- 1School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai, China
- 2State Key Laboratory of Vegetable Germplasm Innovation, Tianjin, China
The development of trichomes (spines) on cucumber fruits is an important agronomic trait. It has been reported that two MYB family members, CsMYB6 (Csa3G824850) and CsTRY (Csa5G139610) act as negative regulators of trichome or fruit spine initiation. To further study the functions of these two genes, we overexpressed them in tobacco, and found that the flowers and seed coats of transformants overexpressing CsTRY displayed an unexpected defect in pigmentation that was not observed in plants overexpressing CsMYB6. Moreover, the expression of key genes in the flavonoid synthesis pathway was repressed in CsTRY overexpressing plants, which resulted in the decrease of several important flavonoid secondary metabolites. In addition, CsTRY could interact with the AN1 homologous gene CsAN1 (Csa7G044190) in cucumber, which further confirmed that CsTRY not only regulates the development of fruit spines, but also functions in the synthesis of flavonoids, acting as the repressor of anthocyanin synthesis.
Introduction
Cucumber (Cucumis sativus L.) is a horticultural crop that is consumed worldwide (Huang et al., 2009; Yundaeng et al., 2015), and trichomes (spines) on the fruit are considered as an important commodity trait (Zhang et al., 2010; Yang et al., 2014; Li et al., 2015; Pan et al., 2015). The cucumber fruit, a pepo that develops from the ovary and receptacle, is covered with a thick cuticle, tubercules and trichomes (spines) (Roth, 1977; Wang et al., 2015). In the model plant Arabidopsis thaliana, trichome developments is initiated by a ternary complex (GL1-GL3/EGL1-TTG1) from epidermal cells, which leads to the expression of GL2 and TRY (Oppenheimer et al., 1991; Galway et al., 1994; Walker, 1999; Payne et al., 2000; Szymanski et al., 2000; Larkin et al., 2003; Zhang et al., 2003). The TRY protein moves into neighboring cells, where it competes with GL1 for binding to GL3/EGL3 and prevents differentiation of the cells into trichomes (Schellmann et al., 2002; Esch et al., 2003; Zhang et al., 2003). Further, the transcriptional complex can also regulate anthocyanin biosynthesis genes to mediate anthocyanin biosynthesis, including NADPH-dependent dihydroflavonol reductase (DFR), leucoanthocyanidin dioxygenase (LDOX), and UDP-Glc:flavonoid 3′- O-glucosyltransferase (UF3GT). In Artemisia annua, an MYB family member, AaMIXTA1, can promotes trichome development and regulates cuticle biosynthesis. These reports suggest that, the genes that regulate trichome developments usually function in secondary metabolite biosynthesis (Yan et al., 2018).
In cucumber, csgl1/mict/tbh mutants produced microtrichomes, which means the GL1 gene may be related to the development of trichomes. csgl3/tril mutants have a hairless phenotype, which means the GL3 gene is related to the initiation of trichomes (Chen et al., 2014; Li et al., 2015; Pan et al., 2015; Zhao et al., 2015; Wang et al., 2016), and CsTTG is involved in the formation of fruit warts (Chen et al., 2016). It has been reported that CsMYB6 and CsTRY act as negative regulators of trichome initiation, and they can reduce cucumber trichome density (Yang et al., 2018). However, the specific regulatory mechanisms are still unclear.
In this study, we overexpressed CsTRY and CsMYB6 in tobacco (Nicotiana tabacum L.) and found the flowers and seed coats of CsTRY overexpressing transformants displayed an unexpected defect in pigmentation that was not found in CsMYB6 overexpressing plants. Furthermore, the expression of key genes in the flavonoid synthesis pathway were repressed in CsTRY overexpressing plants. In addition, we determined the compound content in the anthocyanin synthesis pathway by LC-MS and found that the content of peonidin and several important flavonoid secondary metabolites was significantly decreased, which is consistent with the gene expression change. CsTRY could interact with the AN1 homologous gene in cucumber. These results suggested that CsTRY not only regulates the development of fruit spines, but also functions in synthesis of flavonoids, acting as the repressor of anthocyanin synthesis.
Materials and Methods
CsTRY and CsMYB6 Construct and Plant Transformation
The full-length CsTRY and CsMYB6 coding region was amplified and inserted in the SacI and PstI sites of the pCambia2300 vector, containing the CaMV 35S promoter. The CsMYB6 and CsTRY overexpression constructs were used for tobacco transformation (Horsch et al., 1985). The primes used are TRY-F(ATGGACAATCATCGT), TRY-R(TCATCCTCTTCTTCT), MYB6-F(ATGGGAAGGTCTCCT), MYB6-R(TCAGAATCTCAGGAA).
Preparation of Nucleic Acids and cDNA Synthesis
Genomic DNA was isolated from N. tabacum young leaf material using a DNeasy plant mini kit (QIAGEN). Total RNAs were extracted from N. tabacum petal material of the wild-type and transgenic plants, respectively, using an RNA extraction kit (TRIzol Reagent, Invitrogen). First-strand cDNA was synthesized from 2 μg total RNA in a 20 μl reaction mixture with 0.5 μg oligo(dT)15, 0.75 mM dNTPs, 10 mM dithiothreitol (DTT), and 100 U SuperScript II RNase H-reverse transcriptase (Invitrogen).
Semi-Quantitative RT-PCR and Real-Time RT-PCR Analysis
For expression analysis of different structural flavonoid genes, the petunia genes of interest, PhCHS, PhCHI, PhF3H, PhF3’H, PhDFR, and PhANS were blasted with N. tabacum EST database. SYBR® Premix Ex Taq from TaKaRa was used for qPCR with an Applied Biosystems 7500 real-time PCR system (Applied Biosystems). The tobacco gene EF1α was used as an internal control (Zhang et al., 2009) in all qPCR reactions. Three biological replicates were performed for each experiment.
