Abstract
Genome editing technologies have progressed rapidly and become one of the most important genetic tools in the implementation of pathogen resistance in plants. Recent years have witnessed the emergence of site directed modification methods using meganucleases, zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindrome repeats (CRISPR)/CRISPR-associated protein 9 (Cas9). Recently, CRISPR/Cas9 has largely overtaken the other genome editing technologies due to the fact that it is easier to design and implement, has a higher success rate, and is more versatile and less expensive. This review focuses on the recent advances in plant protection using CRISPR/Cas9 technology in model plants and crops in response to viral, fungal and bacterial diseases. As regards the achievement of viral disease resistance, the main strategies employed in model species such as Arabidopsis and Nicotiana benthamiana, which include the integration of CRISPR-encoding sequences that target and interfere with the viral genome and the induction of a CRISPR-mediated targeted mutation in the host plant genome, will be discussed. Furthermore, as regards fungal and bacterial disease resistance, the strategies based on CRISPR/Cas9 targeted modification of susceptibility genes in crop species such as rice, tomato, wheat, and citrus will be reviewed. After spending years deciphering and reading genomes, researchers are now editing and rewriting them to develop crop plants resistant to specific pests and pathogens.
Introduction
Plant breeding has been the most successful approach for developing new crop varieties since domestication occurred, making possible major advances in feeding the world and societal development. Crops are susceptible to a large set of pathogens including fungi, bacteria, and viruses, which cause important economic losses (FAO, 2017); the enhancement of plant resistance plays an important role in adjusting crop production to meet global population increases. Approaches to disease control that depend on resistant varieties and agrochemicals are usually highly effective whenever they are deployed. However, due to the high evolutionary potential of many plant pathogens, novel genotypes no longer sensitive to the resistance gene or the phytosanitary product can rapidly emerge via mutation or recombination. When this happens, particular disease control approaches can rapidly be rendered ineffective as the novel genotypes increase in frequency through natural selection and quickly spread to other locations, causing failure of control over large geographic areas.
An understanding of interactions between plants and communities of bacteria, fungi, and other microorganisms has been a major area of investigation for many years. The advent of high-throughput molecular technologies has made a more complete inventory of the pathogens associated with particular crops possible, and provided insight into how these communities may be affected by environmental factors and the crop genotype. Disease involves a complex inter-play between a host plant and a pathogen, and the resistance/susceptibility response can involve several components. Natural and induced mutations may change the interaction and inhibit certain steps in the mechanism of infection (Boyd and O’Toole, 2012; Dracatos et al., 2018).
During pre-genomic years, traditional breeding programs were based on the identification of natural and induced mutant alleles for resistance, and their incorporation into elite genotypes through breeding techniques. These approaches were uncertain and imprecise, leading for instance to the transfer of large genome regions instead of just single gene insertions. Nevertheless, mutation breeding methods have been quite successful in improving disease resistance, and traditional plant breeding has been used to generate new crop varieties for decades. Numerous mutants have been developed through mutation induction, showing enhanced resistance to various diseases. Among the most widely known mutants are those induced at the mildew resistance locus (MLO) in barley for resistance to powdery mildew (Miklis et al., 2007), and mutations conferring resistance to several lettuce diseases (Christopoulou et al., 2015). The mlo mutant is interesting, as the allele has not broken down and has provided unprecedented resistance to mildew for two decades (Panstruga and Schulze-Lefert, 2002). This longevity is due to a gene knockout. In other cases where resistance to specific pathotypes is conferred by a specific host gene allele, mutagenesis needs to be deployed to provide more precise single nucleotide mutations in the target gene sequence.
The revolution driven by the availability of genome and transcriptome sequences offers a new start for plant breeding programs. Association genetics based on single nucleotide polymorphisms (SNPs) and other molecular markers are spreading in plant breeding, creating high throughput data fundamental for the identification of quantitative trait loci (QTL). Major QTL are employed in crops to provide quantitative resistance to pathogens, together with the use of major resistance (R) genes introduced into varieties with superior agronomic characteristics.
New breeding techniques (NBTs) are attracting attention in plant research and concern many different areas, such as developmental biology, abiotic stress tolerance or plant-pathogen resistance (Nelson et al., 2018). NBT include the most recent and powerful molecular approaches for precise genetic modifications of single or multiple gene targets. They employ site-directed nucleases to introduce double stranded breaks at predetermined sites in DNA. These breaks are repaired by different host cell repair mechanisms, resulting either in small insertions or deletions via near homologous end-joining (NHEJ) or micro-homology-mediated end-joining (MMEJ), or in a modified gene carrying predetermined nucleotide changes copied from a repair matrix via homologous recombination (HR). Meganucleases (MNs), zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindrome repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) correspond to the four types of nucleases used in genome editing. The exponential increase in publications reporting the use of CRISPR/Cas9 illustrates the fact that this technology requires less know-how and financial means and has a higher success rate in gene modification compared to the other available nucleases. The application of CRISPR/Cas9 editing has become a powerful tool for future enhancement of agronomic traits in crops (Mohanta et al., 2017).
The objective of this review is to recall the main features of the CRISPR/Cas9 genome editing technique and discuss its application for the enhancement of pathogen resistance in model plants and important crops, with a focus on rice, wheat, and maize.
CRISPR/Cas9: Advances, Limitations, and New Combinations
CRISPR/Cas9 from Streptococcus pyogenes (SpCas9) has rapidly assumed an important role in different application areas of plant research and many other fields (Ding et al., 2018; Liu and Moschou, 2018). In the CRISPR/Cas9 system a single-guide RNA (sgRNA) can bind to Cas9 and target it to specific DNA sequences (Figure 1). The requirement of a protospacer adjacent motif (PAM) limits the possible target sequences in a gene of interest. This limitation is of minor importance if the aim is simply to inactivate a gene by targeted mutagenesis at any position. It has much more importance for genome editing aiming at the precise change of specific nucleotides in a gene. Consequently, major efforts are under way to find Cas9-like proteins with different PAM sequences or to engineer the original Cas9 from S. pyogenes to recognize other PAM sequences. For example, xCas9, an evolved version of SpCas9, has been shown to recognize a broad range of PAM sequences including NG, GAA, and GAT in mammalian cells (Hu et al., 2018). In plants, the most widely explored alternative to SpCas9 is Cpf1 from Prevotella and Francisella with the PAM sequence TTTV, where “V” is A, C, or G (Endo et al., 2016), and an illustrative diagram is shown in Figure 1. Cpf1 is also considerably smaller than Cas9, is capable of RNAse activity to process its guide RNA, and introduces a staggered double break, which can be useful for enhancing homology-directed recombination and generating efficient gene insertion.
FIGURE 1
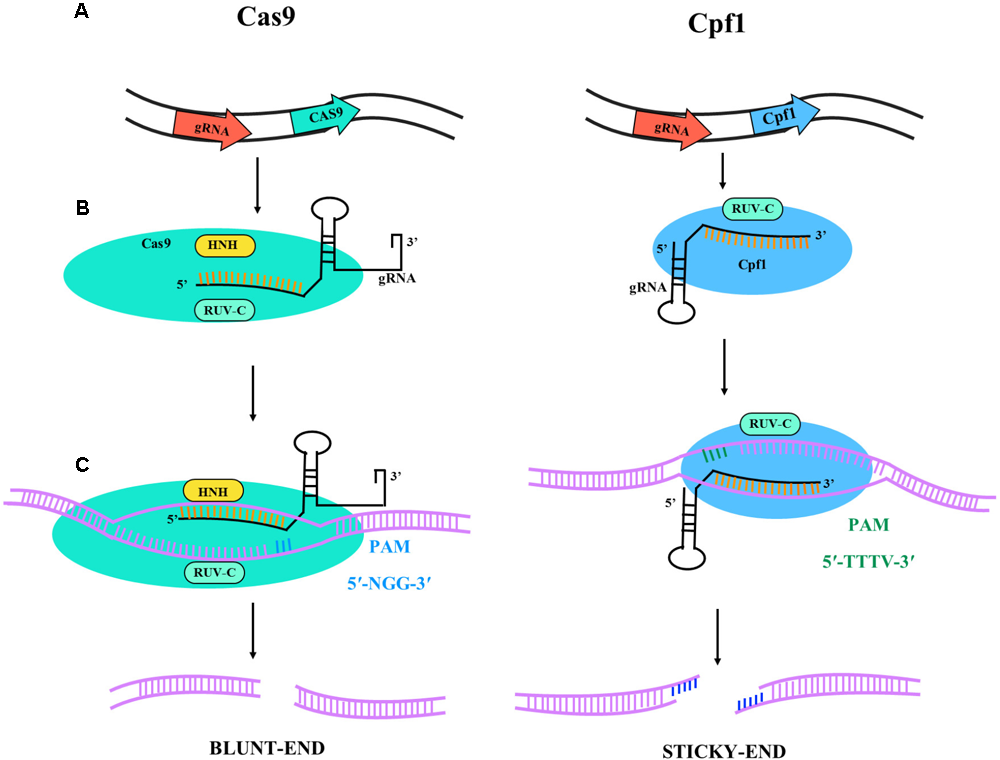
Illustrative diagram of Cas9 and Cpf1 activities. The target specificity is given by the 17-20 nt located at the 5′ end of the sgRNA sequence. (A) Primary transcript and gRNA-nuclease (Cas9 or Cpf1) complex formation. The catalytic domains are RUV-C (light blue) and HNH (yellow) for Cas9 and RUV-C for Cpf1. The Cas9 is colored in light blue and the Cpf1 in dark blue; in black is represented the gRNA for gene targeting. (B) Gene target activity. Cas9 has 5′-NGG-3′ PAM sequence (blue bars) and Cpf1 has 5′-TTTV-3′ PAM sequence (green bars). (C) DNA ends after nuclease activity. Cas9 lead to blunt-end and Cpf1 to sticky-ends.
Multiplex Genome Editing: When Does It Become Useful?
