- 1School of Horticulture Landscape Architecture, Henan Institute of Science and Technology, Xinxiang, China
- 2Henan Province Engineering Research Center of Horticultural Plant Resource Utilization and Germplasm Enhancement, Xinxiang, China
Powdery mildew (PM), which is mainly caused by Podosphaera xanthii, is a serious biotrophic pathogen disease affecting field-grown and greenhouse-grown cucurbit crops worldwide. Because fungicides poorly control PM, the development and cultivation of PM-resistant varieties is critical. A homolog of SGT1 (suppressor of the G2 allele of skp1), which encodes a key component of the plant disease-associated signal transduction pathway, was previously identified through a transcriptomic analysis of a PM-resistant pumpkin (Cucurbita moschata) inbred line infected with PM. In this study, we have characterized this SGT1 homolog in C. moschata, and investigated its effects on biotic stress resistance. Subcellular localization results revealed that CmSGT1 is present in the nucleus. Additionally, CmSGT1 expression levels in the PM-resistant material was strongly induced by PM, salicylic acid (SA) and hydrogen peroxide (H2O2). In contrast, SA and H2O2 downregulated CmSGT1 expression in the PM-susceptible material. The ethephon (Eth) and methyl jasmonate (MeJA) treatments upregulated CmSGT1 expression in both plant materials. The constitutive overexpression of CmSGT1 in Nicotiana benthamiana (N. benthamiana) minimized the PM symptoms on the leaves of PM-infected seedlings, accelerated the onset of cell necrosis, and enhanced the accumulation of H2O2. Furthermore, the expression levels of PR1a and PR5, which are SA signaling transduction markers, were higher in the transgenic plants than in wild-type plants. Thus, the transgenic N. benthamiana plants were significantly more resistant to Erysiphe cichoracearum than the wild-type plants. This increased resistance was correlated with cell death, H2O2 accumulation, and upregulated expression of SA-dependent defense genes. However, the chlorosis and yellowing of plant materials and the concentration of bacteria at infection sites were greater in the transgenic N. benthamiana plants than in the wild-type plants in response to infections by the pathogens responsible for bacterial wilt and scab. Therefore, CmSGT1-overexpressing N. benthamiana plants were hypersensitive to these two diseases. The results of this study may represent valuable genetic information for the breeding of disease-resistant pumpkin varieties, and may also help to reveal the molecular mechanism underlying CmSGT1 functions.
Introduction
The genus Cucurbita is composed of several species, including the cultivated Cucurbita moschata Duch., Cucurbita pepo L., Cucurbita maxima Duch., and several wild species. Pumpkins (C. moschata) are valued for their fruit and seeds. Additionally, they are rich in vitamins, amino acids, flavonoids, phenolics, and carbohydrates, and possess medicinal properties, including anti-diabetic, anti-oxidant, anti-carcinogenic, and anti-inflammatory activities (Wang et al., 2002; Yadav et al., 2010). In China alone, the annual yield of pumpkin, squash, and gourds is 8,051,495 t (i.e., approximately 22.68% of the global yield) from a harvested area of 438,466 ha (i.e., 17.42% of the global area) (Food and Agriculture Organization, 2017)1. Cucurbit powdery mildew (PM) is a serious disease affecting field-grown and greenhouse-grown cucurbit crops worldwide. The disease is mainly caused by Podosphaera xanthii (formerly known as Sphaerotheca fuliginea), which is a biotrophic plant pathogen (Perez-Garcia et al., 2009; Fukino et al., 2013). Fungicide applications poorly control PM and the long-term use of pesticides may lead to increased environmental pollution and the residual chemicals on food crops may be harmful for humans and animals. Therefore, studying the mechanism underlying PM resistance and exploiting the resistance genes to breed resistant varieties represents an effective way to control PM in pumpkin.
To date, there has been relatively little research on pumpkin (2n = 2x = 40), especially at the molecular level, which has seriously hindered developments in the fields of molecular biology and genetics. We previously conducted a RNA sequencing analysis of pumpkin inbred line highly resistant to PM (inbred line “112-2”) and identified 4,716 differentially expressed genes, including genes encoding broad-spectrum disease-resistance/susceptibility proteins [PR protein, ubiquitin, heat shock protein and Mildew Locus O (MLO)], reactive oxygen scavengers (peroxidase, superoxide dismutase, and catalase), signal transduction molecules serine/threonine protein kinase, and transcription factors (WRKY, TGA, and MYB). Additionally, candidate disease-resistance genes, such as WRKY21, MLO3, and SGT1, were identified (Guo et al., 2018). One of the genes cloned from the transcriptome was a homolog of Cucumis melo SGT1 (suppressor of the G2 allele of skp1). The expression of this SGT1 homolog was highly upregulated in PM-resistant material at 6 h after an inoculation with the PM fungus, but not in PM-susceptible material. However, pumpkin SGT1 has not been functionally characterized regarding its potential involvement in the defense response to PM.
The SGT1 protein was originally defined in yeast, in which it interacts with SKP1, which is a component of the Skp1/CDC/F-box protein E3 ubiquitin ligase complex (Kitagawa et al., 1999). The SGT1 protein contains three functional domains, namely the tetratricopeptide repeat (TPR), CHORD SGT1 (CS), and the SGT1-specific sequence (SGS) (Kumar and Kirti, 2015). SGT1 is closely related to the disease resistance mediated by the plant resistance (R) genes. The silencing or mutation of NbSGT1 from Nicotiana benthamiana (N. benthamiana) can lead to reduction of steady-state levels of R proteins and the loss of the mediated resistance, and AtSGT1a overexpression can contribute positively to resistance triggered by the NB-LRR type R proteins, and can complement for loss of AtSGT1b in auxin signaling (Muskett and Parker, 2003; Azevedo et al., 2006). A previous study revealed that NbSGT1 overexpression in N. benthamiana accelerates the development of the hypersensitive response (HR) during R-mediated disease resistance (Wang et al., 2010). Additionally, Hv-SGT1 overexpression in wheat enhances the resistance to PM, which is correlated with increased levels of reactive oxygen intermediates at the pathogen entry sites (Xing et al., 2013). Furthermore, PsoSGT1 from Prunus sogdiana appears to interact with molecular chaperones (RAR1 and HSP90) to activate a nucleotide-binding domain and leucine-rich repeat-containing (NB-LRR)-type protein that confers disease resistance (Zhu et al., 2017). The regulation of NB-LRR-type protein stability or substrate degradation to maintain the balance between the activation and inhibition of plant defense responses (Meldau et al., 2011; Hoser et al., 2013), contributes to the RXLR elicitor-induced HR-related cell necrosis (Xiang et al., 2017).
The recent release of the pumpkin genome provided an opportunity for an additional screening for disease-resistance genes (Sun et al., 2017). In this study, on the basis of the above-mentioned transcriptome analysis that identified differentially expressed genes responsive to PM, we functionally characterized the pumpkin homolog of SGT1 (designated as CmSGT1). The transcription of CmSGT1 in the PM-resistant inbred line “112-2” was strongly induced by PM, salicylic acid (SA), hydrogen peroxide (H2O2), ethephon (Eth), and methyl jasmonate (MeJA). Transgenic N. benthamiana plants that constitutively overexpressed CmSGT1 exhibited increased resistance to PM and were hypersensitive to bacterial wilt and scab.
Results
Cloning of CmSGT1 and Subcellular Localization the Encoded Protein
Pumpkin PM-related candidate genes identified in a transcriptome were reported previously (Guo et al., 2018). One of the isolated clones exhibited 89% identity at the nucleotide level to C. melo SGT1. A full-length clone of this homolog was obtained, and the gene was named CmSGT1 and submitted to GenBank (accession number MH105820). The CmSGT1 gene comprised 1,206 bp, which included a 1,080-bp open reading frame (ORF) encoding 360 amino acids. The predicted polypeptide was basic, with a pI of 5.32, and a molecular mass of 40.3 kDa. An alignment of the deduced CmSGT1 amino acid sequence with homologous sequences is presented in Supplementary Figure S1. At the amino acid level, CmSGT1 was highly similar to the SGT1 from the following plant species: Cucumis melo (CmSGT1, 84.4% identity), and Cucumis sativus (CsSGT1, 82.5% identity), N. benthamiana (NbSGT1.1 and NbSGT1.2, 89.5% identity), and Arabidopsis thaliana (AtSGT1a and AtSGT1b, 75.2% identity). The CmSGT1 sequence contained three conserved domains, namely TPR, CS, and SGS.
The subcellular localization of CmSGT1 was assessed with a CmSGT1-GFP fusion protein that was produced in A. thaliana protoplasts under the control of the 35S CaMV promoter. The GFP signal in protoplasts producing GFP alone was detected in the cytoplasm and nucleus (Figure 1), whereas the signal from the CmSGT1-GFP fusion protein was detected exclusively in the nucleus.
