- 1State Key Laboratory of Crop Stress Biology for Arid Areas, College of Agronomy, Northwest A&F University, Yangling, China
- 2Shaanxi Province Tobacco Company of China National Tobacco Company, Xi’an, China
Cuticular wax accumulation and composition affects drought resistance in plants. Brachypodium distachyon plants subjected to water deficit and polyethylene glycol treatments resulted in a significant increase in total wax load, in which very-long-chain (VLC) alkanes were more sensitive to these treatments than other wax compounds, implying that VLC alkanes biosynthesis plays a more important role in drought resistance in B. distachyon. ECERIFERUM1 (CER1) has been reported to encode a core enzyme involved in VLC alkanes biosynthesis in Arabidopsis (Arabidopsis thaliana), but few corresponding genes are investigated in B. distachyon. Here, we identified eight CER1 homologous genes in B. distachyon, namely BdCER1-1 to BdCER1-8, and then analyzed their sequences feature, expression patterns, stress induction, and biochemical activities. These genes had similar protein structure to other reported CER1 and CER1-like genes, but displayed closer phylogenetic relationship to the rice OsGL1 genes. They were further found to exhibit various tissue expression patterns after being induced by abiotic stresses. Among them, BdCER1-8 gene showed extremely high expression in leaves. Heterologous introduction of BdCER1-8 into the Arabidopsis cer1 mutant rescued VLC alkanes biosynthesis. These results indicate that BdCER1 genes are likely to be involved in VLC alkanes biosynthesis of B. distachyon. Taken together, BdCER1-8 seems to play an explicit and predominant role in VLC alkanes biosynthesis in leaf. Our work provides important clues for further characterizing function of CER1 homologous genes in B. distachyon and also an option to improve drought resistance of cereal crops.
Introduction
The tissue surfaces of higher plants are covered by cuticle consisting of cutin and cuticular wax (Kerstiens, 2006; Pollard et al., 2008; Li and Beisson, 2009). As the major structural constituent in the cuticle, cutin is covered or embedded by cuticular wax (Pollard et al., 2008; Li and Beisson, 2009). Cuticular wax is a complex mixture of the very-long-chain fatty acids (VLCFAs, ≥C20) and their derivatives, such as primary alcohols, aldehydes, alkanes, alkenes, branched alkanes, secondary alcohols, unsaturated fatty alcohols, ketones, wax esters, and cyclic compounds (Samuels et al., 2008; Lee and Suh, 2013). In addition to its primary function in restraining uncontrolled non-stomatal water loss in plants, the cuticle also plays roles in helping plants against excess UV radiation, insects, and bacterial and fungal pathogens (Eigenbrode and Espelie, 1995; Solovchenko and Merzlyak, 2003; Shepherd and Griffiths, 2006). Surprisingly, an unexpected function of wax was found to participate in proper pollen–stigma interaction (Preuss et al., 1993).
Cuticular wax biosynthesis occurs in epidermal cells (Kunst and Samuels, 2009). First, the de novo synthesis of C16 or C18 fatty acid is processed in the plastid of epidermal cells, which are catalyzed into C16 or C18 fatty acyl-CoAs by the fatty acid synthase complex. Then these products are elongated to VLCFAs mainly with 26 to 32 carbons by fatty acid elongase complex in the endoplasmic reticulum (Samuels et al., 2008; Lee and Suh, 2013; Kunst and Samuels, 2009). After elongation, VLCFAs are converted into primary alcohols and wax esters by the alcohol-forming pathway or alkanes and their derivatives by the alkane-forming pathway, respectively (Samuels et al., 2008; Kunst and Samuels, 2009). Among these wax components, VLC alkanes are predominant in Arabidopsis, accounting for 50–80% of the total wax contents (Bourdenx et al., 2011). They are increased by 93% under water deficit, suggesting that VLC alkanes biosynthesis plays an important role in drought resistance (Kosma et al., 2009).
Several genes involved in VLC alkanes biosynthesis have been identified through the phenotypic characterizations of the Arabidopsis cer1 and cer3 mutants. Compared with the wild-type plants, the cer1 mutant displays glossy green stems, significantly decreased contents of alkanes and their derivatives, and slightly increased contents of aldehydes, indicating that CER1 encodes an aldehyde decarboxylase responsible for the conversion of aldehydes to alkanes (Aarts et al., 1995; Bourdenx et al., 2011). In the cer3 mutant, the stems also exhibit glossy green, but the contents of alkanes, secondary alcohols, ketones, and aldehydes go decreased dramatically, suggesting that CER3 encodes an enzyme critical for production of aldehydes using VLC acyl-CoAs as substrates (Chen et al., 2003; Kurata et al., 2003; Rowland et al., 2007). Further, Bernard et al. (2012) used yeast as a heterologous expression system to build a new pathway of VLC alkanes biosynthesis: CER1 physically interacts with CER3 and CYTB5s to convert VLC acyl-CoAs to VLC alkanes, during which CER1/CER3 complex is the core component of this process, while CYTB5s function to improve CER1 efficiency as CER1-specific cofactors (Bernard et al., 2012). As another member of the CER1 family in Arabidopsis, CER1-like1 also plays a vital role in VLC alkanes biosynthesis by using the same mechanism as that of CER1, but the CER1-like1/CER3 complex prefers to produce shorter chain alkanes than the CER1/CER3 complex, suggesting that alkane-forming complexes have distinct chain-length specificities and are coexisted in plants (Pascal et al., 2019).
The biological functions of CER1 and CER3 have been well characterized in Arabidopsis. The CER1 gene was found to be mainly expressed in the epidermis of aerial organs and could also be induced by abiotic stresses (Bourdenx et al., 2011). T-DNA insertional mutants of CER1 are devoid of VLC alkanes and their derivatives, whereas the contents of odd carbon-numbered alkanes and iso-branched alkanes are significantly increased in the CER1-overexpressing plants (Bourdenx et al., 2011). The cer1 mutant exhibits male sterility under low humidity, whereas CER1-overexpressing plants show reduced cuticle permeability, slow response to soil water deficit, and sensitive response to bacterial and fungal pathogens (Bourdenx et al., 2011). On the other hand, CER3 transcript is detected in silique, leaf, stem, and flower, but is absent in root. Loss-of-function mutation in CER3 alters cuticle membrane and wax synthesis. Moreover, the cer3 mutant shows post-genital organ fusions, decreased fertility under low humidity, and reduced stomatal index (Chen et al., 2003; Rowland et al., 2007). A recent study shows that mutation of CER1 or CER3 can also reduce the amounts of branched alkanes in leaf and flower, implying that they may partially be involved in branched chain wax component synthesis as well (Busta and Jetter, 2017).
Several orthologs of CER1 and CER3 genes have also been identified in other plants. As a homologous gene of CER1, BnCER1 transcript is most severely suppressed among wax biosynthetic genes in a glossy mutant (BnaA.GL) of Brassica napus, suggesting that BnCER1 may be a candidate gene involved in alkane-forming pathway (Pu et al., 2013). CsCER1 and CsWAX2 in Cucumber (Cucumis sativus) are respective homologs of CER1 and CER3, both of which are involved in VLC alkanes biosynthesis and affect pollen fertility (Wang et al., 2015a, Wang et al., 2015b). The maize (Zea mays) CER3 homologous gene GLOSSY1 (GL1) is involved in cuticular wax accumulation in seedling leaves (Sturaro et al., 2005). A BLAST search in rice database using GL1 protein further retrieved 11 putative GL1 paralogous genes, named OsGL1-1 to OsGL1-11 (Islam et al., 2009). Among them, OsGL1-1, OsGL1-2, and OsGL1-3 are more closely related to CER3 and GL1, while OsGL1-4, OsGL1-5, OsGL1-6, and OsGL1-7 are more closely related to CER1 (Islam et al., 2009). Despite of their large family members, these CER1 and CER3 homologs in rice exhibit partially divergent functions during plant growth, pollen development, and cuticular wax biosynthesis. OsGL1-1 was found to be expressed ubiquitously, involved in leaf cuticular wax and cuticle membrane formations in rice (Qin et al., 2011). OsGL1-2, OsGL1-3, and OsGL1-6 show varying expression levels in various organs and the disruption of their functions result in different wax productions and drought resistance levels (Islam et al., 2009; Qin et al., 2011; Zhou et al., 2013; Zhou et al., 2015). As homologs of CER1, Wax-deficient Anther 1 (WDA1) and OsCER1 affect wax production and pollen development in rice (Jung et al., 2006; Ni et al., 2018). However, as an outstanding referencing model plant for studying functional genomics of grasses and cereals such as wheat (Triticum aestivum L.) and barley (Hordeum vulgare L.) (Draper et al., 2001; Vogel et al., 2010), Brachypodium distachyon is still a bit far from being well investigated on VLC alkanes biosynthesis. Only few studies have been carried out including one recent report showing that BdWAX2 is required for cuticular wax biosynthesis (Hsia et al., 2017). But the alkane-forming pathway in B. distachyon remains unknown. To address this concern, a full picture of the CER1 homologs involved in VLC alkanes biosynthesis in B. distachyon need to be investigated.
In this article, we systematically examined the influence of drought stress on leaf cuticular wax of B. distachyon, and found that VLC alkanes may play a critical role against drought stress. Then, sequence features and expression profiles of CER1 homologous genes that are possibly involved in VLC alkanes biosynthesis in B. distachyon were analyzed in details. Further, we generated BdCER1-8 heterologous expression in the Arabidopsis cer1 mutant to unveil its role in wax biosynthesis. Our results show that VLC alkanes are more sensitive to drought stress than other wax compounds. It was found that there were eight CER1 homologous genes identified in B. distachyon that were likely involved in VLC alkanes biosynthesis. Among them, BdCER1-8 rescued wax production defect in the cer1 mutant by participating in the VLC alkanes biosynthesis.
