- Centre for Functional Ecology, Department of Life Sciences, University of Coimbra, Coimbra, Portugal
Plant regeneration is a well-known capacity of plants occurring either in vivo or in vitro. This potential is the basis for plant micropropagation and genetic transformation as well as a useful system to analyse different aspects of plant development. Recent studies have proven that RNA species with no protein-coding capacity are key regulators of cellular function and essential for cell reprogramming. In this review, the current knowledge on the role of several ncRNAs in plant regeneration processes is summarized, with a focus on cell fate reprogramming. Moreover, the involvement/impact of microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and small-interfering RNAs (siRNAs) in the regulatory networks of cell dedifferentiation, proliferation and differentiation is also analysed. A deeper understanding of plant ncRNAs in somatic cell reprogramming will allow a better modulation of in vitro regeneration processes such as organogenesis and somatic embryogenesis.
Introduction
Plant propagation through in vitro culture techniques, usually known as micropropagation, is a useful biotechnological set of tools that can contribute to satisfying the food and aesthetical needs of the growing world population, mainly, for horticultural plants, such as fruits, vegetables and ornamentals. However, most underlying mechanisms remain unclear and achieving an effective in vitro regeneration, via somatic embryogenesis (SE) or organogenesis, is still challenging for many species. Besides the huge natural variation in plant regeneration capacity between and within species and/or genotypes (Lardon and Geelen, 2020), many obstacles and difficulties prevent reaching suitable protocols to induce plant cell reprogramming and effective regeneration. These bottlenecks may be related to more technical aspects such as contamination or browning of the explants, and/or to different settings requirements (nutrients, plant growth regulators, solidifying agents, pH, osmotic stress, light and temperature) (Abdalla et al., 2022). But they can also be related to the in vitro regeneration process itself, for instance, the difficulty of in vitro establishment and propagation, the recalcitrance or low explant responsivity, which can depend on the plant donor age and/or type, on the oxidative state of the explant, on callus proliferation without organogenic or embryogenic ability, on the difficulty in maintaining and proliferating embryogenic cultures or on the formation of abnormal somatic embryos (Correia et al., 2012a; Martínez et al., 2019). Such constraints cause in vitro regeneration processes to be frequently based on trial-and-error experiments, which reduces their effectiveness. Both organogenesis and SE can be induced directly in the explant or through a two-step process in which a callus phase first occurs followed by shoot or somatic embryo formation from the callus cells (Long et al., 2022). In any of the regeneration pathways, a complex series of events occurs that is controlled by genetic and epigenetic mechanisms that are still poorly understood (Ikeuchi et al., 2019). Moreover, either organogenesis or SE involve cell proliferation and further differentiation with the formation of shoot meristems or embryos that go through different morphological stages until a complete plant could be obtained. For example, in the case of SE, totipotency must be acquired, embryo development must go through different morphological phases (globular, hearth-shaped, torpedo and cotyledonary), and maturation must be achieved before somatic embryos germinate into plantlets (Méndez-Hernández et al., 2019). This complex series of interconnected events must be precisely controlled for the regeneration process to be successful. Among these factors are physiological, cellular, biochemistry and genetic factors. Understanding the molecular framework of these processes is crucial for improving protocols and overcoming those hindrances for several crops. In this review, we focused on a field of investigation that has been assuming an increasing relevance in the understanding of the mechanisms involved in developmental transitions, more specifically the regulatory role of non-coding RNAs (ncRNAs) on cell reprogramming and in vitro plant regeneration. This field has undergone important advances in animal cell systems, but in plants it remains little explored.
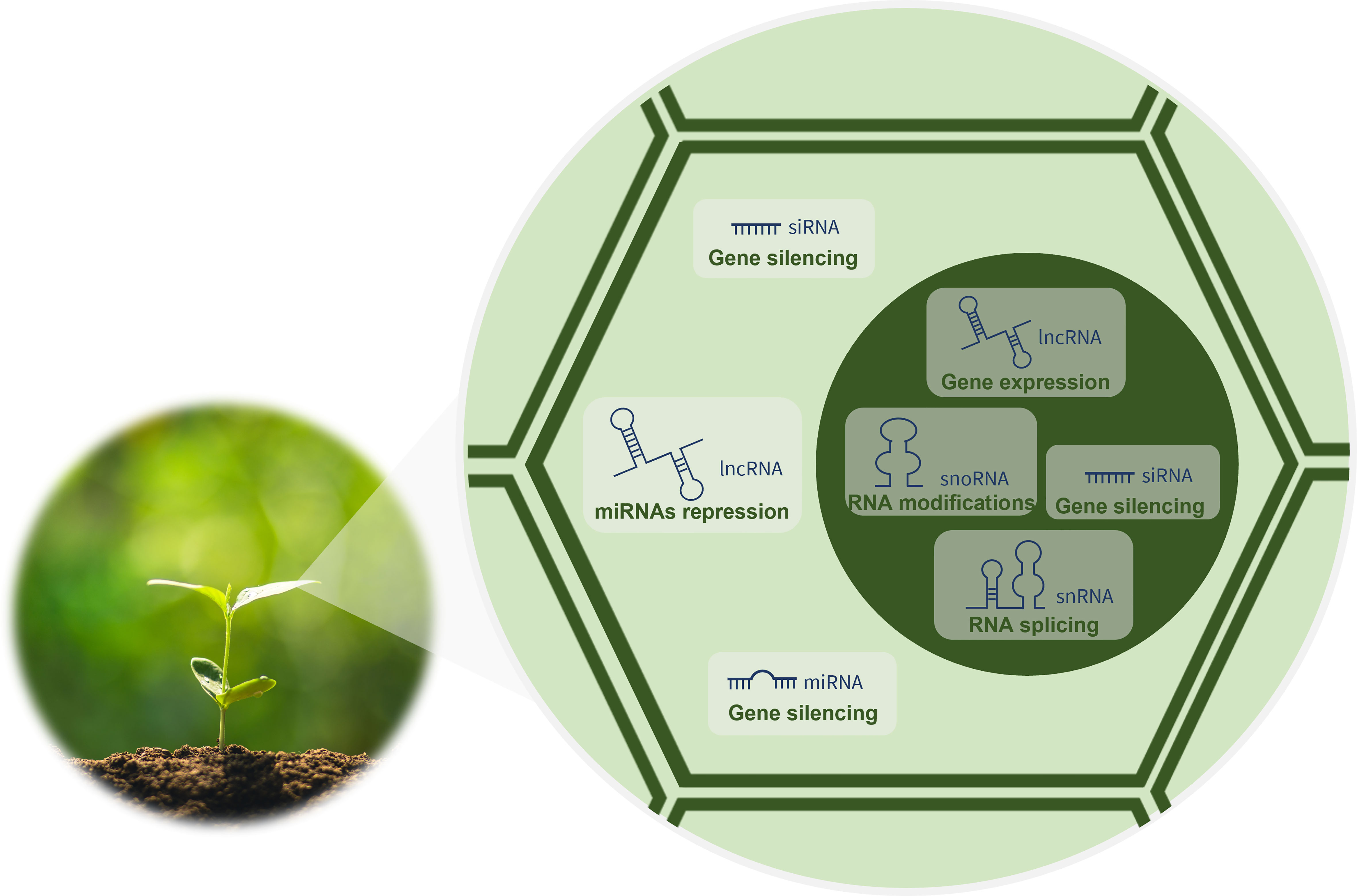
The development of advanced technologies for sequencing and characterizing RNA has revealed key players in the biology of organisms hitherto unknown. Currently, it is known that the eukaryotic cell transcriptome encompasses a wide variety of RNA forms that differ in their biogenesis, mode of action and function. Messenger RNAs (mRNAs) are very well-known RNAs, as they are the templates for protein synthesis. However, these RNAs represent only around 2% of the 90% of the eukaryotic genome that is transcribed into RNA (Rai et al., 2019). The remaining 98% of the transcriptome has no protein-coding capacity and is transcribed from what was previously called junk DNA (Ariel et al., 2015). Nevertheless, it is now known that these genetic information-containing molecules are functional and grouped into two categories (Waititu et al., 2020): the housekeeping and the regulatory ncRNAs. The housekeeping, also called infrastructural or constitutive ncRNAs, are abundant in all cell types and include ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs). Being constitutively expressed, these ncRNAs regulate generic basic cellular functions maintaining them (Zhang et al., 2019a). In turn, the regulatory ncRNAs modulate gene expression at transcriptional, post-transcriptional and epigenetic levels (Zhang et al., 2019a) as indicated in Figure 1. They include transcripts longer than 200 nucleotides (nt), the long ncRNAs (lncRNAs), but also RNAs shorter than 30 nt in length, the small RNAs (sRNAs). In plants, sRNAs, such as microRNAs (miRNAs), small-interfering RNAs (siRNAs) and phased siRNAs (phasiRNAs), have been recently discovered and characterized (Borges and Martienssen, 2015).
