- 1Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 2Sustainability Department, Traditional Medicinals, Sebastopol, CA, United States
- 3Institute of Botany, Chinese Academy of Sciences, Beijing, China
- 4Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing, China
Artemisinin is the first-line drug for anti-malaria recommended by the World Health Organization (WHO). As the sole natural plant source of artemisinin, ecotypes of Artemisia annua L. vary widely in artemisinin content between nations, and China is the main producing area of A. annua. Here we present a three-dimensional evaluation on ecotypic diversity of A. annua from 12 main producing areas in China using high-performance liquid chromatography coupled with evaporative light scattering detection (HPLC-ELSD) method, DNA barcoding and ecological analyses. The results indicated that A. annua exhibited high ecotypic diversity. A. annua grown in the South of the Qinling Mountains-Huaihe River Line had a high artemisinin content, whereas the northern ones were low. Similar pattern was noted in the genetic diversity. The southern A. annua had high intraspecific variation in contrast to the northern A. annua. In terms of ecological analyses, humidity and sunshine time could be the major limiting ecological factors that affect the accumulation of artemisinin. This is the first reported three-dimensional evaluation integrating chemical, molecular and ecological analyses of the ecotypic diversity of A. annua. The work will facilitate exploring the genetic basis of chemical variations and developing strategies for the breeding and cultivation of high quality A. annua.
Introduction
Artemisinin, an endoperoxide sesquiterpene lactone, is effective against drug-resistant malaria. The ‘high efficacy, fast-action and low-toxicity’ artemisinin-based combination therapy (ACT) is recommended by World Health Organization (WHO) as the best choice for acute malaria (Douglas et al., 2010). Researches show that ACT has a cure rate up to 97% against severe falciparum malaria (Dondorp et al., 2005; WHO, 2012). Artemisia annua L. (sweet or annual wormwood, member of Asteraceae family) is the only plant that synthesizes and accumulates artemisinin (Chinese Pharmacopoeia Commission, 2015; Su and Miller, 2015). A. annua is native of Asia, and now it is widely dispersed throughout the world, including America, Australia, Africa, Bulgaria, France, Argentina, Spain and Hungary (Knudsmark Jessing et al., 2014). The plant has been established as crop in agriculture after the statement of WHO, as a valuable component of combinatorial therapy for malaria (Ferreira, 2007). A. annua is at present the sole commercial source for artemisinin production since the chemical synthesis and biosynthesis is complicated and uneconomical (Corsello and Garg, 2015). Similar to many other secondary metabolites, trace amounts of artemisinin at a range of 0.01 to 1.4% of dry leaf weight of A. annua result in a low ratio of production to demand of this effective drug (Liu et al., 2006). Previous researches show that the Chinese germplasm has higher artemisinin contents than those from Europe, North America, Australia and East Africa (Jain et al., 1996; Zhang et al., 2016).
Chinese scientist Youyou Tu was awarded the 2015 Nobel Prize in Physiology or Medicine for her contribution to artemisinin discovery, which is regarded as a significant breakthrough in 20th century tropical medicine and an important health improvement for people of tropical developing countries (Su and Miller, 2015). Tu screened more than 2,000 Chinese herbal remedies to search for drugs with antimalarial activity. An extract from the wormwood plant A. annua proved especially effective while other species of Artemisia did not contain artemisinin. In Tu’s artemisinin isolation trials, artemisinin content of samples from Beijing Province, Shandong Province, Liaoning Province, and Anhui Province of China, were low, whereas samples from the Sichuan Province, Hunan Province, Chongqing Province of China had high artemisinin contents (Tu, 2009; Zhang et al., 2016). The result suggested that the artemisinin content was variable in A. annua from different geographic origins.
It has been long recognized that plants in general exhibit ecotypic population diversification that reflects influence of environment on a variety of morphological, physiological and chemical traits (Oleksyn et al., 1992; Hancock et al., 2011; Reich et al., 2014). The study of diversification and ultimately speciation is central to evolution and relevant for biology conservation (Weissing et al., 2011). Biogeographic variation has been extensively documented. For instance, Asian cultivated rice consists of two important ecotypes, upland and irrigated, that have, respectively, adapted to either dry land or irrigated cultivation. Upland rice contains abundant drought-related adaptation genes (Lyu et al., 2014). Moringa oleifera Lam. ecotypes show a diverse growth performance and leaf mass production (Forster et al., 2015). Our previous research shows that Panax quinquefolium L. producing in China can be divided into two chemoecotypes: ginsenoside Rb1-Re from outside Shanhaiguan, and ginsenoside Rg2-Rd from inside Shanhaiguan (Huang et al., 2013). However, although evidence shows that ecotypic variation occurs is abundant (Adams et al., 2016; Perreault-Payette et al., 2017), comprehensive characterization of ecologically important intraspecific variation across biogeographic gradients is lacking, greatly hindering progress in the breeding and cultivation of quality varieties. As the demand for artemisinin remains high worldwide, exploring the chemical and genetic intraspecific variation especially knowledge about its genetic mechanisms for high-yielding become more and more important. Public interest has grown in recent decades; however, the market is not always regulated. Consequently, it is also important to develop accurate and efficient authentication methods of differentiating A. annua.
In the present paper, we aim to (i) estimate the chemical variation pattern of A. annua from different regions in China using high-performance liquid chromatography coupled with evaporative light scattering detection (HPLC-ELSD), (ii) reveal intraspecific variation of A. annua based on DNA barcoding analysis using four regions (ITS2, psbA-trnH, matK and rbcL), and differentiate A. annua from its adulterants, and (iii) find possible ecological factors that affect the artemisinin yield. The results will facilitate exploring the genetic basis of chemical variations and developing strategies for the utilization and conservation of A. annua.
