- School of Forest Resources and Environmental Science, Michigan Technological University, Houghton, MI, United States
Growth and architectural traits in trees are economically and environmentally important and thus of considerable importance to the improvement of forest and fruit trees. These traits are complex and result from the operation of a number of molecular mechanisms. This review will focus on the regulation of crown architecture, secondary woody growth and adventitious rooting. These traits and processes have significant impact on deployment, management, and productivity of tree crops. The majority of the described work comes from experiments in model plants, poplar, apple, peach, and plum because these species allow functional analysis of the involved genes and have significant genomics resources. However, these studies convincingly show conserved mechanisms for elaboration of specific growth and architectural traits. The conservation of these mechanisms suggest that they can be used as a blueprint for the improvement of these traits and processes in phylogenetically diverse tree crops. We will specifically consider the involvement of flowering time, transcription factors and hormone-associated genes. The review will also discuss the impact of recent technological advances as well as the challenges to the dissection of these traits in trees.
Introduction
Intensive forest plantation can alleviate the harvesting pressure on native forests via allowing production of the same or larger amount of wood on a much smaller land base (Paquette and Messier, 2010). Improved genetics through breeding is one, if not the leading factor in this increased productivity (Fenning and Gershenzon, 2002; Ruotsalainen, 2014). However, tree breeding is slow due to long generation times, traits that need a long time to evaluate and complex genetic architecture of these traits (Fenning and Gershenzon, 2002). Understanding the involved genetic mechanism could significantly accelerate the process through both conventional breeding and genetic engineering.
Here we review the current knowledge about the molecular mechanisms that underpin three developmental processes in trees with significant impact on intensive plantation deployment, management and growth. The review will focus on mechanisms and genes that can provide positive effects and thus are of breeding value rather than exhaustively discuss progress in the dissection of each process. Where available, the reader will be pointed to reviews that deal with these processes in a more comprehensive manner.
Manipulation of Crown Architecture
Crown architecture is a compound trait resulting from the position, size, periodicity, angle and density of the branches. Crown characteristics affect plantation density, interception of photosynthetic light and quality of the derived wood. Depending on the plantation purpose, the direction and extent to which these characteristics need to be changed can vary. Branches originate in axillary meristems (AMs) and thus establishment and outgrowth of AMs has a profound effect on branch characteristics and crown architecture. AM initiation is exclusively characterized in herbaceous plants and there is no information about the effect of these genes in trees. We therefore will not cover here these developments. Excellent reviews on AM initiation discuss in detail these genes and mechanisms (Janssen et al., 2014; Yang and Jiao, 2016).
Branch Outgrowth
Once established, the AM outgrowth is typically suppressed, a phenomenon known as apical dominance. Auxin is central to the establishment and maintenance of apical dominance (Figure 1). The regulatory roles of auxin in apical dominance are indirect and are explained by the canalization and secondary messengers’ models (Domagalska and Leyser, 2011; Teichmann and Muhr, 2015). However, only the latter provides genes and mechanisms manipulated in trees and is thus covered here. According to this model, auxin synthesized in the shoot apex, moves basipetally to the roots to generate a secondary signal that travels acropetally to regulate bud outgrowth. Cytokinin was the first candidate for a second messenger because it has a strong positive effect on axillary bud outgrowth when exogenously applied (Figure 1). However, cytokinin acropetal transport was not able to activate bud outgrowth (Faiss et al., 1997). This led to the discovery of the shoot branching hormone strigolactones (SLs). SLs have strong negative effects on bud outgrowth (Figure 1), are synthesized in roots, acropetally transported to shoots and biosynthetic genes are positively regulated by auxin. SLs metabolic and signaling genes are strong regulators of bud outgrowth in a number of plant species including several trees (Domagalska and Leyser, 2011; Muhr et al., 2016; Foster et al., 2018). RNAi knockdown of poplar and apple orthologs of SLs biosynthetic genes resulted in increased sylleptic branching (branches developed from lateral buds that have not undergone dormancy) (Muhr et al., 2016; Foster et al., 2018).
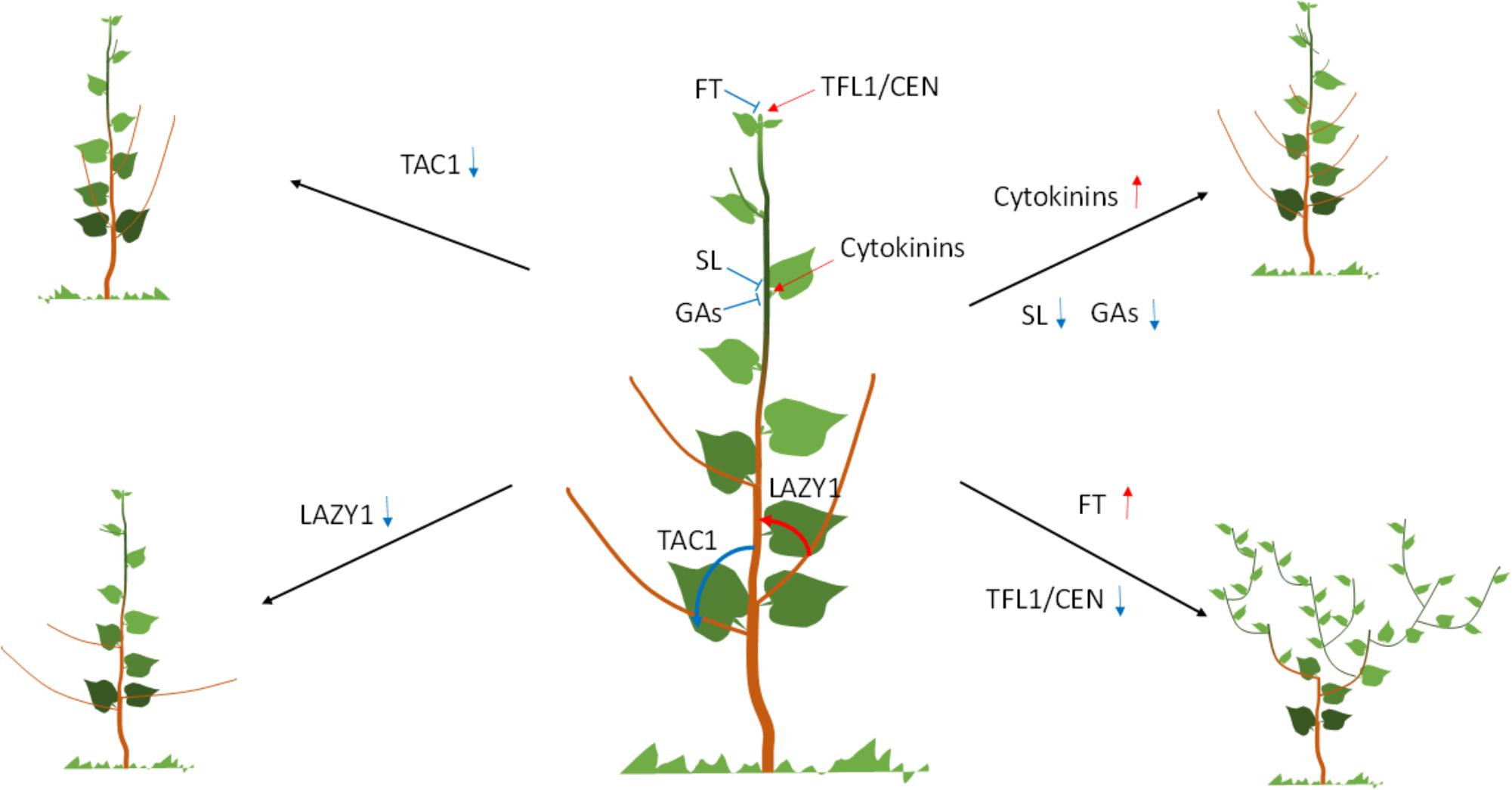
FIGURE 1. Regulation of crown architecture in trees. Diagram in the middle shows a WT tree with the different regulators and their putative roles indicated. The tree drawings to the left and right shows the effect of the different regulators on the crown architecture. Arrows show positive while blunted line negative effect. Vertical up red arrow indicates upregulation, while blue down arrow downregulation of the gene.
