- Department of Plant Pathology, Washington State University, Pullman, WA, United States
Over the last decades, medical research has utilized DNA altering procedures in cancer treatments with the objective of killing cells or suppressing cell proliferation. Simultaneous research related to enhancing disease resistance in plants reported that alterations in DNA can enhance defense responses. These two opposite perspectives have in common their effects on the center for gene transcription, the nuclear chromatin. A review of selected research from both anticancer- and plant defense-related research provides examples of some specific DNA altering actions: DNA helical distortion, DNA intercalation, DNA base substitution, DNA single cleavage by DNases, DNA alkylation/methylation, and DNA binding/exclusion. The actions of the pertinent agents are compared, and their proposed modes of action are described in this study. Many of the DNA specific agents affecting resistance responses in plants, e.g., the model system using pea endocarp tissue, are indeed anticancer agents. The tumor cell death or growth suppression in cancer cells following high level treatments may be accompanied with chromatin distortions. Likewise, in plants, DNA-specific agents activate enhanced expression of many genes including defense genes, probably due to the chromatin alterations resulting from the agents. Here, we propose a hypothesis that DNA damage and chromatin structural changes are central mechanisms in initiating defense gene transcription during the nonhost resistance response in plants.
Introduction
Features of DNA-specific agents and their actions on cancer cells may share modes of action related to those inducing disease resistance in plants. The objective of cancer treatments is mainly to selectively stop cancer growth with little collateral damage to healthy cells. Some of the same DNA-specific compounds (Hendry et al., 2007) have been shown to activate defense response genes, termed pathogenesis-related (PR) genes (Hartney et al., 2007; Hadwiger, 2009). Research on the plant side is aimed at stopping fungal growth. The characterization of DNA damage-induced protein synthesis in plants is variable and involves traits ranging from DNA damage-related repair proteins to defensins (peptides) that are directly toxic to fungal pathogens (Chiang and Hadwiger, 1991; Almeida et al., 2006).
Hypothesis Defined
Pathogenesis-related genes are major contributors to the plant’s nonhost resistance to pathogens (Hadwiger, 2015a). In addition, the DNA-specific signals for activation of these genes can be initiated by “elicitors” or “effectors” of pathogen origin (Jones and Dangl, 2006; Boller and Felix, 2009). The transcription of these defense genes is ultimately coded by the DNA within the chromatin of the nucleus. Based primarily on the accumulated data on defense gene activation in pea endocarp tissue we are hypothesizing that multiple DNA-specific agents can activate PR genes and stimulate secondary metabolic pathways (e.g., producing antifungal compounds called phytoalexins) by generating direct effects on chromatin conformation. In a manner similar to how effectors can initiate signals (via cascading routes) to engage the transcription factors and positively affect stalled genes, the DNA/chromatin-specific agents can increase transcription via direct conformational changes (Hadwiger, 2008).
This paper assembles mechanistic information from current and previously published literature on transcription initiation (Hager et al., 2009). Because of the complexity of chromatin, the understanding of its ability to determine how and when the appropriate genes within are suppressed or expressed, is a challenge for all eukaryotic research. The RNA polymerase complex that transcribes the DNA code is confronted by a tightly packed genomic DNA in a nucleosome structure. Thus, gene transcription requires that a single DNA strand transit the DNA polymerase II enzyme in an environment of tight DNA helixes and attached nuclear proteins (Ma et al., 2013). Transcription benefits from removal of DNA helices and temporarily dissociating DNA from histones and other nuclear proteins (Yaniv, 2014). The genes coding PR and other defense gene products are apparently silent, stalled or partially suppressed prior to contact with a fungal pathogen. The suppressed environment of sensitive DNA regions (Teves and Henikoff, 2014) can be changed by: DNA intercalators, DNA base substitution, thymidine dimerization, DNA minor groove insertion, histone modification or removal, DNA strand cleavage and other chromatin-specific effects– to a transcription positive state. Within these agent actions are the eliciting agents, chitosan oligomers (Kendra et al., 1989) and a single strand cleaving DNase known to be released by pathogens (Hadwiger and Polashock, 2013) and transferred to the host nucleus in the pea nonhost resistance response.
Our hypothesis is that these general conformational changes occur within sensitive regions present in multiple chromosomes since the genomic mapping of the pea genome locates PR genes in multiple chromosomes (Pilet-Nayel et al., 2002; Ramirez-Prado et al., 2018). We also realize that DNA/chromatin changes can also stimulate some genes not directly involved in disease resistance. The following paragraphs detail the data upon which the hypothesis was derived.
DNA Damage: Insights Into the DNA Targets of Anticancer Agents and Phytoalexin Elicitors
Specific DNA altering actions including DNA intercalation, DNA distortion, DNA base substitution, DNA single and double strand cleavage, alkylation and methylation, DNA binding and exclusion in cancer related research (Martinez and Cha′con-Garcia, 2005) compare with the action of many of the same agents affecting resistance responses investigated primarily in the model endocarp tissue system of pea plants (Pisum sativum) (Hadwiger, 2015a). Early research on disease resistance in pea tissue revealed alterations in nuclear DNA that enhance defense responses (Hadwiger and Schwochau, 1971). These two opposite perspectives have in common their effects on the center for gene transcription, nuclear chromatin (Nair and Kumar, 2012). The similarities of action at the chromatin level in both systems are based on the degree of interaction.
The chromatin/DNA perspective presented herein by-passes a different interpretation of the signaling events that involve the plant receptor-like kinases as initiators of disease resistance or plant defense that is reviewed elsewhere (Nürnberger et al., 2004; Boller and Felix, 2009; Antolin-Llovera et al., 2014). Briefly, such signaling between an elicitor PAMPs (pathogen-associated molecular patterns) via receptor-mediated transfer to specific defense response genes within chromatin or intact pea tissue has been observed but primarily with high levels of two PAMPs (Hadwiger and Chang, 2015). These high PAMP concentrations were also associated with DNA damage and thus have commonality with the DNA-specific agents discussed herein.
Cell death or suppression in cancer following high-intensity treatments may be accompanied by chromatin distortions capable of activating the expression of less-desirable collateral genes. Likewise, in the pea endocarp, high-level treatments of DNA-specific agents can cause cell death, while low-level chromatin alterations activate the defense genes associated with immunity, i.e., nonhost resistance (Hadwiger et al., 1974; Choi et al., 2001; Hartney et al., 2007; Isaac et al., 2009). Some of the anticancer drugs remaining in use today are DNA damaging agents, and those that have been used to the best advantage in the past are being re-visited (Gurova, 2009). These agents have the potential to target the DNA of tumor cells, resulting in their destruction. However, their clinical use can result in adverse side effects, and since some are also carcinogenic, their continued use can promote secondary cancers.
DNA Damage, Damage Repair, and Chromatin Alterations in Cancer and Age-Related Diseases of Humans
DNA repair contributes to innate and acquired immunity (Song et al., 2014). DNA damage triggers the activation of DNA repair pathways and DNA repair protects against oxidized DNA damage generated by infectious and inflammatory diseases. Thus, DNA damage is involved in innate and adaptive immunity (Fontes et al., 2014). At the transcriptional level there is the regulation of cytokines and other genes involved in the inflammatory response. Chemical modifications to DNA and the histone components of chromatin potentiate gene expression. As an example, chromatin must become accessible to allow activation-induced cytidine deaminase (AID)-mediated deamination of cytosines in DNA (Daniel and Nussenzweig, 2013). In response to DNA damage there is a removal of DNA lesions. In the arousal of the immune system there can be an expression of antimicrobial peptides and development of ligands for receptors found on immune cells. Components that can arouse include DNA damage sensors, transducer kinases, and effectors (Nakad and Schumacher, 2016). Some progress has been reported in distinguishing which molecular and cellular pathways of the DNA damage activate immune signaling (Kastan and Bartek, 2001).
Induction of Pea Defense Responses
Investigations into the induction of plant defense responses by DNA-specific compounds in peas have occurred in parallel over multiple decades (Figure 1). Messenger RNA from pea tissue treated with DNA-specific agents was subsequently translated in vitro. This technique identified the total array of newly expressed gene products as characteristic protein patterns in 2-D electrophoretic separations. These patterns enhanced by the DNA specific anti-cancer actinomycin D in the plant host were similar to those induced following inoculation with fungal pathogens (Loschke et al., 1983). Both treatments also promoted the production of the anti-fungal phytoalexin, pisatin (Schwochau and Hadwiger, 1968; Hartney et al., 2007; Hadwiger and Tanaka, 2017a).
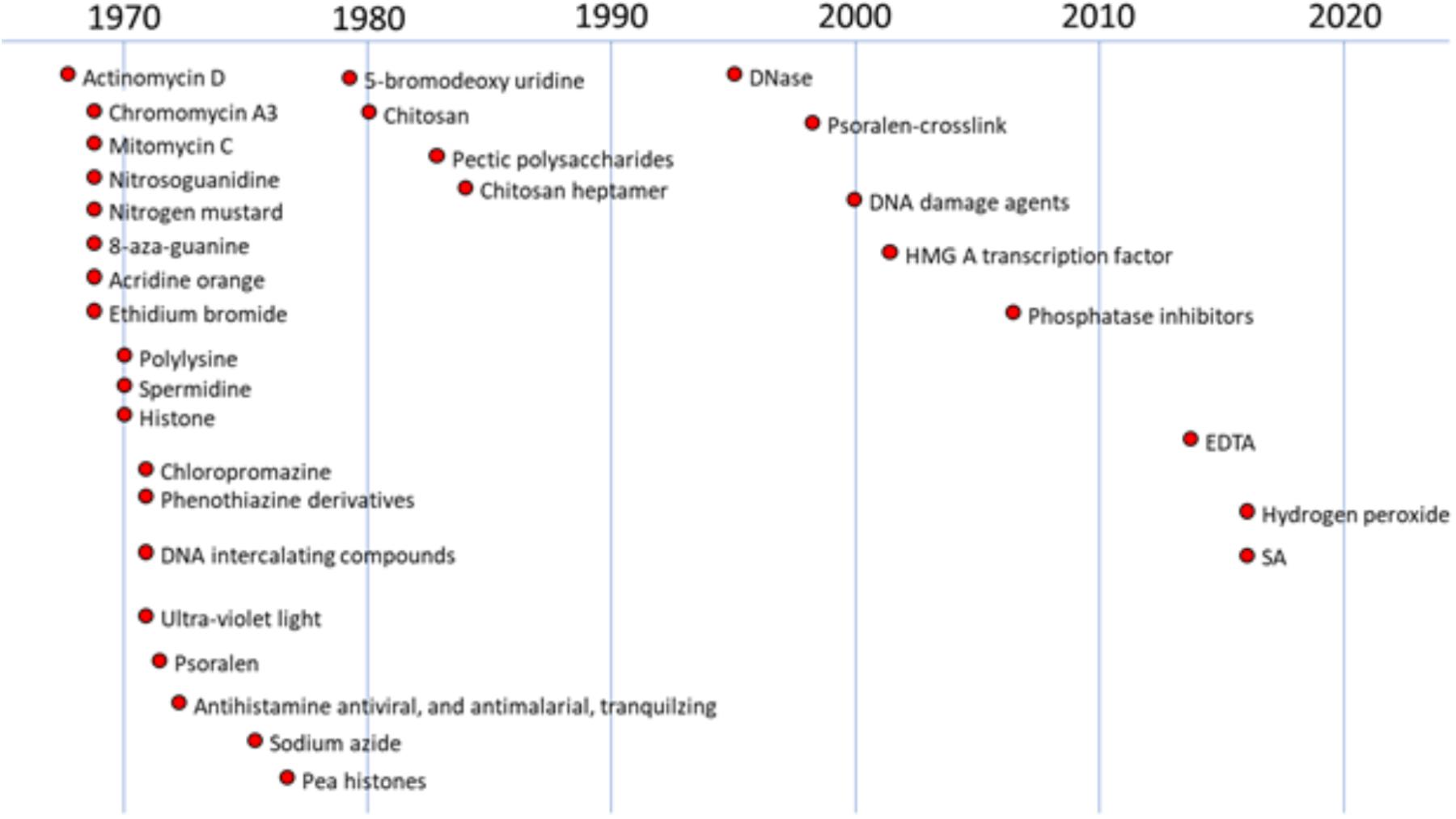
FIGURE 1. History of studies regarding anti-cancer and other compounds on DNA damage in plants. The figure were created based on the following references (chronological order): Schwochau and Hadwiger, 1968, 1969; Hadwiger and Schwochau, 1970, 1971; Hadwiger and Martin, 1971; Hess and Hadwiger, 1971; Hadwiger, 1972a,b; Hadwiger et al., 1976, 1977, 1995; Sander and Hadwiger, 1979; Hadwiger and Beckman, 1980; Walker-Simmons et al., 1983; Kendra et al., 1989; Parsons and Hadwiger, 1998; Choi et al., 2001; Klosterman et al., 2003; Hartney et al., 2007; Hadwiger and Tanaka, 2015; Hadwiger and Tanaka, 2017b; Tanaka and Hadwiger, 2017.
