- 1Institute of Botany, Heinrich Heine University, Düsseldorf, Germany
- 2Center of Excellence on Plant Sciences (CEPLAS), Germany
Membrane identity and dynamic processes, that act at membrane sites, provide important cues for regulating transport, signal transduction and communication across membranes. There are still numerous open questions as to how membrane identity changes and the dynamic processes acting at the surface of membranes are regulated in diverse eukaryotes in particular plants and which roles are being played by protein interaction complexes composed of peripheral and integral membrane proteins. One class of peripheral membrane proteins conserved across eukaryotes comprises the SEC14-like phosphatidylinositol transfer proteins (SEC14L-PITPs). These proteins share a SEC14 domain that contributes to membrane identity and fulfills regulatory functions in membrane trafficking by its ability to sense, bind, transport and exchange lipophilic substances between membranes, such as phosphoinositides and diverse other lipophilic substances. SEC14L-PITPs can occur as single-domain SEC14-only proteins in all investigated organisms or with a modular domain structure as multi-domain proteins in animals and streptophytes (comprising charales and land plants). Here, we present an overview on the functional roles of SEC14L-PITPs, with a special focus on the multi-domain SEC14L-PITPs of the SEC14-nodulin and SEC14-GOLD group (PATELLINs, PATLs in plants). This indicates that SEC14L-PITPs play diverse roles from membrane trafficking to organism fitness in plants. We concentrate on the structure of SEC14L-PITPs, their ability to not only bind phospholipids but also other lipophilic ligands, and their ability to regulate complex cellular responses through interacting with proteins at membrane sites.
Highlights
• SEC14-like phosphatidylinositol transfer proteins (SEC14L-PITPs) can bind phospholipids and other lipophilic ligands.
• The occurrence of multi-domain SEC14L-PITPs in higher eukaryotes including land plants underlines their functional roles.
• Plant SEC14 proteins function as cellular regulators via protein-protein and/or protein-lipid interaction and lipid transfer to achieve chloroplast functioning, cell polarity and development, and to control the response to environmental stimuli and iron nutrition.
1 Introduction
Cells are surrounded by membranes, which function as hydrophobic permeable barriers regulating the exchange of molecules and the flow of information. Within the cell, membranes have different compositions resulting in their specific identity and allowing them to fulfill specific tasks (Watson, 2015; Heilmann, 2016; Mamode Cassim et al., 2019). The plasma membrane, for example, is involved in uptake of molecules from the environment, cell-to-cell communication or cell shape changes (Cooper, 2000; Luschnig and Vert, 2014). The thylakoid membrane, on the other hand, is essential for photosynthesis and uses an electron gradient to generate ATP (O'Connor and Adams, 2010). In addition to building the basic membrane backbone, lipids may have regulatory roles (Stevenson et al., 2000). Minor changes in lipid composition and structure can result in major modifications to essential cellular processes (Harayama and Riezman, 2018). Especially the phospholipid composition of a membrane has significant effects on the regulation of cellular and tissue functions (Heilmann, 2016). A crucial group of regulatory phospholipids are the phosphorylated derivatives of phosphatidylinositol (PI), which are the phosphoinositides (PIPs) PI(3)P, PI(4)P, PI(5)P, PI(3,4)P2, PI(3,5)P2, PI(4,5)P2 and PI(3,4,5)P3 (Irvine, 2016). PI and PIPs provide cues to membrane identity, although they make up less than 1% of membrane lipids (Simon et al., 2014; Simon et al., 2016; Gerth et al., 2017; Noack and Jaillais, 2020). They are key players controlling growth, development and polarization as well as influencing multiple processes through binding to a great number of interaction partners in adaptation to, e.g., environmental changes (Heilmann, 2016; Roman-Fernandez et al., 2018). It is among the open compelling questions in plant cell biology how membranes are remodeled and controlled and how identity and dynamics of membrane systems are determined, maintained or changed (Roeder et al., 2022). Here, we review one class of peripheral membrane proteins, namely SEC14 domain-containing lipid transfer proteins, that are promising candidates to alter the phospholipid identity and membrane dynamics in their functions as lipid transfer, lipid-binding and membrane-associated proteins with protein-protein interaction capabilities.
2 Overview of phosphatidylinositol transfer proteins
Lipids can be distributed in the cell through vesicle-independent trafficking, which includes spontaneous lipid transfer, flip-flop exchange within bilayers, lateral diffusion or single-lipid transfer by lipid-transfer proteins, that act, for example at membrane contact sites (Lev, 2010; Peretti et al., 2019). On the other hand, trafficking of proteins, lipids and other metabolites between cell compartments is achieved by the highly regulated and coordinated vesicular trafficking mechanism, in which macromolecules are transported within membrane vesicles (Tokarev et al., 2013; Goring and Di Sansebastiano, 2017). Generally, single-lipid transfer and membrane vesicular trafficking are controlled by regulatory proteins in response to developmental cues and external stimuli. It is not yet well investigated how this is controlled in plant cells. One group of proteins able to link lipid recognition, metabolism and signaling are phosphatidylinositol transfer proteins (PITPs). PITPs can be clustered in two independent protein families with distinctly separated biological functions (Wirtz, 1991).
The first group is simply named the phosphatidylinositol transfer protein (PITP)-superfamily, defined through its e.g. phosphatidylinositol transfer protein and Lipin/Ned1/Smp2 (PITP/LNS2) domain (InterPro accession number (IPR): IPR031315). Such a domain is thought to promote the exchange of phospholipids at the membrane contact sites of the endoplasmic reticulum (ER) and the plasma membrane by non-vesicular lipid transport (Cockcroft and Raghu, 2018). Proteins containing a PITP/LNS2 domain can be found in mammals, invertebrates and plants (Hsuan and Cockcroft, 2001; Cockcroft, 2012). Defects in PITP proteins can lead to for example neurodegenerative diseases (Hsuan and Cockcroft, 2001; Cockcroft, 2012). In Arabidopsis thaliana (Arabidopsis) the PITP/LNS2 domain can for example be found in two phosphatidate phosphohydrolase proteins involved in galactolipid synthesis and necessary to maintain membrane structure by lipid remodeling due to phosphate starvation (Nakamura et al., 2009; Yoshitake et al., 2017). These examples show that PITPs have profound functions in cellular and physiological integrity of higher eukaryotes.
The second PITP-superfamily and subject of this review is defined through its SEC14 domain (IPR001251), named the SEC14-like phosphatidylinositol transfer protein (SEC14L-PITP)-superfamily. SEC14L-PITPs are able to recognize, bind, exchange and transfer small lipophilic molecules between membranes by non-vesicular transport (Figures 1A–F) (Bankaitis et al., 1990; Cleves et al., 1991). Additionally, they are involved in regulation of membrane trafficking within a cell (Bankaitis et al., 2010). SEC14 proteins are found in yeast, plants, invertebrates, and mammals, suggesting a conserved and essential role (Ren et al., 2011; Aravind and Iyer, 2012). In the following sections, we will review their general characteristics, distinguish functions of single- and multi-domain SEC14-PITPs and highlight recent work describing their roles in plants, focusing on Arabidopsis thaliana.
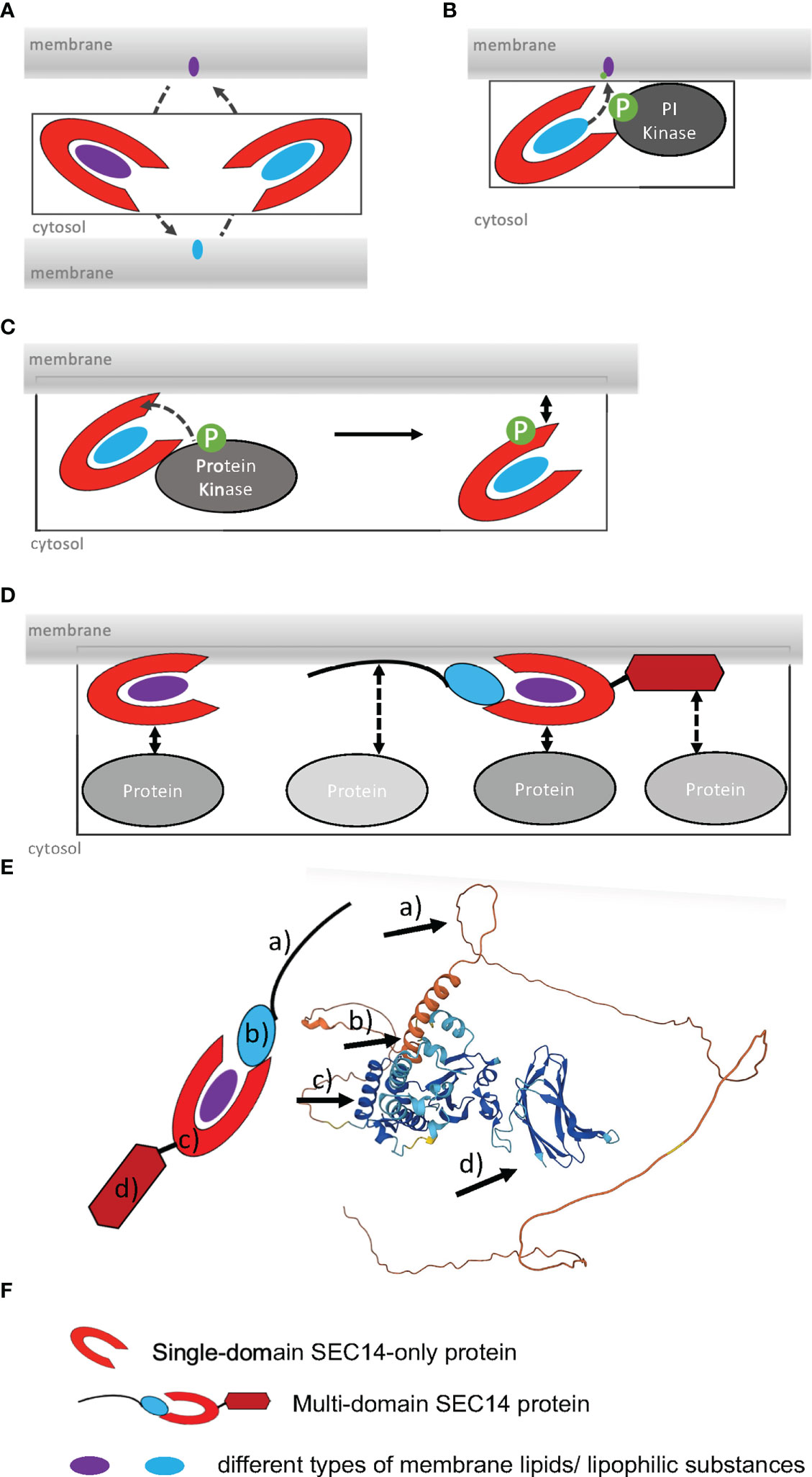
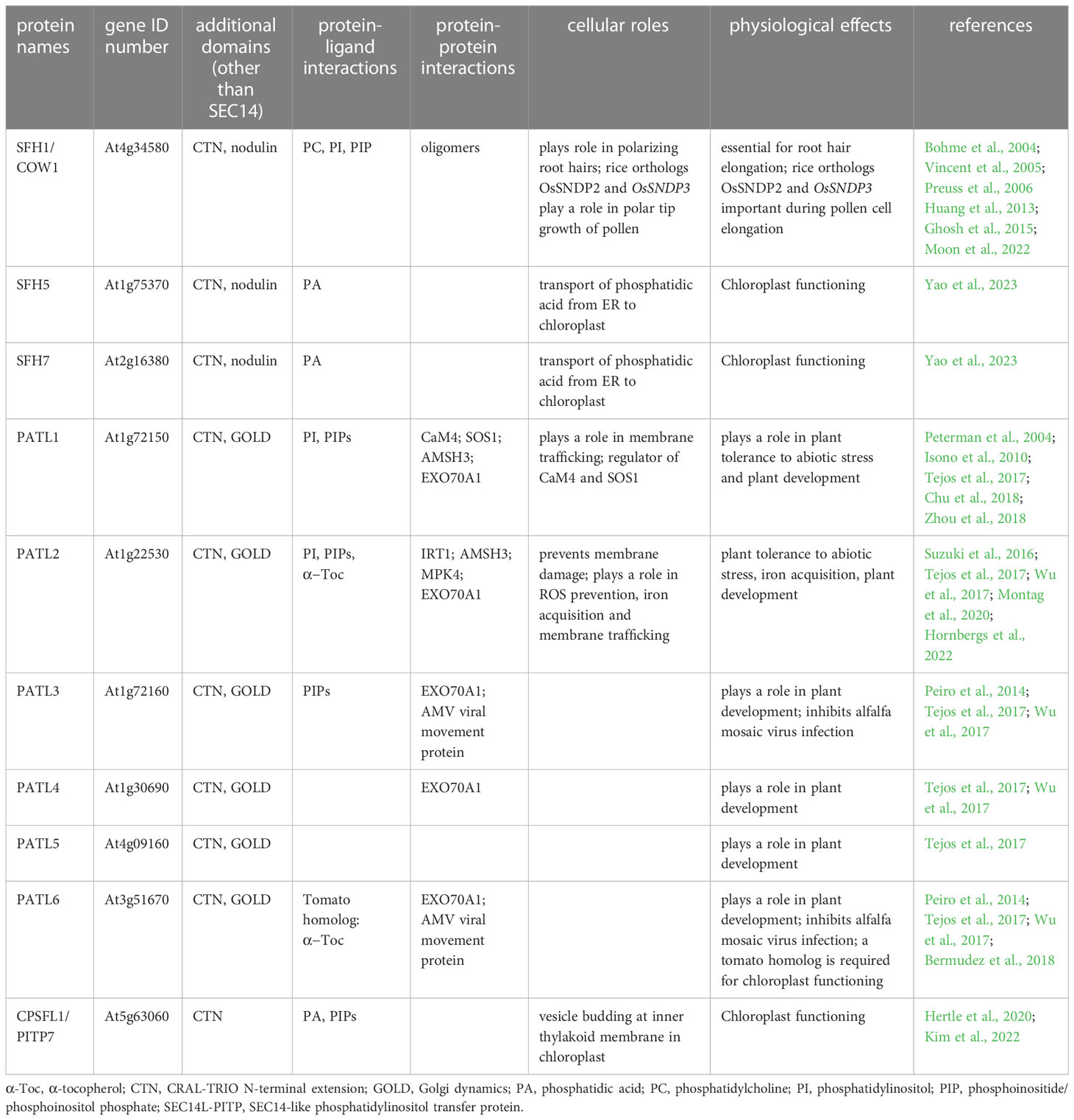
Figure 1 Functions and regulation modes of SEC14L-PITPs. (A), SEC14 protein-mediated lipid transfer and heterotypic exchange of lipids between two membranes. (B), Lipid presentation model, interaction of SEC14 protein with PI kinase and phosphorylation of lipid during transfer (de Campos and Schaaf, 2017). (C), Regulation of membrane binding by phosphorylation of SEC14 domain by a protein kinase. (D), Increase in the number of potential protein-protein interactions of multi-domain versus single-domain SEC14L-PITPs. (E), Alphafold model of the multi-domain SEC14 protein PATL2 (At1g22530). The arrows point to a) intrinsically disordered N-terminal region; b) CTN domain; c) SEC14 domain with lipid-binding site, gate and anchor helices; d) GOLD domain. Alphafold was used, as described (Jumper et al. 2021; Varadi et al. 2021). (F), Symbols used in (A–D).