The regulatory functions of plant ncRNAs in several biological processes are arousing a growing interest (Böhmdorfer and Wierzbicki, 2015; Yu et al., 2019; Bhogireddy et al., 2021; Song et al., 2021). In turn, in animals, ncRNAs are known as regulators of embryogenesis since they are responsible for developmental transitions and have a pivotal role in cell fate determination, controlling the specification and differentiation of cell types and organ morphogenesis (Pauli et al., 2011; Kanwal et al., 2020). Moreover, ncRNAs are proving to be indispensable in somatic cell reprogramming, specifically in the acquisition and maintenance of pluripotency (Luginbühl et al., 2017).
The ability of some organisms to return some cell types to a meristematic state and regenerate new tissues, organs or whole organisms during post-embryonic development is particularly remarkable in plants (Ikeuchi et al., 2016; Kareem et al., 2016). Their developmental plasticity is used by some species as a reproductive strategy or to survive damage for instance, by the replacement of a lost part after wounding (Perianez-Rodriguez et al., 2014; Ikeuchi et al., 2016). In vitro, plant regeneration can be achieved by culturing differentiated tissues or organs in a medium with appropriate nutrients and plant growth regulators. This can occur through three mechanisms: 1) SE, a process in which a somatic cell gives rise to an embryo that later develops into a complete organism (expression of totipotency); 2) organogenesis, which involves the formation of new organs from mature non-meristematic tissues (reflecting pluripotency) and 3) proliferation of axillary shoot meristems (Kumar, 2011). Thus, induced SE and organogenesis processes are noteworthy systems to study the molecular mechanisms underlying plant cell reprogramming, including ncRNA-mediated regulation.
Being sessile organisms, cell reprogramming is required by plants to cope with the multitude of edaphic-climatic stresses they experience during post-embryonic development and for the expression of totipotency and/or pluripotency. In induced regeneration processes, cell reprogramming is also required and can be achieved directly, with no dedifferentiation stage, or indirectly, involving major reprogramming, depending on culture conditions (Paul et al., 2011; Acanda et al., 2013; Sánchez and Dallos, 2018; Zhang et al., 2021). (Atta et al., 2009; Sugimoto et al., 2010; Chandler, 2011; Perianez-Rodriguez et al., 2014). Furthermore, changes in cell fate are accompanied by deep changes in metabolic pathways (Kanwal et al., 2020). In animal cells, for example, a general metabolic switch from oxidative phosphorylation to glycolysis is required for reprogramming and to acquire the ability to proliferate indefinitely (Wu et al., 2016). Likewise, Araucaria cell lines with high embryogenic potential were associated with the presence of glycolysis-related proteins (dos Santos et al., 2016). Reinforcing the need for energy metabolism for the achievement of embryogenic potential, glucose metabolism-related proteins were predominantly expressed during the SE of tamarillo (Correia et al., 2012b). Moreover, glycosylases are involved in DNA demethylation (Zhao and Chen, 2014), necessary for cell reprogramming as described later in this review.
Despite being reported as key regulators in plant SE (Chen et al., 2018; López-Ruiz et al., 2019a; Siddiqui et al., 2019; Alves et al., 2021), the ncRNAs’ involvement in plant cell plasticity is still far from being fully understood. Nevertheless, the increasing evidence of the importance of the ncRNAs-mediated regulation in cell pluripotency and regeneration ability brings to light important questions to discuss, such as: (1) To what extent do ncRNAs impact plant regeneration processes? (2) Which regulatory networks are most involved/affected? (3) How can this knowledge be used to modulate induced regeneration processes? Thus, in this review, we highlight the current knowledge on the role of several ncRNAs on the intrinsic plant pluripotency and regeneration capacity, focusing mainly on the regulatory roles of miRNAs, siRNAs and lncRNAs.
miRNAs
miRNAs are the most abundant class of sRNAs and are encoded by MIR genes and transcribed by RNA POLYMERASE II (Pol II). Despite its tiny length of only 20-22 nt, these molecules are involved in gene expression regulation, acting post-transcriptionally by cleaving their target mRNA transcripts or causing translational repression (Yu et al., 2017). miRNAs are involved in almost every intracellular event, despite being highly tissue type-dependent and, therefore, their expression patterns and timings vary among species and throughout development. Although essential for plant meristem maintenance, growth and proliferation control, miRNAs are also emerging as key players in somatic to embryonic cell reprogramming, given their capacity in modulating gene expression (López-Ruiz et al., 2019a).
Kanwal et al. (2020) showed that variation in miRNAs expression can enhance or inhibit the reprogramming towards diverse cell types. Also, numerous studies in a wide range of plant species have pointed out differential miRNAs expression in tissues undergoing not only dedifferentiation but also further regeneration (Figure 2). During the dedifferentiation phase, plant tissues present a general increase in miRNAs levels (Luo et al., 2006; Szyrajew et al., 2017). Nevertheless, when the callus phase is reached, some miRNAs exhibit low expression levels, as is the case of miR156, miR160, miR166, miR395, miR396 and miR397, which might be a response to the dedifferentiation stimulus (Gao et al., 2019; Juárez-González et al., 2019). Tissues with high embryogenic potential show high miRNAs levels compared to less embryogenic ones, mainly miR156, miR164 and miR394 in maize (López-Ruiz et al., 2019b). This suggests that these miRNAs are likely enhancers of the embryogenic response and modulation of their expression in non-embryogenic tissues could overcome recalcitrance. Besides the low miRNAs levels in non-embryogenic callus, some of them were reported as up-regulated in this type of calluses such as miR169, miR172, miR390, miR395 and miR408 (Zhang et al., 2010; Wu et al., 2015). Silencing these miRNAs in this kind of calluses or tissues could enhance embryogenic competency and ultimately its expression. When hormone depletion occurs in plant regeneration, there is a slight decrease in the miRNAs amount followed by a new accumulation (López-Ruiz et al., 2019b). Surprisingly, tissues in either the callus or plant regeneration phase show a high accumulation of miRNA targets, similarly to what happens with miRNA levels (López-Ruiz et al., 2019b).
