- 1Department of Agronomy and Plant Genetics, University of Minnesota, St. Paul, MN, United States
- 2Department of Horticultural Science, University of Minnesota, St. Paul, MN, United States
- 3Department of Biology, Virginia Commonwealth University, St. Paul, MN, United States
- 4Department of Plant and Microbial Biology, University of Minnesota, St. Paul, MN, United States
Until the mid-1950s, it was believed that genetic crossovers did not occur within genes. Crossovers occurred between genes, the “beads on a string” model. Then in 1956, Seymour Benzer published his classic paper describing crossing over within a gene, intragenic recombination. This result from a bacteriophage gene prompted Oliver Nelson to study intragenic recombination in the maize Waxy locus. His studies along with subsequent work by others working with maize and other organisms described the outcomes of intragenic recombination and provided some of the earliest evidence that genes, not intergenic regions, were recombination hotspots. High-throughput genotyping approaches have since replaced single gene intragenic studies for characterizing the outcomes of recombination. These large-scale studies confirm that genes, or more generally genic regions, are the most active recombinogenic regions, and suggested a pattern of crossovers similar to the budding yeast Saccharomyces cerevisiae. In S. cerevisiae recombination is initiated by double-strand breaks (DSBs) near transcription start sites (TSSs) of genes producing a polarity gradient where crossovers preferentially resolve at the 5′ end of genes. Intragenic studies in maize yielded less evidence for either polarity or for DSBs near TSSs initiating recombination and in certain respects resembled Schizosaccharomyces pombe or mouse. These different perspectives highlight the need to draw upon the strengths of different approaches and caution against relying on a single model system or approach for understanding recombination.
Introduction
Recombination is the exchange of genetic information between chromosomes. Meiotic recombination is a major contributor to genetic diversity and facilitates selection by nature and breeders. A large share of our current understanding of recombination is based on work studying intragenic recombination (recombination within genes) in model fungal species, especially the budding yeast (Saccharomyces cerevisiae). Conclusions from genetic fungal studies have been supported by recent molecular and genomic approaches, providing a relatively detailed, although still incomplete, picture of recombination (reviewed in Keeney et al., 2014; Gray and Cohen, 2016). Beginning in the early 2000s, studies in the plant model system Arabidopsis thaliana have supported a similar picture of recombination (Wang and Copenhaver, 2018). Maize has been a genetic model organism since the early 1900s, and there is an extensive history of intragenic recombination studies in maize. Maize studies identified genes as recombination hotspots with crossovers distributed approximately evenly across many genes, which conflicts with the discrete hotspots and polarity found in S. cerevisiae. Our purpose here is to review the maize intragenic recombination work, and place this work in context with results from genomic studies of maize recombination and work in fungal and animal model systems.
To fully comprehend what intragenic as well as large-scale genomics studies can tell us about recombination, we recapitulate and reconcile knowledge from historic and more recent studies. Figure 1 depicts the approaches for gene-scale and genomic-scale to illustrate their data origins and differences. To facilitate a smooth and easy understanding of the information in this review, we first clarify the following terms which are frequently used:
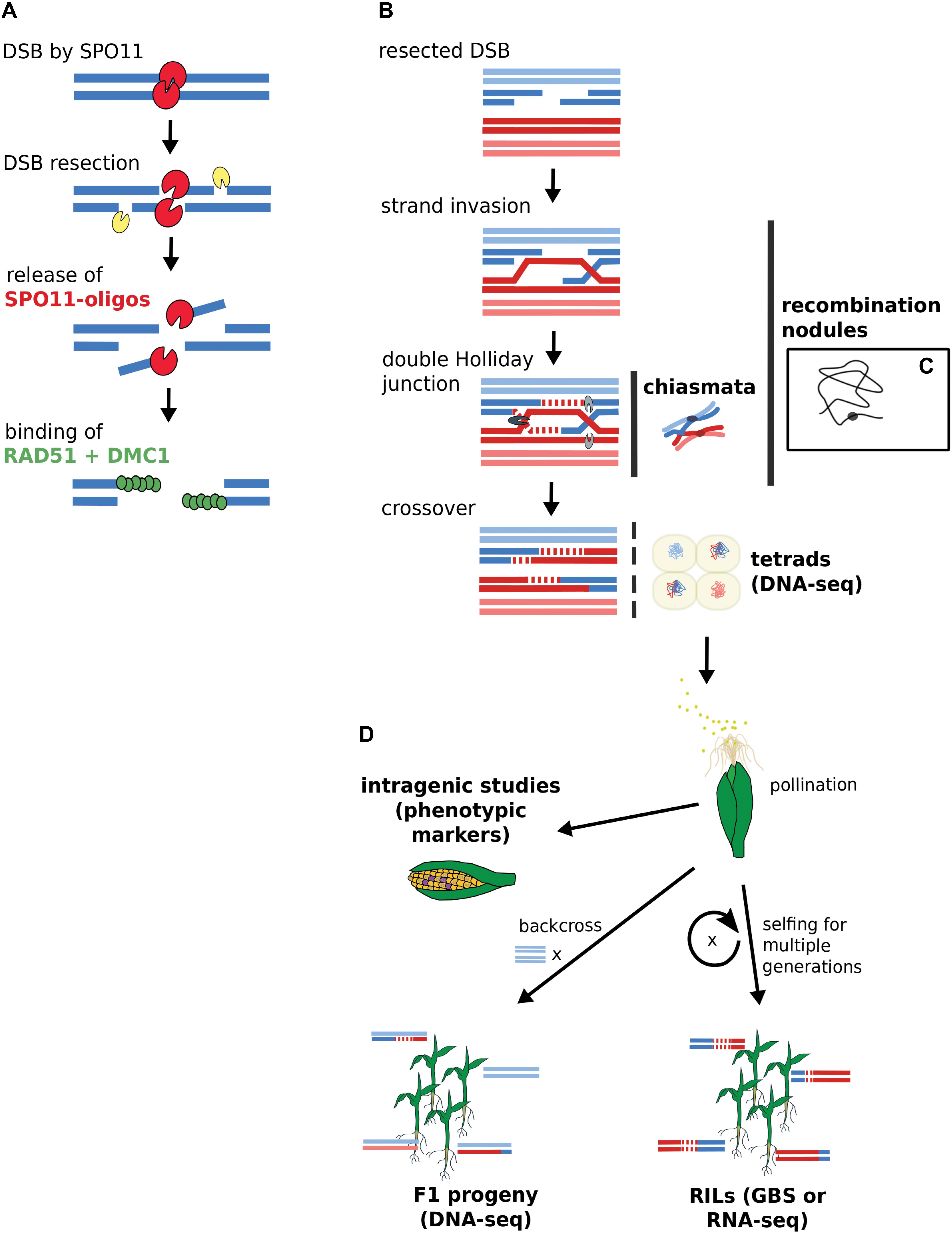
FIGURE 1. Acquisition of DSB and CO data by gene-scale and genome-scale approaches. (A) DSB generation by SPO11 with subsequent binding of RAD51 and DMC1. DSB data derives from SPO11-oligos or RAD51-bound fragments via ChIP-seq. (B) CO generation via double Holliday junction. (C) Chiasmata and recombination nodules are visible via microscopy. A single recombination nodule on a chromosome is illustrated. (D) Mapping recombinants. Sequencing approaches rely on isolated microspores from tetrads or progeny lines. Intragenic studies directly score recombination via visible kernel markers. Terms in bold indicate data source options. RILs, recombinant inbred lines; GBS, genotyping by sequencing.
• “Recombination” is a term used for mechanisms of somatic DNA repair as well as for exchange of genetic information during meiosis. While underlying mechanisms and involved proteins overlap, there are profound differences between somatic and meiotic recombination. Meiotic recombination can refer to crossovers (COs) which are due to exchange of whole chromosome parts, to non-crossovers (NCOs) which are locally restricted, or to both. Both COs and NCOs can result in local genetic changes via gene conversion.
• “Gene conversion” (GC) is the non-reciprocal transfer of information. Resolution of double Holliday structures into COs produces a gene conversion tract, as do pathways leading to NCOs. The latter is often used interchangeably with GC although it does not cover all GC instances.
• “Intragenic recombination” refers to both NCOs and COs within genes. Some intragenic recombination studies look at short regions containing several genes, albeit mostly with emphasis on the outcome within genes. These loci or genes are often recombination hotspots.
• “Hotspots” are genomic regions with elevated levels of recombination-related events, and can refer to the meiotic double-strand breaks (DSBs) initiating recombination, COs or general recombination including both COs and NCOs. There is no standard definition of hotspots regarding their strength or size. The amount of events distinguishing hotspots from cold regions is arbitrary, and the definition and identification depends strongly on the respective study.
• “Polarity” exists when there is a gradient of recombination, e.g., higher recombination rates at the 5′ or 3′ end of a gene. The 5′ end is defined here as the transcription start site (TSS), and the 3′ end as the transcription termination site (TTS). Other definitions have been used such as the promoter region for the 5′ end. However, in practice, COs are localized to intervals defined by available sequence polymorphisms which may not coincide with the defined 5′ and 3′ ends.
We have two goals for this review. First, we hope to show how the study of individual genes may influence our interpretation of genomic studies of recombination. Second, we describe characteristics in several model systems to illustrate the variation present in nature, and to argue that recombination in maize shares some, but not all, properties of each of these systems.
A History of Maize Intragenic Recombination
The classical conception of genes posited that genes were indivisible units and recombination occurs between genes (reviewed in Green, 1955; Portin, 1993). Recombination within genes, intragenic recombination, was not believed to exist, especially since several apparent exceptions turned out to be recombination between duplicated gene copies in a complex locus. An alternative position was supported by several prominent geneticists who viewed genes as having multiple sites where crossing over could occur (Pontecorvo, 1955). Arthur Chovnick’s Perspectives article in Genetics provides a historical overview (Chovnick, 1989).
Today, Seymour Benzer’s papers demonstrating intragenic recombination in bacteriophage are often seen as the experimental work changing our understanding of recombination and genes (Benzer, 1955). At that time however, it was not clear. Several explanations for the contrasting results from bacteriophage versus Drosophila melanogaster and other familiar genetic systems were proposed (Green, 1955). One possibility was that recombination was different in bacteriophage and eukaryotes. Alternatively, detecting intragenic recombination might require screening very large populations.
Nelson (1957) proposed testing intragenic recombination at the maize Waxy (Wx) locus. Wx encodes a starch synthase required for amylose in the kernel endosperm and pollen. A recombination event between two mutant alleles would create a non-mutant Wx allele giving a revertant Wx pollen grain. Non-mutant Wx pollen contains a mixture of amylose and amylopectin starches and is stained a dark black by potassium iodine, while mutant wx pollen contains only amylose and stains reddish. This pollen phenotype is readily scored under a microscope and allows screening of very large numbers of meiotic products. Using this pollen assay, Nelson was able to detect intragenic recombination in a higher eukaryote (Nelson, 1959). A second study incorporated genetic markers flanking the Wx locus to connect recombination within the Wx locus with the exchange of flanking markers (Nelson, 1962).
