- 1Department of Genetics and Genome Biology, Adrian Building, University of Leicester, Leicester, United Kingdom
- 2Genetics, Diversity and Ecophysiology of Cereals, Unité Mixte de Recherche (UMR) 1095, The Institut National de la Recherche Agronomique (INRAE), Université Clermont Auvergne, Clermont-Ferrand, France
During meiosis, the chromosome axes and synaptonemal complex mediate chromosome pairing and homologous recombination to maintain genomic stability and accurate chromosome segregation. In plants, ASYNAPSIS 1 (ASY1) is a key component of the chromosome axis that promotes inter-homolog recombination, synapsis and crossover formation. Here, the function of ASY1 has been cytologically characterized in a series of hypomorphic wheat mutants. In tetraploid wheat, asy1 hypomorphic mutants experience a reduction in chiasmata (crossovers) in a dosage-specific manner, resulting in failure to maintain crossover (CO) assurance. In mutants with only one functional copy of ASY1, distal chiasmata are maintained at the expense of proximal and interstitial chiasmata, indicating that ASY1 is required to promote chiasma formation away from the chromosome ends. Meiotic prophase I progression is delayed in asy1 hypomorphic mutants and is arrested in asy1 null mutants. In both tetraploid and hexaploid wheat, single asy1 mutants exhibit a high degree of ectopic recombination between multiple chromosomes at metaphase I. To explore the nature of the ectopic recombination, Triticum turgidum asy1b-2 was crossed with wheat-wild relative Aegilops variabilis. Homoeologous chiasmata increased 3.75-fold in Ttasy1b-2/Ae. variabilis compared to wild type/Ae. variabilis, indicating that ASY1 suppresses chiasma formation between divergent, but related chromosomes. These data suggest that ASY1 promotes recombination along the chromosome arms of homologous chromosomes whilst suppressing recombination between non-homologous chromosomes. Therefore, asy1 mutants could be utilized to increase recombination between wheat wild relatives and elite varieties for expediting introgression of important agronomic traits.
Introduction
The majority of sexually reproducing eukaryotes undergo meiosis, a specialized cell division required to produce haploid gametes from diploid progenitor cells. Meiosis is characterized by the homologous recombination of genetic material between chromosomes that is necessary to ensure accurate chromosome segregation as well as create new combinations of alleles. In wheat, meiotic recombination is initiated by ~2,000 programmed DNA double-strand breaks (DSBs) (Gardiner et al., 2019), catalyzed by SPO11-1/SPO11-2 (Benyahya et al., 2020; Da Ines et al., 2020; Hyde et al., 2022). DSBs are repaired as crossovers (COs) when a reciprocal exchange of DNA takes place between homologous chromosomes (that are cytologically detected as chiasmata), or non-crossovers (NCOs) when DSBs are repaired by non-reciprocal exchange of DNA, via either the sister chromatid or homologous chromosome as a template. In plants, ~85% of COs form via the class I pathway that ensures every chromosome pair receives at least one “obligate CO” so that homologous chromosomes are tethered together at metaphase I and accurately segregate during meiosis II (Higgins et al., 2004; Higgins et al., 2008b; Osman et al., 2011). Class I COs are sensitive to interference and therefore more likely to be spaced apart than by random chance (Jones and Franklin, 2006). The class II pathway accounts for ~15% of COs and is insensitive to interference (Berchowitz et al., 2007; Higgins et al., 2008a; Lambing et al., 2017; Wang and Copenhaver, 2018; Desjardins S. D. et al., 2020). In wheat, the FANCM helicase promotes class I COs as well as suppressing class II CO formation, suggesting that the two CO pathways are intimately linked (Desjardins et al., 2022).
In plants, inter-homolog recombination and the obligate CO are promoted by the synaptonemal complex (SC), which also imposes CO interference (Higgins et al., 2005; Sanchez-Moran et al., 2007; Sanchez-Moran et al., 2008; Ferdous et al., 2012; Chambon et al., 2018; Capilla-Perez et al., 2021; France et al., 2021). The SC is an evolutionary conserved tripartite proteinaceous structure that assembles and disassembles during meiotic prophase I (Page and Hawley, 2004; Hughes and Hawley, 2020). The SC is composed of two chromosome axes that mature into lateral elements upon installation of the transverse filament proteins (Page and Hawley, 2004; Gao and Colaiacovo, 2018). The core components of the chromosome axes are ASYNAPSIS 1 (ASY1)/PAIR2, ASY3/PAIR3, and ASY4 (Armstrong et al., 2002; Nonomura et al., 2004; Yuan et al., 2009; Ferdous et al., 2012; Chambon et al., 2018) as well as the transverse filament proteins ZYP1/ZEP1 (Higgins et al., 2005; Wang et al., 2010; Barakate et al., 2014). ASY1 possesses a conserved HORMA domain that is predicted to bind to chromatin along with its interacting partners p31COMET, ASY3, and ASY4 (Caryl et al., 2000; Armstrong et al., 2002; Sanchez-Moran et al., 2007; Ferdous et al., 2012; Chambon et al., 2018; Balboni et al., 2020). ASY1 also acts as a gene dosage-dependent antagonist of telomere-led recombination in Arabidopsis, thereby promoting interfering COs (Lambing et al., 2020), although ASY1 immunoprecipitation experiments in wheat suggest that the protein is more abundant toward the chromosome ends (Tock et al., 2021).
Wheat is an allopolyploid crop in which COs predominantly form toward the chromosome ends (Saintenac et al., 2009; Osman et al., 2021; Higgins et al., 2022). It has evolved a meiotic program in which homoeologous chromosomes rarely recombine due to the Pairing homoeologous (Ph) 1 and 2 loci (Riley and Chapman, 1958; Mello-Sampayo, 1971). TaZIP4-B2 gene in the Ph1 locus is required for both promotion of homologous COs and restriction of homoeologous COs in wheat/Aegilops variabilis hybrids (Rey et al., 2017; Rey et al., 2018), while TaMSH7-3D in the Ph2 locus is necessary for recombination partner selection (homologous vs. homoeologous) by likely increasing the instability of homoeologous recombination in wheat/Ae. variabilis hybrids (Serra et al., 2021). In addition, reduced expression of ASY1 by RNAi in hexaploid wheat generated high levels of multiple chromosome configurations at metaphase I, implying loss of CO control and elevated homoeologous recombination (Boden et al., 2009).
Here, cytological analysis of hypomorphic wheat asy1 TILLING (Targeting Induced Local Lesions In Genomes) mutants has revealed a delay in meiotic progression, loss of the obligate chiasma, and ectopic recombination between multiple chromosomes. Tetraploid wheat asy1 mutants crossed with wheat-wild relative Ae. variabilis exhibit an increase in chiasma formation, indicating that ASY1 is dosage-sensitive for promoting accurate homologous recombination while suppressing non-homologous recombination during meiosis.
Experimental procedures
Plant material and greenhouse conditions
Triticum turgidum ‘Kronos’ and Triticum aestivum ‘Cadenza’ were used as wild-type controls for experiments involving TILLING mutant lines received from www.SeedStor.ac.uk. The Ensembl Plants database (http://plants.ensembl.org) was used to identify ASY1 genes: TtASY1-5A, TRITD5Av1G167820; TtASY1-5B, TRITD5Bv1G159710; TaASY1-5A, TraesCS5A02G286500; TaASY1-5B, TraesCS5B02G285800; and TaASY1-5D, TraesCS5D02G294100. TILLING mutants were screened by BLAST search on the Wheat TILLING database (http://www.wheat-tilling.com/): Ttasy1a, K0706; Ttasy1b-1, K0157; Ttasy1b-2, K2071 (Krasileva et al., 2017); and Taasy1b, C0971 (Appels et al., 2018). To create hypomorphic mutants, homozygous lines were crossed (K0706 Ttasy1a × K0157 Ttasy1b-1 and K0706 Ttasy1a × K2071 Ttasy1b-2), while heterozygous individuals from the F1’s (AaBb) were self-pollinated to create F2’s. Wild-type Kronos and the Ttasy1b-2 mutant line (K2071) were crossed with Ae. variabilis (accession no. 26248, https://www6.clermont.inrae.fr/umr1095_eng/Organisation/Experimental-Infrastructure/Biological-Resources-Centre; UUSS, 2n = 4x = 28) to produce Kronos/Ae. variabilis haploid hybrids (ABUS, n = 28). Briefly, Kronos inflorescences were emasculated and pollinated with fresh Ae. variabilis pollen. Inflorescences were then bagged to avoid cross-pollination, and seeds were collected when mature. Plants were grown in soil-based compost (Levington Advance Pot and Bedding M1 Compost) under greenhouse conditions with a photoperiod of 16-h days light cycle at a constant temperature of 22°C (day)/16°C (night) and relative humidity ~60%.
Validating SNP mutations
To validate that the point mutations induced by ethyl methanesulfonate treatment would be transcribed into mRNA, total RNA was extracted from tetraploid wheat T. turgidum ‘Kronos’ and hexaploid T. aestivum ‘Cadenza’ inflorescences using the ISOLATE II RNA Mini Kit (https://www.bioline.com/). cDNA was synthesized using the Tetro cDNA Synthesis Kit (https://www.bioline.com/), followed by PCR with Q5® High-Fidelity DNA Proofreading Polymerase (https://www.neb.uk.com/) with primers TaASY1cDNAF and TaASY1cDNAR (Supplementary Table 1). PCR amplicons were ligated into pDrive (https://www.qiagen.com/) and Sanger sequenced (https://eurofinsgenomics.eu/). Following validation, single-nucleotide polymorphism (SNP)-specific primers were designed to amplify individual TILLING lines for genotyping optimized by gradient PCR.
