- 1Department of Ecological and Environmental System, Kyungpook National University, Sangju, Republic of Korea
- 2Department of Forest Science, Chungbuk National University, Cheongju, Republic of Korea
Natural hybridization between Asplenium incisum and A. ruprechtii has been observed in Northeast Asia and its allotetraploid species, A. castaneoviride, was reported. However, the hybridization process between the parental species and the origin of the allotetraploid taxon remains obscure. Additionally, the systematic affinities of the recently described hybrid A. bimixtum, considered to have originated from the hybridization of A. ruprechtii, A. trichomanes, and A. incisum, is unresolved owing to its similarity to A. castaneoviride. The goals of this study were to (1) investigate the hybridization between A. ruprechtii and A. incisum; (2) verify the origin of A. castaneoviride occurring in Korea, whether it independently arose from 2x sterile hybrids; and (3) elucidate the reliability of identifying A. bimixtum. Three genotypes, A. incisum, A. ruprechtii, and their hybrid, were identified based on the nuclear gene pgiC sequence and finally divided them into six types by ploidy levels: diploid A. incisum, A. ruprechtii, and four hybrid types (diploid A. × castaneoviride, triploid A. × castaneoviride, allotetraploid A. castaneoviride, and A. bimixtum). In the analyses of plastid DNA, all hybrids had an A. ruprechtii-type rbcL gene. In addition, the four plastomes of A. ruprechtii and the hybrids had high pairwise sequence identities greater than 98.48%. They increased up to 99.88% when a large deletion of A. x castaneoriviride (2x) collected from Buramsan populations was ignored. Notably, this large deletion was also found in triploid A. × castaneoviride and allotetraploid A. castaneoviride in the same populations. Sequence data of the nuclear and plastid genes showed that hybridization is unidirectional, and A. ruprechtii is the maternal parent. The large deletion of rpoC2-rps2 commonly found in the different ploidy hybrids of the Buramsan population suggests that the allotetraploid A. castaneoviride can be created independently from sterile hybrids. We assume that both polyploidization driving allopolyploidy and minority cytotype exclusion took place independently in the population, since A castaenoviride co-occurs with A. ruprechtii in small populations. Furthermore, it was also observed that an enlarged noncoding region in fern organelle (ENRIFO) of the plastome was found in the genus Asplenium.
Introduction
Among the processes involved in plant evolution, polyploidization plays an important role in species formation (Stebbins, 1940). Especially in ferns, most homosporous species are likely to be derived from ancient polyploidy for their high basic chromosome number (Wagner and Wagner, 1980). Comparison of chromosome numbers shows that polyploidization affects speciation, and frequent successive polyploidization was regarded to have occurred in ferns more than angiosperms (Wood et al., 2009).
Asplenium Lis is a widely distributed subcosmopolitan fern consisting of more than 700 species (Lin and Viane, 2013). Species in this genus have various ploidy levels caused by the sexual reproductive ability of unreduced gametophytes, which form not only new autopolyploids such as A. sarelii (2x) → A. pekinense (4x) but also allopolyploids like A. sarelli (2x) + A. tenuicaule (2x) → A. anogrammoides (4x) (Liang et al., 2021). To understand the polyplodization event in the Asplenium, investigations using molecular data have been done and revealed a reticulate relationship among the species of this genus (Dyer et al., 2012; Chang et al., 2013; Ohlsen et al., 2014; Chang et al., 2018).
Asplenium castaneoviride Baker is an allotetraploid species derived from Asplenium ruprechtii Sa. Kurata and Asplenium incisum Thunb. and is distributed across China, Japan, and Korea (Lin and Viane, 2013). The fertile allotetraploid A. castaneoviride is close to A. ruprechtii and exhibits a sympatric distribution together with A. incisum in Korea. Based on the diploid hybrid between A. ruprechtii and A. incisum found in the natural population, we probed the origin of the allotetraploid A. castaneovirids. To seek answers regarding the origin of this allotetraploid species, we made the following assumptions:
If diploid hybrids are generated between A. ruprechtii and A. incisum, it can independently become a fertile allotetraploid via chromosome doubling (Figure 1A) to overcome its sterility or very low fertility, with low or identical genetic variation between progenitors and hybrids. The genetic variation in allotetraploid species may be distinguished from their progenitors if chromosome doubling occurs in certain diploid hybrids, spread with a significant divergence time throughout Northeast Asia (Figure 1B).
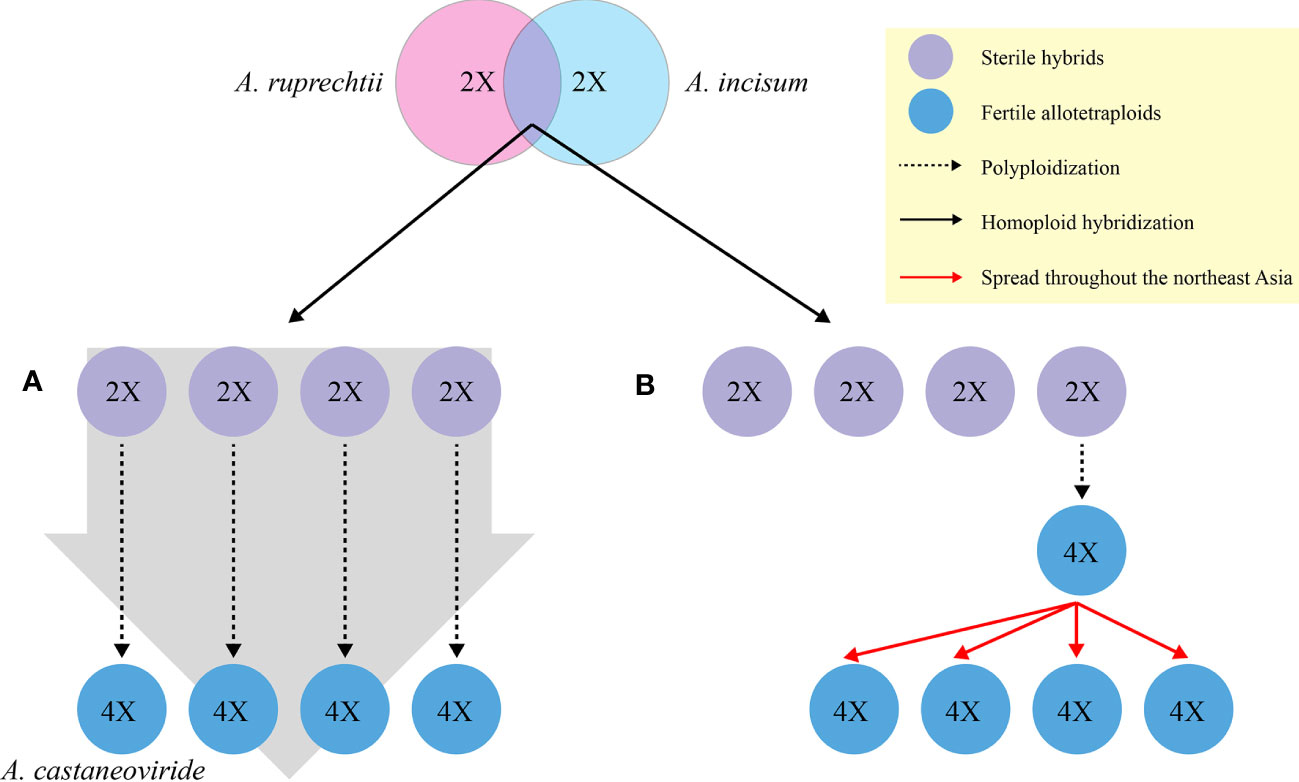
Figure 1 Hypotheses for the formation of allotetraploid A castaneoviride in Korea. (A) Allotetraploid A castaneoviride independently arises from duplication of sterile hybrids. (B) Allotetraploid A castaneoviride arises from certain sterile hybrids and spreads throughout Northeast Asia after polyploidization.
Gender-biased hybridization in ferns has captivated researchers for several years. Stein and Barrington (1990) reported an equal distribution of parental species, even though the sample size was too small to be statistically significant. In contrast, Vogel et al. (1998b) showed predominantly unidirectional hybridization between A. septentrionale and A. trichomanes, and Xiang et al. (2000) discussed how population-specific gender-biased hybridization occurs in Dryopteris × triploidea. In diploid and tetraploid hybrids of A. ruprechtii and A. incisum, it was assumed that A. ruprechtii was the maternal parent because it was found alongside hybrids in the field. However, this assumption is empirical and has not been experimentally proven to date. Therefore, the hybridization producing progenitors of A. castaneoviride remains unclear.
