- Division of Plant Molecular Biology, Rajiv Gandhi Centre for Biotechnology, Thiruvananthapuram, India
MADS box transcription factors have been studied extensively in flowering plants but remain less studied in non-seed plants. MADS box is one such example of a gene which is prevalent across many classes of plants ranging from chlorophyta to embryophyta as well as fungi and animals. MADS box transcription factors are of two types, Type I and Type II. Type II transcription factors (TF) that consist of a MADS domain, I region, K domain, and C terminal domain are discussed in this review. The Type II/ MIKC class is widespread across charophytes and all major lineages of land plants but unknown in green and red algae. These transcription factors have been implicated in floral development in seed plants and thus the question arises, “What is their role in non-seed plants?” From the studies reviewed here it can be gathered that unlike seed plants, MIKCC genes in non-seed plants have roles in both gametophytic and sporophytic generations and contribute to the development of both vegetative and reproductive structures. On the other hand as previously observed in seed plants, MIKC* genes of non-seed plants have a conserved role during gametophyte development. With respect to evolution of MIKC genes in non-seed plants, the number of common ancestors is probably very few at each branch. The expansion of this gene family in seed plants and increased plant complexity seem to be correlated. As gradually the genomes of non-seed plants are becoming available it is worthwhile to gather the existing information about MADS box genes in non-seed plants. This review highlights various MIKC MADS box genes discovered so far in non-seed plants, their possible roles and an insight into their evolution.
Introduction
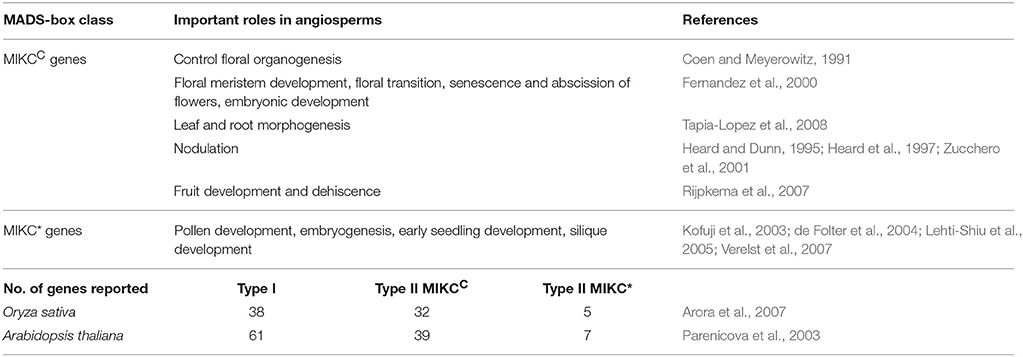
The main principle of evo-devo (evolutionary developmental genetics) proposes that development and evolution are mutually interrelated processes (Gilbert et al., 1996). Since development is under genetic control, the genes which regulate the development of an organ may play a major role in their evolution (Theissen and Saedler, 1995). MADS-box gene family (MCM1/AGAMOUS/DEFICIENS/SRF) is one such important gene family which has been extensively explored because it is widely present in eukaryotes like plants, animals, fungi and might have originated at least 1 billion years ago (Purugganan et al., 1995; Theissen et al., 1996, 2000). MADS domain proteins are homeotic transcription factors with a DNA-binding MADS domain and play a major role during reproductive development (Schwarz-Sommer et al., 1990). They are broadly classified into two major classes based on their structure and phylogeny, Type I and Type II class (Alvarez-Buylla et al., 2000). Type I consists of Mα, Mβ, and Mγ subgroups that contain a MADS domain and a variable C terminal domain and Type II class genes have two subfamilies MIKCC and MIKC* that are characterized by a MADS domain, I region, K domain and C terminal domain (Ma et al., 1991; Henschel et al., 2002; Becker and Theissen, 2003). The I region may have a role in providing specificity to protein dimer formation, the K domain promotes protein dimerization and C terminal domain may function in transcriptional activation and in the formation of ternary and quaternary protein complexes (Ma et al., 1991; Davies and Schwarz-Sommer, 1994; Shore and Sharrocks, 1995; Riechmann and Meyerowitz, 1997; Cho et al., 1999; Egea-Cortines et al., 1999; Honma and Goto, 2001; Theissen and Saedler, 2001).
MIKC Type MADS-Box Genes in Charophycean Green Algae
Charophycean green algae are the closest relatives of the extant land plants (Graham et al., 2000). Tanabe et al. (2005) isolated and characterized MADS-box genes from three Charophycean green algae, CgMADS1 from the stonewort Chara globularis, CsMADS1 from the coleochaete Coleochaete scutata and CpMADS1 from the desmid Clostridium peracerosum-strigosum-littorale complex. The sequence analysis of these genes when compared with the already reported MIKCC and MIKC* type genes revealed that all the four M, I, K, and C domains were present in the isolated algal genes. Expression analysis suggests that these genes may have a putative role in haploid reproductive cell differentiation and during the course of evolution they were recruited into a diploid generation. The MIKC type genes in land plants fall into several groups with diverse functions and expression patterns, whereas in Charophycean green algae, only one MIKCC type gene was reported in each of the three charophycean taxa (Tanabe et al., 2005). This extensive diversification of MIKC type genes may have played an important role in the development of sophisticated developmental systems in land plants (Tanabe et al., 2005).
MIKC Type MADS-Box Genes in Bryophytes
Non-vascular land plants liverworts, mosses, and hornworts form the bryophyte group. According to current information the phylogeny of liverworts, mosses, and hornworts with respect to each other and its relation to land plants remains unresolved as there are multiple hypotheses for their positions (Szövényi, 2016).
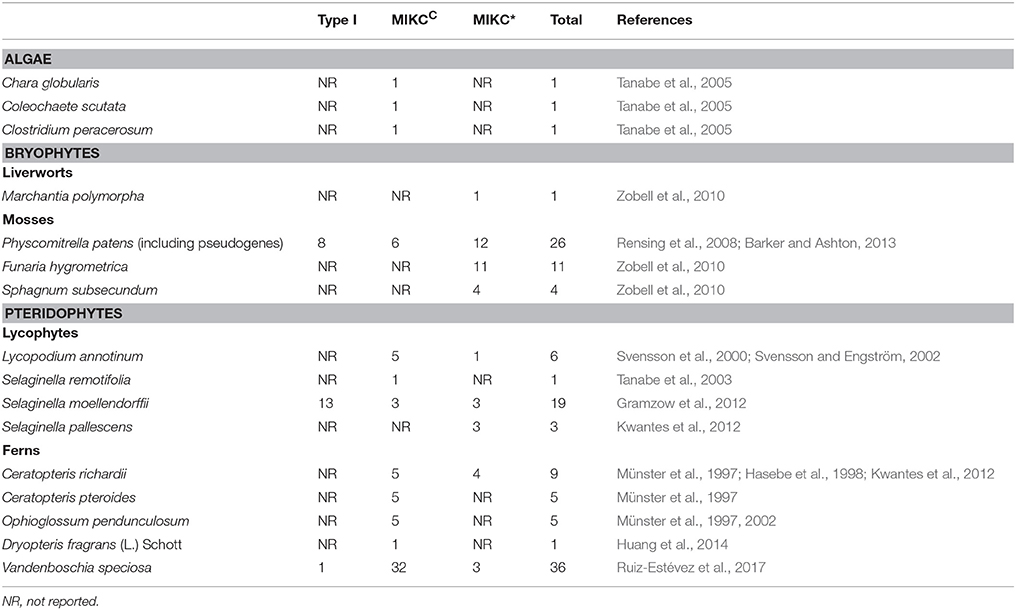
Both Type I and Type II class of MADS-box genes have been reported from various bryophyte taxa. The pseudo-chromosomal genome assembly of the moss Physcomitrella patens confirmed the presence of 26 MADS-box genes. It has 6 MIKCC type genes (PPM1- 2, PpMADS1 and S, PPMC5, PPMC6), 11 MIKC* type genes (PPM3-4, PPM6-7, PpMADS2-3, PPMA 8-12), 2 pseudogenes (PPMA5, PPTIM6) and the rest are Type I (PPTIM1-5,7,8) where PPTIM2,3 are Type I Mα genes and PPTIM6-8 are Type I Mβ-γ genes (Krogan and Ashton, 2000; Henschel et al., 2002; Hohe et al., 2002; Riese et al., 2005; Quodt et al., 2007; Singer et al., 2007; Rensing et al., 2008; Singer and Ashton, 2009; Barker and Ashton, 2013, 2016).
