- 1Department of Agricultural Food Forest Sciences, University of Palermo, Palermo, Italy
- 2Council for Agricultural Research and Economics (CREA), Research Centre for Plant Protection and Certification (CREA-DC), Bagheria, Italy
- 3Hyris Limited, Milan, Italy
- 4Department of Plant Sciences, University of California, Davis, Davis, CA, United States
Olive Quick Decline Syndrome (OQDS)
Xylella fastidiosa subsp. pauca (Xfp) is a gram-negative pathogenic bacteria responsible for serious diseases (Purcell, 2013) that inflicts considerable economic loss (Li et al., 2007; Luvisi et al., 2017). The pathogen has been linked to olive quick decline syndrome (OQDS). This devastating olive disease was first observed in Salento (Apulia, southeastern Italy) in 2009. Infected trees respond to Xfp infection with scattered desiccation of twigs and small branches in the upper crown, which extend to the rest of the canopy, showing the characteristic blight effect. The disease causes tree death within a few years from the onset of symptoms (Martelli, 2016). The primary agronomic procedure for counteracting the infection is by heavy pruning to stimulate new growth (Martelli et al., 2016). However, this does not prevent the withering and desiccation of upper vegetation in the infected tree. Lignin deposition increases the tolerance of some hosts to Xylella fastidiosa. Elevated concentration of quinic acid, a lignin precursor, less concentration of hydroxytyrosolglucoside and the up-regulation of cinnamoyl-CoA reductase and polyphenol oxidase were observed in the most tolerant olive cultivar, Leccino (Sabella et al., 2018).
In this opinion article, we explore the use of a portable instrument to detect OQDS, based on the host responses at the transcript level. This approach was proposed previously to detect Huanglongbing, a severe disease affecting Citrus worldwide (Dandekar et al., 2010; Martinelli et al., 2014b). These innovative methods of plant disease detection had been reviewed recently (Martinelli et al., 2014a).
A Portable Instrument To Detect Early Olive Quick Decline Syndrome
Molecular tools for early in-field detection of Xylella fastidiosa infections have been developed (Baldi and La Porta, 2017; Chiriacò et al., 2018). A new portable instrument, the bCUBE, marketed by Hyris, Ltd., can effectively detect plant diseases. We believe that this instrument could also be used to detect olive responses to Xylella fastidiosa infections. The portable instrument analyzes mRNA using in-field qRT-PCR (Figure 1). Analysis of expression could be conducted on leaf, fruit, or bark tissues, targeting genes linked with OQDS syndrome. RNA can be extracted in the field with a quick procedure developed to detect other plant pathogens with bCUBE. Results are provided in ~ 45 min. The already-developed software and hardware can be used to optimize the instrument to detect expression of host Xfp-regulated genes. A bioinformatics analysis can be conducted through meta-analysis of published gene expression data related to Xf-host interactions. There is already a wide analysis of transcriptomic responses to this pathogen in different crops. Raw data deposited in public databases can be re-analyzed using the same pipeline to determine which differentially regulated genes are most commonly expressed. This analysis would show how much currently available information can be used to develop a portable instrument that detects these Xfp-regulated biomarkers. Implementation and validation tests would analyze the same tissues simultaneously in the field (with the portable instrument) and in the lab (traditional qRT-PCR) to verify the consistency of the field results provided by the instrument and to validate and optimize its use. We believe that this instrument should be tested in an olive grove where OQDS syndrome is already present by analyzing trees at different disease stages: healthy, apparently healthy, and both asymptomatic and symptomatic tissues of symptomatic trees.
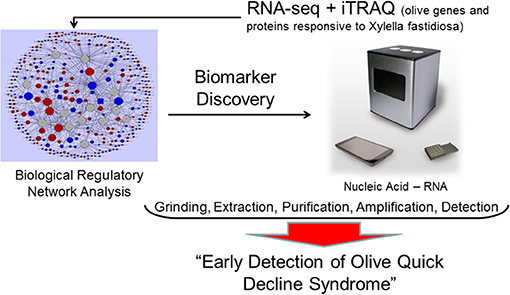
Figure 1. Multi-omic approach to detect early olive quick decline syndrome with portable instruments in the field. RNA-seq and proteomic iTRAQ approaches can identify host and pathogen biomarkers that can be analyzed directly in the field by a portable instrument for rapid in-field diagnosis of Xylella fastidiosa infections.