Another allotetraploid, A. bimixtum putatively derived from A. trichomanes, A. ruprechtii, and A. incisum, has been reported in Korea. Based on morphological features, multiple hybridization has been suggested for the development of A. bimixtum (Lee et al., 2015). After the first report of this hybrid, Lee et al. (2019) argued the origin of this new species based on morphological characteristics, genome size, and phylogenetic relationships. However, the origin and status of A. bimixtum require reconsideration since morphological features of A. bimixtum are almost identical to those of A. castaneoviride. In their cytometric study, some of the genome size data had a high standard deviation, since they ran the sample without any reference plant having the standard genome size, which generally runs together with the target sample for measuring accuracy (Lee et al., 2019). Additionally, the authors described that A. bimixtum has two different types of plastid genes originating from A. ruprechtii and A. incisum and suggested a multi-maternal hypothesis to explain their results. However, this is questionable because Vogel et al. (1998a) reported the maternal transmission of chloroplasts in the genus Asplenium, and different plant materials were used for each dataset of rbcL and rps4-trnS in their phylogenetic analysis.
The plastomes of angiosperms are normally conserved in sequence, gene content, and organization (Ruhlman and Jansen, 2014) except in certain lineages (Funk et al., 2007; McNeal et al., 2007; Cai et al., 2008; Guisinger et al., 2011; Knox, 2014). However, the plastomes of ferns have undergone more genomic mutations during evolution than those of the seed plants. Generally, genomic inversions, IR expansions/contractions, and gene duplications/deletions occur frequently throughout the fern lineages (Wolf et al., 2010; Gao et al., 2013; Grewe et al., 2013; Kim et al., 2014; Labiak and Karol, 2017). Recently, foreign DNAs, located within the plastid genome but did not match any plastome sequence, have been found in fern species at different locations (Kim and Kim, 2018; Robison et al., 2018; Lehtonen and Cardenas, 2019). These could be used as molecular markers to distinguish closely related taxa (Kim and Kim, 2020). In addition, nucleotide substitution rates for fern plastome genes are significantly higher than those for seed plant plastomes (Wolf et al., 2011). Consequently, the levels of sequence and indel divergence between intraspecific taxa in ferns are frequently higher than those of closely related intraspecific and interspecific taxa of seed plants (Kim and Kim, 2014). Owing to these dynamic variations, complete plastome sequences allow us to design molecular markers for distinguishing populations or closely related taxa in ferns.
The goals of this study were to (1) investigate the hybridization pattern between A. ruprechtii and A. incisum, (2) verify whether allotetraploid A. castaneoviride occurred independently originating from 2x hybrids, and (3) clarify the reliability of recognizing another allotetraploid, A. bimixtum, recently reported in Korea. For this purpose, we first applied two methods of genome size measurement using flow cytometry analysis and spore viability check in the manner of distinguishing a sterile hybrid with abortive spores and fertile hybrids with normal viable spores (Wagner et al., 1986) to identify the ploidy and sterility of hybrids, respectively. The complete plastome sequences of four species (A. bimixtum, A. castaneoviride, A. incisum, and A. ruprechtii) and 2x sterile hybrids between A. incisum and A. ruprechtii were compared to design a molecular marker to identify the maternal inheritance of each individual. A codominant nuclear DNA marker pgiC was also used to identify paternal hybrids, as well as to trace the origin of the allotetraploid A. castaneoviride. Hereafter, sterile hybrids of A. incisum and A. ruprechtii were named A. × castaneoviride with ploidy levels (e.g., 2x or 3x) to distinguish them from A. castaneoviride (allotetraploid hybrid species) in the present study.
Materials and methods
Sampling and DNA extraction
Four species (A. incisum, A. ruprechtii, A. castaneoviride, and A. bimixtum) and sterile hybrids were collected from 14 populations throughout Korea and transplanted into a greenhouse at Chungbuk National University (Supplementary Table 1). All voucher specimens were deposited in the herbarium of the Chungbuk National University (CBNU) and their accession numbers were described in Supplementary Table 2. Genomic DNAs were extracted from fresh leaves using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol.
Genome size estimation
After checking the ploidy levels of all plant materials, the genome sizes of A. incisum, A. ruprechtii, and their hybrid progenies (2x, 3x, and 4x) were measured using live materials selected from the sampled populations, which includes all hybrids (Supplementary Figure 1, Supplementary Table 2). 4x Solanum tuberosum with a 1c-value of 2.1 (Bennett and Smith, 1976) was used as the standard reference for calculating the genome size. Only 1 × 1 mm2 of the reference leaf and one-tenth leaf of Asplenium species were chopped together with 500 μl of nuclei extraction buffer using a CyStain UV Precise P kit (Sysmex Partec, Germany) in a Petri dish placed on ice. The debris was filtered using a non-sterile CellTrics filter Green 30 μm (Sysmex Partec), and nuclei stained with the staining buffer in CyStain UV Precise P kit (Sysmex Partec). The particle size was measured at least three times per sample using a CyFlow Cube 6 (Sysmex Partec).
Plastome sequencing, assembly, and annotation
NGS libraries for four high-quality DNAs were constructed (Macrogen, Seoul, Korea) and sequenced using Illumina HISeq X Ten. The raw reads were trimmed by trimmomatic 0.36 (Bolger et al., 2014) using the following options: leading, 10; trailing, 10; sliding window, 4:20; and minlen, 50. The assembly method was identical to that of Kim et al. (2015) using A. nidus (NC_045119.1) and A. prolongatum (NC_035838) as reference sequences. Genes in plastome sequences were annotated and compared with the genes of A. nidus and A. prolongatum using Geneious 10.2.6 (Kearse et al., 2012) and confirmed using BLAST search (Altschul et al., 1990) and tRNAscan-SE (Lowe and Eddy, 1997).
Amplification of pgiC, rbcL, and rpoC2-rps2 regions
The gene encoding cytosolic phosphoglucose isomerase, pgiC, merits phylogenetic reconstruction in seed plants (Gottlieb and Ford, 1996) and is generally translated from a single copy gene, except in certain lineages (Gottlieb, 1982; Thomas et al., 1993). A codominant nuclear DNA marker using pgiC for homosporous ferns was developed to estimate mating and to study population genetics (Ishikawa et al., 2002) and has been applied to various fern lineage studies (Ebihara et al., 2005; de Groot et al., 2011; Dyer et al., 2012; Sessa et al., 2012).
We used a pair of primer sets for pgiC designed by Ishikawa et al. (2002) and rbcL designed by Schuettpelz and Pryer (2007) and de Groot et al. (2011) to verify the hybridization pattern between A. incisum and A. ruprechtii. Only individuals confirmed to have both types of pgiC were tested for the rbcL. To detect large deletions (2,139 bp) between rpoC2 and rps2 in A. ruprechtii-type plastomes, we designed a pair of primers (ARuRPS2: AGTGGATTCCTGCTGCCATC, ARuRPOC2: TGAGGGATTGAGTCGGCAAC) and applied them to all A. bimixtum, A. ruprechtii, and hybrids between A. incisum and A. ruprechtii.
The polymerase chain reaction (PCR) conditions for these loci were as follows: a 5-min denaturation step at 95°C, followed by 35 cycles at 95°C for 45 s, 50–53°C for 30–45 s, and 72°C for 60–75 s, followed by a 5-min final extension step at 72°C. The PCR products were purified using a PCR purification kit (Geneall, Seoul, Korea) and sequenced using an ABI 3730×l System (Macrogen).
Phylogenetic analysis
A total of 68 rbcL sequences from A. incisum (17), A. ruprechtii (28), and their hybrids, including A. bimixtum (23), and 5 from A. oligophlebium, A. boreale, A. normale, A. tripteropus, and A. trichomanes were aligned using MAFFT (Katoh et al., 2002). All the rbcL sequence data generated in the present study were deposited in the NCBI database (Supplementary Table 3). Phylogenetic analysis was performed by the maximum likelihood method using IQ-TREE (Nguyen et al., 2015). The best-fit model for base substitution was selected by Bayesian information criterion using ModelFinder (Kalyaanamoorthy et al., 2017), and branch supports were accessed using UFBoot2 (Hoang et al., 2018).
Identification of enlarged noncoding regions in the plastomes of A. incisum and A. ruprechtii
The rpoC2-rps2 regions of the two plastomes, excluding from −1 to −100 upstream of rps2, were blasted against nucleotide collection using BLASTn (Altschul et al., 1990) with an e-value of 1e−3 and a word size of 11.
Results
Ploidy analyses of individuals based on the nuclear genotype and genome size measurement
Based on leaf morphology, the plant samples were first separated into four groups (Figure 2, Supplementary Table 1): A. incisum (17 individuals), A. ruprechtii (28), hybrid (18), and A. bimixtum (5). All individuals morphologically identified as A. incisum and A. ruprechtii had homozygous nuclear genotypes. Meanwhile, sequencing traces of pgiC from hybrids showed a mixed template pattern with two peaks found at the same location. For an additional check of the correct sequence at these sites, seven PCR products (including an A. bimixtum individual) from 23 hybrids were cloned and confirmed to have both nuclear genotypes of A. incisum and A. ruprechtii.

Figure 2 Plants and their natural habitats analyzed in the present study. (A) A incisum; (B) A ruprechtii; (C) A castaneoviride; (D) A bimixtum.