The ease of multiplexing, i.e., the simultaneous targeting of several genes with a single molecular construct, is one of the major advantages of CRISPR/Cas9 technology with respect to MN, ZFN, or TALEN. For example, the simultaneous mutation of 14 different genes by a single construct has been demonstrated in Arabidopsis (Peterson et al., 2016). In crops, several multiplex genome editing (MGE) strategies were reported early on (Ma et al., 2014; Xing et al., 2014; Zhou et al., 2014; Xu et al., 2016), which were all based on a common strategy, i.e., the assembly of multiple gRNAs under the control of a U3 or U6 promoter into a single construct. In maize, the ISU Maize CRISPR platform (Char et al., 2017) permits the cloning of up to four gRNAs for multiplex gene targeting.
More recent multiplex systems exploit self-cleavage capacity of RNA molecules containing tRNA sequences. Constructs alternating sgRNA and tRNA sequences under the control of a single U3 or U6 promoter permit reduction of the size of the construct and limit the risk of silencing due to direct repetitions of promoter sequences. The use of such a strategy employing polycistronic tRNA-gRNA (PTG) to generate hereditable mutation in TaLpx-1 and TaMLO genes has been reported in hexaploid wheat (Wang et al., 2018); the PTG system is described in Figure 2. Starting from a previous study on gene silencing of TaLpx-1, which encodes the wheat 9-lipoxygenase resistance gene to Fusarium graminearum (Nalam et al., 2015), the editing of homologs in wheat was tested. The PTG system containing gRNA activity was validated in wheat confirming gene editing efficacy and providing an effective tool for rapid trait pyramiding in breeding programs.
FIGURE 2
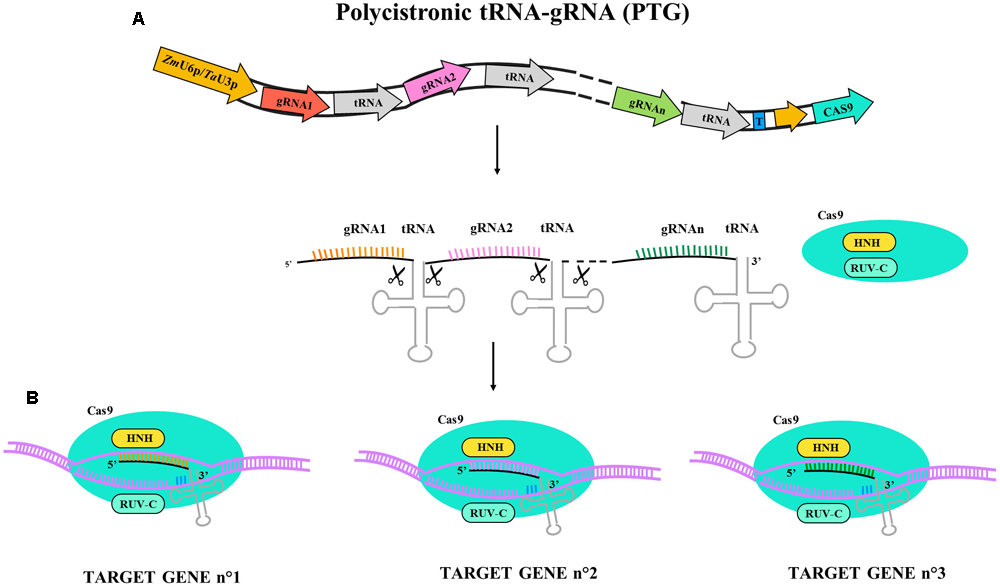
Illustrative diagram of polycistronic tRNA-gRNA (PTG) gene construct and targeting activity for Cas9. PTG is composed of t-RNA-gRNA repeats and is upregulated by ZmU6 promoter or TaU3 promoter according the experimental design as different terminator region (T) are adopted. (A) PTG primary transcript. Endogenous endonuclease cuts the tRNA ends and let each tRNA-gRNA targeting the corresponding gene sequence. (B) In PTG system more sequence targets are available (n° gene targets) and the different gRNA are represented in different colors (orange, pink, and green).
Recently, an alternative approach for MGEs based on PTG has been reported in rice, where crRNA transcription was obtained from introns inserted into Cpf1 and Cas9 sequences (Ding et al., 2018). Multiplex PTG/Cas9 systems can help with multigene family analysis, as reported for the closely related mitogen-activated protein kinase MPK1 and MPK6 in rice (Minkenberg et al., 2017). 67% of all lines were double mutants for MPK genes with a high frequency of biallelic mutations on multiple target sites. The possibility of programming the PTG/Cas9 to delete chromosomal fragments could be adopted to remove genes and regulatory elements in order to produce transgene free plants.
Off-Target Mutations: Frequency and Limitations
High specificity is frequently put forward as a major argument in favor of CRISPR/Cas9 technology, for example in comparison to chemically or irradiation-induced mutagenesis. This raises the question of to what extent a gRNA targets only fully complementary genomic DNA sequences, and to what extent other genomic regions (off-target regions) can also be recognized and cleaved by the CRISPR/Cas9 tool, provoking potentially unwanted damage. Two types of off-target effects are evoked by scientists and regulatory agencies: (i) expected off-target in genome regions with high sequence similarity to the target and (ii) unexpected off-target in unrelated genome regions. The former is generally addressed by PCR amplification and sequencing of regions known to be similar to the target, the latter by whole genome sequencing (Feng et al., 2016).
Genome sequence information is necessary for the prediction of expected off-target effects. The search focuses on the 20 bp target sequence involved in base pairing with the gRNA but excludes the PAM 5′-NGG-3′. The PAM functions as a recognition site outside of the targeted element and does not give specificity for nuclease cleavage (Shah et al., 2013). Moreover, the CRISPR/Cas9 system accepts at least three mismatches in the 20 bp DNA target sequence. Most CRISPR/Cas9 design tools take this into account and propose only specific gRNA designs that do not bind theoretical off-target sites with more than 17 bp identity anywhere else in the genome. Such state-of-the art design is effortless if the gene is unique in the genome, but it becomes rather challenging if the gene has one or more paralogs. This also means that the design is generally easier for diploid genomes without recent duplications than for recently duplicated or polyploid genomes. In silico genome analysis of potential target sequences in dicots and monocots has confirmed that, as expected, larger genomes contain more PAMs and more potential targets (Bortesi et al., 2016). High specificity of between 87.3% and 94.3% was observed in relatively simple genomes of Arabidopsis, rice, tomato, and soybean, whereas maize, a recent allotetraploid with high levels of repetitive DNA, revealed only 29.5% specific targeting (Bortesi et al., 2016).
Analysis of expected off-target sites, with only one to several mismatches with the primary target, has revealed that the position of the mismatches in the sequence is significant. Mismatches in the seed sequence (“seed” is defined as the 12 bp close to the PAM) are generally not supported or poorly supported by the sgRNA/Cas9 complex (Tsai et al., 2015), causing mutation less frequently at off-target sites, although in some cases mutations have been observed, as in barley (Lawrenson et al., 2015), soybean (Jacobs et al., 2015), and rice (Xie and Yang, 2013). Unwanted off-target mutations become more frequent when mismatches are located far from the seed region (Zhang et al., 2014).
To clarify the off-targeting issue in crops, recent investigations have screened progenies of CRISPR/Cas9 knockout in polyploid species. A study of CRISPR mutation frequency and mutation heritability of TaGW2, TaLpx-1, and TaMLO genes in the allohexaploid wheat was conducted (Wang et al., 2018). The results were different for the three genes: highly conserved for TaGW2 (target sequence was specific for all three genomes), moderate for TaLpx-1 (target sequence specific in two genomes), and low for TaMLO. The study showed the flexibility of CRISPR/Cas9 technology for implementing complex gene editing where the majority of genes have more than three homologous copies. Also, the gene editing process was investigated across generations: new mutant variants were recovered across multiple gene targets suggesting the transgenerational activity of CRISPR/Cas9 (Wang et al., 2018).
Another study on target accuracy and efficiency was performed in rice on paralogs OsBEIIb and OSBEIIa (Baysal et al., 2016). The study reveals the discrepancy in gRNA prediction and mutagenesis efficiency, confirming that gRNA with low predicted efficiency can achieve high mutation frequency even though the prediction suggested different targets with high mutagenesis scores. Empirical testing seems necessary in order to avoid putative gRNA inefficiency. Moreover, the authors also investigated off-target mutagenesis, reporting no mutation in the OSBEIIa paralog when only OsBEIIb was targeted, confirming the high accuracy of the strategy. CRISPR accuracy has been shown also in tomato (Čermák et al., 2015; Pan et al., 2016; Nekrasov et al., 2017).
To conclude, the CRISPR/Cas9 complex can bind with lower efficiency sequences with one to three mismatches. Therefore, expected off-target mutations do occur but can be avoided by rigorous design of the CRISPR/Cas9 tool. Unexpected off-target mutations do not occur at a frequency above the spontaneous mutation rate of plants.
Plant Transformations: Conventional and Alternative Techniques
The bottleneck in the application of CRISPR/Cas9 technology to a wide range of crops is clearly the regeneration of fertile plants from the cells into which the CRISPR/Cas9 tool has been introduced (Altpeter et al., 2016). Consequently, the efficiency of the entire process remains very species- and genotype-dependent, meaning that in many crop species only a few lab varieties are accessible to CRISPR/Cas9 technology. Other important parameters are the quality of the design of the CRISPR/Cas9 tool and the method chosen for its introduction into the plant cell. As in conventional transgenesis, the introduction of the CRISPR/Cas9 tool can be achieved by the Agrobacterium-mediated and biolistic transformation of explants, or by direct transformation of protoplasts. The latter two systems have the advantage that not only can the DNA coding for Cas9 and the sgRNA be transferred, but this also applies for ribonucleoproteins (RNPs), i.e., an in vitro assembled complex of Cas9 protein with an sgRNA (Malnoy et al., 2016; Svitashev et al., 2016; Liang et al., 2017), or intermediate versions such as a DNA or RNA coding for Cas9 and an RNA representing the sgRNA (Svitashev et al., 2015; Zhang et al., 2016). In addition, both biolistics and direct DNA transfer permit an increase in the ratio of repair matrix DNA over DNA encoding Cas9 and sgRNA readily, thereby favoring HR over NHEJ/MMEJ.
