- Structure and Evolution of Plant Genomes Group, Centre for Research in Agricultural Genomics, CSIC-IRTA-UAB-UB, Campus UAB, Edifici CRAG, Barcelona, Spain
LTR-retrotransposons share a common genomic organization in which the 5′ long terminal repeat (LTR) is followed by the gag and pol genes and terminates with the 3′ LTR. Although GAG-POL-encoded proteins are considered sufficient to accomplish the LTR-retrotransposon transposition, a number of elements carrying additional open reading frames (aORF) have been described. In some cases, the presence of an aORF can be explained by a phenomenon similar to retrovirus gene transduction, but in these cases the aORFs are present in only one or a few copies. On the contrary, many elements contain aORFs, or derivatives, in all or most of their copies. These aORFs are more frequently located between pol and 3′ LTR, and they could be in sense or antisense orientation with respect to gag-pol. Sense aORFs include those encoding for ENV-like proteins, so called because they have some structural and functional similarities with retroviral ENV proteins. Antisense aORFs between pol and 3′ LTR are also relatively frequent and, for example, are present in some characterized LTR-retrotransposon families like maize Grande, rice RIRE2, or Silene Retand, although their possible roles have been not yet determined. Here, we discuss the current knowledge about these sense and antisense aORFs in plant LTR-retrotransposons, suggesting their possible origins, evolutionary relevance, and function.
Introduction
LTR-retrotransposons are transposable elements (TEs) characterized by the presence of two long direct repeats (long terminal repeats, LTRs) flanking an internal region that contains the gag and pol genes encoding proteins required for transposition (Figures 1A,B). Long terminal repeats provide the promoters and terminators associated with the transcription of the LTR-retrotransposon by RNA polymerase II (Kumar and Bennetzen, 1999). The internal region contains the primer binding site (PBS) and the polypurine tract (PPT), both used during the retrotransposition process. The PBS is a 10–20-nucleotide sequence located next to the 5′ LTR that can partly base-pair with the 3′ end of a cytoplasmic tRNA. The PBS is used to prime the synthesis of the first DNA strand during the retrotranscription process. The PPT is a short stretch of purine-rich DNA (8–49 nt) located in the internal region next to the 3′ LTR and is used to prime the synthesis of the second DNA strand during retrotranscription. The internal region also contains the gag and pol genes, which encode all the proteins necessary for the retrotranscription and integration processes not provided by the cell. Gag encodes the structural proteins, including capsid (CA) and nucleocapsid (NC), that assemble into virus-like particles (VLPs) Dodonova et al., 2019). Pol encodes the proteins that provide the enzymatic machinery for reverse transcription and integration into the host genome: aspartic proteinase (AP), reverse transcriptase (RT), RNase H (RH), and integrase (INT) Kumar and Bennetzen, 1999).
