- 1Engineering Research Center of Ecology and Agricultural Use of Wetland, Ministry of Education/Hubei Key Laboratory of Waterlogging Disaster and Agricultural Use of Wetland, Yangtze University, Jingzhou, China
- 2State Key Laboratory of Crop Stress Adaptation and Improvement, Henan University, Kaifeng, China
- 3State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Science and Technology, Guangxi University, Nanning, China
- 4School of Science, Western Sydney University, Penrith, NSW, Australia
- 5Hawkesbury Institute for the Environment, Western Sydney University, Penrith, NSW, Australia
Toxic heavy metals and metalloids in agricultural ecosystems are crucial factors that limit global crop productivity and food safety. Industrial toxic heavy metals and metalloids such as cadmium, lead, and arsenic have contaminated large areas of arable land in the world and their accumulation in the edible parts of crops is causing serious health risks to humans and animals. Plants have co-evolved with various concentrations of these toxic metals and metalloids in soil and water. Some green plant species have significant innovations in key genes for the adaptation of abiotic stress tolerance pathways that are able to tolerate heavy metals and metalloids. Increasing evidence has demonstrated that phytohormone abscisic acid (ABA) plays a vital role in the alleviation of heavy metal and metalloid stresses in plants. Here, we trace the evolutionary origins of the key gene families connecting ABA signaling with tolerance to heavy metals and metalloids in green plants. We also summarize the molecular and physiological aspects of ABA in the uptake, root-to-shoot translocation, chelation, sequestration, reutilization, and accumulation of key heavy metals and metalloids in plants. The molecular evolution and interaction between the ABA signaling pathway and mechanisms for heavy metal and metalloid tolerance are highlighted in this review. Therefore, we propose that it is promising to manipulate ABA signaling in plant tissues to reduce the uptake and accumulation of toxic heavy metals and metalloids in crops through the application of ABA-producing bacteria or ABA analogues. This may lead to improvements in tolerance of major crops to heavy metals and metalloids.
Introduction
Toxic mineral elements, such as metals and metalloids, are ubiquitous in the Earth's crust. The amount of toxic metals and metalloids in water and soil is increasingly attributed to anthropogenic activities (Bowell et al., 2014; Zhu et al., 2014; Zhao et al., 2015). These contaminates affect agricultural productivity and ecosystem function and also threat human health, posing a great risk to global economic growth (Landrigan et al., 2018). Moreover, toxic metals and metalloids accumulated in edible parts of plants, such as cereals and vegetables, as well as in pasture for animals, should be tightly controlled to reduce health risks. Therefore, urgent actions are required to mitigate the serious problems from heavy metals and metalloids' contamination.
Since the evolution of land plants from ancestral green algae (Cheng et al., 2019; Zhao et al., 2019; Wang et al., 2020), land plants have been indispensable to the biosphere and our daily life. In addition to feeding the world's population, many plant species have also been employed to alleviate the increasing pollution of heavy metals and metalloids through bioremediation. Over 700 plant species have been identified as hyperaccumulators of trace metals, metalloids, and nonmetals. The hyperaccumulator species are from 52 families belonging to angiosperms and petridophyta (Reeves et al., 2017). For instance, one plant used is a hardy, versatile, fast-growing brake fern (Pteris vittata) with extreme efficiency in extracting and translocating soil arsenic (As) to the above-ground biomass, which can significantly remove arsenic from contaminated soils (Ma et al., 2001; Yan et al., 2019). Therefore, exploring the early-divergent plant species for their tolerance mechanisms is essential to utilize them as potential hyperaccumulators for heavy metals and metalloids.
Among the toxic minerals, Arsenic, cadmium (Cd), and lead (Pb) were ranked as the top hazardous substances (Clemens and Ma, 2016) due to their toxicity, prevalence, and potential for human exposure. During the last decade, considerable advances in As and Cd accumulation and detoxification mechanisms in angiosperms, in particular the model plant species rice (Oryza sativa) and Arabidopsis (Arabidopsis thaliana) (Clemens and Ma, 2016; Singh et al., 2016; Deng et al., 2019; Deng et al., 2020; Zhao and Wang, 2020) have been made. Although genetic engineering is a powerful strategy for generating ideal plants for food safety and phytoremediation (Shim et al., 2013; Deng et al., 2018), it's not widely used in agriculture at present due to its controversy in potential risk to human health and agricultural ecosystems (Andersen et al., 2015). Therefore, understanding the molecular mechanisms of elements absorption and root-to-shoot translocation make it possible to promote the efficiency of phytoremediation instead of genetic engineering.
Abscisic acid (ABA) plays vital roles in plant responses to a range of abiotic stresses such as drought, salinity, high light, nutrient deficiency, and heavy metals (Chen Z. H. et al., 2016; Cai et al., 2017; Liu X. et al., 2017; Vishwakarma et al., 2017; Wang F. et al., 2017; Kuromori et al., 2018; Babla et al., 2019; Mak et al., 2019; Mega et al., 2019; Feng et al., 2020; Shabala et al., 2020) and its agonist and antagonist were considered as candidate compounds to overcome these stresses (Joshi-Saha et al., 2011; Kitahata and Asami, 2011; Miyakawa et al., 2013; Park et al., 2015; Gupta et al., 2020). ABA is one of the foremost phytohormone driving plant resistance to toxic metals and metalloids such as As, Cd, and Pb (Maestri and Marmiroli, 2012; Chmielowska-Bak et al., 2014; Vishwakarma et al., 2017; Amir et al., 2018; Pál et al., 2018; Shi et al., 2019; Zhang P. et al., 2019; Zhang W. et al., 2019; Pan et al., 2020). Mechanisms of ABA in response to heavy metals and metalloids stresses in non-angiosperm plant lineages is still limited; we took a comparative genomic evolutionary approach to shed some light on the insights of ABA and tolerance to heavy metals and metalloids.
There have been many excellent reviews on ABA and plant stress tolerance in recent years (Osakabe et al., 2014; Mittler and Blumwald, 2015; Zhu, 2016; Hauser et al., 2017; Kuromori et al., 2018; Chen et al., 2020). Here, we summarize three ABA-activated pathways that contribute to heavy metals detoxification in angiosperms using rice and Arabidopsis as model species. We also attempt to trace the origin and evolution of the core components linking ABA and tolerance to toxic metals and metalloids involved in the processes.
Overview of ABA Signaling Network
Abscisic acid (ABA) is a vital phytohormone that regulates many developmental processes in plants and in the response to abiotic stresses including drought, cold, salinity, and heavy metals (Chmielowska-Bak et al., 2014; Mittler and Blumwald, 2015; Chen et al., 2017; Hauser et al., 2017; Chen et al., 2019; Zhang P. et al., 2019; Zhao et al., 2019). The biosynthesis, catabolism, transport, signal perception and transduction, downstream response, and modulation of ABA have been extensively investigated in angiosperms, in particular in Arabidopsis thaliana (Hauser et al., 2011; Cai et al., 2017; Hauser et al., 2017; Chen et al., 2020).
ABA is primarily synthesized from carotenoids, which are catalyzed by various enzymes including β-carotene hydroxylases, zeaxanthin epoxidase (ZEP, ABA1), 9-cis-epoxycarotenoid dioxygenase (NCEDs), short-chain alcohol dehydrogenase/reductases (SDRs, such as ABA2), abscisic aldehyde oxidases (AAOs), molybdenum cofactor sulfurase (MOCO, ABA3), and ABA4, which is required for neoxanthin synthesis (Nambara and Marion-Poll, 2005; North et al., 2007; Finkelstein, 2013; Cai et al., 2017; Hauser et al., 2017). The hydroxylation and esterification of ABA are two major pathways for regulating ABA levels mediated by four CYP707As and eight glucosyltransferases (UGTs). The inactivated ABA-glucosyl ester (ABA-GE) conjugation is a storage or transport form of ABA and the site can be cleaved by β-glucosidases (BGLUs) (Finkelstein, 2013). The mobility of ABA from vascular tissues to target tissues is transported by three groups of membrane-localized proteins: G-type ATP binding cassette transporters (ABCG22, ABCG25, ABCG30, ABCG31, ABCG40), ABA-Importing Transporters (AIT1~4) belonging to the Nitrate Transporter 1/Peptide Transporter (NRT1/PTR, NPF) family, and a member of DTX/Multidrug and Toxic Compound Extrusion (MATE) family DTX50 in Arabidopsis (Kang et al., 2010; Kuromori et al., 2010; Kuromori et al., 2011; Kanno et al., 2012; Zhang et al., 2014; Kang et al., 2015).
The core components of ABA perception and transduction consist of intracellular Pyrabactin Resistance 1 (PYR1)/PYR-Like (PYL)/Regulatory Component of ABA receptors (RCARs), clade A Protein Phosphatases PP2C (ABA Insensitive 1/2, ABI1/2, and Hypersensitive to ABA1/2, HAB1/2), sucrose non-fermenting-1-related protein kinase 2 family members (SnRK2s) (Ma et al., 2009; Park et al., 2009; Umezawa et al., 2009). SnRK2.2, -2.3, and -2.6 (Open Stomata 1, OST1) are strongly activated in the presence of ABA (McLoughlin et al., 2012). SnRK2.6 is a key regulator of stomatal closure by enhancing Cl-, K+, malate2- efflux from guard cells mediated by S-type anion channel 1 (SLAC1), K+ uptake transporter 6 (KUP6), and Aluminum-activated malate transporter (ALMT12), and inhibiting the activity of Potassium channel 1 (KAT1) to reduce K+ influx (Finkelstein, 2013; Chen et al., 2020). ABA also enables the activation of guard cell membrane-localized transporters through phosphorylation mediated by calcium dependent kinases (CDPKs) and other kinases, indicating alternative stomatal regulatory pathways independent of SnRK2s (Cai et al., 2017; Pornsiriwong et al., 2017). Most recently, subgroup B Raf-like kinases have been identified as upstream regulators of SnRK2s for ABA signal transduction and response to osmotic and drought stresses in Arabidopsis (Saruhashi et al., 2015; Katsuta et al., 2020; Lin et al., 2020; Soma et al., 2020; Takahashi et al., 2020). Furthermore, it's revealed that this regulatory module is evolutionarily conserved in land plants, at least for conferring protection against drought (Katsuta et al., 2020). The other biological regulations induced by ABA are mostly implemented through transcriptional processes mediated by ABA Insensitive 3/4 (ABI3/4) and 9 ABA-response element (ABRE) binding factors (ABFs) (Chen et al., 2020). ABI3 and ABI4 genes encode B3-type and APETALA2 domain a transcription factors, respectively, while ABFs consisting of ABF1~4, ABI5, bZIP15, bZIP67, and EEL (Enhanced Em Level) belong to group A subfamily of bZIP (basic region/leucine zipper) transcription factors (Choi et al., 2000; Finkelstein, 2013; Huang et al., 2016; Skubacz et al., 2016; Chen et al., 2020). All the listed components are candidates for the role of ABA in response to toxic metals and metalloids stresses.
ABA Alleviates Toxic Metals and Metalloids Stresses in Plants
First of all, the biosynthesis and signaling pathways of ABA are affected by heavy metal stresses. Elevated endogenous ABA content was detected in rice, potato (Solanum tuberosum), oilseed rape (Brassica napus), Malus hupehensis, Sedum alfredii, and other plants exposed to Cd, partially due to the upregulation of genes for ABA biosynthesis (Hsu and Kao, 2003; Stroiński et al., 2013; Yan et al., 2016; Shi et al., 2019; Zhang W. et al., 2019; Lu et al., 2020b). In rice, the expression levels of OsNCED3, OsNCED4, and OsNCED5 were upregulated by Cd (Tan et al., 2017). In addition, Cd-induced rapid ABA production was more significant in the leaves and roots of Cd-tolerant rice cultivar than those in the Cd-sensitive genotype (Hsu and Kao, 2003), indicating the positive correlation between endogenous ABA content and Cd tolerance.
In Arabidopsis, enhanced Cd sensitivity and increased Cd accumulation was observed in three ABA-deficient mutants (aba-1, aba-3, aba-4, nced3) and two ABA-insensitive mutants (abi2-1, abi3-1) (Sharma and Kumar, 2002; Zhang W. et al., 2019). These genes are involved in ABA synthesis (ABA1, ABA3, ABA4, nced3) and signal transduction (ABI2, ABI3), respectively. Similarly, the Arabidopsis mutants bglu10 and bglu18 with reduced root cytoplasmic ABA levels were more sensitive to Cd stress compared to the wild type (Table 1) (Wang et al., 2018). Ectopic expression of Malus hupehensis NCED3 (MhNCED3) in Arabidopsis increased ABA content and reduced Cd accumulation in both root and leaves (Zhang W. et al., 2019). Moreover, in Cd-contaminated soil, greater biomass and lower Cd concentrations were determined in Arabidopsis and Brassica chinensis inoculated with ABA-generating bacteria, Azospirillum brasilense or Bacillus subtilis (Xu et al., 2018; Pan et al., 2019). By contrast, decreasing the endogenous ABA amount by inoculation with an ABA-catabolizing bacteria, Rhodococcus qingshengii, significantly increased Cd content in Arabidopsis shoots by 47% (Lu et al., 2020a). Compared to the wild type, higher root Cd concentration was detected in ABA-deficient Slsit tomato mutant (Table 1) (Pompeu et al., 2017). Application of ABA further enhanced Cd tolerance and accumulation activity in the Cd-hyperaccumulating ecotype (HE) of Sedum alfredii (Lu et al., 2020b). Interestingly, elevated levels of endogenous ABA accompanied by up-regulated SaNCED and SaABA2 was observed in the non-hyperaccumulating ecotype (NHE) subjected to Cd treatment compared to those in HE, restricting radial transport of Cd toward root vascular tissues (Tao et al., 2019; Lu et al., 2020b).
Elevated ABA content was increased in both roots and shoots of As-exposed Indian mustard (Brassica juncea), which is a potential As accumulator for phytoremediation (Srivastava et al., 2013). Furthermore, ABA-related genes were regulated predominately in As-tolerant ecotype Col-0, however, all the 25 genes involved in ABA biosynthesis, receptor, and signaling pathways detected in sensitive ecotype Ws-2 were unaltered by the treatment of As (Fu et al., 2014). In addition to the upregulated OsNCED1, OsNCED2, OsNCED3, and OsABA4 responsible for ABA biosynthesis, the expression levels of genes probably involved in ABA signaling including OsPP2Cs, OsbZIP10 (OsABI5), OsbZIP12 (OsABF1), OsbZIP66 (OsABF5), and OsbZIP72 (OsABF4) were also elevated when rice plants were exposed to As (Huang et al., 2012; Yu et al., 2012). Moreover, increased endogenous ABA levels were detected in germinating chickpea (Cicer arietinum) and leaves of pea (Pisum sativum) exposed to Pb (Parys et al., 1998; Atici et al., 2005). Compared to that of control, ABA concentration was increased by 107% in the leaves of Gray Poplar (Populus × canescens) with Pb exposure, while application of exogenous ABA alleviated Pb toxicity (Shi et al., 2019). Recently, comparative transcriptomic analyses between Arabidopsis (Pb sensitive) and Hirschfeldia incana (Pb tolerant) revealed that genes involved in ABA biosynthesis were upregulated in the roots and shoots of H. incana subjected to Pb (Auguy et al., 2016).
