- The Key Laboratory of Integrated Crop Pest Management of Shandong Province, College of Plant Health and Medicine, Qingdao Agricultural University, Qingdao, China
Black shank caused by Phytophthora nicotianae is one of the most devastating diseases in tobacco production. In this study, we characterized a novel cytochromic resistance gene, SoCYP85A1, from spinach, which was upregulated in response to P. nicotianae infection. Overexpression of SoCYP85A1 in tobacco resulted in remarkable resistance to pathogen inoculation, with diverse resistance levels in different transgenic lines. Meanwhile, a significant accumulation of castasterone (CS) was detected in transgenic plants when challenged with the pathogen. Moreover, activities of antioxidant enzymes were enhanced by SoCYP85A1 in the transgenic lines as compared to those in the wild types inoculated with P. nicotianae. In addition, the alteration of CS content resulted in interference of phytohormone homeostasis. Overall, these results demonstrate that SoCYP85A1 can participate in the defense response to P. nicotianae through the involvement of defense enzymes and by interaction with certain phytohormones. Our findings suggest that SoCYP85A1 could be used as a potential candidate gene for improving resistance to black shank disease in tobacco and other economic crops.
Introduction
Members of oomycetes are destructive pathogens that cause damages to important economic crops (Kamoun et al., 2015). Among the oomycetes that are pathogenic to plants, Phytophthora nicotianae is one of the most devastating for tobacco (Nicotiana tabacum L.) production (Gallup et al., 2018); it causes serious disease in the roots and stems of tobacco in many tobacco-growing regions around the world (Johnson et al., 2002). Plants have evolved defense mechanisms to resist pathogen infections. One such mechanism involves regulation of the biosynthesis of phytohormones, such as abscisic acid (ABA), gibberellic acid (GA), jasmonic acid (JA), and salicylic acid (SA) (Vlot et al., 2009; Fang et al., 2016; Gao et al., 2016; Tang et al., 2017).
Brassinosteroids (BRs) are one of the most important phytohormones and involve in various physiological processes (Choudhary et al., 2012). In addition of their roles in plant growth and development (Arteca and Arteca, 2001; Yu et al., 2004; Gruszka, 2013), BRs have been implicated in tolerance to pathogens (Nakashita et al., 2003; Albrecht et al., 2012). However, most of the previous studies focusing on the regulation of plant response to pathogens by BRs have been performed using exogenously applied BRs (Nakashita et al., 2003; Divi et al., 2010; Albrecht et al., 2012). The genetic mechanisms underlying the role of endogenous BRs in regulating the response of plants to pathogens remain poorly understood.
Over the past few years, the biosynthesis pathway of BRs has been elucidated, and key enzymes involved in the pathway have been identified (Shimada et al., 2003; Kim et al., 2005; Ohnishi et al., 2012). Among these, CYP85A1 catalyzes the conversion of intermediate metabolites to castasterone (CS) (Shimada et al., 2001), which is the precursor of brassinolide (BL), the end-product of BRs biosynthesis pathway. However, the rate of conversion of CS to BL is extremely low (Noguchi et al., 2000). CS possesses its own function and performs the same biological activity as BL (Kim et al., 2005). In a previous work, we observed that overexpression of SoCYP85A1 (GenBank Accession: KT900949), a spinach cytochrome P450 enhanced drought tolerance in transgenic tobacco by increasing the CS content. CS was assumed to be a bioactive BR and displayed the same biological function (Duan et al., 2017). However, whether endogenous CS participates in plant response to pathogen infection remains unknown.
To explore the function CS in plants subjected to pathogen invasion, we analyzed tobacco seedlings overexpressing SoCYP85A1 infected with P. nicotianae. It was observed that overexpression of SoCYP85A1 enhanced black shank tolerance of transgenic tobacco seedlings compared to that of wild type seedlings. Furthermore, this enhanced tolerance was demonstrated to be through the upregulation of activities of defense enzymes at the transcriptional level. In addition, CS was identified as a bioactive component in tobacco, which was induced by the P. nicotianae infection. The alteration of CS content resulted in interference of phytohormone homeostasis. This study provides new information for the disease control network and a new strategy for disease resistance breeding.
Materials and Methods
Plant Materials and Growth Conditions
The genotype of tobacco that was genetically engineered was N. tabacum cv. Xanthi-nc. T3 seeds from the transgenic lines overexpressing SoCYP85A1 driven by the CAMV35S promoter were produced and confirmed as described in our previous study (Duan et al., 2017), in which the SoCYP85A1 gene has been stable inheritance and expression. T3 seeds were grown and developed to T4 transgenic soybean plants for further analysis. The wild type (WT) and transgenic lines were grown in growth chambers under a photoperiod of 16 h light/8 h dark and maintained at 30°C and 60% relative humidity for 1 month.
Genomic PCR and GFP Analysis of Transgenic Plants
Genomic DNA was extracted from tobacco leaves using Plant DNA Kit (Omega Bio-tek, Norcross, GA, United States). The primers (Forward: 5′-ATGGCCGTTTTTATGGTGGTTTTTGCTGT-3′ and Reverse: 5′-CTAATAACTCGAAACTCGAATGC-3′) were used to amplify the open reading frame (ORF) of the SoCYP85A1 gene. For green fluorescent protein (GFP) analysis, fluorescence signal was detected with the NEWTON 7.0 smart imaging system (Vilber Lourmat, France).
Stress Treatments
For P. nicotianae treatment, 10 seedlings per type in each treatment were infected with the pathogen following the method described by Gao et al. (2010). The plants treated with the same amount of sterile water in the same way served as controls. The tested seedlings were transplanted in another growth chamber under a 16 h light/8 h dark cycle at 35°C and 90% relative humidity. Each treatment was performed in three biological replicates, in which there were three technical replicates.
Assays of Resistance of Transgenic Tobacco Against P. nicotianae
The disease symptoms on each seedling were monitored at 2, 3, 4, and 7 days post inoculation (dpi), respectively. The severity of disease was graded according to GB/T 23222-2008 (National Standardization Management Committee, 2009). Disease index (DI) was calculated using the following formula: DI (%) = [Σ (DGi × ni)/(DGi max ×n)] × 100, where DGi is the value of the disease grade, ni is the number of plants with each disease grade, DGi max is the maximum value of disease grades, and n is the total number of inoculated plants. The unifoliate leaves were harvested at 0 and 4 dpi, immediately frozen in liquid nitrogen, and stored at -80°C to investigate the function of SoCYP85A1.
Quantitative Real-Time RT-PCR of SoCYP85A1
Total RNA was extracted from WT and transgenic tobacco seedlings with Plant RNA Kit (Omega Bio-tek, Norcross, GA, United States). Reverse transcription was performed using Prime ScriptTM RTase Kit (TaKaRa, Dalian, China) in accordance with the manufacturer’s instructions. The relative expression level of SoCYP85A1 was examined by SYBR Premix Ex Taq II Kit (TaKaRa, Dalian, China) before and after the inoculation of P. nicotianae. The PCR program was as follows: 95°C for 5 min followed by 40 cycles of 30 s at 95°C, 30 s at 60°C, and 30 s at 72°C. Spinacia oleracea actin gene (GenBank: JN987183.1) served as an internal control to determine the relative transcript levels of SoCYP85A1. The relative gene expression level was calculated using the ΔΔCt method (Livaka and Schmittgenb, 2001). qRT-PCR was performed in three biological replicates and each reaction was proceeded three times.
Extraction and Quantification of Endogenous BRs
Both extraction and quantification of endogenous BRs were conducted according to Ding et al. (2013).