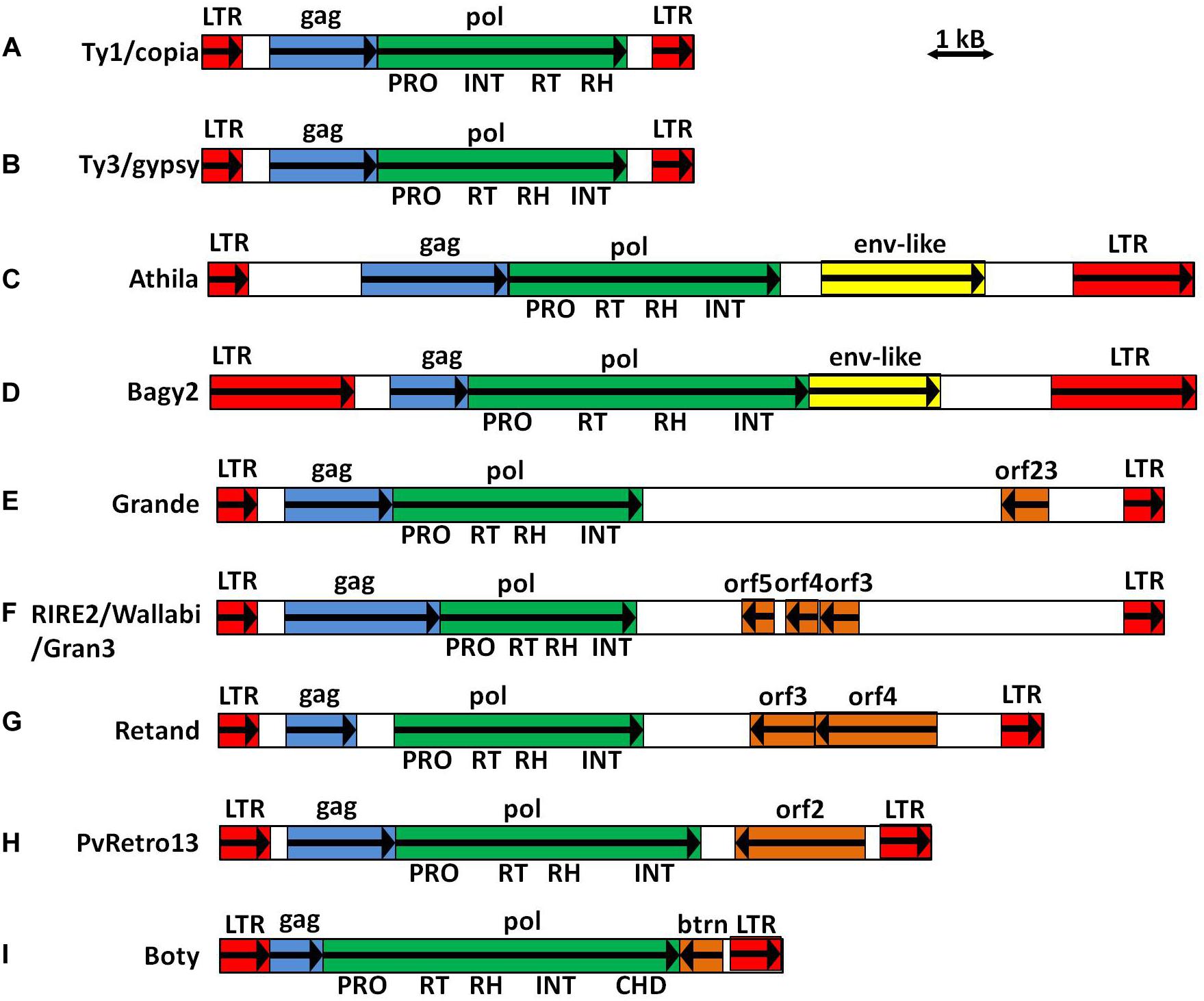
Figure 1. Schematic representation of LTR-retrotransposons. (A) Model Ty1/copia element. (B) Model Ty3/gypsy element. (C) Arabidopis thaliana Athila (Wright and Voytas, 2002). (D) Barley Bagy2 (Vicient et al., 2001). (E) Maize Grande (Gómez-Orte et al., 2013). (F) Rice RIRE2 (Ohtsubo et al., 1999), Wallabi and Gran3 (Wicker et al., 2007). (G) Silene latifolia Retand (Kejnovsky et al., 2006). (H) Phaseolus vulgaris PvRetro13 (Gao et al., 2014). (I) Botrytis cinerea Boty (Zhao et al., 2011).
As sequence data accumulate, the recognizing of sequences encoding for additional proteins (aORFs) in the internal region of plant LTR-retrotransposons seems to be more frequent (Neumann et al., 2019). The aORF can be found in LTR-retrotransposon families with low or high copy numbers, in sense or antisense orientation with respect to the gag-pol genes, and upstream or downstream of them (Steinbauerová et al., 2011). Among them, those located between pol and 3′ LTR in sense or in antisense with respect to the gag and pol are the most frequent.
