- Department of Plant Pathology and Microbiology, The Robert H. Smith Faculty of Agriculture, Food and Environment, The Hebrew University of Jerusalem, Rehovot, Israel
Beneficial rhizobacteria dwell in plant roots and promote plant growth, development, and resistance to various stress types. In recent years there have been large-scale efforts to culture root-associated bacteria and sequence their genomes to uncover novel beneficial microbes. However, only a few strains of rhizobacteria from the large pool of soil microbes have been studied at the molecular level. This review focuses on the molecular basis underlying the phenotypes of three beneficial microbe groups; (1) plant-growth promoting rhizobacteria (PGPR), (2) root nodulating bacteria (RNB), and (3) biocontrol agents (BCAs). We focus on bacterial proteins and secondary metabolites that mediate known phenotypes within and around plants, and the mechanisms used to secrete these. We highlight the necessity for a better understanding of bacterial genes responsible for beneficial plant traits, which can be used for targeted gene-centered and molecule-centered discovery and deployment of novel beneficial rhizobacteria.
Introduction
The term rhizosphere was first defined by Hiltner, who described it as the soil compartment influenced by the root (Hiltner, 1904). The rhizosphere differs from the surrounding bulk soil and the plant endophytic compartment in microbial diversity (Hacquard et al., 2015) and its members influence the release of root exudates. Root exudates are responsible for shaping the microbial community structure, including attraction of beneficial microbes (Clark, 1949; Zhalnina et al., 2018; Korenblum et al., 2020). After successfully colonizing plant roots, beneficial microbes secrete proteins and secondary metabolites, relevant for nutrient acquisition, improved plant fitness, and inhibition of pathogen colonization (Pieterse et al., 2014; Bakker et al., 2018; Yu et al., 2019a). Beneficial microbes are subdivided in a coarse manner into plant growth promoting rhizobacteria (PGPR), biocontrol agents (BCAs), and root-nodulating bacteria (RNB; Berendsen et al., 2012). PGPR directly or indirectly induce plant growth via secretion of secondary metabolites, which are in turn involved in plant hormone synthesis and nutrient acquisition from soil (Lugtenberg and Kamilova, 2009). RNB are also referred to as biofertilizers. They interact with legume roots as mutualists. Nodules allow the energetically expensive process of nitrogen fixation. The ammonia produced in the nodules as part of this process is transported into the plant cells in exchange for carbon required for bacterial growth. BCAs or biopesticides in the roots act by eliminating phytopathogens and pests, either indirectly by induction of the plant immune response through induced or acquired systemic resistance, or directly by producing and releasing antimicrobial and pesticidal toxins or by physical niche occupation (Kuc and Tuzun, 1992; Van Wees et al., 1997; Zamioudis and Pieterse, 2012; Bernal et al., 2018).
The effectiveness of beneficial microbes is frequently dependent on secretion systems. Some secretion systems allow translocation of proteins, called effectors, directly from one cell into another without being degraded or utilized by another organism. Other secretion systems and efflux pumps release proteins and secondary metabolites into the medium, respectively. The secreted proteins and metabolites play roles in root colonization, as well as in interactions with the plant immune response and the surrounding prokaryotic and eukaryotic organisms (Figure 1; Lugtenberg and Kamilova, 2009; Pieterse et al., 2014; Wu et al., 2018; Jamali et al., 2020). The goal of this minireview is to describe important bacterial secreted effectors, secondary metabolites and secretion systems which play a role in the interactions of beneficial microbes with plants and surrounding microbes, including bacteria and fungi.
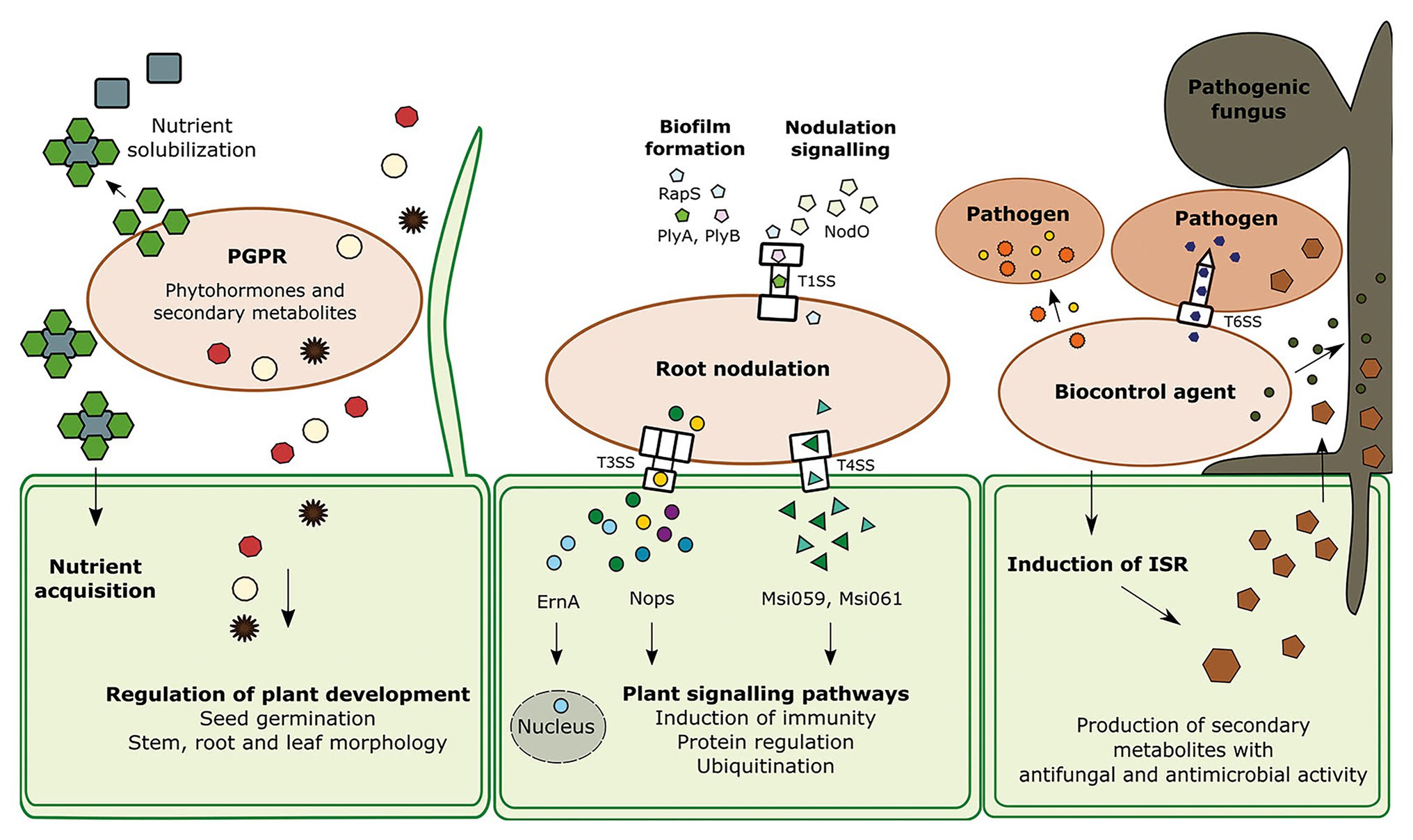
Figure 1. The interaction of the three groups of beneficial bacteria with other species; plant growth promoting rhizobacteria (PGPR), root nodulating bacteria (RNB), and biocontrol agents (BCAs) and their neighboring cells. PGPR produce various secondary metabolites including phytohormones which are regulating several processes in the plant development such as seed germination, stem, leaf, and root morphology. Another feature of PGPR is the solubilization of nutrients. RNB contain several secretion systems that can transport effectors directly into the host cell to regulate certain processes. Type I protein secretion system (TISS) of Rhizobium leguminosarum bv. Viciae is responsible for biofilm formation via the effectors PlyA, PlyB, and Rhizobium-adhering proteins (RapS). The T1SS is recognizable due to the outer membrane protein TolC. Type III secretion system (T3SS) and type IV secretion system (T4SS) are secreting effectors which can trigger protein regulation and induce plant immune responses. At least one effector travels into the plant cell nucleus. BCAs produce antibacterial and antifungal protein toxins and small molecules. Proteinaceous toxins are transferred through the Type VI secretion system (T6SS), a powerful nanoweapon, into the host cell. Specific antimicrobials can kill phytopathogens like fungi, oomycetes, and bacteria. In addition, biocontrol agents can trigger the plant immunity pathway induced systemic resistance (ISR), which leads to the production of antimicrobials which can eliminate a broad spectrum of organisms.
Plant Growth Promoting Rhizobacteria
Plant growth promoting rhizobacteria can improve the plant growth in multiple ways. They can indirectly promote growth by forming a biofilm that serves as a protective layer against pathogens or as an enhanced surface for nutrient acquisition from the surrounding soil (Weselowski et al., 2016). They can also produce and secrete growth phytohormones or their intermediates, which directly increase the root surface area, and promote plant development, growth and health (Figure 1; Spaepen et al., 2014). Additionally, PGPR increase abiotic stress tolerance in crops.
Plant growth promoting rhizobacteria secrete organic acids and other secondary metabolites that solubilize macronutrients and micronutrients and increase their bioavailability for plants. Nitrogen and phosphorus are two of the essential macronutrients for plant growth. Nitrogen fixation will be discussed in the next section. Phosphorus in soil is highly unavailable for plants. Beneficial microbes mobilize phosphorus via organic chelators like citric, gluconic and malic acid. The secretion of these acids leads to a decrease of the soil pH and production of plant bioavailable HPO42− (Rodríguez and Fraga, 1999; Ivanova et al., 2006; Yasmin et al., 2009). Some members of the Bacillus, Pseudomonas and Enterobacter genera are very efficient phosphorus solubilizing bacteria and have been shown to improve yield and growth of crops (Jha et al., 2012; Goswami et al., 2014).
Iron is an essential micronutrient for plant growth and development and iron-deficient plants suffer from yellow stripe chlorosis in young leaves (Abadía et al., 2002). The application of Alcaligenes 637Ca and Staphylococcus MFDCa1 to pear and apple roots, respectively, increased the foliar enzymatic activity of a plant enzyme that is responsible for Fe3+ reduction and helps plants uptake iron under iron deprivation (İpek et al., 2017; Aras et al., 2018). Some Pseudomonas spp. PGPR secrete siderophores like carboxylates, catecholate, and hydroxamate for Fe acquisition in Zea mays. Siderophores also have antimicrobial properties against the phytopathogens Rhizoctonia solani and Sclerotium rolfs (Sharma and Johri, 2003; Yu et al., 2011; Scagliola et al., 2016; Trapet et al., 2016; Mullins et al., 2019).
