- 1State Key Laboratory of Crop Gene Exploration and Utilization in Southwest China, Sichuan Agricultural University, Chengdu, China
- 2Ministry of Education Key Laboratory for Crop Genetic Resources and Improvement in Southwest China, Sichuan Agricultural University, Chengdu, China
Wheat is a significant source of protein and starch worldwide. The defective kernel (Dek) mutant AK-3537, displaying a large hollow area in the endosperm and shrunken grain, was obtained through ethyl methane sulfonate (EMS) treatment of the wheat cultivar Aikang 58 (AK58). The mode of inheritance of the AK-3537 grain Dek phenotype was determined to be recessive with a specific statistical significance level. We used bulked segregant RNA-seq (BSR-seq), BSA-based exome capture sequencing (BSE-seq), and the ΔSNP-index algorithm to identify candidate regions for the grain Dek phenotype. Two major candidate regions, DCR1 (Dek candidate region 1) and DCR2, were identified on chromosome 7A between 279.98 and 287.93 Mb and 565.34 and 568.59 Mb, respectively. Based on transcriptome analysis and previous reports, we designed KASP genotyping assays based on SNP variations in the candidate regions and speculated that the candidate gene is TraesCS7A03G0625900 (HMGS-7A), which encodes a 3-hydroxy-3-methylglutaryl-CoA synthase. One SNP variation located at position 1,049 in the coding sequence (G>A) causes an amino acid change from Gly to Asp. The research suggests that functional changes in HMGS-7A may affect the expression of key enzyme genes involved in wheat starch syntheses, such as GBSSII and SSIIIa.
Introduction
Wheat (Triticum aestivum L.) is a major global crop, providing approximately 20% of the total caloric intake for the world’s population (Ogbonnaya et al., 2013; Ma et al., 2022). Therefore, maintaining high grain quality and yield of wheat is essential for food security (Shiferaw et al., 2013). Wheat grain, the reproductive and storage organ, plays a crucial role in wheat propagation, spread, and yield and primarily consists of the embryo, endosperm, and seed coat (Ebrahimnejad and Rameeh, 2016). In wheat, grain filling refers to the process of starch biosynthesis and accumulation in the endosperm (Ahmed et al., 2015). Starch is the main component of wheat grain, accounting for 65%–70% of dry grain weight (Housley et al., 1981), and it significantly impacts wheat flour quality (Mancebo et al., 2015).
Starch synthesis in wheat grain starts after fertilization and continues until approximately 35 days, when the grain matures and dries (Khatun and Ahmed, 2015). When starch is synthesized in the endosperm, sucrose produces through leaf photosynthesis and enters the cytoplasm, serving as the carbon source for starch synthesis in wheat (Guo et al., 2015). Multiple factors influence wheat grain development, including wheat tissue organs (Barneix, 2007), starch synthesis-related enzymes, plant hormones, and environmental factors. Relevant tissue organs include leaves and stem sheaths. At the same time, starch synthesis-related enzymes encompass sucrose synthase (SuSy), ADP-glucose pyrophosphorylase (AGPase), granule-bound starch synthase (GBSS), starch synthase (SS), starch branching enzyme (SBE), starch debranching enzyme (DEB), starch phosphorylase (SP), and sucrose convertase (SC), among others (Wang et al., 2014). Plant hormones affecting wheat grain development include ethylene (ET), brassinosteroid (BR), gibberellin (GA), and abscisic acid (ABA), among others (Liu et al., 2013; Xiong et al., 2022). Environmental factors such as temperature, light, and soil moisture also play a role (Liu et al., 2016; Jiang et al., 2017; Djanaguiraman et al., 2020; Mirosavljević et al., 2021).
Defective kernel (Dek) mutants exhibit shrunken grains, and grain filling in crop Dek mutants is drastically impaired (Wang et al., 2017). Generally, the Dek phenotype reduces grain weight and significantly affects grain appearance and seed vigor (Li et al., 2017; Fu et al., 2019; Qi et al., 2019). Many Dek mutants have been identified in maize and rice, and numerous genetic loci regulating grain fullness have also been discovered. Dek mutants, such as Dek10, Dek35, Dek36, Dek37, Dek39, Dek40, and Dek42 (Chen et al., 2017; Qi et al., 2017; Wang et al., 2017; Dai et al., 2018; Li et al., 2018; Ren et al., 2019; Zuo et al., 2019), display germinated mutant kernels that are lethal during the seedling stage; Dek15, Dek38, Dek41, and Dek44 (Lid et al., 2002; Garcia et al., 2017; He et al., 2019; Qi et al., 2019; Zhu et al., 2019) seeds cannot germinate at all, resulting in lethal embryo mutations. For example, the Dek15 gene encodes sister chromatid cohesion protein 4 (SCC4), and mutation of this gene disrupts the cell cycle and nuclear replication, leading to the complete failure of seed germination (He et al., 2019). The Dek38 gene encodes TEL2-interaction protein 2 (TTI2) molecular chaperone protein, which affects the development of male germ cells (Garcia et al., 2017). The Dek1 gene, located in the 47.1 to 47.4 Mb region on chromosome 1 in maize, is involved in the differentiation and development of maize aleurone cells. A mutation of this gene leads to embryo lethality and affects the development of the aleurone layer and the accumulation of endosperm gliadin content (Lid et al., 2002; Song et al., 2020). At present, only a few Dek-related studies have been reported in Triticeae. Three QTLs, QDek.Caas-3BS.1, QDek.Caas-3BS.2, and QDek.Caas-4AL, associated with wheat grain Dek were identified using wheat mutant groups, explaining 14.78%–18.17%, 16.61%–21.83%, and 19.08%–28.19% of phenotypic variances, respectively (Fu et al., 2019). The loss-of-function mutation of the sex6 (SSIIa) gene on chromosome 7H in barley causes amylopectin synthesis to decrease to less than 20% of the wild-type level. Simultaneously, the mutation also affects the binding of starch synthetases SSI, SBEIIa, and SBEIIb to starch granules and ultimately causes barley grain to become shrunken (Dek) (Morell et al., 2003). The barley’s sex6 (SSIIa) mutant was crossed with the amo1 (SSIIIa) mutant to generate the sex6amo1 double mutant, which produces high-amylose starch. The level of granule-bound starch synthase I (GBSSI) protein in starch granules increased, and starch synthase I (SSI), SSIIa, starch branching enzyme IIa (SBEIIa), and SBEIIb also significantly increased in the starch granules. The double mutant’s Dek phenotype was restored to a normal grain phenotype, indicating that changes in starch synthase function in cereal crops can also lead to shrunken grains (Li et al., 2011). These genes are crucial for synthesizing and accumulating starch and protein in the endosperm. Therefore, excavating DeK-related genes will be conducive to improving crop grain yield and quality.
This study analyzed Aikang58 (AK58) and its Dek mutant line AK-3537 to investigate grain characteristics in different environments over several years. AK-3537 exhibited poor grain filling, collapsed abdominal grooves, and shrunken grains. Transcriptome analysis using RNA-seq was performed on the grains of AK58 and AK-3537, and an F2 population with 130 individuals was constructed using AK-3537 and AK58. Two high-confidence candidate regions, DCR1 (Dek candidate region 1) and DCR2, regulating wheat grain Dek, were mapped using BSE-seq (Bulked Segregant Exome Capture Sequencing) and BSR-seq (Bulked Segregant RNA-seq). We identified TraesCS7A03G0625900 in the DCR1 region as a candidate gene for the wheat grain Dek phenotype using KASP (Kompetitive Allele-Specific Polymerase Chain Reaction) markers, the Chinese Spring reference genome (RefSeq 2.1), and the AK58 genome. This study will contribute to a deeper understanding of the regulatory mechanisms underlying wheat grain morphology and provide new insights to improve wheat yield through breeding.
Materials and methods
Plant materials
The EMS (ethyl methane sulfonate) mutant Dek line AK-3537 originated from the wheat variety AK58. The Dek phenotype was stably inherited after eight generations of self-pollination. For genetic analysis and mapping, AK-3537 was crossed with AK58 to produce an F2 population of 130 individuals. Wheat materials were cultivated in experimental fields and greenhouses at Sichuan Agricultural University, Chengdu Chongzhou (103° 38’ E, 30° 32’ N), Wenjiang (103° 51’ E, 30° 43’ N), and Xishuangbanna (99° 56’ E, 21° 08’ N), China. All field trials were well irrigated and managed following local standard practices, and all AK-3537 × AK58 F2 plants were grown in a greenhouse at 20°C with a 16-h/8-h light/dark cycle.
