- 1Yunnan Key Laboratory for Wild Plant Resources, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 2University of Chinese Academy of Sciences, Beijing, China
As an endoplasmic reticulum (ER) stress sensor, inositol-requiring enzyme 1 (IRE1) splices the bZIP60 mRNA, and produces an active bZIP60 transcription factor that regulates genes involved in the unfolded protein response (UPR) during ER stresses. This IRE1-bZIP60 pathway is conserved in plant species and recently implicated in plant-pathogen interaction. However, it is unclear whether this IRE1-bZIP60 pathway is involved in Nicotiana attenuata resistance to necrotic fungal pathogen, Alternaria alternata. In this study, transcriptional levels of chaperone protein genes, including luminal binding protein (BiP), protein disulfide isomerase (PDI), calnexin 1-like (CNX 1-like), and calreticulin (CRT), and genes involved in IRE1-bZIP60 pathway, were all significantly induced in N. attenuata leaves after A. alternata inoculation. Silencing IRE1 or bZIP60 led to N. attenuata plants more susceptible to A. alternata, which were associated with reduced gene expressions of Feruloyl-CoA 6′-hydroxylase 1 (F6′H1), a gene encoding a key enzyme for phytoalexin scopoletin and scopolin biosynthesis. Further, electromobility shift assays (EMSA) indicated that bZIP60 protein of spliced form could directly bind to the promoter region of F6′H1 in vitro. JA signaling pathway is required for N. attenuata resistance to A. alternata. Interestingly, the fungus-elicited transcriptional levels of BiP, PDI, CNX 1-like, CRT, IRE1, and bZIP60(s) were all significantly decreased in JA-deficient or JA-insensitive plants. Meanwhile, those genes were significantly induced by methyl jasmonate (MeJA) when applied exogenously. However, the transcriptional levels of JA-regulated genes allene oxide synthase (AOS) and lipoxygenease 3 (LOX3) were not affected in plants impaired with IRE1-bZIP60 pathway. Thus, it is concluded that IRE1-bZIP60 pathway is required for N. attenuata resistance to A. alternata, and JA signaling pathway plays an important role in the elicitation of chaperone protein genes and IRE1-bZIP60 pathway.
Introduction
The endoplasmic reticulum (ER) is a major protein-folding organelle in cells (Ellgaard and Helenius, 2003; Trombetta and Parodi, 2003). During stresses, the protein-folding process is disturbed, thus a considerable amount of misfolded proteins are accumulated in the ER, leading to a situation known as ER stress (Gething and Sambrook, 1992; Vitale and Boston, 2008). The signaling network called unfolded protein response (UPR) plays an essential role in rescuing misfolded proteins to maintaining ER homeostasis, by enhancement of protein-folding and degradation of misfolded proteins, or by delaying new proteins synthesis. If UPR fails to mitigate ER stress, it will lead to programmed cell death and autophagy (Liu et al., 2012; Ruberti et al., 2015).
To facilitate folding and ensure quality of the produced protein, many factors including chaperones, folding factors, and enzymes, such as luminal binding protein (BiP), calnexin (CNX), calreticulin (CRT), and protein disulfide isomerase (PDI), are employed during ER stress (Siffroi-Fernandez et al., 2002; Freitas et al., 2016). The gene expressions of these factors, in many cases, also serve as molecular markers of the elicitation of UPR (Denecke et al., 1995; Klein et al., 2006).
There are two main branches of plant UPR to rehabilitate the ER capabilities, IRE1-bZIP60 and bZIP28-S1P/S2P pathways (Moreno and Orellana, 2011). IRE1-bZIP60 pathway is mediated by inositol-requiring enzyme 1 (IRE1) through a conserved mechanism. As a membrane kinase and ribonuclease, IRE1 serves as a stress sensor in eukaryotes by catalyzing the unconventional splicing of the mRNA of basic leucine zipper (bZIP) transcription factors when ER stress occurs. Plant bZIP60 has two stem loop structures, IRE1 recognizes these structures and cleaves two phosphodiester bonds, removes the coding region for a transmembrane domain (TMD), releasing the active form of bZIP60 (Deng et al., 2011). The transcription factor subsequently enters into the nucleus regulating the target genes for the restoration of ER proteostasis (Moreno and Orellana, 2011; Wan and Jiang, 2016). Another branch of plant UPR involves bZIP28. It is an ER membrane-associated transcription factor, migrates to Golgi during ER stress. bZIP28 is then cleaved off the transmembrane anchor by two Golgi-localized proteases S1P and S2P, and transported to the nucleus to regulate UPR-related genes (Wan and Jiang, 2016).
Heat, salt, drought, extreme osmotic and heavy metals stresses can activate the UPR in plant cell (Gao et al., 2008; Deng et al., 2011; Liu et al., 2011; Wang et al., 2011; Zhang et al., 2017; Li et al., 2018). There are also some reports linking pathogen attack and the activation of UPR (Tateda et al., 2008; Moreno et al., 2012; Ye et al., 2012; Sun et al., 2013). In Arabidopsis, ire1a mutant plants showed enhanced susceptibility to bacterial pathogen Pseudomonas syringae (Moreno et al., 2012). In addition, rice black-streaked dwarf virus P10 and potato virus X TGBp3 elicited the UPR responses in N. benthamiana (Sun et al., 2013; Ye et al., 2013). The IRE1/bZIP60 pathway suppressed systemic accumulation of potyviruses and potexviruses in Arabidopsis and N. benthamiana (Gaguancela et al., 2016). However, it is unclear whether UPR is involved in plant resistance to necrotrophic fungal pathogens.
The necrotrophic fungal pathogen Alternata alternata (tobacco pathotype) causes brown spot disease, which is one of the most common and destructive fungal diseases of Nicotiana species. In response to this fungal infection, phytoalexins scopoletin and scopolin and transcripts of their key enzyme gene feruloyl-CoA 6′-hydroxylase 1 (F6′H1) are highly elicited in N. attenuata leaves around the attacked sites (Sun et al., 2014; Li and Wu, 2016). Phytoalexins are low-molecular mass antimicrobial secondary metabolites in response to pathogen infection, including camalexin, the major one in Arabidopsis, kauralexin, zealexin DIMBOA, and HDMBOA in maize, and scopoletin and capsidiol in Nicotiana species (Gnonlonfin et al., 2012). Scopoletin is a phenolic coumarin deriving from the phenylpropanoid pathway, playing an important role in N. attenuata-A. alternata interaction (Sun et al., 2014).
Jasmonate (JA) signaling pathway is usually associated with the defense against necrotrophic pathogens. Plants impaired with JA productions or JA perceptions are highly susceptible to A. alternata in N. attenuata (Sun et al., 2014). Importantly, phytoalexin scopoletin and scopolin biosyntheses are completely dependent on JA signaling, as their production and F6′H1 transcripts are abolished in JA-deficient (irAOC) and JA-insensitive (irCOI1) plants (Sun et al., 2014, 2017). It is currently unknown whether IRE1-bZIP60 pathway is involved in the regulation of scopoletin and scopolin by JA signaling pathway.
Here, we investigated whether UPR was activated in N. attenuata plants in response to A. alternata inoculation. The role of IRE1-bZIP60 pathway in plant resistance was tested in plants silenced with either IRE1 or bZIP60 via virus-induced gene silencing (VIGS), and the relation among IRE1-bZIP60 pathway, F6′H1, and JA signaling pathway was also explored in this study.
Materials and Methods
Plant and Fungal Material
Seeds of the N. attenuata were used as the wild-type (WT) genotype in all experiments. Stably transformed lines of irAOC and irCOI1 were previously generated (Paschold et al., 2007; Kallenbach et al., 2012) and used as plants that were silenced in the expression of AOC (the gene encoding the key enzyme of JA biosynthesis, allene oxide cyclase) and COI1 (the gene encoding the JA-Ile receptor, COI1). Seed germination and plant growth were conducted as described by Krugel et al. (2002). Sterilized N. attenuata seeds were treated with 1:50 (v/v) liquid smoke (House of herbs, Passaic, NY, USA) and 1 mM gibberellic acid (GA3, www.sigmaaldrich.com) to break dormancy, and were sown on agar with Gamborg B5. After 10 days, seedlings were planted into soil in Teku pots for another 10 days, seedlings of similar size were then transferred into 1-L pots in greenhouse with day/night temperatures of 25/19°C. Plants around 35-day-old before bolting were used for experiments.
Alternaria alternata were grown and used for inoculation as described (Sun et al., 2014). In brief, the source-sink transition leaves (0 leaves) were detached and inoculated with 4 PDA plugs (3 mm diameter) containing actively growing hyphae of A. alternata, which had been grown on PDA plates for 6–8 days, and then placed in a transparent 12 × 12 cm square Petri dish with 100% humidity at 25°C with a 16-h light/8-h dark cycle. Detached leaf without PDA plugs was served as mock control. Lesion diameters were measured after inoculation for 5–7 days.