In contrast to the indirect effect of solubilization of nutrients on plant growth, phytohormone production by PGPR directly interfaces with plant growth signaling networks. The five main phytohormones are produced by PGPR: auxins, cytokinins, gibberellins (GAs), ethylene (ET), and abscisic acid. One of the most studied PGPRs is the Azospirillum genus. Azospirillum. brasilense consists of four different pathways to produce IAA, which is the most common phytohormone from the auxin class. The indole-3-pyruvate pathway produces the highest amount of IAA (Kloepper et al., 1989; Puente et al., 2004). The IAA biosynthesis pathways are well understood in bacteria, but the reason for the existence of those pathways in bacteria is still unknown (Molina et al., 2018). The indole-3-pyruvate pathway is highly dependent on the key enzyme indole-3-pyruvate decarboxylase encoded by the gene ipdC (Spaepen et al., 2014). ipdC alone is responsible for induction of root hair formation as was shown by laboratory inoculation experiments of Arabidopsis thaliana with A. brasilense SP245 strain (Cohen et al., 2008; Rivera et al., 2018). However, field experiments are inconsistent and did not reproduce the results (Díaz-Zorita and Fernández-Canigia, 2009). Generally, field experiments inoculated with Azospirillum showed an inconsistent increase in grain yield (Dobbelaere et al., 1999; Vande Broek et al., 1999). Azospirillum has many features, in addition to auxin production, which could lead to plant growth promotion including nitrogen fixation, siderophore production, and phosphate solubilization. Hence, there might be a growth promotion as an additive or a synergistic combination of the various pathways (Spaepen et al., 2008). The technological progress in the field allows us to combine large-scale ecological studies with a reductionist genetic approach that reveals bacterial genes that promote growth. A recent study using a 185-member bacterial community showed that this community causes Arabidopsis root growth inhibition. Interestingly, several strains of the Variovorax genus were found to maintain root development. Further genetic approach identified that the Variovorax genomes encode an auxin degrading operon that is necessary and sufficient for causing this beneficial phenotype even in such a complex ecosystem (Finkel et al., 2020).
Another example for phytohormone production in rhizobacteria is of the phytohormones gibberellins. GAs are involved in many developmental processes in plants, such as flowering regulation, seed germination, stem and leaf elongation, and pollen maturation (Achard et al., 2007; Ariizumi and Steber, 2007). Biosynthesis of GA was found in many bacteria such as Bacillus pumilus, Bacillus licheniformis, and Leifsonia soli (Kang et al., 2016; Kim et al., 2017). Leifsonia soli SE134 has been shown to enhance plant growth of the GA deficient Waito-C rice dwarf mutant cultivar and can extend shoot length, plant weight, and seed germination in cucumber, and tomato under greenhouse conditions, which may be due to GA synthesis (Kang et al., 2014).
Cytokinins are another group of growth-stimulating phytohormones that are responsible for cell division, plant senescence, seed germination, flower and fruit development, and apical dormancy (Akhtar et al., 2020). Pseudomonas fluorescens G20-18 and 6–8 strains produces cytokinins (García de Salamone et al., 2001; Pallai et al., 2012). It has been shown that canola inoculated with G20-18 and 6–8 strains had greater root length than the non-inoculated control in a gnotobiotic assay (Pallai et al., 2012). Großkinsky et al. (2016) revealed by constructing various knock-out and gain of function mutants of G20-18, that cytokinins have a protective role against Pseudomonas syringae pv. tomato DC3000 and can suppress disease response in A. thaliana (Großkinsky et al., 2016).
The aforementioned studies showed that PGPR can secrete multiple molecules which lead to various phenotypes. Exactly which conditions favor release of these beneficial growth-promoting molecules is still poorly understood. Knowing these conditions is important given that abiotic and biotic stressors can affect phytohormone production (Díaz-Zorita and Fernández-Canigia, 2009).
Ethylene is an important plant growth hormone that ameliorates harmful effects of abiotic stress conditions in plants (Glick, 2014). Its precursor is 1-aminocyclopropane-1-carboxylate (ACC). PGPR can improve plant tolerance to abiotic stressors via the production of ACC deaminase, which cleaves ET and produces α-ketobutyrate and ammonia. ACC deaminase indirectly counteract saline plant growth inhibitory effects in plants, hence plants are more salt stress tolerant (Orozco-Mosqueda et al., 2019, 2020). Pseudomonas putida UW4 carrying the acdS gene that encode for ACC deaminase was able to restore 66% of canola shoot fresh mass when grown in cold temperatures under high salt levels. Remarkably, the ΔacdS strain yielded only 20% of shoot fresh mass under the same conditions, demonstrating the importance of this single bacterial gene in promoting plant growth (Cheng et al., 2007). Trehalose, a non-reducing disaccharide, is especially induced in bacteria under saline stress and reduces osmotic, ionic and saline stress responses, by interacting with ABA, volatile compounds and exopolysaccharides (EPS; Avonce et al., 2006). Recently the synergetic effect of trehalose accumulation and ACC-deaminase production has been discovered in Pseudomonas sp. UW4 protecting tomato plants under saline stress. The plants were unable to survive the abiotic stress when the UW4 acdS and treS (trehalose synthesis) genes where knocked out (Orozco-Mosqueda et al., 2019). More detailed information about the synergistic effect of rhizobacteria produced ACC deaminase and plant compounds were reviewed recently by Forni et al. (2017). ACC deaminase also plays a role in synergetic function with other soil living-organisms such as in rhizobacteria for induced nodulation. Pseudomonas fluorescens YsS6 promotes the growth of Rhizobia tropici CIAT899, leading to an induced growth of Phaseolus vulgaris. The plant growth induction was only observed when YsS6 expressed acdS (Nascimento et al., 2019).
Root-Nodulating Bacteria
Root-nodulating bacteria have developed an impressive and complex symbiosis with their legume host. One of the first steps in this relationship, is the secretion of flavonoids by the host plant that diffuse across the membrane of the Rhizobia and induce synthesis of the NodD protein which activates transcription of other genes involved in nodulation including Nod factor (NF) production (Wang et al., 2012). NFs are primary signal molecules produced by bacteria and detected by the plant to induce nodule organogenesis (Nelson and Sadowsky, 2015). In addition to NFs, other molecules and proteins mediate other aspects of the rhizobia-legume symbiosis such as root colonization, symbiont recognition and suppression of the plant immune system. To perform all of these tasks, Rhizobia make use of special secretion systems that translocate effectors to their host. These include type I, type III and IV secretion systems (Cianciotto, 2005; Schmeisser et al., 2009; Nelson and Sadowsky, 2015).
Type I protein secretion system (TISS) of Rhizobium leguminosarum bv. viciae is encoded by the prsD and prsE genes. This T1SS is responsible for secretion of the EPS-glycanases PlyA and PlyB (Russo et al., 2006). These enzymes play a key role in biofilm formation; by cleaving the EPS chains they modulate the structure and maturation of the biofilm. Mutations in prsD and prsE greatly suppress the formation of biofilm on glass surfaces (Russo et al., 2006). Biofilm formation is an important step in root colonization and in symbiotic interaction formation. Once rhizobia attach to root hairs, they aggregate and form a biofilm, which is encased in a structure called a cap that is made of cellulose and EPS (Smit et al., 1987; Downie, 2010). Some proteins such as Rhizobium-adhering proteins (Raps) are required for stability of the cap, and are exported through the PrsDE T1SS (Smit et al., 1987; Russo et al., 2006; Krehenbrink and Allan, 2008; Poole et al., 2018). RapA1 is a calcium-binding Rap located at the cell pole (Poole et al., 2018). RapA1 overexpression in R. leguminosarum bv. trifolii R200 increased attachment to red clover roots by up to 5-fold and its overexpression in Rhizobium etli enhanced the capability of attachment to common bean roots (Mongiardini et al., 2009; Frederix et al., 2014). TISS also secretes NodO, a well-studied protein from R. leguminosarum, that is critical for signaling during nodulation (Finnie et al., 1997).
TolC is an integral membrane protein that is part of the outer membrane component of T1SS. TolC from Sinorhizobium meliloti functions in the symbiotic relationship with Medicago sativa (Cosme et al., 2008). S. meliloti tolC mutant showed an 8-fold reduction in the number of nodules compared with the wild type and presented an ineffective nitrogen fixation in the roots of M. sativa (Cosme et al., 2008). TolC may participate in the efflux of antimicrobial compounds produced by the host plant, resistance to osmotic or oxidative stress, polysaccharide biosynthesis, and the secretion of proteins or other molecules relevant for the symbiosis, such as NFs, that can affect directly or indirectly the formation of nodules in the roots of M. sativa (Srinivasan et al., 2015; Mergaert, 2018).
Other secretion systems, such as the type III secretion system (T3SS), are employed for effector translocation into the host plants. T3SS is mostly studied for its role in plant disease. The effectors can interfere with plant signaling and plant cell recognition. Transcriptional studies have shown expression of T3SS genes at different stages of the Plant-Rhizobium interaction such as root colonization, infection and nodulation. The T3SS of Bradyrhizobium japonicum USDA110 is expressed in infection threads and developing nodules of soybean (Zehner et al., 2008). Several T3SS genes of Rhizobium sp. NGR234 are expressed in mature nodules of Cajanus cajan and Vigna unguiculate (Viprey et al., 1998; Perret et al., 1999; Tampakaki, 2014). Regulatory analyses of the T3SS of Rhizobium sp. NGR234 showed that it is activated after Nod factors generation and its activity continues for at least 24 h (Kobayashi et al., 2004; Marie et al., 2004). These results indicate that effector secretion through T3SS concurs with development of the infection thread. T3SS is strongly regulated after sensing potential plant hosts.
