- 1Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Department of Plant Pathology and Ministry of Agriculture Key Laboratory for Plant Pathology, China Agricultural University, Beijing, China
- 3School of Medical Science, Chifeng University, Chifeng, China
- 4College of Life Sciences, Beijing Normal University, Beijing, China
Meloidogyne incognita is highly specialized parasite that interacts with host plants using a range of strategies. The effectors are synthesized in the esophageal glands and secreted into plant cells through a needle-like stylet during parasitism. In this study, based on RNA-seq and bioinformatics analysis, we predicted 110 putative Meloidogyne incognita effectors that contain nuclear localization signals (NLSs). Combining the Burkholderia glumae–pEDV based screening system with subcellular localization, from 20 randomly selected NLS effector candidates, we identified an effector MiISE6 that can effectively suppress B. glumae-induced cell death in Nicotiana benthamiana, targets to the nuclei of plant cells, and is highly expressed in early parasitic J2 stage. Sequence analysis showed that MiISE6 is a 157-amino acid peptide, with an OGFr_N domain and two NLS motifs. Hybridization in situ verified that MiISE6 is expressed in the subventral esophageal glands. Yeast invertase secretion assay validated the function of the signal peptide harbored in MiISE6. Transgenic Arabidopsis thaliana plants expressing MiISE6 become more susceptible to M. incognita. Inversely, the host-derived RNAi of MiISE6 of the nematode can decrease its parasitism on host. Based on transcriptome analysis of the MiISE6 transgenic Arabidopsis samples and the wild-type samples, we obtained 852 differentially expressed genes (DEGs). Integrating Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses, we found that expression of MiISE6 in Arabidopsis can suppress jasmonate signaling pathway. In addition, the expression of genes related to cell wall modification and the ubiquitination proteasome pathway also have detectable changes in the transgenic plants. Results from the present study suggest that MiISE6 is involved in interaction between nematode-plant, and plays an important role during the early stages of parasitism by interfering multiple signaling pathways of plant. Moreover, we found homologs of MiISE6 in other sedentary nematodes, Meloidogyne hapla and Globodera pallida. Our experimental results provide evidence to decipher the molecular mechanisms underlying the manipulation of host immune defense responses by plant parasitic nematodes, and transcriptome data also provide useful information for further study nematode–plant interactions.
Introduction
Meloidogyne incognita, the southern root knot nematode (RKN), is an economically important, sedentary nematode that infests roots of a wide range of crop plants (Alioto et al., 2015). During parasitism, RKNs establish a close relationship with their hosts and induce the formation of giant cells (GCs) that provide essential nutrition for nematode parasitic life stages. The formation of GCs are mediated through secretions (referred to as effectors) from the esophageal gland (dorsal and subventral) that are injected into root cells via the stylet, resulting in physiological and morphological changes in root cells (Abad et al., 2003). The esophageal glands of M. incognita are composed of two subventral gland (SvG) cells and one dorsal gland cell. The SvG cells are active in the pre-parasitic and parasitic stages, producing effectors required for root invasion. The dorsal gland cell is active during the sedentary stages, synthesizing proteins involved in feeding site development and maintenance (Davis et al., 2000). Once secreted from esophageal glands into root cells, effectors may be localized to different cellular compartments where they may assume diverse cellular functions to increase parasitism. The manipulation of host cellular functions, such as host transcription, chromatin remodeling, and immune responses, involves targeting of the host nucleus by secreted effectors (Quentin et al., 2013).
It is hypothesized that effector proteins participate in a diverse range of host processes during nematode parasitism. Nematode effectors work by interacting with host proteins or mimicking host proteins, and are capable of degrading and modifying plant cell walls (Smant et al., 1998); suppressing host defense responses; and targeting plant signaling pathways, such as those involved in the shikimate pathway and the ubiquitin-proteasome pathway (Chronis et al., 2013; Chen et al., 2015; Niu et al., 2016). In addition to triggering the development of GCs, effectors play a central role in suppressing host defense responses to facilitate infection (Haegeman et al., 2012).
A growing number of studies have revealed that some secreted effectors can target the host cell nucleus and manipulate essential host cellular processes such as transcription, chromatin remodeling, and histone modification (Kay et al., 2007; Clapier and Cairns, 2009). Many effectors contain nuclear localization signals (NLSs). The functional characterization of nuclear-targeted effectors from bacteria is well documented. For instance, the VirE2 of Agrobacterium, which contains a NLS, was involved in the transfer of T-DNA to the plant cell nucleus to modulate histone gene expression and exert infection (Tzfira et al., 2001; Citovsky et al., 2004). Another TAL effector AvrXa7 of Xoo can activate the accumulation of OsSWEET14, which can induce a sugar efflux to feed bacteria in the xylem and apoplasm (Chen et al., 2010). In Meloidogyne nematode, previous studies have identified one nuclear located effector, 7H08, through subcellular localization verification among 13 candidate NLS effectors that are expressed in the esophageal gland (Zhang et al., 2015). The M. javanica effector MJ-NULG1a was found to contain two NLSs, and immunolocalization analysis showed that MJ-NULG1a was localized in the nuclei of giant cells during nematode parasitism, and that it played a central role in M. javanica parasitism (Lin et al., 2013). In Heterodera schachtii, the NLS-containing effector 10A07 can localize to the nucleus and then specifically interact with the nuclear-localized transcription factor IAA16, leading to interference of auxin signaling. Overexpression of H. schachtii 10A07 in Arabidopsis thaliana increased sensitivity to H. schachtii infection (Hewezi et al., 2015). Mounting evidence suggests that nucleus-targeted effectors play important roles in weakening host resistance or enhancing pathogen virulence during plant–pathogen interactions. However, further studies on additional related effectors are required to elucidate the molecular mechanisms behind this process.
Benefits from advances in genomic, transcriptomic, and bioinformatic approaches, have led to progress in identification and functional characterization of some nematode effectors, but most effectors are still unidentified. In this study, by transcriptome analysis, we predict 110 candidate NLS effectors from M. incognita. Of 20 randomly selected effector candidates, three can strongly suppress B. glumae-caused fast, localized HR responses in non-host N. benthamiana. Two of these effectors are verified as localizing to the nucleus. Of which, MiISE6 is highly expressed in the early parasitism stages, and has a functional signal peptide. Utilizing transgenic technology and host-derived RNAi approaches, in addition to comparative transcriptome analysis, we try to determine the roles of MiISE6 in M. incognita parasitism.
Materials and Methods
Nematodes, Plants and Growth Conditions
Meloidogyne incognita nematodes were propagated on 3-week-old pepper (Capsicum annuum, Qiemen) plants in an artificial environment from a single female egg mass after isolated from the Sijiqing farm (Beijing, China). Three lines (Avir-1, Avir-2, and Avir-3) of the nematodes that each from a female egg mass were continually reproduced for 30 generations to form stable populations and then used for experiments. Infective, second stage juveniles were collected by hatching the handpicked egg masses at 26°C.
Nicotiana benthamiana was used for plant hypersensitive response (HR) assay and subcellular localization observation. Arabidopsis thaliana (Col-0 ecotype) was used for transgene, plant-nematode interaction and parasitism analyses. Both plants were cultivated in a growth chamber at 23°C.
Secreted NLSs Effector Prediction
Nematode genomes and transcripts used to predict effector proteins containing nuclear localization signals (NLSs) and for sequence homologue analysis in were downloaded from the National Center for Biotechnology Information (NCBI) (Supplementary Table S1). Signal peptide predictions were performed using SignalP 4.11, predisi2, and phobius3. Subcellular location of proteins was predicted by TargetP4, and the proteins located in mitochondria were removed. The prediction of transmembrane domain and NLS were performed using TMHMM5 and NucPred (Brameier et al., 2007) based on a sequence score ≥ 0.5. Domains were predicted in local PuTTY based on HMMER 3.1b1 (Wong et al., 2015) and the active conserved domains were blasted to the Pfam database (Finn et al., 2014). A phylogenetic analysis of homologues was carried out using the maximum likelihood method with MEGA 5 (Tamura et al., 2011).
Cell Death Suppression
The suppression of cell death elicited by B. glumae (from a gift of Dr. Sun, Chinese Agricultural University) on N. benthamiana leaves was performed as described in previous study (Sharma et al., 2013). Candidate effectors and GFP coding sequences were cloned into the pEDV vector to generate pEDV::effector and pEDV::GFP constructs, respectively. All constructs were introduced into B. glumae by electroporation, and the transformed bacteria were then suspended in 0.9% NaCl at 600 nm (OD600 = 0.4). Burkholderia glumae cells carrying pEDV::effector were infiltrated into the right side of N. benthamiana leaves using a 1 ml syringe. As controls, B. glumae cells carrying the pEDV::GFP were infiltrated into the left side of N. benthamiana leaves. Three leaves on 5 plants were used for each biological replicate, and at least three independent experiments were performed. Plants were kept at 25°C. Cell death was observed at 4 days post-inoculation. All primers were synthesized by Sangon and were listed in Supplementary Table S2.
Subcellular Localization
For subcellular localization assays, MiISE2Δsp, MiISE10Δsp, MiISE6Δsp, MiISE6Δsp_Δ109_157, MiISE6Δsp_Δ33_108 and MiISE6Δsp_MΔ109_118 were amplified by PCR from the M. incognita cDNA, using I-5TM 2 × High-Fidelity Master Mix (MCLAB, United States). PCR products were digested with Nco I and Spe I and inserted into pCAMBIA1302 digested with the same enzymes. The resulting plasmids harbored the effector gene fused with the GFP gene. The constructs were transformed into Agrobacterium tumefaciens GV3101. After inoculation in LB followed by horizontal shaking for approximately 30 h, A. tumefaciens cultures were diluted to an OD600 of 0.8 using infiltration medium (10 mM MES, 10 mM MgCl2 and 0.1 mM AS). Leaves at the four-leaf stage of N. benthamiana were infiltrated with bacterial inocula using a 1 ml syringe with no needle. After 24–48 h, the infiltrated zones of the leaf were observed under a confocal microscope (Zeiss LSM700, Germany).
Quantitative RT-PCR (q-PCR) Assays
RNA samples were extracted from 300 M. incognita at different life stages (pre-J2, parasitic (par)-J2, par-J3, par-J4 and female) for developmental expression analysis using RNAprep pure Tissue Kit (Tiangen, China). The 18S gene (Genbank accession no. U81578) was used as an internal control.
In order to confirm the results of RNA-seq analysis, Arabidopsis plants grow under same condition as those used for the RNA-seq analysis were collected for q-PCR assays. RNA samples were extracted from 100 mg frozen plant tissue using RNAprep pure Plant Kit (Tiangen, China). The Actin2 gene (AT3G18780) was used as an internal control.
Then, 1 μg RNA was used for cDNA synthesis using PrimeScriptTM RT reagent Kit with gDNA Eraser (Perfect Real Time) (Takara, Japan) following the manufacturer’s protocol. Each 25 μl reaction mixture was prepared using SYBR Premix Ex TaqTM||(TaKaRa, Japan) on a BIO-RAD CFX96 (BIO-RAD, United States) following the manufacturer’s protocol. Cycling conditions for quantitative PCR were: 95°C for 5 min and 40 cycles of 95°C for 30 s and 60°C for 30 s. Gene expression changes were calculated using the 2-ΔΔCT method. At least three independent experiments were performed, with four technical replicates for each reaction.
In Situ Hybridization
Parasitic stage J2s of M. incognita were processed for in situ hybridization to confirm the expression site of MiISE6 as previously described (de Boer et al., 1998). Digoxigenin-labeled sense and antisense cDNA probes were synthesized using the primers DIG-MiISE6-F and DIG-MiISE6-R. Hybridization signals within the nematode were detected with alkaline phosphatase-conjugated anti-digoxigenin antibody and substrate, and observed with an Olympus IX53 microscope.
Fluorescence in situ hybridization was performed using a 28 bp specific cDNA probe (CTTTCTTCAATTCCTT TCATCAACATCC) whose 5′ end was labeled with fluorescein isothiocyanate (FITC, Sangon Biotech) as previously described with some modification (Moter and Gobel, 2000), and the hybridization signals were observed under a confocal microscope (Zeiss LSM700, Germany).
