- 1Department of Molecular Genetics and Department of Biological Chemistry and Pharmacology, The Ohio State University, Columbus, OH, United States
- 2Plant Cellular and Molecular Biology Graduate Program, The Ohio State University, Columbus, OH, United States
In bacteria and energy generating organelles, c-type cytochromes are a class of universal electron carriers with a heme cofactor covalently linked via one or two thioether bonds to a heme binding site. The covalent attachment of heme to apocytochromes is a catalyzed process, taking place via three evolutionarily distinct assembly pathways (Systems I, II, III). System II was discovered in the green alga Chlamydomonas reinhardtii through the genetic analysis of the ccs mutants (cytochrome c synthesis), which display a block in the apo- to holo- form conversion of cytochrome f and c6, the thylakoid lumen resident c-type cytochromes functioning in photosynthesis. Here we show that the gene corresponding to the CCS2 locus encodes a 1,719 amino acid polypeptide and identify the molecular lesions in the ccs2-1 to ccs2-5 alleles. The CCS2 protein displays seven degenerate amino acid repeats, which are variations of the octatricopeptide-repeat motif (OPR) recently recognized in several nuclear-encoded proteins controlling the maturation, stability, or translation of chloroplast transcripts. A plastid site of action for CCS2 is inferred from the finding that GFP fused to the first 100 amino acids of the algal protein localizes to chloroplasts in Nicotiana benthamiana. We discuss the possible functions of CCS2 in the heme attachment reaction.
Introduction
Energy-transducing membranes are specialized membranes in archaea, bacteria, mitochondria, and chloroplasts, which rely on electron carriers to generate the proton gradient necessary for ATP synthesis. In energy-transducing membranes, the c-type cytochromes, also generically referred to as cytochromes c, are a class of structurally diverse metalloproteins with one or more covalently linked heme(s) (ferro-protoporphyrin IX) that participate in electron transfer reactions (Thony-Meyer, 1997; Bonnard et al., 2010; Verissimo and Daldal, 2014). On the positive side (p-side) of the energy-transducing membrane (i.e., bacterial or archaeal periplasm, mitochondrial intermembrane space, thylakoid lumen), a group of cytochromes c occur as both soluble and membrane proteins with the heme moiety typically attached via a thioether linkage to two cysteine sulfhydryl(s) in a characteristic motif in the apocytochrome c. The consensus motif, also referred to as the heme-binding site, is CXnCH where n is usually equal to two and X can be any residue except cysteine in naturally occurring cytochromes c. The histidine in this motif acts as a ligand of ferroheme (Bowman and Bren, 2008; Allen et al., 2009). Heme attachment to the heme-binding site is a catalyzed reaction in vivo and requires heme transport across at least one biological membrane and covalent linkage of ferroheme to free sulfhydryls of the heme binding cysteines in the CXnCH motif (Bonnard et al., 2010; Mavridou et al., 2013).
To date, three Systems (I, II, and III) have been identified as being necessary for the post-translational assembly of cytochromes c located on the p-side of the membrane, where these molecules function. These systems have been identified through extensive investigation of both bacterial and eukaryotic models (Kranz et al., 2009; Allen, 2011; de Vitry, 2011; Mavridou et al., 2013; Verissimo and Daldal, 2014). In plastids, the heme attachment reaction occurs on the luminal side of the thylakoid membrane and is under the control of System II. This system is composed of multiple pathways and occurs in plastids of all photosynthetic eukaryotes, cyanobacteria, and most proteobacteria of the β-, δ-, and 𝜀-group (Hamel et al., 2009; Bonnard et al., 2010; Simon and Hederstedt, 2011).
System II first emerged through genetic screens for photosynthesis-impaired ccs mutants (ccs for cytochrome c synthesis) in the green alga Chlamydomonas reinhardtii (Hamel et al., 2009; Simon and Hederstedt, 2011; Gabilly and Hamel, 2017). The ccs mutants were isolated on the basis of photosynthetic deficiency due to loss of plastid c-type cytochromes, namely membrane-bound cytochrome f and soluble cytochrome c6 (Howe and Merchant, 1992). These two plastid cytochromes c reside in the thylakoid lumen and function in photosynthesis. The defect is specific to plastid cytochromes c, as abundance of mitochondrial cytochromes c was unaffected in the ccs mutants (Howe and Merchant, 1992). Pulse-chase analyses revealed that precursor forms of apocytochrome f and c6 are synthesized, translocated to the lumen, and therein cleaved by the thylakoid peptidase. However, they are not converted to their respective holoforms. This indicates that the CCS loci control the heme attachment reaction in the lumen (Howe and Merchant, 1992, 1993, 1994; Xie et al., 1998; Gabilly et al., 2010). At least seven loci, plastid ccsA and nuclear CCS1 to CCS6, were uncovered through genetic analysis of the ccs mutants isolated via several UV and insertional mutageneses (Xie et al., 1998; Page et al., 2004; Gabilly et al., 2010).
Functional analysis of plastid CcsA and Ccs1 led to the proposal that these two proteins act together to relay heme from its site of synthesis, the stroma, to its site of function, the lumen, possibly in an assembly complex alongside other CCS factors (Xie and Merchant, 1996; Inoue et al., 1997; Dreyfuss et al., 2003; Hamel et al., 2003). Biochemical studies supported this model with evidence that bacterial CcsA and Ccs1 operate as a functional unit with a dual activity in heme transport across the membrane and heme ligation to apocytochromes c in the periplasmic space (Frawley and Kranz, 2009).
In addition to heme delivery and attachment, the maintenance of the cysteine sulfhydryls (reduced vs. oxidized) within the heme binding site is a prerequisite for covalent linkage of heme (Bonnard et al., 2010; Sanders et al., 2010; Verissimo and Daldal, 2014). The proposed model is that the cysteines are first oxidized by disulfide bond forming enzymes on the p-side of the membrane and must subsequently be reduced to provide free sulfhydryls for the heme ligation reaction. In plastids and bacteria, this requirement is fulfilled by the operation of a disulfide reducing pathway, which conveys electrons across the energy-transducing membrane via thiol-disulfide exchanges in order to reduce the disulfide bond formed between the heme-linking cysteines in the heme binding motif of apocytochromes c (Karamoko et al., 2013; Verissimo and Daldal, 2014; Gabilly and Hamel, 2017).
In bacteria, the CcsA/Ccs1-dependent heme delivery/attachment pathway and the disulfide-reducing pathway are all that is required for cytochrome c assembly (Beckett et al., 2000; Le Brun et al., 2000; Feissner et al., 2005). However, screening of ccs mutants in Chlamydomonas reinhardtii has revealed four additional CCS loci (CCS2, CCS3, CCS4, and CCS6), an indication that the assembly of cytochromes c in the plastid is a more complicated process (Xie et al., 1998; Page et al., 2004). While we have proposed that CCS4 regulates the disulfide reducing pathway in the plastid (Gabilly et al., 2011), the gene products for CCS2, CCS3, and CCS6 remain unknown.
In this article, we report the molecular identification of the CCS2 locus by functional complementation of the ccs2 mutant. The CCS2 gene encodes a plastid-localized, 170 kDa protein that was previously identified by bioinformatics to be a member of the octatricopeptide repeat (OPR) family (Eberhard et al., 2011).
Materials and Methods
Strains and Culture Condition
The ccs2-1 through ccs2-5 strains were used in complementation experiments (Xie et al., 1998). Mutants ccs2-1 and ccs2-2 were crossed to wild-type strain 3A+ (mt+ arg7-8) and 4C- (mt- arg7-8), respectively, to generate the ccs2-1 arg7-8 and ccs2-2 arg7-8 recipient strains. The ccs2-1 arg7-8 mutant was then crossed to wild-type strain CC425 (arg7-8 cw15) to generate the ccs2-1 arg7-8 cw15 mutant used for detection of the HA-tagged CCS2 protein. The MCA1-HA expressing strain is described in Raynaud et al. (2007). Strains were maintained at 25°C on Tris-acetate phosphate (TAP) liquid or solid medium supplemented with 400 mg/mL arginine (Harris, 1989) at 0.6 μE/m2/s. Complemented ccs2 strains were assessed for restoration of photoautotrophic growth on minimal medium (Min) (Harris, 1989) or acetate containing (1.7 mM) minimal medium. For protein extraction, wild-type and complemented strains were grown in liquid TAP supplemented with arginine under 50 μE/m2/s illumination while ccs mutants were grown under 0.6 μE/m2/s illumination. Cell wall-less mutants (cw15) were cultured in liquid and on solid media supplemented with 50 mM sorbitol. Copper-free media for induction of cytochrome c6 was prepared as described previously (Howe and Merchant, 1992; Quinn and Merchant, 1998).
Molecular Cloning of the CCS2 Gene
An indexed cosmid library of Chlamydomonas genomic DNA was used for transformation by electroporation as described by Shimogawara et al. (1998) with the following exceptions: the vector backbone was the cosmid pCB412 containing the ARG7 marker and the transformation required a 30 min incubation in autolysin (to digest the cell wall) followed by electroporation of 5 μg of DNA per cosmid pool on a Biorad Micropulser at 1300 V. Transformants were selected under 50 μE/m2/s light on minimal media supplemented with 1.7 mM acetate. Plasmid pMOL+8.2 kb was generated by cloning the 8.2 kb BamHI/HindIII CCS2 genomic fragment isolated from complementing cosmid (c8G6), into BamHI/HindIII digested pMOLUC (Cha et al., 2002).
Assembly of the CCS2 cDNA
Chlamydomonas RNAs were extracted and retro-transcribed using a bacterial reverse transcriptase from the Roche Transcriptor High Fidelity cDNA Synthesis Kit and the CCS2 specific primers CCS2.30 and CCS2.66STP. Overlapping fragments were amplified with the following primer pairs: CCS2.69 and CCS2.66STP, CCS2.19 and CCS2.54, CCS2.21 and CCS2.18, CCS2.79 and CCS2.02, CCS2.27 and CCS2.28, CCS2.51 and CCS2.26, and, finally, CCS2.81ATG and CCS2.70 using DV Ready Mix (Sigma) and the total retro-transcribed RNAs as a template. Amplified fragments were isolated after electrophoresis in agarose gel, purified, re-amplified and AT-cloned into pGEM-T Easy for sequencing. All primer sequences are listed in Supplementary Table S1.
