- 1College of Resources and Environmental Sciences, Nanjing Agricultural University, Nanjing, China
- 2Shanghai Center for Plant Stress Biology, National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, Shanghai, China
- 3Flower Research Institute, Yunnan Academy of Agricultural Sciences, Kunming, China
Aluminum (Al) can target multiple sites of root cells for toxicity, including the cell wall, the plasma membrane and symplastic components. Previous work revealed that the cell cycle checkpoint regulator (ATR) Ataxia Telangiectasia-mutated and Rad3-related is required for Al toxicity-induced root growth inhibition in als3 and that the symplastic component DNA is an important target site of Al for the toxicity. However, whether monitoring DNA integrity through ATR-regulated pathway is required for Al-induced root growth inhibition in other Al-sensitive mutants remains unknown. In this study, we demonstrated that the atr mutation could also rescue the Al hypersensitivity and Al-induced cell cycle arrest in star1, which supports the hypothesis that ALS3 and STAR1 function together to be involved in the detoxification of Al in Arabidopsis. However, mutation of ATR could not rescue the Al-sensitive phenotype of almt1 or stop1, both of which are defective in external detoxification mechanisms of Al. We further showed that the Al hypersensitivity and Al-induced quiescent center (QC) differentiation in als1 could also be rescued by the atr mutation. Therefore, our results suggest that ATR-regulated pathway is involved in the modulation of internal Al toxicity-mediated root growth inhibition in Arabidopsis.
Introduction
Aluminum (Al) comprises about 7% of the earth’s crust and is the most abundant metallic element. In neutral or alkaline soils, Al exists as insoluble aluminosilicates or oxides, which are non-toxic to plants. However, in acid soils with a pH of 5.5 or lower, solubilization of Al is enhanced and phytotoxic forms of Al are released into soil to levels that affect root growth. As a consequence, Al toxicity on acid soils becomes one of the most severe global problems since these soils comprise approximately 50% of the world’s potentially arable land (von Uexkull and Mutert, 1995; Kochian et al., 2004).
In acidic soils, Al exists as the octahedral hexahydrate Al(H2O)63+, which is more commonly referred to as Al3+. The phytotoxic Al3+ is the hardest Lewis acid, which is characterized by a low covalent and a high ionic index. Hard metal ions have strong interactions with organic molecules bearing oxygen groups (Poschenrieder et al., 2008). Therefore, Al3+ preferentially binds to phosphate, sulfate, and carboxyl groups for toxicity. Considering the components of a plant cell, Al is believed to target multiple sites for toxicity, including the cell wall, the plasma membrane and inside the cells. Cell walls and intercellular spaces are the first sites of the root in contact with Al when the roots are exposed to Al. Many studies have shown that most of the Al is bound to the cell wall. The ratio of cell wall Al to the total Al has been reported to range from 85 to 99.9% (Ma, 2007). Al can also bind to the plasma membrane and alter the membrane fluidity and surface potential (Kinraide, 2001), block ion channel activity (Pineros and Kochian, 2001), and induce the reactive oxygen species (ROS) as well as lipid peroxidation on the plasma membrane (Yamamoto et al., 2001). Furthermore, a small portion of Al can enter the symplasm rapidly and may interact with a number of symplastic targets (Lazof et al., 1996; Silva et al., 2000). For example, Al disrupts the cytoskeleton by interacting with both microtubules and actin filaments (Grabski and Schindler, 1995; Blancaflor et al., 1998), and blocks signal transduction pathways, particularly in Ca2+ homeostasis and signaling (Jones and Kochian, 1995; Jones et al., 1998; Zhang and Rengel, 1999). Al can also interact with DNA (Karlik et al., 1980; Karlik and Eichhorn, 1989), which is expected to have serious effects on gene expression and chromosome structure.
To cope with Al toxicity, plants have evolved Al-resistance mechanisms, including external and internal detoxification of Al (Ma et al., 2001; Kochian et al., 2004). In Arabidopsis thaliana, external detoxification of Al is primarily achieved through AtALMT1-mediated secretion of malate to form a non-toxic form of Al-malate in the apoplast (Hoekenga et al., 2006), and the citrate transport AtMATE play a minor role in the external detoxification of Al (Liu et al., 2009). STOP1, a C2H2 transcription factor, is involved in the detoxification of Al mainly through the regulation of AtALMT1 expression (Iuchi et al., 2007). For the internal detoxification of Al, the tonoplast-localized ATP-binding cassette (ABC) transporter ALS1 is required, which tolerates Al presumably via the transport of cytosolic Al into vacuoles (Larsen et al., 2007). STAR1 and STAR2/ALS3 encode a nucleotide-binding domain and transmembrane domain of a bacterial-type ABC transporter, respectively, and are suggested to be involved in Al tolerance through modification of cell wall or redistribution of Al from Al-sensitive root tips to other less Al-sensitive tissues (Larsen et al., 2005; Huang et al., 2009, 2010). Recently, Dong et al. (2017) reported that unlike rice STAR1 and STAR2, Arabidopsis ALS3 interacts with AtASTAR1 to be localized to the tonoplast, suggesting that AtSTAR1/ALS3 might be also required for the internal detoxification of Al.
Through the screening of the suppressors of the Al hypersensitivity of als3 mutant, Gabrielson et al. (2006) identified a dozen of suppressor mutants, and two of them had different mutations on the same gene ATR (Rounds and Larsen, 2008). ATR (Ataxia Telangiectasia-mutated and Rad3-related) is a cell cycle checkpoint regulator that functions in detecting DNA damage and then halting cell division (Culligan et al., 2004). atr mutant is hypersensitive to clastogenic and genotoxic stresses, but shows increased tolerance to Al because of failure to halt cell cycle progression. Together with the recovery of the Al hypersensitivity of als3 by the atr mutation, the results suggest that Al acts as a mild genotoxic agent and can target DNA to arrest root growth through ATR-regulated pathway (Rounds and Larsen, 2008).
In this study, to determine whether ATR-dependent pathway is required for the Al hypersensitivity in all Al-sensitive mutants, we created a series of double mutants between Al-sensitive mutants and atr mutant and then evaluated their sensitivity to Al in Arabidopsis. Our results revealed that the atr mutation could rescue the Al-sensitive phenotype of als3, star1 and als1, but not that of almt1 and stop1. These findings suggest that ATR-regulated pathway is required for internal Al toxicity-induced root growth inhibition.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis thaliana (Columbia ecotype, Col-0) was used for all the control experiments. The T-DNA insertion lines atr (SALK_032841C), star1 (GABI_762A06), als3 (SALK_004094), stop1 (SALK_114108), almt1 (SALK_00962) and the mutant als1-1 (CS3847) were all derived from uNASC1. Plants were grown in a growth chamber or controlled room at 22–25°C with 14 h of light and 10 h of darkness.
