- 1School of Crop Protection, College of Post Graduate Studies in Agricultural Sciences, Central Agricultural University (Imphal), Meghalaya, Imphal, India
- 2Director of Research, Central Agricultural University (Imphal), Imphal, India
- 3School of Natural Resource Management, College of Post Graduate Studies in Agricultural Sciences, Central Agricultural University (Imphal), Imphal, India
- 4Sarat Chandra Singha (SCS) College of Agriculture, Assam Agricultural University (Jorhat), Dhubri, Assam, India
- 5Krishi Vigyan Kendra (KVK)-Tinsukia, Assam Agricultural University (Jorhat), Tinsukia, Assam, India
- 6Krishi Vigyan Kendra (KVK)-Sepahijala, Central Agricultural University (Imphal), Tripura, Sepahijala, India
- 7Department of Plant Pathology, Dr Rajendra Prasad Central Agricultural University, Bihar, Samastipur, India
- 8Department of Agronomy, Central Agricultural University (Imphal), Pasighat, India
- 9Department of Mycology and Microbiology, Tea Research Association, North Bengal Regional, R & D Center, Jalpaiguri, West Bengal, India
Trichoderma spp. (Hypocreales) are used worldwide as a lucrative biocontrol agent. The interactions of Trichoderma spp. with host plants and pathogens at a molecular level are important in understanding the various mechanisms adopted by the fungus to attain a close relationship with their plant host through superior antifungal/antimicrobial activity. When working in synchrony, mycoparasitism, antibiosis, competition, and the induction of a systemic acquired resistance (SAR)-like response are considered key factors in deciding the biocontrol potential of Trichoderma. Sucrose-rich root exudates of the host plant attract Trichoderma. The soluble secretome of Trichoderma plays a significant role in attachment to and penetration and colonization of plant roots, as well as modulating the mycoparasitic and antibiosis activity of Trichoderma. This review aims to gather information on how Trichoderma interacts with host plants and its role as a biocontrol agent of soil-borne phytopathogens, and to give a comprehensive account of the diverse molecular aspects of this interaction.
1 Introduction
Agriculture is an economic activity that deals with the scientific production of crops to address world hunger. Food is a fundamental right of humans; therefore, it is the utmost concern of all countries to increase their agricultural production, as the global population is expected to reach nearly 10 billion by 2050 (Gill and Garg, 2014; Dutta et al., 2022a). However, one of the major challenges encountered by agriculture today is the sustainable production of high-quality food in a sufficient quantity to meet the needs of the producer and consumer. Among the various biotic and abiotic factors contributing to the economic yield loss of crops, destruction due to diseases caused by filamentous fungi is of foremost importance (Singh, 2014). Soil-borne plant pathogens lead to a significant reduction in crop yield by causing diseases such as die-back, wilting, and root rot. They usually target the roots to enter into the plant system and directly influence water and nutrient uptake capacity. Soil-borne diseases therefore have a direct negative impact on plant growth and development (Dignam et al., 2022). The management of soil-borne plant diseases is a cumbersome task. Large amounts of chemical pesticides are administered early in the farming process to counteract these phytopathogens. The use of chemical pesticides, however, has a negative impact on the environment, such as residual toxicity and soil pollution. Therefore, the biological management of plant diseases with different bacterial and fungal biocontrol agents is considered a safer option. Trichoderma, a soil-inhabiting ascomycete fungus, is widely used for its versatile plant growth-promoting (PGP) and biocontrol activity. First described by Persoon (1794), the genus Trichoderma outraces phytopathogens in the competition for space, nutrients, antibiosis, and mycoparasitism (Mukherjee et al., 2013). Furthermore, Trichoderma is also known to colonize plant roots, to enhance plants’ systemic defenses, viz., systemic acquired resistance (SAR) and induced systemic resistance (ISR), and to promote plant growth by modulating the phytohormonal blend. To do so, Trichoderma needs to interact and establish a good relationship with the host plant. Proteins or peptides are the communicating molecule in any plant–fungus interaction. Plants and fungi communicate and perceive their surroundings via the secretion and perception of different peptides. Understanding the soluble secretome of Trichoderma will shed light on the mechanisms of molecular crosstalk between plant roots and Trichoderma, and will explain the mechanism behind PGP and biocontrol activities.
2 Trichoderma: role as a biocontrol agent
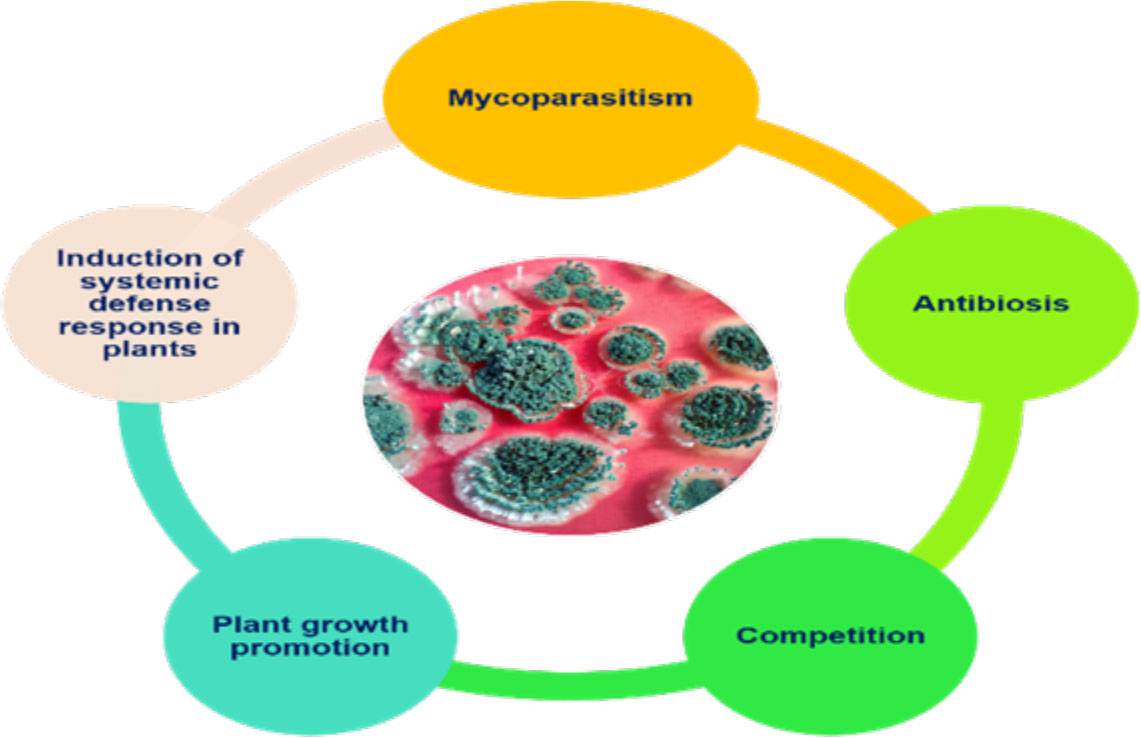
In the early 1930s, the biological control potential of Trichoderma was realized. Weindling, while working with T. lignorum and Rhizoctonia solani, observed that the mycelial growth of R. solani was inhibited by the profuse mycelial growth of Trichoderma. Microscopic observation led to the discovery of a new phenomenon, whereby the hyphae of T. lignorum coil around the hyphae of the phytopathogen and penetrate them, which subsequently leads to complete dissolution of the host cytoplasm. The mechanism of parasitization of another fungus by Trichoderma was named mycoparasitism by Weindling (1932). The discovery of the mycoparasitic nature of Trichoderma led a great volume of work on the subject by many researchers. The studies carried out led to the discovery of different biocontrol mechanisms exhibited by fungi from the genus Trichoderma. Genome profiling of three Trichoderma species, viz., T. virens, T. atroviride, and T. reesei, by Mukherjee et al. (2012) has opened avenues for understanding the molecular mechanism behind their advantageous biocontrol activities. Mycoparasitism, competition with other soil inhabitants/invaders, and antibiosis are the major modes of action for the biocontrol activity of Trichoderma (Zhang et al., 2021). Synchronization between mycoparasitism and antibiosis is necessary for the proper functioning of this biocontrol agent (BCA) (Keswani et al., 2014). Moreover, the host defense activation triggered by Trichoderma is a key part of its ability to protect plants against several phytopathogens. Therefore, it can be said that a combination of competitive exclusion, antibiosis, mycoparasitism, and induced systemic resistance (Figure 1) is crucial for Trichoderma-mediated disease suppression/management (Sharma et al., 2017). Trichoderma is a very fast-growing BCA that rapidly colonizes the spermosphere and/or rhizosphere, thereby providing protection to germinating seeds against major soil-borne, seed-borne, and air-borne plant diseases (Mukherjee et al., 2022).
3 Major biological control strategies of Trichoderma
3.1 Mycoparasitism: Trichoderma’s deadly weapon for the management of phytopathogens
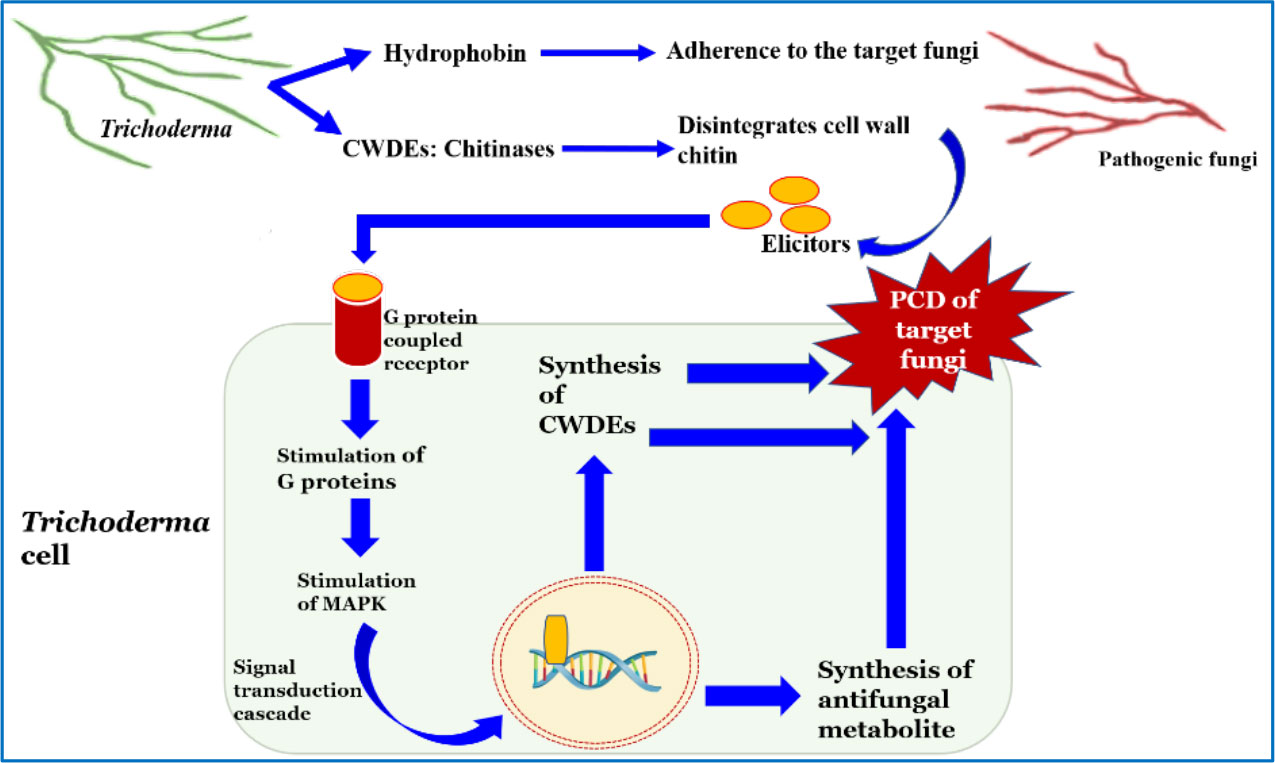
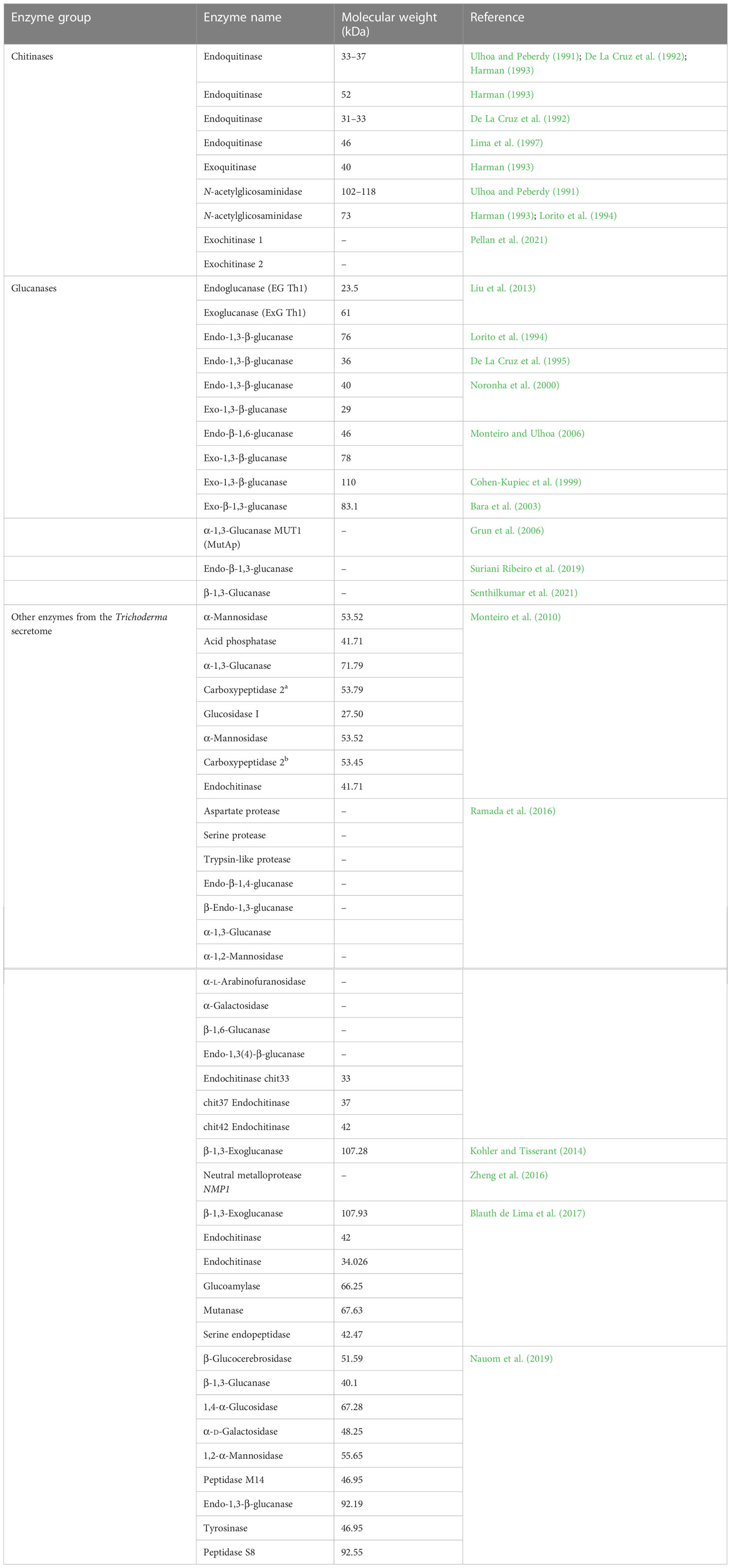
The phenomenon of mycoparasitization by Trichoderma, as first reported by Weindling (1932), is a complex process involving sequential events. Direct confrontation with the fungal pathogen and the secretion of cell wall-degrading enzymes (CWDEs) is followed by the penetration and subsequent killing of the fungal phytopathogen (Woo and Lorito, 2007). The primary identification and attachment between Trichoderma and the prey fungi is mediated by the binding of the cell wall carbohydrates of Trichoderma to the lectin of the target fungi, which is followed by hyphal coiling. The adhesion of Trichoderma to the mycelium of the host fungi is facilitated by hydrophobins, which is evident from the expression of the Vel1 gene of T. virens encoding hydrophobins (Viterbo and Chet, 2006). Penetration into the target hyphae occurs via the development of appressoria containing a high concentration of osmotic solutes such as glycerol, which is necessary for exerting mechanical pressure to invade the hyphal wall. The use of fungitoxic CWDEs, such as chitinases, glucanases, and proteases, by Trichoderma combined with the mechanical strength exerted by the appressorium is crucial to the successful penetration of the host hyphae. Following penetration into the lumen of the target hyphae, the cumulative effect of the CWDEs dissolves the host cell wall, which ultimately results in parasitization and facilitates the assimilation of cell wall content, leading to the subsequent killing of the target fungus (Howell, 2003; Harman et al., 2004; Sood et al., 2020). In addition, Trichoderma disarms the target fungi by deactivating the enzymes necessary for pathogenic fungi to colonize and penetrate the plant tissue (Harman et al., 2004). There are approximately 75 species of Trichoderma reported to exhibit mycoparasitic activity against a wide range of phytopathogens (Verena et al., 2011; Harwoko et al., 2021). Different studies have revealed the significant effect of several strains/species of Trichoderma in the management of phytopathogens such as Fusarium oxysporum, F. culmorum, Gaeumannomyces graminis var. tritici, Pythium aphanidermatum, R. solani, and Sclerotium rolfsii in both greenhouse and field conditions (Das et al., 2006; Dutta et al., 2008; Gajera et al., 2013). The hyperparasitization of F. graminearum by Trichoderma takes place by the Trichoderma clutching and coiling around the target mycelium, interpenetration, and other mechanisms, resulting in deformed mycelium of F. graminearum that eventually disappear (Tian et al., 2018). Chitinase secreted by T. hamatum plays an important role in promoting disintegration of fungal cell wall, chitin assimilation, mycelial autolysis, mycoparasitism, and impeding mycelial growth, spore germination, and spore formation (Saravanakumar et al., 2017). Similarly, T. koningiopsis exhibits mycoparasitic activity against S. sclerotiorum by invading the host hyphae, which it achieves by attaching to and wrapping around the targeted hyphae and then breaking them into small fragments until it completely disintegrates (Shaw et al., 2016). Trichoderma perceives the presence of target fungi in its surroundings via seven transmembrane G protein-coupled receptors, e.g., Grp1 (Omann et al., 2012). When pathogen ligands bind to the receptor, it causes a downstream signaling cascade by stimulating G proteins and mitogen-activated protein kinases (MAPKs). There are three MAPKs known in different Trichoderma species, viz., MAPKKK, MAPKK, and MAPK. Signal transduction via these pathways may have an important role in the mycoparasitization and biocontrol of phytopathogens. Furthermore, the synthesis and secretion of pathogenesis-related enzymes, viz., CWDE and fungitoxic secondary metabolites such as peptaibols, is an extremely useful chemical resource used by Trichoderma to eradicate pathogens (Figure 2) (Omann et al., 2012; Gajera et al., 2013; Dutta et al., 2022b). Although Trichoderma spp. are traditionally known as necrotrophic mycoparasites, an extensive scientific study (Mukherjee et al., 2022) has also revealed hemibiotrophic nature. Hemibiotrophic Trichoderma causes minor damage to the cell wall of the host fungi and is reported to exist intracellularly for a notable period of time.