Generation of VIGS Plants
Specific cDNA fragments of the bZIP60, IRE1a, and IRE1b were amplified by primers (XZ97 and XZ98 for bZIP60, XZ159 and XZ160 for IRE1a, XZ163 and XZ164 for IRE1b; all primers used in this study are included in Supplementary Table 1) and cloned into pTV00 respectively (Ratcliff et al., 2001). Agrobacterium tumefaciens GV3101 carrying above constructs were combined with those cells carrying pBINTRA, and then were inoculated into 3-week-old N. attenuata leaves, generating bZIP60-, IRE1a-, and and IRE1b-silenced plants (VIGS bZIP60, VIGS IRE1a, and VIGS IRE1b plants). To monitor the progress of VIGS, phytoene desaturase (PDS) was silenced, which would lead to a visible bleaching of tissues 2–3 weeks after inoculation (Saedler and Baldwin, 2004; Wu et al., 2008). When the leaves of PDS-silenced plants began to bleach, the source-sink transition leaves of VIGS plants and empty vector-inoculated plants (EV plants) were selected for further experiments. Around 25 plants were inoculated for each construct, and all VIGS experiments were repeated another two times.
Real-Time PCR Assay
Total RNA of five biological-replicate samples was extracted from a 1.5 × 1.5 cm2 area of leaf lamina with the inoculation site at the center. cDNA was synthesized from 500 ng of total RNA with reverse transcriptase (Thermo Scientific, http://www.thermoscientifcbio.com). Real-time PCR was performed as described (Sun et al., 2014) on a CFX Connect qPCR System (Bio-Rad) with iTaq Universal SYBR Green Supermix (Bio-Rad) and gene-specific primers according to the manufacturer’s instructions. The transcript abundance of Actin II was not altered in leaves either inoculated with A. alternata at 1 or 3 dpi or treated with MeJA and thus was used as an internal standard (Xu et al., 2018). All primers used in this study are included in Supplementary Table 1.
Electromobility Shift Assays (EMSA)
The full-length coding sequence of bZIP60 (spliced form) was cloned in frame into the EcoR I-Xho I sites of the pET28a (+), His- bZIP60(s) were expressed and purified with Ni-NTA agarose (QIAGEN). Biotin-labeled probes FBP1 (5′-aattttttttACGTaaatgattttcttttaccTGAAAAtttcccttga-3′), FBP2 (5′-taaggggcgTGAAAAgttcaaagt-3′), and FBP3 (5′-ttttgcactCAGTCAggctaaaatcg-3′) were designed from F6′H1 promoter and synthesized from Sangon Biotech (Shanghai). The detection of the binding of the recombinant protein and the probes (300 ng of recombinant protein and 30 ng labeled probe) was carried out with a chemiluminescent EMSA kit (Beyotime Biotechnology) according to the protocol suggested by the manufacturer.
Methyl Jasmonate (MeJA) Treatments
MeJA treatment is the most commonly used means of eliciting JA-regulated responses in plants; the chemical will be quickly de-methylated into jasmonic acid by plant MeJA-esterase when applied (Wu et al., 2008). A solution of 1 mM MeJA1 was prepared by distilled water, and sprayed directly on 0 leaves of 35-day-old rosette staged N. attenuata plants until run off with a fine mist according to Xu et al. (2018). The treated plants were immediately covered with a plastic bag, and harvested after 1, 3, 6, and 12 h. Distilled water was sprayed and used as a control treatment.
Accession Numbers
Sequence data from this article can be found in the GeneBank data library under accession numbers: XM_019373376.1 (luminal binding protein); XM_019393813.1 (protein disulfide isomerase); XM_019379334.1 (Calnexin 1-like); XM_019387223.1 (Calreticulin); XM_019409728.1 (IRE1a); XM_019389310.1 (IRE1b) and XM_019396471.1 (bZIP60); AY133805.1 (AtbZIP60); NM_127306.4 (AtIRE1a); and NM_001203454.1 (AtIRE1b).
Statistical Analyses
Statistical differences in gene expression between mock and fugus infection were analyzed with the Student’s t-test (p < 0.05). The lesion diameter was analyzed with Student’s t-test (p < 0.05). All analyses were performed using SPASS statistical software.
Result
Elicitation of Chaperone Protein Genes and Genes Involved in IRE1-bZIP60 Pathway in N. attenuata After A. alternata Inoculation
To test whether unfolded protein response was involved in N. attenuata resistance to A. alternata, we detected gene expression of chaperone proteins, which were usually up-regulated to accelerate protein folding during ER stress. All four chaperone protein genes tested in this study, BiP, PDI, CNX 1-like, and CRT, were significantly induced after 1 day post inoculation (dpi) (Figure 1). This result indicated that ER stress occurred in N. attenuata plants after A. alternata infection and host plants tried to alleviate this stress by activation of UPR.
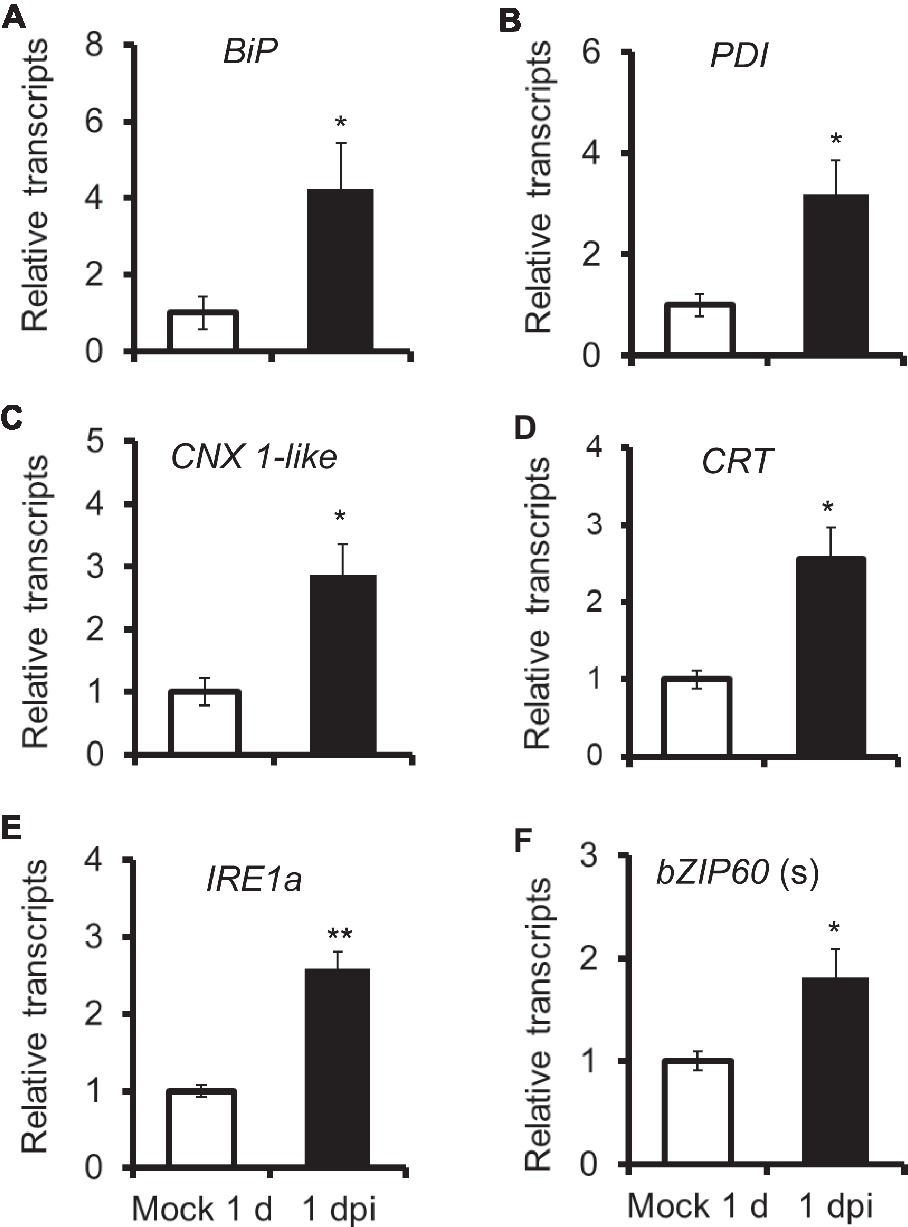
Figure 1. Elicitation of chaperone protein genes and genes involved in IRE-bZIP60 pathway after A. alternata inoculation. BiP (A), PDI (B), CNX 1-like (C), CRT (D), IRE1a (E), and bZIP60(s) (F) transcripts were measured by real-time PCR in source-sink transition leaves treated with mock infection of with A. alternata at 1 day post inoculation (dpi). All transcriptional levels were normalized with a housekeeping gene Actin II. Values are means ±SE for five biological replicates. Asterisks indicate the level of significant difference between mock and infected leaves (Student’s t-test: *, p < 0.05; **, p < 0.01, n = 5). All those experiments were repeated three times.
