- 1International Rice Research Institute, South Asia Hub, International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Patancheru, India
- 2International Rice Research Institute, Metro Manila, Philippines
Dry direct-seeded rice (DSR) is an alternative crop establishment method with less water and labor requirement through mechanization. It provides better opportunities for a second crop during the cropping season and therefore, a feasible alternative system to transplanted lowland rice. However, lodging is one of the major constraints in attaining high yield in DSR. Identification of QTLs for lodging resistance and their subsequent use in improving varieties under DSR will be an efficient breeding strategy to address the problem. In order to map the QTLs associated with lodging resistance, a set of 253 BC3F4 lines derived from a backcross between Swarna and Moroberekan were evaluated in two consecutive years. A total of 12 QTLs associated with lodging resistance traits [culm length (qCL), culm diameter (qCD), and culm strength (qCS)] were mapped on chromosomes 1, 2, 6, and 7 using 193 polymorphic SNP markers. Two major and consistent effect QTLs, namely qCD1.1 (with R2 of 10%) and qCS1.1 (with R2 of 14%) on chromosome 1 with id1003559 being the peak SNP marker (flanking markers; id1001973-id1006772) were identified as a common genomic region associated with important lodging resistance traits. In silico analysis revealed the presence of Gibberellic Acid 3 beta-hydroxylase along with 34 other putative candidate genes in the marker interval region of id1001973-id1006772. The positive alleles for culm length, culm diameter, and culm strength were contributed by the upland adaptive parent Moroberekan. Our results identified significant positive correlation between lodging related traits (culm length diameter and strength) and grain yield under DSR, indicating the role of lodging resistant traits in grain yield improvement under DSR. Deployment of the identified alleles influencing the culm strength and culm diameter in marker assisted introgression program may facilitate the lodging resistance under DSR.
Introduction
Lodging in cereal crops is the result of the combined effects of the plants morphological traits and adverse weather conditions such as heavy winds and rains. Reduced crop yields, quality deterioration, and minimized harvesting efficiency are significantly associated with lodging (Kono, 1995). In recent years, due to limited water supply and escalating labor costs, direct-seeded rice (DSR) has become a major alternative among farmers in tropical countries (Sinniah et al., 2012). However, the large-scale cultivation of DSR is also prone to lodging, along with other problems such as uncontrolled weeds, yield reductions, and poor nutrient uptake (Nguyen and Ferrero, 2006). Moreover, lodging has been found to be more severe in DSR than in transplanted rice (Setter et al., 1997). Three types of lodging exist in cereal crops, namely: root lodging, stem bending, and stem breakage (Kono, 1995). Dry DSR is most susceptible to root lodging due to its shallow rooting system (Kobayashi et al., 2006; Hirano et al., 2014). Lowland rice is generally affected by the bending type of lodging due to the bending pressure of the upper internodes during strong winds and rains. Stem breakage occurs in the lower internodes of culms having a thin diameter and weak tensile strength (Islam et al., 2007).
Lodging resistance is a complex trait and influenced by many interacting agro-morphological traits. Different techniques have been used to measure the lodging effects in various crops (Kono, 1995; Watanabe, 1997), the most common of which is pushing the resistance of the lower part of the plant (Terashima et al., 1992; Won et al., 1998; Berry et al., 2003).
A number of investigations reported the culm diameter and thickness as the major factors contributing to lodging resistance (Ookawa and Ishihara, 1992; Zuber et al., 1999; Zhu et al., 2008; Ookawa et al., 2010). Various reports have found the correlation of culm strength with culm length, culm diameter, and yield in many cereal crops such as rice (Kashiwagi et al., 2008; Zhu et al., 2008), wheat (Keller et al., 1999; Zuber et al., 1999; Kelbert et al., 2004; Verma et al., 2005), maize (Flint-Garcia et al., 2003) and barley (Sameri et al., 2009).
The short stature of plants has earlier been the main target in improving the lodging resistance and harvest index of rice (Keller et al., 1999; Peng and Khush, 2003). However, statements cannot be generalized as the susceptibility of rice plant to lodging varies among cultivars with short plant height (Terashima et al., 1992). Reducing the plant's height also reduces its photosynthetic capacity and leads to a decrease in total biomass production, thus, restricting the plant's potential for further yield increase (Flintham et al., 1997; Murai et al., 2002; Okuno et al., 2014). The dwarfing gene (sd1) associated with short plant height has been used to show the negative pleiotropic effects on culm morphology in rice (Yano et al., 2015). Therefore, moderate plant height, large stem diameter, thick stem walls, and high lignin deposition have been recommended as the corrected preferential traits for the improvement of lodging resistance under DSR (Mackill et al., 1996).
Considering the current yield constraints under DSR conditions due to lodging, the identification of quantitative trait loci (QTLs) and candidate genes will increase our understanding of the regulatory mechanism of lodging resistance and will help breeders to improve lodging resistance under this system. A number of QTL mapping studies for culm length, strength, and thickness related to lodging resistance have been carried out using different rice segregating populations (Kashiwagi and Ishimaru, 2004; Mu et al., 2004; Kashiwagi et al., 2008; Zhu et al., 2008; Ookawa et al., 2010; Yano et al., 2015). By keeping all these points in mind, the present study aimed to (1) identify the genomic regions associated with lodging resistance, (2) examine the correlations of lodging resistance traits with grain yield, and (3) look into the possible candidate genes within the identified genetic regions.
Materials and Methods
Plant Materials and Field Experiments
The field experiments were conducted during the wet seasons (WS) of 2014 and 2015 at the International Rice Research Institute – South Asia Hub (IRRI-SAH), ICRISAT, Hyderabad, India (78° 16′longitude, 17° 32′ latitude with 540 m above the sea level). The experimental material consisted of a set of 253 lines derived from a backcross mapping population of Swarna*3/Moroberekan (Dixit et al., 2014). Genomic regions associated with early vigor and direct seeded rice related traits have been reported in same background in our previous study (Singh et al., 2017). The field trials were laid out in alpha-lattice design in two replications in the year 2014 and in augmented RCBD in the year 2015. Seeds were direct seeded in leveled, unpuddled soil under aerobic conditions by dibbling at a depth of 3 cm. Single row plot of 4 m were planted with row spacing of 20 cm. The pre-emergence herbicide Pendimethalin @1.5 ml/l was applied within 2 days after seeding (DAS) at surface moisture level. Manual weeding was followed regularly with 2 sprays of the post-emergence spray bispyribac-sodium (Nominee Gold) at 2 ml/l. The appropriate dosages of fertilizers and nutrients were administered during the critical stages of growth as recommended under the DSR system (Dingkuhn et al., 1990; Pal et al., 2008). The trials were irrigated once in a week throughout the cropping period. The tensiometers were installed across the field at 30 cm soil depth to measure soil moisture levels.
