- 1Department of Plant Physiology, University of Rostock, Rostock, Germany
- 2Centre of Advanced Study in Botany, Banaras Hindu University, Varanasi, India
- 3Department of Plant and Environmental Sciences, The Hebrew University of Jerusalem, Jerusalem, Israel
- 4Applied Metabolome Analysis, Department of Molecular Physiology, Max Planck Institute of Molecular Plant Physiology, Potsdam, Germany
- 5Department Life, Light and Matter, University of Rostock, Rostock, Germany
Photorespiratory phosphoglycolate (2PG) metabolism is essential for cyanobacteria, algae, and plants. The first enzyme of the pathway, 2PG phosphatase (PGPase), is known from plants and algae but was scarcely investigated in cyanobacteria. In silico analysis revealed four candidate genes (slr0458, slr0586, sll1349, and slr1762) in the genome of the model cyanobacterium Synechocystis sp. PCC 6803 that all belong to the 2-haloacid dehalogenase (HAD) superfamily and could possibly encode PGPase proteins. However, in contrast to known algal and plant PGPases, the putative cyanobacterial PGPases belong to another HAD subfamily implying that PGPases in eukaryotic phototrophs did not originate from cyanobacterial PGPases. To verify their function, these four genes were inactivated both individually and in combination. A mild high-CO2-requiring (HCR) growth phenotype typical for photorespiratory mutants was observed only in Δsll1349. Combinatorial inactivation enhanced the HCR phenotype in specific double and triple mutants. Heterologous expression of the putative cyanobacterial PGPases in E. coli led to higher PGPase activities in crude cell extracts, but only the purified Slr0458 protein showed PGPase activity. Hence, we propose that a consortium of up to four photorespiratory PGPases may initiate photorespiratory 2PG metabolism in Synechocystis. We suggest that redundancy of this essential enzyme activity could be related to the highly adaptive lifestyle of cyanobacteria such as Synechocystis sp. PCC 6803, which allows them to grow under very diverse conditions.
Introduction
All oxygenic phototrophs use the enzyme ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) for carbon assimilation. However, Rubisco catalyzes two competing reactions, carboxylation and oxygenation of ribulose-1,5-bisphosphate (RuBP), the ratio of which mainly depends on the concentrations of CO2 and O2. Oxygenation of RuBP produces 2PG, a compound that inhibits Calvin-Benson cycle enzymes (e.g., triosephosphate isomerase, Husic et al., 1987; sedoheptulose 1,7-bisphosphate phosphatase, Flügel et al., 2017), lowering the rate of photosynthesis. Therefore, 2PG is rapidly metabolized by the photorespiratory pathway (Tolbert, 1997; Bauwe et al., 2010), which recovers one molecule of 3-phosphoglycerate from two molecules of 2PG while one carbon atom is lost as photorespiratory CO2. Moreover, the conversion of two molecules of glycine into serine releases NH3. In addition to these functions, detoxification and organic carbon salvage, it has been suggested that photorespiration provides glycine and serine to cellular metabolism and helps protecting from photoinhibition and oxidative damage (e.g., Wingler et al., 2000).
It has been established that there is no oxygenic photosynthesis without photorespiration (Bauwe et al., 2012). Accordingly, the inactivation of photorespiration-related genes in C3 plants typically produces mutants that show a HCR phenotype (Somerville, 2001; Timm et al., 2012). To deal with limiting CO2 amounts, cyanobacteria, many algae, and C4 plants evolved inorganic carbon concentrating mechanisms (CCM; Giordano et al., 2005), which were believed to make photorespiration superfluous. However, Chlamydomonas and maize mutants with intact CCM but defective photorespiration also showed the HCR photorespiratory phenotype (Nakamura et al., 2005; Zelitch et al., 2009). Photorespiration is also essential for cyanobacteria despite their CCM (Eisenhut et al., 2008b). All studied cyanobacteria, even those with small genome size as the picoplanktonic Prochlorococcus and Synechococcus spp., have preserved complete and active sets of photorespiratory genes (Kern et al., 2011). Among cyanobacteria, most of the functional analysis of photorespiration has been performed in the model strain Synechocystis sp. PCC 6803 (hereafter Synechocystis), which possesses three different routes for 2PG metabolism: plant-like C2 pathway, bacterial glycerate pathway, and complete decarboxylation of glyoxylate (Eisenhut et al., 2008b). The photorespiratory intermediate glycolate is produced in Synechocystis WT cells even at 5% CO2 (Huege et al., 2011) suggesting that operation of the CCM is not sufficient to completely suppress RuBP oxygenation. Due to the similarity of photorespiratory enzymes in plants and cyanobacteria and the essential relation between photorespiration and photosynthesis, we hypothesized that the photorespiratory pathway might have evolved very early among cyanobacteria, before the primary endosymbiosis leading to the first photosynthetic eukaryotes (Eisenhut et al., 2008b).
Many enzymes of the cyanobacterial photorespiratory metabolism have been studied during the last few years (e.g., Eisenhut et al., 2008b; Hackenberg et al., 2011), but PGPase (EC 3.1.3.18) and the corresponding gene(s) were only scarcely investigated (Haimovich-Dayan et al., 2015). PGPase catalyses the entry reaction into photorespiratory 2PG metabolism: dephosphorylation of 2PG to glycolate. The enzyme belongs to the HAD protein superfamily that possesses three conserved motifs: motif I, DX(D/T/Y)X(T/V)(L/V); motif II, (S/T); and motif III, K(G/S)(D/S)XXX(D/N). An aspartate (D) at position three in motif I is characteristic for phosphatases. During catalysis, motif I is self-phosphorylated, motif II forms hydrogen bonds with the substrate, and motif III interacts with divalent metal ions at the active site (Kim et al., 2004). PGPases have been purified from plants such as tobacco, spinach, maize, pea; and the green alga Chlamydomonas as well as the cyanobacterium Coccochloris (Kerr and Gear, 1974; Husic and Tolbert, 1984; Hardy and Baldy, 1986; Norman and Colman, 1991). Pellicer et al. (2003) compared the structural and functional features of PGPases of different origins and found similarity of the sequences, implying the use of the same catalytic mechanism. However, the bacterial proteins belong to the dehr (dehalogenase-related)-like protein family, whereas PGPases from eukaryotic phototrophs are members of the NagD (ribonucleotide monophosphatase)-like protein family among the HAD protein superfamily (Burroughs et al., 2006). PGPase mutants have been isolated from Arabidopsis (Somerville and Ogren, 1979; Schwarte and Bauwe, 2007) and Chlamydomonas (Suzuki et al., 1999; Ma et al., 2013). All of them were reported to exhibit the characteristic HCR phenotype (Timm et al., 2012; Flügel et al., 2017).
Here, we report on the identification of Synechocystis PGPases. By using the basic local alignment search tool (BLAST), we found four candidate genes (slr0458, slr0586, sll1349, and slr1762). The four candidate cyanobacterial genes encode for proteins of the HAD superfamily (Supplementary Figure S1) but contrary to known algal and plant PGPases, the putative cyanobacterial PGPases belong to another HAD subfamily implying that PGPases in eukaryotic phototrophs did not originate from the cyanobacterial proteins. Their function was studied in vitro after expression in E. coli and by the analysis of single and combined Synechocystis mutants. We conclude that a consortium of up to four photorespiratory PGPases may initiate photorespiratory 2PG metabolism in Synechocystis.
Materials and Methods
Strains and Culture Conditions
The glucose-tolerant Synechocystis sp. strain PCC 6803 served as WT. Axenic cultures were grown photo-autotrophically in batch cultures (3 cm glass vessels with 5 mm glass tubes) by bubbling with air (flow rate 5 mL min-1) enriched with CO2 (5% CO2, defined as HC) at 29°C under continuous illumination of 100 μmol photons m-2 s-1 (warm white light; Osram L58 W32/3) in BG11 medium (Rippka et al., 1979) buffered with TES-KOH (20 mM final concentration) at pH 8.0. Cultures at an optical density at 730 nm (OD730) of approximately 1.0 (equivalent to approximately 109 cells mL-1) was used for the experiments. Shifts to ambient air conditions (low CO2 of 0.04%, LC) were performed by switching the bubbling from CO2-supplemented to air as described previously (Hackenberg et al., 2009).
Construction of Single and Multiple Mutants
To generate single and multiple mutations in the selected genes, interposon mutagenesis was applied by insertion of antibiotic resistance cassettes into the coding sequences at selected restriction sites (e.g., Bryant et al., 1990). The primers were designed for the coding sequences and neighboring sequences (approximately 2 kb length) using the complete genome sequence of Synechocystis (Kaneko et al., 1996). Supplementary Table S1 includes the respective list of antibiotic cassettes used for each gene and the primers. Total DNA from Synechocystis was isolated according to Hagemann et al. (1997).