Materials and Methods
Plant Growth and Treatments
B. distachyon ‘Bd21’ seeds were germinated and grown in the greenhouse with a 14-h light/10-h dark cycle. Two-month-old seedlings were used for all stress treatments. For water deficit treatment, plants were stopped watering until lower leaves were wilted, which needed 6 days. For polyethylene glycol (PEG) treatment, plants were watered with 20% (w/v) PEG6000 three times over a 6-day period. Plants watered as needed with tap water were used as control plants. All leaves were harvested at seventh day after treatments for extracting cuticular wax. To investigate the effects of stresses on the expressions of BdCER1 genes, 2-month-old seedlings were incubated in water for 2 days and then transferred to water containing 200 mM NaCl, 20% (w/v) PEG6000, or 100 µM ABA. For drought stress evaluation, surface water of roots was dried gently by napkins, and then seedlings were transferred to filter papers without water supply. Plant leaves under all treatments were sampled at 0, 2, 4, 6, 8, 12, and 24 h. To examine tissue-specific expression profiles of the BdCER1 genes, four tissues (leaves, stems, nodes, and roots) were collected.
Seeds of T3 homozygous BdCER1-8 transgenic lines, the cer1 mutant (SALK_008544) and wild-type Arabidopsis Col-0 (WT) plants were germinated on MS medium. After 2 weeks, seedlings were transferred into Arabidopsis seedling substrate with moderate vermiculites and grown in the illumination incubator at 23°C, 14-h-light/10-h-dark photoperiod. Two-month-old inflorescence stems and 4-week-old rosette leaves were collected to analyze the expression level of BdCER1-8, wax crystal, and cuticular wax, respectively. Four-week-old rosette leaves were further used to check chlorophyll efflux and water loss rates. For soil water deficit experiment, 4-week-old different lines were deprived of water until the cer1 mutant wilted. All materials for expression analysis were immediately frozen in liquid nitrogen and stored at −80°C for further analysis.
Identification and Sequence Analyses of CER1 Homologous Genes in B. distachyon
To identify putative CER1 homologous genes in B. distachyon, the CER1 protein sequence (GenBank accession: NP_001184890) was subjected to do BLASTP search against B. distachyon genome by using the Plant Genomics Resource database (https://phytozome.jgi.doe.gov/). All putative non-redundant protein sequences were manually examined with Pfam program (http://pfam.xfam.org/search) to confirm the presence of the fatty acid hydroxylase (FAH) superfamily (accession no. PF04116). Finally, these potential protein sequences that harbor the FAH superfamily were used as candidate members of CER1 homologous proteins and were further analyzed. The reported CER1 homologous genes in Arabidopsis, cucumber, rape, rice, and maize were obtained from the NCBI database. All sequences of CER1-homologous proteins were listed in Supplementary Data Set 1.
The amino acid sequences of CER1 homologous proteins were aligned by ClustalW and then a phylogenetic tree was constructed using MEGA5 software based on the neighbor-joining method (Saitou and Nei, 1987; Tamura et al., 2011). The molecular weight (MW) and theoretical isoelectric point (pI) of BdCER1s were obtained using the ProtParam tool (http://web.expasy.org/protparam/) (Bjellqvist et al., 1994). The potential motifs in protein sequences were analyzed using the MEME program (http://meme-suite.org/tools/meme) (Bailey et al., 2009). To confirm whether the FAH superfamily of BdCER1 proteins has three His-rich motifs or not, the amino acid sequences of FAH superfamily from different plants were aligned by using ClustalX program with default parameters (Thompson et al., 1997).
Cuticular Wax Extraction and Chemical Characterization
Bd21 leaves and Arabidopsis leaves or stems were immersed in chloroform for 60 s to extract cuticular wax, respectively. N-Tetracosane (C24) was used as an internal standard in each sample. Each sample was transferred to GC vials and dried under nitrogen gas, and then they were derivatized with 30 µl pyridine (Alfa Aesar) and 30 µl BSTFA [bis-N,O-(trimethylsilyl) trifluoroacetamide] (Fluka) at 70°C for 60 min. After derivatization, all samples were dried again under nitrogen gas, and re-dissolved in 500 µl chloroform. Gas chromatography–mass spectrometry (GC–MS) (GCMS-QP2010, SHIMADZU, Japan) and GC with flame ionization detector (GC-FID) (GC-2010Plus, SHIMADZU, Japan) were used to do qualification and quantification analysis, respectively. The surface areas were calculated using digital images and ImageJ software. The wax contents were expressed per unit of surface area.
RNA Extraction and Transcript Level Analysis
The total RNA was extracted using the HiPure Plant RNA Mini Kit (Magen, Guangzhou, China) following standard protocol. Before reverse transcription, about 1–2 µg total RNA was treated with gDNA Eraser to remove genomic DNA, and then the first-strand cDNA was synthesized by PrimeScript RT Enzyme Mix I (Takara, Dalian, China) according to the manufacturer’s instruction. Finally, the products were diluted 10-fold with sterile distilled water.
Quantitative real-time PCR (qRT-PCR) was carried out by using an ABI StepOnePlus instrument (Applied Biosystems). Each reaction volume of 25 µl contained 12.5 µl SYBR® Green I Mix (ToYoBo, Osaka, Japan), 2.5 µl cDNA, 1 µl 10 µM forward primer, 1 µl 10 µM reverse primer, and 8 µl ddH2O. The PCR cycle used was as follows: 95°C for 1 min, 40 cycles of 95°C for 15 s, 58°C for 30 s, and 72°C for 45 s. Data were collected during the extension step. Dissociation curve analysis was performed as follows: 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. Specific primers of BdCER1 genes were listed in Supplementary Table 7. BdACTIN gene (Bradi_1g10630) was selected as an endogenous control. To check expression levels of BdCER1-8 in leaves and stems of different transgenic Arabidopsis lines, both semi-quantitative RT-PCR and qRT-PCR assays were performed using AtACT8 gene (At1g49240) as an endogenous control (corresponding primers also listed in Supplementary Table 7). The relative expression levels were analyzed using the 2−ΔΔCT method (Livak and Schmittgen, 2001). Expression levels of the BdCER1 homologous genes under abiotic stresses were analyzed using the software package of Heatmap on the BMKCloud platform (www.biocloud.net).
Arabidopsis Transformation
The full-length CDS of BdCER1-8 was isolated from leaves of 2-month-old plants using gene-specific primers (Supplementary Table 7) and cloned into pCXSN vector under the control of 35S promoter. The construct was introduced into Agrobacterium tumefaciens strain GV3101 by freeze–thaw method. Two cer1 mutant lines: cer1-1 (SALK_008544) and cer1-2 (SALK_014839) have been previously characterized in Bourdenx et al. (2011). The full-length transcript of CER1 is not detected in the cer1-1 allele, but a weak CER1 band can still be detected in the cer1-2 allele by RT-PCR (Bourdenx et al., 2011), which may interfere the transgene effect. Thus, as a null-allele and due to its severest phenotypes among all alleles, we picked cer1-1 (SALK_008544) as the background material for examining the transgene effect of BdCER1-8 following the Arabidopsis transformation protocol. The cer1-1 (SALK_008544) seeds were obtained from the Arabidopsis Biological Resource Centre (ABRC). As a result after transformation, we obtained over 30 T1 independent transgenic lines that exhibited various levels of transgene expression and morphological phenotypes. Genomic DNAs were extracted from the rosette leaves of transgenic plants, and PCR was performed to confirm positive transformants by standard PCR protocol using specific primers (Supplementary Table 7). qPCR was further carried out to examine the relative BdCER1-8 expression among all individual positive transformants. As a result, 11 ones with the highest expressions were chosen for further selection of BdCER1-8 transgene homozygotes on MS medium containing 50 mg/L hygromycin. The resulting homozygotes in T3 generation coming from independent T1 lines were retained for subsequent analyses.
Phenotyping and Measurements of Transgenic Plants
Chlorophyll efflux, water loss, and soil water deprivation experiments were performed to measure the influence of BdCER1-8 overexpression on cuticle permeability in the cer1 mutant. These three assays were conducted as described previously (Kosma et al., 2009). To investigate the influence of BdCER1-8 overexpression on wax crystal formation, scanning electron microscopy (SEM) was conducted as described previously (Wang et al., 2017).
