- 1Hunan Provincial Key Laboratory of Plant Resources Conservation and Utilization, College of Biology and Environmental Sciences, Jishou University, Jishou, China
- 2State Key Laboratory of Plant Genomics, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China
- 3Key Laboratory for the Creation Cotton Varieties in the Northwest, Ministry of Agriculture, Join Hope Seeds Corporation, Ltd., Changji, China
- 4Research Base, State Key Laboratory of Cotton Biology, Zhengzhou University, Zhengzhou, China
Verticillium wilt, caused by Verticillium dahliae, seriously limits cotton production. It is difficult to control this pathogen damage mainly due to the complexity of the molecular mechanism of plant resistance to V. dahliae. Here, we identified three homologous cotton Walls Are Thin (WAT) genes, which were designated as GhWAT1, GhWAT2, and GhWAT3. The GhWATs were predominantly expressed in the roots, internodes, and hypocotyls and induced by infection with V. dahliae and treatment with indole-3-acetic acid (IAA) and salicylic acid (SA). GhWAT1-, GhWAT2-, or GhWAT3-silenced plants showed a comparable phenotype and level of resistance with control plants, but simultaneously silenced three GhWATs (GhWAT123-silenced), inhibited plant growth and increased plant resistance to V. dahliae, indicating that these genes were functionally redundant. In the GhWAT123-silenced plants, the expression of SA related genes was significantly upregulated compared with the control, resulting in an increase of SA level. Moreover, the histochemical analysis showed that xylem development was inhibited in GhWAT123-silenced plants compared with the control. However, lignin deposition increased in the xylem of the GhWAT123-silenced plants compared to the control, and there were higher expression levels of lignin synthesis- and lignifications-related genes in the GhWAT123-silenced plants. Collectively, the results showed that GhWATs in triple-silenced plants acts as negative regulators of plant resistance against V. dahliae. The potential mechanism of the WATs functioning in the plant defence can modulate the SA biosynthesis and lignin deposition in the xylem.
Introduction
Cotton verticillium wilt caused by Verticillium dahliae is a soil-borne fungal vascular disease, leading to annual yield losses of more than 30% and a serious economic loss of approximately 250–310 million dollars in China (Gong et al., 2017). However, it is difficult to control the pathogen damage to cotton plants, although many attempts have been made, including attempts with fungicides and cultural approaches (Mohamed and Akladious, 2017; Wei and Yu, 2018). In general, to protect plants from pathogen damage, disease resistance cultivars should be widely planted. Although some genes/proteins have been characterised in cotton plant defence against pathogens, few effective candidate genes have been updated for use in disease resistance breeding (Cai et al., 2009; Zeng et al., 2016; Gong et al., 2017). Therefore, the molecular mechanism of plant resistance to V. dahliae and the functional dissection of defence-related genes need to be further explored.
The molecular mechanisms of cotton plant defence against V. dahliae are complex. Recently, SA mediated resistance and lignin, as the major physical barriers of cell wall, were reported to play an important role in cotton plant defence against V. dahliae (Pieterse et al., 2012; Seyfferth and Tsuda, 2014; Yan and Dong, 2014; Zhu et al., 2018). In SA-related resistance, the SA signalling pathway mainly participates in Arabidopsis plant defence through some hub components, including non-expressor of pathogenesis-related genes 1 (NPR1), NPR3/NPR4, TGAs and pathogenesis-related gene (PR) proteins (Shah, 2003; Fu et al., 2012; Moreau et al., 2012; Seyfferth and Tsuda, 2014; Yan and Dong, 2014; Ding et al., 2018). In cotton, many SA signalling-related genes have been characterised in plant resistance to V. dahliae (Chen et al., 2016; Yang et al., 2017; Gong et al., 2018). For instance, the expression of cotton ribosomal protein L18 (GaRPL18) was upregulated by SA treatments, and silencing GaRPL18 significantly reduced plant resistance to V. dahliae (Gong et al., 2017). The GhSNAP33 mediates cotton resistance to V. dahliae infection by SA treatments (Wang et al., 2018). Additionally, the SA synthesis pathways are also involved in plant defence (Durrant and Dong, 2004; Seyfferth and Tsuda, 2014; Zhou et al., 2018). Such as in Arabidopsis, isochorismate synthase 1 (ICS1) is the major required gene for SA synthesis; enhanced disease susceptibility 1 (EDS1) and phytoalexin deficient 4 (PAD4) positively regulate SA biosynthesis by a feedback loop and the mobilisation of salicylic acid defence (Wildermuth et al., 2001; Rietz et al., 2011). In cotton, ectopic expressions of GhPGIP1 can upregulate the expression levels of ICS1, EDS1, and PAD4, enhancing resistance to Verticillium (Liu et al., 2017). Cotton EDS1 can promote SA biosynthesis to enhance plant defence against V. dahliae (Su et al., 2014; Zhang et al., 2016, 2017). Together, the SA synthesis and signalling response participate in cotton resistance to V. dahliae.
Recently, an increasing number of reports have shown that lignin accumulation in the secondary cell walls of cotton plants enhanced plants sensitivity to V. dahliae pathogen infection and colonisation (Xu et al., 2011; Shi et al., 2012; Gao et al., 2013). For example, the ectopic expression of the cotton laccase gene GhLAC15 in Arabidopsis increased cell wall lignifications, strengthening resistance against V. dahliae (Zhang Y. et al., 2018). The lignin contents in different cotton varieties were positively correlated with plant resistance against V. dahliae, resulting from increasing lignifications and cross-linking of lignin components (Xu et al., 2011). An ethylene response-related factor GbERF1-like gene contributed to the resistance to V. dahliae through positively regulating lignin synthesis-related genes and enhancing the lignin content (Guo et al., 2016). Interestingly, GhUMC1 was identified to be involved in cotton resistance to V. dahliae through both the SA signalling pathway and lignin synthesis (Zhu et al., 2018). Additionally, other hosts of Verticillium pathogens can increase their resistance through lignin deposition in the secondary cell wall or vascular tissue. For instance, Verticillium infection triggers de novo xylem formation in Arabidopsis, which depends on NAC domain 7 to maintain plant growth under stress (Reusche et al., 2012). Lignin composition and accumulation in plants contributed to resisting V. dahliae colonisation by strengthening the cell walls in pepper (Novo et al., 2017). The tomato R gene Ve participated in the plant resistance response to V. dahliae by increasing lignin- and PAL-related gene expression (Gayoso et al., 2010). Together, these studies documented that SA and lignin play important roles in plant defence against Verticillium pathogen infection. However, some novel SA- and/or lignin-related genes remain to be mined for cotton disease resistance breeding.
Arabidopsis walls are thin 1 (WAT1), which is a Medicago truncatula NODULIN 21 (MtN21) homologue gene, was identified as a member of the drug/metabolite transporter (DMT) superfamily (Gamas et al., 1996; Ranocha et al., 2010). WAT1 functions in the regulation of auxin polar transport, secondary cell wall deposition and cell elongation (Ranocha et al., 2010). WAT1 is localised in the tonoplast and mediates auxin transport across the tonoplast from the vacuole to external space, regulating the homeostasis of auxin intracellular metabolism (Ranocha et al., 2010, 2011, 2013). Recent reports have shown that WAT1 participates in plant broad-spectrum resistance to vascular pathogens by regulating SA metabolism and signalling transduction through affecting auxin polar transport (Ranocha et al., 2010, 2013; Denancé et al., 2013). Thus, WAT1 participates in plant defence against vascular pathogens, possibly involved in SA synthesis and secondary cell wall lignifications.
In this study, we identified three cotton WAT genes, GhWAT1, GhWAT2, and GhWAT3, and dissected their function in plant defence against V. dahliae. The knockdown of three GhWATs by a virus-induced gene silencing (VIGS) method can increase plant resistance. According to molecular, biochemical and histological analyses, the results showed that simultaneously silencing three GhWATs promoted the expression of SA synthesis-related genes and increase SA level. However, the knockdown of GhWATs increases lignin deposition in the xylem sections. Collectively, the data showed that GhWATs as negative regulators increases disease resistance in silenced-plants infected by V. dahliae, possibly resulting from modulating SA synthesis and lignin accumulation.
Materials and Methods
Plant Materials, Growth Conditions, and Hormone Treatment
G. hirsutum cv. BD18 seeds were soaked in tap water at 37°C overnight, and then the imbibed seeds were placed on wet tissue to germinate in the dark at 28°C for 2 days. Similar germinating seeds were transferred into a sterilised soil mixture (a complex of organic matter soil and vermiculite) or to hydroponic conditions (Hoagland nutrient solution) in a growth chamber at 25°C with a 16/8 h light/dark photoperiod and 75% relative humidity.
N. benthamiana was cultivated in a sterilised soil mixture with the same conditions as described for G. hirsutum growth for the subcellular location analysis.
To analyse the gene response to phytohormones, 3-week-old seedlings were transferred to Hoagland nutrient solution. IAA or SA was added into culture solution. IAA and SA were adjusted to 50 μM and 2 mM, respectively (Shi et al., 2010; Hu et al., 2018). The seedlings were mock-treated with the corresponding concentration of ethanol at the same time. Then roots were harvested for RNA extraction at 0, 0.25, 0.5, 1, 4, 6, and 12 h after hormone and mock treatment. Over 10 seedlings were collected at each time point. The experiments were performed with three biological replicates.
Phylogenetic Analysis
The three homologous GhWATs were retrieved in the Cottongen database1. The functional domains of GhWATs and the transmembrane helices were predicted using UniProt database2. Protein hydrophobicities analyses were performed by DNAMAN 7.0. BlastP was employed to analyse GhWATs homologous sequences in NCBI3. The amino acid sequence alignment and phylogenetic relationship analyses were performed on ClustalX 2.1 and MEGA5.0, respectively.
VIGS Mediated by Agrobacterium tumefaciens
Virus-induced gene silencing technology has been successfully reported to determine cotton gene function (Gao et al., 2013; Pang et al., 2013). Specific fragments of the GhWAT1, GhWAT2, GhWAT3 and positive control GhPDS genes were amplified by PCR, were digested with BamH I and Kpn I and then cloned into the tobacco rattle virus (TRV) vector pYL156, generating pYLW1, pYLW2, pYLW3, and pYLPDS vectors, respectively. In addition, we cloned shorter fragments of GhWAT1, GhWAT2, and GhWAT3 by PCR with corresponding specific primers, and these were tandemly inserted into pYL156 to form the pYLW123 vector. Then, all pYL-candidate gene vectors and the auxiliary vector pYL192 were transformed into A. tumefaciens strains GV3101 by electroporation. A. tumefaciens containing the indicated vectors was cultured in LB medium with 50 μg/mL kanamycin and 40 μg/mL rifampicin at 28°C overnight. The Agrobacterium cells were collected and then resuspended in MMA solution (10 mM N-morpholino ethanesulfonic acid, 10 mM MgCl2, and 200 mM acetosyringone). Every suspension concentration was adjusted to a 1.2 value of OD600. The Agrobacterium cells containing the aforementioned vectors were equally mixed with those containing pYL192 and were incubated at room temperature for 3 h in the dark. Finally, the mixed Agrobacterium cells were injected into the fully expanded cotyledons of 7-day-old seedlings by a 1-mL needleless syringe. The experiment was performed three times. Each sample comprised of more than 50 cotyledons. The primers used in the experiment are shown in Supplementary Table S1.