Next, we investigated transcriptional level of genes involved in IRE1-bZIP60 pathway. The N. attenuata genome comprises two IRE1s, IRE1a (XM_019409728.1), and IRE1b (XM_019389310.1), and one bZIP60 (XM_019396471.1). Both IRE1a and IRE1b shared 47% amino acid identity (Supplementary Data 1). Specific primers were designed to distinguish the bZIP60 spliced form (bZIP60 (s)) and unspliced form (bZIP60 (u); Supplementary Data 3). Our results indicated that IRE1a and bZIP60 (s) were both significantly elicited in N. attenuata plants at 1 dpi (Figure 1).
IRE1-bZIP60 Pathway Is Required for N. attenuata Resistance to the Fungal Pathogen A. alternata
To study the role of IRE1-bZIP60 pathway in N. attenuata resistance to A. alternata, IRE1a, IRE1b, and bZIP60 were silenced via virus-induced gene silencing (VIGS) individually. The transcriptional levels of IRE1a were reduced by more than 80% in plants transformed with IRE1a-silenced construct (VIGS IRE1a) when compared with plants transformed with empty vector (EV) at 1 dpi, and decreased by 66% at 3 dpi, suggesting that IRE1a was successfully silenced in VIGS IRE1a plants (Figure 2A). Similarly, VIGS IRE1b plants and VIGS bZIP60 plants were also successfully generated (Figure 2A). Notably, significantly bigger lesions were observed in source-sink transition leaves of plants silenced with IRE1a, IRE1b, or bZIP60 at 6 dpi when compared with EV plants (Figure 2B, Supplementary Data 4). These results indicate that plants impaired with IRE1-bZIP60 pathway are more susceptible to A. alternata than EV plants.
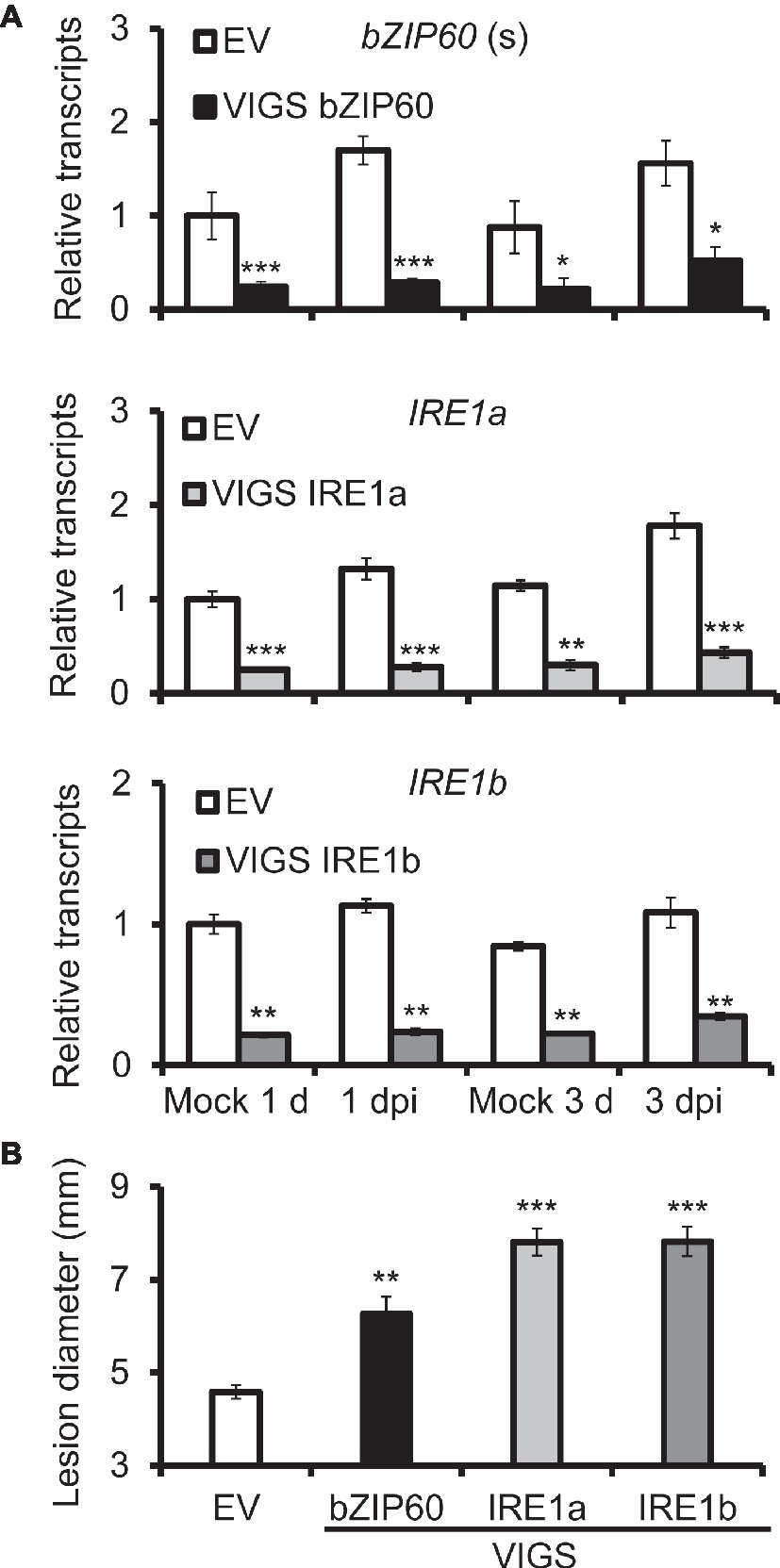
Figure 2. Silencing bZIP60 and IRE1s led to plants susceptible to A. alternata. (A) Mean (±SE) bZIP60(s), IRE1a, and IRE1b transcripts were measured by real-time PCR in six biological replicates of source-sink transition leaves of EV, VIGS bZIP60, VIGS IRE1a, and VIGS IRE1a plants at 1 and 3 dpi (n = 6). (B) Mean (±SE) diameter of necrotic lesions of 20 biological replicates of source-sink transition leaves of EV, VIGS bZIP60, VIGS IRE1a, and VIGS IRE1b plants at 6 dpi (n = 20). Asterisks indicate the level of significant differences between EV and VIGS plants with the same treatments (Student’s t-test: *, p < 0.05; **, p < 0.01; ***, p < 0.005). All those experiments were repeated four times.
As scopoletin and scopolin are important phytoalexins produced in N. attenuata against A. alternata (Sun et al., 2014; Li and Wu, 2016), we also investigated the gene expression level of the key enzyme gene for their biosynthesis, feruloyl-CoA 6′-hydroxylase 1 (F6′H1). F6′H1 was highly elicited in EV plants after inoculation, but its expression was dramatically reduced in VIGS IRE1a, VIGS IRE1b, or VIGS bZIP60 plants at 1 dpi (Figure 3A), suggesting that scopoletin-based defense was impaired in VIGS plants.
