- 1State Key Laboratory of Crop Gene Exploration and Utilization in Southwest China, Wenjiang, China
- 2Triticeae Research Institute, Sichuan Agricultural University, Wenjiang, China
Fluorescence in situ hybridization karyotypes have been widely used for evolutionary analysis on chromosome organization and genetic/genomic diversity in the wheat alliance (tribe Triticeae of Poaceae). The karyotpic diversity of Aegilops umbellulata, Ae. markgrafii, Ae. comosa subsp. comosa and subsp. subventricosa, and Ae. uniaristata was evaluated by the fluorescence in situ hybridization (FISH) probes oligo-pSc119.2 and pTa71 in combination with (AAC)5, (ACT)7, and (CTT)12, respectively. Abundant intra- and interspecific genetic variation was discovered in Ae. umbellulata, Ae. markgrafii, and Ae. comosa, but not Ae. uniaristata. Chromosome 7 of Ae. umbellulata had more variants (six variants) than the other six U chromosomes (2–3 variants) as revealed by probes oligo-pSc119.2 and (AAC)5. Intraspecific variation in Ae. markgrafii and Ae. comosa was revealed by oligo-pSc119.2 in combination with (ACT)7 and (CTT)12, respectively. At least five variants were found in every chromosome of Ae. markgrafii and Ae. comosa, and up to 18, 10, and 15 variants were identified for chromosomes 2 of Ae. markgrafii, 4 of Ae. comosa subsp. comosa, and 6 of Ae. comosa subsp. subventricosa. The six Ae. uniaristata accessions showed identical FISH signal patterns. A large number of intra-specific polymorphic FISH signals were observed between the homologous chromosomes of Ae. markgrafii and Ae. comosa, especially chromosomes 1, 2, 4, and 7 of Ae. markgrafii, chromosome 4 of Ae. comosa subsp. comosa, and chromosome 6 of Ae. comosa subsp. subventricosa. Twelve Ae. comosa and 24 Ae. markgrafii accessions showed heteromorphism between homologous chromosomes. Additionally, a translocation between the short arms of chromosomes 1 and 7 of Ae. comosa PI 551038 was identified. The FISH karyotypes can be used to clearly identify the chromosome variations of each chromosome in these Aegilops species and also provide valuable information for understanding the evolutionary relationships and structural genomic variation among Aegilops species.
Introduction
The genus Aegilops serves as a valuable genetic resource for expanding the genetic basis of cultivated bread wheat, as it is closely related to Triticum and has played a pivotal role in the evolution of bread wheat (Schneider et al., 2008). Aegilops contains 11 diploid species that harbor different nuclear and cytoplasmic genomes (Van Slageren, 1994). The diploid Aegilops species include seven basic genomes, namely U, C, M, N, D, S, and T (Kimber and Tsunewaki, 1988). Ae. umbellulata, Ae. markgrafii, Ae. comosa, and Ae. uniaristata are diploid donor species of polyploid Aegilops harboring the U, C, M, and N genomes, respectively.
In addition to the evolutionary contribution to polyploid Aegilops species, the diploid Aegilops species including those with U, M, C, and N genomes also harbor abundant beneficial genes for the genetic improvement of bread wheat with respect to, for instance, resistance to leaf and stripe rust (Sears, 1956; Riley et al., 1968; Riar et al., 2012; Toor et al., 2016; Bansal et al., 2017; Liu et al., 2019) and powdery mildew (Gill et al., 1985; Zhu et al., 2006; Weidner et al., 2012), tolerance to salt (Gorham, 1990) and aluminum stress (Miller et al., 1995), accumulation of zinc and iron (Wang et al., 2011; Neelam et al., 2012), high efficiency in zinc uptake (Cakmak et al., 1999), as well as high protein content and gluten content (Gong et al., 2017; Wang et al., 2017).
Aegilops species are distributed from the Mediterranean via Southwest Asia to central Asia and contain sufficient genetic diversity for adaptation to various environments. Several methods, including morphological observation (Kawahara, 2002; Tahernezhad et al., 2010), C-banding karyotype (Badaeva et al., 1996a), biochemical markers (Rodríguez-Quijano et al., 2001; Dai et al., 2015), and molecular markers (Sasanuma et al., 2004; Tahernezhad et al., 2010; Thomas and Bebeli, 2010), have been adopted to assess the genetic diversity and evolutionary relationships of Aegilops species. The karyotypes of some Aegilops species have been established with a C-banding technique (Teoh et al., 1983; Friebe et al., 1992, 1995, 1996b). Hybridization signals of the (CTT)n probe on Aegilops chromosomes are often consistent with their C-banding patterns (Ruban and Badaeva, 2018). Furthermore, some cloned repeats and oligo-nucleotide sequences have been used as probes to establish fluorescence in situ hybridization (FISH) karyotypes rather than C-banding karyotypes due to improved efficiency and easier operation.
Fluorescence in situ hybridization is a valid tool for the direct physical mapping of DNA sequences on chromosomes and is often utilized in evolutionary and speciation studies as well as for the assessment of genetic diversity among and within species (Badaeva et al., 2002, 2004, 2015). Several probes, such as pSc119 or oligo-pSc119.2, Afa family, (AAC)5, (GAA)n, oligo-pTa535, and oligo-pTa71, have been used for the FISH karyotyping of diploid Aegilops species, including Ae. umbellulata, Ae. markgrafii, Ae. comosa, and Ae. uniaristata, and their introgression lines with wheat (Badaeva et al., 1996a, 2011; Iqbal et al., 2000; Molnár et al., 2011, 2015, 2016; Kwiatek et al., 2013; Mirzaghaderi et al., 2014; Danilova et al., 2017; Liu et al., 2019; Song et al., 2019).
The goatgrasses, including Ae. umbellulata, Ae. markgrafii, Ae. comosa, and Ae. uniaristata, are necessary germplasms for the genetic improvement of cultivated hexaploid wheat and exhibit rich genetic diversity at the chromosome level. In previous studies, the FISH karyotypes were mainly used for chromosome identification and, to a lesser extent, for the analysis of genomic diversity, chromosome organization, and evolutionary patterns. Currently, FISH karyotype-based chromosome organization and the evolutionary patterns of these Aegilops species are still insufficiently. Therefore, the main objectives of the present study were to elucidate the genetic diversity of the four diploid Aegilops species using FISH karyotypes and to understand their chromosomal organization. In the present investigation, six FISH probes were tested for the selection of suitable probes for evaluating the diversity of four diploid Aegilops species with U, C, M, and N chromosomes. These data provide useful information for understanding the genome evolution and differentiation as well as the genetic diversity of Aegilops species.
Materials and Methods
Plant Materials
A collection of 145 accessions belonging to Ae. umbellulata (2n = 2x = 14, UU, 47 accessions), Ae. markgrafii (2n = 2x = 14, CC, 44 accessions), Ae. comosa (2n = 2x = 14, MM, 13 accessions of subsp. comosa and 35 accessions of subsp. subventricosa), and Ae. uniaristata (2n = 2x = 14, NN, six accessions) were subjected to FISH karyotyping (Table 1). These materials were supplied from the USDA-ARS germplasm bank1.
FISH Karyotyping
Ten randomly selected seeds from each accession were germinated on Petri dishes lined with double-layer moist filter papers at 4°C for ∼24 h and then incubated in a container under a 16 h photoperiod (light/dark temperature 22/16°C). Root tips were excised when the roots reached 1–2 cm and were treated with 1.0 MPa nitrous oxide (NO) gas for 2 h. Then, the root tips were fixed in glacial acetic acid for at least 5 min before storing in 70% ethanol for slide preparation (Kato, 1999). Root tips were suspended in cellulase/pectinase enzyme solution (4: 2) before dropping onto slides (Komuro et al., 2013). The FISH procedure was the same as described by Hao et al. (2013). Ten microliters of hybridization mixture solution was added to each slide. Each slide was then placed at 37°C for at least 1 h. DAPI (4′, 6-diamidino-2-phenylindole) was used to counter-stain the slides for visualizing the FISH signals. The chromosomal observations of hybridization signals were conducted with an Olympus BX-63 epifluorescence microscope, and the photographs were recorded with a Photometric SenSys Olympus DP80 CCD camera (Olympus, Tokyo, Japan). After capturing the images, the coverslips of each slide were removed and the slides were washed for the next FISH (Komuro et al., 2013). Original images were processed using Photoshop V7.0 (Adobe Systems Incorporated, United States). At least five metaphase cells of each accession were observed in order to count chromosome variants.
DNA Probes and Chromosome Identification
Six probes, namely oligo-pSc119.2, oligo-pTa71, oligo-pTa713, (AAC)5, (CTT)12, and (ACT)7, which are suited for use in identifying the chromosomes of common wheat and Aegilops species (Cuadrado and Jouve, 2010; Tang et al., 2014; Zhao et al., 2016) were used in the current investigation. The chromosomes of the four diploid Aegilops species were identified and classified based on the FISH patterns of these DNA probes combined with the C-banding karyotypes in previous investigations (Friebe et al., 1995, 1996a; Badaeva et al., 1996a; Danilova et al., 2017; Liu et al., 2019; Song et al., 2019). The 5′-ends of oligo-pSc119.2 and oligo-pTa71 were labeled with 6-carboxyfluorescein (6-FAM), and the remaining four probes were labeled with 6-carboxy tetramethylrhodamine (Tamra). All of these probes were synthesized by Sangon Biotech (Shanghai, China).
Results
FISH Markers for Each Species
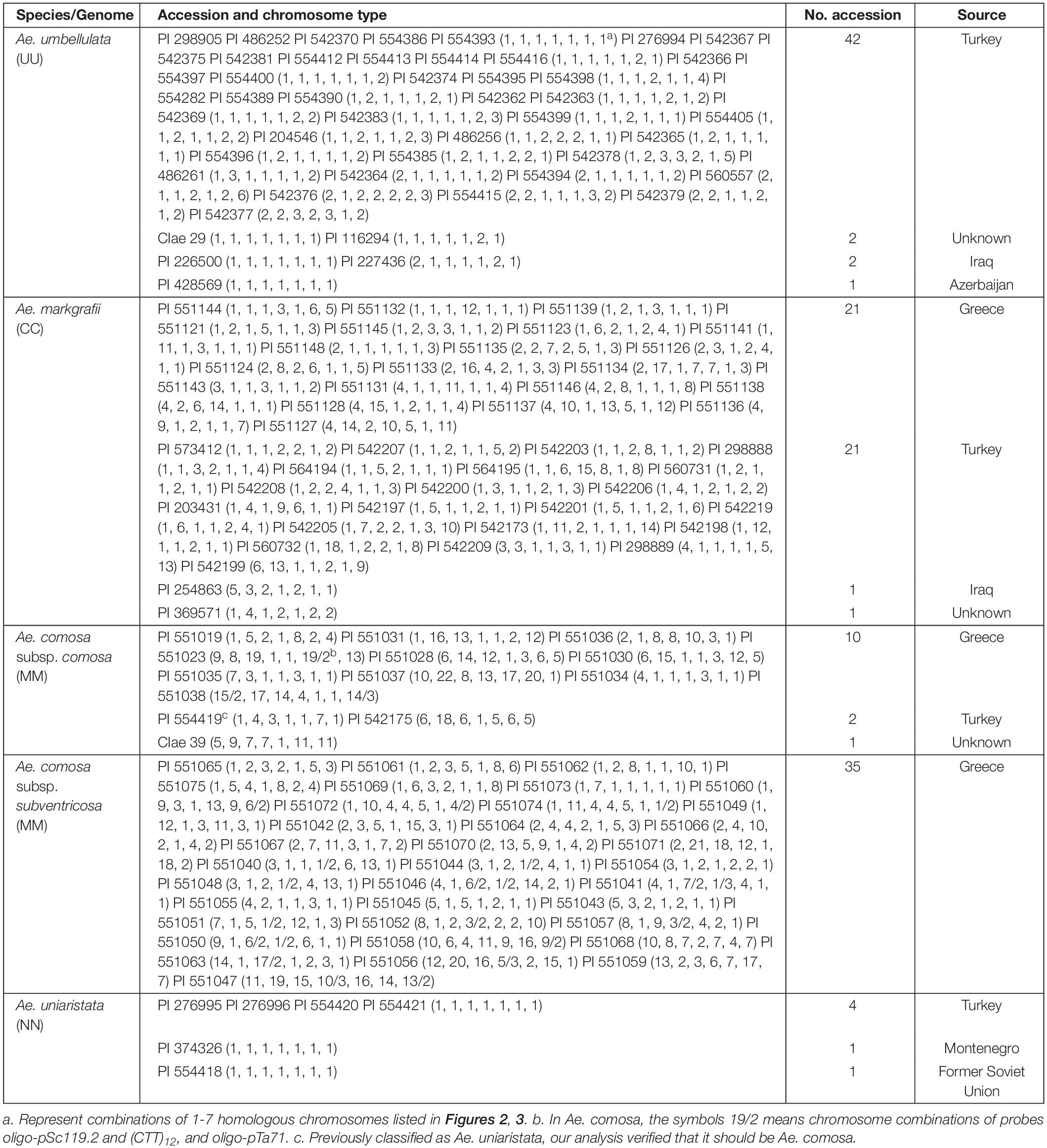
Six probes were initially screened for one accession each from four diploid species, namely Ae. umbellulata (accession CIae 29), Ae. markgrafii (PI 542197), Ae. comosa subsp. subventricosa (PI 551068), and Ae. uniaristata (PI 554418) (Figure 1).