Construction of HA-Tagged CCS2 and CCS2-GFP Expressing Constructs
Versions of the CCS2 gene carrying an internal HA tag were created by cutting pMOL+8.2 kb by BsiWI or BspEI and inserting the HA-tag sequence via In-Fusion® (CloneTech). The HA-tag sequence was generated using PCR based fill-in of primers CCS2-HA_BsiW1-F and CCS2-HA_BsiW1-R or CCS2-HA_BspEI-F and CCS2-HA_BspEI-R. Introduction of an internal HA tag sequence at the BspEI site (residue 298) abolished CCS2 function while the tag at BsiWI (residue 1672) had no impact (not shown).
To generate the series of “p8” plasmids used in Figure 5, a 606 bp sequence was synthesized (Genscript) and cloned into pUC57 (pUC57+ccs2bit). This sequence contained two distinct fragments that were modified from the Chlamydomonas genomic DNA. The first was a 207 bp fragment corresponding to the 3′ end of CCS2 genomic DNA that both changed the stop codon in the CCS2 ORF into an alanine and added the restriction sites XbaI, SwaI, and SpeI downstream of the stop codon. The second was a 394 bp fragment encompassing the 5′ end of CCS2 genomic DNA designed to remove the native CCS2 promoter and introduce restriction sites BglII and XhoI upstream of the ATG.
Carboxy-terminus 3xHA tags were generated by cutting pMOL+8.2 kb by BsiWI (146 bp upstream the stop codon) and PshA1 (570 bp downstream of the stop codon in the 3′ UTR) and cloning the 207 bp fragment, which was amplified by primers CCS2-BsiWI-F and CCS2-PshAI-R using pUC57+ccs2bits as a template. The 207 bp fragment was cloned using In-Fusion® and the resulting plasmid is p8-noS. The p8-noS plasmid was then digested by XbaI and SpeI and a sequence corresponding to a 3xHA tag (and including a stop), created through PCR fill-in of primers 8.3xHA-f and 8.3xHA-r, was inserted via In-Fusion®. This created vector p8-3xHA expressing CCS2-HA from its native promotor. The plasmids expressing CCS2 or CCS2-HA under the PSAD promoter were generated in a similar manner. The 394 bp fragment was amplified from pUC57+ccs2bits using primers CCS2-XcmI.R and CCS2-XcmI.F and inserted via In-Fusion® after digestion of pMOL+8.2 kb and p8-3xHA by XcmI (322 bp from the initiation codon). This created plasmids p8-noP and p8-3xHAnoP. These constructs were then used to generate plasmids p8-PROM and p8-3xHA+P. To create these vectors, the PSAD promoter and 5′ UTR were amplified from plasmid pSL18 (Pollock et al., 2004), using primers 8.PROM-f and 8.PROM-r, and inserted via In-Fusion® at the introduced restriction sites BglII and XhoI.
The plasmid pGWB5/ccs2target, expressing the CCS2-GFP fusion protein, was constructed from pUC57+ccs2targeting, which contains the first 300 bp from CCS2 ORF, corresponding to the first 100 amino acids of CCS2, cloned into pUC57. This 300 bp sequence was codon optimized for expression in tobacco and synthesized by GenScript. Using primers CCS2.t1 and CCS2.t2, the sequence was amplified from the pUC57+ccs2targeting template and then inserted in frame with the GFP reporter in the expression vector pGWB5 (Nakagawa et al., 2007) using entry vector pENTR/SD/TOPO (Invitrogen) via TOPO cloning. The GFP-NLS/GFP-NES expressing construct in pK7WGF2 (Karimi et al., 2002) is a gift from Dr. I. Meier (Ohio State University). The NLS (nuclear localization sequence) is from the SV40 large T-antigen. The NES (nuclear export signal) is from the HIV-1 Rev response element. Plasmids were transferred to Agrobacterium tumefaciens GV3101 by electroporation.
Protein Preparation and Analysis
Cytochrome f and c6 detection was performed as in Howe and Merchant (1992). Soluble fractions for cytochrome c6 detection were obtained by freeze-thaw fractionation of cells grown in copper deficient conditions. Fractions were electrophoretically separated and cytochromes c revealed by immunodetection or a heme-staining procedure (Howe and Merchant, 1992). Anti-HA immunoblotting analysis was performed on whole cells extracts, prepared as follows: cells were grown to early logarithmic phase under 0.6 μE/m2/s illumination and moved into 30 μE/m2/s light for 5 h. Cells were then pelleted and re-suspended in 10 mM NaPO4 buffer with protease inhibitors (4 mM benzamine, 0.4 mM 6-amino-n-hexanoic acid, 2 mM PMSF, 10 μM leupeptin, 1 μM pepstatin, 1 mM ortho-phenanthroline, 40 μg/mL chymostatin, 10 μM E-64) (Kurvari et al., 1998) to a final concentration of 3 × 108 cells. Laemmli Buffer was then added to a final concentration of 1.2 × 108 cells and 0.1 M DTT. Cells were placed in a sonication bath for 120 s and the solubilized protein sample was denatured for 20 min at 70°C before electrophoresis. Polyclonal antisera raised against Chlamydomonas cytochrome c6, cytochrome f GST-fusion protein, and CF1 were used for immunodetection by alkaline phosphatase-conjugated secondary antibodies. The rat monoclonal anti-HA antibody (clone 3F10) (Roche) was used for immunodetection by peroxidase-conjugated secondary antibody.
Fluorescence Rise and Decay Kinetics
Fluorescence rise and decay kinetics were taken as described in Gabilly et al. (2010) except that strains were grown in liquid medium overnight and measurements were recorded on 20 μL culture aliquots against a white background. Fluorescence transients were measured using Handy Fluorcam from Photon System Instruments. The fluorescence is in arbitrary units (A.U.) and recorded over a 3-s illumination period.
GFP Fluorescence and Imaging
Nicotiana benthamiana was transformed with Agrobacterium carrying the GFP-CCS2 or NLS-GFP/NES-GFP expressing construct via infiltration after a 1 h incubation in Induction Medium (10 mM MgCl2, 10 mM 2-[N-Morpholino]ethanesulfonic acid, 100 μM Acetosyringone). After 3 days, protoplasts were extracted from infiltrated leaves by a 30-min incubation in a Digestion Buffer (1.5% cellulose, 0.4% macerozyme, 0.4 M mannitol, 20 mM KCl, 20 mM 2-[N-Morpholino]ethanesulfonic acid -KOH pH 5.5, 10 mM CaCl2, 0.1% Bovine serum albumin) and concentrated with a 1 s spin at 100 rpm. Supernatant was removed and protoplasts were re-suspended in 100 μL Digestion Buffer before imaging. GFP-dependent fluorescence was taken at 515 nm and chlorophyll auto-fluorescence at 650 nm on a Nikon Cl confocal microscope (Eclipse C90i) using a medium aperture. Images were processed using the NIS-Elements software.
Analysis of OPR Motifs in Proteins
Information presented in Figure 4 was generated by the MEME algorithm (Bailey and Elkan, 1994). Individual protein sequences (Supplementary Figure S1) were entered into the program and base settings were altered to search for any number of repetitions, one motif, between 70 and 600 sites, and between 28 and 50 amino acids wide. The OPR motifs identified by the MEME program (Supplementary Figure S2) were fed into Weblogo 3.3 (Crooks et al., 2004).
Results
Cloning of the CCS2 Gene by Functional Complementation of the ccs2-2 Mutant
To gain further insights into plastid cytochrome c assembly, we sought to clone the gene corresponding to the CCS2 locus, defined by the ccs2-1 to -5 alleles (Xie et al., 1998). All ccs mutants display a b6f-deficient phenotype due to loss of cytochrome f assembly and hence are unable to grow photoautotrophically (Howe and Merchant, 1992; Xie et al., 1998). Using the photosynthetic deficient phenotype of the ccs mutants, we cloned the CCS2 gene via transformation of the ccs2-2 arg7-8 double mutant with an indexed cosmid library (Purton and Rochaix, 1994; Zhang et al., 1994). Transformants were selected for restored growth on acetate-containing minimal medium under standard illumination (50 μE/m2/s). Two cosmids (c8G6 and c5D9) were isolated based on their ability to restore photosynthetic growth to the ccs2-2 mutant. Sequence analysis revealed that c8G6 contains a 30.2 kb region from chromosome 19 while c5D9 appears to have rearranged. The complementing activity in c8G6 could be narrowed down to an 8.2 kb BamHI-HindIII fragment. Both c8G6 and the cloned 8.2 kb BamHI-HindIII fragment complemented strains ccs2-1 through ccs2-5, restoring photosynthetic growth (Figure 1A).