Pathogen Infection and Plant Disease Assays
The highly aggressive defoliating fungal pathogen V. dahliae strain V991 was provided by the Institute of Plant Protection, CAAS, Beijing, China. And then it was cultured on potato dextrose agar plates for 3 days at 25°C in the dark. Then, hyphae were transferred into Czapek’s medium (NaNO3, 0.3% w/v; MgSO4, 0.1% w/v; KH2PO4, 0.1% w/v; FeSO4, 0.0002% w/v; KCl, 0.1% w/v; and sucrose, 3% w/v; pH 6.0) for harvesting the fungal conidia. Plants were inoculated with V. dahliae by the root dip method previously described (Zhang et al., 2012). In brief, the 3-week-old plants were dipped into a fungal conidia suspension (1 × 106 conidia/mL) for 1 min and transferred into a growth chamber with soil culture to allow the disease to progress. The conditions of cultivation were consistent with the plants’ growth conditions. To investigate the disease resistance of silenced plants to V. dahliae, the disease index (DI), fungal recovery assay and fungal biomass quantification assay were performed. The DI was calculated according to the method reported by Wang et al. (2004) with minor modification. In brief, the degree of plant infection by the fungi was divided into four grades according to disease scores ranging from 1 and 4 (1: healthy plants, or <10% of the leaves show yellowing or yellow spots; 2: 10–<40% of the leaves show yellow spots, a few leaves fall off; 3: 40–<70% of the leaves show yellow or brown spots, some leaves fall off; 4: >70% of the leaves showed brown spots, many leaves fall off, even the death of the whole plant). The DI was calculated following formulae: DI = [(Σ disease grade × number of infected plants)/total checked plants × 4] × 100%. The fungal recovery experiment was performed according to the method previously described by Fradin et al. (2009). The fungal biomass quantification assay was performed according to the method described by Wang et al. (2017). The β-tubulin gene of V. dahliae was used to measure the fungal biomass by qPCR. GhUB-7 was used to normalise the DNA quantity of the plant. The same experiments were performed with three biological replicates.
DNA/RNA Extraction and qRT-PCR
For fungal growth biomass, fungal DNA was extracted using five cotton stems through the CTAB method at 3 weeks after V. dahliae infection (Muhammad et al., 2013). Total RNA from cotton samples, including the roots, internodes, leaves, cotyledons, and hypocotyls, was extracted using a plant total RNA extraction kit (Sangon, Shanghai, China) following the manufacturer’s protocol. First-strand cDNA was synthesised from 3 μg of total RNA using the Goldenstar RT6 cDNA Synthesis kit Ver. 2 (Tsingke, Beijing, China) to analyse the expression levels of related genes by qRT-PCR. Quantitative detection was performed using the 2 × T5 Fast qPCR mix kit (Tsingke, Beijing, China) in a real-time quantitative PCR instrument (Bio-Rad, Foster City, CA, United States) and the relative quantification was calculated using the comparative Ct method (Livak and Schmittgen, 2001; Li et al., 2005). To normalise gene expression, GhUB-7 was used as an internal control. The experiments were performed with three technical replicate and three biological replicates (n ≥ 3). All primers used in the experiments are shown in Supplementary Table S1.
Subcellular Location
The coding sequences of GhWAT1, GhWAT2, and GhWAT3 were amplified from G. hirsutum cDNA with the following specific primer pairs: WAT1-F/R, WAT2-F/R, and WAT3-F/R, respectively. The tonoplast marker gene AtTIP2 was amplified with TIP2-F/R primer pairs from Arabidopsis cDNA. The pUC-mGFP4 and pUC-mCherry vectors were double digested with Kpn I and Xba I (TAKARA, Dalian, China). Then, the amplified products of GhWATs and AtTIP2 were inserted into the pUC-mGFP4 and pUC-mCherry vectors, respectively, by using a homologous recombination kit (Vazyme, Nanjing, China) according to the manufacturer’s guidelines, generating the GhWATs:GFP and AtTIP2:mCherry vectors. The primers used in PCR are shown in Supplementary Table S1.
To observe the subcellular location of GhWATs, the aforementioned plasmids were extracted using the QIAGEN Plasmid Midi Kit (QIAGEN, Germany). The N. benthamiana mesophyll protoplasts were isolated and transformed according to methods reported previously (Schweiger and Schwenkert, 2014; Lei et al., 2015). Then, protoplasts that had been incubated for 12–18 h at room temperature under a low level of light were observed under a confocal laser scanning microscope (TCS SP2; Leica, Germany).
Histochemical Staining and Lignin Content Analysis
Histochemical staining with Wiesner reagent was used to visualise xylem development and lignin deposition (Xu et al., 2011). More than five cotton internodes and hypocotyls of the GhWAT123-silenced and control plants were crosscut into sections by hand and directly dipped in phloroglucinol solution (2% phloroglucinol dissolved in 95% ethanol) to incubate for 10 min, followed by treatment with 18% HCl for 20 min. The stained sections were observed and photographed using a stereomicroscope (DM2500; Leica, Germany). The lignin contents of hypocotyls from the silenced and control plants were determined by the Klason method as previously described (Ronald et al., 1994; Abdullah et al., 2006). The same experiment was conducted with three biological replicates, and each replicate comprised more than five individual seedlings.
Paraffin Sectioning
To observe the xylem vessels of the different samples, a paraffin sectioning was carried out according to the method described by Kong et al. (2017) with minor modifications. The five young stems from the silenced and control plants were fixed with FAA fixation solution (95% ethanol, 5% glacial acetic acid, 37% formalin and DDW) for 24 h. The paraffin sections were stained with safranin solution (1% safranin dissolved in 50% ethanol) overnight, which stains lignified tissues, such as the xylem. The stained paraffin sections were rinsed with 95% ethanol and observed using an inverted microscope (Axiovert 200 M; Zeiss, Germany) (Vazquez-Cooz and Meyer, 2002; De Micco and Aronne, 2007; Bond et al., 2008).
SA Extraction and Measurement
The SA extraction was performed as described by Sun et al. (2014). In brief, 100–200 mg root samples were ground to powder in liquid nitrogen and homogenised twice with the cold extraction buffer (methanol:ddH2O:glacial acetic acid = 80:19:1) for oscillation overnight in the dark at 4°C. The extraction was evaporated dry with N2. The dry powder was dissolved with 300 μL methanols; and the solution was filtered by 0.22 μm filters. SA content was measured by HPLC-MS/MS performed by AB SCIEX QTRAP 4500 system (AB SCIEX, Foster City, CA, United States) as described by Zhou et al. (2018). The experiment was repeated three times, with samples containing more than three seedlings.
Results
Identification and Characterisation of G. hirsutum WAT1
Recently, reports have shown that many genes associated with lignin deposition in the cell wall and xylem vessels participate in cotton plant defence against V. dahliae (Xu et al., 2011; Gao et al., 2013). Arabidopsis WAT1 functions in secondary wall formation in interfascicular and xylary fibres, which participates in plant resistance to Ralstonia solanacearum, V. dahliae, and other vascular pathogens (Denancé et al., 2013, 2014). Thus, we were prompted to explore cotton WAT functions in plant defence and lignin synthesis. Three homologous WATs in G. hirsutum were identified by blasting Arabidopsis WAT1 in the available genomic sequence of G. hirsutum4, and they were designated GhWAT1 (Gh_A05G1621 and Gh_D05G1805 located in the A and D subgenomes, respectively), GhWAT2 (Gh_A06G1836), and GhWAT3 (Gh_A01G0922 and Gh_D01G0964). Both GhWAT1 and GhWAT3 genes contain six exons and five introns, while GhWAT2 only has five exons and four introns. GhWAT1, GhWAT2, and GhWAT3 encode 384, 386, and 392 amino acid peptides, respectively (Figure 1A). They encode proteins belonging to the EamA-like transporter (DOMAINS OF UNKNOWN FUNCTION 6, DUF6) family (Figure 1B). According to the WAT hydrophobicity profile analysis, two DUF6s possess ten potential alpha helix transmembrane-spanning domains (Figure 1B), which is consistent with the structure of Arabidopsis WAT1 (Ranocha et al., 2010). Amino acid sequence alignment analysis suggested that GhWATs exhibited sequence highly identifies with Arabidopsis WAT1 and Medicago sativa N21, reaching 88.4 and 88.8%, respectively. The protein similarities of the three GhWATs reached 92.9%, indicating that there could be functional redundancy among them (Supplementary Figure S1).
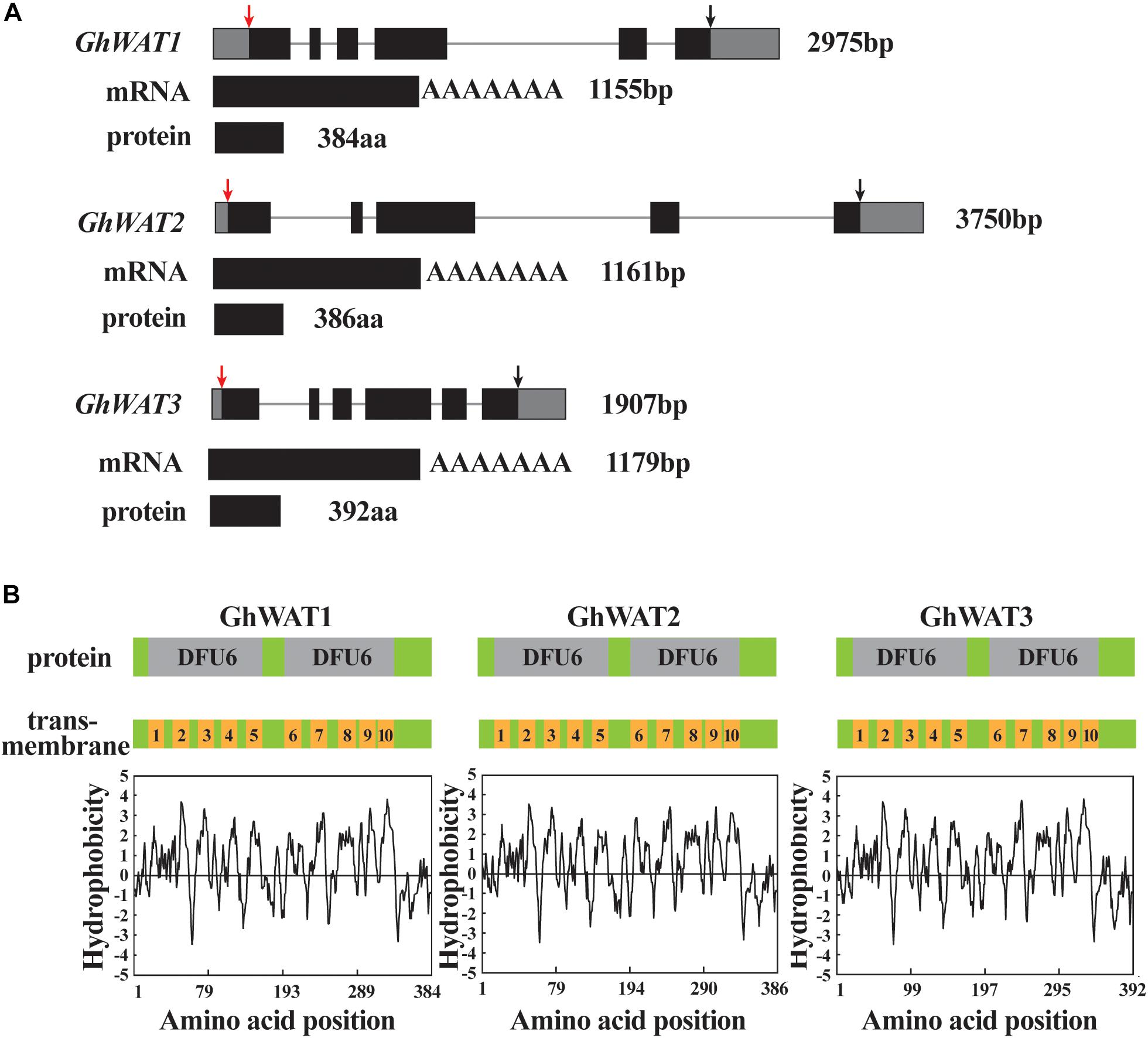
Figure 1. Three GhWATs genes structural characteristics and their corresponding predicted proteins. (A) GhWATs gene structure, mRNA and deduced proteins. Lines represent introns, black boxes represent exons in gene structure. The red arrow points to ATG and the black points to stop codon. (B) The protein domains and transmembrane domains of GhWATs according to predictions of hydrophobicity regions. The GhWATs contain two DUF6 domains; each of the DFU6 domains encompasses five transmembrane-spanning domains.