Measurement of Culm Strength and Other Traits
At fully ripened stage, a prostrate tester (Daiki Rika Kogyou Co., Tokyo, Japan) was set perpendicular to the middle of the plant from 20 to 25 cm above the ground and the plant was pushed at an angle of 45° to measure the pushing resistance (Kashiwagi and Ishimaru, 2004). Culm strength (CS) was then estimated using the following formula (Hai et al., 2005):
The culm strength was averaged over three plants for each line of the mapping population. Culm diameter (CD) in mm was measured using a sliding vernier caliper in the field from the same three plants at a height of 30 cm above ground level. Culm length (CL) in cm was measured as plant height minus panicle length (Mu et al., 2004). The other morphological traits viz., days to 50% flowering, plant height (cm), tiller number, panicle length (cm), and grain yield (kg ha−1) were recorded in both the years of 2014 and 2015. Days to 50% flowering (DTF) was recorded as when 50% of the panicles across the plot have emerged. At maturity plant height (cm) of three randomly chosen plants per plot was measured from ground level to the tip of the highest panicle and then averaged for analysis. The grain were harvested from each plot at physiological maturity, dried to moisture content ~14%, and then weighed to calculate the grain yield in kg ha−1. Tiller numbers were counted manually and panicle length was measured using a centimeter scale.
Statistical Analysis
Genotype means for each trial were estimated in the first stage analysis using the REML option of Proc Mixed (Littell et al., 2006), taking lines as fixed and replicates and blocks as random. In the second stage, a mixed model is fitted to the genotype-environment table of means and the weights estimated in the first stage (Frensham et al., 1997; Möhring and Piepho, 2009). The weights are the reciprocal of the variance of genotype means. The experimental error of each genotype-year combination is weighted by the variance of mean of the particular year. The mixed model is
Where γij is the adjusted mean of the ith genotype in the jth environment, μj is the main effect of the jth environment, gi is the main effect of the ith genotype and ωij is the interaction of the ith genotype with the jth environment. Note that ωij subsumes residual error of the adjusted mean (Piepho, 2009). The residual variance is set to unity.
Year-wise correlations of lodging-associated traits with yield and other morphological traits such as days to 50% flowering, plant height (cm), tiller number, panicle length (cm), with grain yield (kg ha−1) were calculated using SAS PROC CORR (SAS Institute, 1992).
Construction of Linkage Map and QTL Identification for Lodging Resistance in Rice
A total of 193 polymorphic SNPs were used to construct the linkage map by covering all 12 chromosomes spanning 1,525 cM with an average interval of 7.86 cM (Dixit et al., 2014) Phenotypic data on lodging associated traits were subject to QTL analysis to identify the genetic regions using QGene ver 4.3.10 (Joehanes and Nelson, 2008). The stepwise cofactor selection and default values were used for setting the CIM procedure in QGene. The genome scan interval was set to 1 cM and Window size was set at 10 cM. A LOD (logarithm of odds) score of 2.5 with 1,000 permutations was used for confirming the presence of a putative QTL. The QTLs responsible for phenotypic variance of more than 10% were considered as major-effect QTLs. The putative candidate genes for lodging-associated traits were identified based on the available literature and on the RAP database (http://rapdb.dna.affrc.go.jp/).
Results and Discussion
Phenotypic Evaluations of Culm Traits with Lodging Resistance
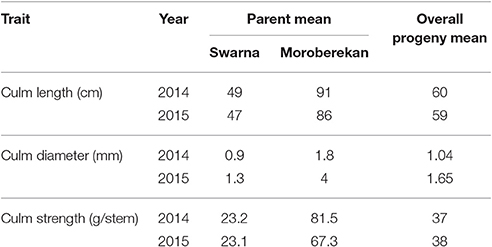
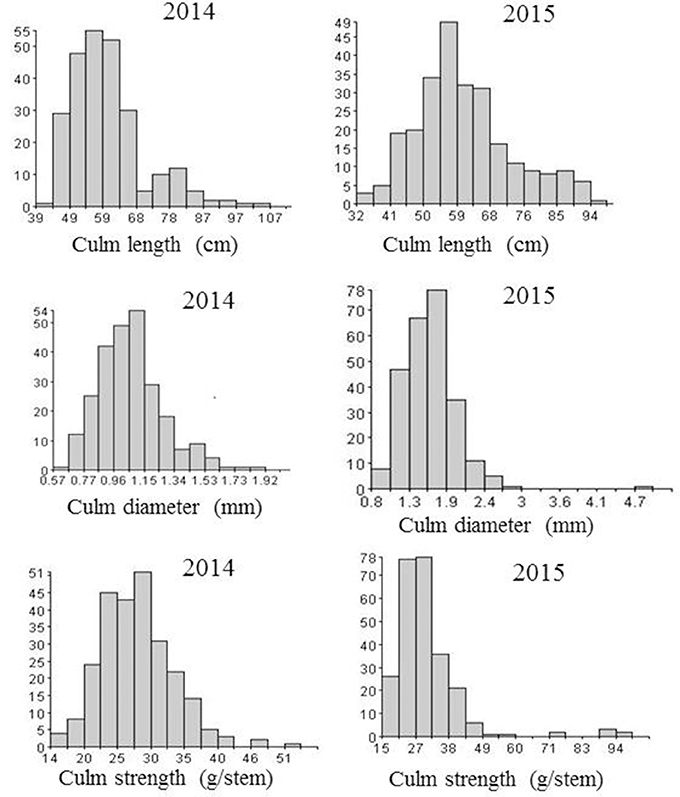
Three culm traits associated with lodging resistance and other morphological traits such as plant height, days to 50% flowering, number of tillers, panicle length and grain yield were investigated. Analysis of variance revealed significant difference (p < 0.01) between the two parents (Swarna and Moroberekan) in all traits measured in both years. The overall progeny means for lodging-related traits were: culm length (59 ± 10.9 cm), culm diameter (1.65 ± 0.20 mm), and culm strength (28 ± 7.7 g/stem) with Moroberekan on the higher side in both years (Table 1). No transgressive segregant with a thicker diameter and higher culm strength than Moroberekan was observed. The phenotypic distribution of lodging-associated traits (CL, CD, and CS) was nearly normal in year 2014, while in 2015, CD and CS were skewed toward the Swarna (Figure 1). The culm strength of the progenies was classified from infirmness to good strength (good strength ≥ 40.1 gram/stem, medium 20.1~40 gram/stem, infirmness ≤ 20 gram/stem) on the basis of instrument reading (prostrate tester) and visual observation of plant lodging tendency correlated with the stem strength reading.