Photometric Determination of Anthocyanins
Petals of mature flowers were harvested, ground in liquid nitrogen to produce a fine powder, and then immediately freeze-dried, and stored at −80°C until use. Anthocyanins were detected as previously described (Zhang et al., 2009). All samples were measured as triplicates in three independent biological replicates. Error bars represent +SE.
Yeast Two-Hybrid Screen
We cloned the cDNA sequences of CsTRY (full-length) and fused it into the pGADT7 vectors. The ORF of CsAN1, a homolog gene of the Arabidopsis PhANTHOCYANIN1 (AN1) (Spelt, 2000) was cloned and fused into the pGBKT7. All recombinant constructs were separately transformed into the yeast strain AH109. At least three independent experiments were performed, and the result of one representative experiment is shown.
Scanning Electron Microscopy
N. tabacum young leaf samples were fixed, washed, post fixed, dehydrated, coated (Chen et al., 2014), and observed using a Hitachi S-4700 scanning electron microscope with a 2-kV accelerating voltage.
Bimolecular Fluorescence Complementation (BiFC) Assay
To generate the BiFC constructs, the full-length cDNA sequences of CsTRY and CsAN1 were cloned and fused with the pXY104 and pXY106 vectors (Yu et al., 2008; Liu and Howell, 2010). Tobacco (N. tabacum) leaves were used for co-expression studies as previously described (Schütze et al., 2009). The fluorescence signal was detected 2 to 4 days after infiltration, using an Olympus BX 51 fluorescence microscope to acquire fluorescent images. YFP (yellow fluorescent protein) imaging was performed at an excitation wavelength of 488 nm. CFP served as the internal control in all BiFC analyses. At least three independent replicates were performed, and the result of one representative experiment is shown.
Metabolite Profiling
The petals of transgene plants were grounded into a fine powder. Each 20 mg of fine powder was used for metabolite extraction prior to UHPLC-Q-TOF-MS analysis. The metabolite extraction was analyzed as previously described (Hu et al., 2018). The metabolites were annotated by searching the Personal Compound Database and Library (PCD/PCDL) (Hu et al., 2015), and by comparing the MS and MS/MS of the compounds in the Metlin database (Want, 2005) and the Massbank database (Horai et al., 2010). Data acquisition, metabolite annotation and peak area extraction were performed with the Agilent softwares (Agilent Technologies Inc., Palo Alto, CA, USA), of MassHunter Acquisition 7.0, MassHunter Qualitative 7.0 and Mass Profinder 8.0, respectively. All measurements were performed in three replicates per genotype.
Results
1. Flower Pigmentation and Trichome Distribution Were Affected in Transgenic Tobacco Plants Overexpressing CsTRY, but Not in Plants Overexpressing CsMYB6