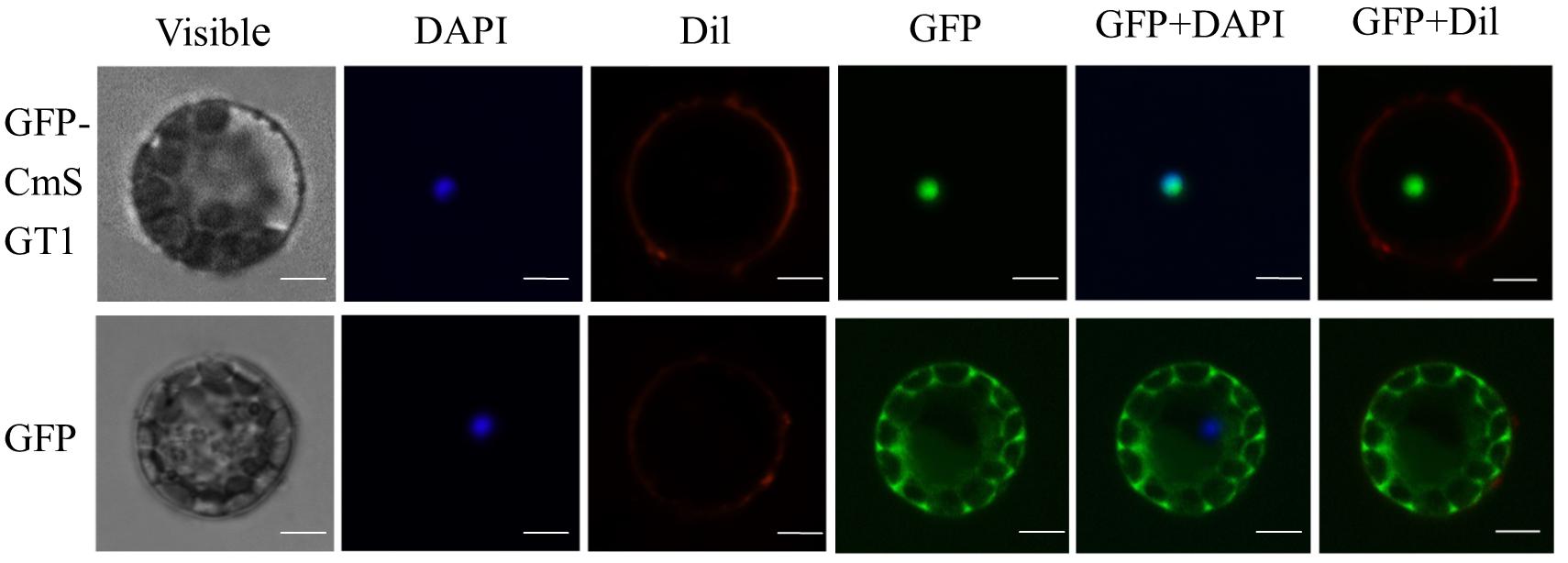
Figure 1. The subcellular localization of pumpkin CmSGT1. The fused pBI221-GFP- CmSGT1 and pBI221-GFP constructs were introduced into Arabidopsis protoplast by polyethylene glycol (PEG)-mediated protoplast transformation. The fluorescent signals were detected using a confocal fluorescence microscope. Scale bars = 5 μm.
Expression of CmSGT1 in Pumpkin Seedlings in Response to PM and Exogenous Treatments
A qRT-PCR assay was completed to analyze the CmSGT1 expression patterns in PM-resistant inbred line “112-2” and PM-susceptible cultivar “JJJD” treated with PM, H2O2, SA, abscisic acid (ABA), Eth, and MeJA (Figure 2). The expression data were normalized against that of the β-actin gene and were recorded relative to the CmSGT1 transcript level in the water-sprayed control “JJJD” plants at 0 hour post-inoculation (hpi).
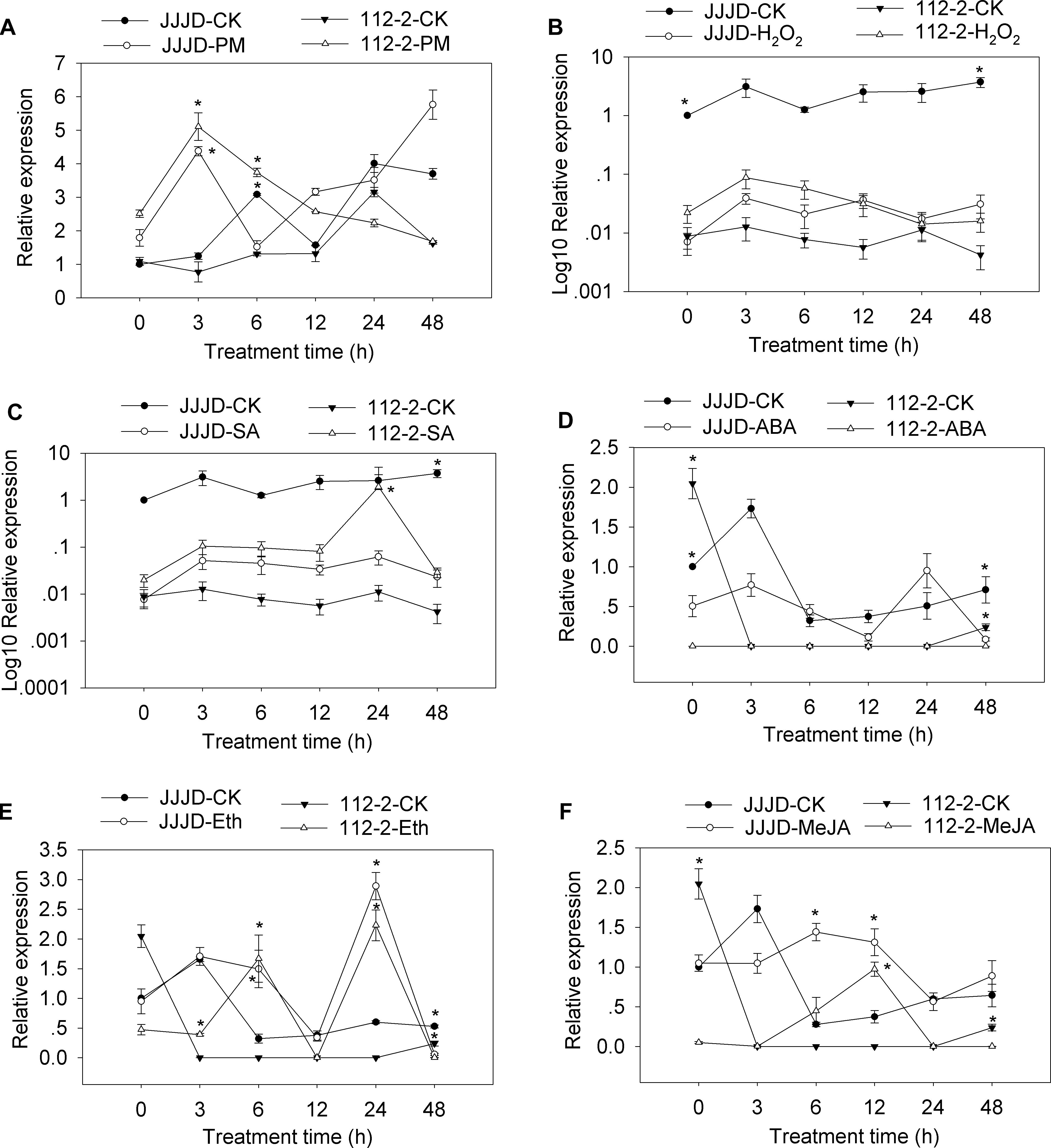
Figure 2. CmSGT1 expression in response to powdery mildew and exogenous hormones. The pumpkin seedlings were sprayed with a spore suspension (A), exogenous H2O2 (B), SA (C), ABA (D), Eth (E) and MeJA (F). The pumpkin β-actin gene was used as an internal reference gene for qRT-PCR. The transcript level of CmSGT1 in the susceptible cultivar “JJJD” at 0 h is used as control (quantities of calibrator) and was assumed as 1. The relative gene expression in B and C (Y-axis) was transformed to a log10 scale. The values are the means ± SEs of three biological replicates. Data between treatments (112-2-treatment vs. 112-2-CK and JJJD- treatment vs. JJJD-CK) were analyzed by one-way ANOVA and * denotes statistical significance at p < 0.05.