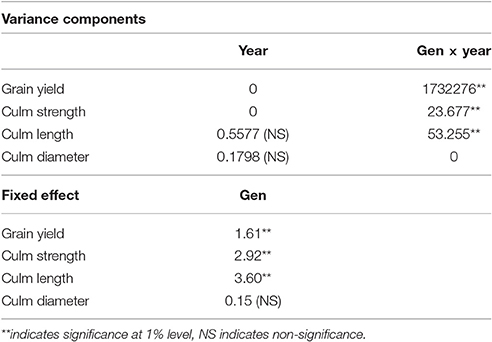
A two-stage analysis was used as the experimental design varied between the 2 years. The analysis indicated significant genotype × year interaction for grain yield, culm length and culm strength (Table 2). However the trial variance was 0 in the case of culm strength, culm length and grain yield and the genotype × year interaction was 0 in the case of culm diameter. The differences between genotypes were non-significant in the case of culm diameter. The best entries for grain yield, culm length and culm strength performing across the years are shown in Supplementary Table 1.
Phenotypic Correlation between Culm Strength and Other Related Traits
In both years, yield exhibited a moderate positive significant correlation with culm diameter and culm length however; it had a weak positive correlation with culm strength (Table 3). At the same time, culm strength and culm diameter were positively and significantly correlated (r = 0.682, p < 0.01 and r = 0.722, p < 0.01 in year 2014 and 2015, respectively). Earlier reports on various cereals including rice (Duan et al., 2004; Kashiwagi et al., 2008; Zhu et al., 2008), wheat (Li et al., 2000; Tripathi et al., 2003; Wang et al., 2006) and barley (Sameri et al., 2009) have reported significant associations of lodging resistance with culm diameter and the wall thickness of basal internodes. These findings indicate that increasing the culm diameter can improve the resistance of cereals to lodging and subsequently increase yields. It was also observed that the lines with yields greater than 5,000 kg ha−1 possessed intermediate culm length (59–70 cm) in 2014 and moderate to large culm length (51–90 cm) in 2015. The high yielding lines also possessed moderate measurements for culm length, strength, and diameter (Table 4).
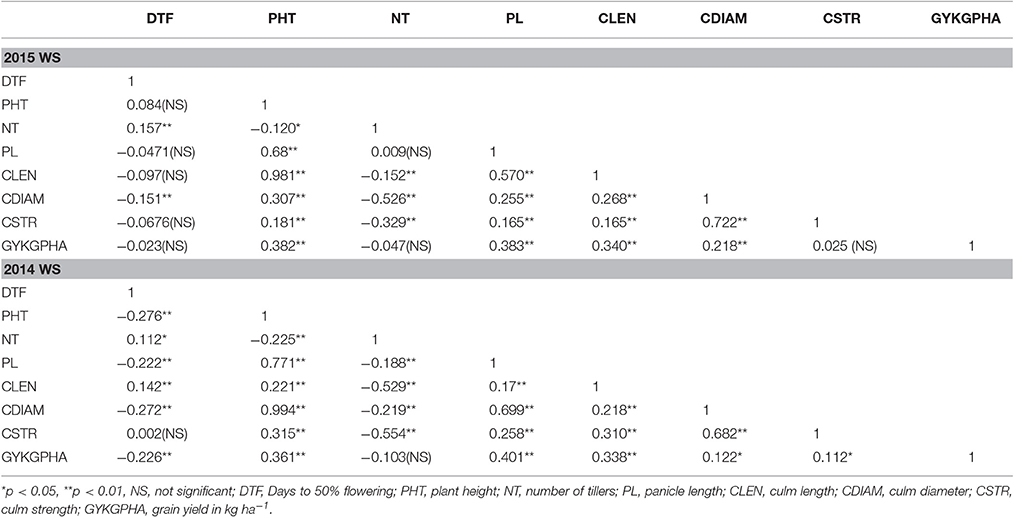
Table 3. Correlation coefficients between 8 agro-morphological characters in 253 genotypes evaluated in 2 years (2014 and 2015).
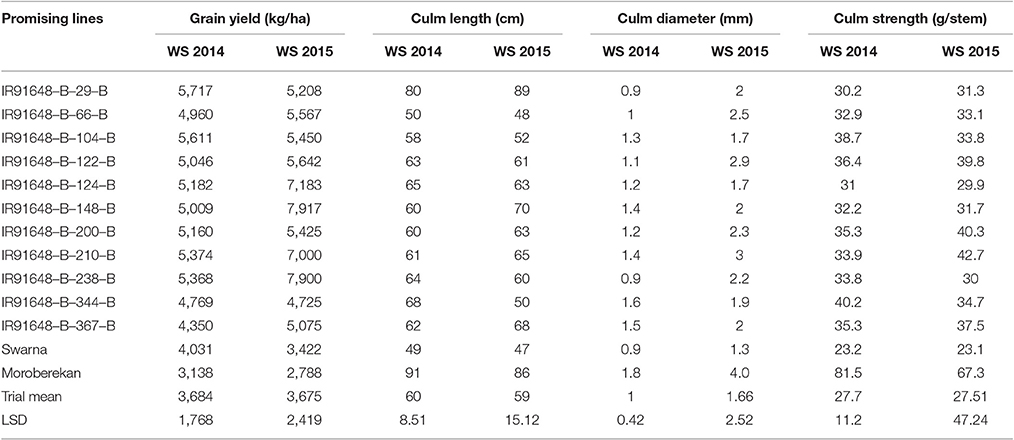
Table 4. Means of promising lines for culm length, culm diameter, and culm strength with grain yield in both the years 2014 and 2015.