T3SS genes called rhc (Rhizobium conserved), encode different nodulation outer proteins (Nops) that can be divided into two groups. The first group is composed of the core components of T3SS pilus that spans the plant cell wall (Saad et al., 2008; Deakin and Broughton, 2009; López-Baena et al., 2016). NopA and NopB are the major and minor subunits, respectively. NopX likely polymerizes to form a transmembrane pore (the translocon) through which other effectors enter the plant cytoplasm (Deakin and Broughton, 2009; López-Baena et al., 2016). The second group is composed of the effectors that are injected through T3SS machinery into the host cytoplasm. Several Sinorhizobial proteins secreted through the T3SS have been identified. These include NopL and NopP that may interfere with plant signaling pathways, as both can be phosphorylated by plant kinases and have shown to be responsible for optimal nodulation of host plants Flemingia congesta and Tephrosia vogelii (Bartsev et al., 2004; Skorpil et al., 2005; Gourion et al., 2015). NopL was shown to interfere with mitogen-activated protein kinase (MAPK) that is involved in pathogen recognition in both basal plant defense and R-mediated resistance (Pedley and Martin, 2005; Zhang et al., 2011). NopM belongs to the IpaH-SspH-YopM family of effectors found in animal pathogens, which are known to be involved in targeting nuclei of host cells and ubiquitination process (Bartsev et al., 2004; Skorpil et al., 2005; Rohde et al., 2007). A later study indicated a possible role for NopM as a functional E3 ubiquitin ligase domain in Rhizobium sp. strain NG234 (Xin et al., 2012). In the same study it was further mentioned that when expressed in Nicotiana benthamiana, NopM reduced reactive oxygen species (ROS) and induced plant defense gene expression (Xin et al., 2012). NopT effector has homology with the avirulence protein AvrPphB of the phytopathogen P. syringae and YopT of Yersinia spp. which are known to possess a protease activity. NopT mutants of NGR234 affected nodulation either positively (P. vulgaris cv. Yudou No. 1; T. vogelii) or negatively (Crotalaria juncea; Dai et al., 2008). NopM and NopT have shown to have either negative or positive effects in nodulation in a host dependent manner (Dai et al., 2008; Kambara et al., 2009)
Another effector, NopD in Sinorhizobium fredii HH103, has been predicted to be a C48 cysteine peptidase (Rodrigues et al., 2007). The C48 cysteine peptidase family contains the protein XopD, a T3SS effector from the plant pathogen Xanthomonas campestris (Hotson et al., 2003). It functions in planta to target SUMO-conjugated proteins (Hotson et al., 2003). XopD interferes with the plant’s ability to regulate the expression of specific proteins (Nelson and Sadowsky, 2015). NopC is a T3SS-dependent effector that lacks homologues in pathogenic bacteria but its function in plants is still unknown (Jiménez-Guerrero et al., 2015). NopJ acts as acetyltransferase that prevents phosphorylation of MAP kinases by acetylating the phosphorylation sites, thereby inactivating the MAP kinases (Mukherjee et al., 2006). Recently, a conserved T3SS effector, ErnA, was described in Bradyrhizobium (Teulet et al., 2019). Interestingly, this effector is targeted to the plant nucleus and may bind nucleic acids in the plant nuclei. Gain and loss of function experiments demonstrated the direct involvement of ErnA for nodule formation. All T3SS effectors and their predicted function are described in Table 1.
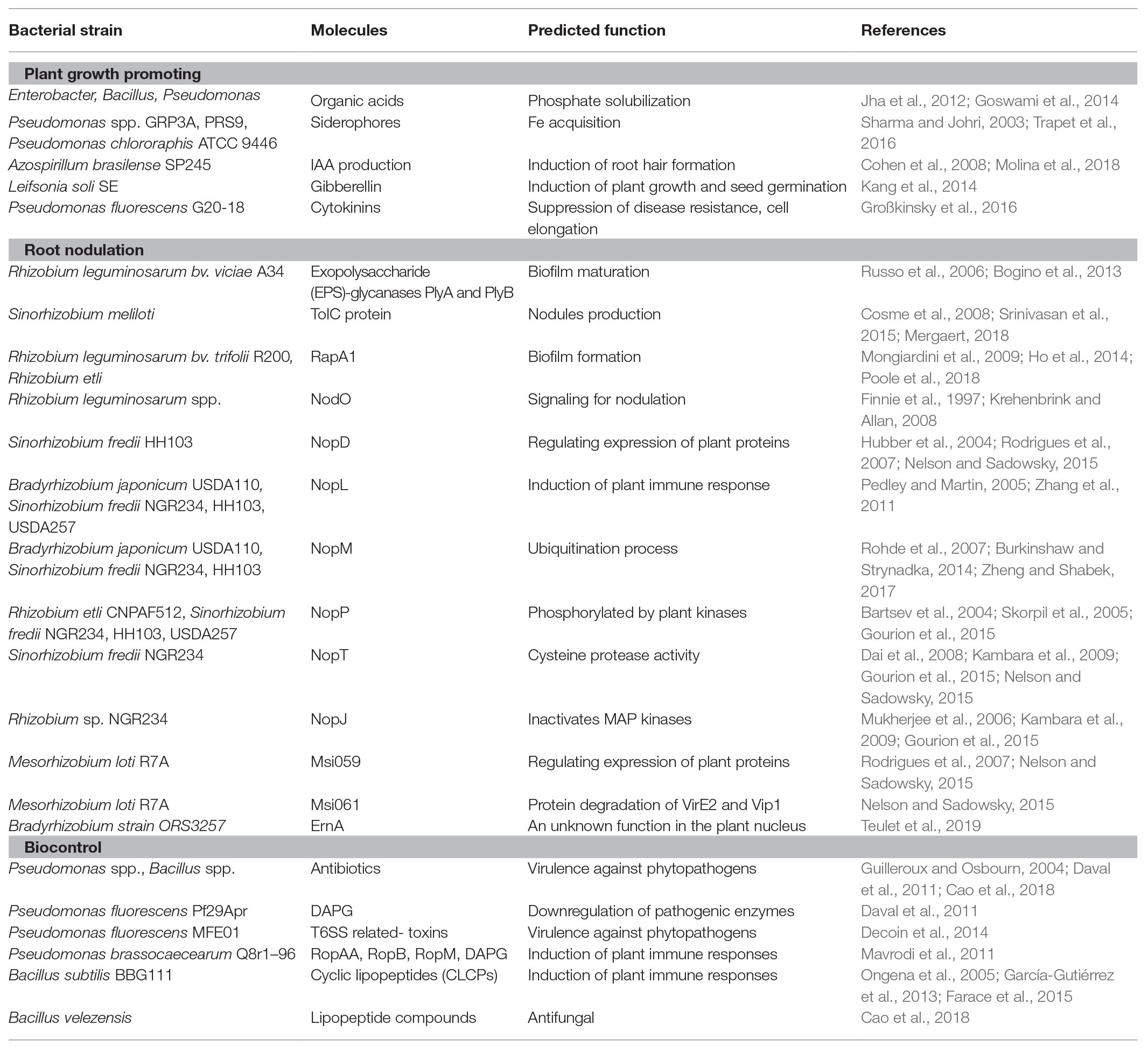
Table 1. Summary of all discussed bacteria, predicted function, and secreted molecules in this review. Some molecules are secreted from different bacteria.
The bacterial type IV secretion systems (T4SS) is a unique system in its ability to transfer large nucleic acid molecules, in addition to proteins, across the cell envelope (Christie and Cascales, 2005; Sgro et al., 2019). Rhizobial T4SS shares strong homology to the VirB/VirD4 subunits found in Agrobacterium (Sullivan et al., 2002; Christie et al., 2014). The T4SS in Agrobacterium tumefaciens, is used for translocation of both T-DNA and effector proteins (Kuldau et al., 1990; Zupan and Zambryski, 1995). T4SS has been identified in rhizobia such as Mesorhizobium loti R7A (Hubber et al., 2007; Miwa and Okazaki, 2017) and R. etli CFN42 (Lacroix and Citovsky, 2016). T4SS could be involved in the nodulation process in Rhizobium in early stages. M. loti T4SS mutants delayed nodulation on Lotus corniculatus and allows effective nodulation on Leucaena leucocephala (Hubber et al., 2004, 2007). R. etli encodes a T4SS locus (vir) and is able to mediate transfer and integration of DNA into plant cell genome when provided with a T-DNA (Lacroix and Citovsky, 2016). However, a T-DNA-like sequences in R. etli was not identified, suggesting that Rhizobium-mediated plant transformation does not occur in nature, although it cannot be ruled out that other Rhizobium strains, not yet sequenced, harbor a T-DNA.
Thus far, only two T4SS candidate effector proteins were identified in rhizobia. These are Msi059 and Msi061 from M. loti R7A (Nelson and Sadowsky, 2015). Msi059 shares a partial protein sequence similarity to the XopD C48 cysteine peptidase (Rodrigues et al., 2007; Nelson and Sadowsky, 2015). The other T4SS effector Msi061, shares protein similarity with A. tumefaciens effector VirF (Tzfira et al., 2004). VirF interacts with the host Skp1 protein to facilitate protein degradation of effector proteins VirE2 and Vip1 leading to unbinding of the T-DNA after entry into the host cell (Tzfira et al., 2004). The specific role of the Msi059 and Msi061 in RNB remains unidentified, but the latest evidence suggests that they modulate protein expression levels in planta (Nelson and Sadowsky, 2015).
Type VI Secretion System (T6SS) contractile nanoweapons allows bacteria to inject toxins directly into prey cell membranes, periplasm or cytoplasm, leading to cell growth arrest. In rhizobia, T6SS sequence have been found in several species such as R. leguminosarum, B. japonicum, M. loti, Sinorhizobium saheli, and S. fredii (Bladergroen et al., 2003). T6SS was related to the prevention of nodulation on Pisum sativum cv. Rondo (Bladergroen et al., 2003). Recently, it was reported that R. etli Mim1 T6SS mutant produced plants with lower dry weight and smaller nodules than the wild-type strain, suggesting for the first time a positive role of T6SS in Rhizobium-legume symbiosis (Salinero-Lanzarote et al., 2019). The rhizobacterium Azorhizobium caulinodans ORS571 utilizes its T6SS to outcompete other strains during infection of its host Sesbania rostrata (Lin et al., 2018). However, the researchers could not show involvement in inter-bacterial competition in vitro. The nitrogen fixing bacteria Azoarcus olearius BH72 encodes two T6SS operons, one of which is strongly up-regulated when nitrogen is absent (Jiang et al., 2019). Kosakonia strains are endophytic nitrogen fixers involved in plant growth promotion in rice (Bertani et al., 2016). T6SS of Kosakonia KO348 is important for rhizoplane and endosphere colonization but it is not clear exactly how (Mosquito et al., 2019). One possibility is that the microbes use the T6SS to facilitate colonization by inhibiting competitors in the rhizosphere.
Although different secretion systems and effectors have been identified in RNB, their specific role in symbiosis and nodulation is still unclear. Further molecular and biochemical work should be done to characterize the molecular mechanisms leading to secretion of proteins and other molecules and their effects in planta.