After the initial observation of intragenic recombination in maize, the Wx locus was used for further studies focusing on exploring the recombination process. Nelson’s studies provided early evidence for a non-crossover recombination pathway, by using lines where the wx-C allele was located inside a chromosome inversion or a complex chromosomal rearrangement (Nelson, 1975). Single-crossovers between these wx-C alleles on a rearranged chromosome and the wx-90 allele on a normal chromosome produce inviable gametes unless there was a second crossover within the inversion (Figure 2). When both wx-C and wx-90 were on normal chromosomes crossover events accounted for approximately 65% of the Wx revertants based on the segregation of flanking markers. There was crossing over between the flanking markers in 35% of the Wx revertants when wx-C was located within a pericentric inversion. A portion of these crossovers occurred outside of the inversion and accompanied a NCO event between the wx alleles. When wx-C was within a complex rearrangement the few revertants arose through non-crossover events.
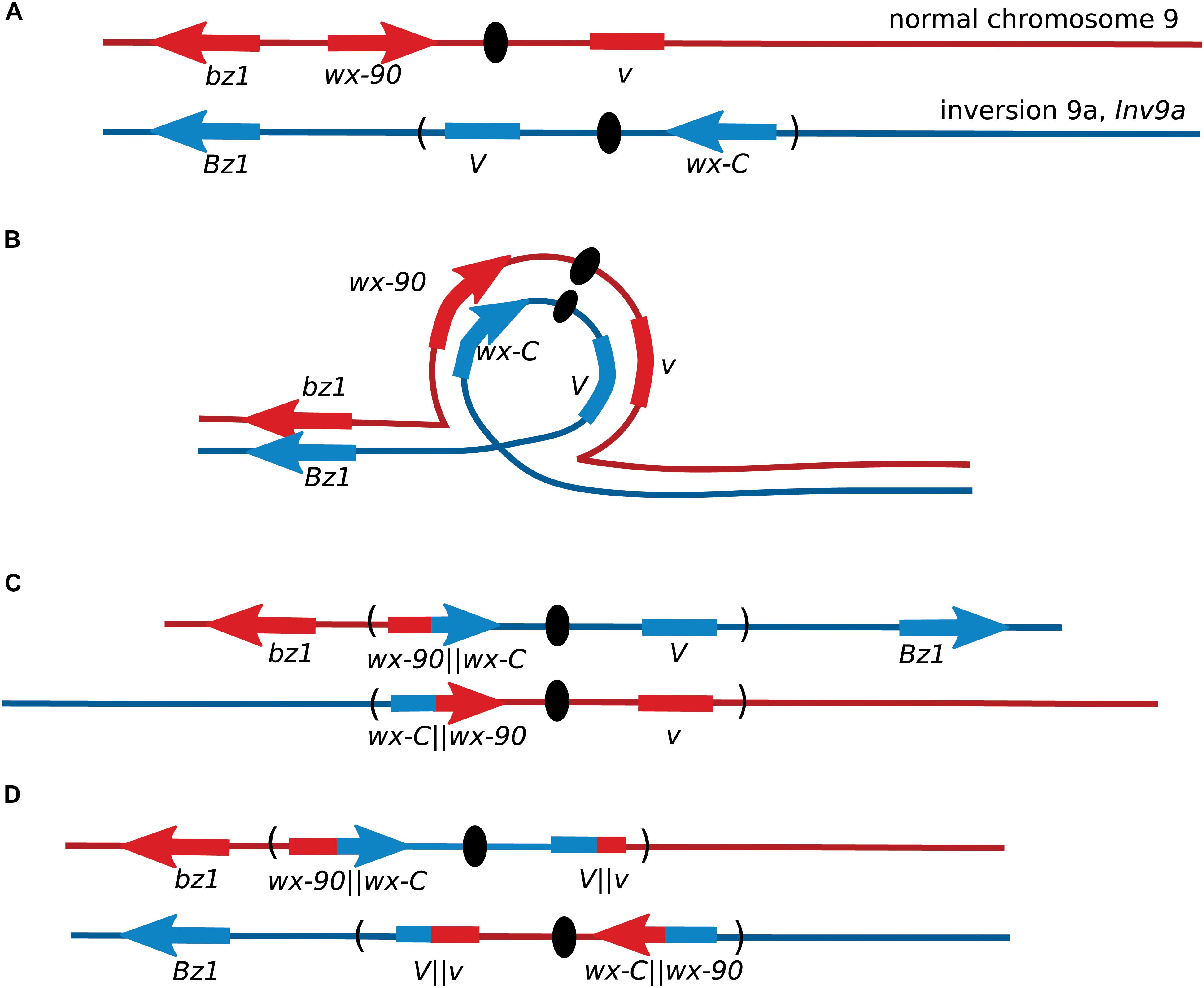
FIGURE 2. Recovery of maize Wx recombinants from an inversion heterozygote. (A) Locations of the Wx locus and the flanking markers Bronze1 (Bz1) and Virescent (V) on the normal chromosome 9, and their locations on the pericentric inversion chromosome 9. The orientation of V is not known. (B) Chromosome pairing during meiosis. (C) Gametes from meiosis with crossovers within a pericentric inversion are generally inviable because one centromere carries with it both short arms and the other centromere carries both long arms. Non-crossover events may produce non-mutant Wx kernels. (D) Crossovers between wx-C and wx-90 will produce inviable pollen unless there is a second crossover within the inversion. Most Wx revertant pollen will be from non-crossover events as double crossovers are rare in short genetic intervals. This illustration shows the second crossover occurring within V.
The Wx pollen system was also used to explore whether the distance of a locus from the centromere altered recombination frequency (Yu and Peterson, 1973). Using chromosome translocation lines with wx alleles at different distances from the centromere they showed that distance of a locus from the centromere is correlated with recombination frequency. Other studies by Peterson examined the effects of chemical treatments on recombination and noted the effect of environment on recombination at the Wx locus (Sukhapinda and Peterson, 1980).
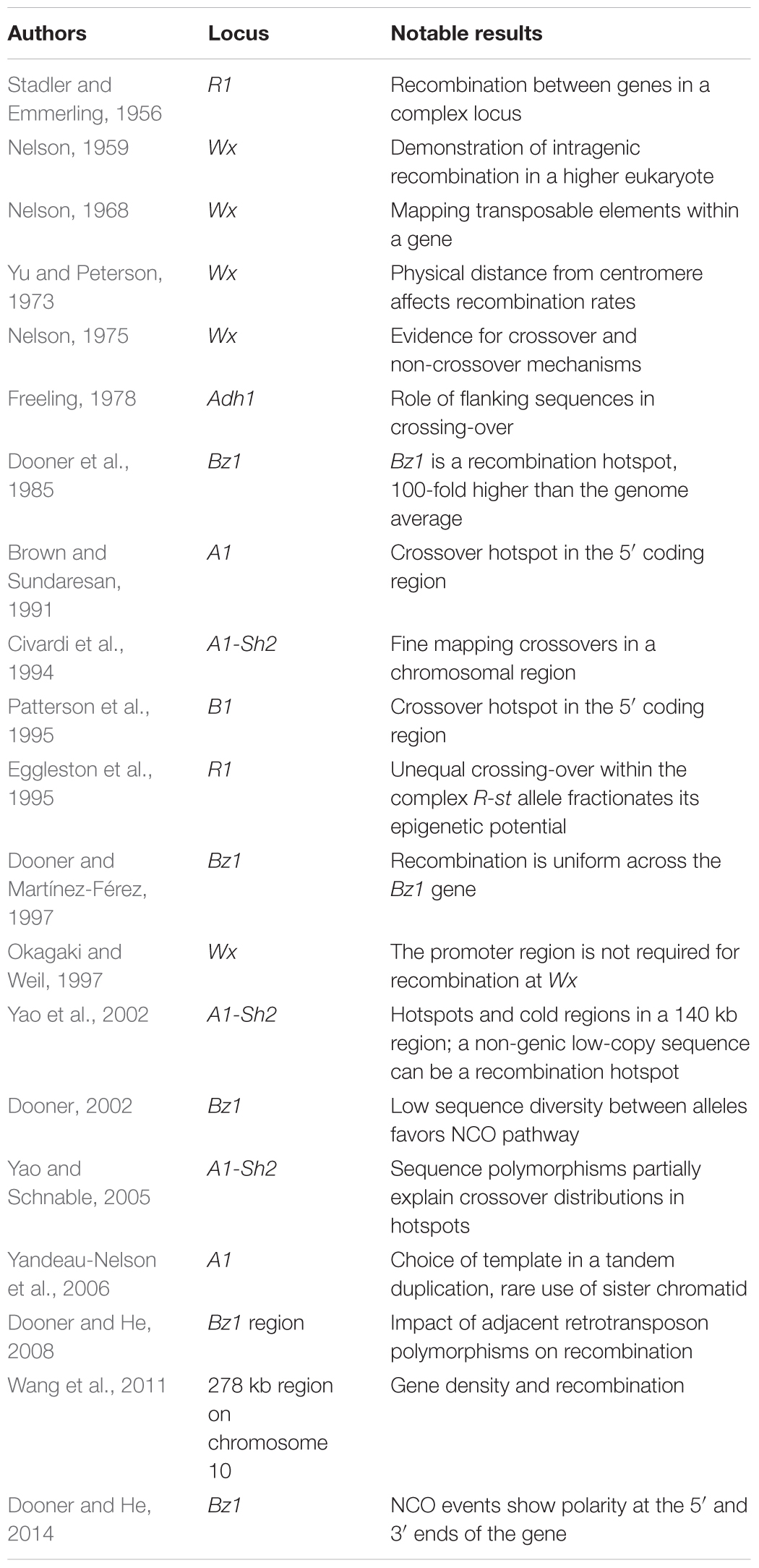
Pollen phenotyping procedures were developed for other genes to study intragenic recombination. Mike Freeling described an odd situation where alcohol dehydrogenase1 (adh1) alleles derived from the same progenitor allele showed intragenic recombination, but adh1 alleles derived from different progenitor alleles did not recombine (Freeling, 1978). One possibility suggested at the time was that local structural differences between progenitor alleles inhibited synapsis and recombination. This conjecture was supported by subsequent molecular findings of little similarity between regions flanking most, but not all, parental Adh1 alleles (Johns et al., 1983; Sachs et al., 1986). In general, maize intragenic studies focused on genes with an easily scored phenotype, and most genes studied proved to be hotspots. A partial list of key results from intragenic studies is presented in Table 1.
With the cloning and sequencing of maize genes, it became possible to compare the frequency of recombination within genes to the genome average. Hugo Dooner, studying the Bronze1 (Bz1) gene estimated that the ratio of genetic to physical distance within Bz1 was 100-fold higher than the genome average (Dooner, 1986). This result was consistent with the conjecture that recombination is restricted to genes (Thuriaux, 1977), and stands in complete contrast to the initial view of recombination occurring only between genes.
