- 1Plantain and Banana Improvement Program, International Institute of Tropical Agriculture, Ibadan, Nigeria
- 2Department of Plant Sciences (Plant Breeding), University of the Free State, Bloemfontein, South Africa
- 3Food and Nutrition Sciences Laboratory, International Institute of Tropical Agriculture, Ibadan, Nigeria
- 4International Institute of Tropical Agriculture, The Nelson Mandela African Institution of Science and Technology, Arusha, Tanzania
- 5Laboratory of Tropical Crop Improvement, Bioversity International, Heverlee, Belgium
- 6Department of Biosystems, KU Leuven, Heverlee, Belgium
Bananas (Musa spp.), native to South East Asia, have spread worldwide and are integrated into the diets of millions of people in tropical regions. Carotenoid content varies dramatically between different banana genotypes, providing an opportunity for vitamin A biofortification. Polyploidization is a useful tool for crop improvement with potential to generate new diversity, especially in a polyploid crop like bananas. Ten induced tetraploids generated from six diploid banana genotypes were evaluated for their agronomic attributes and fruit carotenoid content in comparison to their diploid progenitors. Tetraploids had distinct plant morphology, but generally displayed inferior vegetative and yield characteristics with 20% lower bunch weights than their original diploids. Similarly, a 50% decrease in fruit provitamin A carotenoids (α-carotene, 13-cis β-carotene, 9-cis β-carotene, trans-β-carotene) accompanied by a corresponding increase in lutein was recorded in induced tetraploids in comparison to their original diploids. Additionally, all lines were subjected to pollen viability tests to assess their fertility. Pollen viability tests indicated over 70% viability for induced tetraploids and diploid controls, suggesting their possible use in crosses. These findings provide a basis for the application of induced polyploidization in bananas to generate useful genetic material for integration in hybridization programmes aiming to produce vitamin A enriched triploids valuable to malnourished populations.
Introduction
Micronutrient deficiency, also known as “hidden hunger,” is a major public health concern, caused by inadequate dietary intake of essential nutrients or minerals. One in three people worldwide are malnourished, with two billion people lacking key micronutrients, like iron and vitamin A (Global Nutrition Report, 2017). Traditional strategies, such as mineral supplementation and food fortification, have failed to completely eradicate micronutrient deficiencies due to lack of economic, social, and cultural mechanisms for efficient implementation. Recently, crop improvement efforts have focused on producing nutrient-rich high yielding crops through biofortification, as a sustainable solution to address micronutrient malnutrition (Bouis and Welch, 2010; Garg et al., 2018). Biofortification involves the use of conventional breeding or transgenic approaches to enhance the nutritional value of crops. Conventional breeding, which exploits natural genetic variability has been the preferred method for total carotenoids with vitamin A activity (pVAC) enhancement of sweet potato, maize, and cassava, with biofortified varieties already available in farmers’ fields (Bouis and Saltzman, 2017; Garg et al., 2018).
Bananas (Musa spp.), native to South East Asia, have spread worldwide and are integrated into the diets of millions of people in tropical regions. Most banana cultivars are derived from intra- and inter-specific hybridization of two wild diploid ancestral species, Musa acuminata Colla (A genome) and Musa balbisiana Colla (B genome), although Musa schizocarpa (S genome) and Musa textilis (T genome) are also reported to have contributed to the origin of some cultivars (Heslop-Harrison and Schwarzacher, 2007; Häkkinen, 2013; Christelová et al., 2017; Němečková et al., 2018). Based on ploidy and genome distribution, bananas are classified as diploid (AA and AB), triploid (AAA, AAB, and ABB), or tetraploid (AAAA, AAAB, and AABB) (Heslop-Harrison and Schwarzacher, 2007). Most important cultivars for food security are of the triploid category comprising dessert bananas (AAA and AAB), plantains (AAB), East African highland bananas (EAHB) (AAA-EA) and the Bluggoe and Pisang awak cooking types (ABB), hence the focus of banana breeding programs.
Besides being a principal source of calories, some bananas are a good source of micronutrients such as vitamin A, which can be enhanced by biofortification. Banana cultivars exhibit considerable genetic variability for pVACs, with some cultivars accumulating up to 35 µg g−1 FW of fruit pulp (Davey et al., 2009; Borges et al., 2014; Heng et al., 2017; Amah et al., 2019), but this variability has not been fully exploited in breeding. Banana breeding strategies rely on sexual hybridization across ploidy categories and phenotypic selection from viable offspring. The predominant breeding scheme involves crosses between 3x cultivars and 2x wild or improved accessions, selecting 4x and 2x hybrids from intermediate products and crossing selected 4x and 2x hybrids to generate sterile 3x hybrids (Tenkouano et al., 2011; Ortiz and Swennen, 2014; Brown et al., 2017). However, this process is relatively slow and is further complicated by parthenocarpy, polyploidy, and associated irregular meiotic behavior, low fertility, low seed viability, long generation times, diverse genome configurations, and the narrow genetic base (Ortiz, 2013; Brown et al., 2017; Batte et al., 2019). Nevertheless, triploid hybrids have been efficiently obtained using the 4x–2x scheme making tetraploids vital to 3x breeding.
Polyploidization has been advocated as a tool for genetic improvement in several crops with the aim of creating new polyploid varieties or increasing fertility of interspecific hybrids (Dhooghe et al., 2011; Sattlser et al., 2016; Denaeghel et al., 2018; Parsons et al., 2019). Polyploid induction has been proposed as an alternative breeding approach for bananas. This strategy aims to create triploids directly from doubled ancestral diploid cultivars through reconstitutive breeding (Bakry et al., 2009; Tenkouano et al., 2011). Polyploid induction in Musa spp. is achieved through treatment of highly proliferating totipotent explants with anti-mitotic compounds (colchicine or oryzalin), which interfere with microtubule formation, followed by plantlet regeneration and isolation of induced tetraploids through flow cytometry (Hamill et al., 1992; Van Duren et al., 1996; Ganga and Chezhiyan, 2002; Bakry et al., 2007; Goigoux et al., 2013; Salles Pio et al., 2014; Do Amaral et al., 2015). Although there are several reports on induction of polyploidization in banana aiming to generate tetraploid breeding lines towards triploid breeding, most studies are limited to the establishment of in vitro polyploidization systems for specific cultivars and isolation of induced tetraploids. There are currently no reports on characterization of induced tetraploids for their use in triploid breeding. Following the creation of induced tetraploids, there is a need for the evaluation of their agronomic/morphological characteristics, fruit quality, and fertility to ascertain their suitability for triploid breeding.
The triploid status, limited fertility, and narrow genetic variability of most cultivated bananas necessitate the use of diploid bananas as a source of diversity for genetic improvement. Wild M. acuminata (AA) diploids are quite diverse and have been differentiated into four sub-species (Perrier et al., 2009). Recently, AA wild and cultivated diploids have been further differentiated into several distinct clusters based on cytological and molecular characterization, reflecting evolutionary relationships as well as geographical origins (Christelová et al., 2017). Particularly, diploid cultivars implicated in the evolutionary process of present cultivated varieties may constitute a source of diversity with relevance for banana improvement (Bakry et al., 2009; Brown et al., 2017; Perrier et al., 2018). For example, the AA cultivars of the ssp. banksii cluster from Papua New Guinea (PNG), which may have contributed to the A genome in plantain cultivars (Perrier et al., 2009; Hippolyte et al., 2012; Christelová et al., 2017), could be of relevance to plantain breeding. AA diploid cultivars with origins from PNG have also been reported to have high pVAC content with potential for vitamin A biofortification (Fungo and Pillay, 2011; Amah et al., 2019). Such valuable diploid germplasm can serve as progenitors for induced polyploidization to generate tetraploids for triploid breeding. Induced polyploidization is a useful breeding tool for creating novel genetic variation for cultivar improvement and could offer possibilities to utilize existing pVAC diversity within the diploid genepool for biofortification in Musa spp.
The aim of this study was to assess the effect of in vitro polyploidization on agronomic attributes, pVAC content, and pollen fertility on induced tetraploids derived from diploid banana genotypes. These findings provide a basis for the potential application of polyploid induction as a breeding strategy towards banana biofortification.
Materials and Methods
Plant Material
Six diploid AA genotypes (Table 1), were subjected to in vitro polyploidization to generate tetraploids. In vitro polyploidization was carried out as described by Bakry et al. (2007) using oryzalin (PESTANAL®, Sigma-Aldrich). Proliferating shoot clusters were established from in vitro shoot tips on Murashige and Skoog (MS) basal medium (Sigma-Aldrich) supplemented with 4 mg l−1 6-benzylaminopurine (BAP), 30 g l−1 sucrose and 2.0 g l−1 Gelrite (Gelzan™, Sigma-Aldrich) with pH adjusted to 5.8 ± 0.1. Selected bud clusters were subjected to oryzalin treatment (45 µM; 15.6 mg l−1) for 48 hours on a gyratory shaker at 90 rpm in liquid MS medium, after which they were rinsed in sterile distilled water for 24 hours at 90 rpm. Clusters were transferred to semi-solid MS medium with 1 mg l−1 BAP. and surviving plants were sub-cultured at 30-day intervals four times to allow chimera dissociation and plantlet development. Young leaf tissue was obtained from selected, fully developed plantlets for ploidy analysis. Ploidy level of each accession was estimated by flow cytometry according to Dolezel et al. (2007), using a reference AA diploid, Calcutta 4 as external standard.