In maize, ISU Maize CRISPR is a high efficiency public platform using Agrobacterium-mediated transformation (Char et al., 2017). The main genotypes used for immature embryo transformation are A188, A634, H99, W117 (Ishida et al., 2007), B104 and the hybrid Hi-II (Char et al., 2017). Private companies seem to prefer biolistic transformation to Agrobacterium-mediated transformation in the case of gene editing with donor template (Shi et al., 2017), particularly where multiple copies of donor template DNA molecules can be delivered (Svitashev et al., 2015). Even though both transformation processes have decent efficiencies nowadays, they remain limited to the above genotypes with poor agronomic traits. This limitation has recently been overcome by the overexpression of Baby boom (Bbm) and Wuschel2 (Wus2) genes, which stimulated callus growth and increased the overall transformation frequency in maize, including in recalcitrant genotypes. Proof of concept has also been provided for enhanced transformation in sorghum (Lowe et al., 2016).
In rice, most genotypes can easily be transformed both via Agrobacterium-mediated transformation and by biolistic methods. In order to achieve CRISPR-mediated HR the DNA template is normally introduced via the biolistic method to increase its copy number in the host (Baysal et al., 2016). As for maize, but involving a higher number of studies, protoplast transient assay is becoming an efficient tool for testing CRISPR-target before starting the transformation of embryos or scutellum derived calli by Agrobacterium or particle bombardment (Gao et al., 2013; Jiang et al., 2013; Xie and Yang, 2013; Zhou et al., 2014; Lowder et al., 2015; Li et al., 2016; Luo et al., 2016; Wang et al., 2016). Regeneration of rice protoplasts is still very challenging, but important optimization efforts may render it feasible in the near future. In wheat, although very high Agrobacterium-mediated transformation efficiencies of up to 90% have been reported for specific wheat genetic backgrounds (Ishida et al., 2015a,b), particle bombardment has been more widely accepted as the standard method in wheat genetic transformation (Hakam et al., 2015; Wang et al., 2018). Remarkable success has been achieved by particle bombardment of both immature embryos and callus cells to obtain transient expression of the CRISPR/Cas9 DNA, and transgene-free homozygous mutant T0 plants have been generated in the absence of any selection (Zhang et al., 2016). Three studies have reported CRISPR mutagenesis in barley by using Agrobacterium-mediated transformation of immature embryos (Lawrenson et al., 2015; Holme et al., 2017; Kapusi et al., 2017), while in Kapusi et al. (2017) a comparison with particle bombardment was carried out. Higher numbers of mutants were reported with the Agrobacterium-mediated compared to the biolistic transformation approach.
In conclusion, although preferences for certain delivery methods exist for certain species, efficiency is not only linked to the technique itself, but also to the know-how of a given lab as regards a given technique. Polyethylene glycol (PEG) or electroporation-mediated DNA transient expression in protoplasts have proven very useful for the evaluation of the efficiency of CRISPR/Cas9 designs (Malnoy et al., 2016). The importance of preliminary screens will certainly increase with the foreseeable shift from targeted mutagenesis to repair matrix based genome editing, which will increase the number of events to analyze due to lower efficiency. RNP technology has been established in plants and may help toward exemption from regulatory oversight, but its efficiency needs to be improved to make it a routine tool.
CRISPR/Cas-Based Strategies Conferring Biotic Resistance
Biotic stresses including viral, fungal, and bacterial diseases are responsible for losses ranging from 20% to 40% of global agricultural productivity (Savary et al., 2012). Conferring host plant resistance to pathogens can reduce the impact of disease on crop development and yield, thereby addressing the challenge of feeding the world’s growing population.
Advances in genome editing tools have opened new ways to achieve the improvement of resistance in crops. In recent years, the CRISPR/Cas system has been employed to respond to several agricultural challenges, including the achievement of improved biotic stress resistance (Arora and Narula, 2017). The application of CRISPR/Cas tools has mainly been explored against virus infection, followed by efforts to improve fungal and bacterial disease resistance. Recent studies demonstrating the power of the CRISPR/Cas technology in establishing resistance to these pathogen categories will be further discussed below.
Virus Resistance via CRISPR/Cas
Plant viruses are a serious threat to many economically important staple and specialty crops. Based on their genome nature, they are classified into six major groups: double-stranded DNA (dsDNA) viruses with no plant viruses in this group, single-stranded DNA (ssDNA), reverse-transcribing viruses, double-stranded RNA (dsRNA), negative sense single-stranded RNA (ssRNA-), and positive sense single-stranded RNA (ssRNA+) viruses (Roossinck et al., 2015). Most studies involving CRISPR-edited plants for virus resistance have targeted ssDNA geminivirus genomes (Ali et al., 2015, 2016; Baltes et al., 2015; Ji et al., 2015) (Table 1).
Table 1
| Plant species | Virus | Target gene | Gene function | Strategy | Reference |
|---|---|---|---|---|---|
| Nicotiana benthamiana and Arabidopsis thaliana | BeYDV | CP, Rep, and IR | RCA mechanism | Agrobacterium-mediated transformation of leaves with Cas9/gRNA expression plasmid vectors | Ji et al., 2015 |
| Nicotiana benthamiana | BSCTV | LIR and Rep/RepA | RCA mechanism | Agrobacterium-mediated transformation of leaves with Cas9/gRNA expression plasmid vectors | Baltes et al., 2015 |
| Nicotiana benthamiana | TYLCV BCTV MeMV | CP, Rep and IR | RCA mechanism | Agrobacterium-mediated transformation of leaves with a TRV vector in Cas9 overexpressing plants | Ali et al., 2015 |
| Nicotiana benthamiana | CLCuKoV MeMV TYLCV | CP, Rep, and IR | RCA mechanism | Agrobacterium-mediated transformation of leaves with a TRV vector in Cas9 overexpressing plants | Ali et al., 2016 |
| Nicotiana benthamiana | TuMV | GFP1, GFP2, HC-Pro, CP | Replication mechanism | Agrobacterium-mediated transformation of leaves with a TRV vector in Cas13a overexpressing plants | Aman et al., 2018 |
| Nicotiana benthamiana and Arabidopsis thaliana | CMV TMV | ORF1, 2, 3, CP and 3′UTR | Replication mechanism | Agrobacterium-mediated transformation of leaves with FnCas9/gRNA expression binary vectors Floral dipping for Arabidopsis | Zhang et al., 2018 |
| Cucumis sativus | CVYV ZYMV PRSV-W | eIF4E | Host factor for RNA viruses translation | Agrobacterium-mediated transformation of cut cotyledons (without embryo) with Cas9/gRNA binary vectors | Chandrasekaran et al., 2016 |
| Arabidopsis thaliana | TuMV | eIF(iso)4E | Host factor for RNA viruses translation | Agrobacterium-mediated transformation with Cas9/gRNA recombinant plasmid binary vectors (floral dipping) | Pyott et al., 2016 |
| Oryza sativa L. japonica | RTSV | eIF4G | Host factor for RNA viruses translation | Agrobacterium-mediated transformation of immature embryos with Cas9/gRNA expression plasmid vectors | Macovei et al., 2018 |
CRISPR/Cas9 applications for virus resistance.
BeYDV, bean yellow dwarf virus; BSCTV, beet severe curly top virus; TYLCV, tomato yellow leaf curl virus; BCTV, beet curly top virus; MeMV, Merremia mosaic virus; TRV, tobacco rattle virus; CLCuKoV, cotton leaf curl Kokhran virus; TuMV, turnip mosaic virus; CMV, cucumber mosaic virus; TMV, tobacco mosaic virus; CVYV, cucumber vein yellowing virus; ZYMV, zucchini yellow mosaic virus; PRSV-W, papaya ring spot mosaic virus-W; PVX, potato virus X; TCV, turnip crinkle virus; CMV, cucumber mosaic virus; RTSV, rice tungro spherical virus; CP, coat protein; Rep, replication association protein; IR, intergenic region; RCA, rolling-circle amplification; LIR, long intergenic region; GFP1, green fluorescent protein 1; GFP2, green fluorescent protein 2; HC-Pro, helper component proteinase silencing suppressor; ORF, open reading frame; UTR, untranslated terminal repeat; eIF4E, eukaryotic translation initiation factor 4E; eIF4G, eukaryotic translation initiation factor 4G.
Geminiviridae is a large family of plant viruses causing worldwide crop losses among several important families, such as Cucurbitaceae, Euphorbiaceae, Solanaceae, Malvaceae, and Fabaceae (Zaidi et al., 2016). The virus genome is replicated through a rolling-circle amplification mechanism via a dsDNA replicative form, or by recombination-mediated replication (Hanley-Bowdoin et al., 2013). The most important genus of geminiviruses in economic terms is Begomovirus. Begomoviruses infect dicotyledonous plants via the sweet potato/tobacco/silverleaf whitefly (Bemisia tabaci) and are mainly found associated to the phloem of infected plants (Gilbertson et al., 2015). Their genome is organized in one (A, monopartite) or two (A and B, bipartite) components containing a common region of ∼220 bp (Fondong, 2013).
The first two studies focusing on resistance to geminiviruses, beet severe curly top virus (BSCTV) and bean yellow dwarf virus (BeYDV) in model plants N. benthamiana and Arabidopsis were reported by Baltes et al. (2015) and Ji et al. (2015) (Table 1). Ji et al. (2015) screened 43 candidate sgRNA/Cas9 target sites in coding and non-coding regions of the BSCTV genome. All the sgRNA/Cas9 constructs reduced virus accumulation in inoculated leaves at varying levels, but a greater resistance to virus infection was observed in Nicotiana and Arabidopsis plants showing the highest levels of expression of Cas9 and sgRNAs. Similar findings were described by Baltes et al. (2015), who employed 11 sgRNAs targeting Rep motifs, Rep-binding sites, hairpin, and the nonanucleotide sequence of BeYDV, and reported up to 87% reduction in the targeted viral load in N. benthamiana.
