- State Key Laboratory of Crop Stress Biology for Arid Areas, College of Agronomy, Northwest A&F University, Yangling, China
Late blight, caused by the pathogen Phytophthora infestans, is one of the most devastating diseases of potato. Here, we describe a new single dominant resistance gene, Rpi2, from the Mexican diploid wild species Solanum pinnatisectum that confers high level and broad spectrum resistance to late blight. The Rpi2 locus confers full resistance to complex isolates of P. infestans, for which race specificity has not yet been demonstrated. This new gene, flanked by the RFLP-derived marker SpT1756 and AFLP-derived marker SpAFLP2 with a minimal genetic distance of 0.8 cM, was mapped to potato chromosome 7. Using the genomic sequence data of potato, we estimated that the physical distance of the nearest marker to the resistance gene was about 27 kb. The map location and other evidence indicated that this resistance locus was different from the previously reported resistance locus Rpi1 on the same chromosome from S. pinnatisectum. The presence of other reported resistance genes in the target region, such as Gro1-4, I-3, and three NBS-LLR like genes, on a homologous tomato genome segment indicates the Rpi2-related region is a hotspot for resistance genes. Comparative sequence analysis showed that the order of nine markers mapped to the Rpi2 genetic map was highly conserved on tomato chromosome 7; however, some rearrangements were observed in the potato genome sequence. Additional markers and potential resistance genes will promote accurate location of the site of Rpi2 and help breeders to introduce this resistance gene into different cultivars by marker-aided selection.
Introduction
Late blight caused by Phytophthora infestans is one of the most devastating diseases of potato worldwide. Under favorable conditions for the pathogen, complete defoliation of a potato plant can occur in just a few weeks. The development and deployment of cultivars with genetic resistance is the most economical and eco-friendly approach for reducing yield losses due to late blight. Wild potato species are a valuable genetic pool for finding late blight resistant genes. The first paradigm came from the hexaploid Mexican wild species Solanum demissum. Eleven resistance (R) genes, named R1 to R11, were identified in this wild species and introduced into S. tuberosum(Black, 1951; Black et al., 1953; Malcolmson and Black, 1966). However, these R genes conferred race-specific resistance and those that were introgressed into potato varieties were quickly overcome by the pathogen because of its high genetic variability (Wastie, 1991). Hence, new sources of resistance are required, especially those conferring race non-specific resistance to late blight.
The co-evolution of the pathogen and wild species in Central America indicated the possibility of finding resistance in species from Mexico such as S. bulbocastanum, S. pinnatisectum, and S. trifidum. A set of late blight resistance genes has already been identified in these species. Notably, in S. bulbocastanum, four different loci with broad spectrum late blight resistance have been identified, namely Rpi-Blb1/RB (Helgeson et al., 1998), Rpi-blb2 (van der Vossen et al., 2005), Rpi-blb3 (Park et al., 2005a), and Rpi-apbt (Park et al., 2005b). Recently, several other wild Solanum species have been reported as potential sources of resistance, such as S. mochiquense (Jones et al., 2013), S. chacoense (Vossen et al., 2011), and S. × edinense (De Vetten et al., 2014). Sustainable breeding efforts using these resistance sources have resulted in several new potato cultivars (Jo et al., 2014; Haesaert et al., 2015).
Extensive investigations have shown that the molecular basis of R gene resistance is a gene family characterized by two domains, the nucleotide binding site (NBS) and leucine-rich repeat (LRR) domains (Martin et al., 2003). The conserved nature of the motifs within these domains has been exploited to search for new resistance gene-like sequences or resistance gene analogs (RGAs) using a homology-dependent PCR-based approach (Kanazin et al., 1996; Leister et al., 1996; Chen et al., 1998; Hayes and Maroof, 2000). Many RGAs have been mapped to genomic positions containing known resistance specificities, and RGAs have been shown to be derived from known resistance genes (Collins et al., 1999). Thus, RGAs represent candidates for functional resistance genes. NBS-LRR genes can generally be divided into two distinct groups: one encoding an N-terminal domain with Toll/Interleukin-1 Receptor homology (TIR-NBS-LRR) and the other with an N-terminal coiled-coil motif (CC-NBS-LRR) (Martin et al., 2003). So far, over 20 late blight resistance genes, such as R1, R2, R3a, R3b, RB, Rpi-blb2, Rpi-blb3, Rpi-abpt, Rpi-sto1, Rpi-pta1, Rpi-vnt1.1, and Rpi-vnt1.3, which all belong to the CC-NBS-LRR class, have been cloned (Ballvora et al., 2002; Song et al., 2003; van der Vossen et al., 2003; Huang et al., 2005; Vleeshouwers et al., 2008; Foster et al., 2009; Lokossou et al., 2009; Pel et al., 2009). The publishing of the potato genome sequence derived from the S. tuberosum Group Phureja clone DM1-3 516 R44 (DM) accelerated the identification of 438 NB-LRR type genes from ∼39,000 potato gene models, and will increase the velocity of functional NB-LRR gene cloning from Solanum species (Jupe et al., 2012).
High level and broad spectrum late blight resistance has also been observed in the Mexican diploid wild species S. pinnatisectum (Menke et al., 1996; Chen et al., 2003). Compared with S. bulbocastanum, S. pinnatisectum has received less attention in late blight research. Kuhl et al. (2001) screened 13 accessions of S. pinnatisectum and found that most were resistant to late blight. Chen et al. (2003) revaluated the late blight resistance of S. pinnatisectum (PI275233) and found that it showed broad-spectrum resistance against various known P. infestans strains including the R9 isolate. They also found different levels of resistance among different accessions of S. pinnatisectum, suggesting the presence of different resistance genes. To date, only Kuhl et al. (2001) have reported the genetic analysis and identification of a single dominant resistance locus in S. pinnatisectum (PI253124), Rpi1, which was mapped to chromosome 7 in an interval of 14.6 cM between two RFLP markers, CP56 and TG20A.
The hybridization barrier between S. pinnatisectum and cultivated potatoes had been overcome by the combination of the Sli gene and chromosome-doubling techniques (Sanetomo et al., 2014). Therefore, the wild species S. pinnnatisectum should receive more attention as a resource for potato late blight resistance breeding. The objective of this study was to characterize and map a late blight resistance gene from S. pinnatisectum (PI275233) through genetic linkage analysis and collinearity analysis. This gene may be useful for developing potato cultivars with broad spectrum resistance.
Materials and Methods
Plant Materials
A backcross population was developed by crossing the susceptible diploid S. cardiophyllum (PI186548) as the male parent with the resistant diploid S. pinnatisectum (PI275233). Several clones of a single resistant F1 individual, propagated through in vitro culture, were then backcrossed with the susceptible parent to generate a backcross mapping population.