Validation of the Functionality of Signal Peptide
We used the yeast secretion system based on the yeast signal sequence trap vector pSUC2T7M13ORI (Jacobs et al., 1997) to validate the function of MiISE6 signal peptide. PCR products were digested with EcoRI and XhoI and inserted into pSUC2T7M13ORI to generate pSUC2::MiISE6. The primer pairs used for PCR amplification were pSUC2_MiISE6_F and pSUC2_ MiISE6_R. The constructs were transformed into the invertase negative yeast strain YTK12 using the Frozen-EZ Yeast Transformation II kit (Zymo Research, United States). After transformation, yeast was plated on CMD-W (minus Trp) plates for 3 days. Single colonies of YTK12 were replicated onto raffinose-containing YPRAA plates (with raffinose instead of sucrose). Invertase enzymatic activity was detected with TTC as described by Oh et al. (2009).
Generation of Transgenic Arabidopsis Plants
For MiISE6 overexpression in Arabidopsis, the coding sequence of the MiISE6 gene without signal peptide was cloned into the vector pEGAD, and for host-derived gene silencing, the coding sequence of MiISE6 was inserted into the pFGC5941 vector in both sense and antisense orientations. There was an CHSA intron between sense and antisense fragments. Both pEGAD vector and pFGC5941 vector habor the BASTA selectable marker gene. The transformation of Arabidopsis was performed as previously described (Martínez-Zapater and Salinas, 1998). Transformed T1 seeds were sown on soil and were screened by spraying the herbicide BASTA 2 weeks later to select for transgenic plants. The spraying was repeated three times. Homozygous T3 seeds were used in this research. The expression level of MiISE6 in different transgenic line were confirmed by RT-PCR, RNAi transgenic lines was confirmed by checking the CHSA intron fragment using primer CHSA-F/CHSA-R.
Nematode Infection Assays
Four-week-old Arabidopsis were inoculated using 400 J2 of M. incognita per plant. 42 days after inoculation, the number of females was counted.
At least 15 plants were used for each treatment, and three independent experiments were performed. Statistically significant differences between treatments were determined by independent samples t-test (P < 0.05) using SPSS.
RNA Sequencing and Transcriptome Analysis
For M. incognita transcriptome: 50 mg of freshly collected nematodes were used for RNA extraction using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. At least 20 mg of total RNA was sent to the Shenzhen Genomics Institute for Solexa sequencing (commercial service, Shenzhen China), and three independent replicates (Avir-1, Avir-2, and Avir-3) were sequenced. The full genome sequences and gene annotation of M. incognita were downloaded from Wormbase as a reference genome (Abad et al., 2008). The RNA-seq reads were mapped to the reference genome using Tophat and subsequently assembled using cufflinks (Trapnell et al., 2010). The expression abundance of each gene was estimated with fragments per kilobase of transcript per million fragments mapped (FPKM) (Anders et al., 2015).
For A. thaliana transcriptome: The 14-day transgenic Arabidopsis and WT Arabidopsis grown on Murashige and Skoog (MS) medium were used for RNA extraction. RNA samples were extracted from 100 mg frozen plant tissue using RNAprep pure Plant Kit (Tiangen, China). Library construction and RNA-Seq were performed at BerryGenomics (Beijing, China), and four independent replicates were sequenced. The cDNA libraries with average insert sizes of 280 bp were sequenced by Illumina technology. The adaptor sequences and low-quality reads were filtered from the raw data. Tophat2 tools were used to map mRNA reads to the Arabidopsis genome (Langdon, 2015), and htseq-count was then used to calculate the expected FPKM mapped (Anders et al., 2015), and the edgeR package in R was applied to identify DEGs of 2 groups of samples with a threshold criterion of (FDR < 0.01) and |log2FC|≥ 1. We used AgriGO6 and KOBAS (Xie et al., 2011) to perform GO functional enrichment analysis and KEGG enrichment analysis with default parameters.
Results
Transcriptome Sequence Assembly and Prediction of Effectors Containing Secreted Nuclear Localization Signals (NLSs)
In total, 8.22 billion clean base pairs were generated from three independent replicates of M. incognita wild type population, with Illumina/Solexa sequencing reads of ∼70x of M. incognita genome. Of these, 13398 genes (95.90% of total) were shared by the three replicates (Supplementary Figure S1); detailed information is shown in Supplementary Datasheet S1. By a series bioinformatic analysis, such as searching out the sequences with signal peptides and discarding the sequences with putative transmembrane-spanning regions, analyzing subcellular location and removing proteins located in mitochondria, NLS motif predicting, totally 110 potential NLS effectors are predicted from M. incognita, and they are taken as candidate effectors with putative immune function for further analysis. Detailed information is shown in Supplementary Datasheets S1, S2.
Immune Suppression Studies on Putative Effectors
Previous studies have shown that PPN effectors can suppress the plant hypersensitive response (HR) to facilitate parasitism (Niu et al., 2016; Chen et al., 2017). Here we used a pEDV-based type III secretion system (T3SS) in which B. glumae is used to deliver effectors to plant cells to verify the function of 20 randomly selected novel effector candidates in plant immune suppression. Burkholderia glumae is a bacterial rice pathogen that can cause a fast, localized cell death in non-host N. benthamiana. We also cloned a green fluorescent protein gene (GFP) into the vector as a negative control, and HR symptoms were monitored within 3 days after agroinfiltration. Screening results 3 out of 20 candidate effectors had strong HR suppression ability, including MiISE2 (Minc04520), MiISE10 (Minc08615), and MiISE6 (Minc06775) (Figure 1).

FIGURE 1. Putative M. incognita effectors suppress B. glumae-induced HR in N. benthamiana. The N. benthamiana leaves were photographed within 4 days after B. glumae inoculation to observe the cell death symptoms. The left half leaf sections were injected with B. glumae carrying pEDV::GFP and the right half sections were injected with B. glumae carrying pEDV::effector. The volume of bacteria solution injected in both side were equivalent. Numbers on the pictures, for example 19/25, indicate that 19 right leaf sections out of 25 inoculated leaves showed no or significantly less severe symptoms as compared with left sections.
Subcellular Localization of Three Candidate Effectors
To investigate the subcellular localization of the three candidate effectors in plant cells, the green fluorescent protein (GFP) gene was fused with the effector (without signal peptide and stop codon) at its C terminus with the 35 S promoter, and the effector proteins were then transiently expressed in N. benthamiana. Confocal microscopy revealed distinct subcellular localization of the 3 candidate effectors. MiISE2 shows nuclear-cytoplasmic localization; MiISE10 appears to localize in the cytoplasm; and MiISE6 is enriched in the nucleus (Figure 2A). The subcellular localization of MiISE2 and MiISE6 are consistent with the predictions by NucPred. Moreover, we used q-PCR to quantify the expression levels of three effectors during pre-parasitic J2 (pre-J2), parasitic J2 (par-J2), J3 and Female stages. The candidate effectors MiISE2 and MiISE10 have a high expression level in late parasitism stages (J3 and Female), but they exhibit a very low expression level in early parasitism stages. Conversely, the candidate effector MiISE6 is upregulated in early sedentary stages, reaching a peak in the early par-J2 at 1 dpi, and then undergoing a dramatic reduction in the J3 parasitism stage (Figure 2B). The expression pattern of MiISE6 is similar to other published nematode effectors which play central roles in nematode parasitism (Chronis et al., 2013; Lin et al., 2016; Chen et al., 2017), so we focus on MiISE6 and speculate it may play an important role during the initial establishment of the plant-nematode interaction.
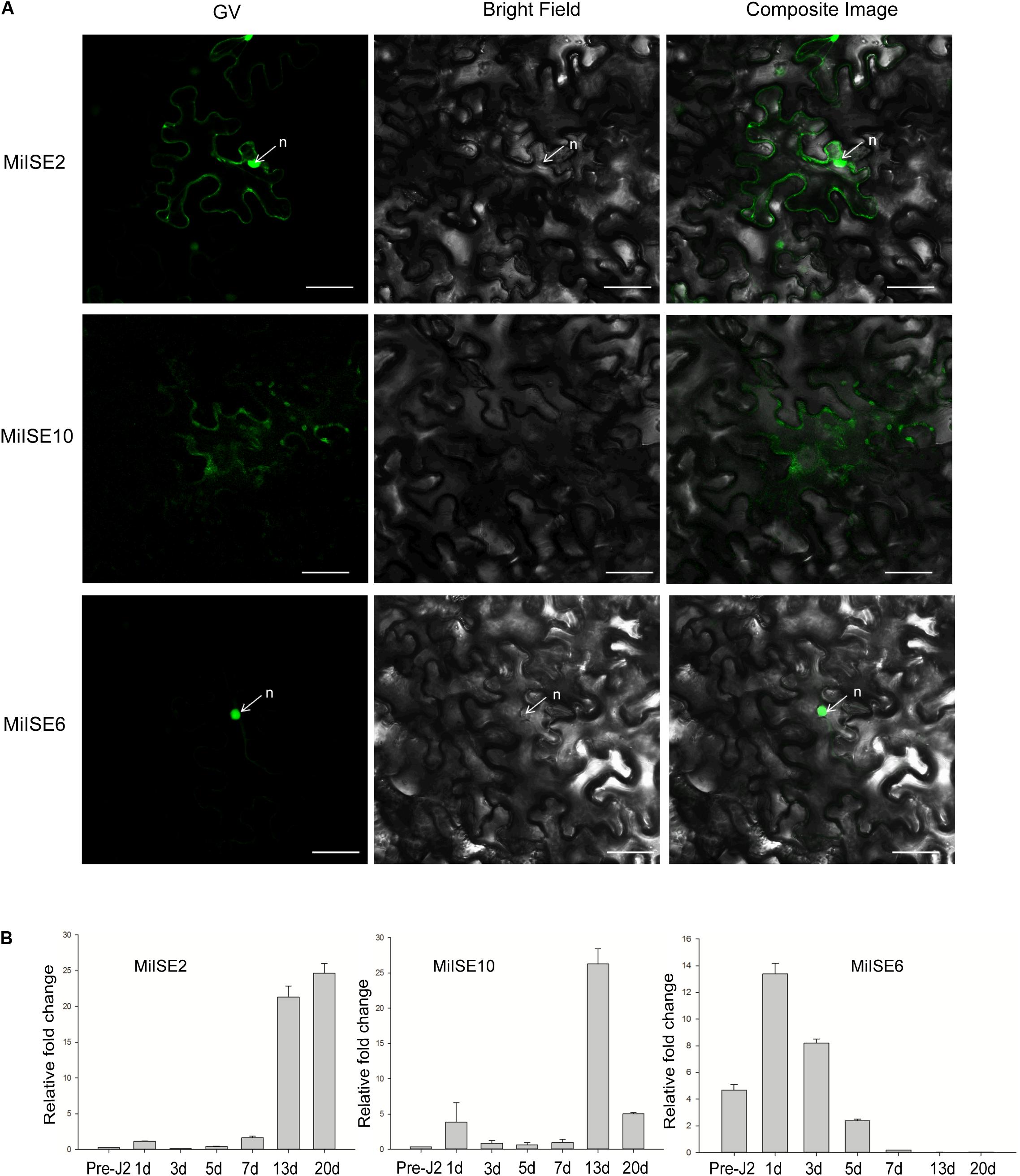
FIGURE 2. (A) Subcellular localization of C-terminally GFP-tagged candidate NLS effectors in N. benthamiana leaves. N. benthamiana leaves were monitored using confocal microscopy 24–48 h after transfection with effector::GFP construct. (B) The developmental expression of these effectors. The relative expression of effectors was quantified using q-PCR in four different M. incognita life stages: pre-J2s, par-J2s (1–3 days), J3s (7–13 days), and J4s (20 days). Each bar represents the mean plus SD of three biological replicates; with four technical replicates for each independent experiment. The 18S gene was used as an internal control. n, nucleus. Scale bars = 10 μm.
Sequence and Homolog Analysis of MiISE6 From M. incognita
The M. incognita MiISE6 cDNA encodes a 158 amino acid protein with a predicted N-terminal signal peptide (SP, residues 1–32) for secretion; an OGFr_N region (residues 35–62); and two NLS motifs (residues 89–98, 109–118) (Supplementary Figure S2). A search of homologues of MiISE6 from 16 nematodes species (including 4 free living nematodes, 6 animal or insect parasitic nematodes, and 6 plant parasitic nematodes) (Supplementary Table S1) was performed with BlastP (E ≤ 1e-10), and we obtained 20 homologues (Supplementary Table S3). These homologs were not only distributed in plant parasitic nematodes, but also in animal or insect parasitic nematodes and free-living nematodes. Based on the amino acid sequences of MiISE6, a phylogenetic tree was constructed in MEGA5 using the maximum likelihood method with 1000 bootstrap replicates. MiISE6 was closest to the homologue from M. hapla, and they clustered with MiISE6 homologues from Globodera pallida (Figure 3).
