transcription factors, which act as integrators of JA and ethylene signaling pathways. Overexpression of ERF1 and ORA59 caused increased resistance, and ora59 mutant enhanced susceptibility to B. cinerea (Berrocal-Lobo et al., 2002; Pre et al., 2008). However, the transcript levels of DEWAX were elevated neither by JA nor by ethephon1, although DEWAX is classified with ERF1 and ORA59 into the group IX of Arabidopsis AP2/ERF gene family (Nakano et al., 2006). Based on the previous reports that Arabidopsis defense responses to herbivores and pathogens are regulated by circadian rhythms (Wang et al., 2011; Goodspeed et al., 2012), it would be interesting to examine whether upregulation of defense-related genes, including PDF1.2 by the diurnally controlled DEWAX may be important in circadian clock-mediated plant innate immunity. In addition, JA and salicylic acid are known to play a role as an antagonist in plant innate immunity (Thaler et al., 2012). The induction of the PDF1.2 gene in DEWAX-overexpressing Arabidopsis indicates that JA-associated defense responses are activated, and this may cause the suppression of SA-related signaling. This hypothesis supports that DEWAX-overexpressing plants are more resistant to the necrotropic fungal pathogen, B. cinerea, but are more susceptible to the semi-biotropic bacterial pathogen, Pto DC3000 compared with WT.
Although C. sativa is considered to be resistant to many diseases, it is reportedly sensitive to some fungal and bacterial pathogens causing wilting, root rot, clubroot, white rust, and downy mildew diseases (Séguin-Swartz et al., 2009; Zakharchenko et al., 2013). Zakharchenko et al. (2013) reported that transgenic C. sativa overexpressing the antimicrobial peptide cecropin P1 exhibits enhanced resistance to Erwinia carotovora and Fusarium sporotrichioides. In this study, the Arabidopsis dewax mutant showed increased resistance to Pto DC3000, whereas overexpression of DEWAX conferred elevated resistance to B. cinerea in Arabidopsis and C. sativa, suggesting the potential for developing Brassica lines resistant to various diseases. In conclusion, our study revealed that DEWAX is a transcriptional activator that upregulates the expression of defense-related genes, and overexpression of DEWAX enhances the resistance to B. cinerea in Arabidopsis and C. sativa. We provide that DEWAX-mediated defense mechanisms may be applicable in the improvement of crops with enhanced resistance to fungal and bacterial pathogens.
Author Contributions
SJ, YG, and MS conceived and designed research. SJ, YG, and HC conducted experiments. SJ, JP, and MS analyzed data. SJ, JP, and MS wrote the manuscript. All authors read and approved the manuscript.
Funding
This research was supported by grants from the Cooperative Research Program for Agriculture Science and Technology Development (Next-Generation BioGreen 21 Program PJ011052) of the Rural Development Administration, the National Research Foundation (NRF-2016R1A2B2010068) of South Korea, and Korea Institute of Planning and Evaluation for Technology (IPET) of Ministry for Food, Agriculture, Forestry and Fisheries (312033-05), South Korea.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01210/full#supplementary-material
Footnotes
References
