- 1Tobacco Research Institute of Chinese Academy of Agricultural Sciences, Qingdao, China
- 2State Key Laboratory of Rice Biology, China National Rice Research Institute, Hangzhou, China
- 3Zhejiang Provincial Key Laboratory of Biometrology and Inspection and Quarantine, College of Life Sciences, China Jiliang University, Hangzhou, China
- 4Department of Horticulture, College of Agriculture and Biotechnology, Zhejiang University, Hangzhou, China
Wild rice (Zizania spp.), an aquatic grass belonging to the subfamily Gramineae, has a high economic value. Zizania provides food (such as grains and vegetables), a habitat for wild animals, and paper-making pulps, possesses certain medicinal values, and helps control water eutrophication. Zizania is an ideal resource for expanding and enriching a rice breeding gene bank to naturally preserve valuable characteristics lost during domestication. With the Z. latifolia and Z. palustris genomes completely sequenced, fundamental achievements have been made toward understanding the origin and domestication, as well as the genetic basis of important agronomic traits of this genus, substantially accelerating the domestication of this wild plant. The present review summarizes the research results on the edible history, economic value, domestication, breeding, omics research, and important genes of Z. latifolia and Z. palustris over the past decades. These findings broaden the collective understanding of Zizania domestication and breeding, furthering human domestication, improvement, and long-term sustainability of wild plant cultivation.
1 Introduction
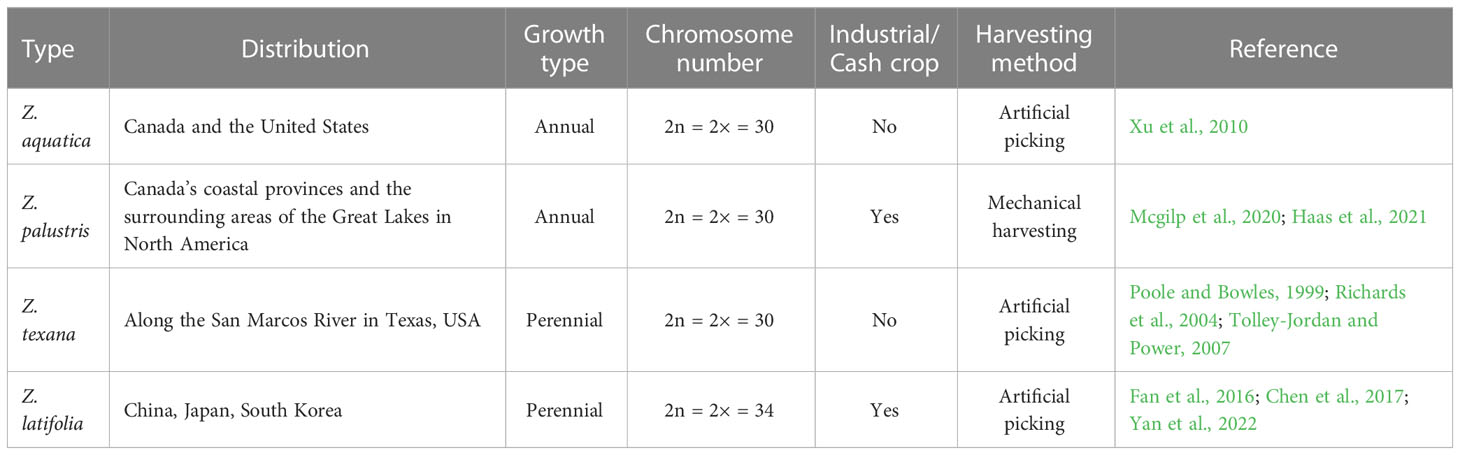
The genus Zizania (family Poaceae) is a type of wild rice that includes four species: Z. aquatica, Z. palustris, Z. texana, and Z. latifolia (Table 1) (Makela et al., 1998; Lu et al., 2022). Zizania diverged from Oryza approximately 26–30 million years ago (mya), whereas Z. palustris and Z. latifolia diverged from one another approximately 6–8 mya (Haas et al., 2021). Due to the differences in geographic distribution and ecological environments, a significant variability has been observed between the morphology and reproductive cycle of species in East Asia (Z. latifolia) and those in North America (Z. aquatica, Z. palustris, and Z. texana). Z. aquatica is found in Southern Ontario, Quebec, the East Coast of Canada, and the Atlantic and Gulf Coasts of the US (Florida and Louisiana) (Xu et al., 2010). Z. palustris is widely distributed across the Canadian prairie provinces (Alberta, Manitoba, and Saskatchewan), along the Great Lakes region, and across the prairies in North America (Mcgilp et al., 2020; Haas et al., 2021). Z. texana, a rare species of Zizania, is limited to a small region along the San Marcos River in Texas. Its small range and limited population size have led to it being classified as endangered by the US federal government (Poole and Bowles, 1999; Richards et al., 2004; Tolley-Jordan and Power, 2007). In numerous places along the Great Lakes, areas with abundant Zizania populations are already endangered. The growing body of research on Zizania, including genetics, domestication, breeding, etc., has greatly enhanced our ability to conserve this important plant species (Kennard et al., 2002; Lu et al., 2005; Kahler et al., 2014; Haas et al., 2021). The morphology of East Asian wild rice differs from its North American counterpart (Table 1). Z. latifolia, also known as East Asian wild rice, is believed to have originated in China and is found in various water bodies including rivers, lakes, ditches, ponds, and paddy fields throughout the country, especially in the Yangtze and Huaihe River basins (Yan et al., 2018). This species is not limited to China and can also be found in other parts of East Asia, such as Japan, South Korea, and Southeast Asia (Fan et al., 2016; Chen et al., 2017; Yan et al., 2018).
Wild rice, classified as a whole grain, is a caryopsis with a seed coat that is thinner and longer in shape than conventional rice (Oryza sativa). The grain has acuminose ends, black-brown and glossy cortex, and a milky white and brittle endosperm (Surendiran et al., 2014). Zizania is a highly nutritious food, rich in protein, dietary fiber, vitamins, and minerals, with a low glycemic index, low fat content, and reasonable amino acid composition (Zhai et al., 2001; Yu et al., 2020). Additionally, wild rice contains various biologically active substances such as phytosterol, γ-oryzanol, γ-aminobutyric acid, and phenolic compounds, which have potential health benefits (Yu et al., 2022). Several studies have shown that whole grain consumption helps reduce the risk of chronic diseases, such as cardiovascular disease, obesity, cancer, and diabetes (Juan et al., 2017). Various international projects have been initiated in the 21st century aimed at increasing the benefits and content of plant functional components in whole grain for lowering cholesterol, and regulating blood glucose metabolism in dietary fiber, in addition to reducing the risk of cardiovascular and cerebrovascular diseases (Ward et al., 2008; Juan et al., 2017; Guo et al., 2022).
Northern wild rice (NWR, Z. palustris) serves as a traditional food among American Indians and has gradually been incorporated into the diets of various cultures across the world (Qiu et al., 2010). Following its “nonshattering” domestication in the late 1960s, Z. palustris gained extremely high commercial value. Currently, large-scale commercial planting of Z. palustris is concentrated primarily in the US (Minnesota and California) (Mcgilp et al., 2020). Z. latifolia has a long history in ancient China; however, due to the human population increases in Southern China following the Tang and Song Dynasties, subsequent agricultural development, and the man-made drying of lakes for rice cultivation, the prevalence of Z. latifolia has drastically reduced (Yan et al., 2018). Moreover, the low yield of Z. latifolia caused by the difficulty in its harvesting (seeds are prone to fall off after ripening) is another contributing factor. In addition, with the widespread cultivation of rice, the yield of rice has increased, reducing the importance of human reliance on Chinese wild rice (CWR) as a staple food. Therefore, CWR has gradually faded out of people’s lives (Yan et al., 2018). In contrast, wild rice remains an expensive and popular food in North America (Surendiran et al., 2014), while it is not often consumed in modern China.
The global population is expected to exceed 9 billion by 2050; thus, higher agricultural output is required to meet the growing demand for food. However, the acceleration of urbanization and reduction of resources (land, water, and human resources available for agriculture) have presented major challenges to production requirements (Zhang et al., 2022). For example, Z. latifolia, a known perennial crop that does not require replanting after each harvest, unlike annuals such as Z. palustris, whose cultivation mandates substantial seed input, increased investment in agricultural machinery, greater water and soil loss, soil nutrient depletion, as well as other socio-economic and ecological challenges (Lobell et al., 2011). Therefore, breeding perennial food crops (e.g., Z. latifolia) can serve as a novel approach to tackling food security and environmental challenges (Glover et al., 2010). Zizania breeding and domestication have recently become a necessary direction in large-scale commercial farming and can bring favorable economic gains to farmers. Currently, genomics approaches are widely used for crops such as rice and have provided new directions for crop domestication and utilization of heterosis (Huang et al., 2012; Huang et al., 2016; Yu and Li, 2022). In particular, the multi-omics methods, including genomics, transcriptomics, proteomics, and metabolomics, have been successfully utilized for the genetic improvement of crops, achieving more efficient and accurate breeding through molecule design (Zeng et al., 2017; Wu et al., 2021). This article reviews the research progress on the edible history, economic value, domestication, breeding, omics research, and functional verification of important genes of Z. latifolia and Z. palustris over the past few decades, providing strong theoretical support for greater domestication of this genus with high-yield, -quality, -stress resistance, and -nutrient utilization.
2 Edible history and economic value of Z. latifolia and Z. palustris
Zizania spp., particularly Z. palustris and Z. latifolia, have extremely high ecological and economic values, as they provide habitat for wild animals, in addition to their use as grains and vegetables. Furthermore, wild rice is used as a raw material in paper-making and a genetic resource for expanding the rice gene pool; it is also used in controlling water eutrophication and possesses certain medicinal values (Figure 1).