Materials and Methods
Materials
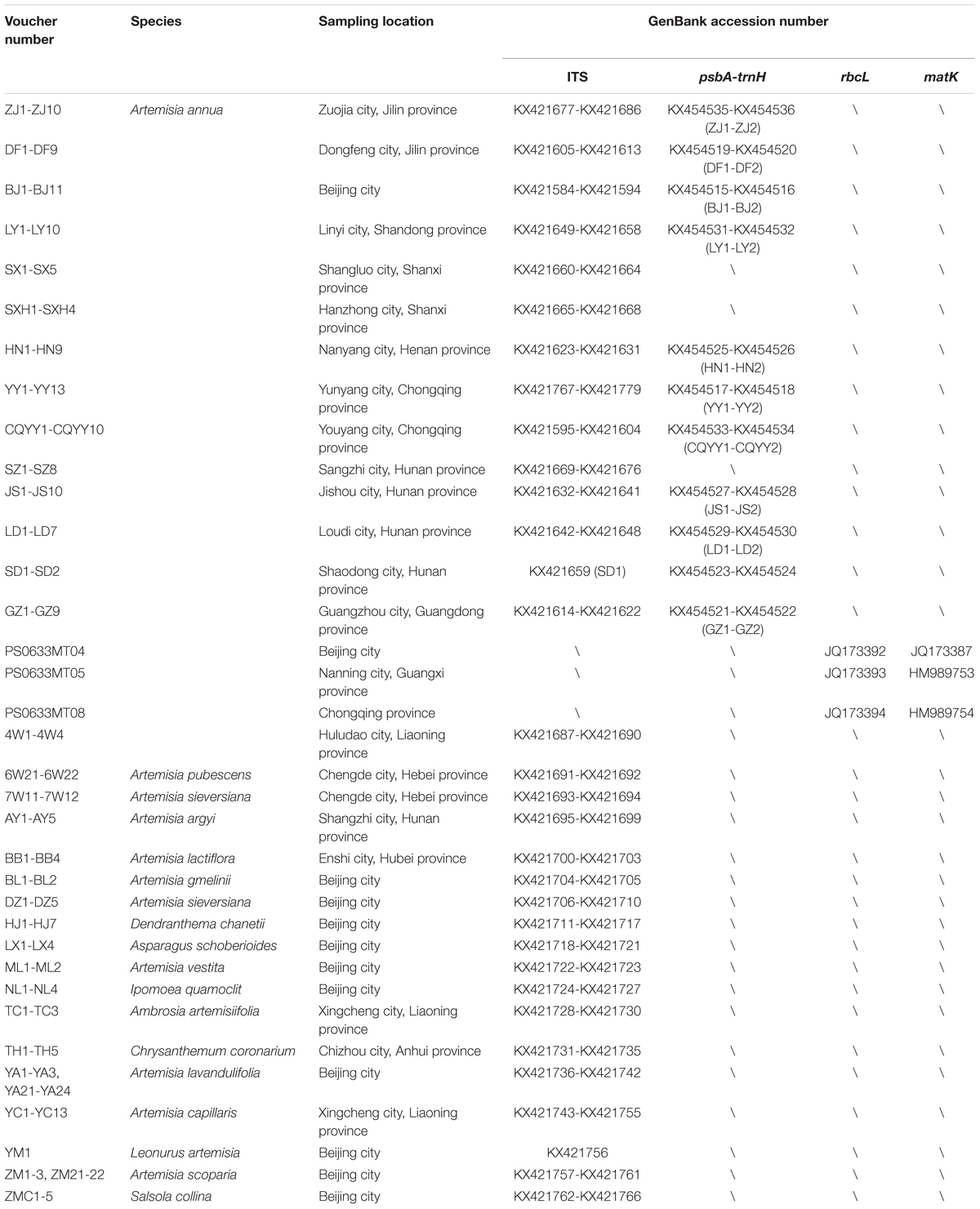
A total of 102 A. annua samples for chemical detection were collected from the following 12 main producing areas in China: (1) North of Qinling Mountains-Huaihe River Line: Zuojia city, Jilin province; Dongfeng city, Jilin province; Beijing city; Linyi city, Shandong province; Shangluo city, Shaanxi province; Hanzhong city, Shaanxi province; (2) South of Qinling Mountains-Huaihe River Line: Sangzhi city, Hunan province; Jishou city, Hunan province; Shaodong city, Hunan province; Loudi city, Hunan province; Yunyang city, Chongqing province and Guangzhou city, Guangdong province. The 116 A. annua samples for genetic variation analysis were collected from Nanyang city, Henan province, Youyang city, Chongqing province and all the above sampling sites (Nanyang and Youyang are all located in the South of Qinling Mountains-Huaihe River Line). We also downloaded 3 rbcL and 3 matK sequences of A. annua from GenBank. The 80 adulterant samples for the identification analysis included 18 species belonging to 8 genera (Artemisia, Dendranthema, Asparagus, Quamoclit, Ambrosia, Chrysanthemum, Leonurus, Salsola). The samples were all cultivars collected in 2015 by Prof. Linfang Huang. The details of these samples are listed in Table 1. All the samples were authenticated by Prof. Bing Liu at the Institute of Botany, the Chinese Academy of Sciences (IB-CAS). All corresponding voucher specimens were deposited to the Herbarium of the Institute of Medicinal Plant Development (IMPLAD) at the Chinese Academy of Medical Sciences in Beijing, China.
Artemisinin Determination
In this study, The artemisinin in A. annua was quantified by high-performance liquid chromatography coupled with evaporative light scattering detection (HPLC-ELSD). Acetonitrile and methanol (HPLC grade) for HPLC analysis were bought from Fisher (Fair Lawn, NJ, United States). Deionized water was obtained from Millipore Milli-Q purification system (Millipore, Bedford, MA, United States). HPLC-ELSD analyses were performed with a 1525 HPLC System (Waters, United States) equipped with an autosampler, a binary pump, a vacuum degasser, a thermostated column compartment and a evaporative light scattering detection (Alltech, United States).
The dry plant material of A. annua were grounded to fine particles and milled to 60 mesh powder before extraction. About 1 g A. annua powder sample was extracted in a sealed flask containing 25 mL methanol under room temperature for a given time. Then the suspensions were sonicated with ultrasonic apparatus in a pre-setting procedure for 20 min under 300 W. After extraction, the power was switched off and then the extracts were allowed to cool to room temperature. The extracts were filtered through 0.45 μm nylon membranes prior to injection into HPLC (Hui et al., 2006).
The C18-RP HPLC column (250 mm × 4.6 mm ID; 5.0 μm pore size) was maintained at room temperature. The mobile phase consisted of (A) water-methanol (1:1, v/v) and (B) acetonitrile. The HPLC conditions in the C18 column included the (A):(B) (1:1 v/v) mixture as mobile phase at a flow rate of 1 mL/min. ELSD conditions were optimized at nebulizer-gas flow rate of 2.0 L/min and drift tube temperature of 70°C under the impactor off-mode, and the gain was set at 2 (Avery et al., 1999; Liu C.Z. et al., 2007; Liu J.-L. et al., 2007; Lapkin et al., 2009).
DNA Extraction, PCR Amplification, and Sequencing
The material specimens were dried by natural methods, and 20 mg of dried plant material was used for DNA extraction. Samples were rubbed for 2 min at a frequency of 30 r/s in a FastPrep bead mill (Retsch MM400, Germany). Genomic DNA was isolated using the Plant Genomic DNA Kit (Tiangen Biotech Co., China), according to the DNA extraction protocol. We made the following modifications to the protocol: chloroform was diluted with isoamyl alcohol (24:1), and the buffer solution GP2 was diluted with isopropanol (same volume). The powdered sample was mixed with 700 μL of 65°C GP1 and 1 μL of β-mercaptoethanol for 10–20 s before the mixture was incubated for 60 min at 65°C. Then, 700 μL of a chloroform: isoamyl alcohol mixture was added, and the solution was centrifuged for 5 min at 12, 000 rpm (∼13400 × g). After removed and transferred into a new tube, the supernatant was added 700 μL isopropanol and mixed for 15–20 min. The mixture was centrifuged in CB3 spin columns for 40 s at 12,000 rpm. The filtrate was discarded and 500 μL GD (adding quantitative anhydrous ethanol before use) was added before centrifugation at 12,000 rpm for 40 s. The filtrate was discarded, and 700 μL PW buffer (quantitative anhydrous ethanol was added before use) was used to wash the membrane before centrifugation for 40 s at 12,000 rpm. This step was repeated with 500 μL PW, followed by a final centrifugation step for 2 min at 12,000 rpm to remove the residual wash buffer. The spin column was dried at room temperature for 3–5 min and then centrifuged for 2 min at 12,000 rpm to obtain the total DNA.
General PCR reaction conditions and universal DNA barcode primers were used for the ITS barcodes, as presented in Supplementary Table S1. PCR amplification was performed on 25 μL reaction mixtures containing approximately 20–50 ng of genomic DNA, 8.5 μL of distilled deionized water, 12.5 μL of 2 × Taq PCR Master Mix (Beijing TransGen Biotech Co., China), and 1/1 μL of the forward/reverse (F/R) primers (2.5 μM). The reaction mixtures were amplified in a 9700 GeneAmp PCR system (Applied Biosystems, United States). The amplified products were purified using electrophoresis on 1% agarose gels. The purified PCR products were sequenced in both directions by an ABI 3730XL sequencer (Applied Biosystems, United States).