Branch Angle
Significant progress has been made in trees in elucidating the mechanism underpinning branch angle characteristics. Using an innovative sequencing approach and a distinct peach mutant with acute branch angle, the causative gene was isolated to be TILLER ANGLE CONTROL1 (TAC1) (Dardick et al., 2013). TAC1 was originally found to control tiller angle in rice (Yu et al., 2007). TAC1 belongs to a small family of genes. All the genes in the family, characterized to date in several plant species, including trees (poplar and plum) control branch or lateral root angles (Figure 1; Xu et al., 2017; Hollender et al., 2018). Depending on presence of a conserved domain, members of the family can increase (TAC1) or decrease (LAZY1) branch angles (Figure 1; Dardick et al., 2013; Hollender and Dardick, 2015; Xu et al., 2017).
Roles of Gibberellins
Gibberellins control stem elongation, but can also regulate crown characteristics (Figure 1). Gibberellin 2-oxidase (GA2ox) overexpression leads to low levels of bioactive GAs, and proliferation of long sylleptic branches at a wide, almost perpendicular angle to the main stem (Mauriat et al., 2011; Zawaski et al., 2011). After 2 years in the field, GA2ox overexpressors produced a wide oval crown (Zawaski et al., 2011). A similar effect was also observed in turf grass and rice (Agharkar et al., 2007; Lo et al., 2008). These effects are possibly mediated via the GAs regulation of PIN auxin efflux carrier abundance (Willige et al., 2011; Lofke et al., 2013; Mauriat et al., 2014). In contrast, modifications of GA signaling via DELLA domain proteins produces a highly compact crown consisting of short branches with narrow acute angle (Zawaski et al., 2011). The effect of DELLA domain proteins on branching may be due their interactions with the transcription factor BRANCHED1 (Daviere et al., 2014).
Flowering and Crown Architecture
The determinacy of the meristem is genetically programmed, heritable and significantly affects plant architecture, including crown characteristics in trees (McGarry and Ayre, 2012). Indeterminate meristems typically produce monopodial growth characterized by a pronounced primary stem. In contrast, plants with determinate meristems show sympodial growth, a process of repeated loss of the shoot apical meristem (SAM) through terminal differentiation and lateral outgrowth from the axillary meristem resulting in a compound shoot architecture. Monopodial and sympodial growth types result from differences in the expression of genes and localization of proteins from the CENTRORDIALIS/TERMINAL FLOWER/SELF PRUNING (CETS) family that control flowering (McGarry and Ayre, 2012). CETS genes form a small gene family in Arabidopsis and other plant species. Very small (few amino acid) changes in the sequence of the proteins can reverse their function (Hanzawa et al., 2005). For example, FT promotes, while a close family member, TFL1 inhibits flowering (Hanano and Goto, 2011). FT is a mobile signal originating in the leaf that moves through the phloem stream to reach the shoot or axillary meristems and initiates terminal flower development (Pin and Nilsson, 2012). TFL1 plays an antagonistic role to FT in the SAM (Kobayashi et al., 1999). Low and high FT/TFL1 ratio in the SAM results in indeterminate and determinate growth respectively, (McGarry and Ayre, 2012; Figure 1). This model has been confirmed through transgenic overexpression of FT orthologs in several tree species (Hsu et al., 2006, 2011; Srinivasan et al., 2012; Klocko et al., 2016). FT overexpression leads to early flowering and highly branched, sympodial growth. Increase in FT/TFL1 balance via downregulation of TFL1/CEN genes in apple leads to similar effects as with FT overexpression (Kotoda et al., 2006; Flachowsky et al., 2012). RNAi downregulation of two TFL1/CEN-like homologs in poplar (PopCEN1 and PoCEN2) produced similar but more moderate flowering and architectural phenotypes (Mohamed et al., 2010). Interacting factors and regulators of FT and TFL1/CEN, can also produce changes in tree architecture. Overexpression of CsRAV1, a chestnut ortholog of TEMPRANILLO, a regulator of FT (Castillejo and Pelaz, 2008), led to upregulation of a poplar ortholog of FT (PttFT2) expression (Triozzi et al., 2018) and consequently to increased branching both under greenhouse and field conditions (Moreno-Cortes et al., 2012, 2017). Under field conditions, increased branching led to increased biomass (Moreno-Cortes et al., 2017). Similarly, overexpression of a poplar ortholog of GIGANTEA, a positive regulator of FT, upregulated PttFT2 and increased sylleptic branching in poplar (Ding et al., 2018). FT interacts with FD to promote flowering and overexpression of poplar FD homolog led to precocious flowering and sympodial, highly branched growth (Parmentier-Line and Coleman, 2015).
