- 1Unit of Polar Genomics, Korea Polar Research Institute, Incheon, South Korea
- 2Department of Systems Biology, Yonsei University, Seoul, South Korea
- 3Polar Science, University of Science & Technology, Daejeon, South Korea
Few plant species can survive in Antarctica, the harshest environment for living organisms. Deschampsia antarctica is the only natural grass species to have adapted to and colonized the maritime Antarctic. To investigate the molecular mechanism of the Antarctic adaptation of this plant, we identified and characterized D. antarctica C-repeat binding factor 4 (DaCBF4), which belongs to monocot CBF group IV. The transcript level of DaCBF4 in D. antarctica was markedly increased by cold and dehydration stress. To assess the roles of DaCBF4 in plants, we generated a DaCBF4-overexpressing transgenic rice plant (Ubi:DaCBF4) and analyzed its abiotic stress response phenotype. Ubi:DaCBF4 displayed enhanced tolerance to cold stress without growth retardation under any condition compared to wild-type plants. Because the cold-specific phenotype of Ubi:DaCBF4 was similar to that of Ubi:DaCBF7 (Byun et al., 2015), we screened for the genes responsible for the improved cold tolerance in rice by selecting differentially regulated genes in both transgenic rice lines. By comparative transcriptome analysis using RNA-seq, we identified 9 and 15 genes under normal and cold-stress conditions, respectively, as putative downstream targets of the two D. antarctica CBFs. Overall, our results suggest that Antarctic hairgrass DaCBF4 mediates the cold-stress response of transgenic rice plants by adjusting the expression levels of a set of stress-responsive genes in transgenic rice plants. Moreover, selected downstream target genes will be useful for genetic engineering to enhance the cold tolerance of cereal plants, including rice.
Introduction
Plants are sessile organisms and are constantly exposed to adverse environmental conditions, to which they have adapted (Lata and Prasad, 2011; Claeys and Inzé, 2013). The maritime Antarctic climate has low temperatures, high soil salinity, and a water deficit, which hamper the survival of angiosperms. Deschampsia antarctica Desv. (Poaceae) is one of two flowering plants that naturally inhabit Antarctica (Alberdi et al., 2002). This species has developed various adaptive mechanisms to survive in the Antarctic.
The adaptations that enable D. antarctica to survive in the harsh environment of the maritime Antarctic include changes in the leaf anatomy and the physiology of the photosynthetic apparatus (Giełwanowska et al., 2005; Sáez et al., 2017). Increased levels of nonstructural carbohydrates, apoplastic antifreeze proteins, and ice recrystallization inhibition proteins (biochemical cryoprotectants) reportedly enable plants to flourish under Antarctic conditions (Bravo and Griffith, 2005; John et al., 2009; Pastorczyk et al., 2014). In addition, transcriptome analysis of D. antarctica under abiotic stress demonstrated changes in the expression levels of stress-responsive genes (Lee et al., 2013a). However, the signaling pathways that mediate activation of the expression of the stress-related genes responsible for the abiotic stress tolerance of this plant are unclear.
In Arabidopsis, C-repeat binding factor (CBF)/dehydration-responsive element-binding protein (DREB) is the major transcription factor responsible for induction of cold tolerance (Thomashow, 2010). Also, CBF overexpression in monocot plants resulted in enhanced tolerance to diverse abiotic stresses including cold and freezing. When overexpressed in barley, TaDREB3, TaCBF14, and TaCBF15 from wheat and HvCBF2a from barley resulted in enhanced frost tolerance by increasing the transcript levels of downstream target genes such as COR14b and DHN5 (Kovalchuk et al., 2013; Soltész et al., 2013; Jeknić et al., 2014). Moreover, TaCBF3 overexpression enhanced tolerance to frost and cold in wheat and barley (Morran et al., 2011). Expression of cotton GhDREB in barley also resulted in enhanced cold tolerance (Gao et al., 2009). Indeed, overexpression of the CBF genes from rice (OsDREB1), barley (HvCBF4), and maize (ZmCBF3) resulted in enhanced cold tolerance in rice (Ito et al., 2006; Oh et al., 2007; Xu et al., 2011).
Followed by the discovery of AtCBF1 as the major transcriptional regulator responsible for inducing the expression of downstream COR genes in Arabidopsis (Jaglo-Ottosen et al., 1998), the CBF proteins of monocot cereal plants have been discovered genome-wide (Stockinger, 2009; Tondelli et al., 2011). Moreover, Badawi et al. (2007) divided monocot CBF proteins into groups I–IV, and group V was recently identified (Byun et al., 2015).
However, few studies have aimed to identify the downstream target genes responsible for the cold tolerance in cereal plants. Overexpression of the rice CBF homolog OsDREB1A, have shown enhanced tolerance of transgenic rice to drought, high-salt, and low-temperature stresses. A microarray analysis of 21,500 rice genes together with northern blot analysis confirmed that 12 genes, including dehydrins and a protease inhibitor, were upregulated in OsDREB1-overexpressing rice plants (Ito et al., 2006). Rice plants continuously overexpressing HvCBF4 from barley showed improved tolerance to cold, drought, and high-salt stresses. A microarray analysis revealed that the transcript levels of 15 genes, including Bowman Birk trypsin inhibitor 1 (BBTI1), were upregulated in HvCBF4 overexpressing rice plants under normal condition (Oh et al., 2007).
In previous work, we generated transgenic rice plants overexpressing DaCBF7, a CBF gene from D. antarctica, and analyzed the expression levels of downstream genes. DaCBF7 overexpression resulted in enhanced cold tolerance compared to wild-type plants (Byun et al., 2015). In this study, we found that the D. antarctica CBF4 gene (DaCBF4) encodes a homolog of cereal CBF group IV. The expression of this gene was induced in D. antarctica plants under cold and drought stress. Overexpression of DaCBF4 in rice resulted in a cold-specific phenotype similar to that of the DaCBF7-overexpressing plants. Thus, we identified the genes responsible for increasing the cold tolerance of rice by screening differentially regulated genes common to DaCBF7- and DaCBF4-overexpressing rice plants. Based on these results, we suggest that the Antarctic hairgrass DaCBF4 gene plays a crucial role in the cold tolerance in transgenic rice plants. Selected downstream target genes common to Ubi:DaCBF4 and Ubi:DaCBF7 will be useful for genetic engineering to enhance the cold-stress tolerance of cereal plants, including rice.
Materials and Methods
Phylogenetic Analysis
The amino acid sequences of DaCBF4 and other CBF/DREB homologs from monocot crops were retrieved from the GenBank database and proofread. All downstream analyses were performed using the MEGA7 software (Kumar et al., 2016). Phylogenetic trees were constructed from the data sets by the neighbor-joining method based on the JTT matrix-based model. All positions with <95% site coverage were eliminated. Fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. Supports for internal branches were tested by bootstrap analyses of 1,000 replications. The accession numbers of CBF homologs from five monocot species—Antarctic hairgrass (D. antarctica), barley (Hordeum vulgare), rice (Oryza sativa), hexaploid wheat (Triticum aestivum), and einkorn wheat (Triticum monococcum)—are presented in Supplementary Table 1.
Subcellular Localization Experiment
The synthetic green fluorescent protein (sGFP)-coding region was fused in-frame to the 3′ end of the full-length DaCBF4-coding region and inserted into the pEarleyGate 100 binary vector. The vector was transformed into Agrobacterium tumefaciens strain GV3101 by electroporation. Tobacco (Nicotiana benthamiana) leaves were co-infiltrated using A. tumefaciens that contained the 35S:DaCBF4-sGFP or 35S:sGFP constructs. A 35S:nuclear localizing signal-monomeric red fluorescent protein (35S:NLS-mRFP) construct was used as a control nuclear protein. Two days after infection, protoplasts were extracted from the tobacco leaves and visualized by fluorescence microscopy (BX51, Olympus).
Plant Materials and Stress Conditions
Deschampsia antarctica was collected near the King Sejong Antarctic Station (62°14′29″S; 58°44′18″W) on the Barton Peninsula of King George Island in January 2007. The plants were cultured in vitro in tissue culture medium [Murashige and Skoog (MS) medium; 2% sucrose and 0.8% phytoagar (pH 5.7)] under a 16-h light/8-h dark photoperiod with a light intensity of 150 μmol m−2 s−1 at 15°C. For cold-stress conditions, plants grown at 15°C were transferred to a climate chamber at 4°C for the indicated periods. For drought-stress conditions, plants were transferred to filter paper and dried at 15°C. For high-salt conditions, plants were transferred to MS medium supplemented with 150 mM NaCl and incubated at 15°C. RNA was extracted from leaves and analyzed at various times after the imposition of stress. All the samplings for expression analysis was conducted at the same time to avoid variation by circadian rhythm.
Total RNA Extraction and Reverse Transcription-Quantitative Polymerase Chain Reaction
Total RNA was isolated from leaves of five D. antarctica plants with four tillers each (longest tiller, 5 cm) using an RNeasy Plant Mini Kit in conjunction with the RNase free DNase set (Qiagen) according to the manufacturer's instructions. The quantity and quality of total RNA were determined by spectroscopic measurements at 230, 260, and 280 nm using an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE), and RNA integrity was checked by electrophoresis in a 2% agarose gel. First-strand cDNA was synthesized from 2 μg of total RNA using Superscript III (Invitrogen). Real-time reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis was performed in 20-μL reaction mixtures that included 1 μL of a 1:15 diluted cDNA template, 2 μM of each primer, and 10 μL of QuantiFast SYBR Green PCR kit (Qiagen). Amplified signals were monitored continuously using the Mx3000P qPCR System (Stratagene). The amplification protocol was as follows: 5 min of denaturation and enzyme activation at 95°C; followed by 40 cycles at 95°C for 5 s, 58°C for 20 s, and 72°C for 15 s. The DaEF1a gene was used as an internal control (Lee et al., 2010). The DNA sequences of primers used for PCR amplification are listed in Supplementary Table 2.
