- 1Department of Pharmacognosy, Faculty of Pharmacy, Mahidol University, Bangkok, Thailand
- 2Graduate School of Biological Science, Nara Institute of Science and Technology, Ikoma, Japan
- 3Max-Planck-Institute of Molecular Plant Physiology, Potsdam, Germany
Nuts, such as peanut, almond, and chestnut, are valuable food crops for humans being important sources of fatty acids, vitamins, minerals, and polyphenols. Polyphenols, such as flavonoids, stilbenoids, and hydroxycinnamates, represent a group of plant-specialized (secondary) metabolites which are characterized as health-beneficial antioxidants within the human diet as well as physiological stress protectants within the plant. In food chemistry research, a multitude of polyphenols contained in culinary nuts have been studied leading to the identification of their chemical properties and bioactivities. Although functional elucidation of the biosynthetic genes of polyphenols in nut species is crucially important for crop improvement in the creation of higher-quality nuts and stress-tolerant cultivars, the chemical diversity of nut polyphenols and the key biosynthetic genes responsible for their production are still largely uncharacterized. However, current technical advances in whole-genome sequencing have facilitated that nut plant species became model plants for omics-based approaches. Here, we review the chemical diversity of seed polyphenols in majorly consumed nut species coupled to insights into their biological activities. Furthermore, we present an example of the annotation of key genes involved in polyphenolic biosynthesis in peanut using comparative genomics as a case study outlining how we are approaching omics-based approaches of the nut plant species.
Introduction
Nuts, such as chestnut and hazelnut, are oil-rich seeds comprising of an edible fruit with a hard outer shell attached to a cupule. Additionally, drupe seeds such as almond, peanut, pistachio, walnut, macadamia, pecan, and cashew nut also contain a hard shell and are thus referred to as “nuts.” Since nuts contain precious types of phytonutrients which exhibit beneficial health-promoting properties, they are regarded as one of the most valuable culinary crops (Ros, 2010; Bodoira and Maestri, 2020). Indeed, consuming nuts provides a rich source of nutritional components, including fatty acids, minerals, vitamins, proteins, and fibers (Ros, 2010; Chen C. Y. O. et al., 2019). In addition, nuts were also found as a rich source of “plant-specialized (secondary) metabolites” which are a vast array of bioactive compounds. Plant-specialized compounds tend to act as stress protectants against biotic and abiotic stresses from the external environment (Tohge et al., 2013b, 2016; Kessler and Kalske, 2018; Pais et al., 2018; Scossa et al., 2019). These metabolites broadly correspond to important physiological and ecological functions, for example, attracting insects for pollination by volatiles and color; as antifeedants and allelochemicals against herbivores; for visible pigmentation; and for lightening from stress conditions, e.g., ultraviolet radiation, elicitors, temperature, and water deficiency (Bodoira and Maestri, 2020; Corso et al., 2020; Yuan and Grotewold, 2020).
Plant-specialized metabolites are allocated to three main categories, namely, terpenoids, alkaloids, and polyphenols, classified on their chemical core skeletons and biosynthetic pathways (Tohge et al.,2013a,b, 2014; Chen C. Y. O. et al., 2019; Bodoira and Maestri, 2020; Yuan and Grotewold, 2020). Polyphenols are ubiquitously present among fruits, vegetables, and seeds, including nuts, with multiple claims of these compounds presenting human health beneficial effects on human health (Lepiniec et al., 2006; Tohge and Fernie, 2017; Hano and Tungmunnithum, 2020; Alseekh et al., 2020a). Numerous research publications investigating polyphenols in nuts have been published (Bolling et al., 2011; Alasalvar and Bolling, 2015; Bodoira and Maestri, 2020). According to our current update, polyphenols in nut plant species are classified into six major groups, including phenols, flavonoids, tannins, stilbenoids, lignans, and coumarins (Figure 1). Phenols including phenolic acids (C6–C1 skeleton) and hydroxycinnamic acids (C6–C3) are the compounds containing an aromatic ring attached with at least one hydroxyl moiety (Figure 1A; Bodoira and Maestri, 2020; Hano and Tungmunnithum, 2020). Flavonoids (C6–C3–C6), one of the largest classes of specialized metabolites, are distributed extensively in the plant kingdom (Figure 1B; Tohge et al.,2013a,b; Mathesius, 2018). Flavonoids reported in seeds of ten major nut plant species are subdivided into eight subclasses, i.e., flavanols, flavones, flavonols, flavanones, flavanonols, isoflavones, anthocyanins, and proanthocyanidins (Figures 1B,C; Tohge et al., 2013b; Saigo et al., 2020). Tannins, comprising of hydrolyzable and condensed tannins, are astringent and bitter plant substances being particularly numerous in nuts seeds (Landete, 2011). Stilbene compounds are comprised of two aromatic rings connecting with an ethylene bridge (C6–C2–C6 backbone) and commonly found as monomers and oligomers in both aglycone and glycoside forms (Figure 1D; Akinwumi et al., 2018; Hano and Tungmunnithum, 2020). Oxidative dimerization of two or more phenylpropanoid molecules produces lignans for lignin synthesis, and as such important precursors for the development of plant vessels and cell walls (Figure 1E; Barker, 2019). Coumarins are plant benzo-α-pyrone compounds generated by the reaction of pyran-benzene ring condensation (Figure 1F; Küpeli Akkol et al., 2020).
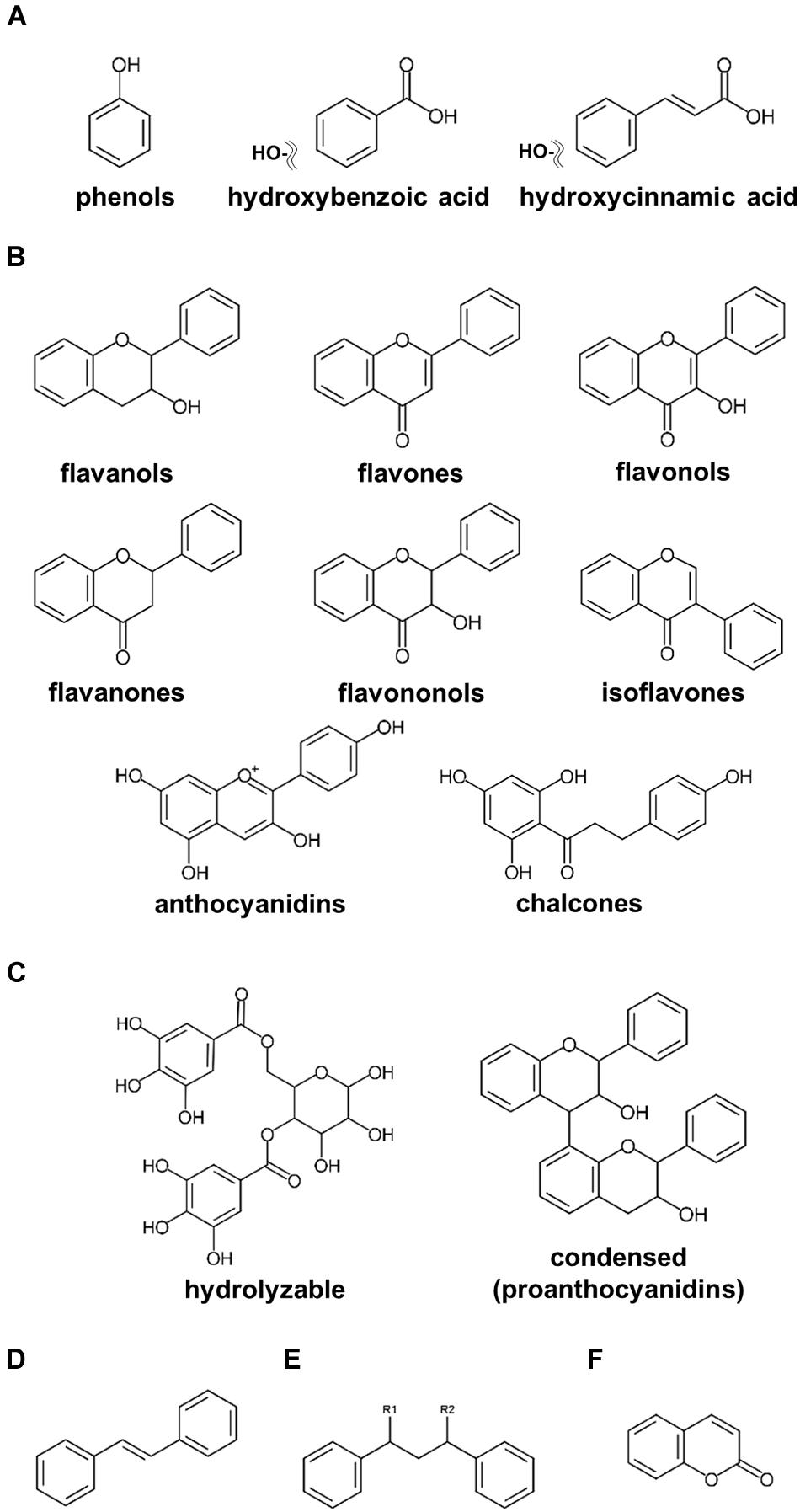
Figure 1. Core structures of polyphenolics found in major nut plant species. (A) phenols and phenolic acids. (B) flavonoids. (C) tannins. (D) stilbenoids. (E) lignans. (F) coumarins.
With respect to biological or pharmaceutical activities, polyphenols have been found to be beneficial components in both human health promotion and disease prevention. They were shown to exhibit antioxidative, anticancer, cardio-protective, antibacterial, anti-inflammatory, and immune system-promoting properties and to exert protection for skin against UV radiation, against neurodegenerative diseases, chronic diseases, obesity, and diabetes, and against the current pandemic coronavirus disease (COVID-19), being reported (Nayak et al., 2015; Tohge and Fernie, 2017; Puksasook et al.,2017a,b; Cory et al., 2018; Tungmunnithum et al., 2018; Renaud and Martinoli, 2019; Ngwa et al., 2020). Several review articles focus on the health benefits of nut consumption (Ros, 2010; Bolling et al., 2011; Alasalvar and Bolling, 2015), with the major groups of polyphenols present in nut seeds being characterized (Bolling et al., 2011; Alasalvar and Bolling, 2015; Bodoira and Maestri, 2020). Currently, the chemical diversity of specialized metabolites and metabolic polymorphisms have specifically highlighted the decoration of polyphenols, with such decorations being found to be a key factor in the enhancement of bioactivity of specialized compounds (Tohge et al., 2016; Peng et al., 2017). In fact, most of the aforementioned biological activities of polyphenols are derived from not only the aglycone form but also the decorated form. The capacity of biological activities such as antioxidant capacity significantly depends on the chemical structures, since these dramatically affect the bioavailability and especially absorption of the compounds (Cipolletti et al., 2018; Gulcin, 2020). Although the core biosynthetic pathways of many polyphenols are conversed among genetically and taxonomically distant plant species, these species often accumulate polyphenols in a tissue-specific manner. In the case of seed-specific specialized metabolites, polyphenols are assumed to be involved in environmental stress protection during seed desiccation and dormancy. As such, the elucidation and understanding of physiological functions of such tissue-specific specialized metabolites are highly valuable.
Current technological and theoretical development of omics-based approaches has enabled that the genome-wide characterization of biosynthetic genes can be carried out, representing an important route by which phenol and polyphenol production could be enhanced in plants (Butelli et al., 2008; Tohge and Fernie, 2010; Zhang et al., 2015; Fernie and Tohge, 2017; Alseekh et al., 2020b). Notably, almost all genes encoding enzymes responsible for structure decoration remain ambiguous. However, as yet, genomic data is only available for peanut and almond; therefore, studies concerning nut plant polyphenolics are not as extensive as they could have been. As such, it is important to update and synthesize the collective information concerning chemical diversity in nut plant species. Given the presence of some species-specific polyphenolics in nuts, it is likely that such a compendium will prove a useful resource for biological activity investigations.
In this review, the current knowledge of polyphenolic compounds in major nut plant species is summarized in terms of their biological activities, chemical diversity, and biosynthetic genes. Ten eminently consumed nut plant species, namely, groundnut/peanut (Arachis hypogaea), almond (Prunus dulcis), pistachio (Pistacia vera), Japanese chestnut (Castanea crenata), Chinese chestnut (Castanea mollissima), walnut (Juglans regia), hazelnut (Corylus avellana), macadamia (Macadamia integrifolia), pecan (Carya illinoinensis), and cashew nut (Anacardium occidentale), are presented (Figure 2 and Table 1). We additionally propose a future perspective for generating an integrative omics approach for functional genomics utilizing polyphenolic biosynthesis in nut plant species as a case study.

Figure 2. Phylogenetic relationship of ten nut plant species presented in this review. The phylogenetic tree was constructed by MEGA X (Kumar et al., 2018) by using the sequences of ITS (internal transcribed spacer) gene of each species which were retrieved from the NCBI database. P. sativum was considered as an outgroup. The parameters for maximum-likelihood analysis were shown as followed: Kimura 2-parameter model, complete deletion, and bootstrap (1000 replicates). Values presented on the branches demonstrated bootstrap support in percentage.
