- 1Plant Gene Expression Center, United States Department of Agriculture – Agricultural Research Service, Albany, CA, United States
- 2Department of Plant and Microbial Biology, University of California, Berkeley, Berkeley, CA, United States
Multicellular organisms rely on the precise and consistent regulation of gene expression to direct their development in tissue- and cell-type specific patterns. This regulatory activity involves arrays of DNA-binding transcription factors and epigenetic factors that modify chromatin structure. Among the chromatin modifiers, trithorax (trxG) and Polycomb (PcG) group proteins play important roles in orchestrating the stable activation and repression of gene expression, respectively. These proteins have generally antagonistic functions in maintaining cell and tissue homeostasis as well as in mediating widespread transcriptional reprogramming during developmental transitions. Plants utilize multiple trxG factors to regulate gene transcription as they modulate their development in response to both endogenous and environmental cues. Here, I will discuss the roles of trxG factors and their associated proteins in post-embryonic plant development.
Introduction
The development of multicellular organisms is driven by precise patterns of gene transcription that are tightly regulated in a spatial and temporal manner. Establishing and sustaining specific transcription states at gene loci are complex, multi-step processes. They require repertoires of sequence-specific DNA-binding transcription factors as well as epigenetic factors that alter chromatin structure and thereby affect accessibility by the transcriptional machinery. Epigenetic regulators classified as trithorax group (trxG) and Polycomb group (PcG) factors are critical for maintaining the stable transcription patterns at developmental regulatory loci by organizing chromatin in an active or inactive state, respectively (Schwartz and Pirrota, 2007). trxG and PcG factors generally act in large, multi-component complexes that function antagonistically to generate and maintain a balanced state of gene expression (Piunti and Shilatifard, 2016).
TrxG genes were first identified in Drosophila as positive regulators of PcG developmental target genes (Ingham, 1988; Kennison, 1995), and their protein products operate in multiple complexes that affect gene expression on a global scale (Piunti and Shilatifard, 2016). Because transcription activation involves numerous steps, trxG factors are heterogeneous and fall into several functional categories: chromatin remodeling proteins, histone modifying methyltransferase and demethylase proteins, and DNA-binding and accessory proteins (Xiao et al., 2016) (Table 1). The chromatin remodeling proteins include members of the SWI/SNF, ISWI, and CHD families that utilize ATP to alter nucleosome assembly and distribution (Gentry and Hennig, 2014). The histone modifying enzymes deposit H3K4me2/3 and/or H3K36me2/3 marks associated with transcription activation to counteract the activity of PcG complexes such as POLYCOMB REPRESSIVE COMPLEX 2 (PRC2) that deposit H3K27me3 as the major repressive mark for transcription (Piunti and Shilatifard, 2016).
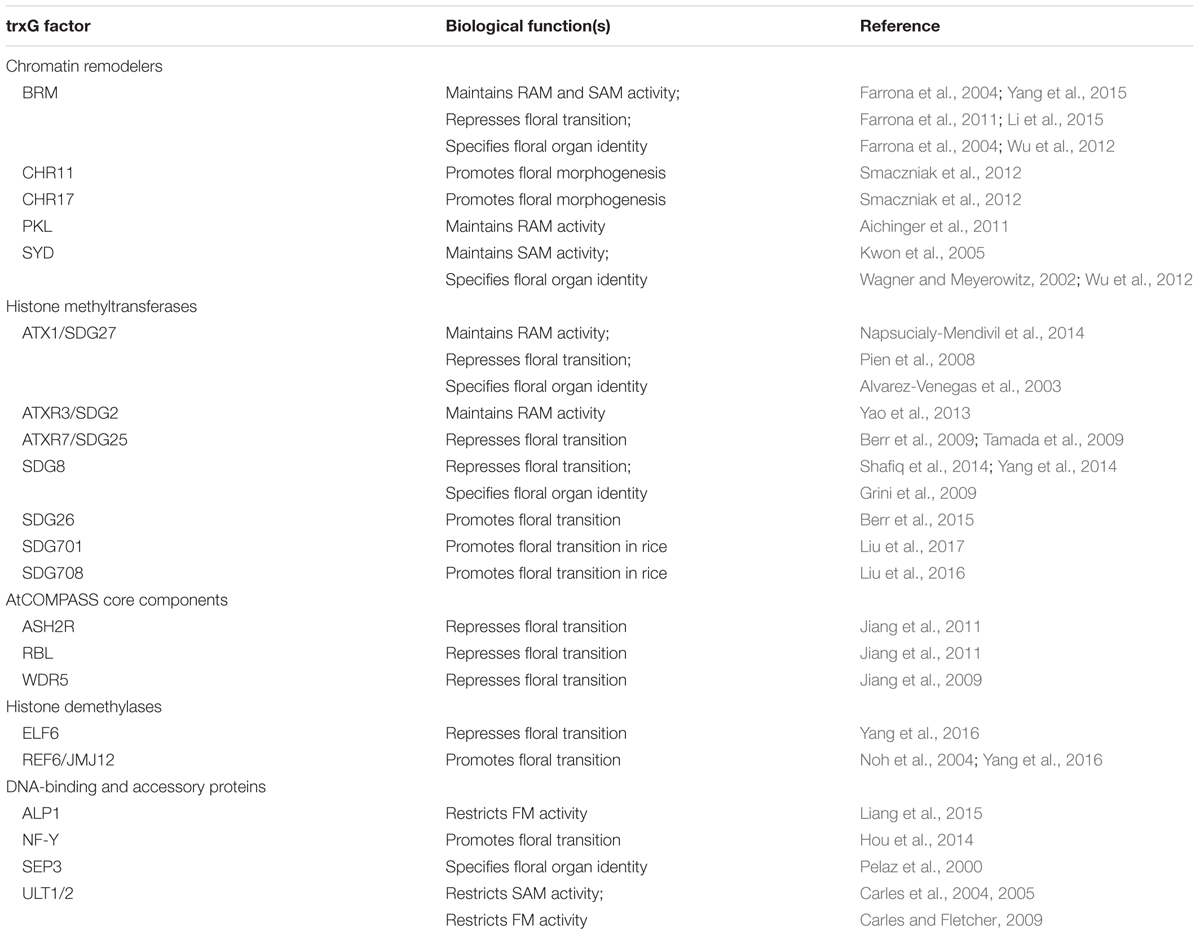
TABLE 1. Biological functions of the trxG factors and their accessory proteins in post-embryonic development.
