Abstract
Glutaredoxins (GRXs) have at least three major identified functions. In apoforms, they exhibit oxidoreductase activity controlling notably protein glutathionylation/deglutathionylation. In holoforms, i.e., iron–sulfur (Fe–S) cluster-bridging forms, they act as maturation factors for the biogenesis of Fe–S proteins or as regulators of iron homeostasis contributing directly or indirectly to the sensing of cellular iron status and/or distribution. The latter functions seem intimately connected with the capacity of specific GRXs to form [2Fe–2S] cluster-bridging homodimeric or heterodimeric complexes with BOLA proteins. In yeast species, both proteins modulate the localization and/or activity of transcription factors regulating genes coding for proteins involved in iron uptake and intracellular sequestration in response notably to iron deficiency. Whereas vertebrate GRX and BOLA isoforms may display similar functions, the involved partner proteins are different. We perform here a critical evaluation of the results supporting the implication of both protein families in similar signaling pathways in plants and provide ideas and experimental strategies to delineate further their functions.
Introduction
Many cellular reactions and biological processes require metalloproteins, among which those containing iron (Fe) cofactors such as mononuclear and dinuclear (non-heme) Fe centers, hemes and iron–sulfur (Fe–S) clusters, are particularly crucial. Unlike other metals such as copper or zinc, there is no universal Fe chaperone described and so far, only poly rC-binding proteins (PCBPs) were shown to coordinate Fe entry in the cytosol and serve for the metalation of non-heme Fe enzymes in mammals (Philpott et al., 2017). In contrast, the synthesis/assembly of hemes and Fe–S clusters requires more complex and universally conserved pathways (Couturier et al., 2013; Barupala et al., 2016). The machineries dedicated to the maturation of Fe–S proteins present in mitochondria and chloroplasts, named ISC (iron–sulfur cluster) and SUF (sulfur mobilization), respectively, are also found in bacteria (Lill, 2009). On the other hand, cytosolic and nuclear Fe–S proteins are maturated via the eukaryote-specific cytosolic iron–sulfur cluster assembly (CIA) machinery, which is, however, dependent on the mitochondrial ISC machinery for sulfur supply (Lill, 2009). Hence, given the high cellular demand for iron, sophisticated systems exist to control Fe uptake and intracellular distribution due to its potential toxicity. Strikingly, the Fe sensing systems and associated transcription factors generally differ in bacteria, yeast/fungi, mammals, and plants, but might include common actors such as glutaredoxins (GRXs) and BOLAs (Couturier et al., 2015).
Two GRX classes, I and II, are present in most organisms whereas additional classes are specific to some species/genus/kingdoms (Alves et al., 2009; Couturier et al., 2009). GRXs of the first class are involved in redox regulation, reducing protein disulfides or glutathione-protein mixed disulfides. GRXs from class II participate in the regulation of Fe homeostasis (Mühlenhoff et al., 2010; Haunhorst et al., 2013) and in the maturation of Fe–S proteins owing to their capacity to ligate and exchange [2Fe–2S] clusters with partner proteins (Table 1; Rodríguez-Manzaneque et al., 2002; Bandyopadhyay et al., 2008). They are also referred to as monothiol GRXs or CGFS GRXs owing to their conserved CGFS active site signature.
Table 1
| Organism | Protein names | Mutant phenotype(s) | References | |
|---|---|---|---|---|
| Mono-domain (organellar) GRXs | Saccharomyces cerevisiae | Grx5 | Defaults in Fe–S cluster assembly | Rodríguez-Manzaneque et al., 2002; Mühlenhoff et al., 2003 |
| Schizosaccharomyces pombe | Grx5 | Defaults in Fe–S cluster assembly, decreased amount of mitochondrial DNA, reduced growth, and sensitivity toward oxidants | Chung et al., 2005; Kim et al., 2010 | |
| Danio rerio | GRX5 | Embryo lethal | Wingert et al., 2005 | |
| Homo sapiens | GLRX5 | Defaults in Fe–S cluster assembly leading to sideroblastic anemia | Camaschella et al., 2007; Ye et al., 2010 | |
| Trypanosoma brucei | 1-C-Grx1 | Lethal | Comini et al., 2008 | |
| Sinorhizobium meliloti | Grx2 | Defaults in Fe–S cluster assembly, deregulation of RirA transcriptional activity, increased intracellular iron content, modified nodule development | Benyamina et al., 2013 | |
| Escherichia coli | Grx4 | Sensitivity to iron depletion, defect in respiratory complex I | Yeung et al., 2011; Burschel et al., 2019 | |
| Arabidopsis thaliana | GRXS14 | Sensitivity to prolonged darkness | Rey et al., 2017 | |
| Arabidopsis thaliana | GRXS15 | Lethal, decreased amounts of lipoate synthase and of lipoic acid dependent H subunits of the glycine cleavage system in RNAi lines | Moseler et al., 2015; Ströher et al., 2016 | |
| Arabidopsis thaliana | GRXS16 | None described for co-suppressed and RNAi lines | Rey et al., 2017 | |
| Multi-domain (cytosolic) GRXs | Saccharomyces cerevisiae | Grx3 | Impaired regulation of Aft1/2 and iron homeostasis | Ojeda et al., 2006; Pujol-Carrion et al., 2006 |
| Saccharomyces cerevisiae | Grx4 | Impaired regulation of Aft1/2 and iron homeostasis | Ojeda et al., 2006; Pujol-Carrion et al., 2006 | |
| Saccharomyces cerevisiae | Grx3–Grx4 | Lethal in some background. Impaired iron trafficking and assembly of Fe–S proteins, heme, and iron-containing proteins | Pujol-Carrion et al., 2006; Mühlenhoff et al., 2010 | |
| Schizosaccharomyces pombe | Grx4 | Lethal | Chung et al., 2005 | |
| Cryptococcus neoformans | Grx4 | Slow growth upon iron deprivation | Attarian et al., 2018 | |
| Danio rerio | GRX3 | Impaired heme synthesis and Fe–S protein maturation | Haunhorst et al., 2013 | |
| Homo sapiens | GLRX3/PICOT | Decreased activities of cytosolic Fe–S proteins | Haunhorst et al., 2013 | |
| Arabidopsis thaliana | GRXS17 | Growth defects (meristem arrest) upon elevated temperature and long photoperiod. No decrease in cytosolic Fe–S protein activity | Cheng et al., 2011; Knuesting et al., 2015; Yu et al., 2017 | |
| BOLA | Saccharomyces cerevisiae | Bol1 | No growth defect and no decrease in Fe–S enzyme activity | Melber et al., 2016; Uzarska et al., 2016 |
| Saccharomyces cerevisiae | Bol3 | Slightly decreased complex II (SDH) activity | Melber et al., 2016; Uzarska et al., 2016 | |
| Saccharomyces cerevisiae | Bol1–Bol3 | Decreased activity of lipoic acid-dependent enzymes, aconitase, and respiratory complex II | Melber et al., 2016; Uzarska et al., 2016 | |
| Saccharomyces cerevisiae | Bol2/Fra2 | Impaired regulation of Aft1/2 and iron homeostasis | Kumánovics et al., 2008; Uzarska et al., 2016 | |
| Schizosaccharomyces pombe | BolA2/ Fra2 | Impaired regulation of the Fep1 transcription factor | Jacques et al., 2014 | |
| Homo sapiens | BOLA1 | Oxidation of the mitochondrial GSH pool | Willems et al., 2013 | |
| Homo sapiens | BOLA2 | None described for siRNA lines | Frey et al., 2016 | |
| Homo sapiens | BOLA3 | Defect in lipoic acid-dependent enzymes and in respiratory complexes I and II | Cameron et al., 2011 | |
| Escherichia coli | BolA | Partial defect in respiratory complex I assembly | Burschel et al., 2019 | |
| Escherichia coli | IbaG | None described | Burschel et al., 2019 | |
| Escherichia coli | BolA – IbaG | Decreased complex II activity | Burschel et al., 2019 | |
| Salmonella typhimurium | BolA | Decreased resistance to acidic and oxidative stresses and decreased virulence | Mil-Homens et al., 2018 | |
| Arabidopsis thaliana | BOLA2 | None described under control conditions, increased resistance to oxidative conditions | Qin et al., 2015 | |
Iron-related phenotypes of bolA and glutaredoxin mutants from various sources.