Increase of Secondary Woody Growth
Secondary growth originates in a lateral meristem known as vascular cambium, which in trees shows exaggerated and perennial activity, compared to herbaceous plants, resulting in production of massive amounts of conductive and supportive tissues, referred to as wood (Helariutta and Bhalerao, 2003; Elo et al., 2009; Barra-Jimenez and Ragni, 2017). Bifacial periclinal division of the cambium cells, followed by growth and differentiation results in production of phloem/bark to the outside and xylem/wood to the inside of the tree trunk (Helariutta and Bhalerao, 2003; Zhang et al., 2014). Excellent reviews comprehensively discuss the process (Groover, 2005; Demura and Fukuda, 2007; Groover et al., 2010; Spicer and Groover, 2010; Mizrachi and Myburg, 2016). Here we focus on genes and mechanisms that have positive effects on secondary woody growth and thus are of potential breeding/improvement value (Figure 2A). The only exception would be the genes that affect bark development, Bark is typically considered as waist byproduct and thus decrease of bark production would be favored. However, notable exceptions where bark increase would be the goal would be special plantation for production of cork as well as breeding for resistance to pests, fires and drought.
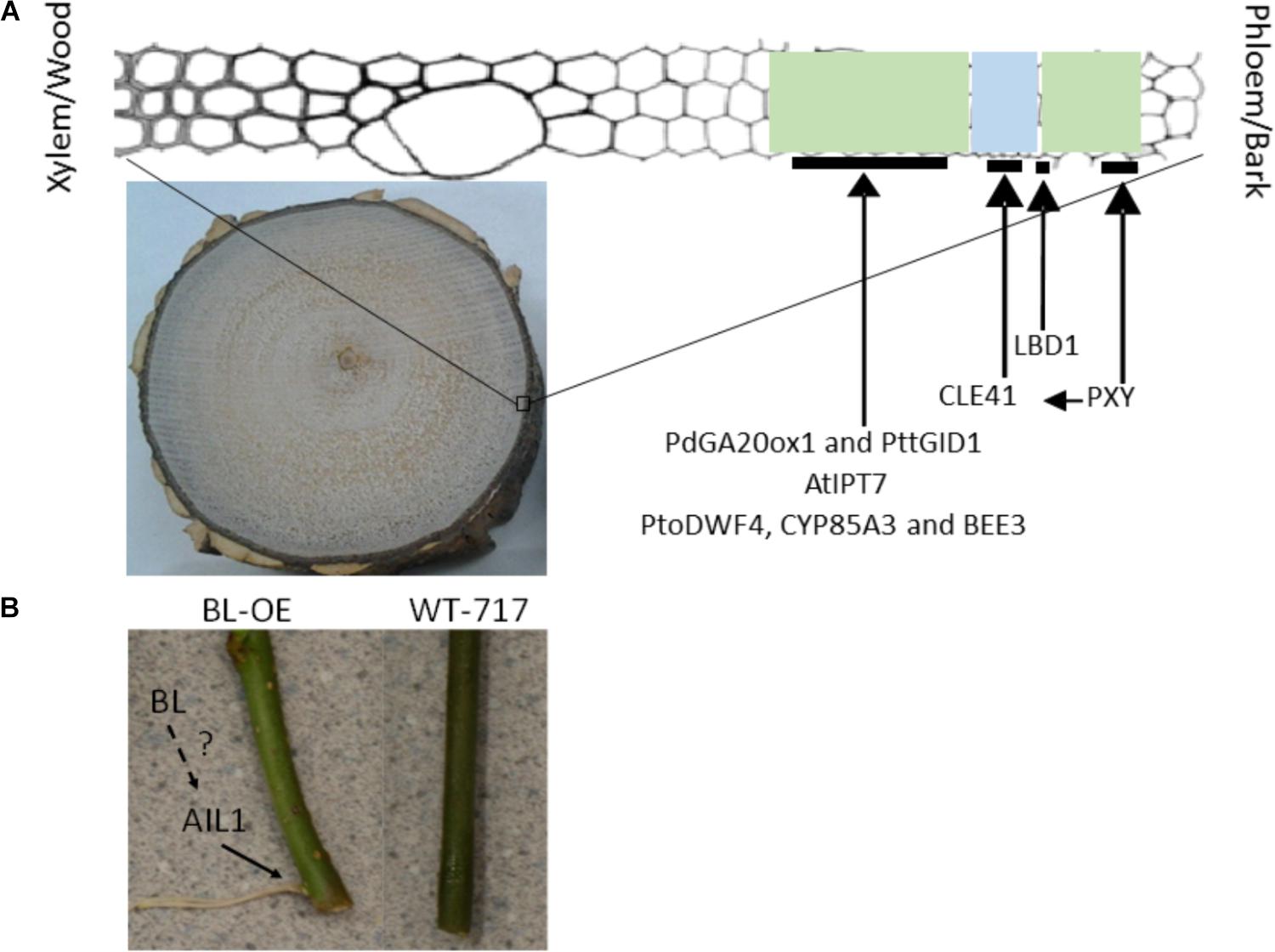
FIGURE 2. Genes increasing secondary woody growth (A) and adventitious rooting (B) in trees. (A) Picture on the bottom shows a cross section from a from a 3-year-old poplar tree, approximately 10 cm in diameter, showing an extensive wood production in the middle and bark at the periphery. The diagram on the top shows the cambium zone with the developing xylem and phloem. Green highlighted area indicates developing xylem and phloem. Blue highlight indicates cambium zone. The black lines show the approximate presumed location where the positive regulators of secondary growth play critical roles. (B) Picture to the left is a transgenic plant overexpressing BL (see text for detail) and to the right WT-717. See Yordanov et al., 2017 and the text for further details. The dotted line shows the presence of additional unknown regulators, indicated by a question mark.
Gibberellins
The first demonstration of increased secondary growth was via transgenic modifications of gibberellin biosynthesis (Eriksson et al., 2000). Overexpression of the Arabidopsis GA-20 oxidase (GA20ox), a key biosynthetic enzyme, resulted in significant (2-fold) increase in wood production (Figure 2). Similarly, overexpression of pine PdGA20ox1 in poplar resulted in nearly 3-fold increase in woody biomass (Jeon et al., 2016; Figure 2A). In addition, overexpression of the poplar orthologs of the GA receptor PttGIBBERELLIN-INSENSITIVE DWARF1 (PttGID1) resulted in similar wood biomass enhancement (Mauriat and Moritz, 2009; Figure 2A). Increase in bioactive gibberellins also increased fiber length and cellulose/xylan content (Eriksson et al., 2000; Jeon et al., 2016). Increase in GA signaling, however, did not result in changes in fiber length. The increased GA synthesis and signaling in the transgenics poplars, had negative effect on root development, decreased expression of defense-related genes and resulted in poor leaf development (Eriksson et al., 2000; Mauriat et al., 2014; Jeon et al., 2016). These negative pleiotropic effects resulting from the constitutive overexpression were mitigated by using a xylem-specific promoter (Jeon et al., 2016). The xylem specific expression resulted in similar increases of wood biomass (Jeon et al., 2016). However, tissue-specific upregulation of PttGID1 using a different xylem-specific promoter did not result in any increases in woody biomass (Mauriat and Moritz, 2009), suggesting that different promoter::gene combination can have specific effects. Cisgenic modifications of several poplar GA 20-oxidase genes led to no pleiotropic effects and increased wood biomass and fiber length but the gains were more modest than these obtained with the constitutive and xylem-specific promoters (Han et al., 2011).