Specific concentrations of actinomycin provided resistance against Fusarium solani f. sp. pisi (Fspi) a true pathogen in pea (Figure 2). The variation of resistance that is concentration-related, probably due to the progression of DNA changes as more actinomycin molecules become involved. Actinomycin D 1 μg/ml applied 1 h prior to the pathogen spores (Figure 2B) there gave no cytologically detectible induction of resistance allowing the pathogen to proceed as it did following the water treatment (Figure 2A) in the absence of the hypersensitive host response. At 3 μg/ml (Figure 2C) the presence of actinomycin induces a resistance that is a plant disease resistance response rather than a direct antifungal action.
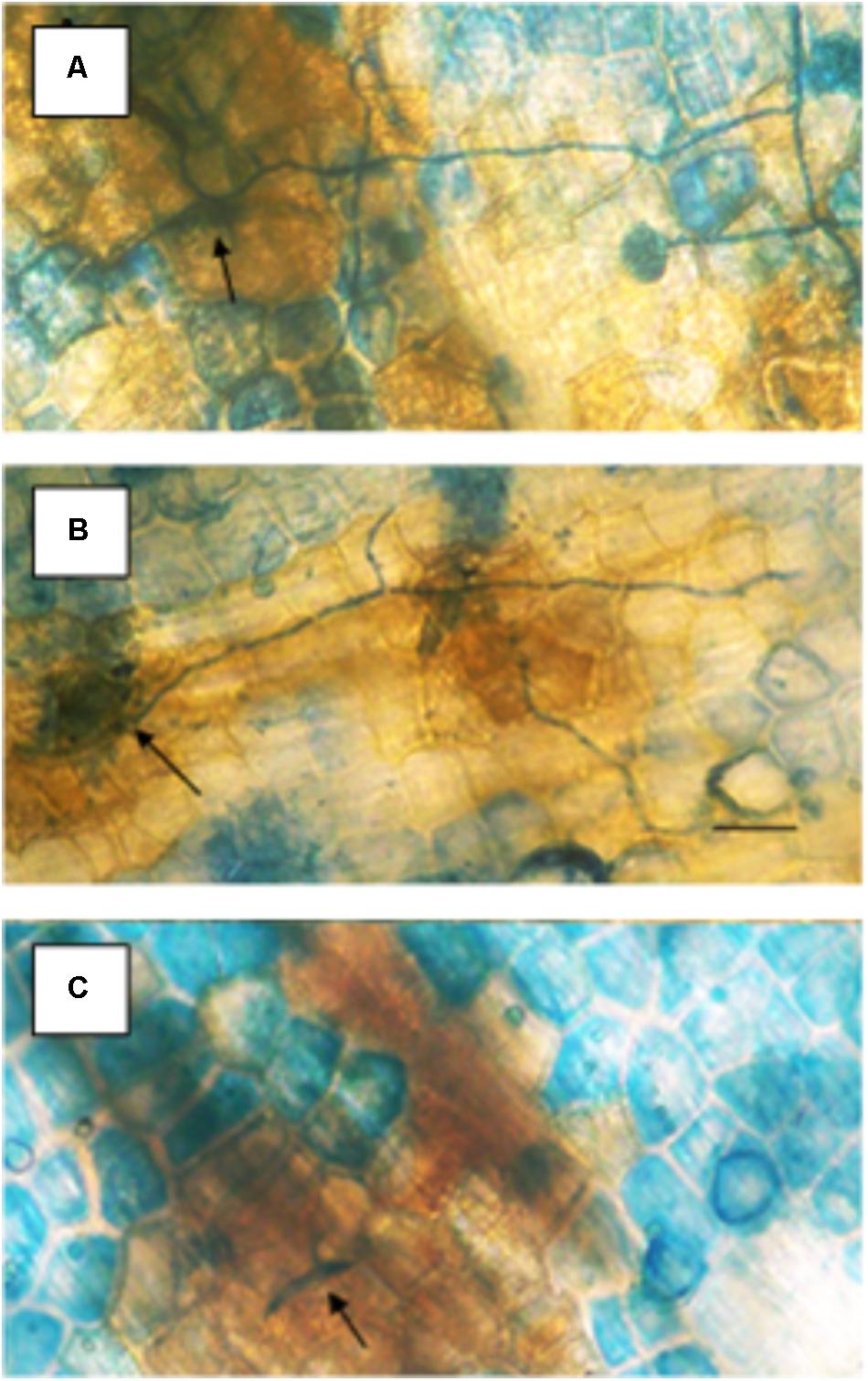
FIGURE 2. Effect of different concentrations of actinomycin D applied 1 h prior to inoculation on the susceptibility of pea endocarp tissue to the true pathogen, Fusarium solani f. sp. pisi (Fspi). The resistance responses of pea endocarp tissue against a pea pathogen (Fspi) are sharply influenced by different concentrations of actinomycin D applied 1 h prior to inoculation. Concentrations in photos are as follows: A = H2O; B = 1 μg/ml; and C = 3 μg/ml. Arrows indicate the inoculated spore. Bar = 50 microns.
Follow-up research utilized the chemical properties of other DNA-specific agents to investigate the basis of defense gene induction in plants, which may relate to DNA conformations or chromatin alterations.
The actions of compounds such as actinomycin D that specifically target DNA base sequences were valued for use because of the available background of physical and chemical information. The DNA intercalating property was first thought to primarily inhibit RNA synthesis; however, there were reports that it super-induced certain genes in other eukaryotes (Steinberg et al., 1975). Actinomycin D was also found to increase mRNA for specific pea genes. Examination of chromatin spreads from pea cells injected with labeled uridine indicated that regions of the chromatin are unraveled by actinomycin D, and unraveled chromatin supports hot spots of RNA synthesis (Hadwiger, 2015a). The action of actinomycin D demonstrates the complexity of DNA damage-related changes. The defense response induction by DNA-specific agents in plants was obtained with low actinomycin concentration levels. Actinomycin D was widely utilized in biological research for its ability to complex intimately (Reich and Goldberg, 1964) with DNA by intercalating the planer ring structure between base pairs and subsequently suppressing mRNA production (Flamm et al., 1966). It was noted that the binding of actinomycin D to the DNA in chromatin was restricted by the chromosomal proteins, and thus the binding of actinomycin D to chromatin could be a measure of the amount of DNA not masked by the chromosomal proteins (Beato et al., 1970). Alternately, this measure was used in plant systems to determine how much externally applied actinomycin D was transferred to the nucleus and to evaluate the open regions of DNA in pea cells; and the increased template activity that developed following treatment with elicitors and fungal challenges (Hadwiger et al., 1974).
These results demonstrated that both anticancer agents and defense gene activators can influence the structure and function of chromatin. Why is actinomycin D not inhibiting RNA synthesis in pea? In bacterial cells, actinomycin D is able to intercalate DNA at a rate of 1 molecule per 1000 base pairs and successfully suppress mRNA production (Hyman and Davidson, 1970). Alternately, the optimal induction of pea defense responses occurs when less than 1 molecule of actinomycin D inserts per 10,000 DNA base pairs (Hadwiger et al., 1974), a level that does not significantly suppress RNA synthesis. Thus, there is an apparent difference in action between plants and other systems based on the degree of intercalation. The activation of defense genes in pea tissue is proposed to occur by direct action on chromatin structure (Isaac et al., 2009) and is often accompanied by DNA damage. This disruption can be observed by electron microscopy (Hadwiger and Adams, 1978). The regions of disrupted chromatin structure have been shown to be regions of intense labeling with RNA precursors (Hadwiger, 2015a). The chromatin alteration hypothesis has been further tested in pea endocarp tissue and is compared with a series of compounds with well-researched modes of action (Hartney et al., 2007). Chromosome dynamics can also be influenced by inherent cytoskeleton polymers such as actin filaments, microtubules and intermediate filaments that connect to the nuclear envelope (Figure 3) (Spichal and Gabre, 2017). The smaller of these molecules can enter the nucleus and act as chromatin remodelers.
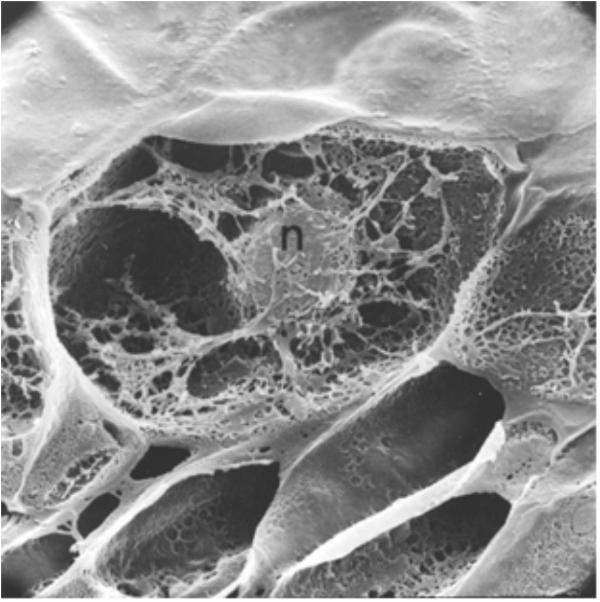
FIGURE 3. A cross-section of a pea endocarp cell, viewed in a scanning electron microscope, showing the intimate association of the pea nucleus with the cytoskeleton. Reproduced from a previous publication (Hadwiger and Adams, 1978).
In pea, an assay for detecting agents initiating the transcription of defense responses monitors a secondary pathway that culminates in part with the production of the anti-fungal isoflavonoid, pisatin. Compounds that are elicitor-positive in this assay were further examined to determine whether similar changes occur in the elicitation of total disease resistance in pea by a bean pathogen (nonhost resistance) or in furthering susceptibility to both pea and bean-specific pathogens (Hartney et al., 2007). Additional assays of pea tissue involved cell fractionation and cytological preparations that specifically examined DNA damage (Isaac et al., 2009), nuclear protein modification (Klosterman et al., 2003), and nuclear diameters changes (Tanaka and Hadwiger, 2017). As indicated, the accumulation of phytoalexin, pisatin, and PR gene activation are responses that are associated with the defense response of pea.
Variation in DNA-Specific Agents Action
The modes of action of selected compounds on DNA in vitro are defined in Table 1 and their relative effects on the accumulations of the phytoalexin, pisatin are presented in Table 2. A large number of cyclic molecules have the potential to intercalate between the base pairs of DNA. Many derivatives of acridine have been shown to positively induce pisatin production (Hadwiger, 1972a). This action is shared by the compounds with planar three ring structures (e.g., in ethidium bromide). A positively charged nitrogen in the azole ring or on the side chain presumably attracts the negatively charged phosphate groups of DNA (Schwochau and Hadwiger, 1968). Unfortunately, many medically important compounds, including antihistamines, antimalarials, decongestants, chelators, etc., are also capable of intercalating DNA (Hadwiger, 1972a). Not all DNA intercalators are cytotoxic. Some small molecule drugs have now been shown to have a wide range of biological activities: i.e., vitamins, hormones, hormone antagonists, antipsychotics, antidepressants, and antihistamines. The DNA helix is flexible and can be readily wound or unwound. When unwound cavities appear between the base pairs, the space approximates that of small molecule natural products. For example, the shape of the steroid hormone estradiol is a good fit between base pairs of unwound DNA (Hendry et al., 2007). The plant hormone gibberellic acid fits into the intercalation site 5′-dTdG-3′ 5′-dTdA-3′ (Witham et al., 1978). Other natural products, such as caffeine, vitamin D and riboflavin, fit into unwound DNA (Hendry et al., 1977). The specific sequences in DNA into which ligands best intercalated were found in the consensus sequences of genes activated by nuclear receptors, indicating that intercalation was central to their mode of action.