To explore the function of CsTRY and CsMYB6, we used 35S promoter to regulate these two genes and overexpress them in tobacco by genetic transformation (Figure 1A). In transgenic 35S:CsTRY tobacco, there were clear phenotypic changes in petal pigmentation, which resulted in a complete loss of pigmentation and pure white petals. Moreover, there was a decrease in seed pigmentation, which was lighter than the wild type (Figure 1B).
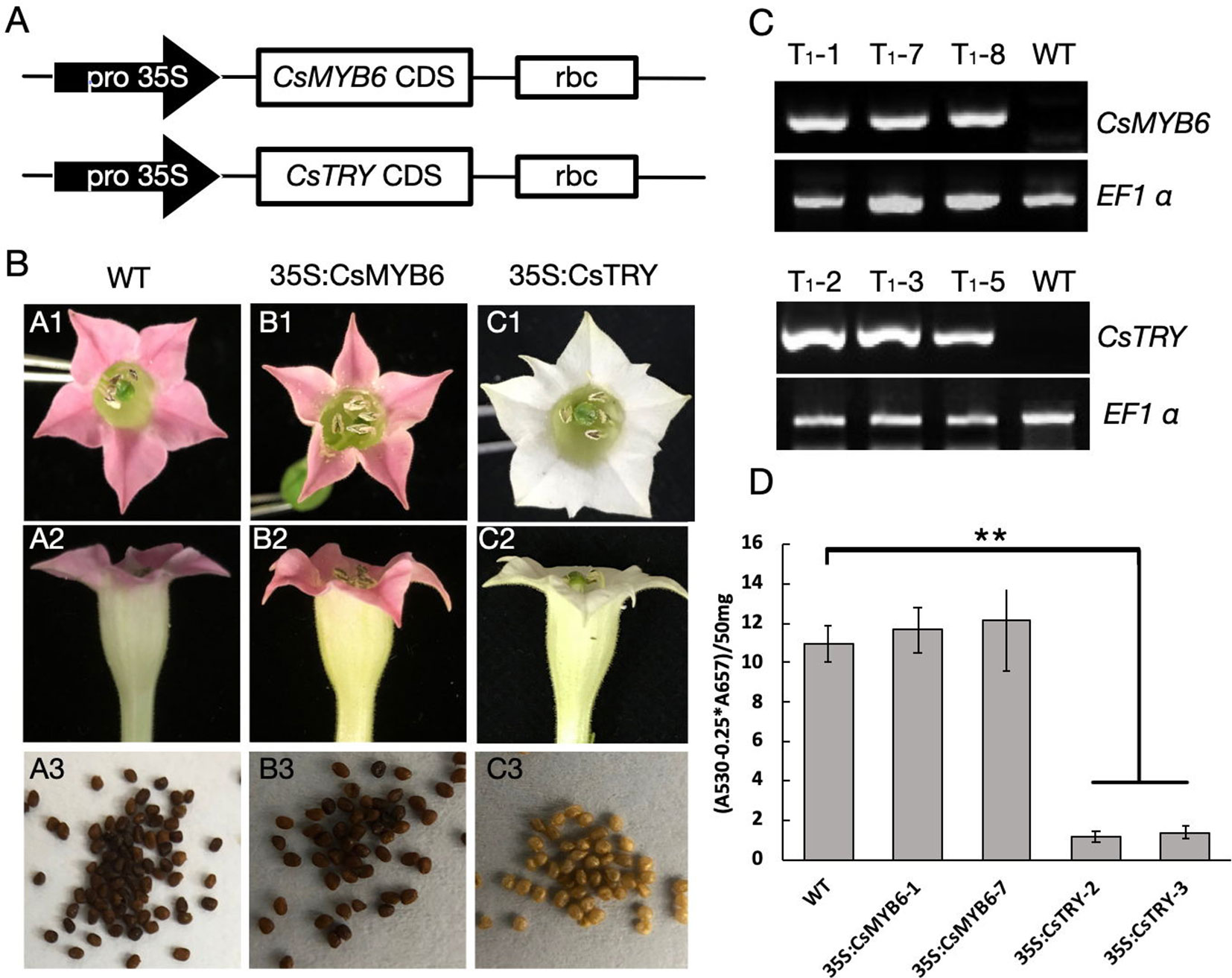
Figure 1 The gene construct for ectopic expression of CsTRY and CsMYB6 and changes in flower and seed pigmentation phenotype of transgenic tobacco. (A) Structure of the gene construct used to express. (B)CsTRY and CsMYB6 expressing lines exhibiting different phenotype characteristics. A1–A3 is the petal and seeds of WT; B1–B3 is the petal and seeds of 35S:CsMYB6 transgenic tobacco plants; and C1–C3 is of 35S:CsTRY transgenic plants (C) Semi-quantitative RT-PCR analysis of CsTRY and CsMYB6 expression levels in mature leaves of T1 generation plants. EF1α transcript abundance was used as a control. (D) Photometric determination of anthocyanin content in methanolic extracts of petals in tobacco lines 35S:CsTRY (35S:CsTRY-2, 35S:CsTRY-3), 35S:CsMYB6 (35S:CsMYB6-1, 35S:CsMYB6-7) and the wild-type. A530, absorption at 530 nm; A657, absorption at 657 nm. Error bars represent +SE. Significant differences were determined according to Duncan’s multiple range test (P < 0.05) or Student’s t-test (**P < 0.01).
Anthocyanin quantification results measured by spectrophotometer revealed that anthocyanin accumulation in the petals of 35S:CsTRY transgenic tobacco plants was clearly reduced, indicating that CsTRY may be negatively regulating the synthesis of tobacco anthocyanin (Figure 1D).
In addition, the morphology and quantity of glandular hairs of transgenic lines were observed by scanning election microscopy (Figure 2). It was found that the number of long stalked glandular hairs and the density of glandular hairs decreased, which is consistent with the known negative regulation of the epidermis. However, overexpression of CsMYB6 in tobacco showed no difference in phenotypes, such as flower pigment and glandular density.
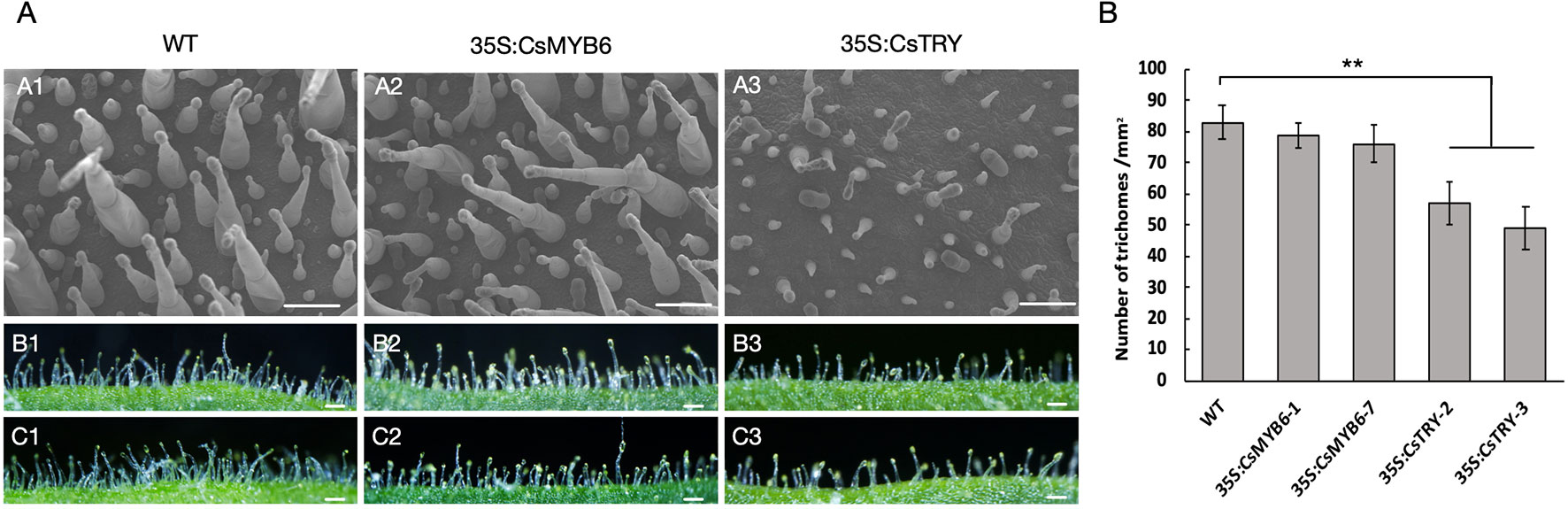
Figure 2 The morphology and quantity of trichome in wild type and 35S:CsTRY, 35S:CsMYB6 tobacco lines (A) Patterns of trichome distribution on the leave surface of T1 generation plants of CsTRY and CsMYB6 overexpression transgenic tobacco. Bars = 100 µm. (B) Number of trichomes on 1 mm−2 adaxial surface of the leave of T1 generation plants of CsTRY and CsMYB6 overexpression transformed lines and the wild-type. Error bars represent +SE. Significant differences were determined according to Duncan’s multiple range test (P < 0.05) or Student’s t-test (**P < 0.01).
