- 1Department of Life Science (BK21 Program), Chung-Ang University, Seoul, South Korea
- 2Department of Integrative Plant Science, Chung-Ang University, Anseong, South Korea
Abscisic acid (ABA) is a plant hormone that plays a critical role in the response to environmental stress conditions, especially regulation of the stomatal aperture under water-deficit conditions. The signal transduction occurring during the stress response is initiated by transcription of defense-related genes. Here, we isolated the pepper ethylene-responsive transcription factor CaAIEF1 (Capsicum annuum ABA Induced ERF 1). The CaAIEF1 gene was significantly induced after exposure to ABA, drought, and high salinity. Fusion of the acidic domain in the C-terminal region of CaAIEF1 to the GAL4 DNA-binding domain had a transactivation effect on the reporter gene in yeast. Further, the CaAIEF1-GFP fusion constructs localized in the nucleus. We used CaAIEF1-silenced plants and CaAIEF1-overexpressing (OX) plants to elucidate the biological function of CaAIEF1 in response to ABA and drought stress. CaAIEF1-silenced pepper plants and CaAIEF1-OX Arabidopsis plants displayed drought-sensitive and -tolerant phenotypes, respectively, which were characterized by regulation of transpirational water loss and stomatal aperture. In drought stress condition, quantitative RT-PCR analyses revealed that the expression levels of pepper stress-related genes were higher in CaAIEF1-silenced pepper plants than control plants. Moreover, expression levels of Arabidopsis stress-related genes were significantly reduced in CaAIEF1-OX plants compared with control plants in drought stress condition. Our findings suggest that CaAIEF1 positively regulates the drought stress response and the ABA signaling.
Introduction
Plants are sessile organisms, and therefore, their growth and development is affected by abiotic stresses, including drought, high salinity, and extreme temperatures. Drought is a key abiotic stress that leads to loss of crop yield. Drought stress limits photosynthesis, reduces growth, and influences hormonal balance (Zhu, 2002). To adapt to drought stress conditions, plants have evolved various defense strategies to enhance water retention in cells via minimizing transpiration from the stomata and maximizing water uptake from the roots (Apse and Blumwald, 2002; Yamaguchi-Shinozaki and Shinozaki, 2006). The physiological mechanisms functioning under drought stress conditions have been intensively examined; however, the plant defense response is a complex phenomenon, and the precise adaptive mechanisms induced by drought stress remain elusive (Lee and Luan, 2012; Murata et al., 2015).
The major phytohormone abscisic acid (ABA) regulates many plant growth and development processes and abiotic stress responses (Cutler et al., 2010). Under drought stress conditions, ABA is biosynthesized in various tissues and accumulated in the leaf tissue, and this leads to induction of stress-adaptive mechanisms—including induction of defense-related genes, promotion of stomatal closure, and accumulation of various protective metabolites (Tan et al., 2003; Lee and Luan, 2012). A number of studies have indicated that the plant defense response to drought stress involves various mechanisms—from transcription to post-translational modification—and these mechanisms are influenced by the ABA signal transduction pathway (Lee and Luan, 2012).
Under stress conditions, defense-related genes are induced by the interaction between specific cis-acting elements and transcription factors (Thatcher et al., 2012; Llorca et al., 2014; Muller and Munne-Bosch, 2015). Transcription factors control the majority of defense-related genes and have several characteristics—such as transcriptional activation or repression and nuclear localization—that are essential for inducing or preventing target gene expression (Eulgem et al., 2000; Jakoby et al., 2002; Licausi et al., 2013; Llorca et al., 2014). In Arabidopsis, more than 1,800 transcription factors have been identified, and many of these transcription factors are involved in adaptation to stress conditions, including pathogens, drought and high salinity (Gong et al., 2004; Lee et al., 2006; Froidure et al., 2010; Mizoi et al., 2012; Fu and Dong, 2013). In particular, members of the APETALA2/Ethylene-Responsive Factor (AP2/ERF) transcription factor family are conservatively widespread in plants and are classified into five subfamilies according to the similarity and number of DNA-binding domains (Sakuma et al., 2002; Licausi et al., 2013). Previous studies have demonstrated that AP2/ERF transcription factors function as positive or negative regulators in the response to abiotic stress, and this facilitates plant adaptation to stress conditions. Thus, AP2/ERF-overexpressing (OX) plants exhibit tolerant or sensitive phenotypes after exposure to different stresses. For example, ERF1-OX Arabidopsis plants showed enhanced tolerance to drought and high salinity; these plants had increased levels of ABA and proline, which contribute to stress tolerance (Cheng et al., 2013). Activation of AtERF7 inhibited ABA-responsive genes; hence, AtERF7-OX plants exhibited an ABA-hyposensitive and drought-sensitive phenotype (Song et al., 2005). The involvement of the AP2/ERF transcription factors in the plant adaptive response to abiotic stress is well established (Xu et al., 2011; Licausi et al., 2013; Muller and Munne-Bosch, 2015).
In this study, we characterized the drought-tolerant ERF transcription factor CaAIEF1 from Capsicum annuum. We used GAL4 fusion proteins in yeast to investigate the in vitro function of CaAIEF1 as a transacting factor. We evaluated the in vivo function of CaAIEF1 by examining the expression profiles of CaAIEF1 in CaAIEF1-silenced pepper plants and CaAIEF1-overexpressing (OX) Arabidopsis plants in response to abiotic stresses. CaAIEF1-silenced pepper plants exhibited a drought-sensitive phenotype characterized by decreased stomatal closure and increased water loss. In contrast, CaAIEF1-OX plants displayed an drought-tolerant and ABA-hypersensitive phenotypes characterized by increased stomatal closure and low levels of transpirational water loss. Our findings imply that the CaAIEF1 transcription factor functions as a positive regulator of the defense response to drought stress.
Materials and Methods
Plant Materials
Seeds of pepper (C. annuum), Arabidopsis thaliana (ecotype Col-0), and tobacco (Nicotiana benthamiana) were sown in a soil mix, including sand, loam soil, vermiculite, perlite, peat moss (10:10:5:3:2, v:v:v:v:v). These plants were grown in a fixed condition at 24 ± 1°C (16 h/8 h, light/dark).
Transactivation Analysis of CaAIEF1
The coding region of the CaAIEF1 gene was cloned into the pGBKT7 vector, which includes a nuclear localization and GAL4 DNA-binding domains. We used Saccharomyces cerevisiae strain AH109 for CaAIEF1 transactivation analysis, which have two reporter genes (Ade1 and His3) with GAL4 promoters. The AH109 cells were transformed with pGBKT7 vector that carried GAL4-CaAIEF1 fusion genes. The transformed yeast cells were selected into SD-adenine-histidine-leucine-tryptophan medium to confirm transcriptional activation.
Virus-Induced Gene Silencing
For the loss-of function analysis of CaAIEF1, virus-induced gene silencing (VIGS) was conducted in pepper plants, as described by Lim et al. (2015b). Briefly, pTRV1 and pTRV2:CaAIEF1 or pTRV2:00 transformed to Agrobacterium tumefaciens strain GV3101. The Agrobacterium cells were infiltrated by syringe to the pepper cotyledons (OD600 = 0.2 for each construct). Plants were grown in pepper growth condition described previously for spreading the virus.
Generation of CaAIEF1-Overexpressing Arabidopsis Plants
For the elucidation of biological function of CaAIEF1, CaAIEF1 overexpressing (OX) Arabidopsis was generated. The full-length coding region of CaAIEF1 (accession no. KY652734) was inserted into pENTR/D-TOPO vectors (Invitrogen, Carlsbad, CA, United States). Through the LR reaction, the inserted genes were introduced into pK2GW7 to constitutively express each gene with the cauliflower mosaic virus (CaMV) 35S promoter (Karimi et al., 2002); the generated construct was transformed into Agrobacterium tumefaciens strain GV3101. The floral dip method was applied for transformation of Arabidopsis with the CaAIEF1 gene (Clough and Bent, 1998). Overexpressing plants were selected by germinating putative transformed seeds on Murashige and Skoog (MS) plates supplemented with 50 μg⋅mL-1 of kanamycin as selection marker. Seeds of T3 plants were harvested from second-generation transgenic plants showing a 3:1 segregation ratio on MS plates supplemented with the same antibiotic.