Cytokinins
The regulatory role(s) of cytokinins during secondary woody growth has been known (Nieminen et al., 2008). However, it was only recently demonstrated that modification of cytokinin biosynthesis can have a positive effect on secondary woody growth (Immanen et al., 2016). Transgenic poplars transformed with the Arabidopsis AtIPT7 (key cytokinin biosynthetic gene) driven by a xylem-specific promoter, showed significant (nearly 2-fold) increases in secondary growth and no negative pleiotropic effects (Immanen et al., 2016) (Figure 2A).
Brassinosteroids
Recent evidence suggests that both brassinosteroid (BR) biosynthesis and signaling has a positive effect on woody biomass production (Noh et al., 2015; Jin et al., 2017; Shen et al., 2018; Figure 2). Overexpression of key biosynthetic genes (PtoDWF4 and CYP85A3) led to increased brassinosteroid concentrations and woody biomass (Jin et al., 2017; Shen et al., 2018). The productivity gains however, were much smaller than, these observed with the manipulations of GA and cytokinin biosynthesis/signaling. The increase in brassinosteroids led to longer fibers and no or little impact on cell wall chemistry (Jin et al., 2017; Shen et al., 2018). Similar results were obtained with manipulation of brassinosteroid signaling. Overexpression of a poplar ortholog of BEE3 (Brassinosteroid Enhanced Expression 3), a transcription factor involved in BR signaling increased stem, leaf and root biomass. As with the enhancement of BR biosynthesis, gains in wood biomass were less than these observed with GA and cytokinin and ranged between 25 and 50%. The modifications of both BRs biosynthesis and signaling did not cause any negative pleiotropic effects, despite the strong constitutive promoters used in both studies.
Small Protein Signaling in the Cambium
Cambial cell division in Arabidopsis is controlled by a protein ligand receptor complex (Fisher and Turner, 2007; Hirakawa et al., 2008; Etchells and Turner, 2010). The ligand is the small CLE41 protein, produced in the phloem and transported into the cambium, where it interacts with the PXY receptor to stimulate cambium cell division (Fisher and Turner, 2007; Hirakawa et al., 2008; Etchells and Turner, 2010). Recently, constitutive expression of aspen orthologs of the ligand and receptor in transgenic poplar trees resulted in highly pleiotropic and negative effects on growth and tissue organization (Etchells et al., 2015). However, when the ligand and receptor were simultaneously upregulated in their native tissue domains employing tissue specific promoters, not only that the negative effects were completely mitigated, but also the double transgenic plants showed a nearly double increases in wood production (Figure 2A).
Secondary Phloem and Bark Development
Secondary growth also yields secondary phloem and bark. Using activation tagging in poplar, the first gene that regulates secondary phloem development was discovered (Yordanov et al., 2010). The gene encodes a transcription factor of the LATERAL ORGAN BOUNDARIES (LBD) gene family that is a positive regulator of secondary phloem development (Yordanov et al., 2010; Yordanov and Busov, 2011). Transgenic plants overexpressing the gene produced more, while dominant negative modification of the protein produced less secondary phloem (Figure 2A).
Genes Promoting Adventitious Rooting
Adventitious rooting (AR) is root formation from organs and tissues that typically do not produce roots. The process is most important in forestry and horticulture for clonal propagation and deployment of elite germplasm. The cellular and molecular events underlying AR has been reviewed elsewhere (Diaz-Sala, 2014; Legue et al., 2014; Pacurar et al., 2014). Here we focus on several genes that have been functionally characterized in trees and provide strong positive effects on AR formation.
Controls of Cell Proliferation Provide Points for AR Manipulation
AINTEGUMENTA (ANT) and ANT-like (AIL) genes are a group of eight AP2 transcription factors in Arabidopsis with important functions in regulation of meristem establishment and maintenance as well as organ growth and size (Horstman et al., 2014). One of the members of the AIL family from poplar (AIL1), showed induction during AR primordia activation (Rigal et al., 2012) and overexpression of the gene caused increase (Figure 2B), while RNAi downregulation decrease in the number of ARs. AIL1 transcriptionally regulates Cyclin D3.1 by binding to its promoter (Karlberg et al., 2011). Thus, AIL1 promotes AR at least in part via activation of cell proliferation.
The BIG LEAF/STERILE APETALA (BL/SAP) gene from poplar has a positive effect on AR formation when ectopically expressed (Yordanov et al., 2017; Figure 2B). BL is an F box protein that regulates leaf size in poplar and Arabidopsis through control of cell proliferation (Wang et al., 2016; Li et al., 2018). BL/SAP targets proteins for degradation that negatively regulate AIL genes (PLETHORA 1 and 2) (Horstman et al., 2014; Wang et al., 2016; Li et al., 2018). Thus, BL likely regulates AR formation through promoting degradation of a repressor(s)of the AIL-like genes (Figure 2B), which has a positive effect on cell proliferation and meristem organization.
Both AIL1 and BL, when overexpressed have significant pleiotropic effects (Rigal et al., 2012; Yordanov et al., 2017) and to serve as biotechnological tool for increased AR formation, will require inducible or tissue-specific upregulation.
Gibberellins
GAs inhibit AR likely thought interfering with polar auxin transport (Mauriat et al., 2014). Increase and decrease in GA biosynthesis and signaling leads to decreased and increased AR (Busov et al., 2006; Gou et al., 2010; Elias et al., 2012). As mentioned earlier, GAs have strong positive effects on secondary woody growth and thus the decrease of AR may present an impediment for the clonal propagation of transgenics with increased GAs biosynthesis. Alternatively, decrease in bioactive GAs and block of signaling, which promotes AR formation, leads to various levels of dwarfism. Dwarfism is a desirable trait in fruit and ornamental tree crops and the increased AR formation would provide an additional benefit for the propagation of these genotypes. In forestry, however, extreme dwarfism may lead to loss in biomass productivity and thus, this effect can be either mitigated via increased girth growth using gene stacking with other transgenes that promote radial expansion (see above) or selection of semi-dwarfism genotypes (Elias et al., 2012).
