- 1National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, China
- 2University of Chinese Academy of Sciences, Beijing, China
Transient gene expression systems using protoplasts have been widely used for rapid functional characterization of genes and high-throughput analysis in many model and crop species. Here, we describe a simplified and highly efficient root protoplast isolation and transient expression system in the model legumes Lotus japonicus and Medicago truncatula. Firstly, we presented an efficient protocol for isolating protoplasts from L. japonicus and M. truncatula roots. We then established an efficient transient expression system in these legumes root protoplasts. Using this protocol, the subcellular localization of two symbiosis related proteins (SYMRK and ERN1) were visualized in the plasma membrane and nuclei, respectively. Collectively, this efficient protoplast isolation and transformation protocol is sufficient for studies on protein subcellular localization, and should be suitable for many other molecular biology applications.
Introduction
Legumes are the third largest group of angiosperms and include many very important food, feed and biofuel crops. Moreover, legumes have evolved a symbiotic interaction with rhizobia bacteria to efficiently fix nitrogen (Masson-Boivin et al., 2009), this nutrient source not only can satisfy its own growth, but also provides nitrogen for other plants grown in rotation or intercropped with legume plants. Nitrogen is one of the most important limiting factors to plant growth and crop yield. Modern agriculture relies heavily on industrial fertilizers to achieve high yields. Industrial nitrogen fixation is a highly energy consuming process. In addition, applying chemical fertilizers is a largely inefficient process, as more than half the fertilizers are lost to leaching, resulting in significant environmental problems, such as eutrophication (Galloway et al., 2008). Therefore, the nitrogen fixation ability of legumes provides them with a distinct advantage over other plant species in agriculture. Detailed investigation of the molecular mechanisms of the symbiosis may help us understand the essence of this phenomenon and provide scientific knowledge for exploring non-legume nitrogen fixation thus reduce the use of industrial fertilizers in future (Perrine-Walker et al., 2007; Rogers and Oldroyd, 2014).
The genome sequence of several model legume species, such as Medicago truncatula, Lotus Japonicus, and Glycine max been published recently (Sato et al., 2008; Schmutz et al., 2010; Young et al., 2011). Several mutant collections have been established by EMS chemical induction (Perry et al., 2009), LORE1 (Fukai et al., 2012; Urbanski et al., 2012) or Tnt1 insertion (Tadege et al., 2008) and fast neutron bombardment (FNB) mutagenesis (Chen et al., 2017). Many of the key components involved in symbiosis signal transduction, rhizobial infection and nodule organogenesis have been successively identified in these models (Svistoonoff et al., 2014). However, due to long life cycle of the plants, and the time consuming and low efficiency of the stable genetic transformation systems in these model legumes, the progress of the molecular mechanisms studies of these symbiotic proteins has been greatly limited. Some heterologous systems such as tobacco and Arabidopsis have been used for functional characterization of legume proteins, however, the expression of proteins in heterologous systems has certain limitations. For example, the encoded proteins of some Arabidopsis genes introduced into rice for subcellular localization have been shown to mis-localize (Zhang et al., 2011). In addition, the nodulation process occurs in legume roots and is a very host-specific process. Rhizobia secrete signal molecules, such as Nod factor (Valera and Alexander, 1965), which are very important for rhizobial infection and nodule organogenesis (Oldroyd and Downie, 2008), and heterologous systems cannot respond properly to this signal molecules. The plant protoplast system provides a complementary or alternative stable transgenic system for many types of gene functional studies, such as protein subcellular localization, in vivo protein-protein interaction etc. However, an efficient isolation and transient expression system in legumes root protoplast has not yet been established.
Because of the presence of cell walls, many routine molecular biology techniques in animal cells are difficult to perform in plant cells. However, plant protoplasts without cell walls behave similarly to animal cells in vitro thus offer a physiological and versatile cell-based experimental system. It is the first report on successful isolation of tomato protoplast in 1960 by Cocking (Cocking, 1960). By using different methods such as PEG-calcium fusion, microinjection, and electroporation, macromolecules including DNA, RNA and proteins can be delivered to protoplasts. By using tobacco mesophyll protoplasts, Fischer and Hain performed the first successful experiment for the transfection into protoplasts (Fischer and Hain, 1995). Subsequently, efficient systems for transient gene expression using protoplasts have been successfully established in different model and crop species including Arabidopsis (Yoo et al., 2007), tobacco (Fischer and Hain, 1995), maize (Sheen, 2001), rice (Zhang et al., 2011), perennial ryegrass (Yu et al., 2017) and Brachypodium (Hong et al., 2012). However, efficient systems for transient gene expression mostly focused on leaf mesophyll protoplasts. Root system are important for nutrient intake and response to biotic or abiotic environment. Nitrogen fixation between rhizobia and legume occurs in the host plant roots. However, the root protoplast isolation and transfection protocol in model legumes have not been established. We are trying to isolate legume root protoplasts for functional study of genes related to symbiotic nitrogen fixation and it will be helpful for other studies as well.
In this study, we report a simplified and highly efficient transient gene expression system using root protoplasts from L. japonicus and M. truncatula. And we also applied this system to get correct subcellular localization of classic genes in nodulation. The application of the new technique in legumes will significantly shorten the time for characterization of gene function.