Measurement of the Activities of Antioxidant Enzymes and Their Corresponding Gene Expression Levels
The activities of defense-related enzymes, including superoxide dismutase (SOD), peroxidase (POD), polyphenol oxidase (PPO), and catalase (CAT), were spectrophotometrically detected using different kits (obtained from Jiancheng, Nanjing, China; catalog number A001-4 for SOD, A084-3 for POD, A136 for PPO, and A003-3 for CAT) in accordance with the manufacturer’s instructions.
qRT-PCR Analysis of Defense-Related Genes
qRT-PCR was employed to examine the relative expression levels of defense-related genes, which included four genes encoding the enzymes mentioned above (NtSOD, NtAPX, NtPPO, and NtCAT), ABA biosynthesis gene (NtNCED1), SA-responsive genes (NtPR1a/c and NtPR2), JA-responsive gene (NtPR1b), and HR-associated gene (NtHSR203J). The sequences of the all the primers used for analysis were listed in Table 1. The NtActin gene served as the internal control. The analysis was performed for three biological replicates of each sample.
Extraction and Quantification of Endogenous ABA, GA3, JA, and SA
Extraction and quantification of ABA, GA3, and JA were performed as described previously (Duan et al., 2017). The procedure for the measurement of SA was slightly modified from the method described Duan et al. (2017). The extract of SA was vacuum evaporated to remove the solvent at 40°C and dissolved in 0.5 mL Reagent 3 for HPLC analysis. The flowing phase was detected at a wave-length of 240 nm for 30 min. Each sample was monitored using HPLC (e2695, Waters, United States) equipped with a reversed phase Kromasil C18 column.
Statistical Analysis
The experimental design was a completely randomized block design with three replicates. Each replicate contained three plants. Statistical comparisons were implemented using the SPSS statistical software (ver.16.0, SPSS Inc., Chicago, IL, United States) by one-way analysis of variance. A Tukey’s test was conducted to evaluate treatment efforts. Error bars represent standard deviations (SD) with different letters at P < 0.05 significant level.
Results
Molecular Confirmation of Transgenic Tobacco Plants Overexpressing SoCYP85A1
The SoCYP85A1 gene was inserted in pB7WG2D,1-GFP to create the overexpressing vector. T4 generation of transgenic tobacco plants was confirmed by PCR analysis (Figure 1A) and fluorescence detection of GFP protein (Figure 1B). The 1,392-bp fragment of the SoCYP85A1 gene was amplified in the transgenic lines. Further analysis by semi-quantitative RT-PCR confirmed successful integration of SoCYP85A1 in all the tested transgenic lines (Figure 1C), among which, the transgenic line 8 (L8) had the highest level of the overexpressed transcript.
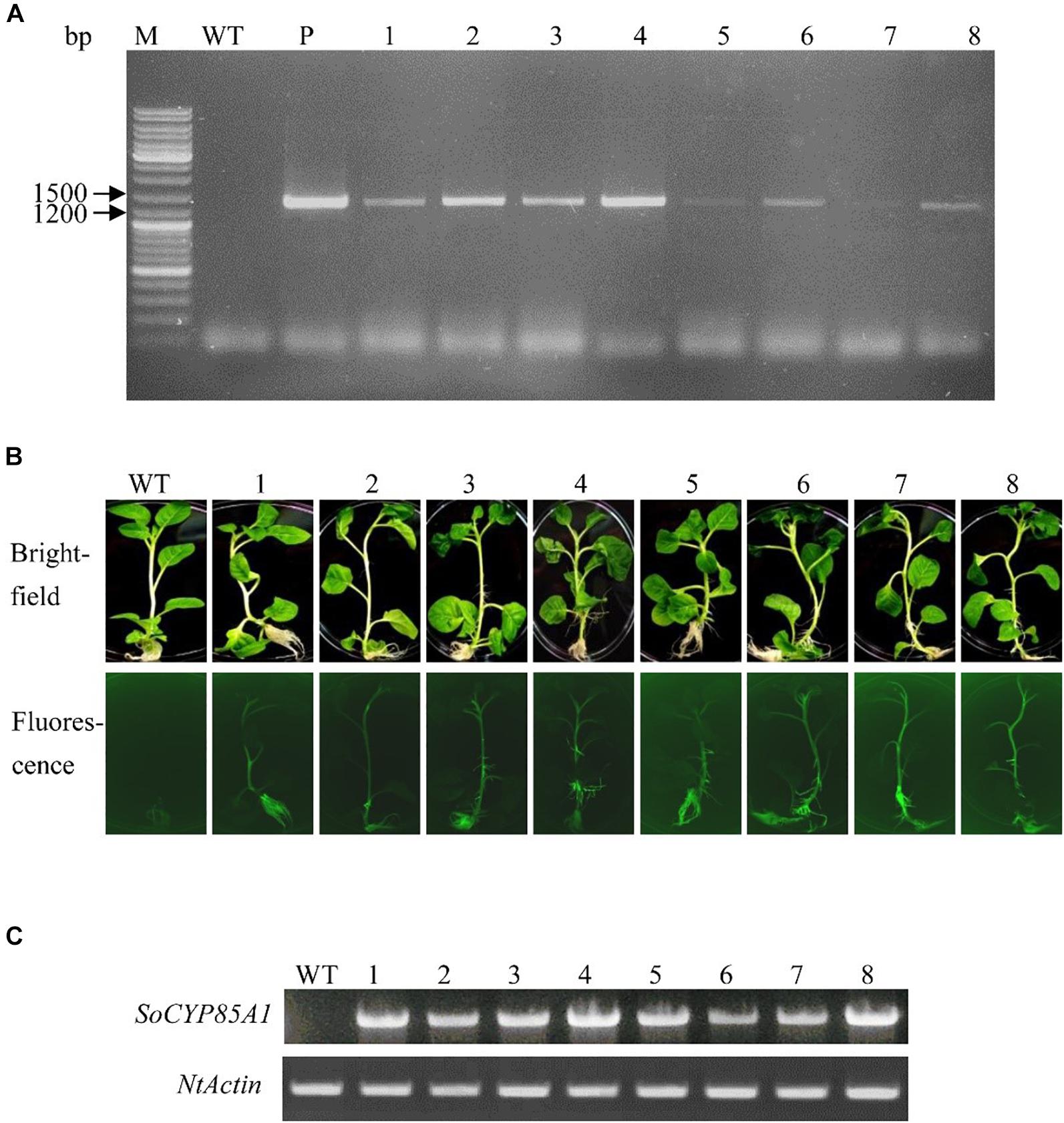
Figure 1. Molecular identification of transgenic tobacco lines. (A) PCR analysis of genomic DNA with specific primers for the ORF of SoCYP85A1. (B) GFP analysis of transgenic lines. (C) RT-PCR analysis of WT and different transgenic lines. M, Molecular marker; WT, Wild type; P, Plasmid; 1–8: transgenic lines.
Overexpression of SoCYP85A1 Enhances the Resistance of Tobacco Plants to P. nicotianae
To investigate whether the overexpression of SoCYP85A1 could enhance the resistance to P. nicotianae, the T4 transgenic tobacco plants were inoculated with P. nicotianae. All the transgenic lines exhibited enhanced but different levels of resistance to the pathogen (Table 2). Among the eight transgenic lines, L4 and L8 possessed higher resistance, with both lower disease rate and disease index, whereas the stem of WT plants displayed severe necrosis, with 7.23% disease index and 75.56% rate at 7 dpi. As shown in Figure 2A, the infection of pathogen resulted in a slightly necrotic stem and wilting of leaves in the WT plants, whereas no symptoms were observed at 4 dpi in the L8 line, which accumulated the highest level of the SoCYP85A1 transcript. Extremely severe wilting, necrosis, and lodging symptoms appeared in the WT seedlings at 7 dpi, whereas only faint wilting symptoms were observed in L8 plants (Figure 2B). The above results indicated that the overexpression of SoCYP85A1 notably enhanced the resistance against P. nicotianae in transgenic tobacco plants. Based on the results, L4 and L8 were selected as the representative lines for further analyses.