A Transdisciplinary Approach to Identify Olive Xfp-Responsive Genes as Biomarkers for In-Field Detection
A broad range of environmental stress biomarkers (transcripts, miRNAs, proteins, or metabolites) have been identified through integrated omic approaches (Natali et al., 2007; Martinelli and Tonutti, 2012; Tosetti et al., 2012). As the pathogen is not evenly distributed in an infected plant's tissues, identifying specific genes induced by the pathogen in a range of tissues is necessary to discover tissue-specific host biomarkers (Giovino et al., 2015). The pathogen's presence is perceived by the infected plant before the pathogen DNA concentration exceeds the detection limit of traditional real-time systems. Such analyses will clarify pathogenetic disease mechanisms by clarifying the relationships between cause and effect. It is still unclear how the pathogen contributes to disease manifestation and how other environmental factors contribute. The proposed approach would also provide molecular targets to develop resistant genotypes as it refines existing technology into a novel portable instrument for real-time disease detection in orchards. It links genomics to technology that enhances the discovery of disease-specific biomarkers and closes the gaps in conventional disease scouting and molecular diagnostic methods. Disease-specific biomarkers will be discovered through genomic and network analysis of host and pathogen responses. Close integration of different activities will combine technology and research. Graft-infected olive plants should be inoculated. Five to ten plants should be sampled in each category and tissues should be collected from the top branches at 7, 21, 35, and 60 days after infection. When symptoms appear, infection status should be verified by PCR. In addition to the longitudinal experiment, a transcriptomic analysis of naturally infected trees at different disease stages should be performed. The first category of trees (healthy) might be available in a nearby olive grove where the pathogen has not yet been detected. If it is subjected to similar climatic, agronomic and pedological conditions to infected trees, these trees can represent control uninfected conditions. The second category of trees is present in the infected field, but is apparently healthy and without detectable pathogen. Since we cannot exclude the presence of the pathogen, these trees provide a different control because later diagnostic analysis might detect infection. The third category of trees is clearly symptomatic, PCR-positive or ELISA-positive. The discovery of potential markers linked with early olive response to Xfd infection may be performed using RNA-seq approaches and Illumina technology (Hiseq 2000). Some transcriptomic studies at different symptomatic stages have been conducted in Vitis and Citrus crops infected by Xylella fastidiosa (Rodrigues et al., 2013; Zaini et al., 2018). Some genes are commonly expressed in Vitis and Citrus responses to Xylella fastidiosa (Table 1). Interestingly, some of these genes are differentially regulated by Xylella fastidiosa in susceptible and tolerant genotypes, highlighting their possible use as biomarkers for tolerance. These data provide a good starting point to test the instrument's ability to identify olive orthologs of these Xf-regulated genes. Our meta-analysis identified some good candidate genes: a leucine-rich repeat protein kinase family protein, MYB66, chitinase A, a trypsin inhibitor family protein, expansin A4 and some auxin-responsive and gibberellin-regulated genes (Table 1). Xf-regulated genes commonly present in the two transcriptomic works conducted in Vitis and olive might be also of interest (Giampetruzzi et al., 2016; Zaini et al., 2018; Table 2). Some key cell wall modification genes that are potentially involved in pathogen signaling responses were observed: a pectin-lyase, a laccase and a poligalacturonase. This latter gene was commonly regulated in transcriptomic data obtained from all three species (Citrus, Vitis, and olive). This preliminary meta-analysis does not replace a longitudinal RNA-seq analysis focusing on early asymptomatic stages of infection, required to render the instrument highly sensitive and reliable. RNA-seq analysis cannot be exploited fully unless the quality of the olive genome sequence is improved. At the moment, only a draft version of the Farga genotype is available (Cruz et al., 2016). However, a more advanced version of the olive genome is expected soon due to ongoing international initiatives by an Italian-Spanish consortium that began in 2009. With the progressive reduction of sequencing costs, an improved olive genome is expected soon, allowing improved RNA-seq approaches. Indeed, we believe that the concept of an in-field device to detect host transcriptomic responses to Xfp will push efforts to obtain a high-quality olive genome sequence.