Materials and Methods
Reagents
• Cellulase Onozuka R-10 (Yakult Pharmaceutical Ind. Co., Japan);
• Macerozyme Onozuka R-10 (Yakult Pharmaceutical Ind. Co., Japan);
• D-Sorbitol (Sigma, cat. no. S3889);
• Potassium chloride (KCl; Sigma-Aldrich, cat. no. P3911);
• Calcium chloride dehydrate (CaCl2; Sigma, cat. no. C7902);
• Sodium chloride (NaCl; Sigma-Aldrich, cat. no. S9888);
• Magnesium chloride hexa-hydrate (MgCl2. 6 H2O; Sigma-Aldrich, cat. no. 9272);
• BSA (Shanghai Bo AO Biotech Co., Ltd., China);
• MES hydrate (Sigma, cat. no. M8250);
• Cell-wall degrading enzyme mix viscozyme (Sigma, cat. no. V2010);
• PEG4000 (Sigma-Aldrich, cat. no. 81240);
• Plasmid Maxi Kit (QIAGEN, cat. no. 12163);
• Gamborg B-5 Basal Medium (Phytotechnology, cat. no. G398);
• T4 DNA Ligase (Takara, cat. no. 2011A);
• Bam HI-HF (NEB, cat. no. R3136S);
• Stu I (NEB, cat. no. R0187S).
Reagents Setup
• Digestion solution (always prepare freshly):
• Prepare 10 mM MES (pH 5.7) containing 1.5% (wt/vol) cellulase R-10, 2% (wt/vol) macerozyme R-10, 0.4M D-sorbitol and 10 mM CaCl2. Warm the solution at 55°C for 10 min and cool it slowly to room temperature (25°C). At the last, add 5% cell-wall degrading enzyme mix viscozyme and 1% BSA.
• W5 solution: 154 mM NaCl, 125 mM CaCl2 and 5 mM KCl and 10 mM MES (pH 5.7). The prepared W5 solution can be stored in the refrigerator for several weeks.
• WI solution: 0.4 M D-sorbitol, 20 mM KCl and 4 mM MES (pH 5.7). The prepared WI solution can be stored at room temperature.
• DMG solution: 0.4 M D-sorbitol, 15 mM MgCl2 and 4 mM MES (pH 5.7). The prepared DMG solution can be stored at room temperature.
• PEG–calcium transfection solution: 20% (wt/vol) PEG4000, 0.2 M D-sorbitol and 100 mM CaCl2.
Equipment
• Fluorescence microscope (Nikon ECLIPSE Ni; Nikon);
• Confocal laser scanning microscope (Leica TCS SP8; Leica);
• Millex-HV filter: Unit-33 mm-0.45 μm (Merck, cat. no. SLHV033RB);
• Cell Strainer, 300 Screen Mesh, Hand Handle (56 μm, Sangon biotech, cat. no. F513442-0001);
• 6-well TC Treated Plates (Sangon biotech, cat. no. F603201-0001);
• Petri dishes 40 × 9 mm (Sangon biotech, cat. no. F611001-0001);
• pH meter (Five Easy pH; Mettler-Toledo);
• Syringe 5 ml (Shanghai Misawa Medical Industry Co., Ltd., China);
• 10-ml round-bottomed Centrifuge tube (Sangon biotech, cat. no. F600889-0001);
• Microscope Cover glassed 22 × 22-1 (Fisherbrand, lot no. 12-542-B);
• Hemacytometer (Shanghai Qiujing Biochemical Reagent Instrument Co., Ltd., China).
Stepwise Procedures
To establish an efficient transient expression protoplast system for legumes root, we evaluated the protocol established for protoplast isolation from L. japonicus MG20 root by modifying various experimental parameters including enzyme digestion time and concentrations, solution recipes, culture medium etc. (Figure 1; Den Herder et al., 2012). The entire procedure is divided into six main stages from A to F. (A) Plant growth, (B) Vector construction and plasmid preparation, (C) Isolation of protoplasts from legumes roots, (D) DNA-PEG-calcium transfection, (E) Protoplast culture and harvest, and (F) Images are obtained by laser scanning confocal microscopy.