2. CsTRY Negatively Regulates the Synthesis of Anthocyanins by Suppressing the Expression of Genes in the Flavonoid Metabolic Pathway
To elucidate the molecular mechanisms involved in the marked decrease of anthocyanins in 35S:CsTRY transformants, the transcript levels of seven key genes encoding the enzymes of the flavonoid pathway from the first stage to the third were measured in flowers by real-time RT-PCR analyses (Figure 3).
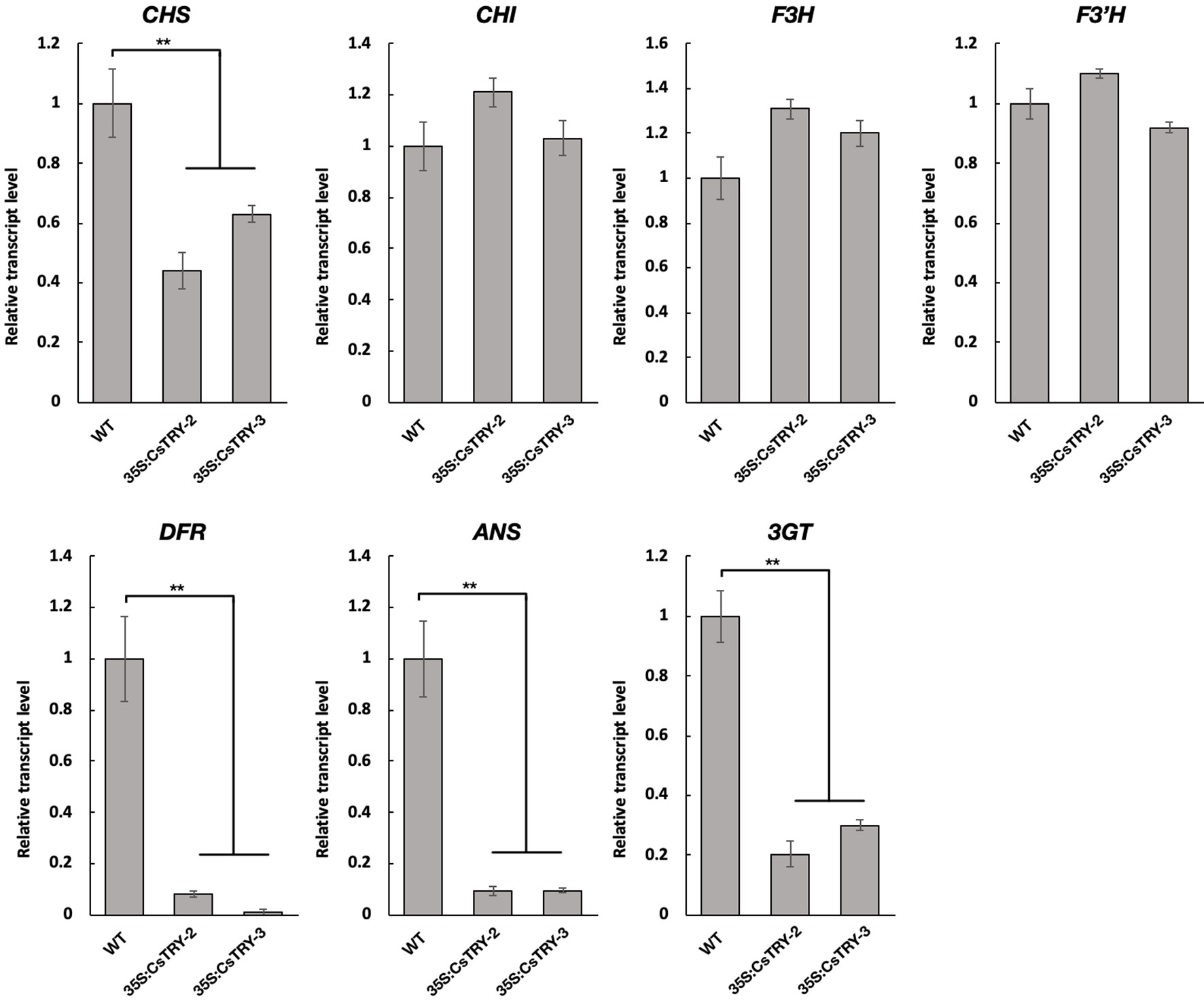
Figure 3 Relative transcript levels of flavonoid structural genes in the petals of tobacco lines 35S:CsTRY (35S:CsTRY-2, 35S:CsTRY-3) and wild type, with the EF1α gene as an internal control. Error bars represent +SE. Significant differences were determined according to Duncan’s multiple range test (P < 0.05) or Student’s t-test (**P < 0.01).
Based on the different responses of the seven genes to CsTRY, we divided them into three categories. The first type is Chalcone synthase (CHS) located upstream of the second stage of the flavonoid metabolic pathway, and its expression level is significantly inhibited; the second type contains Chalcone isomerase (CHI), flavanone 3-hydroxylas (F3H), and flavonoid 3′-hydroxylase (F3′H) located downstream of the second stage of the flavonoid metabolic pathway, and its expression level is upregulated to varying degrees. The third category is dihydroflavonol4-reductas (DFR), Anthocyanidin Synthase (ANS) and Anthocyanin 3-0-g1ucosyltransferase (3GT), which is directly related to the synthesis of anthocyanins and is clearly strongly inhibited.