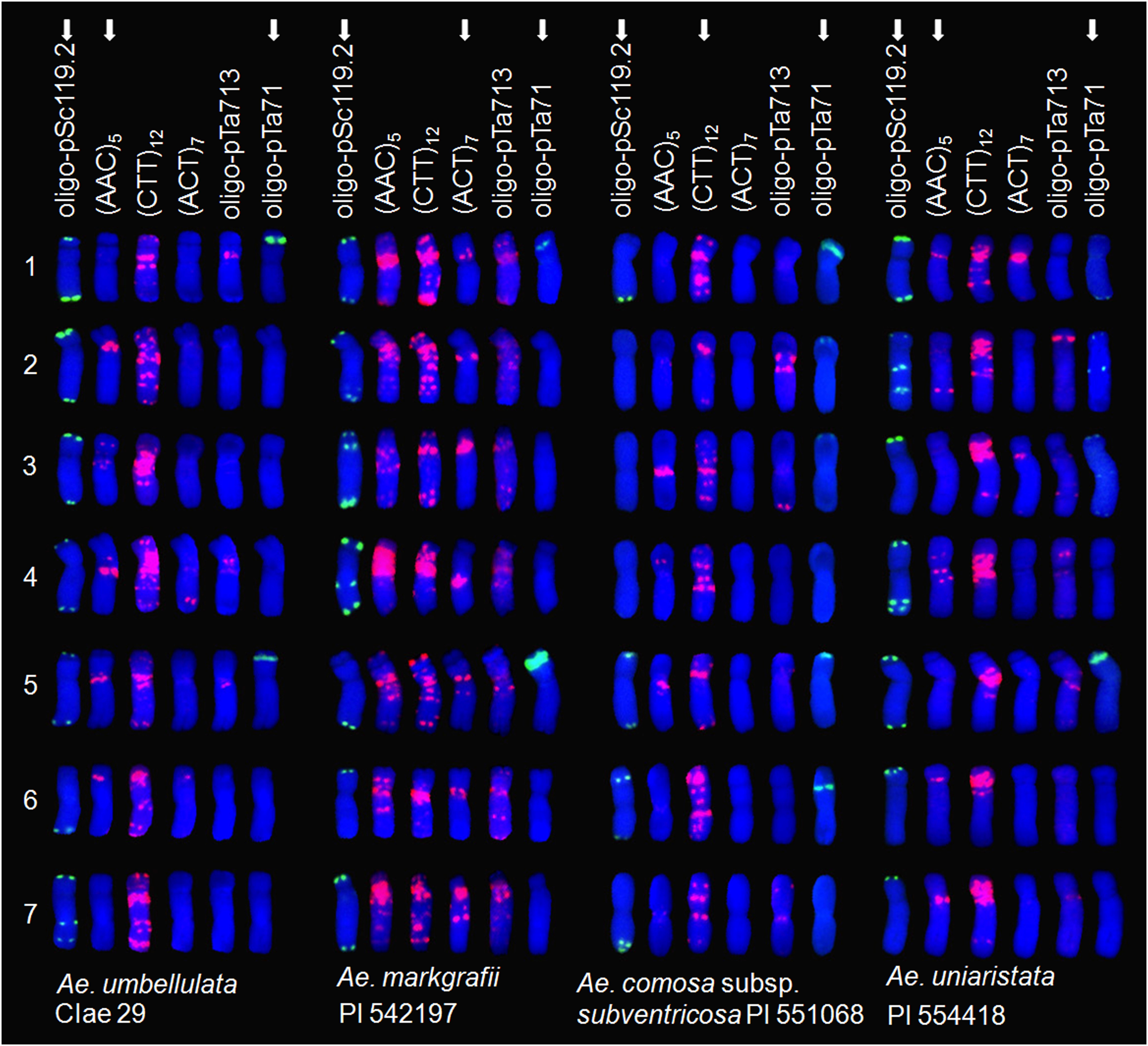
Figure 1. FISH karyotyping of the chromosomes of four diploid Aegilops species using six DNA probes. The probes eventually used for polymorphism analysis of each diploid Aegilops species are shown by white arrowheads (see text for details).
Three [oligo-pSc119.2, (AAC)5 and (CTT)12] of the six probes showed hybridization signals on nearly all U chromosomes of Ae. umbellulata, while the remaining three probes had hybridization signals on some U chromosomes, including 1U, 4U, and 5U. The (CTT)12 probe hybridized to many sites on every U chromosome and stretched throughout the whole chromosome, whereas (ACT)7 and oligo-pTa713 harbored a few signals on the pericentromeric region of chromosomes 1U, 4U, and 5U and chromosome arms 4UL and 6US. The probes oligo-pSc119.2 and (AAC)5 hybridized to the telomeric regions of every U chromosome and the pericentromeric regions of chromosomes 2U, 3U, 4U, 5U, and 6U, and their combinations showed hybridization signals on every U chromosome and could distinguish 1U, 3U, 4U, 6U, and 7U. Although oligo-pTa71 only hybridized to the nucleolar organizing regions of chromosome arms 1US and 5US, it was helpful to differentiate chromosomes 1U and 5U from 2U, 3U, and 4U.
Five of the six probes (all except for oligo-pTa71) hybridized to all C chromosomes of Ae. markgrafii. The oligo-pSc119.2 probe hybridized to the telomeric regions of chromosomes 1C, 2C, 3C, 4C, and 7C and chromosome arms 6CS and 5CL, as well as the central region of chromosome arm 4CL. The hybridization sites of probes (AAC)5, (ACT)7, (CTT)12, and oligo-pTa713 were found on every C chromosome, while those of oligo-pTa71 hybridized to the nucleolar organizing regions of chromosome arms 1CS and 5CS. The probe combinations oligo-pSc119.2/(ACT)7 and oligo-pTa71 could clearly differentiate all C chromosomes.
All six probes other than (ACT)7 harbored hybridization sites on the M chromosomes of Ae. comosa. The hybridization sites of the oligo-pTa713 probe were located on the telomeric region of chromosome arm 3ML, near the telomeric region of chromosome arm 7MS, and the pericentromeric regions of chromosomes 2M and 7M, while those of (AAC)5 were located on the pericentromeric regions of chromosomes 4M to 7M. The oligo-pSc119.2 probe hybridized to the telomeric region of chromosome arms 1ML and 7ML and chromosomes 5M and 6M. However, the hybridization sites (CTT)12 mainly targeted the pericentromeric region and telomeric region of chromosomes 1M to 7M. The oligo-pTa71 probe hybridized to sites on chromosome arm 1MS and chromosomes 2M to 6M. The probes oligo-pSc119.2/(CTT)12 and oligo-pTa71 had the ability to distinguish each M chromosome. The remaining three probes, (AAC)5, (ACT)7, and oligo-pTa713, were not further used as they lacked sufficient hybridization sites on the M chromosome.
All six probes hybridized to sites on the N chromosomes of Ae. uniaristata. The hybridization signals of the (AAC)5 probe were distributed on the pericentromeric regions of chromosomes 1N, 4N, 6N, and 7N and chromosome arm 2NL. The oligo-pSc119.2 hybridized to the telomeric regions on chromosome arms 2NS, 3NS, 6NS, 7NS, and chromosomes 1N, 4N, and 5N, as well as the central region on chromosome arm 2NL. The hybridization signal sites (CTT)12 were mainly on the pericentromeric regions of every N chromosome, while those of (ACT)7 were on the pericentromeric regions of chromosomes 1N and 3N. The oligo-pTa713 hybridized to pericentromeric regions on chromosomes 4N, 5N, and 7N and near the telomeric region on chromosome arm 2NS and the middle region of chromosome arm 3NL. The oligo-pTa71 probe exhibited strong hybridization signals on nucleolar organizing regions 5NS and weak signals on the pericentromeric regions of chromosomes 1N and 2N and the telomeric regions of chromosome arms 1NL and 2NS and chromosome 3N. The probes oligo-pSc119.2/(AAC)5 could clearly differentiate every N chromosome with the aid of oligo-pTa71.
Among the selected accession in each species, the oligo-pSc119.2 probe showed strong signals on all seven chromosomes of Ae. umbellulata, Ae. markgrafii, and Ae. uniaristata, as well as on the four chromosomes (1M, 5M, 6M, and 7M) of Ae. comosa. The (AAC)5 signals were distributed on every chromosome of Ae. markgrafii, on five chromosomes of Ae. umbellulata (2–6U) and Ae. uniaristata (1N, 2N, 4N, 6N, and 7N), and on six chromosomes (except for 1M) of Ae. comosa. The (ACT)7 signals of Ae. markgrafii were presented on all seven chromosomes, while only a few or no such signal was detected from the other three Aegilops species. Of the six probes, (CTT)12 had the most signal points, while oligo-pTa713 and oligo-pTa71 exhibited the fewest signal points. The (CTT)12 probe showed strong signals on all of the seven chromosomes of the four Aegilops species, but the signals were mainly concentrated on the pericentromeric regions. There were more (CTT)12 signals in Ae. umbellulata and Ae. markgrafii than in Ae. comosa and Ae. uniaristata.
Polymorphic Variants for Each Species
Ae. umbellulata
All Ae. umbellulata accessions showed diverse hybridization signals for probes oligo-pSc119.2 and (AAC)5 and only one signal pattern for the oligo-pTa71 probe (Table 1 and Figure 2). The number of polymorphic variants for every U chromosome ranged from two to six (Figure 2).