Climatic Data Collection and Data Analysis
The climatic data were collected from the China meteorological data sharing service system (Website)1. The data of 21 climatic factors from 1981 to 2010 were included in our analysis. These climatic factors are listed below: (1) average temperature in January; (2) average temperature in February; (3) average temperature in March; (4) average temperature in April; (5) average temperature in May; (6) average temperature in June; (7) average temperature in July; (8) average temperature in August; (9) average temperature in September; (10) average air pressure; (11) average annual temperature; (12) relative humidity; (13) total annual precipitation; (14) wind speed; (15) average surface temperature; (16) average sunshine time; (17) average vapor pressure; (18) average date of frost; (19) evaporation capacity; (20) longitude; (21) latitude.
The one-way analysis of variance (ANOVA) for the artemisinin content was conducted by SPSS for Windows (release version 22.0; SPSS Institute, Cary, NC, United States). PCA for climatic factors and correlation analysis between artemisinin content and climatic factors were also performed with the SPSS. The level of statistical significance was set at p-value < 0.05. The DNA sequencing peak diagrams were obtained and proofread before the contigs were assembled with CodonCode Aligner 5.0.1 (CodonCode Co., United States). All of the sequences were aligned using ClustalW. The NJ trees were constructed based on the Kimura 2-parameter model (Kimura, 1980), and bootstrap tests were conducted with 1,000 repeats to assess the confidence of the phylogenetic relationships by MEGA 6.0 software (Tamura et al., 2013).
Results
Chemical Analysis
The contents of artemisinin in A. annua obtained from different locations were determined and analyzed. As shown in Figure 1, A. annua grown in the South of the Qinling Mountains-Huaihe River Line (SA) contained an average artemisinin content of 1.52%. In contrast, A. annua grown in the North of the Qinling Mountains-Huaihe River Line (NA) had an extremely low artemisinin yield which was below the detection limit. As the center of Wuling Mountain region, samples from Sangzhi and Jishou showed greater artemisinin contents of 1.89 and 1.85%, respectively, forming a distribution pattern that the artemisinin reduced from the Wuling Mountain region to the periphery. A one-way analysis of variance (ANOVA) was employed to perform statistical analysis of the result. It indicated that Group SA and Group NA were significantly different in their content of artemisinin (p < 0.05). The results were presented in Supplementary Table S2.
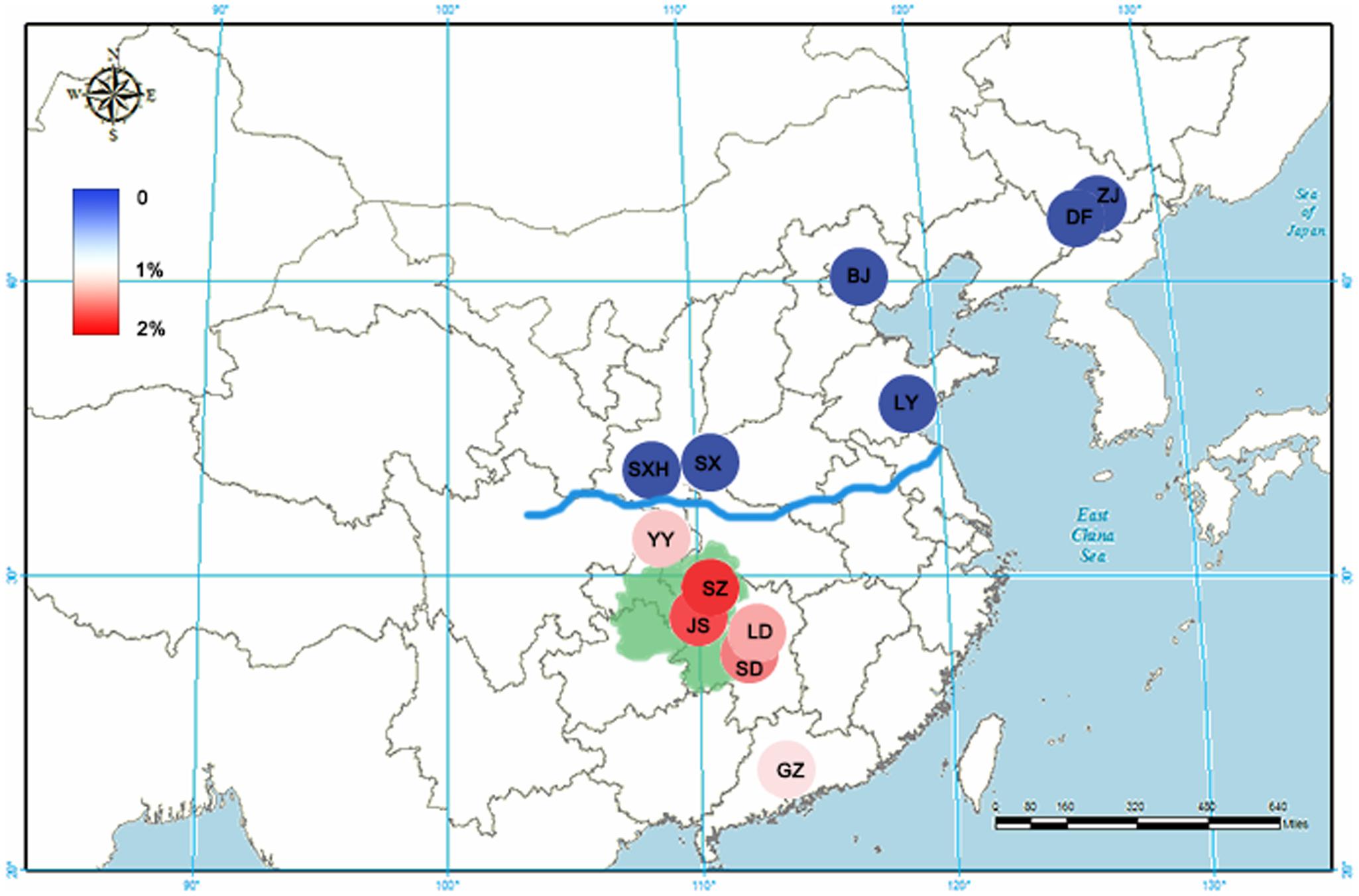
FIGURE 1. The artemisinin content for Artemisia annua from 12 main producing areas in China. The artemisinin contents are depicted with an increasingly stronger blue or red color. The blue line represents the Qinling Mountains-Huaihe River Line. The green zone represents the Wuling Mountain region. ZJ, Zuojia city; DF, Dongfeng city; BJ, Beijing city; LY, Linyi city; SX, Shangluo city; SXH, Hanzhong city; SZ, Sangzhi city; JS, Jishou city; SD, Shaodong city; LD, Loudi city; YY, Yunyang city; GZ, Guangzhou city.
