- Department of Botany, University of British Columbia, Vancouver, BC, Canada
Salicylic acid (SA) is a key signaling molecule in plant immunity. Two types of SA receptors, NPR1 and NPR3/NPR4, were reported to be involved in the perception of SA in Arabidopsis. SA is also synthesized in the non-vascular moss Physcomitrella patens following pathogen infection. Sequence analysis revealed that there is only one NPR1/NPR3/NPR4-like protein in P. patens. This agrees with the phylogenetic study that showed the divergence of NPR1 and NPR3/NPR4 from the same ancestor during the evolution of higher plants. Intriguingly, expression of the P. patens NPR1/NPR3/NPR4-like gene in Arabidopsis does not complement the constitutive defense phenotype of the npr3 npr4 double mutant, but can partially rescue the mutant phenotypes of npr1-1, suggesting that it functions as an NPR1-like positive regulator of SA-mediated immunity and P. patens does not have an SA receptor functioning similarly as NPR3/NPR4. Future characterization of the P. patens NPR1-like protein and analysis of its functions through knockout and biochemical approaches will help us better understand how SA is perceived and what its functions are in P. patens.
Systemic acquired resistance (SAR) is a plant immune response induced after primary infection, which leads to enhanced resistance to a broad spectrum of pathogens in distal tissue (Durrant and Dong, 2004). Early studies of SAR identified increased accumulation of salicylic acid (SA) and induction of PATHOGENESIS RELATED (PR) gene expression as two key features of SAR (Malamy et al., 1990; Metraux et al., 1990; Rasmussen et al., 1991; Ward et al., 1991; Yalpani et al., 1991; Uknes et al., 1992). Clear evidence showed that SA accumulation is necessary for SAR, as blocking SA accumulation by expressing a bacterial salicylate hydroxylase in transgenic plants results in low SA levels and loss of SAR (Gaffney et al., 1993). In addition, two SA-deficient mutants, sid2 and eds5, display enhanced susceptibility to pathogens and compromised SAR (Nawrath and Metraux, 1999; Wildermuth et al., 2001). On the other hand, exogenous application of SA or its analogs is sufficient to induce PR gene expression and resistance to pathogen infection (White, 1979; Metraux et al., 1991; Gorlach et al., 1996). These studies suggest that SA is a key signaling molecule in plant immunity.
Several genetic screens conducted in Arabidopsis to identify signaling components downstream of SA led to the isolation of a large number of npr1 alleles (Cao et al., 1994; Delaney et al., 1995; Shah et al., 1997). In npr1 mutants, SA-induced PR gene expression and resistance to pathogens are blocked, suggesting that NPR1 is a key immune regulator downstream of SA. NPR1 encodes a protein containing two protein–protein interaction motifs: a BTB/POZ (broad-complex, tram track, and bric-a-brac/poxvirus, zinc finger) domain at the N-terminus and an ankyrin-repeat domain in the middle of the protein (Cao et al., 1997; Ryals et al., 1997). NPR1 binds to SA in vitro, suggesting that it may function as a receptor for SA (Wu et al., 2012; Manohar et al., 2014). Yeast two-hybrid screens identified several TGA transcription factors as interactors of NPR1 (Zhang et al., 1999; Despres et al., 2000; Zhou et al., 2000). TGAs bind to the SA-responsive element (as-1) in the PR1 promoter that are required for the induction of PR gene expression by SA, suggesting that they are important for regulating PR1 expression (Zhang et al., 1999). Knockout analysis of TGA2/TGA5/TGA6 further revealed that induction of PR1 expression and pathogen resistance by the SA analog INA was abolished in the tga2 tga5 tga6 triple mutant, but unaffected in tga6 or tga2 tga5 mutant plants, suggesting that TGA2/TGA5/TGA6 function redundantly as positive regulators of SA-induced defense responses (Zhang et al., 2003).