Aharoni, A., Dixit, S., Jetter, R., Thoenes, E., van Arkel, G., and Pereira, A. (2004). The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16, 2463–2480. doi: 10.1105/tpc.104.022897
Almagro, L., Gomez Ros, L. V., Belchi-Navarro, S., Bru, R., Ros Barcelo, A., and Pedreno, M. A. (2009). Class III peroxidases in plant defence reactions. J. Exp. Bot. 60, 377–390. doi: 10.1093/jxb/ern277
An, D., and Suh, M. C. (2015). Overexpression of Arabidopsis WRI1 enhanced seed mass and storage oil content in Camelina sativa. Plant Biotechnol. Rep. 9, 137–148. doi: 10.1007/s11816-015-0351-x
An, G. (1987). Binary Ti vectors for plant transformation and promoter analysis. Methods Enzymol. 9, 137–148. doi: 10.1186/1472-6750-10-67
Bednarek, P., Pislewska-Bednarek, M., Svatos, A., Schneider, B., Doubsky, J., Mansurova, M., et al. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323, 101–106. doi: 10.1126/science.1163732
Beisson, F., Li-Beisson, Y., and Pollard, M. (2012). Solving the puzzles of cutin and suberin polymer biosynthesis. Curr. Opin. Plant Biol. 15, 329–337. doi: 10.1016/j.pbi.2012.03.003
Bernard, A., and Joubes, J. (2013). Arabidopsis cuticular waxes: advances in synthesis, export and regulation. Prog. Lipid Res. 52, 110–129. doi: 10.1016/j.plipres.2012.10.002
Berrocal-Lobo, M., Molina, A., and Solano, R. (2002). Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29, 23–32. doi: 10.1046/j.1365-313x.2002.01191.x
Bessire, M., Borel, S., Fabre, G., Carraca, L., Efremova, N., Yephremov, A., et al. (2011). A member of the PLEIOTROPIC DRUG RESISTANCE family of ATP binding cassette transporters is required for the formation of a functional cuticle in Arabidopsis. Plant Cell 23, 1958–1970. doi: 10.1105/tpc.111.083121
Bessire, M., Chassot, C., Jacquat, A. C., Humphry, M., Borel, S., Petetot, J. M. C., et al. (2007). A permeable cuticle in Arabidopsis leads to a strong resistance to Botrytis cinerea. EMBO J. 26, 2158–2168. doi: 10.1038/sj.emboj.7601658
Bird, D., Beisson, F., Brigham, A., Shin, J., Greer, S., Jetter, R., et al. (2007). Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J. 52, 485–498. doi: 10.1111/j.1365-313X.2007.03252.x
Breeden, L., and Nasmyth, K. (1985). Regulation of the yeast HO gene. Cold. Spring Harb. Symp. Quant. Biol. 50, 643–650. doi: 10.1101/SQB.1985.050.01.078
Cameron, K. D., Teece, M. A., and Smart, L. B. (2006). Increased accumulation of cuticular wax and expression of lipid transfer protein in response to periodic drying events in leaves of tree tobacco. Plant Physiol. 140, 176–183. doi: 10.1104/pp.105.069724
Catinot, J., Huang, J. B., Huang, P. Y., Tseng, M. Y., Chen, Y. L., Gu, S. Y., et al. (2015). ETHYLENE RESPONSE FACTOR 96 positively regulates Arabidopsis resistance to necrotrophic pathogens by direct binding to GCC elements of jasmonate - and ethylene-responsive defence genes. Plant Cell Environ. 38, 2721–2734. doi: 10.1111/pce.12583
Cessna, S. G., Sears, V. E., Dickman, M. B., and Low, P. S. (2000). Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. Plant Cell 12, 2191–2200. doi: 10.1105/tpc.12.11.2191
Cevik, V., Kidd, B. N., Zhang, P. J., Hill, C., Kiddle, S., Denby, K. J., et al. (2012). MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol. 160, 541–555. doi: 10.1104/pp.112.202697