QTL Detection
The introduction of the semi-dwarfing genes rht1 in wheat (Peng et al., 1999) and sd1 in rice (Sasaki et al., 2002) improved lodging resistance and increased yield significantly. However, to further improve yield, an increase of 10–20 cm in the height of the presently cultivated semi-dwarf rice varieties has been suggested as a viable alternative (Kumar et al., 1999). Therefore, identifying QTLs related to culm traits, together with increased plant height, is needed for improving lodging resistance and yield (Chen et al., 2014).
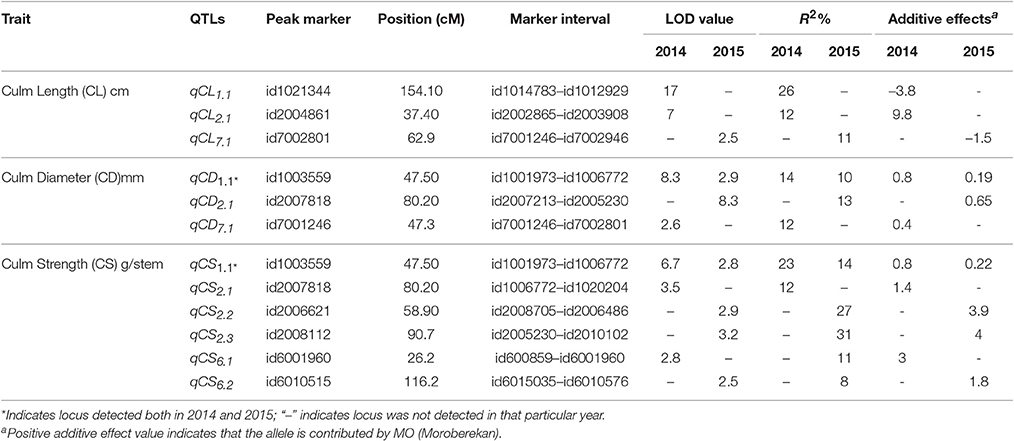
In the present study, 12 QTLs controlling culm length, culm diameter, and culm strength were detected over four different chromosomes in the backcross population of Swarna*3/Moroberekan (Figure 2, Table 5). Chromosome 1 harbored the maximum number of QTLs for lodging-associated traits. A total of three culm length QTLs (LOD>3.0) were detected on chromosomes 1, 2, and 7 (qCL1.1, qCL2.1, and qCL7.1) over the 2 years. The allele for higher culm length for qCL2.1 was from the taller parent Moroberekan while the semi-dwarf parent Swarna contributed favorable alleles to the other two loci (qCL1.1 and qCL7.1). The explained phenotypic variance ranges from 11 to 26.0%. Previously, Mu et al. (2004) identified six additive QTLs associated with culm length on chromosomes 2, 3, 4, 5, and 6 with a phenotypic variation of 39% collectively. Matsubara et al. (2016) mapped four QTLs for culm length on chromosome 1, 2, 5, and 6. In present study, the QTL qCL1.1 identified at physical position of 36.1 Mbp on chromosome 1 with 26% of phenotypic variance was 4 Mbp closer from large effect QTL for culm length (with R2 of 73%) reported by Matsubara et al. (2016).
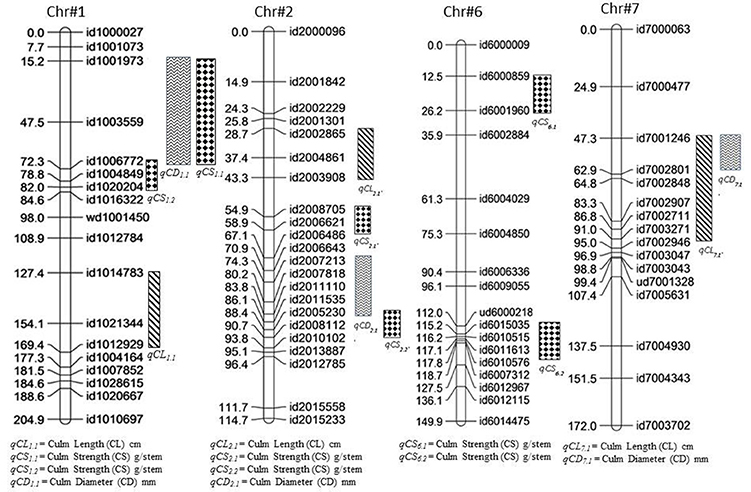
Figure 2. Positions of the QTLs identified for culm length (qCL), culm diameter (qCD) and culm strength (qCS) derived from backcross mapping population (Swarna*3/Moroberekan).
Three QTLs that affected culm diameter were detected on chromosomes 1 (qCD1.1), 2 (qCD2.1), and 7 (qCD7.1) with phenotypic variance explained by 10–14%. The alleles for culm diameter (qCD1.1, qCD2.1, and qCD7.1) were contributed by Moroberekan. The QTL qCD1.1 was consistent in both 2014 and 2015 with a phenotypic variance of 14 and 10%, respectively. Many researchers have detected QTLs for higher culm diameter on chromosome 1 at physical interval of 6Mbp-11 Mbp using various mapping populations (Kashiwagi and Ishimaru, 2004; Mu et al., 2004; Yano et al., 2015). The culm diameter was reported to have positive effects on the section modulus (SM) and bending moment at breaking (M), leading to lodging resistance without any yield loss (Ookawa et al., 2010). Keller et al. (1999) identified two QTLs for culm thickness associated with lodging resistance in a population between wheat and spelt. The previous reports and present findings demonstrate the role of culm diameter in enhancing lodging resistance in rice.
