- 1Key Laboratory of Biology and Genetic Improvement of Oil Crops, Ministry of Agriculture and Rural Affairs, Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences, Wuhan, China
- 2Hubei Hongshan Laboratory, Wuhan, China
- 3Hubei Key Laboratory of Food Crop Germplasm and Genetic Improvement, Key Laboratory of Crop Molecular Breeding, Ministry of Agriculture and Rural Affairs, Institute of Food Crops, Hubei Academy of Agricultural Sciences, Wuhan, China
Diverse molecular processes regulate the interactions between insect herbivores and their host plants. When plants are exposed to insects, elicitors induce plant defenses, and complex physiological and biochemical processes are triggered, such as the activation of the jasmonic acid (JA) and salicylic acid (SA) pathways, Ca2+ flux, reactive oxygen species (ROS) burst, mitogen-activated protein kinase (MAPK) activation, and other responses. For better adaptation, insects secrete a large number of effectors to interfere with plant defenses on multiple levels. In plants, resistance (R) proteins have evolved to recognize effectors and trigger stronger defense responses. However, only a few effectors recognized by R proteins have been identified until now. Multi-omics approaches for high-throughput elicitor/effector identification and functional characterization have been developed. In this review, we mainly highlight the recent advances in the identification of the elicitors and effectors secreted by insects and their target proteins in plants and discuss their underlying molecular mechanisms, which will provide new inspiration for controlling these insect pests.
Introduction
Plants are constantly being attacked by various insects. Nearly half a million insect species live on plants (Wu and Baldwin, 2010). The vast majority of herbivorous insects feed on plants from a single taxonomic family or a few closely related plant species specifically, while only 10% of them establish intimacy with multiple plant species (Schoonhoven et al., 2005). In addition to the direct damage caused by feeding, insects can also injure plants indirectly by transmitting viral, bacterial, and fungal pathogens. The main strategy for crop protection against insects over the past several decades was the application of chemical insecticides. However, due to the emergence of insect resistance to pesticides and the negative effect on the environment, the use of such compounds has declined in recent years (Du et al., 2020). Scientists have begun to unravel the molecular mechanisms underpinning the interactions between plants and insects in order to find better ways to control these pests.
Over the years, evidence has been accumulated during the long-term interaction and evolution of plants and insects, and both host plants and insect herbivores have obtained diverse sophisticated mechanisms to adapt to each other. In general, the perception of insect attack is the first step of plant defenses. Insect elicitors are the biologically active molecules from insects’ saliva or gut regurgitant; they are recognized by plants and subsequently induce plant defenses (Chen and Mao, 2020; Snoeck et al., 2022). These elicitors are also called herbivore-associated molecular patterns (HAMPs) (Snoeck et al., 2022). The elicitor-induced defenses include depolarization of the plasma trans-membrane potential, activation of JA and SA pathways, ROS burst, callose deposition, Ca2+ influx, MAPK activation, etc. (Erb and Reymond, 2019; Ye et al., 2019; Li et al., 2019a). For successful infestation, insect herbivores secret salivary molecules into plant cells to weaken their defense responses; these active molecules are called effectors (Mutti et al., 2008; Bos et al., 2010; Hogenhout and Bos, 2011; Naessens et al., 2015; Rodriguez et al., 2017). Effectors that suppress the plant’s responses can be recognized by their corresponding resistance proteins, inducing a second layer of defense, the effector-triggered immunity (ETI) (Jones and Dangl, 2006; Takken and Tameling, 2009). Notably, the second layer of defense response is much more fierce than the first layer. In summary, the active molecules from insect secretion have a significant impact on plant immunity. The molecules that can trigger plant defense responses are defined as elicitors, while those that weaken plant defenses are called effectors (Chen and Mao, 2020). In this review, we mainly discuss the recent advances in research on elicitors and effectors secreted by insects and their roles in the interactions between insects and their host plants. Dissecting the plant host factors and pathways targeted by these active insect molecules will facilitate the characterization of the molecular mechanisms of plant-insect interactions.
Herbivore feeding behaviors
To obtain nutrients from the hosts, insects employ diverse feeding strategies upon landing. Based on the different mouthparts and feeding habits, herbivorous insects can be divided into two groups: chewing and piercing-sucking insects (Schoonhoven et al., 1998; Walling, 2000). The insect species that cause damage with mouthparts evolved for chewing, snipping, or tearing belong to chewing insects, like leaf-eating beetles, caterpillars, or cotton bollworms. Chewing insects have a chewing type of mouth, which consists of the labrum, mandibles, first maxillae, second maxillae, hypopharynx, and epipharynx. The rectangular flap-like labrum is in the middle. The mandibles are paired and bear toothed edges at their inner surfaces; they masticate food using two sets of muscles transversely. The first maxillae and second maxillae are paired. The first maxillae are responsible for holding food and the second maxillae are responsible for pushing masticated food into the mouth. The hypopharynx has a single median tongue-like process, and the opening of the salivary duct lies under the hypopharynx. The epipharynx with taste buds is a single small membranous piece at the base of the labrum (Kahl, 1982; Felton et al., 1999; Stotz et al., 1999). Oral secretion (OS, consisting of regurgitant and saliva) of chewing insects contains active molecules that have a big impact on plant defense responses that are distinguishable from general mechanical damage (Hogenhout and Bos, 2011; Chen and Mao, 2020).
Piercing-sucking herbivorous insects, such as aphids, whiteflies, and planthoppers feed on plants through specially adapted mouthparts known as stylets, which they use to puncture the plant surface to access the phloem sap. The mouthparts of piercing-sucking insects are composed of the labrum, the labium, and the stylet. Among them, the stylet is used for piercing and sucking phloem sap from plants (Sogawa, 1982; Backus, 1988). The feeding strategies of piercing-sucking insects are mainly divided into three major phases, labial exploration, stylet penetration, and phloem-sap sucking (Spiller, 1990; Hao et al., 2008; Cheng et al., 2013b; Will et al., 2013). During their initial encounter with their host plants, insects walk rapidly and dab repeatedly on the plant’s surface to find a suitable feeding site, which is essential for the survival of the insects (Sogawa, 1982; Backus, 1988; Walling, 2008). Rice leaf sheath surface is featured in units and subunit structures, including the silico-phellem block, stomate block, large tubercle block, and vein, which are often covered with tubercle papicles, little papicles, glochids, and tenuous hairs. Recent research has shown that the brown planthopper (Nilaparvata lugens Stål, BPH), the most destructive pest of rice, preferentially selects the smooth long-cell block to probe their stylets into the leaf sheaths (Shi et al., 2021). Using an Atomic Force Microscope (AFM), Shi et al. (2021) found that the surface hardness of the long-cell block was much lower than that of the other cells. Sensilla basiconica, arranged symmetrically in two separate areas at the distal segment of the labium, was speculated to have a mechano-receptive function (Sogawa, 1982; Backus, 1988). Thus, we suppose that the labium may guide the stylets to find the suitable feeding site by sensing the mechanical heterogeneity of different structures on the plant surface.
Piercing-sucking insects penetrate plants with their stylet and move the stylet toward the phloem (Will et al., 2013). Along the stylet track, different types of cells are regularly penetrated (Will et al., 2013). Sucrose and pH are suggested to be indicators of phloem penetration (Hewer et al., 2010; Hewer et al., 2011). During the penetration process, piercing-sucking insects secrete both gelling and watery saliva from their salivary glands into the plant cells, and the protein compositions of the two types of saliva were shown to have some overlap (Walling, 2008; Huang et al., 2016). The secreted gelling saliva quickly solidifies and forms a continuous salivary sheath along its stylets for providing mechanical stability and protection (Wang et al., 2008). Some secretary proteins have been proven to be the key factors for forming the salivary sheath (Will and Vilcinskas, 2015; Huang et al., 2016; Huang et al., 2017; Shangguan et al., 2018; Huang et al., 2023). Watery saliva contains many active molecules that are involved in the induction or suppression of defenses against insect attack, i.e., the elicitors and effectors (Kaloshian and Walling, 2016; Chen and Mao, 2020).
Multi-omics approach to identifying elicitors and effectors
Saliva is a complex mixture of biomolecules with potential roles in encounters with plant immune responses (Miles, 1999; Will et al., 2013). Functional approaches such as proteomics and transcriptomics have facilitated the high-throughput identification of elicitors/effectors in regurgitant or saliva from various insect species (Harmel et al., 2008; Bos et al., 2010; Cooper et al., 2011; Nicholson et al., 2012; Ji et al., 2013; Nicholson and Puterka, 2014; Huang et al., 2016; Liu X. et al., 2016; Huang et al., 2018; Rao et al., 2019; Huang et al., 2023). The majority of the reported elicitors or effectors discussed below were identified using multi-omics approaches. Here, we take the salivary proteome and transcriptome of N. lugens as examples. Through comparative transcriptome analysis of the salivary glands of TN1 and Mudgo populations, 352 genes were predicted to encode secretory proteins (Ji et al., 2013). Among them, endo-β-1,4-glucanase (NlEG1) and NlSEF1 play important roles in rice-BPH interactions (Ji et al., 2017; Ye et al., 2017). Huang et al. (2016) performed proteomic analyses combined with genomic and transcriptomic analysis and identified 202 secreted salivary proteins in N. lugens. RNA interference revealed that salivap-3 is required for forming the salivary sheath, while annexin-like5 and carbonic anhydrase are indispensable for BPH survival (Huang et al., 2016). Recently, 1140 protein-coding genes were predicted in the secretome of N. lugens by Rao et al. (2019). Sequence analysis and homology searches revealed the presence of both conserved and rapidly evolving salivary proteins. Furthermore, six N. lugens secreted elicitors (Nl12, Nl16, Nl28, Nl32, Nl40, and Nl43) were identified by a series of predictions and functional analysis, as discussed below (Rao et al., 2019). The high-throughput identification of these secreted salivary proteins provides the possibility of understanding some aspects of plant-insect molecular interaction mechanisms and identifying potential targets for pest management.