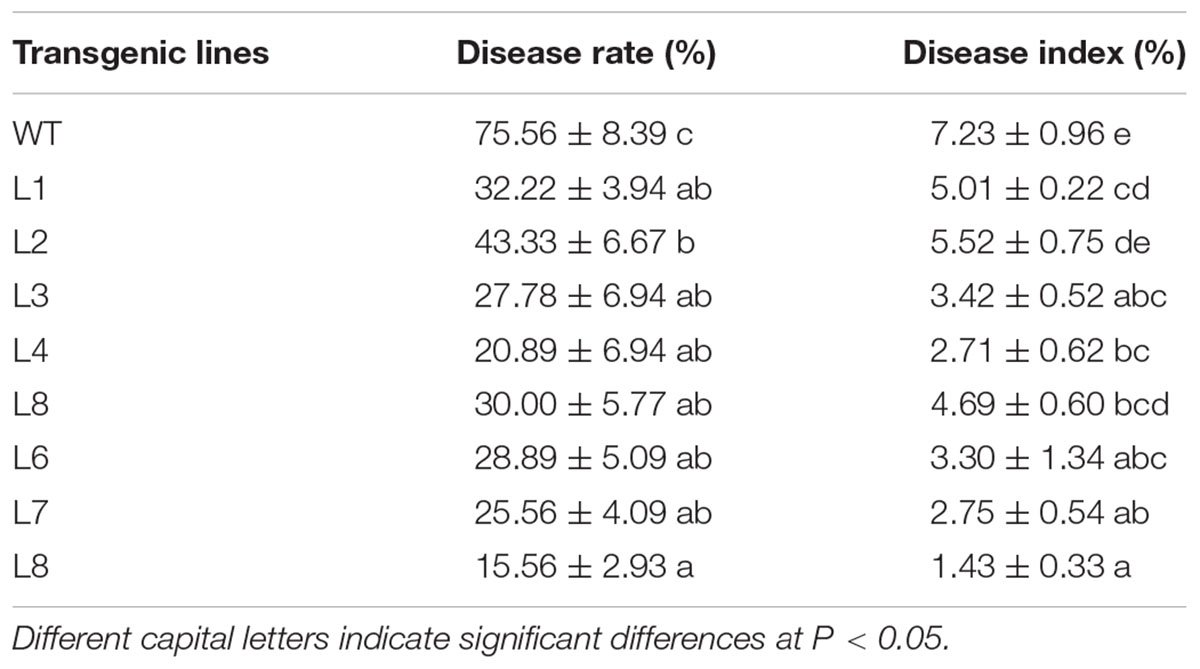
Table 2. Disease rate and disease index of wild-type (WT) and different transgenic lines after inoculation with Phytopthora nicotianae.

Figure 2. Tobacco plants inoculated with Phytopthora nicotianae. (A) Phenotypes of wild-type (WT) and L8 line infected with P. nicotianae at 4 days post inoculation (dpi) and (B) 7 dpi, respectively.
Overexpression of SoCYP85A1 Increases the Accumulation of SoCYP85A1 Transcript and CS After Pathogen Inoculation
The qRT-PCR results showed that the transcription of SoCYP85A1 was not detectable in the un-infected or infected WT plants. In contrast, pathogen infection induced a significant increase in the transcript level of SoCYP85A1 in the transgenic lines (Figure 3A).
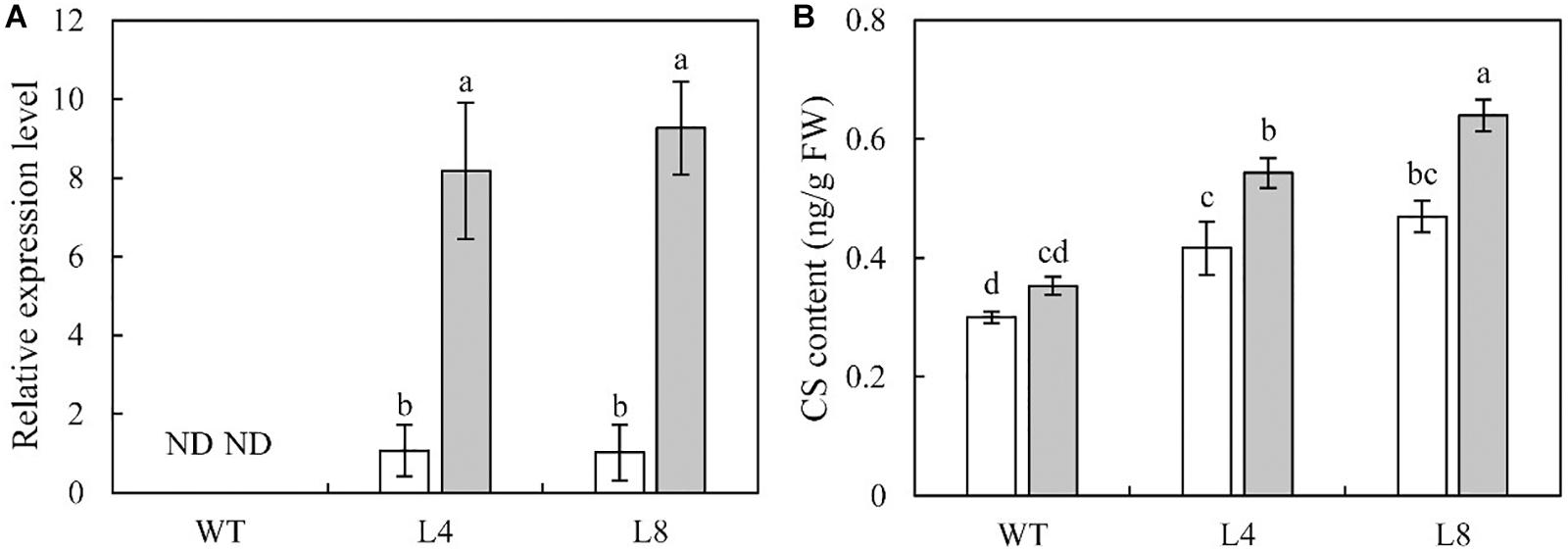
Figure 3. Effect on castasterone (CS) content in wild-type (WT), and transgenic lines 4 and 8 inoculated with P. nicotianae. (A) Expression level of SoCYP85A1 and (B) CS content in WT and transgenic lines 4 and 8 before and after infection with P. nicotianae at 4 days post inoculation (dpi). Different capital letters indicate significant differences at P < 0.05.
To understand whether SoCYP85A1 indeed functions in BRs biosynthesis, the endogenous BRs in the WT and transgenic plants were examined before and after the inoculation of the pathogen. The intermediates in BR biosynthetic pathway, which include teasterone (TE), typhasterol (TY), 6-deoxo castasterone (6-deoxoCS), and 28-norcastasterone (28-norCS) were undetectable in both WT and transgenic lines under the absence and presence of the pathogen. No significant changes were observed in the content of BL. However, compared to WT plants, an obvious increase in the content of CS was detected in L4 and L8 plants. Furthermore, when subjected to pathogen infection, WT plants showed a slight increase in the content of CS, whereas L4 and L8 showed a remarkable increase (Figure 3B). These results suggested that overexpression of SoCYP85A1 enhanced the content of CS in the transgenic tobacco plants. Moreover, pathogen infection resulted in a significant increase in CS content in the transgenic plants.
