- 1National Innovation Alliance of Wintersweet, Henan Academy of Forestry, Zhengzhou, China
- 2Scientific Research Department, Scientific Research Department, Henan Colorful Horticulture Co., Ltd, Zhengzhou, China
- 3Research Institute of Subtropical Forestry, Chinese Academy of Forestry, Hangzhou, Zhejiang, China
Magnoliids are the largest flowering plant clades outside of the eudicots and monocots, which are distributed worldwide and have high economic, ornamental and ecological values. Eudicots, monocots and magnoliids are the three major clades of Mesangiospermae, and their phylogenetic relationship is one of the most interesting issues. In recent years, with the continuous accumulation of genomic information, the evolutionary status of magnoliids has become a hot spot in plant phylogenetic research. Although great efforts have been made to study the evolution of magnoliids using molecular data from several representative species such as nuclear genome, plastid genome, mitochondrial genome, and transcriptome, the results of current studies on the phylogenetic status of magnoliids are inconsistent. Here, we systematically describe the current understanding of the molecular research on magnoliid phylogeny and review the differences in the evolutionary state of magnoliids. Understanding the research approaches and limitations of magnoliid phylogeny can guide research strategies to further improve the study of the phylogenetic evolution of magnoliids.
Introduction
Angiosperms, also known as flowering plants, are the highest and most diverse category of the plant kingdom and have a significant dominance on Earth (Tang et al., 2014; Yang L. et al., 2020; Yang Y. et al., 2020). It has been reported that there are over 35,2000 species of angiosperms (http://www.theplantlist.org/), which are essential sources of oxygen, food, fiber, medicines and other materials for humans and animals (Judd et al., 1999; Tilman et al., 2002). Darwin referred to the phenomenon of rapid origin and species diversity of angiosperms in a relatively short geological period as an “abominable mystery” (Davies et al., 2004; Crepet and Niklas, 2009; Friedman, 2009; Buggs, 2017; Chen et al., 2017). Phylogenetic relationships among organisms are fundamental to evolutionary biology and many other disciplines (Zhang et al., 2012). The establishment of a classification system that truly reflects plant phylogeny has been an important goal of botanical and evolutionary biology research for the largest group in the plant kingdom, angiosperms (Hsu, 1984; Cronquist, 1988; Takhtajan, 1997).
Angiosperms have long been classified into two major groups: monocotyledons and dicotyledons, according to the four major classification systems of Cronquist, Takhtajan, Engler, and Hutchinson (Engler, 1964; Hutchinson, 1973; Cronquist, 1988; Takhtajan, 1997). With the development of molecular biology, the phylogenetic studies of angiosperms have made amazing progress and the taxonomic perspective of angiosperms has undergone revolutionary changes. The Angiosperm Phylogeny Group (APG), composed of several scholars, proposed the APG system based on molecular data in 1998 (Bremer et al., 1998), which is a new classification system of angiosperms based on cladistics and molecular systematics in three revised versions (Bremer et al., 2003; The APG, 2009; Chase et al., 2016). The APG system has changed the traditional phylogenetic research based on fossil records, species morphology and physiological characteristics, and has had a significant impact on the phylogenetic study of angiosperms. Today, the APG system has become a widely used classification system for angiosperms. In the most recent APG IV classification system, angiosperms are classified as basal angiosperms and Mesangiospermae (Chase et al., 2016). The basal angiosperms (ANA clade) include Amborellales, Nymphaeales and Austrobaileyales (Qiu et al., 1999), and Mesangiospermae include five branches: eudicots, monocots, magnoliids, ceratophyllales, and chloranthales. Among them, eudicots and monocots are the two most abundant groups, accounting for about 75% and 20% of angiosperm species, respectively (Zeng et al., 2014). Magnoliids are the third largest branch with more than 10,000 species, accounting for less than 3% of angiosperm species (http://www.theplantlist.org/). Chloranthales and ceratophyllales are few in number and rare in morphology, with only 77 and 6 species, respectively (Zeng et al., 2014).
Many species of magnoliids are early diverging lineages and play an important role in the study of plant evolution and phylogeny, which can be used to better understand the evolution of extant angiosperms (Massoni et al., 2015; Chen et al., 2019; Shang et al., 2020; Liu et al., 2020; Wu et al., 2021). Moreover, many species have high economic, ornamental and ecological values and are widely distributed worldwide (Massoni et al., 2014; Zeng et al., 2014; Massoni et al., 2015; Dong et al., 2021; Shen et al., 2021). Therefore, magnoliids are of great interest to botanists and plant breeders. Nevertheless, the evolutionary relationships between eudicots, monocots, and magnoliids remain unclear, and differences in topology may reveal the phylogenetic complexity behind the rapid radiation of angiosperms (Soltis and Soltis, 2019). In this paper, the phylogenetic research of magnoliids is reviewed and the potential reasons for differences in the evolutionary state of magnoliids are summarized and discussed, with a view to providing guidance for future research.
Overview of magnoliids
The majority of phylogenetic findings support Magnoliales, Laurales, Canellales and Piperales as a branch of Mesangiospermae with early and rapid differentiation (Soltis et al., 1999; Soltis et al., 2007, Cai et al., 2006; Cantino et al., 2007; Moore et al., 2010; Qiu et al., 2010; Moore et al., 2011; Soltis et al., 2011; Ruhfel et al., 2014), but this branch differs from the Magnoliidae, as defined in the Takhtajan classification system (Takhtajan, 1997) or the Cronquist classification system (Cronquist, 1988). Giulietti et al. (2005) and Cantino et al. (2007) associated the name of Magnoliidae with this branch, while in the APG system (Bremer et al., 2003; The APG, 2009; Chase et al., 2016), with this branch being referred to as magnoliids. This paper follows the name of magnoliids in the APG system, which is equivalent to Magnoliidae in some of the literature (Cantino et al., 2007; Massoni et al., 2014; Massoni et al., 2015).
Magnoliids are the next clades of angiosperms after eudicots and monocots, including some of the “earliest angiosperms” defined in earlier studies (Zeng et al., 2014). Magnoliids have played an important role in the development of human society, with species such as black pepper (Piper nigrum), avocado (Persea americana), Litsea cubeba and Chimonanthus salicifolius having high economic value, while other species such as C. praecox, Yulania denudata, Magnolia grandiflora, and Liriodendron chinense have high ornamental value. Besides, many organisms (including various butterfly and beetle groups) are highly dependent on this group for feeding or reproduction, which is an important part of the forest ecosystem (Massoni et al., 2015). Magnoliids have morphological characteristics of both eudicots and monocots (Tang et al., 2014). For example, the L. chinense flower has three cardinal numbers with single pore pollen grains and exhibits typical monocot characteristics, while the cotyledons and roots show typical eudicot characteristics (Chen et al., 2019).