Plant trxG factors have been identified either by homology to known trxG factors in animals or by genetic characterization based on their ability to suppress PcG mutant phenotypes. Given their fundamental roles in the epigenetic regulation of gene expression states, mutations in plant trxG genes often cause pleiotrophic developmental phenotypes, including defects in seedling growth, anther and ovule formation, and gametophyte development (Grini et al., 2009; Guo et al., 2010; Carter et al., 2016; Chen et al., 2017). They also play key roles during developmental transitions when widespread gene reprogramming occurs. Here I will summarize our current understanding of trxG protein function in plant meristems, which drive post-embryonic development.
Role of trxG Factors in Root and Shoot Apical Meristem Maintenance
Plants are sessile organisms that grow continuously and alter their development in response to changes in their environment. Organogenesis occurs throughout the life cycle from specialized structures at the growing shoot and root tips, called apical meristems (Steeves and Sussex, 1989). Both the root and shoot apical meristems (SAMs) contain small reservoirs of stem cells that constantly replenish themselves as well as provide progeny cells for continuous organ formation. The flexible regulation of gene expression via chromatin remodeling is essential for maintaining these pluripotent stem cell populations whose progeny can assume different fates. Animal stem cells possess special chromatin signatures (Bernstein et al., 2006) that permit plasticity in stem cell dynamics, and much is known about the epigenetic factors and mechanisms involved (Spivakov and Fisher, 2007). In contrast, the roles of epigenetic factors in regulating plant stem cell activity are only beginning to be revealed.
Recent genetic studies have uncovered roles for multiple trxG factors in root apical meristem (RAM) maintenance. The RAM generates the entire underground root system and has a stereotypical organization. Four rarely dividing cells known as the quiescent center (QC) act as a niche (van den Berg et al., 1997) that maintains the surrounding cells as stem cells, aka initial cells, which undergo asymmetric cell divisions to generate the distinct root cell lineages. A gradient of the hormone auxin across the root tip exists due to the activity of members of the PIN-FORMED (PIN) family of auxin transport proteins (Blilou et al., 2005; Grieneisen et al., 2007). The auxin concentration maximum coincides with the QC and promotes the expression of the PLETHORA (PLT) AP2 domain transcription factor (TF) genes, which are essential for root stem cell niche maintenance (Aida et al., 2004).
Two H3K4 histone methyltransferase trxG factors have been implicated in Arabidopsis RAM maintenance (Figure 1A). The SET domain protein SET DOMAIN GROUP 2 (SDG2) is the major H3K4 trimethyltransferase in Arabidopsis and is necessary for genome-wide H3K4me3 deposition (Guo et al., 2010). In the RAM SDG2 is required to maintain the auxin gradient and QC maximum, and to sustain cell identity and stem cell activity in the QC and surrounding initial cells (Yao et al., 2013). These functions correlate with a requirement for SDG2 to promote PLT1 expression and global H3K4me3 deposition in root cells (Yao et al., 2013). The ARABIDOPSIS HOMOLOG OF TRITHORAX1 (ATX1/SDG27) protein contributes ∼15% of genome-wide H3K4 trimethylation (Alvarez-Venegas and Avramova, 2005). ATX1 is needed for TATA binding protein (TBP) and RNA Polymerase II recruitment to its target promoters (Ding et al., 2011) and is also critical for H3K4me3 deposition associated with transcription elongation (Ding et al., 2012). Like SDG2, ATX1 is necessary for normal RAM organization, but also restricts the expression of QC markers such as WOX5 to the stem cell niche in an auxin-independent fashion (Napsucialy-Mendivil et al., 2014), indicating that the two H3K4 histone methyltransferases have distinct as well as shared roles in RAM maintenance.
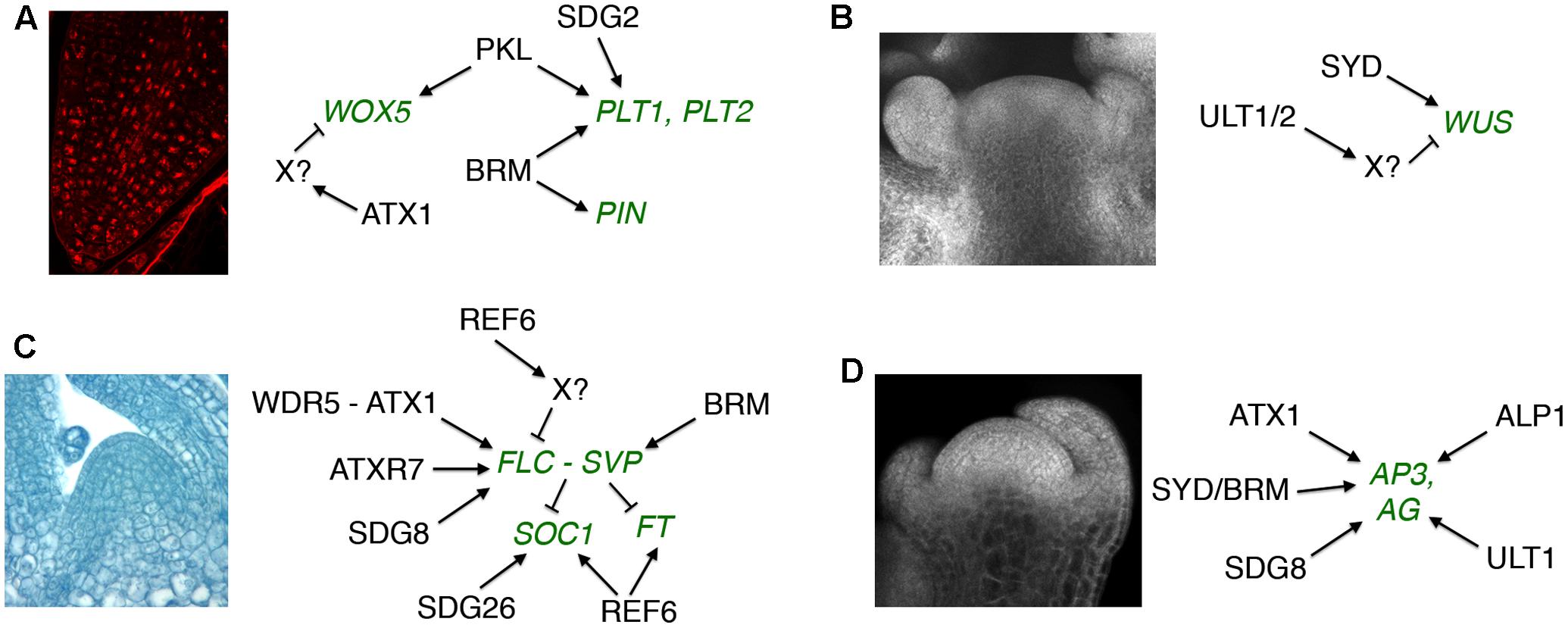
FIGURE 1. Regulatory targets of trxG factors in post-embryonic development. (A) Root apical meristem maintenance. (B) Shoot apical meristem maintenance. (C) Vegetative to reproductive meristem transition. (D) Floral meristem patterning. Gene targets shown in green type. Arrows indicate positive and bars indicate negative regulatory interactions. The SAM image in (B) is reprinted from Fiume et al. (2010). No permission is required for its reproduction.