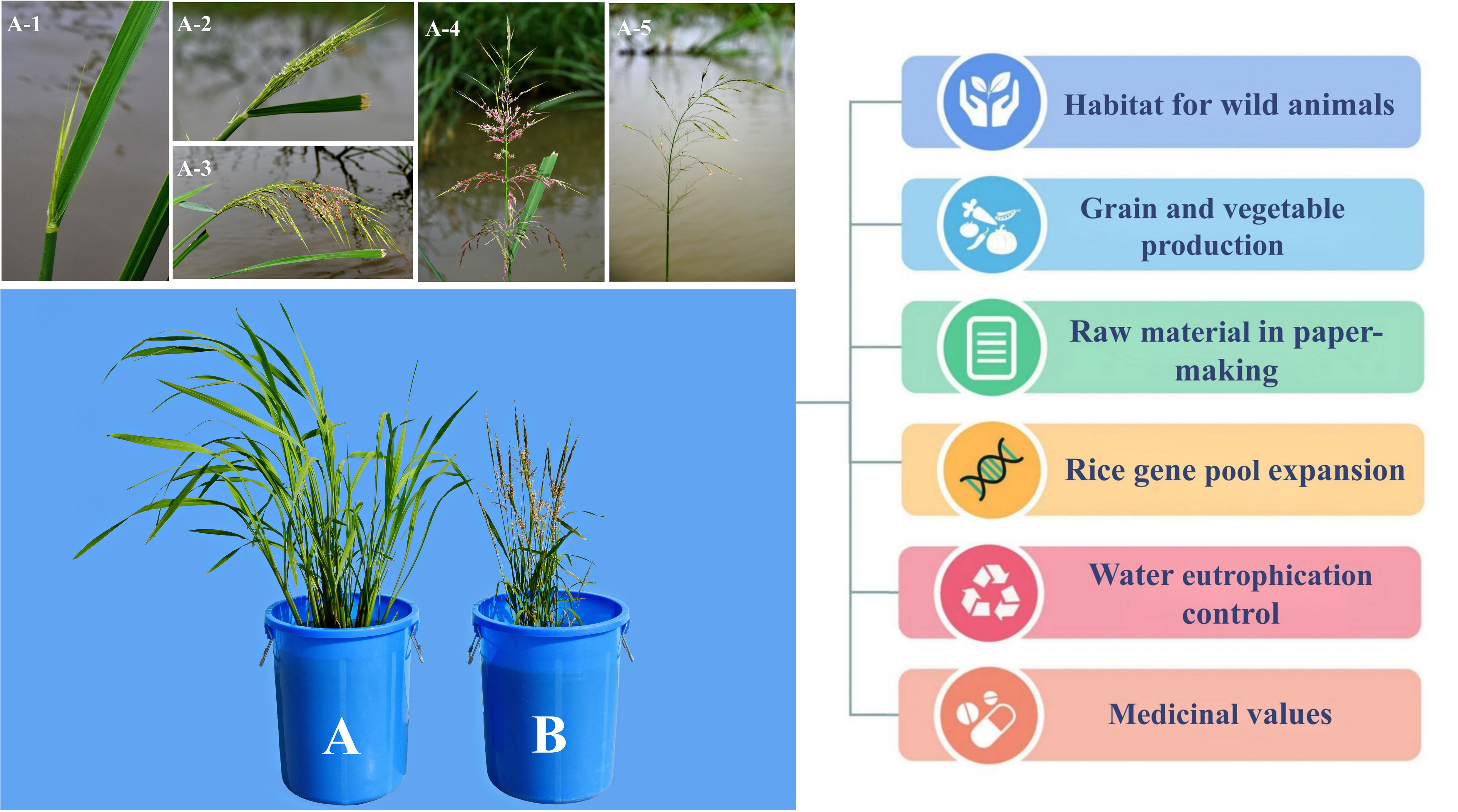
Figure 1 Application value of Zizania latifolia (A) and Zizania palustris (B). A1–A5 represent the inflorescences of Z. latifolia at different stages after flowering.
2.1 Edible history and economic value of Z. palustris
Z. palustris, also known as wild rice, is an annual, outcrossing, aquatic species in the Poaceae family and native to North America (Hayes et al., 1989; Zaitchik et al., 2000; Kennard et al., 2002). Currently, Z. palustris, a nutritious grain, is the only successfully domesticated and widely planted wild rice. Z. palustris is typically found in shallow lakes, rivers, and coastal areas within the north-central US and southern Canada (Oelke, 2004). This plant provides habitats for various birds, mammals, fish, and invertebrates. In addition, Z. palustris participates in the water nutrient cycle and helps stabilize the sediment of coastal river wetlands (Meeker, 1996), playing a significant role in the local food network and wetland ecology (Drewes and Silbernagel, 2012). For centuries, American Indians have collected Z. palustris from the lakes and rivers of the Great Lakes region (Hayes et al., 1989). Z. palustris is considered a high-value crop and has become a cash crop in recent years. Z. palustris is classified as a whole grain food containing > 75% carbohydrates, 6.2% dietary fiber, 14.7% protein, and 1.1% lipids (Yan et al., 2018; Yu et al., 2021). In addition, the antioxidant activity of Z. palustris is 10–15 times higher than that of rice, while its protein and essential amino acid content are doubled, and dietary fiber content is 5 times greater, with a low-fat content, most of which are essential unsaturated fatty acids, including ω-6 (35.0–37.8%) and ω-3 (20.0–31.5%) (Wiser, 1975; Bunzel et al., 2002; Aladedunye et al., 2013; Kahler et al., 2014; Surendiran et al., 2014; Timm and Slavin, 2014). History has documented the economic position of wild rice. Early Native Americans, especially the Ojibway, Menomini, and Cree peoples, consider Z. palustris a traditional food (Lorenz and Lund, 1981). In the 17th century, Europeans successively entered the Great Lakes Region of North America, where a priest named Hennepin was the first to observe the collection of Z. palustris by American Indians, describing it as an abundant aquatic oat that grows in the lakes without any cultivation (Steeves, 1952). Hennepin believed that aquatic oats were an inadequate translation and revised it to wild rice (Horne and Kahn, 1997).
Currently, the indigenous populations hand-harvest and sell Z. palustris, representing an important component of their income (Drewes and Silbernagel, 2012). In northern Michigan and Wisconsin, as well as most of northeastern Minnesota, tribes have preserved their methods of Z. palustris processing, a right protected by law (Walker et al., 2006). Although American Indians have processed and harvested Z. palustris for food over many centuries, researchers first proposed the domestication of Z. palustris as a cultivated crop in the mid-18th century (Porter, 2019). However, in the 1950s, paddy field planting and harvesting of Z. palustris began in earnest (Porter, 2019). Z. palustris seeds are large and can produce considerable yield, being highest in Minnesota and California (Oelke, 2004). However, this crop has relatively fragile seeds and inconsistent ripening periods, making it difficult to achieve full domestication (Oelke and Albrecht, 1978; Drewes and Silbernagel, 2012). Currently, Canada, Hungary, and Australia have implemented commercialized production of Z. palustris (Qiu et al., 2009; Small, 2012); however, Z. aquatica and Z. texana seeds are fragile, have low yields, while their distribution ranges and population sizes are substantially smaller than that of Z. palustris, restricting their collection for consumption. Moreover, relatively little research and domestication efforts have been dedicated to these species (Oxley et al., 2008).
2.2 Edible history and economic value of Z. latifolia
The history of harvesting and utilizing wild rice by Chinese ancestors dates to the Zhou Dynasty 3000 years ago (Yan et al., 2018). Wild rice was a precious food material offered to royalty in Ancient China, and its historical position exceeds that of other crops (Yan et al., 2018). The economic value of Z. latifolia primarily rests on two aspects: CWR is a nutritious whole-grain food that is being increasingly accepted, and Z. latifolia is the source of the domesticated, cultivated species Jiaobai (Yu et al., 2020). Approximately 2000 years ago, Z. latifolia infected by U. esculenta resulted in culm enlargement and the formation of a fleshy edible gall—Jiaobai. Based on this process, Chinese ancestors domesticated it into a vegetable (Zhao et al., 2018; Tu et al., 2019). Jiaobai is a famous aquatic vegetable that has also been introduced to Japan, South Korea, and across Southeast Asia (Xu et al., 2010). Currently, the total planting area of Jiaobai in China is > 60,000 hm2, second only to lotus as the most cultivated aquatic vegetable in China (Yan et al., 2018). The cultivation of this vegetable is an important economic resource for many households in Southern China (Guo et al., 2007; Yan et al., 2013a). Jiaobai is a widely enjoyed fresh, tender, crisp, and sweet vegetable that provides sugar, protein, vitamins, minerals, and essential amino acids (Yan et al., 2018). In addition, enzyme-treated Z. latifolia extract (ETZL) can clear free radicals and block elevated triglyceride and malondialdehyde levels in the liver (Chang et al., 2021). Moreover, ETZL significantly decreases t-BHP-induced HepG2 cytotoxicity and active oxygen generation and improves liver damage induced by excessive alcohol consumption via upregulating antioxidant defense mechanisms to prevent alcoholism (Chang et al., 2021; Gao et al., 2022). ETZL and its main compound, tricin, can inhibit the production of metalloproteinases in the extracellular matrix of human skin following UV exposure, thereby helping the skin resist ultraviolet radiation (Park et al., 2019).
The caryopsis of Z. latifolia is classified as one of the “six grains”, along with rice, broomcorn millet, panicled millet, wheat, and beans, in Ancient China. As a grain, Z. latifolia primarily exists in a wild state and has not been artificially domesticated (Yan et al., 2022). Although the edible history of Z. latifolia traces back to the Zhou Dynasty (3 kya), Z. latifolia was gradually replaced by cultivated rice due to its strong seed shattering and low yield characteristics (Yan et al., 2022). Z. latifolia was subsequently used as a traditional Chinese medicinal crop that was categorized as a treatment for diabetes and gastrointestinal diseases in the Compendium of Materia Medica of Li Shizhen during the Ming Dynasty (Yu et al., 2022). Z. latifolia is classified as a whole grain with high nutritional value. In particular, it is rich in protein, essential amino acids, fatty acids, vitamins, and microelements (Yan et al., 2018). Due to its high nutritional value, developing germplasms resistant to seed shattering remains a top priority in the domestication and breeding of Z. latifolia (Xie et al., 2022). As a graminaceous crop closely related to rice, Z. latifolia boasts myriad beneficial traits that rice lacks, including rice blast resistance, thick stalks, strong tillering, low-temperature resistance, flood resistance, rapid grouting maturity, high biological yields, and optimizing protein and lysine contents in seeds (Chen et al., 2012; Ye et al., 2016; Yan et al., 2022). Hence, it is capable of overcoming the narrow bottleneck of genetic resources for rice breeding and providing important materials for optimizing genetic traits. For example, a Z. latifolia and rice hybrid exhibited increased rice blast resistance (Wang et al., 2013). However, the genomes of hybrid rice strains comprise a small proportion of Z. latifolia DNA sequences, and inserting exogenous DNA fragments introduces extensive cytosine methylation variation and transposon activation. The resulting sequence variation may serve as the primary driver of character variations within introgressive hybrid lines (Liu et al., 2004; Shan et al., 2005; Wang et al., 2005). Meanwhile, given the advantageous characteristics of Z. latifolia, it may represent a source of genes for future rice molecular breeding. As such, the introduction of Z. latifolia as a perennial food crop may prove ecologically beneficial and sustainable, thus providing a means to address food security and environmental challenges. Indeed, appropriate domestication of Z. latifolia across large planting areas can fulfill the demand of an increasing population for a nutritional food source while alleviating the environmental challenges driven by the reduction of cultivated land and resources.
Previous studies have shown that Z. latifolia contains phytoliths, showing considerable carbon sequestration potential (Li et al., 2022). In addition, this species exhibits excellent sequestration under nitrogen and phosphorus eutrophication conditions compared to other plants. Planting Z. latifolia in highly eutrophicated areas with seasonal harvesting and clearing may effectively control pollution through plant absorption and microbial degradation (Liu et al., 2007; Tang et al., 2013). During industrial production, Z. latifolia has found further application as pulp for paper making, which can be prepared by sulfate treatment of the residual stems and leaves after Jiaobai harvesting. Physical performance evaluations have shown that the tear, tensile, and rupture strengths of the resulting paper are not significantly different from those of conventional old corrugated container paper (Chen et al., 2022).