Plant LTR-Retrotransposons with Conserved aORFs in Sense Orientation Between pol and 3′ LTR
Retroviruses and LTR-retrotransposons share many structural features, and the main difference is that all retroviruses contain a third coding domain in their internal region called env that is located between pol and 3′ LTR. Env encodes for proteins involved in interacting with cellular receptors and mediate fusion of the host and viral membranes (Nisole and Saïb, 2004). Since retrotransposons have an intracellular retrotransposition cycle, it was initially thought that they do not need the ENV protein and, in fact, most do not have it. However, the presence of coding domains in addition to the gag and pol genes between pol and 3′ LTR in the same sense as gag and pol (aORFs-3S) have been described in some LTR-retrotransposons in insects (Song et al., 1994) and in plants (Vicient et al., 2001; Wright and Voytas, 2002; Laten and Gaston, 2012). They encode proteins with certain similarities to ENV, suggesting that they may also exist in, at least, some LTR-retrotransposons (Figures 1C,D).
One of the conserved characteristics of the retroviral env domains is that they code for proteins with transmembrane domains, a characteristic that some of these aORFs-3S also have, which may suggest some functional similarity (Vicient et al., 2001). On the other hand, the retroviral mRNA is usually spliced to give rise to a message capable of expressing the envelope protein. Similar splicing events have been reported for some plant retrotransposons like barley Bagy2 (Vicient et al., 2001). Together, the data suggest that aORFs-3S could encode proteins with functional similarities to retroviral ENV and therefore they are often called env-like domains (Carvalho et al., 2010). However, an ENV-like function in plants remains controversial because the plant cell wall may represent a barrier to the interaction of ENV with cellular receptors present in the plasma membrane. As a consequence, it has been proposed that ENV-like proteins in plants may have a different function than in retroviruses as, for example, to modify the molecular size exclusion limit of plasmodesmata (Carrington et al., 1996) or to serve as chaperone proteins to facilitate replication (Havecker et al., 2004).
Plant LTR-Retrotransposons with Conserved aORFs in Antisense Orientation Between pol and 3′ LTR
The presence of coding domains in addition to the gag and pol genes between pol and 3′ LTR in antisense with respect to gag and pol (aORFs-3R) has been described in different plant LTR-retrotransposons. Some examples are maize Grande1, rice RIRE2, Retand from Silene latifolia, and PvRetro13 from Phaseolus vulgaris (Martínez-Izquierdo et al., 1997; Ohtsubo et al., 1999; Kejnovsky et al., 2006; Figures 1E–H). They are also present in non-plant species as, for example, in Boty from Botrytis cinerea and Sclerotinia sclerotiorum (Zhao et al., 2011; Figure 1I). The function of these aORF-3Rs is not known, but their presence in most of the copies of a family, with a degree of sequence conservation similar to that of other retrotransposon-encoded proteins, suggests that they may be important for the retrotransposition process (Ohtsubo et al., 1999; Gómez-Orte et al., 2013).
Although a complete analysis of the presence, species distribution, and types of these aORF-3Rs is not yet available, the current data indicates that some of them are distributed in several species. For example, the sequences of the aORF-3R of RIRE2, Wallabi, and Gran3, from different species of the genus Oryza, show similarities with the aORF-3R of Grande from Zea species (Ohtsubo et al., 1999). Retand aORF-3Rs from S. latifolia show homology with sequences from other species, and sequences similar to PvRetro13 aORF-3R were detected in other species (Gao et al., 2014). These homologies indicate an ancient origin, at least for some of the aORF-3R.