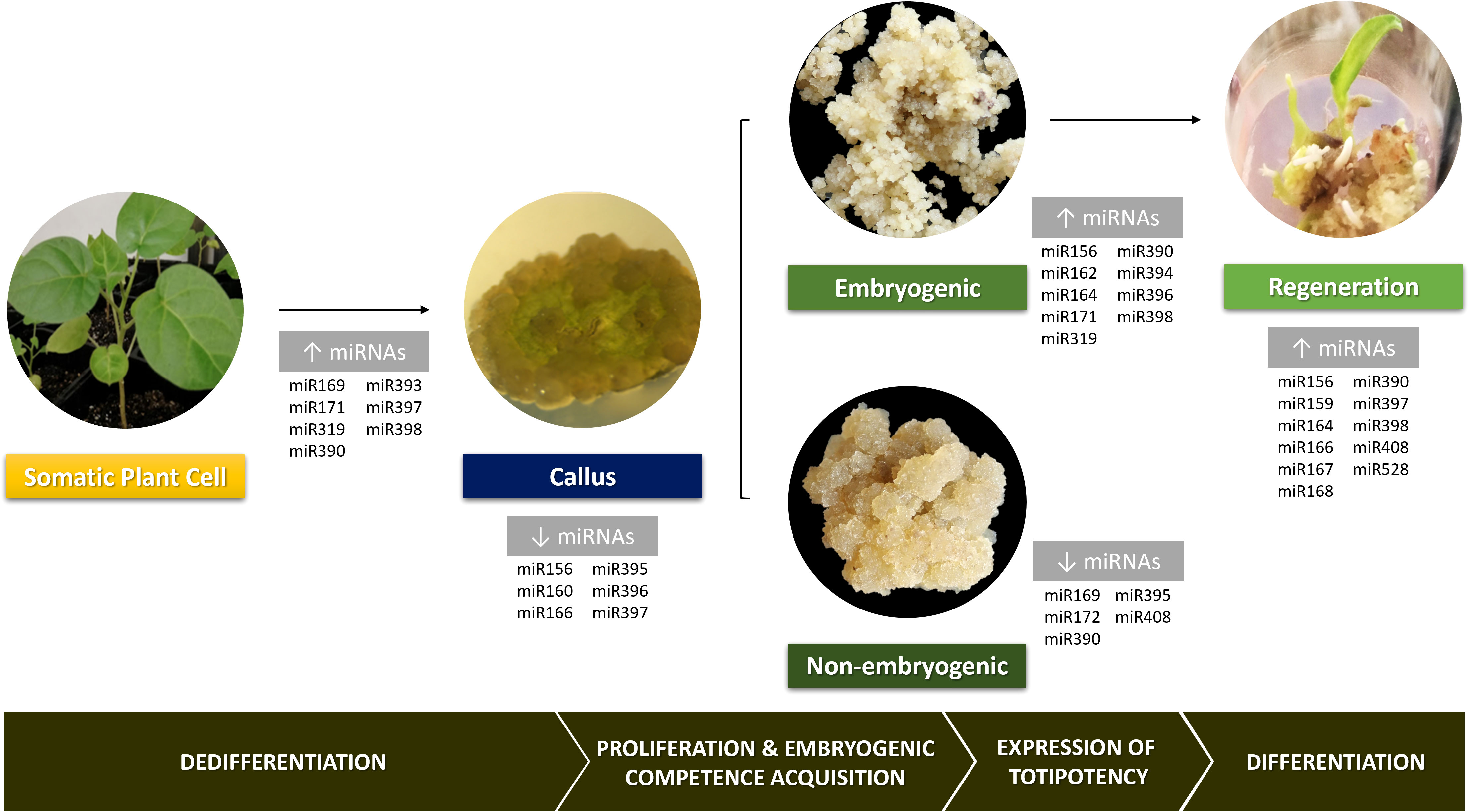
Figure 2 General overview of the involvement of several miRNAs families in various stages of plant cell reprogramming during in vitro indirect somatic embryogenesis. Pictures of Solanum beataceum illustrate key stages of the regeneration process.
Recently, in explants going through later developmental stages and, hence, with lower embryogenic potential, it was found a decrease in the levels of development-related miRNAs and an accumulation of stress- and nutrient- transport-related miRNAs (Juárez-González et al., 2019). This suggests that these last miRNAs are blocking embryogenic potential acquisition, preventing totipotency achievement, probably due to imposed stress and hormone concentration during dedifferentiation. Indeed, unlike moderate stress, which is beneficial for dedifferentiation, excessive stress causes loss of totipotence (Peng et al., 2020).
Cell dedifferentiation goes along with cell wall loosening, which might be associated with miR397-mediated down-regulation of laccases, enzymes involved in cell wall lignification and thickness (Constabel et al., 2000; Sun et al., 2018). Indeed, the levels of expression of miR397 increase during the initial phases of the SE process (López-Ruiz et al., 2019a). In callus tissue, miR397 expression undergoes a slight decline allowing accumulation of laccases and hence lignin deposition, which can be explained by the fact that this tissue contains lignified parenchyma cells (Cao et al., 2020). During plant regeneration phases, miR397 accumulates continually, reaching the highest levels in the adventitious shoots, suggesting a major regulatory role in morphogenesis during advanced differentiation (Liu et al., 2014). Curiously, in conifer, miR397 predominate in somatic embryos but greatly decreased in zygotic ones (Rodrigues et al., 2019).
As miR397, miR160 and miR166 were also defined as key repressors of callus initiation, by modulating the interplay between auxin and cytokinin during organogenesis and through regulation of auxin biosynthesis and response genes, respectively (Cao et al., 2020). Nevertheless, miR160 was proven to be engaged in the embryogenic transition targeting AUXIN RESPONSE FACTORS (ARF) 10 and 16 and contributing to the LEAFY COTYLEDON2-mediated auxin-related pathway induced in Arabidopsis thaliana SE (Wójcik et al., 2017). Also contributing to this pathway is the miR65/166 that targets PHABULOSA/PHAVOLUTA and is involved in meristem maintenance (Wójcik et al., 2017). Despite the decrease in the expression levels of these three miRNAs (miR160, miR166 and miR397) during the change from differentiated cells to callus stages, they increase their expression at further differentiation stages (López-Ruiz et al., 2019b).
miR156, highly conserved in plants and targeting SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes, was confirmed to be involved in callus formation by regulating the cytokinin signalling pathway in citrus (Long et al., 2018; Cao et al., 2020), in modulating redifferentiation during induction of organogenesis in Acacia crassicarpa (Liu et al., 2014), in in vivo regeneration in tomato (Cao et al., 2020), and also in the ability to form somatic embryos from citrus embryogenic callus (Chen and Zhang, 2016).
Accumulation of miR159a, miR319a, miR162, miR171q, miR390a and miR396 seems to be required for embryogenic callus formation in A. crassicarpa organogenesis (Liu et al., 2014) and rice SE (Luo et al., 2006). miR159, which was reported as the most abundantly expressed in tissues under regeneration (Cao et al., 2020), also modulates gene expression during embryogenic callus differentiation in several species such as A. crassicarpa, larch and longan (Zhang et al., 2012; Lin and Lai, 2013; Liu et al., 2014).
After hormone depletion, miR156, miR164, miR168, miR397, miR398, miR408 and miR528 tend to increase, regardless of photoperiod absence/presence, demonstrating the great influence of hormone on specific miRNA expression during plant regeneration (Chávez-Hernández et al., 2015). However, photoperiod seems to influence miR164a, miR167a and miR168a expression levels that increase when callus is cultured in the light as well as some miRNAs targets (Chávez-Hernández et al., 2015). The increase of those miRNAs reveals their stage-specific functions in bud formation (Wu et al., 2011; Liu et al., 2014).
Long non-coding RNAs
lncRNAs are transcripts longer than 200 nt that are transcribed by Pol II and can be grouped in linear and circular lncRNAs (lincRNAs and circRNAs, respectively) and sub-grouped according to the genome region from which they arise, i.e., intergenic, intronic and coding region (Yu et al., 2019). These poorly conserved RNA molecules function as precursors of miRNAs and other sRNAs (Liu et al., 2015) and regulate chromatin remodelling, transcription process and post-transcriptional processing (Mercer et al., 2009).