The SWI2/SNF2 chromatin remodeling complex ATPase genes PICKLE (PKL) and BRAHMA (BRM) also regulate RAM activity in Arabidopsis. PKL acts antagonistically to the PRC2 PcG factor CURLY LEAF (CLF) to maintain RAM stem cell activity (Aichinger et al., 2011). PKL does not induce the activity of the root stem cell niche by affecting auxin accumulation. Instead, PKL elevates the expression levels of and limits CLF-mediated H3K27me3 deposition at PLT1, PLT2, and WOX5 (Aichinger et al., 2011). BRM likewise maintains the RAM stem cell niche by promoting expression of PLT1 and PLT2 (Yang et al., 2015). However, BRM unlike PKL affects auxin accumulation in the root tip by directly binding to and up-regulating the transcription of five PIN loci (Yang et al., 2015). Thus the evidence to date suggests that the auxin-dependent and auxin-independent regulatory pathways utilize distinct trxG factors to sustain RAM activity.
Maintenance of stem cell reservoirs in Arabidopsis shoot and floral meristems occurs via a spatial negative feedback loop mediated by the WUSCHEL (WUS)-CLAVATA (CLV) signal transduction pathway. The homeobox TF gene WUS is expressed in the core of the meristem and confers stem cell identity on the overlying cells (Laux et al., 1996). The stem cell-inducing activity of WUS is antagonized by the secreted polypeptide CLV3 (Fletcher et al., 1999; Rojo et al., 2002), which is produced by the stem cells and activates a signal transduction pathway in the meristem interior that limits the accumulation of WUS-expressing cells (Brand et al., 2000). This feedback loop mediates stem cell homeostasis, balancing the loss of stem cells to organ formation with their replenishment via cell division.
Although little is known about trxG activity in the SAM, the ratio of H3K4me3 active to H3K27me3 repressive marks is known to be important for reproductive SAM development in rice (Liu et al., 2015). The SWI2/SNF2 trxG factors BRM and SYD both act to sustain SAM activity in Arabidopsis (Farrona et al., 2004; Kwon et al., 2005), with SYD shown to bind to the WUS promoter and elevate its transcription in the SAM (Kwon et al., 2005) (Figure 1B). In contrast, the SAND domain-containing proteins ULTRAPETALA1 (ULT1) and ULT2 restrict shoot and floral stem cell activity by limiting the size of the WUS expression domain (Carles et al., 2004, 2005). The SAND domain occurs in a small number of eukaryotic proteins including the human AIRE transcriptional regulator that is implicated in autoimmune diseases (Abramson and Goldfarb, 2016). ULT1 antagonizes the repressive activity of PRC1 and PRC2 PcG complex components (Carles and Fletcher, 2009; Pu et al., 2013), and physically associates with the H3K4 methyltransferase ATX1 (Carles and Fletcher, 2009) as well as the transcription factors KANADI1 (KAN1), KAN2 and ULTRAPETALA1 INTERACTING FACTOR (UIF1) (Pires et al., 2014; Moreau et al., 2016). Because both ATX1 and the AIRE protein form complexes with RNA Pol II and RNA-processing components (Abramson et al., 2010; Ding et al., 2012), the ULT proteins may coordinate chromatin mark modification at transcriptionally priming loci with basal transcription machinery recruitment by stage- and tissue-specific transcription factors (Carles and Fletcher, 2010; Engelhorn et al., 2014a).
Roles of trxG Factors in Floral Induction at the Shoot Apical Meristem
The floral induction process directs the SAM to transition from generating vegetative organs (leaves) to reproductive organs (flowers). The timing of this dynamic meristem cell fate switch is critical for plant reproductive success and occurs in response to endogenous pathways such as the age, GA and autonomous pathways (APs) as well as environmental cues including photoperiod, vernalization and temperature (Amasino, 2010; Srikanth and Schmid, 2011; Andres and Coupland, 2012). The MADS domain TF FLOWERING LOCUS C (FLC) is the main floral repressor in Arabidopsis seedlings (Michaels and Amasino, 1999) and is a key target of both endogenous and environmental signaling pathways (Sheldon et al., 2000; Michaels and Amasino, 2001). During vegetative development FLC directly represses the transcription of the flowering time integrators FLOWERING LOCUS T (FT), FLOWERING LOCUS D (FD) and SOC1, which promote the transition to flowering (Michaels and Amasino, 2001). FT protein is produced in the rosette leaves and moves to the SAM, where it interacts with the FD protein. The FT-FD complex then induces the floral transition by activating the expression of TF genes such as SOC1, LEAFY (LFY) and APETALA1 (AP1), which confer floral meristem (FM) identity on the primordia that form on the flanks of the primary SAM (Irish, 2010).
In Arabidopsis, chromatin modifications at key regulatory loci such as FLC, SOC1 and FT are crucial to the timing of the floral transition, and a number of trxG factors are involved in these processes (Figure 1C). Because the role of epigenetic factors in FLC regulation during vernalization has been extensively reviewed (Jarillo and Pineiro, 2011; Andres and Coupland, 2012; He, 2012; Hepworth and Dean, 2015), I will focus here on the control of flowering through other pathways. To prevent premature flowering during vegetative growth, the FLC locus is maintained in a transcriptionally active state marked by H3K36 tri-methylation (Zhao et al., 2005; Yang et al., 2014), which inhibits accumulation of H3K27me3 repressive marks (Shafiq et al., 2014). SDG8 is the major H3K36me3 methyltransferase in Arabidopsis (Yang et al., 2014) and is required for H3K36me3 deposition at the FLC locus (Shafiq et al., 2014; Yang et al., 2014). SDG8 associates with components of the transcription machinery, including RNA Pol II and PAF1, as well as with the H3K27 demethylase EARLY FLOWERING 6 (ELF6) (Yang et al., 2016). These physical associations (Figure 2) couple removal of repressive histone marks with deposition of active marks and transcription initiation/elongation to sustain high levels of FLC expression. ATXR7/SDG25 also represses the floral transition by binding to the FLC promoter and augmenting both H3K36me3 and H3K4me3 accumulation (Berr et al., 2009; Tamada et al., 2009), but whether SDG8 and ATXR7 function in concert to induce FLC transcription is unknown.