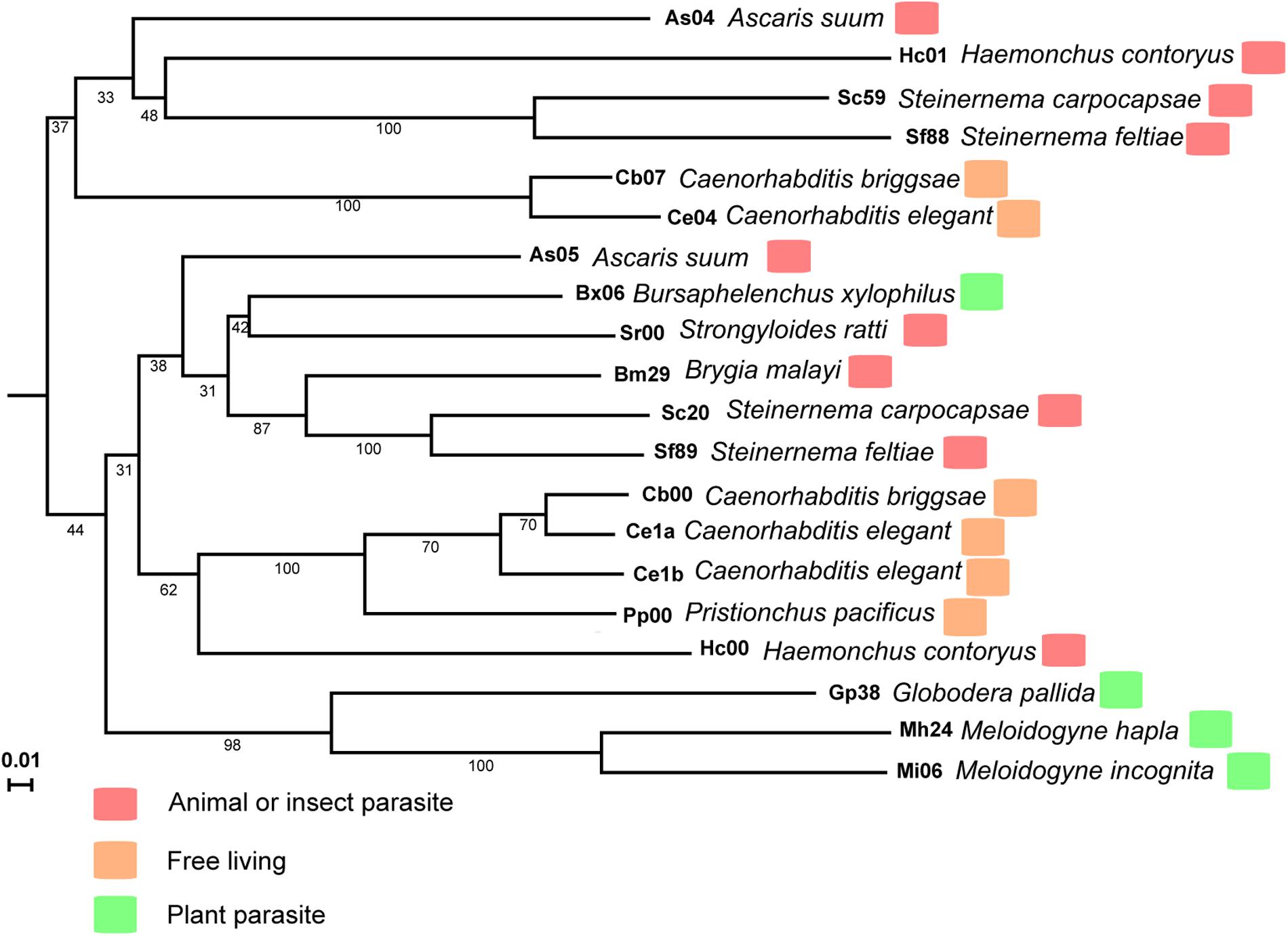
FIGURE 3. The phylogenetic tree of MiISE6 homologues inferred using the maximum likelihood method. The bootstrap value is given on each node.
Identification of Nuclear Localization Domains in MiISE6
Sequence analysis revealed that MiISE6 harbors two putative NLSs including 89GMQKRKRALD98 and 109FDRKRRALDM118. To test which NLS play function in nuclear localization, we firstly generate two constructs MiISE6Δsp_Δ109_157:GFP and MiISE6Δsp_Δ33_108:GFP, which contain NLS89-98 and NLS109-118, respectively. As shown in Figure 4, the MiISE6Δsp_Δ109_157:GFP localized to the cytoplasm (Figure 4A), whereas MiISE6Δsp_Δ33_108:GFP localized to the nucleus in N. benthamiana leaves (Figure 4B). The results indicate that the predicted NLS89-98 motif may not responsible for the nuclear localization of MiISE6. To determine whether NLS109-118 motif play role in nuclear localization, we used site-directed mutagenesis to generate MiISE6Δsp_MΔ109_118:GFP, and the fluorescence accumulation was observed in cytoplasm, and no fluorescence was observed in the nucleus (Figure 4C). These results indicate that the NLS109-118 lead to the nuclear localization of MiISE6.
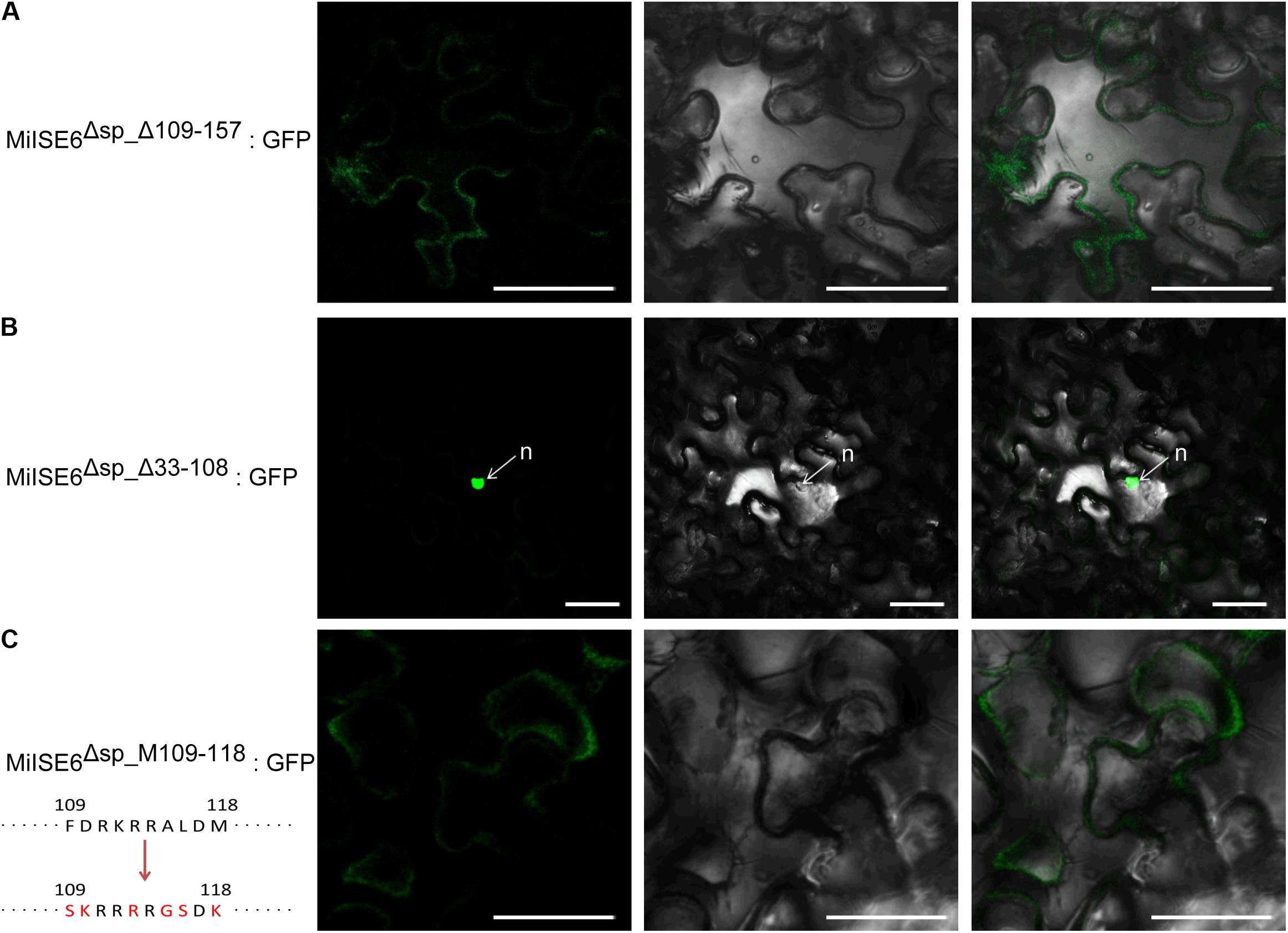
FIGURE 4. Identification of nuclear localization domains in MiISE6. C-terminally GFP-tagged MiISE6Δsp_Δ33_108, MiISE6Δsp_MΔ109_118, and MiISE6Δsp_MΔ109_118 were transiently expressed in N. benthamiana leaves, respectively. N. benthamiana leaves were monitored using confocal microscopy 24–48 h after transfection. (A) Cytoplasmic localization of MiISE6Δsp_Δ109_157:GFP. (B) Nuclear localization of MiISE6Δsp_Δ33_108:GFP. (C) Cytoplasmic localization of MiISE6Δsp_MΔ109_118:GFP. There was 5 amino acid substitution (red colored) in the NLS109-118 of MiISE6Δsp_MΔ109_118compared to MiISE6Δsp. n, nucleus. Scale bars = 10 μm.
MiISE6 Is Expressed in Subventral Esophageal Gland Cells
The tissue localization of MiISE6 transcripts in the nematode was verified using in situ hybridization. With digoxigenin-labeled antisense cDNA probes of MiISE6, there is a strong signal within the subventral gland cells of the pre-parasitic J2s. No signals are detected using the sense cDNA probes. Consistent with the result of DIG-labeled in situ mRNA hybridization, strong fluorescent signals are observed in the subventral esophageal gland by fluorescence in situ hybridization, and there is no tissue specificity when dyed with DAPI (Figure 5).
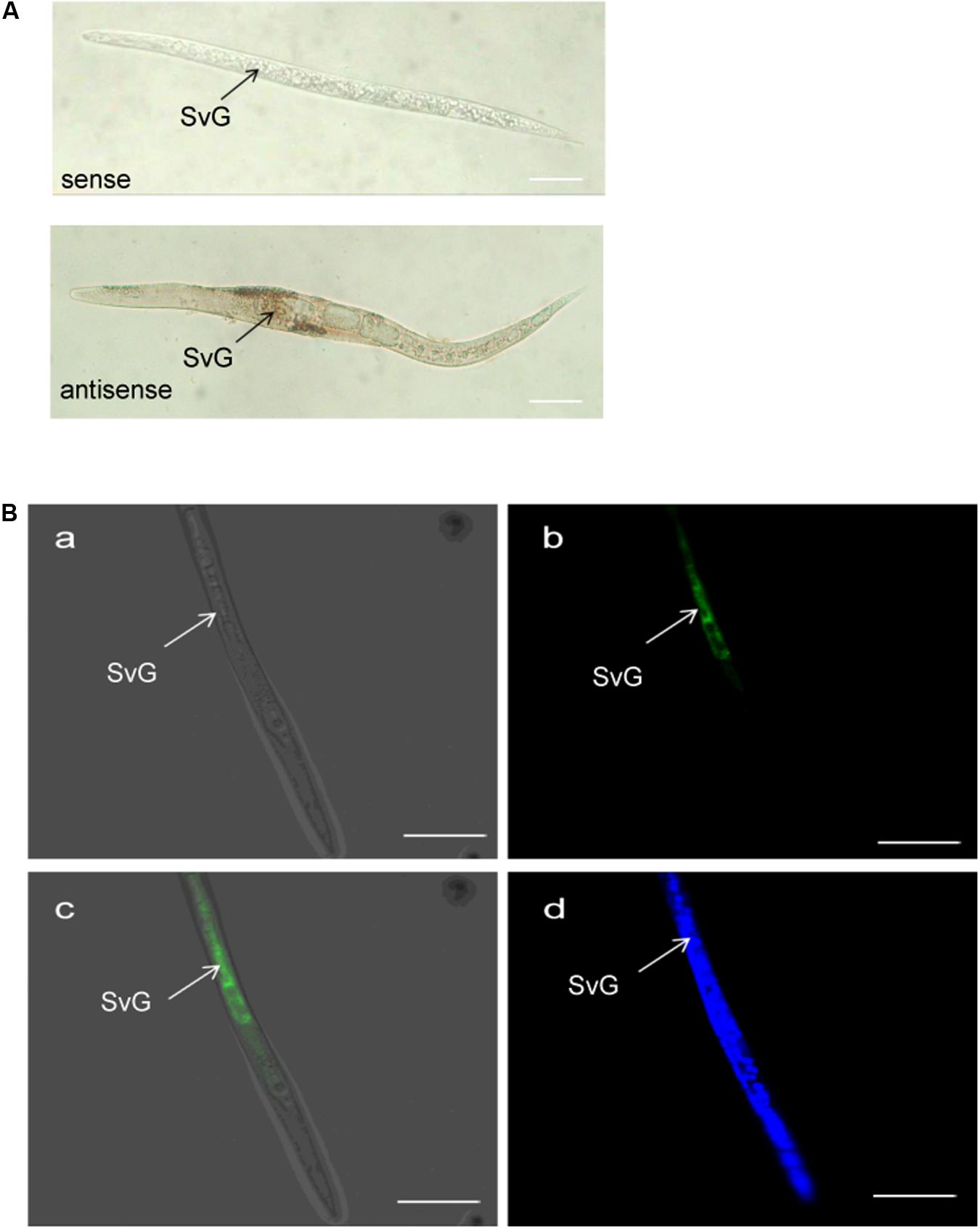
FIGURE 5. Spatial expression of MiISE6. (A) Digoxigenin-labeled in situ mRNA hybridization. (B) FITC-labeled in situ mRNA hybridization. Both digoxigenin-labeled antisense MiISE6 cDNA probe and 5′ end labeled with FITC cDNA probe localized MiISE6 transcripts within the subventral gland cells. (B,a) Bright Field. (B,b) GFP channel. (B,c) Composite Image. (B,d) DAPI channel.