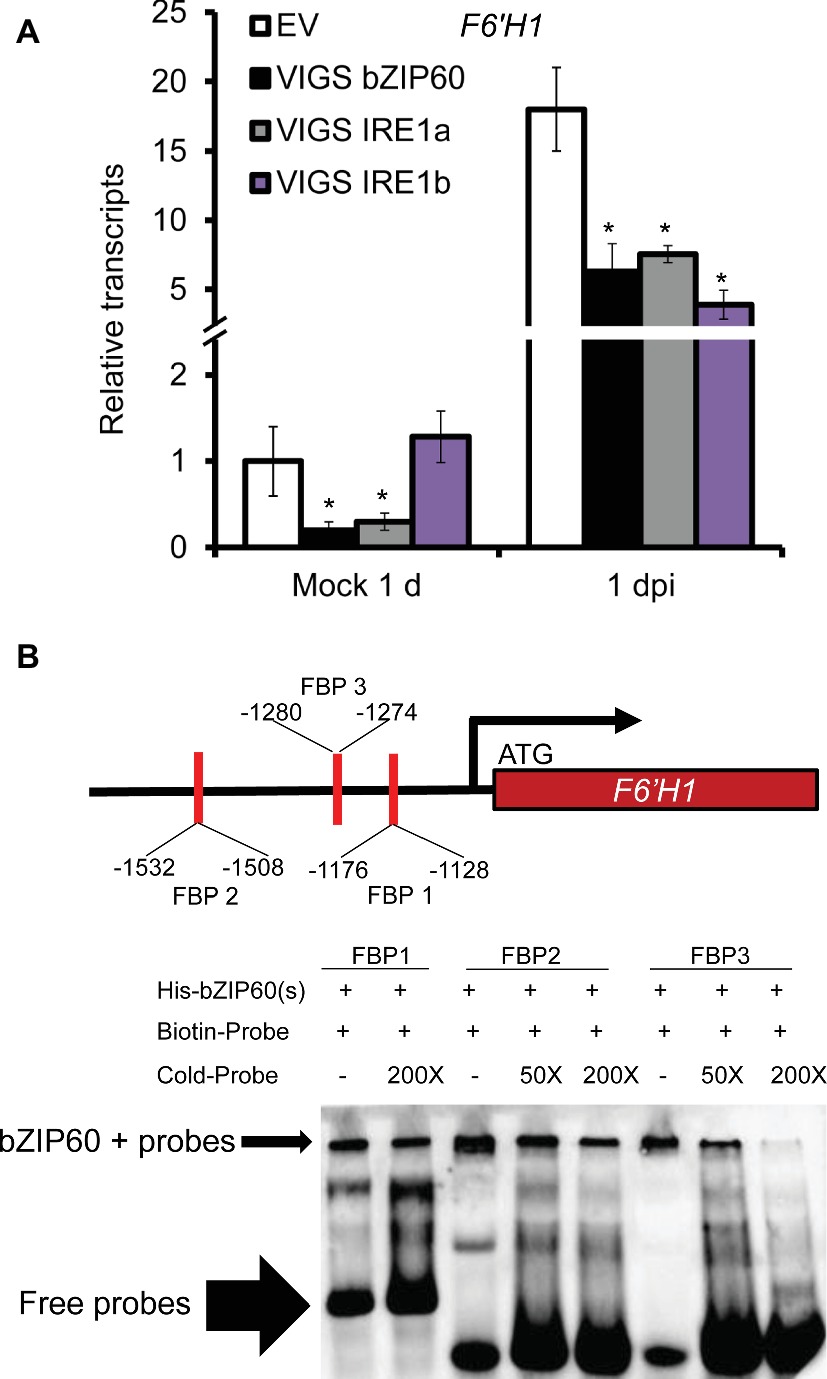
Figure 3. Reduction of A. alternata-elicited F6′H1 expression levels in plants impaired with IRE1-bZIP60 pathway and binding of bZIP60(s) proteins to the probes designed from F6′H1 promoter. (A) Mean (±SE) F6′H1 transcripts were measured by real-time PCR in five biological replicates of source-sink transition leaves treated with mock infection of A. alternata at 1 and 3 dpi. Asterisks indicate the level of significant difference between EV and VIGS plants with the same treatments (Student’s t-test: *, p < 0.05; **, p < 0.01, n = 5). All those experiments were repeated three times. (B) The spliced form of bZIP60 (His-bZIP60(s)) was purified from Ni-NTA agarose (QIAGEN) beads and incubated with biotin-labeled FBP1, FBP2, and FBP3 designed from F6′H1 promoter. Unlabeled FBP1, FBP2, and FBP3 were used as the competitor to show binding specificity. The arrows indicate the position of the binding bands and free probes. All those experiments were repeated three times.
By promoter motif scanning, we found three candidate binding sites of bZIP proteins, namely FBP1, FBP2, and FBP3 in F6′H1 promoter region, according to TGACTGR, TACGTA, and TGAAAA motifs which were identified previously (Jakoby et al., 2002; Iwata et al., 2008). We investigated whether bZIP60(s) protein could bind to those probes by EMSA. Our results indicated that bZIP60(s) protein could directly bind to all the three biotin-labeled probes (Figure 3B). To see whether the binding was specific or not, excessive unlabeled probes were added respectively and shown to be an effective competitor for each binding (Figure 3B).
Requirement of JA Signaling in A. alternata-Elicited Chaperone Protein Genes and Genes Involved in IRE1-bZIP60 Pathway
As JA-deficient (irAOC, plants silenced with allene oxide cyclase) and JA-insensitive (irCOI1, plants silenced with JA-Ile receptor COI1) plants were highly susceptible to A. alternata (Sun et al., 2014), we tried to investigate the role of JA signaling in this A. alternata-induced UPR. Compared with WT plants, all four chaperone protein genes decreased their expression levels significantly in irAOC and irCOI1 plants at 1 dpi (Figure 4). Furthermore, the fungus-elicited expression levels of IRE1a and bZIP60(s) were abolished in irAOC and irCOI1 plants at 1 dpi (Figure 5). Although the gene expression of IRE1b was not affected after fungal inoculation, its level was significantly reduced in irAOC and irCOI1 plants at 1 dpi (Figure 5). Those data suggest that JA signaling is required for the elicitation of chaperone protein genes and genes involved in IRE1-bZIP60 pathway.
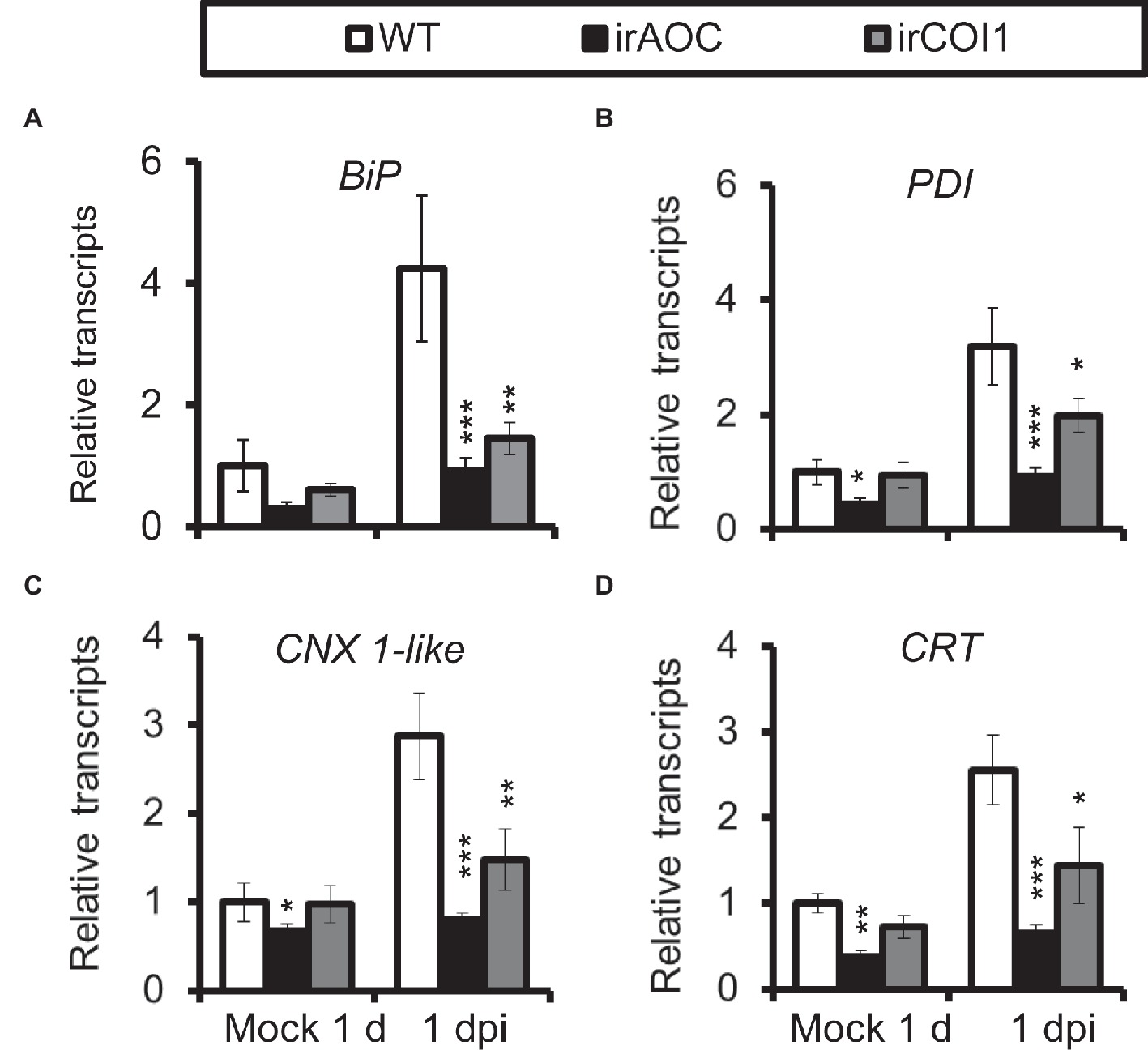
Figure 4. Transcriptional levels of A. alternata-induced chaperone protein genes were decreased in JA-deficient or JA-insensitive plants. Mean (±SE) BiP (A), PDI (B), CNX 1-like (C), and CRT (D) transcripts were measured by real-time PCR in five biological replicates of source-sink transition leaves treated with mock infection of A. alternata at 1 dpi. Asterisks indicate the level of significant difference between WT and transgenic plants with the same treatments (Student’s t-test: *, p < 0.05; **, p < 0.01; ***, p < 0.005, n = 5). All those experiments were repeated three times.