3.1.1 Evolution of mycoparasitism from a genomic perspective
The ability of the fungi to grow indefinitely as hyphae with great metabolic diversity and their ability to interact with other living components of the ecosystem account for their evolutionary success (Naranjo-Ortiz and Gabaldon, 2019). Mycoparasitic associations were found in the oldest fungal fossil, aged 410 million years (Hass et al., 1994). Comparison of the genome sequences of T. atroviride, T. reesei, and T. virens revealed information about the common ancestral mycotrophic lifestyle of these species (Kubicek et al., 2011; Schmoll et al., 2016; Karlsson et al., 2017). Their mycotrophic lifestyle further evolved to slowly colonize dead wood, plants, animals, and immunocompromised humans, providing new ecological niches for their growth and development (Druzhinina et al., 2018). It is speculated that the ancestor of Trichoderma had limited cellulolytic capacity and fed on either fungi or arthropods, and lateral gene transfer (LGT) is to some extent considered responsible for the formation of this genus. Nearly half the genes for plant CWDEs [belonging to the group called carbohydrate active enzymes (CAZymes)] found in genome profiling of Trichoderma were from plant-associated Ascomycota, indicating a competitive advantage for the mycoparasite in colonizing and feeding on these fungi. However, LGT is not reported in Trichoderma for the mycoparasitism of unrelated fungi such as basidiomycetes and oomycetes (Druzhinina et al., 2018; Mukherjee et al., 2022). A comparative analysis of the presence in 12 Trichoderma spp. of peculiar gene families that are not shared by other fungi might be useful in explaining their ability to deconstruct host/prey cells. An extremely high number of genes gained by Trichoderma belong to different domains, viz., heterokaryon incompatibility (HET), ankyrin repeat, and the major facilitator superfamily (MFS) transporter families. In comparison with other fungi, Trichoderma genomes have been found to possess a higher number of gene families encoding CAZymes, secondary metabolism-related genes, and transcription factors. Moreover, as well as heterokaryon incompatibility, the HET genes may play a important role in sensing the prey fungus (Kubicek et al., 2019).
The study of the transcriptomics of Trichoderma’s interactions with host fungi revealed that the strategies taken up against prey fungi differ among different Trichoderma species. Atanasova et al. (2013) conducted a transcriptomic study on the specific interactions of three Trichoderma spp., viz., T. atroviride, T. virens, and T. reesei, with R. solani, which revealed the different strategies adopted by these BCAs. Th results revealed that T. atroviride uses diverse strategies that include the up-regulation of the biosynthesis of secondary metabolites and enzymes such as GH16 β-glucanases, proteases, and small secreted proteins. T. reesei was found to increase the expression of cellulases, hemicellulases, and transporter-encoding genes, whereas T. virens relied strongly on its toxic secondary metabolite, expressing primarily the genes responsible for gliotoxin biosynthesis. Expression of genes encoding β-1,3- and β-1,6-endoglucanase were observed in the transcriptome of T. harzianum during co-culture with host fungi. Similar results were obtained for T. asperellum during co-culture with mycophytopathogens (Mukherjee et al., 2022). Further research on transcriptomics revealed the up-regulation of genes encoding proteases in Trichoderma transcriptomes, indicating that proteolysis is a prominent process in this BCA’s mycoparasitism (Morán-Diez et al., 2019). Zapparata et al. (2021) studied the interactions of T. gamsii with F. graminearum, with a special focus on transcriptomic changes in both organisms during the sensing phase. The findings revealed an increase in the expression of genes for ferric reductase in T. gamsii, which are essential for iron competition among both fungi; similarly, the expression of defensive genes such as genes encoding killer toxin and transporters was upregulated by F. graminearum.
3.2 Antibiosis and role of secondary metabolites of Trichoderma
Study of the mycoparasitic activity of T. lignorum on R. solani (Weindling, 1932) via coiling of hyphae, coagulation of protoplasts, and loss of vacuolated structures led to the discovery of a lethal principle with the ability to suppress R. solani in pot cultures (Weindling and Emerson, 1936; Weindling and Fawcett, 1936). The lethal principle was later identified as gliotoxin—a highly antimicrobial secondary metabolite of Trichoderma (Weindling, 1934). Trichoderma secretes a wide range of chemically divergent secondary metabolites with broad-spectrum antimicrobial activity in the vicinity of their niche, which in turn inhibit the growth or spore germination of mycopathogens, a process known as antibiosis (Keswani et al., 2014). Owing to their biochemical nature, the secondary metabolites of Trichoderma perform antibiosis by acting as the metabolic inhibitor of translational pathways, thereby blocking protein synthesis and promoting mycoparasitism by helping in the penetration of the hyphae of prey fungus, as well as inhibiting cell wall synthesis, growth, reproduction, sporulation, nutrient uptake, and metabolite production by target pathogens. Moreover, the antibiosis mediated by the secondary metabolites of Trichoderma is greatly influenced by the species and strain of the fungal agent (Khan et al., 2020). A pot assay targeting R. solani and P. debaryanum in cucumber and pea revealed that the suppression of targeted pathogens was attributed to the antibiosis mechanism of T. virens (Allen and Haenseler, 1935). Diverse studies conducted since then have revealed considerable antifungal/antimicrobial activities exhibited by several secondary metabolites of Trichoderma against a wide range of phytopathogens (Table 1). Viridiol is another antifungal compound released by Trichoderma spp. that also inhibits the enzyme activity of 5′-hydroxyaverantin dehydrogenase, which is necessary for aflatoxin biosynthesis in Aspergillus flavus and A. parasiticus, and, therefore, affects aflatoxin biosynthesis (Sakuno et al., 2000; Wipf and Kerekes, 2003). In addition to antibiosis, these metabolites could also play a significant role in competition, mycoparasitism, and stimulating the plant’s immune system (Zeilinger et al., 2016). It is, therefore, very difficult to study mycoparasitism in isolation. A comparative assessment of sclerotial parasitism, hyphal parasitism, and antibiosis exhibited by T. virens (P strain), led Mukherjee et al. (1995), concluded that sclerotial parasitism is the major mechanism used in controlling S. rolfsii and R. solani in soil (Dutta et al., 2013; Dutta, 2018). In a mutant of T. virens (developed using gamma ray-induced mutagenesis) with upregulated genes for plant interaction, the production of secondary metabolites was found to provide excellent protection against collar rot in lentil and chickpea in both greenhouse and on-farm trials (Mukherjee et al., 2019). A UV-induced mutant of T. virens deficient in genes for mycoparasitism was observed to be an equally efficient biocontrol agent against R. solani as its parental type, and gliotoxin-deficient mutants exhibited a similar result (Howell, 1987; Howell, 2003); however, sclerotial parasitism remained unexamined by these experiments. Therefore, this case raised the question of the mechanism used by Trichoderma for efficient control of phytopathogens, and the probable role of induced systemic resistance in host plant was emphasized. Tu (1980) observed that the sclerotia of Sclerotinia sclerotiorum were readily parasitized by T. virens, with extensive hyphal growth of the mycoparasite inside the colonized sclerotia; however, they could not find any conidia inside. A study conducted by Liu et al. (2009) revealed that, during symbiotic colonization of plant roots by Trichoderma, Trichoderma secretes a higher number of secondary metabolites, such as pachybasin and chrysophanol, with a lower degree of oxidation and less antimicrobial activity. Interestingly, when the host plant is encountered by any phytopathogens, the reactive oxygen species (ROS) released by the plant convert these weakly antimicrobial metabolites to highly antimicrobial oxidized secondary metabolites, viz., 1,5-dihydroxy-3-hydroxymethyl-9,10-anthraquinone, 1,7-dihydroxy-3-hydroxymethyl-9,10-anthraquinone, and emodine. They perform dual beneficial roles by both acting as a powerful antifungal agent that helps in promoting the competitive efficiency of Trichoderma and escalating the immune response of the host plant to other phytopathogens. This finding indicates that, in a tripartite interaction of plant–Trichoderma–pathogen, the plant has been previously well equipped with a reservoir of anthraquinone secondary metabolites of Trichoderma that engage in antagonistic activity only when the plant encounters a pathogenic invasion. A study of the genomics of T. virens revealed that NRPS Tex2 (non-ribosomal peptide synthetase-encoding gene Tex2) is responsible for the assemblage of 11- and 14-module peptaibols (Mukherjee et al., 2011), which elicit strong antimicrobial effects. Trichokonin VI, a peptaibol isolated from T. pseudokoningii, is reported to cause programmed cell death in F. oxysporum via formation of voltage-gated channels in the pathogen membrane. Similarly, SMF2-derived trichokonin VI in T. pseudokoningii was reported to exhibit antimicrobial activity against wide range of fungal phytopathogens by stimulating wide-ranging apoptotic programmed cell death (PCD) (Tijerino et al., 2011; Shi et al., 2012; Sood et al., 2020). Gliotoxin and gliovirin are the polyketides synthesized by the P and Q group strains of Trichoderma and have a significant role in managing deadly soil-borne phytopathogens. Interestingly, the T. virens P group strain is highly antagonistic to P. ultimum, but not to R. solani. Similarly, the Q group strain adversely affects R. solani (Howell et al., 2000). Further research into the genomic perspective of the secondary metabolism of Trichoderma revealed that the T. virens gene veA ortholog vel1 encoded the VELVET protein, which is responsible for regulation of both the biosynthesis and biocontrol activity of gliotoxin, and also adjusts the expression of other genes involved in the secondary metabolism (Mukherjee et al., 2012).
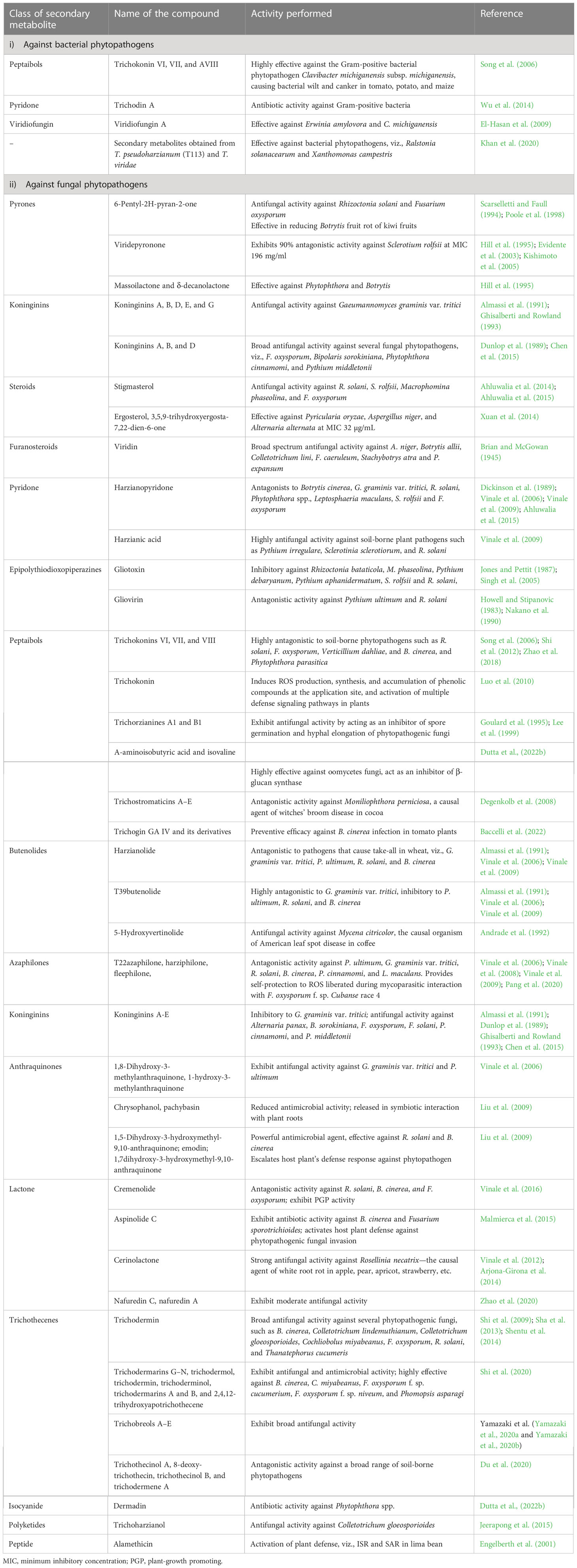
Table 1 Diverse secondary metabolites secreted by Trichoderma spp. and their functions in plant disease management.
3.3 Competitive exclusion of phytopathogens by Trichoderma
Trichoderma is known as an aggressive colonizer of plant roots that competes for space, nutrients, water, or oxygen by mobilizing immobile soil nutrients, thereby eliminating other micro-organisms that inhabit their niche (Elad et al., 2000; Dutta et al., 2022b due to the diversified composition of root exudates secreted by plants. Competition among micro-organisms is a strategy to utilize the nutrient hotspots present in the rhizosphere by eliminating other competitors (Guzmán-Guzmán et al., 2023). Therefore, to be an effective colonizer of plant roots, those organisms must have metabolic versatility and the competitive capacity to occupy the nutrient hotspots. In this regard, Trichoderma can be considered as an aggressive competitor because it has the capacity to secrete a plethora of chemically diverse secondary metabolites that have an antagonistic effect on other micro-organisms (i.e., competitive capacities) and it also exhibits rapid growth and colonization strategies (indicating metabolic versatility) that enable it to occupy space in rhizosphere, enhance plant growth, and restrict further growth of potentially pathogenic micro-organisms (Saravanakumar et al., 2017). The presence of ATP-binding cassettes transporters (ABC transporters) in Trichoderma ensures enhanced competitive ability by conferring resistance to toxic metabolites secreted by other micro-organisms (Harman et al., 2004). Moreover, Trichoderma is compatible with sublethal doses of chemical fertilizers such as urea and muriate of potash, and many chemical pesticides such as thiamethoxam, methomyl, imidacloprid, and methyl bromide, which is attributed to the presence of ABC transporters in Trichoderma (Chet et al., 1997; Gajera et al., 2013; Dutta et al., 2017). Trichoderma releases certain iron chelators, i.e., siderophores, which become bound to iron present in soil. Iron is a key micronutrient for the viability of fungi, and therefore the release of iron-chelating siderophores by Trichoderma is detrimental to the growth of other fungi. This is one of the main reasons for the biocontrol potential of Trichoderma against soil-borne phytopathogens such as Pythium, Fusarium, and Botrytis, which is inversely proportional to the concentration of nutrients in soil (Tjamos et al., 1992). The discovery of Gtt1 (high-affinity glucose transporter) in T. harzianum CECT 2413 raised questions about the probable role of glucose transporters during competition by Trichoderma. Delgado-Jarana et al. (2003) observed that Gtt gene expression is upregulated when T. harzianum CECT 2413 is subjected to growth in nutrient-deficient media. Moreover, a mutant of Trichoderma with an additional copy of the glucose transporter gene performed strongly, with a two- to threefold increase in glucose uptake. Glucose metabolism is essential in the assimilation of enzymes and permeases, as well as proteins involved in membrane and cell wall modifications (Delgado-Jarana et al., 2003).
3.4 Impact of Trichoderma colonization on plant defense and growth promotion
The sessile lifestyle of plants depends on their ability to adapt to the challenges presented by the outside environment in terms of pathogen attack, nutrient starvation, and exposure to toxins and contaminants which are detrimental to its growth. Owing to the abiotic and biotic stresses faced by plants, growth–defense trade-offs take place, which prioritize the acquisition and use of resources (Hacquard et al., 2016). The production of ROS under such conditions determines important developmental processes and cross-kingdom relationships (Segal and Wilson, 2018).
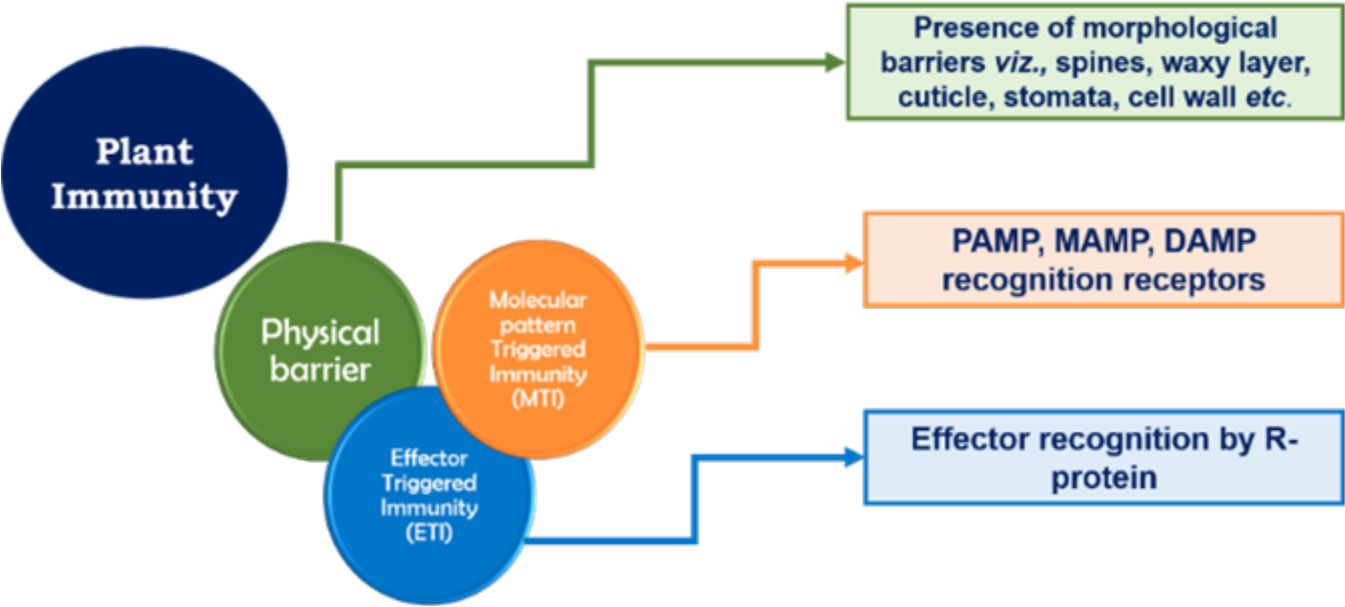
Plant immunity consists of a robust three-layered protection (Figure 3). The first layer of defense safeguards them from foreign invasion and takes the form of physical barriers such as wax, a cuticle layer, stomata, and the cell wall (Lawry, 2016). The second and third layers are based on molecular pattern recognition. All micro-organisms, irrespective of whether they are pathogenic or beneficial, possess unique molecular patterns known as microbe-associated molecular patterns (MAMPs). The unique molecular patterns present in pathogenic microbes are known as pathogen-associated molecular patterns (PAMPs). The second layer of plant immunity consists of different enzymes and pattern recognition receptors (PRRs) that recognize the MAMPs/PAMPs, which leads to the activation of active immune responses. Another possible element of this layer of plant immunity is damage-associated molecular pattern (DAMP) recognition. DAMP recognition receptors help the plant in recognizing any damage caused to the plant system due to invasion by a micro-organism. Therefore, the innate or basal plant immunity comprises three components, viz., MAMP, PAMP, and DAMP recognition receptors, and together this layer of plant immunity is known as molecular pattern-triggered plant immunity (MTI) (Jones and Dangl, 2006; Benedetti et al., 2015). The third layer is based on effector recognition and is known as effector-triggered immunity (ETI). Effectors (previously known as avirulence or Avr proteins) are the molecules released by pathogens/micro-organisms to escape the MTI of plants. Effectors released by micro-organisms help them to counter MAMP-triggered immunity by, for example, scavenging MAMPs, degrading proteases released by plant, and/or deregulating the primary and secondary signaling pathways of plant host. Containing a nucleotide-binding site and leucine-rich repeats (NBS-LRR), the resistance protein (R protein) present in the plant responds to the effectors and triggers a systemic resistance response (i.e., SAR) due to the accumulation of salicylic acid. However, the effector-triggered interaction is always under a tremendous selection pressure that would enable the pathogen/micro-organism to overcome plant immunity and the plant host to retain its immunity (Lawry, 2016). These two layers of plant immunity (viz., MTI and ETI) greatly influence the plant’s response to invading microbes and trigger strong systemic resistance reactions in the plant. Therefore, in order to successfully enter the plant roots and colonize them, Trichoderma need to breach these layers of plant immunity by establishing molecular dialogs with the host plant.
