- 1FFoQSI GmbH – Austrian Competence Centre for Feed and Food Quality, Safety and Innovation, Tulln, Austria
- 2Institute of Applied Botany and Pharmacognosy (IAB), Veterinary University of Vienna, Vienna, Austria
- 3Center for Health & Bioresources, Bioresources Unit, AIT Austrian Institute of Technology GmbH, Tulln, Austria
The modes of interactions between plants and plant-associated microbiota are manifold, and secondary metabolites often play a central role in plant-microbe interactions. Abiotic and biotic (including both plant pathogens and endophytes) stress can affect the composition and concentration of secondary plant metabolites, and thus have an influence on chemical compounds that make up for the taste and aroma of fruit. While the role of microbiota in growth and health of plants is widely acknowledged, relatively little is known about the possible effect of microorganisms on the quality of fruit of plants they are colonizing. In this work, tomato (Solanum lycopersicum L.) plants of five different cultivars were grown in soil and in hydroponics to investigate the impact of the cultivation method on the flavor of fruit, and to assess whether variations in their chemical composition are attributable to shifts in bacterial microbiota. Ripe fruit were harvested and used for bacterial community analysis and for the analysis of tomato volatiles, sugars and acids, all contributing to flavor. Fruit grown in soil showed significantly higher sugar content, whereas tomatoes from plants under hydroponic conditions had significantly higher levels of organic acids. In contrast, aroma profiles of fruit were shaped by the tomato cultivars, rather than the cultivation method. In terms of bacterial communities, the cultivation method significantly defined the community composition in all cultivars, with the bacterial communities in hydroponic tomatoes being more variable that those in tomatoes grown in soil. Bacterial indicator species in soil-grown tomatoes correlated with higher concentrations of volatiles described to be perceived as “green” or “pungent.” A soil-grown specific reproducibly occurring ASV (amplicon sequence variants) classified as Bacillus detected solely in “Solarino” tomatoes, which were the sweetest among all cultivars, correlated with the amount of aroma-relevant volatiles as well as of fructose and glucose in the fruit. In contrast, indicator bacterial species in hydroponic-derived tomatoes correlated with aroma compounds with “sweet” and “floral” notes and showed negative correlations with glucose concentrations in fruit. Overall, our results point toward a microbiota-related accumulation of flavor and aroma compounds in tomato fruit, which is strongly dependent on the cultivation substrate and approach.
Introduction
Tomato (Solanum lycopersicum L.) is the highest value fruit crop worldwide (Tieman et al., 2017) and major dietary source of valuable vitamins, minerals and antioxidants. Based on the increased global demand of fresh produce in the recent years, sustained efforts have been pursued in order to increase yield and fruit size, minimize pest susceptibility and therefore provide the consumer with year-round fresh produce. Such efforts include breeding programs which have extensively improved certain fruit qualities at the expense of aroma, taste, and nutritional value (Maul et al., 2000; Tieman et al., 2017; Wang and Seymour, 2017). For example, due to the deterioration of the commercial tomato flavor, which goes hand-in-hand with reduced consumer acceptance in the recent years (Klee, 2010), a growing trend in research emerged aiming at elucidating the causes underlying this phenomenon.
Flavor is described as a complex interaction between volatile and non-volatile compounds, which are perceived by retronasal olfaction and by the gustatory receptors on the tongue, respectively. These interactions and perceptions can be enhanced by orthonasal olfaction (“smelling”) as well as by visual and textural signals in the brain (Spence, 2015). Fresh tomatoes possess a characteristic sweet-sour flavor due to the presence of a complex mixture of volatile compounds that interact with sugars and acids in the fruit (Baldwin et al., 2008). Sugars are important compounds in regard to fruit yield, quality and organoleptic properties (Quinet et al., 2019), as they provide sweetness and are key in the generation of turgor pressure to promote cell expansion, ultimately affecting fruit size (Kanayama, 2017). In tomato fruit, sugars (primarily fructose and glucose) as well as acids (malic and citric acid) are the main contributors to taste (Tieman et al., 2012). Accordingly, high sugar and relatively high acid content are required for a favorable taste and several studies have shown that sweeter tomatoes are more acceptable (Malundo et al., 1995).
With respect to aroma, up to 400 organic volatiles have been identified in the tomato fruit, but only a fraction correlated with consumer liking and/or were associated with flavor intensity (Tieman et al., 2017). These volatiles derive from essential dietary nutrients like fatty acids, amino acids and colored carotenoids (Goff and Klee, 2006; Zanor et al., 2009). Furthermore, several studies demonstrated that the amount of some volatiles enhance the perception of sweetness in tomato fruit, increasing their likability (Baldwin et al., 2008; Tieman et al., 2012).
Genetic factors are responsible for the variability in the volatile profiles of tomato fruit. It was recently demonstrated that modern commercial cultivars show poor flavor compared to heirloom and wild accessions (Tieman et al., 2017). Intriguingly, despite the low rate of DNA sequence diversity among modern commercial tomato cultivars, Tieman et al. (2012) observed a variation in volatile content of as much as 3,000-fold across cultivars, possibly reflecting deviations in cultivation methods and post-harvest practices among lots (Tieman et al., 2012).
Hydroponic cultivation in greenhouses is considered effective in increasing productivity by unit land, as it allows extending the duration of the cropping season while minimizing the use of resources and negative environmental impacts (FAO, 2013). However, it is a widespread notion among consumers that tomatoes grown in hydroponics have bland flavor and aroma compared to those cultivated in soil (Padilla et al., 2007).
In the recent years, plant-associated microbial communities have taken a spotlight in research for their potential to impact plant health and, consequently, crop productivity and quality (Coleman-Derr and Tringe, 2014). Microorganisms are an integral part of the composition of fruits and vegetables and are found as epiphytes on the surfaces as well as endophytes within tissues (Droby and Wisniewski, 2018). Besides their roles in nutrient acquisition, stress alleviation as well as pest and disease control, the activities of plant microbiota can also influence the composition of secondary metabolites in their hosts (Berg et al., 2016), which include precursors of flavor-relevant compounds in fruit such as flavonoids and carotenoids (Robards and Antolovich, 1997; Kochevenko et al., 2012). This microbial-mediated modulation of fruit flavor has been demonstrated with methylotrophic bacteria in strawberry (Verginer et al., 2010) and with rhizobacteria in Basmati rice (Deshmukh et al., 2016). Furthermore, it has been proposed to be key in the taste of grapes and ultimately, the organoleptic characteristics of wine (Gilbert et al., 2014; Bokulich et al., 2016). Recently. changes in fruit quality and aroma have been linked to shifts in the fruit-associated bacterial community composition in raspberry (Sangiorgio et al., 2021). Compared to soil, hydroponic greenhouse cropping systems are an ecosystem with low microbiological complexity. Consequently, the microbial communities of tomato plants in hydroponics is less diverse than of those cultivated in soil (Rosberg et al., 2014). Therefore, along with the intrinsically impaired flavor of modern tomato cultivars, it is possible that the cultivation method could indirectly play a crucial role in the flavor of tomato fruit.
In this study, we intended to shed light on the impact of the cultivation method in the bacterial assemblages of ripe tomato fruit and how these communities correlate with tomato aroma and flavor profiles. With this aim, we cultivated five different commercial tomato cultivars (‘Ardiles,’ ‘Campari,’ ‘Cappricia,’ ‘Savantas’ and ‘Solarino’) in soil and in hydroponics. Ripe fruit were harvested and used for bacterial community analysis by 16S rRNA gene amplicon sequencing and HS-SPME GC/MS (Headspace Solid-Phase Micro-Extraction coupled with Gas Chromatography and a Mass Spectrometer) analytics for tomato aroma components, and GC-MS for flavor compounds. We described the various aroma, flavor as well as the bacterial community profiles for each cultivar-cultivation method combination and identified associations of microbial taxa and fruit organoleptic characteristics.
Materials and Methods
Cultivation and Harvesting of Tomato Fruit
Seeds of five tomato (Solanum lycopersicum L.) cultivars with different organoleptic properties, namely ‘Ardiles,’ ‘Campari,’ ‘Cappricia,’ ‘Savantas’ and ‘Solarino’ were provided by the Viennese LGV Sonnengemüse cooperative (Figure 1 and Supplementary Table 1). To obtain the fruit material needed for this study, the tomato plants were cultivated under two different approaches: (1) in soil and (2) in hydroponics.
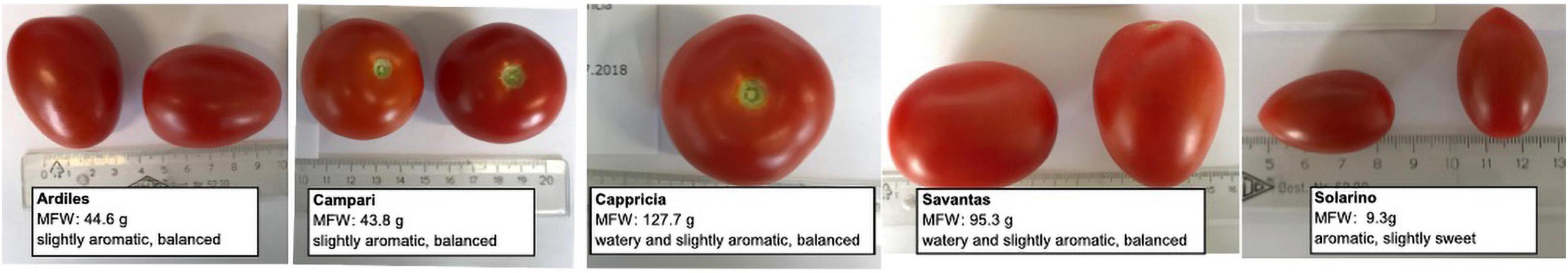
Figure 1. Fruit of five tomato cultivars ‘Ardiles,’ ‘Campari,’ ‘Cappricia,’ ‘Savantas’ and ‘Solarino.’ Shown are the mean fresh weight (MFW) and the scoring obtained from a trained tasting panel regarding the level of aroma perception and sugar/acid ratio. Scoring details are listed in Supplementary Table 1.
Cultivation in soil was started by sowing seeds in trays filled with potting substrate (Profi Substrat, Einheitserde special, Sinntal-Altengronau, Germany). After three weeks, seedlings were planted into 1.5 L pots filled with potting substrate, treated with organic-mineral NPK (5% + 4% + 8%) fertilizer (Gärtner Exclusiv Bio-Tomatendünger, gpi green partners international GmbH & Co. KG, Gladbeck, Germany) in a concentration of 2.2 kg/10 m2 and further kept in the greenhouse. After another three weeks, the tomato plants were transplanted into larger pots (12 L) filled with potting substrate placed in a wire-house, allowing for cultivation in semi-field conditions. Plants were watered whenever needed with tap water. Beginning nine weeks after sowing we treated each plant every two weeks with 1 L calcium nitrate greenhouse grade fertilizer (Haifa Cal CC) (Haifa Group, Israel) in a concentration of 33 g/L to prevent blossom end rot. Fourteen weeks after sowing, we fertilized the plants a second time with organic-mineral NPK (5% + 4% + 8%) solution.
For cultivation in hydroponics, seeds were sown on autoclaved rockwool plugs (2.5 cm × 2.5 cm × 4 cm) (Grodan, Roermond, Netherlands), which were pre-soaked in a diluted Complete Nutrient Solution for young plants (CNS, recipe in Supplementary Material) and placed into plastic Stervient High Containers (10.7 cm × 9.4 cm × 9.6 cm), with a plastic cover to prevent drying, and maintained at a temperature of 25°C in a Memmert Climatic Chamber until germination. After three days the containers were put into the greenhouse and rockwool plugs were watered daily with CNS. Two weeks after sowing, tomato plants in rockwool plugs were inserted into autoclaved rockwool cubes (10 cm × 10 cm × 6.5 cm) previously soaked in CNS and three cubes each with plants of the same tomato cultivar were placed on a tray to prevent exchange of microbiota between cultivars. Automated irrigation with a drip system was installed by inserting one drip into each rockwool cube and plants were drip irrigated with CNS for young plants. At this stage, plants had 3–4 leaves and were 15–20 cm high. Forty days after sowing, the rock wool cubes were placed on cocopeat mats (GBC-Österreich e. Gen. GartenBauCentrum, Wels, Austria), which were prior drip irrigated with nutrient solution for three days to achieve the necessary increase in volume (100 cm × 15 cm × 10 cm). From then on plants were irrigated with CNS for cocopeat mats.