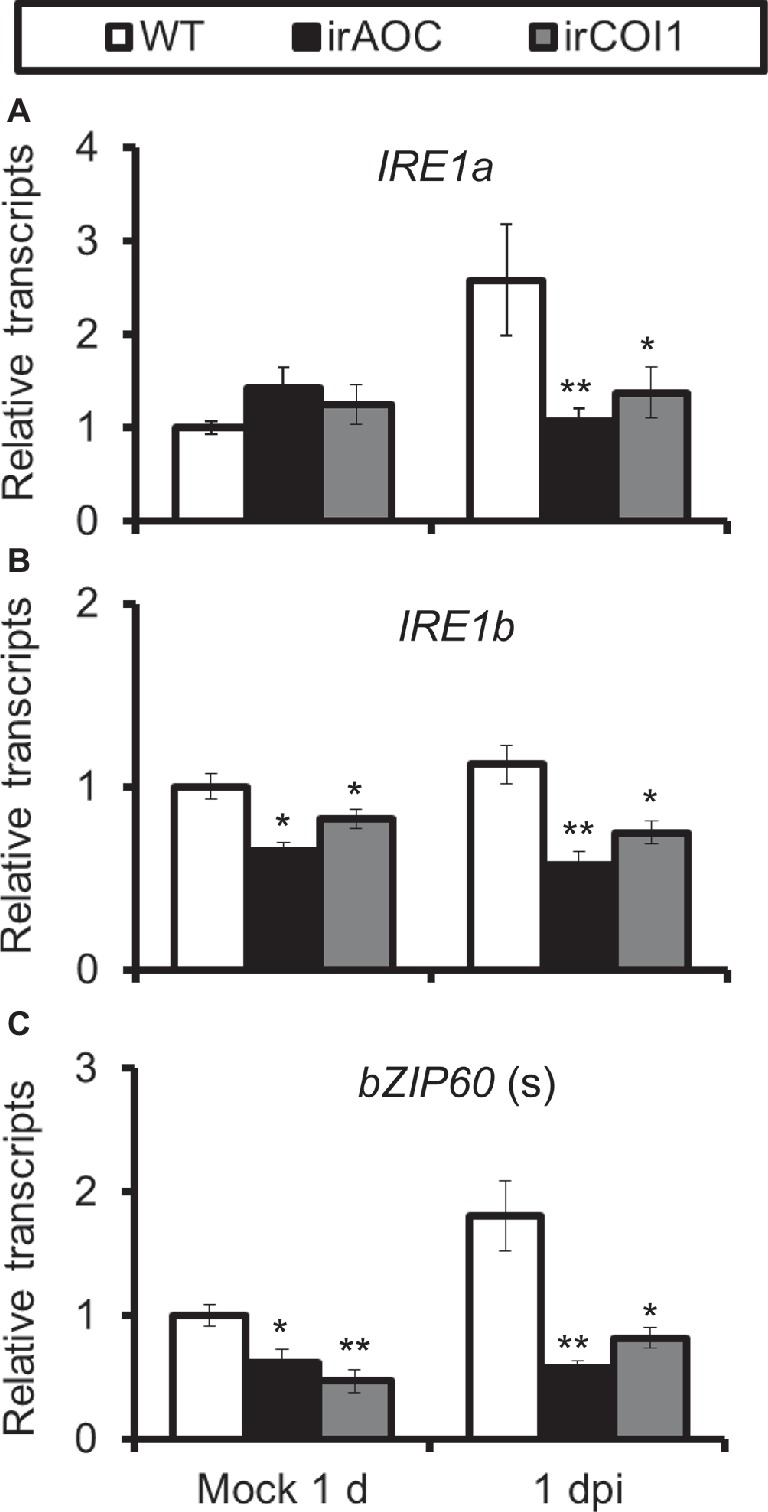
Figure 5. Transcriptional levels of A. alternata-induced genes involved in IRE1-bZIP60 pathway were reduced in JA-deficient or JA insensitive plants. Mean (±SE) IRE1a (A), IRE1b (B), and bZIP60(s) (C) transcripts were measured by real-time PCR in five biological replicates of source-sink transition leaves treated with mock infection of A. alternata at 1 dpi. Asterisks indicate the level of significant difference between WT and transgenic plants with the same treatments (Student’s t-test: *, p < 0.05; **, p < 0.01, n = 5). All those experiments were repeated three times.
When treated with methyl jasmonate (MeJA) exogenously, all genes were significantly induced in N. attenuata leaves. All four chaperone protein genes reached their peaks at 3-h treatment, while the highest level of IRE1a transcripts was detected after treatment with 1 h, and bZIP60(s) at 12 h (Figure 6). These results indicated that MeJA treatment itself is sufficient for elicitation of chaperone protein genes and genes involved in IRE1-bZIP60 pathway.
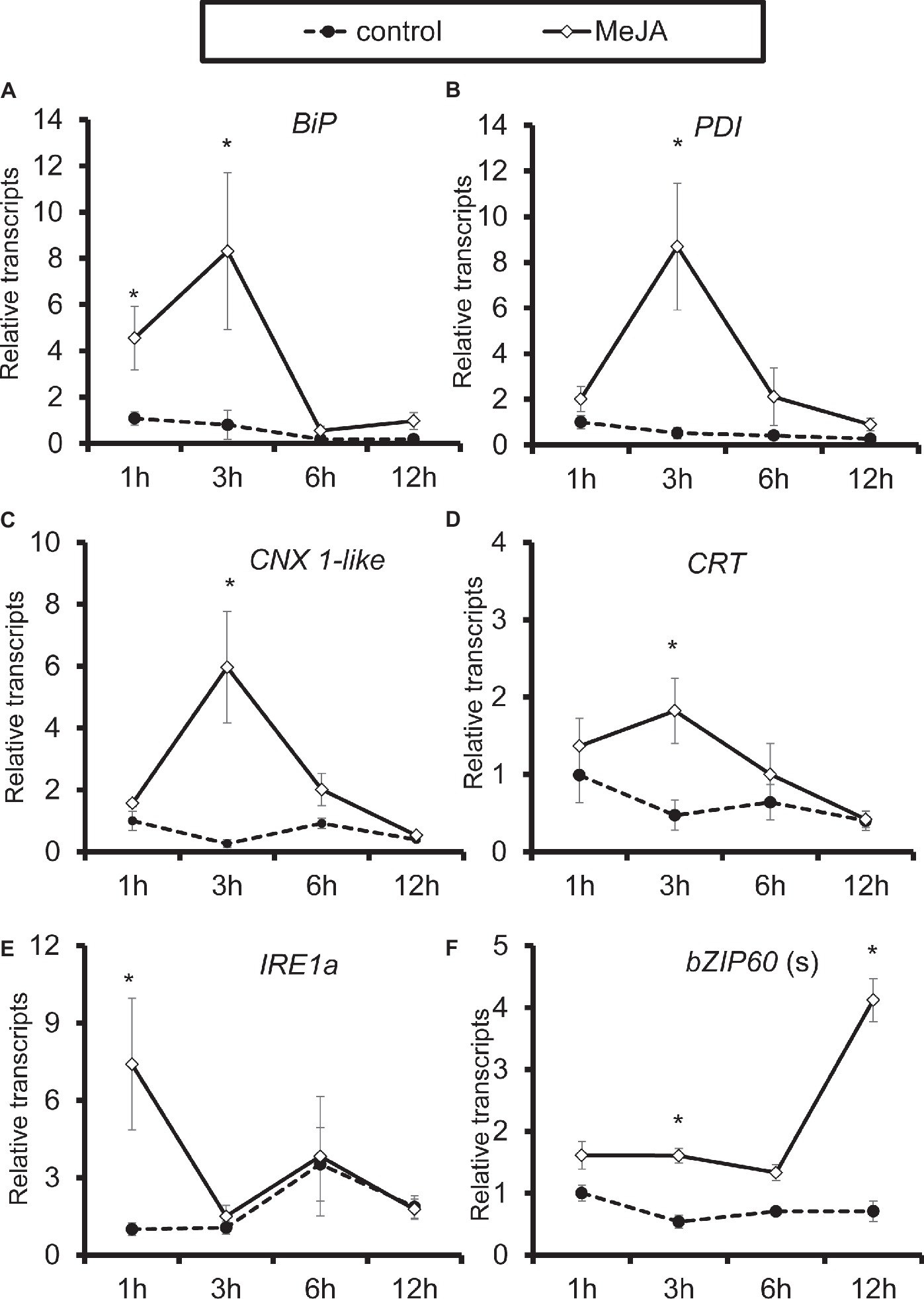
Figure 6. Elicitation of chaperone protein genes and genes involved in IRE-bZIP60 pathway by methyl jasmonate (MeJA). Mean (±SE) BiP (A), PDI (B), CNX 1-like (C), CRT (D), IRE1a (E), and bZIP60(s) (F) transcripts were measured by real-time PCR in five biological replicates of source-sink transition leaves treated with mock infection of MeJA at 1, 3, 6, and 12 h. Asterisks indicate the level of significant difference between mock or MeJA treatment (Student’s t-test: *, p < 0.05, n = 5). All those experiments were repeated three times.