The F1 and BC1 populations were maintained vegetatively from tubers following their first propagation from true seed. A total of 931 clones from the backcross population were selected to analyze the genetics of resistance to late blight using detached leaf methods at the first clonal generation (Chen et al., 2003).
Detached Leaf Assay for Evaluating Late Blight Resistance
An inoculum was prepared from the P1801C.16 strain of P. infestans (US-8/A2 mating type) and diluted to a final concentration of 30,000 sporangia per ml. Inoculation and the detached leaf assay were performed according to Chen et al. (2003). Three compound leaves were excised for the late blight test, including the third to fifth leaves from the top on each plant’s main branch. Each compound leaf with five leaflets was inserted into prepared moist vermiculite in a plastic tray. The inoculum was sprayed onto the surface of all leaflets. Trays with inoculated compound leaves were incubated in a growth cabinet under an 18/6 h and 20/18°C day/night regime for about 15 days. The susceptible parent was inoculated as a susceptible check. Plant resistance was evaluated after 8 and 15 days. Disease severity was estimated using mean disease severity values (DSVs) of three compound leaves based on the percentage of leaf area with symptoms of late blight. Severity values were scored using a scale of 0–5 where 0 = no disease to <3%; 1 = 3–24%; 2 = 25–49%; 3 = 50–74%; 4 = 75–94%; and 5 = 95–100% infection. Plants with a DSV of 0 were classified as resistant and those with DSVs of 2–5 were classified as susceptible.
AFLP Analysis
DNA was extracted from 100 mg of young leaves for each potato plant using a Genomic DNA Purification Kit (Promega, Fitchburg, WI, United States). Bulked segregant analysis (BSA) was used to screen for molecular markers associated with late blight resistance (Michelmore et al., 1991). Two susceptible bulks were constructed from 10 highly susceptible individuals and a resistant bulk was developed with equal amounts of DNA from 10 highly resistant individuals among the BC1 population. DNA markers were screened for the two susceptible bulks, the resistant bulk and the resistant parent plant.
AFLP analysis was performed as described by Vos et al. (1995) using EcoRI and MseI as rare- and frequent-cutter enzymes, respectively. Genomic DNA digestion and ligation were conducted using an AFLP Core Reagent Kit (Invitrogen, Carlsbad, CA, United States) according to the instructions. A pre-amplification was carried out with 1-bp extension primer combinations (EcoRI+A/MseI+C and EcoRI+A/MseI+A) and the PCR products were diluted at a ratio of 1:30 with TE buffer. Selective amplification using primer combinations of EcoRI+3 and MseI+3 was conducted and the products were separated on a 6% PAGE sequencing gel run at 100 W for 2.5 h after pre-electrophoresis for 30 min. The gel was stained by the silver-staining method (Bassam et al., 1991).
DNA Sequencing and Analysis
AFLP fragments were excised from the dried silver-stained polyacrylamide gel and placed into microfuge tubes containing 30 μl distilled water. The samples were boiled for 10 min and centrifuged, and then 3 μl of the supernatant was used for PCR under the same conditions as those used for AFLP analysis. The PCR products were then inserted into the pGEM-T easy vector (Promega, Fitchburg, WI, United States) and sequenced. A search for sequences homologous to the AFLP fragments was conducted using the GenBank website1, and Clustal W22 was used to compare the sequence homology.
RGA Markers
The digestion of genome DNA were performed by two restriction enzymes, EcoRI and MseI, according to the instructions of AFLP Core Reagent Kit. The primer combination EcoRI/MseI was used to generate pre-amplification products. Then, the second amplification step was carried out with five primer combinations. The primer combinations were respectively combined by five P-loop based RGAs primers, S1 (5′-GGTGGGGTTGGGAAGACAACG-3′), S2 (5′-GGIGGIGTIGGIAAIACIAC-3′), AS1 (5′-CAACGCTAGTGGCAATCC-3′), AS2 (5′-IAAIGCIAGIGGIAAICC-3′) and AS3 (5′-IAGIGCIAGIGGIAGICC-3′) (Leister et al., 1996), with the EcoRI AFLP primer. PCR conditions were somewhat different from the standard AFLP procedure; 30 s DNA denaturation at 94°C, 30 s primer annealing at 55°C and a 1 min elongation step at 72°C (35 cycles). Prior to the cycling, the template DNA was denatured for 1 min at 94°C and the PCR was finalized by applying an extra 5 min elongation step at 72°C. The procedures for running the gel and fragment extraction were the same as described for AFLP section.
Locus-Specific Marker Development
Locus-specific markers on chromosome 7 of potato and tomato3 were selected to develop PCR-based markers. Generally, the RFLP probe sequences were used as queries to search ESTs using the BLASTn program (Altschul et al., 1997). Then, the ESTs and RFLP probes were assembled with a criterion of more than 95% identity over a stretch of 40 nucleotides using SeqMan II (LASERGENE, DNASTAR, Madison, WI, United States). Primers were designed according to the assembled sequences guided by intron finder4 to amplify regions spanning introns and avoid placing primers in exon–intron boundaries. The PCR products were separated after digestion with one of the 4-bp cutter restriction enzymes TaqI, Trul1, Msp1, Rsa1, and Tail1.
The PCR amplification reactions were conducted in 20 μl reaction mixtures containing 10 mM TRIS-HCl, pH 8.3, 50 mM KCl, 2 mM MgCl2, 100 μM of each dNTP, 200 nM primers, approximately 20 ng template DNA and 1 Unit Taq DNA polymerase. The cycling program consisted of an initial 3 min denaturation step at 94°C, followed by 35 cycles of 94°C (30 s), 55°C (30 s), and 72°C (50 s), and a final 5 min extension step at 72°C. The PCR products were size-separated on a 2% agarose gel, stained with ethidium bromide, and visualized on a Gel Imaging system (Bio-Rad, San Diego, CA, United States).
Linkage Analysis
Linkage analysis was performed using the software package MAPMAKER V3.0 (Lander et al., 1987). Markers and their corresponding distances (cM) were included in the framework map only if the LOD value for the ripple was >3. The Kosambi mapping function was employed to convert recombination frequencies to map distances in cM (Kosambi, 1943). Collinearity analysis results were visualized using Circos-0.67 (Krzywinski et al., 2009).
Results
The BC1 generation produced 440 resistant plants and 491 susceptible plants. The segregation ratio fit a monogenic Mendelian inheritance model of 1:1 (resistant:susceptible) in the population (χ2 = 2.794, P = 0.095). This result suggested that a single dominant locus controlled the late blight resistance in S. pinnatisectum (PI275233). Subsequently, 164 susceptible and 101 resistant plants with extreme phenotypes were chosen for mapping.
AFLP and AFLP-Derived Markers
In an attempt to find AFLP markers linked to the resistance locus, 324 EcoRI+3/MseI+3 (196 E-A/M-C and 128 E-A/M-A) AFLP primer combinations were screened in the bulk material using a BSA strategy. Ten putative AFLP fragments were identified and segregation analysis in the BC1 population confirmed that seven of them were associated with the resistance locus. The two closest markers, EAGCMCGA-450 and EACAMAGG-330, were determined to be linked to the resistance locus at distances of 1.2 and 0.8 cM, respectively.