The inactivated genes for slr0458 and sll1349 were amplified via PCR from the previously used mutants of Synechocystis (Eisenhut et al., 2006) and ligated into pGEMT (Promega). To inactivate slr0586, the coding sequence and flanking DNA was obtained from the Synechocystis WT and ligated into pGEMT. The coding sequence was cut with MunI and a blunt-ended erythromycin resistance cartridge was inserted. A clone harboring the resistance gene in the same transcriptional orientation as slr0586 was used for the transformation of the Synechocystis WT. The coding sequence of gene slr1762 together with 800 bp flanking sequence was amplified from WT DNA and ligated into pGEMT. The central part of the coding sequence was deleted by restriction with EcoO109I and after generating blunt ends it was replaced by the spectinomycin resistance cassette.
All mutation constructs were verified by restriction analysis and sequencing (Seqlab, Germany). Positive constructs were used for transforming Synechocystis as described previously (Hagemann and Zuther, 1992). Transformation, selection, and segregation of mutants were done in the presence of 5% CO2 to decrease selective pressure from photorespiratory activity. Mutant colonies were screened against the corresponding antibiotics added to the petri dishes containing BG11 with 0.8% Kobe agar. Drug-resistant colonies were transferred on fresh plates to allow complete segregation of the Synechocystis genome. The constructs for generating single mutations in slr0458, slr0586, and sll1349 were subsequently used to transform the Synechocystis mutantΔslr1762 for the second and third transformations raising double and triple gene mutants, respectively.
All mutants were further stored and cultivated under HC conditions. To show alterations in the genotype, PCR with gene-specific oligonucleotides (Supplementary Table S1) was carried out by using the Taq-PCR Master Mix (Qiagen) as described in Eisenhut et al. (2006). Mutant colonies showing complete segregation of the target gene, only DNA fragment with sizes corresponding to the mutated genes in the absence of the respective WT gene bands (see Supplementary Figure S2), were used for further experiments.
Phenotypic Characterization of Synechocystis Mutants
For physiological and molecular phenotyping, exponentially growing cultures (OD730 = 0.5) were pre-cultured at HC conditions as described by Hackenberg et al. (2009). These cells were then further grown in the Multi-Cultivator MC 1000 (Photon System Instruments, Czech Republic) by bubbling with ambient air (LC) at 150 μmol photons m-2 s-1 light and 30°C. The MC1000 allows an automated measurement of growth as optical density of cultures at 730 nm (OD730). The optical density was recorded every 30 min during a growth period of 5 days. Periods of exponential growth were used to calculate specific growth rates. In each experiment, the WT and selected mutants were grown in duplicates to compare the specific growth rates. The experiments were repeated at least 2-times. Statistically significant differences in the growth rates between the WT and the mutant strains were analyzed using 1-way ANOVA. Cultures, pre-grown in 5% CO2 with OD730 = 0.5 were also used for drop dilution assay tests to search for the HCR phenotype. WT and mutant cultures were serially diluted 10, 100, 1000, and 10,000 fold; 2 μl of these dilutions were spotted on BG11-Kobe agar plates, allowed to dry and incubated in ambient air at similar temperature and light conditions as used for the cultivation of liquid cultures.
Metabolite Profiling for Selected Mutants
Cells grown in BG11 liquid media under LC culture conditions were harvested by fast filtration and immediately frozen in liquid nitrogen as described by Eisenhut et al. (2008a). The mutants were grown in duplicates and at least three replicates from each culture were analyzed in all cases. Steady state contents of metabolites, e.g., 2PG and glycolate, in cyanobacterial cells were determined using non-targeted metabolite profiling by gas chromatography-electron impact-time of flight-mass spectrometry (GC-EI-TOF-MS). Amounts of metabolites were normalized to biomass using the OD730 of each sample (Eisenhut et al., 2008a; Huege et al., 2011) and to the internal standard U13C-sorbitol using the normalization procedure described by Lisec et al. (2011). Analytes were identified by at least 3 specific mass fragments per compound and a retention index deviation < 1.0% (Hummel et al., 2010). Metabolites were assessed by relative changes expressed as response ratios, i.e., x-fold factors of mutant cells in comparison to the WT that was arbitrarily set to 1. Statistical analysis was performed using the heteroscedastic Student’s t-test in Excel 2010. The complete data set is shown in Supplementary Table S3.
Cloning of Genes for Overexpression in E. coli
For overexpression of the candidate genes, the pET28a Expression System (Novagen) was used. Open reading frames of all the four genes (slr0458, slr0586, sll1349, and slr1762) were amplified from Synechocystis WT chromosomal DNA by PCR using Taq polymerase master mix (Qiagen) and elongase enzyme mix (Thermo Fisher Scientific). Gene-specific primers (Supplementary Table S1) contained added NdeI and EcoRI restriction sites at the 5′ and 3′ ends, respectively, to facilitate cloning of cyanobacterial genes in frame with the N-terminal His-tag. Before the ligation with pET28a, the PCR fragments were cloned into pGEMT (Promega) and the inserts were verified by sequencing. Correct inserts were then excised with NdeI and EcoRI, ligated into pET28a and transferred into E. coli strain BL21 DE3 (Novagen). The cells were cultured at 37°C at 170 rpm shaking in LB medium until the suspension reached an OD600 of 0.5. Cultures with empty vector were simultaneously maintained as control. Gene expression was induced by addition of 0.5 mM IPTG and the cells were then cultured overnight at room temperature. Soluble proteins were extracted from E. coli by sonication with homogenization buffer (20 mM Tris buffer, 500 mM NaCl, pH 7.8) and centrifugation (40,000 ×g, 4°C). His-tagged recombinant proteins were purified by affinity chromatography on Ni-NTA columns (ProBond, Thermo Fisher Scientific) and eluted with gradually increasing imidazole concentrations. The purity of recombinant proteins was confirmed by SDS-PAGE (see Supplementary Figure S4). For comparison, the recombinant PGLP1 (At5g36700) from Arabidopsis was obtained as described by Schwarte and Bauwe (2007).
Protein Quantification and PGPase Activity
His-tag-purified recombinant proteins and soluble protein extracts from E. coli were used for the PGPase assay. Total protein concentrations of all samples were determined according to Bradford (1976) and enzymatic assays (N = 3) performed as described in Schwarte and Bauwe (2007). In short, assays were incubated for 10 min at 37°C with 2 mM 2PG (Sigma-Aldrich). Aliquots taken every 2 min were assayed for inorganic phosphate via the formation of reduced phosphomolybdate (Ames, 1966). A set of different Na2HPO4 concentrations was used for calibration.
Phylogenetic Analyses
The sequences used for the alignment were identified via BlastP (Altschul et al., 1990) using the Arabidopsis PGPase (At5g36700; Schwarte and Bauwe, 2007) and the Synechocystis PGPase candidate proteins Slr0586, Slr1762, Sll1349 or Slr0458 as template. The BlastP searches were limited to specific taxonomic groups (e.g., cyanobacteria). In addition to plant and cyanobacterial proteins, we also included verified PGPases from the bacteria Ralstonia eutropha, Escherichia coli and Rhodobacter sphaeroides (Schäferjohann et al., 1993; Gibson and Tabita, 1997; Lyngstadaas et al., 1999). The sequences were aligned using the high accuracy algorithm Probcons (Do et al., 2005) implemented at the MPI TOOLKIT platform (Biegert et al., 2006). Thereafter, putative chloroplast target peptides and other miss-aligned regions in the C- or N-terminus were removed. According to the Akaike information criterion calculated via ProtTest 3.2 (Darriba et al., 2011), the best fitting model is the LG+G+I (amino acid replacement matrix from Le and Gascuel, 2008; including a proportion of invariant sites and a gamma model of rate heterogeneity). After visual inspection, the alignment was used for phylogenetic reconstruction using Bayesian interference implemented in MrBayes 3.2 (Ronquist et al., 2012) running 2,000,000 generations, whereas the first 25% of samples were disregarded as burn-in. Additionally, Maximum Likelihood with 1000 bootstrap replicates implemented in RAxML (Stamatakis, 2014) was applied. The phylogenetic tree was visualized using FigTree v1.4.21. The alignments can be found as Supplementary Figure S5. Species names and accession numbers are as follows: Microcystis aeruginosa PCC 9701 (CCI35500.1, WP_002803420.1, CCI39026.1), Microcoleus vaginatus FGP-2 (WP_006635473.1, EGK86926.1, EGK83991.1), Synechocystis sp. PCC 6803 (Slr0586, Slr1349, Slr1762, Slr0458; CyanoBase), Nostoc sp. PCC 7120 (Alr3258, Alr4944, All0135; CyanoBase), Synechocystis sp. PCC 6714 (WP_028946651.1), E. coli O157:H7 (P58422.1), R. eutropha (AAA20197), R. sphaeroides ATCC 17025 (A4WW31.1), Arabidopsis thaliana (At5g36700; TAIR), Physcomitrella patens (XP_001758778), Zea mays (ACF84012). If not otherwise mentioned sequences were retrieved from NCBI.