In the field, a total of 20 plants from AK58 (10 individuals) and AK-3537 (10 individuals) were randomly selected to investigate agronomic traits, including plant height (PH), tiller number (TN), heading date (HD), flag leaf length (FLL), and flag leaf width (FLW), using the method reported in previous studies. Thousand kernel weight (TKW), grain length (GL), and grain width (GW) were also measured using previously reported methods (Liu et al., 2017). Excel 2019 (Microsoft, Redmond, WA, USA) was used to calculate the phenotypic data. Analysis of variance was conducted, and individuals were ranked through Duncan’s test and plotted using GraphPad Prism V9.0.0. R software (version 4.2.1) was used as a plotting tool to calculate the wheat Dek phenotypic data.
Identification of Dek phenotype
After maturity, wheat grains were harvested and threshed by hand. One hundred grains were randomly sampled from each individual, and the sampling was repeated three times. The wheat grains were visually examined for the Dek phenotype, and the incidence (percentage of grain Dek phenotype) was calculated. To better observe the wheat grain Dek phenotype, the grains of AK58 and AK-3537 were stored at 4°C in FAA (Formalin-Aceto-Alcohol) fixative (ensuring the kernels did not float on the surface of the fixative solution) during the wheat grain filling stage. Subsequently, the cell structures of the normal phenotype and Dek phenotype were observed using frozen and free-hand sections (Leica CM1860) and x-ray computed tomography (Micro-CT).
The BSE-seq and BSR-seq for rapid map Dek gene
(AK-3537 × AK58) F1 grains were observed at the mature grain period in the natural field. F2 populations were used for genetic analysis. We then performed a chi-square test (χ2) to test phenotypic data (grain Dek phenotype) for a goodness of fit to the ratio of 3:1 expected for a single gene (or semi-dominant) genetic basis in Excel 2019 by CHISQ.TEST function (p > 0.05 means no deviation from expectations of 3:1). Using the combined approaches of BSE-seq and BSR-seq, AK58, AK-3537, and the F2 population (grains with normal phenotype and Dek phenotype, with a mix of 30 random individuals each) were selected for DNA and RNA segregant pools. Leaves were collected for DNA extraction using the Plant Genomics DNA Extraction Kit (BIOFIT®, DN32-100, Chengdu, China), and grains (10–20 days post-anthesis, DPA) were sampled to extract total RNA using the Plant RNA Extraction Kit (BIOFIT®, RN34050, Chengdu, China). RNA sequencing generating 150 bp paired-end reads was performed on the Illumina HiSeq™ × platform. Clean RNA-seq data were mapped onto the Chinese Spring reference genome (RefSeq 2.1) and the AK58 genome (Jia J et al., 2021) using the software Bowtie2, and SNP calling was performed with the SAMtools software. The newly designed exome capture probe panel (Dong et al., 2020) and ΔSNP-index algorithm (Abe et al., 2012) were used to map the grain Dek gene in wheat rapidly. High-quality reads were aligned to the Chinese Spring reference genome (RefSeq 2.1) and the AK58 genome (Jia J et al., 2021) with default parameters. The parental AK58 sequencing data (DNA and RNA sequencing data) were used as a “background” to identify the causal mutation based on the assumption. Calculations were analyzed on PlantGmap (Zhang et al., 2021) (http://183.223.252.63:3333/).
Lastly, considering the characteristics of EMS mutagenesis, certain variations were filtered (Dong et al., 2020). Only the candidate regions identified in BSE-seq and BSR-seq were considered. Then, the Chinese Spring reference genome (RefSeq 2.1) was used to obtain the candidate gene, gene sequence, and gene annotation. Homologous analysis and gene expression patterns were evaluated on WheatOmics 1.0 (http://202.194.139.32/). Arabidopsis (TAIR10) and rice (IRGSP-1.0) genomes were used for comparative genomics analyses. A phylogenetic tree was constructed using MEGA11, and Geneious was employed to assemble high-quality reads.
Differentially expressed gene analysis
Grains of AK-3537 and AK58 during the grain filling stage (10 DPA) were collected. RNA extraction, library construction, and RNA sequencing were performed as described previously. Differentially expressed gene (DEG) analysis, Gene Ontology (GO) annotation, and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were adopted to confirm the putative biological functions and biochemical pathways of DEGs (AK-3537 and AK58) on the OmicsShare Tools (https://www.omicshare.com/).
Validation of KASP markers and quantitative reverse transcription polymerase chain reaction
DNA extraction was performed as previously described, and KASP markers were used for genotyping in parents and the F2 population. The experimental method for KASP markers was referenced from previous reports (Li et al., 2020). The quantitative reverse transcription polymerase chain reaction (qRT-PCR) experiment was conducted to detect the expression levels of key genes involved in the wheat starch synthesis pathway, including GBSSI, GBSSII, SBEI, SBEII, SSI, SSII, SSIIIa, SSIV, TaAGPL1, TaAGPS1a, TaBEI, TaBEIIa, TaBEIIb, and TaBEIII. The experimental method for qRT-PCR was referenced from previous reports (Li et al., 2022). Actin was selected as the reference gene (Wang et al., 2014), and the relative quantification formula (2−ΔΔCt) ± standard error of the mean (SEM) was used to evaluate quantitative variation further. Three biological replicates were tested for each sample. All primers used in this study are listed in Supplementary Table 1.
Results
Characterization of the Dek mutant phenotype
In a screening of wheat grain, we identified a Dek mutant, AK-3537, with grain shrunken from an ethyl methyl sulfide (EMS) mutant library in the AK58 background (Figure 1A). The grains of the wild-type AK58 were plump and normal (AK58 type), whereas AK-3537 showed a 100% grain Dek phenotype rate across all environments (AK-3537 type) (Figure 1). To further establish the genetic basis of this phenotype, a cross was performed between AK-3537 and AK58. All F1 plants exhibited a normal grain phenotype, suggesting the presence of a recessive gene controlling the wheat grain Dek phenotype. To assess AK-3537 mutation segregation, (AK-3537 × AK58) F1 plants were self-pollinated and an F2 segregating population (130 individuals) was developed. The F2 population was segregated into two categories: 95 plants exhibited an AK58-type phenotype, and 35 plants showed an AK-3537-type phenotype (Table 1). The results were in agreement with a 3:1 segregation ratio [χ2 = 0.26 < χ2 (0.05, 1) = 3.84, p = 0.61], suggesting that a single gene regulates grain Dek.
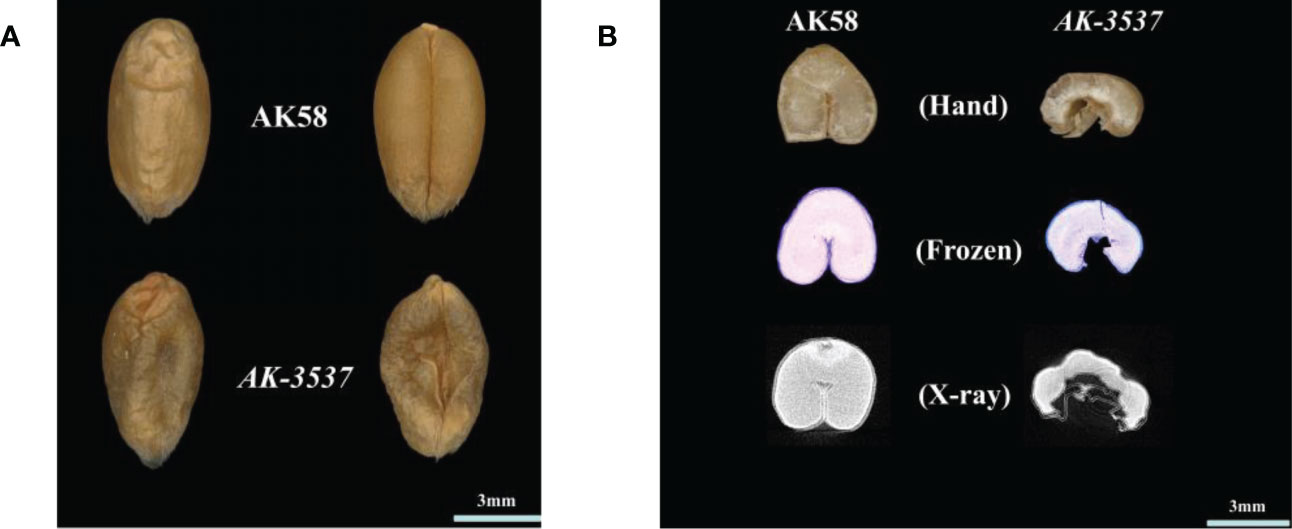
Figure 1 Wheat grain Dek phenotype and the internal structure of AK58 and AK-3537. (A) Dek phenotype of AK-3537 mature grain compared to AK58 normal grain. (B) The internal structure of AK58 and AK-3537 mature grains was observed by free-hand section (Hand), frozen section (Frozen), and x-ray tomography (X-ray).