Polyphenolics in Nuts and Their Bioactivities Associated With Health-Promoting Benefits
Several popular nut plant species are consumed as snacks and food supplements, since they are rich in phytonutrients especially fatty acids, protein, minerals, and polyphenolics (Ros, 2010; Vinson and Cai, 2012). To date, research in food chemistry has suggested several biological activities of nut extracts with studies on the antioxidant activity being particularly prominent. From a health beneficial perspective, antioxidants are responsible for the elimination of reactive oxygen species (ROS) or free radical molecules such as superoxide, nitric oxide, and hydrogen peroxide radicals from the human body in order to prevent the generation of hazardous substances which underlie many chronic diseases (Gulcin, 2020). Polyphenols are one of the best-known and major sources of natural antioxidants due to their effective scavenging activity resulting from the presence of several hydroxyl groups present on the structures, especially those on the ortho- and para- positions of the aromatic ring (Shahidi and Ambigaipalan, 2015; Gulcin, 2020). On the basis of their total polyphenolic quantity, the highest antioxidant activities were found in raw walnut and roasted almond (Vinson and Cai, 2012). The biological activities of nut polyphenol antioxidants against major stress such as oxidative stress, aging, and age-related disease prevention were summarized in previous reviews (Ros, 2010; Bodoira and Maestri, 2020; Hano and Tungmunnithum, 2020). Nut consumption has been suggested to play a key role in cardio-protection by reducing cardiovascular risk factors, including coronary heart disease, hypertension, and blood cholesterol levels (Ros, 2010; De Souza et al., 2017). In a recent study, almond skin extract was found to show antimicrobial and antiviral activities against Staphylococcus aureus and herpes simplex virus type I, respectively (Musarra-Pizzo et al., 2019). Due to such indication of nut consumption and health-promoting benefits, polyphenols contained in culinary nuts have been focused and studied with quantification of known health-promoting polyphenols in food chemistry research. Bioactivities of major and specific polyphenolics in nut plant species are summarized in Table 2. Given that health beneficial components such as resveratrols, chlorogenic acids, catechins, and rutin are detected in nuts, health-promoting benefits of culinary nuts are considered with bioactivities and concentrations of these polyphenols. Additionally, nut-specific polyphenolics such as cardanols and anacardic acid were found as cashew nut-specific antioxidant compounds (Table 2). Ellagic acid, which is present in several nuts including almond, walnut, pecan, Japanese chestnut, and hazelnut, has been reported as an inhibitor of inflammatory mediator molecules such as cyclooxygenase and nuclear factor κB, providing anti-inflammatory activity (El-Shitany et al., 2014). Anacardic acid from cashew nut, ellagic acid from walnut and pecan, genistein from peanut and hazelnut, and resveratrol from peanut have demonstrated anticancer properties with numerous molecular targets (Falasca et al., 2014). Resveratrol and its prenylated derivatives in peanut have been reported to mitigate against neurodegenerative diseases such as Alzheimer’s and Parkinson’s disease via their antioxidant, anti-β-amyloid aggregation, anti-β-secretase, neuroprotective, and neuritogenicity properties (Puksasook et al.,2017a,b; Navarro et al., 2018). Captivatingly, myricetin which is found in pistachio and hazelnut as well as resveratrol in peanut, almond, and pistachio are recently claimed as potential phytochemical compounds that could counteract the current COVID-19 pandemic (Han et al., 2020; Ngwa et al., 2020).
Chemical Structural Diversity of Polyphenolics Among Seeds of Nut Plant Species
The chemo-diversity of plant metabolism is a highly important factor affecting plant ecological processes and plant metabolic evolution (Kessler and Kalske, 2018). Furthermore, the various characteristics of phytochemical structures show diverse modes of action with regard to the prevention and treatment of human diseases given differences in their physicochemical properties. Recent overviews focusing on plant structural diversity in anthocyanins/proanthocyanidins (Saigo et al., 2020), glucosinolates (Blažević et al., 2020), and diterpenoid alkaloids (Shen et al., 2020) have been published. With this regard, for example, some decorations such as oxidation for enhancing acidity, methylation and acylation for reducing polarity, and glycosylation for stability and solubility are considered as the potential factors corresponding to diversification of biological functions. Moreover, the updated plant chemo-diversity database is a powerful tool for enthusing new pharmaceutical drug discovery (Lautié et al., 2020). To illustrate the diversity of chemical structures of polyphenolics found in seeds of nut plant species, both raw and processed nut seeds were included in our chemical diversity analysis from the renowned literature-based phytochemical database KNApSAcK (http://kanaya.naist.jp/KNApSAcK/, searched by plant scientific names in July, 2020; Afendi et al., 2011). Furthermore, several current phytochemical reports were included to illustrate the structural diversity of nut polyphenols. A list of all 214 polyphenols is provided in Supplementary Table 1, while the structural diversity of nut polyphenolic compounds is presented in Figure 3.
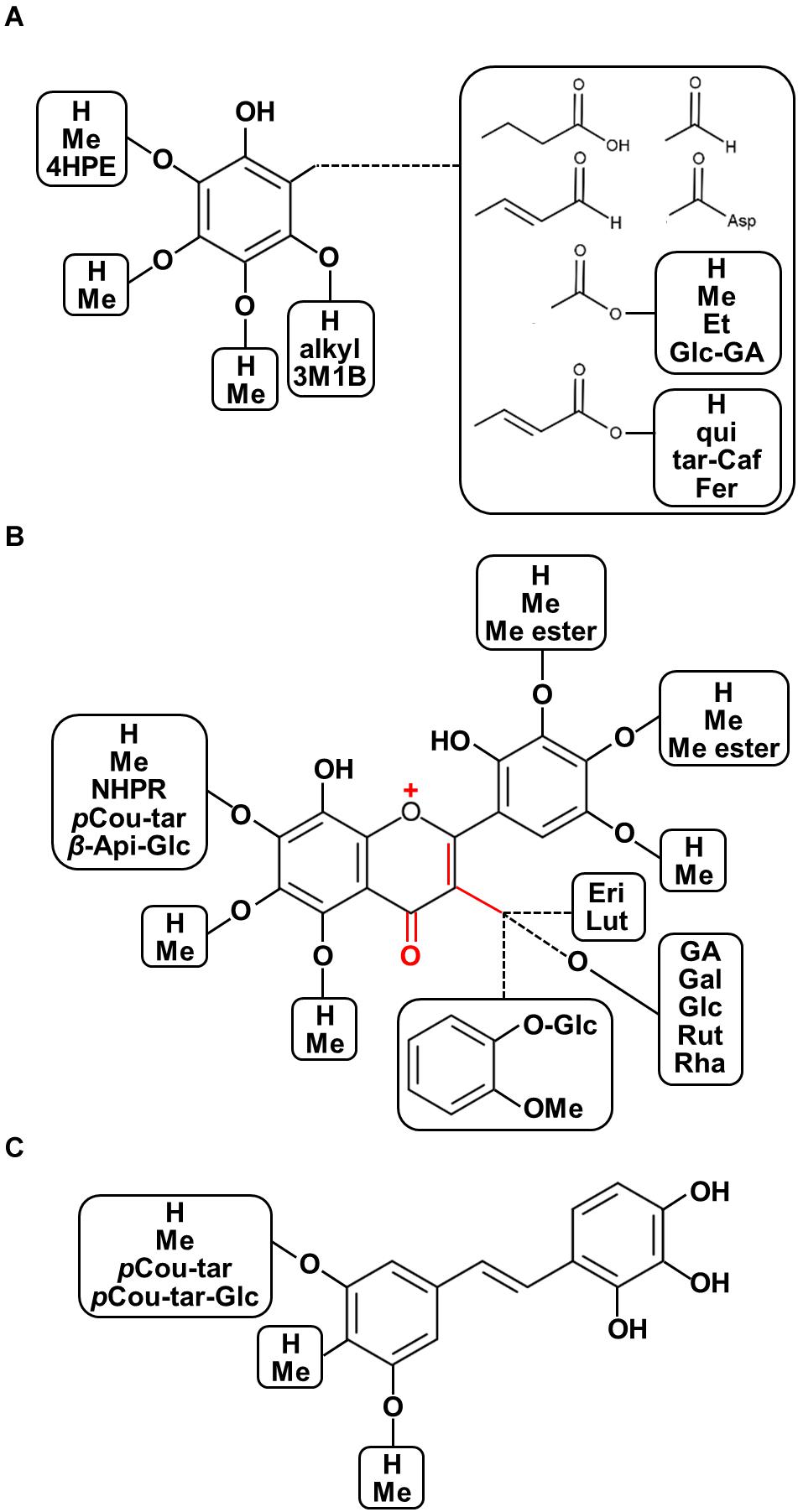
Figure 3. Diversity of chemical structures of polyphenolics found in seeds of nut plant species. (A) Phenols and phenolic acid derivatives, (B) flavonoids, and (C) stilbenoids. Abbreviations: OMe, methoxy; 4HPE, 4-(hydroxypheny)ethylene; Asp, aspartate; Me, methyl; Et, ethyl; qui, quinic acid; tar, tartaric acid; fer, ferulic acid; 3M1B, 3-methyl-1-butenyl; pCou-tar, p-coumaroyl-tartarate; pCou-tar-Glc, p-coumaroyl-tartarate-glucoside; Pre, isoprenyl group; Me ester, methyl ester; NHPR, neohesperidose; Api-Glc, -apiosyl-glucose; Eri, eriodictyol; Lut, luteolin; GA, gallic acid; Gal, galactose; Glc, glucose; Rut, rutinose; and Rha, rhamnose.
Phenols and Phenolic Acids
Several types of phenols have been found in nut plant species (Table 2 and Figure 3A). Four chemical isomers, alpha-, beta-, gamma-, and delta- of tocopherol, were detected in pistachio kernel, walnut kernel, and whole cashew nut (Horvath et al., 2006; Ballistreri et al., 2009; Trox et al., 2011). Cardanols and their derivatives were reported only in the kernel of the cashew nut (Trevisan et al., 2006; Alvarenga et al., 2016; Bodoira and Maestri, 2020). In pecan and walnut kernel, many glycosylated and methylated ellagic acid derivatives are present at high abundance (De La Rosa et al., 2011; Grace et al., 2014; Regueiro et al., 2014; Robbins et al., 2014; Jia et al., 2018). Gallic, p-coumaric, chlorogenic, and p-hydroxybenzoic acids are the most abundant phenolic acids found in the kernel and skin of seeds of ten nut plant species. In addition, several phenolic acids were specifically presented in some of the nut plant species. Anacardic acid and derivatives were present in cashew nut and pistachio kernel (Trevisan et al., 2006; Alvarenga et al., 2016; Bodoira et al., 2019; Salehi et al., 2019; Bodoira and Maestri, 2020). Coutaric and fertaric acids are constituents in peanut and hazelnut skin (Ma et al., 2014; Pelvan et al., 2018). Chinese chestnut skin contains high levels of gentisic acid and 2,3,4-trihydroxybenzoic acid (Xu et al., 2020). In fact, Bodoira and Maestri have mentioned that phenolic acids of peanut are found only in the skin (Bodoira and Maestri, 2020). Nevertheless, nut phenolic acids, such as phloretic acid and dihydroxybenzoic acid, were found not only in the skin but also in the kernel of peanut (Bisby, 1994). Apart from common phenolic acids, numerous derivatives of gallic, hydroxybenzoic, and hydroxycinnamic acids, such as esterification with other phenolic acids, hydroxylated and methylated derivatives, are also conspicuously reported in nut seeds (Bodoira and Maestri, 2020). Polymerization of tartaric acid with two other molecules of phenolic acids is produced only in peanut skin reported by Ma et al. (2014).