3 Domestication and breeding of Z. latifolia and Z. palustris
Crop domestication is the foundation of modern agriculture and comprises long and complex evolutionary processes (Pickersgill, 2007; Vaughan et al., 2007). Improving the cultivation of wild plants entails changing their morphology and physiology to accommodate human and production needs, which is fundamental to agricultural development (Tang et al., 2013). Although the current growth of global crop yield is stable, botanists face an enormous challenge of food security in addition to climate change and the increasing growth of the global population (Ren et al., 2005; Ma et al., 2015).
3.1 Domestication and breeding of Z. palustris
Compared to rice, maize, wheat, and other major food crops, Z. palustris has a shorter breeding and commercial production time. Hundreds of years ago, American Indians processed and harvested Z. palustris for food; however, only in 1853, Z. palustris was proposed as a crop for planting (Porter, 2019). In 1950, farmers in Northern Minnesota successfully harvested the first batch of manually planted Z. palustris. In 1962, Uncle Ben’s company signed contracts to purchase and sell Z. palustris with planters in Minnesota. In 1972, farmers planted Z. palustris in a paddy field; however, they could not harvest seeds due to its strong seed-shattering tendencies. Z. palustris planters consulted the University of Minnesota Agronomy and Phytogenetics Center for improvements, pursuing a cultivar suitable for large-scale planting. Accordingly, the University of Minnesota developed a special scientific research team to conduct the artificial domestication of Z. palustris, which has been ongoing for 40 years until now (Kahler et al., 2014). Currently, the primary objective of breeders for Z. palustris domestication is the improvement of various growth traits, including reducing seed shattering following maturation and increasing the seed maturation consistency (Grombacher et al., 1996). Seed shattering is an important trait of wild plants to adapt to natural environments and maintain reproduction levels (Zhang et al., 2009); further, it is considered a direct morphological basis for identifying the domestication of wild plants (Ray and Chakraborty, 2018). Although strong seed shattering facilitates self-breeding and population conservation of Z. palustris, the reduction of seed shattering can promote an effective collection of seeds after maturation, thus sustaining production and yield (Yan et al., 2015; Lv et al., 2018). During Z. palustris planting, excessive seed shattering can cause harvest loss within 24 h amounting to 10–20% of the total harvest, whereas the loss of seed maturity can reach 70% during the harvest season (Elliott and Perlinger, 1977; Kennard et al., 2002). Recent collinearity analyses of the Z. palustris and rice genomes screened 20 orthologous genes with rice seed shattering genes qSH1, SH1, SHAT1, SH4, SH5, and OsLG1 as potential candidate genes to improve seed shattering (Haas et al., 2021).
A major component of the University of Minnesota Z. palustris breeding program centers around using conventional breeding methods for Z. palustris, selecting superior phenotypes that can significantly improve variety yield and production efficiency (Grombacher et al., 1996). Progress in crop improvement has been successful using the phenotypic mass recurrent selection method of plant breeding and has led to the introduction of improved wild rice varieties (Kennard et al., 2000). Certain Z. palustris hybrids are currently under commercial cultivation, among which “Itasca-C12” was released in 2007 by the Minnesota Cultivated Wild Rice Council as a variety with lower seed shattering that is suitable for mechanized harvesting in large planting areas. Therefore, “Itasca-C12” is a standard in the Z. palustris industry and is used for genome sequencing (Haas et al., 2021) and research on seed dormancy (Mcgilp et al., 2022) of Z. palustris. These artificially selected Z. palustris varieties are primarily grown in Minnesota and California (Haas et al., 2021). Z. palustris seeds have moderate tolerance to recalcitrance and dehydration, limiting their survival in off-site storage to 1–2 years. Hence, save for annual seed maintenance, Z. palustris cannot be stored in seed banks or repositories, posing challenges to Z. palustris conservation and breeding plans (Mcgilp et al., 2022). Seed dormancy refers to the lack of germination under generally favorable environmental conditions, i.e., sufficient water, oxygen, temperature, and light (Hilhorst, 1995). Z. palustris exhibits a minimum three-month dormancy period (Cardwell, 1978), which is affected by temperature, as well as the stability of seed pericarps and phytohormones, such as abscisic acid and gibberellic acid (Grombacher et al., 1996). Within Poaceae, 98.6% of species are classified as orthodox or desiccation tolerant, while only 0.8% and 0.6% are considered recalcitrant and intermediate, respectively (Dickie and Pritchard, 2002). Z. palustris is generally considered recalcitrant; however, its water resistance is impacted by storage temperature, degree of dryness, and metabolic activity (Probert and Longley, 1989; Kovach and Bradford, 1992; Berjak and Pammenter, 2008). Z. palustris seeds can survive drying under specific conditions, with embryonic water content reaching as low as 6% (Kovach and Bradford, 1992). Therefore, maintaining the vitality of wild rice seeds is essential for Z. palustris breeding (Kennard et al., 1999; Jin et al., 2005; Porter, 2019).
3.2 Domestication and breeding of Z. latifolia
The economic value of Z. latifolia is associated primarily with the aquatic vegetable Jiaobai and CWR (Yan et al., 2018). Chinese ancestors domesticated Z. latifolia as an aquatic vegetable Jiaobai approximately 2000 years ago (as noted in the Chinese first dictionary book “Erya” in the Qin Dynasty, 207–221 BC) (Guo et al., 2007; Guo et al., 2015). During its long-term symbiosis with the endophytic fungus U. esculenta, domesticated Z. latifolia Jiaobai has lost its flower structures and sexual propagation ability, only undergoing asexual reproduction through underground rhizomes (Guo et al., 2015). U. esculenta primarily colonizes the underground root and rhizome of the Jiaobai plant, completing its whole life cycle within the host (Yan et al., 2013b; Jose et al., 2016). Currently, China has > 100 regional cultivars of Jiaobai, and it has become an important economic source for many rural households in southern China (Guo et al., 2007). Domesticated Jiaobai exhibits significant divergence from Z. latifolia based on plant type, means of reproduction, and swollen gall metamorphosis. Under natural conditions, the fleshy stem of the plant formed after infection by U. esculenta, some of them are smaller and full of black teliospores, called “gray Jiaobai” (Figure 2). Jiaobai cultivars are compact, vertical, have swollen, fleshy stems, and lack or only possess rudimentary teliospores (“normal Jiaobai”; Yan et al., 2013a). Notably, the genetic variation and relationships of Z. latifolia and Jiaobai varieties exhibit obvious population structures (Zhao et al., 2019). However, the levels of genetic variation within Jiaobai are extremely low, suggesting that Jiaobai arose from a single domestication (Xu et al., 2008; Zhao et al., 2019). The domestication of Jiaobai and the diversity of its cultivars are mainly affected by the genetic variation of U. esculenta (Zhao et al., 2019). Specifically, U. esculenta strains in normal and gray Jiaobai resulting from atavistic mutation exhibit differences in morphology and internal transcribed spacers, with marked differentiation in the morphology and genetics between the two forms (You et al., 2011).
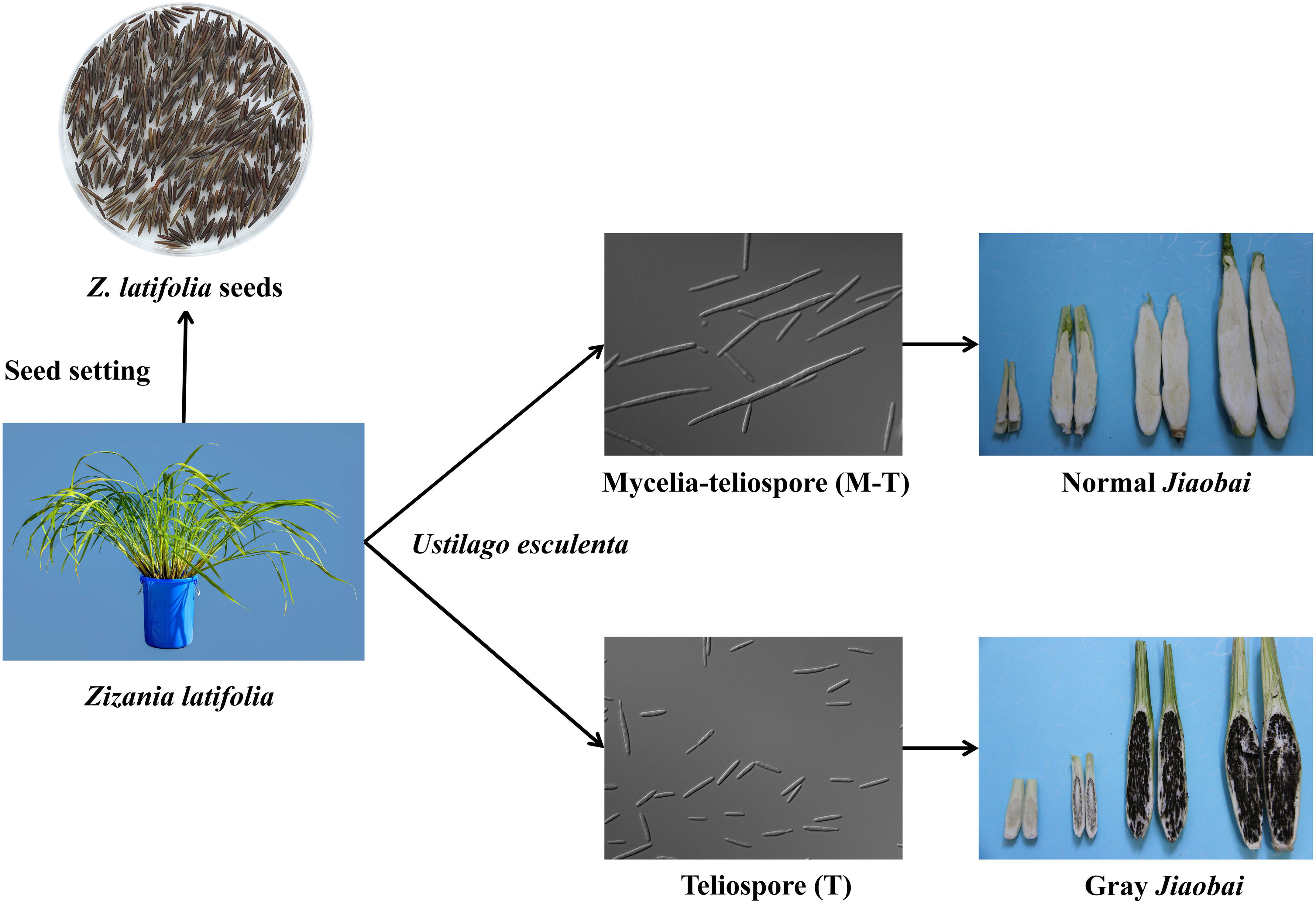
Figure 2 Jiaobai (normal and gray Jiaobai) formed from the infection of Zizania latifolia with Ustilago esculenta. In the images, U. esculenta is a haploid strain, the mycelia-teliospore (M-T) has a long haploid phase with multifocal budding, while the teliospore (T) has a short haploid phase with normal budding and generally does not exhibit multifocal budding.