The phylogeny of magnoliids, monocots and eudicots is related to the early origin and evolution of angiosperms. Clarifying the phylogenetic status of magnoliids will provide a new direction for phytogenetic studies and promote the interpretation of evolutionary mysteries and the disclosure of earth history (Zhang et al., 2022). In recent years, the evolutionary status of magnoliids has become a hot spot for plant phylogenetic studies. Among the existing published studies on the phylogeny of magnoliids, most researchers have tried to explain the phylogenetic status of magnoliids using nuclear genomes and other molecular data (plastid genomes, mitochondrial genomes, transcriptomes, etc.) of several representative species, but the conclusions are inconsistent. In general, the topological structure of the evolutionary relationships between the three clades include the following three types: (1) magnoliids + (eudicot + monocot); (2) monocot + (eudicot + magnoliids); (3) eudicot + (magnoliids + monocot).
Phylogeny based on the sequencing of magnoliids own nuclear genome
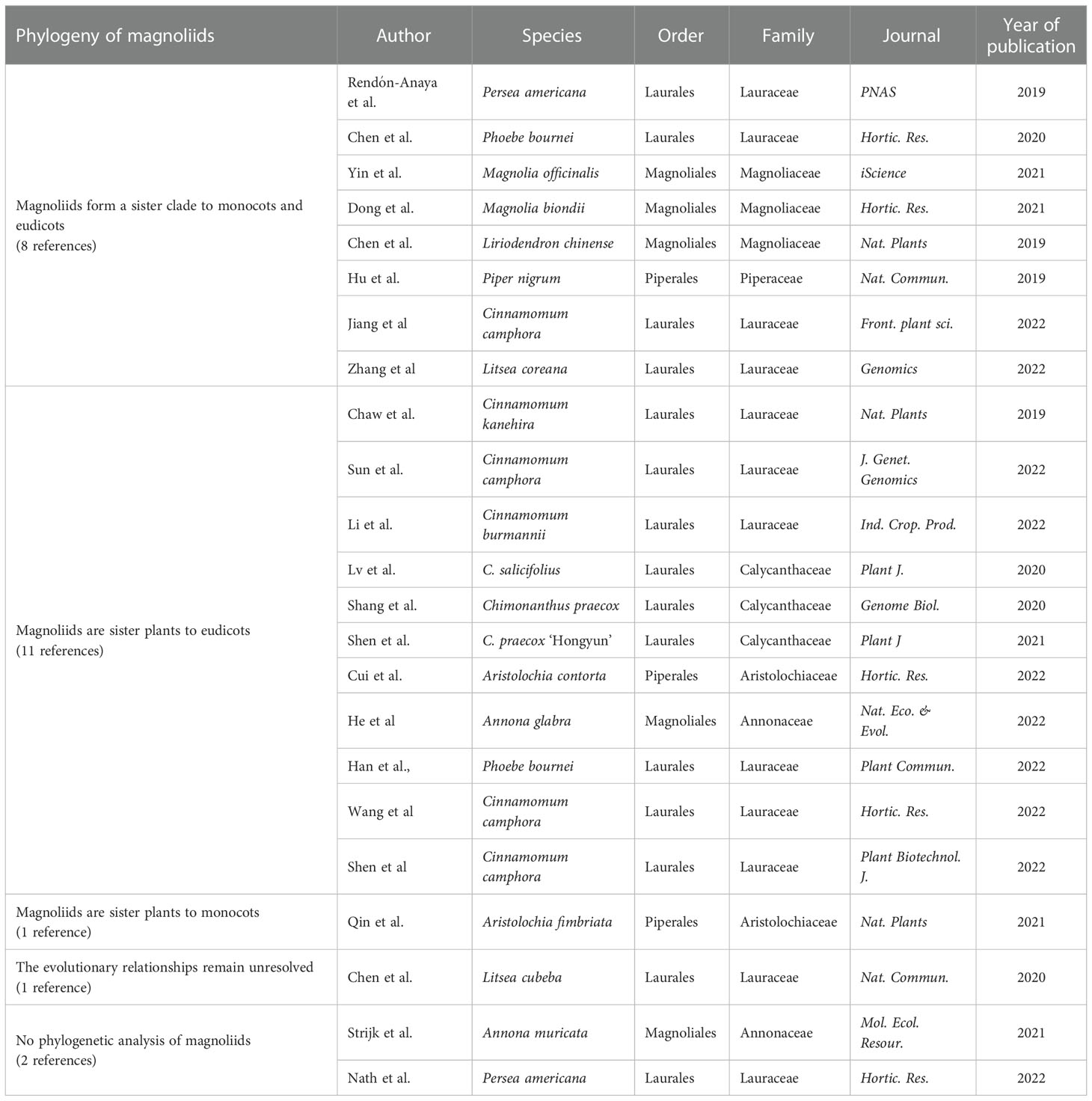
Plant cells contain three sets of genomes: the nuclear genome, the plastid genome and the mitochondrial genome. The nuclear whole-genome sequences contain rich genetic information and have greater application potential in phylogenetic research, with potent capabilities to decipher complex phylogenetic models and evolutionary processes, which can deepen our understanding of plant phylogeny and evolution. The reported species of magnoliids with sequenced nuclear genomes include Magnoliales, Laurales, and Piperales (17 species, 23 references in total). Examples include Laurales: Cinnamomum kanehirae (Chaw et al., 2019), avocado (Persea americana) (Rendón-Anaya et al., 2019; Nath et al., 2022), Litsea cubeba (Chen Y. C. et al., 2020), Phoebe bournei (Chen S. P. et al., 2020; Han et al., 2022), Cinnamomum camphora (Jiang et al., 2022; Shen et al., 2022; Sun et al., 2022; Wang et al., 2022), C. burmannii (Li et al., 2022), Litsea coreana (Zhang et al., 2022) of Lauraceae, Chimonanthus praecox (Shang et al., 2020; Shen et al., 2021) and C. salicifolius (Lv et al., 2020) of Calycanthaceae; Magnoliales: L. chinense (Chen et al., 2019), Magnolia biondii (Dong et al., 2021), M. officinalis (Yin et al., 2021); Annona muricata (Strijk et al., 2021), A. glabra (He et al., 2022); Pepperales: Piper nigrum (Hu et al., 2019), Aristolochia fimbriata (Qin et al., 2021) and A. contorta (Cui et al., 2022). Twenty of the 23 references in the genome analysis magnoliids discussed the evolutionary status of magnoliids, providing new insights into the early evolution of angiosperms, but the results of the analysis are inconsistent.