Generation of DaCBF4-Overexpressing Transgenic Rice Plants
To generate Ubi:DaCBF4 transgenic rice plants, the 651-bp DaCBF4-coding region was inserted into the pGA2897 vector. The binary vector was transformed into A. tumefaciens strain LBA4404 by electroporation and used for rice transformation as described previously (Byun and Kim, 2014). Callus tissue was induced from wild-type rice (O. sativa L. japonica variety “Dong-Jin”) seeds, co-cultivated with A. tumefaciens, and selected on callus induction medium containing antibiotics (40 mg/L hygromycin B and 250 mg/L carbenicillin). Selected callus tissue was transferred to a regeneration medium. Transgenic T0 plants were transplanted to soil in a greenhouse and T2 plants were used for phenotypic analysis.
RT-PCR and DNA Gel-Blot Analysis
Total RNA was isolated from mature leaves of wild-type and Ubi:DaCBF4 transgenic rice plants using TRIzol reagent [38% equilibrated phenol, pH 4.3, 1 M guanidine thiocyanate, 1 M ammonium thiocyanate, 0.3 M sodium acetate, pH 5.2, and 5% glycerol (v/v)]. First-strand cDNA was synthesized from 2 μg of total RNA using oligo(dT) primers and TOPscript Reverse Transcriptase (Enzynomics). To compare the DaCBF4 expression levels of the transgenic lines, DaCBF4 was amplified using a gene-specific primer set (Supplementary Table 2). To distinguish the independent lines of Ubi:DaCBF4 transgenic plants, genomic Southern blotting was performed. Total genomic DNA was isolated from mature rice leaves using cetyltrimethylammonium bromide (CTAB) solution, digested with BamHI, and separated in a 0.8% agarose gel. The gel was treated sequentially with depurifying, denaturing, and neutralizing solutions, and the DNA was transferred to Hybond-N nylon membranes. The blot was hybridized with a 32P-labeled hygromycin B phosphotransferase (hph) probe under high-stringency conditions.
Rice Plant Materials and Stress Conditions
Dry rice seeds were sterilized with 0.4% NaClO solution for 30 min and washed several times with sterilized water. Seeds of wild-type and Ubi:DaCBF4 transgenic rice plants were germinated on half-strength MS medium containing vitamins (Duchefa Biochemie), 3% sucrose, and 0.7% phytoagar. Seedlings were grown for 2 weeks at 28°C under a 16-h light/8-h dark photoperiod and then transplanted to soil in a greenhouse as described by Byun et al. (2017). To investigate the effect of DaCBF4 on stress tolerance, 6-week-old plants grown in the growth room were subjected to various abiotic stresses, observed, and photographed. For cold-stress conditions, wild-type and DaCBF4-overexpressing plants (independent lines #1 and #2) grown at 28°C were transferred to 4°C for 5 days, and then returned to 28°C.
RNA-Sequencing
RNA-sequencing (RNA-seq) was carried out using total RNA from 6-week-old wild-type and DaCBF4-overexpressing line #1 plants under normal and cold-stress conditions (1 or 6 days after transfer to 4°C). Total RNAs were extracted from leaves of each genotype and each treatment using TRIzol reagent, treated with DNase I to remove contaminant genomic DNA, and purified using the RNeasy Mini kit (Qiagen) following the manufacturer's instructions. Three different biological replicates were prepared. The integrity and concentration of RNA were determined using a Bioanalyzer system (RIN > 6) (Agilent Technologies) and a Qubit RNA broad-range assay kit (Life Technologies), respectively. To construct the sequencing library, 1.5 μg of total RNA from each sample was used as input for the TruSeq RNA sample prep kit ver. 2 (Illumina). The libraries were validated, quantified using the Bioanalyzer and the library qPCR quantification method, multiplexed with equal ratios, and loaded into the flow cell of the Illumina MiSeq Reagent Kit ver. 3 (2 × 75 runs). Sequencing was performed using a MiSeq Sequencer system (Illumina) and a total of 3 Gb (40 M paired-end reads) of sequencing data were generated (Q30 > 98%). The RNA-Seq data have been deposited to the Sequence Read Archive (SRA) under accession number SRP134977.
Transcriptomic Data Analysis
All analyses were performed using the CLC Genomics Workbench ver. 8 module (Qiagen). After quality and adapter trimming, the raw reads were mapped to the rice reference genome using a gene model annotation file from the Michigan State University rice genome annotation project database ver. 7 (http://rice.plantbiology.msu.edu/). The gene expression levels were determined in units of fragments per kilobase of exon model per million mapped reads (FPKM) normalized values (Mortazavi et al., 2008). For statistical analysis, t-tests and Baggerley's tests were performed on the original and normalized read counts. In addition, several relevant values for analysis, such as p-values, corrected p-values for multiple correction, and test statistics, were calculated in the “multi-group comparison” option. Differentially expressed genes were those with a p < 0.05, corrected p-value of false discovery rate (FDR) < 0.05, and absolute fold-change value > 1.5 from pair-wise comparisons of FPKM values among different samples.
Recombinant Protein Expression and Gel Retardation Assay
Recombinant protein expression was conducted as described previously (Byun et al., 2015) with minor modifications. Full-length coding region of DaCBF4 was inserted into the pProEx-HTa protein expression vector (Invitrogen, Carlsbad, CA, USA). Recombinant protein expressed in E. coli Rosetta (DE3) cells was purified by affinity chromatography using Ni-NTA agarose (Qiagen) according to the manufacturer's protocols. Purified protein was concentrated with Amicon Ultra-15 Centrifugal Filter Units (Merck). To investigate the characteristics of DaCBF4 as a transcription factor, gel retardation assays was conducted. The DNA fragments containing the CRT/DRE core repeat or low-temperature responsive element (LTRE) sequence were labeled with 32P-dCTP and incubated with recombinant DaCBF4 proteins in binding buffer [10 mM Tris HCl, pH 8.0, 50 mM NaCl, 1 mM EDTA, 1 mM DTT, and 5% glycerol (v/v)]. After incubation for 10 min on ice, components of the reaction mixtures were separated on 6% non-denatured polyacrylamide gels in 0.5× Tris-borate EDTA buffer. Gel was dried and visualized by autoradiography.
Results
Identification and Characterization of DaCBF4
As the overexpression of barley HvCBF4 and wheat TaCBF14 (group IV) increased the cold-stress tolerance of transgenic rice and barley plants, respectively (Oh et al., 2007; Soltész et al., 2013), we cloned the D. antarctica cDNA sequence encoding the CRT-DREB-binding factor protein based on its sequence homology with HvCBF4 (Lee et al., 2013b). The gene was designated DaCBF4, and the sequence was submitted to GenBank (KM978992).
DaCBF4 encodes a protein of 216 amino acids with a conserved AP2 DNA-binding domain. To examine its phylogenetic relationships with CBF proteins from other monocot plants, we compared the amino acid sequence of DaCBF4 with those of 37 hexaploid wheat, 9 einkorn wheat, 17 barley, 9 rice, and 1 D. antarctica CBF homologs (Badawi et al., 2007; Byun et al., 2015). All members of the monocot CBF family were divided into five groups; DaCBF4 belonged, as expected, to group IV (Supplementary Figure 1). In the AP2 domain and flanking regions, DaCBF4 shares 70–75, 76–78, 67–79, 75–91, and 56–65% sequence identities with the members of groups I, II, III, IV, and V, respectively.
DaCBF4 Is a Nuclear-Localized Transcription Factor
Transcription factors must localize to the nucleus to induce the expression of downstream target genes. To examine the cellular localization of DaCBF4, an in vivo cell targeting experiment was performed. The sGFP gene was fused in-frame to the 3′ end of the DaCBF4-coding region under the control of the CaMV 35S promoter. DaCBF4-sGFP and NLS-mRFP (a nuclear marker) constructs were co-expressed in tobacco (N. benthamiana) leaves using the Agrobacterium-mediated infiltration method. The protoplasts were extracted and visualized by fluorescence microscopy. The fluorescence signal of sGFP was uniformly distributed throughout the cell (Figure 1). In contrast, the DaCBF4-sGFP fusion protein was found primarily in the nucleus, and its signal merged with that of NLS-mRFP, indicating that DaCBF4 is targeted to the nucleus of plant cells.
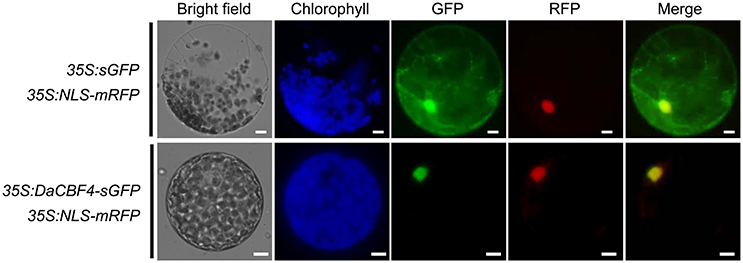
Figure 1. Subcellular localization of DaCBF4. Tobacco (Nicotiana benthamiana) leaves were co-infiltrated using Agrobacterium that contained the 35S:DaCBF4-sGFP or 35S:sGFP construct. NLS-mRFP was used as a nuclear marker protein. Scale bars = 5 μm.
Induction of DaCBF4 Expression in D. antarctica by Cold and Drought Stresses
The expression of numerous monocot CBF genes is reportedly induced in response to abiotic stresses in plants (Dubouzet et al., 2003; Vágújfalvi et al., 2005; Byun et al., 2015). To investigate the effects of abiotic stresses on the DaCBF4 transcript level in Antarctic hairgrass, DaCBF4 mRNA levels were analyzed by RT-qPCR using total RNA isolated from leaves of D. antarctica plants subjected to cold (4°C), salt (150 mM NaCl), or drought (air-dried on filter paper) stress. The DaCBF4 transcript level increased in response to cold-stress treatment. DaCBF4 expression was induced from 30 min after cold stress, peaked at 4 h, and then declined but remained high until 48 h. Drought stress resulted in a similar pattern of DaCBF4 expression. In contrast, salt stress did not activate the transcription of DaCBF4 (Figure 2). The DaIRIP, DaP5CS, and DaDHN1 genes were used as positive controls for the cold, salt, and drought stresses, respectively.
