- 1Graduate School of Science, Kobe University, Kobe, Japan
- 2Faculty of Science and Engineering, Konan University, Kobe, Japan
- 3School of Food and Nutritional Sciences, University of Shizuoka, Shizuoka, Japan
The shape of plant nuclei varies among different species, tissues, and cell types. In Arabidopsis thaliana seedlings, nuclei in meristems and guard cells are nearly spherical, whereas those of epidermal cells in differentiated tissues are elongated spindle-shaped. The vegetative nuclei in pollen grains are irregularly shaped in angiosperms. In the past few decades, it has been revealed that several nuclear envelope (NE) proteins play the main role in the regulation of the nuclear shape in plants. Some plant NE proteins that regulate nuclear shape are also involved in nuclear or cellular functions, such as nuclear migration, maintenance of chromatin structure, gene expression, calcium and reactive oxygen species signaling, plant growth, reproduction, and plant immunity. The shape of the nucleus has been assessed both by labeling internal components (for instance chromatin) and by labeling membranes, including the NE or endoplasmic reticulum in interphase cells and viral-infected cells of plants. Changes in NE are correlated with the formation of invaginations of the NE, collectively called the nucleoplasmic reticulum. In this review, what is known and what is unknown about nuclear shape determination are presented, and the physiological significance of the control of the nuclear shape in plants is discussed.
Introduction
The cell nucleus (hereinafter referred to as the nucleus) of eukaryotes consists of chromatin, other nuclear contents (molecules forming/not forming nuclear bodies, such as nucleolus), and surrounding nuclear envelope (NE; Dreger et al., 2001). Usually, the nucleus is observed by labeling chromatin, a nuclear protein, or a NE protein by staining or expressing a fluorescent protein fusion. Although nuclei of most cells are either spherical/round or ellipsoid/oval in shape, alterations of the nuclear shape and size occur, and the alterations are associated with differentiation, aging, and diseases (Webster et al., 2009). Therefore, nuclear shapes can be indicators or diagnostic markers of differentiation stages and pathological states (Zink et al., 2004; Skinner and Johnson, 2017), although the link between the nuclear shape and the physiological function of the nucleus of an organism is unclear.
Plants have several cell types with different shape, such as polygon-shaped meristematic cells, cylindrical-shaped root cells, and jigsaw puzzle-like-shaped leaf epidermal pavement cells. In meristematic cells, nuclei are small and become larger during differentiation. The nuclei in differentiated cells are often appressed to the plasma membrane by a large vacuole. Regardless of the shape or the differentiation state of the cells, the shape of the nuclei is usually either spherical/round or ellipsoid/oval. However, the vegetative nucleus (VN) of pollen is irregularly shaped and often lobulated in various plant species (Dumas et al., 1985; Heslop-Harrison and Heslop-Harrison, 1989a,b,c; Borges et al., 2012). Additionally, the sperm cell nuclei (SCNs) of plants are often elongated (Borg and Twell, 2011). In the first model plant Arabidopsis thaliana, the shape of nuclei in epidermal cells, except for guard cells, changes from spherical to spindle shape during differentiation (Figure 1; Tamura and Hara-Nishimura, 2011; Meier et al., 2016).
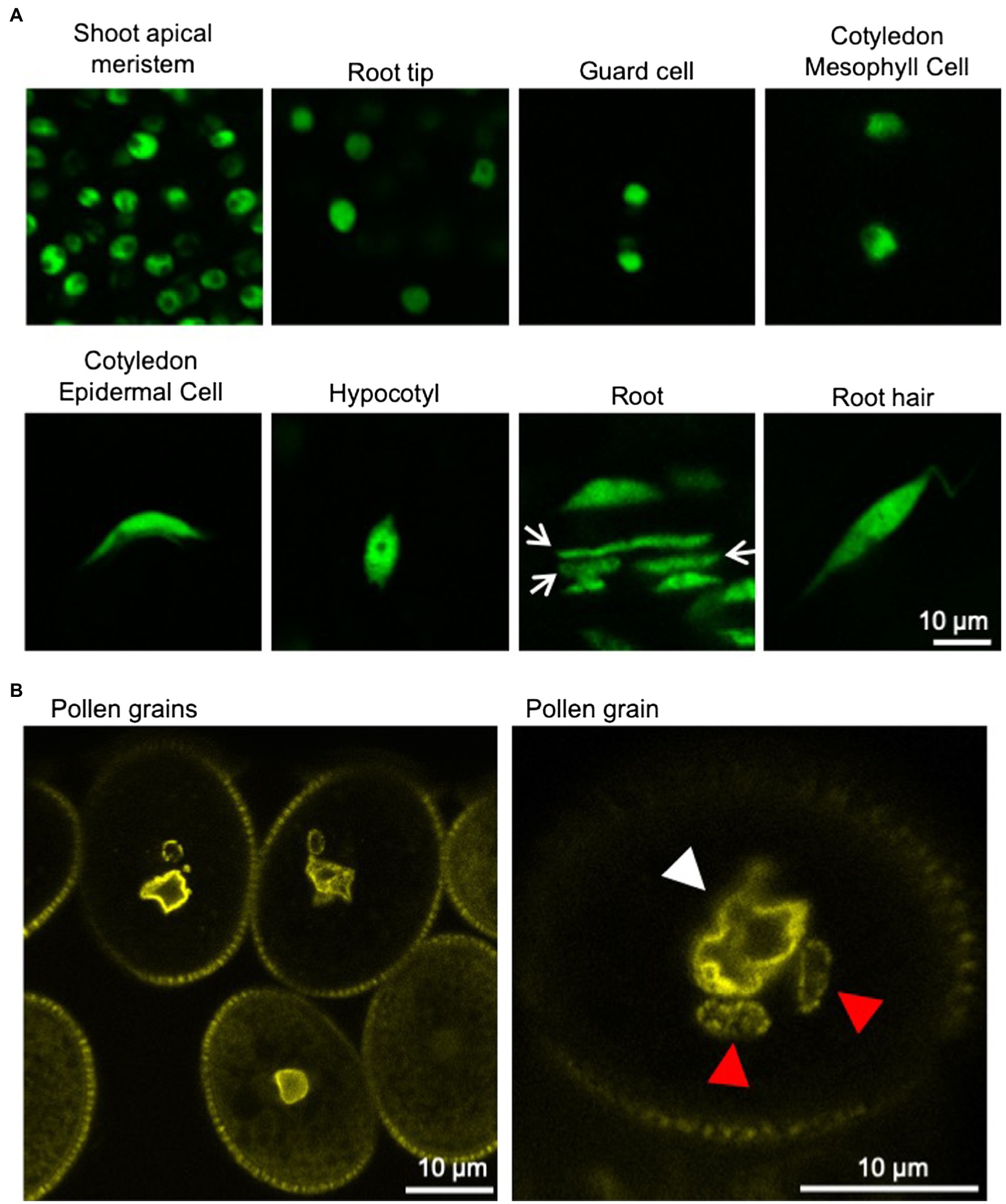
Figure 1. Nuclei in various tissues of Arabidopsis thaliana. (A) Confocal microscope images of nuclei in various tissues of 6-day-old A. thaliana seedlings stably expressing GFP-tagged histone protein (HTA8-GFP) under the control of 35S promoter. Spherical-shaped nuclei are observed in shoot apical meristems, root tips, guard cells, and cotyledon mesophyll cells (upper panels). Spindle-shaped nuclei are observed in cotyledon epidermal cells, hypocotyls, root cells, and root hairs (lower panels). Rod-shaped nuclei (arrow) are also observed in roots. (B) Confocal microscope images of nuclei in pollen grains of A. thaliana stably expressing YFP-tagged nucleoporin protein (RAE1-EYFP) under the control of 35S promoter (Goto et al., 2020). Fluorescently labeled NE clearly shows the irregular shapes of the VNs (white arrowhead). Sperm cell nuclei (red arrowhead) are often elongated.
Studies of nonplant organisms have identified the factors that influence the nuclear shape, such as NE proteins, cytoskeletons, vesicle trafficking, lipid biosynthesis, chromatin structure, and mitosis. Such NE proteins include lamins, which are intermediate filament proteins that form the scaffold called nuclear lamina (Dechat et al., 2008; Hampoelz and Lecuit, 2011), some lamin-binding proteins, such as lamin B receptor (Ellenberg et al., 1997; Schirmer and Foisner, 2007; Wilson and Foisner, 2010; Hampoelz and Lecuit, 2011), the inner nuclear membrane (INM) protein in Drosophila called Kugelkern (Brandt et al., 2006; Polychronidou et al., 2010), other INM proteins called Sad1/UNC-84 (SUN) domain proteins (Starr and Fridolfsson, 2010), outer nuclear membrane (ONM) proteins called Klarsicht/ANC-1/Syne/homology (KASH) domain proteins (Starr and Fridolfsson, 2010), and the nucleoporin Nup153 (Zhou and Panté, 2010). Whereas the actin cytoskeleton pulls nuclei, microtubules (MTs) exert compressive forces on the nucleus in metazoans (Mazumder and Shivashankar, 2010; Hampoelz and Lecuit, 2011). Vesicle trafficking is also reported to maintain the nuclear shape (Kimata et al., 1999; Webster et al., 2009). In yeast, the deletion of certain genes affecting lipid biosynthesis (e.g., SPO7, NEM1, or PAH1) or early protein secretion pathways (SEC31, SEC53, and SAR1) leads to both peripheral ER and NE expansion (Walters et al., 2012). The SWI/SNF chromatin-remodeling enzyme ATPase, BRG1, has been shown to regulate nuclear shape (Imbalzano et al., 2013). Chromatin histone modification and rigidity affect nuclear morphology (Stephens et al., 2018). In Aspergillus nidulans, a mitotic NE tether for Gle1 also affects nuclear and nucleolar architecture (Chemudupati et al., 2016).
Plants have no homologous lamin genes (Meier, 2007). Instead, NE proteins that regulate nuclear shape in plants have been identified one after another in the last 15 years (Meier et al., 2016). In addition, it has been suggested that NE proteins or the nuclear shape controlled by them affects nuclear function (migration of the nucleus, chromatin regulation, gene expression, and meiosis) and plant physiological functions (viability, reproduction, plant defense, and salicylic acid response; Evans et al., 2020; Groves et al., 2020). This review introduces what is known and what is unknown about the factors involved in controlling nuclear shape in plants, and discusses about the role of the factors or the nuclear shape in plant physiology.
Structure and Visualization of the Nucleus
Structure of the Nucleus
The nucleus has a structure in which the chromatin and the other components containing nuclear bodies, such as the nucleolus, are surrounded by the NE (Lanctôt et al., 2007). The NE consists of INM, ONM, nuclear lamina, and nuclear pore complexes (NPCs) in metazoans (Dultz and Ellenberg, 2007) and plants (Graumann et al., 2013; Ciska and de la Espina, 2014). The nuclear lamina is a fibrillar meshwork structure underlying the INM, of which the structure is observed in metazoan (Aebi et al., 1986) and plant (Fiserova et al., 2009) cells by electron microscopy. In yeasts, nuclear lamina-like structure is not observed (Cardenas et al., 1990), although two lamina proteins are immunologically identified (Galcheva-Gargova and Stateva, 1988). INM and ONM can be discriminated by transmission electron microscopy (TEM; Starr and Fridolfsson, 2010), and the space between the INM and the ONM is called the perinuclear space (Prunuske and Ullman, 2006). When the NE is observed with a light microscope, INM and ONM can be discriminated between by labeling them with fluorescent markers specific to each membrane (Xu et al., 2007b). INM and ONM are fused to form what is sometimes referred to as the pore membrane (Prunuske and Ullman, 2006), at which an NPC that consists of ~30 nucleoporins is embedded (D’angelo and Hetzer, 2008; Grossman et al., 2012; Tamura and Hara-Nishimura, 2013). ONM is contiguous with the ER (Starr and Fridolfsson, 2010), and a fluorescent protein that localizes in the ER lumen labels both ER and NE (Figure 2A). Although the NE is a type of ER sheet (Schwarz and Blower, 2016), the NE has specific proteins that interact with chromatin (Mattaj, 2004; Zuleger et al., 2011).