Regarding the BOLA family, an extensive phylogenetic analysis allowed delineating four groups, namely BOLA1–BOLA4 (Couturier et al., 2014). BOLA1s are present in both bacteria and eukaryotes, BOLA2s and BOLA3s in eukaryotes and BOLA4s in photosynthetic organisms, archaea, and bacteria. Pioneer works revealed functions for Escherichia coli BolA in the regulation of cell morphology, possibly as a transcriptional regulator (Aldea et al., 1989), for Saccharomyces cerevisiae cytosolic Bol2/Fra2 (Fe repressor of activation 2) in the regulation of iron homeostasis (Lesuisse et al., 2005; Kumánovics et al., 2008), and for mitochondrial BOLAs (human BOLA3 and yeast Bol1, Bol3) in the maturation of Fe–S clusters (Table 1; Cameron et al., 2011; Melber et al., 2016; Uzarska et al., 2016).
A very close relationship between class II GRXs and BOLAs was initially evident from genome (gene co-occurrence and clustering, existence of fusion proteins) and large-scale interactomic analyses in various organisms (reviewed in Przybyla-Toscano et al., 2017). Then, the molecular and structural determinants of the complexes were investigated in detail using mutational, spectroscopic and structural analyses on recombinant proteins. This led to demonstrate that class II GRXs and BOLAs form both apo- and holo-heterodimers bridging a [2Fe–2S] cluster, usually more stable than the [2Fe–2S] cluster-bridging GRX homodimers, and to identify the residues serving as ligands (Li and Outten, 2012; Couturier et al., 2015; Przybyla-Toscano et al., 2017). In GRX-BOLA holo-heterodimers, the [2Fe–2S] cluster is ligated using the GRX conserved cysteine, a cysteine from glutathione (as in GRX holo-homodimers), and, on the BOLA side, using a C-terminally located conserved histidine and an histidine or a cysteine in the β1–β2 loop, referred to as [H/C] loop (Figure 1A; Li et al., 2011, 2012; Roret et al., 2014; Dlouhy et al., 2016; Nasta et al., 2017).
FIGURE 1
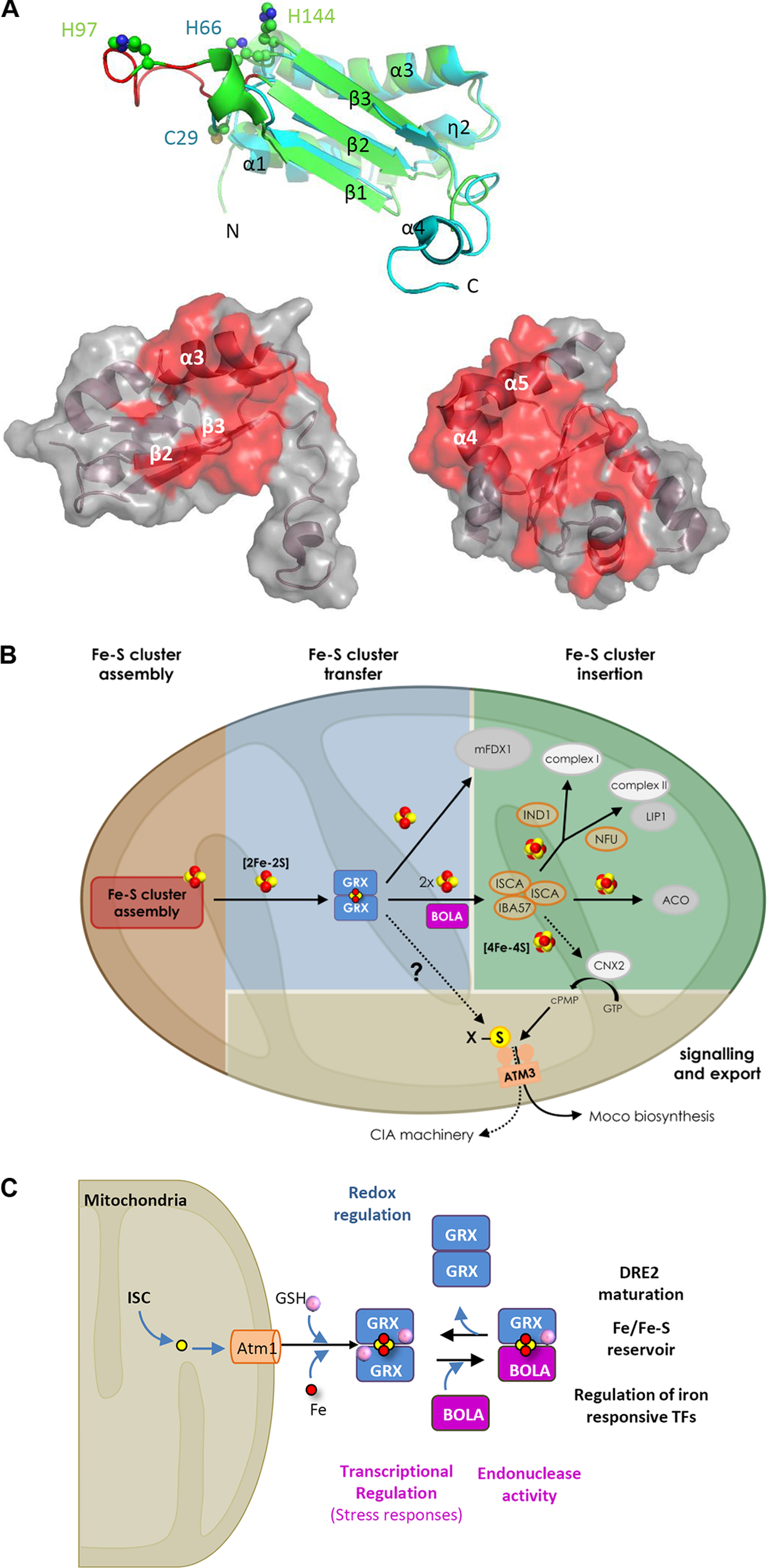
Properties and hypothetical roles of the class II GRX-BOLA couple in plants. (A) Tridimensional structures of plastidial Arabidopsis thaliana BOLA1 and GRXS14 proteins highlighting the residues involved in the interactions. On the top, superimposition of AtBOLA1 (green) and AtBOLA2 (blue) structures. Both proteins have a α/β-structure made of four helices and three strands with an α1β1β2η2α3β3α4 (η: 310-helix) topology (Roret et al., 2014). The β-strands form a central three-stranded β-sheet. In addition to the extended C-terminal part in AtBOLA2, both proteins differ by the length of the β1–β2 loop (in red in BOLA1), referred to as [H/C] loop, and which contains the histidine (His97 in AtBOLA1) or cysteine (Cys29 in AtBOLA2) residues provided by BOLA proteins for Fe–S cluster bridging together with the His66 (AtBOLA2) or His144 (AtBOLA1). The putative DNA binding site in BOLAs is formed by the η2 and α3 helices, the loop containing a specific FXGX signature (type II β-turn), the α3 helix containing a positively charged RHR motif and the β3 strand. Below, from left to right, AtBOLA2 structure showing 18 residues (mainly part of the β2 and β3 strands and α3 helix) identified by NMR titration as involved in the interaction with apo-AtGRXS14; and AtGRXS14 structure showing 32 residues (many present in the C-terminal α3 and α4 helices) identified by NMR titration as involved in the interaction with AtBOLA2 (Roret et al., 2014). These residues, plus some additional ones, are also involved in the formation of the [2Fe–2S] cluster-bridged heterodimer as determined using human proteins (Nasta et al., 2017). (B) Hypothetical model for the role of GRXS15 and BOLA4 in plant mitochondria. By analogy with the yeast system, GRXS15 (shortened as GRX) should receive a [2Fe–2S] cluster synthesized de novo by a multi-protein assembly complex (details about the proteins involved in the early steps of Fe–S cluster assembly and transfer have been omitted). GRXS15 is supposed to transfer its [2Fe–2S] cluster to client proteins as the mitochondrial ferredoxin 1 (mFDX1) (Moseler et al., 2015) or to ISCA proteins for the reductive conversion of two [2Fe–2S] clusters into a [4Fe–4S] cluster (as shown with human proteins) and its subsequent delivery to client proteins bearing such cluster. In the absence of genetic analysis about bola4 mutants, the contribution of BOLA4 (shortened as BOLA) for the respective roles of GRXS15 is unclear, but the confirmed interaction between both proteins (Couturier et al., 2014) prompted us to include BOLA at this step as yeast Bol1/3 proteins are only required for the maturation of [4Fe–4S] proteins. The specific defects observed for aconitase (ACO) and lipoic-acid dependent proteins in the GRXS15 mutant lines indicate a direct or indirect role for GRXS15 in the maturation of both lipoate synthase (LIP1) and aconitase. Finally, whether GRXS15 is required for the maturation and activity of cytosolic and nuclear Fe–S proteins by fueling the CIA machinery as shown for yeast Grx5, or by indirectly contributing to the synthesis of molybdenum cofactor, that is present in several cytosolic Fe–S proteins, is unknown. (C) Roles associated with the various oligomeric forms involving nucleo-cytosolic GRXs and BOLAs irrespective of the organisms considered. The color code is as follows: in blue, functions associated with apo-dimeric GRX forms, in purple those associated with apo-BOLA and in black those associated with the GRX homodimeric or GRX-BOLA heterodimeric forms bridging a [2Fe–2S] cluster.