Phylogenetic analysis reveals that MIKCC genes from P. patens clustered into a separate clade from the MIKCC type genes of other taxa suggesting that angiosperm orthologs are not found in mosses. The MIKC* genes clustered within a larger cluster containing Arabidopsis MIKC* genes. The Type I genes formed three clusters, one which was exclusively for P. patens, the second with Mα genes of Arabidopsis and third with Mβ-γ genes of Arabidopsis (Riese et al., 2005; Barker and Ashton, 2013). This suggests that at least 4 different types of MADS box genes existed in the most recent common ancestor (MRCA) of extant mosses and vascular plants about 450 million years ago (MYA) (Gramzow and Theissen, 2010; Barker and Ashton, 2013).
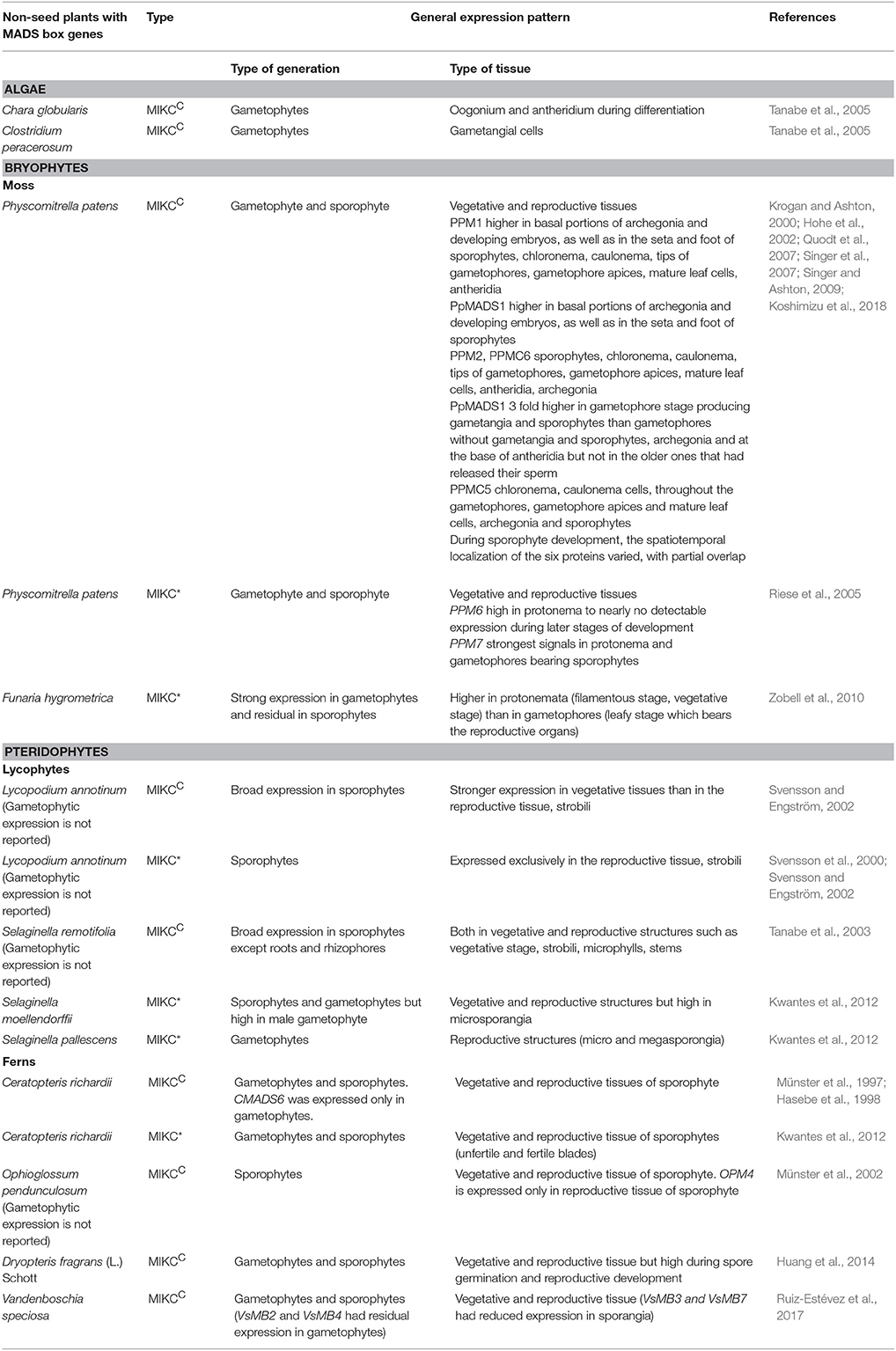
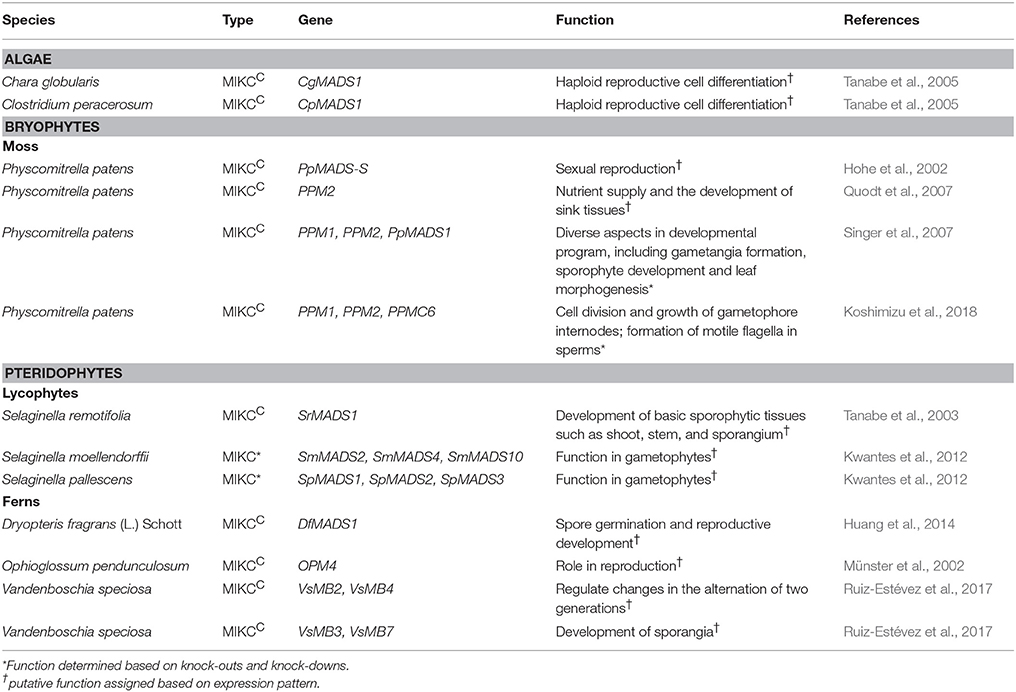
PPM1 and PpMADS1expression was detected in both vegetative and reproductive tissues in a study carried out using protein fused with GUS reporter. Though their expression pattern was broad, the protein was found at increased levels at the basal portions of archegonia and developing embryos, as well as in the seta and foot of sporophytes (Singer et al., 2007). Another protein localization study of PPM1 fused with reporters (GUS and citrine) expressed in both vegetative and reproductive tissues like chloronema, caulonema, tips of gametophores, gametophore apices, mature leaf cells, antheridia, and sporophytes (Koshimizu et al., 2018). PPM2 was reported to have a weak ubiquitous expression with increased expression in male and female gametangia and in the basal parts of the developing sporophytes (Quodt et al., 2007). This increased expression suggests that probably PPM2 has a role in defining the sink tissues for proper development of the organs required for transition from gametophyte to sporophyte (Quodt et al., 2007). In another study, PPM2 was found to be localized in different developmental stages like chloronema, caulonema, tips of gametophores, gametophore apices, mature leaf cells, antheridia, archegonia, and sporophytes. In the study by Quodt et al. (2007) PPM2 expression was detected in the basal part of the developing sporophyte whereas in the recent study by Koshimizu et al. (2018) expression is seen toward the apical part. It should be noted that this difference in expression pattern has not been discussed by Koshimizu et al. (2018). Another MIKCC protein PPMC6 was also found to be localized in different tissues similar to PPM2 (Koshimizu et al., 2018). PpMADS S expression was found to be 3 fold higher in gametophore stage producing gametangia and sporophytes than gametophores without gametangia and sporophytes (Hohe et al., 2002). In localization studies carried out by Koshimizu et al. (2018) PpMADS S was detected in archegonia and at the base of antheridia but not in the older ones that had released their sperm. These studies suggest it may have a role during sexual reproduction of the moss (Hohe et al., 2002). PPMC5 localization was detected in chloronema, caulonema cells, throughout the gametophores, gametophore apices and mature leaf cells, archegonia, and sporophytes (Koshimizu et al., 2018). During sporophyte development, the spatiotemporal localization of the six proteins varied, with partial overlap (Koshimizu et al., 2018). These studies show that the MIKCC genes may redundantly function in various moss developmental stages.