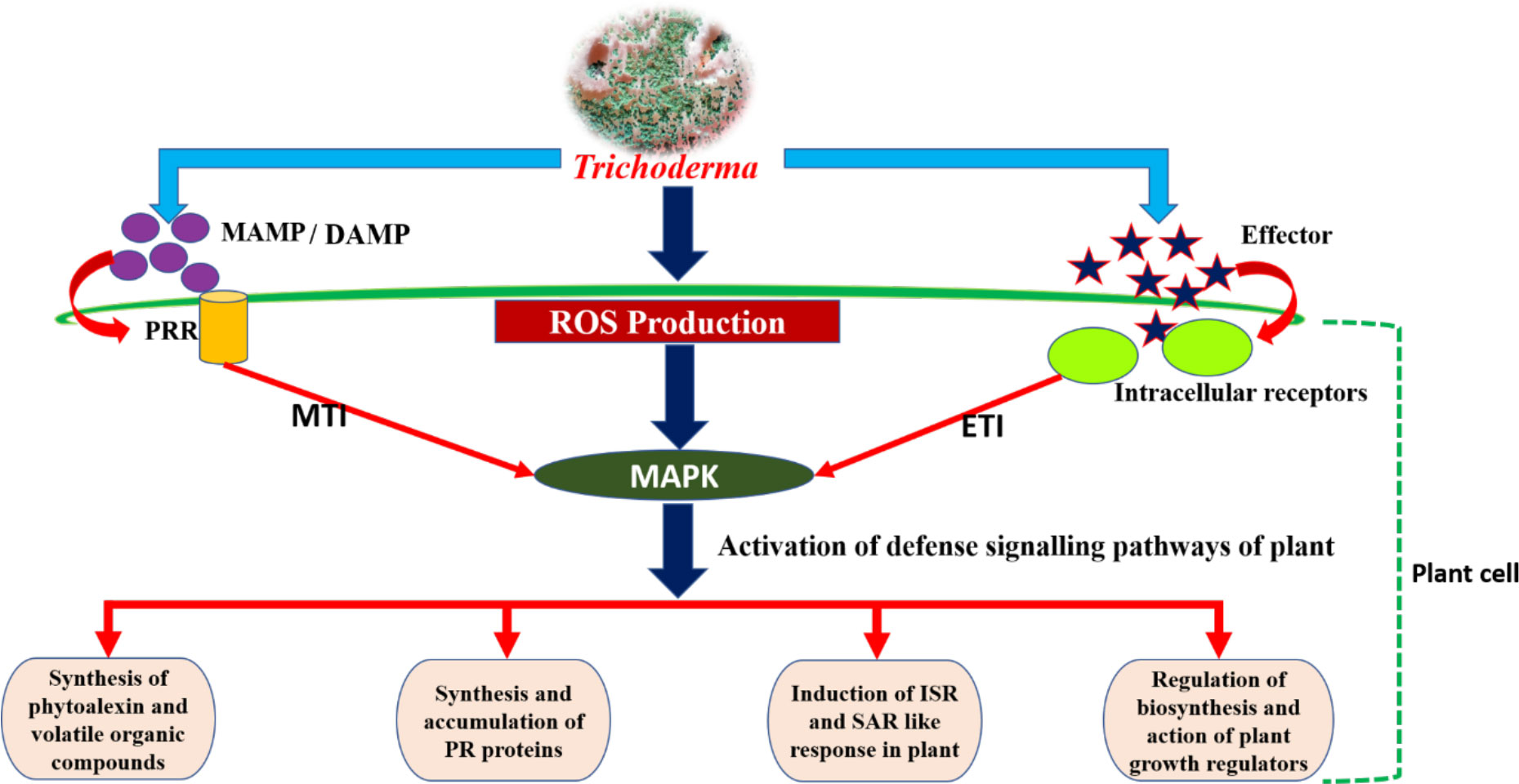
The soil-inhabiting fungi Trichoderma are mainly found to be root colonizers. They can establish themselves in the plant as an endophyte after a process of molecular crosstalk that brings a plethora of positive changes for the host. The sucrose-rich plant root exudates act as an attractant for Trichoderma, causing root colonization by Trichoderma, which activates the plant defense responses and enhances leaf photosynthesis (Vargas et al., 2009). The colonization of plant roots involves Trichoderma’s ability to recognize and adhere to roots, penetrate them, and withstand the toxic metabolites produced by the plant in response to invasion. Activation of plant defense takes place through MTI and ETI, which leads to the production of ROS such as H2O2, , and hydroxyl radical. The ROS further act as a signaling molecule in signal transduction via mitogen-activated protein kinase (MAPK), thereby stimulating different pathways of plant defense, such as activation of the phenylalanine ammonia lyase (PAL) enzyme, which is essential in phytoalexin production and synthesis and the accumulation of pathogenesis-related (PR) proteins, and activates the host’s defense responses (Mendoza-Mendoza et al., 2017; Figure 4). NADPH oxidase (Nox) is the key enzyme that regulates the production of ROS. Studies have revealed that Nox proteins, particularly NoxR and Nox1, greatly influence the molecular dialog between plant roots and Trichoderma during their interaction (Villalobos-Escobedo et al., 2020). The defense responses exhibited by plants to any microbial invasion are energy consuming, and are expressed at the cost of the plants’ own growth and development. Therefore, Trichoderma elicits plant growth and development alongside the induction of a strong immune response in plants (Hermosa et al., 2013). In this context, the Nox protein plays a significant role. In a study conducted with a Trichoderma atroviride mutant expressing the NoxR protein, co-culture with Arabidopsis produced a decrease in feeder root proliferation and phytostimulation when compared with the wild-type strain. However, this also caused an exacerbated response of jasmonic acid-mediated systemic resistance response in the plant when compared with treatment with wild-type T. atroviride (Villalobos-Escobedo et al., 2020). Reduction in lateral growth and development in plants when cultured with a Trichoderma NoxR mutant may be due to the overactivation of jasmonic acid-mediated responses, leading to a shortage of carbon/other energy resources in the plant that are required for the development of lateral root primordia (Guo et al., 2018).
Invasion and colonization by Trichoderma lead to the synthesis and accumulation of different phytohormones, viz., salicylic acid (SA), jasmonic acid (JA), and ethylene (ET). Due to their ability to modulate plant immune responses, these phytohormones are known as the central players of plant defense. The timing, composition, and quantity of the phytohormonal blend produced by plants in response to microbial invasion is greatly influenced by the strain, time, and/or inoculum concentration of the microbe (Pieterse et al., 2009). A study conducted with cucumber and Trichoderma in a hydroponic system by Segarra et al. (2007) revealed that, 4 h post inoculation of cucumber roots with Trichoderma, the plants exhibited a SAR-like response via up-regulated activity of peroxidase and SA. Furthermore, application of a higher inoculum density of Trichoderma induced a systemic increase in SA and JA levels within the plant system. This may be due to the expression of ETI causing oxidative bursts in the plant cells, leading to a hypersensitive response in the plant system that ultimately results in the activation of SAR via an SA-mediated pathway. These defense responses of plants due to the activation of SA-dependent pathways can be overcome by Trichoderma by increasing the JA/ET and auxin responses in plant, which can act as antagonists to SA (Hermosa et al., 2012). When Trichoderma was inoculated into the roots of the model plant Arabidopsis by Morán-Diez et al. (2012), a decrease in plant defense mediated by SA and JA was observed after 24 h; however, over a longer period, the level of plant defense increased both locally and systematically. They suggested that the lower level of plant defense during the initial 24 h of Trichoderma inoculation could be because the plant did not consider the Trichoderma as hostile at that point in time. Over time, Trichoderma colonizes the epidermal and cortical cells of roots, which plants perceive as a threat, and subsequently the plants try to limit Trichoderma from entering into their vascular system by activating plant defenses both locally and systematically via upregulation of genes mediating ISR and SAR. Therefore, it can be said that the SA-mediated response of plants is essential in limiting the root colonization by Trichoderma to the first two layers of root cortical cells, preventing further invasion into the vascular system (Segarra et al., 2007). Moreover, the ability of Trichoderma strains to withstand the highly oxidizing, toxic environment created within the plant system is also a determinant of the degree to which they are effective in colonizing the plant roots (Chen et al., 2011).
The phytohormonal blend produced by plants in response to root colonization by Trichoderma also plays an important role in determining plant growth and development. A balanced trade-off between growth and defense in plants upon colonization by Trichoderma can be explained in terms of cross-communication among phytohormones, viz., ET, SA, and JA (the central players of defense); abscisic acid (ABA), which is related to abiotic stress and plant growth; indole acetic acid (IAA), which is commonly associated with plant growth and lateral root growth of plants; and gibberellins, which modulate plant growth and defense responses via degradation of the DELLA protein (D, aspartic acid; E, glutamic acid; L, leucine; L, leucine; A, alanine) (Hermosa et al., 2012). During Trichoderma–plant interactions, the 1-aminocyclopropane-1-carboxylate deaminase (ACCD) activity of Trichoderma reduces ET production by lowering the availability of the substrate 1-aminocyclopropane 1-carboxylic acid (ACC), which is necessary for ET biosynthesis. As a result, ABA biosynthesis decreases and the activation of gibberellin signaling takes place via degradation of the DELLA protein, which results in an increase in PGP activities. Moreover, JA- and SA-mediated defense responses in plants are also modulated by gibberellin through regulation of DELLA protein degradation. Furthermore, IAA and ET can reciprocally regulate biosynthesis of each other (Stepanova et al., 2007) and, according to this finding, ABA biosynthesis is regulated by exogenous auxin-stimulated ET biosynthesis via ACC synthase (Hermosa et al., 2012). A decrease in ABA biosynthesis is inversely proportional to stomatal conductance, thereby ensuring a higher rate of photosynthesis, and vice versa. Tucci et al. (2011) observed an increase in PGP activity in tomato plants subjected to treatments containing T. atroviride and T. harzianum (Dutta and Das, 1999; Dutta and Das, 2002). The probable reasons for a reduction in ET production were suggested to be either a decrease in the precursor ACC through microbial degradation of IAA in the rhizosphere or the presence of ACCD activity in Trichoderma (Tucci et al., 2011). T. asperellum mutants with RNA interference (RNAi) silencing of the ACCD gene showed an inability to promote root elongation in treated canola seedlings, suggesting the important role of ACCD in root elongation and development (Viterbo et al., 2010; Kubicek et al., 2011). Exogenous production of IAA by Trichoderma stimulates ET biosynthesis through ACC synthase (Figure 5). Liu et al. (2021) conducted an experiment to identify the growth-promoting effect of the T. guizhouense NJAU 4742 strain on cucumber seedlings in a hydroponic study. They observed a significant increase in plant biomass and the modification of lateral root architecture, with a 64.7% increase in lateral root tips of treated plants compared with control. Further study on in situ biosynthesis of auxin by T. guizhouense during interaction with cucumber roots revealed a gradual increase of auxin in the growing media, which was 1.15 and 0.5 times more than the control and IAA-containing treatments (external source) at 30 days post inoculation. These findings indicate that, after interaction with host roots, the exogenous production of IAA by Trichoderma increased considerably, which could be the underlying reason behind plant growth promotion. Similarly, Dutta et al. (2021) observed that groundnut plants treated with a T. harzianum-based bioformulation, made from a native isolate of Meghalaya, were not only protected from tikka disease but also exhibited enhanced plant growth parameters along with increased lateral root growth and root nodulation. Thus, the role of the phytohormonal blend resulting from Trichoderma colonization in determining plant growth promotion and immune response cannot be denied. Therefore, it can be said that plant root colonization by Trichoderma and the existence of Trichoderma within the plant as an endophyte (Zheng et al., 2021), which stimulate the plant’s immunity responses, constitute a complex yet profitable relationship that enables the plant to withstand subsequent biotic and abiotic stresses.
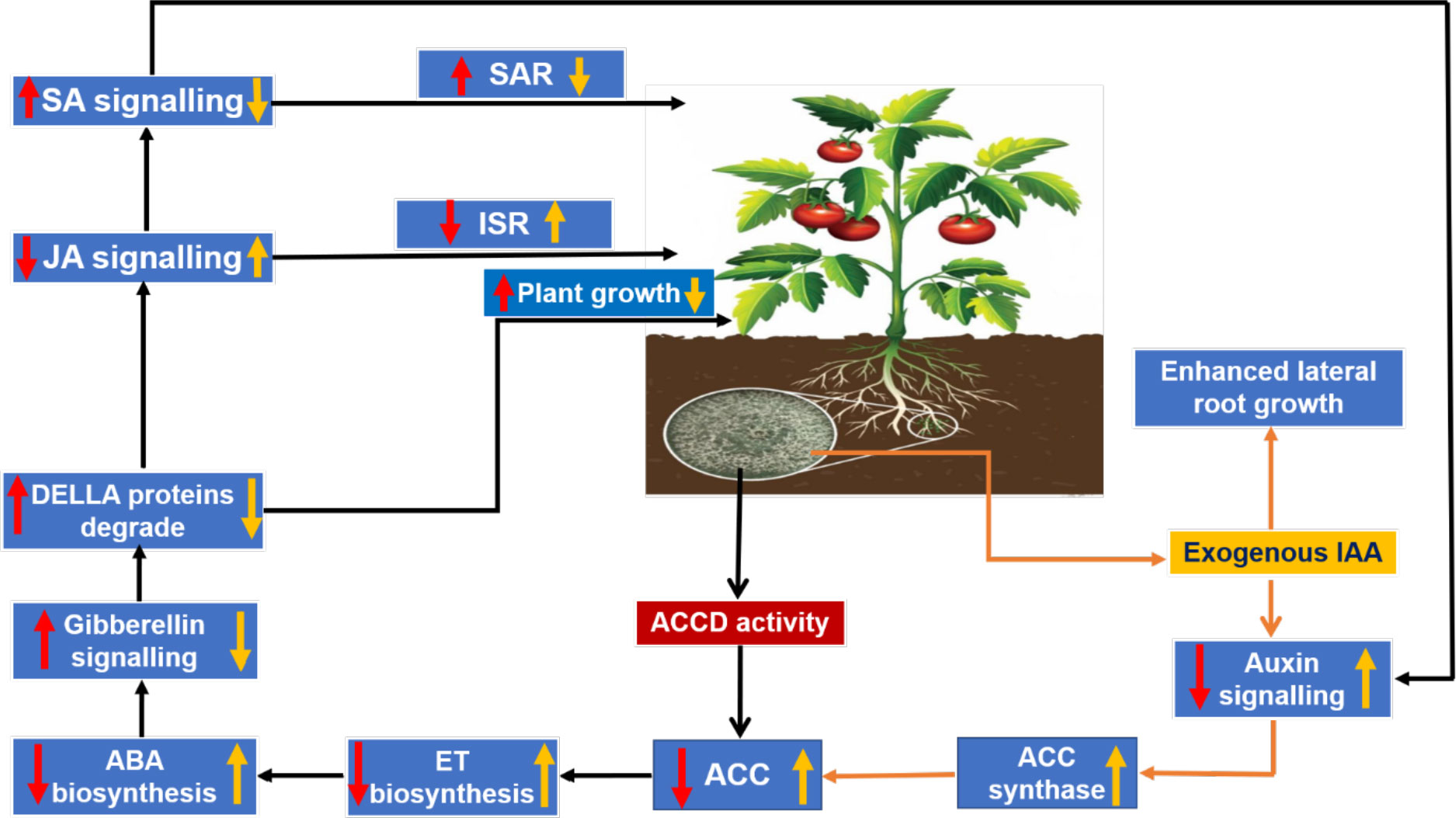
Figure 5 Trichoderma–plant cross-communication model via regulation of the phytohormonal blend: red arrows indicate the effects on the plant due to 1-aminocyclopropane-1-carboxylate deaminase (ACCD) activity in Trichoderma; and yellow arrows indicate the modulation in phytohormonal concentration due to exogenous production of indole acetic acid (IAA) by Trichoderma.
4 Soluble secretome of Trichoderma: role in host–plant interaction and biological control of plant diseases
The emergence of the era of molecular science in the 1940s and 1950s, and its subsequent progress, with the development of different biotechnology tools, made it possible for scientists to isolate, study, and determine the chemical composition of individual genes present in any organism, and ultimately paved the way for whole-genome sequencing. The ability to map and study genes present in a genome made it easier for scientists to understand how genes are assembled in a genome and how they perform their function. In this context, the development of evolutionary trees was also fine-tuned by the detailed knowledge obtained from the understanding of genomics (Merrill and Mazza, 2006). The comprehensive study or global assessment of a set of molecules is referred to as “omics”. Next-generation sequencing of genetic materials and the development of other high-throughput technologies has led to availability of omics data worldwide. The first omics to appear was genomics, which deals with the study of the whole genome of an organism (Hasin et al., 2017). Different omics, viz., genomics, transcriptomics, proteomics, and metabolomics, contribute to the wealth of omics data available publicly across the globe (Figure 6). Trichoderma reesei was the first species of Trichoderma to have its whole genome sequenced (Martinez et al., 2008). Subsequently, the complete genome sequencing of many species of Trichoderma was carried out, and the genomic information is available from the NCBI (National Center for Biotechnology Information) GenBank database (Table 2).