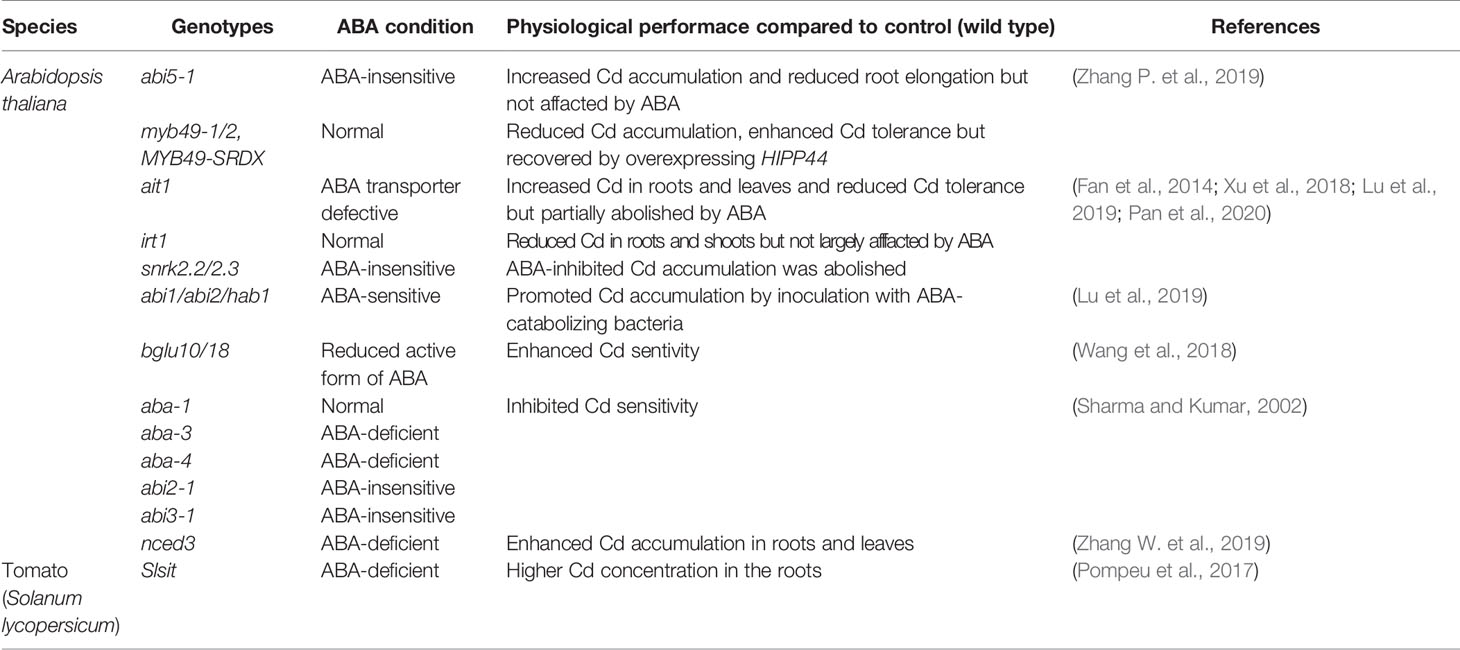
These imply a positive role of ABA in alleviating accumulation and toxicity of heavy metals and metalloids. Consistently, the application of exogenous ABA to angiosperms subjected to Cd, As, or Pb could alleviate the stresses. Three major pathways involved in the detoxification of toxic metals and metalloids can be triggered by ABA, inhibiting the uptake, altering the translocation from root to shoot, and promoting the conjugation with chelators (Figure 1).
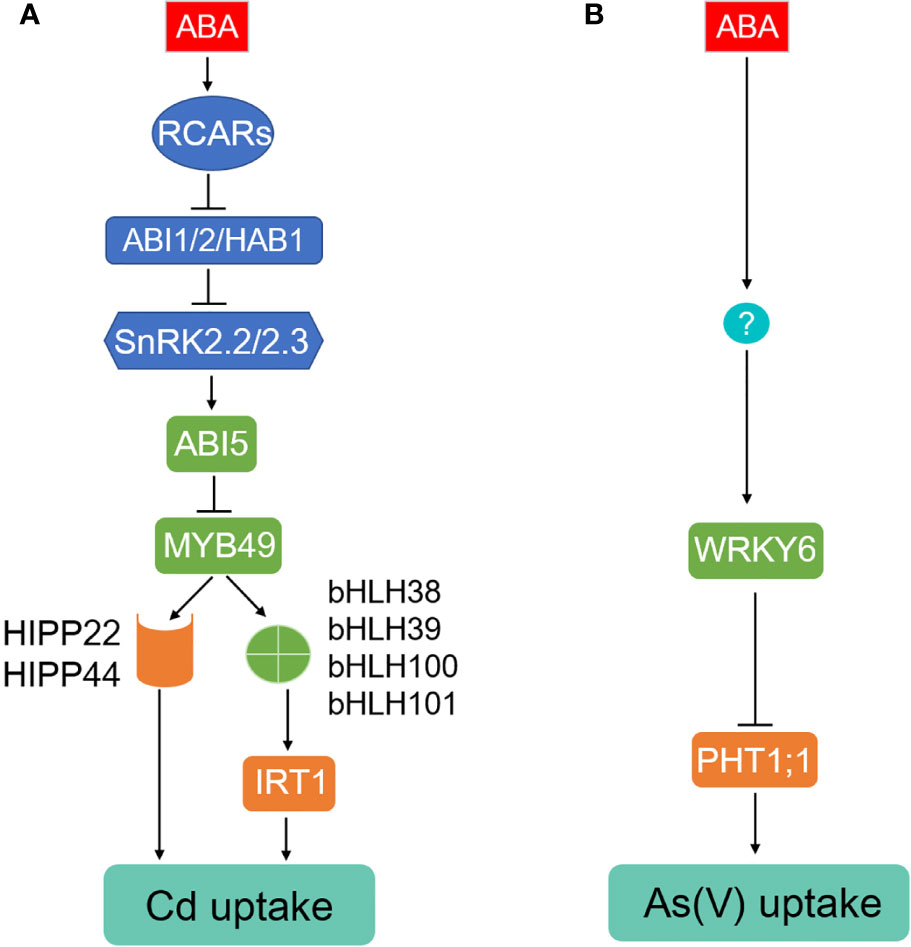
Figure 1 ABA inhibits Cd and As(V) uptake in Arabidopsis. (A) Cd uptake and accumulation mediated by IRT1 and HIPP proteins is activated by a transcriptional factor MYB49, which can be inactivated by interaction with ABA-induced ABI5. (B) Arsenate uptake mediated by PHT1;1 is repressed by ABA-induced transcriptional factor WRKY6.
ABA Limits Toxic Heavy Metals and Metalloids Uptake
In Arabidopsis, Iron Regulated Transporter 1 (IRT1) functions as a primary transporter for Cd uptake from the rhizosphere (Lux et al., 2011). The expression of IRT1 is tightly regulated by FIT (FER-like Deficiency Induced Transcripition Factor) and Ib subgroup of the bHLH (basic helix-loop-helix) transcriptional factors including bHLH38, bHLH39, bHLH100, and bHLH101 in Arabidopsis (Wu et al., 2012; Wang et al., 2013). Application of low concentrations of ABA (0.1~1.0 μM) or inoculation with ABA-generating bacteria strains decreased Cd uptake through inhibiting transcription of IRT1 and thereby alleviating Cd-induced growth inhibition (Fan et al., 2014; Zhang P. et al., 2019; Pan et al., 2020). By contrast, an increase was observed in expression level of IRT1 and its homologous genes such as ZIP1 (Zinc Regulated Transporter/IRT-like Protein 1) and ZIP4 with Cd transporting-activity in the roots inoculated with ABA-catabolizing bacteria (Xu et al., 2018; Pan et al., 2019; Lu et al., 2020a). These positive effects were reduced in the ABA-Importing Transporter 1 (AIT1)-deficient mutant ait1 but enhanced in the AIT1-overexpressing transgenic plants (Pan et al., 2020). Furthermore, the loss-of-function Arabidopsis mutant of ABI5 and ABA-insensitive double mutant of snrk2.2/2.3 abolished the reduction of Cd accumulation induced by exogenously applied ABA or ABA-generating bacteria. The elevated IRT1 transcription was diminished in snrk2.2/2.3 mutant but further enhanced in the roots of ABA-sensitive triple PP2Cs mutant abi1/hab1/abi2 lines, suggesting the involvement of AIT1, ABI5, SnRK2s, and PP2Cs in Cd absorption mediated by IRT1 (Fan et al., 2014; Xu et al., 2018; Pan et al., 2019; Zhang P. et al., 2019; Lu et al., 2020a). Additionally, ABI5 can be directly phosphorylated by SnRK2.2/2.3 and inactivated via dephosphorylation of ABI1/HAB1/ABI2 (Fujii et al., 2009; Skubacz et al., 2016). ABI5 could directly interact with Cd-induced transcriptional factor MYB49 to inhibit its binding to the promoter regions of bHLH38 and bHLH101, which are required for IRT1 expression (Wu et al., 2012; Wang et al., 2013; Zhang P. et al., 2019). As a result, ABI5 is a negative regulator of IRT1 and Cd uptake. Moreover, MYB49 directly induces the expression of HIPP22 and HIPP44, members belonging to Heavy metal-associated Isoprenylated Plant Proteins family (HIPPs) involved in Cd accumulation (Tehseen et al., 2010; Zhang P. et al., 2019). The expression of Arabidopsis HIPP22 and HIPP44 are positively correlated with Cd accumulation (Tehseen et al., 2010; Zhang P. et al., 2019). It is noteworthy that tobacco (Nicotiana tabacum) and Arabidopsis SnRK2s are transiently activated by Cd exposure, knockout of AtSnRK2.4 enhanced Cd tolerance. However, the induced SnRK2.4 is ABA-independent but probably influences ROS accumulation (Kulik et al., 2012).
Cd absorption was also observed to be inhibited in lettuce (Lactuca sativa) and oilseed rape (Brassica napus), however, whether this is a conserved route is not clear (Shen et al., 2017; Tang et al., 2020). On the other hand, Cd absorption was not affected by ABA in rice, Indian mustard, and A. helleri (Salt et al., 1995; Hsu and Kao, 2003; Zhao et al., 2006), which could be due to a different Cd uptake system in these species. For instance, the major transporters for Cd uptake in rice is OsNramp5 (Sasaki et al., 2012) with less contribution from OsIRT1 and OsIRT2 (Nakanishi et al., 2006). OsNramp5 is a member belonging to Natural Resistance-Associated Macrophage Protein family and polarly localized at the distal side of both root exodermis and endodermis cells (Sasaki et al., 2012). Knockout of OsNramp5 almost abolished the ability to take up Cd (Ishikawa et al., 2012; Sasaki et al., 2012). In summary, we propose that ABA alleviates Cd uptake potentially through the ABI5-MYB-bHLHs-IRT1 and ABI5-MYB49-HIPPs pathways in Arabidopsis (Figure 1A), and the ABA core components including AIT1, ABI1/HAB1/ABI2, and SnRK2.2/2.3 are involved in these processes.
In As stress, Phosphate Transporters (PHTs, PTs) and Nodulin 26-like Intrinsic membrane Proteins (NIPs) are two major families responsible for arsenate (AsV) and arsenite (AsIII) uptake, respectively (Lindsay and Maathuis, 2017; Deng et al., 2020). In Arabidopsis, As(V) uptake is mainly mediated via PHT1;1 and PHT1;4 (Shin et al., 2004). An As(V)-responsive transcription factor WRKY6 was identified as a negative regulator for As(V) uptake through directly repressing expression of PHT1;1 and removal of PHT1;1 from the plasma membrane (Figure 1B) (Castrillo et al., 2013). The expression of WRKY6 was induced by ABA (Song et al., 2016), knockout of WRKY6 resulted in ABA insensitivity, while WRKY6-overexpressing lines showed ABA-hypersensitive phenotypes during seed germination (Huang et al., 2016), indicating that WRKY6 functions as a positive regulator in ABA signaling. Therefore, we speculate that ABA may inhibit As(V) uptake through WRKY6-PHT1;1 route in Arabidopsis, which requires future investigation. It's also worthy to isolate and functionally characterize rice and brake fern homologs of AtWRKY6 in As(V) uptake because OsPT1, OsPT4, OsPT8, PvPHT1, PvPHT1;3, and PvPHT1;4 confer to As(V) accumulation and toxicity in rice and P. vittata, respectively, most likely via their role in As(V) uptake in roots (Kamiya et al., 2013; DiTusa et al., 2016; Wang et al., 2016; Cao et al., 2017; Ye et al., 2017; Cao et al., 2019; Sun et al., 2019). Arabidopsis NIP1;1, NIP3;1, NIP3;2, and NIP7;1 function in As(III) uptake and accumulation (Lindsay and Maathuis, 2017; Deng et al., 2020), however, the regulation of them being mediated by ABA is still elusive.
Tremendous progress has been made recently in dissecting the entry of Cd and As into plant cells (Clemens and Ma, 2016; Lindsay and Maathuis, 2017; Zhao and Wang, 2020), and the involvement of ABA in Cd and As(V) uptake is strongly supported by experimental evidence. By contrast, the molecular understanding of Pb uptake pathways is limited. In Pb stress, the plasma membrane-localized G-type ABC members AtABCG36 (PDR8, for Cd and Pb), AtABCG40 (PDR12, for Pb) function as extrusion pumps conferring Pb resistance by limiting their accumulation in Arabidopsis (Lee et al., 2005; Kim et al., 2007; Fu et al., 2019; Wang et al., 2019). Most recently, it has been reported that the expression of PcABCG40 were stimulated by exogenous ABA in Pb-exposed Gray Poplar (Shi et al., 2019), while the transcription of AtABCG40 is activated by Pb-Sensitive 1 (AtPSE1), which is a cytoplasmic protein conferring to Pb tolerance in Arabidopsis (Fan et al., 2016). Besides, Arabidopsis ABCG36 is positively regulated by WRKY13 through directly binding to its promoter (Sheng et al., 2019). Therefore, further studies are required for uncovering the involvement of ABA in Pb uptake, such as the role of ABA in Pb extrusion from root, potentially via ABCGs, PSE1, and WRKY13.
ABA Alters Toxic Metals and Metalloids Distribution Between Root and Shoot
Exogenous ABA tends to hinder metal ion and metalloid translocation to the shoot via inhibiting transpiration. Xylem loading is a limiting step for metal ion and metalloid accumulation in the above-ground tissues; the activity is largely dependent on transpiration activity and membrane transporters (Uraguchi et al., 2009; Clemens and Ma, 2016; Deng et al., 2018; Deng et al., 2019). For Cd stress, pre-treatment with ABA dramatically reduced Cd accumulation in the leaves of Indian mustard (Salt et al., 1995) and less Cd accumulation was detected in parallel with a reduced transpiration rate and stomatal closure with up to 100 μM ABA treatment in rice and Arabidopsis helleri (Salt et al., 1995; Hsu and Kao, 2003; Zhao et al., 2006; Uraguchi et al., 2009). Besides, involvement of transpiration capacity in the ABA-induced reduction of Cd allocation from roots to leaves is yet to be validated in lettuce (Lactuca sativa) and field mustard (Brassica campestris) (Shen et al., 2017; Tang et al., 2020).