The CmSGT1 expression level in “112-2” plants were upregulated by PM (except at 24 h), H2O2, and SA treatments, with the PM-induced expression level at 3 hpi upregulated by 6.62-fold. In contrast, the ABA treatment essentially had no effect. The expression of CmSGT1 in “JJJD” plants was inhibited by H2O2, SA, and ABA (except at 24 h) treatments, with irregular expression-level changes induced by PM. After the Eth treatment, the CmSGT1 expression level was significantly higher in the “112-2” and “JJJD” seedlings than in the water-treatment control (CK) seedlings (except at 48 h), with peak levels occurring at 24 h (i.e., 10.2-fold and 4.5-fold increases in “112-2” and “JJJD” seedlings, respectively). In response to the MeJA treatment, CmSGT1 expression levels were higher in “112-2” (except at 48 h) and “JJJD” (except at 3 h) seedlings than in CK seedlings. The results indicated that SA, and H2O2 upregulated CmSGT1 expression in inbred line “112-2” (PM-resistant material), but downregulated CmSGT1 expression in cultivar “JJJD” (PM-sensitive material). Moreover, Eth and MeJA induced CmSGT1 expression regardless of PM susceptibility.
Improved PM Resistance of CmSGT1-Overexpressing N. benthamiana Plants
Firstly, transcript level of the high homology (NbSGT1) modulated by the overexpression of CmSGT1 under normal conditions was determined by qRT-PCR. Compared to wild type (WT) plants, the expression of the homologous genes was not basically altered in transgenic plants when grown in normal condition (Figure 3), indicating that overexpression of pumpkin CmSGT1 gene has no obvious effect on NbSGT1 transcript in N. benthamiana. The CmSGT1 gene was not detected in WT plants.
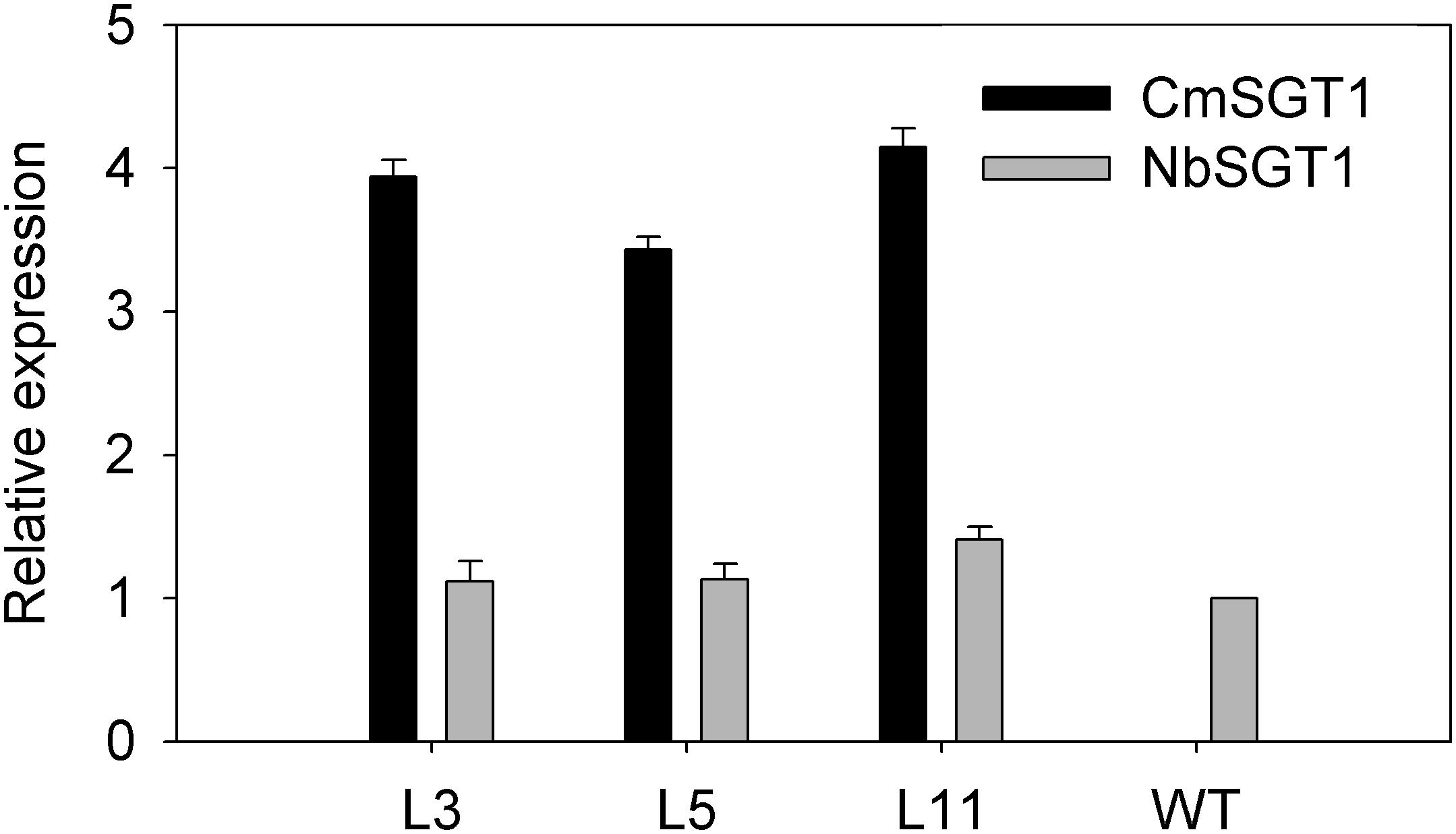
Figure 3. Relative expression of CmSGT1 and NbSGT1 in transgenic or wild-type plants under normal growth conditions. N. benthamiana encodes NbSGT1 isoform. L3, L5, and L11 are three independent transgenic lines that overexpress CmSGT1. Three biological triplicates were averaged and Bars indicate standard error of the mean. Data (NbSGT1 expression in transgenic lines vs. NbSGT1 expression in wild-type plants) were analyzed by one-way ANOVA and unmarked * denotes no statistical differences at p < 0.05.
The disease severity at 10 days post-inoculation (dpi) was 78% lower for the transgenic plants than for the CK plants (Table 1). Powdery mildew symptoms were detectable on infected WT seedlings at 7 dpi, and the infected spots were chlorotic at 28 dpi. In contrast, the PM symptoms were undetectable and relatively slight at 7 and 28 dpi, respectively, in the CmSGT1-overexpressing transgenic plants (Figure 4A). A comparison of the leaves of PM-infected WT and transgenic plants revealed more blue spots displayed by the trypan blue staining, on the transgenic leaves than on the WT leaves at 4 dpi, although blue spots were developing on the WT leaves. The blue spots on the transgenic leaves expanded at 5 and 7 dpi, and were bigger than those of WT leaves, implying that the overexpression of CmSGT1 in transgenic plants accelerated cell death following a PM infection (Figure 4B). Moreover, there were more brown spots manifested by the DAB staining, on the transgenic leaves than on the WT leaves at 1 dpi. These brown spots reflected the accumulation of H2O2, and were darker brown and larger at 3 dpi. At 5 dpi, the brown spots lightened in the infected plants, but were more intense in the transgenic plants than the spots in the WT plants. The results indicated that the overexpression of CmSGT1 promoted the accumulation of H2O2 in transgenic plants infected with PM (Figure 4C).