These two AFLP fragments were cloned and sequenced. BLAST analysis showed that the sequence of EAACMATC-330 had no similarity to any known sequence in GenBank, whereas EAGCMCGA-450 hit four potato ESTs (BQ509088, BG600948, DV623421, DV623416). These four ESTs and two other potato ESTs were assembled into a 1720-bp contig with a complete coding region that showed high similarity to the Arabidopsis gene GLUCAN SYNTHASE-like 7 (1e-139) in Blastx5) analysis. Based on this assembled sequence, a CAPS marker of EAGCMCGA-450, SpAFLP1, was developed (Figures 1A,D). In addition, EAACMATC-330 was converted into the CAPS marker SpAFLP2 (Figures 1B,E).
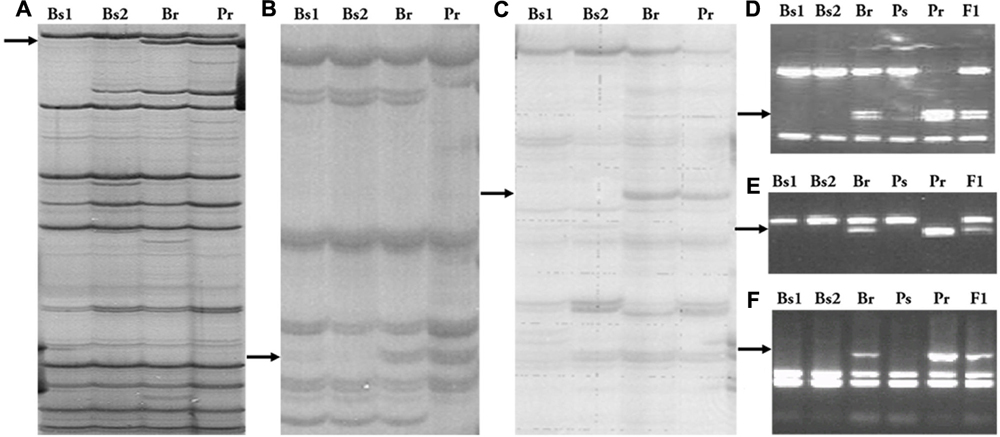
FIGURE 1. AFLP and RGA markers and their conversion into PCR-based markers. (A) Marker EAGC/MCGA-450 and (D) its SCAR marker SpAFLP1. (B) Marker EAAC/MATC-330 and (E) its SCAR marker SpAFLP2. (C) The RGA1 marker and (F) its SCAR marker SpGrol-1. Bs1, Susceptible bulk 1; Bs2, susceptible bulk 2; Br, resistant bulk; Ps, susceptible parent; Pr, resistant parent.
Integration of Rpi2 into the SGN Map
The 1720-bp contig of EAGCMCGA-450 was used to search the high-throughput genomic sequence (HTGS) database with BLASTn6 and a tomato BAC (C07HBa0116M01) was identified (e-112). Annotation of this BAC (C07HBa0116M01) revealed a partial VPS16-like gene in the 3′ terminus. This partial gene sequence was used as a query to search the EST database with BLASTn and 23 matching ESTs were identified. All 23 ESTs were assembled to a 2.4-kb contig, named cEST1. BLAST analysis showed that this sequence had homology to an RFLP marker, TG572 (e-120), which was mapped to tomato chromosome 7. Subsequently, a CAPS maker named SpTG572 was developed according to this sequence and was shown to co-segregate with SpAFLP1 (Figure 2).
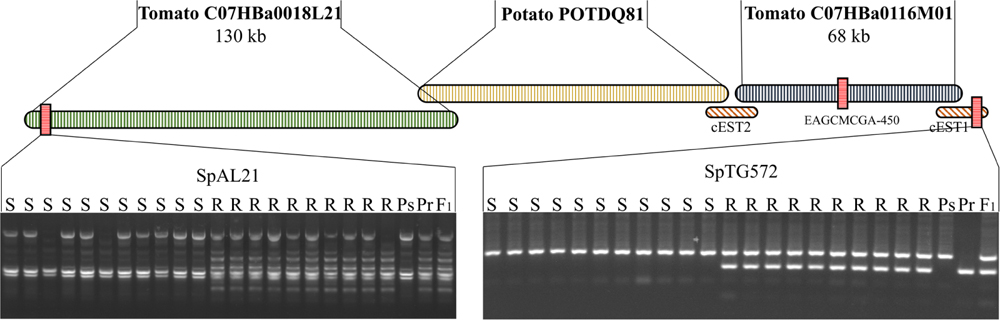
FIGURE 2. A contig built near the Rpi2 locus. Three BACs (C07HBa0018L21, POTDQ81, and C07HBa0116M01) and two EST assembles (cEST1 and cEST2) overlapped and were assembled into a contig. Two markers located at either end of the contig were developed, SpAL21 and SpTG572. Ps, Susceptible parent; Pr, resistant parent.
The 5′ terminal sequence of BAC C07HBa0116M01 was used to build a 2-kb contig with 10 ESTs, named cEST2. This contig hit a potato BAC end sequence, POTDQ81TR. Using the other end sequence POTDQ81TF as a query, we identified a tomato BAC, C07HBa0018L21. A PCR marker, SpAL21, was developed based on the left end sequence of BAC C07HBa0018L21 and recombination was found between this marker and SpTG572 (Figure 2).
TG572 was near I-3, a gene for fusarium wilt resistance from the wild tomato species Lycopersicon pennellii, with a genetic distance less than 0.3 cM (Hemming et al., 2004). Two additional markers closely linked to I-3, CT226, and Got2, were converted to SCAR markers in our mapping population, and named SpCT226 and SpGot2, respectively. Segregation analysis indicated SpCT226 and SpGot2 were proximal and distal with genetic distances of 2.8 and 3.2 cM, respectively (Figure 3B).