Tomato fruit were harvested, when a single branch of a plant yielded at least 150 g of ripe fruit material. Fruit were considered ripe when they developed full red color. The fruit were then picked with gloved hands, put into sterile plastic bags and immediately further processed in the lab. Tomato fruit of five individual plants per tomato cultivar and cultivation condition were sampled. This resulted in 50 tomato samples [5 cultivars × 5 replicates × 2 cultivation conditions (in soil or in hydroponics)].
Sample Preparation
Harvested fruit were processed to obtain a homogenized tomato juice using a modified version of the method presented by Xiao et al. (2015). Briefly, 150 g of fruit per replicate were mixed to puree using a household blender (Waring Pro BB90E, Silva Homeline, Salzburg, Austria) that was sterilized and subsequently washed three times with sterilized water. Two 2-mL aliquots of the puree were transferred to sterile reaction tubes by using a disposable transfer pipette (VWR, Vienna, Austria) and directly frozen at −20°C for later DNA isolation. The remaining puree was then transferred to sterile tubes and centrifuged at 4°C and 14,000 × g for 15 min. The supernatant (fresh tomato juice) was recovered and further processed to determine volatile, sugar and acid concentrations.
Measurement and Identification of Volatile Compounds in Tomato Fruit
For organic volatile analyses, 8 g of fresh tomato juice were combined with 1.5 g of sodium chloride, 100 μL EDTA/NaOH (1M, pH = 7.5) and 8 μL 1-octen-3-ol (100 mg/L) as internal standard. Samples and reagents were mixed thoroughly in a 20 mL headspace vial equipped with a PTFE/butyl septum (References: 5188-2753 and 5188-2759; Agilent Technologies, United States). Samples were conditioned by incubating each vial at 60°C for 15 min on a thermoblock stirring at 250 rpm. Subsequently, the sample headspace was exposed to the solid-phase micro extraction (SPME) fiber (65 μm PDMS/DVB, Reference: 57298-U; Supelco, United States) for further 50 min under the same conditions. Desorption of the analytes into the injector port of the gas chromatography (GC) apparatus was carried out at 270°C for 15 min with a 3:1 split ratio. Headspace conditioning, sampling and injection into the GC was performed with CombiPAL (CTC Analytics, Switzerland). Volatile compounds in the headspace of the tomato juice were separated on the Agilent 7890A GC. The gas flow was equally split after separation with a deans switch (Agilent Technologies) into two detectors, an Agilent 5975C mass selective (MS) detector for identification and a flame ionization detector (FID) for quantification. Chromatographic separation of the analytes took place in a VF-5ms (30 m × 250 μm × 0.25 μm) column (Agilent Technologies). The oven temperature was set to hold at 60°C for 6 min, followed by a ramp of 5°C/min to 280°C. Helium was used as gas carrier at a flow rate of 1 mL/min. The area of each peak was calculated by the ChemStation software (Agilent Technologies). To ensure that no peaks derived from previous runs or any instrumentation, blank runs with pure water were run every 12 samples. Identification of the volatile compounds was based on comparing mass-fragmented patterns with mass spectra in the NIST Database or with previously reported Kovats indices (KI) in the literature. Kovats indices were calculated by using retention data of n-alkanes (C7-C24) along with retention data of the analytes. Furthermore, the concentrations of the volatile compounds in each sample were quantified based on the peak area of the internal standard.
Assessment of Sugar Content in Tomato Fruit
For the estimation of both sugar and acid content in the tomato fruit, the protocol presented by Agius et al. was employed (Agius et al., 2018). Briefly, 2 mL of freshly prepared tomato juice were filtered through a 0.22 μm membrane (Reference: 8-7066; Ahlstrom ReliaPrep, Finland). Subsequently, 400 μL of the filtrate were combined with 100 μL of both internal standards, namely tricarballylic acid (10 g/L) and lactose (100 g/L), as well as with 400 μL acetonitrile (ACN, 100%). The mixture was then centrifuged at 4°C and 14,000 × g for 5 min. Sugars and acids in the samples were separated by solid phase extraction (SPE) as indicated by the authors using one Supelclean™ LC-NH2 SPE 100 mg tube per sample (Reference: 504483, Sigma Aldrich). Sugar analyses were performed using high pressure liquid chromatography (HPLC) coupled with a diode array detector (DAD) by transferring 200 μL of sugar eluate to an autosampler vial together with 8 μL ACN 90%. Chromatographic separation of the sugars in the samples took place in a Shodex Asahipak NH2P-50 4E 125 mm × 4 mm column preceded by a Shodex Asahipak NH2P-50G 4A 10 mm × 4.6 mm precolumn (Showa Denko Europe, Germany). The column oven temperature was set to 40°C. 20 μL of the sample was injected. For elution, a mixture of 75% ACN and NaOH (pH = 12) was used at a flow rate of 1.1 ml/min. Quantification of sugar concentrations was based on the results obtained from a standard mix containing fructose, glucose and lactose at 5 concentration levels (Agius et al., 2018).
Estimation of Acid Content in Tomato Fruit
Hundred microliters of eluate (see previous section) containing the acidic compounds of the tomato fruit was evaporated in a desiccator overnight. The next day, 120 μL of derivatization reagent (2M (trimethylsilyl) diazomethane in diethyl-ether together with 900 μL methanol) was added until the mixture remained yellow. The reaction was incubated at 25°C and 800 rpm for 5 min in a thermomixer. 5 μL of 1M acetic acid (in methanol) were added until the yellow color disappeared. The reactions were subsequently centrifuged at 14,000 × g for 5 min at room temperature. 50 μL of the reaction were transferred to an autosampler vial for analyses on GC-MS using the previously described column. Helium served as carrier gas at a flow rate of 1 mL/min. Injection was performed at 220°C in splitless mode. The temperature program was set to hold one minute at 45°C, then to increase at 15°C/min until 210°C and finally to reach 300°C at a rate of 30°C/min. The transfer line was operated at 280°C, the ion trap at 160°C and the manifold at 40°C. Quantification of acid concentrations was based on standard curves of citric and malic acids run at five concentrations (Agius et al., 2018).
DNA Extraction
DNA was isolated from 0.5 mL tomato puree with the FastDNA® SPIN Kit for Soil (MP Biomedicals, Solon, OH, United States) and following the manufacturers’ protocol using the beat-beating method. To test the efficacy of the protocol in extracting bacterial DNA from tomato puree, selected samples were mixed with 75 μL of thawed mock community suspension (ZymoBIOMICS™ Microbial Community Standard, Zymo Research, United States). In addition, DNA was isolated directly from the pure mock community using 75 μL to provide control DNA samples to test whether there are substances in the DNA extractions from tomato puree, which inhibit PCR. DNA (5 μL) was separated and visually tested for quality by electrophoresis (80 V) on 1% (w/v) agarose gels. In addition, DNA concentration was measured with a NanoDrop ND-1000 spectrophotometer (Thermo Fischer Scientific, DE, United States).
16S rRNA Library Preparation
To analyze the bacterial communities of tomato fruit the V5-V7 region of the 16S rRNA gene was amplified in a two-step PCR approach as described previously in comparable studies (Escobar Rodríguez et al., 2018). In the first step, PCR was carried out with the PCR primers 799f (5′- AACMGGATTAGATACCCKG -3′) and 1175r (5′- ACGGGCGGTGTGTRC -3′) in 25 μL reaction volume containing 5–10 ng DNA, 1 × KAPA buffer, 0.3 mM dNTPS, 0.3 μM forward and reverse primer each, 0.5 U KAPA polymerase (Kapa Biosystems, Boston, MA, United States) and PCR grade water. Amplifications were performed in a Thermocycler (peqSTAR 96x HPL, VWR International, Vienna, Austria) and cycling conditions were as follows: 95°C for 3 min, followed by 25 cycles of 30s at 95°C, 30s at 55°C and 30s at 72°C with a final extension step at 72°C for 5 min. Reactions were performed in triplicate and pooled. Pools were loaded onto a 2% agarose gel (w/v) in sterile-filtrated TAE (Biozym Biotech Trading, Vienna, Austria), and the bacterial 16S rRNA gene amplicons were separated from mitochondrial 18S rRNA gene amplicons by electrophoresis for 80 min at 110 V and excised from the gel using X-tracta Gel Extraction Tools (Sigma Aldrich, Vienna, Austria). The gel pieces were then placed inside the top part of a cut sterile filter tip placed inside a 2 mL Eppendorf tube and DNA collected by centrifugation at 10,000 rpm for 1 min in an Eppendorf™ 5424 Microcentrifuge (Eppendorf, Hamburg, Germany). Then, the filter tip was carefully removed with sterile tweezers, and 2 μL of the eluate were used for the second round of PCR with the primers 799f and 1175r that had specific indices (Supplementary Table 2) attached for sample recognition in sequencing. The concentration of PCR amplicons was estimated from the intensity of the amplicon bands on agarose gels with Image Lab™ Software 6.0 (BioRad, Hercules, CA, United States) using the bands of the DNA marker as a point of reference. For library preparation, 48 amplicons were pooled in equimolar amounts, and libraries purified with the Agencourt® AMPure® XP system (New England Biolabs, Ipswich, MA, United States). Total DNA in the library was quantified with Quant-iT™ PicoGreen® following the instructions of the manufacturer in a plate reader (BioTek, Winooski, VT, United States). For sequencing, pooled and purified libraries were subjected to Illumina adapter ligation and sequencing using 2 × 250 bp MiSeq v2 sequencing at LGC Genomics (Berlin, Germany). Controls included in this analysis were: (1) the controls for DNA isolation efficacy described above, (2) the negative control for each PCR reaction, for which water was used instead of DNA and (3) pieces of the agarose gel used for the isolation of the bacterial amplicons.
Raw Sequence Data Processing
For removing the Illumina inherent Phi X contamination raw reads were filtered with Bowtie2 v.2.3.4.3 (Langmead and Salzberg, 2012) and the overall quality was checked with FastQC v.0.11.8 (Andrews, 2010). Demultiplexing and stripping the adapters and primers was done with Cutadapt v.1.18 (Martin, 2018). The DADA2 v. 1.12.1 Bioconductor R package (Callahan et al., 2016) was then applied to quality filter, trim, denoise and merge paired-reads while also screening for chimeras. Using Metaxa2 v.2.2 (Bengtsson-Palme et al., 2015) unique Amplicon sequence variants (ASVs) stemming from the V5-V7 region of the 16S rRNA gene of bacteria and archaea were identified. Those were then assigned to taxa by using the RDP classifier (Wang et al., 2020) re-implemented in DADA2 against the SILVA SSU v.132 database (Quast et al., 2013) and finally an ASV count matric was generated for the statistical analysis. Prior to any further analyses, ASVs deriving from control samples were removed from the dataset.
Bacterial Community Analyses
Representative bacterial communities in our samples were obtained by considering bacterial amplicon sequence variants (ASVs) with a relative abundance greater than 0.01%. A further filtering step consisted in extracting reproducibly-occurring ASVs (rASVs), namely those observed in at least three of five replicates. Alpha diversity values (total ASV count and Simpson’s Index) were calculated using the rtk R package (Saary et al., 2017) averaging the results of 999 iterations. The rarefaction minimum depth was 151 for reproducibly-occurring communities. For beta diversity analyses, a cumulative sum scaling (CSS) normalization was applied (Paulson et al., 2013). Indicator genera for all cultivars in each cultivation method was calculated fitting a multivariate generalized linear model to the rASV table (mvabund:manyglm).
Statistical Analyses
Processed sequence and chemical data were analyzed in R v3.6.3 software. We investigated the role of cultivation method, tomato cultivar and interaction thereof (genotype x environment, GxE) in the concentrations of total volatile compounds, aroma-relevant volatiles and most abundant compounds, as well as in the amount of sugars (glucose and fructose) and acids (malic and citric) of the tomato fruit using analysis of variance (ANOVA). Volatile compound variation across samples was assessed employing principal component analysis (PCA) and visualized with factoextra:fviz_pca, depicting only those volatiles with a contribution greater than 10 in the loadings plot (Kassambara and Mundt, 2020). The effect of cultivation method, tomato cultivar, and GxE (genotype x environment) interaction in the diversity and composition of the microbial communities of the fruit was assessed using the Phyloseq (McMurdie and Holmes, 2013) and vegan (Oksanen et al., 2020) packages by testing with permutational multivariate analysis of variance (PERMANOVA, 9999 permutations). Pairwise comparisons among cultivars were conducted using the permwise.perm.manova of the RVAideMemoire package (Hervé, 2021), where p values were adjusted using false discovery rate controlling procedures. Unless otherwise indicated, statistical significance threshold was set at α = 0.05. Spearman correlation analyses between abundances of rASVs and absolute volatile, sugar and acid compound concentrations was conducted using microbiome:associate and depicted with the heat command considering significant adjusted p-values (Lahti et al., 2017).
