- 1Key Laboratory of Soybean Molecular Design Breeding, Northeast Institute of eography and Agroecology, The Innovative Academy of Seed Design, Chinese Academy of Sciences, Changchun, China
- 2College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing, China
Thiamine is an essential cofactor in several enzymatic reactions for all living organisms. Animals cannot synthesize thiamine and depend on their diet. Enhancing the content of thiamine is one of the most important goals of plant breeding to solve the thiamine deficiency associated with the low-thiamin staple crops. In this study, a Glycinemaxpalegreenleaf 1 (Gmpgl1) mutant was isolated from the EMS mutagenized population of soybean cultivar, Williams 82. Map-based cloning of the GmPGL1 locus revealed a single nucleotide deletion at the 292th nucleotide residue of the first exon of Glyma.10g251500 gene in Gmpgl1 mutant plant, encoding a thiamine thiazole synthase. Total thiamine contents decreased in both seedlings and seeds of the Gmpgl1 mutant. Exogenous application of thiazole restored the pale green leaf phenotype of the mutant. The deficiency of thiamine in Gmpgl1 mutant led to reduced activities of the pyruvate dehydrogenase (PDH) and pyruvate decarboxylase (PDC), and decreased contents of six amino acids as compared to that in the wild type plants. These results revealed that GmPGL1 played an essential role in thiamine thiazole biosynthesis.
Introduction
Thiamine is a water-soluble vitamin essential to all organisms. Thiamine is widely available in green vegetables, beans, cereal embryos and nuts. However, humans and animals are incapable of synthesizing thiamine, and depend entirely on plant sources for this vitamin (Goyer, 2010). Thiamine deficiency leads to various chronic diseases in humans, such as edema, neurological disorders and beriberi (Rapala-Kozik, 2011). Thiamin esters are available as thiamin, thiamin monophosphate (TMP) and thiamine diphosphate (TPP) (Gangolf et al., 2010). TPP is the functional form of thiamine and takes part as a cofactor in plant central energy metabolizing enzymes (Mimura et al., 2016), which are pyruvate dehydrogenase (PDH) of glycolysis, α-ketoglutarate dehydrogenase (α-KGDH) of the tricarboxylic acid cycle, transketolase (TK) of the Calvin Benson cycle. It is also a cofactor in some enzymatic reactions, such as (i) in aerobic energy metabolism, (ii) carbohydrate catabolism, (iii) the pentose phosphate pathway and (iv) branched-chain amino acid biosynthesis (Linka and Weber, 2010; Bocobza et al., 2013; Khozaei et al., 2016). The major cereals, such as wheat, rice and corn, contain inadequate levels of this vitamin after processing and can lead to thiamine deficiencies. Vitamins has been selected as the target in the fight against micronutrient malnutrition in humans (Strobbe and Van Der Straeten, 2018).
The thiamine biosynthetic pathway in plants is similar to that in prokaryotes (Jurgenson et al., 2009). Thiamine is assembled from pyrimidine and thiazole moieties, which are synthesized independently (Goyer, 2010). The THIC (4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate synthase) protein catalyzes synthesis of pyrimidine moiety (4-amino-2-methyl-5-hydro-xymethylpyrimidine monophosphate, HMP-P) of thiamine from 5-aminoimidazole ribonucleotide (AIR). THIC is localized to the stroma of chloroplasts (Kong et al., 2008). THI1 catalyzes the synthesis of thiazole moiety (4-methyl-5- (2-hydroxyethyl)-thiazole phosphate, HET-P) from glycine, NAD+ and sulfur. THI1 is the only enzyme involved in thiazole biosynthesis (Papini-Terzi et al., 2003; Yazdani et al., 2013). In plants, thiamine deficiency leads to development of chlorotic leaves (Ajjawi et al., 2007a; Rapala-Kozik et al., 2007).
The THI1 gene was identified from Arabidopsis thaliana, which complemented bacterial defects in DNA repair (Machado et al., 1996). In thi1 mutants, the rosette leaves are yellow and requires on exogenous thiamine application for its survival (Papini-Terzi et al., 2003). THI1 gene is highly expressed in dividing tissues and regulates organ formation (Woodward et al., 2010). THI1 protein is typically modified at the N-terminus with signals for targeting to chloroplast and mitochondria (Chabregas et al., 2001). In Arabidopsis thaliana, th1, th2, th3, py and tz mutants requiring thiamine for survival have been isolated. In th1, th2 and th3, leaves are white and they require thiamine supplementation to become normal; whereas, py and tz can be rescued with an exogenous supply of HMP and HET, respectively (Feenstra, 1964; Redei, 1965). The three-dimensional structure of the Arabidopsis THI1 protein was resolved by Godoi et al. (2006). The protein assembles as an octamer, and the protein complex is a potential intermediate of thiazole biosynthesis. The octamer is tightly packed as a two-layer ring torus structure from four dimers. Proteomics studies have indicated that THI1 contains two to three conserved cysteine residues and is a potential target of chloroplast thioredoxin (Lemaire et al., 2004). THI1 encodes HET-P synthase, which is essential for thiamine biosynthesis. HET-P synthase has been detected in chloroplast and mitochondria in Z. mays and Arabidopsis (Belanger et al., 1995; Chabregas et al., 2003). HET-P synthase targeted to mitochondrial protect any DNA damage, while HET-P synthase targeted to chloroplasts is used in biosynthesis of thiamine (Ajjawi et al., 2007b). Vitamins can reinforce plant disease resistance and environmental stress tolerance, as well as act directly as exert antimicrobial activities (Hong et al., 2016; Huang et al., 2016). And it was recently reported that TPP also played a role in metabolic acclimation to the photoperiod (Rosado-Souza et al., 2019).
Here, we report the characterization of the Glycinemaxpalegreenleaf 1 (Gmpgl1) mutant. The GmPGL1 gene encodes a thiamine thiazole synthase (THI1). Pale green leaves of Gmpgl1 mutant seedling are rescued by supplementation with thiazole. Activities of TPP-dependent enzymes pyruvate dehydrogenase and pyruvate decarboxylase decreased in the Gmpgl1 mutant as compared to the wild type Williams 82. The metabolic network of amino acid synthesis and carbohydrate synthesis strongly affected in Gmpgl1. The presented results suggested that GmPGL1 gene was involved in biosynthesis of thiamine in soybean.
Materials and Methods
Plant Materials and Growth Conditions
We mutagenized 10,000 dry seeds of Williams 82 with 0.6% ethyl methyl sulfonate (EMS) in April 2013. The mutagenized seeds were grown in the Chang-Chun experiment filed of Northeast Institute of Geography and Agroecology, CAS in May 2013 and 4,354 M2 lines were harvested in October 2013. Twenty progeny of each M2 line were sowed in the field for phenotyping during May to October 2014. The screened mutant plants were backcrossed for four generations to purify the genetic background from July 2014 to October 2016 in both greenhouse and filed as described before (Chen et al., 2016). One of pale green leaf mutant, named as Glycinemaxpalegreenleaf 1 (Gmpgl1), was selected to mapping the mutation locus and function study.
Thiazole was exogenously applied as described by earlier (Hsieh et al., 2017) with minor modification. The seeds were germinated with H2O for two days in dark at 25°C, and then transferred into Hoagland nutrient solution, supplemented thiazole (Shanghai Macklin Biochemical Co. Ltd) with 15 mM for five days in a growth chamber under a 16 h/8 h light dark cycle at 30–25°C. Control plants were treated with H2O instead of thiazole. Three plants were investigated in each treatment.
Mapping of GmPGL1 Using Bulked-Segregant Analysis
F2 plants derived from a cross between the Gmpgl1 mutant and a Chinese local cultivar, Hedou12, were used to map the GmPGL1 gene (Song et al., 2015). DNA from 32 F2 individuals with the Gmpgl1 mutant phenotype and 50 F2 individuals with wild-type phenotype were bulked into mutant and wild-type pools, respectively. These DNA samples were used to construct libraries that were subjected to whole-genome sequencing using the Illumina HiSeq™ 2500 platform. Single-nucleotide polymorphisms (SNPs) and small InDels were calculated between the Gmpgl1 and Williams 82 bulked DNA samples by aligning the sequence reads of individual bulked DNA samples to the Glycine max Wm82.a2.v1 reference genome (https://phytozome.jgi.doe.gov/pz/portal.html). The candidate genomic regions were identified by QTL-seq method using bulked segregants from F2 population (Takagi et al., 2013). Sliding-window analysis with a 1 Mb window size and with an 100 kb increment for all chromosomes of soybean genome was conducted to develop the SNP index plots. The allele frequencies were calculated based on the formula SNP index = SNP Williams 82/(SNP Williams 82 + SNP Hedou12), and △(SNP-index) = (SNP-index of mutant-bulk)-(SNP-index of wild type-bulk). The genomic region with △(SNP-index) > 0.5 was selected as candidate area.
