- 1College of Life Sciences, Shaanxi Normal University, Xi’an, China
- 2Department of Plant and Microbial Biology, University of Minnesota, Saint Paul, MN, United States
- 3National Engineering Laboratory for Resource Developing of Endangered Chinese Crude Drugs in Northwest of China, Shaanxi Normal University, Xi’an, China
Brassinosteroid insensitive 1 (BRI1) is a multidomain plant leucine-rich repeat receptor-like kinase (LRR-RLK), belongs to the LRR X subfamily. BRI1 perceives plant hormone brassinosteroids (BRs) through its extracellular domain that constitutes of LRRs interrupted by a 70 amino acid residue island domain (ID), which activates the kinase domain (KD) in its intracellular domain to trigger BR response. Thus, the KD and the ID of BRI1 are highly conserved and greatly contribute to BR functions. In fact, most bri1 mutants are clustered in or surrounded around the ID and the KD. However, the role of the less conserved LRR domains, particularly the first few LRRs after the signal peptide, is elusive. Here, we report the identification of a loss-of-function mutant bri1-235 that carries a mutation in the less conserved fourth LRR of BRI1 extracellular domain in Arabidopsis. This mutant had a base alteration from C to T, resulting in an amino acid substitution from serine to phenylalanine at the 156th position of BRI1. Compared with the wild-type plants, bri1-235 exhibited round leaves, prolonged life span, shorter stature, and approximately normal fertility under light conditions. The bri1-235 mutant was less sensitive to exogenous brassinolide under normal conditions. Importantly, both wild-type BRI1 expression and a sbi1 mutant that activates BRI1 rescued bri1-235 and resembled the wild type. Furthermore, bri1-235 protein was localized in endoplasmic reticulum rather than plasma membrane, suggestive of a cause for reducing BR sensitive in bri1-235. Taken together, our findings provide an insight into the role of the less conserved LRRs of BRI1, shedding light on the role of LRRs in a variety of LRR-RLKs that control numerous processes of plant growth, development, and stress response.
Introduction
In the complex and changing environment, plants have developed multiple strategies to sense various signals from their environment. These signals are first perceived at the cell surface and then transmitted to the cell’s interior. These events critically rely on cell-cell recognition and communication mediated by receptor-like kinases (RLKs). The structure organization of RLKs includes an extracellular domain, a single-pass transmembrane segment, and a cytoplasmic serine/threonine protein kinase domain (Shiu and Bleecker, 2001). Based on their different extracellular domains, RLKs can be divided into 13 subfamilies, of which the largest one is leucine-rich repeat (LRR) subfamily (Shiu and Bleecker, 2003). The LRRs in the extracellular domain of RLKs vary in number and distribution patterns, enabling them to have diverse functions. CLAVATA 1 (CLV1) with 21 LRRs controls meristem development (Clark et al., 1997). ERECTA (ER) and HAESA with 20 LRRs affect stomatal patterning and floral organ abscission, respectively (Stone et al., 1994; Jinn et al., 2000; Hunt and Gray, 2009). EXCESS MICROSPOROCYTES1 (EMS1) with 30 LRRs decides anther cell differentiation (Zhao et al., 2002). FLAGELLIN-SENSING 2 (FLS2) and EF-TU RECEPTOR (EFR), processing 28 LRRs and 21 LRRs, respectively, are involved in various responses to biotic stresses for innate defense in Arabidopsis (Gomez-Gomez et al., 2001; Zipfel et al., 2006). PHYTOSULFOKINE RECEPTOR 1 (PSKR1), with 21 LRRs interrupted by a 36-amino-acid island domain between 17th and 18th LRR, is involved in plant growth through elevating cell expansion and enhancing pathogen response (Matsubayashi et al., 2002; Amano et al., 2007; Mosher and Kemmerling, 2013). The co-receptor BRI1-ASSOSIATE KINASE1 (BAK1), with only five LRRs, directly associates with BRI1 in brassinosteroid signaling pathway and with FLS2 to mediate immune response (Nam and Li, 2002; Chinchilla et al., 2007). BAK1 also can interact with several other LRR-RLKs such as ER, EMS1, and PSKR1 (Zhao et al., 2002; Meng et al., 2015; Wang et al., 2015).
The crystal structure of a few LRR-RLKs have shown a right-handed superhelix of twisted LRRs (Di Matteo et al., 2003; Hothorn et al., 2011; She et al., 2011) or an anti-β-sheet (Wang et al., 2015). Variation in secondary structures on the convex side affects the curvature of the LRR domain, and this in turn permits interaction with an enormous diversity of ligands including hormone, peptide, and the entire protein. Many small signaling peptides have been identified, and typical examples are CLAVATA3 (CLV3) for CLV1 (Schoof et al., 2000), Phytosulfokine (PSK) for PSKR1 (Matsubayashi et al., 2002; Wang et al., 2015), Inflorescence deficient in abscission (IDA) for HAESA AND HAESA-LIKE2 (Butenko et al., 2003), and EPIDERMAL PATTERNING FACTORS (EPF) for ER (Shpak et al., 2005; Hara et al., 2007). The small protein, bacterial flagellin and TAPETUM DETERMINANT1 (TPD1) are perceived by FLS2 and EMS1, respectively (Gomez-Gomez and Boller, 2000; Yang et al., 2003). Among all LRR-RLKs in Arabidopsis, BRASSINOSTEROID INSENSITIVE 1 (BRI1) and BRI1-likes (BRLs) are the only examples that directly bind to phytohormone brassinosteroid (BR) (Cano-Delgado et al., 2004; Kinoshita et al., 2005). The newly formed BRI1-brassinolide (BL) surface can interact directly with extracellular residues of BAK1 via several hydrogen bonds (Santiago et al., 2013; Sun et al., 2013). The BRI1-BL-BAK1 complex can then initiate early BR signaling events and activate its downstream signaling cascade (Wang et al., 2008).
BRI1, with 25 LRRs and a 70-amino-acid island domain between 21st and 22nd LRR, regulates male fertility, flowering time, leaf senescence, vascular differentiation, dark-grown phenotype, and stress resistance (Clouse et al., 1996; Noguchi et al., 1999; Friedrichsen et al., 2000; Wang et al., 2001; Shang et al., 2011; Belkhadir et al., 2012; Gou et al., 2012). Three other BRI1 homologs, BRI1-LIKE 1 (BRL1), BRI1-LIKE 2 (BRL2), and BRI1-LIKE 3 (BRL3), were identified in Arabidopsis (Cano-Delgado et al., 2004; Zhou et al., 2004). Upon the expression of these homologs in Arabidopsis bri1 mutants, their phenotypes were rescued by BRL1 and BRL3, but not BRL2, showing that BRL1 and BRL3 but not BRL2 can interact with BL of high affinity (Cano-Delgado et al., 2004; Kinoshita et al., 2005). Yet, BRL2 regulated vascular development (Ceserani et al., 2009). Based on these observations, BRI1 and its three homologs have specific functions in cell growth and vascular differentiation in Arabidopsis (Wang et al., 2001; Cano-Delgado et al., 2004; Kinoshita et al., 2005). Since the discovery of BRI1, more than 30 unique bri1 mutants have been identified in Arabidopsis (Jiang et al., 2013). Their mutations are mainly clustered in the ID or the LRRs surrounding the ID in the extracellular region and the KD in the cytoplasmic region (Vert et al., 2005; Jiang et al., 2013). These mutants helped to determine the significance of the ID and KD of BRI1. However, the role of the less conserved LRR domains, particularly the first few LRRs after the signal peptide, remains unclear. The possible reason might be that this region is considered less important for BRI1 full function or may be neglected by researchers due to the lack of observable phenotype or systematic studies. Therefore, further studies are required to comprehensively understand the function of these less conserved LRR regions in different LRR-RLKs.
Here, we report the identification of a new mutation in the less conserved LRR regions of BRI1. This mutation significantly altered the growth and development of Arabidopsis plants. We described the isolation and characterization of this new bri1 allele and named it bri1-235, which showed a weak morphological phenotype and a reduced sensitivity to BRs. The point mutation from C to T in the region encoding the 4th LRR of BRI1 resulted in the conversion of Ser to Phe at the 156th position, leading to mislocalization of BRI1 and less BR sensitivity. Our findings on bri1-235 may provide insight to elucidate detailed functions of less conserved LRRs not only in BR receptors but also among numerous LRR-RLKs that control plant growth, development, and stress response.
Materials and Methods
Plant Materials and Growth Conditions
The ecotype Columbia (Col-0) of Arabidopsis thaliana was used as wild-type control. The bri1-235 was selected in the Col-0 background by ethyl methane sulfonate (EMS)-mutagenesis. bri1-5 mutant is in the Wassilewskija (Ws) background. The sbi1 (suppressor of bri1) is an A. thaliana bri1 suppressor mutant, which was identified from EMS-mutagenized bri1-5 (Wu et al., 2011). bri1-235 sbi1 was selected by crossing bri1-235 and sbi1 mutant. The seeds of Arabidopsis were surface-sterilized with 70% (v/v) ethanol for 3 min and 50% sodium hypochlorite for 8 min, followed by washing three times with sterilized water. After pre-incubated at 4°C in the dark for 3 days, the sterilized seeds were plated on 1/2 Murashige and Skoog (MS) medium containing 0.8% agar. After 7 days, seedlings were transferred to the moistened soil. Plants were grown under long daylight conditions (16-h light/8-h dark cycles) at 23°C.