Compared to humans and other animals, only a few plant lncRNAs have been studied (Zhang et al., 2019b). Despite the limited functional characterization of most lncRNAs, studies so far have uncovered a wide range of possible functions and molecular mechanisms mediated by plant lncRNA activities (Kim and Sung, 2012; Datta and Paul, 2019). Acting in cis or trans modes, lncRNAs regulate the expression of neighbouring or distant genes, respectively, during different plant developmental processes (Yu et al., 2019). Although recent studies have exposed the role of lncRNAs in the control of cell differentiation and pluripotency maintenance in animal stem cells (Guttman et al., 2011; Chen and Zhang, 2016), similar evidence was not yet fully described for plant cells. Nevertheless, an increasing number of functional studies suggest the contributions of lncRNAs as essential modulators in plant responses to abiotic stresses (Urquiaga et al., 2021).
Indeed, a few reports describe the role of lncRNAs during the induction of SE, a stress-induced plant regeneration process. During SE in longan, some lncRNAs were found differentially expressed and revealed to be involved in gene expression regulation during the process by regulatory networks with miRNAs and mRNAs (Chen et al., 2018). For instance, some lncRNAs function as blockers of miRNA cleavage, which greatly affect the regulation of plant cell differentiation and the development process (Chen et al., 2018). This is the case of the lncRNA INDUCED BY PHOSPHATE STARVATION1 (IPS1) that acts as a target-mimic of miR399 and averts the cleavage of its target mRNA (Franco-Zorrilla et al., 2007). Also, in white spruce, the stress-induced SE was reported to be regulated by a lncRNA-miRNA-mRNA network (Gao et al., 2022). In this study, three lncRNAs (MSTRG.33602.1, MSTRG.505746.1, and MSTRG.1070680.1) were found to positively regulate target genes involved in stress response, auxin signal transduction and early somatic embryo development. Having a protective, or sponge effect, for miRNAs targeting these genes, these lncRNAs are therefore involved in SE and ultimately contribute to the embryogenicity of mature somatic embryos (Gao et al., 2022).
As cell reprogramming inherent to plant regeneration can be understood as a stress-responsive event, the increasing number of lncRNAs described with a regulatory role in plant development and response to various stresses can also be understood as being related to plant developmental plasticity. For instance, the lncRNA LONG DAY-SPECIFIC MALE-FERTILITY-ASSOCIATED RNA (LDMAR) was found to be essential for rice fertility (Ding et al., 2012). As a lncRNA involved in pollen development, LDMAR could also be involved in other developmental transitions such as the ones that occur during plant regeneration. Another example is the case of asHSFB2a, which controls the expression of the heat shock factor HSFB2a after heat stress with influence on vegetative and gametophytic development in Arabidopsis (Wunderlich et al., 2014). With a validated role in development transitions, more studies in the asHSFB2a action could bring to light its participation in cell reprogramming. BoNR8 (AtR8 lncRNA homolog in Brassica oleracea) negatively affects seed germination, seedling root growth and silique development in Arabidopsis (Wu et al., 2019) Localized in the epidermal tissues of the root elongation zone and induced by abiotic stresses and auxins, this lncRNA is a potential regulator of in vitro-induced regeneration processes.
The lncRNAs involved in specific developmental transitions are possible candidates to study as cell reprogramming regulators as well. The lncRNA ALTERNATIVE SPLICING COMPETITOR (ASCO) is an alternative splicing competitor in Arabidopsis, that modulates the splicing patterns of some genes and introduces changes during specific developmental transitions, such as the ability to form lateral roots from pericycle cells (Bardou et al., 2014). In the same way, the lncRNAs related to flowering, such as COLD-INDUCED LONG ANTISENSE INTERGENIC NON-CODING RNA (COOLAIR) (Chen and Penfield, 2018), may also affect the developmental transitions that occur during pluripotency acquisition and expression.
Since plant regeneration is an auxin-dependent process, the lncRNAs involved in plant development through auxin transport and signalling may also be important players during this process. Accordingly, five lncRNAs (LTCONS-00006334, LTCONS-00008111, LTCONS-00025525, LTCONS-00030223 and LTCONS-00055024) targeting ARFs were found differentially expressed during longan early SE (Chen et al., 2018). Another lncRNA involved in auxin signalling is AUXIN-REGULATED PROMOTER LOOP (APOLO) which is involved in the modulation of PINOID (PID), a key regulator of polar auxin transport during root growth in Arabidopsis (Ariel et al., 2014) (Figure 3).
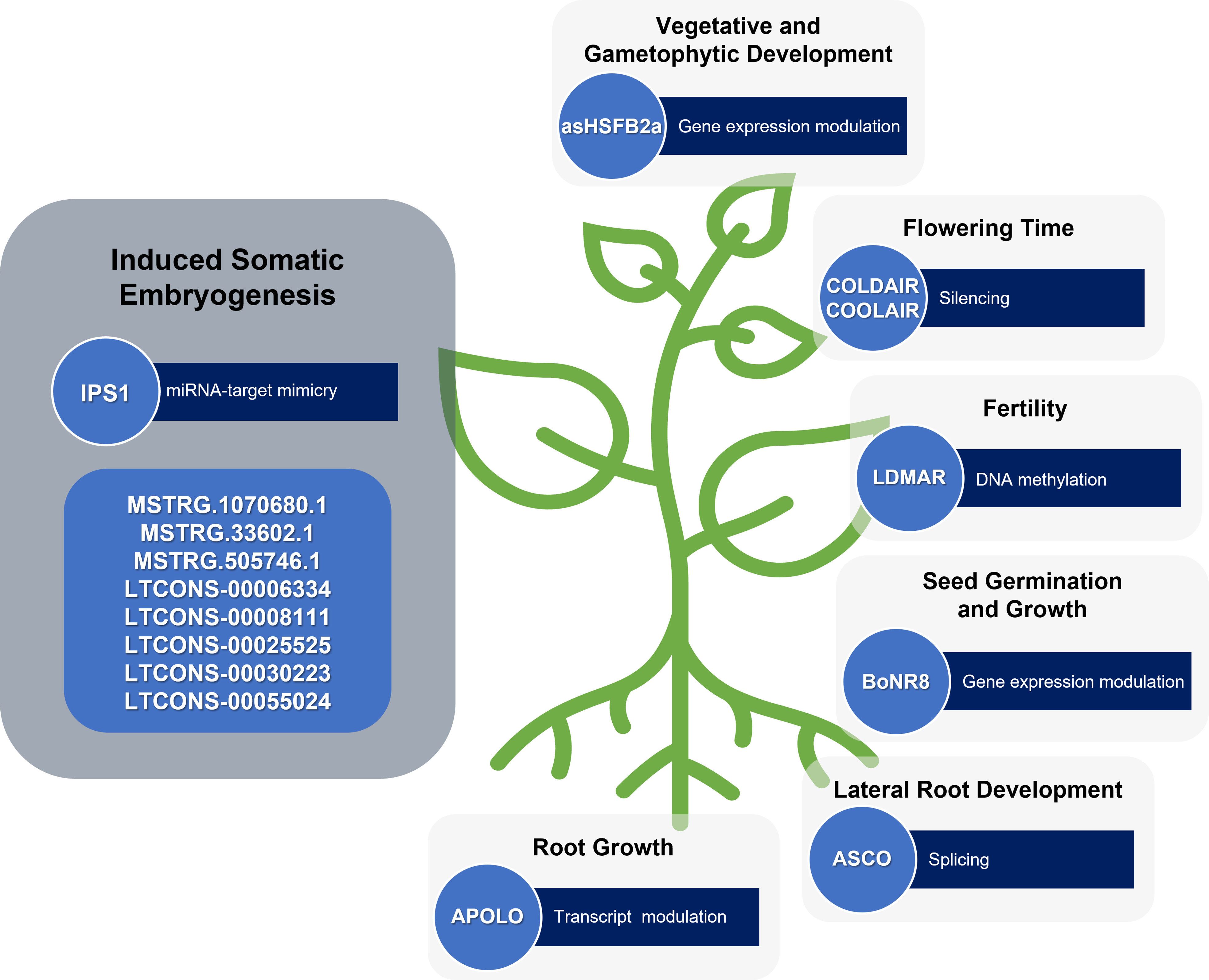
Figure 3 lncRNAs involved in induced plant somatic embryogenesis and other developmental processes. Some lncRNAs still require functional validation.