Analyses of GhWAT Expression Profiles
To investigate the expression pattern of GhWAT1, GhWAT2, and GhWAT3 in plant tissues, the GhWAT mRNA levels of samples from roots, internodes, leaves, cotyledons, and hypocotyls were analysed by qPCR. As shown in Supplementary Figure S2, GhWAT1, GhWAT2, and GhWAT3 were predominantly expressed in the roots, internodes, and hypocotyls but were slightly expressed in the leaves and cotyledons. Since Arabidopsis WAT1 participates in plant defence through regulating SA synthesis and IAA transport (Denancé et al., 2013), we evaluated the GhWATs expression levels of plants infected with V. dahliae and treated with either IAA or SA. The results showed that the expression levels of GhWAT1 exhibited an increasing trend over 0–72 h ranges in the treated roots (0 h-treated samples collected from the parallel plants without any treatment as a control) and, specifically, that at 48 h the expression level significantly increased compared with other time points. The GhWAT2 expression levels showed significantly decreased at 12 and 36 h among all time points. GhWAT3 expression was significantly upregulated at 48 and 72 h in the treated roots among all time points. These data suggested that the three homologous GhWATs had different expression responses to V. dahliae infection (Figure 2A).
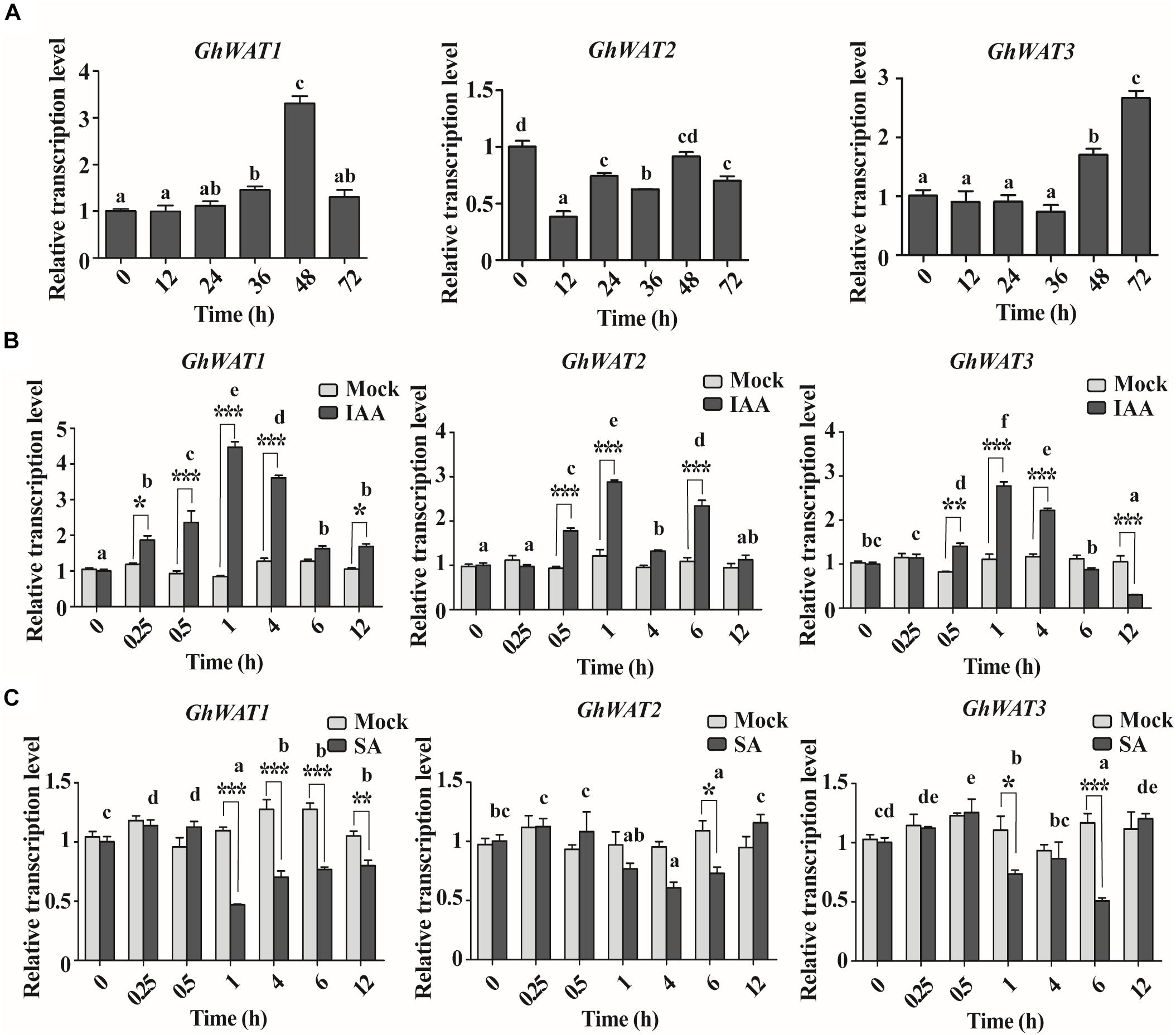
Figure 2. The pathogen and hormone-induced expression patterns of three GhWATs. (A) The expression analysis of GhWAT1, GhWAT2, and GhWAT3 in cotton roots in response to V. dahliae infection. Relative expression levels were normalised to the levels at 0 h of infection. GhUB-7 was used as a reference gene. The error bars represent the standard error of the mean (SEM) of three biological replicates. Different letters represent statistically different means at P < 0.05 (one-way ANOVA with a Duncan post hoc test). (B,C) The expression levels of three GhWATs in plants treated with 50 μm IAA (B), 2 mM SA (C) and the corresponding mock treatments. Relative expression levels were normalised to the levels at 0 h of infection. GhUB-7 was used as a reference gene. The error bars represent the standard error of the mean (SEM) of three biological replicates. As compared with the corresponding time of the mock treatment, two-way ANOVA with a Bonferroni post-test: ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. Comparing all the time points, different letters represent statistically different means at P < 0.05 (two-way ANOVA followed by Duncan post hoc test).
The IAA treatment was first performed to investigate GhWATs expression profiles in hormone-induced plants. As shown in Figure 2B, GhWAT1 exhibited significantly upregulated expression in 0.25, 0.5, 1, 4, and 12 h-treated roots compared with the mock treatment; among all time points, the GhWAT1 at 0.5, 1, and 4 h were significantly upregulated expression. The expression level of GhWAT2 was significantly upregulated in 0.5, 1, and 6 h-treated roots compared with the mock treatment, and the three time points showed significant upregulation in GhWAT2 expression among all time points. The GhWAT3 expression levels significantly increased in 0.5, 1, and 4 h-treated roots and then significantly decreased at the 12 h-treated samples compared with the mock treatment; the expression levels of GhWAT3 were significantly upregulated at 0.5, 1, and 4 h among all time. Then, GhWATs expression levels were checked under SA treatment. The Compared with mock treatment samples, expression levels of GhWAT1 significantly decreased in 1, 4, 6, and 12 h samples, those of GhWAT2 significantly downregulated in 6 h samples and GhWAT3 significantly decreased expression in 1 and 6 h samples. Among all the time points, the expression of GhWAT1, GhWAT2, and GhWAT3 at various time points showed significant differences (Figure 2C). These results suggested that the expression of the three GhWATs was induced by IAA treatments and repressed by SA treatments.
The Knockdown of GhWATs Increases Plant Resistance to V. dahliae
To characterise the function of GhWATs in plant defence, the VIGS method was used to generate GhWAT1-, GhWAT2-, and GhWAT3-silenced plants. And the vectors of pYL156, pYLW1, pYLW2, pYLW3, and pYLW123 construction showed in Figure 3A. Seven-day-old plants were injected with A. tumefaciens strain GV3101 cells containing the indicated vectors using a 1-mL syringe without a needle. At 14 days post-Agro-infiltration, the positive control plants that contained silenced phytoene dehydrogenase (GhPDS) genes showed white bleached leaves (Figure 3B), which indicated that the TRV-VIGS system worked well in cotton plants. At this time point, the corresponding GhWATs expression levels of the silenced plant leaves and roots were detected by qPCR to analyse the silencing efficiency. As shown in Supplementary Figure S3, the expression levels of GhWAT1, GhWAT2, and GhWAT3 significantly decreased in the silenced leaves and roots compared with the those in the control plants injected with the pYL156 vector (as the control named TRV:00), and the expression levels of GhWAT1, GhWAT2, and GhWAT3 reduced, exhibiting 0.13-, 0.35-, and 0.17-fold of the control in leaves and 0.3-, 0.04-, and 0.09-fold in roots, respectively. Expectedly, the expression of GhWAT1, GhWAT2, and GhWAT3 was greatly reduced in the GhWAT123-silenced plants (Supplementary Figure S3). At 21 days post-Agro-infiltration, the GhWAT1-, GhWAT2-, and GhWAT3-silenced plants showed comparable phenotypes with the control plants, while the GhWAT123-silenced plants exhibited smaller phenotypes than control plants with significantly lower biomass (Supplementary Figures S4A,B). As shown in Figures 3C,D, the plant height and root length of the GhWAT123-silenced plants were clearly shorter than those in the control. The first internode growth in the GhWAT123-silenced plants was significantly inhibited compared with that in the control, with the length reduced by 39% of that of the control (Figure 3E). Similar results were detected for the root growth of the GhWAT123-silenced plants, which showed a reduction of 25% of that of the control (Figure 3F).