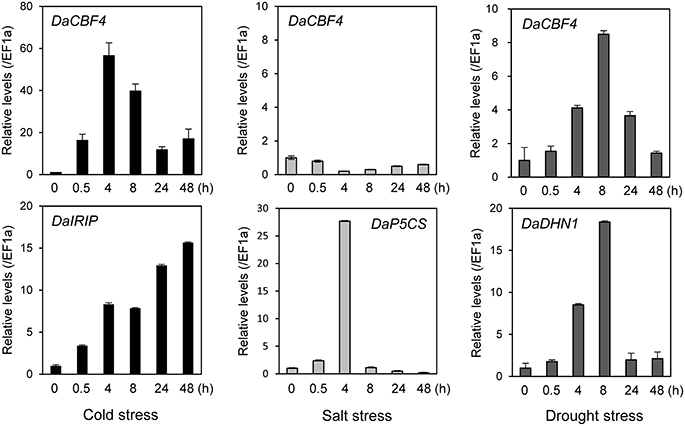
Figure 2. Expression of DaCBF4 in D. antarctica under various abiotic stress conditions. The DaCBF4 expression levels were measured by RT-qPCR with total RNA from D. antarctica leaves under cold (4°C), salt (150 mM NaCl), or drought (air dried on a filter paper at 15°C) stress. The D. antarctica EF1a gene was used as an internal control for normalization. The expression level of DaCBF4 grown on normal MS at 15°C was used as a control (calibrator for quantification) and was assumed as 1. Error bars represents standard deviation of means (n = 3). The DaIRIP, DaP5CS, and DaDHN1 genes were positive controls for cold, salt, and drought stresses, respectively. DNA sequences of gene-specific primers used for RT-qPCR are shown in Supplementary Table 2.
Generation and Molecular Analysis of Ubi:DaCBF4 Transgenic Rice Plants
Genetic transformation is widely used to investigate the functional roles of specific genes in plants. However, transformation of genes into D. antarctica is not yet possible. Therefore, we overexpressed DaCBF4 in rice, a monocot model plant of the same family as D. antarctica.
We generated transgenic rice plants constitutively overexpressing DaCBF4 under the control of the maize ubiquitin (Ubi) promoter (Figure 3A). Semi-quantitative RT-PCR confirmed that two different Ubi:DaCBF4 transgenic plants overexpressed DaCBF4 (Figure 3B). DNA gel-blot analysis indicated that these Ubi:DaCBF4 transgenic plants were independent lines (Figure 3C). Transgenic Arabidopsis plants overexpressing AtCBF1, AtCBF2, or AtCBF3 at high levels exhibit the dwarf phenotype (small stature and slow growth) under normal conditions (Gilmour et al., 2004). Moreover, in cereals, transgenic rice overexpressing OsDREB1 and transgenic barley overexpressing TaCBF14, TaCBF15, or HvCBF2A, showed moderate retarded development under normal condition compared to wild-type plants (Ito et al., 2006; Soltész et al., 2013; Jeknić et al., 2014). In contrast, the two independent Ubi:DaCBF4 transgenic lines (#1 and #2) exhibited wild-type vegetative growth under our experimental conditions (Figure 3D).
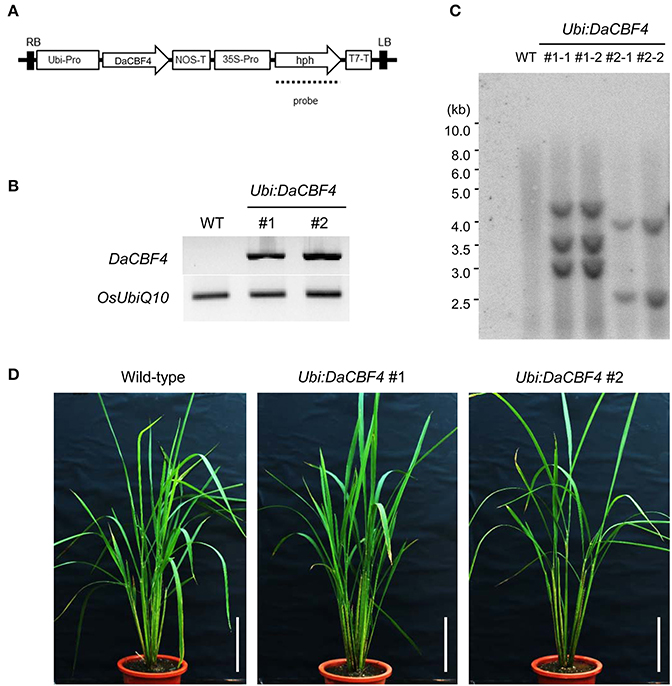
Figure 3. Generation and molecular characterization of Ubi:DaCBF4 transgenic rice plants. (A) Schematic representation of the binary vector construct used for DaCBF4 overexpression under the control of the maize ubiquitin promoter. RB, right border; Ubi-Pro, ubiquitin promoter; NOS-T, terminator sequence from nopaline synthase gene; 35S-Pro, CaMV 35S promoter; hph, hygromycin B phosphotransferase; T7-T, T7 terminator; LB, left border. (B) Semi-quantitative RT-PCR analysis of 6-week-old wild-type and two independent Ubi:DaCBF4 T2 transgenic plants (lines #1 and #2). (C) Genomic Southern blot analysis. Total leaf genomic DNA was isolated from wild-type and T2 Ubi:DaCBF4 transgenic rice plants. DNA was digested by BamHI and hybridized with 32P-labeled hph probe under high-stringency conditions. (D) Overall morphology of 2-month-old wild type and T2 Ubi:DaCBF4 transgenic (independent lines #1 and #2) rice plants. Rice plants were grown under greenhouse conditions. Scale bars = 10 cm.
Effect of DaCBF4 Overexpression on Abiotic Stress Tolerance in Transgenic Rice Plants
Overexpression of heterologous CBF genes in rice results in tolerance to cold (Byun et al., 2015), drought and high salt (Oh et al., 2005; Cui et al., 2011), or cold, drought, and high salt (Oh et al., 2007; Xu et al., 2011). To investigate whether overexpression of DaCBF4 is correlated with tolerance to abiotic stresses in transgenic plants, we assessed the effect of cold stress on Ubi:DaCBF4 transgenic rice plants. Wild-type and Ubi:DaCBF4 transgenic plants were grown at 28°C for 6 weeks in the growth room and transferred to the cold room (4°C). After 5 days of cold-stress conditions, wild-type plants showed wilting and did not recover after removal from the cold-stress conditions, while most of the Ubi:DaCBF4 transgenic rice plants recovered, appeared healthy, and continued to grow (Figure 4A). The survival rate of the wild-type rice plants was 11.8% (12 of 101), and the survival rates of Ubi:DaCBF4 lines #1 and #2 were 63.6% (35 of 55) and 41.3% (19 of 46), respectively (Figure 4B).
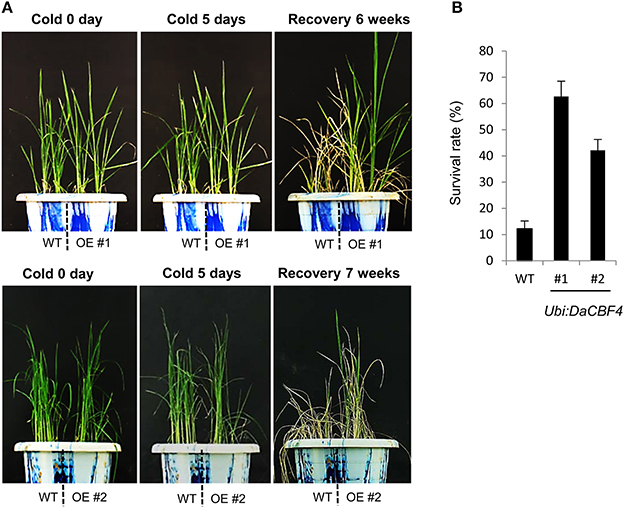
Figure 4. Comparison of cold-stress tolerance between wild-type (WT) and Ubi:DaCBF4 transgenic plants. (A) Six-week-old wild type (WT) and T2 Ubi:DaCBF4 (line #1 and #2) rice plants were subjected to cold treatment at 4°C for 5 days, recovered to normal condition at 28°C, and then compared their survival rates.(B) Survival rates of WT and two independent transgenic lines under cold-stress conditions. Results are expressed as means from four independent experiments and 11 plants were used for each experiment. Error bars represents standard deviation of means of independent experiments.
As DaCBF4 expression was also induced by drought (Figure 2), we assessed the tolerance of wild-type and Ubi:DaCBF4 rice plants to drought stress. Six-week-old wild-type and Ubi:DaCBF4 plants were grown without a water supply for 9 days. Both wild-type and DaCBF4-overexpressing plants were severely wilted, and most plants did not recovered and died even after re-watering. Next, we compared the salt tolerance of the wild-type and Ubi:DaCBF4 transgenic lines. Six-week-old plants were irrigated with water supplemented with 200 mM NaCl for 10 days, re-watered with tap water, and their growth patterns were observed. Under this condition, both wild-type and transgenic plants were almost entirely bleached and unable to recover. Therefore, constitutive expression of DaCBF4 increased the tolerance of rice to cold stress, but had no effect on the tolerance to dehydration or salt stress.