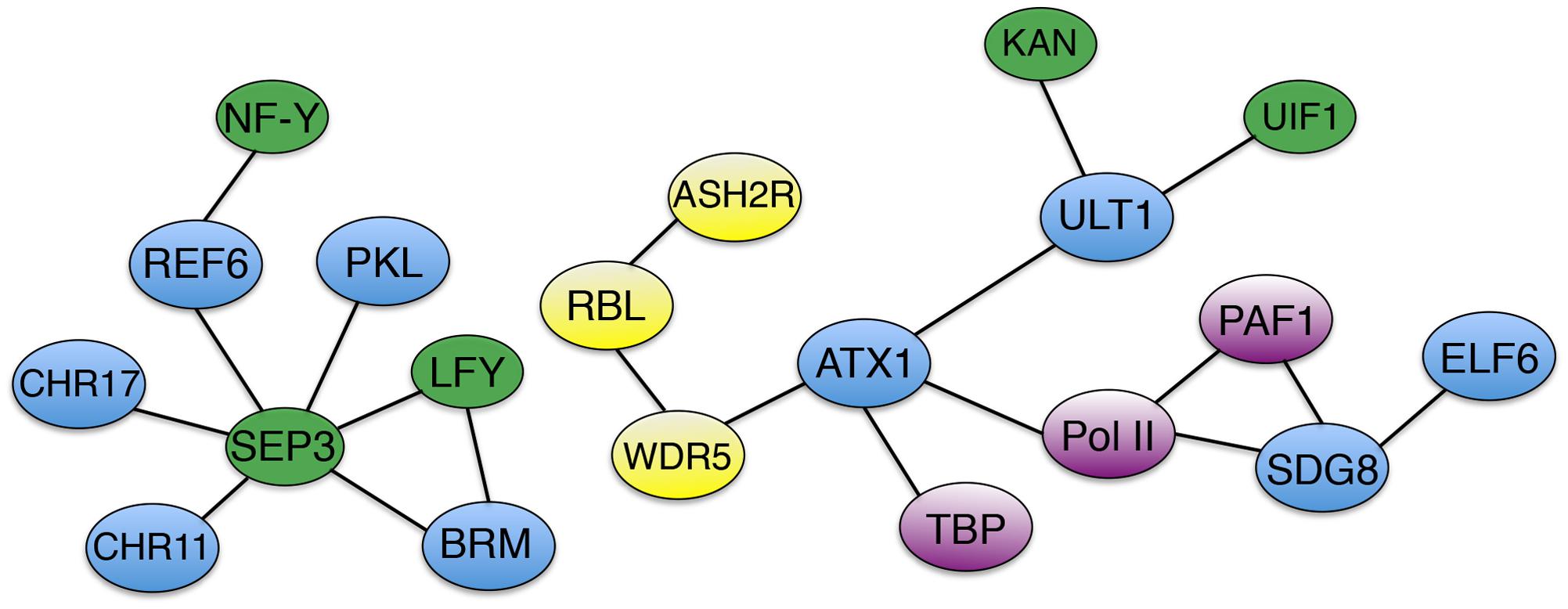
FIGURE 2. Association networks of trxG factors and interacting proteins. Solid bars designate physical associations between trxG factors (blue), transcription factors (green), AtCOMPASS components (yellow), and transcription machinery components and accessory proteins (purple).
ATXR7/SDG25 does act independently of the H3K4me3 methyltransferase ATX1 in repressing the flowering transition (Tamada et al., 2009), although ATX1 also directly binds to the FLC locus to deposit H3K4me3 and elevate expression of the floral repressor gene (Pien et al., 2008). ATX1 physically associates with WDR5a (Figure 2), a core component of the Arabidopsis COMPASS-like complex (AtCOMPASS) that also binds the FLC locus and promotes its expression by elevating H3K4me3 levels (Jiang et al., 2009). Two other core AtCOMPASS components, ASH2R and RbBP5-LIKE (RBL), also repress the floral transition by enhancing FLC expression, with ASH2R availability apparently being the rate-limiting factor in H3K4 tri-methylation at FLC and other target loci (Jiang et al., 2011). Therefore both H3K36me3 and H3K4me3 contribute to the maintenance of active FLC transcription in Arabidopsis.
The chromatin remodeling ATPase BRM prevents premature flowering by regulating FLC and FLC-related gene expression (Farrona et al., 2011; Li et al., 2015). Loss of function brm seedlings exhibit elevated H3K4me3 levels and reduced H3K27me3 levels in the FLC promoter, indicating that BRM imposes a repressive chromatin configuration at the FLC locus (Farrona et al., 2011). In addition, BRM directly activates the expression of the FLC-related MADS box TF gene SHORT VEGETATIVE PHASE (SVP) (Li et al., 2015). SVP forms a complex with FLC to repress flowering under non-inductive conditions (Fujiwara et al., 2008; Li et al., 2008). SVP expression is regulated by the AP, GA and temperature pathways, and directly represses SOC1 and FT transcription (Li et al., 2008). BRM represses flowering largely by inducing SVP transcription in vegetative tissues, binding to the SVP locus where it limits H3K27me3 accumulation by restricting CLF occupancy and activity (Li et al., 2015). Thus the early flowering phenotype of brm mutants can be accounted for by a reduction in SVP mRNA levels leading to lower abundance of the FLC-SVP repressor complex, resulting in elevated FT transcript levels that induce precocious flowering.
Repression of FLC transcription is crucial for the transition from the vegetative to the reproductive state. The JmjC domain-containing H3K27 demethylase REF6/JMJ12 promotes flowering independently of photoperiod by repressing FLC transcription (Noh et al., 2004). Because REF6 acts antagonistically to CLF by removing repressive H3K27me2/3 marks (Lu et al., 2011), binding its target genes in a sequence-specific fashion via its C2H2 zinc-finger domains (Cui et al., 2016) and facilitating recruitment of BRM (Li et al., 2016), the repression of FLC by REF6 is likely to be indirect (Yang et al., 2016). REF6 also induces transcription of the floral activator genes SOC1 and FT in an FLC-independent fashion (Noh et al., 2004; Lu et al., 2011). It is recruited by the nuclear factor Y (NF-Y) transcription factor complex to demethylate the SOC1 locus in response to the photoperiod and GA pathways (Hou et al., 2014), indicating that REF6 is a component of both endogenous and environmental signaling modules. Like REF6, the SDG26 histone methyltransferase also binds to and induces SOC1 transcription, augmenting the deposition of both H3K4me3 and H3K36me3 at the locus to promote the floral transition (Berr et al., 2015).
Finally, two methyltransferases that accelerate the floral transition independent of photoperiod have recently been characterized in rice. SDG708 encodes a methyltransferase that deposits up to three methyl groups on H3K36, and promotes flowering by catalyzing H3K36 methylation at the key flowering time regulatory genes H3Da and RFT1, which are closely related homologs of Arabidopsis FT, and Ehd1 (Liu et al., 2016). SDG701 encodes an H3K4 di- and tri-methyltransferase that likewise promotes flowering by depositing H3K4me3 to elevate the expression of H3Da and RFT1 (Liu et al., 2017).