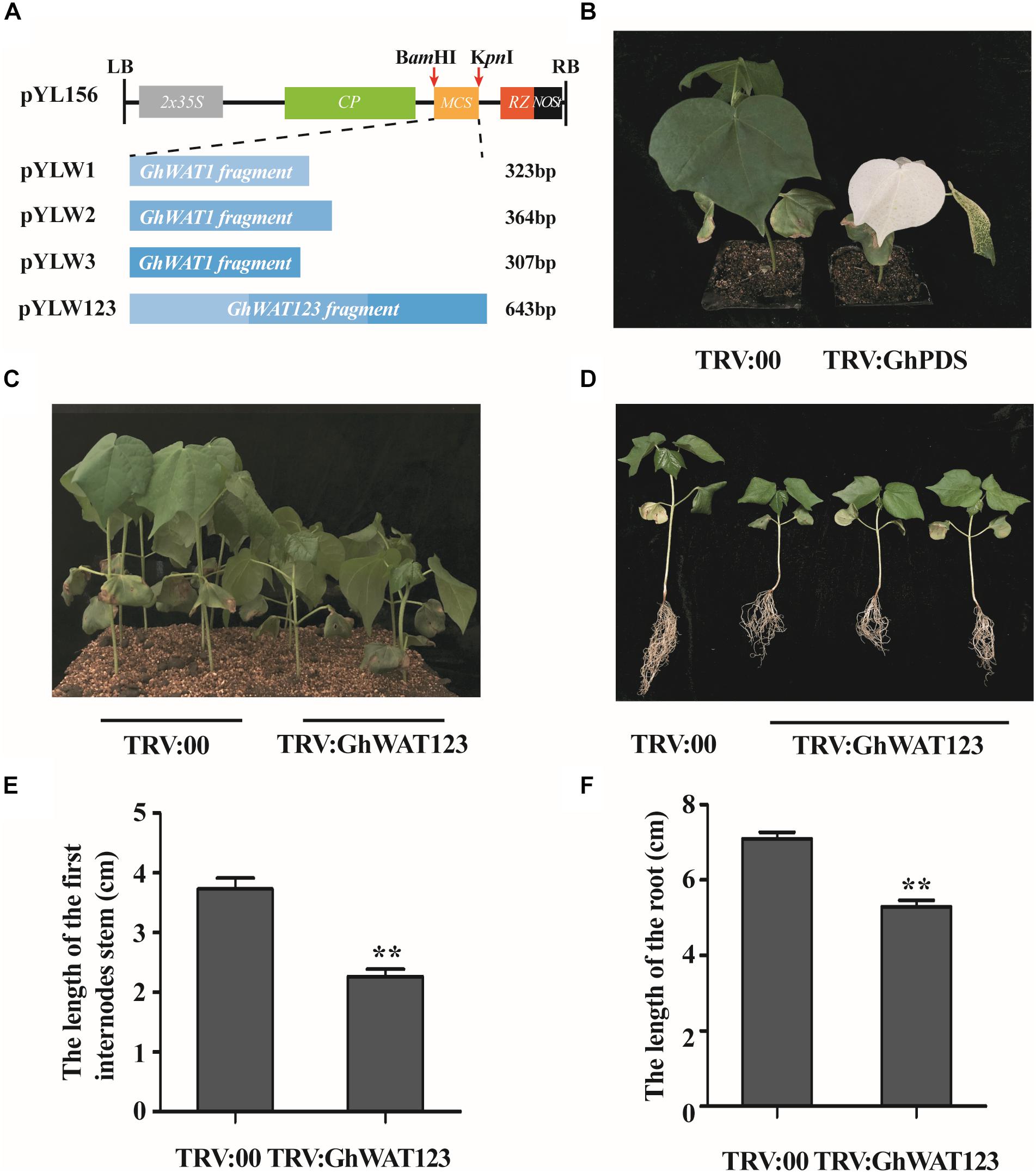
Figure 3. Development of GhWATs-silenced plants and their phenotypes by VIGS. (A) Schematic diagram of the pYL156, pYLW1, pYLW2, pYLW3, and pYLW123 vectors. LB and RB, left and right borders of T-DNA; 2 × 35S, CaMV35S promoter with the duplicated enhancers; CP, coat protein; MCS, multiple cloning sites; RZ, self-cleaving ribozyme; Nos, NOS terminator. (B) Photobleaching phenotypes of GhPDS-silenced plant leaves (positive control) at 2 weeks post-Agro-infiltration. (C,D) Phenotypic characteristics of GhWAT123-silenced and control plants grown in plots with soil and exposed to Hoagland nutrient solution. (E,F) The length of the first internode (E) and the root length (F) of silenced plants and the control plants. TRV:00 is control plants injected with the pYL156 vector. Statistical significance was determined by Student’s t-test: ∗∗P < 0.01. The error bars represent the SEM of three biological replicates.
Previous reports showed that Arabidopsis WAT1 mutation conferred broad-spectrum resistance to vascular pathogens, such as V. dahliae (Denancé et al., 2013). The GhWAT1-, GhWAT2-, GhWAT3-, and, GhWAT123-silenced plants and the control plants were inoculated with V. dahliae through the root dip method. Three weeks after inoculation, the single-silenced plants and the control showed a similarly high level of susceptibility to the pathogen, with disease symptoms including plant wilt, stunted growth, leaf chlorosis and even defoliation (Figure 4A). However, the triple-silenced plant, GhWAT123-silenced plant, showed a stronger level of resistance to V. dahliae compared with the single-silenced plant and the control (Figure 4A). As observed in the corresponding longitudinal stem sections, there were darker brown vascular vessels in the single-silenced plants and the control plants than in the GhWAT123-silenced plants (Figure 4B). The disease index of the GhWAT123-silenced plants was significantly reduced compared with the GhWAT1-, GhWAT2-, and, GhWAT3-silenced plants and the control, which showed decreases of 14.6, 18.4, 23.1, and 19.1%, respectively (Figure 4C). The fungal biomass results in the infected stems showed that the relative fungal DNA levels of the three single GhWAT-silenced plants were comparable with the controls, while that of the GhWAT123-silenced plants was significantly lower than those of the controls (Figure 4D). To further investigate the effects of GhWATs on plant resistance, hyphae recovery growth from cutting sections of the infected stem was assayed. We found that most of the stem sections from the single-silenced plants and the control could grow mycelia, while significantly fewer fungal colonies were observed in the cutting sections from the GhWAT123-silenced plants than in those of the other plants (Figures 4E,F). Altogether, these results showed that GhWAT1, GhWAT2, and, GhWAT3 are functionally redundant and together participate in plant defence against V. dahliae.
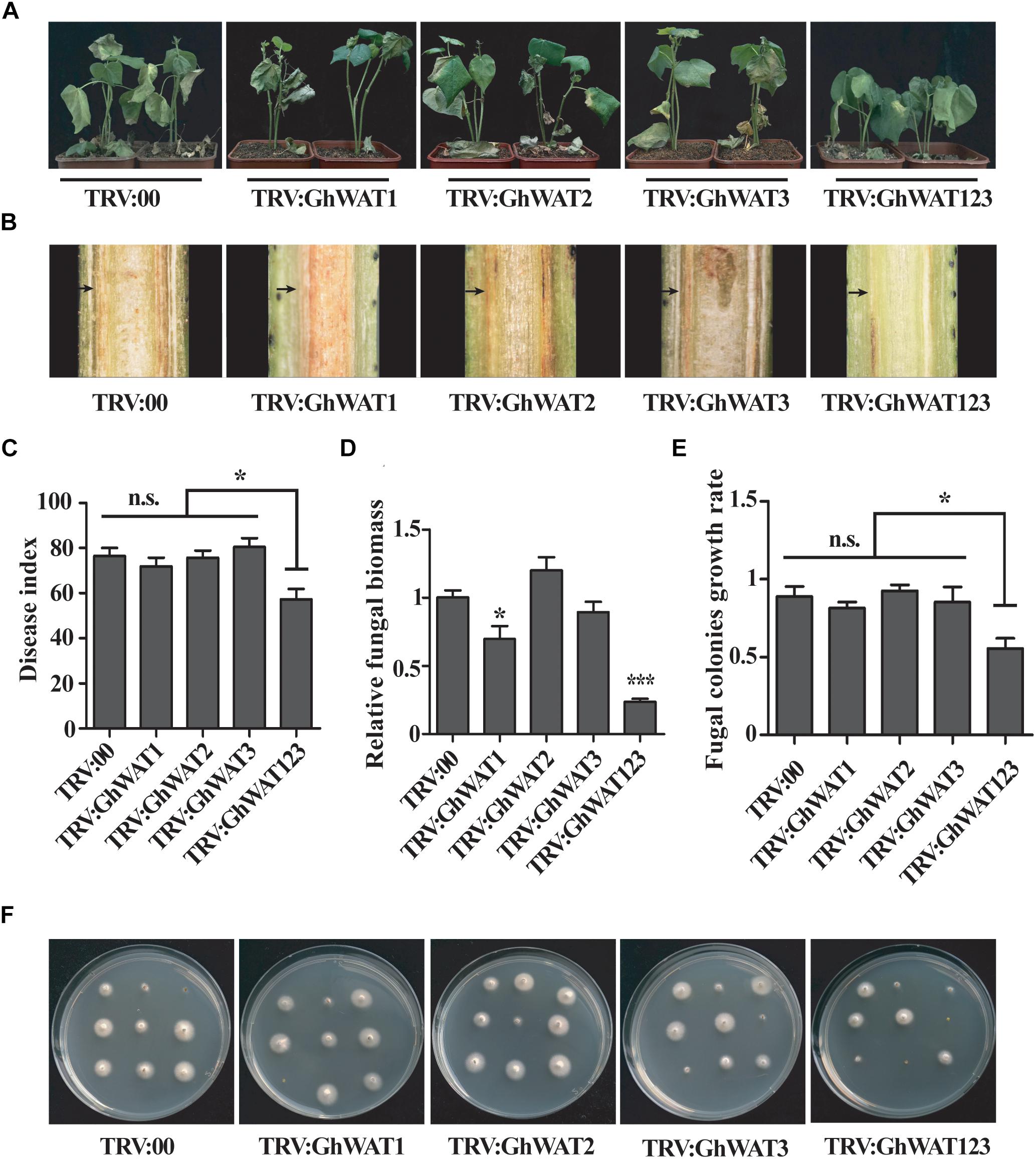
Figure 4. The simultaneous knockdown of three GhWATs increased plant resistance to V. dahliae. (A) Disease symptoms of the three single GhWAT-silenced plants, the GhWAT123-silenced plants and the control plants inoculated with V. dahliae (106 conidia/mL) at 21 days. (B) The browning phenotypes in the xylem of longitudinal cross-sections of silenced hypocotyls at 21 days after infection. The black arrow points to the browning xylem. (C) The disease index of the control, three single GhWATs- and GhWAT123-silenced plants 21 days after inoculation. The star represents P < 0.05 (one-way ANOVA with a Duncan post hoc test). n.s. = 0.5536. (D) The relative V. dahliae fungal biomass in the infected stems was quantified by qRT-PCR. The DNA contents of the V. dahliae β-tubulin gene in the V. dahliae-inoculated plants were analysed by qPCR. The GhUB-7 DNA levels were normalised to the pathogen DNA contents of the β-tubulin gene. The statistically significant differences compared with the GhWAT123-silenced plants were determined by Student’s t-test (∗P < 0.05, ∗∗∗P < 0.001). Student’s t-test was used to determine significant differences between the TRV:00 control and other silenced plants. TRV:00 is injected with the pYL156 vector. Error bars represent the SEM of three biological replicates. (E) Statistics of the fungal colonies. The number of fungal colonies of each plate means the rate of recovery growth rate. The star represents P < 0.05 (one-way ANOVA with a Duncan post hoc test). n.s. = 0.3942. (F) A fungal recovery assay. The first internode of the plants inoculated with V. dahliae for 21 days was cultured on potato dextrose agar medium for 3 days.
GhWATs Subcellular Localisation and the Expression Response of Auxin-Related Genes
To determine the subcellular localisation of the GhWATs to characterise their function, three vectors containing the GhWATs:mGFP4 fusion protein driven by the CaMV35S promoter were constructed and transiently expressed in Nicotiana benthamiana mesophyll protoplasts. A tonoplast protein AtTIP2 (AT3G26520):mCherry vector was constructed for use as a co-localised tonoplast maker. When GhWATs:mGFP4 and AtTIP2:mCherry were co-expressed in protoplasts, the GFP signal of GhWATs:mGFP4 was merged with the image of AtTIP2:mCherry, indicating that the three GhWATs probably localised in the tonoplasts (Figure 5A).