Flavonoids
A multitude of flavonoids including flavanols, flavones, flavonols, flavanones, flavanonols, isoflavones, anthocyanins, and proanthocyanidins have been found nut plant species (Table 2 and Figures 1B, 3B). When the hydroxyl group is connected to carbon position three of the C ring, the molecule belongs to the flavanol subclass. Flavonols represent the principal subgroups of flavonoids since they display a rich diversity of derivatives and are also a basic structure of proanthocyanidins (Tohge et al., 2017; Saigo et al., 2020). Catechin and epicatechin are visibly rich in seeds of almost all of the ten major nut plant species. Moreover, their derivatives via esterification with gallic acid, epicatechin-gallate, gallocatechin-gallate, epigallocatechin, and epigallocatechin gallate, were also determined to be abundant in these species being found in whole almond seed (Bolling, 2017), kernel and skin of cashew nut (Salehi et al., 2019), pecan, walnut, Chinese chestnut kernel (Regueiro et al., 2014; Jia et al., 2018; Zhang Y. et al., 2020), and hazelnut skin (Del Rio et al., 2011; Pelvan et al., 2018). Luteolin and apigenin are typical flavones found in peanut skin (Bodoira et al., 2017), almond kernel (Čolić et al., 2017), kernel and the skin of pistachio (Tomaino et al., 2010; Fabani et al., 2013), Chinese chestnut kernel (Zhang Y. et al., 2020), and Japanese chestnut skin (Tuyen et al., 2017). Predominantly among studied nut plant species, whole almond seeds comprise the most diverse types of flavonols, including quercetin, kaempferol, isorhamnetin, and their O-glycoside derivatives (Milbury et al., 2006; Monagas et al., 2007; Bolling et al., 2010; Valdés et al., 2015; Bolling, 2017; Čolić et al., 2017). Flavonol derivatives are frequently glycosylated at the hydroxyl group at position three. Aside from major flavonols, in Chinese and Japanese chestnut kernel, minor flavonols such as rhamnetin and morin have been detected (Tuyen et al., 2017; Zhang Y. et al., 2020). Metabolites containing the saturated C ring belong to the flavanone subgroup of flavonoids. Eriodictyol is the main flavanone detected among nut seeds. As for flavonols, several types of flavanones and their derivatives are characterized in whole almond seeds (Milbury et al., 2006; Čolić et al., 2017; Bodoira and Maestri, 2020) with glycosylated derivatives generally displaying glycosylation on the hydroxyl group at position seven. Particularly, naringin (naringenin-7-O-neohesperidoside) is a major flavanone derivative in kernels of almond, walnut, and Chinese chestnut as well as in pistachio kernel and skin (Tomaino et al., 2010; Čolić et al., 2017; Vu et al., 2018; Zhang Y. et al., 2020). By contrast, flavanonols and hydroxylated derivatives of flavanones are only minor constituents of nuts; for example, aromadendrin and taxifolin are reported in whole almond seeds (Monagas et al., 2007; Bolling, 2017; Vu et al., 2018). The core structure of isoflavones differs from other flavonoids by linkage of the phenyl ring to position three of ring C supplemented with the ketone group at position four. Isoflavones are not broadly found in nut seeds. However, genistein and its glucosides, daidzein, and daidzin, are present in peanut kernel and skin, whole almond, and pistachio kernel (Ballistreri et al., 2009; Tomaino et al., 2010; Bolling, 2017; Bodoira and Maestri, 2020). Anthocyanidins (aglycone form) and anthocyanins (glycoside form) are extensively known as plant pigments in seeds, flowers, and fruits (Saigo et al., 2020). Cyanidin and its glycoside derivatives, i.e., glucoside and galactoside, were reported as constituents in whole almond seeds (Bolling, 2017) and pistachio kernel and skin (Ballistreri et al., 2009; Tomaino et al., 2010; Fabani et al., 2013; Bodoira and Maestri, 2020). Chalcones or the so-called open-chain flavonoids are a very unique flavonoid subclass in the plant kingdom. Phloretin and its glucoside derivative, phlorizin, are the two chalcones found in the almond kernel and hazelnut skin (Del Rio et al., 2011; Čolić et al., 2017).
Tannins
Both tannin subgroups, i.e., hydrolyzable and condensed tannins, are abundant in peanut skin, whole almond, and walnut kernel. Hydrolyzable tannins are further allocated into two subgroups gallotannins and ellagitannins (Soares et al., 2020). The latter including strictinin, pedunculagin, tellimagrandin, glasrin, rugosin, casuarinin, and praecoxin (Fukuda et al., 2003; Grace et al., 2014; Regueiro et al., 2014; Robbins et al., 2014; Jia et al., 2018) are more prominent than the former. Brown or non-visible colors of plant seed, peel, and bark are caused by proanthocyanidins or condensed tannins (Saigo et al., 2020). As mentioned above, proanthocyanidin is synthesized by combining flavonol molecules, i.e., catechin, epicatechin, or their galloylated derivatives; polymerization started from dimers by several types of inter flavan linkage (Soares et al., 2020). Proanthocyanidins have been characterized in seed testa of nut plant species, especially in peanut, almond, pistachio, and hazelnut (Lou et al., 1999, 2004; Yu et al., 2006; Monagas et al., 2007; Del Rio et al., 2011; Sarnoski et al., 2012; Fabani et al., 2013; Grace et al., 2014; Slatnar et al., 2015; Bodoira et al., 2017, 2019; Bolling, 2017; Pelvan et al., 2018; Bodoira and Maestri, 2020). As stated in the current review article (Saigo et al., 2020), proanthocyanidins and tannins are arduous to study, since their condensed structures are very complex due to high molecular weight and various types of chemical bond configuration.
Stilbenoids
Typically, stilbene compounds are rarely found in the seeds of nut plant species. Resveratrol, one of the most well-known stilbenes ubiquitously found in grape, Vitis vinifera L. (Salehi et al., 2018), was detected in peanut skin, whole almond seeds, and pistachio kernel (Ballistreri et al., 2009; Ballard et al., 2010; Xie and Bolling, 2014; Čolić et al., 2017; Bodoira and Maestri, 2020). Moreover, various types of stilbene derivatives were also found. Two prenylated resveratrols, arachidin I and II, were found in the peanut kernel (Bisby, 1994). In whole almond seed, the glycosylated resveratrol named polydatin is the most prominently detected stilbenoid along with small amounts of a methylated and two hydroxylated resveratrols called pterostilbene, piceatannol, and oxyresveratrol, respectively (Xie and Bolling, 2014).
Lignans and Coumarins
Although lignans are not well-investigated in nut plant species, several major plant lignans and their hydroxylated derivatives have been reported in the nut species that we are reviewing in this article, such as lariciresinol, matairesinol, secoisolariresinol, cyclolariciresinol, and 7-hydroxymatairesinol, whereas cashew nut contains the highest total lignan contents (Bolling, 2017; Rodríguez-García et al., 2019). The same is true of lignans in nut species; coumarins in seeds of nut plant species have not been characterized well. Some simple and pyrone-substituted coumarin compounds, e.g., aesculin, aesculetin, umbelliferone, and coumestrol, are mostly found in whole almond, Japanese chestnut skin, and Chinese chestnut kernel (Bolling, 2017; Čolić et al., 2017; Tuyen et al., 2017; Chang et al., 2020).
Recent Updates of the Polyphenolic Biosynthetic Framework in Seeds of Nut Plant Species
An overview of the known polyphenolic biosynthetic framework is summarized in Figure 4 (Lushchak and Semchuk, 2012; Cheynier et al., 2013; Anantharaju et al., 2016; Valanciene et al., 2020). Even though the enzymatic genes regarding polyphenolic biosynthesis are elucidated and well characterized in model plants and crop species, such as Arabidopsis thaliana, Zea mays, and Camellia sinensis (Falcone Ferreyra et al., 2012; Jiang et al., 2013), numerous key genes are largely uncharacterized in seeds of nut plant species, probably due to the lower sequence similarity of genes between major model plants and model nut plant species. Currently, the information of biosynthetic genes has been reported only in peanut and pecan. Peanut chalcone isomerase (CHI) types I and II have been identified (Wang et al., 2012; Liu et al., 2015), and stilbene synthase (AhSTS), the most vital enzyme for resveratrol biosynthesis, is functionalized (Condori et al., 2009). In the pecan kernel, three chalcone synthases (CHS) were isolated and properly characterized (Zhang et al., 2019b). Moreover, RNA-Seq also provided several other gene candidates of flavonoid biosynthesis, including PAL, C4H, 4CL, CHI, F3H, F3′H, DFR, ANS, LAR, ANR, and MYB transcription factor (Zhang et al., 2019a). However, those candidates have been well identified by Huang and colleagues (Huang et al., 2019). In 2014, Chen et al. (2014) performed the first MYB gene family in peanut investigation and found that AhMYB15 is related to flavonol biosynthesis. Aside from seeds, gallate glucosyltransferase (GGT) I and II are important for β-glucogallin, the intermediate compound for hydrolyzable tannin biosynthesis, which were identified in walnut leaves (Martínez-García et al., 2016). Stilbenoid prenyltransferases named AhR4DT-1 and AhR3′DT-1 involved in prenylated resveratrol production in peanut kernel were explicated in peanut hairy root (Yang et al., 2018). According to current knowledge, it is apparent that the responsible key genes for polyphenolics in the seed of nut plant species remain largely undetermined, especially in the case of decoration enzymatic genes, which are largely responsible for producing chemical structure diversification. Identification of these genes thus represents an important priority for future research.
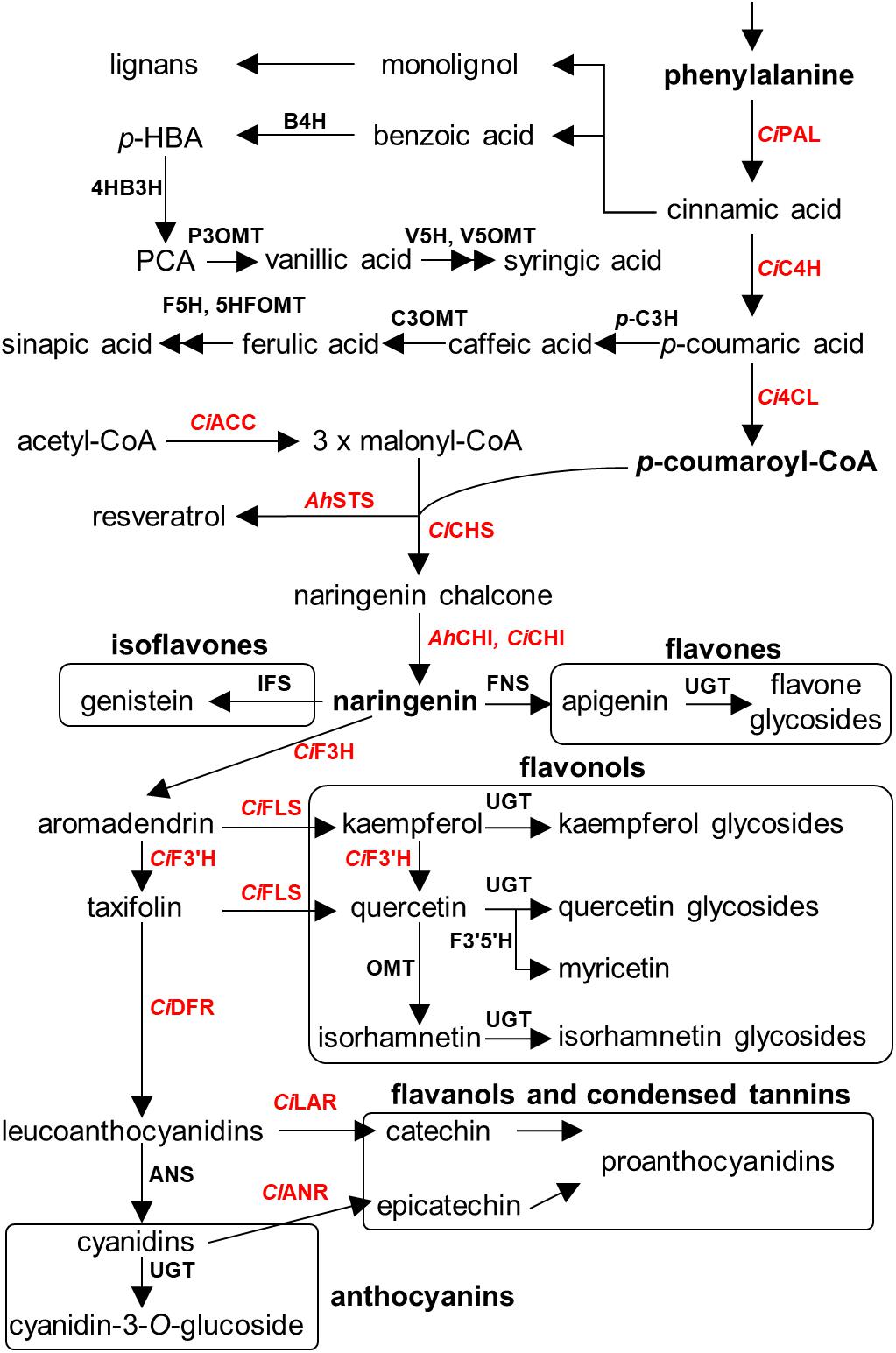
Figure 4. Overview of polyphenolics biosynthesis in major nut plant species. Genes involved in each biosynthetic step are displayed. Characterized genes are shown in red color. Abbreviations used. E4P, erythrose-4-phosphate; PEP, phosphoenolpyruvate; DAHPS, 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase; DHQS, 3-dehydroquinate synthase; DHQ, 3-dehydroquinate; DHD, 3-dehydroquinate dehydratase; DHS, 3-dehydroshikimate; JrGGT, gallate 1-β-glucosyltransferase from walnut; SDH, shikimate dehydrogenase; SK, shikimate kinase; EPSPS, 5-enolpyruvylshikimate 3-phosphate synthase; CS, chorismate synthase; CM, chorismate mutase; HPPD, p-hydroxyphenylpyruvate dioxygenase; PAT, prephenate aminotransferase; AD, arogenate dehydratase; CiPAL, phenylalanine ammonia lyase of pecan; B4H, benzoic acid 4-hydroxylase; 4HB3H, 4-hydroxybenzoic acid 3-hydroxylase; P3OMT, protocatechuic acid-3-O-methyltransferase; V5H, vanillic acid 5-hydroxylase; V5OMT, vanillic acid 5-O-methyltransferase; CiC4H, cinnamic acid-4-hydroxylase from pecan; p-C3H, p-coumaric acid 3-hydroxylase; C3OMT, caffeic acid 3-O-methyltransferase; F5H, ferulic acid 5-hydroxylase; 5HFOMT, 5-hydroxyferulic acid O-methyltransferase; Ci4CL, 4-coumaroyl-CoA ligase from pecan; CiACC, acetyl coenzyme A carboxylase from pecan; AhSTS, stilbene synthase from peanut; CiCHS, chalcone synthase from pecan; AhCHI, CiCHI, chalcone isomerase from peanut and pecan; IFS, isoflavone synthase; FNS, flavone synthase; CiF3H, flavanone 3-hydroxylase from pecan; CiFLS, flavonol synthase from pecan; CiF3′H, flavonoid 3′-hydroxylase from pecan; CiF3′5′H, flavanoid 3′5′-hydroxylase from pecan; CiDFR, dihydroflavonol reductase from pecan; ANS, anthocyanidin synthase; CiLAR, leucoanthocyanidin reductase; CiANR, anthocyanidin reductase from pecan; and UGT, uridine diphosphate glycosyltransferase.