Results
Changes in Cuticular Wax Accumulation of B. distachyon Leaves Under Drought Stress
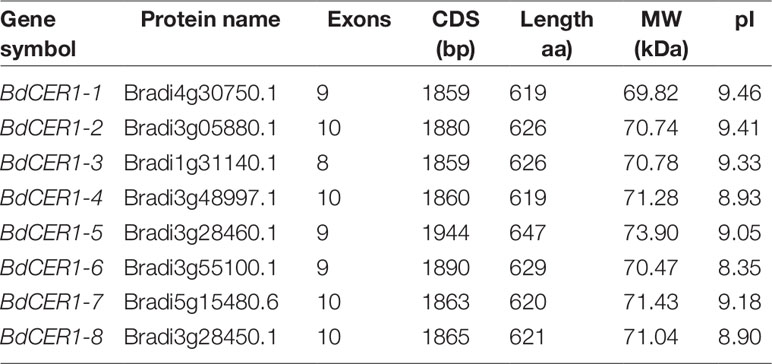
Cuticular wax of B. distachyon ‘Bd21’ leaves under water deficit and 20% PEG6000 treatments were measured to assess the effect of drought stress on wax composition. In the control leaves, total wax content was 9.04 µg/cm2, which was increased by 64.0% to 14.83 µg/cm2 under water deficit treatment and by 61.1% to 14.57 µg/cm2 with 20% PEG6000 treatment, respectively (Figure 1A, Supplementary Table 1). Compared to the control leaves, the primary alcohols content under the water deficit and 20% PEG6000 treatments was increased by 60.5% and 60.6%, respectively (Figure 1A, Supplementary Table 1). The alkanes content was increased by 108.9% and 73.6%, respectively (Figure 1A, Supplementary Table 1). The aldehydes content was increased by 61.7% and 39.7%, respectively (Figure 1A, Supplementary Table 1). The fatty acids content was increased by 50.4% and 50.0%, respectively (Figure 1A, Supplementary Table 1). However, wax composition was unchanged by all treatments, but the contents of different chain length wax components exhibited different elevation levels (Figure 1B). Significant increases in primary alcohols C24 (223.8% and 267.5%), C26 (56.4% and 55.7%), C28 (39.8% and 39.3%), and C32 (172.3% and 102.2%); alkanes C27 (164.4% and 107.3%), C29 (150.9% and 101.0%), and C33 (271.8% and 196.3%); aldehydes C24 (69.0% and 66.8%), C26 (84.2% and 62.6%), and C30 (61.2% and 95.3%); and fatty acids C20 (79.8% and 23.0%), C22 (162.0% and 176.0%), C24 (75.9% and 60.6%), and C26 (140.9% and 200.8%) were detected under the water deficit and 20% PEG6000 treatments, respectively (Figure 1B, Supplementary Table 2). These data showed that alkanes were more sensitive to drought stress than other wax compounds, implying that alkanes biosynthesis may play an important role in drought resistance of B. distachyon.
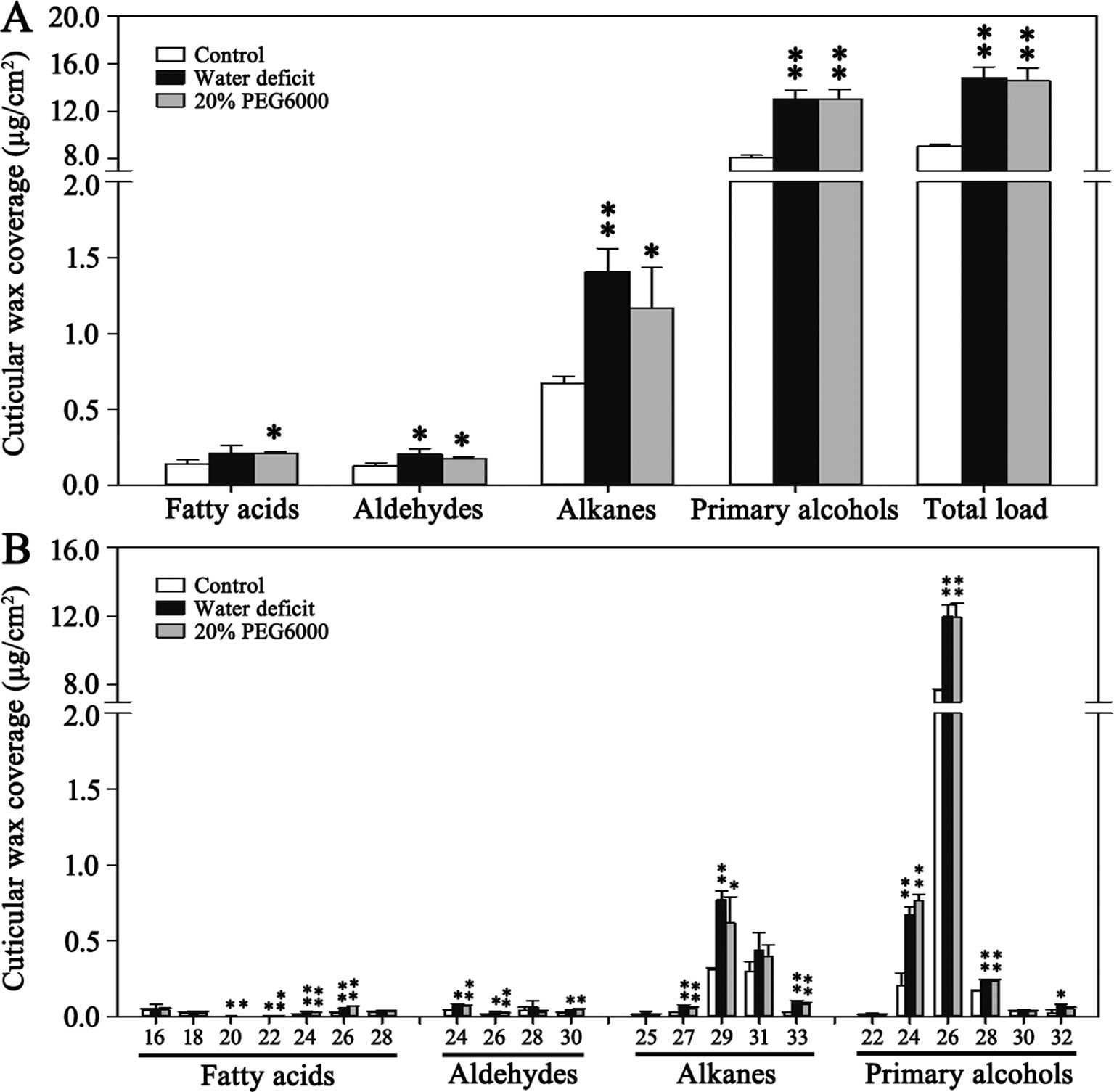
Figure 1 Cuticular wax accumulation and chain length distribution of the individual wax constituents on Bd21 leaves under various treatments. (A) Changes in wax components and total load under various treatments. (B) Changes in chain length of the individual wax constituents under various treatments. The results show averages of three biological replicates, and error bars indicate ± SD. Asterisks indicate significant differences from control (t-test: * for p < 0.05; ** for p < 0.01). Numbers on the x-axis of (B) indicate carbon numbers of the compounds.
Identification of CER1 Homologous Genes in B. distachyon
The Arabidopsis CER1 protein was used to search against B. distachyon genome, and eight putative CER1 homologous genes located on four different chromosomes were found, which were separately named as BdCER1-1 to BdCER1-8 (Table 1). Although the number of exons varied from 8 to 10, there were no significant differences in coding DNA sequences (CDS), amino acid lengths, pIs, and MWs (Table 1). To determine evolutionary relationships among the BdCER1s, their amino acid sequences together with other 15 reported orthologous CER1s were used to construct a phylogenetic tree. Based on the phylogenetic tree topology, all proteins were characterized into two main clades (I and II), of which clade I and clade II mainly contained the CER1-related and the CER3-related members, respectively (Figure 2A), suggesting that function diversification between CER1 and CER3 seemed to have taken place before monocot/dicot separation. As a result, CER1 and CER3 might represent most of their individual ancestor functions, respectively. In B. distachyon, BdCER1-4, -5, -6, -7, and -8 were classified into clade I, while BdCER1-1, -2, and -3 were clustered into clade II (Figure 2A), implying that the duplication events of BdCER1s happened after the separation of dicot from monocot, indicating that the formation of BdCER1s was independent and their functions might also be diverse to some extent.

Figure 2 Phylogenetic and protein structure analyses on homologous genes of CER1 in various plants. (A) Phylogenetic tree. Black triangles point to candidate genes in this study. Two significant clades separately belong to homologous genes of CER1 (Clade I) and CER3 (Clade II). The abbreviations of diverse species: At, Arabidopsis thaliana; Bn, Brassica napus; Cs, Cucumis sativus; Zm, Zea mays; Os, Oryza sativa; Bd, Brachypodium distachyon. (B) Five putative motifs in CER1 homologous genes identified by MEME. Different color rectangles indicate putative motifs
By using the MEME program, five putative motifs were identified and annotated in BdCER1s protein sequences (Figure 2B, Supplementary Figure 1). In the FAH superfamily, three His-rich motifs (H-X3-H, H-X2-HH, and H-X2-HH, where X represents any amino acid) were present in all BdCER1s members (Supplementary Figure 2), which are very important for CER1 and CER1-like1 genes to function in VLC alkanes biosynthesis (Bourdenx et al., 2011; Pascal et al., 2019). These analyses suggested that BdCER1s may be candidate genes involved in VLC alkanes biosynthesis in B. distachyon.
Expression Profiles of BdCER1 Genes in Various Tissues and Under Different Stresses
The expression levels of BdCER1 genes in four tissues of Bd21 including leaves, stems, nodes, and roots were examined by qRT-PCR (Figure 3A). BdCER1-1 exhibited relatively high expressions in all tissues. The expression of BdCER1-2 and -7 were detected in all tissues except in roots. The high expression levels of BdCER1-3, -4, and -6 were detected in stems and nodes. Relatively high expression was detected in stems, leaves, and nodes for BdCER1-5. Interestingly, BdCER1-8 was preferentially expressed in leaves and its expression level was the highest among all BdCER1 genes examined, which was in agreement with the electronic expression levels of BdCER1 genes in leaves according to the Plant Genomics Resource database (Supplementary Figure 3). These analyses showed that BdCER1 genes may be involved in wax accumulation of different tissues.