3 General characteristics of SEC14L-PITPs
The SEC14 domain forms a characteristic hydrophobic phospholipid-binding pocket at its carboxy (C)-terminus (Sha et al., 1998). Yeast Sec14p (304 AA) is the prototype for the SEC14 domain ([37-279AA] 12x α - helices, 6x β-strands, 8x 310-helices; 2x distinct domains) (Sha et al., 1998) and was initially identified in a screen for secretory mutants (termed “SEC”) (Novick et al., 1980). An identical phospholipid-binding pocket was observed in several mammalian proteins, including the CELLULAR RETINAL-BINDING PROTEIN (CRALBP), TRIO and α-TOCOPHEROL-TRANSFER PROTEIN (α-TTP) (Crabb et al., 1998; Min et al., 2003). That is why the SEC14 domain is also known as CRAL-TRIO domain (Panagabko et al., 2003). A unique feature of the SEC14 domain is that the lipophilic ligand is bound and enclosed as a whole molecule in the hydrophobic lipid-binding pocket (Min et al., 2003; Schaaf et al., 2006), while other lipid-binding domains, like FYVE or PH, only bind lipid headgroups (Stahelin, 2009). The alpha helical amino (N)-terminus of Sep14p is defined as CRAL-TRIO-N-terminal extension (CTN) (IPR011074) and cannot be identified in all SEC14L-PITPs (Saito et al., 2007a). The ability to open and close the SEC14 lipid-binding pocket by structural changes seems to be essential for domain activity and the biological function of this domain (Ryan et al., 2007; Schaaf et al., 2008; Schaaf et al., 2011; Kono et al., 2013). The open status is believed to be the membrane-attached structure, while the closed conformation, the one binding a substrate, is understood as the cytosolic version of the SEC14 domain (Tripathi et al., 2014). This fits the observation that the CTN-SEC14 module is crucial for membrane association of SEC14L-PITPs, since loss of the module leads to the accumulation of the protein in the cytosol (Skinner et al., 1993; Sirokmany et al., 2006; Sun et al., 2006; Saito et al., 2007a; Saito et al., 2007b; Montag et al., 2020). The lipid-presentation model of SEC14L-PITP function is based on results indicating that the SEC14 domain is essential for promoting membrane trafficking by supporting PI(4)P-OH kinase activity (Strahl and Thorner, 2007) (Figure 1B). For example, the SEC14 domain can recognize membrane-bound phosphatidylcholine (PC) and present PI bound in the lipid-binding pocket of the SEC14 domain to a PI(4)P-OH kinases for phosphorylation, as proposed for Arabidopsis SEC14 protein named SFH1 (Figure 1C), according to (Kf de Campos and Schaaf, 2017). Phosphorylation of SEC14 protein may regulate the association of SEC14 proteins with membranes (Suzuki et al., 2016) (Figure 1C). It is remarkable that unicellular organisms have SEC14-only single-domain SEC14L-PITPs, whereas multicellular organisms of the animal and plant lineage have single-domain and multi-domain SEC14L-PITPs (Figures 1D, 2) (Montag et al., 2020). The SEC14 domain represents a hydrophobic pocket-like lipid-binding site, in which a hydrophobic lipid substrate can bind (Sha et al., 1998). This site was found to be flanked by helical regions termed the anchor helix and gate helix. The anchor helix confers association with the membrane. The amphipathic gate helix keeps the lipid-binding site in an open or close conformation (Sha et al., 1998; Schaaf et al., 2008; Sugiura et al., 2021; Hornbergs et al., 2022; Yao et al., 2023) (Figure 1E). Additional residues may favor orientation either towards the negatively charged membrane or the cytosol, and they may also steer specificity for the lipid substrate (Sha et al., 1998; Schaaf et al., 2008; Sugiura et al., 2021; Hornbergs et al., 2022; Yao et al., 2023). Upon a heterotypic lipid exchange, an intermediate with two different lipophilic substrate-binding sites can form (Schaaf et al., 2008).
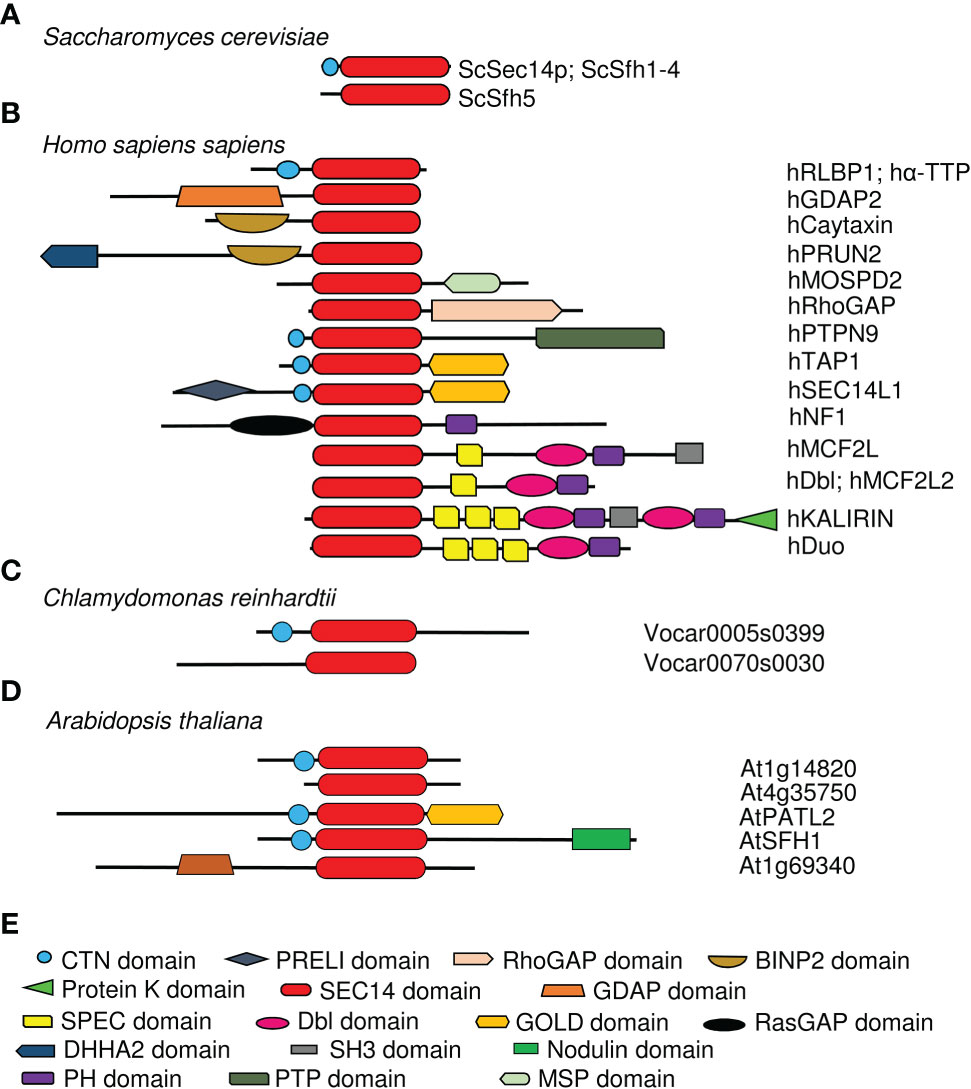
Figure 2 Schematic structures of representative SEC14L-PITPs showing the differing complexity between single- and multi-domain SEC14L-PITPs in unicellular and multicellular species. SEC14-PITPs in (A), yeast Saccharomyces cerevisiae; (B), Homo sapiens, (C), Chlamydomonas reinhardtii; (D), Arabidopsis thaliana. The presence of different types of domains is indicated in colors, and example names are provided on the right. (E) Symbols used in (A–D). Unicellular eukaryotes have simple single-domain SEC14L-PITPs, while multicellular eukaryotes have single- and various multi-domain proteins with various types of additional domains attached. Some multidomain proteins have independently evolved in the animal and plant lineage, e.g. SEC14-GOLD domain proteins, while others are unique in one of the lineages, e.g. SEC14-nodulin proteins in land plants.
While the functions of several single-domain SEC14-only proteins are studied well, only a few multi-domain SEC14L-PITPs are characterized. At the same time, their abundance in higher eukaryotes highlights their importance. This raises several fundamental questions: Which are the pathways that single- and multi-domain SEC14L-PITPs are integrated in? What are the functions of the different domains within the multi-domain SEC14L-PITPs? Which protein-protein and protein-ligand interactions are relevant for the functions of SEC14L-PITPs? Herein, we review SEC14L-PITPs and focus on structure and function of single- and multi-domain SEC14-PITPs, especially in plants. We highlight particularly the subgroup of SEC14-GOLD proteins. We analyze which role might be played by subdomains to allow binding with phospholipids of the membrane and protein-protein interaction.