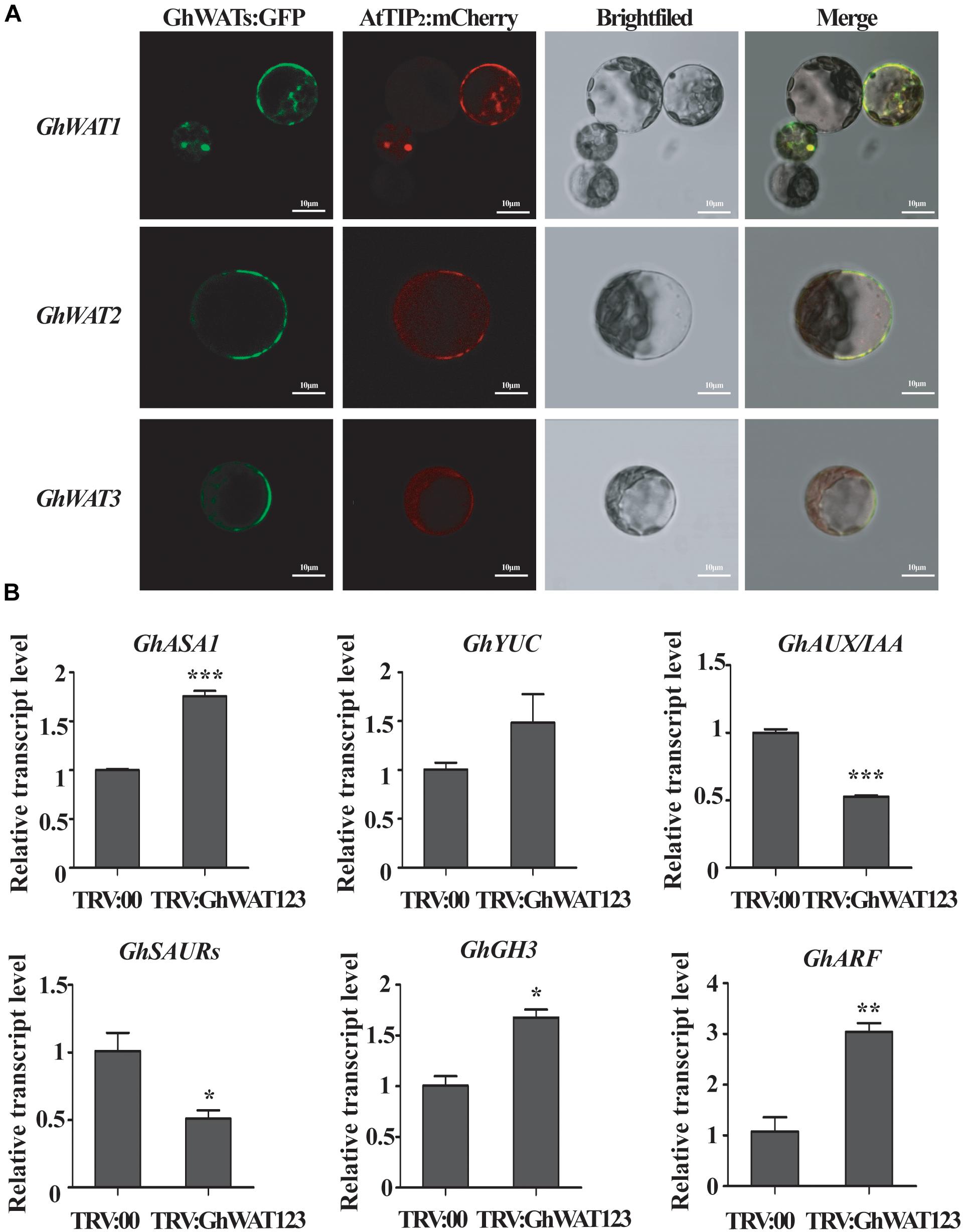
Figure 5. Subcellular location of GhWATs and the expression levels of auxin-related genes. (A) The three GhWATs:GFP fusion proteins co-localised with AtTIP2:mCherry in the tonoplast in N. benthamiana mesophyll protoplasts. AtTIP2 is a tonoplast protein maker. Scale bars: 10 μm. (B) The expression levels of auxin-related genes. TRV:00 is control plants injected with the pYL156 vector, which relative expression level was normalised to ‘1’, and GhUB-7 was used as a reference gene. Student’s t-test were performed in the same experiments (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001). Error bars represent the SEM of three biological replicates.
In wat1 mutant roots, indole metabolism was repressed, and the expression of IAA-related genes was reprogrammed (Denancé et al., 2013; Ranocha et al., 2013). In our studies, the expression levels of two auxin biosynthesis-related genes, anthranilate synthase α1 (ASA1) and Flavin Monooxygenase-Like Enzyme YUCCA1 (YUC1), were increased in the silenced plants compared with the control plants (Figure 5B). The expression levels of four auxin signalling response-related genes, auxin/indole-3-acetic acid (AUX/IAA) and small auxin-up RNAs (SAUR), were significantly downregulated than the control plants, while the expression levels of auxin-responsive Gretchen Hagen 3 (GH3) and AUXIN RESPONSE FACTOR (ARF) were significantly upregulated (Figure 5B). These data suggested that the knockdown of GhWATs affected auxin biosynthesis and signalling response possibly by disrupting auxin polar transport across the tonoplast.
GhWATs Participate in Regulation of SA Synthesis- and Signal Response-Related Genes
In both IAA and SA biosynthesis, chorismate acted is shared as a common substrate (Jones and Dangl, 2006; Vlot et al., 2009; Ranocha et al., 2010, 2013; Denancé et al., 2013; Fu and Dong, 2013). Thereby, the SA content in the roots was measured to evaluate the GhWATs role in SA biosynthesis. The SA content in triple-silenced plants was significantly higher than the control (Figure 6A). The expression of ICS1, SA synthesis-required gene, was twofold higher in the GhWAT123-silenced plants than in the control plants. The expression levels of EDS1 and PAD4, which contribute to the positive feedback loop involved in SA biosynthesis, were significantly increased in the GhWAT123-silenced plants compared with the control plants (Figure 6). These results indicated that the knockdown of GhWATs can activate SA synthesis and positive feedback loop process, which increases plant resistance to pathogens.
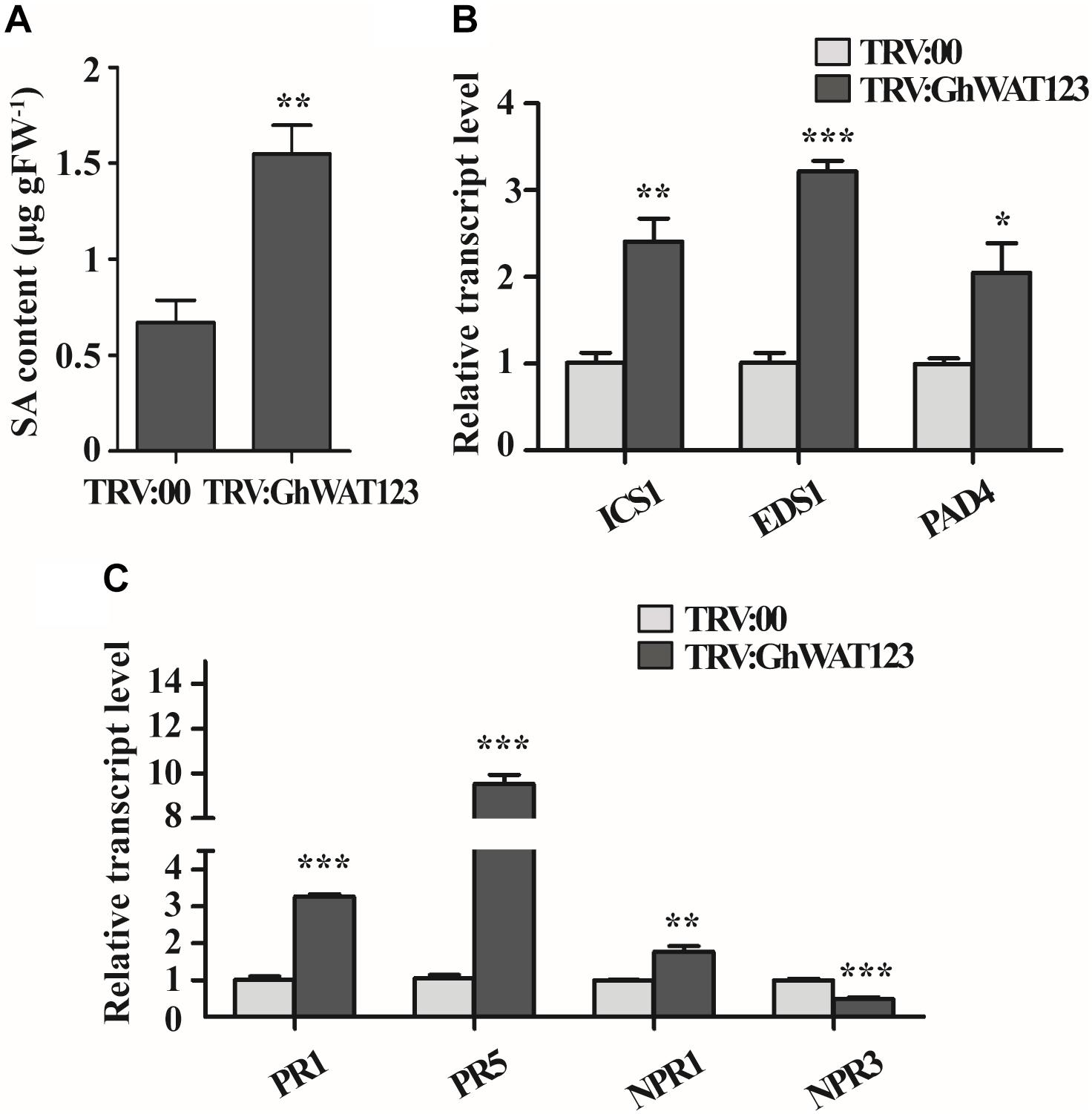
Figure 6. SA contents and expression patterns of SA-related genes. (A) SA content in the root of GhWAT123-silenced and control plants. FW, fresh weight. (B) The expression levels of three SA biosynthesis-related genes, ICS1, EDS1, and PAD4, in GhWAT123-silenced and control plants. (C) The expression levels of PR1, PR5, NPR1, and NPR3 in GhWAT123-silenced and control plants. TRV:00 is controlled plants injected with the pYL156 vector and the relative expression levels were normalised to ‘1’. GhUB-7 was used as the reference gene. The statistically significant differences were run by Student’s t-test (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001). Error bars represent the SEM of three technical replicates.
To investigate whether the activation of SA synthesis- and positive feedback loop-related genes could increase the expression of defence-related genes, four genes, including SA signal component factors and defence-related genes, were monitored at the transcript level (Denancé et al., 2013). As shown in Figure 6C, the expression level of NPR1 in the GhWAT123-silenced plants was significantly higher than that in the control, while that of NPR3 was lower. PR1 and PR5 expression was upregulated in the GhWATs knockdown plants compared with the control and were 3.2- and 9-fold higher than control plants, respectively. These results showed that the knockdown of GhWATs can reprogram the transcription of SA defence-related genes.