Activation Tagging Discovery of AR-Involved Genes
Using activation tagging (AT), the poplar gene ETHYLENE RESPONSE FACTOR 3 (ERF003) was shown to have positive effect on AR formation (Trupiano et al., 2013). In addition to ERF003, several other AT mutants affected in AR and associated with ethylene signaling and biosynthesis were also discovered (Trupiano et al., 2013). These genes however, have not been recapitulated through re-transformation experiments and thus their involvement and utility in manipulation of AR formation is still tentative.
Future Outlook
Improvements in Transformation Technologies
Transformation is the golden standard for asserting gene function and preferred method of choice in delivering advanced editing tools like CRIPSR/Cas9 system (Busov et al., 2005; Altpeter et al., 2016; Ran et al., 2017). However, transformation technologies are slow, inefficient, require cumbersome tissue culture processes and remain largely genotype-specific (Busov et al., 2005; Altpeter et al., 2016; Baltes et al., 2017), even in genera considered as ‘easy-to-transform’ like poplars. Thus, major strides in understanding and improving transformation technologies are needed (Altpeter et al., 2016).
Understanding Promoter Architecture
The need for research in isolation and engineering artificial promoters for precise targeting of transgenic manipulations has been known and well-recognized. However, research in this area has been lagging behind. The advances in gene editing and synthetic technologies would further necessitate better understanding promoter architecture in order to being able to effectively modify and design level and specificity of promoter activities.
Application of CRISPR Technology
New technological advances in gene editing technologies like CRISPR/Cas9 promise to revolutionize tree improvement (Tsai and Xue, 2015). The CRISPR/Cas9 was successfully implemented in a poplar tree (Fan et al., 2015; Zhou et al., 2015). CRISPR/Cas9 compared to RNAi produced stronger and more uniform phenotypic effects when the same gene was targeted (Zhou et al., 2015). Although now, the majority of the CRISPR/Cas9 applications involve generation of knock-outs via non-homologous end joining, a significant progress is also made in the application of CRISPR/Cas9 for gene editing through homologous recombination (Schaeffer and Nakata, 2015). However, the latter is still in developmental stages for plants. CRIPSR/Cas9 can also alleviate the regulatory burdens associated with field-testing because, in some countries CRISPR/Cas9-modified genotypes are considered as a non-GMO type of modification.
Using Induced and Natural Mutants
Application of natural or induced mutants in tree research has been rare. However, significant strides have been made in both approaches (Busov et al., 2010; Dardick et al., 2013). As described above, using activation tagging in poplar, genes important for secondary growth (Yordanov et al., 2010, 2014, 2017) and AR formation (Trupiano et al., 2013) were discovered. In addition to induced mutants, many natural tree mutants exist. The new sequencing technologies allow efficient mapping of the causative mutations, (Dardick et al., 2013). These approaches can be further used for identifying genes affecting various aspects of tree growth and development.
Understanding Integrative System Controls
A significant progress has been made in identification of individual genes and pathways regulating different traits. However, it has been long known at an organismal level that the various processes are highly coordinated at tissue and organismal level but also in response to various environmental cues. Identification of the coordinating genes, signals and mechanisms can lead to more integrative manipulation of one or several traits.
Author Contributions
The author confirms being the sole contributor of this work and approved it for publication.
Funding
This research was supported by grants from the Plant Feedstock Genomics for Bioenergy: A Joint Research Program of USDA and DOE (2009-65504-05767 and DE-SC0008462), USDA McIntire Stennis Fund (Grant 1001498), and USDA BRAG (grant 2016-33522-25626).
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Agharkar, M., Lomba, P., Altpeter, F., Zhang, H., Kenworthy, K., and Lange, T. (2007). Stable expression of AtGA2ox1 in a low-input turfgrass (Paspalum notatum Flugge) reduces bioactive gibberellin levels and improves turf quality under field conditions. Plant Biotechnol. J. 5, 791–801. doi: 10.1111/j.1467-7652.2007.00284.x
Altpeter, F., Springer, N. M., Bartley, L. E., Blechl, A. E., Brutnell, T. P., Citovsky, V., et al. (2016). Advancing crop transformation in the era of genome editing. Plant Cell 28, 1510–1520. doi: 10.1105/tpc.16.00196
Baltes, N. J., Gil-Humanes, J., and Voytas, D. F. (2017). Genome engineering and agriculture: opportunities and challenges. Prog. Mol. Biol. Transl. Sci. 149, 1–26. doi: 10.1016/bs.pmbts.2017.03.011
Barra-Jimenez, A., and Ragni, L. (2017). Secondary development in the stem: when Arabidopsis and trees are closer than it seems. Curr. Opin. Plant Biol. 35, 145–151. doi: 10.1016/j.pbi.2016.12.002
Busov, V., Meilan, R., Pearce, D. W., Rood, S. B., Ma, C., Tschaplinski, T. J., et al. (2006). Transgenic modification of gai or rgl1 causes dwarfing and alters gibberellins, root growth, and metabolite profiles in Populus. Planta 224, 288–299. doi: 10.1007/s00425-005-0213-9
Busov, V., Yordanov, Y., Gou, J., Meilan, R., Ma, C., Regan, S., et al. (2010). Activation tagging is an effective gene tagging system in Populus. Tree Gen. Genom. 7, 91–101. doi: 10.1007/s11295-010-0317-7
Busov, V. B., Brunner, A. M., Meilan, R., Filichkin, S., Ganio, L., Gandhi, S., et al. (2005). Genetic transformation: a powerful tool for dissection of adaptive traits in trees. New Phytol. 167, 9–18. doi: 10.1111/j.1469-8137.2005.01412.x
Castillejo, C., and Pelaz, S. (2008). The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 18, 1338–1343. doi: 10.1016/j.cub.2008.07.075
Dardick, C., Callahan, A., Horn, R., Ruiz, K. B., Zhebentyayeva, T., Hollender, C., et al. (2013). PpeTAC1 promotes the horizontal growth of branches in peach trees and is a member of a functionally conserved gene family found in diverse plants species. Plant J. 75, 618–630. doi: 10.1111/tpj.12234
Daviere, J. M., Wild, M., Regnault, T., Baumberger, N., Eisler, H., Genschik, P., et al. (2014). Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr. Biol. 24, 1923–1928. doi: 10.1016/j.cub.2014.07.012
Demura, T., and Fukuda, H. (2007). Transcriptional regulation in wood formation. Trends Plant Sci. 12, 64–70. doi: 10.1016/j.tplants.2006.12.006
Diaz-Sala, C. (2014). Direct reprogramming of adult somatic cells toward adventitious root formation in forest tree species: the effect of the juvenile-adult transition. Front. Plant Sci. 5:310. doi: 10.3389/fpls.2014.00310
