- State Key Laboratory of Crop Stress Biology for Arid Area, College of Life Sciences, Northwest A&F University, Yangling, China
The single-celled trichomes of Arabidopsis thaliana have long served as an elegant model for elucidating the mechanisms of cell differentiation and morphogenesis due to their unique growth patterns. To identify new components in the genetic network that governs trichome development, we carried out exhaustive screens for additional Arabidopsis mutants with altered trichome morphology. Here, we report one mutant, aberrantly branched trichome1-1 (abt1-1), with a reduced trichome branching phenotype. After positional cloning, a point mutation in the SPIKE1 (SPK1) gene was identified in abt1-1. Further genetic complementation experiments confirmed that abt1-1 is a new allele of SPK1, so abt1-1 was renamed as spk1-7 according to the literatures. spk1-7 and two other spk1 mutant alleles, covering a spectrum of phenotypic severity, highlighted the distinct responses of developmental programs to different SPK1 mutations. Although null spk1 mutants are lethal and show defects in plant stature, trichome and epidermal pavement cell development, only trichome branching is affected in spk1-7. Surprisingly, we found that SPK1 is involved in the positioning of nuclei in the trichome cells. Lastly, through double mutant analysis, we found the coordinated regulation of trichome branching between SPK1 and two other trichome branching regulators, ANGUSTIFOLIA (AN) and ZWICHEL (ZWI). SPK1 might serve for the precise positioning of trichome nuclei, while AN and ZWI contribute to the formation of branch points through governing the cMTs dynamics. In summary, this study presented a fully viable new mutant allele of SPK1 and shed new light on the regulation of trichome branching and other developmental processes by SPK1.
Introduction
The differentiation of distinct cell types is one of the most fundamental features of multicellular organisms (Lee and Schiefelbein, 2002). In higher plants, plant trichomes have served as an amazingly effective paradigm for the investigation of cell differentiation due to their conspicuous presence and defined developmental patterns (Hülskamp et al., 1994; Tominaga-Wada et al., 2011; Grebe, 2012; Ivakov and Persson, 2013; Pattanaik et al., 2014). In model plant Arabidopsis thaliana, leaf trichomes are unicellular structures usually bearing three or four branches (Hülskamp et al., 1994), and numerous pathways that control trichome branching have been identified in the last two decades (Folkers et al., 1997; Hülskamp, 2004; Guimil and Dunand, 2007; Ishida et al., 2008; Yang and Ye, 2013).
Generally, the number of trichome branches is closely linked with plant cell cycle control (Schnittger and Hülskamp, 2002; Kasili et al., 2010; Yang and Ye, 2013). During Arabidopsis trichome development, once the cell fate is determined, trichome progenitor cells switch from mitotic division to endoreplication and typically undergo 4 rounds of endoreplication, giving the trichome cells an average of 32C (C equals haploid DNA content per nucleus) in the nuclear DNA content (Folkers et al., 1997; Hülskamp, 2004). A myriads of mutants including kaktus, spindly, polycomb, triptychon, rastifari, and rpt2a showed extended endoreplication and elevated ploidy, and thus displayed hyperbranched trichomes, suggesting a certain degree of correlation between trichome branching and endoreplication (Szymanski and Marks, 1998; Perazza et al., 1999; Sonoda et al., 2009). However, the correlation between branching and ploidy and nuclear DNA content cannot be extrapolated to all trichome branching mutants, implicating that additional pathways are involved in the elaboration of trichome branching (Ilgenfritz et al., 2003).
Pharmacological and molecular evidences suggest that the organization and dynamics of cortical microtubules (cMTs) are also intimately related with trichome branching determination (Oppenheimer et al., 1997; Mathur and Chua, 2000; Kirik et al., 2002a,b; Abe et al., 2004). During trichome morphogenesis, the arrangement of cMTs changes dramatically at branching points (Mathur and Chua, 2000). Mutations in genes involved in the formation of α/β-tubulin heterodimers or cMTs dynamics often lead to altered trichome branching (Oppenheimer et al., 1997; Burk et al., 2001; Kirik et al., 2002a,b; Abe et al., 2004; Abe and Hashimoto, 2005). Dominant-negative mutations in α-tubulin 4 (TUA4) and TUA6 genes display abnormal arrangement and destabilization of cMTs and show a reduction in trichome branching (Thitamadee et al., 2002; Abe et al., 2004). In contrast, certain mutant forms of TUA6 render cMTs more polymerization-prone and promote more branched trichomes formed (Abe and Hashimoto, 2005). Additional factors that are functionally associated with cMTs also play roles in trichome branching determination. ANGUSTIFOLIA (AN) encodes a C-terminal binding proteins/brefeldin ADP-ribosylated substrates (CtBP/BARS) related protein (Folkers et al., 2002; Kim et al., 2002), and the Arabidopsis an mutant showed an abnormal organization of cMTs and predominantly two-branched trichomes (Folkers et al., 2002). ZWICHEL (ZWI) encodes a member of the unique Kinesin-like Calmodulin-Binding Protein (KCBP) family that can directly bind to cMTs (Reddy et al., 1996; Oppenheimer et al., 1997). Loss of function of ZWI leads to the plants with two-branched and short stalk trichomes. Interestingly, ZWI could interact genetically and physically with AN, suggesting a role for AN in cMTs mediated intracellular trafficking (Reddy et al., 1996; Oppenheimer et al., 1997; Folkers et al., 2002). Recent results showed that ZWI may serve as a hub to integrate and coordinate cMTs and actin cytoskeleton to achieve the cytoskeletal configuration necessary for trichome development (Tian et al., 2015).
Despite the tremendous progress in our understanding of the process, the full regulatory network of plant trichome development has not yet to be established. To identify additional genes and processes that regulate trichome development, we carried out large-scale forward genetic screening in A. thaliana and identified a mutant which we named abt1-1. Through map-based cloning, we cloned the ABT1 locus and identified a G to A mutation in the SPK1 gene. Genetic and phenotypic analyses further confirmed that ABT1 is SPK1 gene, which encodes a member of the CDM (named after the Caenorhabditis elegans CED-5, human DOCK180, and Drosophila melanogaster myoblast city) family proteins that functions as a guanine nucleotide exchange factor (GEF) (Kiyokawa et al., 1998; Bompard and Caron, 2004). Taking advantage of the three SPK1 mutant alleles with varied severity, we demonstrated that the developmental processes regulated by SPK1 show differential responses to these mutations, revealing a previous unknown mode of plant responses to SPK1. More importantly, we determined that SPK1 is involved in the regulation of nucleus positioning in the trichome cell. Genetic interaction analyses established that SPK1 interact genetically with AN and ZWI in the regulation of trichome branch formation. In summary, our findings provide new insight in the functions of SPK1 and the modes of regulation of plant growth and development by SPK1.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis thaliana materials used in this study are all in the Columbia-0 (Col) background unless indicated otherwise. Ethyl methanesulfonate (EMS)-mutagenesis was carried out in the abs3-1D mutant background following previously described procedures (Kim et al., 2006). abs3-1D has been described by Wang et al. (2015). The spk1-7 line described in this study was isolated in the M2 mutant pool. The abs3-1D mutant background does not influence the trichome branching defects of spk1-7 and was removed by backcrossing the original mutant with Col wild type. Two additional rounds of backcross were performed prior to further analysis of spk1-7. T-DNA insertional lines for SPK1 (SALK_136776C, spk1-5; SAIL_520H04, spk1-6), AN (SALK_026489, an), ZWI (SALK_017886, zwi), and a marker line for microtubule arrays GFP-TUB6 (CS6550) were obtained from the Arabidopsis Biological Resource Center (ABRC). The T-DNA insertion sites and homozygous mutants for these lines were confirmed by PCR and sequencing. All primers used in this study are listed in Supplementary Table S1. Arabidopsis seeds were sowed on commercial soil mix (Pindstrup, Denmark) and grown at 22°C under continuous illumination (∼100 μmol m-2 s-1).
Phenotypic Characterization of SPK1 Alleles
The trichome branching phenotypes were analyzed as described in An et al. (2011). In brief, the third and fifth rosette leaves of two-week-old Arabidopsis plants were examined with a SZ61sterescope (Olympus). Numbers of branches of all the trichomes on each leaf were counted and recorded. For each genotype, at least 16 plants were used in the statistical analysis. Student’s t-test was used to assess the difference between wild type and mutants. The experiments were repeated as least twice.
For scanning electron microscopy (SEM), fresh leaves were taped on the sample stages, and directly viewed and photographed using a tabletop SEM TM3030 (Hitachi, Japan).
To visualize the outlines of pavement cells, cotyledons of one-week-old seedlings were stained in propidium iodide (PI) (1 mg⋅ml-1 in H2O) for 5 min and washed multiple times in deionized water. Stained cotyledons were mounted in water and examined with confocal microscopy (Nikon A1) using the following setting: Ex, 561 nm; Em: 595/50 nm.
Map-Based Cloning
Map-based cloning was conducted according to Lukowitz et al. (2000). spk1-7 was crossed with Landsberg erecta (Ler) to generate an F2 mapping population. Bulked segregant analysis first mapped spk1-7 mutation to a region near the Indel markers FCA2#1 and F17L22#1 on chromosome IV. Additional molecular markers were used to fine map the spk1-7 mutation. Detailed information of molecular markers used in this study has been described in Liu et al. (2010). Candidate genes in the final interval were sequenced to determine the mutation sites.
RNA Extraction and Real-Time Quantitative RT-PCR
Total RNA was isolated from pooled samples of one-week-old seedlings using the TRIzol reagent (Invitrogen) following the manufacturer’s instructions. First strand cDNAs were synthesized from 1 μg of DNase I treated total RNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche). Real-time PCRs were performed with the SYBR green PCR mix (Roche), and running on a Bio-Rad CFX96 Real-Time PCR system. Relative expression levels of the target genes were calculated with 2-ΔCt. The ACTIN2 gene was used as the internal control. Primers for real-time quantitative RT-PCR analysis were listed in Supplementary Table S1.
Trichome Nuclei Position Analysis
The third and fifth rosette leaves from three-week old wild type and mutant plants were used in trichome cell nuclei position analysis. Leaves were fixed and cell nuclei were stained by fluorescent dye 4′,6-Diamidino-2-phenylindole dihydrochloride (DAPI), then the nuclei of the DAPI stained trichome cells were observed under the a DM5000B fluorescence microscope (Leica), and the nuclei positions of the cells were captured and recorded. Statistical analyses were performed based on the observation and at least 30 independent trichome cells of each genotype were used.
Construction of Double Mutants
To generate the spk1-7 an and spk1-7 zwi double mutants, spk1-7 was crossed with an and zwi, respectively. Genotypes of the F1 and F2 progeny were determined by PCR using gene specific primers. Trichome phenotypes were assesses at F2 generation after genotyping. The same strategies were used to construct the spk1-5 an, spk1-5 zwi, spk1-6 an, spk1-6 zwi double mutants, and spk1-7 an zwi triple mutants, respectively.