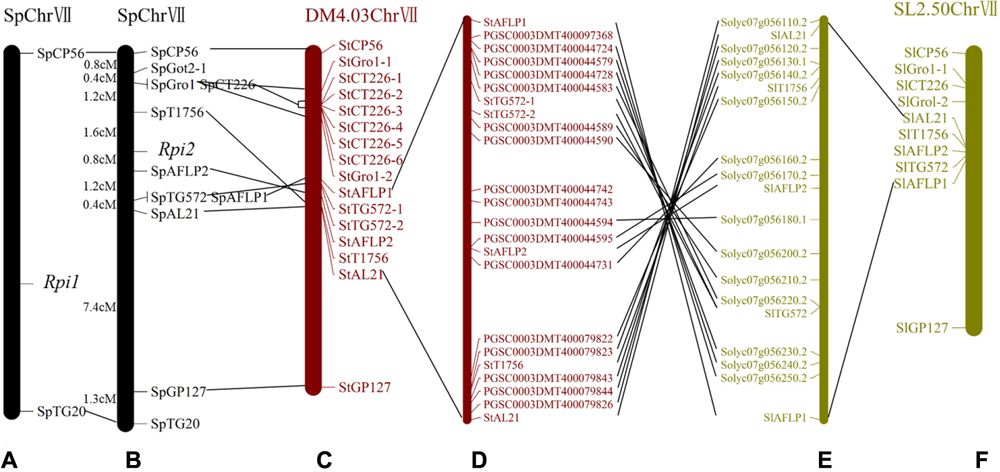
FIGURE 3. Genetic maps of the late blight resistance gene Rpi2 and their correspondence to comparative genomic maps of DM and tomato. (A) A genetic map of Rpi1 established by Kuhl et al. (2001). (B) A genetic map of Rpi2 established in this study. (C) Homologous markers on Solanum tuberosum group Phureja DM1-3(DM) chromosome 7. (D) Annotated genes on DM chromosome 7 between StAFLP1 and StAL21. (E) Annotated genes on Solanum lycopersicum (SL2.50) chromosome 7 between SlAFLP1 and SlAL21. (F) Homologous markers on S. lycopersicum chromosome 7. Collinear loci are indicated by black lines.
The flanking markers TG572, T0810, and T1756 in the SGN map7 were developed into PCR-based markers and tested in the mapping population. The results delimited the resistance locus to the interval between StAFLP2 and SpT1756 on potato chromosome 7 (Figure 3B).
An RGA Flanks the Resistance Locus
The RGA fingerprinting technique was used to identify functionally relevant markers linked to the resistance for late blight. An RGA fragment, RGA1, amplified by the AS2 primer (Leister et al., 1996) in combination with the E00 AFLP primer, did not exist in the two susceptible bulks but appeared in the resistant bulk and the resistant parent (Figure 1C). Sequence analysis of this 320-bp long fragment revealed homology to an RGA sequence previously mapped to the long arm of potato chromosome 7, Gro1-5 (Leister et al., 1996). Therefore, this fragment was named SpGrol-1. A PCR marker was developed and segregation analysis indicated that SpGrol-1 was 2.8 cM from the resistance locus (Figure 1F).
Genetic Relationship between Rpi1 and Rpi2
Previously, the late blight resistant locus Rpi1, also derived from S. pinnatisectum, was assigned to chromosome 7 flanked by two RFLP markers, TG20 and CP56 (Kuhl et al., 2001). To compare the map positions of Rpi1 and our target resistance locus, the RFLP markers TG20, CP56 and their interval marker GP1278 were converted into PCR-based markers. The marker information is listed in Supplementary Table S1. Segregation analysis of these converted PCR-based markers showed a link between the late blight resistance loci. The genetic distance between CP56 and TG20 was 15.1 cM, similar to the map of Kuhl et al. (2001) (14.6 cM). However, there was an obvious difference in the genetic distance between the markers linked to the resistance genes. CP56 and TG20 were 4.0 and 11.1 cM away from our target gene, respectively, which was different to the genetic distances of Rpi1 to these two markers (9.4 and 5.2 cM, respectively) (Figures 3A,B). Therefore, our resistance gene was called Rpi2.
Although the two resistance genes were derived from the same species, they came from two different accessions; Rpi1 from PI235214 and Rpi2 from PI 275233. To further identify genetic differences between the accessions PI275233 and PI235214, we screened all of the markers and found that the two accessions had different genotypes at two loci, SpAFLP2 and SpCT226.
Collinearity Analysis of Target Chromosome Regions between Potato and Tomato
The molecular marker sequences were used as queries to search for homologous loci in the genome sequence databases of potato and tomato. The majority of markers linked to Rpi2 showed homology to STChr7 and SLChr7, and were included in two orthologous genomic regions spanning 2.9 and 2.4 Mb, respectively (Figure 4B). Specifically, 9 of the 11 molecular markers mapped in the Rpi2 genetic linkage map generated hits to 16 homologous loci in STChr7 and 10 loci in SLChr7 (Figures 3C,F). Because cloning and sequencing failed, homologous loci of SpGot2-1 and SpTG20 were not found.
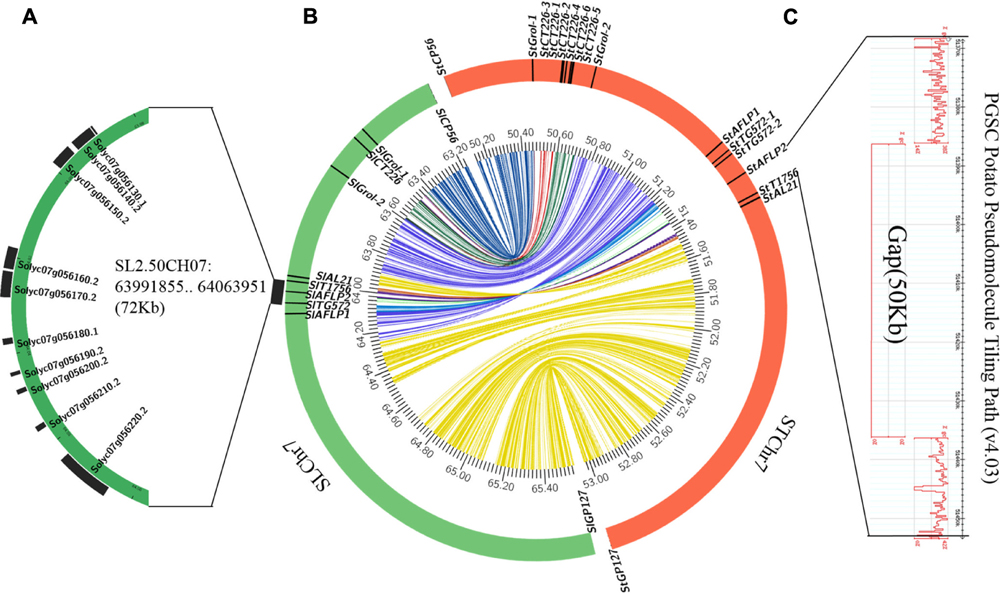
FIGURE 4. Collinearity analysis of target chromosome regions between potato and tomato. (A) The chromosome region between SlT1756 and SlTG572, which contains 10 annotated genes in the tomato genome. (B) Collinearity analysis of Rpi2-related chromosome regions between potato (DM v4.03) and tomato (SL2.50). (C) A 50-kb gap between StT1756 and StAFLP2 in the potato genome.