Roles of trxG Factors in Patterning the Floral Meristem
Flowering signals induce reproductive development in plants by reprogramming the SAM into an inflorescence meristem (IM) that produces floral meristems (FMs) instead of leaves. A small suite of floral homeotic transcription factor genes then specifies the identity of each floral organ – sepals, petals, stamens, and carpels – from the outside to the inside of the flower (Coen and Meyerowitz, 1991). The activation of the floral homeotic genes at the onset of flower patterning requires counteracting the PcG-mediated repressive state that has persisted throughout vegetative development (Pu and Sung, 2015), an activity that is associated with increases in H3K4me3 levels at PcG target genes (Engelhorn et al., 2017) and involves multiple trxG factors.
The plant specific TF LFY and the MADS domain TF SEPALLATA3 (SEP3) play crucial roles in activating the expression of MADS box-containing floral homeotic genes such as APETALA3 (AP3) and AGAMOUS (AG) that specify petal, stamen, and carpel identity in the developing flower (Weigel and Meyerowitz, 1993; Pelaz et al., 2000). SYD and BRM physically associate with the LFY and SEP3 proteins (Figure 2), which recruit SYD to the AP3 and AG loci (Wu et al., 2012). At the onset of flower patterning, SYD and BRM redundantly regulate floral organ identity specification (Wagner and Meyerowitz, 2002; Farrona et al., 2004) (Figure 1D) by activating AP3 and AG expression, antagonizing CLF activity at the two loci by reducing H3K27me3 deposition and enhancing H3K4me3 deposition (Wu et al., 2012). ATX1 and SDG8 also specify floral organ identity by maintaining floral homeotic gene expression levels (Alvarez-Venegas et al., 2003; Grini et al., 2009), although the mechanistic details are as yet unknown. Finally, REF6 and PKL as well as two ISWI-type chromatin remodelers, CHR11 and CHR17, occur in floral MADS domain protein complexes and affect floral organ morphogenesis (Smaczniak et al., 2012). SEP3 and several other floral homeotic TFs bind their target genes prior to detectable increases in DNA accessibility (Pajaro et al., 2014), suggesting that they work closely with epigenetic factors to facilitate transcription initiation during early flower development by modulating chromatin accessibility at target loci.
The ULT1 trxG protein induces AG transcription in the center of the FM at stage 3 of flower formation (Alvarez-Venegas et al., 2003; Carles and Fletcher, 2009), binding directly to the locus to limit CLF-mediated H3K27me3 deposition and enhance H3K4me3 deposition (Carles and Fletcher, 2009). ULT1 thus sets the timing of the transition of the AG locus from a repressed to an active state, helping trigger a molecular pathway that ultimately terminates FM activity (Carles and Fletcher, 2009; Engelhorn et al., 2014b; Cao et al., 2015; Sun and Ito, 2015). Mutations in the domesticated transposase gene ANTAGONIST OF LIKE HETEROCHROMATIN PROTEIN1 (ALP1) enhance ult1 FM phenotypes, and ALP1 promotes floral organ identity gene expression in the absence of LFY (Liang et al., 2015). ALP1 antagonizes CLF function, acting genetically as a trxG factor, and is required for the activity of PcG target genes such as AP3 and AG. Notably, the ALP1 protein complex lacks known trxG factors but consists of core components of PRC2 and accessory factors such as EMF1 and LHP1. ALP1 is therefore proposed to antagonize PcG activity by blocking the interaction between PRC2 and accessory factors that stimulate its activity (Liang et al., 2015).
Conclusion
A variety of trxG factors exist in plants that carry out diverse biochemical activities to promote active gene expression states: chromatin-remodeling ATPases, histone methyltransferases, AtCOMPASS core components, histone demethylases, as well as DNA-binding and accessory proteins (Table 1). It is becoming clear from recent research that members of all of these categories of proteins play important roles in regulating landmark post-embryonic developmental processes such as meristem maintenance and floral induction. Moreover some trxG factors, such as BRM and ATX1, mediate multiple developmental processes during plant growth whereas others appear to have more restricted roles. The coupling of a core set of reiteratively used trxG components together with stage-, tissue- or process-specific trxG components may provide a flexible mechanism for tailoring the basic process of transcription activation to discrete gene networks in response to changing endogenous and environmental signals during the life cycle.
Although significant progress has been made in determining the roles of trxG factors in plant development, many gaps in our understanding remain. It still remains to be determined how many trxG complexes exist in plants, not to mention their full composition and whether that composition is static or changes depending on the developmental stage or tissue. The DNA binding proteins that recruit trxG factors to developmental regulatory loci are only beginning to be identified, while elucidating the chromatin signatures of plant stem cell populations can provide a valuable starting point for determining how tissue- and stage-specific epigenetic states are ultimately achieved during development. Finally, much work remains to decipher how developmental switches between trxG and PcG activities are implemented at individual loci as well as broadly across the genome to coordinate widespread transcriptional reprogramming. Further investigation in these areas will provide a more complete picture of how plants are able to maintain and as necessary adjust their gene expression programs during development in response to a wealth of endogenous and environmental cues.
Author Contributions
The author confirms being the sole contributor of this work and approved it for publication.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author apologizes for references omitted due to space limitations. This work was supported by research grants from the USDA (CRIS 2030-21000-041-00D) and the NSF (IOS-105-2050).