Origin of Antisense aORFs
Retroviruses have the potential to capture complete or parts of cellular genes in a process known as gene transduction. Gene transduction events have also been described in some Class I TEs. For example, human L1 retrotransposons can capture gene fragments by transduction (Goodier et al., 2000). There are also some described examples in plants. Bs1, a maize LTR-retrotransposon, has transduced sequences from different host genes (Elrouby and Bureau, 2010). A total of 400 genes have been identified as transposon-captured genes in maize (Schnable et al., 2009), 672 in rice and 1343 in sorghum (Jiang and Ramachandran, 2013). In maize, the majority of TE-captured genes were from Helitron elements, in rice from Pack-MULEs, and in sorghum from LTR-retrotransposons, and a high percentage of LTR-retrotransposon-captured genes are still functional in sorghum (Jiang and Ramachandran, 2013). However, retrotransposon-transduced gene sequences are usually in the same sense as the retrotransposon gag and pol genes. Moreover, the lack of gene sequences similar to that of aORFs outside the TEs suggests that aORFs are not transduced gene sequences. Another possible origin of the aORF is the insertion of a TE that, once inserted, became part of the element, losing part of its structure. However, although nested insertions of TEs are relatively frequent in plant genomes (SanMiguel and Bennetzen, 1998), the lack of similarity of the aORF sequences with that of other TEs or viruses does not support this hypothesis. In consequence, the origin of most of these aORFs remains unknown.
Function of Antisense aORFs
No clear similarities with other proteins in databases have been described for any of the proteins encoded by aORFs-3R. However, some of these peptides localize in the nucleus, as the one encoded by Grande (GENE23; Gómez-Orte et al., 2013), and the one encoded by PvRetro13 (ORF2) contains a conserved SMC domain (structural maintenance of chromosomes) that binds DNA and acts in organizing and segregating chromosomes for partitioning (Marchler-Bauer et al., 2011). These results suggest that these proteins may fulfill some nuclear function. Moreover, the aORF-3R protein encoded by Retand (ORF4) contains a transposase 28 domain (pfam04195), suggesting a possible role in the retrotransposition process. During retrotransposition, the pre-integration complex (PIC) produced in the cytoplasm must translocate to the nucleus (McLane et al., 2008). In some retrotransposons, like the fission yeast Tf1, the nuclear localization signal is provided by GAG (Kim et al., 2005), but in retroviruses some accessory proteins are involved (Vogt, 1997; Kogan and Rappaport, 2011). Many retroviruses encode more ORFs in addition to gag, pol, and env, called accessory factors. These ORFs could be in antisense orientation and be located between pol and 3′ LTR. The Accessory factors encode for structural and enzymatic proteins essential for the regulation of transcription (Tat), the transport of unspliced and partially spliced viral RNAs from the nucleus into the cytoplasm (Rev), and others (Vif, Vpr, Vpu, Vpx, and Nef) (Sauter and Kirchhoff, 2018). All these suggest that the proteins encoded by aORFs-3R in LTR-retrotransposons may play a role similar to some of the retroviral accessory proteins, regulating the retrotransposition process.
Transcription of aORFs-3R
The transcription of gag-, pol-, and env-like genes in LTR-retrotransposons is directed by a promoter located in the 5′ LTR. In maize Grande, the region corresponding to gene23 (aORF-3R) is ubiquitously transcribed in a relatively high level in antisense with respect to the gag-pol genes. This transcription is directed by a promoter located in the upstream region of gene23 (Gómez et al., 2006; Vicient, 2010; Gómez-Orte et al., 2013). A weak ubiquitous expression was also detected for Retand aORFs-3R (Kejnovsky et al., 2006). Antisense promoters are not unusual in retrovirus. For example, HTLV has an antisense promoter located inside the 3′ LTR (Barbeau and Mesnard, 2011). Antisense promoters have been also identified in other retrotransposons as, for example, the apple Mdoryco1-1 (Wang et al., 2017), Arabidopsis thaliana AtRE1 (Kato et al., 2005), and Drosophila hydei micropia (Lankenau et al., 1994). However, in the case of micropia, it transcribes two antisense RNAs of 1.0 and 1.6 kb with no protein coding capacity. Antisense non-coding transcripts have also been described for the Drosophila non-LTR TART element (Danilevskaya et al., 1999) and in mouse IAP elements that have both sense and antisense promoter activities in their LTRs (Druker et al., 2004). In view of these last examples, we cannot rule out that, at least in some cases, the production of antisense mRNAs has itself some regulatory roles. Antisense transcripts may produce double-stranded RNAs (dsRNAs) when they hybridize with the genomic RNA of the LTR-retrotransposon, generating 21–24-nt siRNAs, which can act as inhibitors of the retrotransposition by a dsRNA-mediated silencing mechanism. These two possible functions based on the generation of a protein or on the generation of dsRNA can be true simultaneously and represent a fine regulation of the LTR-retrotransposition mechanism.