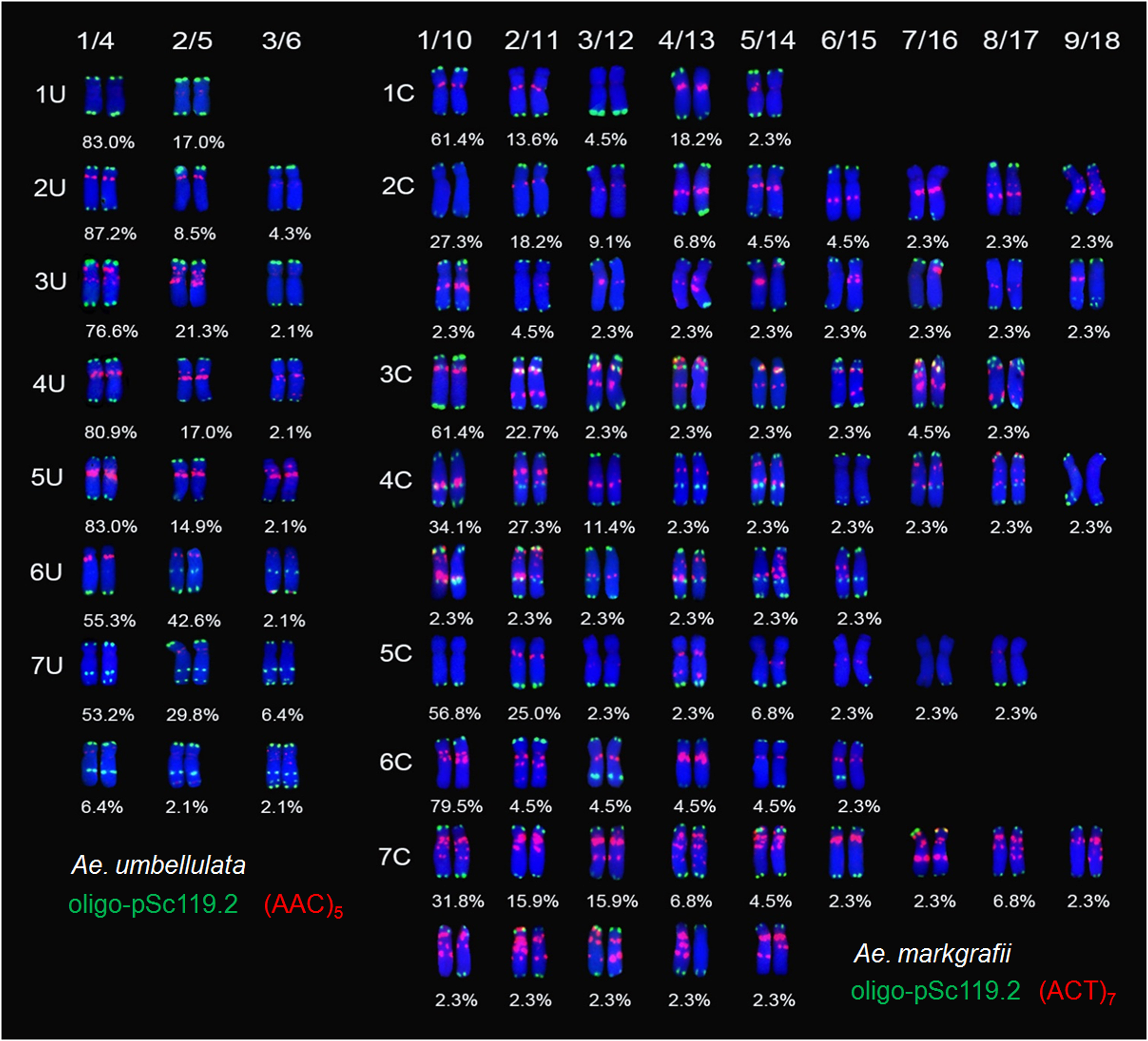
Figure 2. Chromosome variants and frequency analysis of Ae. umbellulata and Ae. markgrafii with the FISH probes oligo-pSc119.2/(AAC)5 and oligo-pSc119.2/(ACT)7. Numbers indicate variants (numbered consecutively); in case these variants are shown in two rows (e.g., 7U, 2C), the number before the slash refers to the variant shown in the upper row, the number after the slash refers to the variant shown in the lower row.
Chromosome 1U had two variants for probe (AAC)5 and only one signal pattern for oligo-pSc119.2. Variant 1 has no (AAC)5 signal on the entire 1U chromosome, while variant 2 showed weak signals on the short arm (Figure 2). Each of the chromosomes 2U, 3U, 4U, 5U, and 6U had three variants for oligo-pSc119.2 and (AAC)5. The hybridization signals of the oligo-pSc119.2 probe on chromosomes 2U and 3U occurred on the telomeric regions of both the long and short arms (for variants 1 and 3) or the short arms (for variant 2). The 3U (AAC)5 hybridization signals varied in intensity (variants 1 and 2 vs. 3: strong vs. weak signal), whereas those of 2U varied in both the intensity and location (strong signal on central 2US for variants 1 and 2 and weak signal on the near-centromeric regions of 2US for variant 3). Among the three 4U variants, a major difference occurred for (AAC)5 rather than oligo-pSc119.2. All three 4U variants shared similar (AAC)5 signal sites on the pericentromeric regions and dissimilar signals on the short or long arm. Compared to 4U variant 1, variants 2 and 3 had an additional pair of signals on either the short or long arm. Chromosome 5U had three variants varying for the hybridization sites of both the oligo-pSc119.2 and (AAC)5 probes. Variant 3 lacked a pair of oligo-pSc119.2 signals that were specific to telomeric region of the short arm of variants 1 and 2. All three 5U variants shared strong (AAC)5 signals on the pericentromeric regions. Additionally, 5U variants 2 and 3 had an additional pair of signals on the short arms in comparison with variant 1. For the three 6U variants, variants 2 and 3 had a pair of oligo-pSc119.2 hybridization signals on the pericentromeric regions that were absent in variant 1. On the other side, variant 3 had an extra (AAC)5 telomeric signal on the long arms as compared to variants 1 and 2. Chromosome 7U had six variants. These 7U variants exhibited different hybridization sites for both the oligo-pSc119.2 and (AAC)5 probes. For example, an additional pair of oligo-pSc119.2 signals on the telomeric regions of the long arms was unique to variants 3 and 6, and a pair of oligo-pSc119.2 signals on the telomeric regions of the long arms was lacking in variant 4 in comparison with the other variants. The (AAC)5 signals of variants 2, 4, 5, and 6 were weak and appeared on the pericentromeric regions. Furthermore, weak (AAC)5 signals were also present on the telomeric regions of the long arms for variant 5.
A total of 25 FISH banding patterns were identified for the 47 Ae. umbellulata accessions (Table 1), of which the pattern harbored by PI 542367 (1, 1, 1, 1, 1, 2, 1) was the dominant variant (9 accessions, 19.1%). Seventeen of the 24 patterns were rare, with one accession for each of them.
Ae. markgrafii
All the Ae. markgrafii accessions showed polymorphic variants for probes oligo-pSc119.2 and (ACT)7 and identical signals for oligo-pTa71. The polymorphic variants for each C chromosome varied from 5 to 18 (Figure 2). Interestingly, heteromorphismin the homologous chromosomes was detected for every C chromosome.
Chromosome 1C had five variants, but only three variants (variants 1–3) showed identical signals between homologous chromosomes. Variants 2, 3, and 4 lacked a pair of oligo-pSc119.2 hybridization signals on the telomeric regions of the short arms in two (variants 2 and 3) or one (variant 4) 1C chromosome in comparison with the other two variants. Similarly, three of the five (except for variant 5 in one of the two 1C and variant 3) 1C variants showed (ACT)7 signal sites on the pericentromeric regions. A total of 18 variants for chromosome 2C were found, of which nine variants (variants 10–18) showed heteromorphismin the homologous chromosomes. All of the 2C variants showed oligo-pSc119.2 hybridization signals on the telomeric regions of both the long and short arms, except for variants 7 and 17, which lacked hybridization signals on the telomeric regions of the short arms. The telomeric signals of oligo-pSc119.2 on chromosome arm 2CL showed high diversity and could be arranged in four groups (I–IV; variants showing similar signal patterns were defined as a group, and the same meaning applies here after for groups), consisting of nine (variants 1, 2, 4, 7, 9, 11, 15, 16, and 17), three (variants 3, 6, and 8), three (variants 12, 13, and 14), and three variants (variants 5, 10, and 18). Group I had a pair of telomeric signals that was absent from group II. Both groups III and IV showed signals on one of the two 2C chromosomes; however, extra signal sites occurred on one of the two 2C chromosomes for group IV. All 18 variants except for variant 1 showed (ACT)7 hybridization signals on chromosome 2C. Heteromorphism in the homologous chromosomes 2C mainly occurred on the long arm. Chromosome 3C had eight variants, of which five variants (variants 1–5) shared similar (ACT)7 signals between homologous chromosomes. Seven of the eight 3C variants (except for variant 5) shared similar oligo-pSc119.2 signals. On chromosome arm 3CS, variant 5 had a pair of oligo-pSc119.2 signals, while the other seven variants had two pairs of signals. The eight variants also exhibited different (ACT)7 hybridization signals. For example, the (ACT)7 signals of variant 1 were located on the short arms, whereas an additional pair of signals was present on the long arms of variants 2, 3, and 5, and two further pairs of signals were present on both the long and short arms of variant 4. Chromosome 4C had 15 variants, but only eight variants (variants 1–8) showed consistent signals between homologous chromosomes. The oligo-pSc119.2 signal patterns of chromosome 4C could be divided into six groups, consisting of seven (variants 1, 2, 4, 5, 8, 14, and 15), two (variants 7 and 10), two (variants 6 and 9), two (variants 11 and 13), one (variant 3), and one (variant 12) variants. Group I had three pairs of signals on the telomeric regions on both the long and short arms and the central region of the long arms, while group II lost the signal on the telomeric region of the long arms. Group III showed three pairs of signals on the telomeric regions of both the long and short arms and proximal telomeric regions of the long arms. Group IV lost signals on the telomeric region of the long arms in one of the two 4C chromosomes when compared with group I. Group V had two pairs of signals on the telomeric regions of both chromosome arms, while group VI had an additional signal site on one of the two 4C chromosomes. Of the 15 4C variants, 14 variants (except for variant 6) showed (ACT)7 hybridization signals in the middle of the chromosome and extended from the long arm to the short arm. Chromosome 5C had eight variants. Of them, four variants (variants 1–4) shared similar hybridization signal sites between homologous chromosomes. All of the eight variants had a pair of oligo-pSc119.2 signals on the telomeric region of the long arms, except for variants 6 and 7, which lacked such hybridization signals on one of the two 5C. Six of the eight 5C variants (except for variants 1 and 7) had (ACT)7 signals distributed in different positions. Chromosome 6C had six variants, of which two variants (variants 5 and 6) showed heteromorphism in the homologous chromosomes. All of the six variants shared similar oligo-pSc119.2 signals on the telomeric region of the short arms, while an additional pair of oligo-pSc119.2 signals was present on the long arms of two 6C for variants 3 and 4, and one 6C for variants 5 and 6. All of the six variants had (ACT)7 signals spread around the pericentromeric regions. Chromosome 7C had 14 variants. Of them, seven variants (variants 8–14) exhibited heteromorphism in the homologous chromosomes. All of the 14 variants showed oligo-pSc119.2 signals on the telomeric regions of both chromosome arms, except for variants 10 and 14, which lacked a pair of signals on the telomeric regions of the long arms of one 7C for the former and double 7C for the latter. All of the 14 variants showed (ACT)7 signals mainly distributed on the pericentromeric regions of the long arms, except for one 7C in variant 13 that had three pairs of (ACT)7 signals fewer.
The 44 Ae. markgrafii accessions showed 43 different chromosome variants for the seven pairs of chromosomes (Table 1), suggesting that abundant FISH diversity had occurred within this species.
Ae. comosa
The 48 Ae. comosa accessions showed different signal patterns for the seven chromosomes as revealed by probes oligo-pSc119.2, (CTT)12, and oligo-pTa71 (Table 1). Biosystematically, Ae. comosa contains two subspecies, subsp. comosa and subsp. subventricosa. The main results of each subspecies were as follows.
Subspecies comosa
The 13 accessions of subsp. comosa showed 5–10 variants for probes oligo-pSc119.2 and (CTT)12 in every M chromosome and two variants for probe oligo-pTa71 for each of three M chromosomes (1M, 6M, and 7M) (Figure 3A).

Figure 3. Chromosome variants and frequency analysis of Ae. comosa subspecies comosa (A) and subventricosa (B) with FISH probes oligo-pSc119.2, (CTT)12, and oligo-pTa71. Numbering of variants as explained for Figure 2.