Membrane transporters play critical roles in loading minerals toward root vascular tissues and subsequent accumulation in plants (Song et al., 2014a; Clemens and Ma, 2016; Deng et al., 2019; Zhao and Wang, 2020). For example, the plasma membrane-localized heavy metal ATPases (HMAs), such as AtHMA2, AtHMA4, and OsHMA2, confer to loading Cd into the stele and subsequent accumulation in shoot (Hussain et al., 2004; Mills et al., 2005; Yamaji et al., 2013), while the tonoplast-localized AtHMA3 and OsHMA3 limit Cd allocation to the stem, leaves, and grain through sequestration Cd into the vacuole of root cells (Morel et al., 2009; Ueno et al., 2010). As a result, the mutant plants without functional AtHMA2, AtHMA4, or OsHMA2 accumulate less Cd compared to the wild type (Hussain et al., 2004; Mills et al., 2005; Yamaji et al., 2013). Knockout of AtHMA3 or OsHMA3 increased Cd accumulation in aerial organs (Morel et al., 2009; Ueno et al., 2010), whereas overexpressing OsHMA3 enhanced Cd tolerance and produced Cd-free rice lines (Ueno et al., 2010; Sasaki et al., 2014; Lu C. et al., 2019). Similar functions of their homologs have been reported in other plants, including barley (Hordeum vulgare), wheat (Triticum aestivum), soybean (Glycine max), cucumber (Cucumis sativus), and Cd hyperaccumulators Sedum plumbizincicola, Noccaea caerulescens, and Arabidopsis halleri (Hanikenne et al., 2008; Miyadate et al., 2011; Ueno et al., 2011; Mills et al., 2012; Wang et al., 2012; Tan et al., 2013; Migocka et al., 2015; Liu H. et al., 2017). Natural variations in the HMA3 genes are key determinants of Cd translocation to and accumulation in the shoot of Arabidopsis, rice, soybean, and Brassica rapa (Chao et al., 2012; Wang et al., 2012; Liu et al., 2019; Sui et al., 2019; Zhang L. et al., 2019). In addition, some members belonging to NRAMPs also transport free Cd ions (Clemens and Ma, 2016; Zhang et al., 2018). For instance, the expression of HMA3, Nramp1, Nramp3, and Nramp4 were upregulated in Arabidopsis incubation with ABA-catabolizing bacteria (Lu et al., 2020a), but overexpression of MhNCED3 in Arabidopsis inhibited the expression of IRT1, Nramp1, and HMA2, leading to reduced Cd uptake and root-to-shoot translocation (Zhang W. et al., 2019). Application of ABA promotes Cd resistance and mobility from root to shoot in the Cd-hyperaccumulating ecotype (HE) of Sedum alfredii by inducing the transcription of SaHMA2, SaHMA3, and SaHMA4 (Lu et al., 2020b). Interestingly, more endogenous ABA was generated in the non-hyperaccumulating ecotype (NHE) subjected to Cd treatment and induced the expression of lignin- and suberin-related biosynthetic enzymes in NHE roots to limit Cd radial transport towards the stele as well as accumulation in the shoot (Tao et al., 2019). Moreover, some membrane transporters are indirectly involved in ABA-related Cd tolerance. In Arabidopsis, nitrate transporter NRT1.5 plays a vital role in the root-to-shoot translocation of nitrate (Lin et al., 2008), while NRT1.8 is responsible for removing nitrate from xylem vessels and also confers tolerance to CD in a nitrate-dependent manner (Li et al., 2010). Exogenous ABA inhibits the expression of NRT1.5 but has no effect on the transcripts of NRT1.8, leading to increased accumulation of nitrate in the roots and thus enhances Cd resistance (Wang et al., 2018). Vacuolar proton pumps V-ATPase and V-PPase are able to resist Cd through enhanced compartmentation activity into root vacuoles (Wang et al., 2018). Knockout of BGLU10 or BGLU18 reduced endogenous active ABA level, resulting in higher levels of NRT1.5 and lower V-ATPase and V-PPase activities, resulting in higher Cd accumulation and sensitivity (Wang et al., 2018). However, the direct regulators of these transporters response to ABA remains to be discovered and functionally characterized.
For As stress tolerance and translocation, many of the fundamental studies were conducted in rice. For instance, a plasma membrane-localized ABC transporter OsABCC7, highly expressed in the root xylem parenchyma cells, is involved in the root-to-shoot translocation of glutathione (GSH)- and Phytochelatins (PCs)-conjugated As (Tang Z. et al., 2019). As(III) uptake and loading to shoot are predominantly accomplished by plasma membrane-polar localized OsLsi1 and OsLsi2 in rice (Ma et al., 2008). OsLsi1 is a member of NIPs required for As(III) uptake from soil into the root cells while OsLsi2 is responsible for the subsequent As(III) transport out of epidermal and endodermal cells toward the stele (Ma et al., 2008). Knockout of OsLsi1 reduced As uptake, while mutation of OsLsi2 decreased As accumulation in rice straw and grain (Ma et al., 2008). OsLsi6, a homolog of OsLsi1 highly expressed in rice nodes, was implicated as a transporter required for As distribution from leaf and node to panicle (Yamaji et al., 2015; Deng et al., 2020). The expression levels of OsLsi1, OsLsi2, and OsLsi6 were negatively regulated by Arsenite-Responsive MYB 1 (OsARM1) through the direct binding to the promoters or genomic regions of the three key As transporters (Figure 2) (Wang F.-Z. et al., 2017). Knockout of OsARM1 improved tolerance to As(III) and increased As accumulation in shoot and the upmost node, while As concentrations in these organs and the tolerance to As(III) were reduced in OsARM1-overexpressing plants compared to those of wild-type plants (Wang F.-Z. et al., 2017). Most interestingly, it was found that the expression of OsARM1 was repressed by exogenous ABA treatment (Sato et al., 2011; Guo et al., 2016). Therefore, we propose that ABA may enhance As tolerance by promoting As accumulation in above-ground tissues, which is partially dependent on the OsARM1-OsLsi1/Lsi2/Lsi6 pathway. The direct components for the ABA-inhibited transcription of OsARM1 require further investigations.
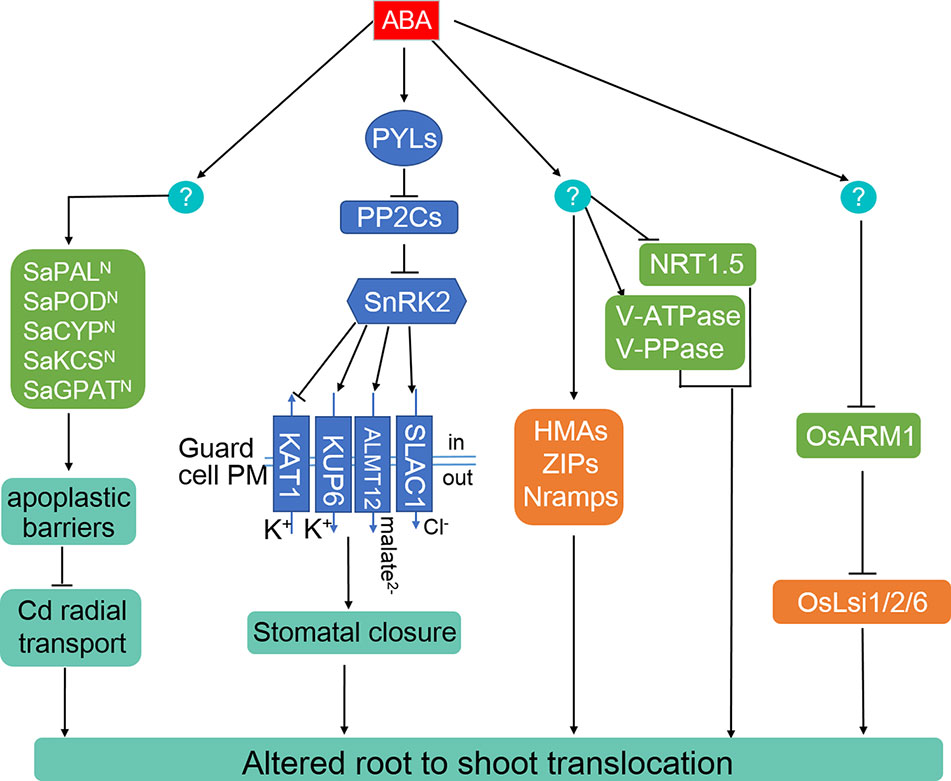
Figure 2 ABA alters the root-to-shoot translocation of Cd, As, and Pb through multiple routes. ABA is able to promote the generation of root apoplastic barriers to inhibit Cd radial transport toward stele in the non-hyperaccumulating ecotype of Sedum alfredii. ABA-induced stomatal closure repress the allocation of Cd, As, and Pb from root to shoots. ABA can change the distribution of heavy metals by inducing the expression of certain HMAs, ZIPs, and Nramps transporter genes in various plant species. ABA change Cd distribution between root and shoots through increasing the activates of vacuolar protons, V-ATPase, and V-PPase and inhibiting nitrate movement from root to shoot mediated by NRT1.5. In rice, As uptake, distribution, and tolerance mediated by OsLsi1, OsLsi2, and OsLsi6 is repressed by the ABA-inhibited transcriptional factor, OsARM1.
For Pb stress tolerance, stomatal closure and a decreased transpiration ratio were observed in the pea leaves with Pb exposure, accompanied with elevated amount of endogenous ABA (Parys et al., 1998). Moreover, the mobility of Pb from root to shoot in rice and Gray Poplar (Populus × canescens) was also affected by exogenous ABA in a dose-dependent manner. When subjected to 10 μM exogenous ABA, stimulated Pb uptake and vascular loading in the roots was observed in Gray Poplar (Shi et al., 2019). In rice, a low concentration of ABA (0.38 μM) decreased Pb translocation from roots to shoots, whereas a high concentration of ABA (3.8 and 38 μM) resulted in antipodal Pb distribution (Zhao et al., 2009). However, the genetic evidence underlying the ABA-affected Pb distribution is still lacking.
In summary, ABA is important in regulating the root-to-shoot translocation ratio of heavy metals and metalloids. Many physiological investigations suggest that ABA-induced stomatal closure and a reduced transpiration rate limit the long-distance transport of heavy metals and metalloids from root to shoot. Numerous transporters for Cd, As, and Pb distribution have been identified in Arabidopsis and rice, potentially supporting the participation of ABA in these processes.
ABA Promotes Chelation and Vacuolar Sequestration of Toxic Metals and Metalloids
The cytosolic Cd, As, and Pb can be conjugated by thiol-containing chelators, GSH, and/or PCs and then compartmented into vacuoles. These processes play pivotal roles in the detoxification of toxic metals and metalloids by restricting their mobility (Clemens et al., 1999; Gong et al., 2003; Kim et al., 2006; Song et al., 2010; Park et al., 2012; Song et al., 2014b; Hayashi et al., 2017; Deng et al., 2018; Deng et al., 2019). It is well documented that GSH is synthesized through γ-glutamylcysteine synthetase (GSH1 or γ-ECS) and glutathione synthetase (GSH2), while PCs are polymerized from GSH by PCS (PCS1, PCS2) in Arabidopsis (Cobbett, 1999; Vatamaniuk et al., 1999; Cobbett, 2000).
In Arabidopsis, the transcription of GSH1, GSH2, PCS1, and PCS2 are positively regulated by a Cd-induced Cysteine-2/Histidine-2 type zinc-finger (C2H2) transcriptional factor, ZAT6 (Chen J. et al., 2016). Overexpressing ZAT6 significantly enhanced Cd tolerance, whereas loss of function of ZAT6 led to Cd sensitivity (Chen J. et al., 2016). In a systematic study of the ABA transcriptional regulatory network, the expression of ZAT6 was upregulated by exogenous ABA treatment (Song et al., 2016). In addition to activating the transcription of ABCG40, genes involved in PCs synthesis are also activated by PSE1, which confers Pb tolerance in Arabidopsis (Figure 3) (Fan et al., 2016). WRKY12 negatively regulates Cd tolerance by repressing the expression of PCs synthesis genes (Han et al., 2019). Recently, OsPCS1 and OsPCS2 were identified and played crucial roles in the detoxification and accumulation of As and Cd in rice (Hayashi et al., 2017; Uraguchi et al., 2017; Yamazaki et al., 2018). Loss-of-function of OsPCS1 increased As allocation from node to seed, while overexpressing OsPCS1 significantly reduced grain As content (Hayashi et al., 2017). Overexpression of wheat TaPCS1, Morus notabilis MnPCS1, and MnPCS2 or Populus tomentosa PtPCS in Arabidopsis and/or tobacco enhanced Cd tolerance (Figure 3) (Fan et al., 2018). In addition, a plastid envelope membrane-localized CRT-like transporter, OsCTL1, is required for As and Cd detoxification through exporting γ-glutamylcysteine and GSH from plastids to the cytoplasm, where PCS synthesis takes place (Yang et al., 2016). Treatment of potato (Solanum tuberosum) plants with ABA clearly enhanced StPCS1 transcript level, PCS activity, and PCs content in roots, while application of the ABA biosynthesis inhibitor, fluridone, limited the Cd-induced PCS activity (Stroiński et al., 2010; Stroiński et al., 2013). ABA-induced expression of StPCS1 was in parallel with an elevated level of StbZIP, encoding a potential ABF on the upstream of StPCS1 (Figure 3) (Stroiński et al., 2010; Stroiński et al., 2013). Similarly, the transcript levels of PcECS1 and PcPCS1.1 genes, encoding rate-limiting enzymes for GSH and PCs synthesis, were upregulated in the roots of Gray Poplar treated with ABA compared with the control, irrespective of Pb treatments (Figure 3) (Shi et al., 2019).
Figure 3 ABA-dependent and -independent regulation of heavy metals and metalloids chelation and compartmentation. The generation of GSH and PCs can be promoted by the ABA-induced transcriptional factors AtZAT6 in Arabidopsis, bZIP members in different plant species. AtWRKY12 functions as an ABA-independent negative regulator of AtGSH1, while PCS expression is enhanced by PSE1. Moreover, PcABCC1.1 is activated by ABA and may participate in Pb sequestration in Gray Polar.
The members of the ABC transporter family mediate the transport of GSH- or PCs-conjugated toxic metals and metalloids for detoxification (Song et al., 2010; Park et al., 2012; Song et al., 2014b; Brunetti et al., 2015). The vacuolar membrane-localized Arabidopsis ABCC1 and ABCC2 mediate tolerance to both Cd and As by sequestrating the complexes into vacuoles, while ABCC3 only confers Cd detoxification (Song et al., 2010; Park et al., 2012; Brunetti et al., 2015). Knockout of OsABCC1 resulted in enhanced As sensitivity and largely increased As accumulation in rice grain, and ectopic expression of OsABCC1, ScYCF1 (Saccharomyces cerevisiae yeast cadmium factor 1, homolog of ABCC1 in yeast), and γ-ECS reduced grain As content by 70% compared to that of control by trapping AS-PCs into vacuoles of root cortical cells (Song et al., 2014b; Deng et al., 2018), indicating the critical role of OsABCC1 in As tolerance and reducing As distribution to grains. Overexpression of ScYCF1 in Arabidopsis enhanced tolerance and accumulation of Cd and Pb (Song et al., 2003). The mitochondrial transporter ABCB25 (ATM3) contributes to Cd and Pb resistance, possibly by transporting glutamine synthetase (GS)-conjugated complexes across the mitochondrial membrane (Kim et al., 2006). In addition to the upregulated expression of genes for PCs synthesis, the mRNA levels of the PcABCC1.1 was increased by exogenous ABA in Pb-exposed Gray Poplar (Figure 3) (Shi et al., 2019), indicating a possible enhanced Pb compartmentation activity of plants with ABA treatment.
Taken together, ABA alleviates heavy metals and metalloids' toxicity partially through increasing the generation of GSH and PCs as well as the vacuolar sequestration capacity of the conjugated complexes. PSE1 and transcriptional factors bZIPs, C2H2, and WRKYs are the likely candidates responsible for the ABA-induced GSH and PCs production. However, the evolutionary origin and conservation of this regulatory pattern in land plants and algae needs further evidence.
