- 1Molecular Biotechnology of Plants and Micro-Organisms, Institute of Botany and Microbiology, KU Leuven, Leuven, Belgium
- 2National Crops Resources Research Institute, National Agriculture Research Organization, Kampala, Uganda
- 3M’bé Research Station, Africa Rice Center (AfricaRice), Bouaké, Côte d’Ivoire
- 4Breeding Innovations Platform, International Rice Research Institute, Metro Manila, Philippines
- 5Graduate School of Agriculture, Tokyo University of Agriculture, Tokyo, Japan
Rice is the main food crop for people in low- and lower-middle-income countries in Asia and sub-Saharan Africa (SSA). Since 1982, there has been a significant increase in the demand for rice in SSA, and its growing importance is reflected in the national strategic food security plans of several countries in the region. However, several abiotic and biotic factors undermine efforts to meet this demand. Rice yellow mottle virus (RYMV) caused by Solemoviridae is a major biotic factor affecting rice production and continues to be an important pathogen in SSA. To date, six pathogenic strains have been reported. RYMV infects rice plants through wounds and rice feeding vectors. Once inside the plant cells, viral genome-linked protein is required to bind to the rice translation initiation factor [eIF(iso)4G1] for a compatible interaction. The development of resistant cultivars that can interrupt this interaction is the most effective method to manage this disease. Three resistance genes are recognized to limit RYMV virulence in rice, some of which have nonsynonymous single mutations or short deletions in the core domain of eIF(iso)4G1 that impair viral host interaction. However, deployment of these resistance genes using conventional methods has proved slow and tedious. Molecular approaches are expected to be an alternative to facilitate gene introgression and/or pyramiding and rapid deployment of these resistance genes into elite cultivars. In this review, we summarize the knowledge on molecular genetics of RYMV-rice interaction, with emphasis on host plant resistance. In addition, we provide strategies for sustainable utilization of the novel resistant sources. This knowledge is expected to guide breeding programs in the development and deployment of RYMV resistant rice varieties.
Introduction
Food security is a primary concern for many economies around the world because the human population is increasing, and food production must increase to keep pace (Food and Agriculture Organization of the United Nations, 2010; Boyd et al., 2013; Savary et al., 2019). According to Valin et al. (2014), a 74% increase in food demand is expected by 2050, which would affect consumers worldwide. Meeting this demand is a daunting task, and will require major improvements in agricultural production systems, including improved strategies to minimize crop losses to biotic and abiotic stresses (Das and Rao, 2015). Cereal crops account for more than 60% of food intake and will continue to contribute significantly to global food security. This contribution is reflected in projected increases in the consumption of wheat, corn, and rice by an average of 53, 106, and 47%, respectively, by 2050 (Valin et al., 2014; FAOSTAT, 2018). Of these three main crops, rice is the most important food crop for people in low- and lower-middle-income countries. In sub-Saharan Africa (SSA), where most of the population falls within the category of low incomes, the demand for rice has increased considerably since 1982. This increased demand is due to improving economies, rising household incomes, and growing population and urbanization that have led to increased consumption of rice as a major staple (Balasubramanian et al., 2007). To meet the increased demand, the region is continuously pushing for coordinated efforts to increase rice self-sufficiency in several rice-producing countries (Seck et al., 2013). However, the prospects for rice self-sufficiency are being subverted by abiotic and biotic factors, including plant diseases. Several important rice diseases have emerged recently in SSA (Séré et al., 2013). The most notorious of these has been Rice yellow mottle virus (RYMV), which is restricted to only Africa (Bakker, 1974; Kouassi et al., 2005; Savary et al., 2019).
RYMV was first identified in East Africa in 1966 (Bakker, 1974), from where it is believed to have spatially spread westward into West Africa (Pinel et al., 2000). The virus has now spread to all rice-growing countries in the region, and as such is described as an emerging disease (Fargette et al., 2006). The natural hosts of RYMV are limited to Oryza sativa, Oryza glaberrima, and wild rice, including Oryza logistiminata and Oryza barthi (Bakker, 1970; Allarangaye et al., 2007). Recently, some wild accessions from the primary gene pool (AA genome), including Oryza glumaepatula, Oryza breviligulata, Oryza meridionalis, Oryza rufipogon, and Oryza nivara, have also shown susceptibility in screening experiments (Allarangaye et al., 2007; Odongo et al., 2019), which indicate that RYMV is capable of infecting several species of Oryzae. Clear symptoms of RYMV include severe leaf mottling, yellow-green streaking, decreased tillering, and stunting of plants during the vegetative stage. Reproductive stage symptoms include poor emergence of panicles and panicle sterility (Bakker, 1970). RYMV yield losses range from 10 to 100%, depending on the time of infection, ecology, viral strain, and the rice genotype. Plants that display severe symptoms during seedling and early vegetative stages often result in plant death (Bakker, 1974; Kouassi et al., 2005). In lowland ecologies, susceptible genotypes can be severely damaged in a short period of time during periods of intense disease activity. This has been observed in susceptible varieties grown in West African countries. For example, in Nigeria and Côte d’Ivoire, yield losses above of 90% have been reported in susceptible cultivars, Bouake 189 and FARO 29 (Onwughalu et al., 2011; Soko et al., 2016), while in Sierra Leone, losses of 82% were reported on varieties PN 623-3, TOX 516-12-SLR, and ROK 3 (Taylor et al., 1990). Most recently in Burkina Faso, losses of 84% were recorded on popular varieties, FKR56N, FKR62N, and TS2 (Traoré et al., 2015).
The prevalence of RYMV varies between rice-producing countries, probably because of multiple factors including transitions from subsistence production to large-scale intensive rice production, and the associated increase in the diversity of vectors involved in virus transmission. For example, the incidence and severity of RYMV in most of Uganda’s rice-growing areas is between 50 and 75% under rainfed lowlands (Ochola and Tusiime, 2010), while in Burkina Faso, the incidence of the disease tends to be lower at 28% (Traoré et al., 2015). In Côte d’Ivoire, the incidence is higher during the dry season than during the rainy season while the opposite is the case in Nigeria (Heinrichs et al., 1997). In Niger, RYMV is generally sporadic and field infections are often found in patches (Sarra and Peters, 2003). High incidence of RYMV has been also reported in Burundi, Zimbabwe, Ethiopia, the Central African Republic, the Democratic Republic of Congo, and Zanzibar (Traoré et al., 2001; Abubakar et al., 2003; Hubert et al., 2013). The spread and severity of the disease in the Democratic Republic of Congo is attributed to changing agricultural patterns with most farmers increasingly cultivating rice in irrigated lowlands. Such changes create a conducive environment for RYMV vectors to multiply (Hubert et al., 2013), a trend that could increase transmission and create a risk of severe infections.
The response from the scientific community to the RYMV threat has been positive and has made progress. Yield losses have been addressed through adapting RYMV control measures to local situations. This includes crop improvement toward local production systems and utilization of specific agronomic practices including the use of varietal mixtures and pesticide sprays, early planting, destruction of previous plants, ratoons and volunteer crops, removal of infected plants, crop rotation, and optimum fertilizer application. These practices aim at disrupting the life cycle of the disease and improving crop health (Traoré et al., 2009). However, their use is limited and still ineffective, especially when the disease occurs in epidemic proportions. Genetic resistance is the best feasible option for economical and sustainable long-term RYMV management. Recent advances in genetics and molecular biology have contributed to the identification of RYMV resistance genes and quantitative trait loci (QTLs) that have been mapped and some cloned from O. sativa and O. glaberrima (Ndjiondjop et al., 1999; Ioannidou et al., 2003; Albar et al., 2006; Rakotomalala et al., 2008; Thiémélé et al., 2010; Pidon et al., 2017). However, Traoré et al. (2006b) and Fargette et al. (2008a) have highlighted the nuisance associated with RYMV resistance management. The virus evolves rapidly, and resistance-breaking variants have been observed across SSA (Traoré et al., 2006a; Fargette et al., 2008a,b). So much work remains to be done to identify the best strategies to limit the prevalence and yield losses caused by RYMV. In this review, we present an update of RYMV genetics in relation to virus-rice interactions and discuss some interventions focusing on natural genetic resistance. In addition, we highlight the implications of using known genetic resistance in breeding for durable resistance to the virus in SSA.
RYMV Genome Structure, Gene Function and Diversity
RYMV is a member of the genus Sobemovirus in the Solemoviridae family (Bakker, 1974; Hébrard et al., 2021). The virus capsid consists of 180 copies of 26 kDa coat protein (CP) subunits assembled in a T = 3 icosahedral structure with a positive-sense single-stranded RNA (+ssRNA) particle size of 25–28 nm in diameter (Bakker, 1974; Opalka et al., 2000; Qu et al., 2000). RYMV encodes a small genome of approximately 4,500 nucleotides, of these, roughly, 330 nucleotides are in non-coding regions (Yassi et al., 1994) (Figure 1). Additionally, the virus encapsidates a non-coding viroid like satellite RNA (satRNA) of about 220 nucleotides, which depends on a helper virus for replication. The satRNA plays no role during infection process (Tamm and Truve, 2000; Sõmera et al., 2015; Hébrard et al., 2021). The genome is a polycistronic RNA harboring five overlapping open reading frames (ORFs): ORF1, ORFx, ORF2a, ORF2b, and ORF3 (Yassi et al., 1994; Ling et al., 2013). ORF1 and ORF3 are more variable while ORF2a and ORF2b are quite conserved. Because of this, the genome size may vary depending on nucleotide diversity of ORFs. For instance, the Mali strain is about 4,450 nucleotides, and some Nigerian isolates are reported to be about 4,452 nucleotides. In ORF3, a few insertion-deletions involving basic amino acids occur depending on the strain (Fargette et al., 2004). Moreover, two non-coding regions of about 80 and 289 nucleotides, respectively, are adjacent to the furthest 5' and 3' ends of the genome (Yassi et al., 1994; Hébrard et al., 2008a). ORF1 is at the 5' end, and encodes the 17.8 kDa P1 protein of 157 amino acids. This protein is involved in viral accumulation and post-transcriptional gene silencing by suppressing the mechanisms of host RNA silencing (Siré et al., 2008; Lacombe et al., 2010). P1 is dispensable for the local movement and replication of RYMV but is essential for systemic infection (Brugidou et al., 1995; Bonneau et al., 1998; Nummert et al., 2017). At the 5' terminus of ORF1 is a viral genome-linked protein (VPg) instead of a cap while the 3' end of the viral genome is not polyadenylated (Hull, 1977; Yassi et al., 1994). ORFx, a newly assigned ORF, is embedded beneath the 5' end of ORF2. It encodes a replication protein, named protein X. Unlike in other Sobemoviruses, ORFx does not overlap with ORF1. Its translation initiates at a highly conserved CUG codon and proceeds through a leaky scanning and ribosomal frameshift mechanism (Ling et al., 2013). The next ORFs are the two overlapping ORF2a and ORF2b. ORF2a expresses VPg in the first 134 amino acids (Yassi et al., 1994; Fargette et al., 2004), which is pinpointed to influence the virulence against resistance conferred by two major genes (RYMV1 and RYMV2) (Hébrard et al., 2008b; Pinel-Galzi et al., 2016). ORF2a also encodes a polyprotein protease P2a, and two additional proteins, P10 and P8, the functions of which have yet to be investigated. ORF2b encodes the (P2b) RNA-dependent RNA polymerase (RdRP) protein. ORF3 encodes the viral coat protein (CP) translated from the sub genomic RNA. The coat protein is responsible for cell-to-cell movement, virus packaging, and stability (Yassi et al., 1994; Bonneau et al., 1998; Opalka et al., 1998).