Deletion mutants and overexpression lines of these three MIKCC genes (PPM1, PPM2, PPMC6) revealed that, these genes redundantly and negatively regulate the cell division and growth of the gametophore internodes thereby influencing the external water conduction which is necessary for the sperms to swim in order to reach and fertilize the eggs. They were also found to redundantly regulate the formation of motile flagella in sperms. Therefore, the authors suggest that both functions are necessary for fertilization to occur (Koshimizu et al., 2018). Earlier, the PPM1 knockdowns created by antisense technology resulted in multifaceted mutant phenotype which may be due to the variation in the expressivity of the antisense PPM1 gene or homology of the PPM1 antisense RNA molecules to other P. patens MIKCC genes (Singer et al., 2007). The knockdowns showed aberrant gametangia formation with significantly fewer antheridia and no archegonia. These lines also produced fewer sporophytes and sporophyte morphogenesis was abnormal as they were pale green, swollen and larger than control sporophytes (Singer et al., 2007). Single knock out mutants of MIKCc genes in Physcomitrella patens were phenotypically normal when compared to the wildtype (Singer et al., 2007). The expression and functional studies in P. patens indicate that MIKCC genes have broad expression pattern and play a role in both gametophyte and sporophyte generation.
Additionally in bryophytes MIKC* genes are also present. Two MIKC* type genes (PPM6 and PPM7) were isolated from P. patens (Riese et al., 2005). Zobell et al. (2010) isolated MIKC*genes from liverwort, Marchantia polymorpha (MpMADS1) and the mosses Sphagnum subsecundum (SsMADS1-4), and Funaria hygrometrica (FhMADS1-11). Their attempts to isolate MIKC* genes from the hornworts Anthoceros agrestis and Anthoceros formosae was unsuccessful. The genes were either lost in the hornwort lineage or escaped identification due to unavailability of the whole genome (Zobell et al., 2010). But a recent article dealing with draft genome of A. agrestis mentions it has MADS box genes (details not mentioned) though the number is dramatically reduced when compared to Physcomitrella (Szövényi, 2016). It should be noted that completion of genome sequencing of Anthoceros punctatus will throw light on the status of MADS box genes in hornworts.
The liverwort gene MpMADS1 was found to be the sister of all moss homologs thus suggesting that there was only one MIKC* MADS-box gene before the separation of the major bryophyte lineages. Sphagnum subsecundum has fewer MIKC* genes than P. patens and F. hygrometrica and formed a separate monophyletic clade suggesting expansion in the former was lesser in comparison to the latter. MIKC* genes of P. patens and F. hygrometrica were grouped under one clade which is consistent as these two genera belong to the same family Funariales (Zobell et al., 2010).
Transcript level analysis showed ubiquitous expression of PPM7 (MIKC*) with strongest signals in protonema and gametophores bearing sporophytes. PPM6 (MIKC*) expression decreased from high in protonema to nearly undetectable in later stages of sporophytic development providing evidence for spatial and temporal regulation of this gene (Riese et al., 2005). MpMADS1 was found to be a functional transcription factor with the ability to bind to DNA and localize in the nucleus. The protein was also able to form homo-dimers. This gene was able to partially restore pollen germination in an Arabidopsis MIKC* mutant (Zobell et al., 2010). FhMADS genes expression was strong in gametophytes and only residual in sporophytes. The expression was higher in protonema as compared to the gametophore indicating a role for FhMADS genes during vegetative development rather than promoting onset of reproductive development during the haploid phase (Zobell et al., 2010). The expression studies in P. patens and F. hygrometrica and functional study in M. polymorpha thus reveal that MIKC* genes may play an important role during gametophyte development. Further studies to confirm the presence/absence of MIKCC genes among these bryophyte species (Funaria, Marchantia, and Sphagnum) may be useful to understand the origin and evolution of MADS-box genes.
MIKC Type MADS-Box Genes in Lycophytes
Lycophytes (clubmosses and spikemosses) are the most basal vascular plants and they appeared roughly 400 MYA (Kenrick and Crane, 1997). Surveying the MADS-box genes in these taxa may give us a better understanding of the evolutionary history of MADS-box genes and the evolution of advanced organs in flowering plants.
As seen in bryophytes, the lycophytes also have both Type I and Type II class MADS box genes. Lycopodium annotinum is a clubmoss in which 5 MIKCC (LAMB2-6) MADS-box genes and 1 MIKC* (LAMB1) MADS-box gene are reported so far (Svensson et al., 2000; Svensson and Engström, 2002). Tanabe et al. (2003) reported a MIKCC type MADS-box gene, SrMADS1 from the spike moss Selaginella remotifolia. Genome analysis of Selaginella moellendorffii revealed the presence of 19 putative MADS-box genes (Gramzow et al., 2012). It involves 13 type I genes (SmMADS5-9, 11-15, 17-20) where SmMADS14,15 are of α type and SmMADS5,7,17 are of β-γ origin and rest are Selaginella type. This lycophyte also has 3 MIKCC genes (SmMADS1, 3, 6), and 3 MIKC* genes (SmMADS 2, 4, 10) (Gramzow et al., 2012).
Phylogenetic analysis showed that LAMB2-6 form a separate clade and does not show any orthology to moss and fern MIKCC proteins (Svensson and Engström, 2002). SrMADS1 forms a separate clade with the LAMB2 group (LAMB2, 4, 6) in the phylogenetic analysis. So SrMADS1 may be sister to LAMB2 group (Tanabe et al., 2003). SmMADS MIKCC were not monophyletic but SrMADS1 and SmMADS1 clustered together. These genes did not form a monophyletic group with LAMB2,4,6 which was earlier reported by Tanabe et al. (2003) probably due to difficulties in determining the deep branching of widely divergent taxa (Gramzow et al., 2012). Notably no orthology was found between the MIKCC of S. moellendorffii and angiosperms which suggests that the common ancestor of mosses and vascular plants had a single ancestral MIKCC gene (Gramzow et al., 2012).
LAMB2, 4 and 6 were similar to MADS-K-box genes and were grouped under MIKCC class of genes. The other two genes (LAMB3 and 5) did not encode for a K domain. LAMB3 has only M, I domains and has a region with homology to partial K box which may be a truncated protein. LAMB5 has only M domain and ends 8 amino acid residues downstream of the M domain (Svensson and Engström, 2002). In SrMADS1, in addition to M, I, K, and C domains, SrMADS1 have additional amino acid residues in the N terminal of the MADS domain which was not reported in other genes of the LAMB2 group (Tanabe et al., 2003). SmMADS1,3,6 contain the same number of exons as seen in Arabidopsis MIKCC genes (Gramzow et al., 2012).
LAMB2, 4, 5, 6 have broad expression patterns in sporophytic tissues such as roots and apices (Svensson and Engström, 2002). Expression of SrMADS1 was found in all sporophytic tissues except in root and rhizophores. The expression pattern of SmMADS1,3, 6 is currently not available. These findings hypothesize that MIKCC genes may be involved in the development of sporophytic tissues like shoot, stem, sporogium in lycophytes (Tanabe et al., 2003).
Svensson et al. (2000) reported the first MIKC type gene LAMB1 from L. annotinum. Although it has all the four domains – M, I, K, and C domains which is a prerequisite feature of classical MIKC genes, it differed from others in both sequence and structure as the intervening region is unusually longer and it also has a longer C region. These unusual characteristics of LAMB1 made it different from the classical MIKC type genes (Svensson et al., 2000). LAMB1 was thus hypothesized to be a primitive MIKC class gene, representing an ancestral state though they mentioned that the opposite possibility was also open (Svensson et al., 2000). In the following years after the discovery of MIKC* class genes, phylogenetic analysis showed that LAMB1 clustered with the MIKC* clade (Henschel et al., 2002; Gramzow et al., 2012; Kwantes et al., 2012). Thus LAMB1 was the first MIKC* gene to be discovered (Henschel et al., 2002). In the other lycophyte studied, S. moellendorffii, there are three MIKC* genes namely SmMADS2, 4, 10 which has similar exon-intron structure to that of Arabidopsis MIKC* (Gramzow et al., 2012; Kwantes et al., 2012). Phylogenetic analysis revealed that the relationship of MIKC* proteins of the lycophyte S. moellendorffii and LAMB1 of L. annotinum to the other clades of MIKC* proteins or amongst themselves remained unresolved (Kwantes et al., 2012).