Figure 6 Omics strategies for a better understanding of molecular dialogues in Trichoderma–plant/phytopathogen interaction.
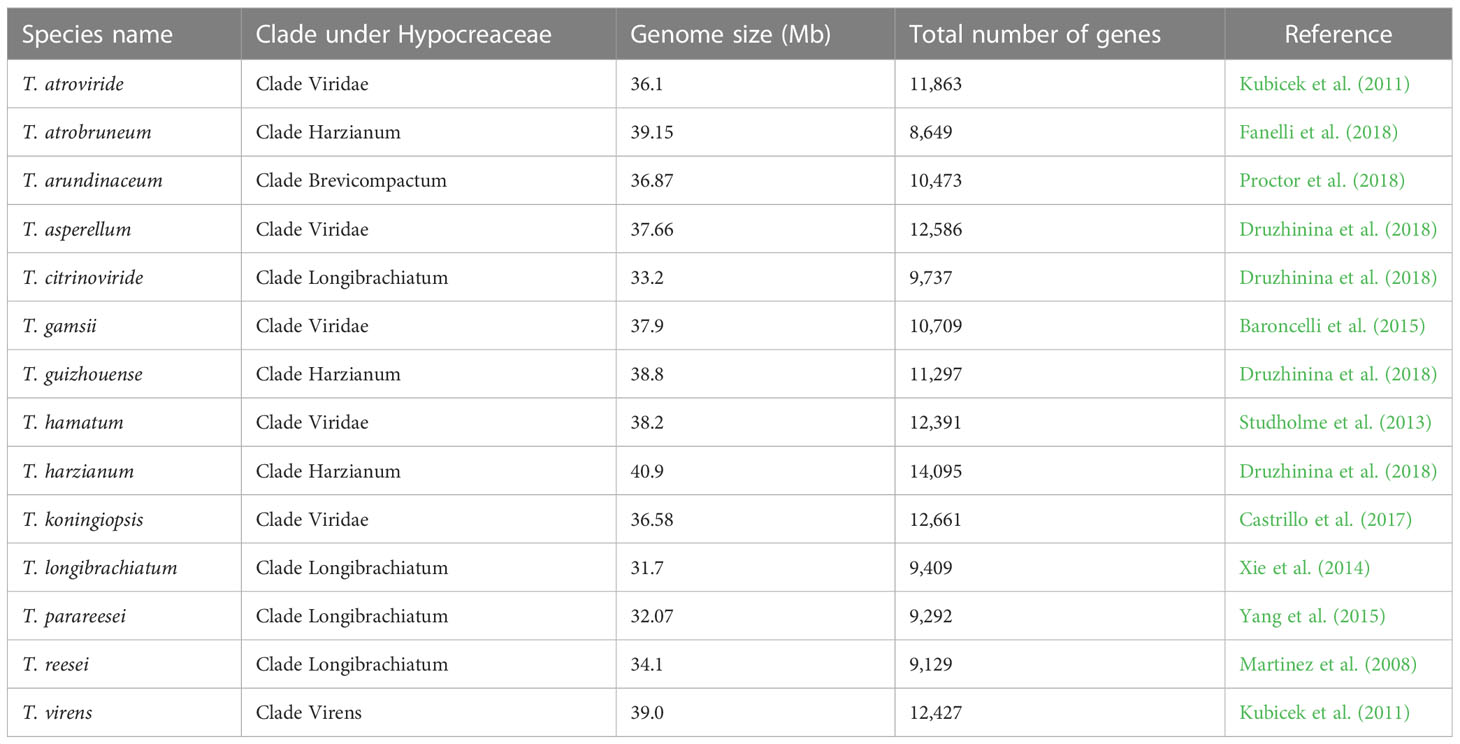
Table 2 Genomic information for different Trichoderma species available from the NCBI GenBank database.
Sanger expressed sequence tag (EST) projects have made transcriptomics studies of Trichoderma–plant interactions easier (Steindorff et al., 2012; Silva et al., 2019). In addition, a transcriptomic study of Trichoderma genes present in the fungal cell wall can be obtained using high-throughput techniques such as suppression subtractive hybridization (SSH) (Vieira et al., 2013). Furthermore, a combined omics strategy can be adopted for an in-depth study of Trichoderma–plant/pathogen interactions (Figure 6).
5 Omics in determining the adaptation behavior of Trichoderma to host root colonization
Trichoderma, being a beneficial microbe, has attracted the attention of researchers, who have studied how it adapts to plant root colonization. The upregulation of genes responsible for the formation of infection structures was observed in an early transcriptomic study of Trichoderma colonizing tomato roots in a hydroponic system (Samolski et al., 2009). Similarly, Lawry (2016) observed the presence of an appressorium-like structure in a Trichoderma virens–maize hydroponic system that helped the biocontrol agent penetrate the plant cell wall and form an intercellular infection peg. The infection pegs possess structures similar to a haustorium, thereby indicating intracellular colonization of maize roots by T. virens. Moreover, forced penetration, by pushing away the outer cell layer of the epidermis, was observed in maize grown in T. virens-inoculated soil, and may be considered as another root colonization mechanism of Trichoderma. Rubio et al. (2014) reported a decrease in nutrient and carbohydrate metabolism activity in plants in response to hyphal attachment of Trichoderma. After 20 h of interaction between Trichoderma and tomato grown in a hydroponic system, Rubio et al. (2012) observed the differential regulation of genes in Trichoderma as a response to the host’s fluctuating behavior. It revealed an upregulation of Trichoderma genes involved in carbohydrate metabolism, nutrient exchange with the plant, the generation of building blocks, and cell wall synthesis, and a downregulation of genes that indicate a sufficient availability of nitrogen. Furthermore, the upregulation or downregulation of genes in Trichoderma during root colonization of either tomato or maize plants is greatly regulated by the host itself. In a T. virens–maize co-culture system, the genes that are mostly upregulated belong to classes such as the glycosyl hydrolases (GHs), oxidoreductases, and small secreted proteins, and the symbiosis-related invertase TvInv (Vargas et al., 2009). The gene most preferentially expressed in T. virens during its interaction with tomato was revealed to be a secreted quino-protein, glucose dehydrogenase. Several studies on the secretome of different species of Trichoderma co-cultured with plants have reported an increased expression of genes encoding glycosidases and peptidases during the initial phase of their interaction (Lamdan et al., 2015; González-López et al., 2021). Reduction in tomato root colonization by a T. harzianum mutant with a silenced thpg1 gene encoding endopolygalactouronase indicates the probable role of fungal glycosidases and peptidases in plant–Trichoderma interactions. Therefore, it can be said that the lytic enzymes released by Trichoderma during the initial phase of their interaction with plants is essential for disintegrating cell wall components to aid in the successful colonization of plant roots (González-López et al., 2021). Furthermore, during the first 24 h of interaction, the genes encoding a group of antioxidant enzymes were found to be exclusively expressed, which is necessary for ROS detoxification, and therefore demonstrates Trichoderma’s strategy of adaptation to the highly reactive environment of plants (González-López et al., 2021). However, there is still a need to perform more function-oriented experiments to obtain a clear understanding of the biochemical significance of host specificity at a transcriptome level. Overall, it can be summarized that the host and Trichoderma appear to coordinate their counterattacks, and Trichoderma has several genes for secreted proteins that apparently play a key role in determining its ability to survive in the complex rhizosphere ecosystem.
Proteomic study of the secretome of Trichoderma revealed that 3.5%–6.0% of total proteins are released via the type II secretion system into the apoplastic space of the plant cell, which is also known as the soluble secretome of Trichoderma. Gene ontology studies of the soluble secreted proteins of Trichoderma have revealed the range of different functions exhibited by these proteins, which are primarily CWDEs, cell wall adherence proteins (such as adhesins, hydrophobins, and tandem repeats), effector-like proteins, proteins determining host surface attachment and recognition, and proteins involved in secondary metabolisms (Mendoza-Mendoza et al., 2017). An effort is made here to briefly discuss the literature available on the soluble secretome of Trichoderma with regard to plant root colonization and its role in the biological control of plant diseases.
6 Cell wall-degrading enzymes
In every plant–microbe interaction, the host cell wall is at the forefront and so is the basal defense. Therefore, it is essential that the micro-organisms digest cell walls by releasing CWDEs in order to break through to the host system. Plant CWDEs are considered a major source of communication molecules and belong to CAZymes groups. GHs essentially digest plant cell walls and facilitate fungal entry into the host tissue. A study conducted with different species of Trichoderma revealed that T. harzianum and T. guizhouense secrete a higher number of CAZyme modules than other Trichoderma species. This may be indicative of their larger genome size and/or their particular behavior in the competitive environment of soil and when interacting with plants and pathogens (Li et al., 2013). In soil, Trichoderma releases a lignocellulolytic enzyme that helps it to live a saprophytic life. However, in the presence of host root exudates they respond differently, synthesizing and secreting CWDEs (Cragg et al., 2015). Harman et al. (2004) reported two glucosyltransferases (GTs) from the soluble secretome of mycoparasitic fungi T. virens and T. atroviride.
The soluble secretome of Trichoderma contains a significant amount of CWDEs. They are responsible for alerting the plant immune system to the presence of an invader. As reported by Avni et al. (1994), inactive cellulase and xylanase were the first MAMPs obtained from Trichoderma. Furthermore, CWDEs cause damage to the plant cell wall, thereby generating DAMP signals. A study of T. harzianum–root interaction in Arabidopsis and tomato led to the discovery of the first DAMP, which corresponds to the oligogalactouronides produced by the enzymatic activity of CWDE endopolygalacturonase ThPG1 in T. harzianum, which was found to be capable of inducing systemic defense in the plants (Moran-Diez et al., 2009). In a study conducted by Baroncelli et al. (2016), the expression of two endo-polygalcturonase genes (viz., TvPg1 and TvPg2) from T. virens I10 was examined during the interaction with tomato roots. The results revealed that, while interacting with, in particular, host roots or pectin, expression of TvPg1 was induced, whereas TvPg2 was later expressed constitutively. According to Sarrocco et al. (2017), this constitutively produced endopolygalacturonase was responsible for eliciting ISR in the host. Similarly, the direct activity of plant chitinases or the mycotrophic nature of Trichoderma against rhizospheric fungi yields chito-oligosaccharides, which can also function as DAMPs in the activation of systemic plant immunity (Woo et al., 2006). Apart from inducing plant immunity by generating MAMP and DAMP signals, CWDEs also play an important role in determining the efficient colonization of roots by Trichoderma. They achieve this by either increasing the plasticity of the host cell wall or causing irreversible deterioration of the cell wall structure.
As the fungal cell wall comprises mainly chitin, glucan, and proteins, mycoparasitism by Trichoderma involves the extensive use of CWDEs, viz., chitinases, glucanases, and proteases (Table 3). Use of a Trichoderma microarray to study the transcriptomic changes of genes in T. atroviride overgrown on a Verticillium dahliae colony revealed that there was total 143 differentially regulated genes (almost 98%) that belonged to the T. atroviride genome. The upregulated genes were all from classes of CAZymes and proteases, viz., serine, aspartic acid, and metallopeptidases genes that are crucial in weakening and disintegrating the fungal host cell wall (Morán-Diez et al., 2019). Therefore, it can be said that the differentially regulated genes of T. atroviride are unequivocally associated with the mycoparasitic and antagonistic activity against the targeted pathogen.
7 Enzymes for chitin degradation
It has been reported that the Trichoderma genome harbors a greater number of genes encoding chitinolytic enzymes, which is attributed to Trichoderma’s mycoparasitic nature. Fungal chitinases belong to the GH18 and GH20 families. GH18 chitinases can be further categorized into subfamilies A, B, and C. It has been observed that genes encoding chitinases from GH18 are significantly expanded in T. atroviride, T. asperellum, T. atrobrunneum, T. gamsii, T. harzianum, and T. virens (Kubicek et al., 2011). Chitin and chitosan (a partial or complete deacetylated derivative of chitin) comprise the chitinous layer of fungal cell wall. Chitosan in mycoparasitic fungi such as Trichoderma plays an important role in the scavenging of ROS produced by parasitized fungi. Kappel et al. (2020) observed that out of six genes encoding chitin deacetylase, the deletion of genes cda1 or cda5 in T. atroviridae led to severely impaired mycoparasitic ability. This result indicates that a decrease in or absence of chitin deacetylase enzymes results in a low level of chitosan in Trichoderma, and, therefore, the Trichoderma is not protected from ROS (Mukherjee et al., 2022). Further study led to the discovery of cell wall remodeling in T. atroviridae during mycoparasitic interaction via the upregulation of all six genes encoding chitosanase, especially toward the later stage of interaction. One interesting finding made during this study by Kappel et al. (2020) was CHS8, which is called as a hybrid synthase due to its similarity to both chitin synthases and hyaluronan synthases, and can utilize both UDP-N-acetylglucosamine and UDP-d-glucuronate as substrates. The authors, therefore, speculated that CHS8, along with CDA1, forms a chitin glycol-polymer layer that protects the Trichoderma cell wall during mycoparasitic interactions (Kappel et al., 2020).
8 Glucan degradation
The enzymes α- and β-glucanase are necessary for the deconstruction of the glucan layer. Fungal α-1,3-glucanases are members to the GH71 family. T. harzianum and T. asperellum explicitly secrete the exo-α-1,3-glucanases AGN13.1 and AGN13.2, respectively, in the presence of the Botrytis cinerea cell wall. Enzyme AGN13.1 is found to possess lytic properties against fungal cell walls and exhibit antifungal activity (Mukherjee et al., 2022).
β-1,3-Glucanases are classified into GH families, viz., 16, 17, 55, 64, and 81. The mycoparasitic Trichoderma genomes comprise a large number of genes encoding GH55 and GH64 family members (Kubicek et al., 2011). They play a significant role in the mycoparasitization of oomycete fungi (which have a cell wall composed of cellulose and β-1,3- and β-1,6-glucans). A study of T. virens mutants in which the bgn3 gene, encoding β-1,6-glucanase, is overexpressed found that such mutants exhibited enhanced antagonism toward Globisporangium ultimum, whereas mutants overexpressing the genes for both β-1,3-glucanase and β-1,6-glucanase were found to exhibit enhanced inhibition of G. ultimum (Djonovic et al., 2007).
9 Protein degradation
Proteases are an important group of enzymes released by Trichoderma in the event of mycoparasitism. The differential regulation of several protease genes of Trichoderma is reported during mycoparasitism (Mukherjee et al., 2022). Overexpression of the T. atroviride prb1 gene (encoding protease) reportedly provides increased protection against R. solani (Pozo et al., 2004). Cortes et al. (1998) observed that expression of the prb gene was induced before contact with the fungal host. Further study of gene behavior in nitrogen-limited conditions led to the finding that the promoter region of the prb gene contains a binding site for transcriptional activator of nitrogen catabolite-repressed genes, viz., ARE1 (Olmedo-Monfil et al., 2002; Mukherjee et al., 2022). These findings caused Druzhinina et al. (2011) to hypothesize that, in the early stage of mycoparasitic interaction, the activity of proteolytic enzymes results in host-derived nitrogenous products, which are responsible for the activation of mycoparasitism-relevant genes via their binding to nitrogen sensors present on the Trichoderma cell surface.
10 Small cysteine-rich proteins as effectors of Trichoderma
Effectors released during Trichoderma’s interactions with plants or fungi participate in ROS scavenging, chitinase and glucanase production, fungal cell wall masking, protease inhibition, and the prevention of defense alarm activation in neighboring cells colonized by the invader (Rabe et al., 2013; Lanver et al., 2017). A study conducted by Mendoza-Mendoza et al. (2017) revealed the presence of 70–123 effector proteins in the soluble secretome of Trichoderma. However, not all these effectors possess a clear functional domain. Some of the effector proteins with known functional domains are discussed briefly herein.
11 Common in fungal extracellular membrane domain proteins
Common in fungal extracellular membrane is a protein domain containing eight cysteines, which distinguishes it characteristically from other known cysteine-rich proteins. CFEM domain proteins were first discovered in rice blast pathogen Magnaporthe grisea (Kulkarni et al., 2003) and are known to play important roles in fungal pathogenicity. The functions of CFEM domain proteins include plant-surface sensing, appressorium development, asexual development (Sabnam and Barman, 2017), iron assimilation (Nasser et al., 2016), and redox homoeostasis (Kou et al., 2017). Fifty soluble secreted proteins of Trichoderma have so far been found to contain CFEM domains.
In an experiment on a Trichoderma–maize co-culture conducted in a hydroponic system, a decreased abundance of several CFEM-containing secreted proteins was observed. On developing deletion mutants for two genes encoding CFEM domain proteins with IDs 92810 and 111486 (Joint Genome Institute (JGI) v2.0), Lamdan et al. (2015) observed an increased ISR response against necrotic phytopathogens. They suggested that an increased degradation or sequestering of CFEM domain proteins by host roots could be the reason for their loss of abundance in a Trichoderma–maize co-culture system. However, further study of CFEM domain proteins is needed to reveal their modeof action in Trichoderma–plant or Trichoderma–phytopathogen interactions.
12 LysM-like putative effectors
Chitin, a homopolymer of N-acetyl-d-glucosamine, represents the second most abundant organic matter after cellulose. Chitin is widely distributed in fungi as a major component of the cell wall, but is absent in plants. The presence of chitin in the plant system is recognized by specific lysin motif (LysM)-containing pattern recognition receptors (PRRs) in the plant cell surface, which trigger an innate immune response in plants (Marshall et al., 2011). The absence of these PRRs compromises the plant’s defense against fungal pathogens. Therefore, the plant’s ability to perceive chitin is very important in recognizing phytopathogenic fungi. However, successful plant colonizers have evolved strategies that overcome chitin-induced defense in plants. Alteration of cell wall composition and release of LysM-like effector proteins are some of the strategies adopted by micro-organisms. LysM-like effectors released by plant colonizers bind to the free chitin released in the plant apoplastic space during fungal growth and mask the colonizer’s presence. In this way, they overcome chitin-induced plant defense. Genomic study of mycoparasitic and endophytic Trichoderma has revealed that they contain an increased number of genes encoding LysM-containing secreted and non-secreted proteins as well as chitinases. These proteins help in the penetration and establishment of Trichoderma within the plant system by binding themselves to the fungal chitin and thereby avoiding ligand–PRR binding (Hermosa et al., 2013). Moreover, it has also been suggested that proteins containing a LysM domain may provide a mechanism of self-protection against the Trichoderma’s own chitinases (Gruber and Seidl-Seiboth, 2012).