Crossing Over and Polarity
Lessons From S. cerevisiae and A. thaliana
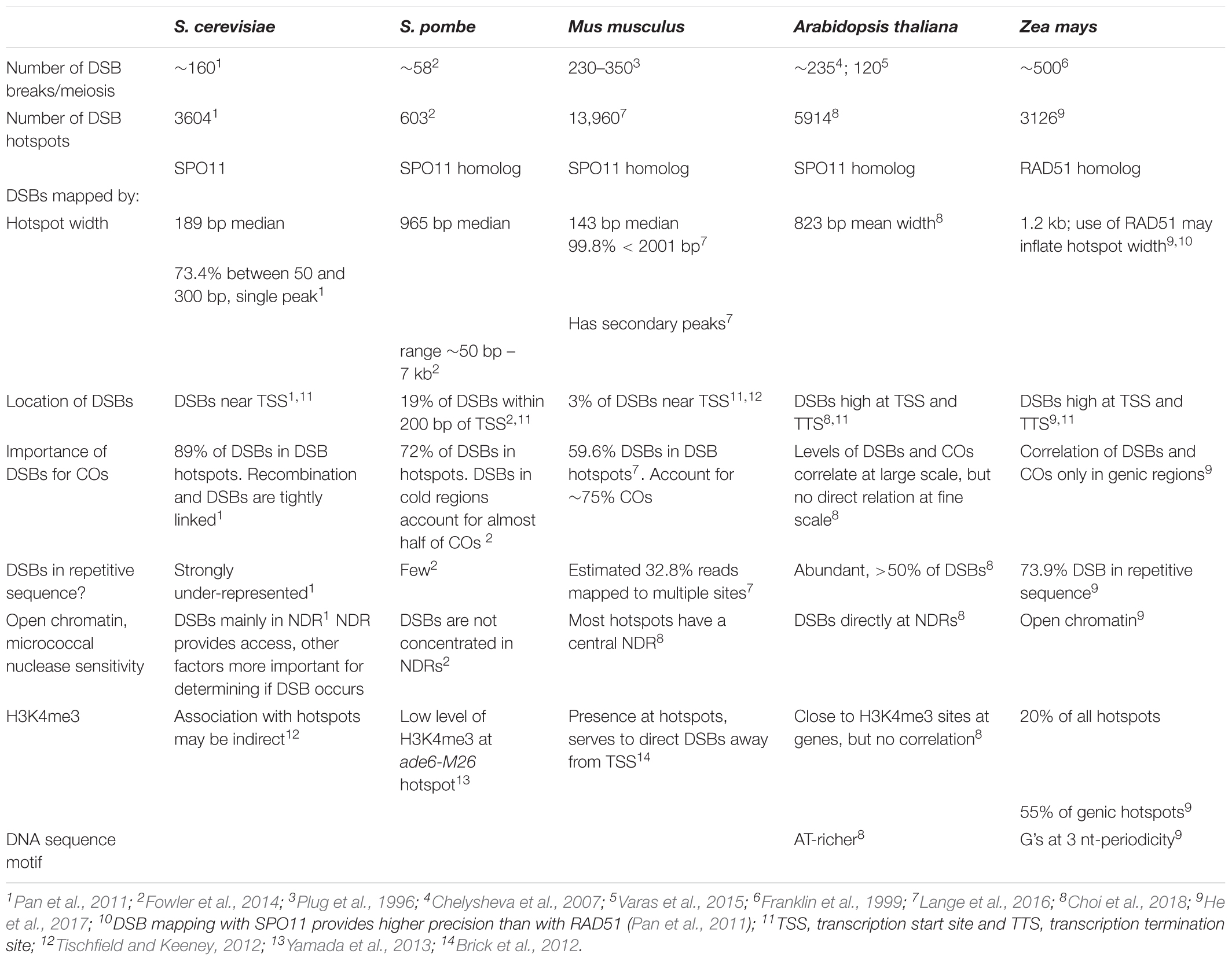
The foundation for our understanding of recombination is built upon intragenic recombination studies in fungal systems, particularly the budding yeast, S. cerevisiae (Nicolas and Petes, 1994; Gray and Cohen, 2016). These studies established a picture of recombination initiating at DSB hotspots which were usually found near TSSs (Petes, 2001). Later, this model was confirmed by genomic studies on the genome wide distribution of DSBs and meiotic recombination (Figure 1). DSB break maps, produced by capturing and sequencing SPO11-bound oligonucleotides released during initial DSB resection, confirmed that DSBs occur mainly near TSSs in S. cerevisiae (Pan et al., 2011). High resolution mapping of COs and NCOs placed 84% of recombination hotpots overlapping promoters near TSSs (Mancera et al., 2008). The agreement of genome-wide DSB maps and high-resolution recombination maps provides a clear picture of the general recombination pattern in S. cerevisiae (Table 2).
In Arabidopsis, DSB hotspots also localize to gene promoters, additionally to terminators, as well as introns (Choi et al., 2018). Though only a fraction of DSBs is resolved into COs in Arabidopsis, DSB and CO levels were shown to correlate strongly at the chromosome scale, though varying along arms (Choi et al., 2018) (Table 2).
The recombination machinery has been extensively described and reviewed in general as well as in plants (Pradillo et al., 2014; Lambing et al., 2017). Briefly, recombination initiates with a DSB. Resection creates a 3′-overhang that invades a homologous DNA region and pairs with its complementary sequence, binding it as a repair template. DNA synthesis can then proceed from the exposed 3′-end. From here, the invading strand plus newly synthesized sequence may dissociate from the complementary strand giving non-crossover events through SDSA (synthesis-dependent strand annealing). Alternatively, a double Holliday junction structure may form which can resolve into a crossover.
Both COs and NCOs give rise to a short region with non-reciprocal transfer of genetic information known as a gene conversion tract. The length of gene conversion tracts depends on DSB resection, synthesis from the exposed 3′-end, and migration of Holliday junctions. Median gene conversion tract lengths in S. cerevisiae have been measured at 2.0 kb for COs and 1.8 kb for NCOs (Mancera et al., 2008). NCO conversion tracts reached up to 40.8 kb in length, but 97% were less than 5 kb in length. Some CO and NCO conversion tracts had complex tracts arising via template switching between the parental alleles (Marsolier-Kergoat et al., 2018). In general, crossovers in S. cerevisiae are located close to the position of initiating DSBs.
Gene conversion tracts in Arabidopsis wild type have been far more difficult to detect and characterize, and are in general fewer and shorter than in budding yeast. For COs, gene conversion tracts were detected first at a maximal median length of ∼1.1 kb, for NCOs in the range of 1 bp to ∼6.6 kb (Lu et al., 2012). In another study, Arabidopsis NCO gene conversion ranged from mean tracts of 1 bp to ∼0.5 kb, the longest at ∼3.0 kb (Drouaud et al., 2013). The marker resolution underlies the precision at which gene conversion tracts can be defined, and might explain the even shorter estimates of CO-associated tracts of ∼0.3–0.4 kb and NCO-associated tracts of 25–50 bp (Wijnker et al., 2013).
Polarity for recombination is seen in many S. cerevisiae genes. The small discrete DSB hotspots located near TSSs concentrate recombination events at the 5′ end of many genes. A DSB hotspot at the 5′ end of a gene can give polarity near the 3′ end of a neighboring gene. Variation in gene conversion tract length and mismatch repair both contribute to polarity (Nicolas and Petes, 1994). Polarity can also be seen in Arabidopsis genes where recombination peaks near TSS, then decreases toward the end of genes (Hellsten et al., 2013; Choi et al., 2016). In maize, the relative importance of polarity in recombination is one of the questions arising between intragenic recombination studies and high-throughput genotyping studies.
Distribution of Crossovers in Maize Intragenic Studies
In a series of studies beginning in 1985, and continuing today, Hugo Dooner described a number of properties of maize recombination using the Bz1 locus (Table 1). For genetic crossovers, there is no polarity within Bz1. The ratio of physical distance to genetic distance (kb/cM) at the 5′ and 3′ ends of Bz1 are similar (Dooner and Martínez-Férez, 1997). This absence of polarity extends beyond Bz1 into adjacent sequences. Upstream, the genetic distance from a marker within Bz1 to the upstream gene Mkk1 100 kb away was less than the genetic length of Bz1 (Dooner, 1986; Fu et al., 2002). Similarly, no crossovers were detected in the downstream interval between Bz1 and the adjacent gene, Stc1 (He and Dooner, 2009). There is, however, polarity for NCOs at both ends of the Bz1 gene. Point mutations at both ends of Bz1 are converted more frequently than point mutations in the middle of the gene. 5′ flanking sequences are required for polarity at the 5′ end; the requirement for 3′ flanking sequences have not been tested (Dooner and He, 2014). To summarize, the number of NCOs peak at the ends of Bz1, but COs are evenly distributed across the entire Bz1 coding region and are rare in upstream and downstream regions.
Outside of Bz1 the most extensive intragenic recombination data comes from the Wx locus. Here too there is no evidence for polarity of COs. Nelson fine-mapped 29 wx alleles, and a number of these mutations were sequenced in Sue Wessler’s lab. Looking at recombination between pairs of alleles with a variety of genetic backgrounds and mutational lesions found no indication of polarity at Wx (Okagaki and Weil, 1997). Though limited to only four alleles, results at the rice Wx locus are in agreement (Inukai et al., 2000). Similarly, at the maize Stc1 locus, COs are found across the length of the gene with similar numbers of crossovers at the 5′ and 3′ ends of the gene (Dooner and He, 2008; He and Dooner, 2009).
Contrasting with the absence of polarity at the Wx locus is the strong polarity for COs at the maize A1, B1, and R1 genes. 5′-polarity was seen at A1 and B1. Thirty-three of 35 crossovers in B1 mapped to a 620 bp interval overlapping the start codon (Patterson et al., 1995. The A1 gene has a 377 bp hotspot beginning in exon 1 and spanning exon 2 (Xu et al., 1995). Recombination at R1 showed a polarity gradient with highest levels of recombination at the 3′-end of R1 that declined to low levels in the middle of the gene, a distance of approximately 3.5 kb (Eggleston et al., 1995; Dietrich, 1998; Kermicle, personal communication).
Intragenic recombination studies have also looked at recombination within small genetic intervals. Since high recombination rates measured within genes suggests that little recombination happens in intergenic regions this work directly asks whether crossing over can take place outside of genes. Studying recombination in the Al – Sh2 interval, Patrick Schnable’s group identified three CO hotspots in the 130–140 kb interval (Yao et al., 2002; Yao and Schnable, 2005). Two of the four genes in the region were CO hotspots, and the third hotspot was in a unique non-genic sequence. Only four of the 101 COs mapped outside of the three hotspots (Yao et al., 2002). The genic region surrounding Bz1 presents a similar pattern with a majority of the genes in the region functioning as CO hotspots (Fu et al., 2002; He and Dooner, 2009). The large block of repetitive sequence upstream of Bz1 is heavily methylated consistent with methylation suppressing recombination as has been seen in Arabidopsis (Melamed-Bessudo and Levy, 2012; Mirouze et al., 2012; Yelina et al., 2015). Haplotype structure and local sequence differences locally suppressing recombination provides an additional mechanism for modifying crossover frequencies (Yao and Schnable, 2005; Dooner and He, 2008).
In summary, the key results from intragenic recombination studies in maize are as follows: (1) both crossover and non-crossover events are detected; (2) many maize genes are recombination hotspots; (3) some but not all genes show polarity that may be punctate or have a gradient; (4) some recombination hotspots are in non-genic low-copy sequences; (5) sequence differences between parental chromosomes affect the distribution of recombination events. However, the number of studies with adequate data is small and conclusions about relative frequency of genes showing polarity should not be drawn.