Model Nuts: The State of the Art
At present, nut plant species, including peanut (Bertioli et al., 2019), almond (Sánchez-Pérez et al., 2019), pistachio (Zeng et al., 2019), walnut (Martínez-García et al., 2016), and hazelnut (Lucas et al., 2020), have been genome sequenced (Table 1). Furthermore, draft genome sequences of some chestnut cultivars have been currently deposited in the genome database. Such information provides the advantages of those plant species which became “model nut” species for elucidation of physiological and biological functions of key metabolic genes via omics-based approaches. The genome sequence of the cultivated peanut, known as allotetraploids, was elucidated after sequencing its diploid ancestors, A. duranensis and A. ipaensis, providing helpful hints for peanut domestication (Bertioli et al., 2016). Currently, using peanut genomics data, lipid metabolism (Chen X. et al., 2019) and the genes involved in size and lipid content in seeds, leaf disease resistance, and nitrogen fixing capacity (Zhuang et al., 2019) were investigated and annotated. Importantly, PacBio and chromosome conformation capture (Hi-C) technologies were performed in order to improve data reading quality and a complete peanut genome sequence was lately reported (Bertioli et al., 2019), resulting in a very high-quality genome.
The walnut genome which was first reported in 2016 revealed some of the genes involved in polyphenolic transformations (Martínez-García et al., 2016). The first walnut reference genome was used to generate high-density 700-K single-nucleotide polymorphism (SNP) arrays (Marrano et al., 2019). Importantly, this tool was used to identify gene candidates responsible for flowering process disclosure (Bernard et al., 2020). However, given that the early walnut genome remained at the scaffold level, multi-omics studies for unraveling biological function and regulation were obstructed. For this reason, the de novo assembly of the complete walnut genome was attempted by various techniques. The hybridization of walnut species sequencing by single-molecule or PacBio long-read sequencing and optical genome mapping technologies demonstrated a high quality of parental genome sequence (Wu and Gmitter, 2019; Zhu et al., 2019). Recently, nanopore long-read sequencing supplemented with Hi-C technology has been utilized for a high-quality walnut chromosome level genome assembly (Marrano et al., 2020). Illumina sequencing coupled with Hi-C data is also found to provide high-quality genome data and uncover differences (Zhang J. et al., 2020). Almond is also recently described to have a complete genome sequence. The complete genome sequence of almond was initially reported to be coupled with forty-six kilobases of the gene cluster encoding five basic helix–loop–helix (bHLH) transcription factors (Sánchez-Pérez et al., 2019). Fascinatingly, bHLH2 is identified to be involved in amygdalin biosynthesis (Sánchez-Pérez et al., 2019). Alioto and colleagues similarly performed almond genome sequencing and determined transposon elements related to amygdalin biosynthesis and diversification in peach (Alioto et al., 2020). With an ever-increasing number of high-quality nut plant genomes, gene conservation of functional genes is one of the important topics deserving further investigation of plant metabolism. Hazelnut is the most recently reported complete genome sequence by a hybrid sequencing strategy combining short reads, long reads, and proximity ligation methods (Lucas et al., 2020). The European hazelnut (Corylus avellana L. cv. Tombul) was sequenced focusing on gene families encoding hazelnut allergens and the pathogen-resistance locus proteins that are an important for crop improvement in C. avellana.
Biosynthetic Gene Conservation in Nut Genomes
In spite of the fact that flavonoids are highly diverse in seeds of nut plant species, the enzymatic gene involved in flavonoid biosynthesis named CHS is frequently mentioned as one of the most conserved key enzymes. In addition, stilbene synthase (STS), the key enzyme of stilbene biosynthesis which resulted in resveratrol as a first product, is discovered as a homolog of CHS since they play a similar function and contain a conserved cysteine residue; hence, STS is described as belonging to the CHS family (Schröder and Schröder, 1990; Lanz et al., 1991). In the nut species presented in this article, AhSTS1 (arahy.QVKQ5Y, XM_025790597.1, ABY86219.1) in A. hypogaea has been characterized (Condori et al., 2009). Both CHS and STS belong to the type III polyketide synthase (PKS) family and have been occurred during functional diversification of PKS (Tohge et al., 2013b). In current informatics research, the phylogenomic synteny network combined with phylogenetic analyses of whole-genomic data of 126 plant species has been developed and focused on the macroevolution of diversification of the PKS family (Naake et al., 2020). In this review, we performed the comparative genomics of CHS and STS between nuts and closely taxonomic-related species by phylogenomic synteny analysis. The total diverged genomic synteny regions of CHS and STS in eight legume plant species are comprised in twenty regions (Figures 5A,B). Three regions (b, k, and l) are commonly found in all studied plant species, whereas m, n, o, and p regions are specific to peanut species. Additionally, the numerous tandem gene duplication is prominently presented in regions b, c, d, I, l, m, n, and p. Even though CHS and STS genes are generally noticed in every mentioned plant species, the flavonoid and/or stilbenoid are still not reported providing a research gap for further metabolite investigation. Based on CHS and STS syntenic regions, a phylogenetic analysis demonstrates the clearly visible separation of region p in wild and cultivated peanut (Arachis ipaensis and A. hypogaea, respectively), soybean (Glycine max), red clover (Trifolium pretense), and medicago (Medicago truncatula) from the others; it suggests that those uncharacterized genes may function as CHSs (Figure 5C). Interestingly, two characterized grapevine STSs show a separated relationship from PhSTS with high bootstrap support. This suggests that these two enzymes evolved from different ancestors may be caused by neo-functionalization of species-specific tandem gene duplications. Nevertheless, a major part of the two displays low bootstrap percentage, which suggests that the CHS and STS phylogeny of the current studied plant species is highly assorted and complex; as such, further investigation is imperative. Apart from genomics investigations, transcriptomic studies are also important for metabolomics studies and crop improvement. The gene expression profiles of various peanut tissues have been revealed (Clevenger et al., 2016). Thus, we attempted to link our synteny analysis with this previously published gene expression data. phSTS (XP_025646382.1, ABY86219.1) is expressed in fruits of peanuts. Obviously, XP_025642791 (gene ID. Arahy.0FI6RG) which is pinpointed on region b is mainly expressed during the development of peanut seeds (Figure 5D). Furthermore, a co-expression network analysis of these genes is presented in Figure 6. phSTS shows a highly correlated expression with many tandem gene-duplicated CHSs, genes encoding CHI, and O-methyltransferase (OMT). On the other hand, Arahy.0FI6RG is correlated with genes related to several types of enzymes, including hydrolases, oxidoreductases, epimerases, and kinases. Moreover, some of these genes are involved in the regulation of transcription factors and transporters. The functions of these candidate genes need to be evaluated experimentally; however, our example of the in silico omics-based approach provides insights for future researches into the elucidation of key genes involved in nut polyphenols for metabolomics-assisted breeding approaches aimed at enhancing health-beneficial components.
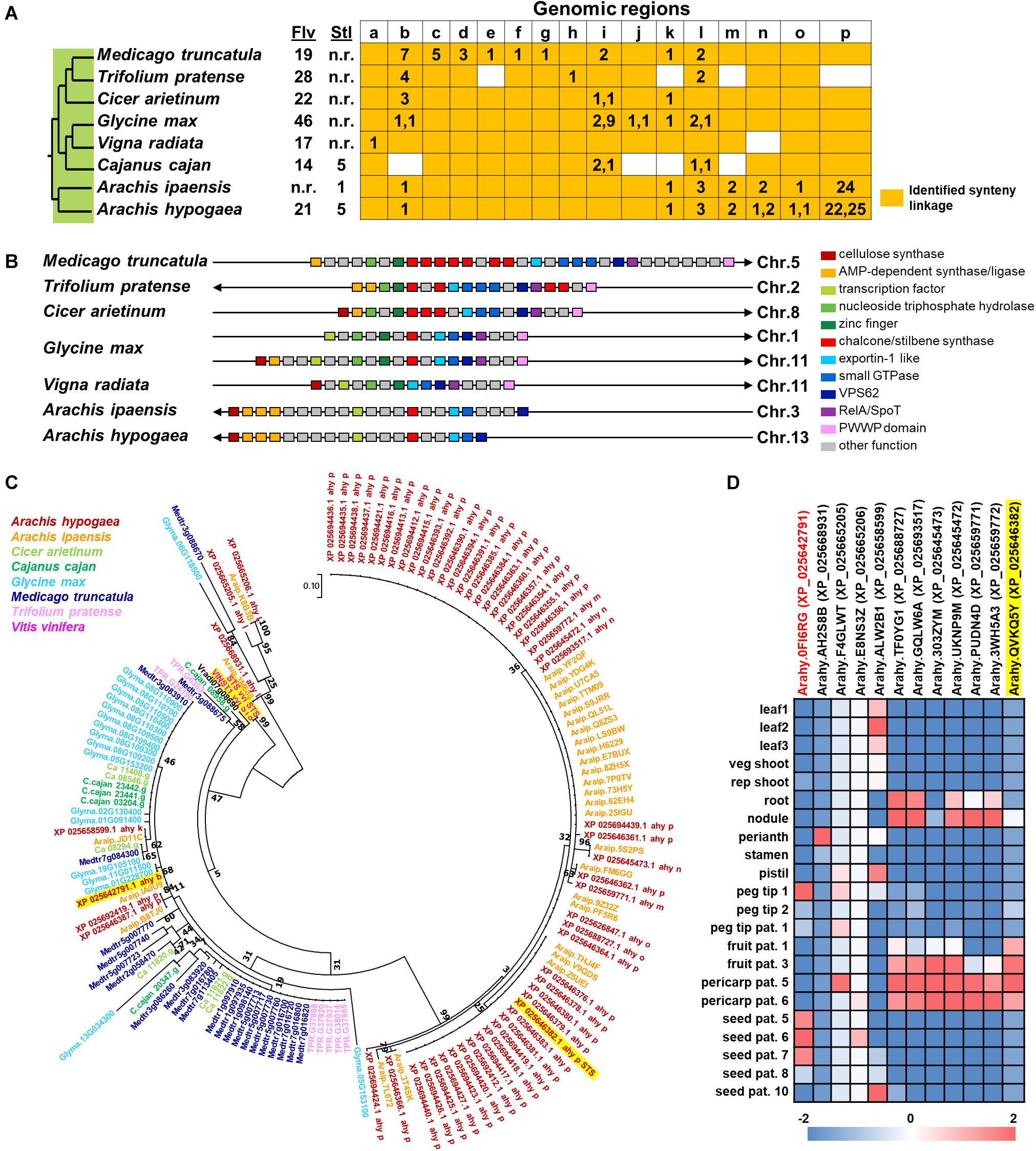
Figure 5. Comparative genomics of CHS and STS in eight legume plant species. (A) Genomic synteny analysis of CHS and STS. Gene lists of six legume plant species were retrieved from Plaza database (Dicot 4.5; http://bioinformatics.psb.ugent.be/plaza/). Almond and peanut genome sequences were obtained from the NCBI database. Numbers in each region indicate the number of tandem duplicated genes. Numbers in each region indicate the number of tandem duplicated genes and intra synteny genes. Abbreviations used: Flv, flavonoids; and Stl, stilbenoids. (B) Genomic structure of synteny regions b presented in Figure 1A. (C) Phylogenetic relationship of CHS and STS located in genome synteny analysis of thirteen plant species. Amino acid sequences were attained from the Plaza database (Dicot 4.5; http://bioinformatics.psb.ugent.be/plaza/) coupled with the NCBI database. The phylogenetic trees were constructed with aligned protein sequences by MEGA7 (Kumar et al., 2016) using the neighbor-joining method with the following parameters: Poisson correction, complete deletion, and bootstrap (1000 replicates, random seed). The protein sequences were aligned by MUSCLE implemented in MEGA. Values on the branches indicate bootstrap support in percentages. The tree with the highest log likelihood (–1287.37) is shown. Colors of circle indicate plant species. (D) Gene expression profile of peanut tissues. Each gene is connected with the syntenic region. Gene expression data; part and stage of peanuts are described by previous work of Clevenger and colleagues (Clevenger et al., 2016).
