- 1Institut de Biologie Moléculaire des Plantes (IBMP), UPR2357 CNRS, Université de Strasbourg, Strasbourg, France
- 2State Key Laboratory of Subtropical Silviculture, Zhejiang A&F University, Hangzhou, China
- 3College of Bioscience and Biotechnology, International Associated Laboratory of CNRS-Fudan-HUNAU on Plant Epigenome Research, Hunan Agricultural University, Changsha, China
The H2A/UBIQUITIN-binding proteins AtZRF1a/b have been reported as key regulators involved in multiple processes of Arabidopsis plant growth and development. Yet, the cellular and molecular mechanisms underlying the mutant phenotype remain largely elusive. Here we show that loss-of-function of AtZRF1a/b causes defective root elongation and deformed root apical meristem organization in seedlings. The premature termination of the primary root in the atzrf1a;atzrf1b double mutant is associated with an advanced onset of endoreduplication and subsequent consumption of reservoir stem cells. Cytological analyses using cell type-specific markers and florescent dyes indicate that AtZRF1a/b are involved in maintenance of proper cell layer organization, determinacy of cell identity, and establishment of auxin gradient and maximum at the root tip. During embryogenesis AtZRF1a/b act dominantly in regulating the maintenance of ground tissue initial cells and production of lateral root cap. Lastly, quantitative real-time polymerase chain reaction analysis shows mis-expression of some key genes involved in regulating cell patterning, cell proliferation and/or hormone pathways. Our results provide important insight into AtZRF1a/b function in cell fate determinacy and in establishment and maintenance of proper stem cell reservoir during embryonic and post-embryonic root development.
Introduction
The Arabidopsis root has a well-organized structure with simple longitudinal organization and few well-defined cell lineages, providing an excellent model system to investigate asymmetric cell division and cell fate determinacy. Root development relies on the root apical meristem (RAM), which not only maintains stem cell self-renewal but also provides different types of daughter cells, which subsequently undergo expansion to form elongation zone and then differentiation to form root hair zone (maturation zone). The balance between cell division and cell differentiation determines RAM size. The RAM consists of the proliferation domain with high dividing-cell content and the transition domain with low dividing-cell content along the longitudinal axis (Ivanov and Dubrovsky, 2013). In Arabidopsis (Arabidopsis thaliana) root proliferation domain, four types of initial stem cells surrounding approximately four quiescent center (QC) cells (Scheres et al., 1994), together constitute the stem cell niche (SCN). At the distal region, columella cells are generated from anticlinal divisions of the columella stem cell initials, and epidermal cell and lateral root cap are derived from sequentially anticlinal and periclinal divisions of their common epidermal/lateral root cap initials. At the proximal region, cortex and endodermis are derived from the periclinal division of their common ground tissue initials, and stele is formed from the stele stem cell initials. Within the root SCN, QC cells with slowly mitotic activity provide a reservoir for maintenance and replenishment of the surrounding initial stem cells, which exhibit high frequency of cell divisions (Heyman et al., 2013). These ascribed basic RAM cell pattern has been originally established during embryogenesis and maintained during postembryonic primary root growth (Dolan et al., 1993; Scheres et al., 1994).
Transcription factors and phytohormone auxin play critical roles in regulating RAM maintenance and stem cell homeostasis (Drisch and Stahl, 2015). ETHYLENE RESPONSE FACTOR115 (ERF115) as a rate-limiting factor of QC divisions is expressed in dividing QC cells, but it is usually restrained through proteolysis by the APC/CCCS52A2 ubiquitin ligase in normal condition (Heyman et al., 2013). WUSCHEL-RELATED HOMEOBOX 5 (WOX5) as one of the most important root stem cell regulatory factor is specifically expressed in QC cells, necessary for the maintenance of undifferentiated state of surrounding stem cells (Sarkar et al., 2007). The CLAVATA3/EMBRYO SURROUNDING REGION (CLE) peptide CLE40 from columella cells is perceived via its receptors ARABIDOPSIS CRINKLY4 (ACR4) and CLAVATA1 (CLV1) to modulate the expression level and positioning of WOX5 (CLE-WOX5 feedback loop), consequently regulating columella stem cell fates (Stahl et al., 2009; Stahl et al., 2013). It is known that auxin signal and distribution are widely involved in root patterning and polarity, SCN maintenance, and distal stem cell identity (Sabatini et al., 1999; Friml et al., 2002; Ding and Friml, 2010). In fact, auxin signal and transcriptional factor usually function in a coordinate way. For instance, WOX5 action is balanced through the activity of indole-3-acetic acid 17 (IAA17) auxin response repressor, together forming WOX5–IAA17 feedback circuit, essential for the maintenance of auxin gradient in RAM and the auxin-mediated columella stem cell differentiation (Tian et al., 2013; Tian et al., 2014).
Polycomb Group (PcG) proteins are conserved in animals and plants, mainly constitute Polycomb Repressive Complex1 (PRC1) and PRC2, acting as key epigenetic regulators during organism growth and developmental processes (reviewed in (Hennig and Derkacheva, 2009; Xiao and Wagner, 2015; Forderer et al., 2016). In Arabidopsis, PcG proteins play essential roles in root development. PRC2 components CLF, SWN, EMF2, VRN2, and FIE are involved in root meristem development and vascular cell proliferation in the maturation zone (Aichinger et al., 2011; de Lucas et al., 2016). CLF also associates with EMF2 to repress founder cell establishment during lateral root initiation associated with down-regulation of root auxin maxima (Gu et al., 2014). Additionally, PRC2 deficiency gives rise to mitotic reactivation and somatic embryogenesis in terminally differentiated root hairs (Ikeuchi et al., 2015). Consistently, the PRC1 subunits AtRING1a/b and AtBMI1a/b/c inhibit the formation of pkl-type root-phenotype displaying embryonic traits in primary root mainly through preventing an ectopic expression of embryonic master regulators (Bratzel et al., 2010; Chen et al., 2010). The AtBMI1-interacting factors VAL1/2 also have a similar function (Hoppmann et al., 2011; Yang et al., 2013).
The ZUOTIN-RELATED FACTOR (ZRF) proteins in eukaryotes constitute a novel clade of HSP40 family, which in general serves as co-chaperone of HSP70s to assist protein translation, folding, unfolding, translocation and degradation (Chen et al., 2014). However, the human ZRF1 was found to compete with and replace PRC1 RING1B from chromatin via competitively binding H2Aub1 mark, and to favor H2Aub1 removal via recruiting the specific deubiquitinase USP21, consequently leading to repressive-to-active chromatin state switch (Richly et al., 2010). In Arabidopsis, two ZRF1 homologs, AtZRF1a and AtZRF1b, showed redundant functions. AtZRF1b can bind ubiquitin in vitro and pull-down H2Aub1 and H2A from plant protein extracts (Feng et al., 2016), which is in agreement with the human ZRF1 acting as a H2Aub1 reader. The AtZRF1a/b genes display broad expression pattern, but with higher levels in the dividing cell-enriched tissues, e.g. meristem, floral bud, and developing embryo. Loss-of-function of AtZRF1a/b causes pleiotropic abnormalities including delayed seed germination, plant dwarfism, formation of multiple ectopic meristems, and defects in flower development and gametophyte transmission as well as embryogenesis (Feng et al., 2016; Guzman-Lopez et al., 2016). The atzrf1a;atzrf1b mutant displays severely disrupted root developmental phenotype; yet, the underlying mechanism is far from clear.
In this study, we investigated in detail the root developmental defects of the atzrf1a;b mutant. We demonstrated that AtZRF1a/b play crucial roles in regulating stem cell activity, cell layer organization, cell fate determinacy and cell division orientation in RAM during embryonic and post-embryonic root development.
Materials and Methods
Plant Materials and Growth Conditions
The atzrf1a-1;atzrf1b-1 and atzrf1a-2;atzrf1b-1 double mutants (Feng et al, 2016) and some marker lines CYCB1;1:Dbox-GUS (Colon-Carmona et al., 1999), WOX5::erGFP (Blilou et al., 2005), SCR::SCR-YFP (Heidstra et al., 2004), CO2::H2B-YFP (Heidstra et al., 2004), DR5rev::GFP (Friml et al., 2003) have been described previously. The Haseloff enhancer trap GFP lines J1092 (N9147) and J2341 (N9118) were obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org) or the European Arabidopsis Stock Centre (http://arabidopsis.info). The different reporter lines were respectively introgressed by crossing and homogenized by backcrossing two generations with atzrf1a-1;atzrf1b-1 double mutant. The genotyping of atzrf1a-1 or atzrf1b-1 mutation sites was performed as described (Feng et al, 2016). The existences of reporter genes were detected by directly GUS staining or fluorescent observation. Homozygous atzrf1a-1;atzrf1b-1 mutant harboring each reporter gene was used for further analysis, and WT harboring the corresponding reporter was set as control. Seeds were surface sterilized (70 and 95% ethanol for 10 min, respectively) and plated on MS medium (MS salts, 0.9% sucrose, pH 5.7, 0.9% bactoagar). After stratification at 4°C for 2 days in the dark, seeds were transferred to a growth chamber at 22°C in a 16/8 h light/dark regime. After 2 weeks, seedlings were transferred to soil at the same condition.
Phenotypic Analysis
For root length comparison, mutant and control plants were grown side by side on a same plate. Root length was measured from the root tip to the root/hypocotyl border of vertically grown seedlings via ImageJ software. The lengths of RAM, proliferation domain and transition domain were measured according to the defined criteria (Napsucialy-Mendivil et al., 2014). In mature embryo, the number of cortex cells was counted in a cell file extending from QC to hypocotyl rootward border; the maximum number of columella cell layers was counted in the columella cell cap including initials. All the above experiments were repeated three times (mean ± SE), each repeat containing at least 15 plants.
Histology and Microscopy
GUS staining assay was performed as described (Chen et al., 2010). Briefly, whole transgenic marker seedlings were sequentially treated by fixative cold acetone for 30min, and GUS staining buffer (100 mM phosphate buffer, pH 7.4, 2 mM ferricyanide, and 0.5 mM ferrocyanide, and 4 mM X-Gluc) at 37°C in the dark for 4 h. Samples were cleared overnight in 90% lactic acid and were photographed with a differential interference contrast microscope (Leica). For whole-mount visualization, the roots were directly cleared in chloral hydrate solution (chloral hydrate/glycerol/H2O, 8/2/1, m/v/v). For starch granule staining, roots were stained with Lugol solution (Chen et al, 2010). For GFP/YFP observation, the roots were counterstained with 20 µg/ml of propidium iodide. For RAM observation, mature embryos and root tips were stained via the mPS-PI method (Truernit et al., 2008).