Six QTLs (qCS1.1, qCS2.1, qCS2.2, qCS2.3, qCS6.1, and qCS6.2) for culm strength were detected on three chromosomes in the present study. The QTLs with positive effect for culm strength were contributed by the Moroberekan parent. Previously, a number of QTLs viz., prl5, SCM1, SCM2, SCM3, and SCM4 were reported for culm strength (Kashiwagi and Ishimaru, 2004; Ookawa et al., 2010; Yano et al., 2015). However, some of the identified QTLs region was quite large for its utilization in marker-assisted selection; fine mapping, sequencing and allele mining of the identified regions could be useful in further deployment. The QTL qCS1.1 identified in present study was reproducible in both 2014 and 2015 with a phenotypic variance of 23 and 14%, respectively and located on chromosome 1. Mu et al. (2004) also reported QTL for culm strength on same chromosome in marker interval of RM5-RM302. The QTLs qCS6.1 and qCS6.2 were identified on chromosome 6 with phenotypic variance of 11 and 8%, respectively. The QTL qCS6.2 for culm strength identified in the present study was in agreement with earlier reported QTL STRONG CULM2 (SCM2) at physical position of 27 Mbp (Ookawa et al., 2010). The detailed analysis of SCM2 through positional cloning and sequencing reveals that the gene is similar to APO1 favorably leads to increase in the spikelet number and enhanced culm strength by increasing the cell proliferation rate (Ookawa et al., 2010). The QTL-hotspot for early vigor, early uniform emergence (qEV6.1, qEUE6.1) in interval of 11.7-27.6 cM under DSR conditions reported by Singh et al. (2017) was located near to QTL for culm strength (qCS6.1) identified in present study at position of 26.2 cM. The observed direct relationship among the seedling establishment and lodging resistant traits in term of co-location of genetic regions could be an important landmark for various desirable traits under DSR conditions.
Co-localization of QTLs
In this study, the stable and consistent effect QTL region between id1001973-id1006772 markers reported to be associated with the traits culm diameter and culm strength indicating the pleiotropic effect of the identified genomic region or linkage of loci controlling both the traits. Current finding in the present study are in agreement with the previous report of lodging resistant traits in rice (Kashiwagi and Ishimaru, 2004; Mu et al., 2004). The identified shared loci can be fine mapped and cloned to resolve the underlying gene functions constituting the QTL region (Eshed and Zamir, 1995; Ashikari et al., 2005; Pelgas et al., 2011). Though the genetic distance of this co-localized region is of considerable length (47.5–72.3 cM), it may provide an important clue for future efforts to explore the region by fine mapping to better understand the mechanism of this complex trait.
Identification of Putative Candidate Genes
The genomic regions within the consistent QTL for culm diameter (qCD1.1) and culm strength (qCS1.1) were analyzed in silico for the presence of possible candidate genes previously reported for lodging resistance. A total of 35 putative genes were found within the stable QTL region identified for culm-associated traits (Table 6). Many genes within the QTL were responsible for phytohormones synthesis, hypothetical and expressed protein, heat shock protein, transcriptional factors, and precursors for various biochemical and metabolic pathways. Among them, a putative candidate gene Gibberellic Acid (GA) 3 beta-hydroxylase with single copy 4003659-4004946 bp was identified. This gene is the final precursor in the biosynthetic pathway of the plant hormone Gibberellin (Itoh et al., 1999). Another report (Okuno et al., 2014) highlighted the effect of the plant hormone GA on lodging resistance in rice and on increased total biomass signifying the positive impact of over-expression of GA on lodging resistance due to increased culm diameter and lignin deposition. GA was also found to increase biomass yield. A few other findings reported that higher lignin content is responsible for improved varietal resistance to the bending type of lodging (Ookawa and Ishihara, 1993; Biemelt et al., 2004). Recently, Ookawa et al. (2016) also discussed the effect of high GA level results in enlargement of culm diameter by increasing the number of parenchymatous cells in the culm. Further studies needed to confirm the positive regulation of GA on culm thickness in the genomic region on chromosome 1. Such candidate genes need to be further characterized to be used as potential targets for marker-assisted breeding for strong/thick culm providing lodging resistance in rice.
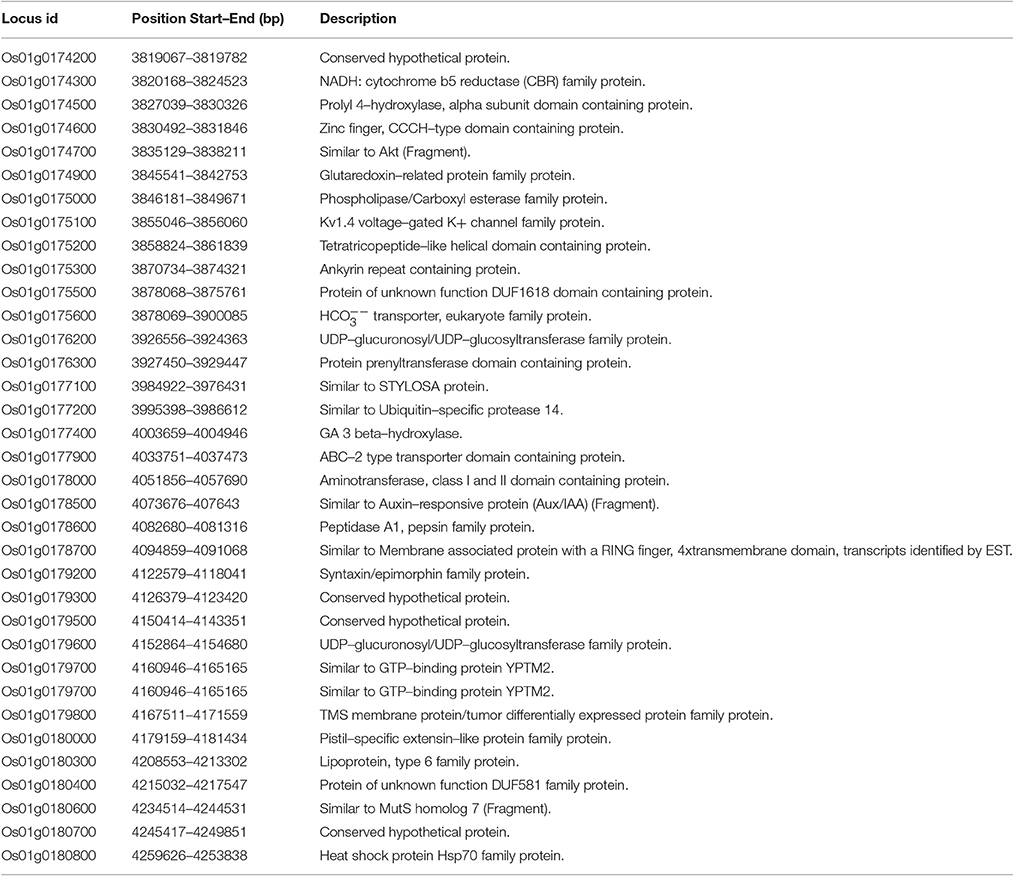
Table 6. List of putative genes found within the stable QTL on chromosome1 in the marker interval of id1001973–id1006772.
