- College of Horticulture, Northwest A&F University, Yangling, China
Fruit set and development occur following successful fertilization. Parthenocarpy, a valuable trait in some self-incompatible species, produces seedless fruit without fertilization. Gibberellin (GA) is a crucial hormone in fruit-set regulation and development. While investigating the development of parthenocarpy in pear (Pyrus bretschneideri Rehd.), we determined that GA 20-oxidases (GA20ox) may play key roles in seedless pear fruit development. Sequence analysis revealed three PbGA20ox genes: PbGA20ox1, PbGA20ox2, and PbGA20ox3. We analyzed the expression patterns of candidate genes and found that PbGA20ox2 levels significantly changed in pollinated fruits. Tissue-specific expression assays revealed that PbGA20ox2 is highly expressed in young fruit and leaves. Subcellular localization assays showed it was located in the cytoplasm, nucleus, and plasma membrane. Overexpressed PbGA20ox2 tomato plants were taller and had longer hypocotyls and internodes, and the emasculated flowers produced parthenocarpic fruit. In pear, the transient overexpression of PbGA20ox2 promoted fruit development and delayed the drop of nonpollinated fruit. Furthermore, the fruit of PbGA20ox2-overexpressing tomato and transient PbGA20ox2-overexpressing pear had increased GA4 (but not GA3 and GA1) contents. This result provided evidence that PbGA20ox2 was necessary for GA4-dependent pear fruit development. Our study revealed that PbGA20ox2 altered the GA biosynthetic pathway and enhanced GA4 synthesis, thereby promoting fruit set and parthenocarpic fruit development.
Introduction
Fruit set, a key developmental process that occurs following successful fertilization in higher plants, is the first step in fruit development (Joldersma and Liu, 2018). Fruit set and development represent the commitment of an angiosperm plant to the development of mature fruit (Joldersma and Liu, 2018). In many fruiting plants, pollination and fertilization are necessary for fruit set and development, with these events leading to the development of seeds that promote cell division and fruit development in a synchronized manner (McAfee et al., 2013).
‘Dangshansu’ (Pyrus bretschneideri Rehd.), a pear cultivar with fleshy fruits, is highly studied because of its considerable economic importance. In most fruiting species, fruit set and development are triggered by pollination and fertilization. However, many fruit species, including pear, exhibit natural self-incompatibility. Parthenocarpy, which induces seedless fruit, is independent of pollination and fertilization and is a valuable trait for some self-incompatible species.
Plant hormones play key roles in parthenocarpic fruit growth. Parthenocarpy can be induced by the exogenous application of plant hormones, such as gibberellins (GAs), auxins, and melatonin (Cong et al., 2018; Liu et al., 2018a; Liu et al., 2018b), as well as the regulation of endogenous hormone contents (Mesejo et al., 2010; Ding et al., 2013; Sugiyama et al., 2014; Niu et al., 2015). Application of GAs can be used for this purpose in many horticultural plants, such as apple (Watanabe et al., 2008), loquat (Aslmoshtaghi and Shahsavar, 2013) and pear (Liu et al., 2018b). Genes related to GA biosynthetic or signaling pathways may also have functions in parthenocarpic fruit development. In tomato, the silencing of the GA 2-oxidase (GA2ox) gene, which encodes a deactivating enzyme in the GA biosynthetic pathway, induces parthenocarpic fruit and inhibits lateral branching (Martínez-Bello et al., 2015). The silencing of the DELLA gene, which negatively regulates GA signaling, induces facultative parthenocarpy in tomato fruit (Martí et al., 2007). In pea, the overexpression of GA3ox, which converts inactive GA to active GA, induces parthenocarpic fruit (Reinecke et al., 2013).
In ‘Dangshansu’ pear, exogenous application of GA4+7 (Gibberellin A4 and A7), but not GA3, can induce parthenocarpy (Liu et al., 2018b). GA plays important roles in regulating diverse developmental processes (Hedden and Sponsel, 2015), such as shoot elongation, germination, flowering, fruit set, and growth (Hedden and Kamiya, 1997; Olszewski et al., 2002). The GA metabolic pathways have previously been described (Singh et al., 2010; Hedden and Thomas, 2012). GAs are synthesized from trans-geranylgeranyl pyrophosphate and metabolized to GA12 and/or GA53. The two precursors are converted to active GA4 and GA1 by GA20ox and GA3ox (García-Hurtado et al., 2012). GA20ox plays a key role in this pathway and is involved in the successive oxidation of C-20, leading to its removal and the formation of C19-GAs (GA9 and GA20) (Rieu et al., 2008a). GA4 and GA1 are synthesized from GA9 and GA20, respectively. In brief, two parallel pathways exist that include consecutively acting GA20ox: non-13-hydroxylation and early-13-hydroxylation pathways (Supplementary Figure S1). GA20ox has been cloned and characterized in many plants. For example, five GA20ox genes are present in Arabidopsis, six in grape and at least four in tomato (Serrani et al., 2008; Rieu et al., 2008b; Giacomelli et al., 2013). In these species, GA20ox is involved in diverse developmental processes, including vegetative and reproductive growth.
Here, we reported that PbGA20ox2 regulated fruit set and development by increasing the contents of specific active GAs during an early stage of fruit development in pear. Using quantitative real-time PCR (qRT-PCR), tissue-specific expression analyses and subcellular location assays, we investigated the expression patterns of PbGA20ox2. Furthermore, we investigated the functions of PbGA20ox2 in fruit set and development using a stable genetic overexpression assay in tomato and a transient overexpression assay in pear.
Materials and Methods
Plant Material, Growth Conditions and Treatments
Experiments were carried out in a pear orchard located in Meixian, Shaanxi Province, China (34.29°N, 107.76°E; 514 m above sea level). The average annual precipitation at this location was 574.6 mm, and the average annual temperature was 12.7°C. During anthesis, the average temperature was 12°C.
‘Dangshansu’ pear, which was grafted onto P. betulifolia Bge. rootstock, was used for the study. Two days before anthesis, all treated and control plants were bagged to prohibit pollination. Healthy and uniform plants were subjected to three treatments: (i) application of water to unpollinated flowers, serving as the nonpollination treatment (Control); (ii) spraying of a solution of 50 mg L−1 GA4+7 (GA) [the concentration previously determined by Liu et al. (2018b)] on unpollinated ‘Dangshansu' flowers at anthesis followed by immediately bagged, and (iii) hand pollination (P), performed at the same time. The three treatments were carried out on three trees, and three branches were selected on each three. Three branches from each treatment were used as three replicates, and each treatment contained about 150 flowers. The whole fruits were collected at 0, 3, 9, and 14 days after treatment (DAT), and each sample consisted of 60 fruits at every time point. Ovules were separated from 30 fruits which were collected at 0, 3, 9 DAT and used for qRT-PCR. Pear is a kind of pseudocarp, and the hypanthiums develop into fruit. The information of pear floral organ was shown in Figure S2, and the sample of pear labeled as fruit included hypanthiums, ovules, and ovaries in this study. For a tissue-specific expression assay, total RNA was isolated from the whole fruits (including hypanthiums, ovules, and ovaries), pedicels, sepals, styles, young leaves, and young stems at 6 DAA (days after anthesis) respectively. Each sample was frozen in liquid nitrogen and stored at −80°C for further analyses.
Tomato (Solanum lycopersicum L. ‘Micro-Tom') was used in the transgenic experiments. Plants were placed in pots (120 × 100 mm) containing a mixture of peat: vermiculite (1:1 v/v) in a growth chamber at 25°C under 16-h/8-h light/dark conditions and irrigated daily with Hoagland's nutrient solution (García-Hurtado et al., 2012).
Isolation of PbGA20ox Family Genes
GA20ox genes were identified from the pear-genome coding DNA sequence (CDS) database (Wu et al., 2013) (http://www.ncbi.nlm.nih.gov/genome/?term = pyrus). Hidden Markov Model profiles of GA20-ox family gene conserved domains (PF14226 and PF03171) were obtained from the Pfam database (Pan et al., 2017) and used as queries for searching against the pear genome database. To confirm the existence of a corresponding highly similar EST sequence for each predicted gene, sequences of candidate genes were queried against the pear genome EST database (Zhai et al., 2016) using BLASTN. Alignment of amino acid sequences of the candidate genes was performed using ClustalW (Thompson et al., 1997).
Transcriptome Analysis
Transcriptome data generated in our previous study (Liu et al., 2018b) were used to analyze fruit development-related genes. The gene expression data were obtained from samples of unpollinated fruits, unpollinated fruits subjected to GA4+7 treatment, and pollinated fruits collected at three different stages: 3, 9, and 14 days after anthesis.