LAMB1 expression was restricted to the reproductive structure strobili found in sporophyte during sporogenesis (Svensson et al., 2000). SmMADS2, 4, 10 was shown to be expressed in microsporangia which have the male gametophyte- containing microspores (Kwantes et al., 2012). Thus in Selaginella evidence for strong gametophytic expression was shown whereas in Lycopodium only sporophytic expression was found. This shows that MIKC* genes were also recruited in the sporophytes in early vascular plants but they did not have a conserved function in this generation (Kwantes et al., 2012).
MIKC Type MADS-Box Genes in Ferns
Ferns are non-seed vascular plants with simple reproductive structures and form spores in the naked sporangium present on the abaxial side of the leaf (Gifford and Foster, 1989; Stewart and Rothwell, 1993; Hasebe et al., 1998).
The role of MADS box genes in the development of simple reproductive structures was looked into where, 2 MIKCC genes (CRM1, CRM3) from the leptosporangiate fern, Ceratopteris richardii and 4 MIKCC genes (CRM2, CRM4, CRM5, CRM6) from Ceratopteris pteroides was reported (Münster et al., 1997). Five MADS-box genes (CMADS1-4, and 6) were isolated from Ceratopteris richardii by Hasebe et al. (1998) in which CMADS3 and CMADS6 were identical to the previously reported CRM1 and CRM3 genes. Münster et al. (1997) also isolated a MIKCC gene (OPM1) from Ophioglossum pendunculosum, a eusporangiate fern thereby showing that MADS-box genes are present in other groups of ferns as well (Münster et al., 1997). They followed it up by discovering five cDNAs belonging to MIKCC type MADS-box OPM1-OPM5, where OPM1 and OPM2 may represent closely related genes or alleles (Münster et al., 2002). In Dryopteris, a new MIKCC MADS box gene DfMADS1 was isolated which shared homology to other pteridophyte MADS-box genes (Huang et al., 2014). Thirty six putative MADS-box genes were reported in the endangered fern, Vandenboschia speciosa by the analysis of Next Generation Sequencing assembled transcriptome data. Among the reported 36 putative MADS-box genes, 1 gene was found to be type I, 32 were MIKCC and 3 were MIKC* type (Ruiz-Estévez et al., 2017).
Phylogenetic reconstruction resulted in 3 subfamilies of CRM proteins (CRM1-, CRM6-, and CRM3-like sequences) interspersed among spermatophyte clades (Münster et al., 1997). CMADS1, CMADS2/3/4, and CMADS6 fall under CRM6, CRM1, and CRM3 groups respectively (Hasebe et al., 1998). OPM1/2 and OPM5 may belong to the CRM6 family whereas the other proteins did not group into any specific clade of either the seed plants or the ferns (Münster et al., 2002). DfMADS1 groups with the CRM1 family of fern MADS domain proteins (Huang et al., 2014). VsMB2, 5, 7 proteins group with CRM1 subfamily and VsMB3 and VsMB6 proteins groups with the CRM3 subfamily and VsMB4 to the CRM6-like subfamily (Ruiz-Estévez et al., 2017). The results discussed here show that the leptosporangiate ferns studied till now have genes representing three clades CRM1, CRM3, and CRM6 whereas Ophioglossum has three CRM6 like genes along with 2 unique genes which may be characteristic to the eusporangiate ferns. Further work on eusporangiate ferns will be required to validate the origin of these unique genes and whether these genes are specific to eusporangiate ferns. In the phylogenetic trees discussed, Münster et al. (1997, 2002) show that CRM3 clade is closer to the AG clade whereas Hasebe et al. (1998) and Huang et al. (2014) show that CRM6 clade is closely related to the AG clade. The latter trees seem to be clearer since the CRM6 proteins (CMADS1, CerMADS2, CerMADS3) have additional N terminal amino acids preceding the MADS domain which is similar to the AG protein. Another variation noticeable is that, according to Hasebe et al. (1998) CRM3 and CRM6 clades are closely associated whereas in all the other studies CRM1 and CRM3 are phylogenetically close. Thus the position of CRM3 within the fern clades remains unclear but CRM1 and CRM6 always formed distinct clades. Hence according to the studies discussed, the MRCA of ferns and seed plants probably had at least 2 MIKCC type genes. Further gene duplication events might have led to lineage specific diversification and expansion of MIKCC class in extant ferns and seed plants.
Northern blot analysis showed CRM1 and CRM3 were expressed in the gametophytic (haploid) as well as the sporophytic (diploid) phase of the fern life cycle (Münster et al., 1997). CMADS1 (CRM6), CMADS 2-4 (CRM1) expression was detected in both vegetative and reproductive tissues of the sporophyte and CMADS6 (CRM3) was detected in gametophytic tissues but not in sporophytic tissues. Further spatiotemporal survey of mRNA expression of these MADS genes has revealed that they may be involved in regulating cell division during early organ development and playing an unknown role in the differentiated vasculature(Hasebe et al., 1998). OPM1, OPM3, OPM5 were expressed in both trophophore and sporophore at an almost same level of expression, whereas OPM4 was detected only in sporophore. Thus this indicated that OPM4 has a role specific to spore development whereas the other genes may have a universal role. The ubiquitous expression of OPM1, OPM3, and OPM5 are similar to the genes of Ceratopteris, which is characteristic to the MADS-box genes of the fern family but not an absolute feature (Münster et al., 2002). Though DfMADS1 was expressed in both sporophytes and gametophyte, it was expressed at very high levels in the spores and in the young prothallus indicating that this particular MADS-box gene may be involved in spore germination and reproductive development (Huang et al., 2014). In V. speciosa, the expression level of 6 MIKCC type MADS-box genes (VsMB2, VsMB3, VsMB4, VsMB5, VsMB6, VsMB7) was analyzed in sporophytes, gametophytes and sporangia. All the six genes (VsMB2-7) and most of the genes (VsMB3, VsMB5, VsMB6, VsMB7) were found to have a broad expression pattern in sporophytes and gametophytes respectively. VsMB2 and VsMB4 expression level in gametophytes was found to be residual when compared to their expression level in sporophytes. These genes may have a specialized role in the changes occurring during the alternation of the two major phases in the life cycle of V. speciosa. Also, reduced expression of VsMB3 and VsMB7 genes was reported in the sporangium which is a reproductive structure. The down-regulation of these genes may be important for the development of the reproductive structure, sporangia (Ruiz-Estévez et al., 2017).
Four MIKC* type MADS-box genes namely CRM13-16 were reported from Ceratopteris richardii (Kwantes et al., 2012). Phylogenetic analysis showed that, CRM13 and CRM16 grouped into the P clade whereas CRM14 and CRM15 into the S clade thus indicating that there were two different types of MIKC* genes in the ancestor of ferns and spermatophytes (Kwantes et al., 2012). Expression analysis revealed high expression of all the genes in roots and both S-clade (CRM14) and P-clade (CRM16) genes in male and hermaphroditic gametophytes. In other tissues such as fertile and unfertile blades, even though the S-clade genes expressed the expression of P-clade genes were found to be dominating (Kwantes et al., 2012). P-clade members formed homodimers and heterodimers which might function in both sporophytic and gametophytic tissues, but heterodimers between the members of the S-(CRM14) and P-clades (CRM16) were shown to be typical for the gametophytes and roots of Ceratopteris (Kwantes et al., 2012).