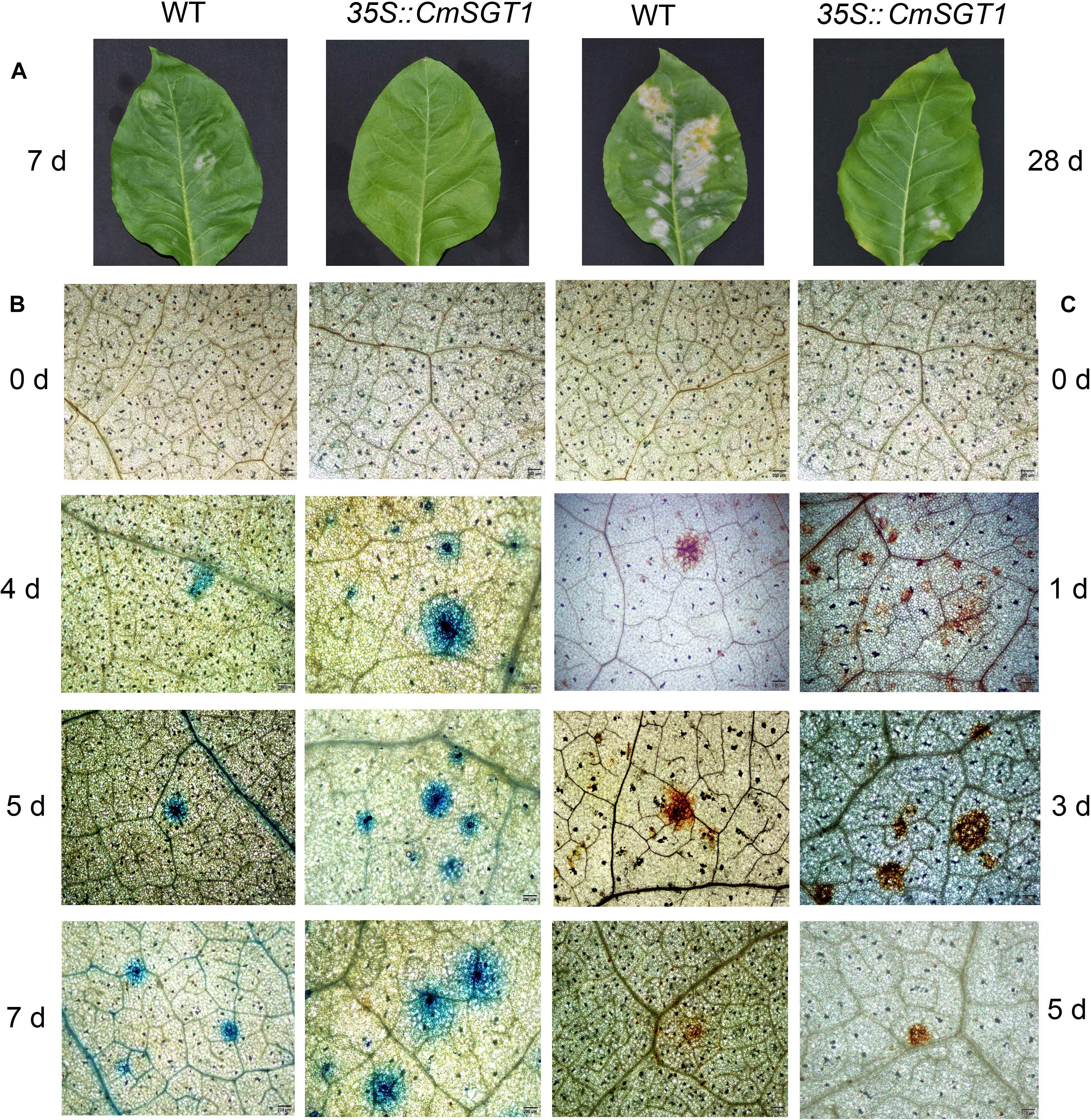
Figure 4. The pathogenic symptoms, trypan blue and DAB staining of N. benthamiana leaves treated with powdery mildew. The pathogenic symptoms of transgenic N. benthamiana and WT at 7 and 28 days post-inoculation (A); trypan blue staining was performed to visualize cell death (B); DAB staining was performed to visualize H2O2 accumulation (C). Scale bars = 200 μm.
Expression of Signal-Related Genes in Transgenic N. benthamiana Plants
To investigate the signal transduction pathways affected by CmSGT1 during plant defense responses to PM, the expression levels of five signaling-associated genes (NPR1, PR5, PR1a, PAL, and PDF1.2) in the SA, JA, and ET signal transduction pathways were analyzed by qRT-PCR for the CmSGT1-overexpressing transgenic and WT N. benthamiana plants, with water-treated WT plants at 0 hpi serving as the control samples (Figure 5). The NPR1, PAL (except at 120 h), and PR5 expression levels were lower in the transgenic and WT plants infected with PM than in the CK plants, implying that PM inhibited the expression of these genes. The PR1a expression level was higher in the PM-infected transgenic plants than in the CK plants (i.e., 68.3-fold at 120 h), whereas the PR1a level was lower in the PM-infected WT plants than in the CK plants. The PDF1.2 expression levels in the PM-infected transgenic plants were lower at 12 hpi and higher at 48 hpi than in the CK plants, with no differences thereafter. Thus, in response to the PM infection, the NPR1, PAL (except at 24 and 48 h), and PDF1.2 (except at 48 h) expression levels were significantly lower in the transgenic plants than in the WT plants, whereas the opposite pattern was observed for the PR1a, and PR5 expression levels. These results implied that the overexpression of CmSGT1 in N. benthamiana upregulates the expression of PR1a, and PR5, which are involved in SA defense signal transduction. Furthermore, the increased PM resistance of the transgenic N. benthamiana plants appears to be related to the upregulated expression of these genes.
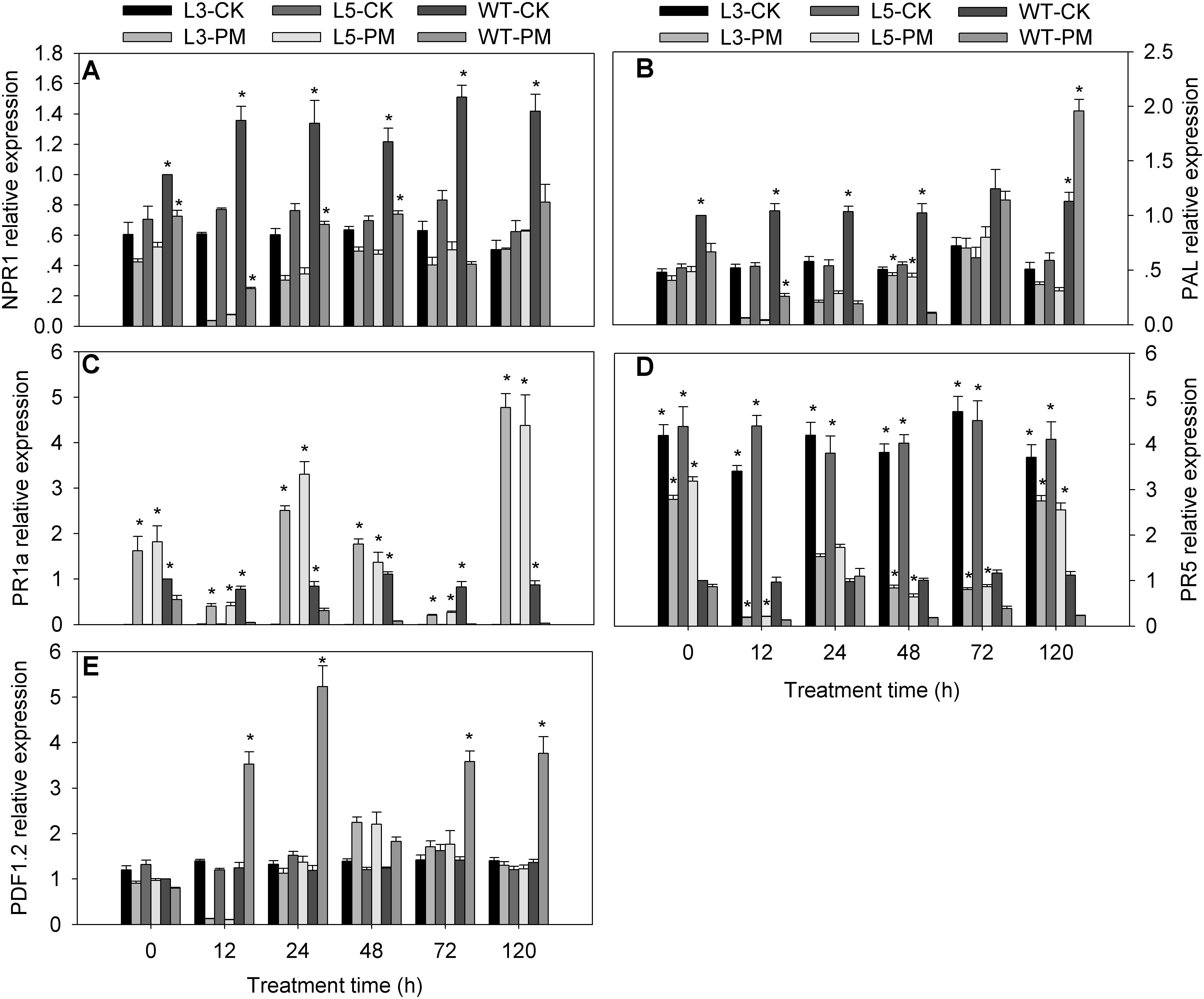
Figure 5. Expression of signal-related genes in transgenic and wild-type plants treated with powdery mildew. The samples of two genetically modified N. benthamiana lines were used to analyze the genes expression by qRT-PCR. (A) NtNPR1; (B) NtPAL; (C) NtPR1a; (D) NtPR5; (E) NtPDF1.2. L3/5-CK represents transgenic N. benthamiana L3 or L5 line grown under normal conditions; L3/5-PM represents transgenic N. benthamiana L3 or L5 line infected with powdery mildew; WT-CK represents wild-type plants grown under normal conditions; WT-PM represents wild-type plants infected with powdery mildew. N. benthamiana NtEF1-α gene (AF120093) was used as an internal control for normalization of different cDNA samples. The expression levels of signal-related genes in wild-type plants at 0 h were used as control (quantities of calibrator) and were assumed as 1. Three biological triplicates per line were averaged and Bars indicate standard error of the mean. Data between transgenic plants and WT plants (L3/5-PM vs. WT-PM and L3/5-CK vs. WT-CK) were analyzed by one-way ANOVA and “*” denotes statistical significance at p < 0.05.