Transcriptome Analysis of Ubi:DaCBF4 Transgenic Plants Under Cold-Stress Conditions
As DaCBF4-overexpressing rice plants were more tolerant to cold stress than wild-type plants, we next performed a transcriptomic analysis of Ubi:DaCBF4 and wild-type plants to identify the downstream target genes of DaCBF4 responsible for the enhanced cold tolerance. We conducted RNA-seq analysis using the total RNA from 6-week-old leaves of wild-type and Ubi:DaCBF4 (transgenic line #1) plants grown under normal (before cold treatment) and cold-stress (1 and 6 days at 4°C) conditions. Transgenic line #1 was selected for RNA-seq analysis because it displayed a higher survival rate after cold stress than line #2 (Figure 4). The raw reads were mapped to the rice reference genome. The expression values were measured in FPKM and the differentially expressed genes (DEGs) were determined.
We reported previously that constitutive expression of DaCBF7, one of the CBF genes of D. antarctica, enhanced the cold-stress tolerance of transgenic rice plants by modulating the expression levels of downstream genes (Byun et al., 2015). Because the DaCBF4-overexpressing plants showed a cold-specific phenotype similar to that of the DaCBF7-overexpressing plants, we identified the genes responsible for the increased the cold tolerance by screening for genes up- or down-regulated in both transgenic rice plants.
Genes Upregulated Under Normal Conditions
Under normal growth conditions, 33 genes were upregulated in Ubi:DaCBF4 line #1 compared to wild-type rice plants (Figure 5 and Supplementary Table 3). This indicated that constitutive expression of DaCBF4 influenced the gene expression profile of transgenic plants even before cold treatment. Among them, nine genes were also upregulated in Ubi:DaCBF7 plants compared to wild-type plants (Table 1), including those encoding a protease inhibitor family protein (BBTI12, Os01g04050) and non-specific lipid transfer proteins (LTPL114, Os03g01300; LTPL12, Os12g02320). The expression of three genes annotated as expressed proteins was highly induced in both transgenic plants. Furthermore, the induction of the expression of the genes encoding dirigent (Os11g10850), thaumatin (Os12g43380), and ribonuclease T2 family domain-containing protein (Os09g36680) differed between the two transgenic plants under normal conditions.
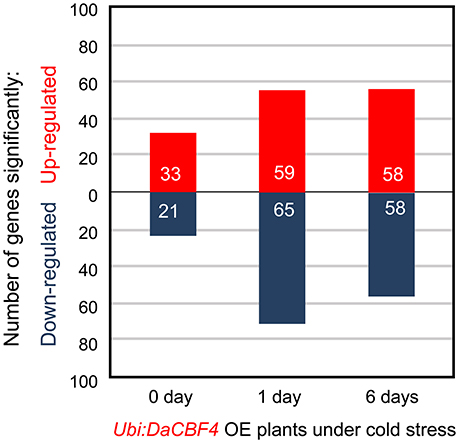
Figure 5. Transcriptome analysis with RNA-seq of the cold-stress response of Ubi:DaCBF4 transgenic plants. Number of differentially expressed genes in Ubi:DaCBF4 line #1 relative to the expression level of wild-type (WT) at each time point (0, 1, and 6 days) of cold treatment (p < 0.05, corrected p-value of FDR < 0.05).
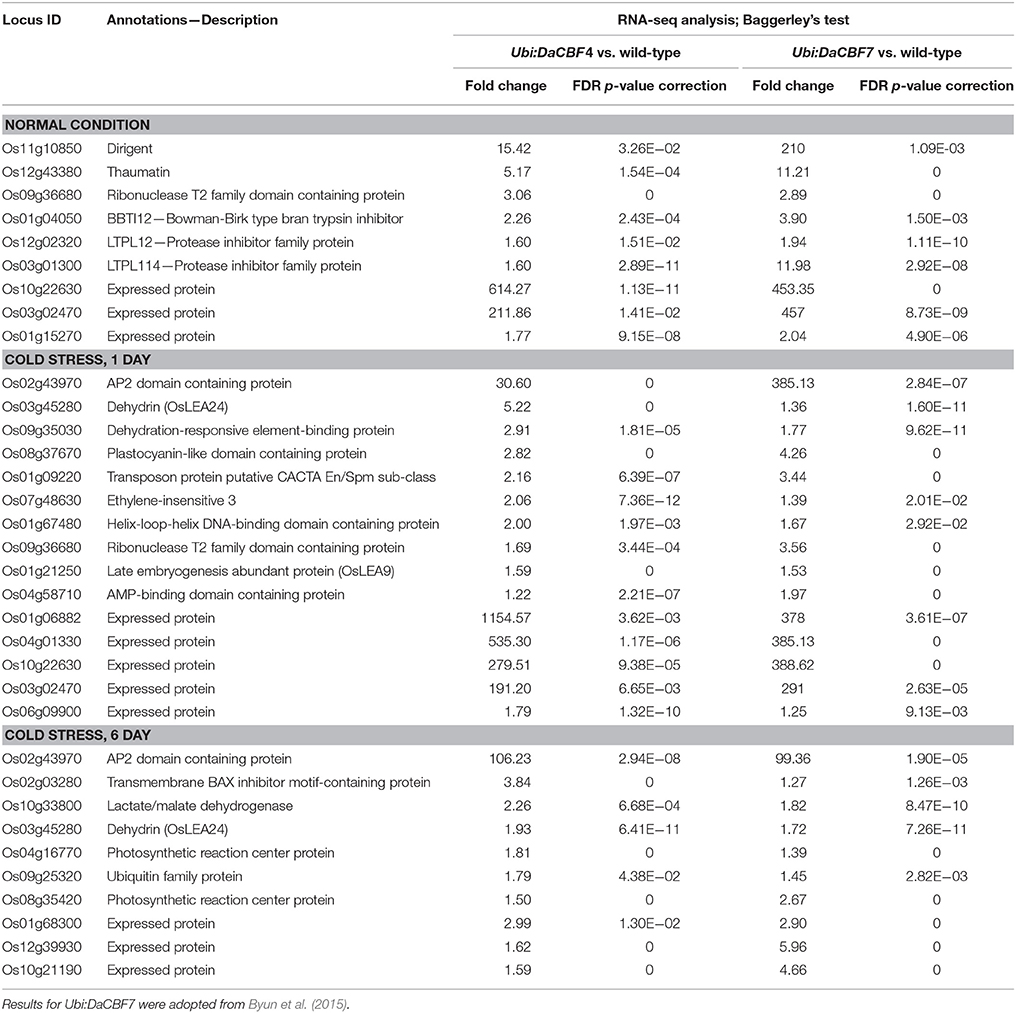
Table 1. List of differentially induced genes in both transgenic rice plants Ubi:DaCBF4 and Ubi:DaCBF7 under normal control or cold stress conditions when compared to wild type plants.
Genes Upregulated Under Cold-Stress Conditions
After 1 day of cold-stress conditions, 59 genes were upregulated in the Ubi:DaCBF4 transgenic rice plants compared to the wild type (Figure 5 and Supplementary Table 4). Of these, 15 were upregulated in both Ubi:DaCBF4 and Ubi:DaCBF7 after 1 day of cold-stress conditions (Table 1). The gene encoding AP domain-containing protein (Os02g43970) exhibited the greatest fold-change in expression in both transgenic lines compared to wild-type plants, except for five genes annotated as expressed proteins. This indicates that activation of the expression of downstream COR genes in transgenic plants is a multi-step process. Three transcription factor-encoding genes, dehydration-responsive element-binding protein (Os09g35030), ethylene-insensitive 3 (Os07g48630), and helix-loop-helix DNA-binding domain containing protein (Os01g67480), were upregulated, suggesting a role in the cold tolerance of rice. In addition, the expression of two late embryogenesis-abundant protein (LEA) genes encoding OsLEA9 (Os01g21250) and OsLEA24 (Os03g45280) were induced under cold-stress conditions. The expression of OsLEA24 was induced after 1 and 6 days of cold-stress conditions, but OsLEA9 expression was induced only after 6 days. The expression levels of OsBI (transmembrane BAX inhibitor motif-containing protein; Os02g03280) and OsMDH (lactate/malate dehydrogenase; Os10g33800) were increased after 6 days of cold-stress conditions.
Genes Downregulated in Transgenic Rice Plants
In total, 130 genes were downregulated in Ubi:DaCBF4 under normal and cold-stress conditions (Supplementary Table 5). To identify enriched Gene Ontology (GO) terms for these genes, over-represented GO categories were analyzed using AgriGO (Fisher's exact test p < 0.05) (Tian et al., 2017).
In the Biological Process category, the GO terms photosynthesis, generation of precursor metabolites and energy, and translation were significantly enriched in downregulated genes in Ubi:DaCBF4 plants; the most highly enriched GO term was photosynthesis with the lowest corrected p-value of FDR, 2.21e−15 (Supplementary Figure 2). Six of the 14 genes included in the GO term photosynthesis were significantly downregulated in Ubi:DaCBF7 plants under cold-stress compared to normal conditions. These comprised the genes encoding chlorophyll A-B binding protein (Os01g41710 and Os07g37240), photosynthetic reaction center protein (Os08g35420), photosystem II 10 kDa polypeptide chloroplast precursor (Os08g10020), photosystem II 44 kDa reaction center protein (Os04g16874), and photosystem II D2 protein (Os04g16872) (Table 2).
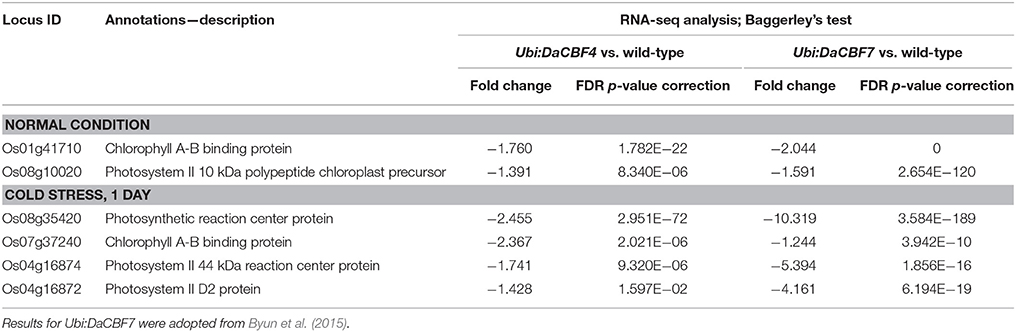
Table 2. List of differentially down-regulated genes belonging to GO term “Photosynthesis” (GO:0015979) in both transgenic rice plants Ubi:DaCBF4 and Ubi:DaCBF7under normal control or cold stress condition when compared to wild type plants.