Induced tetraploids were isolated from separate doubling events for each genotype for further evaluation (Table 1). Tetraploids and their diploid progenitors were established on proliferation medium to generate clones, and plantlets were rooted on MS medium devoid of BAP and supplemented with 1 mg l−1 1-naphtaleneacetic acid (NAA), for 40 days, after which they were simultaneously acclimatized in a screenhouse for field establishment.
Research Site and Experimental Design
Experimental plants were established at the IITA Ibadan research station (3° 54’ E, 7° 30’ N, at 240 m a.s.l.), in the sub humid-derived savanna agro-ecology where the soil type is predominantly a ferric luvisol. The rainfall pattern is bimodal, with two distinct rainy seasons, the first from April to July, and the second from August to November. Fully acclimatized plants of all six diploid progenitors and 10 induced tetraploids (Table 1) were planted in a completely randomized design with nine replications. All plants were grown under standard field conditions at a spacing of 3 m × 2 m at a density of 1,666 plants ha−1. Plants were evaluated between the months of April 2017 and May 2018.
Agronomic Assessment
Agronomic characteristics (vegetative and yield-related traits) were evaluated at flowering and at harvest. Plant height (PHT), pseudostem girth at 100 cm from soil surface (PGT) and number of suckers (NSF) were recorded at flowering. Bunch weight (BWT), number of hands or clusters (NH), total number of fingers or fruits (NF), fruit weight (FWT), fruit length (FLT) and fruit circumference (FC) were recorded at harvest when fruits were filled or when a fruit showed signs of yellowing. Fruit measurements were taken from the middle fruit of the third hand. Flowering date was recorded upon emergence of the flag leaf and harvest date was recorded at harvest. Days to flowering (DF) was recorded as the number of days between planting and flowering. Days to fruit maturity (DFM) were recorded as number of days between flowering and harvesting.
Carotenoid Quantification
Sample preparation, carotenoid extraction, and quantification through high-performance liquid chromatography (HPLC) were carried out as previously described (Amah et al., 2019). Briefly, carotenoids were extracted from finely macerated ripe fruit samples (10 g) using acetone as a solvent and partitioned into petroleum ether via a separatory funnel. Petroleum ether extracts were then filtered through anhydrous sodium sulphate, dried under nitrogen gas, and reconstituted in dichloromethane/methanol (v/v) for analysis.
Separation and quantification of individual carotenoids was carried out on a Waters Alliance e2695 HPLC system (Waters Corporation, Milford, MA, USA) equipped with a polymeric YMCTM C30 column (4.6 × 250 mm, 5-µm particle size) and a photodiode array detector. The UV spectrum was observed at 200 to 600 nm, and carotenoids were detected at 450 nm. Data collection and processing were conducted using Empower software (Waters Corporation, Milford, MA, USA). Lutein, α-carotene, trans-β-carotene (trans-BC), 13-cis-β-carotene (13-cis-BC), and 9-cis-β-carotene (9-cis-BC) were identified through an external standard method based on the calibration curve established from pure standards and verification of absorption spectrum and co-elution with authentic commercial standards (β-carotene, α-carotene, and lutein).
Total carotenoids (TC) with provitamin A activity was computed as: pVACs (µg g−1 of fresh weight) = α-carotene +13-cis-BC + 9-cis-BC + trans-BC and TC computed as TC (µg g−1) = total pVACs + lutein. Provitamin A content expressed in terms of β-carotene equivalents (BCE) was also computed as BCE (µg g−1) = 0.5 (α-carotene +13-cis-BC + 9-cis-BC) + trans-BC.
Pollen Viability Assessment
Pollen viability tests were carried out using 1% 2,3,5-triphenyltetrazolium chloride (TTC) stain diluted in Tris buffer (hydrochloric acid 0.15 M, pH 7.8) as described by Soares et al. (2015). Male flowers were collected from each genotype at anthesis (7.30–10.30 am) and immediately conveyed to the laboratory for analysis. Pollen grains were collected from two anthers of the same flower and manually spread on a glass slide, after which a drop of TTC stain was added. The preparation was covered with a cover slip and allowed to stand for two hours to allow uptake of the stain. Two slides were prepared of each genotype (four flowers from each individual plant) and preparations were observed under bright field illumination using a light microscope (Olympus BX51). Pollen grains stained with TTC appears light pink or red when viable and remains transparent when non-viable. Viable and non-viable pollen grains were recorded from observation of three selected microscopic fields of each slide. The percentage of stained pollen grain was calculated from the pollen counts and expressed as percentage pollen viability.
Statistical Analysis
All data were collected, summarized, and analyzed using SAS software version 9.4 for Windows Copyright © 2002–2015 by SAS Institute Inc., Cary, NC, USA. The PROC GLM statement was used for one-way ANOVA followed by Student-Newman-Keuls test and Duncan’s Multiple Range Test to detect significant differences among means for agronomic and carotenoid traits, respectively, and the statistical significance level was set to 0.05. Heatmap correlation (genotypes versus variables) was performed using “heatmap.2” function through gplots v3.0.1.1 library implemented in R (R Core Team, 2018).
Results
Agronomic Characteristics
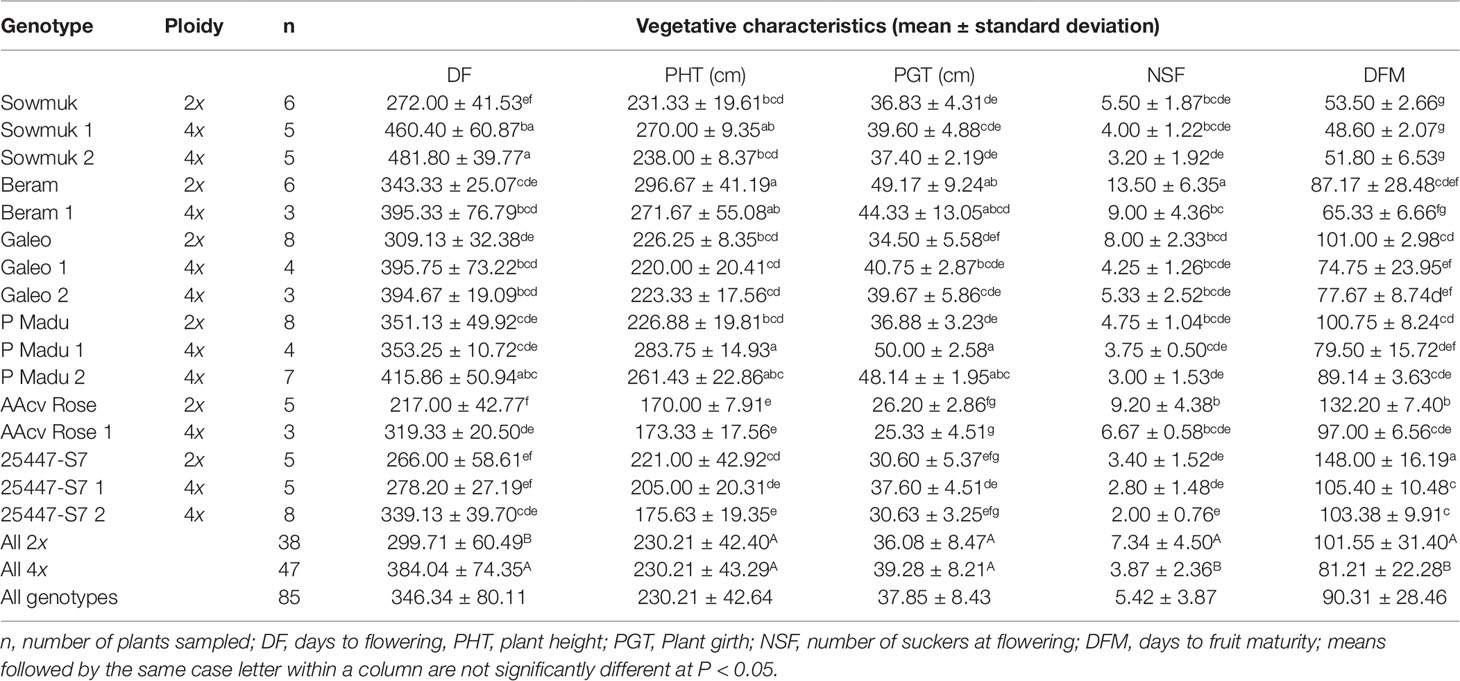
Results for agronomic characteristics are only presented for plants harvested at full maturity, as some plants suffered wind damage. Significant phenotypic differences were observed between diploid plants and their induced tetraploids for vegetative and yield traits. Tetraploid plants had longer, curved, and drooping leaves, while diploid plants had shorter and more erect leaves (Figure 1). Data for vegetative characteristics are presented in Table 2 and illustrated in Figure 2. Generally, number of days to flowering (384.04) was significantly longer for induced tetraploids plants than for diploids (299.71) but the fruit maturity time was significantly shorter for induced tetraploids (81.21) than for original diploids (101.55). Plant height and plant girth did not differ significantly for both groups but the number of suckers at flowering was significantly higher for diploids (7.34) than for induced tetraploids (3.87).