Magnoliids form a sister clade to monocots and eudicots
The diversity of rapid formation or differentiation of common ancestors of magnoliids, monocots and eudicots leads to differences in the topological results of phylogenetic trees (incomplete lineage sorting, ILS). Chen et al. (2019) sequenced the nuclear genome of L. chinense and deduced three topological structures based on 502 low-copy (no more than two per species) gene trees from 17 species. The species tree was further constructed by the amino acid coalescence approach, and further topological analysis of 78 chloroplast genes and gene families specific to monocots and eudicots in the Liriodendron genome was performed, respectively, which all supported that magnoliids are sister plants to monocots and eudicots. Hu et al. (2019), based on nuclear genome sequencing of P. nigrum and 82 single-copy genes identified in 21 species, used the amino acid concatenation approach to support that magnoliids form a sister clade to monocots and eudicots. Rendón-Anaya et al. (2019) sequenced the nuclear genome of avocado and determined 176 single-copy genes from 19 species. Based on protein sequences, avocado is considered to be sister to monocots and eudicots; based on CDS sequences, avocado is sister to monocots; also, based on 4,694 low-copy genes, avocado is considered to be sister to eudicots. Besides, a neighbor-joining tree was generated based on modal dissimilarity scores from thousands of syntenically validated ortholog pairs, indicating that avocado is sister plants to monocots and eudicots. By evaluating the three positions of avocado, the authors concluded that the three different positions of avocado in angiosperms may be indistinguishable for purely biological reasons. However, according to the Akaike information criterion (AIC) comparison based on the free rate (FR) model, avocado is preferred as sister plants to monocots and eudicots. Chen S. P. et al. (2020) sequenced the nuclear genome of P. bournei and constructed five evolutionary trees based on 292 single-copy genes from 18 species. Among them, three trees (Bayesian tree, coalescent and concatenation trees based on amino acid sequences) support that magnoliids form a sister clade to monocots and eudicots; two trees (coalescent and concatenation trees based on nucleotide sequences) support that magnoliids are sister to monocots. The authors support the Bayesian tree. Zhang et al. (2022) sequenced the nuclear genome of L. coreana. Using 71 single-copy genes of the nuclear genome from 13 species (Amborella trichopoda as an outgroup), nucleic acid sequence-based and amino acid sequence-based concatenation trees were constructed to support that magnoliids are sister to eudicots; the constructed amino acid sequence-based coalescent tree supports that magnoliids form a sister clade to monocots and eudicots and that black pepper is closer to monocots and eudicots. The authors concluded that Magnoliids are more likely to be the basal species of angiosperms due to the possibility of ILS. Consistently, the results of nuclear genome sequencing analysis of M. officinalis, M. biondii, C. camphora, etc. support that magnoliids form a sister clade to monocots and eudicots (Dong et al., 2021; Yin et al., 2021; Jiang et al., 2022).
Magnoliids are sister to eudicots
Chaw et al. (2019) sequenced the nuclear genome of C. kanehirae. Using 211 single-copy genes determined from 13 species, amino acid sequence-based coalescent and concatenation trees support the idea that magnoliids are sister to eudicots. Meanwhile, this topology is also supported by transcriptome data from 22 magnoliids species (although the BS is somewhat low). Similarly, in the genome sequencing analysis of C. salicifolius, Lv et al. (2020) constructed an amino acid sequence-based concatenation tree of 103 single-copy gene sets and coalescent tree of 1,420 low-copy gene sets from 17 species, all of which support that magnoliids are sister plants to eudicots. In addition, Shang et al. (2020) used two software (OrthoMCL and SonicParanoid) to identify two single-copy gene sets and construct amino acid sequence-based concatenation and coalescent trees, respectively. A total of four trees support that magnoliids are sister plants to eudicots. However, based on 38 chloroplast single-copy genes from 26 species, the results of amino acid sequence-based concatenation tree support that magnoliids form a sister clade to monocots and eudicots. The authors suggest that the phylogenetic inconsistency between chloroplast genomes and nuclear genomes may be caused by the ILS effect. Furthermore, a concatenation phylogenetic tree was constructed using nucleic acid sequences of 2,420 gene sets from 29 plants (including transcriptome data), again demonstrating that magnoliids are sister plants to eudicots. Therefore, the authors believe that it is relatively accurate that magnoliids are the sister plants to eudicots in the current data set. In the nuclear genomic analysis of the red flower wintersweet, taking into full consideration various factors that may affect the evolutionary position of magnoliids, Shen et al. (2021) constructed concatenation and coalescent trees using nucleic acid and amino acid sequences of 70 single-copy gene families from 25 genomes, as well as phylogeny trees of 123 plants (47 transcripts, 76 genomes) based on the nucleotide sequences of selected low-copy nuclear ortholog groups. The results suggest that magnoliids are more likely to form a sister clade to eudicots, which is supported by more phylogenetic trees. Consistently, the results of nuclear genome sequencing analysis of C. camphora, A. glabra, P. bournei, A. contorta, C. burmannii, etc. support that magnoliids are sister plants to eudicots (Cui et al., 2022; Han et al., 2022; He et al., 2022; Li et al., 2022; Shen et al., 2022; Sun et al., 2022; Wang et al., 2022).
Magnoliids are sister to monocots or the evolutionary relationships remain unresolved
Qin et al. (2021) compared the genome structure of A. fimbriata and representative species of major angiosperm groups and placed magnoliids as sister groups of monocots. Chen Y. C. et al. (2020) sequenced the L. cubeba nuclear genome, obtained 160 common single-copy gene families of 34 angiosperms from the BUSCO database, and constructed concatenation and coalescent trees using nucleic acid and amino acid sequences. Among them, the amino acid sequence-based coalescent tree supports that magnoliids are sister plants to monocots, and the other three trees support that magnoliids are sister plants to eudicots. Analysis by ASTRAL software suggested that a possible ILS effect on the rapid differentiation of early Mesangiospermae. Based on this, the authors conclude that the evolutionary relationships between magnoliids, monocots, and eudicots remain unresolved.