Two paralogs of NPR1, NPR3 and NPR4, also interact with TGA2/TGA5/TGA6, but they function as negative regulators of PR gene expression and pathogen resistance (Zhang et al., 2006). The npr3 npr4 double mutants exhibit elevated basal PR gene expression and enhanced resistance against pathogens. Similar to NPR1, NPR3 and NPR4 also bind to SA, suggesting that they also function as SA receptors (Fu et al., 2012). They were proposed to negatively regulate plant immunity by degrading NPR1 in response to SA (Fu et al., 2012).
Physcomitrella patens is the first bryophyte to have its whole genome sequenced and has been widely used in studying evolutionary changes during the evolution of land plants (Rensing et al., 2008). Few studies have been carried out on the immune system of P. patens. It was shown that SA levels in P. patens increase rapidly after inoculation with Botrytis cinerea (Ponce De Leon et al., 2012). Exogenous application of SA also strongly induces the expression of a PAL gene in P. patens (Ponce De Leon et al., 2012), suggesting that SA is synthesized and perceived during pathogen infection.
To better understand how SA is perceived in P. patens, we carried out BLAST searches to look for homologs of NPR1 in P. patens. Search of the NCBI database found that Pp3c19_7560 encodes an NPR1 homolog. Search of the JGI Phytozome database revealed that another predicted protein encoded by Pp3c21_7570 also has high similarity with Arabidopsis NPR1. To determine the actual coding sequences of these two NPR1-like genes in P. patens, we sequenced their cDNAs amplified by RT-PCR. Analysis of the cDNA sequences revealed that the predicted Pp3c19_7560 gene model is incorrect and there is an early frame shift in the cDNA, resulting in the truncation of the encoded protein, suggesting that Pp3c19_7560 is likely a pseudogene. The cDNA sequence of Pp3c21_7570 is also different from the predicted mRNA sequence. The first exon is larger than predicted.
The predicted Pp3c21_7570 protein based on the cDNA sequence shows 34% identity and 53% similarity with Arabidopsis NPR1, 37% identity and 55% similarity with NPR3, and 36% identity and 56% similarity with NPR4. Similar to NPR1 and NPR3/NPR4, it contains a conserved N-terminal BTB/POZ domain, a central ankyrin-repeat domain and a C-terminal domain with nuclear localization signals (Figure 1A). In Arabidopsis NPR1, Cys521, and Cys529 in the C-terminal domain were reported to be required for SA-binding, but they are not universally conserved in NPR1 orthologs (Wu et al., 2012). These two residues are not conserved in Pp3c21_7570 and Arabidopsis NPR3/NPR4 either. Most likely SA-binding in SA receptors involves in additional amino acid residues. Phylogenetic analysis of proteins in the NPR1 family from different plant species revealed that NPR1 and NPR3/NPR4 diverged from Pp3c21_7570 during evolution of higher plants (Figure 1B). It is not obvious whether Pp3c21_7570 is more related to Arabidopsis NPR1 or NPR3/NPR4 simply from the phylogenetic tree.