Isolation and Mapping of bri1-235
The seeds of ecotype Col-0 were mutagenized with 0.4% ethyl methanesulfonate (EMS) (Sigma). The homozygous M0 seeds grown in the greenhouse were pollinated with Landsberg erecta (Ler) pollens to propagate M1 seeds, which germinated and allowed self-pollination to generate a segregating M2 mapping population. The dwarf seedlings of M2 were selected and grown under normal light to produce M3 seeds, which were subsequently subjected to the second-step screening for those showing a normal etiolated phenotype when grown in the dark.
Root and Hypocotyl Growth Assay
To determine the sensitivity of the plants to exogenous BR and BR biosynthesis inhibitor, the seeds of wild type, bri1-235, and transgenic plants were surface-sterilized, cold-stratified, germinated, and lined in petri plates containing 1/2 MS supplemented with 24-epibrassinolide (24-eBL, Sigma) at the 0, 0.1, 1, 10, 100, and 1,000 nM under light conditions or with propiconazole (PCZ) at the 0 and 5 μM under dark conditions. PCZ is a specific BR biosynthesis inhibitor in plants. PCZ binds to the BR biosynthesis enzyme CYP90D1, which is responsible for side chain C23 hydroxylation of BR, and inhibits the steps of BR biosynthesis from campestanol (CN) and teasterone (TE) (Hartwig et al., 2012; Oh et al., 2016). When the seedlings grew vertically in plate for 7 days, 20 seedlings were photographed and the hypocotyl length and root length were measured with Image J software. The measurement was repeated three times.
Endo H Treatment
The leaves from 4-week-old soil-grown plants were homogenized in “Tris, NaCl, EDTA, and Triton X (TSEX) buffer” (50 mM Tris pH 8.0, 100 mM NaCl, 5 mM EDTA, 0.2% Triton X-100, 10% glycerol, and protease inhibitors). After boiling for 5 min, the leaf samples were centrifuged for 10 min at 10,000 g. After centrifugation, the protein extracts were denatured at 95°C for 10 min in denaturing buffer and incubated with or without 1,000 U of endoglycosidase H (Endo H) (New England Biolabs) in the G5 buffer for 1 h at 37°C. The immunoprecipitated proteins were separated by 7% SDS/PAGE and immunoblotted with anti-BRI1 antibody. The anti-BRI1 antibody was purchased from Agrisera. Horseradish peroxidase-linked anti-rabbit antibodies were used as secondary antibodies, and the signal was detected by Western blotting. The experiment was repeated three times.
Confocal Microscopy
The localization patterns of BRI1-GFP, bri1-5-GFP, and bri1-235-GFP were examined by imaging the root tips of 5-day-old seedlings of BRI1-GFP, bri1-5-GFP, and bri1-235-GFP bri1-235 double transgenic lines with a Leica TCSSP8 confocal microscope using 100× water immersion objective. GFP signal was acquired on conditions of excitation at 488 nm with emission at 505–530 nm.
Generation of Transgenic Plants
Map-based cloning approach was used in cloning of pBRI1:BRI1-GFP, pBRI1:bri1-235-GFP, and pBRI1:bri1-5-GFP into expression. The full-length CDS fragment of BRI1 was amplified and cloned into a T-Vector PMD19. The cloned gene was sequence verified, digested, and cloned into the expression vector to get the construct pBRI1:BRI1-GFP, which was used for generating BRI1-GFP transgenic plants. The BRI1-GFP was transformed into bri1-235 mutant via the Agrobacterium tumefaciens (GV3101)–mediated floral dip method (Clough and Bent, 1998). To confirm the transformation, the transformed seeds were germinated and selected on 1/2 MS medium containing kanamycin (50 mg L−1).
Transcript Analysis by RT-PCR
The 2-week-old seedlings of Col-0, bri1-235, and BRI1-GFP/bri1-235 grown on 1/2 MS plates were collected. Total RNAs were extracted from whole plant seedlings by using RNeasy Plant Mini Kit (Qiagen). The mRNA concentration was estimated by spectrophotometer. First-strand cDNA was synthesized from 2 mg of total RNA by utilizing M-MLV, a reverse transcriptase (Invitrogen), following the manufacturer’s instruction. The resulting cDNAs corresponding to 100 ng of total RNAs were amplified by using StepOnePlus Real-Time PCR System (Applied Biosystems) with gene-specific primers for DWF4, CPD, BAS1, and ACT2 at an annealing temperature of 55°C for 30 cycles (Tanaka et al., 2005). Primers used for RT-PCR are given in the following paragraph.
DWF4 (forward: 5′-CGAAGGAAGGCTCTTTGAATG-3′ and reverse: 5′-CTTCAACGGCTTTAGGGCAA-3′); CPD (forward: 5′-GTTCTTATCCTGCTTCCATTTG-3′ and reverse: 5′-AGCCACTCGTAGCGTCTCATT-3′); BAS1 (forward: 5′-GTTCAGGACATTGTGGAGGAG-3′ and reverse: 5′-GGATAAAGCAACATAAGGACG-3′); and ACT2 (forward: 5′-ACTCTCCCGCTATGTATGTCG-3′ and reverse: 5′-TGGACCTG CCTCATCATACTC-3′). RT-PCR experiment was repeated three times.
Sequences Alignment and Phylogenetic Analysis
The amino acid sequences of BRI1, BRL1, BRL2, BRL3, and EMS1 of Arabidopsis thaliana were downloaded from the Arabidopsis thaliana TAIR1. The BRI1 protein sequences of Brachypodium distachyon, Oryza sativa, Setaria italica, Sorghum bicolor, Zea mays, Solanum lycopersicum, Solanum tuberosum, Populus trichocarpa, and Phaseolus vulgaris were retrieved by blast searches in the genome-wide sequenced plant databases from Phytozome v12.12. EMS1 was defined as outgroup. The multiple sequence alignment was made by DNAman software. The full-length protein sequences were aligned with ClustalX and manually adjusted. The maximum likelihood tree was calculated with MEGA5.2.2 for 1,000 bootstrap repetitions. Branch lengths are proportional to the number of substitutions per 100 residues (indicated by the bar below the tree), and the branch point values indicate the percentage bootstrap support.
Results
Isolation and Characterization of a New bri1 Allele, bri1-235
Using EMS mutagenesis and genetic screening, we isolated a weak morphological mutant when the mutants grew under normal light conditions. The mutants were dwarfed and displayed round leaves, short petioles, prolonged life span, reduced rosette size, and approximate normal fertility under the light conditions (Figures 1A,B). Compared to the wild-type Col-0, bri1-235 mutants of Arabidopsis exhibited reduced root elongation under light conditions and reduced hypocotyl elongation under dark conditions (Figures 1C–E). These results suggest that the mutation in this mutant results in reduced BR response in Arabidopsis.
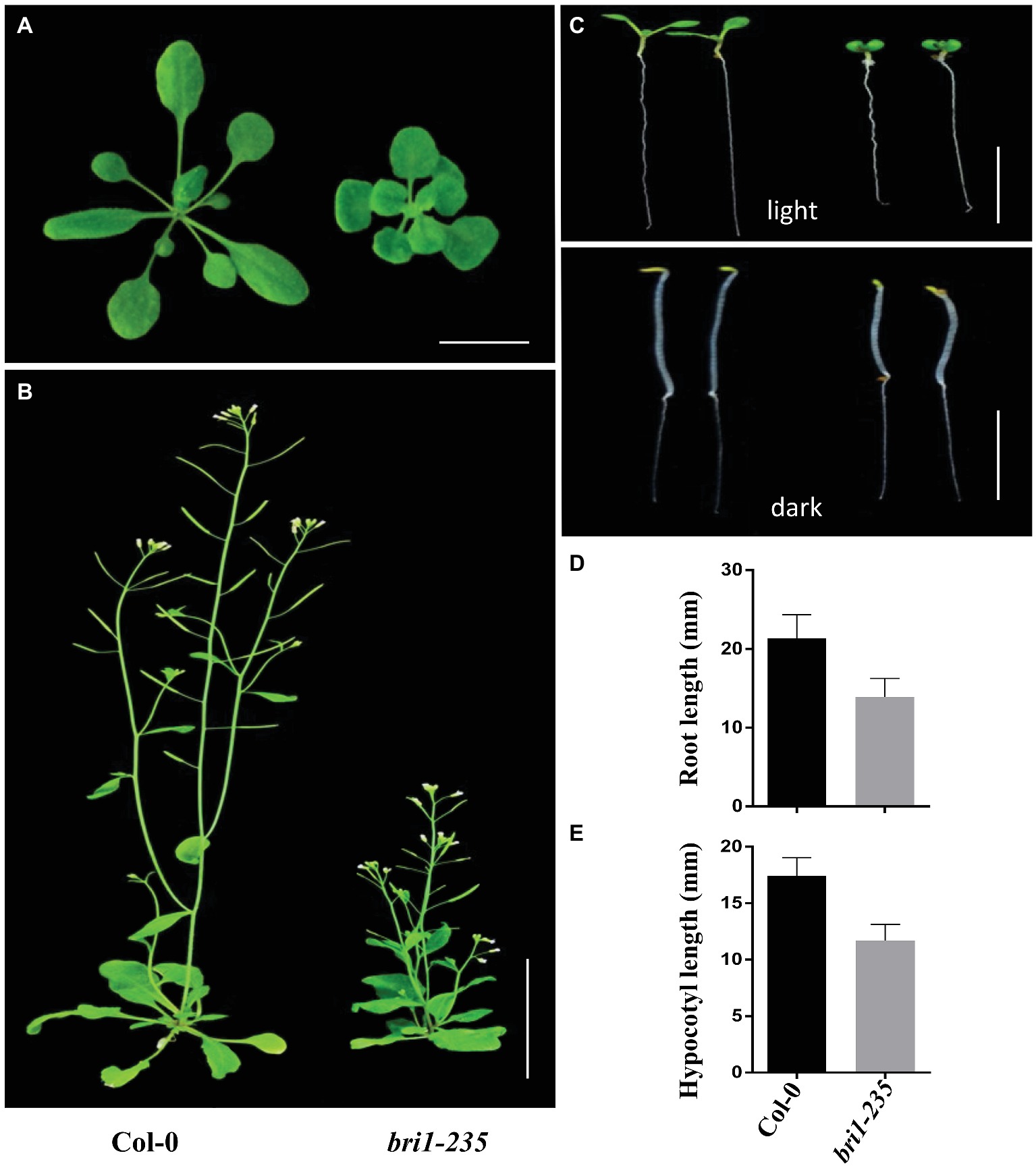
Figure 1. Phenotypes of bri1-235 mutant. (A) One-month-old wild type and bri1-235 grown under normal light at 23°C. Scale bar = 1 cm. (B) Two-month-old wild type and bri1-235 grown under normal light at 23°C. Scale bar = 5 cm. (C) Seven-day-old wild-type and bri1-235 seedlings grown on ½ MS plates under normal light or in the dark. Scale bar = 1 cm. (D) Quantitative measurements of root length of 7-day-old seedlings grown under normal light. (E) Quantitative measurements of hypocotyl length of 7-day-old seedlings grown in the dark. Data are presented the mean ± standard deviation from three independent biological replicates (n = 20 seedlings).
