Abstract
Multiplex genome-editing (MGE) technologies are recently developed versatile bioengineering tools for modifying two or more specific DNA loci in a genome with high precision. These genome-editing tools have greatly increased the feasibility of introducing desired changes at multiple nucleotide levels into a target genome. In particular, clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (Cas) [CRISPR/Cas] system-based MGE tools allow the simultaneous generation of direct mutations precisely at multiple loci in a gene or multiple genes. MGE is enhancing the field of plant molecular biology and providing capabilities for revolutionizing modern crop-breeding methods as it was virtually impossible to edit genomes so precisely at the single base-pair level with prior genome-editing tools, such as zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs). Recently, researchers have not only started using MGE tools to advance genome-editing applications in certain plant science fields but also have attempted to decipher and answer basic questions related to plant biology. In this review, we discuss the current progress that has been made toward the development and utilization of MGE tools with an emphasis on the improvements in plant biology after the discovery of CRISPR/Cas9. Furthermore, the most recent advancements involving CRISPR/Cas applications for editing multiple loci or genes are described. Finally, insights into the strengths and importance of MGE technology in advancing crop-improvement programs are presented.
Introduction
Plant scientists have long been searching for biological tools to generate site-specific mutations in the plant genome for research. Until the emergence of genome-editing tools, it was almost impossible to recognize and alter specific genomic sequences at a chosen locus. Recently, nucleases have been engineered that recognize and cleave DNA at target genomic sites, introducing double strand breaks (DSBs) that are subsequently repaired by either error-prone non-homologous end joining (NHEJ) or homology-directed repair (HDR) pathways. During the repair process, precise DNA fragment deletion, insertion, or replacement can occur, resulting in gene editing, mutation, or replacement (Hsu et al., 2013; Miglani, 2017; Mishra et al., 2018).
Using nucleases to simultaneously and precisely introduce two or more DSBs at specific loci within a single genome in a single round of mutagenesis will hereafter be called multiplex genome editing (MGE) (Cong et al., 2013). MGE generates rapid desirable changes in specific but different genes or quantitative trait loci (QTLs), resulting in the development of new plant genotypes carrying multiple mutations in one generation. The capability to create such genomic changes will allow researchers to understand and study the interactions between members of a gene family that have redundant functions, and it may provide an opportunity to analyze epistatic relationships among closely related genetic pathways (Xing et al., 2014). Furthermore, these multiple genomic modifications or edits could provide the most practical platform for metabolic pathway engineering in plants. MGE can be used to facilitate many desirable crop improvements, such as the production of seedless plant species and better disease resistance (Naim et al., 2018; Tripathi et al., 2019), as well as to skip extensive segregation in vegetatively propagating crop plants, such as sugarcane (Kannan et al., 2018).
Unlike traditional random mutagenesis, MGE tools are designed to target specific genomic regions and to generate specific mutations or variations with much more precision (Belhaj et al., 2015). Accomplishing multiple gene mutations in an elite cultivar formerly required many years and several cycles of crossing and selection. Moreover, the traditional approach for conventional breeding has sometimes been more complicated because of linkage drag, which impedes the transmission of good traits aside from deleterious genes. For instance, developing high-yielding genotypes with premium grain quality traits has always been considered a challenge for plant breeders utilizing conventional breeding approaches (Abdelrahman and Zhao, 2020). However, many researchers have reported using MGE tools to improve traits with such complications (Wang et al., 2014; Li W. et al., 2016; Kannan et al., 2018). MGE tools have become more user-friendly, particularly after the development and adaptation of the CRISPR-Cas system (Jinek et al., 2012; Cho et al., 2013; Cong et al., 2013; Mali et al., 2013). Among genome-editing technologies, including zinc-finger nucleases (ZFNs) (Bibikova et al., 2002; Lloyd et al., 2005; Ainley et al., 2013; Qi et al., 2014) and transcription activator-like effector nucleases (TALENs) (Christian et al., 2010; Li et al., 2011), the CRISPR/Cas system uniquely depends on base complementarity between the guide RNA (gRNA) and the target DNA for recognition, which greatly simplifies the genome-editing process (Jinek et al., 2012). Due to its simplicity for designing multiple target genetic loci, researchers have preferred the CRISPR/Cas system for genome editing in recent years. TALENs have also been successfully utilized for MGE in polyploid plants, including wheat, tobacco, and sugarcane (Wang et al., 2014; Li W. et al., 2016; Kannan et al., 2018). However, while using the multiplex TALENs approach, each unit must be designed separately, which increases the difficulty in delivering different units simultaneously to a plant cell. The CRISPR/Cas system has many advantages over TALENs and ZFNs: it is more efficient in editing target loci, and multiple single-guide RNAs (sgRNAs) can be designed to be expressed at once for simultaneous editing of multiple loci (Cong et al., 2013). Adding more gRNAs to the transformation construct is far simpler than engineering new proteins to improve the trait of choice in plants. By using just one Cas protein with multiple gRNAs, multiple gene mutations have been achieved in various plants, including rice, wheat, maize, tomato, and Arabidopsis (Li et al., 2013; Upadhyay et al., 2013; Ma et al., 2015; Yan et al., 2016; Rothan et al., 2019).
Further investigations and developments are ongoing to improve MGE applications for the genetic improvement of crops. To date, MGE using CRISPR/Cas has been efficiently conducted to enhance crop yield, quality, and stress resistance (Xu et al., 2016; Kaur et al., 2018; Li Y. et al., 2019; Zhou et al., 2019). The results of different experiments have shown that different applications and procedures are progressively appearing. Immense technical progress has recently occurred in this research field, utilizing more complex genetic, epigenetic, and transcriptional manipulations for research needs. Recently, Kannan et al. (2018) reported targeted mutagenesis of more than 100 alleles in sugarcane to improve saccharification efficiency without affecting biomass yield. In this review, we discuss different emerging applications that have been developed and adapted for MGE and their potency and associated experiments for crop-improvement programs. We further summarize and provide recent information about the progress on MGE in plants, emphasizing its role in developing plant molecular biology and plant breeding.
Tools for MGE
TALENs for Targeting Gene Families
When MGE tools were first developed, before there was a CRISPR/Cas system, Wang et al. (2014) reported using a single TALEN pair to edit three homoeoalleles encoding mildew resistance locus proteins (MLOs) in hexaploid bread wheat (Triticum aestivum L., 2n = 42, AABBDD). The goal was to develop broad-spectrum and durable Blumeria graminis f. sp. tritici (Bgt) resistant wheat lines. The single TALEN pair recognition sequences were strictly conserved at the MLO-B1 and MLO-D1 target sites and has one nucleotide mismatch at the MLO-A1 target site; they had mutations at the three target sites. The results revealed that of the 27 T0 plants, four mutants carried two edited targets in either homozygous or heterozygous forms, and only one plant harbored triple mutations. Plants with triple mutations at the three different targets showed resistance, with no apparent fungal growth on the leaves, although abundant fungal growth was found on the leaves of wild-type plants, and the plants with combinations of other mutations.
Multiplex editing using TALENs was also employed to produce highly potent biopharmaceutical glycoprotein products. In an attempt to increase their activity, stability, and immunogenicity in plants, Li J. et al. (2016) reported that two α (1,3)-fucosyltransferase (FucT1 and FucT2) and two β (1,2)-xylosyltransferase (XylT1 and XylT2) genes within Nicotiana benthamiana were successfully edited, resulting in plants with improved capacity to produce glycoproteins devoid of plant-specific residues. Conserved sequences in the first exon of each gene family were analyzed, and one sequence-specific TALEN pair for each gene family was designed accordingly. The researchers tried to knockout the two-gene family members separately and together by cotransformation of the two TALEN pairs. Both FucT and XylT mutant plants had significant changes in their N-glycan profiles. Notably, this was the only trial of multiplex editing using two different modules of TALEN pairs to target two divergent gene families in one genotype.
To date, the highest record for simultaneous targeting genome editing in the same plant genotype has been reported in sugarcane crops. Kannan et al. successfully edited 107 of 109 gene copies of the sugarcane caffeic acid O-methyltransferase (COMT) gene family by introducing a single TALEN pair (Kannan et al., 2018). A conserved region of COMT was targeted with a single TALEN pair for multiallelic mutagenesis to modify lignin biosynthesis in sugarcane. This improved saccharification efficiency by up to 43.8% and significantly reduced lignin content in the mutant, without significant differences in biomass production compared with the wild type.
These studies made it clear that TALEN-based genome editing allows for efficient multiplex editing of gene families with conserved sequences among different gene copies, and this has been successfully demonstrated in polyploid genomes. However, Li et al. succeeded in editing two different gene families with two different cotransformed TALEN pairs; the editing efficiency of several TALEN pairs for each target was tested before conducting the experiment (Li J. et al., 2016). Furthermore, the procedure is more complicated if more gene families are targeted simultaneously because TALEN technologies require the engineering of DNA-binding domains for individual targeting applications (Lowder et al., 2015).
The CRISPR/Cas9 System Offers Excellent MGE Ability
The CRISPR/Cas9 is much easier and more efficient than other genome-editing tools, particularly for the editing of multiple unrelated genes. Using multiple sgRNAs and expressing only one Cas protein significantly simplifies and enhances the editing efficiency of multiple genes. The ease of plasmid construction in CRISPR/Cas9 experiments further allows the function of multiple associated genes to be investigated that either has negative effects or that improve plant-specific characteristics.
Attempts have been made to improve the CRISPR/Cas system to further simplify and enhance the efficiency of editing multiple genes. The core focus has been to develop an easy and fast toolbox to insert multiple sgRNA expression cassettes into one binary vector. Several sequential rounds of regular cloning steps can be utilized to insert different expression cassettes containing sgRNAs for different targets into a single construct, but this is very laborious and there is a limited number of assembled sgRNAs (Li et al., 2013; Mao et al., 2013; Zhou et al., 2014; Yan et al., 2015). A more advanced cloning method, called Golden Gate cloning, depends on the use of type IIS restriction enzymes in the DNA assembly. Golden Gate cloning has been used to generate non-palindromic sticky ends in DNA fragments carrying sgRNAs. This technique facilitates the cloning of different constructs into binary vectors based on the different toolkits available for CRISPR/Cas9-mediated genome editing. The methods investigated to enhance the cloning of different gRNAs into a single binary vector can be divided based on the technique used to assemble the sgRNAs together in the single delivery construct. These strategies are either based on assembling multiple individual gRNA expression cassettes, each transcribed from RNA polymerase III promoters, such as U6 or U3 (Xing et al., 2014; Lowder et al., 2015; Ma et al., 2015; Wang C. et al., 2015; Yan et al., 2015; Vazquez-Vilar et al., 2016; Zhang et al., 2016; Zhao et al., 2016; Char et al., 2017; Ordon et al., 2017; Tian et al., 2017), or through a single transcript of polycistronic mRNAs that are cleaved into individual gRNAs posttranscriptionally by the CRISPR-associated RNA endoribonuclease Csy4 from Pseudomonas aeruginosa (Tsai et al., 2014); the tRNA processing enzymes, naturally present in the host cells (Xie et al., 2015); or self-processing ribozymes (Gao and Zhao, 2014; Tang et al., 2016).