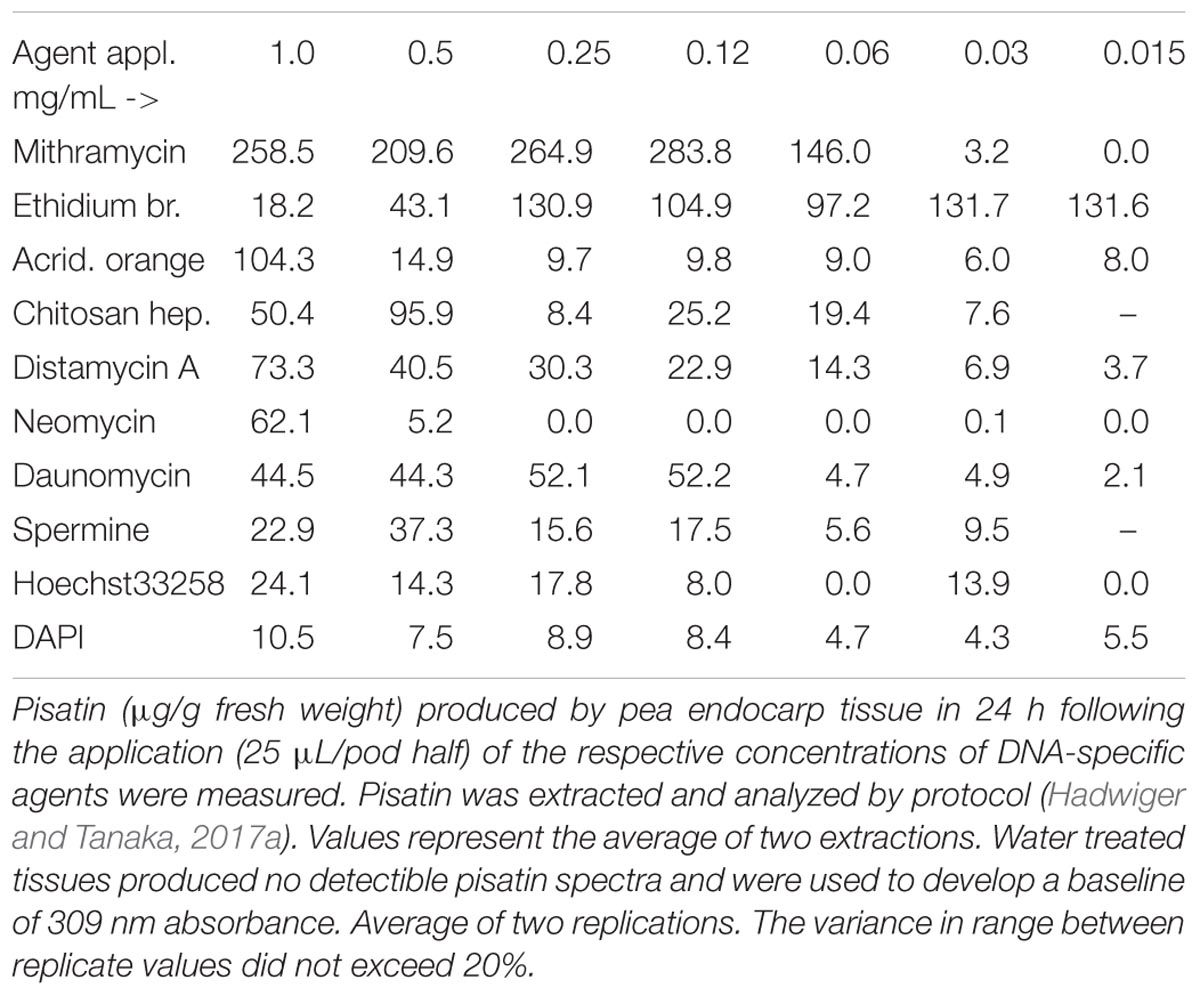
TABLE 2. Pisatin production in pea endocarp tissue 24 h after treatment with DNA-specific compounds, capable of DNA intercalation or minor groove localization.
The intercalator modes of action are also likely to occur by altering the DNA torsions (unwinding) that can affect the transcription of some genes (Ma et al., 2013). The mechanics by which transcription is affected by DNA intercalators have been investigated. Although there are multiple interpretations, the following actions and conditions are well understood (Pruss and Drlica, 1989):
The packaging of DNA into the cell is assisted by histones and supercoiling, often causing negative supercoiling of the DNA. The supercoiling of the DNA in advance of the polymerase transcription complex must be removed, and the polymerase action itself is accompanied by supercoiling (Figure 4). As the region in front of the polymerase is unwound, there is compensatory positive supercoiling well ahead of the complex (Gilbert and Allan, 2014). Alternately, the DNA behind the complex is rewound with the development of compensating negative supercoils. DNA intercalators can twist DNA, thus affecting the supercoiling independent of the aid from a protein. Topoisomerases and DNA gyrases can relieve some of the stress. Some SWI/SNF genes code for gyrase enzymes. Additionally, SWI/SNF complexes can cause a bulge mechanism that may cause the dissociation of DNA at the edge of the nucleosome, followed by re-association of the DNA inside the nucleosome (Tang et al., 2010). Such complexes can function as tumor suppressors.
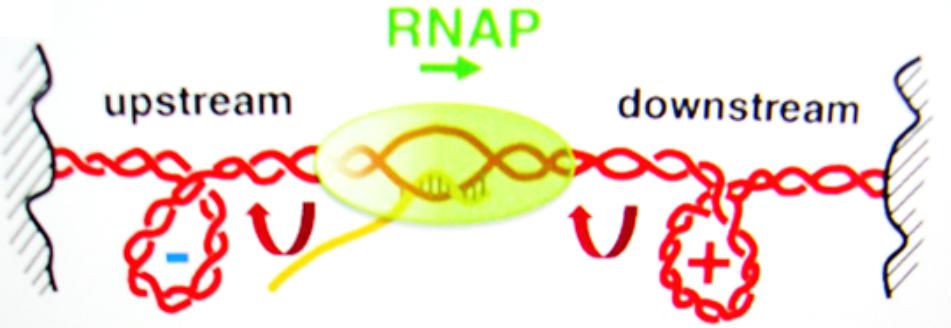
FIGURE 4. The supercoiling of DNA is a removable barrier to the RNA polymerase complex (RNAP) transcription of genes. Reproduced with copyright permission (Ma et al., 2013).
As indicated earlier, extensive research on the DNA-specific intercalator actinomycin D indicates the diversity of action in vivo. Actinomycin D was found to be a super inducer of the synthesis of some animal genes (Chatterjee et al., 1979). In an early screening of intercalators, we found actinomycin D and other intercalators to be strong activators of plant defense responses (Schwochau and Hadwiger, 1968). Actinomycin D action in pea tissue, in contrast to mRNA inhibition, has been explained in various ways, such as suppression of the production of transcription factors or suppression of RNase activity. However, as a general conclusion of the action of DNA intercalators in pea tissue, we propose that the torsional effect on the DNA helix is a major factor in promoting transcription, as indicated in cancer research (Teves and Henikoff, 2014). Additionally, because of the many similarities of plant and animal chromatin structure and the effect of such DNA-specific compounds on plant chromatin, concentrations below the lethal action are likely acting on varying levels of transcription enhancement and thus on the differential activation of genes.
Substitution of DNA Bases and Helixes
Externally applied base analogs, such as 5-bromo deoxyuridine and 5-iododeoxyuridine, can activate the pisatin pathway in pea (Sander and Hadwiger, 1979). The base analog must be incorporated into pea DNA before any induction occurs. The nuclei undergo condensation just prior to the detection of the induced increase in phenylalanine ammonia lyase (PAL) activity. The mode of action involved the insertion of a base analog into the DNA, and the transcriptional increase was likely due to a change in the DNA helical structure during the removal of the aberrant abduct.
DNA Cross-Linking Agents
The alteration of the DNA helix that developed from a cross-linked psoralen activates phytoalexin (pisatin) production in pea endocarp tissue and is likely triggered during the DNA repair process that would remove this aberration (Parsons and Hadwiger, 1998). In humans, such cross-linking may be general along the genome as the associated symptoms are extensive. The effect of cross-linking DNA by psoralen compounds was first reported as an environmental hazard on celery harvesters. Psoralen compounds develop when celery plants are infected by the fungal pathogen Sclerotinia sclerotiorum (Floss et al., 1969). The hazardous effect on workers occurred when the psoralen entered the skin of their hands. The additional environmental action came from the UV content of sunlight that enables the compound to cross-link DNA strands. The psoralen is activated to form covalent bonds. The subsequent human symptom was tumorous growth on the workers’ hands. Psoralen compounds have also been shown to activate multiple plant defense responses in pea endocarp tissue (Parsons and Hadwiger, 1998). Prior to the development of other molecular assays, the psoralen cross-linkage was also utilized to locate DNA segments within open reading frames of the PR genes, which provided evidence that the DNA abduct had occurred in the vicinity of the defense gene. This site-specific adduct was detected on southern blotting analyses run on alkaline gels (the cross-link of DNA slowed the electrophoretic of cross-linked segments and not the migration of alkali separated DNA segments). The precise effect of crosslinking in activating the pea defense response is not known; however, the removal of this adduct, such as the removal of other adducts, renders the DNA free to unwind or modify as the repair is undertaken.
The DNA within chromatin can be negatively or positively helically coiled; thus, the presence of these supercoils can be obstructive to the progression of the RNA polymerase complex. The progression of this complex along the DNA molecule during transcription requires an absence of obstruction, as well as a separation of the strands, as shown in the drawing (Figure 5). The loosening of the nucleosome structure by a single strand cleaving DNase can occur both by freeing a single strand and exposing DNA for enzyme access and by allowing a release of the negative helix of the supercoiled DNA.
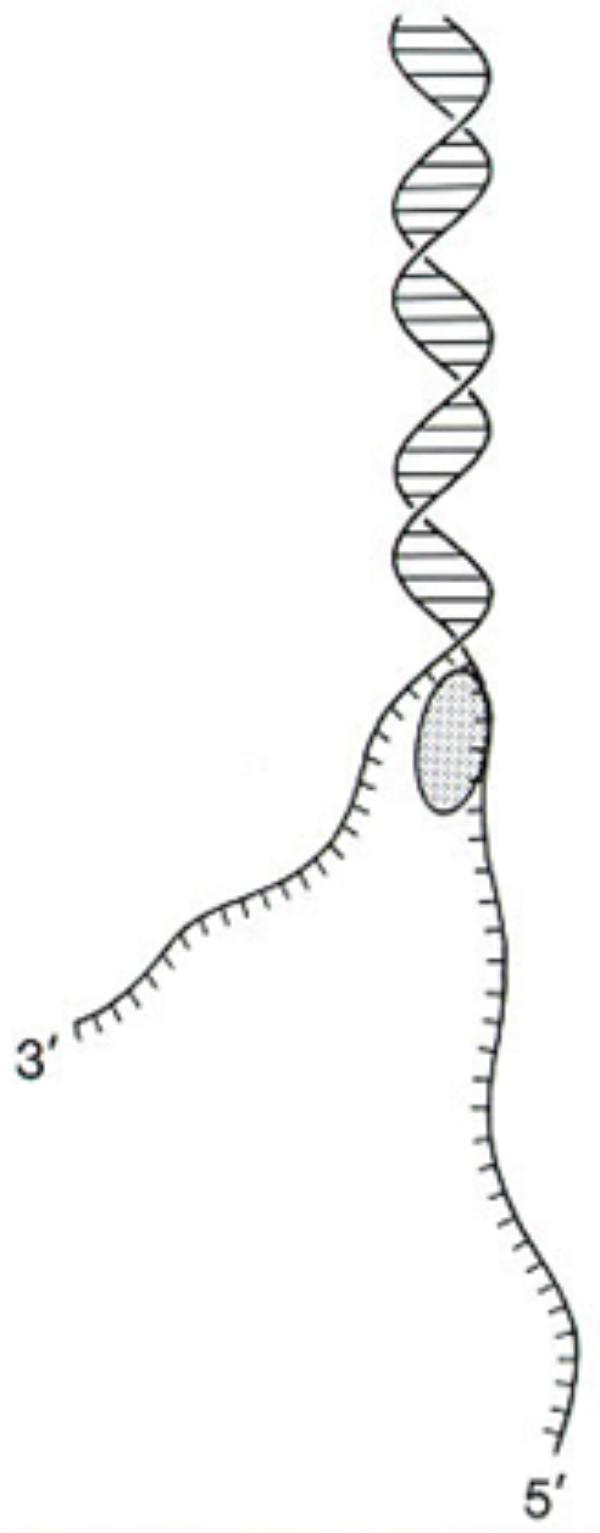
FIGURE 5. DNA single strands can develop during DNA repair or by DNase I-like or gyrase-like enzymes. Reproduced from a previous publication (Neigeborn and Carlson, 1984).
Biotic DNA Targeting Agents
Fungus-related DNase function, in support of the growing fungal mycelium, was thought to occur as a means to break down DNA as a nutritional source of nucleic acid bases. A fungal DNase capable of cleaving single DNA strands is synthesized in most fungi (Hadwiger and Polashock, 2013) with an N-terminal signal peptide that enables it to cross membranes (Klosterman et al., 2001). However, as an inadvertent occurrence, the immediate plant defense response slows fungal growth. The resultant DNase accumulation that normally occurs in old mycelia for the purpose of digesting and recovering DNA components for reuse now occurs in the hyphal tip. It appears to accumulate close to the growing tip and effectively cleaves the single strands of the DNA that must remain intact for cell division. In the absence of a functional nucleus, fungal growth is terminated (Hadwiger, 2015c). All of the genomes of fungi sequenced thus far contain the DNA coding sequence for this mitochondrial DNase (Hadwiger and Polashock, 2013). The universality of the DNA strand cleaving function in eliciting a defense response is likely a major contribution to the development of “nonhost resistance” that protects plants from all but their true pathogens. The growth of a true pathogen is not so severely suppressed by the pea plant defense response, and mycelial tips can retain some viable nuclei. Mycelia with viable nuclei can continue growth on the plant tissue (Hadwiger, 2015c), as the major defense response subsides.