Evolution of MADS Box Genes in Non-seed Plants
Genome analysis of the green algae Chlamydomonas reinhardtii (Tanabe et al., 2005), Ostreococcus tauri (Derelle et al., 2006), Ostreococcus lucimarinus (Palenik et al., 2007) and the red algae Cyanidioschyzon merolae (Matsuzaki et al., 2004) reported no MIKCC or MIKC* type genes although a single gene with MADS box lacking I,K,C region in C. reinhardtii and C. merolae and lacking K domain in Ostreococcus sp's was found which resembled the MEF2 type MADS domain (Type II like) (Kaufmann et al., 2005; Tanabe et al., 2005). Hence the MRCA of chlorophytes and streptophytes contained a protein with MADS domain similar to Type II approximately 1,000 million years ago (MYA) representing the ancestral MADS domain protein (Kaufmann et al., 2005). One MIKCC type MADS-box gene was reported in each of the three Charophycean green algal species, which are representatives of charophytes that are believed to be the common ancestor of land plants (Tanabe et al., 2005). These findings suggest that the MIKC type MADS-box genes evolved by the addition of a K domain to the ancestral MADS-box gene in the charophycean—land plant lineage after its divergence from the Chlamydomonas lineage at least 700 MYA (Figure 1) (Kaufmann et al., 2005; Tanabe et al., 2005; Gramzow and Theissen, 2010). There were at least 2 types of ancestral MIKC genes (1 MIKCC and 1 MIKC*) and 2 types of Type I genes (α and β-γ) in the common ancestor leading to bryophytes (450 MYA), lycophytes (400 MYA) and higher vascular plants (Figure 1) (Gramzow and Theissen, 2010; Gramzow et al., 2012; Barker and Ashton, 2013). It is in the common ancestor of bryophytes and vascular plants that MIKC diverged into MIKCC and MIKC*, no further diversification seems to have taken place in lineage leading to lycophytes (Figure 1)(Gramzow et al., 2012; Barker and Ashton, 2013). The MRCA of monilophytes/ferns and seed plants (380 MYA) had at least 2 MIKCC type and 2 MIKC* type (S and P-type) genes (Figure 1) (Münster et al., 1997, 2002; Hasebe et al., 1998; Huang et al., 2014; Ruiz-Estévez et al., 2017). The information on Type I genes in ferns is currently very limited, only 1 gene reported in V. speciosa (Ruiz-Estévez et al., 2017). In each of the taxa (bryophytes, lycophytes, and ferns) discussed, there has also been lineage specific diversification and expansion of the MADS box genes which have led to the unique body plan of these non-seed plants (Gramzow et al., 2012; Barker and Ashton, 2013; Ruiz-Estévez et al., 2017). Thus it can be hypothesized that the diversification and duplication of Type II MIKC genes accelerated in the common ancestor of monilophytes and seed plants and continued to expand extensively in the spermatophytes.
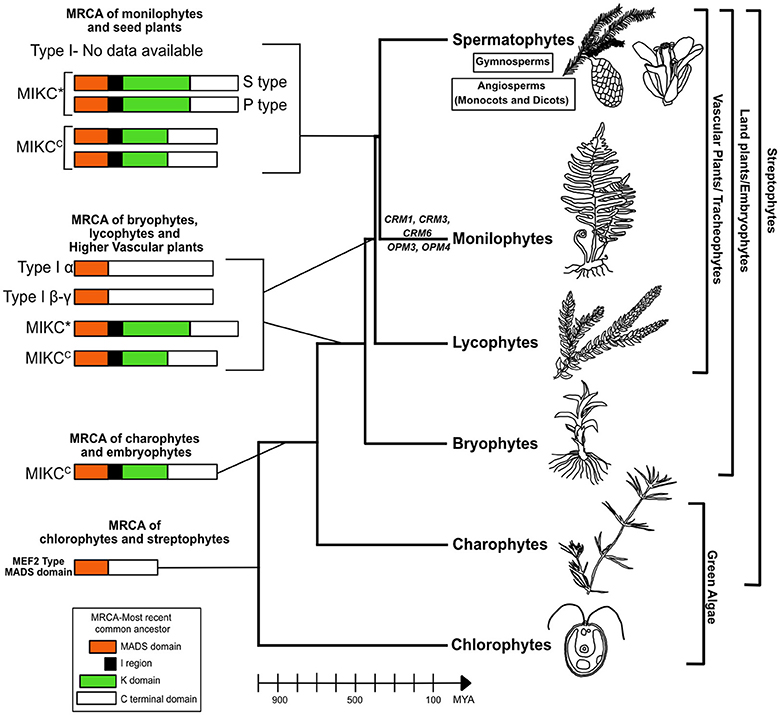
Figure 1. Model for evolution of MADS box genes in non-seed plants. MIKC-type MADS proteins seem to have evolved in streptophytes (700 MYA) by addition of K domain from a MEF2-like ancestor in chlorophytes. A gene duplication event in the common ancestor of bryophytes and tracheophytes (450 MYA) led to MIKCC and MIKC* proteins where MIKC*-type proteins have an elongated K domain. The origin of Type I is still not clear which lacks the I region and K domain but in the MRCA of bryophytes, lycophytes and tracheophytes there are two subtypes M alpha and M beta-gamma. The information on Type I genes in the MRCA of monilophytes and spermatophytes is not available. The MRCA of monilophytes/ferns and seed plants (380 MYA) had at least 2 MIKCC type and 2 MIKC* type (S and P clades) genes. Further diversification and expansion took place independently in MIKCC type genes in the lineages leading to extant ferns and spermatophytes. This gave rise to the large number of MIKCC genes in present day ferns and seed plants. In leptosporangiate ferns, there are genes belonging to three clades CRM1, CRM3, and CRM6 whereas in Ophioglossum (a eusporangiate fern) in addition to the three CRM6-like genes there are two unique genes OPM3 and OPM4 which may be specific to eusporangiate ferns. It should also be noted that in each taxa from bryophytes onwards there has been lineage specific diversification and expansion of MIKC genes. It is a possibility that with increase in number of MIKC genes there has been increase in complexity (from single celled algae to multicellular algae to multicellular non-vascular plants to early vascular plants and finally seed plants) which needs further evidence. MYA, million years ago.
Structurally the MIKC genes of non-seed plants and seed plants are very similar and the proteins function in a similar mode by forming homodimers and heterodimers as inferred from the reports available for MIKC* proteins (Tanabe et al., 2005; Zobell et al., 2010; Kwantes et al., 2012). Some of the CRM6 clade proteins (CMADS1, CerMADS2, and CerMADS3) have additional amino-terminal amino acids like AGAMOUS (AG), they seem to be the most closely related to the AG clade in the seed plants than other MADS box genes (Hasebe et al., 1998). One MIKCC gene from S. remotifolia (SrMADS1), one MIKCC (SmMADS1) and four Type I genes (SmMADS12,18,19,20) from S. moellendorffii and all the MIKCC genes from P.patens also have an N-terminal region preceding the MADS box (Tanabe et al., 2003; Gramzow et al., 2012; Barker and Ashton, 2013). This shows that the N-terminal region preceding the MADS box region was retained in AGAMOUS genes as well as some other genes in extant seed plants (Mizukami et al., 1996). It has been shown that this region in AG is not necessary for DNA binding in-vitro (Mizukami et al., 1996). Unlike AG, the N terminal region of SRF has been shown to be involved in DNA binding and affinity in-vitro (Sharrocks et al., 1993; Nurrish and Treisman, 1995), hence the function of the N-terminal region in MADS domain proteins of non-seed plants remains to be explored.
Origin of MIKC* Type MADS-Box Genes
Henschel et al. (2002) proposed two hypotheses regarding the difference between MIKC* and MIKCC genes. According to the authors there were differences in the length of I domain and K domain between MIKC* and MIKCC genes. According to the first hypothesis during evolution the ancestral I domain might have elongated in the MIKC* genes. The second hypothesis states that the difference in length upstream of C domain between MIKCC and MIKC* type genes is due to de novo insertion within K domain and not due to elongation of I domain. Sequence analysis and exon-intron structure analysis of various MIKC-type genes favored longer I domain hypothesis wherein the I region of MIKC* was found to be encoded by 4 or 5 exons whereas in MIKCC it is encoded by just one exon (Henschel et al., 2002). Later Kwantes et al. (2012) redefined the existing hypotheses which seems more reliable that states, MIKCC type gene was found in the common ancestor of charophycean-land plant lineage and later duplication events in the first part of the Keratin-like region resulted in the evolution of MIKC* group genes which distinguished it from classical MIKCC type gene (Kwantes et al., 2012).