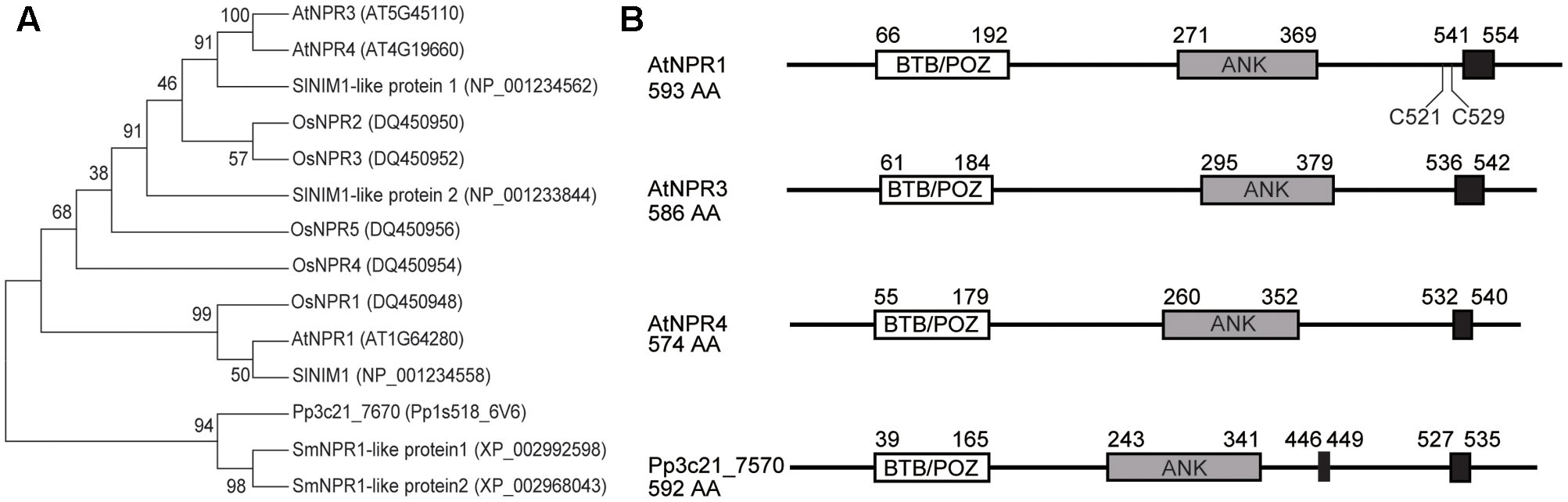
FIGURE 1. Pp3C21_7570 encodes an NPR1-like protein in Physcomitrella patens. (A) Phylogenetic analysis of Pp3C21_7570, Arabidopsis NPR1/NPR3/NPR4 and NPR1/NPR3/NPR4-like proteins in rice (OsNPR1, OsNPR2, OsNPR3, OsNPR4, and OsNPR5), tomato (SlNIM1, SlNIM1-like protein 1, and SlNIM1-like protein 2) and the lycophyte Selaginella moellendorffii (SmNPR1-like protein 1 and SmNPR1-like protein 2). The protein sequences were aligned by MEGA version 7.0.26, and the maximum likelihood tree was generated using the Maximum Likelihood method. Bootstrap replication (500 replications) was used for statistical support for the nodes in the phylogenetic tree. (B) Predicted protein structures of Arabidopsis NPR1/NPR3/NPR4 and Pp3C21_7570. BTB/POZ domain (white box), Ankyrin-repeat (gray box), and nuclear localization signals (black box) are shown.
To determine whether Pp3c21_7570 has similar functions as NPR3/4 or NPR1, we generated a construct expressing Pp3c21_7570 driven by the 35S promoter and transformed it into Arabidopsis npr3-2 npr4-2 and npr1-1 mutants to test whether it can complement the mutant phenotype of npr3-2 npr4-2 or npr1-1. As shown in Figure 2A, the 35S::Pp3c21_7570 transgenic plants in the npr3-2 npr4-2 double mutant background still exhibit elevated basal PR1 expression, and the expression levels of PR1 in the transgenic lines are actually higher than in npr3-2 npr4-2. Similarly, the enhanced resistance to H.a. Noco2 in npr3-2 npr4-2 is unaffected in the 35S::Pp3c21_7570 transgenic lines (Figure 2B). These data suggest that the 35S::Pp3c21_7570 transgene does not complement the mutant phenotypes of npr3-2 npr4-2.