Two recent works have also employed a CRISPR/Cas9 approach for achieving resistance to begomoviruses (Ali et al., 2015, 2016) (Table 1). Both studies were based on the strategy of expressing the CRISPR/Cas9 system in the host cell nucleus to target and cleave the virus genome during replication. Ali et al. (2015) developed sgRNA molecules delivered via a tobacco rattle virus (TRV) vector into N. benthamiana plants stably overexpressing the Cas9 endonuclease. SgRNAs were specific for different tomato yellow leaf curl virus (TYLCV) coding and non-coding sequences, targeting the viral capsid protein (CP), the RCRII motif of the replication protein (Rep) and the intergenic region (IR). All sgRNAs were able to interfere with TYLCV genome sequences, but targeting the stem-loop invariant sequence contained in the IR caused a more significant reduction of viral replication and accumulation. The same CRISPR/Cas9 system was tested for targeting simultaneously the monopartite beet curly top virus (BCTV) and the bipartite Merremia mosaic virus (MeMV), geminiviruses that share the same stem-loop sequence in the IR. The results showed attenuated symptoms for both viruses, demonstrating that mixed infection immunity can be developed via a single sgRNA specific for conserved sequences of multiple viral strains.
Furthermore, Ali et al. (2016) analyzed not only the targeting efficiencies of the CRISPR/Cas9 tool but also the emergence of mutated viruses capable of replication and systemic movement. The CRISPR/Cas9 tool was designed to interfere with different coding and non-coding sequences of cotton leaf curl Kokhran virus (CLCuKoV), MeMV, and different severe and mild strains of TYLCV. The work revealed that when the sgRNA/Cas9 complex edited sites in the coding regions of all viruses, virus variants were generated capable of replicating and moving to escape the CRISPR/Cas9 machinery. Conversely, no novel variants were detected in N. benthamiana plants carrying sgRNAs addressing the IR sequences. Even though the NHEJ machinery repaired the double strand breaks caused by the Cas9 protein, the IR-repaired variants generated virus genomes unable to replicate, thus providing a better overall interference with the viral life cycle.
Protection against RNA viruses has seemed more difficult to achieve, since the classical SpCas9 from Streptococcus pyogenes only recognizes dsDNA. However, the search for and characterization of related nucleases has led to the discovery of enzymes that can bind to and cut RNA, such as FnCas9 from Francisella novicida or LwaCas13a from Leptotrichia wadei. A first report demonstrating resistance to RNA viruses (Zhang et al., 2018) (Table 1) expressed FnCas9 and RNA-targeting sgRNAs specific for cucumber mosaic virus (CMV) and tobacco mosaic virus (TMV) in N. benthamiana and Arabidopsis plants. Transgenic plants showed CMV and TMV accumulation reduced by 40–80% compared with control plants. Furthermore, the resistance obtained by expressing the sgRNA-FnCas9 system was quite stable and still active in the T6 generation. Importantly, Zhang et al. (2018) observed that the endonuclease activity of FnCas9 was not required for interference with the CMV genome, whereas its RNA-binding activity was essential, meaning that this particular application of FnCas9 can be considered as a CRISPR interference (CRISPRi) tool, similar to catalytically inactive SpCas9 proteins programmed to mitigate gene expression (Larson et al., 2013). The use of a catalytically inactive variant of FnCas9 has the advantage of limiting the onset of mutated viral variants capable of escaping CRISPR/Cas9. Moreover, in contrast with the previously described interference with geminivirus replication in the nucleus, no nuclear localization signal is necessary for FnCas9, which interferes with the RNA viruses in the cytoplasm.
Similar work has been carried out with Cas13a. Aman et al. (2018) exploited this RNA-guided ribonuclease to manipulate the turnip mosaic virus (TuMV) RNA genome (Table 1). Four different viral genomic regions were targeted: two targets in the green fluorescent protein (GFP) region, one in the helper component proteinase silencing suppressor (HC-Pro), and one in the coat protein (CP). The most efficient virus interference was observed with CRISPR RNA editing HC-Pro and GFP2 genes and resulted in a reduced replication and spread of TuMV in tobacco leaves. Furthermore, due to the innate ability of Cas13 to process pre-CRISPR RNA into functional CRISPR RNA, the multiplex targeting of several viral mRNA could be markedly improved through this alternative system (Aman et al., 2018).
All the systems aiming at protection against viruses described so far require the maintenance of a transgene for Cas9 and sgRNA in the genome of the crop plants, rendering them subject to genetically modified organism (GMO) regulation. A second strategy for the achievement of viral disease resistance consists in modifying plant genes that will generate virus resistance traits, to segregate the CRISPR/Cas9 tool and to release non-transgenic mutants in the field (Chandrasekaran et al., 2016; Pyott et al., 2016; Macovei et al., 2018) (Table 1). Plant host factors are required by RNA viruses to maintain their life cycle, including the eukaryotic translation initiation factors eIF4E, eIF(iso)4E and eIF4G (Sanfacon, 2015). Chandrasekaran et al. (2016) developed cucumber plants resistant to potyviruses by mutating independently two different sites of the host susceptibility gene eIF4E by CRISPR/Cas9. Non-transgenic Cucumis eif4e mutant plants were obtained by segregation of the CRISPR/Cas9 tool by three generations of backcrossing, making these plants safe for human consumption and for release into the environment, according to the authors. When challenged with viruses from the Potyviridae family, cucumber vein yellowing virus (CVYV), zucchini yellow mosaic virus (ZYMV), and papaya ring spot mosaic virus-W (PRSV-W), homozygous eif4e mutants showed immunity. Conversely, heterozygous knockout plants and non-mutant plants did not reveal any resistance to these viruses.
A similar editing approach was utilized by Pyott et al. (2016) in order to introduce site-specific mutations at the closely related eIF(iso)4E locus in Arabidopsis plants. Both 1 bp insertions and 1 bp deletions in eIF(iso)4E conferred complete resistance to the single-stranded RNA potyvirus (+ssRNA) TuMV and no off-target modification was detected in this study. Furthermore, homozygous T3 eIF(iso)4E mutants did not significantly differ in growth and development compared to wild-type plants.
Recently, Macovei et al. (2018) have developed new sources of resistance to rice tungro spherical virus (RTSV) through mutagenesis of eIF4G alleles in rice plants. The RTSV-resistant T2 plants obtained did not show any detectable mutation in the off-target sites and were negative when tested for the presence of Cas9. Furthermore, after inoculation with RTSV, agronomic parameters such as plant height and grain yield were enhanced in the edited rice plants compared to their wild-type counterparts under glasshouse conditions.
The advantage of knocking out host susceptibility genes is that this is a relatively simple method that renders following the mutation easy. The loss of a host factor required for the viral life cycle is a form of recessive resistance that should be more durable than that of dominant R genes because viruses undergo a lower selective pressure, preventing their evolution to hinder host defense mechanisms. A possible disadvantage of the knockout strategy is that it may negatively influence plant vigor, supporting the selection of virus variants breaking the resistance, as already observed in nature (Abdul-Razzak et al., 2009). Pyott et al. (2016) and Macovei et al. (2018) did not observe any significant difference in growth defects between mutants and normal plants, although further investigations should be carried out in order to test the durability of this edited recessive resistance.
Resistance to Fungi Through CRISPR/Cas
Fungal pathogens are responsible for numerous diseases such as mildew, smut, rust, rot and many more. These diseases not only cause dramatic yield losses annually throughout the world but also compromise the quality of the harvested products. Moreover, mycotoxigenic fungi represent a serious concern due to the production of secondary metabolites known as mycotoxins, which cause severe health problems in humans and animals after exposure to contaminated food and feed. Several strategies have been evolved to enhance fungal resistance in plant species based on the current knowledge of molecular mechanisms implicated in plant-pathogen interaction. Potential candidate genes and gene products involved in plant resistance against fungi have been described, and nowadays these are prime targets for editing through the CRISPR/Cas9 approach.
As previously partially discussed, MLO loci have been targeted by RNA-guided Cas9 endonuclease in three different plant species: bread wheat, tomato, and grapevine (Wang et al., 2014; Malnoy et al., 2016; Nekrasov et al., 2017) (Table 2). MLO encodes a protein with seven transmembrane domains localized in the plasma membrane and is ubiquitously present in monocots and dicots (Acevedo-Garcia et al., 2014). It had previously been reported that MLO were susceptibility (S) genes and that homozygous loss-of-function mutants had significantly increased resistance to powdery mildew in barley, Arabidopsis and tomato (Piffanelli et al., 2004; Consonni et al., 2006; Bai et al., 2008). Bread wheat plants mutated by CRISPR/Cas9 in one (TaMLO-A1) of the three MLO homeoalleles showed improved resistance to Blumeria graminis f. sp. tritici infection, a finding that once again demonstrated the important role of TaMLO genes in powdery mildew disease (Wang et al., 2014). In tomato, SlMlo1, previously identified as the most important of 16 SlMlo genes, was targeted at two sites and a deletion of 48 bp was obtained. The edited plants were self-pollinated in order to generate CRISPR/Cas cassette-free individuals. This new non-transgenic variety, “Tomelo,” was fully resistant to Oidium neolycopersici. Furthermore, off-target analysis did not reveal any effect on the genomic regions outside the SlMlo1 locus (Nekrasov et al., 2017). In grapevine, the molecular feasibility of VvMLO7 knockout has been demonstrated through CRISPR/Cas9 RNP in protoplasts, but no plants have been regenerated (Malnoy et al., 2016). Parallel experiments with RNAi plants showed that the loss of VvMLO7 reduced susceptibility to Erysiphe necator in grapevine (Pessina et al., 2016).