Based on these three nuclear genotypes of pgiC (homozygous A. incisum, homozygous A. ruprechtii, and heterozygous genotypes of both) and the different genome sizes detected among the heterozygotes, they were grouped into six types. The average genome sizes of A. incisum and A. ruprechtii known as diploid were 3.75 pg and 2.79 pg, respectively, compared to the reference standard of S. tuberosum, (Figure 3, Table 1, Supplementary Table 2). Among the hybrids, the average genome size of 11 individuals in Buramsan and one in Bukhansan was 6.51 pg, which was close to the sum of the two parental species. These individuals were considered as allotetraploid A. castaneoviride because they had normal viable spores. In contrast, the average genome size of 10 hybrid individuals collected from five different populations was 3.28 pg, which is approximately close to the mean genome size of A. incisum and A. ruprechtii. These individuals were considered sterile diploid hybrid A. × castaneoviride (described as A. × castaneoviride 2x) because they had abortive spores. Only one triploid hybrid collected from Buramsan had a genome size of 4.62 pg, resembling the mean genome size of an allotetraploid A. castaneoviride and a diploid-origin A. ruprechtii. This triploid also had abortive spores, and was considered to be A. × castaneoviride 3x. The four individuals collected from Seongsan, known as A. bimixtum, had an average genome size of 6.32 pg with normal spores.
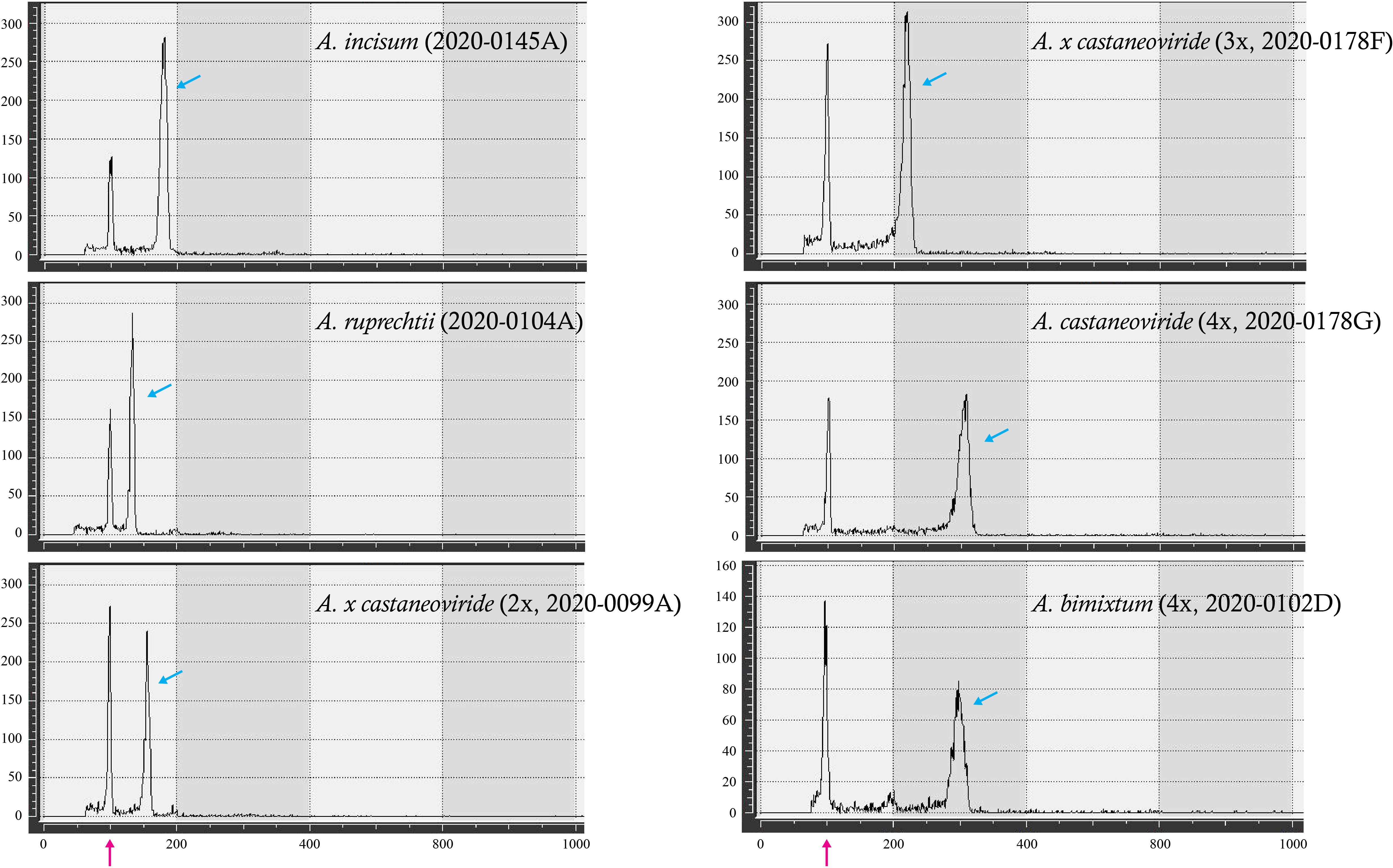
Figure 3 Examples of flow cytometry histograms of A. incisum, A. ruprechtii, and their hybrids. The X- and Y-axes show a relative fluorescence intensity and number of particles, respectively. Red and blue arrows refer to reference (S. tuberosum) and sample peaks, respectively.
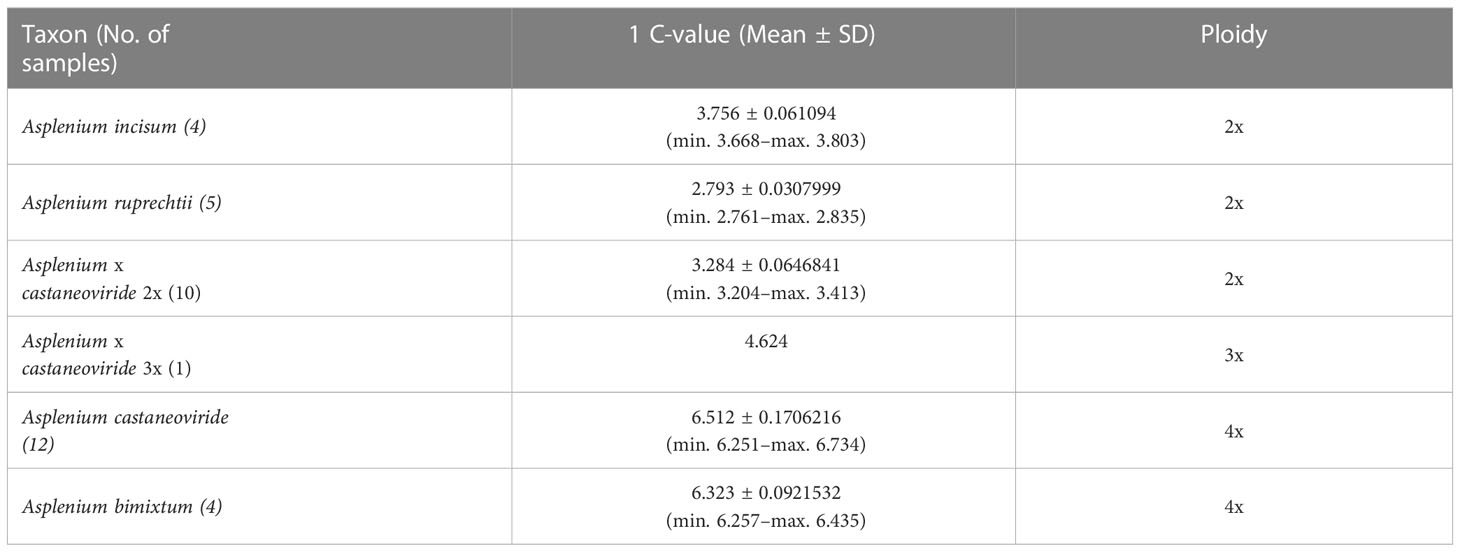
Table 1 Summary of genome size comparison for A. incisum, A. ruprechtii, and their hybrid progenies.