Nucleotide Sequence Accession Numbers
Sequence data are available in the NCBI SRA database under the BioProject number PRJNA513967.
Results
Fruit Volatile Profile Is Mainly a Function of the Tomato Cultivar, While Sugar and Acids Levels Depend on the Cultivation Method
A total of 34 volatile organic compounds (VOCs) were detected by HS-SPME-GC-MS analysis of the tomato juice, including 13 aldehydes, 9 alcohols, four ketones, four esters and four other compounds (Table 1). Among all cultivars, VOCs with the highest measured concentrations included hexanal, E-2-hexenal, 2-isobutylthiazole and methyl salicylate. Sixteen of all detected compounds in this study either had log odor units > 0 (e.g., beta-damascenone), or were listed by Tieman and colleagues to be linked with overall liking by the consumer, sweetness perception and/or flavor intensity: isovaleraldehyde, benzaldehyde, E-2-heptenal, E-2-hexenal, nonyl-aldehyde, phenylacetaldehyde, guaiacol, 2-methylbutanol, eugenol, Z-3-hexen-1-ol, 6-methyl-5-hepten-2-one, beta-ionone, hexyl-acetate, 1-nitro-methylbutane, and 2-isobuthylthiazole (Tieman et al., 2012, 2017). There were no significant differences in total volatile amounts between tomatoes grown in soil and those from hydroponic cultures, while the cultivar and the GxE interaction significantly determined the total VOCs concentrations (Anova, F-values = 9.9 and 3.9; p < 0.001, respectively; Figure 2A). In fact, the concentrations of 20 individual VOCs were found to differ significantly among tomato cultivars, whereas only nine were affected by the cultivation method and GxE interactions (Table 1 and Supplementary Figure 1). PCA analysis of the VOC concentrations explained a total of 75.7% of the variance in the first two principal components (PC) and revealed clustering of samples belonging to the same fruit cultivar (Figure 2B). When grown in hydroponics, the cultivars ‘Savantas’ and ‘Ardiles’ were characterized mainly by their high hexanal contents together with less pronounced amounts of 2-butyl-octenal, eugenol and geranyl-acetone. The fruit of the soil-grown cultivars ‘Campari’ and ‘Ardiles’ were rich in methyl salycilate, E-2-hexenal and 2-isobutylthiazole. The cultivar ‘Solarino,’ soil-grown, however, showed characteristic amounts of 3-hexen-1-ol (Figures 2B,D). It was noteworthy that groupings within cultivation methods were not observed (Figure 2C). Prevalent volatiles among samples are displayed as variables and their loadings in Figures 2B–D, respectively as well as in Supplementary Figure 1.
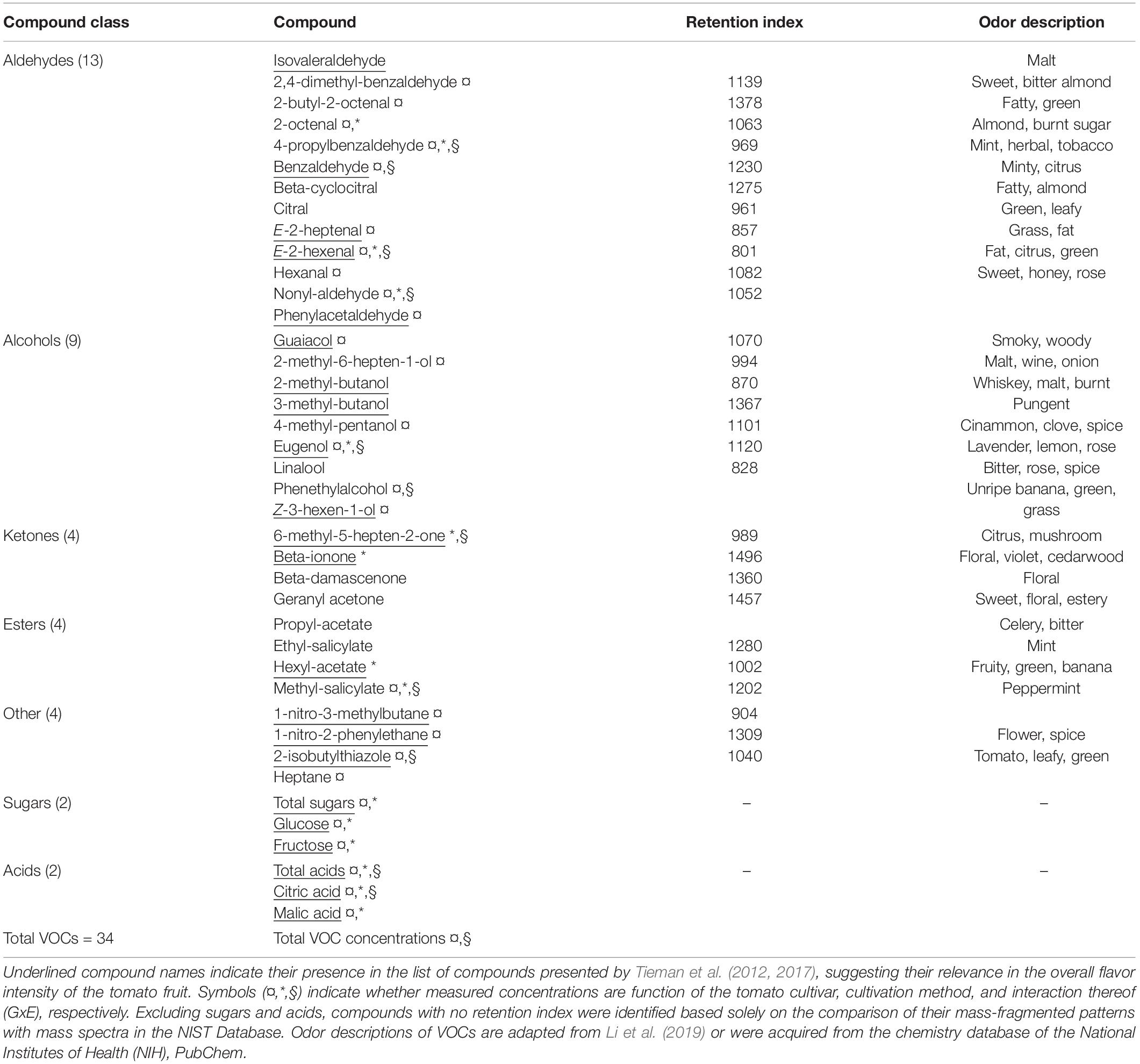
Table 1. List of measured sugars and acids as well as volatile compounds (VOCs) detected in the headspace of juice made from tomato fruit of five different cultivars.
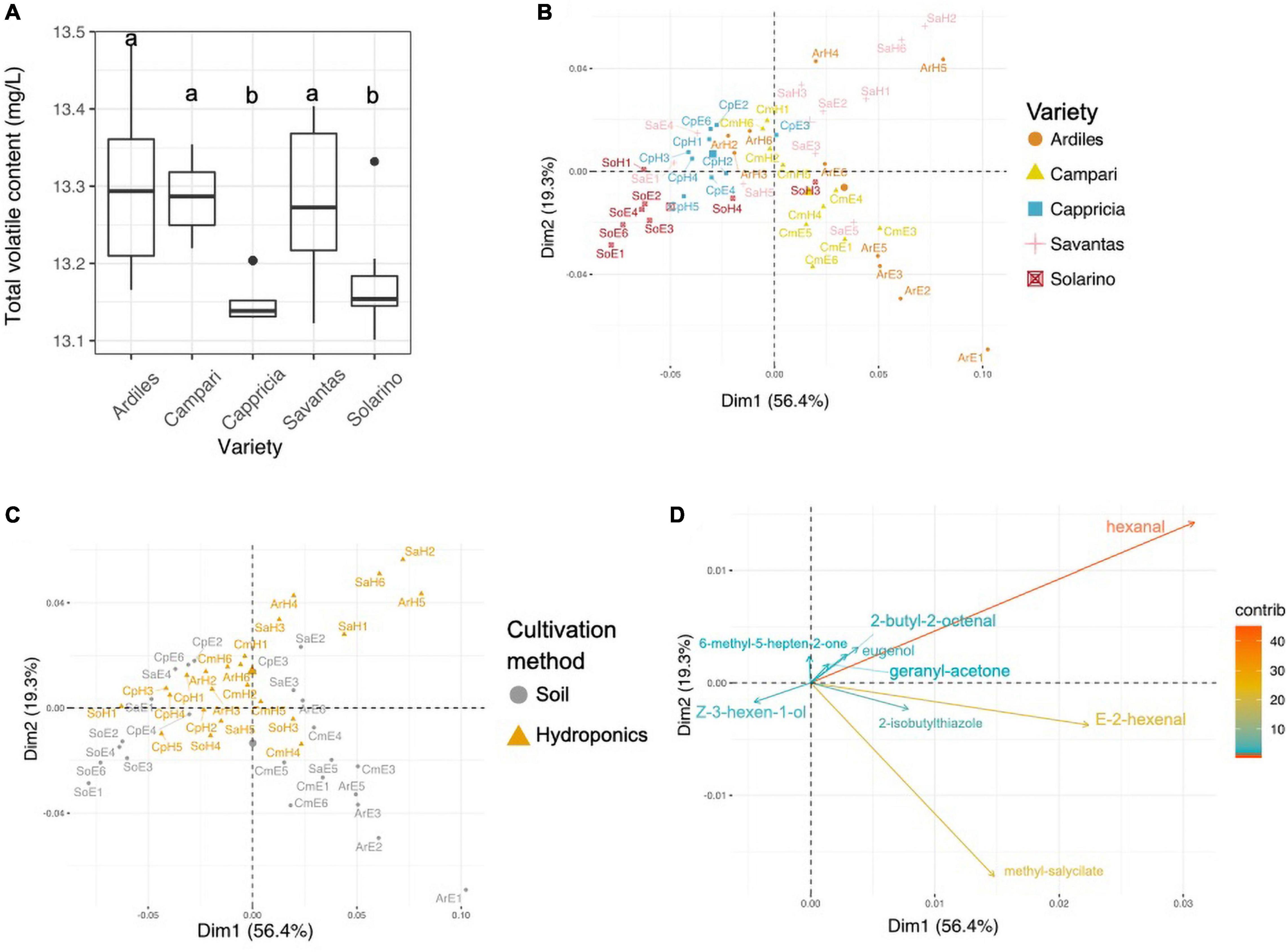
Figure 2. (A) Total concentration of organic volatile compounds (VOCs) in tomato fruit of five different cultivars expressed in mg/L. Letters over each box indicate members of homologous groups according to a Tukey HSD test for pairwise comparisons among tomato cultivars. (B) Principal component analyses (PCA) showing groupings of individual samples colored by tomato cultivar. (C) PCA with individual samples colored according to the employed cultivation method. (D) PCA loadings of VOCs with a contribution >10.
Tomato fruit grown in soil showed significantly higher concentrations of glucose and fructose than those grown in hydroponics (Figure 3A). Fruit of the cultivar ‘Solarino’ had significantly more sugar than the other cultivars, with over 3.5% mean sugar content (Figure 3B). In contrast, amounts of the main organic acids were significantly higher in tomatoes grown in hydroponics (Figure 3C). Citric acid predominated in tomato fruit reaching up to 5 g/L and its concentration was significantly higher in the cultivars ‘Ardiles,’ ‘Campari’ and ‘Solarino’ grown in hydroponics. The content of malic acid, on the other hand, reached a maximum of 750 mg/L and was significantly higher in fruit of the cultivar ‘Cappricia’ (Figure 3D). Overall, the major contributors to the total and individual sugar content were mainly the cultivation method and secondly, the tomato cultivar (F-values = 24.2, 9.2; p < 0.001 respectively). All three factors significantly shaped the total acid concentration, with the main contributor being the cultivation method, followed by the cultivar and lastly, the GxE interaction (F-values = 12.05, 6.72 and 2.9, p < 0.01) respectively. Amounts of citric acid were mainly a function of the tomato cultivar, followed by the cultivation method and lastly the GxE interaction (F-values = 11.6, 10.8 and 3.9; p < 0.01, respectively), while the cultivar and the cultivation method were the main contributors to malic acid concentrations in the tomato fruit (F-values = 46.8, 8.3; p < 0.001, respectively) (Table 1).