RNA Isolation and Real-Time Quantitative Polymerase Chain Reaction Analyses
Total RNA was extracted from the soybean tissue samples using Trizol reagent (Tiangen) according to the manufacturer's instructions. The first strand of cDNA was performed using the PrimeScript™ RT reagent kit (Takara, RR014) following the manufacture's methods. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using a SYBR® Premix Ex Taq™ Kit (Takara, RR420) on Stratagene MX3005P Real-Time PCR System (Agilent Technologies, Inc., Santa Clara, CA) according to the manufacturer's instructions. The qRT-PCR procedure was conducted as follows: 10 min at 95°C, followed by 40 cycles of 95°C for 10 s, 58°C for 20 s, and 72°C for 20 s. Three independent biological replications were performed for each sample to calculate the relative expression level using the 2−ΔΔCt method after normalization to Cons4 (ATP binding cassette transporter gene, Glyma.12G020500) (Libault et al., 2008). The primer pairs are listed in Table S1.
Plasmid Construction and Soybean Transformation
To generate GmPGL1 knock out plants, CRISPR/Cas system for targeted genome modification of crop plants was used (Shan et al., 2013). A pair of 23 bp 5′-ATTGAGCAGTCCGTGAGCCCCGG-3′, 5′-AACCCGGGGCTCACGGACTGCTC-3′ oligonucleotides specific to GmPGL1 was annealed and cloned into the modified VK005-04-soU6-2-GmUbi3 vector in the BspQI endonuclease restriction site (Du et al., 2016). The knock out construct was introduced into Agrobacterium tumefaciens EHA105, which was then use to transform cotyledonary explants of the 'Dongnong 50' (Zhao et al, 2016; Dai et al, 2018).
Database Searching and Phylogenetic Analysis
GmPGL1 homologues were identified by running the BLASTP program in the phytozome gbrowser (https://phytozome.jgi.doe.gov/pz/portal.html) for GmPGL1. GmPGL1 amino acid and its homologues were aligned with the ClustalW2 (version 2.0.8). Evolutionary relationships were deduced using the neighbor-joining algorithm (Saitou and Nei, 1987). Bootstrapping was performed using the PHYLIP program (version 3.6.7) with 1,000 replicates. The unrooted phylogenetic tree was constructed using the MEGA7 phylogenetic program (Kumar et al., 2016). Protein motifs of GmPGL1-like genes are profiled by MEME (version 5.0.5, Seattle, WA, USA).
Analysis of GmPGL1-GFP Subcellular Localization
The full-length CDS of the GmPGL1 gene is amplified using primers OL8155 and OL8156 that carry NdeI and SacI endonuclease restriction sites at their 5'-ends, respectively. The GmPGL1 CDS was cloned into the modified pUC19-2x35S-GFP vector carrying the NdeI and SacI sites (Wang et al., 2015) and a recombinant plasmid carrying the 2×35S: GFP-GmPGL1 fusion gene was obtained. The 2×35S: GFP-GmPGL1 plasmid was transiently introduced into Arabidopsis (Col-0) mesophyll protoplasts (Gao et al., 2017). The GFP signals were observed using a Nikon confocal microscope C2 (Japan).
Thiamin, Pigment, Amino Acids, Oxaloacetic Acid, Pyruvate, and α-Ketoglutarate Contents Determination
The thiamine was determined by HPLC as described earlier (Dong et al., 2015). The level of pigment contents of the first true leaves were harvested from Williams 82 and Gmpgl1, their 8-day-old seedlings were analyzed using a spectrophotometer as described earlier (Arnon, 1949). Contents of amino acids from 8-day-old leaves were determined using a previously described method (Schwarz et al., 2005; Gheshlaghi et al., 2008). The oxaloacetic acid, pyruvate and α-ketoglutarate from the leaves were extracted using a commercial chemical assay service from Jiangsu Comin Biotechnology Institute (Suzhou, China) according a previously described method (Petrarulo et al., 1990). Three biological replications were examined for both wild type and mutant plants for all above measurements.
Pyruvate Dehydrogenase, α-Ketoglutarate Dehydrogenase, and Pyruvate Decarboxylase Activity Assay
PDH, α-KGDH and PDC of leaves were determined by a commercial chemical assay service from Jiangsu Comin Biotechnology Institute (Suzhou, China) according the previous described method (Park et al., 2000; Sterk et al., 2003; Kikuchi et al., 2014; Eram and Ma, 2016). The activity assays were repeated from three biological replications.
Transmission Electron Microscopy
Transmission electron microscopy (TEM) was performed according to a previously described method (Kwon and Cho, 2008). Unifoliate leaves of the 8-day-old Williams 82 and Gmpgl1 seedlings were observed under a transmission electron microscopy (Hitachi, H-7650) at an accelerating voltage of 100 kV. Ten samples were investigated for both wild type and mutant plants.
Results
Isolation and Phenotypic Characterization of the Gmpgl1 Mutant
To investigate the genetic mechanisms affecting thiamine accumulation in soybean, Glycinemaxpalegreenleaf1 (Gmpgl1) mutant was isolated from an M2 population induced by EMS. The pale green leaf phenotype of Gmpgl1 mutant appeared yellower in the early development stage of first true leaves, as the leaf developed, the defective phenotype gradually disappeared, and phenotype appeared in the early development of ternately compound leaf (Figure 1A). The content of chlorophyll and carotenoid were determined in the Williams 82 and Gmpgl1 mutant because the content of pigment is associated with changes in leaf color. The total content of chlorophyll, chlorophyll a, chlorophyll b and carotenoid of the Gmpgl1 mutant was 0.67 ± 0.01, 0.44 ± 0.01 and 0.22 ± 0.01 mg.g-1 fresh weight, respectively. While these values of Williams 82 are 1.72 ± 0.12, 1.13 ± 0.08, and 0.59 ± 0.12 mg.g-1 fresh weight respectively. These indicated that the pigment contents of Gmpgl1 mutant are much less than those of wild type (Figure 1B).