Results
Phylogenetic Analysis of PGPase Proteins
BlastP searches with the Arabidopsis PGPase (PGLP1, At5g36700) in the Synechocystis genome identified Slr0458, Slr0586, Sll1349, and Slr1762 as candidates for putative cyanobacterial PGPases. These proteins possess HAD family domains, a characteristic feature of functional PGPases (Supplementary Figure S1). Notably, the four proteins showed only a low degree of similarity (48–65%) to each other and with At5g36700 (23–32%; Supplementary Table S2). Among the Synechocystis PGPase candidate genes, only sll1349 is preliminarily annotated as PGPase in CyanoBase2, whereas the other three proteins are annotated as hypothetical or unknown proteins. Within the HAD superfamily, all cyanobacterial proteins belong to the dehr (dehalogenase-related)-like protein family, whereas PGLP1 of Arabidopsis is a member of the NagD (ribonucleotide monophosphatase)-like protein family (Burroughs et al., 2006).
In the phylogenetic tree (Figure 1), all bacterial proteins belonging to the dehr-like protein family grouped into on large clade, which is clearly separated from the plant proteins of the NagD-like family. The four candidate proteins from Synechocystis are separated into four different subclades. The Slr0458 protein forms one clade together with the bacterial CbbZ clade, which comprises well characterized PGPases from bacteria such as E. coli, R. sphaeroides, and R. eutropha H16. The Slr1762–like cyanobacterial homologs are situated next to this clade. The Sll1349 homolog in Synechococcus PCC 7942 is also annotated as cbbZ3, however, Sll1349-like proteins cluster separately from the CbbZ-Slr048-Slr1762 clades. The Slr0586 protein and homologs from other cyanobacterial strains build a third clade (Figure 1). The plant PGPase proteins, e.g., PGLP1 (At5g36700) of Arabidopsis, form a separate group together with PGPases from algae and were used as outgroup (Figure 1).
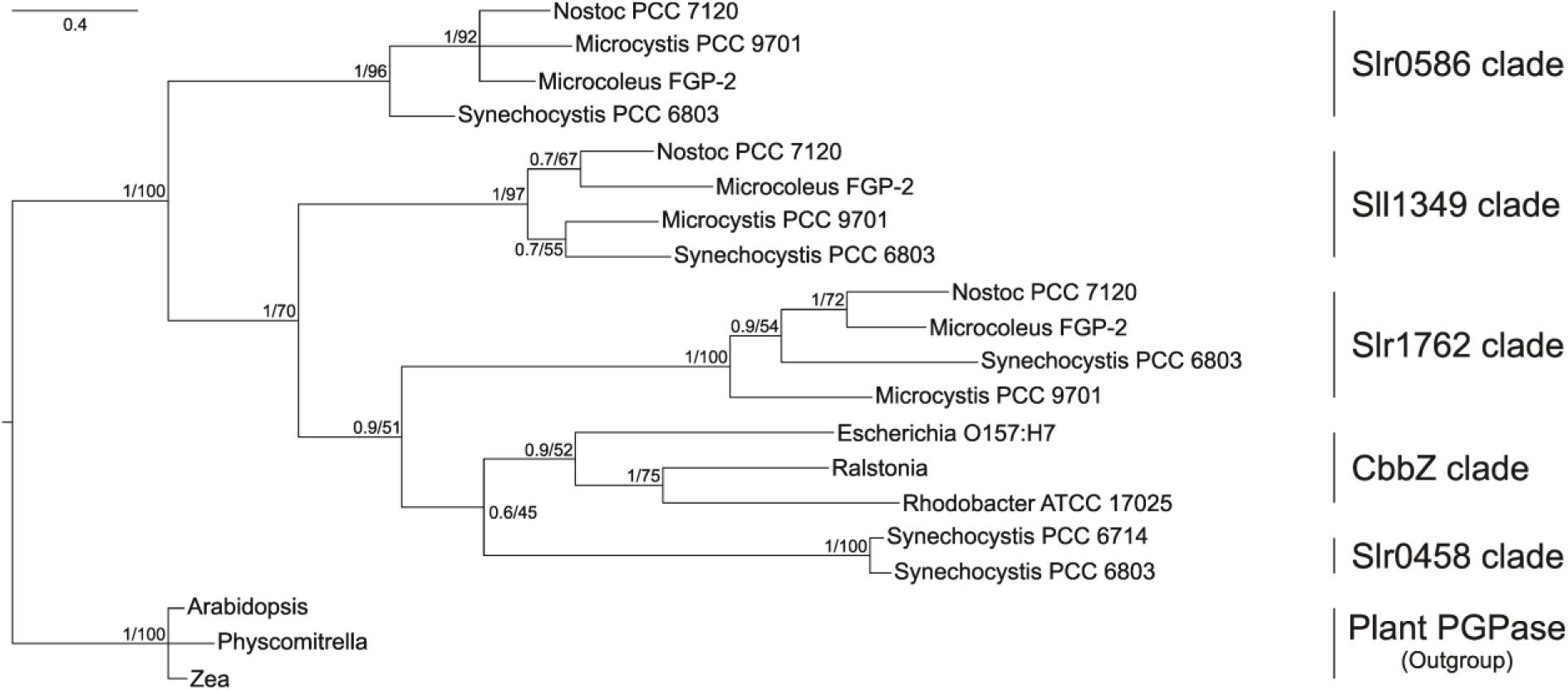
FIGURE 1. Phylogenetic tree of cyanobacterial PGPase candidate proteins showing relationship of Synechocystis PGPases with proteins from other cyanobacteria, bacteria, and plants. The tree was constructed using Bayesian interference running 2,000,000 generations whereas the first 25% were sort as burn-in. Numbers at the node indicate Bayesian post probabilities and Maximum likelihood bootstrap support values, respectively. Species names and accession number are given in the Material and methods section.
BLAST searches against complete cyanobacterial genomes listed in CyanoBase4 revealed that Slr0586, Slr1762, and Sll1349 proteins have homologs in many other cyanobacteria (80–60 other strains), which all are annotated as HAD-like proteins. Remarkably, the closest CbbZ-homolog Slr0458 is only found in the genome of Synechocystis and its closely related strain Synechocystis sp. PCC 6714 (Kopf et al., 2014). It needs to be mentioned that we did not find similar proteins in any of the genomes from strains of the marine Synechococcus/Prochlorococcus clades.
Thus, the cyanobacterial PGPase candidates are not homologous to the well characterized enzyme from Arabidopsis (Schwarte and Bauwe, 2007). To verify whether these proteins are PGPases, we generated insertion mutants affected in single and multiple genes, which were characterized regarding their ability to grow and accumulate 2PG under LC conditions. The candidate genes were also cloned into E. coli expression vectors to obtain recombinant proteins to verify the PGPase activity in vitro.
Generation of Single, Double, and Triple Synechocystis Mutations
To analyze the function of the four candidate proteins, we first inactivated each coding sequence via insertion of different drug resistance cartridges, which allowed the subsequent combination of the gene defects. PCR analyses showed that all WT copies of the respective genes were inactivated (Supplementary Figure S2). The complete segregation of the mutants revealed that under HC (5% CO2 in air) conditions none of the genes is essential for viability particularly since glycolate, the immediate product of PGPase, was also detected under HC (Huege et al., 2011). The mutants were then subjected to LC (0.04% CO2) conditions, where all of the single mutants were able to grow. The growth was comparable to WT in drop dilution assays on solid media (Supplementary Figure S3). Comparing specific growth rates in liquid media (Figure 2), three single mutants showed no significant changes compared to WT at LC, whereas mutant Δsll1349 showed slower growth, i.e., has a weak HCR phenotype. This finding indicates that more than one functional PGPase exists in Synechocystis. Hence, we generated a set of various double mutants. Again, it was possible to obtain completely segregated clones of Synechocystis mutants in which different pairs of putative PGPase-encoding genes were disrupted under HC conditions. All generated double mutants were able to grow at LC on plates (Supplementary Figure S3). However, the estimated growth rates in liquid cultures revealed differences in the performances under LC conditions. Compared with the WT, the double mutant Δsll1349/slr0458 showed a 60%, Δsll1349/slr0586 a 41%, Δsll1349/slr1762 a 34%, and Δslr0586/slr1762 a 17% decrease in specific growth rates in LC (Figure 2). An even stronger HCR phenotype was observed with the triple mutants Δslr1762/slr0586/sll1349 and Δsll1349/slr0586/slr0458. These mutants also seem to grow ∼50% slower than WT at HC conditions, because it took more than 3 weeks before clones of triple mutants appeared on agar plates, whereas other clones already appeared after 1 week. At LC conditions, a clear decline of growth was observed for the triple mutants. The growth rate of Δslr1762/slr0586/sll1349 was dropped by approximately 70% of WT when cultivated at LC. A similar growth decline was observed for another triple mutant, Δsll1349/slr0586/slr0458. Notably, several attempts to generate the respective quadruple mutant failed, we could not obtain mutant colonies even under HC conditions.