Chassot, C., Nawrath, C., and Metraux, J. P. (2007). Cuticular defects lead to full immunity to a major plant pathogen. Plant J. 49, 972–980. doi: 10.1111/j.1365-313X.2006.03017.x
Clay, N. K., Adio, A. M., Denoux, C., Jander, G., and Ausubel, F. M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323, 95–101. doi: 10.1126/science.1164627
Enjalbert, J.-N., Zheng, S., Johnson, J. J., Mullen, J. L., Byrne, P. F., and McKay, J. K. (2013). Brassicaceae germplasm diversity for agronomic and seed quality traits under drought stress. Ind. Crops Prod. 47, 176–185. doi: 10.1016/j.indcrop.2013.02.037
Gachon, C., and Saindrenan, P. (2004). Real-time PCR monitoring of fungal development in Arabidopsis thaliana infected by alternaria brassicicola and Botrytis cinerea. Plant Physiol. Biochem. 42, 367–371. doi: 10.1016/j.plaphy.2004.04.001
Gendrel, A. V., Lippman, Z., Martienssen, R., and Colot, V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2, 213–218. doi: 10.1038/nmeth0305-213
Go, Y. S., Kim, H., Kim, H. J., and Suh, M. C. (2014). Arabidopsis cuticular wax biosynthesis is negatively regulated by the DEWAX gene encoding an AP2/ERF-type transcription factor. Plant Cell 26, 1666–1680. doi: 10.1105/tpc.114.123307
Goodspeed, D., Chehab, E. W., Min-Venditti, A., Braam, J., and Covington, M. F. (2012). Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc. Natl. Acad. Sci. U.S.A. 109, 4674–4677. doi: 10.1073/pnas.1116368109
Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. doi: 10.1007/BF00014672
Horemans, N., Raeymaekers, T., Beek, K. V., Nowocin, A., Blust, R., Broos, K., et al. (2007). Dehydroascorbate uptake is impaired in the early response of Arabidopsis plant cell cultures to cadmium. J. Exp. Bot. 58, 4307–4317. doi: 10.1093/jxb/erm291
Huang, M., and Geng, M. (2017). Exploiting histone deacetylases for cancer therapy: from hematological malignancies to solid tumors. Sci. China Life Sci. 60, 94–97. doi: 10.1007/s11427-016-0300-y
Hutcheon, C., Ditt, R. F., Beilstein, M., Comai, L., Schroeder, J., Goldstein, E., et al. (2010). Polyploid genome of Camelina sativa revealed by isolation of fatty acid synthesis genes. BMC Plant Biol. 10:233. doi: 10.1186/1471-2229-10-233
Jenks, M. A., Andersen, L., Teusink, R. S., and Williams, M. H. (2001). Leaf cuticular waxes of potted rose cultivars as affected by plant development, drought and paclobutrazol treatments. Physiol. Plant. 112, 62–70. doi: 10.1034/j.1399-3054.2001.1120109.x
Katagiri, F., Thilmony, R., and He, S.-Y. (2002). “Arabidopsis thaliana-Pseudomonas syringae interaction,” in The Arabidopsis Book, eds C. Somerville and E. M. Meyerowitz (Rockville, MD: American Society of Plant Biologists).
Kim, H., Choi, D., and Suh, M. C. (2017). Cuticle ultrastructure, cuticular lipid composition, and gene expression in hypoxia-stressed Arabidopsis stems and leaves. Plant Cell Rep. 36, 815–827. doi: 10.1007/s00299-017-2112-5
Kim, H., Park, J. H., Kim, D. J., Kim, A. Y., and Suh, M. C. (2016). Functional analysis of diacylglycerol acyltransferase1 genes from Camelina sativa and effects of CsDGAT1B overexpression on seed mass and storage oil content in C. sativa. Plant Biotechnol. Rep. 10, 141–153. doi: 10.1007/s11816-016-0394-7
Kolattukudy, P. E. (1985). Enzymatic penetration of the plant cuticle by fungal pathogens. Annu. Rev. Phytopathol. 23, 223–250. doi: 10.1146/annurev.py.23.090185.001255