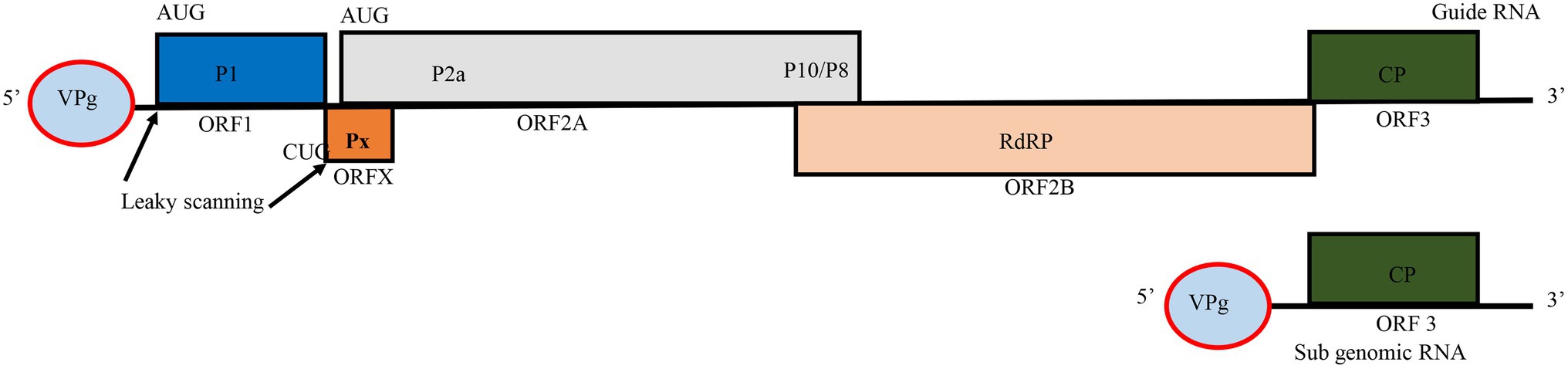
Figure 1. Genome organization of Rice yellow mottle virus (RYMV). Boxes represent open reading frames (ORFs) and the encoded proteins in each ORF. VPg, viral genome-linked protein; P1, gene silencing suppressor protein; P2a, viral protease; P10/P8, polyproteins with no assigned function; RdRP, RNA dependent RNA polymerase; CP, major coat protein. The figure was adapted from Ling et al. (2013).
RYMV has a distinct geographic diversity (Fargette et al., 2004, 2008b), and the highest diversity is established in East Africa–the putative center of ancestry and diversification (Pinel-Galzi et al., 2015). Five serotypes (Ser1–Ser5) across Africa are distinguished based on serological typing (Konate et al., 1997; N’guessan et al., 2000). Molecular typing of the CP of these serotypes recognizes six strains including S1, S2, and S3 in West Africa and S4, S5, and S6 in East Africa. Nucleotide and amino-acid difference between East African and West African strains can be up to 11%; however, at the nucleotide level, low variation within strain and within isolate is found (Pinel et al., 2000). Polymorphisms of amino-acids in the bipartite nuclear focusing motif of the R domain of the CP, and near the conserved position 151–154 of the S domain presumably determine differences and aggressiveness among the strains and isolates (Pinel et al., 2000). For instance, new Ugandan isolates were serotyped and grouped into Ser 4, based on polymorphisms in the amino acid sequences of CP gene (Uke et al., 2016). Recently, other variants in the S4, S6, and S1 subtypes have been identified (Adego et al., 2018). These serotypes are possibly segregated by polymorphisms in two amino acids: alanine vs threonine at position 115 and valine vs threonine at 191. These positions possibly localize in the antigenic sites and share an epitope placed within a conserved region. Thus, these polymorphisms may elucidate the cross-reactivity between RYMV isolates (Fargette et al., 2002a).
RYMV-Host Plant Interaction
RYMV is transmitted by about 12 insect vectors, which accelerate viral spread in most localities (Sere et al., 2008; Koudamilore et al., 2014). Chrysomelid beetles are the primary vectors that transmit the virus from its reservoirs in a semi-persistent manner (Bakker, 1970; Abo et al., 2000; Allarangaye et al., 2006). The virus can be found in the seed but no direct seed transmission has been definitively established in host plants (Konate et al., 2001; Allarangaye et al., 2006). It is unclear whether this is due to endogenous viral elements that have been reported to constitute footprints of previous infections by existing or ancient viruses (Maumus et al., 2014). Experimentally, RYMV is mechanically transmissible through sap inoculation. Other secondary forms of RYMV transmission include farming implements, wind, irrigation water, and animals (Traoré et al., 2009). Seedbed nurseries and infected crop stubble also serve as auxiliary sources of the virus (Traoré et al., 2006b; Uke et al., 2014).
Viral entry is facilitated through natural wounds caused by insect vectors and/or intercellular transfer as virion or viral ribonucleoproteins (Schoelz et al., 2011; Wang et al., 2015; Garcia-Ruiz, 2018). For compatible interaction, RYMV, like all plant viruses, must achieve both local and systemic infection in their hosts (van Loon, 1987). This infection process involves a sophisticated molecular interaction between the plant host factors and virus proteins (Figure 2). Compatible host proteins facilitate encapsidation, replication, translation, movement, and assembly. After the viral (+) RNA is released in the cytoplasm, replication is initiated. Like other Sobemoviruses, the mechanism of replication initiation of plus-and minus-strands in RYMV is still ambiguous, and needs to be further investigated (Tamm and Truve, 2000). However, recent studies suggest that the viral replication complex (VRC) replicates the nascent complementary negative sense RNA (−) using the primary +ssRNA. Subsequent (−) RNAs undergo translation and replication cycles yielding more (+) mRNAs. These are then used to produce new viral proteins and more (−) RNA and finally, the genome is enscapsidated to produce new virus particles (Souza, 2020). The new viral particles are transported cell-to-cell via the plasmodesmata to the vascular tissues for systemic spread (Mandadi and Scholthof, 2013; Heinlein, 2015). The systemic spread of RYMV occurs through xylem vessels in which large amounts of viral RNA accumulate and are then transported with solutes to new cells. RYMV disrupts the pit membranes in the xylem vessels, thus enabling the movement of virus particles into new cells (Opalka et al., 1998). To facilitate the active passage of virus particles to neighboring cells, the plasmodesmata must be modified with the help of host proteins. ORF1 and ORF3 play a key role in the modification of plasmodesmata (Bonneau et al., 1998; Nummert et al., 2017). Generally, two hypotheses explain plasmodesmata modification during viral transport (Schoelz et al., 2011; Reagan and Burch-Smith, 2020): the size exclusion limit and the removal of desmotubules and endoplasmic reticulum membranes (Schoelz et al., 2011; Reagan and Burch-Smith, 2020). RYMV uses the size exclusion limit strategy for movement to new cells (Opalka et al., 1998; Kouassi et al., 2006). RYMV particles can be localized in various plant cells, such as epidermis, nucleus vacuole, mesophyll, vesicles, bundle sheath, and vascular parenchymal cells, within 3–6 days post-infection (dpi; Opalka et al., 1998; Ndjiondjop et al., 2001), as well as in the chloroplast (Brugidou et al., 2002). Intracellularly, these viral particles occur in three isoforms described based on the presence of divalent ions and pH. The first is a stable compact form that is pH independent and has Ca2+, the second is the stable transitional form dependent on acidic pH but devoid of Ca2+, and the third is the unstable swollen form dependent on basic pH and lacks Ca2+ (Opalka et al., 1998; Brugidou et al., 2002). The viral particle stability depends on the stage of infection in the host, for instance, the transitional and swollen isoforms are more abundant during early infection, while the compact isoforms increase during late stages of infection (Brugidou et al., 2002).
In plant cells, the virus triggers host defense reactions that involve differential expression of genes that are regulated by various signaling pathways (Whitham et al., 2006). Transcriptomic studies using ESTs and cDNA-AFLP revealed differential activation of defense, metabolic, and photosynthesis pathways (Ventelon-Debout et al., 2003, 2008). In the partially resistant cultivar Azucena, in comparison with the susceptible IR64, RYMV induces expression of defense and stress related genes. Proteomic studies, using 2D-DIGE and LC-MS/MS to compare infected cell suspensions of the resistant cultivar IR64 with those of the partially resistant cultivar Azucena, further revealed protein profiles involved in metabolism, defense, and stress-related protein translation and synthesis (Ventelon-Debout et al., 2004). Delalande et al. (2005) identified three candidate proteins belonging to multigenic families including a phenylalanine ammonia-lyase, a mitochondrial chaperonin-60, and an aldolase C; however, the role of these proteins in RYMV infection remains to be validated. Further studies by Brizard et al. (2006), using SDS-PAGE and nano-LC-MS/MS identified 223 differentially regulated proteins that were categorized into functional pathways comparable to those observed using 2D-DIGE. In an incompatible interaction, RYMV induces proteins in glycolysis pathway, defense-related proteins including superoxide dismutase (SOD) (Ventelon-Debout et al., 2004; Brizard et al., 2006). In contrast, there is a marked downregulation of SOD in susceptible cultivars (Ventelon-Debout et al., 2004). In addition, studies also reveal that there is an increased accumulation of HSP 70 in the susceptible cultivar IR64 during early RYMV stress compared to partially resistant Azucena (Ventelon-Debout et al., 2004). Expression of heat shock protein 70 (HSP70) is a common incidence in response to plant viral infections, though the function of HSP70 during viral infection is poorly understood (Aparicio et al., 2005). RYMV also induces translation and protein synthesis like ribosomal proteins, translation initiation, and elongation factors, protein disulfide isomerases, chaperone proteins, and proteins involved in protein turnover such as the 20S proteasome (Ventelon-Debout et al., 2004; Brizard et al., 2006). The role of these proteins during RYMV infection remains to be validated functionally.
While these studies provide a good view of the genes and proteins expressed during RYMV-rice interactions, no recent high throughput genome-based technologies, such as RNA-seq, have been used to study RYMV-rice interactions. To clarify the different transcriptomic responses between compatible and incompatible RYMV-rice interactions and to describe more genes involved in this process, RNA-seq based approaches ought to be applied.