Microscopy and Image Processing
For the fluorescence observation of the cMTs, a marker line CS6550 which carries a GFP-TUB6 construct was crossed to the different mutant background and the double mutants were selected by genomic PCR and GFP fluorescence detection. Seeds were sowed into the soil and the cotyledons of 4-day-old seedlings were observed under a spinning disk confocal microscope equipped with lasers for GFP (Nikon A1). Images were stored and processed with the Adobe Photoshop 7.0 program. For the quantitative analysis of the cMTs organization, the microtubule angles were measured by Image J software according to Yao et al. (2008). The value of microtubules angles of that parallel to the cell’s longitudinal axis were defined as 0°, while the value of those were perpendicular to the cell’s longitudinal axis were defined as 90°. The cMTs anisotropy was evaluated according to Boudaoud et al. (2014), and the following convention was used: the anisotropy score 0 for no order (purely isotropic arrays) and 1 for perfectly ordered (purely anisotropic arrays).
Results
The Isolation of the abt1-1 Mutant
In our ongoing effort to identify new factors that define trichome morphology, we carried out large-scale EMS-mutagenesis and a trichome branching defective mutant, designated abt1-1 was isolated. The overall growth and development of abt1-1 was not significantly altered compared with the wild type, but displayed a conspicuous trichome branching defect (Figures 1A–C). The majority of leaf trichomes had three branches in wild type while two-branched trichomes became the predominant form in abt1-1 leaves (Figures 1B–E). Notably, the reduced trichome branching phenotype in abt1-1 was more pronounced in the fifth rosette leaves than in the third rosette leaves as we observed an average of 68% of two-branched trichomes in the third leaves (Figure 1D) versus an average of 76% of two-branched in the fifth leaves (Figure 1E). This observation suggests that the trichome hypobranching defect in abt1-1 was developmentally associated. Consistent with the leaf trichome branching phenotype, trichomes on the main inflorescence stem also had fewer branches in abt1-1 compared to those in the wild type (Figure 1F). Genetic analysis indicated that abt1-1 behaved as a single recessive mutation. These data suggest that ABT1 functions as a positive regulator of trichome branching.
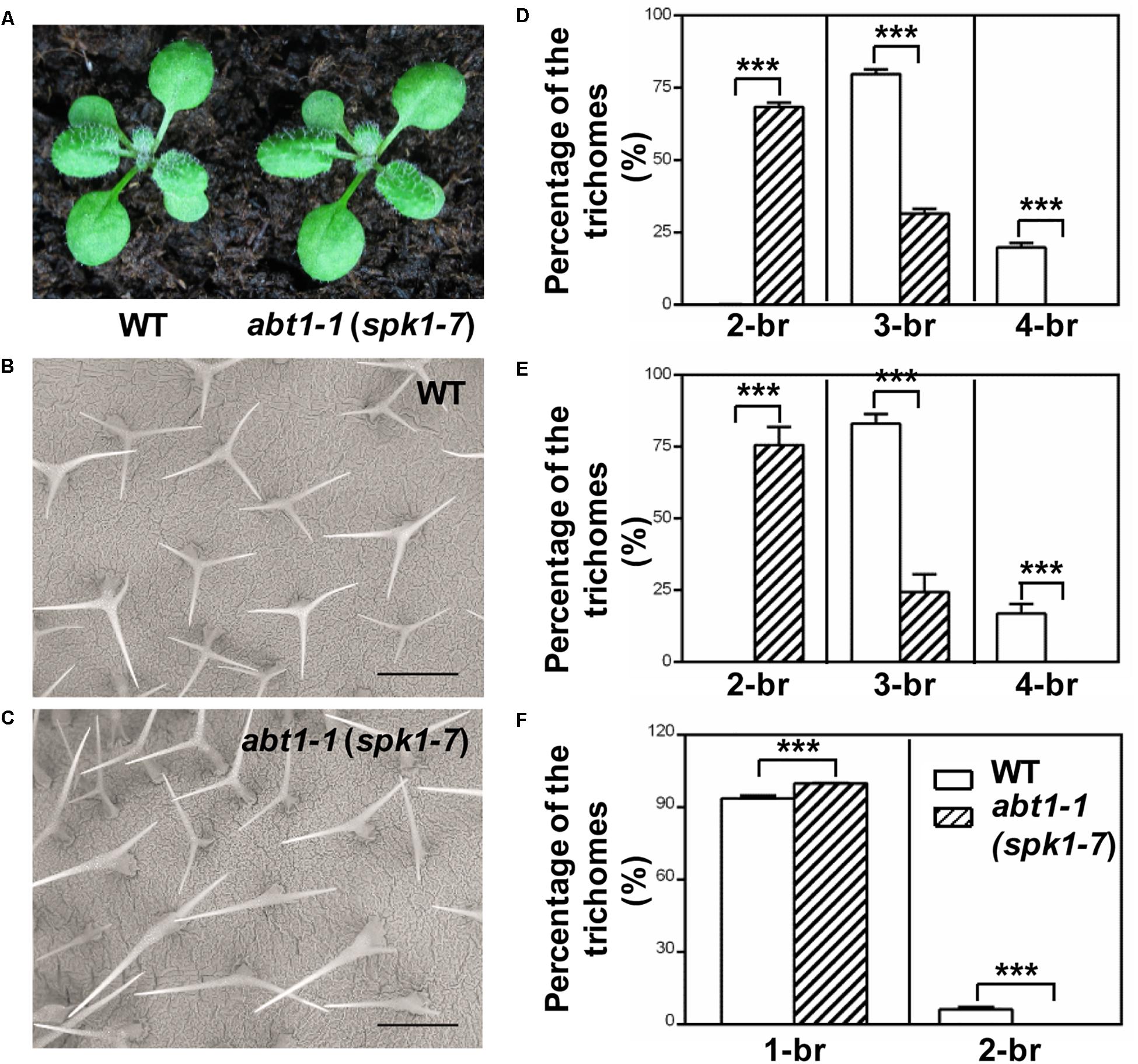
Figure 1. Phenotypes of WT and abt1-1 (spk1-7) plants. (A) The overall growth phenotypes of two-week-old wild type (WT) and abt1-1 (spk1-7). (B,C) Examination of the trichome branching phenotypes on the fifth rosette leaves of three-week-old WT (B) and abt1-1 (spk1-7) (C) by SEM. Bars are 250 μM. (D–F) Quantification of the trichome branching phenotypes of WT and abt1-1 (spk1-7) on the third rosette leaves (D), the fifth rosette leaves (E) and the main inflorescence stems (F). Data were presented as mean ± SD of three biological replicates. ∗∗∗p < 0.001. 2/3/4-br for two/three/four-branched trichomes, respectively.
abt1-1 Is a New Mutant Allele of SPK1
To further investigate the function of ABT1 on regulating trichome branching, we carried out map-based cloning. Initial bulked segregant analyses placed the ABT1 locus near the molecular markers FCA2#1 and F17L22#1 on chromosome IV (Figure 2). Further fine mapping narrowed down the physical interval harboring the abt1-1 mutation to a ∼200 kb region between molecular markers FCA5#12 and FCA6#1 (Figure 2). We examined all the annotated genes in this interval to identify known loci in which mutations could lead to reduced trichome branching phenotype. Interestingly, T-DNA insertional mutants of one locus, SPK1/At4g16340, was reported to have predominantly one-branched trichomes (Qiu et al., 2002; Basu et al., 2008). To test whether the mutation in SPK1 caused the trichome branching defects of abt1-1, genomic region of the SPK1 locus was amplified and sequenced, and a single G to A transition mutation was identified at the +5,893 position (from the ATG start codon) in the SPK1 locus in abt1-1 mutant background (Figure 2). Theoretically, this mutation would convert a deduced glycine codon (GGA, Gly760) to a codon for glutamate (GAA, Glu760) (Figure 2).
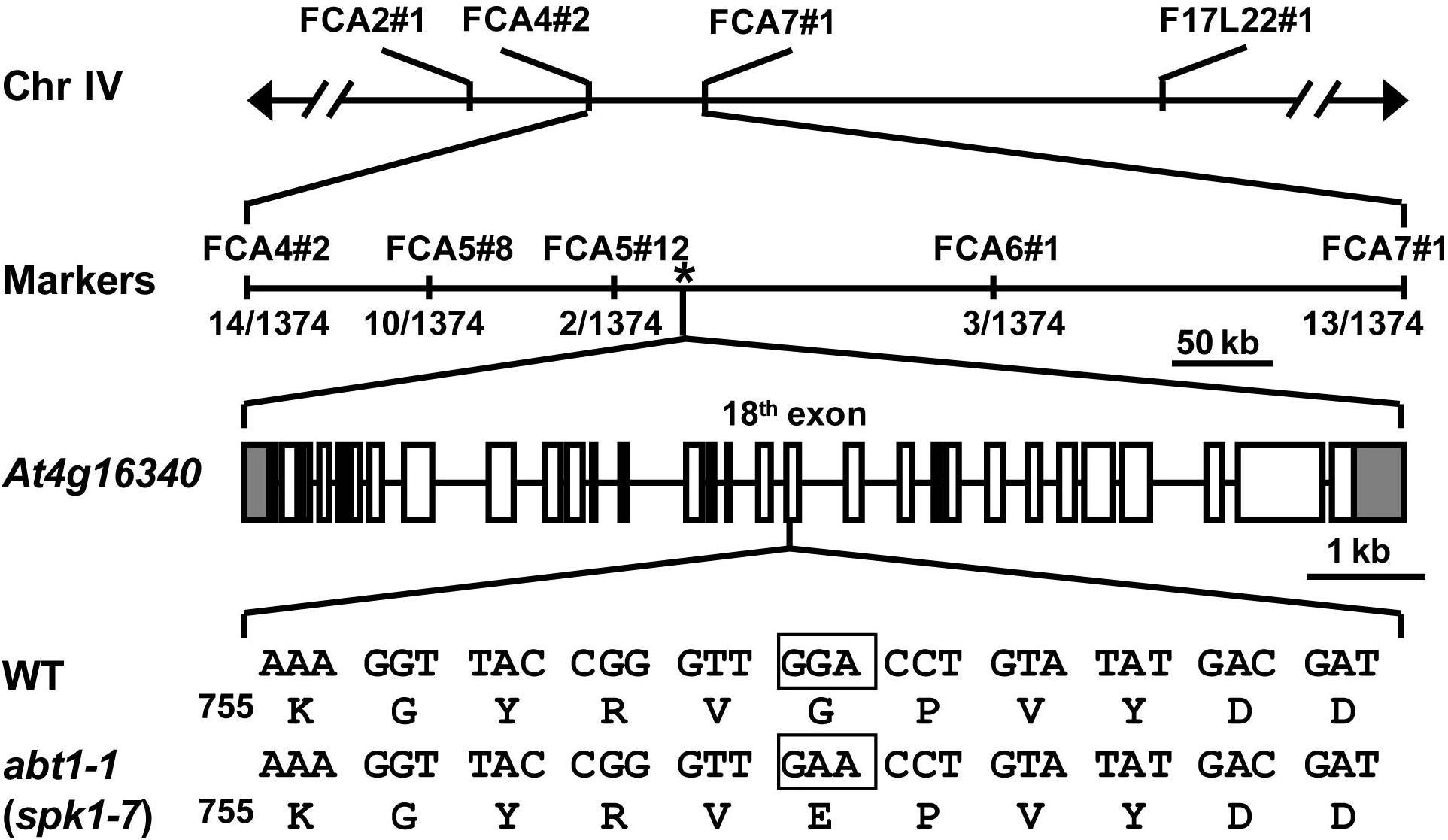
Figure 2. Map-based cloning of abt1-1 (spk1-7). Schematic representation of the map-based cloning of abt1-1 (spk1-7) as described in Methods. abt1-1 (spk1-7) mutation was linked to markers FCA2#1 and F17L22#1 on chromosome IV. Numbers of recombinants were shown under each marker. The asterisk indicated the position of SPK1 gene, At4g16340. In the gene model, boxes and solid lines represent the exons and introns, respectively. The 5′ and 3′ untranslated regions were shown as shaded boxes. The boxed nucleotides below the gene model indicated the exact position of the mutation site.