4 Single-domain SEC14-only PITPs
In addition to Sec14p and its homologs in yeast (Bankaitis et al., 1990; Cleves et al., 1991; Schnabl et al., 2003; Griac, 2007), single-domain SEC14-only proteins can be identified in Chlamydomonas and in higher eukaryotes, either with or without CTN (Figure 2) (Saito et al., 2007a; Montag et al., 2020). While all SEC14-only proteins in yeast are well characterized by demonstrating their roles in different aspects of the phospholipid metabolism, for example organization of the actin cytoskeleton, activation of phospholipase D or prevention of saturated fatty-acid accumulation (Li et al., 2000; Desfougeres et al., 2008; Yakir-Tamang and Gerst, 2009), only few SEC14-only proteins have been studied till now in higher multicellular eukaryotes. In spite of this, the studies of human SEC14-only proteins have increased the knowledge about the functions of SEC14L-PITPs and their essential roles within organisms. For example, human CRALBP is able to transport 11-cis retinaldehyde, the photosensitive component of rhodopsin, in its SEC14 lipid-binding pocket (Crabb et al., 1998; Fishman et al., 2004). This feature makes CRALBP essential for photoreceptor function. Mutations in its SEC14 domain can be the causes of neurodegenerative diseases affecting the eyesight by photoreceptor involution (Maw et al., 1997; Burstedt et al., 2001; Eichers et al., 2002). Another important human SEC14-only protein is α-TTP found to be most abundant in liver cells, where it is involved in vitamin E secretion, especially as α-tocopherol (α-Toc), but it is also expressed in mammalian uterine and placental cells during embryogenesis (Sato et al., 1993; Arita et al., 1997; Miller et al., 2012). Vitamin E is known to be an important antioxidant neutralizing reactive oxygen species (ROS) and radical formation involving membrane lipids (Nukala et al., 2018). Important for the biological function and localization of α-TTP is its ability to not only bind α-Toc (in its lipid-biding pocket) but also PIPs (at the entrance of the lipid-binding pocket) (Kono et al., 2013; Chung et al., 2016). PIP binding mediates the release of α-Toc at membranes by inducing the conformational change of the SEC14 binding pocket from closed to open (Meier et al., 2003; Kono et al., 2013). Mutations in α-TTP lead to the neurodegenerative disease AVED (ataxia, with vitamin E deficiency) caused by dramatic vitamin E deficiency, which results in disturbance of muscle activity (Ouahchi et al., 1995; Min et al., 2003). These two examples of human single-domain SEC14-only proteins show their critical roles in binding and transporting additional lipophilic ligands besides PI, PIPs and PC. Generally, the presence of SEC14-only proteins in unicellular and multicellular eukaryotes demonstrates the importance of regulating lipophilic-substance transport within a cell.
Out of the 15 described single-domain SEC14-only proteins of A. thaliana, a functional role and structural characteristics are known for CPSFL1, also known as PITP7 (Figure 2; Table 1) (Montag et al., 2020). This chloroplast-localized single-domain SEC14 protein is involved in formation of vesicles from the inner thylakoid membrane, where it may transfer phosphatidic acid and PIPs (Hertle et al., 2020; Kim et al., 2022). Interestingly, a similar single-domain SEC14 protein function was described for Chlamydomonas reinhardtii, where CPSFL1 defects also caused chloroplast dis-functioning, light sensitivity and low carotene contents in plastoglobuli and eyespot (García-Cerdán et al., 2020). Chlamydomonas CPSFL1 is able to bind besides phosphatidic acid also carotene and precursor substrates (García-Cerdán et al., 2020). Hence, this CPSFL1 single-domain SEC14 proteins seem to function in the development of chloroplasts by transporting and transferring relevant lipophilic substances inside chloroplasts.
5 Multi-domain SEC14L-PITPs
The number and modular complexity of SEC14L-PITPs increases in multicellular eukaryotes (Figure 2) (Montag et al., 2020). Through the presence of one or more additional domains the functions of SEC14L-PITPs are extended, the functions of different domains are better coordinated with respect to each other in a cell and upon loss of function the risk of second-site dominant effects is reduced (Figure 1D). Multi-domain SEC14L-PITPs are not only regulators of lipophilic substance transport but they may have the ability to function, e.g. as proteins with enzymatic functions, guanine exchange factors (GEFs) or GTPase-activating proteins (GAPs). For example, the human multi-domain SEC14L-PITP TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 9 (PTPN9) has an additional protein phosphatase catalytic (PTP) domain (IPR000242) and functions as a tyrosine phosphatase (Denu and Dixon, 1998). The CTN-SEC14 module of PTPN9 is responsible for protein localization to the outer surface of secretory vesicles binding either phosphatidylserine (PS) or PI(3,4,5)P3 (and other PIPs) (Kruger et al., 2002; Krugmann et al., 2002; Huynh et al., 2003; Zhao et al., 2003; Saito et al., 2007a; Saito et al., 2007b). The membrane targeting function of the (CTN)-SEC14 domain and its role in intracellular vesicle trafficking could also be recognized in human multi-domain SEC14L-PITPs functioning as GEFs and GAPs and thereby regulating the Ras/Raf-signaling pathway (Ueda et al., 2004; Sirokmany et al., 2006; Sun et al., 2006). For example, human SEC14L-PITPs with a GAP function are KALIRIN, Dou or RhoGAP, while MCF2 or MCF2L are functioning as GEFs. Mutations in all these proteins are linked to neurodegenerative diseases or cancer. Defects in human NEUROFIBROMIN 1 (NF1), a putative negative regulator of the Ras-signaling pathway, are disease-associated, especially when occurring in the double domain structure of SEC14-PH (Rad and Tee, 2016). The Pleckstrin homology (PH) domain (IPR001849) has a phospholipid-binding specificity for PI(4,5)P2 and seems to be involved in protein recruitment to membranes (Hyvonen et al., 1995; Lemmon and Ferguson, 1998; Lemmon, 2007). Patients with an NF1 mutation are developing the Recklinghausen disease/Watson syndrome and have a significantly increased cancer risk (Rasmussen and Friedman, 2000; D'Angelo et al., 2006; Yap et al., 2014; Peltonen et al., 2017). Another example of SEC14L-PITPs playing an important role in human health is the prostate cancer suppressor PROTEIN Prune Homolog 2 With BCH Domain (PRUNE2) containing the additional BINP2 domain and DHHA2 domain (IPR004097) at its N-terminus (Salameh et al., 2015). The human multi-domain SEC14L-PITP GANGLIOSIDE-INDUCED DIFFERENTIATION-ASSOCIATED PROTEIN 2 (GDAP2) contains a GDAP macro domain (IPR035793) at its N- terminus, which possibly binds ADP-ribose, and is localized to the lysosomal membrane (Martzen et al., 1999; Hassa et al., 2006). Its exact function is unknown, but recently homologs were identified in plants, suggesting a conserved function (Montag et al., 2020).
5.1 SEC14-nodulin proteins (plant-specific)
A plant-specific subfamily of multi-domain SEC14L-PITPs are SEC14-nodulin proteins exhibiting an additional C-terminal nodulin domain, present in this combination in seed plants (Figure 2; Table 1) (Kapranov et al., 2001; Denancé et al., 2014; Montag et al., 2020). The nodulin name is derived from nodulin genes and proteins highly expressed and accumulating during the nitrogen-fixing symbiosis of legume plants at the root hair invasion, infection and nodule developmental and nitrogen-fixing site (Denancé et al., 2014). It was therefore very interesting to find that one function of SEC14-nodulin proteins is to be basic regulators in polarizing membrane trafficking (Vincent et al., 2005; Huang et al., 2013; Ghosh et al., 2015). A well-studied member of this protein family in this context is A. Thaliana SFH1, also known as CAN OF WORMS1 (COW1), which is involved in root hair biogenesis by controlling the tip-directed gradient of PI(4,5)P2 (and PI(4)P) (Bohme et al., 2004; Vincent et al., 2005; Preuss et al., 2006; Ghosh et al., 2015). Sfh1 loss-of-function mutant plants have short root hairs defective in elongation (Ghosh et al., 2015). Essential for this are both the SEC14 domain and the nodulin domain including the C-terminal poly-lysine motif stretch of the nodulin domain (Ghosh et al., 2015). The SEC14 domain lipid-binding activity is important, since mutations leading to reduced phosphatidylinositol transfer in vitro do not complement the root hair phenotype of sfh1 mutants (Huang et al., 2016). The poly-lysine stretch is presumably required for PI(4,5)P2 binding and assembly of the SEC14-nodulin protein at the plasma membrane since mutants devoid of such a stretch do not complement the sfh1 phenotype, do not locate at the plasma membrane in yeast cells and have an altered oligomerization behavior. According to the working model proposed by Ghosh et al., (2015), oligomers of SFH1 present PI contained inside the SEC14 domains at membrane sites to phosphatidylinositol-4 or -5-phosphate kinase, thereby changing the phosphatidylinositol landscape and properties of the plasma membrane. Subsequently, oligomeric SFH1 complexes may form at the plasma membrane. These events may trigger root hair cell elongation. A similar function is proposed for rice OsSNDP2 and OsSNDP3 during pollen tube elongation (Moon et al., 2022). Two further SEC14-nodulin proteins from Arabidopsis have been recently studied in physiological contexts, which are SFH5 and SFH7 (Yao et al., 2023). These two proteins were found to localize at ER and chloroplast membranes. SFH5 and SFH7 were able to bind phosphatidic acid and transfer it between membranes in vitro. Additionally, it was shown that double knockout mutants had aberrant thylakoid membrane structures in chloroplasts. Interestingly, phosphatidic acid is the precursor to several lipids produced in chloroplasts and required for thylakoid assembly, and the sfh5 sfh7 double mutants had reduced amounts of such lipids. Hence, these findings strongly suggest that the SEC14-nodulin proteins SFH5 and SFH7 mediate the transport of phosphatidic acid from ER to chloroplast perhaps at interorganellar contact sites (Yao et al., 2023). SEC14-nodulin proteins therefore play roles in different organs (roots and leaves) and are involved in different processes requiring lipid transfer, such as polarity control and organellar contacts.
5.2 SEC14-GOLD proteins
A well-studied family of multi-domain SEC14L-PITPs is the SEC14-GOLD family. All SEC14-GOLD proteins contain a Golgi dynamics (GOLD) domain (IPR009038) at their C-terminus (Figures 2, 3; Table 1). They can be found in insects and vertebrates. In the green lineage, SEC14-GOLD proteins were identified in bryophytes Marchantia polymorpha and vascular plants (Montag et al., 2020). It has been shown that the GOLD domain functions in membrane trafficking along the secretory pathway by mediating diverse protein-protein and protein-membrane interactions (Sohda et al., 2001; Anantharaman and Aravind, 2002; Carney and Bowen, 2004; Pastor-Cantizano et al., 2016; Pastor-Cantizano et al., 2018). The GOLD domain is either present in single-domain proteins or co-occurs with other domains, which are all involved in lipid-binding (Anantharaman and Aravind, 2002; McPhail et al., 2017; Pastor-Cantizano et al., 2016).
Figure 3 Overview of human and Arabidopsis thaliana SEC14-GOLD proteins. Phylogenetic trees of (A), human and (B), Arabidopsis SEC14-GOLD proteins and their modular architecture. [Phylogenetic analysis, domain identification and alignments were performed as described in Montag et al., 2020]. (C), Symbols used in (A, B) Plant PATLs have differing N-terminal regions, while human SEC14-GOLD proteins have either no extensive N-terminal region or a N-terminal region with PRELI domain. CTN, cellular retinal-binding protein and TRIO protein N-terminal extension; PRELI, PRELI/MSF resembling human PRELI protein; SEC14, secretory mutant 14 protein; GOLD, Golgi dynamics.