Genome structure and gene contents of plastomes in Asplenium species
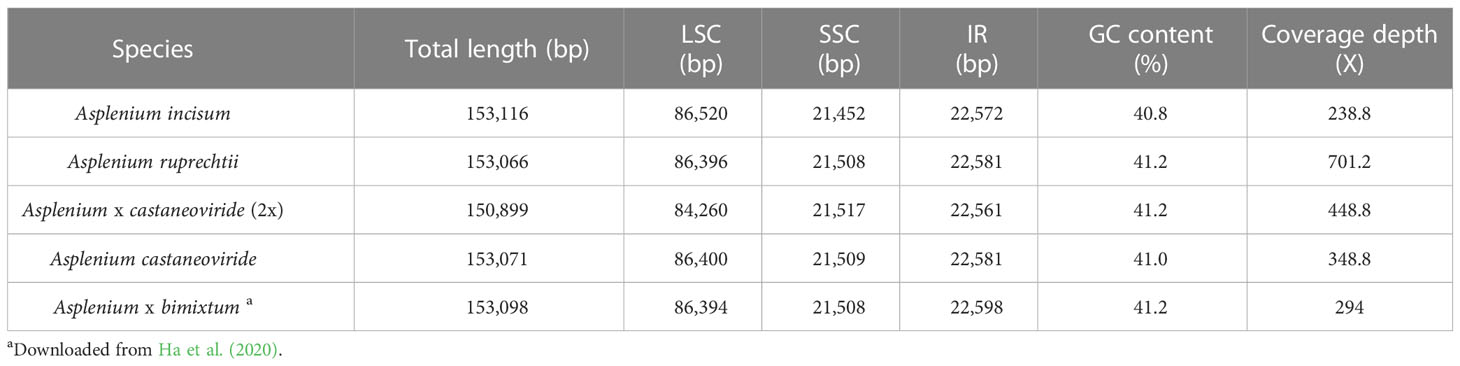
The four plastomes of Asplenium species newly reported in the present study were from 150,899 (A. × castaneoviride 2x) to 153,116 bp (A. incisum) in length with a GC content of 40.8%–41.2% (Table 2). The length variation among the five plastomes including A. bimixtum was higher in the large single-copy (LSC) region than in the other regions because of the 2,139-bp deletion found in the plastome of A. × castaneoviride 2x. The LSC-IR boundaries were highly conserved, but the SSC-IR boundary of A. incisum was slightly different from that of the others because of the 22-bp IR expansion into chlL (Supplementary Figure 2).
Asplenium castaneoviride, A. bimixtum, and A. × castaneoviride (2x) have A. ruprechtii-type plastomes, whereas A. incisum always has its own plastome. Pairwise comparison of the whole plastome sequences showed that A. ruprechtii-type plastomes had high identities to each other with more than 98.48%, and increasing up to 99.88% when a large deletion that occurred in A. × castaneoriviride (2x) was excluded (Supplementary Table 4). In addition, two small inversions and a tandem repeat were found among A. ruprechtii-type plastomes (Supplementary Figure 3).
In contrast to the results of Lee et al. (2019), the plastome of A. bimixtum was clearly A. ruprechtii-type and not belonging to A. incisum-type. Therefore, the rbcL regions of four individuals of A. bimixtum were additionally sequenced and confirmed that they were identical to the A. ruprechtii-type rbcL without any exception.
New finding of the enlarged noncoding region in the plastomes
The enlarged noncoding regions of rpoC2-rps2 in the plastomes of A. incisum and A. ruprechtii and their hybrids were newly found compared to the previously reported plastomes in other ferns (Kim and Kim, 2020). Although 100 bp upstream of rps2 was highly conserved within the genus, the intergenic spacer of rpoC2-rps2 of A. incisum, A. ruprechtii, and their hybrids was 1,870–4,277 bp longer than that of other species with non-expanded plastomes (Table 3). Among the enlarged regions in rpoC2-rps2 of A. incisum, A. reprechtii, and their hybrids, a large deletion of 2,139 bp was detected in the plastome of A. × castaneoviride 2x. We checked the presence/absence of this large deletion in all collected samples and confirmed that this event also occurred in other individuals from the Buramsan populations: five out of nine in A. ruprechtii, five out of six in A. × castaneoviride (2x), one in A. × castaneoviride (3x), and one out of five in A. castaneoviride.
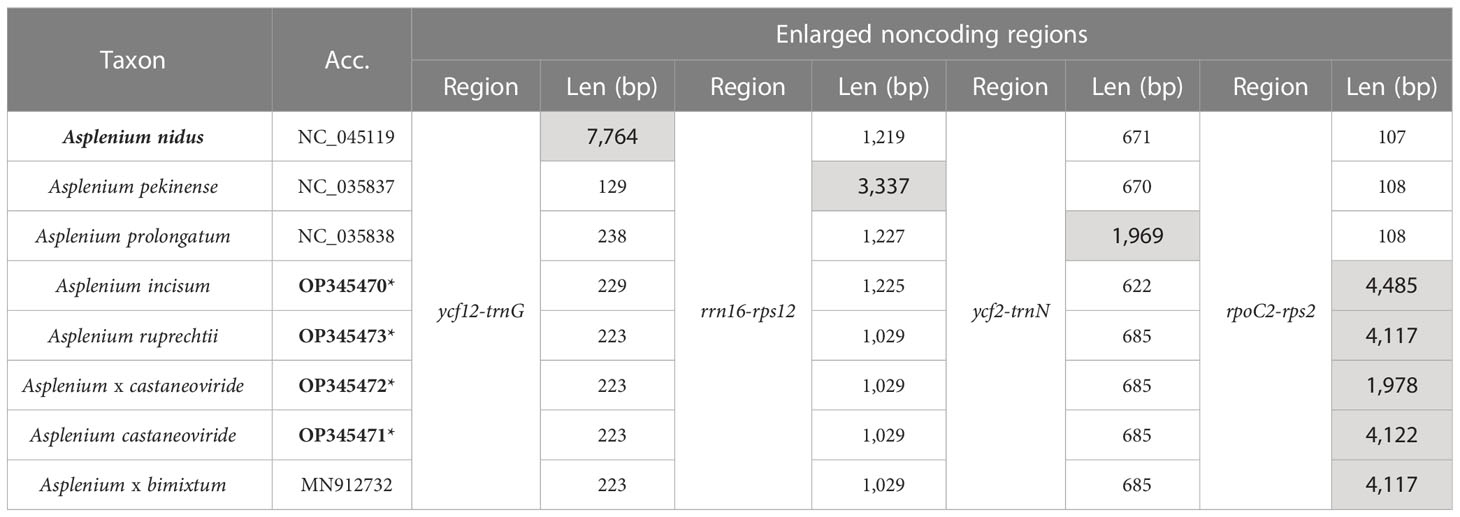
Table 3 Enlarged noncoding regions (gray cells) in Asplenium plastomes. (*Bold accession numbers indicate the plastomes that were newly determined in the present study).
The enlarged DNAs of A. incisum and A. ruprechtii were originated from other sources even though they are very closely related taxa. The BLAST results for the A. ruprechtii rpoC2-rps2 region (from −101 to −4,108 upstream of rps2) showed that most regions were hit at least once in 48 organisms tested consisting of 47 ferns and one fungi (Figure 4A and Supplementary Table 5); only the 87-bp region had no hits against the nucleotide collection. In contrast to the A. ruprechtii rpoC2-rps2 region, the 1,021-bp region (from −101 to −1,121 bp upstream of rps2) of the A. incisum rpoC2-rps2 region was only hit in the fern plastomes. Although the 49-bp region positioned at −3,440 bp upstream of rps2 hit the mitochondrial genome of the fungus, this region was new in this plastome (Figure 4B).