Overexpression of SoCYP85A1 Reduces the Accumulation of ABA, GA3, JA, and SA After P. nicotianae Infection
To further explore the potential function of SoCYP85A1 upon P. nicotianae infection, HPLC analysis of leaf extracts from WT and transgenic plants was performed to quantify the endogenous contents of ABA, GA3, JA, and SA. No significant differences in the contents of the phytohormones were detected between the WT and transgenic seedlings before pathogen treatment, with the exception of SA, whose accumulation was much higher in WT than in the transgenic lines (Figure 4D). Significantly, WT plants had higher contents of the four phytohormones compared to those in transgenic plants after pathogen infection. It was worth noting that the contents of GA3 and SA decreased sharply in L4 and L8 lines after the pathogen attack (Figures 4B,D). Conversely, transgenic plants overexpressing SoCYP85A1 showed a higher accumulation of JA upon pathogen infection (Figure 4C). However, the ABA content was increased slightly in the transgenic seedlings. These results highlighted that alteration of CS content could alter the abundance of other phytohormones.
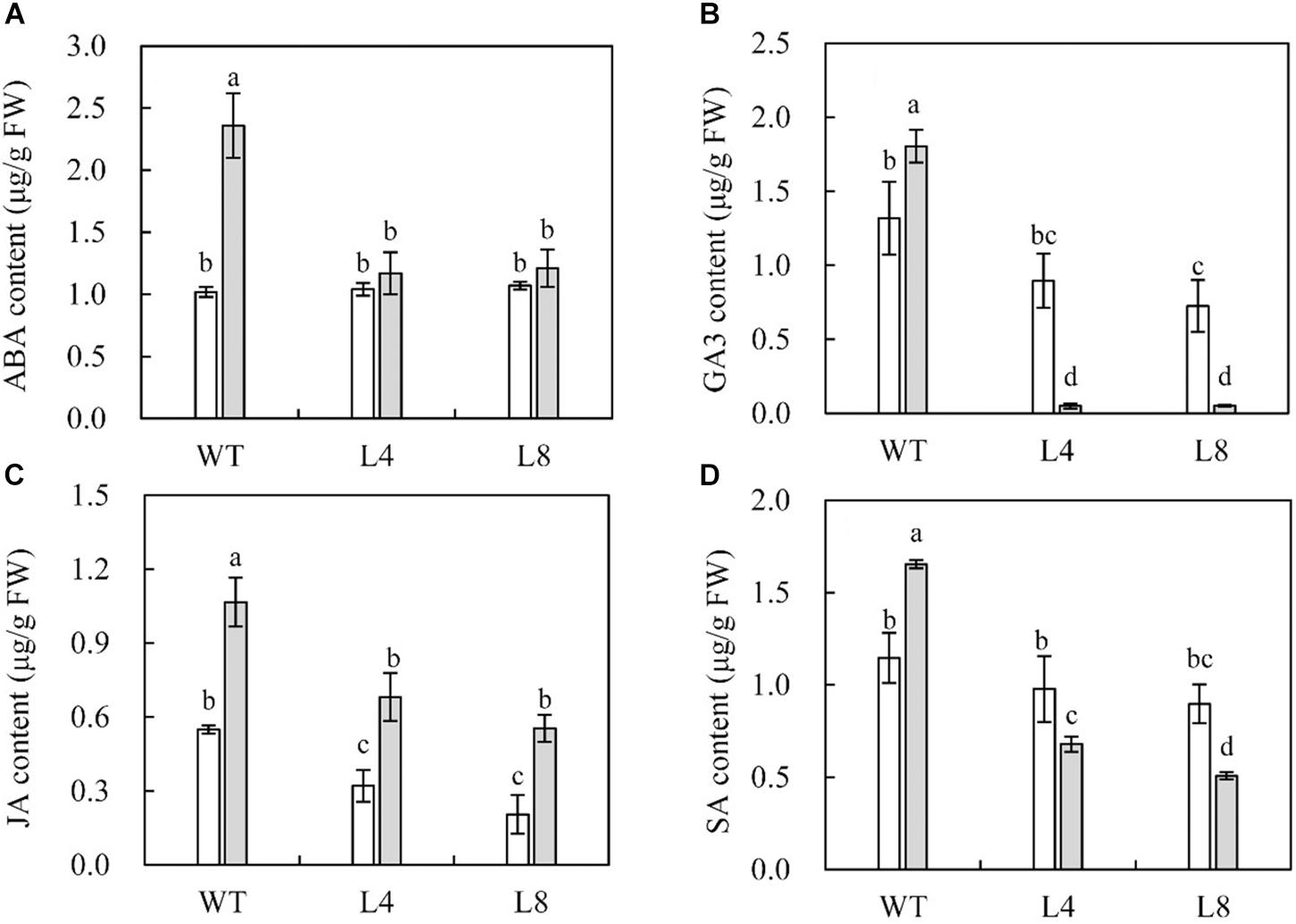
Figure 4. Contents of endogenous (A) abscisic acid (ABA), (B) gibberellic acid (GA3), (C) jasmonic acid (JA), and (D) salicylic acid (SA) in wild-type (WT) and transgenic plants under control and P. nicotianae inoculation conditions at 4 days post inoculation (dpi). Different capital letters indicate significant differences at P < 0.05.
Overexpression of SoCYP85A1 Increases the Activities of Some Defense Enzymes After P. nicotianae Infection
To examine whether SoCYP85A1 overexpression would cause physiological changes in transgenic lines, the activities of four key defense enzymes (SOD, POD, PPO, and CAT) were determined in the WT and transgenic plants with and without pathogen inoculation. The results demonstrated that before inoculation the activities of these enzymes were not much different between the WT and the two transgenic lines (Figure 5). However, when exposed to the pathogen, all the treated seedlings displayed an obvious increase in the activities of SOD, POD, PPO, and CAT, which were significantly higher in L8, showing 58, 51, 78, and 76% increase, respectively, compared to those in the WT seedlings (Figure 5). These results demonstrated that overexpression of SoCYP85A1 could enhance the activities of defense enzymes in transgenic seedlings upon challenge with P. nicotianae.