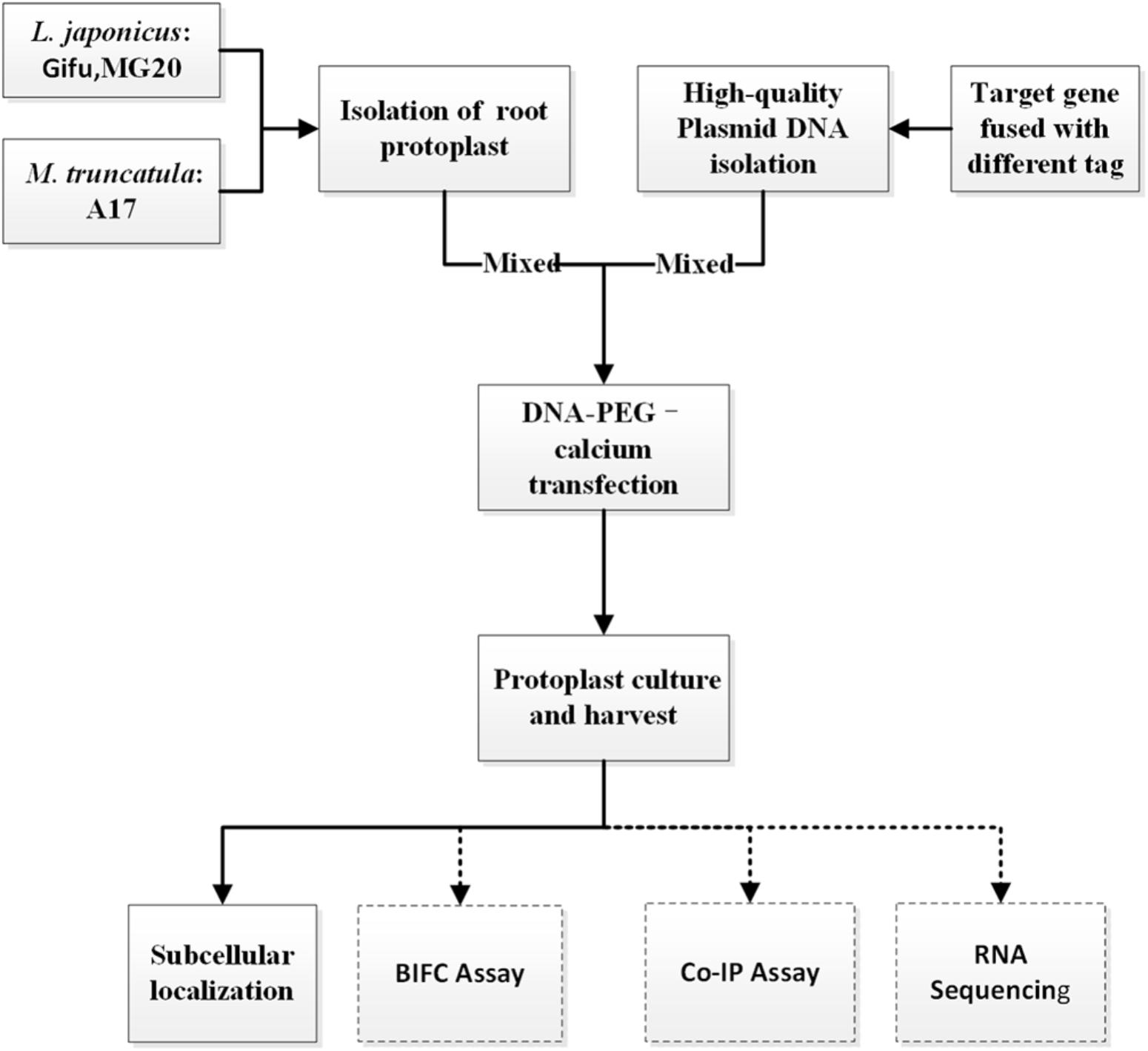
FIGURE 1. An overview flowchart describing transient gene expression systems using legume root protoplasts.
CRITICAL: Except for different plant growth conditions, this protocol (solution recipes and step-by-step) has also applied to Gifu and M. truncatula A17.
Stage A. Plant Growth
Time: 6–10 day
Lotus japonicus ecotype Gifu and Miyakojima (MG20), M. truncatula ecotype A17 were used. The seeds were scarified with sandpaper or immersed for 5–7 min in concentrated H2SO4 (for M. truncatula), then surfaced sterilized with 10% sodium hypochlorite solution for 5–10 min, washed 5 times in sterile H2O and kept overnight in water. Imbibed seeds were transferred to 0.8% DWA (Distilled Water Agar) plate and germinated in dark. M. truncatula seeds were placed in 4 degree freezers for 3 days before germinated in dark. Then the germinated seedlings were transferred into 1 × Gamborg B5 square plates with a sterilized filter paper placed on the top of the medium as to cover half of the petri dish, germinated seeds are placed on the border of the filter paper so that emerging roots will stay attached to it. The plates were vertical positioned in a controlled growth chamber (For Gifu, 2 days in the dark and 7–8 days in 16 h light/8 h dark; For MG20, 1 day in the dark and 5–6 days in 16 h light/ 8 h dark. For M. truncatula A17, seeds were germinated overnight at room temperature, then 5–6 days in 16 h light/8 h dark growth chamber).
CRITICAL: The growth status of the plant is very important for root protoplasts isolation and transfection. Healthy plant growing on the 1 × B5 medium instead of 1/2 × B5 can be recommend isolating root protoplasts. Please avoid isolating protoplasts from stressed or older root tissue.
Stage B. Vector Construction and Plasmid Preparation
Time: 2 days
To generate 35S::SYMRK-GFP and 35S::ERN1-GFP construct, the SYMRK coding region (CDS) and ERN1 CDS was PCR-amplified with cDNA from L. japonicus seedlings using 5′- ggggggatcc ATGATGGAGTTACCAGCTA-3′ and 5′- ggggaggcctTCTCGGCTGTGGGTGAGAC -3′ for SYMRK, 5′- ggggggatcc ATGGAGATTCAATTCCAGC -3′ and 5′- ggggaggcct ACAGAACAATGAGCACAAGG -3′ for ERN1. The PCR fragment was fused to N termini of GFP in vector pUC19 digested with Bam H I /Stu I. The constructs were confirmed by sequencing.
For protoplast transfection, high concentration and purity of plasmid DNA is required. Using the QIAGEN MAXI Kit, we were able to isolate sufficiently high-quality plasmid using the manufacturer’s suggested protocols. The extracted plasmid DNA quality was verified using a nanodrop machine and by running a sample on an agarose gel.
PAUSE POINT: High concentration and purity of plasmids can be stored at a -20°C refrigerator for several months.
CRITICAL: We recommend using small size plasmids, because transfection efficiency is higher than large size binary vectors.