ABA, Dehydration, and NaCl Treatments
To analyze the expression patterns of CaAIEF1 in pepper plants, which had been treated with ABA, NaCl, and dehydration, leaf samples were prepared as described by Lim et al. (2015b). For the ABA and NaCl treatment, the six-leaf-stage pepper plants were treated with 100 μM ABA, irrigated with a salt solution (200 mM). Whole plants were dried on 3-mm paper (Whatman, Clifton, United Kingdom), or the roots were removed and the aerial parts of plants were dried. After treatment, the third and fourth leaves were harvested at the several time points.
For the dehydration phenotype analysis, 5-week-old pepper plants and 3-week-old Arabidopsis plants were randomly placed and subjected to dehydration treatment by withholding watering for 12 and 11 days, respectively. After re-watering, the survival rate was calculated by counting the plant number with resumed growth. To determine the dehydration tolerance in a quantitative manner, rates of water loss were measured by drying leaves detached from gene-silenced pepper plants and Arabidopsis transgenic plants.
For quantitative real-time transcription-polymerase chain reaction (qPCR) analysis, 4-week-old wild-type and CaAIEF1-OX Arabidopsis were treated with dehydration stress, and harvested at the several time points.
Measurement of Stomatal Aperture and Leaf Temperature
The measurements of stomatal pore size and leaf temperature were performed as described previously (Lim and Lee, 2016). Briefly, pepper and Arabidopsis leaf peels were floated in stomatal opening buffer (SOS: 50 mM KCl and 10 mM MES-KOH, pH 6.15, 10 mM CaCl2) in the light condition for 2.5 h. Stomatal closure was induced by replacing the buffer with fresh SOS containing various concentrations of ABA. After an additional 2.5 h of incubation in each buffer, 100 stomata per each sample were observed under a Nikon Eclipse 80i microscope. An infrared camera (FLIR systems; T420) were used for obtaining the thermal images, and FLIR Tools+ ver 5.2 software measured the leaf temperatures.
Quantitative Real-time Transcription-Polymerase Chain Reaction
RNA samples were digested with RNA-free DNase to inhibit contamination of genomic DNA. The cDNA were synthesized using a Transcript First Strand cDNA Synthesis kit (Roche, Indianapolis, IN, United States). For qPCR analysis, the specific primers and CFX96 TouchTM Real-Time PCR detection system (Bio-Rad, Hercules, CA, United States) were used (Supplementary Table S1). The PCR was programmed as follows: 95°C for 5 min; 45 cycles each at 95°C for 20 s and 60°C for 20 s; and 72°C for 20 s. To determine relative expression level, we used the ΔΔCt method (Livak and Schmittgen, 2001). For the normalization, the Arabidopsis AtACT8 and pepper CaACT1 genes were used.
Subcellular Localization of CaAIEF1
CaAIEF1-GFP proteins were expressed in the leaves of N. benthamiana epidermis cells by using infiltration of Agrobacterium tumefaciens strain GV3101 with strain carrying p19 suppressor (1:1 ratio; OD600 = 0.5). The GFP signal was observed 2 days after infiltration, using a confocal microscope (510 UV/Vis Meta; Zeiss, Oberkochen, Germany).
Results
Isolation of Ethylene-Responsive Transcription Factor CaAIEF1
To identify the novel ABA-induced transcription factor, we conducted an RNA-seq assay using control and ABA treated pepper leaves; we identified the putative pepper ABA-induced ERF. We designated gene name CaAIEF1 (C. annuum ABA Induced ERF 1) by domain analysis (Figure 1A and Supplementary Figure S1). CaAIEF1 encoded 180 amino acids. The predicted protein consisted of an AP2/ERF domain with 65 amino acids (59–123) in the central region and an acidic domain (AD) consisting of 21 amino acids (160–180); these domains specifically bind to promoter regions and activate target genes (Figure 1A) (Licausi et al., 2013). Moreover, a putative nuclear localization signal (NLS) was detected in the AP2/ERF domain. Amino acid sequence alignments of CaAIEF1 with ERF proteins revealed that these ERF protein members have a highly conserved AP2/ERF domain. CaAIEF1 shares identity with other ERF proteins of Solanum tuberosum (accession no. XP_006367196.1, 87.9% identity), N. tomentosiformis (accession no. XP_009619682.1, 81.3% identity), N. tabacum (accession no. XP_016487471.1, 80.7% identity), Solanum lycopersicum (accession no. XP_001233987.1, 79.1% identity) and A. thaliana (accession no. NP_188965.1, 32.9% identity). However, CaAIEF1 does not share any identity with pepper ERF proteins (Supplementary Figure S2).
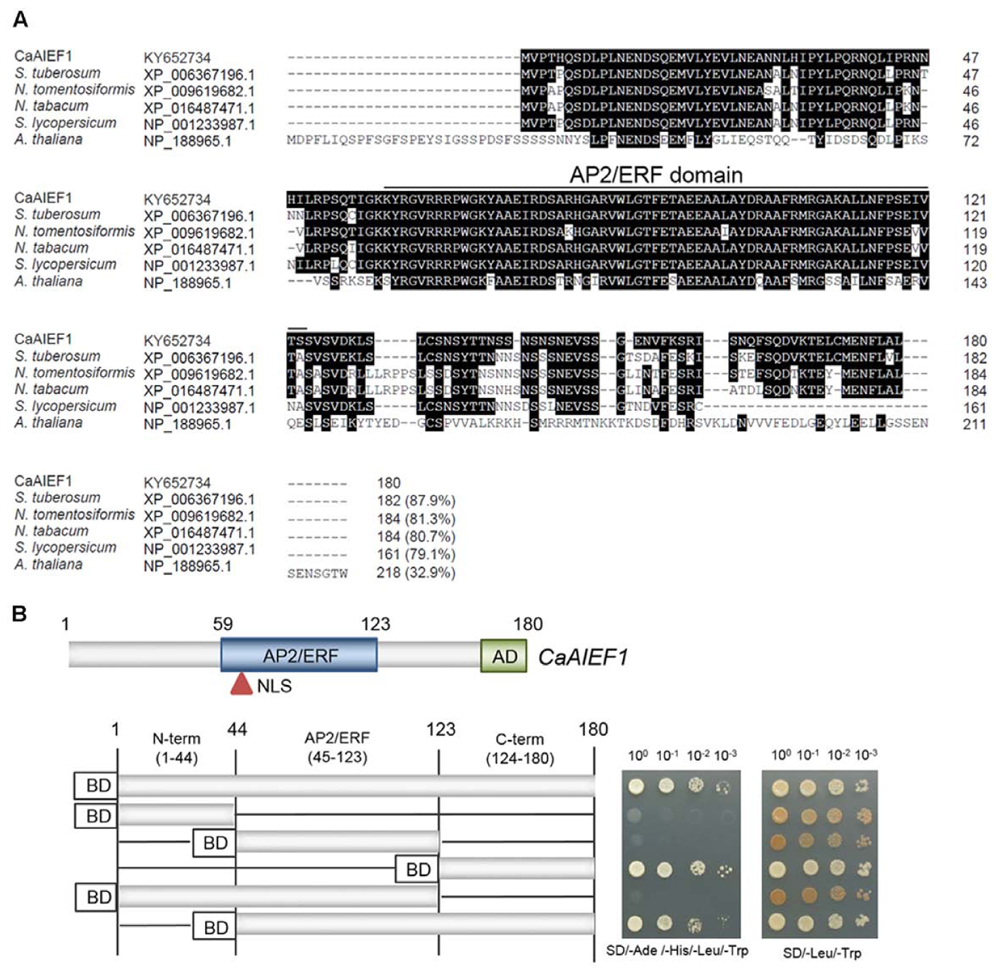
FIGURE 1. Molecular characterization of the CaAIEF1 (Capsicum annuum ABA Induced ERF 1) protein. (A) Comparisons of the deduced amino acid sequence of the CaAIEF1 protein with those of the Solanum tuberosum (accession no. XP_006367196.1), Nicotiana tomentosiformis (accession no. XP_009619682.1), N. tabacum (accession no. XP_016487471.1), Solanum lycopersicum (accession no. XP_001233987.1) and Arabidopsis thaliana (accession no. NP_188965.1) proteins. Identical amino acid residues are highlighted in black, and the upper line indicates the AP2/ERF domain. (B) Transactivation of GAL4-responsive transcription by the full-length and truncated forms of the CaAIFF1 gene using a GAL4 yeast system. Derivatives of S. cerevisiae strain AH109 harboring plasmids that encode GAL4-CaAIFF1 were grown on SD medium lacking adenine, histidine, leucine, and tryptophan (SD-adenine-histidine-leucine-tryptophan; left) or leucine and tryptophan (SD-leucine-tryptophan, right). Plates were incubated at 28°C for 5 days. pGBKT7 indicates the vector used in this experiment; this vector expressed GAL4 BD.