Clean reads were mapped using Bowtie 1.12 to generate read alignments for each sample (Langmead et al., 2009). Gene expression levels were calculated as fragments per kilobase of exon model per million mapped reads (FPKM) (Wapinski et al., 2013). RNA-seq data of unpollinated fruits were used as the controls. In order to avoid missing important genes, we did not set the minimum value of FPKM. The pairwise comparison of FPKM values includes nine combinations, P vs UP, P vs GA, GA vs UP at 3 DAA, 9 DAA, and 14 DAA, respectively. The results were used as references to screen candidate genes. Three independent biological replications were sequenced and analyzed.
Phylogenetic Analysis
Sequences of designated pear GA20ox genes were downloaded from the pear genome database (Wu et al., 2013) (http://www.ncbi.nlm.nih.gov/genome/?term = pyrus) and aligned with GA20ox genes from Arabidopsis, apple, tomato, citrus, rice, and grape using ClustalW. The phylogenetic analysis was performed using the minimum-evolution method under the JTT model in MEGA 5.0. Information on the analyzed genes was listed in Supplementary Table S5.
Cloning of the PbGA20ox2 Gene
The CDS of PbGA20ox2 (LOC103960493) was cloned from cDNA of ‘Dangshansu' pear using primer sets (Supplementary Table S3) designed from the sequence data in the pear genome database (http://www.ncbi.nlm.nih.gov/genome/?term = pyrus). The methods of RNA extraction and RNA reverse-transcription are described below. The PCR amplification was carried out using the following protocol: 40 cycles at 98°C for 10 s, 55°C for 15 s and 72°C for 10 s. The primers are based on CDS and described in Table S3.
Expression Analysis by qRT-PCR
Total RNA was extracted using a RNAprep Pure Plant kit (Tiangen, Beijing, China). RNA concentration and quality were assessed by UV spectrophotometry and on a 0.8% agar ethidium bromide-stained gel, respectively. Next, 1 μg of total RNA was reverse-transcribed to cDNA using a PrimeScript RT reagent kit with gDNA Eraser (Takara, Dalian, China). qRT-PCR amplifications were performed on an ABI instrument (Thermo Fisher Scientific, Massachusetts, USA) using a SYBR Premix Ex Taq kit (Takara) and selected gene primers, which were designed with Primer Premier 5.0 software (Liu et al., 2018b). qRT-PCR amplifications were carried out using the following protocol: initial incubation at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. All reactions were performed with three biological repeats. Data were collected by AB StepOne Plus software (Thermo Fisher Scientific, Massachusetts, USA) and we used the ΔΔCT algorithm to analyze the results (Livak and Schmittgen, 2001). The primers for actin genes of pear and tomato, PbGA20ox1, PbGA20ox2, and PbGA20ox3 were described in Table S3.
Subcellular Localization of PbGA20ox2
The full-length CDS of GA20ox2 was cloned into a pCambia2300–green fluorescent protein (GFP) to form a translation fusion with the N-terminus of the GFP. The vector was kindly provided by Professor Xu Yan, Northwest A&F University. Agrobacterium tumefaciens strain EHA105, containing either a pCambia 2300 vector with 35S::GFP or the GA20ox2—35S::GA20ox2-GFP, was grown at 28°C in Luria–Bertani medium containing 50 mg L−1 kanamycin and 25 mg L−1 rifampicin. After 24 h, the Agrobacterium cells were harvested and resuspended in infiltration buffer [10 mM MgCl2, 10 mM MES (pH 5.6) and 150 mM acetosyringone] to a final OD600 of 0.8. The resuspended cells were shaken for 4 h at room temperature and then subjected to infiltration by a syringe. The methods of infection were based on Hellens et al. (2005). The leaves were incubated in the dark at 22°C for 12 h and then placed in a growth chamber (25°C, 16-h/8-h day/night) for 4 to 5 days. Fluorescence Microscopic (BX51 + PD72 + IX71, OLYMPUS, Japan) imaging system was used to observe the anatomical images. Excitation and emission wavelengths were 498 nm and 516 nm respectively.
Production of Transgenic Tomato Plants
The complete CDS of PbGA20ox2 was fused to the CaMV 35S promoter of a pCambia 1301 binary vector (Supplementary Figure S3). The construct 35S::GA20ox2 was transformed into tomato using Agrobacterium tumefaciens strain EHA105 as described in García-Hurtado et al. (2012). Briefly, the tomato first leaf sections (from 20-day-old seedlings grown under sterile conditions) were cultured for 2 days in the dark in Petri dishes containing solidified preculture (PC) medium [MS salts supplemented with vitamins, 3% (w/v) sucrose, 100 mg/L myo-inositol, 4 mg/L indole acetic acid (IAA), 4 mg/L kinetin, and 0.8% (w/v) agar]. Then they were immersed in bacterial suspensions (OD600nm = 0.4) containing 200 µM acetosyringone for 10 min. Blotted explants were cultured in the dark for 2 days in solidified PC medium with 200 µM acetosyringone, washed in washing medium [MS salts, 2% (w/v) sucrose, 100 mg/L myo-inositol, and 500 mg/L cefotaxime], and cultured for 2 days in the dark in solidified PC medium. Then the explants were transferred to PC medium containing 1 mg/L zeatin, 300 mg/L cefotaxime, and 100 mg/L kanamycin. Explants developing resistant calli produced shoots, which were excised and placed on rooting medium [MS salts, 2% (w/v) sucrose, 100 mg/L myo-inositol, 1 mg/L thiamine, 0.1 mg/L IAA, and 0.8% (w/v) agar]. Rooted explants were cultured in pots containing vermiculite, watered with Hoagland's solution, and conditioned in a growth chamber before transferring to the greenhouse.
RNA was extracted from young leaves, and used for qRT-PCR to identify the positive transgenic lines. The specific primers: 5′-CAATGGCACTCCATTAGCCC-3′ (sense) and 5′-TTCACTGTTCACCACTGCCCT-3′ (antisense) were used. The method of qRT-PCR was described as above. Progeny from the transgenic plants were obtained by selfing under controlled conditions. According to the phenotypes, we designed three experiments of self-cross. Then we obtained homozygous lines in the third generation, which were used for the study and replicated each transgenic line in tissue culture.
Tomato Germination and Growth Conditions
Tomato seeds from transgenic lines and wild-type (WT) plants were imbibed in 50°C water for 4–5 h. The seeds were then bagged in wet gauze, placed in 90-mm-diameter plastic Petri dishes and sprayed with double-deionized water every 3 h at 25°C in the dark. After germination for 48 h, the seedlings were transferred to pots (120 × 100 mm) containing a mixture of peat: vermiculite (1:1, v/v) and cultured in a growth chamber (25°C, relative air humidity of 70%–80%, 16-h/8-h day/night photoperiod and photon flux of 115 μ mol m–2 s–1). Seedlings were irrigated daily with Hoagland's solution, and the natural light was supplemented with Osram lamps (Powerstar HQI-BT, 400W) to provide 16 h of daylight (Martínez-Bello et al., 2015).
Transgenic Tomato Phenotypes
For each line, we recorded the following phenotypes: (i) hypocotyl length (mean length from bottom of the stem to cotyledon); (ii) height to first inflorescence (mean length from soil surface to first inflorescence); (iii) internode length (average of three internode lengths between first to fourth leaves); (iv) stem diameter (stem diameter of third internode); (v) leaves to first inflorescence; (vi) root length (measured 5 days after seed germination in the growth chamber); and (vii) days to anthesis (time to first open flower).
Parthenocarpic Capacity of Transgenic Tomato
We analyzed the parthenocarpic capacity of transgenic tomato in two independent assays, each involving four independent lines (WT, 10-1, -6 and -8). Two or three trusses and three to four emasculated flowers were left on at least nine plants per line in two independent experiments. Flower emasculation was carried out 2 days before anthesis to prevent self-pollination, and all non-selected flowers were removed. The percentage of developed fruits and their weights were determined at maturity.
Transient Overexpression Assay in Pear Inflorescences
The complete CDS of PbGA20ox2 was fused to the CaMV 35S promoter of a pCambia 1301 binary vector (35S::GA20ox2) (Supplementary Figure S3). Agrobacterium cell growth and resuspension methods were the same as those used in the subcellular localization assay described above. Flowers at 3 DAA and at same position (the center flowers of the inflorescence) were used. Plant infiltration was performed as follows: a syringe with the needle removed was fixed on the inflorescence. 3–4 flowers on each inflorescence for the infiltration and the unpollinated inflorescence was dipped in the Agrobacterium infiltration buffer. Figure S4 is a simplified diagram of the experiment. During dipping, negative pressure was maintained for 10 min by pull syringe piston. We then removed the syringe, and the inflorescence was bagged to prohibit pollination. The negative controls were infiltrated with Agrobacterium containing empty pCambia 1301 vectors (Supplementary Figure S3). At 5 DAT, We randomly selected fruits to analyze PbGA20ox2 expression levels in the control and the two lines overexpressing PbGA20ox2 by qRT-PCR. Starting at 11 DAT, we measured fruit size, including fruit length and diameter, every 3 days until fruit drop. The measuring of the fruit size was carried out in three biological repeats, each with 7 fruits at 11 DAT and 14 DAT and the sample randomly selected fruit in the middle of inflorescence.