Distribution of Maize Crossovers by High-Throughput Genotyping
At the genome-wide scale, maize COs form a particular U-shape pattern, with COs increasing strongly toward chromosome ends (Anderson et al., 2003; Li et al., 2015; Rodgers-Melnick et al., 2015; Kianian et al., 2018). Maize chromosomes have rather big pericentromeric heterochromatin regions that cover more than half of them (Baucom et al., 2009; Wei et al., 2009). Heterochromatin is thus negatively correlated with COs at large scale, but we want to keep the focus on the gene-scale data to allow comparison between traditional intragenic studies in maize and the newer cohort of sequencing-technology-driven studies.
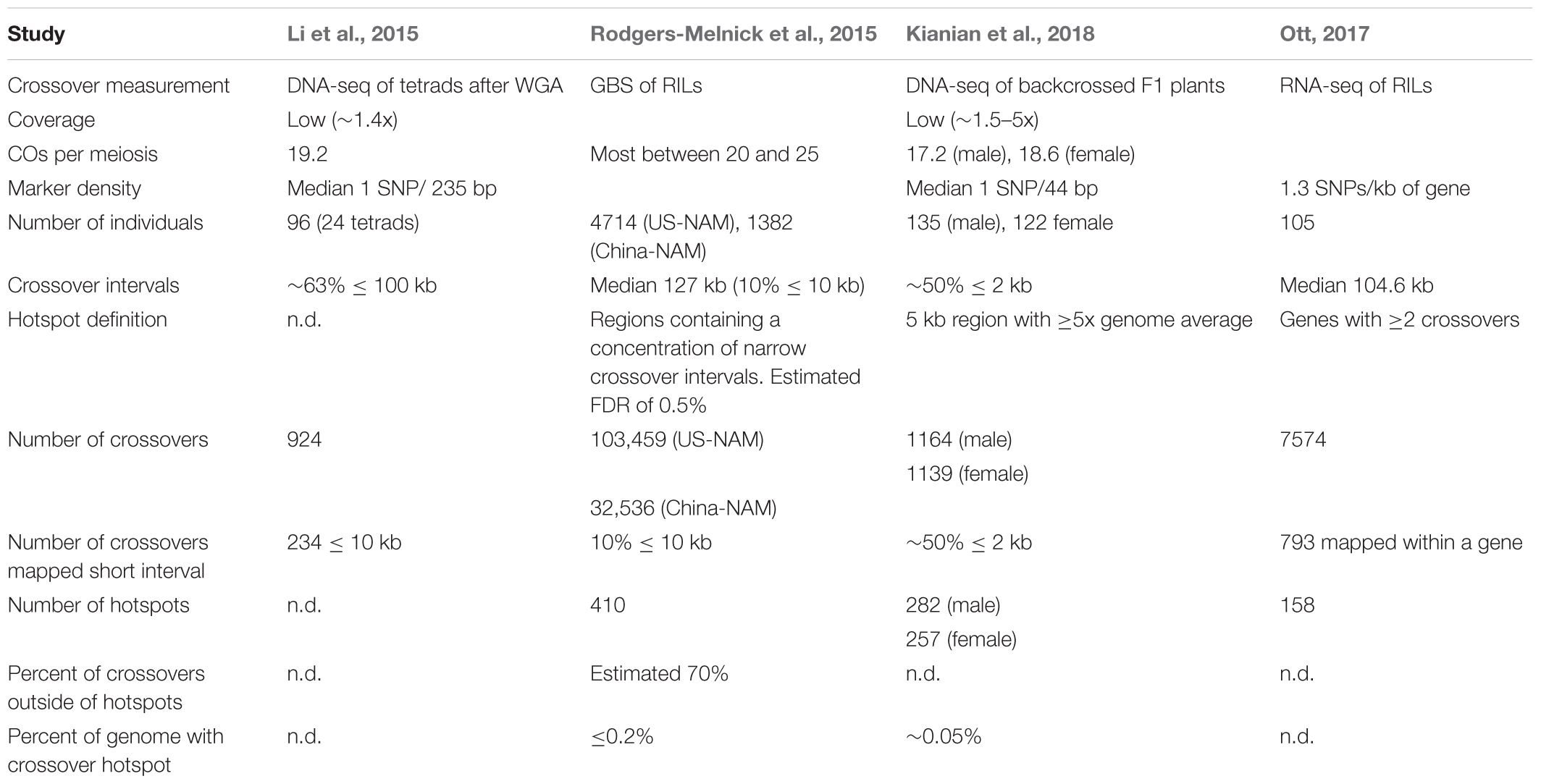
Four studies based on next-generation sequencing have mapped recombination events in maize (Table 3). Three studies have been published (Li et al., 2015; Rodgers-Melnick et al., 2015; Kianian et al., 2018). Data from the fourth study was reported in Alina Ott’s Ph.D. dissertation, and a manuscript is in preparation (Ott, 2017). High marker density is critical for studying polarity and other questions. Three maize genome-wide studies reported polarity for crossovers, with COs most frequent in the 5′ region of genes and low in the central region of genes (Li et al., 2015; Ott, 2017; Kianian et al., 2018). The resolution of one study was generally not sufficient to address this question (Rodgers-Melnick et al., 2015). Crossovers were mapped with sufficient precision to identify polarity for approximately 50% (Kianian et al., 2018) and approximately 10% of crossovers placed (Li et al., 2015; Ott, 2017). Two of the studies reported evidence for high crossover frequency at the 3′ end of the gene (Li et al., 2015; Kianian et al., 2018). Recombination polarity at the 5′ and often 3′ ends of genes has been reported to be the common pattern in several plant species (reviewed in Choi and Henderson, 2015).
Crossover hotspots were identified in three studies (Rodgers-Melnick et al., 2015; Ott, 2017; Kianian et al., 2018). Kianian’s study defined hotspots as 5 kb regions with CO rates fivefold higher than the genome average; there were 282 and 257 hotspots in the male and female parents of the population respectively (Kianian et al., 2018). Using the 793 COs mapped within a gene, Ott identified 158 genes with more than one CO event in her population; many of these genes are statistically likely to be CO hotspots (Ott, 2017). These two studies relied on relatively small populations. Using a much larger population, Rodgers-Melnick’s study found 410 hotspots (Rodgers-Melnick et al., 2015). Not all of the genic hotspots described by intragenic recombination studies were identified in these studies. Sampling depth may be a limiting factor in detecting CO hotspots, but the variable number and locations of CO hotspots suggests we need to think carefully about the meaning of hotspots.
Conclusions drawn from high-throughput genotyping studies emphasized polarity with COs concentrated at 5′ and 3′ ends of genes (Li et al., 2015; Ott, 2017; Kianian et al., 2018). Although this differs at first glance from intragenic studies which reported genes with and without polarity, the data actually agrees. Intragenic studies reporting on individual genes found a mix of genes showing polarity and others that do not. What is reported in genome-wide studies is an accumulated pattern from 100s of genes. The polarity found in these studies could be a result of a generalized polarity at most genes or the result of a mix of genes with and without CO polarity as seen in intragenic studies. While high-throughput studies emphasize increased CO rates at 5′ and 3′ ends of genes, intragenic studies report on individual genes, exposing the mix of genes with large diffuse hotspots or localized hotspots (Figure 3). Similarly, in Arabidopsis, it has been shown that the level of polarity-underlying DSBs at the TSS and TTS are independent from each other (Choi et al., 2018).
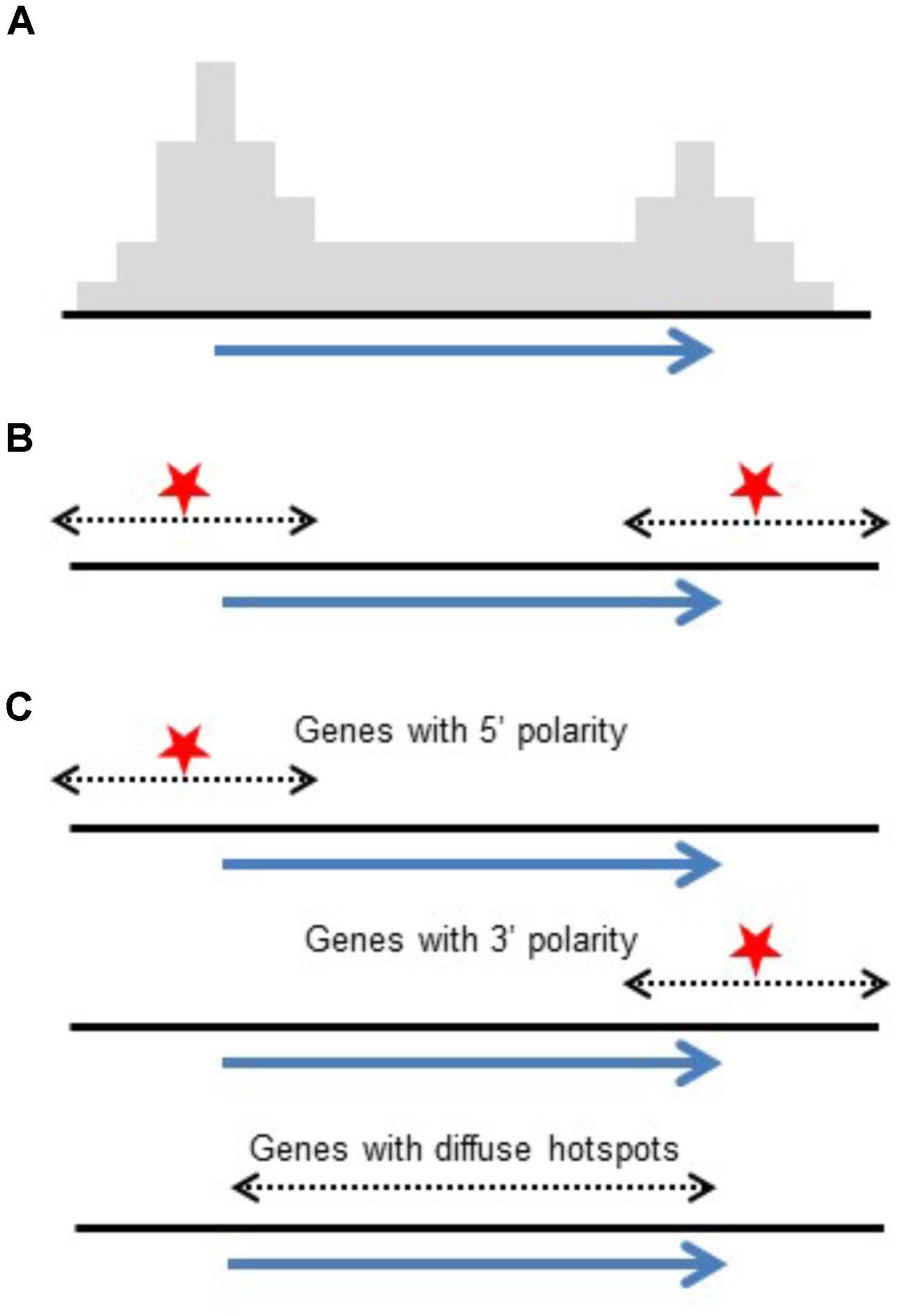
FIGURE 3. Reconciling polarity gradients as seen in high-throughput genotyping versus intragenic studies. (A) Histogram representing crossover polarity seen in high-throughput genotyping studies. (B) DSB hotspots at both ends of genes could produce this distribution. (C) Alternatively, a mix of genes having DSB hotspots at their 5′ ends, their 3′ ends, and genes that are diffuse hotspots give the same pattern of crossovers.