Furthermore, we detected a variety of flavonoids in transgenic tobacco petals by LC-MS (Figure 4). We found that a number of secondary metabolites related to anthocyanins, such as (kaempferol-3-O-rhamnoside-7-O-rhamnoside and kaempferol-3-O-rutinoside-7-O-rhamnoside), naringenin hexoside, chalcone 2″-O-glucoside, anthocyanin (peonidin di-hexoside I, II, and III), were significantly reduced in transgenic plants.
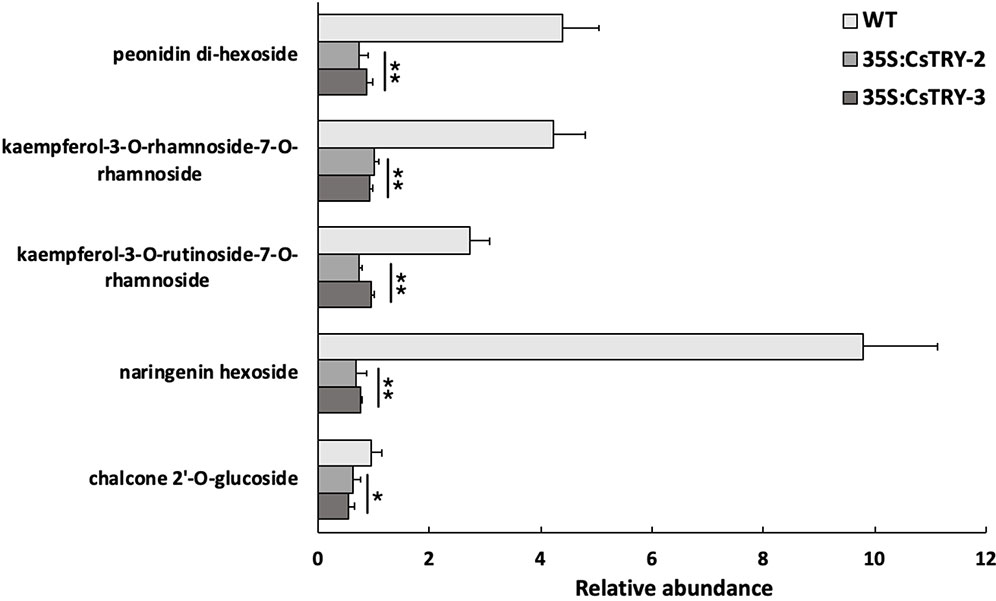
Figure 4 Determination of the relative abundance of flavonoids in flowers of CsTRY overexpressing transgenic tobacco lines (35S:CsTRY-2, 35S:CsTRY-3). Error bars represent +SE. Significant differences were determined according to Duncan’s multiple range test (P < 0.05) or Student’s t-test (*P < 0.05, **P < 0.01).
This is consistent with the expression of genes. In the secondary stage of Anthocyanin synthesis, CHS catalyzes the stepwise condensation of three acetate units from malonyl-COA with p-coumaroyl-COA to yield tetrahydroxychalcone. CHI then catalyzes the stereospecific isornerization of the yellow-colored tetrahydroxychalcone to the colorless naringenin. Naringenin is converted to dihydrokaempferol by F3H. Thus, the decrease in the expression of CHS and the increase in the expression of CHI, F3H reduced the content of chalcone and trihydroxyflavanone. In the last stage of Anthocyanin synthesis, the dihydroflavonol is reduced to the flavan 9,4 cis-diol (leucoanthocyanidins) by dihydroflavone-4-reductase (DFR). Then Anthocyanin formed by catalysis of ANS and 3GT. Owing to the inhibited of the expression three genes, DFR, ANS, and 3GT, the decrease of Anthocyanin is reasonable.
3. The CsTRY Protein Interacts With the Known Anthocyanin Synthesis Regulator CsAN1
The PhAN1 protein has previously been shown to be essential for anthocyanin synthesis in petunia. To further verify whether CsTRY can interact with known modulators of anthocyanin metabolism, we cloned the homologous gene of PhAN1 in cucumber, CsAN1, through yeast two-hybrid and BiFC and found that CsTRY can interact with CsAN1 (Figure 5).
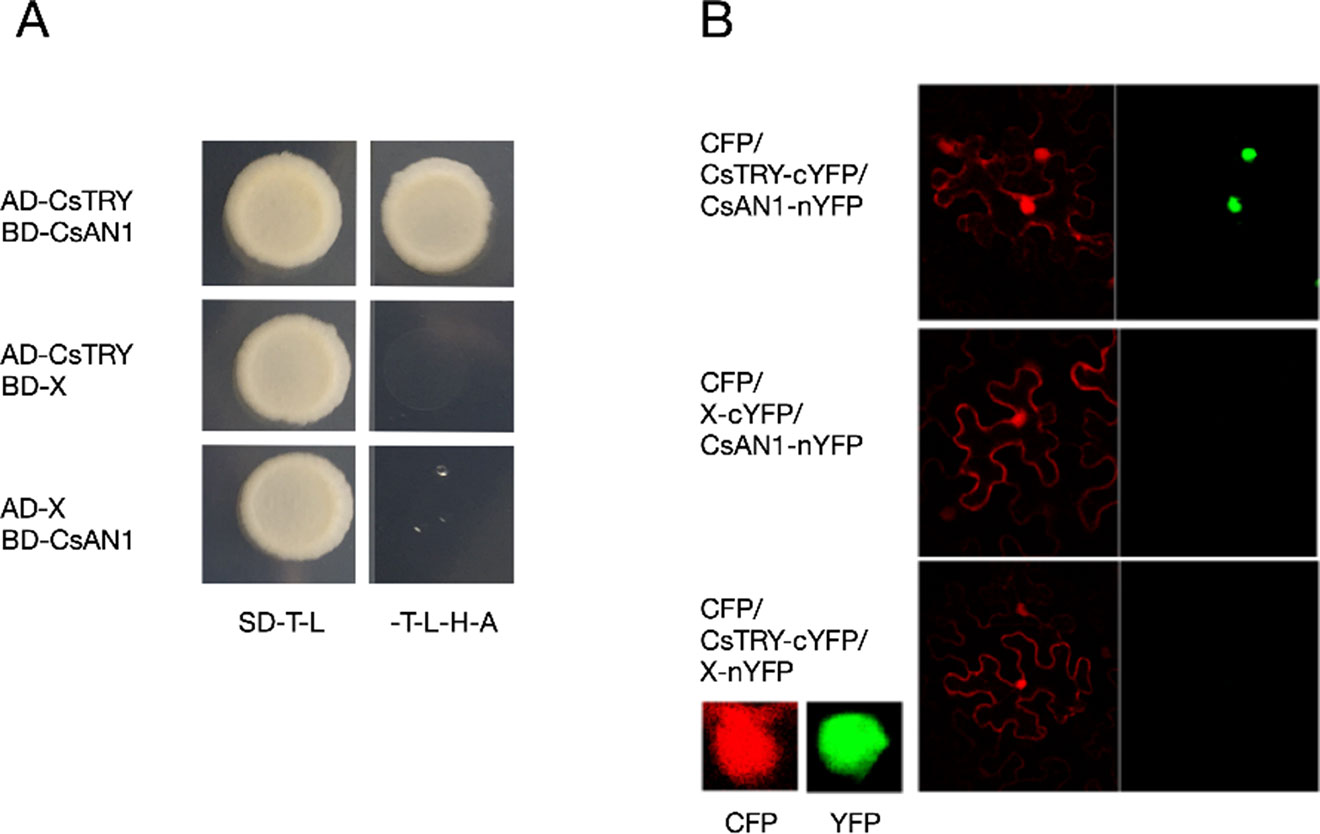
Figure 5 Yeast two-hybrid analysis and BiFC (A) Activation activity of CsTRY to CsAN1 in yeast. (B) BiFC analysis of the physical interaction between CsAN1 (fused with the N-terminal fragment of YFP) and CsTRY (fused with the C-terminal fragment of YFP).
Discussion
Trichomes are generally considered biofactories that produce secondary metabolites. The genes regulating unicellular trichome developments are usually related to anthocyanin synthesis (Jin and Martin, 1999). In A. annua, which has multicellular trichomes, genes involved in the development of trichomes also regulate the synthesis and transportation of secondary metabolites, such as artemisinin (Yan et al., 2018). In the cucumber trichome development mutants csgl3/tril and csgl1/mict/tbh, the genes that regulate anthocyanin synthesis are also differentially expressed, suggesting that although trichomes morphology and related genes differ between multicellular and unicellular