SPK1 genomic region spans13,133 bp, consisting of 30 exons and encodes a predicted protein of 1,830 amino acids. To verify that abt1-1 indeed represents a new mutant allele of SPK1, two SPK1 alleles, SALK_136776C and SAIL_520H04, were obtained from ABRC. Through PCR and genomic sequencing, we confirmed that the T-DNAs were inserted in the 27th intron and the in the ninth exon of the SPK1 gene in SALK_136776C and SAIL_520H04, respectively, (Figure 3A). SEM examination and quantification of trichome branching on the third and fifth rosette leaves of homozygous SALK_136776C and SAIL_520H04 lines clearly showed that they also display trichome hypobranching phenotypes similar to those of abt1-1 (Figures 3B–D). SALK_136776C showed a slightly stronger branching defect than abt1-1. The percentage of two-branched trichomes is higher in SALK_136776C compared to that of abt1-1 (Figures 3C,D). Occasionally, unbranched trichomes could be also observed in SALK_136776C (Figures 3C,D). SAIL_520H04 showed the strongest trichome branching defect, with an average of ∼30% unbranched trichomes (Figures 3C,D). These observations were consistent with the trichome branching defects in other spk1 alleles reported in previous studies (Qiu et al., 2002; Basu et al., 2008). Moreover, the extent of trichome branching reduction observed in abt1-1, SALK_136776C and SAIL_520H04 agrees in general with the natures of the three mutations (Figures 3E,F).
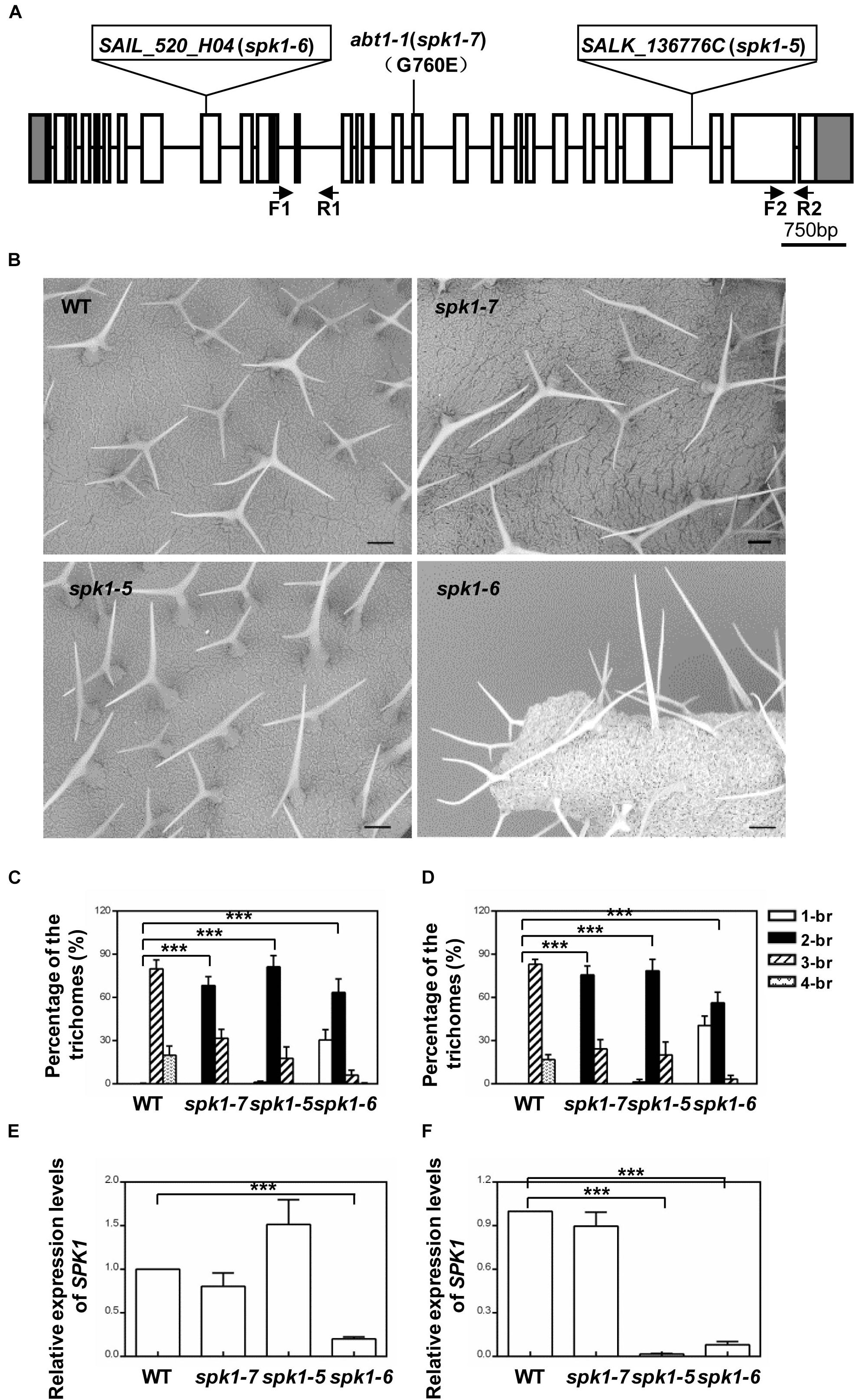
Figure 3. Analysis of the trichome branching phenotypes of various SPK1 alleles. (A) Schematic representation of the mutation sites in abt1-1 (spk1-7), spk1-5 (SALK_136776C), and spk1-6 (SAIL_520H04). (B) Trichome branching phenotypes on the fifth rosette leaves of three-week-old WT, spk1-7, spk1-5, and spk1-6 examined by SEM. Bars are 250 μM. (C,D) Quantification of the trichome branching phenotypes of WT, spk1-7, spk1-5, and spk1-6 on the third rosette leaf (C) and the fifth rosette leaf (D). (E,F) Real-time quantitative RT-PCR analysis of transcript accumulations of SPK1 in different SPK1 alleles with primers F1 coupled with R1 spanning the 11th–13th exon (E), and primers F2 coupled with R2 covering the 29th exon (F). Data were presented as mean ± SD of three biological replicates. ∗∗∗p < 0.001. 1/2/3/4-br represented one/two/three/four-branched trichomes, respectively.
Next, we carried out genetic crosses to further assess whether abt1-1 is allelic to SALK_136776C and SAIL_520H04. To this end, abt1-1 was crossed with SALK_136776C and SAIL_520H04, respectively, and trichome phenotypes of the F1 progeny were examined. While wild type plants scarcely show two-branched trichomes, two-branched trichomes were the most prevalent type found on rosette leaves of the F1 progeny from both crosses (Figures 4A–C). On the third rosette leaves, F1 plants from the cross between abt1-1 and SALK_136776C showed an average of 85.8% two-branched trichomes, while those from the cross between abt1-1 and SAIL_520H04 showed an average of 90.6% two-branched trichomes (Figure 4B). Moreover, an average of 13.9% and 9.3% three-branched trichomes were observed in the abt1-1 SALK_136776C F1 progeny and the abt1-1 SAIL_520H04 F1 progeny, respectively, in contrast to the 79.8% in wild type on the fifth rosette leaves (Figure 4C). These observations indicate that abt1-1 failed to complement SALK_136776C or SAIL_520H04. ABT1 and SPK1 likely represent the same genetic locus. In addition, these data showed that the reduction of trichome branching was more severe in the F1 plants from the cross between abt1-1 and SAIL_520H04, than from the cross between abt1-1 and SALK_136776C, agreeing with the severity of trichome phenotypes of individual alleles. Taken together, we conclude that ABT1 is SPK1, and abt1-1 is a new mutant allele of SPK1. Hence, we designated SALK_136776C as spk1-5, SAIL_520H04 as spk1-6, and abt1-1 as spk1-7 respectively, according to the literatures.
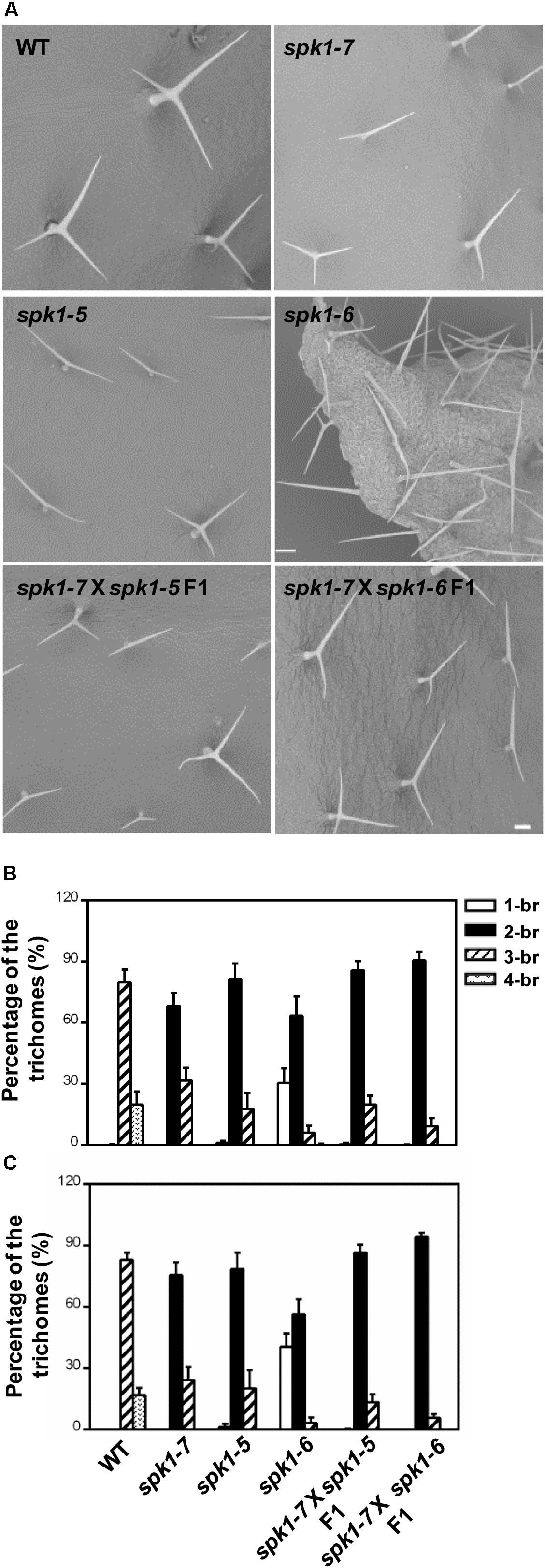
Figure 4. abt1-1 (spk1-7), spk1-5, and spk1-6 are allelic to one another. (A) Trichome branching phenotypes on the fifth rosette leaves of WT, spk1-7, spk1-5, spk1-6, spk1-7 × spk1-5 F1, and spk1-7 × spk1-6 F1 examined by SEM. Bars are 250 μM. (B,C) Quantitative analysis of the trichome branching phenotypes on the third (B) and fifth (C) rosette leaves of indicated genotypes. Data were presented as mean ± SD of three biological replicates. 1/2/3/4-br represented one/two/three/four-branched trichomes, respectively.