Cytological procedures
Anther sizes were measured with a Nikon SMZ 745 dissecting microscope and 10 mm/0.1 mm graticule. Chromosome spreads were stained with DAPI and examined by light microscopy as previously described (Higgins, 2013; Desjardins S. et al., 2020). Nikon Ni-E and Eclipse Ci fluorescence microscopes equipped with NIS elements software were used to image chromosomes. The following primary antibodies were used for immunolocalization: anti-TaASY1 guinea pig, 1:500 (Desjardins S. D. et al., 2020); and anti-AtZYP1 rabbit 1:500 (Osman et al., 2018). Secondary antibodies: goat anti-guinea pig Alexa Fluor 488 (https://www.abcam.com/) and goat anti-rabbit DyLight 594 (https://www.2bscientific.com/) were used at 1:200. Chiasma counts were performed using NIS software, and significance (p adj < 0.01) was established using pairwise Wilcoxon rank sum tests adjusted with Bonferroni correction method (RStudio v1.2.5033). The karyology of Kronos/Ae. variabilis hybrids was checked by aceto-carmine chromosome spreads as previously described (Serra et al., 2021).
Statistical analysis
A chi-square test for analysis of meiotic progression in asy1 hypomorphic mutants was performed to test the association between meiotic prophase I stages and anther lengths, and a significant p-value was set less than 0.05. A statistical analysis of seed counts per plant comparing the primary inflorescence was performed on Minitab 20 with a t-test two-sample distribution.
Results
Identification of wheat ASY1
ASY1 is a component of the meiotic chromosome axis that is highly expressed in anthers during prophase I of meiosis (Boden et al., 2009; Alabdullah et al., 2019; Tock et al., 2021; Jiang et al., 2023). The wheat ASY1 coding sequences were cloned and sequenced from tetraploid ‘Kronos’ and hexaploid ‘Cadenza’ cDNA (Supplementary Figure 1). A wheat consensus ASY1 protein sequence derived from the clones shares a high level of sequence similarity to PAIR2 in Oryza sativa (80%), ASY1 in Arabidopsis thaliana (54%), and ASY1 in Brassica oleracea (51%). The polyploid wheat ASY1 homoeologous sequences share >96% nucleotide identity and >94% amino acid identity (Supplementary Table 2). ASY1 is located on the long arm of chromosome 5 in tetraploid wheat and hexaploid wheat. A Phyre2 structural analysis (Kelley et al., 2015) predicts that the wheat ASY1 proteins contain a conserved N-terminal HORMA domain (100% prediction at residues 6–236 for 5A and 3–231 for 5B and 5D) and a winged helix DNA binding domain (96% prediction at residues 339–457) (Figure 1 and Supplementary Table 3). ASY1-5A and ASY1-5B are predicted to contain a Set3 PhD finger H3K4me3 domain (85%–91% prediction for ASY1-5A and 21%–42% ASY1-5B at residues 317–400) but not detected in ASY1-5D (Supplementary Table 3). The predicted domains and immunoprecipitation experiments (Tock et al., 2021) indicate that ASY1 binds to DNA and chromatin at the chromosome axis during wheat meiosis.
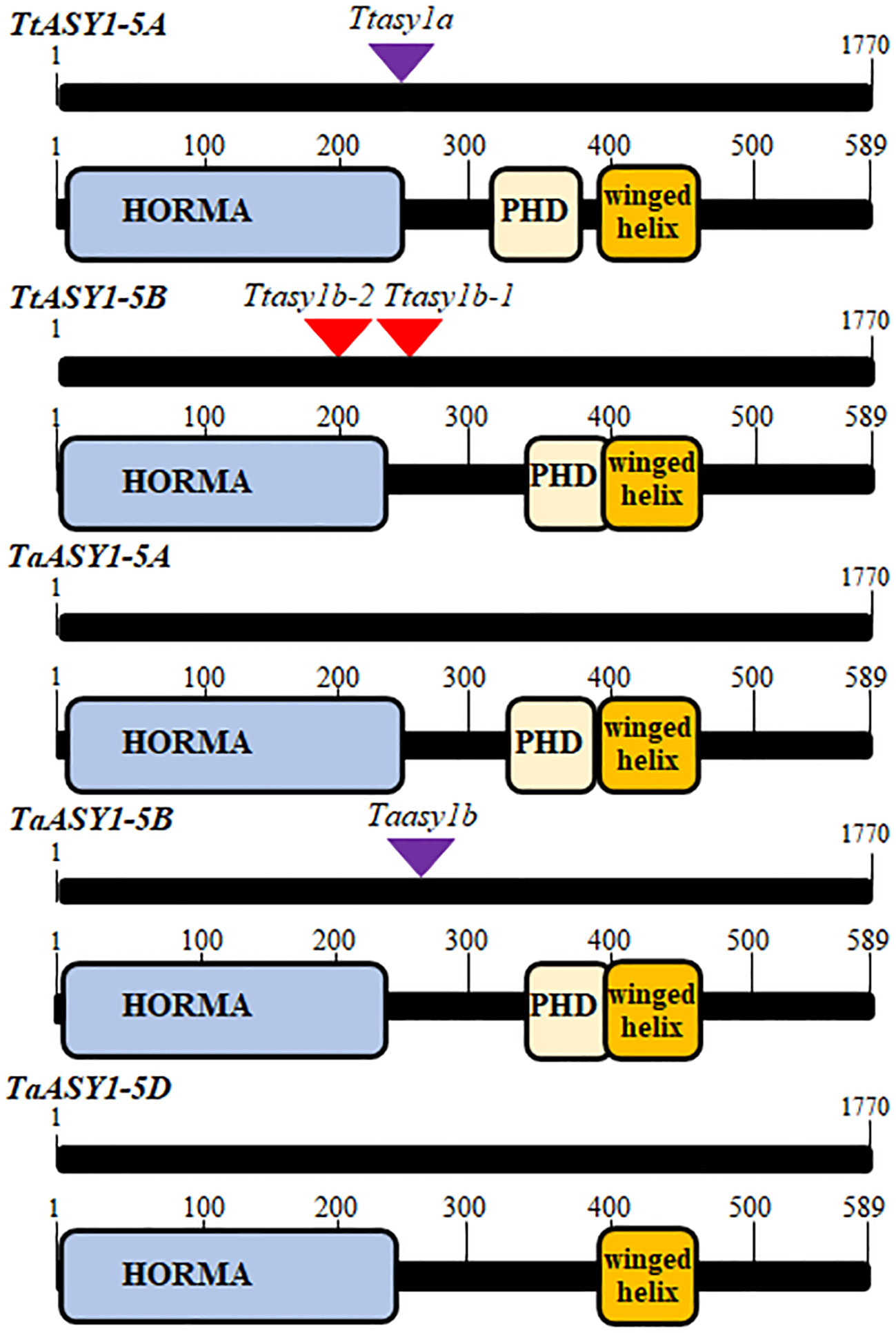
Figure 1 Schematic representation of wheat ASY1 coding regions and altered proteins from TILLING mutations. The coding regions of ASY1 and the Phyre2 predicted protein domains are shown relative to the TILLING mutations. The purple triangle represents a mutated splice donor site that retains an intron, whereas the red triangles represent a stop codon.
Wheat asy1 TILLING mutants
T. turgidum ‘Kronos’ and T. aestivum ‘Cadenza’ asy1 mutants were identified in the wheat TILLING populations (Krasileva et al., 2017; Appels et al., 2018). Two Kronos lines possessing a premature STOP codon and one line containing a mutation at a splice donor site that retained an intron and subsequent STOP codon were sequenced and verified. The mutations are predicted to truncate and create non-functional ASY1 proteins (Ttasy1b-1, 785 C > T, Q 307 > STOP; Ttasy1b-2, 148 G > A, W 156 > STOP; Ttasy1a, 714 G > A, V 231 > STOP) (Figure 1 and Supplementary Figures 2–4). In Cadenza, only one asy1 mutant on chromosome 5B was identified, and this is predicted to disrupt the protein function due to a splice donor site mutation and intron retention that led to a STOP codon ((Taasy1b (C0971), 1195 G > A, P 254 > STOP)) (Figure 1 and Supplementary Figure 5). The asy1 transcripts were sequenced from the TILLING lines to confirm that the mutations in the genomic DNA led to stop codons in the coding sequences (Supplementary Figures 2–5). As tetraploid Kronos contains four ASY1 copies, a phenotypic analysis could be performed on hypomorphic mutants: Ttasy1a (aaBB), Ttasy1b-1 (AAbb), Ttasy1b-2 (AAbb), Ttasy1Ab (Aabb), Ttasy1aB (aabB), Ttasy1_1 (aabb), and Cadenza Taasy1b (AAbbDD). Seed-set per plant significantly decreased from 22 ± 3 SD per plant in wild-type Kronos (n = 10) to 16 ± 4 SD in Ttasy1a (n = 10), 15 ± 3 SD (n = 10) in Ttasy1a/b, 8 ± 1 SD in Ttasy1Ab/aB (n = 10), and 0 in Ttasy1_1 (n = 10) as well as from 41 ± 1 SD in wild-type Cadenza (n = 10) to 35 ± 3 SD (n = 10) in Taasy1b (p < 0.001 Mann–Whitney) (Supplementary Table 4). Since fertility is affected and ASY1 is a known meiosis gene, this suggests that meiosis may be disturbed in the mutants leading to infertile gametes. We therefore analyzed the meiotic behavior of these mutants.