Kolattukudy, P. E., Rogers, L. M., Li, D. X., Hwang, C. S., and Flaishman, M. A. (1995). Surface signaling in pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 92, 4080–4087. doi: 10.1073/pnas.92.10.4080
Kosma, D. K., Bourdenx, B., Bernard, A., Parsons, E. P., Lu, S., Joubes, J., et al. (2009). The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol. 151, 1918–1929. doi: 10.1104/pp.109.141911
Kunst, L., and Samuels, L. (2009). Plant cuticles shine: advances in wax biosynthesis and export. Curr. Opin. Plant Biol. 12, 721–727. doi: 10.1016/j.pbi.2009.09.009
Kurdyukov, S., Faust, A., Nawrath, C., Bar, S., Voisin, D., Efremova, N., et al. (2006). The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. Plant Cell 18, 321–339. doi: 10.1105/tpc.105.036079
Lay, F. T., and Anderson, M. A. (2005). Defensins–components of the innate immune system in plants. Curr. Protein Pept. Sci. 6, 85–101. doi: 10.2174/1389203053027575
Lee, H. W., Kim, M. J., Kim, N. Y., Lee, S. H., and Kim, J. (2013). LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis. Plant J. 73, 212–224. doi: 10.1111/tpj.12013
Lee, S. B., Go, Y. S., Bae, H. J., Park, J. H., Cho, S. H., Cho, H. J., et al. (2009). Disruption of glycosylphosphatidylinositol-anchored lipid transfer protein gene altered cuticular lipid composition, increased plastoglobules, and enhanced susceptibility to infection by the fungal pathogen Alternaria brassicicola. Plant Physiol. 150, 42–54. doi: 10.1104/pp.109.137745
Lee, S. B., Kim, H., Kim, R. J., and Suh, M. C. (2014). Overexpression of Arabidopsis MYB96 confers drought resistance in Camelina sativa via cuticular wax accumulation. Plant Cell Rep. 33, 1535–1546. doi: 10.1007/s00299-014-1636-1
Lee, S. B., and Suh, M. C. (2013). Recent advances in cuticular wax biosynthesis and its regulation in Arabidopsis. Mol Plant. 6, 246–249. doi: 10.1093/mp/sss159
Lee, S. B., and Suh, M. C. (2015). Advances in the understanding of cuticular waxes in Arabidopsis thaliana and crop species. Plant Cell Rep. 34, 557–572. doi: 10.1007/s00299-015-1772-2
L’Haridon, F., Besson-Bard, A., Binda, M., Serrano, M., Abou-Mansour, E., Balet, F., et al. (2011). A permeable cuticle is associated with the release of reactive oxygen species and induction of innate immunity. PLoS Pathog. 7:e1002148. doi: 10.1371/journal.ppat.1002148
Li, Y., Beisson, F., Koo, A. J., Molina, I., Pollard, M., and Ohlrogge, J. (2007). Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proc. Natl. Acad. Sci. U.S.A. 104, 18339–18344. doi: 10.1073/pnas.0706984104
Li-Beisson, Y., Shorrosh, B., Beisson, F., Andersson, M. X., Arondel, V., Bates, P. D., et al. (2013). Acyl-lipid metabolism. Arabidopsis Book 11:e0161. doi: 10.1199/tab.0161
Liu, X., Brost, J., Hutcheon, C., Guilfoil, R., Wilson, A. K., Leung, S., et al. (2012). Transformation of the oilseed crop Camelina sativa by Agrobacterium-mediated floral dip and simple large-scale screening of transformants. In Vitro Cell Dev. Biol. 48, 462–468. doi: 10.1007/s11627-012-9459-7
Lorenzo, O., Piqueras, R., Sanchez-Serrano, J. J., and Solano, R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15, 165–178. doi: 10.1105/tpc.007468
Lu, C., and Kang, J. (2008). Generation of transgenic plants of a potential oilseed crop Camelina sativa by Agrobacterium-mediated transformation. Plant Cell Rep. 27, 273–278. doi: 10.1007/s00299-007-0454-0