For young embryo observation, embryos at different stages were dissected from developing seeds with tweezers and fine syringe needles, and either were stained with newly-developed cell-wall Dye SCRI renaissance 2200 (SR2200) as described (Musielak et al., 2015) or were cleared by chloral hydrate solution for differential interference contrast observation.
All the above experiments were repeated three times, each repeat containing at least 10 plants.
Quantitative Real-Time Polymerase Chain Reaction
Total RNA was isolated from 7-day-old roots using the NucleoSpin RNA Plant kit (Macherey-Nagel, Germany). First strand complementary DNA synthesis was carried out using Improm-II reverse transcriptase (Promega). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed on a light cycler 480II (Roche) according to the manufacturer’s instructions. Reaction volumes (10 µl) were comprised of 5 µl of 2× LightCycler 480 SYBR Green I Master (Roche), 2 µl of primer mix, 1 µl of template complementary DNA, and 2 µl of ddH2O. Reactions were performed at 95°C for 10 min, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. The specific PCR product was validated through melting curve analysis. PP2A and EXP was used as internal reference. Data represent mean values of three technical replicates on three independent biological repeats. The primers for qRT-PCR are listed in Supplementary Table S1.
Flow Cytometry
Nuclei were prepared from roots of 1-week-old plants and analyzed on an Attune™ Acoustic Focusing Cytometer (Applied Biosystems, USA). Typically, 10,000 nuclei per sample from at least 100 WT roots and 200 mutant roots were analyzed. Data represent mean values of three technical replicates on three independent biological repeats. Endoreduplication index (EI) which represents the average number of endocycles undergone by a typical nucleus (EI = [0 · n2C + 1 · n4C + 2 · n8C + 3 · n16C]/[n2C + n4C + n8C + n16C]) was calculated as published (Barow and Meister, 2003; Vanstraelen et al., 2009).
Results
Loss of AtZRF1a/b Causes Primary Root Growth Arrest
The atzrf1a-1;atzrf1b-1 and atzrf1a-2;atzrf1b-1 double mutants displayed the same defective root phenotype, whereas WT and atzrf1 single mutants displayed the normal root growth (Supplementary Figure S1). Thus, the atzrf1a-1;atzrf1b-1 (briefly as atzrf1a;b) mutant was chosen for further analysis in this study. The atzrf1a;b mutant showed a drastically reduced root growth rate (Figure 1A), leading to an extremely short-root phenotype, e.g. only ∼2 mm in length for the mutant roots as compared to ∼20 mm for the WT roots at 7 days after stratification (DAS). The mutant root displayed mature zone characteristically covered by root hairs that arise in close proximity to the root tip (Figure 1B), indicating a drastic reduction of the meristem and elongation zone as well as developmentally advanced cell differentiation. The mutant primary roots ceased to grow as early as at 14 DAS (Figure 1D), whereas the WT primary roots continuously grew and produced lateral roots (Figure 1C). Later on, the mutant plants produced many adventitious roots (Figures 1E–G), which sustain plant growth in water and nutrition acquisition.
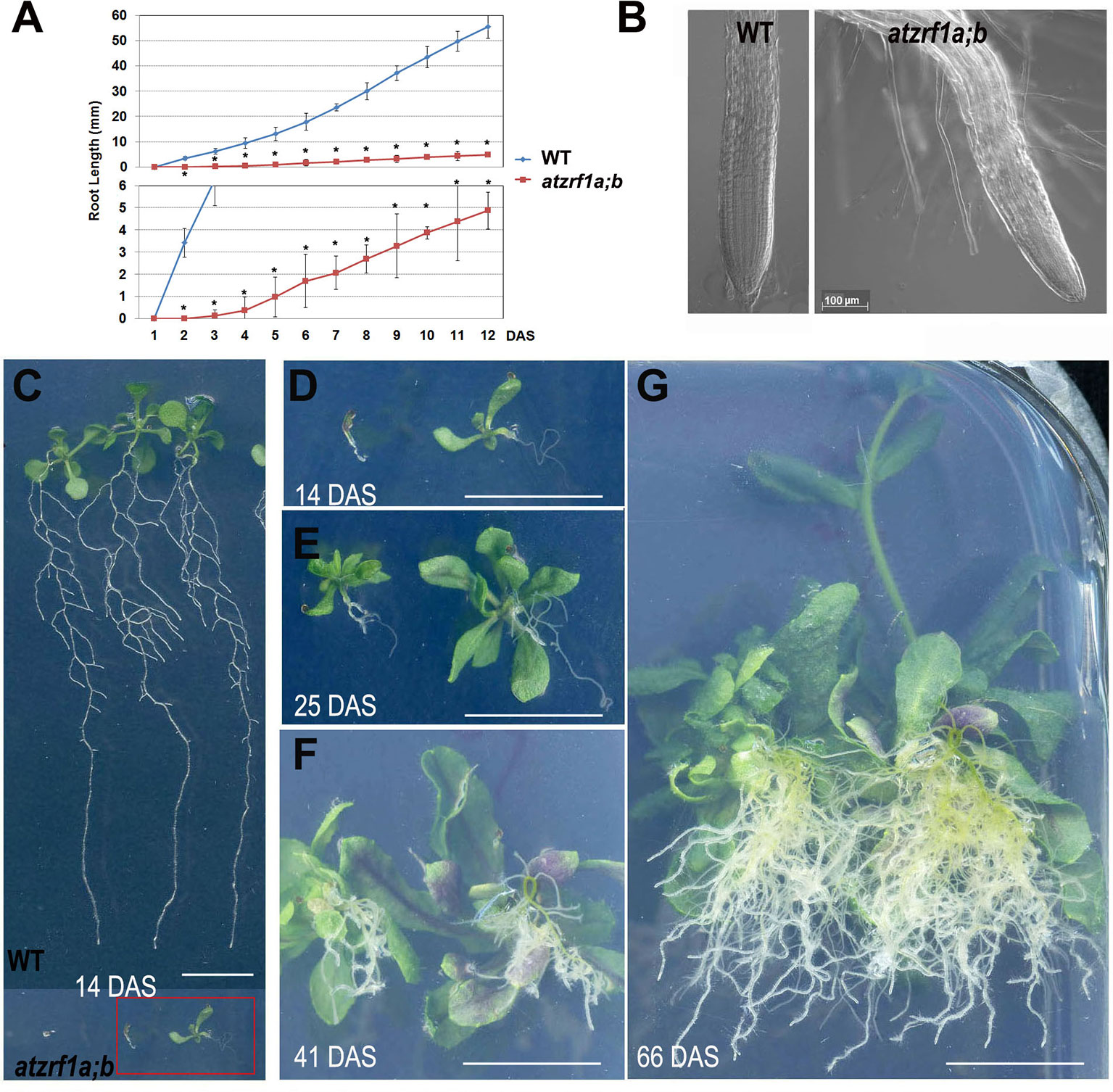
Figure 1 Defective primary root development in the atzrf1a;b mutant. (A) Comparison of primary root growth between the wild type (WT) and the atzrf1a;b mutant plants during 12 days after stratification (DAS). Histogram at the bottom shows a magnification of the Y-axe scale to better view the time course of the atzrf1a;b mutant growth. Data represent mean values of three technical replicates on three independent biological repeats. Asterisk indicates student’s t-test statistically significant differences at P < 0.01. (B) Representative images of the WT root tip and the atzrf1a;b mutant root tip at 5 DAS. (C) Representative WT seedlings showing continuous primary root growth and proliferation of lateral roots at 14 DAS. (D–G) Representative seedlings of atzrf1a;b showing primary root growth arrest and adventitious root development at 14, 25, 41, 66 DAS. Bars = 50 μm in (B), and 1 cm in (C) to (G).
The atzrf1a;b Mutant Root Exhibits Cell Division Arrest and Precocious Cell Differentiation
Hereinafter we focused on primary roots to investigate AtZRF1a/b function. Root growth largely depends on the RAM activity in which cells undergo mitotic cell division, cell expansion and cell differentiation. RAM size is relatively fixed in WT and constantly maintained by the dynamic balance between cell proliferation and cell differentiation. We found that RAM including proliferation domain and transition domain in atzrf1a;b is significantly shorter than that in the WT control (Figures 2A, C and Supplementary Figure S2). Sometimes, the cell arrangement was largely disorganized in atzrf1a;b RAM, so that it was hardly distinguished among different cell types (Figure 2C and Supplementary Figure S3). The root diameters in mutant became evidently narrow mainly due to thinner stele (Figures 2B, C). Moreover, the average height to width ratio of the RAM cortical cells in mutant (1.3, n = 30) was higher than that in WT (0.7, n = 30), suggesting cell elongation in advance in mutant RAM. In 3-week-old seedling, the RAM region in mutant was almost completely consumed (Supplementary Figure S4). These results indicated that RAM cells in atzrf1a;b mutant were undergoing premature differentiation.