Naturally occurring proteins/peptides and synthesized polymers rich in the basic amino acids arginine (A) and lysine (K) were found to be capable of producing pisatin in peas. Protamine, histones, spermidine, spermine and some basic enzyme protein domains present in RNase and snake venom elicit pisatin production. All are rich in basic amino acids or basic charges (Hadwiger and Schwochau, 1970). The synthetic peptides poly-L-lysine and poly-L-arginine are elicitors but are unlikely to be natural pisatin elicitors. However, these basic peptides provide clues regarding the potential of natural protein segments rich in arginine or lysine to act in this capacity (Brunner et al., 2002). These synthetic proteins (peptides) can be mimicked by carbohydrates that are also strongly positively charged. Chitosan is a basic polymer of glucosamine and is a signaling component in the pea/Fusarium interaction (Hadwiger et al., 1981; Hadwiger, 2015b). Chitosan shares the DNA affinity property of basic peptides and can activate the same responses in pea endocarp tissue as the bean pathogen, F. solani f. sp. phaseoli (Fsph) (Loschke et al., 1983). Furthermore, a large group of microbes contain chitin (polymers of β-linked N-acetyl glucosamine) and chitosan (polymers of β linked glucosamine). Chitosan heptamers of seven sugars or more represent optimal-sized elicitors (Kendra et al., 1989). Although chitin structure has similarities to chitosan, the added acetyl group negates the positive charge of the amino groups, rendering it less effective as an elicitor unless there is a chitin receptor to carry forth the signal (Hadwiger and Chang, 2015). A computer analysis indicates that the chitosan heptamer (seven glucosamine residues) fits into the minor groove of the DNA molecule (Hadwiger et al., 1989).
Other Minor Groove Targeting Agents
The minor groove of DNA is a target of anticancer drugs (Figure 6). These include distamycin A and mithramycin (Figure 7), and Hoechst 33258 (pibenzimol), 4′,6-diamidino-2-phenylindole (DAPI) and neptropsin, which are topoisomerase poisons or helicase inhibitors, preferring an AT-tract duplex DNA (Varqiu et al., 2008). Chitosan has had only limited evaluations as an anticancer agent; however, chitosan, actinomycin D, and camptothecin all activate the production of p53, a tumor-suppressing protein, in the mouse pre-neoplastic mammary cell line CL-S1 (Hadwiger et al., 1997). All three agents activate defense genes in pea (Isaac et al., 2009), and although each agent is capable of altering chromatin structure within the nucleosome (Figure 8), they reportedly have differing specific modes of action. The concentration of positive charges on chitosan may compete with pea histones that function to compact the cellular DNA in the nucleus (Hadwiger, 2008; Isaac et al., 2009). Chitosan’s action on chromatin is able to loosen the compaction of the nucleosome structure, allowing stalled genes to resume transcription (Hadwiger, 2015a). Messenger RNA from chitosan-treated pea tissue when transcribed in an in vitro protein synthesis system also produces protein 2-D patterns closely related to mRNA from pea tissue that responds to a bean pathogen, Fusarium solani f. sp. phaseoli (Fsph) (Loschke et al., 1983).
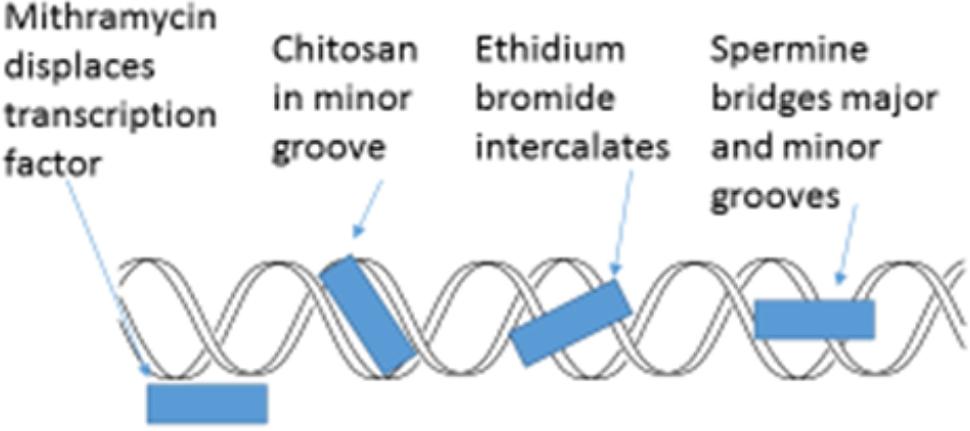
FIGURE 6. Schematic representation of four major actions on the helical condition of DNA. Mithramycin as a minor groove targeting agent displaces the Sp2 transcription factor (Neigeborn and Carlson, 1984; Miskovic et al., 2013). Chitosan resides in the DNA minor groove (Hadwiger et al., 1989). Ethidium bromide intercalates between DNA base pairs (Lenglet and David-Cordonnier, 2010). Spermine can both enter the minor groove and the major groove in a manner that enables the bridging of both (Bryson and Greenall, 2000).
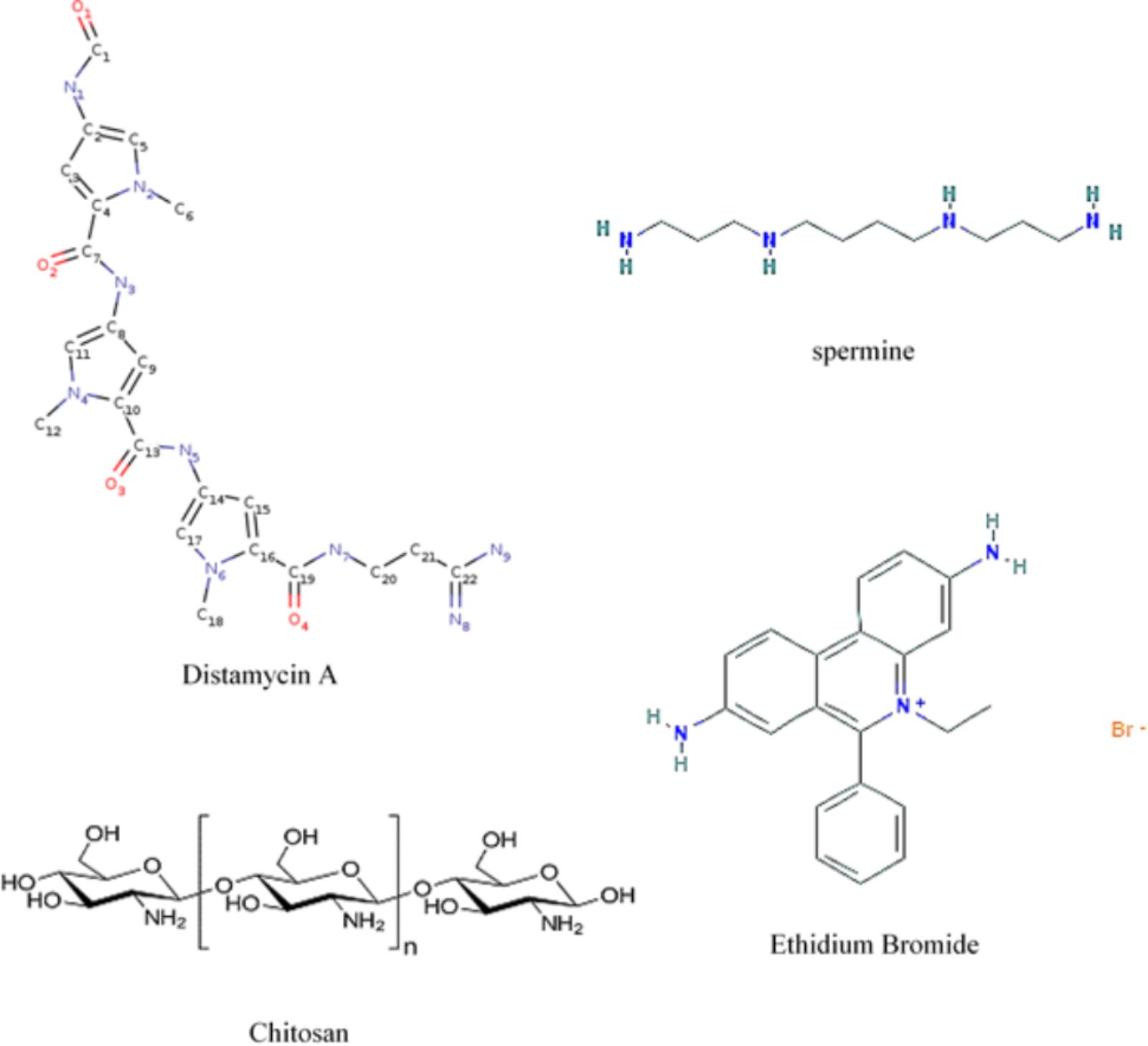
FIGURE 7. Formulas showing molecular locations of nitrogens when present and planar ring structures. Distamycin A, spermine, and ethidium bromide (by permission from NIH – PubChem and Cayman Chemical) and chitosan.
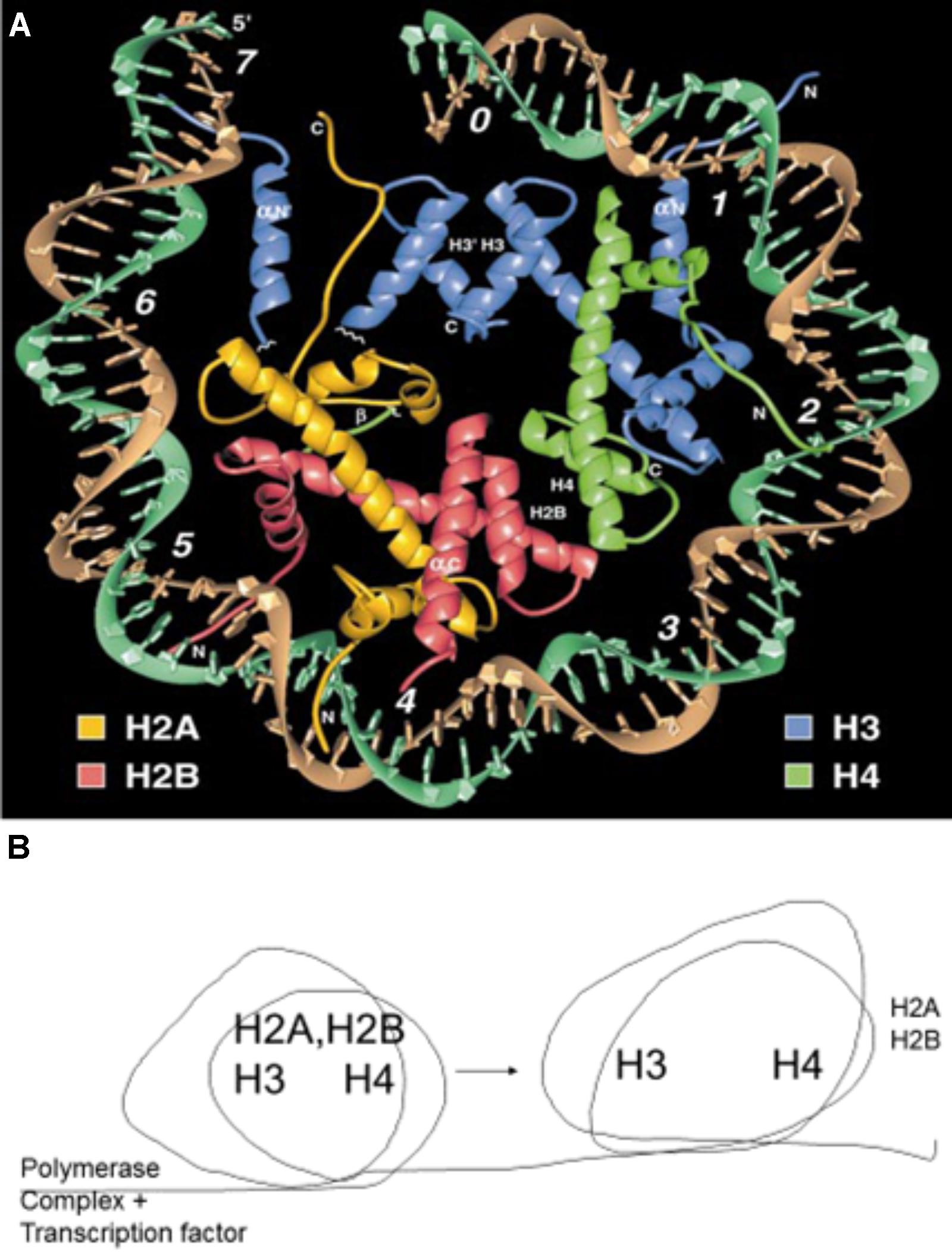
FIGURE 8. (A) Crystal structure of a nucleosome. The DNA helix circles the attached histones, H2A, H2B, H3, and H4. Reproduced with copyright permission (Luger et al., 1997). (B) Schematic description of the nucleosome rolling action that results in the temporary removal of histones H2A and H2B (Gerasimova et al., 2016). This temporary removal is essential for the RNA polymerase complex to transcribe genes and the histones are reassembled following the successful passing of the complex.