Genetic Variation Analysis of A. annua
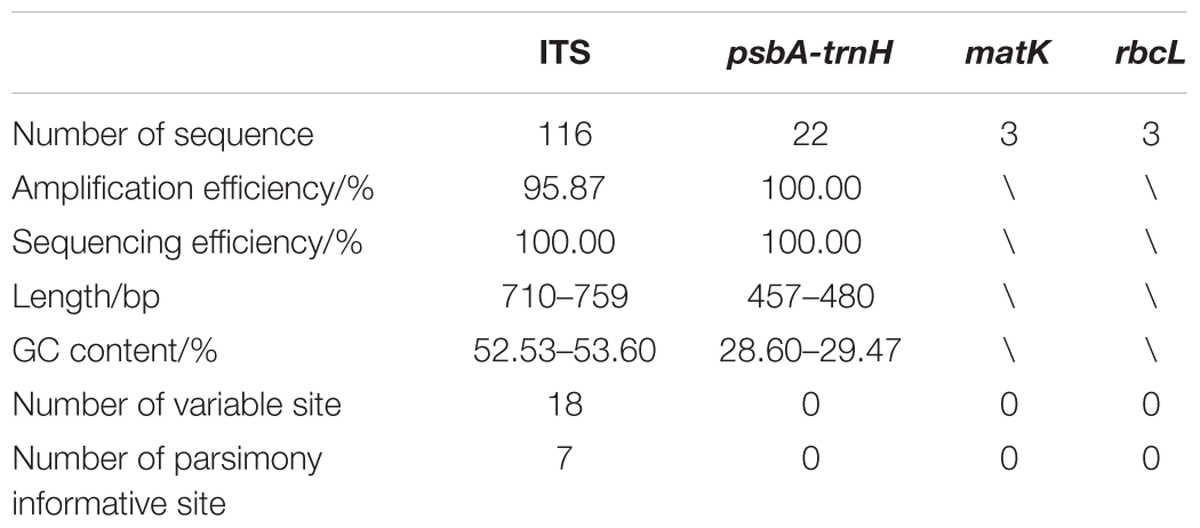
To reveal the intraspecific polymorphisms in A. annua from different producing areas, the ITS2 and psbA-trnH sequences were successfully amplified and sequenced. In addition, another two regions, rbcL and matK, were downloaded from GenBank. The characteristics of the four barcodes were listed in Table 2.
ITS
A total of 116 high-quality bidirectional ITS sequences of A. annua were aligned. There were 18 variable sites and 7 parsimony-informative sites in ITS region that segregated as 13 sequence haplotypes. The variations were generally nucleotide C to nucleotide T, though few variations were A nucleotides to nucleotide G (Figure 2A). The main variants with occurrences greater than 2 were listed in Figure 2B. For SA, samples from Wuling Mountain region (Yunyang city, Jishou city, and Youyang city) had high intraspecific variation, which was consistent with the distribution pattern of artemisinin. A1 and A3 represented approximately 85% of the haplotypes, indicating them may be the major ITS haplotype of SA. For NA, a low intraspecific divergence was observed. However, a single nucleotide polymorphism (SNP) sites was found in NA, which was a T nucleotide variation at position 148. The haplotype A2, which represents this variation, coupled with A1 were the dominant haplotype of NA.
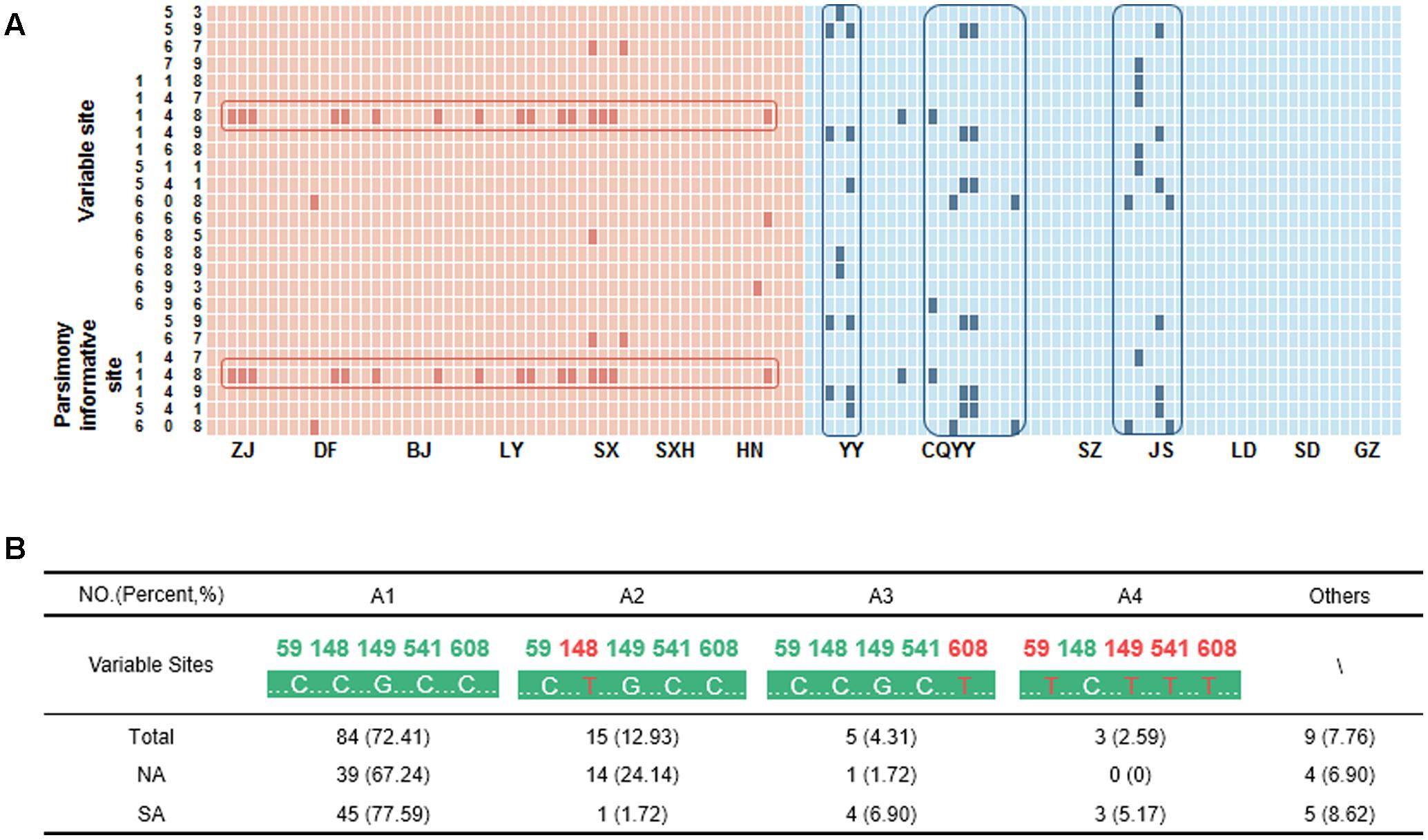
FIGURE 2. The variable sites and haplotypes for A. annua. (A) The pink and blue block represent nucleotides in A. annua from the South (SA) and North (NA) of the Qinling Mountains-Huaihe River Line, respectively. The deep pink and deep blue block represent the variable sites. HN, Nanyang city; CQYY, Youyang city. (B) The main haplotypes of A. annua. The pink nucleotides represent the variable sites.
psbA-trnH, matK, and rbcL
Regions of the psbA-trnH spacer were successfully amplified and sequenced from 22 A. annua samples (100%). The results showed that the psbA-trnH regions of all the individuals within the A. annua were completely conserved. No variable sites were found among them. The same result was observed upon examination of matK and rbcL sequences of A. annua obtained from GenBank, which indicated that the matK and rbcL genes had a low intraspecific divergence in this species as well.