Evolution of Gene Families for ABA-Responsive Heavy Metals and Metalloids Detoxification
Growing evidence has revealed the evolutionarily conserved roles of ABA and its biosynthesis and signaling systems from bryophytes to angiosperms that cause them to survive and thrive in terrestrial conditions (Sakata et al., 2014; Cuming and Stevenson, 2015; Shinozawa et al., 2019). However, our understating of ABA signaling processes leading to heavy metals detoxification in non-angiosperms is still limited. Using a range of available tools in bioinformatics and plant evolutionary biology (Leebens-Mack et al., 2019; Zhao et al., 2019; Adem et al., 2020), we were able to glimpse the evolution of gene families for ABA-responsive heavy metals and metalloids' detoxification and trace the origin and co-evolution of ABA signaling and tolerance to metals and metalloids in plants.
ABA Biosynthesis and Signaling Network
As described above, physiological and genetic evidence reveals the involvement of ABA in plants resistance to Cd, As, and Pb stresses. Certain members belonging to ZEPs, NCEDs, AAOs, MOCOs, BGLUs, AITs (NPF4s), SnRK2s, and ABFs (bZIPs) are positive regulators, while PP2Cs play negative roles. Bioinformatics analyses were performed to identify the predicted gene families responsible for ABA biosynthesis, catabolism, transport, signal perception, and transduction in these 10 gene families across 41 species, including chlorophyte and streptophyte algae, red algae, and plants (Figure 4). All ZEPs, NCEDs, MOCOs, BGLUs, ABC transporters, and DTX transporters have been identified across most tested land plant and algal species (Figure 4), which showed the same pattern to those of SnRK and PP2C protein families (Cai et al., 2017; Chen et al., 2017). The orthologs of SDRs and AAOs were mainly identified in fern Azolla filiculoides and seed plants but not in any algae. NPFs were found in Chlorophyta Volvox carteri, all tested Streptophyte, and land plants, but not in red algae (Figure 4). Molecular and genetic evidence revealed that core ABA signaling networkz consisting of PYR/PYL/RCARs, PP2Cs, and SnRK2s of early land plants is comparable to that of Arabidopsis (Tougane et al., 2010; Takezawa et al., 2015; Bowman et al., 2017; Briskine et al., 2017; Cai et al., 2017; Eklund et al., 2018; Jahan et al., 2019; Shinozawa et al., 2019). As a result, the ABA signaling network evolved before the land plants.
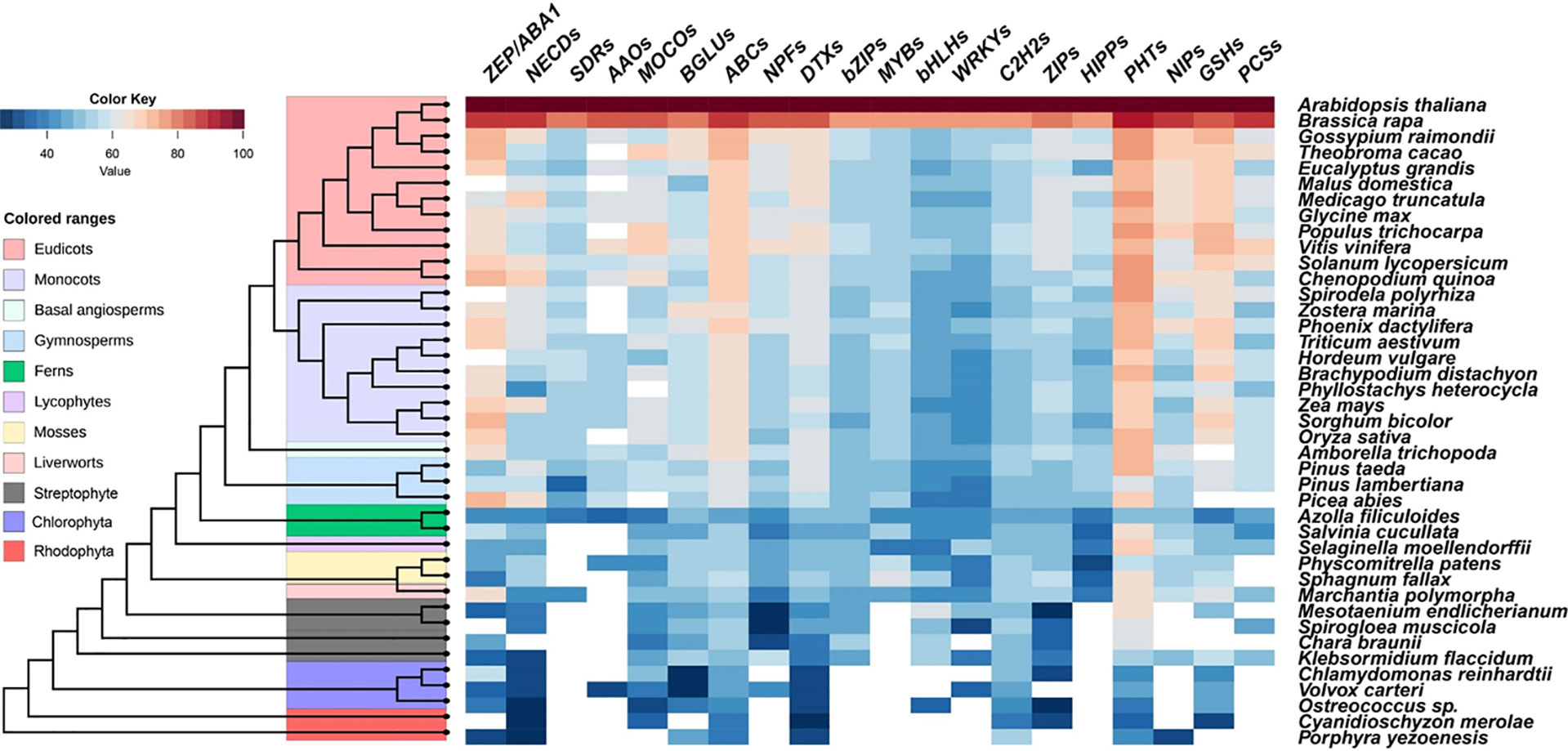
Figure 4 Similarity heat map of ABA signaling components and ABA-responsive heavy metals and metalloids-related proteins in different species. Candidate protein sequences were selected by BLASTP searches which satisfied E value <10-10 and query coverage >50%. Colored squares indicate protein sequence similarity from zero (Blue) to 100% (red). White squares indicate proteins that satisfied neither of the selection. ZEP, zeaxanthin epoxidase; NCEDs, 9-cis-epoxycarotenoid dioxygenase; SDRs, short-chain alcohol dehydrogenase/reductases; AAOs, abscisic aldehyde oxidases; MOCOs, molybdenum cofactor sulfurase; BGLUs, β-glucosidases; ABCs, ATP-binding cassette transporters; NPFs, Nitrate Transporter 1/Peptide Transporter; DTXs, DTX/Multidrug and Toxic Compound Extrusion (MATE); bZIPs, Basic region/leucine zipper proteins; MYBs, MYB transcriptional factors; bHLHs, basic helix-loop-helix transcriptional factors; WRKYs, WRKY transcriptional factors; C2H2s, Cysteine-2/Histidine-2 type zinc-finger transcriptional factors; ZIPs, Zinc Regulated Transporter/Iron Regulated Transporter-like Proteins; HIPPs, Heavy metal-associated Isoprenylated Plant Proteins; PHTs, Phosphate transporters; NIPs, Nodulin 26-like Intrinsic membrane Proteins; GSHs, glutathione synthetases; PCSs, Phytochelatin synthetases.
The above summarized experimental evidence revealed that ABFs belonging to bZIPs are the main regulators for Cd, As, and Pb uptake, distribution, and detoxification regulated by ABA signaling. The origin of land plant gene families that are relevant to ABA and metal and metalloid tolerance can be traced to Streptophyte algae with 7 members of bZIP transcriptional factors in the basal Streptophyta species Klebsormidium flaccidum and Chara braunii (Figure 4) (Cai et al., 2017; Chen et al., 2017; Zhao et al., 2019; Adem et al., 2020), indicating an early evolution of these gene families in Viridiplantae (green plants). The bZIPs gene family has since evolved and diversified into multiple members based on genome assembly of recently sequenced Streptophyte algae Spirogloea muscicola (Figure 4) in the Zygnematophyceae family (Cheng et al., 2019), which includes mosses, liverworts, lycophytes, ferns, gymnosperms, and angiosperms. For instance, there are 78 genes in the bZIP family in Arabidopsis thaliana (Jakoby et al., 2002) and the evolution of orthologues of bZIPs can be identified in the basal lineage of mosses: Sphagnum fallax in Sphagnopsida and Physcomitrella patens in Bryopsida (Figure 4).
Regulation of stomata and transpiration are vital for plants' tolerance to heavy metals and metalloids by reducing their uptake. Rapid stomatal responsiveness to fluctuant environmental stimulus is an essential adaptation to terrestrial plant life (Cai et al., 2017; Chen et al., 2017; Zhao et al., 2019). The opening and closing of stomata is predominantly dependent on ABA, which can be induced in plants under heavy metals and metalloids' stress (Amir et al., 2018; Chen et al., 2020). In flowering plants, stomata are directly regulated by ion flux mediated by several guard cell membrane-localized transporters, which are triggered by ABA in the Ca2+-dependent and Ca2+-independent pathway (Cai et al., 2017; Chen et al., 2017; Chen et al., 2019; Zhao et al., 2019; Chen et al., 2020). Comparative genomics and transcriptomics revealed that the ABA reception complex protein families including RCARs, PP2Cs, and SnRK2s, guard cell transporter families consisting of SLACs, KATs, and ALMTs, and kinases CDPKs and CIPKs critical for ABA-induced stomatal closure have been identified across the land plant species with stomata (Lind et al., 2015; Chater et al., 2016; Cai et al., 2017; Chen et al., 2017; Zhao et al., 2019). The physiological roles of certain members from liverworts, mosses, and ferns have been verified through genetic complementation tests (Tougane et al., 2010; Chater et al., 2011; Ruszala et al., 2011; Komatsu et al., 2013; Lind et al., 2015; Cai et al., 2017). The results reveal that the molecular mechanism underlying ABA-regulated stomatal aperture tends to be similar across plant lineages, but the stomata of vascular plants are generally more responsive to ABA than those in early plant species (Chen et al., 2017). In addition to ABA-regulated stomata responsiveness, the emergence of functional root and vascular tissues, which connect the various organs of plants and are essential for the long-distance transport of minerals, is indispensable for the Cd, As, and Pb distribution among tissues driven by ABA. Therefore, we propose that ABA is able to regulate heavy metals and metalloids' distribution in all vascular plant species, which of course remain to be investigated by the plant research community.
Regulation of ABA-Responsive Heavy Metals and Metalloids Detoxification
Some proteins responsible for the uptake, distribution, chelation, and compartmentation of Cd, As, and Pb are directly or indirectly regulated by ABA. Here, we identified the orthologues of transporter families including ZIPs, HIPPs, PHTs, NIPs, NPFs, and ABCs, transcriptional factor families consisting of bZIPs, MYBs, bHLHs, WRKYs, and C2H2s, enzymes for GSH and PCs synthesis from 41 plant and algal genomes (Figure 4).
In Arabidopsis, ABA is able to inhibit Cd uptake and accumulation mediated by IRT1, HIPP22, and HIPP44 at transcriptional levels (Zhang P. et al., 2019), while reducing endogenous ABA content increased the expression of IRT1 and its homologs, ZIP1 and ZIP4 (Lu et al., 2020a). IRT1 is a member of the ZIP family with board substrates including iron (Fe), zinc (Zn), cobalt, manganese (Mn), and Cd (Rogers et al., 2000; Vert et al., 2002). Homologs of ZIPs were identified in almost all examined algae and land plants, except in Rhodophyta Porphyra yezoenesis and Chlodophyta Volvox carteri (Figure 4), indicating the early arising of ZIPs prior to the evolution of land plants. There are 17 ZIPs in Arabidopsis, and IRT1-like members are only found in angiosperms (Lo et al., 2016). For example, five and four ZIPs were isolated from M. polymorpha and P. patens respectively; they were clustered into an IRT3-like group encoding Fe/Zn transporters in Arabidopsis and ZIP2-like subgroup transporting Zn, Fe, and Mn but not Cd (Lo et al., 2016). The activation of IRT1 by MYB49 requires Ib subgroup members of bHLHs to act as the bridge regulators, while the expression of HIPP22 and HIPP44 are directly regulated by MYB49 (Zhang P. et al., 2019). The origin of bHLHs can be traced to Chlorophyta, but MYBs and HIPPs appear to be land plant specific (Figure 4). Therefore, the analysis implicated that the ABI5-MYB49-bHLH-IRT1 pathway is likely to be conserved in land plants.
Arsenate uptake mediated by PHT1;1 can be restricted by WRKY6 transcriptional factor in Arabidopsis (Castrillo et al., 2013), while As(III) take up and distribution mediated by NIPs can be repressed by MYB protein OsARM1, whose expression is downregulated by ABA (Wang F.-Z. et al., 2017). Both As(V) and phosphate (P), which is an essential macro element for all organisms, are the substrates of PHTs (Lindsay and Maathuis, 2017). Four, eight, and twelve PHT orthologues are identified in Porphyra yezoenesis, Chlorophyta Ostreococcus sp., and Klebsormidium flaccidum, but the numbers are rapidly expanded to 25 in moss Physcomitrella patens, 26 in rice, and 19 in Arabidopsis (Figure 4). In addition, the PHT proteins show high similarity (over 60%) among all the land plants (Figure 4). Transcription factors WRKYs tend to be widespread in all the species examined, except Rhodophyta (Figure 4). The putative NIPs are found in the most ancient Rhodophyta Porphyra yezoenesis, basal Streptophyta Klebsormidium flaccidum, and all land plants examined (Figure 4). The origin of NIPs is found from horizontal gene transfer of bacterial aquaporin group with As efflux activity, and NIPs from Charophytes, mosses, and angiosperms are permeable to As (Pommerrenig et al., 2020). Moreover, Arabidopsis NPFs are divided into 8 subgroups (Léran et al., 2014), 4 NPF4s are ABA-importing transporters (Pan et al., 2020), while 2 NPF7s indirectly participate in ABA-altered Cd distribution in roots and shoots (Wang et al., 2018). Using 53 Arabidopsis NPFs as reference, we identified 1,990 orthologs from 36 representative genomes consisting of Chlorophyta Volvox carteri, Streptophyte algae, and land plants (Figure 4). Among the 2,398 putative NPFs identified from 33 genomes, 350 members were clustered into NPF4s and 206 belonged to NPF7s (Léran et al., 2014), which may be the candidates with ABA transport activity and tolerance to Cd induced by ABA in different plant species, respectively.
The ubiquitous thiol-containing small peptide, PCs, protect cells against the toxic effects of heavy metals and metalloids (Clemens, 2006) and the synthesis of PCs is catalyzed by PCS using reduced GSH and related thiols as substrates (Cobbett, 1999). Genes encoding GSHs can be traced to an ancestral streptophyte alga Klebsormidium flaccidum (also found in Cyanidioschyzon merolae, an unicellular red alga the phylum Rhodophyta), and the similarity among the members from monocots and dicots is over 60% (Figure 4). The potential PCS orthologs are found in the Klebsormidium flaccidum, Spirodela polyrhiza, liverwort Marchantia polymorpha, and most vascular plants. However, the activities and responsiveness to various metals of PCS are divergent in various kinds of plants. In general, the PCSs of basal plants appear to be less active compared with Arabidopsis PCS (Degola et al., 2014; Petraglia et al., 2014). Expression of GSHs and PCSs could be activated by ABA-induced C2H2 transcriptional factor AtZAT6 and StbZIP but repressed by AtWRKY12 (Stroiński et al., 2013; Chen J. et al., 2016; Han et al., 2019). We found that the C2H2 member family shows the same evolutionary origin as that of GSHs, whereas WRKYs may be originated from Chlorophyta (Figure 4). Transcriptional factors belonging to the bZIP subfamily activates PCS transcription in potato and Gray Poplar subjected to exogenous ABA (Stroiński et al., 2010; Stroiński et al., 2013; Shi et al., 2019), which is consistent with the parallel evolution of bZIPs and PCSs from Streptophyte algae (Figure 4). Further experiments have to be conducted to test the conservation of these regulatory modules in different plant species.