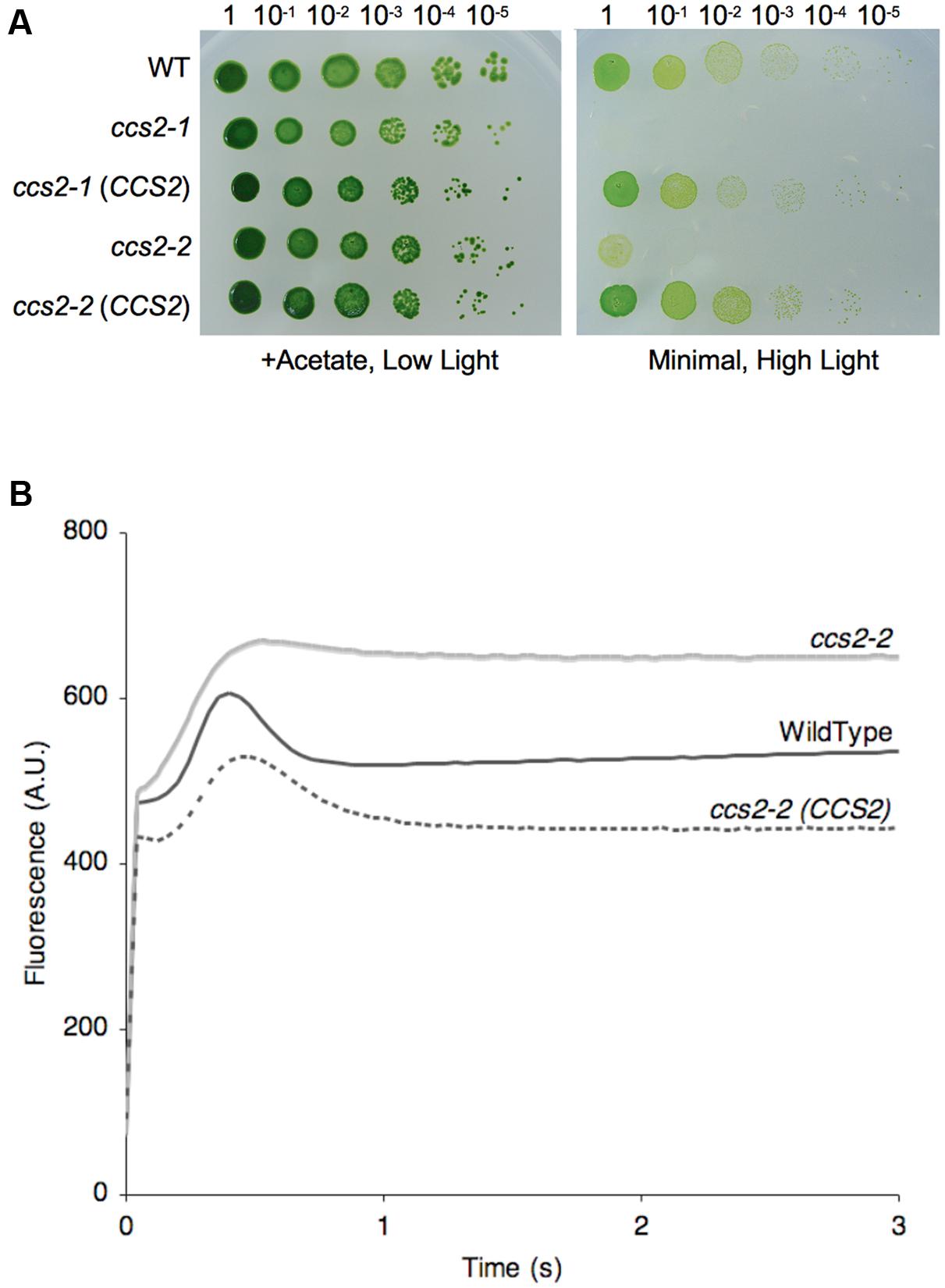
FIGURE 1. The CCS2 gene restores photoautotropic growth to the ccs2 mutants. For (A,B), wild-type CC124 (WT), ccs2-1 and ccs2-2 strains, and CCS2 complemented mutants were used. (A) Ten-fold dilution series of algal cultures were plated on solid medium, with or without acetate, and incubated under low light (0.6 μE/m2/s) or high light (50 μE/m2/s), respectively, for 1 week at 25°C. The ccs2-3, ccs2-4, and ccs2-5 mutants are also restored for photoautotrophic growth upon transformation with the CCS2 gene (not shown). (B) Representative fluorescence rise and decay kinetics indicate restoration of the cytochrome b6f complex in the complemented ccs2 mutant. While all complemented mutants were restored, only the ccs2-2 strain is shown. The fluorescence is in arbitrary units (A.U.) and recorded over a 3-s illumination period after a dark adaption period.
To test if the cytochrome f assembly was restored in the complemented transformants, we performed analyses of fluorescence rise and decay kinetics (Figure 1B). In such experiments, the emitted fluorescence of excited chlorophyll in photosystem II is taken as an indication of the functionality of the b6f complex, which receives electrons from photosystem II. As seen in Figure 1B, the rise and plateau curve for ccs2 is characteristic of a specific block in electron transfer at the level of the cytochrome b6f complex because of its impaired assembly in the absence of membrane-bound holocytochrome f. When the energy absorbed by the chlorophyll cannot be utilized, in this case as a result of a block in photosynthetic transfer through cytochrome b6f, an increase in the chlorophyll fluorescence is observed. In wild-type and complemented strains, the decay phase corresponds to the re-oxidation of the quinone pool, the primary electron acceptor of the photosystem II, by the cytochrome b6f complex. This indicates holocytochrome f assembly and cytochrome b6f functionality is restored in the complemented strains. In accord with this result, we also showed that both holocytochrome f and cytochrome c6 accumulation is restored to wild-type levels in the complemented strains (Figures 2A,B).
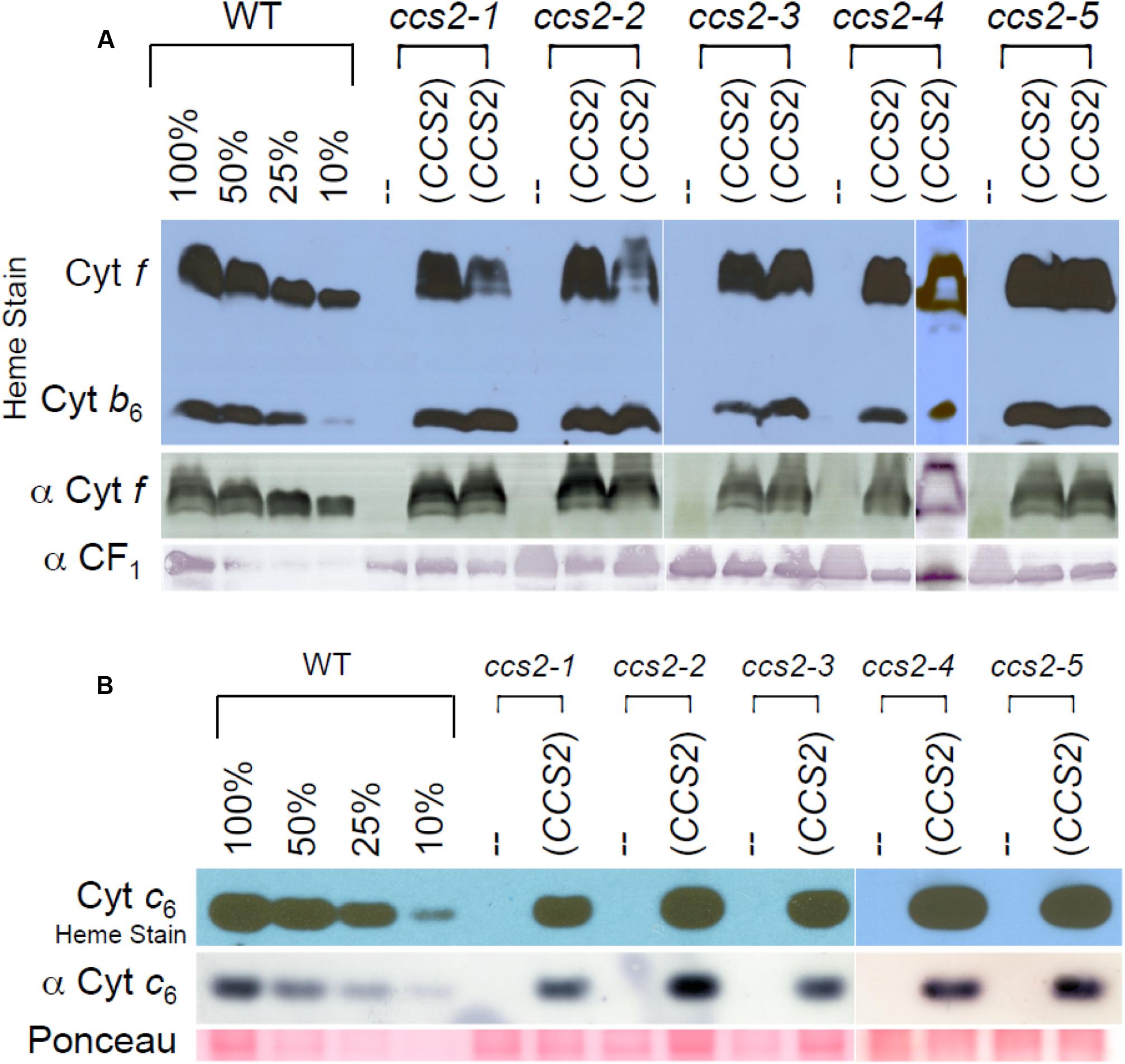
FIGURE 2. Restoration of plastid cytochrome c assembly in the CCS2 complemented strains. For (A,B), the ccs2-1 to ccs2-5 mutants were complemented with the 8.2 kb genomic fragment containing the CCS2 gene. Dilutions of the wild-type sample serve to estimate the cytochrome f (A) and cytochrome c6 (B) abundance. (A) The wild-type CC124 strain (WT), ccs2 mutants, and two independently complemented ccs2 transformants for each ccs2 allele (CCS2) were analyzed for cytochrome f accumulation via heme stain and immunoblotting. Detection of the CF1 of the ATPase is shown as a loading control. Note that heme staining also reveals the presence of covalently attached heme ci in holocytochrome b6. As evidenced from the heme stain, holocytochrome b6 accumulates to a lower level due to loss of cytochrome f assembly in the ccs2 mutants. Different levels of holocytochrome f accumulation in the transformants might reflect differential expression of the introduced CCS2 gene due to position effect of non-homologous integration of the construct in the chromosome. (B) Same as in (A), except only one transformant was analyzed for cytochrome c6 accumulation by heme staining and immunoblotting. Ponceau staining is shown as a loading control. The white lines in (A,B) indicate assembly from independent immunoblots.
The CCS2 Gene Encodes a Protein of the OPR Family
Because of the large size, low abundance, and high GC content (74%) of the CCS2 mRNA, the corresponding full-length cDNA proved difficult to amplify. Instead, overlapping cDNA fragments, approximately 2 kb in length, were amplified and the full-length transcript was extrapolated by aligning the sequenced fragments with the CCS2 genomic DNA. Sequence comparison of the assembled cDNA with the current gene model Cre03.g213201 extended the 5′ end of exon 1 and identified four introns within the CCS2 gene (Figure 3A). The Chlamydomonas transcriptome from the Joint Genome Institute, University of California (JGI-UCLA) and Genoscope1 corroborates this transcript sequence under the previous unique gene ID Cre19.g757200. We assigned the start codon in exon 1 based on the fact that this is the 5′ most ATG preceded by stop codons in all three reading frames.