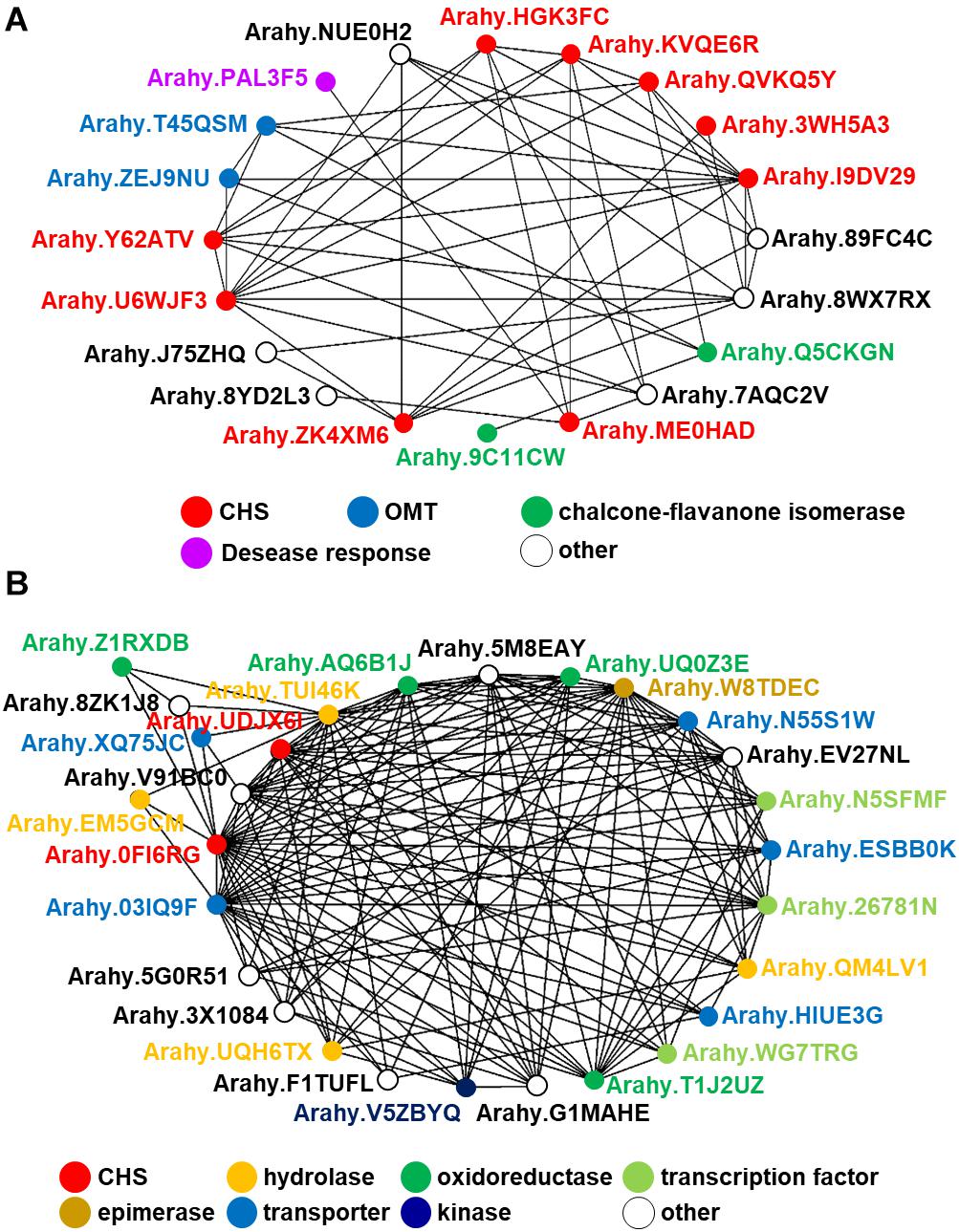
Figure 6. In silico co-expression gene network analysis of the peanut STS gene putatively annotated by comparative genomics. (A) AhSTS, (B) Arahy.0FI6RG.
Summary and Future Perspective
Nuts are regarded as treasured food crops due to their high contents of potential bioactive components which are able to promote human health benefit. In our summary of the chemical diversity of nut polyphenols, flavonoids are found as the major structurally diversified polyphenols in both aglycone and decorated forms among seeds of nut plant species. With regard to the latter, the glycoside is the main category of polyphenolic derivatives. Diversification of chemical structures results in different effectiveness of biological activities, particularly antioxidants. Although the polyphenolic biosynthetic pathway is widely known, genes of nut plant species encoding enzymes responsible for each step remain uncharacterized. Genome synteny analysis of CHS and STS provides a strategic example for understanding the evolution and conservation of these two enzymes in seeds of nut plant species. Notably, there are several research gaps for nut plant species since much of our knowledge is fragmentary and considerable further investigation is required. Deciphering the multi-omics (genomics, transcriptomics, proteomics, and metabolomics) of nut plant species will provide fundamental data for their physiological function and potential for crop improvement, including increasing crop yield, stress, and disease tolerance, as well as enhance the production of human health beneficial specialized metabolites.
Author Contributions
CA and TT wrote the manuscript. CA, MW, and TT conceived, designed, and conceptualized the outline of the manuscript. TS, CA, and TN performed comparative genome analysis. VS, SB, MW, AF, and TT supervised and edited the manuscript. All authors have read and approved the final manuscript.
Funding
Research activity of TT was additionally supported by the JSPS KAKENHI Grant-in-Aid for Scientific Research B (19H03249) and C (19K06723).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
CA would like to specially thank Mahidol Medical Scholars Program (MSP), Mahidol University, Thailand, for the opportunity to work on this review article at Graduate School of Biological Science, Nara Institute of Science and Technology (NAIST), Ikoma, Japan. SB and VS thank Faculty of Pharmacy, Mahidol University. TN and AF would like to thank the funding from the Max-Planck-Society. TT, MW, and TS gratefully acknowledge the partial support by the NAIST.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.642581/full#supplementary-material
References
Abdulkhaleq, L. A., Assi, M. A., Noor, M. H. M., Abdullah, R., Saad, M. Z., and Taufiq-Yap, Y. H. (2017). Therapeutic uses of epicatechin in diabetes and cancer. Vet. World 10, 869–872. doi: 10.14202/vetworld.2017.869-872
Afendi, F. M., Okada, T., Yamazaki, M., Hirai-Morita, A., Nakamura, Y., Nakamura, K., et al. (2011). KNApSAcK family databases: integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 53:e1. doi: 10.1093/pcp/pcr165
Akinwumi, B., Bordun, K.-A., and Anderson, H. (2018). Biological activities of stilbenoids. Int. J. Mol. Sci. 19:792. doi: 10.3390/ijms19030792
Alasalvar, C., and Bolling, B. W. (2015). Review of nut phytochemicals, fat-soluble bioactives, antioxidant components and health effects. Br. J. Nutr. 113, S68–S78. doi: 10.1017/s0007114514003729
Alioto, T., Alexiou, K. G., Bardil, A., Barteri, F., Castanera, R., Cruz, F., et al. (2020). Transposons played a major role in the diversification between the closely related almond and peach genomes: results from the almond genome sequence. Plant J. 101, 455–472. doi: 10.1111/tpj.14538
Alseekh, S., Ofner, I., Liu, Z. Y., Osorio, S., Vallarino, J., Last, R. L., et al. (2020a). Quantitative trait loci analysis of seed-specialized metabolites reveals seed-specific flavonols and differential regulation of glycoalkaloid content in tomato. Plant J. 103, 2007–2024. doi: 10.1111/tpj.14879
Alseekh, S., Perez de Souza, L., Benina, M., and Fernie, A. R. (2020b). The style and substance of plant flavonoid decoration; towards defining both structure and function. Phytochemistry 174:112347. doi: 10.1016/j.phytochem.2020.112347
Alvarenga, T. A., De Oliveira, P. F., De Souza, J. M., Tavares, D. C., Andrade, E., Silva, M. L., et al. (2016). Schistosomicidal activity of alkyl-phenols from the cashew anacardium occidentale against Schistosoma mansoni adult worms. J. Agric. Food Chem. 64, 8821–8827. doi: 10.1021/acs.jafc.6b04200
Anand David, A. V., Arulmoli, R., and Parasuraman, S. (2016). Overviews of biological importance of Quercetin: a bioactive Flavonoid. Pharmacogn. Rev. 10, 84–89. doi: 10.4103/0973-7847.194044
Anantharaju, P. G., Gowda, P. C., Vimalambike, M. G., and Madhunapantula, S. V. (2016). An overview on the role of dietary phenolics for the treatment of cancers. Nutr. J. 15:99. doi: 10.1186/s12937-016-0217-2
Bae, J., Kim, N., Shin, Y., Kim, S.-Y., and Kim, Y.-J. (2020). Activity of catechins and their applications. Biomed. Dermatol. 4:8. doi: 10.1186/s41702-020-0057-8
Bajpai, V. K., Shukla, S., Paek, W. K., Lim, J., Kumar, P., Kumar, P., et al. (2017). Efficacy of (+)-Lariciresinol to control bacterial growth of Staphylococcus aureus and Escherichia coli O157:H7. Front. Microbiol. 8:804. doi: 10.3389/fmicb.2017.00804
Ballard, T. S., Mallikarjunan, P., Zhou, K., and O’Keefe, S. (2010). Microwave-assisted extraction of phenolic antioxidant compounds from peanut skins. Food Chem. 120, 1185–1192. doi: 10.1016/j.foodchem.2009.11.063
Ballistreri, G., Arena, E., and Fallico, B. (2009). Influence of ripeness and drying process on the Polyphenols and Tocopherols of Pistacia vera L. Molecules 14, 4358–4369. doi: 10.3390/molecules14114358
Bernard, A., Marrano, A., Donkpegan, A., Brown, P. J., Leslie, C. A., Neale, D. B., et al. (2020). Association and linkage mapping to unravel genetic architecture of phenological traits and lateral bearing in Persian walnut (Juglans regia L.). BMC Genom. 21:203. doi: 10.1186/s12864-020-6616-y
Bernatova, I. (2018). Biological activities of (–)-epicatechin and (–)-epicatechin-containing foods: focus on cardiovascular and neuropsychological health. Biotechnol. Adv. 36, 666–681. doi: 10.1016/j.biotechadv.2018.01.009
Bertioli, D. J., Cannon, S. B., Froenicke, L., Huang, G., Farmer, A. D., Cannon, E. K. S., et al. (2016). The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 48, 438–446. doi: 10.1038/ng.3517
Bertioli, D. J., Jenkins, J., Clevenger, J., Dudchenko, O., Gao, D., Seijo, G., et al. (2019). The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 51, 877–884. doi: 10.1038/s41588-019-0405-z
Blažević, I., Montaut, S., Burčul, F., Olsen, C. E., Burow, M., Rollin, P., et al. (2020). Glucosinolate structural diversity, identification, chemical synthesis and metabolism in plants. Phytochemistry 169:112100. doi: 10.1016/j.phytochem.2019.112100
Bodoira, R., and Maestri, D. (2020). Phenolic compounds from nuts: extraction, chemical profiles, and bioactivity. J. Agric. Food Chem. 68, 927–942. doi: 10.1021/acs.jafc.9b07160
Bodoira, R., Rossi, Y., Montenegro, M., Maestri, D., and Velez, A. (2017). Extraction of antioxidant polyphenolic compounds from peanut skin using water-ethanol at high pressure and temperature conditions. J. Supercrit. Fluids 128, 57–65. doi: 10.1016/j.supflu.2017.05.011
Bodoira, R., Velez, A., Rovetto, L., Ribotta, P., Maestri, D., and Martínez, M. (2019). Subcritical fluid extraction of antioxidant phenolic compounds from pistachio (Pistacia vera L.) Nuts: experiments, modeling, and optimization. J. Food Sci. 84, 963–970. doi: 10.1111/1750-3841.14507
Bolling, B. W. (2017). Almond polyphenols: methods of analysis, contribution to food quality, and health promotion. Compr. Rev. Food Sci. Food Saf. 16, 346–368. doi: 10.1111/1541-4337.12260
Bolling, B. W., Chen, C. Y. O., McKay, D. L., and Blumberg, J. B. (2011). Tree nut phytochemicals: composition, antioxidant capacity, bioactivity, impact factors. A systematic review of almonds, Brazils, cashews, hazelnuts, macadamias, pecans, pine nuts, pistachios and walnuts. Nutr. Res. Rev. 24, 244–275. doi: 10.1017/s095442241100014x
Bolling, B. W., Dolnikowski, G., Blumberg, J. B., and Chen, C. Y. O. (2010). Polyphenol content and antioxidant activity of California almonds depend on cultivar and harvest year. Food Chem. 122, 819–825. doi: 10.1016/j.foodchem.2010.03.068
Butelli, E., Titta, L., Giorgio, M., Mock, H. P., Matros, A., Peterek, S., et al. (2008). Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26, 1301–1308. doi: 10.1038/nbt.1506
Chang, X., Liu, F., Lin, Z., Qiu, J., Peng, C., Lu, Y., et al. (2020). Phytochemical profiles and cellular antioxidant activities in chestnut (Castanea mollissima BL.) kernels of five different cultivars. Molecules 25:178. doi: 10.3390/molecules25010178
Chen, C. Y. O., Milbury, P. E., and Blumberg, J. B. (2019). Polyphenols in almond skins after blanching modulate plasma biomarkers of oxidative stress in healthy humans. Antioxidants 8:95. doi: 10.3390/antiox8040095
Chen, X., Lu, Q., Liu, H., Zhang, J., Hong, Y., Lan, H., et al. (2019). Sequencing of cultivated peanut, Arachis hypogaea, yields insights into genome evolution and oil improvement. Mol. Plant 12, 920–934. doi: 10.1016/j.molp.2019.03.005