4 Omics research of Z. latifolia and Z. palustris
4.1 Genome research of Z. latifolia and Z. palustris
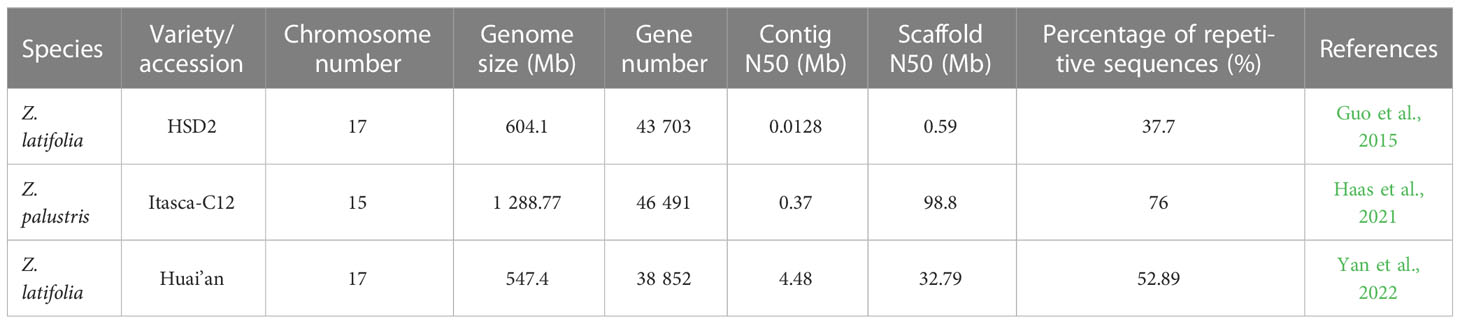
A reference genome is the foundation of genomic and gene function research (Jansen et al., 2007; Moore et al., 2010). Alignment of molecular markers and reference genomes can provide researchers with genes of interest and functional relationships between characteristics, important physiological mechanisms, and the structures of intraspecies genetic diversity. However, as of 2020, the genome resources of Z. palustris were limited to only a few studies on molecular markers, including isozyme (Lu et al., 2005), restrictive fragment length polymorphism (Kennard et al., 1999; Kennard et al., 2002), simple sequence repeats (Kahler et al., 2014), and single nucleotide polymorphism (Shao et al., 2020). In 2021, Haas et al. (2021) assembled the high-quality genome of the ‘Itasca-C12’ NWR cultivar and anchored 98.53% of sequences to 15 chromosomes, with an assembly length of 1.29 Gb and high reproducibility (~76.0%; Table 2). This genome assembly and annotation provide an important reference to the comparative genomics of the rice tribe (Oryzeae) and lays a foundation for future NWR protection and breeding work (Haas et al., 2021). Estimates of divergence times revealed that the Zizania genus diverged from Oryza by approximately 26–30 mya, whereas Z. palustris and Z. latifolia diverged from one another by approximately 6–8 mya (Haas et al., 2021).
Given that Z. latifolia lacks a high-quality genome, research on the genetics and genes related to various characteristics of Z. latifolia remains considerably stunted compared to that of rice. Zizania is also a monoecious outcrosser with severe inbreeding depression, which increases the difficulty of genetic mapping research. The genome of Z. latifolia chloroplasts comprises typical circular double-stranded DNA molecules with genome size, overall structure, gene number, and gene sequence highly conserved with most terrestrial plant genomes (Zhang et al., 2016). The genome size is 136,501 bp, and its sequence was derived from the direct purification of chloroplast DNA (Zhang et al., 2016). Guo et al. (2015) used second-generation sequencing technology to build the Z. latifolia accession “HSD2” genome sequence for the first time. They performed transcriptome analysis of the molecular mechanism of the Jiaobai swollen culm formed following the infection of Z. latifolia by U. esculenta. Due to technical limitations and a lack of genetic linkage maps, the Z. latifolia genome remains relatively scattered at a scaffolding level, with a contig N50 of only 13 kb (Table 2). With third-generation technology, sequencing of the Z. latifolia genome revealed a total genome length of 545.36 Mb distributed over 17 chromosomes, accounting for 99.63%, and 300 corresponding sequences were obtained (the longest and shortest being 49.61 Mb and 17.01 Mb, respectively). In addition, the assembly of the Z. latifolia genome (Contig N50 = 4.48 Mb) is 343.62-fold longer than the previously completed “HSD2” genome (Contig N50 = 13kb). Through genome annotation, 289.5 Mb (52.89%) of the repetitive sequence was identified in the assembled genome, which is significantly higher than the 227.50 Mb (37.70%) reported in the previously assembled version (Yan et al., 2022).
4.2 Transcriptome research of Z. latifolia
Transcriptomics is the systematic study of global gene expression patterns to identify the molecular mechanisms of complex biological pathways and trait regulation networks (Ponting et al., 2009). These studies have revealed that the fungal pathogenicity-related genes of U. esculenta, as well as those associated with phytohormone biosynthesis, may cause culm enlargement of Z. latifolia (Wang et al., 2017; Li et al., 2021; Zhang et al., 2021). Moreover, two Cys2-His2 (C2H2) zinc finger proteins, GME3058_g and GME5963_g, from U. esculenta may impact fungal growth and infection at the initial stage of swollen culm formation (Zhang et al., 2021). Numerous U. esculenta genes related to effectors and teliospore formation have been shown to exhibit marked upregulation (Wang et al., 2020) and stage-specific expression patterns during the initial and subsequent enlargement of culm (Wang et al., 2017). For instance, Wang et al. (2020) reported that the expression of the melanin biosynthesis gene of the teliospore (T) strain is upregulated compared with that in the mycelia-teliospore (M-T) strain of U. esculenta; the T strain exhibits stronger pathogenicity and teliospore-forming properties than the M-T strain. Indeed, two types of gene regulatory networks contribute to the formation of different types of swollen culm (normal and grey) (Wang et al., 2020). During the process of culm swelling, many genes related to the synthesis, metabolism, and signal transduction of hormones of the host plant are stimulated and exhibit specific expression patterns. In particular, the expression of ZlYUCCA9—a flavin monooxygenase that serves as the key enzyme in the indole-3-acetic acid biosynthesis pathway—is markedly upregulated (Zhang et al., 2021). Moreover, host plant genes become differentially expressed before and after U. esculenta infection, some of which are primarily involved in plant hormone signal transduction and cell wall–loosening factors (Li et al., 2021). Although “a hormone–cell wall loosening model” was proposed to explain the symbiotic mechanism in culm enlargement (Li et al., 2021), cytokinins appear to play a more important role (Wang et al., 2017; Li et al., 2021).
4.3 Proteome research of Z. latifolia
Proteomics is the study of changes in cells, tissues, or protein composition of organisms, with the proteome functioning as the object of study (Singh et al., 2016). Isobaric tags for relative and absolute quantification (iTRAQ) have been used in recent years for high-throughput screening in quantitative proteomics and exhibit good quantitative results along with satisfactory reproducibility (Owiti et al., 2011; Chu et al., 2019). Based on the pattern of phenolic compound change during CWR germination, researchers selected the representative phases (germinated for 36 and 120 h) for further proteomics analysis of the mechanism of phenolic compound accumulation during CWR germination using iTRAQ. The results showed that the differentially expressed proteins in these two phases were primarily associated with metabolic pathways, including biosynthesis of secondary metabolites and phenylpropanoid biosynthesis (Chu et al., 2019). Other researchers performed two-dimensional electrophoresis of total proteins in CWR and rice seeds; the information obtained from peptide mass fingerprinting indicated that a glutelin precursor, caffeoyl coenzyme A O-methyltransferase, and putative bithoraxoid-like protein could provide good gene sources for improving rice seed quality (Jiang et al., 2016). U. esculenta evades host defenses through at least seven metabolic pathways and five biological processes to successfully reproduce in Z. latifolia. Observing the proteins extracted from the topmost internodal region below the apical meristematic tissue deriving from the infected and uninfected parts of Z. latifolia through transmission electron and fluorescence microscopy showed that U. esculenta hyphal morphological transitions and movement occurred both inter- and intracellularly, while sporulation occurred only intracellularly in selective cells (Jose et al., 2019). This study revealed why differentially expressed proteins in U. esculenta allow the inflorescence organization to be replaced by a swollen fleshy stem and why Z. latifolia develops resistance to infection by U. esculenta (Jose et al., 2019).
4.4 Metabolome research of Z. latifolia and Z. palustris
Plant metabolomics is the study of the relationship between gene function and phenotype through analysis of the metabolites in plant tissues during specific changes in the external environment (Oliver, 1997). Plant metabolic networks are complex and are typically classified as primary or secondary metabolisms; however, there is no clear boundary between these two types as they are closely related. Currently, ~200,000 metabolites have been found in plants (Wink, 1988). Some scientists have studied Jiaobai through metabolo- and other omic approaches (Luo et al., 2012; Luo et al., 2019; Bata Gouda et al., 2022). Jiaobai is of substantial economic value; however, its shelf-life and quality during post-harvest storage are reduced due to respiratory disorders, shell etiolation, surface browning, transpiration, and tissue hollowness (Luo et al., 2012, 2019). Therefore, researchers believe that the physiological, biochemical, and molecular processes involved in the post-harvest aging of Jiaobai, as well as post-harvest treatment methods to ameliorate aging and improve storage quality, are worth investigating (Bata Gouda et al., 2022; Qian et al., 2023). The mechanisms of Jiaobai aging during storage at 25°C were investigated using integrated methods of transcriptomics and metabolomics. The results showed that Z. latifolia aging is closely associated with reactive oxygen species (ROS) accumulation, ethylene biosynthesis, energy metabolism consumption caused by cell membrane degradation, and abiotic stress (Bata Gouda et al., 2022). Aging may also be weakly associated with ornithine decarboxylase, polyamine oxidase, transcription factor A, jasmonic acid-amino synthetase, coronatine insensitive protein 1, brassinosteroid insensitive 1 kinase inhibitor 1, mitogen-activated protein kinase, calmodulin, and catalase genes, as well as lower organic acid, l-alanine, and γ-linolenate contents (Bata Gouda et al., 2022). Following 1-MCP treatment, these genes, metabolism products, and enzyme activities changed, thereby delaying Jiaobai post-harvest aging (Bata Gouda et al., 2022). Recently, Qian et al. (2023) found that the lignification of Jiaobai during cold storage was regulated by respiratory burst oxidase homolog-mediated ROS signaling. These studies improved our understanding on the mechanisms of Jiaobai post-harvest aging and serve as important references for maintaining post-harvest quality, as well as extending the shelf life of Jiaobai.