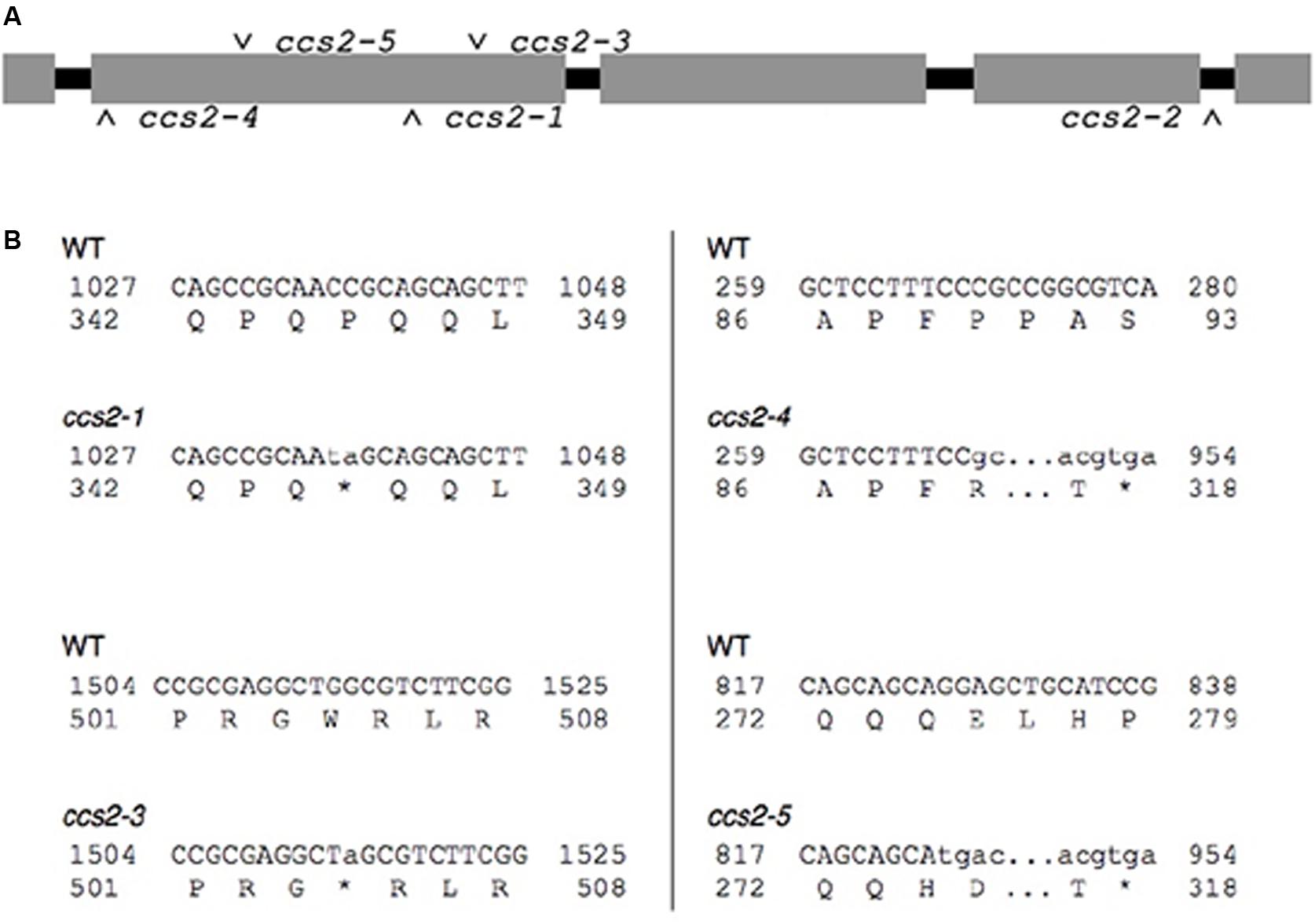
FIGURE 3. Molecular identification of the ccs2-1 to ccs2-5 mutations. (A) Schematic representation of the CCS2 immature transcript. Introns are shown in black and exons in gray. The position of the molecular lesions in the ccs2 alleles is indicated. (B) Identified mutations in the ccs2 alleles. The top line corresponds to the nucleotide sequence of the CCS2 ORF and numbers displayed to either side refer to position of the nucleotides within the ORF. The lower-case letters indicate the nucleotide sequence due to change(s) induced by the UV mutagenesis. The lower line shows the corresponding amino acid sequence and the change within the protein sequence resulting from the molecular lesion(s). The asterisk indicates a stop in the protein sequence. The ccs2-1 allele is a CC to TA mutation at position 1036-1037 and the ccs2-3 allele is a G to A change at position 1514. In the ccs2-4, deletion of a single C occurred at position 269. The ccs2-5 mutation combines a G to T mutation at position 826 and deletion of a single G at position 829. The GenBank accession number for the CCS2 nucleotide sequence is KC292647.
Sequencing of the genomic DNA in the ccs2-1 to ccs2-5 mutants revealed molecular lesions in the CCS2 coding sequence (Figure 3A). All lesions introduced nonsense (ccs2-1 and ccs2-3) or frameshift (ccs2-4 and ccs2-5) mutations, which result in truncations within the first 500 amino acids of the protein except for the ccs2-2 allele (Figure 3B). In the ccs2-2 mutant, a single guanine has been deleted from a stretch of 11 guanines in intron 4, 10 bp downstream of the 3′ end of exon 4. It is possible that this change impairs the splicing of intron 4 in the CCS2 transcript. The identification of the molecular lesions confirms that the complementing sequence we isolated corresponds to the wild-type CCS2 gene rather than an extragenic suppressor of the ccs2 mutation.
The CCS2 gene encodes a 1,719 amino acid protein with a predicted molecular weight of 171,753 Da. The most striking feature of the CCS2 protein is the presence of several 38–40 amino acid repeats occurring between residues 720 and 1610 (Figure 4AB). Such motifs, named OPR for octatricopeptide repeats were first defined in TBC2 (translation factor for chloroplast psbC mRNA), a nuclear encoded protein required for the translation of psbC RNA in the chloroplast (Auchincloss et al., 2002). OPRs are also recognized in other factors controlling translation (TDA1, TAB1), maturation (RAT2, RAA1, RAA8, RAP), or stability (MCG1,MBI1) of chloroplast transcripts (Balczun et al., 2005; Merendino et al., 2006; Eberhard et al., 2011; Rahire et al., 2012; Kleinknecht et al., 2014; Marx et al., 2015; Wang et al., 2015).
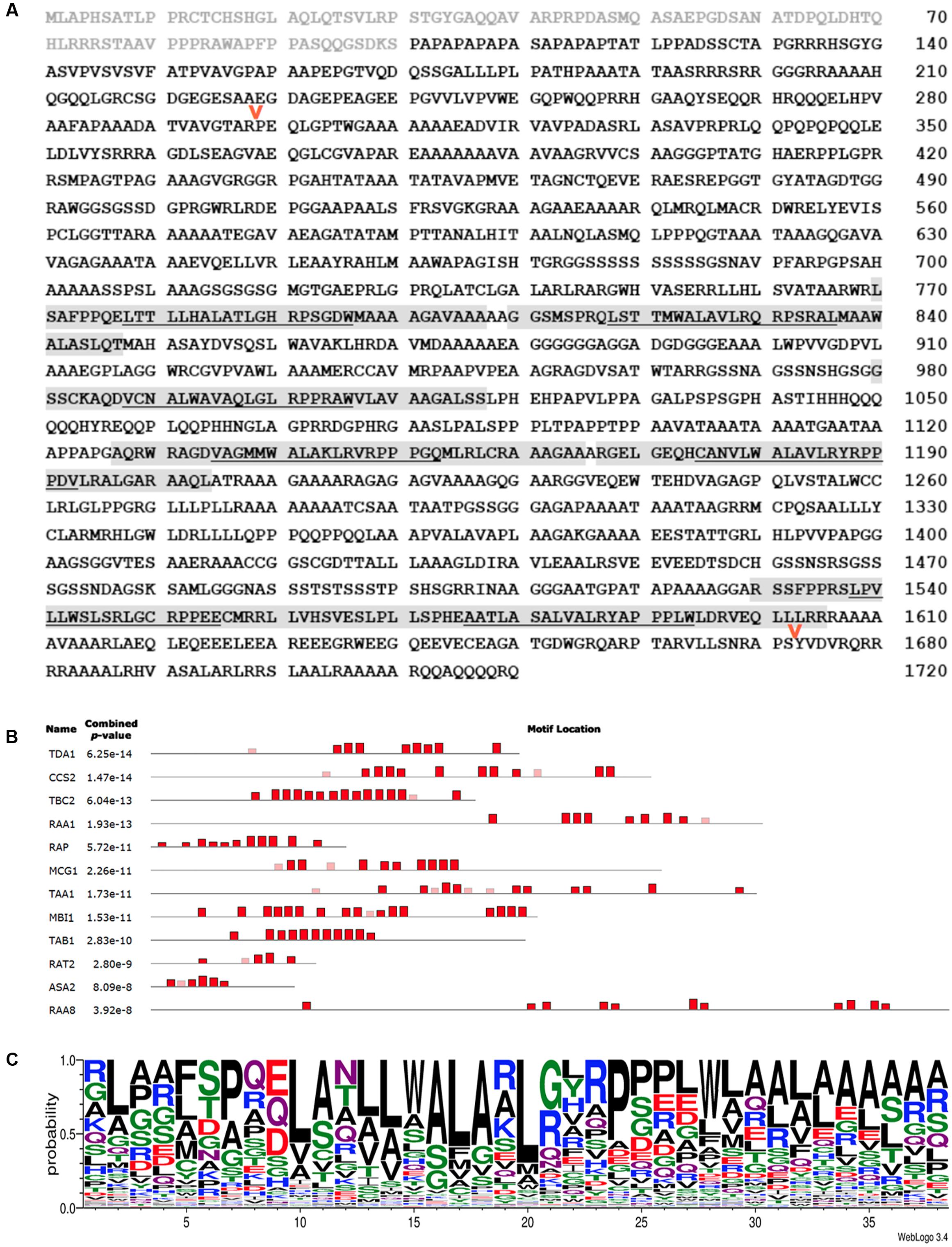
FIGURE 4. CCS2 is an OPR protein. (A) Amino acid sequence of the deduced CCS2 protein. Light gray text indicates the protein sequence that was fused to GFP for targeting experiments (Figure 6), light gray highlighting indicates OPRs recognized with p-values of at least 1e-10, as shown in (B). The underlined portion indicates the location of the previously recognized “PPPEW” motif. The red arrows indicate the positions of internal HA tags that were introduced to test neutrality of the tag with respect to CCS2 function (see Materials and Methods). (B) MEME-generated distribution of OPR motifs within the Chlamydomonas OPR proteins that have been functionally characterized. Heights of blocks indicate relative proportion of the p-value 1e-10. Red boxes represent motifs found by the MEME program while faded boxes represent motifs found by other programs within the MEME Suite. Sequences of Chlamydomonas OPR proteins CCS2 (KC292647), ASA2 (EDP00850.1), TBC2 (CAD20887.1), TDA1 (CCA62914.20), RAA1 (CAE53330.1), TAB1 (ADY68544.1), RAT2 (EDP02536.1), TAA1 (Cre06.g262650), RAA8 (Cre10.g440000), MCG1 (Cre10.g429400), MBI1 (Cre06.g272450), and Arabidopsis RAP (OAP08625.1) were used (see Supplementary Figure S1). (C) WebLogo consensus sequence of OPRs identified by the MEME program depicted in red in Figure 4B (Bailey and Elkan, 1994; Crooks et al., 2004). Letter height indicates the probability of a particular amino acid (y-axis) at a given position within the 38 amino acid repeat (x-axis). OPR sequences used to calculate this consensus sequence are found in Supplementary Figure S2.