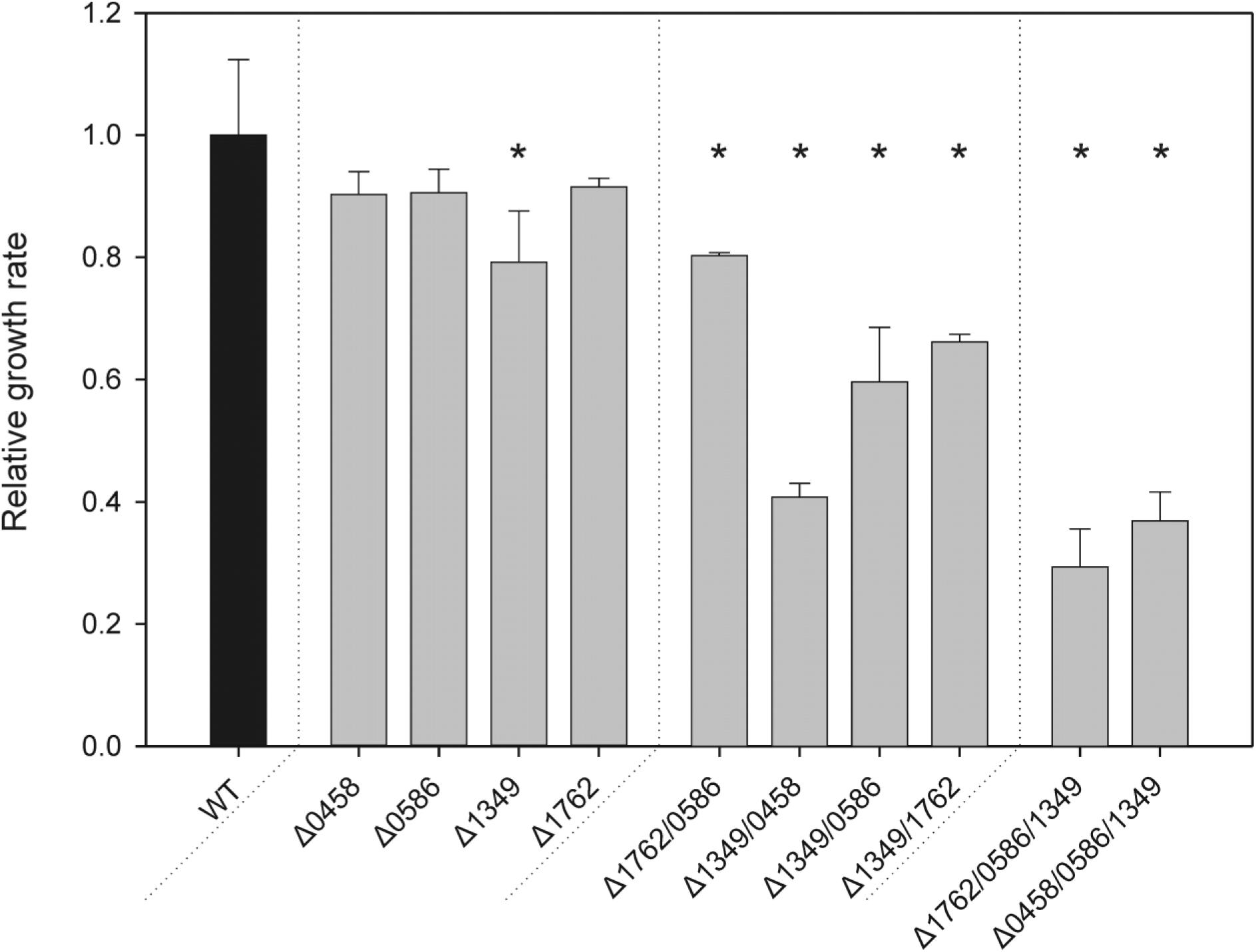
FIGURE 2. Specific growth rates of PGPase mutants as compared to the wild type (WT). Different sets of single, double, or triple mutants (gray bars) were grown side by side with the WT(black bar) under LC conditions. Given values are means ± SD (n = 3) and asterisks indicate values statistically different from the control (∗p < 0.05, 1-way ANOVA). The growth of the WT was set to 1 and corresponds to a growth rate of 0.017 h-1.
Accumulation of 2PG in Mutant Cells
The growth experiments showed gradually decreasing growth rates, starting from single over double to triple mutants, which could be associated with increased 2PG levels if the enzymes have PGPase activity. Thus, mutant gene combinations Δsll1349, Δslr1762/slr0586, Δsll1349/slr1762, Δsll1349/slr0458, Δsll1349/slr0586, and Δsll1349/slr0586/slr1762 were selected for profiling of the intracellular concentrations of 2PG and glycolate after cultivation at LC conditions (Figure 3). No major changes in 2PG levels were observed in the cells of the tested single mutant Δsll1349. Cells of the double mutant Δslr1762/slr0586 and Δsll1349/slr1762 showed 55 and 77% increased accumulation of 2PG compared to WT cells, whereas the double mutant Δsll1349/0458 only slightly increased the internal 2PG amounts (Figure 3A). Highest accumulation of 2PG, 86% increase compared to the WT was recorded for the triple mutant Δslr1762/slr0586/sll1349 (Figure 3A) In contrast to 2PG, only small amounts of glycolate were found in all cells, which are largely unaltered in all mutants compared to WT (Figure 3B). Generally, the 2PG accumulation is consistent with the previous findings of declined growth of double and triple mutants at LC conditions.
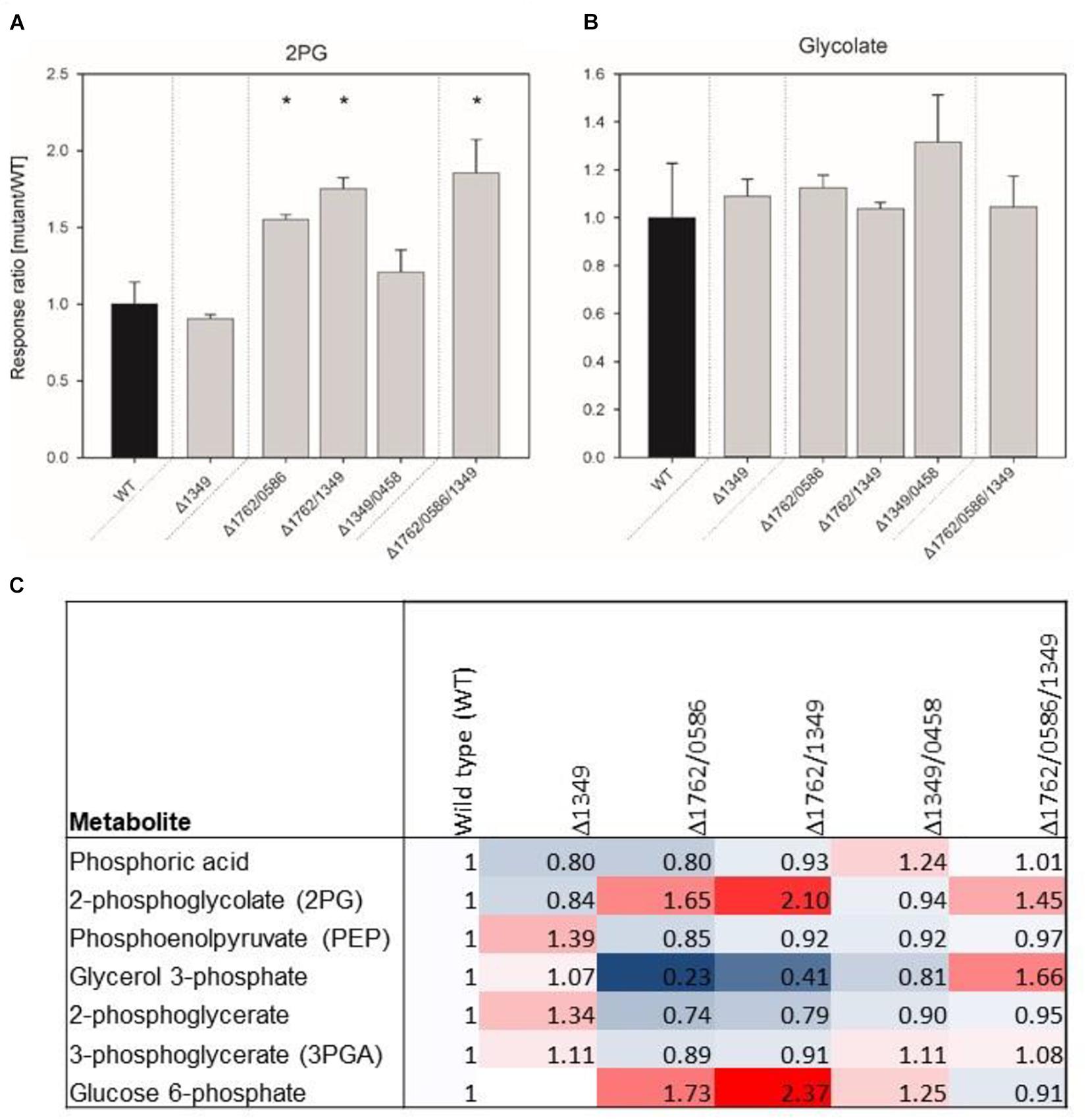
FIGURE 3. Relative contents of 2-phosphoglycolate (2PG, A), glycolate (B) and several other phosphorylated compounds (C) in mutant and wild type (WT) cells of Synechocystis. The data represent response ratios and correspond to averaged x-fold values of three biological replicates measured by at least two technical replicates. Factors are calculated relative to WT. Error bars represent standard error. Significant differences to the WT are marked by asterisks (∗p < 0.05; Student’s t-test).