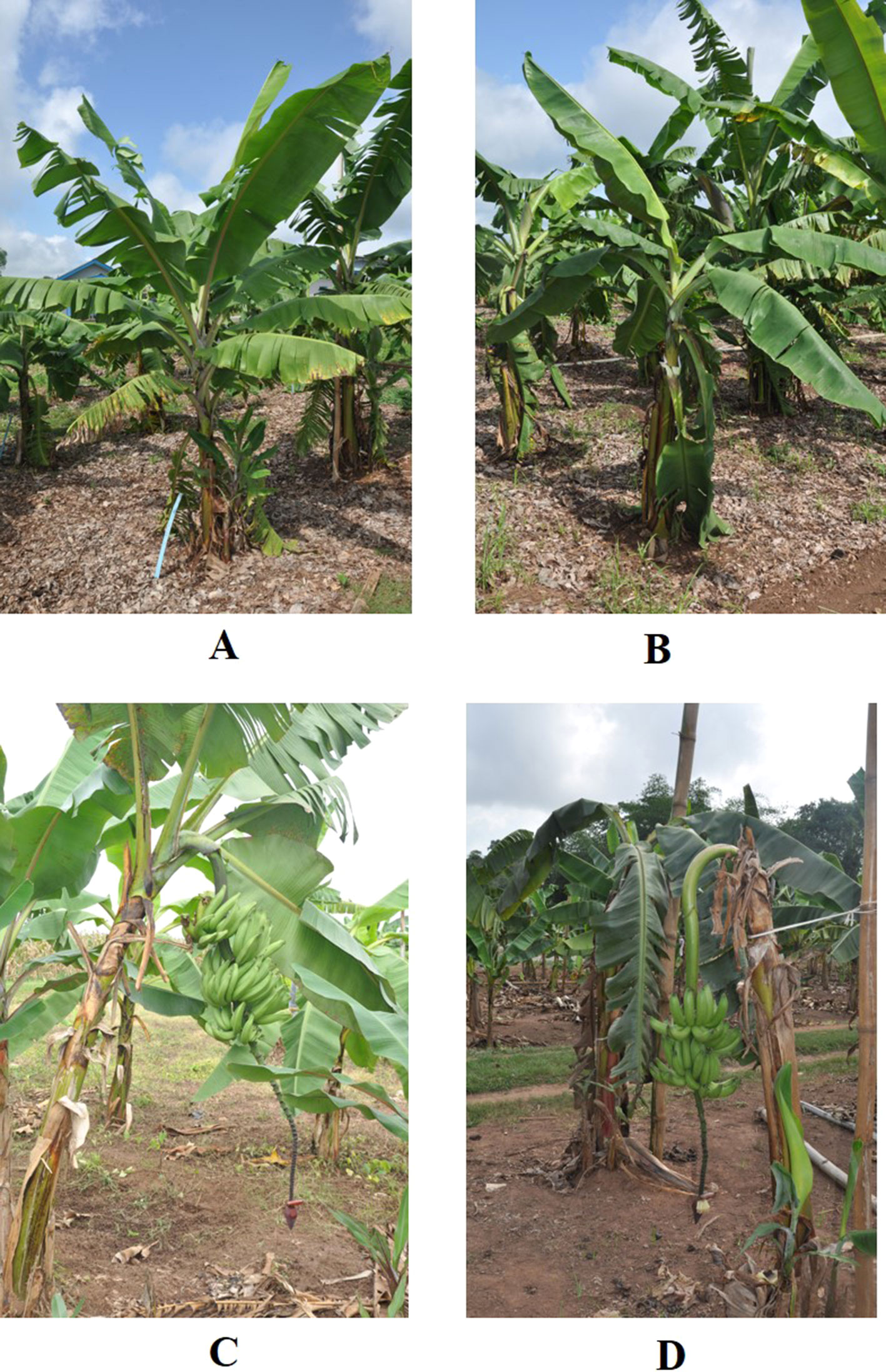
Figure 1 Leaf characteristics of diploid and induced tetraploid banana cultivar Galeo. Pre-flowered plants of diploid (A) and induced tetraploid (B) and mature plants for diploid (C) and induced tetraploid (D) of cultivar Galeo.
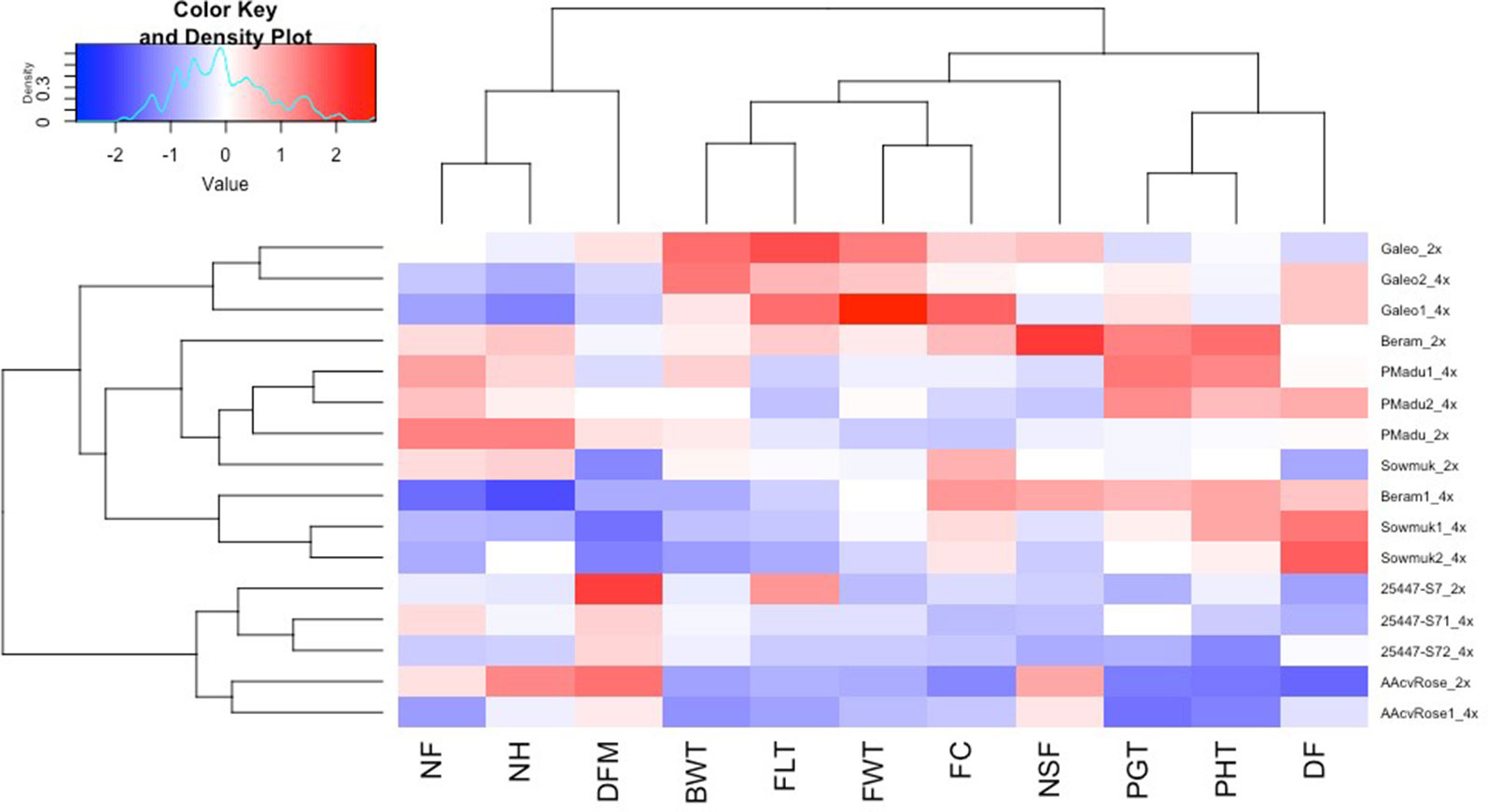
Figure 2 Heatmap and hierarchical clustering for agronomic traits in diploid and induced tetraploid bananas. Genotype names on the vertical axis followed by 2x and 4x indicates diploids and tetraploid lines respectively. DF, days to flowering; PHT, plant height; PGT, Plant girth; NSF, number of suckers at flowering; DFM, days to fruit maturity; BWT, bunch weight; NH, number of hands; NF, number of fruits; FLT, fruit length; FC, fruit circumference; FWT fruit weight. Bright blue indicates lowest values while bright red indicates highest values for each trait.