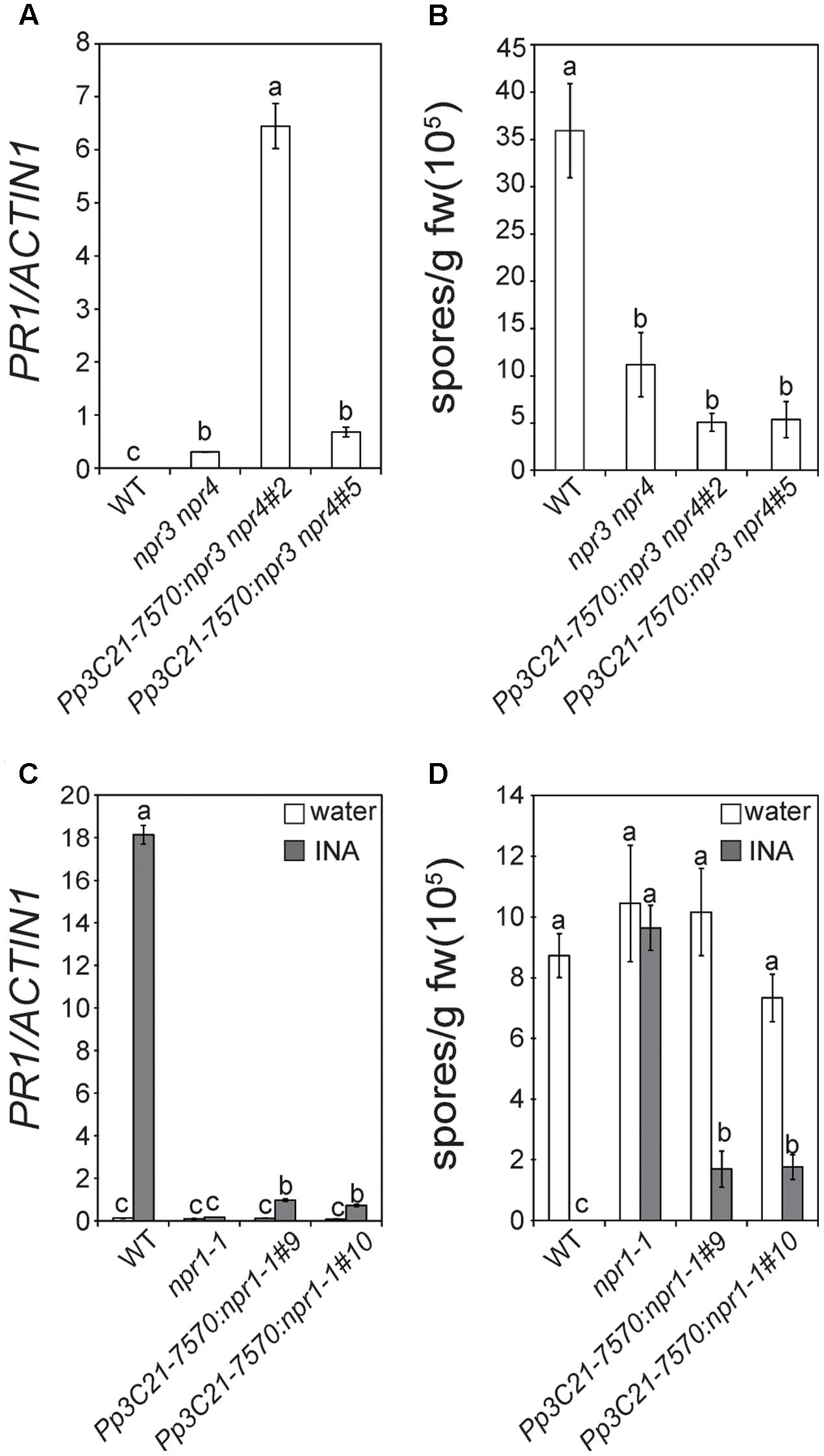
FIGURE 2. Characterization of transgenic plants expressing Pp3C21_7570 in the Arabidopsis npr3-2 npr4-2 and npr1-1 mutants. (A) Expression of PR1 in 2-week-old seedlings of wild type (WT), npr3-2 npr4-2 and 35S::Pp3C21_7570 transgenic lines in npr3-2 npr4 background (line #2 and #5). Statistical differences among the samples are labeled with different letters (P < 0.01, one-way ANOVA; n = 3). (B) Growth of H.a. Noco2 on WT, npr3-2 npr4-2 and 35S::Pp3C21_7570 transgenic plants in npr3-2 npr4-2 background (line #2 and #5). Two-week old seedlings were sprayed with H.a. Noco2 spores at a concentration of 50,000 spores/ml. Statistical differences among the samples are labeled with different letters (P < 0.01, one-way ANOVA; n = 4). (C) Expression level of PR1 in WT, npr1-1 and 35S::Pp3C21_7570 transgenic lines in npr1-1 background (line #9 and #10) before and after INA treatment. 11-day-old seedlings were sprayed with 0.33 mM INA. Samples were collected before treatment and 48 h after treatment. Statistical differences among the samples are labeled with different letters (P < 0.01, one-way ANOVA; n = 3). (D) Growth of H.a. Noco2 on WT, npr1-1 and 35S::Pp3C21_7570 transgenic lines in npr1-1 background (line #9 and #10). Twelve-day-old plants were sprayed with water or 0.65 mM INA 3 days before spraying with H.a. Noco2 spores at a concentration of 20,000 spores/ml. Statistical differences among the samples are labeled with different letters (P < 0.01, one-way ANOVA; n = 4). The coding region of Pp3C21_7570 was amplified by RT-PCR and cloned into a modified pCambia1300 vector with a 35S promoter. 35S::Pp3C21_7570 transgenic lines were generated by transforming Arabidopsis npr3-2, npr4-2, and npr1-1 mutant plants with Agrobacteria carrying the construct. Expression levels of PR1 in (A,C) were determined by quantitative RT-PCR. Values were normalized by the expression levels of Actin1. Quantitative analysis of H.a. Noco2 growth in (B,D) was carried out as previously described (Bi et al., 2010). After spraying with H.a. Noco2 spores, plants were grown at 18°C under 12 h day/12 h night cycle for 7 days before the conidiospores on the plants were quantified.