For GUS staining, plant materials were stained with 5-bromo-4-chloro-3-indolyl glucuronide at 37°C for 12 h as described in Fillatti et al. (1987).
Phytohormone Analysis
Gibberellins levels were determined using high-performance liquid chromatography–tandem mass spectrometry (AB SCIEX TripleTOF 5600+, Darmstadt, IN, USA). Approximately 0.5 g of fruits was ground in liquid nitrogen and extracted using solvent A (1 L 80% methanol containing 0.5 g citric acid and 0.2 g butylated hydroxytoluene). The sample was extracted with 4 ml of solvent A, and the extract was shaken at 4°C for 12 h. After centrifuging the sample at 10,000 ×g and 4°C for 15 min, the supernatant was collected. Next, 2 ml of solvent A was added to the sample, which was shaken again at 4°C for 1 h. The sample was centrifuged at 10,000 ×g at 4°C for 15 min, and the supernatant was collected. This step was repeated. All supernatant aliquots were mixed and dried under nitrogen gas, dissolved in 250 µl methanol and passed through a 0.22-µm filter membrane. The high-performance liquid chromatography analysis was performed using an ACQUITY UPLC HSS T3 (1.8 μm, Waters, USA) column (2.1 × 100 mm). The mobile phase solvent was the same as in Balcke et al. (2012), and the injection volume was 2 µl. The mass spectrometry conditions were as follows: a spray voltage of 4,500 V, and air curtain, nebulizer, and auxiliary gas pressures of 15, 65, and 70 psi, respectively. The atomizing temperature was 400°C. Each sample consisted of three replicates from independent experiments.
Statistical Analyses
Data were subjected to analysis of variance and tested for significant (*P < 0.05) treatment differences using Duncan's test. Results were presented as the means ± standard deviation (SD) of three replicate samples.
Results
Isolation and Identification of GA20ox Genes in Pear
In our previous study, we found that GA4+7 induced seedless pear fruit (Liu et al., 2018b); to investigate the mechanism by which GA induces parthenocarpy in pear, we analyzed the RNA-seq data, which showed that pollination induced the expression of gibberellin biosynthesis pathway related genes, such as, PbGA3ox, Ent-Kaurene Synthase, Ent-Kaurene Oxidase, and PbGA20ox genes, and the expression of PbGA20ox2 was increased dramatically. The expression of PbGA2ox2, which transfers active GAs to inactive GAs, was downregulated by pollination at 9 DAA (Supplementary Table S1).These results indicated that pollination promoted GAs biosynthesis. GA20ox, as a rate-limiting enzyme, plays a key role in the GA synthesis pathway, so we inferred that it may be related to fruit development. On the basis of conserved domains of the GA20ox subfamily, we identified three PbGA20ox genes in pear, PbGA20ox1 (LOC103942611), PbGA20ox2 (LOC103960493), and PbGA20ox3 (LOC103957121). Amino acid sequence alignment uncovered two conserved domains, DIOX_N and 2OG-Fell_Oxy, and indicated that the sequence ‘LPWKET’ was highly conserved in all PbGA20ox genes (Figure 1A). Phylogenetic analysis revealed that PbGA20ox1 and PbGA20ox2 are most closely related to MdGA20ox1c and MdGA20ox1a, while PbGA20ox3 is similar to MdGA20ox5 and VvGA20ox1, 3, 5 (Supplementary Figure S5). To explore PbGA20ox functions, we quantitatively assessed the expression profiles of the three PbGA20ox genes (Figure 1B). Hand pollination increased the expression of PbGA20ox2 dramatically, whereas PbGA20ox2 expression was reduced in GA-treated nonpollinated fruits compared with nonpollinated ones (Figure 1B). PbGA20ox1 and PbGA20ox3 gene expression levels were reduced in pollinated fruits at 9 DAA; their expression levels after pollination were lower than in fruit treated with exogenous GA at 14 DAA, although the expression of PbGA20ox3 increased at 3 DAA (Figure 1B). We also quantitatively assessed the three genes in ovules. The result showed PbGA20ox2 increased at 3 DAA and 9 DAA in the pollinated ovules, but the PbGA20ox1 and 3 didn't increase in pollinated ovules (Supplementary Figure S6). On the basis of these results, PbGA20ox2 was considered to be a candidate gene for further study.
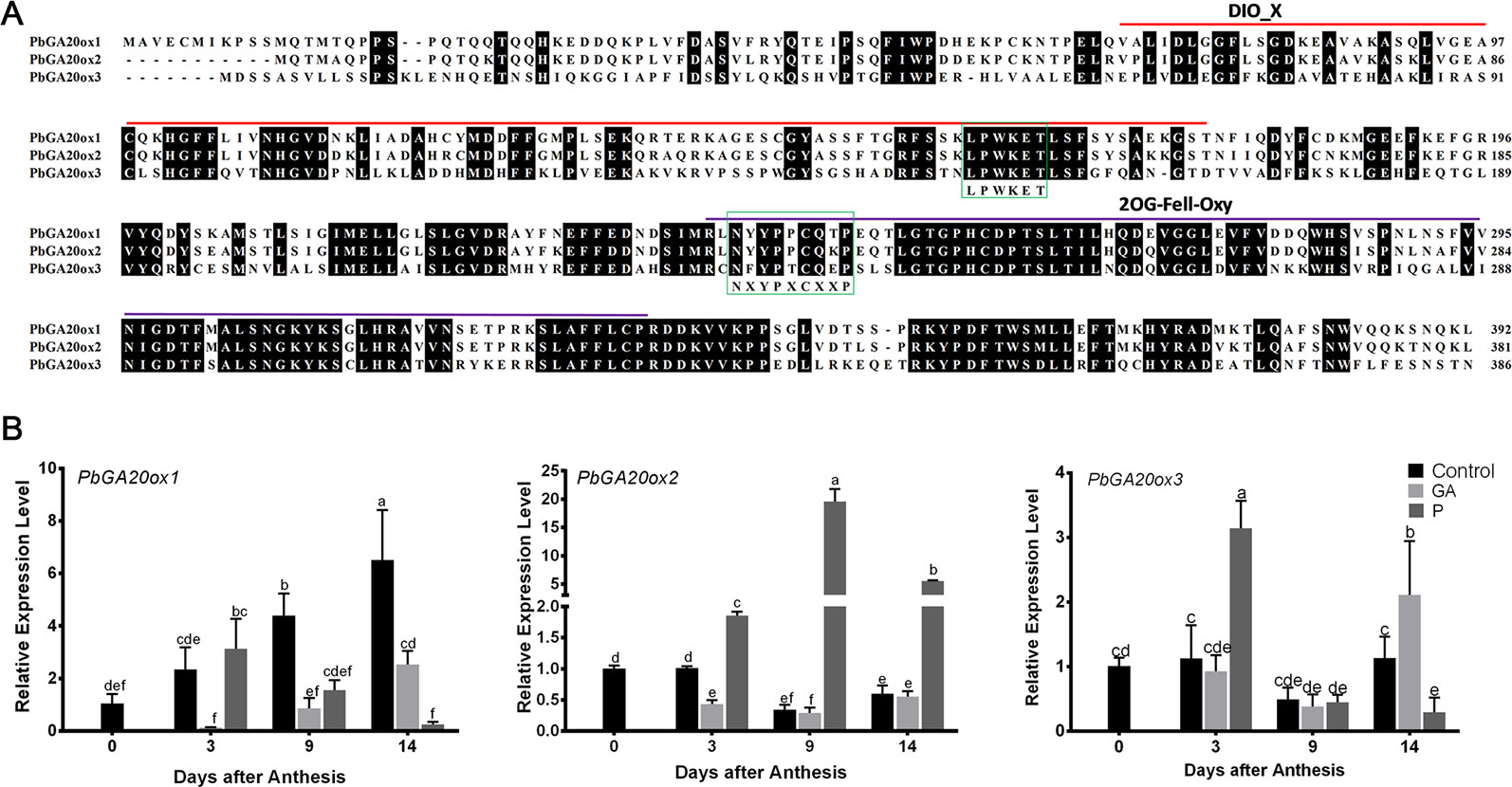
Figure 1 Alignment of amino acid sequences of PbGA20oxs and their expression models after different treatments. (A) Alignment of the three amino acid sequences. DIO_X and 2OG-Fell-Oxy domains are marked with bold lines, and identical residues are shaded in black. The proposed conserved sequences ‘NXYPXCXXP’ and ‘LPWKET’ are indicated by green boxes. (B) Relative expression levels of PbGA20ox genes. Relative transcript quantities were determined by qRT-PCR. The expression level of each gene in fruits under control conditions at 0 DAT was normalized to 1.0. The results shown are means ± SD (n = 3). Different letters between bars indicate significant differences at P < 0.05 (Duncan's range test). Control, nonpollinated; GA, gibberellin treatment; HP, hand pollination.