Mutant Genotyping
To select homozygous mutants of atr, als3, star1, stop1, and almt1, primer pairs flanked each T-DNA insertion were used as follows: ATR (5′-ACTGCATGCCAT TTACTCCTAC-3′ and 5′-GATCAGCTTGATCATCCAAACT-3′), ALS3 (5′- CAA TGTTCTTGCTCGTCCTCCT-3′ and 5′-TGGTTCACGTAGTGGGCCATCG-3′), STAR1 (5′-TCGTAGAGTTGGAATGCTTTTTC-3′ and 5′-GTTGAAGAAACCTCTGTGCCATT-3′), ALMT1 (5′-TTGAGAGAGCTGAGTGACCA-3′ and 5′-ACAAC GATATCAGCGCGAAC-3′), and STOP1 (5′-TCTTAAAGCGGCCATTGGTG-3′ and 5′-TTAGAGACTAGTATCTGAAACAGACTCAC-3′). For als1-1 mutant, a dCAPS (derive Cleaved Amplified Polymorphic sequences) marker was developed by using a primer pair (5′-TGTGAAACAGTTTGGTCGCT-3′ and 5′-TGCGTTTAGTCCTCCGAAGA-3′) and a restriction endonuclease TfiI. To generate double or triple mutants, crosses were made between atr and each Al-sensitive mutant or between als3atr and star1 and then the derived F2 plants were genotyped and selected. For genotyping of CyclinB1;1 and QC46 marker lines, a primer pair for the GUS gene was used (5′-ATGTTACGTCCTGTAGAAACC-3′ and 5′-TCATTGTTTGCCTCCC TGCTGC-3′).
RNA Isolation and Expression Analysis
Seeds were sterilized and stratified at 4°C for 2 days and then sowed on a 0.3% Gellan gum (G1910; Sigma–Aldrich) nutrient medium consisting of 1 mM KNO3, 0.2 mM KH2PO4, 2 mM MgSO4, 0.25 mM (NH4)2SO4, 1 mM Ca(NO3)2, 1 mM CaSO4, 1 mM K2SO4, 1 μM MnSO4, 5 μM H3BO3, 0.05 μM CuSO4, 0.2 μM ZnSO4, 0.02 μM NaMoO4, 0.1 μM CaCl2, 0.001 μM CoCl2 and 1% sucrose. After 7 days growth, the seedlings were transferred to a 0.5 mM CaCl2 solution for 6 h pretreatment at pH 4.8 and then exposed to a 0.5 mM CaCl2 solution (pH 4.8) with or without 20 μM AlCl3 for 12 h. Total RNA was extracted using TaKaRa MiniBEST plant RNA Extraction Kit (Cat # 9769). Around one microgram total RNA was first digested with DNase I and then subjected for the synthesis of first-strand cDNAs by using HiScript® 1st Strand cDNA Synthesis Kit (Vazyme Biotech Co., Ltd., Nanjing, China). One twentieth of the cDNA products and the SYBR® Green Master Mix kit (Vazyme Biotech Co., Ltd., Nanjing, China) were used for RT-PCR and real-time RT-PCR analysis. The primers for RT-PCR analysis of ATR, ALS3, STAR1, ALMT1, and STOP1 were same to those primers for genotyping as shown above. The primers for real-time RT-PCR analysis were as follows: ATR (5′-CTGACTGAGGACTGTGGTCTGGT-3′ and 5′-GACGGTCACCAAGCCCAACA-3′), ALS3 (5′-CGTATCTCTTCATGGTCTCTGTCG-3′ and 5′-GTAACTCCGGTGACGGTCATG-3′), STAR1 (5′-TTCAAGGGACTGTTGCGGATA-3′ and 5′-AAGAGCACTTGTTGGTTCATCG-3′), ALS1 (5′-GCCTCACAGTTGGTTCATCGG-3′ and 5′-GTCGTTTTTCCTCCACCGCT-3′), ALMT1 (5′-TGCAAGCTGCGTTGTCGAC-3′ and 5′-CAAAATCTTGAAGGAAGTGGGAG-3′) and STOP1 (5′-TCACATAGCTCTGTTCCAGGGA-3′ and 5′-ATCAGTCATTCCAGGCTGTGT-3′). UBQ10 was used as an internal control and the forward and reverse prime sequences of UBQ10 are 5′-CGTCTTCGTGGTGGTTTCTAA-3′ and 5′-GGATTATACAAGGCCCCAAAA-3′, respectively.
Evaluation of Sensitivity to Al
For assessment of Al sensitivity in hydroponic conditions, we referred to a previous method with slight modifications (Huang et al., 2010). Briefly, seeds of each line were stratified at 4°C for 2 days and then sowed on a plastic mesh floating on a 1/30 strength Hoagland nutrient solution (NH4H2PO4 omitted) plus 1 mM CaCl2 and different concentrations of AlCl3 at pH 5.0 for 7 days. The solution was renewed every 3 days. After the treatment, the seedlings were photographed and root length was measured by ImageJ. Relative root growth expressed as (root length with Al treatment/root length without Al) × 100 was used to evaluate the Al sensitivity. For soaked gel experiments, we adopted the method developed by Larsen et al. (2005). Nutrient agar medium was first prepared, which consisted of 50 ml of 1 mM KNO3, 0.2 mM KH2PO4, 2 mM MgSO4, 0.25 mM (NH4)2SO4, 1 mM Ca(NO3)2, 1 mM CaSO4, 1 mM K2SO4, 1 μM MnSO4, 5 μM H3BO3, 0.05 μM CuSO4, 0.2 μM ZnSO4, 0.02 μM NaMoO4, 0.1 μM CaCl2, 0.001 μM CoCl2, 1% sucrose, and 0.3% Gellan gum (G1910; Sigma–Aldrich). The agar medium was then soaked with 25 ml of the same nutrient medium containing 0, 0.5, 0.75, or 1 mM AlCl3. After 2 days soaking, the solution was removed and seeds were grown on the agar medium plates for 7 days. The seedlings were then pictured and compared and the root length was measured by ImageJ.
GUS Activity Assay
To investigate the effect of Al on Cyclin B1;1 accumulation, seeds of CycB1;1:GUS –containing WT, atr, star1, and star1atr were grown on a soaked gel medium containing 0 or 0.5 mM AlCl3 for 7 days. The seedlings were then stained with a commercialized GUS staining solution (161031; O’Biolab Co., Ltd., Beijing, China) for 2 h at 37°C. For determination of the status of the quiescent center (QC) after Al treatment, seeds of QC46 (GUS-based QC marker)-containing WT, atr, als1, and als1atr were grown on a soaked gel medium containing 0 or 1.5 mM AlCl3. After growth for 7 days, the seedlings were stained with the GUS staining solution overnight at 37°C. Stained tissues were observed and photographed with a microscope (Olympus BX53F, Japan).