Compromised Resistance of Transgenic N. benthamiana Plants to Bacterial Diseases
To analyze the effect of CmSGT1 on other plant diseases, two common bacterial pathogens causing bacterial wilt (Ralstonia solanacearum) and scab (Xanthomonas euvesicatoria) were injected into N. benthamiana plants (Figure 6). At 6 days after inoculation with R. solanacearum and X. euvesicatoria, the chlorosis and yellowing of the sixth leaf veins was greater for transgenic N. benthamiana plants than for the WT plants. Additionally, there were 5.94- and 21.1-times more the concentration of bacterial wilt and scab bacteria, respectively, in the transgenic plants than in the WT plants. Yellowing was also observed between the veins of the 12th leaf in transgenic N. benthamiana plants, and there were 13.3- and 8.28-times more R. solanacearum and X. euvesicatoria bacteria, respectively, in the transgenic plants than in the WT plants. These observations suggested that the overexpression of CmSGT1 in N. benthamiana decreases the resistance to bacterial wilt and scab.
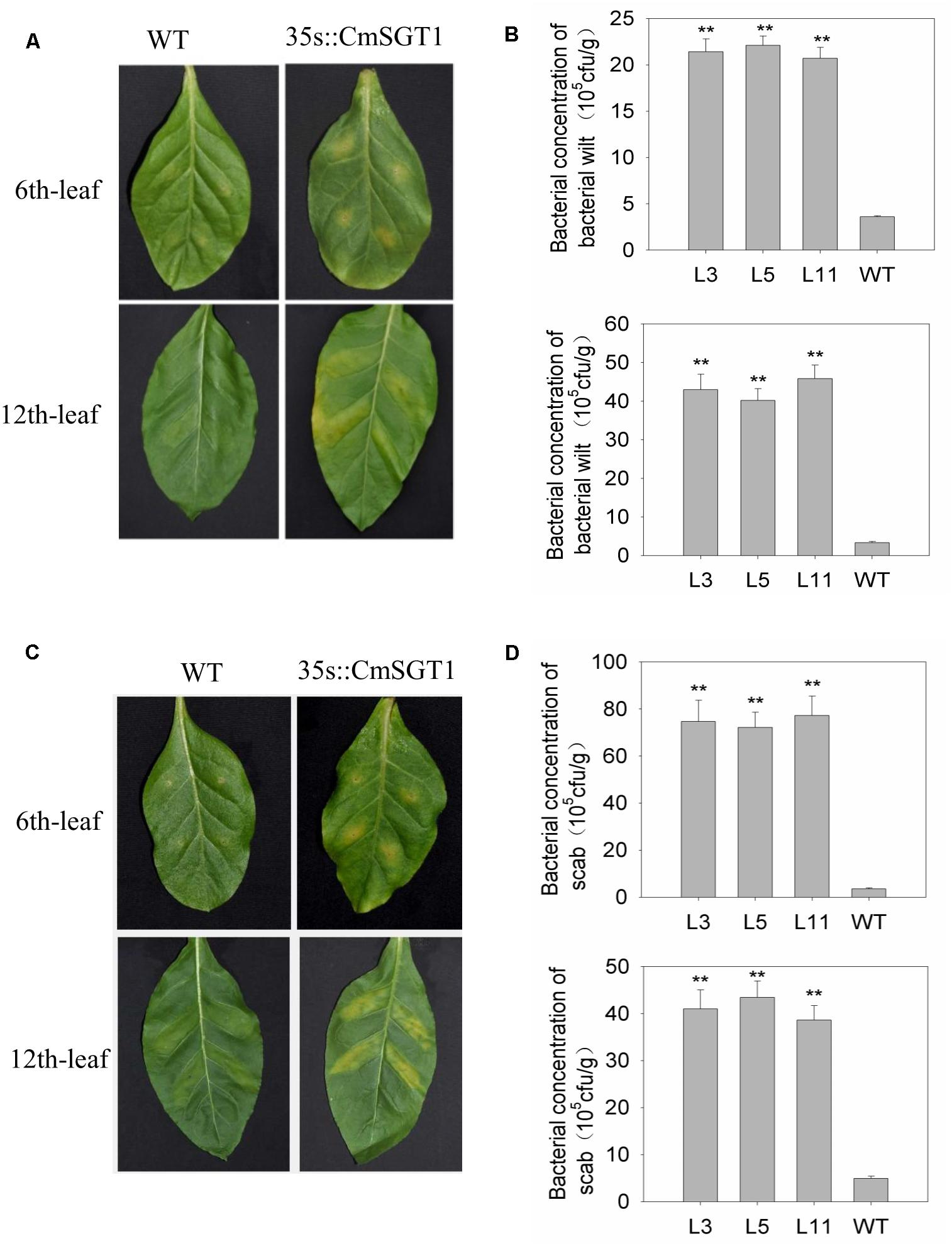
Figure 6. The resistance of CmSGT1 in transgenic and wild-type plants to N. benthamiana bacterial wilt and scab. (A) pathogens symptoms of the 6th-upper and 12th-upper leaf injection sites was injected with bacterial wilt bacteria with a needle-removed syringe. (B) concentration bacteria of the 6th-upper and 12th-upper leaf injection sites was injected with bacterial wilt bacteria with a needle-removed syringe. (C) pathogens symptoms of the 6th-upper and 12th-upper leaf injection sites was injected with scab bacteria with a needle-removed syringe. (D) concentration bacteria of the 6th-upper and 12th-upper leaf injection sites was injected with scab bacteria with a needle-removed syringe. “∗∗” denotes significant differences between WT and transgenic plants at p < 0.01.
Discussion
In this study, we isolated a novel pumpkin SGT1 gene, which was designated as CmSGT1. The predicted amino acid sequence was 89 and 82% identical to the NbSGT1.1 and CsSGT1 sequences, respectively. In A. thaliana and N. benthamiana, SGT1 encodes three functional domains (TPR, CS, and SGS) that are essential for SGT1 activity (Peart et al., 2002; Noël et al., 2007). In this study, the CmSGT1-GFP fusion protein was localized to the nucleus in A. thaliana protoplasts, which was inconsistent with the results of earlier studies involving SGT1 in other plant species (Xing et al., 2013; Liu et al., 2016). This inconsistency may be related with low identity (Haynaldia villosa HvSGT1, 64.0% identity; pepper CaSGT1, 63.7% identity) or the number of transformed cells examined. Arabidopsis SGT1b fused to Cerulean localized to the cytosol, but could be seen in nuclei of 25% of 55 transformed cells examined (Noël et al.,2007), and translocation of the SGT1/SRC2-1 complex from the plasma membrane and cytoplasm to the nuclei is required in pepper upon the inoculation of Phytophthora capsici (Liu et al., 2016), suggesting the movement of SGT1 between the cytosol and nucleus.
The interplay among complex signaling networks, including various pathways regulated by phytohormones, such as SA, JA, ethylene (ET) and ABA, considerably influences plant resistance to diseases. An earlier investigation proved that HvSGT1 expression levels substantially increase following treatments with H2O2 and MeJA, slightly increase following exposure to ET or ABA, and are unchanged in response to SA (Xing et al., 2013). In the current study, SA and H2O2 treatments considerably upregulated CmSGT1 expression in the PM-resistant inbred line “112-2”, but downregulated CmSGT1 expression in the PM-susceptible material. The expression of CmSGT1 in both materials was upregulated by Eth and MeJA treatments. Recent reports indicated that SGT1 expression may be induced by phytopathogens, such as Blumeria graminis in wheat and P. capsici in pepper (Xing et al., 2013; Liu et al., 2016). In this study, the CmSGT1 expression in inbred line “112-2” was induced by PM. Additionally, the overexpression of CmSGT1 in transgenic N. benthamiana plants decreased the disease index by 78%, accelerated cell necrosis, and increased the accumulation of H2O2. These results indicated that the PM resistance of the transgenic N. benthamiana plants was enhanced, likely because of the changes to the HR-related cell necrosis and H2O2 accumulation. Our findings are consistent with the results of an earlier study that revealed that H2O2 accumulation and the subsequent cell death usually lead to the resistance to diseases caused by biotrophic pathogens (Li et al., 2011). Moreover, SGT1 helps mediate cell death during compatible and incompatible plant–pathogen interactions, suggesting that SGT1 is an essential component of common signaling pathways responsible for cell death (Cuzick et al., 2009; Uppalapati et al., 2010; Wang et al., 2010). The silencing of the pepper SGT1 gene adversely affects HR-related cell death, prevents H2O2 accumulation, and downregulates HR-related and SA/JA-dependent marker gene expression levels, and influences the PcINF1/SRC2-1-induced pepper defense response by SGT1 interacting with SRC2-1 (Liu et al., 2016).