CRITICAL: High concentration and purity of plasmids DNA are required for protoplasts transfection, and we extract plasmids using the CsCl gradient if necessary.
Stage C. Isolation of Protoplasts From Legumes Roots
Time: 8–10 h
(1) The roots of 6-day-old MG20 seedlings were used for protoplast isolation.
(2) Vigorously growing roots from 6-day-old seedling were used.
(3) Young root tissues from 100 seedlings were cut into 1 mm slices and then dipped into 10 ml of digestion solution. At this step, the amount of digestion solution should be enough to soak all root tissues.
CRITICAL: It is difficult to distinguish root protoplasts because color of root tissue is similar to digestion solution. Hypocotyl can be cut in part to show green as an indicator for root protoplasts.
(4) Incubate for 8–12 h in the digestion solution without shaking at 23°C in the dark. After digestion, the solution should display a strong brown color, which indicates the release of protoplasts. If the incubation time is too long, the protoplasts will be stressed.
CRITICAL: Older roots may need longer digestion time, and there are obvious small bubbles in the tissue indicating that the digestion is complete.
CRITICAL: Due to root tissue are denser than leaves, we gently clip root tissues by a large pair of tweezers to push protoplasts out by mechanical force.
(5) Check for the release of root protoplasts in the solution under the microscope; the size of MG20 root protoplasts is approximately 30–50 μm. To remove undigested root tissue, the protoplasts were collected by filtration through 50 μm nylon meshes and diluted with an equal volume of pre-chilled W5 solution. Our method generated approximately 1 × 105 cells varying from 10 to 50 μm in size.
CRITICAL: The size of root protoplasts is smaller than leaf, so we filter cells by 300 screen mesh with smaller aperture. To get cleaner root protoplasts, we can filter it twice.
(6) Centrifuge at 400 RCF 4°C for 2 min in a swinging-bucket rotor. After centrifugation, discard as much supernatant as possible and re-suspend the protoplasts in the 10 ml pre-chilled W5 solution.
CRITICAL: Use the swinging-bucket rotor instead of the corner rotor.
(7) Pellet the protoplasts by centrifugation at 400 RCF 4°C for 2 min in a swinging–bucket rotor. Add 5 ml pre-chilled W5 solution and re-suspend the protoplasts in a 10-ml round-bottomed tube.
CRITICAL: We suggest using round-bottomed tube in the whole experiments.
(8) Keep protoplasts on ice for 30–60 min. After 15 min protoplasts should begin to settle at the bottom of the tube by gravity. Remove as much of the W5 solution as possible without touching the protoplast pellet. We diluted a suitable concentration of the protoplasts for efficient transfection in DMG solution by using hemacytometer.
CRITICAL: The suitable concentration of the protoplasts is very critical for transfection efficiency.
Stage D. DNA-PEG-Calcium Transfection
Time: 30 min
(1) Add 20 μL plasmid DNA (plasmid DNA concentration about 1 μg/μL, size from 5 to 10 kb) to a 5-mL-round-bottom tube.
(2) Mix 200 μL protoplasts (104 protoplasts) and plasmid DNA gently.
(3) Add 220 μL with an equal volume of PEG solution, and then mix well by gently tapping the tube. Incubate for 5 min in the room temperature.
(4) Terminate transfection process by adding 880–900 μL W5 solution at room temperature and mix completely by gently rocking or inverting the tube.
(5) Centrifuge at 400 RCF 4°C for 2 min using a swinging-bucket rotor and discard supernatant.
(6) Re-suspend protoplasts gently with 2 mL WI solution and transfer mixture to each well of a 6-well tissue culture plate.
CRITICAL: WI solution recipes can only maintain the basic living of root protoplasts.
Stage E. Protoplast Culture and Harvest
Time: 8–20 h
(1) Incubate protoplasts at room temperature (20 °C -25°C) for 12-20h in the dark.
CRITICAL: Because roots belong to underground tissues, we culture protoplast in the darkness.
(2) Harvest protoplasts by centrifugation at 400 RCF 4°C for 2 min a swinging-bucket rotor.
(3) Discard the supernatant and re-suspend protoplasts for further analysis.
Stage F. Images Are Obtained by Laser Scanning Confocal Microscopy
Time: 1 day
Root protoplasts were visualized using SP8 confocal microscope (Leica TCS 8 SP8). The filter sets used for excitation (Ex) and emission (Em) were as follows: GFP, 488 nm (Ex)/from 498 to 550 nm (Em); bright field, 633 nm. Signals were captured in multi-channel mode. All fluorescence experiments were independently repeated at least three times.
Timing
Stage A: 6–10 days;
Stage B: 2 days;
Stage C: 8–10 h;
Stage D: 30 min;
Stage E: 8–20 h;
Stage F: 1 day;
Anticipated Results
Highly Protoplast Isolation Efficiency
For L. japonicus MG20, our protocol generated approximately 1 × 105 cells varying from 10 to 50 μm in size (Figure 2A). Above 90% of isolated root protoplasts were healthy by fluorescein diacetate (FDA) staining (Figure 2B and Table 1). We diluted the suitable concentration of the protoplasts (1 × 104 cells per transfection) for efficient transfection by using a hemacytometer under a microscope.