Genetic Determinants and Mechanisms of Qualitative Resistance to RYMV
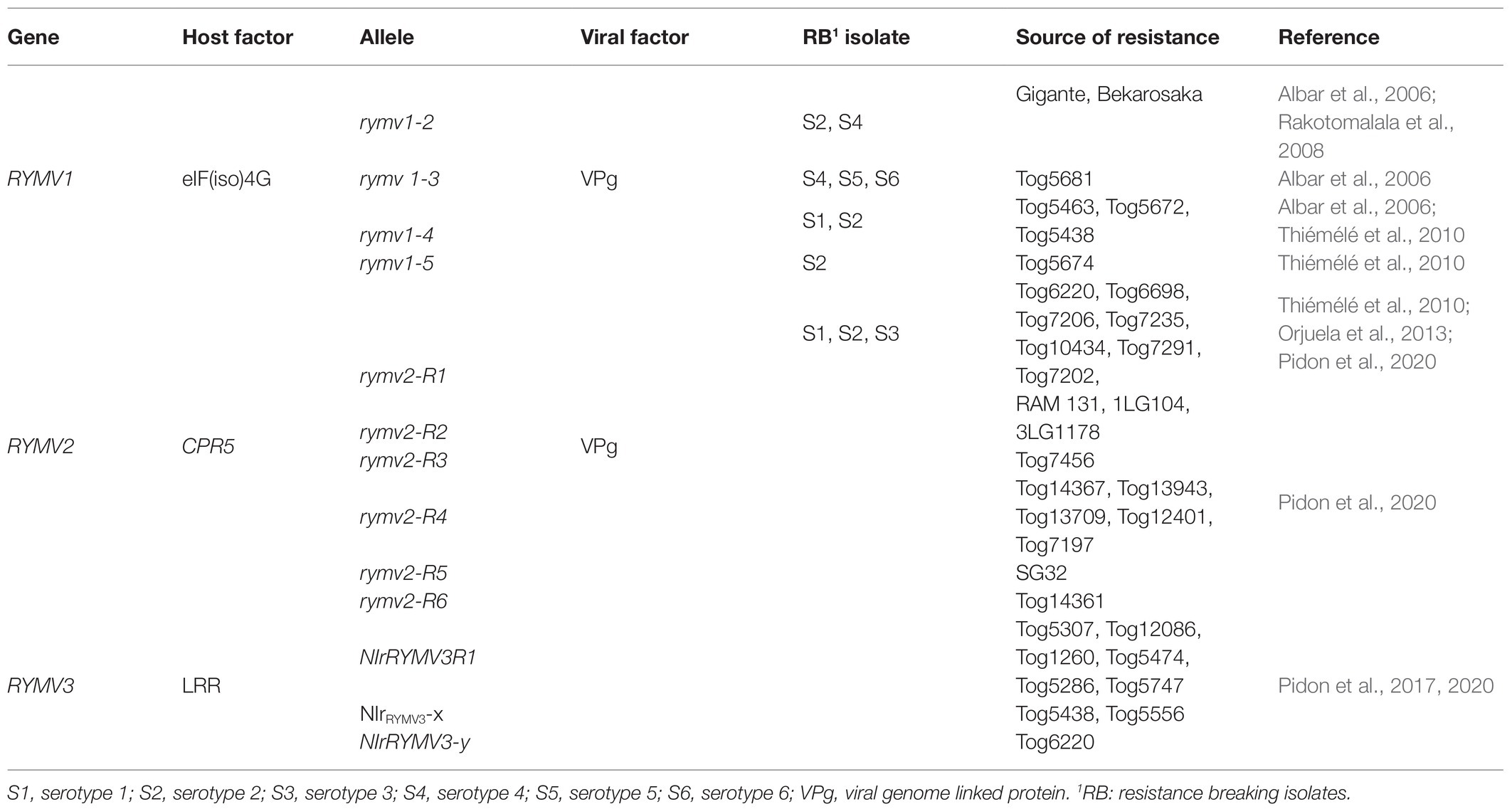
Qualitative disease resistance genes show monogenic or near complete resistance and are thus called major genes (Nelson et al., 2018). Qualitative disease resistance can be described based on two models: gene-for-gene and the matching allele model (Fraile and García-Arenal, 2010). Gene-for-gene resistance involves activation of resistance proteins such as nucleotide-binding domain leucine-rich repeat proteins (NB-LRR), which play a role in the perception of pathogen invasion and activation of plant defense mechanisms. Resistant plants usually encode NB-LRR proteins which are normally genetically dominant (R genes) (Fraile and García-Arenal, 2010; Nelson et al., 2018) (Figure 2). The matching allele interaction/resistance is conferred by the absence of host factors (susceptibility factors) required for completion of the virus infection cycle (Moffett, 2009; Fraile and García-Arenal, 2010; Wang and Krishnaswamy, 2012). Some of the genes expressed in the matching allele model induce recessive resistance. Examples of recessive host factors include the eukaryotic-translation initiation factors (eIFs) such as eIF4E and eIF4G and their isoforms. Recessive resistance is the predominant form of resistance against plant viruses (Wang and Krishnaswamy, 2012; Hashimoto et al., 2016), and can be as a result of loss-of-function of susceptibility (S genes) (Wang and Krishnaswamy, 2012; Sanfaçon, 2015; Hashimoto et al., 2016; Garcia-Ruiz, 2018). Most of the genes conferring resistance against RYMV fall in this category. These include RYMV1 (Resistance to the yellow mottle virus1) and RYMV2 (Albar et al., 2006; Thiémélé et al., 2010; Orjuela et al., 2013; Pidon et al., 2017, 2020).
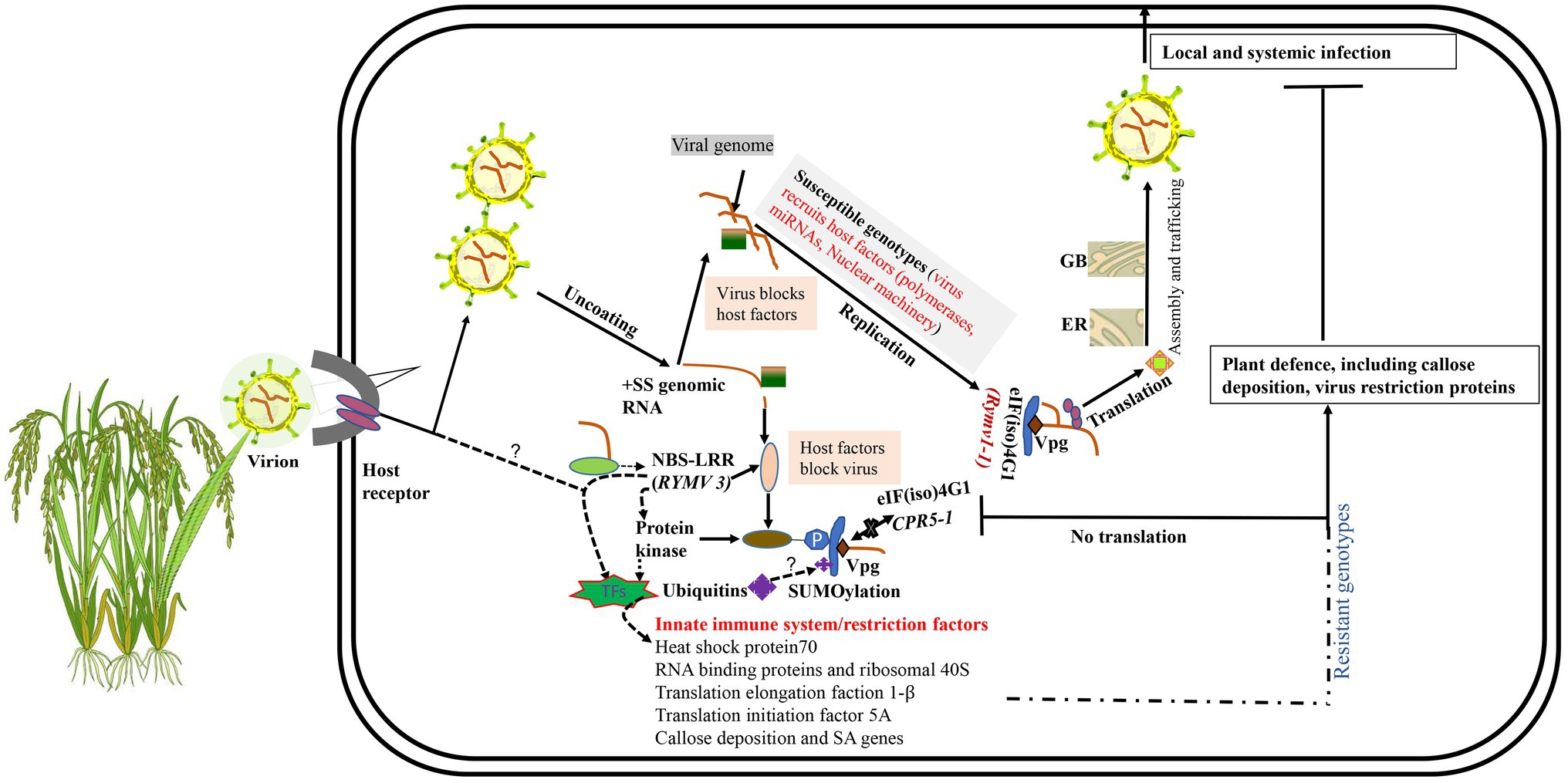
Figure 2. Mechanisms of rice-RYMV interactions. Viruses rely on numerous interactions with the host cell to replicate and transmit. GB, Golgi body; ER, endoplasmic reticulum. Unknown hypotheses are indicated as “?.” In a susceptible interaction, a virus attaches, un-coats, replicates, and expresses proteins, and then assembles and egresses. eIF(iso)4G1 encoded by rymv1-1 (susceptibility allele) possesses altered sites that facilitates viral RNA Vpg binding for successful translation of viral proteins. In a resistant reaction, we hypothesize that when the virus enters the plant cell, viral RNAs or its associated molecules in the cytoplasm are recognized by NBS-LRR or unknown receptors. The signal is transmitted from NBS-LRR via transcription factors (TFs) to the nucleus where induced resistance genes that limit viral replication, translation, and movement are transcribed. NBS-LRR proteins could be activated by direct recognition of the viral RNA. The signal could also be transmitted via either protein kinase monomers or dimers upon binding to viral RNA, which phosphorylates eIF(iso)4G1, thereby leading to translation suppression. Ubiquitination and SUMOylation could also play a role in translation regulation of eIF(iso)4G1.
The first major gene to be found and most studied is RYMV1. RYMV1 located on chromosome four and encodes eIF(iso)4G1. In a pro-viral interaction, eIF(iso)4G is recruited and directly interacts with VPg of RYMV for successful infection (Albar et al., 2006). In antiviral interactions, nonsynonymous mutations in eIF(iso)4G interfere with direct interaction with the VPg causing a resistant phenotype. These mutations may have occurred independently in O. sativa and O. glaberrima, each affecting the specificity of the allele/isolate interaction (Hébrard et al., 2010). The RYMV1 locus harbors four alleles with point mutation or small deletions; including rymv1-2, the only allele identified in O. sativa, and rymv1-3, rymv1-4, and rymv1-5, the three alleles reported in O. glaberrima (Albar et al., 2006; Thiémélé et al., 2010). The resistance conferred by rymv1-2 and rymv1-4 is due to a substitution of a glutamic acid (E) for a lysine (K) at amino acid positions 309 and 321 of eIF(iso)4G, whereas, the resistance conferred by rymv1-3 is characterized by a deletion of three amino acids at positions 322–324 in the central domain of eIF(iso)4G (Albar et al., 2006). A deletion in positions 313–315 in the central domain of eIF(iso)4G is responsible for the resistance conferred by rymv1-5 (Thiémélé et al., 2010). Some rice accessions harboring RYMV1 resistance exhibit complete resistance, and these are generally described as highly resistant accessions (Table 1).