DNA Groove-Binding Architectural Proteins
Chromatin architectural proteins is a major group of nuclear proteins that impact chromatin structure and function. TATA-box-containing protein and high mobility group HMG A protein complexes with DNA can have sequence-specific recognition (Bewley et al., 1998; Klosterman and Hadwiger, 2002). Both bind in the minor groove of DNA and make conformational changes in the DNA. Both occur widely in eukaryotic organisms, including plants. Some chemicals that binds the minor groove of DNA such the bis-benzimidazoles (Hoechst 33258) and DAPI have been used for cancer therapy (Baraldi et al., 2004). These compounds interact physically with DNA and cause reversible inhibition of DNA-dependent functions. Hoechst 33258 (but not DAPI) was found to be an elicitor (weak) of phytoalexin production in pea tissue. These two compounds, along with chitosan, reportedly enter the DNA minor groove (Baraldi et al., 2004). The strong induction by the chitosan preparation (Table 1) with heptamer-sized polymers may benefit from molecular lengths large enough to outperform the smaller spermine and Hoechst 33258 compounds in initiating the pisatin induction, using this single parameter for comparison. Given information on the mechanism of action of DNA-specific abiotic compounds in altering the DNA within chromatin and activating a defense response, the information should be useful to understand the mechanism of the biotic DNA-specific action of chitosan that also occurs in the minor groove of DNA. Similarly, a comparison of the multiple compounds utilized in cancer therapy would be useful in determining which groove-binding abiotic compound was most active or inactive in inducing collateral gene activation responses. The pisatin assay was utilized to evaluate the optimal accumulations that could be generated by the compounds listed in Table 2.
In calf thymus tissue, the non-histone proteins HMG 1 and HMG 2 are capable of unwinding the DNA double helix. Pea tissue also possesses a HMG A protein (Klosterman et al., 2003) that is reduced in the chromatin material during the initiation of the pea defense response (Isaac et al., 2009). HMG A is considered an architectural transcription factor with a wide array of actions in both stabilizing and altering chromatin structure. Its action is reportedly influenced by the associated salt solution of the assay (Javaherian and Sadeghi, 1979).
There is an alteration of nuclear structure that occurs in the early minutes of pea/Fusarium solani formae species interactions. Interactions at 5 h were globally more intense in the compatible interaction than in the resistance reaction (Isaac et al., 2009). Western analyses, mass spectrometry, and [32P] techniques were used to follow the disappearance of the architectural transcription factor HMG A and histones H2A/H2B. Of more specific interest, at 5 h, these nuclear proteins were also observed to be less abundantly complexed in the vicinity of two PR genes, DRR206 and the β-glucanase gene, utilizing chromatin immunoprecipitation analyses. There is an early ubiquitination of HMG A and some histones (Isaac et al., 2009). This suggests that the DNA breaks and the removal of nuclear proteins may assist the progression of stalled genes that had previously been obstructed. Some of the specific defense genes become activated as nuclear proteins (histone/HMG A) are removed.
DNA Damage and Repair Aspects From Cancer Therapeutic Research
DNA damage is an early event the pea endocarp/fungal pathogen interaction (Tanaka and Hadwiger, 2017) and occurs following other DNA inducing treatments. DNA damage is also a linking mechanism in animal immunity development (Brzostek-Racine et al., 2011; Nakad and Schumacher, 2016). The DNA damage activates immune signaling through molecular and cellular pathways and drives chronic inflammation in humans. The DNA damage response can also induce interferon production.
Some of the chemotherapy-induced DNA damage responses include genes for DNA repair (Woods and Turchi, 2013). Ataxia telangiectasia mutated (ATM) kinases are activated and phosphorylate many substrates, including proteins involved in checkpoint activation, DNA replication and DNA damage repair. It remains uncertain whether ATM binds directly to DNA. As indicated above, the transcription complex movement is facilitated in the absence of helical blocks and histone attachments to the DNA. Additional effects on the helical structure are possible by non-histone proteins such as HMG A (Isaac et al., 2009), which can unwind the double helix, and by nucleases such as ribonuclease and gyrases, which can cause destabilizing effects on DNA helical structure (Felsenfeld et al., 1963). The development of anticancer drugs needs to take these direct actions on DNA into consideration. Alternately, the defense response of pea tissue is strongly affected by these additional proteins. Ribonuclease A strongly induces the accumulation of the pea phytoalexin pisatin. This activity is diminished by half if the ribonuclease is autoclaved prior to application, and reportedly, ribonuclease S loses half of its pisatin-inducing potential if only the non-enzymatic portion of the “S” molecule is applied, indicating that the action is a combination of enzyme activity and non-enzymatic cationic proteins. Additionally, the digestion of the RNA content may have a functional role in chromatin structural change. A number of other basic proteins also induce pisatin production without any obvious nuclease activity (Hadwiger et al., 1974).
Reactive Oxygen Species (ROS): Potential Signals in Cancer and Disease Resistance Via DNA Damage
Reactive oxygen species (ROS) production is a mechanism shared by all non-surgical therapeutic approaches for cancers, including chemotherapy radiotherapy and photodynamic therapy. ROS are usually increased in cancer cells due to oncogene activation and are involved in the initiation, progression and metastasis of cancers. Thus, ROS are considered oncogenic (Wang and Yi, 2008). Oxidative stress has a significant impact on the progression of cancer and other human pathologies. It has a global influence on chromatin structure, mediating a number of cellular changes, including gene expression. This makes the targeting of oxidative stress pathways important in the control of cancer (Kreuz and Fischle, 2016). ROS in eukaryotic tissue cause multiple DNA base changes, such as from thymine to thymine glycol (Dizdaroglu and Jaruga, 2012) and 5-hydroxy methyl-2-deoxy uridine (Chaung and Boorstein, 1997). Most of these changes cause mismatches during DNA replication, leading to mutagenesis. ROS are capable of directly altering plant DNA. Application of hydrogen peroxide to pea endocarp tissue increases DNA fragmentation and activates defense genes (PR genes) (Tanaka and Hadwiger, 2017). Although direct effects on the DNA are detectable in pea, it is likely that other damage to the pea chromatin is involved and that the induction of repair responses may occur as it does in animals. ROS mediate a systemic signal network for developing plant immunity (Alvarez et al., 1998). A part of this network is the DNA damage inflicted by ROS. In pea tissue, this damage is associated with the post-treatment period during which PR genes are activated (Tanaka and Hadwiger, 2017).
Reactive oxygen species are induced in mammalian tissue as an antimicrobial defense. Their importance is based on the observation that individuals with deficiencies in generating ROS are highly susceptible to infection by a broad range of microbes. A likely mode of defense occurs following damage to mitochondrial DNA. Interestingly, DNA repair mechanisms were required to resist killing by ROS. Although ROS play a role, direct killing may not be the key mechanism. ROS may affect ROS-dependent signaling controls, such cytokine production (Deffert et al., 2014). Excessive ROS can damage cellular proteins, lipids and DNA, leading to fatal lesions in cells that contribute to carcinogenesis. Low levels of ROS facilitate cancer cell survival. High levels of ROS can suppress tumor growth through the sustained activation of cell-cycle inhibitors and the induction of cell death (Ramsey and Sharpless, 2006). A cancer cell can die in three ways: apoptosis, necrosis and autophagy. The cytotoxic nature of ROS is the driving force behind apoptosis, but with even higher amounts, ROS can result in both apoptosis and necrosis, a form of uncontrolled cell death in cancer cells (Hampton and Orrenius, 1997).
Other Potential Signals in Disease Resistance Via DNA Damage
It has been reported other potential signals for nonhost disease resistance via DNA damage as shown in Figure 1 (Yan et al., 2013; Hadwiger and Tanaka, 2015, 2017b). Another potential signal for speculation is damage-associated molecular patterns (DAMPs; Tanaka et al., 2014). For examples, extracellular DNA that trigger plant immunity are in addition to the hypothesized PAMPs discussed above. These signals are unique in that only self-DNA fragments (Barbero et al., 2016; Duran-Flores and Heil, 2018) and are active and maybe differ from the PAMPs in that the likely target is host DNA rather than a pattern recognition receptor located in the vicinity of the cell membrane. The specificity of the self-DNA requirement and the rapidity of the response were demonstrated by developing immunity in common bean with extracellular DNA from other same species plants and unsuccessfully from DNA from unrelated species. The fragments all less than 700 bp suggest that the molecules reach host DNA and that like other introduced DNA can quickly find homologous regions in the genome. The mechanism of action has not been unequivocally determined (Mazzoleni et al., 2015a,b) but the presence of a homologous third strand fragment is likely destructive or at least competitive to chromatin organization in the homologous region. As indicated previously seemingly minor changes in chromatin organization can effect transcription. Interestingly, these externally applied DNAs can also affect plant growth.
Essence of the Compiled Information on DNA Damage in Disease Resistance in Plants and Cancer Development in Animals
Multiple black boxes of unknown regulatory components are prevalent within chromatin (Dekker et al., 2013). Such variations in chromatin structures were demonstrated in the 1950s by a cytologist looking at the bands within giant chromosomes of Drosophila salivary glands. Regions sensitive to treatments with hormones, DNA intercalators, heat, etc. were observed to puff out from certain bands of the chromatin within the giant chromosome. The multi-action damage of ROS to plant nuclei activating defense responses may be more global in comparison to activation by actinomycin D, which recognizes specific DNA sequences. This actinomycin-DNA specificity results in more direct action on sensitive areas of the chromatin (Lewis et al., 1975), as visualized in the puffing effects on giant chromosomes of Drosophila (Watson et al., 1987). In general, actinomycin D prefers GpC regions. It binds to DNA by intercalating its phenoxazine ring at a GpC step such that the two cyclic pentapeptides of the drug area are located in the DNA minor groove (Sobell and Jain, 1972). As the base sequence becomes more deviant, there can be more radical changes. For example, actinomycin D induces nucleotide flipping out, sharp bends and a left-handed twist in CGG triplet repeats. Heat denaturation, circular dichroism and surface plasmon resonance analyses indicate that adjacent GpC sequences flanking a G:G mismatch are preferred actinomycin D binding sites (Lo et al., 2013). The detection of sensitive regions within chromatin regions of pea chromosomes has been defined genetically as QTLs. The mapping of these regions in pea detects some of the induced defense genes residing within QTLs and thus may characterize special features of the pea chromosome (Pilet-Nayel et al., 2002).
Condensed State of DNA in Plant Chromatin
Nucleosomes help condense the almost 1-m length of DNA within a pea cell, and similarly in many other eukaryotic cells, into the small volume of the nucleus of ∼10 microns in diameter (Hadwiger and Adams, 1978). This fete is accomplished in part in co-operation with nucleosomes. Each turn of the nuclear DNA strand may contain six nucleosomes, as shown in the Figure 8A, each composed of two molecules of the following histone molecules: H3, H4, H2A, and H2B. This structure is stable because of the electrostatic interaction between the negatively charged DNA and the basic histones (Yaniv, 2014).
The presence of histones and the condensed structure of chromatin restricts the access of specific proteins to DNA sequences except when appropriate for transcription, repair, etc. (Petesch and Lis, 2008; Pang et al., 2013; Mao et al., 2014). Both RNA and DNA polymerases must separate the strands of DNA. This can be accomplished enzymatically in eukaryotic cells by helicases. Helicase motifs have been found in genetic complexes (SNF genes in yeast that act in control of certain other genes) (Neigeborn and Carlson, 1984). Re-expression of the helicase in human cells previously lacking a helicase strongly increased the expression of a glucocorticoid hormone receptor (GR) (Muchardt and Yaniv, 1993). These examples suggest a helicase function in DNA strand separation that assists the transcription process.
The broad implication of complexes containing SNF genes in cancer is that the loss or change in this activity can result in a multitude of re-regulated cellular programs affecting cell survival and cell death of malignant transformation and may relate to the strand separating function of helicases. In plants, the re-regulation of genes due to abnormal insults to organized chromatin via pathogen invasion can also affect transcription patterns for cell viability and cell death. Fortunately, there are windows in this array in which previously suppressed genes can become beneficial to the immune response of the plant tissue.
The signaling of such new gene expression levels with respect to time after inoculation of an “inappropriate” pathogen has been defined as what is termed a nonhost resistance response (Hadwiger, 2015c). This early response is almost universally observed as more excessive than the plant’s response to pathogens considered to be in the range of that particular plant species. Plants have obviously diverted evolutionarily from animal systems; however, there is a conservation of similarity in the transcriptional machinery within plant and animal cells. It was recognized early that the amino acid sequences of an array of histone proteins in both plant and animal cells were highly conserved. Additionally, there is some similarity in certain transcription factors such as HMG A, which is regarded as an architectural transcription factor with AT-hook motives within the protein specific to AT-rich regions of the DNA (Klosterman et al., 2000; Klosterman et al., 2003; Reeves, 2010). HMG A is retained in both plant and animal systems. Plants and animals also have SNF/SWI-like complexes that affect transcription (Bezhani et al., 2007; Jerzmanowski, 2007; Sarnowska et al., 2016).
A portion of the pea and cancer cell chromatin contain genes with various states of activity ranging from open expression to “stalled” (Nelson et al., 2007). Stalled genes have obstructions that are related to the state of transcription factors, DNA helical obstructions and nuclear protein content. Therefore, it is likely that chromatin modifications from agents with slightly different modes of action can assume multiple changes increasing (or suppressing) gene expression. Furthermore, because the agents can possess differing base-sequence preferences, their proximity to the genes expressed will also be an influencing factor. Conformational states of chromatin in the vicinity of the PR genes may effect transcription enhancement via nucleosome disassembly or histone H2A/H2B releases similar to that found in other eukaryotic systems (Adkins and Tyler, 2006; Weake and Workman, 2008).