The Differential Regulation of Multiple Developmental Processes by SPK1
SPK1 has been shown to regulate several aspects of plant growth and development processes including the overall plant stature, trichome and epidermal pavement cell morphogenesis, as well as leaf, petal and root development (Qiu et al., 2002; Basu et al., 2008; Zhang et al., 2010, 2013; Lin et al., 2012; Ren et al., 2016). Quantification of the trichome branching phenotypes of three SPK1 alleles clearly showed that different mutations in SPK1 gene confer varied degree of trichome branching reduction (Figures 3B–D). To explore the potential differential regulation of these processes by SPK1, we examined the other two processes regulated by SPK1. First, we compared the overall plant stature and found that spk1-7 resembled the wild type at both seedling and mature stages (Figures 1A, 5A,B). spk1-5 showed a modest reduction of plant stature and the cotyledon development was also slightly affected (Figures 5A,B). Similar to previously reported strong alleles of SPK1 (Qiu et al., 2002; Basu et al., 2008), spk1-6 showed the most severe developmental defects, with small and narrow cotyledons, a gross reduction of plant size and was unable to reach reproductive stage when grown on the soil (Figures 5A,B).
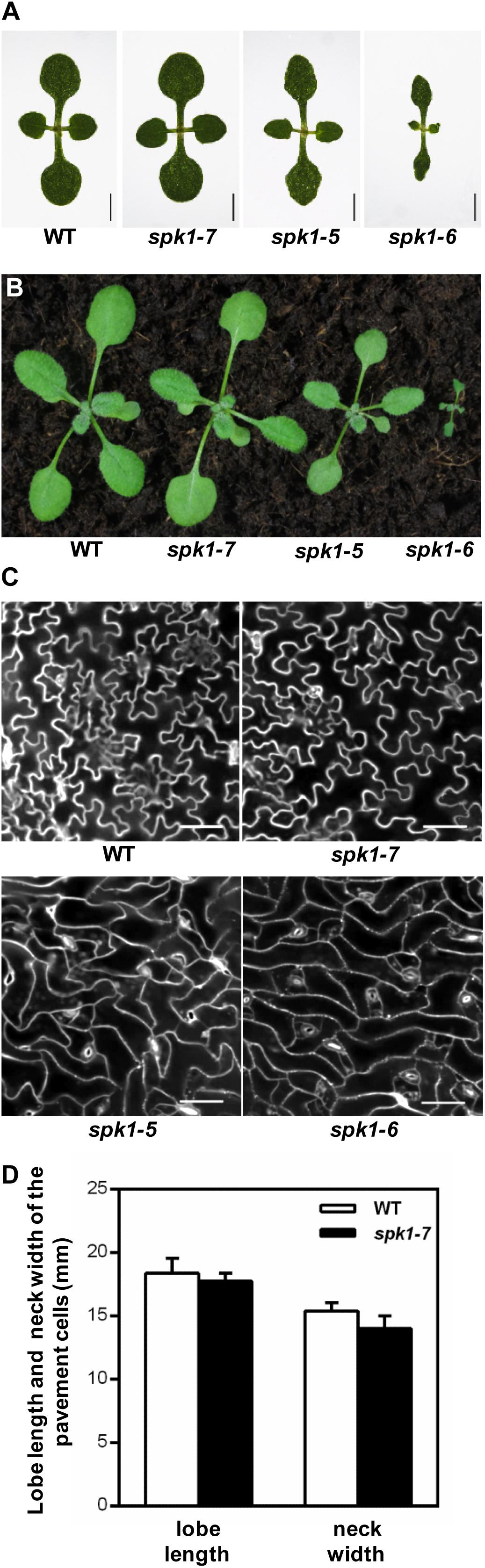
Figure 5. Three different SPK1 alleles showed varied developmental phenotypes. (A,B) Overall plant stature of WT, spk1-7, spk1-5, and spk1-6 at one-week-old (A) and two-week-old (B). (C) Cotyledon pavement cells morphology of WT, spk1-7, spk1-5, and spk1-6. The outlines of pavement cells were stained by PI and examined with confocal microcopy. Bars are 20 μM. (D) Quantification of the lobe length and the neck width of the cotyledon pavement cell of WT plant and spk1-7 mutant.
Next, we examined the cotyledon pavement cell morphology in three SPK1 mutant alleles. In wild type, cotyledon epidermal pavement cells developed pronounced lobes and indentations, showing a characteristic “jigsaw puzzle” arrangement (Figure 5C). In line with other previously reported spk1 alleles, the outgrowth of pavement cell lobes was greatly repressed and the interdigitation of pavement cells was almost lost in spk1-5 and spk1-6 (Figure 5C). Surprisingly, the “jigsaw puzzle” arrangement of pavement cell in spk1-7 resembled those of the wild type (Figure 5C), and further statistics on the length of lobes and the width of necks of the pavement cells suggested that spk1-7 mutation didn’t affect pavement cells morphogenesis (Figure 5D). These results indicate that different SPK1 alleles can lead to differential displays of mutant phenotypes, suggesting that these developmental processes show distinct responses to the activities of SPK1in different mutant backgrounds.
Mutations of SPK1 Affect Nuclei Positioning in Trichome Cells
The numbers of trichome branches are closely correlated with nuclear DNA content (Tominaga-Wada et al., 2011; Yang and Ye, 2013). Typically, elevated nuclear DNA contents are associated with more trichome branches, while reduced DNA ploidy results in fewer trichome branches. The reduced trichome branching phenotype of spk1-7 prompted us to test whether SPK1 is involved in the regulation of endoreduplication and further nuclear DNA ploidy. To this end, we firstly checked the transcript levels of cell cycle related genes in wild type and spk1-7 mutant alleles. These genes include CYCD3;1 for the G1 phase; CDC6a, CDC6b, CDT1a, CDT1b, HISH4, and CYCA3;1 for the S phase; CDKB1;1, CYCB1;1, KRP1, and KRP2 for the G2/M phase (Sonoda et al., 2009). No significant differences were observed for the expression levels of these genes in the spk1-7 mutant compared to those of the wild type (Supplementary Figure S1). Then, we examined the trichome DNA content based on the quantitative fluorescence measurements of DAPI stained nuclei, and found that the relative nuclear DNA content of spk1-7 trichomes were not remarkably changed compared with that in the wild type (Supplementary Figure S1). These results indicate that reduced trichome branching in the spk1-7 might not be accompanied with the changes in the trichome nuclear DNA content. SPK1 may promote trichome branching via a cell cycle independent pathway.
Coincidentally, during the investigation of plant nuclei DNA contents, we observed a high frequency of the abnormally positioned nuclei in two branched trichomes in spk1-5 and spk1-7 mutants (Figure 6A). In wild type plants, trichome nuclei were often situated just below the primary branch point in the trichome stalk (Folkers et al., 1997; Figures 6A,B). However, the nuclei of the spk1-5 and spk1-7 mutants trichome cells were frequently found in the trichome branches (Figures 6A,B). For the three branched trichomes in spk1-5 and spk1-7, the nuclei position of which was similar to that in the wild type plants (Supplementary Figure S2). In addition, we also examined the nuclei location of the epidermal pavement cells in spk1-5 and spk1-7 mutants respectively, and found that there were no remarkable differences among that in wild type and in mutants background. All of the nuclei of the pavement cells examined were resided closed to the cell edges (Supplementary Figure S3). The migration of nuclei during trichome development is a well-known phenomenon but the molecular mechanism underlining this process is poorly understood (Hülskamp et al., 1994; Folkers et al., 1997; Griffis et al., 2014). Our results suggest that SPK1 might contribute to the correct positioning of trichome nuclei.
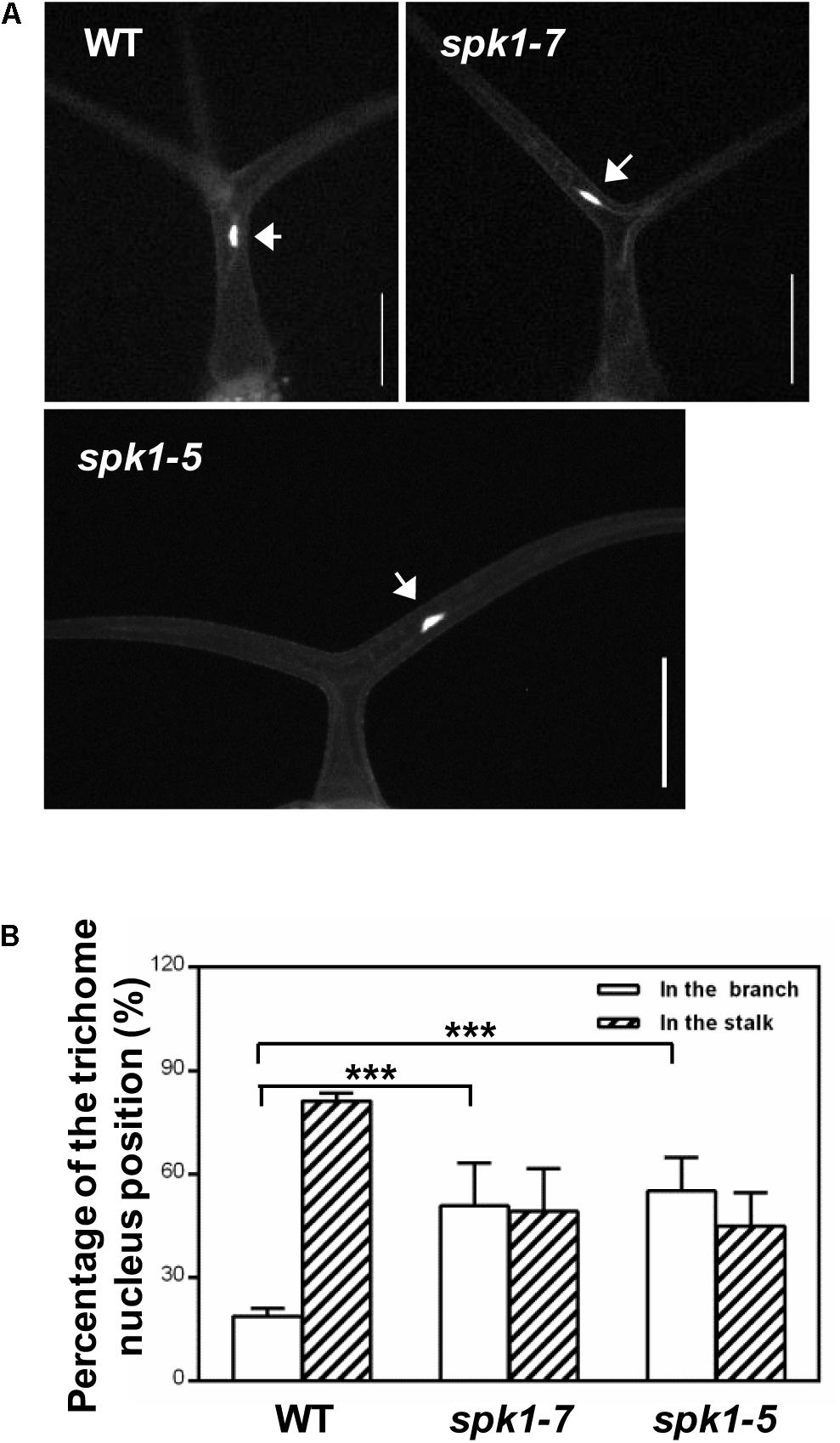
Figure 6. Analysis of trichome nuclei positions in WT and SPK1 mutants.(A) Representative leaf trichomes of WT, spk1-7 and spk1-5. White arrows point to the DAPI-stained trichome nuclei. Images were captured by fluorescence microscopy. Bars are 100 μM. (B) Quantification of trichome nuclei positions in WT, spk1-7 and spk1-5. Data were presented as mean ± SD of three biological replicates. ∗∗∗p < 0.001.