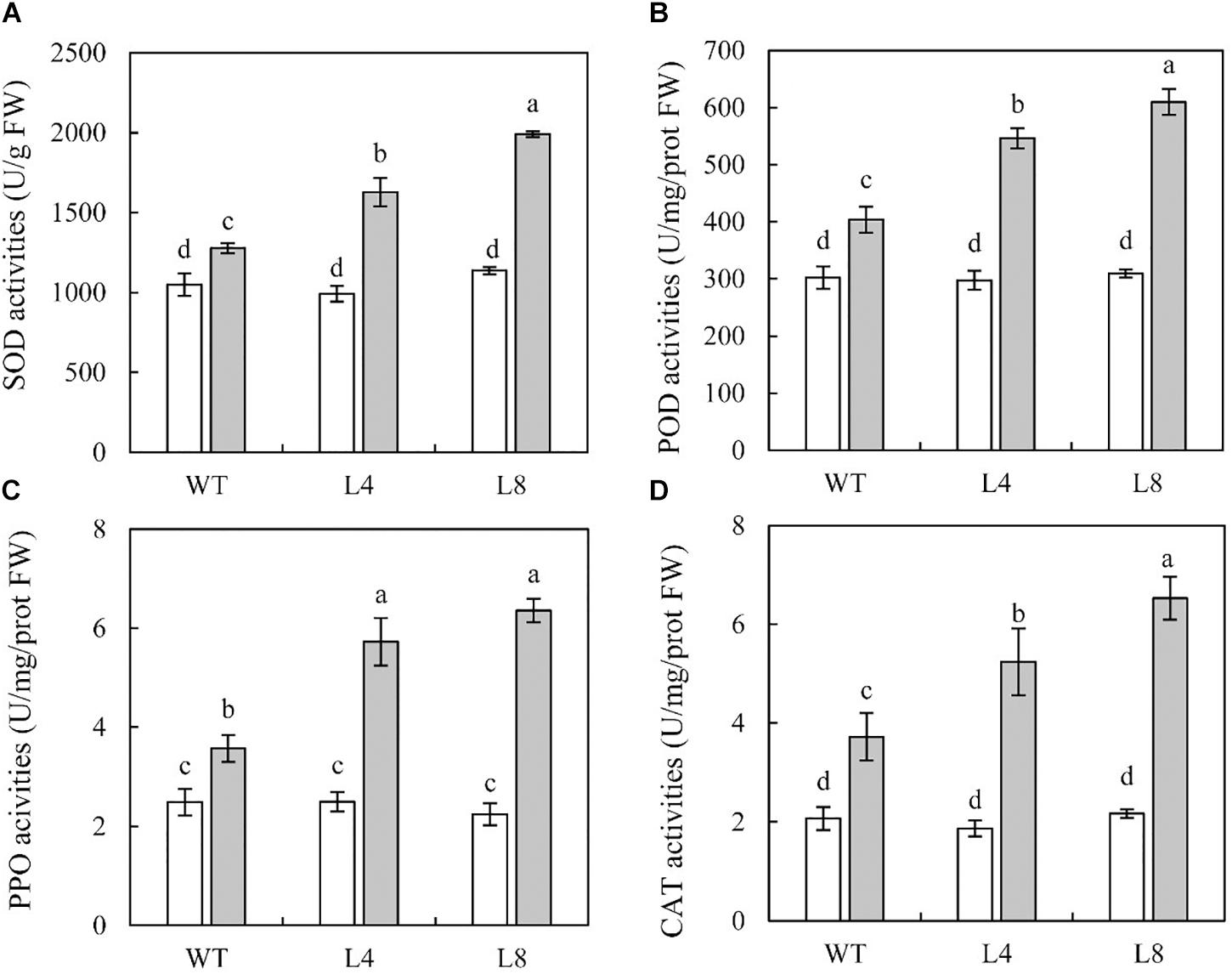
Figure 5. Activities of defense-related enzymes in wild-type (WT) and transgenic lines inoculated with P. nicotianae at 4 days post inoculation (dpi). (A) superoxide dismutase (SOD), (B) peroxidase (POD), (C) polyphenol oxidase (PPO), and (D) catalase (CAT). Different capital letters indicate significant differences at P < 0.05.
Overexpression of SoCYP85A1 Increases the Transcript Levels of Genes Encoding Defense Enzymes After P. nicotianae Infection
For deciphering the molecular mechanism through which the overexpression of SoCYP85A1 could enhance the resistance to P. nicotianae, qRT-PCR analysis was carried out to detect the transcript levels of genes in both WT and transgenic plants before and after the pathogen infection. In the absence of pathogen infection, no obviously differences were detected in the transcript levels of genes in WT and transgenic seedlings, with the exception of NtPR2, which exhibited a slight or significant decrease in L4 and L8 lines, respectively (Figure 6). However, when subjected to pathogen treatment, the transcript levels of NtSOD, NtAPX, NtPPO, and NtCAT were notably enhanced in transgenic plants compared to those in the WT plants, while the transcript levels of NtNCED1, NtPR1b, and NtHSR203J were lower in the transgenic lines than those in the WT plants (Figure 6). Pathogen infection resulted in a decrease in the abundance of NtPR1a/c and NtPR2 transcripts in both the L4 and L8 lines. This result, together with those mentioned above, demonstrate that overexpression of SoCYP85A1 could enhance the resistance of tobacco plants to P. nicotianae infection through upregulation of the expression of genes encoding defense enzymes at the transcriptional level.
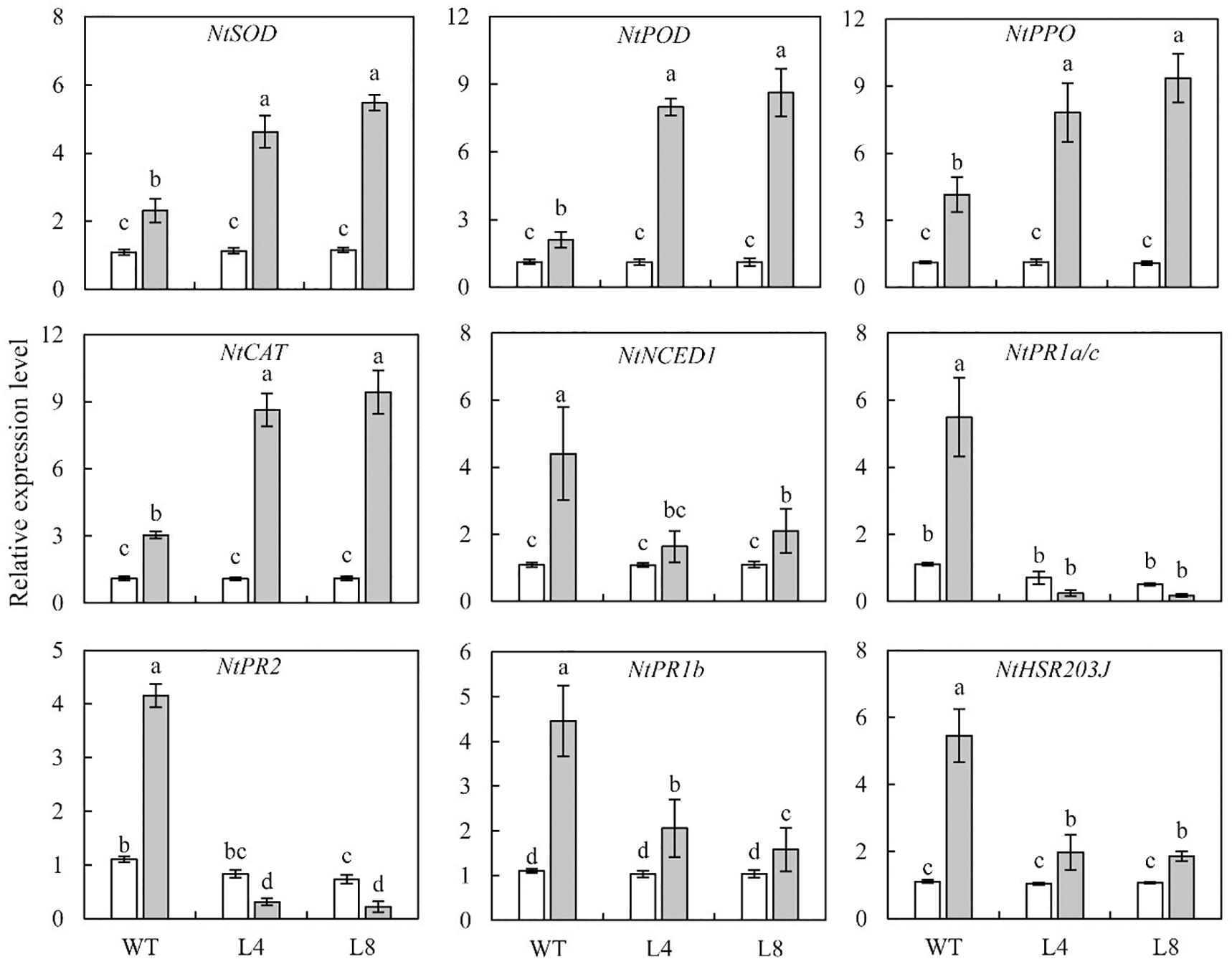
Figure 6. Relative transcript levels of genes involved in the resistance of wild-type (WT) and transgenic lines to P. nicotianae infection at 4 days post inoculation (dpi). Different capital letters indicate significant differences at P < 0.05.