Insect-associated elicitors
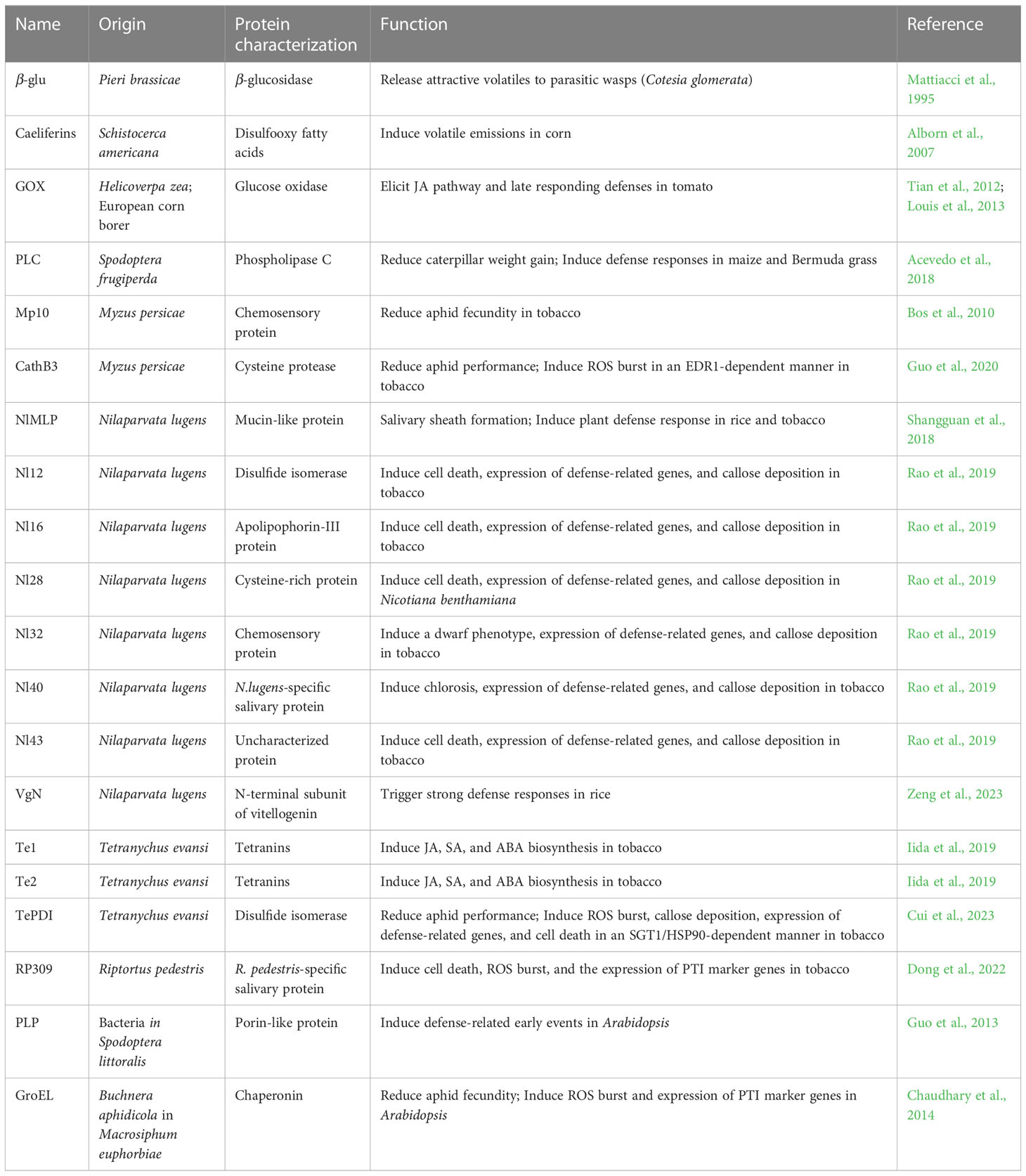
In general, plants can recognize elicitors and produce a complex series of defenses. The first reported elicitor β-glucosidase was isolated from the regurgitant of the white butterfly (Pieri brassicae). Leaves treated with β-glucosidase enhanced the emission of volatiles that are highly attractive to the parasitic wasp (Mattiacci et al., 1995). The glucose oxidase (GOX) present in the saliva extracted from Noctuid caterpillars (Helicoverpa zea) and European corn borer (Ostrinia nubilalis) upregulates the expression of genes related to the JA biosynthesis pathway and the late responding defense, such as proteinase inhibitor 2 (Pin2) in tomato (Tian et al., 2012; Louis et al., 2013). External spraying of phospholipase C (PLC), a salivary protein from fall armyworm (Spodoptera frugiperda), activates defense responses in maize and Bermuda grass and reduces caterpillar weight gain (Acevedo et al., 2018).
Except for the elicitors isolated from chewing insects, some elicitors were identified in piercing-sucking insects. Mp10 and Mp42 were two elicitors that were identified using a functional genomics approach in aphids. Aphid fecundity decreased when feeding on plants over-expressing Mp10 and Mp42. In addition, Mp10 specifically induced chlorosis in N. benthamiana leaves in a SUPPRESSOR OF G2 ALLELE OF skp1 (SGT1)-dependent manner (Bos et al., 2010; Rodriguez et al., 2014). Cysteine protease Cathepsin B3 (CathB3) was also recognized as a potential elicitor protein, which suppresses aphid feeding by triggering ROS through interacting with an ENHANCED DISEASE RESISTANCE 1-like (EDR1-like) protein (Guo et al., 2020). The mucin-like salivary protein (NlMLP) is a dual-functional protein both for insects and plants. In BPH, NlMLP is required for the formation of salivary sheath. In plants, NlMLP induces cell death, the expression of defense-related genes, and callose deposition (Shangguan et al., 2018). When BPH feed or oviposit, the small N-terminal subunit of vitellogenins (VgN) induces strong defenses, such as ROS burst and other responses in rice (Zeng et al., 2023). The ectopic expression of six secreted salivary proteins from BPH (Nl12, Nl16, Nl28, Nl32, Nl40, and Nl43) could induce cell death, chlorosis, or a dwarf phenotype, respectively in N. benthamiana leaves (Rao et al., 2019). Some salivary proteins from other piercing-sucking insects were also identified as the elicitors, like Tet1, Tet2, disulfide isomerase (TetPDI) from spider mite (Tetranychus evansi) (Iida et al., 2019; Cui et al., 2023), and RP309 from Fabricius (Riptortus pedestris) (Dong et al., 2022). It is noteworthy that although elicitor-induced plant defenses impair the performance of insects on plants, RNA interference (RNAi) experiments have revealed that elicitors are still essential for the survival of insects (Shangguan et al., 2018; Guo et al., 2020; Cui et al., 2023; Zeng et al., 2023).
In addition to the elicitors coming from insects themselves, some elicitors are generated from the microbes they carry. Buchnera aphidicola is the endosymbiont of potato aphids (Macrosiphum euphorbiae). Over-expression of Buchnera GroEL in Arabidopsis plants induces ROS burst and PTI, which is associated with the BRASSINOSTEROID INSENSITIVE1-ASSOCIATED RECEPTOR KINASE 1 (BAK1), thus reducing the fecundity of the aphid (Chaudhary et al., 2014). A porin-like protein (PLP) from bacteria in oral secretions of Spodoptera littoralis larvae induces Ca2+ flux in vitro and upregulates the calmodulin-like CML42 (Guo et al., 2013).
Some elicitors are relatively conserved in their ability to induce responses across a range of plant species. Both Nl32 in planthopper and MP10 in aphids are chemosensory proteins (CSPs), small water-soluble proteins with an OS-D domain that are conserved among different insects (Pelosi et al., 2005; Bos et al., 2010; Rao et al., 2019). Eleven CSPs (NlCSP-1 to -11) were previously identified in BPH, and six out of the eleven CSPs induced similar effects on N. benthamiana to those caused by Nl32 and Mp10 (Bos et al., 2010; Zhou et al., 2015; Rao et al., 2019). Nl12 and TetPDI, deriving from planthopper and spider mite, respectively, are the two members of the conserved disulfide isomerase family in eukaryotic organisms (Rao et al., 2019; Cui et al., 2023). Moreover, PDIs from phylogenetically distinct herbivorous and non-herbivorous arthropods could induce plant immunity in an SGT1/HSP90-dependent way (Cui et al., 2023). GOX was also conserved among caterpillar species (Tian et al., 2012; Louis et al., 2013). In Figure 1 and Table 1, we summarize the reported insect-associated elicitors from different species and their respective roles.