Other ncRNAs
snRNAs are a class of ncRNAs with 100–200 nt in length. These uridine-rich sRNAs constitute the spliceosome, the molecular machinery that catalyzes pre-mRNA splicing (Will and Lührmann, 2011). It was demonstrated that to achieve in vitro cell dedifferentiation and organogenesis, higher levels of snRNAs are required than those for seedling development (Ohtani et al., 2015). This suggests the critical role of the splicing capacity in cell pluripotency expression.
siRNAs are double-stranded 20-24 nt long RNA molecules. In plants, they include heterochromatic siRNAs (hc-siRNAs), natural antisense transcript siRNAs (nat-siRNAs), trans-acting siRNAs (tasiRNAs), repeat-associated siRNAs (ra-siRNAs) and long siRNAs (lsiRNAs) (Zhang et al., 2019a). siRNAs lead to gene silencing acting either at the transcriptional level, by inducing epigenetic modifications such as DNA and/or histone methylation, or at the posttranscriptional level, inducing mRNA degradation (Borges and Martienssen, 2015). In vitro cultured plant cells undergo extensive epigenetic reprogramming which results in changes in the DNA methylation patterns (Zhang et al., 2018). The increase or decrease of 24 nt siRNAs is correlated with hyper or hypomethylation of the corresponding target genes, respectively (Matzke et al., 2009). Thus, several studies have pointed out a relation between the amount of 24 nt siRNAs and the tissue embryogenic potential, by their impact on genetic reprogramming (Elhiti et al., 2013). In maize, a decrease in the 24 nt siRNAs population is associated with the establishment of embryogenic callus (Alejandri-Ramírez et al., 2018) and, in turn, an increase was observed in fewer embryogenic tissues (Juárez-González et al., 2019). On the contrary, in citrus, a lower abundance of 24 nt siRNAs in non-embryogenic callus has been reported (Wu et al., 2015). Likewise, in grapevine, larger amounts of 24 nt siRNAs were found in embryogenic samples compared to non-embryogenic (Dal Santo et al., 2022). Also, an overrepresentation of 24 nt siRNAs was associated with SE synchronism (Zhang et al., 2014). Thus, as noted by Dal Santo et al. (2022), a readjustment of 24 nt siRNAs appears to be essential for embryogenic commitment.
The multigenerational protection against invasive transposable elements has been described as an important function of 24 nt ncRNAs, particularly at late seed set in conifers, altering developmental programs (Liu and El-Kassaby, 2017).
Recently, López-Ruiz et al. (2020) revealed a dynamic tasiRNAs regulation mediated by ARFs in in vitro plant regeneration. The authors concluded that miR390 and tasiRNAs-ARFs contribute differently to the embryogenic potential depending on the plant species and the in vitro culture process (López-Ruiz et al., 2020). For instance, in cotton embryogenic callus, three tasiRNAs (TAS3a, TAS3b, and TAS3d) were found up-regulated while TAS3c was down-regulated (Yang et al., 2013), pointing to a role of these RNAs in SE regulation.
Recently, chromatin-enriched ncRNAs (cheRNAs) were identified as positive regulators of genes related to somatic cell reprogramming, being required for cell dedifferentiation and plant regeneration ability in rice (Zhang et al., 2022).
Despite not being essential for the restart of cell division during tissue culture, rRNA transcription is upregulated and required during the dedifferentiation of plant cells (Ohtani, 2015).
Conclusions
As important genetic and epigenetic regulators, ncRNAs have proven to be essential to achieve pluripotency, and hence, plant regeneration capacity, through gene expression regulation and chromatin remodelling. Indeed, stress-enhanced embryogenicity is achieved through ncRNAs networks that modulate the epigenetic state of the somatic cell towards pluripotency. As stated in this review, an important class of ncRNAs, miRNAs, are proven as key elements in plant regeneration and totipotency, and an increasing amount of evidence integrates their action with lncRNAs that target important genes in such developmental mechanisms. Nevertheless, and despite the growing number of works that demonstrate differential expression of lncRNAs in regeneration processes, there are no functional studies on lncRNAs in induced plant cell reprogramming and regeneration.
Once ncRNAs are gene expression modulators, variation of their expression, by overexpression or downregulation, alters the expression of specific key genes of cell reprogramming which in turn could improve in vitro regeneration processes. For instance, overexpression of embryogenic-related miRNAs, such as miR156, would increase explants’ regeneration capacity. In the same way, downregulation of stress- and nutrient-transport-related miRNAs could unlock embryogenic potential acquisition.
Despite appreciable success in understanding the role of miRNAs in plant development, the functions and biological mechanisms of other emerging ncRNAs are still unclear. Intensive efforts are needed to ascertain the functional and regulatory role of ncRNAs in plant cell reprogramming. This is crucial to modulate and improve in vitro-induced regeneration processes.
Studies have demonstrated multiple mechanisms of lncRNAs in animals, such as lncRNA-encoded peptides and interaction with DNA or RNA methylases and demethylases; however, the mechanism of action of lncRNAs to regulate RNA methylation of mRNA and protein expression has not been reported in plants. Although plant lncRNAs involved in these functions have not been reported yet, the increasing amount of evidence from other biological systems gives reasons to believe that many of the unknown functions of lncRNAs will be realized through these nodes. The rapid evolution in the integration of data generated by different omics tools is allowing better correlations between coding and non-coding transcriptomes, particularly in the interaction between mRNA-miRNA-lncRNA, revealing new functional interactions whose validation will be crucial for a full understanding of the regulatory mechanisms and their possible modulation. These mechanisms of ncRNA action in plants can provide directions for future research, and more functions of plant ncRNAs can be determined.