Biocontrol Agents
Biocontrol agents secrete a broad spectrum of secondary metabolites and proteins which can serve as antibacterial and antifungal compounds, such as enzymes which are able to degrade different compartments of various organisms (Mullins et al., 2019; Vesga et al., 2020). Some BCAs employ secretion systems to penetrate the neighboring cells and inject toxins into them. Pseudomonas spp., and Bacillus spp. are two of the most studied organisms in the BCA field. The most important and most studied secondary metabolites are antibiotics such as Phenazines, Phloroglucinols, Dialkylresorcinols, Pyrolnitrin, Pyoluteorin, Mupirocin, Peptide antibiotics, Hyrdogen cyanide, Rhizoxins, and Oxyvinylglycines (Raaijmakers et al., 2002; Weller, 2007; Mavrodi et al., 2011). Bacillus velezensis strains isolated from tomato rhizosphere strongly inhibit growth of Ralstonia solanacearum and Fusarium oxysporum under both laboratory and greenhouse conditions (Cao et al., 2018). This is done by production of different lipopeptide compounds whose production is stimulated during the BCA interaction with R. solanacearum. Recently, a survey of bacteria isolated from the phyllosphere of A. thaliana revealed novel antibiotics, with possible novel modes of actions (Helfrich et al., 2018). Antibiotics can be identified via HPLC and then tested for their antagonistic effect against different pathogens (Shahid et al., 2017). Many toxins (proteins or secondary metabolites) which are produced by beneficial bacteria have been studied beyond their antimicrobial/antifungal activity. For example, the BCA P. fluorescens Pf29Arp downregulates relevant pathogenicity enzymes (laccasses, exogluanases, and mitogen-activated kinases) in the fungus Gaeumannomyces graminis var. tritici, the causing agent of take-all disease (Guilleroux and Osbourn, 2004; Daval et al., 2011). Other assays can be used for profiling secondary metabolites, such as the use of LC-MS on crude extracts from BCA strains, in silico screening of antagonistic potential on pathogenic genes, and finally in vitro screening against specific pathogens (Jinal and Amaresan, 2020). Burkholderia ambifaria, a biocontrol agent was screened for its antimicrobial metabolites which led to detection of Cepacin A via LC-MS. Mutants for Cepain A production in B. ambifaria have a significantly reduced inhibition activity against Pythium ultimum in a pea infection model (Mullins et al., 2019).
Disease-suppressive soils prevent establishment of pathogens or lead to minor plant disease. The Raaijmakers group was able to demonstrate the involvement of beneficial bacteria from Burkholderiaceae family in disease-suppressive activity against R. solani (Chapelle et al., 2016; Carrión et al., 2018). They isolated representative Burkholderiaceae strains and uncovered genes involved in in vitro and in situ antifungal activity via the production of sulfurous volatile compounds (Carrión et al., 2018). Recently, they showed that an endophytic consortium of Chitinophaga and Flavobacterium consistently inhibited Rhizoctonia solani infection (Carrión et al., 2019). Moreover, they showed that the fungal infection enriched the root metagenome for chitinase genes and candidate biosynthetic gene clusters that likely produce antifungals. Finally, site-directed mutagenesis revealed a new NRPS-PKS gene cluster from Flavobacterium that is essential for disease suppression by the consortium. This is a fine example of how years of research revealed first specific BCA strains and later on their molecular mechanism that underlies a reproducible root microbiome that mediated plant protection.
As discussed already in the root nodulation section, Gram-negative bacteria can be equipped with different secretion systems. The T6SS translocates toxins into the neighboring cells that are killed if they do not have the matching immunity protein (Hood et al., 2010). T6SS genes were found, for examples, in P. fluorescens strain MFE01. Different T6SS effectors are injected by this strain. However, those toxins are not virulent against eukaryotic cells, but against a broad spectrum of pathogenic bacteria (Decoin et al., 2014). Bernal and colleges identified in P. putida KT2440 three T6SS clusters and 10 T6SS effector-immunity pairs. One of the T6SS loci is responsible to bactericidal activity against phytopathogens in vitro and in planta on N. benthamiana, although the in planta effect was mild (Bernal et al., 2017).
Biocontrol agents can also induce plant responses by secreting secondary metabolites. Often this results in an induction of the plant immune response called induced systemic resistance (ISR), which is regulated by the plant hormones jasmonic acid (JA) and ET (Berendsen et al., 2012; Pieterse et al., 2014). ISR is a response which is known to be triggered by rhizobacteria and leads to secretion of antimicrobial secondary metabolites from plants (Pieterse et al., 2012; Yu et al., 2019b). Pseudomonas fluorescens Q8r1–96 contains T3SS effectors RopAA, RopB, and RopM. In N. benthamiana these effectors suppress two plant immune pathways after leaf infection with P. syringae DC3000; the hypersensitive response and the production of reactive oxygens species (Mavrodi et al., 2011). Q8r1-96 also produces DAPG, which suppresses the take-all disease in wheat (Brazelton et al., 2008; Mavrodi et al., 2011; Kwak et al., 2012; Yang et al., 2020). Bacillus spp. is a well-established ISR elicitor. Bacillus subitilis BBG111 releases cyclic lipopeptides (CLPs), which are magnifying the plant microbe-associated molecular patterns (MAMPs) triggered immunity (MTI). The MTI recognizes microbe derived compounds, such as flagellins, lipopolysaccharides, and chitin that trigger the ISR pathway in rice against R. solani (Chandler et al., 2015; Lastochkina et al., 2019). This induction of ISR does not necessarily lead to the resistance against one phytopathogen since it is not species-specific (Chandler et al., 2015). Bacillus spp. and Pseudomonas spp. increase ISR in different kinds of crops (tomato, melon, and bean) against different organisms including fungi, bacteria, and nematodes (Ongena et al., 2005; García-Gutiérrez et al., 2013; Farace et al., 2015). The induction of ISR is very powerful, however its broad-spectrum activity may lead to killing of beneficial bacteria.
Often the combination of both PGPR and BCA can ensure both plant protectiveness and growth induction. Both traits can be tested in vitro on specific media. Liu et al. (2017) screened 196 PGPR strains based on their disease suppression for broad-spectrum antagonistic activity. In a second screen selected strains were tested for PGPR traits in vitro. For example, nitrogen fixation was tested on nitrogen-free semisolid medium and phosphate solubilization on media with different phosphate sources. In advanced screens, the PGPR strains were tested in planta for biological control of multiple plant diseases and most of them significantly reduced at least two tested diseases. Gene encoding antimicrobials were predicted but have not been experimentally validated (Liu et al., 2017).
The root nodules are also sites of active antimicrobial production. Brevibacillus brevis is an accessory species which resides near dominant rhizobia species. An untargeted in planta metabolomics study of this strain led to identification of nonribosomal peptides, Britacidin and gramicidin. Sequencing of the strain’s genome led to assignment of these antimicrobials to their cognate biosynthetic gene clusters. It is yet unknown whether these antimicrobials are used in competition between the natural nodule microbiome or protect it from pathogen infection (Hansen et al., 2020).
Discussion
Much research has been conducted regarding PGPR, BCA, and RNB and many effectors are known and are not mentioned in this review. Despite the knowledge of those secreted molecules, their functionality in planta remains unclear (Bai et al., 2020; Kumar and Dubey, 2020; Zhou et al., 2020). The importance of the RNB secretion system in nodule formation and symbiosis between rhizobia and legumes is known; however, direct interactions of effectors and plant proteins and the specific processes regulated by the effectors are not understood. Many hypotheses have been postulated but were not confirmed experimentally (Sachs et al., 2018). High-quality ecological studies revealed the function of specific rhizobacteria in protecting plants against bacterial, fungal, and oomycete pathogens but did not reveal the compounds responsible for this effect (Duran et al., 2018; Kwak et al., 2018). High-density transposon screens coupled with in planta phenotyping can uncover the genes responsible for these antagonistic phenotypes. Another approach that should be applied is systematic gain of function approach to uncover the secondary metabolome encoded by the biosynthetic gene clusters of beneficial rhizobacteria. This can be done by using large scale operon cloning, induction of operons in organisms such as Escherichia coli, and applying the lysates on plants to couple microbial metabolites with beneficial functions.
We believe that genetic, metagenomics, transcriptomics, proteomics (secretome), and metabolomics analyses should increase our knowledge about the effectors and small molecules injected by rhizobacteria into the host, nearby pathogens, or released into the surrounding soil (Levy et al., 2018a). Identification of the specific genes, proteins and molecules responsible for growth promotion and protection against pathogens will allow a more accurate identification of beneficial strains and engineering of plant supportive microbiomes. We think that the entire field will gain important basic and applied insights by moving from identification of beneficial strains through extensive phenotype screening toward molecule-centered or gene-centered phenotypic associations. Identification of new genes and molecules that underlie a beneficial phenotype will allow accurate discovery of novel beneficial strains based on their genetic and chemical features identified from metagenome and metabolome surveys. Downstream functional analysis such as random mutagenesis of beneficial microbes coupled with identification of phenotypes in planta, protein binding assays to identify the binding partners of effectors in plant cells, or cell-based assays to show translocation of effectors into plants could improve molecular understanding of beneficial bacterial interaction with plants. Specifically, very little is known on the interaction of proteins and small molecules from beneficial microbes with the different branches of the plant immune system.
In addition to the lack of functional studies revealing the molecular basis for a beneficial microbial phenotype in plants, the understanding of bacteria communities in soil is also very partially understood. Recently, more studies include synthetic communities, revealing that certain assemblies of rhizobacteria are having a positive influence on plant fitness and health (Finkel et al., 2017; Helfrich et al., 2018). Sequenced and annotated genomes of those bacterial communities are available, however functional analysis lags behind (Finkel et al., 2017; Helfrich et al., 2018; Levy et al., 2018a,b).
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
ML is supported by the Landtagsstipendium from the Ministry of Science, Research and Arts from Baden-Württemberg in Germany. MC is supported by a scholarship from IFARHU. AL is supported by Alon Fellowship of the Israeli council of higher education. This research was supported by the Israel Science Foundation (Grant No. 1535/20).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Omri Finkel and Alexander Martin Geller for critical reading and editing of the manuscript.