Results
Mutation of ATR Rescued the Al-Sensitive Phenotype of Both als3 and star1 Mutants
To confirm the previous observation that mutation of ATR could rescue the Al-sensitive phenotype of als3 (Rounds and Larsen, 2008), we generated als3atr double mutant through a genetic cross between atr and als3 single mutants. RT-PCR analysis revealed that ATR and ALS3 were knocked out in respective single or double mutants (Figure 1A). We evaluated the tolerance of WT, atr, als3, and als3atr mutants to Al in both hydroponic and soaked gel conditions. Consistent with previous results, atr mutant showed more tolerance to Al than WT, and the atr mutation was able to reduce the sensitivity of als3 to Al at all Al concentrations (Figures 1C,E,F). Nevertheless, mutation of ATR was not able to fully rescue the Al-sensitive phenotype of als3, especially at high Al concentrations (Figures 1C,E,F), suggesting that other Al toxicity mechanisms are also required for Al-induced growth inhibition in als3 mutant. As STAR1 interacts with ALS3 to be involved in the regulation of Al tolerance in Arabidopsis (Huang et al., 2010; Dong et al., 2017), we investigated whether the atr mutation could also rescue the Al-sensitive phenotype of star1. We generated star1atr double mutant through crossing and genotyping and RT-PCR analysis confirmed that both STAR1 and ATR were knocked out in the double mutant (Figure 1B). Evaluation of Al tolerance in the double mutant showed that star1atr was more tolerant to Al than star1 at all Al concentrations (Figures 1D,G,H), indicating that ATR is required for Al-induced growth inhibition in star1 mutant. Additionally, similar to that in als3atr mutant, mutation of ATR did not fully rescue the Al-sensitive phenotype of star1 (Figures 1D,G,H). We also generated star1als3 and star1als3atr mutants to further investigate whether mutation of ATR could rescue the Al sensitivity in star1als3 double mutant. Results showed that the Al-sensitive phenotype of star1als3 could also be rescued by the introduction of the atr mutation (Figure 1I). Together, these results confirm that STAR1 and ALS3 regulate Al tolerance through the same pathway and indicate that ATR-dependent pathway is also required for Al-induced growth inhibition in star1 mutant.
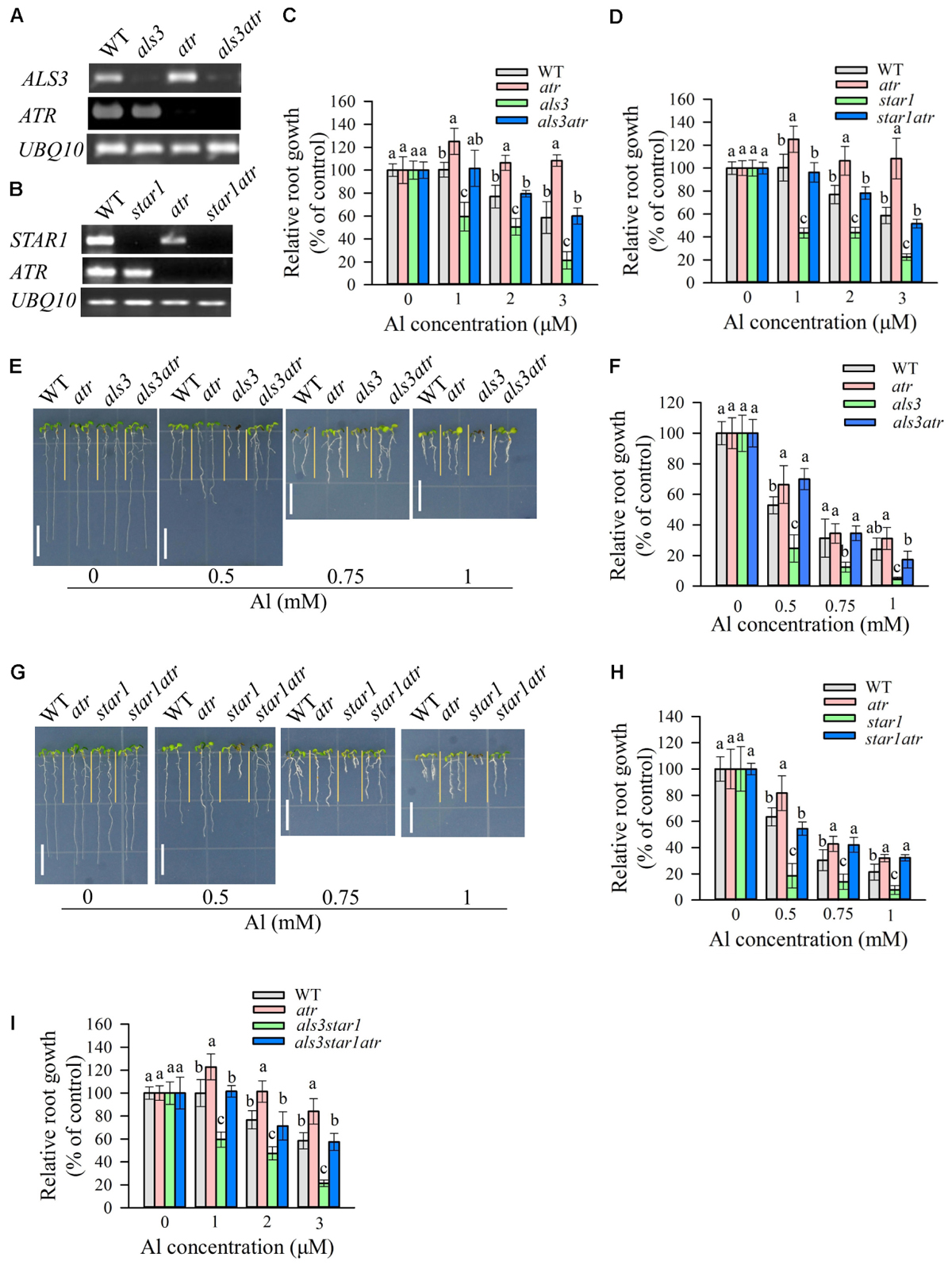
FIGURE 1. Rescue of the Al-sensitive phenotype of als3 and star1 by atr mutation. (A,B) RT-PCR analysis of ATR, ALS3, or STAR1 in WT and different single or double mutants. UBQ10 was used as internal control. (C,D) Evaluation of Al tolerance in als3 (C) or star1 (D)-related mutants in hydroponic conditions. Seedlings were grown on a nutrient solution containing 0, 1, 2, or 3 μM Al at pH 5.0 for 7 days and then root length was measured and compared. Data are means ± SD (n = 15–20). (E–H) Evaluation of Al tolerance in soaked gel conditions. Seedlings were grown on a soaked gel medium containing 0, 0.5, 0.75, or 1 mM Al for 7 days. Data are means ± SD (n = 10–15). (E,F) Rescue of the Al-sensitive phenotype of als3 by atr. (G,H) Rescue of the Al-sensitive phenotype of star1 by atr. (I) Rescue of the Al-sensitive phenotype of als3star1 by atr in hydroponic conditions. Means with different letters are significantly different (P < 0.05, Tukey’s test). Scale bar = 1 cm.
Al-induced inhibition of root growth was correlated with the increase in the number of cells trapped in the G2 stage, which causes the hyperaccumulation of Cyclin B1;1 in root tips (Rounds and Larsen, 2008). To examine the effect of Al on the accumulation of Cyclin B1;1 in star1 mutant background, we introduced CycB1;1:GUS into atr, star1 and star1atr through crossing. In the absence of Al, GUS expression was detected at relatively low levels in all the materials (Figure 2). After exposure to a low toxic level of Al, while GUS activity was slightly increased in WT, GUS expression in star1 was dramatically increased in root tips, suggesting that cell cycle progression was halted in star1 (Figure 2). In star1atr, GUS activity was detected at similar low levels to that in WT and atr, which suggested that the arrest of cell cycle progression in star1 was rescued by the atr mutation. The Cyclin B1;1 expression results support the conclusion that knockout of ATR is able to rescue the Al hypersensitivity in star1.