Tissue-Specific Expression Analysis of PbGA20ox2
The biosynthesis of active GAs is a complex, multistep process, and GA biosynthetic genes are differentially expressed among different tissues, developmental stages, and cell types (Binenbaum et al., 2018). We analyzed the expression of PbGA20ox2 in different tissues using qRT-PCR. PbGA20ox2 was highly transcribed in fruits, which contain (hypanthium, ovary, and ovules), and leaves, with comparatively lower expression levels in pedicels, styles, and stems but no expression in sepals (Figure 2). PbGA20ox1 and PbGA20ox3 were highly transcribed in fruits (Supplementary Figure S7).
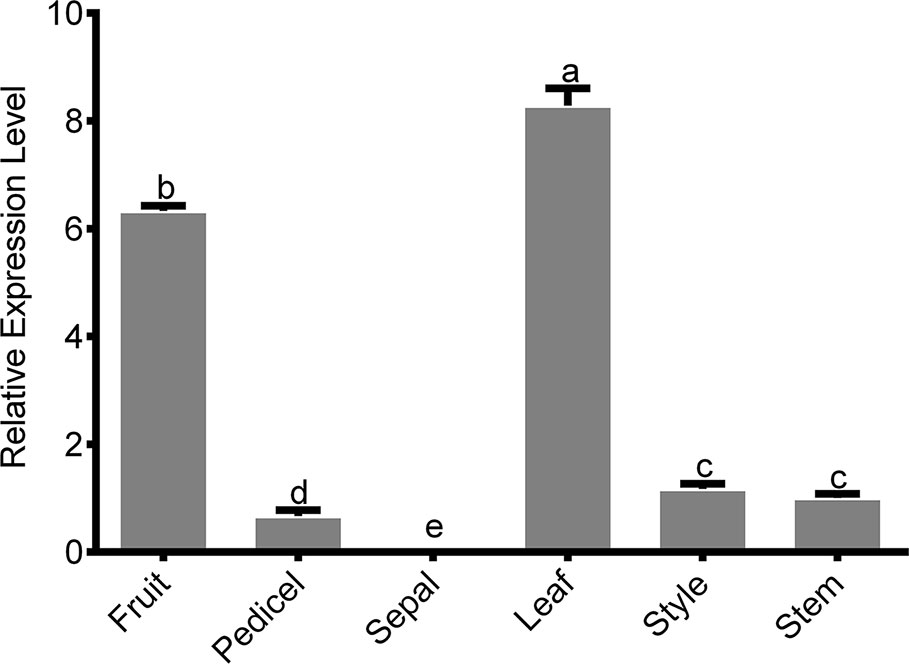
Figure 2 Tissue-specific expression of PbGA20ox2. A qRT-PCR analysis was used to determine the relative expression level of PbGA20ox2 in various tissues and organs. Values shown are means ± SD (n = 3). Significant differences (P < 0.05) among different tissues as determined by Duncan's test are indicated using different lowercase letters.
Subcellular Localization of PbGA20ox2
To determine the precise location of PbGA20ox2 expression, we fused the coding sequence of PbGA20ox2 to a GFP protein under the control of the CaMV 35S promoter (35S::PbGA20ox2-GFP), with the empty vector (EV, 35S::GFP) used as a control. We independently transiently expressed these constructs in tobacco leaves. Four days after transformation, the GFP fluorescence signal was examined using a fluorescence microscope. Merged bright and green fluorescence images are presented and the results suggested that PbGA20ox2 is localized in the cytoplasm, nucleus, and plasma membrane (Figure 3).
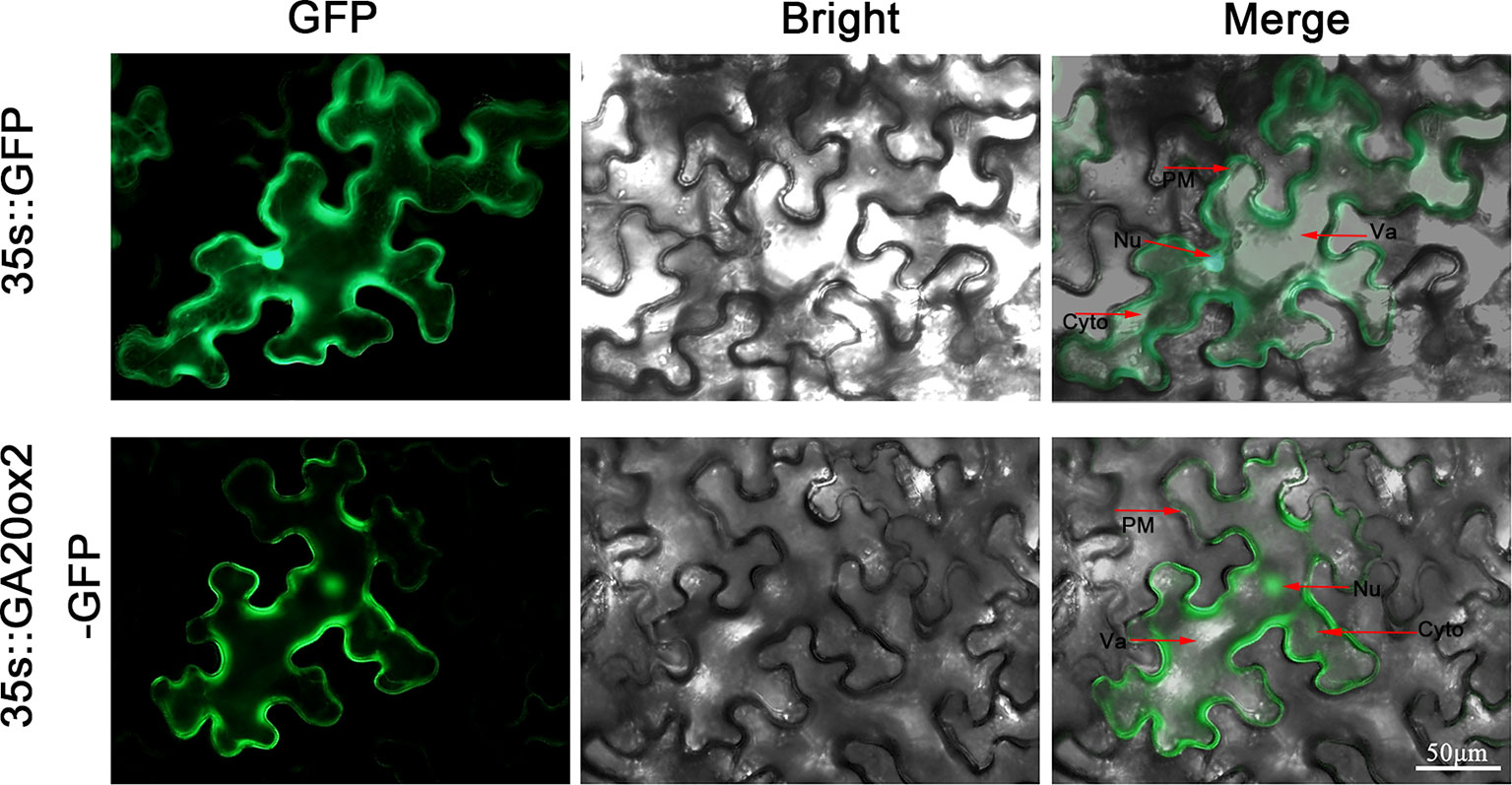
Figure 3 Subcellular localization of PbGA20ox2 in tobacco leaves. The fusion protein (PbGA20ox2-GFP) and positive control (GFP) were independently transiently expressed in tobacco leaves. PbGA20ox2 was detected in the cytoplasm, nucleus, and plasma membrane. PM, plasma membrane; Nu, nucleus; Cyto, cytoplasm; Va, vacule.
Production of Parthenocarpic Fruit in PbGA20ox2-Overexpressing Tomato
To investigate the function of PbGA20ox2 in fruit set and development, we obtained transgenic tomato lines overexpressing the PbGA20ox2 gene (GA20ox2-OE). Compared with the controls, plants of the transgenic lines were taller and had longer hypocotyls and roots, thinner stems and longer internodes (Figures 4A, B; Supplementary Table S2). The leaves of transgenic lines had smooth edges, whereas the WT had the characteristic serrated borders (Figure 4C). In addition, the flowering time and number of leaves before the first inflorescence were also changed in transgenic tomato lines (Supplementary Table S2). We obtained 10 positive lines, 3 of which (10-1, -6 and -8) were selected for analysis. A qRT-PCR assay verified the expression of PbGA20ox2 in young leaves of three lines (Figure 4E), while a phytohormone analysis revealed significant increases in the content of GA4, but not of GA3 and GA1, in the fruitlets of three lines (Figures 4F–H).