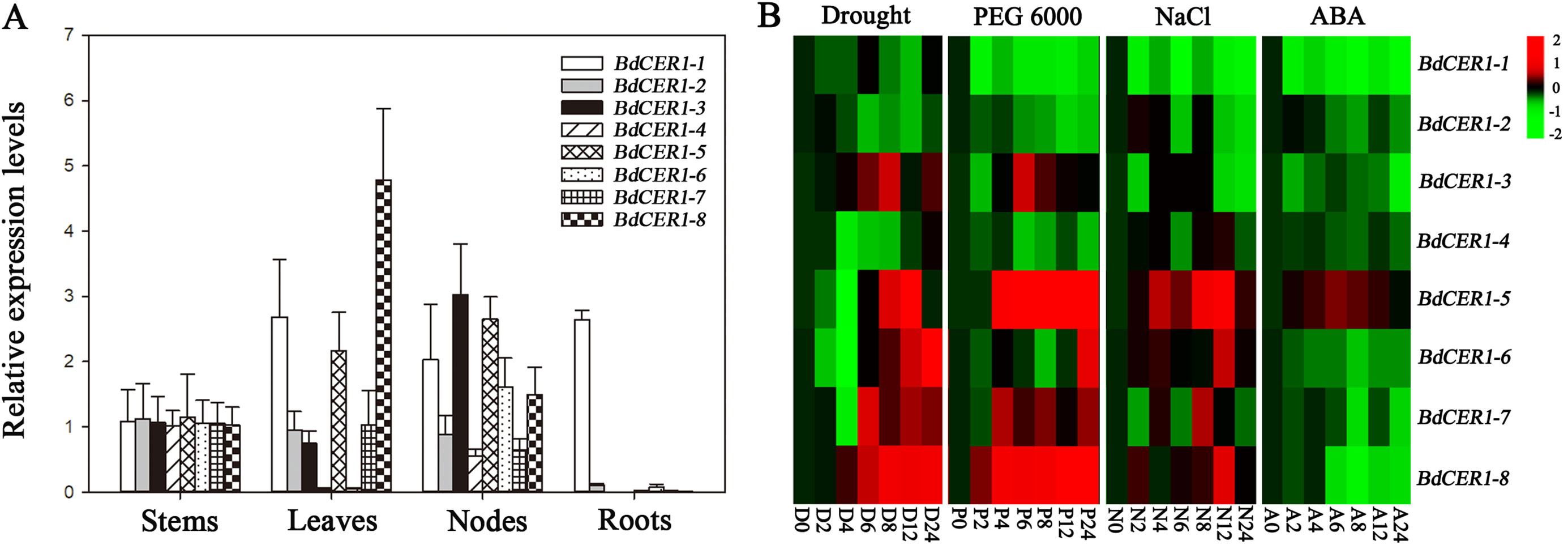
Figure 3 Gene expression analysis of BdCER1s. (A) Tissue expression analysis. (B) Expression analysis under drought (D), PEG (P), ABA (A), and NaCl (N) treatments. Results are presented as relative expression levels. The data represent the means ± standard deviations of three biological replicates.
To investigate the roles of BdCER1 genes in stress resistance, the transcript levels of BdCER1 genes under drought, PEG6000, ABA, and NaCl treatments were analyzed using qRT-PCR (Figure 3B). Among them, expressions levels of five genes (BdCER1-3, -5, -6, -7, and -8) under drought and PEG6000 treatments were gradually increased throughout the time course. However, expressions of BdCER1-5, -6, -7, and -8 with NaCl treatment showed diversified patterns. The expression level of BdCER1-5 was constitutively increased throughout the time course. The expression levels of BdCER1-6 and -8 peaked at 12 h after the treatment and then became decreased, whereas the BdCER1-7 only showed strong induction at 8 h after the treatment. Expression of only one gene (BdCER1-5) was induced by ABA treatment, and its expression level was gradually increased within 12 h after the treatment and then gradually went decreased. These analyses indicated that BdCER1 genes may participate in stress responses and they may play diverse roles in different stress pathways.
Molecular and Phenotypic Characterizations of BdCER1-8 Transgenic Plants
Due to its unique features including high expression level in leaf where wax synthesis is active and intense responses to osmotic stresses, BdCER1-8 was selected as representation of BdCER1 family genes for functional study by introducing its full-length CDS into Arabidopsis cer1 mutant. Eleven T3 independent transgenic lines were used to detect a 3:1 separation ratio. Among these, five homozygous transgenic lines were obtained and further analyzed. Semi-quantitative RT-PCR showed that the BdCER1-8 expression was detected in all five transgenic lines but not detected in the cer1 mutant and WT plant (Figure 4A), which was further confirmed by qRT-PCR analysis (Figure 4B). The Arabidopsis CER1 gene was expressed at very low levels in both the cer1 mutant and the transgenic lines compared to that in WT plant. Whereas the BdCER1-8 transgene was extremely highly expressed in transgenic lines and was absent in the WT and cer1 plants (Figure 4B, Supplementary Figure 4), indicating that BdCER1-8 gene had been successfully expressed in the cer1 mutant without interfering the normal remain of CER1 function in the cer1 mutant.
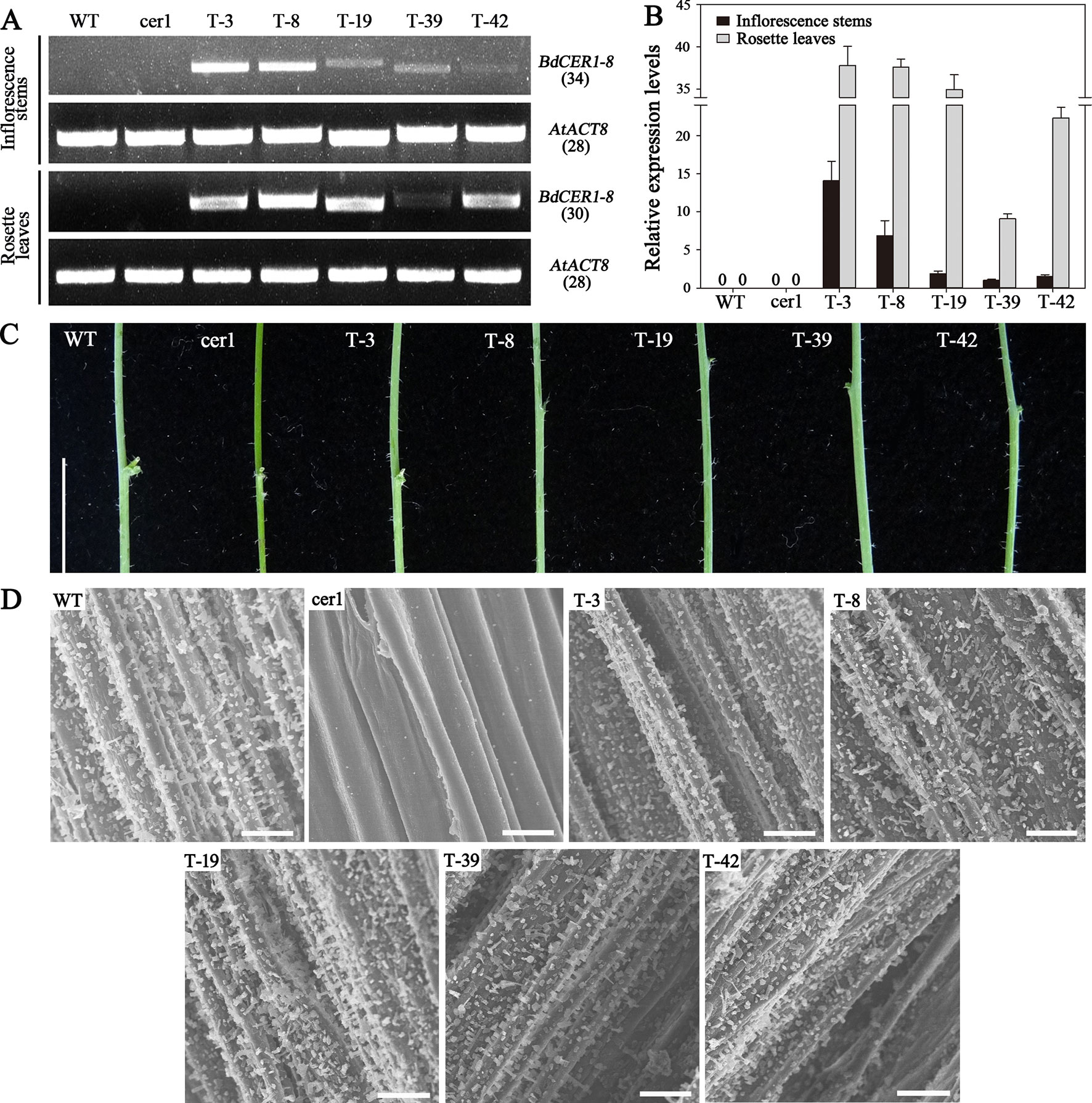
Figure 4 Molecular and phenotypic characterizations of BdCER1-8 transgenic plants. (A) Semi-quantitative RT-PCR analysis of BdCER1-8 transcripts in WT plant, the cer1 mutant, and the transgenic lines with BdCER1-8 transgene. (B) qRT-PCR analysis of BdCER1-8 gene expression in the WT plant, the cer1 mutant, and the transgenic lines with BdCER1-8 transgene. The AtACT8 gene is used as a constitutively expressed control. Results are presented as relative expression levels of T-39 transgenic line in stem. The data represent the means ± standard deviations of three technical replicates. (C) Stem color comparison among WT, cer1, and transgenic plants. Bars = 1 cm. (D) Epicuticular wax crystal of stems from WT, cer1, and transgenic plants observed by SEM at 1,000 × magnification. Bars = 10 μm
To examine the biological function of BdCER1-8, its heterologous expression in the cer1 mutant could be reflected by viewing the stem color (Figure 4C). Compared with WT plant, the stem surface of the cer1 mutant exhibited a glossy green phenotype. However, the BdCER1-8 heterologous expression in the cer1 mutant could prevent this glossy green phenotype and displayed a similar glaucous color to the WT plant. To confirm these observations in details, the stem surfaces of different lines were observed by SEM (Figure 4D). Indeed, compared with the WT plant, the stem surface of the cer1 mutant did not have wax crystal formation, whereas the BdCER1-8 heterologous expression in the cer1 mutant restored its wax crystal formation, suggesting that the stem wax formation defect in the cer1 mutant could be rescued by BdCER1-8.