Author contributions
DC and SC conceived the idea of the review and prepared the initial outline. DC wrote the first draft. JC and SC were involved in manuscript refinement. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Foundation for Science and Technology (Portugal) through a doctoral research fellowship (SFRH/BD/136925/2018) awarded to DC and was carried out at the R&D Unit Centre for Functional Ecology - Science for People and the Planet (CFE), with reference UIDB/04004/2020, financed by FCT/MCTES through national funds (PIDDAC).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdalla, N., El-Ramady, H., Seliem, M. K., El-Mahrouk, M. E., Taha, N., Bayoumi, Y., et al. (2022). An academic and technical overview on plant micropropagation challenges. Horticulturae 8, 677. doi: 10.3390/HORTICULTURAE8080677
Acanda, Y., Prado, M. J., González, M. V., Rey, M. (2013). Somatic embryogenesis from stamen filaments in grapevine (Vitis vinifera l. cv. mencía): changes in ploidy level and nuclear DNA content. In Vitro Cell. Dev. Biol. - Plant 49, 276–284. doi: 10.1007/s11627-013-9499-7
Alejandri-Ramírez, N. D., Chávez-Hernández, E. C., Contreras-Guerra, J. L., Reyes, J. L., Dinkova, T. D. (2018). Small RNA differential expression and regulation in tuxpeño maize embryogenic callus induction and establishment. Plant Physiol. Biochem. 122, 78–79. doi: 10.1016/j.plaphy.2017.11.013
Alves, A., Cordeiro, D., Correia, S., Miguel, C. (2021). Small non-coding RNAs at the crossroads of regulatory pathways controlling somatic embryogenesis in seed plants. Plants 10, 504. doi: 10.3390/plants10030504
Ariel, F., Jegu, T., Latrasse, D., Romero-Barrios, N., Christ, A., Benhamed, M., et al. (2014). Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol. Cell 55, 383–396. doi: 10.1016/j.molcel.2014.06.011
Ariel, F., Romero-Barrios, N., Jégu, T., Benhamed, M., Crespi, M. (2015). Battles and hijacks: noncoding transcription in plants. Trends Plant Sci. 20, 362–371. doi: 10.1016/j.tplants.2015.03.003
Atta, R., Laurens, L., Boucheron-Dubuisson, E., Guivarc’h, A., Carnero, E., Giraudat-Pautot, V., et al. (2009). Pluripotency of arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J. 57, 626–644. doi: 10.1111/j.1365-313X.2008.03715.x
Bardou, F., Ariel, F., Simpson, C. G., Romero-Barrios, N., Laporte, P., Balzergue, S., et al. (2014). Long noncoding RNA modulates alternative splicing regulators in arabidopsis. Dev. Cell 30, 166–176. doi: 10.1016/j.devcel.2014.06.017
Bhogireddy, S., Mangrauthia, S. K., Kumar, R., Pandey, A. K., Singh, S., Jain, A., et al. (2021). Regulatory non-coding RNAs: A new frontier in regulation of plant biology. Funct. Integr. Genomics 21, 313–330. doi: 10.1007/S10142-021-00787-8
Böhmdorfer, G., Wierzbicki, A. T. (2015). Control of chromatin structure by long noncoding RNA. Trends Cell Biol. 25, 623–632. doi: 10.1016/j.tcb.2015.07.002
Borges, F., Martienssen, R. A. (2015). The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 16, 727–741. doi: 10.1038/nrm4085
Cao, H., Zhang, X., Ruan, Y., Zhang, L., Cui, Z., Li, X., et al. (2020). miRNA expression profiling and zeatin dynamic changes in a new model system of in vivo indirect regeneration of tomato. PLoS One 15, e0237690. doi: 10.1371/journal.pone.0237690
Chandler, J. W. (2011). Founder cell specification. Trends Plant Sci. 16, 607–613. doi: 10.1016/j.tplants.2011.08.005
Chávez-Hernández, E. C., Alejandri-Ramírez, N. D., Juárez-González, V. T., Dinkova, T. D. (2015). Maize miRNA and target regulation in response to hormone depletion and light exposure during somatic embryogenesis. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00555
Chen, Y., Li, X., Su, L., Chen, X., Zhang, S., Xu, X., et al. (2018). Genome-wide identification and characterization of long non-coding RNAs involved in the early somatic embryogenesis in Dimocarpus longan lour. BMC Genomics 19, 805. doi: 10.1186/s12864-018-5158-z
Chen, M., Penfield, S. (2018). Feedback regulation of COOLAIR expression controls seed dormancy and flowering time. Sci. (1979) 360, 1014–1017. doi: 10.1126/science.aar7361
Chen, L., Zhang, S. (2016). Long noncoding RNAs in cell differentiation and pluripotency. Cell Tissue Res. 366, 509–521. doi: 10.1007/s00441-016-2451-5
Constabel, C. P., Yip, L., Patton, J. J., Christopher, M. E. (2000). Polyphenol oxidase from hybrid poplar. cloning and expression in response to wounding and herbivory. Plant Physiol. 124, 285–295. doi: 10.1104/pp.124.1.285
Correia, S., Cunha, A. E., Salgueiro, L., Canhoto, J. M. (2012a). Somatic embryogenesis in tamarillo (Cyphomandra betacea): approaches to increase efficiency of embryo formation and plant development. Plant Cell Tissue Organ Cult 109, 143–152. doi: 10.1007/s11240-011-0082-9
Correia, S., Vinhas, R., Manadas, B., Lourenço, A. S., Veríssimo, P., Canhoto, J. M. (2012b). Comparative proteomic analysis of auxin-induced embryogenic and nonembryogenic tissues of the solanaceous tree Cyphomandra betacea (tamarillo). J. Proteome Res. 11, 1666–1675. doi: 10.1021/pr200856w
Dal Santo, S., de Paoli, E., Pagliarani, C., Amato, A., Celii, M., Boccacci, P., et al. (2022). Stress responses and epigenomic instability mark the loss of somatic embryogenesis competence in grapevine. Plant Physiol. 188, 490–508. doi: 10.1093/plphys/kiab477
Datta, R., Paul, S. (2019). Long non-coding RNAs: fine-tuning the developmental responses in plants. J. Biosci. 4, 1–11. doi: 10.1007/S12038-019-9910-6
Ding, J., Shen, J., Mao, H., Xie, W., Li, X., Zhang, Q. (2012). RNA-Directed DNA methylation is involved in regulating photoperiod-sensitive male sterility in rice. Mol. Plant 5, 1210–1216. doi: 10.1093/mp/sss095
dos Santos, A. L. W., Elbl, P., Navarro, B. V., de Oliveira, L. F., Salvato, F., Balbuena, T. S., et al. (2016). Quantitative proteomic analysis of Araucaria angustifolia (Bertol.) kuntze cell lines with contrasting embryogenic potential. J. Proteomics 130, 180–189. doi: 10.1016/j.jprot.2015.09.027
Elhiti, M., Stasolla, C., Wang, A. (2013). Molecular regulation of plant somatic embryogenesis. In Vitro Cell. Dev. Biol. - Plant 49, 631–642. doi: 10.1007/s11627-013-9547-3
Franco-Zorrilla, J. M., Valli, A., Todesco, M., Mateos, I., Puga, M. I., Rubio-Somoza, I., et al. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39, 1033–1037. doi: 10.1038/ng2079
Gao, Y., Cui, Y., Zhao, R., Chen, X., Zhang, J., Zhao, J., et al. (2022). Cryo-treatment enhances the embryogenicity of mature somatic embryos via the lncRNA-miRNA-mRNA network in white spruce. Int. J. Mol. Sci. 23, 1111. doi: 10.3390/ijms23031111
Gao, Y., Li, D., Zhang, L. L., Borthakur, D., Li, Q. S., Ye, J. H., et al. (2019). MicroRNAs and their targeted genes associated with phase changes of stem explants during tissue culture of tea plant. Sci. Rep. 9, 20239. doi: 10.1038/s41598-019-56686-3
Guttman, M., Donaghey, J., Carey, B. W., Garber, M., Grenier, J. K., Munson, G., et al. (2011). LincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300. doi: 10.1038/nature10398