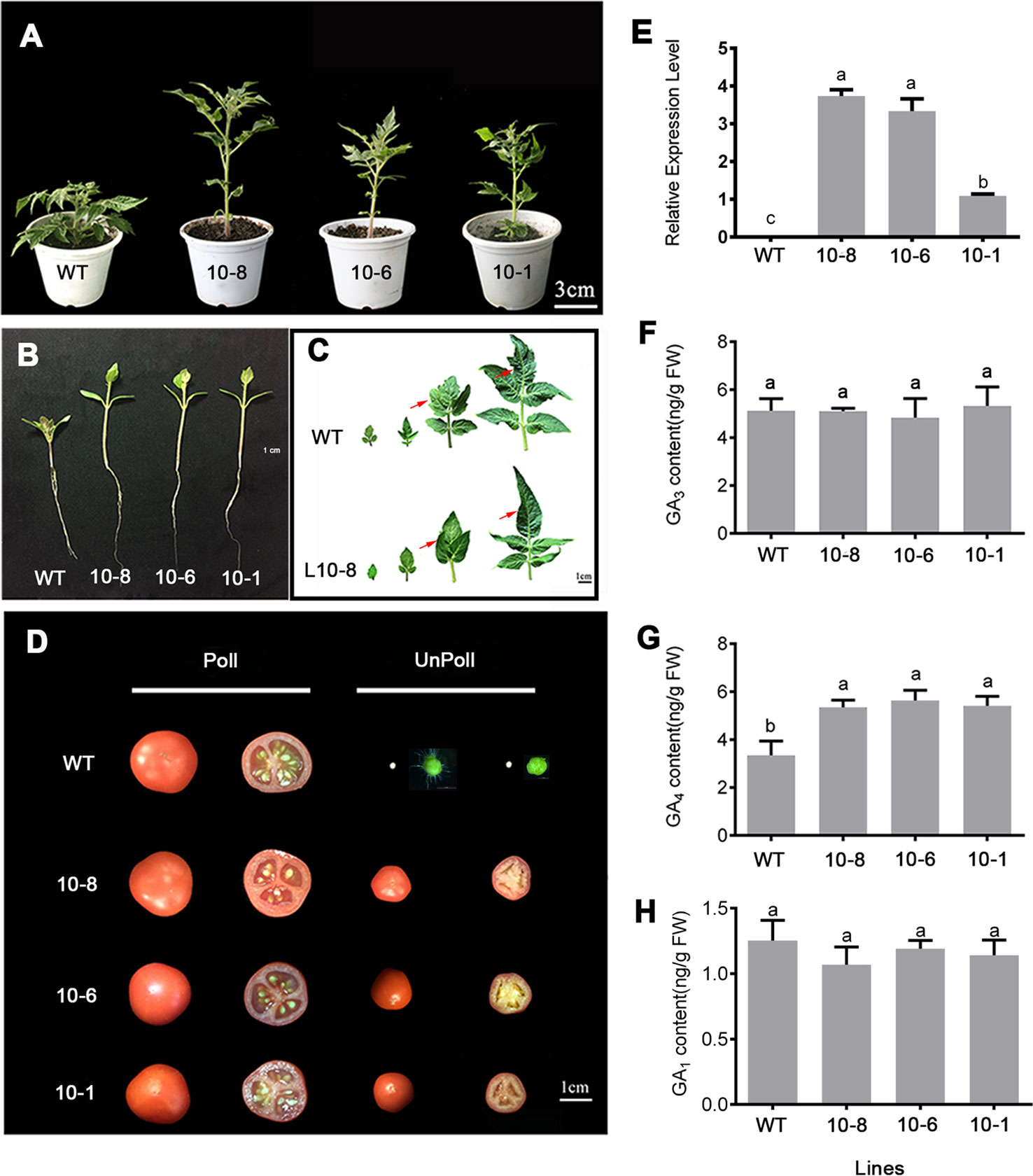
Figure 4 Phenotypes of wild-type (WT) and PbGA20ox-OE lines (10-8, -6, and -1) of tomato. (A) Plant before flowering. (B) Roots of a representative 10-day-old seeding. (C) Leaves at different positions on the plant (first to fourth, left to right). The difference points were marked with a red arrow. (D) Fruits of WT and GA20ox-OE lines. (E) Expression levels of PbGA20ox2 in young leaves of WT and transgenic lines as assessed by qRT-PCR. Line 10-1 was used as control. Ten-day-old fruits were used for phytohormone analysis. (F) GA3 content. (G) GA4 content. (H) GA1 content. Data are means ± SD (n = 3). Different letters between bars indicate significant differences at P < 0.05 (Duncan's range test). Poll, pollinated; Unpoll, nonpollinated.
To determine the function of PbGA20ox2, we analyzed fruit development in transgenic tomato lines and analyzed the parthenocarpic capacity of WT and PbGA20ox2-OE lines (10-1, -6 and -8) in two independent experiments. In total, 10.0–15.5% of nonpollinated fruits developed and produced parthenocarpic fruit in transgenic lines (Figure 4D, Table 1). In contrast, the rate of parthenocarpic fruit was 0 in WT lines unpollinated (Figure 4D, Table 1). In regard to fruit set and weight, the pollinated fruits of transgenic and control lines were not significantly different: the fruit-set rate was 100% with pollination, and the weights per fruit were ~4–5 g in the two types of lines (Table 1). In addition, the average weight of a parthenocarpic fruit was 0.8–0.9 g (Table 1). No mature fruits were generated from nonpollinated fruits of WT plants (Figure 4D). Consequently, PbGA20ox2 altered vegetative growth and induced parthenocarpic fruit development in tomato.
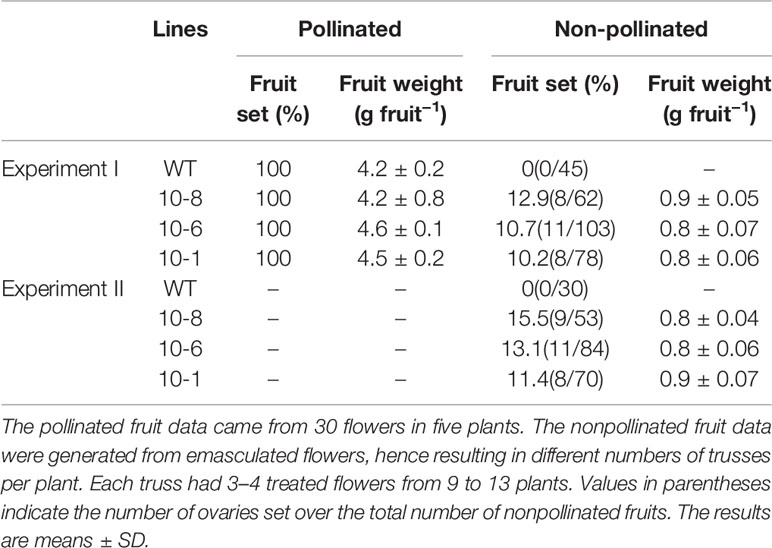
Table 1 Fruit-set capacity of wild-type (WT) and PbGA20ox2-OE lines (10-8, -6, and -1) in two independent experiments.
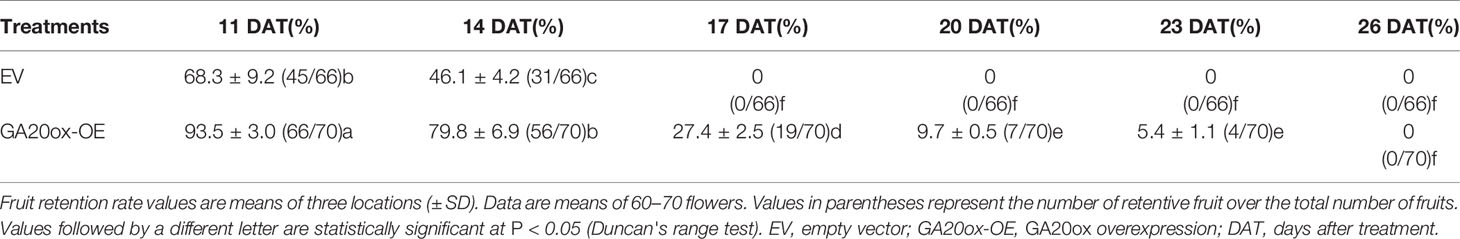
Delayed Fruit Drop in PbGA20ox2-Overexpressing Pear
To further determine the functions of PbGA20ox2 in fruit development, we overexpressed the construct 35S::PbGA20ox2 in pear inflorescences (GA20ox-OE), with the EV used as the control. Fruit infection was validated by monitoring GUS signals (Supplementary Figure S8). Fruit retention rate percentages were respectively 68.3 and 93.5% in control and GA20ox2-OE at 11 DAT. In addition, 53.9% of control fruit had dropped at 14 DAT, but 79.8% of fruits were set in GA20ox-OE. At 17 DAT, all control fruits had dropped, while GA20ox-OE fruit dropped at 26 DAT (Figure 5A, Table 2). We analyzed the expression of PbGA20ox2 at 5 DAT by qRT-PCR, which revealed an increased expression of PbGA20ox2 under the control of the CaMV 35S promoter in fruits compared with the control (infiltrated EV) (Figure 5B). Overexpression of PbGA20ox2 also increased the expression of GA 3-oxidase and genes associated with cell division and expansion in fruits (Supplementary Figure S9). We also measured the fruit size of PbGA20ox2-transiently overexpressed and control (EV) fruits and found significant increases in PbGA20ox2-overexpressed fruit compared with the control (Supplementary Table S4). Furthermore, we determined that a significant change in GA4 content, but not GA1 and GA3 content, occurred at 5 DAT (Figures 5C–E). To conclude, PbGA20ox2 promoted fruit development and delayed unpollinated fruit drop in pear by enhancing GA4 content.