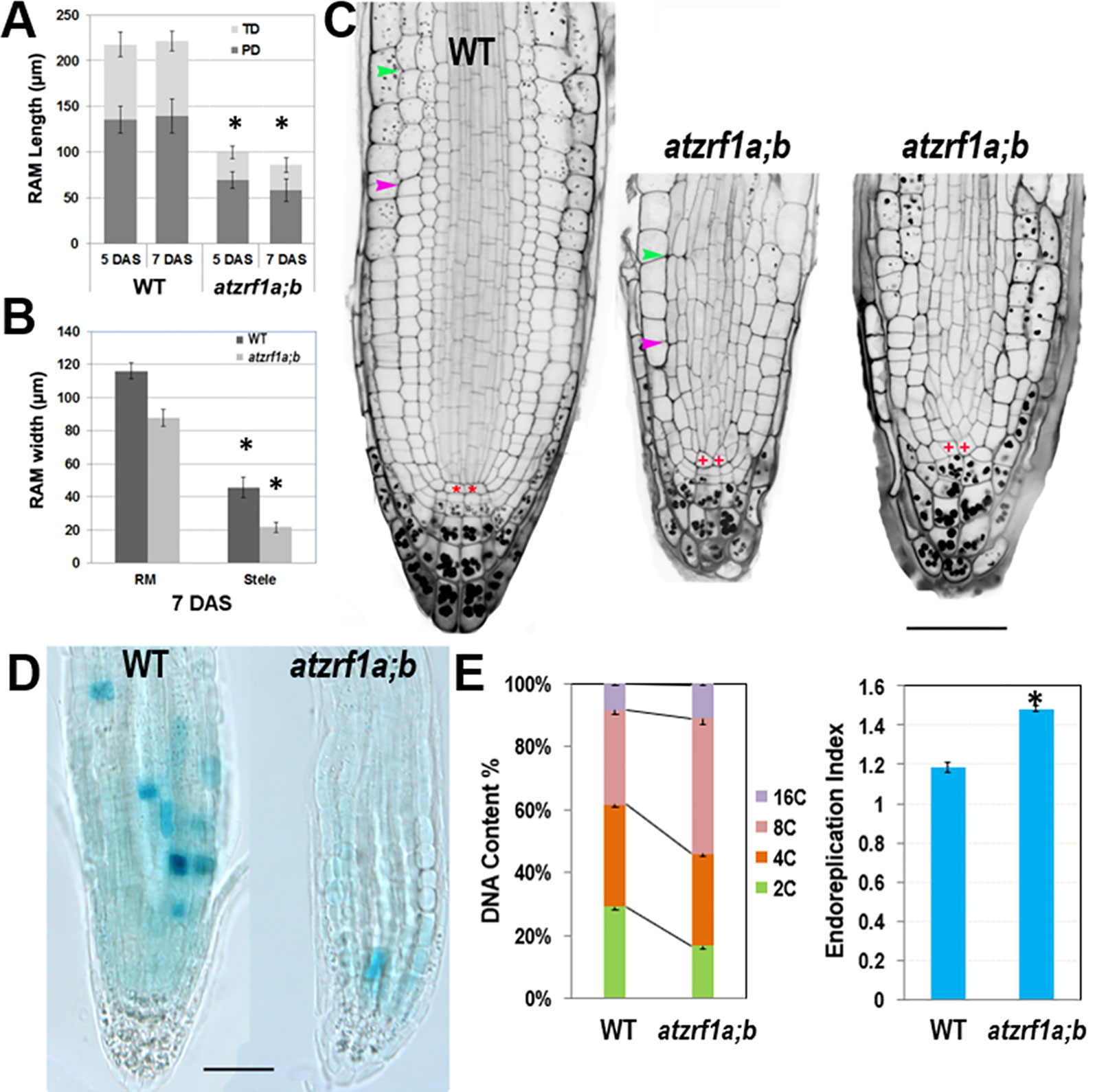
Figure 2 Defective root development is associated with cell division arrest and precocious cell differentiation in the atzrf1a;b mutant. (A, B) Comparison of meristem size between the wild-type (WT) control and the atzrf1a;b mutant at 5 or 7 days after stratification (DAS). The length of RAM including proliferation domain (PD) and transition domain (TD) indicates the vertical distance from QC to PD distal border, and finally to TD distal border. RAM width indicates the diameter at TD distal position. (C) 5-day-old RAM in WT and atzrf1a;b after mPS-propidium iodide staining. Starch granule is visible in dark and accumulates in the root cap. Pink and green arrowheads indicate the distal borders of PD and TD, respectively. Asterisk indicates position of QC cells. Bar = 50 µm. (D) GUS activity of CYCB1;1::Dbox-GUS reporter in 5-day-old WT and atzrf1a;b mutant. Blue staining indicates for positive GUS activity. Bar = 50 µm. (E) Ploidy analysis in 7-day-old WT and atzrf1a;b mutant roots. Percentages of 2C, 4C, 8C, and 16C DNA content nuclei are shown. Data show means ± SE from three biological repeats. Asterisk indicates student’s t-test statistically significant differences at P < 0.01.
In order to investigate the underlying mechanism, we introgressed into the atzrf1a;b mutant the CYCB1;1::Dbox-GUS reporter which marks the cells at the G2-to-M transition of the cell cycle (Colon-Carmona et al., 1999). Compared to WT, the mutant RAM had reduced GUS staining (Figure 2D), which indicates attenuated mitotic activity. On the other hand, we investigated the level of root endopolyploidy, which is associated with cell differentiation. In line with the reduced mitotic activity, EI in atzrf1a;b was significantly increased, mainly due to the greatly elevated proportion of 8C and to a less degree of 16C cells (Figure 2E), indicating for an early mitosis-to-endocycle transition. Taken together, our data suggest that the decreased mitotic cell division capacity and the advanced onset of endoreduplication lead to reduced RAM size in the atzrf1a;b mutant.
AtZTf1a/b Are Required for Organization and Maintenance of Root Stem Cell Niche
To gain insight about cell fate determinacy, root cell-type specific markers were introgressed into the atzrf1a;b mutant. Consistent with previous report (Blilou et al., 2005), WOX5::erGFP showed specifically expression in QC cells in WT (Figure 3A). In contrast, it was found expressed in QC often with aberrant morphology as well as in adjacent cortex/endodermis initial cells in the atzrf1a;b mutant (Figures 3B–D). The abnormal pattern observed in the mutant might provide inappropriate position cues to surrounding stem cells, as reflected by irregular SCN formation in the mutant (Figure 2C). The J2341 enhancer trap marker carrying ER-tethered GFP (Kim et al., 2005) was found expressed in approximately four columella initials in WT (Figure 3E), but was found only expressed in one cell below the QC in the atzrf1a;b mutant (Figure 3F), indicating weakened columella initial stem cell activity in the mutant. The SCR::SCR-YFP endodermis marker (Heidstra et al., 2004) was found expressed in endodermis, cortex/endodermis initials and QC in both WT (Figure 3G) and the atzrf1a;b mutant (Figure 3H). Remarkably, in the mutant, additionally SCR::SCR-YFP showed weak but significant expression in cells normally corresponding to the cortex cells (Figure 3H). To further verify this mutant defect, we examined expression pattern of CO2::H2B-YFP, a marker specific for cortex cells (Heidstra et al., 2004). As expected, CO2::H2B-YFP was found expressed specifically in cortex layer cells in WT (Figure 3I). In the atzrf1a;b mutant, however, only a few cells from the cortex layer showed roughly normal level of CO2::H2B-YFP expression whereas the other cells showed low or absence of CO2::H2B-YFP expression (Figure 3J). These observations using both SCR::SCR-YFP and CO2::H2B-YFP indicate that the atzrf1a;b mutant is impeded in establishment and maintenance of the cortex cell fate during root development. The J1092 enhancer trap line (Blilou et al., 2002) displayed strong GFP signal in the lateral root cap initial cells and to a lesser extent in the columella root cap initial cells in WT (Figure 3K), but showed rather uniform expression level throughout the root cap initial cells in the atzrf1a;b mutant (Figure 3L), implying a weakened distinction between the two types of root cap initial cells in the mutant. Taken together, our analyses using cell fate markers indicate that AtZRF1a/b are required for whole SCN architecture, including QC localization, columella stem cell maintenance, separation between cortex and endodermis identities as well as stable maintenance of cortex cell fate, and distinction between the columella and lateral root cap initials.
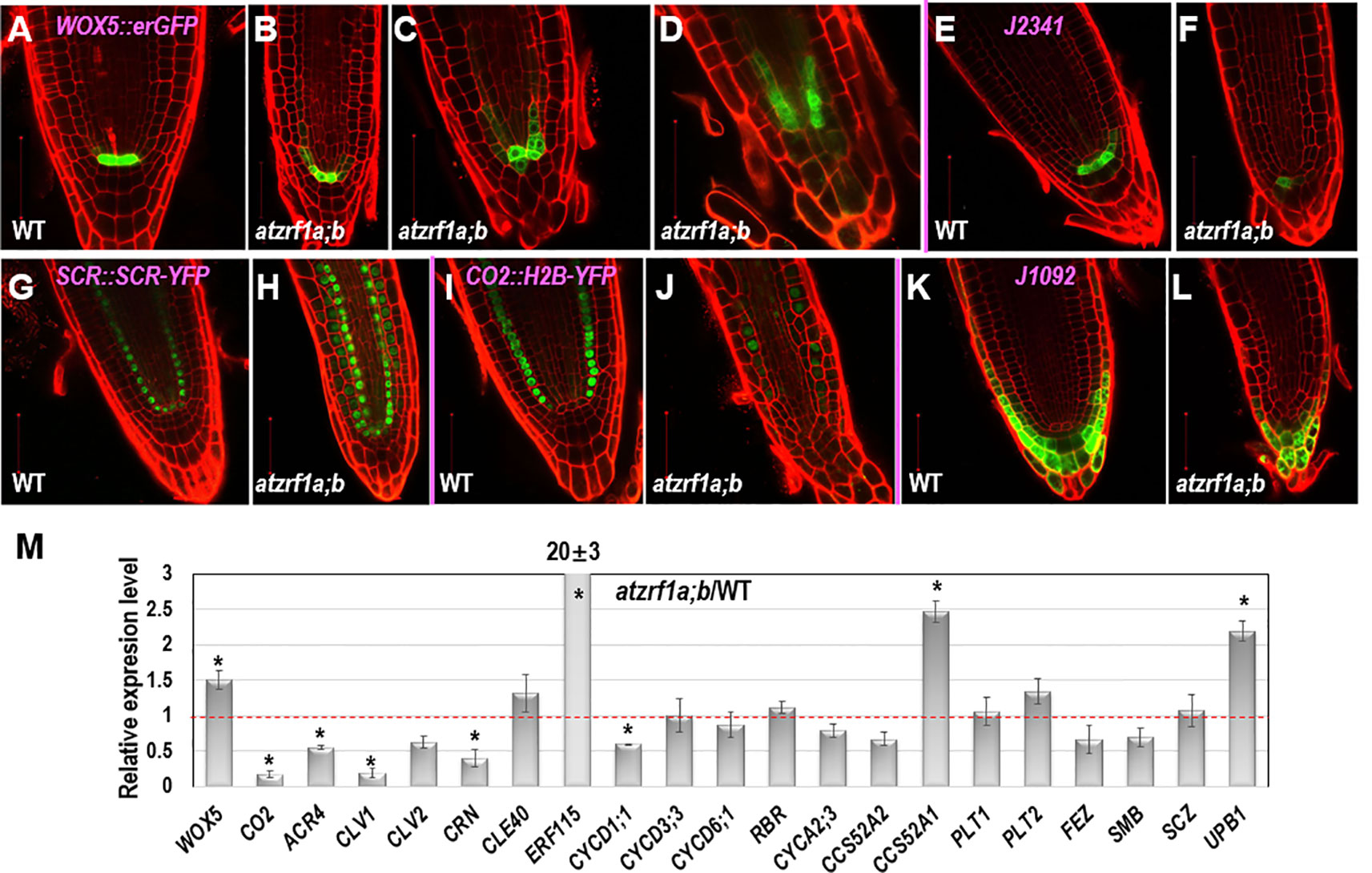
Figure 3 The expression patterns of RAM specific markers in WT and atzrf1a;b roots. 5-day-old root was counterstained with propidium iodide and observed with confocal microscopy. (A–D) WOX5::erGFP markers in WT and atzrf1a;b. (E–F) J2341 markers. (G–H) SCR::SCR-YFP markers. (I–J) CO2::H2B-YFP markers. Inset indicates the relative expression level of CO2 in 7-day-old root of atzrf1a;b compared with that of WT. (K–L) J1092 markers. (M) Relative expression levels of some RAM-regulating genes in atzrf1a;b compared with WT (set as 1) examined by quantitative real-time polymerase chain reaction. Data show means ± SE from three biological repeats. Asterisk indicates student’s t-test statistically significant differences at P < 0.01. Bars = 50 μm.