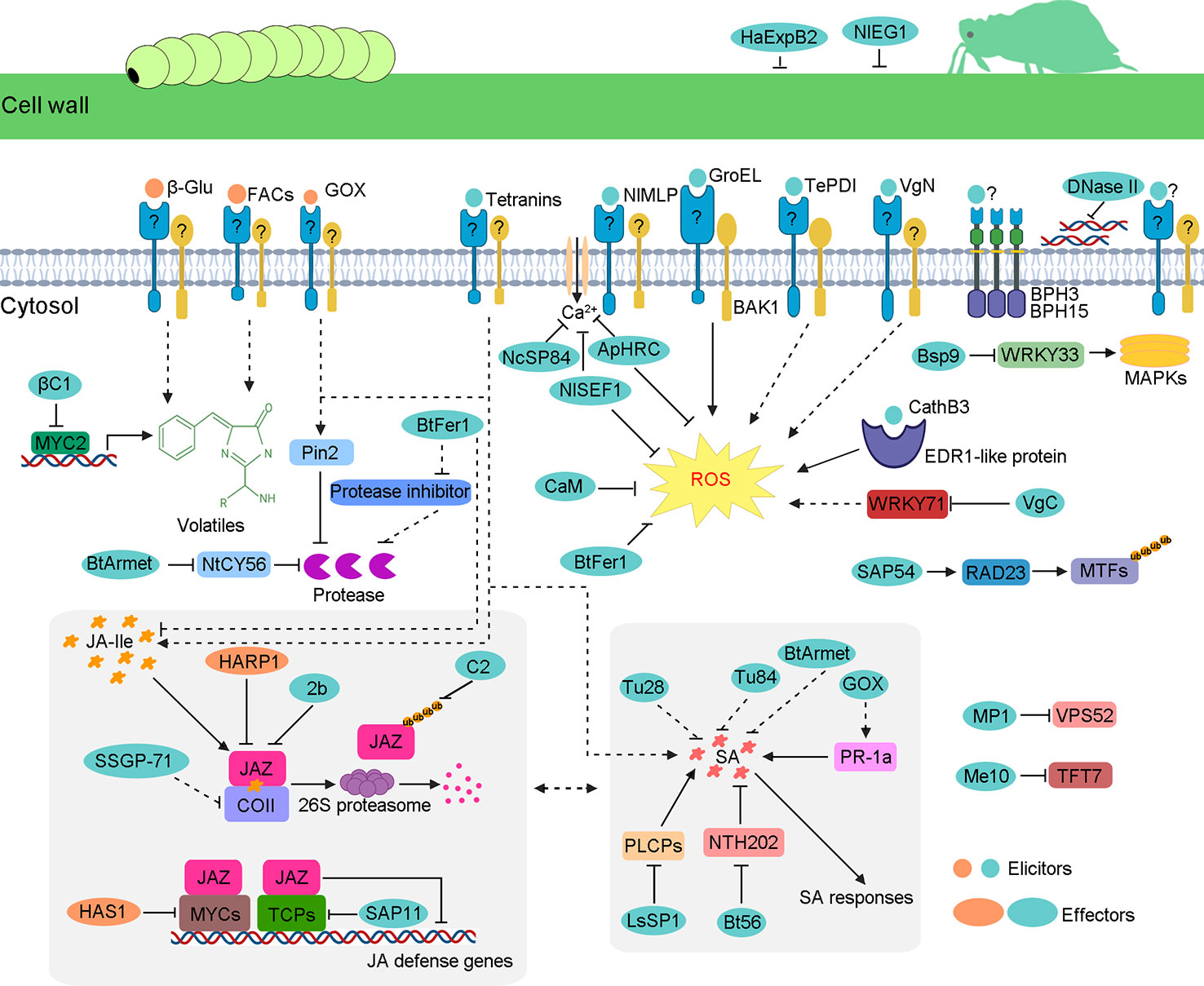
Figure 1 Schematic model of insect-secreted elicitors and effectors regulating plant defenses. When insects feed on plants, elicitors induce a complex series of plant defenses, such as ROS burst; upregulation of JA, SA and some volatile; and other unknown responses. However, insects secrete effectors to suppress these defense responses. Some effectors weaken JA pathways, including HARP1, HAS1, 2b, C2, βC1, SAP11, SSGP-71, and BtFer1. Some effectors interfere with SA pathways, such as BtArmet, Bt56, GOX, and LsSP1. The same effector protein can participate in diverse defense pathways. For example, as well as the reduction of the Ca2+ influx, ApHRC and NlSEF1 also suppress ROS burst. The B. tabaci effector BtFer1 not only reduces the accumulation of ROS and JA but also weakens protease inhibitor activity, thus increasing the content of protease to help whitefly feed better. Similar to BtFer1, effector BtArmet interacts with protease inhibitor NtCY56 to block the inhibition of whitefly protease. Mp1 and Me10 target plant proteins VPS52 and TFT7, respectively, which are required for insect resistance. The DNase II targets the extracellular DNA that is released by damaged cells. NlEG1 and HaExPB2 enable the insect’s stylet to reach the phloem by degrading celluloses in host plant cell walls.
Effectors involved in plant-insect interactions
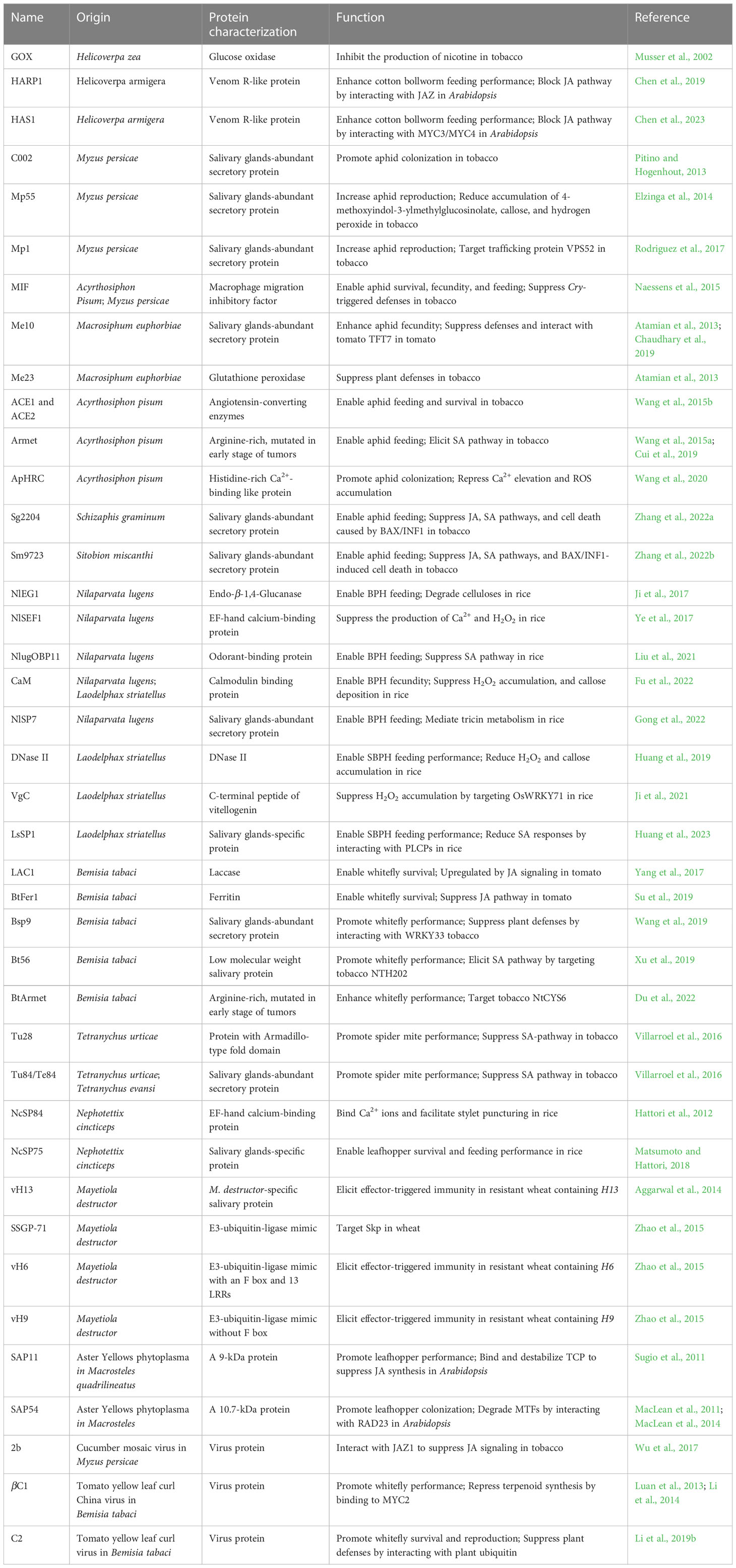
To adapt to their host plants, insects secrete a repertoire of effectors to disturb host plant defense responses (Figure 1 and Table 2). GOX from caterpillar H. zea was the first reported insect effector. The nicotine accumulation was suppressed significantly by GOX in tobacco (Musser et al., 2002; Musser et al., 2005). Interestingly, the same GOX was characterized as an elicitor in the ‘Insect-associated elicitors’ section because it induces plant responses in tomato (Tian et al., 2012; Louis et al., 2013). These results indicate that the same protein can act as the effector or as the elicitor when encountering different host plants. The cotton bollworm (Helicoverpa armigera) is a destructive lepidopteran insect widely existing in agriculture. Chen et al. (2019) identified an effector, a venom-like protein termed HARP1, from the OS of H. armigera. HARP1 stabilizes JAZ degradation and blocks wound-induced JA signaling transduction by forming a protein complex with JAZ. The weight of H. armigera larvae was increased significantly on transgenic plants with high-level HARP1 (Chen et al., 2019). HAS1 is another effector of H. armigera. Plants over-expressing HAS1 exhibit more susceptibility to insect herbivores accompanied by the suppressed JA pathway due to the interactions between HAS1 and JASMONATE-ZIM-domain repressors MYC3/MYC4 (Chen et al., 2023). These results indicate that interfering with the JA pathway is a common strategy of effectors in chewing insects.