Identification Analysis of A. annua and Its Adulterants
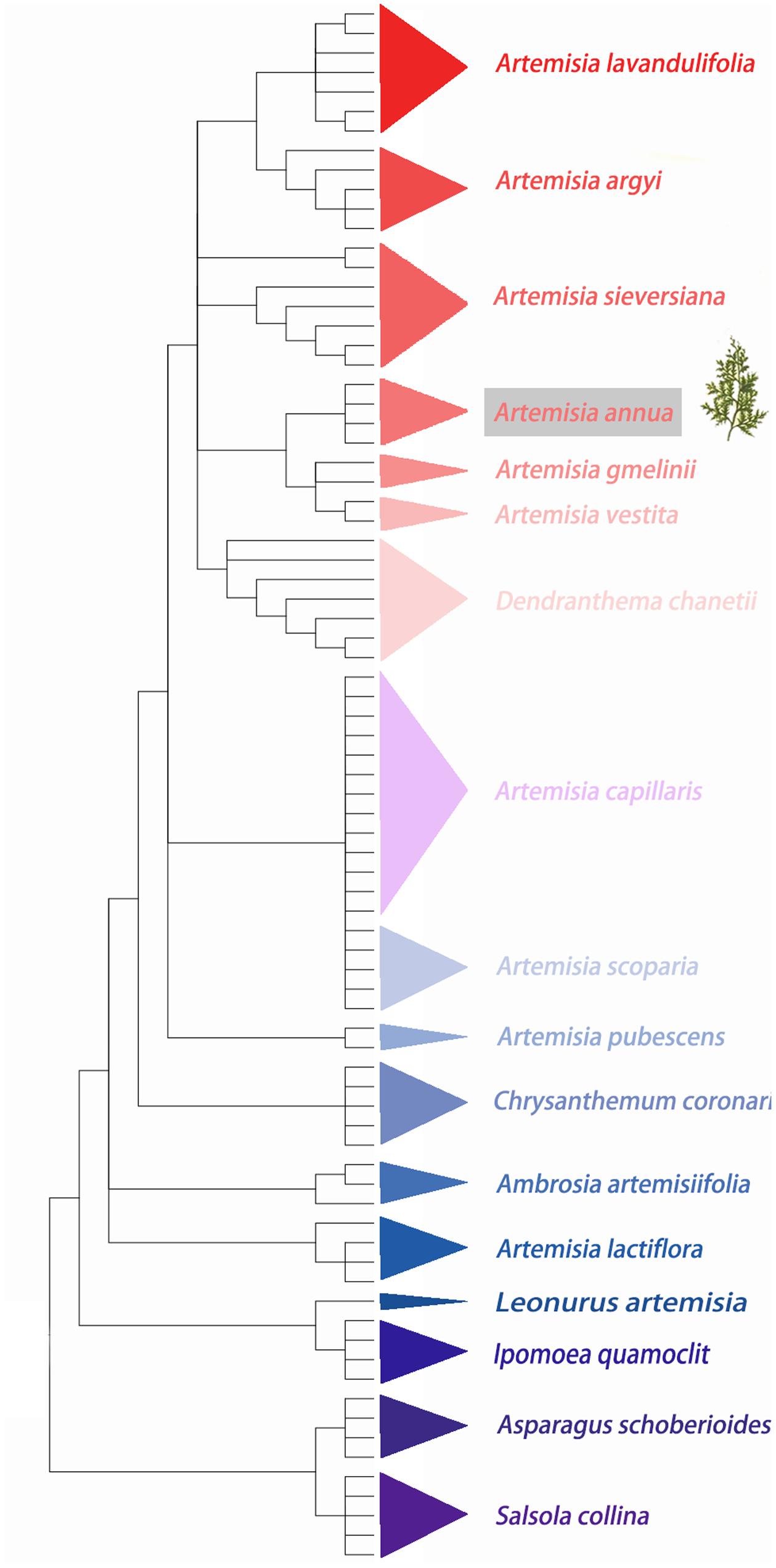
A total 80 ITS sequences from 18 species that belonged to 8 genera (Artemisia, Dendranthema, Asparagus, Quamoclit, Ambrosia, Chrysanthemum, Leonurus, and Salsola) were analyzed. The ITS sequences varied in length from 509 to 731 bp, and the GC content varied from 51.4 to 66.6%. Nearest genetic distance, BLAST and neighbor-joining (NJ) tree methods were used to estimate the capability of species authentication by ITS. Sequence divergences represented by six parameters (Table 3) showed that the minimum inter-specific distance was much more than the coalescent depth (maximum intra-specific distance) (Table 3), indicating that A. annua can be easily distinguished from these 17 adulterants based on the ITS sequences by K2P distance method. NJ tree result also showed that every species clustered into their own clade and A. annua can be discriminated from other included taxa (Figure 3). Moreover, the successful identification of differences at the species level was 86.3% for BLAST1 and 43.8% for the nearest distance method.

TABLE 3. Analyses of intra- and inter-specific divergence and identification efficiency of ITS sequences.
Ecological Factor Analysis
To explore the differences in ecology among the sampling sites, a principal component analysis (PCA) was employed. Figure 4A showed that the three-component PCA model cumulatively accounted for 97.049% of the variation (PC1, 66.869%; PC2, 18.870%; PC3, 11.310). PC1 was mainly characterized by the average temperature in May, average temperature in June, average temperature in September, average temperature in April. PC2 was correlated with evaporation capacity and relative humidity. Thus, PC1 and PC2 can be defined as temperature factor and humidity factor, respectively. In the score plot (Figure 4B), twelve sampling areas were divided into two groups according to their ecological factors. The SA group fell into a large cluster to the top right, whereas the NA group appeared more scattered at the left bottom. The SA group had a higher temperature and humidity than the NA group. The result matched the natural conditions. According to the correlation analysis results (Figure 4C), relative humidity demonstrated a significantly positive correlation with artemisinin content, whereas the average sunshine time had a crucially negative influence on the artemisinin content. Furthermore, average temperature in August, average temperature in July, average vapor pressure and latitude were also important ecological factors. Generally, humidity and sunshine time were crucial factors that influenced the artemisinin content.
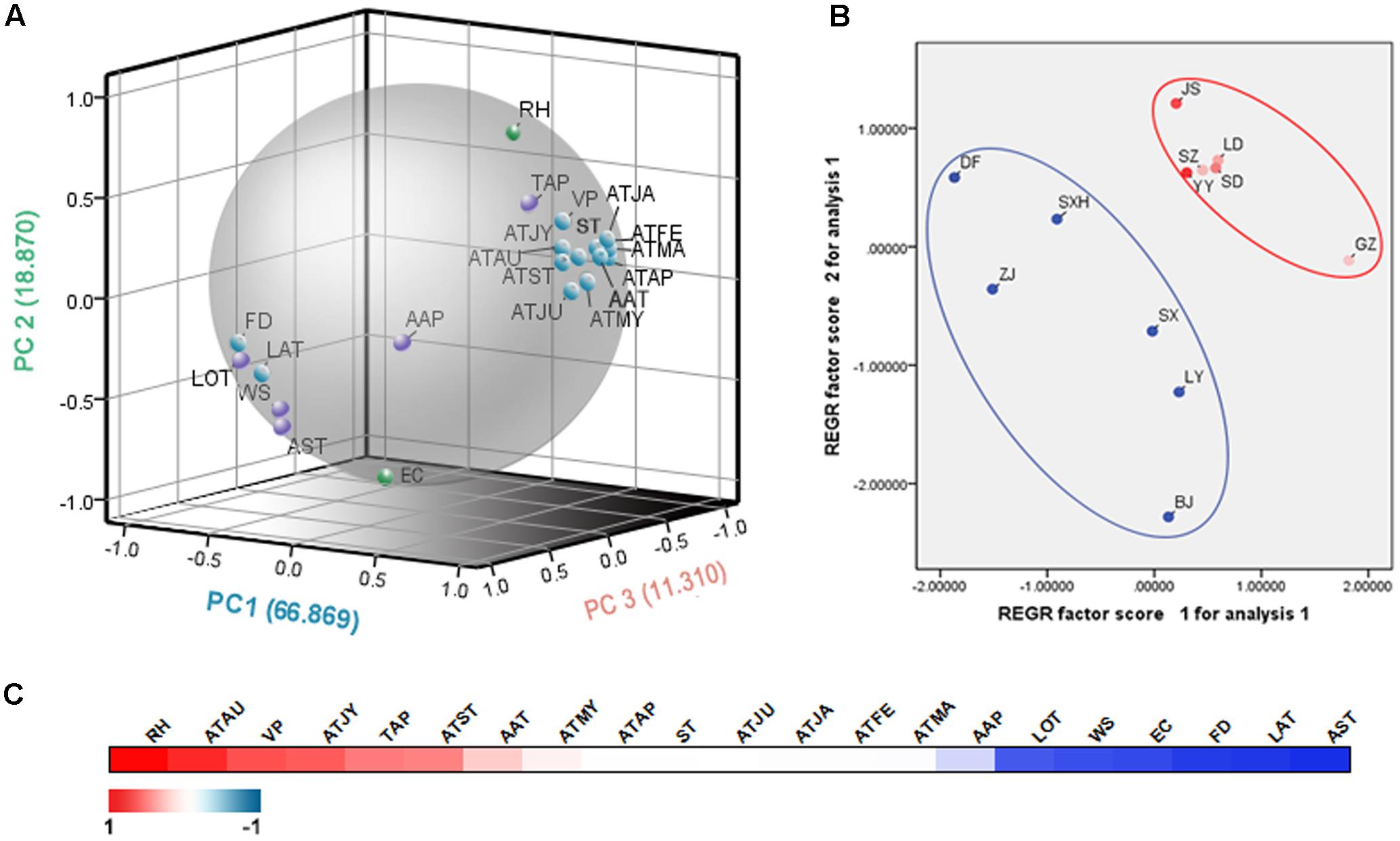
FIGURE 4. The ecological factor analysis of the 12 main producing areas of A. annua. (A) The PCA loading plot of the ecological factors. ATJA, average temperature in January; ATFE, average temperature in February; ATMA, average temperature in March; ATAP, average temperature in April; ATMY, average temperature in May; ATJU, average temperature in June; ATJY, average temperature in July; ATAU, average temperature in August; ATST, average temperature in September; AAP, average air pressure; AAT, average annual temperature; RH, relative humidity; TAP, total annual precipitation; WS, wind speed; ST, average surface temperature; AST, average sunshine time; VP, average vapor pressure; FD, average date of frost; EC, evaporation capacity; LOT, longitude; LAT, latitude. (B) The PCA score plot of the sampling sites. (C) The correlation analysis of ecological factors and artemisinin contents. The correlation coefficients are depicted with an increasingly stronger blue or red color.