MiISE6 Contains Functional Signal Peptide
We used a yeast system requiring invertase secretion to grow on raffinose media to validate the function of the signal peptide of MiISE6. The predicted signal peptide sequence and the subsequent 24 amino acids of MiISE6 was firstly fused with the SUC2 minus the signal peptide and then inserted in vector pSUC2T7M13ORI (Jacobs et al., 1997). The final construct enables the successful secretion of invertase, and then the invertase mutant yeast strain YTK12 can hydrolyze raffinose and grow on YPRAA media. The invertase secretion was confirmed with 2,3,5-triphenyltetrazolium chloride (TTC), which reacts with monosaccharides to form the insoluble (red colored) triphenylformazan. Compared with the negative control, the fusion of signal peptide of Avrblb2 and MiISE6 to the SUC2 enabled the secretion of invertase, which lead to the yeast cells growing on YPRAA media (Figure 6). These results revealed that the MiISE6 protein habor a functional signal peptide.
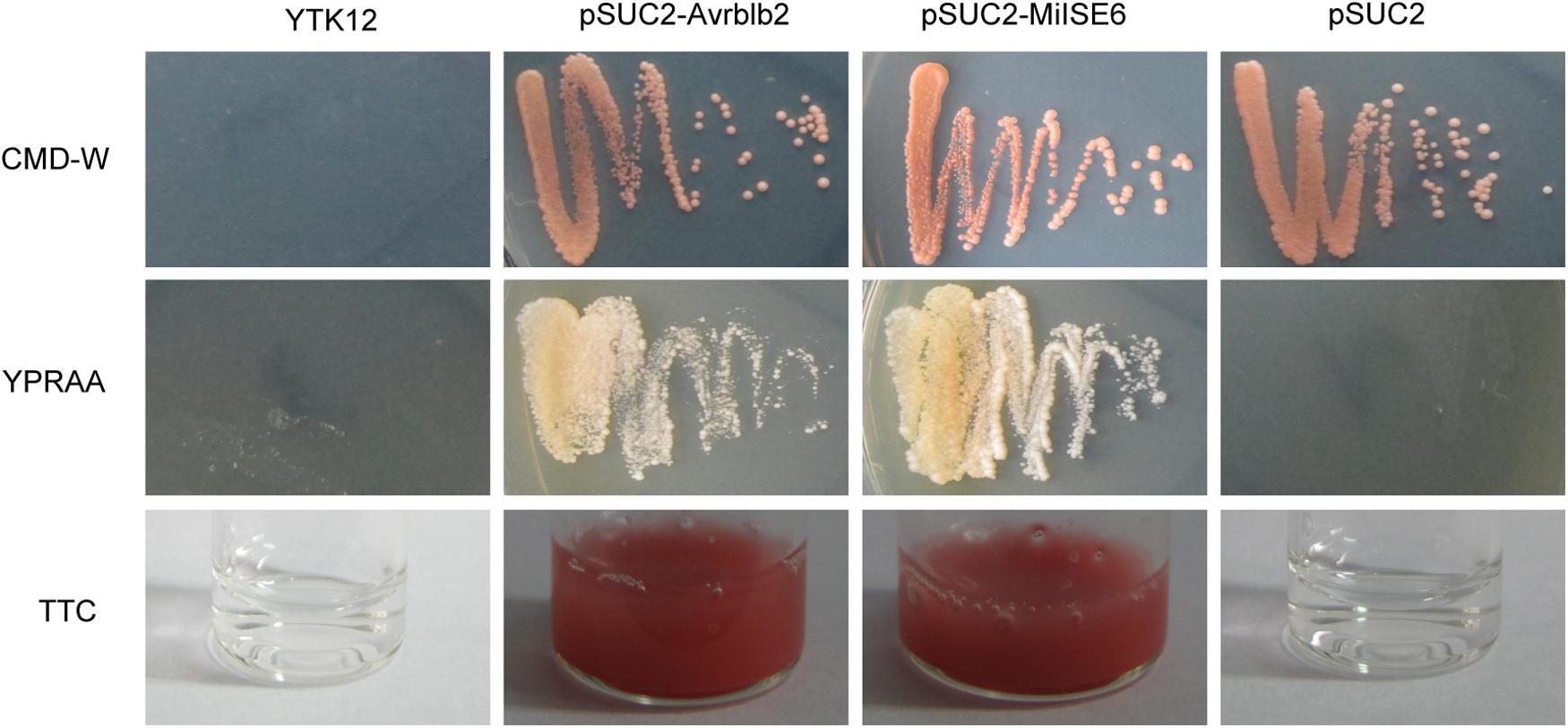
FIGURE 6. Functional validation of the signal peptides of MiISE6. Functional validation of the signal peptide of MiISE6 was performed using the yeast invertase secretion assays. Yeast YTK12 strain carrying the Avrblb2 and MiISE6 signal peptide, which expresses two signal peptide fragments fused in frame to the mature invertase gene SUC2, is able to grow in both the CMD-W media and YPRAA media (with raffinose instead of sucrose), as well as react with TTC and turn into red formazan, indicating secretion of invertase. YTK12 carrying the pSUC2 vector was used as negative control.
MiISE6 Increases M. incognita Parasitism
To determine whether the expression of MiISE6 in Arabidopsis can change plant phenotype and nematode susceptibility, we generated transgenic Arabidopsis lines expressing ΔspMiISE6 driven by the CaMV35S promoter. Three independent transgenic lines (T_1, T_2, T_8) were selected for phenotype observation and the nematode inoculation. We firstly detected the expression of transcripts in independent transgenic lines by RT-PCR. The photo (Figure 7A and Supplementary Figure S3) shows that MiISE6 gene is really expressed in Arabidopsis plants. Then, we counted the number of root-knots in each transgenic plant, and compared the difference between each transgenic line and the wild-type plants by statistical significance test. The results show that there are more root-knots in each transgenic line plants than those in wild-type plants (mean 23–25 vs. 18–19) (Supplementary Figure S4). The difference is significant at a statistic level (all of the three tests with P < 0.001, Supplementary Figure S5). It indicates that all three transgenic lines are more susceptible to M. incognita than control, and there is an increase in females with an average rate of 25.15, 35.26, and 38.80%, respectively (Figure 7B). To further assess the role of MiISE6 in nematode parasitism, we used a host-derived gene silencing method to knock down the expression of MiISE6 in M. incognita during the nematode infection. Three RNAi lines (RNAi1, RNAi5, RNAi11), with at least 15 plants for each treatment, and three independent experiments. By RT-qPCR assays, it is shown that the expression of MiISE6 in M. incognita collected from inoculated on RNAi plants at 3 dpi is significant reduced compared with those from the wild-type (WT) plants (Supplementary Figure S6 and Figure 8A). We also counted the number of root-knots in each RNAi plant and wild-type plant. Statistical analysis showed that there is an obvious reduction on RNAi plants (mean 13-15 knots) (Supplementary Figure S7), compared to those on wild type plants (mean 21). There was a 29.16, 35.26, and 30.86% reduction of females in three RNAi plants, respectively (Figure 8B). The results of t-tests show that the difference is significant (P < 0.001, Supplementary Figure S8). These data indicate that MiISE6 is an important effector in mediating parasitic success of M. incognita.
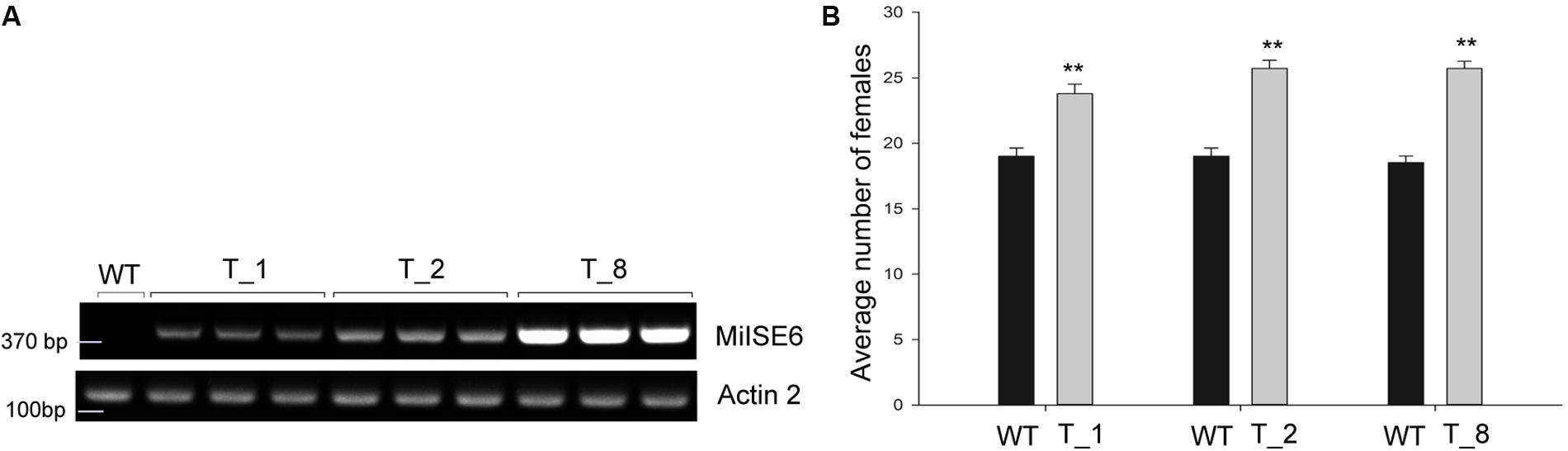
FIGURE 7. The overexpression of MiISE6 in Arabidopsis promotes nematode susceptibility. (A) RT-PCR assays were used to determine the expression of MiISE6 in three independent transgenic lines with Actin2 as reference. Upper image, agarose gel image of MiISE6; Lower image, agarose gel image of Actin2. (B) There was an increase of females in the T_1, T_2, and T_8 transgenic lines compared with WT Arabidopsis (Col-0). Independent samples t-tests were used for significance test between treatments, with a 95% confidence interval. Error bars represent standard Error (SED), and the mean values significantly different from the control are demoted by ∗∗ (∗∗P < 0.01). The experiments were performed with three independent replicates.