trichomes, the mechanism of secondary metabolites coupled with epidermal hair development is conserved. Overexpression of CsTRY in tobacco can affect the flowering and seed coat color, but overexpression of CsMYB6 does not. This result suggests that although both genes can regulate the density of trichomes in cucumber, the specific mechanism and range of the regulation may not be the same. In addition, CsMYB6 was significantly downregulated in the cucumber hairless mutants tril and mict, but the expression of CsTRY did not change, which also indicated that the regulation patterns of CsTRY and CsMYB6 may be different (Chen et al., 2014; Li et al., 2015; Zhao et al., 2015).
In previous study, overexpression CsTRY or CsMYB6 in cucumber can decrease the density of fruit trichome and CsTRY is directly regulated by CsMYB6. However, overexpression of CsMYB6 inhibited rather than promoted, the expression of CsTRY, which means the relationship between CsTRY and CsMYB6 is not simple (Yang et al., 2018). In this study, overexpression of CsTRY in tobacco can affect the number of trichome, but overexpression of CsMYB6 does not. It can be inferred that CsTRY, a R3 MYB transcription factor in the relative downstream, was more directly related to glandular trichomes and metabolites, while CsMYB6, a R2R3 MYB transcription factor in the relative upstream, might be affected by other proteins in tobacco. Interestingly, overexpression CsTRY or CsMYB6 in cucumber can decrease the density of fruit trichome rather than other organs (Yang et al., 2018), which means the regulation mechanism of fruit trichome is different from that of other organs. SlMIXTA-like, a R2R2MYB transcription factor of tomato, regulates trichome formation on fruit surface, which also indicates that the formation of fruit trichome may be different from that of other organs (Ying et al., 2019). Therefore, overexpress CsMYB6 in tobacco did not change the phenotype, suggesting that CsMYB6 only plays a role in the regulation of cucumber fruit trichome, not other organs.
At present, there are three main ways to regulate anthocyanin synthesis: MYB-bHLH protein binary complex, MYB-WD40 protein binary complex, which is independent of bHLH transcription factor, and MYB-bHLH-WD40 protein ternary complex. The anthocyanin pathway in most plants is activated by MYB-bHLH-WD40 ternary complex (Broun, 2005; Antoine et al., 2010). In this study, we found that CsTRY can interact with the bHLH protein CsAN1. AN1 is an important regulator involved in anthocyanin synthesis in Petunia hybrida. It is a homology of the structural gene DFR and can directly regulate the expression of DFR (Spelt, 2000). Without the research of WD40 protein, we speculate that CsTRY may function by forming a ternary complex of MYB-bHLH-WD40 protein or a binary complex of MYB-bHLH protein in tobacco. Moreover, CsTRY can interact with CsAN1, a homolog of the Arabidopsis AN1 gene, indicating that it also acts as a negative regulator of anthocyanin synthesis in cucumber and has regulatory mechanisms similar to Arabidopsis.