Chromosome 1M had seven variants for probes oligo-pSc119.2 and (CTT)12 and two variants for probe oligo-pTa71. The hybridization signals of oligo-pSc119.2 were located on the telomeric region of the long arms, while those of the (CTT)12 were distributed on the entire 1M chromosome (variants 2, 3, 4, and 6) or only on the pericentromeric regions (variants 1, 5, and 7). The 1M variant 2 of probe oligo-pTa71 lacked a pair of intensity signals on the subtelomeric regions of the short arms in one of the two chromosomes when compared with variant 1. Chromosome 2M had six variants for probes oligo-pSc119.2 and (CTT)12. Four variants (variants 1, 2, 4, and 5) lacked the oligo-pSc119.2 signal on both the long and short arms, while the other two variants had a pair of oligo-pSc119.2 signals on the short arm of two 2M (variant 3) and on the long arm of one 2M (variant 6). The hybridization signals of (CTT)12 were mainly distributed on the pericentromeric and telomeric regions of the long arms (such as variant 1) or only on the pericentromeric regions (such as variant 5). Chromosome 3M had seven variants for probes oligo-pSc119.2 and (CTT)12. Four variants (variants 1–4) lacked the oligo-pSc119.2 signal on the entire chromosome, while variants 5 and 6 had a pair of signals on the long and short arms, respectively. Variant 7 showed heteromorphism between two 3M homologous chromosomes for both the oligo-pSc119.2 and (CTT)12 signals. The (CTT)12 signals of chromosome 3M could be divided into three groups according to their signals on the short arm and pericentromeric regions (variants 2 and 3), on the long arm and pericentromeric regions (variants 5, 6, and 7), or on the whole chromosomes (variants 1 and 4). Chromosome 4M had 10 variants for probes oligo-pSc119.2 and (CTT)12, and the telomeric signals of probe oligo-pSc119.2 could be divided into five groups (I–V). All of the variants in groups I (variants 1, 2, 6, and 9) and II (variant 5) had a pair of telomeric signals located on the long and short arms, respectively. Two variants (variants 3 and 7) belonged to group III and lacked the oligo-pSc119.2 signal on both 4M chromosomes, and the variant in group V (type 10) lacked such a signal on one 4M. Two variants (variants 4 and 8) in group IV had a pair of subtelomeric signals on both the long and short arms. The (CTT)12 signals of chromosome 4M could be divided into two groups (I and II). The signals of the seven variants (variants 1, 2, 5, 6, and 8–10) in group I were distributed on the entire chromosome, while those of the three variants in groups II (variants 3, 4, and 7) were on the pericentromeric regions. Chromosome 5M had five variants for probes oligo-pSc119.2 and (CTT)12. Variants 1 and 4 had a pair of telomeric signals for the oligo-pSc119.2 probe on both the long and short arms, while variants 2 and 3 had a pair of oligo-pSc119.2 signals on either the short or long arms. Variant 5 showed heteromorphism between two 5M homologous chromosomes, with one 5M lacking a pair of oligo-pSc119.2 signals. The (CTT)12 signals were distributed on the entire chromosomes (variant 1), on the long arms (variant 2), or on both the long and short arms (variants 3, 4, and 5). Chromosome 6M had eight variants for probes oligo-pSc119.2 and (CTT)12, and the hybridization signals of oligo-pSc119.2 could be divided into three groups (I–III). The variants among the three groups showed difference in a pair of oligo-pSc119.2 signals. Three variants (variants 3, 4, and 5) in group I and four variants (variants 1, 2, 6, and 7) in group II had signals on both chromosome arms and only on the long arms, respectively, while the sole variant (variant 8) in group III lacked a pair of signals on one of the two 6M chromosomes. The (CTT)12 signals were located on the short arm and pericentromeric regions (variants 2, 4, 6, and7), on the long arms and pericentromeric regions (variants 3 and 5), or on the long and short arms (variant 1). Chromosome 6M had two variants for probe oligo-pTa71. Both variants 1 and 2 shared strong oligo-pTa71 signals on the nucleolar organizer regions, whereas variant 2 had an additional pair of oligo-pTa71 signals on the telomeric regions of the short arm on one of the two 6M chromosomes. Chromosome 7M had seven variants for probes oligo-pSc119.2 and (CTT)12 and two variants for probe oligo-pTa71. All of these variants shared two pairs of oligo-pSc119.2 telomeric signals on the long arms, except for variant 2, which had lost a pair of such signals. The (CTT)12 signals were located on the short arm and pericentromeric regions (variants 2, 4, 6, and 7), on the long arms and pericentromeric regions (variants 3 and 5), or on the long and short arms (variant 1). The 7M variant 2 of probe pTa71 differed from variant 1 by the existence of a pair of weak signals on the telomeric regions of both 7M and a pair of strong signals on the subtelomeric region of one of the two 7M chromosomes.
Subspecies subventricosa
The 35 accessions of subspecies subventricosa showed 10–15 variants for probes oligo-pSc119.2 and (CTT)12 in all of the seven M chromosomes, 2–3 variants in three (3M, 5M, and 7M) of seven chromosomes, and only one signal pattern for the remaining four chromosomes for probe oligo-pTa71 (Figure 3B).
Chromosome 1M had 12 variants for probes oligo-pSc119.2 and (CTT)12 and two variants for probe oligo-pTa71. All of the variants shared oligo-pSc119.2 signals on the telomeric regions, but the signals of most variants (10 variants) were located on the long arms, while those of the remaining two variants (variant 6 and one of the two 1M in variant 10) were on the short arms. The (CTT)12 signals of 1M could be divided into four groups (I–IV), comprising seven (variants 2, 4, 6, 7, 8, 9, and 12), one (variant 5), two (variants 3 and 10), and two (variants 1 and 11) variants, respectively. The hybridization signals of groups I, II, III, and IV were distributed on the entire 1M chromosome, on the pericentromeric regions, on both the long and short arms and pericentromeric regions, and on the pericentromeric regions and the short arms, respectively. Both variants 1 and 2 shared pTa71 signals on the telomeric regions of the short arms, but such a signal was lacking in one of the 1M homologous chromosomes of variant 2. Chromosome 2M had 14 variants for probes oligo-pSc119.2 and (CTT)12 and only one signal pattern for probe oligo-pTa71. All of these variants lacked the oligo-pSc119.2 signal, but their hybridization signals of (CTT)12 showed obvious differences, as they were mainly distributed on the pericentromeric regions and the long arms (such as variant 1) or only on the pericentromeric regions (such as variants 6 and 7). Chromosome 3M had 14 variants for probes oligo-pSc119.2 and (CTT)12 and two variants for probe oligo-pTa71. The oligo-pSc119.2 signals of chromosome 3M could be classified into three groups (I–III). Nine variants (variants 1–7, 11, and 12) in group I lacked the oligo-pSc119.2 signal on the entire chromosome. Four variants (variants 8, 9, 10, and 14) in group II had a pair of signals on the long or short arms, and the sole variant (variant 12) in group III had signals on one of the two 3M chromosomes. The (CTT)12 signals of this chromosome could be divided into four groups based on the distribution of the signals on the short arms and pericentromeric regions (variants 7 and 11), on the long arms and pericentromeric regions (variants 2, 3, 5, 8, and 12), on the pericentromeric regions (variants 4,9, 10, 13 and 14), or on the entire chromosomes (variants 1 and 6). Both the two variants of the pTa71 probe shared telomeric signals on the short arms, while variant 2 had two additional pairs of signals on the near-centromeres and long arms. Chromosome 4M had 14 variants for probes oligo-pSc119.2 and (CTT)12 and only one signal pattern for probe oligo-pTa71. The signal pattern of probe oligo-pSc119.2 could be classified into five groups (I–V). Seven variants (variants 1, 3, 4, 5, 8, 12, and 14) in group I and three variants (variants 6, 9, and 10) in group II had a pair of telomeric signals on the long and short arms, respectively. The two variants (variants 2 and 7) in group III lacked the oligo-pSc119.2 signal. The sole variant in group IV (variant 11) had a pair of subtelomeric signals on both the long and short arms, and the only variant (variant 13) in group V did not possess a pair of oligo-pSc119.2 signals on one of the two 4M chromosomes. Moreover, the (CTT)12 signals of chromosome 4M could be divided into three groups (I–III). The signals of the seven variants (variants 1, 2, 4, 5, 10, 12, and 13) in group I were distributed on the entire chromosome, while those of the two variants each in group II (variants 3, 6, 7, 9, 11, and 14) and the sole variant (variant 8) in group III had lacked the (CTT)12 signal on the short and long arms, respectively. Chromosome 5M had 10 variants for probes oligo-pSc119.2 and (CTT)12 and three variants for probe oligo-pTa71. The 10 variants had different (CTT)12 signals that were located on the entire chromosome (variants 1, 5, and 9), on the short arms (variant 4), on the pericentromeric regions and long arms (variant 3, 7, 8, and 10), or on both the long and short arms (variants 2 and 6). Chromosome 5M had three variants for probe oligo-pTa71. Variant 1 had a pair of oligo-pTa71 signals on the telomeric regions of the short arms, and variant 2 had a pair of signals on the middle of the long arms and on the telomeric regions of the short arms. One of the two 5M chromosomes in variant 3 had lost a pair of oligo-pTa71 signals that were located in the middle of the long arms in variant 2. Chromosome 6M had 15 variants for probes oligo-pSc119.2 and (CTT)12 and only one variant for probe oligo-pTa71. The hybridization signals of probe oligo-pSc119.2 could be divided into three groups (I–IV). Seven variants (variants 4, 6–9, 14, and 15) in group I, five variants (types 1–3, 11, and 12) in group II, and the sole variant (variant 5) in group III had a pair of signals located on both the long and short arms, on the long arms, and on the short arms, respectively. Two variants (types 10 and 13) in group IV lacked a pair of signals on one of the two 6M homologous chromosomes. The (CTT)12 signals of these variants were distributed on entire chromosomes (variants 4, 5, 13, and 15), the short arms and pericentromeric regions (variant 7), the long arms and pericentromeric regions (variants 1–3, 9–12, and 14), and both chromosome arms (variants 6 and 8). Chromosome 7M had 10 variants for probes oligo-pSc119.2 and (CTT)12 and three variants for probe oligo-pTa71. These variants had different (CTT)12 signals that were located on the short arm and pericentromeric regions (variants 3, 5 and 7), on the long arms and pericentromeric regions (variants 4 and 8), on the pericentromeric regions (variant 2), or on the entire chromosomes (variants 1, 6, 9, and 10). Chromosome 7M had three variants for probe oligo-pTa71, of which variant 1 lacked such a signal, while variants 2 and 3 shared a pair of telomeric signals on the short arms, and variant 3 had an additional pair of strong oligo-pTa71 signals on the subtelomeric regions in one of the two 7M chromosomes.
Ae. uniaristata
Fluorescence in situ hybridization karyotyping of the six Ae. uniaristata accessions was conducted with probes oligo-pSc119.2, (AAC)5, and oligo-pTa71, but no variant was found (Figures 1, 4). Using PI 554418 as an example (Figures 4A1–A3), the hybridization signals of oligo-pSc119.2 were located on the telomeric regions of chromosome arms 2NS, 3NS, 6NS, and 7NS and chromosomes 1N, 2N, and 4N. Additionally, a pair of oligo-pSc119.2 signals was also present in the middle of chromosome arm 2NL and the subtelomeric region of chromosome arm 4NL. The hybridization signals of probe (AAC)5 existed on five (except for 3N and 5N) of the seven N chromosomes. The signals of chromosome 4N were distributed on the pericentromeric regions, middle of the short arms, and near the pericentromeric regions of the long arms, and those of 1N and 7N, and 6N were located on the pericentromeric regions and the middle of the short arms, respectively.
Figure 4. Comparison of the hybridization patterns of probes oligo-pSc119.2 (A1–C1), (AAC)5 (A2–C2), and oligo-pTa71 (A3–C3) on the metaphase chromosomes of Ae. uniaristata PI 554418 (A1–A3), PI 554419 (formerly Ae. uniaristata, now Ae. comosa,B1–B3), and Ae. comosa PI 551066 (C1–C3). The different hybridization signals of oligo-pSc119.2 between PI 554419 and PI 551066 on 3M and 4M are indicated by green arrowheads.
Comparison of the FISH Pattern of PI 554419 in Ae. uniaristata and Ae. comosa
Previously, PI554419 was classified as Ae. uniaristata1. Many differences were detected between the FISH karyotypes of PI 554419 and Ae. uniaristata PI554418; however, similar FISH karyotypes were found between PI 554419 (Figures 4B1–B3) and Ae. comosa PI 551066 (Figures 4C1–C3) as revealed by probes oligo-pSc119.2, (AAC)5, and oligo-pTa71. Thus, based on the FISH karyotypes, PI 554419 should be treated as Ae. comosa, although a minor difference was detected between PI 554419 and PI 551066. For example, PI 554419 had no weak telomeric signals on chromosome arms 3ML and 4ML in comparison with PI 551066. Furthermore, PI554419 was verified as Ae. comosa by comparison with the herbarium specimens of both species2.
Heteromorphism in the Homologous Chromosomes of Ae. markgrafii and Ae. comosa
Some Ae. markgrafii and Ae. comosa accessions showed heteromorphism in the homologous chromosomes (viz., two 1M or two 1C, etc.).