The second S gene, RYMV2, was identified in the O. glaberrima accession Tog7291. RYMV2 is predicted to be localized on chromosome 1, encodes a homolog of the Arabidopsis CONSTITUTIVE EXPRESSION OF PATHOGENESIS RELATED GENE-5-1 (CPR5-1) (Thiémélé et al., 2010; Orjuela et al., 2013; Pidon et al., 2020). In rice, two Arabidopsis CPR5 homologs are reported to be present, CPR5-1 and CPR5-2. Resistance conferred by CPR5 is linked to a point or nonsense mutation in the CPR5-1 sequence (Orjuela et al., 2013). This gene encodes a transmembrane nucleoprotein in Arabidopsis and is known to regulate effector-triggered immunity by controlling the cell cycle and defense mechanisms. In response to the activation of immunoreceptors, CPR5 undergoes a conformational switch from oligomer to monomer. In this process, loss of function occurs and leads to the release of cyclin dependent kinase inhibitors and the permeabilization of the nuclear pore complex, which activates constitutive resistance to several pathogens (Gu et al., 2016). Despite resistance against pathogens, this loss of function mutants usually come with detrimental phenotypes that are undesirable for crop improvement (Bowling et al., 1997; Pidon et al., 2020). Six frameshift or truncated alleles have been identified in O. glaberrima accessions harboring RYMV2 (Table 1), interestingly, none of them display undesirable traits. Their activity is attributed to a partial functional redundancy of CPR5-1, suggesting that this gene is not a functional homolog of Arabidopsis CPR5. The third major gene and the only R gene is NLRRYMV3, or RYMV3, found in the O. glaberrima accessions Tog5307 and Tog5672. RYMV3 maps on chromosome 11 and encodes for a NB-LRR (Pidon et al., 2017, 2020). The LRR proteins represent one of the most diverse and abundant classes of R-gene families in plants and can be variable even among closely related plants due to presence or absence of polymorphism (Dodds and Rathjen, 2010; Luo et al., 2012; Tan and Wu, 2012; Marone et al., 2013). At the RYMV3 locus, three candidate alleles have been mapped: two alleles, NirRYMV3R1 and NirRYMV3-x, involve a substitution of one amino acid at position K779R and A823V, respectively. The resistance conferred by the third allele, NIrRYMV3-y, is a result of a truncated protein in the LRR domain with 11 amino acid substitutions (Pidon et al., 2020). The molecular basis of NB-LRR gene resistance to virus infections is expressed in two forms: first, the hypersensitivity reaction restricts the virus to the primary infection site, and second, the extreme reaction, in which the intercellular movement of the virus is completely stopped (Dodds and Rathjen, 2010). Resistance mediated by RYMV3 displays features of an extreme reaction with no symptoms expressed after infection (Pidon et al., 2020).
Collectively high genetic diversity for resistance has been identified among O. glaberrima genotypes compared to the commercially important species O. sativa (Albar et al., 2006; Thiémélé et al., 2010; Orjuela et al., 2013; Pidon et al., 2020) (Table 1). Among O. sativa accessions, only one major resistance gene has been found with two resistance alleles, while three major resistance genes, each with multiple alleles have been discovered in O. glaberrima accessions. Recessive genes (RYMV1 and RYMV2) have less polymorphisms, which is attributed to conservative selection leading to low mutation rates. For instance, five and 10 non-synonymous mutations have been identified in RYMV1 and RYMV2, respectively, whereas the R gene (RYMV3) is highly polymorphic with about 35 non-synonymous mutations identified (Pidon et al., 2020). These high mutation rates have been observed in all LRR genes due to selective pressure, which promotes the evolution of new receptors to counteract pathogens effectors (Mondragón-Palomino et al., 2002; Marone et al., 2013). This feature might be responsible for high allelic diversity in resistance loci of O. glaberrima (Pidon et al., 2020). A shorter evolutionary interaction between RYMV and O. glaberrima is however predicted as RYMV could have emerged only a few decades (Pinel-Galzi et al., 2015; Trovao et al., 2015; Pidon et al., 2020). Thus, the high variability of RYMV3 gene is interesting, and might confer resistance to other pathogens.
The use of genetic resistance is an early prevention method to reduce the impact of viral diseases (Todorovska et al., 2009; Marone et al., 2013). However, the deployment of novel varieties harboring major genes is affected by the ability of pathogens to rapidly evolve and overcome such resistance. Most R genes are known to be race specific and only give resistance to a single or limited strain(s) of a given a pathogen (Burdon et al., 2014). The mechanisms of RYMV evolution responsible for its genetic diversity are presumably 2-fold: mutations (N’guessan et al., 2000) and recombination (Ochola et al., 2015; Ndikumana et al., 2017). Recombination analyses found that two new isolates, Ke101 and Ke105, evolved from homologous recombination between strains S4lv and S4ug recognized as emerging types from Western Kenya (Adego et al., 2018), whereas mutations in the central domains of the VPg or different regions of polyprotein P2a contribute to emergence RYMV variants that overcome major resistance genes (Hébrard et al., 2008b, 2010, 2018; Poulicard et al., 2014; Pinel-Galzi et al., 2016; Pidon et al., 2020).
Mutations in the VPg are predicted to develop in three sequential ways (Pinel-Galzi et al., 2007; Traoré et al., 2010; Poulicard et al., 2014). Pathway I occurs via the sequential substitution of arginine (A) with glycine (G) and glutamic acid (E); pathway II involves sequential mutations to valine (V) but coexists with isoleucine (I); and pathway III involves sequential mutations substituting arginine with tryptophan (W). Pathway I is more frequent and efficient, whereas II and III are isolate specific (Pinel-Galzi et al., 2007; Traoré et al., 2010). In addition, polymorphism at VPg codon 49 which involve either glutamic (E) or threonic (T) acid residues (E/T polymorphism) influences the virulence of RYMV isolates (Poulicard et al., 2012). This polymorphism involves three pathotypes. The first are the T-pathotypes, in which isolates with threonine (T) at codon 49 only break resistance in O. glaberrima accessions. T-pathotypes with substitutions at codons 41 and 52 of the VPg overcome resistance conferred by rymv1-3 and rymv1-5 alleles, respectively, while substitutions from threonine to alanine at position 43 overcome resistance of the rymv1-4 allele. The T-pathotypes with substitutions at codon 804, exchanging phenylalanine for leucine in polyprotein P2a overcome RYMV2-mediated resistance (Pinel-Galzi et al., 2016) and RYMV3 encoded resistance (Pidon et al., 2017), although the mechanism of resistance breakdown is not fully understood. The second are the E-pathotypes with a glutamic acid at codon 49 which overcome rymv1-2 resistance in O. sativa. These pathotypes are polymorphic at codon 48 of VPg, and in avirulent isolates, a conserved arginine is present in this region. Substitutions of R48E and E309K in the VPg of E-pathotypes and eIF(iso)4G, respectively, provide direct interaction between the viral VPg and eIF(iso)4G (Pinel-Galzi et al., 2007; Hébrard et al., 2008b, 2010; Traoré et al., 2010). The third is a highly virulent pathotype suspected to overcome all known sources of resistance. The Hypervirulent T9-pathotype contains a mutation in the central domain of the VPg and was recently identified in West-Central Africa (Hébrard et al., 2018).
Geographically, the E-pathotypes are prevalent in East Africa while the T-pathotypes are common in West Africa. Similarly, the geographic clustering of rice accessions harboring resistance alleles in RYMV1, RYMV2, and RYMV3 loci has been recently demonstrated (Pidon et al., 2020). Thus, the geographic specificity of isolates and resistance genes clearly indicates a pattern of adaptation which would guide the development and deployment of rice breeding products in SSA. Resistance coffered by rymv1-5 and RYMV3 are quite durable in both regions (Hébrard et al., 2018; Pidon et al., 2020), whereas resistance mediated by rymv 1-3, rymv 1-4 and RYMV2 might be effective in only East Africa. However, the high durability of rymv1-4 in Tog5672 than in Tog5438 suggests epistatic control of resistance.
Genetic Determinants and Mechanisms of Quantitative Resistance to RYMV
Quantitative disease resistance is controlled by multiple genes (minor genes) with small effects or quantitative trait loci (QTLs). Quantitative resistance is characterized by a reduced virus load in the plant tissues and a slow progression of disease development due to the weakened movement of the virus. Thus, because it does not show the dramatic breakdown observed with major R gene deployment, quantitative resistance is usually viewed as durable and consequently often used in a broad range of crops (Parlevliet, 2002; Burdon et al., 2014).
Quantitative resistance to RYMV is limited to the 1st week of infection, beyond which the disease progresses, virus titer increases, and symptoms reach levels comparable to those of susceptible varieties but with less loss of yield (Albar et al., 1998; Ioannidou et al., 2000). Quantitative resistance to RYMV has been identified mostly in tropical upland rice varieties such as Azucena and Moroberekan (Japonica subtypes) (Ghesquière et al., 1997; Ioannidou et al., 2003). Fifteen QTLs have been uncovered on seven chromosomal fragments. Using an Azucena × IR64 double haploid population, genetic mapping identified these QTLs on chromosomes 1, 2, 4, 7, 8, 9, and 12 (Boisnard et al., 2007). However, most of these QTLs co-localize with growth and development QTLs, except for those on chromosome 12, and are, therefore, difficult to target. The RYMV QTL on chromosome 12 is considered to be major (Ghesquière et al., 1997; Albar et al., 1998; Ioannidou et al., 2003; Boisnard et al., 2007). Epistatic interactions between QTLs on chromosomes 7 and 12 presumably play a vital role in the regulation of partial resistance to RYMV in Azucena (Pressoir et al., 1998; Ahmadi et al., 2001; Ioannidou et al., 2003), which might limit their usefulness in marker assisted selection (MAS) taken into consideration. Other partial resistance QTLs are probably in similar positions with the major RYMV resistance genes (Boisnard et al., 2007). For example, Orjuela et al. (2013) reported on the co-localization of QTL1 in a 151-kb interval of the RYMV2 gene, hence this QTL may combine complete and partial resistance. Genome-wide association analysis study (GWAS) involving various accessions of O. glaberrima also revealed considerable relationships between quantitative resistance and two known major resistance genes (Cubry et al., 2020). In the same study, several single nucleotide polymorphisms (SNPs) close to two major resistance genes RYMV1 and RYMV3 were identified. However, the SNPs associated with resistance conferred by RYMV1 and RYMV3 were not found in regions flanking any of the identified partial resistance QTLs indicating differences in genes and pathways of resistance (Cubry et al., 2020) suggesting the complex mechanism of partial resistance to RYMV in rice.
Breeding for RYMV Durable Resistance
Breeding for disease resistance has been the most sustainable way of crop improvement to avert potential crop yield losses due to plant viruses (Burdon et al., 2014; Mundt, 2014; Fuchs, 2017). Genetic resistance is durable if it is effective over time when deployed in an environment that is favorable for disease development (Johnson, 1981; Kobayashi et al., 2014). Resistance durability of a gene depends on the nature of resistance, pathogen variability, and environmental factors (Maule et al., 2007; Gómez et al., 2009; Fonseca and Mysore, 2019). These factors presumably influence the epidemiology of RYMV and complicate the deployment of control strategies across Africa (Traoré et al., 2009; Trovao et al., 2015).
The context of durable resistance depends on a given breeding programs, for instance, lasting resistance is critical when new varieties are released less frequently compared to programs that often release novel resistant varieties (Nelson et al., 2018). Generation of broad-spectrum resistance has been the focus of most breeding programs for RYMV resistance. Preliminary focus of breeding for RYMV resistance has been on the deployment of resistance derived from Gigante (rymv1-2), given its O. sativa background and high F1 hybrid fertility in cross combinations. Recently, rymv1-2 allele was introduced into some elite cultivars, and several near-isogenic rice lines (NILs) were generated (Ndjiondjop et al., 2013). These NILs combine the rymv1-2 allele and agronomically important traits. Efforts to pyramid RYMV major resistance genes and resistance QTLs through conventional breeding methods have proven futile (Ndjiondjop et al., 2013). This has been attributed to the recessive nature of resistance conferred by most RYMV genes. If a recessive gene is introduced into an elite variety by backcrossing, the theoretical segregation ratio between the heterozygotes with recessive resistance and homozygotes with no resistance gene is 1:1 in the first generation (Griffiths et al., 2012). At this stage, it is difficult to discriminate which individuals are the heterozygotes. The heterozygotes must be tested in the second generation, which requires more time and resources. However, the use of marker assisted breeding (MAB) would allow effective selection of rymv recessive alleles in the heterozygous state. No selfing or test crossing is needed to detect rymv alleles in populations, thus saving time and accelerating breeding progress.