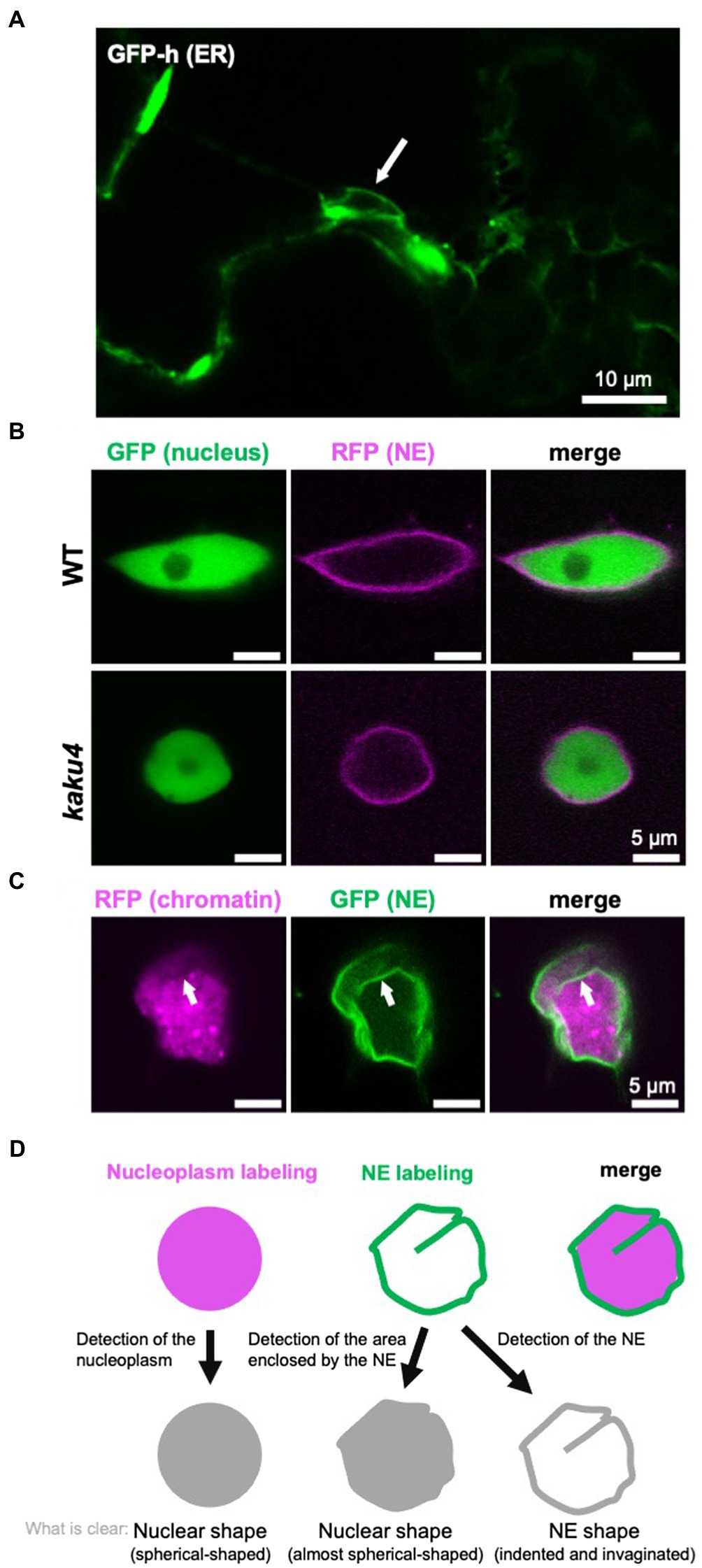
Figure 2. Detection and definition of the nuclear shape and the NE shape. (A) A confocal microscope image of A. thaliana cotyledon epidermal cells of 7-day-old GFP-h (Mitsuhashi et al., 2000; Hayashi et al., 2001; Nakano et al., 2009), in which ER is fluorescently labeled by GFP. The NE, contiguous with ER, is also visualized. The white arrow indicates a nucleus. (B) Confocal images of A. thaliana WT and kaku4 root cells stably coexpressing Nup50a-GFP (Tamura et al., 2010, 2013; Goto et al., 2014) and SUN2-TagRFP (Tamura et al., 2013), each under the control of the 35S promoter. The nucleus, except for the nucleolus, is fluorescently visualized by Nup50a-GFP, and the NE is fluorescently visualized by SUN2-TagRFP. (C) Confocal images of Nicotiana tabacum leaf cells transiently coexpressing H2B-TagRFP and KAKU4-GFP (Goto et al., 2014), each under the control of the 35S promoter. The chromatin is fluorescently visualized by H2B-GFP, and the NE is fluorescently visualized by KAKU4-GFP. The NE invagination (white arrow) is clearer when the NE is visualized. (D) Diagram depicting “nuclear shape” and “NE shape.” In this review, the shapes outlined by both nucleoplasm labeling and NE labeling are called the nuclear shape, whereas shapes outlined by only the labeled NE are called the NE shape.
The NE breaks down and reassembles during mitosis in metazoans and plants (Boettcher and Barral, 2013; Pradillo et al., 2019). This type of mitosis is called “open” mitosis. In contrast, the NE is intact during “closed” mitosis in budding yeast, in which the spherical metaphase nucleus transforms into an elongated dumbbell shape in anaphase (Boettcher and Barral, 2013). After mitosis, in some cell types, nuclear shape and size change as the cell differentiates (Dauer and Worman, 2009; Tamura and Hara-Nishimura, 2011; Meier et al., 2016).
Visualization of the Nucleus That Makes Nuclear Shape and NE Shape Recognizable
When cells are observed under a light microscope, nuclei are visible in the bright field without staining regardless of species. However, nuclei are often fluorescently labeled and observed to obtain a clearer image. For fluorescent labeling of nuclei, 4',6-diamidino-2-phenylindole (DAPI; e.g., Collings et al., 2000) and Hoechst 33342 (e.g., Goto et al., 2014) are typically used, which stain chromatin by intercalating into the minor grooves of DNA (Paul and Bhattacharya, 2012). Other DNA-binding reagents can also be used for the visualization of nuclei. In plants, it is effective to fix the cells to allow the staining solution to penetrate the cell wall. As cells are killed by the fixation, a fluorescent protein fusion of chromatin protein [e.g., H2B-green fluorescent protein (GFP)] or nuclear protein (e.g., NLS-GFP) is used to trace the nuclear shape and/or nuclear movement in living cells. Not only the nucleus but also the NE is often labeled by an NE protein fused with a fluorescent protein (e.g., SUN2-GFP). In many cases, the shape and size of the visualized nucleus are very similar between when the nucleus (chromatin or whole nucleus, excluding the NE) is fluorescently labeled and when the NE is fluorescently labeled. However, differences in the labeling targets result in differences in nuclear shape and size, as below: (1) Labeling chromatin cannot visualize the nucleoplasm, except for chromatin, whereas the labeled NE encloses all contents of the nucleus. (2) The nuclear periphery is not always smooth; the NE itself can deform and result in invaginations into the nucleoplasm or nuclear blebs extruded from the nucleus; invaginations of the NE are collectively called the nucleoplasmic reticulum (NR; see section “NE Shapes Under Normal State”). (3) The deformations of the NE are not always accompanied by consistent changes in the shape of the nucleus visualized by labeling the chromatin or nucleoplasm. For example, in the Arabidopsis mutant kaku1 with a mutation in myosin XI-i, nuclear shapes look different depending on the labeling targets (Tamura et al., 2013). The nuclear surface is smooth when the kaku1-1 nuclei are visualized by the expression of Nup50a-GFP that diffuses throughout the nucleus. In contrast, the kaku1-1 NE visualized by an NE marker SUN2-TagRFP is irregularly and intricately invaginated, not detected by labeling the nucleoplasm. Other examples of the difference in the appearance of the nucleus due to the difference in the visualization targets (nucleus/NE) are also shown in Figures 2B,C.
The nuclear shape determined by the nucleoplasm and the nuclear shape determined by the NE enclosing nucleoplasm are very similar but strictly not the same, as discussed above. The contents of the nucleus can show morphological changes not revealed by membrane imaging, and vice versa, under light microscope. When focusing on the NE, NE deformations may be classified into two types for convenience. One is a relatively large NE deformation with nucleoplasm deformation. In this type, both NE and nucleoplasm deformations can be detected under both light and electron microscopes. The other is local NE deformation without most of the nucleoplasm deformation. In this type, relatively small NE deformations can be detected under an electron microscope but hardly under a light microscope, and the shape of nucleoplasm changes little regardless of the microscope for detection. The above two types are sometimes not completely distinguishable. However, the classification of NE deformation as above could help understand the mechanism of change in the nuclear shape. For example, the very small and local changes in nuclear shape by the proliferation of the NE are classified into the latter type of NE deformation above, in which “change in NE shape” or “nuclear membrane growth” may be more proper than “change in nuclear shape” to explain the morphological phenotype of the nucleus.
As mentioned above, the nuclear shape can differ depending on labeling targets and microscopes used for observation. In this review, the shapes of chromatin or nucleoplasm and shapes outlined by the NE that are expected to be consistent with the shape of nucleoplasm are tentatively referred to as the “nuclear shape.” In contrast, the overall or local shapes that can be visualized with the labeled NE but hardly with the labeled nucleoplasm are tentatively referred to as the “NE shape” (Figure 2D).
Quantification of the Nuclear Shape
Nuclear shape and size are sometimes quantified and statistically analyzed. The circularity index and the nucleus area in a cross-sectional image are often calculated to quantify the morphological characterization of the nucleus. In case the nuclear shape is oval, the major and minor axes could be good indexes to quantify the nuclear shape. To quantify the nuclear shape and size more precisely, it is necessary to obtain and analyze a stack of three-dimensional images. NucleusJ, an ImageJ plugin, a useful tool to analyze nuclear morphology and chromatin organization, was released (Poulet et al., 2015) and updated (Dubos et al., 2020). The quantification of the nuclear shape is not only convincingly showing that the nucleus of some mutants is abnormal compared to the wild-type (WT) nucleus but also used to analyze the nuclear mechanical properties (Hampoelz and Lecuit, 2011).
Nuclear Shape in Plants
Various Types of Nuclear Shapes in Plants
In many plants, the nuclear shape is spherical/round or ellipsoid/oval. Nuclei in differentiated epidermal cells in leaves of Nicotiana tabacum and Allium cepa are spherical/round or ellipsoid/oval. In A. thaliana, the nuclear shape differs depending on cell or tissue types (nicely depicted in Meier et al., 2016). In 6-day-old Arabidopsis seedlings, nuclei are almost spherical in the root meristem, shoot apical meristem, and guard cells, whereas nuclei in the epidermal tissue of other parts in the root, hypocotyl, and cotyledon are spindle-shaped (Figure 1A; Tamura and Hara-Nishimura, 2011; Goto et al., 2014; Meier et al., 2016). Rod-like nuclei were located within vascular tissues (Chytilova et al., 1999). Recently, it was reported that nucleus compression occurs at the organ-meristem boundary (Fal et al., 2021).
The VN of pollen is irregularly shaped and often lobulated throughout various plant species (Dumas et al., 1985; Heslop-Harrison and Heslop-Harrison, 1989a,b,c; Borges et al., 2012). In mature pollen of Arabidopsis, the VN is highly invaginated (Figure 1B; Borges et al., 2012; Meier et al., 2016). The VN moves around in the pollen grain while changing its shape and then enters the pollen tube, in which the VN forms an elongated shape (Zhou and Meier, 2014). Additionally, the SCNs of angiosperm are often elongated (Borg and Twell, 2011). Interestingly, the nucleus becomes elongated with coiling during spermatogenesis in bryophytes (Renzaglia and Duckett, 1987). In most bryophytes, sperm nuclei form an elongated shape with slightly sinistral coiling, except for hornworts, of which nuclei are dextral (Renzaglia and Duckett, 1989). In vertebrates, nonround nuclei are observed in neutrophils, neurons, and spermatozoa (Dauer and Worman, 2009; Webster et al., 2009; Skinner and Johnson, 2017). There seems to be a tendency that nuclei form an elongated or irregular shape when the nucleus itself or the cell containing the nucleus migrates through narrow spaces.