In this study, the overexpression of CsTRY in tobacco greatly affected the synthesis of anthocyanin, indicating that CsTRY may have conserved function in cucumber. A number of transcription factors (e.g.,CsGl1/Mict/tbh, CsGl3/Tril, Tu and Ts) play key roles in cucumber trichome (spine) differentiation and development. However, the functions of their homologues in Arabidopsis are irrelevant to trichome development (Yang et al., 2014; Zhao et al., 2015; Guo et al., 2018). Moreover, CsTRY can complement the Arabidopsis try mutant, whereas CsMYB6 cannot complement the gl1 mutant. These results suggest that the regulation network of multicellular-trichome development may be partially consistent with unicellular-trichome development, but key genes, such as CsGl3/Tril and CsGl1/Mict/tbh, may have independent evolutionary pathways in the two different trichome types.
Data Availability Statement
All datasets generated for this study are included in the manuscript/supplementary files.
Author Contributions
HH conceptualized the research. JuP designed experiments. JiP participated in writing, editing, and revising the manuscript. RC conceptualized the research, designed and performed experiments. GW performed experiments and prepared figures. LZ performed experiments, analyzed the data, and wrote the manuscript. HD prepared the figures.
Funding
This work was supported by the National Key R&D Program of China (Grant No. 2018YFD0100701), National Natural Science Foundation of China (31471156), Shanghai Agriculture Applied Technology Development Program (Grant No.G2015060402).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the reviewers for critically reading the manuscript. We also thank Hanfan Wen and Yue Chen for technical assistance and data analysis of the experiments.
References
Antoine, B., Heim, M. A., Bertrand, D., Michel, C., Bernd, W., LoC, L. (2010). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39 (3), 366–380. doi: 10.1111/j.1365-313X.2004.02138.x
Broun, P. (2005). Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr. Opin. Plant Biol. 8 (3), 272–279. doi: 10.1016/j.pbi.2005.03.006
Chen, C., Liu, M., Jiang, L., Liu, X., Zhao, J., Yan, S., et al. (2014). Transcriptome profiling reveals roles of meristem regulators and polarity genes during fruit trichome development in cucumber (Cucumis sativus L.). J. Exp. Bot. 65 (17), 4943–4958. doi: 10.1093/jxb/eru258
Chen, C., Yin, S., Liu, X., Liu, B., Yang, S., Xue, S., et al. (2016). The WD-repeat protein CsTTG1 regulates fruit wart formation through interaction with the homeodomain-leucine zipper I protein Mict. Plant Physiol. 171 (2), 1156–1168. doi: 10.1104/pp.16.00112
Esch, J. J., Chen, M., Sanders, M., Hillestad, M., Ndkium, S., Idelkope, B., et al. (2003). A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development 130, 5885–5894. doi: 10.1242/dev.00812
Galway, M. E., Masucci, J. D., Lloyd, A. M., Walbot, V., Davis, R. W., Schiefelbein, J. W. (1994). The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166 (2), 0–754. doi: 10.1006/dbio.1994.1352
Guo, C., Yang, X., Wang, Y., Nie, J., Yang, Y., Sun, J., et al. (2018). Identification and mapping of ts (tender spines), a gene involved in soft spine development in Cucumis sativus. Theor. Appl. Genet. 131 (1), 1–12. doi: 10.1007/s00122-017-2954-9
Horai, H., Arita, M., Kanaya, S., Nihei, Y., Ikeda, T., Suwa, K., et al. (2010). Massbank: a public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 45 (7), 703–714. doi: 10.1002/jms.1777
Horsch, R., Hoffmann, J., Eichholtz, D., Rogers, S., Fraley, R. (1985). A simple and general method for transferring genes into plants. Science 227 (4691), 1229–1231. doi: 10.1126/science.227.4691.1229
Hu, C., Tohge, T., Chan, S. A., Song, Y., Rao, J., Cui, B., et al. (2015). Identification of conserved and diverse metabolic shifts during rice grain development. Sci. Rep. 6, 20942. doi: 10.1038/srep20942
Hu, C., Zhao, H., Wang, W., Xu, M., Shi, J., Nie, X., et al. (2018). Identification of conserved and diverse metabolic shift of the stylar, intermediate and peduncular segments of cucumber fruit during development. Int. J. Mol. Sci. 19 (1). doi: 10.3390/ijms19010135
Huang, S., Li, R., Zhang, Z., Li, L., Gu, X., Fan, W., et al. (2009). The genome of the cucumber, Cucumis sativus L. Nat. Genet. 41 (12), 1275–1281. doi: 10.1038/ng.475
Jin, H., Martin, C. (1999). Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 41 (5), 577–585. doi: 10.1023/A:1006319732410
Larkin, J. C., Brown, M. L., Schiefelbein, J. (2003). How do cells know what they want to be when they grow up? Lessons from epidermal patterning in Arabidopsis. Annu. Rev. Plant Biol. 54, 403–430. doi: 10.1146/annurev.arplant.54.031902.134823
Li, Q., Cao, C., Zhang, C., Zheng, S., Wang, Z., Wang, L., et al. (2015). The identification of Cucumis sativus Glabrous 1 (CsGL1) required for the formation of trichomes uncovers a novel function for the homeodomain-leucine zipper I gene. J. Exp. Bot. 66 (9), 2515–2526. doi: 10.1093/jxb/erv046
Liu, J. X., Howell, S. H. (2010). bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22 (3), 782–796. doi: 10.1105/tpc.109.072173
Oppenheimer, D. G., Herman, P. L., Sivakumaran, S., Esch, J., Marks, M.D. (1991). A MYB gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67, 483–493. doi: 10.1016/0092-8674(91)90523-2