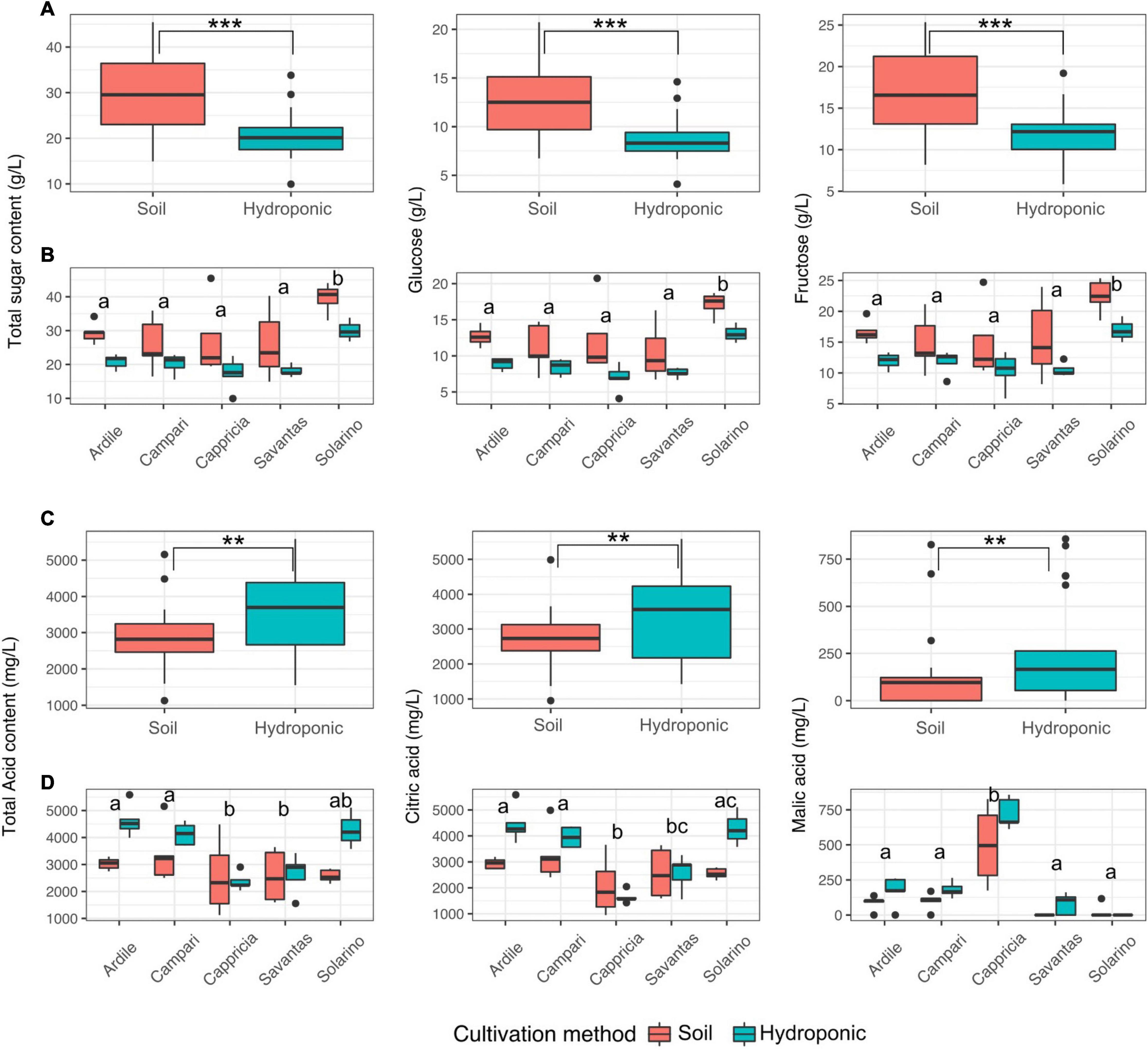
Figure 3. (A) Total sugar as well as glucose and fructose concentrations in juices prepared from tomatoes grown in soil and in hydroponics. (B) Sugar concentrations in juices of five different tomato cultivars grown in soil and hydroponics. (C) Total acid as well as malic and citric acid concentrations in juices prepared from tomatoes grown in soil and in hydroponics. (D) Acid concentrations in juices of five different tomato cultivars grown in soil and in hydroponics. Letters over each box indicate members of homologous groups according to a Tukey HSD test for pairwise comparisons among tomato cultivars. Significance values computed by ANOVA for each factor are indicated as (**) for p > 0.01 and (***) for p > 0.001.
The Cultivation Method Affects the Bacterial Diversity in Tomato Fruit of Some Cultivars and Significantly Shapes the Community Structure
Sequencing of the V5 to V7 region of the 16S rRNA genes present in 50 tomato puree samples (5 cultivars × 5 replicates × 2 cultivation conditions resulted in a total of 983,497 high-quality merged reads, corresponding to an average of 19,669.94 ± 11,641.69 reads per sample (n = 50) and a mean length of 375 bp. ASVs with relative abundances greater than 0.01% were used for further analyses in R 3.6.3, resulting in a total of 851 ASVs.
We investigated the role of both, cultivation method and tomato cultivar, in the diversity and composition of the reproducibly occurring bacterial communities of the fruit. We could not ascertain any significant differences in the numbers of total rASVs or diversity between fruit grown in soil and those from hydroponic cultures (Figure 4A). However, there were significant effects observed in the GxE interaction [F = 21.52, Pr (> F) = 0.0001]: numbers of identified rASVs were significantly higher in hydroponic “Cappricia” and “Savantas” tomatoes compared to their soil-derived counterparts, whereas soil cultivated “Ardiles” fruit harbored more bacterial rASVs than those that were grown in hydroponics. The same trend was observed in the Simpson indices (Figure 4B). In contrast, no significant differences in alpha diversity among tomato cultivars were observed. In terms of beta diversity, the cultivation method, tomato cultivar and the interaction thereof significantly shaped the bacterial community composition of the tomato fruit (Table 2). The largest amount of variation among samples was observed in the communities of hydroponic “Solarino” fruit as indicated by the scattering of samples across the second axis of the constrained ordination plot (CAP) depicted in Figure 5. Moreover, the effect of the cultivar on the bacterial community structures of the fruit was significantly stronger in soil-grown tomatoes than from those of hydroponic cultures: while communities of all soil-derived tomatoes differed significantly from each other and from their hydroponic counterparts, no significant differences were identified among hydroponic fruit of “Solarino” and the rest of cultivars, as well as between “Cappricia” and “Savantas” (Supplementary Table 3).
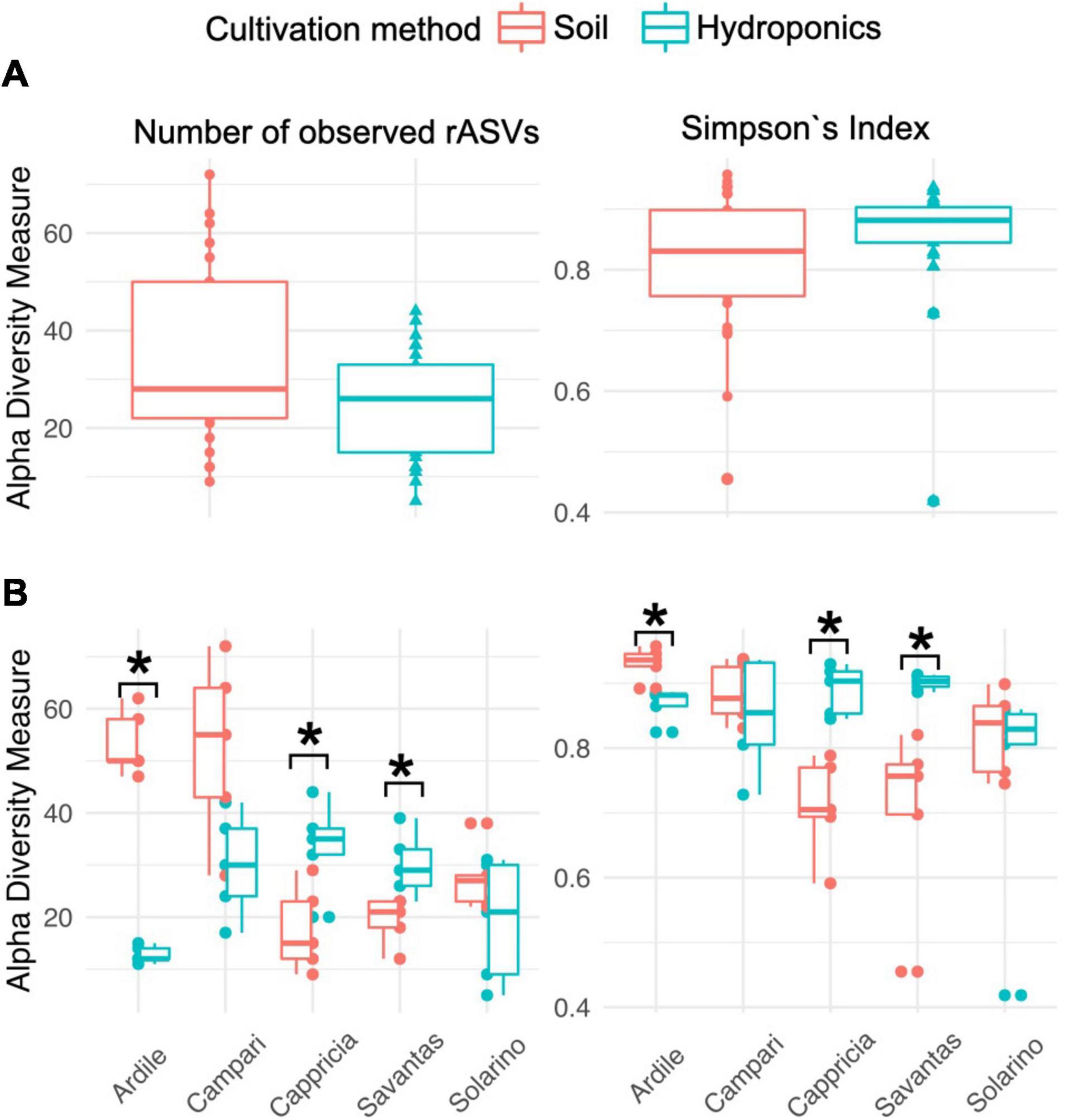
Figure 4. (A) Total observed rASVs and diversity (Simpsons Index) of tomato fruit grown in soil and in hydroponics. (B) Alpha diversity measures considering the effect of cultivation method in each tomato cultivar. Significance values computed by ANOVA are indicated as (*) for p > 0.05.
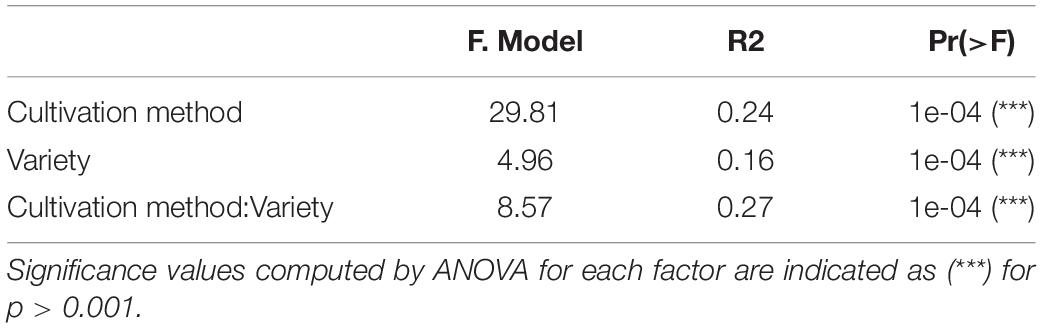
Table 2. Roles of cultivation method and variety in the bacterial community structures of tomato fruits.