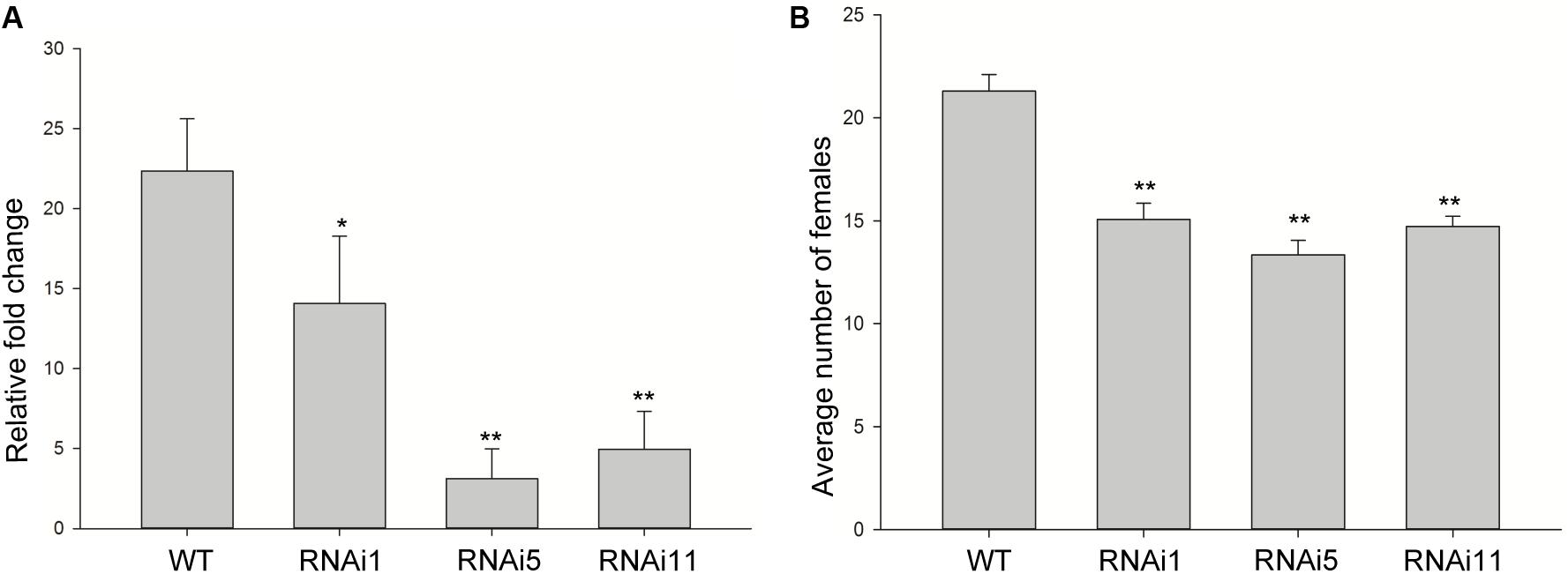
FIGURE 8. Host-derived MiISE6 silencing effect on nematode parasitism. (A) q-PCR assays were used to determine the expression level of MiISE6 in WT and three independent RNAi lines. (B) There was a reduction of females in three RNAi lines (RNAi1, RNAi5, RNAi11) compared with control plants. Independent samples t-tests were used for significance test between treatments, with a 95% confidence interval. Error bars represent standard Error (SED), and the mean values significantly different from the control are demoted by ∗ (∗P < 0.05, ∗∗P < 0.01). The experiments were performed with three independent replicates.
Identification of Differentially Expressed Genes (DEGs) Between MiISE6 Transgenic Plants and WT Plants
The difference in expression level of DEGs between WT and MiISE6 transgenic Arabidopsis plants was calculated according to the FPKM method. Under the criteria of |log2FC|≥ 1 and FDR ≤ 0.01, a total of 852 genes were identified in MiISE6-overexpressing samples compared with WT samples, of which 772 were upregulated and 80 were downregulated. Detailed information is provided in Supplementary Datasheets S3, S4.
GO and KEGG Enrichment Analysis of the DEGs
To functionally classify the DEGs, gene ontology (GO) terms were assigned to each DEG using AgriGO based on the TAIR 10 database. DEGs are categorized into three groups: Biological Process (BP), Cellular Component (CC), and Molecular Function (MF). The majority of the DEGs are assigned to the BP category, such as cellular process, metabolic process, and response to stimulus. In addition, we find that DEGs are significantly involved in binding, catalytic activity, and transcription regulator activity in the MF category. The results also show that in the CC category, the majority of the DEGs are significantly related with cell part and organelle (Figure 9). The expression of MiISE6 in Arabidopsis leads to changes in multiple plant immune responses, including hormones signaling pathway, ubiquitination proteasome pathway, and calcium signaling pathway (Supplementary Table S4). In “regulation of defense response (GO:0031347)”, most enriched genes work as negative regulator of plant defense responses, such as WRKY40, WRKY18, and NUDT7(Yang et al., 2006; Ge et al., 2007; Pandey et al., 2010). In “protein ubiquitination (GO:0016567),” most enriched genes are plant U-BOX E3 ubiquitin ligases (PUBs). The U-Box domain mediates the binding to the conjugating enzyme (E2) during ubiquitination process. Previous study showed that there was a reduced bacteria infection in pub22/pub24 double mutant and pub22/pub23/pub24 triple mutant compared to control. Although the molecular mechanism is not well clarified, previous studies indicated that those genes may act as negative regulator of PAMP-triggered immunity by altering the activity of 26S proteasome (Stegmann et al., 2012; Cho et al., 2015). In “response to hormone stimulus,” genes involved in modulating multiple hormone signaling pathway, including JA signaling (GO:0009867), SA signaling (GO:0009751) and auxin signaling (GO:0009733). Moreover, we find that a number of genes are enriched in “cell wall modification (GO:0042545),” such as xyloglucan endotransglucosylase-hydrolase (XTHs) and expansin-like gene (EXLA). Those genes involved in the reorganization or loosening of cell walls (Cosgrove, 2000; Olsen and Krause, 2017). Recent studies have suggested that diverse pathogen acquired and exploited these plant derived enzymes for successful parasitism (Balestrini et al., 2005; Nikolaidis et al., 2014; Olsen and Krause, 2017). Our result shows that most of those genes exhibit a significant higher expression level in the MiISE6 transgenic plants compared to control. Based on the present data, we draw a model illustrating the interplay between potential DEGs involved in host defense responses (Figure 10).
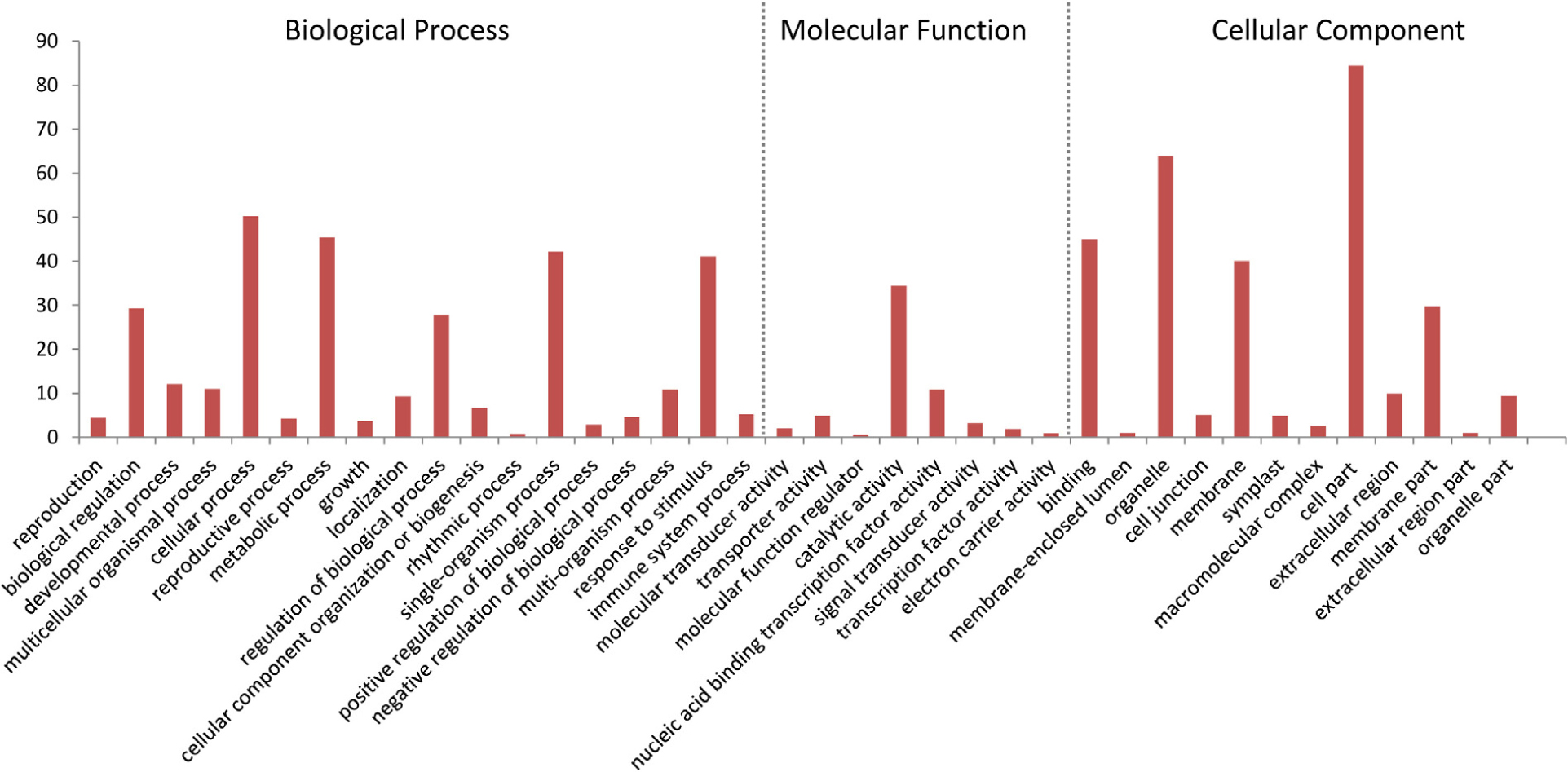
FIGURE 9. Gene ontology (GO) distribution of DEGs. All of the DEGs were assigned to three categories: cellular component, molecular function, and biological process.
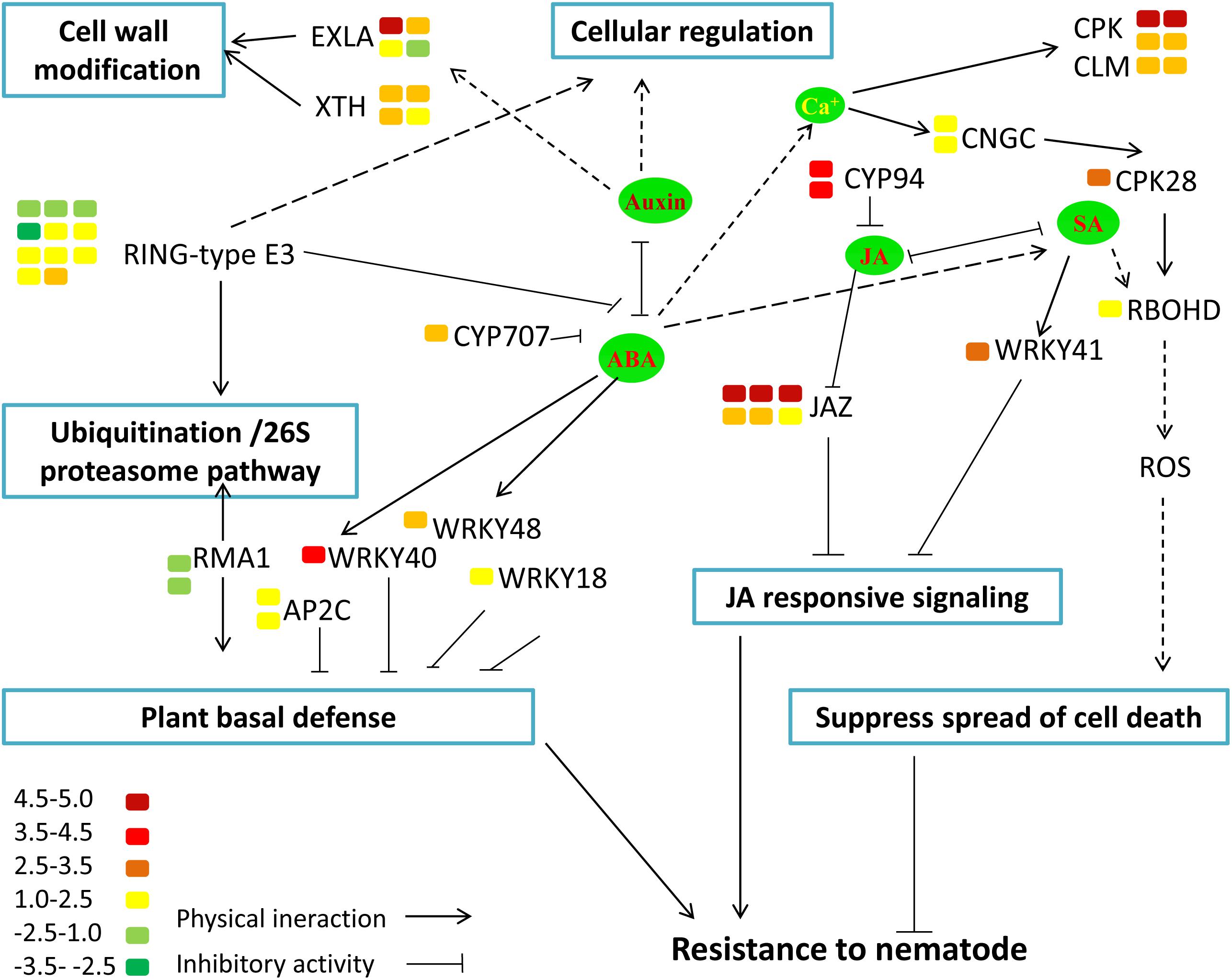
FIGURE 10. Model illustrating the interplay between potential DEGs involved in host defense responses. The color bar represents the expression level (log2FC) and number of DEGs.