BdCER1-8 Overexpression Rescues Wax Biosynthesis in the Arabidopsis cer1 Mutant
Cuticular wax accumulation on stems and leaves from different lines including the WT plant, the cer1 mutant, and three transgenic lines with relatively high BdCER1-8 expressions (T-3, T-8, and T-19) was analyzed in details (Figure 5). Compared with the cer1 mutant, the total wax contents on stems was increased by 66.1%, 67.7%, and 70.5% in T-3, T-8, and T-19 lines, respectively. This change was largely due to an increased content of alkanes, ketones, and secondary alcohols that are produced through the alkane-forming pathway (Figure 5A, Supplementary Table 3). Further, the most prominent changes were contributed by C29 alkane, C29 ketone, and C29 secondary alcohol (98.9%, 99.4%, and 99.5% increases, respectively) (Figure 5B, Supplementary Table 4). On the contrary, the contents of other wax components were very slightly changed (Figure 5A, Supplementary Table 3). The heterologous expression of BdCER1-8 restored stem wax content of the cer1 mutant to 74.2–85.5% of that of the WT plant, but the stem total wax contents between transgenic lines and WT plant did not reach significant differences (Figure 5A, Supplementary Table 3), suggesting that BdCER1-8 alone might be sufficient for the restoration of wax loss in the cer1 stem.
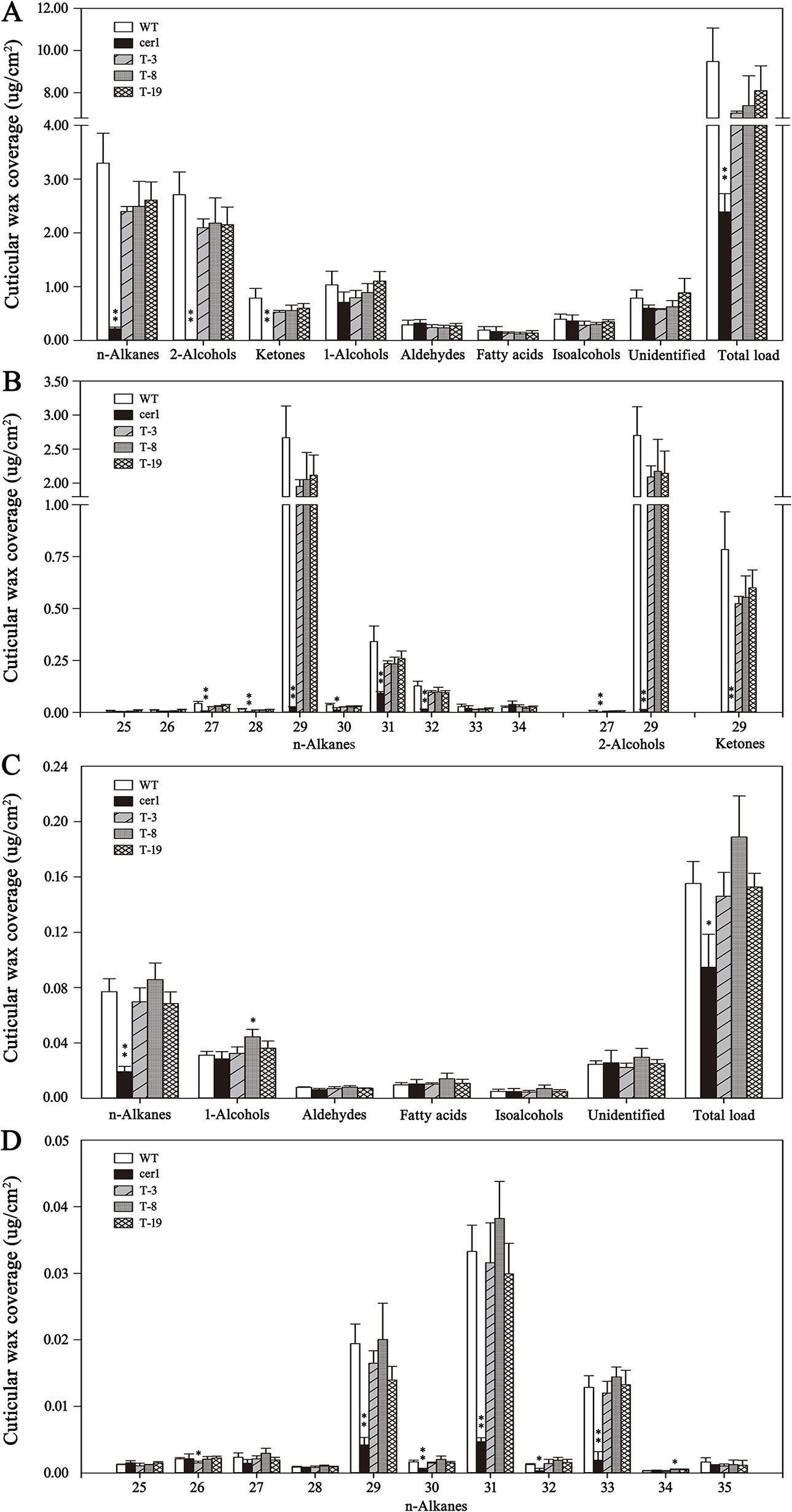
Figure 5 Cuticular wax amount and composition in inflorescence stems and rosette leaves of WT, cer1, and transgenic plants. (A and B) are cuticular wax loads and compositions in the inflorescence stems of different lines, respectively. (C and D) are cuticular wax loads and compositions in the rosette leaves of different lines, respectively. The results show averages of three biological replicates, and error bars indicate ± SD. Asterisks indicate significant differences from WT plant (t-test: * for p < 0.05; ** for p < 0.01). Numbers on the x-axis of (B and D) indicate carbon numbers of the compounds.
In leaves, the total wax contents of T-3, T-8, and T-19 lines were also significantly increased compared with those in the cer1 mutant (35.2%, 49.9%, and 38.0% increases, respectively), which was due to the increases of alkane amounts (around 74.0% increases) (Figure 5C, Supplementary Table 5). More precisely, this modification was largely contributed by the increases of the C29, C31, and C33 alkane contents (74.5%, 85.7%, and 85.4% increases, respectively) (Figure 5D, Supplementary Table 6). Nevertheless, there were only slight changes for other wax components (Figure 5C, Supplementary Table 5). The heterologous expression of BdCER1-8 in the cer1 mutant restored leaf total wax content to 91.1–117.9% of that in WT plant (Figure 5C, Supplementary Table 5). All these results suggested that BdCER1-8 rescued wax production through its participation in the alkane-forming pathway in the transgenic lines.
BdCER1-8 Overexpression Changes Cuticle Properties and Susceptibility to Soil Water Deficit of the cer1 Mutant
Many studies have indicated that change in wax content is generally related to altered cuticle permeability of plants (Kosma et al., 2009; Bourdenx et al., 2011; Xu et al., 2014; Wang et al., 2015a, Wang et al., 2015b; Zhou et al., 2015; Ni et al., 2018). Thus, we conducted chlorophyll leaching and water loss assays in WT plant, cer1 mutant, and T-3, T-8, and T-19 transgenic lines to assess whether the impact of BdCER1-8 over-expression would affect cuticle permeability of the cer1 mutant (Figure 6A, B). The rates of both water loss and chlorophyll leaching in the transgenic lines were lower than those in the cer1 mutant but similar to those in the WT plant at all examined times, indicating that the BdCER1-8 heterologous expression changed cuticle properties of the cer1 mutant. To further test responses of the transgenic lines to soil water deficit, 4-week-old lines were not watered until the cer1 mutant became wilted (Figure 6C). After 10 days of water deprivation treatment, the wilted cer1 mutant showed a relative water content (RWC) of 25.1%. In contrast, transgenic plants showed a less sensitive response. Compared with the cer1 mutant, the transgenic lines maintained a high RWC of 46.3%, 52.1%, and 42.4% in T-3, T-8, and T-19 lines, respectively (Figure 6D). These data suggested that cuticle changes induced by BdCER1-8 decreased the susceptibility of the cer1 mutant to soil water deficit.