Validation of DEGs in Ubi:DaCBF4 Transgenic Plants
To validate the DEG results, 12 genes that were differentially expressed in Ubi:DaCBF4 transgenic plants under normal and cold-stress conditions were subjected to RT-qPCR (Figure 6A). Under normal conditions, the expression patterns of nine genes—including BBTI12 (Os01g04050), RNase T2 (Os09g36680), LTPL114 (Os03g01300), Thaumatin (Os12g43380), LTPL12 (Os12g02320), Dirigent (Os11g10850)—and three unknown genes—Os01g15270, Os03g02470, and Os10g22630—were in overall agreement with those determined by RNA-seq. The fold-changes in expression determined by RNA-seq and RT-qPCR differed slightly for LTPL114, Thaumatin, and Os10g22630, but their expression was increased in Ubi:DaCBF4 line #1 and line #2 compared to wild-type plants. Under cold-stress conditions, the expression patterns of five genes—including OsLEA24 (Os03g45280), OsBI (Os02g03280), OsMDH (Os10g33800)—and two unknown genes—Os03g02470 and Os10g22630—were in agreement with those determined by RNA-seq, and the expression of these genes was higher in independent two lines of Ubi:DaCBF4 compared to wild-type plants.
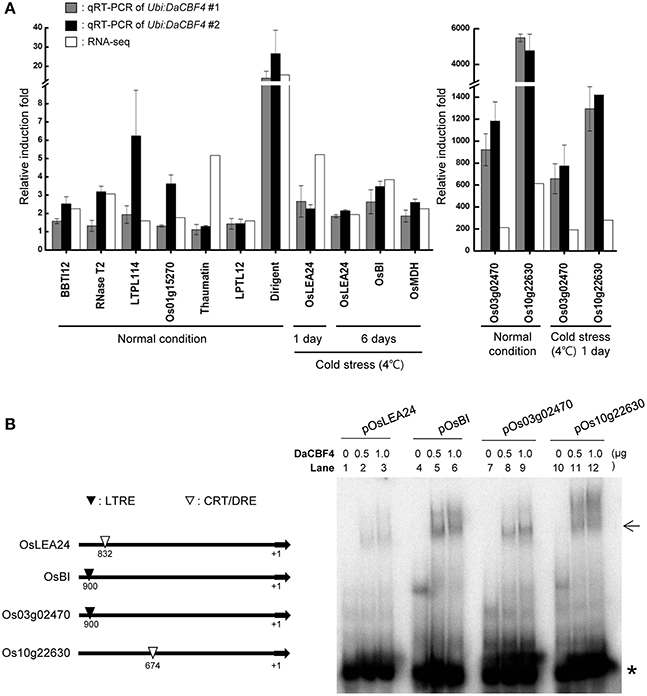
Figure 6. Validation of the gene expression profiles obtained in RNA-seq by RT-qPCR and gel retardation analysis. (A) Light-grown, 6-week-old wild-type and Ubi:DaCBF4 plants were subjected to cold (4°C) stress for 1 day and 6 days, respectively. DEGs commonly up-regulated in Ubi:DaCBF4 and Ubi:DaCBF7 transgenic plants under normal condition or cold stress condition were confirmed by RT-qPCR using the gene-specific primers listed in Supplementary Table 2. Data represent the fold induction of each gene in Ubi:DaCBF4 #1 and #2 plants relative to wild type plants. Gray bars show relative gene expression of Ubi:DaCBF4 #1 normalized to the level of OsActin mRNA as an internal control, which indicate means ± SD from three biological replicates. In addition, black bars indicate relative induction fold of Ubi:DaCBF4 #2, which indicate means ± SD from biological duplicates. White bars represent fold change of the selected genes in RNA-seq. (B) Gel retardation assay. (Left panel) Schematic presentation of four DaCBF4-induced target genes. +1 indicates a transcription initiation site. Putative CRT/DRE and LTRE motifs are indicated by ∇ and ▼, respectively. (Right panel) Gel retardation assay was performed with the 32P-radiolabeled promoter sequence of OsLEA24 (lanes 1-3), OsBI (lanes 4-6), Os03g02470 (lanes 7-9), Os10g22630 (lanes 10-12). Each set of lanes contained 0, 0.5, or 1.0 μg of bacterially-expressed, full-length His-DaCBF4 recombinant protein, respectively. Arrow and asterisk (*) indicate positions of DNA-DaCBF4 complex and free probe, respectively.
Promoter analysis represented that OsLEA24 and Os10g22630 contain putative CRT/DRE cis-acting elements in their promoter regions (Figure 6B). In addition, the upstream regions of the OsBI and Os03g02470 have a putative low temperature responsive element (LTRE). To investigate the function of DaCBF4 as a transcription factor, we conducted gel retardation analysis with 32P-labeled promoter regions of the target gene promoter. As shown in Figure 6B, the His-DaCBF4 protein bound to DNA and formed nucleoprotein complex. These results are in agreement with the fact that the above-mentioned genes are regulated by ectopic expression of DaCBF4 in rice plants.
Differential Expression of AP2 Genes in Ubi:DaCBF4 and Ubi:DaCBF7
In addition to searching for genes responsible for increased cold tolerance, investigating the specificity of DaCBF4 to its downstream targets is important for understanding the molecular mechanism of regulation for downstream genes by the group IV CBF. We could make a list of 72 genes which were upregulated exclusively in Ubi:DaCBF4 under cold stress condition, compared to Ubi:DaCBF7. When over-represented GO categories were analyzed using AgriGO (Fisher's exact test p < 0.05) (Tian et al., 2017), the GO term regulation of metabolic process was the only significantly enriched one (corrected p-value of FDR = 0.022). Notably, seven genes included in this group were encoding AP2 domain containing protein (Supplementary Table 6). For comparative analysis, we also identified 7 more AP2 genes, which were induced exclusively in Ubi:DaCBF7 under cold stress. As a result of promoter analysis, we could identify 9 DRE/CRT and 4 LTRE elements from DaCBF4 induced 7 AP2 genes, and 5 DRE/CRT and 8 LTRE elements from DaCBF7 induced 7 AP2 genes, which imply that different CBF homolog protein may prefer different type of cis-element in promoters of target genes.
Discussion
Cloning and Characterization of DaCBF4 From D. antarctica
We isolated DaCBF4 from the Antarctic plant D. antarctica (Poaceae; Pooideae). Phylogenetic analysis revealed that DaCBF4 is grouped within the cereal CBF group IV (Badawi et al., 2007). Because cereal CBF group IV is found only in the Pooideae, members of this group are considered to have evolved during the recent translocation of these plants from tropical to temperate habitats, suggesting possible roles in adaptation to low temperature (Badawi et al., 2007). RT-qPCR analyses revealed that the expression levels of DaCBF4 in D. antarctica under cold and drought stresses were increased, and cold stress resulted in an increase in expression of greater magnitude than did drought stress (Figure 2). Induction of CBF IV group gene expression by cold stress also occurs in barley, wheat, and Brachypodium distachyon (Kume et al., 2005; Skinner et al., 2005; Chen et al., 2016). Together with the cellular localization of 35S:DaCBF4-sGFP in the nucleus of tobacco protoplasts (Figure 1), DNA binding affinity of DaCBF4 (Figure 6B) represented possible roles of this protein as a transcription factor in Antarctic hairgrass. The considerable induction of DaCBF4 expression under cold stress suggests that DaCBF4 regulates development of cold tolerance in D. antarctica during adaptation to the low-temperature environment of the Antarctic.
Phenotype of Ubi:DaCBF4 Plants Under Normal and Cold-Stress Conditions
We generated CBF-overexpressing transgenic plants to assess the roles of these genes in tolerance to abiotic stresses. Most of the transgenic plant lines exhibited enhanced tolerance to cold, drought, and salt stresses; this was mediated by modulating the expression of downstream stress-responsive genes (Lata and Prasad, 2011). Constitutive expression of CBF transcription factors generally enhances stress tolerance, but in some cases results in retarded normal growth (dwarf phenotype) and delayed flowering (Ito et al., 2006; Wei et al., 2017). In contrast, other CBF-overexpressing transgenic plants displayed no differences in phenotype under normal growth conditions (Oh et al., 2007; Xu et al., 2011; Byun et al., 2015). In this study, constitutive expression of DaCBF4 from D. antarctica enhanced cold-stress tolerance without affecting development. The distinct effects of CBF overexpression on vegetative growth may be due to different generation of transgenic plants analyzed (Oh et al., 2007). However, the molecular mechanisms underlying the different growth phenotypes of the various transgenic plant lines remain to be elucidated.
Comparative RNA-Seq Analysis of Ubi:DaCBF4 and Ubi:DaCBF7
Overexpression of CBF reportedly enhances the cold tolerance of barley, rice, and wheat (Ito et al., 2006; Oh et al., 2007; Gao et al., 2009; Morran et al., 2011; Xu et al., 2011). However, only a few studies have performed high-throughput transcriptome analysis of downstream genes regulated by overexpressed CBF transcription factors, all of them in rice (Oh et al., 2005, 2007; Ito et al., 2006). We reported previously that the overexpression of DaCBF7, a CBF homolog from D. antarctica, resulted in enhanced tolerance to cold in rice, similar to DaCBF4 in this study. Moreover, the transcriptome of Ubi:DaCBF7 was analyzed by RNA-seq to identify downstream genes involved in promoting cold tolerance in transgenic rice plants (Byun et al., 2015). Therefore, in this study we focused on downstream target genes regulated by both DaCBF7 and DaCBF4 that enhance the cold tolerance of rice.