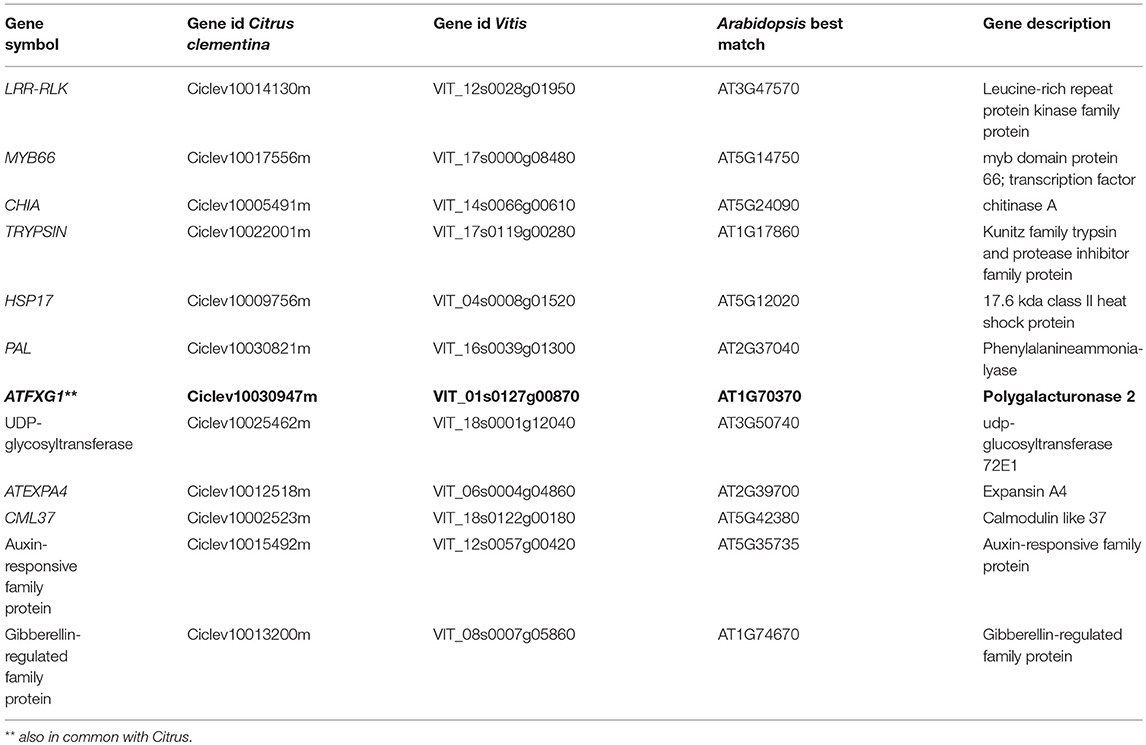
Table 1. Common Citrus and Vitis differentially-expressed genes in response to Xylella fastidiosa infection (Rodrigues et al., 2013; Zaini et al., 2018).