The QTL for culm strength (qCS6.2) identified in the present study was also explained by Ookawa et al., 2010 by identifying candidate gene SCM2 within the marker interval of RM 20546–RM 20562 at physical location of 27 Mbp. This gene was found to be responsible for the physical strength of the culm and for the increase in spikelet number because of pleiotropic gene action. This finding points to the authenticity of the QTL hot spot region for lodging resistance genes and will be an important landmark in future breeding programs for improved lodging resistance in rice.
Conclusion
Culm diameter and culm strength are potential target traits for breeding rice varieties with enhanced lodging resistance under DSR conditions. The stable and consistent genetic region within the marker interval of id1001973-id1006772 could be an important source of alleles responsible for enhancing lodging resistance in rice. The polymorphic SNP markers in these QTL regions may be utilized for future marker-assisted breeding programs. Functional validation of the candidate gene SCM2 as well as genes for GA biosynthesis with precursors of various metabolic pathways associated with culm QTLs (qCS6.2, qCS1.1, and qCD1.1) will be a valuable future step to clearly understand the mechanism of lodging resistance.
Author Contributions
SY was involved in conducting the experiment, recording observations and drafting the article; US, SN, CV, and PR helped in experimental work and contributed to the manuscript modification; KR, NS was involved in experimental analysis, interpretation of data and revising the manuscript; AK was involved in the design of the experiment and in the critical revision of the manuscript. All authors approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research work is conducted under Department of Biotechnology, Government of India grant (BT/AB/01/IRRI India/ 2012 dt.01.07.2013) to the corresponding author. Authors thank DBT for financial support to conduct this study.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01431/full#supplementary-material
Abbreviations
DSR, Direct seeded rice; CL, Culm length; CD, Culm diameter; CS, Culm strength; SNP, Single nucleotide polymorphism; CIM, Composite Interval Mapping; PCA, Principal component analysis; SAS, Statistical Analysis System.
References
Ashikari, M., Sakakibara, H., Lin, S., Yamamoto, T., Takashi, T., and Nishimura, A. (2005). Cytokinin oxidase regulates rice grain production. Science 309, 741–745. doi: 10.1126/science.1113373
Berry, P. M., Spink, J. H., Gay, A. P., and Craigon, J. A. (2003). Comparison of root and stem lodging risks among winter wheat cultivars. J. Agric. Sci. 141, 191–202. doi: 10.1017/S002185960300354X
Biemelt, S., Tschiersch, H., and Sonnewald, U. (2004). Impact of altered gibberellin metabolism on biomass accumulation, lignin biosynthesis, and photosynthesis in transgenic tobacco plants. Plant. Physiol. 135, 254–265. doi: 10.1104/pp.103.036988
Chen, W. Y., Liu, Z. M., Deng, G. B., Pan, Z. F., Liang, J. J., Zeng, X. Q., et al. (2014). Genetic relationship between lodging and lodging components in barley (Hordeum vulgare) based on unconditional and conditional quantitative trait locus analyses. Genet. Mol. Res. 13, 1909–1925. doi: 10.4238/2014.March.17.19
Dingkuhn, M., Schnier, H. F., De Datta, S. K., Dorffling, K., and Javellana, C. (1990). Nitrogen fertilization of direct-seeded flooded vs. transplanted rice. II. Interactions among canopy properties. Crops. Sci. 30, 1284–1292. doi: 10.2135/cropsci1990.0011183X003000060025x
Dixit, S., Huang, B. E., Sta Cruz, M. T., Maturan, P. T., Ontoy, J. C. E., and Kumar, A. (2014). QTLs for tolerance of drought and breeding for tolerance of abiotic and biotic stress: an integrated approach. PLoS ONE 9:e109574. doi: 10.1371/journal.pone.0109574
Duan, C. R., Wang, B. C., Wang, P. Q., Wang, D. H., and Cai, S. X. (2004). Relationship between the minute structure and the lodging resistance of rice stems. Colloids Surf. 35, 155–158. doi: 10.1016/j.colsurfb.2004.03.005
Eshed, Y., and Zamir, D. (1995). An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141, 1147–1162.