Comparative genomic analysis revealed that 14 of the 17 annotated genes in potato between StAFLP1 and StAL21 had similarity to corresponding regions in tomato (Figures 3D,E), again revealing high levels of collinearity in the Rpi2 region between potato and tomato. Furthermore, the order of these similar genes was highly conserved but reversed. Segmental inversion, which was a reasonable explanation for the reverse order of the conserved genes, was observed on STChr7 between StAFLP1 and StAL21 compared with the Rpi2 linkage map and SLChr7.
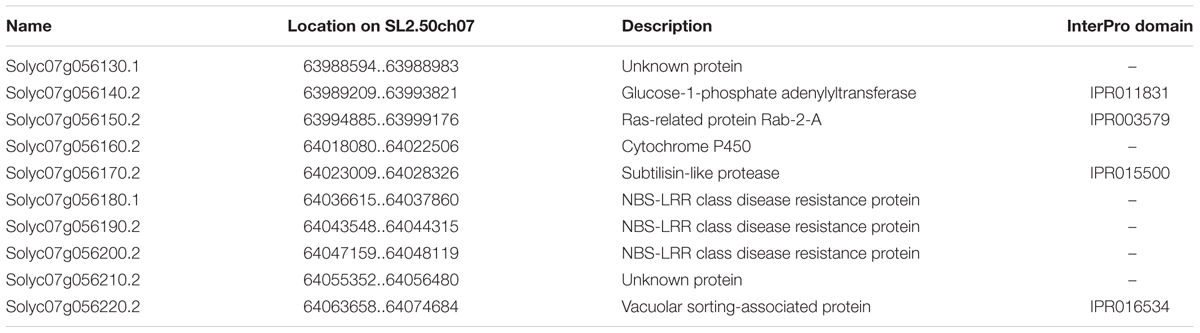
Scanning the Spud DB Genome Browser for Potato (Solanum tuberosum group Phureja DM1-3) PGSC v4.03 Pseudomolecules9 suggested the physical distance between StAFLP2 and StT1756 was about 84 kbp. Consequently, we estimated that the physical distance between Rpi2 and SpAFLP2 was about 28 kbp by referring to their genetic distance. Unfortunately, there was a 50-kbp gap in this potato genome region that potentially contained the homologous gene of Rpi2 (Figure 4C). However, the homologous segment in the tomato genome was assembled completely, in which 3 of 10 loci Table 1 were annotated as NBS-LRR class disease resistance proteins (Accession nos. Solyc07g056180.1, Solyc07g056190.2, and Solyc07g056200.2) (Figure 4A).
Discussion
The short-lived R genes from S. demissum prompted potato breeders and geneticists to look for resistance genes in other wild Solanum species (Van Soest et al., 1984; Colon and Budding, 1988; Douches et al., 2001). High-level resistance has been found in several diploid Mexican species, including S. bulbocastanum and S. pinnatisectum (Helgeson et al., 1998; Kuhl et al., 2001; Chen et al., 2003). These species may have adapted to coexist with highly complex and dynamic P. infestans populations (Niederhauser, 1953; Niederhauser et al., 1954). Genetic mapping studies indicated that the resistance in both S. bulbocastanum and S. pinnatisectum might be conferred by a single gene or a few dominant genes (Naess et al., 2000; Kuhl et al., 2001). Here, we identified a single dominant late blight resistance gene from the wild potato species S. pinnatisectum (PI 275233) and mapped it to an interval of 2.4 cM on the long arm of chromosome 7.
A Hotspot of Resistance Genes on Potato Chromosome 7
Accumulated evidence has suggested that resistance loci are not distributed randomly along chromosomes. Several hotspots for resistance genes have been described in Solanum species. For instance, at least five R genes against diverse pathogens have been mapped to the GP21–GP179 interval on chromosome 5 in different genetic backgrounds; Gpa and Grp1 conferring resistance to potato cyst nematodes (Kreike et al., 1994; van der Voort et al., 1998), Nb and Rx2 conferring resistance to potato virus X (Ritter et al., 1991; DeJong et al., 1997), and R1 conferring resistance to P. infestans (Leonards-Schippers et al., 1992). In the current study, we found that Rpi2 was located in a major cluster on the long arm of chromosome 7 in which several R genes have been mapped, including Rpi1 conferring resistance to P. infestans, Gro1-4 conferring resistance to Globodera rostochiensis, and I-3 conferring resistance to Fusarium oxysporum (Bournival et al., 1989; Ballvora et al., 1995; Kuhl et al., 2001; Paal et al., 2004; Ruggieri et al., 2014; Catanzariti et al., 2015). Clearly, this region is another hotspot for resistance genes, and can be expected to contain more resistance genes.
Resistance loci regions are usually enriched in NBS-LRR homologs. For instance, there are at least 13 TIR-NBS-LRR sequences clustered across more than 400 kb in the locus Gro1 (Paal et al., 2004) and one of them, Gro1-4, has been shown to be responsible for a resistance trait. In this research, we used the RGA profiling strategy to identify an RGA fragment, SpGrol-1, linked to the resistance locus. Sequence analysis showed that SpGrol-1 belonged to the TIR-NBS-LRR family, and that the most similar sequence was Gro1-5, a gene at the Gro1 locus. However, mapping analysis showed this RGA was proximal to Rpi2 with a genetic distance of 2.8 cM.
Resistance gene analogs are generally clustered in the genome (Meyers et al., 1998; Michelmore and Meyers, 1998). Clusters of R genes can be tightly organized or spaced over several megabases of sequence (Meyers et al., 1998; Noël et al., 1999). We thought that similar Gro1-like sequences might be present in our resistance locus. Hence, we designed a set of primers according to an alignment of 13 Gro1 sequences and developed three PCR markers. However, all of these markers co-localized to SpGrol-1 (data not shown). This indicated that there was more than one Gro1-like sequence and that the Rpi2 gene might not be a Gro1-like gene.
A similar observation was also described for the resistance gene I-3 from the wild tomato L. pennellii (Hemming et al., 2004). I-3 co-segregated with RGA St332; however, RGA St332 was ruled out as a candidate gene for I-3 because it was a single-copy pseudo gene in L. pennellii. I-3 was flanked by two RFLP markers TG572 and CT226 in an interval of 0.6 cM. That Rpi2 and I-3 share the flanking markers CT226 and TG572 supports that these two genes are in a syntenic region.
Comparative Sequence Analysis of the Rpi2 Region
Comparative genomics between potato and tomato facilitated the mapping and isolation of the late-blight R genes R3a and Rpi-blb2 from potato in a previous study, as these genes were mapped to regions of the potato genome that were syntenic to previously cloned gene loci (I2 and Mi, respectively) in tomato (Huang et al., 2005; van der Vossen et al., 2005). Recently, both the potato and tomato genomes have been sequenced (Xu et al., 2011; Sato et al., 2012). This sequence information should greatly accelerate the cloning of the Rpi2 gene through comparative genomics.