Table 2
| Plant species | Fungus | Target gene | Gene function | Strategy | Reference |
|---|---|---|---|---|---|
| Triticum aestivum | Powdery mildew (Blumeria graminis f. sp. tritici) | MLO-A1 | Susceptibility (S) gene involved in powdery mildew disease | Particle bombardment of immature wheat embryos with Cas9/gRNA expression plasmid vectors | Wang et al., 2014 |
| Solanum lycopersicum | Powdery mildew (Oidium neolycopersici) | MLO1 | Major responsible for powdery mildew vulnerability | Agrobacterium-mediated transformation of cotyledons with Cas9/gRNA expression plasmid vectors | Nekrasov et al., 2017 |
| Vitis vinifera | Powdery mildew (Erysiphe necator) | MLO-7 | Susceptibility (S) gene involved in powdery mildew disease | PEG-mediated protoplast transformation with CRISPR ribonucleoproteins | Malnoy et al., 2016 |
| Vitis vinifera | Gray mold (Botrytis cinerea) | WRKY52 | Transcription factor involved in response to biotic stress | Agrobacterium-mediated transformation of proembryonal masses with Cas9/gRNA expression binary vectors | Wang et al., 2018 |
| Theobroma cacao | Black pod disease (Phytophthora tropicalis) | NPR3 | Regulator of the immune system | Agrobacterium-mediated transient transformation of stage C leaves with Cas9/gRNA expression binary vectors | Fister et al., 2018 |
| Oryza sativa L. japonica | Rice blast disease (Magnaporthe oryzae) | SEC3A | Subunit of the exocyst complex | Protoplast transformation with Cas9/gRNA expression binary vectors | Ma et al., 2018 |
| Oryza sativa L. japonica | Rice blast disease (Magnaporthe oryzae) | ERF922 | Transcription factor implicated in multiple stress responses | Agrobacterium-mediated transformation of embryogenic calli with Cas9/gRNA expression binary vectors | Wang et al., 2016 |
CRISPR/Cas9 applications for fungal resistance.
MLO, MILDEW RESISTANT LOCUS; NPR3, non-expressor of pathogenesis-related 3; ERF922, ethylene responsive factor.
The RNP approach has also been used for editing DIPM-1, DIPM-2, and DIPM-4 genes in apple protoplasts in order to confer resistance to fire blight disease (Malnoy et al., 2016). Again, only the molecular analysis attesting mutations has been carried out, not disease assay on regenerated plants. In perennial crops such as grapevine and apple, which take several years to flower, the transient introduction of genome editing tools in protoplasts is particularly interesting, since the segregation of stably integrated CRISPR/Cas9 cassettes by backcrosses would take a lot longer than in annual crops with generation times of only a few months. Secondly, the delivery of Cas9/sgRNA complex as RNP is a rapid approach, making possible the achievement of transformed protoplasts and the evaluation of sgRNA efficiency within 1 or 2 days. Thirdly, no foreign DNA is integrated into the genome and the Cas9/sgRNA complexes can be degraded rapidly during the cell culture regeneration process. Furthermore, even in transient approaches, the employment of plasmids can sometimes cause their undesired integration into the host genome, and the prolonged presence of CRISPR/Cas9 tools in the genome increases the risk of off-target mutations, while the CRISPR/Cas9 RNP shows improved on-target specificity. The drawback of this approach is the need to optimize plant regeneration protocols in order to apply this technology.
An example of the successful protection of grapevine by the CRISPR/Cas9 system is the VvWRKY52 transcription factor, which was targeted by four gRNAs (Wang et al., 2018) (Table 2). About 21% of the transgenic plants showed biallelic mutations and were more resistant to Botrytis cinerea compared to the monoallelic mutants. No marked difference was observed in phenotype between wild-type and biallelic mutant plants, confirming the efficiency of the CRISPR/Cas9 strategy in woody plants with long reproductive cycles.
A further strategy to expedite genome editing application in slow generation tree crops is the employment of transient leaf transformation coupled to disease assays as demonstrated in Theobroma cacao (Fister et al., 2018) (Table 2). The authors reported for the first time the transient introduction of CRISPR/Cas9 components into cacao leaves targeting the Non-Expressor of Pathogenesis-Related 3 (NPR3) gene, a suppressor of the immune system, and obtained leaves with increased resistance to Phytophthora tropicalis. This new system of in vivo mutagenesis in adult cacao trees is a fast and useful technique for validating sgRNA design and observing CRISPR mutagenized phenotypes. It encouraged the authors to regenerate genome-edited somatic embryos to validate the observed results at whole-plant level.
Plants resistant to rice blast disease were generated through CRISPR/Cas9 by disrupting OsERF922 and OsSEC3A genes in rice (Wang et al., 2016; Ma et al., 2018) (Table 2). Ossec3a mutant plants disrupted in a putative subunit of a complex involved in exocytosis, revealed a pleiotropic phenotype including improved resistance against Magnaporthe oryzae, higher levels of salicylic acid (SA) content and up-regulation of pathogenesis- and SA-related genes, but also dwarf stature (Ma et al., 2018). In contrast, no alteration of a number of agronomic traits was observed in T1 and T2 transgene free plants mutated in the ethylene responsive factor (ERF)922, a transcription factor implicated in multiple stress responses. The mutant plants had a reduced number of blast lesions at both seedling and tillering stages (Wang et al., 2016). Overall, these results demonstrate the powerful and advantageous application of the CRISPR/Cas9 system for crop improvement as regards fungal disease resistance.
Resistance to Bacteria Through CRISPR/Cas
Among the bacterial species living on earth, just a few hundred are involved in crop damage, which often reveals multiple symptoms of disease (Schloss and Handelsman, 2004). Phytopathogenic bacteria are difficult to control, mainly because of undetected asymptomatic infections and the lack of suitable agrochemicals. Generally speaking, bacteriological plant control is based on prevention and exclusion of the pathogen by using genetic resistance, agronomic practices, and biocontrol agents (Kerr, 2016).
Phytopathogenic bacteria can be classified as crop specific, such as Clavibacter michiganensis, which is the causal agent of tomato bacterial ring rot; polyphagous specific, such as Ralstonia solanacearum, which causes disease in multiple monocot and dicot species; and “kingdom crosser,” such as Dickeya dadantii, an entomo-phytopathogen, which can affect plants and animals.
Relatively few studies (Table 3) have been published on the application of the CRISPR/Cas system to counteract crop bacterial diseases. CRISPR/Cas9 mutagenesis of OsSWEET13 has been performed in rice to achieve resistance to bacterial blight disease caused by γ-proteobacterium Xanthomonas oryzae pv. oryzae (Zhou et al., 2015). OsSWEET13 is a susceptibility (S) gene encoding a sucrose transporter involved in plant-pathogen interaction. X. oryzae produces an effector protein, PthXo2, which induces OsSWEET13 expression in the host and the consequent condition of susceptibility. In a previous work concerning OsSWEET14 promoter mutagenesis adopting a TALEN approach, the disruption of this gene rendered the X. oryzae effector unable to bind OsSWEET14 and ultimately resulted in disease resistance (Li et al., 2012). Similarly, Zhou et al. (2015) obtained a null mutation in OsSWEET13 in order to better explore PthXo2-dependent disease susceptibility, and resultant mutants were resistant to bacterial blight. Further genome editing strategies for multiplexed recessive resistance using a combination of the major effectors and other resistance (R) genes will be the next step toward achieving bacterial blight resistance.
Table 3
| Plant species | Fungus | Target gene | Gene function | Strategy | Reference |
|---|---|---|---|---|---|
| Oryza sativa | Bacterial blight (Xanthomonas oryzae pv. oryzae) | SWEET13 | Sucrose transporter gene | Agrobacterium-mediated transformation of embryogenic callus with Cas9/gRNA expression plasmid vectors and TALEN | Li et al., 2012; Zhou et al., 2015 |
| Citrus paradisi | Citrus canker (Xanthomonas citri subspecies citric) | LOB1 | Susceptibility (S) gene promoting pathogen growth and pustule formation | Agrobacterium-mediated transformation of epicotyl with Cas9/gRNA expression plasmid vectors | Jia et al., 2016 |
| Citrus sinensis Osbeck | Citrus canker (Xanthomonas citri subspecies citric) | LOB1 | Susceptibility (S) gene promoting pathogen growth and pustule formation | Agrobacterium-mediated transformation of epicotyl with Cas9/gRNA expression plasmid vectors | Peng et al., 2017 |
| Malus domestica | Fire blight (Erwinia amylovora) | DIPM-1 DIPM-2 DIPM-4 | Susceptibility factor involved in fire blight disease | PEG-mediated protoplast transformation with CRISPR ribonucleoproteins | Malnoy et al., 2016 |
CRISPR/Cas9 applications for bacterial resistance.
CsLOB1, Lateral Organ Boundaries 1; DIPM, DspE-interacting proteins of Malus.
Two recent works have reported the employment of CRISPR/Cas9 with the aim of producing citrus plants resistant to citrus bacterial canker (CBC). CBC is caused by Xanthomonas citri subsp. citri (Xcc) and is the most widespread disease among commercial cultivars. In the first work, Jia et al. (2016) generated canker resistant mutants by editing the PthA4 effector binding elements in the promoter of the Lateral Organ Boundaries 1 (CsLOB1) gene in Duncan grapefruit. Mutated lines showed a decrease in typical canker symptoms 4 days post inoculation with Xcc, and no further phenotypic alterations were detectable. Furthermore, no potential off-target mutations in other LOB family genes were found by PCR-sequencing. The second work, by Peng et al. (2017), confirmed the link between CsLOB1 promoter activity and CBC disease susceptibility in Wanjincheng orange (Citrus sinensis Osbeck). The complete deletion of the EBEPthA4 sequence from both CsLOB1 alleles induced resistance enhancement to CBC. Moreover, no alteration in plant development was observed after CsLOB1 promoter modification. Additional efforts will be required to generate non-transgenic canker-resistant citrus varieties for facilitating their agronomic application in CBC prevention.