The PDF1.2 gene is important for the JA/ET-dependent signaling pathway. The disease-associated NPR1 gene (non-expresser of PR1) affects various disease-resistance signal transduction pathways, and encodes one of the important transcription factors downstream of SA. The SA-dependent disease-resistance signal transduction pathway can be divided into NPR1-dependent and NPR1-independent transduction pathways (Gao and Zhang, 2012). The PAL, PR1a, and PR5 expression levels are markers of the SA signaling pathway. In wheat, HvSGT1 activates PM resistance mechanisms through JA-dependent defense pathways and suppresses the activities of SA-dependent defense pathways (Xing et al., 2013). In the current study, following a PM infection, the NPR1, PAL, and PDF1.2 expression levels were lower in the transgenic plants than in the WT plants, whereas the opposite pattern was observed for the PR1a and PR5 expression levels. This suggests that in the SA pathway, the transactivation of NPR1 is unaffected by CmSGT1, whereas the transactivation of PR1a, and PR5 is dependent on CmSGT1. Additionally, CmSGT1 does not directly affect the JA/ET-dependent defense pathway to regulate PDF1.2 expression. We propose that CmSGT1 activates stress-resistance mechanisms through SA-dependent defense pathways without inducing NPR1 and PAL expression levels, but suppresses the activities of JA/ET-dependent defense pathways. Therefore, we speculate that CmSGT1 positively regulates the H2O2 and SA pathways. Moreover, H2O2 might directly transfer the SA signal to regulate the expression of downstream response genes in the CmSGT1-overexpressing transgenic plants infected with PM. In NPR1-dependent SA signal transduction pathways, the activation of PR genes needs NPR1 and TGA transcription factor binding to identify its promoters (Fan and Dong, 2002; Spoel and Dong, 2012). The equivalent mutation in NPR1 abolishes its ability to bind SA and promotes SA-induced defense gene expression (Ding et al., 2018). We speculated that whether there is the relation of the downregulation of NPR1 and SA-mediated transcriptional activation of PR genes or not. Notably, the phenotypes and genes in CmSGT1-overexpressing transgenic N. benthamiana plants infected with N. benthamiana PM might not be exactly the same as those regulated by CmSGT1 in C. moschata in response to cucurbit PM. Further studies will be necessary to reveal biological functions for CmSGT1 in C. moschata infected with PM.
Two globally important diseases that affect N. benthamiana, bacterial wilt and scab, are caused by the necrotrophic R. solanacearum and the hemi-biotrophic X. euvesicatoria, respectively. There are differences in the resistance mechanisms and patterns of fungal development for biothophic (P. xanthii) and necrotrophic (R. solanacearum) pathogens. Recent studies confirmed that SGT1 promotes the resistance to biotrophic pathogens, while suppressing the resistance to necrotrophic and hemibiotrophic pathogens. Silencing of SGT1 compromised resistance to the barley and H. villosa biothoph B. graminis (Shen et al., 2003; Xing et al., 2013) and the wheat biotroph Puccinia striiformis (Scofield et al., 2005), while enhancing the resistance of N. benthamiana to the necrotrophic pathogen Botrytis cinerea (El Oirdi and Bouarab, 2007). A recent study involving N. benthamiana indicated silencing of NbSGT1 compromised the protective effect of systemic acquired resistance-induced plants on neighboring plants against bacteria wilt caused by R. solanacearum (Cheol Song et al., 2016). In the current study, the chlorosis and yellowing of the infection sites on leaves were greater in transgenic N. benthamiana plants than in WT plants at 6 dpi. Additionally, there were substantially more concentration of bacterial wilt and scab bacteria in the transgenic plants than in the WT plants, indicating that the overexpression of CmSGT1 in N. benthamiana adversely affects the resistance to bacterial wilt and scab, which differs from the effects of CmSGT1 overexpression on the resistance to PM. Consequently, SGT1 may function differently depending on the plant–pathogen combinations with diverse effectors and R proteins.
In conclusions, the results of this study indicate that the overexpression of CmSGT1 may increase the PM resistance, but decrease bacterial wilt and scab resistance, in transgenic N. benthamiana plants. Additionally, CmSGT1 overexpression may improve PM resistance by enhancing HR-related cell death and H2O2 accumulation and upregulating the expression of SA-mediated defense-response genes. Further analyses of the CmSGT1 gene may be useful for characterizing biotic stress signaling pathways and for the genetic engineering of novel pumpkin cultivars. The results described herein may be relevant for future biotechnology-based investigations and the molecular breeding of pumpkins and related plant species.
Materials and Methods
Plant Materials and Treatments
Pumpkin (C. moschata) inbred line “112-2” and cultivar “Jiujiangjiaoding” (abbreviated “JJJD”), which are resistant and susceptible to PM, respectively, were provided by the Henan Institute of Science and Technology, Xinxiang, Henan, China (Zhou et al., 2010). Pumpkin seeds were germinated and the resulting seedlings were grown as previously described (Guo et al., 2018). Seedlings at the three-leaf stage were treated as follows. The PM infection was initiated as described by Guo et al. (2018). Specifically, conidia were collected from the leaves of pumpkin plants naturally infected with PM in a local greenhouse. Seedlings were sprayed with a freshly prepared spore suspension (106 spores/ml) or an exogenous signaling molecule or hormone, including 1.5 mM H2O2, 100 μM SA, 100 μM ABA, 100 μM MeJA, 0.5 g/L Eth. Moreover, water alone was used for the control treatment (CK). The treated seedlings were maintained in a growth chamber with a 15-h light (28°C)/9-h dark (18°C) cycle (5,500 lux light intensity) and harvested after 0, 3, 6, 12, 24, and 48 h to examine the CmSGT1 expression pattern. At each time point, two young leaves were collected from the upper parts of four seedlings (i.e., one sample), wrapped in foil, frozen in liquid nitrogen, and stored at −80°C. The treatments were arranged in a randomized complete block design, with three biological replicates.
Isolation of CmSGT1 cDNA Clone and Sequence Analysis
The SGT1 homolog expressed sequence tag (GenBank accession number SRR5369792) was identified in a PM-resistant pumpkin seedling transcriptome by Guo et al. (2018). The full-length ORF of the SGT1 homolog was obtained with the cDNA fragment of this homolog used as a probe, as previously described (Guo et al., 2014). The theoretical molecular weight (Mw) and isoelectric point (pI) were calculated with the ExPASy Compute pI/Mw tool (Bjellqvist et al., 1993). Sequence data were analyzed with the ClustalW program (Thompson et al., 1994). The NCBI databases were screened for homologous sequences with the default parameters of the BLAST program2 (Altschul et al., 1997).
Subcellular Localization Analysis of CmSGT1
The CmSGT1 ORF (without the termination codon) was ligated into the pBI221-GFP vector for the subsequent production of a green fluorescent protein (GFP)-tagged CmSGT1 fusion protein. Polyethylene glycol was used during the transformation of A. thaliana protoplasts with the recombinant plasmid (Lee et al., 2013). The subcellular localization of CmSGT1 was determined based on the GFP signal, which was detected with the confocal fluorescence microscope (UltraVIEW VoX, Olympus, Japan) under excitation wavelength 488 nm and captured light wavelength range 448–508 nm. The cell membrane and nucleus were stained with 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (Dil) and 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI), respectively.
Generation of CmSGT1-Overexpressing Transgenic N. benthamiana Plants
Pumpkin are known to be one of the plants most refractory for transformation. To date, only two reports on transformation in C. moschata existed using a combined method of vacuum infiltration and Agrobacterium infection (Nanasato et al., 2011; Nanasato and Tabei, 2015). So, we choose a heterologous overexpression assay in N. benthamiana instead of generating C. moschata transgenic plants. Forward and reverse primers with an added BamH I site and Kpn I site, respectively, were used to amplify CmSGT1. The amplified sequence was then inserted into the pVBG2307 vector for the subsequent expression of CmSGT1 under the control of the 35S cauliflower mosaic virus (CaMV) promoter. The recombinant plasmid was introduced into Agrobacterium tumefaciens GV3101 cells as previously described (Guo et al., 2014). The resulting A. tumefaciens cells were used to transform N. benthamiana plants according to a previously described leaf disk method (Li et al., 2012). The transgenic N. benthamiana plants were confirmed by examining the segregation ratio of the kanamycin selectable marker and by PCR analysis of NPTII and CmSGT1. T2 lines that produced 100% kanamycin-resistant plants in the T3 generation were considered as homozygous transformants. In each experiment, T2 generations of homozygous transgenic lines (L3, L5 and L11) were selected for further analysis. Similar phenotypes and results used for this study were observed in more than three independent lines of transgenic plants.
Primer Design
Details regarding all of the primers designed and used in this study are provided in Supplementary Table S1.