Physiological Contexts of Nuclear Shape in Plants
In Arabidopsis seedlings, the amount of DNA increases due to endoreduplication but at the same time changes the nucleus from circular to spindle shape (Jovtchev et al., 2006; Sliwinska et al., 2012). However, endoreduplication or the increase of DNA content does not seem to be the decisive factor in inducing a change in the shape of the nucleus to a spindle shape as the nucleus remains spherical even after endoduplication in Barbarea stricta (Jovtchev et al., 2006). The nuclear shape differs depending on cell or tissue types as mentioned above, which suggests that nuclear morphogenesis, in which spherical nuclei change to the bigger spindle-shaped nuclei, seems to occur during cell differentiation (Tamura and Hara-Nishimura, 2011). Nuclei maintain the ability to change their shape even after they accomplish their morphogenesis as seen in roots (Chytilova et al., 2000; Tamura et al., 2013) and pollen grains (Zhou and Meier, 2014), in which nuclei flexibly change their shapes as they migrate. In contrast, nuclei maintain the spindle shape formed by the morphogenesis after their isolation and each treatment of reagent to break down the NE, actin filaments, and MTs (Sakamoto and Takagi, 2013). The formation and maintenance of the nuclear shape in plants seem to be regulated mainly by NE proteins (see also section “Factors Involved in the Regulation of the Nuclear Shape”). The links between the nuclear shape and NE proteins in plants are revealed by a molecular genetic approach using Arabidopsis.
Shape of the NE in Plants
NE Shapes Under Normal State
The surface of the nucleus is wrinkled rather than smooth. In nuclei of A. cepa stained with DAPI, nuclear grooves and invaginations are observed and also confirmed in nuclei labeled by nucleus-targeted GFP (N-GFP) or ER-targeted GFP (ER-GFP; Collings et al., 2000). Nuclear grooves and invaginations are also observed in cultured cells from N. tabacum expressing ER-GFP (Collings et al., 2000; Gupton et al., 2006). F-actin is present in nuclear grooves and surrounds the nucleus in cells of A. cepa (Collings et al., 2000). Nuclear grooves and invaginations appear to associate preferentially with nucleoli in epidermal cells of A. cepa (Collings et al., 2000). Invaginations of the NE, collectively called the NR, have been reported in various normal and abnormal cells from the plant and animal kingdoms (Malhas et al., 2011). The name nucleoplasmic reticulum (NR) was suggested for the convolutions of the NE, including deep, branching invaginations by morphological comparisons with the endoplasmic reticulum (ER). Common to plants and animals, under normal conditions, NE deformation or NR formation appears to be prominent, especially in reproductive cells and surrounding cells. For example, fluorescent labeling of the NE shows that pollen VNs are irregularly shaped, with invagination of the NE, which is an example of the prominent NE deformation (Figure 1B; Zhou and Meier, 2014; Zhou et al., 2015b). It is also observed that only INM invaginates into the nucleus in the meiocyte of Marsilea vestita (Sheffield et al., 1979) and placental cells of Lilium regale (Singh et al., 1998). Evaginations of only ONM and invaginations of the NE are also observed in microspores of Podocarpus macrophyllus (Aldrich and Vasil, 1970). One of the well-studied animal NRs is a “nucleolar channel system” that develops in postovulation human endometrial cells, and the transient presence of the nucleolar channel system has been associated with human fertility (Kittur et al., 2007). The characteristic NE shape in each tissue may imply the physiological role of the NE specific to that tissue.
Changes in the NE Shape by a Viral Infection
The deformation of the NE or formation of NR is observed under not only normal but also pathological conditions of plants and animals. It is observed under a TEM that many vesicles are formed in the perinuclear space of pea enation mosaic virus-infected plant cells (De Zoeten et al., 1972), which is reminiscent of the primary enveloped virion in the perinuclear cleft of herpesvirus-infected animal cells (Mettenleiter et al., 2006). TEM analysis also reveals that the infection of cowpea severe mosaic virus and bean yellow mosaic virus induces intranuclear inclusions (Carr and Kim, 1983). Not only TEMs but also fluorescence microscopes detect NE deformations induced by a viral infection. Infection of Nicotiana benthamiana with Potato yellow dwarf virus (PYDV) or Sonchus yellow net virus (SYNV) induces the accumulation of membranes within the nuclei (Goodin et al., 2005). The M protein of PYDV can induce the intranuclear accumulation of the INM in the absence of any other viral protein (Bandyopadhyay et al., 2010). A complex containing M protein of SYNV is discovered that appears to bud from the nucleus and that moves on ER membranes (Goodin et al., 2007). This budding of the M protein implies the budding of mature virions from the perinuclear space, of which the mechanism would be akin to the “bud-in, bud-out” envelopment and de-envelopment of herpesvirus particles at the NE (Goodin et al., 2007). It is proposed that herpesviruses have hijacked a nuclear export pathway employed by endogenous RNPs (Speese et al., 2012).
Factors Involved in the Regulation of the Nuclear Shape
Overview of Nuclear Shape Determination in Plants
Thus far, molecular genetic analyses have revealed that several NE proteins and cytoskeleton-associated proteins are involved in controlling the nuclear shape in plants (Figure 3). Single or multiple mutants deficient in nuclear lamina constituent protein candidates (CRWNs and KAKU4), plant LINC complex components (SUNs, WIPs, and WITs), myosin XI-i, or nucleoporins (Nup136/Nup1 and Nup160) have more spherical nuclei compared with WT. Among them, KAKU4, CRWN4, and Nup136/Nup1 promote NE deformation when overexpressed. Unlike the above, gip1 gip2 mutant has an abnormally shaped nucleus with jagged edges at the root tip. These findings were summarized by gene in previous reviews (Tamura et al., 2015; Meier et al., 2016). This section briefly reviews the factors involved in controlling nuclear shape after categorization based on their effect on the nuclear shape. It includes recent updates and introduces factors that have been found to control nuclear shapes in animals but with unclear effects on plant nuclear shapes (section “Other Possible Factors That Can Be Involved in Nuclear Shape Determination”). The last part of this section discusses how nuclear shapes in plants are controlled (section “Regulation of the Nuclear Shape by GIP, SUN-WIP-WIT-Myosin XI-i, and CRWN-KAKU4”).
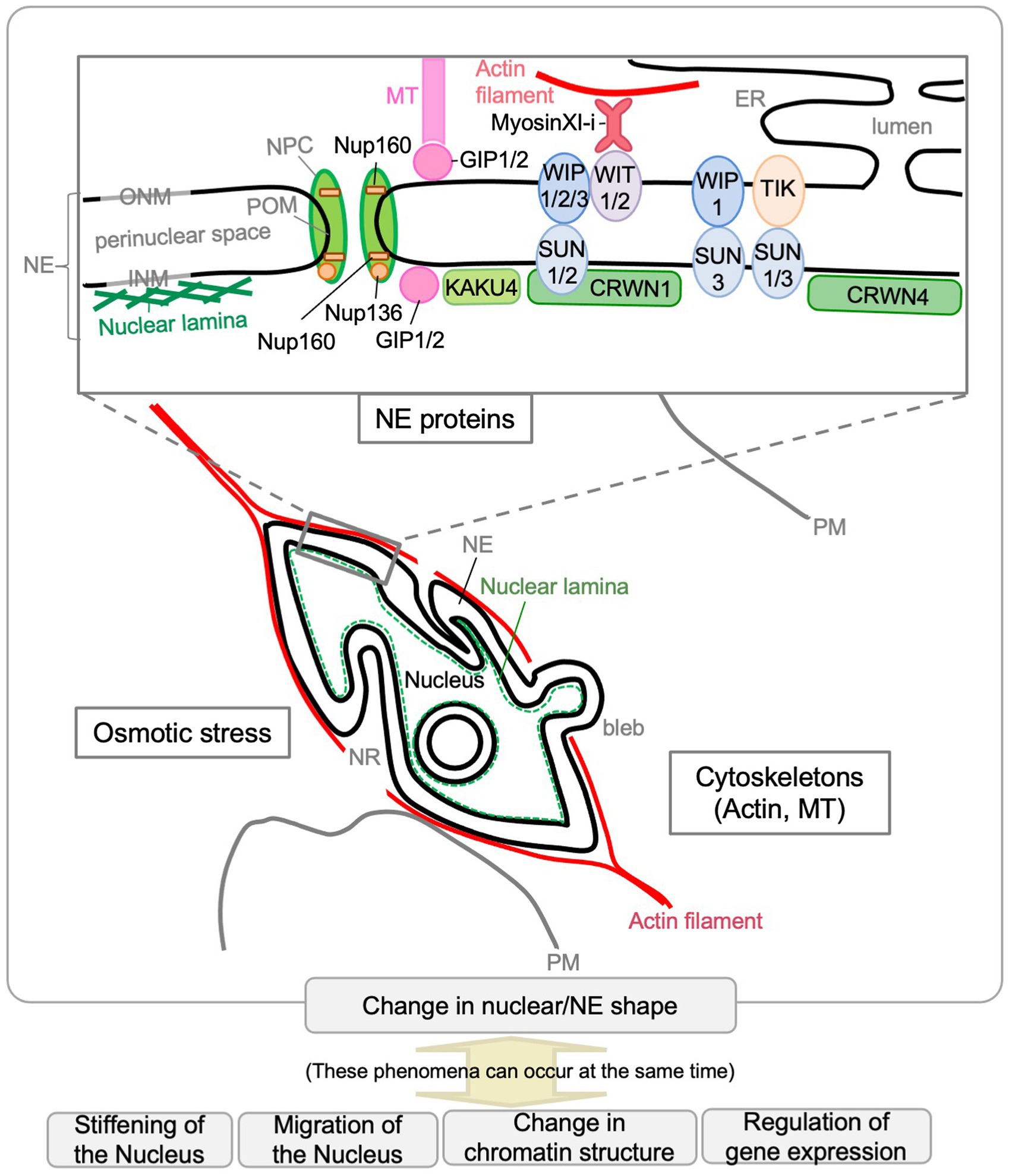
Figure 3. Factors that affect the nuclear shape in plants. Scaffolding or other functions by NE proteins and cytoskeletons (actin and MTs) are involved in controlling the nuclear shape in plants. Osmotic stress affects nuclear shape. The Arabidopsis NE proteins that affect nuclear shape are depicted in the enlarged diagram of the NE structure. The phenomena shown at the bottom of the diagram can occur during nuclear shape changes.
NE Proteins of Which the Deficiency Results in Small Spherical Nuclei in Arabidopsis
Many, but not all, NE proteins are involved in the formation and/or maintenance of nuclear shape (Figure 3). While nuclei become bigger and spindle-shaped during differentiation in WT A. thaliana, nuclei remained spherical in the following single or multiple mutants: crwn1 crwn2, crwn1, crwn4, kaku4, sun1 sun2, sun3, wip1 wip2 wip3, wit1 wit2, tik, myosin XI-i, nup136/nup1, and nup160.
Candidate Components of the Nuclear Lamina-Like Structure: NMCPs/CRWNs and KAKU4
The CROWDED NUCLEI1 (CRWN1) gene was originally named as LITTLE NUCLEI1 (LINC1; Dittmer et al., 2007), for the Arabidopsis homolog of the gene encoding the insoluble protein nuclear matrix constituent protein 1 (NMCP1), isolated from carrots (Masuda et al., 1997). LINC1 was renamed CRWN1 in 2013 (Wang et al., 2013). A. cepa NMCP1 (Ciska et al., 2013) and Arabidopsis CRWN1 (Sakamoto et al., 2020) have been suggested to be localized within the nuclear lamina. The small spherical nuclei in crwn1 crwn2 (Dittmer et al., 2007) are the first reported nuclei with a shape different to that of the WT. The CRWN family is plant specific, and Arabidopsis has three NMCP1 homologs (CRWN1-3) and an NMCP2 homolog (CRWN4; Ciska et al., 2013). Whereas crwn1 and crwn4 clearly show spherical nuclei, nuclei in crwn2 and crwn3 are less different to those in WT (Sakamoto and Takagi, 2013).