The use of MAB would also provide opportunities to combine R and or S genes with QTLs that could provide durable and more resilient forms of resistance to RYMV. Thus, it is important to understand the impact of each of the RYMV major resistance genes alone or in combination with other resistance genes in the field before deployment. Combining the candidate QTLs with RYMV major resistance genes in a single genetic background would substantially enhance the durability of resistance as the partial resistance would delay the breakdown of the major resistance gene (Mundt, 2014; Fuchs, 2017). Thus, more QTLs for partial resistance should be identified and included in the breeding program in SSA to enhance the durability of rymv1-2 and other major genes. Additional ways to improve durability of a major resistance gene is to combine them with another major gene (Rimbaud et al., 2018), especially those that interrupt the interaction with the conserved domains of the virus genome during the infection cycle. This would reduce the genetic adaptation of virulent variants to their environments and hosts (Fuchs, 2017). Pyramiding the recessive (RYMV1) and the dominant (RYMV3) resistance genes may provide much desired broad-spectrum resistance against RYMV and other pathogens in rice.
Integration of New Breeding Techniques for RYMV Resistance
The continuous improvements in genomics and bioinformatics has a potential to speed up crop breeding (Batley and Edwards, 2016). Advances in genomic-assisted selection have become essential to facilitate the introduction of traits that may not be done using conventional breeding techniques. Molecular markers can be used to facilitate the selection of rare traits at early stages without phenotypic analysis, thus speeding up plant breeding and crop improvement (Varshney et al., 2005; Batley and Edwards, 2016).
The scope of MAS breeding for targeted introgression of RYMV resistance genes has been successfully demonstrated in SSA (Ndjiondjop et al., 2013). A number of SNP markers linked to rymv1-2, rymv1-3, rymv1-4, and rymv1-5 alleles are now available (Albar et al., 2003; Thiémélé et al., 2010). Improvements in high-throughput genotyping have enhanced the identification of QTLs through GWAS. For instance, Cubry et al. (2020) observed a total of 2,199 SNPs, close to chromosomes 4 and 11 associated with RYMV1 and RYMV3 resistance, respectively. However, adoption and utilization of genomics-selection approaches in many breeding programs has remained low due to less know-how on genomic tools, and the cost involved in MAS (Ndjiondjop et al., 2013). However, given the current developments in refining SNPs associated with all RYMV major resistance genes at the International Rice Research Institute (IRRI), Institut de Recherche pour le Développement (IRD), and AfricaRice, it is likely that introgression of these genes through MAS will be more successful. These genes could also be pyramided with other biotic and abiotic stress resistance genes using anther culture techniques. Recently, successful crosses between O. glaberrima and Milyang23 (O. sativa) for resistance to multiple pathogens, such as RYMV, Bacterial leaf blight, Rice blast and Bacterial leaf streak, was demonstrated using anther culture techniques. Hybrids with combined resistance to these diseases were shown to be highly resistant to all four pathogens (Lamo et al., 2015).
Genetic engineering is another important strategy in crop improvement against plant viruses, considering the limited sources of natural resistance and the constraints related to the introduction of these genes to popular cultivars. RNA interference (RNAi) is one of the methods that has been explored to develop transgenic virus-resistant rice (Pinto et al., 1999; Sasaya et al., 2014; Kreuze and Valkonen, 2017). This approach involves the expression of hairpin RNA constructs in the host plant (Sanford and Johnston, 1985; Baulcombe, 1996; Brodersen and Voinnet, 2006), and it has shown success in generating RYMV resistant transgenic plants targeting the conserved P2a protein in the susceptible cultivar IR64 (Pinto et al., 1999). Additionally, genome editing is another approach with enormous potential to improve breeding for RYMV resistance. Genome editing tools, such as CRISPR-Cas9, are powerful techniques to facilitate crop improvement. CRISPR-Cas9 is based on introducing targeted mutations at specific sites by utilizing non-homologous end-joining to repair induced double-strand breaks (Belhaj et al., 2013; Haque et al., 2018; Kalinina et al., 2020). Full implementation of CRISPR-Cas9 technology is dependent on the identification of plant host factors which can be manipulated to limit virus proliferation (Kalinina et al., 2020). Some of these host factors have been demonstrated to interact directly with the viral RNA (Hyodo et al., 2014; Van Schie and Takken 2014; Garcia-Ruiz, 2018), although only a few of them have been identified to date. In rice, only eIF(iso)G4, CPR-5 (Albar et al., 2006; Thiémélé et al., 2010), is known to interact with RYMV. This limited number of plant-interacting proteins, with experimental evidence, makes targeting specific host genes for gene modification difficult. Therefore, to provide more breeding options for RYMV resistance, additional susceptibility genes need to be identified and validated. Once validated, these genes can be modified using CRISPR-Cas9 to disrupt the interaction with viral proteins. This brings the susceptible host plant outside the host range of the virus, to confer resistance.
Conclusion
RYMV continues to cause losses to rice production in SSA due to lack of resistant elite cultivars. Considering the high adaptive potential of this virus and frequent resistance breakdown in rice cultivars, breeding for resistance and its durability should be given top priority in SSA. Encouraging news comes from recent knowledge on rice-RYMV interactions, which has enabled identification and isolation of three major resistance genes (RYMV1, RYMV2, and RYMV3), and the subsequent definition of geographic specificities of these genes and RYMV strains. There is need to build on these successes by further identifying the genomic localization of desired SNPs linked to the major resistance genes for use in genomic-assisted breeding. The speedy development of homozygous lines through double haploid breeding when coupled with MAS would be more effective for developing and deployment of durable RYMV resistant varieties. Parallel efforts are needed to identify additional genes and extensively screen host-virus protein interactors to identify and validate additional host factors that assist or suppress the virus. With these interaction factors identified, a reverse genetics approach could be used to identify novel host S genes that could be modified by genome editing to impair susceptibility. These short and long-term strategies will provide immediate products and build a large germplasm base and knowledge necessary to respond to future RYMV attacks.
Author Contributions
PO wrote the first draft of the manuscript. GO helped to write and edited the manuscript. PO and GO drew the figures and tables. KG edited, supervised, and supported the writing. KN, TA, and OR edited all versions of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by Belgian Directorate General for Development and Humanitarian Aid (DGD) and implemented by VLIR-UOS under grant number BE2017GMUKULA101.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully thank Alice Kennedy, Molecular Biotechnology of Plants and Micro-organisms, KU Leuven, for her help in language editing.
References
Abo, M. E., Alegbejo, M. D., Sy, A. A., and Misari, S. M. (2000). An overview of the mode of transmission, host plants and methods of detection of Rice yellow mottle virus. J. Sustain. Agric. 17, 19–36. doi: 10.1300/J064v17n01_04
Abubakar, Z., Ali, F., Pinel, A., Traoré, O., N’Guessan, P., Notteghem, J. L., et al. (2003). Phylogeography of Rice yellow mottle virus in Africa. J. Gen. Virol. 84, 733–743. doi: 10.1099/vir.0.18759-0
Adego, A. K., Poulicard, N., Pinel-Galzi, A., Mukoye, B., Fargette, D., Wéré, H. K., et al. (2018). Full-length genome sequences of recombinant and nonrecombinant sympatric strains of Rice yellow mottle virus from western Kenya. Genome Announc. 6, e01508–e01517. doi: 10.1128/genomeA.01508-17
Ahmadi, N., Albar, L., Pressoir, G., Pinel, A., Fargette, D., and Ghesquière, A. (2001). Genetic basis and mapping of the resistance to Rice yellow mottle virus. III. Analysis of QTL efficiency in introgressed progenies confirmed the hypothesis of complementary epistasis between two resistance QTLs. Theor. Appl. Genet. 103, 1084–1092. doi: 10.1007/s001220100642
Albar, L., Bangratz-Reyser, M., Hébrard, E., Ndjiondjop, M. N., Jones, M., and Ghesquière, A. (2006). Mutations in the eIF(iso)4G translation initiation factor confer high resistance of rice to Rice yellow mottle virus. Plant J. 47, 417–426. doi: 10.1111/j.1365-313X.2006.02792.x
Albar, L., Lorieux, M., Ahmadi, N., Rimbault, I., Pinel, A., Sy, A. A., et al. (1998). Genetic basis and mapping of the resistance to rice yellow mottle virus. I. QTLs identification and relationship between resistance and plant morphology. Theor. Appl. Genet. 97, 1145–1154. doi: 10.1007/s001220051003
Albar, L., Ndjiondjop, M.-N. N., Esshak, Z., Berger, A., Pinel, A., Jones, M., et al. (2003). Fine genetic mapping of a gene required for Rice yellow mottle virus cell-to-cell movement. Theor. Appl. Genet. 107, 371–378. doi: 10.1007/s00122-003-1258-4
Allarangaye, M. D., Traoré, O., Traoré, E. V. S., Millogo, R. J., Guinko, S., and Konaté, G. (2007). Host range of rice yellow mottle virus in Sudano-sahelian savannahs. Pak. J. Biol. Sci. 10, 1414–1421. doi: 10.3923/pjbs.2007.1414.1421
Allarangaye, M. D., Traoré, O., Traoré, E. V. S., Millogo, R. J., and Konaté, G. (2006). Evidence of non-transmission of Rice yellow mottle virus through seeds of wild host species. J. Plant Pathol. 88, 309–315. doi: 10.4454/jpp.v88i3.877
Aparacio, F., Thomas, C. L., Lederer, C., Niu, Y., Wang, D., and Maule, A. J. (2005). Virus induction of heat shock protein 70 reflects a general response to protein accumulation in the plant cytosol. Plant Physiol. 138, 529–536. doi: 10.1104/pp.104.058958
Bakker, W. (1970). Rice yellow mottle, a mechanically transmissible virus disease of rice in Kenya. Neth. J. Plant Pathol. 76, 53–63. doi: 10.1007/BF01974433
Bakker, W. (1974). Characterization and ecological aspects of rice yellow mottle virus in Kenya. Agric. Res. Rep. 829, 1–152.