Transcription factors have an activation domain, which functions in activation of the target gene. The activation domain is enriched in glutamine, proline, or acidic amino acids (Schwechheimer et al., 1998a,b; Jakoby et al., 2002; Cantin et al., 2003). Transactivation of acidic domains has universal functions in eukaryotic transcription factors (Hahn, 1993; Schwechheimer et al., 1998a). CaAIEF1 contains an acidic domain in the C-terminal region; hence, we used deletion constructs of the CaAIEF1 gene to determine whether the CaAIEF1 protein functions as a transactivation factor in yeast. The deletion constructs were inserted into the pGBKT7 vector carrying the GAL4 DNA-binding domain, expressed in S. cerevisiae strain AH109 (Figure 1B). Yeast cells carrying the C-terminal region (containing the acidic domain) grew well in the selection medium, suggesting activation of the reporter genes. Our results indicate that the C-terminal region of CaAIEF1 act as a potential transcriptional activator.
Expression of the CaAIEF1 Gene and Subcellular Localization of the CaAIEF1 Protein
To examine whether CaAIEF1 is expressed in response to several stresses, including drought, ABA, and NaCl, we performed quantitative RT-PCR analysis (Figure 2A). In drought-treated pepper leaves, the expression level of CaAIEF1 was up-regulated at 2 h and thereafter gradually decreased. ABA is a key abiotic stress-related hormone involved in stress signal transduction—especially under drought stress conditions (Lee and Luan, 2012). In ABA-treated pepper leaves, CaAIEF1 was induced within 6–24 h. CaAIEF1 transcripts started to induce at 2 h and reached peak levels after 12 h by NaCl treatment. The CaAIEF1 is ethylene responsive factor; hence, we checked whether CaAIEF1 is induced by ethylene. As shown in Supplementary Figure S3A, the transcription level of CaAIEF1 was up-regulated by ethylene. Moreover, the CaAIEF1 expression was also regulated by developmental stages (Supplementary Figure S3B).
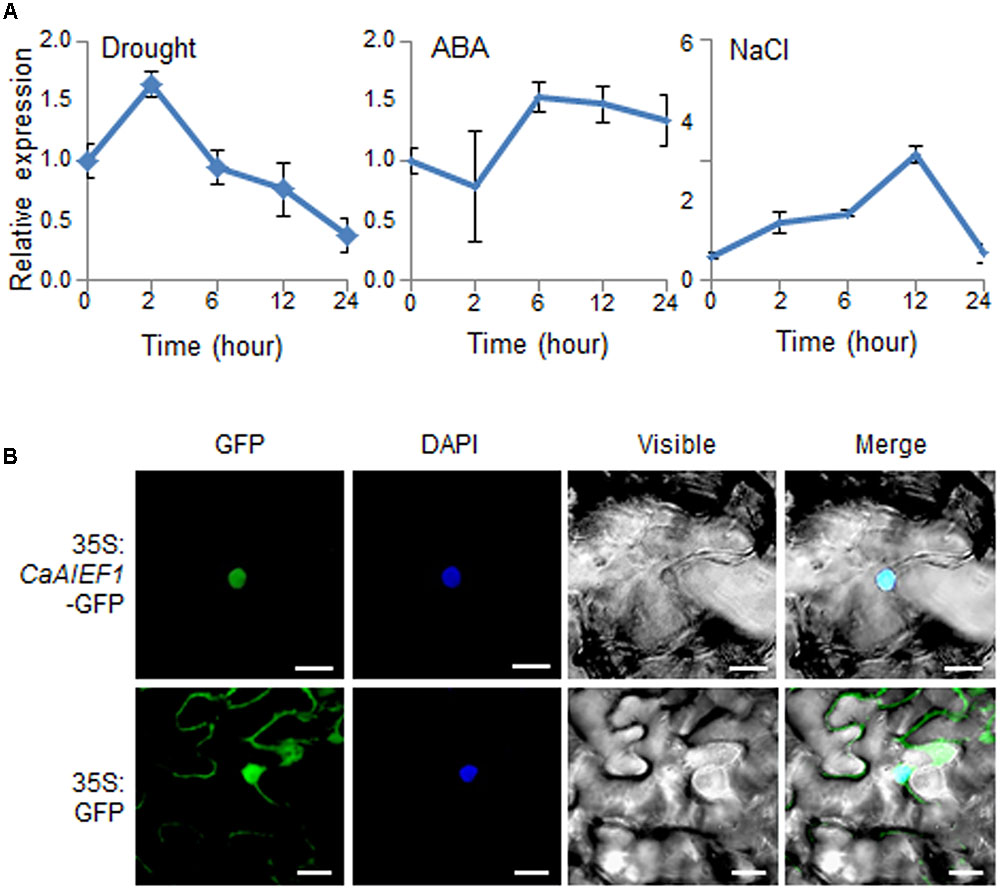
FIGURE 2. Expression of CaAIEF1 and subcellular localization of CaAIEF1. (A) The expression pattern of the CaAIEF1 gene was analyzed in the leaves of pepper plants after treatment with 100 μM abscisic acid (ABA), drought, or 200 mM NaCl. The pepper Actin1 gene was used as an internal control. (B) Subcellular localization of the 35S: CaAIEF1-GFP fusion protein in N. benthamiana epidermal cells. The 35S: CaAIEF1-GFP construct was expressed via agroinfiltration of N. benthamiana leaves and observed using a confocal laser-scanning microscope. 4′,6-Diamidino-2-phenylindole (DAPI) staining was used as a marker for the nucleus. The scale bar represents 20 μm.
CaAIEF1 has an NLS in the AP2/ERF domain of the central region (amino acids 202–219); hence, we predicted that CaAIEF1 localizes in the nucleus. To confirm the subcellular localization of the CaAIEF1 protein (Figure 2B), we used the CaAIEF1 coding region with green fluorescent protein (GFP) cDNA. GFP fluorescence signals were detected in the nucleus of N. benthamiana cells, indicating that CaAIEF1 functions in the nucleus.
Decreased Tolerance of CaAIEF1-Silenced Pepper Plants in Response to Drought Stress
The expression level of CaAIEF1 was induced by ABA and abiotic stresses; hence, we investigated the in vivo function of CaAIEF1 using virus-induced gene silencing (VIGS) in pepper plants and Arabidopsis transgenic plants. Semi-quantitative RT-PCR analysis revealed that CaAIEF1 transcripts were not expressed in CaAIEF1-silenced pepper plants in normal and drought stress conditions (Figure 3A). We investigated the biological function of CaAIEF1 in drought stress condition (Figure 3B). Under well-grown conditions, we did not observe any phenotypic differences in both plants (Figure 3B, upper panel). However, when we subjected pepper plants to dehydration for 12 days and rehydration for 3 days, CaAIEF1-silenced pepper plants showed a more wilted phenotype than control plants (Figure 3B, middle and lower panels). In addition, after rehydration, the 80% of control and only 10.0% of CaAIEF1-silenced pepper plants resumed their growth. To examine whether the drought-sensitive phenotype was cause by rapid transpirational water loss, the fresh weight of rosette leaves were measured during 8 h after detachment (Figure 3C). The higher transpirational water loss was detected in CaAIEF1-silenced plants than in control plants. Under dehydrated conditions, ABA is synthesized in several tissues and accumulates in the leaves; this leads to stomatal closure via regulation of guard cell turgor pressure (Lee and Luan, 2012). To examine the biological role of CaAIEF1 in drought stress-induced ABA signaling, we measured the stomatal apertures and leaf temperatures of control and CaAIEF1-silenced pepper plants with or without ABA (Figures 3D–G). In the absence of ABA, we did not observe differences between control and CaAIEF1-silenced pepper plants. However, after exposure to 10 and 20 μM ABA, the stomatal pore sizes were large in CaAIEF1-silenced pepper plants compared with control plants (Figures 3D,E). Moreover, the leaf temperatures were significantly lower in CaAIEF1-silenced plants than in control pepper plants, implying that increased evaporative cooling was caused by decreased stomatal closure (Figures 3F,G). Our data indicated that the decreased ABA sensitivity of CaAIEF1-silenced pepper plants leads to increased water loss and a drought-sensitive phenotype.