The OPR family is further characterized by the presence of low complexity regions, which are regions containing little diversity in their amino acid composition (Balczun et al., 2005; Merendino et al., 2006; Eberhard et al., 2011; Rahire et al., 2012; Marx et al., 2015; Wang et al., 2015). Indeed, we noted the occurrence of several stretches of three or more alanine, glycine, serine, or glutamine repeats in CCS2 (Figure 4A). CCS2 has a high content of alanine (24.2%), glycine (10.7%), proline (8.7%), and leucine (8.7%), a feature shared by other OPR proteins (Auchincloss et al., 2002; Balczun et al., 2005; Merendino et al., 2006; Eberhard et al., 2011; Rahire et al., 2012; Marx et al., 2015; Wang et al., 2015). OPR motifs found in CCS2 are highlighted in Figure 4B and a consensus OPR motif from all OPR containing proteins that have been functionally identified to date, with the exception of NCC1 and NCC2 (Boulouis et al., 2015), can be seen in Figure 4C. The relative locations of the motifs in the proteins used to generate the consensus motif in Figure 4C can be seen in Figure 4B.
Immunodetection of the CCS2 Protein
In order to detect CCS2 and assess its subcellular localization, a series of constructs expressing HA tagged proteins were engineered. The tagged proteins were either expressed under the control of the native CCS2 promoter or the PSAD promoter, which allows increased expression of Chlamydomonas genes (Fischer and Rochaix, 2001). We saw no difference in the ability of the two HA-tagged CCS2 constructs to complement the ccs2-1 mutation as compared to the WT gene (Figure 5A).
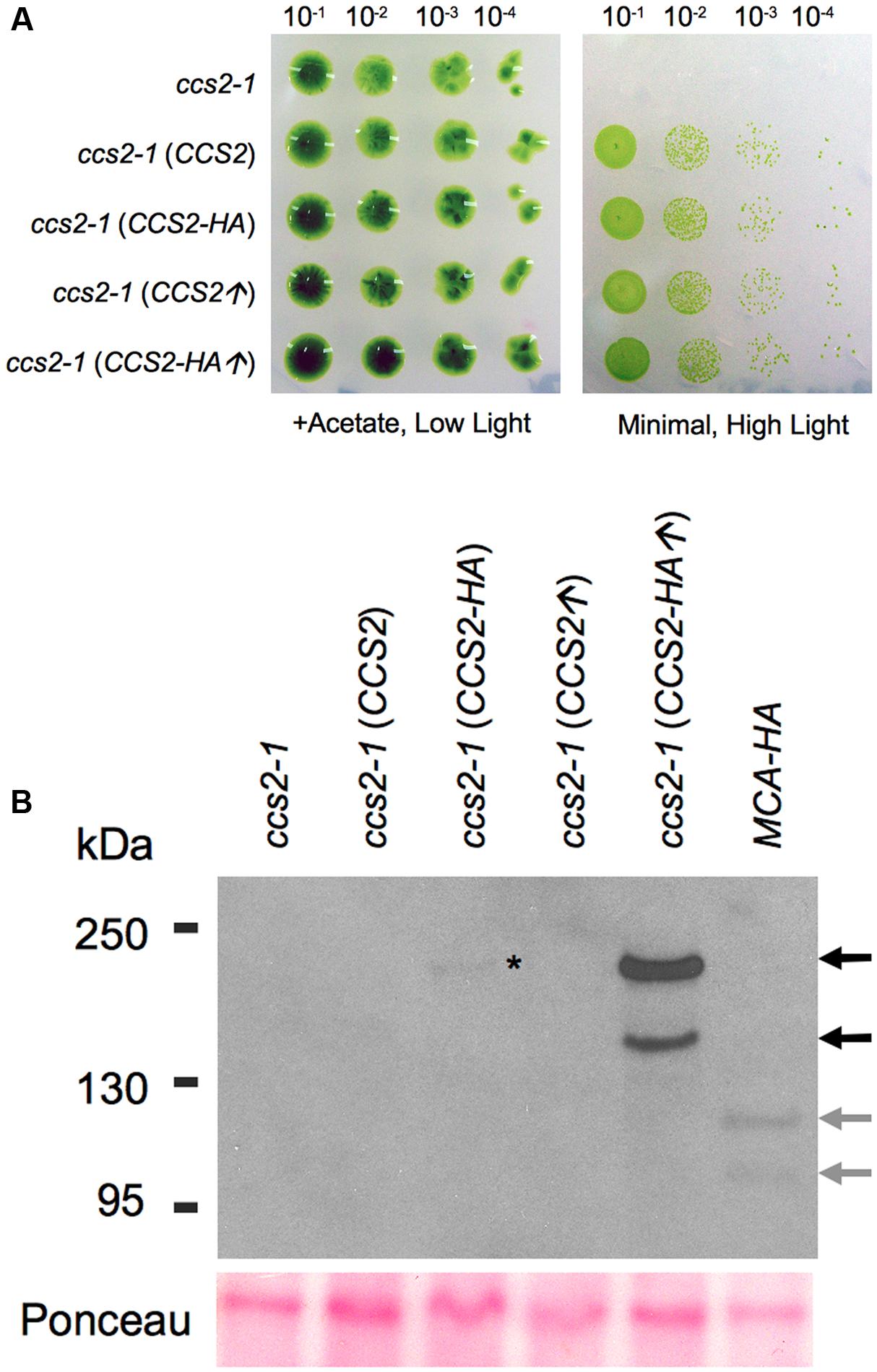
FIGURE 5. Immunodetection of CCS2-HA in whole cells. For (A,B), a cell wall minus ccs2-1 strain was transformed with plasmids carrying the 8.2 kb genomic fragment (CCS2), the 8.2 kb genomic fragment expressing HA tagged CCS2 (CCS2-HA), the CCS2 coding sequence under the PSAD promoter (CCS2↑), and HA tagged CCS2 coding sequence under the PSAD promoter (CCS2-HA↑), respectively. One representative transformant, selected on minimal medium, was used for the analysis. (A) Ten-fold dilution series of algal cells on acetate containing (0.6 μE/m2/s) or minimal media (50 μE/m2/s), incubated for 1 week at 25°C. (B) Anti-HA immunodetection using whole cell extracts. The HA-tagged MCA1 strain is used as a control (Raynaud et al., 2007). MCA1 is a plastid-localized PPR protein (PPR for pentatricopeptide repeat). Black arrows indicate the two CCS2 species, gray arrows indicate the two MCA1 species. The lower bands correspond to the expected molecular mass for CCS2 (∼176 kDa) and MCA1 (∼105 kDa). The asterisk indicates the CCS2 species of high molecular weight that is detected in the ccs2 strain expressing CCS2-HA under its native promoter. Ponceau staining serves as a loading control.
Immunoblotting analysis against whole cell extracts from transformants over-expressing CCS2-HA revealed that the protein occurs as two species, one of which appears to migrate at the expected size (≃176 kDa) (Figure 5B). A similar pattern was also seen in cells expressing HA-tagged MCA1, a PPR (PPR for pentatricopeptide repeat) protein with plastid localization (Raynaud et al., 2007; Boulouis et al., 2011). This suggests that the immunoreactive species with higher mobility may be a result of the extraction conditions and/or the denaturation step needed to immunodetect CCS2 (see Materials and Methods). Detection of CCS2-HA was only possible from freshly grown cultures when a cocktail of protease inhibitors was used immediately followed by denaturing at 70°C (instead of 100°C), an indication that the protein is likely highly sensitive to proteolysis. Interestingly, Rahire et al. (2012) noted that the OPR protein TDA1 was also very susceptible to proteolysis.