References
Abramson, J., Giraud, M., Benoist, C., and Mathis, D. (2010). Aire’s partners in the molecular control of immunological tolerance. Cell 140, 123–145. doi: 10.1016/j.cell.2009.12.030
Abramson, J., and Goldfarb, Y. (2016). AIRE: from promiscuous molecular partnerships to promiscuous gene expression. Eur. J. Biochem. 46, 22–33. doi: 10.1002/eji.201545792
Aichinger, E., Villar, C. B., Di Mambro, R., Sabatini, S., and Kohler, C. (2011). The CHD3 chromatin remodeler PICKLE and Polycomb Group proteins antagonistically regulate meristem activity in the Arabidopsis root. Plant Cell 23, 1047–1060. doi: 10.1105/tpc.111.083352
Aida, M., Beis, D., Heidstra, R., Willemsen, V., Blilou, I., Galinha, C., et al. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119, 109–120. doi: 10.1016/j.cell.2004.09.018
Alvarez-Venegas, R., and Avramova, Z. (2005). Methylation patterns of histone H3 Lys 4, Lys 9 and Lys 27 in transcriptionally active and inactive Arabidopsis genes and in atx1 mutants. Nucleic Acids Res. 33, 5199–5207. doi: 10.1093/nar/gki830
Alvarez-Venegas, R., Pien, S., Sadder, M., Witmer, X., Grossniklaus, U., and Avramova, Z. (2003). ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr. Biol. 13, 627–637. doi: 10.1016/S0960-9822(03)00243-4
Amasino, R. (2010). Seasonal and developmental timing of flowering. Plant J. 61, 1001–1013. doi: 10.1111/j.1365-313X.2010.04148.x
Andres, F., and Coupland, G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639. doi: 10.1038/nrg3291
Bernstein, B. E., Mikkelsen, T. S., Xie, X., Kamal, M., Huebert, D. J., Cuff, J., et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326. doi: 10.1016/j.cell.2006.02.041
Berr, A., Shafiq, S., Pinon, V., Dong, A., and Shen, W.-H. (2015). The trxG family histone methyltransferase SET DOMAIN GROUP 26 promotes flowering via a distinctive genetic pathway. Plant J. 81, 316–328. doi: 10.1111/tpj.12729
Berr, A., Xu, L., Gao, J., Cognat, V., Steinmetz, A., Dong, A., et al. (2009). SET DOMAIN GROUP 25 encodes a histone methyltransferase and is involved in FLC activation and repression of flowering. Plant Physiol. 151, 1476–1485. doi: 10.1104/pp.109.143941
Blilou, I., Xu, J., Wildwater, M., Willemsen, V., Paponov, I., Friml, J., et al. (2005). The PIN auxin efflux carrier facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44. doi: 10.1038/nature03184
Brand, U., Fletcher, J. C., Hobe, M., Meyerowitz, E. M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619. doi: 10.1126/science.289.5479.617
Cao, X., He, Z., Guo, L., and Liu, X. (2015). Epigenetic mechanisms are critical for the regulation of WUSCHEL in floral meristems. Plant Physiol. 168, 1189–1196. doi: 10.1104/pp.15.00230
Carles, C. C., Choffnes-Inada, D., Reville, K., Lertpiriyapong, K., and Fletcher, J. C. (2005). ULTRAPETALA1 encodes a putative SAND domain transcription factor that controls shoot and floral meristem activity in Arabidopsis. Development 132, 897–911. doi: 10.1242/dev.01642
Carles, C. C., and Fletcher, J. C. (2009). The SAND domain protein ULTRAPETALA1 acts as a trithorax group factor to regulate cell fate in plants. Genes Dev. 23, 2723–2728. doi: 10.1101/gad.1812609
Carles, C. C., and Fletcher, J. C. (2010). Missing links between histones and RNA Pol II arising from SAND? Epigenetics 5, 381–385. doi: 10.4161/epi.5.5.11956
Carles, C. C., Lertpiriyapong, K., Reville, K., and Fletcher, J. C. (2004). The ULTRAPETALA1 gene functions early in Arabidopsis development to restrict shoot apical meristem activity and acts through WUSCHEL to regulate floral meristem determinacy. Genetics 167, 1893–1903. doi: 10.1534/genetics.104.028787
Carter, B., Henderson, J. T., Svedin, E., Fiers, M., McCarthy, K., Smith, A., et al. (2016). Cross-talk between sporophyte and gametophyte generations is provided by CHD3 chromatin remodelers in Arabidopsis thaliana. Genetics 203, 817–829. doi: 10.1534/genetics.115.180141
Chen, L.-Q., Luo, J.-H., Cui, Z.-H., Xue, M., Wang, L., Zhang, X.-Y., et al. (2017). ATX3, ATX4 and ATX5 encode putative H3K4 methyltransferases and are critical for plant development. Plant Physiol. 174, 1795–1806. doi: 10.1104/pp.16.01944
Coen, E. S., and Meyerowitz, E. M. (1991). The war of the whorls - genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1038/353031a0
Cui, X., Lu, F., Qiu, Q., Zhou, B., Lu, G., Zhang, S., et al. (2016). REF6 recognizes a specific DNA sequence to demethylate H3K27me3 and regulate organ boundary formation in Arabidopsis. Nat. Genet. 48, 694–699. doi: 10.1038/ng.3556
Ding, Y., Avramova, Z., and Fromm, M. H. (2011). Two distinct roles of ARABIDOPSIS HOMOLOG OF TRITHORAX1 (ATX1) at promoters and within transcribed regions of ATX1-related genes. Plant Cell 23, 350–363. doi: 10.1105/tpc.110.080150
Ding, Y., Ndamukong, I., Xu, Z., Lapko, H., Fromm, M. H., and Avramova, Z. (2012). ATX1-generated H3K4me3 is required for efficient elongation of transcription, not initiation, at ATX1-regulated genes. PLOS Genet. 8:e1003111. doi: 10.1371/journal.pgen.1003111
Engelhorn, J., Blanvillain, R., and Carles, C. C. (2014a). Gene activation and cell fate control in plants: a chromatin perspective. Cell Mol. Life Sci. 71, 3119–3137. doi: 10.1007/s00018-014-1609-0
Engelhorn, J., Blanvillain, R., Kroner, C., Parrinello, H., Rohmer, M., Pose, D., et al. (2017). Dynamics of H3K4me3 chromatin marks prevails over H3K27me3 for gene regulation during flower morphogenesis in Arabidopsis thaliana. Epigenomes 1:8. doi: 10.3390/epigenomes1020008
Engelhorn, J., Moreau, F., Fletcher, J. C., and Carles, C. C. (2014b). ULTRAPETALA1 and LEAFY pathways function independently in specifying identity and determinacy at the Arabidopsis floral meristem. Ann. Bot. 114, 1497–1505. doi: 10.1093/aob/mcu185
Farrona, S., Hurtado, L., Bowman, J. L., and Reyes, J. L. (2004). The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development 131, 4965–4975. doi: 10.1241/dev.01363