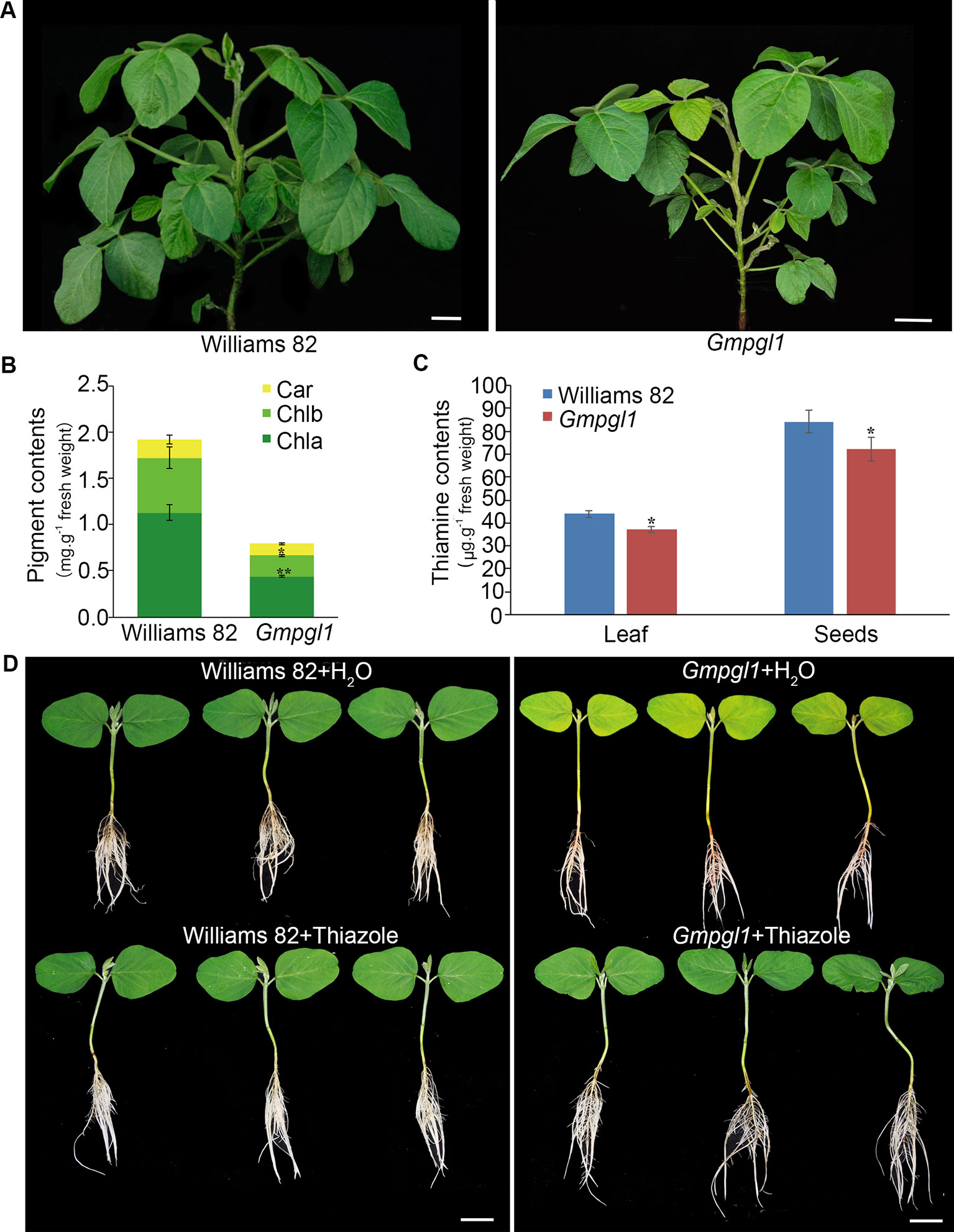
Figure 1 (A) The phenotypes of Williams 82 and Gmpgl1 mutant plant. Scale bars, 4 cm. (B) Pigment contents in the unifoliate leaves of 8-day old Williams 82 and Gmpgl1 seedlings. Chla, chlorophyll a; Chlb, chlorophyll b; Car, carotenoids. (C) Thiamine contents in unifoliate leaves of the 8-day-old seedlings, and mature seeds. Values are means from three biological replications. Asterisks indicate a stastically significant difference between the data of Williams 82 and Gmpgl1 determined by student's t-test (**P < 0.01, *P < 0.05) and the error bars represent standard deviations. (D) The 8-day old seedlings of Williams 82 and Gmpgl1 supplemented with water and 15 mM thiazole. Scale bars, 3 cm.
To determine if Gmpgl1 mutant influences thiamine accumulation, we quantified the total thiamine content of leaves of 8-day-old seedlings and mature seeds in the Williams 82 and Gmpgl1 mutant. The content of thiamine in unifoliate leaves of the 8-day-old Gmpgl1 mutant seedlings was only 74.6% of the corresponding value in the Williams 82 unifoliate leaves, and the content of thiamine in mature seeds decreased by 9.12% compared with that in the Williams 82 (Figure 1C).
Feeding of Gmpgl1 mutant and wild type seedlings with thiazole (15mM) restored the normal green unifoliate leaf phenotype in the Gmpgl1 mutant (Figure 1D). The complementation of the pale green unifoliate phenotype of the Gmpgl1 with thiazole confirmed that Gmpgl1 is a thiazole-deficient mutant.
Genetic Mapping of the Gmpgl1 Mutation Locus from F2 Population
The Gmpgl1 mutant was crossed to Hedou12 to generate a segregation population for mapping the GmPGL1 gene. The F1 plants were normal, but a 3:1 (WT: mutant = 108:32, χ2 test, p = 0.67) segregation ratio was observed in the F2 generation, indicating that the Gmpgl1 mutation is single and recessive. Based on the phenotype of Gmpgl1, the DNA from 32 F2 individuals carrying the GmPGL1 mutation in homozygous condition and 50 F2 individuals of wild-type phenotype were pooled into a Gmpgl1 bulk and a Williams 82 bulk for further bulked segregant analysis (BSA) analysis, respectively. From this sequence comparison, 1,208,350 SNPs and InDels were identified that distinguish the mutant bulk and wild type-bulk genotypes after filtering from 1,434,460 SNPs and InDels. One major statistically significant peak with the highest △ (SNP-index) value was identified in the intervals of 46.90-48.90 Mb region of the Chromosome 10 and was considered as the GmPGL1 region (Figures 2A, B). In this region, three SNPs and two InDels were identified from 3,250 SNPs and 814 InDels related to mutation locus (Table 1, Table S2). Among them, the C to T transversion of Glyma.10G259100 at 48,503,156 bp resulted in Asp to Asn conversion in its protein of this gene, and a T-deletion at 47,970,082 bp of Glyma.10G251500 caused its ORF frameshift; the rest three mutations were not in the coding region. The T-deletion of Glyma.10G251500.1 was at 292 nucleotide position of its CDS, which resulted in a truncated protein with a premature stop codon at the 357th nucleotide position (Figure 2C).
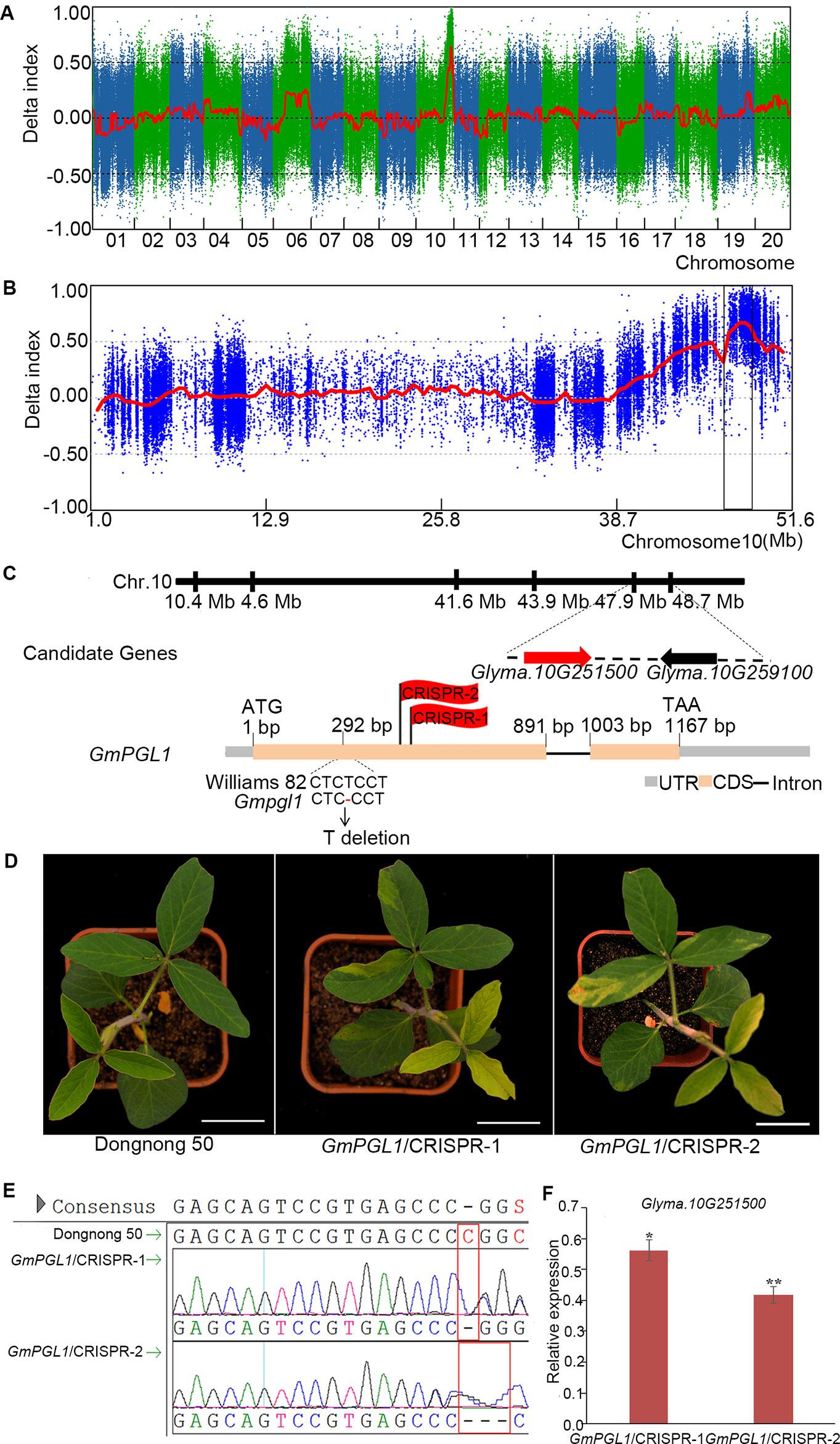
Figure 2 Bulked segregant analysis (BSA) mapping of Gmpgl1. (A) SNP index plot on all chromosomes of F2. (B) SNP index plots on chromosomes 10 for Gmpgl1 mutant from F2 population. The interval between 46.90 - 48.90 Mb of chromosome 10 with a highest peak is the candidate region for Gmpgl1, in which the ΔSNP-index was greater than mean value (0.5). (C) The candidate gene GmPGL1 (Glyma.10G251500.1) is shown in the red arrow, the others in the black arrows. ATG and TAA are the start and stop codons, respectively. One thymidine nucleotide is deleted at 292 bp in the first exon of GmPGL1 gene. (D) Knockout of GmPGL1 in wild-type background phenocopied the Gmpgl1 mutant. Scale bars, 3 cm. (E) The sequences comparison of CRISPR plants and Dongnong 50 (WT) in the knock down regions. (F) The relative expression of GmPGL1 gene in CRISPR plants compared with Dongnong 50. Asterisks indicate a statistically significant difference determined by student's t-test (**P < 0.01, *P < 0.05) and the error bars represent standard deviations.