Hereafter, based on the most recent results and known roles in non-photosynthetic organisms, we discuss the putative or confirmed functions of GRX and BOLA, alone or in complex, in photosynthetic organisms.
The Class II GRX and Bola Couple Proteins Present in Bacteria or in Eukaryote Organelles Are Involved in the Synthesis of Fe–S Clusters
The first evidence about GRX involvement in the biogenesis of Fe–S proteins were obtained from a S. cerevisiae mutant for the mitochondrial Grx5 (Table 1; Rodríguez-Manzaneque et al., 2002; Mühlenhoff et al., 2003). Orthologs of this single domain-containing GRX are found in bacteria, archaea and plant plastids. The current view is that Grx5 is required for the maturation of all types of Fe–S clusters in mitochondria, receiving a [2Fe–2S] cluster from ISCU-type scaffold proteins and transferring it to ISCA-type transfer proteins for subsequent maturation of [4Fe–4S] proteins (Figure 1B). Grx5 is also required for the maturation of nucleo-cytosolic Fe–S proteins and the activation of the Aft1 transcription factor, pointing to its key position in S. cerevisiae (see below) (Uzarska et al., 2013). Yeast Bol1 and Bol3, which have the capacity to form heterodimers with Grx5, were later shown to be required for a specific set of mitochondrial [4Fe–4S] proteins, without affecting de novo synthesis of [2Fe–2S] proteins (Uzarska et al., 2016). So far, human BOLA3, but not BOLA1, has been demonstrated as required for the maturation of specific Fe–S proteins (Table 1; Cameron et al., 2011; Willems et al., 2013). The client proteins are notably the succinate dehydrogenase/complex II and lipoate synthase. Moreover, the fact that bol1–bol3Δ mutants are neither affected in the CIA machinery, nor in Aft1 activation, indicates that Grx5 has physiological roles independent of Bol1 and Bol3 (Uzarska et al., 2016). Additional studies suggested that Bol1 indeed acts early in the ISC pathway in concert with Grx5 (possibly only for [4Fe–4S] proteins) whereas Bol3 may preferentially act with NFU1, a late Fe–S cluster transfer protein, to preserve the [4Fe–4S] center found in some specific mitochondrial client proteins, as lipoate synthase, from oxidative damage (Figure 1B; Melber et al., 2016).
Concerning bacteria, the sole Grx isoform (Grx4/D) and both BolAs (BolA and IbaG) from E. coli were recently shown as implicated in the maturation of the respiratory complexes I and II, but the effects are only visible when multiple genes are mutated (Burschel et al., 2019). This role in maturating Fe–S proteins is consistent with (i) the synthetic lethality of the Grx4 gene with genes present in the ISC operon (Butland et al., 2008), (ii) the interaction in vitro of Grx4 with the MiaB Fe–S protein (Boutigny et al., 2013), and (iii) the capacity of Grx4-BolA and Grx4-IbaG to form the usual [2Fe–2S] cluster-bridging heterodimers (Yeung et al., 2011; Dlouhy et al., 2016). In Sinorhizobium meliloti, deletion of the sole class II GRX also leads to impaired maturation of Fe–S proteins and increased intracellular iron content (Benyamina et al., 2013).
In plants, the corresponding mitochondrial GRX is named GRXS15. Knockout Arabidopsis mutants are lethal due to defective embryo development (Moseler et al., 2015). Plants expressing a mutated GRXS15 form modified for its ability to coordinate an Fe–S cluster exhibit severely reduced growth and impaired aconitase activity (Moseler et al., 2015). Additionally, Arabidopsis GRXS15 down-regulated lines display slowed growth and impaired activity of enzymes dependent on lipoic acid, the synthesis of which is ensured by the Fe–S cluster-containing lipoyl synthase (Ströher et al., 2016). Whether GRXS15 fulfills its function in concert with BOLA4, the sole mitochondrial BOLA, remains to be explored, but their interaction was demonstrated in yeast and in planta (Figure 1B; Couturier et al., 2014). Plants also have class II GRXs (GRXS14 and S16) and mono-domain BOLAs (BOLA1, BOLA4) in plastids (Couturier et al., 2013). So far, in planta evidence for their implication in the biogenesis of Fe–S proteins are scarce (Table 1). GRXS14-deficient Arabidopsis plants exhibit accelerated chlorophyll loss upon prolonged darkness, a treatment also leading to a decreased abundance of proteins acting in Fe–S cluster metabolism (Rey et al., 2017). Nevertheless, the demonstration that Arabidopsis and/or poplar GRXS14 and GRXS16 interact both with BOLA1 and BOLA4 (Couturier et al., 2014), bind Fe–S clusters alone or in complex (Bandyopadhyay et al., 2008; Dhalleine et al., 2014; Roret et al., 2014) and transfer it to partner proteins (Mapolelo et al., 2013) give strong credence to such a role. Even more importantly, all plant GRX and BOLA genes complement totally or partially (GRXS15) the corresponding yeast grx5 and bol1–bol3 mutants, indicating that they possess similar structural and functional determinants (Bandyopadhyay et al., 2008; Moseler et al., 2015; Uzarska et al., 2018).