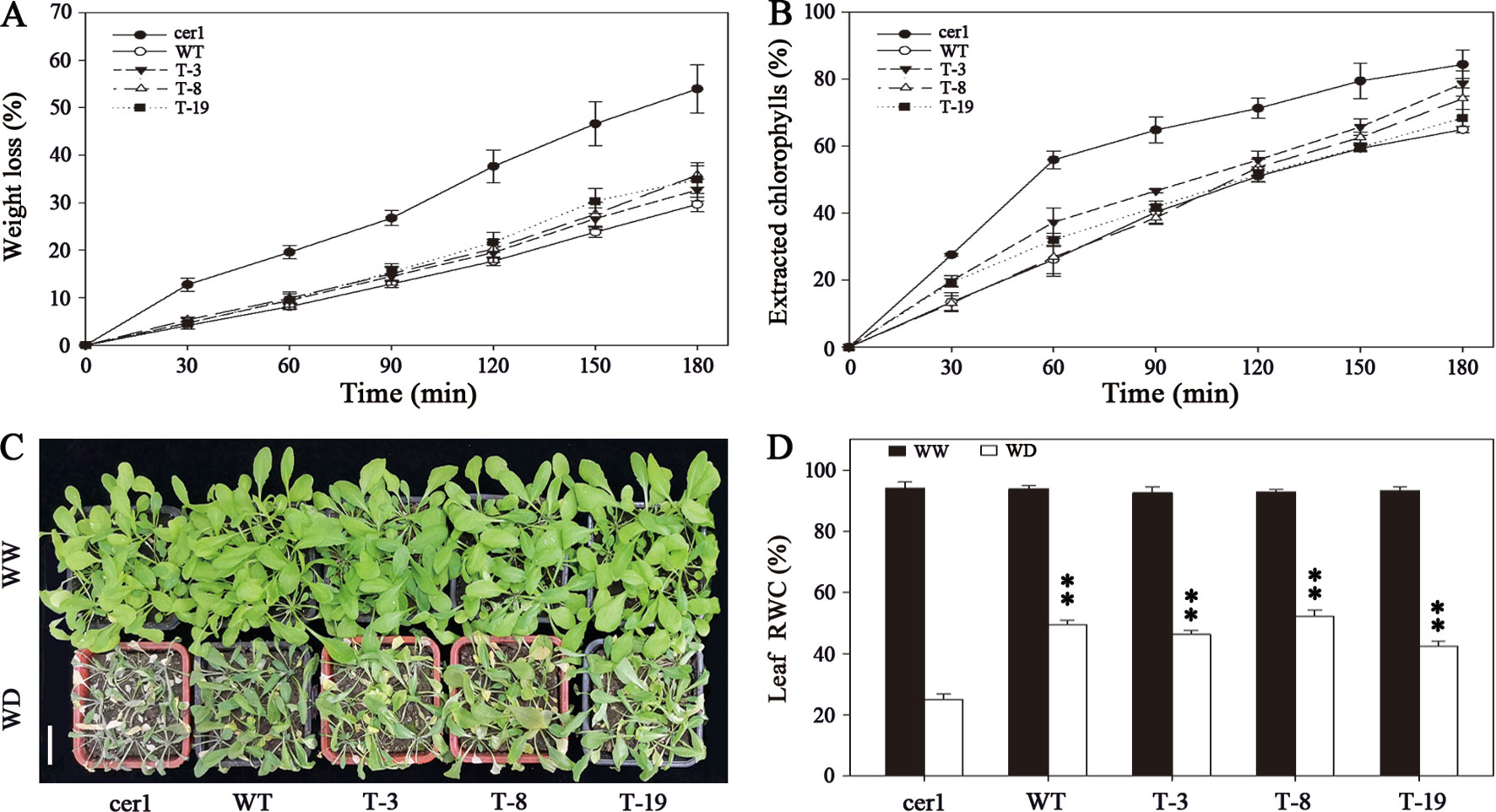
Figure 6 BdCER1-8 alters cuticle properties and response to soil water deficit of the cer1 mutant. (A) Water-loss rates of isolated rosette leaves from WT, cer1, and transgenic plants. (B) Chlorophyll leaching rates of isolated rosette leaves from different lines as indicated above. (C) Soil water deprivation experiment. The abbreviations of WW and WD indicate well-watered plants and water-deprived plants, respectively. Bar = 3 cm. (D) Leaf RWC of well-watered and water-deprived plants. Data are shown by mean ± SD of four biological replicates. Asterisks indicate significant differences from the cer1 mutant (t-test: * for p < 0.05; ** for p < 0.01).
Discussion
In this study, we systematically investigated the influence of drought stress on the leaf cuticular wax accumulation in B. distachyon. Our data indicate that alkanes were more sensitive to drought stress than other wax compounds. Then, we identified eight putative homologous genes of CER1 involved in the VLC alkane biosynthesis of B. distachyon, and further suggested that BdCER1-8 was predominant in VLC alkanes production of leaf due to its relatively high expression levels in leaf and its capability to have nearly fully rescued Arabidopsis cer1 mutant phenotype.
The accumulation of leaf cuticular wax is an important physiological process in plants, which is also seriously affected by environmental stresses (Jenks et al., 2002). Drought stress often results in significant increase of cuticular wax in many plants (Cameron and Smart, 2006; Kosma et al., 2009; Xu et al., 2014; Xu et al., 2016). In Arabidopsis, the leaf wax content was increased by 75% after drought stress, primarily due to increases in wax alkanes (Kosma et al., 2009). Additionally, the content of alkanes was also the most significant change in the Yukon ecotype of saltwater cress and Populus euphratica after water deficit (Xu et al., 2014; Xu et al., 2016). Above results imply that the synthesis of alkanes plays a key role in drought stress response. In our study, to assess the influence of drought stress on leaf cuticular wax accumulation in B. distachyon, both water deficit and PEG treatments were performed. Results showed that the cuticular wax of control plants mainly contained primary alcohols, alkanes, aldehydes, and fatty acids, in which primary alcohols were dominant components with few contents of other compounds (Figure 1), which is consistent with the results of a recent study that the leaf cuticular wax of B. distachyon mainly consists of primary alcohols with less contents of aldehydes, alkanes, and fatty acids throughout leaf development (Wang et al., 2018). All treatments resulted in significant increases of total wax load and individual compound without changing wax composition (Figure 1). However, the change of alkanes content was mostly obvious among all wax compounds, suggesting that alkanes biosynthesis may play an indispensable role in drought resistance of B. distachyon.
The Arabidopsis CER1 gene encodes an essential enzyme involved in the VLC alkane biosynthesis with the presence of FAH superfamily and WAX2 C-terminal domain (Bourdenx et al., 2011; Bernard et al., 2012). The FAH superfamily is functionally involved in zeaxanthin synthesis, cholesterol biosynthesis, and plant cuticular wax biosynthesis (Arthington et al., 1991; Aarts et al., 1995; Bard et al., 1996), in which three His-rich motifs are essential for CER1 and CER1-like1 to function in VLC alkane biosynthesis (Bernard et al., 2012; Pascal et al.,2019). By analyzing FAH superfamily of CER1-related proteins from monocots and dicots (Supplementary Figure 2), we observed that the three His-rich motif core is a shared common feature, indicating that homologous genes of CER1 could be functionally similar and are likely evolved from the same ancestor with His-rich motifs. But according to the phylogenetic tree, they were separately characterized into the CER1-related and the CER3-related groups, respectively (Figure 2A), suggesting that their functional diversification seems to have taken place before monocot/dicot separation, causing subsequent gains of new functions during the long evolutionary history. Additionally in rice, the disruptions in functions of OsGL1s including OsGL1-1, OsGL1-2, OsGL1-3, and OsGL1-6 affect cuticular wax biosynthesis and drought resistance capability in leaf, whereas others including OsGL1-4/OsCER1 and OsGL1-5/WDA1 are involved in VLC alkanes biosynthesis and pollen development in anther (Jung et al., 2006; Islam et al., 2009; Qin et al., 2011; Zhou et al., 2013; Zhou et al., 2015; Ni et al., 2018). These OsGL1s were phylogenetically more closely related to BdCER1s, and their corresponding orthologs were also identified to be the BdCER1 family genes (Figure 2A), indicating that though BdCER1s have multiple copies (five CER1-like genes and three CER3-like genes) and shared core motifs (H-X3-H, H-X2-HH, and H-X2-HH, where X represents any amino acid), they have experienced extensive neofunctionalization since their duplication events because their independent gene birth allow diversification during this long history.
The neofunctionalization consequences may also be related with the differential expression profiles of BdCER1 genes in various tissues (Figure 3A), which is similar to what their rice homologous genes (OsGL1s) behave in different organs to affect wax accumulation in leaf or anther (Jung et al., 2006; Islam et al., 2009; Qin et al., 2011; Zhou et al., 2013; Zhou et al., 2015; Ni et al., 2018). Thus, in consideration of the tissue expression profiles of BdCER1 family genes (Figure 3A), we further inferred that BdCER1-1, -3, and -5 might play key roles in cuticular wax biosynthesis of node where BdCER1-1 might also play an important role in wax production of root. In contrast, BdCER1-8 mainly affected cuticular wax accumulation in leaf. In Arabidopsis, although CER1 and CER1-like1 show different tissues expression levels, they separately interact with CER3 and CYTB5-B to produce VLC alkanes in yeast (Bourdenx et al., 2011; Bernard et al., 2012; Pascal et al., 2019). As such, we provide a model that CER1-related genes (BdCER1-4, -5, -6, -7, and -8) maybe separately form alkane-forming complexes with CER3-related genes (BdCER1-1, -2, and -3) to participate in VLC alkanes accumulation of different tissues; while as specific cofactors of CER1-related genes, CYTB5-B family genes function to improve their efficiencies in this process. This model needs further bench work confirmation in the future study.
Heterologous expression of BdCER1-8 in the Arabidopsis cer1 mutant mainly resulted in increased contents of alkanes and their derivatives in leaf or stem, whereas other wax compounds were only slightly changed (Figure 5), indicating that BdCER1-8 is involved in the alkane-forming pathway in the cer1 mutant. Even though B. distachyon genome encodes multiple CER1-homologous proteins, our analysis suggests that BdCER1-8 may play a more predominant role in alkane production of leaf than its paralogs because it has the highest expression level in leaf among all BdCER1s and it along can nearly completely restored the cer1 defects. This explicit and dominant function of BdCER1-8 may suggest extensive functional diversification existing among BdCER1s, allowing optimized spatial and temporal regulation of wax synthesis in different tissues.