We also tested whether or not JA signaling was altered in plants impaired with IRE1-bZIP60 pathway. The transcripts of the key enzyme genes for JA synthesis LOX3 and AOS, which were usually up-regulated after JA elicitation, were not altered in bZIP60-silenced plants (Figure 7), suggesting that JA signaling pathways were not affected in IRE1-bZIP60-silenced plants.
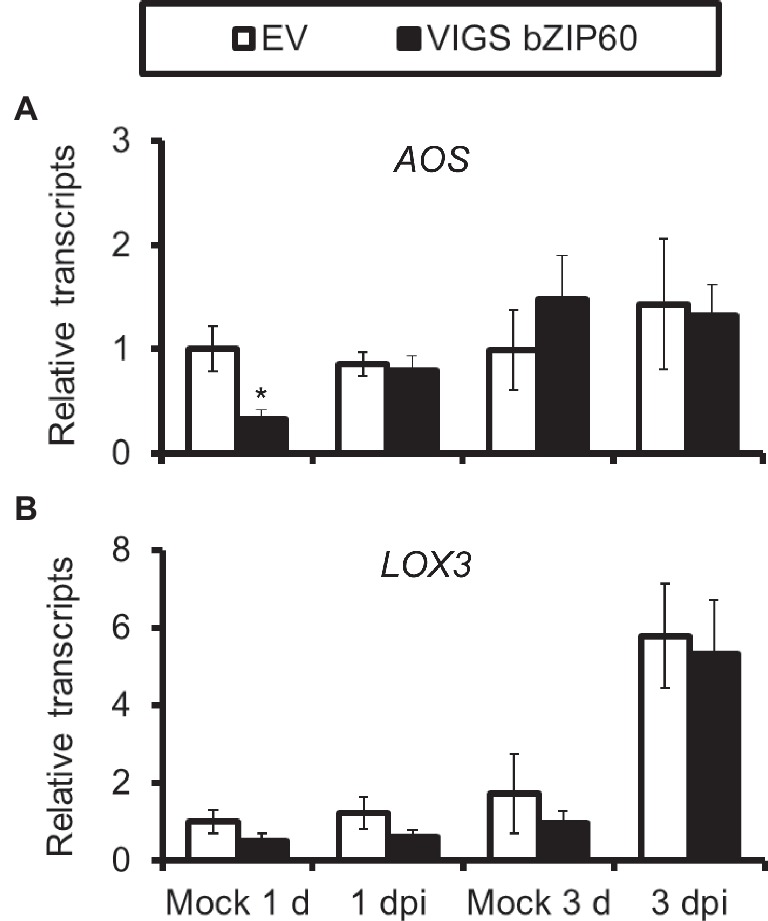
Figure 7. AOS and LOX3 expressions are not affected in VIGS bZIP60 plants. Mean (±SE) AOS (A) and LOX3 (B) transcripts were measured by real-time PCR in five biological replicates of source-sink transition leaves treated with mock infection of A. alternata at 1 dpi and 3 dpi. All those experiments were repeated three times (Student’s t-test: *, p < 0.05; n=5).
Discussion
The UPR signaling pathway plays an important role in adaptation to adverse environmental stresses and growth in eukaryotes. It is generally considered that IRE1-bZIP60 branch is conserved throughout metazoans and plants. The Arabidopsis genome encodes two IRE1s, AtIRE1a and AtIRE1b that share 41% amino acid identity (Noh et al., 2002). Both AtIRE1a and AtIRE1b are required for the splicing of bZIP60 mRNA (Nagashima et al., 2011), indicating they are functionally redundant. Similarly, two AtIRE1 homologs, IRE1a and IRE1b, were identified in N. attenuata genome. They shared 70 and 44% identity to that of Arabidopsis, respectively (Supplementary Data 1). The bZIP60 in N. attenuata shared 48% amino acid sequence identity to AtbZIP60 (Supplementary Data 1). Although we did not have the direct evidence of this IRE1a and IRE1b catalyzing the unconventional splicing of the mRNA of bZIP60, we did observe the existence of spliced form of bZIP60 by PCR with the specific primers (Supplementary Data 3), which was also confirmed by sequencing. Moreover, the abundance of bZIP60 (s) expression in either VIGS IRE1a or VIGS IRE1b plants was significantly reduced (Supplementary Data 3). These results indicated that the IRE1a and IRE1b in N. attenuata also functioned reluctantly in splicing bZIP60 mRNA.
Recent reports indicate that IRE1-bZIP60 pathway is involved in plant resistance to pathogens. For example, the expression of bZIP60 in N. benthamiana is up-regulated upon infection with non-host bacterial pathogen (Tateda et al., 2008). IRE1 and its substrate bZIP60 function as a strictly cognate enzyme-substrate pair to control viral pathogenesis in Arabidopsis and N. benthamiana (Zhang et al., 2015; Gaguancela et al., 2016). In this study, we showed that UPRs were also activated in N. attenuata when inoculated with A. alternata, as chaperone genes BiP, PDI, CNX 1-like, and CRT were all significantly up-regulated after inoculation (Figure 1). Both IRE1a and the spliced form of bZIP60 were also significantly elicited (Figure 1), and clearly the IRE1-bZIP60 pathway was required for plant resistance to this fungal pathogen, as significantly bigger lesions were observed in plants impaired with IRE1-bZIP60 pathway when compared with EV plants (Figure 2).
Why plants impaired with IRE1-bZIP60 pathway were more susceptible to A. alternata? It might be due to the delayed up-regulation of chaperone genes which are important to restore proper protein-folding and release ER stress to avoid cell death when the stresses are overwhelming. Among the four up-regulated chaperone genes, PDI and CNX 1-like showed reduced gene expression levels in VIGS bZIP60 plants at 1 dpi, but they were elicited to levels similar to that of EV plants at 3 dpi (Supplementary Data 2); while BiP and CRT were induced to the same levels in VIGS bZIP60 and EV plants at both 1 and 3 dpi (Supplementary Data 2). In addition, F6′H1, the key enzyme gene of phytoalexins scopoletin and scopolin previously shown to be crucial for N. attenuata resistance to A. alternata (Sun et al., 2014; Li and Wu, 2016), was found significantly less elicited in plants impaired with IRE1-bZIP60 pathway (Figure 3). Further EMSA experiments indicated that bZIP60(s) could directly bind to all the three biotin-labeled probes designed from F6′H1 promoter. Thus, we concluded that the IRE1-bZIP60 pathway was not only required for releasing the stresses in ER but also regulated chemical defense by direct binding of bZIP60(s) proteins to F6′H1 promoter.
JA signaling pathway is essential for plant resistance to A. alternata (Sun et al., 2014). It regulates a series of defense-related genes; although some of them play a negative role in resistance, like the recently reported patatin-like protein in N. attenuata (Cheng et al., 2019), many of them are required for plant resistance to A. alternata, for example the pleiotropic drug resistance transporters NaPDR1 and NaPDR1-like (Xu et al., 2018), and F6′H1 (Sun et al., 2014; Li and Wu, 2016). However, whether JA plays a role in UPR is unclear. In this study, we showed that the elicitations of all the four chaperone genes used in this study, IRE1a, IRE1b, and bZIP60 by the fungus were dramatically reduced in JA-deficient (irAOC) and JA-insensitive (irCOI1) plants (Figures 4 and 5), suggesting that JA signaling pathway is required for the mounting of UPR. Furthermore, chaperone genes BiP, PDI, CNX 1-like, CRT, IRE1a, and bZIP60(s) were all significantly induced when N. attenuata leaves were treated with MeJA (Figure 6). In contrast, the transcripts of the key enzyme genes for JA synthesis, LOX3 and AOS, were not reduced in bZIP60-silenced plants (Figure 6), indicating that JA levels are not affected in these plants. Thus our data strongly suggest that JA signaling is located upstream of IRE1-bZIP60 pathway (Figure 8).