Chen, N., Yang, Q., Pan, L., Chi, X., Chen, M., Hu, D., et al. (2014). Identification of 30 MYB transcription factor genes and analysis of their expression during abiotic stress in peanut (Arachis hypogaea L.). Gene 533, 332–345. doi: 10.1016/j.gene.2013.08.092
Cheynier, V., Comte, G., Davies, K. M., Lattanzio, V., and Martens, S. (2013). Plant phenolics: recent advances on their biosynthesis, genetics, and ecophysiology. Plant Physiol. Biochem. 72, 1–20. doi: 10.1016/j.plaphy.2013.05.009
Cho, J.-Y., Moon, J.-H., Seong, K.-Y., and Park, K.-H. (1998). Antimicrobial activity of 4-hydroxybenzoic acid and trans 4-hydroxycinnamic acid isolated and identified from rice hull. Biotechnol. Biochem. 62, 2273–2276. doi: 10.1271/bbb.62.2273
Choi, S.-W., Park, K.-I., Yeon, J.-T., Ryu, B. J., Kim, K.-J., and Kim, S. H. (2014). Anti-osteoclastogenic activity of matairesinol via suppression of p38/ERK-NFATc1 signaling axis. BMC Complement. Altern. Med. 14:35. doi: 10.1186/1472-6882-14-35
Cipolletti, M., Solar Fernandez, V., Montalesi, E., Marino, M., and Fiocchetti, M. (2018). Beyond the antioxidant activity of dietary polyphenols in cancer: the modulation of estrogen receptors (ERs) signaling. Int. J. Mol. Sci. 19:2624. doi: 10.3390/ijms19092624
Clevenger, J., Chu, Y., Scheffler, B., and Ozias-Akins, P. (2016). A developmental Transcriptome map for Allotetraploid Arachis hypogaea. Front. Plant Sci. 7:1446. doi: 10.3389/fpls.2016.01446
Čolić, S. D., Fotirić Akšić, M. M., Lazarević, K. B., Zec, G. N., Gašić, U. M., and Dabić Zagorac, D. Č, et al. (2017). Fatty acid and phenolic profiles of almond grown in Serbia. Food Chem. 234, 455–463. doi: 10.1016/j.foodchem.2017.05.006
Condori, J., Medrano, G., Sivakumar, G., Nair, V., Cramer, C., and Medina-Bolivar, F. (2009). Functional characterization of a stilbene synthase gene using a transient expression system in planta. Plant Cell Rep. 28, 589–599. doi: 10.1007/s00299-008-0664-0
Corso, M., Perreau, F., Mouille, G., and Lepiniec, L. (2020). Specialized phenolic compounds in seeds: structures, functions, and regulations. Plant Sci. 296:110471. doi: 10.1016/j.plantsci.2020.110471
Cory, H., Passarelli, S., Szeto, J., Tamez, M., and Mattei, J. (2018). The role of polyphenols in human health and food systems: a mini-review. Front. Nutr. 5:87. doi: 10.3389/fnut.2018.00087
De La Rosa, L. A., Alvarez-Parrilla, E., and Shahidi, F. (2011). Phenolic compounds and antioxidant activity of kernels and shells of mexican pecan (Carya illinoinensis). J. Agric. Food Chem. 59, 152–162. doi: 10.1021/jf1034306
De Souza, R. G. M., Schincaglia, R. M., Pimentel, G. D., and Mota, J. F. (2017). Nuts and human health outcomes: a systematic review. Nutrients 9:1311. doi: 10.3390/nu9121311
Del Rio, D., Calani, L., Dall’Asta, M., and Brighenti, F. (2011). Polyphenolic composition of hazelnut skin. J. Agric. Food Chem. 59, 9935–9941. doi: 10.1021/jf202449z
Deng, Z., Hassan, S., Rafiq, M., Li, H., He, Y., Cai, Y., et al. (2020). Pharmacological activity of Eriodictyol: the major natural Polyphenolic Flavanone. Evid. Based Complement. Altern. Med. 2020:6681352. doi: 10.1155/2020/6681352
Dong, H., Yang, X., He, J., Cai, S., Xiao, K., and Zhu, L. (2017). Enhanced antioxidant activity, antibacterial activity and hypoglycemic effect of luteolin by complexation with manganese(ii) and its inhibition kinetics on xanthine oxidase. RSC Adv. 7, 53385–53395. doi: 10.1039/C7RA11036G
El-Shitany, N. A., El-Bastawissy, E. A., and El-Desoky, K. (2014). Ellagic acid protects against carrageenan-induced acute inflammation through inhibition of nuclear factor kappa B, inducible cyclooxygenase and proinflammatory cytokines and enhancement of interleukin-10 via an antioxidant mechanism. Int. Immunopharmacol. 19, 290–299. doi: 10.1016/j.intimp.2014.02.004
Fabani, M. P., Luna, L., Baroni, M. V., Monferran, M. V., Ighani, M., Tapia, A., et al. (2013). Pistachio (Pistacia vera var Kerman) from argentinean cultivars. A natural product with potential to improve human health. J. Funct. Foods 5, 1347–1356. doi: 10.1016/j.jff.2013.05.002
Falasca, M., Casari, I., and Maffucci, T. (2014). Cancer chemoprevention with nuts. J. Natl. Cancer Inst. 106:dju238. doi: 10.1093/jnci/dju238
Falcone Ferreyra, M. L., Rius, S. P., and Casati, P. (2012). Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 3:222. doi: 10.3389/fpls.2012.00222
Fernie, A. R., and Tohge, T. (2017). The genetics of plant metabolism. Annu. Rev. Genet. 51, 287–310. doi: 10.1146/annurev-genet-120116-024640
Fukuda, T., Ito, H., and Yoshida, T. (2003). Antioxidative polyphenols from walnuts (Juglans regia L.). Phytochemistry 63, 795–801. doi: 10.1016/s0031-9422(03)00333-9
Ganeshpurkar, A., and Saluja, A. K. (2017). The pharmacological potential of Rutin. Saudi Pharm. J. SPJ Off. Publ. Saudi Pharm. Soc. 25, 149–164. doi: 10.1016/j.jsps.2016.04.025
Gao, Z., Xu, H., and Huang, K. (2002). Effects of rutin supplementation on antioxidant status and iron, copper, and zinc contents in mouse liver and brain. Biol. Trace Elem. Res. 88, 271–279.
Gil, M., and Wianowska, D. (2017). Chlorogenic acids - their properties, occurrence and analysis. Ann. UMCS Chem. 72:61. doi: 10.17951/aa.2017.72.1.61
Grace, M. H., Warlick, C. W., Neff, S. A., and Lila, M. A. (2014). Efficient preparative isolation and identification of walnut bioactive components using high-speed counter-current chromatography and LC-ESI-IT-TOF-MS. Food Chem. 158, 229–238. doi: 10.1016/j.foodchem.2014.02.117
Gulcin, İ (2020). Antioxidants and antioxidant methods: an updated overview. Arch. Toxicol. 94, 651–715. doi: 10.1007/s00204-020-02689-3
Han, Y.-J., Ren, Z.-G., Li, X.-X., Yan, J.-L., Ma, C.-Y., Wu, D.-D., et al. (2020). Advances and challenges in the prevention and treatment of COVID-19. Int. J. Med. Sci. 17, 1803–1810. doi: 10.7150/ijms.47836
Hano, C., and Tungmunnithum, D. (2020). Plant Polyphenols, more than just simple natural antioxidants: oxidative stress, aging and age-related diseases. Medicines 7:26. doi: 10.3390/medicines7050026
Heleno, S. A., Martins, A., Queiroz, M. J. R. P., and Ferreira, I. C. F. R. (2015). Bioactivity of phenolic acids: metabolites versus parent compounds: a review. Food Chem. 173, 501–513. doi: 10.1016/j.foodchem.2014.10.057
Horvath, G., Wessjohann, L., Bigirimana, J., Jansen, M., Guisez, Y., Caubergs, R., et al. (2006). Differential distribution of tocopherols and tocotrienols in photosynthetic and non-photosynthetic tissues. Phytochemistry 67, 1185–1195. doi: 10.1016/j.phytochem.2006.04.004
Huang, Y., Xiao, L., Zhang, Z., Zhang, R., Wang, Z., Huang, C., et al. (2019). The genomes of pecan and Chinese hickory provide insights into Carya evolution and nut nutrition. Gigascience 8:giz036. doi: 10.1093/gigascience/giz036
Hwang, B., Cho, J., Hwang, I., Jin, H.-G., Woo, E.-R., and Lee, D. G. (2011). Antifungal activity of lariciresinol derived from Sambucus williamsii and their membrane-active mechanisms in Candida albicans. Biochem. Biophys. Res. Commun. 410, 489–493. doi: 10.1016/j.bbrc.2011.06.004
Isemura, M. (2019). Catechin in human health and disease. Molecules 24:528. doi: 10.3390/molecules24030528
Jia, X., Luo, H., Xu, M., Zhai, M., Guo, Z., Qiao, Y., et al. (2018). Dynamic Changes in phenolics and antioxidant capacity during pecan (Carya illinoinensis) kernel ripening and its phenolics profiles. Molecules 23:435. doi: 10.3390/molecules23020435
Jiang, X., Liu, Y., Li, W., Zhao, L., Meng, F., Wang, Y., et al. (2013). Tissue-specific, development-dependent phenolic compounds accumulation profile and gene expression pattern in tea plant [Camellia sinensis]. PLoS One 8:e62315. doi: 10.1371/journal.pone.0062315
Kahkeshani, N., Farzaei, F., Fotouhi, M., Alavi, S. S., Bahramsoltani, R., Naseri, R., et al. (2019). Pharmacological effects of gallic acid in health and diseases: a mechanistic review. Iran. J. Basic Med. Sci. 22, 225–237. doi: 10.22038/ijbms.2019.32806.7897
Kawahara, S., Iwata, I., Fujita, E., Yamawaki, M., Nishiwaki, H., Sugahara, T., et al. (2010). IgE-suppressive activity of (À)-matairesinol and its structure-activity relationship. Biosci. Biotechnol. Biochem. 74, 1878–1883. doi: 10.1271/bbb.100275
Kessler, A., and Kalske, A. (2018). Plant secondary metabolite diversity and species interactions. Annu. Rev. Ecol. Evol. Syst. 49, 115–138. doi: 10.1146/annurev-ecolsys-110617-062406
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Küpeli Akkol, E., Genç, Y., Karpuz, B., Sobarzo-Sánchez, E., and Capasso, R. (2020). Coumarins and coumarin-related compounds in pharmacotherapy of cancer. Cancers 12:1959. doi: 10.3390/cancers12071959
Kuršvietienė, L., Stanevičienė, I., Mongirdienė, A., and Bernatonienė, J. (2016). Multiplicity of effects and health benefits of resveratrol. Medicina 52, 148–155. doi: 10.1016/j.medici.2016.03.003
Landete, J. M. (2011). Ellagitannins, ellagic acid and their derived metabolites: a review about source, metabolism, functions and health. Food Res. Int. 44, 1150–1160. doi: 10.1016/j.foodres.2011.04.027
Lanz, T., Tropf, S., Marner, F. J., Schröder, J., and Schröder, G. (1991). The role of cysteines in polyketide synthases. Site-directed mutagenesis of resveratrol and chalcone synthases, two key enzymes in different plant-specific pathways. J. Biol. Chem. 266, 9971–9976.