Multiple Functions in the Regulation of Iron Homeostasis of the Class II GRX and Bola Couple in the Cytosol/Nucleus of Eukaryotes
Eukaryote cytosolic class II GRXs are multidomain proteins formed by an N-terminal thioredoxin-like domain fused to one to three GRX domains (Couturier et al., 2009). Most organisms have a single GRX of this type and also a single cytosolic BOLA isoform, referred to as BOLA2/Bol2/Fra2. The pioneering studies showing the involvement of class II GRXs and BOLAs in Fe homeostasis have been conducted in S. cerevisiae mutants deregulated in Grx3, Grx4, and Bol2/Fra2 genes (Table 1; Lesuisse et al., 2005; Ojeda et al., 2006; Pujol-Carrion et al., 2006). In yeast, the regulation of Fe concentration is achieved at the transcriptional level by low-(Aft1 and Aft2) and high-level (Yap5) sensing transcription factors and at the post-transcriptional level by mRNA-binding proteins (Outten and Albetel, 2013). Both types of transcription factors bind [2Fe–2S] clusters allowing them to perceive the cellular Fe or Fe–S cluster status (Poor et al., 2014; Rietzschel et al., 2015). Whereas Grx4 expression is regulated by Yap5, it is not documented whether Yap5 localization or activity is controlled by a GRX/BOLA complex. Regarding Aft1/Aft2, their subcellular (nuclear vs. cytosolic) localization is controlled by a Fra2-Grx3/4 inhibitory complex (possibly requiring also the aminopeptidase Fra1) (Kumánovics et al., 2008). The current view is that the presence of an Fe–S cluster in the Fra2-Grx3/4 complex is synonymous of iron-replete conditions and of a correct functioning of the ISC machinery (Figure 1C; Kumánovics et al., 2008). By transferring a cluster to Aft1/2, the GRX-BOLA complex should either retain them in the cytosol or promote their dissociation from DNA if in the nucleus (Ueta et al., 2012; Poor et al., 2014).
Some aspects of Fe homeostasis in other yeasts and fungi are also controlled by GRX and/or BOLA. In Cryptococcus neoformans, Fe repletion promotes Grx4 relocation from the nucleus to the cytoplasm allowing the regulation of Cir1, a master regulator of Fe-responsive genes (Attarian et al., 2018). In Schizosaccharomyces pombe, Fe metabolism is regulated by two transcriptional repressors, the GATA-type iron sensing Fep1 and the CCAAT-binding factor complex subunit Php4 (Brault et al., 2015). Their localization and/or DNA binding activity are regulated by Grx4 and/or Fra2 (reviewed in Outten and Albetel, 2013; Brault et al., 2015). The binding of a [2Fe–2S] cluster between Grx4 and Php4 may promote Php4 release from the CCAAT-binding complex at the DNA targets and suppress its inhibitory effect on the expression of Fe storage genes (Dlouhy et al., 2017). Unlike Php4, the regulation of which does not involve Fra2, the formation of a [2Fe–2S]-Grx4/Fra2 heterodimeric complex is required for regulating Fep1 activation (Jacques et al., 2014; Encinar del Dedo et al., 2015).
In mammals, the regulation of Fe metabolism and homeostasis is ensured by IRP1/2 and RNA-binding proteins (Rouault and Maio, 2017). Under Fe limitation, both IRPs bind to the so-called Iron Responsive Elements (IREs) in untranslated regions of mRNAs coding for proteins implicated in Fe assimilation and homeostasis (Rouault and Maio, 2017). Doing so, they control either mRNA stabilization or translational blocking. Whereas IRP2 release from IREs is mediated by proteasomal degradation (Guo et al., 1995), IRP1 function may depend on GLRX3/PICOT (but also on mitochondrial GLRX5) as it relies on the binding of an Fe–S cluster. Under Fe sufficiency, IRP1 binds a [4Fe–4S] cluster and acts as an aconitase whereas under Fe limitation the protein turns into an apoform binding to IREs. Consequently, IRP1 requires functional mitochondrial and cytosolic Fe–S cluster assembly machineries. Having two GRX domains, human GLRX3 forms homodimers or heterotrimers with two BOLA2 molecules bridging two [2Fe–2S] clusters (Li et al., 2012; Banci et al., 2015b; Frey et al., 2016). It also binds a [4Fe–4S] cluster and transfers it in vitro to an apo-IRP1 (Xia et al., 2015). GLRX3 silencing in human HELA cells decreases the activity of several cytosolic Fe–S proteins, including IRP1 (Table 1; Haunhorst et al., 2013). In zebrafish, GLRX3 deletion impairs heme biosynthesis during embryo development (Haunhorst et al., 2013). All of this indicates important functions of vertebrate GLRX3 in Fe metabolism.
In addition to an Fe sensing function, an Fe or Fe–S cluster trafficking function was proposed for yeast Grx3/4 and the human GLRX3-BOLA2 complex to ensure proper assembly of several types of Fe-containing centers. In fact, most multidomain GRXs are able to rescue the Fe–S cluster maturation defects of the yeast grx5 mutant (Molina et al., 2004; Bandyopadhyay et al., 2008; Knuesting et al., 2015) suggesting that they have the capacity of exchanging Fe–S clusters. Accordingly, both human GLRX3 homodimers and GLRX3-BOLA2 trimeric complexes bridging two [2Fe–2S] clusters can deliver their clusters to the anamorsin/CIAPIN/DRE2 protein (Banci et al., 2015a,b). From the observation that the maturation of yeast Grx3/4 and human GLRX3-BOLA2 heterodimers requires the mitochondrial ISC machinery but not CIA components (Mühlenhoff et al., 2010; Frey et al., 2016), it is concluded that cytosolic class II GRXs should build their cluster from a sulfur compound exported by the mitochondrial ATM transporter (Figure 1C). In yeast grx3/4Δ, the Fe or Fe-cofactor insertion in various proteins present in cytosol [catalase, ribonucleotide reductase (RNR)], and mitochondria (complexes II and III, aconitase, Coq7 mono-oxygenase) is altered (Mühlenhoff et al., 2010; Zhang et al., 2011). Moreover, the respective increased and decreased Fe levels in cytosol and mitochondria of Grx3/4 depleted cells pointed to impaired Fe distribution (Mühlenhoff et al., 2010). These additional functions of yeast Grx3/4 are well exemplified in the case of RNR di-iron cofactor biogenesis because Grx4 provides the Fe atoms, but also serves for the maturation of holo-Dre2, that provides the required electrons (Li et al., 2017). A contribution of yeast Bol2 for these functions is unclear even though a general role in cytosolic Fe–S protein maturation is excluded (Uzarska et al., 2016). In human, GLRX3-BOLA2 trimeric complexes bridging two [2Fe–2S] clusters were proposed to constitute a reservoir for delivering Fe or Fe–S cluster to some Fe-containing target proteins based notably on the six–eightfold increased abundance observed in response to elevated iron (Frey et al., 2016).