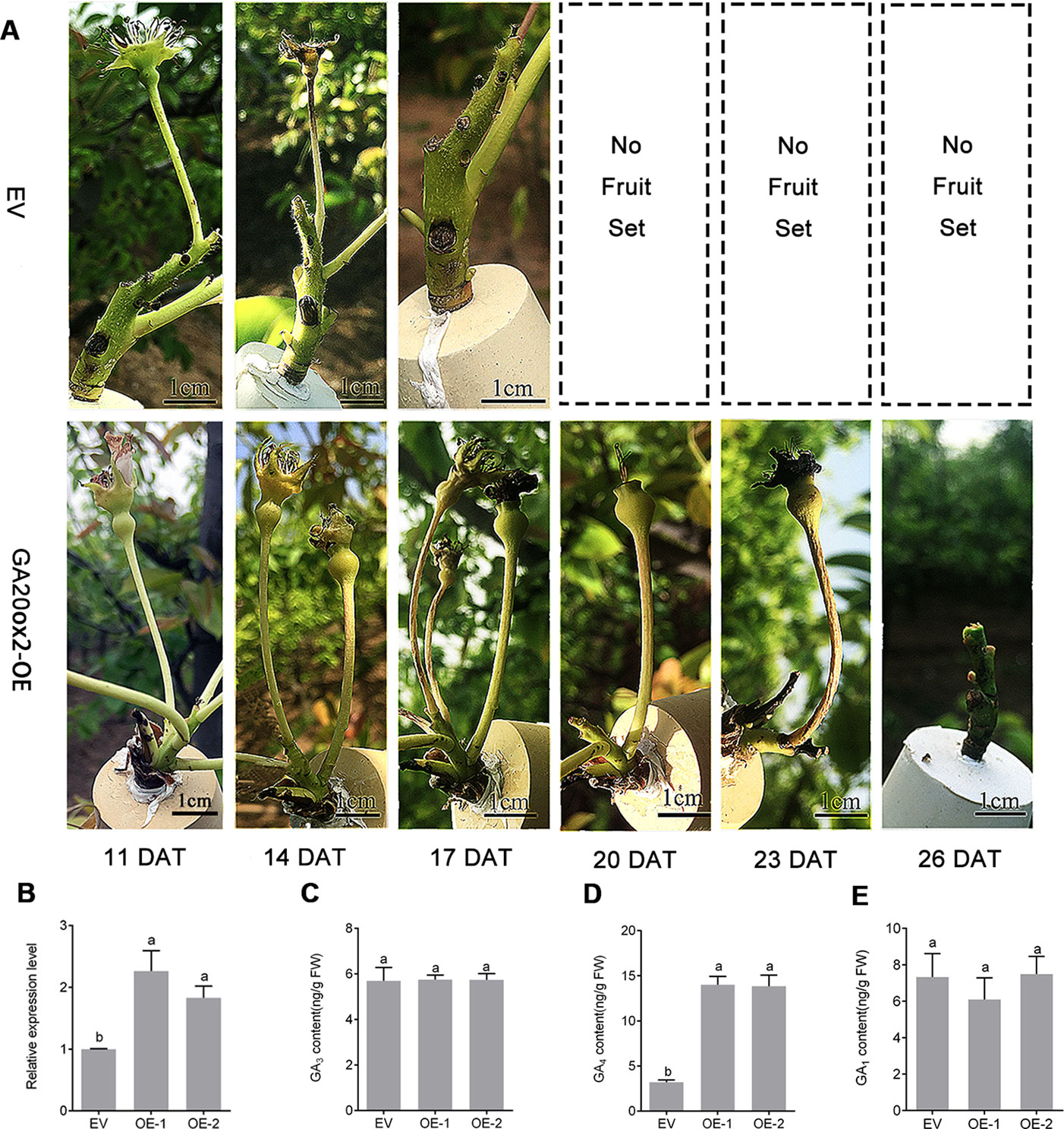
Figure 5 Transient overexpression assay of PbGA20ox2 in ‘Dangshansu' pear fruits. (A) Images of inflorescence development on different days after the transient overexpression of PbGA20ox2 in ‘Dangshansu' pear. (B) Relative expression levels of PbGA20ox2 in control (empty vector, EV) and two independent overexpressed lines. The expression level of the gene in fruits transformed with the EV was normalized to 1.0. (C–E) GA3, GA4 and GA1 contents in pear fruits at 5 DAT. OE-1 and OE-2 represent two biological repeats with overexpressing PbGA20ox2. Results are means ± SD (n = 3). Different letters between bars indicate significant differences at P < 0.05 (Duncan's range test). DAT, days after treatment.
Discussion
Fruit set in higher plants requires pollination. In fruiting plants, ovarian cell division is temporarily stopped at anthesis until pollination and fertilization occur (Reig et al., 2018). During this process, GA biosynthetic pathways are active in ovules (Dorcey et al., 2009). In this study, we isolated PbGA20ox genes, which may be related to fruit development. On the basis of conserved domains of the GA20ox family, we identified three PbGA20ox genes (Figure 1A). As assessed by a qRT-PCR assay, PbGA20ox2 was significantly increased in pollinated fruits and was therefore selected as a candidate gene (Figure 1B). The expression profile of PbGA20ox2 was similar between fruit and ovules, but the expression profile of PbGA20ox3 showed a difference between fruits and ovules, so we inferred that the different expression patterns of PbGA20ox3 may be related to its low expression in pear at the early stage (Figure 1B; Supplementary Figure S6). We next analyzed the location of PbGA20ox2 expression. The biosynthesis of active GAs is a complex, multistep process, and GA biosynthetic genes are differentially expressed among different tissues, developmental stages, and cell types (Binenbaum et al., 2018). In rice flowers, GA biosynthetic genes are extremely highly expressed in the tapetum cells of anthers (Hedden and Phillips, 2000). In Arabidopsis thaliana, GA20ox1 is expressed in the receptacle, and GA20ox3 is expressed in the leaves, roots, hypocotyls, and internodes, with the five AtGA20ox genes mainly expressed in different locations (Plackett et al., 2012). In our study, PbGA20ox2 was mainly expressed in leaves and fruits, with relatively lower expression detected in pedicels, styles, and stems and none in sepals (Figure 2). We have confirmed PbGA20ox2 was related with fruit development, and we inferred that PbGA20ox2 may also play important roles in the leaf. Additionally, our subcellular localization assay revealed that PbGA20ox2 was localized in the nucleus, cytosol, and plasma membrane (Figure 3).
To determine the function of PbGA20ox2 in fruit development, we overexpressed PbGA20ox2 in tomato. We detected significant alterations in the architecture and vegetative growth of transgenic tomato (Figure 4, Supplementary Table S2). The similar alterations have been observed in diverse plants overexpressing GA20ox genes of different species, such as aspen (Eriksson et al., 2000), tobacco (Vidal et al., 2001), and switchgrass (Do et al., 2016). The morphological changes in these species were consistent with the higher expression of GA20ox and bioactive GA levels.
In transgenic tomato, emasculated flowers produced parthenocarpic fruit (Figure 4D), but the percentage of seedless fruit set was lower, and fruit size was smaller than that of pollinated fruit (Table 1). The limited parthenocarpic capacity of transgenic tomato was probably due to the relatively lower increase in GA4 in fruits compared with that resulting from the exogenous application of GAs to tomato (García-Hurtado et al., 2012). Furthermore, we overexpressed PbGA20ox2 in pear inflorescences through a transient expression assay. Fruit drop was delayed approximately 9 days in GA20ox-OE compared with the control (Figure 5A, Table 2), and GA4 contents increased in transgenic tomato and pear fruits (Figures 4G and 5D). Following the expression of PbGA20ox2, the PbGA3ox gene was expressed at a higher level in transgenic pear fruits relative to nontransformed ones (Supplementary Figure S9). In our case, however, the different pear fruit development patterns between GA20ox-OE and control may have been due to the greater viability of transgenic fruits and therefore was an indirect effect of PbGA20ox2 overexpression. During pollination and fertilization, GA synthesis is activated in ovules, and fruit development begins during this process (Dorcey et al., 2009). In previous studies, transgenic tomato in which CsGA20ox was overexpressed (García-Hurtado et al., 2012) or SlGA2ox was silenced (Martínez-Bello et al., 2015) was found to induce parthenocarpy, and the active GA content increased. GA also negatively regulates the formation of the abscission layer to prevent fruit drop (Olsson and Butenko, 2018). Therefore, we confirmed that the overexpression of PbGA20ox2 induced parthenocarpic fruit and delayed fruit drop by increasing GA4 content.