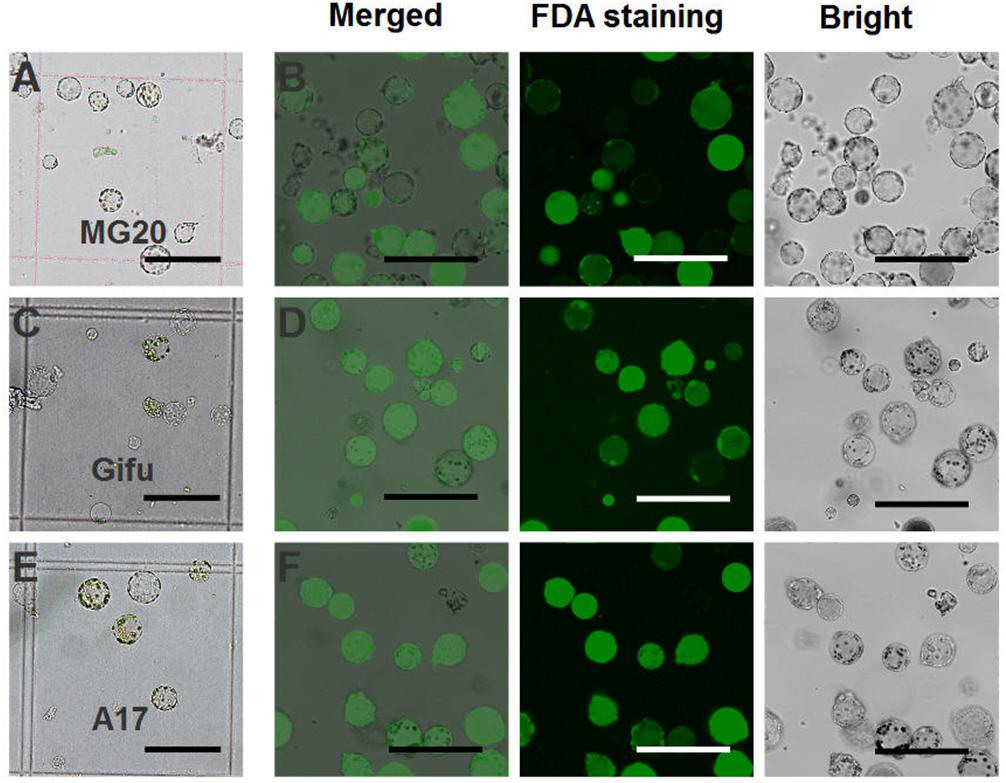
FIGURE 2. Isolation of protoplasts from legumes roots. Isolated root protoplasts from (A) 6-day-old L. japonicus MG20 seedling. (C) 8-day-old L. japonicus Gifu seedling. (E) 5-day-old M. truncatula A17 seedling. FDA staining of root protoplasts from (B) MG20. (D) Gifu. (F) A17. Image of protoplasts obtained under a Nikon microscope with a 20 × objective. Scale bar = 100 μm.
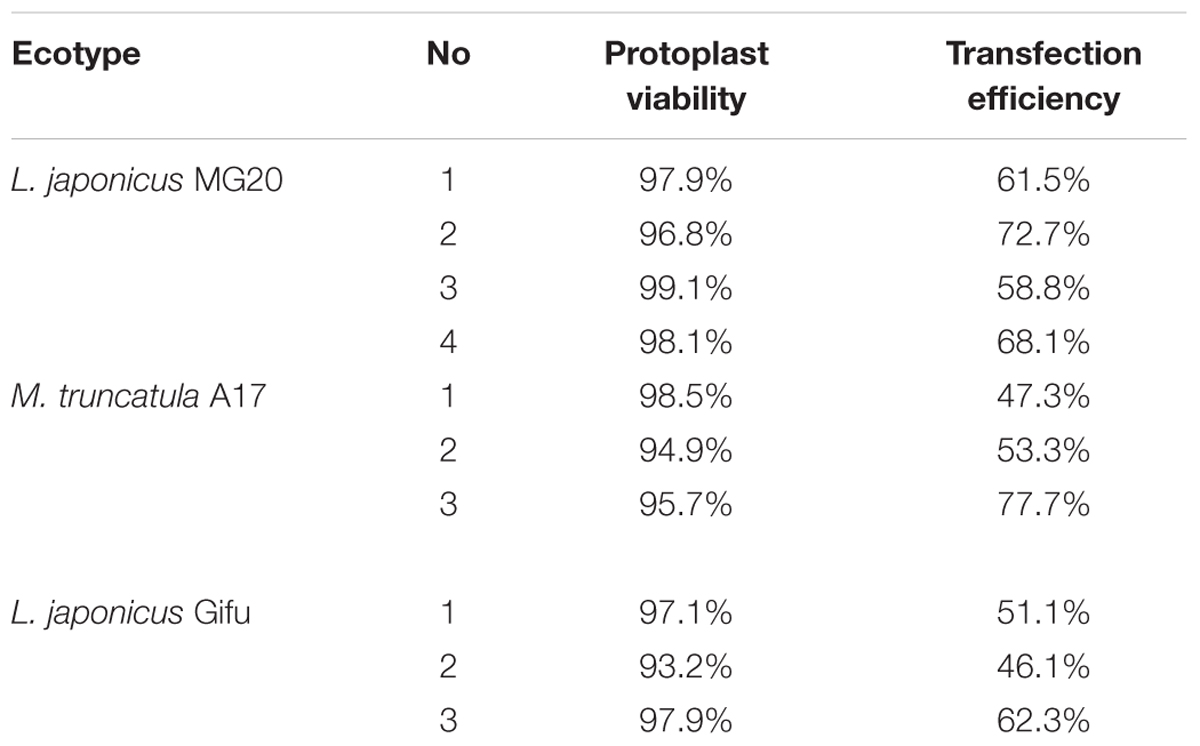
TABLE 1. Viability and transfection efficiency of root protoplasts by expression of vector 2 × 35S::GFP-pUC19.