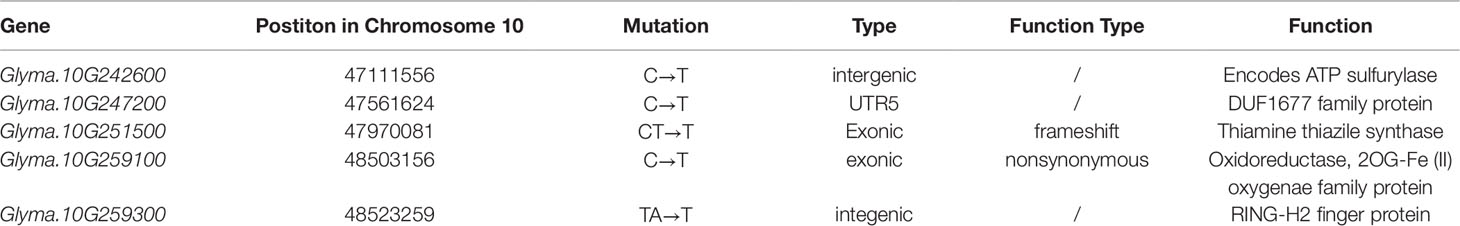
Table 1 Three SNPs and two InDels in the candidate region between 46.90 Mb and 48.90 Mb on chromosome 10 in Gmpgl1 mutant.
We compared the expression level of two candidate genes in the leaves of wild-type and Gmpgl1 mutant. The relative expression of Glyma.10G251500 is 2.57 ± 0.52 in Gmpgl1 mutant, and 13.21 ± 2.23 in Williams 82. However, the expression of Glyma.10G259100 is 0.19 ± 0.018 in Gmpgl1 mutant and 0.22 ± 0.04 in Williams 82 (Figure S1). The expression of the former is much lower in Gmpgl1 mutant compared with wild-type, while the latter is no significant difference. This indicated that Glyma.10G251500 might be more likely linked to mutation phenotype. And the independent mutation screening of Glyma.10G259100 mutation from our lab also proved its mutation could not bring the pale green yellow phenotype.
To confirm whether Glyma.10G251500.1 was the GmPGL1 gene, loss-of-function T1 heterozygosis transgenic lines were generated by inducing mutations in the Glyma.10G251500.1 gene using a CRISPR/Cas9 system. The CRISPR/Cas9-induced mutations in Glyma.10G251500.1 in two independent mutants caused development of the Gmpgl1-specific mutant phenotype (Figures 2D, E), the expression level of Glyma.10G251500.1 decreased in GmPGL1/CRISPR-1 and GmPGL1/CRISPR-2 (Figure 2F), and supplying thiazole could reduce the CRISPR mutants' phenotype to wild type (Figure S2). Above results suggested that Glyma.10G251500.1 is the GmPGL1 gene.
GmPGL1 Encoded Thiamine Thiazole Synthase
Blast searches revealed that GmPGL1 was highly homologous to the Arabidopsis THI1 (AT5G54770) protein (69.5% identical amino acids), which encodes a HEP-P synthase. In soybean, the most homologous gene of GmPGL1 is Glyma.20G142000.1, which exhibits 98.3% high-sequence identity with GmPGL1, and 67.5% with Arabidopsis THI1. Both of their proteins are 351 amino acid in length. Amino acid sequence comparison generated a phylogenetic tree with Glycine max THI1 family and its homologs from dicot plant species and monocotyledon plant species, Phaseolus vulgaris, Arabidopsis thaliana, Medicago, Lotus corniculatus, Oryza.sativa L, Zea mays L, and Brassica napus L (Figure 3A). It suggests that two THI1 homologues are evolutionarily conserved among plant kingdom, which share the common genomic structure in observed plant species.
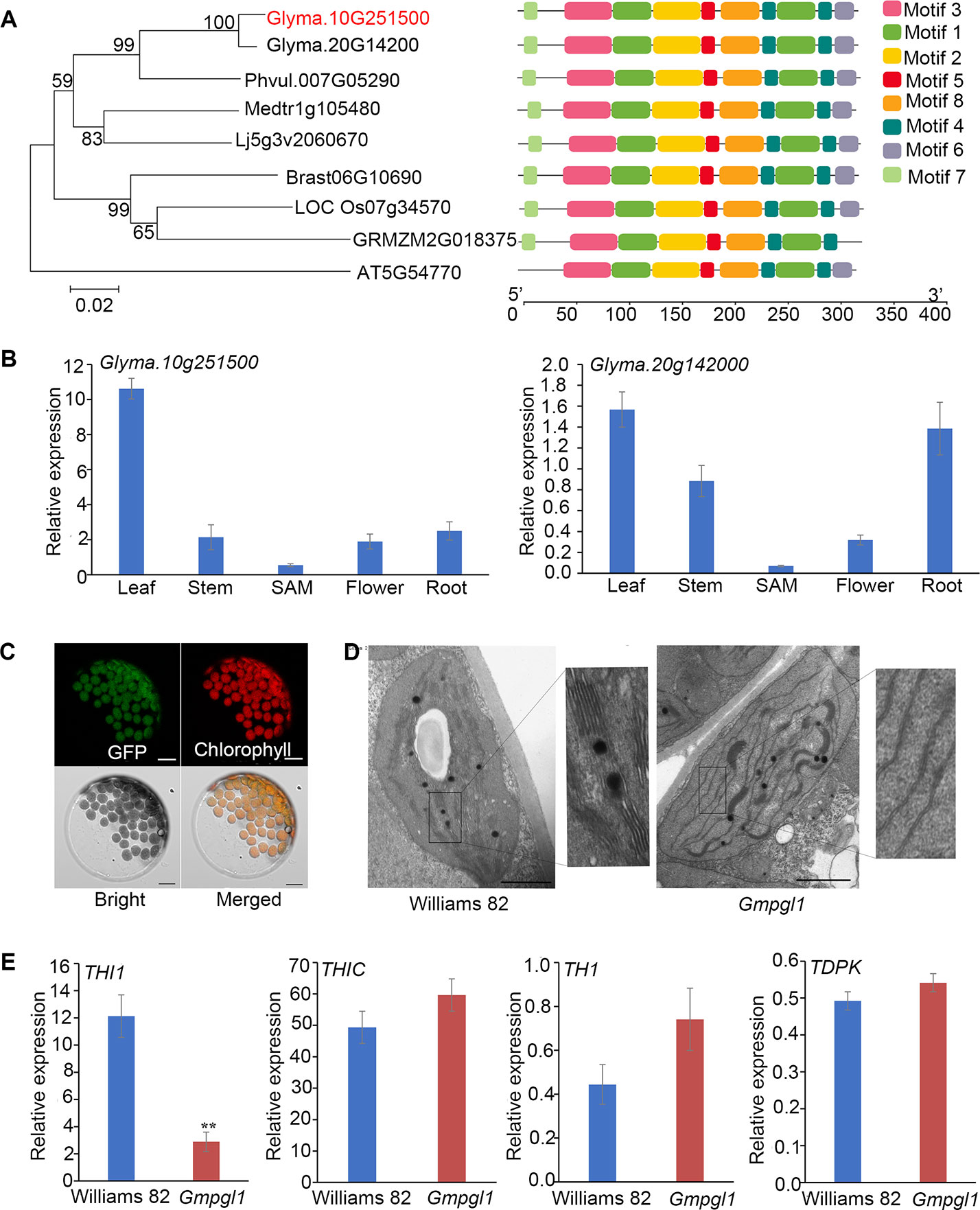
Figure 3 (A) Phylogenetic tree of GmPGL1 and its homologous proteins from Phaseolus vulgaris, Arabidopsis thaliana, Medicago, Lotus corniculatus, Oryza.sativa L, Zea mays L and Brassica napus L. The 8 conserved motifs are represented by colored boxes. The scale of protein length is given below the schematic diagram. (B) Expression levels of GmPGL1 and Glyma.20G142000.1 gene in different tissues of wild-type Williams 82 plant. (C) Transient expresison of GFP-GmPGL1 fusion proteins in Arabidopsis protoplasts. GFP, fluorescence signals of the fusion proteins; Chlorophyll, autofluorescence signals of chloroplasts; Bright, the protoplasts under the bright field; Merged, Merged images of the GFP-GmPGL1 fusion protein, chlorophyll, and bright field images. Scale bars, 10 μm. (D) Transmission electron micrographs of chloroplasts from the 8-day old leaves of Williams 82 and Gmpgl1 mutant. Scale bars, 10 μm. (E) The relative expression levels of the genes involved in thiamine biosynthesis between the Williams 82 and Gmpgl1 mutant unifoliate leaves. The reference gene is Cons4 of soybean, values from three independent biological replicates. Asterisks indicate a stastically significant difference between the data of Williams 82 and Gmpgl1 determined by Student's t-test (**P < 0.01) and the error bars represent standard deviations.