Genetic Interactions Between SPK1, AN and ZWI
It is well-known that the migration of the nuclei in the plants cells is tightly correlated with the cMTs status. The abnormal nuclei positions of trichome cells in the spk1-5 and spk1-7 mutants reminded us to speculate whether the regulation of trichome branching of SPK1 depend on the regulation of arrangement of the cMTs cytoskeleton. To investigate this possibility, we tested genetic interactions between SPK1 and AN and ZWI, which are characterized to define trichome branching through affecting cMTs dynamics (Reddy et al., 1996; Oppenheimer et al., 1997; Folkers et al., 2002; Kim et al., 2002; Tian et al., 2015). Moreover, loss of functions of either AN or ZWI displays two-branched trichome phenotypes. We obtained and confirmed T-DNA insertion mutants for an (SALK_026489) and zwi (SALK_017886) and generated spk1-7 an, spk1-7 zwi double mutants, and examined the trichome branching phenotypes of the double mutants, respectively. In contrast to the predominantly two-branched trichomes in spk1-7 and an single mutant, almost all the trichomes (97.3%) in spk1-7 an double mutants only had a single branch (Figures 7A,B). In spk1-7 zwi double mutants, although not as dramatic as that in the spk1-7 an double mutants, a significant increase in the percentage of single-branched trichomes (54.3%) was observed compared to either of the single mutants (Figures 7A,B). These data implicate that SPK1 might act coordinately with AN and ZWI to promote trichome branching. To validate this hypothesis, we also generated double mutants of spk1-5 an, spk1-5 zwi, spk1-6 an, and spk1-6 zwi, and detected the trichome phenotypes, respectively. Similar to the trichome phenotypes that we observed in spk1-7 an double mutants, about 74.5% of the trichomes on the fifth rosette leaves displayed one branched in spk1-5 an double mutants (Figures 7A,C). In spk1-5 zwi double mutants, although not as conspicuous as that in the spk1-5 an double mutants, the number of the one branched trichomes are also remarkably increased (Figures 7A,C). The statuses of trichome phenotypes of the spk1-6 an and spk1-6 zwi double mutants were the same as those in spk1-7 an and spk1-7 zwi double mutants, respectively, the trichome branching events were largely prevented (Figures 7A,D).
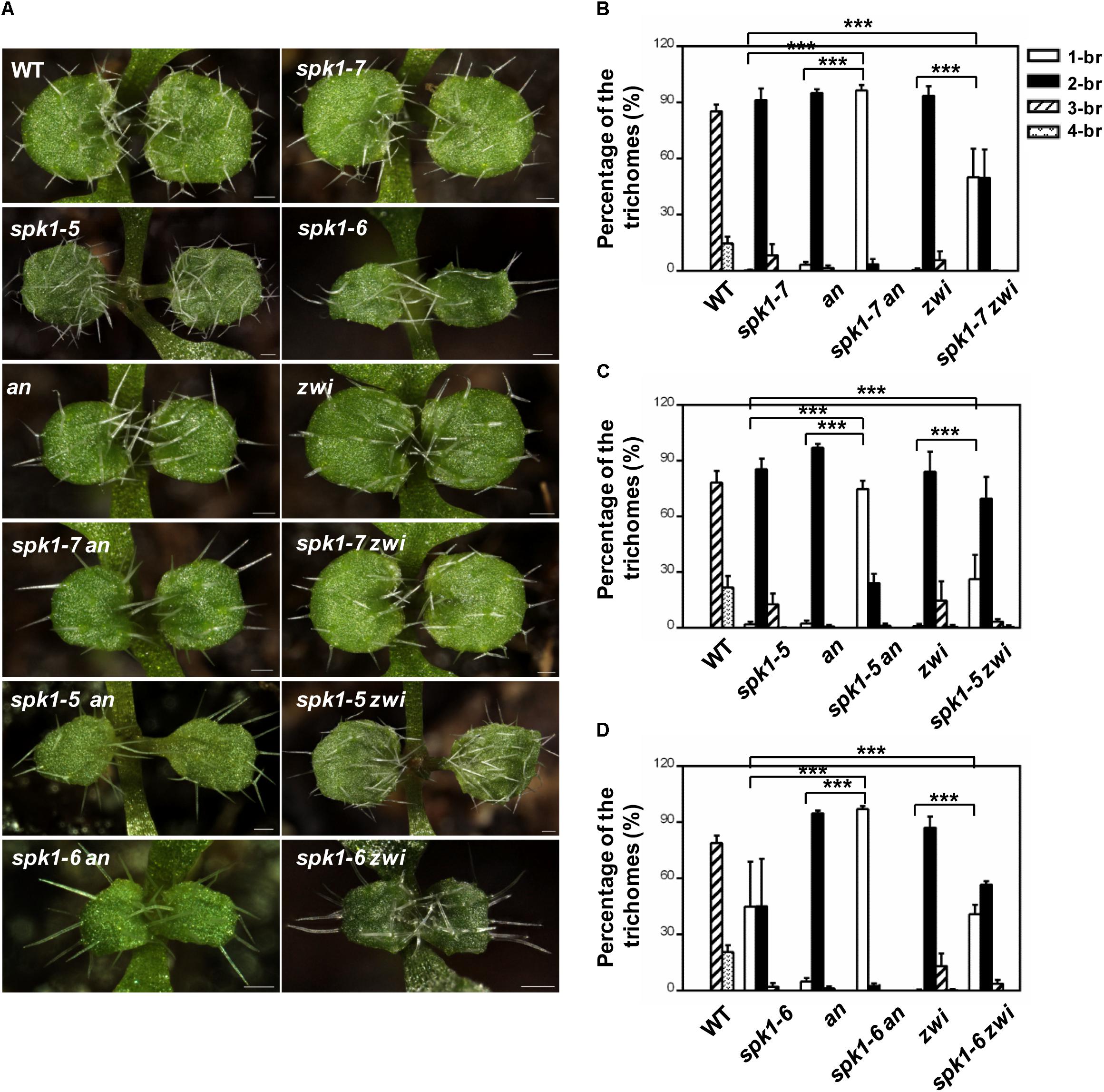
Figure 7. Analysis of the trichome branching phenotype of the double mutants. (A) Trichome branching phenotypes on the first pair of rosette leaves of WT, spk1-7, spk1-5, spk1-6, an, zwi, spk1-7 an, spk1-7 zwi, spk1-5an, spk1-5 zwi, spk1-6 an, and spk1-6 zwi double mutants. Bars are 100 μM. (B–D) Quantitative analysis of the trichome branching phenotypes on the third rosette leaves of indicated genotypes. Data were presented as mean ± SD of three biological replicates. ∗∗∗p < 0.001. 1/2/3/4-br represented one/ two/ three/ four -branched trichomes, respectively.
Considering the genetic interaction among AN, ZWI, and SPK1, we were interested in further understanding whether the regulation of epidermal pavement cell development by SPK1 is also associated with the cMTs arrays dynamics. We detected the cMTs arrangements in the pavement cells of cotyledons of spk1-5, spk1-6, and spk1-7 mutants, respectively. As shown in Figure 8A, the configuration of cMTs arrays in spk1-7 mutant was observed to similar to that in the wild type (CS6550), but in the spk1-5 and spk1-6 background, the cMTs arrays were assembled more orderly, which was largely different from that in the wild type, suggesting a certain degree of correlation between the pavement cell shape and the cMTs arrangements. The regulation of SPK1 on the pavement cell development might due to its influences on the cMTs dynamics. To validate this point, we measured the cMTs angles and quantitatively analyzed their distribution. In consistent with our observation, the quantitative data showed in wild type and spk1-7 mutant, the cMTs angles displayed a relative even distribution in different orientations, while in the spk1-5 and spk1-6 mutants, a significant peak of cMTs angles were observed between 80°-100° (Figure 8B). We also examined the anisotropy of the cMTs in wild type, spk1-7, spk1-5 and spk1-6 mutants, and found that the mean of the anisotropy value in spk1-5 and spk1-6 mutants were about 0.29 ± 0.10 and 0.35 ± 0.11, respectively, compared to that of about 0.18 ± 0.09 in wild type and 0.15 ± 0.04 in spk1-7 mutant (Figure 8C), implicating the isotropic arrangements of cMTs in spk1-5 and spk1-6 mutants comparing to that in wild type plant. We detected the organization of the cMT arrays in the pavement cells in the spk1-7 zwi double mutants background at the same time, and found that the status of the cMTs arrays resembled to that in zwi single mutants (Figures 8A–C). These data indicate that SPK1 might affect cMTs cytoskeleton dynamics through AN and ZWI during plant development.