Lautié, E., Russo, O., Ducrot, P., and Boutin, J. A. (2020). Unraveling plant natural chemical diversity for drug discovery purposes. Front. Pharmacol. 11:397. doi: 10.3389/fphar.2020.00397
Lee, M., Rho, H. S., and Choi, K. (2019). Anti-inflammatory effects of a p-coumaric acid and Kojic acid derivative in LPS-stimulated RAW264.7 macrophage cells. Biotechnol. Bioprocess Eng. 24, 653–657. doi: 10.1007/s12257-018-0492-1
Lepiniec, L., Debeaujon, I., Routaboul, J.-M., Baudry, A., Pourcel, L., Nesi, N., et al. (2006). Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 57, 405–430. doi: 10.1146/annurev.arplant.57.032905.105252
Li, W., Du, Q., Li, X., Zheng, X., Lv, F., Xi, X., et al. (2020). Eriodictyol inhibits proliferation, metastasis and induces apoptosis of glioma cells via PI3K/Akt/NF-κB signaling pathway. Front. Pharmacol. 11:114. doi: 10.3389/fphar.2020.00114
Lin, Y., Shi, R., Wang, X., and Shen, H.-M. (2008). Luteolin, a flavonoid with potential for cancer prevention and therapy. Curr. Cancer Drug Targets 8, 634–646. doi: 10.2174/156800908786241050
Liu, Y., Zhao, S., Wang, J., Zhao, C., Guan, H., Hou, L., et al. (2015). Molecular cloning, expression, and evolution analysis of type II CHI gene from peanut (Arachis hypogaea L.). Dev. Genes Evol. 225, 1–10. doi: 10.1007/s00427-015-0489-0
Lou, H., Yamazaki, Y., Sasaki, T., Uchida, M., Tanaka, H., and Oka, A. (1999). A-type proanthocyanidins from peanut skins. Phytochemistry 51, 297–308. doi: 10.1016/S0031-9422(98)00736-5
Lou, H., Yuan, H., Ma, B., Ren, D., Ji, M., and Oka, S. (2004). Polyphenols from peanut skins and their free radical-scavenging effects. Phytochemistry 65, 2391–2399. doi: 10.1016/j.phytochem.2004.06.026
Lucas, S. J., Kahraman, K., Avşar, B., Buggs, R. J. A., and Bilge, I. (2020). A chromosome-scale genome assembly of European hazel (Corylus avellana L.) reveals targets for crop improvement. Plant J. 105, 1413–1430. doi: 10.1111/tpj.15099
Luo, Y., Shang, P., and Li, D. (2017). Luteolin: a flavonoid that has multiple cardio-protective effects and its molecular mechanisms. Front. Pharmacol. 8:692. doi: 10.3389/fphar.2017.00692
Lushchak, V. I., and Semchuk, N. M. (2012). Tocopherol biosynthesis: chemistry, regulation and effects of environmental factors. Acta Physiol. Plant 34, 1607–1628. doi: 10.1007/s11738-012-0988-9
Ma, Y., Kosińska-Cagnazzo, A., Kerr, W. L., Amarowicz, R., Swanson, R. B., and Pegg, R. B. (2014). Separation and characterization of phenolic compounds from dry-blanched peanut skins by liquid chromatography-electrospray ionization mass spectrometry. J. Chromatogr. 1356, 64–81. doi: 10.1016/j.chroma.2014.06.027
Maia, F. J. N., Ribeiro, F. W. P., Rangel, J. H. G., Lomonaco, D., Luna, F. M. T., de Lima-Neto, P., et al. (2015). Evaluation of antioxidant action by electrochemical and accelerated oxidation experiments of phenolic compounds derived from cashew nut shell liquid. Ind. Crops Prod. 67, 281–286. doi: 10.1016/j.indcrop.2015.01.034
Marrano, A., Britton, M., Zaini, P. A., Zimin, A. V., Workman, R. E., Puiu, D., et al. (2020). High-quality chromosome-scale assembly of the walnut (Juglans regia L.) reference genome. Gigascience 9:giaa050. doi: 10.1093/gigascience/giaa050
Marrano, A., Martínez-García, P. J., Bianco, L., Sideli, G. M., Di Pierro, E. A., Leslie, C. A., et al. (2019). A new genomic tool for walnut (Juglans regia L.): development and validation of the high-density AxiomTM J. regia 700K SNP genotyping array. Plant Biotechnol. J. 17, 1027–1036. doi: 10.1111/pbi.13034
Martínez-García, P. J., Crepeau, M. W., Puiu, D., Gonzalez-Ibeas, D., Whalen, J., Stevens, K. A., et al. (2016). The walnut (Juglans regia) genome sequence reveals diversity in genes coding for the biosynthesis of non-structural polyphenols. Plant J. 87, 507–532. doi: 10.1111/tpj.13207
Mathesius, U. (2018). Flavonoid functions in plants and their interactions with other organisms. Plants 7:30. doi: 10.3390/plants7020030
Milbury, P. E., Chen, C.-Y., Dolnikowski, G. G., and Blumberg, J. B. (2006). Determination of Flavonoids and Phenolics and their distribution in almonds. J. Agric. Food Chem. 54, 5027–5033. doi: 10.1021/jf0603937
Monagas, M., Garrido, I., Lebrón-Aguilar, R., Bartolome, B., and Gómez-Cordovés, C. (2007). Almond (Prunus dulcis(Mill.) D.A. Webb) skins as a potential source of bioactive polyphenols. J. Agric. Food Chem. 55, 8498–8507. doi: 10.1021/jf071780z
Morais, S. M., Silva, K. A., Araujo, H., Vieira, I. G. P., Alves, D. R., Fontenelle, R. O. S., et al. (2017). Anacardic acid constituents from cashew nut shell liquid: nmr characterization and the effect of unsaturation on its biological activities. Pharmaceuticals 10:31.
Muroi, H., and Kubo, I. (1996). Antibacterial activity of anacardic acid and totarol, alone and in combination with methicillin, against methicillinresistant Staphylococcus aureus. J. Appl. Bacteriol. 80, 387–394. doi: 10.1111/j.1365-2672.1996.tb03233.x
Musarra-Pizzo, M., Ginestra, G., Smeriglio, A., Pennisi, R., Sciortino, M. T., and Mandalari, G. (2019). The antimicrobial and antiviral activity of polyphenols from almond (Prunus dulcis L.) Skin. Nutrients 11:2355. doi: 10.3390/nu11102355
Naake, T., Maeda, H. A., Proost, S., Tohge, T., and Fernie, A. R. (2020). Kingdom-wide analysis of the evolution of the plant type III polyketide synthase superfamily. Plant Physiol. kiaa086. doi: 10.1093/plphys/kiaa086
Navarro, G., Martínez-Pinilla, E., Ortiz, R., Noé, V., Ciudad, C. J., and Franco, R. (2018). Resveratrol and related Stilbenoids, Nutraceutical/dietary complements with health-promoting actions: industrial production, safety, and the search for mode of action. Compr. Rev. Food Sci. Food Saf. 17, 808–826. doi: 10.1111/1541-4337.12359
Nayak, B., Liu, R. H., and Tang, J. (2015). Effect of processing on phenolic antioxidants of fruits, vegetables, and grains—a review. Crit. Rev. Food Sci. Nutr. 55, 887–918. doi: 10.1080/10408398.2011.654142
Ngwa, W., Kumar, R., Thompson, D., Lyerly, W., Moore, R., Reid, T.-E., et al. (2020). Potential of flavonoid-inspired Phytomedicines against COVID-19. Molecules 25:2707. doi: 10.3390/molecules25112707
Pais, A. L., Li, X., and Jenny Xiang, Q.-Y. (2018). Discovering variation of secondary metabolite diversity and its relationship with disease resistance in Cornus florida L. Ecol. Evol. 8, 5619–5636. doi: 10.1002/ece3.4090
Palazzolo, G., Horvath, P., and Zenobi-Wong, M. (2012). The flavonoid isoquercitrin promotes neurite elongation by reducing RhoA activity. PLoS One 7:e49979. doi: 10.1371/journal.pone.0049979
Pelvan, E., Olgun, E. Ö, Karadağ, A., and Alasalvar, C. (2018). Phenolic profiles and antioxidant activity of Turkish Tombul hazelnut samples (natural, roasted, and roasted hazelnut skin). Food Chem. 244, 102–108. doi: 10.1016/j.foodchem.2017.10.011
Peng, M., Shahzad, R., Gul, A., Subthain, H., Shen, S. Q., Lei, L., et al. (2017). Differentially evolved glucosyltransferases determine natural variation of rice flavone accumulation and UV-tolerance. Nat. Commun. 8:1975. doi: 10.1038/s41467-017-02168-x
Pugazhendhi, D., Pope, G. S., and Darbre, P. D. (2005). Oestrogenic activity of p-hydroxybenzoic acid (common metabolite of paraben esters) and methylparaben in human breast cancer cell lines. J. Appl. Toxicol. 25, 301–309. doi: 10.1002/jat.1066
Puksasook, T., Kimura, S., Tadtong, S., Jiaranaikulwanitch, J., Pratuangdejkul, J., Kitphati, W., et al. (2017a). Erratum to: semisynthesis and biological evaluation of prenylated resveratrol derivatives as multi-targeted agents for Alzheimer’s disease. J. Nat. Med. 71, 683–684. doi: 10.1007/s11418-017-1110-9
Puksasook, T., Kimura, S., Tadtong, S., Jiaranaikulwanitch, J., Pratuangdejkul, J., Kitphati, W., et al. (2017b). Semisynthesis and biological evaluation of prenylated resveratrol derivatives as multi-targeted agents for Alzheimer’s disease. J. Nat. Med. 71, 665–682. doi: 10.1007/s11418-017-1097-2
Regueiro, J., Sánchez-González, C., Vallverdú-Queralt, A., Simal-Gándara, J., Lamuela-Raventós, R., and Izquierdo-Pulido, M. (2014). Comprehensive identification of walnut polyphenols by liquid chromatography coupled to linear ion trap-Orbitrap mass spectrometry. Food Chem. 152, 340–348. doi: 10.1016/j.foodchem.2013.11.158
Renaud, J., and Martinoli, M.-G. (2019). Considerations for the use of polyphenols as therapies in neurodegenerative diseases. Int. J. Mol. Sci. 20:1883. doi: 10.3390/ijms20081883
Robbins, K. S., Ma, Y., Wells, M. L., Greenspan, P., and Pegg, R. B. (2014). Separation and characterization of phenolic compounds from U.S. pecans by liquid chromatography-tandem mass spectrometry. J. Agric. Food Chem. 62, 4332–4341. doi: 10.1021/jf500909h
Rodríguez-García, C., Sánchez-Quesada, C., Toledo, E., Delgado-Rodríguez, M., and Gaforio, J. (2019). Naturally Lignan-rich foods: a dietary tool for health promotion? Molecules 24:917. doi: 10.3390/molecules24050917
Saigo, T., Wang, T., Watanabe, M., and Tohge, T. (2020). Diversity of anthocyanin and proanthocyanin biosynthesis in land plants. Curr. Opin. Plant Biol. 55, 93–99. doi: 10.1016/j.pbi.2020.04.001
Salehi, B., Gültekin, ÖM., Kırkın, C., Özçelik, B., Morais, B., Carneiro, F. C., et al. (2019). Anacardium plants: chemical, nutritional composition and biotechnological applications. Biomolecules 9:465. doi: 10.3390/biom9090465
Salehi, B., Mishra, A., Nigam, M., Sener, B., Kilic, M., Sharifi-Rad, M., et al. (2018). Resveratrol: a double-edged sword in health benefits. Biomedicines 6:91. doi: 10.3390/biomedicines6030091
Sánchez-Pérez, R., Pavan, S., Mazzeo, R., Moldovan, C., Aiese Cigliano, R., Del Cueto, J., et al. (2019). Mutation of a bHLH transcription factor allowed almond domestication. Science 364, 1095–1098. doi: 10.1126/science.aav8197
Sarnoski, P. J., Johnson, J. V., Reed, K. A., Tanko, J. M., and O’Keefe, S. F. (2012). Separation and characterisation of proanthocyanidins in Virginia type peanut skins by LC-MSn. Food Chem. 131, 927–939. doi: 10.1016/j.foodchem.2011.09.081
Sato, Y., Itagaki, S., Kurokawa, T., Ogura, J., Kobayashi, M., Hirano, T., et al. (2011). In vitro and in vivo antioxidant properties of chlorogenic acid and caffeic acid. Int. J. Pharm. 403, 136–138. doi: 10.1016/j.ijpharm.2010.09.035
Schneider, B. U. C., Meza, A., Beatriz, A., Pesarini, J. R., Carvalho, P. C., de Mauro, M., et al. (2016). Cardanol: toxicogenetic assessment and its effects when combined with cyclophosphamide. Genet. Mol. Biol. 39, 279–289.