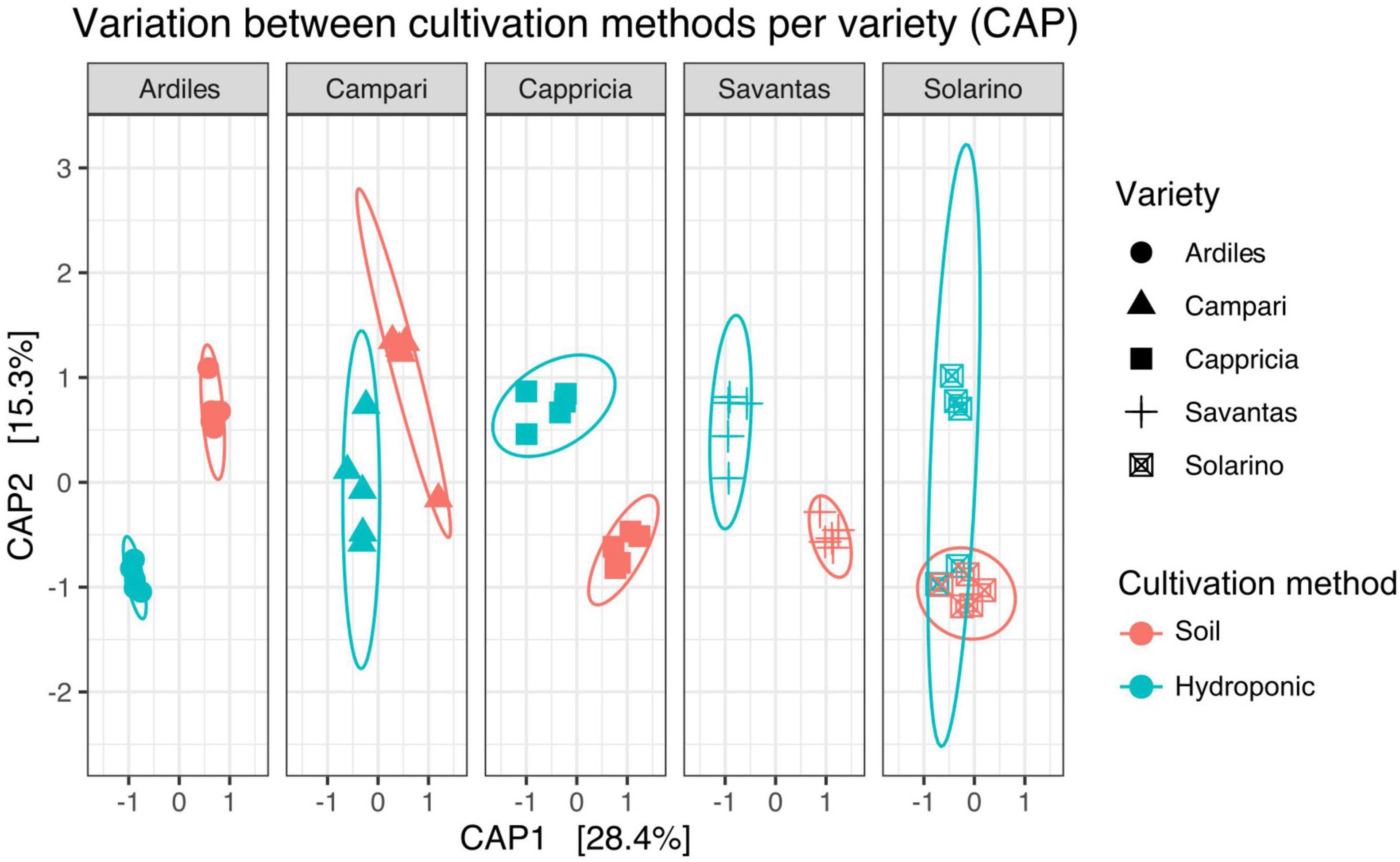
Figure 5. Variation across samples of both cultivation methods is visible for each tomato cultivar (CAP).
The Taxonomic Composition of Bacterial Communities Colonizing Tomato Fruit From Cultivation in Soil and in Hydroponics
Overall, a total of 851 ASVs were classified among 77 bacterial orders of 18 classes, with an average of 17.02 ASVs per sample (Supplementary Data 1). ASVs classified as Proteobacteria were predominant among tomato fruit, with a relative abundance of over 30% of total reads, followed by Firmicutes, Actinobacteria and Bacteroidetes with around 5% prevalence each. Most represented were orders of the Gammaproteobacteria (Enterobacteriales and Pseudomonadales, Betaproteobacteriales), and Alphaproteobacteria (Rhizobiales and Sphingomonadales). At the genus level, Pseudomonas, Burkholderia-Caballeronia-Paraburkholderia, Raoultella, Sphingomonas, Massilia and Paenibacillus were among the most abundant taxa detected in the tomato fruit in this study (Supplementary Figure 2). In order to identify representative bacterial communities (“core”) within tomatoes from both cultivation methods, we extracted reproducibly occurring ASVs (rASVs) from the data set for further investigation. One hundred and twenty-seven (127) rASVs occurred in at least 3 of 5 replicates and were classified among 22 bacterial orders. These reproducibly occurring communities represented a higher portion of the overall bacterial abundance in hydroponic fruit (up to 80%) compared to those from soils (around 60%). Differences between reproducibly occurring communities of tomatoes from both cultivation methods were minimal in taxonomic composition but evident through the lack of members of the Flavobacteriales and Kineosporiales in hydroponic tomatoes (Figure 6A). Furthermore, of the remaining 55 “accessory” bacterial orders (i.e., which did not occur in a reproducible fashion), 21 were identified exclusively in soil-grown tomatoes and eleven were specific to fruit from plants grown in hydroponics (Figure 6B). Accessory bacterial orders that were specific to any of the cultivation methods occurred in low relative abundances (Figure 6D). Twenty-three orders occurred in fruit grown in both soil and hydroponics (Supplementary Data 1 and Figures 6C,D).
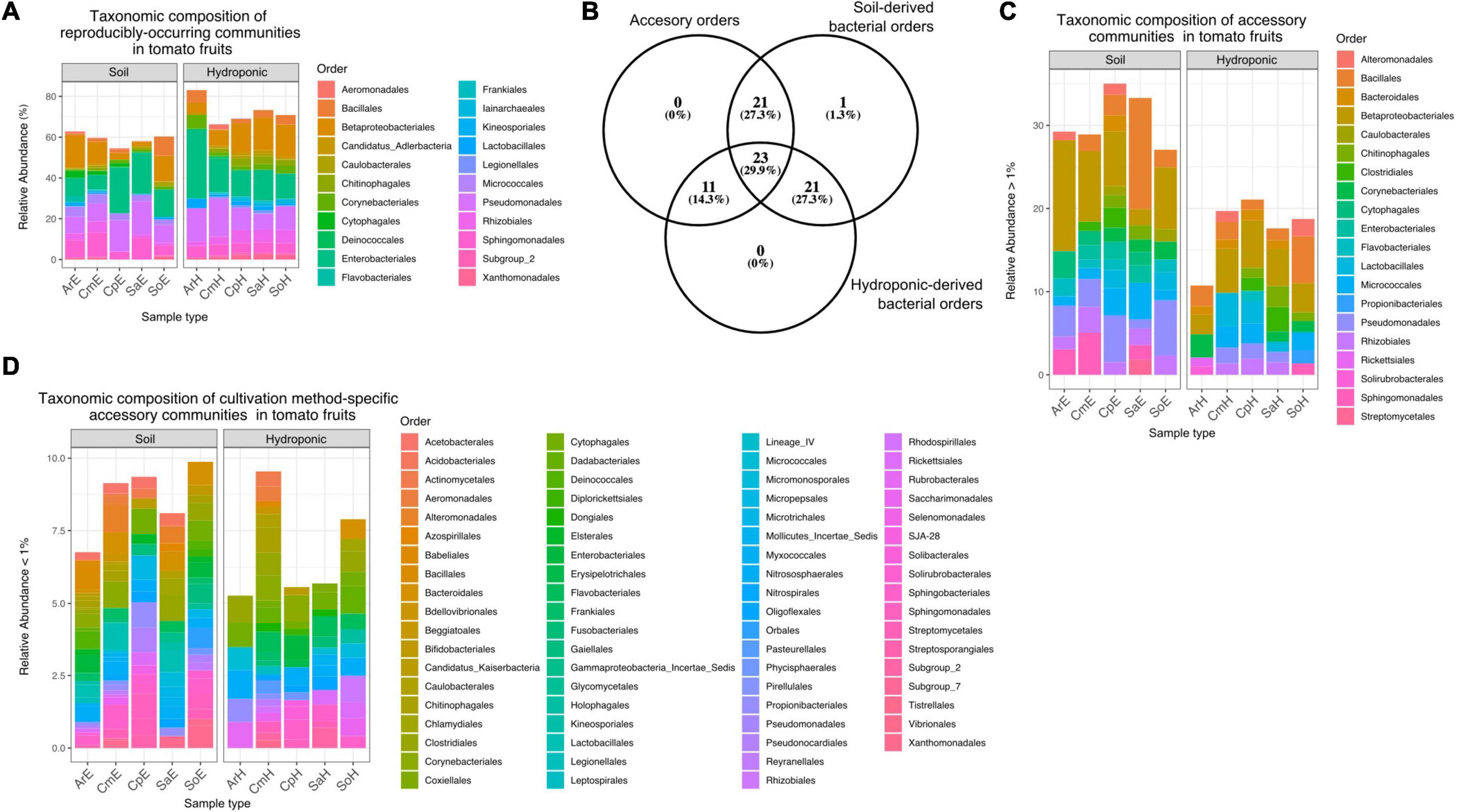
Figure 6. (A) Relative abundance of core orders in soil- and hydroponic grown tomato fruit. rASVs represent a higher portion of the overall communities in hydroponic tomatoes (65–82%) as compared to soil derived fruit (∼60%). Reproducibly occurring communities of hydroponic tomatoes lack member rASVs from Flavobacteriales and Kineosporiales. (B) Number of accessory bacterial orders (accessory) that are specific to soil-grown tomatoes or hydroponic-grown fruit. (C) Abundance of 20 predominant accessory bacterial orders detected in tomato fruit. None of these orders were specific to any cultivation method, but rather common among them. (D) Accessory bacterial orders that were specific to each cultivation method occurred in low relative abundance.
Indicator Genera of Cultivation Method Correlate With the Abundance of Specific Volatile and Non-volatile Compounds
Relative abundances of 99 rASVs correlated with the concentration of at least one volatile or non-volatile compound detected in this study. Among these, 46 rASVs belonging to 23 bacterial genera were identified as indicator species in at least one sample type (cultivar/cultivation method combinations) (Supplementary Figure 3A). Indicator rASVs of soil-grown tomatoes included members of Pseudomonas, Pantoea, Sphingomonas, Massilia, Methylobacterium and Hymenobacter, whose relative abundances correlated significantly with higher concentrations of methyl salicylate, nonyl aldehyde, 2-isobutylthiazole, E-2-hexenal and lower levels of 2-octenal and 6-methyl-5-hepten-2-one. Furthermore, soil derived “Solarino” tomatoes were characterized by higher abundances of a Bacillus (rASV_135) which was depleted in their hydroponic counterparts and not detectable in other tomato cultivars. This Bacillus rASV_135 correlated with higher abundances of fructose and glucose, as well as increased concentration of important aroma volatiles such as Z-3-hexen-1-ol, E-2-heptenal and benzaldehyde. Moreover, its abundance negatively correlated with lower concentrations of geranylacetone, E-2-hexenal and hexanal. On the other hand, indicator communities of tomatoes grown in hydroponics were represented by members of Burkholderia-Caballeronia-Paraburkholderia, Dyella, Mesorhizobium, Raoultella, Acinetobacter and Novosphingobium. The relative abundance of the former three genera correlated with higher levels of geranylacetone while the latter three correlated with concentrations of beta-ionone, 6-methyl-5-hepten-2-one and hexylacetate. Furthermore, relative abundances of indicator genera from hydroponic tomatoes correlated with decreased sugar levels. Moreover, an rASV classified as Raoultella (rASV_126) was characteristic of “Savantas,” “Campari” and “Cappricia” tomatoes grown in hydroponics and correlated positively with citric acid concentrations (Figure 7 and Supplementary Figure 3B).