Pan, Y., Bo, K., Cheng, Z., Weng, Y. (2015). The loss-of-function GLABROUS 3 mutation in cucumber is due to LTR-retrotransposon insertion in a class IV HD-ZIP transcription factor gene CsGL3 that is epistatic over CsGL1. BMC Plant Biol. 15, 302. doi: 10.1186/s12870-015-0693-0
Payne, C. T., Zhang, F., Lloyd, A. M. (2000). Gl3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156 (3), 1349–1362. doi: 10.1016/S1474-6670(17)69558-2
Roth, I. (1977). Fruits of Cucurbitaceae. Encyclopedia Plant Anat. 10, 471–477. doi: 10.1016/S1474-6670(17)69558-2
Schellmann, S., Schnittger, A., Kirik, V., Wada, T., Okada, K., Beermann, A., et al. (2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO (Eur. Mol. Biol. Organ.) J. 21 (19), 5036–5046. doi: 10.1093/emboj/cdf524
Schütze, K., Harter, K., Chaban, C. (2009). Bimolecular fluorescence complementation (BiFC) to study protein–protein interactions in living plant cells. Methods Mol. Biol. 479, 189–202. doi: 10.1007/978-1-59745-289-2_12
Spelt, C. (2000). Anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural Anthocyanin genes. Plant Cell 12 (9), 1619–1632. doi: 10.1105/tpc.12.9.1619
Szymanski, D. B., Lloyd, A. M., Marks, M. D. (2000). Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis. Trends Plant Sci. 5 (5), 214–219. doi: 10.1016/S1360-1385(00)01597-1
Walker, A. R. (1999). The transparent testa glabra1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell Online 11 (7), 1337–1350. doi: 10.1105/tpc.11.7.1337
Wang, W., Zhang, Y., Xu, C., Ren, J., Liu, X., Black, K., et al. (2015). Cucumber ECERIFERUM1 (CsCER1), which influences the cuticle properties and drought tolerance of cucumber, plays a key role in VLC alkanes biosynthesis. Plant Mol. Biol. 87 (3), 219–233. doi: 10.1007/s11103-014-0271-0
Wang, Y. L., Nie, J. T., Chen, H. M., Guo, C. L., Pan, J., He, H. L., et al. (2016). Identification and mapping of Tril, a homeodomain-leucine zipper gene involved in multicellular trichome initiation in Cucumis sativus. Theor. Appl. Genet. 129 (2), 305–316. doi: 10.1007/s00122-015-2628-4
Want, E. J. (2005). Metlin: a metabolite mass spectral database. Ther. Drug Monit. 27 (6), 747. doi: 10.1097/01.ftd.0000179845.53213.39
Yan, T., Li, L., Xie, L., Chen, M., Shen, Q., Pan, Q., et al. (2018). A novel HD-ZIP IV/MIXTA complex promotes glandular trichome initiation and cuticle development in Artemisia annua. New Phytol. 218 (2), 567–578. doi: 10.1111/nph.15005
Yang, S., Cai, Y., Liu, X., Dong, M., Zhang, Y., Chen, S., et al. (2018). A CsMYB6-CsTRY module regulates fruit trichome initiation in cucumber. J. Exp. Bot. 69 (8), 1887–1902. doi: 10.1093/jxb/ery047
Yang, X., Zhang, W., He, H., Nie, J., Bie, B., Zhao, J., et al. (2014). Tuberculate fruit gene Tu encodes a C2 H2 zinc finger protein that is required for the warty fruit phenotype in cucumber (Cucumis sativus L.). Plant J. 78 (6), 1034–1046. doi: 10.1111/tpj.12531
Ying, S. Y., Su, M., Wu, Y., Zhou, L., et al. (2019). Trichome regulator SlMIXTA-like directly manipulates primary metabolism in tomato fruit. Plant Biotechnol. J., 1–10. doi: 10.1111/pbi.13202
Yu, X., Li, L., Li, L., Guo, M., Chory, J., Yin, Y. (2008). Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 105 (21), 7618–7623. doi: 10.1073/pnas.0802254105
Yundaeng, C., Somta, P., Tangphatsornruang, S., Chankaew, S., Srinives, P. (2015). A single base substitution in BADH/AMADH is responsible for fragrance in cucumber (Cucumis sativus L.), and development of SNAP markers for the fragrance. Theor. Appl. Genet. 128 (9), 1881–1892. doi: 10.1007/s00122-015-2554-5
Zhang, F., Gonzalez, A., Zhao, M., Payne, C. T., Lloyd, A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130 (20), 4859–4869. doi: 10.1242/dev.00681
Zhang, W., He, H., Guan, Y., Du, H., Yuan, L., Li, Z., et al. (2010). Identification and mapping of molecular markers linked to the tuberculate fruit gene in the cucumber (Cucumis sativus L.). Theor. Appl. Genet. 120 (3), 645–654. doi: 10.1007/s00122-009-1182-3
Zhang, W., Ning, G., Lv, H., Liao, L., Bao, M. (2009). Single MYB-type transcription factor AtCAPRICE: a new efficient tool to engineer the production of anthocyanin in tobacco. Biochem. Biophys. Res. Commun. 388 (4), 742–747. doi: 10.1016/j.bbrc.2009.08.092
Keywords: cucumber, trichome, anthocyanin, CsTRY, CsMYB6, tobacco
Citation: Zhang L, Pan J, Wang G, Du H, He H, Pan J and Cai R (2019) Cucumber CsTRY Negatively Regulates Anthocyanin Biosynthesis and Trichome Formation When Expressed in Tobacco. Front. Plant Sci. 10:1232. doi: 10.3389/fpls.2019.01232
Received: 14 June 2019; Accepted: 05 September 2019;
Published: 08 October 2019.
Edited by:
Feishi Luan, Northeast Agricultural University, ChinaReviewed by:
Xuehao Chen, Yangzhou University, ChinaYuhong Li, Northwest A&F University, China
Huazhong Ren, China Agricultural University (CAU), China
Copyright © 2019 Zhang, Pan, Wang, Du, He, Pan and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junsong Pan, anNwYW43MUBzanR1LmVkdS5jbg==; Run Cai, Y2FpcnVuQHNqdHUuZWR1LmNu
†These authors have contributed equally to this work
 Leyu Zhang
Leyu Zhang Jian Pan
Jian Pan Gang Wang1
Gang Wang1 Junsong Pan
Junsong Pan