KAKU4 was novelly identified from mutant screening with a focus on nuclear shape (Goto et al., 2014). Mutant screening also resulted in the isolation of kaku1 (myosin XI-i; Tamura et al., 2013) and kaku2 (another allele of crwn1; Goto et al., 2014). KAKU4 is localized near the INM, which suggests its localization to the nuclear lamina (Goto et al., 2014). KAKU4 interacts with CRWN1 and CRWN4 (Goto et al., 2014) and is conserved in seed-bearing plants (Goto et al., 2014; Poulet et al., 2017b). Two KAKU4-like genes have been identified in some plant species, including Glycine max and Zea mays (Poulet et al., 2017b; Gumber et al., 2019a), whereas A. thaliana has only one KAKU4 gene (Goto et al., 2014; Poulet et al., 2017b). Almost all nuclei in kaku4 seedlings were spherical, similar to those in crwn1 (Goto et al., 2014). KAKU4 is also required to form irregularly shaped VNs in pollen grains (Goto et al., 2020). Based on their localization and effects on the nuclear shape, CRWN1 and KAKU4 are expected to be functional analogs of the intermediate filament proteins lamin and its binding proteins of metazoans that make up the nuclear lamina (Guo and Fang, 2014; Tamura et al., 2015; Meier et al., 2017; Groves et al., 2020).
Components of Plant LINC Complexes and the Myosin: SUNs, WIPs, WITs, Myosin XI-i, TIK, and MLKS2
Analysis of the localization, FRET, and FRAP of the protein fusions suggests that CRWN1 interacts with SUN1 and SUN2 (Graumann, 2014), reminiscent of animal lamins interacting with the INM protein SUNs (Starr and Fridolfsson, 2010). A. thaliana SUN1 and SUN2 were identified via a protein BLAST search with the Caenorhabditis elegans SUN domain protein UNC-84 sequence as the query (Graumann et al., 2010). Subsequently, a further SUN domain search identified SUN3, SUN4, and SUN5 in Arabidopsis (Graumann et al., 2014). These SUNs are classified into two subfamilies based on the position of the SUN domain within the protein: SUN1 and SUN2 are referred to as Cter-SUN, whereas SUN3, SUN4, and SUN5 are referred to as mid-SUN (Graumann et al., 2014). The nuclei of root hairs were rounder in sun1sun2 than in WT (Oda and Fukuda, 2011; Zhou et al., 2012). Statistical analysis of circularity suggests that sun3 also has a rounder nucleus compared to the WT (Graumann et al., 2014). The nuclear shape in sun4 and sun5 was similar to that in the WT (Graumann et al., 2014).
In metazoans and fungi, SUN domain proteins interact with KASH domain proteins in the perinuclear space to form LINC complexes (Starr and Fridolfsson, 2010). The LINC complex interacts with the nuclear lamina on the nucleoplasmic side and the cytoskeleton on the cytoplasmic side, and is involved in controlling the nuclear shape, migration of the nucleus, and mechanotransduction from the cell membrane to the nucleus (Dahl et al., 2008; Starr and Fridolfsson, 2010). In plants, some components of the LINC complexes are involved in controlling nuclear morphology: Arabidopsis SUN-WIP-WIT complex (Zhou et al., 2012; Tamura et al., 2013), Arabidopsis SUN3-TIK complex (Graumann et al., 2014), and Z. mays SUN2-MLKS2 complex (Gumber et al., 2019b). In Arabidopsis, SUN proteins (SUNs) interact with WIP proteins (WIPs), WIPs interact with WIT proteins (WITs), and WITs interact with myosin XI-i (Zhou et al., 2012; Tamura et al., 2013). The SUN-WIP-WIT complex is expected to have functions equivalent to the LINC complex in metazoans and fungi (Tamura et al., 2013). WIPs were biochemically identified as WPP-interacting factors (Xu et al., 2007a), and WITs were biochemically identified as WIP-interacting factors (Zhao et al., 2008). The C-terminal tail of WIP1, which is expected to be located in the perinuclear space and to interact with SUNs, has a low degree of similarity to the Opisthokont KASH domains (Zhou et al., 2012). The triple mutant wip1wip2wip3 (Zhou et al., 2012) and the double mutant wit1wit2 (Tamura et al., 2013) have spherical nuclei in the cell types that have spindle-shaped nuclei in WT plants. Myosin XI-i, whose anchoring to the nuclear membrane requires WITs, was identified as the gene responsible for the kaku1 mutation (Tamura et al., 2013). Nuclei in kaku1 are spherical in cells in which WT nuclei are spindle-shaped when visualized with nuclear-localized GFP (Nup50a-GFP; Tamura et al., 2013). Interestingly, it was observed that the NEs of kaku1 are irregularly and intricately invaginated under the stable expression of SUN2-TagRFP or TEM (Tamura et al., 2013). Another putative Arabidopsis KASH domain protein, TIK (TIR-KASH protein), plays a role in nuclear morphology, specifically nuclear size (Graumann et al., 2014). SINEs (SUN-interacting NE proteins) have been identified as other components of plant LINC complexes (Zhou et al., 2014). Arabidopsis SINE1 is required for proper nuclear positioning in guard cells (Zhou et al., 2014). The Z. mays SINE1 homolog MLKS2 interacts with ZmSUN2 and is able to rescue the nuclear phenotype in the Arabidopsis wip1 wip2 wip3 mutant (Gumber et al., 2019b). These results suggest that the Zea SINE-family KASH protein can be a substitute for Arabidopsis WIP-family KASH proteins for nuclear shape control (Gumber et al., 2019b).
Nucleoporins: nup136/nup1 and nup160
The NPC comprises ~30 nucleoporins and is well characterized in vertebrates and yeast (D’angelo and Hetzer, 2008; Hoelz et al., 2011; Grossman et al., 2012). Interactive proteomics has identified 30 nucleoporins in Arabidopsis (Tamura et al., 2010). Nuclei in nup136/nup1 (Tamura et al., 2010) and nup160 (Parry, 2014) are more spherical than those in WT plants. Arabidopsis Nup136 is thought to be a functional homolog of vertebrate Nup153, which is physiologically associated with the nuclear lamina (Tamura et al., 2010; Tamura and Hara-Nishimura, 2011). As in the case of vertebrate Nup153, Arabidopsis Nup136 is a possible key determinant of nuclear structure through the interaction between NPCs and lamin-like structures (Tamura et al., 2010; Tamura and Hara-Nishimura, 2011).
NE-Localized Proteins That Are Involved in the Deformation of the Nucleus/NE in Plants
NE Proteins of Which Overexpression Causes Deformation of the Nucleus/NE: KAKU4, CRWNs, and Nup136/Nup1
Among NE proteins, KAKU4 (Goto et al., 2014), CRWN4 (Sakamoto and Takagi, 2013), and Nup136 (Tamura and Hara-Nishimura, 2011) cause deformations of the nucleus/NE. KAKU4-GFP overexpression causes nuclear membrane growth and NE deformation in whole seedlings of Arabidopsis and leaves of N. tabacum and N. benthamiana (Goto et al., 2014). Indeed, care is required to interpret the phenotypes caused by the overexpression of GFP-fused proteins, as they can impose artificial effects (Groves et al., 2019). However, KAKU4 fused to another tag, FLAG, also causes NE deformation (Goto et al., 2014). The shape of VN in WT mature pollen grains, in which KAKU4 is highly expressed, is irregular and reminiscent of the deformed shape of the nucleus in KAKU4-overexpressing cells, supporting the hypothesis that KAKU4 affects NE shape (Goto et al., 2020). Co-overexpression of FLAG-CRWN1 with GFP-KAKU4 has a greater impact on NE shape in N. tabacum leaves (Goto et al., 2020). CRWN4-GFP overexpression causes nuclear elongation (Sakamoto and Takagi, 2013). Transgenic plants overexpressing Nup136-GFP have elongated nuclei in guard cells, leaf epidermal cells, and trichomes (Tamura and Hara-Nishimura, 2011).
NE-Localized Proteins of Which Deficiency Causes Deformed Nuclei: GIPs and Nup88
Some cytoskeleton-associated proteins also affect the shape of the nucleus/NE. One is the MT-associated protein GIP (Batzenschlager et al., 2014), and the other is a putative actin filament-associated protein, myosin XI-i (Tamura et al., 2013). Perinuclear MTs are nucleated from γ-tubulin complexes (γ-TuCs) located on the surface of the nucleus. The γ-TuC protein 3 (GCP3)-interacting protein 1 (GIP1), a γ-TuC component, was first discovered by a yeast two-hybrid screen (Janski et al., 2008). In Arabidopsis, there is a GIP1 homologous gene, GIP2, which is collectively called GIPs (Janski et al., 2012). At the electron microscopy resolution, GIPs are localized on both sides of the NE (Batzenschlager et al., 2013). Nuclei of gip1gip2 mutant cells exhibit an increased size and are highly deformed in root tips (Batzenschlager et al., 2013). The gip1gip2 nuclei detected by DAPI staining are irregularly shaped and highly deformed (Batzenschlager et al., 2013), whereas the kaku1/myosin XI-i nuclei detected by Hoechst staining are spherical and hardly deformed, although the NE of kaku1/myosin XI-i is extensively invaginated (Tamura et al., 2013). It has also been shown that the gip1gip2 mutant exhibits constitutive hyperosmotic stress response with stiffer and deformed nuclei (Goswami et al., 2020). In N. benthamiana plants, RNA silencing of Nup88 causes shrunken nuclei in leaf cells (Ohtsu et al., 2014).
Other Possible Factors That Can Be Involved in Nuclear Shape Determination
Although chromatin involvement seems to have relatively little effect on nuclear shape through animals and plants, it is reported that nuclei of BY-2 cells are elongated on long-term treatment with aphidicolin, an inhibitor of DNA polymerase (Yasuhara and Kitamoto, 2014). Recently, it is reported that hyperosmotic stress by 0.3 M mannitol treatment affects the nuclear shape in Arabidopsis root tips (Goswami et al., 2020). In yeast and metazoans, lipid biosynthesis and vesicle trafficking affect the shape of the nucleus/NE (Webster et al., 2009; Walters et al., 2012). Although Arabidopsis has genes PAH1 and PAH2 that are homologous to yeast PAH1, which is involved in lipid biosynthesis and affects the nuclear shape, unlike the yeast pah1-delta mutant, the volume of the nucleus was not greatly enlarged in cells of pah1pah2-1 leaves, and its shape was not distorted (Eastmond et al., 2010). In Scenedesmus acutus, significant NE deformation and ONM budding were observed after treatment of brefeldin A, which is an inhibitor of vesicle transport (Noguchi et al., 1998). This implies that direct and/or indirect involvement of membrane traffic factors in the NE shape.
Regulation of the Nuclear Shape by GIP, SUN-WIP-WIT-Myosin XI-i, and CRWN-KAKU4
In Arabidopsis, the nucleus changes from spherical shape to spindle shape at the same time as it grows in volume. Many reports mentioned above suggested that proteins localized in the NE, including cytoskeleton-related factors, contribute to the morphogenesis and maintenance of the nucleus and the NE. On the NE of Arabidopsis, the SUN-WIP-WIT complex connects the CRWN1-KAKU4 complex expected to form nuclear lamina with the cytoplasmic myosin XI-i, which is expected to be associated with the actin cytoskeleton (Figure 3). The intricate NE in the kaku1/myosin XI-i mutant suggests that the myosin pulls the NE under normal conditions (Tamura et al., 2013), which is consistent with the indication that the actin cytoskeleton pulls nuclei in metazoans (Mazumder and Shivashankar, 2010; Hampoelz and Lecuit, 2011). In contrast to the actin cytoskeleton, it is indicated that MTs exert compressive forces on the nucleus in metazoans (Mazumder and Shivashankar, 2010; Hampoelz and Lecuit, 2011), which may be related to that only gip1gip2 shows a different phenotype on the nuclear shape compared to other mutants, including kaku1/myosin XI-i (Batzenschlager et al., 2013). As nuclei under hyperosmotic stress or those in gip1gip2 shrink and become stiffer, nuclear shape and stiffness of the nucleus could be related (Goswami et al., 2020). The phenotype on the nuclear shape of the mutant lines defective in nuclear morphogenesis, except for gip1gip2, is similar in that nuclei are spherical in the cells in which nuclei are spindle-shaped in WT plants. However, the shape of the NE is different among mutant lines. The NE morphology of crwn1-1 is smooth, different from the invaginated nuclei of sun1-KO sun2-KD and wit2-1 (Zhou et al., 2015a). In addition, loss of CRWN1 does not affect the localization of the SUN-WIP-WIT2 complex, suggesting that CRWN1 acts independently of the SUN-WIP-WIT complex (Zhou et al., 2015a). CRWN1 has a synergistic effect on NE deformation when coexpressed with KAKU4 (Goto et al., 2014). Thus, CRWN1 may have the function of nuclear membrane growth and the NE deformation as KAKU4 does. The NE shape in KAKU4-overexpressing cells is reminiscent of the NE shape in Drosophila kugelkern-overexpressing cells (Brandt et al., 2006; Polychronidou et al., 2010). Drosophila kugelkern is the INM protein required for nuclear elongation during cellularization in Drosophila embryo and induces nuclear membrane growth (Brandt et al., 2006). About Drosophila kugelkern, a model has been established in which the farnesylated C-terminus affects the nuclear shape by directly interacting with the lipid bilayer (Polychronidou and Großhans, 2011). In contrast, KAKU4 and CRWN1 have no predicted site for lipid modification, and the mechanism by which they change the shape of nuclear membranes is unclear. In the metazoans and yeasts, nucleoskeleton-cytoskeleton interactions, chromatin conformational changes, and altered lipid biosynthesis are involved in shaping the nuclear membrane (Polychronidou and Großhans, 2011). In Arabidopsis, at least nucleoskeleton (CRWNs and KAKU4) and cytoskeleton (myosin XI-i and GIP) near the NE seem to play a major role in regulating the nuclear shape (Tamura et al., 2015; Meier et al., 2016, 2017; Evans et al., 2020).