The function of GRXS17, the sole nucleo-cytosolic class II GRX in plants, has been explored using several approaches. Tandem affinity purification using a tagged GRX form expressed in Arabidopsis cell cultures and seedlings pointed to the association of GRXS17 with CIA components and BOLA2 (Iñigo et al., 2016). The interactions with DRE2 and BOLA2 have been confirmed in vivo by binary yeast two-hybrid and BiFC and/or in vitro by co-expression in E. coli (Couturier et al., 2014; Dhalleine et al., 2014; Iñigo et al., 2016). As GRXs interact with Dre2/Anamorsin in yeast and human cells (Zhang et al., 2011; Banci et al., 2015b), the only direct CIA partner of GRXS17 might be DRE2 and the other proteins part of a complex. Besides, the binding of GRXS17 with putative Fe–S client proteins involved in purine salvage (xanthine dehydrogenase 1) or tRNA modification (thiouridylase subunits 1 and 2) was shown (Iñigo et al., 2016). Thus, one would expect that plants deficient in GRXS17 display a marked phenotype in relation with Fe metabolism, but the analysis of Arabidopsis grxs17 plants led to relatively complex data. Indeed, their development is only mildly affected under standard growth conditions, but gets severely impaired (elongated leaves, modified shoot apical meristem structure, and altered auxin response) at high temperature or under long photoperiod (Cheng et al., 2011; Knuesting et al., 2015). It is not yet clear whether a redox- and/or an Fe-related function of GRXS17 is responsible for these alterations. In fact, in vitro pull-downs performed using the recombinant protein allowed recovering many non-Fe–S proteins including the NF-YC11 transcriptional regulator (Knuesting et al., 2015). Moreover, there is no variation in the Fe content in mature leaves and only a slight increase in seeds (Yu et al., 2017) of grxs17 plants that exhibit no or minor decreases in the activity of three Fe–S containing enzymes: aconitase, aldehyde oxidase and xanthine dehydrogenase (Knuesting et al., 2015; Iñigo et al., 2016). On the other hand, GRXS17-deficient lines exhibit a slightly increased sensitivity to genotoxic stress which is reminiscent of mutants compromised in the CIA pathway (Iñigo et al., 2016). Finally, when GRXS17-deficient lines are exposed to Fe deficiency, the primary root growth reduction, that is already visible under standard conditions, is exacerbated and ROS levels are elevated (Iñigo et al., 2016; Yu et al., 2017). Whether plant GRXS17 and BOLA2 act in concert remains unclear. The Arabidopsis bola2 (incorrectly named bola3) mutant displays no phenotype under control conditions and no change in the activity of typical Fe–S enzymes (Qin et al., 2015). Surprisingly, this line is more tolerant to oxidative stress generated by an Fe excess (Qin et al., 2015). In conclusion, bola2 and grxs17 plants exhibit relatively mild phenotypes, visible mostly under stress conditions, compared to those described for human and yeast orthologs and to the embryo-lethality of most Arabidopsis mutants defective for early acting CIA components (Bernard et al., 2013). This raises some questions about the exact functions of BOLA2 and GRXS17 in the regulation of Fe homeostasis in plant cells and about the existence of an alternative system, notably for delivering Fe–S clusters to DRE2, whose function is essential.
Roadmap Toward the Understanding of the Roles of GRX/Bola Couples in Plants
In this section, we propose some ideas and experimental strategies that should warrant deciphering the functions associated to GRX/BOLA couples in plants.
Evidence obtained so far indicate that the class II mono-domain GRXs and BOLAs present in mitochondria of non-plant eukaryotes and in bacteria act as maturation factors for the biogenesis of Fe–S proteins. A similar role seems true for the plant mitochondrial GRXS15, but it is now mandatory to examine whether it also contributes to the maturation of extra-mitochondrial proteins. Another challenge will be to understand why it is essential in plants unlike in yeast. Also, the physiological consequences of BOLA4 depletion must be investigated to see whether this fits with a function connected to GRXS15. Concerning plastidial proteins (GRXS14, GRXS16, BOLA1, and BOLA4), a role in the maturation of Fe–S proteins still needs to be demonstrated in planta, despite they can functionally substitute to their mitochondrial yeast counterparts.
With regard to the cytosolic multi-domain GRXs and BOLAs, a role in Fe metabolism seems evolutionary conserved, but their contribution and partners differ. In yeast, their primary function is to regulate Fe-responsive transcription factors. Additional functions are to ensure a proper Fe distribution toward all types of Fe cofactors (including heme and non-heme Fe centers) and/or to serve for Dre2 maturation, thus contributing to the correct functioning of the CIA machinery. In this case, Grx3/4 have an exclusive or predominant role because the corresponding mutant is lethal or strongly affected, unlike the bol2/fra2 mutant. Experimental evidence indicate that the involvement of GRX and/or BOLA in DRE2 maturation is likely also true in mammals and plants, but evidence supporting other functions are scarce.
A first prerequisite to future molecular and physiological analyses is to generate the missing single knock-out lines but also multiple knock-out lines for possibly redundant proteins. This would be particularly important to obtain lines combining mutations for GRXS14 and GRXS16, for BOLA1 and BOLA4, but also for GRXS17 and the only other Fe–S ligating GRXs reported so far in the cytosol, namely GRXC1 (Rouhier et al., 2007), or BOLA2. In case the single or multiple mutants are lethal, an option for obtaining viable lines would be to generate RNAi lines as for GRXS15, but also dominant negative mutant lines expressing mutated versions of GRX or BOLA unable for instance to ligate the Fe–S cluster, i.e., mutated for the catalytic cysteine of GRXs or the conserved histidine residue of BOLA.
At the physiological level, the growth of these plants should be analyzed under standard conditions, but also under environmental constraints as the shoot phenotypes of grxs17 mutants are only visible in specific conditions. For the BOLA2-GRXS17 couple, understanding their connection and discriminating between Fe- or redox-related functions will require in particular to assess the phenotypes of the corresponding mutants in the same experimental setup and conditions. Considering the described importance of GSH for ligating Fe–S cluster in GRX homodimer or GRX-BOLA heterodimer and for the maturation of cytosolic Fe–S proteins (Sipos et al., 2002), crossing some of these mutants with mutants having an altered GSH homeostasis would certainly be informative.
In other respects, an obvious strategy is to measure the abundance/activity of representative Fe–S proteins in these lines. However, performing quantitative proteomic and metabolomic approaches may be more informative and help obtaining a broader view of the molecular and cellular mechanisms affected and of the compensations established. It may also rapidly point to metabolic differences existing among mutants.
In all cases, determining the identity of the direct and indirect targets of both GRXs and BOLAs would represent a mandatory information. For instance, the proteins involved in the Fe–S cluster maturation process may act at different steps. Various approaches complementary to quantitative proteomics proved valuable even for detecting supposedly transient interactions among Fe–S cluster donors and acceptors (Touraine et al., 2019). Hence, it is possible to combine it to another non-targeted approach such as co-immunoprecipitation or to binary yeast two-hybrid experiments which has the advantage for instance to allow studying rapidly sequence requirements by mutational analysis.
In summary, the combination of genetic approaches, omics analyses and conventional biochemical tools should in principle allow better delineating the roles and specificities of GRX/BOLA couples in the maintenance of Fe homeostasis in plants.
Statements
Data availability statement
All datasets analyzed for this study are included in the manuscript and the Supplementary Files.
Author contributions
All authors wrote the text and approved the final version of the manuscript.
Funding
The UMR1136 is supported by a grant overseen by the French National Research Agency (ANR) as part of the “Investissements d’Avenir” program (ANR-11-LABX-0002-01, Lab of Excellence ARBRE). The work on plant GRX and BOLA proteins was supported by the Agence Nationale de la Recherche (Grant No. 2010BLAN1616).