The analyses of SEC14-GOLD proteins revealed two subgroups of this family in humans (Figure 3A). Next to the two defining domains, SEC14-LIKE1 (SEC14L1) and SEC14-LIKE5 (SEC14L5) have an additional N-terminal PRELI/MSF1 domain (IPR009038/IPR006797) (Anantharaman and Aravind, 2002). Similar SEC14-GOLD proteins with additional PRELI domain can also be identified in other higher eukaryotes like Zebrafish, Drosophila melanogaster, Mus musculus, and Caenorhabditis elegans, but not in any of the checked plant and yeast species. It is assumed that the PRELI/MSF1 domain could function in protein association to membranes as well as in transferring lipids (Anantharaman and Aravind, 2002; Yu et al., 2015), due to its involvement in mitochondrial protein sorting and phosphatidylethanolamine metabolism (Nakai et al., 1993; Hall et al., 2011). For example, hSEC14L1 is able to associate with two transporters, the VESICULAR ACETYLCHOLIN TRANSPORTER (VACht) and the CHOLINE TRANSPORTER 1 (CHT1), on synaptic vesicles (Ribeiro et al., 2007). This indicates that hSEC14L1 may play a fundamental role in intracellular vesicle trafficking (Ribeiro et al., 2007). Additionally, it has a negative regulatory function on RETINOIC ACID-INDUCIBLE GENE I (RIG-I), important for antiviral immunity response. RIG-I interaction with other proteins is inhibited through its interactions with hSEC14L1, via its PRELI domain and SEC14 domain (Li et al., 2013). hSEC14L5 was characterized as a potential target for post-traumatic stress disorder (PTSD) found in a study to identify molecular and genetic key players in this disease (Chitrala et al., 2016). Human TOCOPHEROL-ASSOCIATED PROTEINs (TAPs), TAP1 (SPF/SEC14L2), TAP2 (p45/SEC14L3) and TAP3 (SFP2/SEC14L4), have a SEC14-GOLD domain combination with no additional N-terminal extensions (termed “N region”) (Figure 3A). They have the ability to bind α-Toc, α-Toc derivatives, squalene, phosphatidylglycerol, phosphatidylcholine (PC), PI and PIPs (Chin and Bloch, 1985; Kempna et al., 2003; Stocker and Baumann, 2003). TAP proteins have a Rab-like small GTPase activity (Habermehl et al., 2005; Gong et al., 2017). Furthermore, the ability to bind α-Toc and its derivatives indicates a role of human TAPs in preventing lipid peroxidation. This is supported by the observation that phosphorylated hTAP1 is able to stimulate cellular cholesterol biosynthesis, since protecting low density lipoproteins from oxidation may inhibit cholesterol uptake (Neuzil et al., 1997; Shibata et al., 2001; Stocker and Baumann, 2003). Another hint to that assumption is that the presence of hTAP1 is able to increase vitamin E-mediated membrane protection from lipid peroxidation, which positively influences RNA replication of the hepatitis C virus in cell cultures (Saeed et al., 2015; Li et al., 2018). Additionally, hTAP1 and its functional orthologue Cgr-1 in C. elegans are playing a conserved role in the Ras/Raf pathway by being regulators of the Raf-signal activation and thereby suppressing its oncogenic capacity (Johnson and Kornfeld, 2010). Here again its ability to bind α-Toc positively influences health by regulating the uptake of α-Toc into cancer cells to stop cell growth and amplification. But hTAP1 is not only involved in tumor suppression by mediating α-Toc uptake and lipid protection, it also contributes to the regulation of PI(3)P Kinase γ (PI3Kγ) activity, either by blocking its subunit interaction or starting its activity, which then leads to VASCULAR ENDOSOMAL FACTOR (VEGF) expression (Ni et al., 2005; Wang et al., 2009; Zingg et al., 2014; Zingg et al., 2015; Zingg et al., 2017). Another fact linking hTAP1 to carcinogenesis is the observation that it is highly expressed in breast and prostate tissue, but downregulated in prostate and breast cancer cell lines, as well as in human breasts with invasive breast carcinomas (Ni et al., 2005; Wang et al., 2009). In zebrafish, TAP2 is crucial for the hydrolysis of PI(4,5)P2 by phospholipase C (Gong et al., 2017). Rat (Rattus norvegicus) p45, a hTAP2 homolog, especially binds PI(3,4,5)P3 in vitro and localizes with it in secretory vesicles, the cytoplasm and the extracellular space (Merkulova et al., 2005). Deletion of the SEC14 domain leads to inhibited secretion into the extracellular space, indicating that the SEC14 domain is essential for secretion (Merkulova et al., 2005). The expression of rat SPF2, a homolog of hTAP3, is mainly observed in skin and respiratory tissue (Merkulova et al., 1999; Kempna et al., 2003). Recombinant SPF2 is able to stimulate the monooxygenase but not as efficient as TAP1 (Mokashi et al., 2004). Its activity is thereby stronger dependent on regulation by protein kinase A phosphorylation, guanine nucleotides and α-Toc, than TAP1 (Mokashi et al., 2004). An alternative splicing pattern was obtained for human TAP3 (Kempna et al., 2003). Due to this, reduced levels of biologically active hTAP3 could increase the risk of disease outbreak associated with the secretory capability of tissues/cells (Zingg et al., 2008; Kempna et al., 2010), underlining possible roles of TAPs as tumor suppressors. Thus, the data on animal SEC14-GOLD proteins demonstrate SEC14-GOLD protein roles in intracellular vesicle trafficking by interaction with PIPs. Additionally, it shows their function as negative regulators via protein-protein interactions and demonstrates their oncogenic role. Furthermore, the data highlights their possible function as tumor suppressers, e.g. by mediating vitamin E transport and by preventing cellular damage by ROS and radicals.
In Arabidopsis and other plants, SEC14-GOLD proteins are called PATELLINs (PATLs), named after patella, the Latin word for small plate, referring to PATL1 localization at the developing cell plate (Peterman et al., 2004). Analysis of the SEC14L-PITP superfamily in Arabidopsis revealed six PATL proteins, with a CTN-SEC14 and GOLD domain but no other N-terminal domains (Figure 3B; Table 1) (Peterman et al., 2004). PATLs display a variable N region of unknown structure, however different small motifs (coiled coil- and PXXP motifs) can be found (Peterman et al., 2004; Neduva and Russell, 2006; Diella et al., 2008; Montag et al., 2020). The N regions of PATLs vary in amino-acid sequences and are unique for each protein (Montag et al., 2020). PATLs, except of PATL6, show an overall acidic N region due to repeats of glutamate (E) (Peterman et al., 2004). But they also show a pattern of lysines (K) surrounding the E repeats in PATL1, PATL2 and PATL4 (Montag et al., 2020). Independently of the N regions, plant SEC14-GOLD proteins form three clades with subgroup-specific amino acid substitutions in the GOLD domain, which may define different functional categories (Peterman et al., 2006; Bermudez et al., 2018; Montag et al., 2020). Expression analyses of PATLs uncovered overlapping and clade-specific clusters, and together with studies on multiple knock-out plants, this indicates partial redundancy within the family (Tejos et al., 2017; Montag et al., 2020). Multiple patl mutants demonstrated the essential role of PATLs in Arabidopsis patterning and polarity by revealing auxin response phenotypes and developmental defects due to decreased polarization of the auxin transporter PIN-FORMED 1 (PIN1) (Tejos et al., 2017). The role of PATLs during plant development can be confirmed by the observation that PATL1 expression was increased in developing leaves and vascular tissues and by cellular localization of PATL1 at the plasma membrane and the cell plate during cell division (Peterman et al., 2004). Distinct and overlapping localization patterns, were found for all other PATLs. PATLs are peripheral membrane proteins found to localize at the plasma membrane, at the cell plate and/or were found to be cytosolic. PATL genes were expressed in leaf epidermis cells, vascular tissues, during embryogenesis, during development of lateral-root primordia and during differentiation of the root apical meristem (Suzuki et al., 2016; Tejos et al., 2017; Wu et al., 2017). Protein localization during development and differentiation links PATLs closely to membrane trafficking supported through the observation that PATL1, PATL2 and PATL3 bind PIPs (Peterman et al., 2004; Suzuki et al., 2016; Wu et al., 2017). PATLs are involved in a number of different protein interactions at membrane sites (Figure 4). Although PATL1 preferentially binds PI(5)P, PI(3)P and PI(4,5)P2 and AtPATL3 mainly binds PI(4)P and PI(4,5)P2, both still have the ability to associate with all other PIPs (Peterman et al., 2004; Wu et al., 2017). All domains of PATL2 contributed to PIP association (Montag et al., 2020) (Figure 4). The CTN-SEC14 module of PATL2 was found to govern membrane association of the protein, and the GOLD domain to specify plasma membrane localization, presumably by recognizing PI(4,5)P2 maybe through its lysine motif (Montag et al., 2020). A hint linking PATLs to plasma membrane protein regulation and membrane trafficking is the observation that PATL1 and PATL2 were able to interact with plasma membrane proteins (Chu et al., 2018; Zhou et al., 2018; Hornbergs et al., 2022) (Figure 4). Through its GOLD domain PATL1 interacted with CALMODULIN-4 (CaM4), a multifunctional sensor for Ca2+, and via its N region it interacted with SALT OVERLY-SENSITIVE 1 (SOS1), a Na+/H+ antiporter localized at the plasma membrane (Chu et al., 2018; Zhou et al., 2018). Its closest homologue PATL2 interacted through its N region with IRON-REGULATED TRANSPORTER 1 (IRT1), an essential protein for iron acquisition by roots in soil (Hornbergs et al., 2022) (Figure 4B). PATL1 and PATL2 contribute to stress tolerance by affecting plant responses to cold, salt and iron nutrition-related stress. In addition, both seem to be involved in preventing damage caused by reactive oxygen species (ROS) and radicals (Zhou et al., 2018; Hornbergs et al., 2022). patl2 loss-of-function mutants exhibited enhanced iron reduction activity in roots, a response required for iron acquisition via IRT1 in Arabidopsis, as well as enhanced lipid peroxidation phenotypes (Hornbergs et al., 2022). Interestingly, interactome analysis of tagged PATL2 retrieved ROS response/metabolism proteins and, under iron deficiency, endomembrane trafficking regulators. PATL2 protein was found to bind the antioxidant α-Toc in the lipid-binding SEC14 domain (Hornbergs et al., 2022). Vitamin E deficiency also caused iron utilization phenotypes. Taken together, PATL2 may recruit a ROS response interactome to IRT1 sites and present or transfer α-Toc to IRT1 membrane sites. Since IRT1 mediates uptake of reactive iron and other metal ions, vitamin E compounds may protect from potential oxidative stress due to lipid peroxidation catalyzed in the presence of these reactive Fenton metal ions. Subsequently, endomembrane trafficking may affect the regulation of IRT1 (Hornbergs et al., 2022). In this context, it is interesting to note that phosphorylation of the N region was identified under Fe deficiency (Lan et al., 2011) and during salt stress, in response to oligogalacturonides, and brassinosteroid signaling (Tang et al., 2008; Hsu et al., 2009; Chang et al., 2012; Mattei et al., 2016), indicating that protein interaction of PATL2 with IRT1 may be under control of protein phosphorylation of the interacting N region. PATL1 and PATL2 co-immunoprecipitated with ASSOCIATED MOLECULE WITH THE SH3 DOMAIN OF STAM3 (AMSH3), a deubiquitinating enzyme required for intracellular trafficking and vacuole biogenesis in Arabidopsis, next to other proteins with reported or expected function in intracellular trafficking processes (Isono et al., 2010). PATL2 is phosphorylated by MAP KINASE4 (MPK4) within the SEC14 domain, which might be important for the release of PATL2 from the membrane (Suzuki et al., 2016) (Figure 4A). Additionally, phosphorylation of the SEC14 domain of PATL2 could be detected after short- term cytokinin treatment and sugar stress (Niittyla et al., 2007; Cerny et al., 2011). This indicates that PATL2 might undergo dynamic post-translational regulation in response to plant stress. PATL3 recruitment to the plasma membrane depended on interaction of its GOLD domain with EXO70A1, a subunit of the exocyst complex participating in intracellular vesicle transport (He and Guo, 2009; Fendrych et al., 2013; Wu et al., 2017). Interestingly, all other PATLs except of PATL5, were able to interact with EXO70A1 (Wu et al., 2017). Moreover, PATL3 and PATL6 inhibited stem infection spread of the alfalfa mosaic virus by interfering with virus movement through interaction with a PLASMODESMATA TARGETING MOVEMENT PROTEIN (AMV MP) and thereby preventing subcellular targeting (Peiro et al., 2014) (Figure 4A). Tomato TOCOPHEROL BINDING PROTEIN (SlTBP) is a homologue of Arabidopsis PATL6 and clusters together with other plant PATLs due to a plastid-targeting signal (Bermudez et al., 2018). The SlTBP gene is mainly expressed in photosynthetic active tissues and the protein is localized to plastids. Its potential ability to bind α-Toc makes it a key player in controlling possibly vitamin E movement between plastids and the ER, which affects lipid metabolism within these organelle (Bermudez et al., 2018). Additionally, SlTBP is involved in maintaining chloroplast membrane structure, affecting its lipid profile (Bermudez et al., 2018). Interestingly, PATL1, PATL2 and PATL5 were identified as putative cargo receptors in proteomic studies searching for components of the chloroplast vesicle transport pathway. The proteins were identified localizing to the chloroplast envelope (Kleffmann et al., 2004; Ferro et al., 2010; Khan et al., 2013).