Next, we performed qRT-PCR to compare between the atzrf1a;b mutant and the WT control for relative expression levels of some of the above described genes as well as others genes known in previous publications as important regulators of root development. As shown in Figure 3M, the expression of WOX5 was upregulated whereas that of CO2 was drastically downregulated in the atzrf1a;b mutant roots. This is in agreement with the WOX5::erGFP and CO2::H2B-YFP expression pattern described above. Consistent with the CLE-WOX5 feedback repressive pathway, the expression levels of several CLE-reception component genes, i.e. ACR4, CLV1, CLV2, and CORYNE (CRN) (Miwa et al., 2008; Stahl et al., 2009; Meng and Feldman, 2010; Stahl et al., 2013), were downregulated albeit CLE40 itself was upregulated in the atzrf1a;b mutant roots (Figure 3M). In addition, expression of ERF115, which is associated with dividing QC cells (Heyman et al., 2013), was found drastically upregulated in atzrf1a;b, further indicating defects of QC regulation in the mutant. We then examined expression of several cell cycle regulatory genes known as being involved in root development, including the G1-phase D-type cyclins (CYCD1;1, CYCD3;3 and CYCD6;1), the G1-S transition inhibitor RETINOBLASTOMA-RELATED (RBR), the S-phase A-type cyclin CYCA2;3, and the endocycle switch regulators CCS52A1/FZR2 and CCS52A2/FZR1 (Vanstraelen et al., 2009; Sozzani et al., 2010; Cruz-Ramirez et al., 2012; Forzani et al., 2014). It was found that CYCD1;1, CCS52A2/FZR1 and to a less degree CYCA2;3 were downregulated whereas CCS52A1/FZR2 was upregulated and the other ones remained unchanged in the atzrf1a;b mutant roots (Figure 3M). The downregulation of CYCD1;1 correlates with the high level of WOX5 in atzrf1a;b, which is in agreement with CYCD1;1 being repressed by WOX5 (Forzani et al., 2014). Lastly, we checked expression of several root-patterning transcription factor genes, including PLETHORA1 (PLT1) and PLT2 involved in auxin-dependent axial patterning (Aida et al., 2004), FEZ and SOMBRERO (SMB) that antagonistically regulate asymmetric cell division of epidermal and lateral cap initials as well as columella stem cells (Willemsen et al., 2008; Bennett et al., 2014), SCHIZORIZA (SCZ) that is required for the specification of cortex identity and the separation of cell fates in surrounding RAM layers (Pernas et al., 2010; ten Hove et al., 2010), and UPBEAT1 (UPB1) that functions in the maintenance of cell proliferation-differentiation balance by controlling ROS production (Tsukagoshi et al., 2010). It was found that UPB1 and to a less degree PLT2 were upregulated whereas FEZ and SMB were downregulated in atzrf1a;b (Figure 3M and Supplementary Table S2), implying defects in the regulation of cell fate determinacy and homeostasis between cell proliferation and cell differentiation in the mutant. Taken together, our data indicate that loss of AtZRF1a/b perturbs expression of multiple sets of genes involved in diverse pathways in the regulation of postembryonic root development.
AtZRf1a/b Are Required for Proper Auxin Regulation of Root Development
Generation and maintenance of auxin gradients and regional maxima in root tip is crucial for normal root development (reviewed in Overvoorde et al., 2010). To survey whether the atzrf1a;b mutant abnormal root development is related to any impaired auxin regulation, we first tested plant growth sensitivity to auxin treatment. We found that the root growth of both WT and mutant was seriously inhibited by exogenous 1-naphthalene acetic acid (NAA) increasingly with higher concentrations (Figure 4A). The mutant root growth became most completely blocked at the presence of 2.5 µM NAA. When grown at 5 µM NAA for 3 weeks, all the mutant roots and about 10% (N = 50) of whole seedlings developed into callus-like structures with root hairs appeared on the surface whereas WT seedlings never showed callus formation (Figure 4B). These data indicate that the atzrf1a;b mutant is more sensitive to auxin treatment than WT.
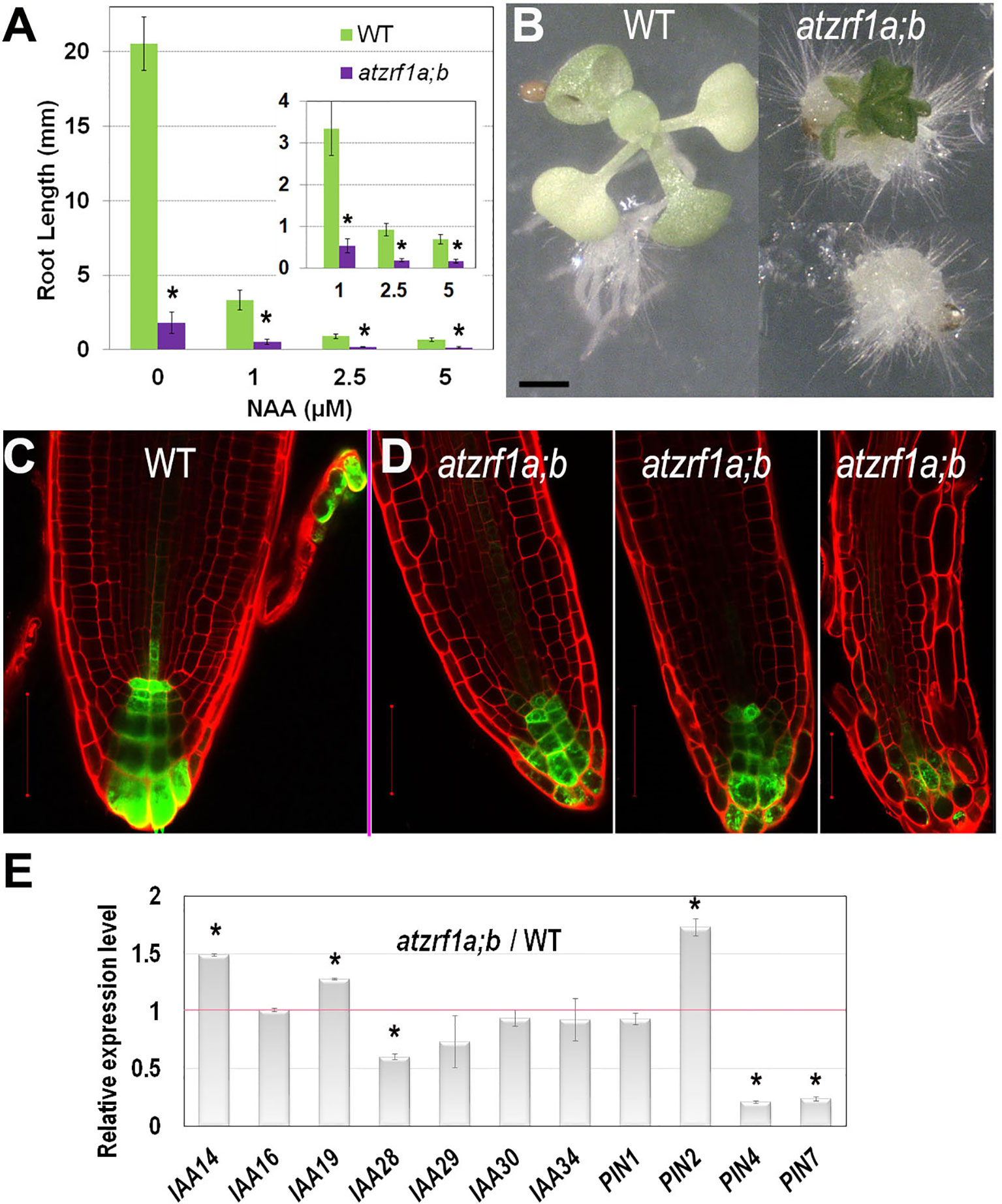
Figure 4 Effect of auxin treatment on atzrf1a;b double mutant. (A) Effects of exogenous NAA on root growth of WT and atzrf1a;b seedlings. Seeds were sown on MS medium containing the indicated concentration of NAA for 1 week. (B) WT and atzrf1a;b plants grown on the plate containing 5μM NAA in 3 weeks. (C–D) DR5rev::GFP markers in 5-day-old seedlings of WT (C) and atzrf1a;b (D). Propidium iodide was used to stain the cell wall. (E) Relative expression levels of auxin-responsive genes in 7-day-old atzrf1a;b compared with WT. Data show means ± SE from three biological repeats. Asterisk indicates student’s t-test statistically significant differences at P < 0.01. Bars = 1 mm in (B), 50 μm in (C) and (D).
To investigate whether auxin distribution is disturbed in the atzrf1a;b mutant RAM, we introgressed into the mutant the auxin-response reporter DR5rev::GFP (Friml et al., 2003). In WT, DR5rev::GFP was expressed in QC, columella stem cells and columella, displaying auxin gradient maxima in QC and distal columella cells (Figure 4C). In the atzrf1a;b mutant, the number of cells expressing DR5rev::GFP was reduced at varied degrees in individual roots and the auxin gradient maxima at both QC and distal columella cells were lost or weakened (Figure 4D). Actually in the cotyledon, a main vein and several secondary veins unite into almost four areoles in WT, but retain unclosed in atzrf1a;b (Supplementary Figure S5), in consistent with the damaged auxin distribution in mutant. Furthermore, we carried out qRT-PCR to analyze the expression levels of auxin-responsive genes (IAA14, IAA16, IAA19, IAA28-IAA30, IAA34) and polar auxin transporter genes (PIN1, PIN2, PIN4, PIN7), which play important roles in root development (Blilou et al., 2005; Overvoorde et al., 2010). It was found that expression of IAA14, IAA19, and PIN2 was increased whereas that of IAA28, IAA29, PIN4, and PIN7 was decreased in the atzrf1a;b mutant roots (Figure 4E and Supplementary Table S3). Collectively, our data indicate that defects in auxin signaling, transport and/or cell type-specific distribution contribute partly to the atzrf1a;b mutant root phenotype.