Acknowledgments
Ms. Anna Moseler is greatly acknowledged for the artwork on Figure 1.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
AldeaM.GarridoT.Hernández-ChicoC.VicenteM.KushnerS. R. (1989). Induction of a growth-phase-dependent promoter triggers transcription of bolA, an Escherichia coli morphogene.EMBO J.83923–3931. 10.1002/j.1460-2075.1989.tb08573.x
2
AlvesR.VilaprinyoE.SorribasA.HerreroE. (2009). Evolution based on domain combinations: the case of glutaredoxins.BMC Evol. Biol.9:66. 10.1186/1471-2148-9-66
3
AttarianR.HuG.Sánchez-LeónE.CazaM.CrollD.DoE.et al (2018). The monothiol glutaredoxin grx4 regulates iron homeostasis and virulence in Cryptococcus neoformans.mBio9:e02377-18. 10.1128/mBio.02377-18
4
BanciL.CamponeschiF.Ciofi-BaffoniS.MuzzioliR. (2015a). Elucidating the molecular function of human BOLA2 in GRX3-dependent anamorsin maturation pathway.J. Am. Chem. Soc.13716133–16143. 10.1021/jacs.5b10592
5
BanciL.Ciofi-BaffoniS.GajdaK.MuzzioliR.PeruzziniR.WinkelmannJ. (2015b). N-terminal domains mediate [2Fe-2S] cluster transfer from glutaredoxin-3 to anamorsin.Nat. Chem. Biol.11772–778. 10.1038/nchembio.1892
6
BandyopadhyayS.GamaF.Molina-NavarroM. M.GualbertoJ. M.ClaxtonR.NaikS. G.et al (2008). Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe-2S] clusters.EMBO J.271122–1133. 10.1038/emboj.2008.50
7
BarupalaD. P.DzulS. P.Riggs-GelascoP. J.StemmlerT. L. (2016). Synthesis, delivery and regulation of eukaryotic heme and Fe-S cluster cofactors.Arch. Biochem. Biophys.59260–75. 10.1016/j.abb.2016.01.010
8
BenyaminaS. M.Baldacci-CrespF.CouturierJ.ChibaniK.HopkinsJ.BekkiA.et al (2013). Two Sinorhizobium meliloti glutaredoxins regulate iron metabolism and symbiotic bacteroid differentiation.Environ. Microbiol.15795–810. 10.1111/j.1462-2920.2012.02835.x
9
BernardD. G.NetzD. J. A.LagnyT. J.PierikA. J.BalkJ. (2013). Requirements of the cytosolic iron-sulfur cluster assembly pathway in Arabidopsis.Philos. Trans. R. Soc. Lond. B. Biol. Sci.368:20120259. 10.1098/rstb.2012.0259
10
BoutignyS.SainiA.BaidooE. E. K.YeungN.KeaslingJ. D.ButlandG. (2013). Physical and functional interactions of a monothiol glutaredoxin and an iron sulfur cluster carrier protein with the sulfur-donating radical S-adenosyl-L-methionine enzyme MiaB.J. Biol. Chem.28814200–14211. 10.1074/jbc.M113.460360
11
BraultA.MourerT.LabbéS. (2015). Molecular basis of the regulation of iron homeostasis in fission and filamentous yeasts.IUBMB Life67801–815. 10.1002/iub.1441
12
BurschelS.Kreuzer DecovicD.NuberF.StillerM.HofmannM.ZupokA.et al (2019). Iron-sulfur cluster carrier proteins involved in the assembly of Escherichia coli NADH:ubiquinone oxidoreductase (complex I).Mol. Microbiol.11131–45. 10.1111/mmi.14137
13
ButlandG.BabuM.Díaz-MejíaJ. J.BohdanaF.PhanseS.GoldB.et al (2008). eSGA: E. coli synthetic genetic array analysis.Nat. Methods5789–795. 10.1038/nmeth.1239
14
CamaschellaC.CampanellaA.De FalcoL.BoschettoL.MerliniR.SilvestriL.et al (2007). The human counterpart of zebrafish shiraz shows sideroblastic-like microcytic anemia and iron overload.Blood1101353–1358. 10.1182/blood-2007-02-072520
15
CameronJ. M.JanerA.LevandovskiyV.MackayN.RouaultT. A.TongW.-H.et al (2011). Mutations in iron-sulfur cluster scaffold genes NFU1 and BOLA3 cause a fatal deficiency of multiple respiratory chain and 2-oxoacid dehydrogenase enzymes.Am. J. Hum. Genet.89486–495. 10.1016/j.ajhg.2011.08.011
16
ChengN.-H.LiuJ.-Z.LiuX.WuQ.ThompsonS. M.LinJ.et al (2011). Arabidopsis monothiol glutaredoxin, AtGRXS17, is critical for temperature-dependent postembryonic growth and development via modulating auxin response.J. Biol. Chem.28620398–20406. 10.1074/jbc.M110.201707
17
ChungW.-H.KimK.-D.RoeJ.-H. (2005). Localization and function of three monothiol glutaredoxins in Schizosaccharomyces pombe.Biochem. Biophys. Res. Commun.330604–610. 10.1016/j.bbrc.2005.02.183
18
CominiM. A.RettigJ.DirdjajaN.HanschmannE.-M.BerndtC.Krauth-SiegelR. L. (2008). Monothiol glutaredoxin-1 is an essential iron-sulfur protein in the mitochondrion of African trypanosomes.J. Biol. Chem.28327785–27798. 10.1074/jbc.M802010200
19
CouturierJ.JacquotJ.-P.RouhierN. (2009). Evolution and diversity of glutaredoxins in photosynthetic organisms.Cell. Mol. Life Sci.662539–2557. 10.1007/s00018-009-0054-y
20
CouturierJ.Przybyla-ToscanoJ.RoretT.DidierjeanC.RouhierN. (2015). The roles of glutaredoxins ligating Fe-S clusters: sensing, transfer or repair functions?Biochim. Biophys. Acta18531513–1527. 10.1016/j.bbamcr.2014.09.018
21
CouturierJ.TouraineB.BriatJ.-F.GaymardF.RouhierN. (2013). The iron-sulfur cluster assembly machineries in plants: current knowledge and open questions.Front. Plant Sci.4:259. 10.3389/fpls.2013.00259
22
CouturierJ.WuH.-C.DhalleineT.PégeotH.SudreD.GualbertoJ. M.et al (2014). Monothiol glutaredoxin-BolA interactions: redox control of Arabidopsis thaliana BolA2 and SufE1.Mol. Plant7187–205. 10.1093/mp/sst156
23
DhalleineT.RouhierN.CouturierJ. (2014). Putative roles of glutaredoxin-BolA holo-heterodimers in plants.Plant Signal. Behav.9:e28564. 10.4161/psb.28564
24
DlouhyA. C.BeaudoinJ.LabbéS.OuttenC. E. (2017). Schizosaccharomyces pombe Grx4 regulates the transcriptional repressor Php4 via [2Fe-2S] cluster binding.Met. Integr. Biometal Sci.91096–1105. 10.1039/c7mt00144d
25
DlouhyA. C.LiH.AlbetelA.-N.ZhangB.MapoleloD. T.RandeniyaS.et al (2016). The Escherichia coli BolA protein IbaG forms a histidine-Ligated [2Fe-2S]-bridged complex with Grx4.Biochemistry556869–6879. 10.1021/acs.biochem.6b00812
26
Encinar del DedoJ.GabrielliN.CarmonaM.AytéJ.HidalgoE. (2015). A cascade of iron-containing proteins governs the genetic iron starvation response to promote iron uptake and inhibit iron storage in fission yeast.PLoS Genet.11:e1005106. 10.1371/journal.pgen.1005106
27