Discussion
In previous studies, the effects of BRs on plants were mostly investigated using exogenous application of BRs (Nemhauser et al., 2006). Thus, the role of the endogenous BRs in the regulation of plant diseases remained elusive and required further experimentation. Although the function of endogenous BRs in the growth and development of plants was amply highlighted in previous research (Pérez-España et al., 2011; Li et al., 2016a,b), their involvement in plant’s response to pathogen invasion needed to be verified. Here, we provided evidence that an increase in the content of endogenous BRs could enhance the resistance of tobacco plants overexpressing SoCYP85A1 to P. nicotianae by enhancing the activities of defense enzymes. Our results provided a potential approach for enhancing black shank tolerance of tobacco plants through the regulation of genes involving in the biosynthetic pathway of BRs.
SoCYP85A1 Was First Identified to Enhance the Resistance to P. nicotianae in Tobacco Transformants
Over the past few years, molecular biology studies have been conducted to investigate the interaction of tobacco plants with P. nicotianae. Lipoxygenase (LOX) was the first proved enzyme during this interaction. The expression of antisense LOX gene could suppress the incompatible interaction between tobacco and this pathogen, which suggested the crucial role of LOX in the defense of host plants (Rancé et al., 1998). Subsequently, several defense-related genes had been identified to play a role in the response to black shank disease in tobacco, among which, the expressions of glutathione S transferase (GST), hsr203J, and RING finger protein genes, were reported to be highly induced during compatible and incompatible interactions with the pathogen (Chacón et al., 2009). On the contrary, functional analysis indicated that silencing of GST resulted in a significant increase in the resistance of plants to black shank disease as a result of upregulation of some defense genes (Hernández et al., 2009). These inconsistent results might sound reasonable because different GST genes were found to respond to fungal infection in different ways (Dean et al., 2005). Another type of defense-related genes includes serine/threonine protein kinase genes, which encode receptor proteins that mediate signal transduction in plant defense response. The overexpression of NrSTK significantly enhanced resistance to black shank in tobacco (Gao et al., 2015). In the present study, a novel category of the gene conferring resistance to black shank in tobacco was characterized as a cytochrome P450 gene, named as SoCYP85A1. Transgenic tobacco plants overexpressing SoCYP85A1 showed enhanced resistance to P. nicotianae, although different transgenic lines exhibited different degrees of resistance to this pathogen. Among them, the L8 showed the highest resistance to black shank disease, which could result from the greatest induction at the transcriptional level. These results demonstrated that SoCYP85A1 could participate in conferring resistance against P. nicotianae in plants.
CS Is a Bioactive Component Which Is Induced in Tobacco Plants Upon P. nicotianae Infection
To dissect the elaborate biological function of SoCYP85A1, we overexpressed this gene in tobacco plants and examined the endogenous content of BRs. The result showed that most of the intermediates involved in the BR biosynthetic pathway were undetectable, with the exception of CS, which was accumulated remarkably. Similar results were obtained in tomato and poplar plants overexpressing PtCYP85A3 (Jin et al., 2017). However, the endogenous level of CS in the 35S-CYP85A1 Arabidopsis was almost the same as that in wild-type, whereas high levels of 28-norCS, CS, and BL were detected in transgenic Arabidopsis plants overexpressing AtCY85A2 (Kim et al., 2005). The dissimilarity could be explained by the fact that the CYP85 family members function uniquely in BR metabolism in different species. Taken together, SoCYP85A1 might be a functional allele of PtCYP85A3 and AtCY85A2.
In addition, transgenic plants overexpressing SoCYP85A1 showed significant accumulation of CS, especially when subjected to P. nicotianae infection, which implied that CS might be a pathogen-infection-induced compound. Besides, the increased content of CS conferred enhancement of black shank resistance, indicating that CS might act as a bioactive component functioning in resistance to pathogen invasion. As another bioactive component among the BRs, BL is one of the most potent steroid hormones in plants. In Arabidopsis, an interplay between the plant immune system and BL was elucidated that BL biosynthesis could modulate the immunity of plants through BRI1-Associated Kinase 1 (BAK1) (Belkhadir et al., 2012). Therefore, BL was demonstrated to be involved in the response of Arabidopsis plants to pathogen attack. However, unexpectedly, in the present study, the BL content displayed no significant changes in both the WT and transgenic tobacco plants before and after pathogen treatment and rather CS was confirmed to be involved in the response of plants to infection. Regarding these inconsistencies, it appeared reasonable to assume that BRs might exert their biological effects through different intermediates or through the final product in different species. This assumption was also supported by the fact that 28-nor CS was identified as the bioactive compound in tomato (Li et al., 2016a).
Alteration in the Content of CS Interferes With Homeostasis of Phytohormones
During the last decade, various phytohormones have been identified to be involved in the modulation of plant–pathogen interactions (Zhou et al., 2005; Fan et al., 2009; Vlot et al., 2009; De et al., 2012; Nagel et al., 2017). BRs are important phytohormones involved in various aspects of plant development (Kim et al., 2005; Li et al., 2016a,b). Recently, a number of evidences have indicated that BRs can also participate in the resistance of plants to pathogens (Albrecht et al., 2012; Belkhadir et al., 2012; Prince et al., 2014). However, the interplay between BRs and other phytohormones has remained unresolved.
In this research, we studied the interaction between CS and four other phytohormones (ABA, GA, JA, and SA) by taking advantage of the fact that the concentrations of CS are highly regulated in transgenic seedlings overexpressing SoCYP85A1, especially when attacked by P. nicotianae. In the absence of the pathogen, the contents of ABA, GA, and JA in transgenic seedlings displayed no obvious changes, compared to the levels in the WT plants. We speculated that this was because of the fact that although CS was significantly accumulated, this increase did not have an effect on the three phytohormones. When CS was accumulated up to a certain level, it was worth noting that the concentrations of the four hormones in transgenic plants were much less than those in the WT plants upon challenge with the pathogen. Based on the result, the antagonistic interaction between CS and the three hormones could not take place until its content reached a critical level; however, even lower content of CS could reduce the biosynthesis of SA. Therefore, upon pathogen challenge, CS could act antagonistically on ABA, GA, JA, and SA by upregulating the CYP85A1 expression in tobacco plants. A similar interaction was observed between BRs and JA, wherein JA was shown to interrupt BR signaling by downregulating the expression of CYP90B1 during the response of plants to pathogens (Kim et al., 2013).
Activities of Antioxidant Enzymes Are Upregulated by SoCYP85A1 at Transcriptional Level
To resist pathogen invasion, plants have evolved defense mechanisms, in which different enzymes play the most important roles (Lebeda et al., 2001). In our study, we observed that the activities of SOD, POD, PPO, and CAT were highly regulated in L8 compared to those in the WT plants, which indicated that transgenic lines possessed more efficient detoxification system against phytopathogen attack. It was reported that a combination of these antioxidant enzymes in plants might contribute to greater resistance of plants to the pathogens (Hafez et al., 2012; Leite et al., 2014).