To identify the sequence alteration that resulted in bri1-235 phenotype, we amplified the genomic region of BRI1 using PCR and then performed DNA sequencing analysis on bri1 allele, bri1-235. We found a single base alteration by comparing the genomic DNA sequences of the bri1-235 mutant with the wild-type BRI1. The single base substitution from C to T in the 4th LRRs of BRI1 resulting in an amino acid substitution from serine to phenylalanine at the 156th amino acid position of BRI1 in the bri1-235 (Figure 2A). Sequence alignment of the 4th LRR showed that S residue in position 156 is highly conserved among different species. Apart from the serine at position 156, the other highly conserved residue in the 4th LRR is the asparagine at position 154, while most of its surrounding residues are not conversed among BRI1 in different species (Figure 2B).
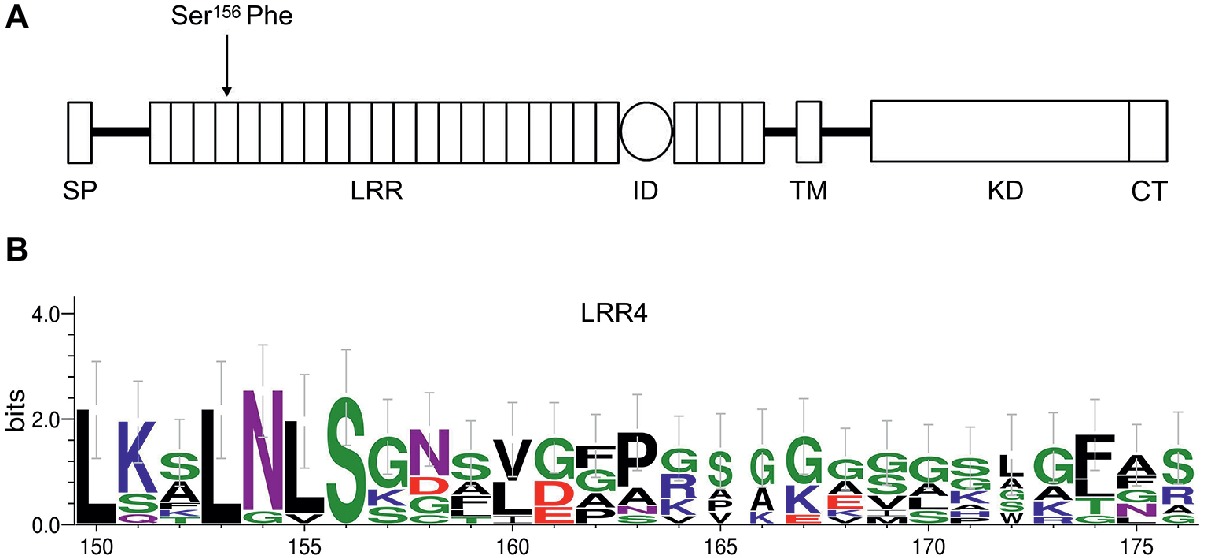
Figure 2. An amino acid substitution in bri1-235 from Ser to Phe at the 156th position of BRI1. (A) The full-length BRI1 protein with a defined signal peptide (SP) domain, leucine-rich repeat (LRR) domain, island domain (ID), transmembrane domain (TM), kinase domain (KD), and C-terminal (CT). bri1-235 has a point mutation in the region encoding the 4th LRR of BRI1, resulting in a change from Ser to Phe. (B) Sequence alignment of the 4th LRR among different species. S residue is highly conserved, and N at position 154 is the other highly conserved residue; however, other residues are less conversed among different species.
Most Amino Acids From the 1st to 10th LRR Are Variable Among BRI1 of Different Species
Among BRI1’s three homologs, BRL1 and BRL3 can also perceive and bind brassinosteroid; however, the ligand for BRL2 has not been found yet (Cano-Delgado et al., 2004). Phylogenetic analysis of BRI1 and its homologs in plants showed that BRI1 falls into the same clade with BRL1 and BRL3 but not with BRL2 (Figure 3A). Sequences of different plant species including five monocots (Oryza sativa, Brachypodium distachyon, Setaria italic, Sorghum bicolor, and Zea mays) and five dicots (Arabidopsis thaliana, Solanum lycopersicum, Solanum tuberosum, Populus trichocarpa, and Phaseolus vulgaris) were compared in similarity plots to study the conservation of the protein in this domain. Multiple sequence alignment of ectodomain BRI1 showed that the 11th–25th LRRs were highly conserved in the extracellular domains of BRI1 from different plant species. However, the 1st–10th LRR amino acid region of BRI1 extracellular domain was less conserved and highly variable (Figure 3B). In addition, there was insertion or deletion variation between the 1st and 10th LRR among different species. Similar alteration was found in between BRI1 and its homologs, BRL1, BRL3, and BRL2 from Arabidopsis thaliana.
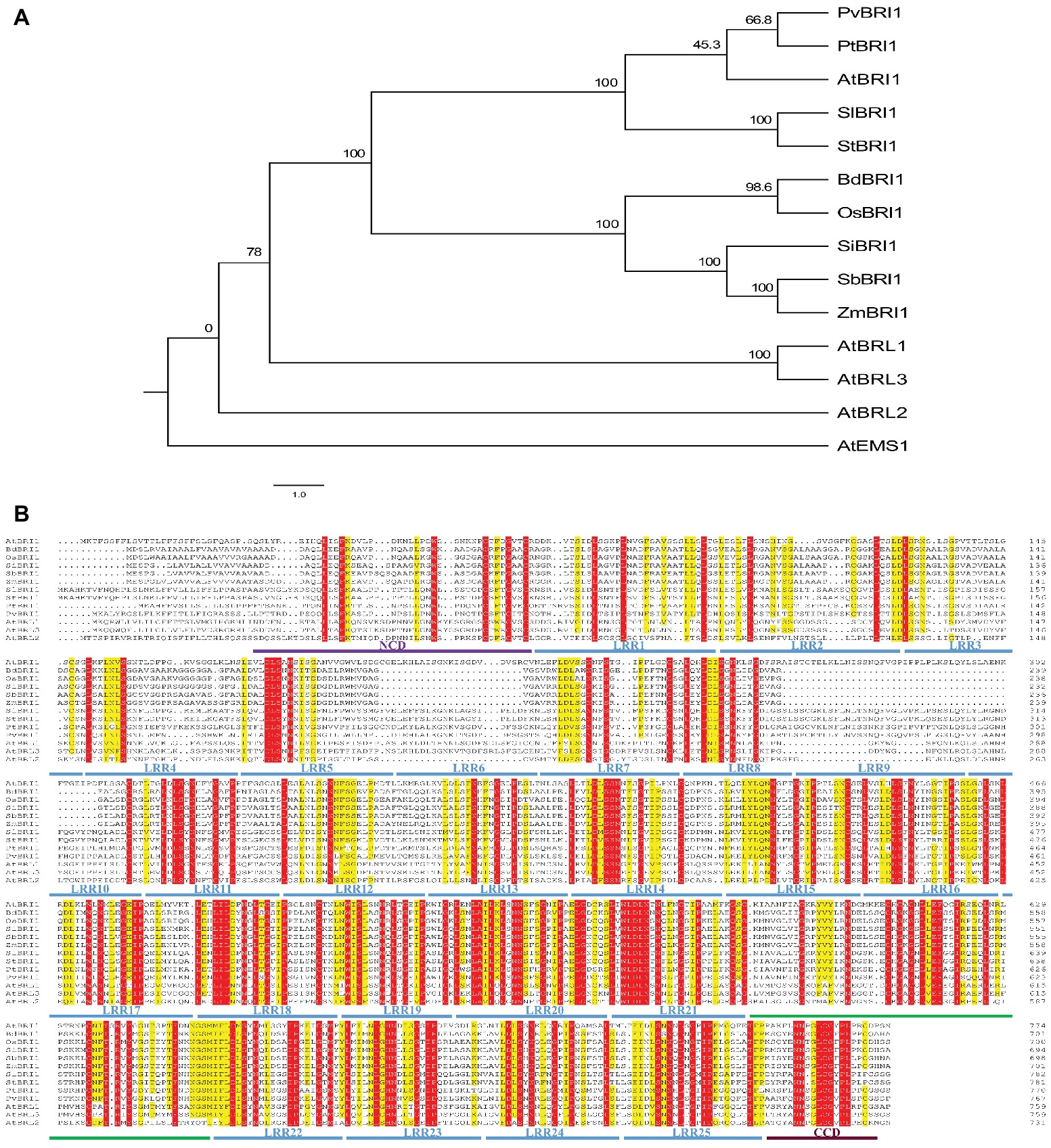
Figure 3. The phylogenetic relationship among BRI1’s homologs and sequence alignment of ectodomain BRI1 among different species. (A) The phylogenetic relationship among BRI1’s homologs. The full-length protein sequences were aligned with ClustalX and manually adjusted. The maximum likelihood tree was calculated with MEGA5.2.2 for 1,000 bootstrap repetitions. Branch lengths are proportional to the number of substitutions per 100 residues (indicated by the bar below the tree), and the branch point values indicate the percentage bootstrap support. (B) Conserved and similar residues are highlighted with red and yellow grounds, respectively. “NCD” and “CCD” represent the N-terminal and C-terminal capping domain, respectively. In (A) and (B), “At,” “Bd,” “Os,” “Si,” “Zm,” “Sl,” “St,” “Pt,” and “Pv” represent Arabidopsis thaliana, Brachypodium distachyon, Oryza sativa, Setaria italica, Zea mays, Solanum lycopersicum, Solanum tuberosum, Populus trichocarpa, and Phaseolus vulgaris, respectively.