Comparing the homologous regions in the potato and tomato genomes, the genetic linkage map of Rpi2 showed high uniformity except that a chromosome inversion had occurred in the sequenced DM genome (Figure 3). Although this inversion may be a result of chromosomal variation during evolution, incorrect sequencing or assembly could equally have led to the observed recombination because short reads, a large amount of repetitive sequence, the sequence GC composition and other effects can impede uniform and complete sequencing coverage along the genome (Maiti and Bouvagnet, 2001; van Hijum et al., 2005; Aird et al., 2011; Schatz et al., 2012; Berlin et al., 2015). The gap between SpAFLP2 and SpT1756 on StChr7 indicated the accuracy of the assembly in the Rpi2-related region was not sufficiently high. In other words, the fragment inversion observed by comparative analysis was not sufficient evidence to demonstrate chromosome inversion. Furthermore, the lack of sequence information between the flanking markers prevented us from obtaining candidate genes from the DM genome data. Therefore, constructing a higher quality genome assembly for the Rpi2-related region requires enhanced approaches.
At present, an effort to introgress disease resistance genes from S. pinnatisectum into potato is being carried out to develop resistant cultivars. Because of the ploidy level barrier and endosperm balance number incompatibility, it is difficult to transfer resistance traits from S. pinnatisectum to cultivated potato. Fortunately, the hybridization barrier between S. pinnatisectum and cultivated potatoes can be overcome by embryo rescue, protoplast fusion, and chromosome-doubling techniques (Chen et al., 2008; Sanetomo et al., 2014). Our molecular markers could help breeders to introduce this resistance gene into different cultivars by marker-assisted selection.
Author Contributions
LY and DW: Conducted the experiments, analyzed the data, and wrote the manuscript. YX: Identified the resistant parental line and made the cross. LW and XC: Participated in detecting molecular makers and contributed to the genotyping experiment. HZ: Assisted in analyzing the data. YC and QC: Conceived and directed the project and revised the manuscript.
Funding
This study was financially supported by Key Research and Development Program of Shaanxi Province, China (grant no. 2017ZDXM-NY-004) and the Research Funding for Special Professor of Northwest Agriculture and Forestry University (grant no. Z111021202).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Robbie Lewis, M.Sc., from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01729/full#supplementary-material
TABLE S1 | Information of molecular markers involved in this study.
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/genbank/
- ^ http://www.ebi.ac.uk/Tools/msa/clustalw2/
- ^ https://solgenomics.net/cview/index.pl
- ^ http://ftp.sgn.cornell.edu/tools/intron_detection/find_introns.pl
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastx&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome
- ^ https://solgenomics.net/cview/index.pl
- ^ https://solgenomics.net/marker/SGN-M16437/details
- ^ http://solanaceae.plantbiology.msu.edu/cgi-bin/gbrowse/potato/
References
Aird, D., Ross, M. G., Chen, W. S., Danielsson, M., Fennell, T., Russ, C., et al. (2011). Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 12:R18. doi: 10.1186/gb-2011-12-2-r18
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Ballvora, A., Ercolano, M. R., Weiss, J., Meksem, K., Bormann, C. A., Oberhagemann, P., et al. (2002). The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. Plant J. 30, 361–371. doi: 10.1046/j.1365-313X.2001.01292.x
Ballvora, A., Hesselbach, J., Niewohner, J., Leister, D., Salamini, F., and Gebhardt, C. (1995). Marker enrichment and high-resolution map of the segment of potato chromosome VII harbouring the nematode resistance gene Gro1. Mol. Gen. Genet. 249, 82–90. doi: 10.1007/BF00290239
Bassam, B. J., Caetano-Anolles, G., and Gresshoff, P. M. (1991). Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal. Biochem. 196, 80–83. doi: 10.1016/0003-2697(91)90120-I
Berlin, K., Koren, S., Chin, C. S., Drake, J. P., Landolin, J. M., and Phillippy, A. M. (2015). Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol. 33, 623–630. doi: 10.1038/nbt.3238
Black, W. (1951). XVII.—inheritance of resistance to blight (Phytophthora infestans) in potatoes: inter-relationships of genes and strains. Proc. R. Soc. Edinb. 64, 312–352. doi: 10.1017/S0080455X00009905
Black, W., Mastenbroek, C., Mills, W. R., and Peterson, L. C. (1953). A proposal for an international nomenclature of races of Phytophthora infestans and of genes controlling immunity in Solanum demissum derivatives. Euphytica 2, 173–179. doi: 10.1007/bf00053724
Bournival, B. L., Scott, J. W., and Vallejos, C. E. (1989). An isozyme marker for resistance to race 3 of Fusarium oxysporum f. sp. lycopersici in tomato. Theor. Appl. Genet. 78, 489–494. doi: 10.1007/BF00290832
Catanzariti, A. M., Lim, G. T., and Jones, D. A. (2015). The tomato I-3 gene: a novel gene for resistance to Fusarium wilt disease. New Phytol. 207, 106–118. doi: 10.1111/nph.13348
Chen, Q., Kawchuk, L. M., Lynch, D. R., Goettel, M. S., and Fujimoto, D. K. (2003). Identification of late blight, Colorado potato beetle, and blackleg resistance in three Mexican and two south American wild 2x (1EBN) Solanum species. Am. J. Potato Res. 80, 9–19. doi: 10.1007/BF02854552
Chen, Q., Li, H. Y., Shi, Y. Z., Beasley, D., Bizimungu, B., and Goettel, M. S. (2008). Development of an effective protoplast fusion system for production of new potatoes with disease and insect resistance using Mexican wild potato species as gene pools. Can. J. Plant Sci. 88, 611–619. doi: 10.4141/CJPS07045
Chen, X. M., Line, R. F., and Leung, H. (1998). Genome scanning for resistance-gene analogs in rice, barley, and wheat by high-resolution electrophoresis. Theor. Appl. Genet. 97, 345–355. doi: 10.1007/s001220050905
Collins, A., Milbourne, D., Ramsay, L., Meyer, R., Chatot-Balandras, C., Oberhagemann, P., et al. (1999). QTL for field resistance to late blight in potato are strongly correlated with maturity and vigour. Mol. Breed. 5, 387–398. doi: 10.1023/A:1009601427062
Colon, L. T., and Budding, D. J. (1988). Resistance to late blight (Phytophthora infestans) in ten wild Solanum species. Euphytica 39, 77–86. doi: 10.1007/bf00043370
De Vetten, N. C. M. H., Verzaux, E. C., Vossen, J. H., Rietman, H., and Jacobsen, E. (2014). Cloning and Exploitation of a Functional R-Gene from Solanum x edinense. US Patent WO 2011152722 A3.