13 Hydrophobins
Hydrophobins are small, unique, surface-active fungal proteins with the ability to form an amphipathic membrane at the interface of hydrophilic and hydrophobic environments. Their β-structured core is composed of eight highly conserved cysteine residues linked by four disulfide bridges. Hydrophobin proteins have a large exposed hydrophobic area, which explains their high surface activity (Linder et al., 2005; Bayry et al., 2012). Class I hydrophobin molecules form rodlet layers on the fungal cell wall by organizing themselves into a highly insoluble amphipathic membrane at the junction of the hydrophilic fungal cell wall and the hydrophobic environment. Class II hydrophobins form micro-aggregates to give rise to dimers and tetramers in a rodlet-like structure (Mendoza-Mendoza et al., 2017). Kubicek et al. (2008) carried out a comparative evolutionary study on class II hydrophobins produced by the ascomycetes group of fungi and noted that the genus Trichoderma ranked first in number and diversity of class II hydrophobins. Guzman-Guzman et al. (2017) reported that a class II hydrophobin, viz., TVHYDII1 of T. virens, contributes to the antagonistic activity of T. virens against R. solani and promotes Arabidopsis root colonization by Trichoderma. Huang et al. (2015) observed an up-regulation in hydrophobin synthesis and secretion in T. asperellum when placed in a 1% Alternaria alternata cell wall and 5% A. alternata fermentation broth, which is indicative of hydrophobins’ role in mycoparasitism. Microarray analysis of T. virens T87 genes revealed that genes encoding hydrophobins were largely downregulated during Trichoderma–tomato interaction (Rubio et al., 2012) and found to have a negative effect on the growth and development of tomato plants in in vitro conditions. This result may be indicative of limited root attachment of T. virens T87 due to fewer hydrophobins, affecting its interaction with tomato plants. Interestingly, Przylucka et al. (2017) have observed upregulated HFB7 genes of T. virens in interactions with tomato. T. harzianum secretes QID74, which is a hydrophobin-like cell wall protein with a high molecular mass. It is particularly involved in fungal cell wall protection, adherence to the host cell, and modification of the host root architecture by increasing lateral roots growth, which, in turn, ensures increased nutrient uptake. Effective utilization of these nutrients results in increased plant biomass, and promising results have been obtained in cucumber and tomato plants (Samolski et al., 2012).
14 Ceratoplatanin family proteins
Ceratoplatanins (CPs) are non-enzymatic unique fungal proteins similar to plant expansin proteins. Trichoderma CPs bind to chitin (Pazzagli et al., 2014), which may be helpful in opening the physical spaces of the parasitized fungal cell wall. Similarly, during colonization of the host plant, the behavior of CP proteins might be helpful in masking fungal cell wall chitin from detection by the host plant’s receptors (Quarantin et al., 2016). CPs are also known as eliciting plant response-like proteins (EPLs) due to their role in the induction of SAR in plants (Gaderer et al., 2014). According to Gao et al. (2020), the number of EPL-encoding genes in Trichoderma species is either three or four. During T. harzianum–S. sclerotiorum interaction, the role of EPL1 was found to be significant for the expression of genes related to mycoparasitism and coiling around S. sclerotiorum. Moreover, EPL-encoding genes found to downregulate the expression of virulence genes present in B. cinerea necessary for botrydial biosynthesis. Therefore, EPLs have two major functions in Trichoderma–pathogen interaction, viz., the expression of mycoparasitism-related genes and protection of Trichoderma from secondary metabolites produced defensively by pathogens (Mukherjee et al., 2022).
In Trichoderma, the Sm1 gene encodes a small, secreted protein belonging to the CP family. Sm1 gene abundantly expressed throughout fungal development. Specific growth conditions modulate the expression of the Sm1 gene in Trichoderma. Studies conducted by Vargas et al. (2011) concluded that the detection of sucrose-rich plant root exudates and their trafficking activates the expression of Sm1 in T. virens. Djonovic et al. (2006) demonstrated the role of Sm1 in T. virens as a non-enzymatic effector of plant defense. In addition, purified Sm1 protein was observed to trigger ROS production, thereby eliciting local and systemic resistance in the plant. Furthermore, these proteins are found to have no phytotoxic or antimicrobial function. Trichoderma has a wide host range, and therefore its interactions with its hosts have diverse consequences. Salas-Marina et al. (2015) observed that Sm1- and Epl1-deleted mutants of T. virens and T. atroviride led to decreased systemic resistance in treated tomato plants, and overexpression of these genes resulted in enhanced protection against phytopathogens. In a tripartite interaction involving T. virens Gv29-8, maize, and Colletotrichum graminearum, Crutcher et al. (2015) observed an enhanced expression of Sm2 protein by T. virens. The use of mutants developed by deletion of the Sm2-encoding gene revealed that they were able to induce same level of ISR in maize plants; however, the root colonization ability of T. virens was found to decrease significantly. Conversely, a study on the interaction of Sm1-deleted mutants of T. virens, I10 maize, and Cochliobolus heterostrophus revealed a decreased level of ISR, and an Sm1- and Sm2-deleted mutant caused a more severe reduction in the plant’s defense response (Gaderer et al., 2015). The diverse responses obtained from these tripartite interactions could be due to the different lifestyles of the phytopathogens used for the study. The plant’s immune response to biotrophs, hemibiotrophs, and necrotrophs upon infection could determine which Trichoderma elicitors/effectors are deployed. Indeed, the activity of the same Trichoderma-derived effector could be altered by the host plant’s response, thereby explaining some of the distinct actions of Sm1 and its paralogs depending on the three-way interaction.
15 Swollenin
Swollenin is a soluble secreted protein first described in T. reesei. Swollenin and its orthologs are structurally characterized by the fungal carbohydrate-binding domain (CBD) in their N-terminal, which is followed by a region with domains 1 and 2, similar to plant expansin (Saloheimo et al., 2002). The presence of CBD in their N-terminal helps them to bind to the carbohydrate molecules present in the plant cell wall, and facilitates access to and colonization of the plant system. Brotman et al. (2008) observed that TasSwo—a swollenin protein secreted by T. asperellum—recognizes and binds to the cellulose present in the plant cell wall via the CBD and alters the architecture of the plant cell wall in favor of root colonization by Trichoderma. Furthermore, the authors demonstrated that the CBD present in these proteins acts as a MAMP by inducing plant-innate immunity in cucumber to phytopathogens such as Botrytis cinerea and Pseudomonas syringae.
16 Wall stress responsive-component domain protein
Genomic study of Trichoderma has revealed the presence of several proteins with the cell wall stress-responsive component (WSC) domain. Although no direct information is yet available on how these proteins help Trichoderma in their interactions with plants or pathogens, similar proteins encoded in Trichoderma genome are also reported in other plant-beneficial endophytes. For instance, in Piriformospora indica, the WSC domain protein FGB1 performs the function of plant immunity suppressor by altering its cell wall composition and properties, and therefore aids its establishment within the host plant (Wawra et al., 2016). Moreover, as reported by Tong et al. (2016), these proteins may have a role in promoting cellular resistance, cell wall disruption, high osmolarity, the production of metal ions (Mg2+, Zn2+, Fe2+, Ca2+, Mn2+, and K+), and oxidation (Silva et al., 2019). In addition, under stressed conditions, WSC domain proteins, viz., FGB1 and WSC3, may be involved in β-glucan remodeling in the fungal cell wall (Wawra et al., 2019). A comparative secretome analysis of Trichoderma under salt stress conditions revealed that, in the presence of its plant host, the expression of WSC domain proteins in Trichoderma decreases, which may be indicative of the benefits derived by the fungus from its symbiont (i.e., the plant) through intensified root colonization (Rouina et al., 2022).
Other than the above-discussed secreted proteins, there are certain proteins identified in Trichoderma genomes, the function of which are not yet known. The presence of genes for different proteins, e.g., necrosis-inducing polypeptides (NPP1), killer-like toxins, GLEYA adherence proteins, and fungal ribonucleases (RNAses), are reported in different Trichoderma genomes. The expression of some killer-like toxin protein-encoding genes, such as KP4, hinders plant growth (Allen et al., 2008); however, as reported by Allen et al. (2011), the inclusion of this gene in transgenic plants is effective in protecting the plants from phytopathogens. Similarly, across Trichoderma species, the number of secreted GLEYA adhesin protein differs. According to previous studies, T. guizhouense harbors the maximum number of such proteins, i.e., three; T. atroviride, T. gamsii, and T. parareesei secrete two proteins; and T. harzianum, T. virens, and T. reesei contain only one such protein. The presence of necrosis-inducing proteins (NPP1) in both mycoparasitic and saprophytic Trichoderma genomes and their role in plant interaction is still not clear. However, it has been speculated that NPP1 genes and their expression are not always related to necrosis in the host plant, but may also play a role in fungal growth and sporulation (Santhanam et al., 2013). Recently, the use of RNA-interacting proteins and RNA by fungi has been reported in the establishment of successful interactions with the host plant (Spanu, 2015). Among three different families of RNAses present in fungi, viz., non-specific RNases, RNase T1, and RNase T2, Trichoderma genomes are reported to express two RNAse families, i.e., RNase T2 and non-specific RNases. Although the T2 family RNases are known to perform functions such as nutrient acquisition, phosphate solubilization, defense against phytopathogens, self-incompatibility, and senescence (Deshpande and Shankar, 2002); however, their role in establishing interaction with plants is still not known.
17 Conclusion
Trichoderma is widely used across the globe due to its biocontrol and plant growth-promoting abilities. The interactions of Trichoderma spp. with host plants and pathogens at a molecular level will provide insights on the mechanisms that make Trichoderma a superior biocontrol agent. Mycoparasitism by Trichoderma is a complex process; therefore, a comprehensive study at the gene level is important to understand how the BCA safeguards itself from the defense strategies adopted by the parasitized fungi. Moreover, knowledge of secondary metabolites secreted by Trichoderma during their interaction with either the plant host or fungal host may be helpful in formulating effective bioactive molecule-based formulations that can provide enhanced protection to plants for a longer time. An understanding of the molecular dialogues between the host plant/fungus and Trichoderma is important to realizing the full potential of Trichoderma as a biocontrol agent.
Author contributions
MM prepared the original draft, and PD reviewed and edited the article. All authors contributed to the article and approved the submitted version.
Funding
The authors are thankful to the Department of Science and Technology, Government of India, New Delhi, for the DST INSPIRE Junior Research Fellowship (IF-210048) given to MM, and the Department of Biotechnology, Government of India, for providing grants BT/KIS/123/SP45224/2022 and BT/NER/143/SP42744/2021 to PD.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahluwalia, V., Kumar, J., Rana, V. S., Sati, O. P., Walia, S. (2015). Comparative evaluation of two Trichoderma harzianum strains for major secondary metabolite production and antifungal activity. Nat. Prod. Res. 29 (10), 914–920. doi: 10.1080/14786419.2014.958739
Ahluwalia, V., Walia, S., Sati, O. P., Kumar, J., Kundu, A., Shankar, J., et al. (2014). Isolation, characterisation of major secondary metabolites of the Himalayan Trichoderma koningii and their antifungal activity. Arch. Phytopathol. Plant Prot. 47 (9), 1063–1071. doi: 10.1080/03235408.2013.829715
Allen, M. C., Haenseler, C. M. (1935). Antagonistic action of Trichoderma on Rhizoctonia and other soil fungi. Phytopathol 25, 244–252.
Allen, A., Islamovic, E., Kaur, J., Gold, S., Shah, D., Smith, T. J. (2011). Transgenic maize plants expressing the totivirus antifungal protein, KP4, are highly resistant to corn smut. Plant Biotechnol. J. 9, 857–864. doi: 10.1111/j.1467-7652.2011.00590.x
Allen, A., Snyder, A. K., Preuss, M., Nielsen, E. E., Shah, D. M., Smith, T. J. (2008). Plant defensins and virally encoded fungal toxin KP4 inhibit plant root growth. Planta 227, 331–339. doi: 10.1007/s00425-007-0620-1
Almassi, F., Ghisalberti, E. L., Narbey, M. J., Sivasithamparam, K. (1991). New antibiotics from strains of Trichoderma harzianum. J. Nat. Prod. 54 (2), 396–402. doi: 10.1021/np50074a008
Andrade, R., Ayer, W. A., Mebe, P. P. (1992). The metabolites of Trichoderma longibrachiatum. part 1. isolation of the metabolites and the structure of trichodimerol. Can. J. Chem. 70 (10), 2526–2535. doi: 10.1139/v92-320
Arjona-Girona, I., Vinale, F., Ruano-Rosa, D., Lorito, M., López-Herrera, C. J. (2014). Effect of metabolites from different Trichoderma strains on the growth of Rosellinia necatrix, the causal agent of avocado white root rot. Eur. J. Plant Pathol. 140, 385–397. doi: 10.1007/s10658-014-0472-z
Atanasova, L., Crom, S. L., Gruber, S., Coulpier, F., Seidl-Seiboth, V., Kubicek, C. P., et al. (2013). Comparative transcriptomics reveals different strategies of trichoderma mycoparasitism. BMC Genomics 14 (1), 1–15. doi: 10.1186/1471-2164-14-121
Avni, A., Bailey, B. A., Mattoo, A. K., Anderson, J. D. (1994). Induction of ethylene biosynthesis in nicotiana tabacum by a trichoderma viride xylanase is correlated to the accumulation of 1- aminocyclopropane-1-carboxylic acid (ACC) synthase and ACC oxidase transcripts. Plant Physiol. 106, 1049–1055. doi: 10.1104/pp.106.3.1049
Baccelli, I., Luti, S., Bernardi, R., Favaron, F., De Zotti, M., Sella, L. (2022). Water-soluble trichogin GA IV -derived peptaibols protect tomato plants from botrytis cinerea infection with limited impact on plant defenses. Front. Plant Sci. 13.
Bara, M. T. F., Lima, A. L., Ulhoa, C. J. (2003). Purification and characterization of an exo-b-1,3-glucanase produced by trichoderma asperellum. FEMS Microbiol. Lett. 219 (1), 81–85. doi: 10.1016/S0378-1097(02)01191-6
Baroncelli, R., Matarese, F., Moncini, L., Vannacci, G., Vergara, M. (2016). Two endopolygalacturonase genes in trichoderma virens: in silico characterization and expression during interaction with plants. J. Phytopathol. 164, 18–28. doi: 10.1111/jph.12414
Baroncelli, R., Piaggeschi, G., Fiorini, L., Bertolini, E., Zapparata, A., Pe, M. E., et al. (2015). Draft whole-genome sequence of the biocontrol agent Trichoderma harzianum T6776. Genome Announc. 3 (3), e00647–15. doi: 10.1128/genomeA.00647-15
Bayry, J., Aimanianda, V., Guijarro, J. I., Sunde, M., Latge, J. P. (2012). Hydrophobins–unique fungal proteins. PloS Pathog. 8 (5), e1002700. doi: 10.1371/journal.ppat.1002700
Benedetti, M., Pontiggia, D., Raggi, S., Cheng, Z., Scaloni, F., Ferrari, S., et al. (2015). Plant immunity triggered by engineered in vivo release of oligogalacturonides, damage-associated molecular patterns. Proc. Nat. Acad. Sci. 112 (17), 5533–5538. doi: 10.1073/pnas.1504154112
Blauth de Lima, F., Felix, C., Osorio, N., Alves, A., Vitorino, R., Domingues, P., et al. (2017). Trichoderma harzianum T1A constitutively secretes proteins involved in the biological control of Guignardia citricarpa. Biol. Control. 106, 99–109. doi: 10.1016/j.biocontrol.2017.01.003
Brian, P. W., McGowan, J. G. (1945). Viridin: a highly fungistatic substance produced by trichoderma viride. Nat 156 (3953), 144–145. doi: 10.1038/156144a0
Brotman, Y., Briff, E., Viterbo, A., Chet, I. (2008). Role of swollenin, an expansin-like protein from trichoderma, in plant root colonization. Plant Physiol. 147 (2), 779–789. doi: 10.1104/pp.108.116293
Castrillo, M. L., Bich, G. A., Modenutti, C., Turjanski, A., Zapata, P. D., Villalba, L. L. (2017). First whole-genome shotgun sequence of a promising cellulase secretor, trichoderma koningiopsis strain POS7. Genome Announc. 5 (37), e00823–e00817. doi: 10.1128/genomeA.00823-17
Chen, J. L., Liu, K., Miao, C. P., Guan, H. L., Zhao, L. X., Sun, S. Z. (2015). Chemical constituents with siderophores activities from Trichoderma koningiopsis YIM PH30002. Nat. Prod. Res. Dev. 27, 1878–1883.
Chen, L. L., Yang, X., Raza, W., Li, J., Liu, Y., Qiu, M., et al. (2011). Trichoderma harzianum SQR-T037 rapidly degrades allelochemicals in rhizospheres of continuously cropped cucumbers. Appl. Microbiol. Biotechnol. 89, 1653–1663. doi: 10.1007/s00253-010-2948-x
Chet, I., Inbar, J., Hadar, I. (1997). Fungal antagonists and mycoparasites. In: Charnley, A.K., Wicklow, D.T., Soderstrom, B. (Eds.) The mycota IV: environmental and microbial relationships, Springer-Verlag, Berlin, pp.165–184.
Cohen-Kupiec, R., Broglie, K. E., Friesem, D., Broglie, R. M., Chet, I. (1999). Molecular characterization of a novel b-1,3-exoglucanase related to mycoparasitism of trichoderma harzianum. Gene. 226(2): 147–154. doi: 10.1080/15389588.2014.999857
Corke, A. T. K., Hunter, T. (1979). Biocontrol of Nectaria galligena infections of pruning wounds of apple shoots. J. Horticul. Sci. 54, 47. doi: 10.1080/00221589.1979.11514847
Cortes, C., Gutierrez, A., Olmedo, V., Inbar, J., Chet, I., Herrera-Estrella, A. (1998). The expression of genes involved in parasitism by Trichoderma harzianum is triggered by a diffusible factor. Mol. Gen. Genet. 260, 218–225. doi: 10.1007/s004380050889
Cragg, S. M., Beckham, G. T., Bruce, N. C., Bugg, T. D. H., Distel, D. L., Dupree, P., et al. (2015). Lignocellulose degradation mechanisms across the tree of life. Curr. Opin. Chem. Biol. 29, 108–119. doi: 10.1016/j.cbpa.2015.10.018
Crutcher, F. K., Moran-Diez, M. E., Ding, S., Liu, J., Horwitz, B. A., Mukherjee, P. K., et al. (2015). A paralog of the proteinaceous elicitor SM1 is involved in colonization of maize roots by trichoderma virens. Fungal Biol. 119, 476–486. doi: 10.1016/j.funbio.2015.01.004
Das, B. C., Das, B. K., Dutta, P., Sarmah, D. K. (2006). Bioformulation of Trichoderma harzianum rifai for management of soyabean stem-rot caused by Rhizoctonia solani Kuhn. J. Biol. Control. 20 (1), 57–64.