RNA Polymerase III Promoters Drive the Expression of Multiple sgRNAs
Two sets of CRISPR/Cas9 constructs have been developed by Xing et al. (2014) and Ma et al. (2015), where the gRNAs are driven by small RNA polymerase III (Figure 1A) and designed for cloning into a binary vector with prepared sgRNA expression cassettes in a single round of Golden Gate cloning using Type IIS restriction enzymes, such as BsaI. This approach has allowed for simultaneous knockout of multiple genes in Arabidopsis and rice. Both sets of CRISPR/Cas9 constructs are the most commonly used tools of this type, and both require a specific promoter to drive each sgRNA. Similarly, Lowder et al. (2015) designed an approach for MGE that uses RNA polymerase III to drive sgRNA expression in a successive three-step procedure, which is as follows: an intermediate vector carrying the sgRNAs of interest is constructed first, followed by Type IIS restriction enzyme digestions, recovery of the sgRNA cassettes, and then Golden Gate cloning into a binary vector.
Figure 1
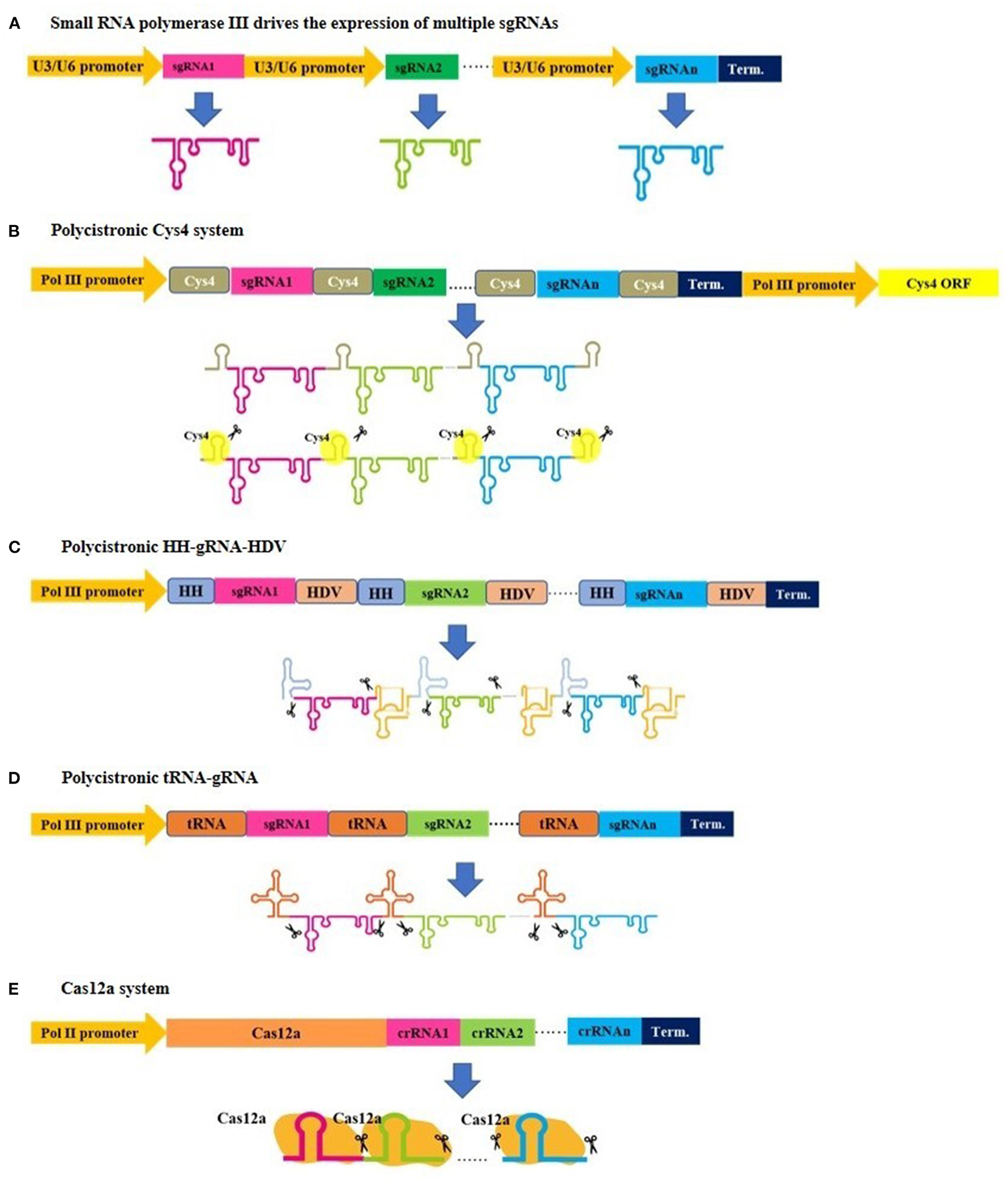
An illustration demonstrating various strategies for expressing multiplex gRNA cassettes in plants. (A) Small gRNAs are cloned after U3 or U6 promoters and derived by small RNA polymerase III to generate individual gRNAs. (B–D) Small gRNAs are cloned to be transcribed into a single transcript, and subsequent posttranscriptional processing is needed for gRNA separation, where Csy4, tRNAs, and hammerhead ribozyme regulate this separation. Similarly, a single transcript is generated in the (E) Cas12a system, but this system has a gRNA self-cleaving feature and does not require additional elements for posttranscriptional processing.
Single Transcript System for the Expression of Multiple sgRNAs
Although the abovementioned method in section RNA Polymerase III Promoters Drive the Expression of Multiple sgRNAs is frequently used for MGE, one limitation is the size of the final construct, which can sometimes be too large and repetitive due to repeating promoter sequences. These repeats can induce frequent recombination events (Vidigal and Ventura, 2015; Ding et al., 2020) and cause plasmid instability in E. coli and Agrobacterium. This might be why this method has been reported to result in the assembly of two to eight gRNA expression cassettes. Recently, Stuttmann et al. (2021) reported a new CRISPR system to allow multiplexing of up to 24 gRNAs in Arabidopsis; they observed that processing polycistronic single transcripts containing multiple gRNAs has a major advantage because they can be produced by a polymerase II promoter, which can produce longer transcripts than a polymerase III promoter (Arimbasseri et al., 2013).
In an early attempt to have a polycistronic CRISPR/Cas9 system for MGE, Tsai et al. developed a vector in which multiple gRNAs can be assembled, each flanked by cleavage sites for the Csy4 RNase of Pseudomonas aeruginosa (Haurwitz et al., 2010; Tsai et al., 2014). Based on this, each gRNA must be designed to have a 3′ end with a Csy4 recognition site to allow the coexpressed Csy4 to cleave the long transcript, thereby releasing the gRNAs (Figure 1B).
Mimicking the native tRNA-snoRNA43 dicistron (Kruszka et al., 2003) in cells, Xie et al. further designed a polycistronic tRNA-gRNA (PTG) system for simultaneous production of single gRNA transcripts (Figure 1C), in which the gRNAs are transcribed by RNA polymerase III and linked together by their tRNA sequences (Xie et al., 2015). The tRNAs are then recognized and processed by endogenous RNases (e.g., RNase P and RNase Z in plants) that excise the individual gRNAs from the transcript. These gRNAs direct Cas9 (driven by RNA polymerase II) to their targets for genome editing. This application has resulted in highly efficient genome editing, although Xie and his group observed a reduction in the efficiency of PTGs with an increasing number of gRNAs, possibly due to multiple sgRNAs competing for binding to Cas9.
In an alternative approach, Tang et al. designed a CRISPR/Cas system to allow the expression of both Cas9 and sgRNAs from a single Pol II promoter using a self-processing hammerhead ribozyme (Haseloff and Gerlach, 1988; Tang et al., 2016). The Cas9 and sgRNAs were linked by ribozyme (RZ) cleavage sites, allowing transcripts to be cleaved by the cis-acting ribozyme (Figure 1D). The approach was tested using six target sites in three rice genes, and the mutation frequencies in T0 plants ranged from 53.8 to 100% with a majority of biallelic mutations.
In the same context, Cermák et al. designed several multiplexed gene-editing tools based on the posttranscriptional processing of a single polycistronic mRNA using the CRISPR-associated Csy4 RNA endoribonuclease from Pseudomonas aeruginosa (Tsai et al., 2014; Cermák et al., 2017), the tRNA processing enzymes naturally present in the host cells (Xie et al., 2015), and ribozymes (Gao and Zhao, 2014). Furthermore, Cermák and colleagues compared the editing efficiency of these systems for targeting two sites in the AUXIN RESPONSE FACTOR 8A (ARF8A) gene of tomato, and found that Csy4 had the best efficiency, followed by the tRNA system. The trial results were confirmed with eight gRNAs and in four different tomato genes. The efficiency of the Csy4 system was further demonstrated using up to 12 gRNAs for targeted gene deletions.
The CRISPR/Cas12a (Cpf1) System: An Emerging Tool for MGE
For the aforementioned CRISPR/Cas9 systems, the size of the final construct is one of the most critical limitations in multiplexing experiments. Moreover, the requirement of GC-rich protospacer-adjacent motif (PAM) sequences is a drawback of using Cas9, especially when the number of genes to be edited is large. Furthermore, even in cases where single polycistronic multiple gRNA methods are employed, additional sequences must be added for further processing after transcription. These limitations may be overcome by a newer member of the Cas protein family, Cas12a, formerly known as Cpf1 (Zetsche et al., 2015). Cas12a has a relatively small size, its required PAM sequence allows T-rich regions to be targeted, and it requires a short crRNA (42 nt) which facilitates construction and multiplexing (Figure 1E). Moreover, the Cas12a multiplexing system has major advantages over other DNA editing systems. The Cas12a system does not require either the addition of several pol III promoters (only one Pol III promoter is enough to drive several small crRNAs) or the addition of specific sequences for multiple crRNA constructs because it can sufficiently process and mature crRNAs using its ribonuclease activity (Zetsche et al., 2015, 2017; Fonfara et al., 2016). These features make Cas12a an ideal system for MGE. Moreover, Cas12a cleaves the DSB at cohesive ends distal from the PAM site, which allows continuous cleavage of DNA (Zetsche et al., 2015). Swarts and Jinek compared the structure function relationships of Cas9 and Cas12a and their implications in genome editing in detail (Swarts and Jinek, 2018). Importantly, Cas12a was found to be very specific in plant MGE using whole-genome sequencing (Tang et al., 2018). Similar findings were reported while using CIRCLE-seq (Tsai et al., 2017; Lee et al., 2019).
Inactive Cas Proteins for MGE
Mutating specific amino acids in Cas9 and Cas12a abolishes the DNA cleavage activity of these proteins, converting them into inactive (or dead) Cas (dCas). Fusion of dCas with cytosine deaminase (CBE) or adenosine deaminase (ABE) and sgRNAs induces point mutations at the target site. Editing a single nucleotide may be sufficient to alter gene structure to obtain a desirable function (Komor et al., 2016; Eid et al., 2018; Mishra et al., 2020). Shimatani et al. used such technology to induce multiple herbicide-resistance point mutations in rice (Shimatani et al., 2017). Recently, Xu et al. increased the efficiency of this technique by 2- to 5-fold by developing a tool kit that uses high-fidelity dCas9 variants fused to a tandemly arrayed tRNA-modified sgRNA architecture in rice (Xu et al., 2019). Furthermore, oriented transcriptional regulation was achieved by the fusion of dCas enzymes to effector domains, including CRISPR-mediated inhibitor (CRISPRi) and activator (CRISPRa) (Gilbert et al., 2013; Qi et al., 2013). Lowder et al. (2017) described a toolbox for constructing and cloning multiplex CRISPR/Cas9 transcriptional activation and the repression of Arabidopsis genes.