Physiological Significance of Controlling the Nuclear Shape
Overview of the Effects of the Nuclear Shape on Plant Physiology
There are two phenomena in which nuclear shape itself may be deeply involved: control of the order of migration of VN and SCs in pollen tubes (section “Migration of the Nucleus in Pollen and Roots”) and gene expression regulation (section “Regulation of Gene Expression”). In Arabidopsis, the absence of nuclear morphogenesis does not appear to be a fatal defect because plant growth is normal in mutants with aberrant nuclear shapes, such as crwn1 (Wang et al., 2013) and kaku4 (Goto et al., 2014). On the other hand, some nuclear shape mutants have defects in migration of the nucleus, maintenance of chromatin structure, meiosis, immunity, salicylic acid response, Ca2+ and reactive oxygen species (ROS) signaling, pollen development, and survival (section “Other Plant Phenotypes Affected by NE Proteins Involved in the Control of the Nuclear Shape”). Little is known about the direct effects of the NE deformation, including the formation of NR, on plant physiology, but studies in other organisms may be useful for the future development of plant research (section “Physiological Significance of NE Deformation”).
Migration of the Nucleus in Pollen and Roots
When moving through the pollen tube, the VN precedes SCs. Time-lapse imaging of in vitro elongated pollen tubes has shown that the VN precedes sperm cells when they enter the pollen tube from pollen grains, and the migration order is mostly maintained during their migration in pollen tubes (Zhou and Meier, 2014), which implies that the positions of the VN and sperm cells when they enter the pollen tube are important in determining the migration order. The VN within pollen grains is irregularly shaped throughout various plant species, and the VN envelope is invaginated. Based on cases where the VN precedes and cases where SCs precede appear randomly in pollen tubes of kaku4 of which mature pollen grains have the VN with less invaginated NE (Goto et al., 2020), the irregular shape of VNs is expected to help the VN slides into the pollen tube before the membrane-enclosed mass of sperm cells enter the pollen tube. Mutations in the genes encoding of NE proteins WIPs, WITs, or SUNs reverse the order of the migration of VN and SCs (Zhou and Meier, 2014; Zhou et al., 2015b). Recently, it became apparent that wit1 wit2 VNs are shorter and more circular than WT VNs (Moser et al., 2020), which supports that the VN shape contributes to the control of the migration order of the VN and SCs. The possibility that nuclear mobility is more important than nuclear shape to determine the migration order of VN and SCs cannot be excluded, although kaku1-4, a myosin XI-i null mutant, exhibits a normal migration order (Zhou and Meier, 2014). The correct migration order of VN and SCs and/or keeping VNs at a certain distance from pollen tubes seems to be necessary for maintaining fertilization efficiency (Goto et al., 2014), Ca2+ and ROS signaling (Moser et al., 2020), pollen tube reception and seed production (Zhou and Meier, 2014; Zhou et al., 2015b).
In roots, the nucleus moves actively while changing its shape, suggesting that nuclear shape and nuclear movement can be linked (Chytilova et al., 2000). The kaku1/myosin XI-i with spherical nuclei moves much more slowly than WT spindle-shaped nuclei (Tamura et al., 2013). The flexibility of the nucleus to change its shape may affect the migration of the nucleus and the cell that harbors the nucleus. To be consistent with this idea, the neutrophils that migrate through tissues in mammalian cells have multilobed nuclei, typically exhibiting three or four lobes connected by thin DNA-containing filaments (Webster et al., 2009). Cells with less lobulated nuclei derived from the Pelger-Huet anomaly show a lower migration rate than normal control (Park et al., 1977).
Regulation of Gene Expression
When mammalian cells are placed in small holes of various sizes and artificially changed their shapes, the nuclear shape also changes (Thomas et al., 2002). In this experimental system, collagen I synthesis correlates directly with the cell shape and nuclear shape index (Thomas et al., 2002). This report raises the possibility that the nuclear shape itself affects gene expression. Reduction of the chromocenter number and induction of expression of the gene encoding a linker histone (H1.3) are found in the organ-meristem boundary of Arabidopsis, in which the nuclear shape changes (Fal et al., 2021). In addition, NE proteins GIPs and CRWNs affect both the nuclear shape (section “NE-Localized Proteins That Are Involved in the Deformation of the Nucleus/NE in Plants”) and gene expression (as below). Transcriptomic analysis of gip1gip2 compared to WT plants shows that 2092 genes containing many touch response genes are upregulated in gip1gip2 (Goswami et al., 2020). Altered transcriptional profiles are detected in three single mutants crwn1, crwn2, and crwn4 (Choi et al., 2019) and two double mutants crwn1crwn2 (van Zanten et al., 2011; Choi et al., 2019) and crwn1crwn4 (Choi et al., 2019; Sakamoto et al., 2020). More than 2000 differentially expressed genes were identified in the crwn1crwn4 double mutant (Sakamoto et al., 2020). It is indicated that CRWNs could tether the locus of copper-associated genes to the nuclear periphery (Sakamoto et al., 2020), which suggests that gene expressions are regulated by tethering chromatin to the NE rather than a change in the nuclear shape. As GIP1 localizes not only the cytoplasmic side but also the nucleoplasmic side of the NE and CRWN1 localizes at the nucleoplasmic side of the NE, it is possible that they affect gene expression through binding of the proteins to the chromatin independently of the control of the nuclear shape.
In metazoans, it is well known that the tethering of chromatin to the NE mediates gene regulation (Czapiewski et al., 2016). Also in plants, it is suggested that association with the nuclear periphery is involved in the regulation of gene expression. Smith et al. showed that tethering of a reporter gene with nucleoporins affects the level of its expression in Arabidopsis (Smith et al., 2015). This suggests that plant NE proteins influence transcriptional activity by tethering genes to the nuclear periphery. It is shown that 10–20% of the regions on the chromosome arms are anchored at the nuclear periphery in plants (Bi et al., 2017). In Arabidopsis, CRWN1 is a key component of the lamina-chromatin network (Hu et al., 2019). CRWN1 suppresses the expression of the PATHOGENESIS-RELATED1 (PR1) gene by enhancing the binding of NAC WITH TRANSMEMBRANE MOTIF1-LIKE9 (NTL9), an NAC transcription factor involved in plant immunity, to the PR1 promoter (Guo et al., 2017). The crwn1crwn2 double mutant shows enhanced resistance against the virulent pathogen (Guo et al., 2017). Loss of CRWN proteins induces the expression of the salicylic acid biosynthetic gene ISOCHORISMATE SYNTHASE1, which leads to spontaneous defense responses in crwn1crwn2 and crwn1crwn4 mutants (Choi et al., 2019). In addition to CRWNs, other NE proteins called NE-associated proteins (NEAPs) may connect to chromatin as NEAPs interact with a putative transcription factor called AtbZIP18 (Pawar et al., 2016). Tethering chromatin to the NE is associated with chromatin structure as well as the regulation of gene expression. DNA density is increased crwn1crwn2 (Dittmer et al., 2007), and the chromocenter organization is altered in crwn1crwn2 and crwn4 (Wang et al., 2013). The absence of SUN1 and SUN2 proteins leads to a delay in meiotic progression and defects in synapsis and recombination (Varas et al., 2015). In wifi, the quintuple mutants deficient of WIPs and WITs, and sun1sun4sun5, chromocenters are further decondensed compared to WT (Poulet et al., 2017a). The absence of MLKS2 leads to multiple meiotic defects in Z. mays. The Arabidopsis gip1gip2 mutants exhibit centromeric cohesion defects (Batzenschlager et al., 2015).
Other Plant Phenotypes Affected by NE Proteins Involved in the Control of the Nuclear Shape
Some mutant lines that are deficient in genes affecting the nuclear shape show defects in seed germination, plant growth, survival, immune response, reproduction, and signal transduction. Whereas crwn1crwn2 shows similar dormancy levels to WT (van Zanten et al., 2011), crwn1crwn3 shows a low germination rate and hypersensitiveness to abscisic acid (Zhao et al., 2016). Whereas crwn single mutant lines have rosette leaves with a size similar to that of WT, some crwn double and triple mutant lines have smaller rosette leaves, and a mutant combining alleles in all four CRWN genes cannot be isolated (Wang et al., 2013). The crwn1crwn2 double mutant shows enhanced resistance against the virulent pathogen as mentioned above (section “Regulation of Gene Expression”). Seed production is reduced in wip1wip2wip3, wit1wit2, and wifi (Zhou and Meier, 2014). It is shown that loss of WITs or WIPs impairs the pollen tube reception and SC-to-ovule migration (Zhou and Meier, 2014). wit1wit2 and wifi pollen tubes are hyposensitive to exogenous hydrogen peroxide (H2O2), which induce pollen tube (Moser et al., 2020). Nuclear Ca2+ peaks observed after ROS (H2O2) treatment in growing pollen tubes are disrupted in the wit1wit2 mutant (Moser et al., 2020). nup136 mutants exhibited an early flowering phenotype (Tamura et al., 2010). NUP1 (also known as NUP136) deficiency affects both male and female gametophyte development, resulting in reduced seed production (Bao et al., 2019). In addition to the plant phenotype of Arabidopsis mutants above, the Z. mays mlks2 mutants have defects in male meiosis, pollen viability, and stomatal complex development (Gumber et al., 2019b).
Physiological Significance of NE Deformation
What is the physiological significance of NE deformation, including NR formation? NR is the intranuclear network formed by invaginations of the NE (section “NE Shapes Under Normal State”; Malhas et al., 2011). It has been suggested based on metazoan researches that the presence of an NR increases the surface-to-volume ratio within the nucleus, thereby facilitating both the entry and exit of Ca2+ (Bootman et al., 2009). It is also discussed that the cytoplasmic core of NR might also facilitate the translation of mRNA generated adjacent to the invagination, with NPCs within it permitting the prompt translocation to the cytoplasm of the NR core (Malhas et al., 2011). The recent findings of Ca2+ fluctuations in the nucleus in Arabidopsis pollen tubes, which are altered in wit1wit2 (Moser et al., 2020), might be a clue to elucidate the relation between NR and Ca2+ signaling in plants. It is common between metazoans and plants that the invaginated NE often associates with nucleoli, which might facilitate mRNA export to the cytoplasm (Collings et al., 2000).
Nuclear egress of herpesviruses in vertebrates (Mettenleiter et al., 2006) and NE budding for ribonucleoprotein particle export during synaptic Wnt signaling in Drosophila (Speese et al., 2012) are accompanied by small and local NE deformation. Although similar changes in the NE shape during viral infections are also observed in plants (Malhas et al., 2011; mentioned in section “Changes in the NE Shape by a Viral Infection”), little is known about the mechanism underlying NE budding in plants. Nucleophagy (Kvam and Goldfarb, 2007; Park et al., 2009; Nezis et al., 2010) and NE rupture (Shah et al., 2017), both accompanied by a change in the NE shape, do not seem to have been elucidated yet in plants.