Analysis of Transgenic N. benthamiana Plants Infected With Powdery Mildew
Conidia were collected from N. benthamiana leaves naturally infected with Erysiphe cichoracearum DC., which causes PM. Transgenic N. benthamiana seedlings at the five-leaf stage were used for phenotypic analyses. Specifically, the petiole of the second leaf from the top of the seedlings was wrapped with cotton moistened with water. The leaf was placed on a porcelain tray containing filter paper, after which it was sprayed with a spore suspension (106 spores/ml). The tray was then covered with film to maintain humidity. At 10 dpi, disease severity of leaves in vitro was calculated as [(5A + 4B+ 3C+ 2D+ E)/5F] × 100 according to Ishii et al. (2001). Additionally, the first fully expanded true leaf of the transgenic and WT plants were sprayed with the above-mentioned spore suspension and sampled after 0, 12, 24, 48, 72, and 120 hpi, frozen in liquid nitrogen, stored at −80°C and used for extraction of total RNA. On the other hand, these leaves were harvested symmetrically along the sides of the main vein after 0, 1, 3, 4, 5, and 7 days to examine of cell death and H2O2 accumulation. The treatments were arranged in a randomized complete block design with three replicates.
Leaves from the transgenic N. benthamiana seedlings were stained with 3, 3′-diaminobenzidine (DAB) and trypan blue staining as previously described (Choi et al., 2012) to analyze H2O2 accumulation and cell death. For DAB staining, whole leaves were immersed in DAB solution (1 mg/ml, pH 5.7) and incubated overnight (almost 8 h) in darkness. Leaves were then de-stained three times with 95% ethanol. Regarding trypan blue staining, whole leaves were boiled in a lactophenol-ethanol trypan blue solution (10 ml lactic acid, 10 ml glycerol, 10 g phenol, 30 ml absolute ethanol, and 10 mg trypan blue dissolved in 10 ml distilled water) for 10 min and then maintained at room temperature overnight. Leaves were de-stained with 2.5 g/ml chloral hydrate in distilled water.
Analysis of Transgenic N. benthamiana Plants Infected With Bacterial Wilt and Scab
The sixth and twelfth leaf from the top of transgenic N. benthamiana seedlings were inoculated with bacterial solutions (108 cfu/ml). Specifically, the bacterial solutions were injected into the underside of leaves between the lateral veins with a syringe lacking a needle. Each leaf was inoculated in four places. The petiole of leaves placed on a porcelain plate was wrapped with water-saturated degreased cotton, after which the plate was covered with film to maintain humidity and then incubated at 28°C with a 16-h light/8-h dark cycle. Water was periodically added to the cotton to maintain an appropriate moisture level. Experiments were done in triplicate for each line. After a 6-day incubation, the concentration of bacteria at the injection sites was determined as follows. The injection sites were sampled with a circular perforator (1 cm diameter) and ground in aseptic water (0.1 g added to 0.9 ml water). The ground material was serially diluted to produce the 102-fold, 103-fold, and 104-fold diluents. A 0.1-ml aliquot of the 103-fold, 104-fold, and 105-fold diluents were added to a petri dish containing NA medium. The medium was incubated upside down at 28°C. Plates with 30–300 colonies were considered ideal. The bacterial concentration was calculated with the following formula: colony average × 10 × dilution/g.
Quantitative Real-Time PCR Analysis
The RNA extraction, first-strand cDNA synthesis and quantitative real-time (qRT-PCR) were completed as described by Guo et al. (2015). Relative gene expression levels were determined with the 2–ΔΔCT method. Total RNA was extracted from the leaves of pumpkin seedlings treated with various stresses or distilled water for 0, 3, 6, 12, 24, or 48 h as described above. The β-actin gene was used as an internal control, because it was confirmed as a suitable reference gene for normalizing of gene expression levels in pumpkin (Wu and Cao, 2010). On the other hand, total RNA was extracted from CmSGT1-overexpressing transgenic and WT N. benthamiana seedlings to examine the expression of five hormone-related genes (NtNPR1, NtPR1a, NtPR5, NtPDF1.2, and NtPAL) and N. benthamiana isoform (NbSGT1). The N. benthamiana NtEF1-α gene (AF120093) was included in the assays as an internal control.
Statistical Analyses
Values were expressed as the mean ± standard error of three independent determinations. Data were compared by analysis of variance (ANOVA) using a one-way ANOVA, and differences between WT and transgenic plants were tested by a post hoc comparison test (Student–Newman–Keuls) at p < 0.05 with SPSS 19.0 for Windows (SPSS Inc., Chicago, IL, United States).
Data Availability
The datasets generated for this study can be found in NCBI, MH105820.
Author Contributions
W-LG and Y-YG conceived and designed the experiments. W-LG and B-HC performed the experiments. J-YM and H-LY analyzed the data. Y-LW, J-GZ, and X-ZL contributed reagents, materials, and analysis tools. W-LG wrote the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (No. 31401876), Henan High-level Talent Introduction and Training Special Foundation (No. 103020218003/008), and Natural Science Foundation of Henan Province (No. 162300410110).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Guizhou Academy of Tobacco Sciences researcher Hancheng Wang for providing tobacco E. cichoracearum and Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac) for editing the English text of a draft of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00955/full#supplementary-material
FIGURE S1 | Amino acid sequences alignment of pumpkin CmSGT1 with others. The three conserved domains (TPR, CS, and SGS) are shown by thin underlines. The genes included are CmSGT1 (Cucumis melo L., XP_008439299.1) and CsSGT1 (Cucumis sativus L., XP_004140745.1) AtSGT1A (Arabidopsis, AT4G23570.3) and AtSGT1B (Arabidopsis, AT4G11260.1), NbSGT1.1 (N. benthamiana, AF516180) and NbSGT1.2 (N. benthamiana, AF516181).
TABLE S1 | Primers used in this investigation.
Footnotes
References
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., and Miller, W. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Azevedo, C., Betsuyaku, S., Peart, J., Takahashi, A., Noël, L., Sadanandom, A., et al. (2006). Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J. 25, 2007–2016. doi: 10.1038/sj.emboj.7601084
Bjellqvist, B., Hughes, G. J., Pasquali, C., Paquet, N., Ravier, F., Sanchez, J. C., et al. (1993). The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 14, 1023–1031. doi: 10.1002/elps.11501401163
Cheol Song, G., Sim, H. J., Kim, S. G., and Ryu, C. M. (2016). Root-mediated signal transmission of systemic acquired resistance against above-ground and below-ground pathogens. Ann. Bot. 118, 821–831. doi: 10.1093/aob/mcw152
Choi, D. S., Hwang, I. S., and Hwang, B. K. (2012). Requirement of the cytosolic interaction between PATHOGENESIS-RELATED PROTEIN10 and LEUCINE-RICH REPEAT PROTEIN1 for cell death and defense signaling in pepper. Plant Cell 24, 1675–1690. doi: 10.1105/tpc.112.095869
Cuzick, A., Maguire, K., and Hammond-Kosack, K. E. (2009). Lack of the plant signaling component SGT1b enhances disease resistance to Fusarium culmorum in Arabidopsis buds and flowers. New Phytol. 181, 901–912. doi: 10.1111/j.1469-8137.2008.02712.x
Ding, Y., Sun, T., Ao, K., Peng, Y., Zhang, Y., Li, X., et al. (2018). Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 6, 173–203. doi: 10.1016/j.cell.2018.03.044
El Oirdi, M., and Bouarab, K. (2007). Plant signalling components EDS1 and SGT1 enhance disease caused by the necrotrophic pathogen Botrytis cinerea. New Phytol. 175, 131–139. doi: 10.1111/j.1469-8137.2007.02086.x
Fan, W., and Dong, X. (2002). In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid–mediated gene activation in Arabidopsis. Plant Cell 14, 1377–1389. doi: 10.1105/tpc.001628
Fukino, N., Yoshioka, Y., Sugiyama, M., Sakata, Y., and Matsumoto, S. (2013). Identification and validation of powdery mildew (Podosphaera xanthii)-resistant loci in recombinant inbred lines of cucumber (Cucumis sativus L.). Mol. Breed. 32, 267–277. doi: 10.1007/s11032-013-9867-3
Gao, L. J., and Zhang, Y. X. (2012). Effects of salicylic acid on the expression of SOD, PPO Isozymes and NPR1 in Pear. Acta Hortic. Sin. 40, 41–48.