Conclusion
Additional open reading frames located between the pol gene and the 3′ LTR are present in some plant LTR-retrotransposon families. Sense aORFs show some functional and structural characteristics similar to the env genes in retroviruses, although their possible roles in retrotransposition remain unclear. Antisense aORFs are also present in different retrotransposon families, but their functions are yet unknown. The nuclear localization identified in some cases and the comparison with the antisense genes of retroviruses suggest they may play a regulatory role in retrotransposition. Antisense transcription may also play a regulatory role itself, through a dsRNA-mediated silencing mechanism. In conclusion, we believe that it is necessary to pay more attention to the presence of this type of additional ORFs in the annotations of the TEs. We also think that it is necessary to look at the possible presence of antisense and spliced transcripts. Finally, we think it would be interesting to carry out research efforts on the possible functions that the transcripts and the proteins they encode could perform.
Author Contributions
CV drafted the manuscript with contributions of JC. Both CV and JC revised and approved the manuscript.
Funding
This work was funded by grants AGL2016-78992-R from FEDER/Ministerio de Ciencia, Innovación y Universidades-Agencia Estatal de Investigación (Spain) and by the CERCA Programme of the Generalitat de Catalunya. We also acknowledge financial support from the Spanish Ministerio de Economía y Competitividad through the “Severo Ochoa Programme for Centres of Excellence in R&D” 2016-2019 (SEV-2015-0533).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Barbeau, B., and Mesnard, J. M. (2011). Making sense out of antisense transcription in human T-cell lymphotropic viruses (HTLVs). Viruses 3, 456–468. doi: 10.3390/v3050456
Carrington, J. C., Kasschau, K. D., Mahajan, S. K., and Schaad, M. C. (1996). Cell-to-cell and long-distance transport of viruses in plants. Plant Cell 8, 1669–1681. doi: 10.1105/tpc.8.10.1669
Carvalho, M., Ribeiro, T., Viegas, W., Morais-Cecilio, L., and Rocheta, M. (2010). Presence of env-like sequences in Quercus suber retrotransposons. J. Appl. Genet. 51, 461–467. doi: 10.1007/BF03208875
Danilevskaya, O. N., Traverse, K. L., Hogan, N. C., DeBaryshe, P. G., and Pardue, M. L. (1999). The two Drosophila telomeric transposable elements have very different patterns of transcription. Mol. Cell Biol. 19, 873–881. doi: 10.1128/mcb.19.1.873
Dodonova, S. O., Prinz, S., Bilanchone, V., Sandmeyer, S., and Briggs, J. A. G. (2019). Structure of the Ty3/Gypsy retrotransposon capsid and the evolution of retroviruses. Proc. Natl. Acad. Sci. U.S.A. 116, 10048–10057. doi: 10.1073/pnas.1900931116
Druker, R., Bruxner, T. J., Lehrbach, N. J., and Whitelaw, E. (2004). Complex patterns of transcription at the insertion site of a retrotransposon in the mouse. Nucleic Acids Res. 32, 5800–5808. doi: 10.1093/nar/gkh914