We further examined the expression patterns of the GmPGL1 and its homologue Glyma.20G142000. GmPGL1 expressed higher than Glyma.20G142000 in all tested tissues, and GmPGL1 also especially highly expressed in leaves compared that in stem, root and flower (Figure 3B). This result suggests that GmPGL1 might be dominant functional paralogue in soybean.
In the early development of leaves, the relative expression level of GmPGL1 reduced in Gmpgl1 mutant compared with Williams 82, however, the relative expression level in Glyma.20G142000 increased in Gmpgl1 mutant (Figure S1). It suggested the expression of GmPGL1 reduction caused the lowing of thiamine contents in Gmpgl1 mutant, the elevated Glyma.20G142000 expression might compensate for the loss of function of the former.
GmPGL1 Localized in Chloroplasts and Influenced Chloroplast Development
It has been reported that THI1 protein had two distinct functions due to different locations in cells (Chabregas et al., 2003; Ajjawi et al., 2007b). Targeting to chloroplasts, THI1 protein is involved in thiamine biosynthesis; while in mitochondria, it is involved in protecting DNA damages in Arabidopsis. GmPGL1 is predicted to locate in the chloroplast with TargetP program (http://www.cbs.dtu.dk/services/TargetP/). Transient expression of the GFP-GmPGL1 fusion gene revealed localization of the GFP-GmPGL1 fusion protein with the auto-fluorescent signals of chlorophyll (Figure 3C). This result indicated that GmPGL1 was located in the chloroplasts.
In order to make sure whether GmPGL1 influences chloroplast development, TEM analysis was performed. The chloroplasts of Williams 82 cells have abundant and well-ordered thylakoids and stacked membranes, whereas chloroplasts in the Gmpgl1 mutant had fewer lamellae per chloroplast and fewer lamellae per granum (Figure 3D). Because of the complementation of the Gmpgl1 mutant phenotype with exogenous application of thiazole, it was most likely due to thiamin deficiency but not a direct effect of GmPGL1 that led to the reduction in chlorophyll content and abnormal chloroplast development in Gmpgl1 mutant.
To resolve the function of GmPGL1 in thiamine biosynthesis pathway, we examined the expression differences of key genes of the thiamine biosynthetic pathway between wild type and Gmpgl1 mutant including THI1 (GmPGL1, Glyma.10g251500), THIC (Glyma.18g065700), TH1 (Glyma.08g18200) and TDPK (Glyma.03g213500). The relative expression levels of THI1, THIC, TH1 and TPK1 comparisons with Cons4 are 2.89 ± 0.71, 59.65 ± 10.38, 0.74 ± 0.14, 0.54 ± 0.03 in Gmpgl1 mutant respectively, while these values of Williams 82 are 12.13 ± 1.56, 49.37 ± 6.89, 0.44 ± 0.09 and 0.49 ± 0.06 respectively. The expression of THI1 (GmPGL1) was down-regulated in the Gmpgl1 mutant as compared to Williams 82, while the other genes were slightly up-regulated (Figure 3E).
Mutation in GmPGL1 Led to Altered Primary Metabolism
Thiamine is a co-factor in several enzymes in key cellular metabolic pathways, such as, PDC, PDH and α-KGDH. PDC is the first enzyme for all homo-fermentative ethanol pathways, and PDH and α-KGDH are two central enzymes in energy metabolism. The activity of α-KGDH, PDH and PDC were 87.79 ± 1.45, 0.90 ± 0.004 and 0.17 ± 0.004 µmol.min-1.g-1 fresh weight respectively in Gmpgl1 mutant, while 79.54 ± 0.81, 2.88 ± 0.02 and 0.23 ± 0.006 µmol.min-1.g-1 fresh weight in Williams 82 (Figure 4A). The activity of α-KGDH increased in the Gmpgl1 mutant as compared to Williams 82. Whereas, the activities of PDH and PDC were significantly reduced in Gmpgl1 mutant, as compared to that in Williams 82. The changes of above enzymes' activities in the Gmpgl1 mutant were consistent with the trends of substrate accumulations, such as the reduction of α-ketoglutaric acid, and increment of pyruvic acid in mutant plant (Figure 4B).
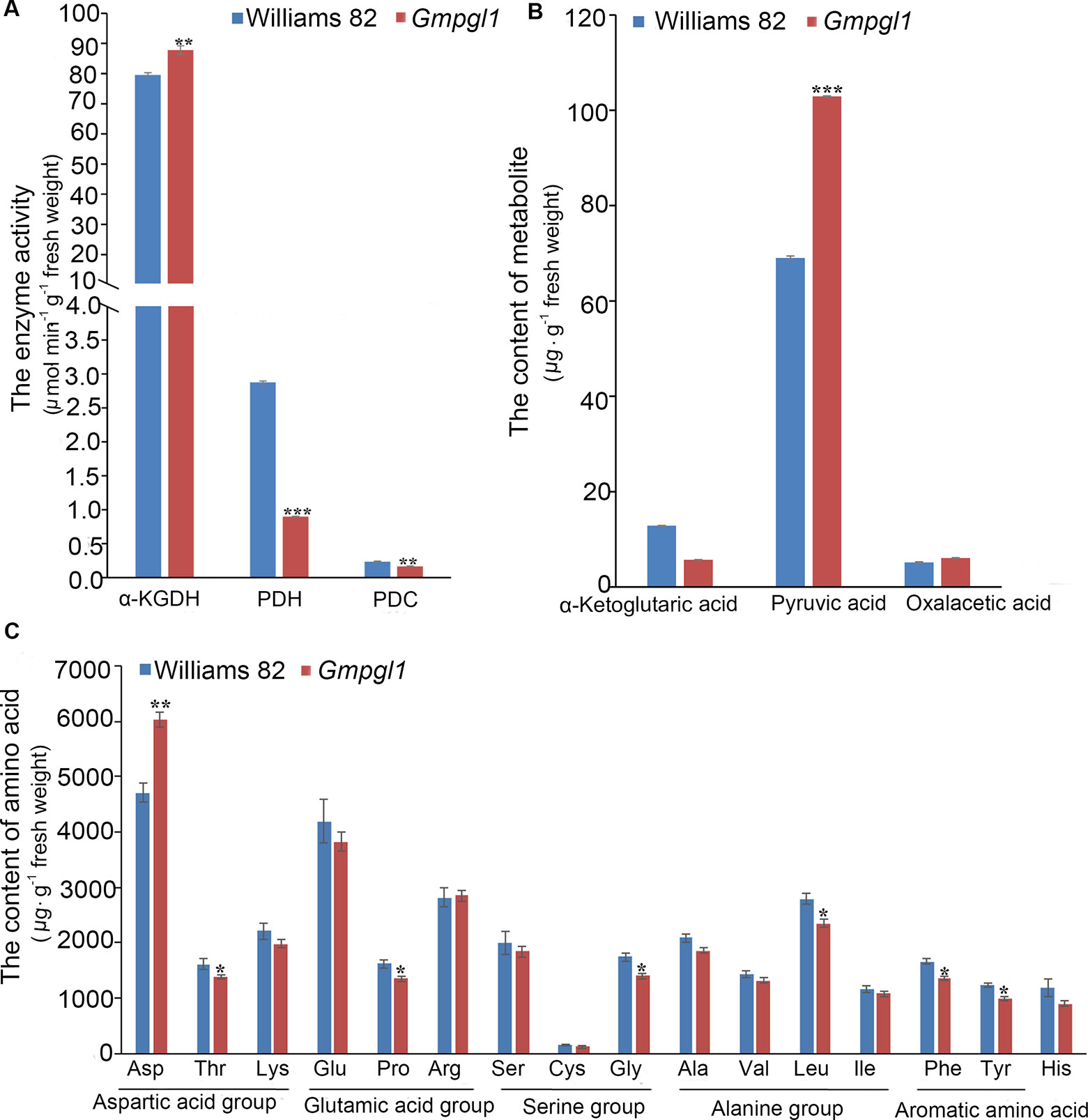
Figure 4 Some enzymes and metabolites changes between Williams 82 and Gmpgl1 mutant in the unifoliate leaves of 8-day-old seedlings. (A) The enzyme activities of α-KGDH, PDH and PDC. (B) The contents of α-ketoglutaric, pyruvic acid and oxalacetic acid. (C) The contents of 16 amino acid. Values are means ± SD of three replicates, asterisks indicate the statistically significant differences between the Williams 82 and Gmpgl1 mutant. Student's t-test: ***P ρ < 0.001, **P ρ <0.01, *P ρ < 0.05.