Ikeuchi, M., Favero, D. S., Sakamoto, Y., Iwase, A., Coleman, D., Rymen, B., et al. (2019). Molecular mechanisms of plant regeneration. Annu. Rev. Plant Biol. 70, 377–406. doi: 10.1146/annurev-arplant-050718-100434
Ikeuchi, M., Ogawa, Y., Iwase, A., Sugimoto, K. (2016). Plant regeneration: cellular origins and molecular mechanisms. Development 143, 1442–1451. doi: 10.1242/dev.134668
Juárez-González, V. T., López-Ruiz, B. A., Baldrich, P., Luján-Soto, E., Meyers, B. C., Dinkova, T. D. (2019). The explant developmental stage profoundly impacts small RNA-mediated regulation at the dedifferentiation step of maize somatic embryogenesis. Sci. Rep. 9, 1–14. doi: 10.1038/s41598-019-50962-y
Kanwal, S., Guo, X., Ward, C., Volpe, G., Qin, B., Esteban, M. A., et al. (2020). Role of long non-coding RNAs in reprogramming to induced pluripotency. Genomics Proteomics Bioinf. 18, 16–25. doi: 10.1016/j.gpb.2019.06.003
Kareem, A., Radhakrishnan, D., Sondhi, Y., Aiyaz, M., Roy, M., Sugimoto, K., et al. (2016). De novo assembly of plant body plan: a step ahead of deadpool. Regeneration 3, 182–197. doi: 10.1002/reg2.68
Kim, E. D., Sung, S. (2012). Long noncoding RNA: unveiling hidden layer of gene regulatory networks. Trends Plant Sci. 17, 16–21. doi: 10.1016/j.tplants.2011.10.008
Kumar, N. (2011). In vitro plant propagation: a review. J. For. Environ. Sci. 27, 61–72. doi: 10.7747/jfs.2011.27.2.1
Lardon, R., Geelen, D. (2020). Natural variation in plant pluripotency and regeneration. Plants 9, 1261. doi: 10.3390/PLANTS9101261
Lin, Y., Lai, Z. (2013). Comparative analysis reveals dynamic changes in miRNAs and their targets and expression during somatic embryogenesis in longan (Dimocarpus longan lour.). PLoS One 8, e60337. doi: 10.1371/journal.pone.0060337
Liu, Y., El-Kassaby, Y. A. (2017). Landscape of fluid sets of hairpin-derived 21-/24-nt-long small RNAs at seed set uncovers special epigenetic features in Picea glauca. Genome Biol. Evol. 9, 82–92. doi: 10.1093/gbe/evw283
Liu, X., Hao, L., Li, D., Zhu, L., Hu, S. (2015). Long non-coding RNAs and their biological roles in plants. Genomics Proteomics Bioinf. 13, 137–147. doi: 10.1016/j.gpb.2015.02.003
Liu, W., Yu, W., Hou, L., Wang, X., Zheng, F., Wang, W., et al. (2014). Analysis of miRNAs and their targets during adventitious shoot organogenesis of Acacia crassicarpa. PLoS One 9, e93438. doi: 10.1371/journal.pone.0093438
Long, J. M., Liu, C. Y., Feng, M. Q., Liu, Y., Wu, X. M., Guo, W. W. (2018). miR156-SPL modules regulate induction of somatic embryogenesis in citrus callus. J. Exp. Bot. 69, 2979–2993. doi: 10.1093/jxb/ery132
Long, Y., Yang, Y., Pan, G., Shen, Y. (2022). New insights into tissue culture plant-regeneration mechanisms. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.926752
López-Ruiz, B. A., Juárez-González, V. T., Gómez-Felipe, A., de Folter, S., Dinkova, T. D. (2020). tasiR-ARFs production and target regulation during in vitro maize plant regeneration. Plants 9, 849. doi: 10.3390/plants9070849
López-Ruiz, B. A., Juárez-González, V. T., Luján-Soto, E., Dinkova, T. D. (2019a). “The role of small RNAs in plant somatic embryogenesis,” in Epigenetics in plants of agronomic importance: Fundamentals and applications. Eds. Alvarez-Venegas, R., De-la-Peña, C., Casas-Mollano, J. (Cham: Springer), 311–338. doi: 10.1007/978-3-030-14760-0_12
López-Ruiz, B. A., Juárez-González, V. T., Sandoval-Zapotitla, E., Dinkova, T. D. (2019b). Development-related miRNA expression and target regulation during staggered in vitro plant regeneration of tuxpeño VS-535 maize cultivar. Int. J. Mol. Sci. 20, 2079. doi: 10.3390/ijms20092079
Luginbühl, J., Sivaraman, D. M., Shin, J. W. (2017). The essentiality of non-coding RNAs in cell reprogramming. Noncoding RNA Res. 2, 74–82. doi: 10.1016/j.ncrna.2017.04.002
Luo, Y. C., Zhou, H., Li, Y., Chen, J. Y., Yang, J. H., Chen, Y. Q., et al. (2006). Rice embryogenic calli express a unique set of microRNAs, suggesting regulatory roles of microRNAs in plant post-embryogenic development. FEBS Lett. 580, 5111–5116. doi: 10.1016/j.febslet.2006.08.046
Martínez, M. T., San-José, M., del, C., Arrillaga, I., Cano, V., Morcillo, M., et al. (2019). Holm Oak somatic embryogenesis: current status and future perspectives. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00239
Matzke, M., Kanno, T., Daxinger, L., Huettel, B., Matzke, A. J. (2009). RNA-Mediated chromatin-based silencing in plants. Curr. Opin. Cell Biol. 21, 367–376. doi: 10.1016/j.ceb.2009.01.025
Méndez-Hernández, H. A., Ledezma-Rodríguez, M., Avilez-Montalvo, R. N., Juárez-Gómez, Y. L., Skeete, A., Avilez-Montalvo, J., et al. (2019). Signaling overview of plant somatic embryogenesis. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00077
Mercer, T. R., Dinger, M. E., Mattick, J. S. (2009). Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10, 155–159. doi: 10.1038/nrg2521
Ohtani, M. (2015). Regulation of RNA metabolism is important for in vitro dedifferentiation of plant cells. J. Plant Res. 128, 361–369. doi: 10.1007/s10265-015-0700-4
Ohtani, M., Takebayashi, A., Hiroyama, R., Xu, B., Kudo, T., Sakakibara, H., et al. (2015). Cell dedifferentiation and organogenesis in vitro require more snRNA than does seedling development in Arabidopsis thaliana. J. Plant Res. 128, 371–380. doi: 10.1007/s10265-015-0704-0
Paul, S., Dam, A., Bhattacharyya, A., Bandyopadhyay, T. K. (2011). An efficient regeneration system via direct and indirect somatic embryogenesis for the medicinal tree Murraya koenigii. Plant Cell Tissue Organ Cult 105, 271–283. doi: 10.1007/s11240-010-9864-8
Pauli, A., Rinn, J. L., Schier, A. F. (2011). Non-coding RNAs as regulators of embryogenesis. Nat. Rev. Genet. 12, 136–149. doi: 10.1038/nrg2904
Peng, C., Gao, F., Wang, H., Shen, H., Yang, L. (2020). Physiological and biochemical traits in Korean pine somatic embryogenesis. Forests 11, 577. doi: 10.3390/f11050577
Perianez-Rodriguez, J., Manzano, C., Moreno-Risueno, M. A. (2014). Post-embryonic organogenesis and plant regeneration from tissues: two sides of the same coin? Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00219
Rai, M. I., Alam, M., Lightfoot, D. A., Gurha, P., Afzal, A. J. (2019). Classification and experimental identification of plant long non-coding RNAs. Genomics 111, 997–1005. doi: 10.1016/j.ygeno.2018.04.014
Rodrigues, A. S., Chaves, I., Costa, B. V., Lin, Y. C., Lopes, S., Milhinhos, A., et al. (2019). Small RNA profiling in Pinus pinaster reveals the transcriptome of developing seeds and highlights differences between zygotic and somatic embryos. Sci. Rep. 9, 11327. doi: 10.1038/s41598-019-47789-y
Sánchez, C., Dallos, P. (2018)Histological study of direct and indirect somatic embryogenesis in mango (Mangifera indica l.) cultivar “hilacha”. In: Proceeding of the international association for plant biotechnology congress. Available at: https://www.researchgate.net/publication/325965284_Histological_study_of_direct_and_indirect_somatic_embryogenesis_in_mango_Mangifera_indica_L_cultivar_hilacha (Accessed October 24, 2022).