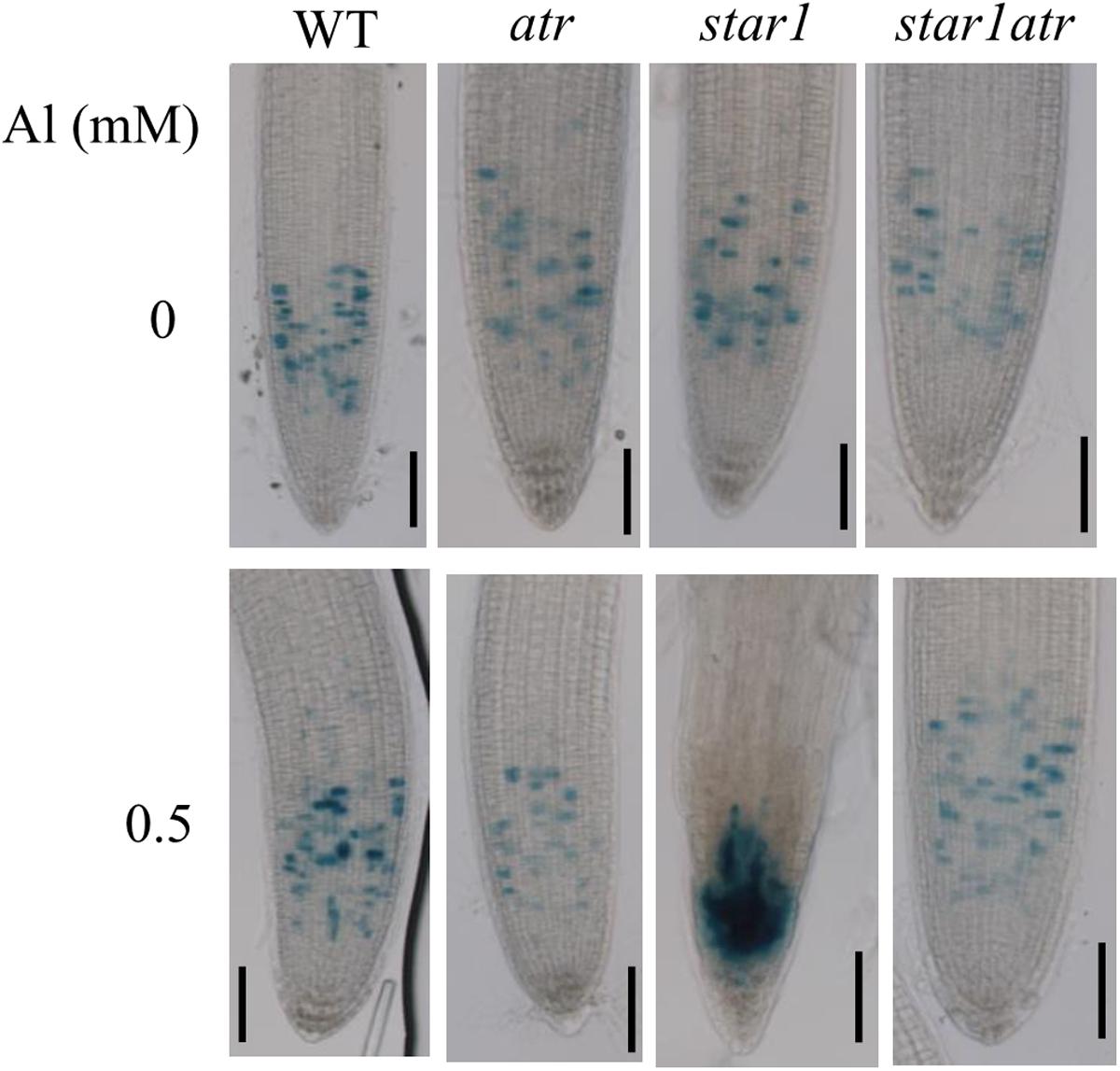
FIGURE 2. Rescue of cell cycle progression defects in star1 by atr mutation under Al stress conditions. Seedlings of WT, atr, star1, and star1atr harboring CycB1;1:GUS marker were grown on a soaked gel medium containing 0 or 0.5 mM Al for 7 days and the roots were stained and observed under a microscope. Scale bar = 50 μm.
The atr Mutation Could Not Rescue the Al Hypersensitivity in Either almt1 or stop1 Mutants
To investigate whether mutation of ATR could rescue the hypersensitivity of almt1 and stop1 to Al, we introduced the atr mutation into stop1 and almt1 mutants by crossing and genotyping, respectively. RT-PCR analysis confirmed that ALMT1 or STOP1 were knocked out in the corresponding mutants (Figures 3A,B). Phenotypic analysis of Al tolerance showed that the tolerance of almt1atr to Al did not differ from that of almt1 at all Al concentrations in both hydroponic and soaked gel conditions (Figures 3C,D,G,H), indicating that mutation of ATR could not rescue Al-sensitive phenotype of almt1. Similarly, Al tolerance in stop1atr was also not different from that in stop1 under all Al treatment (Figures 3E,F), demonstrating that the atr mutation was not able to rescue the Al-sensitive phenotype of stop1 either. These results suggest that ATR is not required for Al-induced growth inhibition in those Al-sensitive mutants that are defective in the external detoxification of Al.