Future Prospects
In an era marked by political and societal pressure to reduce the use of pesticides, crop protection by genetic improvement provides a promising alternative with no obvious impact on human health or the environment. Genome editing is one of the genetic levers that can be adopted, and disease resistance is frequently cited as the most promising application of CRISPR/Cas9 technology in agriculture. There are three main reasons for this: firstly, scientific knowledge of the molecular mechanisms underlying numerous pathosystems is sufficiently advanced to enable the proposal of genes to be edited in order to achieve resistance. Secondly, disease resistance can frequently be achieved by the modification of a single gene, which is technically less challenging. This is similar to the modification of metabolic pathways, where the editing of a single gene can also have an all-or-nothing effect, but different from abiotic stress tolerance, where generally numerous genes have to be modified in a coordinated fashion to achieve incremental improvements. Thirdly, targeted mutagenesis, the only use of CRISPR/Cas9 technology at present mastered with respect to crops, is readily applicable to disease resistance, since the inactivation of susceptibility genes leads to protection. For other agriculturally interesting traits the achievement of positive effects by the loss-of-function of genes is a more delicate matter. However, acting as the spearhead of genome editing in crops also puts a certain responsibility on plant pathologists.
The first challenge is to demonstrate that the promises made by proofs of concept in confined environments can be maintained under field conditions. It is one thing to show that the population of a pathogen or the size of disease lesions is reduced in a greenhouse and another to protect a crop year after year under varying environmental conditions. Field tests are also necessary for correct evaluation of the agronomic fitness of the edited crops. Most of the genes inactivated by CRISPR/Cas9 technology in order to obtain disease resistance are likely to have roles in the physiology of the plant other than that linked to the life cycle of the pathogen. For example, triple knockouts of wheat TaMLO were not only resistant to powdery mildew but also showed leaf chlorosis (Wang et al., 2014), whereas EMS-induced triple mutants with non-conservative point mutations in TaMLO did not show obvious pleiotropic phenotypes (Acevedo-Garcia et al., 2017). Therefore, encouraging greenhouse observations of plant development or measurements of key parameters such as height, leaf area or grain weight absolutely must be confirmed under field conditions by multi-environmental yield trials in order to measure the relative importance of negative side effects. A final limitation of many published proofs of concept is that they involve lab varieties, which can easily be regenerated after the introduction of Cas9 and sgRNA, but which often have only a limited agronomic value. It remains to be shown that the phenotypic effects are maintained in elite lines under field conditions.
The second challenge is the durability of the disease resistances, and their agronomic management. This challenge needs to be dealt with seriously, in order to convince a public often hostile to this technology. Durability is not a specific aspect of resistance genes obtained by genome editing, and the answers are the same as for introgressed resistance genes discovered in the genetic variability of the species: (i) the stacking of several resistance genes, preferably with different modes of action, (ii) a focus on systems other than NBS-LRR receptor kinases known to break down rapidly, and (iii) good agronomic practices, including, in particular, crop rotation and the concomitant use of biocontrol agents. An example of two independent CRISPR/Cas9-derived resistances against the same disease are the knockouts of TaMLO (Wang et al., 2014) and TaEDR1 (Zhang et al., 2017), both conferring resistance to powdery mildew in wheat. Beyond the creation of novel alleles conferring protection, CRISPR/Cas9 technology can also be helpful in the stacking process itself. In contrast with the introgression of conventional resistance genes, the technology not only avoids genetic drag on neighboring regions with potentially negative impacts on agronomic performance, but also permits the simultaneous creation of multiple resistances in a single generation by multiplexing, i.e., the parallel use of several sgRNAs targeting different genes. Admittedly, multiplexing becomes more challenging with increasing ploidy levels, and in the above example in hexaploid wheat (A, B, and D genome), three TaMLO genes and three TaEDR1 genes would need to be modified in parallel.
The third challenge is to overcome the present technical limitation regarding targeted mutagenesis and to implement true genome editing in crop plants. Targeted mutagenesis introduces random mutations (generally short insertions or deletions) at a predetermined site of a given gene, leading generally to loss-of-function, whereas true genome editing introduces predetermined base changes at one or several specific positions in a gene. For example, the elongation initiation factor 4E (eIF4E) is necessary for the translation of RNA into protein for both the host cell and single-stranded RNA viruses of the Potyviridae family. As described above, loss-of-function of eIF4E by targeted mutagenesis has been achieved in several model and crop species, consistently conferring resistance to potyviruses but also impacting the host plants to varying degrees. The specific modification of amino acids known to be important for the translation of viral but not host proteins would permit driving resistance to potyviruses without affecting plant physiology (Bastet et al., 2017). The expression of a transgene carrying a synthetic allele with six mutations in an Arabidopsis eif4e mutant validated the concept (Bastet et al., 2018), demonstrating indirectly the potential benefit of genome editing over targeted mutagenesis. However, at present true genome editing by HR is still hampered by very low efficiencies in plants, although it has recently become routine in many animal species. Continued efforts to improve its efficiency, for example by the use of lig4 (Endo et al., 2016) or polQ mutations (Saito et al., 2017), or a copy number increase of the repair matrix by virus vectors (Čermák et al., 2015), are crucial to increasing the range of tools available to plant pathologists. Base editing, to date permitting C to T and A to G transitions in plants, is more limited in scope but has recently emerged as a readily available alternative for certain editing projects (Zhang et al., 2017; Hua et al., 2018).
The long term success of CRISPR/Cas9 technology in plant protection is dependent on new scientific knowledge. CRISPR/Cas9 technology can only be used if one knows which gene(s) to modify and which modification(s) to carry out in these genes in order to render plants resistant to disease. When pathogen resistance is achieved by the knock-out of one or several genes, inactivating mutations can easily be provoked by CRISPR-mediated specific DNA break and activation of the cell’s error prone DNA repair, based on NHEJ. In this case, CRISPR can be used to target and inactivate a single gene or large gene families, both through single gRNA which matches several targets, or by multiplexing the system by introducing several gRNAs simultaneously. On the contrary, when specific allelic variants are involved in resistance, CRISPR-DNA break can be coupled with the less frequent cell repair mechanism based on HDR. The DNA template for HDR should be introduced into the cell together with the effector nuclease. This permits the introduction of a custom-designed sequence into the genome. The use of HDR, compared to NHEJ, can indefinitely expand the possibility of the type of mutations inserted by CRISPR, as any sequence can be inserted into a site of choice. Nevertheless, HDR is still technically challenging due to its low efficiency, the difficulty of having a selective marker and the lack of multiplexing protocols. These are aspects that will need to be improved if CRISPR applications are to expand in plant breeding. Despite the recent judgment of the Court of Justice of the European Union issued that organisms created using genome editing techniques are to be regulated as GMOs (Callaway, 2018), anyhow continuous efforts in plant pathology are necessary, in order to identify and characterize the genes involved in plant pathogen interactions. For example, the past decade was marked by the discovery of hundreds of effector molecules that are synthesized by different classes of pathogens and transferred into the host cell. A major challenge is to identify the host proteins targeted by these effectors and to characterize the underlying genes, which are one of many possible targets for future genome editing approaches. New knowledge does not necessarily have to stem from the crop species of interest. For example, the targeted mutagenesis of wheat TaMLO was based on knowledge of another crop, barley, where Hvmlo mutant varieties have provided good protection against powdery mildew that has not yet broken down, and the modification of TaERF1 exploited knowledge from the model species Arabidopsis. These examples perfectly illustrate the added value of genome editing, which permits the enlargement of the gene pool of a crop species beyond all the available natural variability, by means of the transfer of knowledge acquired in other crops or model species.
Statements
Author contributions
VGB contributed by writing and editing the major part of the review. AL, AM, and PR organized and prepared some of the parts of this review. VB and PR critically revised the manuscript. AM and AL contributed to the design of the work’s layout and were responsible for obtaining final approval from the other contributors.
Funding
VGB was supported by the Doctoral School on the Agro-Food System (Agrisystem) of Università Cattolica del Sacro Cuore (Italy). PR declares (i) a pending patent application involving CRISPR/Cas9 as one of many biotechnologies to obtain haploid inducing maize lines, (ii) funding by the biotechnology company Meiogenix for research on targeting meiotic recombination to specific genome regions by CRISPR/Cas9 technology, and (iii) funding by the seed company Limagrain for research on haploid induction in maize.
Acknowledgments
We acknowledge funding by the Investissement d’Avenir Program of the French National Agency of Research for the project GENIUS (ANR-11-BTBR-0001_GENIUS).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
Abdul-RazzakA.GuiraudT.PeypelutM.WalterJ.HouvenaghelM. C.CandresseT.et al (2009). Involvement of the cylindrical inclusion (CI) protein in the overcoming of an eIF4E- mediated resistance against Lettuce mosaic potyvirus.Mol. Plant Pathol.10109–113. 10.1111/j.1364-3703.2008.00513.x
2
Acevedo-GarciaJ.KuschS.PanstrugaR. (2014). Magical mystery tour: MLO proteins in plant immunity and beyond.New Phytol.204273–281. 10.1111/nph.12889
3
Acevedo-GarciaJ.SpencerD.ThieronH.ReinstadlerA.Hammond KosackK.PhillipsA. L.et al (2017). Mlo-based powdery mildew resistance in hexaploid bread wheat generated by a non-transgenic TILLING approach.Plant Biotechnol. J.15367–378. 10.1111/pbi.12631
4
AliZ.AbulfarajA.IdrisA.AliS.TashkandiM.MahfouzM. M. (2015). CRISPR/Cas9-mediated viral interference in plants.Genome Biol.16:238. 10.1186/s13059-015-0799-6
5
AliZ.AliS.TashkandiM.ShanS.ZaidiA.MahfouzM. M. (2016). CRISPR/Cas9-mediated immunity to geminiviruses: differential interference and evasion.Sci. Rep.6:26912. 10.1038/srep26912
6
AltpeterF.SpringerN. M.BartleyL. E.BlechlA. E.BrutnellT. P.CitovskyV.et al (2016). Advancing crop transformation in the era of genome editing.Plant Cell281510–1520. 10.1105/tpc.16.00196
7
AmanR.AliZ.ButtH.MahasA.AljedaaniF.KhanM. Z.et al (2018). RNA virus interference via CRISPR/Cas13a system in plants.Genome Biol.19:1. 10.1186/s13059-017-1381-1
8
AroraL.NarulaA. (2017). Gene editing and crop improvement using CRISPR-Cas9 system.Front. Plant Sci.8:1932. 10.3389/fpls.2017.01932
9
BaiY.PavanS.ZhengZ.ZappelN. F.ReinstädlerA.LottiC.et al (2008). Naturally occurring broad-spectrum powdery mildew resistance in a central american tomato accession is caused by loss of Mlo function.Mol. Plant Microbe Interact.2130–39. 10.1094/MPMI-21-1-0030
10
BaltesN. J.HummelA. W.KonecnaE.CeganR.BrunsA. N.BisaroD. M.et al (2015). Conferring resistance to geminiviruses with the CRISPR–Cas prokaryotic immune system.Nat. Plants.1:15145. 10.1038/nplants.2015.145
11
BastetA.LedererB.GiovinazzoN.ArnouxX.German-RetanaS.ReinboldC.et al (2018). Trans-species synthetic gene design allows resistance pyramiding and broad-spectrum engineering of virus resistance in plants.Plant Biotechnol. J.10.1111/pbi.12896[Epub ahead of print].