References
Abadía, J., López-Millán, A. F., Rombolà, A., and Abadía, A. (2002). Organic acids and Fe deficiency: a review. Plant Soil 241, 75–86. doi: 10.1023/A:1016093317898
Achard, P., Liao, L., Jiang, C., Desnos, T., Bartlett, J., Fu, X., et al. (2007). DELLAs contribute to plant photomorphogenesis. Plant Physiol. 143, 1163–1172. doi: 10.1104/pp.106.092254
Akhtar, S. S., Mekureyaw, M. F., Pandey, C., and Roitsch, T. (2020). Role of cytokinins for interactions of plants with microbial pathogens and pest insects. Front. Plant Sci. 10:1777. doi: 10.3389/fpls.2019.01777
Aras, S., Arıkan, Ş., İpek, M., Eşitken, A., Pırlak, L., Dönmez, M. F., et al. (2018). Plant growth promoting rhizobacteria enhanced leaf organic acids, FC-R activity and Fe nutrition of apple under lime soil conditions. Acta Physiol. Plant. 40:120. doi: 10.1007/s11738-018-2693-9
Ariizumi, T., and Steber, C. M. (2007). Seed germination of GA-insensitive sleepy1 mutants does not require RGL2 protein disappearance in arabidopsis. Plant Cell 19, 791–804. doi: 10.1105/tpc.106.048009
Avonce, N., Mendoza-Vargas, A., Morett, E., and Iturriag, G. (2006). Insights on the evolution of trehalose biosynthesis. BMC Evol. Biol. 6, 1–15. doi: 10.1186/1471-2148-6-109
Bai, Y. -C., Chang, Y. -Y., Hussain, M., Lu, B., Zhang, J. -P., Song, X. -B., et al. (2020). Soil chemical and microbiological properties are changed by long-term chemical fertilizers that limit ecosystem functioning. Microorganisms 8:694. doi: 10.3390/microorganisms8050694
Bakker, P. A. H. M., Pieterse, C. M. J., de Jonge, R., and Berendsen, R. L. (2018). The soil-borne legacy. Cell 172, 1178–1180. doi: 10.1016/j.cell.2018.02.024
Bartsev, A. V., Deakin, W. J., Boukli, N. M., McAlvin, C. B., Stacey, G., Malnoë, P., et al. (2004). NopL, an effector protein of rhizobium sp. NGR234, thwarts activation of plant defense reactions. Plant Physiol. 134, 871–879. doi: 10.1104/pp.103.031740
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Bernal, P., Allsopp, L. P., Filloux, A., and Llamas, M. A. (2017). The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J. 11, 972–987. doi: 10.1038/ismej.2016.169
Bernal, P., LlamBernal, P., Llamas, M. A., and Filloux, A. (2018). Type VI secretion systems in plant-associated bacteria. Environ. Microbiol. 20, 1–15. doi: 10.1111/1462-2920.13956
Bertani, I., Abbruscato, P., Piffanelli, P., Subramoni, S., and Venturi, V. (2016). Rice bacterial endophytes: isolation of a collection, identification of beneficial strains and microbiome analysis. Environ. Microbiol. Rep. 8, 388–398. doi: 10.1111/1758-2229.12403
Bladergroen, M. R., Badelt, K., and Spaink, H. P. (2003). Infection-blocking genes of a symbiotic rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant-Microbe Interact. 16, 53–64. doi: 10.1094/MPMI.2003.16.1.53
Bogino, P. C., Oliva, M. D. L. M., Sorroche, F. G., and Giordano, W. (2013). The role of bacterial biofilms and surface components in plant-bacterial associations. Int. J. Mol. Sci. 14, 15838–15859. doi: 10.3390/ijms140815838
Brazelton, J. N., Pfeufer, E. E., Sweat, T. A., McSpadden Gardener, B. B., and Coenen, C. (2008). 2,4-Diacetylphloroglucinol alters plant root development. Mol. Plant-Microbe Interact. 21, 1349–1358. doi: 10.1094/MPMI-21-10-1349
Burkinshaw, B. J., and Strynadka, N. C. (2014). Assembly and structure of the T3SS. Biochim. Biophys. Acta 1843, 1649–1663. doi: 10.1016/j.bbamcr.2014.01.035
Cao, Y., Pi, H., Chandrangsu, P., Li, Y., Wang, Y., Zhou, H., et al. (2018). Antagonism of two plant-growth promoting bacillus velezensis isolates against Ralstonia solanacearum and Fusarium oxysporum. Sci. Rep. 8, 1–14. doi: 10.1038/s41598-018-22782-z
Carrión, V. J., Cordovez, V., Tyc, O., Etalo, D. W., de Bruijn, I., de Jager, V. C. L., et al. (2018). Involvement of Burkholderiaceae and sulfurous volatiles in disease-suppressive soils. ISME J. 12, 2307–2321. doi: 10.1038/s41396-018-0186-x
Carrión, V. J., Perez-Jaramillo, J., Cordovez, V., Tracanna, V., De Hollander, M., Ruiz-Buck, D., et al. (2019). Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 366, 606–612. doi: 10.1126/science.aaw9285
Chandler, S., Van Hese, N., Coutte, F., Jacques, P., Höfte, M., and De Vleesschauwer, D. (2015). Role of cyclic lipopeptides produced by Bacillus subtilis in mounting induced immunity in rice (Oryza sativa L.). Physiol. Mol. Plant Pathol. 91, 20–30. doi: 10.1016/j.pmpp.2015.05.010
Chapelle, E., Mendes, R., Bakker, P. A. H. M., and Raaijmakers, J. M. (2016). Fungal invasion of the rhizosphere microbiome. ISME J. 10, 265–268. doi: 10.1038/ismej.2015.82
Cheng, Z., Park, E., and Glick, B. R. (2007). 1-Aminocyclopropane-1-carboxylate deaminase from pseudomonas putida UW4 facilitates the growth of canola in the presence of salt. Can. J. Microbiol. 53, 912–918. doi: 10.1139/W07-050
Christie, P. J., and Cascales, E. (2005). Structural and dynamic properties of bacterial type IV secretion systems (review). Mol. Membr. Biol. 22, 51–61. doi: 10.1080/09687860500063316
Christie, P. J., Whitaker, N., and González-Rivera, C. (2014). Mechanism and structure of the bacterial type IV secretion systems. Biochim. Biophys. Acta-Mol. Cell Res. 1843, 1578–1591. doi: 10.1016/j.bbamcr.2013.12.019
Cianciotto, N. P. (2005). Type II secretion: A protein secretion system for all seasons. Trends Microbiol. 13, 581–588. doi: 10.1016/j.tim.2005.09.005
Clark, F. E. (1949). Soil microorganisms and plant roots. Adv. Agron. 1, 241–288. doi: 10.1016/S0065-2113(08)60750-6
Cohen, A. C., Bottini, R., and Piccoli, P. N. (2008). Azospirillum brasilense Sp 245 produces ABA in chemically-defined culture medium and increases ABA content in arabidopsis plants. Plant Growth Regul. 54, 97–103. doi: 10.1007/s10725-007-9232-9
Cosme, A. M., Becker, A., Santos, M. R., Sharypova, L. A., Santos, P. M., and Moreira, L. M. (2008). The outer membrane protein TolC from Sinorhizobium meliloti affects protein secretion, polysaccharide biosynthesis, antimicrobial resistance, and symbiosis. Mol. Plant-Microbe Interact. 21, 947–957. doi: 10.1094/MPMI-21-7-0947
Dai, W. J., Zeng, Y., Xie, Z. P., and Staehelin, C. (2008). Symbiosis-promoting and deleterious effects of NopT, a novel type 3 effector of rhizobium sp. strain NGR234. J. Bacteriol. 190, 5101–5110. doi: 10.1128/JB.00306-08
Daval, S., Lebreton, L., Gazengel, K., Boutin, M., Guillerm-Erckelboudt, A. Y., and Sarniguet, A. (2011). The biocontrol bacterium Pseudomonas fluorescens Pf29Arp strain affects the pathogenesis-related gene expression of the take-all fungus Gaeumannomyces graminis var. tritici on wheat roots. Mol. Plant Pathol. 12, 839–854. doi: 10.1111/j.1364-3703.2011.00715.x
Deakin, W. J., and Broughton, W. J. (2009). Symbiotic use of pathogenic strategies: Rhizobial protein secretion systems. Nat. Rev. Microbiol. 7, 312–320. doi: 10.1038/nrmicro2091
Decoin, V., Barbey, C., Bergeau, D., Latour, X., Feuilloley, M. G. J., Orange, N., et al. (2014). A type VI secretion system is involved in Pseudomonas fluorescens bacterial competition. PLoS One 9:e89411. doi: 10.1371/journal.pone.0089411
Díaz-Zorita, M., and Fernández-Canigia, M. V. (2009). Field performance of a liquid formulation of Azospirillum brasilense on dryland wheat productivity. Eur. J. Soil Biol. 45, 3–11. doi: 10.1016/j.ejsobi.2008.07.001
Dobbelaere, S., Croonenborghs, A., Thys, A., Vande Broek, A., and Vanderleyden, J. (1999). Phytostimulatory effect of Azospirillum brasilense wild type and mutant strains altered in IAA production on wheat. Plant Soil 212, 155–164. doi: 10.1023/A:1004658000815
Downie, J. A. (2010). The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. FEMS Microbiol. Rev. 34, 150–170. doi: 10.1111/j.1574-6976.2009.00205.x
Duran, P., Thiergart, T., Garrido-Oter, R., Agler, M., Kemen, E., and Schulze-Lefert, P. (2018). Microbial Interkingdom interactions in roots promote Arabidopsis survival. Cell 175, 973–983.e14. doi: 10.1016/j.cell.2018.10.020
Farace, G., Fernandez, O., Jacquens, L., Coutte, F., Krier, F., Jacques, P., et al. (2015). Cyclic lipopeptides from Bacillus subtilis activate distinct patterns of defence responses in grapevine. Mol. Plant Pathol. 16, 177–187. doi: 10.1111/mpp.12170
Finkel, O. M., Castrillo, G., Herrera Paredes, S., Salas González, I., and Dangl, J. L. (2017). Understanding and exploiting plant beneficial microbes. Curr. Opin. Plant Biol. 38, 155–163. doi: 10.1016/j.pbi.2017.04.018
Finkel, O. M., Salas-González, I., Castrillo, G., Conway, J. M., Law, T. F., Teixeira, P. J. P. L., et al. (2020). A single bacterial genus maintains root development in a complex microbiome. bioRxiv [Preprint] doi: 10.1101/645655
Finnie, C., Hartley, N. M., Findlay, K. C., and Downie, J. A. (1997). The Rhizobium leguminosarum prsDE genes are required for secretion of several proteins, some of which influence nodulation, symbiotic nitrogen fixation and exopolysaccharide modification. Mol. Microbiol. 25, 135–146. doi: 10.1046/j.1365-2958.1997.4471803.x
Forni, C., Duca, D., and Glick, B. R. (2017). Mechanisms of plant response to salt and drought stress and their alteration by rhizobacteria. Plant Soil 410, 335–356. doi: 10.1007/s11104-016-3007-x
Frederix, M., Edwards, A., Swiderska, A., Stanger, A., Karunakaran, R., Williams, A., et al. (2014). Mutation of praR in Rhizobium leguminosarum enhances root biofilms, improving nodulation competitiveness by increased expression of attachment proteins. Mol. Microbiol. 93, 464–478. doi: 10.1111/mmi.12670
García-Gutiérrez, L., Zeriouh, H., Romero, D., Cubero, J., de Vicente, A., and Pérez-García, A. (2013). The antagonistic strain Bacillus subtilis UMAF6639 also confers protection to melon plants against cucurbit powdery mildew by activation of jasmonate- and salicylic acid-dependent defence responses. Microb. Biotechnol. 6, 264–274. doi: 10.1111/1751-7915.12028
García de Salamone, I. E., Hynes, R. K., and Nelson, L. M. (2001). Cytokinin production by plant growth promoting rhizobacteria and selected mutants. Can. J. Microbiol. 47, 404–411. doi: 10.1139/w01-029
Glick, B. R. (2014). Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 169, 30–39. doi: 10.1016/j.micres.2013.09.009
Goswami, D., Dhandhukia, P., Patel, P., and Thakker, J. N. (2014). Screening of PGPR from saline desert of Kutch: growth promotion in Arachis hypogea by Bacillus licheniformis A2. Microbiol. Res. 169, 66–75. doi: 10.1016/j.micres.2013.07.004
Gourion, B., Berrabah, F., Ratet, P., and Stacey, G. (2015). Rhizobium-legume symbioses: the crucial role of plant immunity. Trends Plant Sci. 20, 186–194. doi: 10.1016/j.tplants.2014.11.008
Großkinsky, D. K., Tafner, R., Moreno, M. V., Stenglein, S. A., De Salamone, I. E. G., Nelson, L. M., et al. (2016). Cytokinin production by Pseudomonas fluorescens G20-18 determines biocontrol activity against pseudomonas syringae in Arabidopsis. Sci. Rep. 6:23310. doi: 10.1038/srep23310
Guilleroux, M., and Osbourn, A. (2004). Gene expression during infection of wheat roots by the “take-all” fungus Gaeumannomyces graminis. Mol. Plant Pathol. 5, 203–216. doi: 10.1111/j.1364-3703.2004.00219.x
Hacquard, S., Garrido-Oter, R., González, A., Spaepen, S., Ackermann, G., Lebeis, S., et al. (2015). Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 17, 603–616. doi: 10.1016/j.chom.2015.04.009
Hansen, B. L., Pessotti, R.d. C., Fischer, M. S., Collins, A., El-Hifnawi, L., Liu, M. D., et al. (2020). Cooperation, competition, and specialized metabolism in a simplified root nodule microbiome. MBio 11, 1–20. doi: 10.1128/mBio.01917-20
Helfrich, E. J. N., Vogel, C. M., Ueoka, R., Schäfer, M., Ryffel, F., Müller, D. B., et al. (2018). Bipartite interactions, antibiotic production and biosynthetic potential of the arabidopsis leaf microbiome. Nat. Microbiol. 3, 909–919. doi: 10.1038/s41564-018-0200-0
Hiltner, L. (1904). Uber nevere Erfahrungen und Probleme auf dem Gebiet der Boden Bakteriologie und unter besonderer Beurchsichtigung der Grundungung und Broche. Arbeit. Deut. Landw. Ges. Berlin 98, 59–78.