AtZRf1a/b Regulate Cell Division and Cell Patterning in Embryonic Root
To trace the embryonic origin of defective RAM formation in the atzrf1a;b mutant, embryos at different developmental stages were analyzed using the cell-wall fluorescent dye SR2200, which had been previously demonstrated to be powerful for investigation of early stages of embryogenesis (Musielak et al., 2015). In globular stage, the extra-proembryo-derived hypophysis located at the uppermost suspensor cell underwent an asymmetric division to produce an upper lens-shaped QC cell and a lower columella initial in both WT and atzrf1a;b without showing significant difference (Supplementary Figures S6A–D). In triangular stage, ground tissue initial in WT performed a typical anticlinal division to maintain self-renewal and at the same time create a daughter cell which subsequently underwent a periclinal asymmetric division (first formative division) to generate a cortical initial and an endodermis initial (Figure 5A). However, the potential ground tissue initial in atzrf1a;b seemingly bypassed the former anticlinal division and directly underwent the first periclinal division to give rise to the presumptive cortex and endodermis (Figure 5B) and absence of obvious ground tissue initial after this division (Figures 5C–I and Supplementary Figures S6E–G). In late heart stage, an additional cortical layer arises from the secondary formative divisions of endodermal cells in WT (Figure 5A), demarcating the boundary between root and hypocotyl (Bougourd et al., 2000). Similarly, another periclinal division in mutant also happened in the inner ground tissue cell (Figure 5F). In the early torpedo stage, the lowest of the protoderm cells in WT formally served as epidermal/lateral root cap initials characterized by the emergence of lateral root cap due to the periclinal division (Figure 5A. However, the corresponding protoderm cells in mutant lacking the hallmarking periclinal division failed to achieve the cell fate transition and to generate lateral root cap (Figures 5F–I). These indicate AtZRF1a/b are necessary for the formative cell division giving rise to epidermal/lateral root cap initials and lateral root cap. QC cells in WT are mitotically quiescent and were transversely aligned in the center of embryonic RAM throughout the whole embryogenesis (Figure 5A). In comparison, QC cells in mutant were also easily recognized prior to the heart stage even their morphology gradually growing abnormal from trapezoid to triangle, and then inclined to become atypical in later stages due to their active and irregular divisions giving rise to ill-organized patterning (Figures 5G–I). Consistently, the columella cell and columella initial exhibited anatomic defects to a different extent in atzrf1a;b mutant (Figures 5H, I and Supplementary Figures S6E–G). In mutant mature embryo, columella initial and columella cell in atzrf1a;b mutant displayed distorted and reduced number of cell layers, with 82% of mutants showing three layers instead of four in WT (Figures 5A, I, J). Consistently, 3-day-old atzrf1a;b seedlings still have three layers of columella cells, less than WT with five layers (Figure 5K). Additionally, the embryonic radicle length in atzrf1a;b mutant (∼10 µm) was only about one seventh of that in WT during mature embryo stage, giving rise to a round end phenotype (Figure 5I, L). Correspondingly, the mutant radicle had only ∼2 cells in a longitudinal cortex file, much fewer than WT with ∼10 (Figure 5M), indicating that AtZRF1a/b promote cell division during embryonic root morphogenesis. Suspensor is comprised of a single file of about seven cells, bridges the embryo proper to surrounding endosperm tissues, and transports nutrients and growth regulators to the embryo (Kawashima and Goldberg, 2010). As embryo grows up, suspensor gradually degenerates till to disappear. But in mutant, suspensor cells were usually arranged into two files at the basal part (Figure 5B and Supplementary Figure S6H), and sometimes proliferated into a cell mass (Supplementary Figures S6I, J). Even in mature embryos, 32% (n = 50) of suspensors were still visible (Supplementary Figure S6K). In summary, AtZRF1a/b participate in the proper radicle cell patterning, maintenance of QC and surrounding stem cell identity, promotion of cell division, and normal degradation of suspensor cell during embryonic radicle formation.
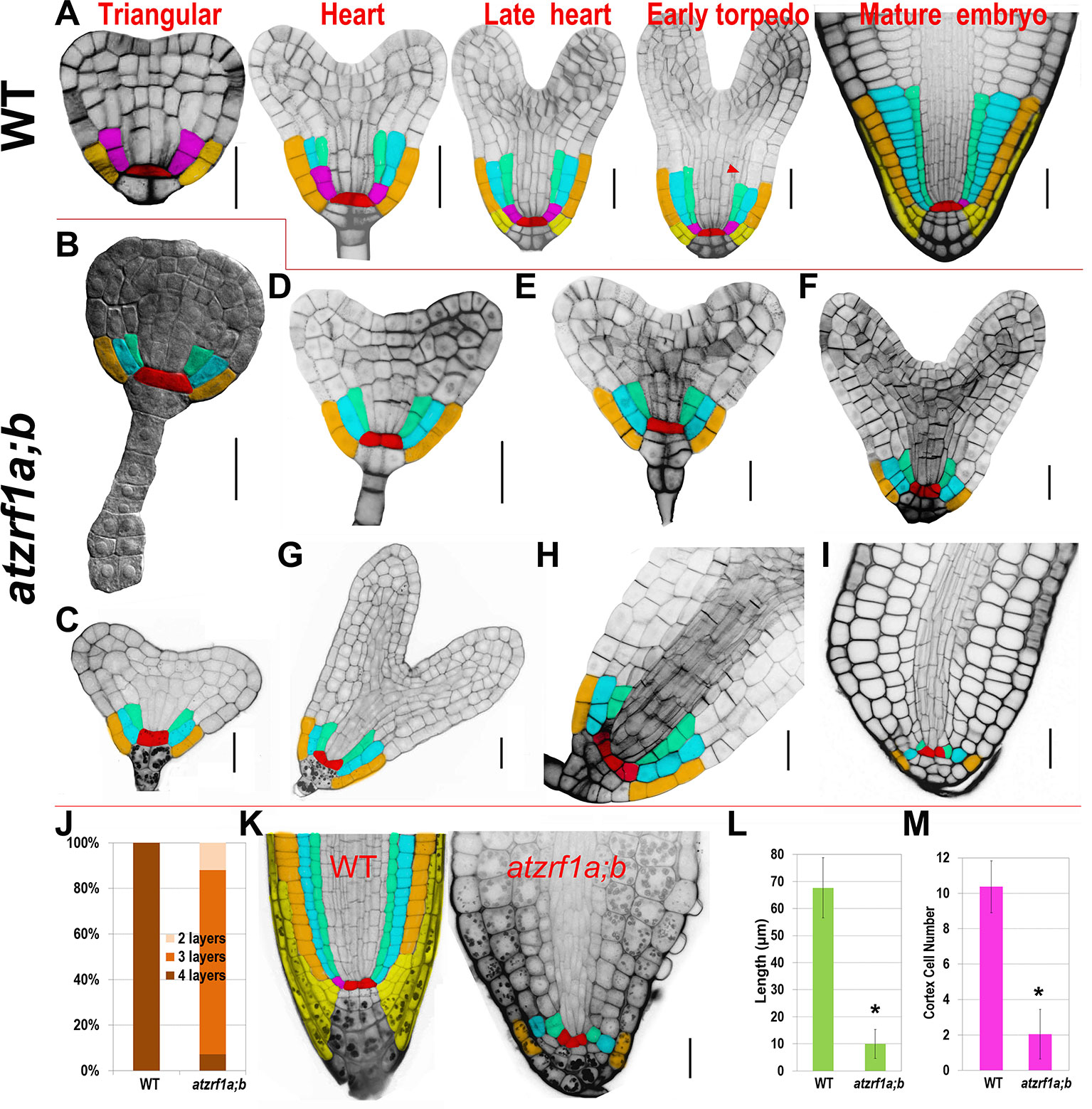
Figure 5 Defective embryogenesis in atzrf1a;b mutant. (A) Representative embryogenetic stages in WT according to previously reported (Scheres et al., 1994). (B – I) Embryogenesis in atzrf1a;b. (B) in triangular stage, (C–E) in different heart-stages, (F–H) in torpedo stages, and (I) in mature embryo stage. (J) Columella cell layers of mature embryos in WT and atzrf1a;b. (K) Three-day-old RAM in WT and atzrf1a;b mutant observed via mPS-propidium iodide method. (L) Length of mature embryonic roots in WT and atzrf1a;b. (M) Cell numbers in a single file of cortex of mature embryos in WT and atzrf1a;b. (A) and (D–I) were stained with SR2200 for confocal observation (Musielak et al., 2015); (B) was cleared by chloral hydrate solution for differential interference contrast observation; (K) was obtained by the mPS-propidium iodide method (Truernit et al., 2008). Red, QC; pink, ground tissue initial; cyan, cortex; green, endodermis; brown, epidermis; yellow, lateral root cap and its initials. Arrowhead, the second formative cell division giving rise to the second cortex file. Data show means ± SE from three biological repeats. Asterisk indicates student’s t-test statistically significant differences at P < 0.01. Bars = 20 µm.
AtZRf1a/b Are Required for Embryonic Root Cell Fate Establishment
To gain insight into the embryonic root cell identity in atzrf1a;b, some aforementioned marker lines were investigated during embryogenesis. QC-specific marker WOX5::erGFP also displayed significantly diffused expression in adjacent initials from torpedo stage onward in mutant embryo (Figures 6A–C, G–I), consistent with the WOX5 performance in the postembryonic root. SCR::SCR-YFP marked QC, ground tissue initial, and endodermis in embryo as it is in seedling stage in WT. In mutant, SCR signal firstly appeared in the inner ground tissue layer from the first unusual periclinal division of ground tissue initial in triangular-stage mutant embryo, confirming the ground tissue inner layer adapted the endodermis cell fate. Subsequently, signal was also observed in the fourth layer from the second periclinal division in mutant embryo, corresponding to the same endodermis layer in WT hypocotyl (Figures 6D–F, J–L). CO2::H2B-YFP marker was firstly observed in the cortex cells in the upper part of torpedo-stage WT embryo but excluding embryonic root region, and subsequently in all the cortex cells in later stages (Figures 6M, N). However, in atzrf1a;b mutant, YFP signal was not detected in torpedo-stage, but later was sporadically found in some cortex cells excluding embryonic root region (Figures 6S, T), indicating AtZRF1a/b are required for determination of cortex cell identity in embryonic root. J1092 marker specified root cap in the embryos of WT and atzrf1a;b mutant, though mutant had no lateral root cap or significantly reduced lateral root cap region in later stage (Figures 6O, P, U, V). DR5rev::GFP marker had the strongest expression in the nearest suspensor cell attaching to embryo proper in WT and mutant, and subsequently auxin distribution gradients were established from columella cells to QC in WT, but almost absent in mutant (Figures 6Q, R, W, X). These data suggested AtZRF1a/b are necessary for cell identity maintenance of QC and different initials and the formation of auxin gradients during embryogenesis.