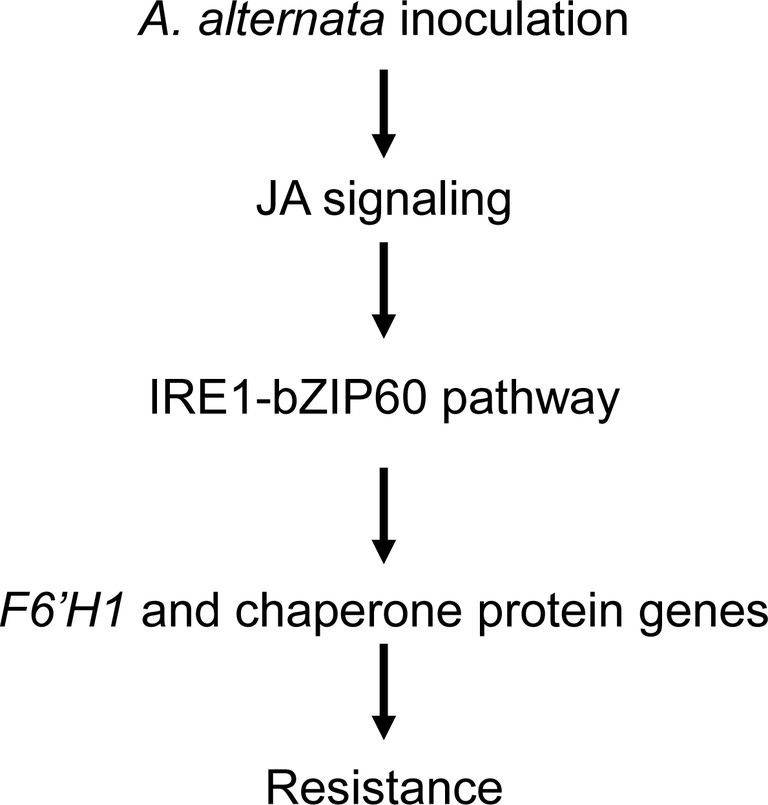
Figure 8. Proposed working model of IRE-bZIP60 pathway in N. attenuata resistance to A. alternata. After A. alternata inoculation, JA signaling pathway is activated, which subsequently regulates F6′H1 and chaperone protein genes by IRE-bZIP60 pathway.
Taken together, our study demonstrated that UPR is activated after A. alternata inoculation in N. attenuata, and the UPR branch, IRE1-bZIP60 pathway, played an important role in plant defense against A. alternata. Furthermore, we provided evidence that JA signaling is required for the elicitation of gene expressions of chaperone proteins and genes involved in IRE1-bZIP60 pathway after fungal inoculation.
Author Contributions
ZX and JW contributed to the conception and design of the study. ZX and JW organized the database. ZX and JW performed the statistical analysis. NS proved the cDNA of irAOC and irCOI1, and cDNA of plants exogenously treated with methyl jasmonate (MeJA), and ZX and JW wrote the manuscript. All authors contributed to manuscript revision, read and approved the submitted version.
Funding
This project is supported by NSFC Grant (No. 31670262), the Key Project of Applied Basic Research Program of Yunnan (2014FA040), and 100-Oversea-Top-Talents Recruitment plan of Yunnan to JW for ZX’s stipend and most of the research. NSFC Grant (No. 31700231) and CAS “Light of West China” Program to LM for greenhouse services and part of the research. NSFC Grant (No. 31670262) provided open access publication fees.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Ian T. Baldwin (Max-Planck Institute for Chemical Ecology, Jena, Germany) for providing all the transgenic Nicotiana attenuata seeds and Biological Technology Open Platform of Kunming Institute of Botany (CAS) for greenhouse and instrument services.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00263/full#supplementary-material
SUPPLEMENTARY DATA 1 | Alignment of the Arabidopsis thaliana IRE1a and IRE1b (a), and bZIP60 (b) with those of N. attenuata protein sequences.
SUPPLEMENTARY DATA 2 | CRT and BiP expressions were not affected but PDI and CNX 1-like (b) were reduced at 1 dpi in VIGS bZIP60 plants. Mean (±SE) CRT, PDI, CNX 1-like and BiP transcripts were measured by real-time PCR in five replicates of source-sink transition leaves treated with mock or A. alternata at 1 and 3 dpi. (Student’s t-test: *, p < 0.05; **, n = 5) All those experiments were repeated twice.
SUPPLEMENTARY DATA 3 | Splicing of bZIP60 mRNA by IRE1a and IRE1b. (a) Mean (±SE) bZIP60 (s) and bZIP60 (u) transcripts were measured by real-time PCR in five replicates of source-sink transition leaves treated with mock or A. alternata at 1 and 3 day post inoculation (dpi). All transcriptional levels were normalized with a housekeeping gene Actin II. Asterisks indicate the level of significant difference between EV and VIGS plants with the same treatments (Student’s t-test: *, p < 0.05; **, p < 0.01, n = 5). (b) Detection of bZIP60 spliced form. Unspliced and spliced forms of bZIP60 were amplified by RT-PCR with primers (XZ141 and XZ142; Supplementary Table 1) from RNA sample extracted from A. alternata inoculated leaves at 1 dpi. The PCR products were run on the gel. The band of spliced form of bZIP60 was pointed by the arrow, and confirmed by sequencing.
SUPPLEMENTARY DATA 4 | Photographs were taken of leaves inoculated with A. alternata at 5 days post inoculation. Two leaves of EV, VIGS bZIP60, VIGS IRE1a and VIGS IRE1b were provided. Black arrows indicate lesions.
SUPPLEMENTARY TABLE 1 | All primers used in this study.
Footnotes
References
Cheng, J., Song, N., and Wu, J. (2019). A patatin-like protein synergistically regulated by jasmonate and ethylene signaling pathways plays a negative role in Nicotiana attenuata resistance to Alternaria alternata. Plant Diversity. 41, 7–10. doi: 10.1016/j.pld.2018.12.001
Denecke, J., Carlsson, L. E., Vidal, S., Hoglund, A. S., Ek, B., Vanzeijl, M. J., et al. (1995). The tobacco homolog of mammalian calreticulin is present in protein complexes in-vivo. Plant Cell 7, 391–406. doi: 10.1105/tpc.7.4.391
Deng, Y., Humbert, S., Liu, J. X., Srivastava, R., Rothstein, S. J., and Howell, S. H. (2011). Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 7247–7252. doi: 10.1073/pnas.1102117108
Ellgaard, L., and Helenius, A. (2003). Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4, 181–191. doi: 10.1038/nrm1052
Freitas, C. P., Clemente, I., Mendes, T., and Novo, C. (2016). Trichinella spiralis: genome database searches for the presence and immunolocalization of protein disulphide isomerase family members. J. Helminthol. 90, 62–67. doi: 10.1017/S0022149X14000807
Gaguancela, O. A., Zuniga, L. P., Arias, A. V., Halterman, D., Flores, F. J., Johansen, I. E., et al. (2016). The IRE1/bZIP60 pathway and Bax Inhibitor 1 suppress systemic accumulation of Potyviruses and Potexviruses in Arabidopsis and Nicotiana benthamiana plants. Molecular Plant-Microbe Interactions 29, 750–766. doi: 10.1094/Mpmi-07-16-0147-R
Gao, H. B., Brandizzi, F., Benning, C., and Larkin, R. M. (2008). A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 105, 16398–16403. doi: 10.1073/pnas.0808463105
Gething, M. J., and Sambrook, J. (1992). Protein folding in the cell. Nature 355, 33–45. doi: 10.1038/355033a0
Gnonlonfin, G. J. B., Sanni, A., and Brimer, L. (2012). Review scopoletin - a coumarin phytoalexin with medicinal properties. Crit. Rev. Plant Sci. 31, 47–56. doi: 10.1080/07352689.2011.616039
Iwata, Y., Fedoroff, N. V., and Koizumi, N. (2008). Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 20, 3107–3121. doi: 10.1105/tpc.108.061002
Jakoby, M., Weisshaar, B., Droge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. doi: 10.1016/S1360-1385(01)02223-3
Kallenbach, M., Bonaventure, G., Gilardoni, P. A., Wissgott, A., and Baldwin, I. T. (2012). Empoasca leafhoppers attack wild tobacco plants in a jasmonate-dependent manner and identify jasmonate mutants in natural populations. Proc. Natl. Acad. Sci. U.S.A. 109, E1548–E1557. doi: 10.1073/pnas.1200363109
Klein, E. M., Mascheroni, L., Pompa, A., Ragni, L., Weimar, T., Lilley, K. S., et al. (2006). Plant endoplasmin supports the protein secretory pathway and has a role in proliferating tissues. Plant J. 48, 657–673. doi: 10.1111/j.1365-313X.2006.02904.x
Krugel, T., Lim, M., Gase, K., Halitschke, R., and Baldwin, I. T. (2002). Agrobacterium-mediated transformation of Nicotiana attenuata, a model ecological expression system. Chemoecology 12, 177–183. doi: 10.1007/PL00012666