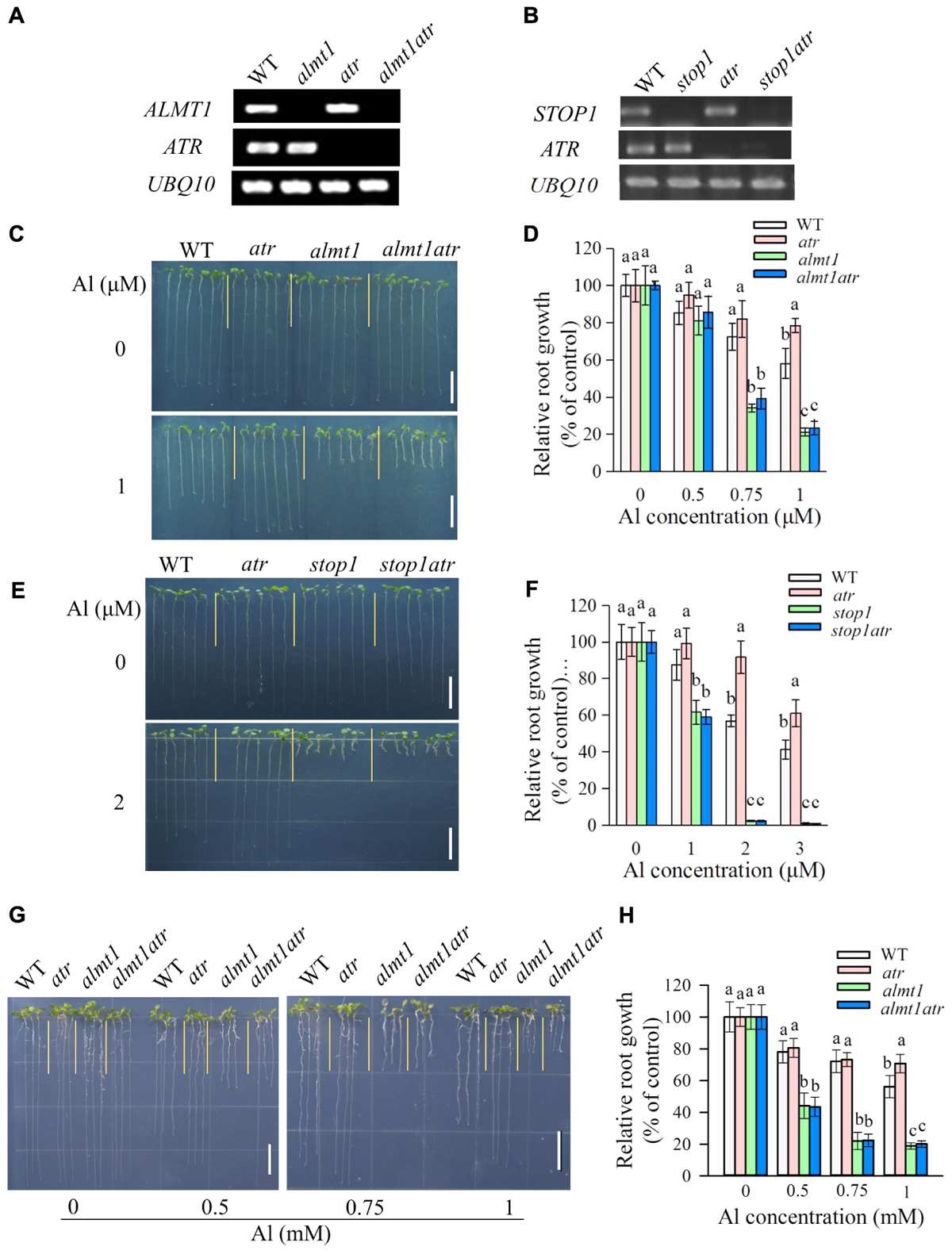
FIGURE 3. The Al hypersensitivity defects in almt1 and stop1 could not be rescued by the atr mutation. (A,B) RT-PCR analysis of ATR, ALMT1, or STOP1 in WT and different single or double mutants. UBQ10 was used as internal control. (C–F) Evaluation of Al tolerance in almt1 (C,D) or stop1 (E,F)-related mutants in hydroponic conditions. Seedlings were grown on a nutrient solution with different concentrations of Al at pH 5.0 for 7 days and then root length was measured and compared. Data are means ± SD (n = 15–20). (G,H) Evaluation of Al tolerance in almt1-related mutants in soaked gel conditions. Seedlings were grown on a soaked gel medium containing 0, 0.5, 0.75, or 1 mM Al for 7 days. Data are means ± SD (n = 10–15). Means with different letters are significantly different (P < 0.05, Tukey’s test). Scale bar = 1 cm.
The Al-Sensitive Phenotype of als1 Could Also Be Rescued by the atr Mutation
Since ATR is localized in the nucleus and required for Al-induced halting cell division in als3 or star1 (Figures 1, 2), there are two possibilities that ATR might detect general internal Al toxicity signal or star1/als3-specific Al toxicity signal. To distinguish these two, we utilized another Al-sensitive mutant als1, which is deficient in the sequestration of Al into vacuoles (Larsen et al., 2007). Introduction of atr mutation into als1 mutant could also rescue its Al-sensitive phenotype at various Al concentrations (Figures 4A,B). These results imply that ATR is required for internal Al toxicity-mediated root growth inhibition.
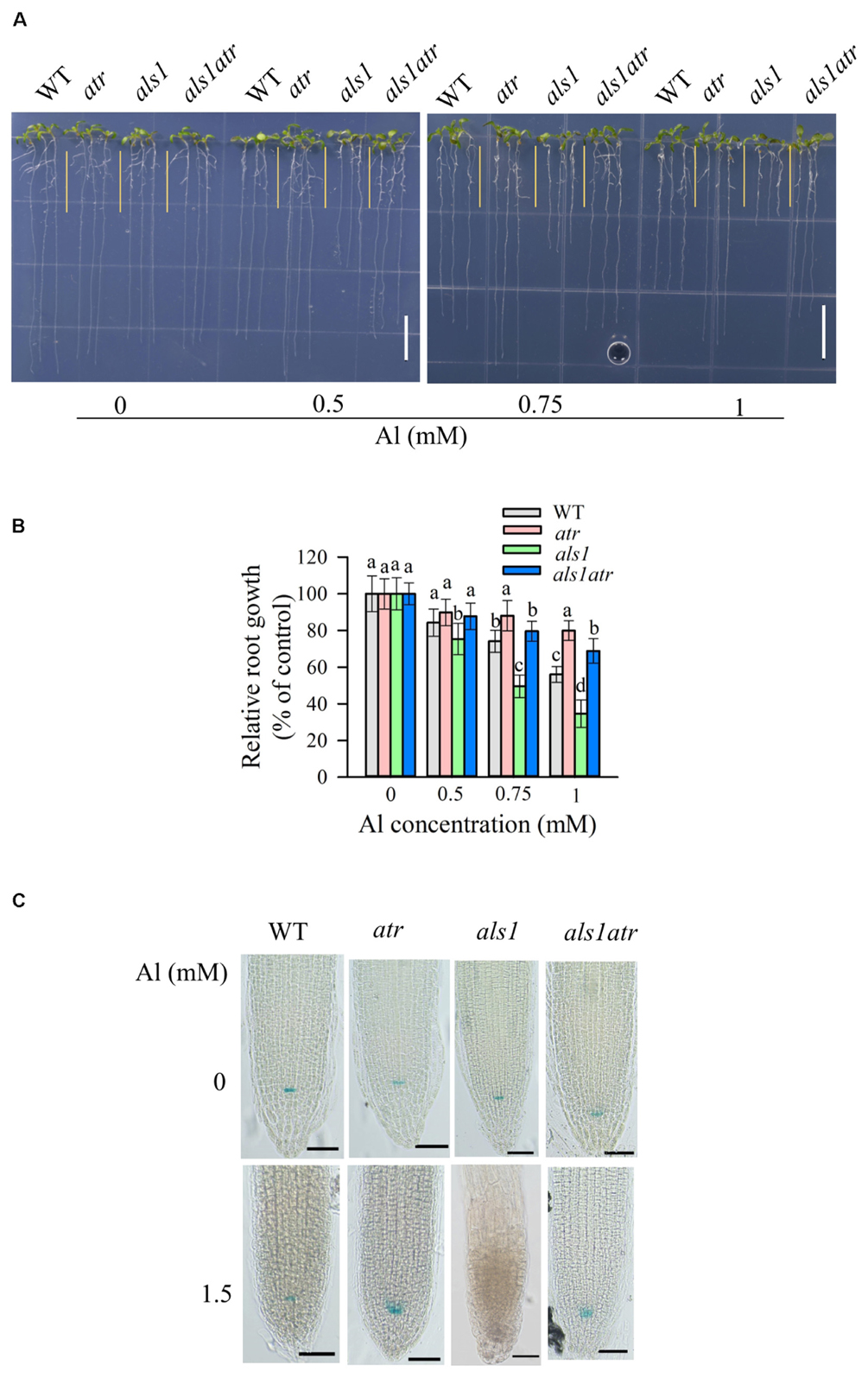
FIGURE 4. Rescue of the Al-sensitive phenotype of als1 by atr mutation. (A,B) Seedlings of WT, atr, als1, and als1atr were grown on a soaked gel medium containing 0, 0.5, 0.75, or 1 mM Al for 7 days. Data are means ± SD (n = 10–15). Means with different letters are significantly different (P < 0.05, Tukey’s test). Scale bar = 1 cm. (C) Rescue of QC differentiation of als1 by atr mutation. Seedlings of WT, atr, als1, and als1atr harboring QC46 (QC-specific marker) were grown on a soaked gel medium containing 0 or 1.5 mM Al for 7 days and the roots were stained with GUS staining solution and observed under a microscope. Scale bar = 50 μm.