Siddiqui, Z. H., Abbas, Z. K., Ansari, M. W., Khan, M. N. (2019). The role of miRNA in somatic embryogenesis. Genomics 111, 1026–1033. doi: 10.1016/j.ygeno.2018.11.022
Song, L., Fang, Y., Chen, L., Wang, J., Chen, X. (2021). Role of non-coding RNAs in plant immunity. Plant Commun. 2, 100180. doi: 10.1016/j.xplc.2021.100180
Sugimoto, K., Jiao, Y., Meyerowitz, E. M. (2010). Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev. Cell 18, 463–471. doi: 10.1016/j.devcel.2010.02.004
Sun, Q., Liu, X., Yang, J., Liu, W., Du, Q., Wang, H., et al. (2018). microRNA528 affects lodging resistance of maize by regulating lignin biosynthesis under nitrogen-luxury conditions. Mol. Plant 11, 806–814. doi: 10.1016/j.molp.2018.03.013
Szyrajew, K., Bielewicz, D., Dolata, J., Wójcik, A. M., Nowak, K., Szczygieł-Sommer, A., et al. (2017). MicroRNAs are intensively regulated during induction of somatic embryogenesis in arabidopsis. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00018
Urquiaga, M. C., de, O., Thiebaut, F., Hemerly, A. S., Ferreira, P. C. G. (2021). From trash to luxury: the potential role of plant lncRNA in DNA methylation during abiotic stress. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.603246
Waititu, J. K., Zhang, C., Liu, J., Wang, H. (2020). Plant non-coding RNAs: origin, biogenesis, mode of action and their roles in abiotic stress. Int. J. Mol. Sci. 21, 8401. doi: 10.3390/ijms21218401
Will, C. L., Lührmann, R. (2011). Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3, a003707. doi: 10.1101/cshperspect.a003707
Wójcik, A. M., Nodine, M. D., Gaj, M. D. (2017). miR160 and miR166/165 contribute to the LEC2-mediated auxin response involved in the somatic embryogenesis induction in arabidopsis. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02024
Wu, X. M., Kou, S. J., Liu, Y. L., Fang, Y. N., Xu, Q., Guo, W. W. (2015). Genomewide analysis of small RNAs in nonembryogenic and embryogenic tissues of citrus: microRNA- and siRNA-mediated transcript cleavage involved in somatic embryogenesis. Plant Biotechnol. J. 13, 383–394. doi: 10.1111/pbi.12317
Wu, X. M., Liu, M. Y., Ge, X. X., Xu, Q., Guo, W. W. (2011). Stage and tissue-specific modulation of ten conserved miRNAs and their targets during somatic embryogenesis of Valencia sweet orange. Planta 233, 495–505. doi: 10.1007/s00425-010-1312-9
Wu, J., Liu, C., Liu, Z., Li, S., Li, D., Liu, S., et al. (2019). Pol III-dependent cabbage BoNR8 long ncRNA affects seed germination and growth in arabidopsis. Plant Cell Physiol. 60, 421–435. doi: 10.1093/pcp/pcy220
Wunderlich, M., Groß-Hardt, R., Schöffl, F. (2014). Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA. Plant Mol. Biol. 85, 541–550. doi: 10.1007/s11103-014-0202-0
Wu, J., Ocampo, A., Belmonte, J. C. I. (2016). Cellular metabolism and induced pluripotency. Cell 166, 1371–1385. doi: 10.1016/j.cell.2016.08.008
Yang, X., Wang, L., Yuan, D., Lindsey, K., Zhang, X. (2013). Small RNA and degradome sequencing reveal complex miRNA regulation during cotton somatic embryogenesis. J. Exp. Bot. 64, 1521–1536. doi: 10.1093/jxb/ert013
Yu, Y., Jia, T., Chen, X. (2017). The ‘how’ and ‘where’ of plant microRNAs. New Phytol. 216, 1002–1017. doi: 10.1111/nph.14834
Yu, Y., Zhang, Y., Chen, X., Chen, Y. (2019). Plant noncoding RNAs: hidden players in development and stress responses. Annu. Rev. Cell Dev. Biol. 35, 407–431. doi: 10.1146/annurev-cellbio-100818-125218
Zhang, H., Lang, Z., Zhu, J. K. (2018). Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 19, 489–506. doi: 10.1038/s41580-018-0016-z
Zhang, M., Wang, A., Qin, M., Qin, X., Yang, S., Su, S., et al. (2021). Direct and indirect somatic embryogenesis induction in Camellia oleifera Abel. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.644389
Zhang, X., Wang, W., Zhu, W., Dong, J., Cheng, Y., Yin, Z., et al. (2019b). Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int. J. Mol. Sci. 20, 5573. doi: 10.3390/ijms20225573
Zhang, P., Wu, W., Chen, Q., Chen, M. (2019a). Non-coding RNAs and their integrated networks. J. Integr. Bioinform. 16, 20190027. doi: 10.1515/jib-2019-0027
Zhang, J., Zhang, S., Han, S., Wu, T., Li, X., Li, W., et al. (2012). Genome-wide identification of microRNAs in larch and stage-specific modulation of 11 conserved microRNAs and their targets during somatic embryogenesis. Planta 236, 647–657. doi: 10.1007/s00425-012-1643-9
Zhang, J. H., Zhang, S. G., Li, S. G., Han, S. Y., Li, W. F., Li, X. M., et al. (2014). Regulation of synchronism by abscisic-acid-responsive small noncoding RNAs during somatic embryogenesis in larch (Larix leptolepis). Plant Cell Tissue Organ Cult 116, 361–370. doi: 10.1007/s11240-013-0412-1
Zhang, Y. C., Zhou, Y. F., Cheng, Y., Huang, J. H., Lian, J. P., Yang, L., et al. (2022). Genome-wide analysis and functional annotation of chromatin-enriched noncoding RNAs in rice during somatic cell regeneration. Genome Biol. 23, 28. doi: 10.1186/s13059-022-02608-y
Zhang, S., Zhou, J., Han, S., Yang, W., Li, W., Wei, H., et al. (2010). Four abiotic stress-induced miRNA families differentially regulated in the embryogenic and non-embryogenic callus tissues of Larix leptolepis. Biochem. Biophys. Res. Commun. 398, 355–360. doi: 10.1016/j.bbrc.2010.06.056
Keywords: embryogenic potential, lncRNAs, miRNAs, plasticity, pluripotency, somatic embryogenesis, totipotency
Citation: Cordeiro D, Canhoto J and Correia S (2022) Regulatory non-coding RNAs: Emerging roles during plant cell reprogramming and in vitro regeneration. Front. Plant Sci. 13:1049631. doi: 10.3389/fpls.2022.1049631
Received: 20 September 2022; Accepted: 25 October 2022;
Published: 10 November 2022.
Edited by:
Elena Corredoira, Institute of Agrobiological Research of Galicia, (CSIC), SpainReviewed by:
Rita Lourenço Costa, Instituto Nacional Investigaciao Agraria e Veterinaria (INIAV), PortugalManuel Rey, University of Vigo, Spain
Pilar S. Testillano, Margarita Salas Center for Biological Research, (CSIC), Spain
Copyright © 2022 Cordeiro, Canhoto and Correia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniela Cordeiro, ZGFuaWVsYWNvcmRlaXJvQG91dGxsb29rLnB0; Sandra Correia, c2FuZHJhaW1jQHVjLnB0
†Present address: Sandra Correia, InnovPlantProtect CoLAb, Elvas, Portugal
 Daniela Cordeiro
Daniela Cordeiro Jorge Canhoto
Jorge Canhoto Sandra Correia
Sandra Correia