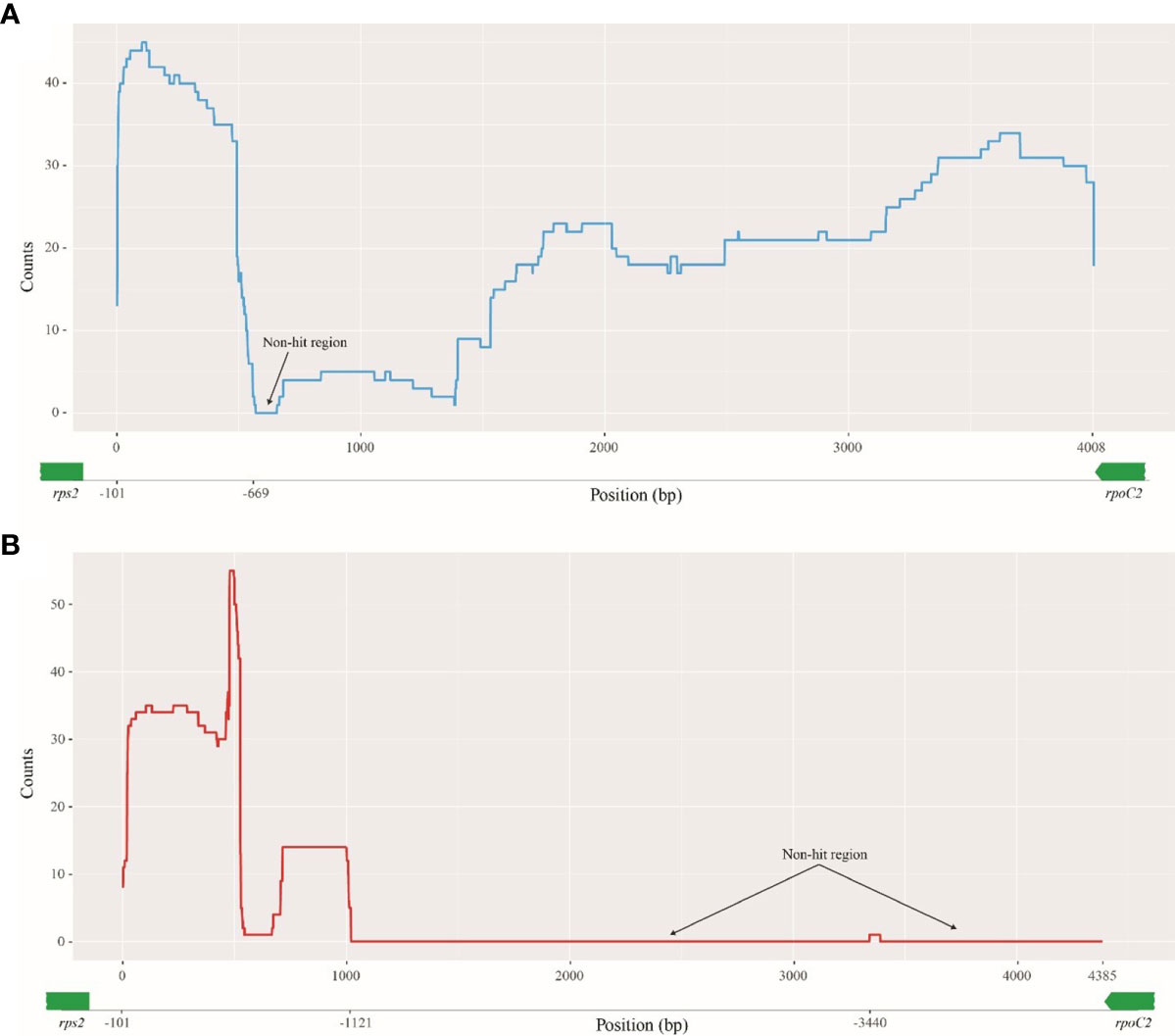
Figure 4 Blast results of enlarged regions against nucleotide collection. The Y-axis shows the number of hits to subject sequences and the X-axis shows the position of rpoC2-rps2 regions. (A) rpoC2-rps2 of A. ruprechtii; (B) rpoC2-rps2 of A. incisum.
Maternal origin of hybrids derived from A. ruprechtii and A. incisum
To confirm the maternal origin of the hybrid progenies between A. ruprechtii and A. incisum, the rbcL sequences of 68 collected samples were compared. All conspecific rbcL sequences of A. ruprechtii (28 individuals) and A. incisum (17 individuals) were almost identical and clearly divided, even though one individual of A. incisum had 1-bp substitution against others. All hybrids, A. × castaneoviride (2x), A. × castaneoviride (3x), A. castaneoviride, and A. bimixtum, had an A. ruprechtii-type rbcL, except for two individuals of A. × castaneoviride (2x), which shared 1-bp substitution against the other A. ruprechtii individuals. In the phylogenetic analysis, they were located in the same clade with their maternal origin A. ruprechtii (Supplementary Figure 4).
Discussion
Hybridization pattern between A. ruprechtii and A. incisum
Asplenium L. tends to hybridize and polyploidize (Dyer et al., 2012; Chang et al., 2018; Liang et al., 2021) in natural populations. Recent experiments based on maternally inherited plastid genes showed that some allotetraploids of the genus (e.g., A. billotii and A. foreziense) have reciprocal parentage. However, other species such as A. adiantum-nigrum and A. balearicum have no evidence of reciprocal origins in the obovatum-adiantum-nigrum group because the former two tetraploids were embedded together in the same clade with respective diploid parents, whereas the latter two tetraploids formed a clade with only maternal diploid of A. onopteris (Sessa et al., 2018). It is possible to assume the path of hybridization using the positions of hybrids and their parental species in the phylogenetic tree, to get insights into the evolution of species over the past, present, and future. For instance, in New Zealand, two octoploids, A. cimmeriorum and A. gracillimum, are reported to result from reciprocal hybridization between two tetraploids, A. bulbiferum and A. hookerianum (Shepherd et al., 2008).
Considering that A. × castaneoviride (2x or 3x) and A. ruprechtii grow side-by-side in Korea, A. ruprechtii is expected to be a maternal parent of A. × castaneoviride. At this point, we have shelved the possibility of allotetraploid formation via a triploid bridge because only one individual of triploid A. × castaneoviride was observed in nature with abortive spores. Our results showed that all hybrids have two types of pgiC originated from A. ruprechtii and A. incisum, but they have only A. ruprechtii-type plastome genes of rbcL. The nuclear single-copy and plastome genes showed completely unidirectional hybrid formation between A. ruprechtii and A. incisum (χ2 = 7, df = 1, p < 0.01). Therefore, A. castaneoviride, a consequence of polyploidization by diploid hybrid A. × castaneoviride, is also expected to have A. ruprechtii-type plastome sequence affecting hybridization between A. ruprechtii and A. incisum.
Gastony and Yatskievych (1992) showed that sexual species play the role of female parent when hybridization occurs between an apogamous and a sexual species in ferns because functional archegonia is lacking in an apogamous species. However, both A. ruprechtii and A. incisum are known to be sexual species, and the sporangium in both species really exhibited 64 spores in our observations. Vogel et al. (1998b) inferred that the predominantly unidirectional fertilization between Asplenium trichomanes and A. septentrionale is related to breeding systems. According to them, the obligately inbreeding taxon may have difficulty playing the role of a maternal parent in the hybridization process because of the synchronous maturity of male and female gametes. Based on the length variation due to the large deletion of rpoC2-rps2 in Buramsan populations of A. ruprechtii and tetraploid A. castaneoviride and their capacity to hybridize with other species in the same genus (Lee et al., 2019), it is suggested that A. ruprechtii is an outbreeding taxon. In contrast, A. incisum has hybridized with species other than A. ruprechtii and acts as a maternal parent (Ebihara, 2016). Therefore, breeding systems might not play an important role in the female parent of A. ruprechtii. If the ontogenetic sequence of gametangia differs between the two parental species, one species beginning with male prothalli may be able to reach the male parent of another species beginning with female prothalli, such as dichogamy in angiosperms. However, gametophytes of A. ruprechtii and A. incisum first produce archegonia, followed by antheridia (Jeong and Lee, 2006).
As mentioned above, A. ruprechtii and A. incisum have the same breeding system of outcrossing, sexual reproductive mode, and diploid level. Nevertheless, it is not known why only A. ruprechtii plays the role of a female parent in this asymmetric hybridization.
One possible scenario for this asymmetric hybridization may be the sex-determination system in ferns. Antheridiogens are pheromones secreted by mature female fern gametophytes, which induce neighboring asexual gametophytes to produce precocious antheridia (Schneller and Ranker, 2008). Hornych et al. (2021) showed that all four species tested in Asplenium responded to conspecific antheridiogen sources, and several species in various lineages responded to these sources. Therefore, it is likely that antheridiogens in genus Asplenium are important for sex determination. If A. incisum gametophytes can inhabit the A. ruprechtii population, but not vice versa, antheridiogens of A. ruprechtii can induce A. incisum gametophytes to grow male gametophytes owing to environmental conditions such as light and moisture. Then, the mature sperm of A. incisum may arrive at the archaegonia of A. ruprechtii to produce a hybrid. Since the available data are insufficient for this scenario, further investigation of asymmetric hybridization between A. ruprechtii and A. incisum is required, including a glasshouse experiment under restricted conditions, because both species have frequently hybridized.
Multiple origins of allotetraploid A. castaneoviride
Asplenium castaneoviride, which is distributed in China, Japan, and Korea (Lin and Viane, 2013) is an allotetraploid species derived from A. ruprechtii and A. incisum. However, the origin of this allotetraploid species has not been discussed.
Multiple allopolyploidization events, in which progenitor species are repeatedly hybridized and independently polyploidized, can produce intraspecific variations within its progenitor species (Sigel, 2016). A large deletion of 2,139 bp that occurred in rpoC2-rps2 was not found in any A. ruprechtii individuals, except for the Buramsan populations (P4) where this mutation may be caused independently. We proposed a hypothesis to induce the allotetraploid A. castaneoviride from the diploid hybrid between A. ruprechtii and A. incisum (Figure 5). A haploid gamete of A. ruprechtii may form a zygote with heterozygous diploid gametes of diploid A. × castaneoviride or A. castaneoviride, and a large deletion of rpoC2-rps2 remains at different ploidy levels (2x, 3x, and 4x) of A. castaneoviride in Buramsan populations. Consequently, the allotetraploid A. castaneoviride was independently produced by more than two maternal origins in the Buramsan population.