On the other hand, three BdFAR genes (BdFAR1, BdFAR2, and BdFAR3) have been demonstrated to be involved in the alcohol-forming pathway and they respond to abiotic stresses in B. distachyon (Wang et al., 2018). These results together suggest that the cuticular wax biosynthetic pathways in B. distachyon are similar to those in Arabidopsis, containing both the alcohol-forming and the alkane-forming pathways. The leaf cuticular wax of B. distachyon consists of primary alcohols, aldehydes, alkanes, and fatty acids, among which primary alcohols are the dominant wax components throughout leaf development, accounting for 82–89% of total wax contents (Wang et al., 2018). Our work also shows that primary alcohols are the dominant wax components, which accounts for 89.6% of total wax load in control plants (Figure 1A, Supplementary Table 1). These results further suggest that the alcohol-forming pathway is a dominant wax-forming pathway in B. distachyon. VLCFAs are common precursors for both the alcohol-forming and the alkane-forming pathways (Samuels et al., 2008; Kunst and Samuels, 2009). Thus, we propose that BdFARs may be predominant in competing VLCFAs from BdCER1s, which results in producing high level of primary alcohols in B. distachyon leaf.
The biological impact of CER1 genes can be interpreted by the water loss rate and chlorophyll leaching rate tests on detached leaves (Figures 5A, B), where water loss rates and chlorophyll leaching rates in transgenic lines showed lower levels than those in the cer1 mutant, but were similar to those in the WT plant at all examined time. Additionally, transgenic plants contained a relatively higher water content than the cer1 mutant after 10 days of water deprivation treatment (Figures 5C, D). All data imply that BdCER1-8 overexpression changes the repelling properties of water loss in the leaf cuticle of the cer1 mutant, which can be caused by the increased alkanes content. Similar effect was also observed in other species. In cucumber, the CsWAX2 overexpression reduced cuticle permeability of the wax2 mutant by rescuing the alkane-forming pathway (Wang et al., 2015a). Further, abnormal expressions of CsCER1 and CsWAX2 in transgenic cucumber plants have significant effects on VLC alkanes biosynthesis, which finally changes resistance to water loss in the transgenic plants from that in the wild type (Wang et al., 2015a, Wang et al., 2015b). These results further supports our conclusion that alkanes biosynthesis plays a crucial role in drought resistance of B. distachyon.
It is also generally known that wax-related genes are involved in regulatory networks of cuticular wax biosynthesis under diverse environments (Lee and Suh, 2013). Many genes, such as CER1 in Arabidopsis (Kosma et al., 2009), OsGL1s in rice (Jung et al., 2006; Islam et al., 2009; Qin et al., 2011; Zhou et al., 2013; Zhou et al., 2015), CsCER1 and CsWAX2 in cucumber (Wang et al., 2015a, Wang et al., 2015b), and BdFARs in B. distachyon (Wang et al., 2018), respond to drought, sodium chloride, cold, and ABA treatments. In our study, qRT-PCR was performed to elucidate the expression patterns of BdCER1 genes under ABA, drought, PEG, and sodium chloride treatments. The results revealed that BdCER1 genes exhibited different responses to these abiotic stresses. Under water deficit and PEG treatments, BdCER1-3, -5, -6, -7, and -8 were significantly up-regulated (Figure 3B), which could explain the obvious alkane induction on B. distachyon, and perhaps provide a drought resistance mechanism for reducing cuticle permeability by increasing wax accumulation. But only BdCER1-5 were significantly up-regulated under ABA treatment (Figure 3B), suggesting that they may be involved in different regulatory networks of wax accumulation under drought stress. In Arabidopsis, the transcript levels of MYB94 or MYB96 are significantly up-regulated in response to ABA-mediated drought signals (Seo et al., 2011; Lee and Suh 2015). Their proteins directly bind to the gene promoters to activate cuticular wax biosynthetic genes, which finally results in cuticular wax accumulation (Seo et al., 2011; Lee and Suh 2015). As a homolog of MYB96 in wheat, TaMYB31 is also a drought-induced transcription factor involved in cuticle biosynthetic pathway (Bi et al., 2016). Therefore, it can be speculated that BdCER1-5 might be involved in ABA-mediated transcription factors activation in B. distachyon to stimulate wax biosynthesis in response to drought stress, possibly through interacting with homologs of MYB94 or MYB96. In contrast, although BdCER1-3, -6, -7, and -8 were significantly up-regulated by drought and PEG treatments, they were down-regulated under ABA treatment (Figure 3B), suggesting that these genes may be involved in other regulatory network to cope with drought stress. Thus, the mechanism how BdCER1-3, -6, -7, and -8 function in this process and their interacting partners remain to be elucidated.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
HW, YC, and LQ contributed to the experimental design. HW, SS, XL, TinL, JW, TiaL, QZ, WS, CL and ZW performed the experiments and analyzed the data. HW and LQ wrote the manuscript and YC revised it. All authors were involved in the revision of the manuscript and approved the final manuscript.
Funding
This research was supported by the Science and Technology Innovation Team Project (2014KCT-25), National Natural Science Foundation of China (31500256), the Key Research and Development Project of Shaanxi Province (No. 2019ZDLNY04-05), and Tobacco Company Project (No. KJ-2016-01) of Shaanxi Province, China.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Yong Wang for sharing the Brachypodium distachyon ‘Bd21’ seeds.
Abbreviations
VLC, very-long-chain; VLCFA, very-long-chain fatty acid; FAH, fatty acid hydroxylase; PEG, polyethylene glycol; CYTB5s, cytochrome b5 isoforms; GC-MS, gas chromatography-mass spectrometry; GC-FID, gas chromatography with flame ionization detector; qRT-PCR, quantitative real-time PCR; RWC, relative water content; SEM, scanning electron microscopy; CDS, coding DNA sequence; bp, base pair; aa, amino acid; MW, molecular weight; kDa, kilodalton; pI, isoelectric point.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01389/full#supplementary-material
References
Aarts, M. G., Keijzer, C. J., Stiekema, W. J., Pereira, A. (1995). Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell 7 (12), 2115–2127. doi: 10.2307/3870155
Arthington, B. A., Bennett, L. G., Skatrud, P. L., Guynn, C. J., Barbuch, R. J., Uibright, C. E., et al. (1991). Cloning, disruption and sequence of the gene encoding yeast c-5 sterol desaturase. Gene 102 (1), 39–44. doi: 10.1016/0378-1119(91)90535-J
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). Meme suite: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bard, M., Bruner, D. A., Pierson, C. A., Lees, N. D., Biermann, B., Frye, L., et al. (1996). Cloning and characterization of ERG25, the Saccharomyces cerevisiae gene encoding c-4 sterol methyl oxidase. Proc. National Acad. Sci. 93 (1), 186–190. doi: 10.1073/pnas.93.1.186
Bernard, A., Domergue, F., Pascal, S., Jetter, R., Renne, C., Faure, J. D., et al. (2012). Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 24 (7), 3106–3118. doi: 10.1105/tpc.112.099796
Bi, H., Luang, S., Li, Y., Bazanova, N., Morran, S., Song, Z., et al. (2016). Identification and characterization of wheat drought-responsive MYB transcription factors involved in the regulation of cuticle biosynthesis. J. Exp. Bot. 67 (18), 5363–5380. doi: 10.1093/jxb/erw298
Bjellqvist, B., Basse, B., Olsen, E., Celis, J. E. (1994). Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis. 15 (1), 529–539. doi: 10.1002/elps.1150150171
Bourdenx, B., Bernard, A., Domergue, F., Pascal, S., Leger, A., Roby, D., et al. (2011). Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol. 156 (1), 29–45. doi: 10.1104/pp.111.172320
Busta, L., Jetter, R. (2017). Structure and biosynthesis of branched wax compounds on wild type and wax biosynthesis mutants of Arabidopsis thaliana. Plant Cell Physiol. 58 (6), 1059–1074. doi: 10.1093/pcp/pcx051
Cameron, K. D., Smart, T. L. B. (2006). Increased accumulation of cuticular wax and expression of lipid transfer protein in response to periodic drying events in leaves of tree tobacco. Plant Physiol. 140 (1), 176–183. doi: 10.1104/pp.105.069724
Chen, X., Goodwin, S. M., Boroff, V. L., Liu, X., Jenks, M. A. (2003). Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 15 (5), 1170–1185. doi: 10.1105/tpc.010926
Draper, J., Mur, L. A., Jenkins, G., Ghosh-Biswas, G. C., Bablak, P., Hasterok, R., et al. (2001). Brachypodium distachyon. a new model system for functional genomics in grasses. Plant Physiol. 127 (4), 1539–1555. doi: 10.1104/pp.010196
Eigenbrode, S. D., Espelie, K. E. (1995). Effects of plant epicuticular lipids on insect herbivores. Ann. Rev. Entomol. 40 (1), 171–194. doi: 10.1146/annurev.en.40.010195.001131
Hsia, M. M., O’Malley, R., Cartwright, A., Nieu, R., Gordon, S. P., Kelly, S., et al. (2017). Sequencing and functional validation of the JGI Brachypodium distachyon T-DNA collection. Plant J. 91 (3), 361–370. doi: 10.1111/tpj.13582
Islam, M. A., Du, H., Ning, J., Ye, H., Xiong, L. (2009). Characterization of Glossy1-homologous genes in rice involved in leaf wax accumulation and drought resistance. Plant Mol.Biol. 70 (4), 443–456. doi: 10.1007/s11103-009-9483-0
Jenks, M. A., Eigenbrode, S. D., Lemieux, B. (2002). “Cuticular waxes of Arabidopsis,” in The Arabidopsis Book. Eds. Somerville, C. R., Meyerowitz, E. M. American Society of Plant Biologists Press.