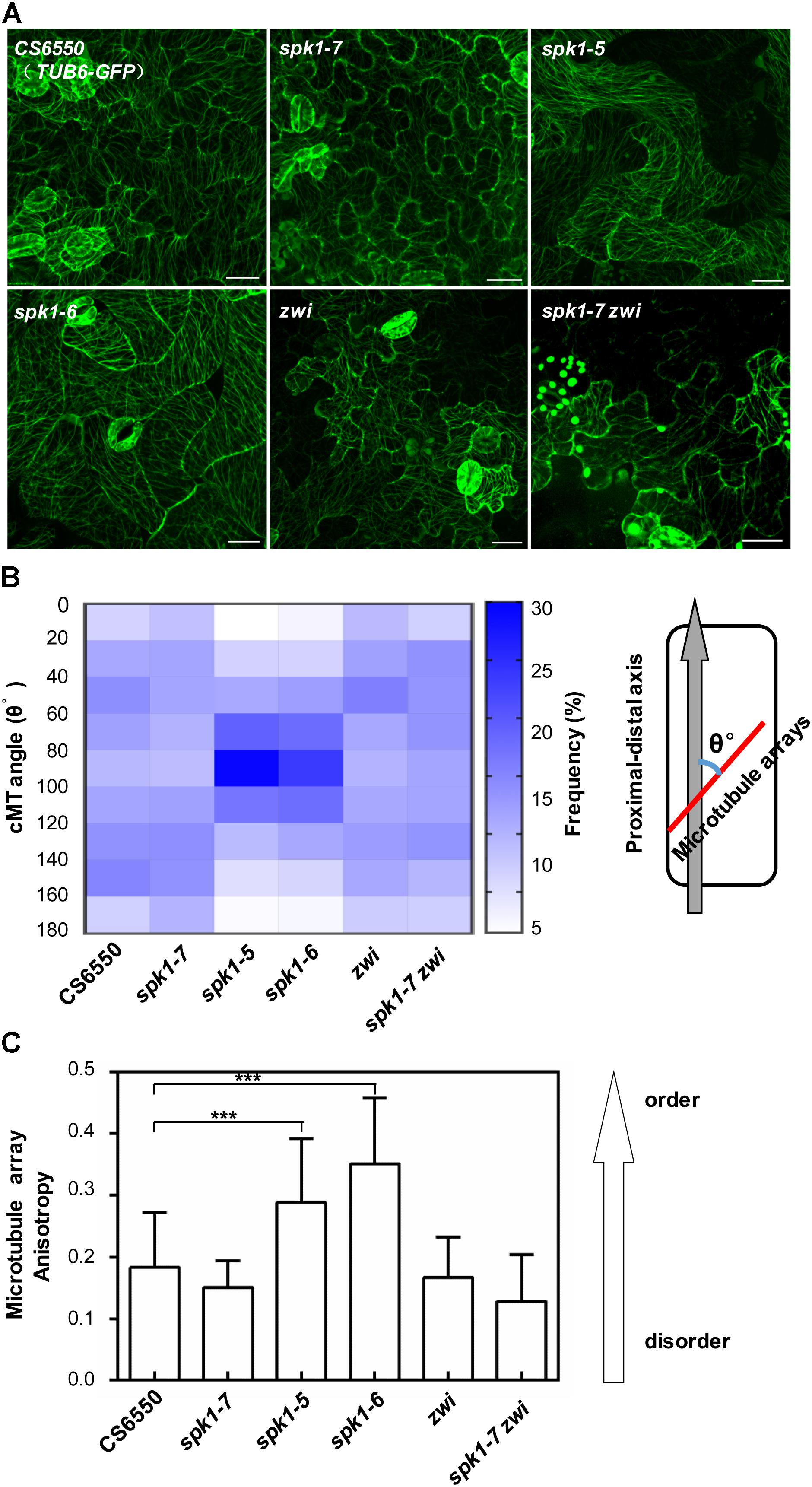
Figure 8. Arrangements of microtubule arrays in the cotyledon pavement cells of spk1, zwi, and spk1-7zwi double mutants. (A) Images of the microtubule arrays in the epidermal pavement cells of 4-day-old cotyledons of the indicated genotypes. Bars are 20 μM for all images. (B,C) Quantitative analysis of the microtubule arrays organization by measurement of cMTs angles (B) and evaluating the cMTs anisotropy (C). The value of microtubules angles of that parallel to the cell’s longitudinal axis were defined as 0°, while the value of those were perpendicular to the cell’s longitudinal axis were defined as 90°. Approximate 1000 microtubule angles were measured for each genotype, and then were divided into nine intervals (0–20°, 20–40°, 40–60°, 60–80°, 80–100°, 100–120°, 120–140°, 140–160°, 160–180°), respectively. The frequency of the microtubule angles in each interval was analyzed, and finally exhibited as a heatmap. For microtubule anisotropy evaluation, about 10 independent cells for each genotype were used. According to Boudaoud et al. (2014), the anisotropy score zero was defined for no order (purely isotropic arrays) and one for perfectly ordered (purely anisotropic arrays). Data were presented as mean ± SD of three biological replicates. ∗∗∗p < 0.001.
To further analysis the interactions among SPK1 and AN and ZWI during plant development, we also investigated the nuclei positions of trichome cells in the spk1-7 an and spk1-7 zwi double mutants, respectively. In an and zwi single mutant, a large portion of the trichome nuclei resided in the stalk, just below the primary branch point (Figures 9A,B), which was similar to that in the wild type, while in spk1-7zwi double mutants about 93.7% of the trichome nuclei were positioned at the branches (Figures 9A,B). When we looked at the trichome branching phenotypes in the spk1-7an double mutants and spk1-7 an zwi triple mutants, found almost all of the trichomes are single-branched (Figure 9A), so we measured the distance between the nuclei and the leaf plane. The data showed that the trichome nuclei were remarkably migrated far away from the leaf plane based on our quantitative analysis (Figure 9C). These data indicate that SPK1 might contribute to the regulation of the migration of the trichome nuclei, while AN and ZWI tend to serve for the cMTs arrays dynamic during trichome branching, and they work together to define the precise trichome branching events.
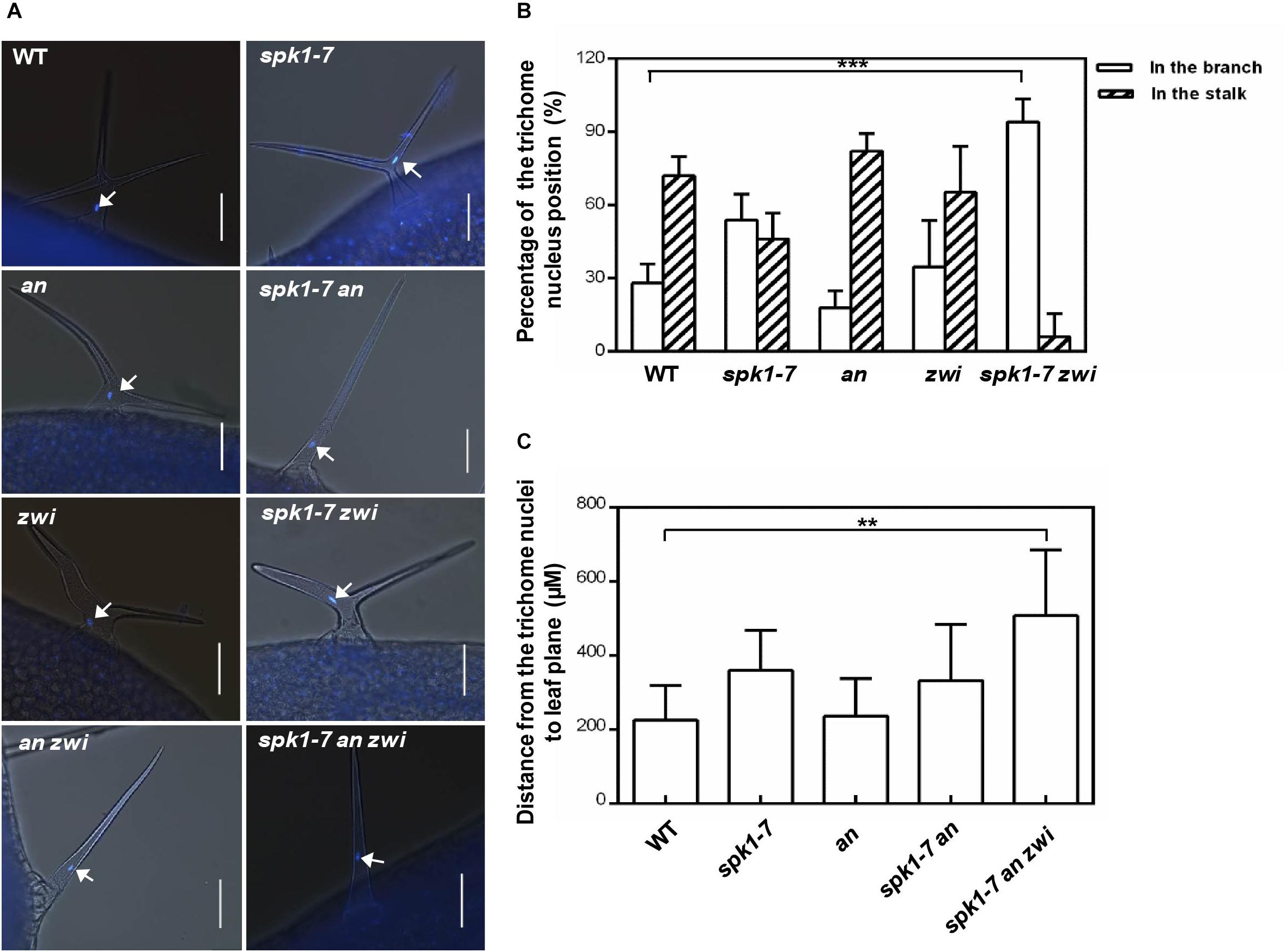
Figure 9. Nuclei positioning statues of the trichome cells in different mutants. (A) Representative trichomes images of the indicated genotype plants. White arrows point to the DAPI-stained trichome nuclei. Bars are 100 μM. (B) Quantification of trichome nuclei positions in WT, spk1-7, an, zwi, and spk1-7 zwi double mutants. (C) Statistic analysis of the distance between the trichome nulei and the leaf plane in WT, an, spk1-7an double mutants, and spk1-7an zwi triple mutants. Data were presented as mean ± SD. ∗∗∗p < 0.001, ∗∗p < 0.005.
Discussion
The Identification of a Novel Allele of SPK1
The differentiation and specification of defined cell types is a hallmark of the development of multi-cellular organisms. In higher plants, trichome cells offer an excellent paradigm for cell differentiation and morphogenesis because of their delicate growth patterns (Hülskamp et al., 1994). The long-term goal of our group is to unravel the molecular mechanisms of trichome cell differentiation, and we have carried out large-scale genetic screens in A. thaliana for mutants with abnormal trichome morphology. In this work, we report the isolation and identification of a novel trichome branching mutant abt1-1. Molecular cloning and genetic analysis revealed that ABT1 is identical to the previously reported locus SPK1 and thus our mutant allele represents a novel allele of SPK1 (Figures 2–4). Therefore, abt1-1 was renamed as spk1-7 according to the previously reported alleles.
In Arabidopsis, previous findings clearly indicate that SPK1 is indispensable for plant development and influences a spectrum of developmental programs including at least three well-defined processes: the overall plant stature, epidermal pavement cell and trichome cell morphogenesis (Qiu et al., 2002). However, further functional and genetic characterizations of SPK1 have been hampered by the seedling lethality of null spk1 mutants. In this work, interestingly, we isolated a fully viable hypomorphic allele of SPK1, spk1-7, which can serve as a valuable tool for future characterizations of SPK1 functions. In contrast to the previously reported seedling lethality of null spk1 mutants, the growth status including the overall plant stature, as well as pavement cell morphology of spk1-7 is not different from that of wild type plants except for the two branched trichomes (Figure 1). Molecular data showed a single G to A transition mutation at the +5893 position (from the ATG start codon) in the SPK1 locus in spk1-7 mutant background (Figure 2). According to the predicted SPK1 amino acid sequences, this mutation would convert a glycine codon (GGA, Gly760) to a codon for glutamate (GAA, Glu760). SPK1 is a large protein with several conserved domains: the pivotal C-terminal CDM domain, other two domains named DHR3/MOD1 and DHR1/MOD2, respectively (Qiu et al., 2002; Basu et al., 2008). The CDM domain has been characterized to confer the SPK1 functioning as a GEF. In this pathway, SPK1 promotes the activities of the conserved Rho-of-Plants (ROP) GTPases by facilitating the exchange of GDP to GTP (Côté and Vuori, 2007; Basu et al., 2008). In turn, activated ROP GTPases regulate the actin cytoskeleton through the SCAR/WAVE-ARP2/3 complexes (Côté and Vuori, 2007; Basu et al., 2008). However, the function of other two domains DHR3/MOD1 and DHR1/MOD2 are still unclear. The mutation site of spk1-7 locates in the middle of the protein, neither in the DHR3/MOD1 nor DHR1/MOD2. This sense mutation doesn’t affect the accumulation of the transcripts of SPK1 in spk1-7 mutant (Figure 3E), and cause only the trichome development impairment, suggesting that this region might specifically interact with the trichome development regulators. The conversion of the glycine codon to the glutamate codon might influence the physical interactions among SPK1 and other factors.