12
BastetA.RobagliaC.GalloisJ. L. (2017). eIF4E resistance: natural variation should guide gene editing.Trends Plant Sci.22411–419. 10.1016/j.tplants.2017.01.008
13
BaysalC.BortesiL.ZhuC.FarréG.SchillbergS.ChristouP. (2016). CRISPR/Cas9 activity in the rice OsBEIIb gene does not induce off-target effects in the closely related paralog OsBEIIa.Mol. Breed.36:108. 10.1007/s11032-016-0533-4
14
BortesiL.ZhuC.ZischewskiJ.PerezL.BassiéL.NadiR.et al (2016). Patterns of CRISPR/Cas9 activity in plants, animals and microbes.Plant Biotechnol. J.142203–2216. 10.1111/pbi.12634
15
BoydC. D.O’TooleG. A. (2012). Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems.Annu. Rev. Cell Dev. Biol.28439–462. 10.1146/annurev-cellbio-101011-155705
16
CallawayE. (2018). CRISPR plants now subject to tough GM laws in European Union.Nature560:16. 10.1038/d41586-018-05814-6
17
ChandrasekaranJ.BruminM.WolfD.LeibmanD.KlapC.PearlsmanM.et al (2016). Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology.Mol. Plant Pathol.171140–1153. 10.1111/mpp.12375
18
CharS. N.NeelakandanA. K.NahampunH.FrameB.MainM.SpaldingM. H.et al (2017). An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize.Plant Biotechnol. J.15257–268. 10.1111/pbi.12611
19
ChristopoulouM.Reyes-ChinW. S.KozikA.McHaleL. K.TrucoM. J.WroblewskiT.et al (2015). Genome-Wide architecture of disease resistance genes in lettuce.G3 (Bethesda)52655–2669. 10.1534/g3.115.020818
20
ConsonniC.HumphryM. E.HartmannH. A.LivajaM.DurnerJ.WestphalL.et al (2006). Conserved requirement for a plant host cell protein in powdery mildew pathogenesis.Nat. Genet.38716–720. 10.1038/ng1806
21
ČermákT.BaltesN. J.ČeganR.ZhangY.VoytasD. F. (2015). High-frequency, precise modification of the tomato genome.Genome Biol.16:32. 10.1186/s13059-015-0796-9
22
DingD.ChenK.ChenY.LiH.XieK. (2018). Engineering introns to express rna guides for Cas9- and Cpf1-mediated multiplex genome editing.Mol. Plant11542–552. 10.1016/j.molp.2018.02.005
23
DracatosP. M.HaghdoustR.SinghD.FraserP. (2018). Exploring and exploiting the boundaries of host specificity using the cereal rust and mildew models.New Phytol.218453–462. 10.1111/nph.15044
24
EndoA.MasafumiM.KayalH.TokiS. (2016). Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida.Sci. Rep.6:38169. 10.1038/srep38169
25
FAO (2017). The Future of Food and Agriculture – Trends and Challenges.Rome: FAO.
26
FengC.YuanJ.WangR.LiuY.BirchlerJ. A.HanF. (2016). Efficient targeted genome modification in maize using CRISPR/Cas9 system.J. Genet. Genomics4337–43. 10.1016/j.jgg.2015.10.002
27
FisterA. S.LandherrL.MaximovaS. N.GuiltinanM. J. (2018). Transient expression of CRISPR/Cas9 machinery targeting TcNPR3 enhances defense response in Theobroma cacao.Front. Plant Sci.9:268. 10.3389/fpls.2018.00268
28
FondongV. N. (2013). Geminivirus protein structure and function.Mol. Plant Pathol.14635–649. 10.1111/mpp.12032
29
GaoZ. Y.ZhaoS. C.HeW. M.GuoL. B.PengY. L.WangJ. J.et al (2013). Dissecting yield-associated loci in super hybrid rice by re- sequencing recombinant inbred lines and improving parental genome sequences.Proc. Natl. Acad. Sci. U.S.A.11014492–14497. 10.1073/pnas.1306579110
30
GilbertsonR. L.BatumanO.WebsterC. G.AdkinsS. (2015). Role of the insect supervectors Bemisia tabaci and Frankliniella occidentalis in the emergence and global spread of plant viruses.Annu. Rev. Virol.267–93. 10.1146/annurev-virology-031413-085410
31
HakamN.VdupaS. M.RobhaA.IbrizM.IraqiD. (2015). Efficient callus induction and plantlets regeneration in bread wheat using immature and mature embryos.Int. J. Biotechnol. Res.31–9.
32
Hanley-BowdoinL.BejaranoE. R.RobertsonD.MansoorS. (2013). Germiniviruses: masters at redirecting and reprogramming plant processes.Nat. Rev. Microbiol.11777–788. 10.1038/nrmicro3117
33
HolmeI. B.WendtT.HumanesJ. G.DeleuranL. C.ColbyG.StarkerC. G.et al (2017). Evaluation of the mature grain phytase candidate HvPAPhy_a gene in barley (Hordeum vulgare L.) using CRISPR/Cas9 and TALENs.Plant Mol. Biol.95111–121. 10.1007/s11103-017-0640-6
34
HuJ. H.MillerS. M.GeurtsM. H.TangW.ChenL.SunN.et al (2018). Evolved Cas9 variants with broad PAM compatibility and high DNA specificity.Nature55657–63. 10.1038/nature26155
35
HuaK.TaoX.YuanF.WangD.ZhuJ. K. (2018). Precise A⋅T to G⋅C base editing in the rice genome.Mol. Plant11627–630. 10.1016/j.molp.2018.02.007
36
IshidaY.HieiY.KomariT. (2007). Agrobacterium-mediated transformation of maize.Nat. Protoc.216144–1621. 10.1038/nprot.2007.241
37
IshidaY.HieiY.KomariT. (2015a). “High efficiency wheat transformation mediated by Agrobacterium tumefaciens,” inAdvances in Wheat Genetics: From Genome to FieldedsOgiharaY.TakumiS.HandaH. (Tokyo: Springer) 10.1007/978-4-431-55675-6_18
38
IshidaY.TsunashimaM.HieiY.KomariT. (2015b). Wheat (Triticum aestivum L.) transformation using immature embryos.Methods Mol. Biol.1223189–198. 10.1007/978-1-4939-1695-5_15
39
JacobsT. B.LaFayetteP. R.SchmitzR. J.ParrottW. A. (2015). Targeted genome modifications in soybean with CRISPR/Cas9.BMC Biotechnol.15:16. 10.1186/s12896-015-0131-2
40
JiX.ZhangH.ZhangY.WangY.GaoC. (2015). Establishing a CRISPR–Cas-like immune system conferring DNA virus resistance in plants.Nat. Plants1:15144. 10.1038/nplants.2015.144
41
JiaH.OrbovicV.JonesJ. B.WangN. (2016). Modification of the PthA4 effector binding elements in Type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccΔpthA4:dCsLOB1.3 infection.Plant Biotechnol. J.141291–1301. 10.1111/pbi.12495
42
JiangL.YuX.QiX.YuQ.DengS.BaiB.et al (2013). Multigene engineering of starch biosynthesis in maize endosperm increases the total starch content and the proportion of amylose.Transgenic Res.221133–1142. 10.1007/s11248-013-9717-4
43
KapusiE.Corcuera-GómezM.MelnikS.StogerE. (2017). Heritable genomic fragment deletions and small indels in the putative ENGase gene induced by CRISPR/Cas9 in barley.Front. Plant Sci.8:540. 10.3389/fpls.2017.00540
44
KerrA. (2016). Biological control of Crown Gall.Australas. Plant Pathol.4515–18. 10.1007/s13313-015-0389-9
45
LarsonM. H.GilbertL. A.WangX.LimW. A.WeissmanJ. S.QiL. S. (2013). CRISPR interference (CRISPRi) for sequence-specific control of gene expression.Nat. Protoc.82180–2196. 10.1038/nprot.2013.132
46
LawrensonT.ShorinolaO.StaceyN.LiC.ØstergaardL.PatronN.et al (2015). Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease.Genome Biol.16:258. 10.1186/s13059-015-0826-7
47
LiQ.ZhangD.ChenM.LiangW.WeiJ.QiY.et al (2016). Development of japonica photo-sensitive genic male sterile rice lines by editing carbon starved anther using CRISPR/Cas9.J. Genet. Genomics43415–419. 10.1016/j.jgg.2016.04.011
48
LiT.LiuB.SpaldingM. H.WeeksD. P.YangB. (2012). High-efficiency TALEN-based gene editing produces disease-resistant rice.Nat. Biotechnol.30390–392. 10.1038/nbt.2199
49
LiangZ.ChenK.LiT.ZhangY.WangY.ZhaoQ.et al (2017). Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes.Nat. Commun.8:14261. 10.1038/ncomms14261
50
LiuC.MoschouP. N. (2018). Phenotypic novelty by CRISPR in plants.Dev. Biol.435170–175. 10.1016/j.ydbio.2018.01.015
51
LowderL. G.ZhangD.BaltesN. J.PaulJ. W.TangX. (2015). A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation.Plant Physiol.169971–985. 10.1104/pp.15.00636
52
LoweK.WuE.WangN.HoersterG.HastingsC.ChoM. J.et al (2016). Morphogenic regulators Baby boom and Wuschel improve monocot transformation.Plant Cell281998–2015. 10.1105/tpc.16.00124
53
LuoM.GilbertB.AyliffeM. (2016). Applications of CRISPR/Cas9 technology for targeted mutagenesis, gene replacement and stacking of genes in higher plants.Plant Cell Rep.351439–1450. 10.1007/s00299-016-1989-8
54
MaJ.ChenJ.WangM.RenY.WangS.LeiC.et al (2018). Disruption of OsSEC3A increases the content of salicylic acid and induces plant defense responses in rice.J. Exp. Bot.691051–1064. 10.1093/jxb/erx458
55
MaY.ZhangL.HuangX. (2014). Genome modification by CRISPR/Cas9.FEBS J.2815186–5193. 10.1111/febs.13110
56
MacoveiA.SevillaN. R.CantosC.JonsonG. B.Slamet-LoedinI.CermakT.et al (2018). Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus.Plant Biotechnol. J.10.1111/pbi.12927[Epub ahead of print].