Li, Z. X., Srivastava, R., Tang, J., Zheng, Z. H., and Howell, S. H. (2018). Cis-effects condition the induction of a major unfolded protein response factor, ZmbZIP60, in response to heat stress in maize. Front. Plant Sci. 9:833. doi: 10.3389/fpls.2018.00833
Li, J., and Wu, J. (2016). Scopolin, a glycoside form of the phytoalexin scopoletin, is likely involved in the resistance of Nicotiana attenuata against Alternaria alternata. J. Plant Pathol. 98, 641–644. doi: 10.4454/JPP.V98I3.006
Liu, Y. M., Burgos, J. S., Deng, Y., Srivastava, R., Howell, S. H., and Bassham, D. C. (2012). Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 24, 4635–4651. doi: 10.1105/tpc.112.101535
Liu, L. J., Cui, F., Li, Q. L., Yin, B. J., Zhang, H. W., Lin, B. Y., et al. (2011). The endoplasmic reticulum-associated degradation is necessary for plant salt tolerance. Cell Res. 21, 957–969. doi: 10.1038/cr.2010.181
Moreno, A. A., Mukhtar, M. S., Blanco, F., Boatwright, J. L., Moreno, I., Jordan, M. R., et al. (2012). IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS One 7:e31944. doi: 10.1371/journal.pone.0031944
Moreno, A. A., and Orellana, A. (2011). The physiological role of the unfolded protein response in plants. Biol. Res. 44, 75–80. doi: 10.4067/S0716-97602011000100010
Nagashima, Y., Mishiba, K., Suzuki, E., Shimada, Y., Iwata, Y., and Koizumi, N. (2011). Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci. Rep. 1:29. doi: 10.1038/srep00029
Noh, S. J., Kwon, C. S., and Chung, W. I. (2002). Characterization of two homologs of Ire1p, a kinase/endoribonuclease in yeast, in Arabidopsis thaliana. Biochim. Biophys. Acta 1575, 130–134. doi: 10.1016/S0167-4781(02)00237-3
Paschold, A., Halitschke, R., and Baldwin, I. T. (2007). Co(i)-ordinating defenses: NaCOI1 mediates herbivore-induced resistance in Nicotiana attenuata and reveals the role of herbivore movement in avoiding defenses. Plant J. 51, 79–91. doi: 10.1111/j.1365-313X.2007.03119.x
Ratcliff, F., Martin-Hernandez, A. M., and Baulcombe, D. C. (2001). Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25, 237–245. doi: 10.1046/j.0960-7412.2000.00942.x
Ruberti, C., Kim, S. J., Stefano, G., and Brandizzi, F. (2015). Unfolded protein response in plants: one master, many questions. Curr. Opin. Plant Biol. 27, 59–66. doi: 10.1016/j.pbi.2015.05.016
Saedler, R., and Baldwin, I. T. (2004). Virus-induced gene silencing of jasmonate-induced direct defences, nicotine and trypsin proteinase-inhibitors in Nicotiana attenuata. J. Exp. Bot. 55, 151–157. doi: 10.1093/jxb/erh004
Siffroi-Fernandez, S., Giraud, A., Lanet, J., and Franc, J. L. (2002). Association of the thyrotropin receptor with calnexin, calreticulin and BiP. Eur. J. Biochem. 269, 4930–4937. doi: 10.1046/j.1432-1033.2002.03192.x
Sun, H., Song, N., Ma, L., Li, J., Ma, L., Wu, J., et al. (2017). Ethylene signalling is essential for the resistance of Nicotiana attenuata against Alternaria alternata and phytoalexin scopoletin biosynthesis. Plant Pathol. 66, 277–284. doi: 10.1111/ppa.12568
Sun, H., Wang, L., Zhang, B., Ma, J., Hettenhausen, C., Cao, G., et al. (2014). Scopoletin is a phytoalexin against Alternaria alternata in wild tobacco dependent on jasmonate signalling. J. Exp. Bot. 65, 4305–4315. doi: 10.1093/jxb/eru203
Sun, Z. T., Yang, D., Xie, L., Sun, L. Y., Zhang, S. L., Zhu, Q. S., et al. (2013). Rice black-streaked dwarf virus P10 induces membranous structures at the ER and elicits the unfolded protein response in Nicotiana benthamiana. Virology 447, 131–139. doi: 10.1016/j.virol.2013.09.001
Tateda, C., Ozaki, R., Onodera, Y., Takahashi, Y., Yamaguchi, K., Berberich, T., et al. (2008). NtbZIP60, an endoplasmic reticulum-localized transcription factor, plays a role in the defense response against bacterial pathogens in Nicotiana tabacum. J. Plant Res. 121, 603–611. doi: 10.1007/s10265-008-0185-5
Trombetta, E. S., and Parodi, A. J. (2003). Quality control and protein folding in the secretory pathway. Annu. Rev. Cell Dev. Biol. 19, 649–676. doi: 10.1146/annurev.cellbio.19.110701.153949
Vitale, A., and Boston, R. S. (2008). Endoplasmic reticulum quality control and the unfolded protein response: insights from plants. Traffic 9, 1581–1588. doi: 10.1111/j.1600-0854.2008.00780.x
Wan, S. C., and Jiang, L. W. (2016). Endoplasmic reticulum (ER) stress and the unfolded protein response (UPR) in plants. Protoplasma 253, 753–764. doi: 10.1007/s00709-015-0842-1
Wang, M. Y., Xu, Q. Y., and Yuan, M. (2011). The unfolded protein response induced by salt stress in Arabidopsis. Methods Enzymol. 489, 319–328. doi: 10.1016/B978-0-12-385116-1.00018-2
Wu, J., Wang, L., and Baldwin, I. T. (2008). Methyl jasmonate-elicited herbivore resistance: does MeJA function as a signal without being hydrolyzed to JA? Planta 227, 1161–1168. doi: 10.1007/s00425-008-0690-8
Xu, Z., Song, N., Ma, L., Fang, D., and Wu, J. (2018). NaPDR1 and NaPDR1-like are essential for the resistance of Nicotiana attenuata against fungal pathogen Alternaria alternata. Plant Diversity 40, 68–73. doi: 10.1016/j.pld.2018.01.001
Ye, C. M., Chen, S. R., Payton, M., Dickman, M. B., and Verchot, J. (2013). TGBp3 triggers the unfolded protein response and SKP1-dependent programmed cell death. Mol. Plant Pathol. 14, 241–255. doi: 10.1111/mpp.12000
Ye, C. M., Kelly, V., Payton, M., Dickman, M. B., and Verchot, J. (2012). SGT1 is Induced by the potato virus X TGBp3 and enhances virus accumulation in Nicotiana benthamiana. Mol. Plant 5, 1151–1153. doi: 10.1093/mp/sss026
Zhang, L. R., Chen, H., Brandizzi, F., Verchot, J., and Wang, A. M. (2015). The UPR branch IRE1-bZIP60 in plants plays an essential role in viral infection and is complementary to the only UPR pathway in yeast. PLoS Genet. 11:e1005164. doi: 10.1371/journal.pgen.1005164
Keywords: unfolded protein response, inositol-requiring enzyme 1 (IRE1), bZIP60, Feruloyl-CoA 6ʹ-hydroxylase 1 (F6ʹH1), jasmonate (JA)
Citation: Xu, Song, Ma and Wu (2019) IRE1-bZIP60 Pathway Is Required for Nicotiana attenuata Resistance to Fungal Pathogen Alternaria alternata. Front. Plant Sci. 10:263. doi: 10.3389/fpls.2019.00263
Edited by:
Péter Poór, University of Szeged,HungaryReviewed by:
Giovanni Stefano, Michigan State University, United StatesXin Deng, Institute of Botany (CAS),China
Copyright © 2019 Xu, Song, Ma and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinsong Wu, amluc29uZ3d1QG1haWwua2liLmFjLmNu
 Zhen Xu1
Zhen Xu1 Jinsong Wu
Jinsong Wu