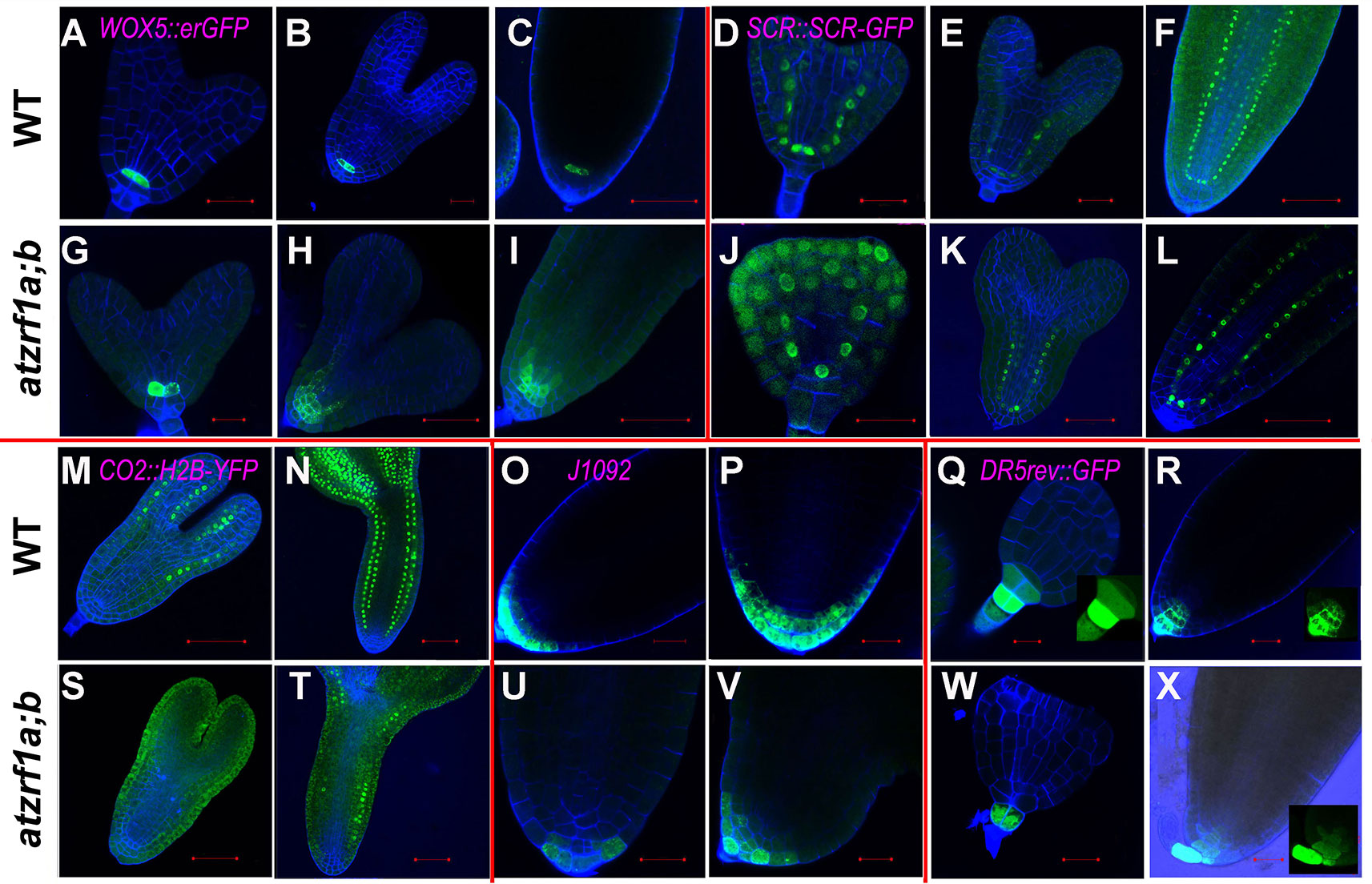
Figure 6 The expression of RAM-specific markers in atzrf1a;b embryos. Different stages of embryos were hand-dissected, counterstained with SR2200 and observed under confocal. (A–C) and (G–I) WOX5::erGFP markers in WT and atzrf1a;b. (D–F) and (J–L) SCR::SCR-YFP markers. (M, N) and (S, T) CO2::H2B-YFP markers. (O, P) and (U, V) J1092 markers. (Q, R) and (W, X) DR5rev::GFP markers. Bars = 20 µm, except 50 µm in (C), (F), (H), (I), (L), (M), and (S).
Discussion
In this study, we provide important insights about the roles of the H2A/UBIQUITIN-binding chromatin regulator genes AtZRF1a/b in embryonic and post-embryonic root development. In the loss-of-function atzrf1a;b mutant, the primary root growth ceased early during seedling growth because RAM became shortened and exhausted due to spoiled balance between cell proliferation and differentiation. The atzrf1a;b RAM displayed low mitotic activities, which was consistent with the very slow root growth. Elevated polyploid levels were detected, indicating an advanced onset of endoreduplication in the mutant roots. In a previous study, endoreduplication levels were also found increased in true leaves of the atzrf1a;b mutant plants (Feng et al., 2016). It thus appears that AtZRF1a/b repress the mitosis-to-endocycle transition in a general rather than an organ-specific manner.
Proper RAM structure organization is crucial in maintaining continuous post-embryonic root development. Our study showed that AtZRF1a/b are crucial in establishment and maintenance of cell fate of various cell types within RAM. The hardly recognizable QC cells together with expanding zone of WOX5::erGFP expression outside QC position indicated that the QC cell fate was drastically impacted in the atzrf1a;b mutant. In addition, the cell identities of surrounding initials and their corresponding descendants were also altered to different extents in the mutant. Columella root cap was frequently found disorganized, correspondingly, columella initial marker J2341 displayed reduced activity in the mutant. lateral root cap was clearly separated from columella root cap in WT, but seemingly became undistinguished from columella root cap or absent in the atzrf1a;b mutant. Ground tissues including cortex and endodermis partially lost their cell identities, which was reflected by diffused SCR::SCR-YFP expression and reduced CO2::H2B-YFP signal in the mutant.
RAM defects in atzrf1a;b could be traced back to early embryogenesis. The first major defect in atzrf1a;b happened in triangular-stage embryo where potential ground tissue initial skipped the first anticlinal division (proliferative division) which was substituted by a periclinal division. The inner ground tissue layer had the endodermis cell identity confirmed by SCR::SCR-YFP marker, which was different from that in seedling, whereas the outer ground tissue layer seemed to loss partial cortex cell identity reflected by CO2::H2B-YFP marker. The second major defect occurred in late heart-stage embryo, in which the potential epidermal/lateral root cap initial failed to perform the periclinal division (formative division), leading to no lateral root cap formation in late stages. Accordingly, J1092 marking root cap has reduced expression domains in mutant. So, the atzrf1a;b root cap mainly results from columella cell cap but not lateral root cap. In fact, the both major defects abovementioned in mutant were characterized by transformation of cell division orientation (anticlinal-to-periclinal or vice versa). So, AtZRF1a/b might be involved in regulating the conversion between proliferative division and formative division rather than specific proliferative or formative division. Additionally, embryonic QC was conspicuous at the beginning, and subsequently conducted a few more divisions even in oblique direction to generate some offspring with similar size and irregular organization, leading to hardly distinguishing from surrounding stem cells in most cases. It seems that AtZRF1a/b are required for repression of cell divisions and maintenance of precise division orientation within QC. WOX5::erGFP also displayed expanded expression during the late embryogenesis, similar to its performance in seedling root.
The phytohormone auxin play key roles in root development. In WT, local auxin maximum as the prerequisite for QC establishment determines the position of the QC, and the auxin gradient is crucial for maintaining columella initial identity (Sabatini et al., 1999; Tian et al., 2014). In the atzrf1a;b mutant, the auxin maximum and/or gradient were perturbed in postembryonic and embryonic roots, where the cell patterning of QC and columella cell was mostly impaired. Accordingly, several IAA genes as auxin signal pathway repressors (Fukaki et al., 2002; De Rybel et al., 2010) and PIN genes as auxin transporters (Blilou et al., 2005) were misregulated in the mutant roots. Exogenous auxin treatment also showed that atzrf1a;b was more sensitive, with enhanced facilities of ectopic callus formation. Future genetic interaction studies may precisely investigate the role and regulatory pathways of auxin in the AtZRF1-regulated root development.
In addition to IAA and PIN genes, several other genes were found deregulated, which likely contributes to the atzrf1a;b mutant root developmental defects. Firstly, AtZRF1a/b may regulate the balance between cell division and differentiation in RAM partially through CCS52A-activating APC/C ubiquitin ligase. CCS52A has two isoforms with antagonistic functions; CCS52A1 expressed in the root elongation zone promotes endocycle onset and mitotic exit through destruction of A2-type cyclin CYCA2;3 (Boudolf et al., 2009), whereas CCS52A2 expressed in the RAM distal region controls QC identity and stem cell maintenance (Vanstraelen et al., 2009) through proteolysis of QC division marker ERF115 (Heyman et al., 2013). In atzrf1a;b, upregulation of CCS52A1 coupled with downregulation of CYCA2;3 was associated with downregulation of CCS52A2 coupled with upregulation of ERF115, which was in line with ccs52a2 mutant root phenotype displaying the consumed and disorganized RAM (Vanstraelen et al., 2009). Secondly, upregulation of UPB1 in atzrf1a;b was consistent with reduced root length and RAM size in UPB1 overexpression line (Tsukagoshi et al., 2010). Lastly, CLE peptide ligands in differentiated columella cells regulate WOX5 expression and columella initial fate through the receptor-like kinases ACR4, CLV1, CRN, and CLV2 (Miwa et al., 2008; Stahl et al., 2009; Meng and Feldman, 2010; Stahl et al., 2013). AtZRF1a/b also regulated RAM organization dependent on CLE-WOX5 pathway inferred from uniformly downregulated expression of CLE receptors in mutant. Correspondingly, ACR4 has an important role in formative cell division and columella cell organization in the root apex (De Smet et al., 2008).