ABC transporters are important for ABA signaling and responses to heavy metals and metalloids. For instance, ABCB25 has a role in Cd and Pb resistance in Arabidopsis and C-type ABC transporters are involved in sequestration of GSH- and PC-conjugated heavy metals and metalloids in various angiosperms. G-type ABC transporters contribute to ABA transport as well as Cd and Pb efflux (Song et al., 2014a; Hwang et al., 2016). We found that ABC transporters are ubiquitous in all 41 genomes used for comparative genetic analysis (Figure 4). There are 69, 125, 130, and 133 ABC transporters in green algae Chlamydomonas reinhardtii, moss Physcomitrella patens, Arabidopsis, and rice, respectively (Hwang et al., 2016). In Rhodophyta Porphyra yezoenesis, 9, 3, and 2 ABC transporter homologs are classified into B-, C-, and G-type subgroups, respectively. The numbers of B-type, C-type, and G-type members increased substantially to 20, 14, and 42, in Physcomitrella paten and 30, 17, and 43 and Arabidopsis, respectively (Hwang et al., 2016). These are the suggested specific proliferations of ABCB, ABCC, and ABCG subfamily members in land plants. Therefore, it is interesting to verify the evolutionary conservation of the functions of ABC transporters for heavy metals and metalloids' detoxification in the early divergent lineages of plant species.
Conclusions and Future Perspectives
In summary, in the key components of the ABA biosynthesis, signaling perception, and transduction pathways, regulatory patterns were evolutionary conserved in land plants but also diversified in different lineages. This may be the case for mechanisms underlying the plant response to toxic metals and metalloids. According to the analyses, we propose that: (1) ABA-repressed Cd uptake mediated by ABI5-MYB49-HIPPs network may be conserved in land plants, (2) ABA-reduced As distribution and detoxification through MYB-NIPs is likely to be conserved in land plants, (3) GSH for metal and metalloid chelation ABA-induced C2H2 transcriptional factor is likely in green plants, and (4) the earliest origin of bZIP-induced PCS can be traced to the Streptophyta (Figure 5). Therefore, future work is suggested to focus on: (1) the discovery of direct regulators of the ABA-responsive transcriptional factors including WRKY6, ZAT6, ARM1, and bZIPs and downstream transporters comprising of ABCs, HMAs, ZIPs, Nramps, NRT1.5, V-ATPase, and V-PPase; (2) characterization of the putative Cd, As, and Pb transporters in major clades of land plants using heterologous expression systems such as yeast and Xenopus laevis; and (3) investigation of the roles of heavy metal and metalloid stress-related and ABA-regulated components of major clades of land plants via genetic complementation of these genes in corresponding mutants of Arabidopsis and rice. The proposed research will shed light on the practices for mitigation the contaminations. For instance, application of ABA or its analogues in crops for diminishing the accumulation of toxic metals and metalloids and their antagonists can be employed in the hyperaccumulators (e.g. algae, plants) for phytoremediation.
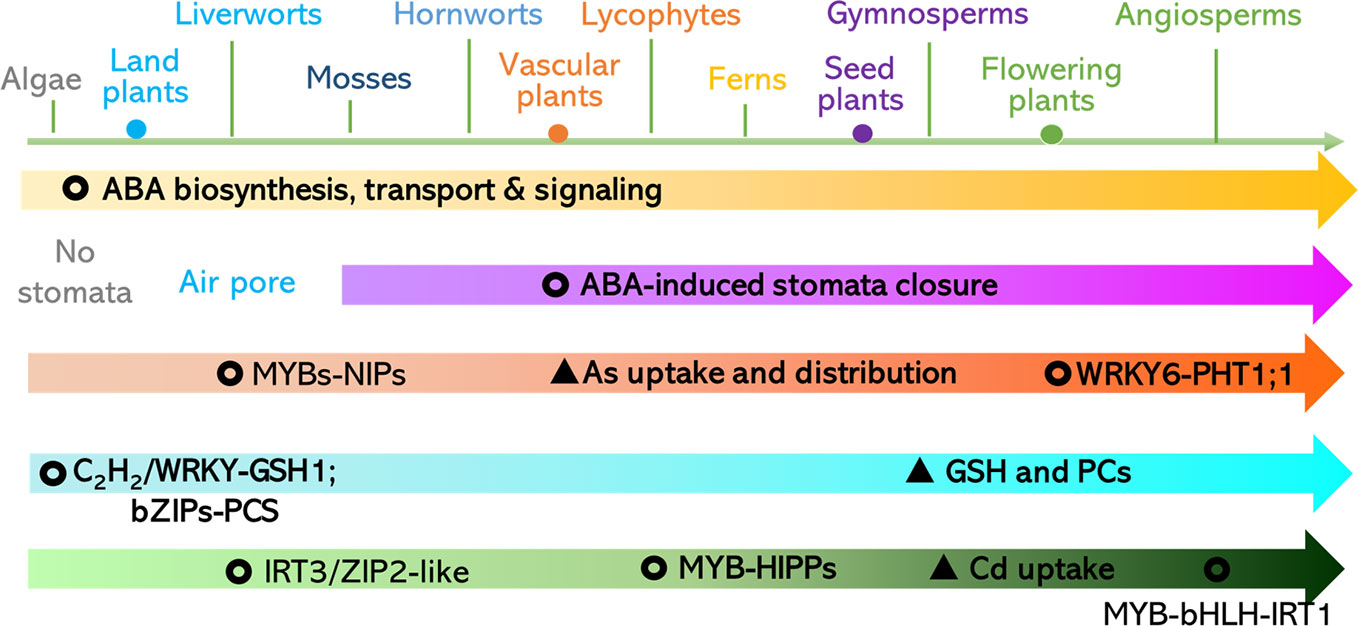
Figure 5 Proposed evolution of ABA-responsive pathways for toxic metals and metalloids uptake, distribution, and detoxification. Please refer to the section of Conclusions and Future Perspectives for a detailed description.
Author Contributions
Z-HC and FD conceptualized the review. FD, BH, and Z-HC wrote the manuscript with support from XC, WG, LL, JX. GC conducted the comparative genomics analyses and prepared Figure 4. FD, BH, and Z-HC did final editing of the manuscript. BH and FD have contributed equally to this work.
Funding
This work was financially supported by the Engineering Research Center of Ecology and Agricultural Use of Wetland, Ministry of Education and Hubei Key Laboratory of Waterlogging Disaster and Agricultural Use of Wetland (KF201908), and funding from the State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources (SKLCUSA-b201910).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adem, G. D., Chen, G., Shabala, L., Chen, Z. H., Shabala, S. (2020). GORK channel: a master switch of plant metabolism? Trends Plant Sci. 25, 434–445. doi: 10.1016/j.tplants.2019.12.012
Amir, R., Taufiq, S., Noor, N., Nauman, I., Munir, F., Keyani, R., et al. (2018). “Stress signaling under metal and metalloid toxicity,” in Plants under metal and metalloid stress: responses, tolerance and remediation. Eds. Hasanuzzaman, M., Nahar, K., Fujita, M. (Singapore: Springer Singapore), 149–184.
Andersen, M. M., Landes, X., Xiang, W., Anyshchenko, A., Falhof, J., Osterberg, J. T., et al. (2015). Feasibility of new breeding techniques for organic farming. Trends Plant Sci. 20, 426–434. doi: 10.1016/j.tplants.2015.04.011
Atici, Ö., Ağar, G., Battal, P. (2005). Changes in phytohormone contents in chickpea seeds germinating under lead or zinc stress. Biol. Plant. 49, 215–222. doi: 10.1007/s10535-005-5222-9
Auguy, F., Fahr, M., Moulin, P., El Mzibri, M., Smouni, A., Filali-Maltouf, A., et al. (2016). Transcriptome changes in Hirschfeldia incana in response to lead exposure. Front. Plant Sci. 6, 1231. doi: 10.3389/fpls.2015.01231
Babla, M., Cai, S., Chen, G., Tissue, D. T., Cazzonelli, C. I., Chen, Z. H. (2019). Molecular evolution and interaction of membrane transport and photoreception in plants. Front. Genet. 10, 956. doi: 10.3389/fgene.2019.00956
Bowell, R. J., Alpers, C. N., Jamieson, H. E., Nordstrom, D. K., Majzlan, J. (2014). The environmental geochemistry of arsenic—An Overview. Rev. Mineral. Geochem. 79, 1–16. doi: 10.2138/rmg.2014.79.1
Bowman, J. L., Kohchi, T., Yamato, K. T., Jenkins, J., Shu, S., Ishizaki, K., et al. (2017). Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171, 287–304. doi: 10.1016/j.cell.2017.09.030
Briskine, R. V., Paape, T., Shimizu-Inatsugi, R., Nishiyama, T., Akama, S., Sese, J., et al. (2017). Genome assembly and annotation of Arabidopsis halleri, a model for heavy metal hyperaccumulation and evolutionary ecology. Mol. Ecol. Resour. 17, 1025–1036. doi: 10.1111/1755-0998.12604
Brunetti, P., Zanella, L., De Paolis, A., Di Litta, D., Cecchetti, V., Falasca, G., et al. (2015). Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin-mediated cadmium tolerance in Arabidopsis. J. Exp. Bot. 66, 3815–3829. doi: 10.1093/jxb/erv185
Cai, S., Chen, G., Wang, Y., Huang, Y., Marchant, D. B., Wang, Y., et al. (2017). Evolutionary conservation of ABA signaling for stomatal closure. Plant Physiol. 174, 732–747. doi: 10.1104/pp.16.01848
Cao, Y., Sun, D., Ai, H., Mei, H., Liu, X., Sun, S., et al. (2017). Knocking out OsPT4 gene decreases arsenate uptake by rice plants and inorganic arsenic accumulation in rice rains. Environ. Sci. Technol. 51, 12131–12138. doi: 10.1021/acs.est.7b03028
Cao, Y., Feng, H., Sun, D., Xu, G., Rathinasabapathi, B., Chen, Y., et al. (2019). Heterologous expression of Pteris vittata phosphate transporter PvPht1;3 enhances arsenic translocation to and accumulation in tbacco shoots. Environ. Sci. Technol. 53, 10636–10644. doi: 10.1021/acs.est.9b02082
Castrillo, G., Sánchez-Bermejo, E., de Lorenzo, L., Crevillén, P., Fraile-Escanciano, A., TC, M., et al. (2013). WRKY6 transcription factor restricts arsenate uptake and transposon activation in Arabidopsis. Plant Cell 25, 2944–2957. doi: 10.1105/tpc.113.114009
Chao, D.-Y., Silva, A., Baxter, I., Huang, Y. S., Nordborg, M., Danku, J., et al. (2012). Genome-wide association studies identify heavy metal ATPase3 as the primary determinant of natural variation in leaf cadmium in Arabidopsis thaliana. PLoS Genet. 8, e1002923. doi: 10.1371/journal.pgen.1002923
Chater, C., Kamisugi, Y., Movahedi, M., Fleming, A., Cuming, A. C., Gray, J. E., et al. (2011). Regulatory mechanism controlling stomatal behavior conserved across 400 million years of land plant evolution. Curr. Biol. 21, 1025–1029. doi: 10.1016/j.cub.2011.04.032
Chater, C. C., Caine, R. S., Tomek, M., Wallace, S., Kamisugi, Y., Cuming, A. C., et al. (2016). Origin and function of stomata in the moss Physcomitrella patens. Nat. Plants 2, 16179. doi: 10.1038/nplants.2016.179
Chen, J., Yang, L., Yan, X., Liu, Y., Wang, R., Fan, T., et al. (2016). Zinc-Finger transcription factor ZAT6 positively regulates cadmium Tolerance through the glutathione-dependent pathway in Arabidopsis. Plant Physiol. 171, 707–719. doi: 10.1104/pp.15.01882
Chen, Z. H., Wang, Y., Wang, J. W., Babla, M., Zhao, C., Garcia-Mata, C., et al. (2016). Nitrate reductase mutation alters potassium nutrition as well as nitric oxide-mediated control of guard cell ion channels in Arabidopsis. New Phytol. 209, 1456–1469. doi: 10.1111/nph.13714
Chen, Z. H., Chen, G., Dai, F., Wang, Y., Hills, A., Ruan, Y. L., et al. (2017). Molecular evolution of grass stomata. Trends Plant Sci. 22, 124–139. doi: 10.1016/j.tplants.2016.09.005
Chen, G., Wang, Y., Wang, X., Yang, Q., Quan, X., Zeng, J., et al. (2019). Leaf epidermis transcriptome reveals drought-Induced hormonal signaling for stomatal regulation in wild barley. Plant Growth Regul. 87, 39–54. doi: 10.1007/s10725-018-0450-0
Chen, K., Li, G. J., Bressan, R. A., Song, C. P., Zhu, J. K., Zhao, Y. (2020). Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 62, 25–54. doi: 10.1111/jipb.12899
Cheng, S., Xian, W., Fu, Y., Marin, B., Keller, J., Wu, T., et al. (2019). Genomes of subaerial Zygnematophyceae provide insights into land plant evolution. Cell 179, 1057–1067.e1014. doi: 10.1016/j.cell.2019.10.019
Chmielowska-Bak, J., Gzyl, J., Rucinska-Sobkowiak, R., Arasimowicz-Jelonek, M., Deckert, J. (2014). The new insights into cadmium sensing. Front. Plant Sci. 5:245. doi: 10.3389/fpls.2014.00245
Choi, H. I., Hong, J. H., Ha, J. O., Kang, J. Y., Kim, S. Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275, 1723–1730. doi: 10.1074/jbc.275.3.1723
Clemens, S., Ma, J. F. (2016). Toxic heavy metal and metalloid accumulation in crop plants and foods. Annu. Rev. Plant Biol. 67, 489–512. doi: 10.1146/annurev-arplant-043015-112301
Clemens, S., Kim, E. J., Neumann, D., Schroeder, J. I. (1999). Tolerance to toxic metals by a gene family of phytochelatin synthases from plants and yeast. EMBO J. 18, 3325–3333. doi: 10.1093/emboj/18.12.3325
Clemens, S. (2006). Evolution and function of phytochelatin synthases. J. Plant Physiol. 163, 319–332. doi: 10.1016/j.jplph.2005.11.010
Cobbett, C. S. (1999). A family of phytochelatin synthase genes from plant, fungal and animal species. Trends Plant Sci. 4, 335–337. doi: 10.1016/S1360-1385(99)01465-X
Cobbett, C. S. (2000). Phytochelatins and their roles in heavy metal detoxification. Plant Physiol. 123, 825–832. doi: 10.1104/pp.123.3.825
Cuming, A. C., Stevenson, S. R. (2015). From pond slime to rain forest: the evolution of ABA signalling and the acquisition of dehydration tolerance. New Phytol. 206, 5–7. doi: 10.1111/nph.13333
Degola, F., De Benedictis, M., Petraglia, A., Massimi, A., Fattorini, L., Sorbo, S., et al. (2014). A Cd/Fe/Zn-responsive phytochelatin synthase is constitutively present in the ancient liverwort Lunularia cruciata (L.) dumort. Plant Cell Physiol. 55, 1884–1891. doi: 10.1093/pcp/pcu117
Deng, F., Liu, X., Chen, Y., Rathinasabapathi, B., Rensing, C., Chen, J., et al. (2020). Aquaporins mediated arsenite transport in plants: Molecular mechanisms and applications in crop improvement. Crit. Rev. Env. Sci. Tec. 50, 1613–1639. doi: 10.1080/10643389.2019.1662704
Deng, F., Yamaji, N., Ma, J. F., Lee, S. K., Jeon, J. S., Martinoia, E., et al. (2018). Engineering rice with lower grain arsenic. Plant Biotechnol. J. 16, 1691–1699. doi: 10.1111/pbi.12905
Deng, F., Yu, M., Martinoia, E., Song, W. Y. (2019). Ideal cereals with lower arsenic and cadmium by accurately enhancing vacuolar sequestration capacity. Front. Genet. 10:322. doi: 10.3389/fgene.2019.00322
DiTusa, S. F., Fontenot, E. B., Wallace, R. W., Silvers, M. A., Steele, T. N., Elnagar, A. H., et al. (2016). A member of the Phosphate transporter 1 (Pht1) family from the arsenic-hyperaccumulating fern Pteris vittata is a high-affinity arsenate transporter. New Phytol. 209, 762–772. doi: 10.1111/nph.13472
Eklund, D. M., Kanei, M., Flores-Sandoval, E., Ishizaki, K., Nishihama, R., Kohchi, T., et al. (2018). An evolutionarily conserved abscisic acid signaling pathway regulates dormancy in the liverwort Marchantia polymorpha. Curr. Biol. 28, 3691–3699.e3693. doi: 10.1016/j.cub.2018.10.018