The Differential Regulation of Overall Plant Stature, Epidermal Pavement Cell and Trichome Cell Differentiation by SPK1
SPK1 has been reported to play essential roles in plant growth and development and null alleles of SPK1 display gross alterations of plant development with dwarfed stature, narrow and distorted cotyledons and rosette leaves, leaf trichome and pavement cell defects, eventually leading to seedling lethality (Qiu et al., 2002). In this work, taking advantage of the three SPK1 mutant alleles of varied severity (Figures 3, 5), we were able to probe the differential regulation of the above-mentioned three developmental processes by SPK1. Based on our results, trichome cell development appears to be the most sensitive to genetic disturbance of SPK1. In spk1-7 mutant, the overall plant development was not visibly affected, yet trichome cell branching was clearly impacted by the spk1-7 mutation (Figure 1). The further reduction of SPK1 activities appears to affect the trichome cell branching and pavement cell morphogenesis more (Figure 3E). As in spk1-5, the pavement cell development was grossly altered, similar to the strongest allele spk1-6 and other reported null mutants of SPK1, yet the overall growth and development was only slightly altered (Figures 5A,B). In strong allele such as spk1-6, all three developmental processes were impacted, similar to previous null mutants (Figures 3, 5; Qiu et al., 2002). spk1-5 and spk1-6 are all T-DNA insertional alleles of SPK1, and the T-DNAs were inserted in the 27th intron and the in the ninth exon of the SPK1 gene in spk1-5 and spk1-6, respectively. These two regions represent the DHR2 domain and the DHR3 domain, respectively, suggesting these two domains might involve in other development processes regulation besides trichome branching control. Moreover, the regulation of SPK1 on cotyledon pavement cell development and trichome branching might refer to cMTs dynamics, as in spk1-5 and spk1-6 mutants which exhibited conspicuous cotyledon pavement cell development impact, the pavement cell cMTs were arranged more orderly, about 25%-30% cMTs oriented to be perpendicular to the cell’s longitudinal axis (Figures 8A–C). In spk1-7 mutant, the pavement cell morphology was undistinguished from that of wild type plants, in consistent, the cMTs organization was similar to that in wild type plants, and the orientation of cMTs was randomly (Figures 8A–C). However, in two-branched trichomes, we found that the distribution of the cMTs were relative isotropic compared that in wild type (Supplementary Figure S4). Taken together, our results raise several interesting scenarios regarding the mode of operation by SPK1. It is possible that different developmental processes may have differential responses to the overall level SPK1 activities. Alternatively, different domains of SPK1may confer differential functions in different developmental processes.
The Genetic Interactions Between SPK1 and AN and ZWI
The discovery of the regulation of the nuclei positioning of trichome cell by SPK1 promoted us to probe how SPK1 interacts genetically with AN and ZWI, two other known regulators of trichome branching determination. Loss of function of AN results in plants with narrow leaves, two branched trichomes and pavement cell shape defects (Tsuge et al., 1996). AN encodes a plant homolog of the C-terminal binding proteins/brefeldin A ADP-ribosylated substrates (CtBP/BARS) protein (Folkers et al., 2002; Kim et al., 2002). In an mutant, the arrangement of cMTs cytoskeleton is disturbed and the abnormal cMTs is thought to cause the mutant phenotypes (Folkers et al., 2002; Kim et al., 2002). Although originally thought to be localized to the nucleus and function as a transcriptional co-repressor, more recent data suggest that AN may partially localize to the trans-Golgi network (TGN) and may exert its functions through the endomembrane system (Minamisawa et al., 2011). In spk1-7 an double mutants, the majority of trichomes are single branched (Figures 7A,B), suggesting that SPK1 and AN interact genetically in regulating trichome branch formation. Considering that SPK1 is associated with ER, it t is likely that endomembrane system including ER and TGN may be involved in the cMTs organization and trichome morphogenesis. Our findings with the double mutant of spk1-7 and zwi further support a functional link between SPK1 and the organization of cMTs. In contrast to an, zwi mutant also shows two branched trichomes with short stalks, but the overall plant growth and development is not significantly affected (Hülskamp et al., 1994). ZWI encodes a plant microtubule associated protein (MAP) that is known as kinesin-like calmodulin-binding protein (KCBP) (Reddy et al., 1996; Oppenheimer et al., 1997; Abdel-Ghany et al., 2005), and could bind to the microtubules (Narasimhulu and Reddy, 1998). spk1-7 zwi double mutants showed increased percentage of single-branched trichomes that are similar to those of spk1-7 an double mutants (Figures 7A,B). Interestingly ZWI/KCBP has been reported to physically interact with AN (Folkers et al., 2002). The similar phenotypes we observed in spk1-7 an and spk1-7 zwi double mutants are consistent with this notion. Moreover, we found that unlike that in an mutant, the gross pavement cell morphology and cMTs organization in zwi as well as in spk1-7 zwi mutant were similar to that in wild type (Figures 8A–C). These data implicated that SPK1 might interact with ZWI to specifically regulate trichome branching.
Another interesting phenomenon we observed is the aberrant placements of the trichome nuclei in spk1 mutants (Figures 6A,B). Nucleus is the core of the eukaryotic cell, and its position is not fixed in the cell. It needs to move to a proper position depending on the cell type, developmental process, and physiological situations (Griffis et al., 2014; Tan et al., 2016; Nakamura et al., 2018). In the mature trichome cells, the nuclei usually are positioned in the stalk, just below the primary branch points (Folkers et al., 1997), and this positioning pattern is considered to depend on the dynamic cMTs, either cMT destabilizing or stabilizing could alter the nuclei positions of trichome cells (Folkers et al., 1997; Mathur and Chua, 2000). However, we found that loss of function of AN seems to have no remarkable effects on nuclei positioning of the trichome cells. In an mutant, most of the nuclei of the trichome cells reside in the stalk (Figures 9A,B), and the distance from the trichome cell nuclei to the leaf plane is about 237.9 μM, which is not different from that in the wild plants (Figure 9C). In the spk1-7 an double mutant, although the positions of the nuclei are elevated, it is not remarkable different from that in spk1-7 plants (Figure 9C). When we investigated the nuclei positions of trichome cells in the spk1-7 an zwi triple mutant, we found that the positions of nuclei are about 508 μM far away from the leaf plane (Figure 9C), which is significantly elevated compared that in spk1-7 plants. These data implicate a coordinative regulation pattern of SPK1 and AN on trichome branching. SPK1 might interact with ZWI to contribute to the regulation of the migration of the trichome nuclei, while AN tends to serve for the cMTs arrays dynamic during trichome branching.
Taken together, based on our data, SPK1, AN and ZWI would all act as positive regulators of trichome branch formation, and their roles in promoting the formation of trichome branches likely involve the microtubule cytoskeleton. It would be interesting in the future to uncover the underlying mechanism of SPK1 on regulating nuclei migration during trichome branching and the functional relationship between SPK1 and the microtubule cytoskeleton.
Author Contributions
LA, FY, and SL designed the experiments. SL, XY, JZ, and JS carried out the experiments. SL and LA wrote the manuscript. XL, YQ, and FY revised the article. MD conducted the additive data in the revised manuscript. All authors agreed to be accountable for the content of the work and gave the final approval for the submission of the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (NSFC) (Grant Nos. 31470290 to LA and 31570267 to FY).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank X. M. Yang and X. Qiao for assistance in scanning electron microscopy and confocal microscopy, respectively.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00016/full#supplementary-material
References
Abdel-Ghany, S. E., Day, I. S., Simmons, M. P., Kugrens, P., and Reddy, A. S. N. (2005). Origin and evolution of kinesin-like calmodulin-binding protein. Plant Physiol. 138, 1711–1722. doi: 10.1104/pp.105.060913
Abe, T., and Hashimoto, T. (2005). Altered microtubule dynamics by expression of modified α-tubulin protein causes right-handed helical growth in transgenic Arabidopsis plants. Plant J. 43, 191–204. doi: 10.1111/j.1365-313X.2005.02442.x
Abe, T., Thitamadee, S., and Hashimoto, T. (2004). Microtubule defects and cell morphogenesis in the lefty1lefty2 tubulin mutant of Arabidopsis thaliana. Plant Cell Physiol. 45, 211–220. doi: 10.1093/pcp/pch026
An, L., Zhou, Z., Su, S., Yan, A., and Gan, Y. (2011). Glabrous Inflorescence Stems (GIS) is required for trichome branching through gibberellic acid signaling in Arabidopsis. Plant Cell Physiol. 53, 457–469. doi: 10.1093/pcp/pcr192
Basu, D., Le, J., Zakharova, T., Mallery, E. L., and Szymanski, D. B. (2008). A SPIKE1 signaling complex controls actin-dependent cell morphogenesis through the heteromeric WAVE and ARP2/3 complexes. Proc. Natl. Acad. Sci. U.S.A. 105, 4044–4049. doi: 10.1073/pnas.0710294105
Bompard, G., and Caron, E. (2004). Regulation of WASP/WAVE proteins: making a long story short. J. Cell Biol. 166, 957–962. doi: 10.1083/jcb.200403127
Boudaoud, A., Burian, A., Borowska-Wykrêt, D., Uyttewaal, M., Wrzalik, R., Kwiatkowska, D., et al. (2014). FibrilTool, an ImageJ plug-in to quantify fibrillar structures in raw microscopy images. Nat. Protoc. 9, 457–463. doi: 10.1038/nprot.2014.024
Burk, D. H., Liu, B., Zhong, R., Morrison, W. H., and Ye, Z. H. (2001). A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13, 807–827. doi: 10.1105/tpc.13.4.807
Côté, J. F., and Vuori, K. (2007). GEF what? Dock180 and related proteins help Rac to polarize cells in new ways. Trends Cell Biol. 17, 383–393. doi: 10.1016/j.tcb.2007.05.001
Folkers, U., Berger, J., and Hülskamp, M. (1997). Cell morphogenesis of trichomes in Arabidopsis: differential control of primary and secondary branching by branch initiation regulators and cell growth. Development 124, 3779–3786.