In multicellular organisms, stem cells can maintain self-renewal and produce the new daughter cells with distinct fates by asymmetric cell divisions or formative divisions, which are coordinated by extrinsic and intrinsic cues (Kajala et al., 2014). asymmetric cell division can be considered as the evolutionary engine, leading to cell differentiation necessary for the innovation of novel organ and the emergence of higher life form. Our study demonstrates that the Arabidopsis AtZRF1a/b are required for formative division giving rise to lateral root cap during embryogenesis. Likely, ZRF1 orthologs have an evolutionarily conserved function in asymmetric cell division. For instance, in the classic animal model Caenorhabditis elegans, DNJ11 is involved in the asymmetric division of the neuroblast via regulating the orientation of the mitotic spindle (Hatzold and Conradt, 2008). In green algae Volvox carteri, Gonidialess A (GlsA) is necessary for separation of germ and somatic cell fate during gonidium formation (Miller and Kirk, 1999).
AtZRF1a and AtZRF1b have similar and broad expression in almost all the young plant organs including root tips, shoot tips, developing leaves, inflorescences, floral buds, and embryos; their expression intensity is positively correlated with dividing activities of the organs (Guzman-Lopez et al., 2016). Consistently, loss of AtZRF1 function results in morphological defects in almost all the developmental phases related to dividing cells and meristematic tissues (Feng et al., 2016; Guzman-Lopez et al., 2016). Recently AtZRF1a/b have been reported to perform both PRC1-related and independent functions in regulating plant growth and development (Feng et al., 2016). Consistently, PRC1 RING-finger proteins functioning as H2Aub1 writers and AtZRF1 as H2Aub1 reader share a set of target genes and partial regulatory pathways (Feng et al., 2016). In addition, PRC1 RING-finger proteins display the similar expression pattern and tendency with ubiquitously organic distribution but high levels in dividing cells (Qin et al., 2008; Chen et al., 2010; Chen et al., 2016). Furthermore, AtRING1a/b and AtBMI1a/b are also widely involved in regulating multiple developmental processes. On the other hand, according to the working model of human ZRF1 (Richly et al., 2010), AtZRF1a/b might also act as a chromatin state switch to remove PRC1 function in the specific developmental context. Future studies are necessary to investigate these different aspects of interplay between AtZRF1a/b and PRC1 complexes in the regulation of gene transcription in the root and other plant organ development.
Conclusion
Our study suggests AtZRF1a and AtZRF1b participate in controlling root development and morphogenesis through regulating cell proliferation and differentiation, cell identity maintenance, cell patterning, and auxin gradient establishment at the root tip during embryonic and post-embryonic stages.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
DC designed the study, performed most of the experiments, analyzed the data, and wrote the draft. QW and JF performed the introduction of gene marks. W-HS conceived the study and supervised the experiments. W-HS, QW, and YR revised the article. All authors have read and approved the final manuscript.
Funding
This work was supported by the French Agence Nationale de la Recherche (ANR-08-BLAN-0200-CSD7, ANR-12-BSV2-0013-02), the National Basic Research Program of China (973 Program, grants no. 2012CB910500), the French Centre National de la Recherche Scientifique (CNRS, LIA PER), and National Natural Science Foundation of China (31870310).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01498/full#supplementary-material
Figure S1 | The representative root phenotype in 5-day-old atzrf1 single and double mutants. The atzrf1a-1;b-1 and atzrf1a-2;atzrf1b-1 double mutants displayed the same extremely short-root root phenotype, whereas WT and atzrf1 single mutants displayed the normal root growth. Bar = 1 cm.
Figure S2 | 7-day-old root phenotype in WT and atzrf1a;b mutant via DIC observation. Bars = 50 µm.
Figure S3 | Lugol staining of 5-day-old RAM in WT and atzrf1a;b mutant via DIC observation. Bars = 20 µm.
Figure S4 | Comparison of RAM development between WT and atzrf1a;b mutant at 21DAS via Lugol staining. Bars = 50 µm.
Figure S5 | Cotyledon vein pattern in 10-day-old seedlings of WT and atzrf1a;b mutant via DIC observation. The samples were cleared in chloral hydrate solution. Bars = 1 mm.
Figure S6 | Other defective embryogenesis in atzrf1a;b mutant. (A-B) Globular stage embryos in WT; (C-K) Embryos of different stages in atzrf1a;b mutant. Bars = 20 µm except 50 µm in (H) to (J).
Table S1 | List of primers used in this study.
Table S2 | Relative expression levels of some RAM-regulating genes in atzrf1a;b compared with WT.
Table S3 | Relative expression levels of auxin-responsive genes in atzrf1a;b compared with WT.
References
Aichinger, E., Villar, C. B., Di Mambro, R., Sabatini, S., Kohler, C. (2011). The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root. Plant Cell 23 (3), 1047–1060. doi: 10.1105/tpc.111.083352
Aida, M., Beis, D., Heidstra, R., Willemsen, V., Blilou, I., Galinha, C. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119 (1), 109–120. doi: 10.1016/j.cell.2004.09.018
Barow, M., Meister, A. (2003). Endopolyploidy in seed plants is differently correlated to systematics, organ, life strategy and genome size. Plant Cell Environ. 26 (4), 571–584. doi: 10.1046/j.1365-3040.2003.00988.x
Bennett, T., van den Toorn, A., Willemsen, V., Scheres, B. (2014). Precise control of plant stem cell activity through parallel regulatory inputs. Development 141 (21), 4055–4064. doi: 10.1242/dev.110148
Blilou, I., Frugier, F., Folmer, S., Serralbo, O., Willemsen, V., Wolkenfelt, H. (2002). The Arabidopsis HOBBIT gene encodes a CDC27 homolog that links the plant cell cycle to progression of cell differentiation. Genes Dev. 16 (19), 2566–2575. doi: 10.1101/gad.237302
Blilou, I., Xu, J., Wildwater, M., Willemsen, V., Paponov, I., Friml, J. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433 (7021), 39–44. doi: 10.1038/nature03184
Boudolf, V., Lammens, T., Boruc, J., Van Leene, J., Van Den Daele, H., Maes, S. (2009). CDKB1;1 forms a functional complex with CYCA2;3 to suppress endocycle onset. Plant Physiol. 150 (3), 1482–1493. doi: 10.1104/pp.109.140269
Bougourd, S., Marrison, J., Haseloff, J. (2000). Technical advance: an aniline blue staining procedure for confocal microscopy and 3D imaging of normal and perturbed cellular phenotypes in mature Arabidopsis embryos. Plant J. 24 (4), 543–550. doi: 10.1046/j.1365-313x.2000.00892.x
Bratzel, F., Lopez-Torrejon, G., Koch, M., Del Pozo, J. C., Calonje, M. (2010). Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr. Biol. 20 (20), 1853–1859. doi: 10.1016/j.cub.2010.09.046
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55 (4), 611–622. doi: 10.1373/clinchem.2008.112797
Chen, D., Molitor, A., Liu, C., Shen, W. H. (2010). The Arabidopsis PRC1-like ring-finger proteins are necessary for repression of embryonic traits during vegetative growth. Cell Res. 20 (12), 1332–1344. doi: 10.1038/cr.2010.151
Chen, D. H., Huang, Y., Liu, C., Ruan, Y., Shen, W. H. (2014). Functional conservation and divergence of J-domain-containing ZUO1/ZRF orthologs throughout evolution. Planta 239 (6), 1159–1173. doi: 10.1007/s00425-014-2058-6
Chen, D., Molitor, A. M., Xu, L., Shen, W. H. (2016). Arabidopsis PRC1 core component AtRING1 regulates stem cell-determining carpel development mainly through repression of class I KNOX genes. BMC Biol. 14 (1), 112. doi: 10.1186/s12915-016-0336-4
Colon-Carmona, A., You, R., Haimovitch-Gal, T., Doerner, P. (1999). Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20 (4), 503–508. doi: 10.1046/j.1365-313x.1999.00620.x
Cruz-Ramirez, A., Diaz-Trivino, S., Blilou, I., Grieneisen, V. A., Sozzani, R., Zamioudis, C. (2012). A bistable circuit involving SCARECROW-RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell 150 (5), 1002–1015. doi: 10.1016/j.cell.2012.07.017
de Lucas, M., Pu, L., Turco, G., Gaudinier, A., Morao, A. K., Harashima, H. (2016). Transcriptional regulation of Arabidopsis polycomb repressive complex 2 coordinates cell-type proliferation and differentiation. Plant Cell 28 (10), 2616–2631. doi: 10.1105/tpc.15.00744
De Rybel, B., Vassileva, V., Parizot, B., Demeulenaere, M., Grunewald, W., Audenaert, D. (2010). A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr. Biol. 20 (19), 1697–1706. doi: 10.1016/j.cub.2010.09.007
De Smet, I., Vassileva, V., De Rybel, B., Levesque, M. P., Grunewald, W., Van Damme, D. (2008). Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322 (5901), 594–597. doi: 10.1126/science.1160158
Ding, Z., Friml, J. (2010). Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc. Natl. Acad. Sci. U.S.A. 107 (26), 12046–12051. doi: 10.1073/pnas.1000672107
Dolan, L., Janmaat, K., Willemsen, V., Linstead, P., Poethig, S., Roberts, K. (1993). Cellular organisation of the Arabidopsis thaliana root. Development 119 (1), 71–84.