Genes Upregulated in Ubi:DaCBF4 and Ubi:DaCBF7 Plants Under Normal Conditions
Nine genes were upregulated in both Ubi:DaCBF4 and Ubi:DaCBF7 transgenic rice plants under normal conditions (Table 1), the majority are abiotic stress-responsive genes in various plant species. LTPL12 (Os12g02320) and LTPL114 (Os03g01300) encode non-specific lipid transfer proteins (LTPs) involved in lipid metabolism. In addition, LTP expression is induced in plant cells upon exposure to abiotic stresses. The bromegrass LTP gene BG-14 was strongly induced during cold acclimation (Wu et al., 2004), and the expression of two barley LTP4 genes, HvLtp4.2, and HvLtp4.3, was increased under cold-stress conditions (Molina et al., 1996). Overexpression of the pearl millet DREB2A transcription factor gene in tobacco and of the rice DREB1A gene in rice plants enhanced abiotic stress tolerance and induced the expression of LTP homologs under cold-stress conditions (Ito et al., 2006; Agarwal et al., 2010). The LTP-mediated resistance of plants to cold is associated with a decrease in thylakoid membrane lipid fluidity (Sror et al., 2003). Dirigent (Os11g10850) encodes a protein that modulates lignin biosynthesis and cell wall metabolism during exposure to abiotic stresses (Paniagua et al., 2017). Dirigent proteins are implicated in modulation of lignification levels upon exposure to abiotic stresses. The expression of several of the DIR-like genes from Brassica rapa and the resurrection plant Boea hygrometrica (BhDIR1) was influenced by water and cold stress (Wu et al., 2009; Arasan et al., 2013). Therefore, modulation of plant cell wall metabolism and fluidity is crucial for the cold-stress tolerance of plants. However, the molecular mechanisms underlying the effects of dirigent proteins on plant abiotic stress tolerance are unclear.
BBTI12 (Os01g04050) belongs to the Bowman-Birk family, which consists of compound inhibitors comprising one to six inhibitor units that target serine proteases (Rawlings, 2010). BBTI1 and BBTI2, rice BBTI homologs, are induced under normal conditions in barley CBF4- or Arabidopsis CBF3-overexpressing transgenic rice plants (Oh et al., 2005, 2007). Proteins are influenced by abiotic stressors and, are the major players in plant responses to stress (Kidrič et al., 2014); thus, BBTI-mediated fine control of protein degradation may be required for survival of plants exposed to abiotic stresses. Expression of OsRNS4, which encodes the ribonuclease T2 family domain-containing protein (Os09g36680) was increased in response to biotic stresses, such as insects and Xanthomonas oryzae, and abiotic stresses, such as wounding and high-salt conditions (Hillwig, 2009). Overexpression of OsRNS4 enhanced tolerance to a high salt concentration, but the effect on cold tolerance was not evaluated (Zheng et al., 2014). Because OsRNS4 is an inactive RNase (Hillwig, 2009), its role in the response to abiotic stresses may be independent of its enzymatic activity. In this study, the expression of Thaumatin, a member of the pathogenesis-related protein 5 (PR5) family, was induced in transgenic rice plants. Moreover, the expression in winter wheat (T. aestivum L.) of TaTLP, which encodes a thaumatin-like protein (TLP), was increased during cold acclimation of wheat seedlings (Kuwabara et al., 2002). Overexpression of ObTLP1, a thaumatin-like protein from basil, in Arabidopsis enhances tolerance of plants to multiple abiotic stresses as well as the phytopathogenic fungi, Scleretonia sclerotiorum and Botrytis cinerea (Misra et al., 2016).
To summarize, DaCBF4 and DaCBF7 enhance cold-stress tolerance by activating the expression of target genes with roles in biotic and abiotic stress responses. Interestingly, most of the genes upregulated in both plants under normal conditions are reportedly responsive to both biotic and abiotic stresses. Therefore, the pathogen resistance of Ubi:DaCBF4 and Ubi:DaCBF7 plants warrants further investigation.
Genes Upregulated in Ubi:DaCBF4 and Ubi:DaCBF7 Under Cold-Stress Conditions
Fifteen and ten genes were upregulated in both transgenic plants in response to 1 day and 6 days of cold stress, respectively (Table 1). Among them, two were LEA (late embryogenesis abundant) genes, which respond to cold, drought, salinity, and ABA stresses during the vegetative stage of growth (Liu et al., 2017). LEA genes are abundant in plant genomes; the Arabidopsis and rice genomes harbor at least 51 and 34 LEA genes, respectively (Wang et al., 2007; Hundertmark and Hincha, 2008). LEA proteins are classified into seven groups according to their motifs (Battaglia et al., 2008). The diverse gene expression levels and protein subcellular distributions suggest that different LEAs are involved in the responses to diverse environmental stimuli (Hundertmark and Hincha, 2008; Candat et al., 2014). LEA proteins may be involved in protection of proteins or endomembrane structures under stress conditions (Koag et al., 2003; Drira et al., 2013). LEA expression enhances stress tolerance in transgenic plants (Kosová et al., 2014). Overexpression of wheat WCI16 or Arabidopsis Cor15A leads to increased freezing tolerance in transgenic plants (Artus et al., 1996; Sasaki et al., 2014). Heterologous expression of individual PmLEAs genes in tobacco conferred enhanced tolerance to cold and drought stress (Bao et al., 2017). OsDhn1-overexpressing plants displayed enhanced drought- and salt-stress tolerance due to increased scavenging of reactive oxygen species (Kumar et al., 2014). Transcriptome analysis of Ubi:DaCBF4 and Ubi:DaCBF7 plants showed specific changes in two LEA proteins, OsLEA9 (group III) and OsLEA24 (group II [dehydrin], characterized by a K-segment). These genes have a DRE/CRT sequence in their upstream regions and may be directly regulated by DaCBF proteins (Byun et al., 2015). Elevated expression levels of OsLEA9 and OsLEA24 are positively correlated with cold-stress tolerance in transgenic rice plants. Overall, these results suggest that the OsLEA9 and OsLEA24 genes, which are downstream of DaCBF, can be used as target genes for genetic engineering of crops with enhanced stress tolerance.
In addition to cold responsive genes, the induction of a rice gene Os02g43970, encoding AP2 domain containing protein, was highly strong under cold condition, but was not significant in the normal condition (Table 1). Moreover, 7 more genes encoding AP2 domain containing protein were significantly upregulated exclusively in Ubi:DaCBF4 under cold condition, but not in Ubi:DaCBF7 under normal or cold conditions (Supplementary Table 6). Based on results that those CBF genes still can exhibit a cold responsive expression pattern, we can assume that their induction in Ubi:DaCBF4 is caused by additive effect of DaCBF4 overexpression and native CBF response to cold treatment. Similar phenomena were also observed in previous studies on transgenic barley overexpressing TaCBF2/3, TaCBF14/15, and HvCBF2A (Morran et al., 2011; Soltész et al., 2013; Jeknić et al., 2014).
For more investigation on difference of target specificities between two transgenes DaCBF4 and DaCBF7, we examined cis-element sequences of two AP2 gene groups, DaCBF4 induced and DaCBF7 induced group. DaCBF4 induced AP2 genes had more DRE/CRT (9) and less LTRE elements (4) than DaCBF7 induced 7 AP2 genes with 5 DRE/CRT and 8 LTRE elements. Obviously further researches on the genome wide analysis of rice AP2 genes and their distribution profile of promoter elements are still necessary, activation of different subset of AP2 genes by DaCBF4 and DaCBF7 can be an indirect evidence of distinctive specificity and biological activity for each CBF protein in plants under abiotic stresses.
Genes Downregulated in Ubi:DaCBF4 and Ubi:DaCBF7 Plants
In total, 130 genes were downregulated in the Ubi:DaCBF4 transgenic line under normal and cold-stress conditions. Photosynthesis was the most enriched GO term (Supplementary Figure 2). Six genes encoding photosystem proteins were significantly downregulated in both Ubi:DaCBF4 and Ubi:DaCBF7 (Table 2). Cold stress generally reduces photosynthetic activity and downregulates the expression of photosynthesis-related genes (Allen and Ort, 2001; Fowler and Thomashow, 2002; Tsonev et al., 2003). In addition, photosynthesis-related genes are downregulated in CBF-overexpressing transgenic plants compared to wild-type plants. However, most studies did not identify the downregulated genes as upregulated genes are more important in terms of stress tolerance. An RNA-Seq study revealed that genes with diverse functions were downregulated in AtCBF2- and AtCBF3-overexpressing Arabidopsis lines, but photosynthesis-related genes were not over-represented among the downregulated genes (Li et al., 2017). However, in this study, the expression of photosynthesis-related genes was decreased in both transgenic rice lines; therefore, the mechanism by which DaCBFs regulate the expression of genes involved in photosynthesis warrants further investigation.
Conclusion
Our results showed that heterologous expression of Antarctic hairgrass DaCBF4 increased the tolerance of transgenic rice plants to low-temperature stress. A comparative transcriptome analysis identified common downstream targets of DaCBF4 and DaCBF7 that resulted in the same phenotype. The genes identified in this study will facilitate genetic engineering of cereal plants with enhanced cold tolerance.