Ding, J., Bohlenius, H., Ruhl, M. G., Chen, P., Sane, S., Zambrano, J. A., et al. (2018). GIGANTEA-like genes control seasonal growth cessation in Populus. New Phytol. 218, 1491–1503. doi: 10.1111/nph.15087
Domagalska, M. A., and Leyser, O. (2011). Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 12, 211–221. doi: 10.1038/nrm3088
Elias, A. A., Busov, V. B., Kosola, K. R., Ma, C., Etherington, E., Shevchenko, O., et al. (2012). Green revolution trees: semidwarfism transgenes modify gibberellins, promote root growth, enhance morphological diversity, and reduce competitiveness in hybrid poplar. Plant Physiol. 160, 1130–1144. doi: 10.1104/pp.112.200741
Elo, A., Immanen, J., Nieminen, K., and Helariutta, Y. (2009). Stem cell function during plant vascular development. Semin. Cell Dev. Biol. 20, 1097–1106. doi: 10.1016/j.semcdb.2009.09.009
Eriksson, M. E., Israelsson, M., Olsson, O., and Moritz, T. (2000). Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat. Biotechnol. 18, 784–788. doi: 10.1038/77355
Etchells, J. P., Mishra, L. S., Kumar, M., Campbell, L., and Turner, S. R. (2015). Wood formation in trees is increased by manipulating PXY-regulated cell division. Curr. Biol. 25, 1050–1055. doi: 10.1016/j.cub.2015.02.023
Etchells, J. P., and Turner, S. R. (2010). The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 137, 767–774. doi: 10.1242/dev.044941
Faiss, M., Zalubilova, J., Strnad, M., and Schmulling, T. (1997). Conditional transgenic expression of the ipt gene indicates a function for cytokinins in paracrine signaling in whole tobacco plants. Plant J. 12, 401–415. doi: 10.1046/j.1365-313X.1997.12020401.x
Fan, D., Liu, T., Li, C., Jiao, B., Li, S., Hou, Y., et al. (2015). Efficient CRISPR/Cas9-mediated targeted mutagenesis in Populus in the first generation. Sci. Rep. 5:12217. doi: 10.1038/srep12217
Fenning, T. M., and Gershenzon, J. (2002). Where will the wood come from? Plantation forests and the role of biotechnology. Trends Biotechnol. 20, 291–296. doi: 10.1016/s0167-7799(02)01983-2
Fisher, K., and Turner, S. (2007). PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr. Biol. 17, 1061–1066. doi: 10.1016/j.cub.2007.05.049
Flachowsky, H., Szankowski, I., Waidmann, S., Peil, A., Trankner, C., and Hanke, M. V. (2012). The MdTFL1 gene of apple (Malus x domestica Borkh.) reduces vegetative growth and generation time. Tree Physiol. 32, 1288–1301. doi: 10.1093/treephys/tps080
Foster, T. M., Ledger, S. E., Janssen, B. J., Luo, Z., Drummond, R. S. M., Tomes, S., et al. (2018). Expression of MdCCD7 in the scion determines the extent of sylleptic branching and the primary shoot growth rate of apple trees. J. Exp. Bot. 69, 2379–2390. doi: 10.1093/jxb/erx404
Gou, J., Strauss, S. H., Tsai, C. J., Fang, K., Chen, Y., Jiang, X., et al. (2010). Gibberellins regulate lateral root formation in Populus through interactions with auxin and other hormones. Plant Cell 22, 623–639. doi: 10.1105/tpc.109.073239
Groover, A. T., Nieminen, K., Helariutta, Y., and Mansfield, S. D. (2010). “Wood formation in Populus,” in Genetics and Genomics of Populus. Plant Genetics and Genomics: Crops and Models, Vol. 8, eds S. Jansson, R. Bhalerao, and A. Groover (New York, NY: Springer), 201–224.
Han, K. M., Dharmawardhana, P., Arias, R. S., Ma, C., Busov, V., and Strauss, S. H. (2011). Gibberellin-associated cisgenes modify growth, stature and wood properties in Populus. Plant Biotechnol. J. 9, 162–178. doi: 10.1111/j.1467-7652.2010.00537.x
Hanano, S., and Goto, K. (2011). Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell 23, 3172–3184. doi: 10.1105/tpc.111.088641
Hanzawa, Y., Money, T., and Bradley, D. (2005). A single amino acid converts a repressor to an activator of flowering. Proc. Natl. Acad. Sci. U.S.A. 102, 7748–7753. doi: 10.1073/pnas.0500932102
Helariutta, Y., and Bhalerao, R. (2003). Between xylem and phloem: the genetic control of cambial activity in plants. Plant Biol. 5, 465–472. doi: 10.1055/s-2003-44780
Hirakawa, Y., Shinohara, H., Kondo, Y., Inoue, A., Nakanomyo, I., Ogawa, M., et al. (2008). Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. U.S.A. 105, 15208–15213. doi: 10.1073/pnas.0808444105
Hollender, C. A., and Dardick, C. (2015). Molecular basis of angiosperm tree architecture. New Phytol. 206, 541–556. doi: 10.1111/nph.13204
Hollender, C. A., Waite, J. M., Tabb, A., Raines, D., Chinnithambi, S., and Dardick, C. (2018). Alteration of TAC1 expression in Prunus species leads to pleiotropic shoot phenotypes. Hortic. Res. 5:26. doi: 10.1038/s41438-018-0034-1
Horstman, A., Willemsen, V., Boutilier, K., and Heidstra, R. (2014). AINTEGUMENTA-LIKE proteins: hubs in a plethora of networks. Trends Plant Sci. 19, 146–157. doi: 10.1016/j.tplants.2013.10.010
Hsu, C. Y., Adams, J. P., Kim, H. J., No, K., Ma, C. P., Strauss, S. H., et al. (2011). FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc. Natl. Acad. Sci. U.S.A. 108, 10756–10761. doi: 10.1073/pnas.1104713108
Hsu, C. Y., Liu, Y., Luthe, D. S., and Yuceer, C. (2006). Poplar FT2 shortens the juvenile phase and promotes seasonal flowering. Plant Cell 18, 1846–1861. doi: 10.1105/tpc.106.041038
Immanen, J., Nieminen, K., Smolander, O. P., Kojima, M., Alonso Serra, J., Koskinen, P., et al. (2016). Cytokinin and auxin display distinct but interconnected distribution and signaling profiles to stimulate cambial activity. Curr. Biol. 26, 1990–1997. doi: 10.1016/j.cub.2016.05.053
Janssen, B. J., Drummond, R. S., and Snowden, K. C. (2014). Regulation of axillary shoot development. Curr. Opin. Plant Biol. 17, 28–35. doi: 10.1016/j.pbi.2013.11.004
Jeon, H. W., Cho, J. S., Park, E. J., Han, K. H., Choi, Y. I., and Ko, J. H. (2016). Developing xylem-preferential expression of PdGA20ox1, a gibberellin 20-oxidase 1 from Pinus densiflora, improves woody biomass production in a hybrid poplar. Plant Biotechnol. J. 14, 1161–1170. doi: 10.1111/pbi.12484
Jin, Y. L., Tang, R. J., Wang, H. H., Jiang, C. M., Bao, Y., Yang, Y., et al. (2017). Overexpression of Populus trichocarpa CYP85A3 promotes growth and biomass production in transgenic trees. Plant Biotechnol. J. 15, 1309–1321. doi: 10.1111/pbi.12717
Karlberg, A., Bako, L., and Bhalerao, R. P. (2011). Short day-mediated cessation of growth requires the downregulation of AINTEGUMENTALIKE1 transcription factor in hybrid aspen. PLoS Genet. 7:e1002361. doi: 10.1371/journal.pgen.1002361
Klocko, A. L., Ma, C., Robertson, S., Esfandiari, E., Nilsson, O., and Strauss, S. H. (2016). FT overexpression induces precocious flowering and normal reproductive development in Eucalyptus. Plant Biotechnol. J. 14, 808–819. doi: 10.1111/pbi.12431
Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. doi: 10.1126/science.286.5446.1960
Kotoda, N., Iwanami, H., Takahashi, S., and Abe, K. (2006). Antisense expression of MdTFL1, a TFL1-like gene, reduces the juvenile phase in apple. J. Am. Soc. Hortic. Sci. 131, 74–81.