Revolutionizing Roles of MGE in Plant Biology
CRISPR/Cas is bringing about a marked transformation in plant biology. MGE using CRISPR/Cas endows scientists with the ability to decipher very difficult biological problems by targeting different genes simultaneously. In this section, of the review, we aim to explore the impacts of the multiplex CRISPR/Cas system. Table 1 summarizes the efforts taken using MGE for revolutionizing plant biology and crop improvement.
Table 1
| MGE application | Target genes/QTLs | Crop | Objective | References |
|---|---|---|---|---|
| Plant biology | ||||
| Knock-out of multiple genes | ||||
| Two targets at ssDNA tomato yellow leaf curl virus (TYLCV) viruses | Tomato | Virus biological control in plants | Ali et al. (2015) | |
| OsGW2, OsGW5, and OsTGW6 | Rice | Larger grain size | Xu et al. (2016) | |
| RAS-PDS1 and RAS-PDS2 | Banana | Demonstration of gene knockout in banana | Kaur et al. (2018) | |
| OsSWEET11 and OsSWEET14 | Rice | Broad-spectrum disease resistance in rice | Xu et al. (2019) | |
| TMS5, Pi21, and Xa13 | Rice | Hybrid rice production and disease resistance | Li S. et al. (2019) | |
| Engineering efficient plant metabolic pathways | ||||
| OsGSTU, OsMRP15, and OsAnP | Rice | Anthocyanin synthesis pathway | Ma et al. (2015) | |
| Three different sites in the OsWaxy | Rice | Decreasing amylose content | Ma et al. (2015) | |
| Two sites in slyPDS | Tomato | Generating photobleached phenotype | Li R. et al. (2018) | |
| GABA-TP1, GABA-TP2, GABA-TP3, CAT9, and SSADH | Tomato | Metabolic engineering and manipulation of the gamma-aminobutyric acid (GABA) | Li R. et al. (2018) | |
| SGR1, LCY-E, Blc, LCY-B1, and LCY-B2 | Tomato | Increase the accumulation of lycopene | Li X. et al. (2018) | |
| GmF3H1, GmF3H2, and GmFNSII-1 | Soybean | Increasing the isoflavone content | Zhang et al. (2020) | |
| Transcriptional regulation | ||||
| Three different sites in the promoter of production of Anthocyanin Pigment1 gene (AtPAP1, encoding a transcription factor) and miR319 (encoding a microRNA) | Arabidopsis | Transcription gene activation | Lowder et al. (2015) | |
| Promoter regions of the WRKY30, RLP23, and CDG1 genes | Arabidopsis | Simultaneous activation of their transcription | Li et al. (2017) | |
| Two sites at locule number (lc) QTL + eight gRNAs targeting the promoters of SlCLV3, S and SP | Tomato | Generating desirable/beneficial regulatory variants | Rodríguez-Leal et al. (2017) | |
| OsWOX11 and OsYUC1 | Rice | Transcription gene activation | Gong et al. (2020) | |
| Chromosomal segment restructuring | ||||
| Two targets flanking a 245 kb segment | Rice | Chromosomal segment deletions | Zhou et al. (2014) | |
| Two targets flanking 1.8 kb segment | Tobacco | Chromosomal segment deletions and inversion | Gao et al. (2015) | |
| Two targets flanking a 100 bp segment | Arabidopsis | Chromosomal segment deletions | Ordon et al. (2017) | |
| 6 targets at a 58 kb region | Medicago | Chromosomal segment deletions | Cermák et al. (2017) | |
| Different genomic regions ranging from 1.7 to 13 kb | Arabidopsis | Chromosomal segment deletions | Wu et al. (2018) | |
| Two targets flanking an 18 (kb) segment | Arabidopsis | Chromosomal inversion | Schmidt et al. (2019) | |
| Multiplex base alterations | ||||
| Three sgRNAs targeting ALS and FTIP1e genes | Rice | Herbicide resistance | Shimatani et al. (2017) | |
| OsACC-T1, OsALS-T1, OsCDC48-T3, OsDEP1-T1, OsDEP1-T2, and OsNRT1.1B-T1. | Rice | Herbicide resistance | Li C. et al. (2018) | |
| AtALS, AtPDS, AtFT, and AtLFY genes and BnALS and BnPDS | Arabidopsis and Rapeseed | System verification | Kang et al. (2018) | |
| Two targets StGBSSI gene | Potato | impaired amylose biosynthesis in potato | Veillet et al. (2019) | |
| ZmALS1 and ZmALS2 | Maize | Herbicide resistance | Li Y. et al. (2019) | |
| Four targets at OsWAXY, OsCDC48, and OsSNB genes | Rice | System optimization | Wang F. et al. (2020) | |
| BnALS1 and BnALS3 | Oilseed rape | Herbicide resistance | Wu et al. (2020) | |
| Multiple sites at OsALS1 gene | Rice | Facilitating the directed evolution of plant | Kuang et al. (2020) | |
| Multiple loci at OsACC | Rice | Herbicide resistance | Liu et al. (2020) | |
| Multiple targets trials | Rice | Prime genome editing optimization | Lin et al. (2020) | |
| Crop improvement | ||||
| Generating diversity and crop domestication | ||||
| Two sites at locule number (Lc) QTL + eight gRNAs targeting the promoters of SlCLV3, S and SP. | Tomato | Wild relative domestication | Rodríguez-Leal et al. (2017) | |
| Six targets at SP, SP5G, SlCLV3 and SlWUS | Tomato | Wild relative domestication | Li T. et al. (2018) | |
| Six targets at SELF PRUNING, OVATE, FRUIT WEIGHT 2.2, LYCOPENE BETA-CYCLASE, MULTIFLORA and FASCIATED/YABBY genes | Tomato | Domestication of wild Solanum pimpinellifolium. | Zsögön et al. (2018) | |
| GS3, GW2, and GN1A | Rice | Wild relative domestication (Oryza glaberrima) | Lacchini et al. (2020) | |
| Multiple | Rice | Wild relative domestication (O. alta) | Yu et al. (2021) | |
| Increasing crop yield potential | ||||
| OsGW2, OsGW5, and OsTGW6 | Rice | Larger grain size | Xu et al. (2016) | |
| OsGS3, OsGW2, and OsGn1a | Rice | Larger grain size | Zhou et al. (2019) | |
| GS3, GW2, and GN1A | Rice | Increasing grain size | Lacchini et al. (2020) | |
| Improving crop product quality | ||||
| 4 alleles of GBSS | Potato | Produce waxy potato | Andersson et al. (2017) | |
| OVATE, FRUIT WEIGHT 2.2 and LYCOPENE BETA CYCLASE | Tomato | Fruit shape | Zsögön et al. (2018) | |
| Several sites at St16DOX | Potato | Reducing steroidal glycoalkaloids (SGAs) | Nakayasu et al. (2018) | |
| Two sites at α-gliadin gene | Wheat | Produce low-gluten wheat | Sánchez-León et al. (2018) | |
| OsGS3, OsGW2, and OsGn1a | Rice | Grain shape | Lacchini et al. (2020) | |
| OsGS3, OsGW2, and OsGn1a | Rice | Grain shape | Lacchini et al. (2020) | |
| Enhancing crop resistance/tolerance to biotic/abiotic stress | ||||
| DIPM-1, DIPM-2, and DIPM-4 | Apple | Disease resistance | Malnoy et al. (2016) | |
| OsSWEET11 and OsSWEET14 | Rice | Generating broad spectrum disease resistance | Xu et al. (2019) | |
| SWEET11, SWEET13 and SWEET14 | Rice | Generating broad spectrum disease resistance | Oliva et al. (2019) | |
| TMS5, Pi21, and Xa13 | Rice | Disease resistance and yield enhancement | Li S. et al. (2019) | |
Summary presentation of the efforts done using multiplex genome editing for revolutionizing plant biology and crop improvement programs.
Knock-Out of Multiple Genes at Once
Loss of function or gene knockout has been widely investigated in plant genome-editing studies mainly because of the activation of the naturally occurring non-homologous end joining (NHEJ) repair pathway after Cas9 protein was reported to cause DSBs at the target DNA site. NHEJ is an error-prone repair mechanism that can generate insertion/deletion (Indels) of nucleotides near the cleavage site (Mao et al., 2013; Xie and Yang, 2013; Ma et al., 2015). These Indels mostly cause a frameshift mutation of the codon. Successful implementation of multiple gene knockouts mainly depends on the precise selection of the target loci.
In this regard, abundant studies have described multiple gene knockouts in different plant species for various purposes. Ali et al. (2015) explained the potential of CRISPR/Cas9 utilization in multiplexing two sgRNAs to knockout ssDNA viruses and expanded the possibilities of virus biological control in plants. In another attempt, Xu et al. generated a multiplex knockout to develop broad-spectrum disease resistance in rice by using a multiplex CRISPR/Cas9 system to alter the effector-binding elements (EBEs) of the bacterial blight TAL effector for two susceptibility genes, OsSWEET11 and OsSWEET14, in rice cv. Kitaake (harboring the defeated OsSWEET13) (Xu et al., 2019). Furthermore, multiple gene knockouts have been involved in metabolite manipulations. In this regard, Kaur et al. designed a single gRNA to target the conserved regions of the two genes encoding RASPDS (Rasthali phytoene desaturase) of the banana cv. Rasthali (Kaur et al., 2018). PDS is one of the carotenoid biosynthesis pathway enzymes that limit the rate of carotenoid synthesis (Cazzonelli and Pogson, 2010; Bai et al., 2016), and its disruption impairs chlorophyll, carotenoid, and gibberellin biosynthesis, thereby producing albino plants. Moreover, multiplex knockout trials indicated the great promise in overcoming redundancy and polyploidy in double- and higher-order knockouts created by multiplex CRISPR/Cas9 in tomatoes (Li W. et al., 2016).
Engineering-Efficient Plant Metabolic Pathways
Reducing or increasing the levels of specific metabolites through modulation of plant endogenous metabolic pathways has become achievable with the application of MGE. MGE has provided opportunities to investigate the roles of multiple genes in synthetic pathways in plants. For instance, using MGE, Ma et al. confirmed the role of OsGSTU, OsMRP15, and OsAnP genes in the anthocyanin synthesis pathway in rice (Ma et al., 2015). Moreover, the authors decreased the amylose content in rice by targeting three different sites in the OsWaxy gene.
Double and higher-order loss-of-function created by CRISPR/Cas9 offers great promise in modulating multiple genes in a single pathway or multiple pathways. Li et al. assessed the efficiency of multiplexing in tomatoes using the robust CRISPR/Cas9 system, as described by Ma et al. (2015) and Li X. et al. (2018) by targeting two loci in exons 6 and 7 of the tomato PDS gene (slyPDS). A strong photobleached phenotype was exhibited among the transgenic lines, and some T0 lines had concurrent homozygous modifications at both target sites. Subsequently, the authors used this system for metabolic engineering and manipulation of the gamma-aminobutyric acid (GABA) shunt metabolic pathway in tomatoes. They targeted five key genes in its pathway, GABA-TP1, GABA-TP2, GABA-TP3, CAT9, and SSADH. The researchers successfully edited five sites, whereas no editing events occurred in the third site, which was thought to be caused by the lack of CG content at this site. Quadruple mutant plants were obtained, which expressed the highest GABA levels in leaves, reaching almost 19-fold higher than that in wild-type plants. Li et al. discussed the relationship between various pathway components and the effects of mutations on certain components in the pathway. These results indicated the power of multiplex CRISPR/Cas9 as a tool for plant metabolic engineering and will improve our understanding of plant polygenic functional outcomes. In a bidirectional strategy, the same group targeted the loss-of-function of five different tomato genes using six gRNAs to increase the accumulation of lycopene while inhibiting its conversion to β- and α-carotene (Li X. et al., 2018).