Compared with that in AK58, a large hollow area was observed in the endosperm of AK-3537 by frozen and free-hand section, and the hollow phenotype was also observed in the Dek grain of AK-3537 by X-ray 3D tomography (Figure 1B). The results showed that AK-3537 grain filling was significantly affected. Finally, we evaluated the agronomic phenotypes of AK58 and AK-3537 in the field. Compared with AK58, FLW, TN, TKW, GL, and GW were all significantly decreased in AK-3537, whereas HD increased considerably (Figures 2A, B).
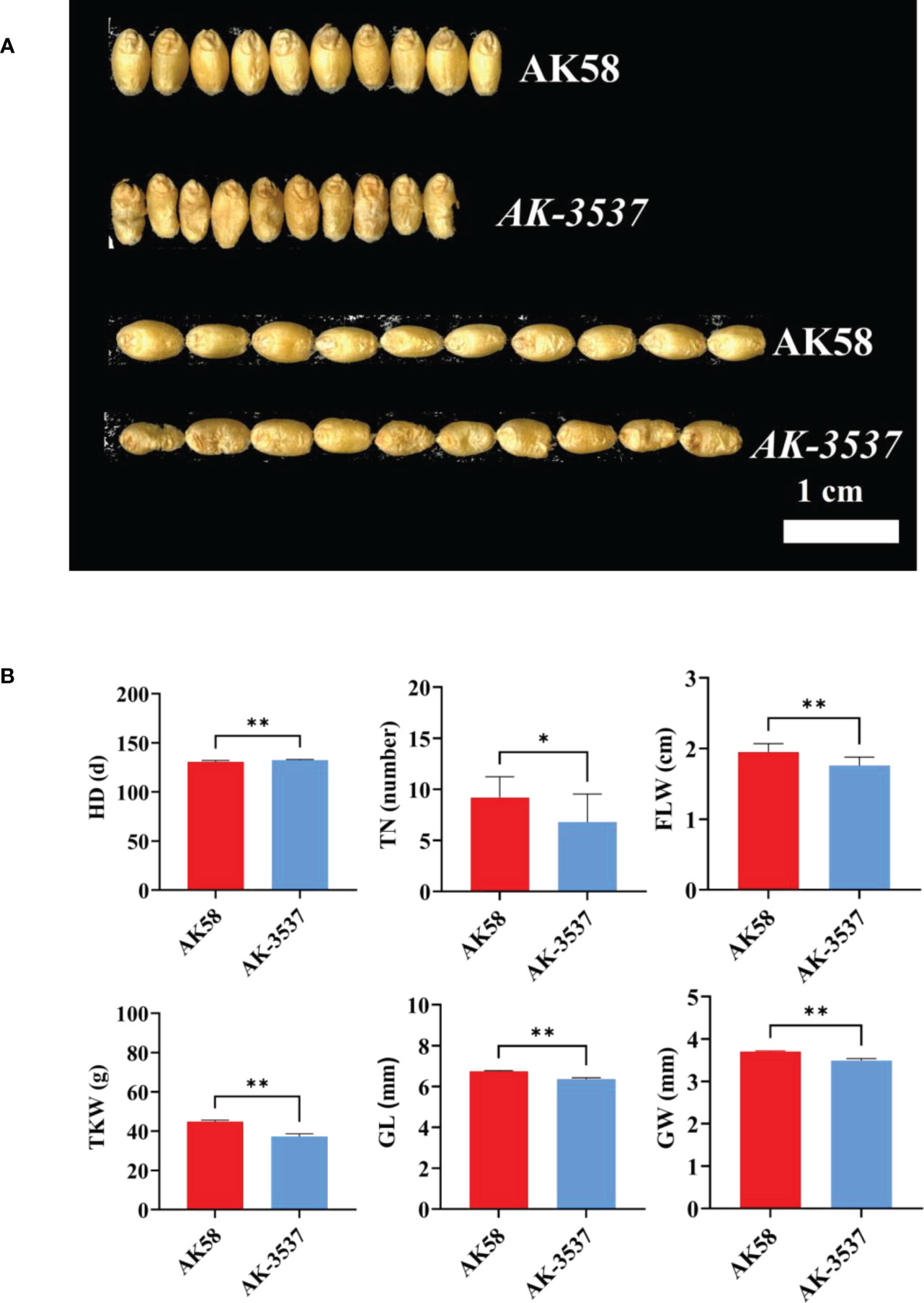
Figure 2 Comparison of grain and field phenotypes between AK58 and AK-3537. (A) Comparison of length and width phenotypes of AK-3537 defective kernel and AK58 normal grain. (B) Statistical analysis of field phenotypes and grain phenotypes of AK58 and AK-3537 including tiller number (TN), heading date (HD), flag leaf width (FLW), thousand kernel weight (TKW), grain length (GL), and grain width (GW). “*” means p < 0.05 and “**” means p < 0.01.
Dek gene mapping by BSE-seq and BSR-seq
For Dek gene mapping, two pooled samples, each comprising 30 normal and Dek phenotype F2 segregants, were constructed. Approximately 20 Gb of sequence data for the DNA pool and approximately 6 Gb for the RNA pool were generated and compared with the Chinese Spring reference genome (RefSeq V2.1) and AK58 genome. Then, we used the ΔSNP-index algorithm (Abe et al., 2012) for Dek gene mapping. ΔSNP-index higher than 0.7 in BSE-seq and BSR-seq Dek gene candidate region (DCR) was defined conservatively as the union of BSA-seq and BSR-seq credible intervals for candidate gene identification. In the AK58 genome, two DCRs, DCR1 and DCR2, were mapped on chromosome 7A by BSE-seq and BSR-seq (Table 2), among which DCR1 was located between 279.98 and 287.93 Mb on Chr7A with two SVs (structural variation) in two genes, and DCR2 was located between the range of 565.34 and 568.58 Mb on Chr7A with two SVs in two genes (Table 2). In the Chinese Spring reference genome (RefSeq V2.1), five DCRs, namely, DCR3, DCR4, DCR5, DCR6, and DCR7, were identified on chromosomes 1B, 7A, and 7B. DCR6 and DCR2 are the same candidate region and contain the same variant genes (Table 2). Because AK58 is a wheat–rye 1B/1R translocation line material, the genetic background of the 1B chromosome is very different between the Chinese Spring reference genome (RefSeq V2.1) and the AK58 genome (Jia J et al., 2021). It has also been reported that wheat chromosome 7B underwent structural rearrangement (Chen et al., 2020). Therefore, we speculated that DCR1 and DCR2 located on chromosome 7A were candidate regions for the wheat grain Dek phenotype, which was predicted to be a moderate functional effect (e.g., missense mutation).
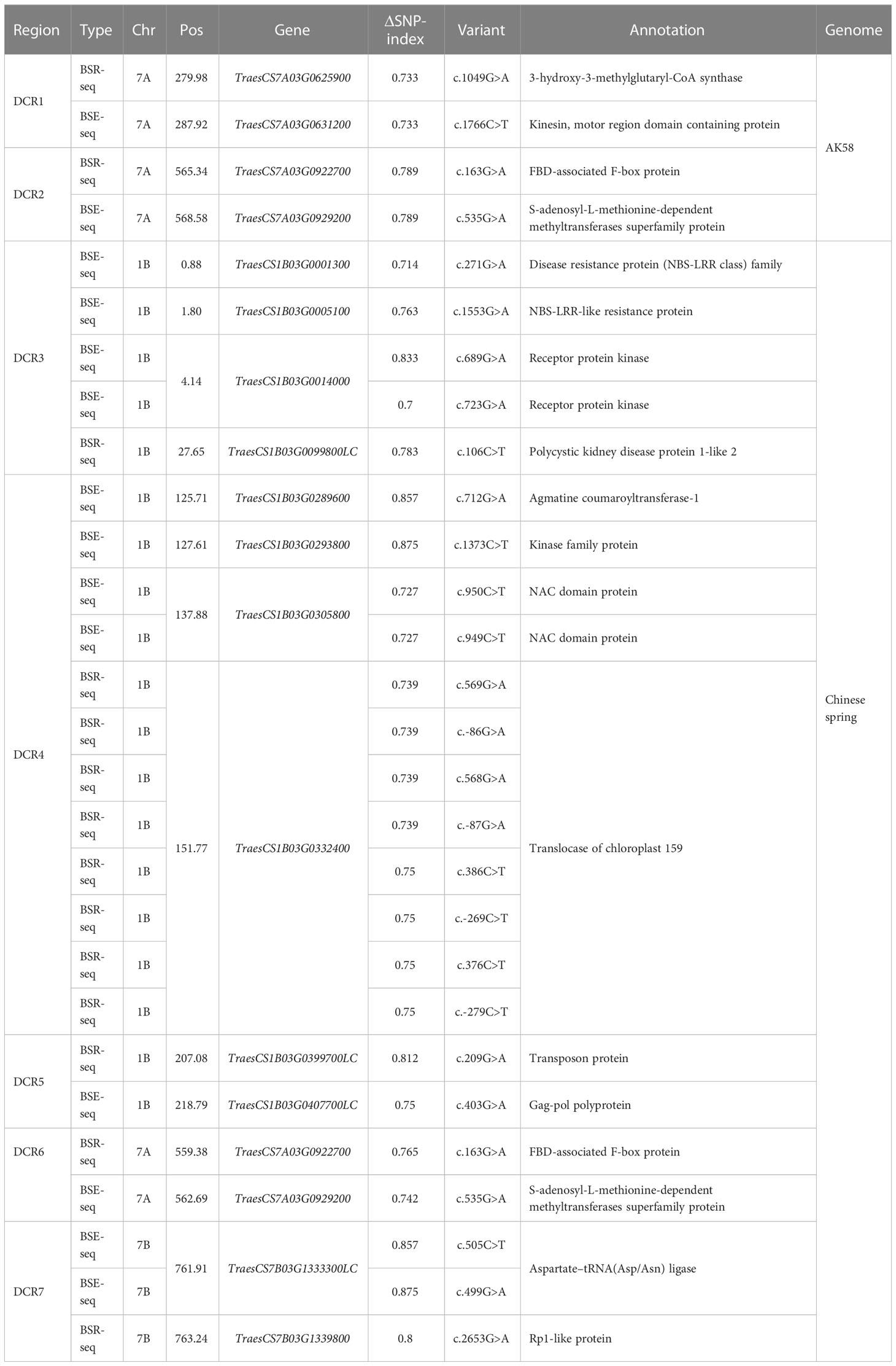
Table 2 The wheat grain Dek candidate region in the AK58 genome and the Chinese Spring reference genome (RefSeq 2.1).