Legue, V., Rigal, A., and Bhalerao, R. P. (2014). Adventitious root formation in tree species: involvement of transcription factors. Physiol. Plant. 151, 192–198. doi: 10.1111/ppl.12197
Li, N., Liu, Z. P., Wang, Z. B., Ru, L. C., Gonzalez, N., Baekelandt, A., et al. (2018). STERILE APETALA modulates the stability of a repressor protein complex to control organ size in Arabidopsis thaliana. PLoS Genet. 14:e1007218. doi: 10.1371/journal.pgen.1007218
Lo, S. F., Yang, S. Y., Chen, K. T., Hsing, Y. L., Zeevaart, J. A. D., Chen, L. J., et al. (2008). A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20, 2603–2618. doi: 10.1105/tpc.108.060913
Lofke, C., Zwiewka, M., Heilmann, I., Van Montagu, M. C., Teichmann, T., and Friml, J. (2013). Asymmetric gibberellin signaling regulates vacuolar trafficking of PIN auxin transporters during root gravitropism. Proc. Natl. Acad. Sci. U.S.A. 110, 3627–3632. doi: 10.1073/pnas.1300107110
Mauriat, M., and Moritz, T. (2009). Analyses of GA20ox- and GID1-over-expressing aspen suggest that gibberellins play two distinct roles in wood formation. Plant J. 58, 989–1003. doi: 10.1111/j.1365-313X.2009.03836.x
Mauriat, M., Petterle, A., Bellini, C., and Moritz, T. (2014). Gibberellins inhibit adventitious rooting in hybrid aspen and Arabidopsis by affecting auxin transport. Plant J. 78, 372–384. doi: 10.1111/tpj.12478
Mauriat, M., Sandberg, L. G., and Moritz, T. (2011). Proper gibberellin localization in vascular tissue is required to control auxin-dependent leaf development and bud outgrowth in hybrid aspen. Plant J. 67, 805–816. doi: 10.1111/j.1365-313X.2011.04635.x
McGarry, R. C., and Ayre, B. G. (2012). Manipulating plant architecture with members of the CETS gene family. Plant Sci. 18, 71–81. doi: 10.1016/j.plantsci.2012.03.002
Mizrachi, E., and Myburg, A. A. (2016). Systems genetics of wood formation. Curr. Opin. Plant Biol. 30, 94–100. doi: 10.1016/j.pbi.2016.02.007
Mohamed, R., Wang, C. T., Ma, C., Shevchenko, O., Dye, S. J., Puzey, J. R., et al. (2010). Populus CEN/TFL1 regulates first onset of flowering, axillary meristem identity and dormancy release in Populus. Plant J. 62, 674–688. doi: 10.1111/j.1365-313X.2010.04185.x
Moreno-Cortes, A., Hernandez-Verdeja, T., Sanchez-Jimenez, P., Gonzalez-Melendi, P., Aragoncillo, C., and Allona, I. (2012). CsRAV1 induces sylleptic branching in hybrid poplar. New Phytol. 194, 83–90. doi: 10.1111/j.1469-8137.2011.04023.x
Moreno-Cortes, A., Ramos-Sanchez, J. M., Hernandez-Verdeja, T., Gonzalez-Melendi, P., Alves, A., Simoes, R., et al. (2017). Impact of RAV1-engineering on poplar biomass production: a short-rotation coppice field trial. Biotechnol. Biofuels 10:110. doi: 10.1186/s13068-017-0795-z
Muhr, M., Prufer, N., Paulat, M., and Teichmann, T. (2016). Knockdown of strigolactone biosynthesis genes in Populus affects BRANCHED1 expression and shoot architecture. New Phytol. 212, 613–626. doi: 10.1111/nph.14076
Nieminen, K., Immanen, J., Laxell, M., Kauppinen, L., Tarkowski, P., Dolezal, K., et al. (2008). Cytokinin signaling regulates cambial development in poplar. Proc. Natl. Acad. Sci. U.S.A. 105, 20032–20037. doi: 10.1073/pnas.0805617106
Noh, S. A., Choi, Y. I., Cho, J. S., and Lee, H. (2015). The poplar basic helix-loop-helix transcription factor BEE3 - Like gene affects biomass production by enhancing proliferation of xylem cells in poplar. Biochem. Biophys. Res. Commun. 462, 64–70. doi: 10.1016/j.bbrc.2015.04.109
Pacurar, D. I., Perrone, I., and Bellini, C. (2014). Auxin is a central player in the hormone cross-talks that control adventitious rooting. Physiol. Plant. 151, 83–96. doi: 10.1111/ppl.12171
Paquette, A., and Messier, C. (2010). The role of plantations in managing the world’s forests in the Anthropocene. Front. Ecol. Environ. 8, 27–34. doi: 10.1890/080116
Parmentier-Line, C. M., and Coleman, G. D. (2015). Constitutive expression of the Poplar FD-like basic leucine zipper transcription factor alters growth and bud development. Plant Biotechnol. J. 14, 260–270. doi: 10.1111/pbi.12380
Pin, P. A., and Nilsson, O. (2012). The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ. 35, 1742–1755. doi: 10.1111/j.1365-3040.2012.02558.x
Ran, Y., Liang, Z., and Gao, C. (2017). Current and future editing reagent delivery systems for plant genome editing. Sci. China Life Sci. 60, 490–505. doi: 10.1007/s11427-017-9022-1
Rigal, A., Yordanov, Y. S., Perrone, I., Karlberg, A., Tisserant, E., Bellini, C., et al. (2012). The AINTEGUMENTA LIKE1 homeotic transcription factor PtAIL1 controls the formation of adventitious root primordia in poplar. Plant Physiol. 160, 1996–2006. doi: 10.1104/pp.112.204453
Ruotsalainen, S. (2014). Increased forest production through forest tree breeding. Scand. J. For. Res. 29, 333–344. doi: 10.1080/02827581.2014.926100
Schaeffer, S. M., and Nakata, P. A. (2015). CRISPR/Cas9-mediated genome editing and gene replacement in plants: transitioning from lab to field. Plant Sci. 240, 130–142. doi: 10.1016/j.plantsci.2015.09.011
Shen, Y., Li, Y., Xu, D., Yang, C., Li, C., and Luo, K. (2018). Molecular cloning and characterization of a brassinosteriod biosynthesis-related gene PtoDWF4 from Populus tomentosa. Tree Physiol. doi: 10.1093/treephys/tpy027 [Epub ahead of print].