Conclusion and Perspectives
Over the last two decades, progress has been made in the study of plant nuclear morphology. NE proteins that connect the cytoplasm to the nucleoplasm have been revealed one after another (summarized by Meier et al., 2017), and many of them are involved in the control of the nuclear shape and size in A. thaliana. Some mutants with an aberrant nuclear shape normally survive, whereas other mutants have defects in nuclear movement, control of migration order of VN and SCs in pollen tubes, maintenance of chromatin structure, meiosis, regulation of gene expression, plant immunity, salicylic acid response, Ca2+ and ROS signaling, pollen development, pollen tube reception, and seed production (Evans et al., 2020; Groves et al., 2020). Little is known about how the shape of the NE, including NR, is regulated. Now that many plant NE proteins have been identified, it could be expected that the molecular mechanisms underlying changes in the NE shape and new physiological phenomena accompanied by changes in the NE shape will be revealed.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by a “Specially Promoted Research” Grant-in-Aid for Scientific Research to IH-N (nos. 22000014 and 15H05776), a Grant-in-Aid for Scientific Research to KT (nos. 26711017 and 18K06283), the Human Frontier Science Program to KT (RGP0009/2018) from the International Human Frontier Science Program Organization, and the Hirao Taro Foundation of KONAN GAKUEN for Academic Research to IH-N.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We appreciate Prof. Fukaki (Kobe University) for encouraging the authors to write the manuscript.
References
Aebi, U., Cohn, J., Buhle, L., and Gerace, L. (1986). The nuclear lamina is a meshwork of intermediate-type filaments. Nature 323, 560–564. doi: 10.1038/323560a0
Aldrich, H. C., and Vasil, I. K. (1970). Ultrastructure of postmeiotic nuclear envelope in microspores of Podocarpus macrophyllus. J. Ultrastruct. Res. 32, 307–315. doi: 10.1016/S0022-5320(70)80011-9
Bandyopadhyay, A., Kopperud, K., Anderson, G., Martin, K., and Goodin, M. (2010). An integrated protein localization and interaction map for Potato yellow dwarf virus, type species of the genus Nucleorhabdovirus. Virology 402, 61–71. doi: 10.1016/j.virol.2010.03.013
Bao, S. G., Shen, G. S., Li, G. C., Liu, Z. K., Arif, M., Wei, Q. Q., et al. (2019). The Arabidopsis nucleoporin NUP1 is essential for megasporogenesis and early stages of pollen development. Plant Cell Rep. 38, 59–74. doi: 10.1007/s00299-018-2349-7
Batzenschlager, M., Herzog, E., Houlné, G., Schmit, A. C., and Chabouté, M. E. (2014). GIP/MZT1 proteins orchestrate nuclear shaping. Front. Plant Sci. 5:29. doi: 10.3389/fpls.2014.00029
Batzenschlager, M., Lermontova, I., Schubert, V., Fuchs, J., Berr, A., Koini, M. A., et al. (2015). Arabidopsis MZT1 homologs GIP1 and GIP2 are essential for centromere architecture. Proc. Natl. Acad. Sci. U. S. A. 112, 8656–8660. doi: 10.1073/pnas.1506351112
Batzenschlager, M., Masoud, K., Janski, N., Houlné, G., Herzog, E., Evrard, J. L., et al. (2013). The GIP gamma-tubulin complex-associated proteins are involved in nuclear architecture in Arabidopsis thaliana. Front. Plant Sci. 4:480. doi: 10.3389/fpls.2013.00480
Bi, X. L., Cheng, Y. J., Hu, B., Ma, X. L., Wu, R., Wang, J. W., et al. (2017). Nonrandom domain organization of the Arabidopsis genome at the nuclear periphery. Genome Res. 27, 1162–1173. doi: 10.1101/gr.215186.116
Boettcher, B., and Barral, Y. (2013). The cell biology of open and closed mitosis. Nucleus 4, 160–165. doi: 10.4161/nucl.24676
Bootman, M. D., Fearnley, C., Smyrnias, I., Macdonald, F., and Roderick, H. L. (2009). An update on nuclear calcium signalling. J. Cell Sci. 122, 2337–2350. doi: 10.1242/jcs.028100
Borg, M., and Twell, D. (2011). “Pollen: structure and development” in eLS (Chichester, UK: John Wiley & Sons, Ltd.).
Borges, F., Gardner, R., Lopes, T., Calarco, J. P., Boavida, L. C., Slotkin, R. K., et al. (2012). FACS-based purification of Arabidopsis microspores, sperm cells and vegetative nuclei. Plant Methods 8:44. doi: 10.1186/1746-4811-8-44
Brandt, A., Papagiannouli, F., Wagner, N., Wilsch-Bräuninger, M., Braun, M., Furlong, E. E., et al. (2006). Developmental control of nuclear size and shape by kugelkern and kurzkern. Curr. Biol. 16, 543–552. doi: 10.1016/j.cub.2006.01.051
Cardenas, M. E., Laroche, T., and Gasser, S. M. (1990). The composition and morphology of yeast nuclear scaffolds. J. Cell Sci. 96, 439–450. doi: 10.1242/jcs.96.3.439
Carr, R. J., and Kim, K. S. (1983). Ultrastructure of mixed plant virus infection: bean yellow mosaic virus with cowpea severe mosaic virus or cowpea mosaic virus in bean. Virology 124, 338–348. doi: 10.1016/0042-6822(83)90350-1
Chemudupati, M., Osmani, A. H., and Osmani, S. A. (2016). A mitotic nuclear envelope tether for Gle1 also affects nuclear and nucleolar architecture. Mol. Biol. Cell 27, 3757–3770. doi: 10.1091/mbc.E16-07-0544
Choi, J., Strickler, S. R., and Richards, E. J. (2019). Loss of CRWN nuclear proteins induces cell death and salicylic acid defense signaling. Plant Physiol. 179, 1315–1329. doi: 10.1104/pp.18.01020
Chytilova, E., Macas, J., and Galbraith, D. W. (1999). Green fluorescent protein targeted to the nucleus, a transgenic phenotype useful for studies in plant biology. Ann. Bot. 83, 645–654. doi: 10.1006/anbo.1999.0866
Chytilova, E., Macas, J., Sliwinska, E., Rafelski, S. M., Lambert, G. M., and Galbraith, D. W. (2000). Nuclear dynamics in Arabidopsis thaliana. Mol. Biol. Cell 11, 2733–2741. doi: 10.1091/mbc.11.8.2733
Ciska, M., and De La Espina, S. M. D. (2014). The intriguing plant nuclear lamina. Front. Plant Sci. 5:166. doi: 10.3389/fpls.2014.00166
Ciska, M., Masuda, K., and De La Espina, S. M. D. (2013). Lamin-like analogues in plants: the characterization of NMCP1 in Allium cepa. J. Exp. Bot. 64, 1553–1564. doi: 10.1093/jxb/ert020
Collings, D. A., Carter, C. N., Rink, J. C., Scott, A. C., Wyatt, S. E., and Allen, N. S. (2000). Plant nuclei can contain extensive grooves and invaginations. Plant Cell 12, 2425–2439. doi: 10.1105/tpc.12.12.2425
Czapiewski, R., Robson, M. I., and Schirmer, E. C. (2016). Anchoring a leviathan: how the nuclear membrane tethers the genome. Front. Genet. 7:82. doi: 10.3389/fgene.2016.00082
Dahl, K. N., Ribeiro, A. J. S., and Lammerding, J. (2008). Nuclear shape, mechanics, and mechanotransduction. Circ. Res. 102, 1307–1318. doi: 10.1161/CIRCRESAHA.108.173989
D’angelo, M. A., and Hetzer, M. W. (2008). Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 18, 456–466. doi: 10.1016/j.tcb.2008.07.009
Dauer, W. T., and Worman, H. J. (2009). The nuclear envelope as a signaling node in development and disease. Dev. Cell 17, 626–638. doi: 10.1016/j.devcel.2009.10.016
De Zoeten, G. A., Gaard, G., and Diez, F. B. (1972). Nuclear vesiculation associated with pea enation mosaic virus-infected plant-tissue. Virology 48, 638–647. doi: 10.1016/0042-6822(72)90148-1
Dechat, T., Pfleghaar, K., Sengupta, K., Shimi, T., Shumaker, D. K., Solimando, L., et al. (2008). Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 22, 832–853. doi: 10.1101/gad.1652708
Dittmer, T. A., Stacey, N. J., Sugimoto-Shirasu, K., and Richards, E. J. (2007). LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. Plant Cell 19, 2793–2803. doi: 10.1105/tpc.107.053231
Dreger, M., Bengtsson, L., Schöneberg, T., Otto, H., and Hucho, F. (2001). Nuclear envelope proteomics: novel integral membrane proteins of the inner nuclear membrane. Proc. Natl. Acad. Sci. U. S. A. 98, 11943–11948. doi: 10.1073/pnas.211201898
Dubos, T., Poulet, A., Gonthier-Gueret, C., Mougeot, G., Vanrobays, E., Li, Y. R., et al. (2020). Automated 3D bio-imaging analysis of nuclear organization by nucleus J 2.0. Nucleus 11, 315–329. doi: 10.1080/19491034.2020.1845012
Dultz, E., and Ellenberg, J. (2007). Nuclear envelope. Curr. Biol. 17, R154–R156. doi: 10.1016/j.cub.2006.12.035
Dumas, C., Knox, R. B., and Gaude, T. (1985). The spatial association of the sperm cells and vegetative nucleus in the pollen grain of Brassica. Protoplasma 124, 168–174. doi: 10.1007/BF01290767
Eastmond, P. J., Quettier, A. L., Kroon, J. T. M., Craddock, C., Adams, N., and Slabas, A. R. (2010). PHOSPHATIDIC ACID PHOSPHOHYDROLASE1 and 2 regulate phospholipid synthesis at the endoplasmic reticulum in Arabidopsis. Plant Cell 22, 2796–2811. doi: 10.1105/tpc.110.221260
Ellenberg, J., Siggia, E. D., Moreira, J. E., Smith, C. L., Presley, J. F., Worman, H. J., et al. (1997). Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 138, 1193–1206. doi: 10.1083/jcb.138.6.1193
Evans, D. E., Mermet, S., and Tatout, C. (2020). Advancing knowledge of the plant nuclear periphery and its application for crop science. Nucleus 11, 347–363. doi: 10.1080/19491034.2020.1838697
Fal, K., Korsbo, N., Alonso-Serra, J., Telesd, J., Liu, M. Y., Refahidi, Y., et al. (2021). Tissue folding at the organ–meristem boundary results in nuclear compression and chromatin compaction. Proc. Natl. Acad. Sci. U. S. A. 118:e2017859118. doi: 10.1073/pnas.2017859118
Fiserova, J., Kiseleva, E., and Goldberg, M. W. (2009). Nuclear envelope and nuclear pore complex structure and organization in tobacco BY-2 cells. Plant J. 59, 243–255. doi: 10.1111/j.1365-313X.2009.03865.x
Galcheva-Gargova, Z., and Stateva, L. (1988). Immunological identification of two lamina-like proteins in Saccharomyces cerevisiae. Biosci. Rep. 8, 287–291. doi: 10.1007/BF01115046
Goodin, M., Yelton, S., Ghosh, D., Mathews, S., and Lesnaw, J. (2005). Live-cell imaging of rhabdovirus-induced morphological changes in plant nuclear membranes. Mol. Plant-Microbe Interact. 18, 703–709. doi: 10.1094/MPMI-18-0703
Goodin, M. M., Chakrabarty, R., Yelton, S., Martin, K., Clark, A., and Brooks, R. (2007). Membrane and protein dynamics in live plant nuclei infected with Sonchus yellow net virus, a plant-adapted rhabdovirus. J. Gen. Virol. 88, 1810–1820. doi: 10.1099/vir.0.82698-0
Goswami, R., Asnacios, A., Milani, P., Graindorge, S., Houlné, G., Mutterer, J., et al. (2020). Mechanical shielding in plant nuclei. Curr. Biol. 30, 1–13. doi: 10.1016/j.cub.2020.03.059