The root protoplasts from other ecotype (L. japonicus Gifu and M. truncatula A17) were isolated following the protocol from L. japonicus MG20. 8-day-old Gifu and 5-day-old A17 seedlings were prepared for protoplast isolation, but compared to MG20, their yield of root protoplasts is lower (Figures 2C,E). FDA staining show that the viability of the cells was no different than root protoplasts from MG20 (Figures 2D,F).
Highly Efficient Transient Transfection
After transfection with the 2 × 35S::GFP plasmid (pUC19-GFP) by using the PEG-mediated transfection approach and incubation for 12 h, the GFP fluorescence was detected both in the cytoplasm and nucleus of the roots protoplasts (Figure 3B). In general, root protoplast transfection efficiencies of approximately 60–70% were achieved with this protocol (Figure 3A and Table 1).
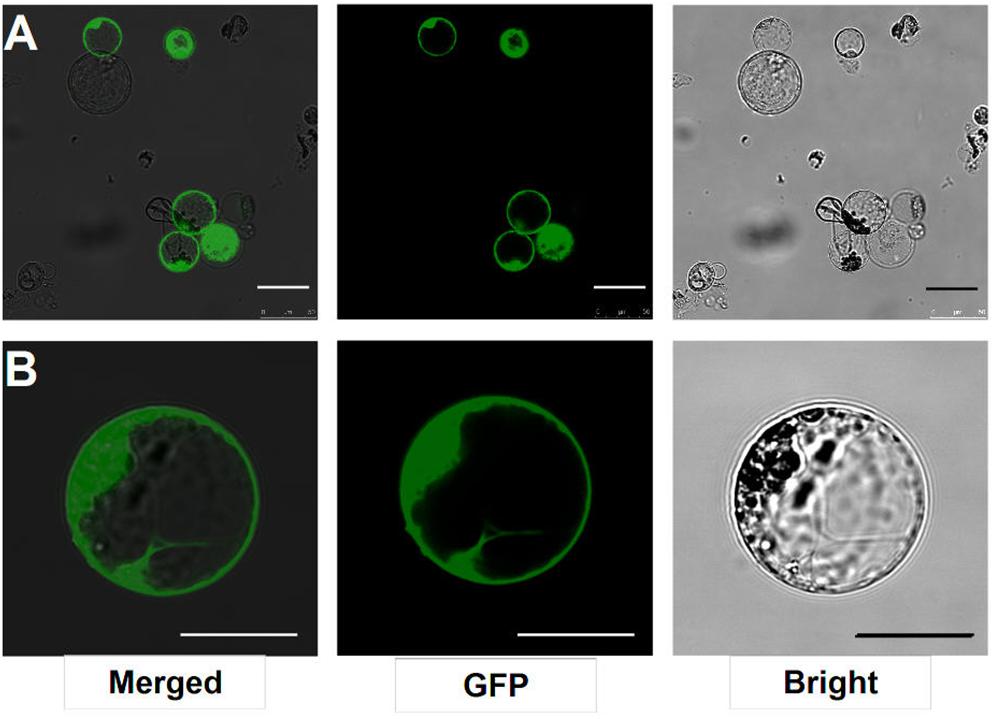
FIGURE 3. Transient expression GFP-pUC19 in L. japonicus MG20 root protoplasts. (A) Detection of transfection efficiency by expression of vector 2 × 35S::GFP-pUC19. More than 60% of root protoplasts were transfected successfully with the GFP reporter. (B) Detail of expression patterns in the root protoplasts transfection by 2 × 35S::GFP-pUC19 are shown under Confocal microscopy (Leica TCS SP8). Scale bars = 25 μm.
SYMRK and ERN1 Protein Subcellular Localization in Lotus Root Protoplasts
Our protocol can be applied to protein subcellular localization. To test this application, we first cloned two symbiosis related genes (SYMRK and ERN1). SYMRK, a symbiosis receptor-like kinase, was previously shown to localize in plasma membrane using hairy root transformation (Stracke et al., 2002; Den Herder et al., 2012). Using the current protocol, we were able to confirm its subcellular localization of plasma membrane in our roots protoplasts system (Figure 4A). ERN1, a AP2/ERF type transcription factor required for nodulation, was shown localized in the nucleus in Lotus root protoplasts by overlap with DAPI DNA stain (Figure 4B) (Andriankaja et al., 2007; Cerri et al., 2017). Its subcellular localization in our homologous protoplast system was consistent with previous reports.