Knockdown of GhWATs Inhibits Xylem Development and Locally Increases Lignin Deposition
In Arabidopsis, WAT1 is required for secondary cell wall deposition and participates in plant resistance to vascular pathogens (Ranocha et al., 2010; Denancé et al., 2013). To investigate secondary cell wall deposition in the GhWATs knockdown plants, cross-sections of hypocotyls stained with phloroglucinol-HCl were observed. The xylem in the vascular tissue showed a red colour with Wiesner reagents, which stain lignin. As shown in Figures 7A,B, compared with the xylem area of the control (1.27 mm2), the xylem area of the GhWAT123-silenced hypocotyls (1.65 mm2) was significantly smaller. However, the red colour density of the xylem area was darker compare with the control. The lignin contents of whole hypocotyls in the silenced plants, which were analysed by the Klason method, were slightly increased compared with those in the control (Supplementary Figure S5A), resulting in lignin deposition was higher in the smaller xylem area of GhWAT123-silenced plants than control. The expression of lignin synthesis-related genes, including Phe:phenylalanine ammonia-lyase (GhPAL), cinnamate 4-hydroxylase (GhC4H), 4-coumarate:CoA ligase (Gh4CL), p-coumarate 3-hydroxylase (GhC3H), Caffeoyl-CoA O-methyltransferase (GhCCoAMT), cinnamyl alcohol dehydrogenase (GhCAD), and cinnamoyl-CoA reductase (GhCCR), was upregulated in the GhWAT123-silenced plants compared with the control (Supplementary Figure S5C). Then the expression levels of three genes involved in the last steps in lignification, laccase 1 (GhLac1), peroxidase 1 (Ghpox1), dirigent 1 (GhDIR1), were detected by qPCR. The expression of GhLac1 was significantly upregulated in silenced-plants compared with the control (Supplementary Figure S5B). These results indicated that silencing GhWATs can repress xylem development but increase lignin accumulation in xylem sections.
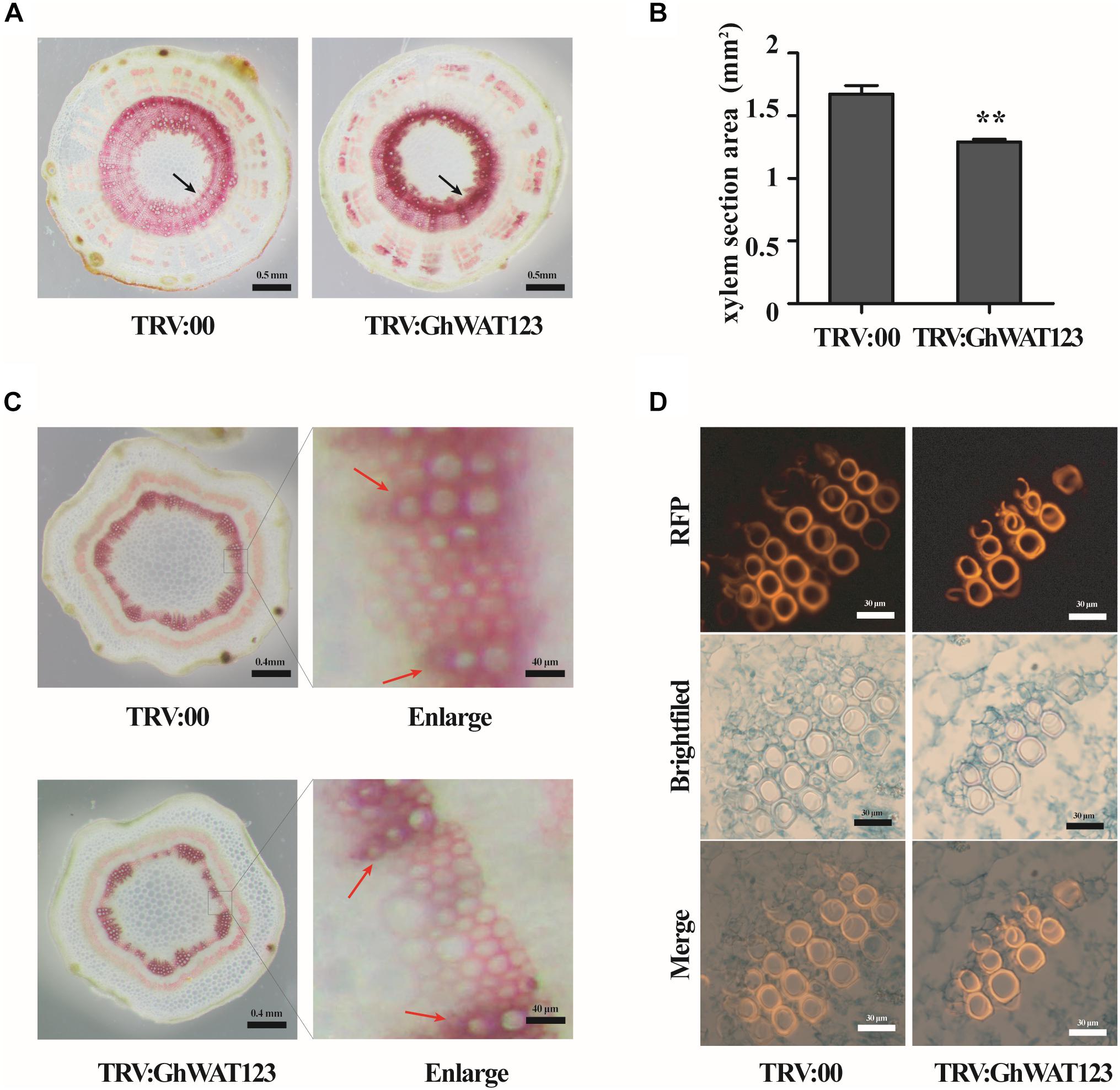
Figure 7. Knockdown of GhWAT123 inhibited xylem development and locally increased lignin deposition. (A) Phloroglucinol-HCl staining of lignin in hypocotyl cross-sections of GhWAT123-silenced and control plants. The xylem sections with lignin were stained red. Scale bars represent 0.5 mm. The black arrow points to the xylem of stained red. (B) The area of the hypocotyl xylem section of silenced and control plants. The xylem section area measurement was performed using Adobe Photoshop CS5. The mean and SEM came from three biological replicates. Double asterisks indicate statistically significant differences compared with the control, which was determined by Student’s t-test (∗∗P < 0.01). (C) Histochemical analysis of transverse sections of young internodes in GhWAT123-silenced and control plants. The lignified tissue and interfascicular fibre were stained red using phloroglucinol-HCl. The red arrow points to the xylary fibres, between the red arrows were interfascicular fibres. Scale bars represent 0.4 mm or 40 μm. The right panels were enlarged from the control or silenced plants by approximately 100-fold. (D) Safranin staining of the paraffin sections of the control and silenced plants. Scale bars represent 30 μm. TRV:00 is control plants. The same experiment was performed three times.
To further evaluate GhWATs function in xylem development, transverse sections of young internodes were stained with phloroglucinol-HCl. The xylem in silenced internodes was stained a slightly purple colour, indicating that the lignin content in the xylem was higher in the GhWATs knockdown internodes than in the control internodes (Figure 7C in enlarging). And the secondary cell wall thickness of interfascicular fibres decreased in the GhWAT123-silenced plants compared with the control plants (Figure 7C). To further observe the xylem structure, paraffin cross-sections were analysed. Safranin was used to stain the lignified tissues to identify lignin-rich cell walls, which exhibited a fluorescent red or orange colour (Vazquez-Cooz and Meyer, 2002; De Micco and Aronne, 2007; Bond et al., 2008). As shown in Figure 7D, a string of xylem vessels in the silenced internodes contained almost two to three layers, while that in the control internodes had three to four layers. The fluorescence of the vessels was stronger in the silenced plants than in the control plants, indicating that more lignin was deposited around xylem vessels in the silenced plants than in the control plants. These data confirmed that the knockdown of GhWATs represses xylem development and locally increases lignin deposition in xylem sections.
Discussion
Verticillium wilt causes devastation to cotton production. Many measures, including the application of tillage, soil solarisation, soil amendments, and biological controls, are not always effective or efficient to control its damage (Gong et al., 2017; Shan et al., 2018). Thus, it is imperative to search for disease-related genes to breed resistant cultivars. Therefore, in order to promote cotton disease resistance breeding, it is vital to mine novel candidate genes/proteins. In this study, we identified three homologous cotton defence-related genes, GhWAT1, GhWAT2, and GhWAT3, which possess redundant functions in negatively regulating plant resistance to V. dahliae.
The GhWATs triple-silenced plants had an increased level of resistance to V. dahliae, however, the three single GhWAT-silenced plants showed a comparable level of sensitivity to V. dahliae with the control plants, suggesting that the three genes redundantly function in plant defence. These results also showed that GhWATs in silenced plants acted as negative regulators of plant defence against pathogens. Genes that negatively regulate plant defence can be directly applied through gene mutations. Thus, it is very important to search for negative regulatory defence genes for plant breeding. To date, some negative regulatory genes have been identified in cotton, including GbJAZ1, GbWRKY1, GhNINJA, Gh14-3-3, and GhCYP82D. The knockout or knockdown of these genes can increase plant resistance against V. dahliae infection (Gao et al., 2013; Li et al., 2014; Sun et al., 2014; Wang et al., 2017; Zhang Z.et al., 2018). Therefore, GhWATs can be used as candidate genes for cotton resistant cultivar breeding.
GhWATs were located in the tonoplast, and their expression levels were induced by IAA treatment, indicating that GhWATs are involved in IAA metabolism and/or IAA transport, which is consistent with results in Arabidopsis (Ranocha et al., 2010, 2013; Denancé et al., 2013). When the GhWATs were silenced, auxin transport was blocked, resulting in a local deficiency of auxin in some tissues. Then, that SA synthesis pathway activity was strengthened and promoted SA levels increasing, which increased plant resistance to V. dahliae infection. The results showed, there is an inverse correlation between SA and auxin levels in cotton, which has been reported in Arabidopsis (Kazan and Manners, 2009; Ludwig-Muller, 2015; Shigenaga et al., 2017). Additionally, SA biosynthesis via two distinct pathways, ICS catalysed chorismate pathway and PAL mediated synthesis pathway (Wildermuth et al., 2001; Shah, 2003; Vlot et al., 2009). In our studies, both ICS pathway-related genes (ICS1, EDS1, and PAD4) and PAL pathway-related gene (PAL) showed significantly upregulated expression in triple-silenced GhWATs plants. Thus the knockdown of GhWATs has promoted the accumulation of SA content and increasing the expression level of SA synthesis- and signalling response-related genes, increasing plant defence against pathogens.
The histological analyses of internodes and hypocotyls showed that the knockdown of three GhWATs depressed xylem development but increased lignin accumulation in xylem sections. In addition, the three GhWATs were predominantly expressed in lignified tissues (roots, internodes, and hypocotyls), indicating that they can involve in tissue lignification. In Arabidopsis, wat1 mutants showed decreased secondary cell wall thickness in their interfascicular fibres (Ranocha et al., 2010). Similar phenotypes were observed in the GhWAT123-silenced plants (Figure 7C). The phenotypes of the GhWAT123-silenced showed a decreased xylem area and increased lignin content in the xylem. The lignin synthesis- and lignifications-related genes in the GhWAT123-silenced plants were upregulated compared with those in the control, while these silenced-plants showed smaller phenotype, suggesting that knockdown of GhWATs increase lignin synthesis and possibly reduces the growth. In Arabidopsis, wat1 mutants were revealed a defect in stems cell elongation resulting in dwarfed phenotypes (Ranocha et al., 2010). Additionally, XIP1 is an important kinase for stem growth and vascular development, in xip1 mutant, the highly lignified cell was aberrant accumulation, and defected in oriented cell division, resulting in a dwarfed habit (Bryan et al., 2012). These results imply that GhWATs regulate xylem development and lignin accumulation in xylem sections.
Many reports have shown that cotton plant defence against V. dahliae involves SA and lignin molecules (Sun et al., 2014; Zhang et al., 2017; Zhu et al., 2018). In the present study, the knockdown of GhWATs promoted accumulation of SA content, activated SA pathway-related gene expression and increased lignin accumulation in xylem sections, which facilitated plant resistance to pathogens. Although the GhWATs-silenced plants were smaller than the controls, the appropriately small plants possessed higher lodging resistance compared with the large plants, suitable for dense planting and mechanised management. Therefore, GhWATs are useful candidate genes for breeding disease resistance cultivars.