Fan, S. K., Fang, X. Z., Guan, M. Y., Ye, Y. Q., Lin, X. Y., Du, S. T., et al. (2014). Exogenous abscisic acid application decreases cadmium accumulation in Arabidopsis plants, which is associated with the inhibition of IRT1-mediated cadmium uptake. Front. Plant Sci. 5, 721. doi: 10.3389/fpls.2014.00721
Fan, T., Yang, L., Wu, X., Ni, J., Jiang, H., Zhang, Q. A., et al. (2016). The PSE1 gene modulates lead tolerance in Arabidopsis. J. Exp. Bot. 67, 4685–4695. doi: 10.1093/jxb/erw251
Fan, W., Guo, Q., Liu, C., Liu, X., Zhang, M., Long, D., et al. (2018). Two mulberry phytochelatin synthase genes confer zinc/cadmium tolerance and accumulation in transgenic Arabidopsis and tobacco. Gene 645, 95–104. doi: 10.1016/j.gene.2017.12.042
Feng, X., Liu, W., Qiu, C.-W., Zeng, F., Wang, Y., Zhang, G., et al. (2020). HvAKT2 and HvHAK1 confer drought tolerance in barley through enhanced leaf mesophyll H+ homoeostasis. Plant Biotechnol. J. 18, 1683–1696. doi: 10.1111/pbi.13332
Finkelstein, R. (2013). Abscisic acid synthesis and response. Arabidopsis book 11, e0166–e0166. doi: 10.1199/tab.0166
Fu, S. F., Chen, P. Y., Nguyen, Q. T., Huang, L. Y., Zeng, G. R., Huang, T. L., et al. (2014). Transcriptome profiling of genes and pathways associated with arsenic toxicity and tolerance in Arabidopsis. BMC Plant Biol. 14, 94. doi: 10.1186/1471-2229-14-94
Fu, S., Lu, Y., Zhang, X., Yang, G., Chao, D., Wang, Z., et al. (2019). The ABC transporter ABCG36 is required for cadmium tolerance in rice. J. Exp. Bot. 70, 5909–5918. doi: 10.1093/jxb/erz335
Fujii, H., Chinnusamy, V., Rodrigues, A., Rubio, S., Antoni, R., Park, S. Y., et al. (2009). In vitro reconstitution of an abscisic acid signalling pathway. Nature 462, 660–664. doi: 10.1038/nature08599
Gong, J. M., Lee, D. A., Schroeder, J. I. (2003). Long-distance root-to-shoot transport of phytochelatins and cadmium in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 100, 10118–10123. doi: 10.1073/pnas.1734072100
Guo, C., Yao, L., You, C., Wang, S., Cui, J., Ge, X., et al. (2016). MID1 plays an important role in response to drought stress during reproductive development. Plant J. 88, 280–293. doi: 10.1111/tpj.13250
Gupta, M. K., Lenka, S. K., Gupta, S., Rawal, R. K. (2020). Agonist, antagonist and signaling modulators of ABA receptor for agronomic and post-harvest management. Plant Physiol. Biochem. 148, 10–25. doi: 10.1016/j.plaphy.2019.12.023
Han, Y., Fan, T., Zhu, X., Wu, X., Ouyang, J., Jiang, L., et al. (2019). WRKY12 represses GSH1 expression to negatively regulate cadmium tolerance in Arabidopsis. Plant Mol. Biol. 99, 149–159. doi: 10.1007/s11103-018-0809-7
Hanikenne, M., Talke, I. N., Haydon, M. J., Lanz, C., Nolte, A., Motte, P., et al. (2008). Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature 453, 391–395. doi: 10.1038/nature06877
Hauser, F., Waadt, R., Schroeder, J. I. (2011). Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 21, 346–355. doi: 10.1016/j.cub.2011.03.015
Hauser, F., Li, Z., Waadt, R., Schroeder, J. I. (2017). SnapShot: abscisic acid signaling. Cell 171, 1708–1708.e1700. doi: 10.1016/j.cell.2017.11.045
Hayashi, S., Kuramata, M., Abe, T., Takagi, H., Ozawa, K., Ishikawa, S. (2017). Phytochelatin synthase OsPCS1 plays a crucial role in reducing arsenic levels in rice grains. Plant J. 91, 840–848. doi: 10.1111/tpj.13612
Hsu, Y., Kao, C. (2003). Role of abscisic acid in cadmium tolerance of rice (Oryza sativa L.) seedlings. Plant Cell Environ. 26, 867–874. doi: 10.1046/j.1365-3040.2003.01018.x
Huang, T. L., Nguyen, Q. T. T., Fu, S.-F., Lin, C.-Y., Chen, Y., Huang, H. J. (2012). Transcriptomic changes and signalling pathways induced by arsenic stress in rice roots. Plant Mol. Biol. 80, 587–608. doi: 10.1007/s11103-012-9969-z
Huang, Y., Feng, C.-Z., Ye, Q., Wu, W.-H., Chen, Y.-F. (2016). Arabidopsis WRKY6 transcription factor acts as a positive pegulator of abscisic acid signaling during seed germination and early seedling development. PLoS Genet. 12, e1005833–e1005833. doi: 10.1371/journal.pgen.1005833
Hussain, D., Haydon, M. J., Wang, Y., Wong, E., Sherson, S. M., Young, J., et al. (2004). P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell 16, 1327–1339. doi: 10.1105/tpc.020487
Hwang, J.-U., Song, W.-Y., Hong, D., Ko, D., Yamaoka, Y., Jang, S., et al. (2016). Plant ABC transporters enable many unique aspects of a terrestrial plant's lifestyle. Mol. Plant 9, 338–355. doi: 10.1016/j.molp.2016.02.003
Ishikawa, S., Ishimaru, Y., Igura, M., Kuramata, M., Abe, T., Senoura, T., et al. (2012). Ion-beam irradiation, gene identification, and marker-assisted breeding in the development of low-cadmium rice. Proc. Natl. Acad. Sci. U. S. A. 109, 19166–19171. doi: 10.1073/pnas.1211132109
Jahan, A., Komatsu, K., Wakida-Sekiya, M., Hiraide, M., Tanaka, K., Ohtake, R., et al. (2019). Archetypal roles of an Abscisic Acid receptor in drought and sugar responses in Liverworts. Plant Physiol. 179, 317–328. doi: 10.1104/pp.18.00761
Jakoby, M., Weisshaar, B., Droge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. doi: 10.1016/s1360-1385(01)02223-3
Joshi-Saha, A., Valon, C., Leung, J. (2011). Abscisic acid signal off the STARting block. Mol. Plant 4, 562–580. doi: 10.1093/mp/ssr055
Kamiya, T., Islam, R., Duan, G., Uraguchi, S., Fujiwara, T. (2013). Phosphate deficiency signaling pathway is a target of arsenate and phosphate transporter OsPT1 is involved in As accumulation in shoots of rice. Soil Sci. Plant Nutr. 59, 580–590. doi: 10.1080/00380768.2013.804390
Kang, J., Hwang, J. U., Lee, M., Kim, Y. Y., Assmann, S. M., Martinoia, E., et al. (2010). PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. U. S. A. 107, 2355–2360. doi: 10.1073/pnas.0909222107
Kang, J., Yim, S., Choi, H., Kim, A., Lee, K. P., Lopez-Molina, L., et al. (2015). Abscisic acid transporters cooperate to control seed germination. Nat. Commun. 6, 8113. doi: 10.1038/ncomms9113
Kanno, Y., Hanada, A., Chiba, Y., Ichikawa, T., Nakazawa, M., Matsui, M., et al. (2012). Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl. Acad. Sci. U. S. A. 109, 9653–9658. doi: 10.1073/pnas.1203567109
Katsuta, S., Masuda, G., Bak, H., Shinozawa, A., Kamiyama, Y., Umezawa, T., et al. (2020). Arabidopsis Raf-like kinases act as positive regulators of subclass III SnRK2 in osmostress signaling. Plant J. doi: 10.1111/tpj.14756
Kim, D.-Y., Bovet, L., Kushnir, S., Noh, E. W., Martinoia, E., Lee, Y. (2006). AtATM3 is involved in heavy metal resistance in Arabidopsis. Plant Physiol. 140, 922–932. doi: 10.1104/pp.105.074146
Kim, D. Y., Bovet, L., Maeshima, M., Martinoia, E., Lee, Y. (2007). The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 50, 207–218. doi: 10.1111/j.1365-313X.2007.03044.x
Kitahata, N., Asami, T. (2011). Chemical biology of abscisic acid. J. Plant Res. 124, 549–557. doi: 10.1007/s10265-011-0415-0
Komatsu, K., Suzuki, N., Kuwamura, M., Nishikawa, Y., Nakatani, M., Ohtawa, H., et al. (2013). Group A PP2Cs evolved in land plants as key regulators of intrinsic desiccation tolerance. Nat. Commun. 4, 2219. doi: 10.1038/ncomms3219
Kulik, A., Anielska-Mazur, A., Bucholc, M., Koen, E., Szymańska, K., Żmieńko, A., et al. (2012). SNF1-related protein kinases type 2 are involved in plant responses to cadmium stress. Plant Physiol. 160, 868–883. doi: 10.1104/pp.112.194472
Kuromori, T., Miyaji, T., Yabuuchi, H., Shimizu, H., Sugimoto, E., Kamiya, A., et al. (2010). ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc. Natl. Acad. Sci. U. S. A. 107, 2361–2366. doi: 10.1073/pnas.0912516107
Kuromori, T., Sugimoto, E., Shinozaki, K. (2011). Arabidopsis mutants of AtABCG22, an ABC transporter gene, increase water transpiration and drought susceptibility. Plant J. 67, 885–894. doi: 10.1111/j.1365-313X.2011.04641.x
Kuromori, T., Seo, M., Shinozaki, K. (2018). ABA transport and plant water stress responses. Trends Plant Sci. 23, 513–522. doi: 10.1016/j.tplants.2018.04.001
Léran, S., Varala, K., Boyer, J.-C., Chiurazzi, M., Crawford, N., Daniel-Vedele, F., et al. (2014). A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 19, 5–9. doi: 10.1016/j.tplants.2013.08.008
Landrigan, P. J., Fuller, R., Acosta, N. J. R., Adeyi, O., Arnold, R., Basu, N., et al. (2018). The Lancet Commission on pollution and health. Lancet 391, 462–512. doi: 10.1016/S0140-6736(17)32345-0
Lee, M., Lee, K., Lee, J., Noh, E. W., Lee, Y. (2005). AtPDR12 contributes to lead resistance in Arabidopsis. Plant Physiol. 138, 827–836. doi: 10.1104/pp.104.058107
Leebens-Mack, J. H., Barker, M. S., Carpenter, E. J., Deyholos, M. K., Gitzendanner, M. A., Graham, S. W., et al. (2019). One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574, 679–685. doi: 10.1038/s41586-019-1693-2
Li, J.-Y., Fu, Y.-L., Pike, S. M., Bao, J., Tian, W., Zhang, Y., et al. (2010). The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22, 1633–1646. doi: 10.1105/tpc.110.075242
Lin, S.-H., Kuo, H.-F., Canivenc, G., Lin, C.-S., Lepetit, M., Hsu, P.-K., et al. (2008). Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20, 2514–2528. doi: 10.1105/tpc.108.060244
Lin, Z., Li, Y., Zhang, Z., Liu, X., Hsu, C. C., Du, Y., et al. (2020). A RAF-SnRK2 kinase cascade mediates early osmotic stress signaling in higher plants. Nat. Commun. 11, 613. doi: 10.1038/s41467-020-14477-9
Lind, C., Dreyer, I., López-Sanjurjo, E. J., von Meyer, K., Ishizaki, K., Kohchi, T., et al. (2015). Stomatal guard cells co-opted an ancient ABA-dependent desiccation survival system to regulate stomatal closure. Curr. Biol. 25, 928–935. doi: 10.1016/j.cub.2015.01.067
Lindsay, E. R., Maathuis, F. J. M. (2017). New molecular mechanisms to reduce arsenic in crops. Trends Plant Sci. 22, 1016–1026. doi: 10.1016/j.tplants.2017.09.015
Liu, H., Zhao, H., Wu, L., Liu, A., Zhao, F.-J., Xu, W. (2017). Heavy metal ATPase 3 (HMA3) confers cadmium hypertolerance on the cadmium/zinc hyperaccumulator Sedum plumbizincicola. New Phytol. 215, 687–698. doi: 10.1111/nph.14622
Liu, X., Fan, Y., Mak, M., Babla, M., Holford, P., Wang, F., et al. (2017). QTLs for stomatal and photosynthetic traits related to salinity tolerance in barley. BMC Genomics 18, 9–9. doi: 10.1186/s12864-016-3380-0
Liu, C. L., Gao, Z. Y., Shang, L. G., Yang, C. H., Ruan, B. P., Zeng, D. L., et al. (2019). Natural variation in the promoter of OsHMA3 contributes to differential grain cadmium accumulation between Indica and Japonica rice. J. Integr. Plant Biol. 62, 314–329. doi: 10.1111/jipb.12794
Lo, J.-C., Tsednee, M., Lo, Y.-C., Yang, S.-C., Hu, J.-M., Ishizaki, K., et al. (2016). Evolutionary analysis of iron (Fe) acquisition system in. Marchantia polymorpha. New Phytol. 211, 569–583. doi: 10.1111/nph.13922
Lu, C., Zhang, L., Tang, Z., Huang, X.-Y., Ma, J. F., Zhao, F.-J. (2019). Producing cadmium-free Indica rice by overexpressing OsHMA3. Environ. Int. 126, 619–626. doi: 10.1016/j.envint.2019.03.004
Lu, Q., Weng, Y., You, Y., Xu, Q., Li, H., Li, Y., et al. (2020a). Inoculation with abscisic acid (ABA)-catabolizing bacteria can improve phytoextraction of heavy metal in contaminated soil. Environ. Pollut. 257, 113497. doi: 10.1016/j.envpol.2019.113497
Lu, Q., Chen, S., Li, Y., Zheng, F., He, B., Gu, M. (2020b). Exogenous abscisic acid (ABA) promotes cadmium (Cd) accumulation in Sedum alfredii Hance by regulating the expression of Cd stress response genes. Environ. Sci.Pollut. Res. 27, 8719–8731. doi: 10.1007/s11356-019-07512-w
Lux, A., Martinka, M., Vaculík, M., White, P. J. (2011). Root responses to cadmium in the rhizosphere: a review. J. Exp. Bot. 62, 21–37. doi: 10.1093/jxb/erq281
Ma, L. Q., Komar, K. M., Tu, C., Zhang, W., Cai, Y., Kennelley, E. D. (2001). A fern that hyperaccumulates arsenic. Nature 409, 579–579. doi: 10.1038/35054664
Ma, J. F., Yamaji, N., Mitani, N., Xu, X.-Y., Su, Y.-H., McGrath, S. P., et al. (2008). Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proc. Natl. Acad. Sci. U. S. A. 105, 9931–9935. doi: 10.1073/pnas.0802361105
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., et al. (2009). Regulators of PP2C phosphatase activity Function as abscisic acid sensors. Science 324, 1064–1068. doi: 10.1126/science.1172408
Maestri, E., Marmiroli, M. (2012). “Genetic and molecular aspects of metal tolerance and hyperaccumulation,” in Metal Toxicity in Plants: Perception, Signaling and Remediation. Eds. Gupta, D. K., Sandalio, L. M. (Berlin, Heidelberg: Springer Berlin Heidelberg), 41–63.