Double-Strand Break Hotspots
Lessons From S. cerevisiae and A. thaliana
In S. cerevisiae, most meiotic DSBs resolve into COs and NCOs. The 140–170 DSBs observed per yeast meiosis (Buhler et al., 2007) closely match the 90.5 COs and 46.5 NCOs counted per meiosis (Mancera et al., 2008). Precise mapping of DSB hotspots via SPO11-bound nucleotides released at resection revealed that almost 90% of DSBs occurred in a described hotspot (Pan et al., 2011) (Table 2). DSBs were underrepresented in repetitive sequences with repetitive DNA comprising 14% of the genome while accounting for only 1.16% of DSB breaks defined by SPO11 reads (Pan et al., 2011). Overall, 95% of DSBs identified by SPO11 oligonucleotides were confined to just 15% of the yeast genome (Marsolier-Kergoat et al., 2018). In Arabidopsis, mapping of SPO11-oligonucleotides also revealed that DSBs are preferentially located in regions with high gene density, and underrepresented in TE dense regions (Choi et al., 2018).
Although there was no consensus target sequence for the SPO11 nuclease responsible for meiotic DSBs, the DNA sequence was non-random with preferred nucleotides at certain positions, and a central AT-rich sequence surrounded by modestly GC-rich sequence. AT-rich motifs were also found at Arabidopsis DSB sites (Choi et al., 2018), likely due to those motifs generally excluding nucleosomes (Segal and Widom, 2009).
Saccharomyces cerevisiae DSBs map preferentially to nucleosome depleted regions (NDRs) around the TSS, a hallmark of open chromatin (Pan et al., 2011). The trimethylated histone H3 lysine 4 (H3K4me3) is a histone modification promoting an open chromatin structure that is found at DSB hotspots (Borde et al., 2009). Noteworthy, however, is that not all NDRs are DSB hotspots and H3K4me3 is absent at some DSB hotspots (Pan et al., 2011; Tischfield and Keeney, 2012). This picture of open chromatin being favored by the DSB machinery is also true in Arabidopsis: here, DSBs were shown to correlate with H3K4me3 and low nucleosome density (Choi et al., 2018).
In summary, the correlation of DSBs and recombination in S. cerevisiae is clear as almost all DSBs lead to recombination and both the initiating DSBs and resolving recombination hotspots are mainly found near TSSs. The picture is different in other systems, including Arabidopsis, where a few 100 DSBs get resolved into only ∼6–12 COs (Giraut et al., 2011; Lu et al., 2012; Salomé et al., 2012; see Table 2).
Location and Distribution of Maize Double-Strand Breaks
Double-strand breaks cannot be directly mapped by intragenic studies, but their possible positions may be deduced by examining recombination in deletion mutations. Homozygous deletions of DSB hotspots reduce recombination and eliminate polarity at the S. cerevisiae HIS4 locus (Detloff et al., 1992). The maize wx-B allele is a 1 kb deletion around the TSS from -459 to +505, while wx-C4 has a smaller deletion within the transcribed region from +257 to +454 (Wessler et al., 1990; Okagaki and Weil, 1997). If DSBs near the TSS contribute substantially to recombination, then more recombinants should be recovered between alleles with intact TSS regions than with alleles containing TSS region deletions. This, however, was not seen when Oliver Nelson measured recombination between wx-B and wx-C4 with downstream alleles (Supplementary Table 1). Second, there is directionality to the repair of meiotic DSBs since DSBs are repaired using sequences on the homologous chromosome. Thus in a line where one allele contains a deletion of its DSB hotspot and the other allele retains its DSB hotspot, recombination preferentially deletes the previously non-mutant sequence (Nicolas et al., 1989). The wx-B1 allele has a deletion from -655 to +299 (Wessler et al., 1990). Recombination between wx-B1 and the wx-I allele, containing a large insertion in the 3′ region, also argued against a single DSB hotspot near the TSS (Okagaki and Weil, 1997). Here, 28 of 29 recombinants were crossovers between wx-B1 and wx-I. The DSBs in this experiment most likely were in the region between wx-B1 and wx-I rather than in the 5′ region.
Indirect evidence against a yeast-like concentration of DSBs near TSSs in maize comes from one whole genome study. A high-throughput recombination study via the maize transcriptome mapped 2634 NCO conversion tracts in a maize RIL population (Ott, 2017). Non-crossover conversion tracts in the 5′, the central, and 3′ regions of maize genes were roughly equally distributed, with a slightly higher number in the middle region of genes (Ott, 2017). According to the original canonical DSB model, gene conversion tracts produced at NCOs and COs will flank or encompass the position of initiating DSBs, thus indirectly mapping DSBs (Szostak et al., 1983). More recent data in S. cerevisiae shows that NCO conversion tracts can be located a short distance from the DSB (Marsolier-Kergoat et al., 2018). The even distribution of NCO conversion tracts across maize genes argues against the concentration of DSBs at the ends of maize genes.
A recent maize genomic DSB study using a genome-wide approach similar to the one used in S. cerevisiae and other model species captured and sequenced single-stranded DNA bound to RAD51 (He et al., 2017). In total, the maize DSB mapping effort identified 3126 hotspots (He et al., 2017), similar to the 3604 DSB hotspots in S. cerevisiae (Pan et al., 2011). The number of defined maize DSB hotspots is conservative. With relaxed stringency and controls the number is considerably larger (He et al., 2017). Although maize DSB hotspots shared some characteristics with their S. cerevisiae counterparts and DSB hotspots from Arabidopsis and other model systems, there are some clear differences (Table 2). The almost 73% of maize DSB hotspots in repetitive sequence contrasts with the under-representation of DSB hotspots in S. cerevisiae repetitive sequences. Maize DSB hotspots were also wider than S. cerevisiae DSB hotspots, 1.2 kb versus 189 bp. This difference might be a consequence of the precision of the SPO11 based approach used in S. cerevisiae versus the lower resolution possible with the RAD51 based approach used in maize, or it could represent physical differences in their DSB hotspots. Perhaps of greater interest is the close relationship between DSB hotspots and COs found in S. cerevisiae may not hold in maize (see section “The Role of Double-Strand Breaks in Maize and Other Model Organisms”). One mapped DSB hotspot was immediately upstream of Bz1. However, as we have seen from intragenic studies, COs at Bz1 do not show polarity, although NCOs show polarity at the 5′ and 3′ ends of Bz1 (Dooner and Martínez-Férez, 1997; Dooner and He, 2014). Thus the importance of a DSB hotspot flanking Bz1 for recombination is unclear.
Approximately, 85% of the maize genome is composed of families of repetitive elements widely distributed in the maize genome (Schnable et al., 2009). Ty1-gypsy-like elements are the most abundant families (Meyers et al., 2001). Over 50% of maize DSB hotspots are in gypsy-like elements (He et al., 2017). Both DNA methylation and sequence polymorphisms are suggested mechanisms for suppressing COs in maize repetitive sequences (Fu et al., 2002; Yao and Schnable, 2005). Repetitive elements are commonly found in blocks separating individual genes or small clusters of genes (Haberer et al., 2005). These blocks are not conserved between maize lines, and two genes may be separated by a short stretch of low-copy sequence in one line and by significant stretches of repetitive sequence resulting from multiple repetitive elements in another line (He and Dooner, 2009). This absence of sequence homology will strongly inhibit recombination. Even when the overall structure of a repetitive block is preserved, nucleotide polymorphisms could locally inhibit crossing over, as seen in the a1-sh2 region (Yao and Schnable, 2005). However, results from one intragenic recombination study argues against sequence polymorphism as the primary mechanism suppressing crossing over in repetitive sequences (Fu et al., 2002). In this study using the Bz1 region, crossing over was compared between a short genic region and a block of repetitive sequence flanking the genes. Because the haplotypes used in this experiment were derived from the same progenitor, there was little if any sequence difference in the region except for the genetic markers. In this region, at least, sequence differences cannot account for the lack of crossing over in repetitive sequence.
The Role of Double-Strand Breaks in Maize and Other Model Organisms
Meiotic DSBs serve two functions, first to promote chromosome pairing and second to produce the crossovers necessary to ensure proper segregation of chromosomes at anaphase (Page and Hawley, 2003). Meiotic DSBs can be visualized on chromosomes as RAD51 foci (Franklin et al., 1999). In mid-zygotene when chromosomes are pairing, there are approximately 500 RAD51 foci decreasing to about 12 RAD51 foci in pachytene (Franklin et al., 1999; Pawlowski et al., 2003). The zygotene foci are distributed across the chromosomes where single-stranded DNA ends produced by DSBs and resection promote chromosome alignment (Smithies and Powers, 1986; Peoples-Holst and Burgess, 2005). The pachytene foci, also known as late recombination nodules when viewed with electron microscopy, represent the sites of crossing-over (Stack and Anderson, 2002). Control of the number of COs and which DSBs are channeled into the crossover pathway is tightly regulated (Lake and Hawley, 2016). There are two types of COs, with interference-sensitive COs (type I) constituting the majority of COs, and a few additionally interspersed interference-insensitive COs (type II) in many species (Gray and Cohen, 2016). Some species lack the type I interference-sensitive pathway (Gray and Cohen, 2016). Though their mechanisms are distinct, their outcomes are treated equally in intragenic as well as whole genome studies.
Double-strand breaks are necessary for recombination, but the importance of DSB hotspots for genetic crossovers is less clear. Compared with S. cerevisiae, the fraction of maize DSBs resolving as crossovers is small. Mapping crossovers using S. cerevisiae tetrads gave an average of 90.5 crossovers per meiosis, with an estimated 160 DSBs per meiosis (Mancera et al., 2008; Pan et al., 2011). In contrast, only a fraction (∼5–10%) of DSBs get resolved into COs in Arabidopsis (Giraut et al., 2011; Lu et al., 2012; Salomé et al., 2012; also see Table 2). Mapping crossovers from maize tetrads determined an average of 19 crossovers per meiosis (Li et al., 2015), similar to the range of cytologically determined CO in maize inbreeds (Sidhu et al., 2015). Thus, less than 4% of maize DSBs resulted in COs versus 56% in S. cerevisiae tetrads. In fact, the majority of DSB hotspots appear unlikely to contribute much to crossing over as almost 73% of hotspots are in repetitive sequence where crossovers are believed to be suppressed (He et al., 2017). It seems reasonable to conclude that a majority of maize DSBs promote chromosome alignment (Peoples-Holst and Burgess, 2005).
The genome-wide maize DSB data identified 3126 high-confidence DSB hotspots, about one-fourth of them in genes. This is a conservative estimate of the number of hotspots based on very stringent criteria (He et al., 2017). What the genomic maize DSB study shows clearly, however, is the concentration of DSBs around TSS and TTS of many genes (He et al., 2017). This is mirrored in other model systems, for example Arabidopsis, with highest DSB levels at TSS and TTS (Choi et al., 2018), and is in general a prerequisite for recombination polarity along a gene body.