Some MIKCC gene orthologs of angiosperms are reported in gymnosperms with functional conservation (Sundstrom and Engstrom, 2002). But no such orthologs are so far identified in non-seed plants (Henschel et al., 2002). This suggests MIKCC class of genes have undergone independent gene duplications and functional diversifications during evolution which resulted in the extant highly diversified MIKCC type MADS-box genes in seed plants (Henschel et al., 2002). This may be a probable reason for the absence of traces of orthology of floral MIKCC type genes in non-seed plants (Henschel et al., 2002). In contrast, MIKC* genes show considerable orthology in ferns, gymnosperms, angiosperms and branched into two monophyletic subgroups S and P (Nam et al., 2004; Verelst et al., 2007; Gramzow et al., 2012; Kwantes et al., 2012; Liu et al., 2013). It should be noted that the MIKC* gene family has a small size in plants across taxa and the S and P clades have been the only reported diversification in the monilophytes and spermatophytes. It is thus hypothesized that conservation of these clades reveals a key role that has been maintained highly similar during the evolution of these plant groups (Kwantes et al., 2012).
Is Size of MIKC MADS-Box Gene Family and Complexity of the Organism Interrelated?
There is an existing hypothesis that size of a gene family involved in development is correlated to the complexity of the organism (Floyd and Bowman, 2007; Quodt et al., 2007). The size of MIKC type MADS-box gene family varies from one in algae, 17 in Physcomitrella to 37 in Rice and 46 in Arabidopsis (Parenicova et al., 2003; Tanabe et al., 2005; Arora et al., 2007; Barker and Ashton, 2013, 2016). This suggests a trend of increasing MIKC numbers during evolution of green plants which implies a correlation between surge in gene numbers and developmental complexity (single celled algae to multicellular algae to multicellular non-vascular plants to early vascular plants and finally seed plants) (Quodt et al., 2007). One such example of the above hypothesis with respect to the MADS box gene family is of the MIKC* clade in bryophytes where the authors speculate that increase in MIKC* genes is related to evolutionary change in growth habit (Zobell et al., 2010). The authors predict that the growth pattern changes from an ancestral predominantly thalloid growth (one gene in M. polymopha) to a condition of transiently thalloid but mainly leafy growth (four or more genes in S subsecundum) or a condition of both filamentous and leafy growth (11 genes in F. hygrometrica and P. patens), can be assigned to increase in gene numbers, however further studies will be required to substantiate this hypothesis (Zobell et al., 2010). Thus it is predicted that MIKC type MADS-box gene family diversified and duplicated extensively after the separation from the charophycean lineage and expansion of this gene family was important for the evolution of advanced morphological features in extant seed plants.
Role of MIKC MADS-Box Genes in Non-seed Plants
In flowering plants, MIKCC and MIKC* genes have more specific expression patterns in sporophytes and gametophytes and its functions are well studied; refer to Table 1D. This specialized expression probably evolved with increase in complexity of the organs in extant seed plants (Münster et al., 1997; Theissen et al., 2000; Theissen and Saedler, 2001; Gramzow and Theissen, 2010). The literature available in non-seed plants, mostly reports the broad expression pattern of MIKCC genes in both sporophytes and gametophytes; refer to Table 1B (Münster et al., 1997, 2002; Hasebe et al., 1998; Huang et al., 2014). This has led to the hypothesis that non-seed plant MIKCC genes may not have any organ specific roles; instead it may have a generalized role in the transcriptional control of different developmental events during its life cycle (Münster et al., 1997; Hasebe et al., 1998; Huang et al., 2014). However there are exceptions reported in some studies which cannot be ignored. Few MIKCC genes were found to have specific expression pattern rather than ubiquitous expression (Münster et al., 2002; Tanabe et al., 2005; Huang et al., 2014; Ruiz-Estévez et al., 2017). DfMADS1 gene was proposed to have role in spore germination and reproductive development of Dryopteris fragrans due to its high expression in spores and prothallus (Huang et al., 2014). VsMB2, VsMB4 may be important in the alternation of the two main phases of V. speciosa life cycle as it is specifically down regulated in the gametophytes (Ruiz-Estévez et al., 2017). OPM4 gene of O. pedunculosum expressed exclusively in the reproductive structures (Münster et al., 2002) while VsMB3, VsMB7 were reported to have reduced expression in the reproductive structures of V. speciosa (Ruiz-Estévez et al., 2017) which shows that differential expression of these genes with respect to the vegetative structures may be critical for the development of the reproductive structures. The MIKCC genes function in the formation of motile flagella sperms in Physcomitrella patens (Koshimizu et al., 2018) and have a putative role in flagellate sperm differentiation in Chara globularis (Tanabe et al., 2005) suggesting they regulate flagellum-related genes in mosses and charophycean green algae. The absence of these homologs in Arabidopsis suggests that they may have been lost during transition from the use of flagellate sperm to pollen tubes for fertilization (Koshimizu et al., 2018). Another function of the MIKCC genes from P. patens seems to be its involvement in gametangia formation i.e., differentiation of reproductive structures from the non-reproductive tissues in the gametophore. This function is analogous to the class C genes in spermatophytes and thus may represent an ancestral function which remained conserved through evolution though the genes have not been shown to be orthologous (Quodt et al., 2007; Singer et al., 2007; Singer and Ashton, 2009).
From the studies discussed above in bryophytes, lycophytes and ferns it seems MIKC* genes have a conserved expression and function with respect to haploid phase of the life cycle (Svensson et al., 2000; Henschel et al., 2002; Zobell et al., 2010; Kwantes et al., 2012). The higher proportion of MIKC* genes than MIKCC genes in bryophytes as compared to vascular plants (Tables 1A,D) indicates that these genes have a role to play in a gametophyte -dominant life cycle (Gramzow et al., 2012). Functional conservation of MIKC* genes across taxa was demonstrated by the partial rescue of pollen in vitro germination defect of Arabidopsis MIKC* mutant pollen by MpMADS1, an MIKC* protein from Marchantia (Zobell et al., 2010; Kwantes et al., 2012). It is possible that perhaps with time their function was restricted to the male gametophyte in the lineage leading to monocots and higher eudicots about 150 million years ago (Verelst et al., 2007; Liu et al., 2013). This kind of conservation has not been reported for MIKCC transcription factors thus suggesting a conserved role of MIKC* transcription factors during gametophytic development (Zobell et al., 2010; Kwantes et al., 2012).
Future Prospects
These limited reports on the specific roles of MIKC genes in non-seed plants are noteworthy. It gives us clues that non-seed plant MIKC genes may also have targeted roles. It questions the existing hypothesis on the generalized function of the non-seed plant MIKC genes. Extensive research has not yet been carried out in non-seed plants as most of the functions assigned to the non-seed plant MIKC genes are putative and on the basis of expression studies (Refer to Table 1C). Knock-outs and overexpression studies in near future to study the exact role of these genes may help us to come up with a clearer picture about the role of MIKC genes in non-seed plants. An initial insight on the hornwort species, Anthoceros agrestis has given a hint that there was a dramatic reduction in the MADS-box gene family size when compared to that of P. patens (Szövényi, 2016). A detailed report on the MADS-box genes in hornworts is still lacking. Hornwort shares some common characteristics with both algae and spermatophytes and is believed to be the immediate sister for all the tracheophytes (vascular plants) (Renzaglia, 1978). Exploring hornwort lineage will be very essential to know the role and evolution of MADS-box gene family in land plants. Information on the MIKCC genes in Marchantia, Funaria, and Sphagnum is also missing hence further work will be required to bring to light their presence or absence in these bryophyte species (Zobell et al., 2010). Like AGAMOUS, some MIKCC genes from Ceratopteris, Selaginella, and Physcomitrella have an N-terminal region preceding the MADS-box, the importance of this region remains to be explored in non-seed plants. Extending functional studies like mutational analysis, gene silencing and targeted genome editing using the CRISPR/Cas9 system to non-seed plants in near future may be useful in understanding the ancestral function of MADS-box genes. Acquiring knowledge on functional and structural aspects of MADS-box gene family in non-seed plants may give us clues on the evolution of advanced organs and developmental processes in extant land plants.
Authors Contributions
SN conceived the idea; GT and SN conceptualized and drafted the review; SN finalized the review.
Funding
SN sincerely thanks the Department of Science and Technology, Government of India for the INSPIRE Faculty Award. We gratefully acknowledge Science and Engineering Board, Department of Science and Technology, Government of India for the Extra Mural Research Grant.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
SN sincerely thanks Prof. M Radhakrishna Pillai and Dr. S Manjula for hosting the INSPIRE Faculty Award at Rajiv Gandhi Centre for Biotechnology, Thiruvananthapuram and providing all necessary facilities and infrastructure. The authors acknowledge the valuable inputs from the reviewers.