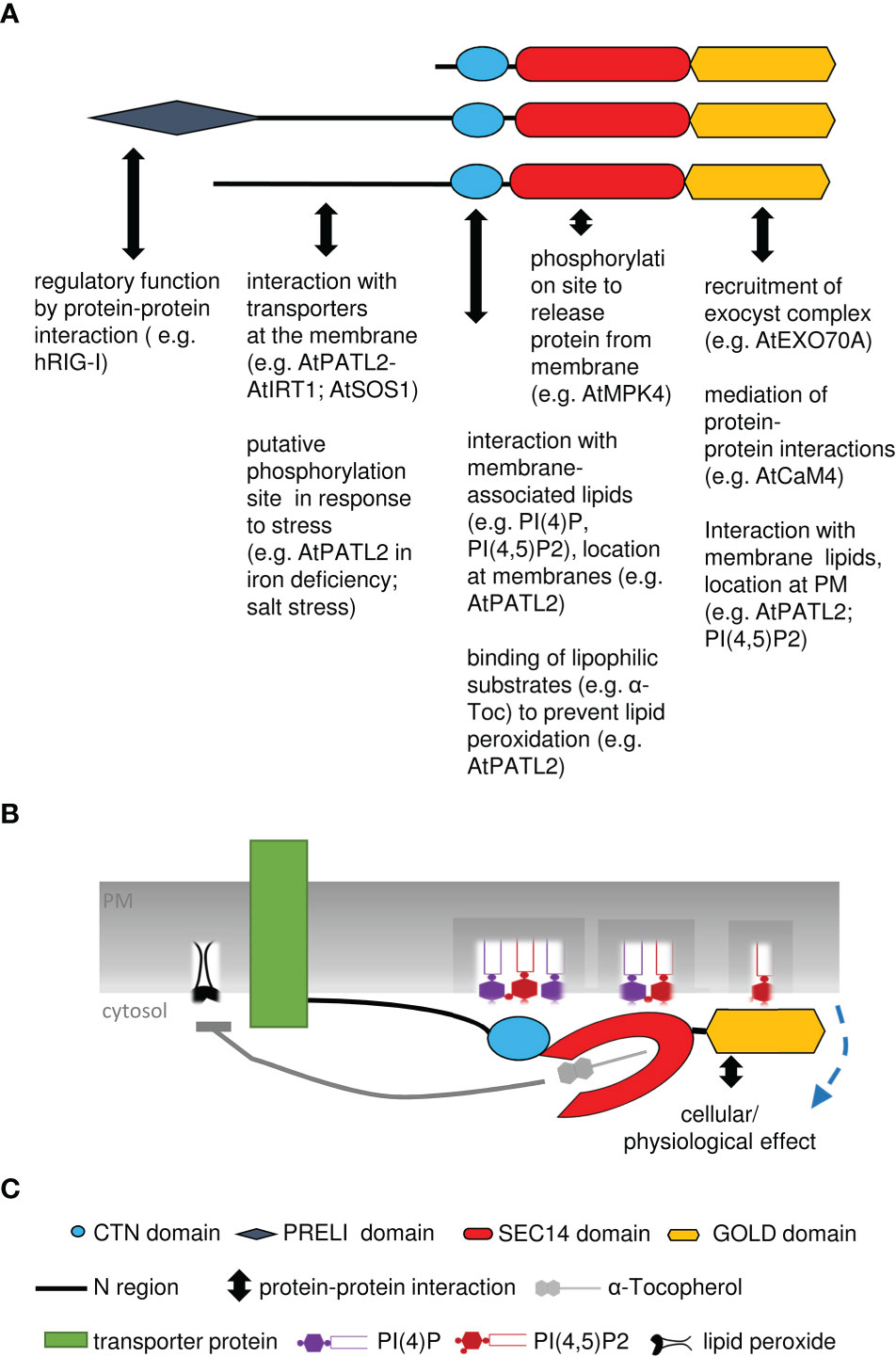
Figure 4 Summary model of SEC14-GOLD protein functions. (A), Summary of various interactions with proteins, lipids or ligands, described for different parts of SEC14-GOLD proteins. (B), Model of AtPATL2, binding to phospholipids and interacting with transport protein in the plasma membrane, leading to prevention of oxidative stress and lipid peroxidation; PATL2 may exchange or present the antioxidant a-tocopherol. The cellular physiological effect may comprise vesicle formation and regulation of transporter abundance and activity. (C), Explanations of symbols used in (A, B).
Taken together, PI/PIP and α-Toc binding, cell plate and membrane localization, phosphorylation, increased expression during stress responses, and interaction with membrane and trafficking proteins indicate that PATLs are basic cell regulators adapting the cell/organism during cell division and growth and to altered environmental influences and in response to external stimuli. As regulatory proteins PATLs may be involved in membrane trafficking, e.g., by initiating vesicle formation. Furthermore, they might play a role in protecting the cell from ROS and radical damage, by offering α-Toc as an antioxidant or through regulating the activity of membrane proteins.
6 Conclusion
The SEC14 domain is the common feature of SEC14 proteins. The domain is capable to recognize, bind, transport, and exchange single lipophilic molecules between membranes inside a lipid-binding site. Unlike other lipid-binding domains, the SEC14 domain not only binds phospholipids, but also other lipophilic substances, e.g., α-Toc, carotenoids. This ability leads to further cellular and physiological effects when these lipid transfer activities are required. This characteristic and the presence of SEC14L-PITPs in higher eukaryotes indicates a conserved function and highlights the need of controlling lipid signaling and membrane trafficking. Especially, the presence of additional domains in multi-domain SEC14L-PITPs of higher multi-cellular eukaryotes indicates an increasing variety of functions due to enhanced possibilities for protein interaction, cellular localization or enzyme activities. These additional domains could be involved in the sensing of the lipid environment at the membrane or in changes of lipid-signaling pathways leading to environmental adaptation. The idea is supported by the findings that multi-domain SEC14L-PITPs are able to influence cell division, vesicle formation, lipid signaling, environment responsiveness and organism development, which directly affects the organism fitness/health. The SEC14 domain is also of special interest, since mutations in the SEC14 domain result in defects in development and in the plant-stress response and tolerance, as well as in neurodegenerative diseases and an increased cancer risk in humans.
Not all complex tasks of SEC14L-PITPs are understood right now and many interesting questions remain to be answered. For example, are SEC14L-PITPs regulated by post- transcriptional regulation? What are the effects of protein phosphorylation? What is the function of the plant-specific regions of SEC14L-PITPs? How do SEC14L-PITPs influence protein activity and metabolic reactions? In which pathways do yet uncharacterized proteins of this kind play a role? Identifying the functions of SEC14L-PITPs on cellular levels can help to understand the adaptation of regulatory pathways to environmental changes.
Author contributions
KM and PB wrote the article and prepared figures. All authors contributed to discussion. PB and RI revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) SFB 1208 (CRC 1208) Project B05 to PB.
Acknowledgments
We sincerely apologize to those authors who were not able to cite their works in this review due to space limitation. We are thankful to Jannik Hornbergs for critical discussions. KM has been member of the MB Train graduate school integrated in the CRC1208.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
α-TOC, α-tocopherol; C-terminal, carboxy-terminal; CTN, CRAL-TRIO-N-terminal extension; ER, endoplasmic reticulum; N-terminal, amino-terminal; N region, N-terminal region of PATLs; PATL, patellin; PC, phosphatidylcholine; PE, phosphatidyl ethanolamine; PI, phosphatidylinositol; PIP, phosphatidylinositol phosphate; PITP, phosphatidyl transfer protein; PS, phosphatidylserine; ROS, reactive oxygen species; SEC14L-PITP, SEC14-like PITP.
References
Anantharaman, V., Aravind, L. (2002). The GOLD domain, a novel protein module involved in golgi function and secretion. Genome Biol. 3 (5), research0023. doi: 10.1186/gb-2002-3-5-research0023
Aravind, L., Iyer, L. M. (2012). The HARE-HTH and associated domains: novel modules in the coordination of epigenetic DNA and protein modifications. Cell Cycle 11 (1), 119–131. doi: 10.4161/cc.11.1.18475
Arita, M., Nomura, K., Arai, H., Inoue, K. (1997). Alpha-tocopherol transfer protein stimulates the secretion of alpha-tocopherol from a cultured liver cell line through a brefeldin a-insensitive pathway. Proc. Natl. Acad. Sci. U.S.A. 94 (23), 12437–12441. doi: 10.1073/pnas.94.23.12437
Bankaitis, V. A., Aitken, J. R., Cleves, A. E., Dowhan, W. (1990). An essential role for a phospholipid transfer protein in yeast golgi function. Nature 347 (6293), 561–562. doi: 10.1038/347561a0
Bankaitis, V. A., Mousley, C. J., Schaaf, G. (2010). The Sec14 superfamily and mechanisms for crosstalk between lipid metabolism and lipid signaling. Trends Biochem. Sci. 35 (3), 150–160. doi: 10.1016/j.tibs.2009.10.008
Bermudez, L., Del Pozo, T., Silvestre Lira, B., de Godoy, F., Boos, I., Romano, C., et al. (2018). A tomato tocopherol-binding protein sheds light on intracellular alpha-tocopherol metabolism in plants. Plant Cell Physiol. 59 (11), 2188–2203. doi: 10.1093/pcp/pcy191
Bohme, K., Li, Y., Charlot, F., Grierson, C., Marrocco, K., Okada, K., et al. (2004). The arabidopsis COW1 gene encodes a phosphatidylinositol transfer protein essential for root hair tip growth. Plant J. 40 (5), 686–698. doi: 10.1111/j.1365-313X.2004.02245.x
Burstedt, M. S., Forsman-Semb, K., Golovleva, I., Janunger, T., Wachtmeister, L., Sandgren, O. (2001). Ocular phenotype of bothnia dystrophy, an autosomal recessive retinitis pigmentosa associated with an R234W mutation in the RLBP1 gene. Arch. Ophthalmol. 119 (2), 260–267.
Carney, G. E., Bowen, N. J. (2004). p24 proteins, intracellular trafficking, and behavior: drosophila melanogaster provides insights and opportunities. Biol. Cell 96 (4), 271–278. doi: 10.1016/j.biolcel.2004.01.004
Cerny, M., Dycka, F., Bobal'ova, J., Brzobohaty, B. (2011). Early cytokinin response proteins and phosphoproteins of arabidopsis thaliana identified by proteome and phosphoproteome profiling. J. Exp. Bot. 62 (3), 921–937. doi: 10.1093/jxb/erq322
Chang, I. F., Hsu, J. L., Hsu, P. H., Sheng, W. A., Lai, S. J., Lee, C., et al. (2012). Comparative phosphoproteomic analysis of microsomal fractions of arabidopsis thaliana and oryza sativa subjected to high salinity. Plant Sci., 185–186. doi: 10.1016/j.plantsci.2011.09.009
Chin, J., Bloch, K. (1985). Stimulation by unsaturated fatty acid of squalene uptake in rat liver microsomes. J. Lipid Res. 26 (7), 819–823. doi: 0.1016/S0022-2275(20)34311-X
Chitrala, K. N., Nagarkatti, P., Nagarkatti, M. (2016). Prediction of possible biomarkers and novel pathways conferring risk to post-traumatic stress disorder. PLoS One 11 (12), e0168404. doi: 10.1371/journal.pone.0168404
Chu, M., Li, J., Zhang, J., Shen, S., Li, C., Gao, Y., et al. (2018). AtCaM4 interacts with a Sec14-like protein, PATL1, to regulate freezing tolerance in arabidopsis in a CBF-independent manner. J. Exp. Bot. 69 (21), 5241–5253. doi: 10.1093/jxb/ery278
Chung, S., Ghelfi, M., Atkinson, J., Parker, R., Qian, J., Carlin, C., et al. (2016). Vitamin e and phosphoinositides regulate the intracellular localization of the hepatic alpha-tocopherol transfer protein. J. Biol. Chem. 291 (33), 17028–17039. doi: 10.1074/jbc.M116.734210
Cleves, A., McGee, T., Bankaitis, V. (1991). Phospholipid transfer proteins: a biological debut. Trends Cell Biol. 1 (1), 30–34. doi: 10.1016/0962-8924(91)90067-j
Cockcroft, S. (2012). “The diverse functions of phosphatidylinositol transfer proteins,” in Phosphoinositides and disease. current topics in microbiology and immunology, vol. 362 . Ed. FALASCA, M. (Dordrech: Springer).
Cockcroft, S., Raghu, P. (2018). Phospholipid transport protein function at organelle contact sites. Curr. Opin. Cell Biol. 53, 52–60. doi: 10.1016/j.ceb.2018.04.011
Cooper, G. M. (2000). The cell: a molecular approach. 2nd edition Vol. 2000 (Sunderland (MA: Sinauer Associates). Available at: https://www.ncbi.nlm.nih.gov/books/NBK9839.