Balasubramanian, V., Sie, M., Hijmans, R. J., and Otsuka, K. (2007). Increasing rice production in sub-Saharan Africa: challenges and opportunities. Adv. Agron. 94, 55–133. doi: 10.1016/S0065-2113(06)94002-4
Batley, J., and Edwards, D. (2016). The application of genomics and bioinformatics to accelerate crop improvement in a changing climate. Curr. Opin. Plant Biol. 30, 78–81. doi: 10.1016/j.pbi.2016.02.002
Baulcombe, D. C. (1996). RNA as a target and an initiator of post-transcriptional gene silencing in trangenic plants. Plant Mol. Biol. 32, 79–88. doi: 10.1007/BF00039378
Belhaj, K., Chaparro-Garcia, A., Kamoun, S., and Nekrasov, V. (2013). Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9:39. doi: 10.1186/1746-4811-9-39
Boisnard, A., Albar, L., Thiéméle, D., Rondeau, M., and Ghesquière, A. (2007). Evaluation of genes from eIF4E and eIF4G multigenic families as potential candidates for partial resistance QTLs to Rice yellow mottle virus in rice. Theor. Appl. Genet. 116, 53–62. doi: 10.1007/s00122-007-0646-6
Bonneau, C., Brugidou, C., Chen, L., Beachy, R. N., and Fauquet, C. (1998). Expression of the rice yellow mottle virus P1 protein in vitro and in vivo and its involvement in virus spread. Virology 244, 79–86. doi: 10.1006/viro.1998.9100
Bowling, S. A., Clarke, J. D., Liu, Y., Klessig, D. F., and Dong, X. (1997). The cpr5 mutant of arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9, 1573–1584. doi: 10.1105/tpc.9.9.1573
Boyd, L. A., Ridout, C., O’Sullivan, D. M., Leach, J. E., and Leung, H. (2013). Plant-pathogen interactions: disease resistance in modern agriculture. Trends Genet. 29, 233–240. doi: 10.1016/j.tig.2012.10.011
Brizard, J. P., Carapito, C., Delalande, F., Van Dorsselaer, A., and Brugidou, C. (2006). Proteome analysis of plant-virus interactome: comprehensive data for virus multiplication inside their hosts. Mol. Cell. Proteomics 5, 2279–2297. doi: 10.1074/mcp.M600173-MCP200
Brodersen, P., and Voinnet, O. (2006). The diversity of RNA silencing pathways in plants. Trends Genet. 22, 268–280. doi: 10.1016/j.tig.2006.03.003
Brugidou, C., Holt, C., Ngon, A., Yassi, M., Zhang, S., Beachy, R., et al. (1995). Synthesis of an infectious full-length cDNA clone of rice yellow mottle virus andmutagenesis of the coat protein. Virology 206, 108–115. doi: 10.1016/S0042-6822(95)80025-5
Brugidou, C., Opalka, N., Yeager, M., Beachy, R. N., and Fauquet, C. (2002). Stability of rice yellow mottle virus and cellular compartmentalization during the infection process in Oryza sativa (L.). Virology 297, 98–108. doi: 10.1006/viro.2002.1398
Burdon, J. J., Barrett, L. G., Rebetzke, G., and Thrall, P. H. (2014). Guiding deployment of resistance in cereals using evolutionary principles. Evol. Appl. 7, 609–624. doi: 10.1111/eva.12175
Cubry, P., Pidon, H., Ta, K. N., Tranchant-Dubreuil, C., Thuillet, A.-C. C., Holzinger, M., et al. (2020). Genome wide association study pinpoints key agronomic QTLs in African rice Oryza glaberrima. bioRxiv [Preprint]. doi: 10.1101/2020.01.07.897298
Das, G., and Rao, G. J. N. (2015). Molecular marker assisted gene stacking for biotic and abiotic stress resistance genes in an elite rice cultivar. Front. Plant Sci. 6:698. doi: 10.3389/fpls.2015.00698
Delalande, F., Carapito, C., Brizard, J. P., Brugidou, C., and Van Dorsselaer, A. (2005). Multigenic families and proteomics: extended protein characterization as a tool for paralog gene identification. Proteomics 5, 450–460. doi: 10.1002/pmic.200400954
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Fargette, D., Konaté, G., Fauquet, C., Muller, E., Peterschmitt, M., and Thresh, J. M. (2006). Molecular ecology and emergence of tropical plant viruses. Annu. Rev. Phytopathol. 44, 235–260. doi: 10.1146/annurev.phyto.44.120705.104644
Fargette, D., Pinel, A., Abubakar, Z., Traoré, O., Brugidou, C., Fatogoma, S., et al. (2004). Inferring the evolutionary history of Rice yellow mottle virus from genomic, phylogenetic, and phylogeographic studies. J. Virol. 78, 3252–3261. doi: 10.1128/JVI.78.7.3252-3261.2004
Fargette, D., Pinel, A., Halimi, H., Brugidou, C., Fauquet, C., and Van Regenmortel, M. (2002a). Comparison of molecular and immunological typing of isolates of Rice yellow mottle virus. Arch. Virol. 147, 583–596. doi: 10.1007/s007050200008
Fargette, D., Pinel, A., Rakotomalala, M., Sangu, E., Traore, O., Sereme, D., et al. (2008b). Rice yellow mottle virus, an RNA plant virus, evolves as rapidly as most RNA animal viruses. J. Virol. 82, 3584–3589. doi: 10.1128/jvi.02506-07
Fargette, D., Pinel, A., Traoré, O., Ghesquière, A., and Konaté, G. (2002b). Emergence of resistance-breaking isolates of Rice yellow mottle virus during serial inoculations. Eur. J. Plant Pathol. 108, 585–591. doi: 10.1023/A:1019952907105
Fargette, D., Pinel-Galzi, A., Sérémé, D., Lacombe, S., Hébrard, E., Traoré, O., et al. (2008a). Diversification of Rice yellow mottle virus and related viruses spans the history of agriculture from the neolithic to the present. PLoS Pathog. 4:e1000125. doi: 10.1371/journal.ppat.1000125
Fonseca, J. P., and Mysore, K. S. (2019). Genes involved in nonhost disease resistance as a key to engineer durable resistance in crops. Plant Sci. 279, 108–116. doi: 10.1016/j.plantsci.2018.07.002
Food and Agriculture Organization of the United Nations (2010). The State of Food Insecurity in the World Addressing food insecurity in protracted crises 2010 Key messages. Available at: http://www.fao.org/docrep/013/i1683e/i1683e.pdf (Accessed April 22, 2021).
Fraile, A., and García-Arenal, F. (2010). “The coevolution of plants and viruses,” in Advances in Virus Research. eds. J P. Carr and Gad Loebenstein (Oxford: Academic Press), 1–32.
Fuchs, M. (2017). Pyramiding resistance-conferring gene sequences in crops. Curr. Opin. Virol. 26, 36–42. doi: 10.1016/j.coviro.2017.07.004
Garcia-Ruiz, H. (2018). Susceptibility genes to plant viruses. Viruses 10:484. doi: 10.3390/v10090484
Ghesquière, A., Albar, L., Lorieux, M., Ahmadi, N., Fargette, D., Huang, N., et al. (1997). A major quantitative trait locus for rice yellow mottle virus resistance maps to a cluster of blast resistance genes on chromosome 12. Phytopathology 87, 1243–1249. doi: 10.1094/PHYTO.1997.87.12.1243
Gómez, P., Rodríguez-Hernández, A. M., Moury, B., and Aranda, M. (2009). Genetic resistance for the sustainable control of plant virus diseases: breeding, mechanisms and durability. Eur. J. Plant Pathol. 125, 1–22. doi: 10.1007/s10658-009-9468-5
Griffiths, A. J. F., Wessler, S. R., Caroll, S. B., and Doebley, J. (2012). Introduction to Genetic Analysis. 10th Edn. New York, NY: W. H. Freeman
Gu, Y., Zebell, S. G., Liang, Z., Wang, S., Kang, B. H., and Dong, X. (2016). Nuclear pore permeabilization is a convergent signaling event in effector-triggered immunity. Cell 166, 1526.e11–1538.e11. doi: 10.1016/j.cell.2016.07.042
Haque, E., Taniguchi, H., Hassan, M. M., Bhowmik, P., Karim, M. R., Śmiech, M., et al. (2018). Application of CRISPR/Cas9 genome editing technology for the improvement of crops cultivated in tropical climates: recent progress, prospects, and challenges. Front. Plant Sci. 9:617. doi: 10.3389/fpls.2018.00617
Hashimoto, M., Neriya, Y., Yamaji, Y., and Namba, S. (2016). Recessive resistance to plant viruses: potential resistance genes beyond translation initiation factors. Front. Microbiol. 7:1695. doi: 10.3389/fmicb.2016.01695
Hébrard, E., Fargette, D., and Konaté, G. (2008a). “Rice yellow mottle virus,” in Encyclopedia of Virology. 3rd Edn. eds. B. W. J. Mahy and H. V. Van Regenmortel (Oxford: Academic Press), 485–490.
Hébrard, E., Pinel-Galzi, A., and Fargette, D. (2008b). Virulence domain of the RYMV genome-linked viral protein VPg towards rice rymv1-2-mediated resistance. Arch. Virol. 153, 1161–1164. doi: 10.1007/s00705-008-0087-9
Hébrard, E., Pinel-Galzi, A., Oludare, A., Poulicard, N., Aribi, J., Fabre, S., et al. (2018). Identification of a hypervirulent pathotype of Rice yellow mottle virus: a threat to genetic resistance deployment in West-Central Africa. Phytopathology 108, 299–307. doi: 10.1094/PHYTO-05-17-0190-R
Hébrard, E., Poulicard, N., Gérard, C., Traoré, O., Wu, H. C., Albar, L., et al. (2010). Direct interaction between the rice yellow mottle virus (RYMV) VPg and the central domain of the rice eIF(iso)4G1 factor correlates with rice susceptibility and RYMV virulence. Mol. Plant-Microbe Interact. 23, 1506–1513. doi: 10.1094/MPMI-03-10-0073
Hébrard, E., Poulicard, N., and Rakotomalala, M. (2021). “Rice yellow mottle virus (Solemoviridae),” in Encyclopedia of Virology. 4th Edn. eds. D. H. Bamford and M. Zuckerman (Oxford: Academic Press), 675–680.