The CCS2 Protein Localizes to the Plastid
A chloroplast targeting sequence is assigned by both ChloroP (Emanuelsson et al., 1999) and WoLF PSORT (Horton et al., 2007) at the N-terminus of CCS2, an indication that CCS2 might reside in the plastid. However, the extraction methods necessary for detection of CCS2-HA precluded the use of subcellular fractionation to determine protein localization. Hence, we tested the ability of the CCS2 N-terminus to direct GFP to the plastid in a heterologous system such as N. benthamiana. This is justified, as targeting sequences from Chlamydomonas nuclear-encoded proteins retain their function as transit peptides for import into chloroplasts in several species of land plants including Nicotiana (Nakazato et al., 2003; Falciatore et al., 2005; Levitan et al., 2005; Li et al., 2016; Yamaoka et al., 2016). To this end, we constructed CCS2-GFP, which encoded a protein consisting of the first 100 amino acids of CCS2, including the putative targeting sequence, translationally fused to the amino-terminus of GFP. This CCS2-GFP expressing construct was introduced into N. benthamiana leaves. Fluorescence microscopy shows clear overlay of chlorophyll auto-fluorescence, which is a feature of the chloroplast, and GFP fluorescence (Figure 6). These results indicate that the first 100 amino acids of CCS2 are sufficient to target GFP to the chloroplast of N. benthamiana and hence the CCS2 protein is likely localized to the plastid of Chlamydomonas.
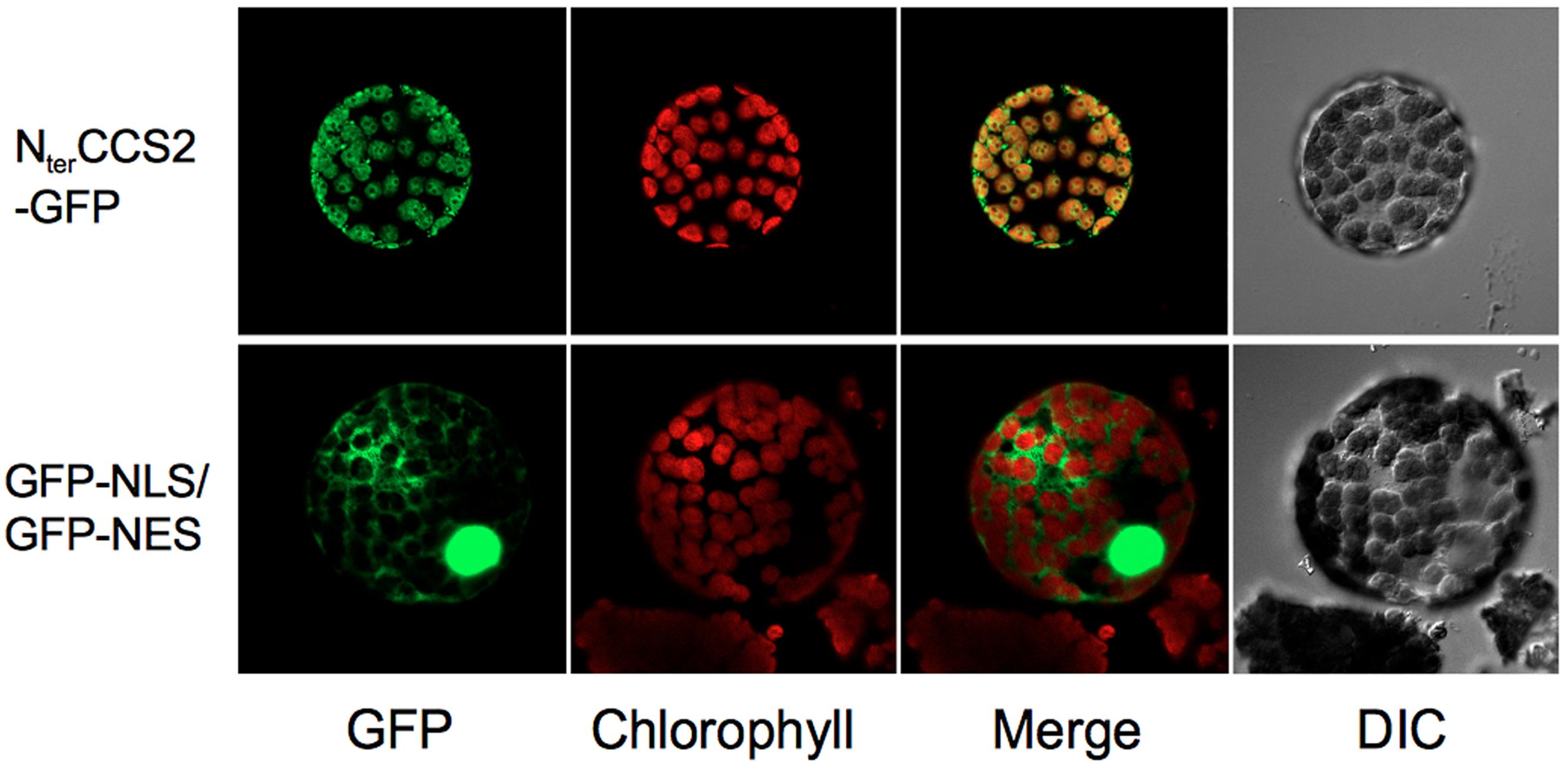
FIGURE 6. Localization of GFP to chloroplast via CCS2 N-terminal sequence. Top row (NterCCS2-GFP): imaging of a protoplast from Nicotiana benthamiana leaves transformed with the pGWB5/ccs2target plasmid expressing the first 100 amino acids of CCS2 fused to GFP. Bottom row (GFP-NLS/GFP-NES): imaging of a protoplast from N. benthamiana leaves transformed with the construct expressing a NLS-GFP/NES-GFP fusion protein shuttling between the nucleus and the cytosol via a nuclear localization signal (NLS) and a nuclear export signal (NES). DIC, differential interference contrast microscopy.
Discussion
Identification of CCS2 as an OPR Protein Controlling Cytochrome c Assembly
Here, we have identified CCS2, a chloroplast localized cytochrome c assembly factor that, based on primary sequence similarity, appears to be unique to specific branches of the chlorophycean algae. This protein has been recognized as a member of the recently designated OPR family, which contains 43 members in Chlamydomonas (Eberhard et al., 2011). OPRs are defined by loosely conserved repeats of 38–40 amino acids (Auchincloss et al., 2002; Eberhard et al., 2011; Rahire et al., 2012).
Via bioinformatics analysis, OPR proteins have been assigned to the ‘α-solenoid’ superfamily, which contains proteins defined by similar tandem repeats, such as TPRs (tetratricopeptide repeat, 34 amino acids) and PPRs (pentatricopeptide repeat, 35 amino acids) (Eberhard et al., 2011; Rahire et al., 2012). Three dimensional structure analyses of TPRs (Das et al., 1998; D’Andrea and Regan, 2003; Zeytuni and Zarivach, 2012) and PPRs (Ringel et al., 2011; Nakamura et al., 2012; Shen et al., 2016) indicate that these motifs result in a series of anti-parallel α-helices. Protein predictions using I-TASSER (Zhang, 2008; Roy et al., 2010) suggest that the OPR proteins are organized in a similar manner (Auchincloss et al., 2002; Eberhard et al., 2011; Rahire et al., 2012). I-TASSER also predicts regions of anti-parallel α-helices in CCS2 (not shown). TPR proteins act as scaffolds to mediate protein–protein interactions and control a wide range of cellular functions such as the cell cycle, transcription in the nucleus, or protein import into mitochondria and peroxisomes (D’Andrea and Regan, 2003). On the other hand, all the PPR proteins described so far were shown to be control maturation, stability, or translation of organellar RNAs, presumably via direct interaction with their target transcript(s) (Barkan and Small, 2014; Manna, 2015).
The PPR family has expanded greatly in the plant lineage, with 450 representatives in Arabidopsis alone, while the typical non-plant, eukaryotic genome encodes for fewer than 40 representatives (Barkan and Small, 2014; Manna, 2015). Similar to the PPR protein family expansion seen in land plants, chlorophycean algae have an expansion in the number of OPR containing proteins (Eberhard et al., 2011; Rahire et al., 2012). The majority of functionally characterized OPR proteins are involved in translation, maturation, or stability of transcripts in the chloroplast of Chlamydomonas. For instance, TBC2, TDA1, and TAB1 are factors involved in the translation of the psbC, atpA, or psaB transcripts, respectively (Auchincloss et al., 2002; Eberhard et al., 2011; Rahire et al., 2012), while RAA1/RAA8 and RAT2 are involved in the maturation of psaA and tscA RNAs (Balczun et al., 2005; Merendino et al., 2006; Marx et al., 2015). In Arabidopsis, RAP is the sole OPR containing protein and loss of RAP function yields a defect in the maturation of the chloroplast 16S ribosomal RNA (Kleinknecht et al., 2014). Recent studies have uncovered the role of MCG1 and MBI1 in stabilizing the petG mRNA, encoding a small subunit of the cytochrome b6f complex and the psbI mRNA, coding for a small subunit of photosystem II, respectively (Wang et al., 2015).
If the role of most OPR containing proteins as transcript-interacting factors was inferred from the phenotypic analysis of loss-of-function mutations, the demonstration that OPR motifs interact directly with their relevant target RNAs was only provided for TAB1 (Rahire et al., 2012). A recent study revealed that gain of function mutations in the OPR motifs of NCC1 and NCC2 confer an ability to recognize chloroplast transcripts as novel targets of action (Boulouis et al., 2015). This suggests that specific amino acids within these OPR motifs must govern nucleotide recognition, but the basis of this specificity still remains to be deciphered.
The molecular mass range of the functionally identified OPR proteins extends from 45 to 269 kDa with an average length of 1428 amino acids and 5–17 OPR motifs. These repeats are found primarily in the central regions of proteins while the carboxyl- and amino-termini are typically characterized by stretches of single amino acid repeats (Auchincloss et al., 2002; Merendino et al., 2006; Rahire et al., 2012). The OPR motifs themselves can be highly divergent, and so the repeat count per protein fluctuates depending on how stringently one defines a single OPR motif (Figure 4B). This divergence is reflected in the fact that OPR repeats in CCS2 are characterized by a LWALAR consensus motif (Figure 4A) while repeats in TBC2, TDA1, and TAB1 contain a PPPEW sequence (Auchincloss et al., 2002; Eberhard et al., 2011; Rahire et al., 2012). While in some instances OPR motifs are serially repeated, the motifs can also be separated by gaps (Figure 4B), often by stretches of amino acids such as alanine. We noted the frequent occurrence of a poly-glutamine stretch upstream of the OPR motif in CCS2. Interestingly, glutamine-rich regions are known to form polar zippers involved in protein interaction but they have also been recognized as motifs for transcriptional activation (Courey and Tjian, 1988; Stott et al., 1995).