Both BRI1 and sbi1 Can Rescue bri1-235 Phenotype
The phenotypic analyses of the bri1-235 and the single-base exchange in the BRI1 gene indicated that the mutant was caused by the loss-of-function mutation in the BRI1 gene. To confirm our hypothesis, we generated transgenic plants that expressed BRI1 or bri1-235 by introducing a BRI1 promoter-driven pBRI1:BRI1-GFP or pBRI1:bri1-235-GFP construct via the Agrobacterium tumefaciens–mediated floral dip method into bri1-235. Consistent with our prediction, the independent transgenic lines exhibited the phenotypes close to wild-type plants in the transgenics with pBRI1:BRI1-GFP, while overexpressing bri1-235 can partially rescue bri1-235 phenotypes. The bri1-235 transgenic plants overexpressing BRI1 showed the nearly normal overall growth patterns with elongated leaves and petiole length as well as total plant stature, similar to those observed in Col-0 as shown in Figures 4A,B. Simultaneously, we obtained bri1-235 sbi1 double mutants by crossing sbi1 to bri1-235. The sbi1 is a loss of function allele of the gene SBI1 encoding a leucine carboxylmethyltransferase that methylated protein phosphatase 2A and controlled its membrane-associated subcellular localization. Plant growth was promoted when SBI1 was mutated in Arabidopsis. Moreover, sbi1 regulates active BRI1 (Wu et al., 2011). Compared with bri1-235, sbi1 partially rescued bri1-235 phenotypes including plant height and rosette width (Figures 4C,D). These results indicate that bri1-235 is a bri1 allele with weak BR response and has kinase activity.
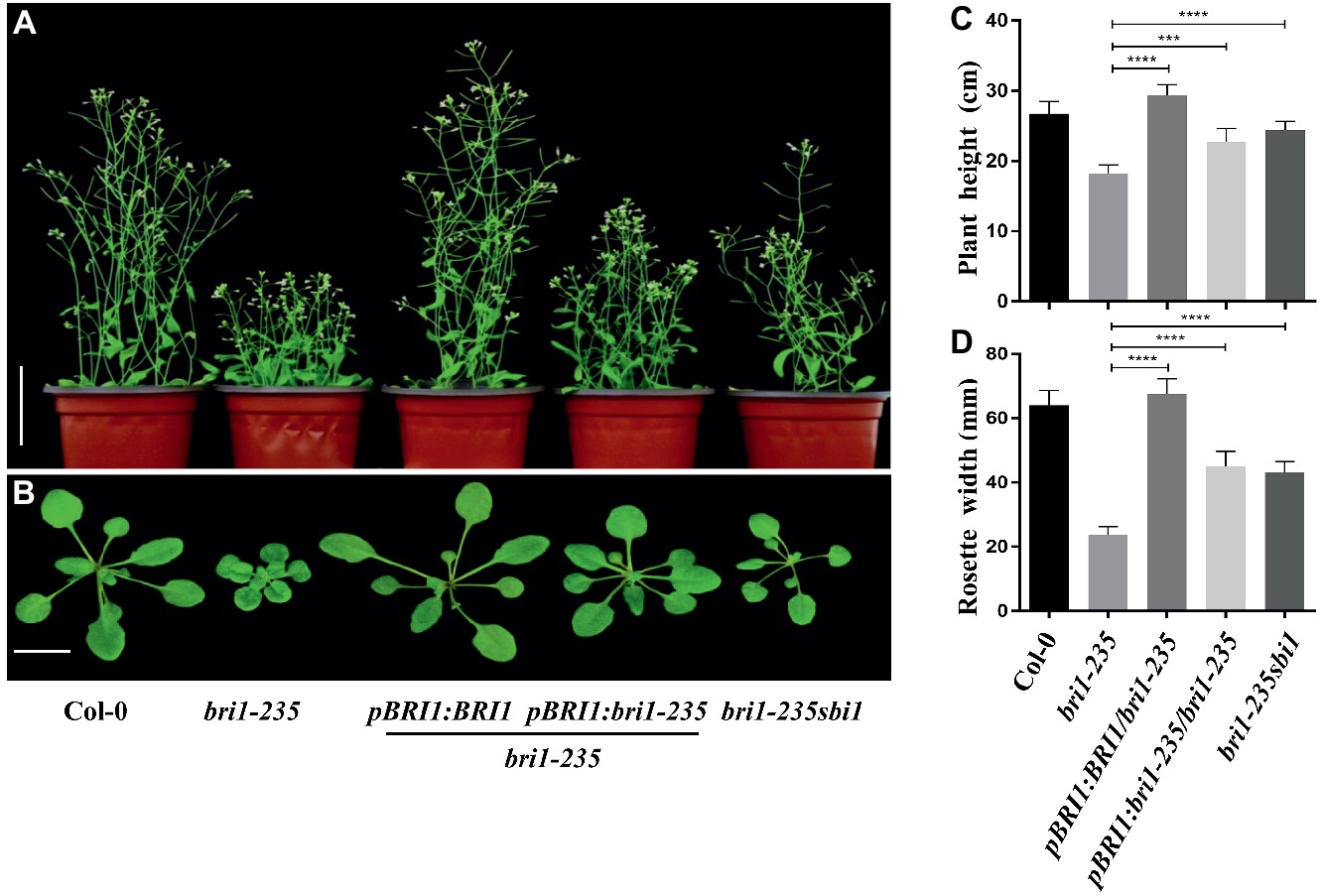
Figure 4. Phenotypes of bri1-235 transgenic lines. (A) Two-month-old transgenic lines expressing pBRI1:BRI1 and pBRI1:bri1-235 in bri1-235, respectively. bri1-235 sbi1 was produced by crossing bri1-235 and sbi1. Scale bar = 1 cm. (B) One-month-old seedlings of the indicated genotypes. Scale bar = 5 cm. (C) Quantitative measurements of plant height grown under normal light. (D) Quantitative measurements of rosette width grown under normal light. Data are presented as the mean ± standard deviation from three independent biological replicates (n = 20 seedlings). *** p < 0.001, **** p < 0.0001 as one-way ANOVA with a Tukey’s test.
The Sensitivity to BR is Reduced in bri1-235
As an essential plant hormone, BR can affect plant morphology at nanomolar level. To check sensitivity of bri1-235 and its transgenic plants to BR and PCZ, seeds were grown in 1/2 MS medium with or without various concentrations of 24-eBL or PCZ. Root growth inhibition analysis showed that the root growth of wild type was sensitive to exogenous 24-eBL treatment in a concentration-dependent manner. However, the roots of bri1-235 seedling were not significantly altered by the application of exogenous applied 24-eBL under light conditions (Figure 5). The transgenic lines, overexpressing BRI1 or bri1-235, restored sensitivity to BR to a certain extent. sbi1 can partially rescue the sensitivity to BR. With regard to the analyses of hypocotyl elongation in dark, the wild-type seedlings responded to exogenous 24-eBL in a concentration-dependent manner and showed reduced hypocotyl elongation, while bri1-235 showed less sensitivity to BL (Figure 5).