To determine if the amino acids metabolic pathways in Gmpgl1 are altered as compared to Williams 82, we conducted metabolic profiling for the amino acids (Figure 4C). The contents of Thr, Pro, Gly, Leu, Phe, and Tyr were decreased in the Gmpgl1 mutant as compared to that in Williams 82, while only Asp was increased. The changes of amino acid related to the contents of their precursors, such as, α-ketoglutaric acid, increment of pyruvic acid and oxaloacetic acid (Figure 4B).
These results show that mutation of GmPGL1 gene effects the thiamine related metabolism enzymes and some primary metabolisms and their precursors.
Discussion
THI1 encoded thiamine thiazole synthase is the essential for thiamine biosynthesis. Arabidopsis tz mutants are chlorotic and die early without thiazole or thiamine supplementation (Redei, 1962). In a tz mutant line, a point mutation in the THI1 gene causes an amino acid substitution in a highly conserved residue (Papini-Terzi et al., 2003), suggesting that the Arabidopsis THI1 gene is essential for thiamine biosynthesis. OsDR8 encodes HET-P synthase and is involved in the synthesis of the thiazole moiety of thiamine in rice (Belanger et al., 1995). In Arabidopsis th1, thi1 and py mutants fail to survive without thiamine supplementation (Redei, 1962; Feenstra, 1964; Li and Redei, 1969); and in maize, blk1-R mutant is rescued by exogenous thiamine supplementation (Woodward et al., 2010). The content of thiamine was decreased in leaves of the young seedlings and mature seeds of the Gmpgl1 mutant. The phenotype of Gmpgl1 can also rescued by thiazole supplementation in our work (Figure 1D). These results indicated that the lost function of THI1 reduced thiamine biosynthesis, and thiamin deficiency caused chloroplast abnormal development and leaf chlorosis.
In the Gmpgl1 mutant, the T-deletion of Glyma.10G251500.1 resulted in a truncated protein with a premature stop codon at the 357th nucleotide position. The expression of the GmPGL1 gene was down-regulated in Gmpgl1 mutant as compared to Williams 82, which might be due to nonsense-mediated mRNA decay (NMD) pathway (Isshiki et al., 2001). However, the relative expression level in GmPGL1 paralogue, Glyma.20G142000, slightly increased in Gmpgl1 mutant, which probably compensate for the loss of function of the Gmpgl1 mutant. The further study of the function of Glyma.20G142000 will help us to comprehensively understand the thiamine biosynthesis pathway in soybean, and the role of thiamine in both abiotic and biotic stresses, soybean development and metabolism.
TPP is the active cofactor form of thiamine required by various enzymes involved in carbohydrate and amino acid metabolism in plants. In Arabidopsis, thiamin level is regulated by a TPP-responsive riboswitch of the THIC pre-mRNA (Bocobza et al., 2007; Wachter et al., 2007). Increasing TMP level in leaves increased extractable PDH, α-KGDH, and TK activities without changing the protein levels of PDH and TK in riboswitch deficiency mutant (Bocobza et al., 2013). However, Hsieh et al. (2017) also reported that transcript levels of the genes encoding TPP-dependent enzymes, such as PDH, α-KGDH, and TK, are up-regulated in the TPP deficiency mutant. These controversial results might result from different measurement condition and organism. The photosynthetic and metabolic phenotypes of TPP riboswitch mutants are photoperiod dependent, appropriate TPP levels are required for acclimation to changes in photoperiod (Rosado-Souza et al., 2019). The thiamin-requiring enzymes' activities might regulate by different TPP availabilities: allosteric regulation may be the main control over under low TPP availability, high TPP availability becomes a major point of control of these enzymes during the dark photoperiod (Bocobza et al., 2013). We observed that activities of PDH, PDC decreased in Gmpgl1 mutant, while that of α-KGDH increased (Figure 4A). Because the enzyme activity is measured by fresh weight per gram in the study, the reduction activities of PDH, PDC in Gmpgl1 mutant might cause by either the protein level of enzyme or the enzyme activity per mole. The difference of alter the difference of altering trend among PDH, PDC and α-KGDH in Gmpgl1 mutant might due to their distributions in plant cells and the binding ability of ThDP cofactor (Hanson et al., 2018; Joshi et al., 2019).
In summary, GmPGL1 encodes thiamine thiazole synthase, which is targeted to chloroplasts and essential for thiamine biosynthesis. Identification of GmPGL1 could improve our understanding of the molecular mechanism for thiamine biosynthesis in soybean.
Data Availability Statement
All datasets for this study are included in the article/Supplementary Material.