Ho, B. T., Dong, T. G., and Mekalanos, J. J. (2014). A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15, 9–21. doi: 10.1016/j.chom.2013.11.008
Hood, R. D., Singh, P., Hsu, F. S., Güvener, T., Carl, M. A., Trinidad, R. R. S., et al. (2010). A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37. doi: 10.1016/j.chom.2009.12.007
Hotson, A., Chosed, R., Shu, H., Orth, K., and Mudgett, M. B. (2003). Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol. Microbiol. 50, 377–389. doi: 10.1046/j.1365-2958.2003.03730.x
Hubber, A. M., Sullivan, J. T., and Ronson, C. W. (2007). Symbiosis-induced cascade regulation of the Mesorhizobium loti R7A VirB/D4 type IV secretion system. Mol. Plant-Microbe Interact. 20, 255–261. doi: 10.1094/MPMI-20-3-0255
Hubber, A., Vergunst, A. C., Sullivan, J. T., Hooykaas, P. J. J., and Ronson, C. W. (2004). Symbiotic phenotypes and translocated effector proteins of the Mesorhizobium loti strain R7A VirB/D4 type IV secretion system. Mol. Microbiol. 54, 561–574. doi: 10.1111/j.1365-2958.2004.04292.x
İpek, M., Aras, S., Arıkan, Ş., Eşitken, A., Pırlak, L., Dönmez, M. F., et al. (2017). Root plant growth promoting rhizobacteria inoculations increase ferric chelate reductase (FC-R) activity and Fe nutrition in pear under calcareous soil conditions. Sci. Hortic. 219, 144–151. doi: 10.1016/j.scienta.2017.02.043
Ivanova, R. P., Bojinova, D. Y., Gruncharov, I. N., and Damgaliev, D. L. (2006). The Solubilization of rock phosphate by organic acids. Phosphorus Sulfur Silicon Relat. Elem. 181, 2541–2554. doi: 10.1080/10426500600758399
Jamali, H., Sharma, A., Roohi, and Srivastava, A. K. (2020). Biocontrol potential of Bacillus subtilis RH5 against sheath blight of rice caused by Rhizoctonia solani. J. Basic Microbiol. 60, 268–280. doi: 10.1002/jobm.201900347
Jha, C. K., Patel, B., and Saraf, M. (2012). Stimulation of the growth of Jatropha curcas by the plant growth promoting bacterium Enterobacter cancerogenus MSA2. World J. Microbiol. Biotechnol. 28, 891–899. doi: 10.1007/s11274-011-0886-0
Jiang, X., Beust, A., Sappa, P. K., Völker, U., Dinse, T., Herglotz, J., et al. (2019). Two functionally deviating type 6 secretion systems occur in the nitrogen-fixing Endophyte Azoarcus olearius BH72. Front. Microbiol. 10:459. doi: 10.3389/fmicb.2019.00459
Jiménez-Guerrero, I., Pérez-Montaño, F., Medina, C., Ollero, F. J., and López-Baena, F. J. (2015). NopC is a rhizobium-specific type 3 secretion system effector secreted by sinorhizobium (ensifer) fredii HH103. PLoS One 10:e0142866. doi: 10.1371/journal.pone.0142866
Jinal, H. N., and Amaresan, N. (2020). Characterization of medicinal plant-associated biocontrol Bacillus subtilis (SSL2) by liquid chromatography-mass spectrometry and evaluation of compounds by in silico and in vitro methods. J. Biomol. Struct. Dyn. 38, 500–510. doi: 10.1080/07391102.2019.1581091
Kambara, K., Ardissone, S., Kobayashi, H., Saad, M. M., Schumpp, O., Broughton, W. J., et al. (2009). Rhizobia utilize pathogen-like effector proteins during symbiosis. Mol. Microbiol. 71, 92–106. doi: 10.1111/j.1365-2958.2008.06507.x
Kang, S. -M., Latif Khan, A., You, Y. -H., Kim, J. -G., Kamran, M., and Lee, I. -J. (2014). Gibberellin production by newly isolated strain Leifsonia soli SE134 and its potential to promote plant growth. J. Microbiol. Biotechnol. 24, 106–112. doi: 10.4014/jmb.1304.04015
Kang, S. M., Asaf, S., Kim, S. J., Yun, B. W., and Lee, I. J. (2016). Complete genome sequence of plant growth-promoting bacterium Leifsonia xyli SE134, a possible gibberellin and auxin producer. J. Biotechnol. 239, 34–38. doi: 10.1016/j.jbiotec.2016.10.004
Kim, M. J., Radhakrishnan, R., Kang, S. M., You, Y. H., Jeong, E. J., Kim, J. G., et al. (2017). Plant growth promoting effect of Bacillus amyloliquefaciens H-2-5 on crop plants and influence on physiological changes in soybean under soil salinity. Physiol. Mol. Biol. Plants 23, 571–580. doi: 10.1007/s12298-017-0449-4
Kloepper, J. W., Lifshitz, R., and Zablotowicz, R. M. (1989). Free-living bacterial inocula for enhancing crop productivity. Trends Biotechnol. 7, 39–44. doi: 10.1016/0167-7799(89)90057-7
Kobayashi, H., Naciri-Graven, Y., Broughton, W. J., and Perret, X. (2004). Flavonoids induce temporal shifts in gene-expression of nod-box controlled loci in rhizobium sp. NGR234. Mol. Microbiol. 51, 335–347. doi: 10.1046/j.1365-2958.2003.03841.x
Korenblum, E., Dong, Y., Szymanski, J., Panda, S., Jozwiak, A., Massalha, H., et al. (2020). Rhizosphere microbiome mediates systemic root metabolite exudation by root-to-root signaling. Proc. Natl. Acad. Sci. U. S. A. 117, 3874–3883. doi: 10.1073/pnas.1912130117
Krehenbrink, M., and Allan, J. A. (2008). Identification of protein secretion systems and novel secreted proteins in rhizobium leguminosarum bv. Viciae. BMC Genomics 9:55. doi: 10.1186/1471-2164-9-55
Kuc, J. A., and Tuzun, S. (1992). Proposed definitions related to induced disease resistance. Biocontrol Sci. Tech. 2, 349–351. doi: 10.1080/09583159209355251
Kuldau, G. A., De Vos, G., Owen, J., McCaffrey, G., and Zambryski, P. (1990). The virB operon of agrobacterium tumefaciens pTiC58 encodes 11 open reading frames. MGG Mol. Gen. Genet. 221, 256–266. doi: 10.1007/BF00261729
Kumar, A., and Dubey, A. (2020). Rhizosphere microbiome: engineering bacterial competitiveness for enhancing crop production. J. Adv. Res. 24, 337–352. doi: 10.1016/j.jare.2020.04.014
Kwak, M. J., Kong, H. G., Choi, K., Kwon, S. K., Song, J. Y., Lee, J., et al. (2018). Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 36, 1100–1116. doi: 10.1038/nbt.4232
Kwak, Y. S., Bonsall, R. F., Okubara, P. A., Paulitz, T. C., Thomashow, L. S., and Weller, D. M. (2012). Factors impacting the activity of 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens against take-all of wheat. Soil Biol. Biochem. 54, 48–56. doi: 10.1016/j.soilbio.2012.05.012
Lacroix, B., and Citovsky, V. (2016). A functional bacterium-to-plant DNA transfer machinery of rhizobium etli. PLoS Pathog. 12:e1005502. doi: 10.1371/journal.ppat.1005502
Lastochkina, O., Seifikalhor, M., Aliniaeifard, S., Baymiev, A., Pusenkova, L., Garipova, S., et al. (2019). Bacillus spp.: efficient biotic strategy to control postharvest diseases of fruits and vegetables. Plan. Theory 8:97. doi: 10.3390/plants8040097
Levy, A., Conway, J. M., Dangl, J. L., and Woyke, T. (2018a). Elucidating bacterial gene functions in the plant microbiome. Cell Host Microbe 24, 475–485. doi: 10.1016/j.chom.2018.09.005
Levy, A., Salas Gonzalez, I., Mittelviefhaus, M., Clingenpeel, S., Herrera Paredes, S., Miao, J., et al. (2018b). Genomic features of bacterial adaptation to plants. Nat. Genet. 50, 138–150. doi: 10.1038/s41588-017-0012-9
Lin, H. -H., Huang, H. -M., Yu, M., Lai, E. -M., Chien, H. -L., and Liu, C. -T. (2018). Functional exploration of the bacterial type VI secretion system in mutualism: Azorhizobium caulinodansORS571– Sesbania rostrata as a research model. Mol. Plant-Microbe Interact. 31, 856–867. doi: 10.1094/MPMI-01-18-0026-R
Liu, K., Newman, M., McInroy, J. A., Hu, C. H., and Kloepper, J. W. (2017). Selection and assessment of plant growth-promoting rhizobacteria for biological control of multiple plant diseases. Phytopathology 107, 928–936. doi: 10.1094/PHYTO-02-17-0051-R
López-Baena, F. J., Ruiz-Sainz, J. E., Rodríguez-Carvajal, M. A., and Vinardell, J. M. (2016). Bacterial molecular signals in the Sinorhizobium fredii-soybean symbiosis. Int. J. Mol. Sci. 17:755. doi: 10.3390/ijms17050755
Lugtenberg, B., and Kamilova, F. (2009). Plant-growth-promoting Rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. doi: 10.1146/annurev.micro.62.081307.162918
Marie, C., Deakin, W. J., Ojanen-Reuhs, T., Diallo, E., Reuhs, B., Broughton, W. J., et al. (2004). TtsI, a key regulator of rhizobium species NGR234 is required for type III-dependent protein secrection and synthesis of rhamnose-rich polysaccharides. Mol. Plant-Microbe Interact. 17, 958–966. doi: 10.1094/MPMI.2004.17.9.958