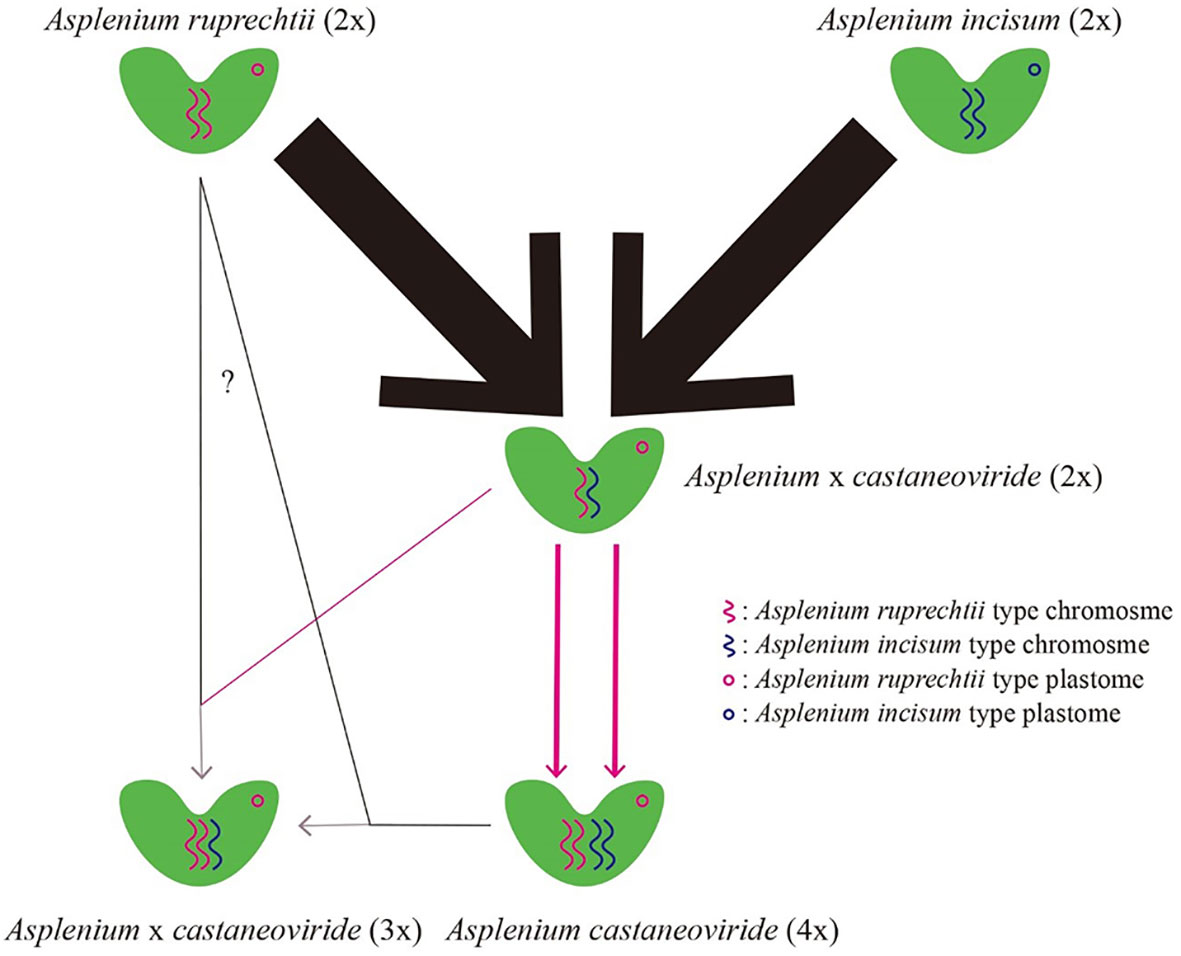
Figure 5 Hypothetical origins of different ploidy levels of diploid, triploid, and tetraploid Asplenium castaneoviride. Red and black lines refer to unreduced diplospory and reduced haplospory, respectively. Line thickness represents the frequency of hybrid individuals found in this study.
Although the allotetraploid A. castaneoviride was fertile, we did not find a population consisting of only A. castaneoviride without the maternal parent of A. ruprechtii in Korea.
Allopolyploids generally have advantages such as heterosis, an adaptive advantage of progeny (Birchler et al., 2010), and gene redundancy masking recessive alleles (Comai, 2005) compared to their progenitors. However, minor cytotypes can be excluded from the population by forming sterile odd-ploidy offspring (Levin, 1975). In Korea, A. ruprechtii and A. incisum are widely distributed, and hybridization frequently occurs between them. During the study, we found four diploid hybrid populations with sympatric distribution of both parental species A. ruprechtii and A. incisum, but allotetraploid species were found in only two populations of Bukhansan (P2) and Buramsan (P4). Although we did not include the plant samples from other ranges of the species like the Chinese one in the present study, we propose that the driving force for the polyploidization of diploid hybrids (A. × castaneoviride) may have been maintained, but restricted to the natural population by a frequency-dependent mating disadvantage (Husband, 2000).
Is Asplenium × bimixtum a hybrid among the three species?
Asplenium x bimixtum C. S. Lee & K. Lee ex Y.H. Ha was first reported by Lee et al. (2015) based on its morphological characteristics and is endemic to Korea. Lee et al. (2015) argued that this species was formed by multiple hybridizations among three species: A. ruprechtii, A. trichmanes, and A. incisum. They attempted to prove the occurrence of these multiple hybridizations based on 1C-value measurements and molecular phylogenetic analyses (Lee et al., 2019). However, they ignored that the standard plant should run together along with running samples to standardize genome size and cover the instability of the equipment. This may explain why the calculated genome sizes of certain species showed a high standard deviation in their study. In addition, they considered all A. castaneotiride individuals to be allotetraploids, but we confirmed the presence of triploid A. × castaneoviride and tetraploid A. castaneoviride from the comparison of 1C-values. Furthermore, the 1C-values of A. bimixtum ranged within those of the allotetraploid A. castaneoviride in their results as well as in this study. Lee et al. (2019) also argued that A. bimixtum has two types of plastid genes: A. ruprechtii-type rbcL and A. incisum-type rps4. However, the complete plastome sequence of A. bimixtum is more similar to A. ruprechtii plastome with 99.96% identity (Supplementary Table 4) and has the same type of rps4-trnS. The four individuals of A. bimixtum used in the present study were all embedded in the A. ruprechtii clade in the phylogenetic tree (Supplementary Figure 4). Although we cannot completely rule out the possibility of overlooking A. x bimixtum in the habitat, this species may be included in the variation of A. castaneoviride, and it will be more apt to treat it as a synonym of allotetraploid A. castaneoviride with the available data. If we recognize A. bimixtum as a member of the alltotetraploid of A. castaneoviride, it could be concluded that A. castaneoviride has derived from more than three origins based on their genetic variation patterns: (1) normal type without large deletion in rpoC2-rps2, (2) with large deletion in rpoC2-rps2, and (3) with a small inversion of trnN-ndhF and more tandem repeats in ycf2 (A. bimixtum type) in Korea, even though they are grouped together in the same clade as their maternal origin.
Enlarged noncoding region in fern organelles found in Asplenium species
Robison et al. (2018) suggested the concept of mobile open reading frames in fern organelles (MORFFO), because several ORFs varying in sequence length, insertion position, and gene arrangement are widely distributed in Pteridaceae plastomes. In contrast, the earliest diverging group with these MORFFOs is in Ophioglossaceae, and six polycistronic genes in the plastome of Mankyua chejuense appear to be the original form of non plastome foreign DNAs in fern plastomes (Kim and Kim, 2018).
Certain flanking regions of MORFFOs are highly divergent or unique though these appear to be conserved throughout the fern lineage if they exist. For instance, the rps12-rrn16 regions of Odontosoria chinensis plastome containing morffo had 94.3% of query coverage hit to nucleotide collection (Robison et al., 2018); however, the trnE-psbM region, which was also expanded by foreign DNAs in the same plastome, had only 45% of query coverage. Similar cases are reported in other fern genera. The trnV-rrn16 regions of Hymenophyllum plastomes were expanded because of foreign DNA insertions (Kim and Kim, 2020); however, only 33.6% of foreign DNAs of H. polyanthos were hit by H. coreanum using BLAST, although these two species are closely related to each other.
In the present study, we also found foreign DNA in the rpoC2-rps2 region of the A. incisum plastome, which has not been reported so far, and confirmed that it originated from a completely different source. To recognize and describe this feature on the plastome, similar to the rpoC2-rps2 of A. incisum, in which no morffo is observed, we propose the term of “Enlarged Noncoding Region in Fern Organelles (ENRIFO),” because many enlarged noncoding regions in fern organelles include unique sequences.