Flint-Garcia, S. A., Jampatong, C., Darrah, L., and McMullen, M. D. (2003). Quantitative trait locus analysis of stalk strength in four maize populations. Crops Sci. 43, 13–22. doi: 10.2135/cropsci2003.0013
Flintham, J. E., Borner, A., Worland, A. J., and Gale, M. D. (1997). Optimizing wheat grain yield: effects of Rht (gibberellin-insensitive) dwarfing genes. J. Agric. Sci. 128, 11–25. doi: 10.1017/S0021859696003942
Frensham, A., Cullis, B., and Verbyla, A. (1997). Genotype by environment variance heterogeneity in a two-stage analysis. Biometrics 53, 1373–1383. doi: 10.2307/2533504
Hai, L., Guo, H., Xiao, S., Jiang, G., Zhang, X., Yan, C., et al. (2005). Quantitative trait loci (QTL) of stem strength and related traits in a doubled-haploid population of wheat (Triticum aestivum L.). Euphytica 141, 1–9. doi: 10.1007/s10681-005-4713-2
Hirano, K., Okuno, A., Hobo, T., Ordonio, R., Shinozaki, Y., Asano, K., et al. (2014). Utilization of stiff culm trait of rice smos1 mutant for increased lodging resistance. PLoS ONE 9:e96009. doi: 10.1371/journal.pone.0096009
Islam, M. S., Peng, S., Visperas, R. M., Ereful, N., Bhiya, S. M. U., and Julfiquar, A. W. (2007). Lodging-related morphological traits of hybrid rice in a tropical irrigated ecosystem. Field Crops Res. 101, 240–248. doi: 10.1016/j.fcr.2006.12.002
Itoh, H., Ueguchi, T., Kawaide, H., Chen, X., Kamiya, Y., and Matsuoka, M. (1999). The gene encoding tobacco gibberellin 3-hydroxylase is expressed at the site of GA action during stem elongation and flower organ development. Plant J. 20, 15–24. doi: 10.1046/j.1365-313X.1999.00568.x
Joehanes, R., and Nelson, J. C. (2008). QGene 4.0, an extensible Java QTL analysis platform. Bioinformatics 24, 2788–2789. doi: 10.1093/bioinformatics/btn523
Kashiwagi, T., and Ishimaru, K. (2004). Identification and functional analysis of a locus for improvement of lodging resistance in rice. Plant Physiol. 134, 676–683. doi: 10.1104/pp.103.029355
Kashiwagi, T., Togawa, E., Hirotsu, N., and Ishimaru, K. (2008). Improvement of lodging resistance with QTLs for stem diameter in rice (Oryza sativa L.). Theor. Appl. Genet. 117, 749–757. doi: 10.1007/s00122-008-0816-1
Kelbert, A. J., Spaner, D., Briggs, K. G., and King, J. R. (2004). The association of culm anatomy with lodging susceptibility in modern spring wheat genotypes. Euphytica 136, 211–221. doi: 10.1023/B:EUPH.0000030668.62653.0d
Keller, M., Karutz, Ch., Schmid, J. E., Stamp, P., Winzeler, M., Keller, B., et al. (1999). Quantitative trait loci for lodging resistance in a segregating wheat × spelt population. Theor. Appl. Genet. 98, 1171–1182. doi: 10.1007/s001220051182
Kobayashi, N., Ebron, L. A., Yanoria, M. J. T., Kato, H., and Atlin, G. (2006). Genetic and breeding study on near isogenic lines of IR64 for yield related traits-resistance to rice blast disease under the water–saving condition in IRRI. Jpn. J. Trop. Agric. 50, 9–10.
Kono, M. (1995). “Physiological aspects of lodging,” in Science of the Rice Plant, Vol. 2, Physiology 2, eds T. Matsuo, K. Kumazawa, R. Ishii, K. Ishihara, and H. Hirata (Tokyo: Food and Agricultural Policy Research Centre), 971–982.
Kumar, A., Tiwari, R. K. S., Parihar, S. S., Pandya, K. S., and Janoria, M. P. (1999). Performance of prototype rice lines from ideotype breeding. Int. Rice. Res. Notes 24, 18–19.
Li, H. B., Bai, K. Z., Kuang, T. Y., Hu, Y. X., Jia, X., and Lin, J. X. (2000). Structural characteristics of thicker-culms in the high-yield wheat cultivars. Acta. Bot. Sin. 42, 1258–1262.
Littell, R. C., Milliken, G. A., Stroup, W. W., Wolfinger, R. D., and Schabenberger, O. (2006). SAS for Mixed Models. Cary, NC: SAS Institute Inc.
Mackill, D. J., Coffman, W. R., and Garrity, D. P. (1996). Rainfed Lowland Rice Improvement. Los Banos, CA: International Rice Research Institute.
Matsubara, K., Yamamoto, E., Kobayashi, N., Ishii, T., Tanaka, J., Tsunematsu, H., et al. (2016). Improvement of rice biomass yield through QTL-based selection. PLoS ONE 11:e0151830. doi: 10.1371/journal.pone.0151830
Möhring, J., and Piepho, H. P. (2009). Comparison of weighting in two-stage analysis of plant breeding trials. Crops. Sci. 49, 1977–1988. doi: 10.2135/cropsci2009.02.0083
Mu, P., Li, Z., Li, C., and Wang, X. (2004). QTL analysis for lodging resistance in rice using a DH population under lowland and upland ecosystems. Acta. Gen. Sin. 31, 717–723.
Murai, M., Takamura, I., Sato, S., Tokutome, T., and Sato, Y. (2002). Effects of the dwarfing gene originating from Dee–geo–woo–gen on yield and its related traits in rice. Breed. Sci. 52, 95–100. doi: 10.1270/jsbbs.52.95
Nguyen, N. V., and Ferrero, A. (2006). Meeting the challenges of global rice production. Paddy. Water Environ. 4, 1–9. doi: 10.1007/s10333-005-0031-5
Okuno, A., Hirano, K., Asano, K., Takase, W., Masuda, R., Morinaka, Y., et al. (2014). New approach to increasing rice lodging resistance and biomass yield through the use of high gibberellin producing varieties. PLoS ONE 9:e86870. doi: 10.1371/journal.pone.0086870
Ookawa, T., and Ishihara, K. (1992). Varietal difference of physical characteristics of the culm related to lodging resistance in paddy rice. Jpn. J. Crop. Sci. 61, 419–425. doi: 10.1626/jcs.61.419
Ookawa, T., and Ishihara, K. (1993). Varietal difference of the cell wall components affecting the bending stress of the culm in relation to the lodging resistance in paddy rice. Jpn. J. Crop. Sci. 62, 378–384. doi: 10.1626/jcs.62.378