Some additional changes were found in the metabolome data set regarding phosphorylated intermediates (Figure 3C). In addition to 2PG other phosphorylated compounds showed changed levels in mutant cells. For example, up to twofold higher amounts of glucose 6-phosphate were found in cells of double mutants, whereas the 3PGA level decreased compared to WT cells in these mutants.
PGPase Activities of Recombinant Proteins
The metabolite profiling data clearly indicated that inactivation of the selected genes results in intracellular accumulation of 2PG. To have direct evidence that the selected Synechocystis genes code for active PGPases, they were expressed in E. coli. First we analyzed the PGPase activities in total cell extracts of recombinant E. coli (Figure 4). Extracts from overexpression strains harboring the cyanobacterial PGPases in the pET28a expression systems showed up to 2 times higher PGPase activity than the extract from the empty vector control (Figure 4). The highest PGPase activity was observed in cells expressing slr0458 followed by cells expressing sll1349. These initial experiments verified that all four candidate proteins from Synechocystis are capable of 2PG dephosphorylation.
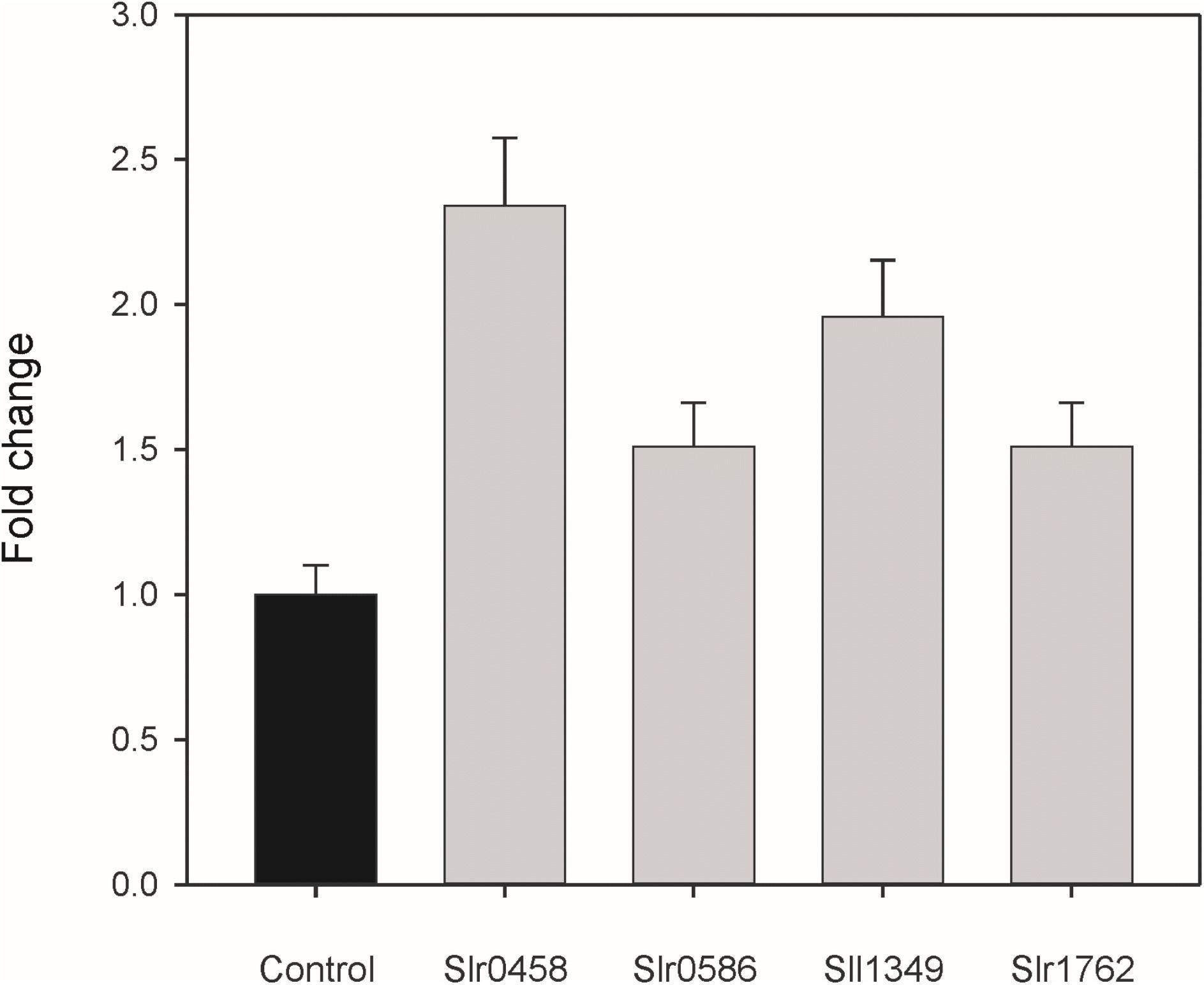
FIGURE 4. PGPase activities of recombinant proteins using 2PG as substrate. The putative PGPase from Synechocystis were expressed as tagged proteins in E. coli BL21 cells. Soluble cell extracts were used for enzyme assays in comparison to cells with the empty expression vector (control). The empty vector control showed PGPase activities of 3.1 nmol Pi min-1 mg-1 protein, which were set to 1 and relative values given are means ± SD.
Next, we went on to isolate purified recombinant proteins. Unfortunately, only three out of the four candidate proteins could be obtained as soluble proteins. Despite many attempts, protein Slr0586 was always found only in the insoluble fraction and therefore could not be tested in vitro. The Slr0458 and Sll1349 proteins were obtained in large amounts; whereas the yield of the Slr1762 protein was significantly lower (Supplementary Figure S4). However, all the purified recombinant proteins showed some instability, which became obvious since they started to precipitated when stored on ice for 1 h. The enzyme assays with 2PG (2 mM final concentration) as substrate revealed low specific PGPase activities for Slr0458 (0.54 ± 0.12 μmol min-1 mg-1) and for Sll1349 (0.23 ± 0.09 μmol min-1 mg-1). Enzymatic active Slr0458 expressed in E. coli was also obtained before by Haimovich-Dayan et al. (2015). The purified recombinant protein Slr1762 showed no PGPase activity at all. For comparison, we included the purified recombinant PGPase from Arabidopsis (PGLP1, At5g36700), which showed high specific PGPase activities of 25.48 ± 1.15 μmol min-1 mg-1. Thus, the purified plant enzyme has a more than 50 fold higher activity compared to the most active cyanobacterial PGPase Slr0458 in these experiments.