To understand the origin of ENRIFO in A. incisum, one should understand how it is completely different from A. ruprechtii. Kim and Kim (2020) suggested that palindromic sequences, including tRNA genes, are strongly related to the genomic instability of fern plastomes, and long foreign DNA is degraded under selective pressure. As the ENRIFOs of Asplenium, except for A. incisum, vary in position in the plastome and share certain sequences, it is possible that a common ENRIFO has changed its position in the plastomes of the genus Asplenium. However, this cannot explain why unidentified sequences are found in the ENRIFOs within the Asplenium plastomes. The simplest explanation for these unidentified ENRIFOs is that these can be easily substituted by foreign DNA, which targets certain regions in the plastome associated with genomic instability. The positions of ENRIFOs generally vary in fern plastomes; however, they are found at the same position in closely related taxa (Kim and Kim, 2020). Considering the low similarity between ENRIFOs in closely related taxa, DNA recombination may have occurred more frequently than expected. To completely understand this genomic feature, further study of the genus Asplenium is needed.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/nuccore/OP345470.1/, https://www.ncbi.nlm.nih.gov/nuccore/OP345473, https://www.ncbi.nlm.nih.gov/nuccore/OP345472, https://www.ncbi.nlm.nih.gov/nuccore/OP345471.
Author contributions
Conceptualization, HK and JK. Methodology, HK and SP. Sequencing, SP. Software and data curation, HK. Resources, SP, HK and JK. Writing—original draft preparation, HK. Writing—review and editing, HK and JK. Visualization, HK. Supervision, JK. Project administration, HK and funding acquisition, HK and JK. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Research Foundation of Korea (NRF) under Grant Nos. 2020R1I1A2053517 and 2021R1I1A3A04037448.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1116040/full#supplementary-material
Supplementary Figure 1 | Map of the location of populations where the plant samples were collected and used to measure the genome size of A. incisum, A. ruprechtii, and their hybrid taxa. The population numbers are identical to the Supplementary Table 1 and the red circles indicate the sympatric populations where the two parental species and their hybrid progenies occupied together.
Supplementary Figure 2 | Comparison the border regions of the LSC, SSC, and IR among five plastomes.
Supplementary Figure 3 | Small inversions and tandem repeats among A. ruprechtii type plastomes. A) Two small inversions found trnN-ndhF and rps15-ycf1, respectively. B) A variable number tandem repeat in ycf2.
Supplementary Figure 4 | Phylogeny of Asplenium incisum, A. ruprechtii and their hybrids constructed by ML method using IQTREE. Numbers on the branch refer to ultrafast bootstrap support value. Number on the base substitution left indicates the position of base substitution on the alignment of rbcL (1283 bp in length).
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 (3), 403–410. doi: 10.1016/S0022-2836(05)80360-2
Bennett, M. D., Smith, J. B. (1976). Nuclear DNA amounts in angiosperms. Philos. Trans. R Soc. Lond B Biol. Sci. 274 (933), 227–274. doi: 10.1098/rstb.1976.0044
Birchler, J. A., Yao, H., Chudalayandi, S., Vaiman, D., Veitia, R. A. (2010). Heterosis. Plant Cell 22 (7), 2105–2112. doi: 10.1105/tpc.110.076133
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30 (15), 2114–2120. doi: 10.1093/bioinformatics/btu170
Cai, Z., Guisinger, M., Kim, H. G., Ruck, E., Blazier, J. C., McMurtry, V., et al. (2008). Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J. Mol. Evol. 67 (6), 696–704. doi: 10.1007/s00239-008-9180-7
Chang, Y., Ebihara, A., Lu, S., Liu, H., Schneider, H. (2018). Integrated taxonomy of the Asplenium normale complex (Aspleniaceae) in China and adjacent areas. J. Plant Res. 131 (4), 573–587. doi: 10.1007/s10265-018-1032-y
Chang, Y. F., Li, J., Lu, S. G., Schneider, H. (2013). Species diversity and reticulate evolution in the Asplenium normale complex (Aspleniaceae) in China and adjacent areas. Taxon 62 (4), 673–687. doi: 10.12705/624.6
Comai, L. (2005). The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6 (11), 836–846. doi: 10.1038/nrg1711
de Groot, G. A., During, H. J., Maas, J. W., Schneider, H., Vogel, J. C., Erkens, R. H. (2011). Use of rbcL and trnL-f as a two-locus DNA barcode for identification of NW-European ferns: an ecological perspective. PloS One 6 (1), e16371. doi: 10.1371/journal.pone.0016371
Dyer, R. J., Savolainen, V., Schneider, H. (2012). Apomixis and reticulate evolution in the Asplenium monanthes fern complex. Ann. Bot. 110 (8), 1515–1529. doi: 10.1093/aob/mcs202
Ebihara, A., Ishikawa, H., Matsumoto, S., Lin, S. J., Iwatsuki, K., Takamiya, M., et al. (2005). Nuclear DNA, chloroplast DNA, and ploidy analysis clarified biological complexity of the vandenboschia radicans complex (Hymenophyllaceae) in Japan and adjacent areas. Am. J. Bot. 92 (9), 1535–1547. doi: 10.3732/ajb.92.9.1535
Funk, H. T., Berg, S., Krupinska, K., Maier, U. G., Krause, K. (2007). Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol. 7, 45. doi: 10.1186/1471-2229-7-45
Gao, L., Wang, B., Wang, Z. W., Zhou, Y., Su, Y. J., Wang, T. (2013). Plastome sequences of Lygodium japonicum and Marsilea crenata reveal the genome organization transformation from basal ferns to core leptosporangiates. Genome Biol. Evol. 5 (7), 1403–1407. doi: 10.1093/gbe/evt099
Gastony, G. J., Yatskievych, G. (1992). Maternal inheritance of the chloroplast and mitochondrial genomes in cheilanthoid ferns. Am. J. Bot. 79 (6), 716–722. doi: 10.2307/2444887
Gottlieb, L. D. (1982). Conservation and duplication of isozymes in plants. Science 216 (4544), 373–380. doi: 10.1126/science.216.4544.373
Gottlieb, L. D., Ford, V. S. (1996). Phylogenetic relationships among the sections of Clarkia (Onagraceae) inferred from the nucleotide sequences of PgiC. Systematic Bot. 21 (1), 45–62. doi: 10.2307/2419562
Grewe, F., Guo, W., Gubbels, E. A., Hansen, A. K., Mower, J. P. (2013). Complete plastid genomes from Ophioglossum californicum, psilotum nudum, and equisetum hyemale reveal an ancestral land plant genome structure and resolve the position of equisetales among monilophytes. BMC Evol. Biol. 13, 8. doi: 10.1186/1471-2148-13-8
Guisinger, M. M., Kuehl, J. V., Boore, J. L., Jansen, R. K. (2011). Extreme reconfiguration of plastid genomes in the angiosperm family geraniaceae: rearrangements, repeats, and codon usage. Mol. Biol. Evol. 28 (1), 583–600. doi: 10.1093/molbev/msq229
Ha, Y.-H., Gil, H.-Y., Kim, D.-K. (2020). Characterization of the complete plastome sequence of Asplenium× bimixtum (Aspleniaceae). Mitochondrial DNA Part B 5 (2), 1854–1856. doi: 10.1080/23802359.2020.1750993
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35 (2), 518–522. doi: 10.1093/molbev/msx281
Hornych, O., Testo, W. L., Sessa, E. B., Watkins, J. E., Jr., Campany, C. E., Pittermann, J., et al. (2021). Insights into the evolutionary history and widespread occurrence of antheridiogen systems in ferns. New Phytol. 229 (1), 607–619. doi: 10.1111/nph.16836
Husband, B. C. (2000). Constraints on polyploid evolution: a test of the minority cytotype exclusion principle. Proc. Biol. Sci. 267 (1440), 217–223. doi: 10.1098/rspb.2000.0990
Ishikawa, H., Watano, Y., Kano, K., Ito, M., Kurita, S. (2002). Development of primer sets for PCR amplification of the PgiC gene in ferns. J. Plant Res. 115 (1117), 65–70. doi: 10.1007/s102650200010
Jeong, J.-A., Lee, C. H. (2006). Gametophyte morphogenesis of 3 fern species belonging to aspleniaceae in vitro. Flower Res. J. 14 (3), 170–175.