Since a single gene regulates the Dek phenotype, we annotated the genes in the two candidate regions based on the wheat Chinese Spring RefSeq v2.1 genome. In DCR1, a gene encoding 3-hydroxy-3-methylglutaryl-CoA synthase (TraesCS7A03G0625900) and a gene encoding a kinesin-like protein (TraesCS7A03G0631200) were annotated. In DCR2, an FBD-associated F-box protein (TraesCS7A03G0922700) and an S-adenosyl-L-methionine-dependent methyltransferase superfamily protein (TraesCS7A03G0929200) were annotated (Figure 3; Table 2). Expression patterns of the candidate genes in the WheatOmics 1.0 (http://202.194.139.32/) database revealed that all four candidate genes were expressed in seeds.
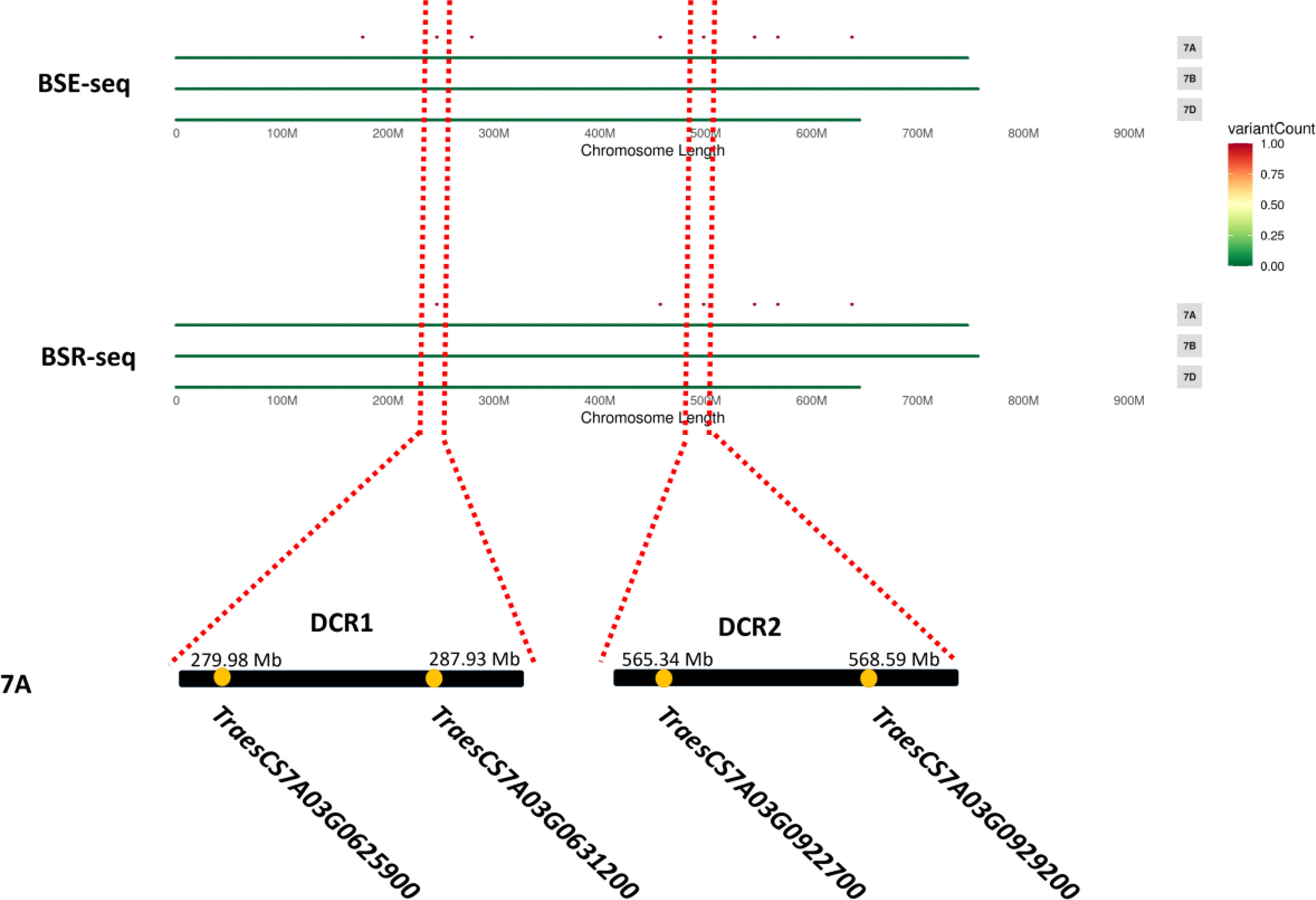
Figure 3 Most likely candidate region for wheat Dek detected via BSE-seq and BSR-seq. “.” means the SNPs associated with wheat grain Dek phenotype after the screening.
To further exclude SNP variations caused by sequence assembly errors, exome capture and RNA-seq data were used to assemble the sequences of the mutated genes in the DCR1 and DCR2 regions. The results showed that the SNP variations at TraesCS7A03G0631200 and TraesCS7A03G0922700 could be detected in both exome capture and RNA-seq data. However, the SNP of TraesCS7A03G0929200 could only be detected in exome capture data, while the SNP of TraesCS7A03G0625900 could only be detected in RNA-seq data (Figure 4; Supplementary Figure 1). To validate the segregation of these SNP variations in the F2 population, KASP markers were developed based on these four SNP variations in this study and validated in the F2 population. The results showed that only the SNP variation at position 1,049 of the TraesCS7A03G0625900 coding region co-segregated with the grain normal and Dek phenotype in the F2 population (Figure 5B). A query of the Chinese Spring RefSeq v2.1 genome found that the coding sequence of the TraesCS7A03G0625900 gene was 1,410 bp in length, and the full-length genome contained 12 exons and 11 introns. Sequencing analysis of the TraesCS7A03G0625900 coding region in AK-3537 revealed a G-to-A mutation at position 1,049, the 11th exon of the coding region, which resulted in an amino acid substitution from glycine (GGC, Gly, G) to aspartic acid (GAC, Asp, D) (Figure 5A). The target gene was tentatively termed HMGS-7A.

Figure 4 Candidate genes influencing wheat grain Dek phenotypes by the assembly of exome capture and RNA-seq sequencing data.