McFarlane, H. E., Shin, J. J., Bird, D. A., and Samuels, A. L. (2010). Arabidopsis ABCG transporters, which are required for export of diverse cuticular lipids, dimerize in different combinations. Plant Cell 22, 3066–3075. doi: 10.1105/tpc.110.077974
Minibayeva, F., Beckett, R. P., and Kranner, I. (2015). Roles of apoplastic peroxidases in plant response to wounding. Phytochemistry 112, 122–129. doi: 10.1016/j.phytochem.2014.06.008
Miura, K., Jin, J. B., Lee, J., Yoo, C. Y., Stirm, V., Miura, T., et al. (2007). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19, 1403–1414. doi: 10.1105/tpc.106.048397
Moser, B. R. (2010). Camelina (Camelina sativa L.) oil as a biofuels feedstock: golden opportunity or false hope?. Lipid Technol. 22, 270–273. doi: 10.1002/lite.201000068
Murashuge, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Nawrath, C. (2006). Unraveling the complex network of cuticular structure and function. Curr. Opin. Plant Biol. 9, 281–287. doi: 10.1016/j.pbi.2006.03.001
Ohta, M., Matsui, K., Hiratsu, K., Shinshi, H., and Ohme-Takagi, M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13, 1959–1968. doi: 10.1105/TPC.010127
Panikashvili, D., Savaldi-Goldstein, S., Mandel, T., Yifhar, T., Franke, R. B., Hofer, R., et al. (2007). The Arabidopsis DESPERADO/AtWBC11 transporter is required for cutin and wax secretion. Plant Physiol. 145, 1345–1360. doi: 10.1104/pp.107.105676
Panikashvili, D., Shi, J. X., Schreiber, L., and Aharoni, A. (2011). The Arabidopsis ABCG13 transporter is required for flower cuticle secretion and patterning of the petal epidermis. New Phytol. 190, 113–124. doi: 10.1111/j.1469-8137.2010.03608.x
Pedreira, J., Herrera, M. T., Zarra, I., and Revilla, G. (2011). The overexpression of AtPrx37, an apoplastic peroxidase, reduces growth in Arabidopsis. Physiol. Plant. 141, 177–187. doi: 10.1111/j.1399-3054.2010.01427.x
Penninckx, I. A., Eggermont, K., Schenk, P. M., Van den Ackerveken, G., Cammue, B. P., and Thomma, B. P. (2003). The Arabidopsis mutant iop1 exhibits induced over-expression of the plant defensin gene PDF1.2 and enhanced pathogen resistance. Mol. Plant Pathol. 4, 479–486. doi: 10.1046/j.1364-3703.2003.00193.x
Pighin, J. A., Zheng, H. Q., Balakshin, L. J., Goodman, I. P., Western, T. L., Jetter, R., et al. (2004). Plant cuticular lipid export requires an ABC transporter. Science 306, 702–704. doi: 10.1126/science.1102331
Piisilä, M., Keceli, M. A., Brader, G., Jakobson, L., Jõesaar, I., Sipari, N., et al. (2015). The F-box protein MAX2 contributes to resistance to bacterial phytopathogens in Arabidopsis thaliana. BMC Plant Biol. 15:53. doi: 10.1186/s12870-015-0434-4
Pollard, M., Beisson, F., Li, Y., and Ohlrogge, J. B. (2008). Building lipid barriers: biosynthesis of cutin and suberin. Trends Plant Sci. 13, 236–246. doi: 10.1016/j.tplants.2008.03.003
Pre, M., Atallah, M., Champion, A., De Vos, M., Pieterse, C. M., and Memelink, J. (2008). The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147, 1347–1357. doi: 10.1104/pp.108.117523
Putnam, D. H., Budin, J. T., Field, L. A., and Breene, W. M. (1993). “Camelina: a promising low-input oilseed,” in New crops, eds J. Janick and J. E. Simon (New York, NY: Wiley), 314–322.