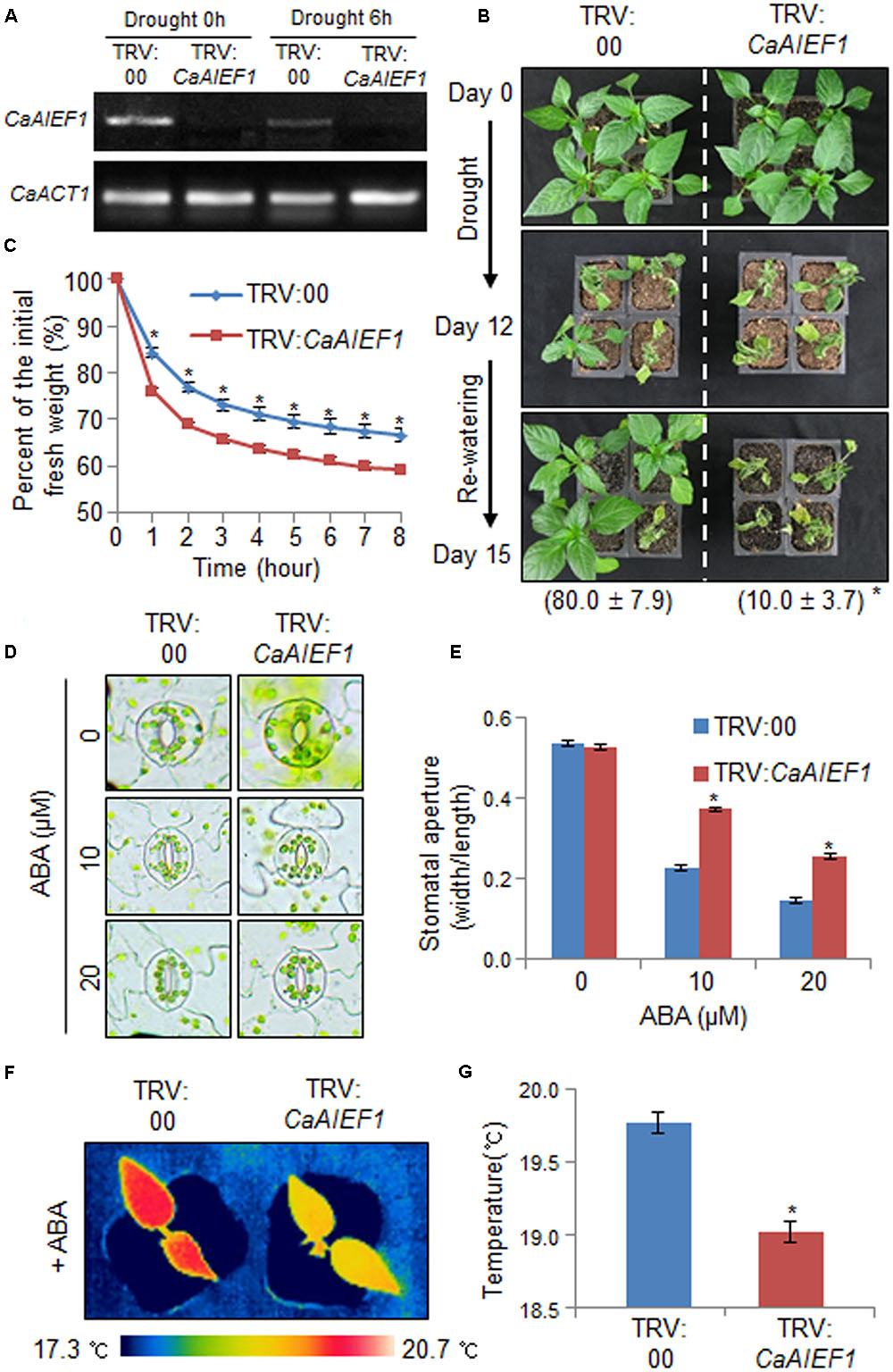
FIGURE 3. Reduced tolerance of CaAIEF1-silenced pepper plants to drought stress. (A) Reverse transcription-polymerase chain reaction (RT-PCR) analysis of CaAIEF1 expression in CaAIEF1-silenced pepper plants in normal and drought stress conditions. The pepper Actin1 (CaACT1) gene was used as an internal control. (B) Drought sensitivity of control (TRV2:00) and CaAIEF1-silenced pepper (TRV2:CaAIEF1) plants. Four-week-old plants of each line were subjected to drought stress by withholding watering for 12 days. The plants were re-watered for 3 days, after which representative images were taken and the percentages of surviving plants were calculated. Data represent the mean ± standard error of three independent experiments, each evaluating 30 plants. (C) Transpirational water loss from the leaves of control and CaAIEF1-silenced pepper plants. Leaves were detached and the fresh weights of each line were measured every hour. Data represent the mean ± standard error of three independent experiments, each evaluating 30 plants. (D,E) ABA-induced stomatal closure in control and CaAIEF1-silenced pepper plants. Leaf peels were incubated with 10 and 20 μM ABA, after which representative images were taken (D), and the stomatal apertures of each line were measured (E). (F,G) Leaf temperatures of control and CaAIEF1-silenced pepper plants after exposure to ABA. Control and CaAIEF1-silenced pepper plants were sprayed with 50 μM ABA. After 6 h, representative thermographic images were taken (F), and the mean leaf temperature was measured using the first and second leaves of each line (n = 18) (G). All data represent the mean ± SE of three independent experiments. Asterisks indicate significant differences (Student’s t-test; P < 0.05).
Hypersensitivity of CaAIEF1-OX Plants in Response to ABA
To further examine the biological function of CaAIEF1, we generated CaAIEF1-OX Arabidopsis plants. Semi-quantitative RT-PCR analysis revealed that CaAIEF1 was only expressed in CaAIEF1-OX plants (Supplementary Figure S4). CaAIEF1 is induced by ABA; hence, we predicted that enhanced expression of CaAIEF1 modulates ABA signaling. To examine whether CaAIEF1 influences the ABA-mediated signaling, we analyzed the ABA sensitivity during the germinative- and post-germinative stages, including seed germination, seedling establishment, and root growth (Figure 4). We did not observe marked differences in germination rates between both plants on growth media without ABA. In the presence of 1.5 μM ABA, CaAIEF1-OX seeds germinated slower than wild-type seeds (Figure 4A). Moreover, CaAIEF1-OX plants were hypersensitive to ABA at seedling stages in seedling establishment (Figures 4B–D) root growth (Figures 4E,F). To determine whether ABA hypersensitivity in root growth and seedling establishment in CaAIEF1-OX plants is caused by late germination, the seedlings at 3 days after germination in the absence of ABA were transferred to medium including ABA. As shown in Supplementary Figure S5, the CaAIEF1-OX plants also exhibited ABA sensitive phenotype. The ABA-hypersensitive phenotype displayed by CaAIEF1-OX plants indicates that CaAIEF1 modulates the ABA-mediated response.