Author Contributions
WTK and HL: conceived and designed the study; MYB, LHC, and AL: performed the experiments; MYB, LHC, JL, and HP: analyzed the data; MYB, LHC, WTK, and HL: discussed the results and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by The Polar Genomics 101 Project: Genome analysis of polar organisms and establishment of application platform (PE18080) to HP, funded by the Korea Polar Research Institute; and a grant from the Next-Generation BioGreen 21 Program for Agriculture and Technology Development (Project Numbers PJ01113801 [NCGC] and PJ01334901 [SSAC]) funded by the Rural Development Administration, Republic of Korea, to WTK.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00601/full#supplementary-material
Abbreviations
CBF, C-repeat binding factor; CRT, C repeat; DEG, differentially expressed gene; DRE, dehydration-responsive element; FDR, false discovery rate; RT-qPCR, Real-time reverse transcription-quantitative polymerase chain reaction; RNA-seq, RNA sequencing; FPKM, fragments per kilobase of exon model per million mapped reads; sGFP, synthetic green fluorescent protein; Ubi, ubiquitin.
References
Agarwal, P., Agarwal, P. K., Joshi, A. J., Sopory, S. K., and Reddy, M. K. (2010). Overexpression of PgDREB2A transcription factor enhances abiotic stress tolerance and activates downstream stress-responsive genes. Mol. Biol. Rep. 37, 1125–1135. doi: 10.1007/s11033-009-9885-8
Alberdi, M., Bravo, L. A., Gutiérrez, A., Gidekel, M., and Corcuera, L. J. (2002). Ecophysiology of Antarctic vascular plants. Physiol. Plant. 115, 479–486. doi: 10.1034/j.1399-3054.2002.1150401.x
Allen, D. J., and Ort, D. R. (2001). Impacts of chilling temperatures on photosynthesis in warm-climate plants. Trends Plant Sci. 6, 36–42. doi: 10.1016/S1360-1385(00)01808-2
Arasan, S. K. T., Park, J. I., Ahmed, N. U., Jung, H. J., Hur, Y., Kang, K. K., et al. (2013). Characterization and expression analysis of dirigent family genes related to stresses in Brassica. Plant Physiol. Biochem. 67, 144–153. doi: 10.1016/j.plaphy.2013.02.030
Artus, N. N., Uemura, M., Steponkus, P. L., Gilmour, S. J., Lin, C., and Thomashow, M. F. (1996). Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc. Natl. Acad. Sci. U.S.A. 93, 13404–13409. doi: 10.1073/pnas.93.23.13404
Badawi, M., Danyluk, J., Boucho, B., Houde, M., and Sarhan, F. (2007). The CBF gene family in hexaploid wheat and its relationship to the phylogenetic complexity of cereal CBFs. Mol. Genet. Genomics 277, 533–554. doi: 10.1007/s00438-006-0206-9
Bao, F., Du, D., An, Y., Yang, W., Wang, J., Cheng, T., et al. (2017). Overexpression of Prunus mume dehydrin genes in tobacco enhances tolerance to cold and drought. Front. Plant Sci. 8:151. doi: 10.3389/fpls.2017.00151
Battaglia, M., Olvera-Carrillo, Y., Garciarrubio, A., Campos, F., and Covarrubias, A. A. (2008). The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 148, 6–24. doi: 10.1104/pp.108.120725
Bravo, L. A., and Griffith, M. (2005). Characterization of antifreeze activity in Antarctic plants. J. Exp. Bot. 56, 1189–1196. doi: 10.1093/jxb/eri112
Byun, M. Y., Cui, L. H., Oh, T. K., Jung, Y. -J., Lee, A., Park, K. Y., et al. (2017). Homologous U-box E3 ubiquitin ligases OsPUB2 and OsPUB3 are involved in the positive regulation of low temperature stress response in rice (Oryza sativa L.). Front. Plant Sci. 8:16. doi: 10.3389/fpls.2017.00016
Byun, M. Y., and Kim, W. T. (2014). Suppression of OsRAD51D results in defects in reproductive development in rice (Oryza sativa L.). Plant J. 79, 256–269. doi: 10.1111/tpj.12558
Byun, M. Y., Lee, J., Cui, L. H., Kang, Y., Oh, T. K., Park, H., et al. (2015). Constitutive expression of DaCBF7, an Antarctic vascular plant Deschampsia antarctica CBF homolog, resulted in improved cold tolerance in transgenic rice plants. Plant Sci. 236, 61–74. doi: 10.1016/j.plantsci.2015.03.020
Candat, A., Paszkiewicz, G., Neveu, M., Gautier, R., Logan, D. C., Avelange-Macherel, M. H., et al. (2014). The ubiquitous distribution of late embryogenesis abundant proteins across cell compartments in Arabidopsis offers tailored protection against abiotic stress. Plant Cell 26, 3148–3166. doi: 10.1105/tpc.114.127316
Chen, L., Han, J., Deng, X., Tan, S., Li, L., Li, L., et al. (2016). Expansion and stress responses of AP2/EREBP superfamily in Brachypodium distachyon. Sci. Rep. 6:21623. doi: 10.1038/srep21623
Claeys, H., and Inzé, D. (2013). The agony of choice: how plants balance growth and survival under water-limiting conditions. Plant Physiol. 162, 1768–1779. doi: 10.1104/pp.113.220921
Cui, M., Zhang, W., Zhang, Q., Xu, Z., Zhu, Z., Duan, F., et al. (2011). Induced over-expression of the transcription factor OsDREB2A improves drought tolerance in rice. Plant Physiol. Biochem. 49, 1384–1391. doi: 10.1016/j.plaphy.2011.09.012
Drira, M., Saibi, W., Brini, F., Gargouri, A., Masmoudi, K., and Hanin, M. (2013). The K-segments of the wheat dehydrin DHN-5 are essential for the protection of lactate dehydrogenase and β-glucosidase activities in vitro. Mol. Biotechnol. 54, 643–650. doi: 10.1007/s12033-012-9606-8
Dubouzet, J. G., Sakuma, Y., Ito, Y., Kasuga, M., Dubouzet, E. G., Miura, S., et al. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt-and cold-responsive gene expression. Plant J. 33, 751–763. doi: 10.1046/j.1365-313X.2003.01661.x
Fowler, S., and Thomashow, M. F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14, 1675–1690. doi: 10.1105/tpc.003483
Gao, S. Q., Chen, M., Xia, L. Q., Xiu, H. J., Xu, Z. S., Li, L. C., et al. (2009). A cotton (Gossypium hirsutum) DRE-binding transcription factor gene, GhDREB, confers enhanced tolerance to drought, high salt, and freezing stresses in transgenic wheat. Plant Cell Rep. 28, 301–311. doi: 10.1007/s00299-008-0623-9
Giełwanowska, I., Szczuka, E., Bednara, J., and Górecki, R. (2005). Anatomical features and ultrastructure of Deschampsia antarctica (Poaceae) leaves from different growing habitats. Ann. Bot. 96, 1109–1119. doi: 10.1093/aob/mci262
Gilmour, S. J., Fowler, S. G., and Thomashow, M. F. (2004). Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 54, 767–781. doi: 10.1023/B:PLAN.0000040902.06881.d4
Hillwig, M. S. (2009). Regulation, Function, and Evolution of T2 RNases. Graduate theses and dissertations. Available online at: http://lib.dr.iastate.edu/etd/11095
Hundertmark, M., and Hincha, D. K. (2008). LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9:118. doi: 10.1186/1471-2164-9-118
Ito, Y., Katsura, K., Maruyama, K., Taji, T., Kobayashi, M., Seki, M., et al. (2006). Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 47, 141–153. doi: 10.1093/pcp/pci230
Jaglo-Ottosen, K. R., Gilmour, S. J., Zarka, D. G., Schabenberger, O., and Thomashow, M. F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280, 104–106. doi: 10.1126/science.280.5360.104
Jeknić, Z., Pillman, K. A., Dhillon, T., Skinner, J. S., Veisz, O., Cuesta-Marcos, A., et al. (2014). Hv-CBF2A overexpression in barley accelerates COR gene transcript accumulation and acquisition of freezing tolerance during cold acclimation. Plant Mol. Biol. 84, 67–82. doi: 10.1007/s11103-013-0119-z
John, U. P., Polotnianka, R. M., Sivakumaran, K. A., Chew, O., Mackin, L., Kuiper, M. J., et al. (2009). Ice recrystallization inhibition proteins (IRIPs) and freeze tolerance in the cryophilic Antarctic hair grass Deschampsia antarctica E. Desv. Plant Cell Environ. 32, 336–348. doi: 10.1111/j.1365-3040.2009.01925.x
Kidrič, M., Kos, J., and Sabotič, J. (2014). Proteases and their endogenous inhibitors in the plant response to abiotic stress. Bot. Serb. 38, 139–158.