In plant cells, the production of pisatin can occur by the amplification of a secondary metabolic pathway that depends on increases in one or more enzymes. The induction of pisatin is usually in synchronization with the activity of PR genes, and both entities possess anti-fungal properties, thus implicating regulatory enhancements in a group of plant genes. The same DNA-specific agents confront similar chromatin structures in animal cancer cells, but the medicinal objectives are intended to negatively affect the viability of actively dividing cancer cells. These negative properties of DNA-specific agents are often acquired with high agent concentrations, it is inevitable that non-targeted peripheral areas will receive diluted concentrations. The results obtained in plant tissue suggest that there is the potential for the lower concentrations of anti-cancer agents to cause a different array of effects.
Perspective Summarization
Following DNA damage within human or plant cells, there is an alteration of the repressed states of some genes encumbered within the respective nuclei. The damage results in actions, such as defense gene activation in plants and suppression of growth in cancer cells with eventual side effects, including programmed cell death (apoptosis). Many of the effects can be duplicated by targeting nuclear DNA by eliciting agents with varying modes of action, such as through DNA intercalation, DNA cleavage, base substitution, nuclear protein modification, etc., that elicit varying responses. This targeting of the sensitive chromatin regions by chemically different agents can produce similar transcriptional changes to those in real biological systems. This abiotic probing provides insight into the biotic changes (Figure 9) experienced by the nucleosomes of the nuclear chromatin of both plant and animal cells. Because of the highly conserved components of chromatin in plants and animal cells, the mechanisms of these changes can have implications that are useful in understanding both systems.
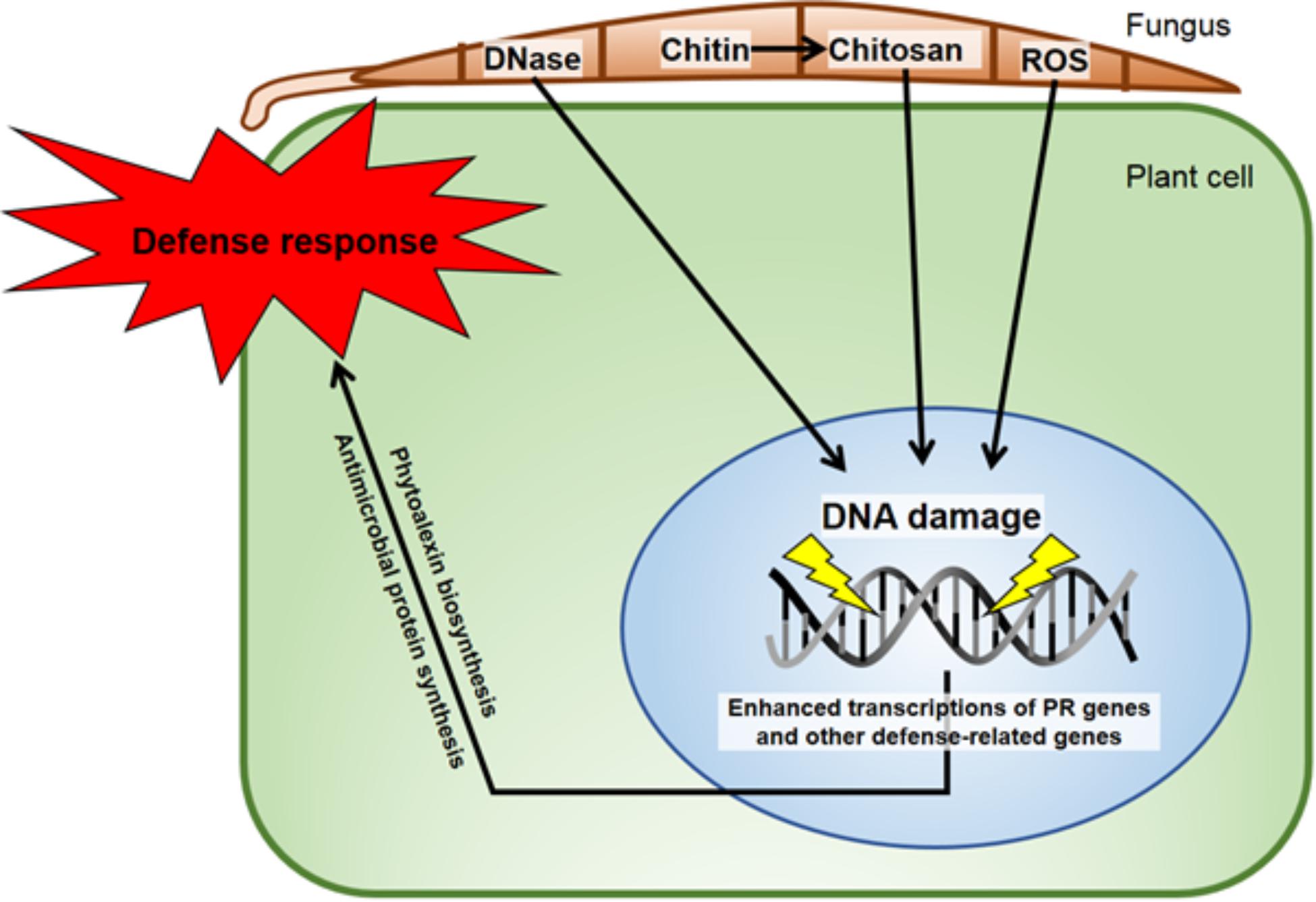
FIGURE 9. Diagram of some DNA-direct effects of biological entities shown capable of activating disease resistance responses in plants. The minor groove-localizing chitosan heptamer is released from the fungus by the plant chitinase and the fungal chitin deacetylase enzymes (Kendra et al., 1989). The DNA torsion/helicity can be affected by small molecule binding/intercalation and single strand cleavage by Fusarium solani f. sp. phaseoli (Fsph)-DNase (Hadwiger and Polashock, 2013). ROS released in the interaction (Tanaka and Hadwiger, 2017) have multiple ways to alter nuclear DNA. The isoflavonoid phytoalexins, e.g., pisatin (Hadwiger, 2015a), and defensin (Almeida et al., 2006) are directly anti-fungal. Fsph DNase accumulation within Fsph terminates fungal growth (Hadwiger, 2015c). PR genes such as DRR49 codes for RNase and DRR230 codes for defensin. Chitinase and β-glucanase that digest fungal chitin and glucan polymers, respectively (Kendra et al., 1989) are present in healthy tissue and increase following fungal challenge. All proteins that transient the host–parasite interface are produced with a N-terminal signal peptide for transfer through membranes (Hadwiger, 2009).
Transcription data over the decades have implicated DNA torsional changes as central to the progression of RNA polymerase complexes through gene open reading frames (Ma et al., 2013). These enhancements of newly expressed genes must remove the barricades of helical stress and nucleosome condensation that restricts the ORF read through RNA polymerase and the subsequent expression of defense and DNA repair genes. A DNA-intercalating scenario may be to insert into proximal DNA, reversing the negatively supercoiled or dispersing nucleosome structure. Another action may be the ubiquitination of histones H2A/H2B and removal from the area downstream from the RNA polymerase complex (Figure 8B). The enhancement of the defense responses in plants can occur in a similar manner. In the latter case, it is the components of the response, the antifungal compounds, that enable resistance. In some plant/bacterial interactions, the complete killing of cells surrounding the lesion is beneficial to resistance as well. The lesson available from the plant responses for cancer therapy is that elicitor-initiated gene activations occurring with low-level treatments in plants may occur randomly at the fringes of the high-level anticancer treatments and may activate genes associated with adverse side effects.
The plant responses that develop from the large number of eliciting agents tested on pea endocarp tissue (Figures 1, Figures 9) indicate that cellular chromatin structural changes relate to the presented chemistry of the agent without respect for what a pharmaceutical company designates as the cellular target. That is, the agent may be designated an antimalarial, antidepressant drug, etc.; however, if there are potential intercalating rings and positive charges exposed, the agent will likely localize next to the negative charges of the DNA and the resulting transcriptional changes will occur based on the chemistry of the interaction.
Author Contributions
LH and KT wrote the manuscript.
Funding
This work was supported by Northwest Potato Research Consortium, NSF (IOS-1557813), USDA NIFA Hatch project (WNP00008, WNP00833, and WNP03847), and CSANR BIOAg Grant Program. PPNS 0759, Department of Plant Pathology, College of Agriculture, Human and Natural Resource Sciences, Agricultural Research Center, Washington State University, Pullman, WA, United States.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Special thanks to Natalia Moroz and Mike Adams for reviewing the manuscript.
References
Adkins, M. W., and Tyler, J. K. (2006). Transcriptional activators are dispensable for transcription in the absence of spt6-mediated chromatin reassembly of promoter regions. Mol. Cell 21, 405–416. doi: 10.1016/j.molcel.2005.12.010
Almeida, M. S., Cabral, K. M., Zingali, R. B., and Kurtenbach, E. (2006). Characterization of two novel defense peptides from pea (Pisum sativum) seeds. Arch. Biochem. Biophys. 378, 278–286. doi: 10.1006/abbi.2000.1824
Alvarez, M. E., Penneli, R. I., Meijer, P.-J., Ishikawa, A., Dixon, R. A., and Lamb, C. (1998). Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92, 773–784. doi: 10.1016/S0092-8674(00)81405-1
Antolin-Llovera, M., Petutsching, E. K., Ried, M. K., Lipka, V., Nurnberger, T., Robatzek, S., et al. (2014). Knowing your friends. New Phytol. 204, 791–802.
Baraldi, P. G., Bovero, A., Fruttarolo, F., Preti, D., Tabrizi, M. A., Pavani, M. G., et al. (2004). DNA minor groove binders as potential antitumor and antimicrobial agents. Med. Res. Rev. 24, 475–528. doi: 10.1002/med.20000
Barbero, F., Guglielmotto, M., Capuzzo, A., and Maffei, M. E. (2016). Extracellular Self-DNA (esDNA), but not heterologous plant or insect DNA (etDNA), induces plasma membrane depolarization and calcium signaling in Lima bean (Phaseolus lunatus) and maize (Zea mays). Int. J. Mol. Sci. 17:1659. doi: 10.3390/ijms17101659
Barcelo, F., Ortiz-Lombardia, M., Martorell, M., Oliver, M., Mendez, C., Salas, J. A., et al. (2010). DNA binding characteristics of mithramycin and chromomycin analogues obtained by combinatorial biosynthesis. Biochemistry 49, 10543–10552. doi: 10.1021/bi101398s
Beato, M., Seifart, K. H., and Sekeris, C. E. (1970). The effect of cortisol on the binding of actinomycin D to the on the template activity of isolated rat liver chromatin. Arch. Biochem. Biophys. 138, 272–284. doi: 10.1016/0003-9861(70)90308-5
Bewley, C. A., Gronenborn, A. M., and Clove, G. M. (1998). Minor groove-binding architectural proteins: structure, function and DNA recognition. Ann. Rev. Biophys. Biomol. Struct. 27, 105–131. doi: 10.1146/annurev.biophys.27.1.105
Bezhani, S., Hershman, L. C., Wagner, J. D., Kennedy, J. F., Kwon, C. S., Pfluger, J., et al. (2007). Unique, shared, and redundant roles for the Arabiddopsis SWI/SNF chromatin remodeling ATPases BRAHMA and SPLAYED. Plant Cell 19, 403–416. doi: 10.1105/tpc.106.048272
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Brunner, F., Rosahl, S., Lee, J., Rudd, J. J., Geiler, C., Kauppinen, S., et al. (2002). Pep-13, a plant defense-inducing pathogen-associated pattern from Phytophthora transglutaminases. EMBO J. 21, 6681–6688. doi: 10.1093/emboj/cdf667
Bryson, K., and Greenall, R. J. (2000). Binding sites of the polymines putrescine, cadaverine, spermidine, and spermine on A- and B-DNA located by simulated annealing. J. Biomol. Struct. Dyn. 18, 393–412. doi: 10.11080/07391102.200010506676
Brzostek-Racine, L., Gordon, C., Van Scoy, S., and Reich, N. C. (2011). The DNA damage response induces IFN. J. Immunol. 187, 5336–5345. doi: 10.4049/jimmunol.1100040
Chatterjee, B., Hopkins, J., Dutchak, D., and Roy, A. K. (1979). Superinduction of α2u globulin by actinomycin D: Evidence for drug-mediated increase in α2u mRNA Proc. Natl. Acad. Sci USA. 76, 1833–1837. doi: 10.1073/pnas.76.4.1833
Chaung, W., and Boorstein, R. J. (1997). Molecular spectrum of mutations induced by 5-hydroxymethyl-2’deoxyuridine in (CHO)-PL61 cells. Mutat. Res. 373, 125–137. doi: 10.1016/S0027-5107(96)00197-2
Chiang, C., and Hadwiger, L. A. (1991). The Fusarium solani-induced expression of a pea gene family encoding high cysteine content proteins. Mol. Plant Microbe Interact. 4, 324–331. doi: 10.1094/MPMI-4-324
Choi, J. J., Klosterman, S. J., and Hadwiger, L. A. (2001). A comparison of the effects of DNA-damaging agents and biotic elicitors on the induction of plant defense genes, nuclear distortion and cell death. Plant Physiol. 125, 752–762. doi: 10.1104/pp.125.2.752
Daniel, J. A., and Nussenzweig, A. (2013). The AID-induced DNA damage response in chromatin. J. Mol. Cell 50, 309–321. doi: 10.1016/j.molcel.2013.04.017
Deffert, C., Cachat, J., and Krause, K. H. (2014). Phagocyte NADPH oxidase, chronic granulomatous disease and mycobacterial infections. Cell. Microbiol. 16, 1168–1178. doi: 10.1111/cmi.12322
Dekker, J., Marti-Renom, M. A., and Mirny, L. A. (2013). Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 14, 390–403. doi: 10.1038/nrg3454
Dizdaroglu, M., and Jaruga, P. (2012). Mechanisms of free radial-induced damage to DNA. Free Radic. Res. 46, 382–419. doi: 10.3109/10715762.2011.653969
Duran-Flores, D., and Heil, M. (2018). Extracellular self-DNA as a damage-associated molecular pattern (DAMP) that triggers self-specific immunity induction in plants. Brain Behav. Immun. 72, 78–88. doi: 10.1016/j.bbi.2017.10.010
Felsenfeld, G., Sandeen, G., and Vonhippel, P. H. (1963). The destabilizing effect of ribonuclease on the helical DNA structure. Proc. Natl. Acad. Sci. U.S.A. 50, 644–648. doi: 10.1073/pnas.50.4.644
Flamm, W. G., Banerjee, M. R., and Counts, W. B. (1966). Topical application of actinomycin D on mouse skin: effect on the synthesis of ribonucleic acid and protein. Cancer Res. 26, 1349–1360.