To sum up, among the 23 references for phylogenetic analysis based on sequencing of magnoliids own nuclear genome, 8 references supported magnoliids as a sister clade to monocots and eudicots, 11 references supported magnoliids as a sister clade to eudicots, and 1 reference supported magnoliids as a sister clade to monocots. In addition, the authors of one reference considered that the evolutionary relationships between magnoliids, monocots and eudicots remain unresolved (Table 1).
Phylogeny of magnoliids based on other molecular data
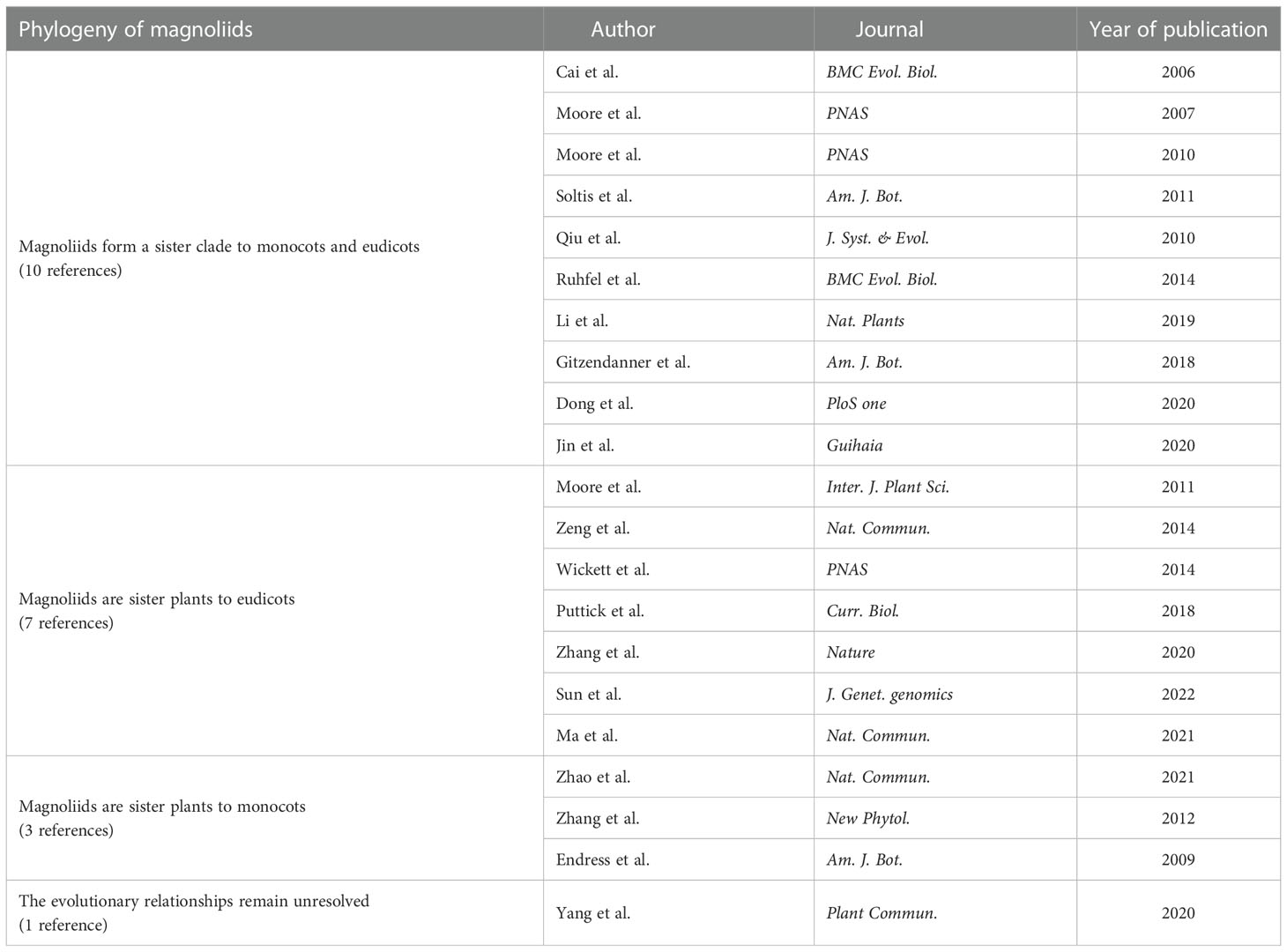
Over the years, researchers have also integrated plastid, mitochondrial, nuclear genome and transcriptome molecular data from multiple species to analyze the phylogeny of magnoliids and the early diversification of angiosperms. Different phylogenetic relationships have also emerged regarding the status of magnoliids.
Magnoliids form a sister clade to monocots and eudicots
In terms of phylogenetic analysis using plastid genomes, Cai et al. (2006); Moore et al. (2007), and Ruhfel et al. (2014) conducted phylogenetic analysis of 61 plastid protein-coding genes from 35 taxa, 61 plastid protein-coding genes from 45 species, and 78 plastid protein-coding data from 360 species, respectively; Moore et al. (2010) conducted a phylogenetic analysis of 83 protein-coding and rRNA genes from 86 seed plant plastid genomes. Gitzendanner et al. (2018) analyzed the phylogenetic tree of 1,827 green plants and 52 outgroups using 78 plastid protein-coding genes. Li et al. (2019) reconstructed the angiosperm phylogeny based on 80 genes from 2,881 plastid genomes, representing 85% of extant families and all orders. In terms of phylogenetic analysis using the mitochondrial genome, Qiu et al. (2010) performed a phylogenetic analysis of 380 species of seed plants based on four mitochondrial gene sequences. Dong et al. (2020) conducted a phylogenetic analysis based on 38 mitochondrial genes from 91 representative angiosperm species. In addition, Soltis et al. (2011) conducted a two-group analysis of 640 plant species from 330 families. The first group included 17 genes representing all three plant genomes (i.e., nucleus, plastid, and mitochondrion); the second group contained 13 genes (representing only the nucleus and plastid). Jin et al. (2020) constructed 20 phylogenetic trees based on nucleotide and amino acid sequences of five gene sets from 89 plants using concatenation and coalescent approaches. The results of all the above analyses support that magnoliids form a sister clade to monocots and eudicots.