Crabb, J. W., Carlson, A., Chen, Y., Goldflam, S., Intres, R., West, K. A., et al. (1998). Structural and functional characterization of recombinant human cellular retinaldehyde-binding protein. Protein Sci. 7 (3), 746–757. doi: 10.1002/pro.5560070324
D'Angelo, I., Welti, S., Bonneau, F., Scheffzek, K. (2006). A novel bipartite phospholipid-binding module in the neurofibromatosis type 1 protein. EMBO Rep. 7 (2), 174–179. doi: 10.1038/sj.embor.7400602
Denancé, N., Szurek, B., Noël, L. D. (2014). Emerging functions of nodulin-like proteins in non-nodulating plant species. Plant Cell Physiol. 55 (3), 469–474. doi: 10.1093/pcp/pct198
Denu, J. M., Dixon, J. E. (1998). Protein tyrosine phosphatases: mechanisms of catalysis and regulation. Curr. Opin. Chem. Biol. 2 (5), 633–641. doi: 10.1016/s1367-5931(98)80095-1
Desfougeres, T., Ferreira, T., Berges, T., Regnacq, M. (2008). SFH2 regulates fatty acid synthase activity in the yeast saccharomyces cerevisiae and is critical to prevent saturated fatty acid accumulation in response to haem and oleic acid depletion. Biochem. J. 409 (1), 299–309. doi: 10.1042/BJ20071028
Diella, F., Haslam, N., Chica, C., Budd, A., Michael, S., Brown, N. P., et al. (2008). Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front. Biosci. 13, 6580–6603. doi: 10.2741/3175
Eichers, E. R., Green, J. S., Stockton, D. W., Jackman, C. S., Whelan, J., McNamara, J. A., et al. (2002). Newfoundland Rod-cone dystrophy, an early-onset retinal dystrophy, is caused by splice-junction mutations in RLBP1. Am. J. Hum. Genet. 70 (4), 955–964. doi: 10.1086/339688
Fendrych, M., Synek, L., Pecenkova, T., Drdova, E. J., Sekeres, J., de Rycke, R., et al. (2013). Visualization of the exocyst complex dynamics at the plasma membrane of arabidopsis thaliana. Mol. Biol. Cell 24 (4), 510–520. doi: 10.1091/mbc.E12-06-0492
Ferro, M., Brugiere, S., Salvi, D., Seigneurin-Berny, D., Court, M., Moyet, L., et al. (2010). AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol. Cell Proteomics 9 (6), 1063–1084. doi: 10.1074/mcp.M900325-MCP200
Fishman, G. A., Roberts, M. F., Derlacki, D. J., Grimsby, J. L., Yamamoto, H., Sharon, D., et al. (2004). Novel mutations in the cellular retinaldehyde-binding protein gene (RLBP1) associated with retinitis punctata albescens: evidence of interfamilial genetic heterogeneity and fundus changes in heterozygotes. Arch. Ophthalmol. 122 (1), 70–75. doi: 10.1001/archopht.122.1.70
García-Cerdán, J. G., Schmid, E. M., Takeuchi, T., McRae, I., McDonald, K. L., Yordduangjun, N., et al. (2020). Chloroplast Sec14-like 1 (CPSFL1) is essential for normal chloroplast development and affects carotenoid accumulation in chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 117 (22), 12452–12463. doi: 10.1073/pnas.1916948117
Gerth, K., Lin, F., Menzel, W., Krishnamoorthy, P., Stenzel, I., Heilmann, M., et al. (2017). Guilt by association: a phenotype-based view of the plant phosphoinositide network. Annu. Rev. Plant Biol. 68, 349–374. doi: 10.1146/annurev-arplant-042916-041022
Ghosh, R., de Campos, M. K., Huang, J., Huh, S. K., Orlowski, A., Yang, Y., et al. (2015). Sec14-nodulin proteins and the patterning of phosphoinositide landmarks for developmental control of membrane morphogenesis. Mol. Biol. Cell 26 (9), 1764–1781. doi: 10.1091/mbc.E14-10-1475
Gong, B., Shen, W., Xiao, W., Meng, Y., Meng, A., Jia, S. (2017). The Sec14-like phosphatidylinositol transfer proteins Sec14l3/SEC14L2 act as GTPase proteins to mediate Wnt/Ca(2+) signaling. Elife 6, e26362. doi: 10.7554/eLife.26362
Goring, D. R., Di Sansebastiano, G. P. (2017). Protein and membrane trafficking routes in plants: conventional or unconventional? J. Exp. Bot. 69 (1), 1–5. doi: 10.1093/jxb/erx435
Griac, P. (2007). Sec14 related proteins in yeast. Biochim. Biophys. Acta 1771 (6), 737–745. doi: 10.1016/j.bbalip.2007.02.008
Habermehl, D., Kempna, P., Azzi, A., Zingg, J. M. (2005). Recombinant SEC14-like proteins (TAP) possess GTPase activity. Biochem. Biophys. Res. Commun. 326 (1), 254–259. doi: 10.1016/j.bbrc.2004.11.021
Hall, B. M., Owens, K. M., Singh, K. K. (2011). Distinct functions of evolutionary conserved MSF1 and late embryogenesis abundant (LEA)-like domains in mitochondria. J. Biol. Chem. 286 (45), 39141–39152. doi: 10.1074/jbc.M111.259853
Harayama, T., Riezman, H. (2018). Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol. 19 (5), 281–296. doi: 10.1038/nrm.2017.138
Hassa, P. O., Haenni, S. S., Elser, M., Hottiger, M. O. (2006). Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol. Mol. Biol. Rev. 70 (3), 789–829. doi: 10.1128/MMBR.00040-05
He, B., Guo, W. (2009). The exocyst complex in polarized exocytosis. Curr. Opin. Cell Biol. 21 (4), 537–542. doi: 10.1016/j.ceb.2009.04.007
Heilmann, I. (2016). Phosphoinositide signaling in plant development. Development 143 (12), 2044–2055. doi: 10.1242/dev.136432
Hertle, A. P., García-Cerdán, J. G., Armbruster, U., Shih, R., Lee, J. J., Wong, W., et al. (2020). A Sec14 domain protein is required for photoautotrophic growth and chloroplast vesicle formation in arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 117 (16), 9101–9111. doi: 10.1073/pnas.1916946117
Hornbergs, J., Montag, K., Loschwitz, J., Mohr, I., Poschmann, G., Schnake, A., et al. (2022). SEC14-GOLD protein PATELLIN2 binds IRON-REGULATED TRANSPORTER1 linking root iron uptake to vitamin e. Plant Physiol, kiac563. doi: 10.1093/plphys/kiac563
Hsu, J. L., Wang, L. Y., Wang, S. Y., Lin, C. H., Ho, K. C., Shi, F. K., et al. (2009). Functional phosphoproteomic profiling of phosphorylation sites in membrane fractions of salt-stressed arabidopsis thaliana. Proteome Sci. 7, 42. doi: 10.1186/1477-5956-7-42
Hsuan, J., Cockcroft, S. (2001). The PITP family of phosphatidylinositol transfer proteins. Genome Biol. 2 (9), REVIEWS3011. doi: 10.1186/gb-2001-2-9-reviews3011
Huang, J., Ghosh, R., Tripathi, A., Lönnfors, M., Somerharju, P., Bankaitis, V. A. (2016). Two-ligand priming mechanism for potentiated phosphoinositide synthesis is an evolutionarily conserved feature of Sec14-like phosphatidylinositol and phosphatidylcholine exchange proteins. Mol. Biol. Cell 27 (14), 2317–2330. doi: 10.1091/mbc.E16-04-0221
Huang, J., Kim, C. M., Xuan, Y. H., Park, S. J., Piao, H. L., Je, B. I., et al. (2013). OsSNDP1, a Sec14-nodulin domain-containing protein, plays a critical role in root hair elongation in rice. Plant Mol. Biol. 82 (1-2), 39–50. doi: 10.1007/s11103-013-0033-4
Huynh, H., Wang, X., Li, W., Bottini, N., Williams, S., Nika, K., et al. (2003). Homotypic secretory vesicle fusion induced by the protein tyrosine phosphatase MEG2 depends on polyphosphoinositides in T cells. J. Immunol. 171 (12), 6661–6671. doi: 10.4049/jimmunol.171.12.6661
Hyvonen, M., Macias, M. J., Nilges, M., Oschkinat, H., Saraste, M., Wilmanns, M. (1995). Structure of the binding site for inositol phosphates in a PH domain. EMBO J. 14 (19), 4676–4685. doi: 10.1002/j.1460-2075.1995.tb00149.x
Irvine, R. F. (2016). A short history of inositol lipids. J. Lipid Res. 57 (11), 1987–1994. doi: 10.1194/jlr.R071712
Isono, E., Katsiarimpa, A., Muller, I. K., Anzenberger, F., Stierhof, Y. D., Geldner, N., et al. (2010). The deubiquitinating enzyme AMSH3 is required for intracellular trafficking and vacuole biogenesis in arabidopsis thaliana. Plant Cell 22 (6), 1826–1837. doi: 10.1105/tpc.110.075952
Johnson, K. G., Kornfeld, K. (2010). The CRAL/TRIO and GOLD domain protein TAP-1 regulates RAF-1 activation. Dev. Biol. 341 (2), 464–471. doi: 10.1016/j.ydbio.2010.03.003
Kapranov, P., Routt, S. M., Bankaitis, V. A., de Bruijn, F. J., Szczyglowski, K. (2001). Nodule-specific regulation of phosphatidylinositol transfer protein expression in lotus japonicus. Plant Cell 13 (6), 1369–1382. doi: 10.1105/tpc.13.6.1369
Kempna, P., Ricciarelli, R., Azzi, A., Zingg, J. M. (2010). Alternative splicing and gene polymorphism of the human TAP3/SEC14L4 gene. Mol. Biol. Rep. 37 (7), 3503–3508. doi: 10.1007/s11033-009-9943-2
Kempna, P., Zingg, J. M., Ricciarelli, R., Hierl, M., Saxena, S., Azzi, A. (2003). Cloning of novel human SEC14p-like proteins: ligand binding and functional properties. Free Radic. Biol. Med. 34 (11), 1458–1472. doi: 10.1016/s0891-5849(03)00173-4
Kf de Campos, M., Schaaf, G. (2017). The regulation of cell polarity by lipid transfer proteins of the SEC14 family. Curr. Opin. Plant Biol. 40, 158–168. doi: 10.1016/j.pbi.2017.09.007
Khan, N. Z., Lindquist, E., Aronsson, H. (2013). New putative chloroplast vesicle transport components and cargo proteins revealed using a bioinformatics approach: an arabidopsis model. PLoS One 8 (4), e59898. doi: 10.1371/journal.pone.0059898
Kim, E. H., Poudyal, R. S., Lee, K. R., Yu, H., Gi, E., Kim, H. U. (2022). Chloroplast-localized PITP7 is essential for plant growth and photosynthetic function in arabidopsis. Physiol. Plant 174 (4), e13760. doi: 10.1111/ppl.13760
Kleffmann, T., Russenberger, D., von Zychlinski, A., Christopher, W., Sjolander, K., Gruissem, W., et al. (2004). The arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 14 (5), 354–362. doi: 10.1016/j.cub.2004.02.039
Kono, N., Ohto, U., Hiramatsu, T., Urabe, M., Uchida, Y., Satow, Y., et al. (2013). Impaired alpha-TTP-PIPs interaction underlies familial vitamin e deficiency. Science 340 (6136), 1106–1110. doi: 10.1126/science.1233508
Kruger, J. M., Fukushima, T., Cherepanov, V., Borregaard, N., Loeve, C., Shek, C., et al. (2002). Protein-tyrosine phosphatase MEG2 is expressed by human neutrophils. localization to the phagosome and activation by polyphosphoinositides. J. Biol. Chem. 277 (4), 2620–2628. doi: 10.1074/jbc.M104550200
Krugmann, S., Anderson, K. E., Ridley, S. H., Risso, N., McGregor, A., Coadwell, J., et al. (2002). Identification of ARAP3, a novel PI3K effector regulating both arf and rho GTPases, by selective capture on phosphoinositide affinity matrices. Mol. Cell 9 (1), 95–108. doi: 10.1016/s1097-2765(02)00434-3
Lemmon, M. A. (2007). Pleckstrin homology (PH) domains and phosphoinositides. Biochem. Soc. Symp 74), 81–93. doi: 10.1042/BSS0740081
Lemmon, M. A., Ferguson, K. M. (1998). Pleckstrin homology domains. Curr. Top. Microbiol. Immunol. 228, 39–74. doi: 10.1007/978-3-642-80481-6_3
Lev, S. (2010). Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat. Rev. Mol. Cell Biol. 11 (10), 739–750. doi: 10.1038/nrm2971
Li, M. T., Di, W., Xu, H., Yang, Y. K., Chen, H. W., Zhang, F. X., et al. (2013). Negative regulation of RIG-i-mediated innate antiviral signaling by SEC14L1. J. Virol. 87 (18), 10037–10046. doi: 10.1128/JVI.01073-13
Li, J., Feng, S., Liu, X., Guo, M., Chen, M., Chen, Y., et al. (2018). Identification of nucleotides in the 5'UTR and amino acids substitutions that are essential for the infectivity of 5'UTR-NS5A recombinant of hepatitis c virus genotype 1b (strain Con1). Virology 518, 253–263. doi: 10.1016/j.virol.2018.03.001