Analysis and utilization of the nutritional components of wild rice are increasingly popular research topics. Enrichment analyses between CWR and NWR showed that the differential metabolites in the phenylpropanoid biosynthesis pathway were significantly enriched, with 357 metabolites forming a significant cluster, among which the relative content of 5 anthocyanin and 4 catechin derivatives differed significantly between CWR and NWR (Yan et al., 2019). In addition, the total phenolic, flavonoid, and proanthocyanidin contents in CWR were significantly higher than those in Indica, Japonica, and red rice (Yu et al., 2021). Using ultra-high performance liquid chromatography coupled to triple quadrupole mass spectrometry-based metabolomic methods, 159 flavonoids were identified in CWR and non-pigment rice, among which 78 exhibited differential expression. KEGG annotation and classification showed that the differentially expressed flavonoids were primarily associated with anthocyanin biosynthesis (Yu et al., 2021). This result is consistent with previous reports of flavonoid accumulation in pigmented rice seeds closely associated with changes in pericarp pigments (Shao et al., 2014). During CWR seed development, the content of total phenols and proanthocyanidins gradually increases. The metabolomic analysis also showed that 57 flavonoids were associated with changes in pericarp color and exhibited gradual increases (Yu et al., 2022). This study also explored the molecular basis of changes in pericarp color during CWR development. The results demonstrated novel perspectives on flavonoid biosynthesis and accumulation research in CWR, while laying the foundation for modern biotechnology to obtain grains with high flavonoid contents.
5 Functional verifications of important genes of Z. latifolia and Z. palustris
As a species transitioning from wildness to domestication, Zizania plants retain the excellent traits lost by many domesticated crops, thus providing a potential source for gene modification of varieties in modern rice breeding and an ideal natural resource for expanding and enriching breeding gene sources (Yan et al., 2022). Guo et al. (2015) and Yan et al. (2022) found that the Z. latifolia and rice genomes are highly collinear. Phylogenetic analysis shows that Z. latifolia is more closely related to Oryza than the other seven plants (Brachypodium distachyon, Hordeum vulgare, Leersia perrieri, Sorghum bicolor, Setaria italica, Zea mays, and Arabidopsis thaliana), with a differentiation time of 19.7–31 mya (Yan et al., 2022). Cross-incompatibility prohibits the direct transfer of these valuable traits to rice (Yan et al., 2022). However, researchers have successfully introduced various high-value genes of interest from Zizania to rice through transgenic technology, allowing for their functional verification and application (Abedinia et al., 2000; Shen et al., 2011; Qi et al., 2023). Abedinia et al. (2000) co-bombarded rice calli with high molecular weight Z. palustris DNA and a pGL2 plasmid encoding the selectable hygromycin resistance gene to generate transgenic plants with NWR grain characteristics for analysis. Amplified fragment length polymorphism analysis showed that this method successfully transferred DNA from large numbers of Z. palustris genes to rice (Abedinia et al., 2000). Following infection of rice with bacterial blight, a disease in which the vascular bundle leaves generally dry out and the non-fruiting rate increases, causing a 1000 grain weight reduction translating to a yield loss of 20–50%, even 100% in severe cases (Lee et al., 2005; Antony et al., 2010). Notably, mining and discovering previously lost beneficial genes in closely related Zizania and rice germplasm resources is one feasible strategy for addressing this situation (Shen et al., 2011). Researchers have designed specific primers based on homologous sequences, screened for a disease resistance gene from the Z. latifolia genome database, and performed alignment analyses on amplified nucleotide sequences. Results showed that ZR1 is part of the nucleotide-binding site domains-C-terminal leucine-rich repeats resistance gene based on its similarity to the P-loop (kinase 1a), kinase 2, kinase 3a, and Gly-Leu-Pro-Leu (GLPL) conserved gene sequences (Shen et al., 2011). Over-expressed transgenic plants with significant resistance to bacterial blight PXO71 were obtained through Agrobacterium-mediated transfer to rice (O. sativa cv. Nipponbare), suggesting that transformation-competent artificial chromosome clones contain ≥ 1 bacterial blight resistance gene—ZlBBR1 (Shen et al., 2011).
The tip of Z. latifolia culm enlarges into a fleshy edible gall after U. esculenta infection, and cytokinin plays a key role in this process. Two-component systems (TCS) connect cytokinin with transcriptional regulation receptors in the nucleus and play a significant role in many bioprocesses (He et al., 2020). Genome-wide identification and transcriptomics were performed on TCS genes in Jiaobai to analyze their expression during culm enlargement. The findings revealed that expression of ZlCHK1, ZlRRA5, ZIRRA9, ZlRRA10, ZlPRR1, and ZlPHYA was associated with Jiaobai culm swelling, among which ARR5, ARR9, and ZlPHYA were rapidly induced by trans-zeatin, supporting that cytokinin signal transduction plays a role in Jiaobai culm enlargement (He et al., 2020). The Jiaobai genome contains 11 chitinase genes (ZlChi1-11), many of which are differentially expressed under abiotic stressors such as salt, extreme temperatures, drought, and the presence of abscisic acid (Zhou et al., 2020). This study provides basic information for analyzing the role of the Z. latifolia chitinase gene family under abiotic stress.
The nutritional composition of CWR is greater than that of normal rice (at 0.26 ± 0.02 g), especially concerning its significantly high lysine content (at 0.62 ± 0.07 g; Zhai et al., 2000). DHDPS encoding the lysine biosynthetic enzyme dihydrodipocolinate synthase plays a significant role in lysine accumulation. Kong et al. (2009) cloned the Z. latifolia DHDPS and named it ZlDHDPS. The ZlDHDPS sequence maintains a high identity with known plant DHDPS in GenBank. RT-PCR analysis showed that ZlDHDPS expression has tissue specificity, as well as high-level expression in rapidly growing and reproductive tissues. Through genome collinearity and homology analysis, ZlRc (Zla16G011250) in Z. latifolia was found to be orthologous to Rc (LOC_Os07g110200) in rice (Qi et al., 2023). Functional validation of ZlRc was performed using subcellular localization and rice transgenes. The results show that ZlRc promotes phenolic compound accumulation and is located within the nucleus (Qi et al., 2023). Moreover, the pericarps of ZlRc-overexpressing rice are brown, while those of wild-type rice are non-pigmented (Qi et al., 2023). The total phenolic, flavonoid, and proanthocyanidin contents, antioxidant activity, as well as enzyme inhibitory effects of ZlRc-overexpressing rice were significantly higher than those of wild-type rice (Qi et al., 2023). These findings show that ZlRc-overexpression promotes phenolic compound accumulation in rice seeds and can bioaugment rice phenolic content. Through genome collinearity and homology analysis, ZlqSH1a (Zla04G033720) and ZlqSH1b (Zla02G027130) in Z. latifolia were found to be orthologous to qSH1 (LOC_Os01g62920) in rice (Yan et al., 2022). Functional validation of ZlqSH1a and ZlqSH1b was performed using subcellular localization and rice transgenes, with the results showing that these genes are involved in regulating the development of the abscission layer and located within the nucleus. Scanning electron and laser confocal microscopy also showed that ZlqSH1a and ZlqSH1b over-expression resulted in a complete abscission layer between the grain and pedicel and significantly enhanced seed shattering following grain maturation in rice (Xie et al., 2022). Studies of gene function related to seed shattering in Z. latifolia have provided a foundation for reducing Z. latifolia seed shattering and thus accelerating its domestication.
6 Conclusion and perspectives
In summary, Zizania is a highly valuable economic crop that provides nutritious grains and vegetables for human use. Laying a strong foundation and utilization of Zizania is currently being done. Recently, genomes of Z. latifolia and Z. palustris have been completed, accelerating the collective understanding of the functional relationships between genes and characteristics, important physiological mechanisms, and genetic diversity of this genus. Hence, the multitude of desirable traits found in Zizania render it an ideal genetic resource for future molecular breeding aimed at enhancing the qualities of rice. Specifically, the domestication of wild plants is beneficial for ecological and sustainable development while providing a novel pathway for addressing food security and environmental challenges; however, current studies on Zizania remain insufficient, with the following research areas demanding further attention:
(1) Collection, identification, and improvement of germplasm resources of Zizania: We should survey, collect, preserve, and identify Zizania germplasm resources and establish a germplasm resource garden. Chemical (ethyl methanesulfonate) or physical (radiation) mutagenesis techniques will be used for improving the germplasm resources of Zizania. Referring to the domestication pathway of Z. palustris, domestication breeding of Z. latifolia will be carried out to screen plants with target traits such as reduced seed shattering, early flowering, consistent maturity, and high seed setting rate.
(2) Analysis of functional components in wild rice: We should use modern separation (solvent extraction), purification (purification of macroporous adsorption resin), and structural identification (spectral and non-spectral methods) techniques to conduct research on the separation and structural identification of bioactive substances from wild rice and accurately analyze the composition, and content of bioactive substances such as amylose, resistant starch, dietary fiber, flavonoids, saponins, anthocyanins, chlorophyll, and phytosterols in wild rice.