57
MalnoyM.ViolaR.JungM. H.KooO. J.KimS.KimJ. S.et al (2016). DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins.Front. Plant Sci.7:1904. 10.3389/fpls.2016.01904
58
MiklisM.ConsonniC.RiyazA. B.VolkerL.Schulze-LefertP.PanstrugaR. (2007). Barley MLO modulates actin-dependent and actin-independent antifungal defense pathways at the cell periphery.Plant Physiol.1441132–1143. 10.1104/pp.107.098897
59
MinkenbergB.XieK.YangY. (2017). Discovery of rice essential genes by characterizing a CRISPR-edited mutation of closely related rice MAP kinase genes.Plant J.89636–648. 10.1111/tpj.13399
60
MohantaT. K.BashirT.HashemA.AllahE.BaeH. (2017). Genome editing tools in plants.Genes8:399. 10.3390/genes8120399
61
NalamV. J.AlamS.KeereetaweepJ.VenablesB.BurdanD.LeeH.et al (2015). Facilitation of Fusarium graminearum infection by 9-Lipoxygenases in Arabidopsis and Wheat.Mol. Plant Microbe Interact.281142–1152. 10.1094/MPMI-04-15-0096-R
62
NekrasovV.WangC.WinJ.LanzC.WeigelD.KamounS. (2017). Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion.Sci. Rep.7:482. 10.1038/s41598-017-00578-x
63
NelsonR.Wiesner-HanksT.WisserR.Balint-KurtiP. (2018). Navigating complexity to breed disease-resistant crops.Nat. Rev. Genet.1921–33. 10.1038/nrg.2017.82
64
PanC.YeL.QinL.LiuX.HeY.WangJ.et al (2016). CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations.Sci. Rep.6:24765. 10.1038/srep24765
65
PanstrugaR.Schulze-LefertP. (2002). Live and let live: insights into powdery mildew disease and resistance.Mol. Plant Pathol.3495–502. 10.1046/j.1364-3703.2002.00145.x
66
PengA.ChenS.LeiT.XuL.HeY.WuL.et al (2017). Engineering canker- resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus.Plant Biotechnol. J.151509–1519. 10.1111/pbi.12733
67
PessinaS.LenziL.PerazzolliM.CampaM.Dalla CostaL.UrsoS.et al (2016). Knockdown of MLO genes reduces susceptibility to powdery mildew in grapevine.Hortic. Res.3:160616. 10.1038/hortres.2016.16
68
PetersonB. A.HaakD. C.NishimuraM. T.TeixeiraP. J.JamesS. R.DanglJ. L.et al (2016). Genome-Wide assessment of efficiency and specificity in CRISPR/Cas9 mediated multiple site targeting in Arabidopsis.PLoS One11:e0162169. 10.1371/journal.pone.0162169
69
PiffanelliP.RamsayL.WaughR.BenabdelmounaA.D’HontA.HollricherK.et al (2004). A barley cultivation-associated polymorphism conveys resistance to powdery mildew.Nature430887–891. 10.1038/nature02781
70
PyottD. E.SheehanE.MolnarA. (2016). Engineering of CRISPR/Cas9- mediated potyvirus resistance in transgene-free Arabidopsis plants.Mol. Plant Pathol.41–13. 10.1111/mpp.12417
71
RoossinckM. J.MartinD. P.RoumagnacP. (2015). Plant virus metagenomics: advances in virus discovery.Phytopathology.105716–727. 10.1094/PHYTO-12-14-0356-RVW
72
SaitoS.MaedaR.AdachiN. (2017). Dual loss of human POLQ and LIG4 abolishes random integration.Nature Commun.8:16112. 10.1038/ncomms16112
73
SanfaconH. (2015). Plant translation factors and virus resistance.Viruses73392–3419. 10.3390/v7072778
74
SavaryS.FickeA.AubertotJ. N.HollierC. (2012). Crop losses due to diseases and their implications for global food production losses and food security.Food Secur.4519–537. 10.1007/s00203-017-1426-6
75
SchlossP. D.HandelsmanJ. (2004). Status of the microbial census.Microbiol. Mol. Biol. Rev.68686–691. 10.1128/MMBR.68.4.686-691.2004
76
ShahS. A.ErdmannS.MojicaF. J.RogerA.GarrettR. A. (2013). Protospacer recognition motifs mixed identities and functional diversity.RNA Biol.10891–899. 10.4161/rna.23764
77
ShiJ.GaoH.WangH.LafitteH. R.ArchibaldR. L.YangM.et al (2017). ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions.Plant Biotechnol. J.15207–216. 10.1111/pbi.12603
78
SvitashevS.SchwartzC.LendertsB.YoungJ. K.CiganA. M. (2016). Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes.Nature Commun.7:13274. 10.1038/ncomms13274
79
SvitashevS.YoungJ. K.SchwartzC.GaoH.FalcoS. C.CiganA. M. (2015). Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA.Plant Physiol.169931–945. 10.1104/pp.15.00793
80
TsaiM.LuY.LiuY.LienH.HuangC.WuJ.et al (2015). Modulation of p53 and met expression by krüppel-like factor 8 regulates zebrafish cerebellar development.Dev. Neurobiol.75908–926. 10.1002/dneu.22258
81
WangF.WangC.LiuP.LeiC.HaoW.GaoY. (2016). Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922.PLoS One11:e0154027. 10.1371/journal.pone.0154027
82
WangW.PanQ.HeF.AkhunovaA.ChaoS.TrickH.et al (2018). Transgenerational CRISPR-Cas9 activity facilitates multiplex gene editing in allopolyploid wheat.CRISPR J.165–74. 10.1089/crispr.2017.0010
83
WangY.ChengX.ShanQ.ZhangY.LiuJ.GaoC.et al (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew.Nat. Biotechnol.32947–952. 10.1038/nbt.2969
84
XieK.YangY. (2013). RNA-guided genome editing in plants using a CRISPR-Cas system.Mol. Plant.61975–1983. 10.1093/mp/sst119
85
XingH. L.DongL.WangZ. P.ZhangH. Y.HanC. Y.LiuB.et al (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants.BMC Plant Biol.14:327. 10.1186/s12870-014-0327-y
86
XuR.YangY.QinR.LiH.QiuC.LiL.et al (2016). Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice.J. Genet. Genomics43529–532. 10.1016/j.jgg.2016.07.003
87
ZaidiS. S.TashkandiM.MansoorS.MahfouzM. M. (2016). Engineering plant immunity: using CRISPR/Cas9 to generate virus resistance.Front. Plant Sci.7:1673. 10.3389/fpls.2016.01673
88
ZhangF.WenY.GuoX. (2014). CRISPR/Cas9 for genome editing: progress, implications and challenges.Hum. Mol. Genet.2340–46. 10.1093/hmg/ddu125
89
ZhangY.BaiY.WuG.ZouS.ChenY.GaoC.et al (2017). Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat.Plant J.91714–724. 10.1111/tpj.13599
90
ZhangY.LiangZ.ZongY.WangY.LiuJ.ChenK.et al (2016). Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA.Nat. Commun.7:12617. 10.1038/ncomms12617
91
ZhangY. Z.ShiM.HolmesE. C. (2018). Using metagenomics to characterize an expanding virosphere.Cell1721168–1172. 10.1016/j.cell.2018.02.043
92
ZhouH.LiuB.WeeksD. P.SpaldingM. H.YangB. (2014). Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice.Nucleic Acids Res.4210903–10914. 10.1093/nar/gku806
93
ZhouJ.PengZ.LongJ.SossoD.LiuB.EomJ. S.et al (2015). Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice.Plant J.82632–643. 10.1111/tpj.12838
Summary
Keywords
CRISPR/Cas9, crop improvement, genome editing, disease resistance, virus, fungus, bacteria
Citation
Borrelli VMG, Brambilla V, Rogowsky P, Marocco A and Lanubile A (2018) The Enhancement of Plant Disease Resistance Using CRISPR/Cas9 Technology. Front. Plant Sci. 9:1245. doi: 10.3389/fpls.2018.01245
Received
11 June 2018
Accepted
06 August 2018
Published
24 August 2018
Volume
9 - 2018
Edited by
Sabrina Sarrocco, Università degli Studi di Pisa, Italy
Reviewed by
Kemal Kazan, Commonwealth Scientific and Industrial Research Organisation (CSIRO), Australia; Kaijun Zhao, Institute of Crop Sciences (CAAS), China
Updates
Copyright
© 2018 Borrelli, Brambilla, Rogowsky, Marocco and Lanubile.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alessandra Lanubile, alessandra.lanubile@unicatt.it
This article was submitted to Plant Microbe Interactions, a section of the journal Frontiers in Plant Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.