Farrona, S., Hurtado, L., March-Diaz, R., Schmitz, R. J., Florencio, F. J., Turck, F., et al. (2011). Brahma is required for proper expression of the floral repressor FLC in Arabidopsis. PLOS ONE 6:e17997. doi: 10.1371/journal.pone.0017997
Fiume, E., Pires, H. R., Kim, J. S., and Fletcher, J. C. (2010). Analyzing floral meristem development. Methods Plant Dev. Biol. 655, 131–142. doi: 10.1007/978-1-60761-765-5_9
Fletcher, J. C., Brand, U., Running, M. P., Simon, R., and Meyerowitz, E. M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914. doi: 10.1126/science.283.5409.1911
Fujiwara, S., Oda, A., Yoshida, R., Niinuma, K., Miyata, K., Tomozoe, Y., et al. (2008). Circadian clock proteins LHY and CCA1 regulate SVP protein accumulation to control flowering in Arabidopsis. Plant Cell 20, 2960–2971. doi: 10.1105/tpc.108.061531
Gentry, M., and Hennig, L. (2014). Remodelling chromatin to shape development of plants. Exp. Cell Res. 321, 40–46. doi: 10.1016/j.yexcr.2013.11.010
Grieneisen, V. A., Xu, J., Maree, A. F. M., Hogeweg, P., and Scheres, B. (2007). Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449, 1008–1013. doi: 10.1038/nature06215
Grini, P. E., Thorstensen, T., Alm, V., Vizcay-Barrena, G., Windju, S. S., Jorstad, T. S., et al. (2009). The ASH1 HOMOLOG 2 (ASHH2) histone H3 methyltransferase is required for ovule and anther development in Arabidopsis. PLOS ONE 4:e7817. doi: 10.1371/journal.pone.0007817
Guo, L., Yu, Y., Law, J. A., and Zhang, X. (2010). SET DOMAIN GROUP2 is the major histone H3 lysine 4 trimethyltransferase in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 107, 18557–18562. doi: 10.1073/pnas.1010478107
He, Y. (2012). Chromatin regulation of flowering. Trends Plant Sci. 17, 556–562. doi: 10.1016/j.tplants.2012.05.001
Hepworth, J., and Dean, C. (2015). Flowering Locus C’s lessons: conserved chromatin switches underpinning developmental timing and adaptation. Plant Physiol. 168, 1237–1245. doi: 10.1104/pp.15.00496
Hou, X., Zhou, J., Liu, C., Liu, L., Shen, L., and Yu, H. (2014). Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat. Commun. 5:4601. doi: 10.1038/ncomms5601
Ingham, P. W. (1988). The molecular genetics of embryonic pattern formation in Drosophila. Nature 335, 25–34. doi: 10.1038/335025a0
Irish, V. (2010). The flowering of Arabidopsis flower development. Plant J. 61, 1014–1028. doi: 10.1111/j.1365-313X.2009.04065.x
Jarillo, J. A., and Pineiro, M. (2011). Timing is everything in plant development. The central role of floral repressors. Plant Sci. 181, 364–378. doi: 10.1016/j.plantsci.2011.06.011
Jiang, D., Gu, X., and He, Y. (2009). Establishment of the winter-annual growth habit via FRIGIDA-mediated histone methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell 21, 1733–1746. doi: 10.1105/tpc.109.067967
Jiang, D., Kong, N. C., Gu, X., Li, Z., and He, Y. (2011). Arabidopsis COMPASS-like complexes mediate histone H3 lysine-4 trimethylation to control floral transition and plant development. PLOS Genet. 7:e1001330. doi: 10.1371/journal.pgen.1001330
Kennison, J. A. (1995). The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu. Rev. Genet. 29, 289–303. doi: 10.1146/annurev.ge.29.120195.001445
Kwon, C. S., Chen, C., and Wagner, D. (2005). WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev. 19, 992–1003. doi: 10.1101/gad.1276305
Laux, T., Mayer, K. F. X., Berger, J., and Jurgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96.
Li, C., Chen, C., Gao, L., Yang, S., Nguyen, V., Shi, X., et al. (2015). The Arabidopsis SWI2/SNF2 chromatin remodeler BRAHMA regulates Polycomb function during vegetative development and directly activates the flowering repressor gene SVP. PLOS Genet. 11:e1004944. doi: 10.1371/journal.pgen.1004944
Li, C., Gu, L., Gao, L., Wei, C.-Q., Chien, C.-W., Wang, S., et al. (2016). Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat. Genet. 48, 687–693. doi: 10.1038/ng.3555
Li, D., Liu, C., SHen, L., Wu, Y., Chen, H., Robertson, M., et al. (2008). A repressor complex governs the integration of flowering signals in Arabidopsis. Dev. Cell 15, 110–120. doi: 10.1016/j.devcel.2008.05.002
Liang, S. C., Hartwig, B., Perera, P., Mora-Garcia, S., de Leau, E., Thornton, H., et al. (2015). Kicking against the PRCs - a domesticated transposase antagonises silencing mediated by Polycomb Group proteins and is an accessory component of Polycomb Repressive Complex 2. PLOS Genet. 11:e1005812. doi: 10.1371/journal.pgen.1005660
Liu, B., Wei, G., Shi, J., Jin, J., Shen, T., Ni, T., et al. (2016). SET DOMAIN GROUP 708, a histone H3 lysine 36-specific methyltransferase, controls flowering time in rice (Oryza sativa). New Phytol. 210, 577–588. doi: 10.1111/nph.13768
Liu, K., Yu, Y., Dong, A., and Shen, W.-H. (2017). SET DOMAIN GROUP701 encodes a H3K4-methyltransferase and regulates multiple key process of rice plant development. New Phytol. 215, 609–623. doi: 10.1111/nph.14596
Liu, X., Zhou, S., Wang, W., Ye, Y., Zhao, Y., Xu, Q., et al. (2015). Regulation of histone methylation and reprogramming of gene expression in the rice inflorescence meristem. Plant Cell 27, 1428–1444. doi: 10.1105/tpc.15.00201
Lu, F., Cui, X., Zhang, Z., Jenuwein, T., and Cao, X. (2011). Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 43, 715–719. doi: 10.1038/ng.854
Michaels, S. D., and Amasino, R. M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. doi: 10.1105/tpc.11.5.949
Michaels, S. D., and Amasino, R. M. (2001). Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13, 935–941. doi: 10.1105/tpc.13.4.935
Moreau, F., Thevenon, E., Blanvillain, R., Lopez-Vidriero, I., Franco-Zorrilla, J. M., Dumas, R., et al. (2016). The Myb-domain protein ULTRAPETALA1 INTERACTING FACTOR 1 controls floral meristem activities in Arabidopsis. Development 143, 1108–1119. doi: 10.1242/dev.127365
Napsucialy-Mendivil, S., Alvarez-Venegas, R., Shishkova, S., and Dubrovsky, J. G. (2014). ARABIDOPSIS HOMOLOG of TRITHORAX1 (ATX1) is required for cell production, patterning, and morphogenesis in root development. J. Exp. Bot. 65, 6373–6384. doi: 10.1093/jxb/eru355