Some plant metabolites, such as isoflavone, are controlled by multiple genes and involve specific pathways content (Li and Zhang, 2017). These compounds not only benefit human health but also play a critical role in the plant-environmental interactions for plant fitness in certain environments. Zhang et al. designed a single CRISPR/Cas9 binary vector to target three genes encoding key enzymes in the competing metabolic isoflavone biosynthesis pathway with the aim of increasing the isoflavone content in soybean plants (Zhang et al., 2020). The triple mutant (T3 homozygous plants) plant leaves had two times the level of isoflavone compared with that of wild-type plant leaves. The results of this study indicated the possibility of using MGE for loss-of-function competing pathway genes to increase the availability of specific plant substances or metabolites for another pathway.
Transcriptional Regulation
Controlling gene expression has long been a goal of plant scientists for deciphering plant gene function and improving crop traits. CRISPR-mediated gene regulation, designed by engineering a nuclease-inactivated Cas9 (dCas9) protein with a transcriptional activation domain (VP64) or repressor domains (SRDX), has been very powerful in activating or repressing the endogenous expression of plant genes of interest (Lowder et al., 2017; Li Z. et al., 2019; Malzahn et al., 2019). With the help of gRNA, the dCas9 fusion protein can specifically bind to the promoter region and manipulate the expression of downstream genes.
To date, MGE-mediated transcription manipulation has mainly been focused on promoters implicated in gene repression, activation, and epigenetic modification. An increase of ~80-, 37-, and 192-fold was reported when targeting the promoter regions of the WRKY30, RLP23, and CDG1 genes for simultaneous activation of their transcription in Arabidopsis (Li et al., 2017). Meanwhile, several gRNAs simultaneously targeting the same promoter in plants using dCas9-VP64 displayed synergistic effects (Lowder et al., 2015, 2018; Vazquez-Vilar et al., 2016). To determine the efficiency of CRISPR/Cas9 in controlling the gene expression of protein-coding and non-coding genes in Arabidopsis, additive transcription activation was observed by Lowder et al., who targeted three different sites in the promoter of the production of anthocyanin pigment1 gene (AtPAP1, encoding a transcription factor) and miR319 (encoding a microRNA; Lowder et al., 2015). Gong et al. (2020) targeted two different genes, OsWOX11 and OsYUC1, involved in crown root development and auxin biosynthesis in rice, respectively. They designed multiple gRNAs to target different sites in the promoter regions upstream of these genes, and the results indicated that targeting two different regions within 350 bp upstream of the TSS is most effective for dCas9-TV-based transcriptional activation.
With the flexibility and expandability of CRISPR/Cas9, it is possible to introduce several regulatory mutations of cis-regulatory elements (CREs), such as promoter enhancers and silencer sequences that regulate gene transcription, which could lead to a range of quantitative transcriptional and phenotypic changes. Rodríguez-Leal et al. (2017) provided a prime example of the utilization of a multiplex CRISPR/Cas9 system to fine-tune gene expression by targeting eight sites in the promoter regions of different tomato genes. The results have demonstrated that alteration of gene expression based on editing the CREs of key developmental transcription factors promises an attractive approach for generating desirable/beneficial regulatory variants, offering great potential for crop molecular breeding.
Chromosomal Segment Restructuring
Chromosomal rearrangements include transposition, duplication, deletion, inversion, or translocation of nucleic acid segments (Gray, 2000). Chromosomal rearrangements occur regularly in plants (Udall et al., 2005; Szinay et al., 2012; Li J. et al., 2016; Zapata et al., 2016; Schmidt et al., 2019). The introduction of two DSBs in a single chromosome by multiple editing mainly leads to deletions (Siebert and Puchta, 2002) and sometimes inversions (Qi et al., 2013; Zhang et al., 2017), whereas two breaks on different plant chromosomes could lead to the occurrence of reciprocal translocation (Pacher et al., 2007).
Induction of deletions, inversions, and translocation using MGE has been employed in mammals and reviewed elsewhere (Cheong et al., 2018). Most of the reports about chromosomal segment restructuring in plants focused on introducing deletions. Deletions smaller than 100 bp were detected at high frequencies in the N. benthamiana T0 and Arabidopsis T2 populations, whereas deletions larger than 120 bp were still feasible, but at lower frequencies in Arabidopsis (Ordon et al., 2017). Durr et al. (2018) aimed to improve the propagation of deletions occurring in somatic cells by direct plant regeneration. Furthermore, different investigators have reported the generation of heritable deletions of genomic regions using CRISPR/Cas9 in Arabidopsis, Medicago, and rice (Zhou et al., 2014; Gao et al., 2015; Cermák et al., 2017; Wu et al., 2018). More recently, a unique study on inversion formation in Arabidopsis was reported, where an 18 kilobase (kb) segment was successfully inverted and transmitted to the next generation (Schmidt et al., 2019). The impact of these chromosome restructuring methods in the field of plant biology is remarkable because it is now feasible to redesign chromosomes according to specific needs (Schmidt et al., 2020).
Multiplex Base Alterations
This application utilizes the CRISPR/Cas system to modify individual bases at multiple sites simultaneously for a gain of function. The establishment of this technology first relied on the fusion of cytosine deaminase or adenosine deaminase to the artificially mutated DNA nickase nCas9, which forms the cytosine editor (CBE) and adenine editor (ABE), respectively. In addition, the use of uracil glycosylase inhibitory protein (UGI) can increase the stability of uracil in DNA, resulting in enhanced editing efficiency. Combined with the multiple-targeting feature of sgRNAs, base editing can be achieved at multiple sites of the genome. Both CBE and ABE have been successfully applied in various model plants and crops (Chen et al., 2019). Most multiplexing base editing researches have focused on herbicide resistance (Shimatani et al., 2017; Kang et al., 2018; Li C. et al., 2018; Li Y. et al., 2019; Veillet et al., 2019; Wu et al., 2020). The reported maximum number of multiplexed sgRNAs in a single construct is four targets in rice, two targets at OsWAXY, and one at OsCDC48 and OsSNB (Wang F. et al., 2020). Shimatani et al. (2017) reported a system with three sgRNAs targeting ALS and FTIP1e that was designed to generate double mutants resulting in the resistance to two herbicides. Moreover, using gRNA libraries, base editors can generate high-density substitutions in target regions, thus facilitating the directed evolution of plant genes (Kuang et al., 2020; Li et al., 2020; Liu et al., 2020). The development of different base-editing platforms was reviewed in detail in Mishra et al. (2020). Ren et al. developed a CRISPR/Cas9 toolkit, extensively enabling efficient base editing at desired sites in the rice genome (Ren et al., 2017). Multiple genomic sites could be targeted for single-base modification. These toolkits were further improved to increase the possibility of generating gain-of-function and loss-of-function mutants for several traits of agronomic importance through base editing (Ren et al., 2018). More recently, Li et al. (2020), created a system that enables the creation of saturated targeted endogenous mutagenesis editors (STEMEs) that allow simultaneous C to T and A to G conversions at specific target sites in plants, where both CBE and ABE are fused together with cas9 nickase and UGI.
A more advanced editing technique that efficiently produces all possible base conversions without the mandate of DSBs or donor DNA was developed, and it is called prime editing (Anzalone et al., 2019). This system uses a catalytically impaired Cas9 endonuclease fused to an engineered reverse transcriptase programmed with a prime editing guide RNA (pegRNA), allowing new genetic information to be written at a specific locus (Lin et al., 2020). Although the efficiency of prime editors is lower than that of base editors, the system is still a versatile and appealing new tool for creating all 12 types of single-point mutations, mixtures of several substitutions, and indels. Gao and her group used this system to develop mutant rice plants carrying G to T mutations, multinucleotide substitutions, and a number of desired nucleotide deletions (Lin et al., 2020). To date, it is very difficult to achieve these types of mutations using any other current editing tool.
MGE in Crop Improvement
Multiplex genome-editing is an innovative approach that accelerates the crop improvement cycle and makes the expected outcome more feasible and achievable. This technology provides effective solutions to overcome the limitations of plant breeding approaches, particularly for providing required variants that cause characteristics desired by breeders at a fast pace without labor-intensive phenotypic and molecular evaluation of breeding germplasm. Although MGE applications are relatively new, the technology has been widely used to improve a diverse range of crops to gain in yield, quality, and nutritional value, herbicide resistance, and biotic and abiotic stress tolerance. In this section, we describe the revolution in crop improvement programs by introgression of this technology for editing plant genotypes to accomplish different breeding objectives.
Generating Diversity and Crop Domestication Using MGE
Plant breeders aiming to develop new genotypes always seek to improve specific plant characteristics, and to achieve their goal, they unintentionally decrease genetic diversity among the crop genotypes. Furthermore, most of the studies on genetic diversity and QTLs depend on naturally occurring mutations that rarely occur in gene-regulatory regions such as promoters and upstream open-reading frames (uORFs) (Rodríguez-Leal et al., 2017), whereas traditional breeding often requires more effort to screen and utilize QTLs carrying favorable mutations.
The CRISPR/Cas9 may allow new genetic diversity to be generated by targeting specific genome loci, which may lead to the identification of new genes. In this context, Rodríguez-Leal et al. demonstrated that genome-editing tools can introduce new cis-regulatory alleles with the aim of identifying new alleles that provide beneficial quantitative traits (Rodríguez-Leal et al., 2017). A CRISPR/Cas9-based system was reported that targets the promoter sequence of genes to regulate three major productivity traits, namely, fruit size, inflorescence branching, and plant architecture in tomatoes. The results demonstrated a wide range of variations that could be achieved by altering the expression of the target genes.
Multiplex genome-editing further accelerates the domestication of crop wild relatives by modifying domestication genes. Domestication genes with marked effects on specific key phenotypes have been reported in barley (Komatsuda et al., 2007), maize (Wang H. et al., 2015), rice (Civán and Brown, 2017), and tomato (Li T. et al., 2018). Zsögön et al. designed a multiplex CRISPR–Cas9 strategy to target six genes in wild Solanum pimpinellifolium that are important for productivity in the present tomato crop breeding lines (Zsögön et al., 2018). The mutants were modified in their fruit number, size, shape, nutrient content, and plant architecture in a single generation and within a single transformation event. In the same context, with the aim of maintaining favorable characteristics of wild ancestors of Solanum pimpinellifolium, Li T. et al. (2018) edited four lines of four wild tomato accessions, all of which were resistant to bacterial spot disease and two of which were salt-tolerant, using the Csy4 multi-gRNA CRISPR/Cas9 system to target coding sequences, cis-regulatory regions, or uORFs of four genes associated with morphology, flower and fruit production, and ascorbic acid synthesis. The generated Cas9-free genotypes had a domesticated phenotype with parental disease resistance and salt tolerance. African rice, Oryza glaberrima, is a wild relative of the cultivated Asian rice Oryza sativa. African rice has specific characteristics to withstand diverse environmental stresses but has low yield ability. Lacchini et al. (2020) disrupted domestication loci of the cultivated African landrace Kabre, which resulted in mutants with significantly improved grain yield. Furthermore, domesticating allotetraploid rice was demonstrated by targeting six agronomically important traits of O. alta (an allotetraploid perennial wild rice) (Yu et al., 2021).