Author Contributions
SY and XZF designed the research. XFF performed the experiments. KT provided bioinformatics analyses. YZ, JL, JM and QW provided the technical assistance. XFF, XZF and SY analyzed the data and wrote the manuscript. All authors declared no conflicting interest on the contents of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professor Madan Bhattacharyya (Iowa State University) for critical reading of the manuscript. The project was funded by the National Key Research and Development Project (Grant Nos. 2016YFD0100401) from the Ministry of Science and Technology of China; and by STS Program of Chinese Academy of Sciences (Grant Nos. KFJ-STS-ZDTP-048).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01546/full#supplementary-material
References
Abe, A., Kosugi, S., Yoshida, K., Natsume, S., Takagi, H., Kanzaki, H., et al. (2012). Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30 (2), 174–178. doi: 10.1038/nbt.2095
Ajjawi, I., Milla, M. A. R., Cushman, J., Shintani, D. K. (2007a). Thiamin pyrophosphokinase is required for thiamin cofactor activation in Arabidopsis. Plant Mol. Biol. 65 (1-2), 151–162. doi: 10.1007/s11103-007-9205-4
Ajjawi, I., Tsegaye, Y., Shintani, D. (2007b). Determination of the genetic, molecular, and biochemical basis of the Arabidopsis thaliana thiamin auxotroph th1 . Arch. Biochem. Biophys. 459 (1), 107–114. doi: 10.1016/j.abb.2006.11.011
Arnon, D. I. (1949). Copper enzymes in isolated chloroplasts-polyphenoloxidase in beta-vulgaris. Plant Physiol. 24 (1), 1–15. doi: 10.1104/pp.24.1.1
Belanger, F. C., Leustek, T., Chu, B. Y., Kriz, A. L. (1995). Evidence for the thiamine biosynthetic pathway in higher-plant plastids and its developmental regulation. Plant Mol. Biol. 29 (4), 809–821. doi: 10.1007/bf00041170
Bocobza, S., Adato, A., Mandel, T., Shapira, M., Nudler, E., Aharoni, A. (2007). Riboswitch-dependent gene regulation and its evolution in the plant kingdom. Genes Dev. 21 (22), 2874–2879. doi: 10.1101/gad.443907
Bocobza, S. E., Malitsky, S., Araujo, W. L., Nunes-Nesi, A., Meir, S., Shapira, M., et al. (2013). Orchestration of thiamin biosynthesis and central metabolism by combined action of the thiamin pyrophosphate riboswitch and the circadian clock in Arabidopsis. Plant Cell 25 (1), 288–307. doi: 10.1105/tpc.112.106385
Chabregas, S. M., Luche, D. D., Farias, L. P., Ribeiro, A. F., van Sluys, M. A., Menck, C. F. M., et al. (2001). Dual targeting properties of the N-terminal signal sequence of Arabidopsis thaliana THI1 protein to mitochondria and chloroplasts. Plant Mol. Biol. 46 (6), 639–650. doi: 10.1023/a:1011628510711
Chabregas, S. M., Luche, D. D., Van Sluys, M. A., Menck, C. F. M., Silva-Filho, M. C. (2003). Differential usage of two in-frame translational start codons regulates subcellular localization of Arabidopsis thaliana THI1 . J. Cell Sci. 116 (2), 285–291. doi: 10.1242/jcs.00228
Chen, W., Gao, J. S., Feng, X. X., Shao, Q., Yang, S. X., Feng, X. Z. (2016). Characterization of dwarf mutants and molecular mapping of a dwarf locus in soybean. J. Integr. Agric. 15, 60345–60347. doi: 10.1016/S2095-3119(15)61312-0
Dai, A., Yang, S., Zhou, H., Tang, K., Li, G., Leng, J., et al. (2018). Evolution and expression divergence of the CYP78A subfamily genes in soybean. Genes (Basel) 9 (12), E611. doi: 10.3390/genes9120611
Dong, W., Stockwell, V. O., Goyer, A. (2015). Enhancement of thiamin content in Arabidopsis thaliana by metabolic engineering. Plant Cell Physiol. 56 (12), 2285–2296. doi: 10.1093/pcp/pcv148
Du, H. Y., Zeng, X. R., Zhao, M., Cui, X. P., Wang, Q., Yang, H., et al. (2016). Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9 . J. Biotechnol. 217, 90–97. doi: 10.1016/j.jbiotec.2015.11.005
Eram, M. S., Ma, K. (2016). Pyruvate decarboxylase activity of the acetohydroxyacid synthase of Thermotoga maritima. Biochem. Biophys. Rep. 7, 394–399. doi: 10.1016/j.bbrep.2016.07.008
Feenstra, W. J. (1964). Isolation of nutritional mutants in Arabidopsis thaliana. Genetica 35 (4), 259–261. doi: 10.1007/bf01804894
Gangolf, M., Czerniecki, J., Radermecker, M., Detry, O., Nisolle, M., Jouan, C., et al. (2010). Thiamine status in humans and content of phosphorylated thiamine derivatives in biopsies and cultured cells. PloS One 5 (10), e13616. doi: 10.1371/journal.pone.0013616
Gao, J., Yang, S., Cheng, W., Fu, Y., Leng, J., Yuan, X., et al. (2017). GmILPA1, encoding an APC8-like protein, controls leaf petiole angle in soybean. Plant Physiol. 174 (2), 1167–1176. doi: 10.1104/pp.16.00074
Gheshlaghi, R., Scharer, J. M., Moo-Young, M., Douglas, P. L. (2008). Application of statistical design for the optimization of amino acid separation by reverse-phase HPLC. Anal. Biochem. 383 (1), 93–102. doi: 10.1016/j.ab.2008.07.032
Godoi, P. H. C., Galhardo, R. S., Luche, D. D., Van Sluys, M. A., Menck, C. F. M., Oliva, G. (2006). Structure of the thiazole biosynthetic enzyme THI1 from Arabidopsis thaliana. J. Biol. Chem. 281 (41), 30957–30966. doi: 10.1074/jbc.M604469200
Goyer, A. (2010). Thiamine in plants: aspects of its metabolism and functions. Phytochemistry 71 (14-15), 1615–1624. doi: 10.1016/j.phytochem.2010.06.022
Goyer, A. (2017). Thiamin biofortification of crops. Curr. Opin. In Biotechnol. 44, 1–7. doi: 10.1016/j.copbio.2016.09.005
Hanson, A. D., Amthor, J. S., Sun, J., Niehaus, T. D., Gregory, J. F., et al. (2018). Redesigning thiamin synthesis: prospects and potential payoffs. Plant Sci. 273, 92–99. doi: 10.1016/j.plantsci.2018.01.019
Hong, J. K., Kim, H. J., Jung, H., Yang, H. J., Kim, D. H., Sung, C. H., et al. (2016). Differential control efficacies of vitamin treatments against bacterial wilt and grey mould diseases in tomato plants. Plant Pathol. J. 32 (5), 469–480. doi: 10.5423/PPJ.OA.03.2016.0076
Hsieh, W. Y., Liao, J. C., Wang, H. T., Hung, T. H., Tseng, C. C., Chung, T. Y., et al. (2017). The Arabidopsis thiamin-deficient mutant pale green1 lacks thiamin monophosphate phosphatase of the vitamin B-1 biosynthesis pathway. Plant J. 91 (1), 145–157. doi: 10.1111/tpj.13552
Huang, W. K., Ji, H. L., Gheysen, G., Kyndt, T. (2016). Thiamine-induced priming against root-knot nematode infection in rice involves lignification and hydrogen peroxide generation. Mol. Plant Pathol. 17 (4), 614–624. doi: 10.1111/mpp.12316
Isshiki, M., Yamamoto, Y., Satoh, H., Shimamoto, K. (2001). Nonsense-mediated decay of mutant waxy mRNA in rice. Plant Physiol. 125 (3), 1388–1395. doi: 10.1104/pp.125.3.1388
Joshi, J., Folz, J. S., Gregory, J. F., McCarty, D. R., Fiehn, O., Hanson, A. D. (2019). Rethinking the PDH bypass and GABA shunt as thiamin-deficiency workarounds. Plant Physiol. 181 (2), 389–393. doi: 10.1104/pp.19.00857
Jurgenson, C. T., Begley, T. P., Ealick, S. E. (2009). The structural and biochemical foundations of thiamin biosynthesis. Annu. Rev. Biochem. 78, 569–603. doi: 10.1146/annurev.biochem.78.072407.102340
Khozaei, M., Fisk, S., Lawson, T., Gibon, Y., Sulpice, R., Stitt, M., et al. (2016). Overexpression of plastid transketolase in tobacco results in a thiamine auxotrophic phenotype. Plant Cell 28 (7), 1752–1754. doi: 10.1105/tpc.16.00527
Kikuchi, D., Minamishima, Y. A., Nakayama, K. (2014). Prolyl-hydroxylase PHD3 interacts with pyruvate dehydrogenase (PDH)-E1 beta and regulates the cellular PDH activity. Biochem. Biophys. Res. Commun. 451 (2), 288–294. doi: 10.1016/j.bbrc.2014.07.114
Kong, D. Y., Zhu, Y. X., Wu, H. L., Cheng, X. D., Liang, H., Ling, H. Q. (2008). AtTHIC, a gene involved in thiamine biosynthesis in Arabidopsis thaliana. Cell Res. 18 (5), 566–576. doi: 10.1038/cr.2008.35
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi: 10.1093/molbev/msw054
Kwon, K. C., Cho, M. H. (2008). Deletion of the chloroplast-localized AtTerC gene product in Arabidopsis thaliana leads to loss of the thylakoid membrane and to seedling lethality. Plant J. 55 (3), 428–442. doi: 10.1111/j.1365-313X.2008.03523.x