Meiotic progression is delayed in asy1 hypomorphic mutants
Meiotic stages in wheat are relatively synchronous and correlate with anther length (Shunmugam et al., 2018). Immunolocalization of ASY1 and ZYP1 was performed on pollen mother cells from wild type and asy1 hypomorphic mutants to determine if meiotic prophase I progression was affected (Figure 2). In the wild type, ASY1 forms linear stretches along the chromosome axes at leptotene, and ZYP1 forms axis-associated foci in anthers 0.7 mm in length (85% nuclei, n = 240) (Figure 2 and Supplementary Figure 6) as previously reported (Sepsi et al., 2017; Osman et al., 2021). At zygotene, ASY1 becomes depleted along the chromosome axes concomitant with ZYP1 polymerization in anthers 0.8 mm in length (90% nuclei, n = 240) (Figure 2 and Supplementary Figure 6) until pachytene when ASY1 is present as a weak, diffuse signal in anthers 0.9 mm in length (90% nuclei, n = 240) (Figure 2 and Supplementary Figure 6). In the Ttasy1a and Ttasy1b mutants, the ASY1 signal appears indistinguishable compared to the wild type at leptotene, although protein quantities were not determined (Figure 2). However, Ttasy1a and Ttasy1b leptotene nuclei were only observed in 0.8-mm anthers (93%, n = 720, χ2 test, p < 0.05, Supplementary Table 5), compared to 0.7 mm in wild type (85%, n = 240, χ2 test, p < 0.05, Supplementary Table 5), suggesting that prophase I progression was delayed. In the wild type, ZYP1 localized as foci or short stretches in 0.7-mm anthers, but equivalent stages were only observed in anthers 0.8 mm in length in Ttasy1a and Ttasy1b (Figure S6). In the minimum ASY1 dose mutants (Ttasy1aB and Ttasy1Ab), leptotene stages were detected in anthers 0.9 mm (95%, n = 480, χ2 test, p < 0.05, Figure S6), indicating a greater delay than the single Ttasy1a and Ttasy1b mutants (Figure 2). Short stretches of ZYP1 were detected in 1.1-mm anthers (n = 480), and in 50% of cells, ZYP1 failed to polymerize, instead forming polycomplexes (Figure 2). Neither ASY1 nor ZYP1 was detected on meiotic chromosomes in 0.7–1.1-mm anthers in Ttasy1_1 (n = 480), suggesting that it was a null asynaptic mutant.
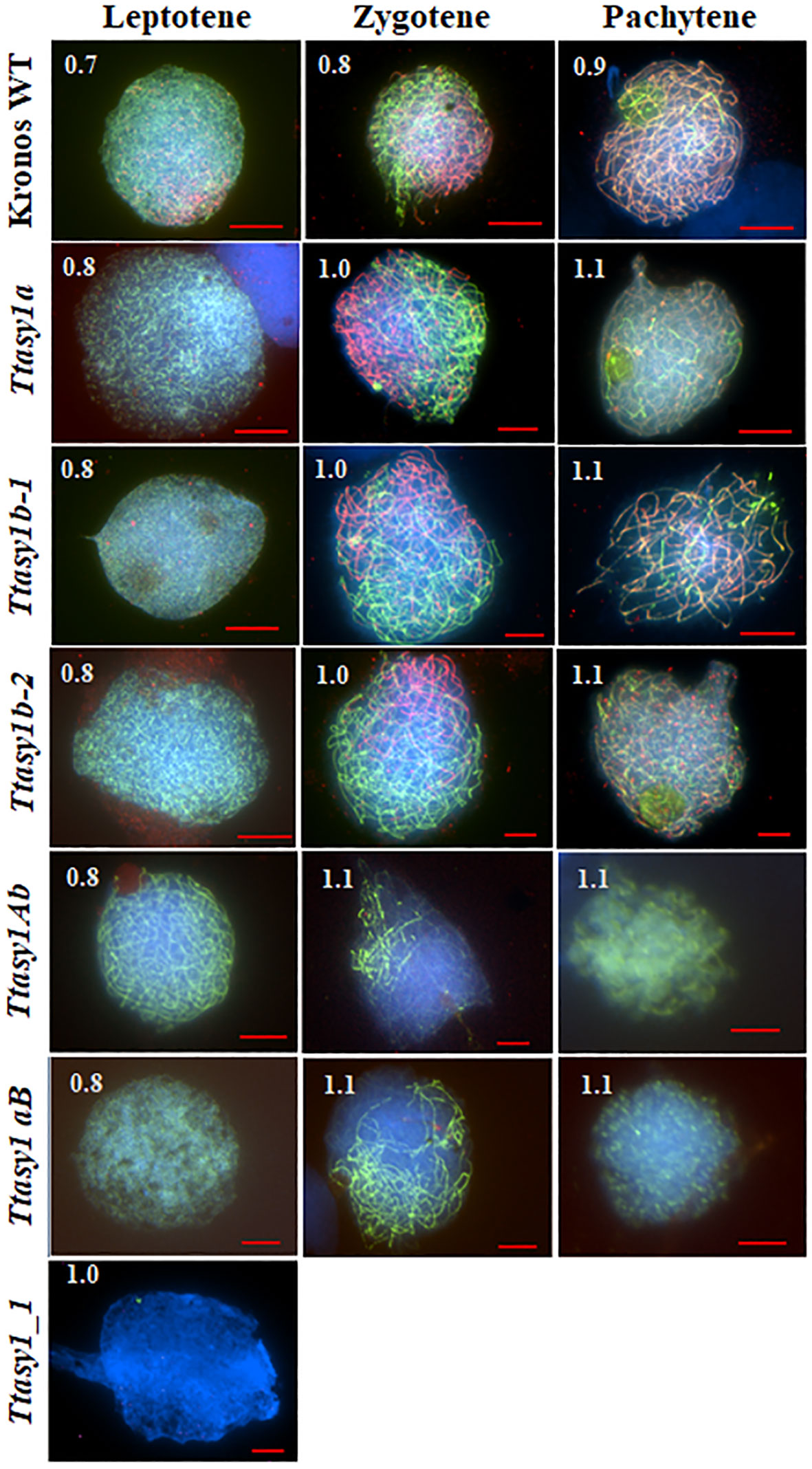
Figure 2 Meiotic prophase I progression in asy1 mutants. Chromosome axes were marked with ASY1 (green), the synaptonemal complex was marked with ZYP1 (red), and chromosomal DNA was counterstained with DAPI (blue). Anther lengths (mm) were measured for each genotype and prophase 1 stage as shown in the top left corner for each image. Scale bar = 10 µm (Kronos wild type, Ttasy1Ab, and Ttasy1aB) and 20 µm (Ttasy1a, Ttasy1b-2, and Ttasy1_1).
ASY1 labeling at leptotene in Cadenza Taasy1b appeared indistinguishable from the wild type, although protein levels were not determined. The A and D copies are expected to be fully functional, although the Set3 PhD finger H3K4me3 domain was not detected in TaASY1-5D, and this could have a detrimental effect (Figure S6). At zygotene, ZYP1 installation in Taasy1b occurred as in the wild type, but polymerization was discontinuous and temporally compromised (1.0 mm in 65% nuclei, n = 240 versus 0.8 mm in 85% wild-type nuclei, n = 240), indicating that a reduced dose of ASY1 delayed meiotic progression in hexaploid wheat (Figure S6).
Correct dosage of ASY1 is required for crossover assurance
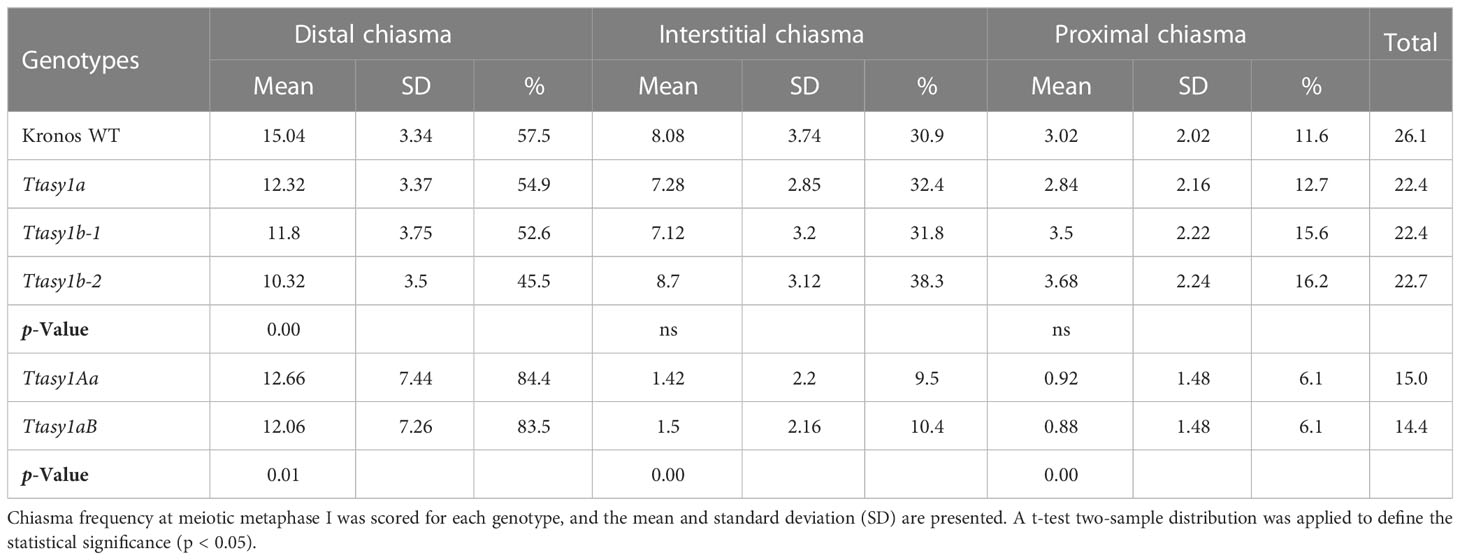
A cytological analysis was performed on the wild type and asy1 Kronos mutants with DAPI-stained metaphase I chromosome spreads. Chiasmata ranged from 21 to 30 per nucleus in wild-type Kronos, with a mean of 26 ± 2.2 (n = 50), and each of the 14 pairs of chromosomes received at least one chiasma (Figure 3A). In Ttasy1a, chiasmata ranged from 16 to 27 per nucleus with a significantly lower mean (22 ± 2.8, n = 50) (pairwise Wilcoxon rank sum test, p < 0.01) (Table 1) compared to the wild type. Similarly, Ttasy1b-1 (n = 50) and Ttasy1b-2 (n = 50) exhibited a mean of 22 ± 2.8 and 23 ± 2.4, respectively (Figures 3A, 4A, 4B and Table 1). Chiasma frequency for the Ttasy1a/b lines was not significantly different from each other (pairwise Wilcoxon rank sum test, p adj > 0.05, Ttasy1a, n = 50; Ttasy1b-1, n = 50; Ttasy1b-2, n = 50), indicating that both A and B sub-genomes provide a similar, non-redundant contribution of ASY1 (Supplementary Tables 6–12). Chiasmata were significantly reduced in Ttasy1Ab (15.0, n = 50) and Ttasy1aB (14.4, n = 50), and no chiasmata were observed in the null mutant Ttasy1_1 (n = 50) (Figure 3A). Similarly, chiasmata were significantly reduced in hexaploid wheat from 39 ± 1.6 (n = 50) in wild type to 37 ± 3.5 (n = 50) in Taasy1b (two-sample t-test, p < 0.001, n = 50 Taasy1b and n = 50 Cadenza wild type) (Figure 4C, D, Table 1, and Supplementary Table 13).