In the 35S::Pp3c21_7570 transgenic lines in the npr1-1 background, INA-induced PR1 expression is partially restored (Figure 2C). Similarly, INA-induced resistance to H.a. Noco2 is largely restored in the 35S::Pp3c21_7570 transgenic lines, although it is not as strong as in wild type (WT) plants (Figure 2D). These data suggest that Pp3c21_7570 can partially complement the mutant phenotypes of npr1-1. Thus Pp3c21_7570 is orthologous to Arabidopsis NPR1 and we named Pp3c21_7570 as PpNPR1.
The partial complementation of the Arabidopsis npr1 mutant phenotypes by PpNPR1 indicates that it functions as a positive regulator of SA-induced defense gene expression, suggesting that similar mechanisms are used to promote defense gene expression and pathogen resistance by SA in P. patens. Since there is only one NPR1-like protein in P. patens, it probably does not use NPR3/NPR4-like SA receptors to negatively regulate defense responses. As NPR3/NPR4 evolved from the same ancestor as NPR1 in higher plants, they probably diverged from NPR1 to take a different role in fine-tuning SA-induced defense responses. Whether P. patens has a simpler immune system which does not need negative regulators like NPR3/NPR4 or it uses an alternative approach to prevent auto-activation of defense responses in the absence of SA remains to be determined.
Bryophytes are remnants of early diverging lineages of embryophytes and occupy an ideal phylogenetic position for reconstruction of ancient evolutionary changes (Rensing et al., 2008). The identification of PpNPR1 as a NPR1-like protein provides a starting point for analyzing SA perception in the bryophyte P. patens. Future analysis of the potential SA-binding activity of PpNPR1 and characterization of the function of PpNPR1 by knockout analysis will help us to better understand its roles in SA perception and defense against pathogens. In Arabidopsis, NPR1 interacts with TGA transcription factors and regulates defense gene expression through TGA2/TGA5/TGA6 (Zhang et al., 1999, 2003; Despres et al., 2000; Zhou et al., 2000). It is likely that PpNPR1 also interacts with TGA transcription factors in P. patens. It will be interesting to determine which TGA transcription factors in P. patens interact with PpNPR1 and analyze their roles in SA-induced defense gene expression.