Magnoliids are sister to eudicots
Moore et al. (2011) analyzed the plastid inversion repeat sequences of 244 plants; Zeng et al. (2014) and Puttick et al. (2018) conducted phylogenetic analysis using transcriptome amino acid sequences from 61 and 103 plants, respectively; Wickett et al. (2014) systematically analyzed 852 protein-coding nuclear genes from 103 plants (92 transcriptomes and 11 nuclear genomes). The results of the above analysis support that magnoliids are sister plants to eudicots. In addition, Zhang et al. (2020) constructed a coalescent tree based on five different low-copy gene sets (comprising 1,167, 834, 683, 602, and 445 genes respectively) from 115 plants (44 nuclear genomes and 71 transcriptomes), most of which support that magnoliids are sister plants to eudicots. Guo et al. (2021) constructed phylogenetic trees based on nuclear genome sequencing of Chloranthus spicatus using four gene sets (257 single-copy genes, 937 single-copy genes, and 2,329 low-copy genes from 18 plants, and 612 single-copy genes from 218 plants, respectively), supporting the idea that magnoliids are sister to eudicots, while the results of chloroplast gene construction support that magnoliids form a sister clade to monocots and eudicots. The analysis suggested that ancient gene flow between monocots and eudicots might have occurred during the early evolution of angiosperms, resulting in inconsistent phylogenetic branches. In addition, Ma et al. (2021) sequenced the nuclear genome of C. sessilifolius and analyzed 1,689 single-copy genes concatenated nucleotide sequences based on nuclear genome data from 14 plants, supporting that magnoliids are sister plants to eudicots. At the same time, the analysis suggests that, in addition to hybridization, ILS may largely explain the observed phylogenetic inconsistencies among gene trees.
Magnoliids are sister to monocots or the evolutionary relationships remain unresolved
Zhao et al. (2021) conducted a phylogenetic analysis based on genome-wide data from 123 plants (covering 31 orders and 52 families); Zhang et al. (2012) constructed a concatenation tree using nucleotide and amino acids based on five low-copy nuclear genes obtained in 94 species; Endress and Doyle (2009) analyzed plastid and morphological data. The results of all these analyses suggest that magnoliids are sister plants to monocots. However, based on 1594, 756, and 296 gene sets from 151 angiosperms (including the five major branches of the core angiosperms), Yang L. et al. (2020) employed both coalescent and concatenation approaches to infer phylogenetic trees of angiosperms. The authors believe that a fully bifurcated species tree may not be the best way to represent the early differentiation of angiosperms.
To sum up, among the 21 references on the phylogenetic analysis of magnoliids, 10 support that magnoliids form a sister clade to monocots and eudicots, 7 support that magnoliids form a sister clade to eudicots, 3 support that magnoliids form a sister clade to monocots, and one believes that the evolutionary relationships between magnoliids, monocots, and eudicots remain unresolved (Table 2).
Summary and perspectives
Magnoliids have important economic, ornamental and ecological values (Massoni et al., 2014; Massoni et al., 2015; Shen et al., 2021; Dong et al., 2021). They are also valuable materials for studying the origin, development and evolution of angiosperms (Massoni et al., 2015; Chen et al., 2019; Liu et al., 2020; Shang et al., 2020; Wu et al., 2021). Despite the large number of studies reporting the phylogenetic status of magnoliids, the evolutionary relationships between eudicots, monocots, and magnoliids remain inconsistent (Tables 1, 2). Long-branch attraction is a major obstacle to phylogenetic reconstruction, which may lead to the wrong inference of distantly related lineages as close relatives (Qu et al., 2017; Shen et al., 2021). Meanwhile, ILS is the result of allele polymorphism in ancestral populations (Chen Y. C. et al., 2020). Many plant species have a century-long growth period, large population sizes, and limited interspecific differences. These factors have generated an important evolutionary network, which is deeply affected by the ILS. In addition, more attention should be paid to methodological choices in phylogenomic analysis, where the same data set may yield conflicting results (Guo et al., 2022). Different tree-building methods may be important factors contributing to the different evolutionary positions of magnoliids (Rendón-Anaya et al., 2019; Chen S.P. et al., 2020; Chen Y. C. et al., 2020; Shen et al., 2021). It is precisely because of the different tree-building methods, the existence of ILS effects, the number of orthologous genes, the limitation of numerical selection in different groups (Bergsten, 2005; Wiens, 2005), and the rapid differentiation of magnoliids in the early evolutionary stage that the results of research on the evolutionary status of magnoliids are different.
A fully resolved and well-supported phylogeny is of great significance for understanding the evolutionary history of magnoliids. Based on the comprehensive analysis of existing research results, how to adopt a more scientific strategy to analyze the phylogeny of magnoliids is a key consideration for future research on the evolution of magnoliids. For a long time, a large number of valuable plant species have not been sequenced due to the cost of sequencing and the complexity of the species’ own genomes. In particular, there are still few genome sequencing samples of magnoliids, which also hinders the in-depth study of these issues to a certain extent. With the rapid development of sequencing technology and the reduction of sequencing cost, an increasing number of plant genome sequencing data will be published, especially more genomic data of magnoliids will be deciphered, and with the more mature means of phylogenetic research, it is believed that in the near future, there will be an industry-recognized result on the phylogenetic status of magnoliids.
Author contributions
ZS was the designer and principal of the project, and drafted the manuscript. XD, JC, and FW were responsible for collecting, sorting, and analyzing some relevant references. HY and MW contributed to the manuscript revision and read the submitted version. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by Henan Key R&D Project, China (Grant No. 221111110700), the Science and Technology Program on Developing Forestry of Henan, China (Grant No. YCHZ[2022]28), and the Fundamental Research Funds of Henan Academy of Forestry, China (Grant Nos. 2021JB01001 and 210612240).