We also determined the status of the QC after Al treatment by introduction of a GUS-based QC marker, QC46 (Sabatini et al., 2003), into atr, als1 and als1atr. Without Al treatment, GUS expression was well detected in all the materials (Figure 4C). However, in the presence of high levels of Al, GUS activity was lost in als1, suggesting that the essential stem cells required for maintenance of root growth was destroyed by Al toxicity in als1 mutant. In contrast, als1atr double mutant displayed normal GUS activity in the QC after Al treatment (Figure 4C). These results indicate that the atr mutation could help als1 mutant to maintain the QC integrity for root growth when exposure to highly toxic levels of Al.
Expression Pattern of ATR and Al-Resistance Genes
To examine whether ATR expression was altered in Al-sensitive mutants, we compared the expression level of ATR between WT and the Al-sensitive mutants. Results showed that there was no significant difference in ATR expression between WT and the mutants in the absence of Al (Figure 5A). Al treatment slightly decreased the expression of ATR, but no significant difference in ATR expression was found in WT and the mutants. This result suggests that increased Al sensitivity of the mutants was not due to altered ATR expression. The expression of Al-resistance genes in atr mutant was also determined. The expression levels of the Al-resistance genes including ALS3, STAR1, ALS1, ALMT1, and STOP1 in atr mutant were similar to those in WT under both –Al and –Al conditions (Figure 5B), suggesting that increased Al tolerance in atr mutant was not caused by elevated expression of Al-resistance genes.
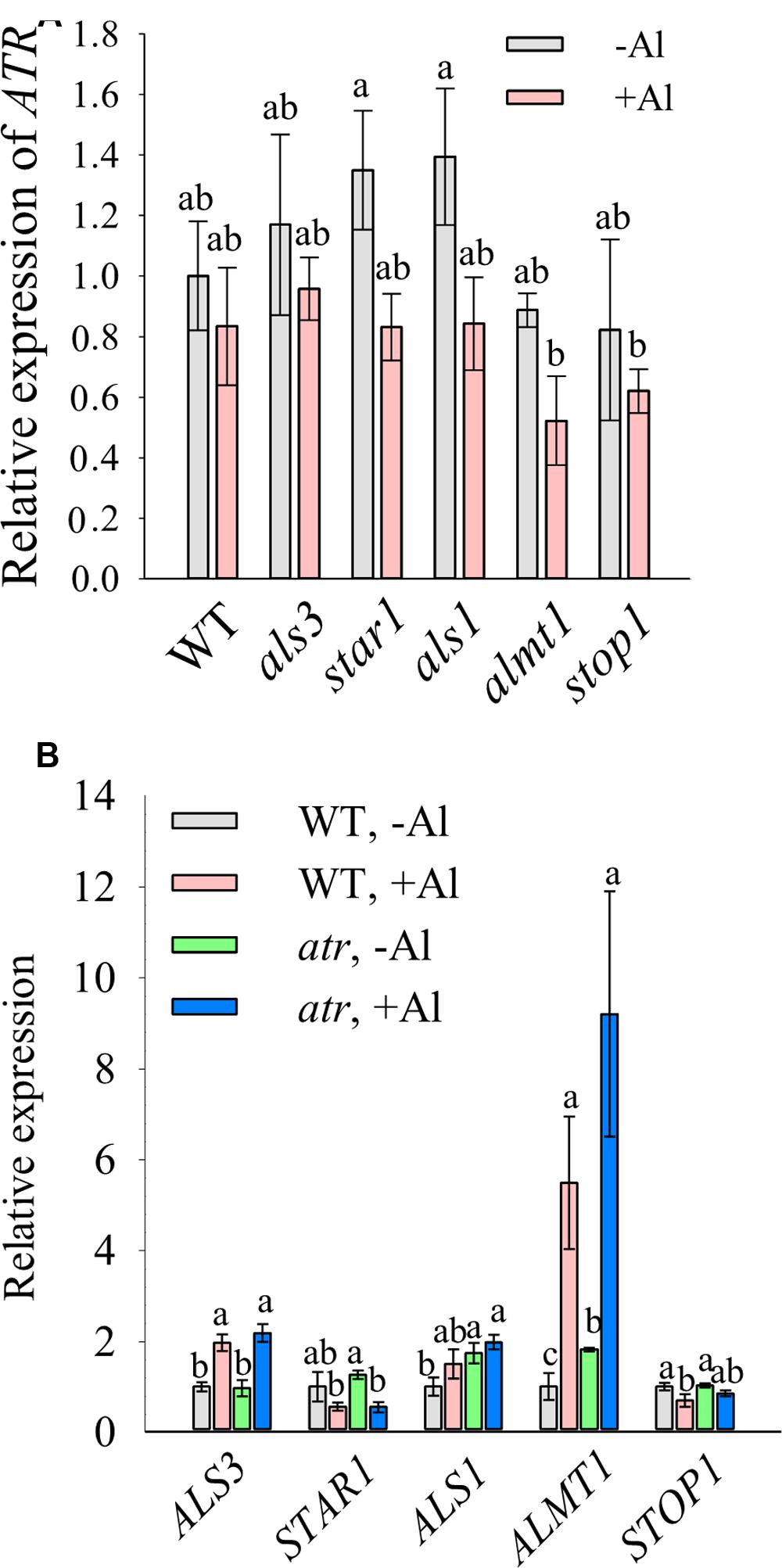
FIGURE 5. Expression analysis of ATR and Al-resistance genes. Seven-day-old seedlings grown on a nutrient agar medium were pretreated with a 0.5 mM CaCl2 solution for 6 h at pH 4.8 and then exposed to the same solution containing 0 or 20 μM Al at pH 4.8 for 12 h. The roots were sampled for expression analysis of ATR (A) or Al-resistance genes (B). Data are means ± SD (n = 3). Means with different letters are significantly different (P < 0.05, Tukey’s test).