Floss, H. G., Guenther, H., and Hadwiger, L. A. (1969). Biosynthesis of furanocoumarins in diseased celery. Phytochemistry 8, 585–588. doi: 10.1016/S0031-9422(00)85404-7
Fontes, F. L., Pinheiro, D. M. L., Sales de Oliveira, A. H., Oliveira, R. K. M., Lajus, T. B. P., and Agnez-Lima, L. F. (2014). Role of DNA repair in host immune response and inflammation. Mutat. Res. 763, 246–257. doi: 10.1016/j.mrrev.2014.11.004
Gerasimova, N. S., Pestov, M. A., and Kulaeva, O. I. (2016). Transcription-induced DNA supercoiling: new roles of intranucleosomal DNA loops in DNA repair and transcription. Transcription 7, 91–95. doi: 10.1080/21541264.2016.1182240
Gilbert, N., and Allan, J. (2014). Supercoiling in DNA and chromatin. Curr. Opin. Genet. Dev. 25, 15–21. doi: 10.1016/j.gde.2013.10.013
Gurova, K. (2009). New hopes from old drugs: revisiting DNA-binding small molecules as anticancer agents. Future Oncol. 5:1685. doi: 10.2217/fon.09.127
Hadwiger, L. A. (1972a). Increased levels of pisatin and phenylalanine ammonia lyase activity in Pisum sativum treated with antihistamine, antiviral, antimalarial, tranquilizing, or other drugs. Biochem. Biophys. Res. Commun. 46, 71–79. doi: 10.1016/0006-291X(72)90631-6
Hadwiger, L. A. (1972b). Induction of phenylalanine ammonia lyase and pisatin by photosensitive psoralen compounds. Plant Physiol. 49, 779–782.
Hadwiger, L. A. (2008). Pea-Fusarium solani interactions, contributions of a system toward understanding disease resistance. Phytopathology 98, 372–379. doi: 10.1094/PHYTO-98-4-0372
Hadwiger, L. A. (2009). Localization predictions for gene products involved in non-host resistance responses in a model plant/fungal pathogen interaction. Plant Sci. 177, 257–265. doi: 10.1016/j.plantsci.2009.06.010
Hadwiger, L. A. (2015a). Anatomy of a nonhost disease resistance response of pea to Fusarium solani: PR gene elicitation via DNase, chitosan and chromatin alterations. Front. Plant Sci. 6:373. doi: 10.3389/fpls.2015.00373
Hadwiger, L. A. (2015b). Chitosan: The preliminary research and the host-parasite system that led to the discovery of its antifungal and gene inducing properties. J Mol. Genet. Med. 9:158. doi: 10.4172/1747-0862.1000158
Hadwiger, L. A. (2015c). Nonhost resistance: self-inflicted DNA damage by fungal DNase accumulation is a major factor in terminating fungal growth in the pea-Fusarium solani f. sp. phaseoli interaction. Physiol. Mole. Plant Pathol. 92, 79–87. doi: 10.1016/j.pmpp.2015.08.003
Hadwiger, L. A., and Adams, M. J. (1978). Nuclear changes associated with the host-parasite interaction between Fusarium solani and peas. Physiol. Plant Pathol. 12, 63–72. doi: 10.1016/0048-4059(78)90019-X
Hadwiger, L. A., and Beckman, J. M. (1980). Chitosan as a component of pea-Fusarium solani interactions. Plant Physiol. 66, 205–211. doi: 10.1104/pp.66.2.205
Hadwiger, L. A., Beckman, J. M., and Adams, M. J. (1981). Localization of fungal components in the pea-Fusarium interaction detected immunochemically with antichitosan and antifungal cell wall antisera. Plant Physiol. 67, 170–175. doi: 10.1104/pp.67.1.170
Hadwiger, L. A., and Chang, M. M. (2015). Low level DNA damage occurs as PAMPs, chitin and flg 22, activates PR genes, and increases pisatin and disease resistance in pea endocarp tissue. New Negat. Plant Sci. 1–2, 6–15. doi: 10.1016/j.neps.2015.04.001
Hadwiger, L. A., Chang, M. M., and Parsons, M. A. (1995). Fusarium solani DNase is a signal for increasing expression of nonhost disease resistance response genes, hypersensitivity and pisatin production. Mol. Plant Microbe Interact. 8, 871–879. doi: 10.1094/MPMI-8-0871
Hadwiger, L. A., Chiang, C., Victory, S., and Horovitz, D. (1989). “The molecular biology of chitosan in plant/pathogen interaction it application in agriculture,” in Chitin and Chitosan, Sources Chemistry, Biochemistry, Physical Properties and Applications, eds G. Skjak-Braek, T. Anthonsen, and P. Sanford (London: Elsevier), 119–131.
Hadwiger, L. A., Jafri, A., von Broembsen, S., and Eddy, R. (1974). Mode of pisatin induction. Increased template activity and dye-binding capacity of chromatin isolated from polypeptide-treated pea pods. Plant Physiol. 53, 52–63. doi: 10.1104/pp.53.1.52
Hadwiger, L. A., Klosterman, S. K., Chang, M. M., Friel, P., and Hosick, H. L. (1997). “Chitosan heptamer alters DNA, induces defense genes in plants and induces the accumulation of gene p53 product in animal cells,” in Advances in Chitin Sciences, Vol. 2, eds A. Domard, G. A. F. Roberts, and K. M. Varum (Lyon: Jacques André Publisher), 102–109.
Hadwiger, L. A., Loschke, D. C., and Teasdale, J. R. (1977). An evaluation of pea histones as disease resistance factors. Phytopathology 67, 755–758. doi: 10.1094/Phyto-67-755
Hadwiger, L. A., and Martin, A. R. (1971). Induced formation of phenylalanine ammonia lyase and pisatin by chlorpromazine and other phenothiazine derivatives. Biochem. Pharmacol. 20, 3255–3261. doi: 10.1016/0006-2952(71)90430-8
Hadwiger, L. A., and Polashock, J. (2013). Fungal mitochondrial DNases. Effectors with the potential to activate plant defenses in nonhost resistance. Phytopathology 103, 81–90. doi: 10.1094/PHYTO-04-12-0085-R
Hadwiger, L. A., Sander, C., Eddyvean, J., and Ralston, J. (1976). Sodium azide-induced mutants of pea that accumulate pisatin. Phytopathology 66, 629–630. doi: 10.1094/Phyto-66-629
Hadwiger, L. A., and Schwochau, M. E. (1970). Induction of phenylalanine ammonia lyase and pisatin in pea pods by poly-lysine, spermidine, or histone fractions Biochem. Biophys. Res. Comm. 38, 683–691. doi: 10.1016/0006-291X(70)90635-2
Hadwiger, L. A., and Schwochau, M. E. (1971). Ultraviolet Light-induced Formation of Pisatin and Phenylalanine Ammonia Lyase. Plant Physiol. 47, 588–590. doi: 10.1104/pp.47.4.588
Hadwiger, L. A., and Tanaka, K. (2015). EDTA a novel inducer of pisatin, a phytoalexin indicator of the non-host resistance in peas. Molecules 20, 24–34. doi: 10.3390/molecules20010024
Hadwiger, L. A., and Tanaka, K. (2017a). A simple and rapid assay for measuring phytoalexin pisatin, an indicator of plant defense response in pea (Pisum sativum L.). Bioprotocol 7:e2362. doi: 10.21769/BioiProtoc.2362
Hadwiger, L. A., and Tanaka, K. (2017b). Nonhost resistance: DNA damage is associated with SA signaling for induction of PR genes and contributes to the growth suppression of a pea pathogen on pea endocarp tissue. Front. Plant Sci. 8:446. doi: 10.3389/fpls.2017.00446
Hager, G. L., McNally, J. G., and Misteli, T. (2009). Transcription dynamics. Mol. Cell 35, 741–753. doi: 10.1016/j.lmolcel.2009.09.005
Hampton, M. B., and Orrenius, S. (1997). Dual regulation of caspase activity by hydrogern peroxide: implications for apoptosis. FEBS Lett. 414, 552–556. doi: 10.1016/S0014-5793(97)01068-5
Hartney, S., Carson, J., and Hadwiger, L. A. (2007). The use of chemical genomics to detect functional systems affecting the non-host disease resistance of pea to Fusarium solani f. sp. phaseoli. Plant Sci. 172, 45–56. doi: 10.1016/j.plantsci.2006.07.014
Hendry, L. B., Mahesh, V. B., Bransome, E. D., and Ewing, D. E. (2007). Small molecule intercalation with double stranded DNA: implications for normal gene regulation and for predicting the biological efficacy and genotoxicity of drugs and other chemicals. Mutat. Res. 623, 53–71. doi: 10.1016/j.mrfmmm.2007.03.009
Hendry, L. B., Witham, F. H., and Chapman, O. L. (1977). Gene regulation: the involvement of stereochemical regulation in DNA-small molecules interactions. Prospect Biol. Med. 21, 120–130. doi: 10.1353/pbm.1977.0018
Hess, S. L., and Hadwiger, L. A. (1971). The induction of phenylalanine ammonia lyase and phaseollin by 9-aminoacridine and other deoxyribonucleic Acid intercalating compounds. Plant Physiol. 48, 197–202. doi: 10.1104/pp.48.2.197
Hyman, R. S., and Davidson, N. (1970). The kinetics of the in vivo inhibition of transcription by actinomycin D. Fed. Proc. 29, 531–532.