FreyA. G.PalencharD. J.WildemannJ. D.PhilpottC. C. (2016). A glutaredoxin ⋅ bola complex serves as an iron-sulfur cluster chaperone for the cytosolic cluster assembly machinery.J. Biol. Chem.29122344–22356. 10.1074/jbc.M116.744946
28
GuoB.PhillipsJ. D.YuY.LeiboldE. A. (1995). Iron regulates the intracellular degradation of iron regulatory protein 2 by the proteasome.J. Biol. Chem.27021645–21651. 10.1074/jbc.270.37.21645
29
HaunhorstP.HanschmannE.-M.BräutigamL.StehlingO.HoffmannB.MühlenhoffU.et al (2013). Crucial function of vertebrate glutaredoxin 3 (PICOT) in iron homeostasis and hemoglobin maturation.Mol. Biol. Cell241895–1903. 10.1091/mbc.E12-09-0648
30
IñigoS.DurandA. N.RitterA.Le GallS.TermatheM.KlassenR.et al (2016). Glutaredoxin GRXS17 associates with the cytosolic iron-sulfur cluster assembly pathway.Plant Physiol.172858–873. 10.1104/pp.16.00261
31
JacquesJ.-F.MercierA.BraultA.MourerT.LabbéS. (2014). Fra2 is a co-regulator of Fep1 inhibition in response to iron starvation.PLoS One9:e98959. 10.1371/journal.pone.0098959
32
KimK.-D.ChungW.-H.KimH.-J.LeeK.-C.RoeJ.-H. (2010). Monothiol glutaredoxin Grx5 interacts with Fe-S scaffold proteins Isa1 and Isa2 and supports Fe-S assembly and DNA integrity in mitochondria of fission yeast.Biochem. Biophys. Res. Commun.392467–472. 10.1016/j.bbrc.2010.01.051
33
KnuestingJ.RiondetC.MariaC.KruseI.BécuweN.KönigN.et al (2015). Arabidopsis glutaredoxin S17 and its partner, the nuclear factor Y subunit C11/negative cofactor 2α, contribute to maintenance of the shoot apical meristem under long-day photoperiod.Plant Physiol.1671643–1658. 10.1104/pp.15.00049
34
KumánovicsA.ChenO. S.LiL.BagleyD.AdkinsE. M.LinH.et al (2008). Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis.J. Biol. Chem.28310276–10286. 10.1074/jbc.M801160200
35
LesuisseE.KnightS. A. B.CourelM.SantosR.CamadroJ.-M.DancisA. (2005). Genome-wide screen for genes with effects on distinct iron uptake activities in Saccharomyces cerevisiae.Genetics169107–122. 10.1534/genetics.104.035873
36
LiH.MapoleloD. T.DingraN. N.KellerG.Riggs-GelascoP. J.WingeD. R.et al (2011). Histidine 103 in Fra2 is an iron-sulfur cluster ligand in the [2Fe-2S] Fra2-Grx3 complex and is required for in vivo iron signaling in yeast.J. Biol. Chem.286867–876. 10.1074/jbc.M110.184176
37
LiH.MapoleloD. T.RandeniyaS.JohnsonM. K.OuttenC. E. (2012). Human glutaredoxin 3 forms [2Fe-2S]-bridged complexes with human BolA2.Biochemistry511687–1696. 10.1021/bi2019089
38
LiH.OuttenC. E. (2012). Monothiol CGFS glutaredoxins and BolA-like proteins: [2Fe-2S] binding partners in iron homeostasis.Biochemistry514377–4389. 10.1021/bi300393z
39
LiH.StümpfigM.ZhangC.AnX.StubbeJ.LillR.et al (2017). The diferric-tyrosyl radical cluster of ribonucleotide reductase and cytosolic iron-sulfur clusters have distinct and similar biogenesis requirements.J. Biol. Chem.29211445–11451. 10.1074/jbc.M117.786178
40
LillR. (2009). Function and biogenesis of iron-sulphur proteins.Nature460831–838. 10.1038/nature08301
41
MapoleloD. T.ZhangB.RandeniyaS.AlbetelA.-N.LiH.CouturierJ.et al (2013). Monothiol glutaredoxins and A-type proteins: partners in Fe-S cluster trafficking.Dalton Trans. Camb. Engl.20033107–3115. 10.1039/c2dt32263c
42
MelberA.NaU.VashishtA.WeilerB. D.LillR.WohlschlegelJ. A.et al (2016). Role of Nfu1 and Bol3 in iron-sulfur cluster transfer to mitochondrial clients.eLife5:e15991. 10.7554/eLife.15991
43
Mil-HomensD.BarahonaS.MoreiraR. N.SilvaI. J.PintoS. N.FialhoA. M.et al (2018). Stress response protein bola influences fitness and promotes Salmonella enterica Serovar Typhimurium virulence.Appl. Environ. Microbiol.84:e02850-17. 10.1128/AEM.02850-17
44
MolinaM. M.BellíG.de la TorreM. A.Rodríguez-ManzanequeM. T.HerreroE. (2004). Nuclear monothiol glutaredoxins of Saccharomyces cerevisiae can function as mitochondrial glutaredoxins.J. Biol. Chem.27951923–51930. 10.1074/jbc.M410219200
45
MoselerA.AllerI.WagnerS.NietzelT.Przybyla-ToscanoJ.MühlenhoffU.et al (2015). The mitochondrial monothiol glutaredoxin S15 is essential for iron-sulfur protein maturation in Arabidopsis thaliana.Proc. Natl. Acad. Sci. U.S.A.11213735–13740. 10.1073/pnas.1510835112
46
MühlenhoffU.GerberJ.RichhardtN.LillR. (2003). Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1p.EMBO J.224815–4825. 10.1093/emboj/cdg446
47
MühlenhoffU.MolikS.GodoyJ. R.UzarskaM. A.RichterN.SeubertA.et al (2010). Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster.Cell Metab.12373–385. 10.1016/j.cmet.2010.08.001
48
NastaV.GiachettiA.Ciofi-BaffoniS.BanciL. (2017). Structural insights into the molecular function of human [2Fe-2S] BOLA1-GRX5 and [2Fe-2S] BOLA3-GRX5 complexes.Biochim. Biophys. Acta18612119–2131. 10.1016/j.bbagen.2017.05.005
49
OjedaL.KellerG.MuhlenhoffU.RutherfordJ. C.LillR.WingeD. R. (2006). Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae.J. Biol. Chem.28117661–17669. 10.1074/jbc.M602165200
50
OuttenC. E.AlbetelA.-N. (2013). Iron sensing and regulation in Saccharomyces cerevisiae: ironing out the mechanistic details.Curr. Opin. Microbiol.16662–668. 10.1016/j.mib.2013.07.020
51
PhilpottC. C.RyuM.-S.FreyA.PatelS. (2017). Cytosolic iron chaperones: proteins delivering iron cofactors in the cytosol of mammalian cells.J. Biol. Chem.29212764–12771. 10.1074/jbc.R117.791962
52
PoorC. B.WegnerS. V.LiH.DlouhyA. C.SchuermannJ. P.SanishviliR.et al (2014). Molecular mechanism and structure of the Saccharomyces cerevisiae iron regulator Aft2.Proc. Natl. Acad. Sci. U.S.A.1114043–4048. 10.1073/pnas.1318869111
53
Przybyla-ToscanoJ.RoretT.CouturierJ.RouhierN. (2017). “FeS Cluster Assembly: Role of Monothiol Grxs and Nfu Proteins,” in Encyclopedia of Inorganic and Bioinorganic Chemistry, edsScottR. A (New York, NY: American Cancer Society), 1–19. 10.1002/9781119951438.eibc2470
54