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14 (6), 587–589. doi: 10.1038/nmeth.4285
Katoh, K., Misawa, K., Kuma, K., Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30 (14), 3059–3066. doi: 10.1093/nar/gkf436
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28 (12), 1647–1649. doi: 10.1093/bioinformatics/bts199
Kim, H. T., Chung, M. G., Kim, K. J. (2014). Chloroplast genome evolution in early diverged leptosporangiate ferns. Mol. Cells 37 (5), 372–382. doi: 10.14348/molcells.2014.2296
Kim, H. T., Kim, K. J. (2014). Chloroplast genome differences between Asian and American Equisetum arvense (Equisetaceae) and the origin of the hypervariable trnY-trnE intergenic spacer. PloS One 9 (8), e103898. doi: 10.1371/journal.pone.0103898
Kim, H. T., Kim, K. J. (2018). Evolution of six novel ORFs in the plastome of Mankyua chejuense and phylogeny of eusporangiate ferns. Sci. Rep. 8 (1), 16466. doi: 10.1038/s41598-018-34825-6
Kim, H. T., Kim, J. S. (2020). The dynamic evolution of mobile open reading frames in plastomes of Hymenophyllum sm. and new insight on Hymenophyllum coreanum nakai. Sci. Rep. 10 (1), 11059. doi: 10.1038/s41598-020-68000-7
Kim, H. T., Kim, J. S., Moore, M. J., Neubig, K. M., Williams, N. H., Whitten, W. M., et al. (2015). Seven new complete plastome sequences reveal rampant independent loss of the ndh gene family across orchids and associated instability of the inverted repeat/small single-copy region boundaries. PloS One 10 (11), e0142215. doi: 10.1371/journal.pone.0142215
Knox, E. B. (2014). The dynamic history of plastid genomes in the campanulaceae sensu lato is unique among angiosperms. Proc. Natl. Acad. Sci. U.S.A. 111 (30), 11097–11102. doi: 10.1073/pnas.1403363111
Labiak, P. H., Karol, K. G. (2017). Plastome sequences of an ancient fern lineage reveal remarkable changes in gene content and architecture. Am. J. Bot. 104 (7), 1008–1018. doi: 10.3732/ajb.1700135
Lee, C. S., Lee, K., Yeau, S. H., Chung, K.-S. (2015). Two new and one unrecorded natural hybrids between Asplenium ruprechtii and related taxa (Aspleniaceae). Korean J. Plant Taxonomy 45 (4), 362–368. doi: 10.11110/kjpt.2015.45.4.362
Lee, C. S., Yeau, S. H., Chung, K.-s. (2019). Exploring natural hybridizations among asplenium ruprechtii and related taxa in Korea. Korean J. Plant Taxonomy 49 (2), 127–139. doi: 10.11110/kjpt.2019.49.2.127
Lehtonen, S., Cardenas, G. G. (2019). Dynamism in plastome structure observed across the phylogenetic tree of ferns. Botanical J. Linn. Soc. 190 (3), 229–241. doi: 10.1093/botlinnean/boz020
Levin, D. A. (1975). Minority cytotype exclusion in local plant populations. Taxon 24 (1), 35–43. doi: 10.2307/1218997
Liang, S. Q., Viane, R. L. L., Zhang, X. C., Wei, R. (2021). Exploring the reticulate evolution in the Asplenium pekinense complex and the A. varians complex (Aspleniaceae). J. Systematics Evol. 59 (1), 125–140. doi: 10.1111/jse.12530
Lin, Y., Viane, R. (2013). “Aspleniaceae,” in Flora of China, vol. 2-3 . Eds. Wu, Z. Y., Raven, P. H., Hong, D. Y. (Beijing: Science Press; St. Louis: Missouri Botanical Garden Press), 267–316.
Lowe, T. M., Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25 (5), 955–964. doi: 10.1093/nar/25.5.955
McNeal, J. R., Kuehl, J. V., Boore, J. L., de Pamphilis, C. W. (2007). Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol. 7, 57. doi: 10.1186/1471-2229-7-57
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32 (1), 268–274. doi: 10.1093/molbev/msu300
Ohlsen, D. J., Perrie, L. R., Shepherd, L. D., Brownsey, P. J., Bayly, M. J. (2014). Investigation of species boundaries and relationships in the Asplenium paleaceum complex (Aspleniaceae) using AFLP fingerprinting and chloroplast and nuclear DNA sequences. Aust. Systematic Bot. 27 (6), 378–394. doi: 10.1071/sb14024
Robison, T. A., Grusz, A. L., Wolf, P. G., Mower, J. P., Fauskee, B. D., Sosa, K., et al. (2018). Mobile elements shape plastome evolution in ferns. Genome Biol. Evol. 10 (10), 2558–2571. doi: 10.1093/gbe/evy189
Ruhlman, T. A., Jansen, R. K. (2014). The plastid genomes of flowering plants. Methods Mol. Biol. 1132, 3–38. doi: 10.1007/978-1-62703-995-6_1
Schneller, J. J. (2008). Antheridiogens. in Biology and Evolution of Ferns and Lycophytes. ed. Ranker, T. A. & Haufler, C. H. (Cambridge: Cambridge University Press). 134–158. doi: 10.1017/CBO9780511541827.006
Schuettpelz, E., Pryer, K. M. (2007). Fern phylogeny inferred from 400 leptosporangiate species and three plastid genes. Taxon 56 (4), 1037–1050. doi: 10.2307/25065903
Sessa, E. B., Vicent, M., Chambers, S. M., Gabriel y Galán, J. M. (2018). Evolution and reciprocal origins in Mediterranean ferns: the asplenium obovatum and A. adiantumnigrum complexes 1. Ann. Missouri Botanical Garden 103 (2), 175–187. doi: 10.3417/2018108
Sessa, E. B., Zimmer, E. A., Givnish, T. J. (2012). Reticulate evolution on a global scale: a nuclear phylogeny for new world Dryopteris (Dryopteridaceae). Mol. Phylogenet Evol. 64 (3), 563–581. doi: 10.1016/j.ympev.2012.05.009
Shepherd, L. D., Perrie, L. R., Brownsey, P. J. (2008). Low-copy nuclear DNA sequences reveal a predominance of allopolyploids in a new Zealand Asplenium fern complex. Mol. Phylogenet. Evol. 49 (1), 240–248. doi: 10.1016/j.ympev.2008.06.015
Sigel, E. M. (2016). Genetic and genomic aspects of hybridization in ferns. J. Systematics Evol. 54 (6), 638–655. doi: 10.1111/jse.12226
Stebbins, G. L. (1940). The significance of polyploidy in plant evolution. Am. Nat. 74 (750), 54–66. doi: 10.1086/280872
Stein, D. B., Barrington, D. S. (1990). Recurring hybrid formation in a population of Polystichum x potteri: evidence from chloroplast DNA comparisons. Ann. Missouri Botanical Garden 77 (2), 334–339. doi: 10.2307/2399548
Thomas, B. R., Ford, V. S., Pichersky, E., Gottlieb, L. D. (1993). Molecular characterization of duplicate cytosolic phosphoglucose isomerase genes in Clarkia and comparison to the single gene in Arabidopsis. Genetics 135 (3), 895–905. doi: 10.1093/genetics/135.3.895
Vogel, J. C., Russell, S. J., Rumsey, F. J., Barrett, J. A., Gibby, M. (1998a). Evidence for maternal transmission of chloroplast DNA in the genus Asplenium (Aspleniaceae, pteridophyta). Botanica Acta 111 (3), 247–249. doi: 10.1111/j.1438-8677.1998.tb00704.x
Vogel, J. C., Russell, S. J., Rumsey, F. J., Barrett, J. A., Gibby, M. (1998b). On hybrid formation in the rock fern Asplenium x alternifolium (Aspleniaceae, pteridophyta). Botanica Acta 111 (3), 241–246. doi: 10.1111/j.1438-8677.1998.tb00702.x
Wagner, W. H., Wagner, F. S. (1980). “Polyploidy in pteridophytes,” in Polyploidy: biological relevance. Ed. Lewis, W. H. (Boston, MA: Springer US), 199–214.
Wagner, W. H., Wagner, F. S., Taylor, W. C. (1986). Detecting abortive spores in herbarium specimens of sterile hybrids. Am. Fern J. 76 (3), 129–140. doi: 10.2307/1547721
Wolf, P. G., Der, J. P., Duffy, A. M., Davidson, J. B., Grusz, A. L., Pryer, K. M. (2011). The evolution of chloroplast genes and genomes in ferns. Plant Mol. Biol. 76 (3-5), 251–261. doi: 10.1007/s11103-010-9706-4
Wolf, P. G., Roper, J. M., Duffy, A. M. (2010). The evolution of chloroplast genome structure in ferns. Genome 53 (9), 731–738. doi: 10.1139/g10-061
Wood, T. E., Takebayashi, N., Barker, M. S., Mayrose, I., Greenspoon, P. B., Rieseberg, L. H. (2009). The frequency of polyploid speciation in vascular plants. Proc. Natl. Acad. Sci. U.S.A. 106 (33), 13875–13879. doi: 10.1073/pnas.0811575106
Keywords: Aplenium, hybridization, polyploidization, genome size, plastome, genotypes
Citation: Kim HT, Park SH and Kim JS (2023) Dynamic hybridization between two spleenworts, Asplenium incisum and Asplenium ruprechtii in Korea. Front. Plant Sci. 14:1116040. doi: 10.3389/fpls.2023.1116040
Received: 05 December 2022; Accepted: 31 May 2023;
Published: 05 July 2023.
Edited by:
Maximilian Weigend, University of Bonn, GermanyReviewed by:
Solange Sotuyo, National Autonomous University of Mexico, MexicoJana Sochorová, Academy of Sciences of the Czech Republic (ASCR), Czechia
Copyright © 2023 Kim, Park and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jung Sung Kim, anVuZ3N1bmdAY2JudS5hYy5rcg==
 Hyoung Tae Kim1
Hyoung Tae Kim1 Jung Sung Kim
Jung Sung Kim