Mavrodi, D. V., Joe, A., Mavrodi, O. V., Hassan, K. A., Weller, D. M., Paulsen, I. T., et al. (2011). Structural and functional analysis of the type III secretion system from Pseudomonas fluorescens Q8r1-96. J. Bacteriol. 193, 177–189. doi: 10.1128/JB.00895-10
Mergaert, P. (2018). Role of antimicrobial peptides in controlling symbiotic bacterial populations. Nat. Prod. Rep. 35, 336–356. doi: 10.1039/c7np00056a
Miwa, H., and Okazaki, S. (2017). How effectors promote beneficial interactions. Curr. Opin. Plant Biol. 38, 148–154. doi: 10.1016/j.pbi.2017.05.011
Molina, R., Rivera, D., Mora, V., López, G., Rosas, S., Spaepen, S., et al. (2018). Regulation of IAA biosynthesis in Azospirillum brasilense under environmental stress conditions. Curr. Microbiol. 75, 1408–1418. doi: 10.1007/s00284-018-1537-6
Mongiardini, E. J., Pérez-Giménez, J., Althabegoiti, M. J., Covelli, J., Quelas, J. I., López-García, S. L., et al. (2009). Overproduction of the rhizobial adhesin RapA1 increases competitiveness for nodulation. Soil Biol. Biochem. 41, 2017–2020. doi: 10.1016/j.soilbio.2009.07.016
Mosquito, S., Bertani, I., Licastro, D., Compant, S., Myers, M. P., Hinarejos, E., et al. (2019). In planta colonization and role of T6SS in two rice Kosakonia endophytes. Mol. Plant-Microbe Interact. 33, 349–363. doi: 10.1094/MPMI-09-19-0256-R
Mukherjee, S., Keitany, G., Li, Y., Wang, Y., Ball, H. L., Goldsmith, E. J., et al. (2006). Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science 312, 1211–1214. doi: 10.1126/science.1126867
Mullins, A. J., Murray, J. A. H., Bull, M. J., Jenner, M., Jones, C., Webster, G., et al. (2019). Genome mining identifies cepacin as a plant-protective metabolite of the biopesticidal bacterium Burkholderia ambifaria. Nat. Microbiol. 4, 996–1005. doi: 10.1038/s41564-019-0383-z
Nascimento, F. X., Tavares, M. J., Franck, J., Ali, S., Glick, B. R., and Rossi, M. J. (2019). ACC deaminase plays a major role in Pseudomonas fluorescens YsS6 ability to promote the nodulation of alpha- and Betaproteobacteria rhizobial strains. Arch. Microbiol. 201, 817–822. doi: 10.1007/s00203-019-01649-5
Nelson, M. S., and Sadowsky, M. J. (2015). Secretion systems and signal exchange between nitrogen-fixing rhizobia and legumes. Front. Plant Sci. 6:491. doi: 10.3389/fpls.2015.00491
Ongena, M., Jacques, P., Touré, Y., Destain, J., Jabrane, A., and Thonart, P. (2005). Involvement of fengycin-type lipopeptides in the multifaceted biocontrol potential of Bacillus subtilis. Appl. Microbiol. Biotechnol. 69, 29–38. doi: 10.1007/s00253-005-1940-3
Orozco-Mosqueda, M.del. C., Duan, J., DiBernardo, M., Zetter, E., Campos-García, J., Glick, B. R., et al. (2019). The production of ACC deaminase and trehalose by the plant growth promoting bacterium pseudomonas sp. UW4 synergistically protect tomato plants against salt stress. Front. Microbiol. 10:1392. doi: 10.3389/fmicb.2019.01392
Orozco-Mosqueda, M.d. C., Glick, B. R., and Santoyo, G. (2020). ACC deaminase in plant growth-promoting bacteria (PGPB): an efficient mechanism to counter salt stress in crops. Microbiol. Res. 235:126439. doi: 10.1016/j.micres.2020.126439
Pallai, R., Hynes, R. K., Verma, B., and Nelson, L. M. (2012). Phytohormone production and colonization of canola (Brassica napus l.) roots by Pseudomonas fluorescens 6-8 under gnotobiotic conditions. Can. J. Microbiol. 58, 170–178. doi: 10.1139/W11-120
Pedley, K. F., and Martin, G. B. (2005). Role of mitogen-activated protein kinases in plant immunity. Curr. Opin. Plant Biol. 8, 541–547. doi: 10.1016/j.pbi.2005.07.006
Perret, X., Freiberg, C., Rosenthal, A., Broughton, W. J., and Fellay, R. M. (1999). High-resolution transcriptional analysis of the symbiotic plasmid of rhizobium sp. NGR234. Mol. Microbiol. 32, 415–425. doi: 10.1046/j.1365-2958.1999.01361.x
Pieterse, C. M. J., Van der Does, D., Zamioudis, C., Leon-Reyes, A., and Van Wees, S. C. M. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. doi: 10.1146/annurev-cellbio-092910-154055
Pieterse, C. M. J., Zamioudis, C., Berendsen, R. L., Weller, D. M., Van Wees, S. C. M., and Bakker, P. A. H. M. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. doi: 10.1146/annurev-phyto-082712-102340
Poole, P., Ramachandran, V., and Terpolilli, J. (2018). Rhizobia: from saprophytes to endosymbionts. Nat. Rev. Microbiol. 16, 291–303. doi: 10.1038/nrmicro.2017.171
Puente, M. E., Li, C. Y., and Bashan, Y. (2004). Microbial populations and activities in the rhizoplane of rock-weathering desert plants II. Growth promotion of cactus seedlings. Plant Biol. 6, 643–650. doi: 10.1055/s-2004-821101
Raaijmakers, J. M., Vlami, M., and De Souza, J. T. (2002). Antibiotic production by bacterial biocontrol agents. Antonie Van Leeuwenhoek 81, 537–547. doi: 10.1023/a:1020501420831
Rivera, D., Mora, V., Lopez, G., Rosas, S., Spaepen, S., Vanderleyden, J., et al. (2018). New insights into indole-3-acetic acid metabolism in Azospirillum brasilense. J. Appl. Microbiol. 125, 1774–1785. doi: 10.1111/jam.14080
Rodrigues, J. A., López-Baena, F. J., Ollero, F. J., Vinardell, J. M., Espuny, M. D. R., Bellogín, R. A., et al. (2007). NopM and NopD are rhizobial nodulation outer proteins: identification using LC-MALDI and LC-ESI with a monolithic capillary column. J. Proteome Res. 6, 1029–1037. doi: 10.1021/pr060519f
Rodríguez, H., and Fraga, R. (1999). Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 17, 319–339. doi: 10.1016/S0734-9750(99)00014-2
Rohde, J. R., Breitkreutz, A., Chenal, A., Sansonetti, P. J., and Parsot, C. (2007). Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe 1, 77–83. doi: 10.1016/j.chom.2007.02.002
Russo, D. M., Williams, A., Edwards, A., Posadas, D. M., Finnie, C., Dankert, M., et al. (2006). Proteins exported via the PrsD-PrsE type I secretion system and the acidic exopolysaccharide are involved in biofilm formation by Rhizobium leguminosarum. J. Bacteriol. 188, 4474–4486. doi: 10.1128/JB.00246-06
Saad, M. M., Staehelin, C., Broughton, W. J., and Deakin, W. J. (2008). Protein-protein interactions within type III secretion system-dependent pili of Rhizobium sp. strain NGR234. J. Bacteriol. 190, 750–754. doi: 10.1128/JB.01116-07
Sachs, J. L., Quides, K. W., and Wendlandt, C. E. (2018). Legumes versus rhizobia: a model for ongoing conflict in symbiosis. New Phytol. 219, 1199–1206. doi: 10.1111/nph.15222
Salinero-Lanzarote, A., Pacheco-Moreno, A., Domingo-Serrano, L., Durán, D., Ormeño-Orrillo, E., Martínez-Romero, E., et al. (2019). Type VI secretion system of rhizobium etli Mim1 has a positive effect in symbiosis. FEMS Microbiol. Ecol. 95:fiz054. doi: 10.1093/femsec/fiz054
Scagliola, M., Pii, Y., Mimmo, T., Cesco, S., Ricciuti, P., and Crecchio, C. (2016). Characterization of plant growth promoting traits of bacterial isolates from the rhizosphere of barley (Hordeum vulgare L.) and tomato (Solanum lycopersicon L.) grown under Fe sufficiency and deficiency. Plant Physiol. Biochem. 107, 187–196. doi: 10.1016/j.plaphy.2016.06.002
Schmeisser, C., Liesegang, H., Krysciak, D., Bakkou, N., Le Quéré, A., Wollherr, A., et al. (2009). Rhizobium sp. strain NGR234 possesses a remarkable number of secretion systems. Appl. Environ. Microbiol. 75, 4035–4045. doi: 10.1128/AEM.00515-09
Sgro, G. G., Oka, G. U., Souza, D. P., Cenens, W., Bayer-Santos, E., Matsuyama, B. Y., et al. (2019). Bacteria-killing type IV secretion systems. Front. Microbiol. 10:1078. doi: 10.3389/fmicb.2019.01078
Shahid, I., Rizwan, M., Baig, D. N., Saleem, R. S., Malik, K. A., and Mehnaz, S. (2017). Secondary metabolites production and plant growth promotion by Pseudomonas chlororaphis and P. aurantiaca strains isolated from cactus, cotton, and Para grass. J. Microbiol. Biotechnol. J. Microbiol. Biotechnol. 27, 480–491. doi: 10.4014/jmb.1601.01021
Sharma, A., and Johri, B. N. (2003). Growth promoting influence of siderophore-producing pseudomonas strains GRP3A and PRS9 in maize (Zea mays L.) under iron limiting conditions. Microbiol. Res. 158, 243–248. doi: 10.1078/0944-5013-00197
Skorpil, P., Saad, M. M., Boukli, N. M., Kobayashi, H., Ares-Orpel, F., Broughton, W. J., et al. (2005). NopP, a phosphorylated effector of Rhizobium sp. strain NGR234, is a major determinant of nodulation of the tropical legumes Flemingia congesta and Tephrosia vogelii. Mol. Microbiol. 57, 1304–1317. doi: 10.1111/j.1365-2958.2005.04768.x