Discussion
As the sole plant producing artemisinin, A. annua has gained much attention during the last several decades. High-quality raw materials face an increase in demand. Information regarding A. annua genetic mechanisms for high-yielding and ecological factors benefiting for producing the plant are central to introduction and cultivation. Given that the variation of secondary metabolite concentration in different plant populations may be largely caused by genetic variation and an interplay of environmental conditions, studies on correlation between chemical constituents variation and genetic diversity can provide better understanding in the ecotypic diversity of medicinal plant (Djabou et al., 2011; Ali et al., 2013; Snyder and Worobo, 2014; Zheng et al., 2014; Xiang et al., 2016a,b). In this paper, a chemical and molecular analysis was combined to reveal the ecotypic population diversification of A. annua.
By analyzing the artemisinin content of 102 A. annua samples from 12 main producing areas in China, we demonstrated that artemisinin yield varied in different areas of China. A. annua grown in the South of the Qinling Mountains-Huaihe River Line had a high artemisinin content, whereas the northern ones had an extremely low artemisinin yields. It can be generally concluded that the artemisinin content reduced from the Wuling Mountain region to the periphery, which was consistent with our previous studies (Huang et al., 2010). In addition, according to the previous research, the artemisinin yield of some provinces located South of the Qinling Mountains-Huaihe River Line, such as Jiangsu province and Zhejiang province were between 0 and 1.00% (Wei et al., 2008; Xiang et al., 2012). And the artemisinin yield of some provinces located North of the Qinling Mountains-Huaihe River Line, such as Shanxi province and Henan province were as low as 0.10% (Liu J.-L. et al., 2007). This proves our conclusion to some extent.
In previous studies, different types of molecular markers have been used to ascertain DNA polymorphism of A. annua. Shi applied RAPD and ISSR to analyze the A. annua germplasm resources, and the results showed that A. annua gathered from Hubei Province of China had a high intraspecific divergence (Shi et al., 2008). RAPD analysis also indicated genetic variation amongst A. annua in the Indian population (Sangwan et al., 1999). By using SRAP markers, Chen et al. found that the A. annua germplasm resources in natural populations of China had an extremely abundant genetic diversity, especially the A. annua germplasm in Chongqing Province (Chen et al., 2011). In EST-SSR analysis, a variety of motifs was detected (Wang et al., 2012). In recent years, DNA barcoding has been suggested as a useful molecular marker technique to complement traditional taxonomic methods for biodiversity inventory construction and fast species identification (Hollingsworth, 2011; Stoeckle et al., 2011; Little and Jeanson, 2013; Nithaniyal et al., 2014; Wu et al., 2015). In the present study, SA contained a high intraspecific divergence in the ITS regions, especially samples from Wuling Mountain region. The NA had a low intraspecific divergence, which was consistent with the distribution pattern of artemisinin. However, a significantly low sequence divergence was observed in the three chloroplast regions investigated (psbA-trnH, matK, and rbcL). One of the possible reasons is that chloroplast genes evolve slowly compared with nuclear genes. The conservation of chloroplast DNA sequences can ensure comparability among groups. Consequently, rbcL sequence is generally applied to the classification of higher taxa above the family level, while matK and psbA-trnH sequences are suitable for identifications at the genus and species level (Hebert and Gregory, 2005; Chase et al., 2007; Kress and Erickson, 2007). The ITS sequence has different copy numbers in different taxa, resulting in high existence of variants in the genome (Alvarez and Wendel, 2003; Nieto Feliner and Rossello, 2007). The abundant variations in ITS can be used in the classification of lower-level taxa (genera, species, subspecies) as far as population level. Our research has confirmed it to a certain extent. Although variations among A. annua from different sampling sites are insufficient to divide into species or subspecies, it reflected an internal variability and DNA polymorphism in A. annua and provide information for the optimal utilization and conservation of A. annua germplasm resource. This work also illustrates the current limitations in the applicability of DNA barcoding to taxonomic surveys. Therefore, further studies are required to protocol improvement and development, particularly design of novel universal primers to extend the barcoding to a broader coverage of plant species.
To identify anti-malaria drugs, A. annua was filtrated from more than 2,000 Chinese herbal remedies. The other species from Artemisia have been proved not to contain artemisinin. After a large-scale national survey, Tu found five main commercial adulterants of A. annua: A. apiacea, A. scoparia, A. capillaries, A. japonica, and A. eriopoda (Dondorp et al., 2005). The adulterants share similar morphological characteristics with A. annua, although they show different pharmacological properties and medicinal values (Xiang et al., 2016b). In the present study, the barcoding ITS effectively identified A. annua from other species of Artemisia, which can significantly reduce the adverse effects on the health of consumers caused by herbal product adulteration and contamination in the traditional herbal medicine market.