Like those in chewing insects, effectors in piercing-sucking insects also disturb plant hormone-related defense pathways. Bt56 from the whitefly (Bemisia tabaci) increases susceptibility to insects by enhancing the accumulation of SA but not JA. Interaction assays have shown that Bt56 interacts directly with a KNOTTED 1-like homeobox transcription factor NTH202 (Xu et al., 2019). The survival rate and fecundity were significantly lower in insects injected with dsBt56 than in those injected with dsGFP (Xu et al., 2019). BtArmet, another effector of the B. tabaci, increased whitefly performance on tobacco plants by suppressing SA accumulation and binding to the cystatin NtCYS6, a protease inhibitor that prevents insects from continuous ingestion and digestion (Du et al., 2022). BtFer1 is a B. tabaci salivary protein with Fe2+ binding ability. The results showed that BtFer1 suppressed the JA-mediated signaling pathway, ROS burst, callose deposition, and accumulation of proteinase inhibitors (Su et al., 2019). The small brown planthopper (Laodelphax striatellus, SBPH) effector LsSP1 not only binds to sheath protein LsMLP to avoid LsMLP protein being recognized by plants but also interacts with rice papain-like cysteine proteases to inhibit SA biosynthesis and SA-related defenses (Huang et al., 2023).
Moreover, some effectors were reported to target other defense-related pathways in plants. The L. striatellus secretes effector protein DNase II to inhibit defense responses by erasing extracellular DNA and reducing hydrogen peroxide (Huang et al., 2019). Interestingly, unlike the VgN, as a reliable elicitor, the C-terminal peptide of vitellogenin (VgC) acts as a novel effector in L. striatellus, which attenuates H2O2-mediated plant defense by interacting directly with the host transcription factor OsWRKY71 for promoting insect performance (Ji et al., 2021; Zeng et al., 2023). Salivary protein 7 (NlSP7), a salivary protein secreted from the brown planthopper, functions as an effector via mediating tricin metabolism in rice plants (Gong et al., 2022). TFT7, 14-3-3 isoform 7, has been proven to be required for aphid resistance in tomato. Macrosiphum euphorbiae saliva-secreted protein Me10 targets the TFT7 as an effective infestation strategy (Chaudhary et al., 2019). Interaction assays have shown that the effector Mp1 from M. persicae associates with the host Vacuolar Protein Sorting Associated Protein52 (VPS52), which has a negative impact on insect infestation (Rodriguez et al., 2017). Effector Bsp9 from B. tabaci interacts with WRKY33 to interfere with the association between WRKY33 and a central regulator in the MAPK cascade, thus inhibiting plant immunity (Wang et al., 2019). The SSGP-71 (Secreted Salivary Gland Proteins-71) family, which has 426 members, has the greatest representation in the salivary proteome of the Hessian fly. Most SSGP-71 genes encode proteins with a signal peptide and an F box domain, which interacts with an Skp1-like protein (Zhao et al., 2015). The host plant cell wall was the first barrier of defense against herbivores (Calderón-Cortés et al., 2012). Both nematode (Heterodera avenae) expansin-like protein (HaEXPB2) and brown planthopper NlEG1 target the cell wall for promoting insect performance (Liu J. et al., 2016; Ji et al., 2017). Some effectors can suppress elicitor/pathogen-associated molecular pattern (PAMP)-triggered immunity. A macrophage migration inhibitory factor (MIF) is secreted from aphid saliva to promote insect feeding. Further study revealed that over-expressing MIF inhibits defense responses caused by the elicitor cryptogein, a 10-kDa protein from the plant pathogen Phytophthora cryptogea (Naessens et al., 2015). Transient overexpression of the salivary effector SG2204 from greenbug (Schizaphis graminum) and Sm9723 from grain aphid (Sitobion miscanthi) could suppress BAX and PAMP INF1-induced cell death (Zhang et al., 2022a; Zhang et al., 2022b). Furthermore, spider mite effectors Te28 and Te84 could also suppress cell death caused by the elicitor TePDI (Cui et al., 2023). However, as yet, the targets or receptors of many effectors in plants have not been identified.
Like elicitors, some effectors also come from insect-borne microbes. Notable examples are the SAP11 and SAP54 from Aster Yellows phytoplasma strain Witches’ Broom (AY-WB). They alter plant development and defense responses by the destabilization of CINCINNATA (CIN)-related TEOSINT BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR (TCP) and MADS domain transcription factors (MTFs) to enhance insect vector reproduction (MacLean et al., 2011; Sugio et al., 2011; MacLean et al., 2014). Other microbe-derived effectors, such as C2 from tomato yellow leaf curl China virus and 2b from cucumber mosaic virus (CMV), promote insect vector infestation by blocking the JA pathway in the plant (Wu et al., 2017; Li et al., 2019b). Together, these examples illustrate that microbe-derived effectors contribute to facilitating the fitness of their insect vectors as an effective strategy for completing their infection cycles.
R gene-mediated plant resistance to insect herbivores
To fight the secreted effectors, host plants have developed resistance proteins. A set of genes in tomato, melon, and rice conferring resistance against insects has been identified and cloned. Two aphid resistance genes, the Mi-1.2 gene characterized in the tomato (Solanum lycopersicum) and the Vat gene characterized in the melon (Cucumis melo) confer resistance to the potato aphid (M. euphorbiae) and the cotton aphid (A.gossypii), respectively (Rossi et al., 1998; Vos et al., 1998; Dogimont et al., 2014). Besides the potato aphid, the Mi-1.2 gene is also resistant to two whitefly biotypes, a psyllid, and three nematode species, suggesting that the Mi-1.2 gene confers a broad-spectrum resistance (Vos et al., 1998). With the availability of genome sequence data and molecular markers in rice, research on BPH-resistance genes has made a spurt of progress. BPH-rice interaction has become an excellent model system for the study of plant-insect interactions and co-evolution (Jing et al., 2017). To date, a total of 17 genes conferring resistance to BPH (Bph1, Bph2, Bph3, Bph6, Bph7, Bph9, Bph10, Bph14, Bph15, Bph18, Bph21, Bph26, bph29, Bph30, Bph32, Bph37, Bph40) have been cloned and characterized in rice plants (Du et al., 2020; Muduli et al., 2021; Shi et al., 2021; Zhou et al., 2021), which has shed a light on the molecular basis of plant-insect interactions.
Bph14, which encodes a typical NLR protein, was the first isolated BPH-resistance gene (Du et al., 2009). Further research has revealed that BPH14 protein stabilizes WRKY46 and WRKY72 to increase the expression of the receptor-like cytoplasmic kinase gene RLCK281 in rice (Hu et al., 2017). Bph9, a BPH-resistance gene mapped on the long arm of rice chromosome 12 (12L), which is allelic with another seven BPH-resistance genes (Bph1, Bph2, Bph7, Bph10, Bph18, Bph21, and Bph26), encodes an unusual NLR protein that confers resistance to BPH by enhancing SA and JA signaling pathways (Zhao et al., 2016). BPH6, an uncharacterized protein that localizes to the exocyst, interacts with the exocyst subunits OsEXO70E1 and OsEXO70H3, increases exocytosis, and participates in cell wall maintenance and reinforcement (Guo et al., 2018; Wu et al., 2022). Recently, a novel dominant BPH-resistance gene, Bph30, was isolated from the short arm of rice chromosome 4 (4S) (Wang et al., 2018; Shi et al., 2021). Bph30 is strongly expressed in sclerenchyma cells and encodes a protein belonging to a novel gene family with two leucine-rich domains (LRDs). A functional study showed that BPH30 enhances cellulose and hemicellulose synthesis, making the cell walls stiffer and sclerenchyma thicker to prevent stylets from penetrating the leaf sheath tissue, thereby conferring broad resistance to BPH and WBPH in rice (Shi et al., 2021). Bph15 encodes a lectin receptor-like kinase (LecRK), which functions in both innate immunity and seed germination in plants (Cheng et al., 2013a). Bph3 consists of a cluster of three genes encoding the plasma membrane-localized LecRKs (OsLecRK1, OsLecRK2, and OsLecRK3), which have a cumulative effect on resistance (Liu et al., 2015). These results indicate the diversity in resistance genes and mechanisms.
Based on our knowledge, except for two lectin receptor-like receptors and a few R proteins with unusual structures, such as BPH6 and BPH30, most isolated BPH-resistance proteins belong to nucleotide-binding and leucine-rich repeat (NLR) proteins, suggesting commonality between the perception of phloem-feeding insects and pathogens by plants. Bph3 and Bph15 encode the LecRKs, which resemble pattern recognition receptors (PRRs). PRRs are activated in response to microbe/pathogen/herbivore-associated molecular patterns or apoplastic effectors (Kaloshian and Walling, 2016; Ngou et al., 2022). The first layer of resistance to BPH may be BPH3 or BPH15, which is activated by the recognition of elicitors or apoplastic effectors. The second layer of resistance to BPH may be BPH6, BPH14, and BPH9 and their alleles, which can specifically recognize their cognate effectors and trigger defense responses (Jing et al., 2017; Du et al., 2020; Zheng et al., 2021).
Effectors recognized in R protein-mediated resistance
Despite the recent insights into the complex repertoire of R proteins, only a few effectors recognized by R proteins have been identified until now. This may be owing to the genetic intractability of the insects. At least 40 brown planthopper-resistant genes have been discovered, but just four corresponding BPH effector loci (Qhp7, Qgr5, Qgr14, and vBph1) were mapped (Jing et al., 2014; Kobayashi et al., 2014). The effectors recognized by R proteins had only been isolated from Hessian fly (Mayetiola destructor) until now (Stuart, 2015). The first Hessian fly virulence gene, virulence to Hessian fly 13 (vH13), was isolated using a map-based cloning strategy (Rider et al., 2002; Aggarwal et al., 2009; Aggarwal et al., 2014). Functional assays have shown that vH13 transcripts are only detected in H13-avirulent larvae and are lost in H13-virulent larvae. RNAi results revealed that the knockdown of vH13 helped some H13-avirulent larvae to escape the resistance triggered by H13 in wheat. Furthermore, vH13 encodes a small modular protein with no sequence similarities to other proteins in the database (Aggarwal et al., 2014).