Smit, G., Kijne, J. W., and Lugtenberg, B. J. (1987). Involvement of both cellulose fibrils and a Ca2+−dependent adhesin in the attachment of Rhizobium leguminosarum to pea root hair tips. J. Bacteriol. 169, 4294–4301. doi: 10.1128/jb.169.9.4294-4301.1987
Spaepen, S., Bossuyt, S., Engelen, K., Marchal, K., and Vanderleyden, J. (2014). Phenotypical and molecular responses of Arabidopsis thaliana roots as a result of inoculation with the auxin-producing bacterium Azospirillum brasilense. New Phytol. 201, 850–861. doi: 10.1111/nph.12590
Spaepen, S., Dobbelaere, S., Croonenborghs, A., and Vanderleyden, J. (2008). Effects of Azospirillum brasilense indole-3-acetic acid production on inoculated wheat plants. Plant Soil 312, 15–23. doi: 10.1007/s11104-008-9560-1
Srinivasan, V. B., Vaidyanathan, V., and Rajamohan, G. (2015). AbuO, a tolc-like outer membrane protein of Acinetobacter baumannii, is involved in antimicrobial and oxidative stress resistance. Antimicrob. Agents Chemother. 59, 1236–1245. doi: 10.1128/AAC.03626-14
Sullivan, J. T., Trzebiatowski, J. R., Cruickshank, R. W., Gouzy, J., Brown, S. D., Elliot, R. M., et al. (2002). Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J. Bacteriol. 184, 3086–3095. doi: 10.1128/JB.184.11.3086-3095.2002
Tampakaki, A. P. (2014). Commonalities and differences of T3SSs in rhizobia and plant pathogenic bacteria. Front. Plant Sci. 5:114. doi: 10.3389/fpls.2014.00114
Teulet, A., Busset, N., Fardoux, J., Gully, D., Chaintreuil, C., Cartieaux, F., et al. (2019). The rhizobial type III effector ErnA confers the ability to form nodules in legumes. Proc. Natl. Acad. Sci. U. S. A. 116, 21758–21768. doi: 10.1073/pnas.1904456116
Trapet, P., Avoscan, L., Klinguer, A., Pateyron, S., Citerne, S., Chervin, C., et al. (2016). The Pseudomonas fluorescens siderophore pyoverdine weakens Arabidopsis thaliana defense in favor of growth in iron-deficient conditions. Plant Physiol. 171, 675–693. doi: 10.1104/pp.15.01537
Tzfira, T., Vaidya, M., and Citovsky, V. (2004). Involvement of targeted proteolysis in plant genetic transformation by agrobacterium. Nature 431, 87–92. doi: 10.1038/nature02857
Van Wees, S. C. M., Pieterse, C. M. J., Trijssenaar, A., Van’t Westende, Y. A. M., Hartog, F., and Van Loon, L. C. (1997). Differential induction of systemic resistance in arabidopsis by biocontrol bacteria. Mol. Plant-Microbe Interact. 10, 716–724. doi: 10.1094/MPMI.1997.10.6.716
Vande Broek, A., Lambrecht, M., Eggermont, K., and Vanderleyden, J. (1999). Auxins upregulate expression of the indole-3-pyruvate decarboxylase gene in Azospirillum brasilense. J. Bacteriol. 181, 1338–1342. doi: 10.1128/jb.181.4.1338-1342.1999
Vesga, P., Flury, P., Vacheron, J., Keel, C., Croll, D., and Maurhofer, M. (2020). Transcriptome plasticity underlying plant root colonization and insect invasion by Pseudomonas protegens. ISME J. 14, 2766–2782. doi: 10.1038/s41396-020-0729-9
Viprey, V., Del Greco, A., Golinowski, W., Broughton, W. J., and Perret, X. (1998). Symbiotic implications of type III protein secretion machinery in rhizobium. Mol. Microbiol. 28, 1381–1389. doi: 10.1046/j.1365-2958.1998.00920.x
Wang, D., Yang, S., Tang, F., and Zhu, H. (2012). Symbiosis specificity in the legume - rhizobial mutualism. Cell. Microbiol. 14, 334–342. doi: 10.1111/j.1462-5822.2011.01736.x
Weller, D. M. (2007). Pseudomonas biocontrol agents of soilborne pathogens: looking back over 30 years. Phytopathology 97, 250–256. doi: 10.1094/PHYTO-97-2-0250
Weselowski, B., Nathoo, N., Eastman, A. W., MacDonald, J., and Yuan, Z. C. (2016). Isolation, identification and characterization of Paenibacillus polymyxa CR1 with potentials for biopesticide, biofertilization, biomass degradation and biofuel production. BMC Microbiol. 16:244. doi: 10.1186/s12866-016-0860-y
Wu, G., Liu, Y., Xu, Y., Zhang, G., Shen, Q., and Zhang, R. (2018). Exploring elicitors of the beneficial Rhizobacterium Bacillus amyloliquefaciens SQR9 to induce plant systemic resistance and their interactions with plant signaling pathways. Mol. Plant-Microbe Interact. 31, 560–567. doi: 10.1094/MPMI-11-17-0273-R
Xin, D. W., Liao, S., Xie, Z. P., Hann, D. R., Steinle, L., Boller, T., et al. (2012). Functional analysis of NopM, a novel E3 ubiquitin ligase (NEL) domain effector of Rhizobium sp. strain NGR234. PLoS Pathog. 8:e1002707. doi: 10.1371/journal.ppat.1002707
Yang, M., Mavrodi, D. V., Mavrodi, O. V., Thomashow, L. S., and Weller, D. M. (2020). Exploring the pathogenicity of pseudomonas brassicacearum Q8r1-96 and other strains of the Pseudomonas fluorescens complex on tomato. Plant Dis. 104, 1026–1031. doi: 10.1094/pdis-09-19-1989-re
Yasmin, F., Othman, R., Sijam, K., and Saad, M. S. (2009). Characterization of beneficial properties of plant growth-promoting rhizobacteria isolated from sweet potato rhizosphere. Afr. J. Microbiol. Res. 3, 815–821.
Yu, K., Pieterse, C. M. J., Bakker, P. A. H. M., and Berendsen, R. L. (2019a). Beneficial microbes going underground of root immunity. Plant Cell Environ. 42, 2860–2870. doi: 10.1111/pce.13632
Yu, K., Tichelaar, R., Liu, Y., Savant, N., Lagendijk, E., Van Kuijk, S., et al. (2019b). Plant-beneficial Pseudomonas spp. Suppress local root immune responses by Gluconic acid-mediated lowering of environmental pH. SSRN Electron. J. 42, 2860–2870. doi: 10.2139/ssrn.3396501
Yu, X., Ai, C., Xin, L., and Zhou, G. (2011). The siderophore-producing bacterium, Bacillus subtilis CAS15, has a biocontrol effect on Fusarium wilt and promotes the growth of pepper. Eur. J. Soil Biol. 47, 138–145. doi: 10.1016/j.ejsobi.2010.11.001
Zamioudis, C., and Pieterse, C. M. J. (2012). Modulation of host immunity by beneficial microbes. Mol. Plant-Microbe Interact. 25, 139–150. doi: 10.1094/MPMI-06-11-0179
Zehner, S., Schober, G., Wenzel, M., Lang, K., and Göttfert, M. (2008). Expression of the Bradyrhizobium japonicum type III secretion system in legume nodules and analysis of the associated tts box promoter. Mol. Plant-Microbe Interact. 21, 1087–1093. doi: 10.1094/MPMI-21-8-1087
Zhalnina, K., Louie, K. B., Hao, Z., Mansoori, N., da Rocha, U. N., Shi, S., et al. (2018). Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3, 470–480. doi: 10.1038/s41564-018-0129-3
Zhang, L., Chen, X. J., Lu, H. B., Xie, Z. P., and Staehelin, C. (2011). Functional analysis of the type 3 effector nodulation outer protein L (NopL) from Rhizobium sp. NGR234: symbiotic effects, phosphorylation, and interference with mitogen-activated protein kinase signaling. J. Biol. Chem. 286, 32178–32187. doi: 10.1074/jbc.M111.265942
Zheng, N., and Shabek, N. (2017). Ubiquitin ligases: structure, function, and regulation. Annu. Rev. Biochem. 86, 129–157. doi: 10.1146/annurev-biochem-060815-014922
Zhou, L., Wang, C., Wang, G. -H., Wei, Z. -W., Fu, Q. -X., Hang, X. -H., et al. (2020). Chemical targeting and manipulation of type III secretion in the Phytopathogen Xanthomonas campestris for control of disease. Appl. Environ. Microbiol. 86, e02349–e02419. doi: 10.1128/AEM.02349-19
Keywords: beneficial bacteria, plant growth promoting bacteria, biocontrol agents, root nodulating bacteria, rhizosphere, effectors, secretion systems
Citation: Lucke M, Correa MG and Levy A (2020) The Role of Secretion Systems, Effectors, and Secondary Metabolites of Beneficial Rhizobacteria in Interactions With Plants and Microbes. Front. Plant Sci. 11:589416. doi: 10.3389/fpls.2020.589416
Edited by:
Christos Zamioudis, Democritus University of Thrace, GreeceReviewed by:
Mika Tapio Tarkka, Helmholtz Centre for Environmental Research (UFZ), GermanyElena A. Dolgikh, All-Russian Research Institute of Agricultural Microbiology of the Russian Academy of Agricultural Sciences, Russia
Copyright © 2020 Lucke, Correa and Levy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Asaf Levy, YWxldnlAbWFpbC5odWppLmFjLmls
†These authors have contributed equally to this work
 Miriam Lucke
Miriam Lucke Mario Gabriel Correa
Mario Gabriel Correa Asaf Levy
Asaf Levy