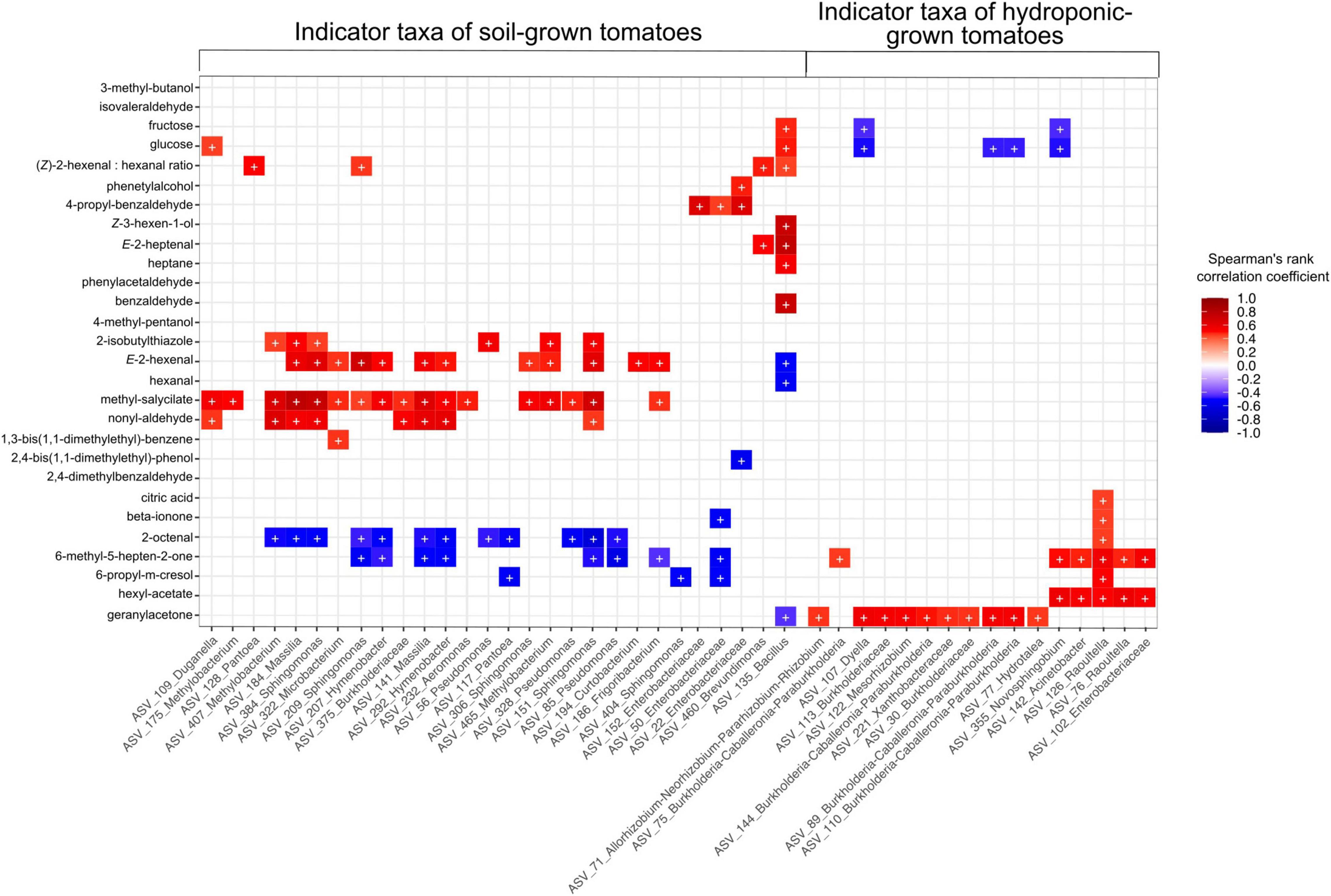
Figure 7. Relative abundance of 46 indicator rASVs for the Cultivation soil × Cultivar (GxE) interaction correlate with concentrations of at least one aroma or flavor compound in this study. Symbols in cells (+) indicate significant correlations (fdr adjusted p > 0.05). Colors in cells with non-significant correlations have been removed for simplification.
Discussion
Aroma Profiles of Fruits Are Characteristic of the Cultivar Whereas the Cultivation Method Affects Amounts of Flavor Compounds
In this study, we asked whether tomatoes from different cultivars grown in soil or in a hydroponic system differ for their emission or content of volatiles, acids, and sugars, and whether such changes in chemical composition may be linked to the bacterial communities in tomato fruit. Volatile profiles in this study were mainly shaped by the tomato cultivar, rather than by the cultivation method. Volatiles related to flavor are derived from essential compounds like fatty acids, amino acids and color carotenoids (Goff and Klee, 2006), which vary in amounts and bioavailability among fruits of wild species and heirlooms and also across commercial hybrids (Rambla et al., 2014). This variability has been attributed to modern breeding processes, which led to the loss of superior alleles for volatiles associated with good aroma (Tieman et al., 2017).
In contrast, the levels of acids and sugars were mainly affected by the cultivation method in this study: soil grown tomatoes showed significantly higher levels of dominant sugars like glucose and fructose, whereas hydroponic tomatoes accumulated higher amounts of citric and malic acids. Numerous studies have shown how the cultivation conditions strongly affect tomato yield, fruit morphology and biochemistry (Kuti and Konuru, 2005; Ortiz et al., 2007; Roselló et al., 2011; Adalid et al., 2012; Panthee et al., 2013; Figàs et al., 2015, 2018): regarding fruit quality, tomato cultivars grown in greenhouse conditions were richer in sugars and antioxidants but poorer in lycopene and ß-carotene than tomatoes from open fields (Kuti and Konuru, 2005; Beckles, 2012; Figàs et al., 2015, 2018), but no difference was found for organic acids under these conditions (Galiana-Balaguer et al., 2006; Figàs et al., 2018). Similarly, grapevine cultivars grown under different conditions result in berries and wines with different biochemical composition and sensory characteristics (Pereira et al., 2005; Son et al., 2009; Robinson et al., 2012).
Several studies have shown that moderate stress conditions may improve tomato fruit quality through increased concentration of flavor compounds in some cultivars including sugars, organic acids, carotenoids and phenolic compounds (Zheng et al., 2013; Albert et al., 2016a,b). While those studies focused on the effects of abiotic stresses during cultivation (e.g., water deficit, salinity and/or heat) on the metabolite production of tomato fruit (reviewed by Quinet et al., 2019), it is possible that tomato plants grown in soil undergo intensified biotic stresses compared to those from hydroponic cultures, which could ultimately lead to changes in the fruit chemistry. Soils contain an extraordinary number and diversity of microorganisms that may interact directly or indirectly with the roots of a nearby plant, e.g., by inducing immune responses and/or altering metabolic pathways (Zamioudis and Pieterse, 2011; Hardoim et al., 2015). In fact, we found that soil grown “Ardiles” and “Campari” tomatoes were characterized by high levels of methyl salicylate, a volatile compound from the phenylpropanoid metabolic pathway known to be involved in plant stress responses (Shulaev et al., 1997; Seskar et al., 1998). This goes hand-in-hand with the findings of Chialva and colleagues, who observed that the microbiota rather than the chemico-physical properties of native soils were crucial in eliciting stress responses in some tomato cultivars, including phenylpropanoid metabolism (Chialva et al., 2018). As for organoleptic perception, methyl salicylate has been described to possess “medicinal” or “peppermint” notes (Zanor et al., 2009), and although it is one of the major contributors to overall tomato flavor, it has been correlated with poor flavor acceptability at higher levels (Vogel et al., 2010). Therefore, biotic stresses related to the microbial complexity of the cultivation substrate may contribute to the altered production of sugars, acids and volatiles in a cultivar-specific manner.
Bacterial Diversity and Community Composition of Tomato Fruits Is Shaped by the Cultivation Method in a Cultivar-Dependent Manner
We further analyzed the bacterial community composition of the tomato fruit and observed no significant differences in total numbers of rASVs nor in alpha diversity measures (Simpson’s index) between tomatoes grown in soil or in hydroponics. However, when we considered the tomato cultivar, the role of the cultivation method in alpha diversity became evident for ‘Ardiles,’ ‘Cappricia’ and ‘Savantas’ tomatoes, while no effect was found in ‘Solarino’ and ‘Campari.’
Plant cultivars are often differently affected by the environment. This phenomenon is known as genotype x environment (GxE) interaction. The tomato plants in this study were grown in controlled hydroponic culture with relatively limited stimuli and environmental variations and, on the other hand, in pots filled with soil inside a wire-house with natural fluctuations in temperature, precipitation and interactions with (micro-)organisms below- and above-ground, providing a myriad of stimuli. It is therefore possible that each of the tomato cultivars reacted differently to these cultivation-dependent stimuli, resulting in fruit with divergent chemical characteristics. These variations in chemical environment within fruit may ultimately provide conditions that support distinct bacterial communities in tomatoes in each cultivar.
Indeed, we observed differences in the composition of the bacterial communities colonizing tomato fruit among the different cultivars and between the cultivation methods. Tomato fruit from all cultivars in this study did not only differ from each other in size (Figure 1), but also in pericarp consistency, locule number and, consequently, water content (not shown). Different tissues of tomato and apple fruit were found to be colonized by distinct bacterial communities (Dong et al., 2019; Wassermann et al., 2019). Therefore, cultivar-specific fruit characteristics like morphology, nutritional availability within the carposphere, size and biomass are likely to be responsible for the observed differences in microbial diversity among cultivars.
In contrast to soils, hydroponic cropping ecosystems are characterized by poor microbial complexities as the microorganisms in these environments derive mainly from plants or seeds, water, insects and personnel (Strayer, 1994). Moreover, plants harbor microbial communities inside, and on every organ. The assembly of these communities depends, among other factors, on the microbial availability in their immediate surroundings. Accordingly, community composition of soil-grown tomatoes of all cultivars differed significantly from their hydroponic counterparts. Interestingly, tomatoes from soil cultures developed “signature” bacterial communities, which were unique for each cultivar and were not observed in hydroponic fruit. Due to their microbial diversity and abundance, soils are considered the primary force driving microbial communities of roots and, albeit less pronounced, those of upper plant organs such as fruit (Zarraonaindia et al., 2015; Müller et al., 2016). Many plant-associated bacteria derive from the rhizosphere environment and may colonize the endospheres by mechanisms that are analogous for both soil and hydroponic cultures: bacteria are attracted to the rhizosphere environment by root exudates and rhizodeposits (Compant et al., 2010). From the roots, bacteria can also migrate to aerial compartments through the xylem of intercellular spaces (Hardoim et al., 2015). Bacteria in fruit may also derive from above-ground sources, like aerosols, pollen or insects (Compant et al., 2011; Glassner et al., 2017; Allard et al., 2018). Root exudate composition shapes microbial communities and have been shown to vary in amount and chemical composition depending on the plant genotype, health and developmental stage (Sasse et al., 2018). Furthermore, irrespective of the point of entry into the plant, microbial colonizers elicit immune responses that are also highly dependent on the above-mentioned host characteristics (Chialva et al., 2018), which shape the community composition by facilitating colonization of some microbial species and discriminating potential pathogens (Brader et al., 2017). Therefore, it is not surprising that fruit of different tomato cultivars harbor distinct microbiotas when provided with a highly diverse substrate like soil.
Bacterial Taxa Correlate With Flavor and Aroma Chemistry of Tomato Fruits
We further aimed to address whether the differences in flavor compounds observed among tomato fruit grown under both cultivation methods are attributable to the abundances of specific members within the fruit microbiota. Indeed, we observed significant correlations among indicator species of soil-grown tomatoes, which included members of genera that have been previously found to be associated with fruit of tomatoes and other plants such as Bacillus, Pseudomonas, Sphingomonas, Massilia, and Methylobacterium (Verginer et al., 2010; Ottesen et al., 2013; Gilbert et al., 2014; Allard et al., 2018). Abundances of these taxa were associated with higher concentrations of volatiles described to contribute to the characteristic pungent, green, and leafy notes of the tomato fruit, namely methyl salicylate, nonyl aldehyde, 2-isobutylthiazole, E-2-hexenal (Klee, 2010; Vogel et al., 2010; Wang et al., 2015; Li et al., 2019). In contrast, indicator species of hydroponic tomatoes included members of Burkholderia-Caballeronia-Paraburkholderia, as well as genera that have been less described in fruit, namely Dyella, Mesorhizobium, Raoultella, Acinetobacter and Novosphingobium. Indicator members of these taxa correlated negatively with sugar levels and positively with levels of citric acid and apocarotenoid volatiles such as geranyl acetone, beta-ionone and 6-methyl-5-hepten-2-one. Apocarotenoid volatiles are products of the oxidative cleavage of various linear and cyclic carotenoids such as lycopene and beta carotene, which fulfill functions as colorants and nutrients in tomatoes. Volatiles derived from carotenoids have generally been described as having fruity and/or floral attributes and contribute to the sweetness perception of tomato fruit (Vogel et al., 2010). Also, reduced levels of beta ionone and 6-methyl-5-hepten-2-one have been associated with poor flavor in tomatoes (Tieman et al., 2017). Therefore, in terms of organoleptic perception, it is possible that the loss of sweetness related to the decreased sugar content observed in hydroponic tomatoes may be compensated to an extent by an increase in the amounts of apocarotenoid volatiles, resulting in an acceptable product for the consumer.