Koag, M. C., Fenton, R. D., Wilkens, S., and Close, T. J. (2003). The binding of maize DHN1 to lipid vesicles. Gain of structure and lipid specificity. Plant Physiol. 131, 309–316. doi: 10.1104/pp.011171
Kosová, K., Vítámvás, P., and Prášil, I. T. (2014). Wheat and barley dehydrins under cold, drought, and salinity - what can LEA-II proteins tell us about plant stress response? Front. Plant Sci. 5:343. doi: 10.3389/fpls.2014.00343
Kovalchuk, N., Jia, W., Eini, O., Morran, S., Pyvovarenko, T., Fletcher, S., et al. (2013). Optimization of TaDREB3 gene expression in transgenic barley using cold-inducible promoters. Plant Biotechnol. J. 11, 659–670. doi: 10.1111/pbi.12056
Kumar, M., Lee, S. C., Kim, J. Y., Kim, S. J., Aye, S. S., and Kim, S.-R. (2014). Over-expression of dehydrin gene, OsDhn1, improves drought and salt stress tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.). J. Plant Biol. 57, 383–393. doi: 10.1007/s12374-014-0487-1
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kume, S., Kobayashi, F., Ishibashi, M., Ohno, R., Nakamura, C., and Takumi, S. (2005). Differential and coordinated expression of Cbf and Cor/Lea genes during long-term cold acclimation in two wheat cultivars showing distinct levels of freezing tolerance. Genes Genet. Syst. 80, 185–197. doi: 10.1266/ggs.80.185
Kuwabara, C., Takezawa, D., Shimada, T., Hamada, T., Fujikawa, S., and Arakawa, K. (2002). Abscisic acid-and cold-induced thaumatin-like protein in winter wheat has an antifungal activity against snow mould, Microdochium nivale. Physiol. Plant. 115, 101–110. doi: 10.1034/j.1399-3054.2002.1150112.x
Lata, C., and Prasad, M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62, 4731–4748. doi: 10.1093/jxb/err210
Lee, H., Kim, J. H., Park, M., Kim, I. C., Yim, J. H., and Lee, H. K. (2010). Reference genes validation for qPCR normalization in Deschampsia antarctica during abiotic stresses. Antarct. Sci. 22, 477–484. doi: 10.1017/S0954102010000428
Lee, J., Noh, E. K., Choi, H. S., Shin, S. C., Park, H., and Lee, H. (2013a). Transcriptome sequencing of the Antarctic vascular plant Deschampsia antarctica Desv. under abiotic stress. Planta 237, 823–836. doi: 10.1007/s00425-012-1797-5
Lee, J., Noh, E. K., Park, H., and Lee, H. (2013b). Transcription factor profile analysis of the Antarctic vascular plant Deschampsia antarctica Desv. (Poaceae). Genes Genom. 35, 575–586. doi: 10.1007/s13258-013-0106-4
Li, A., Zhou, M., Wei, D., Chen, H., You, C., and Lin, J. (2017). Transcriptome profiling reveals the negative regulation of multiple plant hormone signaling pathways elicited by overexpression of C-repeat binding factors. Front. Plant Sci. 8:1647. doi: 10.3389/fpls.2017.01647
Liu, Y., Song, Q., Li, D., Yang, X., and Li, D. (2017). Multifunctional roles of plant dehydrins in response to environmental stresses. Front. Plant Sci. 8:1018. doi: 10.3389/fpls.2017.01018
Misra, R. C., Kamthan, M., Kumar, S., and Ghosh, S. (2016). A thaumatin-like protein of Ocimum basilicum confers tolerance to fungal pathogen and abiotic stress in transgenic Arabidopsis. Sci. Rep. 6:25340. doi: 10.1038/srep25340
Molina, A., Diaz, I., Carbonero, P., García-Olmedo, F., and Vasil, I. K. (1996). Two cold-inducible genes encoding lipid transfer protein LTP4 from barley show differential responses to bacterial pathogens. Mol. Gen. Genet. 252, 162–168. doi: 10.1007/BF02173216
Morran, S., Eini, O., Pyvovarenko, T., Parent, B., Singh, R., Ismagul, A., et al. (2011). Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol. J. 9, 230–249. doi: 10.1111/j.1467-7652.2010.00547.x
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Oh, S. J., Kwon, C. W., Choi, D. W., Song, S. I., and Kim, J. K. (2007). Expression of barley HvCBF4 enhances tolerance to abiotic stress in transgenic rice. Plant Biotechnol. J. 5, 646–656. doi: 10.1111/j.1467-7652.2007.00272.x
Oh, S. J., Song, S. I., Kim, Y. S., Jang, H. J., Kim, S. Y., Kim, M., et al. (2005). Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol. 138, 341–351. doi: 10.1104/pp.104.059147
Paniagua, C., Bilkova, A., Jackson, P., Dabravolski, S., Riber, W., Didi, V., et al. (2017). Dirigent proteins in plants: modulating cell wall metabolism during abiotic and biotic stress exposure. J. Exp. Bot. 18, 3287–3301. doi: 10.1093/jxb/erx141
Pastorczyk, M., Giełwanowska, I., and Lahuta, L. B. (2014). Changes in soluble carbohydrates in polar Caryophyllaceae and Poaceae plants in response to chilling. Acta Physiol. Plant 36, 1771–1780. doi: 10.1007/s11738-014-1551-7
Rawlings, N. D. (2010). Peptidase inhibitors in the MEROPS database. Biochimie 92, 1463–1483. doi: 10.1016/j.biochi.2010.04.013
Sáez, P. L., Bravo, L. A., Cavieres, L. A., Vallejos, V., Sanhueza, C., Font-Carrascosa, M., et al. (2017). Photosynthetic limitations in two Antarctic vascular plants: importance of leaf anatomical traits and Rubisco kinetic parameters. J. Exp. Bot. 68, 2871–2883. doi: 10.1093/jxb/erx148
Sasaki, K., Christov, N. K., Tsuda, S., and Imai, R. (2014). Identification of a novel LEA protein involved in freezing tolerance in wheat. Plant Cell Physiol. 55, 136–147. doi: 10.1093/pcp/pct164
Skinner, J. S., von Zitzewitz, J., Szucs, P., Marquez-Cedillo, L., Filichkin, T., Amundsen, K., et al. (2005). Structural, functional, and phylogenetic characterization of a large CBF gene family in barley. Plant Mol. Biol. 59, 533–551. doi: 10.1007/s11103-005-2498-2
Soltész, A., Smedley, M., Vashegyi, I., Galiba, G., Harwood, W., and Vágújfalvi, A. (2013). Transgenic barley lines prove the involvement of TaCBF14 and TaCBF15 in the cold acclimation process and in frost tolerance. J. Exp. Bot. 64, 1849–1862. doi: 10.1093/jxb/ert050
Sror, H. A., Tischendorf, G., Sieg, F., Schmitt, J. M., and Hincha, D. K. (2003). Cryoprotectin protects thylakoids during a freeze–thaw cycle by a mechanism involving stable membrane binding. Cryobiology 47, 191–203. doi: 10.1016/j.cryobiol.2003.09.005
Stockinger, E. J. (2009). “Winter hardiness and the CBF genes in the Triticeae”, in Plant Cold Hardiness: From the Laboratory to the Field, eds L. V. Gusta, K. K. Tanino, and M. E. Wisniewski (Cambridge: CAB International), 119–130.
Thomashow, M. F. (2010). Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol. 154, 571–577. doi: 10.1104/pp.110.161794
Tian, T., Liu, Y., Yan, H., You, Q., Yi, X., Du, Z., et al. (2017). agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45, W122–W129. doi: 10.1093/nar/gkx382
Tondelli, A., Francia, E., Barabaschi, D., Pasquariello, M., and Pecchioni, N. (2011). Inside the CBF locus in Poaceae. Plant Sci. 180, 39–45. doi: 10.1016/j.plantsci.2010.08.012
Tsonev, T., Velikova, V., Georgieva, K., Hyde, P. F., and Jones, H. G. (2003). Low temperature enhances photosynthetic downregulation in French bean (Phaseolus vulgaris L.) plants. Ann. Bot. 91, 343–352. doi: 10.1093/aob/mcg020
Vágújfalvi, A., Aprile, A., Miller, A., Dubcovsky, J., Delugu, G., Galiba, G., et al. (2005). The expression of several Cbf genes at the Fr-A2 locus is linked to frost resistance in wheat. Mol. Genet. Genomics 274, 506–514. doi: 10.1007/s00438-005-0047-y
Wang, X. S., Zhu, H. B., Jin, G. L., Liu, H. L., Wu, W. R., and Zhu, J. (2007). Genome-scale identification and analysis of LEA genes in rice (Oryza sativa L.). Plant Sci. 172, 414–420. doi: 10.1016/j.plantsci.2006.10.004
Wei, T., Deng, K., Zhang, Q., Gao, Y., Liu, Y., Yang, M., et al. (2017). Modulating AtDREB1C expression improves drought tolerance in Salvia miltiorrhiza. Front. Plant Sci. 8:52. doi: 10.3389/fpls.2017.00052
Wu, G., Robertson, A. J., Liu, X., Zheng, P., Wilen, R. W., Nesbitt, N. T., et al. (2004). A lipid transfer protein gene BG-14 is differentially regulated by abiotic stress, ABA, anisomycin, and sphingosine in bromegrass (Bromus inermis). J. Plant Physiol. 161, 449–458. doi: 10.1078/0176-1617-01259
Wu, R., Wang, L., Wang, Z., Shang, H., Liu, X., Zhu, Y., et al. (2009). Cloning and expression analysis of a dirigent protein gene from the resurrection plant Boea hygrometrica. Prog. Nat. Sci. 19, 347–352. doi: 10.1016/j.pnsc.2008.07.010
Xu, M., Li, L., Fan, Y., Wan, J., and Wang, L. (2011). ZmCBF3 overexpression improves tolerance to abiotic stress in transgenic rice (Oryza sativa) without yield penalty. Plant Cell Rep. 30, 1949–1957. doi: 10.1007/s00299-011-1103-1
Keywords: C-repeat/DRE binding factor, cold tolerance, Deschampsia antarctica, RNA-seq, transgenic plant, rice
Citation: Byun MY, Cui LH, Lee J, Park H, Lee A, Kim WT and Lee H (2018) Identification of Rice Genes Associated With Enhanced Cold Tolerance by Comparative Transcriptome Analysis With Two Transgenic Rice Plants Overexpressing DaCBF4 or DaCBF7, Isolated From Antarctic Flowering Plant Deschampsia antarctica. Front. Plant Sci. 9:601. doi: 10.3389/fpls.2018.00601
Received: 05 February 2018; Accepted: 16 April 2018;
Published: 03 May 2018.
Edited by:
Ju-Kon Kim, Seoul National University, South KoreaReviewed by:
Kazuo Nakashima, Japan International Research Center for Agricultural Sciences, JapanHye Sun Cho, Korea Research Institute of Bioscience & Biotechnology, South Korea
Copyright © 2018 Byun, Cui, Lee, Park, Lee, Kim and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Woo Taek Kim, d3RraW1AeW9uc2VpLmFjLmty
Hyoungseok Lee, c291bGFpZEBrb3ByaS5yZS5rcg==
†These authors have contributed equally to this work.
 Mi Young Byun
Mi Young Byun Li Hua Cui2†
Li Hua Cui2† Andosung Lee
Andosung Lee Woo Taek Kim
Woo Taek Kim Hyoungseok Lee
Hyoungseok Lee