Moreover, the results of KEGG enrichment analysis indicate that a total of 228 annotated DEGs are assigned into 25 pathways. As shown in Supplementary Table S5, these pathways are mainly classified into 8 categories (P ≤ 0.05). The most significantly enriched KEGG pathway is “Plant–pathogen interaction,” and most genes enriched in this pathway are calmodulin-related proteins or calcium dependent kinases, such as CML16, CPK29, CPK28. The expression of CPK28 has a 5.3-fold increase (log2FC = 2.31) in the MiISE6 transgenic plants compared to the WT plants. Previous studies have proven that CPK28 can interact with phosphorylate BIK, and contribute to BIK1 turnover (Monaghan et al., 2014, 2015); the overexpression of CPK28 in Arabidopsis inhibits PTI signaling and immunity (Matschi et al., 2015). Moreover, the NADP oxidase RBOHD is also enriched in the “Plant–pathogen interaction” pathway and presents a higher expression in the transgenic plants, and the expression of SA-responsive genes EDS1, PR1, PR2, and PR5 did not show significant differences in the transgenic plants compared to control. Additionally, in the KEGG pathway “Plant hormone signal transduction,” most enriched genes are JAZ genes, which work as repressors of jasmonate signaling (Chini et al., 2007), and they all showed significantly higher expression in the transgenic plants compared to control. Conversely, the JA-responsive genes PR4 and PDF1.2 showed a significant decrease in the transgenic plants. The expression patterns of these genes were verified by q-PCR, and the results are similar to the transcriptome analysis (Figure 11).
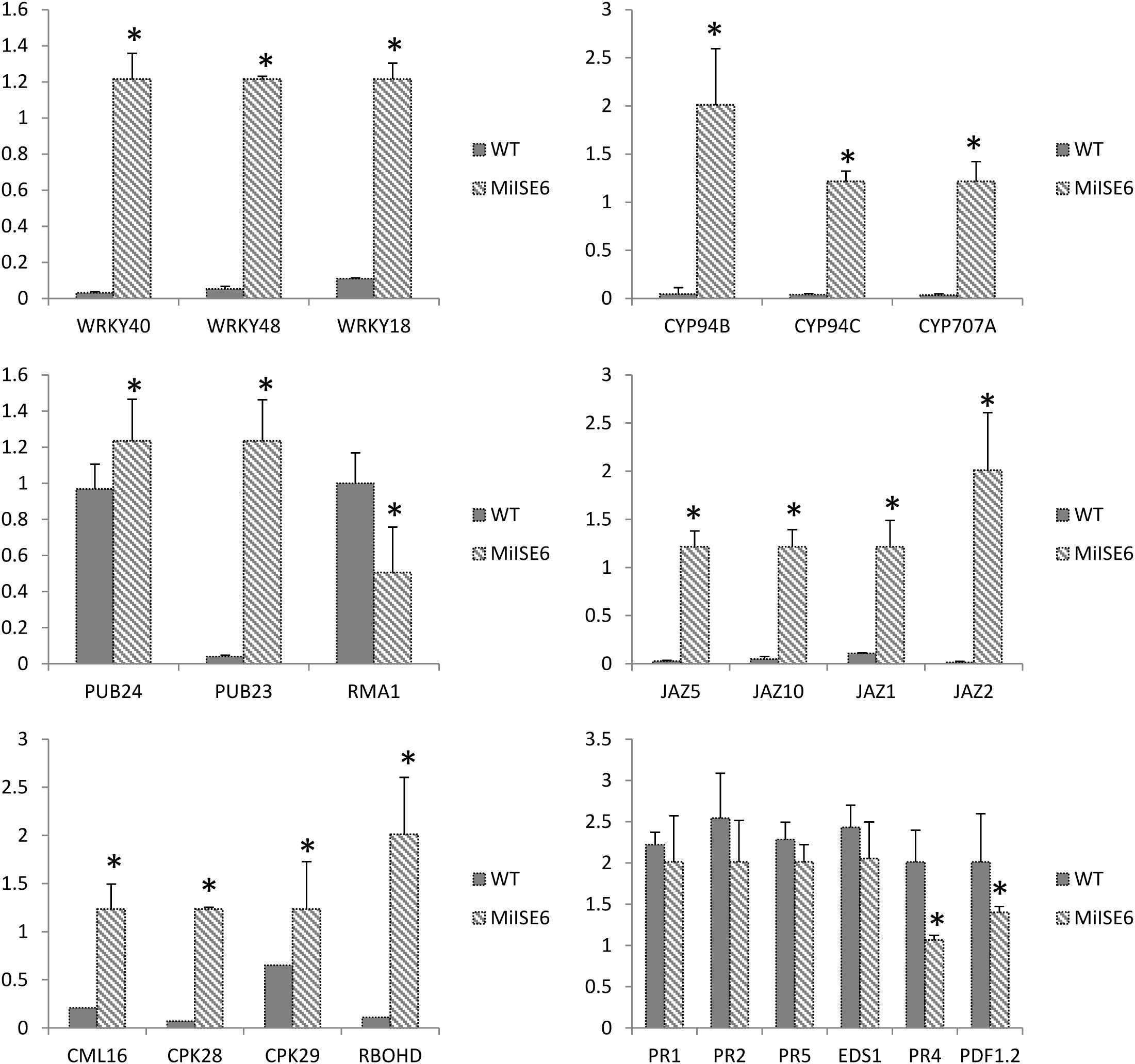
FIGURE 11. Verification of gene expression patterns in WT and MiISE6 transgenic Arabidopsis. 23 DEGs were selected to validate the reliability of the RNA-seq results. Actin2 (AT3G18780) was used as an internal control to normalize the expression of the genes in each corresponding q-PCR samples. Each bar represents the mean plus SD of three biological replicates, with four technical replicates for each independent experiment, and the mean values significantly different from the control are demoted by ∗ as determined by independent samples t-test (P < 0.05).
Discussion
Studies on identification of Meloidogyne spp. pathogenicity-related effectors have been conducted since Meloidogyne genome sequences were first reported. Most of these effectors are synthesized in the esophageal gland and subsequently delivered into plant cells and tissues through the stylet, including cell wall modifying enzymes (Smant et al., 1998; Bera-Maillet et al., 2000), antioxidant enzymes, and some enzymes that function in suppressing host defenses and manipulating plant signaling pathways (Doyle and Lambert, 2003; Dubreuil et al., 2011). Previous studies revealed that Meloidogyne effectors may target different subcellular compartments to counteract plant defenses, and a significant number of these effectors are specifically localized to the plant cell nucleus, such as 7H08, Msp40 and MgGPP (Zhang et al., 2015; Niu et al., 2016; Chen et al., 2017). Increasing evidence suggests that nuclear localized pathogen effectors play a central role in parasitism, by co-opting the host cell nuclear transport system and hijacking the host transcriptional machinery to subvert plant immunity (Kalderon et al., 1984a,b; Garcia-Rodriguez et al., 2006; Kay et al., 2007; Weinthal et al., 2011). In order to screen for more NLS effectors, in this study, based on RNA-seq and bioinformatics analysis, we predicted 110 putative secreted NLS effectors. From 20 randomly selected candidates, we identified three of them (MiISE2, MiISE10, and MiISE6) have strong HR suppression ability by utilizing a pEDV-based type III secretion system. We found the three candidates have distinct subcellular localization in N. benthamiana, and distinct expression in different stages of the nematode. We finally selected MiISE6 for function verification, because it can target the host nucleus, and is predominantly upregulated in the parasitic J2 stage. Our results confirmed MiISE6 as an effector involved in plant-nematode interaction, which can enhance the nematode parasitism by interfering multiple signaling pathways of plants, especially suppression of JA signaling pathway. The other two (MiISE2, MiISE10) are waiting for functional verification. Moreover, in this study, only 20 candidates were randomly selected for functional assay, and a large part of predicted NLS effector candidates are under exploring. We believe more effectors in M. incognita will be identified in the future. Furthermore, transcriptome datasets in our study may provide useful information for further study in nematode–plant interactions.
Sequence analysis of MiISE6 showed that it contains a signal peptide, an OGFr_N domain and two NLS motifs. We have determined the secreted activity of SP and nuclear localization of NLS motif. About OGFr_N domain, it is found in the opioid growth factor receptor (OGFr), which is an integral membrane protein associated with the nucleus and plays important roles in the regulation of cell growth (Zagon et al., 2002). OGFrs are widely reported in the phylum in human, rat and mouse cells (Zagon et al., 1994, 1999; Kren et al., 2015). It was reported that the OGFr was initially expressed on the outer nuclear envelope, and can transport into the nucleus through the nuclear pore (Zagon et al., 2003, 2005; Cheng et al., 2010). Previous studies showed that in sequence of OGFr there is one or more NLS accompanied with OGFr_N domain, which respond for the nuclear transporting of OGFr genes (Cheng et al., 2009). Based on the conserved domain analysis, we found that MiISE6 was a putative OGFr gene, containing an OGFr_N domain and subsequent two NLS motifs, and one motif NLS109-118 is confirmed to work for entering nuclei of plant cells by using sequence deletion variants and site-directed mutagenesis. Sequence analysis showed that identity of protein sequences between MiISE6 and homologs of M. hapla and Globodera pallida are more than 68 and 46%, respectively. We suppose that similar efforts of MiISE6 may also exist in these two nematodes, perhaps it is a common parasitism mechanism in sedentary PPNs.
Sedentary PPNs establish long term relationships with their hosts. Based on the current studies and previous literature, mechanisms of these effectors in parasitism has been elucidated that PPNs can manipulate plant hormone to facilitate successful invasion or establish feeding cells. When plants were invaded by PPN, plant hormones have pivotal roles in the regulation of immune responses toward attack. Nematode induce feeding cells by manipulating many aspects of plant development, which involve auxin transport and plant cell differentiation pathways. It was reported that the effector Hs19C07 of H. schachtii can interact with an auxin influx transporter LAX3 of plant, and the overexpression of this gene in Arabidopsis increased lateral root emergence, indicating a modulating of auxin influx into root cells (Lee et al., 2011). Another H. schachtii effector 10A07 can directly target IAA16 to manipulate auxin responses. Remarkably, the 10A07 transgenic plants showed hypersusceptibility to nematode infection (Hewezi et al., 2015). Auxin is known to involve in cell expansion and cell wall breakdown, accompanied by changes in the expression of plant cell wall modifying proteins (Szakasits et al., 2009), which is important for development of feeding structure. In this study, we found that the expression of some expansin-like genes and xyloglucan endotransglucosylase-hydrolases are obviously upregulated in transgenic Arabidopsis. Expansions are proposed to breakdown the adjacent between xyloglucans and cellulose, resulting in plant cell wall loosing (Cosgrove, 2000). Similar to expansin-like genes, xyloglucan endotransglucosylase-hydrolases also work in loosing cell walls by catalyzing the cleavage and rejoining of the xyloglucans to the primary cell wall (Van Sandt et al., 2007). In addition, the overexpression of XTHs in plants is compatible with a potential role in xyloglucan degradation, which contributing to wall extension (Baumann et al., 2007). We speculate that the expression of MiISE6 in Arabidopsis may accelerate the breakdown of cell wall through interfering the auxin pathway, which help nematode migrating and giant cell formation in plant roots.