Discussion
Chlamydomonas and land pants such as Arabidopsis harbor single genes encoding for chloroplastidial PGPase functioning in the photorespiratory pathway. Additional cytosolic isoforms are not involved in photorespiration (Schwarte and Bauwe, 2007) but serve to destroy toxic side products from mainline carbon metabolism (Collard et al., 2016), which corresponds with our observation that more than one PGPase are present in the non-compartmented cyanobacterial cell of Synechocystis. This conclusion is mainly based on the mutation approach, where two and more genes for the putative PGPases needed to be inactivated before a growth decline and marked 2PG accumulation under LC conditions appeared (see Figures 2, 3). This is in contrast to the finding that inactivation of the single genes encoding for chloroplastidial PGPase of Chlamydomonas or Arabidopsis resulted in HCR phenotypes (Suzuki et al., 1999; Schwarte and Bauwe, 2007).
Only the Synechocystis single mutant Δsll1349 showed a decline in generation time under LC conditions compared to WT. This finding suggests that Sll1349 might represent the most active PGPase in Synechocystis. However, the 2PG level was not significantly changed in this mutant strain, therefore, the involvement of the Sll1349 in another important enzymatic reaction can not be ruled out. Among the double mutants, the combination of sll1349 and slr0458 disruption resulted in the most severe ( > 50%) growth decline pointing at Slr0458 as the second most active PGPase in Synechocystis under the tested growth condition. The protein Sll1349 is already annotated as PGPase in CyanoBase5 due to its similarity to CbbZ in different bacteria. However, our phylogenetic analysis (see Figure 1) revealed that the Sll1349 protein clade is rather distantly related to the CbbZ clade. In contrast, clades of Slr0458 and Slr1762-like proteins from cyanobacteria are situated next to the CbbZ clade. The CbbZ group comprises well-characterized PGPases from R. eutropha, E. coli and R. sphaeroides (Schäferjohann et al., 1993; Gibson and Tabita, 1997; Lyngstadaas et al., 1999). In Rhodobacter, the cbbZ gene is part of the cbbI-operon downstream of the gene for form I Rubisco. Consistent with their functional dependence, the Rubisco and PGPase proteins are co-expressed under photoautotrophic growth conditions (Gibson and Tabita, 1997). The phylogenetically related PGPase of E. coli encoded by gph also clusters in the CbbZ group (Figure 1). However, this likely PGPase is part of the dam containing operon linking it to DNA repair processes in E. coli (Lyngstadaas et al., 1999; Pellicer et al., 2003).
Slr1762, Slr1349, and Slr0586 are the Synechocystis PGPase having homologous proteins in many other cyanobacteria with known genome sequences, where they probably also function as photorespiratory PGPase. Interestingly, the gene for the Slr1762 homolog in Synechococcus elongatus PCC 7942 was found among the LC-induced genes (Schwarz et al., 2011). Photorespiratory genes including the PGPases are also up-regulated by LC in Chlamydomonas (Fang et al., 2012). However, all four PGPase encoding genes of Synechocystis showed similar transcript abundances under different CO2 conditions (Eisenhut et al., 2007). Remarkably, Slr0458, which seems to play an important role in 2PG dephosphorylation in Synechocystis together with Sll1349, is only found in the closely related cyanobacterium Synechocystis sp. PCC 6714 (Kopf et al., 2014).
The absence of a clear photorespiratory phenotype in the Synechocystis double and triple mutants is likely due to the activity of the CCM (Burnap et al., 2015), which minimizes RuBP oxygenation and hence 2PG accumulation. Notably, we were unable to obtain a quadruple mutant, in which all four genes were inactivated even under continuous HC conditions. Probably, the low rate of RuBP oxygenation in cells grown at HC (Huege et al., 2011) already results in 2PG levels that are not tolerated by cells lacking PGPase activity. A very severe HCR phenotype was also observed for the Arabidopsis pglp1 mutant, which does not fully recover at 1% CO2, which is in contrast to other plant mutants affected in photorespiratory enzymes (Timm et al., 2012). The growth decline in the different mutant combinations generally correlated with metabolite profiling data, where a considerable rise in 2PG accumulation was observed from single to double mutants with a maximum in the triple mutant (see Figure 3). These results support the notion that the inactivated genes indeed encode for functional PGPases in vivo that collectively dephosphorylate 2PG. It has been shown that increased 2PG levels due to lack of efficient 2PG recycling reduce growth of plants, because 2PG inhibits triosephosphate isomerase, phosphofructokinase and sedoheptulose 1,7-bisphosphate phosphatase (Anderson, 1971; Kelly and Latzko, 1976; Flügel et al., 2017). However, the slower growth of the single mutant Δsll1349 did not correlate with increased 2PG accumulation. This observation and the alterations in other phosphorylated compounds (see Figure 3C) might indicate that some of these enzymes have broader substrate specificity in vivo. Alternatively, the change in other phosphorylated compounds could also be taken as an indication that 2PG might act as a metabolic regulator.
In contrast to the enhanced values of 2PG in PGPase double and triple mutants, all Synechocystis mutants including the triple mutant contained almost the same amount of glycolate as WT cells (see Figure 3). However, bearing in mind that a significant glycolate accumulation was only detected in LC-grown cells of Synechocystis when the two glycolate dehydrogenases were inactivated (Eisenhut et al., 2008b; Huege et al., 2011), it is likely that the rapid glycolate conversion prevents marked changes in the glycolate pool in the different mutant strains. Hence, we assume that the observed glycolate pool is probably below the affinity of the downstream enzymes, explaining this low and almost constant pool. Nevertheless, the detection of glycolate in all strains supports the notion that more than one PGPase is actively converting 2PG into glycolate in Synechocystis.
To directly verify the enzymatic activity of the candidate proteins, we expressed these genes in E. coli as tagged proteins for biochemical analysis. Enhanced PGPase activity, compared to cells with empty vector, was observed in extracts from cells expressing these cyanobacterial genes (see Figure 4); however, the purified recombinant proteins Slr0458 and Sll1349 showed only low PGPases activity compared to recombinant PGPase of Arabidopsis. The low specific activity is most probably due to the instability of the cyanobacterial proteins, which could have caused an underestimation of the specific activities of cyanobacterial PGPases in this study. A substantial amount of eluted protein precipitated within 1 h of storage on ice indicating incorrect folding of the recombinant proteins. Similar stability problems were reported in previous attempts to purify the PGPase from cyanobacterial cells (Norman and Colman, 1991) or to obtain Slr0458 as recombinant protein (Haimovich-Dayan et al., 2015). The latter study also showed PGPase activity of Slr0458 in crude extracts of E. coli, which was about 4 times higher than the vector control. The instability of enzymes made it impossible to verify the substrate specificity of the four Synechocystis PGPase proteins. Since the levels of some other phosphorylated compounds are also changed in cells of different double mutants (see Figure 3C), it is likely that some of these enzymes act on a number of substrates in vivo.
Conclusion
We demonstrate that Synechocystis PGPases are not homologous to PGPases in eukaryotic phototrophs. The cyanobacterial proteins belong to the previously defined dehr family of HAD family proteins, whereas the plant enzymes are found in the NagD family (Burroughs et al., 2006). In contrast to all other photorespiratory enzymes, the plant PGPase most probably originated from the archaeal host cell (discussed in more detail in Hagemann et al., 2016) during eukaryogenesis (Spang et al., 2015). Only three cyanobacterial strains of the genera Leptolyngbya and Coleofasciculus possess genes for PGPase-like proteins of the NagD family that are closely related to the plant PGPases. However, these genes were most probably obtained via inter-bacterial lateral gene transfer, as the cyanobacterial proteins are situated in-between the bacterial clade (see Hagemann et al., 2016). It is interesting to note that the red alga Galdieria sulphuraria also harbors a Slr0458-like protein (Accession Number: XP_005708578.1) with more than 40% identical amino acids. It should be mentioned that plant-like PGLP1 proteins are also encoded in red algal genomes. The finding of two putative PGPases in the G. sulphuraria genome might indicate that, like cyanobacteria, red algae may use several photorespiratory PGPases, whereas green algae and plants such as Chlamydomonas and Arabidopsis (Suzuki et al., 1999; Schwarte and Bauwe, 2007) use only one photorespiratory PGPases in the chloroplast. The use of multiple PGPases among cyanobacteria could be related to the highly adaptive life-style of cyanobacteria that allows them to grow under diverse conditions. For example, the bloom-forming strain Microcystis aeruginosa PCC 7803 accumulates large amounts of glycolate when exposed to high-light stress (Meissner et al., 2015). The evolution of redundancy of an essential enzyme activity, such as PGPase, protects these organisms from damage by mutations as they frequently occur in non-stable environments. Alternatively, it is possible that at least some of these enzymes (such as Slr0586) have higher activity with substrates other than 2PG in Synechocystis and act as PGPase only when the main enzymes such as Slr0458 and Sll1349 are artificially inactivated. Redundant enzymes were also found for the downstream conversion of glycolate into glyoxylate (Eisenhut et al., 2008b) supporting the notion that the non-compartmented cyanobacteria need to avoid 2PG and glycolate accumulation to a larger extent than plants with their highly compartmented cellular metabolism.