Spicer, R., and Groover, A. (2010). Evolution of development of vascular cambia and secondary growth. New Phytol. 186, 577–592. doi: 10.1111/j.1469-8137.2010.03236.x
Srinivasan, C., Dardick, C., Callahan, A., and Scorza, R. (2012). Plum (Prunus domestica) trees transformed with poplar FT1 result in altered architecture, dormancy requirement, and continuous flowering. PLoS One 7:e40715. doi: 10.1371/journal.pone.0040715
Teichmann, T., and Muhr, M. (2015). Shaping plant architecture. Front. Plant Sci. 6:233. doi: 10.3389/fpls.2015.00233
Triozzi, P. M., Ramos-Sanchez, J. M., Hernandez-Verdeja, T., Moreno-Cortes, A., Allona, I., and Perales, M. (2018). Photoperiodic regulation of shoot apical growth in poplar. Front. Plant Sci. 9:1030. doi: 10.3389/fpls.2018.01030
Trupiano, D., Yordanov, Y., Regan, S., Meilan, R., Tschaplinski, T., Scippa, G. S., et al. (2013). Identification, characterization of an AP2/ERF transcription factor that promotes adventitious, lateral root formation in Populus. Planta 238, 271–282. doi: 10.1007/s00425-013-1890-4
Tsai, C. J., and Xue, L. J. (2015). CRISPRing into the woods. GM Crops Food 6, 206–215. doi: 10.1080/21645698.2015.1091553
Wang, Z., Li, N., Jiang, S., Gonzalez, N., Huang, X., Wang, Y., et al. (2016). SCF(SAP) controls organ size by targeting PPD proteins for degradation in Arabidopsis thaliana. Nat. Commun. 7:11192. doi: 10.1038/ncomms11192
Willige, B. C., Isono, E., Richter, R., Zourelidou, M., and Schwechheimer, C. (2011). Gibberellin regulates PIN-FORMED abundance and is required for auxin transport-dependent growth and development in Arabidopsis thaliana. Plant Cell 23, 2184–2195. doi: 10.1105/tpc.111.086355
Xu, D., Qi, X., Li, J., Han, X., Wang, J., Jiang, Y., et al. (2017). PzTAC and PzLAZY from a narrow-crown poplar contribute to regulation of branch angles. Plant Physiol. Biochem. 118, 571–578. doi: 10.1016/j.plaphy.2017.07.011
Yang, M., and Jiao, Y. (2016). Regulation of axillary meristem initiation by transcription factors and plant hormones. Front. Plant Sci. 7:183. doi: 10.3389/fpls.2016.00183
Yordanov, Y., Regan, S., and Busov, V. (2010). Members of the lateral organ boundaries domain (LBD) transcription factors family are involved in regulation of secondary growth in Populus. Plant Cell 22, 3662–3677. doi: 10.1105/tpc.110.078634
Yordanov, Y. S., and Busov, V. (2011). Boundary genes in regulation and evolution of secondary growth. Plant Signal. Behav. 6, 688–690. doi: 10.4161/psb.6.5.14973
Yordanov, Y. S., Ma, C., Strauss, S. H., and Busov, V. B. (2014). EARLY BUD-BREAK 1 (EBB1) is a regulator of release from seasonal dormancy in poplar trees. Proc. Natl. Acad. Sci. U.S.A. 111, 10001–10006. doi: 10.1073/pnas.1405621111
Yordanov, Y. S., Ma, C., Yordanova, E., Meilan, R., Strauss, S. H., and Busov, V. B. (2017). BIG LEAF is a regulator of organ size and adventitious root formation in poplar. PLoS One 12:e0180527. doi: 10.1371/journal.pone.0180527
Yu, B., Lin, Z., Li, H., Li, X., Li, J., Wang, Y., et al. (2007). TAC1, a major quantitative trait locus controlling tiller angle in rice. Plant J. 52, 891–898. doi: 10.1111/j.1365-313X.2007.03284.x
Zawaski, C., Kadmiel, M., Pickens, J., Ma, C., Strauss, S., and Busov, V. (2011). Repression of gibberellin biosynthesis or signaling produces striking alterations in poplar growth, morphology, and flowering. Planta 234, 1285–1298. doi: 10.1007/s00425-011-1485-x
Zhang, J., Nieminen, K., Serra, J. A., and Helariutta, Y. (2014). The formation of wood and its control. Curr. Opin. Plant Biol. 17, 56–63. doi: 10.1016/j.pbi.2013.11.003
Keywords: hormones, transcription factors, woody biomass growth, molecular mechanisms, crown architecture, adventitious rooting, tree biotechnology
Citation: Busov VB (2018) Manipulation of Growth and Architectural Characteristics in Trees for Increased Woody Biomass Production. Front. Plant Sci. 9:1505. doi: 10.3389/fpls.2018.01505
Received: 29 June 2018; Accepted: 26 September 2018;
Published: 16 October 2018.
Edited by:
Isabel Allona, Universidad Politécnica de Madrid (UPM), SpainReviewed by:
Thomas Teichmann, Georg-August-Universität Göttingen, GermanyRaili Ruonala, University of Helsinki, Finland
Copyright © 2018 Busov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Victor B. Busov, dmJ1c292QG10dS5lZHU=
 Victor B. Busov
Victor B. Busov