Two additional Hessian fly effectors, vH6 and vH9, were identified by the completion of the Hessian fly genome sequencing and gene expression analyses. vH6 and vH9 can overcome the resistance mediated by wheat R protein H6 and H9, respectively (Zhao et al., 2015). Both vH6 and vH9 encode SSGP-71-like proteins. In H6-virulent Hessian flies, an SSGP-71 gene (Mdes009086-RA) is lost, suggesting Mdes009086-RA is the cognate effector of H6. In H9-virulent Hessian flies, two candidate SSGP-71 proteins without F-box domains were perfectly associated with H9 virulence, especially candidate 2 (Mdes015365-RA). These results indicate that the SSGP-71 family may play an essential role in the evolution of Hessian fly biotypes (Zhao et al., 2015). However, no cognate Hessian fly R protein has been cloned successfully, and the recognition mechanism of these Hessian fly effectors by the cognate R protein remains to be explored.
Perspectives and challenges
In recent years, rapid technological progress in the discovery and interrogation of plant and insect genomes, transcriptomes, and proteomes has been made. These developments have provided opportunities for the exploration of molecules delivered by herbivores that activate or suppress plant immunity (Hogenhout and Bos, 2011; Kaloshian and Walling, 2016). Cas9-CRISPR and RNAi technologies can effectively silence host plant/insect genes and, therefore, can help us to reveal the important signal molecules and the key pathways in plant-insect interactions (Ma et al., 2015; Liu et al., 2020; Hough et al., 2022). These discoveries give us an advanced understanding of the plant-insect relationship. In particular, the RNAi tool has made important contributions to the study of the function of insect elicitors and effectors in insect performance and plant immunity. There are several strategies for the delivery of double-stranded RNA (dsRNA), including external spraying, artificial feeding/micro-injection of synthesized dsRNA, and construction of transgenic plant lines with high levels of endogenous dsRNA (Shangguan et al., 2018; Huang et al., 2020; Zhang et al., 2022a). Over the years, RNAi has been considered an effective strategy for the control of insect pests (Liu et al., 2020; Hough et al., 2022). In addition, great progress has been made in the research of insect resistance proteins, especially the resistance mechanism of BPH-resistant genes (Jing et al., 2017; Du et al., 2020; Zheng et al., 2021).
Despite these advances, major gaps in our understanding of interactions between insect herbivores and host plants remain to be filled. Although a large number of elicitors and effectors have been identified, only a few have revealed corresponding host targets. The plant defense pathways interfered with by the majority of elicitors and effectors are obscure. Additionally, little is known about the relationship between cognate insect effector and cognate R protein. On the one hand, no cognate effector of the cloned R genes has been discovered; on the other hand, while three R protein-recognized effectors in Hessian flies were identified, no corresponding Hessian fly R genes have been cloned. Combining map-based cloning and multi-omics approaches may contribute to overcoming these challenges. We believe that these questions will be the priority of research on plant-insect interactions in the next decade, and the answers to these questions will provide more insight into how to control these pests.
Author contributions
WH proposed the idea. HW and SS drafted the manuscript and designed the figure. WH reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This review was supported by grants from the Wuhan Science and Technology Major Project on Key techniques of biological breeding and Breeding of new varieties (2022021302024851), the Natural Science Foundation of Hubei Province (2022CFB832), Hubei Academy of Agricultural Science Foundation (2023NKYJJ01), Hubei Key Laboratory of Food Crop Germplasm and Genetic Improvement Foundation (2022lzjj01), and the National Natural Science Foundation of China (32001921).
Acknowledgments
We acknowledge the BioRender tool that we used to create Figure 1.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Acevedo, F. E., Peiffer, M., Ray, S., Meagher, R., Luthe, D. S., Felton, G. W. (2018). Intraspecific differences in plant defense induction by fall armyworm strains. New Phytol. 218, 310–321. doi: 10.1111/nph.14981
Aggarwal, R., Benatti, T. R., Gill, N., Zhao, C., Chen, M. S., Fellers, J. P., et al. (2009). A BAC-based physical map of the Hessian fly genome anchored to polytene chromosomes. BMC Genomics 2, 293. doi: 10.1186/1471-2164-10-293
Aggarwal, R., Subramanyam, S., Zhao, C. Y., Chen, M. S., Harris, M. O., Stuart, J. J. (2014). Avirulence effector discovery in a plant galling and plant parasitic arthropod, the Hessian fly (Mayetiola destructor). PloS One 9, e100958. doi: 10.1371/journal.pone.0100958
Alborn, H. T., Hansen, T. V., Jones, T. H., Bennett, D. C., Tumlinson, J. H., Schmelz, E. A., et al. (2007). Disulfooxy fatty acids from the American bird grasshopper Schistocerca americana, elicitors of plant volatiles. Proc. Natl. Acad. Sci. U. S. A. 104, 12976–12981. doi: 10.1073/pnas.0705947104
Atamian, H. S., Chaudhary, R., Cin, V. D., Bao, E., Girke, T., Kaloshian, I. (2013). In planta expression or delivery of potato aphid Macrosiphum euphorbiae effectors Me10 and Me23 enhances aphid fecundity. Mol. Plant Microbe In. 26, 67–74. doi: 10.1094/MPMI-06-12-0144-FI
Backus, E. A. (1988). Sensory systems and behaviours which mediate hemipteran plant-feeding: a taxonomic overview. J. Insect Physiol. 34, 151–165. doi: 10.1016/0022-1910(88)90045-5
Bos, J. I., Prince, D., Pitino, M., Maffei, M. E., Win, J., Hogenhout, S. A. (2010). A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid). PloS Genet. 6, e1001216. doi: 10.1371/journal.pgen.1001216
Calderón-Cortés, N., Quesada, M., Watanabe, H., Cano-Camacho, H., Oyama, K. (2012). Endogenous plant cell wall digestion: a key mechanism in insect evolution. Annu. Rev. Ecol. Evol. Syst. 43, 45–71. doi: 10.1146/annurev-ecolsys-110411-160312
Chaudhary, R., Atamian, H. S., Shen, Z., Briggs, S. P., Kaloshian, I. (2014). GroEL from the endosymbiont Buchnera aphidicola betrays the aphid by triggering plant defense. Proc. Natl. Acad. Sci. U. S. A. 111, 8919–8924. doi: 10.1073/pnas.1407687111
Chaudhary, R., Peng, H. C., He, J., MacWilliams, J., Teixeira, M., Tsuchiya, T., et al. (2019). Aphid effector Me10 interacts with tomato TFT7, a 14-3-3 isoform involved in aphid resistance. New Phytol. 221, 1518–1528. doi: 10.1111/nph.15475
Chen, C., Liu, Y., Song, W., Chen, D., Chen, F., Chen, X., et al. (2019). An effector from cotton bollworm oral secretion impairs host plant defense signaling. Proc. Natl. Acad. Sci. U. S. A. 116, 14331–14338. doi: 10.1073/pnas.1905471116
Chen, X., Liu, Y., Wu, M., Yan, L., Chen, C., Mu, Y., et al. (2023). A highly accumulated secretory protein from cotton bollworm interacts with basic helix-loop-helix transcription factors to dampen plant defense. New Phytol. 237, 265–278. doi: 10.1111/nph.18507
Chen, C., Mao, Y. (2020). Research advances in plant-insect molecular interaction. F1000Res 19, F1000 Faculty Rev-198. doi: 10.12688/f1000research.21502.1
Cheng, X., Wu, Y., Guo, J., Du, B., Chen, R., Zhu, L., et al. (2013a). A rice lectin receptor-like that is involved in innate immuneresponses also contributes to seed germination. Plant J. 76, 687–698. doi: 10.1111/tpj.12328
Cheng, X., Zhu, L., He, G. (2013b). Towards understanding of molecular interactions between rice and the brown planthopper. Mol. Plant 6, 621–634. doi: 10.1093/mp/sst030
Cooper, W. R., Dillwith, J. W., Puterka, G. J. (2011). Comparisons of salivary proteins from five aphid (Hemiptera: aphididae) species. Environ. Entomol. 40, 151–156. doi: 10.1603/EN10153
Cui, J., Bing, X., Tang, Y., Liu, F., Ren, L., Zhou, J., et al. (2023). A conserved protein disulfide isomerase enhances plant resistance against herbivores. Plant Physiol. 191, 660–678. doi: 10.1093/plphys/kiac489