Heinlein, M. (2015). Plant virus replication and movement. Virology 479–480, 657–671. doi: 10.1016/j.virol.2015.01.025
Heinrichs, E. A., Sy, A. A., Akator, S. K., and Oyediran, I. (1997). Seasonal occurrence of rice yellow mottle virus in lowland rice in cote d’ivoire. Int. J. Pest Manag. 43, 291–297. doi: 10.1080/096708797228591
Hubert, J. G., Pinel-Galzi, A., Dibwe, D., Cinyabuguma, E., Kaboré, A. D., Fargette, D., et al. (2013). First report of rice yellow mottle virus on rice in the Democratic Republic of Congo. Plant Dis. 97:1664. doi: 10.1094/PDIS-06-13-0650-PDN
Hull, R. (1977). The grouping of small spherical plant viruses with single RNA components. J. Gen. Virol. 36, 289–295. doi: 10.1099/0022-1317-36-2-289
Hyodo, K., Kaido, M., and Okuno, T. (2014). Host and viral RNA-binding proteins involved in membrane targeting, replication and intercellular movement of plant RNA virus genomes. Front. Plant Sci. 5:321. doi: 10.3389/fpls.2014.00321
Ioannidou, D., Lett, J. M., Pinel, A., Assigbétse, K., Brugidou, C., Ghesquière, A., et al. (2000). Responses of Oryza sativa japonica sub-species to infection with rice yellow mottle virus. Physiol. Mol. Plant Pathol. 57, 177–188. doi: 10.1006/pmpp.2000.0292
Ioannidou, D., Pinel, A., Brugidou, C., Albar, L., Ahmadi, N., Ghesquiere, A., et al. (2003). Characterisation of the effects of a major QTL of the partial resistance to Rice yellow mottle virus using a near-isogenic—line approach. Physiol. Mol. Plant Pathol. 63, 213–221. doi: 10.1016/j.pmpp.2003.12.005
Johnson, R. (1981). Durable resistance: definition of, genetic control, and attainment in plant breeding. Phytopathology 71, 567–568. doi: 10.1094/Phyto-71-567
Kalinina, N. O., Khromov, A., Love, A. J., and Taliansky, M. E. (2020). CRISPR applications in plant virology: virus resistance and beyond. Phytopathology 110, 18–28. doi: 10.1094/PHYTO-07-19-0267-IA
Kobayashi, K., Sekine, K.-T. T., and Nishiguchi, M. (2014). Breakdown of plant virus resistance: can we predict and extend the durability of virus resistance? J. Gen. Plant Pathol. 80, 327–336. doi: 10.1007/s10327-014-0527-1
Konate, G., Sarra, S., and Traore, O. (2001). Rice yellow mottle virus is seed-borne but not seed transmitted in rice seeds. Eur. J. Plant Pathol. 107, 361–364. doi: 10.1023/A:1011295709393
Konate, G., Traore, O., and Coulibaly, M. M. (1997). Characterization of rice yellow mottle virus isolates in Sudano-Sahelian areas. Arch. Virol. 142, 1117–1124. doi: 10.1007/s007050050146
Kouassi, N. K., Chen, L., Siré, C., Bangratz-Reyser, M., Beachy, R. N., Fauquet, C. M., et al. (2006). Expression of rice yellow mottle virus coat protein enhances virus infection in transgenic plants. Arch. Virol. 151, 2111–2122. doi: 10.1007/s00705-006-0802-3
Kouassi, N. K., N’Guessan, P., Albar, L., Fauquet, C. M., and Brugidou, C. (2005). Distribution and characterization of Rice yellow mottle virus: a threat to African farmers. Plant Dis. 89, 124–133. doi: 10.1094/PD-89-0124
Koudamilore, A., Nwilene, F. E., Silue, D., Togola, A., Oyetunji, O., Sere, Y., et al. (2014). Identification of insect vectors of Rice yellow mottle virus (RYMV) in Benin. J. Entomol. 11, 153–162. doi: 10.3923/je.2014.153.162
Kreuze, J. F., and Valkonen, J. P. (2017). Utilization of engineered resistance to viruses in crops of the developing world, with emphasis on sub-Saharan Africa. Curr. Opin. Virol. 26, 90–97. doi: 10.1016/j.coviro.2017.07.022
Lacombe, S., Bangratz, M., Vignols, F., and Brugidou, C. (2010). The rice yellow mottle virus P1 protein exhibits dual functions to suppress and activate gene silencing. Plant J. 61, 371–382. doi: 10.1111/j.1365-313X.2009.04062.x
Lamo, J., Cho, G., Jane, I., Dartey, P. K. A., James, E., Ekobu, M., et al. (2015). Developing lowland rice germplasm with resistance to multiple biotic stresses through anther culture in Uganda. J. Korean Soc. Int. Agric. 27, 415–420. doi: 10.12719/KSIA.2015.27.4.415
Ling, R., Pate, A. E., Carr, J. P., and Firth, A. E. (2013). An essential fifth coding ORF in the Sobemoviruses. Virology 446, 397–408. doi: 10.1016/j.virol.2013.05.033
Luo, S., Zhang, Y., Hu, Q., Chen, J., Li, K., Lu, C., et al. (2012). Dynamic nucleotide-binding site and leucine-rich repeat-encoding genes in the grass family. Plant Physiol. 159, 197–210. doi: 10.1104/pp.111.192062
Mandadi, K. K., and Scholthof, K. B. G. (2013). Plant immune responses against viruses: how does a virus cause disease? Plant Cell 25, 1489–1505. doi: 10.1105/tpc.113.111658
Marone, D., Russo, M. A., Laidò, G., De Leonardis, A. M., and Mastrangelo, A. M. (2013). Plant nucleotide binding site-leucine-rich repeat (NBS-LRR) genes: active guardians in host defense responses. Int. J. Mol. Sci. 14, 7302–7326. doi: 10.3390/ijms14047302
Maule, A. J., Caranta, C., and Boulton, M. I. (2007). Sources of natural resistance to plant viruses: status and prospects: review. Mol. Plant Pathol. 8, 223–231. doi: 10.1111/j.1364-3703.2007.00386.x
Maumus, F., Epert, A., Nogué, F., and Blanc, G. (2014). Plant genomes enclose footprints of past infections by giant virus relatives. Nat. Commun. 5:4268. doi: 10.1038/ncomms5268
Moffett, P. (2009). Mechanisms of recognition in dominant R gene mediated resistance. Adv. Virus Res. 75, 1–33. doi: 10.1016/s0065-3527(09)07501-0
Mondragón-Palomino, M., Meyers, B. C., Michelmore, R. W., and Gaut, B. S. (2002). Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res. 12, 1305–1315. doi: 10.1101/gr.159402
Mundt, C. C. (2014). Durable resistance: a key to sustainable management of pathogens and pests. Infect. Genet. Evol. 27, 446–455. doi: 10.1016/j.meegid.2014.01.011
N’guessan, P., Pinel, A., Caruana, M. L., Frutos, R., Sy, A., Ghesquì Ere, A., et al. (2000). Evidence of the presence of two serotypes of rice yellow mottle sobemovirus in Côte d’Ivoire. Eur. J. Plant Pathol. 106, 167–178. doi: 10.1023/A:1008792109954
Ndikumana, I., Pinel-Galzi, A., Fargette, D., and Hébrard, E. (2017). Complete genome sequence of a new strain of Rice yellow mottle virus from Malawi, characterized by a recombinant VPg protein. Genome Announc. 5, e01198–e01217. doi: 10.1128/genomeA.01198-17
Ndjiondjop, M. N., Albar, L., Fargette, D., Fauquet, C., and Ghesquière, A. (1999). The genetic basis of high resistance to rice yellow mottle virus (RYMV) in cultivars of two cultivated rice species. Plant Dis. 83, 931–935. doi: 10.1094/PDIS.1999.83.10.931
Ndjiondjop, M. N., Albar, L., Sow, M., Yao, N., Djedatin, G., Thiemélé, D., et al. (2013). “Integration of molecular markers in rice improvement: a case study on resistance to Rice yellow mottle virus,” in Realizing Africa’s Rice Promise. eds. M. Wopereis, D. Johnson, N. Ahmadi, E. Tollens, and A. Jalloh (Cotonou, Benin: CABI).
Ndjiondjop, M. N., Brugidou, C., Zang, S., Fargette, D., Ghesquiere, A., and Fauquet, C. (2001). High resistance to rice yellow mottle virus in two cultivated rice cultivars is correlated with failure of cell to cell movement. Physiol. Mol. Plant Pathol. 59, 309–316. doi: 10.1006/pmpp.2001.0368
Nelson, R., Wiesner-Hanks, T., Wisser, R., and Balint-Kurti, P. (2018). Navigating complexity to breed disease-resistant crops. Nat. Rev. Genet. 19, 21–33. doi: 10.1038/nrg.2017.82
Nummert, G., Sõmera, M., Uffert, G., Abner, E., and Truve, E. (2017). P1-independent replication and local movement of Rice yellow mottle virus in host and non-host plant species. Virology 502, 28–32. doi: 10.1016/j.virol.2016.12.007
Ochola, D., Issaka, S., Rakotomalala, M., Pinel-Galzi, A., Ndikumana, I., Hubert, J., et al. (2015). Emergence of rice yellow mottle virus in eastern Uganda: recent and singular interplay between strains in East Africa and in Madagascar. Virus Res. 195, 64–72. doi: 10.1016/j.virusres.2014.09.004
Ochola, D., and Tusiime, G. (2010). Survey on incidences and severity of Rice yellow mottle virus disease in eastern Uganda. Int. J. Plant Pathol. 2, 15–25. doi: 10.3923/ijpp.2011.15.25
Odongo, P. J., Kojima, N., Tsuboi, T., Asea, G., and Natsuaki, K. T. (2019). Evaluation of Ugandan rice cultivars for resistance against Rice yellow mottle virus. Trop. Agric. Dev. 63, 47–54. doi: 10.11248/jsta.63.47
Onwughalu, J. T., Abo, M. E., Okoro, J. K., Onasanya, A., and Sere, Y. (2011). Rice yellow mottle virus infection and reproductive losses in rice (Oryza sativa Linn.). Trends Appl. Sci. Res. 6, 182–189. doi: 10.3923/tasr.2011.182.189
Opalka, N., Brugidou, C., Bonneau, C., Nicole, M., Beachy, R. N., Yeager, M., et al. (1998). Movement of rice yellow mottle virus between xylem cells through pit membranes. Proc. Natl. Acad. Sci. U. S. A. 95, 3323–3328. doi: 10.1073/pnas.95.6.3323
Opalka, N., Tihova, M., Brugidou, C., Kumar, A., Beachy, R. N., Fauquet, C. M., et al. (2000). Structure of native and expanded Sobemoviruses by electron cryo-microscopy and image reconstruction. J. Mol. Biol. 303, 197–211. doi: 10.1006/jmbi.2000.4043
Orjuela, J., Thiémélé Deless, E. F., Kolade, O., Chéron, S., Ghesquiére, A., and Albar, L. (2013). A recessive resistance to rice yellow mottle virus is associated with a rice homolog of the CPR5 gene, a regulator of active defense mechanisms. Mol. Plant-Microbe Interact. 26, 1455–1463. doi: 10.1094/MPMI-05-13-0127-R
Parlevliet, J. E. (2002). Durability of resistance against fungal, bacterial and viral pathogens; present situation. Euphytica 124, 147–156. doi: 10.1023/A:1015601731446
Pidon, H., Chéron, S., Ghesquière, A., and Albar, L. (2020). Allele mining unlocks the identification of RYMV resistance genes and alleles in African cultivated rice. BMC Plant Biol. 20:222. doi: 10.1186/s12870-020-02433-0
Pidon, H., Ghesquière, A., Chéron, S., Issaka, S., Hébrard, E., Sabot, F., et al. (2017). Fine mapping of RYMV3: a new resistance gene to Rice yellow mottle virus from Oryza glaberrima. Theor. Appl. Genet. 130, 807–818. doi: 10.1007/s00122-017-2853-0
Pinel, A., N’Guessan, P., Bousalem, M., and Fargette, D. (2000). Molecular variability of geographically distinct isolates of Rice yellow mottle virus in Africa. Arch. Virol. 145, 1621–1638. doi: 10.1007/s007050070080
Pinel-Galzi, A., Dubreuil-Tranchant, C., Hébrard, E., Mariac, C., Ghesquière, A., and Albar, L. (2016). Mutations in rice yellow mottle virus polyprotein P2a involved in RYMV2 gene resistance breakdown. Front. Plant Sci. 7:1779. doi: 10.3389/fpls.2016.01779
Pinel-Galzi, A., Rakotomalala, M., Sangu, E., Sorho, F., Kanyeka, Z., Traoré, O., et al. (2007). Theme and variations in the evolutionary pathways to virulence of an RNA plant virus species. PLoS Pathog. 3:e180. doi: 10.1371/journal.ppat.0030180
Pinel-Galzi, A., Traoré, O., Séré, Y., Hébrard, E., and Fargette, D. (2015). The biogeography of viral emergence: Rice yellow mottle virus as a case study. Curr. Opin. Virol. 10, 7–13. doi: 10.1016/j.coviro.2014.12.002
Pinto, Y. M., Kok, R. A., and Baulcombe, D. C. (1999). Resistance to rice yellow mottle virus (RYMV) in cultivated African rice varieties containing RYMV transgenes. Nat. Biotechnol. 17, 702–707. doi: 10.1038/10917
Poulicard, N., Pinel-Galzi, A., Fargette, D., and Hébrard, E. (2014). Alternative mutational pathways, outside the VPg, of rice yellow mottle virus to overcome eIF(iso)4G-mediated rice resistance under strong genetic constraints. J. Gen. Virol. 95, 219–224. doi: 10.1099/vir.0.057810-0
Poulicard, N., Pinel-Galzi, A., Traoré, O., Vignols, F., Ghesquière, A., Konaté, G., et al. (2012). Historical contingencies modulate the adaptability of rice yellow mottle virus. PLoS Pathog. 8:e1002482. doi: 10.1371/journal.ppat.1002482
Pressoir, G., Albar, L., Ahmadi, N., Rimbault, I., Lorieux, M., Fargette, D., et al. (1998). Genetic basis and mapping of the resistance to rice yellow mottle virus. II. Evidence of a complementary epistasis between two QTLs. Theor. Appl. Genet. 97, 1155–1161. doi: 10.1007/s001220051004
Qu, C., Liljas, L., Opalka, N., Brugidou, C., Yeager, M., Beachy, R. N., et al. (2000). 3D domain swapping modulates the stability of members of an icosahedral virus group. Structure 8, 1095–1103. doi: 10.1016/S0969-2126(00)00508-6
Rakotomalala, M., Pinel-Galzi, A., Albar, L., Ghesquière, A., Rabenantoandro, Y., Ramavovololona, P., et al. (2008). Resistance to Rice yellow mottle virus in rice germplasm in Madagascar. Eur. J. Plant Pathol. 122, 277–286. doi: 10.1007/s10658-008-9282-5
Reagan, B. C., and Burch-Smith, T. M. (2020). Viruses reveal the secrets of plasmodesmal cell biology. Mol. Plant-Microbe Interact. 33, 26–39. doi: 10.1094/MPMI-07-19-0212-FI
Rimbaud, L., Papaïx, J., Rey, J. F., Barrett, L. G., and Thrall, P. H. (2018). Assessing the durability and efficiency of landscape-based strategies to deploy plant resistance to pathogens. PLoS Comput. Biol. 14:e1006067. doi: 10.1371/journal.pcbi.1006067
Sanfaçon, H. (2015). Plant translation factors and virus resistance. Viruses 7, 3392–3419. doi: 10.3390/v7072778
Sanford, J. C., and Johnston, S. A. (1985). The concept of parasite-derived resistance-deriving resistance genes from the parasite’s own genome. J. Theor. Biol. 113, 395–405. doi: 10.1016/S0022-5193(85)80234-4
Sarra, S., and Peters, D. (2003). Rice yellow mottle virus is transmitted by cows, donkeys, and grass rats in irrigated rice crops. Plant Dis. 87, 804–808. doi: 10.1094/PDIS.2003.87.7.804
Sasaya, T., Nakazono-Nagaoka, E., Saika, H., Aoki, H., Hiraguri, A., Netsu, O., et al. (2014). Transgenic strategies to confer resistance against viruses in rice plants. Front. Microbiol. 4:409. doi: 10.3389/fmicb.2013.00409
Savary, S., Willocquet, L., Pethybridge, S. J., Esker, P., McRoberts, N., and Nelson, A. (2019). The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439. doi: 10.1038/s41559-018-0793-y
Schoelz, J. E., Harries, P. A., and Nelson, R. S. (2011). Intracellular transport of plant viruses: finding the door out of the cell. Mol. Plant 4, 813–831. doi: 10.1093/mp/ssr070
Seck, P. A., Touré, A. A., Coulibaly, J. Y., Diagne, A., and Wopereis, M. C. S. (2013). “Africa’s rice economy before and after the 2008 rice crisis,” in Realizing Africa’s Rice Promise. eds. M. Wopereis, D. Johnson, N. Ahmadi, E. Tollens, and A. Jalloh (Cotonou, Benin: CABI), 24–34.