A total of 24 Ae. markgrafii accessions showed heteromorphism in the homologous chromosomes for probes oligo-pSc119.2 and/or (ACT)7, of which nine, eight, four, two, and one accessions exhibited heteromorphism between one to five, respectively, pairs of homologous chromosomes (Figure 5). Some accessions exhibited heteromorphism between a pair of homologous chromosomes in each of the five C chromosomes (1C, 2C, 4C, 6C, and 7C). For example, PI 254863 and PI 551136 showed heteromorphic oligo-pSc119.2 signals between two 1C homologous chromosomes. Six of the seven C chromosomes (all except for 6C) exhibited seven types of heteromorphism [1C with 2C (PI 551128), 4C (PI 551131), and 7C (PI 551146); 2C with 5C (PI 551134) and 7C (PI 542173 and PI 560732); as well as 5C with 3C (PI 551135) and 4C (PI 203431)] between two pairs of homologous chromosomes. Similarly, six of the seven chromosomes (all except for 5C) participated in the formation of three types of heteromorphism [1C with 2C and 7C (PI 542199), 3C and 4C (PI 551138), and 6C and 7C (PI 298889)] among three pairs of homologous chromosomes. Two accessions showed heteromorphism among four pairs of homologous chromosomes [4C, 5C, and 7C with 1C (PI 551137), and 3C (PI 564195)]. Two types of signal patterns were found in different cell division phases from the same root tips of PI551133 at a ratio of nearly 1:1 (Figure 6), which exhibited inconsistent signals between each of the five pairs of chromosomes 2C, 3C, 4C, 6C, and 7C. Chromosome 2C type 1 lacked a pair of (ACT)7 signals on the short arm in one of the two chromosomes as compared to type 2. On the contrary, chromosome 4C type 2 lacked a pair of (ACT)7 signals located on the long arm in one of the two chromosomes 4C in type 1. Similarly, chromosome 6C type 2 lacked a pair of oligo-pSc119.2 signals and a single (ACT)7 signal located on the long arms in one of the two chromosomes 6C and on the pericentromeric regions of two 6C in type 1. Chromosome 3C (type 2) lacked two pairs of (ACT)7 signals on the pericentromeric regions and telomeric regions of the short arm compared to type 1. Chromosome 7C type 2 lacked a single (ACT)7 signal on the near-centromeric regions in one of the two chromosomes as compared to type 1. PI 551127 exhibited heteromorphism among five pairs (1C, 2C, 4C, 5C, and 7C) of homologous chromosomes (Figure 5).
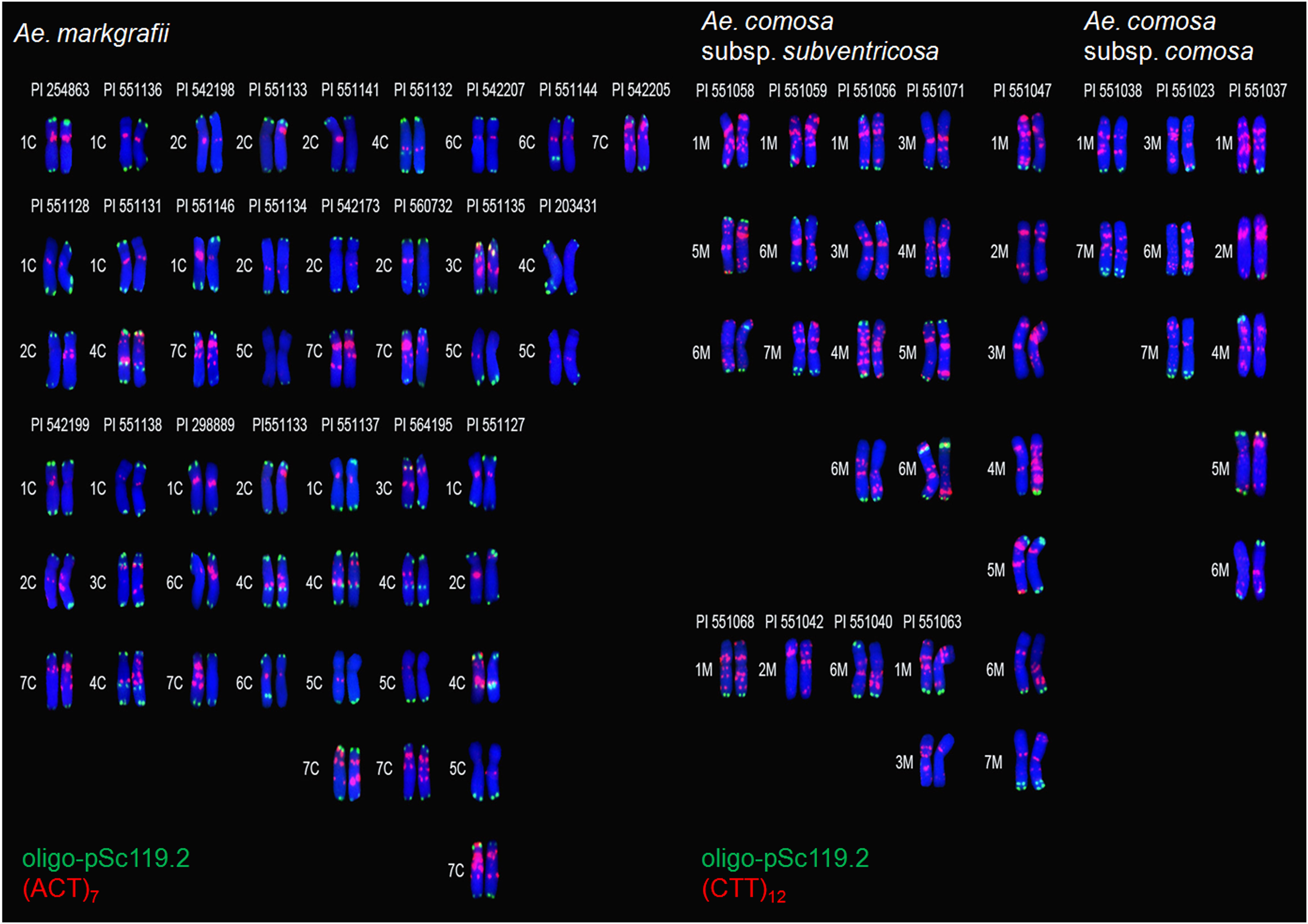
Figure 5. Heteromorphic FISH hybridization signals between the homologous chromosomes of Ae. markgrafii (left) and Ae. comosa (right).
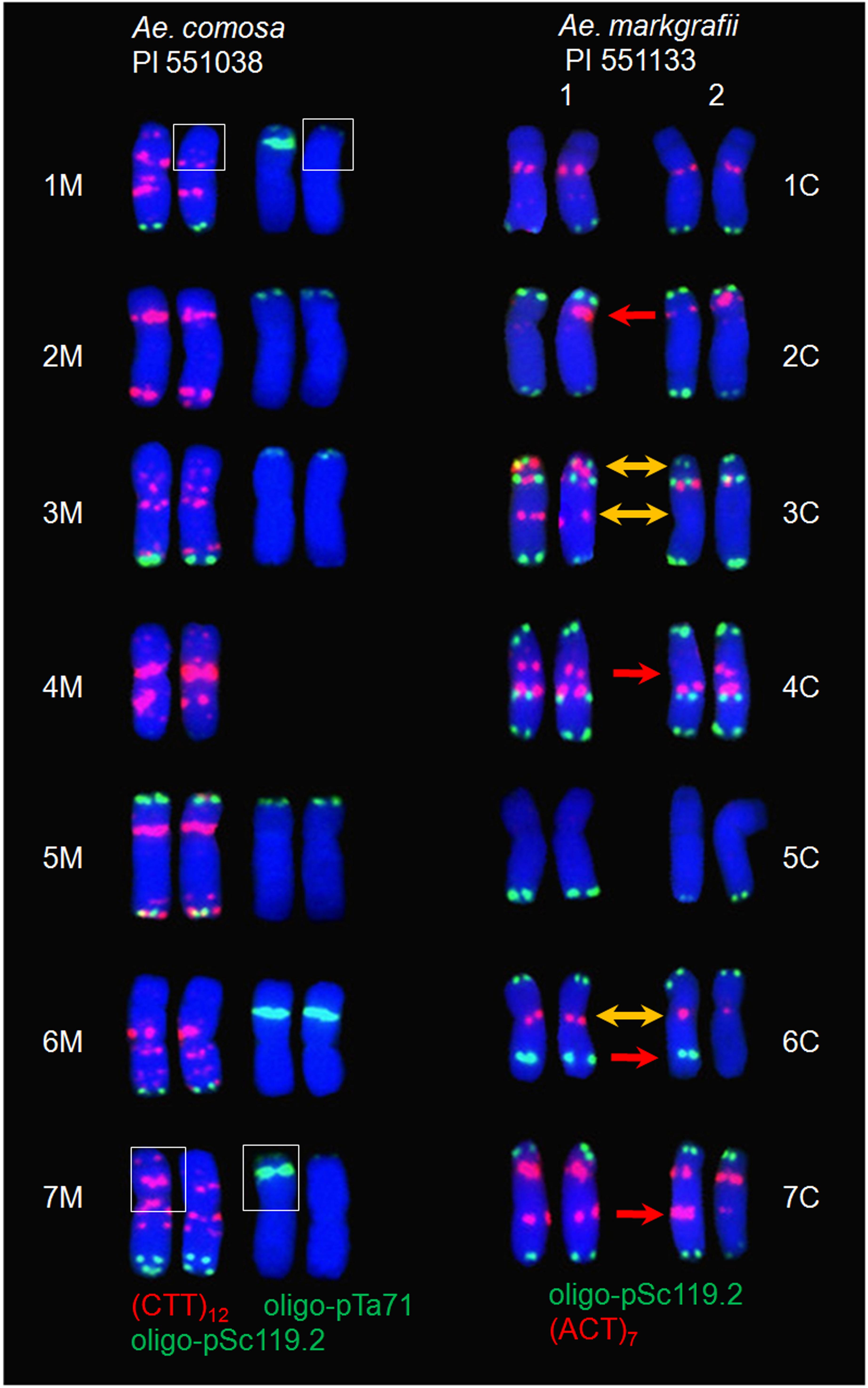
Figure 6. FISH karyotyping of Ae. comosa (PI 551038) and Ae. markgrafii (PI 551133) using DNA probes. The translocation between the chromosome arms 1MS and 7MS of Ae. comosa PI 551038 is shown by white boxes. Two types of reproducible FISH signal patterns were observed within the same metaphase cells of Ae. markgrafii PI551133 at a ratio of 1:1. Signal differences of the same and different types between homologous chromosomes are indicated by red and yellow arrowheads, respectively.
A total of 12 Ae. comosa accessions (three accessions of subsp. comosa and nine of subsp. subventricosa) showed heteromorphic oligo-pSc119.2 and/or (CCT)12 signals between homologous chromosomes, of which three, two, three, two, one, and one accessions showed heteromorphism between one, two, three, four, five and seven, respectively, pairs of homologous chromosomes, respectively (Figure 5). Three M chromosomes (1M, 2M, and 6M) exhibited heteromorphism between a pair of homologous chromosomes. For example, PI 551068, PI 551142, and PI 551140 (all belonged to subsp. subventricosa) showed heteromorphism between two 1M, 2M, and 6M, respectively. PI 551038 (subsp. comosa) also showed heteromorphism between two pairs of 1M and 7M in both the (CTT)12 and oligo-pTa71 signals. Further analysis suggested that a translocation had occurred on the short arms between one of the two chromosome arms 1MS and 7MS (Figure 6). Three chromosomes (1M, 3M, and 7M) were involved in two types of heteromorphism [1M with 3M (PI 551063 subsp. subventricosa) and 7M (PI 551038 subsp. comosa)] between two pairs of homologous chromosomes. Five chromosomes (1M, 3M, 5M, 6M, and 7M) exhibited three types of heteromorphism [6M with 1M and 5M (PI 551058), 1M and 7M (PI 551059) in subsp. subventricosa, and 3M and 7M (PI 551023) in subsp. comosa] among the three pairs of homologous chromosomes. Two accessions of subsp. subventricosa showed heteromorphism among the four pairs of homologous chromosomes [3M, 4M, and 6M with 1M (PI 551056), and 5M (PI 551071)]. One accession (PI 551037 subsp. comosa) exhibited heteromorphism among five pairs (1M, 2M, 4M, 5M, and 6M) of homologous chromosomes. Not expectedly, PI 551047 (subsp. subventricosa) showed heteromorphism for all of the seven pairs of homologous chromosomes.
Discussion
Tandem repeats were previously considered to be junk DNA generated during evolution that lack any biological function (Doolittle and Sapienza, 1980). Now, it is generally accepted that tandem repeats play a pivotal role in chromosome organization, stabilization, and recombination as well as DNA replication (Heslop-Harrison, 2000; Li et al., 2002; Gemayel et al., 2012). Variation in the number and distribution of tandem repeat sequences may be involved in speciation (Flavell et al., 1979; Li et al., 2004; Britten, 2010) and were shown to accelerate the evolution of coding and regulatory sequences (Gemayel et al., 2010). Thus, the diversity of tandem repeat sequences can be used to assess genetic relatedness from the species level to the genome level (Dvorak and Zhang, 1992; Mehrotra and Goyal, 2014).