Furthermore, we observed that the relative abundance of rASV_135, classified as Bacillus (detected in all biological replicates of soil “Solarino” tomatoes but not in other samples) correlated with higher levels of both sugars and important aroma compounds. It is well known that Bacillus spp. are prominent soil inhabitants and typical plant colonizers (Yuan et al., 2015; Amaresan et al., 2019). He and colleagues showed that several Bacillus strains enhanced yield, growth and nutrient uptake of tomato plants when inoculated in the soil (He et al., 2019). A recent study also detected and isolated various Bacillus strains from tomato seeds which displayed multiple positive traits for their hosts including siderophore production and ACC-deaminase activity (Bergna et al., 2018). Accordingly, the authors proposed the seeds as carriers of bacteria into subsequent generations of tomato plants providing beneficial traits. To address the possible origins of potential modulators of tomato flavor such as rASV_135, we looked into bacterial communities of both irrigation water and tomato seedlings germinated under sterile conditions (unpublished). While the irrigation water had the highest overlap of ASVs with fruit in all cultivars, rASV_135 occurred persistently within “Solarino” seedlings, together with other seedling-derived rASVs of Pantoea, Methylobacterium and Burkholderia-Caballeronia and Paraburkholderia, which also correlated with abundance of tomato flavor compounds (Supplementary Data 2). Therefore, it is possible that within the fruit, we have detected seed-derived Bacilli and other microorganisms which may thrive when the seed is cultivated under certain conditions (e.g., soil vs. hydroponics). Although the correlation between the abundance of microbiota members like rASV_135 and the accumulation of flavor-relevant compounds in soil cultivated “Solarino” tomatoes does not necessarily imply causation, the potential role of fruit associated bacteria in the flavor profiles of tomato merits further investigation.
Summarizing, bacterial populations within fruit are shaped by the cultivation method and correlated with the concentrations of chemical compounds known for their implications in the organoleptic quality in these commodities. The tomato cultivar was indeed a key factor shaping the bacterial communities within fruit only when plants were grown in soil, indicating that the microbial diversity of the substrate influences the fruit microbiota. It remains unclear to what extent the correlations between microbial indicator taxa and aroma relevant compounds reflect causal associations. Microbial-mediated modulation of the tomato fruit flavor may occur either directly through the release of metabolites in the fruit or indirectly by enzymatic activities following cell disruption (Bokulich et al., 2016). Further studies investigating the function of the active members of the tomato fruit microbiota will help to disentangle the plant response to key bacterial populations in tomato fruit and are required to understand the role of plant-microbe interactions in fruit flavor. Moreover, the exact pathways involved in the accumulation of aroma-relevant volatiles and their precursors still demand investigation as tomato flavor is not solely based on the amounts of the single volatiles and non-volatile compounds but is rather shaped by the interactions among them, enhancing the perception of some traits while canceling others.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI BioProject – PRJNA782607.
Author Contributions
CE established the chemical analyses of flavor compounds, and mainly contributed in analyses of chemical and microbial data and in the composition of the manuscript. FB and PU contributed in sampling, sample preparation, sequencing and microbial data analyses. MG was mainly involved in tomato plant cultivation and sampling. LB performed analyses of sugars and acids. LA performed the raw data processing of the microbial sequences. JN and BM worked on experimental conception and design, data analyses and discussion of results. All authors contributed to the article and approved the submitted version.
Funding
This work was created within a research project of the Austrian Competence Centre for Feed and Food Quality, Safety and Innovation (FFoQSI). The COMET-K1 competence centre FFoQSI is funded by the Austrian federal ministries BMK, BMDW and the Austrian provinces Lower Austria, Upper Austria and Vienna within the scope of COMET – Competence Centers for Excellent Technologies. The program COMET is handled by the Austrian Research Promotion Agency FFG.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to Harry Bein for ensuring seamless GC analyses and the LGV Sonnengemüse cooperative for providing tomato material and cultivation tips.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.775722/full#supplementary-material
References
Adalid, A. M., Roselló, S., Valcárcel, M., and Nuez, F. (2012). Analysis of the genetic control of β-carotene and l-ascorbic acid accumulation in an orange-brownish wild cherry tomato accession. Euphytica 184, 251–263. doi: 10.1007/s10681-011-0584-x
Agius, C., von Tucher, S., Poppenberger, B., and Rohzon, W. (2018). Quantification of sugars and organic acids in tomato fruit. MethodsX 5, 537–550. doi: 10.1016/j.mex.2018.05.014
Albert, E., Gricourt, J., Bertin, N., Pateyron, S., Tamby, J.-P., Bitton, F., et al. (2016a). Genotype by watering regime interaction in cultivated tomato: lessons from linkage mapping and gene expression. Theor. Appl. Genet. 129, 395–418. doi: 10.1007/s00122-015-2635-5
Albert, E., Segura, V., Gricourt, J., Bonnefoi, J., Derivot, L., and Causse, M. (2016b). Association mapping reveals the genetic architecture of tomato response to water deficit: focus on major fruit quality traits. J. Exp. Bot. 67, 6413–6430. doi: 10.1093/jxb/erw411
Allard, S. M., Ottesen, A. R., Brown, E. W., and Micallef, S. A. (2018). Insect exclusion limits variation in bacterial microbiomes of tomato flowers and fruit. J. Appl. Microbiol. 125, 1749–1760. doi: 10.1111/jam.14087
Amaresan, N., Jayakumar, V., Kumar, K., and Thajuddin, N. (2019). Biocontrol and plant growth-promoting ability of plant-associated bacteria from tomato (Lycopersicum esculentum) under field condition. Microb. Pathog. 136:103713. doi: 10.1016/j.micpath.2019.103713
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed March, 2020).
Baldwin, E. A., Goodner, K., and Plotto, A. (2008). Interaction of volatiles, sugars, and acids on perception of tomato aroma and flavor descriptors. J. Food Sci. 73, 294–307. doi: 10.1111/j.1750-3841.2008.00825.x
Beckles, D. M. (2012). Factors affecting the postharvest soluble solids and sugar content of tomato (Solanum lycopersicum L.) fruit. Postharvest. Biol. Technol. 63, 129–140. doi: 10.1016/j.postharvbio.2011.05.016
Bengtsson-Palme, J., Hartmann, M., Eriksson, K. M., Pal, C., Thorell, K., Larsson, D. G. J., et al. (2015). metaxa2: improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Mol. Ecol. Res. 15, 1403–1414. doi: 10.1111/1755-0998.12399
Berg, G., Rybakova, D., Grube, M., and Köberl, M. (2016). The plant microbiome explored: implications for experimental botany. J. Exp. Bot. 67, 995–1002. doi: 10.1093/jxb/erv466
Bergna, A., Cernava, T., Rändler, M., Grosch, R., Zachow, C., and Berg, G. (2018). Tomato seeds preferably transmit plant beneficial endophytes. Phytobiomes J 2, 183–193. doi: 10.1094/PBIOMES-06-18-0029-R
Bokulich, N. A., Collins, T. S., Masarweh, C., Allen, G., Heymann, H., and Ebeler, S. E. (2016). Associations among wine grape microbiome, metabolome, and fermentation behavior suggest microbial contribution to regional wine characteristics. mBio 7, e00631–e616. doi: 10.1128/mBio.00631-16
Brader, G., Compant, S., Vescio, K., Mitter, B., Trognitz, F., Ma, L.-J., et al. (2017). Ecology and genomic insights into plant-pathogenic and -nonpathogenic endophytes. Annu. Rev. Phytopathol. 55, 61–83. doi: 10.1146/annurev-phyto-080516-035641
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Chialva, M., Salvioli di Fossalunga, A., Daghino, S., Ghignone, S., Bagnaresi, P., and Chiapello, M. (2018). Native soils with their microbiotas elicit a state of alert in tomato plants. New Phytol. 220, 1296–1308. doi: 10.1111/nph.15014
Coleman-Derr, D., and Tringe, S. G. (2014). Building the crops of tomorrow: advantages of symbiont-based approaches to improving abiotic stress tolerance. Front. Microbiol. 5:283. doi: 10.3389/fmicb.2014.00283
Compant, S., Clément, C., and Sessitsch, A. (2010). Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 42, 669–678. doi: 10.1016/j.soilbio.2009.11.024
Compant, S., Mitter, B., Colli-Mull, J. G., Gangl, H., and Sessitsch, A. (2011). Endophytes of grapevine flowers, berries, and seeds: identification of cultivable bacteria, comparison with other plant parts, and visualization of niches of colonization. Microb. Ecol. 62, 188–197.
Deshmukh, Y., Khare, P., and Patra, D. (2016). Rhizobacteria elevate principal basmati aroma compound accumulation in rice variety. Rhizosphere 1, 53–57. doi: 10.1016/j.rhisph.2016.07.001
Dong, C.-J., Wang, L.-L., Li, Q., and Shang, Q.-M. (2019). Bacterial communities in the rhizosphere, phyllosphere and endosphere of tomato plants. PLoS One 14:e0223847. doi: 10.1371/journal.pone.0223847
Droby, S., and Wisniewski, M. (2018). The fruit microbiome: a new frontier for postharvest biocontrol and postharvest biology. Postharvest. Biol. Technol. 140, 107–112. doi: 10.1016/j.postharvbio.2018.03.004
Escobar Rodríguez, C., Mitter, B., Antonielli, L., Trognitz, F., Compant, S., and Sessitsch, A. (2018). Roots and panicles of the C4 model grasses Setaria viridis (L). and S. pumila host distinct bacterial assemblages with core taxa conserved across host genotypes and sampling sites. Front. Microbiol. 9:2708. doi: 10.3389/fmicb.2018.02708
FAO (2013). FAO Plant Production and Protection Paper 2017. Rome: Food and Agriculture Organization.
Figàs, M. R., Prohens, J., Casanova, C., Fernández-de-Córdova, P., and Soler, S. (2018). Variation of morphological descriptors for the evaluation of tomato germplasm and their stability across different growing conditions. Sci. Hortic. 238, 107–115. doi: 10.1016/j.scienta.2018.04.039
Figàs, M. R., Prohens, J., Raigón, M. D., Fita, A., Garcia-Martinez, M. D., Casanova, C., et al. (2015). Characterization of composition traits related to organoleptic and functional quality for the differentiation, selection and enhancement of local varieties of tomato from different cultivar groups. Food Chem. 187, 517–524. doi: 10.1016/j.foodchem.2015.04.083
Galiana-Balaguer, L., Roselló, S., and Nuez, F. (2006). Characterization and selection of balanced sources of variability for breeding tomato (Lycopersicon) internal quality. Genet. Resour. Crop Evol 53, 907–923. doi: 10.1007/s10722-004-6696-6
Gilbert, J. A., van der Lelie, D., and Zarraonaindia, I. (2014). Microbial terroir for wine grapes. Proc. Natl. Acad. Sci. U.S.A. 111, 5–6. doi: 10.1073/pnas.1320471110
Glassner, H., Zchori-Fein, E., Yaron, S., Sessitsch, A., Sauer, U., and Compant, S. (2017). Bacterial niches inside seeds of Cucumis melo L. Plant Soil 422, 101–113. doi: 10.1007/s11104-017-3175-3
Goff, S. A., and Klee, H. J. (2006). Plant volatile compounds: sensory cues for health and nutritional value? Science 311, 815–819. doi: 10.1126/science.1112614
Hardoim, P. R., van Overbeek, L. S., Berg, G., Pirttilä, A. M., Compant, S., Campisano, A., et al. (2015). The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79, 293–320. doi: 10.1128/MMBR.00050-14
He, Y., Pantigoso, H. A., Wu, Z., and Vivanco, J. M. (2019). Co-inoculation of Bacillus sp. and Pseudomonas putida at different development stages acts as a biostimulant to promote growth, yield and nutrient uptake of tomato. J. Appl. Microbiol. 127, 196–207. doi: 10.1111/jam.14273
Hervé, M. (2021). RVAideMemoire: Testing and Plotting Procedures for Biostatistics. R package version 0.9-69-3. Available online at: https://cran.r-project.org/web/packages/RVAideMemoire/RVAideMemoire.pdf (accessed March, 2020).