Cui, N., Lu, H., Wang, T., Zhang, W., Kang, L., Cui, F. (2019). Armet, an aphid effector protein, induces pathogen resistance in plants by promoting the accumulation of salicylic acid. Philos. Trans. R. Soc Lond. B. Biol. Sci. 4, 20180314. doi: 10.1098/rstb.2018.0314
Dogimont, C., Chovelon, V., Pauquet, J., Boualem, A., Bendahmane, A. (2014). The Vat locus encodes for a CC-NBS-LRR protein that confers resistance to Aphis gossypii infestation and A. gossypii-mediated virus resistance. Plant J. 80, 993–1004. doi: 10.1111/tpj.12690
Dong, Y., Huang, X., Yang, Y., Li, J., Zhang, M., Shen, H., et al. (2022). Characterization of salivary secreted proteins that induce cell death from Riptortus pedestris (Fabricius) and their roles in insect-plant interactions. Front. Plant Sci. 4. doi: 10.3389/fpls.2022.912603
Du, B., Chen, R., Guo, J., He, G. (2020). Current understanding of the genomic, genetic, and molecular control of insect resistance in rice. Mol. Breed. 40, 24. doi: 10.1007/s11032-020-1103-3
Du, H., Xu, H., Wang, F., Qian, L., Liu, S., Wang, X. (2022). Armet from whitefly saliva acts as an effector to suppress plant defences by targeting tobacco cystatin. New Phytol. 234, 1848–1862. doi: 10.1111/nph.18063
Du, B., Zhang, W., Liu, B., Hu, J., Wei, Z., Shi, Z., et al. (2009). Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc. Natl. Acad. Sci. U. S. A. 106, 22163–22168. doi: 10.1073/pnas.0912139106
Elzinga, D. A., De Vos, M., Jander, G. (2014). Suppression of plant defenses by a Myzus persicae (green peach aphid) salivary effector protein. Mol. Plant Microbe In. 27, 747–756. doi: 10.1094/MPMI-01-14-0018-R
Erb, M., Reymond, P. (2019). Molecular interactions between plants and insect herbivores. Annu. Rev. Plant Biol. 29, 527–557. doi: 10.1146/annurev-arplant-050718-095910
Felton, G. W., Korth, K. L., Bi, J. L., Wesley, S. V., Huhman, D. V., Mathews, M. C., et al. (1999). Inverse relationship between systemic resistance of plants to microorganisms and to insect herbivory. Curr. Biol. 9, 317–320. doi: 10.1016/s0960-9822(99)80140-7
Fu, J., Shi, Y., Wang, L., Tian, T., Li, J., Gong, L., et al. (2022). Planthopper-secreted salivary calmodulin acts as an effector for defense responses in rice. Front. Plant Sci. 28. doi: 10.3389/fpls.2022.841378
Gong, G., Yuan, L., Li, Y., Xiao, H., Li, Y., Zhang, Y., et al. (2022). Salivary protein 7 of the brown planthopper functions as an effector for mediating tricin metabolism in rice plants. Sci. Rep. 25, 3205. doi: 10.1038/s41598-022-07106-6
Guo, H., Wielsch, N., Hafke, J. B., Svatoš, A., Mithöfer, A., Boland, W. (2013). A porin-like protein from oral secretions of Spodoptera littoralis larvae induces defense-related early events in plant leaves. Insect Biochem. Mol. Biol. 43, 849–858. doi: 10.1016/j.ibmb.2013.06.005
Guo, J., Xu, C., Wu, D., Zhao, Y., Qiu, Y., Wang, X., et al. (2018). Bph6 encodes an exocyst-localized protein and confers broad resistance to planthoppers in rice. Nat. Genet. 50, 297–306. doi: 10.1038/s41588-018-0039-6
Guo, H., Zhang, Y., Tong, J., Ge, P., Wang, Q., Zhao, Z., et al. (2020). An aphid-secreted salivary protease activates plant defense in phloem. Curr. Biol. 30, 4826–4836.e7. doi: 10.1016/j.cub.2020.09.020
Hao, P., Liu, C., Wang, Y., Chen, R., Tang, M., Du, B., et al. (2008). Herbivore-induced callose deposition on the sieve plates of rice: an important mechanism for host resistance. Plant Physiol. 146, 1810–1820. doi: 10.1104/pp.107.111484
Harmel, N., Letocart, E., Cherqui, A., Giordanengo, P., Mazzucchelli, G., Guillonneau, F., et al. (2008). Identification of aphid salivary proteins: a proteomic investigation of Myzus persicae. Insect Mol. Biol. 17, 165–174. doi: 10.1111/j.1365-2583.2008.00790.x
Hattori, M., Nakamura, M., Komatsu, S., Tsuchihara, K., Tamura, Y., Hasegawa, T. (2012). Molecular cloning of a novel calcium-binding protein in the secreted saliva of the green rice leafhopper Nephotettix cincticeps. Insect Biochem. Mol. Biol. 42, 1–9. doi: 10.1016/j.ibmb.2011.10.001
Hewer, A., Becker, A., van Bel, A. J. (2011). An aphid’s odyssey–the cortical quest for the vascular bundle. J. Exp. Biol. 214, 3868–3879. doi: 10.1242/jeb.060913
Hewer, A., Will, T., van Bel, A. J. (2010). Plant cues for aphid navigation in vascular tissues. J. Exp. Biol. 213, 4030–4042. doi: 10.1242/jeb.046326
Hogenhout, S. A., Bos, J. I. (2011). Effector proteins that modulate plant-insect interactions. Curr. Opin. Plant Biol. 14, 422–428. doi: 10.1016/j.pbi.2011.05.003
Hough, J., Howard, J. D., Brown, S., Portwood, D. E., Kilby, P. M., Dickma, M. J. (2022). Strategies for the production of dsRNA biocontrols as alternatives to chemical pesticides. Front. Bioeng Biotechnol. 10. doi: 10.3389/fbioe.2022.980592
Hu, L., Wu, Y., Wu, D., Rao, W., Guo, J., Ma, Y., et al. (2017). The coiled-coil and nucleotide binding domains of BROWN PLANTHOPPER RESISTANCE14 function in signaling and resistance against planthopper in rice. Plant Cell 29, 3157–3185. doi: 10.1105/tpc.17.00263
Huang, H., Cui, J., Xia, X., Chen, J., Ye, Y., Zhang, C., et al. (2019). Salivary DNase II from Laodelphax striatellus acts as an effector that suppresses plant defence. New Phytol. 224, 860–874. doi: 10.1111/nph.15792
Huang, H., Liu, C., Huang, X., Zhou, X., Zhuo, J., Zhang, C., et al. (2016). Screening and functional analyses of Nilaparvata lugens salivary proteome. J. Proteome Res. 15, 1883–1896. doi: 10.1021/acs.jproteome.6b00086
Huang, H., Liu, C., Xu, H., Bao, Y., Zhang, C. (2017). Mucin-like protein, a saliva component involved in brown planthopper virulence and host adaptation. J. Insect Physiol. 98, 223–230. doi: 10.1016/j.jinsphys.2017.01.012
Huang, H., Lu, J., Li, Q., Bao, Y., Zhang, C. (2018). Combined transcriptomic/proteomic analysis of salivary gland and secreted saliva in three planthopper species. J. Proteomics 172, 25–35. doi: 10.1016/j.jprot.2017.11.003
Huang, H., Wang, Y., Li, L., Lu, H., Lu, J., Wang, X., et al. (2023). Planthopper salivary sheath protein LsSP1 contributes to manipulation of rice plant defenses. Nat. Commun. 14, 737. doi: 10.1038/s41467-023-36403-5
Huang, J., Zhang, N., Shan, J., Peng, Y., Guo, J., Zhou, C., et al. (2020). Salivary protein 1 of brown planthopper is required for survival and induces immunity response in plants. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.571280
Iida, J., Desaki, Y., Hata, K., Uemura, T., Yasuno, A., Islam, M., et al. (2019). Tetranins: new putative spider mite elicitors of host plant defense. New Phytol. 224, 875–885. doi: 10.1111/nph.15813
Ji, R., Fu, J., Shi, Y., Li, J., Jing, M., Wang, L., et al. (2021). Vitellogenin from planthopper oral secretion acts as a novel effector to impair plant defenses. New Phytol. 232, 802–817. doi: 10.1111/nph.17620
Ji, R., Ye, W., Chen, H., Zeng, J., Li, H., Yu, H., et al. (2017). A salivary endo-β-1,4-glucanase acts as an effector that enables the brown planthopper to feed on rice. Plant Physiol. 173, 1920–1932. doi: 10.1104/pp.16.01493
Ji, R., Yu, H., Fu, Q., Chen, H., Ye, W., Li, S., et al. (2013). Comparative transcriptome analysis of salivary glands of two populations of rice brown planthopper, Nilaparvata lugens, that differ in virulence. PloS One 8, e79612. doi: 10.1371/journal.pone.0079612
Jing, S., Zhang, L., Ma, Y., Liu, B., Zhao, Y., Yu, H., et al. (2014). Genome-wide mapping of virulence in brown planthopper identifies loci that break down host plant resistance. PloS One 9, e98911. doi: 10.1371/journal.pone.0098911
Jing, S., Zhao, Y., Du, B., Chen, R., Zhu, L., He, G. (2017). Genomics of interaction between the brown planthopper and rice. Curr. Opin. Insect Sci. 19, 82–87. doi: 10.1016/j.cois.2017.03.005
Jones, J. D., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kahl, G. (1982). “Molecular biology of wound healing: the conditioning phenomenon,” in Molecular biology of plant tumors. Eds. Kahl, G., Schell, J. (New York: Academic Press, Inc), 211–267.