Degenkolb, T., Von Doehren, H., Fog Nielsen, K., Samuels, G. J., Brückner, H. (2008). Recent advances and future prospects in peptaibiotics, hydrophobin, and mycotoxin research, and their importance for chemotaxonomy of trichoderma and hypocrea. Chem. Biodivers. 5 (5), 671–680. doi: 10.1002/cbdv.200890064
Delgado-Jarana, J., Moreno-Mateos, M. A., Benítez, T. (2003). Glucose uptake in trichoderma harzianum: role of gtt1. Euk. Cell 2 (4), 708–717.
De La Cruz, J., Hidalgo-Gallego, A., Lora, J. M., Benitez, T., Pintor-Toro, J. A., Llobell, A. (1992). Isolation and characterization of three chitinases from Trichoderma harzianum. Eur. J. Biochem. 206 (3), 859–867. doi: 10.1111/j.1432-1033.1992.tb16994.x
De La Cruz, J., Pintor-Toro, J. A., Benitez, T., Llobell, A., Romero, L. C. (1995). A novel endo-β-1,3-glucanase, BGN13.1, involved in the mycoparasitism of Trichoderma harzianum. J. Bacteriol. 177 (23), 6937–6945. doi: 10.1128/jb.177.23.6937-6945.1995
Deshpande, R. A., Shankar, V. (2002). Ribonucleases from T2 family. Crit. Rev. Microbiol. 28, 79–122. doi: 10.1080/1040-840291046704
Dickinson, J. M., Hanson, J. R., Hitchcock, P. B., Claydon, N. (1989). Structure and biosynthesis of harzianopyridone, an antifungal metabolite of trichoderma harzianum. J. Chem. Soc Perkin Trans. 1 (11), 1885–1887. doi: 10.1039/p19890001885
Dignam, B. E., Marshall, S. D., Wall, A. J., Mtandavari, Y. F., Gerard, E. M., Hicks, E., et al. (2022). Impacts of soil-borne disease on plant yield and farm profit in dairying soils. J. Sustain. Agric. Environ. 1 (1), 16–29. doi: 10.1002/sae2.12009
Djonovic, S., Pozo, M. J., Dangott, L. J., Howell, C. R., Kenerley, C. M. (2006). Sm1, a proteinaceous elicitor secreted by the biocontrol fungus trichoderma virens induces plant defense responses and systemic resistance. Mol. Plant Microbe Interact. 19, 838–853. doi: 10.1094/MPMI-19-0838
Djonovic, S., Vittone, G., Mendoza-Herrera, A., Kenerley, C. M. (2007). Enhanced biocontrol activity of Trichoderma virens transformants constitutively coexpressing b-1,3- and b-1,6- glucanase genes. Mol. Plant Pathol. 8, 469–480. doi: 10.1111/j.1364-3703.2007.00407.x
Druzhinina, I. S., Chenthamara, K., Zhang, J., Atanasova, L., Yang, D., Miao, Y., et al. (2018). Massive lateral transfer of genes encoding plant cell wall-degrading enzymes to the mycoparasitic fungus Trichoderma from its plant-associated hosts. PloS Genet. 14 (4), e1007322. doi: 10.1371/journal.pgen.1007322
Druzhinina, I. S., Seidl-Seiboth, V., Herrera-Estrella, A., Horwitz, B.A., Kenerley, C.M., Monte, E., et al. (2011). Trichoderma: the genomics of opportunistic success. Nat. Rev. Microbiol. 9, 749–759. doi: 10.1038/nrmicro2637
Du, F. Y., Ju, G. L., Xiao, L., Zhou, Y. M., Wu, X. (2020). Sesquiterpenes and cyclodepsipeptides from marine-derived fungus Trichoderma longibrachiatum and their antagonistic activities against soil-borne pathogens. Mar. Drugs 18 (3), 165. doi: 10.3390/md18030165
Dunlop, R. W., Simon, A., Sivasithamparam, K., Ghisalberti, E. L. (1989). An antibiotic from trichoderma koningii active against soilborne plant pathogens. J. Nat. Prod. 52 (1), 67–74. doi: 10.1021/np50061a008
Dutta, P. (2018). Bioformulation of Trichoderma harzianum for the management of soil borne plant diseases. paper presented in ICCPP_2018 at Boston, USA. Phytopathol. S1.274. doi: 10.1094/PHYTO-108-10-S1.240
Dutta, P., Das, B. C. (1999). Effect of seed pelleting and soil application of Trichoderma harzianum in the management of stem rot of soybean. J. Mycol. Plant Pathol. 29 (3), 317–322.
Dutta, P., Das, B. C. (2002). Management of collar rot of tomato by trichoderma spp. and chemicals. Ind. Phytopathol. 55 (2), 235–237.
Dutta, P., Das, B. C., Islam, M. (2008). Eco-friendly strategies for management sclerotinia rot of French bean. J. Biol. Control 22 (2), 405–410.
Dutta, P., Deb, L., Gogoi, J., Mahanta, M., Kumari, A., Yasin, A., et al. (2021). “UmTricho” a liquid bioformulation of indigenous strain of Trichoderma harzianum effectively managed the tikka disease (Cercospora spp.) of groundnut, Arachis hypogea l. under the agroecological condition of meghalaya. Biol. forum-an Int. J. 13 (2), 529–535.
Dutta, P., Deb, L., Pandey, A. K. (2022b). Trichoderma-from lab bench to field application: looking back over 50 years. Front. Agron. 4, 932839. doi: 10.3389/fagro.2022.932839
Dutta, P., Deka, M. K., Pegu, J. R., Das, A. (2013). Efficacy of biocontrol agents against Rhizoctonia solani kuehn. causing banded leaf and sheath blight of maize (Zea mays l.). Pestol XXXVI (6), 10–14.
Dutta, P., Kakati, N., Das, A., Kaushik, H., Boruah, S., Bhowmick, P., et al. (2017). Trichoderma pseudokoningii showed compatibility with certain community used inorganic pesticides, fertilizers and sticker cum spreaders. Int. J. Curr. Microbiol. App. Sci. 6 (2), 140–146. doi: 10.20546/ijcmas.2017.602.020
Dutta, P., Kumari, A., Mahanta, M., Biswas, K. K., Dudkiewicz, A., Thakuria, D., et al. (2022a). Advances in nanotechnology as a potential alternative for plant viral disease management. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.935193
Elad, Y., Freeman, S., Monte, E. (2000). Biocontrol agents: mode of action and interaction with other means of control. IOBC wprs Bull. 24.
El-Hasan, A., Walker, F., Schöne, J., Buchenauer, H. (2009). Detection of viridiofungin a and other antifungal metabolites excreted by Trichoderma harzianum active against different plant pathogens. Eur. J. Plant Pathol. 124, 457–470. doi: 10.1007/s10658-009-9433-3
Engelberth, J., Koch, T., Schüler, G., Bachmann, N., Rechtenbach, J., Boland, W. (2001). Ion channel-forming alamethicin is a potent elicitor of volatile biosynthesis and tendril coiling. cross talk between jasmonate and salicylate signaling in lima bean. Plant Physiol. 125 (1), 369–377. doi: 10.1104/pp.125.1.369
Evidente, A., Cabras, A., Maddau, L., Serra, S., Andolfi, A., Motta, A. (2003). Viridepyronone, a new antifungal 6-substituted 2 h-pyran-2-one produced by trichoderma viride. J. Agric. Food Chem. 51 (24), 6957–6960. doi: 10.1021/jf034708j
Fanelli, F., Liuzzi, V. C., Logrieco, A. F., Altomare, C. (2018). Genomic characterization of Trichoderma atrobrunneum (T. harzianum species complex) ITEM 908: insight into the genetic endowment of a multi-target biocontrol strain. BMC Genomics. 19, 1–18. doi: 10.1186/s12864-018-5049-3
Gaderer, R., Bonazza, K., Seidl-Seiboth, V. (2014). Cerato-platanins: a fungal protein family with intriguing properties and application potential. Appl. Microbiol. Biotechnol. 98, 4795–4803. doi: 10.1007/s00253-014-5690-y
Gaderer, R., Lamdan, N. L., Frischmann, A., Sulyok, M., Krska, R., Horwitz, B. A., et al. (2015). Sm2, a paralog of the trichoderma cerato-platanin elicitor Sm1, is also highly important for plant protection conferred by the fungal-root interaction of trichoderma with maize. BMC Microbiol. 15 (1), 1–9. doi: 10.1186/s12866-014-0333-0
Gajera, H., Domadiya, R., Patel, S., Kapopara, M., Golakiya, B. (2013). Molecular mechanism of trichoderma as bio-control agents against phytopathogen system–a review. Curr. Res. Microbiol. Biotechnol. 1 (4), 133–142.
Gao, R., Ding, M., Jiang, S., Zhao, Z., Chenthamara, K., Shen, Q., et al. (2020). The evolutionary and functional paradox of cerato-platanins in fungi. Appl. Environ. Microbiol. 86 (13), e00696–e00620. doi: 10.1128/AEM.00696-20
Ghisalberti, E. L., Rowland, C. Y. (1993). Antifungal metabolites from trichoderma harzianum. J. Nat. Prod. 56 (10), 1799–1804. doi: 10.1021/np50100a020
Gill, H. K., Garg, H. (2014). Pesticide: environmental impacts and management strategies. Pesticides-toxic aspects 8, 187. doi: 10.5772/57399
Goulard, C., Hlimi, S., Rebuffat, S., Bodo, B. (1995). Trichorzins HA and MA, antibiotic peptides from trichoderma harzianum i. fermentation, isolation and biological properties. J. Antibiot. 48 (11), 1248–1253.
González-López, M. D. C., Jijón-Moreno, S., Dautt-Castro, M., Ovando-Vázquez, C., Ziv, T., Horwitz, B. A., et al. (2021). Secretome analysis of arabidopsis–trichoderma atroviride interaction unveils new roles for the plant glutamate: glyoxylate aminotransferase GGAT1 in plant growth induced by the fungus and resistance against botrytis cinerea. Int. J. Mol. Sci. 22 (13), 6804. doi: 10.3390/ijms22136804
Gruber, S., Seidl-Seiboth, V. (2012). Self versus non-self: fungal cell wall degradation in Trichoderma. Microbiol 158, 26–34. doi: 10.1099/mic.0.052613-0
Grun, C. H., Dekker, N., Nieuwland, A. A., Klis, F. M., Kamerling, J. P., Vliegenthart, J .F., et al. (2006). Mechanism € of action of the endo-(1 / 3)-a-glucanase MutAp from the mycoparasitic fungus trichoderma harzianum. FEBS Lett. 580, 3780–3786. doi: 10.1016/j.febslet.2006.05.062
Guo, Q., Yoshida, Y., Major, I. T., Wang, K., Sugimoto, K., Kapali, G., et al. (2018). JAZ repressors of metabolic defense promote growth and reproductive fitness in arabidopsis. Proc. Natl. Acad. Sci. 115 (45), E10768–E10777. doi: 10.1073/pnas.1811828115
Guzman-Guzman, P., Aleman-Duarte, M. I., Delaye, L., HerreraEstrella, A., Olmedo-Monfil, V. (2017). Identification of effector like proteins in trichoderma spp. and role of a hydrophobin in the plant-fungus interaction and mycoparasitism. BMC Genet. 18 (1), 1–20. doi: 10.1186/s12863-017-0481-y
Guzmán-Guzmán, P., Kumar, A., de los Santos-Villalobos, S., Parra-Cota, F. I., Orozco-Mosqueda, M., Fadiji, A. E., et al. (2023). Trichoderma species: our best fungal allies in the biocontrol of plant diseases–a review. Plants 12, 432. doi: 10.3390/plants12030432
Hacquard, S., Kracher, B., Hiruma, K., Münch, P. C., Garrido-Oter, R., Thon, M. R., et al. (2016). Survival trade-offs in plant roots during colonization by closely related beneficial and pathogenic fungi. Nat. Commun. 7 (1), 11362. doi: 10.1038/ncomms11362
Harman, G. E. (1993). Chitinolytic enzymes of Trichoderma harzianum: purification of chitobiosidase and endochitinase. Phytopathol 83, 313. doi: 10.1094/Phyto-83-313
Harman, G. E., Howell, C. R., Viterbo, A., Chet, I., Lorito, M. (2004). Trichoderma species–opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2 (1), 43–56. doi: 10.1038/nrmicro797
Harwoko, H., Daletos, G., Stuhldreier, F., Lee, J., Wesselborg, S., Feldbrügge, M., et al. (2021). Dithiodiketopiperazine derivatives from endophytic fungi Trichoderma harzianum and Epicoccum nigrum. Nat. Prod. Res. 35 (2), 257–265. doi: 10.1080/14786419.2019.1627348
Hasin, Y., Seldin, M., Lusis, A. (2017). Multi-omics approaches to disease. Genome Biol. 18 (1), 1–15. doi: 10.1186/s13059-017-1215-1
Hass, H., Taylor, T. N., Remy, W. (1994). Fungi from the lower Devonian rhynie chert: mycoparasitism. Am. J. Bot. 81, 29–37. doi: 10.1002/j.1537-2197.1994.tb15405.x
Hermosa, R., Rubio, M. B., Cardoza, R. E., Nicolas, C., Monte, E., Gutierrez, S. (2013). The contribution of trichoderma to balancing the costs of plant growth and defense. Int. Microbiol. 16 (2), 69–80. doi: 10.2436/20.1501.01.181
Hermosa, R., Viterbo, A., Chet, I., Monte, E. (2012). Plant-beneficial effects of Trichoderma and of its genes. Microbiol 158 (1), 17–25. doi: 10.1099/mic.0.052274-0
Hill, R. A., Cutler, H. G., Parker, S. R. (1995). Trichoderma and metabolites as control agents for microbial plant diseases. PCT Int. Appl. 9520879 (10).
Howell, C. R. (1987). Relevance of mycoparasitism in the biological control of Rhizoctonia solani by Gliocladium virens. Phytopathol 77, 992. doi: 10.1094/Phyto-77-992
Howell, C. R. (2003). Mechanisms employed by Trichoderma species in the biological control of plant diseases: the history and evolution of current concepts. Plant Dis. 87, 4–10. doi: 10.1094/PDIS.2003.87.1.4
Howell, C. R., Hanson, L. E., Stipanovic, R. D., Puckhaber, L. S. (2000). Induction of terpenoid synthesis in cotton roots and control of Rhizoctonia solani by seed treatment with Trichoderma virens. Phytopathol 90 (3), 248–252. doi: 10.1094/PHYTO.2000.90.3.248
Howell, C. R., Stipanovic, E. R. D. (1983). Glioviridin, a new antibiotic from Gliocladium virens and its role in the biological control of Pythium ultimum. Can. J. Microbiol. 29, 321–324. doi: 10.1139/m83-053
Huang, Y., Mijiti, G., Wang, Z., Yu, W., Fan, H., Zhang, R., et al. (2015). Functional analysis of the class II hydrophobin gene HFB2-6 from the biocontrol agent Trichoderma asperellum ACCC30536. Microbiol. Res. 171, 8–20. doi: 10.1016/j.micres.2014.12.004
Jeerapong, C., Phupong, W., Bangrak, P., Intana, W., Tuchinda, P. (2015). Trichoharzianol, a new antifungal from Trichoderma harzianum F031. J. Agric. Food Chem. 63 (14), 3704–3708. doi: 10.1021/acs.jafc.5b01258
Jones, J. D. G., Dangl, J. L. (2006). The plant immune system. Nat 444 (7117), 323–329. doi: 10.1038/nature05286
Jones, R. W., Pettit, R. E. (1987). Variation in sensitivity among anastomosis groups of Rhizoctonia solani to the antibiotic gliotoxin. Plant Dis. (USA). 71 (1), 34–36. doi: 10.1094/PD-71-0034
Kappel, L., Münsterkötter, M., Sipos, G., Escobar Rodriguez, C., Gruber, S. (2020). Chitin and chitosan remodeling defines vegetative development and Trichoderma biocontrol. PloS Pathog. 16 (2), e1008320. doi: 10.1371/journal.ppat.1008320
Karlsson, M., Atanasova, L., Jensen, D., Zeilinger, S. (2017). Necrotrophic mycoparasites and their genomes. Microbiol. Spectr. 5, FUNK-0016–2016. doi: 10.1128/microbiolspec.FUNK-0016-2016
Keswani, C., Mishra, S., Sarma, B. K., Singh, S. P., Singh, H. B. (2014). Unraveling the efficient applications of secondary metabolites of various trichoderma spp. Appl. Microbiol. Biotechnol. 98 (2), 533–544. doi: 10.1007/s00253-013-5344-5
Khan, R. A. A., Najeeb, S., Mao, Z., Ling, J., Yang, Y., Li, Y., et al. (2020). Bioactive secondary metabolites from trichoderma spp. against phytopathogenic bacteria and root-knot nematode. Microorg 8 (3), 401. doi: 10.3390/microorganisms8060817
Kishimoto, N., Sugihara, S., Mochida, K. Y. O., Fujita, T. (2005). In vitro antifungal and antiviral activities of γ-and δ-lactone analogs utilized as food flavoring. Biocontrol Sci. 10 (1-2), 31–36. doi: 10.4265/bio.10.31
Kohler, A., Tisserant, E. (2014). Exploring the transcriptome of mycorrhizal interactions. Adv. Bot. Res. 70, 53–78. doi: 10.1016/B978-0-12-397940-7.00002-1
Kou, Y., Tan, Y. H., Ramanujam, R., Naqvi, N. I. (2017). Structure-function analyses of the Pth11 receptor reveal an important role for CFEM motif and redox regulation in rice blast. New Phytol. 214 (1), 330–342. doi: 10.1111/nph.14347
Kubicek, C. P., Baker, S., Gamauf, C., Kenerley, C. M., Druzhinina, I. S. (2008). Purifying selection and birth-and-death evolution in the class II hydrophobin gene families of the ascomycete Trichoderma/Hypocrea. BMC Evol. Biol. 8 (1), 1–16. doi: 10.1186/1471-2148-8-4
Kubicek, C. P., Herrera-Estrella, A., Seidl-Seiboth, V., Martinez, D. A., Druzhinina, I. S., Thon, M., et al. (2011). Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 12 (4), 1–15. doi: 10.1186/gb-2011-12-4-r40
Kubicek, C. P., Steindorff, A. S., Chenthamara, K., Manganiello, G., Henrissat, B., Zhang, J., et al. (2019). Evolution and comparative genomics of the most common Trichoderma species. BMC Genom. 20 (1), 1–24. doi: 10.1186/s12864-019-5680-7
Kulkarni, R. D., Kelkar, H. S., Dean, R. A. (2003). An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. Trends Biochem. Sci. 28 (3), 118–121. doi: 10.1016/S0968-0004(03)00025-2
Lamdan, N. L., Shalaby, S., Ziv, T., Kenerley, C. M., Horwitz, B. A. (2015). Secretome of trichoderma interacting with maize roots: role in induced systemic resistance. Mol. Cell. Proteom. 14, 1054–1063. doi: 10.1074/mcp.M114.046607
Lanver, D., Tollot, M., Schweizer, G., Lo Presti, L., Reissmann, S., Ma, L. S., et al. (2017). Ustilago maydis effectors and their impact on virulence. Nat. Rev. Microbiol. 15 (7), 409–421. doi: 10.1038/nrmicro.2017.33
Lawry, R. (2016). Cross-communication between trichoderma and plants during root colonisation (Lincoln University, New Zealand: Ph.D. Dissertation).