Author contributions
Author contributions are conceptualization, investigation, visualization, writing- original draft preparation: NY, Z-FZ, Y-NX, and Q-QQ; Review and editing W-HL, Y-LL, YZ, H-MW, Y-FZ, Z-HY, D-PG, and QQ. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Youth Innovation Program of the Chinese Academy of Agricultural Sciences (Grant No. Y2023QC34), the Central Public-interest Scientific Institution Basal Research Fund (Grant No. 1610232021006, 1610232022004, and 1610232023003), the Postdoctoral Applied Research Project Fund of Qingdao, and the Agricultural Science and Technology Innovation Program (Grant No. ASTIP-TRIC05-2023 and ASTIP-TRIC-ZD05).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abedinia, M., Henry, R. J., Blakeney, A. B., Lewin, L. G. (2000). Accessing genes in the tertiary gene pool of rice by direct introduction of total DNA from Zizania palustris (wild rice). Plant Mol. Biol. Rep. 18, 133–138. doi: 10.1007/bf02824021
Aladedunye, F., Przybylski, R., Rudzinska, M., Klensporf-Pawlik, D. (2013). γ-oryzanols of north American wild rice (Zizania palustris). J. Am. Oil Chem. Soc 90, 1101–1109. doi: 10.1007/s11746-013-2252-x
Antony, G., Zhou, J. H., Huang, S., Li, T., Liu, B., White, F., et al. (2010). Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene os-11N3. Plant Cell 22, 3864–3876. doi: 10.1105/tpc.110.078964
Bata Gouda, M. H., Peng, S., Yu, R. Y., Li, J. Q., Zhao, G. H., Chen, Y. R., et al. (2022). Transcriptomics and metabolomics reveal the possible mechanism by which 1-methylcyclopropene regulates the postharvest senescence of Zizania latifolia. Food Qual. Saf. 6, 1–13. doi: 10.1093/fqsafe/fyac003
Berjak, P., Pammenter, N. W. (2008). From Avicennia to Zizania: seed recalcitrance in perspective. Ann. Bot. 101, 213–228. doi: 10.1093/aob/mcm168
Bunzel, M., Allerdings, E., Sinwell, V., Ralph, J., Steinhart, H. (2002). Cell wall hydroxycinnamates in wild rice (Zizania aquatica l.) insoluble dietary fibre. Eur. Food Res. Technol. 214, 482–488. doi: 10.1007/s00217-002-0512-3
Cardwell, V. B. (1978). Seed dormancy mechanisms in wild rice (Zizania aquatica). Agron. J. 70, 481–484. doi: 10.2134/agronj1978.00021962007000030029x
Chang, B. Y., Kim, H. J., Kim, T. Y., Kim, S. Y. (2021). Enzyme-treated Zizania latifolia extract protects against alcohol-induced liver injury by regulating the NRF2 pathway. Antioxidants 10, 960. doi: 10.3390/antiox10060960
Chen, Y. Y., Chu, H. J., Liu, H., Liu, Y. L. (2012). Abundant genetic diversity of the wild rice Zizania latifolia in central China revealed by microsatellites. Ann. Appl. Biol. 161, 192–201. doi: 10.1111/j.1744-7348.2012.00564.x
Chen, Y. Y., Liu, Y., Fan, X. R., Li, W., Liu, Y. L. (2017). Landscape-scale genetic structure of wild rice Zizania latifolia: the roles of rivers, mountains, and fragmentation. Front. Ecol. Evol. 5. doi: 10.3389/fevo.2017.00017
Chen, H., Yang, T., Wang, J. H., Song, J. S., Wo, Q. Z., Zheng, B. B., et al. (2022). Study on the application of Zizania latifolia straw in papermaking. Bioresources 17, 5106–5115. doi: 10.15376/biores.17.3.5106–5115
Chu, C., Yan, N., Du, Y. M., Liu, X. M., Chu, M. J., Shi, J., et al. (2019). iTRAQ-based proteomic analysis reveals the accumulation of bioactive compounds in Chinese wild rice (Zizania latifolia) during germination. Food Chem. 289, 635–644. doi: 10.1016/j.foodchem.2019.03.092
Dickie, J. B., Pritchard, H. W. (2002). “Systematic and evolutionary aspects of desiccation tolerance in seeds,” in Desiccation and survival in plants: drying without dying, vol. 239–259. (Wallingford, UK: CAB International). doi: 10.1079/9780851995342.0239
Drewes, A. D., Silbernagel, J. (2012). Uncovering the spatial dynamics of wild rice lakes, harvesters and management across great lakes landscapes for shared regional conservation. Ecol. Modell. 229, 97–107. doi: 10.1016/j.ecolmodel.2011.09.015
Elliott, W. A., Perlinger, G. J. (1977). Inheritance of shattering in wild rice 1. Crop Sci. 17, 851–853. doi: 10.2135/cropsci1977.0011183X001700060008x
Fan, X. R., Ren, X. R., Liu, Y. L., Chen, Y. Y. (2016). Genetic structure of wild rice Zizania latifolia and the implications for its management in the sanjiang plain, northeast China. Biochem. Syst. Ecol. 64, 81–88. doi: 10.1016/j.bse.2015.11.017
Gao, Y., Chen, H. J., Liu, R. L., Wu, W. J., Mu, H. L., Han, Y. C., et al. (2022). Ameliorating effects of water bamboo shoot (Zizania latifolia) on acute alcoholism in a mice model and its chemical composition. Food Chem. 378, 132122. doi: 10.1016/j.foodchem.2022.132122
Glover, J. D., Reganold, J. P., Bell, L. W., Borevitz, J., Brummer, E. C., Buckler, E. S., et al. (2010). Increased food and ecosystem security via perennial grains. Science 328, 1638–1639. doi: 10.1126/science.1188761
Grombacher, A. W., Porter, R. A., Everett, L. E. (1996). Breeding wild rice. Plant Breed Rev. 14, 237–265. doi: 10.1002/9780470650073.ch8
Guo, H. B., Li, S. M., Peng, J., Ke, W. D. (2007). Zizania latifolia turcz. cultivated in China. Genet. Resour. Crop Evol. 54, 1211–1217. doi: 10.1007/s10722-006-9102-8
Guo, L. B., Qiu, J., Han, Z. J., Ye, Z. H., Chen, C., Liu, C. J., et al. (2015). A host plant genome (Zizania latifolia) after a century-long endophyte infection. Plant J. 83, 600–609. doi: 10.1111/tpj.12912
Guo, H., Wu, H., Sajid, A., Li, Z. (2022). Whole grain cereals: the potential roles of functional components in human health. Crit. Rev. Food Sci. Nutr. 62, 8388–8402. doi: 10.1080/10408398.2021.1928596
Haas, M., Kono, T., Macchietto, M., Millas, R., Mcgilp, L., Shao, M. Q., et al. (2021). Whole-genome assembly and annotation of northern wild rice, zizania palustris l., supports a whole-genome duplication in the Zizania genus. Plant J. 107, 1802–1818. doi: 10.1111/tpj.15419
Hayes, P. M., Stucker, R. E., Wandrey, G. G. (1989). The domestication of American wildrice. Econ. Bot. 43, 203–214. doi: 10.1007/bf02859862
He, L. L., Zhang, F., Wu, X. Z., Hu, Y. M., Dong, L. L., Dewitte, W., et al. (2020). Genome-wide characterization and expression of two-component system genes in cytokinin-regulated gall formation in Zizania latifolia. Plants 9, 1409. doi: 10.3390/plants9111409
Hilhorst, H. W. M. (1995). A critical update on seed dormancy. i. primary dormancy1. Seed Sci. Res. 5, 61–73. doi: 10.1017/S0960258500002634
Horne, F., Kahn, A. (1997). Phylogeny of north American wild rice, a theory. Southwestern Nat. 42, 423–434.
Huang, X. H., Kurata, N., Wei, X. H., Wang, Z. X., Wang, A., Zhao, Q., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. doi: 10.1038/nature11532
Huang, X. H., Yang, S. H., Gong, J. Y., Zhao, Q., Feng, Q., Zhan, Q. L., et al. (2016). Genomic architecture of heterosis for yield traits in rice. Nature 537, 629–633. doi: 10.1038/nature19760
Jansen, R. K., Cai, Z. Q., Raubeson, L. A., Daniell, H., Depamphilis, C. W., Leebens-Mack, J., et al. (2007). Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. U.S.A. 104, 19369–19374. doi: 10.1073/pnas.0709121104
Jiang, M. X., Zhai, L. J., Yang, H., Zhai, S. M., Zhai, C. K. (2016). Analysis of active components and proteomics of Chinese wild rice (Zizania latifolia (Griseb) turcz) and Indica rice (Nagina22). J. Med. Food 19, 798–804. doi: 10.1089/jmf.2015.3612
Jin, I. D., Yun, S. J., Matsuishi, Y., Iwaya-Inoue, M. (2005). Changes in the water content and germination rate during seed desiccation and their inter-specific differences among Zizania species. J. Fac. Agric. Kyushu Univ. 50, 573–583. doi: 10.1017/S0021859605005617
Jose, R. C., Bengyella, L., Handique, P. J., Talukdar, N. C. (2019). Cellular and proteomic events associated with the localized formation of smut-gall during Zizania latifolia–Ustilago esculenta interaction. Microb. Pathog. 126, 79–84. doi: 10.1016/j.micpath.2018.10.028
Jose, R. C., Goyari, S., Louis, B., Waikhom, S. D., Handique, P. J., Talukdar, N. C. (2016). Investigation on the biotrophic interaction of Ustilago esculenta on Zizania latifolia found in the indo-Burma biodiversity hotspot. Microb. Pathog. 98, 6–15. doi: 10.1016/j.micpath.2016.06.021
Juan, J., Liu, G., Willett, W. C., Hu, F. B., Rexrode, K. M., Sun, Q. (2017). Whole grain consumption and risk of ischemic stroke results from 2 prospective cohort studies. Stroke 48, 3203–3209. doi: 10.1161/Strokeaha.117.018979
Kahler, A. L., Kern, A. J., Porter, R. A., Phillips, R. L. (2014). Maintaining food value of wild rice (Zizania palustris l.) using comparative genomics (Berlin: Springer Netherlands), 233–248.
Kennard, W. C., Phillips, R. L., Porter, R. A. (2002). Genetic dissection of seed shattering, agronomic, and color traits in American wildrice (Zizania palustris var. interior l.) with a comparative map. Theor. Appl. Genet. 105, 1075–1086. doi: 10.1007/s00122-002-0988-z
Kennard, W., Phillips, R., Porter, R., Grombacher, A. (1999). A comparative map of wild rice (Zizania palustris l. 2n=2x=30). Theor. Appl. Genet. 99, 793–799. doi: 10.1007/s001220051298
Kennard, W. C., Phillips, R. L., Porter, R. A., Grombacher, A. W. (2000). A comparative map of wild rice (Zizania palustris l. 2n= 2x= 30). Theor. Appl. Genet. 101, 677–684. doi: 10.1007/s001220051530
Kong, F. N., Jiang, S. M., Meng, X. B., Song, C. L., Shi, J. F., Jin, D. M., et al. (2009). Cloning and characterization of the DHDPS gene encoding the lysine biosynthetic enzyme dihydrodipocolinate synthase from Zizania latifolia (Griseb). Plant Mol. Biol. Rep. 27, 199–208. doi: 10.1007/s11105-008-0073-0
Kovach, D. A., Bradford, K. J. (1992). Temperature dependence of viability and dormancy of Zizania palustris var. interior seeds stored at high moisture contents. Ann. Bot. 69, 297–301. doi: 10.1093/oxfordjournals.aob.a088345