That CCS2 functions directly in the heme attachment reaction in the thylakoid lumen is very unlikely, considering its large size and the absence of a typical bipartite targeting sequence for luminal localization. Considering its inclusion in the OPR family, one possibility is that CCS2 controls the maturation, stability, or translation of a chloroplast transcript in the stroma involved in cytochrome c assembly. In the chloroplast genome, the ccsA gene encodes a heme delivery factor required for cytochrome c maturation (Xie and Merchant, 1996). While the level of ccsA transcript in the ccs2 mutants was previously shown to be unaltered, indicating that CCS2 is not required to stabilize the ccsA mRNA (Xie et al., 1998), the hypothesis that CCS2 is involved in the translation of ccsA remains plausible. Unfortunately, this was not tested due to the lack of an anti-CcsA antibody. Another likely scenario is that CCS2 functions like a TPR containing protein and mediates the assembly of protein complexes containing other CCS factors. The OPR family also includes ASA2 (ATP Synthase Associated protein), a subunit of the unusual mitochondrial ATP synthase, found only in chlorophycean algae (van Lis et al., 2007; Cano-Estrada et al., 2010) (Figure 4B). While a 200 kDa CCS complex containing ccsA (40 kDa) and Ccs1 (60 kDa) has been identified at the thylakoid membrane (Hamel et al., 2003), it is unlikely that the 170 kDa CCS2 is a component of this complex because of its large size. However, it is conceivable that CCS2 could stabilize the CCS complex via protein-protein interactions or facilitate the recruitment of the components of this complex on the stromal side of the thylakoid membrane.
Author Contributions
SC executed the experiments, interpreted the data and wrote the manuscript; PH designed the experiments, interpreted the data and wrote the manuscript; IL executed experiments.
Funding
This work was primarily supported by a National Science Foundation grant (MCB-0920062) and a U.S. Department of Energy (DOE), Office of Science, Basic Energy Sciences (BES) grant (DE-SC0014562) to PH.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Meier for the gift of the GFP-NLS-GFP-NES expressing construct and Dr. Zhou for technical help with the microscopy experiments. We also wish to thank Dr. O. Vallon for scientific discussion on the OPR proteins and Dr. R. Lamb for her comments on the manuscript. PH thanks Sabeeha Merchant in whose laboratory the project was initiated.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01306/full#supplementary-material
Footnotes
References
Allen, J. W., Sawyer, E. B., Ginger, M. L., Barker, P. D., and Ferguson, S. J. (2009). Variant c-type cytochromes as probes of the substrate specificity of the E. coli cytochrome c maturation (Ccm) apparatus. Biochem. J. 419, 177–184. doi: 10.1042/BJ20081999
Allen, J. W. A. (2011). Cytochrome c biogenesis in mitochondria – systems III and V. FEBS J. 278, 4198–4216. doi: 10.1111/j.1742-4658.2011.08231.x
Auchincloss, A. H., Zerges, W., Perron, K., Girard-Bascou, J., and Rochaix, J. D. (2002). Characterization of Tbc2, a nucleus-encoded factor specifically required for translation of the chloroplast psbC mRNA in Chlamydomonas reinhardtii. J. Cell Biol. 157, 953–962.
Bailey, T. L., and Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36.
Balczun, C., Bunse, A., Hahn, D., Bennoun, P., Nickelsen, J., and Kuck, U. (2005). Two adjacent nuclear genes are required for functional complementation of a chloroplast trans-splicing mutant from Chlamydomonas reinhardtii. Plant J. 43, 636–648.
Barkan, A., and Small, I. (2014). Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65, 415–442. doi: 10.1146/annurev-arplant-050213-040159
Beckett, C. S., Loughman, J. A., Karberg, K. A., Donato, G. M., Goldman, W. E., and Kranz, R. G. (2000). Four genes are required for the System II cytochrome c biogenesis pathway in Bordetella pertussis, a unique bacterial model. Mol. Microbiol. 38, 465–481.
Bonnard, G., Corvest, V., Meyer, E. H., and Hamel, P. P. (2010). Redox processes controlling the biogenesis of c-type cytochromes. Antioxid. Redox Signal. 13, 1385–1401. doi: 10.1089/ars.2010.3161
Boulouis, A., Drapier, D., Razafimanantsoa, H., Wostrikoff, K., Tourasse, N. J., Pascal, K., et al. (2015). Spontaneous dominant mutations in chlamydomonas highlight ongoing evolution by gene diversification. Plant Cell 27, 984–1001. doi: 10.1105/tpc.15.00010
Boulouis, A., Raynaud, C., Bujaldon, S., Aznar, A., Wollman, F. A., and Choquet, Y. (2011). The nucleus-encoded trans-acting factor MCA1 plays a critical role in the regulation of cytochrome f synthesis in Chlamydomonas chloroplasts. Plant Cell 23, 333–349. doi: 10.1105/tpc.110.078170
Bowman, S. E. J., and Bren, K. L. (2008). The chemistry and biochemistry of heme c: functional bases for covalent attachment. Nat. Prod. Rep. 25, 1118–1130. doi: 10.1039/b717196j
Cano-Estrada, A., Vázquez-Acevedo, M., Villavicencio-Queijeiro, A., Figueroa-Martínez, F., Miranda-Astudillo, H., Cordeiro, Y., et al. (2010). Subunit-subunit interactions and overall topology of the dimeric mitochondrial ATP synthase of Polytomella sp. Biochim. Biophys. Acta 1797, 1439–1448. doi: 10.1016/j.bbabio.2010.02.024
Cha, H. J., Dalal, N. G., Pham, M.-Q., Kramer, S. F., Vakharia, V. N., and Bentley, W. E. (2002). Increased efficiency of cloning large DNA fragments using a lower copy number plasmid. Biotechniques 32, 992–998.
Courey, A. J., and Tjian, R. (1988). Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell 55, 887–898.
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190.
D’Andrea, L. D., and Regan, L. (2003). TPR proteins: the versatile helix. Trends Biochem. Sci. 28, 655–662.
Das, A. K., Cohen, P. W., and Barford, D. (1998). The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 17, 1192–1199.
de Vitry, C. (2011). Cytochrome c maturation system on the negative side of bioenergetic membranes: CCB or System IV. FEBS J. 278, 4189–4197. doi: 10.1111/j.1742-4658.2011.08373.x
Dreyfuss, B. W., Hamel, P. P., Nakamoto, S. S., and Merchant, S. (2003). Functional analysis of a divergent System II protein, Ccs1, involved in c-type cytochrome biogenesis. J. Biol. Chem. 278, 2604–2613.
Eberhard, S., Loiselay, C., Drapier, D., Bujaldon, S., Girard-Bascou, J., Kuras, R., et al. (2011). Dual functions of the nucleus-encoded factor TDA1 in trapping and translation activation of atpA transcripts in Chlamydomonas reinhardtii chloroplasts. Plant J. 67, 1055–1066. doi: 10.1111/j.1365-313X.2011.04657.x
Emanuelsson, O., Nielsen, H., and von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984.
Falciatore, A., Merendino, L., Barneche, F., Ceol, M., Meskauskiene, R., Apel, K., et al. (2005). The FLP proteins act as regulators of chlorophyll synthesis in response to light and plastid signals in Chlamydomonas. Genes Dev. 19, 176–187. doi: 10.1101/gad.321305
Feissner, R. E., Beckett, C. S., Loughman, J. A., and Kranz, R. G. (2005). Mutations in cytochrome assembly and periplasmic redox pathways in Bordetella pertussis. J. Bacteriol. 187, 3941–3949.
Fischer, N., and Rochaix, J. D. (2001). The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol. Genet. Genomics 265, 888–894.
Frawley, E. R., and Kranz, R. G. (2009). CcsBA is a cytochrome c synthetase that also functions in heme transport. Proc. Natl. Acad. Sci. U.S.A. 106, 10201–10206. doi: 10.1073/pnas.0903132106
Gabilly, S. T., and Hamel, P. P. (2017). Maturation of plastid c-type cytochromes. Front. Plant Sci. 8:1313. doi: 10.3389/fpls.2017.01313
Gabilly, S. T., Dreyfuss, B. W., Karamoko, M., Corvest, V., Kropat, J., Page, M. D., et al. (2010). CCS5, a thioredoxin-like protein involved in the assembly of plastid c-type cytochromes. J. Biol. Chem. 285, 29738–29749. doi: 10.1074/jbc.M109.099069
Gabilly, S. T., Kropat, J., Karamoko, M., Page, M. D., Nakamoto, S. S., Merchant, S. S., et al. (2011). A novel component of the disulfide-reducing pathway required for cytochrome c assembly in plastids. Genetics 187, 793–802. doi: 10.1534/genetics.110.125369
Hamel, P., Corvest, V., Giege, P., and Bonnard, G. (2009). Biochemical requirements for the maturation of mitochondrial c-type cytochromes. Biochim. Biophys. Acta 1793, 125–138. doi: 10.1016/j.bbamcr.2008.06.017
Hamel, P. P., Dreyfuss, B. W., Xie, Z., Gabilly, S. T., and Merchant, S. (2003). Essential histidine and tryptophan residues in CcsA, a System II polytopic cytochrome c biogenesis protein. J. Biol. Chem. 278, 2593–2603.
Harris, E. H. (1989). The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. San Diego, CA: Academic Press.
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587.
Howe, G., and Merchant, S. (1992). The biosynthesis of membrane and soluble plastidic c-type cytochromes of Chlamydomonas reinhardtii is dependent on multiple common gene products. EMBO J. 11, 2789–2801.
Howe, G., and Merchant, S. (1993). Maturation of thylakoid lumen proteins proceeds post-translationally through an intermediate in vivo. Proc. Natl. Acad. Sci. U.S.A. 90, 1862–1866.
Howe, G., and Merchant, S. (1994). Role of heme in the biosynthesis of cytochrome c6. J. Biol. Chem. 269, 5824–5832.