Conflict of interest
Authors JC and FW were employed by Henan Colorful Horticulture Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bergsten, J. (2005). A review of long-branch attraction. Cladistics Int. J. Willi Hennig Soc. 21 (2), 163–193. doi: 10.1111/j.1096-0031.2005.00059.x
Bremer, B., Bremer, K., Chase, M. W., Reveal, J. L., Soltis, D. E., Soltis, P. S. (2003). An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG II [Review]. Bot. J. Linn. Soc 141 (4), 399–436. doi: 10.1111/j.1095-8339.2009.00996.x
Bremer, K., Chase, M. W., Stevens, P. F., Anderberg, A. A., Backlund, A., Bremer, B. (1998). An ordinal classification for the families of flowering plants. Ann. Mo. Bot. Gard. 85 (4), 531–553. doi: 10.2307/2992015
Buggs, R. J. A. (2017). The deepening of darwin's abominable mystery. Nat. Eco. Evol. 1 (6), 169. doi: 10.1038/s41559-017-0169
Cai, Z., Penaflor, C., Kuehl, J. V., Leebens-Mack, J., Carlson, J. E., dePamphilis, C. W., et al. (2006). Complete plastid genome sequences of drimys, liriodendron, and piper: implications for the phylogenetic relationships of magnoliids. BMC Evol. Biol. 6, 77. doi: 10.1186/1471-2148-6-77
Cantino, P. D., Doyle, J. A., Graham, S. W., Judd, W. S., Olmstead, R. G., Soltis, D. E., et al. (2007). Towards a phylogenetic nomenclature of tracheophyta. Taxon 56 (3), E1–E44. doi: 10.1002/tax.563001
Chase, M. W., Christenhusz, M. J. M., Fay, M. F., Byng, J. W., Judd, W. S., Soltis, D. E., et al. (2016). An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc 181 (1), 1–20. doi: 10.1111/boj.12385
Chaw, S. M., Liu, Y. C., Wu, Y. W., Wang, H. Y., Lin, C. I., Wu, C. S., et al. (2019). Stout camphor tree genome fills gaps in understanding of flowering plant genome evolution. Nat. Plants 5 (1), 63–73. doi: 10.1038/s41477-018-0337-0
Chen, J., Hao, Z., Guang, X., Zhao, C., Wang, P., Xue, L., et al. (2019). Liriodendron genome sheds light on angiosperm phylogeny and species-pair differentiation. Nat. Plants 5 (1), 18–25. doi: 10.1038/s41477-018-0323-6
Chen, F., Liu, X., Yu, C., Chen, Y., Tang, H., Zhang, L. (2017). Water lilies as emerging models for darwin's abominable mystery. Hortic. Res. 4, 17051. doi: 10.1038/hortRes.2017.51
Chen, Y. C., Li, Z., Zhao, Y. X., Gao, M., Wang, J. Y., Liu, K. W., et al. (2020). The Litsea genome and the evolution of the laurel family. Nat. Commun. 11 (1), 1675. doi: 10.1038/s41467-020-15493-5
Chen, S. P., Sun, W. H., Xiong, Y. F., Jiang, Y. T., Liu, X. D., Liao, X. Y., et al. (2020). The phoebe genome sheds light on the evolution of magnoliids. Hortic. Res. 7, 146. doi: 10.1038/s41438-020-00368-z
Crepet, W. L., Niklas, K. J. (2009). Darwin's second 'abominable mystery': Why are there so many angiosperm species? Am. J. Bot. 96 (1), 366–381. doi: 10.3732/ajb.0800126
Cronquist, A. (1988). The evolution and classification of flowering plants. 2nd ed (New York: New York Botanical Garden).
Cui, X., Meng, F., Pan, X., Qiu, X., Zhang, S., Li, C., et al. (2022). Chromosome-level genome assembly of Aristolochia contorta provides insights into the biosynthesis of benzylisoquinoline alkaloids and aristolochic acids. Hortic. Res. 9, uhac005. doi: 10.1093/hr/uhac005
Davies, T. J., Barraclough, T. G., Chase, M. W., Soltis, P. S., Soltis, D. E., Savolainen, V. (2004). Darwin's abominable mystery: Insights from a supertree of the angiosperms. PNAS 101 (7), 1904–1909. doi: 10.1073/pnas.0308127100
Dong, S., Chen, L., Liu, Y., Wang, Y., Zhang, S., Yang, L., et al. (2020). The draft mitochondrial genome of magnolia biondii and mitochondrial phylogenomics of angiosperms. PloS One 15 (4), e0231020. doi: 10.1371/journal.pone.0231020
Dong, S., Liu, M., Liu, Y., Chen, F., Yang, T., Chen, L., et al. (2021). The genome of magnolia biondii pamp. provides insights into the evolution of magnoliales and biosynthesis of terpenoids. Hortic. Res. 8 (1), 38. doi: 10.1038/s41438-021-00471-9
Endress, P. K., Doyle, J. A. (2009). Reconstructing the ancestral angiosperm flower and its initial specializations. Am. J. Bot. 96 (1), 22–66. doi: 10.3732/ajb.0800047
Engler, A. (1964). Syllabus of plant families, volume 2: Angiosperms. (Berlin-Nikolassee: Gebr. Borntraeger).
Friedman, W. E. (2009). The meaning of darwin's 'abominable mystery'. Am. J. Bot. 96 (1), 5–21. doi: 10.3732/ajb.0800150
Gitzendanner, M. A., Soltis, P. S., Wong, G. K., Ruhfel, B. R., Soltis, D. E. (2018). Plastid phylogenomic analysis of green plants: A billion years of evolutionary history. Am. J. Bot. 105 (3), 291–301. doi: 10.1002/ajb2.1048
Giulietti, A. M., Harley, R. M., Queiroz, L., Graas, M. D., Berg, C. (2005). Biodiversity and conservation of plants in brazil. Conserv. Biol. 19 (3), 632–639. doi: 10.1111/j.1523-1739.2005.00704.x
Guo, X., Fang, D., Sahu, S. K., Yang, S., Guang, X., Folk, R., et al. (2021). Chloranthus genome provides insights into the early diversification of angiosperms. Nat. Commun. 12 (1), 6930. doi: 10.1038/s41467-021-26922-4
Guo, C., Luo, Y., Gao, L. M., Yi, T. S., Li, H. T., Yang, J. B., et al. (2022). Phylogenomics and the flowering plant tree of life. J. Integr. Plant Biol. doi: 10.1111/jipb.13415
Han, X., Zhang, J., Han, S., Chong, S. L., Meng, G., Song, M., et al. (2022). The chromosome-scale genome of Phoebe bournei reveals contrasting fates of terpene synthase (TPS)-a and TPS-b subfamilies. Plant Commun. 3 (6), 100410. doi: 10.1016/j.xplc.2022.100410
He, Z., Feng, X., Chen, Q., Li, L., Li, S., Han, K., et al. (2022). Evolution of coastal forests based on a full set of mangrove genomes. Nat. Eco. Evol. 6 (6), 738–749. doi: 10.1038/s41559-022-01744-9