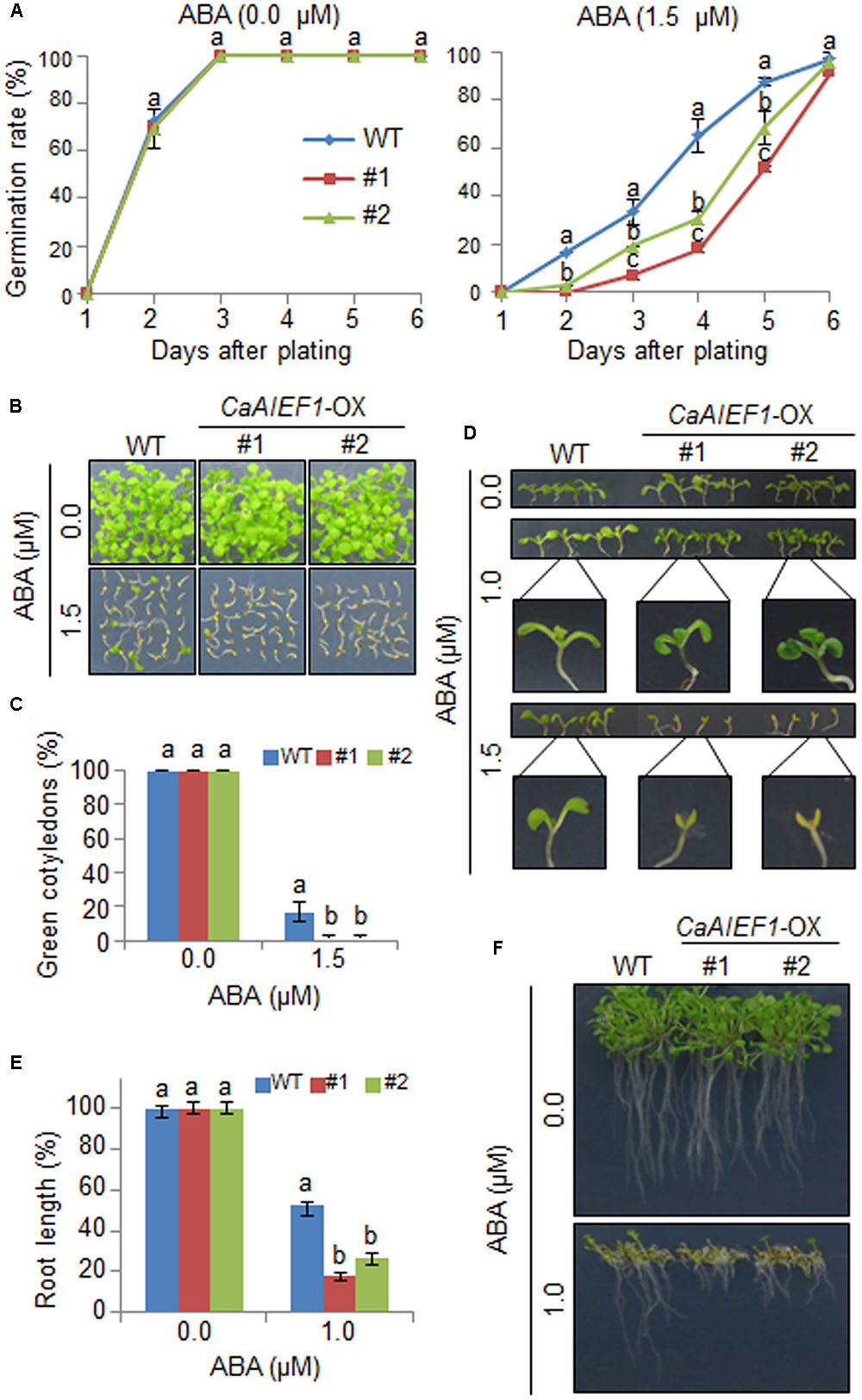
FIGURE 4. Increased sensitivity of CaAIEF1-overexpressing (OX) transgenic Arabidopsis plants to ABA during germination and seedling growth. (A) Seed germination of wild-type (WT) and transgenic lines in response to ABA. Seeds were germinated on 0.5× Murashige and Skoog (MS) agar plates containing 0.0 μM or 1.5 μM ABA. Data represent the mean ± standard error values obtained after evaluating 150 seeds from three independent experiments. (B–D) Growth of WT and transgenic seedlings on 0.5× MS agar plates containing various concentrations of ABA. Representative photographs were taken 12 days after plating (B,C). Quantification of green cotyledons in the wild-type and each mutant line was performed 12 days after plating (D). Data represent the mean ± standard error values obtained after evaluating 72 seeds from three independent experiments. (E,F) Root elongation of WT and transgenic plants in response to ABA. The root length of each plant was measured 9 days after plating. Data represent the mean ± standard error of three independent experiments. Different letters indicate significant differences (ANOVA; P < 0.05).
Increased Tolerance of CaAIEF1-OX Plants in Response to Drought Stress
CaAIEF1-silenced pepper plants showed a drought-sensitive phenotype; hence, we examined whether CaAIEF1-OX plants exhibit drought tolerant phenotype (Figure 5). Under well-grown conditions, we did not observe phenotypic differences in both plants (Figure 5A, left panel). However, when we treated drought stress by dehydration for 11 days and rehydration for 2 days, CaAIEF1-OX plants showed a less wilted phenotype than wild-type plants (Figure 5A, middle and right panels). In addition, the survival rate of CaAIEF1-OX plants (76.6–93.8%) was higher than that of wild-type plants (10.7%). To examine whether different transpirational water loss leads to altered drought tolerance, the leaf fresh weight was measured (Figure 5B). In accordance with the drought tolerance, the transpirational water loss was lower in CaAIEF1-OX plants than in wild-type plants. To examine the biological role of CaAIEF1 in drought stress-induced ABA signaling, we measured the stomatal apertures and leaf temperatures of wild-type and CaAIEF1-OX plants with or without ABA (Figures 5C–E). We did not observe any differences between both plants. ABA treatment led to an increase in stomatal closure in wild-type and transgenic plants. However, after exposure to 10 and 20 μM ABA, the stomatal pore sizes were small in CaAIEF1-OX plants compared with wild-type plants (Figures 5C,D). We measured leaf temperatures, as indicator of stomatal opening/closure via evaporative cooling. The leaf temperatures were significantly higher in CaAIEF1-OX plants than in wild-type plants (Figure 5E). Our results imply that the ABA hypersensitivity displayed by CaAIEF1-OX plants leads to decreased water loss and contributes to a drought-tolerant phenotype.
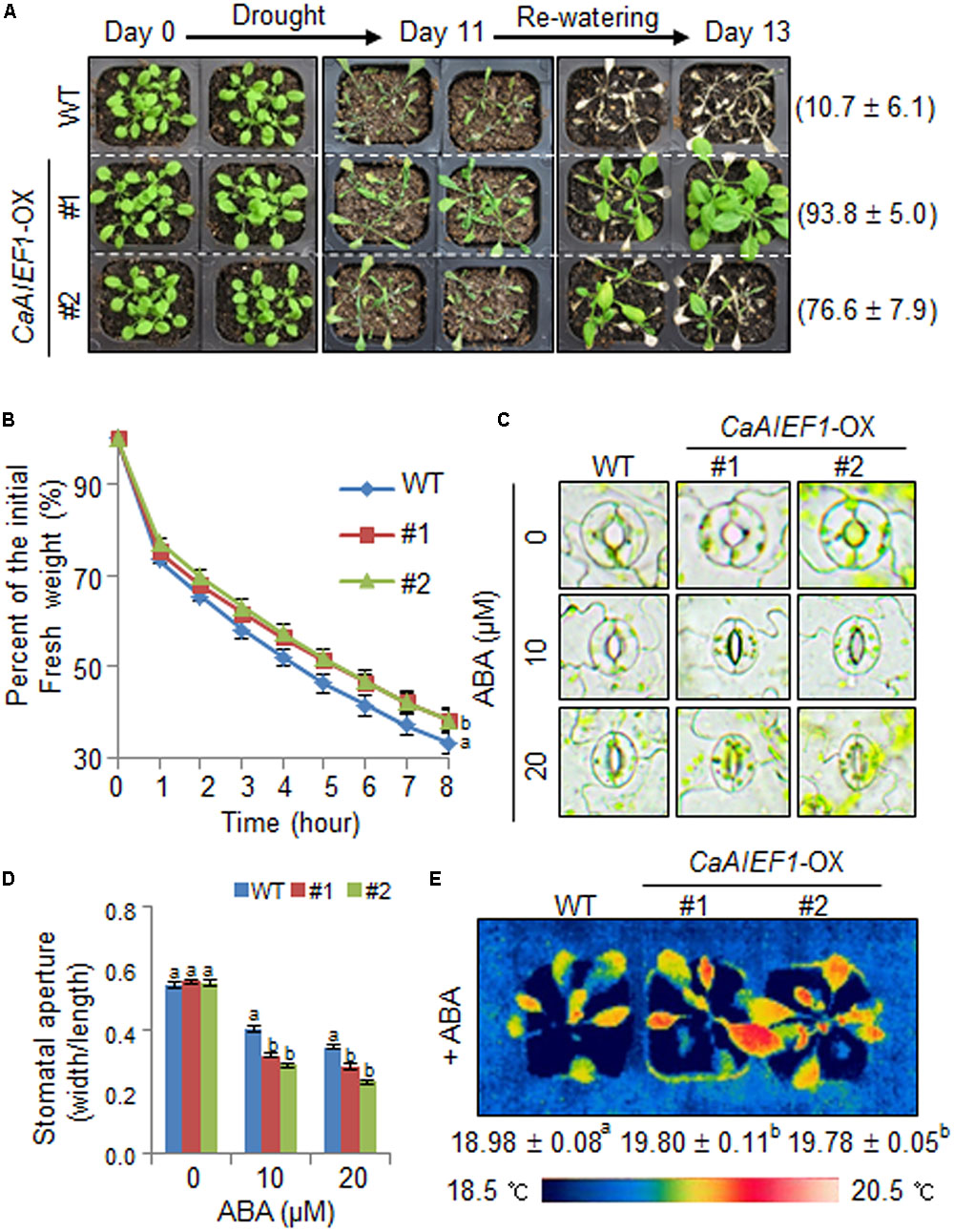
FIGURE 5. Increased tolerance of CaAIEF1-OX plants to drought stress. (A) Drought-tolerant phenotype of CaAIEF1-OX transgenic plants. Four-week-old wild-type (WT) and transgenic plants were subjected to drought stress by withholding watering for 11 days and then re-watering for 2 days. Representative images were taken before (left) and after (middle) drought and after 2 days of re-watering (right). Survival rates of plants were calculated after 2 days of re-watering. Data represent the mean ± standard error of three independent experiments, each evaluating 20 plants. (B) Transpirational water loss from the leaves of WT and transgenic plants at various time points after detachment of rosette leaves. Data represent the mean ± standard error of three independent experiments, each evaluating 50 leaves. (C,D) Stomatal apertures in WT and CaAIEF1-OX plants treated with ABA. Leaf peels were harvested from 4-week-old plants of each line and incubated in stomatal opening solution (SOS) buffer containing the indicated concentrations of ABA. Representative images were taken under a microscope (C) and the stomatal apertures were measured (D). Data represent the mean ± standard error of three independent experiments. (E) Increased leaf temperatures of CaAIEF1-OX plants in response to ABA treatment. Data represent the mean ± standard error of three independent experiments, each evaluating 10 plants. Different letters indicate significant differences (ANOVA; P < 0.05).