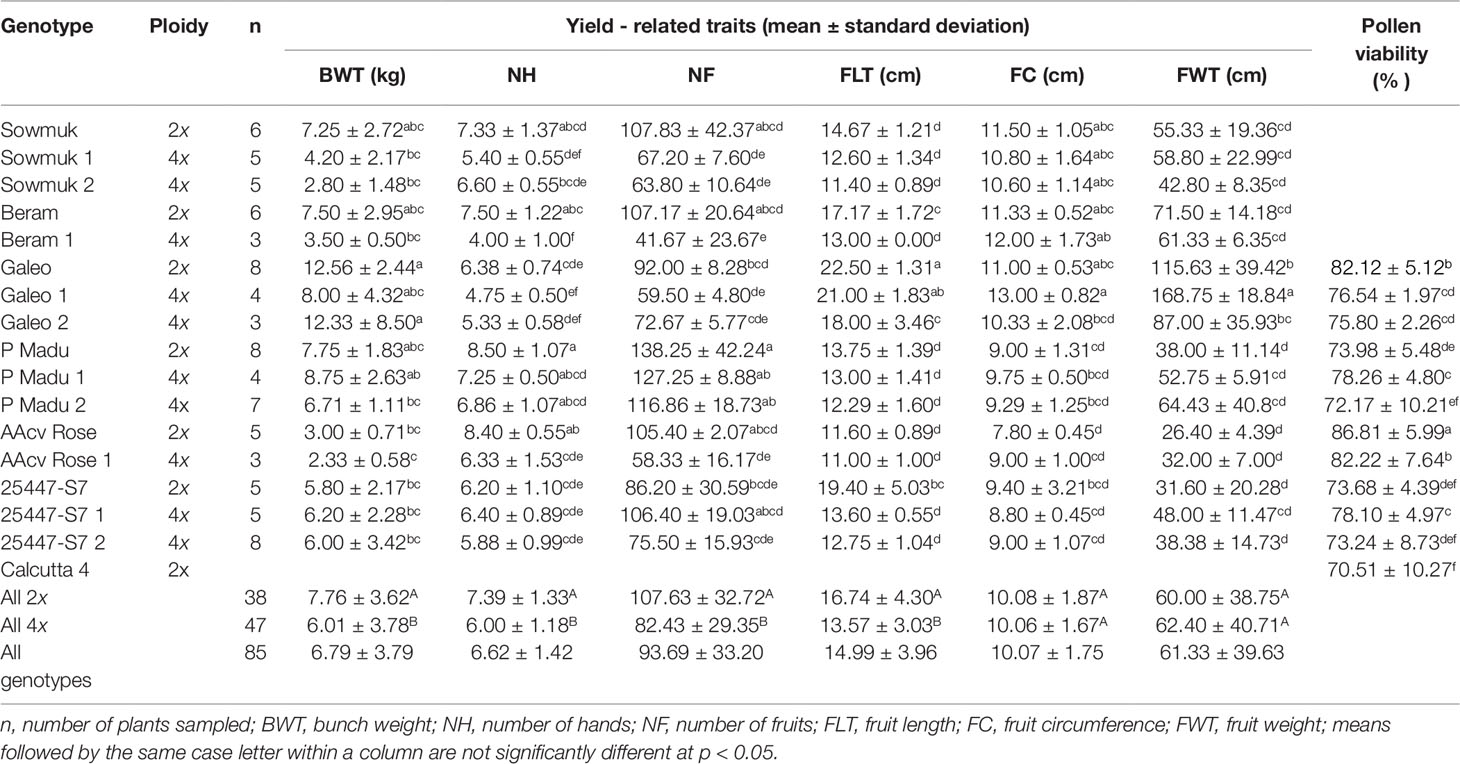
Induced tetraploids generally had significantly lower bunch weights (6.01 kg) than diploids (7.76 kg) but variable changes were observed for individual genotypes (Figure 3; Table 3). Notably, tetraploids from the hybrid 25447-S7 (25447-S7 1 and 2) and Pisang Madu (P Madu 1) had slightly higher bunch weights than their diploid counterparts, while Sowmuk 2 recorded the highest reduction in bunch weight (> 50%) following polyploidization which was significantly different from the diploid version, though these changes in bunch weights were generally not significant (Figure 3, Table 3). Similarly, the number of hands per bunch was higher (7.39) for diploids than for induced tetraploids (6.00), but this difference was only significant for Beram and AAcv Rose (Table 3). Mean number of fruits was significantly higher for diploids (107.63) than tetraploids (82.43) but mean fruit weight and circumference were variable among doubled and non-doubled genotypes.
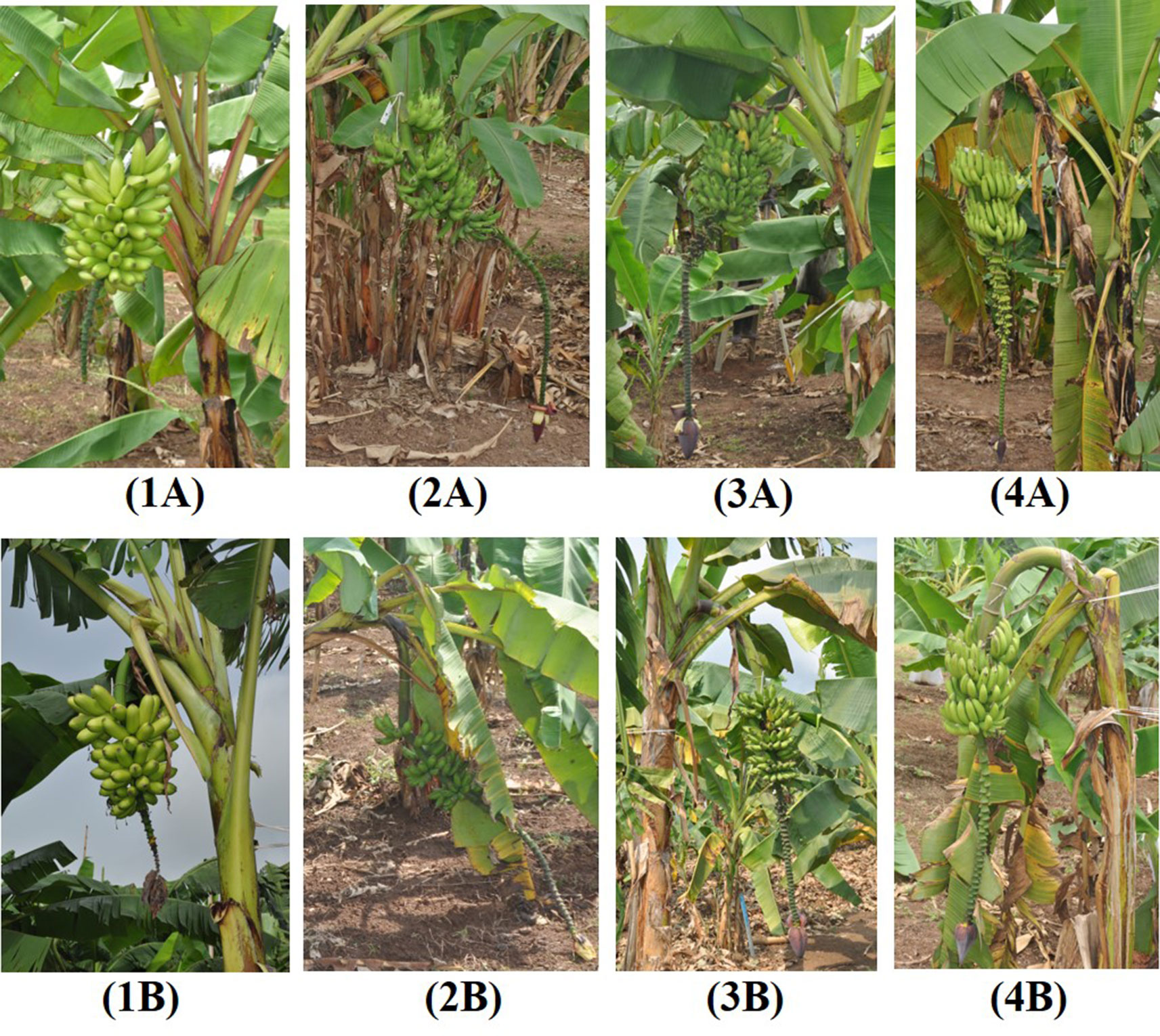
Figure 3 Bunch characteristics of diploid and induced tetraploid bananas. 2x Sowmuk (1A); 4x Sowmuk (1B); 2x AAcv Rose (2A); 4x AAcv Rose (2B); 2x Pisang Madu (3A); 4x Pisang Madu (3B); 2x 25447-S7 (4A); 4x 25447-S7 (4B).
Hierarchical clustering and heat maps of agronomic characteristics of all 16 diploid and induced tetraploid lines were plotted to illustrate the influence of doubling on the variables measured (Figure 2). All entries were clustered in to five main groups based on the 11 agronomic traits measured. The diploid Galeo and its tetraploid forms Galeo 1 and Galeo 2 grouped together in a single cluster showing higher values for bunch weight, fruit length, fruit weight, and fruit circumference, than all other entries. Tetraploids generally had higher days to flowering than diploids and three tetraploids (Beram 1, Sowmuk 1, and Sowmuk 2) with higher days to flowering clustered together. With regard to traits, 11 agronomic traits were grouped into three different groups with plant girth, plant height, and days to flowering clustering together in a first group; bunch weight, fruit length, fruit weight, fruit circumference, and number of suckers at flowering clustering together in a second group and number of fruits, number of hands and days to fruit maturity clustered together in a third group.