Hsu, B. S. (1984). Present day aspects and prospects of the study of angiosperms phylogeny. Acta Bot. Yunnanica 1), 1–10. doi: CNKI:SUN:YOKE.0.1984-01-000
Hu, L., Xu, Z., Wang, M., Fan, R., Yuan, D., Wu, B., et al. (2019). The chromosome-scale reference genome of black pepper provides insight into piperine biosynthesis. Nat. Commun. 10 (1), 4702. doi: 10.1038/s41467-019-12607-6
Jiang, R., Chen, X., Liao, X., Peng, D., Han, X., Zhu, C., et al. (2022). A chromosome-level genome of the camphor tree and the underlying genetic and climatic factors for its top-geoherbalism. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.827890
Jin, X., Cheng, S., Yang, T., Yu, K., Duan, X., Ni, X., et al. (2020). Reconstruction of angiosperm phylogeny based on 5993 nuclear genes. Guihaia 40 (1), 44–59. doi: 10.11931/guihaia.gxzw201905048
Judd, W., Campbell, C., Kellogg, E., Stevens, P. (1999). Plant systematics: A phylogenetic approach. Taxon 49 (1), 137–138. doi: 10.2307/1223950
Li, F., Huang, S., Mei, Y., Wu, B., Hou, Z., Zhan, P., et al. (2022). Genome assembly provided new insights into the Cinnamomum burmannii evolution and d-borneol biosynthesis differences between chemotypes. Ind. Crop Prod. 186, 115181. doi: 10.1016/j.indcrop.2022.115181
Liu, X., Liao, X. Y., Zheng, Y., Zhu, M. J., Yu, X., Jiang, Y. T., et al. (2020). Genome-wide identification of the YABBY gene family in seven species of magnoliids and expression analysis in Litsea. Plants 10 (1), 21. doi: 10.3390/plants10010021
Li, H., Yi, T., Gao, L., Ma, P., Zhang, T., Yang, J., et al. (2019). Origin of angiosperms and the puzzle of the Jurassic gap. Nat. Plants 5 (5), 461–470. doi: 10.1038/s41477-019-0421-0
Lv, Q., Qiu, J., Liu, J., Li, Z., Zhang, W., Wang, Q., et al. (2020). The Chimonanthus salicifolius genome provides insight into magnoliid evolution and flavonoid biosynthesis. Plant J. 103 (5), 1910–1923. doi: 10.1111/tpj.14874
Massoni, J., Couvreur, T. L., Sauquet, H. (2015). Five major shifts of diversification through the long evolutionary history of magnoliidae (angiosperms). BMC Evol. Biol. 15, 49. doi: 10.1186/s12862-015-0320-6
Massoni, J., Forest, F., Sauquet, H. (2014). Increased sampling of both genes and taxa improves resolution of phylogenetic relationships within magnoliidae, a large and early-diverging clade of angiosperms. Mol. Phylogenet. Evol. 70, 84–93. doi: 10.1016/j.ympev.2013.09.010
Ma, J., Sun, P., Wang, D., Wang, Z., Yang, J., Li, Y., et al. (2021). The Chloranthus sessilifolius genome provides insight into early diversification of angiosperms. Nat. Commun. 12 (1), 6929. doi: 10.1038/s41467-021-26931-3
Moore, M. J., Bell, C. D., Soltis, P. S., Soltis, D. E. (2007). Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. PNAS 104 (49), 19363–19368. doi: 10.1073/pnas.0708072104
Moore, M. J., Hassan, N., Gitzendanner, M. A., Bruenn, R. A., Croley, M., Vandeventer, A., et al. (2011). Phylogenetic analysis of the plastid inverted repeat for 244 species: Insights into deeper-level angiosperm relationships from a long, slowly evolving sequence region. Inter. J. Plant Sci. 172 (4), 541–558. doi: 10.1086/658923
Moore, M. J., Soltis, P. S., Bell, C. D., Burleigh, J. G., Soltis, D. E. (2010). Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. PNAS 107 (10), 4623–4628. doi: 10.1073/pnas.0907801107
Nath, O., Fletcher, S. J., Hayward, A., Shaw, L. M., Masouleh, A. K., Furtado, A., et al. (2022). A haplotype resolved chromosomal level avocado genome allows analysis of novel avocado genes. Hortic. Res. 9, uhac157. doi: 10.1093/hr/uhac157
Puttick, M. N., Morris, J. L., Williams, T. A., Cox, C. J., Edwards, D., Kenrick, P., et al. (2018). The interrelationships of land plants and the nature of the ancestral embryophyte. Curr. Biol. 28 (5), 733–745.e2. doi: 10.1016/j.cub.2018.01.063
Qin, L., Hu, Y., Wang, J., Wang, X., Zhao, R., Shan, H., et al. (2021). Insights into angiosperm evolution, floral development and chemical biosynthesis from the Aristolochia fimbriata genome. Nat. Plants 7 (9), 1239–1253. doi: 10.1038/s41477-021-00990-2
Qiu, Y. L., Chen, Z. D., Libo, L. I., Wang, B., Xue, J. Y., Hendry, T. A., et al. (2010). Angiosperm phylogeny inferred from sequences of four mitochondrial genes. J. Syst. Evol. 48 (6), 391–425. doi: 10.1111/j.1759-6831.2010.00097.x
Qiu, Y. L., Lee, J., Bernasconi-Quadroni, F., Soltis, D. E., Soltis, P. S., Zanis, M., et al. (1999). The earliest angiosperms: evidence from mitochondrial, plastid and nuclear genomes. Nature 402 (6760), 404–407. doi: 10.1038/46536
Qu, X. J., Jin, J. J., Chaw, S. M., Li, D. Z., Yi, T. S. (2017). Multiple measures could alleviate long-branch attraction in phylogenomic reconstruction of cupressoideae (Cupressaceae). Sci. Rep. 7, 41005. doi: 10.1038/srep41005
Rendón-Anaya, M., Ibarra-Laclette, E., Méndez-Bravo, A., Lan, T., Zheng, C., Carretero-Paulet, L., et al. (2019). The avocado genome informs deep angiosperm phylogeny, highlights introgressive hybridization, and reveals pathogen-influenced gene space adaptation. PNAS 116 (34), 17081–17089. doi: 10.1073/pnas.1822129116
Ruhfel, B. R., Gitzendanner, M. A., Soltis, P. S., Soltis, D. E., Burleigh, J. G. (2014). From algae to angiosperms-inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol. Biol. 14, 23. doi: 10.1186/1471-2148-14-23
Shang, J., Tian, J., Cheng, H., Yan, Q., Li, L., Jamal, A., et al. (2020). The chromosome-level wintersweet (Chimonanthus praecox) genome provides insights into floral scent biosynthesis and flowering in winter. Genome Biol. 21 (1), 200. doi: 10.1186/s13059-020-02088-y