Several previous studies suggested that the expression of stress-responsive genes influence stress tolerance or stress sensitivity; hence, we examined whether stress-responsive genes in pepper and Arabidopsis are directly or indirectly regulated by the expression level of CaAIEF1 (Figure 6). We subjected plants to dehydration stress by removing the roots from the soil. Contrary to our expectations, q-PCR assay revealed that the induction of stress-responsive genes was higher in CaAIEF1-silenced pepper leaves than in control pepper leaves (Figure 6A). However, the induction of several stress-responsive genes, including NCED3, RD29A, RD29B, COR15A, RAB18, and KIN1, was lower in CaAIEF1-OX leaves than in wild-type leaves (Figure 6B). Our data imply that CaAIEF1 expression negatively affected the expression of stress-responsive genes.
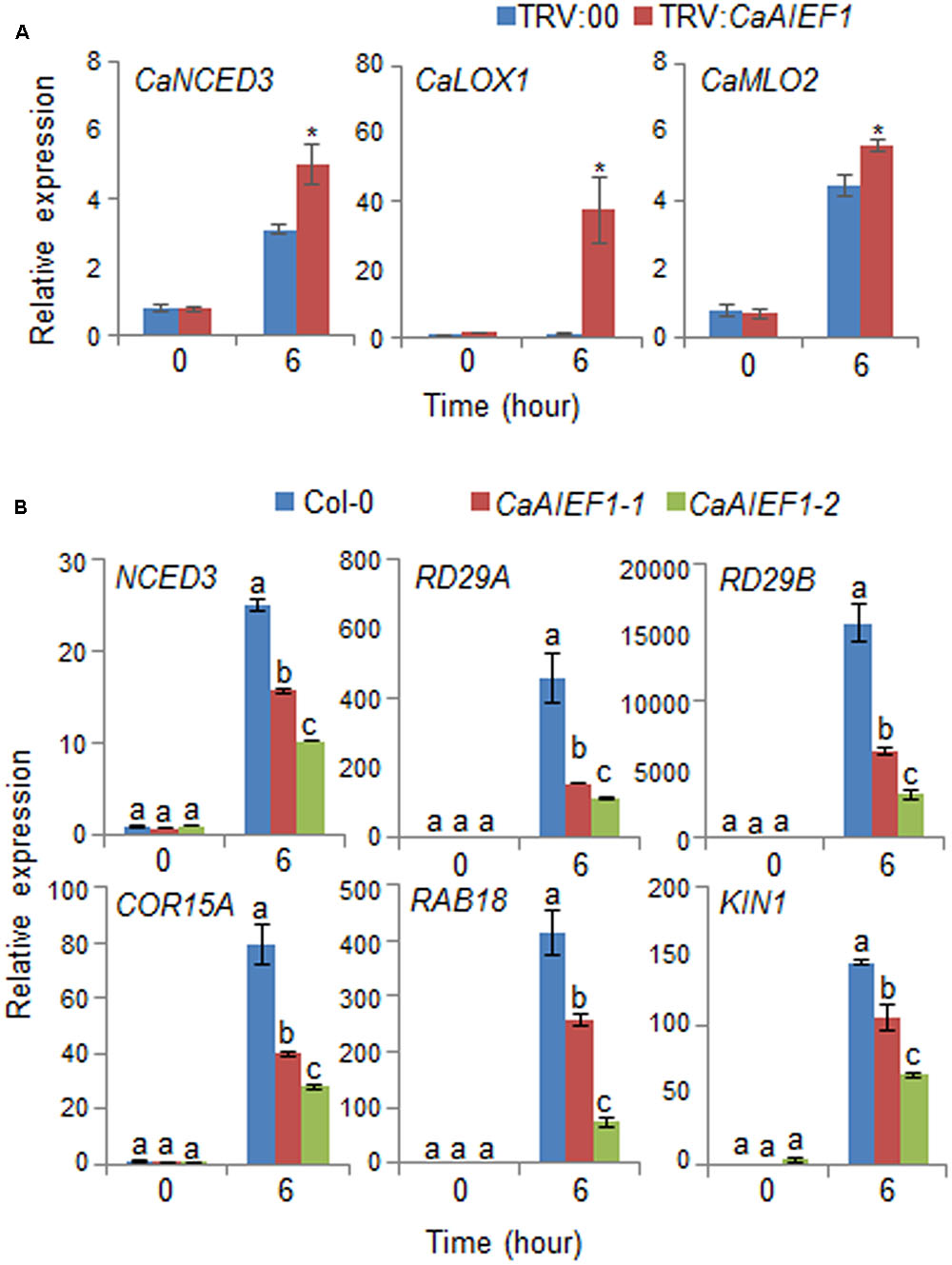
FIGURE 6. Quantitative real-time polymerase chain reaction (qPCR) analysis of stress-responsive genes in the leaves of CaAIEF1-silenced pepper plants (A) and CaAIEF1-OX transgenic Arabidopsis (B) plants exposed to drought stress; analysis was performed 6 h after detachment. The relative expression levels (ΔΔCT) of each gene were normalized to the geometric mean of pepper Actin1 and Arabidopsis Actin8 as internal control genes. Data represent the mean ± standard error of three independent experiments. Different letters indicate significant differences between wild-type and transgenic lines (P < 0.05; ANOVA followed by Fisher’s LSD test).
Discussion
The plant hormone ABA plays a critical role in adaptive response to environmental stresses. Plant response and adaptation to abiotic stress is dependent on the several hormone signals, to regulate the various transcription factor expressions that enable plants to adapt specific stress. The results of our present study suggest that CaAIEF1 is one of ABA signaling component in pepper plants. Many regulatory genes—including transcription factors, protein kinase, and phosphatase—have been identified as essential components of the molecular network in ABA signaling, and the involvement of transcription factors in the stress response is well established (Seo and Koshiba, 2002; Cutler et al., 2010; Wind et al., 2013; Ding et al., 2015; Tian et al., 2015; Li et al., 2016). Several previous studies have demonstrated that the target genes of transcription factors are activated or repressed and that expression or silencing of these genes is the cue for regulation of the stress response. For example, the ABI3—which is upstream of ABI5 in the ABA response—activates ABI5 transcription, leading to acquired osmotic stress tolerance (Lopez-Molina et al., 2002). Ethylene modulates plant growth and development as well as defense response to environmental stresses. In previous studies, several AP2/ERF transcription factors are involved in response to stress signaling (Licausi et al., 2013; Wang et al., 2015a,b). Transcription factors contain at least one DNA-binding domain, which specifically adheres to a distinct promoter region of target genes. In general, plant transcription factors have activation domains, which contain specific amino acids rich region, such as glutamine, proline, and acidic amino acids (Licausi et al., 2013). AP2-containing transcription factors function as transcription activators or repressors, which usually contain activation or repression domain, respectively (Ikeda and Ohme-Takagi, 2009; Licausi et al., 2013). The repression domain accords repression to the transcription factor by binding to the promoter of target genes (Ohta et al., 2001). CaAIEF1 contains an AP2 domain for DNA binding and an acidic domain for transactivation (Figure 1); however, we were unable to find a repression motif. Our results imply that CaAIEF1 functions as a transcription activator in yeast system.