Increasing Crop Yield Potential by MGE
High-yielding ability under different environments is the core trait behind developing new crop genotypes. Therefore, it is necessary to introduce multiple QTLs into an elite genotype to withstand different environmental stresses without yield penalties. These QTLs are generally located in different genetic backgrounds, and there is no certain guarantee that they will perform the best when they are transferred to other genetic backgrounds. Even in conventional crop breeding, this transfer requires multiple rounds of crossing and selection, which cost labor, time, and money, and linkage drag might often interrupt efforts with unfavored genes. MGE allows knocking out of multiple genes with negative effects on yield simultaneously from genotypes with specific resistance to environmental stress in a single generation. Xu et al. (2016) targeted three major genes, namely OsGW2, OsGW5, and OsTGW6, which negatively regulate rice grain weight. Double (gw5tgw6) and triple (gw2gw5tgw6) T1 mutants showed larger grain sizes than wild-type rice. Zhou et al. (2019) targeted negatively regulated genes for grain size, width, weight, and number, namely OsGS3, OsGW2, and OsGn1a, respectively, which directly affected grain yield, in three elite rice varieties. Similar results were obtained by Lacchini et al. (2020) after targeting the same genes (OsGS3, OsGW2, and OsGn1a) in a mutated African rice cv. Kabre with a disrupted HTD1 gene (short stature compared to the wild type).
Improving Crop Product Quality by MGE
Since yield is the most important trait for breeders, the adoption of new genotypes by farmers is strongly driven by crop quality traits (Dalton, 2004; Wang et al., 2012; Abdelrahman and Zhao, 2020). High-yielding varieties with improved quality are more acceptable to consumers, which in turn increases commercial value for the crop. Quality characteristics differ among crops; however, physical properties, eating quality, and nutritional properties are core traits.
Most of the studies that focus on the domestication of crop plants consider quality genes, among others, during planning for multiplex editing of different genes. Zsögön et al. (2018) aimed to create a novel genotype derived from tomato wild-type S. pimpinellifolium by generating loss-of-function mutations in six genes using MGE. Among the edited genes, three genes were related to the quality traits of fruit shape, fruit weight, and lycopene content (OVATE, FRUIT WEIGHT 2.2, and LYCOPENE BETA CYCLASE, respectively). Mutants with loss-of-function in the OVATE gene had an oval fruit shape, while no changes in fruit sizes were reported for FRUIT WEIGHT 2.2 mutants. Meanwhile, mutants of LYCOPENE BETA CYCLASE had 100% more lycopene without a negative effect on β-carotene or lutein compared with the wild type. The level of lycopene, which is a kind of carotenoid, is one of the most important nutritional quality traits of tomato fruit. Li et al. successfully increased lycopene in tomato fruits to ~5.1-fold by using multiplex CRISPR/Cas9 genome editing of five genes that promote the synthesis of lycopene (SGR1 gene) and genes that catalyze the cyclization of lycopene (LCY-E, LCY-B1, and LCY-B2, as well as Blc) (Li X. et al., 2018). The strategy was to promote lycopene biosynthesis while inhibiting its conversion into β-carotene and α-carotene. However, the increased number of mutant genes did not result in a concomitant increase in lycopene (Li X. et al., 2018). Instead, the SGR1 single mutant exhibited the highest lycopene level followed by the plants with triple mutations in SGR1, Blc, and LCY-E.
Multiplex genome-editing has been successfully implemented for altering physical and nutritional quality properties. Lacchini et al. (2020) improved the grain shape of an Oryza glaberrima landrace when multiplexed editing of the OsGS3, OsGW2, and OsGn1a genes was performed. Kim et al. (2017) designed nine crRNAs to simultaneously target two homologous genes, FATTY ACID DESATURASE 2-1A (FAD2-1A, Glyma10g42470) and FAD2-1B (Glyma20g24530), in the soybean genome using CRISPR/Cas12a. FAD2 encodes an enzyme that is responsible for the conversion of monounsaturated oleic fatty acid to a polyunsaturated linoleic fatty acid. Sánchez-León et al. (2018) designed two sgRNAs to target the α-gliadin gene using CRISPR/Cas9 to produce low-gluten wheat.
Moreover, several studies have reported improvements in the eating quality of crops using MGE. In potato (Solanum tuberosum), reducing steroidal glycoalkaloids (SGAs) is a requisite in breeding programs, as the accumulation of SGAs causes unfavorable taste. Nakayasu et al. (2018) multiplexed several sgRNAs to knock out a single gene (St16DOX; encoding a steroid 16α-hydroxylase in SGA biosynthesis) using hairy root culture and succeeded in developing SGA-free hairy roots of tetraploid potato (Nakayasu et al., 2018). In another study aiming to enhance the potato starch quality, Andersson et al. used three sgRNAs to target the four alleles of the potato granule-bound starch synthase (GBSS) gene to produce waxy potato (Andersson et al., 2017). Knocking out the four alleles caused a reduction in GBSS enzyme activity, leading to alteration in starch composition with a concomitant increase in the amylopectin/amylose ratio. Furthermore, mutants with only one functional allele still produced a significant amount of amylose.
Enhancing Crop Resistance/Tolerance to Biotic/Abiotic Stress by MGE
Biotic and abiotic stress threatens food security, causing a reduction in crop production. Abiotic stress is more prominent during global environmental changes. Drought, salinity, and extreme temperatures are the main abiotic stressors that cause biological and biochemical effects on plant growth and development. Most of the gene editing for abiotic stress tolerance has been simplex targeting, i.e., one gene related to one constraint. Zafar et al. reviewed the efforts that have been made for the development of mutations for abiotic stress (Zafar et al., 2020). Ethylene-responsive factors (ERFs) are proteins that enhance plant tolerance to different abiotic stressors by activating stress-responsive genes. However, multiplex gene editing for biotic stress was more feasible because of the ease of targeting diseases susceptibility (S) genes, which confer a negative effect on disease resistance. Recently, Oliva et al. used CRISPR–Cas9-mediated genome editing to introduce mutations in the promoters of three SWEET genes (SWEET11, SWEET13, and SWEET14) of rice cv. Kitaake, IR64, and Ciherang-Sub1 for generating broad-spectrum bacterial blight resistance genotypes (Oliva et al., 2019). With the same objectives, Xu et al. (2019) generated mutations in two EBEs of the S genes OsSWEET11 and OsSWEET14 in rice cv. Kitaake. The resulting mutants were stable and consistently exhibited broad-spectrum resistance to most bacterial blight strains, with few exceptions in two generations of rice lines. Malnoy et al. (2016) mutated DIPM-1, DIPM-2, and DIPM-4 to increase apple resistance to fire blight disease. Disease genes were also multiplexed with other traits to broaden the scope of the multiplexing objectives. Li S. et al. (2019) introduced mutations into the TMS5, Pi21, and Xa13 genes in rice. Triple T1 mutants showed characteristics of thermosensitive genic male sterility (TGMS) with an enhanced blast and bacterial blight resistance. MGE will pave the way toward sustainable development of disease resistance by targeting more S genes in different plant species.
Conclusion and Future Perspectives
Conventional plant breeding has limitations in meeting food security goals for ever-growing populations around the world. However, practical operations of screening germplasm for genetic variations, developing breeding populations, mutation breeding, and hybridization are usually labor-intensive and time-consuming. Genome editing is an innovative technology that is considered an important tool for plant biology studies and for crop improvements. Using MGE to simultaneously target different genomic regions has accelerated both basic and applied researches. Different applications of multiplex technology have been developed that allowed robust, routine CRISPR reagent delivery systems in plants. All these improvements made it easier to utilize MGE technology for enhancing plant biology knowledge by knocking out multiple genes to disrupt gene function and decipher their roles in metabolic pathways. Furthermore, multiplex CRISPR-mediated gene editing provides a gene regulation tool to activate or repress the expression of multiple genes simultaneously. With multiplex CRISPR/Cas9, it is possible to reformat and rearrange chromosomal segments based on the needs of biologists. The utilization of MGE may increase the effect of off-target occurrence as the number of targets increased (McCarty et al., 2020). Several online toolboxes for off-target detection and expectations have been developed and updated for eliminating the percentage of off-target occurrence (Cradick et al., 2014; Kang et al., 2020; Naeem et al., 2020; Störtz and Minary, 2021).
Applications of multiplex base editing have facilitated gain-of-function investigations; however, base editing generates mainly C–T and A–G conversions to date. More recently, prime editing has advanced base editing so that new genetic information can be written into specified DNA loci. This technology will expand the capabilities of MGE that may allow the gain-of-function of multiple genes simultaneously. With the identification and utilization of CRISPR systems, breeding schemes have become faster and more precise. Generating mutations with MGE allows diversity for selection and yield enhancement considering quality aspects. Improving specific traits without any other alteration to others may become possible in the near future.
Multiplex genome-editing will be enhanced by skipping long laborious and complicated plant regeneration and tissue culture steps by allowing CRISPR reagents to be delivered directly into plant meristems or pollen tissues, as reported by Maher et al. (2020), which presented a successful example of tissue culture-free plant gene editing. Furthermore, it is now also possible to write new genetic information to a specific locus in the genome (Lin et al., 2020). Despite diverging social perspectives, it is believed that multiplex CRISPR technology will play an important role in agricultural sustainability by developing resilient germplasm.
Implementation of MGE is necessary for understanding the dynamics of synthetic metabolic pathways. In addition, exploring enzyme functionalities in different pathways and their relationship with other omics information is crucial. Additionally, more attention should be devoted to utilizing this technology for the exploration of interactions among different metabolic pathways and gene regulatory, signal transduction, and protein-protein interaction networks in response to biotic and abiotic stress.
Advanced knowledge regarding plant biology, agronomy, and breeding supports crop improvement programs. Through the utilization of such knowledge and new modeling approaches to predict the performance of the genetic variants (Wang H. et al., 2020), MGE will accelerate the development of elite crop cultivars with precise genomic modifications (Figure 2). MGE will increase and facilitate new plant biology discoveries and/or the development of new elite germplasm.