Drisch, R. C., Stahl, Y. (2015). Function and regulation of transcription factors involved in root apical meristem and stem cell maintenance. Front. Plant Sci. 6, 505. doi: 10.3389/fpls.2015.00505
Feng, J., Chen, D., Berr, A., Shen, W. H. (2016). ZRF1 chromatin regulators have polycomb silencing and independent roles in development. Plant Physiol. 172 (3), 1746–1759. doi: 10.1104/pp.16.00193
Forderer, A., Zhou, Y., Turck, F. (2016). The age of multiplexity: recruitment and interactions of Polycomb complexes in plants. Curr. Opin. Plant Biol. 29, 169–178. doi: 10.1016/j.pbi.2015.11.010
Forzani, C., Aichinger, E., Sornay, E., Willemsen, V., Laux, T., Dewitte, W. (2014). WOX5 suppresses CYCLIN D activity to establish quiescence at the center of the root stem cell niche. Curr. Biol. 24 (16), 1939–1944. doi: 10.1016/j.cub.2014.07.019
Friml, J., Benkova, E., Blilou, I., Wisniewska, J., Hamann, T., Ljung, K. (2002). AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108 (5), 661–673. doi: 10.1016/s0092-8674(02)00656-6
Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., Hamann, T. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426 (6963), 147–153. doi: 10.1038/nature02085
Fukaki, H., Tameda, S., Masuda, H., Tasaka, M. (2002). Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 29 (2), 153–168. doi: 10.1046/j.0960-7412.2001.01201.x
Gu, X., Xu, T., He, Y. (2014). A histone H3 lysine-27 methyltransferase complex represses lateral root formation in Arabidopsis thaliana. Mol. Plant 7 (6), 977–988. doi: 10.1093/mp/ssu035
Guzman-Lopez, J. A., Abraham-Juarez, M. J., Lozano-Sotomayor, P., de Folter, S., Simpson, J. (2016). Arabidopsis thaliana gonidialess A/Zuotin related factors (GlsA/ZRF) are essential for maintenance of meristem integrity. Plant Mol. Biol. 91 (1–2), 37–51. doi: 10.1007/s11103-016-0439-x
Hatzold, J., Conradt, B. (2008). Control of apoptosis by asymmetric cell division. PloS Biol. 6 (4), e84. doi: 10.1371/journal.pbio.0060084
Heidstra, R., Welch, D., Scheres, B. (2004). Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev. 18 (16), 1964–1969. doi: 10.1101/gad.305504
Hennig, L., Derkacheva, M. (2009). Diversity of Polycomb group complexes in plants: same rules, different players? Trends Genet. 25 (9), 414–423. doi: 10.1016/j.tig.2009.07.002
Heyman, J., Cools, T., Vandenbussche, F., Heyndrickx, K. S., Van Leene, J., Vercauteren, I. (2013). ERF115 controls root quiescent center cell division and stem cell replenishment. Science 342 (6160), 860–863. doi: 10.1126/science.1240667
Hoppmann, V., Thorstensen, T., Kristiansen, P. E., Veiseth, S. V., Rahman, M. A., Finne, K. (2011). The CW domain, a new histone recognition module in chromatin proteins. EMBO J. 30 (10), 1939–1952. doi: 10.1038/emboj.2011.108
Ikeuchi, M., Iwase, A., Rymen, B., Harashima, H., Shibata, M., Ohnuma, M. (2015). PRC2 represses dedifferentiation of mature somatic cells in Arabidopsis. Nat. Plants 1, 15089. doi: 10.1038/nplants.2015.89
Ivanov, V. B., Dubrovsky, J. G. (2013). Longitudinal zonation pattern in plant roots: conflicts and solutions. Trends Plant Sci. 18 (5), 237–243. doi: 10.1016/j.tplants.2012.10.002
Kajala, K., Ramakrishna, P., Fisher, A., Bergmann, D. C., De Smet, I., Sozzani, R. (2014). Omics and modelling approaches for understanding regulation of asymmetric cell divisions in Arabidopsis and other angiosperm plants. Ann. Bot. 113 (7), 1083–1105. doi: 10.1093/aob/mcu065
Kawashima, T., Goldberg, R. B. (2010). The suspensor: not just suspending the embryo. Trends Plant Sci. 15 (1), 23–30. doi: 10.1016/j.tplants.2009.11.002
Kim, I., Cho, E., Crawford, K., Hempel, F. D., Zambryski, P. C. (2005). Cell-to-cell movement of GFP during embryogenesis and early seedling development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 102 (6), 2227–2231. doi: 10.1073/pnas.0409193102
Meng, L., Feldman, L. J. (2010). CLE14/CLE20 peptides may interact with CLAVATA2/CORYNE receptor-like kinases to irreversibly inhibit cell division in the root meristem of Arabidopsis. Planta 232 (5), 1061–1074. doi: 10.1007/s00425-010-1236-4
Miller, S. M., Kirk, D. L. (1999). glsA, a Volvox gene required for asymmetric division and germ cell specification, encodes a chaperone-like protein. Development 126 (4), 649–658.
Miwa, H., Betsuyaku, S., Iwamoto, K., Kinoshita, A., Fukuda, H., Sawa, S. (2008). The receptor-like kinase SOL2 mediates CLE signaling in Arabidopsis. Plant Cell Physiol. 49 (11), 1752–1757. doi: 10.1093/pcp/pcn148
Musielak, T. J., Schenkel, L., Kolb, M., Henschen, A., Bayer, M. (2015). A simple and versatile cell wall staining protocol to study plant reproduction. Plant Reprod. 28 (3–4), 161–169. doi: 10.1007/s00497-015-0267-1
Napsucialy-Mendivil, S., Alvarez-Venegas, R., Shishkova, S., Dubrovsky, J. G. (2014). Arabidopsis homolog of trithorax1 (ATX1) is required for cell production, patterning, and morphogenesis in root development. J. Exp. Bot. 65 (22), 6373–6384. doi: 10.1093/jxb/eru355
Overvoorde, P., Fukaki, H., Beeckman, T. (2010). Auxin control of root development. Cold Spring Harb. Perspect. Biol. 2 (6), a001537. doi: 10.1101/cshperspect.a001537
Pernas, M., Ryan, E., Dolan, L. (2010). SCHIZORIZA controls tissue system complexity in plants. Curr. Biol. 20 (9), 818–823. doi: 10.1016/j.cub.2010.02.062
Qin, F., Sakuma, Y., Tran, L. S., Maruyama, K., Kidokoro, S., Fujita, Y. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20 (6), 1693–1707. doi: 10.1105/tpc.107.057380
Richly, H., Rocha-Viegas, L., Ribeiro, J. D., Demajo, S., Gundem, G., Lopez-Bigas, N. (2010). Transcriptional activation of polycomb-repressed genes by ZRF1 . Nature 468 (7327), 1124–1128. doi: 10.1038/nature09574
Sabatini, S., Beis, D., Wolkenfelt, H., Murfett, J., Guilfoyle, T., Malamy, J. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99 (5), 463–472. doi: 10.1016/s0092-8674(00)81535-4
Sarkar, A. K., Luijten, M., Miyashima, S., Lenhard, M., Hashimoto, T., Nakajima, K. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446 (7137), 811–814. doi: 10.1038/nature05703
Scheres, B. J. G., Wolkenfelt, H., Willemsen, V., Terlouw, M., Lawson, E., Dean, C. (1994). Embryonic origin of the Arabidopsis primary root and root meristem initials. Development 120 (9), 2475–2487.
Sozzani, R., Cui, H., Moreno-Risueno, M. A., Busch, W., Van Norman, J. M., Vernoux, T. (2010). Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466 (7302), 128–132. doi: 10.1038/nature09143
Stahl, Y., Wink, R. H., Ingram, G. C., Simon, R. (2009). A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr. Biol. 19 (11), 909–914. doi: 10.1016/j.cub.2009.03.060
Stahl, Y., Grabowski, S., Bleckmann, A., Kuhnemuth, R., Weidtkamp-Peters, S., Pinto, K. G. (2013). Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr. Biol. 23 (5), 362–371. doi: 10.1016/j.cub.2013.01.045
ten Hove, C. A., Willemsen, V., de Vries, W. J., van Dijken, A., Scheres, B., Heidstra, R. (2010). SCHIZORIZA encodes a nuclear factor regulating asymmetry of stem cell divisions in the Arabidopsis root. Curr. Biol. 20 (5), 452–457. doi: 10.1016/j.cub.2010.01.018
Tian, H., Niu, T., Yu, Q., Quan, T., Ding, Z. (2013). Auxin gradient is crucial for the maintenance of root distal stem cell identity in Arabidopsis. Plant Signal Behav. 8 (12), e26429. doi: 10.4161/psb.26429
Tian, H., Wabnik, K., Niu, T., Li, H., Yu, Q., Pollmann, S. (2014). WOX5-IAA17 feedback circuit-mediated cellular auxin response is crucial for the patterning of root stem cell niches in Arabidopsis. Mol. Plant 7 (2), 277–289. doi: 10.1093/mp/sst118
Truernit, E., Bauby, H., Dubreucq, B., Grandjean, O., Runions, J., Barthelemy, J. (2008). High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. Plant Cell 20 (6), 1494–1503. doi: 10.1105/tpc.107.056069
Tsukagoshi, H., Busch, W., Benfey, P. N. (2010). Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143 (4), 606–616. doi: 10.1016/j.cell.2010.10.020
Vanstraelen, M., Baloban, M., Da Ines, O., Cultrone, A., Lammens, T., Boudolf, V. (2009). APC/C-CCS52A complexes control meristem maintenance in the Arabidopsis root. Proc. Natl. Acad. Sci. U.S.A. 106 (28), 11806–11811. doi: 10.1073/pnas.0901193106
Willemsen, V., Bauch, M., Bennett, T., Campilho, A., Wolkenfelt, H., Xu, J. (2008). The NAC domain transcription factors FEZ and SOMBRERO control the orientation of cell division plane in Arabidopsis root stem cells. Dev. Cell 15 (6), 913–922. doi: 10.1016/j.devcel.2008.09.019
Xiao, J., Wagner, D. (2015). Polycomb repression in the regulation of growth and development in Arabidopsis. Curr. Opin. Plant Biol. 23, 15–24. doi: 10.1016/j.pbi.2014.10.003
Keywords: ZRF1, root meristem, embryogenesis, cell layer organization, cell identity
Citation: Chen D, Wang Q, Feng J, Ruan Y and Shen W-H (2019) Arabidopsis ZUOTIN RELATED FACTOR1 Proteins Are Required for Proper Embryonic and Post-Embryonic Root Development. Front. Plant Sci. 10:1498. doi: 10.3389/fpls.2019.01498
Received: 20 August 2019; Accepted: 29 October 2019;
Published: 22 November 2019.
Edited by:
Munetaka Sugiyama, The University of Tokyo, JapanReviewed by:
Tatsuaki Goh, Nara Institute of Science and Technology (NAIST), JapanMitsuhiro Aida, Kumamoto University, Japan
Copyright © 2019 Chen, Wang, Feng, Ruan and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Donghong Chen, ZG9uZ2hvbmcuY2hlbkB6YWZ1LmVkdS5jbg==; Wen-Hui Shen, d2VuLWh1aS5zaGVuQGlibXAtY25ycy51bmlzdHJhLmZy
 Donghong Chen
Donghong Chen Qiannan Wang1
Qiannan Wang1 Jing Feng
Jing Feng Ying Ruan
Ying Ruan Wen-Hui Shen
Wen-Hui Shen