Inoue, K., Dreyfuss, B. W., Kindle, K. L., Stern, D. B., Merchant, S., and Sodeinde, O. A. (1997). CCS1, a nuclear gene required for the post-translational assembly of chloroplast c-type cytochromes. J. Biol. Chem. 272, 31747–31754.
Karamoko, M., Gabilly, S. T., and Hamel, P. P. (2013). Operation of trans-thylakoid thiol-metabolizing pathways in photosynthesis. Front. Plant Sci. 4:476. doi: 10.3389/fpls.2013.00476
Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195.
Kleinknecht, L., Wang, F., Stube, R., Philippar, K., Nickelsen, J., and Bohne, A. V. (2014). RAP, the sole octotricopeptide repeat protein in Arabidopsis, is required for chloroplast 16S rRNA maturation. Plant Cell 26, 777–787. doi: 10.1105/tpc.114.122853
Kranz, R. G., Richard-Fogal, C., Taylor, J.-S., and Frawley, E. R. (2009). Cytochrome c biogenesis: mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control. Microbiol. Mol. Biol. Rev. 73, 510–528. doi: 10.1128/mmbr.00001-09
Kurvari, V., Grishin, N. V., and Snell, W. J. (1998). A gamete-specific, sex-limited homeodomain protein in Chlamydomonas. J. Cell Biol. 143, 1971–1980.
Le Brun, N. E., Bengtsson, J., and Hederstedt, L. (2000). Genes required for cytochrome c synthesis in Bacillus subtilis. Mol. Microbiol. 36, 638–650.
Levitan, A., Trebitsh, T., Kiss, V., Pereg, Y., Dangoor, I., and Danon, A. (2005). Dual targeting of the protein disulfide isomerase RB60 to the chloroplast and the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 102, 6225–6230. doi: 10.1073/pnas.0500676102
Li, Z., Peers, G., Dent, R. M., Bai, Y., Yang, S. Y., Apel, W., et al. (2016). Evolution of an atypical de-epoxidase for photoprotection in the green lineage. Nat. Plants 2:16140. doi: 10.1038/nplants.2016.140
Manna, S. (2015). An overview of pentatricopeptide repeat proteins and their applications. Biochimie 113, 93–99. doi: 10.1016/j.biochi.2015.04.004
Marx, C., Wünsch, C., and Kück, U. (2015). The octatricopeptide repeat protein Raa8 is required for chloroplast trans splicing. Eukaryot. Cell 14, 998–1005. doi: 10.1128/ec.00096-15
Mavridou, D. A. I., Ferguson, S. J., and Stevens, J. M. (2013). Cytochrome c assembly. IUBMB Life 65, 209–216. doi: 10.1002/iub.1123
Merendino, L., Perron, K., Rahire, M., Howald, I., Rochaix, J. D., and Goldschmidt-Clermont, M. (2006). A novel multifunctional factor involved in trans-splicing of chloroplast introns in Chlamydomonas. Nucleic Acids Res. 34, 262–274.
Nakagawa, T., Kurose, T., Hino, T., Tanaka, K., Kawamukai, M., Niwa, Y., et al. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41.
Nakamura, T., Yagi, Y., and Kobayashi, K. (2012). Mechanistic insight into pentatricopeptide repeat proteins as sequence-specific RNA-binding proteins for organellar RNAs in plants. Plant Cell Physiol. 53, 1171–1179. doi: 10.1093/pcp/pcs069
Nakazato, E., Fukuzawa, H., Tabata, S., Takahashi, H., and Tanaka, K. (2003). Identification and expression analysis of cDNA encoding a chloroplast recombination protein REC1, the chloroplast RecA homologue in Chlamydomonas reinhardtii. Biosci. Biotechnol. Biochem. 67, 2608–2613. doi: 10.1271/bbb.67.2608
Page, M. L. D., Hamel, P. P., Gabilly, S. T., Zegzouti, H., Perea, J. V., Alonso, J. M., et al. (2004). A homolog of prokaryotic thiol disulfide transporter CcdA is required for the assembly of the cytochrome b6f complex in Arabidopsis chloroplasts. J. Biol. Chem. 279, 32474–32482. doi: 10.1074/jbc.M404285200
Pollock, S. V., Prout, D. L., Godfrey, A. C., Lemaire, S. D., and Moroney, J. V. (2004). The Chlamydomonas reinhardtii proteins Ccp1 and Ccp2 are required for long-term growth, but are not necessary for efficient photosynthesis, in a low-CO2 environment. Plant Mol. Biol. 56, 125–132.
Purton, S., and Rochaix, J. D. (1994). Complementation of a Chlamydomonas reinhardtii mutant using a genomic cosmid library. Plant Mol. Biol. 24, 533–537.
Quinn, J. M., and Merchant, S. (1998). Copper-responsive gene expression during adaptation to copper deficiency. Methods Enzymol. 297, 263–279.
Rahire, M., Laroche, F., Cerutti, L., and Rochaix, J. D. (2012). Identification of an OPR protein involved in the translation initiation of the PsaB subunit of photosystem I. Plant J. 72, 652–661. doi: 10.1111/j.1365-313X.2012.05111.x
Raynaud, C., Loiselay, C., Wostrikoff, K., Kuras, R., Girard-Bascou, J., Wollman, F. A., et al. (2007). Evidence for regulatory function of nucleus-encoded factors on mRNA stabilization and translation in the chloroplast. Proc. Natl. Acad. Sci. U.S.A. 104, 9093–9098.
Ringel, R., Sologub, M., Morozov, Y. I., Litonin, D., Cramer, P., and Temiakov, D. (2011). Structure of human mitochondrial RNA polymerase. Nature 478, 269–273. doi: 10.1038/nature10435
Roy, A., Kucukural, A., and Zhang, Y. (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738. doi: 10.1038/nprot.2010.5
Sanders, C., Turkarslan, S., Lee, D.-W., and Daldal, F. (2010). Cytochrome c biogenesis: the Ccm system. Trends Microbiol. 18, 266–274. doi: 10.1016/j.tim.2010.03.006
Shen, C., Zhang, D., Guan, Z., Liu, Y., Yang, Z., Yang, Y., et al. (2016). Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins. Nat. Commun. 7:11285. doi: 10.1038/ncomms11285
Shimogawara, K., Fujiwara, S., Grossman, A., and Usuda, H. (1998). High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148, 1821–1828.
Simon, J., and Hederstedt, L. (2011). Composition and function of cytochrome c biogenesis System II. FEBS J. 278, 4179–4188. doi: 10.1111/j.1742-4658.2011.08374.x
Stott, K., Blackburn, J. M., Butler, P. J., and Perutz, M. (1995). Incorporation of glutamine repeats makes protein oligomerize: implications for neurodegenerative diseases. Proc. Natl. Acad. Sci. U.S.A. 92, 6509–6513.
Thony-Meyer, L. (1997). Biogenesis of respiratory cytochromes in bacteria. Microbiol. Mol. Biol. Rev. 61, 337–376.
van Lis, R., Mendoza-Hernandez, G., Groth, G., and Atteia, A. (2007). New insights into the unique structure of the F0F1-ATP synthase from the chlamydomonad algae Polytomella sp. and Chlamydomonas reinhardtii. Plant Physiol. 144, 1190–1199.
Verissimo, A. F., and Daldal, F. (2014). Cytochrome c biogenesis System I: an intricate process catalyzed by a maturase supercomplex? Biochim. Biophys. Acta 1837, 989–998. doi: 10.1016/j.bbabio.2014.03.003
Wang, F., Johnson, X., Cavaiuolo, M., Bohne, A. V., Nickelsen, J., and Vallon, O. (2015). Two Chlamydomonas OPR proteins stabilize chloroplast mRNAs encoding small subunits of photosystem II and cytochrome b6f. Plant J. 82, 861–873. doi: 10.1111/tpj.12858
Xie, Z., Culler, D., Dreyfuss, B. W., Kuras, R., Wollman, F. A., Girard-Bascou, J., et al. (1998). Genetic analysis of chloroplast c-type cytochrome assembly in Chlamydomonas reinhardtii: one chloroplast locus and at least four nuclear loci are required for heme attachment. Genetics 148, 681–692.
Xie, Z., and Merchant, S. (1996). The plastid-encoded ccsA gene is required for heme attachment to chloroplast c-type cytochromes. J. Biol. Chem. 271, 4632–4639.
Yamaoka, Y., Achard, D., Jang, S., Legéret, B., Kamisuki, S., Ko, D., et al. (2016). Identification of a Chlamydomonas plastidial 2-lysophosphatidic acid acyltransferase and its use to engineer microalgae with increased oil content. Plant Biotechnol. J. 14, 2158–2167. doi: 10.1111/pbi.12572
Zeytuni, N., and Zarivach, R. (2012). Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure 20, 397–405. doi: 10.1016/j.str.2012.01.006
Zhang, H., Herman, P. L., and Weeks, D. P. (1994). Gene isolation through genomic complementation using an indexed library of Chlamydomonas reinhardtii DNA. Plant Mol. Biol. 24, 663–672.
Keywords: plastid, photosynthesis, cytochrome c, heme, assembly factor, OPR
Citation: Cline SG, Laughbaum IA and Hamel PP (2017) CCS2, an Octatricopeptide-Repeat Protein, Is Required for Plastid Cytochrome c Assembly in the Green Alga Chlamydomonas reinhardtii. Front. Plant Sci. 8:1306. doi: 10.3389/fpls.2017.01306
Received: 16 April 2017; Accepted: 12 July 2017;
Published: 03 August 2017.
Edited by:
Dario Leister, Ludwig-Maximilians-Universität München, GermanyReviewed by:
Lixin Zhang, Institute of Botany (CAS), ChinaMichael Schroda, Kaiserslautern University of Technology, Germany
Copyright © 2017 Cline, Laughbaum and Hamel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patrice P. Hamel, aGFtZWwuMTZAb3N1LmVkdQ==
†Present address: Sara G. Cline, Department of Mathematics, Computer and Natural Sciences, Athens State University, Athens, AL, United States
 Sara G. Cline1,2†
Sara G. Cline1,2† Isaac A. Laughbaum1
Isaac A. Laughbaum1 Patrice P. Hamel
Patrice P. Hamel