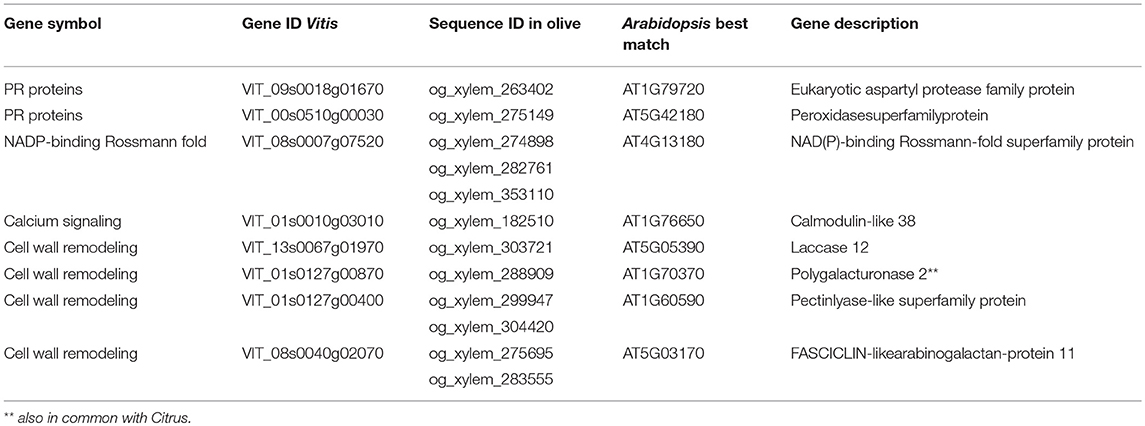
Table 2. Common differentially expressed genes in Vitis (Zaini et al., 2018) and Olea in response to X. fastidiosa infection (Giampetruzzi et al., 2016).
Proteomics may be conducted in parallel with transcriptomics using the iTRAQ method. This approach has been used in Vitis (Chakraborty et al., 2016). The use of proteomics is important to validate transcriptomic markers considering that genes may be regulated by post-transcriptional mechanisms. Proteins can be extracted from powdered olive leaf and bark tissues using a highly efficient phenol extraction procedure (Schuster and Davies, 1983). This protocol can extract protein from tissues that contain highly reactive compounds. The extracted proteins can be precipitated using the ProteoExtractTM Protein Precipitation kit (Calbiochem), dehydrated overnight, then re-suspended and subjected to tryptic digestion. The digested peptides can be analyzed using a QExactive mass spectrometer (Thermo Fisher Scientific) coupled with an Easy-LC (Thermo Fisher Scientific) and a nanospray ionization source. The peptides are loaded onto a trap (100 micron, C18 100Å 5U) and desalted online before separation using a reverse-phased column (75 micron, C18 200Å 3U). Data are acquired using data-dependent ms/ms, which has a full scan range of 300–1600 Da and a resolution of 70,000. Raw data are analyzed using X!Tandem and visualized using Scaffold Proteome Software (Version 3.01). Samples are compared to Uniprot databases appended with the cRAP database, which contains common laboratory contaminants. Reverse decoy databases are also applied to the database prior to the X!Tandem searches. This procedure should identify the secreted virulence factor of Xylella fastidiosa in olive. Some identified pathogen virulence proteins will be selected for in planta expression to validate their virulence phenotype. Synthetic virulence genes should be designed to insure greater in-plant expression. Codons might be optimized by replacing codons that are less frequently used in Citrus by DNAworks software (http://mcl1.ncifcrf.gov/dnaworks/). Signal peptides could be analyzed by SignalP 4.0 server (http://www.cbs.dtu.dk/services/SignalP/). N-Glycosylation sites may be predicted using NetNglyc1.0. More than 4,500 proteins, including several pathogen targets, have been identified in Citrus leaf tissues infected by Huanglongbing disease, comparing tolerant and susceptible genotypes (Martinelli et al., 2016).
Conclusions
Efficient pre-symptomatic diagnosis of OQDS relies on the ability to (i) obtain informative biomarkers from Xylella fastidiosa and host (olive), and then (ii) detect the biomarkers in a rapid, sensitive, and cost-effective manner. Before the onset of disease symptoms, the expression pattern of these biomarkers and their interplay changes at the early stages of infection. A deep transcriptomic analysis using multiple genotypes is highly desirable to define thresholds of biomarker expression and optimize efficiency. Once biomarkers (genes) are determined, the goal is to design a device that integrates sample preparation, nucleic acid enrichment, and final detection. This translational genomic approach will permit the following impacts: (1) develop and deploy inexpensive electronic instruments to identify Xfp infections at early, pre-symptomatic stages, (2) clarify vector-pathogen-plant interaction to monitor the efficacy of field interventions or treatments, (3) enhance disease scouting through new disease detection and diagnosis tools and improved stakeholder outreach, and (4) adapt the detection and diagnostic tools to different specialty crop diseases.