Author Contributions
MH, AK, and HB designed the study. SR performed the most experiments. JK performed the metabolome analysis. SL performed the enzyme assays. RK performed the phylogenetic analysis. SR, JK, AK, HB, and MH analyzed the data. SR and MH wrote the manuscript with contributions from all authors.
Funding
Our work on cyanobacterial PGPases was supported by the Deutsche Forschungsgemeinschaft (German Research Foundation) in the Research Unit FOR 1186 PROMICS (Photorespiration: Origin and Metabolic Integration in Interacting Compartments). The research stay of SR at Rostock University was supported by a DAAD fellowship.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Prof. Dr. Teruo Ogawa (The Bioscience Center, Nagoya University, Chikusa, Nagoya, Japan) was acknowledged for providing the initial mutants defective in slr1349 and sll0458.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01718/full#supplementary-material
Abbreviations
2PG, 2-phosphoglycolate; HAD, 2-haloacid dehalogenase; HC, high CO2; HCR, high-CO2-requiring; LC, low CO2 of ambient air; PCC, Pasteur culture collection; PGPase, 2PG phosphatase; WT, wild type.
Footnotes
- ^http://tree.bio.ed.ac.uk/software/figtree/
- ^http://genome.annotation.jp/cyanobase/Synechocystis/genes/search?q=sll1349&keyword=search&m=gene_symbol%2Cgi_gname%2Cdefinition%2Cgi_pname
- ^http://genome.microbedb.jp/cyanobase/
- ^http://genome.microbedb.jp/CyanoBase
- ^http://genome.annotation.jp/cyanobase/Synechocystis/genes/search?q=sll1349&keyword=search&m=gene_symbol%2Cgi_gname%2Cdefinition%2Cgi_pname
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
Ames, B. N. (1966). Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 8, 115–117.
Anderson, L. E. (1971). Chloroplast and cytoplasmic enzymes. 2. Pea leaf triose phosphate isomerases. Biochim. Biophys. Acta 235, 237–244.
Bauwe, H., Hagemann, M., and Fernie, A. R. (2010). Photorespiration: players, partners and origin. Trend. Plant Sci. 15, 330–336. doi: 10.1016/j.tplants.2010.03.006
Bauwe, H., Hagemann, M., Kern, R., and Timm, S. (2012). Photorespiration has a dual origin and manifold links to central metabolism. Curr. Opin. Plant Biol. 15, 269–275. doi: 10.1016/j.pbi.2012.01.008
Biegert, A., Mayer, C., Remmert, M., Söding, J., and Lupas, A. N. (2006). The MPI bioinformatics toolkit for protein sequence analysis. Nucl. Acids Res. 34, W335–W339.
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254.
Bryant, D. A., de Lorimier, R., Guglielmi, G., and Stevens, S. E. Jr. (1990). Structural and compositional analyses of the phycobilisomes of Synechococcus sp. PCC 7002. Analyses of the wild-type strain and a phycocyanin-less mutant constructed by interposon mutagenesis. Arch. Microbiol. 153, 550–560.
Burnap, R. L., Hagemann, M., and Kaplan, A. (2015). Regulation of CO2 concentrating mechanism in cyanobacteria. Life 5, 348–371. doi: 10.3390/life5010348
Burroughs, M., Allen, K. N., Dunaway-Mariano, D., and Aravind, L. (2006). Evolutionary genomics of the HAD superfamily: understanding the structural adaptations and catalytic diversity in a superfamily of phosphodiesterases and allied enzymes. J. Mol. Biol. 361, 1003–1034.
Collard, F., Baldin, F., Gerin, I., Bolsee, J., Noel, G., Graff, J., et al. (2016). A conserved phosphatase destroys toxic glycolytic side products in mammals and yeast. Nat. Chem. Biol. 12, 601–607. doi: 10.1038/nchembio.2104
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2011). ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165. doi: 10.1093/bioinformatics/btr088
Do, C. B., Mahabhashyam, M. S. P., Brudno, M., and Batzoglou, S. (2005). ProbCons: probabilistic consistency-based multiple sequence alignment. Gen. Res. 15, 330–340.
Eisenhut, M., Kahlon, S., Hasse, D., Ewald, R., Lieman-Hurwitz, J., Ogawa, T., et al. (2006). The plant-like C2 glycolate cycle and the bacterial-like glycerate pathway cooperate in phosphoglycolate metabolism in cyanobacteria. Plant Physiol. 142, 333–342.
Eisenhut, M., Ruth, W., Haimovich, M., Bauwe, H., Kaplan, A., and Hagemann, M. (2008b). The photorespiratory glycolate metabolism is essential for cyanobacteria and might have been conveyed endosymbiontically to plants. Proc. Natl. Acad. Sci. U.S.A. 105, 17199–17204. doi: 10.1073/pnas.0807043105
Eisenhut, M., Huege, J., Schwarz, D., Bauwe, H., Kopka, J., and Hagemann, M. (2008a). Metabolome phenotyping of inorganic carbon limitation in cells of the wild type and photorespiratory mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiol. 148, 2109–2120. doi: 10.1104/pp.108.129403
Eisenhut, M., von Wobeser, E. A., Jonas, L., Schubert, H., Ibelings, B. W., Bauwe, H., et al. (2007). Long-term response toward inorganic carbon limitation in wild type and glycolate turnover mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiol. 144, 1946–1959.
Fang, W., Si, Y., Douglass, S., Casero, D., Merchant, S. S., Pellegrini, M., et al. (2012). Transcriptome-wide changes in Chlamydomonas reinhardtii gene expression regulated by carbon dioxide and the CO2-concentrating mechanism regulator CIA5/CCM1. Plant Cell 24, 1876–1893. doi: 10.1105/tpc.112.097949
Flügel, F., Timm, S., Arrivault, S., Florian, A., Stitt, M., Fernie, A. R., et al. (2017). The photorespiratory metabolite 2-phosphoglycolate regulates photosynthesis and starch accumulation in Arabidopsis. Plant Cell 29, 2537–2551. doi: 10.1105/tpc.17.00256
Gibson, J. L., and Tabita, F. R. (1997). Analysis of the cbbXYZ operon in Rhodobacter sphaeroides. J. Bacteriol. 179, 663–669.
Giordano, M., Beardall, J., and Raven, J. A. (2005). CO2 concentrating mechanisms in algae: mechanisms, environmental modulation, and evolution. Annu. Rev. Plant Biol. 56, 99–131.
Hackenberg, C., Engelhardt, A., Matthijs, H. C. P., Wittink, F., Bauwe, H., Kaplan, A., et al. (2009). Photorespiratory 2-phosphoglycolate metabolism and photoreduction of O2 cooperate in high-light acclimation of Synechocystis sp. strain PCC 6803. Planta 230, 625–637. doi: 10.1007/s00425-009-0972-9
Hackenberg, C., Kern, R., Huege, J., Stal, L. J., Tsuji, Y., Kopka, J., et al. (2011). Cyanobacterial lactate oxidases serve as essential partners in N2 fixation and evolved into photorespiratory glycolate oxidases in plants. Plant Cell 23, 2978–2990. doi: 10.1105/tpc.111.088070
Hagemann, M., Kern, R., Maurino, V. G., Hanson, D. T., Weber, A. P. M., Sage, R. F., et al. (2016). Evolution of photorespiration – from cyanobacteria via algae to land plants. J. Exp. Bot. 67, 2963–2976. doi: 10.1093/jxb/erw063
Hagemann, M., Richter, S., and Mikkat, S. (1997). The gene ggtA encodes for a subunit of the transport system for the osmoprotective compound glucosylglycerol in Synechocystis sp. PCC 6803. J. Bacteriol. 179, 714–720.
Hagemann, M., and Zuther, E. (1992). Selection of mutants of the cyanobacterium Synechocystis sp. PCC-6803 unable to tolerate high salt concentrations. Arch. Microbiol. 158, 429–434.