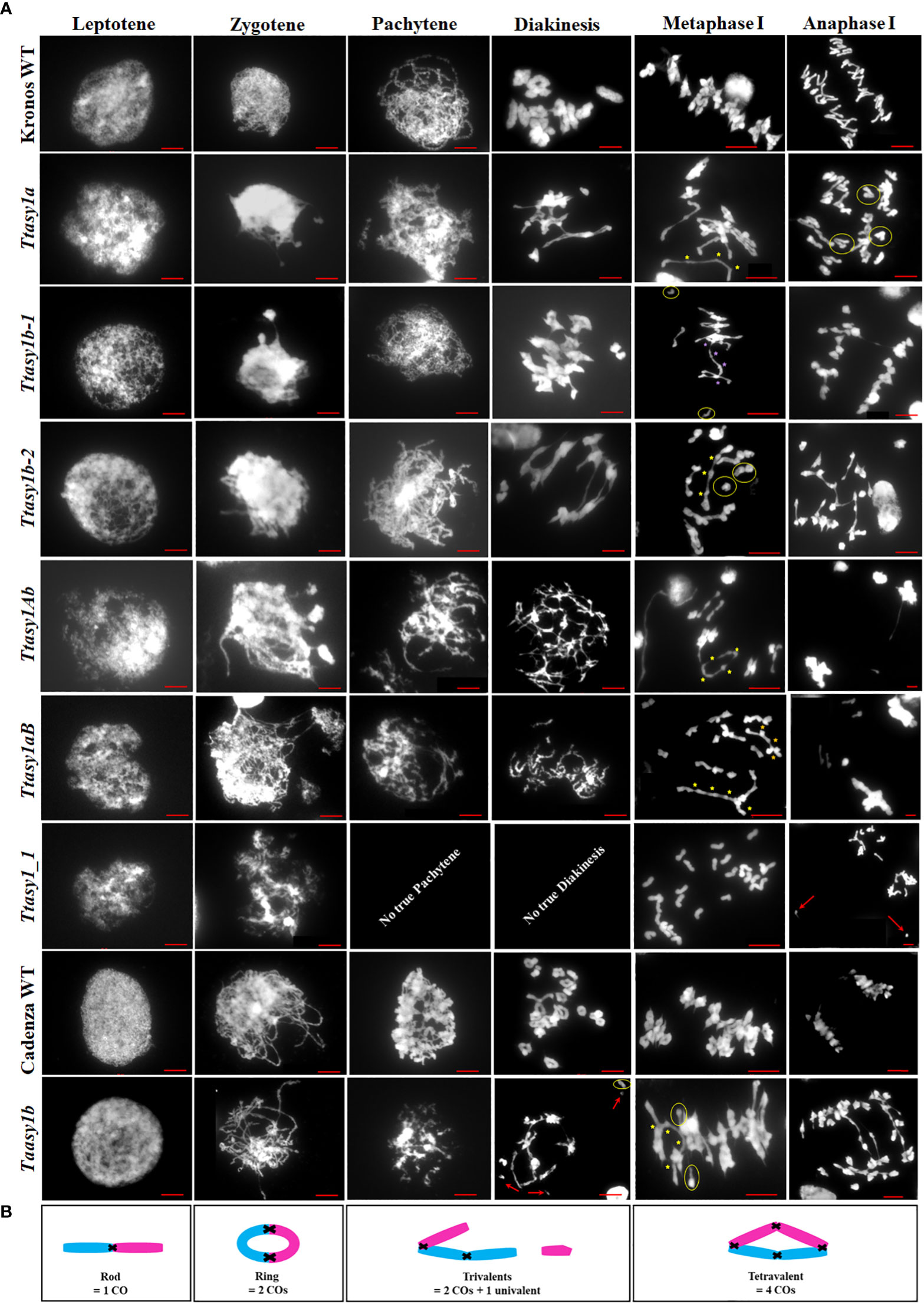
Figure 3 Cytological atlas of asy1 mutants. (A) DAPI-stained meiotic stages from leptotene to anaphase I illustrating phenotypic effects of the asy1 hypomorphic mutants. Yellow circles highlight univalents, yellow stars indicate chiasmata in multivalents, and red arrows highlight lagging chromosomes. Scale bar = 10 µm. (B) Cartoon of chiasma configuration of wheat. The panel depicts bivalent shapes (ring and rod) and trivalent and tetravalent configurations at metaphase I, including points of chiasmata (black crosses) along the chromosomes (blue and pink).
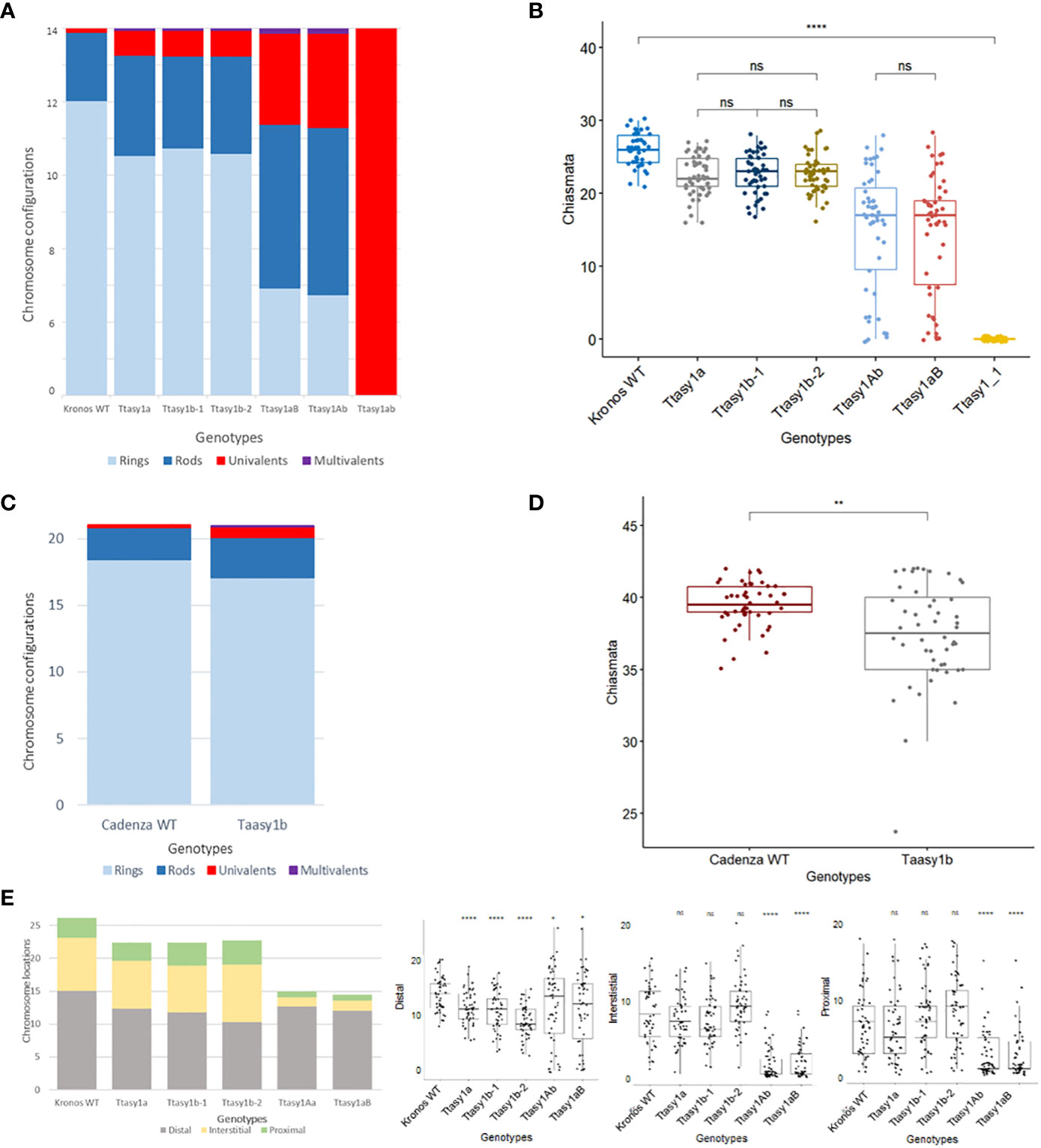
Figure 4 Hypomorphic asy1 mutants display dosage-dependent reduction in chiasma frequency. Bar charts illustrate the mean values of ring bivalents (sky blue), rod bivalents (dark blue), univalents (red), and multivalents (purple) per cell among (A) Kronos wild type and Ttasy1 mutants and (C) Cadenza wild type and Taasy1b mutant. Legend is at the bottom. (B) Box plots exemplify chiasma frequency per male meiocyte among Kronos wild type and Ttasy1 mutants. Significant differences are indicated by pairwise Wilcoxon rank sum test (signif. codes: 0 "****" 0.0001 “***” 0.001 “**” 0.01 “*” 0.1 “ns”). The adjustment methods include the Bonferroni correction. (D) Box plots exemplify chiasma frequency per male meiocyte between Cadenza wild type and Taasy1b mutants. Results of the two-sample t-test are shown (p < 0.001). (E) Position of chiasmata (distal, interstitial, proximal) along the chromosomes in Ttasy1 mutants. Legend is at the bottom. Results of the two-sample t-test are shown (p < 0.001). Significant differences are indicated by pairwise Wilcoxon rank sum test (signif. codes: 0 "****" 0.0001 “***” 0.001 “**” 0.01 “*” 0.1 “ns”).