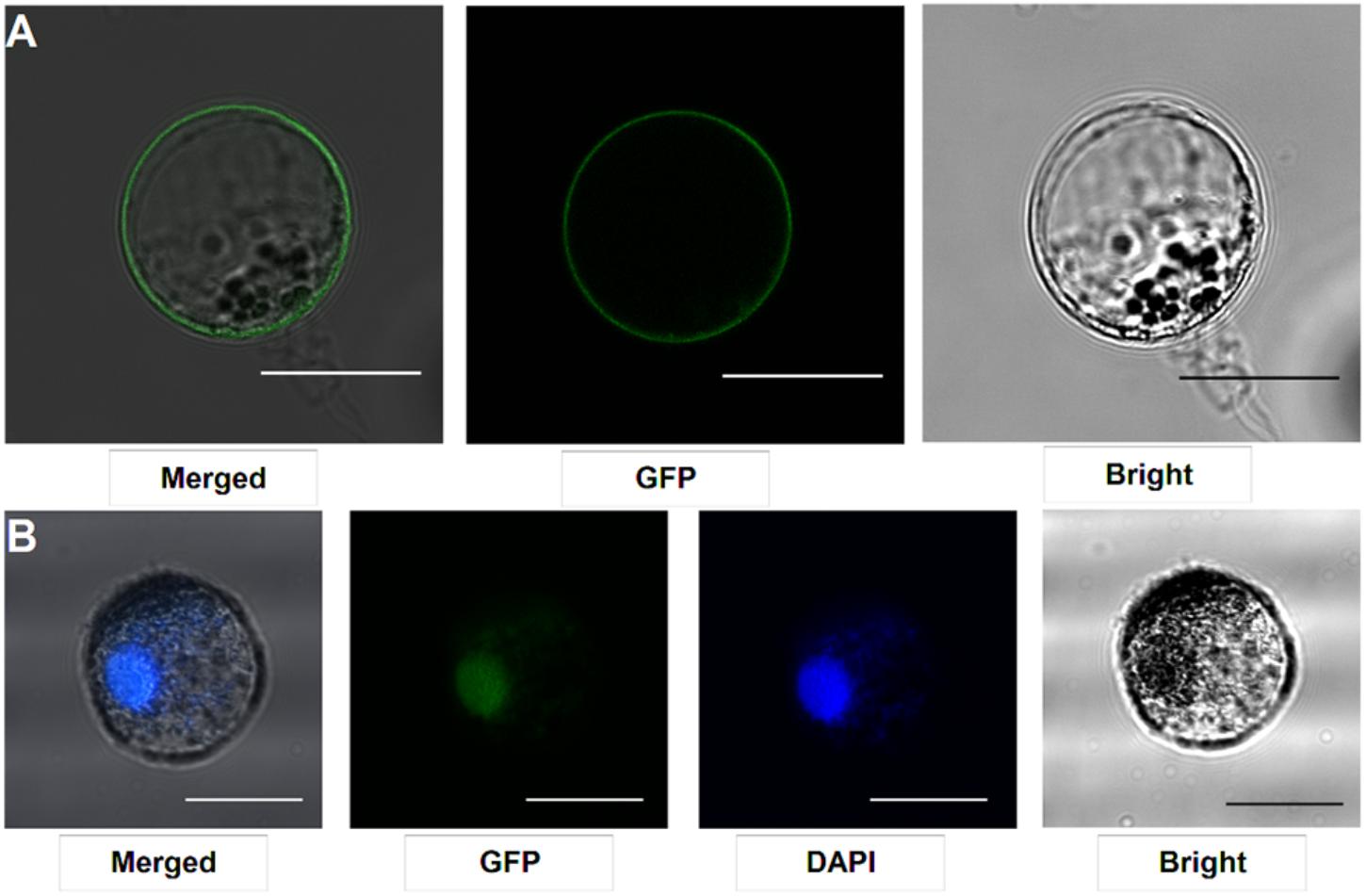
FIGURE 4. Subcellular localization analysis in L. japonicus root protoplasts. (A) SYMRK-GFP targeted to plasma membrane in Gifu root protoplasts. (B) ERN1-GFP nuclear-localized expression patterns in Gifu root protoplasts, the nucleus-localization was merged with DAPI staining. Scale bars = 25 μm. Transfection efficiency (SYMRK) = 10.7%; Transfection efficiency (ERN1) = 13.6%.
Author Contributions
NJ contributed to design of this research, performed the experiments, and wrote the manuscript. YZ performed the research. NJ and FX wrote the paper and all the authors approved the manuscript.
Funding
This work was supported by The National Key R&D Program of China (2016YFA0500500) and the National Natural Science Foundation of China (31700207).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer XL and handling Editor declared their shared affiliation.
Acknowledgments
We thanks Dr. Jeremy Murray for critical reading and modifying of the manuscript, Tongda Xu and Yan Xiong for helping in isolation and transfection protoplasts.