Though the concept is enticing, enrichment of maize DSB hotspots around the TSS and TTS of genes may not exist at all genes, with or without producing polarity. On average, DSBs have an increased tendency to peak at TSS and on both sides of TTS (He et al., 2017), agreeing with CO peaks close to TSS and TTS (Kianian et al., 2018). However, polarity for CO is not seen at Bz1 despite the adjacent DSB hotspot (Dooner and Martínez-Férez, 1997; He et al., 2017). In S. cerevisiae, the tight connection between DSB hotspots and recombination is well-established (Pan et al., 2011; Marsolier-Kergoat et al., 2018), but there are large differences between eukaryotes (Fowler et al., 2014; Stapley et al., 2017). Results from similar studies in the fission yeast Schizosaccharomyces pombe (S. pombe) present a very different picture (Table 2). Less than 20% of S. pombe DSB hotspots are near the TSS. Furthermore, DSBs in S. pombe hotspots are preferentially repaired from the sister chromatid; these events do not contribute to crossing-over. A large fraction of COs are initiated at non-hotspot DSBs in S. pombe (Fowler et al., 2014).
On the other hand, S. pombe DSB hotspots are not strongly correlated with NDRs (Fowler et al., 2014). This contrasts with S. cerevisiae, Arabidopsis and maize.
Mouse DSB hotspots share characteristics of both S. cerevisiae and S. pombe and hint at the complexities underlying hotspots. DSB hotspot widths are similar to S. cerevisiae (Lange et al., 2016). Unlike S. cerevisiae, these hotspots are over-represented within the genic region defined by the start and stop codons, and only 3% are located near the TSS (Smagulova et al., 2011; Brick et al., 2012). The H3K4me3 modification at mouse DSB hotspots is produced by the Prdm9 histone methyltransferase (Baudat et al., 2010). H3K4me3 is present at other sites along mouse chromosomes including TSS. Removing H3K4me3 at DSB hotspots using prdm9 mutant mice blocks DSBs from forming at these hotspots. Instead, DSBs occur at other H3K4me3 sites on the chromosome; many of these are near the TSS (Brick et al., 2012). Of critical importance here is the observation that these mice are defective in DSB repair and chromosome pairing (Hayashi et al., 2005).
Open chromatin is so far the only universal criteria for DSBs and CO locations across different model systems, though caution is needed regarding the scale (Tischfield and Keeney, 2012). The active chromatin mark H3K4me3 for example can be found near DSBs, but overlaps COs even more, arguing that it promotes recombination downstream of DSBs, in yeast and Arabidopsis, while DSB association is merely due to location to promoters (Tischfield and Keeney, 2012; Choi et al., 2018). Rather, chromosomal context and the relationship between the DSB and the synaptonemal complex are important (Medhi et al., 2016). DNA methylation is yet another component underlying the structure of the chromosome, but details on its association with DSB and COs are beyond the scope of this review, which focused on intragenic recombination.
No Model System Explains All
As described here, there are aspects of systems other than S. cerevisiae that provide insight into maize. For example, in S. pombe, DSBs in hotspots contribute far less to COs than expected – most DSBs in hotspots do not resolve as COs. Might this be similar to maize where three-fourths of DSB hotspots are in repetitive sequence? Does the absence of class I COs in S. pombe disqualify S. pombe as a model system for maize recombination, any more than the under-representation of DSB hotspots in S. cerevisiae repetitive sequence disqualifies S. cerevisiae as a model for maize where three-fourths of DSB hotspots are in repetitive sequence? Is there a clear reason why class I versus class II COs are a more important criteria for choosing a reference model system than the weak association between DSB hotspots and COs in maize and S. pombe versus the tight association in S. cerevisiae?
Deleting the promoter region in a yeast gene strongly reduces recombination at the gene (De Massy and Nicolas, 1993; Porter et al., 1993). Deleting the promoter region in maize Bz1 and Wx does not strongly reduce recombination at the gene (see section “Lessons From S. cerevisiae and A. thaliana,” Supplementary Table 1).
Mouse provides the valuable lesson that open chromatin in promoters is not necessarily a target for DSBs. On the other hand, mouse DSB hotspots are mediated by PRDM9 which does not seem to exist in plants. Are plants as the mostly related species not the best system to compare with maize? In spite of different genome architecture of Arabidopsis and maize, many commonalities of DSB and CO hotspots can indeed be found. The agreement of recombination distribution is even better when comparing other large-genome crops with each other, as for example maize, wheat, and barley (Higgins et al., 2012; Darrier et al., 2017). Only when integrating information learned from different model organisms and different approaches do we have a chance of resolving the whole story on DSBs, recombination, polarity, and underlying genome features.
Summary
Maize has been a genetic model genetic system for almost 100 years, and has been used to address questions regarding recombination as fundamental as the connection between cytological crossing over and genetic crossing over (Creighton and McClintock, 1931). However, there is a chasm between what we know based on extensive data, what we think we know, and what is known in other model systems used for studying recombination. Intragenic studies on small genetic regions have characterized most genes as recombination hotspots, but some genes are coldspots and some non-genic regions are recombination hotspots (Yao et al., 2002; He and Dooner, 2009; Wang et al., 2011). Issues may arise when extrapolating results from the handful of maize genes where intragenic recombination has been studied in depth. On the other hand, high-throughput genotyping studies suffer from a lack of resolution or depth, and small sample sizes. The median interval defining crossovers in three of the four studies was over 100 kb (Li et al., 2015; Rodgers-Melnick et al., 2015; Ott, 2017). Greater precision is necessary to minimize the misleading correlations possible with low-resolution mapping (Tischfield and Keeney, 2012).
Where do crossovers occur in maize? At the chromosomal level, intragenic and genome-wide studies identified an association between gene-density and elevated crossover rates (Yao et al., 2002; Wang et al., 2011; Pan et al., 2017). Looking at single genes or small genetic intervals, intragenic studies conclude that most crossovers take place within genes. Genes appear to fall into two categories with crossovers either concentrated at one end of the gene, either 5′ or 3′ polarity, or distributed evenly across the gene (Eggleston et al., 1995; Patterson et al., 1995; Dooner and Martínez-Férez, 1997; Okagaki and Weil, 1997; Yao and Schnable, 2005). High-throughput genotyping studies, drawing parallels with the polarity found in S. cerevisiae genes, emphasize polarity at the 5′ and 3′ ends of genes (Li et al., 2015; Ott, 2017; Kianian et al., 2018). But two of these studies determined that about one-fourth of the crossovers mapped within a gene were in the central region (Li et al., 2015; Ott, 2017). In the third study, 50% of the crossovers did not map to either the 5′ or 3′ regions (Kianian et al., 2018). Intragenic studies look at many recombination events at a few genes while genomic approaches average over many genes with few recombination events each. The two approaches give different snapshots and interpretations of crossing over and point to their relative strengths and weaknesses.
Due to the underlying literature we have focused this review on hotspots for crossover and DSBs. This perspective may be problematic. It seems that hotspots are not the best means to describe crossovers in maize which could be better described as following a chromosome-wide polarity gradient toward the chromosome ends coupled with an avoidance of repetitive sequence in the case of COs. This wider perspective encompasses additional questions including the importance of trans-acting modifiers, frequently genes from recombination pathways (Pan et al., 2017).
In addition, the fraction of crossovers attributed to recombination hotspots is not high. The 410 CO hotspots defined by Rodgers-Melnick accounted for 30.6% crossovers defined to a narrow interval (Rodgers-Melnick et al., 2015). Our interest in hotspots seems to be focusing our attention on local determinants of recombination as a functional unity and away from the larger and possibly more important chromosomal context (Pan et al., 2011). Where a DSB occurs along a chromosome may be just as important in determining the DSB fate as local hotspot features (Serrentino and Borde, 2012). There is now an intense interest and effort in understanding the roles of chromatin features including the synaptonemal axis, loops, and cohesin proteins, which will help refine our views on meiotic recombination mechanisms and patterns (Barrington et al., 2017).
Author’s Note
Results from Alina Ott’s Ph.D. dissertation have now been published in Liu et al. (2018).
Author Contributions
RO: conception of this project. SD-S: re-analyzing recombination data. WE: R1 recombination data. RO and SD-S: drafting of manuscript. RO, SD-S, WBE, and GM: editing of manuscript. All authors approved the final version of the manuscript.
Funding
SD-S thanks Changbin Chen for all his support and the opportunity to work on maize meiosis, supported by the National Science Foundation (IOS-1025881 and IOS-1546792), and a grant-in-aid fund from the University of Minnesota. GM received funding from Endowed Chair in Molecular Genetics Applied to Crop Improvement.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This paper is dedicated to Oliver E. Nelson in recognition for his many contributions to the study of genetics.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01560/full#supplementary-material
References
Anderson, L. K., Doyle, G. G., Brigham, B., Carter, J., Hooker, K. D., Lai, A., et al. (2003). High-resolution crossover maps for each bivalent of Zea mays using recombination nodules. Genetics 165, 849–865.
Barrington, C., Pezic, D., and Hadjur, S. (2017). Chromosome structure dynamics during the cell cycle: a structure to fit every phase. EMBO J. 36, 2661–2663. doi: 10.15252/embj.201798014
Baucom, R. S., Estill, J. C., Chaparro, C., Upshaw, N., Jogi, A., Deragon, J.-M., et al. (2009). Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet. 5:e1000732. doi: 10.1371/journal.pgen.1000732
Baudat, R., Buard, J., Grey, C., Fledel-Alon, A., Ober, C., Przeworski, M., et al. (2010). PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327, 836–840. doi: 10.1126/science.1183439
Benzer, S. (1955). Fine structure of a genetic region in bacteriophage. Proc. Natl. Acad. Sci. U.S.A. 41, 344–354. doi: 10.1073/pnas.41.6.344
Borde, V., Robine, N., Lin, W., Bonfils, S., Géli, V., and Nicolas, A. (2009). Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 28, 99–111. doi: 10.1038/emboj.2008.257
Brick, K., Smagulova, F., Khil, P., Camerini-Otero, R. D., and Petukhova, G. V. (2012). Genetic recombination is directed away from functional genomic elements in mice. Nature 485, 642–645. doi: 10.1038/nature11089
Brown, J. W. S., and Sundaresan, V. (1991). A recombination hotspot in the maize A1 intragenic region. Theor. Appl. Genet. 81, 185–188. doi: 10.1007/BF00215721
Buhler, C., Borde, V., and Lichten, M. (2007). Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol. 5:e324. doi: 10.1371/journal.pbio.0050324
Chelysheva, L., Gendrot, G., Vezon, D., Doutriaux, M.-P., Mercier, R., and Grelon, M. (2007). Zip4/Spo22 is required for Class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet. 3:e83. doi: 10.1371/journal.pgen.0030083
Choi, K., and Henderson, I. R. (2015). Meiotic recombination hotspots – a comparative view. Plant J. 83, 52–61. doi: 10.1111/tpj.12870
Choi, K., Reinhard, C., Serra, H., Ziolkowski, P. A., Underwood, C. J., Zhao, X., et al. (2016). Recombination rate heterogeneity within Arabidopsis disease resistance genes. PLoS Genet. 12:e1006179. doi: 10.1371/journal.pgen.1006179
Choi, K., Zhao, X., Tock, A. J., Lambing, C., Underwood, C. J., Hardcastle, T. J., et al. (2018). Nucleosomes and DNA methylation shape meiotic DSB frequency in Arabidopsis thaliana transposons and gene regulatory regions. Genome Res. 28, 532–546. doi: 10.1101/gr.225599.117
Civardi, L., Xia, Y., Edwards, K. J., Schnable, P. S., and Nikolau, B. F. (1994). The relationship between genetic and physical distances in the cloned a1-sh2 interval of the Zea mays L. genome. Proc. Natl. Acad. Sci. U.S.A. 91, 8268–8272. doi: 10.1073/pnas.91.17.8268
Creighton, H. B., and McClintock, B. (1931). A correlation of cytological and genetical crossing-over in Zea mays. Proc. Natl. Acad. Sci. U.S.A. 17, 492–497. doi: 10.1073/pnas.17.8.492
Darrier, B., Rimbert, H., Balfourier, F., Pingault, L., Josselin, A.-A., Servin, B., et al. (2017). High-resolution mapping of crossover events in the hexaploid wheat genome suggests a universal recombination mechanism. Genetics 206, 1373–1388. doi: 10.1534/genetics.116.196014
De Massy, B., and Nicolas, A. (1993). The control in cis of the position and amount of the ARG4 meiotic double-strand break of Saccharomyces cerevisiae. EMBO J. 12, 1459–1466. doi: 10.1002/j.1460-2075.1993.tb05789.x
Detloff, P., White, M. A., and Petes, T. D. (1992). Analysis of a gene conversion gradient at the HIS4 locus in Saccharomyces cerevisiae. Genetics 132, 113–123.