References
Alvarez-Buylla, E. R., Pelaz, S., Liljegren, S. J., Gold, S. E., Burgeff, C., Ditta, G. S., et al. (2000). An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. U.S.A. 97, 5328–5333. doi: 10.1073/pnas.97.10.5328
Arora, R., Agarwal, P., Ray, S., Singh, A. K., Singh, V. P., Tyagi, A. K., et al. (2007). MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8:242. doi: 10.1186/1471-2164-8-242
Barker, E. I., and Ashton, N. W. (2013). A parsimonious model of lineage-specific expansion of MADS-box genes in Physcomitrella patens. Plant Cell Rep. 32, 1161–1177. doi: 10.1007/s00299-013-1411-8
Barker, E. I., and Ashton, N. W. (2016). Ancestral and more recently acquired syntenic relationships of MADS-box genes uncovered by the Physcomitrella patens pseudochromosomal genome assembly. Plant Cell Rep. 35, 505–512. doi: 10.1007/s00299-015-1898-2
Becker, A., and Theissen, G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29, 464–489. doi: 10.1016/S1055-7903(03)00207-0
Cho, S., Jang, S., Chae, S., Chung, K. M., Moon, Y. H., An, G., et al. (1999). Analysis of the C-terminal region of Arabidopsis thaliana APETALA1 as a transcription activation domain. Plant Mol. Biol. 40, 419–429. doi: 10.1023/A:1006273127067
Coen, E. S., and Meyerowitz, E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1038/353031a0
Davies, B., and Schwarz-Sommer, Z. (1994). “Control of floral organ identity by homeotic MADS-Box transcription factors,” in Plant Promoters and Transcription Factors, ed L. Nover. (Berlin; Heidelberg: Springer Berlin Heidelberg), 235–258.
de Folter, S., Busscher, J., Colombo, L., Losa, A., and Angenent, G. C. (2004). Transcript profiling of transcription factor genes during silique development in Arabidopsis. Plant Mol. Biol. 56, 351–366. doi: 10.1007/s11103-004-3473-z
Derelle, E., Ferraz, C., Rombauts, S., Rouze, P., Worden, A. Z., Robbens, S., et al. (2006). Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. U.S.A. 103, 11647–11652. doi: 10.1073/pnas.0604795103
Egea-Cortines, M., Saedler, H., and Sommer, H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18, 5370–5379. doi: 10.1093/emboj/18.19.5370
Fernandez, D. E., Heck, G. R., Perry, S. E., Patterson, S. E., Bleecker, A. B., and Fang, S. C. (2000). The embryo MADS domain factor AGL15 acts postembryonically. Inhibition of perianth senescence and abscission via constitutive expression. Plant Cell 12, 183–198. doi: 10.1105/tpc.12.2.183
Floyd, S. K., and Bowman, J. L. (2007). The ancestral developmental tool kit of land plants. Int. J. Plant Sci. 168, 1–35. doi: 10.1086/509079
Gifford, E. M., and Foster, A. S. (1989). Morphology and Evolution of Vascular Plants 3rd Edn. New York, NY: W. H. Freeman.
Gilbert, S. F., Opitz, J. M., and Raff, R. A. (1996). Resynthesizing evolutionary and developmental biology. Dev. Biol. 173, 357–372. doi: 10.1006/dbio.1996.0032
Graham, L. E., Cook, M. E., and Busse, J. S. (2000). The origin of plants: body plan changes contributing to a major evolutionary radiation. Proc. Natl. Acad. Sci. U.S.A. 97, 4535–4540. doi: 10.1073/pnas.97.9.4535
Gramzow, L., Barker, E., Schulz, C., Ambrose, B., Ashton, N., Theissen, G., et al. (2012). Selaginella genome analysis - entering the “homoplasy heaven” of the MADS world. Front. Plant Sci. 3:214. doi: 10.3389/fpls.2012.00214
Gramzow, L., and Theissen, G. (2010). A hitchhiker's guide to the MADS world of plants. Genome Biol. 11:214. doi: 10.1186/gb-2010-11-6-214
Hasebe, M., Wen, C. K., Kato, M., and Banks, J. A. (1998). Characterization of MADS homeotic genes in the fern Ceratopteris richardii. Proc. Natl. Acad. Sci. U.S.A. 95, 6222–6227. doi: 10.1073/pnas.95.11.6222
Heard, J., Caspi, M., and Dunn, K. (1997). Evolutionary diversity of symbiotically induced nodule MADS box genes: characterization of nmhC5, a member of a novel subfamily. Mol. Plant Microbe Interact. 10, 665–676. doi: 10.1094/MPMI.1997.10.5.665
Heard, J., and Dunn, K. (1995). Symbiotic induction of a MADS-box gene during development of alfalfa root nodules. Proc. Natl. Acad. Sci. U.S.A. 92, 5273–5277. doi: 10.1073/pnas.92.12.5273
Henschel, K., Kofuji, R., Hasebe, M., Saedler, H., Munster, T., and Theissen, G. (2002). Two ancient classes of MIKC-type MADS-box genes are present in the moss Physcomitrella patens. Mol. Biol. Evol. 19, 801–814. doi: 10.1093/oxfordjournals.molbev.a004137
Hohe, A., Rensing, S. A., Mildner, M., Lang, D., and Reski, R. (2002). Day length and temperature strongly influence sexual reproduction and expression of a novel MADS-box gene in the moss Physcomitrella patens. Plant Biol. 4, 595–602. doi: 10.1055/s-2002-35440
Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409, 525–529. doi: 10.1038/35054083
Huang, Q., Li, W., Fan, R., and Chang, Y. (2014). New MADS-box gene in fern: cloning and expression analysis of DfMADS1 from Dryopteris fragrans. PLoS ONE 9:e86349. doi: 10.1371/journal.pone.0086349
Kaufmann, K., Melzer, R., and Theissen, G. (2005). MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347, 183–198. doi: 10.1016/j.gene.2004.12.014
Kenrick, P., and Crane, P. R. (1997). The origin and early evolution of plants on land. Nature 389, 33–39. doi: 10.1038/37918
Kofuji, R., Sumikawa, N., Yamasaki, M., Kondo, K., Ueda, K., Ito, M., et al. (2003). Evolution and divergence of the MADS-box gene family based on genome-wide expression analyses. Mol. Biol. Evol. 20, 1963–1977. doi: 10.1093/molbev/msg216
Koshimizu, S., Kofuji, R., Sasaki-Sekimoto, Y., Kikkawa, M., Shimojima, M., Ohta, H., et al. (2018). Physcomitrella MADS-box genes regulate water supply and sperm movement for fertilization. Nat. Plants 4, 36–45. doi: 10.1038/s41477-017-0082-9
Krogan, N. T., and Ashton, N. W. (2000). Ancestry of plant MADS-box genes revealed by bryophyte (Physcomitrella patens) homologues. New Phytol. 147, 505–517. doi: 10.1046/j.1469-8137.2000.00728.x
Kwantes, M., Liebsch, D., and Verelst, W. (2012). How MIKC* MADS-box genes originated and evidence for their conserved function throughout the evolution of vascular plant gametophytes. Mol. Biol. Evol. 29, 293–302. doi: 10.1093/molbev/msr200
Lehti-Shiu, M. D., Adamczyk, B. J., and Fernandez, D. E. (2005). Expression of MADS-box genes during the embryonic phase in Arabidopsis. Plant Mol. Biol. 58, 89–107. doi: 10.1007/s11103-005-4546-3
Liu, Y., Cui, S., Wu, F., Yan, S., Lin, X., Du, X., et al. (2013). Functional conservation of MIKC*-Type MADS box genes in Arabidopsis and rice pollen maturation. Plant Cell 25, 1288–1303. doi: 10.1105/tpc.113.110049
Ma, H., Yanofsky, M. F., and Meyerowitz, E. M. (1991). AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 5, 484–495. doi: 10.1101/gad.5.3.484
Matsuzaki, M., Misumi, O., Shin, I. T., Maruyama, S., Takahara, M., Miyagishima, S. Y., et al. (2004). Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428, 653–657. doi: 10.1038/nature02398
Mizukami, Y., Huang, H., Tudor, M., Hu, Y., and Ma, H. (1996). Functional domains of the floral regulator AGAMOUS: characterization of the DNA binding domain and analysis of dominant negative mutations. Plant Cell 8, 831–845. doi: 10.1105/tpc.8.5.831
Münster, T., Faigl, W., Saedler, H., and Theiβen, G. (2002). Evolutionary aspects of MADS-box genes in the eusporangiate fern ophioglossum. Plant Biol. 4, 474–483. doi: 10.1055/s-2002-34130
Münster, T., Pahnke, J., Di Rosa, A., Kim, J. T., Martin, W., Saedler, H., et al. (1997). Floral homeotic genes were recruited from homologous MADS-box genes preexisting in the common ancestor of ferns and seed plants. Proc. Natl. Acad. Sci. U.S.A. 94, 2415–2420. doi: 10.1073/pnas.94.6.2415
Nam, J., Kim, J., Lee, S., An, G., Ma, H., and Nei, M. (2004). Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc. Natl. Acad. Sci. U.S.A. 101, 1910–1915. doi: 10.1073/pnas.0308430100
Nurrish, S. J., and Treisman, R. (1995). DNA binding specificity determinants in MADS-box transcription factors. Mol. Cell. Biol. 15, 4076–4085. doi: 10.1128/MCB.15.8.4076
Palenik, B., Grimwood, J., Aerts, A., Rouze, P., Salamov, A., Putnam, N., et al. (2007). The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. U.S.A. 104, 7705–7710. doi: 10.1073/pnas.0611046104
Parenicova, L., de Folter, S., Kieffer, M., Horner, D. S., Favalli, C., Busscher, J., et al. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15, 1538–1551. doi: 10.1105/tpc.011544
Purugganan, M. D., Rounsley, S. D., Schmidt, R. J., and Yanofsky, M. F. (1995). Molecular evolution of flower development: diversification of the plant MADS-box regulatory gene family. Genetics 140, 345–356.