Raffaele, S., Mongrand, S., Gamas, P., Niebel, A., and Ott, T. (2007). Genome-wide annotation of remorins, a plant-specific protein family: evolutionary and functional perspectives. Plant Physiol. 145, 593–600. doi: 10.1104/pp.107.108639
Schweizer, P., Felix, G., Buchala, A., Müller, C., and Metraux, J. P. (1996). Perception of free cutin monomers by plant cells. Plant J. 10, 331–341. doi: 10.1046/j.1365-313X.1996.10020331.x
Séguin-Swartz, G., Eynck, C., Gugel, R. K., Strelkov, S. E., Olivier, C. Y., Li, J. L., et al. (2009). Diseases of Camelina sativa (false flax). Can. J. Plant Pathol. 31, 375–386. doi: 10.1080/07060660909507612
Seo, P. J., Lee, S. B., Suh, M. C., Park, M. J., Go, Y. S., and Park, C. M. (2011). The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23, 1138–1152. doi: 10.1105/tpc.111.083485
Seo, P. J., and Park, C. M. (2010). MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol. 186, 471–483. doi: 10.1111/j.1469-8137.2010.03183.x
Serrano, M., Coluccia, F., Torres, M., L’Haridon, F., and Metraux, J. P. (2014). The cuticle and plant defense to pathogens. Front Plant Sci. 5:274. doi: 10.3389/fpls.2014.00274
Shepherd, T., and Griffiths, D. W. (2006). The effects of stress on plant cuticular waxes. New Phytol. 171, 469–499. doi: 10.1111/j.1469-8137.2006.01826.x
Solano, R., Stepanova, A., Chao, Q. M., and Ecker, J. R. (1998). Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12, 3703–3714. doi: 10.1101/gad.12.23.3703
Thaler, J. S., Humphrey, P. T., and Whiteman, N. K. (2012). Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 17, 1360–1385. doi: 10.1016/j.tplants.2012.02.010
Tiwari, S. B., Belachew, A., Ma, S. F., Young, M., Ade, J., Shen, Y., et al. (2012). The EDLL motif: a potent plant transcriptional activation domain from AP2/ERF transcription factors. Plant J. 70, 855–865. doi: 10.1111/j.1365-313X.2012.04935.x
Umbach, A. L., Zarkovic, J., Yu, J., Ruckle, M. E., McIntosh, L., Hock, J. J., et al. (2012). Comparison of intact Arabidopsis thaliana leaf transcript profiles during treatment with inhibitors of mitochondrial electron transport and TCA cycle. PLoS ONE 7:e44339. doi: 10.1371/journal.pone.0044339
Veronese, P., Nakagami, H., Bluhm, B., AbuQamar, S., Chen, X., Salmeron, J., et al. (2006). The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18, 257–273. doi: 10.1105/tpc.105.035576
Voisin, D., Nawrath, C., Kurdyukov, S., Franke, R. B., Reina-Pinto, J. J., Efremova, N., et al. (2009). Dissection of the complex phenotype in cuticular mutants of Arabidopsis reveals a role of SERRATE as a mediator. PLoS Genet. 5:e1000703. doi: 10.1371/journal.pgen.1000703
Wang, K., Kang, L., Anand, A., Lazarovits, G., and Mysore, K. S. (2007). Monitoring in planta bacterial infection at both cellular and whole-plant levels using the green fluorescent protein variant GFPuv. New Phytol. 174, 212–223. doi: 10.1111/j.1469-8137.2007.01999.x
Wang, W., Barnaby, J. Y., Tada, Y., Li, H., Tor, M., Caldelari, D., et al. (2011). Timing of plant immune responses by a central circadian regulator. Nature 470, 110–114. doi: 10.1038/nature09766
Waraich, E., Ahmed, Z., Ahmad, R., Ashraf, M., Saifullah Naeem, M., and Rengel, Z. (2013). Camelina sativa, a climate proof crop, has high nutritive value and multiple-uses: a review. Aust. J. Crop Sci. 7, 1551–1559.