Author Contributions
FM conceived the opinion and designed and wrote the article. AM, FPM, TC, AG, ID, and AD approved and contributed to the writing of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Baldi, P., and La Porta, N. (2017). Xylella fastidiosa: host range and advance in molecular identification techniques. Front. Plant Sci. 8:944. doi: 10.3389/fpls.2017.00944
Chakraborty, S., Nascimento, R., Zaini, P. A., Gouran, H., Rao, B. J., Goulart, L. R., et al. (2016). Sequence/structural analysis of xylem proteome emphasizes pathogenesis-related proteins, chitinases and β-1, 3-glucanases as key players in grapevine defense against Xylella fastidiosa. Peer J. 4:e2007. doi: 10.7717/peerj.2007
Chiriacò, M. S., Luvisi, A., Primiceri, E., Sabella, E., De Bellis, L., and Maruccio, G. (2018). Development of a lab-on-a-chip method for rapid assay of Xylella fastidiosa subsp. pauca strain CoDiRO. Sci. Rep. 8:7376. doi: 10.1038/s41598-018-25747-4
Cruz, F., Julca, I., Gomez-Garrido, J., Loska, D., Marcet-Houben, M., Cano, E., et al. (2016). Genome sequence of the olive tree, Olea europea. Gigascience 5:29. doi: 10.1186/s13742-016-0134-5
Dandekar, A. M., Martinelli, F., Davis, C., Bhushan, A., Zhao, W., Fiehn, O., et al. (2010). Analysis of early host responses for asymptomatic disease detection and management of specialty crops. Crit. Rev. Immunol. 30:277–289. doi: 10.1615/CritRevImmunol.v30.i3.50
Giampetruzzi, A., Morelli, M., Saponari, M., Loconsole, G., Chiumenti, M., Boscia, D., et al. (2016). Transcriptome profiling of two olive cultivars in response to infection by the CoDiRO strain of Xylella fastidiosa subsp. pauca. BMC Genom. 17:475. doi: 10.1186/s12864-016-2833-9
Giovino, A., Bertolini, E., Fileccia, V., Al Hassan, M., Labra, M., and Martinelli, F. (2015). Transcriptome analysis of Phoenix canariensis chabaud in response to Rhynchophorusferrugineus Olivier attacks. Front. Plant Sci. 6:817. doi: 10.3389/fpls.2015.00817
Li, Y., Hao, G., Galvani, C. D., Meng, Y., De La Fuente, L., Hoch, H. C., et al. (2007). Type I and type IV pili of Xylella fastidiosa affect twitching motility, biofilm formation, and cell-cell aggregation. Microbiology 153:719–726. doi: 10.1099/mic.0.2006/002311-0
Luvisi, A., Aprile, A., Sabella, E., Vergine, M., Nicolì, F, Nutricati, E., et al. (2017). Xylella fastidiosa subsp. pauca (CoDiRO strain) infection in four olive (Olea europaea L.) cultivars: profile of phenolic compounds in leaves and progression of leaf scorch symptoms. Phytopathol. Mediterr. 56:259. doi: 10.14601/Phytopathol_Mediterr-20578
Martelli, G. P. (2016). The current status of the quick decline syndrome of olive in southern Italy. Phytoparasitica 44, 1–10. doi: 10.1007/s12600-015-0498-6
Martelli, G. P., Boscia, D., Porcelli, F., and Saponari, M. (2016). The olive quick decline syndrome in south-east Italy: a threatening phytosanitary emergency. Eur. J. Plant Pathol. 144, 235–243. doi: 10.1007/s10658-015-0784-7
Martinelli, F., Reagan, R. L., Dolan, D., Fileccia, V., and Dandekar, A. M. (2016). Proteomic analysis highlights the role of detoxification pathways in increased tolerance to Huanglongbing disease. BMC Plant Biol. 16:167. doi: 10.1186/s12870-016-0858-5