DeJong, W., Forsyth, A., Leister, D., Gebhardt, C., and Baulcombe, D. C. (1997). A potato hypersensitive resistance gene against potato virus X maps to a resistance gene cluster on chromosome 5. Theor. Appl. Genet. 95, 246–252. doi: 10.1007/s001220050555
Douches, D. S., Bamberg, J. B., Kirk, W., Jastrzebski, K., Niemira, B. A., Coombs, J., et al. (2001). Evaluation of wild Solanum species for resistance to the US-8 genotype of Phytophthora infestans utilizing a fine-screening technique. Am. J. Potato Res. 78, 159–165. doi: 10.1007/BF02874771
Foster, S. J., Park, T. H., Pel, M., Brigneti, G., Sliwka, J., Jagger, L., et al. (2009). Rpi-vnt1.1, a Tm-2(2) homolog from Solanum venturii, confers resistance to potato late blight. Mol. Plant Microbe Interact. 22, 589–600. doi: 10.1094/MPMI-22-5-0589
Haesaert, G., Vossen, J. H., Custers, R., De Loose, M., Haverkort, A., Heremans, B., et al. (2015). Transformation of the potato variety Desiree with single or multiple resistance genes increases resistance to late blight under field conditions. Crop Prot. 77, 163–175. doi: 10.1016/j.cropro.2015.07.018
Hayes, A. J., and Maroof, M. A. S. (2000). Targeted resistance gene mapping in soybean using modified AFLPs. Theor. Appl. Genet. 100, 1279–1283. doi: 10.1007/s001220051435
Helgeson, J. P., Pohlman, J. D., Austin, S., Haberlach, G. T., Wielgus, S. M., Ronis, D., et al. (1998). Somatic hybrids between Solanum bulbocastanum and potato: a new source of resistance to late blight. Theor. Appl. Genet. 96, doi: 10.1007/s001220050796
Hemming, M. N., Basuki, S., McGrath, D. J., Carroll, B. J., and Jones, D. A. (2004). Fine mapping of the tomato I-3 gene for fusarium wilt resistance and elimination of a co-segregating resistance gene analogue as a candidate for I-3. Theor. Appl. Genet. 109, 409–418. doi: 10.1007/s00122-004-1646-4
Huang, S. W., van der Vossen, E. A. G., Kuang, H. H., Vleeshouwers, V. G. A. A., Zhang, N. W., Borm, T. J. A., et al. (2005). Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato. Plant J. 42, 251–261. doi: 10.1111/j.1365-313X.2005.02365.x
Jo, K. R., Kim, C. J., Kim, S. J., Kim, T. Y., Bergervoet, M., Jongsma, M. A., et al. (2014). Development of late blight resistant potatoes by cisgene stacking. BMC Biotechnol. 14:50. doi: 10.1186/1472-6750-14-50
Jones, J., Foster, S. J., Chu, Z., Park, T. H., and Pel, M. A. (2013). Late Blight Resistance Genes and Methods. US Patent WO 2009013468 A3.
Jupe, F., Pritchard, L., Etherington, G. J., MacKenzie, K., Cock, P. J. A., Wright, F., et al. (2012). Identification and localisation of the NB-LRR gene family within the potato genome. BMC Genomics 13:75. doi: 10.1186/1471-2164-13-75
Kanazin, V., Marek, L. F., and Shoemaker, R. C. (1996). Resistance gene analogs are conserved and clustered in soybean. Proc. Natl. Acad. Sci. U.S.A. 93, 11746–11750. doi: 10.1073/pnas.93.21.11746
Kosambi, D. D. (1943). The estimation of map distances from recombination values. Ann. Eugen. 12, 172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x
Kreike, C. M., Dekoning, J. R. A., Vinke, J. H., Vanooijen, J. W., and Stiekema, W. J. (1994). Quantitatively-inherited resistance to globodera-pallida is dominated by one major locus in Solanum spegazzinii. Theor. Appl. Genet. 88, 764–769. doi: 10.1007/Bf01253983
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kuhl, J. C., Hanneman, R. E., and Havey, M. J. (2001). Characterization and mapping of Rpi1, a late-blight resistance locus from diploid (1EBN) Mexican Solanum pinnatisectum. Mol. Genet. Genomics 265, 977–985. doi: 10.1007/s004380100490
Lander, E. S., Green, P., Abrahamson, J., Barlow, A., Daly, M. J., Lincoln, S. E., et al. (1987). MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1, 174–181. doi: 10.1016/0888-7543(87)90010-3
Leister, D., Ballvora, A., Salamini, F., and Gebhardt, C. (1996). A PCR-based approach for isolating pathogen resistance genes from potato with potential for wide application in plants. Nat. Genet. 14, 421–429. doi: 10.1038/ng1296-421
Leonards-Schippers, C., Gieffers, W., Salamini, F., and Gebhardt, C. (1992). The R1 gene conferring race-specific resistance to Phytophthora infestans in potato is located on potato chromosome V. Mol. Gen. Genet. 233, 278–283. doi: 10.1007/BF00587589
Lokossou, A. A., Park, T. H., van Arkel, G., Arens, M., Ruyter-Spira, C., Morales, J., et al. (2009). Exploiting knowledge of R/Avr genes to rapidly clone a new LZ-NBS-LRR family of late blight resistance genes from potato linkage group IV. Mol. Plant Microbe Interact. 22, 630–641. doi: 10.1094/MPMI-22-6-0630
Maiti, A. K., and Bouvagnet, P. (2001). Assembling and gap filling of unordered genome sequences through gene checking. Genome Biol. 2, rerint0008.0001. doi: 10.1186/gb-2001-2-9-preprint0008
Malcolmson, J. F., and Black, W. (1966). New R genes in Solanum demissum lindl. And their complementary races of Phytophthora infestans (Mont.) de bary. Euphytica 15, 199–203. doi: 10.1007/bf00022324
Martin, G. B., Bogdanove, A. J., and Sessa, G. (2003). Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54, 23–61. doi: 10.1146/annurev.arplant.54.031902.135035
Menke, U., SchildeRentschler, L., Ruoss, B., Zanke, C., Hemleben, V., and Ninnemann, H. (1996). Somatic hybrids between the cultivated potato Solanum tuberosum L and the 1EBN wild species Solanum pinnatisectum Dun: morphological and molecular characterization. Theor. Appl. Genet. 92, 617–626. doi: 10.1007/BF00224566
Meyers, B. C., Chin, D. B., Shen, K. A., Sivaramakrishnan, S., Lavelle, D. O., Zhang, Z., et al. (1998). The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell 10, 1817–1832. doi: 10.1105/tpc.10.11.1817
Michelmore, R. W., and Meyers, B. C. (1998). Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8, 1113–1130. doi: 10.1101/gr.8.11.1113
Michelmore, R. W., Paran, I., and Kesseli, R. V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. U.S.A. 88, 9828–9832. doi: 10.1073/pnas.88.21.9828
Naess, S. K., Bradeen, J. M., Wielgus, S. M., Haberlach, G. T., McGrath, J. M., and Helgeson, J. P. (2000). Resistance to late blight in Solanum bulbocastanum is mapped to chromosome 8. Theor. Appl. Genet. 101, 697–704. doi: 10.1007/s001220051533
Niederhauser, J. S. (1953). Resistance of Solanum species to Phytophthora infestans in Mexico. Phytopathology 43, 456–457.