Wellesen, K., Durst, F., Pinot, F., Benveniste, I., Nettesheim, K., Wisman, E., et al. (2001). Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different roles of fatty acid omega -hydroxylation in development. Proc. Natl. Acad. Sci. U.S.A. 98, 9694–9699. doi: 10.1073/pnas.171285998
Woloshuk, C. P., and Kolattukudy, P. E. (1986). Mechanism by which contact with plant cuticle triggers cutinase gene expression in the spores of Fusariu solani f. sp. pisi. Proc. Natl. Acad. Sci. U.S.A. 83, 1704–1708.
Xia, Y., Gao, Q. M., Yu, K., Lapchyk, L., Navarre, D., Hildebrand, D., et al. (2009). An intact cuticle in distal tissues is essential for the induction of systemic acquired resistance in plants. Cell Host Microbe 5, 151–165. doi: 10.1016/j.chom.2009.01.001
Xia, Y., Yu, K., Gao, Q. M., Wilson, E. V., Navarre, D., Kachroo, P., et al. (2012). Acyl CoA binding proteins are required for cuticle formation and plant responses to microbes. Front. Plant Sci. 3:224. doi: 10.3389/fpls.2012.00224
Xue, Y., Xiao, S., Kim, J., Lung, S. C., Chen, L., Tanner, J. A., et al. (2014). Arabidopsis membrane-associated acyl-CoA-binding protein ACBP1 is involved in stem cuticle formation. J. Exp. Bot. 65, 5473–5483. doi: 10.1093/jxb/eru304
Yant, L., Mathieu, J., Dinh, T. T., Ott, F., Lanz, C., Wollmann, H., et al. (2010). Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22, 2156–2170. doi: 10.1105/tpc.110.075606
Yeats, T. H., and Rose, J. K. (2013). The formation and function of plant cuticles. Plant Physiol. 163, 5–20. doi: 10.1104/pp.113.222737
Yephremov, A., Wisman, E., Huijser, P., Huijser, C., Wellesen, K., and Saedler, H. (1999). Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell 11, 2187–2201. doi: 10.1105/tpc.11.11.2187
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zakharchenko, N. S., Kalyaeva, M. A., and Buryanov, Y. I. (2013). Expression of cecropin P1 gene increases resistance of Camelina sativa (L.) plants to microbial phytopathogens. Genetika 49, 523–529. doi: 10.1134/s102279541305013x
Zarei, A., Korbes, A. P., Younessi, P., Montiel, G., Champion, A., and Memelink, J. (2011). Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol Biol. 75, 321–331. doi: 10.1007/s11103-010-9728-y
Keywords: AP2/ERF-type transcription factor, Arabidopsis thaliana, Botrytis cinerea, Camelina sativa, DEWAX, transcriptional regulation
Citation: Ju S, Go YS, Choi HJ, Park JM and Suh MC (2017) DEWAX Transcription Factor Is Involved in Resistance to Botrytis cinerea in Arabidopsis thaliana and Camelina sativa. Front. Plant Sci. 8:1210. doi: 10.3389/fpls.2017.01210
Received: 22 March 2017; Accepted: 26 June 2017;
Published: 11 July 2017.
Edited by:
Chandrashekhar Pralhad Joshi, Michigan Technological University, United StatesReviewed by:
Aardra Kachroo, University of Kentucky, United StatesTaras P. Pasternak, Albert Ludwig University of Freiburg, Germany
Copyright © 2017 Ju, Go, Choi, Park and Suh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mi Chung Suh, bWNzdWhAY2hvbm5hbS5hYy5rcg==
†Present address: Young Sam Go, Advanced Radiation Technology Institute, Korea Atomic Energy Research Institute, 29 Geumgu-gil, Jeongeup-si Jeollabuk-do, South Korea
 Seulgi Ju
Seulgi Ju Young Sam Go
Young Sam Go Hyo Ju Choi
Hyo Ju Choi Jeong Mee Park
Jeong Mee Park Mi Chung Suh
Mi Chung Suh