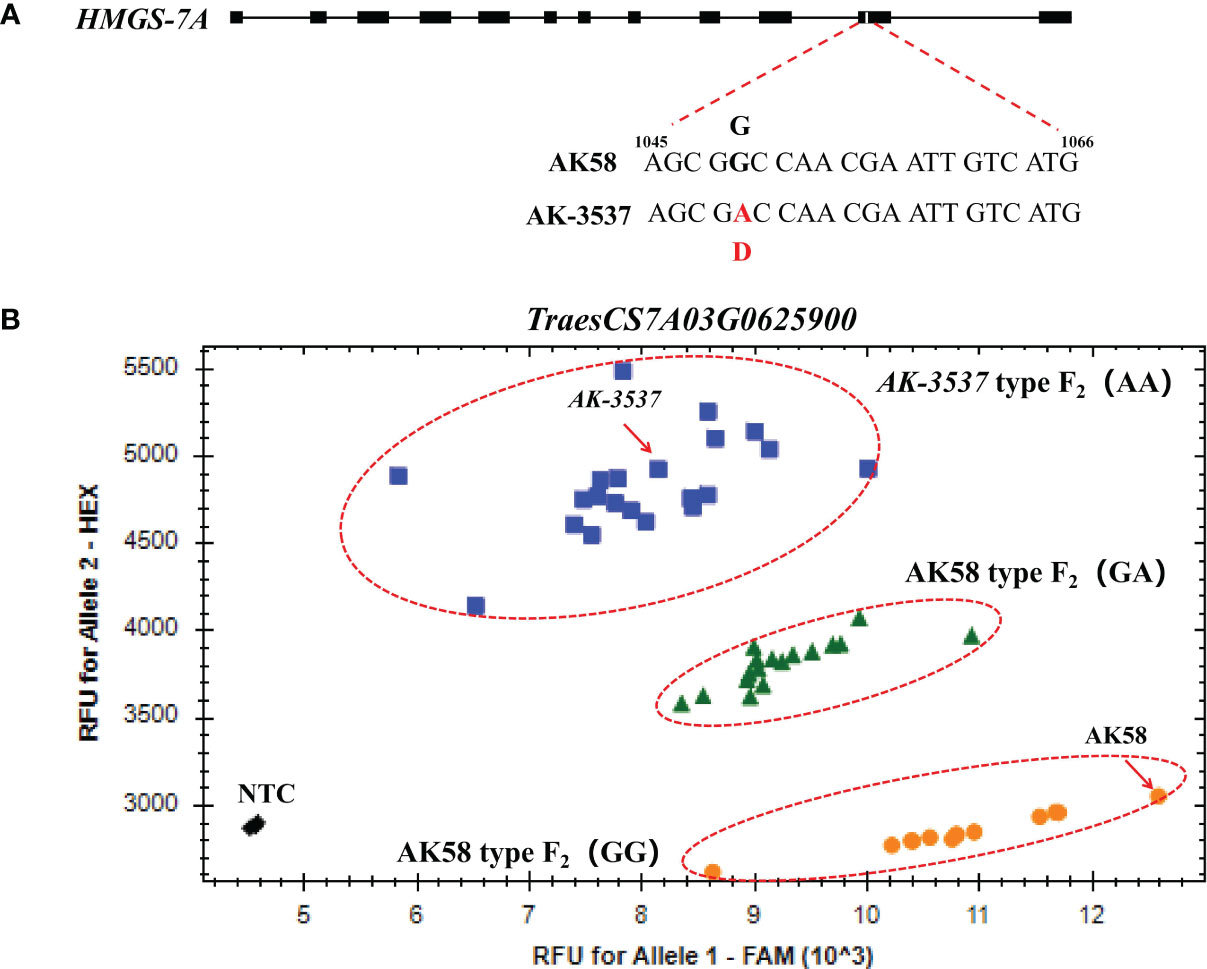
Figure 5 Mutation and marker verification of HMGS-7A gene. (A) Structure and mutation site of HMGS-7A. (B) HMGS-7A1049 was validated by the KASP marker.
Differentially expressed genes between AK58 and AK-3537 grains
In the transcriptome analysis of wheat grains of AK58 and AK-3537, a total of 12,655 DEGs were identified. Enrichment analysis of these DEGs in the wheat grain Dek regulatory pathway showed significant differences in gene expression levels between the mutant AK-3537 and the wild-type AK58, with 6,618 genes downregulated and 6,037 genes upregulated. GO and KEGG analyses were further performed on the screened DEGs to understand the functions and pathways of these DEGs. GO analysis showed that these DEGs were mainly concentrated in processes involved in carbohydrate metabolism (GO:0005975 and GO:0044723) (Figure 6A). KEGG analysis showed significant enrichment (p ≤ 0.05) of energy metabolism and starch synthesis pathway (Ko00500) (Figure 6B). This indicates that, compared to AK58 grains, AK-3537 grains have significant differences in wheat carbon metabolism, photosynthesis product synthesis, and starch synthesis pathways. These DEGs may cause the AK-3537 grain Dek phenotype. In previous reports, starch synthase also regulated similar phenotypes (Morell et al., 2003; Li et al., 2011). Therefore, this study also found that the expression levels of GBSSII and SSIIIa in the Ko00500 pathway were significantly increased in AK-3537 (Figure 7).

Figure 6 Differential expression analysis for AK58 and AK-3537 grain transcriptome. (A) GO analysis for DEGs. (B) KEGG analysis for DEGs.
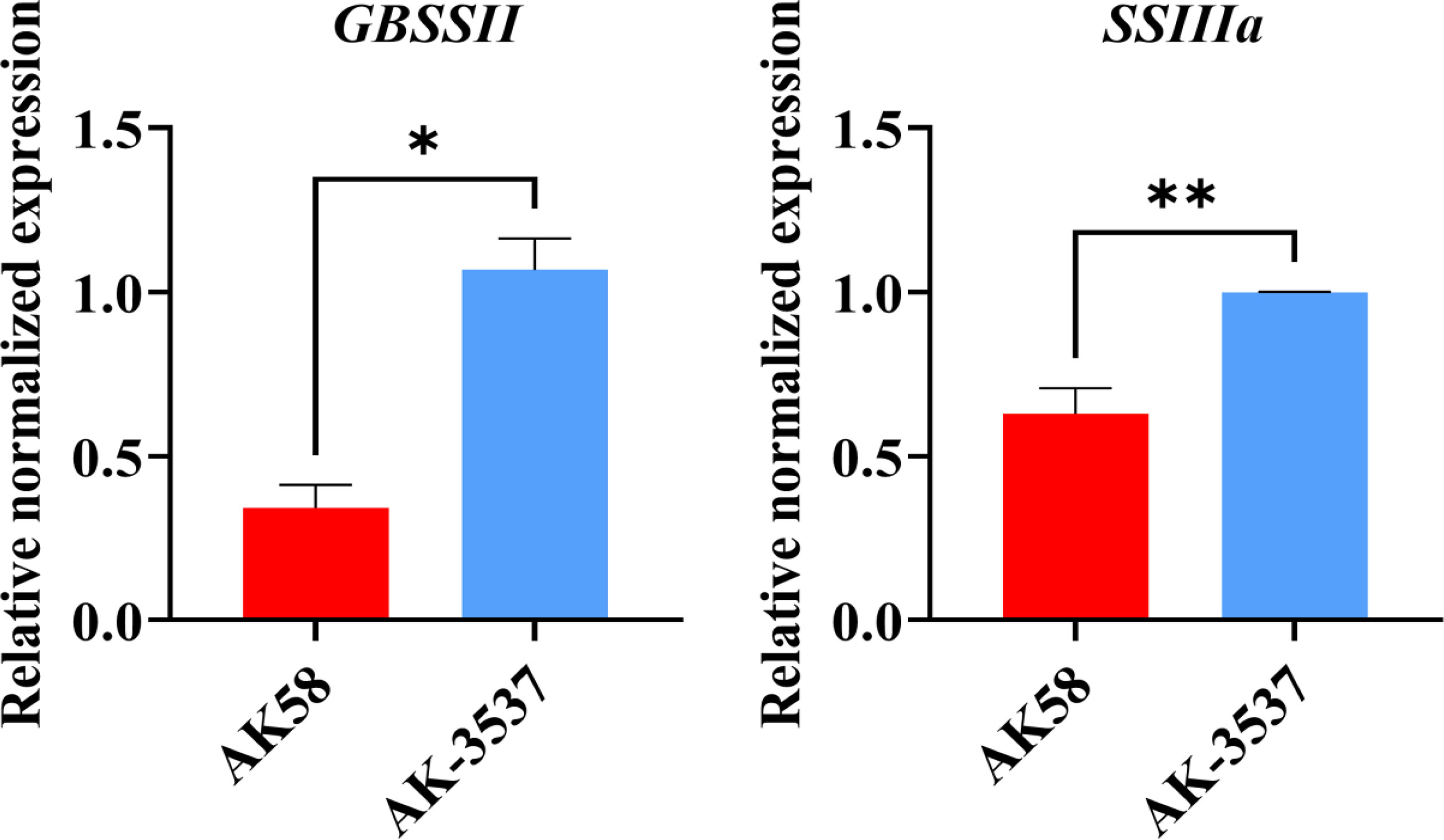
Figure 7 Statistical analysis of gene expression of starch synthetase GBSSII and SSIIIa.* means p < 0.05 and ** means p < 0.01.