Goto, C., Tamura, K., Fukao, Y., Shimada, T., and Hara-Nishimura, I. (2014). The novel nuclear envelope protein KAKU4 modulates nuclear morphology in Arabidopsis. Plant Cell 26, 2143–2155. doi: 10.1105/tpc.113.122168
Goto, C., Tamura, K., Nishimaki, S., Maruyama, D., and Hara-Nishimura, I. (2020). The nuclear envelope protein KAKU4 determines the migration order of the vegetative nucleus and sperm cells in pollen tubes. J. Exp. Bot. 71, 6273–6281. doi: 10.1093/jxb/eraa367
Graumann, K. (2014). Evidence for LINC1-SUN associations at the plant nuclear periphery. PLoS One 9:e93406. doi: 10.1371/journal.pone.0093406
Graumann, K., Bass, H. W., and Parry, G. (2013). SUNrises on the international plant nucleus consortium SEB Salzburg 2012. Nucleus 4, 3–7. doi: 10.4161/nucl.23385
Graumann, K., Runions, J., and Evans, D. E. (2010). Characterization of SUN-domain proteins at the higher plant nuclear envelope. Plant J. 61, 134–144. doi: 10.1111/j.1365-313X.2009.04038.x
Graumann, K., Vanrobays, E., Tutois, S., Probst, A. V., Evans, D. E., and Tatout, C. (2014). Characterization of two distinct subfamilies of SUN-domain proteins in Arabidopsis and their interactions with the novel KASH-domain protein AtTIK. J. Exp. Bot. 65, 6499–6512. doi: 10.1093/jxb/eru368
Grossman, E., Medalia, O., and Zwerger, M. (2012). Functional architecture of the nuclear pore complex. Annu. Rev. Biophys. 41, 557–584. doi: 10.1146/annurev-biophys-050511-102328
Groves, N. R., Biel, A., Moser, M., Mendes, T., Amstutz, K., and Meier, I. (2020). Recent advances in understanding the biological roles of the plant nuclear envelope. Nucleus 11, 330–346. doi: 10.1080/19491034.2020.1846836
Groves, N. R., Mckenna, J. F., Evans, D. E., Graumann, K., and Meier, I. (2019). A nuclear localization signal targets tail-anchored membrane proteins to the inner nuclear envelope in plants. J. Cell Sci. 132:jcs226134. doi: 10.1242/jcs.226134
Gumber, H. K., Mckenna, J. F., Estrada, A. L., Tolmie, A. F., Graumann, K., and Bass, H. W. (2019a). Identification and characterization of genes encoding the nuclear envelope LINC complex in the monocot species Zea mays. J. Cell Sci. 132:jcs221390. doi: 10.1242/jcs.221390
Gumber, H. K., Mckenna, J. F., Tolmie, A. F., Jalovec, A. M., Kartick, A. C., Graumann, K., et al. (2019b). MLKS2 is an ARM domain and F-actin-associated KASH protein that functions in stomatal complex development and meiotic chromosome segregation. Nucleus 10, 144–166. doi: 10.1080/19491034.2019.1629795
Guo, T. T., and Fang, Y. D. (2014). Functional organization and dynamics of the cell nucleus. Front. Plant Sci. 5:378. doi: 10.3389/fpls.2014.00378
Guo, T. T., Mao, X. G., Zhang, H., Zhang, Y., Fu, M. D., Sun, Z. F., et al. (2017). Lamin-like proteins negatively regulate plant immunity through NAC with TRANSMEMBRANE MOTIF1-LIKE9 and NONEXPRESSOR OF PR GENES1 in Arabidopsis thaliana. Mol. Plant 10, 1334–1348. doi: 10.1016/j.molp.2017.09.008
Gupton, S. L., Collings, D. A., and Allen, N. S. (2006). Endoplasmic reticulum targeted GFP reveals ER organization in tobacco NT-1 cells during cell division. Plant Physiol. Biochem. 44, 95–105. doi: 10.1016/j.plaphy.2006.03.003
Hampoelz, B., and Lecuit, T. (2011). Nuclear mechanics in differentiation and development. Curr. Opin. Cell Biol. 23, 668–675. doi: 10.1016/j.ceb.2011.10.001
Hayashi, Y., Yamada, K., Shimada, T., Matsushima, R., Nishizawa, N. K., Nishimura, M., et al. (2001). A proteinase-storing body that prepares for cell death or stresses in the epidermal cells of Arabidopsis. Plant Cell Physiol. 42, 894–899. doi: 10.1093/pcp/pce144
Heslop-Harrison, J., and Heslop-Harrison, Y. (1989a). Actomyosin and movement in the angiosperm pollen tube: an interpretation of some recent results. Sex. Plant Reprod. 2, 199–207. doi: 10.1007/BF00195579
Heslop-Harrison, J., and Heslop-Harrison, Y. (1989b). Conformation and movement of the vegetative nucleus of the angiosperm pollen tube: association with the actin cytoskeleton. J. Cell Sci. 93, 299–308. doi: 10.1242/jcs.93.2.299
Heslop-Harrison, J., and Heslop-Harrison, Y. (1989c). Myosin associated with the surfaces of organelles, vegetative nuclei and generative cells in angiosperm pollen grains and tubes. J. Cell Sci. 94, 319–325. doi: 10.1242/jcs.94.2.319
Hoelz, A., Debler, E. W., and Blobel, G. (2011). The structure of the nuclear pore complex. Annu. Rev. Biochem. 80, 613–643. doi: 10.1146/annurev-biochem-060109-151030
Hu, B., Wang, N., Bi, X. L., Karaaslan, E. S., Weber, A. L., Zhu, W. S., et al. (2019). Plant lamin-like proteins mediate chromatin tethering at the nuclear periphery. Genome Biol. 20:87. doi: 10.1186/s13059-019-1694-3
Imbalzano, K. M., Cohet, N., Wu, Q., Underwood, J. M., Imbalzano, A. N., and Nickerson, J. A. (2013). Nuclear shape changes are induced by knockdown of the SWI/SNF ATPase BRG1 and are independent of cytoskeletal connections. PLoS One 8:e55628. doi: 10.1371/journal.pone.0055628
Janski, N., Herzog, E., and Schmit, A. C. (2008). Identification of a novel small Arabidopsis protein interacting with gamma-tubulin complex protein 3. Cell Biol. Int. 32, 546–548. doi: 10.1016/j.cellbi.2007.11.006
Janski, N., Masoud, K., Batzenschlager, M., Herzog, E., Evrard, J. L., Houlné, G., et al. (2012). The GCP3-interacting proteins GIP1 and GIP2 are required for γ-tubulin complex protein localization, spindle integrity, and chromosomal stability. Plant Cell 24, 1171–1187. doi: 10.1105/tpc.111.094904
Jovtchev, G., Schubert, V., Meister, A., Barow, M., and Schubert, I. (2006). Nuclear DNA content and nuclear and cell volume are positively correlated in angiosperms. Cytogenet. Genome Res. 114, 77–82. doi: 10.1159/000091932
Kimata, Y., Lim, C. R., Kiriyama, T., Nara, A., Hirata, A., and Kohno, K. (1999). Mutation of the yeast ε-COP gene ANU2 causes abnormal nuclear morphology and defects in intracellular vesicular transport. Cell Struct. Funct. 24, 197–208. doi: 10.1247/csf.24.197
Kittur, N., Zapantis, G., Aubuchon, M., Santoro, N., Bazett-Jones, D. P., and Meier, U. T. (2007). The nucleolar channel system of human endometrium is related to endoplasmic reticulum and R-rings. Mol. Biol. Cell 18, 2296–2304. doi: 10.1091/mbc.E07-02-0154
Kvam, E., and Goldfarb, D. S. (2007). Nucleus-vacuole junctions and piecemeal microautophagy of the nucleus in S. cerevisiae. Autophagy 3, 85–92. doi: 10.4161/auto.3586
Lanctôt, C., Cheutin, T., Cremer, M., Cavalli, G., and Cremer, T. (2007). Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat. Rev. Genet. 8, 104–115. doi: 10.1038/nrg2041
Malhas, A., Goulbourne, C., and Vaux, D. J. (2011). The nucleoplasmic reticulum: form and function. Trends Cell Biol. 21, 362–373. doi: 10.1016/j.tcb.2011.03.008
Masuda, K., Xu, Z. J., Takahashi, S., Ito, A., Ono, M., Nomura, K., et al. (1997). Peripheral framework of carrot cell nucleus contains a novel protein predicted to exhibit a long α-helical domain. Exp. Cell Res. 232, 173–181. doi: 10.1006/excr.1997.3531
Mattaj, I. W. (2004). Sorting out the nuclear envelope from the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 5, 65–69. doi: 10.1038/nrm1263
Mazumder, A., and Shivashankar, G. V. (2010). Emergence of a prestressed eukaryotic nucleus during cellular differentiation and development. J. R. Soc. Interface 7, S321–S330. doi: 10.1098/rsif.2010.0039.focus
Meier, I. (2007). Composition of the plant nuclear envelope: theme and variations. J. Exp. Bot. 58, 27–34. doi: 10.1093/jxb/erl009
Meier, I., Griffis, A. H. N., Groves, N. R., and Wagner, A. (2016). Regulation of nuclear shape and size in plants. Curr. Opin. Cell Biol. 40, 114–123. doi: 10.1016/j.ceb.2016.03.005
Meier, I., Richards, E. J., and Evans, D. E. (2017). Cell biology of the plant nucleus. Annu. Rev. Plant Biol. 68, 139–172. doi: 10.1146/annurev-arplant-042916-041115
Mettenleiter, T. C., Klupp, B. G., and Granzow, H. (2006). Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 9, 423–429. doi: 10.1016/j.mib.2006.06.013
Mitsuhashi, N., Shimada, T., Mano, S., Nishimura, M., and Hara-Nishimura, I. (2000). Characterization of organelles in the vacuolar-sorting pathway by visualization with GFP in tobacco BY-2 cells. Plant Cell Physiol. 41, 993–1001. doi: 10.1093/pcp/pcd040
Moser, M., Kirkpatrick, A., Groves, N. R., and Meier, I. (2020). LINC-complex mediated positioning of the vegetative nucleus is involved in calcium and ROS signaling in Arabidopsis pollen tubes. Nucleus 11, 149–163. doi: 10.1080/19491034.2020.1783783
Nakano, R. T., Matsushima, R., Ueda, H., Tamura, K., Shimada, T., Li, L. X., et al. (2009). GNOM-LIKE1/ERMO1 and SEC24a/ERMO2 are required for maintenance of endoplasmic reticulum morphology in Arabidopsis thaliana. Plant Cell 21, 3672–3685. doi: 10.1105/tpc.109.068270
Nezis, I. P., Shravage, B. V., Sagona, A. P., Lamark, T., Bjørkøy, G., Johansen, T., et al. (2010). Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J. Cell Biol. 190, 523–531. doi: 10.1083/jcb.201002035
Noguchi, T., Watanabe, H., and Suzuki, R. (1998). Effects of brefeldin A on the Golgi apparatus, the nuclear envelope, and the endoplasmic reticulum in a green alga, Scenedesmus acutus. Protoplasma 201, 202–212. doi: 10.1007/BF01287416
Oda, Y., and Fukuda, H. (2011). Dynamics of Arabidopsis SUN proteins during mitosis and their involvement in nuclear shaping. Plant J. 66, 629–641. doi: 10.1111/j.1365-313X.2011.04523.x
Ohtsu, M., Shibata, Y., Ojika, M., Tamura, K., Hara-Nishimura, I., Mori, H., et al. (2014). Nucleoporin 75 is involved in the ethylene-mediated production of phytoalexin for the resistance of Nicotiana benthamiana to Phytophthora infestans. Mol. Plant-Microbe Interact. 27, 1318–1330. doi: 10.1094/MPMI-06-14-0181-R
Park, B. H., Dolen, J., and Snyder, B. (1977). Defective chemotactic migration of polymorphonuclear leukocytes in Pelger-Huet anomaly. Proc. Soc. Exp. Biol. Med. 155, 51–54. doi: 10.3181/00379727-155-39743
Park, Y. E., Hayashi, Y. K., Bonne, G., Arimura, T., Noguchi, S., Nonakal, I., et al. (2009). Autophagic degradation of nuclear components in mammalian cells. Autophagy 5, 795–804. doi: 10.4161/auto.8901
Parry, G. (2014). Components of the Arabidopsis nuclear pore complex play multiple diverse roles in control of plant growth. J. Exp. Bot. 65, 6057–6067. doi: 10.1093/jxb/eru346
Paul, A., and Bhattacharya, S. (2012). Chemistry and biology of DNA-binding small molecules. Curr. Sci. 102, 212–231.