Jung, K. H., Han, M. J., Lee, D. Y., Lee, Y. S., Schreiber, L., Franke, R., et al. (2006). Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell 18 (11), 3015–3032. doi: 10.1105/tpc.106.042044
Kerstiens, G. (2006). Water transport in plant cuticles: an update. J. Exp. Bot. 57 (11), 2493–2499. doi: 10.1093/jxb/erl017
Kosma, D. K., Bourdenx, B., Bernard, A., Parsons, E. P., Lu, S., Joubes, J., et al. (2009). The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol. 151 (4), 1918–1929. doi: 10.1104/pp.109.141911
Kunst, L., Samuels, L. (2009). Plant cuticles shine: advances in wax biosynthesis and export. Curr. Opin. Plant Biol. 12 (6), 721–727. doi: 10.1016/j.pbi.2009.09.009
Kurata, T., Kawabata-Awai, Chie, Sakuradani, E., Shimizu, S., Okada, K., Wada, T. (2003). The YORE-YORE gene regulates multiple aspects of epidermal cell differentiation in Arabidopsis. Plant J. 36 (1), 55–66. doi: 10.1046/j.1365-313X.2003.01854.x
Lee, S. B., Suh, M. C. (2013). Recent advances in cuticular wax biosynthesis and its regulation in Arabidopsis. Mol. Plant 6 (2), 246–249. doi: 10.1093/mp/sss159
Lee, S. B., Suh, M. C. (2015). Cuticular wax biosynthesis is up-regulated by the MYB94 transcription factor in Arabidopsis. Plant Cell Physiol. 56 (1), 48–60. doi: 10.1093/pcp/pcu142
Li, Y., Beisson, F. (2009). The biosynthesis of cutin and suberin as an alternative source of enzymes for the production of bio-based chemicals and materials. Biochim. 91 (6), 685–691. doi: 10.1016/j.biochi.2009.03.016
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25 (4), 402–408. doi: doi.org/10.1006/meth.2001.1262
Ni, E., Zhou, L., Li, J., Jiang, D., Wang, Z., Zheng, S., et al. (2018). OsCER1 plays a pivotal role in very-long-chain alkane biosynthesis and affects plastid development and programmed cell death of tapetum in rice (Oryza sativa L.). Front. Plant Sci. 9, 1217. doi: 10.3389/fpls.2018.01217
Pascal, S., Bernard, A., Deslous, P., Gronnier, J., Fournier-Goss, A., Domergue, F., et al. (2019). Arabidopsis CER1-LIKE1 functions in a cuticular very-long-chain alkane-forming complex. Plant physiol. 179 (2), 415–432. doi: 10.1104/pp.18.01075
Pollard, M., Beisson, F., Li, Y., Ohlrogge, J. B. (2008). Building lipid barriers: Biosynthesis of cutin and suberin. Trends Plant Sci. 13 (5), 236–246. doi: 10.1016/j.tplants.2008.03.003
Preuss, D., Lemieux, B., Yen, G., Davis, R. W. (1993). A conditional sterile mutation eliminates surface components from Arabidopsis pollen and disrupts cell signaling during fertilization. Genes and Dev. 7 (6), 974–985. doi: 10.1101/gad.7.6.974
Pu, Y., Gao, J., Guo, Y., Liu, T., Zhu, L., Xu, P., et al. (2013). A novel dominant glossy mutation causes suppression of wax biosynthesis pathway and deficiency of cuticular wax in Brassica napus. BMC Plant Biol. 13, 215. doi: 10.1186/1471-2229-13-215
Qin, B. X., Tang, D., Huang, J., Li, M., Wu, X. R., Lu, L. L., et al. (2011). Rice OsGL1-1 is involved in leaf cuticular wax and cuticle membrane. Mol. Plant 4 (6), 985–995. doi: 10.1093/mp/ssr028
Rowland, O., Lee, R., Franke, R., Schreiber, L., Kunst, L. (2007). The CER3 wax biosynthetic gene from Arabidopsis thaliana is allelic to WAX2/YRE/FLP1. Febs Lett. 581 (18), 3538–3544. doi: 10.1016/j.febslet.2007.06.065
Saitou, N., Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 (4), 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Samuels, L., Kunst, L., Jetter, R. (2008). Sealing plant surfaces: cuticular wax formation by epidermal cells. Annu. Rev.Plant. Biol. 59 (1), 683–707. doi: 10.1146/annurev.arplant.59.103006.093219
Seo, P. J., Lee, S. B., Suh, M. C., Park, M. J., Go, Y. S., Park, C. M. (2011). The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23 (7), 1138–1152. doi: 10.4161/psb.6.7.15606
Shepherd, T., Griffiths, D. W. (2006). The effects of stress on plant cuticular waxes. New Phytol. 171 (3), 469–499. doi: 10.1111/j.1469-8137.2006.01826.x
Solovchenko, A., Merzlyak, M. (2003). Optical properties and contribution of cuticle to UV protection in plants: experiments with apple fruit. Photochem. Photobiol. Sci. 2 (8), 861–866. doi: 10.1039/b302478d
Sturaro, M., Hartings, H., Schmelzer, E., Velasco, R., Salamini, F., Motto, M. (2005). Cloning and characterization of GLOSSY1, a maize gene involved in cuticle membrane and wax production. Plant Physiol. 138 (1), 478–489. doi: 10.1104/pp.104.058164
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28 (10), 2731–2739. doi: 10.1093/molbev/msr121
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic acids res. 25 (24), 4876–4882. doi: 10.1093/nar/25.24.4876
Vogel, J. P., Garvin, D. F., Mockler, T. C., Schmutz, J., Rokhsar, D., Bevan, M. W., et al. (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nat. 463, 763–768. doi: 10.1038/nature08747
Wang, W., Liu, X., Gai, X., Ren, J., Liu, X., Cai, Y., et al. (2015a). Cucumis sativus L. WAX2 plays a pivotal role in wax biosynthesis, influencing pollen fertility and plant biotic and abiotic stress responses. Plant Cell Physiol. 56 (7), 1339–1354. doi: 10.1093/pcp/pcv052
Wang, W., Zhang, Y., Xu, C., Ren, J., Liu, X., Black, K., et al. (2015b). Cucumber ECERIFERUM1 (CsCER1), which influences the cuticle properties and drought tolerance of cucumber, plays a key role in VLC alkanes biosynthesis. Plant Mol.Biol. 87 (3), 219–233. doi: 10.1007/s11103-014-0271-0
Wang, M. L., Wu, H. Q., Xu, J., Li, C. L., Wang, Y., Wang, Z. H. (2017). Five fatty acyl-coenzyme a reductases are involved in the biosynthesis of primary alcohols in Aegilops tauschii leaves. Front. Plant Sci. 8, 1012. doi: 10.3389/fpls.2017.01012
Wang, Y., Sun, Y. L., You, Q. Y., Luo, W. Q., Wang, C., Zhao, S., et al. (2018). Three fatty acyl-coenzyme a reductases, BdFAR1, BdFAR2 and BdFAR3, are involved in cuticular wax primary alcohol biosynthesis in Brachypodium distachyon. Plant Cell Physiol. 59 (3), 527–543. doi: doi.org/10.1093/pcp/pcx211
Xu, X., Feng, J., Lü, S., Lohrey, G. T., An, H., Zhou, Y., et al. (2014). Leaf cuticular lipids on the shandong and yukon ecotypes of saltwater cress, Eutrema salsugineum, and their response to water deficiency and impact on cuticle permeability. Physiol. Plant. 151 (4), 446–458. doi: 10.1111/ppl.12127
Xu, X., Xiao, L., Feng, J., Chen, N., Chen, Y., Song, B., et al. (2016). Cuticle lipids on heteromorphic leaves of Populus euphratica Oliv. growing in riparian habitats differing in available soil moisture. Physiol. Plant. 158 (3), 318–330. doi: 10.1111/ppl.12471
Zhou, L., Ni, E., Yang, J., Zhou, H., Liang, H., Li, J., et al. (2013). Rice OsGL1-6 is involved in leaf cuticular wax accumulation and drought resistance. PLoS One 8 (5), e65139. doi: 10.1371/journal.pone.0065139
Keywords: wax, alkanes, CER1, expression, drought stress, Brachypodium distachyon
Citation: Wu H, Shi S, Lu X, Li T, Wang J, Liu T, Zhang Q, Sun W, Li C, Wang Z, Chen Y and Quan L (2019) Expression Analysis and Functional Characterization of CER1 Family Genes Involved in Very-Long-Chain Alkanes Biosynthesis in Brachypodium distachyon. Front. Plant Sci. 10:1389. doi: 10.3389/fpls.2019.01389
Received: 21 March 2019; Accepted: 08 October 2019;
Published: 01 November 2019.
Edited by:
Basil J. Nikolau, Iowa State University, United StatesReviewed by:
Jay Shockey, Department of Agriculture, United StatesJoaquín J. Salas, Instituto de la Grasa (IG),Spain
Copyright © 2019 Wu, Shi, Lu, Li, Wang, Liu, Zhang, Sun, Li, Wang, Chen and Quan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaofeng Chen, Y2hlbnlmMzgyOEAxMjYuY29t; Li Quan, bHF1YW5AbndhZnUuZWR1LmNu
†These authors have contributed equally to this work
 Hongqi Wu
Hongqi Wu Shandang Shi1†
Shandang Shi1† Jiahuan Wang
Jiahuan Wang Li Quan
Li Quan