Figure 2
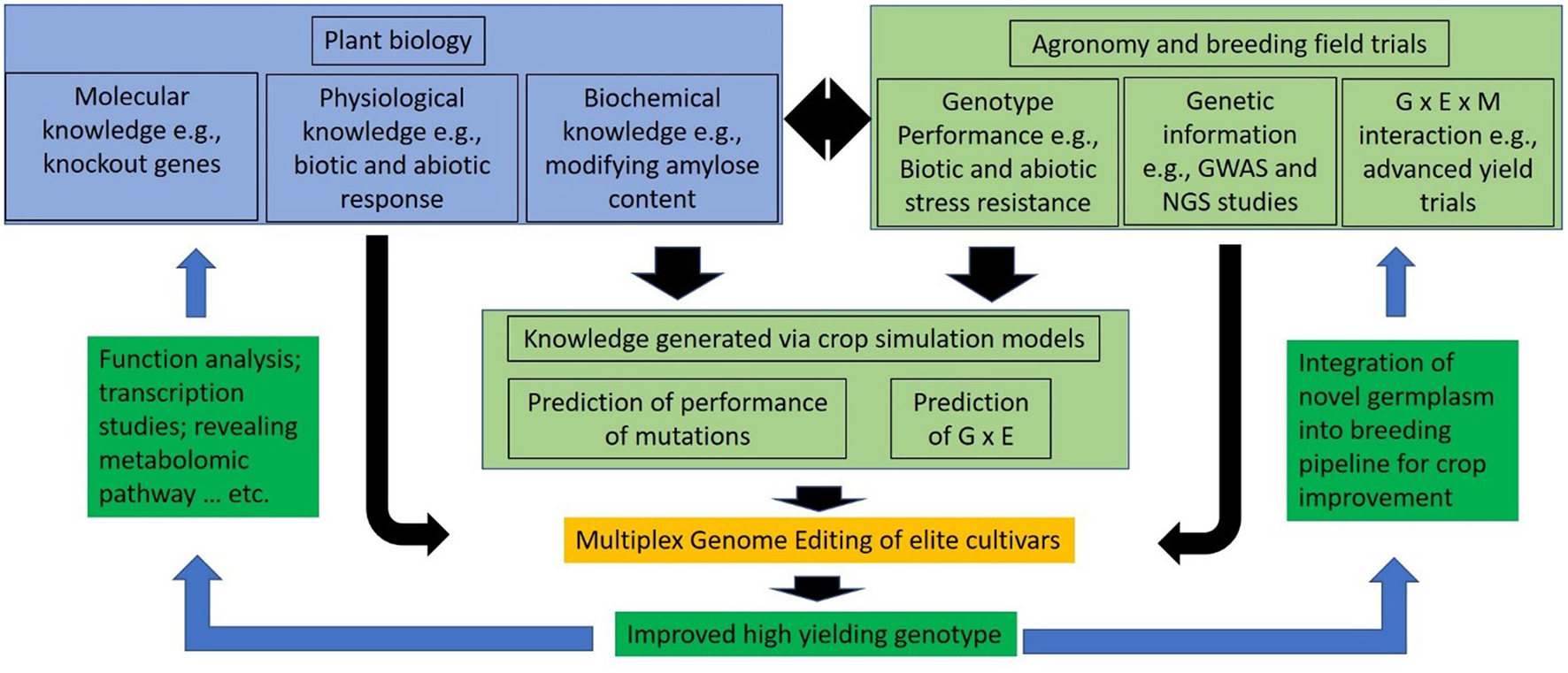
The role of multiplex genome editing (MGE) is explained: the development of novel germplasm is accelerated that either increases knowledge in plant biology or assists crop improvement programs.
Funding
This research was supported by grants from the Key R& D project in Sichuan Province of China (2021YFN0003 to KZ), the National Natural Science Foundation of China (U20A2035 to KZ), and the Talented Young Scientist Program of China (AiJi-18-062 to MA).
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Author contributions
MA and KZ designed and discussed the content. MA and ZW wrote the first draft of the manuscript. JR and KZ critically revised the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
Abdelrahman M. Zhao K. (2020). Genome editing and rice grain quality, in The Future of Rice Demand: Quality Beyond Productivity, eds Costa de OlivieraA.PegoraroC.Ebeling VianaV. (Charm: Springer), 395–422.
2
Ainley W. M. Sastry-Dent L. Welter M. E. Murray M. G. Zeitler B. Amora R. et al . (2013). Trait stacking via targeted genome editing. Plant Biotechnol. J. 11, 1126–1134. 10.1111/pbi.12107
3
Ali Z. Abulfaraj A. Idris A. Ali S. Tashkandi M. Mahfouz M. M. (2015). CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 16:238. 10.1186/s13059-015-0799-6
4
Andersson M. Turesson H. Nicolia A. Fält A. S. Samuelsson M. Hofvander P. (2017). Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep. 36, 117–128. 10.1007/s00299-016-2062-3
5
Anzalone A. V. Randolph P. B. Davis J. R. Sousa A. A. Koblan L. W. Levy J. M. et al . (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature576, 149–157. 10.1038/s41586-019-1711-4
6
Arimbasseri A. G. Rijal K. Maraia R. J. (2013). Transcription termination by the eukaryotic RNA polymerase III. Biochimica Biophys. Acta Gene Regul. Mech. 1829, 318–330. 10.1016/j.bbagrm.2012.10.006
7
Bai C. Capell T. Berman J. Medina V. Sandmann G. Christou P. et al . (2016). Bottlenecks in carotenoid biosynthesis and accumulation in rice endosperm are influenced by the precursor–product balance. Plant Biotechnol. J.14, 195–205. 10.1111/pbi.12373
8
Belhaj K. Chaparro-Garcia A. Kamoun S. Patron N. J. Nekrasov V. (2015). Editing plant genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 32, 76–84. 10.1016/j.copbio.2014.11.007
9
Bibikova M. Golic M. Golic K. G. Carroll D. (2002). Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics161, 1169–1175. 10.1093/genetics/161.3.1169
10
Cazzonelli C. I. Pogson B. J. (2010). Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant Sci.15, 266–274. 10.1016/j.tplants.2010.02.003
11
Cermák T. Curtin S. J. Gil-Humanes J. Cegan R. Kono T. J. Konečná E. et al . (2017). A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell. 29, 1196–1217. 10.1105/tpc.16.00922
12
Char S. N. Neelakandan A. K. Nahampun H. Frame B. Main M. Spalding M. H. et al . (2017). An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotechnol. J. 15, 257–268. 10.1111/pbi.12611
13
Chen K. Wang Y. Zhang R. Zhang H. Gao C. (2019). CRISPR/Cas genome editing and precision plant breeding in agriculture. Ann. Rev. Plant Biol. 70, 667–697. 10.1146/annurev-arplant-050718-100049
14
Cheong T. C. Blasco R. B. Chiarle R. (2018). The CRISPR/Cas9 system as a tool to engineer chromosomal translocation in vivo, in Chromosome Translocation, eds ZhangY. (Singapore: Springer), 39–48.
15
Cho S. W. Kim S. Kim J. M. Kim J. S. (2013). Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31:230. 10.1038/nbt.2507
16
Christian M. Cermák T. Doyle EL Schmidt C. Zhang F. Hummel A. Bogdanove A. J. et al . (2010). Targeting DNA double-strand breaks with TAL effector nucleases. Genetics186, 757–761. 10.1534/genetics.110.120717
17
Civán P. Brown T. A. (2017). Origin of rice (Oryza sativa L.) domestication genes. Genetic Resources Crop Evolution64, 1125–1132. 10.1007/s10722-017-0518-0
18
Cong L. Ran F. A. Cox D. Lin S. Barretto R. Habib N. et al . (2013). Multiplex genome engineering using CRISPR/Cas systems. Science339, 819–823. 10.1126/science.1231143
19
Cradick T. J. Qiu P. Lee C. M. Fine E. J. Bao G. (2014). COSMID: a web-based tool for identifying and validating CRISPR/Cas off-target sites. Mol. Ther. Nucl. Acids3:e214. 10.1038/mtna.2014.64
20
Dalton T. J. (2004). A household hedonic model of rice traits: economic values from farmers in West Africa. Agric. Econ. 31, 149–159. 10.1111/j.1574-0862.2004.tb00253.x
21
Ding T. Huang C. Liang Z. Ma X. Wang N. Huo Y. X. (2020). Reversed paired-gRNA plasmid cloning strategy for efficient genome editing in Escherichia coli. Microbial Cell Factories19, 1–3. 10.1186/s12934-020-01321-4
22
Durr J. Papareddy R. Nakajima K. Gutierrez-Marcos J. (2018). Highly efficient heritable targeted deletions of gene clusters and non-coding regulatory regions in Arabidopsis using CRISPR/Cas9. Sci. Rep. 8:1. 10.1038/s41598-018-22667-1
23
Eid A. Alshareef S. Mahfouz M. M. (2018). CRISPR base editors: genome editing without double-stranded breaks. Biochem. J. 475, 1955–164. 10.1042/BCJ20170793
24
Fonfara I. Richter H. Bratovič M. Le Rhun A. Charpentier E. (2016). The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature532, 517–521. 10.1038/nature17945
25
Gao J. Wang G. Ma S. Xie X. Wu X. Zhang X. et al . (2015). CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol. Biol. 87, 99–110. 10.1007/s11103-014-0263-0
26
Gao Y. Zhao Y. (2014). Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J. Integr. Plant Biol. 56, 343–349. 10.1111/jipb.12152
27
Gilbert L. A. Larson M. H. Morsut L. Liu Z. Brar G. A. Torres S. E. et al . (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell154, 442–451. 10.1016/j.cell.2013.06.044
28
Gong X. Zhang T. Xing J. Wang R. Zhao Y. (2020). Positional effects on efficiency of CRISPR/Cas9-based transcriptional activation in rice plants. aBIOTECH1, 1–5. 10.1007/s42994-019-00007-9
29
Gray Y. H. (2000). It takes two transposons to tango: transposable-element-mediated chromosomal rearrangements. Trends Genet.16, 461–468. 10.1016/S0168-9525(00)02104-1
30
Haseloff J. Gerlach W. L. (1988). Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature334, 585–591. 10.1038/334585a0
31
Haurwitz R. E. Jinek M. Wiedenheft B. Zhou K. Doudna J. A. (2010). Sequence-and structure-specific RNA processing by a CRISPR endonuclease. Science329, 1355–1358. 10.1126/science.1192272
32
Hsu P. D. Scott D. A. Weinstein J. A. Ran F. A. Konermann S. Agarwala V. et al . (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31:827. 10.1038/nbt.2647
33
Jinek M. Chylinski K. Fonfara I. Hauer M. Doudna J. A. Charpentier E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science337, 816–821. 10.1126/science.1225829
34
Kang B. C. Yun J. Y. Kim S. T. Shin Y. Ryu J. Choi M. et al . (2018). Precision genome engineering through adenine base editing in plants. Nat. Plants. 4, 427–431. 10.1038/s41477-018-0178-x
35
Kang S. H. Lee W. J. An J. H. Lee J. H. Kim Y. H. Kim H. et al . (2020). Prediction-based highly sensitive CRISPR off-target validation using target-specific DNA enrichment. Nat. Commun. 11:1. 10.1038/s41467-020-17418-8
36
Kannan B. Jung J. H. Moxley G. W. Lee S. M. Altpeter F. (2018). TALEN-mediated targeted mutagenesis of more than 100 COMT copies/alleles in highly polyploid sugarcane improves saccharification efficiency without compromising biomass yield. Plant Biotechnol. J. 16, 856–866. 10.1111/pbi.12833
37
Kaur N. Alok A. Kaur N. Pandey P. Awasthi P. Tiwari S. (2018). CRISPR/Cas9-mediated efficient editing in phytoene desaturase (PDS) demonstrates precise manipulation in banana cv. Rasthali genome. Funct. Integr. Genomics18, 89–99. 10.1007/s10142-017-0577-5
38
Kim H. Kim S. T. Ryu J. Kang B. C. Kim J. S. Kim S. G. (2017). CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat. Commun. 8, 1–7. 10.1038/ncomms14406
39
Komatsuda T. Pourkheirandish M. He C. Azhaguvel P. Kanamori H. Perovic D. et al . (2007). Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc. Natl. Acad. Sci. U.S.A.104, 1424–1429. 10.1073/pnas.0608580104
40
Komor A. C. Kim Y. B. Packer M. S. Zuris J. A. Liu D. R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature533, 420–424. 10.1038/nature17946
41