Kaloshian, I., Walling, L. L. (2016). Hemipteran and dipteran pests: effectors and plant host immune regulators. J. Integr. Plant Biol. 58, 350–361. doi: 10.1111/jipb.12438
Kobayashi, T., Yamamoto, K., Suetsugu, Y., Kuwazaki, S., Hattori, M., Jairin, J., et al. (2014). Genetic mapping of the rice resistance-breaking gene of the brown planthopper Nilaparvata lugens. Proc. Biol. Sci. 281, 20140726. doi: 10.1098/rspb.2014.0726
Li, P., Liu, C., Deng, W., Yao, D., Pan, L., Li, Y., et al. (2019b). Plant begomoviruses subvert ubiquitination to suppress plant defenses against insect vectors. PloS Pathog. 15, e1007607. doi: 10.1371/journal.ppat.1007607
Li, J., Liu, X., Wang, Q., Huangfu, J., Schuman, M. C., Lou, Y. (2019a). A group d MAPK protects plants from autotoxicity by suppressing herbivore-induced defense signaling. Plant Physiol. 179, 1386–1401. doi: 10.1104/pp.18.01411
Li, R., Weldegergis, B. T., Li, J., Jung, C., Qu, J., Sun, Y., et al. (2014). Virulence factors of geminivirus interact with MYC2 to subvert plant resistance and promote vector performance. Plant Cell 26, 4991–5008. doi: 10.1105/tpc.114.133181
Liu, S., Jaouannet, M., Dempsey, D. A., Imani, J., Coustau, C., Kogel, K. H. (2020). RNA-Based technologies for insect control in plant production. Biotechnol. Adv. 39, 107463. doi: 10.1016/j.biotechadv.2019.107463
Liu, J., Peng, H., Cui, J., Huang, W., Kong, L., Clarke, J. L., et al. (2016). Molecular characterization of a novel effector expansin-like protein from Heterodera avenae that induces cell death in Nicotiana benthamiana. Sci. Rep. 6, 35677. doi: 10.1038/srep35677
Liu, H., Wang, C., Qiu, C., Shi, J., Sun, Z., Hu, X., et al. (2021). A salivary odorant-binding protein mediates Nilaparvata lugens feeding and host plant phytohormone suppression. Int. J. Mol. Sci. 22, 4988. doi: 10.3390/ijms22094988
Liu, Y., Wu, H., Chen, H., Liu, Y., He, J., Kang, H., et al. (2015). A gene cluster encoding lectin receptor kinases confers broad-spectrum and durable insect resistance in rice. Nat. Biotechnol. 33, 301–305. doi: 10.1038/nbt.3069
Liu, X., Zhou, H., Zhao, J., Hua, H., He, Y. (2016). Identification of the secreted watery saliva proteins of the rice brown planthopper, Nilaparvata lugens (Stål) by transcriptome and shotgun LC-MS/MS approach. J. Insect Physiol. 89, 60–69. doi: 10.1016/j.jinsphys.2016.04.002
Louis, J., Peiffer, M., Ray, S., Luthe, D. S., Felton, G. W. (2013). Host-specific salivary elicitor(s) of European corn borer induce defenses in tomato and maize. New Phytol. 199, 66–73. doi: 10.1111/nph.12308
Luan, J., Yao, D., Zhang, T., Walling, L. L., Yang, M., Wang, Y., et al. (2013). Suppression of terpenoid synthesis in plants by a virus promotes its mutualism with vectors. Ecol. Lett. 16, 390–398. doi: 10.1111/ele.12055
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
MacLean, A. M., Orlovskis, Z., Kowitwanich, K., Zdziarska, A. M., Angenent, G. C., Immink, R. G., et al. (2014). Phytoplasma effector SAP54 hijacks plant reproduction by degrading MADS-box proteins and promotes insect colonization in a RAD23-dependent manner. PloS Biol. 12, e1001835. doi: 10.1371/journal.pbio.1001835
MacLean, A. M., Sugio, A., Makarova, O. V., Findlay, K. C., Grieve, V. M., Tóth, R., et al. (2011). Phytoplasma effector SAP54 induces indeterminate leaf-like flower development in arabidopsis plants. Plant Physiol. 157, 831–841. doi: 10.1104/pp.111.181586
Matsumoto, Y., Hattori, M. (2018). The green rice leafhopper, Nephotettix cincticeps (Hemiptera: cicadellidae), salivary protein NcSP75 is a key effector for successful phloem ingestion. PloS One 13, e0202492. doi: 10.1371/journal.pone.0202492
Mattiacci, L., Dicke, M., Posthumus, M. A. (1995). beta-glucosidase: an elicitor of herbivore-induced plant odor that attracts host-searching parasitic wasps. Proc. Natl. Acad. Sci. U. S. A. 92, 2036–2040. doi: 10.1073/pnas.92.6.2036
Miles, P. W. (1999). Aphid saliva. Biol. Rev. Camb. Philos. Soc 4, 41–85. doi: 10.1111/j.1469-185X.1999.tb00181.x
Muduli, L., Pradhan, S. K., Mishra, A., Bastia, D. N., Samal, K. C., Agrawal, P. K., et al. (2021). Understanding brown planthopper resistance in rice: genetics, biochemical and molecular breeding approaches. Rice Sci. 28, 532–546. doi: 10.1016/j.rsci.2021.05.013
Musser, R. O., Cipollini, D. F., Hum-Musser, S. M., Williams, S. A., Brown, J. K., Felton, G. W. (2005). Evidence that the caterpillar salivary enzyme glucose oxidase provides herbivore offense in solanaceous plants. Arch. Insect Biochem. Physiol. 58, 128–137. doi: 10.1002/arch.20039
Musser, R. O., Hum-Musser, S. M., Eichenseer, H., Peiffer, M., Ervin, G., Murphy, J. B., et al. (2002). Herbivory: caterpillar saliva beats plant defences. Nature 416, 599–600. doi: 10.1038/416599a
Mutti, N. S., Louis, J., Pappan, L. K., Pappan, K., Begum, K., Chen, M. S., et al. (2008). A protein from the salivary glands of the pea aphid, Acyrthosiphon pisum, is essential in feeding on a host plant. Proc. Natl. Acad. Sci. U. S. A. 105, 9965–9969. doi: 10.1073/pnas.0708958105
Naessens, E., Dubreuil, G., Giordanengo, P., Baron, O. L., Minet-Kebdani, N., Keller, H., et al. (2015). A secreted MIF cytokine enables aphid feeding and represses plant immune responses. Curr. Biol. 25, 1898–1903. doi: 10.1016/j.cub.2015.05.047
Ngou, B. P. M., Ding, P., Jones, J. D. G. (2022). Thirty years of resistance: zig-zag through the plant immune system. Plant Cell 34, 1447–1478. doi: 10.1093/plcell/koac041
Nicholson, S. J., Hartson, S. D., Puterka, G. J. (2012). Proteomic analysis of secreted saliva from Russian wheat aphid (Diuraphis noxia kurd.) biotypes that differ in virulence to wheat. J. Proteomics 75, 2252–2268. doi: 10.1016/j.jprot.2012.01.031
Nicholson, S. J., Puterka, G. J. (2014). Variation in the salivary proteomes of differentially virulent greenbug (Schizaphis graminum rondani) biotypes. J. Proteomics 105, 186–203. doi: 10.1016/j.jprot.2013.12.005
Pelosi, P., Calvello, M., Ban, L. (2005). Diversity of odorant-binding proteins and chemosensory proteins in insects. Chem. Senses 30, i291–i292. doi: 10.1093/chemse/bjh229
Pitino, M., Hogenhout, S. A. (2013). Aphid protein effectors promote aphid colonization in a plant species-specific manner. Mol. Plant Microbe In. 26, 130–139. doi: 10.1094/MPMI-07-12-0172-FI
Rao, W., Zheng, X., Liu, B., Guo, Q., Guo, J., Wu, Y., et al. (2019). Secretome analysis and in planta expression of salivary proteins identify candidate effectors from the brown planthopper Nilaparvata lugens. Mol. Plant Microbe In. 32, 227–239. doi: 10.1094/MPMI-05-18-0122-R
Rider, S. D., Jr., Sun, W., Ratcliffe, R. H., Stuart, J. J. (2002). Chromosome landing near avirulence gene vH13 in the Hessian fly. Genome 45, 812–822. doi: 10.1139/g02-047
Rodriguez, P. A., Escudero-Martinez, C., Bos, J. I. (2017). An aphid effector targets trafficking protein VPS52 in a host-specific manner to promote virulence. Plant Physiol. 173, 1892–1903. doi: 10.1104/pp.16.01458
Rodriguez, P. A., Stam, R., Warbroek, T., Bos, J. I. (2014). Mp10 and Mp42 from the aphid species Myzus persicae trigger plant defenses in Nicotiana benthamiana through different activities. Mol. Plant Microbe In. 27, 30–39. doi: 10.1094/MPMI-05-13-0156-R
Rossi, M., Goggin, F. L., Milligan, S. B., Kaloshian, I., Ullman, D. E., Williamson, V. M. (1998). The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc. Natl. Acad. Sci. U. S. A. 95, 9750–9754. doi: 10.1073/pnas.95.17.9750
Schoonhoven, L., van Loon, J. J., Dicke, M. (2005). Insect–plant biology. 2nd edn (Oxford: Oxford Univ. Press).