Previous studies have identified the signaling molecules jasmonic acid (JA) and salicylic acid (SA) as important players in induced defense of the plant against pathogens, and the JA and SA signaling can be exploited by pathogens to facilitate parasitism (Gutjahr and Paszkowski, 2009). It was found that a H. glycines effector 10A06 can stimulate polyamine biosynthesis by interacting with Spermidine Synthase 2 (SPDS2), which lead to the disruption of SA-mediated defense signaling (Hewezi et al., 2010). A general view is that SA plays an important role in defense against biotrophic pathogens and JA play role in defense against necrotrophs (Glazebrook et al., 2003). Recent studies have illuminated that JA may play a more dominant role in the plant-pathogen interactions in roots (Fujimoto et al., 2011; Nahar et al., 2011; Mendy et al., 2017). Based on the data from this study, we found that the expression of MiISE6 in Arabidopsis resulted in the upregulation of JAZ genes, which are known as repressors of JA signaling (Chini et al., 2007; Thines et al., 2007; Demianski et al., 2012). It was reported that eight of twelve JAZ genes were induced after DC3000 infection, and the disruption of JAZ10 resulted in the increased susceptibility of Arabidopsis to DC3000 (Demianski et al., 2012). It was hypothesized that JAZ genes attenuate JA signaling by negative feedback regulating the degrading of JAZ proteins by SCFCOL1 (Chico et al., 2008). Moreover, the JA defense response marker genes PR4 and PDF1.2 are downregulated in the transgenic plants, those indicated that the expression of MiISE6 in Arabidopsis can suppress the JA signaling. In addition, we also found that the expression of MiISE6 in Arabidopsis activated the expression of CPK28 and then resulted in the accumulation of RBOHD(Matschi et al., 2015); however, the expression of SA-responsive genes PR1, PR2, PR5, and EDS1 did not show detectable changes in the transgenic plants compared to control. A recent study has demonstrated that parasitic nematodes can restrict infected cell death and promote nurse cell formation by activating the expression of NADPH oxidase RBOHD to produce ROIs (Siddique et al., 2014). As in the transgenic plants, the SA-mediated defense responses were not activated, so we speculated that the concentration of ROIs may be at a low level and may function as signaling molecules to suppress HRs and maintain the development of the feeding site.
All aspects of plant physiology and development are controlled by ubiquitin-proteasome pathway, which function in regulating synthesis of new polypeptides and degradation of existion proteins (Craig et al., 2009). Previous studies have revealed that the ubiquitination system can be exploited by plant pathogens to suppress host immune defense responses (Birch et al., 2009; Dielen et al., 2010). It is reported that a Globodera rostochiensis effector GrUBCEP12 can cleave into free ubiquitin and a CEP12 peptide in planta, which play important role in host immunity suppression and affecting the host 26S proteasome to promote parasitism. In this study, we found that the overexpression of MiISE6 in Arabidopsis lead to the upregulation of PUBs, which act as negative regulator of PAMP-triggered immunity by altering the activity of 26S proteasome, that indicates MiISE6 may also interfering the ubiquitination proteasome pathway to facilitate parasitism.
Conclusion
The overexpression of MiISE6 in host interferes multiple signaling pathways. As different plant signaling may function as networks in host defense responses (Berens et al., 2017), further work such as the target of MiISE6 are required to elucidate the role of MiISE6 during plant-nematode interaction.
Author Contributions
QS and ZM performed the experiments, transcriptome analysis and manuscript. XpZ and XZ did the quantitative real-time PCR. JL and RLin contributed the phylogenetic analysis. RLiu completed the nematode inoculation. YW and YY contributed to the manuscript revision. BX and XC contributed to the study design. All authors read and approved the final manuscript.
Funding
This work was supported by National Key Research & Development (R&D) plan (2017YFD0200600), the National Natural Science Foundation of China (31371923 and 31672010), China Agriculture Research System (CARS-25), the Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-IVFCAAS).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00252/full#supplementary-material
Footnotes
- ^http://www.cbs.dtu.dk/services/SignalP/
- ^http://www.predisi.de/
- ^http://phobius.binf.ku.dk/
- ^http://www.cbs.dtu.dk/services/TargetP/
- ^http://www.cbs.dtu.dk/services/TMHMM-2.0/
- ^http://systemsbiology.cau.edu.cn/agriGOv2/
References
Abad, P., Favery, B., Rosso, M. N., and Castagnone-Sereno, P. (2003). Root-knot nematode parasitism and host response: molecular basis of a sophisticated interaction. Mol. Plant Pathol. 4, 217–224. doi: 10.1046/j.1364-3703.2003.00170.x
Abad, P., Gouzy, J., Aury, J. M., Castagnone-Sereno, P., Danchin, E. G., Deleury, E., et al. (2008). Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 26, 909–915. doi: 10.1038/nbt.1482
Alioto, T. S., Buchhalter, I., Derdak, S., Hutter, B., Eldridge, M. D., Hovig, E., et al. (2015). A comprehensive assessment of somatic mutation detection in cancer using whole-genome sequencing. Nat. Commun. 6:10001. doi: 10.1038/ncomms10001
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Balestrini, R., Cosgrove, D. J., and Bonfante, P. (2005). Differential location of alpha-expansin proteins during the accommodation of root cells to an arbuscular mycorrhizal fungus. Planta 220, 889–899. doi: 10.1007/s00425-004-1431-2
Baumann, M. J., Eklof, J. M., Michel, G., Kallas, A. M., Teeri, T. T., Czjzek, M., et al. (2007). Structural evidence for the evolution of xyloglucanase activity from xyloglucan Endo-transglycosylases: biological implications for cell wall metabolism. Plant Cell 19, 1947–1963. doi: 10.1105/tpc.107.051391
Bera-Maillet, C., Arthaud, L., Abad, P., and Rosso, M. N. (2000). Biochemical characterization of MI-ENG1, a family 5 endoglucanase secreted by the root-knot nematode Meloidogyne incognita. Eur. J. Biochem. 267, 3255–3263. doi: 10.1046/j.1432-1327.2000.01356.x
Berens, M. L., Berry, H. M., Mine, A., Argueso, C. T., and Tsuda, K. (2017). Evolution of hormone signaling networks in plant defense. Annu. Rev. Phytopathol. 55, 401–425. doi: 10.1146/annurev-phyto-080516-035544
Birch, P. R., Armstrong, M., Bos, J., Boevink, P., Gilroy, E. M., Taylor, R. M., et al. (2009). Towards understanding the virulence functions of RXLR effectors of the oomycete plant pathogen Phytophthora infestans. J. Exp. Bot. 60, 1133–1140. doi: 10.1093/jxb/ern353
Brameier, M., Krings, A., and MacCallum, R. M. (2007). NucPred–predicting nuclear localization of proteins. Bioinformatics 23, 1159–1160. doi: 10.1093/bioinformatics/btm066
Chen, C., Liu, S., Liu, Q., Niu, J., Liu, P., Zhao, J., et al. (2015). An ANNEXIN-like protein from the cereal cyst nematode Heterodera avenae suppresses plant defense. PLoS One 10:e0122256. doi: 10.1371/journal.pone.0122256
Chen, J., Lin, B., Huang, Q., Hu, L., Zhuo, K., and Liao, J. (2017). A novel Meloidogyne graminicola effector, MgGPP, is secreted into host cells and undergoes glycosylation in concert with proteolysis to suppress plant defenses and promote parasitism. PLoS Pathog. 13:e1006301. doi: 10.1371/journal.ppat.1006301
Chen, L. Q., Hou, B. H., Lalonde, S., Takanaga, H., Hartung, M. L., Qu, X. Q., et al. (2010). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–532. doi: 10.1038/nature09606
Cheng, F., McLaughlin, P. J., Verderame, M. F., and Zagon, I. S. (2009). Dependence on nuclear localization signals of the opioid growth factor receptor in the regulation of cell proliferation. Exp. Biol. Med. 234, 532–541. doi: 10.3181/0901-RM-16
Cheng, F., McLaughlin, P. J., and Zagon, I. S. (2010). Regulation of cell proliferation by the opioid growth factor receptor is dependent on karyopherin beta and Ran for nucleocytoplasmic trafficking. Exp. Biol. Med. 235, 1093–1101. doi: 10.1258/ebm.2010.010139
Chico, J. M., Chini, A., Fonseca, S., and Solano, R. (2008). JAZ repressors set the rhythm in jasmonate signaling. Curr. Opin. Plant Biol. 11, 486–494. doi: 10.1016/j.pbi.2008.06.003
Chini, A., Fonseca, S., Fernandez, G., Adie, B., Chico, J., Lorenzo, O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. doi: 10.1038/nature06006
Cho, S. K., Bae, H., Ryu, M. Y., Wook Yang, S., and Kim, W. T. (2015). PUB22 and PUB23 U-BOX E3 ligases directly ubiquitinate RPN6, a 26S proteasome lid subunit, for subsequent degradation in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 464, 994–999. doi: 10.1016/j.bbrc.2015.07.030
Chronis, D., Chen, S., Lu, S., Hewezi, T., Carpenter, S. C., Loria, R., et al. (2013). A ubiquitin carboxyl extension protein secreted from a plant-parasitic nematode Globodera rostochiensis is cleaved in planta to promote plant parasitism. Plant J. 74, 185–196. doi: 10.1111/tpj.12125
Citovsky, V., Kapelnikov, A., Oliel, S., Zakai, N., Rojas, M. R., Gilbertson, R. L., et al. (2004). Protein interactions involved in nuclear import of the Agrobacterium VirE2 protein in vivo and in vitro. J. Biol. Chem. 279, 29528–29533. doi: 10.1074/jbc.M403159200
Clapier, C. R., and Cairns, B. R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304. doi: 10.1146/annurev.biochem.77.062706.153223
Cosgrove, D. J. (2000). Loosening of plant cell walls by expansins. Nature 407, 321–326. doi: 10.1038/35030000
Craig, A., Ewan, R., Mesmar, J., Gudipati, V., and Sadanandom, A. (2009). E3 ubiquitin ligases and plant innate immunity. J. Exp. Bot. 60, 1123–1132. doi: 10.1093/jxb/erp059
Davis, E. L., Hussey, R. S., Baum, T. J., Bakker, J., Schots, A., Rosso, M. N., et al. (2000). Nematode parasitism genes. Annu. Rev. Phytopathol. 38, 365–396. doi: 10.1146/annurev.phyto.38.1.365
de Boer, J. M., Yan, Y., Smant, G., Davis, E. L., and Baum, T. J. (1998). In-situ hybridization to messenger RNA in Heterodera glycines. J. Nematol. 30, 309–312.
Demianski, A. J., Chung, K. M., and Kunkel, B. N. (2012). Analysis of Arabidopsis JAZ gene expression during Pseudomonas syringae pathogenesis. Mol. Plant Pathol. 13, 46–57. doi: 10.1111/j.1364-3703.2011.00727.x
Dielen, A. S., Badaoui, S., Candresse, T., and German-Retana, S. (2010). The ubiquitin/26S proteasome system in plant-pathogen interactions: a never-ending hide-and-seek game. Mol. Plant Pathol. 11, 293–308. doi: 10.1111/j.1364-3703.2009.00596.x
Doyle, E. A., and Lambert, K. N. (2003). Meloidogyne javanica chorismate mutase 1 alters plant cell development. Mol. Plant Microbe Interact. 16, 123–131. doi: 10.1094/MPMI.2003.16.2.123
Dubreuil, G., Deleury, E., Magliano, M., Jaouannet, M., Abad, P., and Rosso, M. N. (2011). Peroxiredoxins from the plant parasitic root-knot nematode, Meloidogyne incognita, are required for successful development within the host. Int. J. Parasitol. 41, 385–396. doi: 10.1016/j.ijpara.2010.10.008
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. doi: 10.1093/nar/gkt1223
Fujimoto, T., Tomitaka, Y., Abe, H., Tsuda, S., Futai, K., and Mizukubo, T. (2011). Expression profile of jasmonic acid-induced genes and the induced resistance against the root-knot nematode (Meloidogyne incognita) in tomato plants (Solanum lycopersicum) after foliar treatment with methyl jasmonate. J. Plant Physiol. 168, 1084–1097. doi: 10.1016/j.jplph.2010.12.002
Garcia-Rodriguez, F. M., Schrammeijer, B., and Hooykaas, P. J. (2006). The Agrobacterium VirE3 effector protein: a potential plant transcriptional activator. Nucleic Acids Res. 34, 6496–6504. doi: 10.1093/nar/gkl877
Ge, X., Li, G. J., Wang, S. B., Zhu, H., Zhu, T., Wang, X., et al. (2007). AtNUDT7, a negative regulator of basal immunity in Arabidopsis, modulates two distinct defense response pathways and is involved in maintaining redox homeostasis. Plant Physiol. 145, 204–215. doi: 10.1104/pp.107.103374
Glazebrook, J., Chen, W., Estes, B., Chang, H. S., Nawrath, C., Metraux, J. P., et al. (2003). Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 34, 217–228. doi: 10.1046/j.1365-313X.2003.01717.x
Gutjahr, C., and Paszkowski, U. (2009). Weights in the balance: jasmonic acid and salicylic acid signaling in root-biotroph interactions. Mol. Plant Microbe Interact. 22, 763–772. doi: 10.1094/MPMI-22-7-0763
Haegeman, A., Mantelin, S., Jones, J. T., and Gheysen, G. (2012). Functional roles of effectors of plant-parasitic nematodes. Gene 492, 19–31. doi: 10.1016/j.gene.2011.10.040
Hewezi, T., Howe, P. J., Maier, T. R., Hussey, R. S., Mitchum, M. G., Davis, E. L., et al. (2010). Arabidopsis spermidine synthase is targeted by an effector protein of the cyst nematode Heterodera schachtii. Plant Physiol. 152, 968–984. doi: 10.1104/pp.109.150557
Hewezi, T., Juvale, P. S., Piya, S., Maier, T. R., Rambani, A., Rice, J. H., et al. (2015). The cyst nematode effector protein 10A07 targets and recruits host posttranslational machinery to mediate its nuclear trafficking and to promote parasitism in Arabidopsis. Plant Cell 27, 891–907. doi: 10.1105/tpc.114.135327
Jacobs, K. A., Collins-Racie, L. A., Colbert, M., Duckett, M., Golden-Fleet, M., Kelleher, K., et al. (1997). A genetic selection for isolating cDNAs encoding secreted proteins. Gene 198, 289–296. doi: 10.1016/S0378-1119(97)00330-2
Kalderon, D., Richardson, W. D., Markham, A. F., and Smith, A. E. (1984a). Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311, 33–38.