Carotenoid Content
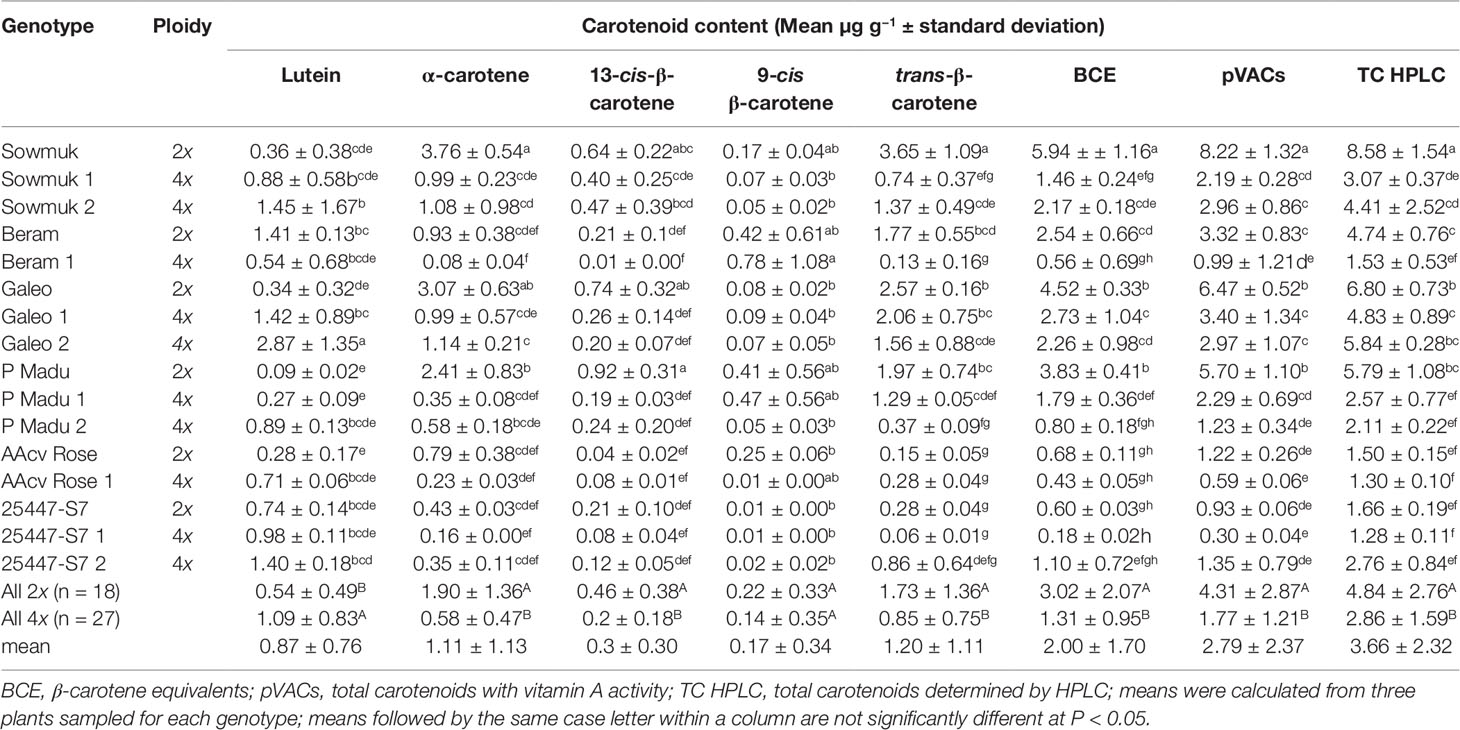
Data for carotenoid traits are presented in Table 4 and illustrated in Figure 4. The predominant carotenoids isolated were pVACs α-carotene, β-carotene (cis and trans versions) and lutein. Total carotenoids determined from HPLC (µg g−1 FW) was highest for diploid Sowmuk (8.58) and Galeo (6.80) and lowest for AAcv Rose (1.50) and 25447-S7 (1.66). Generally, lutein content was higher in tetraploids (1.09 µg g−1 FW) than in diploids (0.54 µg g−1 FW) while all other carotenoids were higher in diploids than in tetraploids except for 25447-S7. Individual genotypes showed increase in lutein content after doubling, but this was only significant for Sowmuk and Galeo, which had the highest carotenoid content at the diploid and tetraploid level. In contrast, induced tetraploids for the genotypes Sowmuk, Beram, Galeo, and P Madu had significantly lower pVACs α-carotene and trans β-carotene with consequently lower BCE and TC content (Table 4). Cis carotenes 13-cis β-carotene and 9-cis β-carotene decreased with doubling in all genotypes, but the decrease was not significant. Hierarchical clustering and heat maps (Figure 4) depicted 3 main clusters for entries based on carotenoid traits measured. The first main cluster contained three (Galeo, P Madu, and Sowmuk) of the five diploids sampled, which were characterized by high values for all carotenoid traits except lutein. The second cluster contained three tetraploids (Galeo 1, Sowmuk 2 and Galeo 2) which showed moderately lower values for the carotenoid traits except for lutein. The third cluster had the highest number of genotypes including 7 of the 10 tetraploids sampled. Genotypes in this cluster were characterized by low carotenoid values. In general, the heatmap shows a decline in carotenoid values of tetraploids when compared to diploids. With regard to clustering of traits, 9-cis-BC and lutein were distinct from the rest of the carotenoid traits measured.
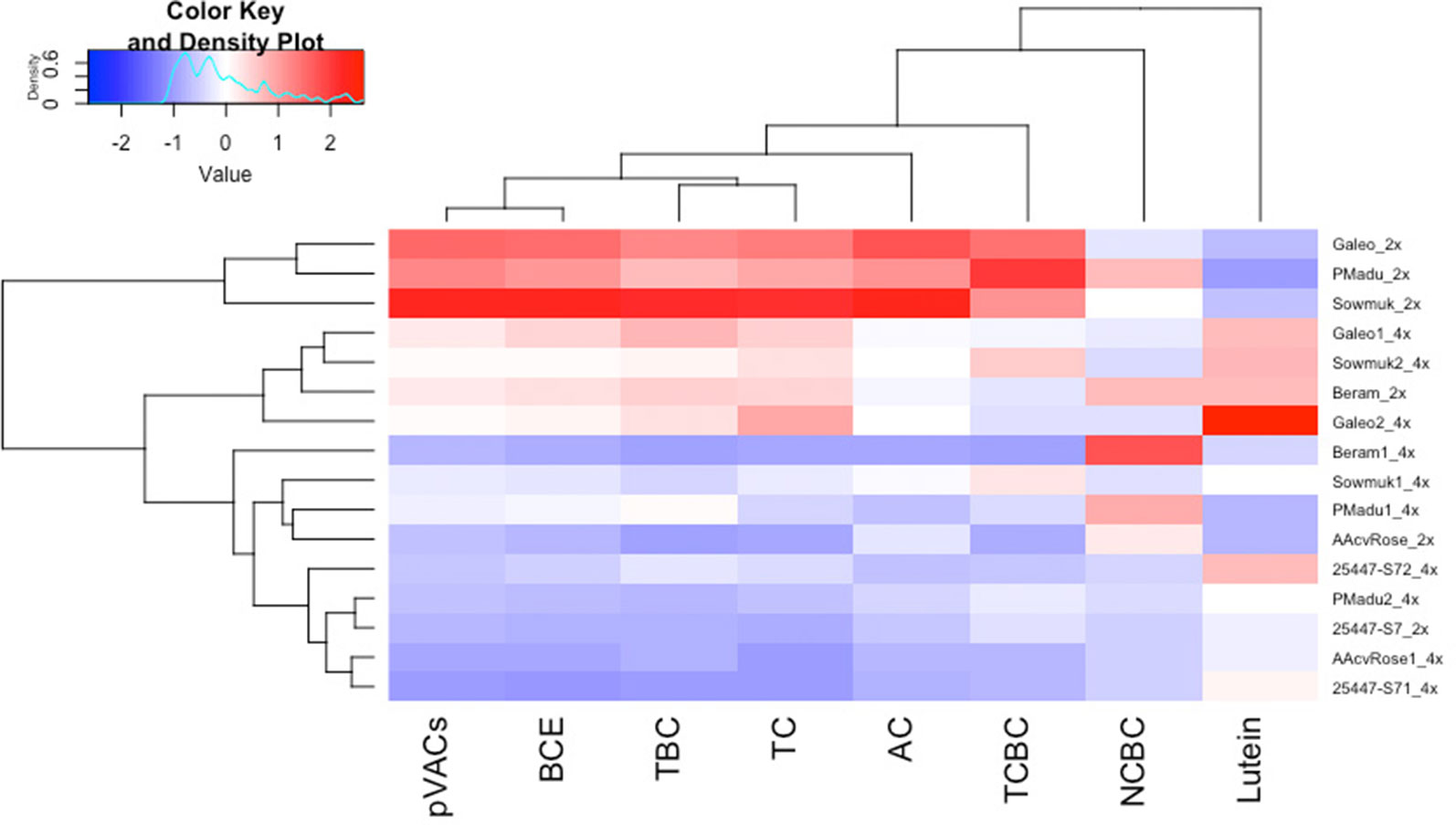
Figure 4 Heatmap and hierarchical clustering for carotenoid traits in diploid and induced tetraploid bananas. Genotype names followed by 2x and 4x indicates diploids and tetraploid lines respectively. AC, α-carotene; TCBC, 13-cis-β-carotene; NCBC, 9-cis β-carotene; TBC, trans-β-carotene; BCE, β-carotene equivalents; pVACs, total carotenoids with vitamin A activity; TC HPLC, total carotenoids determined by HPLC. Bright blue indicates lowest values while bright red indicates highest values for each trait.
Pollen Viability
Pollen viability via staining with TTC was used to estimate male fertility for four diploid genotypes and six induced tetraploids (Figure 5 and Table 3). Genotypes Sowmuk and Beram and their corresponding induced tetraploids were not included in the viability assessment because they recorded very low pollen counts (<10 per field) and exhibited degenerated male buds (Figures 3:1A, 1B). Pollen viability ranged from 70.51% to 86.81% with a mean of 76.95%. All genotypes tested had significantly higher viability than the Calcutta 4 diploid control. For genotypes Galeo and AAcv Rose, the pollen viability of the diploid was significantly higher than that of the induced tetraploids. Genotypes 25447-S7 and P Madu, recorded induced tetraploids with significantly higher viability than the diploid control (Table 3).