Li, X., Routt, S. M., Xie, Z., Cui, X., Fang, M., Kearns, M. A., et al. (2000). Identification of a novel family of nonclassic yeast phosphatidylinositol transfer proteins whose function modulates phospholipase d activity and Sec14p-independent cell growth. Mol. Biol. Cell 11 (6), 1989–2005. doi: 10.1091/mbc.11.6.1989
Luschnig, C., Vert, G. (2014). The dynamics of plant plasma membrane proteins: PINs and beyond. Development 141 (15), 2924–2938. doi: 10.1242/dev.103424
Mamode Cassim, A., Gouguet, P., Gronnier, J., Laurent, N., Germain, V., Grison, M., et al. (2019). Plant lipids: key players of plasma membrane organization and function. Prog. Lipid Res. 73, 1–27. doi: 10.1016/j.plipres.2018.11.002
Martzen, M. R., McCraith, S. M., Spinelli, S. L., Torres, F. M., Fields, S., Grayhack, E. J., et al. (1999). A biochemical genomics approach for identifying genes by the activity of their products. Science 286 (5442), 1153–1155. doi: 10.1126/science.286.5442.1153
Mattei, B., Spinelli, F., Pontiggia, D., De Lorenzo, G. (2016). Comprehensive analysis of the membrane phosphoproteome regulated by oligogalacturonides in arabidopsis thaliana. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01107
Maw, M. A., Kennedy, B., Knight, A., Bridges, R., Roth, K. E., Mani, E. J., et al. (1997). Mutation of the gene encoding cellular retinaldehyde-binding protein in autosomal recessive retinitis pigmentosa. Nat. Genet. 17 (2), 198–200. doi: 10.1038/ng1097-198
Meier, R., Tomizaki, T., Schulze-Briese, C., Baumann, U., Stocker, A. (2003). The molecular basis of vitamin e retention: structure of human alpha-tocopherol transfer protein. J. Mol. Biol. 331 (3), 725–734. doi: 10.1016/s0022-2836(03)00724-1
Merkulova, M. I., Andreeva, S. G., Shuvaeva, T. M., Novoselov, S. V., Peshenko, I. V., Bystrova, M. F., et al. (1999). A novel 45 kDa secretory protein from rat olfactory epithelium: primary structure and localisation. FEBS Lett. 450 (1-2), 126–130. doi: 10.1016/s0014-5793(99)00470-6
Merkulova, M., Huynh, H., Radchenko, V., Saito, K., Lipkin, V., Shuvaeva, T., et al. (2005). Secretion of the mammalian Sec14p-like phosphoinositide-binding p45 protein. FEBS J. 272 (21), 5595–5605. doi: 10.1111/j.1742-4658.2005.04955.x
Miller, G. W., Ulatowski, L., Labut, E. M., Lebold, K. M., Manor, D., Atkinson, J., et al. (2012). The alpha-tocopherol transfer protein is essential for vertebrate embryogenesis. PLoS One 7 (10), e47402. doi: 10.1371/journal.pone.0047402
Min, K. C., Kovall, R. A., Hendrickson, W. A. (2003). Crystal structure of human alpha-tocopherol transfer protein bound to its ligand: implications for ataxia with vitamin e deficiency. Proc. Natl. Acad. Sci. U.S.A. 100 (25), 14713–14718. doi: 10.1073/pnas.2136684100
Mokashi, V., Singh, D. K., Porter, T. D. (2004). Rat supernatant protein factor-like protein stimulates squalene monooxygenase and is activated by protein kinase a. Biochem. Biophys. Res. Commun. 316 (3), 688–692. doi: 10.1016/j.bbrc.2004.02.103
Montag, K., Hornbergs, J., Ivanov, R., Bauer, P. (2020). Phylogenetic analysis of plant multi-domain SEC14-like phosphatidylinositol transfer proteins and structure–function properties of PATELLIN2. Plant Mol. Biol. 104 (6), 665–678. doi: 10.1007/s11103-020-01067-y
Moon, S., Kim, Y. J., Park, H. E., Kim, J., Gho, Y. S., Hong, W. J., et al. (2022). OsSNDP3 functions for the polar tip growth in rice pollen together with OsSNDP2, a paralog of OsSNDP3. Rice (N Y) 15 (1), 39. doi: 10.1186/s12284-022-00586-0
Nakai, M., Takada, T., Endo, T. (1993). Cloning of the YAP19 gene encoding a putative yeast homolog of AP19, the mammalian small chain of the clathrin-assembly proteins. Biochim. Biophys. Acta 1174 (3), 282–284. doi: 10.1016/0167-4781(93)90198-m
Nakamura, Y., Koizumi, R., Shui, G., Shimojima, M., Wenk, M. R., Ito, T., et al. (2009). Arabidopsis lipins mediate eukaryotic pathway of lipid metabolism and cope critically with phosphate starvation. Proc. Natl. Acad. Sci. U.S.A. 106 (49), 20978–20983. doi: 10.1073/pnas.0907173106
Neduva, V., Russell, R. B. (2006). DILIMOT: discovery of linear motifs in proteins. Nucleic Acids Res. 34 (Web Server issue), W350–W355. doi: 10.1093/nar/gkl159
Neuzil, J., Witting, P. K., Stocker, R. (1997). Alpha-tocopheryl hydroquinone is an efficient multifunctional inhibitor of radical-initiated oxidation of low density lipoprotein lipids. Proc. Natl. Acad. Sci. U.S.A. 94 (15), 7885–7890. doi: 10.1073/pnas.94.15.7885
Ni, J., Wen, X., Yao, J., Chang, H. C., Yin, Y., Zhang, M., et al. (2005). Tocopherol-associated protein suppresses prostate cancer cell growth by inhibition of the phosphoinositide 3-kinase pathway. Cancer Res. 65 (21), 9807–9816. doi: 10.1158/0008-5472.CAN-05-1334
Niittyla, T., Fuglsang, A. T., Palmgren, M. G., Frommer, W. B., Schulze, W. X. (2007). Temporal analysis of sucrose-induced phosphorylation changes in plasma membrane proteins of arabidopsis. Mol. Cell Proteomics 6 (10), 1711–1726. doi: 10.1074/mcp.M700164-MCP200
Noack, L. C., Jaillais, Y. (2020). Functions of anionic lipids in plants. Annu. Rev. Plant Biol. 71, 71–102. doi: 10.1146/annurev-arplant-081519-035910
Novick, P., Field, C., Schekman, R. (1980). Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell 21 (1), 205–215. doi: 10.1016/0092-8674(80)90128-2
Nukala, U., Thakkar, S., Krager, K. J., Breen, P. J., Compadre, C. M., Aykin-Burns, N. (2018). Antioxidant tocols as radiation countermeasures (Challenges to be addressed to use tocols as radiation countermeasures in humans). Antioxidants (Basel) 7 (2), 33. doi: 10.3390/antiox7020033
O'Connor, C. M., Adams, J. U. (2010). Essentials of cell biology Vol. 2010 (Cambridge, MA: NPG Education).
Ouahchi, K., Arita, M., Kayden, H., Hentati, F., Ben Hamida, M., Sokol, R., et al. (1995). Ataxia with isolated vitamin e deficiency is caused by mutations in the alpha-tocopherol transfer protein. Nat. Genet. 9 (2), 141–145. doi: 10.1038/ng0295-141
Panagabko, C., Morley, S., Hernandez, M., Cassolato, P., Gordon, H., Parsons, R., et al. (2003). Ligand specificity in the CRAL-TRIO protein family. Biochemistry 42 (21), 6467–6474. doi: 10.1021/bi034086v
Pastor-Cantizano, N., Bernat-Silvestre, C., Marcote, M. J., Aniento, F. (2018). Loss of arabidopsis p24 function affects ERD2 trafficking and golgi structure, and activates the unfolded protein response. J. Cell Sci. 131 (2), 967-985. doi: 10.1242/jcs.203802
Pastor-Cantizano, N., Montesinos, J. C., Bernat-Silvestre, C., Marcote, M. J., Aniento, F. (2016). p24 family proteins: key players in the regulation of trafficking along the secretory pathway. Protoplasma 253 (4), 967–985. doi: 10.1007/s00709-015-0858-6
Peiro, A., Izquierdo-Garcia, A. C., Sanchez-Navarro, J. A., Pallas, V., Mulet, J. M., Aparicio, F. (2014). Patellins 3 and 6, two members of the plant patellin family, interact with the movement protein of alfalfa mosaic virus and interfere with viral movement. Mol. Plant Pathol. 15 (9), 881–891. doi: 10.1111/mpp.12146
Peltonen, S., Kallionpaa, R. A., Peltonen, J. (2017). Neurofibromatosis type 1 (NF1) gene: beyond cafe au lait spots and dermal neurofibromas. Exp. Dermatol. 26 (7), 645–648. doi: 10.1111/exd.13212
Peretti, D., Kim, S., Tufi, R., Lev, S. (2019). Lipid transfer proteins and membrane contact sites in human cancer. Front. Cell Dev. Biol. 7. doi: 10.3389/fcell.2019.00371
Peterman, T. K., Ohol, Y. M., McReynolds, L. J., Luna, E. J. (2004). Patellin1, a novel Sec14-like protein, localizes to the cell plate and binds phosphoinositides. Plant Physiol. 136 (2), 3080–3094. doi: 10.1104/pp.104.045369
Peterman, T. K., Sequeira, A. S., Samia, J. A., Lunde, E. E. (2006). Molecular cloning and characterization of patellin1, a novel sec14-related protein, from zucchini (Cucurbita pepo). J. Plant Physiol. 163 (11), 1150–1158. doi: 10.1016/j.jplph.2006.01.009
Preuss, M. L., Schmitz, A. J., Thole, J. M., Bonner, H. K., Otegui, M. S., Nielsen, E. (2006). A role for the RabA4b effector protein PI-4Kbeta1 in polarized expansion of root hair cells in arabidopsis thaliana. J. Cell Biol. 172 (7), 991–998. doi: 10.1083/jcb.200508116
Rad, E., Tee, A. R. (2016). Neurofibromatosis type 1: fundamental insights into cell signalling and cancer. Semin. Cell Dev. Biol. 52, 39–46. doi: 10.1016/j.semcdb.2016.02.007
Rasmussen, S. A., Friedman, J. M. (2000). NF1 gene and neurofibromatosis 1. Am. J. Epidemiol. 151 (1), 33–40. doi: 10.1093/oxfordjournals.aje.a010118
Ren, J., Schaaf, G., Bankaitis, V. A., Ortlund, E. A., Pathak, M. C. (2011). Crystallization and preliminary X-ray diffraction analysis of Sfh3, a member of the Sec14 protein superfamily. Acta Crystallogr. Sect F Struct. Biol. Cryst Commun. 67 (Pt 10), 1239–1243. doi: 10.1107/S1744309111027096
Ribeiro, F. M., Ferreira, L. T., Marion, S., Fontes, S., Gomez, M., Ferguson, S. S., et al. (2007). SEC14-like protein 1 interacts with cholinergic transporters. Neurochem. Int. 50 (2), 356–364. doi: 10.1016/j.neuint.2006.09.010
Roeder, A. H. K., Otegui, M. S., Dixit, R., Anderson, C. T., Faulkner, C., Zhang, Y., et al. (2022). Fifteen compelling open questions in plant cell biology. Plant Cell 34 (1), 72–102. doi: 10.1093/plcell/koab225
Roman-Fernandez, A., Roignot, J., Sandilands, E., Nacke, M., Mansour, M. A., McGarry, L., et al. (2018). The phospholipid PI(3,4)P2 is an apical identity determinant. Nat. Commun. 9 (1), 5041. doi: 10.1038/s41467-018-07464-8
Ryan, M. M., Temple, B. R., Phillips, S. E., Bankaitis, V. A. (2007). Conformational dynamics of the major yeast phosphatidylinositol transfer protein sec14p: insight into the mechanisms of phospholipid exchange and diseases of sec14p-like protein deficiencies. Mol. Biol. Cell 18 (5), 1928–1942. doi: 10.1091/mbc.e06-11-1024
Saeed, M., Andreo, U., Chung, H. Y., Espiritu, C., Branch, A. D., Silva, J. M., et al. (2015). SEC14L2 enables pan-genotype HCV replication in cell culture. Nature 524 (7566), 471–475. doi: 10.1038/nature14899
Saito, K., Tautz, L., Mustelin, T. (2007a). The lipid-binding SEC14 domain. Biochim. Biophys. Acta 1771 (6), 719–726. doi: 10.1016/j.bbalip.2007.02.010
Saito, K., Williams, S., Bulankina, A., Honing, S., Mustelin, T. (2007b). Association of protein-tyrosine phosphatase MEG2 via its Sec14p homology domain with vesicle-trafficking proteins. J. Biol. Chem. 282 (20), 15170–15178. doi: 10.1074/jbc.M608682200
Salameh, A., Lee, A. K., Cardo-Vila, M., Nunes, D. N., Efstathiou, E., Staquicini, F. I., et al. (2015). PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proc. Natl. Acad. Sci. U.S.A. 112 (27), 8403–8408. doi: 10.1073/pnas.1507882112
Sato, Y., Arai, H., Miyata, A., Tokita, S., Yamamoto, K., Tanabe, T., et al. (1993). Primary structure of alpha-tocopherol transfer protein from rat liver. homology with cellular retinaldehyde-binding protein. J. Biol. Chem. 268 (24), 17705–17710.