We also analyzed the reasons for fruit drop in the transient overexpression assay. Pear undergoes fruit set and development after GA treatment (Liu et al., 2018b). In the transient overexpression assay, all fruit dropped by 26 DAT (Table 2). After overexpression of PbGA20ox2, the increased GA4 content of young transgenic pear fruit did not lead to a fruit set rate as high as that of pollinated fruit (Liu et al., 2018b). We thus infer that the transient overexpression of PbGA20ox2 cannot increase GA4 content to the same extent as that obtained by GA treatment (Liu et al., 2018b), and this observation is correlated with the fruit drop and low parthenocarpic fruit-set rate of transgenic tomato, which was same with García-Hurtado et al. (2012). The GUS staining assay also suggested that the low efficiency of agro-infiltration is directly related to the expression of PbGA20ox2 and the content GA4 in the fruit, which is ultimately and indirectly related to fruit drop. Furthermore, the negative pressure conditions during infiltration in the transient expression assay caused physical injuries to the inflorescence, which might be correlated with fruit drop.
GA1, GA3, GA4 and GA7 are the most common active GAs in higher plants (Ding et al., 2013). In different species, the major active GAs may be different (Dorcey et al., 2009). If a plant has two functional GA forms, then different activity levels may exist, and GA abundance or mode of activity may vary between organs and across developmental stages. In tomato, the main pathway of GA biosynthesis is the early-13-hydroxylation pathway, which can produce GA1 and GA3 (García-Hurtado et al., 2012). In Arabidopsis, GA4 is the major bioactive form and promotes vegetative growth, floral initiation, and maturity (Eriksson et al., 2006). In rice, GA4 is the main active form associated with reproductive growth, while GA1 is the major bioactive form associated with vegetative growth (Kobayashi et al., 1989). In grapes, GA4 content increases during later fruit developmental stages (Giacomelli et al., 2013). Exogenous application of GA4 promotes vegetative and reproductive growth in tomato (García-Hurtado et al., 2012) and cucumber (Qian et al., 2018). Furthermore, the affinity of GA4 to bind GA INSENSITIVE DWARF1 (GID1), a GA signal receptor, is approximately 20 times greater than that of GA3 in rice (Ueguchitanaka et al., 2007). We predicted that GA3 and GA4 may have different functional stages in ‘Dangshansu' pear, an idea that needs further study. GA1 and GA4 represent two parallel GA synthetic pathways. In transgenic tomato fruit and pear fruits, GA4 increased while GA1 and GA3 remained unchanged (Figures 4 and 5). We thus infer that GA4 accumulation in early developmental stage is important for fruit set and development in ‘Dangshansu' pear and that PbGA20ox2 plays key roles in these processes. Combined with the results of tissue specific experiments, we inferred that PbGA20ox2 may mainly catalyze GA12 in fruit at an early stage of fruit development. The involvement of PbGA20ox2 in the functional differences between GA4 and GA3 in pear fruit development also requires further study.
Conclusions
In this study, we demonstrated that PbGA20ox2 plays key roles in fruit set and development. Overexpression of PbGA20ox2 in tomato altered the vegetative phenotype and promoted the production of parthenocarpic fruit. In transient overexpression assays, in addition, the overexpression of PbGA20ox2 delayed pear fruit drop. An analysis of endogenous GAs levels in transgenic tomato and pear revealed that PbGA20ox2 promoted fruit set and development by increasing the accumulation of GA4 (but not GA3) at an early stage of fruit development.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
HW, JL, RZ, and ZW designed the experiments. HW, TW, JL, LC, and YZ performed the experiments. HW analyzed the data. HW, RZ, CY, ZW, FM, and LX wrote and revised the manuscript. All authors have participated in this research and approved the final manuscript.
Funding
This work was supported by the China Agriculture Research System (CARS-28-45) and Weinan Experimental Station foundation of Northwest A&F University.
Conflict of Interest
The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00113/full#supplementary-material
References
Aslmoshtaghi, E., Shahsavar, A. (2013). Study on the induction of seedless loquat. Thai J. Agric. Sci. 77, 83–85. doi: 10.1046/j.1365-2613.1996.00967.x
Balcke, G. U., Handrick, V., Bergau, N., Fichtner, M., Henning, A., Stellmach, H., et al. (2012). An UPLC-MS/MS method for highly sensitive high-throughput analysis of phytohormones in plant tissues. Plant Methods 8, 47. doi: 10.1186/1746-4811-8-47
Binenbaum, J., Weinstain, R., Shani, E. (2018). Gibberellin localization and transport in plants. Trends Plant Sci. 23, 410–421. doi: 10.1016/j.tplants.2018.02.005
Cong, L., Yue, R. R., Wang, H. B., Liu, J. L., Zhai, R., Yang, J., et al. (2018). 2, 4-D-induced parthenocarpy in pear is mediated by enhancement of GA4 biosynthesis. Physiol. Plant. 163, 812–820. doi: 10.1111/ppl.12835
Ding, J., Chen, B., Xia, X., Mao, W., Shi, K., Zhou, Y., et al. (2013). Cytokinin-induced parthenocarpic fruit development in tomato is partly dependent on enhanced gibberellin and auxin biosynthesis. PloS One 8, e70080. doi: 10.1371/journal.pone.0070080
Do, P. T., De Tar, J. R., Lee, H., Folta, M. K., Zhang, Z. J. (2016). Expression of ZmGA20ox cDNA alters plant morphology and increases biomass production of switchgrass (Panicum virgatum L.). Plant Biotechnol. J. 14, 1532–1540. doi: 10.1111/pbi.12514
Dorcey, E., Urbez, C., Blázquez, M. A., Carbonell, J., Perez-Amador, M. A. (2009). Fertilization-dependent auxin response in ovules triggers fruit development through the modulation of gibberellin metabolism in Arabidopsis. Plant J. 58, 318–332. doi: 10.1111/j.1365-313x.2008.03781.x
Eriksson, M. E., Israelsson, M., Olsson, O., Moritz, T. (2000). Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat. Biotechnol. 18, 784–788. doi: 10.1038/77355
Eriksson, S., Böhlenius, H., Moritz, T., Nilsson, O. (2006). GA4 is the active gibberellin in the regulation of leafy transcription and Arabidopsis floral initiation. Plant Cell. 18, 2172–2181. doi: 10.1105/tpc.106.042317
Fillatti, J. J., Kiser, J., Rose, R., Comai, L. (1987). Efficient transfer of a glyphosate tolerance gene into tomato using a binary agrobacterium tumefaciens vector. Biotechnology 5, 726–730. doi: 10.1038/nbt0787-726
García-Hurtado, N., Carrera, E., Ruiz-Rivero, O., Lopez-Gresa, M. P., Hedden, P., Gong, F., et al. (2012). The characterization of transgenic tomato overexpressing gibberellin 20-oxidase reveals induction of parthenocarpic fruit growth, higher yield, and alteration of the gibberellin biosynthetic pathway. J. Exp. Bot. 63, 5803–5813. doi: 10.1093/jxb/ers229
Giacomelli, L., Rota-Stabelli, O., Masuero, D., Acgeampong, A. K., Moretto, M., Caputi, L., et al. (2013). Gibberellin metabolism in Vitis vinifera L. during bloom and fruit-set: functional characterization and evolution of grapevine gibberellin oxidases. J. Exp. Bot. 64, 4403–4419. doi: 10.1093/jxb/ert251
Hedden, P., Kamiya, Y. (1997). Gibberellin biosynthesis: enzymes, genes and their regulation. Annu. Rev. Plant Physiol. Plant MolBiol. 48, 431–460. doi: 10.1146/annurev.arplant.48.1.431
Hedden, P., Phillips, A. L. (2000). Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 5, 523–530. doi: 10.1016/S1360-1385(00)01790-8
Hedden, P., Sponsel, V. (2015). A century of gibberellin research. J. Plant Growth Regul. 34, 740–760. doi: 10.1007/s00344-015-9546-1