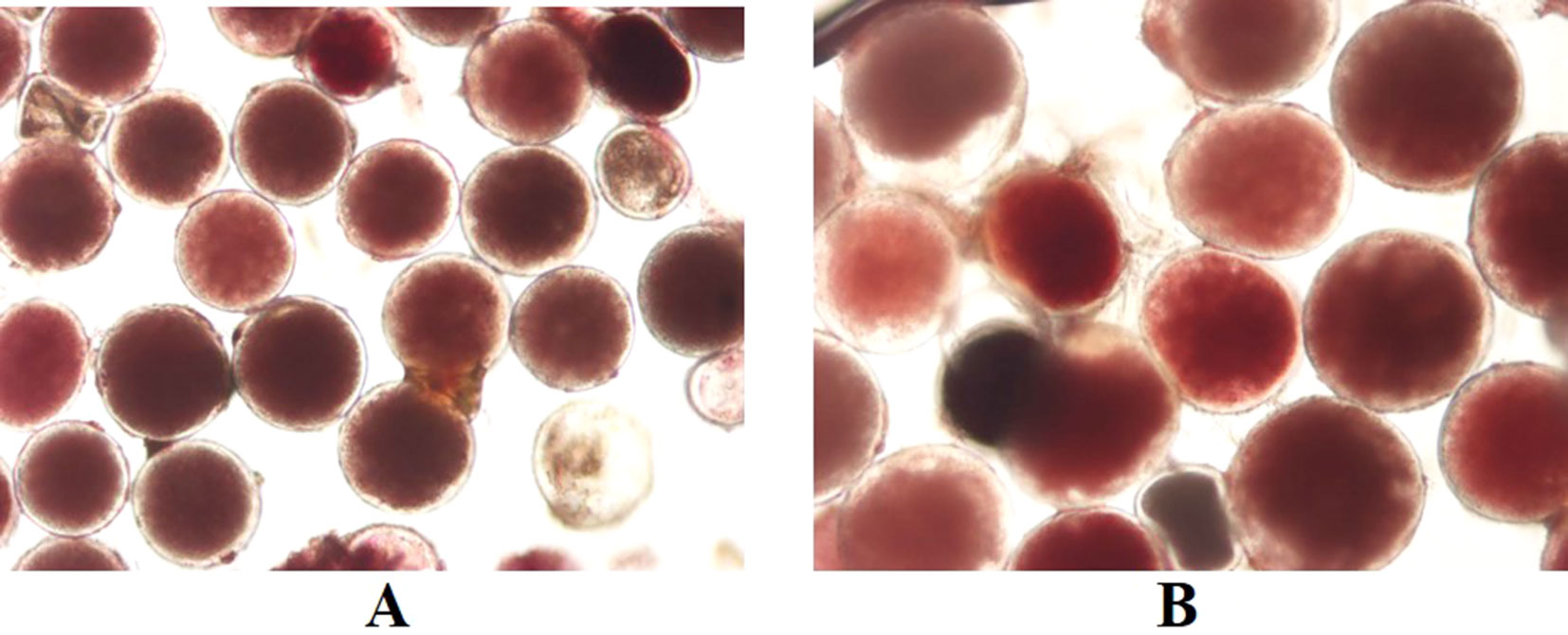
Figure 5 TTC stained pollen from diploid (A) and tetraploid (B) banana cultivar Galeo, showing high viability. Magnification 200x.
Discussion
Polyploidy is believed to impart phenotypic changes such as increased cell nuclei and size, leading to increased plant organs (gigas effect) as a consequence of gene duplications (Hegarty et al., 2013; Sattlser et al., 2016). Other resulting physiological or phenological characteristics may include larger leaves, greater vigor and biomass, increased resistance to biotic and abiotic stress, higher content of chemical substances, altered flowering time, increased fertility, and better post-harvest quality (Dhooghe et al., 2011). Although there are several reports on polyploidy induction in bananas, most reports have focused on developing in vitro polyploidization protocols without a comprehensive report on field evaluation of induced tetraploids. Thus, in this study, tetraploids obtained from six diploid genotypes were characterized for their agronomic attributes, pVAC contents, and fertility, in comparison with their diploid progenitors.
From field observations, induced tetraploids showed distinct morphological characteristics when compared to the diploid progenitors. Induced tetraploids were characterized by long curved leaves with weak petioles resulting in a drooping leaf habit or fallen/broken leaves, as previously observed in banana (Van Duren et al., 1996; Bakry et al., 2007; Do Amaral et al., 2015). Increased leaf size following polyploidization as a result of cell division and expansion has also been reported in several other plant species, with wide applications in the improvement of leaf size and flowers of ornamental plants (Huang et al., 2015; Tamayo-Ordóñez et al., 2016; Denaeghel et al., 2018; Parsons et al., 2019).
With regard to phenology (Table 2, Figure 2), longer days to flowering (28%) were observed in induced tetraploids than original diploids, but once plants had flowered, tetraploids matured faster (25%) than diploids. Plant height and girth at flowering was not significantly affected by doubling, but 47% less suckering was observed in induced tetraploids than in diploid plants. This was in contrast to observations by Do Amaral et al. (2015), who reported that plant height and pseudostem diameter increased while number of suckers decreased with polyploidization in Pisang Lilin. Variable changes in yield attributes following polyploidization, were also recorded. Although tetraploids had larger fruits, this did not necessarily result in increased bunch weight because of the associated decrease in number of hands, number of fruits and fruit length. Faster maturity in tetraploids could also be attributable to the fact that they had fewer fruits but the relationship between both traits is still unclear. Nyine et al. (2017) did not find significant correlation between both traits in bananas when a population consisting of 307 genotypes that included diploid, triploid, and tetraploid plants was assessed. Do Amaral et al. (2015) also observed fewer fruits in the tetraploid Pisang Lilin in the first cycle of evaluation, which later increased in the second cycle. Denaeghel et al. (2018) also reported variable effects of polyploidization in the phenotypes of Escallonia. Polyploidization is known to have played a key role in plant evolution and speciation, leading to better adaptation. While it is expected that induced polyploidization would result in superior traits, this varies among crop species. For example, more vigorous plants with larger flowers were observed in the tetraploid Snowdrop windflower (Zahumenická et al., 2018) while a decreased growth rate or plant vigor was observed in tetraploid apples (Xue et al., 2017). Generally, polyploidy is induced in vitro via the use of toxic anti-tubulin compounds such as oryzalin, which prevent microtubule formation and inhibits mitosis during metaphase, affecting plant growth and vigor. Podwyszyńska et al. (2015) reported that induced tetraploids of daylily exhibited poorer growth than original diploids in the first year but became more vigorous in the second year, possibly attributable to residual toxic effects of anti-mitotic agents.
Carotenoids are widely distributed in nature and specifically pVACs present in edible plant parts are important for human health. Fruit pVAC content is an important factor to consider for banana biofortification. The effect of polyploidy on compositional traits have been studied in some plant species, indicating increased or decreased concentrations (Trojak-Goluch and Skomra, 2013; Zahumenická et al., 2018). However, studies specific to carotenoids are limited. Carotenoids often present in banana fruit pulp are mainly the pVACs α-carotene and β-carotene (cis and trans versions) with smaller quantities of lutein (Davey et al., 2009). Using HPLC, the carotenoid content of banana fruit pulp of tetraploids and diploid plants were assessed to determine the effect of doubling. Polyploidization led to a significant decrease in pVACs in induced tetraploids when compared to diploid progenitors, which was associated with a corresponding increase in contents of the non-pVAC lutein (Figure 4). Trojak-Goluch and Skomra (2013) also reported significantly lower contents of essential oils in tetraploid hops in comparison to their diploid counterparts. On the other hand, Sanwal et al. (2010) reported higher gingerol content and antioxidant activity in tetraploid clones of ginger than that of the original diploids. Parsons et al. (2019) also observed significant changes in secondary metabolites with a 9% increase in cannabidol from tetraploid Cannabis sativa in comparison to their diploid progenitors. The expectation is that whole genome duplication may lead to increased gene expression from increased gene dosage, resulting in enhanced contents of compositional traits like carotenoids. However, this is not always the case as polyploidization may also lead to genome alterations affecting the expression of key genes implicated in hormonal regulation of plant developmental pathways (Sanglard et al., 2017; Xue et al., 2017). Polyploidization may also result in a changed post-transcriptional regulation and translational modification changes of proteins, which play a role in plant biological processes (Wang et al., 2017). Such alterations, possibly leading to the expression of deleterious alleles, may also be associated with poor field establishment and productivity of doubled lines.
The ability to generate progeny depends on gamete fertility, which is known to vary among genotypes. Understanding the pollen viability as an indicator of male fertility or seed set as an indicator of female fertility is critical to ascertain the potential for use of induced tetraploids in hybridizations. The absence of pollen in diploid and tetraploid Sowmuk and Beram (not included in pollen viability studies) is linked to the rapid degeneration of the male bud before maturity. While some diploids may be infertile, fertility may be restored at the tetraploid level. Bakry et al. (2009) reported male and female fertility in tetraploids induced from interspecific AB clones, which were otherwise sterile at the diploid level. In this study, it was observed that diploid and tetraploid versions of genotypes tested had high pollen viability (>70%), which was indeed higher than that of the male fertile diploid Calcutta 4. Notwithstanding, the tetraploid Galeo and AAcv Rose recorded lower pollen viability than their original diploids. However, for P Madu and 25447-S7, pollen viability of tetraploid versions were comparable to, or higher than that of their original diploids. The pollen viability values for induced tetraploids were higher than the 31 to 62.6% viability observed by Soares et al. (2016) on 12 banana tetraploid hybrids assessed for pollen viability using TTC staining, which is a fast and reliable method of accessing pollen viability. Goigoux et al. (2013) also detected viable pollen in doubled diploid Mlali clones Chicame 4x and Paka 4x, which was not significantly different from the diploids, indicating their potential as male parents in banana breeding. The pollen of tetraploid plants generally appeared larger than pollen of diploid plants (Figure 5) possibly also attributable to cell expansion following tetraploidization.