Quodt, V., Faigl, W., Saedler, H., and Munster, T. (2007). The MADS-domain protein PPM2 preferentially occurs in gametangia and sporophytes of the moss Physcomitrella patens. Gene 400, 25–34. doi: 10.1016/j.gene.2007.05.016
Rensing, S. A., Lang, D., Zimmer, A. D., Terry, A., Salamov, A., Shapiro, H., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69. doi: 10.1126/science.1150646
Renzaglia, K. S. (1978). A comparative morphology and developmental anatomy of the Anthocerotophyta. J. Hattori Botanical Lab. 44, 31–90.
Riechmann, J. L., and Meyerowitz, E. M. (1997). Determination of floral organ identity by Arabidopsis MADS domain homeotic proteins AP1, AP3, PI, and AG is independent of their DNA-binding specificity. Mol. Biol. Cell 8, 1243–1259. doi: 10.1091/mbc.8.7.1243
Riese, M., Faigl, W., Quodt, V., Verelst, W., Matthes, A., Saedler, H., et al. (2005). Isolation and characterization of new MIKC*-Type MADS-box genes from the moss Physcomitrella patens. Plant Biol. 7, 307–314. doi: 10.1055/s-2005-865640
Rijpkema, A. S., Gerats, T., and Vandenbussche, M. (2007). Evolutionary complexity of MADS complexes. Curr. Opin. Plant Biol. 10, 32–38. doi: 10.1016/j.pbi.2006.11.010
Ruiz-Estévez, M., Bakkali, M., Martín-Blázquez, R., and Garrido-Ramos, M. A. (2017). Differential expression patterns of MIKCC-type MADS-box genes in the endangered fern Vandenboschia speciosa. PLGENE Plant Gene 12, 50–56. doi: 10.1016/j.plgene.2017.07.006
Schwarz-Sommer, Z., Huijser, P., Nacken, W., Saedler, H., and Sommer, H. (1990). Genetic control of flower development by homeotic genes in Antirrhinum majus. Science 250, 931–936. doi: 10.1126/science.250.4983.931
Sharrocks, A. D., Gille, H., and Shaw, P. E. (1993). Identification of amino acids essential for DNA binding and dimerization in p67SRF: implications for a novel DNA-binding motif. Mol. Cell. Biol. 13, 123–132. doi: 10.1128/MCB.13.1.123
Shore, P., and Sharrocks, A. D. (1995). The MADS-box family of transcription factors. Eur. J. Biochem. 229, 1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x
Singer, S. D., and Ashton, N. W. (2009). MADS about MOSS. Plant Signal. Behav. 4, 111–112. doi: 10.4161/psb.4.2.7479
Singer, S. D., Krogan, N. T., and Ashton, N. W. (2007). Clues about the ancestral roles of plant MADS-box genes from a functional analysis of moss homologues. Plant Cell Rep. 26, 1155–1169. doi: 10.1007/s00299-007-0312-0
Stewart, W. N., and Rothwell, G. W. (1993). Paleobotany and the Evolution of Plants 2nd Edn. Cambridge, UK: Cambridge University Press.
Sundstrom, J., and Engstrom, P. (2002). Conifer reproductive development involves B-type MADS-box genes with distinct and different activities in male organ primordia. Plant J. 31, 161–169. doi: 10.1046/j.1365-313X.2002.01343.x
Svensson, M. E., and Engström, P. (2002). Closely related MADS-box genes in club moss (Lycopodium) show broad expression patterns and are structurally similar to, but phylogenetically distinct from, typical seed plant MADS-box genes. New Phytol. 154, 439–450. doi: 10.1046/j.1469-8137.2002.00392.x
Svensson, M. E., Johannesson, H., and Engstrom, P. (2000). The LAMB1 gene from the clubmoss, Lycopodium annotinum, is a divergent MADS-box gene, expressed specifically in sporogenic structures. Gene 253, 31–43. doi: 10.1016/S0378-1119(00)00243-2
Szövényi, P. (2016). “Chapter 6: The genome of the model species Anthoceros agrestis,” in Advances in Botanical Research, ed S. A. Rensing (Cambridge, MA: Academic Press), 189–211.
Tanabe, Y., Hasebe, M., Sekimoto, H., Nishiyama, T., Kitani, M., Henschel, K., et al. (2005). Characterization of MADS-box genes in charophycean green algae and its implication for the evolution of MADS-box genes. Proc. Natl. Acad. Sci. U.S.A. 102, 2436–2441. doi: 10.1073/pnas.0409860102
Tanabe, Y., Uchida, M., Hasebe, M., and Ito, M. (2003). Characterization of the Selaginella remotifolia MADS-box gene. J. Plant Res. 116, 71–75.
Tapia-Lopez, R., Garcia-Ponce, B., Dubrovsky, J. G., Garay-Arroyo, A., Perez-Ruiz, R. V., Kim, S. H., et al. (2008). An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiol. 146, 1182–1192. doi: 10.1104/pp.107.108647
Theissen, G., Becker, A., Di Rosa, A., Kanno, A., Kim, J. T., Munster, T., et al. (2000). A short history of MADS-box genes in plants. Plant Mol. Biol. 42, 115–149. doi: 10.1023/A:1006332105728
Theissen, G., Kim, J. T., and Saedler, H. (1996). Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol. 43, 484–516. doi: 10.1007/BF02337521
Theissen, G., and Saedler, H. (1995). MADS-box genes in plant ontogeny and phylogeny: Haeckel's ‘biogenetic law’ revisited. Curr. Opin. Genet. Dev. 5, 628–639. doi: 10.1016/0959-437X(95)80032-8
Theissen, G., and Saedler, H. (2001). Plant biology. Floral quartets. Nature 409, 469–471. doi: 10.1038/35054172
Verelst, W., Saedler, H., and Munster, T. (2007). MIKC* MADS-protein complexes bind motifs enriched in the proximal region of late pollen-specific Arabidopsis promoters. Plant Physiol. 143, 447–460. doi: 10.1104/pp.106.089805
Zobell, O., Faigl, W., Saedler, H., and Munster, T. (2010). MIKC* MADS-box proteins: conserved regulators of the gametophytic generation of land plants. Mol. Biol. Evol. 27, 1201–1211. doi: 10.1093/molbev/msq005
Keywords: MADS box, MIKC type, non-seed plants, evolution, algae, bryophytes, lycophytes, ferns
Citation: Thangavel G and Nayar S (2018) A Survey of MIKC Type MADS-Box Genes in Non-seed Plants: Algae, Bryophytes, Lycophytes and Ferns. Front. Plant Sci. 9:510. doi: 10.3389/fpls.2018.00510
Received: 21 December 2017; Accepted: 03 April 2018;
Published: 18 April 2018.
Edited by:
Stefan de Folter, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional, MexicoReviewed by:
Elizabeth Barker, University of Regina, CanadaFederico Valverde, Consejo Superior de Investigaciones Científicas, Spain
Copyright © 2018 Thangavel and Nayar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Saraswati Nayar, c2FyYXN3YXRpbkByZ2NiLnJlcy5pbg==
†These authors have contributed equally to this work.
 Gokilavani Thangavel
Gokilavani Thangavel Saraswati Nayar
Saraswati Nayar