Discussion
ATR functions as a cell cycle checkpoint to detect DNA damage and subsequently prevent cell division (Culligan et al., 2004). Since knockout of ATR is able to rescue the Al hypersensitivity in als3 mutant (Gabrielson et al., 2006; Rounds and Larsen, 2008), two possible mechanisms exist for the increased sensitivity to Al in als3. One possible mechanism is that mutation of ALS3 results in the increased Al accumulation in nucleus and consequently activates ATR-regulated pathway to halt cell division and ultimately inhibit root growth. The other is that Al toxicity-induced specific signal in als3 activates ATR-regulated pathway to cause root growth inhibition. Our results showed that in addition to als3, mutation of ATR can also rescue Al-sensitive phenotype of star1 and als1, indicating that rescue of Al-sensitive phenotype by atr mutation is not specific to als3 mutant. Thus, we prefer the former hypothesis that elevated Al accumulation in the nucleus induces ATR-regulated pathway to inhibit root growth in als3 mutant.
In contrast to its hypersensitivity to clastogenic and genotoxic stresses, atr mutant shows increased tolerance to Al. Al in nucleus might bind to DNA non-covalently and induce a conformational alteration from the B-form to Z-DNA, which affects DNA unwinding during DNA replication (Anitha and Rao, 2002). Nevertheless, unlike other genotoxic stresses, Al is thought to be a mild DNA damage agent and its binding to DNA is likely to be reversible (Rounds and Larsen, 2008; Nezames et al., 2012). This unique interaction of Al with DNA can activate ATR-, ALT2-, and SOG1-regualted transcriptional response to halt cell division and cause the inhibition of root growth (Sjogren et al., 2015). However, it remains unknown about how the interaction of Al with DNA activates the ATR-regulated pathway and what the ATR-regulated downstream transcriptional events that lead to the cease of cell division are.
The inhibition of root growth can be attributed to the disruption of cell division and/or cell elongation. Rapid reduction in root growth suggests an initial impact of Al on cell elongation instead of cell division (Sharp et al., 1988; Kopittke et al., 2015). However, when roots are exposed to Al for a long period of time, inhibition of cell division might also contribute to the reduction of root growth. Al-activated ATR-regulated cease of cell division in als3/star1 or als1 suggests that inhibition of cell division plays a critical role in Al-induced inhibition of root growth in these Al-sensitive mutants. Further work is required to determine whether mutation of atr could rescue the Al-sensitive phenotype of these mutants after a short-term exposure to Al.
Numerous studies have suggested that Al can target multiple sites for toxicity, including apoplastic and symplastic components (Kochian, 1995; Ma, 2007). Nevertheless, it remains debatable about which sites play more important roles in Al-induced inhibition of root growth. We found that the atr mutation could not rescue the Al hypersensitivity in almt1 and stop1, which are defective in the capacity to detoxify Al externally. These results indicate that ATR is not required for Al-induced inhibition of root growth in all Al-sensitive mutants and suggest that both symplastic components such as DNA and apoplastic components including cell wall are important Al target sites that lead to root growth inhibition by Al toxicity. Additionally, our data showed that the atr mutation could not fully rescue the Al hypersensitivity in als3, suggesting that Al also targets other symplastic sites to cause root growth inhibition in als3 mutant.
In rice, OsSTAR1 interacts with OsSTAR2, the rice ortholog of ALS3, to form a functional complex that is suggested to be involved in the modification of cell wall that is required for Al detoxification (Huang et al., 2009). Although Arabidopsis AtSTAR1 can also interact with ALS3 to be involved in the detoxification of Al, AtSTAR1 and ALS3 are localized to tonoplast (Larsen et al., 2005; Huang et al., 2010; Dong et al., 2017), which are different from OsSTAR1 and OsSTAR2 that are localized to vesicle membranes (Huang et al., 2009). We found that in addition to als3, knockout of ATR also rescues the Al-sensitive phenotype of star1. Furthermore, the atr mutation can even rescue Al hypersensitivity in als3star1 double mutant. These results indicate that als3 and star1 share the same mechanism for their hypersensitivity to Al, i.e., ATR-regulated pathway required for Al-induced inhibition of root growth. The results also support the view that STAR1 and STAR2/ALS3 function together to be involved in the same pathway of Al detoxification. We further found that the Al hypersensitivity in als1 was rescued by the atr mutation. als1 has defects in the internal detoxification of Al (Larsen et al., 2007). Together, our results suggest that ATR is required for internal Al toxicity-induced inhibition of root growth and that STAR1 and ALS3 might be involved in the internal detoxification of Al in Arabidopsis. We propose that under Al stress conditions, internal Al detoxification-deficient mutants accumulate high levels of Al in the nucleus, which induces DNA damage and consequently activates ATR-regulated pathway and arrest cell cycle, finally leading to the inhibition of root growth.
Author Contributions
All authors conceived the project. C-FH drafted the manuscript. YZ, JG, MC, LL, and LW performed the experiments. YZ and JG helped to analyze the data and write the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by Jiangsu Science Fund for Distinguished Young Scholars (Grant No. BK20150027), National Natural Science Foundation of China (Grant No. 31570253 to C-FH), the Strategic Priority Research Program (Grant No. XDPB0404) of the Chinese Academy of Sciences, and the Shanghai Center for Plant Stress Biology, Chinese Academy of Sciences.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Philip N. Benfey from Duke University for kindly providing the QC46 marker.