Author Contributions
YP performed the experiments and wrote the paper; TS performed the experiments; YZ designed the experiments and wrote the paper.
Funding
This research is funded by NSERC and CFI grants to YZ.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Rowan van Wersch and Xin Li (University of British Columbia) for discussion and editing of the manuscript. They are grateful for the financial support to YZ from Natural Sciences and Engineering Research Council (NSERC) of Canada and Canada Foundation for Innovation (CFI).
References
Bi, D., Cheng, Y. T., Li, X., and Zhang, Y. (2010). Activation of plant immune responses by a gain-of-function mutation in an atypical receptor-like kinase. Plant Physiol. 153, 1771–1779. doi: 10.1104/pp.110.158501
Cao, H., Bowling, S. A., Gordon, A. S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is non-responsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. doi: 10.1105/tpc.6.11.1583
Cao, H., Glazebrook, J., Clarke, J. D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. doi: 10.1016/S0092-8674(00)81858-9
Delaney, T. P., Friedrich, L., and Ryals, J. A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. U.S.A. 92, 6602–6606. doi: 10.1073/pnas.92.14.6602
Despres, C., DeLong, C., Glaze, S., Liu, E., and Fobert, P. R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12, 279–290. doi: 10.1105/tpc.12.2.279
Durrant, W. E., and Dong, X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209. doi: 10.1146/annurev.phyto.42.040803.140421
Fu, Z. Q., Yan, S., Saleh, A., Wang, W., Ruble, J., Oka, N., et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228–232. doi: 10.1038/nature11162
Gaffney, T., Friedrich, L., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., et al. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261, 754–756. doi: 10.1126/science.261.5122.754
Gorlach, J., Volrath, S., Knauf-Beiter, G., Hengy, G., Beckhove, U., Kogel, K. H., et al. (1996). Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell 8, 629–643. doi: 10.1105/tpc.8.4.629
Malamy, J., Carr, J. P., Klessig, D. F., and Raskin, I. (1990). Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science 250, 1002–1004. doi: 10.1126/science.250.4983.1002
Manohar, M., Tian, M., Moreau, M., Park, S.-W., Choi, H. W., Fei, Z., et al. (2014). Identification of multiple salicylic acid-binding proteins using two high throughput screens. Front. Plant Sci. 5:777. doi: 10.3389/fpls.2014.00777
Metraux, J. P., Ahl-Goy, P., Staub, T., Speich, J., Steinemann, A., Ryals, J., et al. (1991). “Induced resistance in cucumber in response to 2,6-dichloroisonicotinic acid and pathogens,” in Advances in Molecular Genetics of Plant-Microbe Interactions, Vol. 1, eds H. Hennecke and D. P. S. Verma (Dordrecht: Kluwer Academic Publishers), 432–439.