Pujol-CarrionN.BelliG.HerreroE.NoguesA.de la Torre-RuizM. A. (2006). Glutaredoxins Grx3 and Grx4 regulate nuclear localisation of Aft1 and the oxidative stress response in Saccharomyces cerevisiae.J. Cell Sci.1194554–4564. 10.1242/jcs.03229
55
QinL.WangM.ZuoJ.FengX.LiangX.WuZ.et al (2015). Cytosolic bolA plays a repressive role in the tolerance against excess iron and mv-induced oxidative stress in plants.PLoS One10:e0124887. 10.1371/journal.pone.0124887
56
ReyP.BecuweN.TourretteS.RouhierN. (2017). Involvement of Arabidopsis glutaredoxin S14 in the maintenance of chlorophyll content.Plant Cell Environ.402319–2332. 10.1111/pce.13036
57
RietzschelN.PierikA. J.BillE.LillR.MühlenhoffU. (2015). The basic leucine zipper stress response regulator Yap5 senses high-iron conditions by coordination of [2Fe-2S] clusters.Mol. Cell. Biol.35370–378. 10.1128/MCB.01033-14
58
Rodríguez-ManzanequeM. T.TamaritJ.BellíG.RosJ.HerreroE. (2002). Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes.Mol. Biol. Cell131109–1121. 10.1091/mbc.01-10-0517
59
RoretT.TsanP.CouturierJ.ZhangB.JohnsonM. K.RouhierN.et al (2014). Structural and spectroscopic insights into BolA-glutaredoxin complexes.J. Biol. Chem.28924588–24598. 10.1074/jbc.M114.572701
60
RouaultT. A.MaioN. (2017). Biogenesis and functions of mammalian iron-sulfur proteins in the regulation of iron homeostasis and pivotal metabolic pathways.J. Biol. Chem.29212744–12753. 10.1074/jbc.R117.789537
61
RouhierN.UnnoH.BandyopadhyayS.MasipL.KimS.-K.HirasawaM.et al (2007). Functional, structural, and spectroscopic characterization of a glutathione-ligated [2Fe-2S] cluster in poplar glutaredoxin C1.Proc. Natl. Acad. Sci. U.S.A.1047379–7384. 10.1073/pnas.0702268104
62
SiposK.LangeH.FeketeZ.UllmannP.LillR.KispalG. (2002). Maturation of cytosolic iron-sulfur proteins requires glutathione.J. Biol. Chem.27726944–26949. 10.1074/jbc.M200677200
63
StröherE.GrasslJ.CarrieC.FenskeR.WhelanJ.MillarA. H. (2016). Glutaredoxin S15 is involved in Fe-S cluster transfer in mitochondria influencing lipoic acid-dependent enzymes, plant growth, and arsenic tolerance in Arabidopsis.Plant Physiol.1701284–1299. 10.1104/pp.15.01308
64
TouraineB.VignolsF.Przybyla-ToscanoJ.IschebeckT.DhalleineT.WuH.-C.et al (2019). Iron-sulfur protein NFU2 is required for branched-chain amino acid synthesis in Arabidopsis roots.J. Exp. Bot.701875–1889. 10.1093/jxb/erz050
65
UetaR.FujiwaraN.IwaiK.Yamaguchi-IwaiY. (2012). Iron-induced dissociation of the Aft1p transcriptional regulator from target gene promoters is an initial event in iron-dependent gene suppression.Mol. Cell. Biol.324998–5008. 10.1128/MCB.00726-12
66
UzarskaM. A.DutkiewiczR.FreibertS.-A.LillR.MühlenhoffU. (2013). The mitochondrial Hsp70 chaperone Ssq1 facilitates Fe/S cluster transfer from Isu1 to Grx5 by complex formation.Mol. Biol. Cell241830–1841. 10.1091/mbc.E12-09-0644
67
UzarskaM. A.NastaV.WeilerB. D.SpantgarF.Ciofi-BaffoniS.SavielloM. R.et al (2016). Mitochondrial Bol1 and Bol3 function as assembly factors for specific iron-sulfur proteins.eLife5:e16673. 10.7554/eLife.16673
68
UzarskaM. A.Przybyla-ToscanoJ.SpantgarF.ZanniniF.LillR.MühlenhoffU.et al (2018). Conserved functions of Arabidopsis mitochondrial late-acting maturation factors in the trafficking of iron-sulfur clusters.Biochim. Biophys. Acta Mol. Cell Res.18651250–1259. 10.1016/j.bbamcr.2018.06.003
69
WillemsP.WanschersB. F. J.EsselingJ.SzklarczykR.KudlaU.DuarteI.et al (2013). BOLA1 is an aerobic protein that prevents mitochondrial morphology changes induced by glutathione depletion.Antioxid. Redox Signal.18129–138. 10.1089/ars.2011.4253
70
WingertR. A.GallowayJ. L.BarutB.FoottH.FraenkelP.AxeJ. L.et al (2005). Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis.Nature4361035–1039. 10.1038/nature03887
71
XiaH.LiB.ZhangZ.WangQ.QiaoT.LiK. (2015). Human glutaredoxin 3 can bind and effectively transfer [4Fe-4S] cluster to apo-iron regulatory protein 1.Biochem. Biophys. Res. Commun.465620–624. 10.1016/j.bbrc.2015.08.073
72
YeH.JeongS. Y.GhoshM. C.KovtunovychG.SilvestriL.OrtilloD.et al (2010). Glutaredoxin 5 deficiency causes sideroblastic anemia by specifically impairing heme biosynthesis and depleting cytosolic iron in human erythroblasts.J. Clin. Invest.1201749–1761. 10.1172/JCI40372
73
YeungN.GoldB.LiuN. L.PrathapamR.SterlingH. J.WillamsE. R.et al (2011). The E. coli monothiol glutaredoxin GrxD forms homodimeric and heterodimeric FeS cluster containing complexes.Biochemistry508957–8969. 10.1021/bi2008883
74
YuH.YangJ.ShiY.DonelsonJ.ThompsonS. M.SpragueS.et al (2017). Arabidopsis glutaredoxin S17 contributes to vegetative growth, mineral accumulation, and redox balance during iron deficiency.Front. Plant Sci.8:1045. 10.3389/fpls.2017.01045
75
ZhangY.LiuL.WuX.AnX.StubbeJ.HuangM. (2011). Investigation of in vivo diferric tyrosyl radical formation in Saccharomyces cerevisiae Rnr2 protein: requirement of Rnr4 and contribution of Grx3/4 AND Dre2 proteins.J. Biol. Chem.28641499–41509. 10.1074/jbc.M111.294074
Summary
Keywords
BOLA, glutaredoxins, iron–sulfur center, maturation factor, iron homeostasis
Citation
Rey P, Taupin-Broggini M, Couturier J, Vignols F and Rouhier N (2019) Is There a Role for Glutaredoxins and BOLAs in the Perception of the Cellular Iron Status in Plants?. Front. Plant Sci. 10:712. doi: 10.3389/fpls.2019.00712
Received
05 March 2019
Accepted
14 May 2019
Published
04 June 2019
Volume
10 - 2019
Edited by
Thomas J. Buckhout, Humboldt University of Berlin, Germany
Reviewed by
Qingyu Wu, Institute of Agricultural Resources and Regional Planning (CAAS), China; Ping Lan, Institute of Soil Science (CAS), China
Updates
Copyright
© 2019 Rey, Taupin-Broggini, Couturier, Vignols and Rouhier.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicolas Rouhier, Nicolas.Rouhier@univ-lorraine.fr
This article was submitted to Plant Nutrition, a section of the journal Frontiers in Plant Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.