Footnotes
References
Anitha, S., and Rao, K. S. J. (2002). The complexity of aluminum-DNA interactions: relevance to Alzheimer’s and other neurological diseases. Struct. Bond. 104, 79–97. doi: 10.1007/3-540-45425-X_3
Blancaflor, E. B., Jones, D. L., and Gilroy, S. (1998). Alterations in the cytoskeleton accompany aluminum-induced growth inhibition and morphological changes in primary roots of maize. Plant Physiol. 118, 159–172. doi: 10.1104/pp.118.1.159
Culligan, K., Tissier, A., and Britt, A. (2004). ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 16, 1091–1104. doi: 10.1105/Tpc.018903
Dong, J. S., Pineros, M. A., Li, X. X., Yang, H. B., Liu, Y., Murphy, A. S., et al. (2017). An Arabidopsis ABC transporter mediates phosphate deficiency-induced remodeling of root architecture by modulating iron homeostasis in roots. Mol. Plant 10, 244–259. doi: 10.1016/j.molp.2016.11.001
Gabrielson, K. M., Cancel, J. D., Morua, L. F., and Larsen, P. B. (2006). Identification of dominant mutations that confer increased aluminium tolerance through mutagenesis of the Al-sensitive Arabidopsis mutant, als3-1. J. Exp. Bot. 57, 943–951. doi: 10.1093/jxb/erj080
Grabski, S., and Schindler, M. (1995). Aluminum induces rigor within the actin network of soybean cells. Plant Physiol. 108, 897–901. doi: 10.1104/pp.108.3.897
Hoekenga, O. A., Maron, L. G., Pineros, M. A., Cancado, G. M., Shaff, J., Kobayashi, Y., et al. (2006). AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 9738–9743. doi: 10.1073/pnas.0602868103
Huang, C. F., Yamaji, N., and Ma, J. F. (2010). Knockout of a bacterial-type ATP-binding cassette transporter gene, AtSTAR1, results in increased aluminum sensitivity in Arabidopsis. Plant Physiol. 153, 1669–1677. doi: 10.1104/pp.110.155028
Huang, C. F., Yamaji, N., Mitani, N., Yano, M., Nagamura, Y., and Ma, J. F. (2009). A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 21, 655–667. doi: 10.1105/tpc.108.064543
Iuchi, S., Koyama, H., Iuchi, A., Kobayashi, Y., Kitabayashi, S., Ikka, T., et al. (2007). Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc. Natl. Acad. Sci. U.S.A. 104, 9900–9905. doi: 10.1073/pnas.0700117104
Jones, D. L., and Kochian, L. V. (1995). Aluminum inhibition of the inositol 1,4,5-trisphosphate signal transduction pathway in wheat roots: a role in aluminum toxicity? Plant Cell 7, 1913–1922. doi: 10.1105/tpc.7.11.1913
Jones, D. L., Kochian, L. V., and Gilroy, S. (1998). Aluminum induces a decrease in cytosolic calcium concentration in BY-2 tobacco cell cultures. Plant Physiol. 116, 81–89. doi: 10.1104/pp.116.1.81
Karlik, S. J., and Eichhorn, G. L. (1989). Polynucleotide cross-linking by aluminum. J. Inorg. Biochem. 37, 259–269. doi: 10.1016/0162-0134(89)85001-9
Karlik, S. J., Eichhorn, G. L., Lewis, P. N., and Crapper, D. R. (1980). Interaction of aluminum species with deoxyribonucleic acid. Biochemistry 19, 5991–5998. doi: 10.1021/Bi00567a008
Kinraide, T. B. (2001). Ion fluxes considered in terms of membrane-surface electrical potentials. Aust. J. Plant Physiol. 28, 605–616. doi: 10.1071/PP01019
Kochian, L. V. (1995). Cellular mechanisms of aluminum toxicity and resistance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 237–260. doi: 10.1146/annurev.arplant.46.1.237
Kochian, L. V., Hoekenga, O. A., and Pineros, M. A. (2004). How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu. Rev. Plant Biol. 55, 459–493. doi: 10.1146/annurev.arplant.55.031903.141655
Kopittke, P. M., Moore, K. L., Lombi, E., Gianoncelli, A., Ferguson, B. J., Blamey, F. P., et al. (2015). Identification of the primary lesion of toxic aluminum in plant roots. Plant Physiol. 167, 1402–1411. doi: 10.1104/pp.114.253229
Larsen, P. B., Cancel, J., Rounds, M., and Ochoa, V. (2007). Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta 225, 1447–1458. doi: 10.1007/s00425-006-0452-4
Larsen, P. B., Geisler, M. J. B., Jones, C. A., Williams, K. M., and Cancel, J. D. (2005). ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis. Plant J. 41, 353–363. doi: 10.1111/j.1365-313X.2004.02306.x
Lazof, D. B., Goldsmith, J. G., Rufty, T. W., and Linton, R. W. (1996). The early entry of Al into cells of intact soybean roots - A comparison of three developmental root regions using secondary ion mass spectrometry imaging. Plant Physiol. 112, 1289–1300. doi: 10.1104/pp.112.3.1289
Liu, J. P., Magalhaes, J. V., Shaff, J., and Kochian, L. V. (2009). Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 57, 389–399. doi: 10.1111/j.1365-313X.2008.03696.x
Ma, J. F. (2007). Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Int. Rev. Cytol. 264, 225–252. doi: 10.1016/S0074-7696(07)64005-4
Ma, J. F., Ryan, P. R., and Delhaize, E. (2001). Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci. 6, 273–278. doi: 10.1016/S1360-1385(01)01961-6
Nezames, C. D., Sjogren, C. A., Barajas, J. F., and Larsen, P. B. (2012). The Arabidopsis cell cycle checkpoint regulators TANMEI/ALT2 and ATR mediate the active process of aluminum-dependent root growth inhibition. Plant Cell 24, 608–621. doi: 10.1105/tpc.112.095596
Pineros, M. A., and Kochian, L. V. (2001). A patch-clamp study on the physiology of aluminum toxicity and aluminum tolerance in maize. Identification and characterization of Al3+-induced anion channels. Plant Physiol. 125, 292–305. doi: 10.1104/pp.125.1.292
Poschenrieder, C., Gunse, B., Corrales, I., and Barcelo, J. (2008). A glance into aluminum toxicity and resistance in plants. Sci. Total Environ. 400, 356–368. doi: 10.1016/j.scitotenv.2008.06.003
Rounds, M. A., and Larsen, P. B. (2008). Aluminum-dependent root-growth inhibition in Arabidopsis results from AtATR-regulated cell-cycle arrest. Curr. Biol. 18, 1495–1500. doi: 10.1016/j.cub.2008.08.050
Sabatini, S., Heidstra, R., Wildwater, M., and Scheres, B. (2003). SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 17, 354–358. doi: 10.1101/gad.252503
Sharp, R. E., Silk, W. K., and Hsiao, T. C. (1988). Growth of the maize primary root at low water potentials.1. spatial-distribution of expansive growth. Plant Physiol. 87, 50–57. doi: 10.1104/Pp.87.1.50
Silva, I. R., Smyth, T. J., Moxley, D. F., Carter, T. E., Allen, N. S., and Rufty, T. W. (2000). Aluminum accumulation at nuclei of cells in the root tip. Fluorescence detection using lumogallion and confocal laser scanning microscopy. Plant Physiol. 123, 543–552. doi: 10.1104/pp.123.2.543
Sjogren, C. A., Bolaris, S. C., and Larsen, P. B. (2015). Aluminum-dependent terminal differentiation of the Arabidopsis root tip is mediated through an ATR-, ALT2-, and SOG1-regulated transcriptional response. Plant Cell 27, 2501–2515. doi: 10.1105/tpc.15.00172
von Uexkull, H. R., and Mutert, E. (1995). Global extent, development and economic-impact of acid soils. Plant Soil 171, 1–15. doi: 10.1007/BF00009558
Yamamoto, Y., Kobayashi, Y., and Matsumoto, H. (2001). Lipid peroxidation is an early symptom triggered by aluminum, but not the primary cause of elongation inhibition in pea roots. Plant Physiol. 125, 199–208. doi: 10.1104/Pp.125.1.199
Keywords: aluminum toxicity, Arabidopsis thaliana, ATR, cell cycle checkpoint, DNA damage, external, internal
Citation: Zhang Y, Guo J, Chen M, Li L, Wang L and Huang C-F (2018) The Cell Cycle Checkpoint Regulator ATR Is Required for Internal Aluminum Toxicity-Mediated Root Growth Inhibition in Arabidopsis. Front. Plant Sci. 9:118. doi: 10.3389/fpls.2018.00118
Received: 13 November 2017; Accepted: 22 January 2018;
Published: 14 February 2018.
Edited by:
Jian Li Yang, Zhejiang University, ChinaReviewed by:
He Qiang Lou, Zhejiang A & F University, ChinaWei Fan, Yunnan Agricultural University, China
Cuiyue Liang, South China Agricultural University, China
Copyright © 2018 Zhang, Guo, Chen, Li, Wang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chao-Feng Huang, Y2ZodWFuZ0BzaWJzLmFjLmNu
† These authors have contributed equally to this work.
 Yang Zhang1,2†
Yang Zhang1,2† Chao-Feng Huang
Chao-Feng Huang