Pawar, V., Poulet, A., Detourne, G., Tatout, C., Vanrobays, E., Evans, D. E., et al. (2016). A novel family of plant nuclear envelope-associated proteins. J. Exp. Bot. 67, 5699–5710. doi: 10.1093/jxb/erw332
Polychronidou, M., and Großhans, J. (2011). Determining nuclear shape: the role of farnesylated nuclear membrane proteins. Nucleus 2, 17–23. doi: 10.4161/nucl.2.1.13992
Polychronidou, M., Hellwig, A., and Grosshans, J. (2010). Farnesylated nuclear proteins Kugelkern and Lamin Dm0 affect nuclear morphology by directly interacting with the nuclear membrane. Mol. Biol. Cell 21, 3409–3420. doi: 10.1091/mbc.E10-03-0230
Poulet, A., Arganda-Carreras, I., Legland, D., Probst, A. V., Andrey, P., and Tatout, C. (2015). NucleusJ: an ImageJ plugin for quantifying 3D images of interphase nuclei. Bioinformatics 31, 1144–1146. doi: 10.1093/bioinformatics/btu774
Poulet, A., Duc, C., Voisin, M., Desset, S., Tutois, S., Vanrobays, E., et al. (2017a). The LINC complex contributes to heterochromatin organisation and transcriptional gene silencing in plants. J. Cell Sci. 130, 590–601. doi: 10.1242/jcs.194712
Poulet, A., Probst, A. V., Graumann, K., Tatout, C., and Evans, D. (2017b). Exploring the evolution of the proteins of the plant nuclear envelope. Nucleus 8, 46–59. doi: 10.1080/19491034.2016.1236166
Pradillo, M., Evans, D., and Graumann, K. (2019). The nuclear envelope in higher plant mitosis and meiosis. Nucleus 10, 55–66. doi: 10.1080/19491034.2019.1587277
Prunuske, A. J., and Ullman, K. S. (2006). The nuclear envelope: form and reformation. Curr. Opin. Cell Biol. 18, 108–116. doi: 10.1016/j.ceb.2005.12.004
Renzaglia, K. S., and Duckett, J. G. (1987). Spermatogenesis in Blasia-Pusilla - from young antheridium through mature spermatozoid. Bryologist 90, 419–449. doi: 10.2307/3243109
Renzaglia, K. S., and Duckett, J. G. (1989). Ultrastructural studies of spermatogenesis in Anthocerotophyta V. Nuclear metamorphosis and the posterior mitochondrion of Notothylas orbicularis and Phaeoceros laevis. Protoplasma 151, 137–150. doi: 10.1007/Bf01403451
Sakamoto, Y., Sato, M., Sato, Y., Harada, A., Suzuki, T., Goto, C., et al. (2020). Subnuclear gene positioning through lamina association affects copper tolerance. Nat. Commun. 11:5914. doi: 10.1038/s41467-020-19621-z
Sakamoto, Y., and Takagi, S. (2013). LITTLE NUCLEI 1 and 4 regulate nuclear morphology in Arabidopsis thaliana. Plant Cell Physiol. 54, 622–633. doi: 10.1093/pcp/pct031
Schirmer, E. C., and Foisner, R. (2007). Proteins that associate with lamins: many faces, many functions. Exp. Cell Res. 313, 2167–2179. doi: 10.1016/j.yexcr.2007.03.012
Schwarz, D. S., and Blower, M. D. (2016). The endoplasmic reticulum: structure, function and response to cellular signaling. Cell. Mol. Life Sci. 73, 79–94. doi: 10.1007/s00018-015-2052-6
Shah, P., Wolf, K., and Lammerding, J. (2017). Bursting the bubble - nuclear envelope rupture as a path to genomic instability? Trends Cell Biol. 27, 546–555. doi: 10.1016/j.tcb.2017.02.008
Sheffield, E., Cawood, A. H., Bell, P. R., and Dickinson, H. G. (1979). The development of nuclear vacuoles during meiosis in plants. Planta 146, 597–601. doi: 10.1007/BF00388838
Singh, S., Lazzaro, M. D., and Walles, B. (1998). The nuclear reticulum in placental cells of Lilium regale is a part of the endomembrane system. Protoplasma 203, 144–152. doi: 10.1007/BF01279471
Skinner, B. M., and Johnson, E. E. P. (2017). Nuclear morphologies: their diversity and functional relevance. Chromosoma 126, 195–212. doi: 10.1007/s00412-016-0614-5
Sliwinska, E., Mathur, J., and Bewley, J. D. (2012). Synchronously developing collet hairs in Arabidopsis thaliana provide an easily accessible system for studying nuclear movement and endoreduplication. J. Exp. Bot. 63, 4165–4178. doi: 10.1093/jxb/ers099
Smith, S., Galinha, C., Desset, S., Tolmie, F., Evans, D., Tatout, C., et al. (2015). Marker gene tethering by nucleoporins affects gene expression in plants. Nucleus 6, 471–478. doi: 10.1080/19491034.2015.1126028
Speese, S. D., Ashley, J., Jokhi, V., Nunnari, J., Barria, R., Li, Y. H., et al. (2012). Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell 149, 832–846. doi: 10.1016/j.cell.2012.03.032
Starr, D. A., and Fridolfsson, H. N. (2010). Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell Dev. Biol. 26, 421–444. doi: 10.1146/annurev-cellbio-100109-104037
Stephens, A. D., Liu, P. Z., Banigan, E. J., Almassalha, L. M., Backman, V., Adam, S. A., et al. (2018). Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol. Biol. Cell 29, 220–233. doi: 10.1091/mbc.E17-06-0410
Tamura, K., Fukao, Y., Iwamoto, M., Haraguchi, T., and Hara-Nishimura, I. (2010). Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell 22, 4084–4097. doi: 10.1105/tpc.110.079947
Tamura, K., Goto, C., and Hara-Nishimura, I. (2015). Recent advances in understanding plant nuclear envelope proteins involved in nuclear morphology. J. Exp. Bot. 66, 1641–1647. doi: 10.1093/jxb/erv036
Tamura, K., and Hara-Nishimura, I. (2011). Involvement of the nuclear pore complex in morphology of the plant nucleus. Nucleus 2, 168–172. doi: 10.4161/nucl.2.3.16175
Tamura, K., and Hara-Nishimura, I. (2013). The molecular architecture of the plant nuclear pore complex. J. Exp. Bot. 64, 823–832. doi: 10.1093/jxb/ers258
Tamura, K., Iwabuchi, K., Fukao, Y., Kondo, M., Okamoto, K., Ueda, H., et al. (2013). Myosin XI-i links the nuclear membrane to the cytoskeleton to control nuclear movement and shape in Arabidopsis. Curr. Biol. 23, 1776–1781. doi: 10.1016/j.cub.2013.07.035
Thomas, C. H., Collier, J. H., Sfeir, C. S., and Healy, K. E. (2002). Engineering gene expression and protein synthesis by modulation of nuclear shape. Proc. Natl. Acad. Sci. U. S. A. 99, 1972–1977. doi: 10.1073/pnas.032668799
van Zanten, M., Koini, M. A., Geyer, R., Liu, Y. X., Brambilla, V., Bartels, D., et al. (2011). Seed maturation in Arabidopsis thaliana is characterized by nuclear size reduction and increased chromatin condensation. Proc. Natl. Acad. Sci. U. S. A. 108, 20219–20224. doi: 10.1073/pnas.1117726108
Varas, J., Graumann, K., Osman, K., Pradillo, M., Evans, D. E., Santos, J. L., et al. (2015). Absence of SUN1 and SUN2 proteins in Arabidopsis thaliana leads to a delay in meiotic progression and defects in synapsis and recombination. Plant J. 81, 329–346. doi: 10.1111/tpj.12730
Walters, A. D., Bommakanti, A., and Cohen-Fix, O. (2012). Shaping the nucleus: factors and forces. J. Cell. Biochem. 113, 2813–2821. doi: 10.1002/jcb.24178
Wang, H. Y., Dittmer, T. A., and Richards, E. J. (2013). Arabidopsis CROWDED NUCLEI (CRWN) proteins are required for nuclear size control and heterochromatin organization. BMC Plant Biol. 13:200. doi: 10.1186/1471-2229-13-200
Webster, M., Witkin, K. L., and Cohen-Fix, O. (2009). Sizing up the nucleus: nuclear shape, size and nuclear-envelope assembly. J. Cell Sci. 122, 1477–1486. doi: 10.1242/jcs.037333
Wilson, K. L., and Foisner, R. (2010). Lamin-binding proteins. Cold Spring Harb. Perspect. Biol. 2:e000554. doi: 10.1101/cshperspect.a000554
Xu, X. M., Meulia, T., and Meier, I. (2007a). Anchorage of plant RanGAP to the nuclear envelope involves novel nuclear-pore-associated proteins. Curr. Biol. 17, 1157–1163. doi: 10.1016/j.cub.2007.05.076
Xu, X. M., Rose, A., Muthuswamy, S., Jeong, S. Y., Venkatakrishnan, S., Zhao, Q., et al. (2007b). NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/MIp1/MIp2/megator, is involved in mRNA export and SUMO horneostasis and affects diverse aspects of plant development. Plant Cell 19, 1537–1548. doi: 10.1105/tpc.106.049239
Yasuhara, H., and Kitamoto, K. (2014). Aphidicolin-induced nuclear elongation in tobacco BY-2 cells. Plant Cell Physiol. 55, 913–927. doi: 10.1093/pcp/pcu026
Zhao, Q., Brkljacic, J., and Meier, I. (2008). Two distinct interacting classes of nuclear envelope-associated coiled-coil proteins are required for the tissue-specific nuclear envelope targeting of Arabidopsis RanGAP. Plant Cell 20, 1639–1651. doi: 10.1105/tpc.108.059220
Zhao, W. M., Guan, C. M., Feng, J., Liang, Y., Zhan, N., Zuo, J. R., et al. (2016). The Arabidopsis CROWDED NUCLEI genes regulate seed germination by modulating degradation of ABI5 protein. J. Integr. Plant Biol. 58, 669–678. doi: 10.1111/jipb.12448
Zhou, L. X., and Panté, N. (2010). The nucleoporin Nup153 maintains nuclear envelope architecture and is required for cell migration in tumor cells. FEBS Lett. 584, 3013–3020. doi: 10.1016/j.febslet.2010.05.038
Zhou, X., Graumann, K., Evans, D. E., and Meier, I. (2012). Novel plant SUN-KASH bridges are involved in RanGAP anchoring and nuclear shape determination. J. Cell Biol. 196, 203–211. doi: 10.1083/jcb.201108098
Zhou, X., Graumann, K., Wirthmueller, L., Jones, J. D. G., and Meier, I. (2014). Identification of unique SUN-interacting nuclear envelope proteins with diverse functions in plants. J. Cell Biol. 205, 677–692. doi: 10.1083/jcb.201401138
Zhou, X., Groves, N. R., and Meier, I. (2015a). Plant nuclear shape is independently determined by the SUN-WIP-WIT2-myosin XI-i complex and CRWN1. Nucleus 6, 144–153. doi: 10.1080/19491034.2014.1003512
Zhou, X., Groves, N. R., and Meier, I. (2015b). SUN anchors pollen WIP-WIT complexes at the vegetative nuclear envelope and is necessary for pollen tube targeting and fertility. J. Exp. Bot. 66, 7299–7307. doi: 10.1093/jxb/erv425
Zhou, X., and Meier, I. (2014). Efficient plant male fertility depends on vegetative nuclear movement mediated by two families of plant outer nuclear membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 111, 11900–11905. doi: 10.1073/pnas.1323104111
Zink, D., Fischer, A. H., and Nickerson, J. A. (2004). Nuclear structure in cancer cells. Nat. Rev. Cancer 4, 677–687. doi: 10.1038/nrc1430
Keywords: nuclear shape, nuclear envelope, nuclear lamina, nucleoplasmic reticulum, Arabidopsis thaliana
Citation: Goto C, Hara-Nishimura I and Tamura K (2021) Regulation and Physiological Significance of the Nuclear Shape in Plants. Front. Plant Sci. 12:673905. doi: 10.3389/fpls.2021.673905
Edited by:
Marc Libault, University of Nebraska-Lincoln, United StatesReviewed by:
David Edgar Evans, Oxford Brookes University, United KingdomNorman Reid Groves, The Ohio State University, United States
Copyright © 2021 Goto, Hara-Nishimura and Tamura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kentaro Tamura, dGFtdXJhQHUtc2hpenVva2Eta2VuLmFjLmpw
 Chieko Goto
Chieko Goto Ikuko Hara-Nishimura
Ikuko Hara-Nishimura Kentaro Tamura
Kentaro Tamura