Schulz, A. (1998). The phloem. structure related to function. Prog. Bot. 59, 429–475. doi: 10.1007/978-3-642-80446-5_16
Shangguan, X., Zhang, J., Liu, B., Zhao, Y., Wang, H., Wang, Z., et al. (2018). A mucin-like protein of planthopper is required for feeding and induces immunity response in plants. Plant Physiol. 176, 552–565. doi: 10.1104/pp.17.00755
Shi, S., Wang, H., Nie, L., Tan, D., Zhou, C., Zhang, Q., et al. (2021). Bph30 confers resistance to brown planthopper by fortifying sclerenchyma in rice leaf sheaths. Mol. Plant 14, 1714–1732. doi: 10.1016/j.molp.2021.07.004
Snoeck, S., Guayazán-Palacios, N., Steinbrenner, A. D. (2022). Molecular tug-of-war: plant immune recognition of herbivory. Plant Cell 34, 1497–1513. doi: 10.1093/plcell/koac009
Sogawa, K. (1982). The rice brown planthopper: feeding physiology and host plant interactions. Annu. Rev. Entomol. 27, 49–73. doi: 10.1146/annurev.en.27.010182.000405
Spiller, N. J. (1990). An ultrastructural study of the stylet pathway of the brown planthopper Nilaparvata lugens. Entomol. Exp. Appl. 54, 191–193. doi: 10.1111/j.1570-7458.1990.tb01329.x
Stotz, H. U., Kroymann, J., Mitchell-Olds, T. (1999). Plant-insect interactions. Curr. Opin. Plant Biol. 2, 268–272. doi: 10.1016/S1369-5266(99)80048-X
Stuart, J. (2015). Insect effectors and gene-for-gene interactions with host plants. Curr. Opin. Insect Sci. 9, 56–61. doi: 10.1016/j.cois.2015.02.010
Su, Q., Peng, Z., Tong, H., Xie, W., Wang, S., Wu, Q., et al. (2019). A salivary ferritin in the whitefly suppresses plant defenses and facilitates host exploitation. J. Exp. Bot. 70, 3343–3355. doi: 10.1093/jxb/erz152
Sugio, A., Kingdom, H. N., MacLean, A. M., Grieve, V. M., Hogenhout, S. A. (2011). Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 108, E1254–E1263. doi: 10.1073/pnas.1105664108
Takken, F. L. W., Tameling, W. I. L. (2009). To nibble at plant resistance proteins. Science 324, 744–746. doi: 10.1126/science.1171666
Tian, D., Peiffer, M., Shoemaker, E., Tooker, J., Haubruge, E., Francis, F., et al. (2012). Salivary glucose oxidase from caterpillars mediates the induction of rapid and delayed-induced defenses in the tomato plant. PloS One 7, e36168. doi: 10.1371/journal.pone.0036168
Villarroel, C. A., Jonckheere, W., Alba, J. M., Glas, J. J., Dermauw, W., Haring, M. A., et al. (2016). Salivary proteins of spider mites suppress defenses in Nicotiana benthamiana and promote mite reproduction. Plant J. 86, 119–131. doi: 10.1111/tpj.13152
Vos, P., Simons, G., Jesse, T., Wijbrandi, J., Heinen, L., Hogers, R., et al. (1998). The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nat. Biotech. 16, 1365–1369. doi: 10.1038/4350
Walling, L. L. (2000). The myriad plant responses to herbivores. J. Plant Growth Regul. 19, 195–216. doi: 10.1007/s003440000026
Walling, L. L. (2008). Avoiding effective defenses: strategies employed by phloem-feeding insects. Plant Physiol. 146, 859–866. doi: 10.1104/pp.107.113142
Wang, W., Dai, H., Zhang, Y., Chandrasekar, R., Luo, L., Hiromasa, Y., et al. (2015a). Armet is an effector protein mediating aphid-plant interactions. FASEB J. 29, 2032–2045. doi: 10.1096/fj.14-266023
Wang, W., Luo, L., Lu, H., Chen, S., Kang, L., Cui, F. (2015b). Angiotensin-converting enzymes modulate aphid-plant interactions. Sci. Rep. 5, 8885. doi: 10.1038/srep08885
Wang, H., Shi, S., Guo, Q., Nie, L., Du, B., Chen, R., et al. (2018). High-resolution mapping of a gene conferring strong antibiosisto brown planthopper and developing resistant near-isogenic lines in 9311 background. Mol. Breed. 38, 107. doi: 10.1007/s11032-018-0859-1
Wang, Y., Tang, M., Hao, P., Yang, Z., Zhu, L., He, G. (2008). Penetration into rice tissues by brown planthopper and fine structure of the salivary sheaths. Entomol. Exp. Appl. 129, 295–307. doi: 10.1111/j.1570-7458.2008.00785.x
Wang, Q., Yuan, E., Ling, X., Zhu-Salzman, K., Guo, H., Ge, F., et al. (2020). An aphid facultative symbiont suppresses plant defence by manipulating aphid gene expression in salivary glands. Plant Cell Environ. 43, 2311–2322. doi: 10.1111/pce.13836
Wang, N., Zhao, P., Ma, Y., Yao, X., Sun, Y., Huang, X., et al. (2019). A whitefly effector Bsp9 targets host immunity regulator WRKY33 to promote performance. Philos. Trans. R. Soc Lond. B. Biol. Sci. 374, 20180313. doi: 10.1098/rstb.2018.0313
Will, T., Furch, A. C., Zimmermann, M. (2013). How phloem-feeding insects face the challenge of phloem-located defenses. Front. Plant Sci. 29. doi: 10.3389/fpls.2013.00336
Will, T., Vilcinskas, A. (2015). The structural sheath protein of aphids is required for phloem feeding. Insect Biochem. Mol. Biol. 57, 34–40. doi: 10.1016/j.ibmb.2014.12.005
Wu, J., Baldwin, I. T. (2010). New insights into plant responses to the attack from insect herbivores. Annu. Rev. Genet. 44, 1–24. doi: 10.1146/annurev-genet-102209-163500
Wu, D., Guo, J., Zhang, Q., Shi, S., Guan, W., Zhou, C., et al. (2022). Necessity of rice resistance to planthoppers for OsEXO70H3 regulating SAMSL excretion and lignin deposition in cell walls. New Phytol. 234, 1031–1046. doi: 10.1111/nph.18012
Wu, D., Qi, T., Li, W., Tian, H., Gao, H., Wang, J., et al. (2017). Viral effector protein manipulates host hormone signaling to attract insect vectors. Cell Res. 27, 402–415. doi: 10.1038/cr.2017.2
Xu, H., Qian, L., Wang, X., Shao, R., Hong, Y., Liu, S., et al. (2019). A salivary effector enables whitefly to feed on host plants by eliciting salicylic acid-signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 116, 490–495. doi: 10.1073/pnas.1714990116
Yang, C., Guo, J., Chu, D., Ding, T., Wei, K., Cheng, D., et al. (2017). Secretory laccase 1 in Bemisia tabaci MED is involved in whitefly-plant interaction. Sci. Rep. 7, 3623. doi: 10.1038/s41598-017-03765-y
Ye, M., Glauser, G., Lou, Y., Erb, M., Hu, L. (2019). Molecular dissection of early defense signaling underlying volatile-mediated defense regulation and herbivore resistance in rice. Plant Cell 31, 687–698. doi: 10.1105/tpc.18.00569
Ye, W., Yu, H., Jian, Y., Zeng, J., Ji, R., Chen, H., et al. (2017). A salivary EF-hand calcium-binding protein of the brown planthopper Nilaparvata lugens functions as an effector for defense responses in rice. Sci. Rep. 7, 40498. doi: 10.1038/srep40498
Zeng, J., Ye, W., Hu, W., Jin, X., Kuai, P., Xiao, W., et al. (2023). The N-terminal subunit of vitellogenin in planthopper eggs and saliva acts as a reliable elicitor that induces defenses in rice. New Phytol. 238, 1230–1244. doi: 10.1111/nph.18791
Zhang, Y., Liu, X., Francis, F., Xie, H., Fan, J., Wang, Q., et al. (2022a). The salivary effector protein Sg2204 in the greenbug Schizaphis graminum suppresses wheat defence and is essential for enabling aphid feeding on host plants. Plant Biotechnol. J. 20, 2187–2201. doi: 10.1111/pbi.13900
Zhang, Y., Liu, X., Fu, Y., Crespo-Herrera, L., Liu, H., Wang, Q., et al. (2022b). Salivary effector Sm9723 of grain aphid Sitobion miscanthi suppresses plant defense and is essential for aphid survival on wheat. Int. J. Mol. Sci. 23, 6909. doi: 10.3390/ijms23136909
Zhao, C., Escalante, L. N., Chen, H., Benatti, T. R., Qu, J., Chellapilla, S., et al. (2015). A massive expansion of effector genes underlies gall-formation in the wheat pest Mayetiola destructor. Curr. Biol. 25, 613–620. doi: 10.1016/j.cub.2014.12.057
Zhao, Y., Huang, J., Wang, Z., Jing, S., Wang, Y., Ouyang, Y., et al. (2016). Allelic diversity in an NLR gene BPH9 enables rice to combat planthopper variation. Proc. Natl. Acad. Sci. U. S. A. 113, 12850–12855. doi: 10.1073/pnas.1614862113
Zheng, X., Zhu, L., He, G. (2021). Genetic and molecular understanding of host rice resistance and Nilaparvata lugens adaptation. Curr. Opin. Insect Sci. 45, 14–20. doi: 10.1016/j.cois.2020.11.005
Zhou, W., Yuan, X., Qian, P., Cheng, J., Zhang, C., Gurr, G., et al. (2015). Identification and expression profiling of putative chemosensory protein genes in two rice planthoppers, Laodelphax striatellus (Fallén) and Sogatella furcifera (Horváth). J. Asia Pac. Entomol. 18, 771–778. doi: 10.1016/j.aspen.2015.09.006
Keywords: elicitors, effectors, defense responses, multi-omics approach, plant-insect interactions
Citation: Wang H, Shi S and Hua W (2023) Advances of herbivore-secreted elicitors and effectors in plant-insect interactions. Front. Plant Sci. 14:1176048. doi: 10.3389/fpls.2023.1176048
Received: 28 February 2023; Accepted: 31 March 2023;
Published: 19 June 2023.
Edited by:
Shengli Jing, Xinyang Normal University, ChinaReviewed by:
Zhenying Shi, Center for Excellence in Molecular Plant Sciences (CAS), ChinaYan Zhao, Hunan Hybrid Rice Research Center, China
Copyright © 2023 Wang, Shi and Hua. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Hua, aHVhd2VpQG9pbGNyb3BzLmNu
†These authors have contributed equally to this work
 Huiying Wang
Huiying Wang Shaojie Shi2,3†
Shaojie Shi2,3† Wei Hua
Wei Hua