Lee, S. J., Yeo, W. H., Yun, B. S., Yoo, I. D. (1999). Isolation and sequence analysis of new peptaibol, boletusin, from boletus spp. J. Pept. Sci. 5 (8), 374–378.
Li, Q. R., Tan, P., Jiang, Y. L., Hyde, K. D., Mckenzie, E. H., Bahkali, A. H., et al. (2013). A novel Trichoderma species isolated from soil in guizhou. T. guizhouense. Mycol. Prog. 12, 167–172. doi: 10.1007/s11557-012-0821-2
Lima, L. H. C., Ulhoa, C. J., Fernandes, A. P., Felix, C. R. (1997). Purification of a chitinase from trichoderma sp. and its action on sclerotium rolfsii and rhizoctonia solani cell walls. J. Gen. Appl. Microbiol. 43, 31–37. doi: 10.2323/jgam.43.31
Linder, M. B., Szilvay, G. R., Nakari-Setala, T., Penttila, M. E. (2005). Hydrophobins: the protein amphiphiles of filamentous fungi. FEMS Microbiol. Rev. 29, 877–896. doi: 10.1016/j.femsre.2005.01.004
Liu, D., He, X., Li, W., Chen, C., Ge, F. (2013). A b-1,3-glucanase gene expressed in fruit of pyrus pyrifolia enhances resistance to several pathogenic fungi in transgenic tobacco. Eur. J. Plant Pathol. 135, 267–277. doi: 10.1007/s10658-012-0083-5
Liu, S. Y., Lo, C. T., Shibu, M. A., Leu, Y. L., Jen, B. Y., Peng, K. C. (2009). Study on the anthraquinones separated from the cultivation of Trichoderma harzianum strain Th-R16 and their biological activity. J. Agric. Food Chem. 57 (16), 7288–7292. doi: 10.1021/jf901405c
Liu, Q., Tang, S., Meng, X., Zhu, H., Zhu, Y., Liu, D., et al. (2021). Proteomic analysis demonstrates a molecular dialog between Trichoderma guizhouense NJAU 4742 and cucumber (Cucumis sativus l.) roots: role in promoting plant growth. Mol. Plant-Microbe Interact. 34 (6), 631–644. doi: 10.1094/MPMI-08-20-0240-R
Lorito, M., Peterbauer, C., Hayes, C. K., Harman, G. E. (1994). Synergistic interaction between fungal cell wall degrading enzymes and different antifungal compounds enhances inhibition of spore germination. Microbiol. 140 (3), 623–629. doi: 10.1099/00221287-140-3-623
Luo, Y., Zhang, D. D., Dong, X. W., Zhao, P. B., Chen, L. L., Song, X. Y., et al. (2010). Antimicrobial peptaibols induce defense responses and systemic resistance in tobacco against tobacco mosaic virus. FEMS Microbiol. Lett. 313 (2), 120–126. doi: 10.1111/j.1574-6968.2010.02135.x
Malmierca, M. G., Barua, J., McCormick, S. P., Izquierdo-Bueno, I., Cardoza, R. E., Alexander, N. J., et al. (2015). Novel aspinolide production by Trichoderma arundinaceum with a potential role in Botrytis cinerea antagonistic activity and plant defence priming. Environ. Microbiol. 17 (4), 1103–1118. doi: 10.1111/1462-2920.12514
Marshall, R., Kombrink, A., Motteram, J., Loza-Reyes, E., Lucas, J., Hammond-Kosack, et al. (2011). Analysis of two in planta expressed LysM effector homologs from the fungus Mycosphaerella graminicola reveals novel functional properties and varying contributions to virulence on wheat. Plant Physiol. 156, 756–769. doi: 10.1104/pp.111.176347
Martinez, D., Berka, R. M., Henrissat, B., Saloheimo, M., Arvas, M., Baker, S. E., et al. (2008). Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotechnol. 26 (5), 553–560. doi: 10.1038/nbt1403
Mendoza-Mendoza, A., Zaid, R., Lawry, R., Hermosa, R., Monte, E., Horwitz, B. A., et al. (2017). Molecular dialogues between Trichoderma and roots: role of the fungal secretome. Fungal Biol. Rev. 32 (2), 62–85. doi: 10.1016/j.fbr.2017.12.001
Merrill, S. A., Mazza, A. M. (2006). Reaping the benefits of genomic and proteomic research: intellectual property rights, innovation, and public health. Natl. Acad. Press., P. 188.
Monteiro, V. N., do Nascimento Silva, R., Steindorff, A. S., Costa, F. T., Noronha, E. F., Ricart, C. A. O., et al. (2010). New insights in Trichoderma harzianum antagonism of fungal plant pathogens by secreted protein analysis. Curr. Microbiol. 61, 298–305. doi: 10.1007/s00284-010-9611-8
Monteiro, V. N., Ulhoa, C. J. (2006). Biochemical characterization of a b-1,3-glucanase from trichoderma koningii induced by cell wall of rhizoctonia solani. Curr. Microbiol. 52, 92–96. doi: 10.1007/s00284-005-0090-2
Morán-Diez, M. E., Carrero-Carrón, I., Rubio, M. B., Jiménez-Díaz, R. M., Monte, E., Hermosa, R. (2019). Transcriptomic analysis of Trichoderma atroviride overgrowing plant-wilting Verticillium dahliae reveals the role of a new M14 metallocarboxypeptidase CPA1 in biocontrol. Front. Microbiol. 10, 1120. doi: 10.3389/fmicb.2019.01120
Moran-Diez, E., Hermosa, R., Ambrosino, P., Cardoza, R. E., Gutierrez, S., Lorito, M., et al. (2009). The ThPG1 endopolygalacturonase is required for the Trichoderma harzianum-plant beneficial interaction. Mol. Plant Microbe Interact. 22, 1021–1031. doi: 10.1094/MPMI-22-8-1021
Morán-Diez, E., Rubio, B., Domínguez, S., Hermosa, R., Monte, E., Nicolás, C. (2012). Transcriptomic response of Arabidopsis thaliana after 24 h incubation with the biocontrol fungus Trichoderma harzianum. J. Plant Physiol. 169 (6), 614–620. doi: 10.1016/j.jplph.2011.12.016
Mukherjee, P. K., Horwitz, B. A., Herrera-Estrella, A., Schmoll, M., Kenerley, C. M. (2013). Trichoderma research in the genome era. Annu. Rev. Phytopathol. 51, 105–129. doi: 10.1146/annurev-phyto-082712-102353
Mukherjee, P. K., Horwitz, B. A., Kenerley, C. M. (2012). Secondary metabolism in Trichoderma–a genomic perspective. Microbiol 158 (1), 35–45. doi: 10.1099/mic.0.053629-0
Mukherjee, P. K., Mehetre, S. T., Sherkhane, P. D., Muthukathan, G., Ghosh, A., Kotasthane, A. S., et al. (2019). A novel seed-dressing formulation based on an improved mutant strain of trichoderma virens, and its field evaluation. Front. Microbiol. 10, 1910. doi: 10.3389/fmicb.2019.01910
Mukherjee, P. K., Mendoza-Mendoza, A., Zeilinger, S., Horwitz, B. A. (2022). Mycoparasitism as a mechanism of Trichoderma-mediated suppression of plant diseases. Fungal Biol. Rev. 39, 15–33. doi: 10.1016/j.fbr.2021.11.004
Mukherjee, P. K., Mukhopadhyay, A. N., Sarmah, D. K., Shrestha, S. M. (1995). Comparative antagonistic properties of Gliocladium virens and Trichoderma harzianum on Sclerotium rolfsii and Rhizoctonia solani and its relevance to understanding the mechanisms of biocontrol. J. Phytopathol. 143, 275–279. doi: 10.1111/j.1439-0434.1995.tb00260.x
Mukherjee, P. K., Wiest, A., Ruiz, N., Keightley, A., Moran-Diez, M. E., McCluskey, K., et al. (2011). Two classes of new peptaibols are synthesized by a single non-ribosomal peptide synthetase of trichoderma virens. J. Biol. Chem. 286 (6), 4544–4554. doi: 10.1074/jbc.M110.159723
Nakano, H., Hara, M., Mejiro, T., Ando, K., Saito, Y., Morimoto, M. (1990). DC1149B, DC1149R and their manufacture with trichoderma. JP Patent 2218686, 31.
Naranjo-Ortiz, M. A., Gabaldon, T. (2019). Fungal evolution: major ecological adaptations and evolutionary transitions. Biol. Rev. 94, 1443–1476. doi: 10.1111/brv.12510
Nasser, L., Weissman, Z., Pinsky, M., Amartely, H., Dvir, H., Kornitzer, D. (2016). Structural basis of haem-iron acquisition by fungal pathogens. N. Microbiol. 1, 16156. doi: 10.1038/nmicrobiol.2016.156
Nauom, S., da Silva Neto, B. R., Ribeiro, M. S., Pedersoli, W. R., Ulhoa, C. J., Silva, R. N., et al. (2019). Biochemical and molecular study of Trichoderma harzianum enriched secretome protein profiles using lectin affinity chromatography. Appl. Biochem. Biotechnol. 187. 1–13. doi: 10.1007/s12010-018-2795-2
Noronha, E. F., Kipnis, A., Junqueira-Kipnis, A. P., Ulhoa, C. J. (2000). Regulation of 36- kDa b-1,3-glucanase synthesis in trichoderma harzianum. FEMS Microbiol. Lett. 188 (1), 19–22. doi: 10.1016/S0378-1097(00)00205-6
Olmedo-Monfil, V., Mendoza-Mendoza, A., Gomez, I., Cortes, C., Herrera-Estrella, A.. (2002). Multiple environmental signals determine the transcriptional activation of the mycoparasitism related gene prb1 in Trichoderma atroviride. Mol. Genet. Genom. 267, 703–712. doi: 10.1007/s00438-002-0703-4
Omann, M. R., Lehner, S., Rodríguez, C. E., Brunner, K., Zeilinger, S. (2012). The seven-transmembrane receptor Gpr1 governs processes relevant for the antagonistic interaction of trichoderma atroviride with its host. Microbiol 158 (Pt 1), 107. doi: 10.1099/mic.0.052035-0
Pang, G., Sun, T., Yu, Z., Yuan, T., Liu, W., Zhu, H., et al. (2020). Azaphilones biosynthesis complements the defence mechanism of Trichoderma guizhouense against oxidative stress. Environ. Microbiol. 22 (11), 4808–4824. doi: 10.1111/1462-2920.15246
Pazzagli, L., Seidl-Seiboth, V., Barsottini, M., Vargas, W. A., Scala, A., Mukherjee, P. K. (2014). Cerato-platanins: elicitors and effectors. Plant Sci. 228, 79–87. doi: 10.1016/j.plantsci.2014.02.009
Pellan, L., Dieye, C. A. T., Durand, N., Fontana, A., Strub, C., Schorr-Galindo, S. (2021). Biocontrol agents: toolbox for the screening of weapons against mycotoxigenic fusarium. J. Fungi 7 (06), 446. doi: 10.3390/jof7060446
Persoon, C. H. (1794). Neuer versuch einer systematischen einteilung der schwämme. Racodium Römer’s Neues Magazin der Botanik. 1, 123.
Pieterse, C. M., Leon-Reyes, A., van der Ent, S., Van Wees, S. C. (2009). Networking by small molecule hormones in plant immunity. Nat. Chem. Biol. 5 (5), 308–316. doi: 10.1038/nchembio.164
Poole, P. R., Ward, B. G., Whitaker, G. (1998). The effects of topical treatments with 6-pentyl-2-pyrone and structural analogues on stem end postharvest rots in kiwifruit due to botrytis cinerea. J. Sci. Food Agric. 77 (1), 81–86. doi: 10.1002/(SICI)1097-0010(199805)77:1<81::AID-JSFA6>3.0.CO;2-5
Pozo, M. J., Baek, J. M., Garcıa, J. M., Kenerley, C. M. (2004). Functional analysis of tvsp1, a serine protease-encoding gene in the biocontrol agent Trichoderma virens. Fungal Genet. Biol. 41, 336–348. doi: 10.1016/j.fgb.2003.11.002
Proctor, R. H., McCormick, S. P., Kim, H. S., Cardoza, R. E., Stanley, A. M., Lindo, L., et al. (2018). Evolution of structural diversity of trichothecenes, a family of toxins produced by plant pathogenic and entomopathogenic fungi. PloS Pathog. 14 (4), e1006946. doi: 10.1371/journal.ppat.1006946
Przylucka, A., Akcapinar, G. B., Chenthamara, K., Cai, F., Grujic, M., Karpenko, J., et al. (2017). HFB7-a novel orphan hydrophobin of the harzianum and virens clades of Trichoderma, is involved in response to biotic and abiotic stresses. Fungal Genet. Biol. 102, 63–76. doi: 10.1016/j.fgb.2017.01.002
Quarantin, A., Glasenapp, A., Schafer, W., Favaron, F., Sella, L. (2016). Involvement of the Fusarium graminearum ceratoplatanin proteins in fungal growth and plant infection. Plant Physiol. Biochem. 109, 220–229. doi: 10.1016/j.plaphy.2016.10.001
Rabe, F., Ajami-Rashidi, Z., Doehlemann, G., Kahmann, R., Djamei, A. (2013). Degradation of the plant defence hormone salicylic acid by the biotrophic fungus Ustilago maydis. Mol. Microbiol. 89, 179–188. doi: 10.1111/mmi.12269
Ramada, M. H. S., Steindorff, A. S., Bloch, C., Ulhoa, C. J. (2016). Secretome analysis of the mycoparasitic fungus Trichoderma harzianum ALL 42 cultivated in different media supplemented with Fusarium solani cell wall or glucose. Proteomics. 13 (3), 477–490. doi: 10.1002/pmic.201400546
Rouina, H., Tseng, Y. H., Nataraja, K. N., Uma Shaanker, R., Krüger, T., Kniemeyer, O., et al. (2022). Comparative secretome analyses of Trichoderma/Arabidopsis Co-cultures identify proteins for salt stress, plant growth promotion, and root colonization. Front. Ecol. Evol. 9, 933. doi: 10.3389/fevo.2021.808430
Rubio, M. B., Dominguez, S., Monte, E., Hermosa, R. (2012). Comparative study of Trichoderma gene expression in interactions with tomato plants using high-density oligonucleotide microarrays. Microbiol 158, 119–128. doi: 10.1099/mic.0.052118-0
Rubio, M. B., Quijada, N. M., Perez, E., Dominguez, S., Monte, E., Hermosa, R. (2014). Identifying beneficial qualities of Trichoderma parareesei for plants. Appl. Environ. Microbiol. 80, 1864–1873. doi: 10.1128/AEM.03375-13
Sabnam, N., Barman, S. R. (2017). WISH, a novel CFEM GPCR is indispensable for surface sensing, asexual and pathogenic differentiation in rice blast fungus. Fungal Genet. Biol. 105, 37–51. doi: 10.1016/j.fgb.2017.05.006
Sakuno, E., Yabe, K., Hamasaki, T., Nakajima, H. A. (2000). New inhibitor of 5′-hydroxyaverantin dehydrogenase, an enzyme involved in aflatoxin biosynthesis, from Trichoderma hamatum. J. Nat. Prod. 63, 1677–1678. doi: 10.1021/np000194w
Salas-Marina, M. A., Isordia-Jasso, M. I., Islas-Osuna, M. A., Delgado-Sanchez, P., Jimenez-Bremont, J. F., RodriguezKessler, M., et al. (2015). The Epl1 and Sm1 proteins from Trichoderma atroviride and Trichoderma virens differentially modulate systemic disease resistance against different life style pathogens in Solanum lycopersicum. Front. Plant Sci. 6, 77. doi: 10.3389/fpls.2015.00077
Saloheimo, M., Paloheimo, M., Hakola, S., Pere, J., Swanson, B., Nyyssonen, E., et al. (2002). Swollenin, a Trichoderma reesei protein with sequence similarity to the plant expansins, exhibits disruption activity on cellulosic materials. Eur. J. Biochem. 269, 4202–4211. doi: 10.1046/j.1432-1033.2002.03095.x
Samolski, I., de Luis, A., Vizcaino, J. A., Monte, E., Suarez, M. B. (2009). Gene expression analysis of the biocontrol fungus Trichoderma harzianum in the presence of tomato plants, chitin, or glucose using a high-density oligonucleotide microarray. BMC Microbiol. 9, 217. doi: 10.1186/1471-2180-9-217
Samolski, I., Rincon, A. M., Pinzon, L. M., Viterbo, A., Monte, E. (2012). The qid74 gene from Trichoderma harzianum has a role in root architecture and plant biofertilization. Microbiol 158, 129–138. doi: 10.1099/mic.0.053140-0
Santhanam, P., van Esse, H. P., Albert, I., Faino, L., Nurnberger, T., Thomma, B. P. (2013). Evidence for functional diversification within a fungal NEP1-like protein family. Mol. Plant Microbe Interact. 26, 278–286. doi: 10.1094/MPMI-09-12-0222-R
Saravanakumar, K., Li, Y., Yu, C., Wang, Q., Wang, M., Sun, J., et al. (2017). Effect of Trichoderma harzianum on maize rhizosphere microbiome and biocontrol of fusarium stalk rot. Sci. Rep. 7, 1771. doi: 10.1038/s41598-017-01680-w
Sarrocco, S., Matarese, F., Baroncelli, R., Vannacci, G., SeidlSeiboth, V., Kubicek, C. P., et al. (2017). The constitutive endopolygalacturonase TvPG2 regulates the induction of plant systemic resistance by Trichoderma virens. Phytopathol 107, 537–544. doi: 10.1094/PHYTO-03-16-0139-R
Scarselletti, R., Faull, J. L. (1994). In vitro activity of 6-pentyl-α-pyrone, a metabolite of Trichoderma harzianum, in the inhibition of Rhizoctonia solani and Fusarium oxysporum f. sp. lycopersici. Mycol. Res. 98 (10), 1207–1209. doi: 10.1016/S0953-7562(09)80206-2
Schmoll, M., Dattenbock, C., Carreras-Villasenor, N., Mendoza-Mendoza, A., Tisch, D., Alemán, M. I., et al. (2016). The genomes of three uneven siblings: footprints of the lifestyles of three Trichoderma species. Microbiol. Mol. Biol. Rev. 80, 205–327. doi: 10.1128/MMBR.00040-15
Segal, L. M., Wilson, R. A. (2018). Reactive oxygen species metabolism and plant-fungal interactions. Fungal Genet. Biol. 110, 1–9. doi: 10.1016/j.fgb.2017.12.003
Segarra, G., Casanova, E., Bellido, D., Odena, M. A., Oliveira, E., Trillas, I. (2007). Proteome, salicylic acid, and jasmonic acid changes in cucumber plants inoculated with Trichoderma asperellum strain T34. Proteomics 7, 3943–3952. doi: 10.1002/pmic.200700173
Senthilkumar, M., Amaresan, N., Sankaranarayanan, A. (2021). Purification of Trichoderma producing hydrolytic enzyme: β-1, 3 glucanase. in: plant-microbe interactions. Springer Protocols Handbooks. Humana, New York, NY., 215–216. doi: 10.1007/978-1-0716-1080-0_62
Sha, S., Liu, L., Pan, S., Wang, W. (2013). Isolation and purification of antifungal components from Trichoderma harzianum ferment broth by high-speed counter-current chromatography. Chin. J. Biol. Control 29 (1), 83.