Discussion
Wheat grain is the most important component of wheat yield and quality. In this study, we identified a candidate gene, TraesCS7A03G0625900 (HMGS-7A), encoding a 3-hydroxy-3-methylglutaryl-CoA synthase (HMGS), in AK58 and AK-3537 by BSE-seq and BSR-seq, responsible for the Dek phenotype in wheat grain. The G-to-A mutation at position 1,049 of the HMGS-7A coding region leads to amino acid change from glycine (Gly) to aspartic acid (Asp). In previous studies, several genes related to grain size and weight have been mapped and cloned on wheat chromosome 7A, including TaTPP-7A (7A, 135.0 Mb; Liu et al., 2023), TaGASR7 (7A, 176.0 Mb; Zhang et al., 2015), TaTGW-7A (7A, 211.6 Mb; Hu et al., 2016), TaGW8 (7A, 257.3 Mb; Yan et al., 2019), and TaIAA21 (7A, 488.5 Mb; Jia M et al., 2021). The TraesCS7A03G0625900 (HMGS-7A) gene identified in this study does not overlap with these reported grain morphology genes. Therefore, it is speculated to be a novel gene controlling wheat grain Dek phenotype.
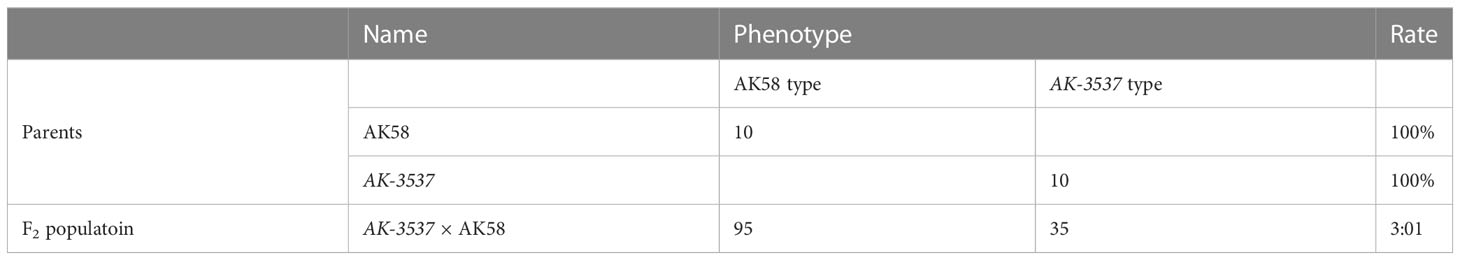
The coding sequences of TraesCS7A03G0625900 (HMGS-7A), and its two homologous genes, TraesCS7B03G0451200 (HMGS-7B) and TraesCS7D02G269600 (HMGS-7D), were obtained from Ensembl plants (https://plants.ensembl.org/index.html) and used to construct a phylogenetic tree with HMGS coding sequences from rice, Arabidopsis, and Brassica (Figure 8). The results showed that the HMGS in wheat is more closely related to that in rice. The importance of HMGS in Arabidopsis has been demonstrated in steroid biosynthesis, pollen fertility, and seed weight (Ishiguro et al., 2010; Bhangu-Uhlmann, 2011; Lange et al., 2015). HMGS is the second key enzyme in the mevalonate (MVA) pathway, significantly affecting plant sterol biosynthesis (Wang et al., 2012; Liao et al., 2014a; Lange et al., 2015; Pérez et al., 2022). Brassinosteroids (BR) is one of the main types of sterols (Zhang et al., 2020), which plays a vital role in the grain-filling process of plants. Transferring the gene encoding C-22 hydroxylase, an enzyme involved in sterol biosynthesis, into rice significantly increased the BR hormone content and the TKW, and the increase in TKW resulted from BR stimulating the transport of photosynthates in rice (Wu et al., 2008). Knockout of TaD11-2A results in dwarfism, a significant decrease in endogenous BR content, and smaller grains in wheat (Xu et al., 2022). TaBRI1 is the BR receptor gene in wheat, and TaBRI1 knockout mutants were insensitive to exogenous BR and significantly reduced TKW (Fang et al., 2020). GW5 is a positive regulator of BR signaling, expressed in various rice organs, considerably affecting the width and weight of rice grains, and is a feasible target for increasing grain yield in rice and other cereal crops through gene editing (Liu et al., 2017). SMG3 and DGS1 regulate the size and weight of rice grains through the BR signaling pathway. Loss of SMG3 or DGS1 function results in smaller grains, while overexpression of SMG3 or DGS1 leads to longer grains (Li et al., 2023).
The sterol content in plant grains may also affect the starch synthesis pathway. A study of sbeIIb mutants in rice showed that many starch synthesis enzyme genes were upregulated, except for genes encoding granule-bound starch synthase, branching enzyme, pullulanase, and starch phosphorylase, which were downregulated. This increased amylose and resistant starch content, in addition to an increase in many other substances such as sugar, fatty acids, amino acids, and plant sterols in the endosperm (Baysal et al., 2020), and the wheat mutant SM482gs, with increased grain size, TKW, and protein content with BR biosynthesis and signal transduction, were significantly upregulated, but AGP-S1, AGP-L2, SSI, SSIIa, SSIIIa, SBEIIa, SBEIIb, and GBSSIa show the lower expression on SM482gs (Zhong et al., 2021), which indicated that plant sterols might be involved in the synthesis of amylose in plant grains. In rice, overexpression of HMGS significantly increased fatty acids, abscisic acid, gibberellins, and lutein in transgenic rice (Pérez et al., 2022), while overexpression of HMGS in mustard significantly increased grain weight (Liao et al., 2014b). In barley, mutations in the starch synthase genes SSIIa and SSIIIa result in grain phenotypes similar to those observed in this study with AK-3537 (Li et al., 2011). Therefore, in this study, we detected the expression levels of key genes involved in starch synthesis in seeds and found that the key gene SSIIIa, which regulates the content of amylose and amylopectin in plants, was highly expressed in AK-3537, indicating that the functional changes of HMGS-7A may affect the expression of key enzyme genes involved in wheat starch synthesis. In the future, we will further analyze HMGS-7A and verify the role of HMGS-7A in wheat grain filling.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
JW and HT designed research; HT performed experiments; HT and MC analyzed the data, ML, ZY, and ZP prepared the plant materials; HT, XG, HD, QC, and JW wrote and revised the paper; JW supervised the project. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Major Program of National Agricultural Science and Technology of China (NK20220607), the National Natural Science Foundation of China (U22A20472), the National Key Research and Development Program of China (2018YFE0112000), the Sichuan Science and Technology Support Project (2021YFH0077, 2021YFYZ0027, and 23NSFSC0770), the Science and Technology Support Project of Chengdu (2021-GH03-00002-HZ), and the open research fund of SKL-CGEUSC (SKL-ZD202212).
Acknowledgments
The authors thank the reviewers and editors for their suggestions to improve this manuscript. The authors thank Professor Jizeng Jia from the Institute of Crop Science, Chinese Academy of Agricultural Science, for sharing the wheat mutation lines.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1173861/full#supplementary-material
Supplementary Figure 1 | Candidate genes influencing wheat grain Dek phenotypes by assembly of exon capture and RNA-seq sequencing data.