Lee, B. M., Park, Y. J., Park, D. S., Kang, H. W., Kim, J. G., Song, E. S., et al. (2005). The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 33, 577–586. doi: 10.1093/nar/gki206
Li, J., Lu, Z. Y., Yang, Y., Hou, J. F., Yuan, L. Y., Chen, G. H., et al. (2021). Transcriptome analysis reveals the symbiotic mechanism of Ustilago esculenta-induced gall formation of Zizania latifolia. Mol. Plant-Microbe Interact. 34, 168–185. doi: 10.1094/Mpmi-05-20-0126-R
Li, W. J., Tan, L., Zhang, N., Chen, H., Fan, X. P., Peng, M., et al. (2022). Phytolith-occluded carbon in residues and economic benefits under rice/single-season Zizania latifolia rotation. Sci. Total Environ. 836, 15504. doi: 10.1016/j.scitotenv.2022.155504
Liu, J. G., Dong, Y., Xu, H., Wang, D. K., Xu, J. K. (2007). Accumulation of cd, Pb and zn by 19 wetland plant species in constructed wetland. J. Hazard. Mater. 147, 947–953. doi: 10.1016/j.jhazmat.2007.01.125
Liu, Z. L., Wang, Y. M., Shen, Y., Guo, W. L., Hao, S., Liu, B. (2004). Extensive alterations in DNA methylation and transcription in rice caused by introgression from Zizania latifolia. Plant Mol. Biol. 54, 571–582. doi: 10.1023/B:PLAN.0000038270.48326.7a
Lobell, D. B., Schlenker, W., Costa-Roberts, J. (2011). Climate trends and global crop production since 1980. Science 333, 616–620. doi: 10.1126/science.1204531
Lorenz, K., Lund, D. (1981). Wild rice: the indian’s staple and the white man’s delicacy. Crit. Rev. Food Sci. 15, 281–319. doi: 10.1080/10408398109527318
Lu, R., Chen, M., Feng, Y., Yuan, N., Zhang, Y., Cao, M., et al. (2022). Comparative plastome analyses and genomic resource development in wild rice (Zizania spp., poaceae) using genome skimming data. Ind. Crop Prod. 186, 115244. doi: 10.1016/j.indcrop.2022.115244
Lu, Y. Q., Waller, D. M., David, P. (2005). Genetic variability is correlated with population size and reproduction in American wild-rice (Zizania palustris var. palustris, poaceae) populations. Am. J. Bot. 92, 990–997. doi: 10.3732/ajb.92.6.990
Luo, H. B., Jiang, L., Zhang, L., Jiang, J., Yu, Z. F. (2012). Quality changes of whole and fresh-cut Zizania latifolia during refrigerated (1°C) storage. Food Bioprocess Technol. 37, 1411–1415. doi: 10.1111/j.1745-4549.2012.00743.x
Luo, H. B., Zhou, T., Kong, X. X., Tao, M. X., Zhang, J. X., Wang, W. H., et al. (2019). iTRAQ-based mitochondrial proteome analysis of the molecular mechanisms underlying postharvest senescence of Zizania latifolia. J. Food Biochem. 43, e13053. doi: 10.1111/jfbc.13053
Lv, S. W., Wu, W. G., Wang, M. H., Meyer, R., Ndjiondjop, M. N., Tan, L. B., et al. (2018). Genetic control of seed shattering during African rice domestication. Nat. Plants 4, 331–337. doi: 10.1038/s41477-018-0164-3
Ma, Y., Dai, X., Xu, Y., Luo, W., Zheng, X., Zeng, D., et al. (2015). Cold1 confers chilling tolerance in rice. Cell 160, 1209–1221. doi: 10.1016/j.cell.2015.01.046
Makela, P., Archibold, O. W., Peltonen-Sainio, P. (1998). Wild rice - a potential new crop for Finland. Agric. Food Sci. Finl. 7, 583–597. doi: 10.23986/afsci.5618
Mcgilp, L., Duquette, J., Braaten, D., Kimball, J., Porter, R. (2020). Investigation of variable storage conditions for cultivated northern wild rice and their effects on seed viability and dormancy. Seed Sci. Res. 30, 1–8. doi: 10.1017/S0960258520000033
Mcgilp, L., Semington, A., Kimball, J. (2022). Dormancy breaking treatments in northern wild rice (Zizania palustris l.) seed suggest a physiological source of dormancy. Plant Growth Regul. 98, 235–247. doi: 10.1007/s10725-022-00859-0
Meeker, J. E. (1996). Wild-rice and sedimentation processes in a lake superior coastal wetland. Wetlands 16, 219–231. doi: 10.1007/Bf03160695
Moore, M. J., Soltis, P. S., Bell, C. D., Burleigh, J. G., Soltis, D. E. (2010). Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci. 107, 4623–4628. doi: 10.1073/pnas.0907801107
Oelke, E. A. (2004). “Rice | wildrice, zizania,” in Encyclopedia of grain science, Amsterdam: Elsevier, 74–85. doi: 10.1016/B0-12-765490-9/00145-2
Oelke, E. A., Albrecht, K. A. (1978). Mechanical scarification of dormant wild rice seed. Agron. J. 70, 691–694. doi: 10.2134/agronj1978.00021962007000040041x
Oliver, S. G. (1997). Yeast as a navigational aid in genome analysis. Microbiol. 143, 1483–1487. doi: 10.1099/00221287-143-5-1483
Owiti, J., Grossmann, J., Gehrig, P., Dessimoz, C., Laloi, C., Hansen, M. B., et al. (2011). iTRAQ-based analysis of changes in the cassava root proteome reveals pathways associated with post-harvest physiological deterioration. Plant J. 67, 145–156. doi: 10.1111/j.1365-313X.2011.04582.x
Oxley, F. M., Echlini, A., Power, P., Tolley-Jordan, L., Alexander, M. L. (2008). Travel of pollen in experimental raceways in the endangered Texas wild rice (Zizania texana). Southwest Nat. 53, 169–174. doi: 10.1894/0038-4909(2008)53[169:Topier]2.0.Co;2
Park, S. H., Lee, S. S., Bang, M. H., Jo, S. K., Jhee, K. H., Yang, S. A. (2019). Protection against UVB-induced damages in human dermal fibroblasts: efficacy of tricin isolated from enzyme-treated Zizania latifolia extract. Biosci. Biotechnol. Biochem. 83, 551–560. doi: 10.1080/09168451.2018.1554424
Pickersgill, B. (2007). Domestication of plants in the americas: insights from mendelian and molecular genetics. Ann. Bot. 100, 925–940. doi: 10.1093/aob/mcm193
Ponting, C. P., Oliver, P. L., Reik, W. (2009). Evolution and functions of long noncoding RNAs. Cell 136, 629–641. doi: 10.1016/j.ce11.2009.02.006
Poole, J., Bowles, D. E. (1999). Habitat characterization of Texas wild-rice (Zizania texana Hitchcock), an endangered aquatic macrophyte from the San marcos river, TX, USA. Aquat. Conserv. 9, 291–302. doi: 10.1002/(SICI)1099-0755(199905/06)9:3<291::AID-AQC349<3.0
Porter, R. (2019). Wildrice (Zizania l.) in north America: genetic resources, conservation, and use. North Am. Crop Wild Relatives 2, 83–97. doi: 10.1007/978-3-319-97121-6_3
Probert, R. J., Longley, P. L. (1989). Recalcitrant seed storage physiology in three aquatic grasses (Zizania palustris, Spartina anglica, and Porteresia coarctata). Ann. Bot. 63, 53–64. doi: 10.1093/oxfordjournals.aob.a087728
Qi, Q., Li, W., Yu, X., Zhang, B., Shang, L., Xie, Y., et al. (2023). Genome-wide analysis, metabolomics, and transcriptomics reveal the molecular basis of ZlRc overexpression in promoting phenolic compound accumulation in rice seeds. Food Front. 1–18 doi: 10.1002/fft2.234
Qian, C., Ji, Z., Sun, Y., Zhang, M., Kan, J., Xiao, L., et al. (2023). Lignin biosynthesis in postharvest water bamboo (Zizania latifolia) shoots during cold storage is regulated by RBOH-mediated reactive oxygen species signaling. J. Agric. Food Chem. 71, 3201–3209. doi: 10.1021/acs.jafc.2c08073
Qiu, Y., Liu, Q., Beta, T. (2009). Antioxidant activity of commercial wild rice and identification of flavonoid compounds in active fractions. J. Agric. Food Chem. 57, 7543–7551. doi: 10.1021/jf901074b
Qiu, Y., Liu, Q., Beta, T. (2010). Antioxidant properties of commercial wild rice and analysis of soluble and insoluble phenolic acids. Food Chem. 121, 140–147. doi: 10.1016/j.foodchem.2009.12.021
Ray, A., Chakraborty, D. (2018). Shattering or not shattering: that is the question in domestication of rice (Oryza sativa l.). Genet. Resour. Crop Evol. 65, 391–395. doi: 10.1007/s10722-017-0586-1
Ren, Z. H., Gao, J. P., Li, L. G., Cai, X. L., Huang, W., Chao, D. Y., et al. (2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37, 1141–1146. doi: 10.1038/ng1643
Richards, C. M., Reilley, A., Touchell, D., Antolin, M. F., Walters, C. (2004). Microsatellite primers for Texas wild rice (Zizania texana), and a preliminary test of the impact of cryogenic storage on allele frequency at these loci. Conserv. Genet. 5, 853–859. doi: 10.1007/s10592-004-1977-3
Shan, X. H., Liu, Z. L., Dong, Z. Y., Wang, Y. M., Chen, Y., Lin, X. Y., et al. (2005). Mobilization of the active MITE transposons mPing and Pong in rice by introgression from wild rice (Zizania latifolia griseb.). Mol. Biol. Evol. 22, 976–990. doi: 10.1093/molbev/msi082
Shao, M. Q., Haas, M., Kern, A., Kimball, J. (2020). Identification of single nucleotide polymorphism markers for population genetic studies in Zizania palustris l. Conserv. Genet. Resour. 12, 451–455. doi: 10.1007/s12686-019-01116-9
Shao, Y. F., Xu, F. F., Sun, X., Bao, J. S., Beta, T. (2014). Phenolic acids, anthocyanins, and antioxidant capacity in rice (Oryza sativa l.) grains at four stages of development after flowering. Food Chem. 143, 90–96. doi: 10.1016/j.foodchem.2013.07.042
Shen, W. W., Song, C. L., Chen, J., Fu, Y. P., Wu, J. L., Jiang, S. M. (2011). Transgenic rice plants harboring genomic DNA from Zizania latifolia confer bacterial blight resistance. Rice Sci. 18, 17–22. doi: 10.1016/S1672-6308(11)60003-6
Singh, S., Parihar, P., Singh, R., Singh, V. P., Prasad, S. M. (2016). Heavy metal tolerance in plants: role of transcriptomics, proteomics, metabolomics, and ionomics. Front. Plant Sci. 6, 1143. doi: 10.3389/fpls.2015.01143
Small, E. (2012). Blossoming treasures of biodiversity. Biodiversity 13, 54–64. doi: 10.1080/14888386.2010.9712667
Steeves, T. A. (1952). Wild rice–Indian food and a modern delicacy. Econ. Bot. 6, 107–142. doi: 10.1007/BF02984871
Surendiran, G., Alsaif, M., Kapourchali, F. R., Moghadasian, M. H. (2014). Nutritional constituents and health benefits of wild rice (Zizania spp.). Nutr. Rev. 72, 227–236. doi: 10.1111/nure.12101
Tang, J. Y., Cao, P. P., Chi, X. U., Liu, M. S. (2013). Effects of aquatic plants during their decay and decomposition on water quality. Chin. J. Appl. Ecol. 24, 83–89.