Kruszka K. Barneche F. Guyot R. Ailhas J. Meneau I. Schiffer S. et al . (2003). Plant dicistronic tRNA–snoRNA genes: a new mode of expression of the small nucleolar RNAs processed by RNase Z. EMBO J. 22, 621–632. 10.1093/emboj/cdg040
42
Kuang Y. Li S. Ren B. Yan F. Spetz C. Li X. et al . (2020). Base-editing-mediated artificial evolution of OsALS1 in planta to develop novel herbicide-tolerant rice germplasms. Mol. Plant28, 565–572. 10.1016/j.molp.2020.01.010
43
Lacchini E. Kiegle E. Castellani M. Adam H. Jouannic S. Gregis V. et al . (2020). CRISPR-mediated accelerated domestication of African rice landraces. PLoS ONE15:e0229782. 10.1371/journal.pone.0229782
44
Lee K. Zhang Y. Kleinstiver B. P. Guo J. A. Aryee M. J. Miller J. et al . (2019). Activities and specificities of CRISPR/Cas9 and Cas12a nucleases for targeted mutagenesis in maize. Plant Biotechnol. J. 17, 362–372. 10.1111/pbi.12982
45
Li C. Zhang R. Meng X. Chen S. Zong Y. Lu C. et al . (2020). Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat. Biotechnol. 13, 1–8. 10.1038/s41587-019-0393-7
46
Li C. Zong Y. Wang Y. Jin S. Zhang D. Song Q. et al . (2018). Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 19:59. 10.1186/s13059-018-1443-z
47
Li J. Stoddard T. J. Demorest Z. L. Lavoie P. O. Luo S. Clasen B. M. et al . (2016). Multiplexed, targeted gene editing in Nicotiana benthamiana for glyco-engineering and monoclonal antibody production. Plant Biotechnol. J. 14, 533–542. 10.1111/pbi.12403
48
Li J. F. Norville J. E. Aach J. McCormack M. Zhang D. Bush J. et al . (2013). Multiplex and homologous recombination–mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691. 10.1038/nbt.2654
49
Li R. Li R. Li X. Fu D. Zhu B. Tian H. et al . (2018). Multiplexed CRISPR/Cas9-mediated metabolic engineering of γ-aminobutyric acid levels in Solanum lycopersicum. Plant Biotechnol. J. 16, 415–427. 10.1111/pbi.12781
50
Li S. Shen L. Hu P. Liu Q. Zhu X. Qian Q. et al . (2019). Developing disease-resistant thermosensitive male sterile rice by multiplex gene editing. J. Integr. Plant Biol. 61, 1201–1205. 10.1111/jipb.12774
51
Li T. Huang S. Jiang W. Z. Wright D. Spalding M. H. Weeks D. P. et al . (2011). TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucl. Acids Res. 39, 359–372. 10.1093/nar/gkq704
52
Li T. Yang X. Yu Y. Si X. Zhai X. Zhang H. et al . (2018). Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 36, 1160–1163. 10.1038/nbt.4273
53
Li W. Challa G. S. Zhu H. Wei W. (2016). Recurrence of chromosome rearrangements and reuse of DNA breakpoints in the evolution of the Triticeae genomes. G3 Genes Genomes Genetics6, 3837–3847. 10.1534/g3.116.035089
54
Li X. Wang Y. Chen S. Tian H. Fu D. Zhu B. et al . (2018). Lycopene is enriched in tomato fruit by CRISPR/Cas9-mediated multiplex genome editing. Front. Plant Sci. 9:559. 10.3389/fpls.2018.00559
55
Li Y. Zhang H. (2017). Soybean isoflavones ameliorate ischemic cardiomyopathy by activating Nrf2-mediated antioxidant responses. Food Function8, 2935–2944. 10.1039/C7FO00342K
56
Li Y. Zhu J. Wu H. Liu C. Huang C. Lan J. et al . (2019). Precise base editing of non-allelic acetolactate synthase genes confers sulfonylurea herbicide resistance in maize. Crop J. 8, 449–456. 10.1016/j.cj.2019.10.001
57
Li Z. Wang F. Li J. F. (2019). Targeted transcriptional activation in plants using a potent dead Cas9–derived synthetic gene activator. Curr. Protocols Mol. Biol. 127:e89. 10.1002/cpmb.89
58
Li Z. Zhang D. Xiong X. Yan B. Xie W. Sheen J. Li J. F. (2017). A potent Cas9-derived gene activator for plant and mammalian cells. Nat. Plant. 3, 930–936. 10.1038/s41477-017-0046-0
59
Lin Q. Zong Y. Xue C. Wang S. Jin S. Zhu Z. et al . (2020). Prime genome editing in rice and wheat. Nat. Biotechnol. 38, 582–585. 10.1038/s41587-020-0455-x
60
Liu X. Qin R. Li J. Liao S. Shan T. Xu R. et al . (2020). A CRISPR-Cas9-mediated domain-specific base-editing screen enables functional assessment of ACCase variants in rice. Plant Biotechnol. J. 27, 1845–1847. 10.1111/pbi.13348
61
Lloyd A. Plaisier C. L. Carroll D. Drews G. N. (2005). Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A.102, 2232–2237. 10.1073/pnas.0409339102
62
Lowder L. G. Zhang D. Baltes N. J. Paul J. W. Tang X. Zheng X. et al . (2015). A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 169, 971–985. 10.1104/pp.15.00636
63
Lowder L. G. Zhou J. Zhang Y. Malzahn A. Zhong Z. Hsieh T. F. et al . (2018). Robust transcriptional activation in plants using multiplexed CRISPR-Act2. 0 and mTALE-Act systems. Mol. Plant11, 245–256. 10.1016/j.molp.2017.11.010
64
Lowder L. G. Paul J. W. Qi Y. (2017). Multiplexed transcriptional activation or repression in plants using CRISPR-dCas9-based systems, in Plant Gene Regulatory Networks, eds KaufmannK.Mueller-RoeberB. (New York, NY: Humana Press), 167–184.
65
Ma X. Zhang Q. Zhu Q. Liu W. Chen Y. Qiu R. et al . (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant8, 1274–1284. 10.1016/j.molp.2015.04.007
66
Maher M. F. Nasti R. A. Vollbrecht M. Starker C. G. Clark M. D. Voytas D. F. (2020). Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 38, 84–89. 10.1038/s41587-019-0337-2
67
Mali P. Aach J. Stranges P. B. Esvelt K. M. Moosburner M. Kosuri S. et al . (2013). CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31:833. 10.1038/nbt.2675
68
Malnoy M. Viola R. Jung M. H. Koo O. J. Kim S. Kim J. S. et al . (2016). DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 7:1904. 10.3389/fpls.2016.01904
69
Malzahn A. Zhang Y. Qi Y. (2019). CRISPR-Act2. 0: an improved multiplexed system for plant transcriptional activation, in Plant Genome Editing With CRISPR Systems, ed QiY. (New York, NY: Humana Press), 83–93.
70
Mao Y. Zhang H. Xu N. Zhang B. Gou F. Zhu J. K. (2013). Application of the CRISPR–Cas system for efficient genome engineering in plants. Mol. Plant6, 2008–2011. 10.1093/mp/sst121
71
McCarty N. S. Graham A. E. Studená L. Ledesma-Amaro R. (2020). Multiplexed CRISPR technologies for gene editing and transcriptional regulation. Nat. Commun. 11, 1–3. 10.1038/s41467-020-15053-x
72
Miglani G. S. (2017). Genome editing in crop improvement: present scenario and future prospects. J. Crop Improvement31, 453–559. 10.1080/15427528.2017.1333192
73
Mishra R. Joshi R. K. Zhao K. (2018). Genome editing in rice: recent advances, challenges, and future implications. Front. Plant Sci. 9:1361. 10.3389/fpls.2018.01361
74
Mishra R. Joshi R. K. Zhao K. (2020). Base editing in crops: current advances, limitations and future implications. Plant Biotechnol. J. 18, 20–31. 10.1111/pbi.13225
75
Naeem M. Majeed S. Hoque M. Z. Ahmad I. (2020). Latest developed strategies to minimize the off-target effects in CRISPR-Cas-mediated genome editing. Cells9:1608. 10.3390/cells9071608
76
Naim F. Dugdale B. Kleidon J. Brinin A. Shand K. Waterhouse P. et al . (2018). Gene editing the phytoene desaturase alleles of Cavendish banana using CRISPR/Cas9. Transgenic Res. 27, 451–460. 10.1007/s11248-018-0083-0
77
Nakayasu M. Akiyama R. Lee H. J. Osakabe K. Osakabe Y. Watanabe B. et al . (2018). Generation of α-solanine-free hairy roots of potato by CRISPR/Cas9 mediated genome editing of the St16DOX gene. Plant Physiol. Biochem. 131, 70–77. 10.1016/j.plaphy.2018.04.026
78
Oliva R. Ji C. Atienza-Grande G. Huguet-Tapia J. C. Perez-Quintero A. Li T. et al . (2019). Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 37, 1344–1350. 10.1038/s41587-019-0267-z
79
Ordon J. Gantner J. Kemna J. Schwalgun L. Reschke M. Streubel J. et al . (2017). Generation of chromosomal deletions in dicotyledonous plants employing a user-friendly genome editing toolkit. Plant J. 89, 155–168. 10.1111/tpj.13319
80
Pacher M. Schmidt-Puchta W. Puchta H. (2007). Two unlinked double-strand breaks can induce reciprocal exchanges in plant genomes via homologous recombination and nonhomologous end joining. Genetics175, 21–29. 10.1534/genetics.106.065185
81
Qi Y. Li X. Zhang Y. Starker C. G. Baltes N. J. Zhang F. et al . (2013). Targeted deletion and inversion of tandemly arrayed genes in Arabidopsis thaliana using zinc finger nucleases. G3 Genes Genomes Genetics3, 1707–1715. 10.1534/g3.113.006270
82
Qi Y. Starker C. G. Zhang F. Baltes N. J. Voytas D. F. (2014). Tailor-made mutations in Arabidopsis using zinc finger nucleases, in Arabidopsis Protocols, eds Sanchez-SerranoJ. J.SalinasJ. (Totowa, NJ: Humana Press), 193–209.