Schröder, G., and Schröder, G. (1990). Stilbene and chalcone synthases: related enzymes with key functions in plant-specific pathways. Z. Naturforsch. C. J. Biosci. 45, 1–8. doi: 10.1515/znc-1990-1-202
Scossa, F., Roda, F., Tohge, T., Georgiev, M. I., and Fernie, A. R. (2019). The hot and the colorful: Understanding the metabolism, genetics and evolution of consumer preferred metabolic traits in pepper and related species. CRC Crit. Rev. Plant Sci. 38, 339–381. doi: 10.1080/07352689.2019.1682791
Shahidi, F., and Ambigaipalan, P. (2015). Phenolics and polyphenolics in foods, beverages and spices: antioxidant activity and health effects - a review. J. Funct. Foods 18, 820–897. doi: 10.1016/j.jff.2015.06.018
Shen, Y., Liang, W.-J., Shi, Y.-N., Kennelly, E. J., and Zhao, D.-K. (2020). Structural diversity, bioactivities, and biosynthesis of natural diterpenoid alkaloids. Nat. Prod. Rep. 37, 763–796. doi: 10.1039/d0np00002g
Shen, Y., Song, X., Li, L., Sun, J., Jaiswal, Y., Huang, J., et al. (2019). Protective effects of p-coumaric acid against oxidant and hyperlipidemia-an in vitro and in vivo evaluation. Biomed. Pharmacother. 111, 579–587. doi: 10.1016/j.biopha.2018.12.074
Slatnar, A., Mikulic-Petkovsek, M., Stampar, F., Veberic, R., and Solar, A. (2015). Identification and quantification of phenolic compounds in kernels, oil and bagasse pellets of common walnut (Juglans regia L.). Food Res. Int. 67, 255–263. doi: 10.1016/j.foodres.2014.11.016
Soares, S., Brandão, E., Guerreiro, C., Soares, S., Mateus, N., and De Freitas, V. (2020). Tannins in food: insights into the molecular perception of astringency and bitter taste. Molecules 25:2590. doi: 10.3390/molecules25112590
Tohge, T., de Souza, L. P., and Fernie, A. R. (2014). Genome-enabled plant metabolomics. J. Chromatogr. B 966, 7–20. doi: 10.1016/j.jchromb.2014.04.003
Tohge, T., de Souza, L. P., and Fernie, A. R. (2017). Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 68, 4013–4028. doi: 10.1093/jxb/erx177
Tohge, T., and Fernie, A. R. (2010). Combining genetic diversity, informatics and metabolomics to facilitate annotation of plant gene function. Nat. Protoc. 5, 1210–1227. doi: 10.1038/nprot.2010.82
Tohge, T., and Fernie, A. R. (2017). An overview of compounds derived from the Shikimate and Phenylpropanoid pathways and their medicinal importance. Mini. Rev. Med. Chem. 17, 1013–1027. doi: 10.2174/1389557516666160624123425
Tohge, T., Watanabe, M., Hoefgen, R., and Fernie, A. R. (2013a). Shikimate and phenylalanine biosynthesis in the green lineage. Front. Plant Sci. 4:62. doi: 10.3389/fpls.2013.00062
Tohge, T., Watanabe, M., Hoefgen, R., and Fernie, A. R. (2013b). The evolution of phenylpropanoid metabolism in the green lineage. Crit. Rev. Biochem. Mol. Biol. 48, 123–152. doi: 10.3109/10409238.2012.758083
Tohge, T., Wendenburg, R., Ishihara, H., Nakabayashi, R., Watanabe, M., Sulpice, R., et al. (2016). Characterization of a recently evolved flavonol-phenylacyltransferase gene provides signatures of natural light selection in Brassicaceae. Nat. Commun. 7:12399. doi: 10.1038/ncomms12399
Tomaino, A., Martorana, M., Arcoraci, T., Monteleone, D., Giovinazzo, C., and Saija, A. (2010). Antioxidant activity and phenolic profile of pistachio (Pistacia vera L., variety Bronte) seeds and skins. Biochimie 92, 1115–1122. doi: 10.1016/j.biochi.2010.03.027
Trevisan, M. T. S., Pfundstein, B., Haubner, R., Würtele, G., Spiegelhalder, B., Bartsch, H., et al. (2006). Characterization of alkyl phenols in cashew (Anacardium occidentale) products and assay of their antioxidant capacity. Food Chem. Toxicol. 44, 188–197. doi: 10.1016/j.fct.2005.06.012
Trox, J., Vadivel, V., Vetter, W., Stuetz, W., Kammerer, D. R., Carle, R., et al. (2011). Catechin and epicatechin in testa and their association with bioactive compounds in kernels of cashew nut (Anacardium occidentale L.). Food Chem. 128, 1094–1099. doi: 10.1016/j.foodchem.2011.04.018
Tungmunnithum, D., Thongboonyou, A., Pholboon, A., and Yangsabai, A. (2018). Flavonoids and other Phenolic compounds from medicinal plants for pharmaceutical and medical aspects: an overview. Medicines 5:93. doi: 10.3390/medicines5030093
Tuyen, P., Xuan, T., Khang, D., Ahmad, A., Quan, N., Tu Anh, T., et al. (2017). Phenolic compositions and antioxidant properties in Bark, Flower, Inner Skin, Kernel and Leaf extracts of Castanea crenata Sieb. et Zucc. Antioxidants 6:31. doi: 10.3390/antiox6020031
Valanciene, E., Jonuskiene, I., Syrpas, M., Augustiniene, E., Matulis, P., Simonavicius, A., et al. (2020). Advances and prospects of phenolic acids production, biorefinery and analysis. Biomolecules 10:874. doi: 10.3390/biom10060874
Valdés, A., Vidal, L., Beltrán, A., Canals, A., and Garrigós, M. C. (2015). Microwave-Assisted extraction of Phenolic compounds from almond skin byproducts (Prunus amygdalus): a multivariate analysis approach. J. Agric. Food Chem. 63, 5395–5402. doi: 10.1021/acs.jafc.5b01011
Valentová, K., Vrba, J., Bancířová, M., Ulrichová, J., and Křen, V. (2014). Isoquercitrin: pharmacology, toxicology, and metabolism. Food Chem. Toxicol. 68, 267–282. doi: 10.1016/j.fct.2014.03.018
Vinson, J. A., and Cai, Y. (2012). Nuts, especially walnuts, have both antioxidant quantity and efficacy and exhibit significant potential health benefits. Food Funct. 3, 134–140. doi: 10.1039/c2fo10152a
Vu, D. C., Vo, P. H., Coggeshall, M. V., and Lin, C.-H. (2018). Identification and characterization of phenolic compounds in black walnut kernels. J. Agric. Food Chem. 66, 4503–4511. doi: 10.1021/acs.jafc.8b01181
Wang, X., Zhang, Y., Xia, H., Yuan, M., Zhao, C., and Li, A. Q. (2012). Cloning and expression analysis of peanut (Arachis hypogaea L.) CHI gene. Electron. J. Biotechnol. 15:5. doi: 10.2225/vol15-issue1-fulltext-6
Wu, G. A., and Gmitter, F. G. (2019). Novel assembly strategy cracks open the mysteries of walnut genome evolution. Hortic. Res. 6:57. doi: 10.1038/s41438-019-0143-5
Xie, L., and Bolling, B. W. (2014). Characterisation of stilbenes in California almonds (Prunus dulcis) by UHPLC-MS. Food Chem. 148, 300–306. doi: 10.1016/j.foodchem.2013.10.057
Xu, D., Hu, M.-J., Wang, Y.-Q., and Cui, Y.-L. (2019). Antioxidant activities of quercetin and its complexes for medicinal application. Molecules 24:1123. doi: 10.3390/molecules24061123
Xu, Z., Meenu, M., Chen, P., and Xu, B. (2020). Comparative study on Phytochemical profiles and antioxidant capacities of chestnuts produced in different geographic area in China. Antioxidants 9:190. doi: 10.3390/antiox9030190
Yamauchi, S., Sugihara, T., Nakashima, Y., Okada, A., Akiyama, K., Kishida, T., et al. (2006). Radical and superoxide scavenging activities of Matairesinol and oxidized Matairesinol. Biosci. Biotechnol. Biochem. 70, 1934–1940. doi: 10.1271/bbb.60096
Yang, T., Fang, L., Sanders, S., Jayanthi, S., Rajan, G., Podicheti, R., et al. (2018). Stilbenoid prenyltransferases define key steps in the diversification of peanut phytoalexins. J. Biol. Chem. 293, 28–46. doi: 10.1074/jbc.ra117.000564
Yu, J., Ahmedna, M., Goktepe, I., and Dai, J. (2006). Peanut skin procyanidins: composition and antioxidant activities as affected by processing. J. Food Comp. Anal. 19, 364–371. doi: 10.1016/j.jfca.2005.08.003
Yuan, L., and Grotewold, E. (2020). Plant specialized metabolism. Plant Sci. 2020:110579. doi: 10.1016/j.plantsci.2020.110579
Zanwar, A. A., Badole, S. L., Shende, P. S., Hegde, M. V., and Bodhankar, S. L. (2014). “Chapter 80 - Role of gallic acid in cardiovascular disorders,” in Polyphenols in Human Health and Disease, eds R. R. Watson, V. R. Preedy, and D. Zibadi (San Diego: Academic Press), 1045–1047. doi: 10.1016/B978-0-12-398456-2.00080-3
Zeng, L., Tu, X. L., Dai, H., Han, F. M., Lu, B. S., Wang, M. S., et al. (2019). Whole genomes and transcriptomes reveal adaptation and domestication of pistachio. Genome Biol. 20:79. doi: 10.1186/s13059-019-1686-3
Zhang, C., Yao, X., Ren, H., Chang, J., and Wang, K. (2019a). RNA-Seq reveals flavonoid biosynthesis-related genes in pecan (Carya illinoinensis) Kernels. J. Agric. Food Chem. 67, 148–158. doi: 10.1021/acs.jafc.8b05239
Zhang, C., Yao, X., Ren, H., Wang, K., and Chang, J. (2019b). Isolation and characterization of three Chalcone synthase genes in pecan (Carya illinoinensis). Biomolecules 9:236. doi: 10.3390/biom9060236
Zhang, J., Zhang, W., Ji, F., Qiu, J., Song, X., Bu, D., et al. (2020). A high-quality walnut genome assembly reveals extensive gene expression divergences after whole-genome duplication. Plant Biotechnol. J. 18, 1848–1850. doi: 10.1111/pbi.13350
Zhang, Y., Yang, Z., Liu, G., Wu, Y., and Ouyang, J. (2020). Inhibitory effect of chestnut (Castanea mollissima Blume) inner skin extract on the activity of α-amylase, α-glucosidase, dipeptidyl peptidase IV and in vitro digestibility of starches. Food Chem. 324:126847. doi: 10.1016/j.foodchem.2020.126847
Zhang, Y., Butelli, E., Alseekh, S., Tohge, T., Rallapalli, G., Luo, J., et al. (2015). Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat. Commun. 6:8635. doi: 10.1038/ncomms9635
Zhu, T., Wang, L., You, F. M., Rodriguez, J. C., Deal, K. R., Chen, L., et al. (2019). Sequencing a Juglans regia×J. microcarpa hybrid yields high-quality genome assemblies of parental species. Hortic. Res. 6:8635. doi: 10.1038/s41438-019-0139-1
Zhuang, W., Chen, H., Yang, M., Wang, J., Pandey, M. K., Zhang, C., et al. (2019). The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 51, 865–876. doi: 10.1038/s41588-019-0402-2
Keywords: flavonoids, chemical diversity, nuts, comparative genomics, polyphenols, health benefits
Citation: Aneklaphakij C, Saigo T, Watanabe M, Naake T, Fernie AR, Bunsupa S, Satitpatipan V and Tohge T (2021) Diversity of Chemical Structures and Biosynthesis of Polyphenols in Nut-Bearing Species. Front. Plant Sci. 12:642581. doi: 10.3389/fpls.2021.642581
Received: 16 December 2020; Accepted: 25 February 2021;
Published: 06 April 2021.
Edited by:
Pedro Mena, University of Parma, ItalyReviewed by:
Wajid Waheed Bhat, Michigan State University, United StatesJoong-Hoon Ahn, Konkuk University, South Korea
Copyright © 2021 Aneklaphakij, Saigo, Watanabe, Naake, Fernie, Bunsupa, Satitpatipan and Tohge. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takayuki Tohge, dG9oZ2VAYnMubmFpc3QuanA=
 Chaiwat Aneklaphakij
Chaiwat Aneklaphakij Tomoki Saigo
Tomoki Saigo Mutsumi Watanabe
Mutsumi Watanabe Thomas Naake
Thomas Naake Alisdair R. Fernie
Alisdair R. Fernie Somnuk Bunsupa
Somnuk Bunsupa Veena Satitpatipan
Veena Satitpatipan Takayuki Tohge
Takayuki Tohge