Haimovich-Dayan, M., Lieman-Hurwitz, J., Orf, I., Hagemann, M., and Kaplan, A. (2015). Does 2-phosphoglycolate serve as an internal signal molecule of inorganic carbon deprivation in the cyanobacterium Synechocystis sp. PCC 6803? Environ. Microbiol. 17, 1794–1804. doi: 10.1111/1462-2920.12638
Hardy, P., and Baldy, P. (1986). Corn phosphoglycolate phosphatase: purification and properties. Planta 168, 245–252. doi: 10.1007/BF00402970
Huege, J., Goetze, J., Schwarz, D., Bauwe, H., Hagemann, M., and Kopka, J. (2011). Modulation of the major paths of carbon in photorespiratory mutants of Synechocystis. PLoS One 6:e16278. doi: 10.1371/journal.pone.0016278
Hummel, J., Strehmel, N., Selbig, J., Walther, D., and Kopka, J. (2010). Decision tree supported substructure prediction of metabolites from GC-MS profiles. Metabolomics 6, 322–333.
Husic, D. W., Husic, H. D., and Tolbert, N. E. (1987). The oxidative photosynthetic carbon cycle or C2 cycle. CRC Crit. Rev. Plant Sci. 5, 45–100.
Husic, H. D., and Tolbert, N. E. (1984). Anion and divalent cation activation of phosphoglycolate phosphatase from leaves. Arch. Biochem. Biophys. 229, 64–72.
Kaneko, T., Sato, S., Kotani, H., Tanaka, A., Asamizu, E., Nakamura, Y., et al. (1996). Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3, 109–136.
Kelly, G. J., and Latzko, E. (1976). Inhibition of spinach-leaf phosphofructokinase by 2-phosphoglycollate. FEBS Lett. 68, 55–58.
Kern, R., Bauwe, H., and Hagemann, M. (2011). Evolution of enzymes involved in the photorespiratory 2-phosphoglycolate cycle from cyanobacteria via algae toward plants. Photosyn. Res. 109, 103–114. doi: 10.1007/s11120-010-9615-z
Kerr, M. W., and Gear, C. F. (1974). Studies on phosphoglycolate phosphatase isolated from pea leaves. Biochem. Soc. Trans. 2, 338–340.
Kim, Y., Yakunin, A. F., Kuznetsova, E., Xu, X., Pennycooke, M., Gu, J., et al. (2004). Structure- and function-based characterization of a new phosphoglycolate phosphatase from Thermoplasma acidophilum. J. Biol. Chem. 279, 517–526.
Kopf, M., Klähn, S., Pade, N., Weingärtner, C., Hagemann, M., Voß, B., et al. (2014). Comparative genome analysis of the closely related Synechocystis strains PCC 6714 and PCC 6803. DNA Res 21, 255–266. doi: 10.1093/dnares/dst055
Le, S. Q., and Gascuel, O. (2008). An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320. doi: 10.1093/molbev/msn067
Lisec, J., Römisch-Margl, L., Nikoloski, Z., Piepho, H. P., Giavalisco, P., Selbig, J., et al. (2011). Corn hybrids display lower metabolite variability and complex metabolite inheritance patterns. Plant J. 68, 326–336. doi: 10.1111/j.1365-313X.2011.04689.x
Lyngstadaas, A., Løbner-Olesen, A., Grelland, E., and Boye, E. (1999). The gene for 2-phosphoglycolate phosphatase (gph) in Escherichia coli is located in the same operon as dam and at least five other diverse genes. Biochim. Biophys. Acta 1472, 376–384.
Ma, Y., Hartman, M. M., and Moroney, J. V. (2013). “Transcriptional analysis of the three phosphoglycolate phosphatase genes in wild type and the pgp1 mutant of Chlamydomonas reinhardtii,” in Proceedings of the 15th Int. Conf. Photos for Photosynthesis Research for Food, Fuel and Future, eds T. Kuang, C. Lu, and L. Zhang (Heidelberg: Springer Nature), 315–318.
Meissner, S., Steinhauser, D., and Dittmann, E. (2015). Metabolomic analysis indicates a pivotal role of the hepatotoxin microcystin in high light adaptation of Microcystis. Environ. Microbiol. 17, 1497–1509. doi: 10.1111/1462-2920.12565
Nakamura, Y., Kanakagiri, S., Van, K., He, W., and Spalding, M. H. (2005). Disruption of the glycolate dehydrogenase gene in the high-CO2-requiring mutant HCR89 of Chlamydomonas reinhardtii. Can. J. Bot. 83, 820–833.
Norman, E. G., and Colman, B. (1991). Purification and characterization of phosphoglycolate phosphatase from the cyanobacterium Coccochloris peniocystis. Plant Physiol. 95, 693–698.
Pellicer, M. T., Nuñez, M. F., Aguilar, J., Badia, J., and Baldoma, L. (2003). Role of 2 phosphoglycolate phosphatase of Escherichia coli in metabolism of the 2-phosphoglycolate formed in DNA repair. J. Bacteriol. 185, 5815–5821.
Rippka, R., Deruelles, J., Waterbury, J. B., Herdman, M., and Stanier, R. Y. (1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111, 1–16.
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Schäferjohann, J., Yoo, J. G., Kusian, B., and Bowien, B. (1993). The cbb operons of the facultative chemoautotroph Alcaligenes eutrophus encode phosphoglycolate phosphatase. J. Bacteriol. 175, 7329–7340.
Schwarte, S., and Bauwe, H. (2007). Identification of the photorespiratory 2-phosphoglycolate phosphatase, PGLP1, in Arabidopsis. Plant Physiol. 144, 1580–1586.
Schwarz, D., Nodop, A., Hüge, J., Purfürst, S., Forchhammer, K., Michel, K. P., et al. (2011). Metabolic and transcriptomic phenotyping of inorganic carbon acclimation in the cyanobacterium Synechococcus elongatus PCC 7942. Plant Physiol. 155, 1640–1655. doi: 10.1104/pp.110.170225
Somerville, C. R. (2001). An early Arabidopsis demonstration. Resolving a few issues concerning photorespiration. Plant Physiol. 125, 20–24.
Somerville, C. R., and Ogren, W. L. (1979). A phosphoglycolate phosphatase-deficient mutant of Arabidopsis. Nature 280, 833–836.
Spang, A., Saw, J. H., Jørgensen, S. L., Zaremba-Niedzwiedzka, K., Martijn, J., Lind, A. E., et al. (2015). Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521, 173–179. doi: 10.1038/nature14447
Stamatakis, A. (2014). RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Suzuki, K., Mamedov, T. G., and Ikawa, T. (1999). A mutant of Chlamydomonas reinhardtii with reduced rate of photorespiration. Plant Cell Physiol. 40, 792–799.
Timm, S., Mielewczik, M., Florian, A., Frankenbach, S., Dreissen, A., Hocken, N., et al. (2012). High-to-low CO2 acclimation reveals plasticity of the photorespiratory pathway and indicates regulatory links to cellular metabolism of Arabidopsis. PLoS One 7:e42809. doi: 10.1371/journal.pone.0042809
Tolbert, N. E. (1997). The C-2 oxidative photosynthetic carbon cycle. Ann. Rev. Plant Physiol. Plant Mol. Biol. 48, 1–25.
Wingler, A., Lea, P. J., Quick, W. P., and Leegood, R. C. (2000). Photorespiration: metabolic pathways and their role in stress protection. Phil. Trans. Roy. Soc. B 355, 1517–1529.
Keywords: cyanobacteria, 2-haloacid dehalogenase, mutant, phosphoglycolate phosphatase, photorespiration, Synechocystis
Citation: Rai S, Lucius S, Kern R, Bauwe H, Kaplan A, Kopka J and Hagemann M (2018) The Synechocystis sp. PCC 6803 Genome Encodes Up to Four 2-Phosphoglycolate Phosphatases. Front. Plant Sci. 9:1718. doi: 10.3389/fpls.2018.01718
Received: 25 September 2018; Accepted: 05 November 2018;
Published: 27 November 2018.
Edited by:
Cornelia Spetea, University of Gothenburg, SwedenReviewed by:
Anja Schneider, Ludwig-Maximilians-Universität München, GermanyCheng-Cai Zhang, Chinese Academy of Sciences, China
Copyright © 2018 Rai, Lucius, Kern, Bauwe, Kaplan, Kopka and Hagemann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martin Hagemann, bWFydGluLmhhZ2VtYW5uQHVuaS1yb3N0b2NrLmRl
†Present address: Ramona Kern, Department of Applied Ecology and Phycology, University of Rostock, Rostock, Germany
 Snigdha Rai
Snigdha Rai Stefan Lucius
Stefan Lucius Ramona Kern
Ramona Kern Hermann Bauwe
Hermann Bauwe Aaron Kaplan
Aaron Kaplan Joachim Kopka
Joachim Kopka Martin Hagemann
Martin Hagemann