Lemaire, S. D., Guillon, B., Le Marechal, P., Keryer, E., Miginiac-Maslow, M., Decottignies, P. (2004). New thioredoxin targets in the unicellular photosynthetic eukaryote Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U. S. A. 101 (19), 7475–7480. doi: 10.1073/pnas.0402221101
Li, S. L., Redei, G. P. (1969). Thiamine mutants of crucifer Arabidopsis. Biochem. Genet. 3 (2), 163–165. doi: 10.1007/bf00520351
Libault, M., Thibivilliers, S., Bilgin, D. D., Radwan, O., Benitez, M., Clough, S. J., et al. (2008). Identification of four soybean reference genes for gene expression normalization. Plant Genome J. 1 (1), 44–54. doi: 10.3835/plantgenome2008.02.0091
Linka, N., Weber, A. P. M. (2010). Intracellular metabolite transporters in plants. Mol. Plant 3 (1), 21–53. doi: 10.1093/mp/ssp108
Lisec, J., Schauer, N., Kopka, J., Willmitzer, L., Fernie, A. R. (2006). Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 1 (1), 387–396. doi: 10.1038/nprot.2006.59
Machado, C. R., Oliveira, R. L. C., Boiteux, S., Praekelt, U. M., Meacock, P. A., Lenck, C. F. M. (1996). Thi1, a thiamine biosynthetic gene in Arabidopsis thaliana, complements bacterial defects in DNA repair. Plant Mol. Biol. 31 (3), 585–593. doi: 10.1007/bf00042231
Mimura, M., Zallot, R., Niehaus, T. D., Hasnain, G., Gidda, S. K., Nguyen, T. N. D., et al. (2016). Arabidopsis TH2 encodes the orphan enzyme thiamin monophosphate phosphatase. Plant Cell 28 (10), 2683–2696. doi: 10.1105/tpc.16.00600
Murashige, T., Skoog, F. (1962). A revised medium for rapid grpwth and bio assay with tobacco tissue cultures. Physiol. Plant. 15 (3), 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Papini-Terzi, F. S., Galhardo, R. D., Farias, L. P., Menck, C. F. M., Van Sluys, M. A. (2003). Point mutation is responsible for Arabidopsis tz-201 mutant phenotype affecting thiamin biosynthesis. Plant Cell Physiol. 44 (8), 856–860. doi: 10.1093/pcp/pcg104
Park, L. C. H., Calingasan, N. Y., Sheu, K. F. R., Gibson, G. E. (2000). Quantitative alpha-ketoglutarate dehydrogenase activity staining in brain sections and in cultured cells. Anal. Biochem. 277 (1), 86–93. doi: 10.1006/abio.1999.4359
Petrarulo, M., Bianco, O., Cosseddu, D., Marangella, M., Pellegrino, S., Linari, F. (1990). Improved high-performance liquid-chromatographic determination of urinary glycolic acid. J. Chromatogr.-Biomed. Appl. 532 (1), 130–134. doi: 10.1016/s0378-4347(00)83759-7
Ping, J. Q., Liu, Y. F., Sun, L. J., Zhao, M. X., Li, Y. H., She, M. Y., et al. (2014). Dt2 is a gain-of-function MADS-domain factor gene that specifies semideterminacy in soybean. Plant Cell 26 (7), 2831–2842. doi: 10.1105/tpc.114.126938
Rapala-Kozik, M., Olczak, M., Ostrowska, K., Starosta, A., Kozik, A. (2007). Molecular characterization of the thi3 gene involved in thiamine biosynthesis in Zea mays: cDNA sequence and enzymatic and structural properties of the recombinant bifunctional protein with 4-amino-5-hydroxymethyl-2-methylpyrimidine (phosphate) kinase and thiamine monophosphate synthase activities. Biochem. J. 408, 149–159. doi: 10.1042/bj20070677
Rapala-Kozik, M. (2011). Vitamin B-1 (Thiamine): A cofactor for enzymes involved in the main metabolic pathways and an environmental stress protectant. Adv. In Botanical Res. 58, 37–91. doi: 10.1016/B978-0-12-386479-6.00004-4
Redei, G. P. (1962). Genetic blocks in thiamine synthesis of angiosperm Arabidopsis. Am. J. Bot. 52 (8), 834–841. doi: 10.2307/2439765
Rosado-Souza, L., Proost, S., Moulin, M., Bergmann, S., Bocobza, S. E., Aharoni, A., et al. (2019). Appropriate thiamin pyrophosphate levels are required for acclimation to changes in photoperiod. Plant Physiol. 180 (1), 185–197. doi: 10.1104/pp.18.01346
Saitou, N., Nei, M. (1987). On the maximum-likelihood method for molecular phylogeny. Japanese J. Genet. 62 (6), 547–548. doi: 10.1007/BF02101285
Schwarz, E. L., Roberts, W. L., Pasquali, M. (2005). Analysis of plasma amino acids by HPLC with photodiode array and fluorescence detection. Clin. Chim. Acta 354 (1-2), 83–90. doi: 10.1016/j.cccn.2004.11.016
Shan, Q. W., Wang, Y. P., Li, J., Zhang, Y., Chen, K. L., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31 (8), 686–688. doi: 10.1038/nbt.2650
Song, X., Wei, H., Cheng, W., Yang, S., Zhao, Y., Li, X., et al. (2015). Development of INDEL markers for genetic mapping based on whole genome resequencing in soybean. G3 (Bethesda) 5, 12, 2793–2799. doi: 10.1534/g3.115.022780
Sterk, J. P., Stanley, W. C., Hoppel, C. L., Kerner, J. (2003). A radiochemical pyruvate dehydrogenase assay: activity in heart. Anal. Biochem. 313 (1), 179–182. doi: 10.1016/s0003-2697(02)00538-9
Strobbe, S., Van Der Straeten, D. (2018). Toward eradication of B-Vitamin deficiencies: considerations for crop biofortification. Front. In Plant Sci. 9, 443. doi: 10.3389/fpls.2018.00443
Takagi, H., Abe, A., Yoshida, K., Kosugi, S., Natsume, S., Mitsuoka, C., et al. (2013). QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 74 (1), 174–183. doi: 10.1111/tpj.12105
Wachter, A., Tunc-Ozdemir, M., Grove, B. C., Green, P. J., Shintani, D. K., Breaker, R. R. (2007). Riboswitch control of gene ex-pression in plants by splicing and alternative 3' end processing of mRNAs. Plant Cell 19 (11), 3437–3450. doi: 10.1105/tpc.107.05364
Wang, S. C., Tiwari, S. B., Hagen, G., Guilfoyle, T. J. (2005). Auxin response factor 7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. Plant Cell 17 (7), 1979–1993. doi: 10.1105/tpc.105.031096
Wang, X. Y., Wang, X. L., Hu, Q. N., Dai, X. M., Tian, H. N., Zheng, K. J., et al. (2015). Characterization of an activation-tagged mutant uncovers a role of GLABRA2 in anthocyanin biosynthesis in Arabidopsis. Plant J. 83 (2), 300–311. doi: 10.1111/tpj.12887
Woodward, J. B., Abeydeera, N. D., Paul, D., Phillips, K., Rapala-Kozik, M., Freeling, M., et al. (2010). A maize thiamine auxotroph is defective in shoot meristem maintenance. Plant Cell 22 (10), 3305–3317. doi: 10.1105/tpc.110.077776
Yamada, T., Watanabe, S., Arai, M., Harada, K., Kitamura, K. (2010). Cotyledonary node pre-wounding with a micro-brush increased frequency of Agrobacterium-mediated transformation in soybean. Plant Biotechnol. 27 (2), 217–220. doi: 10.5511/plantbiotechnology.27.217
Yazdani, M., Zallot, R., Tunc-Ozdemir, M., de Crecy-Lagard, V., Shintani, D. K., Hanson, A. D. (2013). Identification of the thiamin salvage enzyme thiazole kinase in Arabidopsis and maize. Phytochemistry 94, 68–73. doi: 10.1016/j.phytochem.2013.05.017
Zhang, Y., Li, X., Yang, S., Feng, X. (2017). Identification of ZOUPI orthologs in soybean potentially involved in endosperm breakdown and embryogenic development. Front. In Plant Sci. 8, 139. doi: 10.3389/fpls.2017.00139
Keywords: soybean, GmPGL1, thiamine thiazole synthase, carbohydrate synthesis, amino acid synthesis
Citation: Feng X, Yang S, Tang K, Zhang Y, Leng J, Ma J, Wang Q and Feng X (2019) GmPGL1, a Thiamine Thiazole Synthase, Is Required for the Biosynthesis of Thiamine in Soybean. Front. Plant Sci. 10:1546. doi: 10.3389/fpls.2019.01546
Received: 14 August 2019; Accepted: 05 November 2019;
Published: 22 November 2019.
Edited by:
Ling Yuan, University of Kentucky, United StatesReviewed by:
Samuel Edouard Bocobza, Agricultural Research Organization (ARO), IsraelQian Qian, China National Rice Research Institute (CAAS), China
Copyright © 2019 Feng, Yang, Tang, Zhang, Leng, Ma, Wang and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Suxin Yang, eWFuZ3N1eGluQGlnYS5hYy5jbg==
 Xingxing Feng
Xingxing Feng Suxin Yang
Suxin Yang Kuanqiang Tang
Kuanqiang Tang Yaohua Zhang
Yaohua Zhang Jiantian Leng1
Jiantian Leng1 Quan Wang
Quan Wang Xianzhong Feng
Xianzhong Feng