Niederhauser, J. S., Cervantes, J., and Servin, L. (1954). Late blight in Mexico and its implications. Phytopathology 44, 406–408.
Noël, L., Moores, T. L., van der Biezen, E. A., Parniske, M., Daniels, M. J., Parker, J. E., et al. (1999). Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11, 2099–2111. doi: 10.1105/tpc.11.11.2099
Paal, J., Henselewski, H., Muth, J., Meksem, K., Menendez, C. M., Salamini, F., et al. (2004). Molecular cloning of the potato Gro1-4 gene conferring resistance to pathotype Ro1 of the root cyst nematode Globodera rostochiensis, based on a candidate gene approach. Plant J. 38, 285–297. doi: 10.1111/j.1365-313X.2004.02047.x
Park, T. H., Gros, J., Sikkema, A., Vleeshouwers, V. G. A. A., Muskens, M., Allefs, S., et al. (2005a). The late blight resistance locus Rpi-blb3 from Solanum bulbocastanum belongs to a major late blight R gene cluster on chromosome 4 of potato. Mol. Plant Microbe Interact. 18, 722–729.
Park, T. H., Vleeshouwers, V. G. A. A., Hutten, R. C. B., van Eck, H. J., van der Vossen, E., Jacobsen, E., et al. (2005b). High-resolution mapping and analysis of the resistance locus Rpi-abpt against Phytophthora infestans in potato. Mol. Breed. 16, 33–43. doi: 10.1007/s11032-005-1925-z
Pel, M. A., Foster, S. J., Park, T. H., Rietman, H., van Arkel, G., Jones, J. D. G., et al. (2009). Mapping and cloning of late blight resistance genes from Solanum venturii using an interspecific candidate gene approach. Mol. Plant Microbe Interact. 22, 601–615. doi: 10.1094/MPMI-22-5-0601
Ritter, E., Debener, T., Barone, A., Salamini, F., and Gebhardt, C. (1991). RFLP mapping on potato chromosomes of two genes controlling extreme resistance to potato virus X (PVX). Mol. Gen. Genet. 227, 81–85. doi: 10.1007/BF00260710
Ruggieri, V., Nunziata, A., and Barone, A. (2014). Positive selection in the leucine-rich repeat domain of Gro1 genes in Solanum species. J. Genet. 93, 755–765. doi: 10.1007/s12041-014-0458-9
Sanetomo, R., Akino, S., Suzuki, N., and Hosaka, K. (2014). Breakdown of a hybridization barrier between Solanum pinnatisectum Dunal and potato using the S locus inhibitor gene (Sli). Euphytica 197, 119–132. doi: 10.1007/s10681-013-1057-1
Sato, S., Tabata, S., Mueller, L. A., Huang, S., Du, Y., Li, C., et al. (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. doi: 10.1038/nature11119
Schatz, M. C., Witkowski, J., and McCombie, W. R. (2012). Current challenges in de novo plant genome sequencing and assembly. Genome Biol. 13:243. doi: 10.1186/gb4015
Song, J. Q., Bradeen, J. M., Naess, S. K., Raasch, J. A., Wielgus, S. M., Haberlach, G. T., et al. (2003). Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. Proc. Natl. Acad. Sci. U.S.A. 100, 9128–9133. doi: 10.1073/pnas.1533501100
van der Voort, J. R., Lindeman, W., Folkertsma, R., Hutten, R., Overmars, H., van der Vossen, E., et al. (1998). A QTL for broad-spectrum resistance to cyst nematode species (Globodera spp.) maps to a resistance gene cluster in potato. Theor. Appl. Genet. 96, 654–661. doi: 10.1007/s001220050785
van der Vossen, E., Sikkema, A., Hekkert, B., Gros, J., Stevens, P., Muskens, M., et al. (2003). An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant J. 36, 867–882. doi: 10.1046/j.1365-313X.2003.01934.x
van der Vossen, E. A. G., Gros, J., Sikkema, A., Muskens, M., Wouters, D., Wolters, P., et al. (2005). The Rpi-blb2 gene from Solanum bulbocastanum is an Mi-1 gene homolog conferring broad-spectrum late blight resistance in potato. Plant J. 44, 208–222. doi: 10.1111/j.1365-313X.2005.02527.x
van Hijum, S. A., Zomer, A. L., Kuipers, O. P., and Kok, J. (2005). Projector 2: contig mapping for efficient gap-closure of prokaryotic genome sequence assemblies. Nucleic Acids Res. 33, W560–W566. doi: 10.1093/nar/gki356
Van Soest, L. J. M., Schöber, B., and Tazelaar, M. F. (1984). Resistance to Phytophthora infestans in tuber-bearing species of Solanum and its geographical distribution. Potato Res. 27, 393–411. doi: 10.1007/bf02357427
Vleeshouwers, V. G. A. A., Rietman, H., Krenek, P., Champouret, N., Young, C., Oh, S. K., et al. (2008). Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLOS ONE 3:e2875. doi: 10.1371/journal.pone.0002875
Vos, P., Hogers, R., Bleeker, M., Reijans, M., van de Lee, T., Hornes, M., et al. (1995). AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23, 4407–4414. doi: 10.1093/nar/23.21.4407
Vossen, J. H., Nijenhuis, M., and Jacobsen, E. (2011). Cloning and Exploitation of a Functional R-Gene from Solanum chacoense. US Pattern WO 2011034433 A1.
Keywords: potato, late blight, resistance gene, genetic map, collinearity analysis
Citation: Yang L, Wang D, Xu Y, Zhao H, Wang L, Cao X, Chen Y and Chen Q (2017) A New Resistance Gene against Potato Late Blight Originating from Solanum pinnatisectum Located on Potato Chromosome 7. Front. Plant Sci. 8:1729. doi: 10.3389/fpls.2017.01729
Received: 26 July 2017; Accepted: 21 September 2017;
Published: 04 October 2017.
Edited by:
Jacqueline Batley, University of Western Australia, AustraliaReviewed by:
Hussein Shimelis, University of KwaZulu-Natal, South AfricaDavid S. Douches, Michigan State University, United States
Copyright © 2017 Yang, Wang, Xu, Zhao, Wang, Cao, Chen and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qin Chen, Y2hlbnBldGVyMjI4OUBud3N1YWYuZWR1LmNu Yue Chen, MzUzNjA2MTk3QHFxLmNvbQ==
†These authors have contributed equally to this work and should be considered co-first authors.
 Le Yang
Le Yang Dongdong Wang†
Dongdong Wang† Xiaoli Cao
Xiaoli Cao Qin Chen
Qin Chen