The plant content of artemisinin is partly determined by biotic factors, such as germplasm, but also by abiotic factors, such as climate (Hancock et al., 2011). In our PCA analysis, the sampling sites belonging to Wuling Mountain region aggregated closely due to their unique geographical factors. As disclosed in the correlation analysis, humidity and sunshine time were the most distinct positive and negative ecological factors for A. annua, respectively. Temperature, especially temperature in August and July also played an important role in the accumulation of artemisinin. Wuling Mountain region lies in a subtropical monsoon climate that provides suitable temperature and abundant precipitation. Because of the mountains, the region is foggy and lacks sunshine. Consequently, the unique environment facilitates the accumulation of artemisinin. As an annual plant, A. annua blossoms in September. The production of artemisinin usually peaks with flowering. Thus, the plant is very responsive to the temperature in August and July. The result is consistent with our previous study (Huang et al., 2010). Qinling Mountains-Huaihe River Line is one of the most important geographical boundaries in China. The significance of the geographic dividing line is reflected in the boundary of (1) North China and South China; (2) humid areas and semi humid areas; (3) subtropical zones and warm temperate zones; and (4) paddy fields and dry land, namely, rice and wheat main production boundary. Ancient Chinese wisdom indicated that for tangerine plants with similar leaves, tangerine plants grown in the South of Huaihe River would produce tangerines, whereas tangerine plants grown in the North of Huaihe River would produce bitter oranges. China is vast in territory with diversified landforms and mountain ranges and, hence, varying in weather patterns. This diversity of natural ecosystems, climatic conditions and edaphic traits produce abundant diversities among botanical and germplasm resources and contribute to different quality levels of medicinal plants. Environmental stress is an external factor that affects the formation of certain geo-authentic herbs (Zhao et al., 2012). The favorable ecological factors indicated the ecological requirements of A. annua and provided useful guidelines for in vitro and ex situ cultivation.
Conclusion
In this study, a three-dimensional method that integrated chemical detection, molecular identification and ecological analyses was developed for evaluating the ecotypic diversity of A. annua for the first time. By this approach, high ecotypic diversity of A. annua was observed. We demonstrated that the artemisinin content reduced from the Wuling Mountain region to the periphery. We also found high level of intraspecific genetic diversity among A. annua grown in the South of the Qinling Mountains-Huaihe River Line. Humidity and sunshine time were the essential ecological factors influencing the artemisinin content. This work documents the climatic adaptation of A. annua and provides useful guidelines for in vitro and ex situ cultivation. Our intensive research on transcriptomics to test the key enzymes in the biosynthesis of artemisinin are ongoing.
Author Contributions
BL and LH collected and authenticated all the samples. LH, LL, SZ, BJ, and SC contributed to the study design. LL and SZ conducted the experiments. LL analyzed all the data and wrote the manuscript. All authors have read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Funding from the National Natural Science Foundation of China (No. 81473315) and Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (No. 2016-12M-3-015) are gratefully acknowledged.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01225/full#supplementary-material
Footnotes
References
Adams, W. W. I. I. I., Stewart, J. J., Cohu, C. M., Muller, O., and Demmig-Adams, B. (2016). Habitat temperature and precipitation of Arabidopsis thaliana ecotypes determine the response of foliar vasculature, photosynthesis, and transpiration to growth temperature. Front. Plant Sci. 7:1026. doi: 10.3389/fpls.2016.01026
Ali, Z., Ganie, S. H., Narula, A., Sharma, M. P., and Srivastava, P. S. (2013). Intra-specific genetic diversity and chemical profiling of different accessions of Clitoria ternatea L. Ind. Crops Prod. 43, 768–773. doi: 10.1016/j.indcrop.2012.07.070
Alvarez, I., and Wendel, J. F. (2003). Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenet. Evol. 29, 417–434. doi: 10.1016/S1055-7903(03)00208-2
Avery, B. A., Venkatesh, K. K., and Avery, M. A. (1999). Rapid determination of artemisinin and related analogues using high-performance liquid chromatography and an evaporative light scattering detector. J. Chromatogr. B Biomed. Sci. Appl. 730, 71–80. doi: 10.1016/S0378-4347(99)00185-1
Chase, M. W., Cowan, R. S., Hollingsworth, P. M., van den Berg, C., Madriñán, S., Petersen, G., et al. (2007). A proposal for a standardised protocol to barcode all land plants. Taxon 56, 295–299.
Chen, D., Peng, R., Li, L., Cui, G., and Wu, Y. (2011). Genetic diversity of Artemisia annua in natural populations of China revealed by SRAP marker. Chin. Tradit. Herb. Drugs 42, 5.
Chinese Pharmacopoeia Commission (2015). Pharmacopoeia of the People’s Republic of China. Beijing: China Medical Science and Technology Press.
Corsello, M. A., and Garg, N. K. (2015). Synthetic chemistry fuels interdisciplinary approaches to the production of artemisinin. Nat. Prod. Rep. 32, 359–366. doi: 10.1039/c4np00113c
Djabou, N., Battesti, M. J., Allali, H., Desjobert, J. M., Varesi, L., Costa, J., et al. (2011). Chemical and genetic differentiation of Corsican subspecies of Teucrium flavum L. Phytochemistry 72, 1390–1399. doi: 10.1016/j.phytochem.2011.03.024
Dondorp, A., Nosten, F., Stepniewska, K., Day, N., and White, N. (2005). Artesunate versus quinine for treatment of severe falciparum malaria: a randomised trial. Lancet 366, 717–725. doi: 10.1016/s0140-6736(05)67176-0
Douglas, N. M., Anstey, N. M., Angus, B. J., Nosten, F., and Price, R. N. (2010). Artemisinin combination therapy for vivax malaria. Lancet Infect. Dis. 10, 405–416. doi: 10.1016/s1473-3099(10)70079-7
Ferreira, J. F. (2007). Nutrient deficiency in the production of artemisinin, dihydroartemisinic acid, and artemisinic acid in Artemisia annua L. J. Agric. Food Chem. 55, 1686–1694. doi: 10.1021/jf063017v
Forster, N., Ulrichs, C., Schreiner, M., Arndt, N., Schmidt, R., and Mewis, I. (2015). Ecotype variability in growth and secondary metabolite profile in Moringa oleifera: impact of sulfur and water availability. J. Agric. Food Chem. 63, 2852–2861. doi: 10.1021/jf506174v
Hancock, A. M., Brachi, B., Faure, N., Horton, M. W., Jarymowycz, L. B., Sperone, F. G., et al. (2011). Adaptation to climate across the Arabidopsis thaliana genome. Science 334, 83–86. doi: 10.1126/science.1209244
Hebert, P. D., and Gregory, T. R. (2005). The promise of DNA barcoding for taxonomy. Syst. Biol. 54, 852–859. doi: 10.1080/10635150500354886
Hollingsworth, P. M. (2011). Refining the DNA barcode for land plants. Proc. Natl. Acad. Sci. U.S.A. 108, 19451–19452. doi: 10.1073/pnas.1116812108
Huang, L., Xie, C., Duan, B., and Chen, S. (2010). Mapping the potential distribution of high artemisinin-yielding Artemisia annua L. (Qinghao) in China with a geographic information system. Chin. Med. 5, 18. doi: 10.1186/1749-8546-5-18
Huang, L. F., Suo, F. M., Song, J. Y., Wen, M. J., Jia, G. L., Xie, C. X., et al. (2013). [Quality variation and ecotype division of Panax quinquefolium in China]. Yao Xue Xue Bao 48, 580–589.