Author Contributions
JW and QP conceived and designed the experiments. YT, ZZ, and YL performed the experiments. JL, GH, MH, and AC constructed the gene editing vectors and data analysis. YT and JW wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31771848 and 31460066) and sponsored by Funds for Creative Research Groups of China (Grant No. 31621005).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00526/full#supplementary-material
FIGURE S1 | An amino acid alignment of WAT proteins from G. hirsutum, A. thaliana, and Medicago sativa. The complete amino acid sequences of five WATs were retrieved from NCBI database and aligned by DNAMAN 7.0. The two DUF6 domains were noted with black underlines. Go, Gossypium hirsutum; Mt, Medicago sativa; At, Arabidopsis thaliana.
FIGURE S2 | The tissue expression patterns of three GhWATs. Expression levels of three GhWATs in various tissues. Roots, internodes, leaves, cotyledons, and hypocotyls were sampled from 3-week-old seedlings. GhUB-7 was used as the reference gene. The error bars represent the standard error of the mean (SEM) of three biological replicates. More than three individual samples were collected on each of the biological replicates.
FIGURE S3 | The detection of three GhWATs genes expression levels in GhWAT1-, GhWAT2-, GhWAT3-, GhWAT123-silenced, and the control plants. (A,B) The relative expression levels of GhWAT1, GhWAT2, and GhWAT3 were tested in the leaf (A) and root (B) of the corresponding GhWAT1-, GhWAT2-, GhWAT3-silenced plants compared with the control plans, respectively. (C) The relative expression levels of GhWAT1, GhWAT2, and GhWAT3 were tested in the leaves and roots of the GhWAT123-silenced plants compared with the control plants. TRV:00 is control plants injected with the pYL156 vector, which relative expression levels were normalised to ‘1’. GhUB-7 was used as the reference gene. Error bars represent the SEM of three technical replicates. Student’s t-test was used to determine significant differences between corresponding TRV:00 and TRV:GhWAT123 plants (∗∗P < 0.01, ∗∗∗P < 0.001).
FIGURE S4 | The phenotypes and growth of GhWATs-silencing plants. (A) GhWAT1-, GhWAT2-, and GhWAT3-silenced plants showed comparable phenotypes with the control, while GhWAT123-silenced plants exhibited clearly shorter in height than the control at 21 days post-Agro-infiltration. (B) The fresh weight of the control and silenced plants, TRV:00 is control plants injected with the pYL156 vector, Student’s t-test was used to determine significant differences between the control and silenced plants. The error bars represent the standard error of the mean (SEM) of three biological replicates. Each time was collected more than five seedlings (∗P < 0.05).
FIGURE S5 | The lignin content and the expression levels of lignifications- and lignin synthesis-related genes in GhWAT123-silenced and the control plants. (A) The contents of acid-insoluble lignin residues in hypocotyls of GhWAT123-silenced and the control plants by Klason method. The same experiments were performed three times; each time was measured more than five seedlings. (B) The expression levels of lignifications-related genes GhLac1, Ghpox1, GhDIR1 in triple-silenced plants and the control. (C) The relative expression levels of lignin synthesis-related genes in GhWAT123-silenced and the control plants. GhUB-7 was used as the reference gene. The relative expression levels analyses were normalised by control treated data. TRV:00 is control plants injected with the pYL156 vector. The Student’s t-test was performed on statistically significant differences analysis (∗P < 0.05, ∗∗P < 0.01). Error bars represent the SEM of three biological replicates; each time was collected three seedlings.
TABLE S1 | Primer table.
Footnotes
- ^https://www.cottongen.org/icgi/home
- ^https://www.uniprot.org/
- ^https://blast.ncbi.nlm.nih.gov
- ^https://www.cottongen.org/icgi/home
References
Abdullah, N., Ejaz, N., Abdullha, M., Alim-Un-Nisa, and Firdous, S. (2006). Lignocellulosic degradation in solid-state fermentation sugar cane bagasse by termitomyces. Micol. Aplicada Int. 18, 15–19.
Bond, J., Donaldson, L., Hill, S., and Hitchcock, K. (2008). Safranine fluorescent staining of wood cell walls. Biotech. Histochem. 83, 161–171. doi: 10.1080/10520290802373354
Bryan, A. C., Obaidi, A., Wierzba, M., and Tax, F. E. (2012). XYLEM INTERMIXED WITH PHLOEM1, a leucine-rich repeat receptor-like kinase required for stem growth and vascular development in Arabidopsis thaliana. Planta 235, 111–122. doi: 10.1007/s00425-011-1489-6
Cai, Y., He, X., Mo, J., Sun, Q., Yang, J., and Liu, J. (2009). Molecular research and genetic engineering of resistance to Verticillium wilt in cotton: a review. Afr. J. Biotechnol. 8, 7363–7372.
Chen, T., Kan, J., Yang, Y., Ling, X., Chang, Y., and Zhang, B. (2016). A Ve homologous gene from Gossypium barbadense, Gbvdr3, enhances the defense response against Verticillium dahliae. Plant Physiol. Biochem. 98, 101–111. doi: 10.1016/j.plaphy.2015.11.015
De Micco, V., and Aronne, G. (2007). Combined histochemistry and autofluorescence for identifying lignin distribution in cell walls. Biotech. Histochem. 82, 209–216. doi: 10.1080/10520290701713981
Denancé, N., Ranocha, P., Martinez, Y., Sundberg, B., and Goffner, D. (2014). Light-regulated compensation of wat1(walls are thin1) growth and secondary cell wall phenotypes is auxin-independent. Plant Signal. Behav. 5, 1302–1304. doi: 10.4161/psb.5.10.13103
Denancé, N., Ranocha, P., Oria, N., Barlet, X., Riviere, M., Yadeta, K., et al. (2013). Arabidopsis wat1 (walls are thin1)-mediated resistance to the bacterial vascular pathogen, Ralstonia solanacearum, is accompanied by cross-regulation of salicylic acid and tryptophan metabolism. Plant J. 73, 225–239. doi: 10.1111/tpj.12027
Ding, Y., Sun, T., Ao, K., Peng, Y., Zhang, Y., Li, X., et al. (2018). Opposite Roles of Salicylic Acid Receptors NPR1 and NPR3/NPR4 in Transcriptional Regulation of Plant Immunity. Cell 173, 1454–1467. doi: 10.1016/j.cell.2018.03.044
Durrant, W., and Dong, X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209. doi: 10.1146/annurev.phyto.42.040803.140421
Fradin, E., Zhang, Z., Juarez Ayala, J., Castroverde, C., Nazar, R., Robb, J., et al. (2009). Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 150, 320–332. doi: 10.1104/pp.109.136762
Fu, Z., Yan, S., Saleh, A., Wang, W., Ruble, J., Oka, N., et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228–232. doi: 10.1038/nature11162
Fu, Z. Q., and Dong, X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Gamas, P., Niebel, F., Lescure, N., and Cullimore, J. (1996). Use of subtractive hybridization approach to identify new medicago truncatula genes induced during root nodule development. Mol. Plant Microbe. Interact. 9, 233–242.
Gao, W., Long, L., Zhu, L., Xu, L., Gao, W., Sun, L., et al. (2013). Proteomic and virus-induced gene silencing (VIGS) analyses reveal that Gossypol, Brassinosteroids and Jasmonic acid contribute to the resistance of cotton to Verticillium dahliae. Mol. Cell. Proteomics 12, 3690–3703. doi: 10.1074/mcp.M113.031013
Gayoso, C., Pomar, F., Novo-Uzal, E., Merino, F., and de Ilarduya, O. (2010). The Ve-mediated resistance response of the tomato to Verticillium dahliae involves H2O2, peroxidase and lignins and drives PAL gene expression. BMC Plant Biol. 10:232. doi: 10.1186/1471-2229-10-232
Gong, Q., Yang, Z., Chen, E., Sun, G., He, S., Butt, H., et al. (2018). A phi-class glutathione s-transferase gene for verticillium wilt resistance in Gossypium arboreum identified in a genome-wide association study. Plant Cell Physiol. 59, 275–289. doi: 10.1093/pcp/pcx180
Gong, Q., Yang, Z., Wang, X., Butt, H., Chen, E., He, S., et al. (2017). Salicylic acid-related cotton (Gossypium arboreum) ribosomal protein GaRPL18 contributes to resistance to Verticillium dahliae. BMC Plant Biol. 17:59. doi: 10.1186/s12870-017-1007-5
Guo, W., Jin, L., Miao, Y., He, X., Hu, Q., Guo, K., et al. (2016). An ethylene response-related factor, GbERF1-like, from Gossypium barbadense improves resistance to Verticillium dahliae via activating lignin synthesis. Plant Mol. Biol. 91, 305–318. doi: 10.1007/s11103-016-0467-6
Hu, Q., Min, L., Yang, X., Jin, S., Zhang, L., Li, Y., et al. (2018). Laccase GhLac1 modulates broad-spectrum biotic stress tolerance via manipulating phenylpropanoid pathway and jasmonic acid synthesis. Plant Physiol. 176, 1808–1823. doi: 10.1104/pp.17.01628
Jones, J., and Dangl, J. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kazan, K., and Manners, J. (2009). Linking development to defense: auxin in plant-pathogen interactions. Trends Plant Sci. 14, 373–382. doi: 10.1016/j.tplants.2009.04.005
Kong, X., Liu, D., Liao, X., Zheng, J., Diao, Y., Liu, Y., et al. (2017). Comparative analysis of the cytology and transcriptomes of the cytoplasmic male sterility line H276A and its maintainer line H276B of cotton (Gossypium barbadense L.). Int. J. Mol. Sci. 18, 2240–2253. doi: 10.3390/ijms18112240
Lei, R., Qiao, W., Hu, F., Jiang, H., and Zhu, S. (2015). A simple and effective method to encapsulate tobacco mesophyll protoplasts to maintain cell viability. Methods 2, 24–32. doi: 10.1016/j.mex.2014.11.004
Li, C., He, X., Luo, X., Xu, L., Liu, L., Min, L., et al. (2014). Cotton WRKY1 mediates the plant defense-to-development transition during infection of cotton by Verticillium dahliae by activating JASMONATE ZIM-DOMAIN1 expression. Plant Physiol. 166, 2179–2194. doi: 10.1104/pp.114.246694
Li, X. B., Fan, X. P., Wang, X. L., Cai, L., and Yang, W. C. (2005). The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 17, 859–875. doi: 10.1105/tpc.104.029629
Liu, N., Zhang, X., Sun, Y., Wang, P., Li, X., Pei, Y., et al. (2017). Molecular evidence for the involvement of a polygalacturonase-inhibiting protein, GhPGIP1, in enhanced resistance to Verticillium and Fusarium wilts in cotton. Sci. Rep. 7, 39840–39857. doi: 10.1038/srep39840