The transcripts of ABA biosynthesis-, ABA signaling-, and defense responsive-genes are essential for plant adaptation and survival under dehydrated conditions (Zhang et al., 2006; Aubert et al., 2010; Hubbard et al., 2010; Fujita et al., 2011; Lim et al., 2015a). CaAIEF1-silenced pepper plants and CaAIEF1-OX transgenic Arabidopsis plants were used to elucidate the biological function of CaAIEF1. The transformation efficiency was very low in pepper plants; hence, a VIGS analysis in pepper plants and overexpression in Arabidopsis were used to investigate the biological functions of CaAIEF1. CaAIEF1-silenced pepper plants exhibited an ABA-insensitive and drought-sensitive phenotypes characterized by reduced stomatal closure and low leaf temperatures (Figure 3). CaAIEF1-OX Arabidopsis plants displayed a contrasting phenotype to that of CaAIEF1-silenced pepper plants (Figures 4, 5). Under abiotic stress conditions, ABA biosynthesis-related genes—including NCED3—are induced, and ABA is produced in various plant tissues, leading to the defense response (Iuchi et al., 2001). Previous studies have shown that altered expression of defense responsive genes is closely associated with stress tolerance and sensitivity (Verslues and Bray, 2006; Shinozaki and Yamaguchi-Shinozaki, 2007). Therefore, we predicted that under dehydration, the expression levels of defense responsive genes in CaAIEF1-silenced pepper plants and CaAIEF1-OX Arabidopsis plants are down-regulated and up-regulated, respectively. However, contrary to our prediction, the transcription of defense responsive genes in CaAIEF1-silenced pepper and CaAIEF1-OX Arabidopsis plants were up-regulated and down-regulated, respectively. If CaAIEF1-silenced pepper plants do not have the mechanisms to initiate a successful defense response, these plants are not able to alleviate drought stress and therefore cannot adapt to these stress conditions; hence, the drought stress signal is continually transferred to various tissues, leading to defense responsive gene expression. Moreover, this stress signal also induces defense response-related gene expression. Urano et al. (2009) demonstrated that NCED3 positively modulates the expression of defense responsive genes; therefore, up- or down-regulation of NCED3 affects the transcription of defense responsive genes. Moreover, CaAIEF1 may regulate the transcription of different stress-related genes; however, the precise target genes of this transcription factor remain elusive.
Conclusion
The CaAIEF1 protein localizes to the nucleus, where it presumably acts as a transcriptional regulator. Loss-of function of CaAIEF1 in pepper plants led to increased drought sensitivity, whereas ectopic expression of CaAIEF1 in Arabidopsis induced ABA hypersensitivity and drought tolerance. Taken together, our data indicate that CaAIEF1 positively regulates the defense response to drought stress. The precise mechanisms involved in the activation of the defense response by CaAIEF1 remain unclear, and further studies to identify the downstream target genes of this transcription factor are required.
Author Contributions
EH and CWL performed experiments and analyzed the results. S-WH and SCL designed the experiments and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the “Cooperative Research Program for Agriculture & Technology Development (Project No. PJ01100501)” Rural Development Administration and the Research Foundation of Korea funded by the government (NRF-2015R1A2A2A01002674) of the Republic of Korea.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01407/full#supplementary-material
FIGURE S1 | Sequence logo drawn based on the alignment of amino acid sequences of AP2/ERF domain. The overall height of the stack indicates the sequence conservation at that position, while the height of symbols within the stack indicates the relative frequency of each amino at that position.
FIGURE S2 | Comparisons of the deduced amino acid sequence of the CaAIEF1 protein with Capsicum annuum proteins. Identical amino acid residues are highlighted in black, and the upper line indicates the VIGS target region.
FIGURE S3 | Expression of CaAIEF1. (A) The expression pattern of the CaAIEF1 gene was analyzed in the leaves of pepper plants after treatment with ethylene (10 μL/L). (B) The expression pattern of the CaAIEF1 gene was analyzed in the leaves of pepper plants in developmental stages. The pepper Actin1 gene was used as an internal control.
FIGURE S4 | Expression of the CaAIEF1 gene in transgenic Arabidopsis plants. Reverse transcription-polymerase chain reaction (RT-PCR) analysis of CaAIEF1 expression in transgenic Arabidopsis plants overexpressing CaAIEF1. The expression level of CaAIEF1 was analyzed in the leaves of 4-week-old CaAIEF1-OX Arabidopsis plants. The Arabidopsis Actin8 gene was used as an internal control.
FIGURE S5 | Increased sensitivity of CaAIEF1-OX transgenic Arabidopsis plants to abscisic acid (ABA) during post-germinative stage. (A) Root elongation of wild type and transgenic plants exposed to ABA after germination. Three-day-old seedlings grown on 0.5X MS were transferred to 0.5X MS containing 0, 10, or 30 μM ABA. After 9 days, the representative images were taken. (B) Growth of wild-type and transgenic plants exposed to ABA after germination. Two-day-old seedlings grown on 0.5X MS were transferred to 0.5X MS containing 0 or 5 μM ABA. After 4 days, the representative images were taken, and the cotyledon greening in each line was measured. Data represent the mean ± standard error values obtained after evaluating 50 plants from three independent experiments. Different letters indicate significant differences (ANOVA; P < 0.05).