83
Ren B. Yan F. Kuang Y. Li N. Zhang D. Lin H. et al . (2017). A CRISPR/Cas9 toolkit for efficient targeted base editing to induce genetic variations in rice. Sci. China Life Sci. 60, 516–519. 10.1007/s11427-016-0406-x
84
Ren B. Yan F. Kuang Y. Li N. Zhang D. Zhou X. et al . (2018). Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9-guided hyperactive hAID mutant. Mol. Plant11, 623–626. 10.1016/j.molp.2018.01.005
85
Rodríguez-Leal D. Lemmon Z. H. Man J. Bartlett M. E. Lippman Z. B. (2017). Engineering quantitative trait variation for crop improvement by genome editing. Cell171, 470–480. 10.1016/j.cell.2017.08.030
86
Rothan C. Diouf I. Causse M. (2019). Trait discovery and editing in tomato. Plant J. 97, 73–90. 10.1111/tpj.14152
87
Sánchez-León S. Gil-Humanes J. Ozuna C. V. Giménez M. J. Sousa C. Voytas D. F. et al . (2018). Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol. J. 16, 902–910. 10.1111/pbi.12837
88
Schmidt C. Pacher M. Puchta H. (2019). Efficient induction of heritable inversions in plant genomes using the CRISPR/Cas system. Plant J. 98, 577–589. 10.1111/tpj.14322
89
Schmidt C. Schindele P. Puchta H. (2020). From gene editing to genome engineering: restructuring plant chromosomes via CRISPR/Cas. aBIOTECH1, 21–31. 10.1007/s42994-019-00002-0
90
Shimatani Z. Kashojiya S. Takayama M. Terada R. Arazoe T. Ishii H. et al . (2017). Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 35, 441–443. 10.1038/nbt.3833
91
Siebert R. Puchta H. (2002). Efficient repair of genomic double-strand breaks by homologous recombination between directly repeated sequences in the plant genome. Plant Cell. 14, 1121–1131. 10.1105/tpc.001727
92
Störtz F. Minary P. (2021). crisprSQL: a novel database platform for CRISPR/Cas off-target cleavage assays. Nucl. Acids Res. 49, D855–D861. 10.1093/nar/gkaa885
93
Stuttmann J. Barthel K. Martin P. Ordon J. Erickson J. L. Herr R. et al . (2021). Highly efficient multiplex editing: one-shot generation of 8 × Nicotiana benthamiana and 12 × Arabidopsis mutants. Plant J. 106, 8–22. 10.1111/tpj.15197
94
Swarts D. C. Jinek M. (2018). Cas9 versus Cas12a/Cpf1: structure–function comparisons and implications for genome editing. Wiley Interdisciplinary Rev. RNA9:e1481. 10.1002/wrna.1481
95
Szinay D. Wijnker E. van den Berg R. Visser R. G. de Jong H. Bai Y. (2012). Chromosome evolution in Solanum traced by cross-species BAC-FISH. New Phytol. 195, 688–698. 10.1111/j.1469-8137.2012.04195.x
96
Tang X. Liu G. Zhou J. Ren Q. You Q. Tian L. et al . (2018). A large-scale whole-genome sequencing analysis reveals highly specific genome editing by both Cas9 and Cpf1 (Cas12a) nucleases in rice. Genome Biol. 19:84. 10.1186/s13059-018-1458-5
97
Tang X. Zheng X. Qi Y. Zhang D. Cheng Y. Tang A. et al . (2016). A single transcript CRISPR-Cas9 system for efficient genome editing in plants. Mol. Plant9, 1088–1091. 10.1016/j.molp.2016.05.001
98
Tian S. Jiang L. Gao Q. Zhang J. Zong M. Zhang H. et al . (2017). Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Rep. 36, 399–406. 10.1007/s00299-016-2089-5
99
Tripathi J. N. Ntui V. O. Ron M. Muiruri S. K. Britt A. Tripathi L. (2019). CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Commun. Biol. 2:1. 10.1038/s42003-019-0288-7
100
Tsai S. Q. Nguyen N. T. Malagon-Lopez J. Topkar V. V. Aryee M. J. Joung J. K. (2017). CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR–Cas9 nuclease off-targets. Nat. Methods14:607. 10.1038/nmeth.4278
101
Tsai S. Q. Wyvekens N. Khayter C. Foden J. A. Thapar V. Reyon D. et al . (2014). Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 32:569. 10.1038/nbt.2908
102
Udall J. A. Quijada P. A. Osborn T. C. (2005). Detection of chromosomal rearrangements derived from homeologous recombination in four mapping populations of Brassica napus L. Genetics169, 967–979. 10.1534/genetics.104.033209
103
Upadhyay S. K. Kumar J. Alok A. Tuli R. (2013). RNA-guided genome editing for target gene mutations in wheat. G3 Genes Genomes Genetics3, 2233–2238. 10.1534/g3.113.008847
104
Vazquez-Vilar M. Bernabé-Orts J. M. Fernandez-del-Carmen A. Ziarsolo P. Blanca J. Granell A. et al . (2016). A modular toolbox for gRNA–Cas9 genome engineering in plants based on the GoldenBraid standard. Plant Methods12:10. 10.1186/s13007-016-0101-2
105
Veillet F. Chauvin L. Kermarrec M. P. Sevestre F. Merrer M. Terret Z. et al . (2019). The Solanum tuberosum GBSSI gene: a target for assessing gene and base editing in tetraploid potato. Plant Cell Rep. 38, 1065–1080. 10.1007/s00299-019-02426-w
106
Vidigal J. A. Ventura A. (2015). Rapid and efficient one-step generation of paired gRNA CRISPR-Cas9 libraries. Nat. Commun. 6, 1–7. 10.1038/ncomms9083
107
Wang C. Shen L. Fu Y. Yan C. Wang K. (2015). A simple CRISPR/Cas9 system for multiplex genome editing in rice. J. Genet. Genomics42, 703–706. 10.1016/j.jgg.2015.09.011
108
Wang F. Zhang C. Xu W. Yuan S. Song J. Li L. et al . (2020). Developing high-efficiency base editors by combining optimized synergistic core components with new types of nuclear localization signal peptide. Crop J. 8, 408–417. 10.1016/j.cj.2020.01.003e
109
Wang H. Cimen E. Singh N. Buckler E. (2020). Deep learning for plant genomics and crop improvement. Curr. Opin. Plant Biol. 54, 34–41. 10.1016/j.pbi.2019.12.010
110
Wang H. Pandey S. Velarde O. (2012). Synthesis of Key Results and Implications. Patterns of Varietal Adoption and Economics of Rice Production in Asia. Los Baños: International Rice Research Institute, 1–4.
111
Wang H. Studer A. J. Zhao Q. Meeley R. Doebley J. F. (2015). Evidence that the origin of naked kernels during maize domestication was caused by a single amino acid substitution in tga1. Genetics200, 965–974. 10.1534/genetics.115.175752
112
Wang Y. Cheng X. Shan Q. Zhang Y. Liu J. Gao C. et al . (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32:947. 10.1038/nbt.2969
113
Wu J. Chen C. Xian G. Liu D. Lin L. Yin S. et al . (2020). Engineering herbicide-resistant oilseed rape by CRISPR/Cas9-mediated cytosine base-editing. Plant Biotechnol. J. 18, 1857–1859. 10.1111/pbi.13368
114
Wu R. Lucke M. Jang Y. T. Zhu W. Symeonidi E. Wang C. et al . (2018). An efficient CRISPR vector toolbox for engineering large deletions in Arabidopsis thaliana. Plant Methods14:65. 10.1186/s13007-018-0330-7
115
Xie K. Minkenberg B. Yang Y. (2015). Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. U.S.A.112, 3570–3575. 10.1073/pnas.1420294112
116
Xie K. Yang Y. (2013). RNA-guided genome editing in plants using a CRISPR–Cas system. Mol. Plant6, 1975–1983. 10.1093/mp/sst119
117
Xing H. L. Dong L. Wang Z. P. Zhang H. Y. Han C. Y. Liu B. et al . (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 1–2. 10.1186/s12870-014-0327-y
118
Xu R. Yang Y. Qin R. Li H. Qiu C. Li L. et al . (2016). Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice. J. Genet. Genomics43:529. 10.1016/j.jgg.2016.07.003
119
Xu W. Song W. Yang Y. Wu Y. Lv X. Yuan S. et al . (2019). Multiplex nucleotide editing by high-fidelity Cas9 variants with improved efficiency in rice. BMC Plant Biol. 19:1. 10.1186/s12870-019-2131-1
120
Yan L. Wei S. Wu Y. Hu R. Li H. Yang W. et al . (2015). High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol. Plant8, 1820–1823. 10.1016/j.molp.2015.10.004
121
Yan W. Chen D. Kaufmann K. (2016). Molecular mechanisms of floral organ specification by MADS domain proteins. Curr. Opin. Plant Biol. 29, 154–162. 10.1016/j.pbi.2015.12.004
122
Yu H. Lin T. Meng X. Du H. Zhang J. Liu G. et al . (2021). A route to de novo domestication of wild allotetraploid rice. Cell184, 1156–1170. 10.1016/j.cell.2021.01.013
123
Zafar S. A. Zaidi S. S. Gaba Y. Singla-Pareek S. L. Dhankher O. P. Li X. et al . (2020). Engineering abiotic stress tolerance via CRISPR/Cas-mediated genome editing. J. Exp. Botany71, 470–479. 10.1093/jxb/erz476
124
Zapata L. Ding J. Willing E. M. Hartwig B. Bezdan D. Jiao W. B. et al . (2016). Chromosome-level assembly of Arabidopsis thaliana Ler reveals the extent of translocation and inversion polymorphisms. Proc. Natl. Acad. Sci. U.S.A.113, E4052–E4060. 10.1073/pnas.1607532113
125
Zetsche B. Gootenberg J. S. Abudayyeh O. O. Slaymaker I. M. Makarova K. S. Essletzbichler P. et al . (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell163, 759–771. 10.1016/j.cell.2015.09.038
126
Zetsche B. Heidenreich M. Mohanraju P. Fedorova I. Kneppers J. DeGennaro E. M. et al . (2017). Multiplex gene editing by CRISPR–Cpf1 using a single crRNA array. Nat. Biotechnol. 35, 31. 10.1038/nbt.3737
127
Zhang C. Liu C. Weng J. Cheng B. Liu F. Li X. et al . (2017). Creation of targeted inversion mutations in plants using an RNA-guided endonuclease. Crop J. 5, 83–88. 10.1016/j.cj.2016.08.001
128
Zhang P. Du H. Wang J. Pu Y. Yang C. Yan R. et al . (2020). Multiplex CRISPR/Cas9-mediated metabolic engineering increases soya bean isoflavone content and resistance to soya bean mosaic virus. Plant Biotechnol. J. 18, 1384–1395. 10.1111/pbi.13302
129
Zhang Z. Mao Y. Ha S. Liu W. Botella J. R. Zhu J. K. (2016). A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep.35, 1519–1533. 10.1007/s00299-015-1900-z
130
Zhao C. Zhang Z. Xie S. Si T. Li Y. Zhu J. K. (2016). Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 171, 2744–2759. 10.1104/pp.16.00533
131
Zhou H. Liu B. Weeks D. P. Spalding M. H. Yang B. (2014). Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucl. Acids Res. 42, 10903–10914. 10.1093/nar/gku806
132
Zhou J. Xin X. He Y. Chen H. Li Q. Tang X. et al . (2019). Multiplex QTL editing of grain-related genes improves yield in elite rice varieties. Plant Cell Rep. 38, 475–485. 10.1007/s00299-018-2340-3
133
Zsögön A. Cermák T. Naves ER Notini M. M. Edel K. H. Weinl S. Freschi L. et al . (2018). De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 36, 1211–1216. 10.1038/nbt.4272
Summary
Keywords
multiplex genome editing, CRISPR/Cas9, CRISPR/Cas12, plant science, crop improvement
Citation
Abdelrahman M, Wei Z, Rohila JS and Zhao K (2021) Multiplex Genome-Editing Technologies for Revolutionizing Plant Biology and Crop Improvement. Front. Plant Sci. 12:721203. doi: 10.3389/fpls.2021.721203
Received
06 June 2021
Accepted
01 September 2021
Published
06 October 2021
Volume
12 - 2021
Edited by
Maria Raffaella Ercolano, University of Naples Federico II, Italy
Reviewed by
Ulhas S. Kadam, Gyeongsang National University, South Korea; Chonglie Ma, China National Seed Group Co. Ltd., China
Updates
Copyright
© 2021 Abdelrahman, Wei, Rohila and Zhao.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kaijun Zhao zhaokaijun@caas.cn
This article was submitted to Plant Biotechnology, a section of the journal Frontiers in Plant Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.