Metraux, J. P., Signer, H., Ryals, J., Ward, E., Wyss-Benz, M., Gaudin, J., et al. (1990). Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 250, 1004–1006. doi: 10.1126/science.250.4983.1004
Nawrath, C., and Metraux, J. P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11, 1393–1404. doi: 10.2307/3870970
Ponce De Leon, I., Schmelz, E. A., Gaggero, C., Castro, A., Alvarez, A., and Montesano, M. (2012). Physcomitrella patens activates reinforcement of the cell wall, programmed cell death and accumulation of evolutionary conserved defence signals, such as salicylic acid and 12-oxo-phytodienoic acid, but not jasmonic acid, upon Botrytis cinerea infection. Mol. Plant Pathol. 13, 960–974. doi: 10.1111/j.1364-3703.2012.00806.x
Rasmussen, J. B., Hammerschmidt, R., and Zook, M. N. (1991). Systemic induction of salicylic acid accumulation in cucumber after inoculation with Pseudomonas syringae pv syringae. Plant Physiol. 97, 1342–1347. doi: 10.1104/pp.97.4.1342
Rensing, S. A., Lang, D., Zimmer, A. D., Terry, A., Salamov, A., Shapiro, H., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69. doi: 10.1126/science.1150646
Ryals, J., Weymann, K., Lawton, K., Friedrich, L., Ellis, D., Steiner, H. Y., et al. (1997). The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor I kappa B. Plant Cell 9, 425–439. doi: 10.1105/tpc.9.3.425
Shah, J., Tsui, F., and Klessig, D. F. (1997). Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant Microbe Interact. 10, 69–78. doi: 10.1094/MPMI.1997.10.1.69
Uknes, S., Mauch-Mani, B., Moyer, M., Potter, S., Williams, S., Dincher, S., et al. (1992). Acquired resistance in Arabidopsis. Plant Cell 4, 645–656. doi: 10.1105/tpc.4.6.645
Ward, E. R., Uknes, S. J., Williams, S. C., Dincher, S. S., Wiederhold, D. L., Alexander, D. C., et al. (1991). Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3, 1085–1094. doi: 10.1105/tpc.3.10.1085
White, R. F. (1979). Acetylsalicylic acid (aspirin) induces resistance to tobacco mosaic virus in tobacco. Virology 99, 410–412. doi: 10.1016/0042-6822(79)90019-9
Wildermuth, M. C., Dewdney, J., Wu, G., and Ausubel, F. M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565. doi: 10.1038/35107108
Wu, Y., Zhang, D., Chu, J. Y., Boyle, P., Wang, Y., Brindle, I. D., et al. (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 1, 639–647. doi: 10.1016/j.celrep.2012.05.008
Yalpani, N., Silverman, P., Wilson, T. M., Kleier, D. A., and Raskin, I. (1991). Salicylic acid is a systemic signal and an inducer of pathogenesis-related proteins in virus-infected tobacco. Plant Cell 3, 809–818. doi: 10.1105/tpc.3.8.809
Zhang, Y., Cheng, Y. T., Qu, N., Zhao, Q., Bi, D., and Li, X. (2006). Negative regulation of defense responses in Arabidopsis by two NPR1 paralogs. Plant J. 48, 647–656. doi: 10.1111/j.1365-313X.2006.02903.x
Zhang, Y., Fan, W., Kinkema, M., Li, X., and Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. U.S.A. 96, 6523–6528. doi: 10.1073/pnas.96.11.6523
Zhang, Y., Tessaro, M. J., Lassner, M., and Li, X. (2003). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15, 2647–2653. doi: 10.1105/tpc.014894
Zhou, J. M., Trifa, Y., Silva, H., Pontier, D., Lam, E., Shah, J., et al. (2000). NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant Microbe Interact. 13, 191–202. doi: 10.1094/MPMI.2000.13.2.191
Keywords: salicylic acid, NPR1, NPR3, NPR4, plant immunity, SA receptor
Citation: Peng Y, Sun T and Zhang Y (2017) Perception of Salicylic Acid in Physcomitrella patens. Front. Plant Sci. 8:2145. doi: 10.3389/fpls.2017.02145
Received: 08 October 2017; Accepted: 04 December 2017;
Published: 18 December 2017.
Edited by:
Dingzhong Tang, Fujian Agriculture and Forestry University, ChinaReviewed by:
Zhonglin Mou, University of Florida, United StatesZhixiang Chen, Purdue University, United States
Copyright © 2017 Peng, Sun and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuelin Zhang, eXVlbGluLnpoYW5nQHViYy5jYQ==
 Yujun Peng
Yujun Peng Tongjun Sun
Tongjun Sun Yuelin Zhang
Yuelin Zhang