Timm, D. A., Slavin, J. L. (2014). Wild rice: both an ancient grain and a whole grain. Cereal Chem. 91, 207–210. doi: 10.1094/Cchem-08-13-0161-Rw
Tolley-Jordan, L. R., Power, P. (2007). Effects of water temperature on growth of the federally endangered Texas wild rice (Zizania texana). Southwest Nat. 52, 201–208. doi: 10.1894/0038-4909(2007)52[201:Eowtog]2.0.Co;2
Tu, Z. H., Yamada, S., Hu, D., Ito, Y., Iwasaki, T., Yamaguchi, A. (2019). Microbial diversity in the edible gall on white bamboo formed by the interaction between Ustilago esculenta and Zizania latifolia. Curr. Microbiol. 76, 824–834. doi: 10.1007/s00284-019-01693-w
Vaughan, D. A., Balazs, E., Heslop-Harrison, J. S. (2007). From crop domestication to super-domestication. Ann. Bot. 100, 893–901. doi: 10.1093/aob/mcm224
Walker, R. D., Pastor, J., Dewey, B. W. (2006). Effects of wild rice (Zizania palustris) straw on biomass and seed production in northern Minnesota. Can. J. Bot. 84, 1019–1024. doi: 10.1139/B06-058
Wang, Y. M., Dong, Z. Y., Zhang, Z. J., Lin, X. Y., Shen, Y., Zhou, D. W., et al. (2005). Extensive de novo genomic variation in rice induced by introgression from wild rice (Zizania latifolia griseb.). Genet. 170, 1945–1956. doi: 10.1534/genetics.105.040964
Wang, Z. H., Yan, N., Luo, X., Guo, S. S., Xue, S. Q., Liu, J. Q., et al. (2020). Gene expression in the smut fungus Ustilago esculenta governs swollen gall metamorphosis in Zizania latifolia. Microb. Pathog. 143, e104107. doi: 10.1016/j.micpath.2020.104107
Wang, Z. D., Yan, N., Wang, Z. H., Zhang, X. H., Zhang, J. Z., Xue, H. M., et al. (2017). RNA-Seq analysis provides insight into reprogramming of culm development in Zizania latifolia induced by Ustilago esculenta. Plant Mol. Biol. 95, 533–547. doi: 10.1007/s11103-017-0658-9
Wang, Z. H., Zhang, D., Bai, Y., Zhang, Y. H., Liu, Y., Wu, Y., et al. (2013). Genomewide variation in an introgression line of rice-Zizania revealed by whole-genome re-sequencing. PloS One 8, e74479. doi: 10.1371/journal.pone.0074479
Ward, J. L., Poutanen, K., Gebruers, K., Piironen, V., Lampi, A. M., Nystro?M, L., et al. (2008). The healthgrain cereal diversity screen: concept, results, and prospects. J. Agric. Food Chem. 56, 9699–9709. doi: 10.1021/jf8009574
Wink, M. (1988). Plant breeding: importance of plant secondary metabolites for protection against pathogens and herbivores. Theor. Appl. Genet. 75, 225–233. doi: 10.1007/bf00303957
Wiser, E. (1975). Protein and lysine contents in grains of three species of wild-rice (Zizania; gramineae). Bot. Gaz. 136, 312–316. doi: 10.2307/2473618
Wu, L. M., Han, L. Q., Li, Q., Wang, G. Y., Zhang, H. W., Li, L. (2021). Using interactome big data to crack genetic mysteries and enhance future crop breeding. Mol. Plant 14, 77–94. doi: 10.1016/j.molp.2020.12.012
Xie, Y. N., Yang, T., Zhang, B. T., Qi, Q. Q., Ding, A. M., Shang, L. G., et al. (2022). Systematic analysis of bell family genes in Zizania latifolia and functional identification of ZlqSH1a/b in rice seed shattering. Int. J. Mol. Sci. 23, 15939. doi: 10.3390/ijms232415939
Xu, X. W., Ke, W. D., Yu, X. P., Wen, J., Ge, S. (2008). A preliminary study on population genetic structure and phylogeography of the wild and cultivated Zizania latifolia (Poaceae) based on Adh1a sequences. Theor. Appl. Genet. 116, 835–843. doi: 10.1007/s00122-008-0717-3
Xu, X. W., Walters, C., Antolin, M. F., Alexander, M. L., Lutz, S., Ge, S., et al. (2010). Phylogeny and biogeography of the eastern Asian–north American disjunct wild-rice genus (Zizania l., poaceae). Mol. Phylogenet. Evol. 55, 1008–1017. doi: 10.1016/j.ympev.2009.11.018
Yan, N., Du, Y., Liu, X., Chu, C., Shi, J., Zhang, H., et al. (2018). Morphological characteristics, nutrients, and bioactive compounds of Zizania latifolia, and health benefits of its seeds. Molecules 23, 1561. doi: 10.3390/molecules23071561
Yan, N., Du, Y. M., Liu, X. M., Chu, M. J., Shi, J., Zhang, H. B., et al. (2019). A comparative UHPLC-QqQ-MS-based metabolomics approach for evaluating Chinese and north American wild rice. Food Chem. 275, 618–627. doi: 10.1016/j.foodchem.2018.09.153
Yan, H. X., Ma, L., Wang, Z., Lin, Z. M., Su, J., Lu, B. R. (2015). Multiple tissue-specific expression of rice seed-shattering gene SH4 regulated by its promoter pSH4. Rice 8, 1–10. doi: 10.1186/s12284-015-0047-4
Yan, N., Wang, X. Q., Xu, X. F., Guo, D. P., Wang, Z. D., Zhang, J. Z., et al. (2013a). Plant growth and photosynthetic performance of Zizania latifolia are altered by endophytic Ustilago esculenta infection. Physiol. Mol. Plant Pathol. 83, 75–83. doi: 10.1016/j.pmpp.2013.05.005
Yan, N., Xu, X. F., Wang, Z. D., Huang, J. Z., Guo, D. P. (2013b). Interactive effects of temperature and light intensity on photosynthesis and antioxidant enzyme activity in Zizania latifolia turcz. plants. Photosynthetica 51, 127–138. doi: 10.1007/s11099-013-0009-2
Yan, N., Yang, T., Yu, X. T., Shang, L. G., Guo, D. P., Zhang, Y., et al. (2022). Chromosome-level genome assembly of Zizania latifolia provides insights into its seed shattering and phytocassane biosynthesis. Commun. Biol. 5, 36. doi: 10.1038/s42003-021-02993-3
Ye, Z. H., Cui, H. F., An, X. X., Yu, X. P. (2016). Chilling tolerance in Zizania latifolia. Hortic. Bras. 34, 39–45. doi: 10.1590/S0102-053620160000100006
You, W. Y., Liu, Q., Zou, K. Q., Yu, X. P., Cui, H. F., Ye, Z. H. (2011). Morphological and molecular differences in two strains of Ustilago esculenta. Curr. Microbiol. 62, 44–54. doi: 10.1007/s00284-010-9673-7
Yu, X. T., Chu, M., Chu, C., Du, Y., Shi, J., Liu, X., et al. (2020). Wild rice (Zizania spp.): a review of its nutritional constituents, phytochemicals, antioxidant activities, and health-promoting effects. Food Chem. 331, 127293. doi: 10.1016/j.foodchem.2020.127293
Yu, H., Li, J. Y. (2022). Breeding future crops to feed the world through de novo domestication. Nat. Commun. 13, 1171. doi: 10.1038/s41467-022-28732-8
Yu, X. T., Qi, Q. Q., Li, Y. L., Li, N. A., Xie, Y. N., Ding, A. M., et al. (2022). Metabolomics and proteomics reveal the molecular basis of colour formation in the pericarp of Chinese wild rice (Zizania latifolia). Food Res. Int. 162, 112082. doi: 10.1016/j.foodres.2022.112082
Yu, X. T., Yang, T., Qi, Q. Q., Du, Y. M., Shi, J., Liu, X. M., et al. (2021). Comparison of the contents of phenolic compounds including flavonoids and antioxidant activity of rice (Oryza sativa) and Chinese wild rice (Zizania latifolia). Food Chem. 344, 128600. doi: 10.1016/j.foodchem.2020.128600
Zaitchik, B. F., Leroux, L. G., Kellogg, E. A. (2000). Development of male flowers in Zizania aquatica (North American wild-rice; gramineae). Int. J. Plant Sci. 161, 345–351. doi: 10.1086/314268
Zeng, D. L., Tian, Z. X., Rao, Y. C., Dong, G. J., Yang, Y. L., Huang, L. C., et al. (2017). Rational design of high-yield and superior-quality rice. Nat. Plants 3, 17031. doi: 10.1038/nplants.2017.31
Zhai, C. K., Lu, C. M., Zhang, X. Q., Sun, G. J., Lorenz, K. J. (2001). Comparative study on nutritional value of Chinese and north American wild rice. J. Food Compos. Anal. 14, 371–382. doi: 10.1006/jfca.2000.0979
Zhai, C., Zhang, X., Sun, G., Lu, C. (2000). Study on nutrition composition and protein quality of a Chinese wild rice. Wei Sheng Yen Chiu J. Hyg. Res. 29, 375–378.
Zhang, S., Huang, G., Zhang, Y., Lv, X., Wan, K., Liang, J., et al. (2022). Sustained productivity and agronomic potential of perennial rice. Nat. Sustain. 6, 28–38. doi: 10.1038/s41893-022-00997-3
Zhang, D., Li, K., Gao, J., Liu, Y., Gao, L. Z. (2016). The complete plastid genome sequence of the wild rice Zizania latifolia and comparative chloroplast genomics of the rice tribe oryzeae, poaceae. Front. Ecol. Evol. 4. doi: 10.3389/fevo.2016.00088
Zhang, Z. P., Song, S. X., Liu, Y. C., Zhu, X. R., Jiang, Y. F., Shi, L. T., et al. (2021). Mixed transcriptome analysis revealed the possible interaction mechanisms between Zizania latifolia and Ustilago esculenta inducing jiaobai stem-gall formation. Int. J. Mol. Sci. 22, 12258. doi: 10.3390/ijms222212258
Zhang, L. B., Zhu, Q. H., Wu, Z. Q., Ross-Ibarra, J., Gaut, B., Ge, S., et al. (2009). Selection on grain shattering genes and rates of rice domestication. New Phytol. 184, 708–720. doi: 10.1111/j.1469-8137.2009.02984.x
Zhao, Y., Song, Z., Zhong, L., Li, Q., Chen, J., Rong, J. (2019). Inferring the origin of cultivated Zizania latifolia, an aquatic vegetable of a plant-fungus complex in the Yangtze river basin. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01406
Zhao, Y., Zhong, L., Zhou, K., Song, Z. P., Chen, J. K., Rong, J. (2018). Seed characteristic variations and genetic structure of wild Zizania latifolia along a latitudinal gradient in China: implications for neo-domestication as a grain crop. Aob Plants 10, ply072. doi: 10.1093/aobpla/ply072
Keywords: wild rice (Zizania spp.), edible history, economic value, domestication process, breeding objectives, omics research, important genes
Citation: Xie Y-N, Qi Q-Q, Li W-H, Li Y-L, Zhang Y, Wang H-M, Zhang Y-F, Ye Z-H, Guo D-P, Qian Q, Zhang Z-F and Yan N (2023) Domestication, breeding, omics research, and important genes of Zizania latifolia and Zizania palustris. Front. Plant Sci. 14:1183739. doi: 10.3389/fpls.2023.1183739
Received: 10 March 2023; Accepted: 17 May 2023;
Published: 31 May 2023.
Edited by:
Hong An, University of Missouri, United StatesReviewed by:
Xiang Li, University of Massachusetts Amherst, United StatesTing Xiang Tina Neik, University of Nottingham Malaysia Campus, Malaysia
Copyright © 2023 Xie, Qi, Li, Li, Zhang, Wang, Zhang, Ye, Guo, Qian, Zhang and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhong-Feng Zhang, emhhbmd6aG9uZ2ZlbmdAY2Fhcy5jbg==; Ning Yan, eWFubmluZ0BjYWFzLmNu
†These authors have contributed equally to this work and share first authorship
 Yan-Ning Xie
Yan-Ning Xie Qian-Qian Qi1†
Qian-Qian Qi1† Zi-Hong Ye
Zi-Hong Ye De-Ping Guo
De-Ping Guo Qian Qian
Qian Qian Zhong-Feng Zhang
Zhong-Feng Zhang Ning Yan
Ning Yan