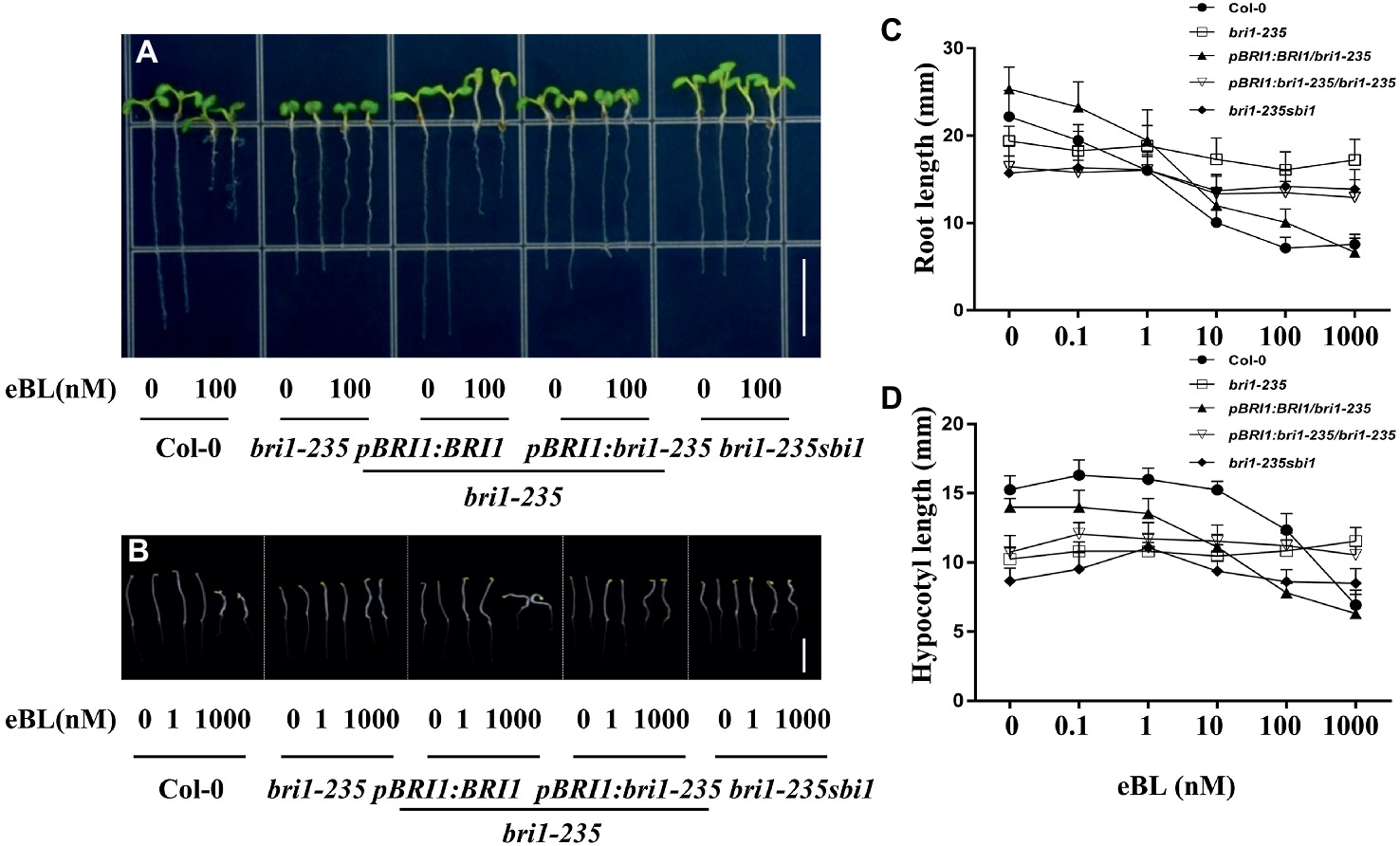
Figure 5. Sensitivity of bri1-235 and its transgenic lines to exogenous eBL. (A) Root phenotype of 5-day-old seedlings under light conditions. Scale bar = 1 cm. (B) Hypocotyl phenotype of 5-day-old seedlings in the dark. Scale bar = 1 cm. (C) Quantitative measurements of root elongation under normal light. (D) Quantitative measurements of hypocotyl elongation in the dark. Data are presented as the mean ± standard deviation from three independent biological replicates (n = 20 seedlings).
In addition, we also tested the sensitivity of this new bri1 allele to PCZ, which is a specific BR biosynthetic inhibitor. As expected, the hypocotyl of this weak mutant and wild-type seedlings exhibited hypersensitivity to PCZ (Figure 6). Similar trend was observed for root length, but it was less sensitive to PCZ as compared to hypocotyl. These findings confirm that the sensitivity of bri1-235 to BR or PCZ altered because of the substitution of serine with phenylalanine in the 4th LRR.
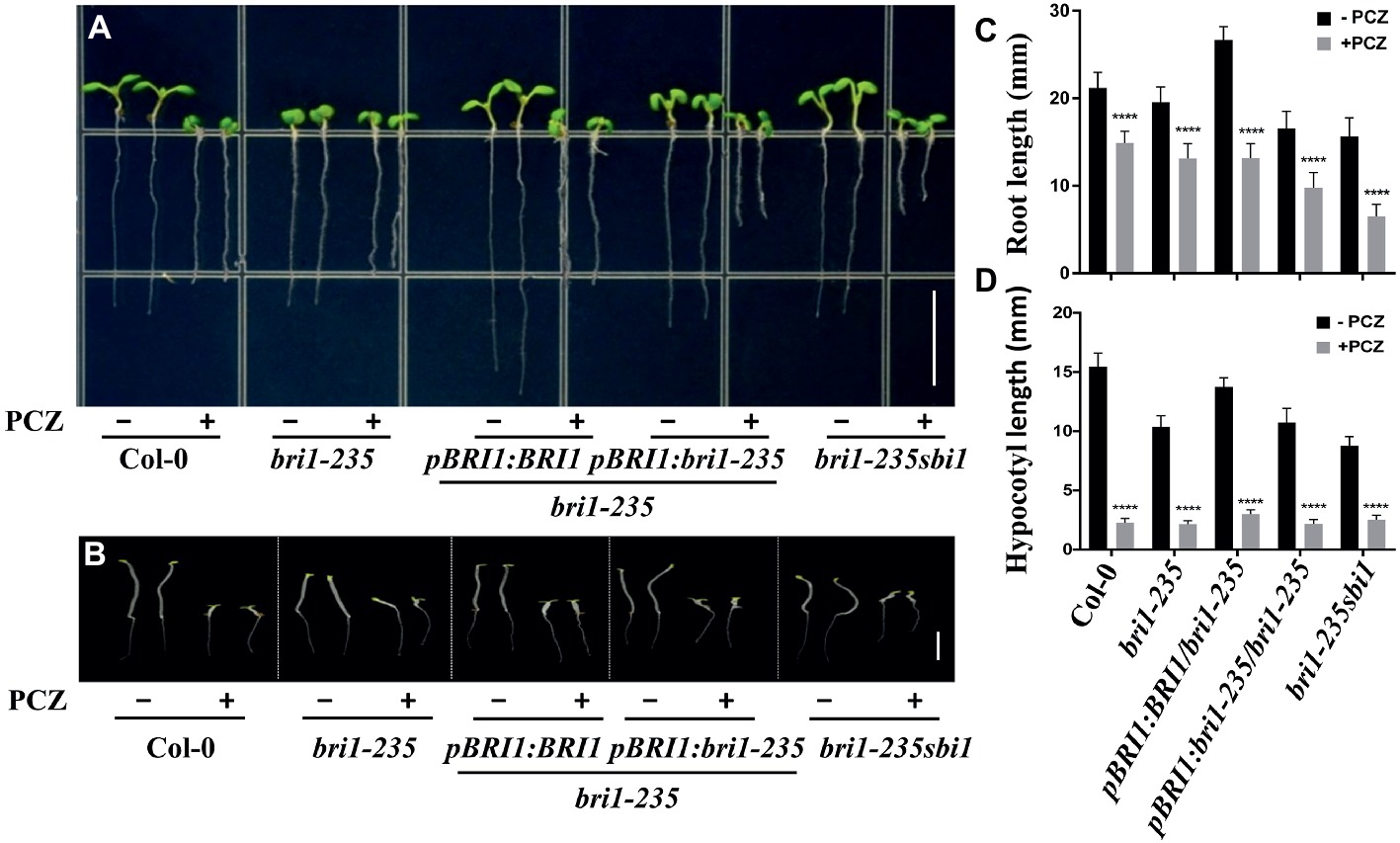
Figure 6. Sensitivity of bri1-235 and its transgenic lines to exogenous PCZ. (A) Root phenotype of 5-day-old seedlings grown on ½ MS plates with or without 5 μM PCZ under light conditions. Scale bar = 1 cm. (B) Hypocotyl phenotype of 5-day-old seedlings grown on ½ MS plates with or without 5 μM PCZ in the dark. Scale bar = 1 cm. (C) Quantitative measurements of root elongation under normal light. (D) Quantitative measurements of hypocotyl elongation in the dark. Data are presented as the mean ± standard deviation from three independent biological replicates (n = 20 seedlings). **** p < 0.0001 as one-way ANOVA with a Tukey’s test.
Several brassinosteroid response genes (i.e., DWF4, CPD, and BAS1) in the brassinosteroid signaling pathway are subjected to feedback regulation upon BL treatment. DWF4 and CPD encode 22α-hydroxylase and C-3 oxidase, respectively, which are involved in BR biosynthesis and are negatively regulated by BRs (Mathur et al., 1998; Gonzalez-Gaitan, 2008). BAS1 encodes a cytochrome P450 that is involved in BR degradation and is positively regulated by BRs (Turk et al., 2003). BR biosynthesis genes, DWF4 and CPD, and BR inactivation genes BAS1 are sensitive markers for enhanced BR signaling (Tanaka et al., 2005; Wang and Chory, 2006; Tang et al., 2008). To further test the altered response of bri1-235 to exogenous 24-eBL, we checked the expression level of these BR responsive genes in the wild type, bri1-235, and the pBRI1:BRI1-GFP/bri1-235 mutants under exogenous 24-eBL. The results show that the expression level of DWF4 and CPD were strongly suppressed by 24-eBL treatment in wild-type but not in bri1-235 mutants (Figure 7). Similar to wild type, the expression level of DWF4 and CPD was strongly suppressed by 24-eBL in the pBRI1:BRI1-GFP/bri1-235. The expression of BAS1 was significantly induced by exogenous 24-eBL in the wild-type plants, but slightly affected in bri1-235 mutant. As expected, similar to the effect of BR on the wild plants, BAS1 was significantly increased by exogenous 24-eBL in the pBRI1:BRI1-GFP/bri1-235 (Figure 7).