Chiasma position was also significantly altered in the asy1 hypomorphic mutants. In the wild type, the majority of chiasmata formed distally to the centromere (57.5%, 15 ± 3), followed by interstitial (31%, 8 ± 4) and proximal (12%, 3 ± 2) (Figure 4E). In Ttasy1a/b, interstitial and proximal chiasmata were reduced by 1% (n = 150), but in Ttasy1Ab/aB, where interstitial and proximal chiasmata decreased from 8 to 1 per nucleus and from 3 to 0.9, respectively, compared to the wild type (n = 100) (Figure 4E). In Ttasy1a/b, the proportions remained similar, although there was a slight reduction in distal chiasmata (53%, t-test, ns) with a minor increase in interstitial (32%, t-test, ns) and proximal chiasmata (15%, t-test, ns). In Ttasy1Ab/aB, distal chiasmata were predominant (84%, t-test, p adj < 0.001), followed by interstitial (10%, t-test, p adj < 0.001), and the remaining 6% of chiasmata were proximal (t-test, p adj < 0.001; Supplementary Tables 6-12) (Figure 4E). These data indicate that ASY1 is required to create a bias for promoting chiasma formation in the centromere proximal and interstitial regions in wheat.
In all Ttasy1a/b and Ttasy1Ab/aB hypomorphic mutants, ring bivalents (where at least one chiasma forms in each chromosome arm) were significantly reduced (by 18%, 9 ± 2, n = 150, t-test, p adj < 0.001; and by 64%, 4 ± 3, n = 100, t-test, p adj < 0.001, respectively). This was accompanied by a 33% (3.5 ± 2, n = 150, t-test, p adj < 0.001) increase in rod bivalents (only one chiasma) in Ttasy1a/b, which further increased by 1.9-fold (6.5 ± 4.2, n = 100, t-test, p adj < 0.001) in Ttasy1Ab/aB (Figure 3A). Lastly, univalents (no chiasma) significantly increased from 0.16 per nucleus in the wild type by fivefold in Ttasy1a/b (1 ± 1, n = 150, t-test, p adj < 0.001) and by 20-fold in Ttasy1Ab/aB (3 ± 5, n = 100, t-test, p adj < 0.001) (Figure 3A), revealing loss of the obligate chiasma and an inability to maintain CO assurance. Loss of the obligate chiasma resulted in chromosome mis-segregation and chromosome bridges at anaphase I (Figure 3A).
ASY1 suppresses non-homologous recombination
Ectopic recombination leading to multiple chromosome associations was observed in all hypomorphic asy1 mutants (Figures 3A, B). In Ttasy1Ab/aB, multiple chromosome associations per nucleus were observed (0.18 ± 0.5, n = 100). These were classified into three groups of which 44% were tetravalents (4 chromosomes), 33% trivalents (3 chromosomes), and 23% multivalents (more than 4 chromosomes). Multiple chromosome associations were twofold more frequent in Ttasy1Ab/aB than in Ttasy1a/b (0.08 ± 0.3, n = 150), of which 100% were tetravalents (n = 150). A meiotic cytological analysis was also performed on the hexaploid wheat at metaphase I, revealing an increase in multivalents from 0 in the wild type to 0.06 ± 0.2 (n = 100) in the Taasy1b mutant (p < 0.005) (Table 2).
To determine if the ectopic recombination observed in Kronos and Cadenza asy1 hypomorphic mutants extended to more divergent genotypes, crosses were made between Ttasy1b-2 and wheat allotetraploid wild-relative Ae. variabilis. Fourteen bivalents would be expected if Kronos and Ae. variabilis were capable of forming the obligate chiasma, and 28 univalents would be expected if chiasmata did not form (Figure 5A). In the Kronos wild type/Ae. variabilis cross, univalents ranged from 22 to 28 with a mean of 26.93 ± 0.12, and bivalents ranged from 0 to 3 with a mean of 0.54 ± 0.06 (n = 155) (Table 3), indicating a low level of CO formation between these divergent wheat genotypes. However, in Ttasy1b-2/Ae. variabilis, the number of univalents decreased in range (16–28, n = 155), with a 3.44-fold increase in the number of bivalents to 1.86 ± 0.1 (p < 0.001 Mann–Whitney) (Figures 5A–C). The mean chiasma frequency significantly increased in Ttasy1b-2/Ae. variabilis by 3.75-fold from 0.55 (n = 155) to 2.06 (n = 152) chiasmata per nucleus with a range from 0 to 7 (p < 0.001 Mann–Whitney) (Figures 5B, C, Table 3 and Supplementary Tables 14–18). This suggests that ASY1 suppresses CO formation between divergent chromosomes in a gene dosage-dependent manner.
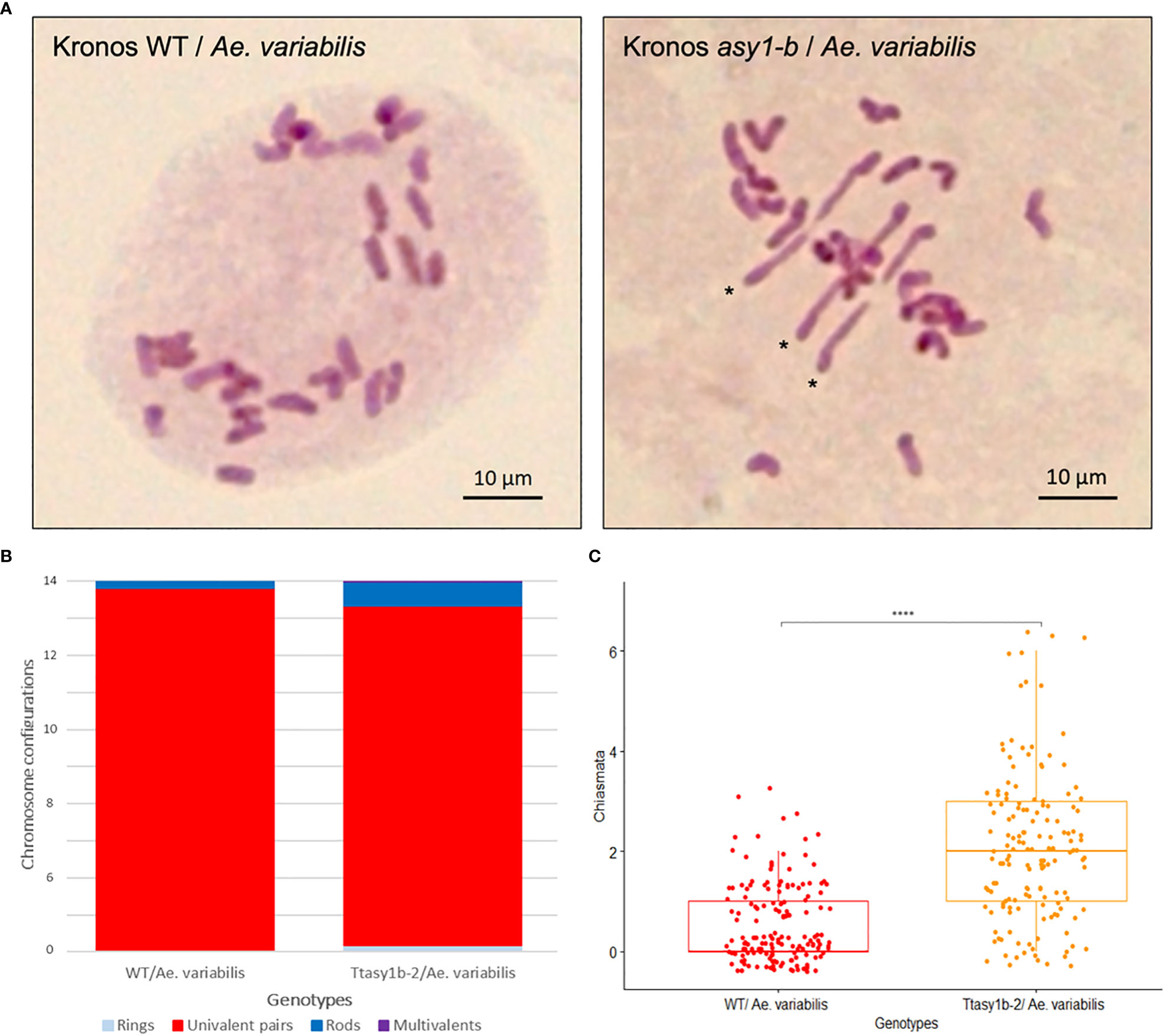
Figure 5 Ttasy1b-2/Aegilops variabilis exhibits increased chiasma frequency at metaphase I. (A) Metaphase I spread of Kronos wild type/Ae. variabilis showing 28 univalents and Ttasy1b-2/Ae. variabilis mutants forming three non-homologous bivalents. Asterisks indicate rod bivalents. Scale bar = 10 µm. (B) Bar chart illustrates the mean values of ring bivalents (sky blue), rod bivalents (dark blue), univalents (red), and multivalents (purple) per cell among Kronos wild type/Ae. variabilis and Ttasy1b-2/Ae. variabilis mutants. Legend is at the bottom. (C) Box plot exemplifies chiasma frequency per male meiocyte among Kronos wild type/Ae. variabilis and Ttasy1b-2/Ae. variabilis mutants. A two-sample t-test was applied to define the statistical significance between wild type/Ae. variabilis and Ttasy1b-2/Ae. variabilis mutant (signif. codes: 0 “****”).