Sharma, V., Salwan, R., Sharma, P. N. (2017). The comparative mechanistic aspects of Trichoderma and probiotics: scope for future research. Physiol. Mol. Plant Pathol. 100, 84–96. doi: 10.1016/j.pmpp.2017.07.005
Shaw, S., Le Cocq, K., Paszkiewicz, K., Moore, K., Winsbury, R., de Torres Zabala, M., et al. (2016). Transcriptional reprogramming underpins enhanced plant growth promotion by the biocontrol fungus Trichoderma hamatum GD12 during antagonistic interactions with Sclerotinia sclerotiorum in soil. Mol. Plant Pathol. 17 (9), 1425–1441. doi: 10.1111/mpp.12429
Shentu, X., Zhan, X., Ma, Z., Yu, X., Zhang, C. (2014). Antifungal activity of metabolites of the endophytic fungus Trichoderma brevicompactum from garlic. Braz. J. Microbiol. 45, 248–254. doi: 10.1590/S1517-83822014005000036
Shi, M., Chen, L., Wang, X. W., Zhang, T., Zhao, P. B., Song, X. Y., et al. (2012). Antimicrobial peptaibols from Trichoderma pseudokoningii induce programmed cell death in plant fungal pathogens. Microbiol 158 (1), 166–175. doi: 10.1099/mic.0.052670-0
Shi, Z. Z., Liu, X. H., Li, X. N., Ji, N. Y. (2020). Antifungal and antimicroalgal trichothecene sesquiterpenes from the marine algicolous fungus Trichoderma brevicompactum a-DL-9-2. J. Agric. Food Chem. 68 (52), 15440–15448. doi: 10.1021/acs.jafc.0c05586
Shi, Y., Shentu, X., Yu, X. (2009). Identification of an endophytic fungus isolated from Ilex cornuta and the biocontrol effects of its secondary metabolite. Acta Phytopathol. Sin. 39 (4), 362–367.
Silva, R. N., Monteiro, V. N., Steindorff, A. S., Gomes, E. V., Noronha, E. F., Ulhoa, C. J. (2019). Trichoderma/pathogen/plant interaction in pre-harvest food security. Fungal Boil. 123 (8), 565–583. doi: 10.1016/j.funbio.2019.06.010
Singh, H. B. (2014). Management of plant pathogens with microorganisms. Proc. Indian Natl. Sci. Acad. 80 (2), 443–454. doi: 10.16943/ptinsa/2014/v80i2/55120
Singh, S., Dureja, P., Tanwar, R. S., Singh, A. (2005). Production and antifungal activity of secondary metabolites of trichoderma virens. Pestic. Res. J. 17 (2), 26–29.
Sivan, A., Chet, I. (1989). The possible role of competition between T. harzianum and F. oxysporum on rhizosphere colonization. Phytopathol 79, 198–203. doi: 10.1094/Phyto-79-198
Song, X. Y., Shen, Q. T., Xie, T. U., Chen, X. L., Sun, C. Y., Zhang, Y. Z. (2006). Broad-spectrum antimicrobial activity and high stability of trichokonins from Trichoderma koningii SMF2 against plant pathogens. FEMS Microbiol. Lett. 260 (1), 119–125. doi: 10.1111/j.1574-6968.2006.00316.x
Sood, M., Kapoor, D., Kumar, V., Sheteiwy, M. S., Ramakrishnan, M., Landi, M., et al. (2020). Trichoderma: the “secrets” of a multitalented biocontrol agent. Plants 9 (6), 762. doi: 10.3390/plants9060762
Spanu, P. D. (2015). RNAeprotein interactions in plant disease: hackers at the dinner table. New Phytol. 207, 991–995. doi: 10.1111/nph.13495
Steindorff, A. S., Silva, R. N., Coelho, A. S. G., Nagata, T., Noronha, E. F., Ulhoa, C. J. (2012). Trichoderma harzianum expressed sequence tags for identification of genes with putative roles in mycoparasitism against Fusarium solani. Biol. Control. 61 (2), 134–140. doi: 10.1016/j.biocontrol.2012.01.014
Stepanova, A. N., Yun, J., Likhacheva, A. V., Alonso, J. M. (2007). Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19 (7), 2169–2185. doi: 10.1105/tpc.107.052068
Studholme, D. J., Harris, B., Le Cocq, K., Winsbury, R., Perera, V., Ryder, L., et al. (2013). Investigating the beneficial traits of Trichoderma hamatum GD12 for sustainable agriculture–insights from genomics. Front. Plant Sci. 4, 258. doi: 10.3389/fpls.2013.00258
Suriani Ribeiro, M., Graciano de Paula, R., Raquel Voltan, A., de Castro, R. G., Carraro, C.B., José de Assis, L., et al. (2019). Endo-b-1,3-glucanase (GH16 family) from trichoderma harzianum participates in cell wall biogenesis but is not essential for antagonism against plant pathogens. Biomolecules 9 (12), 781. doi: 10.3390/biom9120781
Tian, Y., Tan, Y., Yan, Z., Liao, Y., Chen, J., De Boevre, M., et al. (2018). Antagonistic and detoxification potentials of Trichoderma isolates for control of zearalenone (ZEN) producing Fusarium graminearum. Front. Microbiol. 8, 2710. doi: 10.3389/fmicb.2017.02710
Tijerino, A., Cardoza, R. E., Moraga, J., Malmierca, M. G., Vicente, F., Aleu, J., et al. (2011). Overexpression of the trichodiene synthase gene tri5 increases trichodermin production and antimicrobial activity in Trichoderma brevicompactum. Fungal Genet. Biol. 48 (3), 285–296. doi: 10.1016/j.fgb.2010.11.012
Tjamos, E. C., Papavizas, G. C., Cook, R. J. (1992). Biological control of plant diseases- progress and challenges for the future (New York: Plenum Press).
Tong, S. M., Chen, Y., Zhu, J., Ying, S. H., Feng, M. G. (2016). Subcellular localization of five singular WSC domain-containing proteins and their roles in Beauveria bassiana responses to stress cues and metal ions. Environ. Microbiol. Rep. 8, 295–304. doi: 10.1111/1758-2229.12380
Tu, J. C. (1980). Gliocladium virens, a destructive mycoparasite of Sclerotinia sclerotiorum. Phytopathol 70, 670. doi: 10.1094/Phyto-70-670
Tucci, M., Ruocco, M., De Masi, L., De Palma, M., Lorito, M. (2011). The beneficial effect of trichoderma spp. on tomato is modulated by the plant genotype. Mol. Plant Pathol. 12 (4), 341–354. doi: 10.1111/j.1364-3703.2010.00674.x
Ulhoa, C. J., Peberdy, J. F. (1991). Regulation of chitinase synthesis in Trichoderma harzianum. J. Gen. Microbiol. 137 (9), 2163–2169. doi: 10.1371/journal.pone.0038088
Vargas, W. A., Crutcher, F. K., Kenerley, C. M. (2011). Functional characterization of a plant-like sucrose transporter from the beneficial fungus Trichoderma virens. regulation of the symbiotic association with plants by sucrose metabolism inside the fungal cells. New Phytol. 189, 777–789. doi: 10.1111/j.1469-8137.2010.03517.x
Vargas, W. A., Mandawe, J. C., Kenerley, C. M. (2009). Plant-derived sucrose is a key element in the symbiotic association between Trichoderma virens and maize plants. Plant Physiol. 151, 792–808. doi: 10.1104/pp.109.141291
Verena, S. S., Alfredo, H. E., Enrique, M., Susanne, Z. (2011). Trichoderma: the genomics of opportunistic success. Nat. Rev. Microbiol. 9 (10), 749–759. doi: 10.1038/nrmicro2637
Vieira, P. M., Coelho, A. S. G., Steindorff, A. S., de Siqueira, S. J. L., Silva, R. N., Ulhoa, C. J. (2013). Identification of differentially expressed genes from Trichoderma harzianum during growth on cell wall of Fusarium solani as a tool for biotechnological application. BMC Genomics. 14 (1), 1–11. doi: 10.1186/1471-2164-14-177
Villalobos-Escobedo, J. M., Esparza-Reynoso, S., Pelagio-Flores, R., López-Ramírez, F., Ruiz-Herrera, L. F., López-Bucio, J., et al. (2020). The fungal NADPH oxidase is an essential element for the molecular dialog between Trichoderma and Arabidopsis. Plant J. 103 (6), 2178–2192. doi: 10.1111/tpj.14891
Vinale, F., Ghisalberti, E. L., Sivasithamparam, K., Marra, R., Ritieni, A., Ferracane, R., et al. (2009). Factors affecting the production of Trichoderma harzianum secondary metabolites during the interaction with different plant pathogens. Lett. Appl. Microbiol. 48 (6), 705–711. doi: 10.1111/j.1472-765X.2009.02599.x
Vinale, F., Girona, I. A., Nigro, M., Mazzei, P., Piccolo, A., Ruocco, M., et al. (2012). Cerinolactone, a hydroxy-lactone derivative from Trichoderma cerinum. J. Nat. Prod. 75 (1), 103–106. doi: 10.1021/np200577t
Vinale, F., Marra, R., Scala, F., Ghisalberti, E. L., Lorito, M., Sivasithamparam, K. (2006). Major secondary metabolites produced by two commercial Trichoderma strains active against different phytopathogens. Lett. Appl. Microbiol. 43 (2), 143–148. doi: 10.1111/j.1472-765X.2006.01939.x
Vinale, F., Sivasithamparam, K., Ghisalberti, E. L., Marra, R., Barbetti, M. J., Li, H., et al. (2008). A novel role for Trichoderma secondary metabolites in the interactions with plants. Physiol. Mol. Plant Pathol. 72 (1-3), 80–86. doi: 10.1016/j.pmpp.2008.05.005
Vinale, F., Strakowska, J., Mazzei, P., Piccolo, A., Marra, R., Lombardi, N., et al. (2016). Cremenolide, a new antifungal, 10-member lactone from Trichoderma cremeum with plant growth promotion activity. Nat. Prod. Res. 30 (22), 2575–2581. doi: 10.1080/14786419.2015.1131985
Viterbo, A. D. A., Chet, I. (2006). TasHyd1, a new hydrophobin gene from the biocontrol agent Trichoderma asperellum, is involved in plant root colonization. Mol. Plant Pathol. 7 (4), 249–258. doi: 10.1111/j.1364-3703.2006.00335.x
Viterbo, A., Landau, U., Kim, S., Chernin, L., Chet, I. (2010). Characterization of ACC deaminase from the biocontrol and plant growth-promoting agent Trichoderma asperellum T203. FEMS Microbiol. Lett. 305, 42–48. doi: 10.1111/j.1574-6968.2010.01910.x
Wawra, S., Fesel, P., Widmer, H., Neumann, U., Lahrmann, U., Becker, S., et al. (2019). FGB1 and WSC3 are in planta-induced β-glucan-binding fungal lectins with different functions. New Phytol. 222 (3), 1493–1506. doi: 10.1111/nph.15711
Wawra, S., Fesel, P., Widmer, H., Timm, M., Seibel, J., Leson, L., et al. (2016). The fungal-specific β-glucan-binding lectin FGB1 alters cell-wall composition and suppresses glucan-triggered immunity in plants. Nat. Commun. 7 (1), 13188. doi: 10.1038/ncomms13188
Weindling, R. (1932). Trichoderma lignorum as a parasite of other soil fungi. Phytopathol 22, 837–845.
Weindling, R. (1934). Studies on a lethal principle effective in the parasitic action of Trichoderma lignorum on Rhizoctonia solani and other soil fungi. Phytopathol 24, 1153–1179.
Weindling, R., Emerson, O. (1936). The isolation of a toxic substance from the culture filtrate of Trichoderma. Phytopathol 26, 1068–1070.
Weindling, R., Fawcett, H. (1936). Experiments in the control of Rhizoctonia damping-off of citrus seedlings. Hilgardia 10, 1–16. doi: 10.3733/hilg.v10n01p001
Wipf, P., Kerekes, A. D. (2003). Structure reassignment of the fungal metabolite TAEMC161 as the phytotoxin viridiol. J. Nat. Pro. 66 (5), 716–718. doi: 10.1021/np0300277
Woo, S. L., Lorito, M. (2007). Exploiting the interactions between fungal antagonists, pathogens and the plant for biocontrol, In: Novel biotechnologies for biocontrol agent enhancement and management, pp. 107–130. Springer Netherlands.
Woo, S. L., Scala, F., Ruocco, M., Lorito, M. (2006). The molecular biology of the interactions between trichoderma spp., phytopathogenic fungi, and plants. Phytopathol 96, 181–185. doi: 10.1094/PHYTO-96-0181
Wu, B., Oesker, V., Wiese, J., Schmaljohann, R., Imhoff, J. F. (2014). Two new antibiotic pyridones produced by a marine fungus, trichoderma sp. strain MF106. Mar. Drugs 12 (3), 1208–1219. doi: 10.3390/md12031208
Xie, B. B., Qin, Q. L., Shi, M., Chen, L. L., Shu, Y. L., Luo, Y., et al. (2014). Comparative genomics provide insights into evolution of trichoderma nutrition style. Genome Biol. Evol. 6 (2), 379–390. doi: 10.1093/gbe/evu018
Xuan, Q. C., Huang, R., Miao, C. P., Chen, Y. W., Zhai, Y. Z., Song, F., et al. (2014). Secondary metabolites of endophytic fungus trichoderma sp. ym 311505 of Azadirachta indica. Chem. Nat. Compd. 50, 139–141. doi: 10.1007/s10600-014-0891-2
Yamazaki, H., Takahashi, O., Kirikoshi, R., Yagi, A., Ogasawara, T., Bunya, Y., et al. (2020a). Epipolythiodiketopiperazine and trichothecene derivatives from the NaI-containing fermentation of marine-derived Trichoderma cf. brevicompactum. J. Antibiot. 73, 559–567. doi: 10.1038/s41429-020-0314-5
Yamazaki, H., Yagi, A., Takahashi, O., Yamaguchi, Y., Saito, A., Namikoshi, M., et al. (2020b). Antifungal trichothecene sesquiterpenes obtained from the culture broth of marine-derived Trichoderma cf. brevicompactum and their structure activity relationship. Bioorg. Med. Chem. Lett. 30, 127375. doi: 10.1016/j.bmcl.2020.127375
Yang, D., Pomraning, K., Kopchinskiy, A., Karimi Aghcheh, R., Atanasova, L., Chenthamara, K., et al. (2015). Genome sequence and annotation of Trichoderma parareesei, the ancestor of the cellulase producer Trichoderma reesei. Genome Announc. 3 (4), e00885–15. doi: 10.1128/genomeA.00885-15
Zapparata, A., Baroncelli, R., Durling, M. B., Kubicek, C. P., Karlsson, M., Vannacci, G., et al. (2021). Fungal cross-talk: an integrated approach to study distance communication. Fungal Genet. Biol. 148, 103518. doi: 10.1016/j.fgb.2021.103518
Zeilinger, S., Gruber, S., Bansal, R., Mukherjee, P. K. (2016). Secondary metabolism in Trichoderma–chemistry meets genomics. Fungal Biol. Rev. 30, 74–90. doi: 10.1016/j.fbr.2016.05.001
Zheng, J., Bayram Akcapinar, G., Atanasova, L., Rahimi, M. J., Przylucka, A., Yang, D., et al. (2016). The neutral metallopeptidase NMP1 of Trichoderma guizhouense is required for mycotrophy and self-defence. Environ. Microbiol 18, 580–597. doi: 10.1111/1462-2920.12966
Zhang, J. L., Tang, W. L., Huang, Q. R., Li, Y. Z., Wei, M. L., Jiang, L. L., et al. (2021). Trichoderma: a treasure house of structurally diverse secondary metabolites with medicinal importance. Front. Microbiol., 12, 723828. doi: 10.3389/fmicb.2021.723828
Zhao, P., Ren, A., Dong, P., Sheng, Y., Chang, X., Zhang, X. (2018). The antimicrobial peptaibol trichokonin IV promotes plant growth and induces systemic resistance against botrytis cinerea infection in moth orchid. J. Phytopathol. 166 (5), 346–354.
Zhao, D. L., Zhang, X. F., Huang, R. H., Wang, D., Wang, X. Q., Li, Y. Q., et al. (2020). Antifungal nafuredin and epithiodiketopiperazine derivatives from the mangrove-derived fungus Trichoderma harzianum D13. Front. Microbiol. 11, 1495. doi: 10.3389/fmicb.2020.01495
Keywords: soil-borne phytopathogen, Trichoderma, molecular interaction, disease management, host plant
Citation: Dutta P, Mahanta M, Singh SB, Thakuria D, Deb L, Kumari A, Upamanya GK, Boruah S, Dey U, Mishra AK, Vanlaltani L, VijayReddy D, Heisnam P and Pandey AK (2023) Molecular interaction between plants and Trichoderma species against soil-borne plant pathogens. Front. Plant Sci. 14:1145715. doi: 10.3389/fpls.2023.1145715
Received: 16 January 2023; Accepted: 05 April 2023;
Published: 15 May 2023.
Edited by:
Raja Asad Ali Khan, Hainan University, ChinaReviewed by:
Minhui Li, South China Agricultural University, ChinaMiguel J. Beltran-Garcia, Universidad Autónoma de Guadalajara, Mexico
Copyright © 2023 Dutta, Mahanta, Singh, Thakuria, Deb, Kumari, Upamanya, Boruah, Dey, Mishra, Vanlaltani, VijayReddy, Heisnam and Pandey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pranab Dutta, cHJhbmFiZHV0dGE3NEBnbWFpbC5jb20=; Madhusmita Mahanta, bWFkaHVzbWl0YS5tYWhhbnRhMTJAZ21haWwuY29t
 Pranab Dutta
Pranab Dutta Madhusmita Mahanta
Madhusmita Mahanta Soibam Basanta Singh
Soibam Basanta Singh Dwipendra Thakuria
Dwipendra Thakuria Lipa Deb
Lipa Deb Arti Kumari
Arti Kumari Gunadhya K. Upamanya4
Gunadhya K. Upamanya4 Lydia Vanlaltani
Lydia Vanlaltani Dumpapenchala VijayReddy
Dumpapenchala VijayReddy Abhay K. Pandey
Abhay K. Pandey