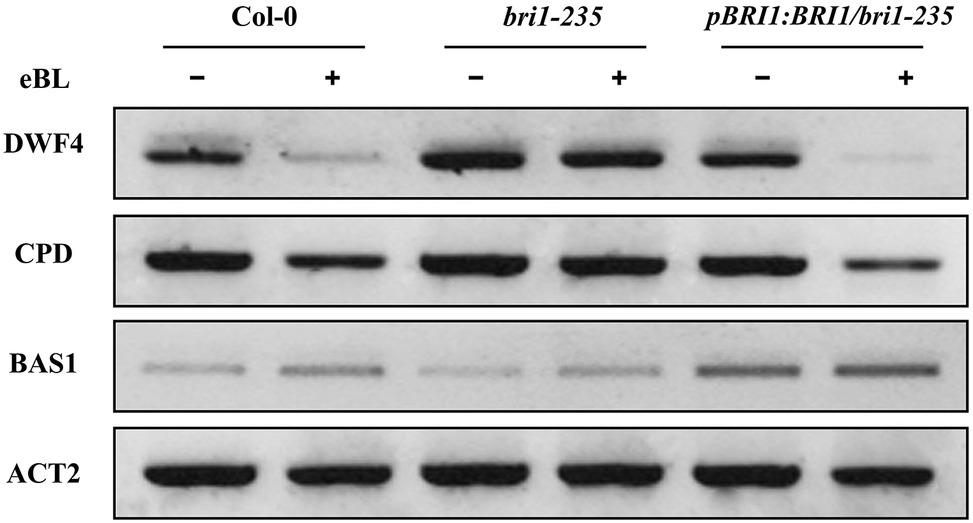
Figure 7. Expression levels of DWF4, CPD, and BAS1 in wild type, bri1-235, and pBRI1:BRI1/bri1-235 seedling grown on ½ MS medium without or with 100 nM eBL in the dark.
The bri1-235 Protein Is Localized in the Endoplasmic Reticulum
Mislocalization of cell surface receptors can lead to their malfunctions. To determine the mechanism of the loss function of bri1-235, we asked the localization of bri1-235 protein in bri1-235. The subcellular localization of bri1-235 was first analyzed by its sensitivity to Endo H that cleaves high mannose-type N-glycans of the endoplasmic reticulum (ER)-localized proteins, whereas it fails to cleave Golgi-processed complex glycans. Thus, Endo H treatment can distinguish the ER-localized BRI1 from the plasma membrane (PM)-localized BRI1. After the total proteins of the wild type, the bri1-235 and bri1-5 seedlings were treated with or without Endo H, the samples were subjected to gel electrophoresis followed by immunoblotting with anti-BRI1 antibody (Mora-Garcia et al., 2004). bri1-5 carrying Cys69Tyr mutation is a proven bri1 mutant whose bri1-5 protein is mainly located in ER (Hong et al., 2008). Here bri1-5 is a positive control to support the ER localization of bri1-235. As shown in Figure 8A, the abundance of bri1-235 was reduced slightly than that of wild type. BRI1 in the wild type was located in the plasma membrane; however, the bri1-235 protein appeared completely sensitive to Endo H digestion, which indicates ER localization (Figure 8B). Confocal study revealed that bri1-235-GFP was localized in endoplasmic reticulum rather than plasma membrane as well (Figure 8C). bri1-9 is another ER-retaining bri1 mutant with Ser662Phe mutation in the 22nd LRR of BRI1 extracellular domain (Jin et al., 2007). Therefore, our results clearly demonstrate that bri1-235 is a new mutant, similar to bri1-5 or bri1-9 mutants, in term of receptor’s mislocalization.
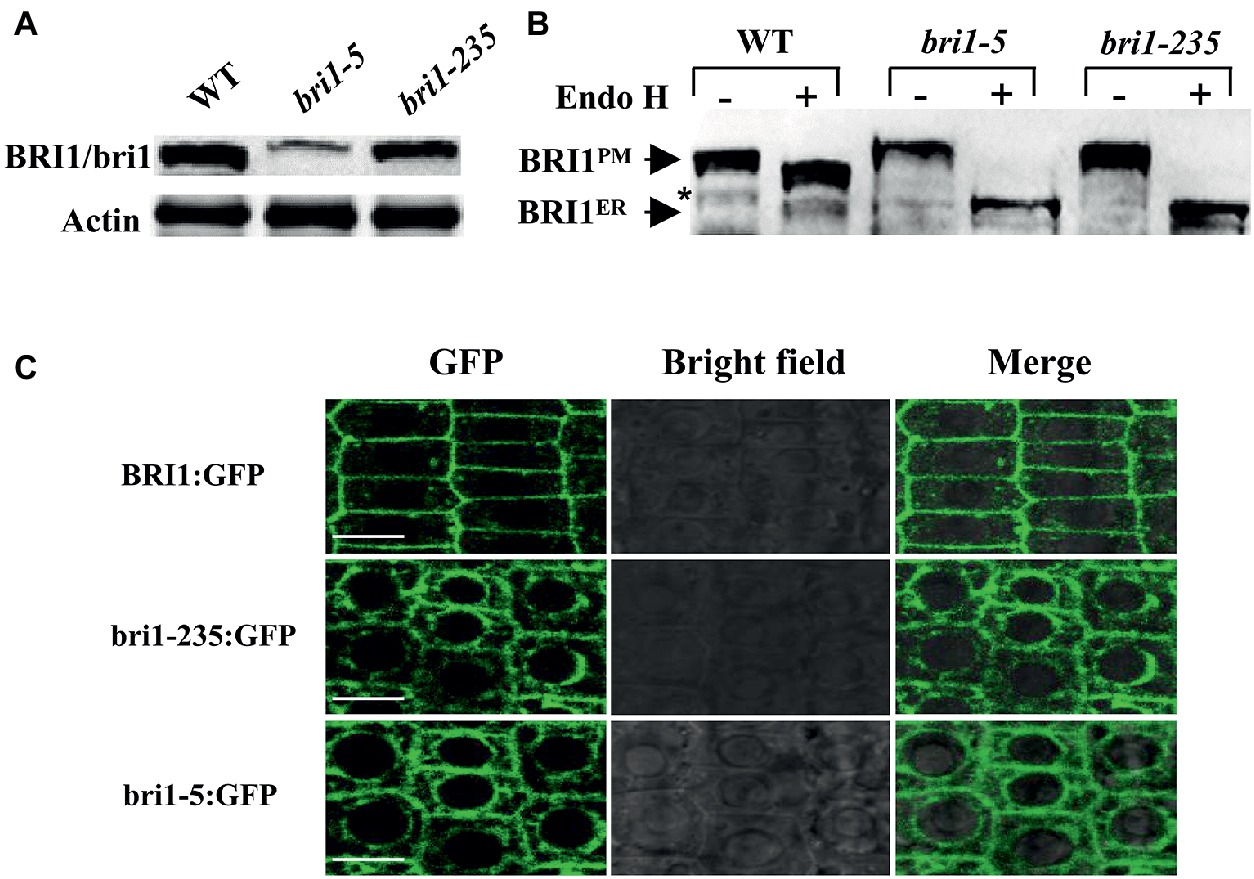
Figure 8. bri1-235 is retained in the endoplasmic reticulum (ER). (A) BRI1 and bri1 were detected by Western blotting. Actin served as the loading control. (B) Endo H assays of bri1-5 and bri1-235. The arrows indicate the positions of bands representing BRI1 in the plasma membrane (BRI1PM) and in the ER (BRI1ER). The asterisk denotes a nonspecific band. (C) Confocal microscopic analysis of BRI1:GFP, bri1-235:GFP, and bri1-5:GFP. Fluorescence patterns of GFP fusion proteins, bright field, and the merged images of green and bright field signals in root tip cells of Arabidopsis are shown from left to right. Scale bar = 5 μm.
Discussion
In the past years, molecular genetics studies have discovered and characterized many BR responsive genes that function in BR biosynthesis, signaling, and inactivation. These studies have provided insight into molecular mechanisms of BR action. DWF4 and CPD, BR biosynthesis genes, were upregulated by exogenous BL in bri1-235. Meanwhile BAS1, a BR inactivation gene, was downregulated in bri1-235 as compared to wild type (Figure 7). This indicates the feedback regulation of BR responsive genes to maintain BR homeostasis and regulate plant development.