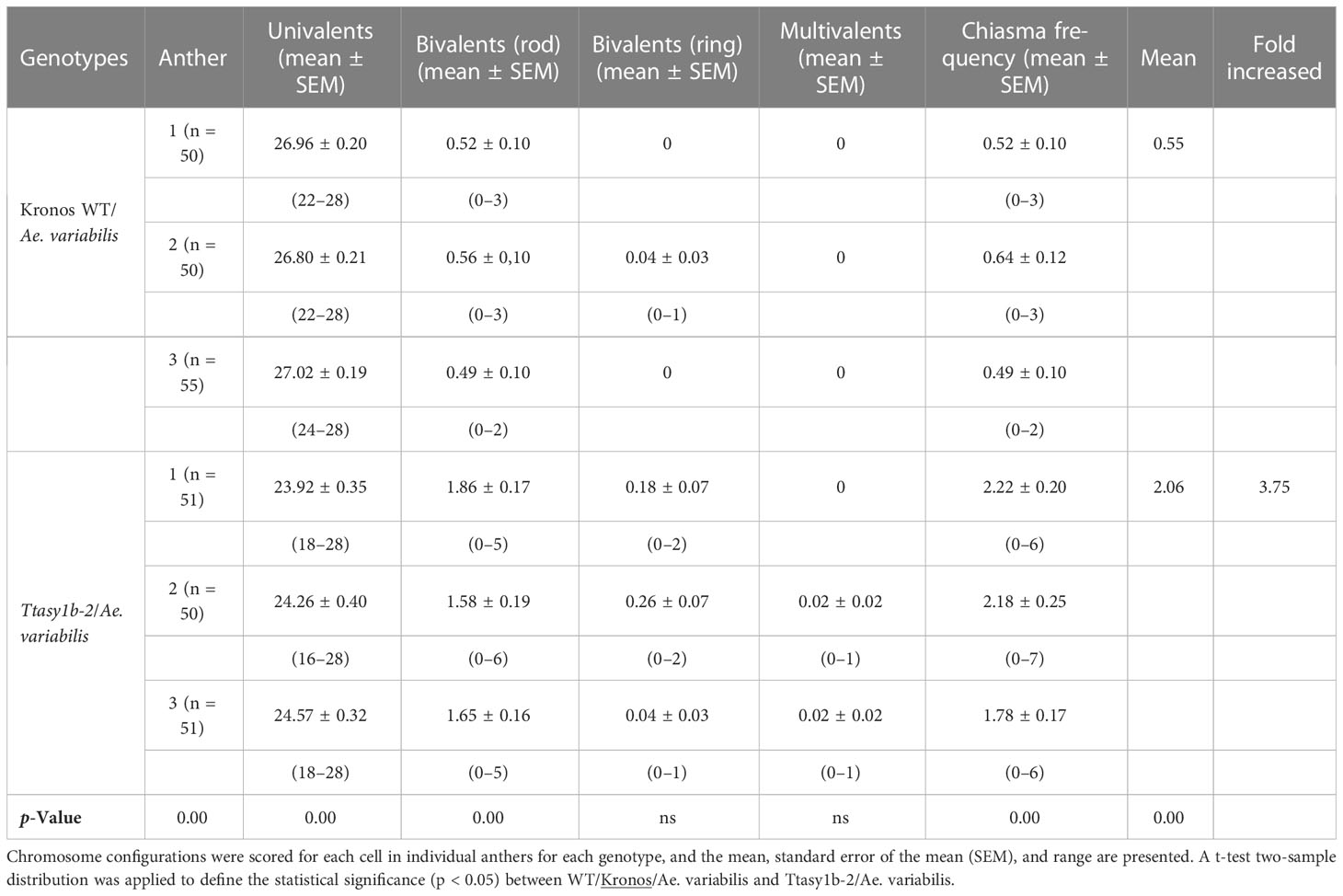
Table 3 Chiasma counts of Kronos wild type/Aegilops variabilis and Ttasy1b-2/Ae. variabilis mutants.
Discussion
ASY1 maintains CO assurance and promotes CO formation away from the chromosome ends
Chiasmata were reduced concomitantly with gene dosage in the Kronos hypomorphic asy1 mutants (WT AABB = 26 chiasmata/cell; AAbb/aaBB =22 chiasmata/cell; Aabb/aaBb = 15 chiasmata/cell; and the null mutant aabb = 0 chiasmata/cell). The asy1 null mutant phenotype is consistent with the ASY1 rice ortholog pair1 mutant where only univalents were observed (Nonomura et al., 2004). Kronos possesses 14 pairs of chromosomes, so the mean number of chiasmata in the hypomorphic mutants is sufficient to ensure the obligate CO, although this is not maintained due to the range around the mean, and also, chiasmata are not equally distributed between the chromosomes. We were unable to determine if this was stochastic or that certain chromosomes were more likely to be affected due to the unreliability of oligonucleotide fluorescence in situ hybridization (FISH) probes to barcode the chromosomes (data not shown).
The cytological data reveal that wheat ASY1 promotes chiasma formation proximal to the centromeres and along the chromosome arms. This is remarkably similar to previous reports in A. thaliana where ASY1 promotes recombination away from the telomeres in a dosage-dependent manner and is essential for the obligate CO (Sanchez-Moran et al., 2007; Lambing et al., 2020; Pochon et al., 2022). In Arabidopsis, barley, and wheat, telomeres cluster during leptotene (Armstrong et al., 2001; Higgins et al., 2012; Sepsi et al., 2017), thus providing an early opportunity for nascent strand invasion events in the sub-telomeric regions to precede those in interstitial regions. These early contacts are likely to bias recombination maturation in the distal regions and prevent further COs from forming in the interstitial regions by CO interference (Higgins et al., 2014). Therefore, ASY1 could alleviate this early bias by forming axial bridges between chromosomes at greater distances to promote strand invasion, thereby enabling CO formation away from the chromosome ends.
Synapsis is dependent on ASY1
ASY1 protein levels were not quantified, but the ASY1 axis signal by immunofluorescence did not appear different in the hypomorphic mutants when compared to the wild type. However, a significant delay in prophase I progression in the hypomorphic mutants associated with a reduction in chiasmata may reflect a lower rate of ASY1 protein production that eventually reached wild-type levels. As no ASY1 protein was detected on the axes in the null asy1 mutant, it is unlikely that truncated forms of the ASY1 proteins would influence the phenotype in these mutants as dominant negatives. The delay in ZYP1 loading at zygotene is also associated with reduced ASY1 dosage in the hypomorphic mutants. Incomplete ZYP1 polymerization in Ttasy1a/b and its total absence in Ttasy1Ab/aB and Ttasy1_1 led to asynchronous meiotic progression that arrested at pachytene and diplotene. In barley, ZYP1 is required for ~85% COs (Barakate et al., 2014), so a delay in synapsis may have had an additive effect in the wheat asy1 mutants on chiasma formation as well as loss of function of ASY1 in promoting interhomolog recombination.
How does ASY1 promote and suppress COs in wheat?
A role for ASY1 in preventing ectopic recombination during meiosis was previously reported (Boden et al., 2009), which is supported by our data. In addition, wheat ASY1 also promotes recombination along the chromosome arms to assuage the telomere-led bias, phenotypically similar to ASY1 in Arabidopsis. Therefore, how does ASY1 promote and suppress COs in wheat? ASY1 may be required to provide a minimum number of interhomolog axial bridge contact points between chromosomes in the pairing process, thereby ensuring accurate fidelity so that the homologous chromosomes can synapse and recombine. It is possible that stronger associations would form between homologous chromosomes rather than homoeologous chromosomes due to the stringency of base pairing and hydrogen bonding of the single-end invasions, promoted by ASY1 and DMC1. If the minimum number of contact points is reduced below a threshold, such as what could happen in the asy1 hypomorphic mutants, then the fidelity of chromosome recognition may be impaired leading to ectopic recombination in the tetraploid/hexaploid mutants and increased chiasmata in asy1/Ae. variabilis. The delay in meiotic progression in the asy1 hypomorphic mutants may reflect a surveillance system performing sub-optimally that is required to monitor accurate pairing and ensure that synapsis initiates between homologous, rather than homoeologous chromosomes.
Does ASY1 share the characteristics of pairing homoeologous loci in wheat?
The pairing homoeologous (Ph) loci in wheat negatively act on recombination between chromosomes of diverged species. Here, we show that the mean chiasma frequency increased by 3.75-fold in Kronos Ttasy1b-2/Ae. variabilis when compared to wild type/Ae. variabilis, indicating that ASY1 suppresses recombination between divergent chromosomes. This is similar to the Ph loci, although to a lesser extent as chiasmata increased by 8.3-fold/cell in Ph1 (zip4 5B) and up to 5.5-fold/cell in Ph2 (msh7-3D) hexaploid wheat mutants when crossed with Ae. variabilis. TaASY1-5B is located on the long arm of chromosome 5, separated by 33.5 Mb of DNA from the class I CO gene ZIP4-5B (Rey et al., 2017; Martín et al., 2018). The novel duplication of ZIP4 on chromosome 5B is indicative of adaptive evolution, whereas there are no obvious hallmarks of adaptation at the ASY1 5B locus (although this requires further investigation). However, phenotypic similarities exist between Ph1 and asy1 such as an increase in homoeologous recombination and a delay in synapsis. Pochon et al. (2022) reported that not all MLH1 foci maturate into COs in Arabidopsis asy1 mutants, reminiscent of the Ph1 phenotype, suggesting a possible association between asy1 and Ph1 (Martín et al., 2014; Martín et al., 2017; Pochon et al., 2022). Moreover, in ph1b, localization of ASY1 was perturbed, adopting a spiral-like pattern during zygotene and pachytene (Boden et al., 2009). Surprisingly, no multivalents were observed in Tazip4-B2 mutant lines, whereas they are observed in ph1b at a low frequency (trivalents 0.2% and tetravalents 0.37%) (Rey et al., 2017) and in the Taasy1 5B mutant line.