Hui, Y., He, J., Yang, C., Kou, Y., and Lu, F. (2006). Determination of artemisinin in Artemisia annua extract by HPLC-ELSD. Chin. Tradit. Herb. Drugs 7, 2. doi: 10.1002/pca.1053
Jain, D. C., Mathur, A. K., Gupta, M. M., Singh, A. K., Verma, R. K., Gupta, A. P., et al. (1996). Isolation of high artemisinin-yielding clones of Artemisia annua. Phytochemistry 43, 993–1001. doi: 10.1016/S0031-9422(96)00369-X
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Knudsmark Jessing, K., Duke, S. O., and Cedergreeen, N. (2014). Potential ecological roles of artemisinin produced by Artemisia annua L. J. Chem. Ecol. 40, 100–117. doi: 10.1007/s10886-014-0384-6
Kress, W. J., and Erickson, D. L. (2007). A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2:e508. doi: 10.1371/journal.pone.0000508
Lapkin, A. A., Walker, A., Sullivan, N., Khambay, B., Mlambo, B., and Chemat, S. (2009). Development of HPLC analytical protocols for quantification of artemisinin in biomass and extracts. J. Pharm. Biomed. Anal. 49, 908–915. doi: 10.1016/j.jpba.2009.01.025
Little, D. P., and Jeanson, M. L. (2013). DNA barcode authentication of saw palmetto herbal dietary supplements. Sci. Rep. 3:3518. doi: 10.1038/srep03518
Liu, C., Zhao, Y., and Wang, Y. (2006). Artemisinin: current state and perspectives for biotechnological production of an antimalarial drug. Appl. Microbiol. Biotechnol. 72, 11–20. doi: 10.1007/s00253-006-0452-0
Liu, C. Z., Zhou, H. Y., and Zhao, Y. (2007). An effective method for fast determination of artemisinin in Artemisia annua L. by high performance liquid chromatography with evaporative light scattering detection. Anal. Chim. Acta 581, 298–302. doi: 10.1016/j.aca.2006.08.038
Liu, J.-L., Li, D.-P., Wei, X., Huang, Y.-L., Lu, F.-L., and Jiang, Y.-S. (2007). Determination of artemisimin in Artemisia annua with RP-HPLC. Guihaia 27, 3. doi: 10.1002/bmc.1719
Lyu, J., Li, B., He, W., Zhang, S., Gou, Z., Zhang, J., et al. (2014). A genomic perspective on the important genetic mechanisms of upland adaptation of rice. BMC Plant Biol. 14:160. doi: 10.1186/1471-2229-14-160
Nieto Feliner, G., and Rossello, J. A. (2007). Better the devil you know? Guidelines for insightful utilization of nrDNA ITS in species-level evolutionary studies in plants. Mol. Phylogenet. Evol. 44, 911–919. doi: 10.1016/j.ympev.2007.01.013
Nithaniyal, S., Newmaster, S. G., Ragupathy, S., Krishnamoorthy, D., Vassou, S. L., and Parani, M. (2014). DNA barcode authentication of wood samples of threatened and commercial timber trees within the tropical dry evergreen forest of India. PLoS ONE 9:e107669. doi: 10.1371/journal.pone.0107669
Oleksyn, J., Tjoelker, M. G., and Reich, P. B. (1992). Growth and biomass partitioning of populations of European Pinus sylvestris L. under simulated 50 degrees and 60 degreesN daylengths: evidence for photoperiodic ecotypes. New. Phytol. 120, 561–574. doi: 10.1111/j.1469-8137.1992.tb01806.x
Perreault-Payette, A., Muir, A. M., Goetz, F., Perrier, C., Normandeau, E., Sirois, P., et al. (2017). Investigating the extent of parallelism in morphological and genomic divergence among lake trout ecotypes in Lake Superior. Mol. Ecol. 26, 1477–1497. doi: 10.1111/mec.14018
Reich, P. B., Rich, R. L., Lu, X., Wang, Y.-P., and Oleksyn, J. (2014). Biogeographic variation in evergreen conifer needle longevity and impacts on boreal forest carbon cycle projections. Proc. Natl. Acad. Sci. U.S.A. 111, 13703–13708. doi: 10.1073/pnas.1216054110
Sangwan, R. S., Sangwan, N. S., Jain, D. C., Kumar, S., and Ranade, S. A. (1999). RAPD profile based genetic characterization of chemotypic variants of Artemisia annua L. Biochem. Mol. Biol. Int. 47, 935–944. doi: 10.1080/15216549900202053
Shi, K., Ai, X., Ding, L., and Zhou, Y. (2008). Study on genetic diversity of Artemisiae annua varieties in the Westen Hubei. Hubei Agric. Sci. 47, 4.
Snyder, A. B., and Worobo, R. W. (2014). Chemical and genetic characterization of bacteriocins: antimicrobial peptides for food safety. J. Sci. Food Agric. 94, 28–44. doi: 10.1002/jsfa.6293
Stoeckle, M. Y., Gamble, C. C., Kirpekar, R., Young, G., Ahmed, S., and Little, D. P. (2011). Commercial teas highlight plant DNA barcode identification successes and obstacles. Sci. Rep. 1, 42. doi: 10.1038/srep00042
Su, X. Z., and Miller, L. H. (2015). The discovery of artemisinin and the Nobel Prize in Physiology or Medicine. Sci. China Life Sci. 58, 1175–1179. doi: 10.1007/s11427-015-4948-7
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Wang, Y., Cai, H., and Jia, W. (2012). Analysis of SSR information in EST resource of sweet wormwood (Artemisia annua) and development of EST-SSR marker. Zhongguo Zhong Yao Za Zhi 37, 570–574.
Wei, S. G., Xiao-Jun, M. A., Feng, S. X., Huang, R. S., Dong, Q. S., Yan, Z. G., et al. (2008). Evaluation on germplasm resources of main production area of Artemisia annua in China. Chin. J. Chin. Mater. Med. 33, 241–244.
Weissing, F. J., Edelaar, P., and van Doorn, G. S. (2011). Adaptive speciation theory: a conceptual review. Behav. Ecol. Sociobiol. 65, 461–480. doi: 10.1007/s00265-010-1125-7
Wu, L., Sun, W., Wang, B., Zhao, H., Li, Y., Cai, S., et al. (2015). An integrated system for identifying the hidden assassins in traditional medicines containing aristolochic acids. Sci. Rep. 5, 11318. doi: 10.1038/srep11318
Xiang, L., Liu, Y., Xie, C., Li, X., Yu, Y., Ye, M., et al. (2016a). The chemical and genetic characteristics of szechuan pepper (Zanthoxylum bungeanum and Z. armatum) cultivars and their suitable habitat. Front. Plant Sci. 7:467. doi: 10.3389/fpls.2016.00467
Xiang, L., Zhang, W., and Chen, S. (2016b). Literature study and DNA barcoding of traditional Chinese medicine Qinghao (Artemisia annua L.). Acta Pharm. Sin. 51, 486–495.
Xiang, W., Li, L., Liu, J., and Yu, B. (2012). Determination of artemisinin in herba Artemisia annua from different locations by UPLC-UV. Chin. Wild Plant Resour. 4, 28–31.
Zhang, X.-B., Zhao, Y.-P., Huang, X.-W., and Huang, L.-Q. (2016). Review on study of Dao-di herbs Artemisiae Annuae Herba. Chin. J. Chin. Mater. Med. 41, 4.
Zhao, Z., Guo, P., and Brand, E. (2012). The formation of daodi medicinal materials. J. Ethnopharmacol. 140, 476–481. doi: 10.1016/j.jep.2012.01.048
Keywords: Artemisia annua, artemisinin, DNA barcoding, ecotype, ecotypic diversity
Citation: Li L, Josef BA, Liu B, Zheng S, Huang L and Chen S (2017) Three-Dimensional Evaluation on Ecotypic Diversity of Traditional Chinese Medicine: A Case Study of Artemisia annua L. Front. Plant Sci. 8:1225. doi: 10.3389/fpls.2017.01225
Received: 08 April 2017; Accepted: 28 June 2017;
Published: 11 July 2017.
Edited by:
Sergio J. Ochatt, INRA – UMR 1347 Agroécologie, FranceReviewed by:
Zhihua Liao, Southwest University, ChinaKexuan Tang, Shanghai Jiao Tong University, China
Copyright © 2017 Li, Josef, Liu, Zheng, Huang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linfang Huang, bGZodWFuZ0BpbXBsYWQuYWMuY24= Shilin Chen, c2xjaGVuQGljbW0uYWMuY24=
 Lin Li
Lin Li Brinckmann A. Josef2
Brinckmann A. Josef2 Linfang Huang
Linfang Huang