Dietrich, W. R. (1998). Determination of Rates and Patterns of Recombination at the Maize Red Color (r1) Locus. MS thesis, Richmond, VA, Virginia Commonwealth University.
Dooner, H. K. (2002). Extensive interallelic polymorphisms drive meiotic recombination into a crossover pathway. Plant Cell 14, 1173–1183. doi: 10.1105/tpc.001271
Dooner, H. K., and He, L. (2008). Maize genome structure variation: interplay between retrotransposon polymorphisms and genic recombination. Plant Cell 20, 249–258. doi: 10.1105/tpc.107.057596
Dooner, H. K., and He, L. (2014). Polarized gene conversion at the bz locus of maize. Proc. Natl. Acad. Sci. U.S.A. 111, 13918–13923. doi: 10.1073/pnas.1415482111
Dooner, H. K., and Martínez-Férez, I. M. (1997). Recombination occurs uniformly within the bronze gene, a meiotic recombination hotspot in the maize genome. Plant Cell 9, 1633–1646. doi: 10.1105/tpc.9.9.1633
Dooner, H. K., Weck, E., Adams, S., Ralston, E., Favreau, M., and English, J. (1985). A molecular genetic analysis of insertions in the bronze locus in maize. Mol. Gen. Genet. 200, 240–246. doi: 10.1007/BF00425430
Drouaud, J., Khademian, H., Giraut, L., Zanni, V., Bellalou, S., Henderson, I. R., et al. (2013). Contrasted patterns of crossover and non-crossover at Arabidopsis thaliana meiotic recombination hotspots. PLoS Genet. 9:e1003922. doi: 10.1371/journal.pgen.1003922
Eggleston, W. B., Alleman, M., and Kermicle, J. L. (1995). Molecular organization and germinal instability of R-stippled maize. Genetics 141, 347–360.
Fowler, K. R., Sasaki, M., Milman, N., Keeney, S., and Smith, G. R. (2014). Evolutionarily diverse determinants of meiotic DNA break and recombination landscapes across the genome. Genome Res. 24, 1650–1664. doi: 10.1101/gr.172122.114
Franklin, A. E., McElver, J., Sunjevaric, I., Rothstein, R., Bowen, B., and Cande, W. Z. (1999). Three-dimensional microscopy of the Rad51 recombination protein during meiotic prophase. Plant Cell 11, 809–824. doi: 10.2307/3870816
Freeling, M. (1978). Allelic variation at the level of intragenic recombination. Genetics 89, 211–224.
Fu, H., Zheng, Z., and Dooner, H. K. (2002). Recombination rates between adjacent genic and retrotransposon regions in maize vary by 2 orders of magnitude. Proc. Natl. Acad. Sci. U.S.A. 99, 1082–1087. doi: 10.1073/pnas.022635499
Giraut, L., Falque, M., Drouaud, J., Pereira, L., Martin, O. C., and Mézard, C. (2011). Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet. 7:e1002354. doi: 10.1371/journal.pgen.1002354
Gray, S., and Cohen, P. E. (2016). Control of meiotic crossovers: from double-strand break formation to designation. Ann. Rev. Genet. 50, 175–210. doi: 10.1146/annurev-genet-120215-035111
Haberer, G., Young, S., Bharti, A. K., Gundlach, H., Raymond, C., Fuks, G., et al. (2005). Structure and Architecture of the Maize Genome. Plant Physiol. 139, 1612–1624. doi: 10.1104/pp.105.068718
Hayashi, K., Yoshida, K., and Matsui, Y. (2005). A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438, 374–378. doi: 10.1038/nature04112
He, L., and Dooner, H. K. (2009). Haplotype structure strongly affects recombination in a maize genetic interval polymorphic for Helitron and retrotransposon insertions. Proc. Natl. Acad. Sci. U.S.A. 106, 8410–8416. doi: 10.1073/pnas.0902972106
He, Y., Wang, M., Dukowic-Schulze, S., Zhou, A., Tiang, C.-L., Shilo, S., et al. (2017). Genomic features shaping the landscape of meiotic double-strand-break hotspots in maize. Proc. Natl. Acad. Sci. U.S.A. 114, 12231–12236. doi: 10.1073/pnas.1713225114
Hellsten, U., Wright, K. M., Jenkins, J., Shu, S., Yuan, Y., Wessler, S. R., et al. (2013). Fine-scale variation in meiotic recombination in Mimulus inferred from population shotgun sequencing. Proc. Natl. Acad. Sci. U.S.A. 110, 19478–19482. doi: 10.1073/pnas.1319032110
Higgins, J. D., Perry, R. M., Barakate, A., Ramsay, L., Waugh, R., Halpin, C., et al. (2012). Spatiotemporal asymmetry of the meiotic program underlies the predominantly distal distribution of meiotic crossovers in barley. Plant Cell 24, 4096–4109. doi: 10.1105/tpc.112.102483
Inukai, T., Sako, A., Hirano, H. Y., and Sano, Y. (2000). Analysis of intragenic recombination at wx in rice: correlation between the molecular and genetic maps within the locus. Genome 43, 589–596. doi: 10.1139/g00-015
Johns, M. A., Strommer, J. N., and Freeling, M. (1983). Exceptionally high levels of restriction site polymorphism in DNA near the maize Adh1 gene. Genetics 105, 733–743.
Keeney, S., Lange, J., and Mohibullah, N. (2014). Self-organization of meiotic recombination initiation: general principles and molecular pathways. Ann. Rev. Genet. 48, 187–214. doi: 10.1146/annurev-genet-120213-092304
Kianian, P. M. A., Wang, M., Simons, K., Ghavami, F., He, Y., Dukowic-Schulze, S., et al. (2018). High-resolution crossover mapping reveals similarities and differences of male and female recombination in maize. Nat. Commun. 9:2370. doi: 10.1038/s41467-018-04562-5
Lake, C. M., and Hawley, R. S. (2016). Becoming a crossover-competent DSB. Semin. Cell Dev. Biol. 54, 117–125. doi: 10.1016/j.semcdb.2016.01.008
Lambing, C., Franklin, F. C. H., and Wang, C.-J. R. (2017). Understanding and manipulating meiotic recombination in plants. Plant Physiol. 173, 1530–1542. doi: 10.1104/pp.16.01530
Lange, J., Yamada, S., Tischfield, S. E., Pan, J., Kim, S., Zhu, X., et al. (2016). The landscape of mouse meiotic double-strand break formation, processing, and repair. Cell 167, 695–708.e16. doi: 10.1016/j.cell.2016.09.035
Li, X., Li, L., and Yan, J. (2015). Dissecting meiotic recombination based on tetrad analysis by single-microspore sequencing in maize. Nat. Commun. 6:6648. doi: 10.1038/ncomms7648
Liu, S., Schnable, J. C., Ott, A., Yeh, C. T., Springer, N. M., Yu, J., et al. (2018). Intragenic meiotic crossovers generate novel alleles with transgressive expression levels. Mol. Biol. Evol. doi: 10.1093/molbev/msy174 [Epub ahead of print].
Lu, P., Han, X., Qi, J., Yang, J., Wijeratne, A. J., Li, T., et al. (2012). Analysis of Arabidopsis genome-wide variations before and after meiosis and meiotic recombination by resequencing Landsberg erecta and all four products of a single meiosis. Genome Res. 22, 508–518. doi: 10.1101/gr.127522.111
Mancera, E., Bourgon, R., Brozzi, A., Huber, W., and Steinmetz, L. M. (2008). High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454, 479–485. doi: 10.1038/nature07135
Marsolier-Kergoat, M.-C., Khan, M. M., Schott, J., Zhu, X., and Llorente, B. (2018). Mechanistic view and genetic control of DNA recombination during meiosis. Mol. Cell 70, 9–20.e6. doi: 10.1016/j.molcel.2018.02.032
Medhi, D., Goldman, A. S. H., and Lichten, M. (2016). Local chromosome context is a major determinant of crossover pathway biochemistry during budding yeast meiosis. eLife 5:e19669. doi: 10.7554/eLife.19669
Melamed-Bessudo, C., and Levy, A. A. (2012). Deficiency in DNA methylation increases meiotic crossover rates in euchromatic but not in heterochromatic regions in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, E981–E988. doi: 10.1073/pnas.1120742109
Meyers, B. C., Tingey, S. V., and Morgante, M. (2001). Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res. 11, 1660–1676. doi: 10.1101/gr.188201
Mirouze, M., Lieberman-Lazarovich, M., Aversano, R., Bucher, E., Nicolet, J., Reinders, J., et al. (2012). Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 5880–5885. doi: 10.1073/pnas.1120841109
Nelson, O. E. (1957). The feasibility of investigating ‘Genetic fine structure’ in higher plants. Am. Nat. 91, 331–332. doi: 10.1086/281997
Nelson, O. E. (1959). Intracistronic recombination in the Wx/Wx region in maize. Science 130, 794–795. doi: 10.1126/science.130.3378.794
Nelson, O. E. (1962). The waxy locus in maize. I. Intralocus recombination frequency estimates by pollen and conventional analysis. Genetics 47, 737–742.
Nelson, O. E. (1968). The WAXY locus in maize. II. The location of the controlling element alleles. Genetics 60, 507–524.
Nelson, O. E. (1975). The waxy locus in maize III. Effect of structural heterozygosity on intragenic recombination and flanking marker assortment. Genetics 79, 31–44.
Nicolas, A., and Petes, T. D. (1994). Polarity of meiotic gene conversion in fungi: contrasting views. Experientia 50, 242–252. doi: 10.1007/BF01924007
Nicolas, A., Treco, D., Schultes, N. P., and Szostak, J. W. (1989). An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature 338, 35–39. doi: 10.1038/338035a0
Okagaki, R. J., and Weil, C. F. (1997). Analysis of recombination sites within the maize waxy locus. Genetics 147, 815–821.