Mak, M., Zhang, M., Randall, D., Holford, P., Milham, P., Wu, F., et al. (2019). Chloride transport at plant-soil Interface modulates barley Cd tolerance. Plant Soil 441, 409–421. doi: 10.1007/s11104-019-04134-6
McLoughlin, F., Galvan-Ampudia, C. S., Julkowska, M. M., Caarls, L., van der Does, D., Laurière, C., et al. (2012). The Snf1-related protein kinases SnRK2.4 and SnRK2.10 are involved in maintenance of root system architecture during salt stress. Plant J. 72, 436–449. doi: 10.1111/j.1365-313X.2012.05089.x
Mega, R., Abe, F., Kim, J.-S., Tsuboi, Y., Tanaka, K., Kobayashi, H., et al. (2019). Tuning water-use efficiency and drought tolerance in wheat using abscisic acid receptors. Nat. Plants 5, 153–159. doi: 10.1038/s41477-019-0361-8
Migocka, M., Papierniak, A., Maciaszczyk-Dziubinska, E. W. A., Posyniak, E., Kosieradzka, A. (2015). Molecular and biochemical properties of two P1B2-ATPases, CsHMA3 and CsHMA4, from cucumber. Plant Cell Environ. 38, 1127–1141. doi: 10.1111/pce.12447
Mills, R. F., Francini, A., Ferreira da Rocha, P. S. C., Baccarini, P. J., Aylett, M., Krijger, G. C., et al. (2005). The plant P1B-type ATPase AtHMA4 transports Zn and Cd and plays a role in detoxification of transition metals supplied at elevated levels. FEBS Lett. 579, 783–791. doi: 10.1016/j.febslet.2004.12.040
Mills, R. F., Peaston, K. A., Runions, J., Williams, L. E. (2012). HvHMA2, a P1B-ATPase from barley, is highly conserved among cereals and functions in Zn and Cd transport. PLoS One 7, e42640. doi: 10.1371/journal.pone.0042640
Mittler, R., Blumwald, E. (2015). The roles of ROS and ABA in systemic acquired acclimation. Plant Cell 27, 64–70. doi: 10.1105/tpc.114.133090
Miyadate, H., Adachi, S., Hiraizumi, A., Tezuka, K., Nakazawa, N., Kawamoto, T., et al. (2011). OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytol. 189, 190–199. doi: 10.1111/j.1469-8137.2010.03459.x
Miyakawa, T., Fujita, Y., Yamaguchi-Shinozaki, K., Tanokura, M. (2013). Structure and function of abscisic acid receptors. Trends Plant Sci. 18, 259–266. doi: 10.1016/j.tplants.2012.11.002
Morel, M., Crouzet, J., Gravot, A., Auroy, P., Leonhardt, N., Vavasseur, A., et al. (2009). AtHMA3, a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiol. 149, 894–904. doi: 10.1104/pp.108.130294
Nakanishi, H., Ogawa, I., Ishimaru, Y., Mori, S., Nishizawa, N. K. (2006). Iron deficiency enhances cadmium uptake and translocation mediated by the Fe2+ transporters OsIRT1 and OsIRT2 in rice. Soil Sci. Plant Nutr. 52, 464–469. doi: 10.1111/j.1747-0765.2006.00055.x
Nambara, E., Marion-Poll, A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56, 165–185. doi: 10.1146/annurev.arplant.56.032604.144046
North, H. M., De Almeida, A., Boutin, J. P., Frey, A., To, A., Botran, L., et al. (2007). The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J. 50, 810–824. doi: 10.1111/j.1365-313X.2007.03094.x
Osakabe, Y., Yamaguchi-Shinozaki, K., Shinozaki, K., Tran, L.-S. P. (2014). ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 202, 35–49. doi: 10.1111/nph.12613
Pál, M., Janda, T., Szalai, G.J.P.G.R. (2018). Interactions between plant hormones and thiol-related heavy metal chelators. Plant Growth Regul. 85, 173–185. doi: 10.1007/s10725-018-0391-7
Pan, W., Lu, Q., Xu, Q. R., Zhang, R. R., Li, H. Y., Yang, Y. H., et al. (2019). Abscisic acid-generating bacteria can reduce Cd concentration in pakchoi grown in Cd-contaminated soil. Ecotoxicol. Environ. Saf. 177, 100–107. doi: 10.1016/j.ecoenv.2019.04.010
Pan, W., You, Y., Shentu, J.-L., Weng, Y.-N., Wang, S.-T., Xu, Q.-R., et al. (2020). Abscisic acid (ABA)-importing transporter 1 (AIT1) contributes to the inhibition of Cd accumulation via exogenous ABA application in Arabidopsis. J. Hazard. Mater. 391, 122189. doi: 10.1016/j.jhazmat.2020.122189
Park, S. Y., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041
Park, J., Song, W. Y., Ko, D., Eom, Y., Hansen, T. H., Schiller, M., et al. (2012). The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant J. 69, 278–288. doi: 10.1111/j.1365-313X.2011.04789.x
Park, S. Y., Peterson, F. C., Mosquna, A., Yao, J., Volkman, B. F., Cutler, S. R. (2015). Agrochemical control of plant water use using engineered abscisic acid receptors. Nature 520, 545–548. doi: 10.1038/nature14123
Parys, E., Romanowska, E., Siedlecka, M., Poskuta, J. W. (1998). The effect of lead on photosynthesis and respiration in detached leaves and in mesophyll protoplasts of Pisum sativum. Acta Physiol. Plant 20, 313. doi: 10.1007/s11738-998-0064-7
Petraglia, A., De Benedictis, M., Degola, F., Pastore, G., Calcagno, M., Ruotolo, R., et al. (2014). The capability to synthesize phytochelatins and the presence of constitutive and functional phytochelatin synthases are ancestral (plesiomorphic) characters for basal land plants. J. Exp. Bot. 65, 1153–1163. doi: 10.1093/jxb/ert472
Pommerrenig, B., Diehn, T. A., Bernhardt, N., Bienert, M. D., Mitani-Ueno, N., Fuge, J., et al. (2020). Functional evolution of nodulin 26-like intrinsic proteins: from bacterial arsenic detoxification to plant nutrient transport. New Phytol. 225, 1383–1396. doi: 10.1111/nph.16217
Pompeu, G. B., Vilhena, M. B., Gratao, P. L., Carvalho, R. F., Rossi, M. L., Martinelli, A. P., et al. (2017). Abscisic acid-deficient sit tomato mutant responses to cadmium-induced stress. Protoplasma 254, 771–783. doi: 10.1007/s00709-016-0989-4
Pornsiriwong, W., Estavillo, G. M., Chan, K. X., Tee, E. E., Ganguly, D., Crisp, P. A., et al. (2017). A chloroplast retrograde signal, 3'-phosphoadenosine 5'-phosphate, acts as a secondary messenger in abscisic acid signaling in stomatal closure and germination. eLife 6, e23361. doi: 10.7554/eLife.23361
Reeves, R. D., Baker, A. J. M., Jaffré, T., Erskine, P. D., Echevarria, G., van der Ent, A. (2017). A global database for plants that hyperaccumulate metal and metalloid trace elements. New Phytol. 218, 407–411. doi: 10.1111/nph.14907
Rogers, E. E., Eide, D. J., Guerinot, M. L. (2000). Altered selectivity in an Arabidopsis metal transporter. Proc. Natl. Acad. Sci. U. S. A. 97, 12356–12360. doi: 10.1073/pnas.210214197
Ruszala, E. M., Beerling, D. J., Franks, P. J., Chater, C., Casson, S. A., Gray, J. E., et al. (2011). Land plants acquired active stomatal control early in their evolutionary Hhstory. Curr. Biol. 21, 1030–1035. doi: 10.1016/j.cub.2011.04.044
Sakata, Y., Komatsu, K., Takezawa, D. (2014). “ABA as a universal plant hormone,” in Progress in Botany: Vol. 75. Eds. Lüttge, U., Beyschlag, W., Cushman, J. (Berlin, Heidelberg: Springer Berlin Heidelberg), 57–96.
Salt, D. E., Prince, R. C., Pickering, I. J., Raskin, I. (1995). Mechanisms of cadmium mobility and accumulation in Indian mustard. Plant Physiol. 109, 1427–1433. doi: 10.1104/pp.109.4.1427
Saruhashi, M., Kumar Ghosh, T., Arai, K., Ishizaki, Y., Hagiwara, K., Komatsu, K., et al. (2015). Plant Raf-like kinase integrates abscisic acid and hyperosmotic stress signaling upstream of SNF1-related protein kinase2. Proc. Natl. Acad. Sci. U. S. A. 112, E6388–E6396. doi: 10.1073/pnas.1511238112
Sasaki, A., Yamaji, N., Yokosho, K., Ma, J. F. (2012). Nramp5 Is a major tansporter responsible for manganese and cadmium uptake in rice. Plant Cell 24, 2155–2167. doi: 10.1105/tpc.112.096925
Sasaki, A., Yamaji, N., Ma, J. F. (2014). Overexpression of OsHMA3 enhances Cd tolerance and expression of Zn transporter genes in rice. J. Exp. Bot. 65, 6013–6021. doi: 10.1093/jxb/eru340
Sato, Y., Antonio, B. A., Namiki, N., Takehisa, H., Minami, H., Kamatsuki, K., et al. (2011). RiceXPro: a platform for monitoring gene expression in japonica rice grown under natural field conditions. Nucleic Acids Res. 39, D1141–D1148. doi: 10.1093/nar/gkq1085
Shabala, S., Chen, G., Chen, Z.-H., Pottosin, I. (2020). The energy cost of the tonoplast futile sodium leak. New Phytol. 225, 1105–1110. doi: 10.1111/nph.15758
Sharma, S. S., Kumar, V. (2002). Responses of wild type and abscisic acid mutants ofArabidopsis thaliana to cadmium. J. Plant Physiol. 159, 1323–1327. doi: 10.1078/0176-1617-00601
Shen, G., Niu, J., Deng, Z. (2017). Abscisic acid treatment alleviates cadmium toxicity in purple flowering stalk (Brassica campestris L. ssp. chinensis var. purpurea Hort.) seedlings. Plant Physiol. Biochem. 118, 471–478. doi: 10.1016/j.plaphy.2017.07.018
Sheng, Y., Yan, X., Huang, Y., Han, Y., Zhang, C., Ren, Y., et al. (2019). The WRKY transcription factor, WRKY13, activates PDR8 expression to positively regulate cadmium tolerance in Arabidopsis. Plant Cell Environ. 42, 891–903. doi: 10.1111/pce.13457
Shi, W. G., Liu, W., Yu, W., Zhang, Y., Ding, S., Li, H., et al. (2019). Abscisic acid enhances lead translocation from the roots to the leaves and alleviates its toxicity in Populus x canescens. J. Hazard. Mater. 362, 275–285. doi: 10.1016/j.jhazmat.2018.09.024
Shim, D., Kim, S., Choi, Y.-I., Song, W.-Y., Park, J., Youk, E. S., et al. (2013). Transgenic poplar trees expressing yeast cadmium factor 1 exhibit the characteristics necessary for the phytoremediation of mine tailing soil. Chemosphere 90, 1478–1486. doi: 10.1016/j.chemosphere.2012.09.044
Shin, H., Shin, H.-S., Dewbre, G. R., Harrison, M. J. (2004). Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 39, 629–642. doi: 10.1111/j.1365-313X.2004.02161.x
Shinozawa, A., Otake, R., Takezawa, D., Umezawa, T., Komatsu, K., Tanaka, K., et al. (2019). SnRK2 protein kinases represent an ancient system in plants for adaptation to a terrestrial environment. Commun. Biol. 2, 55. doi: 10.1038/s42003-019-0312-y
Singh, S., Parihar, P., Singh, R., Singh, V. P., Prasad, S. M. (2016). Heavy metal tolerance in plants: role of transcriptomics, proteomics, petabolomics, and ionomics. Front. Plant Sci. 6, 1143. doi: 10.3389/fpls.2015.01143
Skubacz, A., Daszkowska-Golec, A., Szarejko, I. (2016). The role and regulation of ABI5 (ABA-Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 7, 1884. doi: 10.3389/fpls.2016.01884
Soma, F., Takahashi, F., Suzuki, T., Shinozaki, K., Yamaguchi-Shinozaki, K. (2020). Plant Raf-like kinases regulate the mRNA population upstream of ABA-unresponsive SnRK2 kinases under drought stress. Nat. Commun. 11, 1373. doi: 10.1038/s41467-020-15239-3
Song, W. Y., Sohn, E. J., Martinoia, E., Lee, Y. J., Yang, Y. Y., Jasinski, M., et al. (2003). Engineering tolerance and accumulation of lead and cadmium in transgenic plants. Nat. Biotechnol. 21, 914–919. doi: 10.1038/nbt850
Song, W. Y., Park, J., Mendoza-Cozatl, D. G., Suter-Grotemeyer, M., Shim, D., Hortensteiner, S., et al. (2010). Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proc. Natl. Acad. Sci. U. S. A. 107, 21187–21192. doi: 10.1073/pnas.1013964107
Song, W.-Y., Park, J., Eisenach, C., Maeshima, M., Lee, Y., Martinoia, E. (2014a). “ABC transporters and heavy metals,” in Plant ABC Transporters. Ed. Geisler, M. (Cham: Springer International Publishing), 1–17.