Genetic Diversity Among Four Diploid Aegilops Species With U, C, M, and N Chromosomes
Fluorescence in situ hybridization karyotyping is a valid tool for chromosome authentication, species classification, and evolutionary studies (Badaeva et al., 2002, 2004). It is also reliable for identifying alien chromosome/fragment introgression in wheat-wild distant hybridization crosses when combined with genome in situ hybridization (Wang et al., 2016). A large number of polymorphic FISH karyotypes were detected among and within Ae. umbellulata, Ae. markgrafii, and Ae. comosa, whereas identical karyotypes were observed among Ae. uniaristata accessions due to the limited accessions that were used (Figures 2, 3).
Similar oligo-pSc119.2 signals were mainly presented on the telomeric regions of the long and short arms of the four Aegilops species with C, M, N, and U genomes, while different signals were detected either among species or among seven homologous (Figures 1–3). Polymorphic FISH signals of probe oligo-pSc119.2 were present on 2D, 3D, and 4D of Ae. tauschii (Zhao et al., 2018); 2U, 4U, 5U, and 6U of Ae. umbellulata; and 4M and 6M of Ae. comosa (Schneider et al., 2005). Additionally, polymorphic oligo-pSc 119.2/(AAC)5 signals were also discovered for Ae. umbellulata chromosomes 1U, 6U, and 7U (Song et al., 2019). For example, compared with previous studies, extra oligo-pSc119.2 signals were discovered on the telomeric regions of 7UL in Ae. umbellulata (Schneider et al., 2005), on the telomeric regions of 2CL and 6CL in Ae. markgrafii (Danilova et al., 2017), on the proximal telomeric and subtelomeric regions of 1ML, 3ML, and 3MS in Ae. comosa, as well as on the near-centromeres and near central position of 2N in Ae. uniaristata (Badaeva et al., 1996a; Schneider et al., 2005; Kwiatek et al., 2013). Furthermore, the oligo-pSc119.2 signal was absent on one of the chromosomes arms 5CL of Ae. markgrafii, 6MS of Ae. comosa, and 6NL of Ae. uniaristata (Badaeva et al., 1996a).
The 45S rDNA, a tandem repeat sequence that is located on the nucleolar organizer region of satellite and some non-satellite chromosomes with only a few copy numbers, is represented by the occurrence of the Oligo-pTa71 signals (Long and Dawid, 1980; Mukai et al., 1991; Tang et al., 2014). The pTa71 signals in the present study were mainly located on the telomeric regions of homologous 1 and 5 on the short arms, which is consistent with previous studies (Yamamoto, 1992; Badaeva et al., 1996b; Mirzaghaderi et al., 2014; Song et al., 2019). Additional weak pTa71 signals were present on the telomeric regions of chromosome arm 1NL, the pericentromeric regions of 2N and the telomeric regions of chromosome arm 2NS, and the pericentromeric regions and telomeric regions of chromosome 3N in Ae. uniaristata. Extra strong pTa71 signals were observed on the telomeric regions of chromosome arms 2MS, 3MS, 6MS, and 7MS and on the pericentromeric regions of chromosome 5M in Ae. comosa.
In addition to polymorphic signals revealed by the oligo-pSc119.2 and pTa71 probes, the microsatellite probes (AAC)5, (CTT)12, and (ACT)7 exhibited more hybridization sites on nearly all the chromosomes, and their signals were distributed mainly on the pericentromeric regions and extended to the whole chromosomes according to the probe used. Usually, the (CTT)n-based FISH patterns of some Aegilops species are very similar to their C-banding patterns (Ruban and Badaeva, 2018). The (CTT)12 signals of four diploid Aegilops species are consistent with their C-banding patterns (Figure 1; Friebe et al., 1992, 1995, 1996b), although minor differences occurred in some chromosomes including 1U, 5U, 3C, 6C, 2M, and 6M as well as other chromosomes. For example, the (CTT)12 signal of Ae.comosa showed extensive diversity both between two subspecies and among the seven chromosomes (Figure 3). The present results revealed abundant FISH variants both among and within species.
Intra- and Interspecific Genetic Variations in FISH Patterns and Their Implication in Evolution and Speciation
Intra- and interspecific genetic diversity was detected in the FISH patterns. Slightly higher FISH polymorphisms of oligo-pSc119.2 signals were detected in Ae. comosa than Ae. markgrafii, and both were higher than Ae. umbellulata. Our FISH results are very similar to previous reports where Ae. comosa showed higher genetic diversity than Ae. umbellulata (Resta et al., 1996; Monte et al., 2001; Schneider et al., 2005), where intra- and interspecific genetic diversity was evaluated by using restriction fragment length polymorphism (RFLP), amplified fragment length polymorphism (AFLP), and FISH karyotypes. Similarly, a sequence-tagged site based on molecular markers suggested that the U genome of Ae. umbellulata showed less genetic polymorphism than the M genome of Ae. comosa and C genome of Ae. markgrafii (Chee et al., 1995). On the contrary, a low level of intraspecific variation was discovered with AFLP markers among seven diploid Aegilops species, including Ae. umbellulata and Ae. markgrafii, except for the cross-pollinating Ae. speltoides and Ae. mutica (Sasanuma et al., 2004).
Intraspecific genetic diversity was also found among the diploid Aegilops species with U, C, and M genomes as revealed by oligo-pSc119.2 in combination with a microsatellite probe (AAC)5, (ACT)7, and(CTT)12, respectively. More FISH signal patterns were present in each homologous chromosome of Ae. comosa (5–10 and 10–15 variants for subsp. comosa and subsp. subventricosa, respectively) and Ae. markgrafii (5–18 variants) than those of Ae. umbellulata (two to six variants), and Ae. uniaristata (one variant). The Ae. umbellulata 7U had greater genetic diversity, while 1U had few patterns among the seven U chromosomes (Figure 2). The number of polymorphic FISH patterns among the seven C chromosomes of Ae. markgrafii was ranked as 2C (18 variants) > 4C (15) > 7C (14) > 3C = 5C (8) > 6C (6) > 1C (5) (Figure 2). Meanwhile, the FISH patterns of Ae. comosa subsp. comosa in 13 accessions were richer than that of subsp. subventricosa in 35 accessions and were ranked as 4M (10 variants) > 6M (8) > 1M, 3M and 7M (7) > 2M (6) > 5M (5) and 6M (15 variants) > 2M, 3M and 4M (14) > 1M (12) > 5M and 7M (10) for each subspecies (Figure 3). Intraspecific variation between two subspecies of Ae. comosa was also discovered with the C-banding karyotype, and an obvious difference occurred mainly on the pericentromeric and nucleolar organizing regions (Teoh et al., 1983; Friebe et al., 1996a). Several polymorphic variations for C-banding size and position are present among Ae. markgrafii (Friebe et al., 1992, Ae. umbellulata, Ae. uniaristata, and Ae. comosa accessions (Friebe et al., 1995, 1996b). Considerable genetic diversity among Ae. umbellulata accessions has also been revealed by RNA sequencing analysis (Okada et al., 2018).
Alterations in the number and distribution of tandem repeats are one of the most important manifestations of genetic variation (Gemayel et al., 2010). Currently, abundant intra- and interspecific genetic variations in tandem repeats among four diploid Aegilops species were evaluated by the heterochromatin limited repetitive DNA probe pSc119.2, a tandem repeat sequence 45S rDNA-related DNA probe pTa71, and microsatellite sequence probes (AAC)5, (ACT)7, and (CTT)12. Previous studies have shown tandem repeat variations are involved in speciation and evolution as well as in phenotypic variation (Nagaki et al., 1998; Gemayel et al., 2010). For example, the copy numbers of tandem repeat Afa-family sequences per genome among Triticeae species are highly variable, suggesting that the amplification or deletion of such sequences is related to the evolution and speciation of Triticeae. In hexaploid wheat, the Afa-family sequences between the A and B genomes did not evolve in a concerted manner, and these sequences were amplified all over the chromosomes of the D-genome in a short period (Nagaki et al., 1998). The intraspecific variability of Aegilops speltoides-specific Spelt1 and Aegilops-Triticum-specific Spelt 52 tandem repeats in tetraploid and hexaploid wheat decreased sharply when compared with that of Ae. speltoides, with the exception of Triticum timopheevii Zhuk and T. carthlicum Nevski; both species maintain the amounts of Spelt1 unaltered because they are exceptional in being endemic species with restricted geographical distributions (Pestsova et al., 1998; Salina et al., 2006; Zoshchuk et al., 2009).
Moreover, the number and distribution of the tandem repeat sequences Spelt1 and Spelt 52 in Ae. speltoides exhibited a distinctive geographical gradient, with the number of Spelt 1 in the central population of the species distribution being 12–14 times higher than in marginal populations. The changes in the number of these tandem repeats along an eco-geographical gradient may be ascribed to the depletion of tandem repeats in the marginal populations as a consequence of increased recombination rate under stressful conditions or the accumulation of tandem repeats in conducive climatic/edaphic environments in the central populations (Raskina et al., 2011).
Heteromorphism in Homologous Chromosomes
Heteromorphism in homologous chromosomes has been identified in humans and plants using different cytogenetic methods (Suciu, 1986; Lapitan et al., 1988) and has also been detected in Ae. comosa and Ae. markgrafii as revealed by the FISH probes oligo-pSc119.2 and oligo-pTa71 plus a microsatellite probe (CTT)12 or (ACT)7 (Figures 2, 3, 5). All of the seven chromosomes of Ae. comosa and Ae. markgrafii displayed heteromorphism among one to seven or one to five pairs of homologous chromosomes in each species (Figures 5, 6). Heteromorphism was also discovered in four accessions each of Ae. speltoides as revealed by FISH probes oligo-pSc119.2, between four pairs of homologous chromosomes (1S, 4S, 5S, and 6S) and pAesp_SAT86, and between all seven pairs of homologous chromosomes (Dong et al., 2017; Ruban and Badaeva, 2018). In addition to the heteromorphic FISH signals between homologous pairs, heteromorphic C-banding patterns between two homologous chromosomes of B and D were also reported in one Ae. markgrafii accession (Friebe et al., 1992) and also occurred between the homologous chromosomes of 1R (Alkhimova et al., 1999), and 3R, 4R, 6R, and 7R (Lapitan et al., 1988) of rye (Secale cereale).
Heteromorphism in homologous chromosomes is one manifestation of genetic variation. These genetic variations may have resulted from alterations in chromosome structures, such as chromosome rearrangements, translocations, and inversion between chromosomes within these accessions (Friebe et al., 1992, 1995, 1996b; Badaeva et al., 2004; Schneider et al., 2005). Moreover, the frequent occurrence of heteromorphic chromosomes could be an indicator of open pollination. In Triticeae, Ae. speltoides and rye (Secale cereale) are typical outcrossers with heteromorphism in homologous chromosomes (Lapitan et al., 1988; Ruban and Badaeva, 2018), while Ae. markgrafii is a facultative cross-pollinating plant with a highly asymmetrical karyotype that is indicative of chromosome rearrangements (Kilian et al., 2011; Danilova et al., 2017). Different from Ae. markgrafii, Ae. comosa is a self-pollinating plant (Friebe et al., 1996a). We speculate that the high level of heteromorphic FISH patterns observed in Ae. markgrafii and Ae. comosa is the consequence of hybrid karyotypes, which may be caused by outcrossing within genotypes and/or by chromosomal rearrangements (Tang et al., 2011; Danilova et al., 2017).
In addition to the heteromorphism in homologous chromosomes of FISH karyotypes within Ae. markgrafii and Ae. comosa (Figure 5), a translocation between the 1MS and 7MS of Ae. comosa PI 551038 and two types of FISH signals at a ratio of 1:1 within the same root cells of Ae. markgrafii PI 551133 were another form of genetic variation (Figure 6). A translocation on the 4S of Ae. speltoides may be ascribed to a center inversion, which was detected by the probes (CTT)10 and pAesp_SAT86 (Ruban and Badaeva, 2018). Although so many genetic variations were identified, only a few translocations were identified, which may be because the FISH probes were too small. These variations may have originated from chromosomal rearrangements during the evolutionary process, and this recombination affects the synteny between the homologous chromosomes of Aegilops and Triticum (Devos et al., 1993; Zhang et al., 1998).