Noh, B., Lee, S.-H., Kim, H.-J., Shin, E.-A., Lee, M., Jung, K.-J., et al. (2004). Divergent roles of a pair of homologous Jumonji/Zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16, 2601–2613. doi: 10.1105/tpc.104.025353
Pajaro, A., Madrigal, P., Muino, J. M., Matus, J. T., Jin, J., Mecchia, M. A., et al. (2014). Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. 15:R41. doi: 10.1186/gb-2014-15-3-r41
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E., and Yanofsky, M. F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. doi: 10.1038/35012103
Pien, S., Fleury, D., Mylne, J. S., Crevillen, P., Inze, D., Avramova, Z., et al. (2008). ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. Plant Cell 20, 580–588. doi: 10.1105/tpc.108.058172
Pires, H. R., Monfared, M. M., Shemyakina, E. A., and Fletcher, J. C. (2014). ULTRAPETALA trxG genes interact with KANADI transcription factor genes to regulate Arabidopsis gynoecium patterning. Plant Cell 26, 4345–4361. doi: 10.1105/tpc.114.131250
Piunti, A., and Shilatifard, A. (2016). Epigenetic balance of gene expression by Polycomb and COMPASS families. Science 352:aad9780. doi: 10.1126/science.aad9780
Pu, L., Liu, M.-S., Kim, S. Y., Chen, L.-F. O., Fletcher, J. C., and Sung, Z. R. (2013). EMBRYONIC FLOWER1 and ULTRAPETALA1 act antagonistically on Arabidopsis development and stress response. Plant Physiol. 162, 812–830. doi: 10.1104/pp.112.213223
Pu, L., and Sung, Z. R. (2015). PcG and trxG in plants - friends or foes. Trends Genet. 31, 252–263. doi: 10.1016/j.tig.2015.03.004
Rojo, E., Sharma, V. K., Kovaleva, V., Raikhel, N. V., and Fletcher, J. C. (2002). CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell 14, 969–977. doi: 10.1105/tpc.002196
Schwartz, Y. B., and Pirrota, V. (2007). Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 8, 9–22. doi: 10.1038/nrg1981
Shafiq, S., Berr, A., and Shen, W.-H. (2014). Combinatorial functions of diverse histone methylations in Arabidopsis thaliana flowering time. New Phytol. 201, 312–322. doi: 10.1111/nph.12493
Sheldon, C. C., Rouse, D. T., Finnegan, E. J., Peacock, W. J., and Dennis, E. S. (2000). The molecular basis of vernalization: the central role of FLOWERING LOCUS C. Proc. Natl. Acad. Sci. U.S.A. 97, 3753–3758. doi: 10.1073/pnas.97.7.3753
Smaczniak, C., Immink, R. G. H., Muino, J. M., Blanvillain, R., Busscher, M., Busscher-Lange, J., et al. (2012). Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. U.S.A. 109, 1560–1565. doi: 10.1073/pnas.1112871109
Spivakov, M., and Fisher, A. G. (2007). Epigenetic signatures of stem-cell identity. Nat. Rev. Genet. 8, 263–271. doi: 10.1038/nrg2046
Srikanth, A., and Schmid, M. (2011). Regulation of flowering time: all roads lead to Rome. Cell. Mol. Life Sci. 68, 2013–2037. doi: 10.1007/s00018-011-0673-y
Steeves, T. A., and Sussex, I. M. (1989). Patterns in Plant Development. New York, NY: Cambridge University Press. doi: 10.1017/CBO9780511626227
Sun, B., and Ito, T. (2015). Regulation of floral stem cell termination in Arabidopsis. Front. Plant Sci. 6:17. doi: 10.3389/fpls.2015.00017
Tamada, Y., Yun, J.-Y., Woo, S. C., and Amasino, R. (2009). ARABIDOPSIS TRITHORAX-RELATED7 is required for methylation of lysine 4 of histone H3 and for transcriptional activation of FLOWERING LOCUS C. Plant Cell 21, 3257–3269. doi: 10.1105/tpc.109.070060
van den Berg, C., Willemsen, V., Hendriks, G., Weisbeek, P., and Scheres, B. (1997). Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390, 287–289. doi: 10.1038/36856
Wagner, D., and Meyerowitz, E. M. (2002). SPLAYED, a novel SWI/SNF ATPase homolog, controls reproductive development in Arabidopsis. Curr. Biol. 12, 85–94. doi: 10.1016/S0960-9822(01)00651-0
Weigel, D., and Meyerowitz, E. M. (1993). Activation of floral homeotic genes in Arabidopsis. Science 261, 1723–1726. doi: 10.1126/science.261.5129.1723
Wu, M.-F., Sang, Y., Bezhani, S., Yamaguchi, N., Han, S.-K., Li, Z., et al. (2012). SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc. Natl. Acad. Sci. U.S.A. 109, 3576–3581. doi: 10.1073/pnas.1113409109
Xiao, J., Lee, U.-S., and Wagner, D. (2016). Tug of war: adding and removing histone lysine methylation in Arabidopsis. Curr. Opin. Plant Biol. 34, 41–53. doi: 10.1016/j.pbi.2016.08.002
Yang, H., Howard, M., and Dean, C. (2014). Antagonistic roles for H3K36me3 and H3K27me3 in the cold-induced epigenetic switch at Arabidopsis FLC. Curr. Biol. 24, 1793–1797. doi: 10.1016/j.cub.2014.06.047
Yang, H., Howard, M., and Dean, C. (2016). Physical coupling of activation and derepression activities to maintain an active transcriptional state at FLC. Proc. Natl. Acad. Sci. U.S.A. 113, 9369–9374. doi: 10.1073/pnas.1605733113
Yang, S., Li, C., Zhao, L., Gao, S., Lu, J., Zhao, M., et al. (2015). The Arabidopsis SWI2/SNF2 chromatin remodeling ATPase BRAHMA targets directly to PINs and is required for root stem cell niche maintenance. Plant Cell 27, 1670–1680. doi: 10.1105/tpc.15.00091
Yao, X., Feng, H., Yu, Y., Dong, A., and Shen, W.-H. (2013). SDG2-mediated H3K4 methylation is required for proper Arabidopsis root growth and development. PLOS ONE 8:e56537. doi: 10.1371/journal.pone.0056537
Keywords: trxG, PcG, development, chromatin, histone methylation, transcription, epigenetics, Arabidopsis
Citation: Fletcher JC (2017) State of the Art: trxG Factor Regulation of Post-embryonic Plant Development. Front. Plant Sci. 8:1925. doi: 10.3389/fpls.2017.01925
Received: 25 September 2017; Accepted: 24 October 2017;
Published: 14 November 2017.
Edited by:
Stefan A. Rensing, Philipps University of Marburg, GermanyReviewed by:
Isabel Bäurle, University of Potsdam, GermanyKeqiang Wu, National Taiwan University, Taiwan
Copyright © 2017 Fletcher. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jennifer C. Fletcher, amZsZXRjaGVyQGJlcmtlbGV5LmVkdQ==
 Jennifer C. Fletcher
Jennifer C. Fletcher