The extracellular domains of LRR-RLKs are versatile and diverse, while its kinase domains are conversed in plant. The ID of BRI1 and the LRRs surrounding the ID, are responsible for perceiving BRs (Kinoshita et al., 2005). The crystal structure of BRI1 shows the ID of BRI1 folds back into the interior of the superhelix to bind BL. The superhelix makes the extensive polar and hydrophobic interaction with the 13th LRR–25th LRR (Hothorn et al., 2011; She et al., 2011). Such twisted assemblies of LRRs and superhelix also exist in the plant defence protein PGIP in Phaseolus vulgaris (Di Matteo et al., 2003). Therefore, twisted LRRs in brassinosteroid binding extend to the region from 13th LRR to 25th LRR containing ID. Interestingly, there are significant difference in conservation between the 1st to 10th LRR and 11th to 25th LRR among distinct plant species. The key amino acids of the 11th–25th LRRs were conserved and well matched in the extracellular domain of BRI1 from different plant species including Arabidopsis thaliana, Brachypodium distachyon, Oryza sativa, and Zea mays (Figure 3). However, the amino acid cannot well align in the 1st–10th LRRs of the extracellular domains due to insertion or deletion variation in BRI1 from different species (Figure 3). This indicates the first few LRRs after the signal peptide are less conserved among the BRI1 from distinct species. This high variation in the first 10 LRRs might indicate that the first 10 LRRs are expendable and have only minor or specific role for the receptors’ function. As such, its significance is difficult to uncover, constituting a reason that there is no missense mutants identified in this region so far. In the LRRs of extracellular domain, two mutants, bri1-4 and bri1-120, occur at the 3rd and 13th LRRs, respectively. In bri1-4, a 10-bp segment is deleted, which results in premature termination of the translation resulting in truncated protein. Thus, the bri1-4 gene is a null allele of bri1, whose mutant is comparable to the other null mutant phenotype (Noguchi et al., 1999). The bri1-120 contains a substitution of Ser by Phe at the 399th amino acid in the 13th LRR. This leads to the conformational change affecting the receptor dimerization or reducing the ligand binding capacity (Shang et al., 2011). The finding of bri1-120 showed that the LRR domain plays important role in BR perception. However, no information was known regarding the first few less conserved LRRs after the signal peptide in BRI1, which could also play a critical role in hormone perceiving and receptor activation. Recently, bri1-706 mutants, carried a point mutation Ser-to-Phe in the 8th LRR of the extracellular portion of BRI1, were scanned via Targeted Induced Local Lesions IN Genomes (TILLING) analysis and waiting to be further analyzed (Sun et al., 2017).
In the present investigation, we reported the isolation of a weak bri1 allele, bri1-235. This weak mutant generated by EMS mutagenesis had a single base change from C to T, which led to an amino acid substitution of Ser by Phe at the 156th position. This Ser residue is highly conserved in the 4th LRR of BRI1 (Figure 2). Like other canonical LRR protein, the concave surface of the BRI1 solenoid is composed of a parallel β sheet comprising 25 continuously running parallel β-strands. However, the BRI1 possesses parallel but distorted β-sheets following the inner β-structure in most of the repeats due to plant-specific consensus sequences. The loops of the convex outer surface are stabilized through disulfide bonds formed between consecutive repeats just except the 4th, 9th, and 13th LRR (She et al., 2011). As serine or threonine is able to form an additional hydrogen bond with other part of protein (Choe et al., 2005), the serine residue at the 4th LRR of BRI1 might participate in the formation of the hydrogen bond network, which stabilizes the assembly of repeating LRR motifs. Replacement of this solvent-exposed Ser residue by a bulky hydrophobic Phe reside in bri1-235 mutant might cause a localized structural distortion. However, such a structural perturbation caused only mildly loss of the BRI1 function. When we overexpressed wild-type bri1-235 in bri1-235, its phenotypes were rescued to almost the wild type (Figure 4). Furthermore, we found that sbi1 (suppressor of bri1) also restored bri1-235 to near the wild type (Figure 4). As previously reported, sbi1 only acts on active BRI1 but not inactive BRI1 (Wu et al., 2011). Therefore, our results suggest that bri1-235 is still an active BR receptor. Apart from the serine at position 156, the other highly conserved residue in the 4th LRR is the asparagine (Asn) at position 154 (Figure 2B). According to N-glycosylation sites along the BRI1 superhelix from the C-terminus, this Asn residue is glycosylated and has an important role in structural stabilizes of BRI1 (Hothorn et al., 2011). Together Asn and Ser residues constitute a classic consensus N-glycosylation site (Asn-X-Ser). Mutation of the Ser at position 156 abrogates N-glycosylation at position 154, which may lead to local misfolding or trapping in ER quality control processes, and is the likely cause for the phenotype similar to previous findings for bri1-5 and bri1-9, two best-characterized endoplasmic reticulum-localized bri1 mutants. Thus, mutation of the Ser at position 156 critical for BRI1 folding may be involved to the glycosylation. Subcellular localization of proteins is a vital characteristic that depicts their functions in the cell. This is true for the hormonal receptors because they sense signaling molecules coming from outer space of the cells and transform this signal into the cell interior, thus triggering a precise biological process that affects primary cellular targets in the downstream. BRI1 was reported to undergo a highly conserved ER-mediated protein quality control (ERQC) mechanism before further trafficking to plasma membrane. Among identified BRI1 mutants, bri1-5 and bri1-9 are two typical examples whose proteins are retained in the endoplasmic reticulum. The dwarf phenotype of bri1-9, which carries a Ser-to-Phe mutation in the ligand binding domain of BRI1, is caused by endoplasmic reticulum retention of a structurally imperfect but functionally competent BR receptor via ERQC (Jin et al., 2007). In the endoplasmic reticulum, the stringency of the retention-based ERQC is reduced significantly due to mutations, and the bri1-9 protein was allowed to export to the cell surface for BR perception. Unlike bri1-9, bri1-5 carries a Cys-to-Tyr mutation, which localizes in the amino-terminal pair cysteine before the beginning of the LRRs. The bri1-5 protein is also retained in the endoplasmic reticulum, involving three different retention mechanisms (Hong et al., 2008). Like bri1-5 and bri1-9, the amino acid substitution in bri1-235 occurs in the extracellular domain of BRI1. The susceptibility of bri1-235 to Endo H is similar to that of bri1-5, which retains in the ER (Figure 8). Moreover, the confocal analysis also shows that the bri1-235 is localized to ER instead of PM (Figure 8). Further investigation is required to find out whether bri1-235 is involved in the disturbing of ERQC system or other retention mechanisms. Nevertheless, our study confirms that not only island and the subsequent 13th–25th LRR but also the first few LRRs after the signal peptide are critical for BRI1 folding.
Conclusion
In summary, we demonstrated that the new bri1 mutant, bri1-235, exhibited a weak morphological phenotype and a reduced sensitivity to exogenous BL. The point mutation is found in the first few less conserved LRRs after the signal peptide in BRI1. It was confirmed that the mutation resulted in an amino acid substitution from Ser to Phe at the 156th position of BRI1. Mutation occurred in the 1st–10th LRR regions are slightly conserved and are highly variable among different plant species. bri1-235 can rescue bri1-235 mutant to near wild type. Furthermore, sbi1, which acts on active BRI1, also restores the bri1-235 to almost wild type. Therefore, the bri1-235 is a new mutant having mislocalized functional BR receptor. Our findings likely provide insight into the crucial role of less conserved LRRs in BR receptor BRI1, shedding light on study of many other RLKs.
Data Availability
All datasets generated for this study are included in the manuscript.