To further explore the mechanism through which SoCYP85A1 enhanced black shank resistance at the molecular level, qRT-PCR was conducted to examine the transcript levels of defense-related genes. The transgenic lines overexpressing SoCYP85A1 exhibited significant induction of the four genes encoding antioxidant enzymes at the transcriptional level when attacked by the pathogen. These results correspond with those for enzyme activities, implying that the activities of antioxidant enzymes were remarkably increased because of upregulation of the transcription of related genes by SoCYP85A1. A similar mechanism was also revealed in a previous study that increased transcription of defense enzyme genes conferred resistance to P. nicotianae in transgenic tobacco plants (Hernández et al., 2009).
The pathogen attack, however, did not induce increased transcript accumulation of ABA biosynthetic gene (NtNCED1), SA-responsive genes (NtPR1a/c and NtPR2), JA-responsive gene (NtPR1b), and the HR-associated gene (NtHSR203J) in transgenic lines, in consonance with the results of ABA, JA, and SA quantification (Figure 4). This implied that the above genes were not transcriptionally upregulated by SoCYP85A1 in tobacco plants. Further, we speculated that overexpression of SoCYP85A1 enhanced black shank tolerance in an ABA-, JA-, and SA-independent manner.
Based on the above findings, we concluded that the enhanced resistance of transgenic plants to pathogen infiltration was a result of increased CS accumulation and improved activities of antioxidant enzymes, which were transcriptionally regulated by SoCYP85A1 via an ABA, JA, and SA-independent pathway. However, although the influence of SoCYP85A1 on pathogen resistance was studied in the present study, a comprehensive and elaborate elucidation of the regulatory mechanism underlying the defense response of these transgenic tobacco plants to pathogen invasion would require further investigation.
Author Contributions
WS designed the research and wrote the manuscript. FD characterized the transgenic plants and conducted all the experiments.
Funding
This work was supported by Qingdao Science and Technology Project (16-5-1-54-jch), Research fund for the Doctor Program of Higher Education (663/1111316), and Taishan Scholars Project of Shandong Province.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Albrecht, C., Boutrot, F., Segonzacb, C., Schwessingerb, B., Gimenez-Ibanezb, S., Chinchillac, D., et al. (2012). Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc. Natl. Acad. Sci. U.S.A. 109, 303–308. doi: 10.1073/pnas.1109921108
Arteca, J. M., and Arteca, R. N. (2001). Brassinosteroid-induced exaggerated growth in hydroponically grown Arabidopsis plants. Physiol. Plant 112, 104–112. doi: 10.1034/j.1399-3054.2001.1120114.x
Belkhadir, Y., Jaillais, Y., Epple, P., Balsemão-Pires, E., Dangl, J. L., and Chory, J. (2012). Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc. Natl. Acad. Sci. U.S.A. 109, 297–302. doi: 10.1073/pnas.1112840108
Chacón, O., Hernández, I., Portieles, R., López, Y., Pujol, M., and Borrás-Hidalgo, O. (2009). Identification of defense-related genes in tobacco responding to black shank disease. Plant Sci. 177, 175–180. doi: 10.1016/j.plantsci.2009.05.009
Choudhary, S. P., Yu, J. Q., Yamaguchi-Shinozaki, K., Shinozaki, K., and Tran, L. S. (2012). Benefits of brassinosteroid crosstalk. Trends Plant Sci. 17, 594–605. doi: 10.1016/j.tplants.2012.05.012
De, V. D., Van, B. E., Satoh, K., Balidion, J., Mauleon, R., Choi, I. R., et al. (2012). Brassinosteroids antagonize gibberellin- and salicylate-mediated root immunity in rice. Plant Physiol. 158, 1833–1846. doi: 10.1104/pp.112.193672
Dean, J. D., Goodwin, P. H., and Hsiang, T. (2005). Induction of glutathione s–transferase genes of Nicotiana Benthamiana following infection by colletotrichum destructivum and C. orbiculare and involvement of one in resistance. J. Exp. Bot. 56, 1525–1533. doi: 10.1093/jxb/eri145
Ding, J., Mao, L. J., Wang, S. T., Yuan, B. F., and Feng, Y. Q. (2013). Determination of endogenous brassinosteroids in plant tissues using solidphase extraction with double layered cartridge followed by high-performance liquid chromatography-tandem mass spectrometry. Phytochem. Anal. 24, 386–394. doi: 10.1002/pca.2421
Divi, U. K., Rahman, T., and Krishna, P. (2010). Brassinosteroid-mediated stress tolerance in Arabidopsis shows interactions with abscisic acid, ethylene and salicylic acid pathways. BMC Plant Biol. 10:151. doi: 10.1186/1471-2229-10-151
Duan, F. M., Ding, J., Lee, D., Lu, X. L., Feng, Y. Q., and Song, W. W. (2017). Overexpression of SoCYP85A1, a spinach cytochrome p450 gene in transgenic tobacco enhances root development and drought stress tolerance. Front. Plant Sci. 8:1909. doi: 10.3389/fpls.2017.01909
Fan, J., Hill, L., Crooks, C., and Lamb, D. C. (2009). Abscisic acid has a key role in modulating diverse plant-pathogen interactions. Plant Physiol. 150, 1750–1761. doi: 10.1104/pp.109.137943
Fang, W., Chen, W., Yan, Y., Haihong, J., and Xingqi, G. (2016). Overexpression of cotton Ghmpk11 decreases disease resistance through the gibberellin signaling pathway in transgenic Nicotiana Benthamiana. Front. Plant Sci. 7:00689. doi: 10.3389/fpls.2016.00689
Gallup, C. A., Mccorkle, K. L., Ivors, K., and Shew, H. D. (2018). Characterization of the black shank pathogen, Phytophthora nicotianae, across North Carolina tobacco production areas. Plant Dis. 102, 1108–1114. doi: 10.1094/PDIS-02-17-0295-RE
Gao, S., Guo, W., Feng, W., Liu, L., Song, X., Chen, J., et al. (2016). Ltp3 contributes to disease susceptibility in Arabidopsis by enhancing ABA biosynthesis. Mol. Plant Pathol. 17, 412–426. doi: 10.1111/mpp.12290
Gao, Y., Xu, Z., Jiao, F., Yu, H., Xiao, B., Li, Y., et al. (2010). Cloning, structural features, and expression analysis of resistance gene analogs in tobacco. Mol. Biol. Rep. 37, 345–354. doi: 10.1007/s11033-009-9749-2
Gao, Y. L., Wang, B. W., Xu, Z. L., Li, M. Y., Song, Z. B., Li, W. Z., et al. (2015). Tobacco serine/threonine protein kinase gene NrSTK enhances black shank resistance. Genet. Mol. Res. 14, 16415–16424. doi: 10.4238/2015.December.9.11
Gruszka, D. (2013). Brassinosteroid signaling pathway-new key players and interconnections with other signaling networks crucial for plant development and stress tolerance. Int. J. Mol. Sci. 14, 8740–8774. doi: 10.3390/ijms14058740
Hafez, Y. M., Bacsó, R., Király, Z., Künstler, A., and Király, L. (2012). Up-regulation of antioxidants in tobacco by low concentrations of H2O2 suppresses necrotic disease symptoms. Phytopathology 102, 848–856. doi: 10.1094/PHYTO-01-12-0012-R
Hernández, I., Chacón, O., Rodriguez, R., Portieles, R., Yunior, L., Pujol, M., et al. (2009). Black shank resistant tobacco by silencing of glutathione s-transferase. Biochem. Bioph. Res. Commun. 387, 300–304. doi: 10.1016/j.bbrc.2009.07.003
Jin, Y. L., Tang, R. J., Wang, H. H., Jiang, C. M., Bao, Y., Yang, Y., et al. (2017). Overexpression of Populus trichocarpa CYP85A3 promotes growth and biomass production in transgenic trees. Plant Biotechnol. J. 15, 1309–1321. doi: 10.1111/pbi.12717
Johnson, E. S., Wolff, M. F., Wernsman, E. A., and Rufty, R. C. (2002). Marker-assisted selection for resistance to black shank disease in tobacco. Plant Dis. 86, 1303–1309. doi: 10.1094/PDIS.2002.86.12.1303
Kamoun, S., Furzer, O., Jones, J. D., Judelson, H. S., Ali, G. S., Dalio, R. J., et al. (2015). The Top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 16, 413–434. doi: 10.1111/mpp.12190
Kim, B., Fujioka, S., Mi, K., Jeon, J., and Choe, S. (2013). Arabidopsis brassinosteroid-overproducing gulliver3-D/DWARF4-D mutants exhibit altered responses to jasmonic acid and pathogen. Plant Cell Rep. 32, 1139–1149. doi: 10.1007/s00299-012-1381-2
Kim, T. W., Hwang, J. Y., Kim, Y. S., Joo, S. H., Chang, S. C., Lee, J. S., et al. (2005). Arabidopsis CYP85A2, a cytochrome p450, mediates the baeyer-villiger oxidation of castasterone to brassinolide in brassinosteroid biosynthesis. Plant Cell 17, 2397–2412. doi: 10.1105/tpc.105.033738
Lebeda, A., Luhová, L., Sedláøová, M., and Janèová, D. (2001). The role of enzymes in plant-fungal pathogens interactions. Zeitschrift Für Pflanzenkrankheiten Und Pflanzenschutz 108, 89–111.