Schaaf, G., Betts, L., Garrett, T. A., Raetz, C. R., Bankaitis, V. A. (2006). Crystallization and preliminary X-ray diffraction analysis of phospholipid-bound Sfh1p, a member of the saccharomyces cerevisiae Sec14p-like phosphatidylinositol transfer protein family. Acta Crystallogr. Sect F Struct. Biol. Cryst Commun. 62 (Pt 11), 1156–1160. doi: 10.1107/S1744309106041728
Schaaf, G., Dynowski, M., Mousley, C. J., Shah, S. D., Yuan, P., Winklbauer, E. M., et al. (2011). Resurrection of a functional phosphatidylinositol transfer protein from a pseudo-Sec14 scaffold by directed evolution. Mol. Biol. Cell 22 (6), 892–905. doi: 10.1091/mbc.E10-11-0903
Schaaf, G., Ortlund, E. A., Tyeryar, K. R., Mousley, C. J., Ile, K. E., Garrett, T. A., et al. (2008). Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily. Mol. Cell 29 (2), 191–206. doi: 10.1016/j.molcel.2007.11.026
Schnabl, M., Oskolkova, O. V., Holic, R., Brezna, B., Pichler, H., Zagorsek, M., et al. (2003). Subcellular localization of yeast Sec14 homologues and their involvement in regulation of phospholipid turnover. Eur. J. Biochem. 270 (15), 3133–3145. doi: 10.1046/j.1432-1033.2003.03688.x
Sha, B., Phillips, S. E., Bankaitis, V. A., Luo, M. (1998). Crystal structure of the saccharomyces cerevisiae phosphatidylinositol-transfer protein. Nature 391 (6666), 506–510. doi: 10.1038/35179
Shibata, N., Arita, M., Misaki, Y., Dohmae, N., Takio, K., Ono, T., et al. (2001). Supernatant protein factor, which stimulates the conversion of squalene to lanosterol, is a cytosolic squalene transfer protein and enhances cholesterol biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 98 (5), 2244–2249. doi: 10.1073/pnas.041620398
Simon, M. L., Platre, M. P., Assil, S., van Wijk, R., Chen, W. Y., Chory, J., et al. (2014). A multi-colour/multi-affinity marker set to visualize phosphoinositide dynamics in arabidopsis. Plant J. 77 (2), 322–337. doi: 10.1111/tpj.12358
Simon, M. L., Platre, M. P., Marques-Bueno, M. M., Armengot, L., Stanislas, T., Bayle, V., et al. (2016). A PtdIns(4)P-driven electrostatic field controls cell membrane identity and signalling in plants. Nat. Plants 2, 16089. doi: 10.1038/nplants.2016.89
Sirokmany, G., Szidonya, L., Kaldi, K., Gaborik, Z., Ligeti, E., Geiszt, M. (2006). Sec14 homology domain targets p50RhoGAP to endosomes and provides a link between rab and rho GTPases. J. Biol. Chem. 281 (9), 6096–6105. doi: 10.1074/jbc.M510619200
Skinner, H. B., Alb, J. G., Jr., Whitters, E. A., Helmkamp, G. M., Jr., Bankaitis, V. A. (1993). Phospholipid transfer activity is relevant to but not sufficient for the essential function of the yeast SEC14 gene product. EMBO J. 12 (12), 4775–4784. doi: 10.1002/j.1460-2075.1993.tb06166.x
Sohda, M., Misumi, Y., Yamamoto, A., Yano, A., Nakamura, N., Ikehara, Y. (2001). Identification and characterization of a novel golgi protein, GCP60, that interacts with the integral membrane protein giantin. J. Biol. Chem. 276 (48), 45298–45306. doi: 10.1074/jbc.M108961200
Stahelin, R. V. (2009). Lipid binding domains: more than simple lipid effectors. J. Lipid Res. 50 Suppl, S299–S304. doi: 10.1194/jlr.R800078-JLR200
Stevenson, J., Perera, I., Heilmann, I., Persson, S., Boss, W. (2000). Inositol signaling and plant growth. Trends Plant Sci. 5, 357. doi: 10.1016/S1360-1385(00)01739-8
Stocker, A., Baumann, U. (2003). Supernatant protein factor in complex with RRR-alpha-tocopherylquinone: a link between oxidized vitamin e and cholesterol biosynthesis. J. Mol. Biol. 332 (4), 759–765. doi: 10.1016/s0022-2836(03)00924-0
Strahl, T., Thorner, J. (2007). Synthesis and function of membrane phosphoinositides in budding yeast, saccharomyces cerevisiae. Biochim. Biophys. Acta 1771 (3), 353–404. doi: 10.1016/j.bbalip.2007.01.015
Sugiura, T., Nakao, H., Ikeda, K., Khan, D., Nile, A. H., Bankaitis, V. A., et al. (2021). Biophysical parameters of the Sec14 phospholipid exchange cycle - effect of lipid packing in membranes. Biochim. Biophys. Acta Biomembr 1863 (1), 183450. doi: 10.1016/j.bbamem.2020.183450
Sun, Y. J., Nishikawa, K., Yuda, H., Wang, Y. L., Osaka, H., Fukazawa, N., et al. (2006). Solo/Trio8, a membrane-associated short isoform of trio, modulates endosome dynamics and neurite elongation. Mol. Cell Biol. 26 (18), 6923–6935. doi: 10.1128/MCB.02474-05
Suzuki, T., Matsushima, C., Nishimura, S., Higashiyama, T., Sasabe, M., Machida, Y. (2016). Identification of phosphoinositide-binding protein PATELLIN2 as a substrate of arabidopsis MPK4 MAP kinase during septum formation in cytokinesis. Plant Cell Physiol. 57 (8), 1744–1755. doi: 10.1093/pcp/pcw098
Tang, W., Deng, Z., Oses-Prieto, J. A., Suzuki, N., Zhu, S., Zhang, X., et al. (2008). Proteomics studies of brassinosteroid signal transduction using prefractionation and two-dimensional DIGE. Mol. Cell Proteomics 7 (4), 728–738. doi: 10.1074/mcp.M700358-MCP200
Tejos, R., Rodriguez-Furlan, C., Adamowski, M., Sauer, M., Norambuena, L., Friml, J. (2017). PATELLINS are regulators of auxin-mediated PIN1 relocation and plant development in arabidopsis thaliana. J. Cell Sci 131 (2), jcs204198. doi: 10.1242/jcs.204198
Tokarev, A. A., Alfonso, A., Segev, N. (2013). “Overview of intracellular compartments and trafficking pathways,” in Madame curie bioscience database (Austin (TX: Landes Bioscience), 2000–2013. Available at: https://www.ncbi.nlm.nih.gov/books/NBK7286.
Tripathi, A., Nile, A. H., Bankaitis, V. A. (2014). Sec14-like phosphatidylinositol-transfer proteins and diversification of phosphoinositide signalling outcomes. Biochem. Soc. Trans. 42 (5), 1383–1388. doi: 10.1042/BST20140187
Ueda, S., Kataoka, T., Satoh, T. (2004). Role of the Sec14-like domain of dbl family exchange factors in the regulation of rho family GTPases in different subcellular sites. Cell Signal 16 (8), 899–906. doi: 10.1016/j.cellsig.2004.01.007
Varadi, M., Anyango, S, Deshpande, M, Nair, S, Natassia, C, Yordanova, G, et al. (2021). AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Research 50 (7), D439–D444. doi: 10.1093/nar/gkab1061
Vincent, P., Chua, M., Nogue, F., Fairbrother, A., Mekeel, H., Xu, Y., et al. (2005). A Sec14p-nodulin domain phosphatidylinositol transfer protein polarizes membrane growth of arabidopsis thaliana root hairs. J. Cell Biol. 168 (5), 801–812. doi: 10.1083/jcb.200412074
Wang, X., Ni, J., Hsu, C. L., Johnykutty, S., Tang, P., Ho, Y. S., et al. (2009). Reduced expression of tocopherol-associated protein (TAP/Sec14L2) in human breast cancer. Cancer Invest. 27 (10), 971–977. doi: 10.3109/07357900802392659
Wirtz, K. W. (1991). Phospholipid transfer proteins. Annu. Rev. Biochem. 60, 73–99. doi: 10.1146/annurev.bi.60.070191.000445
Wu, C., Tan, L., van Hooren, M., Tan, X., Liu, F., Li, Y., et al. (2017). Arabidopsis EXO70A1 recruits Patellin3 to the cell membrane independent of its role as an exocyst subunit. J. Integr. Plant Biol. 59 (12), 851–865. doi: 10.1111/jipb.12578
Yakir-Tamang, L., Gerst, J. E. (2009). A phosphatidylinositol-transfer protein and phosphatidylinositol-4-phosphate 5-kinase control Cdc42 to regulate the actin cytoskeleton and secretory pathway in yeast. Mol. Biol. Cell 20 (15), 3583–3597. doi: 10.1091/mbc.E08-10-1073
Yao, H. Y., Lu, Y. Q., Yang, X. L., Wang, X. Q., Luo, Z., Lin, D. L., et al. (2023). Arabidopsis Sec14 proteins (SFH5 and SFH7) mediate interorganelle transport of phosphatidic acid and regulate chloroplast development. Proc. Natl. Acad. Sci. U.S.A. 120 (6), e2221637120. doi: 10.1073/pnas.2221637120
Yap, Y. S., McPherson, J. R., Ong, C. K., Rozen, S. G., Teh, B. T., Lee, A. S., et al. (2014). The NF1 gene revisited - from bench to bedside. Oncotarget 5 (15), 5873–5892. doi: 10.18632/oncotarget.2194
Yoshitake, Y., Sato, R., Madoka, Y., Ikeda, K., Murakawa, M., Suruga, K., et al. (2017). Arabidopsis phosphatidic acid phosphohydrolases are essential for growth under nitrogen-depleted conditions. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01847
Yu, F., He, F., Yao, H., Wang, C., Wang, J., Li, J., et al. (2015). Structural basis of intramitochondrial phosphatidic acid transport mediated by Ups1-Mdm35 complex. EMBO Rep. 16 (7), 813–823. doi: 10.15252/embr.201540137
Zhao, R., Fu, X., Li, Q., Krantz, S. B., Zhao, Z. J. (2003). Specific interaction of protein tyrosine phosphatase-MEG2 with phosphatidylserine. J. Biol. Chem. 278 (25), 22609–22614. doi: 10.1074/jbc.M301560200
Zhou, H., Wang, C., Tan, T., Cai, J., He, J., Lin, H. (2018). Patellin1 negatively modulates salt tolerance by regulating PM Na+/H+ antiport activity and cellular redox homeostasis in arabidopsis. Plant Cell Physiol. 59 (8), 1630–1642. doi: 10.1093/pcp/pcy081
Zingg, J. M., Azzi, A., Meydani, M. (2015). Induction of VEGF expression by alpha-tocopherol and alpha-tocopheryl phosphate via PI3Kgamma/PKB and hTAP1/SEC14L2-mediated lipid exchange. J. Cell Biochem. 116 (3), 398–407. doi: 10.1002/jcb.24988
Zingg, J. M., Azzi, A., Meydani, M. (2017). Alpha-tocopheryl phosphate induces VEGF expression via CD36/PI3Kgamma in THP-1 monocytes. J. Cell Biochem. 118 (7), 1855–1867. doi: 10.1002/jcb.25871
Zingg, J. M., Kempna, P., Paris, M., Reiter, E., Villacorta, L., Cipollone, R., et al. (2008). Characterization of three human sec14p-like proteins: alpha-tocopherol transport activity and expression pattern in tissues. Biochimie 90 (11-12), 1703–1715. doi: 10.1016/j.biochi.2008.07.008
Keywords: lipid binding site, lipid transfer, SEC14, PATELLIN, membrane, phosphatidylinositol, multi-domain, tocopherol
Citation: Montag K, Ivanov R and Bauer P (2023) Role of SEC14-like phosphatidylinositol transfer proteins in membrane identity and dynamics. Front. Plant Sci. 14:1181031. doi: 10.3389/fpls.2023.1181031
Received: 06 March 2023; Accepted: 05 April 2023;
Published: 15 May 2023.
Edited by:
Emily Palm, University of Florence, ItalyReviewed by:
Rosa Laura Lopez-Marques, University of Copenhagen, DenmarkSusanne Hoffmann-Benning, Michigan State University, United States
Armando Albert, Spanish National Research Council (CSIC), Spain
Copyright © 2023 Montag, Ivanov and Bauer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Petra Bauer, cGV0cmEuYmF1ZXJAaGh1LmRl; Rumen Ivanov, cnVtZW4uaXZhbm92QGhodS5kZQ==
 Karolin Montag1
Karolin Montag1 Rumen Ivanov
Rumen Ivanov Petra Bauer
Petra Bauer