Isaac, J., Hartney, S. L., Druffel, K., and Hadwiger, L. A. (2009). The non-host disease resistance response in peas: alterations in phosphorylation and ubiquitination of HMG A and histones H2A/H2B. Plant Sci. 177, 439–449. doi: 10.1016/j.plantsci.2009.07.007
Javaherian, K., and Sadeghi, M. (1979). Non-histone proteins HMG 1 and HMG 2 unwind DNA double helix. Nucleic Acids Res. 6, 3569–3580. doi: 10.1093/nar/6.11.3569
Jerzmanowski, A. (2007). SWI/SNF chromatin remodeling and linker histones in plants. Biochim. Biophys. Acta 1769, 330–345. doi: 10.1016/j.bbaeexp.2006.12.003
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–406. doi: 10.1038/nature05286
Kastan, M. B., and Bartek, J. (2001). Cell-cycle checkpoints and cancer. Nature 432, 316–323. doi: 10.1038/nature03097
Kendra, D. F., Christian, D. A., and Hadwiger, L. A. (1989). Chitosan oligomers from Fusarium solani/pea interactions, chitinase/β-glucanase digestion of sporelings and from fungal wall chitin actively inhibit fungal growth and enhance disease resistance. Physiol. Mol. Plant Pathol. 35, 215–230. doi: 10.1016/0885-5765(89)90052-0
Klosterman, S. J., Chen, J., Choi, J. J., Chinn, E. E., and Hadwiger, L. A. (2001). Characterization of a 20 kDa DNase elicitor from Fusarium solani f. sp. phaseoli and its expression at the onset of induced resistance in Pisum sativum. Mol. Plant Pathol. 2, 147–158. doi: 10.1046/j.1364-3703.2001.00062.x
Klosterman, S. J., Choi, J. J., and Hadwiger, L. A. (2000). Programmed cell death is not mediated by a p53 homolog in Pisum sativum. Physiol. Mol. Plant Pathol. 56, 197–206. doi: 10.1006/pmpp.2000.0267
Klosterman, S. J., Choi, J. J., and Hadwiger, L. A. (2003). Analysis of pea HMG-A expression suggests a role in defense gene regulation Mol. Plant Pathol. 4, 249–258. doi: 10.1046/j.1364-3703.2003.00171.x
Klosterman, S. J., and Hadwiger, L. A. (2002). Plant HMG proteins bearing the AT-hook motif. Plant Sci. 162, 855–866. doi: 10.1016/S0168-9452(02)00056-0
Kreuz, S., and Fischle, W. (2016). Oxidative stress signaling to chromatin in health and disease. Epigenomics 8, 843–862. doi: 10.2217/epi-2016-0002
Lenglet, G., and David-Cordonnier, M. H. (2010). DNA-destabilizing agents as an alternative approach for targeting DNA: mechanisms of action and cellular consequences. J. Nucleic Acids 2010, 1–17. doi: 10.4061/2010/290935
Lewis, M., Helmsing, P. J., and Ashburner, M. (1975). Parallel changes in puffing activity and patterns of protein synthesis in salivary glands of Drosophila. Proc. Nat. Acad. Sci. U.S.A. 72, 3604–3608. doi: 10.1073/pnas.72.9.3604
Lo, Y. S., Tseng, W. H., Chuang, C. Y., and Hou, M. H. (2013). The structural basis of actinomycin D-binding induces nucleotide flipping out, a sharp bend and a left-handed twist in CGG triplet repeats. Nucleic Acids Res. 41, 4284–4294. doi: 10.1093/nar/gkt084
Loschke, D. L., Hadwiger, L. A., and Wagoner, W. (1983). Comparison of mRNA populations coding for phenylalanine ammonia-lyase and other peptides from pea tissue treated with biotic and abiotic phytoalexin inducers. Physiol. Plant Pathol. 23, 163–173. doi: 10.1016/0048-4059(83)90043-7
Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F., and Richmond, T. J. (1997). Crystal structure of the nucleomome core particle at 2.8 Å resolution. Nature 389, 251–260. doi: 10.1038/38444
Ma, J., Bai, L., and Wang, M. D. (2013). Transcription under torsion. Science 340, 1580–1583. doi: 10.1126/science.1235441
Mao, P., Meas, R., Dorgan, K. M., and Smerdon, M. J. (2014). UV damaged-induced RNA polymerase II stalling stimulates H2B deubiquitylation. Proc. Natl. Acad. Sci. U.S.A. 111, 12811–12816. doi: 10.1073/pnas.1403901111
Martinez, R., and Cha′con-Garcia, L. (2005). The search of DNA-intercalators as antitumoral drugs: what worked and what did not. Curr. Med. Chem. 12, 127–151. doi: 10.2174/0929867053363414
Mazzoleni, S., Bonanomi, G., Incerti, G., Chiusano, M. L., Termolino, P., Mingo, A., et al. (2015a). Inhibitory and toxic effects of extracellular self-DNA in litter: a mechanism for negative plant–soil feedbacks? New Phytol. 205, 1195–1210. doi: 10.1111/nph.13121
Mazzoleni, S., Cartenì, F., Bonanomi, G., Senatore, M., Termolino, P., Giannino, F., et al. (2015b). Inhibitory effects of extracellular self-DNA: a general biological process? New Phytol. 206, 127–132. doi: 10.1111/nph.13306
Miskovic, K., Bujak, M., Baus Loncar, M., and Glavas-Obrovac, L. (2013). Antineoplastic DNA-binding compounds: intercalating and minor groove binding drugs. Arh. Hig. Rada Toksikol. 64, 593–602. doi: 10.2478/10004-1254-64-2013-2371
Muchardt, C., and Yaniv, M. A. (1993). A human homologue of S. cerevisiae SNF2/SW12 and Drosophila brm genes co-operates with nuclear hormone receptors in transcriptional activation. EMBO J. 12, 4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x
Nair, S. S., and Kumar, R. (2012). Chromatin remodeling in cancer: a gateway to regulate gene transcription. Mol. Oncol. 6, 611–619. doi: 10.1016/j.molonc.2012.09.005
Nakad, R., and Schumacher, B. (2016). DNA damage response and immune defense: links and mechanisms. Front. Genet. 7:147. doi: 10.3389/fgene.2016.00147
Neigeborn, L., and Carlson, M. (1984). Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics 108, 845–858.
Nelson, S. M., Ferguson, L. R., and Denny, W. A. (2007). Non-covalent ligand/DNA interactions: Minor groove binding agents. Mutat. Res. 623, 24–40. doi: 10.1016/j.mrfmmm.2007.03.012
Nürnberger, T., Brunner, F., Kemmerling, B., and Piater, L. (2004). Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198, 249–266. doi: 10.1111/j.0105-2896.2004.0119.x
Pang, B., Qiao, X., Janssen, L., Velds, A., Groothuis, T., Kerkhoven, R., et al. (2013). Drug-induced histone eviction from open chromatin contributes to the chemotherapeutic effects of doxorubicin (daunomycin). Nat. Commun. 4, 1908–1931. doi: 10.1038/ncomm2921
Parsons, M. A., and Hadwiger, L. A. (1998). Photoactivated psoralens elicit defense genes and phytoalexins production in the pea plant. Photochem. Photobiol. 67, 438–445. doi: 10.1111/j.1751-1097.1998.tb05224.x
Petesch, S. J., and Lis, J. T. (2008). Rapid transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134, 74–84. doi: 10.1016/j.cell2008.05.029
Pilet-Nayel, M. L., Muehlbauer, F. J., McGee, R. J., Kraft, J. M., Baranger, A., and Coyne, C. J. (2002). Quantitative trait loci for partial resistance to Aphanomyces root rot in pea. Theor. Appl. Genet. 106, 28–39. doi: 10.1007/s00122-002-0985-2
Pruss, G. J., and Drlica, K. (1989). DNA supercoiling and prokaryotic transcription. Cell 56, 521–523. doi: 10.1016/0092-8674(89)90574-6
Quigley, G. J., Wang, A. H., Ughetto, G., vander Marel, G., van Boom, J. H., and Rich, A. (1980). Molecular structure of an anticancer drug-DNA complex: daunomycin plus d(CpGpTpApCpG). Proc. Natl. Acad. Sci. U.S.A. 77, 7204–7208. doi: 10.1073/pnas.77.12.7204
Ramirez-Prado, J. S., Piquerez, S. J. M., Bendahmane, A., Hirt, H., Raynaud, C., and Benhamed, M. (2018). Modify the histone to win the battle: chromatin dynamics in plant-pathogen interactions. Front. Plant Sci. 9:355. doi: 10.3389/fpls.2018.00355
Ramsey, M. R., and Sharpless, N. E. (2006). ROS as a tumour suppressor? Nat. Cell Biol. 8, 1213–1215. doi: 10.1038/ncb1106-1213
Reeves, R. (2010). HMG nuclear proteins: linking chromatin structure to cellular phenotype. Biochim. Biophys. Acta 1799, 1–27. doi: 10.1016/j.bbagrm.2009.09.001
Reich, E., and Goldberg, I. H. (1964). “Actinomycin and nucleic acid function,” in Progress in Nucleic Acid Research and Molecular Biology, Vol. 3, eds J. N. Davidson and W. E. Cohn 183–234. doi: 10.1016/S0079-6603(08)60742-4
Sander, C., and Hadwiger, L. A. (1979). L-phenylalanine ammonia-lyase and pisatin induction by 5-bromodeoxyuridine in Pisum sativum. Biochim. Biophys. Acta 563, 278–292. doi: 10.1016/0005-2787(79)90047-9
Sarnowska, E., Gratkowska, D. M., Sacharowski, S. P., Cwiek, P., Tohge, T., Fernie, A. R., et al. (2016). The role of SWI/SNF chromatin remodeling complexes in hormone crosstalk. Trends Plant Sci. 21, 594–608. doi: 10.1016/j.tplants.2016.01.017
Schwochau, M. E., and Hadwiger, L. A. (1968). Stimulation of pisatin production in Pisum sativum by actinomycin D and other compounds. Arch. Biochem. Biophys. 126, 731–733. doi: 10.1016/0003-9861(68)90463-3
Schwochau, M. E., and Hadwiger, L. A. (1969). Regulation of gene expression by actinomycin D and other compounds which change the conformation of DNA. Arch. Biochem. Biophys. 134, 34–41. doi: 10.1016/0003-9861(69)90247-1
Sobell, H. M., and Jain, S. C. (1972). Stereochemistry of actinomycin binding to DNA. II Detailed molecular model of actinomycin-DNA complex and its implications. J. Mol. Biol. 68, 21–34. doi: 10.1016/0022-2836(72)90259-8
Song, J., Keppler, B. D., Wise, R. R., and Bent, A. F. (2014). PARP2 is the predominant poly(ADP-Ribose) polymerase in Arabidopsis DNA damage and immune responses. PLoS Genet. 11:e1005200. doi: 10.1371/journal.pgen.1005200
Spichal, M., and Gabre, E. (2017). The emerging role of the cytoskeleton in chromosome dynamics. Front. Genet. 8:60. doi: 10.339/fgene.2017.00060
Steinberg, R. A., Levenson, B. B., and Tomkins, G. M. (1975). Superinduction of tyrosine aminotransferase by actinomycin D: a reevaluation. Cell 5:29–35. doi: 10.1016/0092-8674(75)90088-4
Tanaka, K., Choi, J., Cao, Y., and Stacey, G. (2014). Extracellular ATP acts as a damage-associated molecular pattern (DAMP) signal in plants. Front. Plant Sci. 5:466. doi: 10.3389/fpls.2014.00446
Tanaka, K., and Hadwiger, L. A. (2017). Nonhost resistance: Reactive oxygen species (ROS) signal causes DNA damage prior to the induction of PR genes and disease resistance in pea tissue. Physiol. Mol. Plant Pathol. 98, 18–24. doi: 10.1016/j.pmpp.2017.01.007
Tang, L., Nogales, E., and Ciferri, C. (2010). Structure and function of SWI/SNF chromatin remodeling complexes and mechanistic implications for transcription. Prog. Biophys. Mol. Biol. 102, 122–128. doi: 10.1016/j.pbiomolbio.2010.05.001
Teves, S. S., and Henikoff, S. (2014). DNA torsion as a feedback mediator of transcription and chromatin dynamics. Nucleus 5, 211–218. doi: 10.4161/nucl.29086
Varqiu, A. V., Ruggerone, P., Magistrato, A., and Carloni, P. (2008). Dissociation of minor groove binders from DNA: Insights from metadynamics simulations. Nucleic Acids Res. 36, 5910–5921. doi: 10.1093/nar/gkn561
Walker-Simmons, M., Hadwiger, L., and Ryan, C. A. (1983). Chitosans and pectic polysaccharides both induce the accumulation of the antifungal phytoalexin pisatin in pea pods and antinutrient proteinase inhibitors in tomato leaves. Biochem. Biophys. Res. Commun. 110, 194–199. doi: 10.1016/0006-291X(83)91279-2
Wang, J., and Yi, J. (2008). Cancer cell killing via ROS: to increase or decrease, that is the question. Cancer Biol. Ther. 7, 1875–1884. doi: 10.4161/cbt.7.12.7067
Watson, J. D., Hopkins, N. H., Roberts, J. W., Steitz, J. A., and Weiner, A. M. (1987). Molecular Biology of the Gene, 4th Edn. Menlo Park, CA: Benjamin/Cummings publishing Co.
Weake, V. M., and Workman, J. L. (2008). Histone ubiquitination: triggering gene activity. Mol. Cell 29, 653–663. doi: 10.1016/j.molcel.2008.02.014
Willis, B., and Arya, D. P. (2006). Recognition of B-DNA by Neomycin-Hoechst 33258 conjugates. Biochemistry 45, 10217–10232. doi: 10.1021/bi0609265
Witham, F. H., Hendry, L. B., and Chapman, O. L. (1978). Chirality and stereochemical recognition in DNA-phytohormone interactions: a model approach. Origins Life 9, 7–15. doi: 10.1007/BF00929709
Woods, D., and Turchi, J. J. (2013). Chemotherapy induced DNA damage response –convergence of drugs and pathways. Cancer Biol. Ther. 14, 379–389. doi: 10.4161/cbt23761
Yan, S., Wang, W., Marqués, J., Mohan, R., Saleh, A., Durrant, W. E., et al. (2013). Salicylic acid activates DNA damage responses to potentiate plant immunity. Mol. Cell. 52, 602–610. doi: 10.1016/j.molcel.2013.09.019
Keywords: nonhost resistance, DNA damage, DNA conformation, chromatin structural changes, anti-cancer agents
Citation: Hadwiger LA and Tanaka K (2018) DNA Damage and Chromatin Conformation Changes Confer Nonhost Resistance: A Hypothesis Based on Effects of Anti-cancer Agents on Plant Defense Responses. Front. Plant Sci. 9:1056. doi: 10.3389/fpls.2018.01056
Received: 03 April 2018; Accepted: 28 June 2018;
Published: 24 July 2018.
Edited by:
Víctor Flors, Universitat Jaume I, SpainReviewed by:
Mónica Meijón, Universidad de Oviedo, SpainMartin Heil, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional (CINVESTAV-IPN), Mexico
Copyright © 2018 Hadwiger and Tanaka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lee A. Hadwiger, Y2hpdG9zYW5Ad3N1LmVkdQ==
 Lee A. Hadwiger
Lee A. Hadwiger Kiwamu Tanaka
Kiwamu Tanaka