References
Abe, A., Kosugi, S., Yoshida, K., Natsume, S., Takagi, H., Kanzaki, H., et al. (2012). Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30, 174–178. doi: 10.1038/nbt.2095
Ahmed, N., Tetlow, I. J., Nawaz, S., Iqbal, A., Mubin, M., Rehman, S., et al. (2015). Effect of high temperature on grain filling period, yield, amylose content and activity of starch biosynthesis enzymes in endosperm of basmati rice. J. Sci. Food Agric. 95, 2237–2243. doi: 10.1002/jsfa.6941
Barneix, A. J. (2007). Physiology and biochemistry of source-regulated protein accumulation in the wheat grain. J. Plant Physiol. 164, 581–590. doi: 10.1016/j.jplph.2006.03.009
Baysal, C., He, W., Drapal, M., Villorbina, G., Medina, V., Capell, T., et al. (2020). Inactivation of rice starch branching enzyme IIb triggers broad and unexpected changes in metabolism by transcriptional reprogramming. Proc. Natl. Acad. Sci. U. S. A. 117, 26503–26512. doi: 10.1073/pnas.2014860117
Bhangu-Uhlmann, A. (2011). “The mevalonate pathway: a monitoring approach in plants by systems biology tools,” in Doctoral dissertation, ETH Zurich (Zurich, Swiss Confederation: ETH Zurich Research Collection). doi: 10.3929/ethz-a-006690303
Chen, X., Feng, F., Qi, W., Xu, L., Yao, D., Wang, Q., et al. (2017). Dek35 encodes a PPR protein that affects cis-splicing of mitochondrial nad4 intron 1 and seed development in maize. Mol. Plant 10, 427–441. doi: 10.1016/j.molp.2016.08.008
Chen, Y., Song, W., Xie, X., Wang, Z., Guan, P., Peng, H., et al. (2020). A collinearity-incorporating homology inference strategy for connecting emerging assemblies in the triticeae tribe as a pilot practice in the plant pangenomic era. Mol. Plant 13, 1694–1708. doi: 10.1016/j.molp.2020.09.019
Dai, D., Luan, S., Chen, X., Wang, Q., Feng, Y., Zhu, C., et al. (2018). Maize Dek37 encodes a p-type PPR protein that affects cis-splicing of mitochondrial nad2 intron 1 and seed development. Genetics 208, 1069–1082. doi: 10.1534/genetics.117.300602
Djanaguiraman, M., Narayanan, S., Erdayani, E., Prasad, P. V. (2020). Effects of high temperature stress during anthesis and grain filling periods on photosynthesis, lipids and grain yield in wheat. BMC Plant Biol. 20, 1–12. doi: 10.1186/s12870-020-02479-0
Dong, C., Zhang, L., Chen, Z., Xia, C., Gu., Y., Wang, J., et al. (2020). Combining a new exome capture panel with an effective varbscore algorithm accelerates bsa-based gene cloning in wheat. Plant Sci. 11. doi: 10.3389/fpls.2020.01249
Ebrahimnejad, S., Rameeh, V. (2016). Correlation and factor analysis of grain yield and some important component characters in spring bread wheat genotypes. Cerce. Agron. Mol. 49, 5–15. doi: 10.1515/cerce-2016-0001
Fang, J., Zhu, W., Tong, Y. (2020). Knock-down the expression of brassinosteroid receptor TaBRI1 reduces photosynthesis, tolerance to high light and high temperature stresses and grain yield in wheat. Plants 9, 840. doi: 10.3390/plants9070840
Fu, C., Du, J., Tian, X., He, Z., Fu, L., Wang, Y., et al. (2019). Rapid identification and characterization of genetic loci for defective kernel in bread wheat. BMC Plant Biol. 19, 483. doi: 10.1186/s12870-019-2102-6
Garcia, N., Li, Y., Dooner, H. K., Messing, J. (2017). Maize defective kernel mutant generated by insertion of a ds element in a gene encoding a highly conserved TTI2 cochaperone. Proc. Natl. Acad. Sci. U. S. A. 114, 5165–5170. doi: 10.1073/pnas.1703498114
Guo, M., Zhao, G., Guo, W., Chang, X., Wang, D., Yang, Y., et al. (2015). Effect of different sowing dates on characteristics of flag leaf photosynthesis and grain filling of wheat cultivars with different gluten. J. Triticeae Crops. 35, 192–197. doi: 10.7606/j.issn.1009-1041.2015.02.07
He, Y., Wang, J., Qi, W., Song, R. (2019). Maize dek15 encodes the cohesin-loading complex subunit SCC4 and is essential for chromosome segregation and kernel development. Plant Cell 31, 465–485. doi: 10.1105/tpc.18.00921
Housley, T. L., Kirleis, A. W., Ohm, H. W., Patterson, F. L. (1981). An evaluation of seed growth in soft red winter wheat. Can. J. Plant Sci. 61, 525–534. doi: 10.4141/cjps81-075
Hu, M. J., Zhang, H. P., Liu, K., Cao, J. J., Wang, S. X., Jiang, H., et al. (2016). Cloning and characterization of TaTGW-7A gene associated with grain weight in wheat via SLAF-seq-BSA. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01902
Ishiguro, S., Nishimori, Y., Yamada, M., Saito, H., Suzuki, T., Nakagawa, T., et al. (2010). The arabidopsis FLAKY POLLEN 1 gene encodes a 3-hydroxy-3-methylglutaryl-coenzyme a synthase required for development of tapetum-specific organelles and fertility of pollen grains. Plant Cell Physiol. 51, 896–911. doi: 10.1093/pcp/pcq068
Jia, M., Li, Y., Wang, Z., Tao, S., Sun, G., Kong, X., et al. (2021). TaIAA21 represses TaARF25-mediated expression of TaERFs required for grain size and weight development in wheat. Plant J. 108 (6), 1754–1767. doi: 10.1111/tpj.15541
Jia, J., Xie, Y., Cheng, J., Kong, C., Wang, M., Gao, L., et al. (2021). Homology-mediated inter-chromosomal interactions in hexaploid wheat lead to specific subgenome territories following polyploidization and introgression. Genome Biol. 22, 1–21. doi: 10.1186/s13059-020-02225-7
Jiang, X., Cheng, H., Jiang, L., Yang, X., Lv, R., Hua, M., et al. (2017). Effect of increasing diffuse radiation fraction under low light condition on the grain-filling process of winter wheat (Triticum aestivum l.). Chin. J. Agrom. 38, 753–760. doi: 10.3969/j.issn.1000-6362.2017.12.001
Khatun, S., Ahmed, J.U (2015). Response of elevated temperature on carbohydrate accumulation and grain yield in different wheat cultivars. Bangladesh J. Agril. Res. 40, 205–215. doi: 10.3329/bjar.v40i2.24558
Lange, I., Poirier, B. C., Herron, B. K., Lange, B. M. (2015). Comprehensive assessment of transcriptional regulation facilitates metabolic engineering of isoprenoid accumulation in arabidopsis. Plant Physiol. 169, 1595–1606. doi: 10.1104/pp.15.00573
Li, J., Fu, J., Chen, Y., Fan, K., He, C., Zhang, Z., et al. (2017). The U6 biogenesis-like 1 plays an important role in maize kernel and seedling development by affecting the 3′ end processing of U6 snRNA. Mol. Plant 10, 470–482. doi: 10.1016/j.molp.2016.10.016
Li, X., Gu, W., Sun, S., Chen, Z., Chen, J., Song, W., et al. (2018). Defective kernel 39 encodes a PPR protein required for seed development in maize. J. Integr. Plant Biol. 60, 45–64. doi: 10.1111/jipb.12602
Li, Z., Li, D., Du, X., Wang, H., Larroque, O., Jenkins, C. L., et al. (2011). The barley amo1 locus is tightly linked to the starch synthase IIIa gene and negatively regulates expression of granule-bound starch synthetic genes. J. Exp. Bot. 62, 5217–5231. doi: 10.1093/jxb/err239
Li, C., Liu, H., Wang, J., Pan, Q., Wang, Y., Wu, K., et al. (2022). Characterization and fine mapping of a lesion mimic mutant (Lm5) with enhanced stripe rust and powdery mildew resistance in bread wheat (Triticum aestivum l.). Theor. Appl. Genet. 135, 1–18. doi: 10.1007/s00122-021-03973-1
Li, C., Tang, H., Luo, W., Zhang, X., Mu, Y., Deng, M., et al. (2020). A novel, validated, and plant height-independent QTL for spike extension length is associated with yield-related traits in wheat. Theor. Appl. Genet. 133, 3381–3393. doi: 10.1007/s00122-020-03675-0
Li, J., Zhang, B., Duan, P., Yan, L., Yu, H., Zhang, L., et al. (2023). An endoplasmic reticulum-associated degradation–related E2–E3 enzyme pair controls grain size and weight through the brassinosteroid signaling pathway in rice. Plant Cell 35, 1076–1091. doi: 10.1093/plcell/koac364
Liao, P., Wang, H., Hemmerlin, A., Nagegowda, D. A., Bach, T. J., Wang, M.. (2014a). Past achievements, current status and future perspectives of studies on 3-hydroxy-3-methylglutaryl-CoA synthase (HMGS) in the mevalonate (MVA) pathway. Plant Cell Rep. 33, 1005–1022. doi: 10.1007/s00299-014-1592-9