Elrouby, N., and Bureau, T. E. (2010). Bs1, a new chimeric gene formed by retrotransposon-mediated exon shuffling in maize. Plant Physiol. 153, 1413–1424. doi: 10.1104/pp.110.157420
Gao, D., Abernathy, B., Rohksar, D., Schmutz, J., and Jackson, S. A. (2014). Annotation and sequence diversity of transposable elements in common bean (Phaseolus vulgaris). Front. Plant Sci. 11:339. doi: 10.3389/fpls.2014.00339
Gómez, E., Schulman, A. H., Martínez-Izquierdo, J. A., and Vicient, C. M. (2006). Integrase diversity and transcription of the maize retrotransposon Grande. Genome 49, 558–562. doi: 10.1139/g05-129
Gómez-Orte, E., Vicient, C. M., and Martínez-Izquierdo, J. A. (2013). Grande retrotransposons contain an accessory gene in the unusually long 3′-internal region that encodes a nuclear protein transcribed from its own promoter. Plant Mol. Biol. 81, 541–551. doi: 10.1007/s11103-013-0019-2
Goodier, J. L., Ostertag, E. M., and Kazazian, H. H. Jr. (2000). Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum. Mol. Genet. 9, 653–657. doi: 10.1093/hmg/9.4.653
Havecker, E. R., Gao, X., and Voytas, D. F. (2004). The diversity of LTR retrotransposons. Genome Biol. 5:225. doi: 10.1186/gb-2004-5-6-225
Jiang, S. Y., and Ramachandran, S. (2013). Genome-wide survey and comparative analysis of LTR retrotransposons and their captured genes in rice and sorghum. PLoS One 8:e71118. doi: 10.1371/journal.pone.0071118
Kato, A., Endo, M., Kato, H., and Saito, T. (2005). The antisense promoter of AtRE1, a retrotransposon in Arabidopsis thaliana, is activated in pollens and calluses. Plant Sci. 168, 981–986.
Kejnovsky, E., Kubat, Z., Macas, J., Hobza, R., Mracek, J., and Vyskot, B. (2006). Retand: a novel family of gypsy-like retrotransposons harboring an amplified tandem repeat. Mol. Genet. Genomics. 276, 254–263. doi: 10.1007/s00438-006-0140-x
Kim, M. K., Claiborn, K. C., and Levin, H. L. (2005). The long terminal repeat-containing retrotransposon Tf1 possesses amino acids in gag that regulate nuclear localization and particle formation. J. Virol. 79, 9540–9555. doi: 10.1128/JVI.79.15.9540-9555.2005
Kogan, M., and Rappaport, J. (2011). HIV-1 accessory protein Vpr: relevance in the pathogenesis of HIV and potential for therapeutic intervention. Retrovirology 8:25. doi: 10.1186/1742-4690-8-25
Lankenau, S., Corces, V. G., and Lankenau, D. H. (1994). The Drosophila micropia retrotransposon encodes a testis-specific antisense RNA complementary to reverse transcriptase. Mol. Cel. Biol. 14, 1764–1775. doi: 10.1128/mcb.14.3.1764
Laten, H. M., and Gaston, G. D. (2012). “Plant Endogenous Retroviruses? A Case of Mysteriuos ORFs,” in Plant Transposable Elements. 2012. Impact on Genome Structure and Function, Topics in Current Genetics 24, eds M. A. Grandbastien and J. M. Casacuberta Berlin: Springer, 89–112.
Marchler-Bauer, A., Lu, S., Anderson, J. B., Chitsaz, F., Derbyshire, M. K., DeWeese-Scott, C., et al. (2011). CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229. doi: 10.1093/nar/gkq1189
Martínez-Izquierdo, J. A., Garcia-Martinez, J., and Vicient, C. M. (1997). What makes Grande1 retrotransposon different? Genetica 100, 15–28.