The prevailing ploidy levels in banana include diploids, triploids, and tetraploids, but triploids are believed to be the optimum ploidy level, since they exhibit better agronomic characteristics and produce non-functional gametes, which ensure seedlessness and edibility. In the banana breeding scheme, tetraploids are synthesized from 3x–2x crosses, and are further crossed with male diploid plants to generate triploids. Therefore, the goal for induced polyploidization in banana breeding is to directly synthesize tetraploids which can be crossed with elite diploids to generate triploids, while keeping the maximum genetic constitution of the diploid at the tetraploid level. Bakry et al. (2009) crossed induced tetraploid clones from Guyod, Galeo, IDN110, and Tjau Lagada with Calcutta 4 and obtained 347 hybrids, which were predominantly (98%) triploids. Oselebe et al. (2010) also pointed out the predominance of triploids in progenies from 4x–2x crosses. Nukaya et al. (2019) also reported the use of induced autotetraploid kumquats (Fortunella spp.) to generate seedless triploids through 4x–2x crosses and reciprocals. Although induced tetraploids generally showed inferior agronomic and pVAC traits in comparison to original diploids, it is expected that these tetraploids will constitute novel diversity for the generation of triploids with superior traits when crossed with elite diploids through transgressive segregation. However, it will be necessary to further evaluate hybrids from crosses with induced tetraploids to ascertain that traits of interests are ultimately expressed in the triploid progeny. Moreover, once tetraploid lines are produced, other desirable traits such as disease and pest resistance may be introgressed by incorporating different diploid cultivars and hybrids in the breeding scheme.
Conclusions
Biofortification through conventional breeding is becoming popular as a trusted approach to tackle micronutrient deficiency and breeding programs are utilizing available diversity to enhance micronutrient levels in staple crops. Although considerable diversity exists for pVACs in Musa genetic resources, genetic enhancement for pVAC content is still limited owing to several challenges with current breeding strategies. The use of in vitro polyploidization was explored as a breeding tool for banana improvement and the effect of polyploidization on agronomic characteristics and pVACs content of induced tetraploids was determined. Induced tetraploid plants showed distinct morphological characteristics, but generally had inferior agronomic characteristics and pVACs when compared to the original diploids. Nevertheless, preliminary fertility assessments indicated that induced tetraploids generated were pollen fertile, hence could provide a significant breeding pool for the synthesis of high pVAC triploid hybrids. While the seemingly negative effect of polyploidization on induced tetraploid is worth noting, polyploidization of diploids offers an opportunity for utilizing useful variability in diploid bananas which may otherwise remain inaccessible for breeding. However, further evaluations are needed to ascertain the (breeding) value of such induced tetraploids in hybridization programs aiming to generate high pVAC triploid hybrids.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
DA, ML, AB, and RS conceived and designed the study. DA conducted the study, performed the statistical analysis, and wrote the first draft of the manuscript. BM-D contributed to the carotenoid analysis. All authors contributed to the data interpretation, writing, and editing of the manuscript.
Funding
This work was part of the Ph.D. research of DA, with financial support from HarvestPlus (www.HarvestPlus.org), a global alliance of agriculture and nutrition research institutions working to increase the micronutrient density of staple food crops through biofortification. The views expressed do not necessarily reflect those of HarvestPlus. This work was also supported by donors through their contributions to the CGIAR Fund (http://www.cgiar.org/who-we-are/cgiar-fund/fund-donors-2/) particularly to the CGIAR Research Program for Roots, Tubers, and Bananas (CRP-RTB).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Amah, D., Alamu, E., Adesokan, M., van Biljon, A., Maziya-Dixon, B., Swennen, R., et al. (2019). Variability of carotenoids in a Musa germplasm collection and implications for provitamin A biofortification. Food Chem. X, 100024. doi: 10.1016/j.fochx.2019.100024
Bakry, F., Carreel, F., Jenny, C., Horry, J. P., (2009). “Breeding plantation tree crops: tropical species,” in Genetic improvement of banana. Ed. Jain, S. M. (New York: Springer), 3–50.
Bakry, F., de la Reberdiere, N. P., Pichot, S., Jenny, C. (2007). In liquid medium colchicine treatment induces non-chimerical doubled-diploids in a wide range of mono-and interspecific diploid banana clones. Fruits 62, 3–12. doi: 10.1051/fruits:2006043
Batte, M., Swennen, R., Uwimana, B., Akech, V., Brown, A., Tumuhimbise, R., et al. (2019). Crossbreeding East African Highland bananas: lessons learnt relevant to the botany of the crop after 21 years of genetic enhancement. Front. Plant Sci. 10, 81. doi: 10.3389/fpls.2019.00081
Borges, C. V., de Oliveira, A. V. B., Ramlov, F., da Silva Ledo, C. A., Donato, M., Maraschin, M., et al. (2014). Characterisation of metabolic profile of banana genotypes, aiming at biofortified Musa spp. cultivars. Food Chem. 145, 496–504. doi: 10.1016/j.foodchem.2013.08.041
Bouis, H. E., Saltzman, A. (2017). Improving nutrition through biofortification: a review of evidence from HarvestPlus, 2003 through 2016. Global Food Secur. 12, 49–58. doi: 10.1016/j.gfs.2017.01.009
Bouis, H. E., Welch, R. M. (2010). Biofortification—a sustainable agricultural strategy for reducing micronutrient malnutrition in the global south. Crop Sci. 50, S21–S32. doi: 10.2135/cropsci2009.09.0531
Brown, A., Tumuhimbise, R., Amah, D., Uwimana, B., Nyine, M., Mduma, H., et al., (2017). “Genetic Improvement of Tropical Crops,” in The genetic improvement of bananas and plantains (Musa spp.). Eds. Campos, H., Caligari, P. D. S. (New York: Springer), 219–240.
Christelová, P., De Langhe, E., Hřibová, E., Čížková, J., Sardos, J., Hušáková, M., et al. (2017). Molecular and cytological characterization of the global Musa germplasm collection provides insights into the treasure of banana diversity. Biodiversity Conserv. 26, 801–824. doi: 10.1007/s10531-016-1273-9
Davey, M. W., Van den Bergh, I., Markham, R., Swennen, R., Keulemans, J. (2009). Genetic variability in Musa fruit provitamin A carotenoids, lutein and mineral micronutrient contents. Food Chem. 115, 806–813. doi: 10.1016/j.foodchem.2008.12.088
Denaeghel, H. E. R., Van Laere, K., Leus, L., Lootens, P., Van Huylenbroeck, J., et al. (2018). The variable effect of polyploidization on the phenotype in Escallonia. Front. Plant Sci. 9, 354. doi: 10.3389/fpls.2018.00354
Dhooghe, E., Van Laere, K., Eeckhaut, T., Leus, L., Van Huylenbroeck, J. (2011). Mitotic chromosome doubling of plant tissues in vitro. Plant Cell Tissue Organ Culture 104, 359–373. doi: 10.1007/s11240-010-9786-5
Do Amaral, C. M., dos Santos-Serejo, J. D., Oliveira e Silva, S., da Silva Ledo, C. A., Amorim, E. P. (2015). Agronomic characterization of autotetraploid banana plants derived from ‘Pisang Lilin’ (AA) obtained through chromosome doubling. Euphytica 202, 435–443. doi: 10.1007/s10681-014-1320-0
Dolezel, J., Greilhuber, J., Suda, J. (2007). Estimation of nuclear DNA content in plants using flow cytometry. Nat. Protoc. 2, 2233–2244. doi: 10.1038/nprot.2007.310
Fungo, R., Pillay, M. (2011). β-Carotene content of selected banana genotypes from Uganda. Afr. J. Biotechnol. 10, 5423–5430. doi: 10.5897/AJB10.1047
Ganga, M., Chezhiyan, N. (2002). Influence of the antimitotic agents colchicine and oryzalin on in vitro regeneration and chromosome doubling of diploid bananas (Musa spp.). J. Hortic. Sci. Biotechnol. 77, 572–575. doi: 10.1080/14620316.2002.11511540
Garg, M., Sharma, N., Sharma, S., Kapoor, P., Kumar, A., Chunduri, V., et al. (2018). Biofortified crops generated by breeding, agronomy, and transgenic approaches are improving lives of millions of people around the world. Front. Nutr. 5, 12. doi: 10.3389/fnut.2018.00012
Global Nutrition Report. (2017). Nourishing the SDGs. Bristol, UK: Development Initiatives. http://globalnutritionreport.org/the-report/ accessed on April 2018.