References
Andriankaja, A., Boisson-Dernier, A., Frances, L., Sauviac, L., Jauneau, A., Barker, D. G., et al. (2007). AP2-ERF transcription factors mediate Nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 19, 2866–2885. doi: 10.1105/tpc.107.052944
Cerri, M. R., Wang, Q., Stolz, P., Folgmann, J., Frances, L., Katzer, K., et al. (2017). The ERN1 transcription factor gene is a target of the CCaMK/CYCLOPS complex and controls rhizobial infection in Lotus japonicus. New Phytol. 215, 323–337. doi: 10.1111/nph.14547
Chen, Y., Wang, X., Lu, S., Wang, H., Li, S., and Chen, R. (2017). An array-based comparative genomic hybridization platform for efficient detection of copy number variations in fast neutron-induced Medicago truncatula Mutants. J. Vis. Exp. 8:129. doi: 10.3791/56470
Cocking, E. C. (1960). A method for the isolation of plant protoplasts and vacuoles. Nature 187, 927–929. doi: 10.1038/187962a0
Den Herder, G., Yoshida, S., Antolin-Llovera, M., Ried, M. K., and Parniske, M. (2012). Lotus japonicus E3 ligase SEVEN IN ABSENTIA4 destabilizes the symbiosis receptor-like kinase SYMRK and negatively regulates rhizobial infection. Plant Cell 24, 1691–1707. doi: 10.1105/tpc.110.082248
Fischer, R., and Hain, R. (1995). Tobacco protoplast transformation and use for functional analysis of newly isolated genes and gene constructs. Methods Cell Biol. 50, 401–410. doi: 10.1016/S0091-679X(08)61046-8
Fukai, E., Soyano, T., Umehara, Y., Nakayama, S., Hirakawa, H., Tabata, S., et al. (2012). Establishment of a Lotus japonicus gene tagging population using the exon-targeting endogenous retrotransposon LORE1. Plant J. 69, 720–730. doi: 10.1111/j.1365-313X.2011.04826.x
Galloway, J. N., Townsend, A. R., Erisman, J. W., Bekunda, M., Cai, Z., Freney, J. R., et al. (2008). Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science 320, 889–892. doi: 10.1126/science.1136674
Hong, S.-Y., Seo, P. J., Cho, S.-H., and Park, C.-M. (2012). Preparation of leaf mesophyll protoplasts for transient gene expression in Brachypodium distachyon. J. Plant Biol. 55, 390–397. doi: 10.1007/s12374-012-0159-y
Masson-Boivin, C., Giraud, E., Perret, X., and Batut, J. (2009). Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes? Trends Microbiol. 17, 458–466. doi: 10.1016/j.tim.2009.07.004
Oldroyd, G. E., and Downie, J. A. (2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59, 519–546. doi: 10.1146/annurev.arplant.59.032607.092839
Perrine-Walker, F. M., Prayitno, J., Rolfe, B. G., Weinman, J. J., and Hocart, C. H. (2007). Infection process and the interaction of rice roots with rhizobia. J. Exp. Bot. 58, 3343–3350. doi: 10.1093/jxb/erm181
Perry, J., Brachmann, A., Welham, T., Binder, A., Charpentier, M., Groth, M., et al. (2009). TILLING in Lotus japonicus identified large allelic series for symbiosis genes and revealed a bias in functionally defective ethyl methanesulfonate alleles toward glycine replacements. Plant Physiol. 151, 1281–1291. doi: 10.1104/pp.109.142190
Rogers, C., and Oldroyd, G. E. (2014). Synthetic biology approaches to engineering the nitrogen symbiosis in cereals. J. Exp. Bot. 65, 1939–1946. doi: 10.1093/jxb/eru098
Sato, S., Nakamura, Y., Kaneko, T., Asamizu, E., Kato, T., Nakao, M., et al. (2008). Genome structure of the legume, Lotus japonicus. DNA Res. 15, 227–239. doi: 10.1093/dnares/dsn008
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Sheen, J. (2001). Signal transduction in maize and Arabidopsis Mesophyll protoplasts. Plant Physiol. 127, 1466–1475. doi: 10.1104/pp.010820
Stracke, S., Kistner, C., Yoshida, S., Mulder, L., Sato, S., Kaneko, T., et al. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417, 959–962. doi: 10.1038/nature00841
Svistoonoff, S., Hocher, V., and Gherbi, H. (2014). Actinorhizal root nodule symbioses: what is signalling telling on the origins of nodulation? Curr. Opin. Plant Biol. 20, 11–18. doi: 10.1016/j.pbi.2014.03.001
Tadege, M., Wen, J., He, J., Tu, H., Kwak, Y., Eschstruth, A., et al. (2008). Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 54, 335–347. doi: 10.1111/j.1365-313X.2008.03418.x
Urbanski, D. F., Malolepszy, A., Stougaard, J., and Andersen, S. U. (2012). Genome-wide LORE1 retrotransposon mutagenesis and high-throughput insertion detection in Lotus japonicus. Plant J. 69, 731–741. doi: 10.1111/j.1365-313X.2011.04827.x
Valera, C. L., and Alexander, M. (1965). Nodulation factor for Rhizobium-Legume symbiosis. J. Bacteriol. 89, 1134–1139.
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Young, N. D., Debelle, F., Oldroyd, G. E., Geurts, R., Cannon, S. B., Udvardi, M. K., et al. (2011). The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524. doi: 10.1038/nature10625
Yu, G., Cheng, Q., Xie, Z., Xu, B., Huang, B., and Zhao, B. (2017). An efficient protocol for perennial ryegrass mesophyll protoplast isolation and transformation, and its application on interaction study between LpNOL and LpNYC1. Plant Methods 13:46. doi: 10.1186/s13007-017-0196-0
Keywords: legumes, root protoplasts, PEG-mediated transformation, transient gene expression, symbiosis
Citation: Jia N, Zhu Y and Xie F (2018) An Efficient Protocol for Model Legume Root Protoplast Isolation and Transformation. Front. Plant Sci. 9:670. doi: 10.3389/fpls.2018.00670
Received: 15 January 2018; Accepted: 02 May 2018;
Published: 04 June 2018.
Edited by:
Deqiang Duanmu, Huazhong Agricultural University, ChinaReviewed by:
Mathias Brault, Université Paris-Saclay, FranceXia Li, Huazhong Agricultural University, China
Copyright © 2018 Jia, Zhu and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fang Xie, eGllZmFuZ0BzaWJzLmFjLmNu
 Ning Jia
Ning Jia Yali Zhu1,2
Yali Zhu1,2 Fang Xie
Fang Xie