Kalderon, D., Roberts, B. L., Richardson, W. D., and Smith, A. E. (1984b). A short amino acid sequence able to specify nuclear location. Cell 39, 499–509.
Kay, S., Hahn, S., Marois, E., Hause, G., and Bonas, U. (2007). A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 318, 648–651. doi: 10.1126/science.1144956
Kren, N. P., Zagon, I. S., and McLaughlin, P. J. (2015). Mutations in the opioid growth factor receptor in human cancers alter receptor function. Int. J. Mol. Med. 36, 289–293. doi: 10.3892/ijmm.2015.2221
Langdon, W. B. (2015). Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) benchmarks. BioData Min. 8:1. doi: 10.1186/s13040-014-0034-0
Lee, C., Chronis, D., Kenning, C., Peret, B., Hewezi, T., Davis, E. L., et al. (2011). The novel cyst nematode effector protein 19C07 interacts with the Arabidopsis auxin influx transporter LAX3 to control feeding site development. Plant Physiol. 155, 866–880. doi: 10.1104/pp.110.167197
Lin, B., Zhuo, K., Chen, S., Hu, L., Sun, L., Wang, X., et al. (2016). A novel nematode effector suppresses plant immunity by activating host reactive oxygen species-scavenging system. New Phytol. 209, 1159–1173. doi: 10.1111/nph.13701
Lin, B., Zhuo, K., Wu, P., Cui, R., Zhang, L. H., and Liao, J. (2013). A novel effector protein, MJ-NULG1a, targeted to giant cell nuclei plays a role in Meloidogyne javanica parasitism. Mol. Plant Microbe Interact. 26, 55–66. doi: 10.1094/MPMI-05-12-0114-FI
Matschi, S., Hake, K., Herde, M., Hause, B., and Romeis, T. (2015). The calcium-dependent protein kinase CPK28 regulates development by inducing growth phase-specific, spatially restricted alterations in jasmonic acid levels independent of defense responses in Arabidopsis. Plant Cell 27, 591–606. doi: 10.1105/tpc.15.00024
Mendy, B., Wang’ombe, M. W., Radakovic, Z. S., Holbein, J., Ilyas, M., Chopra, D., et al. (2017). Arabidopsis leucine-rich repeat receptor-like kinase NILR1 is required for induction of innate immunity to parasitic nematodes. PLoS Pathog. 13:e1006284. doi: 10.1371/journal.ppat.1006284
Monaghan, J., Matschi, S., Romeis, T., and Zipfel, C. (2015). The calcium-dependent protein kinase CPK28 negatively regulates the BIK1-mediated PAMP-induced calcium burst. Plant Signal. Behav. 10:e1018497. doi: 10.1080/15592324.2015.1018497
Monaghan, J., Matschi, S., Shorinola, O., Rovenich, H., Matei, A., Segonzac, C., et al. (2014). The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe 16, 605–615. doi: 10.1016/j.chom.2014.10.007
Moter, A., and Gobel, U. B. (2000). Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J. Microbiol. Methods 41, 85–112. doi: 10.1016/S0167-7012(00)00152-4
Nahar, K., Kyndt, T., De Vleesschauwer, D., Höfte, M., and Gheysen, G. (2011). The jasmonate pathway is a key player in systemically induced defense against root knot nematodes in rice. Plant Physiol. 157, 305–316. doi: 10.1104/pp.111.177576
Nikolaidis, N., Doran, N., and Cosgrove, D. J. (2014). Plant expansins in bacteria and fungi: evolution by horizontal gene transfer and independent domain fusion. Mol. Biol. Evol. 31, 376–386. doi: 10.1093/molbev/mst206
Niu, J., Liu, P., Liu, Q., Chen, C., Guo, Q., Yin, J., et al. (2016). Msp40 effector of root-knot nematode manipulates plant immunity to facilitate parasitism. Sci. Rep. 6:19443. doi: 10.1038/srep19443
Oh, S. K., Young, C., Lee, M., Oliva, R., Bozkurt, T. O., Cano, L. M., et al. (2009). In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell 21, 2928–2947. doi: 10.1105/tpc.109.068247
Olsen, S., and Krause, K. (2017). Activity of xyloglucan endotransglucosylases/hydrolases suggests a role during host invasion by the parasitic plant Cuscuta reflexa. PLoS One 12:e0176754. doi: 10.1371/journal.pone.0176754
Pandey, S. P., Roccaro, M., Schön, M., Logemann, E., and Somssich, I. E. (2010). Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of Arabidopsis. Plant J. 64, 912–923. doi: 10.1111/j.1365-313X.2010.04387.x
Quentin, M., Abad, P., and Favery, B. (2013). Plant parasitic nematode effectors target host defense and nuclear functions to establish feeding cells. Front. Plant Sci. 4:53. doi: 10.3389/fpls.2013.00053
Sharma, S., Hirabuchi, A., Yoshida, K., Fujisaki, K., Ito, A., Uemura, A., et al. (2013). Deployment of the Burkholderia glumae type III secretion system as an efficient tool for translocating pathogen effectors to monocot cells. Plant J. 74, 701–712. doi: 10.1111/tpj.12148
Siddique, S., Matera, C., Radakovic, Z. S., Hasan, M. S., Gutbrod, P., Rozanska, E., et al. (2014). Parasitic worms stimulate host NADPH oxidases to produce reactive oxygen species that limit plant cell death and promote infection. Sci. Signal. 7:ra33. doi: 10.1126/scisignal.2004777
Smant, G., Stokkermans, J. P., Yan, Y., de Boer, J. M., Baum, T. J., Wang, X., et al. (1998). Endogenous cellulases in animals: isolation of beta-1, 4-endoglucanase genes from two species of plant-parasitic cyst nematodes. Proc. Natl. Acad. Sci. U.S.A. 95, 4906–4911. doi: 10.1073/pnas.95.9.4906
Stegmann, M., Anderson, R. G., Ichimura, K., Pecenkova, T., Reuter, P., Zarsky, V., et al. (2012). The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell 24, 4703–4716. doi: 10.1105/tpc.112.104463
Szakasits, D., Heinen, P., Wieczorek, K., Hofmann, J., Wagner, F., Kreil, D. P., et al. (2009). The transcriptome of syncytia induced by the cyst nematode Heterodera schachtii in Arabidopsis roots. Plant J. 57, 771–784. doi: 10.1111/j.1365-313X.2008.03727.x
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., et al. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665. doi: 10.1038/nature05960
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Tzfira, T., Vaidya, M., and Citovsky, V. (2001). VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 20, 3596–3607. doi: 10.1093/emboj/20.13.3596
Van Sandt, V. S., Suslov, D., Verbelen, J. P., and Vissenberg, K. (2007). Xyloglucan endotransglucosylase activity loosens a plant cell wall. Ann. Bot. 100, 1467–1473. doi: 10.1093/aob/mcm248
Weinthal, D. M., Barash, I., Tzfira, T., Gaba, V., Teper, D., Sessa, G., et al. (2011). Characterization of nuclear localization signals in the type III effectors HsvG and HsvB of the gall-forming bacterium Pantoea agglomerans. Microbiology 157, 1500–1508. doi: 10.1099/mic.0.047118-0
Wong, W. C., Yap, C. K., Eisenhaber, B., and Eisenhaber, F. (2015). dissectHMMER: a HMMER-based score dissection framework that statistically evaluates fold-critical sequence segments for domain fold similarity. Biol. Direct 10:39. doi: 10.1186/s13062-015-0068-3
Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. doi: 10.1093/nar/gkr483
Yang, H., Li, Y., and Hua, J. (2006). The C2 domain protein BAP1 negatively regulates defense responses in Arabidopsis. Plant J. 48, 238–248. doi: 10.1111/j.1365-313X.2006.02869.x
Zagon, I. S., Ruth, T. B., Leure-duPree, A. E., Sassani, J. W., and McLaughlin, P. J. (2003). Immunoelectron microscopic localization of the opioid growth factor receptor (OGFr) and OGF in the cornea. Brain Res. 967, 37–47. doi: 10.1016/S0006-8993(02)04172-0
Zagon, I. S., Ruth, T. B., and McLaughlin, P. J. (2005). Nucleocytoplasmic distribution of opioid growth factor and its receptor in tongue epithelium. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 282, 24–37.
Zagon, I. S., Smith, J. P., and McLaughlin, P. J. (1999). Human pancreatic cancer cell proliferation in tissue culture is tonically inhibited by opioid growth factor. Int. J. Oncol. 14, 577–584. doi: 10.3892/ijo.14.3.577
Zagon, I. S., Verderame, M. F., and McLaughlin, P. J. (2002). The biology of the opioid growth factor receptor (OGFr). Brain Res. Brain Res. Rev. 38, 351–376. doi: 10.1016/S0165-0173(01)00160-6
Zagon, I. S., Wu, Y., and McLaughlin, P. J. (1994). Opioid growth factor inhibits DNA synthesis in mouse tongue epithelium in a circadian rhythm-dependent manner. Am. J. Physiol. 267, R645–R652. doi: 10.1152/ajpregu.1994.267.3.R645
Keywords: Meloidogyne incognita, effector, nuclear localization signal (NLS), hypersensitive response (HR), plant–nematode interaction, comparative transcriptome analysis
Citation: Shi Q, Mao Z, Zhang X, Ling J, Lin R, Zhang X, Liu R, Wang Y, Yang Y, Cheng X and Xie B (2018) The Novel Secreted Meloidogyne incognita Effector MiISE6 Targets the Host Nucleus and Facilitates Parasitism in Arabidopsis. Front. Plant Sci. 9:252. doi: 10.3389/fpls.2018.00252
Received: 18 October 2017; Accepted: 12 February 2018;
Published: 23 March 2018.
Edited by:
Vincenzo Lionetti, Sapienza Università di Roma, ItalyReviewed by:
Lei Zhang, Washington State University, United StatesHolger Bohlmann, University of Natural Resources and Life Sciences, Austria
Copyright © 2018 Shi, Mao, Zhang, Ling, Lin, Zhang, Liu, Wang, Yang, Cheng and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bingyan Xie, eGllYmluZ3lhbkBjYWFzLmNu Xinyue Cheng, Y2hlbmd4eUBibnUuZWR1LmNu
†These authors have contributed equally to this work.
 Qianqian Shi
Qianqian Shi Zhenchuan Mao
Zhenchuan Mao Xiaoping Zhang
Xiaoping Zhang Jian Ling
Jian Ling Runmao Lin
Runmao Lin Xi Zhang
Xi Zhang Rui Liu
Rui Liu Yunsheng Wang
Yunsheng Wang Yuhong Yang
Yuhong Yang Xinyue Cheng
Xinyue Cheng Bingyan Xie
Bingyan Xie