Hedden, P., Thomas, S. G. (2012). Gibberellin biosynthesis and its regulation. Biochem. J. 444, 11–25. doi: 10.1042/BJ20120245
Hellens, R. P., Allan, A. C., Friel, E. N., Bolitho, K., Grafton, K., Templeton, M. D., et al. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13. doi: 10.1186/1746-4811-1-13
Joldersma, D., Liu, Z. (2018). The making of virgin fruit: the molecular and genetic basis of parthenocarpy. J. Exp. Bot. 69, 955–962. doi: 10.1093/jxb/erx446
Kobayashi, M., Sakurai, A., Saka, H., Takahashi, N. (1989). Quantitative analysis of endogenous gibberellins in normal and dwarf cultivars of rice. Plant Cell Physiol. 30, 963–969. doi: 10.1093/oxfordjournals.pcp.a077841
Langmead, B., Trapnell, C., Pop, M., Salzberg, S. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. doi: 10.1186/gb-2009-10-3-r25
Liu, J. L., Zhai, R., Liu, F. X., Zhao, Y. X., Wang, H. B., Liu, L. L., et al. (2018a). Melatonin induces parthenocarpy by regulating genes in gibberellin pathways of ‘Starkrimson’ pear (Pyrus communis L.). Front. Plant Sci. 9, 946. doi: 10.3389/fpls.2018.00946
Liu, L. L., Wang, Z. G., Liu, J. L., Liu, F., Zhai, R., Zhu, C. Q., et al. (2018b). Histological, hormonal and transcriptomic reveal the changes upon gibberellin-induced parthenocarpy in pear fruit. Hortic. Res. 5, 1. doi: 10.1038/s41438-017-0012-z
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T) (-Delta Delta C) method. Methods 24, 402–408. doi: 10.1006/meth.2001.1262
Martí, C., Orzáez, D., Ellul, P., Moreno, V., Carbonell, J., Granell, A. (2007). Silencing of DELLA induces facultative parthenocarpy in tomato fruits. Plant J. 52, 865–876. doi: 10.1111/j.1365-313X.2007.03282.x
Martínez-Bello, L., Moritz, T., López-Díaz, I. (2015). Silencing C19-GA 2-oxidases induces parthenocarpic development and inhibits lateral branching in tomato plants. J. Exp. Bot. 66, 5897–5910. doi: 10.1093/jxb/erv300
McAfee, P., Karim, S., Schaffer, R., David, K. (2013). A dynamic interplay between phytohormones is required for fruit development, maturation, and ripening. Front. Plant Sci. 4, 79. doi: 10.3389/fpls.2013.00079
Mesejo, C., Reig, C., Martínez-Fuentes, A., Agustí, M. (2010). Parthenocarpic fruit production in loquat (Eriobotrya japonica Lindl.) by using gibberellic acid. Sci. Hortic. 126, 37–41. doi: 10.1016/j.scienta.2010.06.009
Niu, Q. F., Wang, T., Li, J. Z., Yang, Q. Q., Qian, M. J., Teng, Y. W. (2015). Effects of exogenous application of GA4+7 and N-(2-chloro-4- pyridyl) -N′-phenylurea on induced parthenocarpy and fruit quality in Pyrus pyrifolia ‘Cuiguan'. Plant Growth Regul. 76, 251–258. doi: 10.1007/s10725-014-9995-8
Olsson, V., Butenko, M. A. (2018). Abscission in plants. Curr. Biol. 28, R338–R339. doi: 10.1016/j.cub.2018.02.069
Olszewski, N., Sun, T., Gubler, F. (2002). Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell. 14, S61–S80. doi: 10.1105/tpc.010476
Pan, C., Tian, K. H., Ban, Q. Y., Wang, L. G., Sun, Q. L., He, Y., et al. (2017). Genome-wide analysis of the biosynthesis and deactivation of gibberellin-dioxygenases gene family in camellia sinensis (L.) O. Kuntze. Genes 8, 235. doi: 10.3390/genes8090235
Plackett, A. R. G., Powers, S. J., Fernandez-Garcia, N., Urbanova, T., Takebayashi, Y., Seo, M., et al. (2012). Analysis of the developmental roles of the Arabidopsis Gibberellin 20-Oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell. 24, 941–960. doi: 10.1105/tpc.111.095109
Qian, C., Ren, N., Wang, J., Xu, Q., Chen, X., Qi, X. (2018). Effects of exogenous application of CPPU, NAA and GA4+7 on parthenocarpy and fruit quality in cucumber (Cucumis Sativus L.). Food Chem. 243, 410–413. doi: 10.1016/j.foodchem.2017.09.150
Reig, C., Martinez-Fuentes, A., Mesejo, C., Agusti, M. (2018). Hormonal control of parthenocarpic fruit set in ‘RojoBrillante' persimmon. J. Plant Physiol. 231, 96–104. doi: 10.1016/j.jplph.2018.09.004
Reinecke, D. M., Wickramarathna, A. D., Ozga, J. A., Kurepin, L. V., Jin, A. L., Good, A. G., et al. (2013). Gibberellin 3-oxidase gene expression patterns influence gibberellin biosynthesis, growth, and development in pea. Plant Physiol. 163, 929–945. doi: 10.1104/pp.113.225987
Rieu, I., Eriksson, S., Powers, S. J., Gong, F., Griffiths, J., Woolley, L., et al. (2008a). Genetic analysis reveals that C19-GA 2-oxidation is a major gibberellin inactivation pathway in Arabidopsis. Plant Cell. 20, 2420–2436. doi: 10.1105/tpc.108.058818
Rieu, I., Ruiz-Rivero, O., Fernandez-Garcia, N., Griffiths, J., Powers, S. J., Gong, F., et al. (2008b). The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 53, 488–504. doi: 10.1111/j.1365-313X.2007.03356.x
Serrani, J. C., Ruiz-Rivero, O., Fos, M., García-Martínez, J. L. (2008). Auxin-induced fruit set in tomato is mediated in part by gibberellin. Plant J. 56, 922–934. doi: 10.1111/j.1365-313X.2008.03654.x
Singh, D. P., Filardo, F. F., Storey, R., Jermakow, A. M., Yamaguchi, S. S. (2010). Overexpression of a gibberellin inactivation gene alters seed development, KNOX gene expression, and plant development in Arabidopsis. Physiol. Plant. 138, 74–90. doi: 10.1111/j.1399-3054.2009.01289.x
Sugiyama, K., Kami, D., Muro, T. (2014). Induction of parthenocarpic fruit set in watermelon by pollination with bottle gourd (Lagenariasiceraria (Molina) Standl.) pollen. Sci. Hortic. 171, 1–5. doi: 10.1016/j.scienta.2014.03.008
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., Higgins, D. G. (1997). The Clustal_x windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Ueguchitanaka, M., Nakajima, M., Motoyuki, A., Matsuoka, M. (2007). Gibberellin receptor and its role in gibberellin signaling in plants. Annu. Rev. Plant Biol. 58, 183–198. doi: 10.1146/annurev.arplant.58.032806.103830
Vidal, A. M., Gisbert, C., Talón, M., Primo-Millo, E., López-Díaz, I., García-Martínez, J. L. (2001). The ectopic overexpression of a citrus gibberellin 20-oxidase enhances the non-13-hydroxylation pathway of gibberellin biosynthesis and induces an extremely elongated phenotype in tobacco. Physiol. Plant. 112, 251–260. doi: 10.1034/j.1399-3054.2001.1120214.x
Wapinski, O., Vierbuchen, T., Qu, K., Lee, Q., Chanda, S., Fuentes, D., et al. (2013). Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell 155, 621–635. doi: 10.1016/j.cell.2013.09.028
Watanabe, M., Segawa, H., Murakami, M., Sagawa, S., Komori, S. (2008). Effects of plant growth regulators on fruit set and fruit shape of parthenocarpic apple fruits. J. Jpn. Soc. Hortic. Sci. 77, 350–357. doi: 10.2503/jjshs1.77.350
Wu, J., Wang, Z. W., Shi, Z. B., Zhang, S., Ming, R. R., Khan, M. A., et al. (2013). The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 23, 396–408. doi: 10.1101/gr.144311.112
Keywords: gibberellin, gibberellin 20-oxidase, parthenocarpy, fruit set, fruit development, pear
Citation: Wang H, Wu T, Liu J, Cong L, Zhu Y, Zhai R, Yang C, Wang Z, Ma F and Xu L (2020) PbGA20ox2 Regulates Fruit Set and Induces Parthenocarpy by Enhancing GA4 Content. Front. Plant Sci. 11:113. doi: 10.3389/fpls.2020.00113
Received: 19 July 2019; Accepted: 24 January 2020;
Published: 18 February 2020.
Edited by:
Stefan de Folter, National Polytechnic Institute, MexicoReviewed by:
Concepción Gómez-Mena, Polytechnic University of Valencia, SpainAnn Maureen Callahan, Appalachian Fruit Research Station (ARS-USDA), United States
Copyright © 2020 Wang, Wu, Liu, Cong, Zhu, Zhai, Yang, Wang, Ma and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lingfei Xu, bGluZ2Z4dTIwMTNAc2luYS5jb20=
 Huibin Wang
Huibin Wang Ting Wu
Ting Wu Jianlong Liu
Jianlong Liu Chengquan Yang
Chengquan Yang Zhigang Wang
Zhigang Wang Fengwang Ma
Fengwang Ma Lingfei Xu
Lingfei Xu