Conclusion
Chromosome structural variations of four diploid Aegilops species were evaluated by FISH karyotypes. Extensive intra- and interspecific genetic variation was found in Ae. umbellulata, Ae. markgrafii, and both subspecies of Ae. comosa, but not in Ae. uniaristata. In both subspecies of Ae. comosa and in Ae. markgrafii, frequently occurring heteromorphism in homologous chromosomes constituted an additional component of chromosomal variation. These results will provide important clues for understanding chromosome organization and evolutionary relationships as well as speciation among Aegilops species.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
ZS conducted the experiments and drafted the manuscript. TB, YZ, and QX analyzed the data. JL and GL prepared the plant materials. SD and ZY conceived and designed the study. ZY revised the manuscript.
Funding
The authors would like to acknowledge grants from the Sichuan Science and Technology Program (Nos. 2018HH0130 and 2018HH0113), the Natural National Science Foundation of China (31771783 and U1403185), and the Ministry of Science and Technology of China (2016YFD0100502 and 2017YFD0100903).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Alkhimova, A. G., Heslop-Harrison, J. S., Shchapova, A. I., and Vershinin, A. V. (1999). Rye chromosome variability in wheat–rye addition and substitution lines. Chromosome Res. 7, 205–212. doi: 10.1023/a:1009299300018
Badaeva, E. D., Amosova, A. V., Goncharov, N. P., Macas, J., Ruban, A. S., Grechishnikova, I. V., et al. (2015). A set of cytogenetic markers allows the precise identification of all A-genome chromosomes in diploid and polyploid wheat. Cytogenet. Genome Res. 146, 71–79. doi: 10.1159/000433458
Badaeva, E. D., Amosova, A. V., Muravenko, O. V., Samatadze, T. E., Chikida, N. N., Zelenin, A. V., et al. (2002). Genome differentiation in Aegilops. 3. Evolution of the D-genome cluster. Plant Syst. Evol. 231, 163–190. doi: 10.1007/s006060200018
Badaeva, E. D., Amosova, A. V., Samatadze, T. E., Zoshchuk, S. A., Shostak, N. G., Chikida, N. N., et al. (2004). Genome differentiation in Aegilops. 4. Evolution of the U-genome cluster. Plant Syst. Evol. 246, 45–76. doi: 10.1128/AEM.02052-08
Badaeva, E. D., Dedkova, O. S., Zoshchuk, S. A., Amosova, A. V., Reader, S. M., Bernard, M., et al. (2011). Comparative analysis of the N-genome in diploid and polyploid Aegilops species. Chromosome Res. 19, 541–548. doi: 10.1007/s10577-011-9211-x
Badaeva, E. D., Friebe, B., and Bikram, S. G. (1996a). Genome differentiation in Aegilops. 1. Distribution of highly repetitive DNA sequences on chromosomes of diploid species. Genome 39, 293–306. doi: 10.1139/g96-040
Badaeva, E. D., Friebe, B., and Gill, B. S. (1996b). Genome differentiation in Aegilops. 2. Physical mapping of 5S and 18S-26S ribosomal RNA gene families in diploid species. Genome 39, 1150–1158. doi: 10.1139/g96-145
Bansal, M., Kaur, S., Dhaliwal, H. S., Bains, N. S., Bariana, H. S., Chhuneja, P., et al. (2017). Mapping of Aegilops umbellulata-derived leaf rust and stripe rust resistance loci in wheat. Plant Pathol. 66, 38–44. doi: 10.1111/ppa.12549
Britten, R. J. (2010). Transposable element insertions have strongly affected human evolution. Proc. Natl. Acad. Sci. U.S.A. 107, 19945–19948. doi: 10.1073/pnas.1014330107
Cakmak, I., Tolay, I., Özkan, H., Özdemir, A., and Braun, H. J. (1999). Variation in zinc efficiency among and within Aegilops species. J. Plant Nutr. Soil Sci. 162, 257–262.
Chee, P. W., Lavin, M., and Talbert, L. E. (1995). Molecular analysis of evolutionary patterns of U genome wild wheats. Genome 38, 290–297. doi: 10.1139/g95-036
Cuadrado, Á., and Jouve, N. (2010). Chromosomal detection of simple sequence repeats (SSRs) using nondenaturing FISH (ND-FISH). Chromosoma 119, 495–503. doi: 10.1007/s00412-010-0273-x
Dai, S. F., Zhao, L., Xue, X. F., Jia, Y. N., Liu, D. C., Pu, Z. J., et al. (2015). Analysis of high-molecular-weight glutenin subunits in five amphidiploids and their parental diploid species Aegilops umbellulata and Aegilops uniaristata. Plant Genet. Resour. C 13, 186–189. doi: 10.1017/S1479262114000719
Danilova, T. V., Akhunova, A. R., Akhunov, E. D., Friebe, B., and Gill, B. S. (2017). Major structural genomic alterations can be associated with hybrid speciation in Aegilops markgrafii (Triticeae). Plant J. 92, 317–330. doi: 10.1111/tpj.13657
Devos, K. M., Atkinson, M. D., Chinoy, C. N., Francis, H. A., Harcourt, R. L., Koebner, R. M. D., et al. (1993). Chromosomal rearrangements in the rye genome relative to that of wheat. Theor. Appl. Genet. 85, 673–680. doi: 10.1007/bf00225004
Dong, L., Dong, Q., Zheng, W. L., Hu, X. L., Wang, H. G., and Wang, Y. H. (2017). Karyotypic analysis of Aegilops speltoides revealed by FISH. Sci. Agric. Sin. 50, 1378–1387. doi: 10.3864/j.issn.0578-1752
Doolittle, W. F., and Sapienza, C. (1980). Selfish genes, the phenotype paradigm and genome evolution. Nature 284, 601–603. doi: 10.1038/284601a0
Dvorak, J., and Zhang, H. B. (1992). Reconstruction of the phylogeny of the genus Triticum from variation in repeated nucleotide sequences. Theor. Appl. Genet. 84, 419–429. doi: 10.1007/bf00229502
Flavell, R., O’Dell, M., and Smith, D. (1979). Repeated sequence DNA comparisons between Triticum and Aegilops species. Heredity 42, 309–322. doi: 10.1038/hdy.1979.34
Friebe, B., Badaeva, E. D., Kammer, K., and Gill, B. S. (1996a). Standard karyotypes of Aegilops uniaristata, Ae. mutica, Ae. comosa subspecies comosa and heldreichii (Poaceae). Plant Syst. Evol. 202, 199–210. doi: 10.1007/bf00983382
Friebe, B., Jiang, J., Raupp, W. J., McIntosh, R. A., and Gill, B. S. (1996b). Characterization of wheat alien translocations conferring resistance to diseases and pests, current status. Euphytica 71, 59–87. doi: 10.1007/bf00035277
Friebe, B., Jiang, J., Tuleen, N., and Gill, B. S. (1995). Standard karyotype of Triticum umbellulatum and the characterization of derived chromosome addition and translocation lines in common wheat. Theor. Appl. Genet. 90, 150–156. doi: 10.1007/bf00221010
Friebe, B., Schubert, V., Blüttner, W. D., and Hammer, K. (1992). C-banding pattern and polymorphism of Aegilops caudata and chromosomal constitutions of the amphiploid T. aestivum–Ae. caudata and six derived chromosome addition lines. Theor. Appl. Genet. 83, 589–596. doi: 10.1007/BF00226902
Gemayel, R., Cho, J., Boeynaems, S., and Verstrepen, K. J. (2012). Beyond junk-variable tandem repeats as facilitators of rapid evolution of regulatory and coding sequences. Genes 3, 461–480. doi: 10.3390/genes3030461
Gemayel, R., Vinces, M. D., Legendre, M., and Verstrepen, K. J. (2010). Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu. Rev. Genet. 44, 445–477. doi: 10.1146/annurev-genet-072610-155046
Gill, B. S., Sharma, H. C., and Raupp, W. J. (1985). Evaluation of Aegilops species for resistance to wheat powdery mildew, wheat leaf rust, hessian fly, and greenbug. Plant Dis. 69, 314–316. doi: 10.1094/PD-69-312
Gong, W. P., Han, R., Li, H. S., Song, J. M., Yan, H. F., Li, G. Y., et al. (2017). Agronomic traits and molecular marker identification of wheat-Aegilops caudata addition lines. Front. Plant Sci. 8:1743. doi: 10.3389/fpls.2017.01743
Gorham, J. (1990). Salt tolerance in the Triticeae: K+/Na+ discrimination in Aegilops species. J. Exp. Bot. 41, 615–621. doi: 10.1093/jxb/41.5.615
Hao, M., Luo, J. T., Zhang, L. Q., Yuan, Z. W., Yang, Y. W., Wu, M., et al. (2013). Production of hexaploid triticale by a synthetic hexaploid wheat-rye hybrid method. Euphytica 193, 347–357. doi: 10.1007/s10681-013-0930-2
Heslop-Harrison, J. S. (2000). Comparative genome organization in plants: from sequence and markers to chromatin and chromosomes. Plant Cell 12, 617–636. doi: 10.1016/j.cattod.2009.07.027
Iqbal, N., Reader, S. M., Caligari, P. D. S., and Miller, T. E. (2000). Characterization of Aegilops uniaristata chromosomes by comparative DNA marker analysis and repetitive DNA sequence in situ hybridization. Theor. Appl. Genet. 101, 1173–1179. doi: 10.1007/s001220051594
Kato, A. (1999). Air drying method using nitrous oxide for chromosome counting in maize. Biotech. Histochem. 74, 160–166. doi: 10.3109/10520299909047968
Kawahara, T. (2002). Morphological and isozyme variation in genebank accessions of Aegilops umbellulata Zhuk, a wild relative of wheat. Genet. Resour. Crop Evol. 49, 89–94.
Kilian, B., Mammen, K., Millet, E., Sharma, R., Graner, A., Salamini, F., et al. (2011). “Aegilops,” in Wild Crop Relatives: Genomic and Breeding Resources: Cereals, ed. C. Kole, (New York, NY: Springer), 1–76. doi: 10.1007/978-3-642-14228-4_1
Kimber, G., and Tsunewaki, K. (1988). “Genome symbols and plasma types in the wheat group,” in Proceedings of the 7th International Wheat Genetics Symposium, eds T. E. Miller and R. M. D. Koebner, (Cambridge: Institute of Plant Science Research), 1209–1211.
Komuro, S., Endo, R., Shikata, K., and Kato, A. (2013). Genomic and chromosomal distribution patterns of various repeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome 56, 131–137. doi: 10.1139/gen-2013-0003
Kwiatek, M., Wiśniewska, H., and Apolinarska, B. (2013). Cytogenetic analysis of Aegilops chromosomes, potentially usable in triticale (Triticosecale witt.) breeding. J. Appl. Genet. 54, 147–155. doi: 10.1007/s13353-013-0133-5
Lapitan, N. L. V., Sears, R. G., and Gill, B. S. (1988). Amplification of repeated DNA sequences in wheat x rye hybrids regenerated from tissue culture. Theor. Appl. Genet. 75, 381–388. doi: 10.1007/bf00276739
Li, W., Zhang, P., Fellers, J. P., Friebe, B., and Gill, B. S. (2004). Sequence composition, organization, and evolution of the core Triticeae genome. Plant J. 40, 500–511. doi: 10.1111/j.1365-313x.2004.02228.x
Li, Y. C., Korol, A. B., Fahima, T., Beiles, A., and Nevo, E. (2002). Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol. Ecol. 11, 2453–2465. doi: 10.1046/j.1365-294X.2002.01643.x
Liu, C., Gong, W. P., Han, R., Guo, J., Li, G. R., Li, H. S., et al. (2019). Characterization, identification and evaluation of a set of wheat-Aegilops comosa chromosome lines. Sci. Rep. 9:4773. doi: 10.1038/s41598-019-41219-9
Long, E. O., and Dawid, I. B. (1980). Repeated genes in eukaryotes. Annu. Rev. Biochem. 49, 727–764. doi: 10.1146/annurev.bi.49.070180.003455
Mehrotra, S., and Goyal, V. (2014). Repetitive sequences in plant nuclear DNA: types, distribution, evolution and function. Genomics Proteomics Bioinformatics 12, 164–171. doi: 10.1016/j.gpb.2014.07.003
Miller, T. E., Reader, S. M., Mahmood, A., Purdie, K. A., and King, I. P. (1995). “Chromosome 3N of Aegilops uniaristata-a source of tolerance to high levels of aluminum for wheat,” in Proceedings of the 8th International Wheat Genetics Symposium, eds Z. S. Li, and Z. Y. Xin, (Beijing: China Agricultural Scientech Press), 1037–1042.