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ludwig-Muller, J. (2015). Bacteria and fungi controlling plant growth by manipulating auxin: balance between development and defense. J. Plant Physiol. 172, 4–12. doi: 10.1016/j.jplph.2014.01.002
Mohamed, H., and Akladious, S. (2017). Changes in antioxidants potential, secondary metabolites and plant hormones induced by different fungicides treatment in cotton plants. Pestic. Biochem. Physiol. 142, 117–122. doi: 10.1016/j.pestbp.2017.04.001
Moreau, M., Tian, M., and Klessig, D. (2012). Salicylic acid binds NPR3 and NPR4 to regulate NPR1-dependent defense responses. Cell Res. 22, 1631–1633. doi: 10.1038/cr.2012.100
Muhammad, A., Yusuf, Z., and Shaheen, A. (2013). Silicon carbide whisker-mediated transformation of cotton (Gossypium hirsutum L.). Methods Mol. Biol. 958, 79–92. doi: 10.1007/978-1-62703-212-4_7
Novo, M., Silvar, C., Merino, F., Martinez-Cortes, T., Lu, F., Ralph, J., et al. (2017). Deciphering the role of the phenylpropanoid metabolism in the tolerance of Capsicum annuum L. to Verticillium dahliae kleb. Plant Sci. 258, 12–20. doi: 10.1016/j.plantsci.2017.01.014
Pang, J., Zhu, Y., Li, Q., Liu, J., Tian, Y., Liu, Y., et al. (2013). Development of Agrobacterium-mediated virus-induced gene silencing and performance evaluation of four marker genes in Gossypium barbadense. PLoS One 8:e73211. doi: 10.1371/journal.pone.0073211
Pieterse, C., Van der Does, D., Zamioudis, C., Leon-Reyes, A., and Van Wees, S. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. doi: 10.1146/annurev-cellbio-092910-154055
Ranocha, P., Denance, N., Vanholme, R., Freydier, A., Martinez, Y., Hoffmann, L., et al. (2010). Walls are thin 1 (WAT1), an Arabidopsis homolog of Medicago truncatula NODULIN21, is a tonoplast-localized protein required for secondary wall formation in fibers. Plant J. 63, 469–483. doi: 10.1111/j.1365-313X.2010.04256.x
Ranocha, P., Dima, O., Felten, J., Freydier, A., Hoffmann, L., Ljung, K., et al. (2011). WAT1 (WALLS ARE THIN1) defines a novel auxin transporter in plants and integrates auxin signaling in secondary wall formation in Arabidopsis fibers. BMC Proc. 5(Suppl. 7):O24. doi: 10.1186/1753-6561-5-s7-o24
Ranocha, P., Dima, O., Nagy, R., Felten, J., Corratge-Faillie, C., Novak, O., et al. (2013). Arabidopsis WAT1 is a vacuolar auxin transport facilitator required for auxin homoeostasis. Nat. Commun. 4, 2625–2633. doi: 10.1038/ncomms3625
Reusche, M., Thole, K., Janz, D., Truskina, J., Rindfleisch, S., Drubert, C., et al. (2012). Verticillium infection triggers VASCULAR-RELATED NAC DOMAIN7-dependent de novo xylem formation and enhances drought tolerance in Arabidopsis. Plant Cell 24, 3823–3837. doi: 10.1105/tpc.112.103374
Rietz, S., Stamm, A., Malonek, S., Wagner, S., Becker, D., Medina-Escobar, N., et al. (2011). Different roles of Enhanced Disease Susceptibility1 (EDS1) bound to and dissociated from Phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. New Phytol. 191, 107–119. doi: 10.1111/j.1469-8137.2011.03675.x
Ronald, D., Hans-Joachim, G., John, R., Dwayne, R., and Paul, J. (1994). A comparison of the insoluble residues produced by the Klason lignin and acid detergent lignin procedures. J. Sci. Food Agric. 65, 51–58. doi: 10.1002/jsfa.2740650109
Schweiger, R., and Schwenkert, S. (2014). Protein-protein interactions visualized by bimolecular fluorescence complementation in tobacco protoplasts and leaves. J. Vis. Exp. 85, 51327–51334. doi: 10.3791/51327
Seyfferth, C., and Tsuda, K. (2014). Salicylic acid signal transduction: the initiation of biosynthesis, perception and transcriptional reprogramming. Front. Plant Sci. 5:697. doi: 10.3389/fpls.2014.00697
Shah, J. (2003). The salicylic acid loop in plant defense. Curr. Opin. Plant Biol. 6, 365–371. doi: 10.1016/s1369-5266(03)00058-x
Shan, M., Miao, Y., Ullah, A., Khan, A., Menghwar, H., Khan, A., et al. (2018). Physiological and molecular mechanism of defense in cotton against Verticillium dahliae. Plant Physiol. Biochem. 125, 193–204. doi: 10.1016/j.plaphy.2018.02.011
Shi, H., Liu, Z., Zhu, L., Zhang, C., Chen, Y., Zhou, Y., et al. (2012). Overexpression of cotton (Gossypium hirsutum) dirigent1 gene enhances lignification that blocks the spread of Verticillium dahliae. Acta Biochim. Biophys. Sin. 44, 555–564. doi: 10.1093/abbs/gms035
Shi, J., An, H. L., Zhang, L., Gao, Z., and Guo, X. Q. (2010). GhMPK7, a novel multiple stress-responsive cotton group C MAPK gene, has a role in broad spectrum disease resistance and plant development. Plant Mol. Biol. 74, 1–17. doi: 10.1007/s11103-010-9661-0
Shigenaga, A., Berens, M., Tsuda, K., and Argueso, C. (2017). Towards engineering of hormonal crosstalk in plant immunity. Curr. Opin. Plant Biol. 38, 164–172. doi: 10.1016/j.pbi.2017.04.021
Su, X., Qi, X., and Cheng, H. (2014). Molecular cloning and characterization of enhanced disease susceptibility 1 (EDS1) from Gossypium barbadense. Mol. Biol. Rep. 41, 3821–3828. doi: 10.1007/s11033-014-3248-9
Sun, L., Zhu, L., Xu, L., Yuan, D., Min, L., and Zhang, X. (2014). Cotton cytochrome P450 CYP82D regulates systemic cell death by modulating the octadecanoid pathway. Nat. Commun. 5, 5372–5383. doi: 10.1038/ncomms6372
Vazquez-Cooz, I., and Meyer, R. (2002). A differential staining method to identify lignified and unlignified tissues. Biotech. Histochem. 77, 277–282. doi: 10.1080/bih.77.5-6.277.282
Vlot, A., Dempsey, D., and Klessig, D. (2009). Salicylic Acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Wang, L., Wu, S., Zhu, Y., Fan, Q., Zhang, Z., Hu, G., et al. (2017). Functional characterization of a novel jasmonate ZIM-domain interactor (NINJA) from upland cotton (Gossypium hirsutum). Plant Physiol. Biochem. 112, 152–160. doi: 10.1016/j.plaphy.2017.01.005
Wang, P., Sun, Y., Pei, Y., Li, X., Zhang, X., Li, F., et al. (2018). GhSNAP33, a t-SNARE protein from Gossypium hirsutum, mediates resistance to verticillium dahliae infection and tolerance to drought stress. Front Plant Sci. 9:896. doi: 10.3389/fpls.2018.00896
Wang, Y., Chen, D., Wang, D., Huang, Q., Yao, Z., Liu, F., et al. (2004). Over-expression of Gastrodia anti-fungal protein enhances Verticillium wilt resistance in coloured cotton. Plant Breeding 123, 454–459. doi: 10.1111/j.1439-0523.2004.01005.x
Wei, Z., and Yu, D. (2018). Analysis of the succession of structure of the bacteria community in soil from long-term continuous cotton cropping in Xinjiang using high-throughput sequencing. Arch. Microbiol. 200, 653–662. doi: 10.1007/s00203-018-1476-4
Wildermuth, M., Dewdney, J., Wu, G., and Ausubel, F. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–571.
Xu, L., Zhu, L., Tu, L., Liu, L., Yuan, D., Jin, L., et al. (2011). Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J. Exp. Bot. 62, 5607–5621. doi: 10.1093/jxb/err245
Yan, S., and Dong, X. (2014). Perception of the plant immune signal salicylic acid. Curr. Opin. Plant Biol. 20, 64–68. doi: 10.1016/j.pbi.2014.04.006
Yang, Y., Chen, T., Ling, X., and Ma, Z. (2017). Gbvdr6, a gene encoding a receptor-like protein of cotton (Gossypium barbadense), confers resistance to verticillium wilt in arabidopsis and upland cotton. Front. Plant Sci. 8:2272. doi: 10.3389/fpls.2017.02272
Zeng, H., Chen, R., Luo, X., and Tian, J. (2016). Isolation and anti-Verticillium dahliae activity from Bacillus axarquiensis TUBP1 protein. Process Biochem. 51, 1691–1698. doi: 10.1016/j.procbio.2016.06.014
Zhang, W., Jian, G., Jiang, T., Wang, S., Qi, F., and Xu, S. (2012). Cotton gene expression profiles in resistant Gossypium hirsutum cv. Zhongzhimian KV1 responding to Verticillium dahliae strain V991 infection. Mol. Biol. Rep. 39, 9765–9774. doi: 10.1007/s11033-012-1842-2
Zhang, Y., Wang, X., Rong, W., Yang, J., Li, Z., Wu, L., et al. (2017). Histochemical analyses reveal that stronger intrinsic defenses in Gossypium barbadense than in G. hirsutum are associated with resistance to Verticillium dahliae. Mol. Plant Microbe. Interact. 30, 984–996. doi: 10.1094/MPMI-03-17-0067-R
Zhang, Y., Wang, X., Rong, W., Yang, J., and Ma, Z. (2016). Island cotton Enhanced Disease Susceptibility 1 gene encoding a lipase-like protein plays a crucial role in response to Verticillium dahliae by regulating the SA level and H2O2 accumulation. Front. Plant Sci. 7:1830. doi: 10.3389/fpls.2016.01830
Zhang, Y., Wu, L., Wang, X., Chen, B., Zhao, J., Cui, J., et al. (2018). The cotton laccase gene GhLAC15 enhances Verticillium wilt resistance via an increase in defence-induced lignification and lignin components in the cell walls of plants. Mol. Plant Pathol. 20, 309–322. doi: 10.1111/mpp.12755
Zhang, Z., Gei, X., Luo, X., Wang, P., Fan, Q., Hu, G., et al. (2018). Simultaneous editing of two copies of Gh14-3-3d confers enhanced transgene-clean plant defense against verticillium dahliae in allotetraploid upland cotton. Front. Plant Sci. 9:842. doi: 10.3389/fpls.2018.00842
Zhou, X., Jia, L., Wang, H., Zhao, P., Wang, W., Liu, N., et al. (2018). The potato transcription factor StbZIP61 regulates dynamic biosynthesis of salicylic acid in defense against Phytophthora infestans infection. Plant J. 95, 1055–1068. doi: 10.1111/tpj.14010
Keywords: Gossypium hirsutum L., Verticillium dahliae, verticillium wilt, auxin, salicylic acid, lignin
Citation: Tang Y, Zhang Z, Lei Y, Hu G, Liu J, Hao M, Chen A, Peng Q and Wu J (2019) Cotton WATs Modulate SA Biosynthesis and Local Lignin Deposition Participating in Plant Resistance Against Verticillium dahliae. Front. Plant Sci. 10:526. doi: 10.3389/fpls.2019.00526
Received: 11 December 2018; Accepted: 04 April 2019;
Published: 26 April 2019.
Edited by:
Jens Staal, Flanders Institute for Biotechnology, BelgiumReviewed by:
José Díaz, University of A Coruña, SpainChristina Dixelius, Swedish University of Agricultural Sciences, Sweden
Martijn Rep, University of Amsterdam, Netherlands
Copyright © 2019 Tang, Zhang, Lei, Hu, Liu, Hao, Chen, Peng and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingzhong Peng, cXpwZW5nanN1QDE2My5jb20= Jiahe Wu, d3VqaWFoZUBpbS5hYy5jbg==
 Ye Tang1,2
Ye Tang1,2 Jiahe Wu
Jiahe Wu