Kanayama, Y. (2017). Sugar metabolism and fruit development in the tomato. Hortic. J. 86, 417–425. doi: 10.2503/hortj.OKD-IR01
Kassambara, A., and Mundt, F. (2020). Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. Available online at: https://CRAN.R-project.org/package=factoextra (accessed March, 2020).
Klee, H. J. (2010). Improving the flavor of fresh fruit: genomics, biochemistry, and biotechnology. New Phytol. 187, 44–56. doi: 10.1111/j.1469-8137.2010.03281.x
Kochevenko, A., Araújo, W. L., Maloney, G. S., Tieman, D. M., Phuc, T. D., and Taylor, M. G. (2012). Catabolism of branched chain amino acids supports respiration but not volatile synthesis in tomato fruit. Mol. Plant 5, 366–375. doi: 10.1093/mp/ssr108
Kuti, J. O., and Konuru, H. B. (2005). Effects of genotype and cultivation environment on lycopene content in red-ripe tomatoes. J. Sci. Food Agric 85, 2021–2026. doi: 10.1002/jsfa.2205
Lahti, L., Shetty, S., Blake, T., and Salojarvi, J. (2017). Tools for Microbiome Analysis in R. Version n 1.4.2. Available online at: https://microbiome.github.io/tutorials/ (accessed March, 2020).
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, J., Di, T., and Bai, J. (2019). Distribution of volatile compounds in different fruit structures in four tomato cultivars. Molecules 24, 2594. doi: 10.3390/molecules24142594
Malundo, T. M. M., Shewfelt, R. L., and Scott, J. W. (1995). Flavor quality of fresh tomato (Lycopersicon esculentum Mill.) as affected by sugar and acid levels. Postharvest. Biol. Technol. 6, 103–110. doi: 10.1016/0925-5214(94)00052-T
Martin, M. (2018). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Maul, F., Sargent, S. A., Sims, C. A., Baldwin, E. A., Balaban, M. O., and Huber, D. J. (2000). Tomato flavor and aroma quality as affected by storage temperature. J. Food Sci. 65, 1228–1237. doi: 10.1111/j.1365-2621.2000.tb10270.x
McMurdie, P. J., and Holmes, S. (2013). Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Müller, D. B., Vogel, C., Bai, Y., and Vorholt, J. A. (2016). The plant microbiota: systems-level insights and perspectives. Annu. Rev. Genet. 50, 211–234. doi: 10.1146/annurev-genet-120215-034952
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2020). vegan: Community Ecology Package. R package version 2.5-7. Available online at: https://CRAN.R-project.org/package=vegan
Ortiz, R., Crossa, J., Vargas, M., and Izquierdo, J. (2007). Studying the effect of environmental variables on the genotype × environment interaction of tomato. Euphytica 153, 119–134. doi: 10.1007/s10681-006-9248-7
Ottesen, A. R., González Peña, A., White, J. R., Petengill, J. B., Cong, L., and Allard, S. (2013). Baseline survey of the anatomical microbial ecology of an important food plant: Solanum lycopersicum (tomato). BMC Microbiol. 13:114. doi: 10.1186/1471-2180-13-114
Padilla, M., Oberti, B., Boileau, V., El Jabri, N., and Tekelioglu, Y. (2007). Consumers and hydroponic tomatoes: perception and market place. Acta Hortic. 747, 37–46. doi: 10.17660/ActaHortic.2007.747.2
Panthee, D. R., Labate, J. A., McGrath, M. T., Breksa, A. P., and Robertson, L. D. (2013). Genotype and environmental interaction for fruit quality traits in vintage tomato cultivars. Euphytica 193, 169–182. doi: 10.1007/s10681-013-0895-1
Paulson, J. N., Stine, O. C., Bravo, H. C., and Pop, M. (2013). Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 10, 1200–1202. doi: 10.1038/nmeth.2658
Pereira, G. E., Gaudillere, J.-P., Van Leeuwen, C., Hilbert, G., Lavialle, O., Maucourt, M., et al. (2005). 1H NMR and chemometrics to characterize mature grape berries in four wine-growing areas in Bordeaux, France. J. Agric. Food Chem. 53, 6382–6389. doi: 10.1021/jf058058q
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596. doi: 10.1093/nar/gks1219
Quinet, M., Angosto, T., Yuste-Lisbona, F. J., Blanchard-Gros, R., Bigot, S., Martinez, J.-P., et al. (2019). Tomato fruit development and metabolism. Front. Plant Sci. 10:1554. doi: 10.3389/fpls.2019.01554
Rambla, J. L., Tikunov, Y. M., Monforte, A. J., Bovy, A. G., and Granell, A. (2014). The expanded tomato fruit volatile landscape. J. Exp. Bot. 65, 4613–4623. doi: 10.1093/jxb/eru128
Robards, K., and Antolovich, M. (1997). Analytical chemistry of fruit bioflavonoids. a review. Analyst 122, 11R–34R. doi: 10.1039/A606499J
Robinson, A. L., Adams, D. O., Boss, P. K., Heymann, H., Solomon, P. S., Trengove, R. D., et al. (2012). Influence of geographic origin on the sensory characteristics and wine composition of Vitis vinifera cv. cabernet sauvignon wines from Australia. Am. J. Enol. Vitic. 63, 467–476. doi: 10.5344/ajev.2012.12023
Rosberg, A. K., Gruyer, N., Hultberg, M., Wohanka, W., and Alsanius, B. W. (2014). Monitoring rhizosphere microbial communities in healthy and Pythium ultimum inoculated tomato plants in soilless growing systems. Sci. Hortic. 173, 106–113. doi: 10.1016/j.scienta.2014.04.036
Roselló, S., Adalid, A. M., Cebolla-Cornejo, J., and Nuez, F. (2011). Evaluation of the genotype, environment and their interaction on carotenoid and ascorbic acid accumulation in tomato germplasm. J. Sci. Food Agric. 91, 1014–1021. doi: 10.1002/jsfa.4276
Saary, P., Bork, P., Forslund, K., and Hildebrand, F. (2017). RTK: efficient rarefaction analysis of large datasets. Bioinformatics 33, 2594–2595. doi: 10.1093/bioinformatics/btx206
Sangiorgio, D., Cellini, A., Spinelli, F., Farneti, B., Khomenko, I., Muzzi, E., et al. (2021). Does organic farming increase raspberry quality, aroma and beneficial bacterial biodiversity? Microorganisms 9:1617. doi: 10.3390/microorganisms9081617
Sasse, J., Martinoia, E., and Northen, T. (2018). Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci. 23, 25–41. doi: 10.1016/j.tplants.2017.09.003
Seskar, M., Shulaev, V., and Raskin, I. (1998). Endogenous methyl salicylate in pathogen-inoculated tobacco plants. Plant Physiol. 116, 387–392.
Shulaev, V., Silverman, P., and Raskin, I. (1997). Airborne signalling by methyl salicylate in plant pathogen resistance. Nature 385, 718–721. doi: 10.1038/385718a0
Son, H.-S., Hwang, G.-S., Kim, K. M., Ahn, H.-J., Park, W.-M., van den Berg, F., et al. (2009). Metabolomic studies on geographical grapes and their wines using 1H NMR analysis coupled with multivariate statistics. J. Agric. Food Chem. 57, 1481–1490. doi: 10.1021/jf803388w
Strayer, R. F. (1994). Dynamics of microorganism populations in recirculating nutrient solutions. Adv. Space Res. 14, 357–366. doi: 10.1016/0273-1177(94)90322-0
Tieman, D., Bliss, P., McIntyre, L. M., Blandon-Ubeda, A., Bies, D., Odabasi, A. Z., et al. (2012). The chemical interactions underlying tomato flavor preferences. Curr. Biol. 11, 1035–1039. doi: 10.1016/j.cub.2012.04.016
Tieman, D., Zhu, G., Resende, M. F. R., Lin, T., Nguyen, C., Bies, D., et al. (2017). A chemical genetic roadmap to improved tomato flavor. Science 355, 391–394. doi: 10.1126/science.aal1556
Verginer, M., Siegmund, B., Cardinale, M., Müller, H., Choi, Y., Míguez, C. B., et al. (2010). Monitoring the plant epiphyte Methylobacterium extorquens DSM 21961 by real-time PCR and its influence on the strawberry flavor. FEMS Microbiol. Ecol. 74, 136–145. doi: 10.1111/j.1574-6941.2010.00942.x
Vogel, J. T., Tieman, D. M., Sims, C. A., Odabasi, A. Z., Clark, D. G., and Klee, H. J. (2010). Carotenoid content impacts flavor acceptability in tomato (Solanum lycopersicum). J. Sci. Food Agric. 90, 2233–2240. doi: 10.1002/jsfa.4076
Wang, D., and Seymour, G. B. (2017). Tomato flavor: lost and found? Mol. Plant 10, 782–784. doi: 10.1016/j.molp.2017.04.010
Wang, L., Baldwin, E., Yu, Z., and Bai, J. (2015). The impact of kitchen and food service preparation practices on the volatile aroma profile in ripe tomatoes: effects of refrigeration and blanching. Hortscience 50, 1358–1364. doi: 10.21273/HORTSCI.50.9.1358
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2020). Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wassermann, B., Müller, H., and Berg, G. (2019). An apple a day: which bacteria do we eat with organic and conventional apples? Front. Microbiol. 10:1629. doi: 10.3389/fmicb.2019.01629
Xiao, Z., Zhou, X., Niu, Y., Yu, D., Zhu, J., and Zhu, G. (2015). Optimization and application of headspace-solid-phase micro-extraction coupled with gas chromatography-mass spectrometry for the determination of volatile compounds in cherry wines. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 26, 122–130. doi: 10.1016/j.jchromb.2014.12.006
Yuan, J., Zhang, N., Huang, Q., Li, R., Vivanco, J. M., and Shen, Q. (2015). Organic acids from root exudates of banana help root colonization of PGPR strain Bacillus amyloliquefaciens NJN-6. Sci. Rep. 5:13438. doi: 10.1038/srep13438
Zamioudis, C., and Pieterse, C. M. J. (2011). Modulation of host immunity by beneficial microbes. Mol. Plant Microbe Interact. 25, 139–150. doi: 10.1094/MPMI-06-11-0179
Zanor, M. I., Rambla, J.-L., Chaïb, J., Steppa, A., Medina, A., Granell, A., et al. (2009). Metabolic characterization of loci affecting sensory attributes in tomato allows an assessment of the influence of the levels of primary metabolites and volatile organic contents. J. Exp. Bot. 60, 2139–2154. doi: 10.1093/jxb/erp086
Zarraonaindia, I., Owens, S. M., Weisenhorn, P., West, K., Hampton-Marcell, J., Lax, S., et al. (2015). The soil microbiome influences grapevine-associated microbiota. mBio 6, e002527–e2514. doi: 10.1128/mBio.02527-14
Zheng, J., Huang, G., Jia, D., Wang, J., Mota, M., Pereira, L. S., et al. (2013). Responses of drip irrigated tomato (Solanum lycopersicum L.) yield, quality and water productivity to various soil matric potential thresholds in an arid region of Northwest China. Agric. Water Manag. 129, 181–193. doi: 10.1016/j.agwat.2013.08.001
Keywords: tomato fruit, bacterial microbiota, organoleptic properties, aroma and flavor, hydroponics
Citation: Escobar Rodríguez C, Novak J, Buchholz F, Uetz P, Bragagna L, Gumze M, Antonielli L and Mitter B (2021) The Bacterial Microbiome of the Tomato Fruit Is Highly Dependent on the Cultivation Approach and Correlates With Flavor Chemistry. Front. Plant Sci. 12:775722. doi: 10.3389/fpls.2021.775722
Received: 14 September 2021; Accepted: 24 November 2021;
Published: 24 December 2021.
Edited by:
Brian Farneti, Fondazione Edmund Mach, ItalyReviewed by:
Antonio Cellini, University of Bologna, ItalyAnne Plotto, Horticultural Research Laboratory (USDA-ARS), United States
Copyright © 2021 Escobar Rodríguez, Novak, Buchholz, Uetz, Bragagna, Gumze, Antonielli and Mitter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Johannes Novak, am9oYW5uZXMubm92YWtAdmV0bWVkdW5pLmFjLmF0
 Carolina Escobar Rodríguez
Carolina Escobar Rodríguez Johannes Novak
Johannes Novak Franziska Buchholz
Franziska Buchholz Pia Uetz
Pia Uetz Laura Bragagna
Laura Bragagna Marija Gumze
Marija Gumze Livio Antonielli
Livio Antonielli Birgit Mitter
Birgit Mitter