Martinelli, F., Scalenghe, R., Davino, S., Panno, S., Scuderi, G., Ruisi, P., et al. (2014a). Advanced methods of plant disease detection. A review. Agron. Sustain. Dev. 35, 1–25. doi: 10.1007/s13593-014-0246-1
Martinelli, F., Scalenghe, R., Giovino, A., Pasquale, M., Aksenov, A. A., Pasamontes, A., et al. (2014b). Proposal of a Citrus translational genomic approach for early and infield detection of Flavescence dorée in Vitis. Plant Biosyst. 150, 1–11. doi: 10.1080/11263504.2014.908976
Martinelli, F., and Tonutti, P. (2012). Flavonoid metabolism and gene expression in developing olive (Olea europaea L.) fruit. Plant Biosyst. 146, 164–170. doi: 10.1080/11263504.2012.681320
Natali, L., Giordani, T., Lercari, B., Maestrini, P., Cozza, R., Pangaro, T., et al. (2007). Light induces expression of a dehydrin-encoding gene during seedling de-etiolation in sunflower (Helianthus annuus L.). J. Plant Physiol. 164, 263–273. doi: 10.1016/j.jplph.2006.01.015
Purcell, A. (2013). Paradigms: examples from the bacterium Xylella fastidiosa. Ann. Rev. Phytopathol. 51, 339–356. doi: 10.1146/annurev-phyto-082712-102325
Rodrigues, C. M., de Souza, A. A., Takita, M. A., Kishi, L. T., and Machado, M. A. (2013). RNA-Seq analysis of Citrus reticulata in the early stages of Xylella fastidiosa infection reveals auxin-related genes as a defense response. BMC Genom. 14:676. doi: 10.1186/1471-2164-14-676
Sabella, E., Luvisi, A., Aprile, A., Negro, C., Vergine, M., Nicolì, F., et al. (2018). Xylella fastidiosa induces differential expression of lignification related-genes and lignin accumulation in tolerant olive trees cv. Leccino. J. Plant Physiol. 220, 60–68. doi: 10.1016/j.jplph.2017.10.007
Schuster, A. M., and Davies, E. (1983). Ribonucleic acid and protein metabolism in pea epicotyls: II. Response to wounding in aged tissue. Plant Physiol. 73, 817–821. doi: 10.1104/pp.73.3.817
Tosetti, R., Martinelli, F., and Tonutti, P. (2012). Metabolomics approach to studying minimally processed peach (Prunuspersica) fruit. Acta Hortic. 934, 1017–1022. doi: 10.17660/ActaHortic.2012.934.135
Keywords: in-field detection, olive quick decline syndrome, olive, portable instrument, Xylella fastidiosa
Citation: Martinelli F, Marchese A, Giovino A, Marra FP, Della Noce I, Caruso T and Dandekar AM (2019) In-Field and Early Detection of Xylella fastidiosa Infections in Olive Using a Portable Instrument. Front. Plant Sci. 9:2007. doi: 10.3389/fpls.2018.02007
Received: 08 June 2018; Accepted: 31 December 2018;
Published: 18 January 2019.
Edited by:
Ivan Baccelli, Italian National Research Council, ItalyReviewed by:
Chiaraluce Moretti, University of Perugia, ItalyCopyright © 2019 Martinelli, Marchese, Giovino, Marra, Della Noce, Caruso and Dandekar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Federico Martinelli, ZmVkZXJpY28ubWFydGluZWxsaUB1bmlwYS5pdA==
 Federico Martinelli
Federico Martinelli Annalisa Marchese
Annalisa Marchese Antonio Giovino2
Antonio Giovino2 Francesco Paolo Marra
Francesco Paolo Marra Isabella Della Noce
Isabella Della Noce Tiziano Caruso
Tiziano Caruso Abhaya M. Dandekar
Abhaya M. Dandekar