References
Apse, M. P., and Blumwald, E. (2002). Engineering salt tolerance in plants. Curr. Opin. Biotechnol. 13, 146–150. doi: 10.1016/S0958-1669(02)00298-7
Aubert, Y., Vile, D., Pervent, M., Aldon, D., Ranty, B., Simonneau, T., et al. (2010). RD20, a stress-inducible caleosin, participates in stomatal control, transpiration and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol. 51, 1975–1987. doi: 10.1093/Pcp/Pcq155
Cantin, G. T., Stevens, J. L., and Berk, A. J. (2003). Activation domain-mediator interactions promote transcription preinitiation complex assembly on promoter DNA. Proc. Natl. Acad. Sci. U.S.A. 100, 12003–12008. doi: 10.1073/pnas.2035253100
Cheng, M. C., Liao, P. M., Kuo, W. W., and Lin, T. P. (2013). The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 162, 1566–1582. doi: 10.1104/pp.113.221911
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R., and Abrams, S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. doi: 10.1146/annurev-arplant-042809-112122
Ding, S., Zhang, B., and Qin, F. (2015). Arabidopsis RZFP34/CHYR1, a ubiquitin E3 ligase, regulates stomatal movement and drought tolerance via SnRK2.6-mediated phosphorylation. Plant Cell 27, 3228–3244. doi: 10.1105/tpc.15.00321
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Froidure, S., Canonne, J., Daniel, X., Jauneau, A., Briere, C., Roby, D., et al. (2010). AtsPLA2-alpha nuclear relocalization by the Arabidopsis transcription factor AtMYB30 leads to repression of the plant defense response. Proc. Natl. Acad. Sci. U.S.A. 107, 15281–15286. doi: 10.1073/pnas.1009056107
Fu, Z. Q., and Dong, X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Fujita, Y., Fujita, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124, 509–525. doi: 10.1007/s10265-011-0412-3
Gong, W., Shen, Y. P., Ma, L. G., Pan, Y., Du, Y. L., Wang, D. H., et al. (2004). Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiol. 135, 773–782. doi: 10.1104/pp.104.042176
Hahn, S. (1993). Structure(?) and function of acidic transcription activators. Cell 72, 481–483. doi: 10.1016/0092-8674(93)90064-W
Hubbard, K. E., Nishimura, N., Hitomi, K., Getzoff, E. D., and Schroeder, J. I. (2010). Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev. 24, 1695–1708. doi: 10.1101/gad.1953910
Ikeda, M., and Ohme-Takagi, M. (2009). A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol. 50, 970–975. doi: 10.1093/pcp/pcp048
Iuchi, S., Kobayashi, M., Taji, T., Naramoto, M., Seki, M., Kato, T., et al. (2001). Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 27, 325–333. doi: 10.1046/j.1365-313x.2001.01096.x
Jakoby, M., Weisshaar, B., Droge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. doi: 10.1016/S1360-1385(01)02223-3
Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY((TM)) vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195. doi: 10.1016/S1360-1385(02)02251-3
Lee, S. C., Choi, H. W., Hwang, I. S., Choi, D. S., and Hwang, B. K. (2006). Functional roles of the pepper pathogen-induced bZIP transcription factor, CAbZIP1, in enhanced resistance to pathogen infection and environmental stresses. Planta 224, 1209–1225. doi: 10.1007/s00425-006-0302-4
Lee, S. C., and Luan, S. (2012). ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ. 35, 53–60. doi: 10.1111/j.1365-3040.2011.02426.x
Li, T., Wu, X. Y., Li, H., Song, J. H., and Liu, J. Y. (2016). A dual-function transcription factor, AtYY1, is a novel negative regulator of the Arabidopsis ABA response network. Mol. Plant 9, 650–661. doi: 10.1016/j.molp.2016.02.010
Licausi, F., Ohme-Takagi, M., and Perata, P. (2013). APETALA2/ethylene responsive factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 199, 639–649. doi: 10.1111/nph.12291
Lim, C. W., Han, S. W., Hwang, I. S., Kim, D. S., Hwang, B. K., and Lee, S. C. (2015a). The pepper lipoxygenase CaLOX1 plays a role in osmotic, drought and high salinity stress response. Plant Cell Physiol. 56, 930–942. doi: 10.1093/pcp/pcv020
Lim, C. W., Hwang, B. K., and Lee, S. C. (2015b). Functional roles of the pepper RING finger protein gene, CaRING1, in abscisic acid signaling and dehydration tolerance. Plant Mol. Biol. 89, 143–156. doi: 10.1007/s11103-015-0359-1
Lim, C. W., and Lee, S. C. (2016). Pepper protein phosphatase type 2C, CaADIP1 and its interacting partner CaRLP1 antagonistically regulate ABA signalling and drought response. Plant Cell Environ. 39, 1559–1575. doi: 10.1111/pce.12721
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Llorca, C. M., Potschin, M., and Zentgraf, U. (2014). bZIPs and WRKYs: two large transcription factor families executing two different functional strategies. Front. Plant Sci. 5:169. doi: 10.3389/fpls.2014.00169
Lopez-Molina, L., Mongrand, S., McLachlin, D. T., Chait, B. T., and Chua, N. H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32, 317–328. doi: 10.1046/j.1365-313X.2002.01430.x
Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 86–96. doi: 10.1016/j.bbagrm.2011.08.004
Muller, M., and Munne-Bosch, S. (2015). Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol. 169, 3–41. doi: 10.1104/pp.15.00677
Murata, Y., Mori, I. C., and Munemasa, S. (2015). Diverse stomatal signaling and the signal integration mechanism. Annu. Rev. Plant Biol. 66, 369–392. doi: 10.1146/annurev-arplant-043014-114707
Ohta, M., Matsui, K., Hiratsu, K., Shinshi, H., and Ohme-Takagi, M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13, 1959–1968. doi: 10.1105/tpc.13.8.1959
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Schwechheimer, C., Smith, C., and Bevan, M. W. (1998a). The activities of acidic and glutamine-rich transcriptional activation domains in plant cells: design of modular transcription factors for high-level expression. Plant Mol. Biol. 36, 195–204.
Schwechheimer, C., Zourelidou, M., and Bevan, M. W. (1998b). Plant transcription factor studies. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 127–150. doi: 10.1146/annurev.arplant.49.1.127
Seo, M., and Koshiba, T. (2002). Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 7, 41–48. doi: 10.1016/S1360-1385(01)02187-2
Shinozaki, K., and Yamaguchi-Shinozaki, K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227. doi: 10.1093/jxb/erl164
Song, C. P., Agarwal, M., Ohta, M., Guo, Y., Halfter, U., Wang, P., et al. (2005). Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17, 2384–2396. doi: 10.1105/tpc.105.033043
Tan, B. C., Joseph, L. M., Deng, W. T., Liu, L., Li, Q. B., Cline, K., et al. (2003). Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 35, 44–56. doi: 10.1046/j.1365-313X.2003.01786.x
Thatcher, L. F., Kazan, K., and Manners, J. M. (2012). Lateral organ boundaries domain transcription factors: new roles in plant defense. Plant Signal. Behav. 7, 1702–1704. doi: 10.4161/psb.22097
Tian, M., Lou, L., Liu, L., Yu, F., Zhao, Q., Zhang, H., et al. (2015). The RING finger E3 ligase STRF1 is involved in membrane trafficking and modulates salt-stress response in Arabidopsis thaliana. Plant J. 82, 81–92. doi: 10.1111/tpj.12797
Urano, K., Maruyama, K., Ogata, Y., Morishita, Y., Takeda, M., Sakurai, N., et al. (2009). Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J. 57, 1065–1078. doi: 10.1111/j.1365-313X.2008.03748.x
Verslues, P. E., and Bray, E. A. (2006). Role of abscisic acid (ABA) and Arabidopsis thaliana ABA-insensitive loci in low water potential-induced ABA and proline accumulation. J. Exp. Bot. 57, 201–212. doi: 10.1093/jxb/erj026
Wang, X., Han, H., Yan, J., Chen, F., and Wei, W. (2015a). A new AP2/ERF transcription factor from the oil plant Jatropha curcas confers salt and drought tolerance to transgenic tobacco. Appl. Biochem. Biotechnol. 176, 582–597. doi: 10.1007/s12010-015-1597-z
Wang, X., Liu, S., Tian, H., Wang, S., and Chen, J. G. (2015b). The small ethylene response factor ERF96 is involved in the regulation of the abscisic acid response in Arabidopsis. Front. Plant Sci. 6:1064. doi: 10.3389/fpls.2015.01064
Wind, J. J., Peviani, A., Snel, B., Hanson, J., and Smeekens, S. C. (2013). ABI4: versatile activator and repressor. Trends Plant Sci. 18, 125–132. doi: 10.1016/j.tplants.2012.10.004
Xu, Z. S., Chen, M., Li, L. C., and Ma, Y. Z. (2011). Functions and application of the AP2/ERF transcription factor family in crop improvement. J. Integr. Plant Biol. 53, 570–585. doi: 10.1111/j.1744-7909.2011.01062.x
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803. doi: 10.1146/annurev.arplant.57.032905.105444
Zhang, J. H., Jia, W. S., Yang, J. C., and Ismail, A. M. (2006). Role of ABA in integrating plant responses to drought and salt stresses. Field Crops Res. 97, 111–119. doi: 10.1016/j.fcr.2005.08.018
Keywords: abscisic acid, CaAIEF1, drought stress, pepper, virus-induced gene silencing
Citation: Hong E, Lim CW, Han S-W and Lee SC (2017) Functional Analysis of the Pepper Ethylene-Responsive Transcription Factor, CaAIEF1, in Enhanced ABA Sensitivity and Drought Tolerance. Front. Plant Sci. 8:1407. doi: 10.3389/fpls.2017.01407
Received: 21 June 2017; Accepted: 28 July 2017;
Published: 22 August 2017.
Edited by:
Girdhar Kumar Pandey, University of Delhi, IndiaReviewed by:
Hye Sun Cho, Korea Research Institute of Bioscience and Biotechnology, South KoreaHyong Woo Choi, Boyce Thompson Institute, United States
Du Seok Choi, Pohang University of Science and Technology, South Korea
Copyright © 2017 Hong, Lim, Han and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sang-Wook Han, c3doYW5AY2F1LmFjLmty Sung Chul Lee, c2NsZWUxOTcyQGNhdS5hYy5rcg==
†These authors have contributed equally to this work.
 Eunji Hong1†
Eunji Hong1† Chae Woo Lim
Chae Woo Lim Sung Chul Lee
Sung Chul Lee