The chromosome axis has been implicated in adaptation to meiotic recombination in autotetraploid Arabidopsis arenosa and Arabidopsis lyrata. ASY1 and ASY3 alleles are under selection in these tetraploids that distalize chiasmata to the chromosome ends and reduce their number (Morgan et al., 2020; Seear et al., 2020). This implicates ASY1 as a major gene required to stabilize both allopolyploid and autopolyploid meiotic recombination. It also raises the potential to combine zip4 5B and asy1 (5A, 5B, or 5D) to increase introgression from wheat wild relatives.
In conclusion, this study provides further support for the role of ASY1 in controlling CO number and position as well as CO assurance in plants. The dosage sensitivity of ASY1 in wheat is similar to the haplo-insufficiency reported in Arabidopsis (Lambing et al., 2020), suggesting that ASY1 performs a conserved role in both diploid and polyploid species. The fidelity of accurate chromosome pairing is reduced in the hypomorphic asy1 wheat mutants, leading to ectopic recombination This indicates that ASY1 plays a major role in chromosome recognition and may bias recombination toward the homolog rather than homoeologous chromosomes by monitoring DNA sequence homology during stable strand invasion. Thus, wheat hypomorphic asy1 mutants could provide a tool to enhance the introgression of agronomically important traits from wheat wild relatives into elite varieties.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
JH and PS designed the research performed by CD and HS. JH, PS, CD, and HS analyzed the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The research was funded by the European Union’s Marie Sklodowska-Curie Innovative Training Network MEICOM (H2020-MSCA-ITN-2017 Horizon 2020 grant agreement number 765212).
Acknowledgments
We would like to thank Dr. Stuart Desjardins for technical advice, Dr Neelam Dave for technical support, and the EU for financial support with the MEICOM consortium.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1188347/full#supplementary-material
References
Alabdullah, A. K., Borrill, P., Martin, A. C., Ramirez-Gonzalez, R. H., Hassani-Pak, K., Uauy, C., et al. (2019). A Co-expression network in hexaploid wheat reveals mostly balanced expression and lack of significant gene loss of homeologous meiotic genes upon polyploidization. Front. Plant Sci. 10, 1325. doi: 10.3389/fpls.2019.01325
Appels, R., Eversole, K., Stein, N., Feuillet, C., Keller, B., Rogers, J., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191. doi: 10.1126/science.aar7191
Armstrong, S. J., Caryl, A. P., Jones, G. H., Franklin, F. C. (2002). Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in arabidopsis and brassica. J. Cell Sci. 115, 3645–3655. doi: 10.1242/jcs.00048
Armstrong, S. J., Franklin, F. C., Jones, G. H. (2001). Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in arabidopsis thaliana. J. Cell Sci. 114, 4207–4217. doi: 10.1242/jcs.114.23.4207
Balboni, M., Yang, C., Komaki, S., Brun, J., Schnittger, A. (2020). COMET functions as a PCH2 cofactor in regulating the HORMA domain protein ASY1. Curr. Biol. 30 (21), 4113–4127.e6. doi: 10.1016/j.cub.2020.07.089
Barakate, A., Higgins, J. D., Vivera, S., Stephens, J., Perry, R. M., Ramsay, L., et al. (2014). The synaptonemal complex protein ZYP1 is required for imposition of meiotic crossovers in barley. Plant Cell 26, 729–740. doi: 10.1105/tpc.113.121269
Benyahya, F., Nadaud, I., Da Ines, O., Rimbert, H., White, C., Sourdille, P. (2020). SPO11.2 is essential for programmed double-strand break formation during meiosis in bread wheat (Triticum aestivum l.). Plant J. 104, 30–43. doi: 10.1111/tpj.14903
Berchowitz, L. E., Francis, K. E., Bey, A. L., Copenhaver, G. P. (2007). The role of AtMUS81 in interference-insensitive crossovers in a. thaliana. PloS Genet. 3, e132. doi: 10.1371/journal.pgen.0030132
Boden, S. A., Langridge, P., Spangenberg, G., Able, J. A. (2009). TaASY1 promotes homologous chromosome interactions and is affected by deletion of Ph1. Plant J. 57, 487–497. doi: 10.1111/j.1365-313X.2008.03701.x
Capilla-Perez, L., Durand, S., Hurel, A., Lian, Q., Chambon, A., Taochy, C., et al. (2021). The synaptonemal complex imposes crossover interference and heterochiasmy in arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 118 (12), e2023613118. doi: 10.1073/pnas.2023613118
Caryl, A. P., Armstrong, S. J., Jones, G. H., Franklin, F. C. (2000). A homologue of the yeast HOP1 gene is inactivated in the arabidopsis meiotic mutant asy1. Chromosoma 109, 62–71. doi: 10.1007/s004120050413
Chambon, A., West, A., Vezon, D., Horlow, C., De Muyt, A., Chelysheva, L., et al. (2018). Identification of ASYNAPTIC4, a component of the meiotic chromosome axis. Plant Physiol. 178, 233–246. doi: 10.1104/pp.17.01725
Da Ines, O., Michard, R., Fayos, I., Bastianelli, G., Nicolas, A., Guiderdoni, E., et al. (2020). Bread wheat TaSPO11-1 exhibits evolutionarily conserved function in meiotic recombination across distant plant species. Plant J. 103, 2052–2068. doi: 10.1111/tpj.14882
Desjardins, S., Kanyuka, K., Higgins, J. D. (2020). A cytological analysis of wheat meiosis targeted by virus-induced gene silencing (VIGS). Methods Mol. Biol. 2061, 319–330. doi: 10.1007/978-1-4939-9818-0_22
Desjardins, S. D., Ogle, D. E., Ayoub, M. A., Heckmann, S., Henderson, I. R., Edwards, K. J., et al. (2020). MutS homologue 4 and MutS homologue 5 maintain the obligate crossover in wheat despite stepwise gene loss following polyploidization. Plant Physiol. 183, 1545–1558. doi: 10.1104/pp.20.00534
Desjardins, S. D., Simmonds, J., Guterman, I., Kanyuka, K., Burridge, A. J., Tock, A. J., et al. (2022). FANCM promotes class I interfering crossovers and suppresses class II non-interfering crossovers in wheat meiosis. Nat. Commun. 13, 3644. doi: 10.1038/s41467-022-31438-6
Ferdous, M., Higgins, J. D., Osman, K., Lambing, C., Roitinger, E., Mechtler, K., et al. (2012). Inter-homolog crossing-over and synapsis in arabidopsis meiosis are dependent on the chromosome axis protein AtASY3. PloS Genet. 8, e1002507. doi: 10.1371/journal.pgen.1002507
France, M. G., Enderle, J., Röhrig, S., Puchta, H., Franklin, F. C. H., Higgins, J. D. (2021). ZYP1 is required for obligate cross-over formation and cross-over interference in arabidopsis. Proc. Natl. Acad. Sci. 118, e2021671118. doi: 10.1073/pnas.2021671118
Gao, J., Colaiacovo, M. P. (2018). Zipping and unzipping: protein modifications regulating synaptonemal complex dynamics. Trends Genet. 34, 232–245. doi: 10.1016/j.tig.2017.12.001
Gardiner, L.-J., Wingen, L. U., Bailey, P., Joynson, R., Brabbs, T., Wright, J., et al. (2019). Analysis of the recombination landscape of hexaploid bread wheat reveals genes controlling recombination and gene conversion frequency. Genome Biol. 20, 1–16. doi: 10.1186/s13059-019-1675-6
Higgins, J. D. (2013). Analyzing meiosis in barley. Methods Mol. Biol. 990, 135–144. doi: 10.1007/978-1-62703-333-6_14
Higgins, J. D., Armstrong, S. J., Franklin, F. C. H., Jones, G. H. (2004). The arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in arabidopsis. Genes Dev. 18, 2557–2570. doi: 10.1101/gad.317504
Higgins, J. D., Buckling, E. F., Franklin, F. C. H., Jones, G. H. (2008a). Expression and functional analysis of AtMUS81 in arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J. 54, 152–162. doi: 10.1111/j.1365-313X.2008.03403.x
Higgins, J. D., Osman, K., Desjardins, S. D., Henderson, I. R., Edwards, K. J., Franklin, F. C. H. (2022). Unravelling mechanisms that govern meiotic crossover formation in wheat. Biochem. Soc. Trans. 50, 1179–1186. doi: 10.1042/BST20220405
Higgins, J. D., Osman, K., Jones, G. H., Franklin, F. C. H. (2014). “"Factors underlying restricted crossover localization in barley meiosis,",” in Annual review of genetics, vol. 48 . Ed. Bassler, B. L., 29–47. doi: 10.1146/annurev-genet-120213-092509
Higgins, J. D., Perry, R. M., Barakate, A., Ramsay, L., Waugh, R., Halpin, C., et al. (2012). Spatiotemporal asymmetry of the meiotic program underlies the predominantly distal distribution of meiotic crossovers in barley. Plant Cell 24, 4096–4109. doi: 10.1105/tpc.112.102483
Higgins, J. D., Sanchez-Moran, E., Armstrong, S. J., Jones, G. H., Franklin, F. C. (2005). The arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 19, 2488–2500. doi: 10.1101/gad.354705
Higgins, J. D., Vignard, J., Mercier, R., Pugh, A. G., Franklin, F. C. H., Jones, G. H. (2008b). AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in arabidopsis thaliana, but is not required for synapsis. Plant J. 55, 28–39. doi: 10.1111/j.1365-313X.2008.03470.x