Liao, P., Wang, H., Wang, M., Hsiao, A. S., Bach, T. J., Chye, M. L., et al. (2014b). Transgenic tobacco overexpressing Brassica juncea HMG-CoA synthase 1 shows increased plant growth, pod size and seed yield. PloS One 9, e98264. doi: 10.1371/journal.pone.0108026
Lid, S. E., Gruis, D., Jung, R., Lorentzen, J. A., Ananiev, E., Chamberlin, M., et al. (2002). The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proc. Natl. Acad. Sci. U. S. A. 99, 5460–5465. doi: 10.1073/pnas.042098799
Liu, J., Chen, J., Zheng, X., Wu, F., Lin, Q., Heng, Y., et al. (2017). GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants 3, 1–7. doi: 10.1038/nplants.2017.43
Liu, Y., Gu, D., Wu, W., Wen, X., Liao, Y. (2013). The relationship between polyamines and hormones in the regulation of wheat grain filling. PloS One 8, e78196. doi: 10.1371/journal.pone.0078196
Liu, Y., Liang, H., Lv, X., Liu, D., Wen, X., Liao, Y. (2016). Effect of polyamines on the grain filling of wheat under drought stress. Plant Physiol. Biochem. 100, 113–129. doi: 10.1016/j.plaphy.2016.01.003
Liu, Y., Lin, Y., Gao, S., Li, Z., Ma, J., Deng, M., et al. (2017). A genome-wide association study of 23 agronomic traits in Chinese wheat landraces. Plant J. 91, 861–873. doi: 10.1111/tpj.13614
Liu, H., Si, X., Wang, Z., Cao, L., Gao, L., Zhou, X., et al. (2023). TaTPP-7A positively feedback regulates grain filling and wheat grain yield through T6P-SnRK1 signalling pathway and sugar–ABA interaction. Plant Biotechnol. J., 1–17. doi: 10.1111/pbi.14025
Ma, J., Liu, Y., Zhang, P., Chen, T., Tian, T., Wang, P. (2022). Identification of quantitative trait loci (QTL) and meta-QTL analysis for kernel size-related traits in wheat (Triticum aestivum l.). BMC Plant Biol. 22, 1–18. doi: 10.1186/s12870-022-03989-9
Mancebo, C. M., Merino, C., Martínez, M. M., Gómez, M. (2015). Mixture design of rice flour, maize starch and wheat starch for optimization of gluten free bread quality. J. Food Sci. Technol. 52, 6323–6333. doi: 10.1007/s13197-015-1769-4
Mirosavljević, M., Mikić, S., Župunski, V., Kondić Špika, A., Trkulja, D., Ottosen, C. O., et al. (2021). Effects of high temperature during anthesis and grain filling on physiological characteristics of winter wheat cultivars. J. Agron. Crop Sci. 207, 823–832. doi: 10.1111/jac.12546
Morell, M. K., Kosar-Hashemi, B., Cmiel, M., Samuel, M. S., Chandler, P., Rahman, S., et al. (2003). Barley sex6 mutants lack starch synthase IIa activity and contain a starch with novel properties. Plant J. 34, 173–185. doi: 10.1046/j.1365-313X.2003.01712.x
Ogbonnaya, F. C., Abdalla, O., Mujeebkazi, A., Kazi, A. G., Xu, S., Gosman, N., et al. (2013). Synthetic hexaploids: harnessing species of the primary gene pool for wheat improvement. Plant Breed. Rev. 37, 35–122. doi: 10.1002/9781118497869.ch2
Pérez, L., Alves, R., Perez-Fons, L., Albacete, A., Farré, G., Soto, E., et al. (2022). Multilevel interactions between native and ectopic isoprenoid pathways affect global metabolism in rice. Transgenic Res. 31, 249–268. doi: 10.1007/s11248-022-00299-6
Qi, W., Lu, L., Huang, S., Song, R. (2019). Maize dek44 encodes mitochondrial ribosomal protein l9 and is required for seed development. Plant Physiol. 180, 2106–2119. doi: 10.1104/pp.19.00546
Qi, W., Tian, Z., Lu, L., Chen, X., Chen, X., Zhang, W., et al. (2017). Editing of mitochondrial transcripts nad3 and cox2 by Dek10 is essential for mitochondrial function and maize plant development. Genetics 205, 1489–1501. doi: 10.1534/genetics.116.199331
Ren, R., Lu, X., Zhao, Y., Wei, Y., Wang, L., Zhang, L., et al. (2019). Pentatricopeptide repeat protein DEK40 is required for mitochondrial function and kernel development in maize. J. Exp. Bot. 70, 6163–6179. doi: 10.1093/jxb/erz391
Shiferaw, B., Smale, M., Braun, H. J., Duveiller, E., Reynolds, M., Muricho, G. (2013). Crops that feed the world 10. past successes and future challenges to the role played by wheat in global food security. Food Secur. 5, 291–317. doi: 10.1007/s12571-013-0263-y
Song, X., Hu, S., Zhang, K., Cui, Z., Li, J., Yang, X., et al. (2020). Phenotypic analysis and fine mapping of dek101 in maize. Acta Agron. Sin. 46, 1831–1838. doi: 10.3724/SP.J.1006.2020.03017
Wang, Z., Li, W., Qi, J., Shi, P., Yin, Y. (2014). Starch accumulation, activities of key enzyme and gene expression in starch synthesis of wheat endosperm with different starch contents. J. Food Sci. Technol. 51, 419–429. doi: 10.1007/s13197-011-0520-z
Wang, H., Nagegowda, D. A., Rawat, R., Bouvier-Navé, P., Guo, D., Bach, T. J., et al. (2012). Overexpression of brassica juncea wild-type and mutant HMG-CoA synthase 1 in arabidopsis up-regulates genes in sterol biosynthesis and enhances sterol production and stress tolerance. Plant Biotechnol. J. 10, 31–42. doi: 10.1111/j.1467-7652.2011.00631.
Wang, G., Zhong, M., Shuai, B., Song, J., Zhang, J., Han, L., et al. (2017). E+ subgroup PPR protein defective kernel 36 is required for multiple mitochondrial transcripts editing and seed development in maize and arabidopsis. New Phytol. 214, 1563–1578. doi: 10.1111/nph.14507
Wu, C. Y., Trieu, A., Radhakrishnan, P., Kwok, S. F., Harris, S., Zhang, K., et al. (2008). Brassinosteroids regulate grain filling in rice. Plant Cell 20, 2130–2145. doi: 10.1105/tpc.107.055087
Xiong, M., Feng, G., Gao, Q., Zhang, C., Li, Q., Liu, Q. (2022). Brassinosteroid regulation in rice seed biology. Seed Biol. 1, 1–9. doi: 10.48130/SeedBio-2022-0002
Xu, H., Sun, H., Dong, J., Ma, C., Li, J., Li, Z., et al. (2022). The brassinosteroid biosynthesis gene TaD11-2A controls grain size and its elite haplotype improves wheat grain yields. Theor. Appl. Genet. 135, 2907–2923. doi: 10.1007/s00122-022-04158-0
Yan, X., Zhao, L., Ren, Y., Dong, Z., Cui, D., Chen, F. (2019). Genome-wide association study revealed that the TaGW8 gene was associated with kernel size in Chinese bread wheat. Sci. Rep. 9, 1–10. doi: 10.1038/s41598-019-38570-2
Zhang, L., Dong, C., Chen, Z., Gui, L., Chen, C., Li, D., et al. (2021). WheatGmap: a comprehensive platform for wheat gene mapping and genomic studies. Mol. Plant 14, 187–190. doi: 10.1016/j.molp.2020.11.018
Zhang, X., Lin, K., Li, Y. (2020). Highlights to phytosterols accumulation and equilibrium in plants: biosynthetic pathway and feedback regulation. Plant Physiol. Biochem. 155, 637–649. doi: 10.1016/j.plaphy.2020.08.021
Zhang, D., Wang, B., Zhao, J., Zhao, X., Zhang, L., Liu, D., et al. (2015). Divergence in homoeolog expression of the grain length-associated gene GASR7 during wheat allohexaploidization. Crop J. 3, 1–9. doi: 10.1016/j.cj.2014.08.005
Zhong, X., Lin, N., Ding, J., Yang, Q., Lan, J., Tang, H., et al. (2021). Genome-wide transcriptome profiling indicates the putative mechanism underlying enhanced grain size in a wheat mutant. 3 Biotech. 11, 1–16. doi: 10.1007/s13205-020-02579-6
Zhu, C., Jin, G., Fang, P., Zhang, Y., Feng, X., Tang, Y., et al. (2019). Maize pentatricopeptide repeat protein DEK41 affects cis-splicing of mitochondrial nad4 intron 3 and is required for normal seed development. J. Exp. Bot. 70, 3795–3808. doi: 10.1093/jxb/erz193
Keywords: BSE-seq, BSR-seq, AK58, exome capture, HMGS-7A
Citation: Tang H, Dong H, Guo X, Cheng M, Li M, Chen Q, Yuan Z, Pu Z and Wang J (2023) Identification of candidate gene for the defective kernel phenotype using bulked segregant RNA and exome capture sequencing methods in wheat. Front. Plant Sci. 14:1173861. doi: 10.3389/fpls.2023.1173861
Received: 25 February 2023; Accepted: 03 May 2023;
Published: 05 June 2023.
Edited by:
Reyazul Rouf Mir, Sher-e-Kashmir University of Agricultural Sciences and Technology, IndiaReviewed by:
Tian Li, Chinese Academy of Agricultural Sciences, ChinaYanhao Xu, Hubei Academy of Agricultural Sciences, China
Copyright © 2023 Tang, Dong, Guo, Cheng, Li, Chen, Yuan, Pu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jirui Wang, aXNwaHNjQHNpY2F1LmVkdS5jbg==, d2FuZ2ppcnVpQGdtYWlsLmNvbQ==
 Hao Tang
Hao Tang Huixue Dong1
Huixue Dong1 Zhongwei Yuan
Zhongwei Yuan Zhien Pu
Zhien Pu Jirui Wang
Jirui Wang