Folkers, U., Schöbinger, U., Falk, S., Krishnakumar, S., Pollock, M. A., Oppenheimer, D. G., et al. (2002). The cell morphogenesis gene ANGUSTIFOLIA encodes a CtBP/BARS-like protein and is involved in the control of the microtubule cytoskeleton. EMBO J. 21, 1280–1288. doi: 10.1093/emboj/21.6.1280
Grebe, M. (2012). The patterning of epidermal hairs in Arabidopsis-updated. Curr. Opin. Plant Biol. 15, 31–37. doi: 10.1016/j.pbi.2011.10.010
Griffis, A. H. N., Groves, N. R., Zhou, X., and Meier, I. (2014). Nuclei in motion: movement and positioning of plant nuclei in development, signaling, symbiosis, and disease. Front. Plant Sci. 5:129. doi: 10.3389/fpls.2014.00129
Guimil, S., and Dunand, C. (2007). Cell growth and differentiation in Arabidopsis epidermal cells. J. Exp. Bot. 58, 3829–3840. doi: 10.1093/jxb/erm253
Hülskamp, M. (2004). Plant trichomes: a model for cell differentiation. Nat. Rev. Mol. Cell Biol. 5, 471–480. doi: 10.1038/nrm1404
Hülskamp, M., Miséra, S., and Jürgens, G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76, 555–566. doi: 10.1016/0092-8674(94)90118-X
Ilgenfritz, H., Bouyer, D., Schnittger, A., Mathur, J., Kirik, V., Schwab, B., et al. (2003). The Arabidopsis STICHEL gene is a regulator of trichome branch number and encodes a novel protein. Plant Physiol. 131, 643–655. doi: 10.1104/pp.014209
Ishida, T., Kurata, T., Okada, K., and Wada, T. (2008). A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 59, 365–386. doi: 10.1146/annurev.arplant.59.032607.092949
Ivakov, A., and Persson, S. (2013). Plant cell shape: modulators and measurements. Front. Plant Sci. 4:439. doi: 10.3389/fpls.2013.00439
Kasili, R., Walker, J. D., Simmons, L. A., Zhou, J., De Veylder, L., and Larkin, J. C. (2010). SIAMESE cooperates with the CDH1-like protein CCS52A1 to establish endoreplication in Arabidopsis thaliana trichomes. Genetics 185, 257–268. doi: 10.1534/genetics.109.113274
Kim, G. T., Shoda, K., Tsuge, T., Cho, K. H., Uchimiya, H., Yokoyama, R., et al. (2002). The ANGUSTIFOLIA gene of Arabidopsis, a plant CtBP gene, regulates leaf-cell expansion, the arrangement of cortical microtubules in leaf cells expression of a gene involved in cell-wall formation. EMBO J. 21, 1267–1279. doi: 10.1093/emboj/21.6.1267
Kim, Y., Schumaker, K. S., and Zhu, J. K. (2006). EMS mutagenesis of Arabidopsis. Methods Mol. Biol. 323, 101–103.
Kirik, V., Grini, P. E., Mathur, J., Klinkhammer, I., Adler, K., Bechtold, N., et al. (2002a). The Arabidopsis TUBULIN-FOLDING COFACTOR A gene is involved in the control of the α/β-tubulin monomer balance. Plant Cell 14, 2265–2276.
Kirik, V., Mathur, J., Grini, P. E., Klinkhammer, I., Adler, K., Bechtold, N., et al. (2002b). Functional analysis of the tubulin-folding cofactor C in Arabidopsis thaliana. Curr. Biol. 12, 1519–1523.
Kiyokawa, E., Hashimoto, Y., Kobayashi, S., Sugimura, H., Kurata, T., and Matsuda, M. (1998). Activation of Rac1 by a CrkSH3-binding protein, DOCK180. Genes Dev. 12, 3331–3336. doi: 10.1101/gad.12.21.3331
Lee, M. M., and Schiefelbein, J. (2002). Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell 14, 611–618. doi: 10.1105/tpc.010434
Lin, D., Nagawa, S., Chen, J., Cao, L., Chen, X., Xu, D., et al. (2012). A ROP GTPase-dependent auxinsignaling pathway regulates the subcellular distribution of PIN2 in Arabidopsis roots. Curr. Biol. 22, 1319–1325. doi: 10.1016/j.cub.2012.05.019
Liu, X. Y., Rodermel, S. R., and Yu, F. (2010). A var2 leaf variegation suppressor locus, SUPPRESSOR OF VARIEGATION3, encodes a putative chloroplast translation elongation factor that is important for chloroplast development in the cold. BMC Plant Biol. 10:287. doi: 10.1186/1471-2229-10-287
Lukowitz, W., Gillmor, C. S., and Scheible, W. R. (2000). Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 123, 795–805. doi: 10.1104/pp.123.3.795
Mathur, J., and Chua, N. H. (2000). Microtubule stabilization leads to growth reorientation in Arabidopsis trichomes. Plant Cell 12, 465–477. doi: 10.1105/tpc.12.4.465
Minamisawa, N., Sato, M., Cho, K. H., Ueno, H., Takechi, K., and Kajikawa, M. (2011). ANGUSTIFOLIA, a plant homolog of CtBP/BARS, functions outside the nucleus. Plant J. 68, 788–799. doi: 10.1111/j.1365-313X.2011.04731.x
Nakamura, M., Claes, A. R., Grebe, T., Hermkes, R., Viotti, C., Ikeda, Y., et al. (2018). Auxin and ROP GTPase signaling of polar nuclear migration in root epidermal hair cells. Plant Physiol. 176, 1–11. doi: 10.1104/pp.17.00713
Narasimhulu, S. B., and Reddy, A. S. N. (1998). Characterization of microtubule binding domains in the Arabidopsis kinesin-like calmodulin binding protein. Plant Cell 10, 957–965. doi: 10.1105/tpc.10.6.957
Oppenheimer, D. G., Pollock, M. A., Vacik, J., Szymanski, D. B., Ericson, B., Feldmann, K., et al. (1997). Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 94, 6261–6266. doi: 10.1073/pnas.94.12.6261
Pattanaik, S., Patra, B., Singh, S. K., and Yuan, L. (2014). An overview of the gene regulatory network controlling trichome development in the model plant Arabidopsis. Front. Plant Sci. 5:259. doi: 10.3389/fpls.2014.00259
Perazza, D., Herzog, M., Hülskamp, M., Brown, S., Dorne, A. M., and Bonneville, J. M. (1999). Trichome cell growth in Arabidopsis thaliana can be derepressed by mutations in at least five genes. Genetics 152, 461–476.
Qiu, J. L., Jilk, R., Marks, M. D., and Szymanski, D. B. (2002). The Arabidopsis SPIKE1 gene is required for normal cell shape control and tissue development. Plant Cell 14, 101–118. doi: 10.1105/tpc.010346
Reddy, A. S. N., Narasimhulu, S. B., Safadi, F., and Golovkin, M. (1996). A plant kinesin heavy chain-like protein is a calmodulin-binding protein. Plant J. 10, 9–21. doi: 10.1046/j.1365-313X.1996.10010009.x
Ren, H., Dang, X., Yang, Y., Huang, D., Liu, M., Gao, X., et al. (2016). SPIKE1 activates ROP GTPase to modulate petal growth and shape. Plant Physiol. 172, 358–371. doi: 10.1104/pp.16.00788
Schnittger, A., and Hülskamp, M. (2002). Trichome morphogenesis: a cell-cycle perspective. Philos. Trans. R. Soc. Lond. 357, 823–826. doi: 10.1098/rstb.2002.1087
Sonoda, Y., Sako, K., Maki, Y., Yamazaki, N., Yamamoto, H., Ikeda, A., et al. (2009). Regulation of leaf organ size by the Arabidopsis RPT2a 19S proteasome subunit. Plant J. 60, 68–78. doi: 10.1111/j.1365-313X.2009.03932.x
Szymanski, D. B., and Marks, M. D. (1998). GLABROUS1 overexpression and TRIPTYCHON alter the cell cycle and trichome cell fate in Arabidopsis. Plant Cell 10, 2047–2062. doi: 10.1105/tpc.10.12.2047
Tan, K., Wen, C., Feng, H., Chao, X., and Su, H. (2016). Nuclear dynamics and programmed cell death in Arabidopsis root hairs. Plant Sci. 253, 77–85. doi: 10.1016/j.plantsci.2016.08.006
Thitamadee, S., Tuchihara, K., and Hashimoto, T. (2002). Microtubule basis for left-handed helical growth in Arabidopsis. Nature 417, 193–196. doi: 10.1038/417193a
Tian, J., Han, L., Feng, Z., Wang, G., Liu, W., Ma, Y., et al. (2015). Orchestration of microtubules and the actin cytoskeleton in trichome cell shape determination by plant-unique kinesin. eLife 4:e09351. doi: 10.7554/eLife.09351
Tominaga-Wada, R., Ishida, T., and Wada, T. (2011). New insights into the mechanism of development of Arabidopsis root hairs and trichomes. Int. Rev. Cell Mol. Biol. 286, 67–106. doi: 10.1016/B978-0-12-385859-7.00002-1
Tsuge, T., Tsukaya, H., and Uchimiya, H. (1996). Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development 122, 1589–1600.
Wang, R., Liu, X., Liang, S., Ge, Q., Li, Y., Shao, J., et al. (2015). A subgroup of MATE transporter genes regulates hypocotyl cell elongation in Arabidopsis. J. Exp. Bot. 66, 6327–6343. doi: 10.1093/jxb/erv344
Yang, C., and Ye, Z. (2013). Trichomes as models for studying plant cell differentiation. Cell. Mol. Life Sci. 70, 1937–1948. doi: 10.1007/s00018-012-1147-6
Yao, M., Wakamatsu, Y., Itoh, T. J., Shoji, T., and Hashimoto, T. (2008). Arabidopsis SPIRAL2 promotes uninterrupted microtubule growth by suppressing the pause state of microtubule dynamics. J. Cell Sci. 121, 2372–2381. doi: 10.1242/jcs.030221
Zhang, C., Kotchoni, S. O., Samuels, A. L., and Szymanski, D. B. (2010). SPIKE1 signals originate from and assemble specialized domains of the endoplasmic reticulum. Curr. Biol. 20, 2144–2149. doi: 10.1016/j.cub.2010.11.016
Keywords: branching, cell morphogenesis, cytoskeleton, nucleus positioning, SPIKE1, trichome
Citation: Liang S, Yang X, Deng M, Zhao J, Shao J, Qi Y, Liu X, Yu F and An L (2019) A New Allele of the SPIKE1 Locus Reveals Distinct Regulation of Trichome and Pavement Cell Development and Plant Growth. Front. Plant Sci. 10:16. doi: 10.3389/fpls.2019.00016
Received: 22 August 2018; Accepted: 08 January 2019;
Published: 24 January 2019.
Edited by:
Taku Takahashi, Okayama University, JapanReviewed by:
John Larkin, Louisiana State University, United StatesMitsutomo Abe, The University of Tokyo, Japan
Copyright © 2019 Liang, Yang, Deng, Zhao, Shao, Qi, Liu, Yu and An. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fei Yu, Zmx5ZmVpeXVAZ21haWwuY29t Lijun An, bGlqdW5hbkBud3N1YWYuZWR1LmNu
 Shuang Liang
Shuang Liang Xuying Yang
Xuying Yang Jun Zhao
Jun Zhao Jingxia Shao
Jingxia Shao Yafei Qi
Yafei Qi Xiayan Liu
Xiayan Liu Fei Yu
Fei Yu Lijun An
Lijun An