Shen, Z., Li, W., Li, Y., Liu, M., Cao, H., Provart, N., et al. (2021). The red flower wintersweet genome provides insights into the evolution of magnoliids and the molecular mechanism for tepal color development. Plant J. 108 (6), 1662–1678. doi: 10.1111/tpj.15533
Shen, T., Qi, H., Luan, X., Xu, W., Yu, F., Zhong, Y., et al. (2022). The chromosome-level genome sequence of the camphor tree provides insights into lauraceae evolution and terpene biosynthesis. Plant Biotechnol. J. 20 (2), 244–246. doi: 10.1111/pbi.13749
Soltis, D. E., Smith, S. A., Cellinese, N., Wurdack, K. J., Tank, D. C., Brockington, S. F., et al. (2011). Angiosperm phylogeny: 17 genes, 640 taxa. Am. J. Bot. 98 (4), 704–730. doi: 10.3732/ajb.1000404
Soltis, D. E., Soltis, P. S. (2019). Nuclear genomes of two magnoliids. Nat. Plants 5 (1), 6–7. doi: 10.1038/s41477-018-0344-1
Soltis, P. S., Soltis, D. E., Chase, M. W. (1999). Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature 402 (6760), 402–404. doi: 10.1038/46528
Soltis, D. E., Gitzendanner, M. A., Soltis, P. S. (2007). A 567-taxon data set for angiosperms: the challenges posed by Bayesian analyses of large data sets. Int. J. Plant Sci 168, 137157. doi: 10.1086/509788
Strijk, J. S., Hinsinger, D. D., Roeder, M. M., Chatrou, L. W., Couvreur, T., Erkens, R., et al. (2021). Chromosome-level reference genome of the soursop (Annona muricata): A new resource for magnoliid research and tropical pomology. Mol. Ecol. Resour. 21 (5), 1608–1619. doi: 10.1111/1755-0998.13353
Sun, W. H., Xiang, S., Zhang, Q. G., Xiao, L., Zhang, D., Zhang, P., et al. (2022). The camphor tree genome enhances the understanding of magnoliid evolution. J. Genet. Genomics 49 (3), 249–253. doi: 10.1016/j.jgg.2021.11.001
Takhtajan, A. (1997). Diversity and classification of flowering plants (New York: NY: Columbia Univ. Press).
Tang, H., Lyons, E., Schnable, J. C. (2014). Early history of the angiosperms. Adv. Bot. Res. 69, 195–222. doi: 10.1016/B978-0-12-417163-3.00008-1
The APG (2009). An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG III. Bot. J. Linn. Soc 161 (2), 105–121. doi: 10.1111/boj.12385
Tilman, D., Cassman, K. G., Matson, P. A., Naylor, R., Polasky, S. (2002). Agricultural sustainability and intensive production practices. Nature 418 (6898), 671–677. doi: 10.1038/nature01014
Wang, X. D., Xu, C. Y., Zheng, Y. J., Wu, Y. F., Zhang, Y. T., Zhang, T., et al. (2022). Chromosome-level genome assembly and resequencing of camphor tree (Cinnamomum camphora) provides insight into phylogeny and diversification of terpenoid and triglyceride biosynthesis of Cinnamomum. Hortic. Res. 9, uhac216. doi: 10.1093/hr/uhac216
Wickett, N. J., Mirarab, S., Nguyen, N., Warnow, T., Carpenter, E., Matasci, N., et al. (2014). Phylotranscriptomic analysis of the origin and early diversification of land plants. PNAS 111 (45), E4859–E4868. doi: 10.1073/pnas.1323926111
Wiens, J. J. (2005). Can incomplete taxa rescue phylogenetic analyses from long-branch attraction? Syst. Biol. 54 (5), 731–742. doi: 10.1080/10635150500234583
Wu, J. Y., Xue, J. Y., Van de Peer, Y. (2021). Evolution of NLR resistance genes in magnoliids: Dramatic expansions of CNLs and multiple losses of TNLs. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.777157
Yang, L., Su, D., Chang, X., Foster, C., Sun, L., Huang, C. H., et al. (2020). Phylogenomic insights into deep phylogeny of angiosperms based on broad nuclear gene sampling. Plant Commun. 1 (2), 100027. doi: 10.1016/j.xplc.2020.100027
Yang, Y., Sun, P., Lv, L., Wang, D., Ru, D., Li, Y., et al. (2020). Prickly waterlily and rigid hornwort genomes shed light on early angiosperm evolution. Nat. Plants 6 (3), 215–222. doi: 10.1038/s41477-020-0594-6
Yin, Y., Peng, F., Zhou, L., Yin, X., Chen, J., Zhong, H., et al. (2021). The chromosome-scale genome of Magnolia officinalis provides insight into the evolutionary position of magnoliids. iScience 24 (9), 102997. doi: 10.1016/j.isci.2021.102997
Zeng, L., Zhang, Q., Sun, R., Kong, H., Zhang, N., Ma, H. (2014). Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nat. Commun. 54, 956. doi: 10.1038/ncomms5956
Zhang, L., Chen, F., Zhang, X., Li, Z., Zhao, Y., Lohaus, R., et al. (2020). The water lily genome and the early evolution of flowering plants. Nature 577 (7788), 79–84. doi: 10.1038/s41586-019-1852-5
Zhang, B., Yao, X., Chen, H., Lu, L. (2022). High-quality chromosome-level genome assembly of Litsea coreana l. provides insights into magnoliids evolution and flavonoid biosynthesis. Genomics 114 (4), 110394. doi: 10.1016/j.ygeno.2022.110394
Zhang, N., Zeng, L., Shan, H., Ma, H. (2012). Highly conserved low-copy nuclear genes as effective markers for phylogenetic analyses in angiosperms. New Phytol. 195 (4), 923–937. doi: 10.1111/j.1469-8137.2012.04212.x
Keywords: magnoliids, phylogeny, monocots, eudicots, genome
Citation: Shen Z, Ding X, Cheng J, Wu F, Yin H and Wang M (2023) Phylogenetic studies of magnoliids: Advances and perspectives. Front. Plant Sci. 13:1100302. doi: 10.3389/fpls.2022.1100302
Received: 16 November 2022; Accepted: 28 December 2022;
Published: 16 January 2023.
Edited by:
Xiaoxu Li, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Songlin He, Henan Agricultural University, ChinaWenfang Gong, Central South University Forestry and Technology, China
Yonghua Li, Henan Agricultural University, China
Copyright © 2023 Shen, Ding, Cheng, Wu, Yin and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiguo Shen, aG5zbGt5NjY4QDE2My5jb20=
 Zhiguo Shen
Zhiguo Shen Xin Ding1
Xin Ding1 Hengfu Yin
Hengfu Yin Minyan Wang
Minyan Wang