Mirzaghaderi, G., Houben, A., and Badaeva, E. D. (2014). Molecular-cytogenetic analysis of Aegilops triuncialis and identification of its chromosomes in the background of wheat. Mol. Cytogenet. 7:91. doi: 10.1186/s13039-014-0091-6
Molnár, I., Kubaláková, M., Šimková, H., Cseh, A., Molnár-Láng, M., and Doležel, J. (2011). Chromosome isolation by flow sorting in Aegilops umbellulata and Ae. comosa and their allotetraploid hybrids Ae. biuncialis and Ae. geniculata. PLoS One 6:e27708. doi: 10.1371/journal.pone.0027708
Molnár, I., Vrána, J., Burešová, V., Cápal, P., Farkas, A., Darkó, É., et al. (2016). Dissecting the U, M, S and C genomes of wild relatives of bread wheat (Aegilops spp.) into chromosomes and exploring their synteny with wheat. Plant J. 88, 452–467. doi: 10.1111/tpj.13266
Molnár, I., Vrána, J., Farkas, A., Kubaláková, M., Cseh, A., Molnár-Láng, M., et al. (2015). Flow sorting of C-genome chromosomes from wild relatives of wheat Aegilops markgrafii, Ae. triuncialis and Ae. cylindrica, and their molecular organization. Ann. Bot. 116, 189–200. doi: 10.1093/aob/mcv073
Monte, J. V., De Nova, P. J. G., and Soler, C. (2001). AFLP-based analysis to study genetic variability and relationships in the Spanish species of the genus Aegilops. Hereditas 135, 233–238. doi: 10.1111/j.1601-5223.2001.00233.x
Mukai, Y., Endo, T. R., and Gill, B. S. (1991). Physical mapping of the 18S, 26S rRNA multigene family in common wheat: identification of a new locus. Chromosoma 100, 71–78. doi: 10.1007/BF00418239
Nagaki, K., Tsujimoto, H., and Sasakuma, T. (1998). Dynamics of tandem repetitive Afa-family sequences in Triticeae, wheat-related species. J. Mol. Evol. 47, 183–189. doi: 10.1007/pl00006375
Neelam, K., Rawat, N., Tiwari, V. K., Prasad, R., Tripathi, S. K., Randhawa, G. S., et al. (2012). Evaluation and identification of wheat-Aegilops addition lines controlling high grain iron and zinc concentration and mugineic acid production. Cereal Res. Commun. 40, 53–61. doi: 10.1556/CRC.40.2012.1.7
Okada, M., Yoshida, K., Nishijima, R., Michikawa, A., Motoi, Y., Sato, K., et al. (2018). RNA-seq analysis reveals considerable genetic diversity and provides genetic markers saturating all chromosomes in the diploid wild wheat relative Aegilops umbellulata. BMC Plant Biol. 18:271. doi: 10.1186/s12870-018-1498-8
Pestsova, E. G., Goncharov, N. P., and Salina, E. (1998). Elimination of a tandem repeat of telomeric heterochromatin during the evolution of wheat. Theor. Appl. Genet. 97, 1380–1386. doi: 10.1007/s001220051032
Raskina, O., Brodsky, L., and Belyayev, A. (2011). Tandem repeats on an eco-geographical scale: outcomes from the genome of Aegilops speltoides. Chromosome Res. 19, 607–623. doi: 10.1007/s10577-011-9220-9
Resta, P., Zhang, H. B., Dubcovsky, J., and Dvorak, J. (1996). The origins of the genomes of Triticum biunciale, T. ovatum, T. neglectum, T. columnare and T. rectum (Poaceae) based on variation in repeated nucleotide sequences. Am. J. Bot. 83, 1556–1565. doi: 10.1002/j.1537-2197.1996.tb12813.x
Riar, A. K., Kaur, S., Dhaliwal, H. S., Singh, K., and Chhuneja, P. (2012). Introgression of a leaf rust resistance gene from Aegilops caudata to bread wheat. J. Genet. 91, 155–161. doi: 10.1007/s12041-012-0161-7
Riley, R., Chapman, V., and Johnson, R. (1968). The incorporation of alien disease resistance in wheat by genetic interference with the regulation of meiotic chromosome synapsis. Genet. Res. 12, 199–219. doi: 10.1017/S0016672300011800
Rodríguez-Quijano, M., Nieto-Taladriz, M. T., and Carrillo, J. M. (2001). Polymorphism of high molecular weight glutenin subunits in three species of Aegilops. Genet. Resour. Crop Evol. 48, 599–607. doi: 10.1023/A:1013868629640
Ruban, A. S., and Badaeva, E. D. (2018). Evolution of the S-genomes in Triticum-Aegilops alliance, evidences from chromosome analysis. Front. Plant Sci. 9:1756. doi: 10.3389/fpls.2018.01756
Salina, E. A., Lim, K. Y., Badaeva, E. D., Shcherban, A. B., Adonina, I. G., Amosova, A. V., et al. (2006). Phylogenetic reconstruction of Aegilops section Sitopsis and the evolution of tandem repeats in the diploids and derived wheat polyploids. Genome 49, 1023–1035. doi: 10.1139/g06-050
Sasanuma, T., Chabane, K., Endo, T. R., and Valkoun, J. (2004). Characterization of genetic variation in and phylogenetic relationships among diploid Aegilops species by AFLP: incongruity of chloroplast and nuclear data. Theor. Appl. Genet. 108, 612–618. doi: 10.1007/s00122-003-1485-8
Schneider, A., Linc, G., Molnár, I., and Molnár-Láng, M. (2005). Molecular cytogenetic characterization of Aegilops biuncialis and its use for the identification of 5 derived wheat-Aegilops biuncialis disomic addition lines. Genome 48, 1070–1082. doi: 10.1139/g05-062
Schneider, A., Molnár, I., and Molnár-Láng, M. (2008). Utilisation of Aegilops (goatgrass) species to widen the genetic diversity of cultivated wheat. Euphytica 163, 1–19. doi: 10.1007/s10681-007-9624-y
Sears, E. R. (1956). The transfer of leaf rust resistance from Aegilops umbellulata to wheat. Brookhaven Symp. Biol. 9, 1–22.
Song, Z. P., Dai, S. F., Jia, Y. N., Zhao, L., Kang, L. Z., Liu, D. C., et al. (2019). Development and characterization of Triticum turgidum-Aegilops umbellulata amphidiploids. Plant Genet. Resour. C 17, 24–32. doi: 10.1017/s1479262118000254
Suciu, S. (1986). Constitutive heterochromatin studies in patients with solid tumors. J. Cancer Res. Clin. Oncol. 111, 291–294. doi: 10.1007/BF00389248
Tahernezhad, Z., Zamani, M. J., Solouki, M., Zahravi, M., Imamjomeh, A. A., Jafaraghaei, M., et al. (2010). Genetic diversity of Iranian Aegilops tauschii Coss. using microsatellite molecular markers and morphological traits. Mol. Biol. Rep. 37, 3413–3420. doi: 10.1007/s11033-009-9931-6
Tang, Z. X., Ross, K., Ren, Z. L., Yang, Z. J., Zhang, H. Y., Miftahudin, C. T., et al. (2011). “Chapter 8. Secale,” in Wild Crop Relatives: Genomic and Breeding Resources: Cereals, ed. C. Kole, (New York, NY: Springer), 367–396. doi: 10.1007/978-3-642-14228-4_8
Tang, Z. X., Yang, Z. J., and Fu, S. L. (2014). Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 55, 313–318. doi: 10.1007/s13353-014-0215-z
Teoh, S. B., Miller, T. E., and Reader, S. M. (1983). Intraspecific variation in C-banded chromosomes of Aegilops comosa and Ae. speltoides. Theor. Appl. Genet. 65, 343–348. doi: 10.1007/bf00276575
Thomas, K. G., and Bebeli, P. J. (2010). Genetic diversity of Greek Aegilops species using different types of nuclear genome markers. Mol. Phylogenet. Evol. 56, 951–961. doi: 10.1016/j.ympev.2010.04.041
Toor, P. I., Kaur, S., Bansal, M., Yadav, B., and Chhuneja, P. (2016). Mapping of stripe rust resistance gene in an Aegilops caudata introgression line in wheat and its genetic association with leaf rust resistance. J. Genet. 95, 933–938. doi: 10.1007/s12041-016-0718-y
Van Slageren, M. W. (1994). Wild Wheats: a Monograph of Aegilops L. and Amblyopyrum (Jaub. & Spach) Eig. (Poaceae). Wageningen: Agricultural University Papers.
Wang, J., Wang, C., Zhen, S. M., Li, X. H., and Yan, Y. M. (2017). Low molecular weight glutenin subunits from the 1U genome of Aegilops umbellulata confer superior dough rheological properties and improve breadmaking quality of bread wheat. J. Sci. Food Agric. 98, 2156–2167. doi: 10.1002/jsfa.8700
Wang, S. W., Yin, L. N., Tanaka, H., Tanaka, K., and Tsujimoto, H. (2011). Wheat-Aegilops chromosome addition lines showing high iron and zinc contents in grains. Breed. Sci. 61, 189–195. doi: 10.1270/jsbbs.61.189
Wang, Y. J., Quan, W., Peng, N., Wang, C. Y., Yang, X. F., Liu, X. L., et al. (2016). Molecular cytogenetic identification of a wheat-Aegilops geniculata Roth 7Mg disomic addition line with powdery mildew resistance. Mol. Breed. 36:40. doi: 10.1007/s11032-016-0463-1
Weidner, A., Röder, M. S., and Börner, A. (2012). Mapping wheat powdery mildew resistance derived from Aegilops markgrafii. Plant Genet. Resour. C 10, 137–140. doi: 10.1017/s1479262112000123
Yamamoto, M. (1992). Detection of ribosomal RNA genes in Aegilops by in situ hybridization. Bull. Osaka Private Coll. Assoc. 29, 77–82.
Zhang, H., Jia, J., Gale, M. D., and Devos, K. M. (1998). Relationships between the chromosomes of Aegilops umbellulata and wheat. Theor. Appl. Genet. 96, 69–75. doi: 10.1007/s001220050710
Zhao, L. B., Ning, S. Z., Yi, Y. J., Zhang, L. Q., Yuan, Z. W., Wang, J. R., et al. (2018). Fluorescence in situ hybridization karyotyping reveals the presence of two distinct genomes in the taxon Aegilops tauschii. BMC Genomics 19:3. doi: 10.1186/s12864-017-4384-0
Zhao, L. B., Ning, S. Z., Yu, J. J., Hao, M., Zhang, L. Q., Yuan, Z. W., et al. (2016). Cytological identification of an Aegilops variabilis chromosome carrying stripe rust resistance in wheat. Breed. Sci. 66, 522–529. doi: 10.1270/jsbbs.16011
Zhu, Z. D., Zhou, R. H., Kong, X. Y., Dong, Y. C., and Jia, J. Z. (2006). Microsatellite marker identification of a Triticum aestivum-Aegilops umbellulata substitution line with powdery mildew resistance. Euphytica 150, 149–153. doi: 10.1007/s10681-006-9103-x
Keywords: fluorescence in situ hybridization (FISH), chromosomal variation, karyotypes, diploid Aegilops species, Ae. umbellulata, Ae. markgrafii, Ae. comosa, Ae. uniaristata
Citation: Song Z, Dai S, Bao T, Zuo Y, Xiang Q, Li J, Liu G and Yan Z (2020) Analysis of Structural Genomic Diversity in Aegilops umbellulata, Ae. markgrafii, Ae. comosa, and Ae. uniaristata by Fluorescence In Situ Hybridization Karyotyping. Front. Plant Sci. 11:710. doi: 10.3389/fpls.2020.00710
Received: 25 December 2019; Accepted: 05 May 2020;
Published: 09 June 2020.
Edited by:
Gerald Matthias Schneeweiss, University of Vienna, AustriaReviewed by:
Michał Tomasz Kwiatek, Poznań University of Life Sciences, PolandEkaterina D. Badaeva, Russian Academy of Sciences, Russia
Copyright © 2020 Song, Dai, Bao, Zuo, Xiang, Li, Liu and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shoufen Dai, ZHNmMDIwOEAxNjMuY29t; Zehong Yan, emh5YW4xMDRAMTYzLmNvbQ==
 Zhongping Song1,2
Zhongping Song1,2 Shoufen Dai
Shoufen Dai Zehong Yan
Zehong Yan