Guo, W. L., Chen, B. H., Chen, X. J., Guo, Y. Y., Yang, H. L., Li, X. Z., et al. (2018). Transcriptome profiling of pumpkin (Cucurbita moschata Duch.) leaves infected with powdery mildew. PLoS One 13:e0190175. doi: 10.1371/journal.pone.0190175
Guo, W. L., Chen, R. G., Du, X. H., Zhang, Z., Yin, Y. X., Gong, Z. H., et al. (2014). Reduced tolerance to abiotic stress in transgenic Arabidopsis overexpressing a Capsicum annuum multiprotein bridging factor 1. BMC Plant Biol. 14:138. doi: 10.1186/1471-2229-14-138
Guo, W. L., Wang, S. B., Chen, R. G., Chen, B. H., Du, X. H., Yin, Y. X., et al. (2015). Characterization and expression profile of CaNAC2 pepper gene. Front. Plant Sci. 6:755. doi: 10.3389/fpls.2015.00755
Hoser, R., Zurczak, M., Lichocha, M., Zuzga, S., Dadlez, M., Samuel, M. A., et al. (2013). Nucleocytoplasmic partitioning of tobacco N receptor is modulated by SGT1. New Phytol. 200, 158–171. doi: 10.1111/nph.12347
Ishii, H., Fraaije, B. A., Sugiyama, T., Noguchi, K., Nishimura, K., and Takeda, T. (2001). Occurrence and molecular characterization of Strobilurin resistance in cucumber powdery mildew and downy mildew. Phytopathology 91, 1166–1171. doi: 10.1094/PHYTO.2001.91.12.1166
Kitagawa, K., Skowyra, D., Elledge, S. J., Harper, J. W., and Hieter, P. (1999). SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol. Cell. 4, 21–33. doi: 10.1016/s1097-2765(00)80184-7
Kumar, D., and Kirti, P. B. (2015). Pathogen-induced SGT1 of Arachis diogoi induces cell death and enhanced disease resistance in tobacco and peanut. Plant Biotechnol. J. 13, 73–84. doi: 10.1111/pbi.12237
Lee, M. H., Lee, Y., and Hwang, I. (2013). “In Vivo localization in arabidopsis protoplasts and root tissue,” in G Protein-Coupled Receptor Signaling in Plants. Methods in Molecular Biology (Methods and Protocols), Vol. 1043, ed. M. Running (Totowa, NJ: Humana Press).
Li, A., Zhang, R., Pan, L., Tang, L., Zhao, G., Zhu, M., et al. (2011). Transcriptome analysis of H2O2-treated wheat seedlings reveals a H2O2-responsive ratty acid desaturase gene participating in powdery mildew resistance. PLoS One 6:e28810. doi: 10.1371/journal.pone.0028810
Li, Z., Wang, S., Tao, Q., Pan, J., Si, L., Gong, Z., et al. (2012). A putative positive feedback regulation mechanism in CsACS2 expression suggests a modified model for sex determination in cucumber (Cucumis sativus L.). J. Exp. Bot. 63, 4475–4484. doi: 10.1093/jxb/ers123
Liu, Z. Q., Liu, Y. Y., Shi, L. P., Yang, S., Shen, L., Yu, H. X., et al. (2016). SGT1 is required in PcINF1/SRC2-1 induced pepper defense response by interacting with SRC2-1. Sci. Rep. 6:21651. doi: 10.1038/srep21651
Meldau, S., Baldwin, I. T., and Wu, J. (2011). For security and stability: SGT1 in plant defense and development. Plant Signal. Behav. 6, 1479–1482. doi: 10.4161/psb.6.10.17708
Muskett, P., and Parker, J. (2003). Role of SGT1 in the regulation of plant R gene signaling. Microbes Infect. 5, 969–976. doi: 10.1016/s1286-4579(03)00183-7
Nanasato, Y., Konagaya, K., Okuzaki, A., Tsuda, M., and Tabei, Y. (2011). Agrobacterium-mediated transformation of kabocha squash (Cucurbita moschata Duch) induced by wounding with aluminum borate whiskers. Plant Cell Rep. 30, 1455–1464. doi: 10.1007/s00299-011-1054-6
Nanasato, Y., and Tabei, Y. (2015). Cucumber (Cucumis sativus L.) and kabocha squash (Cucurbita moschata Duch). (eds) Agrobacterium protocols. Methods Mol. Biol. 1223, 299–310. doi: 10.1007/978-1-4939-1695-5_24
Noël, L. D., Cagna, G., Stuttmann, J., Wirthmüller, L., Betsuyaku, S., Witte, C.-P., et al. (2007). Interaction between SGT1 and cytosolic/nuclear HSC70 chaperones regulates Arabidopsis immune responses. Plant Cell 19, 4061–4076. doi: 10.1105/tpc.107.051896
Peart, J. R., Lu, R., Sadanandom, A., Malcuit, I., Moffett, P., Brice, D. C., et al. (2002). Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. U.S.A. 99, 10865–10869. doi: 10.1073/pnas.152330599
Perez-Garcia, A., Romero, D., Fernandez-Ortuno, D., Lopez-Ruiz, F., De-Vicente, A., and Tores, J. A. (2009). The powdery mildew fungus Podosphaera fusca (synonym Podosphaera xanthii), a constant threat to cucurbits. Mol. Plant Pathol. 10, 153–160. doi: 10.1111/j.1364-3703.2008.00527.x
Scofield, S. R., Huang, L., Brandt, A. S., and Gill, B. S. (2005). Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol. 138, 2165–2173. doi: 10.1104/pp.105.061861
Shen, Q. H., Zhou, F., Bieri, S., Haizel, T., and Shirasu, K. (2003). Recognition specificity and RAR1/SGT1 dependency in barley Mla disease resistance alleles to the powdery mildew fungus. Plant Cell 15, 732–744. doi: 10.1105/tpc.009258
Spoel, S. H., and Dong, X. (2012). How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 12, 89–100. doi: 10.1038/nri3141
Sun, H., Wu, S., Zhang, G., Jiao, C., Guo, S., Ren, Y., et al. (2017). Karyotype stability and unbiased fractionation in the paleo-allotetraploid Cucurbita genomes. Mol. Plant 10, 1293–1306. doi: 10.1016/j.molp.2017.09.003
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Uppalapati, S. R., Ishiga, Y., Ryu, C. M., Ishiga, T., Wang, K., Noël, L. D., et al. (2010). SGT1 contributes to coronatine signaling and Pseudomonas syringae pv. Tomato disease symptom development in tomato and Arabidopsis. New Phytol. 189, 83–93. doi: 10.1111/j.1469-8137.2010.03470.x
Wang, K., Uppalapati, S. R., Zhu, X., Dinesh-Kumar, S., and Mysore, K. S. (2010). SGT1 positively regulates the process of plant cell death during both compatible and incompatible plant-pathogen interactions. Mol. Plant Pathol. 11, 597–611. doi: 10.1111/j.1364-3703.2010.00631.x
Wang, P., Liu, J. C., and Zhao, Q. Y. (2002). Studies on nutrient composition and utilization of pumpkin fruit. J. Inner Mongolia Agric. Univ. 23, 52–54.
Wu, T., and Cao, J. (2010). Molecular cloning and expression of a bush related CmV1, gene in tropical pumpkin. Mol. Biol. Rep. 37, 649–652. doi: 10.1007/s11033-009-9505-7
Xiang, J., Li, X., Yin, L., Liu, Y., Zhang, Y., Qu, J. J., et al. (2017). A candidate rxlr effector from plasmopara viticola can elicit immune responses in nicotiana benthamiana. BMC Plant Biol. 17:75. doi: 10.1186/s12870-017-1016-4
Xing, L., Qian, C., Cao, A., Li, Y., Jiang, Z., Li, M., et al. (2013). The Hv-SGT1 gene from Haynaldia villosa contribute to resistances towards both biotrophic and hemi-biotrophic pathogens in common wheat (Triticum aestivum L.). PLoS One 8:e72571. doi: 10.1371/journal.pone.0072571
Yadav, M., Jain, S., Tomar, R., Prasad, G. B. K. S., and Yadav, H. (2010). Medicinal and biological potential of pumpkin: an updated review. Nutr. Res. Rev. 23, 184–190. doi: 10.1017/S0954422410000107
Zhou, J. G., Hu, H. L., Li, X. Z., Zhou, R. J., and Zhang, H. R. (2010). Identification of a resource of powdery mildew resistance in Cucurbita moschata. Acta Hortic. 871, 141–146. doi: 10.17660/actahortic.2010.871.17
Keywords: Cucurbita moschata Duch., powdery mildew, CmSGT1, functional analysis, N. benthamiana
Citation: Guo W-L, Chen B-H, Guo Y-Y, Yang H-L, Mu J-Y, Wang Y-L, Li X-Z and Zhou J-G (2019) Improved Powdery Mildew Resistance of Transgenic Nicotiana benthamiana Overexpressing the Cucurbita moschata CmSGT1 Gene. Front. Plant Sci. 10:955. doi: 10.3389/fpls.2019.00955
Received: 24 April 2019; Accepted: 09 July 2019;
Published: 25 July 2019.
Edited by:
Valentina Fiorilli, University of Turin, ItalyReviewed by:
Yan Xu, Northwest A&F University, ChinaStefano Pavan, University of Bari Aldo Moro, Italy
Copyright © 2019 Guo, Chen, Guo, Yang, Mu, Wang, Li and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin-Zheng Li, bHh6MjI4M0AxMjYuY29t
 Wei-Li Guo
Wei-Li Guo Bi-Hua Chen1,2
Bi-Hua Chen1,2