Ott, A. (2017). Assessment of Genetic Diversity and Recombination in Maize. Graduate Thesis and Dissertations. 16186. Available at: https://lib.dr.iastate.edu/etd/16186
Page, S. L., and Hawley, R. W. (2003). Chromosome choreography: the meiotic ballet. Science 301, 785–789. doi: 10.1126/science.1086605
Pan, J., Sasaki, M., Kniewel, R., Murakami, H., Blitzblau, H. G., Tischfield, S. E., et al. (2011). A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144, 719–731. doi: 10.1016/j.cell.2011.02.009
Pan, Q., Deng, M., and Li, L. (2017). Complexity of genetic mechanisms conferring nonuniformity of recombination in maize. Sci. Rep. 7:1205. doi: 10.1038/s41598-017-01240-2
Patterson, G. I., Kubo, K. M., Shroyer, T., and Chandler, V. L. (1995). Sequences required for paramutation of the maize b gene map to a region containing the promoter and upstream sequences. Genetics 140, 1389–1406.
Pawlowski, W. P., Golubovskaya, I. N., and Cande, W. Z. (2003). Altered nuclear distribution of recombination protein RAD51 in maize mutants suggests the involvement of RAD51 in meiotic homology recognition. Plant Cell 15, 1807–1816. doi: 10.1105/tpc.012898
Peoples-Holst, T. L., and Burgess, S. M. (2005). Multiple branches of the meiotic recombination pathway contribute independently to homolog pairing and stable juxtaposition during meiosis in budding yeast. Genes Dev. 19, 863–874. doi: 10.1101/gad.1293605
Petes, T. D. (2001). Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2, 360–369. doi: 10.1038/35072078
Plug, A. W., Xu, J., Reddy, G., Golub, E. I., and Ashley, T. (1996). Presynaptic association of Rad51 protein with selected sites in meiotic chromatin. Proc. Natl. Acad. Sci. U.S.A. 93, 5920–5924. doi: 10.1073/pnas.93.12.5920
Pontecorvo, G. (1955). Gene structure and action in relation to heterosis. Proc. R. Soc. Lond. B 144, 171–177. doi: 10.1098/rspb.1955.0043
Porter, S. E., White, M. A., and Petes, T. D. (1993). Genetic evidence that the meiotic recombination hotspot at the HIS4 locus of Saccharomyces cerevisiae does not represent a site for a symmetrically-processed double-strand break. Genetics 134, 5–19.
Portin, P. (1993). The concept of the gene: short history and present status. Q. Rev. Biol. 68, 173–223. doi: 10.1086/418039
Pradillo, M., Varas, J., Oliver, C., and Santos, J. L. (2014). On the role of AtDMC1, AtRAD51 and its paralogs during Arabidopsis meiosis. Front. Plant Sci. 5:23. doi: 10.3389/fpls.2014.00023
Rodgers-Melnick, E., Bradbury, P. J., Elshire, R. J., Glaubitz, J. C., Acharya, C. B., Mitchell, S. E., et al. (2015). Recombination in diverse maize is stable, predictable, and associated with genetic load. Proc. Natl. Acad. Sci. U.S.A. 112, 3823–3828. doi: 10.1073/pnas.1413864112
Sachs, M. M., Dennis, E. S., Gerlach, W. L., and Peacock, W. J. (1986). Two alleles of maize alcohol dehydrogenase 1 have 3’ structural and poly(A) addition polymorphisms. Genetics 113, 449–467.
Salomé, P. A., Bomblies, K., Fitz, J., Laitinen, R. A. E., Warthmann, N., Yant, L., et al. (2012). The recombination landscape in Arabidopsis thaliana F2 populations. Heredity 108, 447–455. doi: 10.1038/hdy.2011.95
Schnable, P. S., Ware, D., Fulton, R. S., Stein, J. C., Wei, F., Pasternak, S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115. doi: 10.1126/science.1178534
Segal, E., and Widom, J. (2009). What controls nucleosome positions? Trends Genet. TIG 25, 335–343. doi: 10.1016/j.tig.2009.06.002
Serrentino, M.-E., and Borde, V. (2012). The spatial regulation of meiotic recombination hotspots: are all DSB hotspots crossover hotspots? Exp. Cell Res. 318, 1347–1352. doi: 10.1016/j.yexcr.2012.03.025
Sidhu, G. K., Fang, C., Olson, M. A., Falque, M., Martin, O. C., and Pawlowski, W. P. (2015). Recombination patterns in maize reveal limits to crossover homeostasis. Proc. Natl. Acad. Sci. U.S.A. 112, 15982–15987. doi: 10.1073/pnas.1514265112
Smagulova, F., Gregoretti, I. V., Brick, K., Khil, P., Camerini-Otero, R. D., and Petukhova, G. V. (2011). Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472, 375–378. doi: 10.1038/nature09869
Smithies, O., and Powers, P. A. (1986). Gene conversions and their relation to homologous chromosome pairing. Philos. Trans. R. Soc. Lond. B 312, 291–302. doi: 10.1098/rstb.1986.0008
Stack, S. M., and Anderson, L. K. (2002). Crossing over as assessed by late recombination nodules is related to the pattern of synapsis and the distribution of early recombination nodules in maize. Chromosome Res. 10, 329–345. doi: 10.1023/A:1016575925934
Stadler, L. J., and Emmerling, M. H. (1956). Relation of unequal crossing over to the interdependence of Rr elements (P) and (S). Genetics 41, 124–137.
Stapley, J., Feulner, P. G. D., Johnston, S. E., Santure, A. W., and Smadja, C. M. (2017). Variation in recombination frequency and distribution across eukaryotes: patterns and processes. Philos. Trans. R. Soc. B 372:20160455. doi: 10.1098/rstb.2016.0455
Sukhapinda, K., and Peterson, P. A. (1980). Enhancement of genetic exchange in maize: intragenic recombination. Can. J. Genet. Cytol. 22, 213–222. doi: 10.1139/g80-026
Szostak, J. W., Orr-Weaver, T. L., Rothstein, R. J., and Stahl, F. E. (1983). The double-strand-break repair model for recombination. Cell 33, 25–35. doi: 10.1016/0092-8674(83)90331-8
Thuriaux, P. (1977). Is recombination confined to structural genes on the eukaryotic genome? Nature 268, 460–462. doi: 10.1038/268460a0
Tischfield, S. E., and Keeney, S. (2012). Scale matters. Cell Cycle 11, 1496–1503. doi: 10.4161/cc.19733
Varas, J., Sánchez-Morán, E., Copenhaver, G. P., Santos, J. L., and Pradillo, M. (2015). Analysis of the relationships between DNA double-strand breaks, synaptonemal complex and crossovers using the Atfas1-4 mutant. PLoS Genet. 11:e1005301. doi: 10.1371/journal.pgen.1005301
Wang, G., Xu, J., Tang, Y., Zhou, L., Wang, F., Xu, Z., et al. (2011). Molecular characterization of a genomic interval with highly uneven recombination distribution on maize chromosome 10 L. Genetica 139, 1109–1118. doi: 10.1007/s10709-011-9613-x
Wang, Y., and Copenhaver, G. P. (2018). Meiotic recombination: mixing it up in plants. Annu. Rev. Plant Biol. 69, 577–609. doi: 10.1146/annurev-arplant-042817-040431
Wei, F., Zhang, J., Zhou, S., He, R., Schaeffer, M., Collura, K., et al. (2009). The physical and genetic framework of the maize B73 genome. PLoS Genet. 5:e1000715. doi: 10.1371/journal.pgen.1000715
Wessler, S., Tarpley, A., Purugganan, M., Spell, M., and Okagaki, R. (1990). Filler DNA is associated with spontaneous deletions in maize. Proc. Natl. Acad. Sci. U.S.A. 87, 8731–8735. doi: 10.1073/pnas.87.22.8731
Wijnker, E., James, G. V., Ding, J., Becker, F., Klasen, J. R., Rawat, V., et al. (2013). The genomic landscape of meiotic crossovers and gene conversions in Arabidopsis thaliana. eLife 2:e01426. doi: 10.7554/eLife.01426
Xu, X., Hsia, A. P., Zhang, L., Nikolau, B. J., and Schnable, P. S. (1995). Meiotic recombination break points resolve at high rates at the 5’ end of a maize coding sequence. Plant Cell 7, 2151–2161. doi: 10.1105/tpc.7.12.2151
Yamada, S., Ohta, K., and Yamada, T. (2013). Acetylated Histone H3K9 is associated with meiotic recombination hotspots, and plays a role in recombination redundantly with other factors including the H3K4 methylase Set1 in fission yeast. Nucl. Acids Res. 41, 3504–3517. doi: 10.1093/nar/gkt049
Yandeau-Nelson, M. D., Xia, Y., Li, J., Neuffer, M. G., and Schnable, P. S. (2006). Unequal sister chromatid and homolog recombination at a tandem duplication of the a1 locus in maize. Genetics 173, 2211–2226. doi: 10.1534/genetics.105.05271
Yao, H., and Schnable, P. S. (2005). Cis-effect on meiotic recombination across distinct a1-sh2 intervals in a common Zea genetic genetic background. Genetics 170, 1929–1944. doi: 10.1534/genetics.104.034454
Yao, H., Zhou, Q., Li, J., Smith, H., Yandeau, M., Nikolau, B. J., et al. (2002). Molecular characterization of meiotic recombination across the 140-Kb multigenic a1-sh2 interval of maize. Proc. Natl. Acad. Sci. U.S.A. 99, 6157–6162. doi: 10.1073/pnas.082562199
Yelina, N. E., Lambing, C., Hardcastle, T. J., Zhao, X., Santos, B., and Henderson, I. R. (2015). DNA methylation epigenetically silences crossover hot spots and controls chromosomal domains of meiotic recombination in Arabidopsis. Genes Dev. 29, 2183–2202. doi: 10.1101/gad.270876.115
Keywords: recombination, hotspots, intragenic, polarity, double-strand breaks, maize
Citation: Okagaki RJ, Dukowic-Schulze S, Eggleston WB and Muehlbauer GJ (2018) A Critical Assessment of 60 Years of Maize Intragenic Recombination. Front. Plant Sci. 9:1560. doi: 10.3389/fpls.2018.01560
Received: 25 July 2018; Accepted: 04 October 2018;
Published: 29 October 2018.
Edited by:
Mónica Pradillo, Complutense University of Madrid, SpainReviewed by:
Kyuha Choi, Pohang University of Science and Technology, South KoreaPiotr Andrzej Ziolkowski, Adam Mickiewicz University in Poznań, Poland
Copyright © 2018 Okagaki, Dukowic-Schulze, Eggleston and Muehlbauer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gary J. Muehlbauer, bXVlaGwwMDNAdW1uLmVkdQ==
†These authors have contributed equally to this work
 Ron J. Okagaki
Ron J. Okagaki Stefanie Dukowic-Schulze
Stefanie Dukowic-Schulze William B. Eggleston3
William B. Eggleston3 Gary J. Muehlbauer
Gary J. Muehlbauer