Ookawa, T., Aoba, R., Yamamoto, T., Ueda, T., Takai, T., Fukuoka, S., et al. (2016). Precise estimation of genomic regions controlling lodging resistance using a set of reciprocal chromosome segment substitution lines in rice. Sci. Rep. 6:30572. doi: 10.1038/srep30572
Ookawa, T., Hobo, T., Yano, M., Murata, K., Ando, T., Miura, H., et al. (2010). New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield. Nat. Commun. 1:132. doi: 10.1038/ncomms1132
Pal, S., Datta, S. P., Rattan, R. K., and Singh, A. K. (2008). Diagnosis and amelioration of iron deficiency under aerobic rice. J. Plant. Nutr. 31, 919–940. doi: 10.1080/01904160802043262
Pelgas, B., Bousquet, J., Meirmans, P. G., Ritland, K., and Isabel, N. (2011). QTL mapping in white spruce: gene maps and genomic regions underlying adaptive traits across pedigrees, years and environments. BMC Genomics 12:145. doi: 10.1186/1471-2164-12-145
Peng, J., Richards, D. E., Hartley, N. M., Murphy, G. P., Devos, K. M., Flintham, J. E., et al. (1999). Green revolution' genes encode mutant gibberellin response modulators. Nature 400, 256–261. doi: 10.1038/22307
Peng, S., and Khush, G. S. (2003). Four decades of breeding for varietal improvement of irrigated lowland rice in the International Rice Research Institute. Plant Prod. Sci. 6, 157–164. doi: 10.1626/pps.6.157
Piepho, H. P. (2009). Ridge Regression and Extensions for Genome wide selection in Maize. Crops. Sci. 49, 1165–1176. doi: 10.2135/cropsci2008.10.0595
Sameri, M., Nakamura, S., Nair, S. K., Takeda, K., and Komatsuda, T. (2009). A quantitative trait locus for reduced culm internode length in barley segregates as a Mendelian gene. Theor. Appl. Genet. 118, 643–652. doi: 10.1007/s00122-008-0926-9
Sasaki, A., Ashikari, M., Ueguchi–Tanaka, M., Itoh, H., Nishimura, A., Swapan, D., et al. (2002). Green revolution: a mutant gibberellin–synthesis gene in rice. Nature 416, 701–702. doi: 10.1038/416701a
Setter, T. I., Laureles, E. V., and Mazaredo, A. M. (1997). Lodging reduces yield of rice by self shading and reduction of photosynthesis. Field Crops Res. 49, 95–106. doi: 10.1016/S0378-4290(96)01058-1
Singh, U. M., Yadav, S., Dixit, S., Ramayya, P. J., Devi, M. N., Raman, K. A., et al. (2017). QTL hot spot for early vigor and related traits under Dry- Direct seeded system in rice (Oryza sativa L.). Front. Plant Sci. 8:286. doi: 10.3389/fpls.2017.00286
Sinniah, U. R., Wahyuni, S., Syahputra, B. S., and Gantait, S. (2012). A potential retardant for lodging resistance in direct seeded rice (Oryza sativa L.). Can. J. Plant Sci. 92, 13–18. doi: 10.4141/cjps2011-089
Terashima, K., Ogata, T., and Akita, S. (1992). Eco–physiological characteristics related with lodging tolerance of rice in direct sowing cultivation in comparison of root lodging tolerance among cultivars by the measurement of pushing resistance. Jpn. J. Crop. Sci. 61, 380–387. doi: 10.1626/jcs.61.380
Tripathi, S. C., Sayre, K. D., Kaul, J. N., and Narang, R. S. (2003). Growth and morphology of spring wheat (Triticum aestivum L.) culms and their association with lodging: effects of genotypes, N levels and ethephon. Field Crops Res. 84, 271–290. doi: 10.1016/S0378-4290(03)00095-9
Verma, V., Worland, A. J., Sayers, E. J., and Fish, L. (2005). Identification and characterization of quantitative trait loci related to lodging resistance and associated traits in bread wheat. Plant Breed. 124, 234–241. doi: 10.1111/j.1439-0523.2005.01070.x
Wang, J., Zhu, J. M., Lin, Q. Q., Li, X. J., Teng, N. J., Li, Z. S., et al. (2006). Effects of stem structure and cell wall components on bending strength in wheat. Chin. Sci. Bull. 51, 1–9. doi: 10.1007/s11434-006-0815-z
Watanabe, T. (1997). “Lodging resistance” in Science of the Rice Plant, eds T. Matsuo, Y. Futsuhara, F. Kikuchi, and H. Yamaguchi (Tokyo: Food and Agriculture Policy Research Centre), 567–577.
Won, J. G., Hirahara, Y., Yoshida, T., and Imabayashi, S. (1998). Selection of rice lines using SPGP seedling method for direct seeding. Plant Prod. Sci. 1, 280–285. doi: 10.1626/pps.1.280
Yano, K., Ookawa, T., Aya, K., Ochiai, Y., Hirasawa, T., Ebitani, T., et al. (2015). Isolation of a novel lodging resistance QTL gene involved in strigolactone signaling and its pyramiding with a QTL gene involved in another mechanism. Mol. Plant. 8, 303–314. doi: 10.1016/j.molp.2014.10.009
Zhu, L. H., Zhong, D. B., Xu, J. L., and Yu, S. B. (2008). Differential expression of lodging resistance related QTL in rice (Oryza sativa L.). Plant Sci. 175, 898–905. doi: 10.1016/j.plantsci.2008.09.001
Keywords: direct seeded rice, grain yield, lodging resistance, quantitative trait loci, SNP marker
Citation: Yadav S, Singh UM, Naik SM, Venkateshwarlu C, Ramayya PJ, Raman KA, Sandhu N and Kumar A (2017) Molecular Mapping of QTLs Associated with Lodging Resistance in Dry Direct-Seeded Rice (Oryza sativa L.). Front. Plant Sci. 8:1431. doi: 10.3389/fpls.2017.01431
Received: 21 April 2017; Accepted: 03 August 2017;
Published: 21 August 2017.
Edited by:
Zlatko Satovic, Faculty of Agriculture, University of Zagreb, CroatiaReviewed by:
Habtamu Ayalew, National Agriculture and Food Research Organization (NARO), JapanMarcelo Helguera, Instituto Nacional de Tecnología Agropecuaria (INTA), Argentina
Copyright © 2017 Yadav, Singh, Naik, Venkateshwarlu, Ramayya, Raman, Sandhu and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arvind Kumar, YS5rdW1hckBpcnJpLm9yZw==
 Shailesh Yadav
Shailesh Yadav Uma M. Singh1
Uma M. Singh1 Shilpa M. Naik
Shilpa M. Naik Perumalla J. Ramayya
Perumalla J. Ramayya Nitika Sandhu
Nitika Sandhu Arvind Kumar
Arvind Kumar