Goigoux, S., Salmon, F., Bakry, F. (2013). Evaluation of pollen fertility of diploid and doubled-diploid clones of Mlali and their potential use for banana breeding. Acta Hortic. 986, 195–204. doi: 10.17660/ActaHortic.2013.986.20
Häkkinen, M. (2013). Reappraisal of sectional taxonomy in Musa (Musaceae). Taxon 62, 809–813. doi: 10.12705/624.3
Hamill, S. D., Smith, M. K., Dodd, W. A. (1992). In vitro induction of banana autotetraploids by colchicine treatment of micropropagated diploids. Aust. J. Bot. 40, 887–896. doi: 10.1071/bt9920887
Hegarty, M., Coate, J., Sherman-Broyles, S., Abbott, R., Hiscock, S., Doyle, J. (2013). Lessons from natural and artificial polyploids in higher plants. Cytogenet. Genome Res. 140, 204–225. doi: 10.1159/000353361
Heng, Z., Sheng, O., Yan, S., Lu, H., Motorykin, I., Gao, H., et al. (2017). Carotenoid profiling in the peel and pulp of 36 selected Musa varieties. Food Sci. Technol. Res. 23, 603–611. doi: 10.3136/fstr.23.603
Heslop-Harrison, J. S., Schwarzacher, T. (2007). Domestication, genomics and the future for banana. Ann. Bot. 100, 1073–1084. doi: 10.1093/aob/mcm191
Hippolyte, I., Jenny, C., Gardes, L., Bakry, F., Rivallan, R., Pomies, V., et al. (2012). Foundation characteristics of edible Musa triploids revealed from allelic distribution of SSR markers. Ann. Bot. 109, 937–951. doi: 10.1093/aob/mcs010
Huang, R., Liu, D., Zhao, M., Li, Z., Li, M., Sui, S. (2015). Artificially induced polyploidization in Lobularia maritima (L.) Desv. and its effect on morphological traits. HortScience. 50, 636–639. doi: 10.21273/HORTSCI.50.5.636
Němečková, A., Christelová, P., Čížková, J., Nyine, M., Van den houwe, I., Svačina, R., et al. (2018). Molecular and Cytogenetic Study of East African Highland Banana. Front. Plant Sci. 9, 1371. doi: 10.3389/fpls.2018.01371
Nukaya, T., Sudo, M., Yahata, M., Nakajo, Y., Ohta, T., Yasuda, K., et al. (2019). Characteristics in autotetraploid kumquats (Fortunella spp.) induced by colchicine treatment to nucellar embryos and their utilization for triploid breeding. Sci. Hortic. 245, 210–217. doi: 10.1016/j.scienta.2018.10.015
Nyine, M., Uwimana, B., Swennen, R., Batte, M., Brown, A., Christelová, D., et al. (2017). Trait variation and genetic diversity in a banana genomic selection training population. PloS One 12 (6), e0178734. doi: 10.1371/journal.pone.0178734
Ortiz, R. (2013). Conventional banana and plantain breeding. Acta Hortic. 986, 77–194. doi: 10.17660/ActaHortic.2013.986.19
Ortiz, R., Swennen, R. (2014). From crossbreeding to biotechnology-facilitated improvement of banana and plantain. Biotechnol. Adv. 32, 158–169. doi: doi.10.1016/j.biotechadv.2013.09.010
Oselebe, H. O., Obi, I. U., Uguru, M. I. (2010). Predicting hybrid performances from interploidy crosses in Musa species. Aust. J. Crop Sci. 4, 415–420.
Parsons, J. L., Martin, S. L., James, T., Golenia, G., Boudko, E. A., Hepworth, S. R. (2019). Polyploidization for the genetic improvement of Cannabis sativa. Front. Plant Sci. 10, 476. doi: 10.3389/fpls.2019.00476
Perrier, X., Bakry, F., Carreel, F., Jenny, C., Horry, J. P., Lebot, V., et al. (2009). Combining biological approaches to shed light on the evolution of edible bananas. Ethnobot. Res. Appl. 7, 199–216.
Perrier, X., Jenny, C., Bakry, F., Karamura, D., Kitavi, M., Dubois, C., et al. (2018). East African diploid and triploid bananas: a genetic complex transported from South-East Asia. Ann. Bot. 123, 19–36. doi: 10.1093/aob/mcy156
Podwyszyńska, M., Gabryszewska, E., Dyki, B., Stępowska, A. A., Kowalski, A., Jasiński, A. (2015). Phenotypic and genome size changes (variation) in synthetic tetraploids of daylily (Hemerocallis) in relation to their diploid counterparts. Euphytica 203, 1–16. doi: 10.1007/s10681-014-1212-3
R Core Team. (2018). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org.
Salles Pio, L. A., Pasqual, M., de Oliveira e Silva, S., Souza Rocha, H., Mara Magalhaes, H., de Almeida Santos-Serejo, J. (2014). Inducing and identifying artificially-induced polyploidy in bananas. Afr. J. Biotechnol. 13, 3748–3758. doi: 10.5897/AJB2014.14009
Sanglard, N. A., Amaral-Silva, P. M., Sattler, M. C., de Oliveira, S. C., Nunes, A. C., Soares, T. C., et al. (2017). From chromosome doubling to DNA sequence changes: outcomes of an improved in vitro procedure developed for allotriploid “Híbrido de Timor” (Coffea arabica L.× Coffea canephora Pierre ex A. Froehner). Plant Cell Tissue Organ Culture 131, 223–231. doi: 10.1007/s11240-017-1278-4
Sanwal, S. K., Rai, N., Singh, J., Buragohain, J. (2010). Antioxidant phytochemicals and gingerol content in diploid and tetraploid clones of ginger (Zingiber officinale Roscoe). Sci. Hortic. 124, 280–285. doi: 10.1016/j.scienta.2010.01.003
Sattlser, M. C., Carvalho, C. R., Clarindo, W. R. (2016). The polyploidy and its key role in plant breeding. Planta 243, 281–296. doi: 10.1007/s00425-015-2450-x
Soares, T. L., de Souza, E. H., de Carvalho Costa, M. A., Santos-Serejo, J. A. (2016). Viability of pollen grains of tetraploid banana. Bragantia 75, 145–151. doi: 10.1590/1678-4499.328
Soares, T. L., de Souza, E. H., Sampaio, L. F., de Carvalho Costa, M. A., de Oliveira e Silva, S., dos Santos-Sejero, J. A. (2015). Effect of collection time on the viability of banana pollen grains. Afr. J. Biotechnol. 14, 1207–1214. doi: 10.5897/AJB2015.14449
Tamayo-Ordóñez, M. C., Espinosa-Barrera, L. A., Tamayo-Ordóñez, Y. J., Ayil-Gutiérrez, B., Sánchez-Teyer, L. F. (2016). Advances and perspectives in the generation of polyploid plant species. Euphytica 209, 1–22. doi: 10.1007/s10681-016-1646-x
Tenkouano, A., Pillay, M., Ortiz, R., (2011). “Banana breeding: constraints and progress,” in Breeding techniques. Eds. Pillay, M., Tenkouano, A. (Boca Raton, Florida: CRC Press), 181–202.
Trojak-Goluch, A., Skomra, U. (2013). Artificially induced polyploidization in Humulus lupulus L. and its effect on morphological and chemical traits. Breed. Sci. 63, 393–399. doi: 10.1270/jsbbs.63.393
Wang, Z., Fan, G., Dong, Y., Zhai, X., Deng, M., Zhao, Z., et al. (2017). Implications of polyploidy events on the phenotype, microstructure, and proteome of Paulownia australis. PloS One. 12, e0172633. doi: 10.1371/journal.pone.0172633
Van Duren, M., Morpurgo, R., Dolezel, J., Afza, R. (1996). Induction and verification of autotetraploids in diploid banana (Musa acuminata) by in vitro techniques. Euphytica 88, 25–34. doi: 10.1007/BF00029262
Xue, H., Zhang, B., Tian, J. R., Chen, M. M., Zhang, Y. Y., Zhang, Z. H., et al. (2017). Comparison of the morphology, growth and development of diploid and autotetraploid ‘Hanfu’ apple trees. Sci. Hortic. 225, 277–285. doi: 10.1016/j.scienta.2017.06.059
Zahumenická, P., Fernández, E., Šedivá, J., Žiarovská, J., Ros-Santaella, J. L., Martínez-Fernández, D., et al. (2018). Morphological, physiological and genomic comparisons between diploids and induced tetraploids in Anemone sylvestris L. Plant Cell Tissue Organ Culture 132, 317–327. doi: 10.1007/s11240-017-1331-3
Keywords: banana genetic improvement, biofortification, carotenoid content, diploid bananas, in vitro polyploidization, vitamin A, pollen viability
Citation: Amah D, van Biljon A, Maziya-Dixon B, Labuschagne M and Swennen R (2019) Effects of In Vitro Polyploidization on Agronomic Characteristics and Fruit Carotenoid Content; Implications for Banana Genetic Improvement. Front. Plant Sci. 10:1450. doi: 10.3389/fpls.2019.01450
Received: 22 June 2019; Accepted: 17 October 2019;
Published: 12 November 2019.
Edited by:
Inaki Hormaza, Institute of Subtropical and Mediterranean Hortofruticultura La Mayora (IHSM), SpainReviewed by:
Katarzyna Glowacka, University of Nebraska-Lincoln, United StatesPatrick Ollitrault, Unité Mixte de Recherche Amélioration Génétique et Adaptation des Plantes (UMR Agap) Centre de Coopération Internationale en Recherche Agronomique pour le Développement (CIRAD), France
Copyright © 2019 Amah, van Biljon, Maziya-Dixon, Labuschagne and Swennen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Delphine Amah, RC5BbWFoQGNnaWFyLm9yZw==
 Delphine Amah
Delphine Amah Angeline van Biljon
Angeline van Biljon Bussie Maziya-Dixon3
Bussie Maziya-Dixon3 Maryke Labuschagne
Maryke Labuschagne Rony Swennen
Rony Swennen