Leite, M. E., dos Santos, J. B., Ribeiro, P. M., de Souza, D. A., de Castro Lara, L. A., and de Resende, M. L. V. (2014). Biochemical responses associated with common bean defence against Sclerotinia sclerotiorum. Eur. J. Plant Pathol. 138, 391–404. doi: 10.1007/s10658-013-0341-1
Li, X. J., Chen, X. J., Guo, X., Yin, L. L., Ahammed, G. J., Xu, C. J., et al. (2016a). DWARF overexpression induces alteration in phytohormone homeostasis, development, architecture and carotenoid accumulation in tomato. Plant Biotechnol. J. 14, 1021–1033. doi: 10.1111/pbi.12474
Li, X. J., Guo, X., Zhou, Y. H., Shi, K., Zhou, J., Yu, J. Q., et al. (2016b). Overexpression of a brassinosteroid biosynthetic gene Dwarf enhances photosynthetic capacity through activation of calvin cycle enzymes in tomato. BMC Plant Biol. 16:33. doi: 10.1186/s12870-016-0715-6
Livaka, K. J., and Schmittgenb, T. D. (2001). Analysis of relative gene expression data using real–time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.1262
Nagel, R., Turrini, P. C. G., Nett, R. S., Leach, J. E., Verdier, V., Van Sluys, M. A., et al. (2017). An operon for production of bioactive gibberellin A4 phytohormone with wide distribution in the bacterial rice leaf streak pathogen Xanthomonas oryzae pv. oryzicola. New Phytol. 214, 1260–1266. doi: 10.1111/nph.14441
Nakashita, H., Yasuda, M., Nitta, T., Asami, T., Fujioka, S., Arai, Y., et al. (2003). Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 33, 887–898. doi: 10.1046/j.1365-313X.2003.01675.x
National Standardization Management Committee. (2009). Grade and Investigation Method of Tobacco Diseases and Insect Pests. Available at: http://www.csres.com/detail/205150.html,
Nemhauser, J. L., Hong, F., and Chory, J. (2006). Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126, 467–475. doi: 10.1016/j.cell.2006.05.050
Noguchi, T., Fujioka, S., Choe, S., Takatsuto, S., Tax, F. E., Yoshida, S., et al. (2000). Biosynthetic pathways of brassinolide in Arabidopsis. Plant Physiol. 124, 201–209. doi: 10.1104/pp.124.1.201
Ohnishi, T., Godza, B., Watanabe, B., Fujioka, S., Hategan, L., Ide, K., et al. (2012). CYP90A1/CPD, a brassinosteroid biosynthetic cytochrome P450 of Arabidopsis, catalyzes C-3 oxidation. J. Biol. Chem. 287, 31551–31560. doi: 10.1074/jbc.M112.392720
Pérez-España, V. H., Sánchez-León, N., and Vielle-Calzada, J. P. (2011). CYP85A1 is required for the initiation of female gametogenesis in Arabidopsis thaliana. Plant Signal. Behav. 6, 321–326. doi: 10.4161/psb.6.3.13206
Prince, D. C., Drurey, C., Zipfel, C., and Hogenhout, S. A. (2014). The leucine-rich repeat receptor-like kinase brassinosteroid insensitive1-associated kinase1 and the cytochrome p450 phytoalexin deficient3 contribute to innate immunity to aphids in Arabidopsis. Plant Physiol. 164, 2207–2219. doi: 10.1104/pp.114.235598
Rancé, I., Fournier, J., and Esquerrétugayé, M. T. (1998). The incompatible interaction between Phytophthora nicotianae race 0 and tobacco is suppressed in transgenic plants expressing antisense lipoxygenase sequences. Proc. Natl. Acad. Sci. U.S.A. 95, 6554–6559. doi: 10.1073/pnas.95.11.6554
Shimada, Y., Fujioka, S., Miyauchi, N., Kushiro, M., Takatsuto, S., Nomura, T., et al. (2001). Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol. 126, 770–779. doi: 10.1104/pp.126.2.770
Shimada, Y., Goda, H., Nakamura, A., Takatsuto, S., Fujioka, S., and Yoshida, S. (2003). Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol. 131, 287–297. doi: 10.1104/pp.013029
Tang, Y., Liu, Q., Liu, Y., Zhang, L., and Ding, W. (2017). Overexpression of NtPR-Q up-regulates multiple defense-related genes in Nicotiana tabacum and enhances plant resistance to Ralstonia solanacearum. Front. Plant Sci. 8:1963. doi: 10.3389/fpls.2017.01963
Vlot, A. C., Dempsey, D. A., and Klessig, D. F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Yu, J. Q., Huang, L. F., Hu, W. H., Zhou, Y. H., Mao, W. H., Ye, S. F., et al. (2004). A role for brassinosteroids in the regulation of photosynthesis in Cucumis sativus. J. Exp. Bot. 55, 1135–1143. doi: 10.1093/jxb/erh124
Keywords: black shank disease, transgenic tobacco, SoCYP85A1, castasterone, defense enzymes
Citation: Duan F and Song W (2019) Overexpression of SoCYP85A1 Increases the Accumulation of Castasterone and Confers Enhanced Black Shank Tolerance in Tobacco Through Modulation of the Antioxidant Enzymes’ Activities. Front. Plant Sci. 10:349. doi: 10.3389/fpls.2019.00349
Received: 21 January 2019; Accepted: 07 March 2019;
Published: 22 March 2019.
Edited by:
Stephan Pollmann, Centro de Biotecnología y Genómica de Plantas (CBGP), SpainReviewed by:
Ramsey Steven Lewis, North Carolina State University, United StatesAtanas Ivanov Atanassov, Joint Genomic Center, Bulgaria
Copyright © 2019 Duan and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenwen Song, c29uZ3dlbndlbjIwMDJAMTYzLmNvbQ==
 Fangmeng Duan
Fangmeng Duan Wenwen Song
Wenwen Song