Séré, Y., Fargette, D., Abo, M. E., Wydra, K., Bimerew, M., Onasanya, A., et al. (2013). “Managing the major diseases of rice in Africa,” in Realizing Africa’s Rice Promise. eds. M. Wopereis, D. Johnson, N. Ahmadi, E. Tollens, and A. Jalloh (Cotonou, Benin: CABI), 213–228.
Sere, Y., Sorho, F., Onasanya, A., Jobe, L., Darboe, S., Bojang, Y., et al. (2008). First report of Rice yellow mottle virus in rice in The Gambia. Plant Dis. 92:316. doi: 10.1094/PDIS-92-2-0316B
Siré, C., Bangratz-Reyser, M., Fargette, D., and Brugidou, C. (2008). Genetic diversity and silencing suppression effects of Rice yellow mottle virus and the P1 protein. Virol. J. 5:55. doi: 10.1186/1743-422X-5-55
Soko, D. F., Ayolie, K., Koffi, N. B. C., Tonessia, D. C., Sere, Y., Hilairekouakou, T., et al. (2016). Impact of eight isolates of Rice yellow mottle virus (RYMV) from Gagnoa (Côte d’Ivoire) on rice (Oryza sp) cultivars production. Int. J. Biol. Chem. Sci. 9:2459. doi: 10.4314/ijbcs.v9i5.17
Sõmera, M., Sarmiento, C., and Truve, E. (2015). Overview on Sobemoviruses and a proposal for the creation of the family sobemoviridae. Viruses 7, 3076–3115. doi: 10.3390/v7062761
Souza, P. F. N. (2020). Masters of manipulation: how do positive-sense RNA viruses employ plant proteins to replicate, move from cell to cell, and overcome antiviral immunity? J. Plant Dis. Prot. 127, 425–440. doi: 10.1007/s41348-020-00342-w
Tamm, T., and Truve, E. (2000). Sobemoviruses. J. Virol. 74, 6231–6241. doi: 10.1128/JVI.74.14.6231-6241.2000
Tan, S., and Wu, S. (2012). Genome wide analysis of nucleotide-binding site disease resistance genes in Brachypodium distachyon. Comp. Funct. Genomics 2012:418208. doi: 10.1155/2012/418208
Taylor, D. R., Fofie, A. S., and Suma, M. (1990). Natural infection of rice yellow mottle virus disease (RYMV) on rice in Sierra Leone. Int. Rice Res. newsl. 15:19.
Thiémélé, D., Boisnard, A., Ndjiondjop, M. N., Chéron, S., Séré, Y., Aké, S., et al. (2010). Identification of a second major resistance gene to Rice yellow mottle virus, RYMV2, in the African cultivated rice species, O. glaberrima. Theor. Appl. Genet. 121, 169–179. doi: 10.1007/s00122-010-1300-2
Todorovska, E., Christov, N., Slavov, S., Christova, P., and Vassilev, D. (2009). Biotic stress resistance in wheat—breeding and genomic selection implications. Biotechnol. Biotechnol. Equip. 23, 1417–1426. doi: 10.2478/V10133-009-0006-6
Traoré, V. S. E., Néya, B. J., Camara, M., Gracen, V., Offei, S. K., and Traoré, O. (2015). Farmers’ perception and impact of Rice yellow mottle disease on rice yields in Burkina Faso. Agric. Sci. 6, 943–952. doi: 10.4236/as.2015.69091
Traoré, O., Pinel, A., Fargette, D., and Konaté, G. (2001). First report and characterization of Rice yellow mottle virus in Central Africa. Plant Dis. 85:920. doi: 10.1094/PDIS.2001.85.8.920A
Traoré, O., Pinel, A., Hébrard, E., Dieudonné Gumedzoé, M. Y., Fargette, D., Traoré, A. S., et al. (2006a). Occurrence of resistance-breaking isolates of rice yellow mottle virus in West and Central Africa. Plant Dis. 90, 259–263. doi: 10.1094/PD-90-0259
Traoré, O., Pinel-Galzi, A., Issaka, S., Poulicard, N., Aribi, J., Aké, S., et al. (2010). The adaptation of Rice yellow mottle virus to the eIF(iso)4G-mediated rice resistance. Virology 408, 103–108. doi: 10.1016/j.virol.2010.09.007
Traoré, O., Pinel-Galzi, A., Sorho, F., Sarra, S., Rakotomalala, M., Sangu, E., et al. (2009). A reassessment of the epidemiology of Rice yellow mottle virus following recent advances in field and molecular studies. Virus Res. 141, 258–267. doi: 10.1016/j.virusres.2009.01.011
Traoré, O., Traoré, M. D., Fargette, D., and Konaté, G. (2006b). Rice seedbeds as a source of primary infection by Rice yellow mottle virus. Eur. J. Plant Pathol. 115, 181–186. doi: 10.1007/s10658-006-9004-9
Trovao, N. S., Baele, G., Vrancken, B., Bielejec, F., Suchard, M. A., Fargette, D., et al. (2015). Host ecology determines the dispersal patterns of a plant virus. Virus Evol. 1:vev016. doi: 10.1093/ve/vev016
Uke, A., Asea, G., and Natsuaki, K. (2016). Phylogenic analysis and serotyping of Rice yellow mottle virus strains in Uganda. Trop. Agric. Dev. 75, 81–88. doi: 10.11248/jsta.60.81
Uke, A., Tibanyendela, N., Ikeda, R., Fujiie, A., and Natsuaki, K. T. (2014). Modes of transmission and stability of Rice yellow mottle virus. J. Plant Prot. Res. 54, 363–366. doi: 10.2478/jppr-2014-0054
Valin, H., Sands, R. D., van der Mensbrugghe, D., Nelson, G. C., Ahammad, H., Blanc, E., et al. (2014). The future of food demand: understanding differences in global economic models. Agric. Econ. 45, 51–67. doi: 10.1111/agec.12089
van Loon, L. C. (1987). Disease induction by plant viruses. Adv. Virus Res. 33, 205–255. doi: 10.1016/S0065-3527(08)60319-X
Van Schie, C. C. N., and Takken, F. L. W. (2014). Susceptibility genes 101: how to be a good host. Annu. Rev. Phytopathol. 52, 551–581. doi: 10.1146/annurev-phyto-102313-045854
Varshney, R. K., Graner, A., and Sorrells, M. E. (2005). Genomics-assisted breeding for crop improvement. Trends Plant Sci. 10, 621–630. doi: 10.1016/j.tplants.2005.10.004
Ventelon-Debout, M., Delalande, F., Brizard, J. P., Diemer, H., Van Dorsselaer, A., and Brugidou, C. (2004). Proteome analysis of cultivar-specific deregulations of Oryza sativa indica and O. sativa japonica cellular suspensions undergoing Rice yellow mottle virus infection. Proteomics 4, 216–225. doi: 10.1002/pmic.200300502
Ventelon-Debout, M., Nguyen, T. T. H., Wissocq, A., Berger, C., Laudie, M., Piégu, B., et al. (2003). Analysis of the transcriptional response to Rice yellow mottle virus infection in Oryza sativa indica and japonica cultivars. Mol. Gen. Genomics. 270, 253–262. doi: 10.1007/s00438-003-0903-6
Ventelon-Debout, M., Tranchant-Dubreuil, C., Nguyen, T. T. H., Bangratz, M., Siré, C., Delseny, M., et al. (2008). Rice yellow mottle virus stress responsive genes from susceptible and tolerant rice genotypes. BMC Plant Biol. 8:26. doi: 10.1186/1471-2229-8-26
Wang, A., and Krishnaswamy, S. (2012). Eukaryotic translation initiation factor 4E-mediated recessive resistance to plant viruses and its utility in crop improvement. Mol. Plant Pathol. 13, 795–803. doi: 10.1111/j.1364-3703.2012.00791.x
Wang, Q., Ma, X., Qian, S., Zhou, X., Sun, K., Chen, X., et al. (2015). Rescue of a plant negative-strand RNA virus from cloned cDNA: insights into enveloped plant virus movement and morphogenesis. PLoS Pathog. 11:e1005223. doi: 10.1371/journal.ppat.1005223
Whitham, S. A., Yang, C., and Goodin, M. M. (2006). Global impact: elucidating plant responses to viral infection. Mol. Plant-Microbe Interact. 19, 1207–1215. doi: 10.1094/MPMI-19-1207
Keywords: RYMV, rice-RYMV interaction, resistance mechanisms, durable resistance, rice improvement
Citation: Odongo PJ, Onaga G, Ricardo O, Natsuaki KT, Alicai T and Geuten K (2021) Insights Into Natural Genetic Resistance to Rice Yellow Mottle Virus and Implications on Breeding for Durable Resistance. Front. Plant Sci. 12:671355. doi: 10.3389/fpls.2021.671355
Edited by:
Paul Leslie Guy, University of Otago, New ZealandReviewed by:
Wen-Ming Wang, Sichuan Agricultural University, ChinaMihir Kumar Mandal, Claflin University, United States
Copyright © 2021 Odongo, Onaga, Ricardo, Natsuaki, Alicai and Geuten. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Koen Geuten, a29lbi5nZXV0ZW5Aa3VsZXV2ZW4uYmU=
 Patrick J. Odongo
Patrick J. Odongo Geoffrey Onaga
Geoffrey Onaga Oliver Ricardo
Oliver Ricardo Keiko T. Natsuaki5
Keiko T. Natsuaki5 Titus Alicai
Titus Alicai Koen Geuten
Koen Geuten