Hughes, S. E., Hawley, R. S. (2020). Alternative synaptonemal complex structures: too much of a good thing? Trends Genet. 36, 833–844. doi: 10.1016/j.tig.2020.07.007
Hyde, L., Osman, K., Winfield, M., Sanchez-Moran, E., Higgins, J. D., Henderson, I. R., et al. (2022). Identification, characterisation, and rescue of CRISPR/Cas9 generated wheat SPO11-1 mutants. Plant Biotechnol. J. 21 (2), 405–418. doi: 10.1111/pbi.13961
Jiang, Y., N'diaye, A., Koh, C. S., Quilichini, T. D., Shunmugam, A. S. K., Kirzinger, M. W., et al. (2023). The coordinated regulation of early meiotic stages is dominated by non-coding RNAs and stage-specific transcription in wheat. Plant J. 114 (1), 209–224. doi: 10.1111/tpj.16125
Jones, G. H., Franklin, F. C. (2006). Meiotic crossing-over: obligation and interference. Cell 126, 246–248. doi: 10.1016/j.cell.2006.07.010
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., Sternberg, M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Krasileva, K. V., Vasquez-Gross, H. A., Howell, T., Bailey, P., Paraiso, F., Clissold, L., et al. (2017). Uncovering hidden variation in polyploid wheat. Proc. Natl. Acad. Sci. U.S.A. 114, E913–E921. doi: 10.1073/pnas.1619268114
Lambing, C., Franklin, F. C., Wang, C. R. (2017). Understanding and manipulating meiotic recombination in plants. Plant Physiol. 173, 1530–1542. doi: 10.1104/pp.16.01530
Lambing, C., Kuo, P. C., Tock, A. J., Topp, S. D., Henderson, I. R. (2020). ASY1 acts as a dosage-dependent antagonist of telomere-led recombination and mediates crossover interference in arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 117, 13647–13658. doi: 10.1073/pnas.1921055117
Martín, A. C., Borrill, P., Higgins, J., Alabdullah, A., Ramírez-González, R. H., Swarbreck, D., et al. (2018). Genome-wide transcription during early wheat meiosis is independent of synapsis, ploidy level, and the Ph1 locus. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01791
Martín, A. C., Rey, M.-D., Shaw, P., Moore, G. (2017). Dual effect of the wheat Ph1 locus on chromosome synapsis and crossover. Chromosoma 126, 669–680. doi: 10.1007/s00412-017-0630-0
Martín, A. C., Shaw, P., Phillips, D., Reader, S., Moore, G. (2014). Licensing MLH1 sites for crossover during meiosis. Nat. Commun. 5, 4580. doi: 10.1038/ncomms5580
Mello-Sampayo, T. (1971). Genetic regulation of meiotic chromosome pairing by chromosome 3D of triticum aestivum. Nat. New Biol. 230, 22–23. doi: 10.1038/newbio230022a0
Morgan, C., Zhang, H., Henry, C. E., Franklin, F. C. H., Bomblies, K. (2020). Derived alleles of two axis proteins affect meiotic traits in autotetraploid arabidopsis arenosa. Proc. Natl. Acad. Sci. 117, 8980–8988. doi: 10.1073/pnas.1919459117
Nonomura, K. I., Nakano, M., Murata, K., Miyoshi, K., Eiguchi, M., Miyao, A., et al. (2004). An insertional mutation in the rice PAIR2 gene, the ortholog of arabidopsis ASY1, results in a defect in homologous chromosome pairing during meiosis. Mol. Genet. Genomics 271, 121–129. doi: 10.1007/s00438-003-0934-z
Osman, K., Algopishi, U., Higgins, J. D., Henderson, I. R., Edwards, K. J., Franklin, F. C. H., et al. (2021). Distal bias of meiotic crossovers in hexaploid bread wheat reflects spatio-temporal asymmetry of the meiotic program. Front. Plant Sci. 12, 631323. doi: 10.3389/fpls.2021.631323
Osman, K., Higgins, J. D., Sanchez-Moran, E., Armstrong, S. J., Franklin, F. C. (2011). Pathways to meiotic recombination in arabidopsis thaliana. New Phytol. 190, 523–544. doi: 10.1111/j.1469-8137.2011.03665.x
Osman, K., Yang, J., Roitinger, E., Lambing, C., Heckmann, S., Howell, E., et al. (2018). Affinity proteomics reveals extensive phosphorylation of the brassica chromosome axis protein ASY1 and a network of associated proteins at prophase I of meiosis. Plant J. 93, 17–33. doi: 10.1111/tpj.13752
Page, S. L., Hawley, R. S. (2004). The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 20, 525–558. doi: 10.1146/annurev.cellbio.19.111301.155141
Pochon, G., Henry, I. M., Yang, C., Lory, N., Fernández-Jiménez, N., Böwer, F., et al (2022). The arabidopsis Hop1 homolog ASY1 mediates cross-over assurance and interference. PNAS Nexus. 2 (3), pgac302. doi: 10.1093/pnasnexus/pgac302
Rey, M. D., Martin, A. C., Higgins, J., Swarbreck, D., Uauy, C., Shaw, P., et al. (2017). Exploiting the ZIP4 homologue within the wheat Ph1 locus has identified two lines exhibiting homoeologous crossover in wheat-wild relative hybrids. Mol. Breed 37, 95. doi: 10.1007/s11032-017-0700-2
Rey, M. D., Martin, A. C., Smedley, M., Hayta, S., Harwood, W., Shaw, P., et al. (2018). Magnesium increases homoeologous crossover frequency during meiosis in ZIP4 (Ph1 gene) mutant wheat-wild relative hybrids. Front. Plant Sci. 9, 509. doi: 10.3389/fpls.2018.00509
Riley, R., Chapman, V. (1958). Genetic control of the cytologically diploid behaviour of hexaploid wheat. Nature 182, 713–715. doi: 10.1038/182713a0
Saintenac, C., Falque, M., Martin, O. C., Paux, E., Feuillet, C., Sourdille, P. (2009). Detailed recombination studies along chromosome 3B provide new insights on crossover distribution in wheat (Triticum aestivum l.). Genetics 181, 393–403. doi: 10.1534/genetics.108.097469
Sanchez-Moran, E., Osman, K., Higgins, J. D., Pradillo, M., Cunado, N., Jones, G. H., et al. (2008). ASY1 coordinates early events in the plant meiotic recombination pathway. Cytogenetic Genome Res. 120, 302–312. doi: 10.1159/000121079
Sanchez-Moran, E., Santos, J. L., Jones, G. H., Franklin, F. C. (2007). ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in arabidopsis. Genes Dev. 21, 2220–2233. doi: 10.1101/gad.439007
Seear, P. J., France, M. G., Gregory, C. L., Heavens, D., Schmickl, R., Yant, L., et al. (2020). A novel allele of ASY3 is associated with greater meiotic stability in autotetraploid arabidopsis lyrata. PloS Genet. 16, e1008900. doi: 10.1371/journal.pgen.1008900
Sepsi, A., Higgins, J. D., Heslop-Harrison, J. S., Schwarzacher, T. (2017). CENH3 morphogenesis reveals dynamic centromere associations during synaptonemal complex formation and the progression through male meiosis in hexaploid wheat. Plant J. 89, 235–249. doi: 10.1111/tpj.13379
Serra, H., Svacina, R., Baumann, U., Whitford, R., Sutton, T., Bartos, J., et al. (2021). Ph2 encodes the mismatch repair protein MSH7-3D that inhibits wheat homoeologous recombination. Nat. Commun. 12, 803. doi: 10.1038/s41467-021-21127-1
Shunmugam, A. S., Bollina, V., Dukowic-Schulze, S., Bhowmik, P. K., Ambrose, C., Higgins, J. D., et al. (2018). MeioCapture: an efficient method for staging and isolation of meiocytes in the prophase I sub-stages of meiosis in wheat. BMC Plant Biol. 18, 1–12. doi: 10.1186/s12870-018-1514-z
Tock, A. J., Holland, D. M., Jiang, W., Osman, K., Sanchez-Moran, E., Higgins, J. D., et al. (2021). Crossover-active regions of the wheat genome are distinguished by DMCI, the chromosome axis, H3K27me3, and signatures of adaptation. Genome Res. 31, 1614–1628. doi: 10.1101/gr.273672.120
Wang, Y., Copenhaver, G. P. (2018). Meiotic recombination: mixing it up in plants. Annu. Rev. Plant Biol. 69, 577–609. doi: 10.1146/annurev-arplant-042817-040431
Wang, M., Wang, K., Tang, D., Wei, C., Li, M., Shen, Y., et al. (2010). The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell 22, 417–430. doi: 10.1105/tpc.109.070789
Keywords: chromosomes, chiasma, homoeologous, meiosis, synapsis, introgression
Citation: Di Dio C, Serra H, Sourdille P and Higgins JD (2023) ASYNAPSIS 1 ensures crossover fidelity in polyploid wheat by promoting homologous recombination and suppressing non-homologous recombination. Front. Plant Sci. 14:1188347. doi: 10.3389/fpls.2023.1188347
Received: 20 March 2023; Accepted: 17 April 2023;
Published: 22 May 2023.
Edited by:
Wellington Ronildo Clarindo, Universidade Federal de Viçosa, BrazilReviewed by:
Mariana Cansian Sattler, Universidade Federal de Viçosa, BrazilYuri Shavrukov, Flinders University, Australia
Copyright © 2023 Di Dio, Serra, Sourdille and Higgins. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James D. Higgins, amg1NTVAbGVpY2VzdGVyLmFjLnVr
†Present address: Heïdi Serra, Genetics, Reproduction and Development, CNRS, Inserm, Université Clermont Auvergne, Clermont-Ferrand, France
Chiara Di Dio, Crop Science Centre, Department of Plant Sciences, University of Cambridge, United Kingdom
 Chiara Di Dio
Chiara Di Dio Heïdi Serra
Heïdi Serra Pierre Sourdille2
Pierre Sourdille2 James D. Higgins
James D. Higgins