Author Contributions
GL and QH conceived and designed the work. QH and SS performed the experiments and analyzed the data. HR helped with selecting this mutant. GL prepared the manuscript draft. KA, CB, and GW critically reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31270324), China Postdoctoral Science Foundation (2012M521739), the Fundamental Research Funds for the Central Universities (GK201503046), and the Innovation Team Project in Fundamental Research Funds of the Central Universities (GK201801008).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Amano, Y., Tsubouchi, H., Shinohara, H., Ogawa, M., and Matsubayashi, Y. (2007). Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc. Natl. Acad. Sci. USA 104, 18333–18338. doi: 10.1073/pnas.0706403104
Belkhadir, Y., Jaillais, Y., Epple, P., Balsemao-Pires, E., Dangl, J. L., and Chory, J. (2012). Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc. Natl. Acad. Sci. USA 109, 297–302. doi: 10.1073/pnas.1112840108
Butenko, M. A., Patterson, S. E., Grini, P. E., Stenvik, G. E., Amundsen, S. S., Mandal, A., et al. (2003). Inflorescence deficient in abscission controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 15, 2296–2307. doi: 10.1105/tpc.014365
Cano-Delgado, A., Yin, Y., Yu, C., Vafeados, D., Mora-Garcia, S., Cheng, J. C., et al. (2004). BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131, 5341–5351. doi: 10.1242/dev.01403
Ceserani, T., Trofka, A., Gandotra, N., and Nelson, T. (2009). VH1/BRL2 receptor-like kinase interacts with vascular-specific adaptor proteins VIT and VIK to influence leaf venation. Plant J. 57, 1000–1014. doi: 10.1111/j.1365-313X.2008.03742.x
Chinchilla, D., Zipfel, C., Robatzek, S., Kemmerling, B., Nurnberger, T., Jones, J. D., et al. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500. doi: 10.1038/nature05999
Choe, J., Kelker, M. S., and Wilson, I. A. (2005). Crystal structure of human toll-like receptor 3 (TLR3) ectodomain. Science 309, 581–585. doi: 10.1126/science.1115253
Clark, S. E., Williams, R. W., and Meyerowitz, E. M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. doi: 10.1016/S0092-8674(00)80239-1
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Clouse, S. D., Langford, M., and McMorris, T. C. (1996). A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111, 671–678. doi: 10.1104/pp.111.3.671
Di Matteo, A., Federici, L., Mattei, B., Salvi, G., Johnson, K. A., Savino, C., et al. (2003). The crystal structure of polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein involved in plant defense. Proc. Natl. Acad. Sci. USA 100, 10124–10128. doi: 10.1073/pnas.1733690100
Friedrichsen, D. M., Joazeiro, C. A., Li, J., Hunter, T., and Chory, J. (2000). Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 123, 1247–1256. doi: 10.1104/pp.123.4.1247
Gomez-Gomez, L., Bauer, Z., and Boller, T. (2001). Both the extracellular leucine-rich repeat domain and the kinase activity of FSL2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell 13, 1155–1163. doi: 10.1105/tpc.13.5.1155
Gomez-Gomez, L., and Boller, T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011. doi: 10.1016/S1097-2765(00)80265-8
Gonzalez-Gaitan, M. (2008). The garden of forking paths: recycling, signaling, and degradation. Dev. Cell 15, 172–174. doi: 10.1016/j.devcel.2008.07.001
Gou, X., Yin, H., He, K., Du, J., Yi, J., Xu, S., et al. (2012). Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet. 8:e1002452. doi: 10.1371/journal.pgen.1002452
Hara, K., Kajita, R., Torii, K. U., Bergmann, D. C., and Kakimoto, T. (2007). The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 21, 1720–1725. doi: 10.1101/gad.1550707
Hartwig, T., Corvalan, C., Best, N. B., Budka, J. S., Zhu, J. Y., Choe, S., et al. (2012). Propiconazole is a specific and accessible brassinosteroid (BR) biosynthesis inhibitor for Arabidopsis and maize. PLoS One 7:e36625. doi: 10.1371/journal.pone.0036625
Hong, Z., Jin, H., Tzfira, T., and Li, J. (2008). Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell 20, 3418–3429. doi: 10.1105/tpc.108.061879
Hothorn, M., Belkhadir, Y., Dreux, M., Dabi, T., Noel, J. P., Wilson, I. A., et al. (2011). Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474, 467–471. doi: 10.1038/nature10153
Hunt, L., and Gray, J. E. (2009). The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 19, 864–869. doi: 10.1016/j.cub.2009.03.069
Jiang, J., Zhang, C., and Wang, X. (2013). Ligand perception, activation, and early signaling of plant steroid receptor brassinosteroid insensitive 1. J. Integr. Plant Biol. 55, 1198–1211. doi: 10.1111/jipb.12081
Jin, H., Yan, Z., Nam, K. H., and Li, J. (2007). Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol. Cell 26, 821–830. doi: 10.1016/j.molcel.2007.05.015
Jinn, T. L., Stone, J. M., and Walker, J. C. (2000). HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 14, 108–117. doi: 10.1101/gad.14.1.108
Kinoshita, T., Cano-Delgado, A., Seto, H., Hiranuma, S., Fujioka, S., Yoshida, S., et al. (2005). Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433, 167–171. doi: 10.1038/nature03227
Mathur, J., Molnar, G., Fujioka, S., Takatsuto, S., Sakurai, A., Yokota, T., et al. (1998). Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J. 14, 593–602. doi: 10.1046/j.1365-313X.1998.00158.x
Matsubayashi, Y., Ogawa, M., Morita, A., and Sakagami, Y. (2002). An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science 296, 1470–1472. doi: 10.1126/science.1069607
Meng, X., Chen, X., Mang, H., Liu, C., Yu, X., Gao, X., et al. (2015). Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr. Biol. 25, 2361–2372. doi: 10.1016/j.cub.2015.07.068
Mora-Garcia, S., Vert, G., Yin, Y., Cano-Delgado, A., Cheong, H., and Chory, J. (2004). Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 18, 448–460. doi: 10.1101/gad.1174204
Mosher, S., and Kemmerling, B. (2013). PSKR1 and PSY1R-mediated regulation of plant defense responses. Plant Signal. Behav. 8:e24119. doi: 10.4161/psb.24119
Nam, K. H., and Li, J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212. doi: 10.1016/S0092-8674(02)00814-0
Noguchi, T., Fujioka, S., Choe, S., Takatsuto, S., Yoshida, S., Yuan, H., et al. (1999). Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 121, 743–752. doi: 10.1104/pp.121.3.743
Oh, K., Matsumoto, T., Hoshi, T., and Yoshizawa, Y. (2016). In vitro and in vivo evidence for the inhibition of brassinosteroid synthesis by propiconazole through interference with side chain hydroxylation. Plant Signal. Behav. 11, e1158372–e1158376. doi: 10.1080/15592324.2016.1158372
Santiago, J., Henzler, C., and Hothorn, M. (2013). Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 341, 889–892. doi: 10.1126/science.1242468
Schoof, H., Lenhard, M., Haecker, A., Mayer, K. F., Jurgens, G., and Laux, T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644. doi: 10.1016/S0092-8674(00)80700-X
Shang, Y., Lee, M. M., Li, J., and Nam, K. H. (2011). Characterization of cp3 reveals a new bri1 allele, bri1-120, and the importance of the LRR domain of BRI1 mediating BR signaling. BMC Plant Biol. 11:8. doi: 10.1186/1471-2229-11-8
She, J., Han, Z., Kim, T. W., Wang, J., Cheng, W., Chang, J., et al. (2011). Structural insight into brassinosteroid perception by BRI1. Nature 474, 472–476. doi: 10.1038/nature10178
Shiu, S. H., and Bleecker, A. B. (2001). Plant receptor-like kinase gene family: diversity, function, and signaling. Sci. STKE 2001:re22. doi: 10.1126/stke.2001.113.re22
Shiu, S. H., and Bleecker, A. B. (2003). Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 132, 530–543. doi: 10.1104/pp.103.021964
Shpak, E. D., McAbee, J. M., Pillitteri, L. J., and Torii, K. U. (2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309, 290–293. doi: 10.1126/science.1109710
Stone, J. M., Collinge, M. A., Smith, R. D., Horn, M. A., and Walker, J. C. (1994). Interaction of a protein phosphatase with an Arabidopsis serine-threonine receptor kinase. Science 266, 793–795. doi: 10.1126/science.7973632
Sun, Y. D., Han, Z. F., Tang, J., Hu, Z. H., Chai, C. L., Zhou, B., et al. (2013). Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res. 23, 1326–1329. doi: 10.1038/cr.2013.131
Sun, C., Yan, K., Han, J. T., Tao, L., Lv, M. H., Shi, T., et al. (2017). Scanning for new BRI1 mutations via TILLING analysis. Plant Physiol. 174, 1881–1896. doi: 10.1104/pp.17.00118
Tanaka, K., Asami, T., Yoshida, S., Nakamura, Y., Matsuo, T., and Okamoto, S. (2005). Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol. 138, 1117–1125. doi: 10.1104/pp.104.058040
Tang, W., Kim, T. W., Oses-Prieto, J. A., Sun, Y., Deng, Z., Zhu, S., et al. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321, 557–560. doi: 10.1126/science.1156973
Turk, E. M., Fujioka, S., Seto, H., Shimada, Y., Takatsuto, S., Yoshida, S., et al. (2003). CYP72B1 inactivates brassinosteroid hormones: an intersection between photomorphogenesis and plant steroid signal transduction. Plant Physiol. 133, 1643–1653. doi: 10.1104/pp.103.030882
Vert, G., Nemhauser, J. L., Geldner, N., Hong, F., and Chory, J. (2005). Molecular mechanisms of steroid hormone signaling in plants. Annu. Rev. Cell Dev. Biol. 21, 177–201. doi: 10.1146/annurev.cellbio.21.090704.151241
Wang, X., and Chory, J. (2006). Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313, 1118–1122. doi: 10.1126/science.1127593
Wang, X. F., Kota, U., He, K., Blackburn, K., Li, J., Goshe, M. B., et al. (2008). Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev. Cell 15, 220–235. doi: 10.1016/j.devcel.2008.06.011
Wang, J., Li, H., Han, Z., Zhang, H., Wang, T., Lin, G., et al. (2015). Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature 525, 265–268. doi: 10.1038/nature14858
Wang, Z. Y., Seto, H., Fujioka, S., Yoshida, S., and Chory, J. (2001). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410, 380–383. doi: 10.1038/35066597
Wu, G., Wang, X., Li, X., Kamiya, Y., Otegui, M. S., and Chory, J. (2011). Methylation of a phosphatase specifies dephosphorylation and degradation of activated brassinosteroid receptors. Sci. Signal. 4:ra29. doi: 10.1126/scisignal.2001258
Yang, S. L., Xie, L. F., Mao, H. Z., Puah, C. S., Yang, W. C., Jiang, L., et al. (2003). Tapetum determinant1 is required for cell specialization in the Arabidopsis anther. Plant Cell 15, 2792–2804. doi: 10.1105/tpc.016618
Zhao, D. Z., Wang, G. F., Speal, B., and Ma, H. (2002). The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 16, 2021–2031. doi: 10.1101/gad.997902
Zhou, A., Wang, H., Walker, J. C., and Li, J. (2004). BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. Plant J. 40, 399–409. doi: 10.1111/j.1365-313X.2004.02214.x
Keywords: Arabidopsis, BRI1, less conserved, LRRs, endoplasmic reticulum
Citation: Hou Q, Saima S, Ren H, Ali K, Bai C, Wu G and Li G (2019) Less Conserved LRRs Is Important for BRI1 Folding Front. Plant Sci. 10:634. doi: 10.3389/fpls.2019.00634
Edited by:
Seth Jon Davis, University of York, United KingdomReviewed by:
Xiaofeng Wang, Northwest University, ChinaRichard Strasser, University of Natural Resources and Life Sciences Vienna, Austria
Copyright © 2019 Hou, Saima, Ren, Ali, Bai, Wu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guishuang Li, Z3Vpc2h1YW5nbGlAc25udS5lZHUuY24=
†These authors have contributed equally to this work
 Qiang Hou1†
Qiang Hou1† Guang Wu
Guang Wu Guishuang Li
Guishuang Li