McLane, L. M., Pulliam, K. F., Devine, S. E., and Corbett, A. H. (2008). The Ty1 integrase protein can exploit the classical nuclear protein import machinery for entry into the nucleus. Nucleic Acids Res. 36, 4317–4326. doi: 10.1093/nar/gkn383
Neumann, P., Novák, P., Hoštáková, N., and Macas, J. (2019). Systematic survey of plant LTR-retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mob. DNA. 10:1. doi: 10.1186/s13100-018-0144-1
Nisole, S., and Saïb, A. (2004). Early steps of retrovirus replicative cycle. Retrovirology 1, 9–20. doi: 10.1186/1742-4690-1-9
Ohtsubo, H., Kumekawa, N., and Ohtsubo, E. (1999). RIRE2, a novel gypsy-type retrotransposon from rice. Genes Genet. Syst. 74, 83–91. doi: 10.1266/ggs.74.83
SanMiguel, P., and Bennetzen, J. L. (1998). Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann. Bot. 82, 37–44.
Sauter, D., and Kirchhoff, F. (2018). Multilayered and versatile inhibition of cellular antiviral factors by HIV and SIV accessory proteins. Cytokine Growth Factor Rev. 40, 3–12. doi: 10.1016/j.cytogfr.2018.02.005
Schnable, P. S., Ware, D., Fulton, R. S., Stein, J. C., Wei, F., Pasternak, S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115. doi: 10.1126/science.1178534
Song, S. U., Gerasimova, T., Kurkulos, M., Boeke, J. D., and Corces, V. G. (1994). An env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes Dev. 8, 2046–2057. doi: 10.1101/gad.8.17.2046
Steinbauerová, V., Neumann, P., Novák, P., and Macas, J. (2011). A widespread occurrence of extra open reading frames in plant Ty3/gypsy retrotransposons. Genetica 139, 1543–1555. doi: 10.1007/s10709-012-9654-9
Vicient, C. M. (2010). Transcriptional activity of transposable elements in maize. BMC Genomics 11:601. doi: 10.1186/1471-2164-11-601
Vicient, C. M., Kalendar, R., and Schulman, A. H. (2001). Envelope-containing retrovirus-like elements are widespread, transcribed and spliced, and insertionally polymorphic in plants. Genome Res. 11, 2041–2049. doi: 10.1101/gr.193301
Vogt, P. K. (1997). “Retroviral virions and genomes,” in Retroviruses, eds J. M. Coffin, S. M. Hughes, and H. E. Varmus (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), 27–69.
Wang, L., He, Y., Qiu, H., Guo, J., Han, M., Zhou, J., et al. (2017). Mdoryco1-1, a bidirectionally transcriptional Ty1-copia retrotransposon from Malus×domestica. Sci. Hortic. 220, 283–290.
Wicker, T., Sabot, F., Hua-Van, A., Bennetzen, J. L., Capy, P., Chalhoub, B., et al. (2007). A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8, 973–982. doi: 10.1038/nrg2165
Wright, D. A., and Voytas, D. F. (2002). Athila4 of Arabidopsis and Calypso of soybean define a lineage of endogenous plant retroviruses. Genome Res. 12, 122–131. doi: 10.1101/gr.196001
Keywords: additional ORF, antisense, env, LTR-retrotransposon, retrovirus
Citation: Vicient CM and Casacuberta JM (2020) Additional ORFs in Plant LTR-Retrotransposons. Front. Plant Sci. 11:555. doi: 10.3389/fpls.2020.00555
Received: 03 December 2019; Accepted: 14 April 2020;
Published: 26 May 2020.
Edited by:
Ruslan Kalendar, University of Helsinki, FinlandReviewed by:
Andreas Bachmair, University of Vienna, AustriaAndré Luís Laforga Vanzela, State University of Londrina, Brazil
Copyright © 2020 Vicient and Casacuberta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carlos M. Vicient, Y2FybG9zLnZpY2llbnRAY3JhZ2Vub21pY2EuZXM=
 Carlos M. Vicient
Carlos M. Vicient Josep M. Casacuberta
Josep M. Casacuberta