Song, W. Y., Yamaki, T., Yamaji, N., Ko, D., Jung, K. H., Fujii-Kashino, M., et al. (2014b). A rice ABC transporter, OsABCC1, reduces arsenic accumulation in the grain. Proc. Natl. Acad. Sci. U. S. A. 111, 15699–15704. doi: 10.1073/pnas.1414968111
Song, L., Huang, S.-S. C., Wise, A., Castanon, R., Nery, J. R., Chen, H., et al. (2016). A transcription factor hierarchy defines an environmental stress response network. Science 354, aag1550. doi: 10.1126/science.aag1550
Srivastava, S., Srivastava, A. K., Suprasanna, P., D'Souza, S. F. (2013). Identification and profiling of arsenic stress-induced microRNAs in Brassica juncea. J. Exp. Bot. 64, 303–315. doi: 10.1093/jxb/ers333
Stroiński, A., Chadzinikolau, T., Giżewska, K., Zielezińska, M. (2010). ABA or cadmium induced phytochelatin synthesis in potato tubers. Biol. Plant. 54, 117–120. doi: 10.1007/s10535-010-0017-z
Stroiński, A., Giżewska, K., Zielezińska, M. (2013). Abscisic acid is required in transduction of cadmium signal to potato roots. Biol. Plant. 57, 121–127. doi: 10.1007/s10535-012-0135-x
Sui, F., Zhao, D., Zhu, H., Gong, Y., Tang, Z., Huang, X. Y., et al. (2019). Map-based cloning of a new total loss-of-function allele of OsHMA3 causing high cadmium accumulation in rice grain. J. Exp. Bot. 70, 2857–2871. doi: 10.1093/jxb/erz093
Sun, D., Feng, H.-Y., Li, X.-Y., Ai, H., Sun, S., Chen, Y., et al. (2019). Expression of new Pteris vittata phosphate transporter PvPht1;4 reduces arsenic translocation from the roots to shoots in tobacco plants. Environ. Sci. Technol. 54, 1045–1053. doi: 10.1021/acs.est.9b05486
Takahashi, Y., Zhang, J., Hsu, P. K., Ceciliato, P. H. O., Zhang, L., Dubeaux, G., et al. (2020). MAP3Kinase-dependent SnRK2-kinase activation is required for abscisic acid signal transduction and rapid osmotic stress response. Nat. Commun. 11, 12. doi: 10.1038/s41467-019-13875-y
Takezawa, D., Watanabe, N., Ghosh, T. K., Saruhashi, M., Suzuki, A., Ishiyama, K., et al. (2015). Epoxycarotenoid-mediated synthesis of abscisic acid in Physcomitrella patens implicating conserved mechanisms for acclimation to hyperosmosis in embryophytes. New Phytol. 206, 209–219. doi: 10.1111/nph.13231
Tan, J., Wang, J., Chai, T., Zhang, Y., Feng, S., Li, Y., et al. (2013). Functional analyses of TaHMA2, a P1B-type ATPase in wheat. Plant Biotechnol. J. 11, 420–431. doi: 10.1111/pbi.12027
Tan, M., Cheng, D., Yang, Y., Zhang, G., Qin, M., Chen, J., et al. (2017). Co-expression network analysis of the transcriptomes of rice roots exposed to various cadmium stresses reveals universal cadmium-responsive genes. BMC Plant Biol. 17, 194. doi: 10.1186/s12870-017-1143-y
Tang, Z., Chen, Y., Miller, A. J., Zhao, F. J. (2019). The C-type ATP-binding cassette transporter OsABCC7 is involved in the root-to-shoot translocation of arsenic in rice. Plant Cell Physiol. 60, 1525–1535. doi: 10.1093/pcp/pcz054
Tang, Y., Wang, L., Xie, Y., Yu, X., Li, H., Lin, L., et al. (2020). Effects of exogenous abscisic acid on the growth and cadmium accumulation of lettuce under cadmium-stress conditions. Int. J. Environ. An. Ch. 100, 720–731. doi: 10.1080/03067319.2019.1639686
Tao, Q., Jupa, R., Liu, Y., Luo, J., Li, J., Kováč, J., et al. (2019). Abscisic acid-mediated modifications of radial apoplastic transport pathway play a key role in cadmium uptake in hyperaccumulator Sedum alfredii. Plant Cell Environ. 42, 1425–1440. doi: 10.1111/pce.13506
Tehseen, M., Cairns, N., Sherson, S., Cobbett, C. S. (2010). Metallochaperone-like genes in Arabidopsis thaliana. Metallomics 2, 556–564. doi: 10.1039/C003484C
Tougane, K., Komatsu, K., Bhyan, S. B., Sakata, Y., Ishizaki, K., Yamato, K. T., et al. (2010). Evolutionarily conserved regulatory mechanisms of abscisic acid signaling in land plants: characterization of ABSCISIC ACID INSENSITIVE1-Like Type 2C protein phosphatase in the liverwort Marchantia Polymorpha. Plant Physiol. 152, 1529–1543. doi: 10.1104/pp.110.153387
Ueno, D., Yamaji, N., Kono, I., Huang, C. F., Ando, T., Yano, M., et al. (2010). Gene limiting cadmium accumulation in rice. Proc. Natl. Acad. Sci. U. S. A. 107, 16500–16505. doi: 10.1073/pnas.1005396107
Ueno, D., Milner, M. J., Yamaji, N., Yokosho, K., Koyama, E., Clemencia Zambrano, M., et al. (2011). Elevated expression of TcHMA3 plays a key role in the extreme Cd tolerance in a Cd-hyperaccumulating ecotype of Thlaspi caerulescens. Plant J. 66, 852–862. doi: 10.1111/j.1365-313X.2011.04548.x
Umezawa, T., Sugiyama, N., Mizoguchi, M., Hayashi, S., Myouga, F., Yamaguchi-Shinozaki, K., et al. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 106, 17588–17593. doi: 10.1073/pnas.0907095106
Uraguchi, S., Mori, S., Kuramata, M., Kawasaki, A., Arao, T., Ishikawa, S. (2009). Root-to-shoot Cd translocation via the xylem is the major process determining shoot and grain cadmium accumulation in rice. J. Exp. Bot. 60, 2677–2688. doi: 10.1093/jxb/erp119
Uraguchi, S., Tanaka, N., Hofmann, C., Abiko, K., Ohkama-Ohtsu, N., Weber, M., et al. (2017). Phytochelatin synthase has contrasting effects on cadmium and arsenic accumulation in rice grains. Plant Cell Physiol. 58, 1730–1742. doi: 10.1093/pcp/pcx114
Vatamaniuk, O. K., Mari, S., Lu, Y.-P., Rea, P. A. (1999). AtPCS1, a phytochelatin synthase from Arabidopsis: isolation and in vitro reconstitution. Proc. Natl. Acad. Sci. U. S. A. 96, 7110–7115. doi: 10.1073/pnas.96.12.7110
Vert, G., Grotz, N., Dedaldechamp, F., Gaymard, F., Guerinot, M. L., Briat, J. F., et al. (2002). IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14, 1223–1233. doi: 10.1105/tpc.001388
Vishwakarma, K., Upadhyay, N., Kumar, N., Yadav, G., Singh, J., Mishra, R. K., et al. (2017). Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects. Front. Plant Sci. 8, 161. doi: 10.3389/fpls.2017.00161
Wang, Y., Yu, K.-F., Poysa, V., Shi, C., Zhou, Y.-H. (2012). A single point mutation in GmHMA3 affects cadimum (Cd) translocation and accumulation in soybean seeds. Mol. Plant 5, 1154–1156. doi: 10.1093/mp/sss069
Wang, N., Cui, Y., Liu, Y., Fan, H., Du, J., Huang, Z., et al. (2013). Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol. Plant 6, 503–513. doi: 10.1093/mp/sss089
Wang, P., Zhang, W., Mao, C., Xu, G., Zhao, F. J. (2016). The role of OsPT8 in arsenate uptake and varietal difference in arsenate tolerance in rice. J. Exp. Bot. 67, 6051–6059. doi: 10.1093/jxb/erw362
Wang, F.-Z., Chen, M.-X., Yu, L.-J., Xie, L.-J., Yuan, L.-B., Qi, H., et al. (2017). OsARM1, an R2R3 MYB transcription factor, is involved in regulation of the response to arsenic stress in rice. Front. Plant Sci. 8, 1868. doi: 10.3389/fpls.2017.01868
Wang, F., Chen, Z.-H., Shabala, S. (2017). Hypoxia sensing in plants: on a quest for ion channels as putative oxygen sensors. Plant Cell Physiol. 58, 1126–1142. doi: 10.1093/pcp/pcx079
Wang, T., Hua, Y., Chen, M., Zhang, J., Guan, C., Zhang, Z. (2018). Mechanism enhancing Arabidopsis resistance to cadmium: the role of NRT1.5 and proton pump. Front. Plant Sci. 9, 1892. doi: 10.3389/fpls.2018.01892
Wang, H., Liu, Y., Peng, Z., Li, J., Huang, W., Liu, Y., et al. (2019). Ectopic expression of poplar ABC transporter PtoABCG36 confers Cd tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 20, 3293. doi: 10.3390/ijms20133293
Wang, S., Li, L., Li, H., Sahu, S. K., Wang, H., Xu, Y., et al. (2020). Genomes of early-diverging streptophyte algae shed light on plant terrestrialization. Nat. Plants 6, 95–106. doi: 10.1038/s41477-019-0560-3
Wu, H., Chen, C., Du, J., Liu, H., Cui, Y., Zhang, Y., et al. (2012). Co-overexpression FIT with AtbHLH38 or AtbHLH39 in Arabidopsis-enhanced cadmium tolerance via increased cadmium sequestration in roots and improved iron homeostasis of shoots. Plant Physiol. 158, 790–800. doi: 10.1104/pp.111.190983
Xu, Q., Pan, W., Zhang, R., Lu, Q., Xue, W., Wu, C., et al. (2018). Inoculation with Bacillus subtilis and Azospirillum brasilense produces abscisic acid that reduces IRT1-mediated cadmium uptake of roots. J. Agric. Food Chem. 66, 5229–5236. doi: 10.1021/acs.jafc.8b00598
Yamaji, N., Xia, J., Mitani-Ueno, N., Yokosho, K., Feng Ma, J. (2013). Preferential delivery of zinc to developing tissues in rice is mediated by P-type heavy metal ATPase OsHMA2. Plant Physiol. 162, 927–939. doi: 10.1104/pp.113.216564
Yamaji, N., Sakurai, G., Mitani-Ueno, N., Ma, J. F. (2015). Orchestration of three transporters and distinct vascular structures in node for intervascular transfer of silicon in rice. Proc. Natl. Acad. Sci. U. S. A. 112, 11401–11406. doi: 10.1073/pnas.1508987112
Yamazaki, S., Ueda, Y., Mukai, A., Ochiai, K., Matoh, T. (2018). Rice phytochelatin synthases OsPCS1 and OsPCS2 make different contributions to cadmium and arsenic tolerance. Plant Direct 2, e00034–e00034. doi: 10.1002/pld3.34
Yan, H., Filardo, F., Hu, X., Zhao, X., Fu, D. (2016). Cadmium stress alters the redox reaction and hormone balance in oilseed rape (Brassica napus L.) leaves. Environ. Sci. Pollut. Res. Int. 23, 3758–3769. doi: 10.1007/s11356-015-5640-y
Yan, H., Gao, Y., Wu, L., Wang, L., Zhang, T., Dai, C., et al. (2019). Potential use of the Pteris vittata arsenic hyperaccumulation-regulation network for phytoremediation. J. Hazard. Mater. 368, 386–396. doi: 10.1016/j.jhazmat.2019.01.072
Yang, J., Gao, M.-X., Hu, H., Ding, X.-M., Lin, H.-W., Wang, L., et al. (2016). OsCLT1, a CRT-like transporter 1, is required for glutathione homeostasis and arsenic tolerance in rice. New Phytol. 211, 658–670. doi: 10.1111/nph.13908
Ye, Y., Li, P., Xu, T., Zeng, L., Cheng, D., Yang, M., et al. (2017). OsPT4 contributes to arsenate uptake and transport in rice. Front. Plant Sci. 8, 2197. doi: 10.3389/fpls.2017.02197
Yu, L., Luo, Y., Liao, B., Xie, L., Chen, L., Xiao, S., et al. (2012). Comparative transcriptome analysis of transporters, phytohormone and lipid metabolism pathways in response to arsenic stress in rice (Oryza sativa). New Phytol. 195, 97–112. doi: 10.1111/j.1469-8137.2012.04154.x
Zhang, H., Zhu, H., Pan, Y., Yu, Y., Luan, S., Li, L. (2014). A DTX/MATE-type transporter facilitates abscisic acid efflux and modulates ABA sensitivity and drought tolerance in Arabidopsis. Mol. Plant 7, 1522–1532. doi: 10.1093/mp/ssu063
Zhang, J., Martinoia, E., Lee, Y. (2018). Vacuolar transporters for cadmium and arsenic in plants and their applications in phytoremediation and crop development. Plant Cell Physiol. 59, 1317–1325. doi: 10.1093/pcp/pcy006
Zhang, L., Wu, J., Tang, Z., Huang, X. Y., Wang, X., Salt, D. E., et al. (2019). Variation in the BrHMA3 coding region controls natural variation in cadmium accumulation in Brassica rapa vegetables. J. Exp. Bot. 70, 5865–5878. doi: 10.1093/jxb/erz310
Zhang, P., Wang, R., Ju, Q., Li, W., Tran, L.-S. P., Xu, J. (2019). The R2R3-MYB transcription factor MYB49 regulates cadmium accumulation. Plant Physiol. 180, 529–542. doi: 10.1104/pp.18.01380
Zhang, W., Wang, Z., Song, J., Yue, S., Yang, H. (2019). Cd2+ uptake inhibited by MhNCED3 from Malus hupehensis alleviates Cd-induced cell death. Environ. Exp. Bot. 166, 103802. doi: 10.1016/j.envexpbot.2019.103802
Zhao, F. J., Wang, P. (2020). Arsenic and cadmium accumulation in rice and mitigation strategies. Plant Soil. 46, 1–21. doi: 10.1007/s11104-019-04374-6
Zhao, F. J., Jiang, R. F., Dunham, S. J., McGrath, S. P. (2006). Cadmium uptake, translocation and tolerance in the hyperaccumulator Arabidopsis halleri. New Phytol. 172, 646–654. doi: 10.1111/j.1469-8137.2006.01867.x
Zhao, L., Xiong, J., Li, L. P., Zhu, C. (2009). Low concentration of exogenous abscisic acid increases lead tolerance in rice seedlings. Biol. Plant. 53, 728. doi: 10.1007/s10535-009-0132-x
Zhao, F. J., Ma, Y., Zhu, Y. G., Tang, Z., McGrath, S. P. (2015). Soil contamination in China: current status and mitigation strategies. Environ. Sci. Technol. 49, 750–759. doi: 10.1021/es5047099
Zhao, C., Wang, Y., Chan, K. X., Marchant, D. B., Franks, P. J., Randall, D., et al. (2019). Evolution of chloroplast retrograde signaling facilitates green plant adaptation to land. Proc. Natl. Acad. Sci. U.S.A. 116, 5015–5020. doi: 10.1073/pnas.1812092116
Zhu, Y.-G., Yoshinaga, M., Zhao, F.-J., Rosen, B. P. (2014). Earth abides arsenic biotransformations. Annu. Rev. Earth. Planet Sci. 42, 443–467. doi: 10.1146/annurev-earth-060313-054942
Keywords: plant evolution, comparative genomics, cadmium, arsenic, lead, detoxification
Citation: Hu B, Deng F, Chen G, Chen X, Gao W, Long L, Xia J and Chen Z-H (2020) Evolution of Abscisic Acid Signaling for Stress Responses to Toxic Metals and Metalloids. Front. Plant Sci. 11:909. doi: 10.3389/fpls.2020.00909
Received: 31 March 2020; Accepted: 03 June 2020;
Published: 17 July 2020.
Edited by:
Yoichi Sakata, Tokyo University of Agriculture, JapanReviewed by:
Vijay Pratap Singh, University of Allahabad, IndiaJiaojiao Zhang, Chinese Academy of Sciences (CAS), China
Copyright © 2020 Hu, Deng, Chen, Chen, Gao, Long, Xia and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fenglin Deng, ZGZsQHlhbmd0emV1LmVkdS5jbg==; Zhong-Hua Chen, Wi5DaGVuQHdlc3Rlcm5zeWRuZXkuZWR1LmF1
†These authors have contributed equally to this work
 Beibei Hu
Beibei Hu Fenglin Deng
Fenglin Deng Guang Chen
Guang Chen Xuan Chen
Xuan Chen Wei Gao
Wei Gao Lu Long
Lu Long Jixing Xia
Jixing Xia Zhong-Hua Chen
Zhong-Hua Chen